Abstract
Here, we report the draft genome sequence of Dissulfurirhabdus thermomarina SH388. Improved phylogenetic and taxonomic analysis of this organism using genome-level analyses supports assignment of this organism to a novel family within the phylum Desulfobacterota. Additionally, comparative genomic and phylogenetic analyses contextualize the convergent evolution of sulfur disproportionation and potential extracellular electron transfer in this organism relative to other members of the Desulfobacterota.
Keywords: Deltaproteobacteria, sulfate reduction, comparative genomics, EET
Data Summary
The datasets generated and analysed during the current study are available in the National Center for Biotechnology Information repository under BioProject number PRJNA579145. Raw sequencing data is available in the SRA database under accession number SRR11035951, while the assembled genome is available in the WGS database under accession number JAAGRR000000000.
Impact Statement.
Our understanding of Earth history is largely based on analysis of the rock record, but the preservation of chemical signals in rocks is modulated by the interaction of sediments with microbial metabolisms, many of which are poorly understood. The ways in which microbes are able to breathe insoluble compounds such as elemental sulfur is of particular interest. Dissulfurirhabdus thermomarina is a bacterium in the phylum Desulfobacterota, and is notable for its ability to perform extracellular electron transfer to respire insoluble compounds, as well as its ability to disproportionate elemental sulfur – essentially making a living by fermenting sulfur into sulfide and sulfate in a manner analogous to brewer’s yeast fermenting sugar into alcohol and carbon dioxide. Here, we report the genome of Dissulfurirhabdus thermomarina and show that it evolved its metabolic traits independently from other groups of bacteria capable of similar processes. This has relevance for understanding the biochemical mechanisms, diversity and evolution of both extracellular electron transfer and sulfur disproportionation, enigmatic metabolisms that are important for shaping the preservation of chemical signals in the rock record.
Introduction
The disproportionation of intermediate valence sulfur species (converting substrates such as sulfite, thiosulfate and elemental sulfur into products of sulfate and sulfide) is a poorly understood microbial metabolism that has been implicated in the production and preservation of extremely depleted sulfur isotope signatures in the rock record [1, 2]. Despite its potential significance for understanding paleo-redox proxies [3], this metabolism is not well elucidated [4] and the genetic capacity for sulfur disproportionation is largely indistinguishable from that for sulfate reduction based on sequence data alone [5]. Mechanisms for elemental sulfur disproportionation are poorly understood, yet this process is of significant interest, particularly due to its ability to utilize an insoluble extracellular substrate. Most of the diversity of organisms shown to be capable of disproportionating elemental sulfur are members of the family Desulfobulbaceae of Desulfobacterota (formerly Deltaproteobacteria ) [4]. However, a small number of distantly related organisms are also capable of elemental sulfur disproportionation, although it remains unknown whether these lineages use the same or different pathways for sulfur disproportionation, and whether these pathways have evolved convergently or whether their distribution is the result of a single evolutionary innovation followed by horizontal gene transfer. In order to resolve these possibilities, we report here the draft genome sequence of Dissulfurirhabdus thermomarina SH388. This organism was first isolated from a shallow marine hydrothermal vent and was shown to be capable of disproportionating sulfur compounds (including elemental sulfur and sulfite) [6], but is of uncertain phylogenetic affinity within the Desulfobacterota and is not closely related to elemental sulfur disproportionators within the Desulfobulbaceae . Understanding the genetics and evolutionary history of sulfur metabolism in Dissulfurirhabdus thermomarina , in contrast to members of the Desulfobulbaceae , may therefore provide insight into the diversity, mechanism and evolution of sulfur disproportionation metabolisms more broadly.
Methods
Genome sequencing and analysis followed methods described previously [7, 8], and summarized here. Purified genomic DNA of strain DSM100025 was ordered from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen) and submitted to MicrobesNG for sequencing. Cultures were grown anaerobically at 50 °C on medium 1210a ( Thermosulfurimonas medium) with strain-specific modifications, before genomic DNA extraction with a JetFlex genomic DNA purification kit (Genomed). DNA libraries were prepared with a Nextera XT library prep kit on a Hamilton Microlab Star automated liquid handling system. Libraries were sequenced on an Illumina HiSeq using a 250 bp paired-end protocol. Reads were adapter trimmed using Trimmomatic 0.30 [9] and de novo assembly was performed using SPAdes version 3.7 [10]. Annotation was performed using rast v2.0 [11]. Genome completeness was estimated with CheckM v1.0.12 [12], and likelihood of presence or absence of metabolic pathways was estimated with MetaPOAP v1.0 [13]. The taxonomic assignment of the genome was verified with GTDB-Tk v0.3.2 [14]. Hydrogenase proteins were classified with HydDB [15].
Phylogenetic analyses followed methods described previously [16, 17], and summarized here. Additional Desulfobulbales genomes were downloaded from the National Center for Biotechnology Information (NCBI) GenBank and WGS databases. Protein sequences used in analyses (see below) were identified locally with the tblastn function of blast+ [18], aligned with muscle [19] and manually curated in Jalview [20]. Positive blast hits were considered to be full length (e.g. >90 % the shortest reference sequence from an isolate genome) with E values greater than 1×10−20. Phylogenetic trees were calculated using RAxML [21] on the CIPRES science gateway [22]. Transfer bootstrap support values were calculated by booster [23], and trees were visualized with the Interactive Tree of Life viewer [24]. Taxonomic assignment was confirmed with GTDB-Tk [14]. Histories of vertical versus horizontal inheritance of metabolic genes were inferred by comparison of organismal and metabolic protein phylogenies [25–27].
Results
The Dissulfurirhabdus thermomarina genome was sequenced at ~85× coverage as 508 907 reads. The assembled genome was recovered as 413 contigs (292 >500 nt, 194 >2500 nt) for a total of 2 536 728 nt with 70.82 mol% G+C and an N50 of 14 884. It encodes 2791 coding sequences and 53 RNAs. The genome was estimated by CheckM to be 96.75 % complete.
Discussion
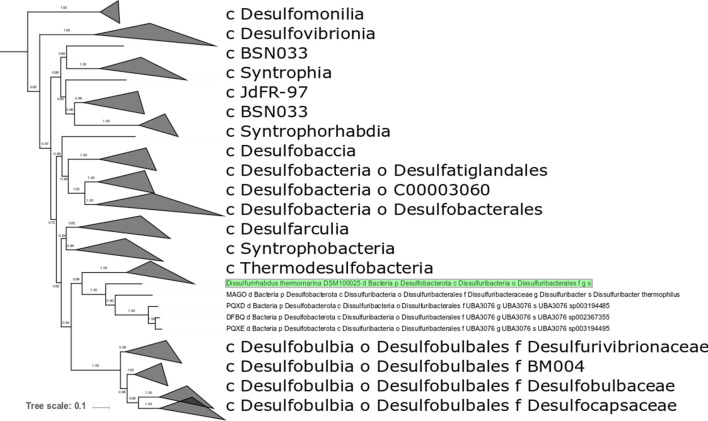
When first isolated, Dissulfurirhabdus thermomarina was placed in the Deltaproteobacteria , but was not assigned at lower taxonomic ranks. Following genome-wide taxonomic analysis with GTDB-Tk, Dissulfurirhabdus thermomarina is robustly placed in the order Dissulfuribacterales (phylum Desulfobacterota, class Dissulfuribacteria), but does not cluster with any characterized families within this order (Fig. 1). Dissulfurirhabdus thermomarina is sufficiently divergent from its closest characterized relative, Dissulfuribacter thermophilus, to suggest that these organisms represent separate families within the Dissulfuribacterales. Therefore, we propose assignment of Dissulfurirhabdus thermomarina as the type species of a novel family, Dissulfurirhabdaceae, within the order Dissulfuribacterales of Desulfobacterota.
Fig. 1.
Phylogeny of the Desulfobacterota, showing the placement of Dissulfurirhabdus thermomarina relative to other lineages, built with concatenated ribosomal proteins following the methods of Hug et al. [38]. Strains are labelled with NCBI WGS database accession numbers and/or taxonomic assignments made with GTDB-Tk [14]. Scale bar represents average number of substitutions per site.
Sulfur disproportionators are expected to encode the same marker genes as sulfate reducers ([5]; consistent with this expectation, Dissulfurirhabdus thermomarina encodes sulfate adenylyltransferase, adenylylsulfate reductase, dissimilatory sulfite reductase and the complex DsrMKJOP, which is associated with sulfite reduction. Known sulfur disproportionators in the family Desulfobulbaceae of Desulfobacterota encode AprB proteins with a truncated tail; this conserved truncation has been proposed as a molecular marker of the capacity for sulfur disproportionation [28]. Dissulfurirhabdus thermomarina encodes an AprB protein with a similar truncation despite encoding an AprB protein that is only distantly related to those of the Desulfobulbaceae (Fig. 2) – this suggests that the AprB truncation evolved independently in Dissulfurirhabdus thermomarina and the Desulfobulbaceae . This reinforces hypotheses for the association between this marker and the capacity for disproportionation as previously proposed [28]. Moreover, the apparently independent acquisition of the AprB truncation and the capacity for disproportionation in Dissulfurirhabdus thermomarina and members of the Desulfobulbaceae suggests that this might be a more widespread trait that has evolved convergently multiple times in ancestrally sulfate-reducing lineages. The relatively small number of physiologically and genomically characterized sulfur disproportionators available at this time limits our ability to confidently test this association, however, and so will necessitate further consideration as more data becomes available.
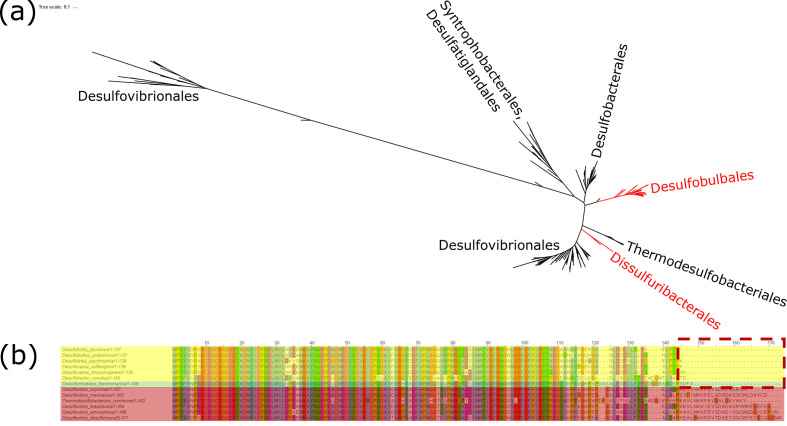
Fig. 2.
(a) Phylogeny of AprB proteins from members of the Desulfobacterota, with sequences from the Dissulfuribacterales (including Dissulfurirhabdus thermomarina ) and members of the Desulfobulbales highlighted in red. These groups include sulfur disproportionators that encode AprB proteins with a truncated tail, but they are not closely related and are separated by many lineages of non-disproportionating bacteria that encode full-length AprB proteins; therefore, it appears that these traits have convergently evolved in the two lineages. Scale bar represents average number of substitutions per site. (b) Multiple sequence alignment of AprB proteins showing the truncated tail (dashed red box) present in members of the Desulfobulbales (highlighted in yellow) and Dissulfurirhabdus thermomarina (highlighted in green), but not in other lineages such as Desulfovibrionales or Thermodesulfobacteriales (highlighted in red).
Dissulfurirhabdus thermomarina is capable of autotrophic growth [6] and encodes CO dehydrogenase/acetyl-CoA synthase, suggesting it makes use of the reductive acetyl-CoA (Wood–Ljungdahl) pathway like many sulfate-reducing bacteria [29]. The genome encodes a hydrogenase annotated by HydDB as a group 1 c NiFe hydrogenase associated with anaerobic respiratory uptake of H2. Dissulfurirhabdus thermomarina does not encode canonical proteins for aerobic respiration or denitrification, consistent with its inability to use O2 or nitrate as electron acceptors in culture [6]. However, this organism encodes a cytochrome bd O2 reductase; while these enzymes can in some cases be coupled to aerobic respiration (e.g. in Nitrospira) [30], they are often found in obligate anaerobes [31] in which they are likely used for O2 detoxification and oxidative stress tolerance [32].
Dissulfurirhabdus thermomarina is capable of disproportionating not only soluble sulfur species such as sulfite, but also insoluble elemental sulfur [6]. The mechanism of elemental sulfur disproportionation in Dissulfurirhabdus thermomarina and other strains capable of this metabolism is not known yet, but likely involves novel extracellular electron transfer pathways. Extracellular multihaem cytochrome proteins are commonly involved in respiratory electron transfer to insoluble mineral substrates [33, 34], and could, therefore, be expected to be involved in disproportionation of elemental sulfur. Analysis of the Dissulfurirhabdus thermomarina genome with CXXCH_finder [33] recovered 81 proteins with haem-binding domains, including one hypothetical protein with 26 haem-binding motifs – on par with proteins from bacteria such as Geobacter and Shewanella , well known for their capacity for extracellular electron transfer and ability to respire extracellular mineral substrates [33]. In addition to haem-binding motifs, this hypothetical protein also encodes seven cytochrome c 3 motifs associated with extracellular iron respiration in Shewanella [35]. This hypothetical protein shows low similarity to other proteins accessible on the NCBI database (<50 % to any sequence), but of all proteins from well-characterized organisms it is most similar to a hypothetical protein from Thermosulfidibacter takaii , a thermophilic bacterium capable of elemental sulfur reduction. The putative extracellular electron transfer protein from Dissulfurirhabdus thermomarina also has notable similarity (~30 %) to extracellular iron oxide respiratory system periplasmic decahaem cytochrome c protein components from Shewanella oneidensis MR-1, including the protein DsmE associated with respiration of extracellular substrates [36]. Therefore, we propose that Dissulfurirhabdus thermomarina utilizes extracellular multihaem cytochrome proteins related to those in dissimilatory iron-reducing bacteria in order to transfer electrons to insoluble substrates such as elemental sulfur. In contrast, members of the Desulfobulbales that are characterized as being capable of elemental sulfur disproportionation (e.g. Desulfobulbus propionicus , Desulfocapsa thiozymogenes ) encode proteins with no more than 11 or 12 CxxCH motifs. However, it remains possible that the capacity for extracellular electron transport has convergently evolved in diverse lineages of sulfur disproportionators, but has so far remained undiagnosed. It has recently been shown that some cable bacteria – members of the Desulfobulbales notable for their capacity for long distance and extracellular electron transfer – may also be capable of sulfur disproportionation [37]. It is therefore likely that the co-occurrence of sulfur disproportionation and extracellular electron transport metabolisms may have evolved independently in multiple lineages of sulfate-reducing bacteria, potentially utilizing diverse mechanisms.
Conclusions
In addition to better constraining the phylogenetic placement and taxonomy of this organism, the draft genome of Dissulfurirhabdus thermomarina provides evidence for the evolutionary history of metabolic pathways it employs. This includes the apparent convergent evolution for the capacity for sulfur disproportionation in different lineages of Desulfobacterota (e.g. Dissulfurirhabdus and members of Desulfobulbaceae ). These organisms appear to have evolved sulfur disproportionation via similar genetic mechanisms independently from an ancestral state of sulfate reduction. The presence of putative multihaem cytochrome proteins encoded by Dissulfurirhabdus thermomarina also suggests that the capacity for elemental sulfur disproportionation may in some cases utilize mechanisms homologous to those used in the respiration of extracellular electron acceptors such as metal oxides, as has been well studied in organisms such as Geobacter and Shewanella , demonstrating the versatility of micro-organisms to adapt proteins encoded in their genomes and acquired via horizontal gene transfer in order to develop novel metabolic traits.
Data Bibliography
Ward, LM, E Bertran, DT Johnston; NCBI BioProject; PRJNA579145 (2020).
Ward, LM, E Bertran, DT Johnston; NCBI WGS; JAAGRR000000000 (2020).
Ward, LM, E Bertran, DT Johnston; NCBI SRA; SRR11035951 (2020).
Funding information
L.M.W. acknowledges support from an Agouron Institute Postdoctoral Fellowship and a Simons Foundation Postdoctoral Fellowship in Marine Microbial Ecology. E.B. acknowledges the National Science Foundation (NSF) (EAR-1149555) and D.T.J. acknowledges the NASA (National Aeronautics and Space Administration) Exobiology (NNX15AP58G) for funding this work.
Acknowledgements
Genomic DNA for strain DSM100025 was acquired from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). Genome sequencing was provided by MicrobesNG (http://www.microbesng.uk), which is supported by the BBSRC (grant number BB/L024209/1).
Author contributions
L.M.W., E.B. and D.T.J. conceived of the study. L.M.W. and E.B. processed samples and analysed data. L.M.W., wrote the manuscript with assistance from E.B. and D.T.J. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviation: NCBI, National Center for Biotechnology Information.
All supporting data, code and protocols have been provided within the article or through supplementary data files.
References
- 1.Canfield DE, Thamdrup B. The production of 34S-depleted sulfide during bacterial disproportionation of elemental sulfur. Science. 1994;266:1973–1975. doi: 10.1126/science.11540246. [DOI] [PubMed] [Google Scholar]
- 2.Canfield DE, Teske A. Late proterozoic rise in atmospheric oxygen concentration inferred from phylogenetic and sulphur-isotope studies. Nature. 1996;382:127–132. doi: 10.1038/382127a0. [DOI] [PubMed] [Google Scholar]
- 3.Johnston DT, Farquhar J, Wing BA, Kaufman AJ, Canfield DE. Multiple sulfur isotope fractionations in biological systems: a case study with sulfate reducers and sulfur disproportionators. Am J Sci. 2005;305:645–660. doi: 10.2475/ajs.305.6-8.645. [DOI] [Google Scholar]
- 4.Finster K. Microbiological disproportionation of inorganic sulfur compounds. J Sulfur Chem. 2008;29:281–292. doi: 10.1080/17415990802105770. [DOI] [Google Scholar]
- 5.Anantharaman K, Hausmann B, Jungbluth SP, Kantor RS, Lavy A, et al. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018;12:1715–1728. doi: 10.1038/s41396-018-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slobodkina GB, Kolganova TV, Kopitsyn DS, Viryasov MB, Bonch-Osmolovskaya EA, et al. Dissulfurirhabdus thermomarina gen. nov., sp. nov., a thermophilic, autotrophic, sulfite-reducing and disproportionating deltaproteobacterium isolated from a shallow-sea hydrothermal vent. Int J Syst Evol Microbiol. 2016;66:2515–2519. doi: 10.1099/ijsem.0.001083. [DOI] [PubMed] [Google Scholar]
- 7.Bertran E, Ward LM, Johnston DT. Draft genome sequence of Acidianus ambivalens DSM 3772, an aerobic thermoacidophilic sulfur disproportionator. Microbiol Resour Announc. 2020;9:e01415-19. doi: 10.1128/MRA.01415-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertran E, Ward LM, Johnston DT. Draft genome sequence of Desulfofundulus thermobenzoicus subsp. thermosyntrophicus DSM 14055, a moderately thermophilic sulfate reducer. Microbiol Resour Announc. 2020;9:e01416-19. doi: 10.1128/MRA.01416-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward LM, Shih PM, Fischer WW. MetaPOAP: presence or absence of metabolic pathways in metagenome-assembled genomes. Bioinformatics. 2018;34:4284–4286. doi: 10.1093/bioinformatics/bty510. [DOI] [PubMed] [Google Scholar]
- 14.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 15.Søndergaard D, Pedersen CNS, Greening C. HydDB: a web tool for hydrogenase classification and analysis. Sci Rep. 2016;6:34212. doi: 10.1038/srep34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward LM, Idei A, Nakagawa M, Ueno Y, Fischer WW, et al. Geochemical and metagenomic characterization of Jinata Onsen, a Proterozoic-analog hot spring, reveals novel microbial diversity including iron-tolerant phototrophs and thermophilic lithotrophs. Microbes Environ. 2019;34:278–292. doi: 10.1264/jsme2.ME19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ward LM, Cardona T, Holland-Moritz H. Evolutionary implications of anoxygenic phototrophy in the bacterial phylum Candidatus Eremiobacterota (WPS-2) Front Microbiol. 2019;10:1658. doi: 10.3389/fmicb.2019.01658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller MA, Pfeiffer W, Schwartz T. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, LA. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees; pp. 1–8. pp. [Google Scholar]
- 23.Lemoine F, Domelevo Entfellner J-B, Wilkinson E, Correia D, Dávila Felipe M, et al. Renewing Felsenstein's phylogenetic bootstrap in the era of big data. Nature. 2018;556:452–456. doi: 10.1038/s41586-018-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doolittle RF. Of URFs and ORFs: a Primer on How to Analyze Derived Amino Acid Sequences. Mill Valley, CA: University Science Books; 1986. [Google Scholar]
- 26.Ward LM, Hemp J, Shih PM, McGlynn SE, Fischer WW. Evolution of phototrophy in the chloroflexi phylum driven by horizontal gene transfer. Front Microbiol. 2018;9:260. doi: 10.3389/fmicb.2018.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward LM, Johnston DT, Shih PM. Phanerozoic radiation of ammonia oxidizing bacteria. bioRxiv. 2020:655399. doi: 10.1038/s41598-021-81718-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertran E. Doctoral Thesis. Harvard University; 2019. Cellular and intracellular insights into microbial sulfate reduction and sulfur disproportionation. [Google Scholar]
- 29.Berg IA. Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–1936. doi: 10.1128/AEM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palomo A, Pedersen AG, Fowler SJ, Dechesne A, Sicheritz-Pontén T, et al. Comparative genomics sheds light on niche differentiation and the evolutionary history of comammox Nitrospira. ISME J. 2018;12:1779–1793. doi: 10.1038/s41396-018-0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ward LM, Hemp J, Pace LA, Fischer WW. Draft genome sequence of Leptolinea tardivitalis YMTK-2, a mesophilic anaerobe from the Chloroflexi class Anaerolineae. Genome Announc. 2015;3:e01356–15. doi: 10.1128/genomeA.01356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forte E, Borisov VB, Vicente JB, Giuffrè A. Advances in microbial physiology. Vol. 71. Academic Press; 2017. Cytochrome bd and gaseous ligands in bacterial physiology; pp. 171–234. [DOI] [PubMed] [Google Scholar]
- 33.McGlynn SE, Chadwick GL, Kempes CP, Orphan VJ. Single cell activity reveals direct electron transfer in methanotrophic consortia. Nature. 2015;526:531–535. doi: 10.1038/nature15512. [DOI] [PubMed] [Google Scholar]
- 34.Shi L, Dong H, Reguera G, Beyenal H, Lu A, et al. Extracellular electron transfer mechanisms between microorganisms and minerals. Nat Rev Microbiol. 2016;14:651–662. doi: 10.1038/nrmicro.2016.93. [DOI] [PubMed] [Google Scholar]
- 35.Gordon EH, Pike AD, Hill AE, Cuthbertson PM, Chapman SK, et al. Identification and characterization of a novel cytochrome c(3) from Shewanella frigidimarina that is involved in Fe(III) respiration. Biochem J. 2000;349:153–158. doi: 10.1042/bj3490153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bewley KD, Firer-Sherwood MA, Mock J-Y, Ando N, Drennan CL, et al. Mind the gap: diversity and reactivity relationships among multihaem cytochromes of the MtrA/DmsE family. Biochem Soc Trans. 2012;40:1268–1273. doi: 10.1042/BST20120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller H, Marozava S, Probst AJ, Meckenstock RU. Groundwater cable bacteria conserve energy by sulfur disproportionation. ISME J. 2020;14:623–634. doi: 10.1038/s41396-019-0554-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]