Abstract
Mobile genetic elements (MGEs) are agents of bacterial evolution and adaptation. Genome sequencing provides an unbiased approach that has revealed an abundance of MGEs in prokaryotes, mainly plasmids and integrative conjugative elements. Nevertheless, many mobilomes, particularly those from environmental bacteria, remain underexplored despite their representing a reservoir of genes that can later emerge in the clinic. Here, we explored the mobilome of the Mycobacteriaceae family, focusing on strains from Brazilian Atlantic Forest soil. Novel Mycolicibacterium and Mycobacteroides strains were identified, with the former ones harbouring linear and circular plasmids encoding the specialized type-VII secretion system (T7SS) and mobility-associated genes. In addition, we also identified a T4SS-mediated integrative conjugative element (ICEMyc226) encoding two T7SSs and a number of xenobiotic degrading genes. Our study uncovers the diversity of the Mycobacteriaceae mobilome, providing the evidence of an ICE in this bacterial family. Moreover, the presence of T7SS genes in an ICE, as well as plasmids, highlights the role of these mobile genetic elements in the dispersion of T7SS.
Keywords: Mycobacterium, Mycobacteriaceae, plasmid, ICE, T7SS, T4SS
Data Summary
Genomic data analysed in this work are available in GenBank and listed in Tables S1 and S2 (available in the online version of this article).
Impact Statement.
In addition to exploring the diversity of soil bacteria isolated from a niche in the Brazilian Atlantic Forest, which is one of the main biodiversity hotspots in the world, we revealed original aspects of Mycobacteriaceae mobilome, as we raised evidence of unique plasmids and the first T4SS-mediated integrative conjugative element (ICE) in this family. Moreover, type-VII secretion systems are part of the accessory genome of these mobilome elements (plasmids and ICE), which contribute to the understanding of Mycobacteriaceae evolution. We also found evidence of horizontal gene transfer within Mycobacteriaceae species in this niche.
Introduction
Bacterial evolution and adaptation are in part facilitated by the acquisition of genetic information by horizontal transfer. In this context, the mobilome represents the set of mobile genetic elements (e.g. integrons, plasmids, insertion sequences, transposons, phages, integrative conjugative elements, among others) that act intra- or inter-cellularly as vectors of gene mobility [1–4].
Conjugative elements, such as conjugative plasmids and integrative conjugative elements (ICEs), mediate their own transfer to other organisms, often carrying cargo genes. ICEs are therefore able to confer novel adaptive traits on their host cells and impact on their evolution [5–7]. These elements have a similar modular structure, clustering genes involved in the same biological function as maintenance, dissemination and regulation [3, 8]. However, while plasmids represent autonomously replicating elements, most ICEs depend on being integrated into a replicon to facilitate their inheritance [3, 9]. ICEs can be classified into two types, T4SS-mediated or AICE (actinomycete ICEs). While the former is characterized by a T4SS apparatus, relaxase and integrase, AICEs rely on a unique TraB protein (FtsK protein family) [10]. Generally, the integration of mobile elements leaves genomic marks, such as direct repeats, flanking the integrated element [11].
Mycobacteria belong to the Mycobacteriaceae family, which encompass ecologically, economically and clinically relevant organisms. Their members can be classified into fast- or slow-growing mycobacteria (the latter group containing the major pathogenic species), depending on the time of growth in the solid medium. Mycobacteria are flexible organisms that can inhabit a wide range of environments, including water bodies, soil, metalworking fluids, animals and humans [12, 13]. Among the mobilome elements of this family, thousands of bacteriophages have been isolated, and over 1700 have been sequenced [14]. Conversely, plasmids are supposed to be rare [15, 16]. So far, only dozens of plasmids have been characterized (https://www.ncbi.nlm.nih.gov/genome/plasmids), most of them in clinical strains [17–30], despite hundreds of species in the Mycobacteriaceae family. Plasmids have primarily been associated with the evolution and dissemination of a specialized secretion system, termed ESX or type-VII secretion system (T7SS), among the mycobacteria. This secretion apparatus consists of a complex of membrane and associated proteins. The T7SS is encoded by six paralogous loci (ESX-1,-2, -3,-4, -5 and -4-bis), each with variations in its genetic organization and function. While ESX-3 and ESX-4 loci are ubiquitously distributed in Mycobacteriaceae , ESX-2 and ESX-5 are only found in slow-growing mycobacteria, and ESX-1 distribution is variable. ESX-1 has been associated with virulence processes in slow-growing mycobacteria, while it has been reported to be involved, along with ESX-4, in horizontal gene transfer in some Mycolicibacterium species. ESX-5 has also been linked to virulence processes, in addition to playing a role in membrane integrity. ESX-3 is considered essential for the survival of mycobacteria, as it is involved with the uptake of iron and zinc. The function of the ESX-2 is still unknown [31–33]. Other mobilome elements, as the integrative conjugative elements, are also rare in this bacterial family. So far, AICEs, but not ICEs, have been reported in few Mycobacterium genomes [9, 34].
Here, we explored the mobilome of Mycobacteriaceae , focusing on plasmids and integrative elements based on metagenomes of soil strains from a low anthropogenic impacted region using in silico approaches. The analyses revealed circular and linear plasmids carrying genes resembling T7SS and T4SS. Moreover, a T4SS-mediated ICE, termed ICEMyc226, was identified and characterized in a Mycolicibacterium sp. strain from Atlantic Forest soil. This ICE harbours genes related to the metabolism of xenobiotics, amino acid and carbohydrate; besides encodes two ESX-systems with distinct origins (plasmid and chromosome).
Methods
Bacterial strains and growing conditions
The bacteria employed in this study encompassed 17 Mycobacteriaceae strains isolated from Atlantic Forest soils (CBMA strains) and deposited in the Bacteria Collection of Environment and Health (CBAS, Fiocruz Institute-Brazil). The CBMA strains were grown in Tryptic Soy Broth (TSB) medium up to 6 days at 23 °C.
Public data set
Representative complete/draft genomes (n=47) and plasmid sequences (n=87) of Mycobacteriaceae family, and Nocardia brasiliensis ATCC 700358 genome (NC_018681.1) were obtained from the National Center for Biotechnology Information (NCBI) public database (July 2019). The accession numbers for these Mycobacteriaceae genomes are supplied in Tables S1 and S2.
CBMA genome sequencing and assembly
In this study, we generated 17 Mycobacteriaceae genomes. The genomic DNA extraction was done using Purelink Genomic DNA Mini Kit (Invitrogen). The genome libraries were constructed using different single/paired-end libraries (Nextera, Truseq and Agilent) following each manufacturer’s instructions. The sequencing was performed on an Illumina Hiseq 2500 for most CBMA strains, generating reads of 100–150 bp length. The strain CBMA 360 was sequenced on 454 GS Junior, generating reads of ~500 bp length. The raw reads were filtered and trimmed (Phred quality score <20 and read length <30) using NGSQCToolkit v.2.3.3 [35] and Quake v.0.3 [36]. The genomes were de novo assembled with SPAdes v.3.5 or v.3.9 [37] and improved using Pilon [38].
Sequence annotation and species phylogeny
The genomes were annotated by Prokka v1.12 [39] and submitted to orthology analysis using GET_HOMOLOGUES v3.0.5 [40] considering a minimum coverage of >=70 % and identity >=40 %. The orthologous genes that represented the core genome of the data set were concatenated and submitted to phylogenetic analysis using RAxML v8.2.12 [41] with 100 bootstrap replicates. The iTOL generated the core genome tree [42] with Nocardia brasiliensis as outgroup. Type-II toxin-antitoxin loci were surveyed in the CBMA mobile elements using TAfinder [43]. Sequence repeats were identified by Unipro UGENE v1.32 with a window size of 100 bp [44], and the figures generated by EasyFig v2.2.2 [45]. Comparative analyses with integrative conjugative elements used the ICEberg database [34]. Functional annotation and assign of KEGG functional categories were performed using the BlastKOALA tool [46]. The metabolic pathway map was generated using iPath3.0: interactive pathways explorer v3 [47] with the KEGG data. The genomic atlas was generated using BRIG [48]. Searches for antibiotic resistance genes were based on The Comprehensive Antibiotic Resistance Database (https://card.mcmaster.ca/) [49]. The prediction of putative horizontal gene transfer (HGT) events was performed by Alien_hunter software v1.7 [50].
In silico approaches for plasmid detection
The plasmid detection in the CBMA genomes included some strategies: (i) blastn searches against NCBI plasmid database using the contigs of the CBMA genomes as queries; (ii) searches against CBMA genomes using hmm profiles of proteins associated with plasmid replication and transfer [9, 51]; (iii) topology inference, using overlapping ends of the contigs together with the paired-end reads that connect both ends of the sequences [52]; (iv) plasmidSPAdes software v.3.13, that predicts plasmids based on sequencing coverage using read sequences [53]; and (v) T7SS phylogeny, since chromosome- and plasmid-borne T7SS sequences evolve independently. Strategy (iii) made it possible to infer the circularity or linearity of the contigs.
T7SS detection and phylogeny
The T7SS core proteins (EccA, EccB, EccC, EccD, EccE and MycP) were searched based on their domains using HMMer package v3.1b2 [54]. ESX loci were selected if at least four core genes were close to each other. The identified protein sequences were aligned with MAFFT v7.310 [55] and the low-quality alignment columns removed using GUIDANCE2 v2.02 [56]. The resulting alignments were concatenated for phylogenetic analysis. A maximum-likelihood tree was generated using PhyML v3.1 [57] with LG+I+G+F substitution model and 100 bootstrap replicates.
Results
Here, to explore and characterize the mobilome of Atlantic Forest Mycobacteriaceae (CBMA strains), we sequenced their genomes and applied a set of in silico approaches.
Genus inference and T7SS phylogeny
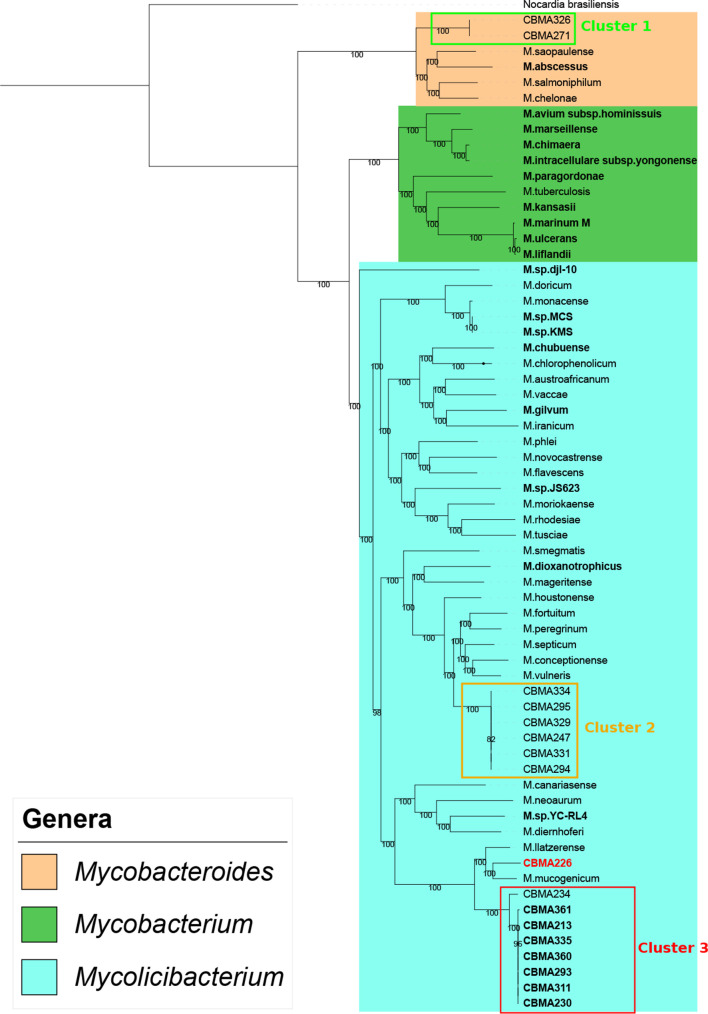
Firstly, we performed a phylogenetic analysis to infer the genus of the Mycobacteriaceae CBMA strains. Based on the Mycobacteriaceae core genome, it was revealed that 15/17 CBMA strains belong to the Mycolicibacterium genus, while the other two, to the Mycobacteroides genus (Fig. 1). With the exception of Mycolicibacterium sp. CBMA 226, the CBMA strains grouped into three clusters, each associated with known Mycobacteriaceae species: cluster 1, Mycobacteroides abscessus complex species; cluster 2, Mycolicibacterium fortuitum complex species; and cluster 3, Mycolicibacterium llatzerense and Mycolicibacterium mucogenicum group. The CBMA genomes of cluster 1 and 2 corresponded to a single lineage in their respective groups, while cluster 3 presented two lineages composed by (i) Mycolicibacterium sp. CBMA 234 and (ii) Mycolicibacterium sp. CBMA 213, 230, 293, 311, 335, 360 and 361 (Fig. 1).
Fig. 1.
Maximum-likelihood core-genome tree based on 408 concatenated genes (totalling 338 kb). The different Mycobacteriaceae genera are depicted by the coloured backgrounds. Genomes harbouring plasmids are in bold. The CBMA genomes are delimited by the clusters, and the CBMA 226 is labelled in red.
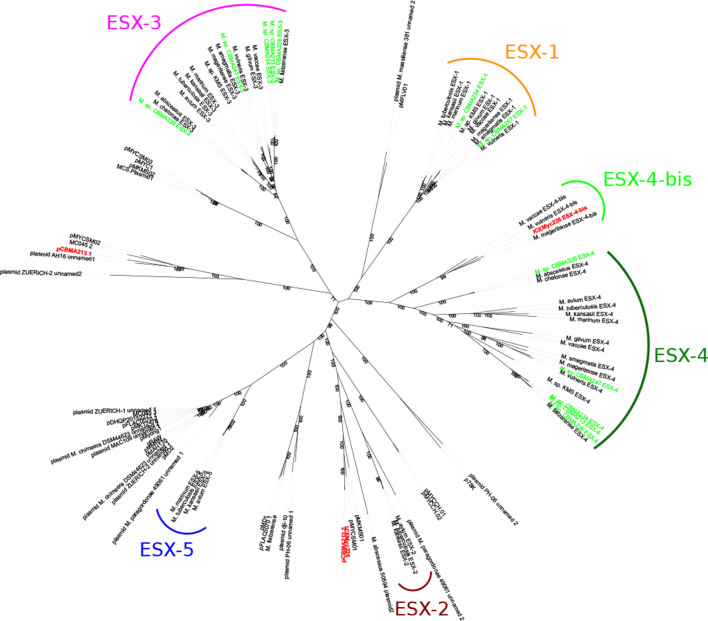
T7SS is a pivotal element in the survivor, communication and virulence of Mycobacteriaceae . Thus, we searched the genes resembling this apparatus in the CBMA genomes using hmm profiles and visual inspection. We identified clusters of ESX (T7SS) core genes in the chromosome of all CBMA strains. An ESX phylogeny was built using representatives of each CBMA lineage ( Mycolicibacterium sp. CBMA 213, 226, 234, 247 and Mycobacteroides sp. CBMA 326), as well as ESX sequences identified in other Mycobacteriaceae species and plasmids (Fig. 2). ESX-3 and ESX-4 were identified in all CBMA chromosomes, while ESX-1 occurred only in Mycolicibacterium sp. CBMA 234 and CBMA 247 (Fig. 2). Another aspect observed in the ESX phylogeny was the separation of chromosome- and plasmid-borne ESX-systems, with the latter branching at the root of the former, indicating independent evolutionary processes.
Fig. 2.
Maximum-likelihood tree of Mycobacteriaceae ESX loci based on concatenated T7SS core proteins. The coloured labels of the ESX types indicate the chromosome-borne ESX-systems, while others are plasmid-borne. The CBMA sequences are labelled in green (chromosome-borne) or red (mobile elements).
Mobilome identification
Using the search strategies, we identified three different plasmids (pCBMA213_1, pCBMA213_2 and pCBMA213_3) among genomes from cluster 3 (Fig. 1) and one integrative conjugative element (ICEMyc226) in Mycolicibacterium sp. CBMA 226 genome. Tables 1 and S3 summarize the major characteristics of each mobile element identified. Since plasmids were first identified in the Mycolicibacterium sp. CBMA 213, they were named referencing this genome and used as representative sequences in later analyses. All CBMA strains from cluster 3 (Fig. 1), except Mycolicibacterium sp. CBMA 234, presented at least one of the three plasmids in distinct combinations (Table 2). Each plasmid was identified as a single contig, except for pCBMA213_2 in Mycolicibacterium sp. CBMA 361, found fragmented into multiple contigs. In that case, we were unable to completely reconstruct the replicon. However, we observed that genes related to replication and mobility (repA and relaxase) showed 100 % identity in comparison to the pCBMA213_2 sequences of the other CBMA genomes, suggesting the presence of pCBMA213_2 in Mycolicibacterium sp. CBMA 361. Although some plasmids are shared by these strains, they are not identical, since there is a small variation in their length (1–6 %) and number of genes (Table S4), which may be due to the genome assembly/sequencing procedure or natural variability. Some details of the different mobile elements are discussed below.
Table 1.
Major features identified in the CBMA mobilome
|
Features |
pCBMA213_1 |
pCBMA213_2 |
pCBMA213_3 |
ICEMyc226 |
|---|---|---|---|---|
|
Size (bp) |
274 124 |
160 489 |
21 616 |
388 440 |
|
GC content |
62 % |
65 % |
64 % |
64 % |
|
# CDS |
328 |
161 |
27 |
364 |
|
Topology |
Linear |
Circular |
Linear |
Circular |
|
repA |
|
X |
X |
|
|
relaxase |
|
X |
|
X |
|
virD4 |
|
X |
|
X |
|
virB4 |
X |
X |
|
X |
|
tcpC |
|
X |
|
X |
|
Helicase |
X |
|
|
X |
|
DNA polymerase |
X |
|
|
|
|
Integrase |
X |
|
|
|
|
Transposase |
X |
X |
|
X |
|
tRNA genes |
X |
|
|
|
|
T7SS |
X |
X |
|
X |
|
Toxin-Antitoxin system |
X |
X |
|
X |
CDS, Coding DNA sequence.
Table 2.
Plasmid distribution in the CBMA strains. The colours represent strains from the same phylogenetic group
|
CBMA strains |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
213 |
226 |
230 |
234 |
247 |
271 |
293 |
294 |
295 |
311 |
326 |
329 |
331 |
334 |
335 |
360 |
361 |
|
|
pCBMA213_1 |
X |
X |
|||||||||||||||
|
pCBMA213_2 |
X |
X |
X |
X |
X* |
||||||||||||
|
pCBMA213_3 |
X |
X |
X |
X |
X |
X |
X |
||||||||||
*Present in multiple contigs.
The pCBMA213_1 plasmid pCBMA213_1 (GenBank accession number MF600313.1) is a megaplasmid composed of 274 124 base pairs with an average GC content of 62 %, presenting a linear topology (Tables 1 and S3). Like other linear plasmids, pCBMA213_1 presented terminal inverted repeats, composed by 145 bp length with 100 % identity (Fig. S1a, b). blast analysis showed that pCBMA213_1 had no extended sequence similarity with other known plasmids, presenting best hit with the unnamed plasmid 1 in Mycobacterium chimaera AH16 strain (coverage of 17 % and identity of 67 %). Most of these shared sequences coded for hypothetical and T7SS genes. Indeed, the T7SS phylogeny of pCBMA213_1 showed that it clustered with the unnamed 1 plasmid of M. chimaera strain, branching at the root of the ESX-3 tree, despite belonging to a different genus, which suggests mobility of this ESX system (Fig. 2). Gene content analysis of pCBMA213_1 revealed sequences that seem to be involved in DNA replication, such as a ribonuclease HI gene (B5P44_p00084), and a 10 kb region (11 to 21 kb) encoding DNA polymerase I (B5P44_p00015), DNA segregation ATPase (FtsK/SpoIIIE family)(B5P44_p00018), and ParA (B5P44_p00022) proteins. Although we could not find a rep gene, pCBMA213_1 also encoded a replicative DNA helicase (DnaB-like) (B5P44_p00269) with high similarity to helicase sequences from Mycolicibacterium sp. JS623 plasmid pMYCSM03 (94 % coverage and 75 % identity) and Mycobacterium chimaera strain ZUERICH-2 plasmid unnamed 2 (89 % coverage and 72 % identity). Interestingly, a ~89 kb region (169 352–258 453 bp), which encodes the 32 tRNA genes, is flanked by transposase genes with 90 % identity (B5P44_p00196 and B5P44_p00348) and inverted repeats (101 bp length with 99 % identity) (Fig. S1c, d). These tRNA genes were explored in a previous study [58]. Moreover, pCBMA213_1 also carries three Toxin-Antitoxin (TA) systems of different type-II TA families (Table S5).
The pCBMA213_2 plasmid pCBMA213_2 (GenBank accession number KY349138.1) is another megaplasmid, composed of 160 489 base pairs with an average GC content of 65 %, and unlike pCBMA213_1, its topology is circular. This plasmid is characterized by the presence of genes related to replication, mobility and conjugation, such as repA, relaxase, transposase, T4SS-like and T7SS genes (Tables 1 and S3). In addition, pCBMA213_2 also carries one TA system (Table S5). blast analysis showed similarity of the pCBMA213_2 sequence with Mycolicibacterium sp. JS623 plasmid pMYCSM01 (21 % coverage and 71 % identity) and Mycolicibacterium sp. KMS plasmid pMKMS01 (22 % coverage and 67 % identity). Like the pCBMA213_1 plasmid, the similarity of pCBMA213_2 with other plasmids is mainly due to the T7SS sequences. This can be seen in the T7SS phylogeny, where pCBMA213_2 branched at the root of the ESX-2 tree (Fig. 2). The analysis of pCBMA213_2 ESX system was presented in a previous study [29].
The pCBMA213_3 plasmid pCBMA213_3 (GenBank accession number MN587875) is the smallest plasmid found in the CBMA genomes, composed of 21 616 base pairs with an average GC content of 64 % (Table 1). Its topology is linear, with terminal inverted repeats of 489 bp length with 100 % identity (Fig. S2). pCBMA213_3 appears to be a cryptic plasmid capable of autonomous replication, since almost all predicted CDS were hypothetical, except for a putative rep gene (pCBMA213_3_00008). blast analysis showed that CBMA213_3 shows similarity to the sequences (encoding hypothetical genes) of two Mycobacterium plasmids: Mycobacterium sp. MOTT plasmid pM90 (17 % coverage and 68 % identity) and Mycobacterium celatum plasmid pCLP (19 % coverage and 69 % identity). These plasmids share similar characteristics with pCBMA213_3, since they are small (18 and 23 kb length), and plasmid pCLP has linear topology [20]. Curiously, pCBMA213_3 shares a gene encoding a DUF4189 domain-containing protein with pCBMA213_1 (respectively, pCBMA213_3_00027 and B5P44_p00076).
Characterization of an integrative conjugative element
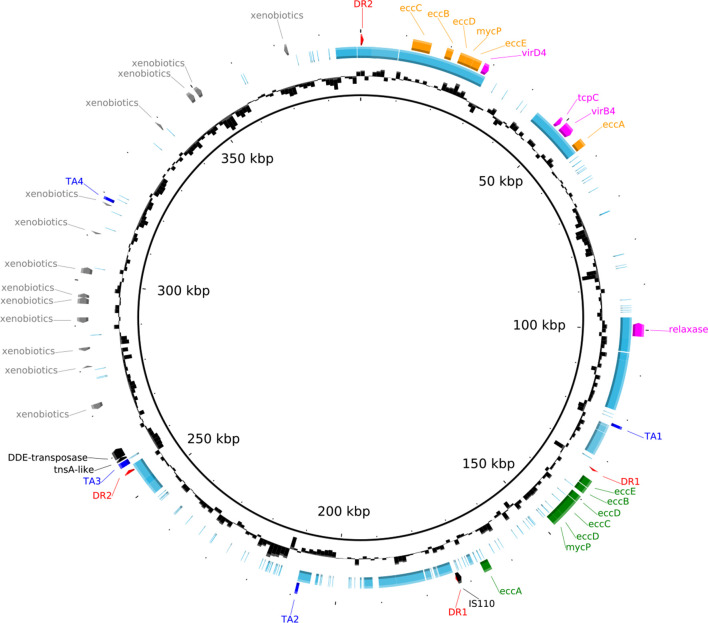
Analysing the Mycolicibacterium sp. CBMA 226 genome, we observed a single contig encoding a set of genes that characterize ICEs, including a relaxase (ICEMyc226_00083), T4SS-like genes (ICEMyc226_00024, ICEMyc226_00036, ICEMyc226_00039) and a DDE-type integrase/transposase/recombinase with the Rve protein domain of retrovirus integrases (ICEMyc226_00243; Tables 1 and S3). ICEMyc226 (GenBank accession number MN587876) is composed of 388 440 base pairs with an average GC content of 64 %, and encodes 364 CDS (Table 1). It was predicted as a circular molecule (Fig. 3), characterizing it as an ICE in its excised form. This element encoded a large number of genes related to partition and maintenance systems, including five genes that encode ParA/B-like family proteins, and four type II TA systems (Table S5). blastp analyses, using all predicted proteins encoded by ICEMyc226 (n=364), against the ICEberg database showed that 101 proteins showed similarities with other ICE proteins, including transposases, VirD4, metabolic and regulatory proteins (22–52 % identity). Interestingly, ICEMyc226 DDE-type integrase/transposase/recombinase showed similarity (~30 % identity) with an integrase of TR2 ICE (154 kb length) from Streptomyces scabiei 87.22, and of other ICEs from Proteobacteria (Fig. S3). No antibiotic resistance genes were identified in ICEMyc226.
Fig. 3.
Genomic map of ICEMyc226 highlighting major features and segments shared with pCBMA213_2 (light blue segments). The external coloured blocks represent the regions that encode for: Toxin-Antitoxin systems (dark blue: TA1, TA2, TA3, and TA4); each end of the direct repeats (red: DR1 and DR2); xenobiotic degrading genes (grey: xenobiotics); transposase elements (black: DDE-transposase, tnsA-like, and IS110); ESX-2-like genes (orange: eccC, eccB, eccD, mycP, eccE and eccA); T4SS-like genes (fuchsia: virD4, tcpC, virB4 and relaxase); ESX-4-bis genes (green: eccE, eccB, eccD, eccC, eccD, mycP, eccA). The second circle, from the inside out, represents the GC content of ICEMyc226.
ICEMyc226 encoded two ESX-systems, one branched at the root of the ESX-2 tree with other plasmid-borne ESX-systems, and the other clustered on ESX-4-bis with chromosome-borne ESX-systems (Fig. 2). When the ESX-4-bis sequences from ICEMyc226 and Mycolicibacterium mageritense were compared, the same ESX gene order was observed: eccE/eccB/eccD/eccC/eccD/mycP/-//-/eccA (eccA located ~17 kb downstream). Interestingly, the ESX-4-bis region of ICEMyc226 (~40 kb) is flanked by direct repeats of 167 bp with 100 % identity (Fig. 3 - DR1), and an IS110 transposase gene is found downstream this ESX locus, adjacent to the direct repeat (Fig. 3). The other ESX locus of ICEMyc226, ESX-2-like, is similar to the ESX of pCBMA213_2 (99 % coverage and 92 % identity). The synteny and identity of the genes of this ESX locus in both elements are conserved in relation to the ESX locus of Mycolicibacterium sp. KMS pMKMS01 plasmid: eccC/eccB/eccD/mycP/eccE/-//-/eccA (eccA downstream) (Fig. 2). In both ICEMyc226 and pCBMA213_2, the region between eccE and eccA encodes several genes, including the T4SS-like genes: virB4, virD4 and tcpC (virB8-like). This region is larger in ICEMyc226 due to an ~14 kb insertion, which encodes two PE-PGRS family protein PE_PGRS18, two-component LuxR family transcriptional regulator, an ATP-binding protein, and a hypothetical protein (Fig. 3).
Since pCBMA213_2 and ICEMyc226 shared a large syntenic block of genes, we performed a comparative analysis of them. pCBMA213_2 is 160 kb in length, and ~70 % of its content (~112 kb) is shared with segments of ICEMyc226 with ~90 % identity, which corresponds to ~30 % of the ICE size. The shared segments include genes associated with the conjugative machinery (T7SS, T4SS-like, and relaxase genes), flanked by direct repeats of 648 bp with 100 % identity (Fig. 3 - DR2) and adjacent to the DDE-type integrase/transposase/recombinase (Fig. 3). Notably, ICEMyc226 lacks the repA gene and 53 mostly hypothetical coding sequences that are present in pCBMA213_2.
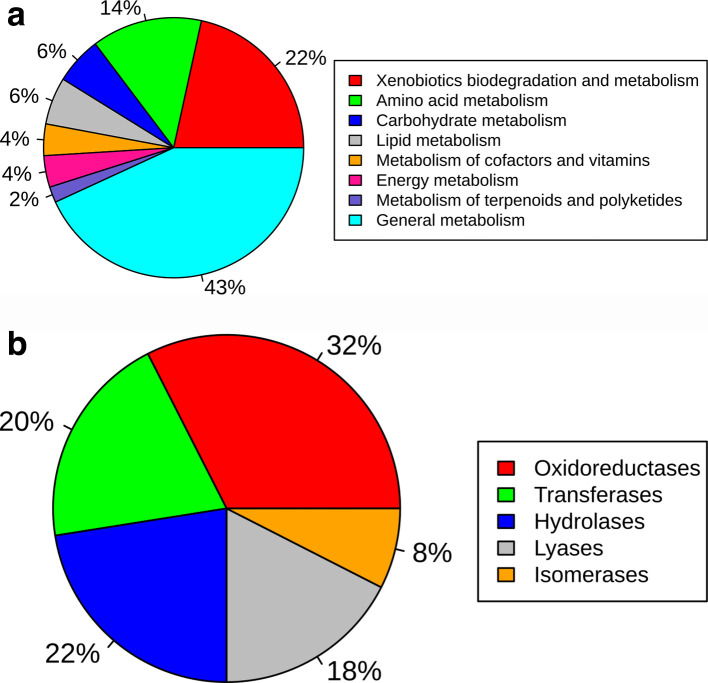
Gene content analysis of ICEMyc226 revealed that ~21 % of the coding sequences (78 CDS) presented similarity with Actinobacteria and Proteobacteria sequences, raising evidence that some genes were horizontally acquired. Moreover, Alien_Hunter software predicted that ~131 kb out of 388 kb (ICEMyc226 length) are related to putative HGT events (Table S6). The functional inference of the ICEMyc226 genome revealed 55 genes related to metabolic processes, such as metabolism of xenobiotics (Fig. 4a) and enzymatic functions (Fig. 4b). Most of the genes associated with xenobiotics metabolism were involved with benzoate (n=4) and styrene (n=3) degradation. A global overview of the metabolic pathways associated with ICEMyc226 genes allowed us to determine the range of processes that this element can influence (Fig. S4). Although ICEMyc226 lacks complete sets of genes needed to metabolize molecules independently of the host, depending on the environment, it could still metabolically complement its host.
Fig. 4.
Distribution of KEGG metabolic categories among ICEMyc226 CDS, including metabolic pathways (a) and enzymatic functions (b).
Discussion
The Mycobacteriaceae mobilome has been extensively explored in relation to bacteriophages [14]. However, to date, there is a lack of information regarding other mobile elements, such as plasmids and genomic islands. Even so, plasmids had a pivotal role in the evolution of this family, as they were involved in the diversification and mobility of T7SS [31–33]. Here, we explored the mobilome of Mycobacteriaceae strains from the soil of the Brazilian Atlantic Forest, which is one of the main biodiversity hotspots in the world and has a prevalence of Mycobacteriaceae [59, 60]. Interestingly, we identified a T4SS-mediated ICE, together with circular and linear plasmids in lineages from the Mycolicibacterium genus. These mobilome elements were restricted to only some of the lineages analysed (Fig. 1, Table 2). So far, only a few AICEs had been reported in genomes of bacteria from this genus [9, 34]. ICEMyc226, like ICEs from other bacterial families, has a backbone of characteristic genes [2, 3], besides two ESX-systems and a set of xenobiotic degrading genes. Interestingly, this is the first evidence of ESX-systems in other mobile elements, besides plasmids.
The CBMA plasmids are distinct from each other, showing differences in size, topology, replication system, conjugative trait, etc. The linear CBMA plasmids, pCBMA213_1 and pCBMA213_3, like that of other linear mycobacteria plasmids, had inverted terminal repeats [18, 25]. However, pCBMA213_1 seems to have a distinct replication system since it does not carry a rep gene [61]. Instead, pCBMA213_1 encoded a DnaB-like replicative helicase, which have been reported to act in the replication of Streptomyces linear plasmids and would characterize this plasmid as a replicon [62, 63]. Interestingly, despite its linear topology, pCBMA213_1 encodes colE1-like elements of theta-type circular plasmids, such as DNA polymerase and RNAse H [64, 65]. A relatively uncommon feature, namely a tRNA array (32 tRNA genes clustered in ~12 kb), had been previously characterized in this replicon [58]. In the case of the circular pCBMA213_2 replicon, it was possible to infer its conjugative nature due to the presence of the relaxase, T4SS and T7SS genes. This set of genes has been shown to play a role in Mycobacterium plasmid conjugation [27]. T7SS is found at one copy in several Actinobacteria plasmids [27, 28, 30–33]. In fact, pCBMA213_1 and pCBMA213_2 encoded ESX-3-like and ESX-2-like, respectively. These two ESX-systems carried by the CBMA plasmids have a close phylogenetic relationship with the ESX-systems of other distinct plasmids that are carried by several species from different niches (Fig. 2). The ESX-3 system is related to metal homeostasis, while the ESX-2 function is still unknown [33]. In addition to the chromosomally encoded ESX-3 and ESX-4 systems, the host strain has an extra ESX-3 copy, which would impact the bacterial fitness in the environment. These results reinforce the hypothesis of the mobilization of the ESX-system through mobile genetic elements, which have driven its evolution and mobility [31–33].
ICEMyc226 encodes an ESX-2-like system that is similar to the one encoded by pCBMA213_2 (Fig. 2). Interestingly, these elements belong to strains from different species (Fig. 1), indicating a horizontal gene transfer event. Both ICE and plasmid also share relaxase and T4SS genes with high identities (~90 %) which, in association with the ESX-2-like system, represent the ICEMyc226 conjugative module. Therefore, we hypothesized an ancestral fusion between pCBMA213_2 and an integrative mobilizable element (IME; e.g. genomic island), with subsequent recombination events, resulting in the current ICEMyc226. IMEs encode genes related to integration and excision and may hijack or subvert the mating apparatus of conjugative elements to promote their own transfer [3, 66–68]. The recombination of some mobile elements, including IMEs and ICEs, rely on DDE-transposases instead of integrases, and this type of enzyme may be associated with the conjugative apparatus of the mobile elements [2, 3, 10, 69–73]. Indeed, plasmids can become ICEs by acquiring integrases from other mobile elements [2, 74], and recombination among mobile elements seems to be frequent during the evolution of plasmids and ICEs [7, 75, 76]. Curiously, DDE-transposases are widespread in Gram-positive bacteria [77].
In addition to carrying the ESX-2-like, the ICEMyc226 also carries an ESX-4-bis. This finding is unique considering the mobile genetic elements of Mycobacteriaceae that harbour ESX-systems, since only one ESX-system was observed in plasmids. The ICEMyc226 ESX-4-bis has a close phylogenetic relationship with chromosome-borne ESX-4-bis of other strains (Fig. 2). This suggests a recent transmission event, probably mediated by IS110, within Mycobacteriaceae , which corroborates the transmissible character of T7SS among replicons [31–33].
Proteobacteria ICEs often carry accessory genes related to antibiotic resistance that impact the clinic [78, 79]. Here, we did not identify antibiotic resistance genes in ICEMyc226; instead, there is a prevalence of genes related to xenobiotics and amino acid metabolism. Indeed, ICEs are more likely to encode metabolism-related genes than antibiotic resistance genes [74]. Moreover, dozens of ICEMyc226 genes presented high identity with genes from non- Mycobacteriaceae bacteria, revealing the occurrence of HGT events between ICEMyc226 host and other soil bacteria genera. Indeed, HGT events have already been observed between Mycobacteriaceae and other genera of Actinobacteria and Proteobacteria [80].
Altogether, this study uncovers an aspect of the underexplored diversity of Mycobacteriaceae mobilome, showing evidence of unique plasmids and an ICE in its repertoire. In particular, the ICE appears to have been the result of a recombination between one of the identified plasmids and another mobile element. Also, this ICE encodes two ESX-systems, one of which presents evidence of mobilization. Thus, besides plasmids, other mobile genetic elements, such as ICEs, could have participated in the spread of T7SS in Mycobacteriaceae . The results presented here reiterate the need for studies with environmental samples to unravel the mobilome diversity of these organisms.
Data Bibliography
1. Raw reads have been deposited at the NCBI under the Bioproject number PRJNA344484.
2. Accession numbers of publicly available genomes used for the core genome multilocus sequence analysis are reported in Table S1.
3. Accession numbers of publicly available plasmids used for the comparative analysis are reported in Table S2.
Supplementary Data
Funding information
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior -Brasil (CAPES) - Finance Code 001, and Oswaldo Cruz Institute grants.
Acknowledgements
We are grateful for support from the Coordination for the Improvement of Higher Education Personnel (CAPES).
Author contributions
S.M.M. performed the in silico analysis, discussed the results and wrote the paper; A.C.P.V. conceived and supervised all steps of the study, discussed the results and wrote the paper.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AICE, actinomycete integrative conjugative element; CBAS, Bacteria Collection of Environment and Health; CBMA, Bacteria Collection of the Atlantic Forest; CDS, coding DNA sequence; DR, direct repeat; HGT, horizontal gene transfer; ICEs, integrative conjugative elements; IME, integrative mobilizable element; IS, insertion sequence; MGEs, mobile genetic elements; NCBI, National Center for Biotechnology Information; TA, toxin-antitoxin; TSB, Tryptic Soy Broth; T4SS, type-IV secretion system; T7SS, type-VII secretion system.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Six supplementary tables and four supplementary figures are available with the online version of this article.
References
- 1.Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 2.Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EPC. The repertoire of ice in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7:e1002222. doi: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guédon G, Libante V, Coluzzi C, Payot S, Leblond-Bourget N. The obscure world of integrative and mobilizable elements, highly widespread elements that Pirate bacterial conjugative systems. Genes. 2017;8:337. doi: 10.3390/genes8110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blesa A, Sánchez M, Sacristán-Horcajada E, González-de la Fuente S, Peiró R, et al. Into the Thermus mobilome: presence, diversity and recent activities of insertion sequences across Thermus spp. Microorganisms. 2019;7:25. doi: 10.3390/microorganisms7010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Getino M, de la Cruz F. Natural and artificial strategies to control the conjugative transmission of plasmids. Microbiol Spectr. 2018;6 doi: 10.1128/microbiolspec.MTBP-0015-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krupovic M, Makarova KS, Wolf YI, Medvedeva S, Prangishvili D, et al. Integrated mobile genetic elements in Thaumarchaeota. Environ Microbiol. 2019;21:2056–2078. doi: 10.1111/1462-2920.14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pesesky MW, Tilley R, Beck DAC. Mosaic plasmids are abundant and unevenly distributed across prokaryotic taxa. Plasmid. 2019;102:10–18. doi: 10.1016/j.plasmid.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 8.Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155:376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Ghinet MG, Bordeleau E, Beaudin J, Brzezinski R, Roy S, et al. Uncovering the prevalence and diversity of integrating conjugative elements in actinobacteria. PLoS One. 2011;6:e27846. doi: 10.1371/journal.pone.0027846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bordeleau E, Ghinet MG, Burrus V. Diversity of integrating conjugative elements in actinobacteria: coexistence of two mechanistically different DNA-translocation systems. Mob Genet Elements. 2012;2:119–124. doi: 10.4161/mge.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 12.Primm TP, Lucero CA. Falkinham JO 3rd. health impacts of environmental mycobacteria. Clin Microbiol Rev. 2004;17:98–106. doi: 10.1128/CMR.17.1.98-106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta RS, Lo B, Son J. Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera. Front Microbiol. 2018;9:67. doi: 10.3389/fmicb.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentile GM, Wetzel KS, Dedrick RM, Montgomery MT, Garlena RA, et al. More evidence of Collusion: a new Prophage-Mediated viral defense system encoded by mycobacteriophage Sbash. mBio. 2019;10:e00196–19. doi: 10.1128/mBio.00196-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shintani M, Sanchez ZK, Kimbara K. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol. 2015;6:242. doi: 10.3389/fmicb.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray TA, Derbyshire KM. Blending genomes: distributive conjugal transfer in mycobacteria, a sexier form of HGT. Mol Microbiol. 2018;108:601–613. doi: 10.1111/mmi.13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labidi A, Mardis E, Roe BA, Wallace RJ. Cloning and DNA sequence of the Mycobacterium fortuitum var fortuitum plasmid pAL5000. Plasmid. 1992;27:130–140. doi: 10.1016/0147-619x(92)90013-z. [DOI] [PubMed] [Google Scholar]
- 18.Picardeau M, Vincent V. Characterization of large linear plasmids in mycobacteria. J Bacteriol. 1997;179:2753–2756. doi: 10.1128/jb.179.8.2753-2756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bachrach G, Colston MJ, Bercovier H, Bar-Nir D, Anderson C, et al. A new single-copy mycobacterial plasmid, pMF1, from Mycobacterium fortuitum which is compatible with the pAL5000 replicon. Microbiology. 2000;146 (Pt 2:297–303. doi: 10.1099/00221287-146-2-297. [DOI] [PubMed] [Google Scholar]
- 20.Le Dantec C, Winter N, Gicquel B, Vincent V, Picardeau M. Genomic sequence and transcriptional analysis of a 23-kilobase mycobacterial linear plasmid: evidence for horizontal transfer and identification of plasmid maintenance systems. J Bacteriol. 2001;183:2157–2164. doi: 10.1128/JB.183.7.2157-2164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirby C, Waring A, Griffin TJ, Grindley NDF, Grindley NDF, et al. Cryptic plasmids of Mycobacterium avium: Tn552 to the rescue. Mol Microbiol. 2002;43:173–186. doi: 10.1046/j.1365-2958.2002.02729.x. [DOI] [PubMed] [Google Scholar]
- 22.Stinear TP, Mve-Obiang A, Small PLC, Frigui W, Pryor MJ, et al. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc Natl Acad Sci U S A. 2004;101:1345–1349. doi: 10.1073/pnas.0305877101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stinear TP, Pryor MJ, Porter JL, Cole ST. Functional analysis and annotation of the virulence plasmid pMUM001 from Mycobacterium ulcerans. Microbiology. 2005;151:683–692. doi: 10.1099/mic.0.27674-0. [DOI] [PubMed] [Google Scholar]
- 24.Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, et al. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One. 2009;4:e5660. doi: 10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabello MCdaS, Matsumoto CK, Almeida LGPde, Menendez MC, Oliveira RSde, et al. First description of natural and experimental conjugation between mycobacteria mediated by a linear plasmid. PLoS One. 2012;7:e29884. doi: 10.1371/journal.pone.0029884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leão SC, Matsumoto CK, Carneiro A, Ramos RT, Nogueira CL, et al. The detection and sequencing of a broad-host-range conjugative IncP-1β plasmid in an epidemic strain of Mycobacterium abscessus subsp. bolletii. PLoS One. 2013;8:e60746. doi: 10.1371/journal.pone.0060746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ummels R, Abdallah AM, Kuiper V, Aâjoud A, Sparrius M, et al. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio. 2014;5:e01744–14. doi: 10.1128/mBio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uchiya K-ichi, Takahashi H, Nakagawa T, Yagi T, Moriyama M, et al. Characterization of a novel plasmid, pMAH135, from Mycobacterium avium subsp. hominissuis. PLoS One. 2015;10:e0117797. doi: 10.1371/journal.pone.0117797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgado SM, Marín MA, Freitas FS, Fonseca EL, Vicente ACP. Complete plasmid sequence carrying type IV-like and type VII secretion systems from an atypical mycobacteria strain. Mem Inst Oswaldo Cruz. 2017;112:514–516. doi: 10.1590/0074-02760160546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim B-J, Cha G-Y, Kim B-R, Kook Y-H, Kim B-J. Insights From the Genome Sequence of Mycobacterium paragordonae, a Potential Novel Live Vaccine for Preventing Mycobacterial Infections: The Putative Role of Type VII Secretion Systems for an Intracellular Lifestyle Within Free-Living Environmental Predators. Front Microbiol. 2019;10:1524. doi: 10.3389/fmicb.2019.01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumas E, Christina Boritsch E, Vandenbogaert M, Rodríguez de la Vega RC, Thiberge J-M, et al. Mycobacterial pan-genome analysis suggests important role of plasmids in the radiation of type VII secretion systems. Genome Biol Evol. 2016;8:387–402. doi: 10.1093/gbe/evw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newton-Foot M, Warren RM, Sampson SL, van Helden PD, Gey van Pittius NC. The plasmid-mediated evolution of the mycobacterial Esx (type VII) secretion systems. BMC Evol Biol. 2016;16:62. doi: 10.1186/s12862-016-0631-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mortimer TD, Weber AM, Pepperell CS. Evolutionary thrift: mycobacteria Repurpose plasmid diversity during adaptation of type VII secretion systems. Genome Biol Evol. 2017;9:398–413. doi: 10.1093/gbe/evx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu M, Li X, Xie Y, Bi D, Sun J, et al. Iceberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 2019;47:D660–D665. doi: 10.1093/nar/gky1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel RK, Jain M. Ngs Qc toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelley DR, Schatz MC, Salzberg SL. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 2010;11:R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;15;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 40.Contreras-Moreira B, Vinuesa P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol. 2013;79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I, Bork P. Interactive tree of life (iTOL) V3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie Y, Wei Y, Shen Y, Li X, Zhou H, et al. TADB 2.0: an updated database of bacterial type II toxin-antitoxin loci. Nucleic Acids Res. 2018;46:D749–D753. doi: 10.1093/nar/gkx1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okonechnikov K, Golosova O, Fursov M, team U. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Darzi Y, Letunic I, Bork P, Yamada T. iPath3.0: interactive pathways explorer V3. Nucleic acids research. 2018;46:W510–W513. doi: 10.1093/nar/gky299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. Blast ring image generator (BRIG): simple prokaryote genome comparisons. BMC genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vernikos GS, Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;15;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Xie Y, Liu M, Tai C, Sun J, et al. oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018;46:W229–W234. doi: 10.1093/nar/gky352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jørgensen TS, Xu Z, Hansen MA, Sørensen SJ, Hansen LH. Hundreds of circular novel plasmids and DNA elements identified in a rat cecum metamobilome. PloS one. 2014;9:e87924. doi: 10.1371/journal.pone.0087924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, et al. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. 2016;32:3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 54.Eddy SR. Accelerated profile HMM searches. PLOS Comp. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K, Standley DM. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 2014;1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- 56.Sela I, Ashkenazy H, Katoh K, Pupko T. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015;43:W7–W14. doi: 10.1093/nar/gkv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 58.Morgado SM, Vicente A. Beyond the limits: tRNA array units in Mycobacterium genomes. Frontiers in microbiology. 2018;9:1042. doi: 10.3389/fmicb.2018.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lambais MR, Crowley DE, Cury JC, Büll RC, Rodrigues RR. Bacterial diversity in tree canopies of the Atlantic forest. Science. 2006;312:1917. doi: 10.1126/science.1124696. [DOI] [PubMed] [Google Scholar]
- 60.Bruce T, Martinez IB, Maia Neto O, Vicente AC, Kruger RH, et al. Bacterial community diversity in the Brazilian Atlantic forest soils. Microb Ecol. 2010;60:840–849. doi: 10.1007/s00248-010-9750-2. [DOI] [PubMed] [Google Scholar]
- 61.Picardeau M, Le Dantec C, Vincent V. Analysis of the internal replication region of a mycobacterial linear plasmid. Microbiology. 2000;146:305–313. doi: 10.1099/00221287-146-2-305. [DOI] [PubMed] [Google Scholar]
- 62.Scherzinger E, Ziegelin G, Bárcena M, Carazo JM, Lurz R, et al. The RepA protein of plasmid RSF1010 is a replicative DNA helicase. J Biol Chem. 1997;272:30228–30236. doi: 10.1074/jbc.272.48.30228. [DOI] [PubMed] [Google Scholar]
- 63.Ahsan S, Kabir MS. Linear plasmids and their replication. Stamford J Microbiol. 2013;2:1–5. [Google Scholar]
- 64.del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lilly J, Camps M. Mechanisms of theta plasmid replication. Microbiology spectrum. 2015;3:PLAS-0029–2014. doi: 10.1128/microbiolspec.PLAS-0029-2014. [DOI] [PubMed] [Google Scholar]
- 66.Antonenka U, Nölting C, Heesemann J, Rakin A. Horizontal transfer of Yersinia high-pathogenicity island by the conjugative RP4 attB target-presenting shuttle plasmid. Mol Microbiol. 2005;57:727–734. doi: 10.1111/j.1365-2958.2005.04722.x. [DOI] [PubMed] [Google Scholar]
- 67.Doublet B, Boyd D, Mulvey MR, Cloeckaert A. The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol. 2005;55:1911–1924. doi: 10.1111/j.1365-2958.2005.04520.x. [DOI] [PubMed] [Google Scholar]
- 68.Douard G, Praud K, Cloeckaert A, Doublet B. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PloS one. 2010;5:e15302. doi: 10.1371/journal.pone.0015302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guérillot R, Siguier P, Gourbeyre E, Chandler M, Glaser P. The diversity of prokaryotic DDE transposases of the mutator superfamily, insertion specificity, and association with conjugation machineries. Genome Biol Evol. 2014;6:260–272. doi: 10.1093/gbe/evu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ambroset C, Coluzzi C, Guédon G DMD, Loux V, et al. New insights into the classification and integration specificity of Streptococcus integrative conjugative elements through extensive genome exploration. Front Microbiol. 2016;6:1483. doi: 10.3389/fmicb.2015.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delavat F, Miyazaki R, Carraro N, Pradervand N, van der Meer JR. The hidden life of integrative and conjugative elements. FEMS Microbiol Rev. 2017;41:512–537. doi: 10.1093/femsre/fux008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sansevere EA, Luo X, Park JY, Yoon S, Seo KS, et al. Transposase-Mediated excision, conjugative transfer, and diversity of ICE6013 elements in Staphylococcus aureus . J Bacteriol Res. 2017;199:e00629–16. doi: 10.1128/JB.00629-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu D, Wan J, Yang Z, Xu J, Wang M, et al. First report of integrative conjugative elements in Riemerella anatipestifer isolates from ducks in China. Front Vet Sci. 2019;6:128. doi: 10.3389/fvets.2019.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cury J, Oliveira PH, de la Cruz F, Rocha E. Host range and genetic plasticity explain the co-existence of integrative and extrachromosomal mobile genetic elements. Mol Biol Evol. 2018;35:2230–2239. doi: 10.1093/molbev/msy123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, et al. Comparative ice genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS genetics. 2009;5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hülter N, Ilhan J, Wein T, Kadibalban AS, Hammerschmidt K, et al. An evolutionary perspective on plasmid lifestyle modes. Curr Opin Microbiol. 2017;38:74–80. doi: 10.1016/j.mib.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 77.Brochet M, Cunha D, V, Couvé E, Rusniok C, Trieu-Cuot P, et al. Atypical association of DDE transposition with conjugation specifies a new family of mobile elements. Mol Microbiol. 2009;71:948–959. doi: 10.1111/j.1365-2958.2008.06579.x. [DOI] [PubMed] [Google Scholar]
- 78.Johnson CM, Grossman AD. Integrative and conjugative elements (ICEs): what they do and how they work. Annual review of genetics. 2015;49:577–601. doi: 10.1146/annurev-genet-112414-055018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clinical microbiology reviews. 2018;31:e00088–17. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Panda A, Drancourt M, Tuller T, Pontarotti P. Genome-Wide analysis of horizontally acquired genes in the genus Mycobacterium. Scientific reports. 2018;8:14817. doi: 10.1038/s41598-018-33261-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.