Abstract
Background
Circular RNA ciRS-7 has been reported to be involved in the progression of various cancers. However, ciRS-7 expression and its role in clear cell renal cell carcinoma (ccRCC) progression remains unclear. This study aimed to investigate the effect of ciRS-7 expression on ccRCC and the related signaling pathway.
Methods
ciRS-7 expression was analyzed using quantitative reverse transcription polymerase chain reaction in 87 pairs of ccRCC and matched adjacent normal tissues. The role of ciRS-7 in ccRCC cell proliferation and invasion was determined using the cell counting kit-8 and invasion assays, respectively. Potential mechanisms underlying the role of ciRS-7 in promoting ccRCC progression were explored by Western blotting. The relationship between the expression of ciRS-7 and features of ccRCC was analyzed by the Chi-square test and progression-free survival was determined using a Kaplan-Meier plot.
Results
ciRS-7 was overexpressed in ccRCC tissues compared with that in matched adjacent normal tissues. In addition, ciRS-7 up-regulation was closely associated with tumor diameter (P = 0.050), clinical stage (P = 0.009), and distant metastasis (P = 0.007). ciRS-7 knockdown in 786O and 769P cells markedly inhibited their proliferative and invasive abilities. In addition, ciRS-7 inhibition reduced phosphorylated epidermal growth factor receptor (p-EGFR) and phosphorylated serine/threonine kinase (p-Akt) levels.
Conclusions
ciRS-7 up-regulation could promote ccRCC cell proliferation and invasion, which may be related with the EGFR/Akt signaling pathway. ciRS-7 might be a potential ccRCC therapeutic target.
Keywords: ciRS-7, Clear cell renal cell carcinoma, Proliferation, Epidermal growth factor receptor, Signaling pathway
Introduction
Clear cell renal cell carcinoma (ccRCC) remains a health burden owing to its tendency to relapse and resistance to chemotherapy and targeted therapy.[1,2] In addition to risk factors such as obesity, tobacco use, and hypertension, genetic mutations and abnormal post-translational protein modifications play crucial roles in the genesis and progression of tumors.[3,4] For instance, von Hippel-Lindau (VHL) gene mutations are common in majority of patients with ccRCC, and these mutations lead to ccRCC metastasis by promoting angiogenesis.[5] Over the past years, although detailed understanding of ccRCC pathogenesis has contributed to the development of novel therapeutic agents, optimization of treatment options, and improvement in patient survival, there remains a lack of efficient systemic therapies for advanced ccRCC. Therefore, a new perspective of research on tumorigenesis will probably shed light on novel therapeutic approaches for this malignancy.
Circular RNA (circRNA) — a type of non-coding RNA — stably exists in cells as it is not easily degraded owing to its specific closed-loop structure.[6] Over the past few years, application of novel technologies has helped discover various circRNAs expressed in different cells and tissues, where they play vital roles in multiple biological processes. For instance, circRNAs have been implicated in cell proliferation and angiogenesis.[7–9] Recently, studies have revealed an increasing number of circRNAs that are abnormally expressed in and linked to many human diseases, including cancer.[10,11] ciRS-7 that also known as cerebellar-degeneration-related protein 1 antisense RNA has been reported to inhibit tumor suppressor miR-7 and act as an oncogene involved in the progression of various cancers.[12–15] However, ciRS-7 expression in ccRCC and its role in cancer progression remain unclear. Thus, in the present study, we assessed ciRS-7 expression and its role in ccRCC and explored the potential mechanisms through which ciRS-7 promotes ccRCC progression.
Methods
Ethical approval
Participants were informed as to the purpose of the study and provided written informed consent before nephrectomy. In addition, the current study was reviewed and verified by the Ethical Reviewing Committee of Qingdao Central Hospital.
Study design
ciRS-7 expression was analyzed using quantitative reverse transcription polymerase chain reaction (qPCR) in 87 pairs of ccRCC and matched adjacent normal tissues. The role of ciRS-7 in ccRCC cell proliferation and invasion was determined using the cell counting kit-8 (CCK-8) and invasion assays, respectively. Potential mechanisms underlying the role of ciRS-7 in promoting ccRCC progression were explored by Western blotting.
Selection and description of participants
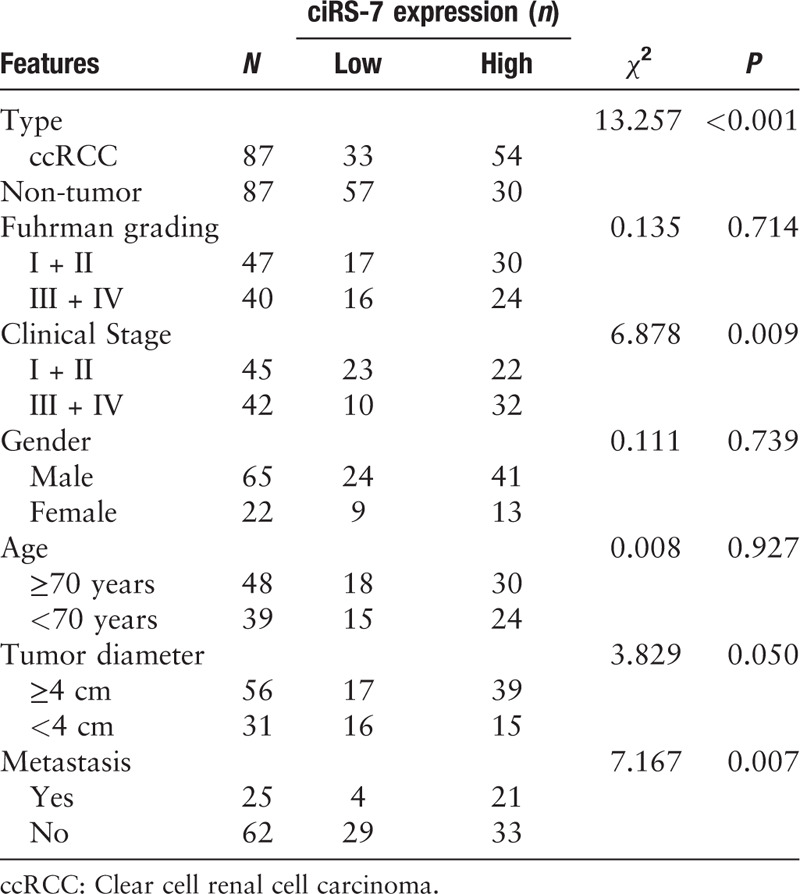
A total of 87 patients diagnosed with ccRCC from 2013 to 2017 in the Qingdao Central Hospital participated in this study. Both ccRCC and matched adjacent normal tissues were immediately collected and stored at –80°C until further analysis. The expression of ciRS-7 was measured in 87 pairs of ccRCC and normal samples. Patient characteristics are summarized in Table 1. Patients were followed up every 6 months by the telephone. The last time of follow-up was October 31, 2019.
Table 1.
Relationship between ciRS-7 expression and clinicopathological features of ccRCC patients.
Cell culture
A normal renal tubular epithelial cell line (HK-2) and two ccRCC cell lines (786O and 769P) were used in the current study and were obtained from the American type culture collection. All cell lines were cultured in complete RPMI-1640 media supplemented with 10% fetal bovine serum (FBS; Invitrogen, Massachusetts, USA) and 100 IU/mL penicillin (Solarbio, Beijing, China). The cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
qPCR
Total RNA was extracted using the TRIzol reagent (Solarbio). A high-capacity complementary DNA (cDNA) reverse transcription kit was then utilized to reverse transcribe the total RNA into cDNA. The tissue and cellular expression of ciRS-7 was analyzed by qPCR using the Fast SYBR Green Master Mix. The cDNA Reverse Transcription Kit and the Fast SYBR Green Master Mix were both purchased from Applied Biosystems (Foster City, CA, USA). For ciRS-7 expression analysis, β-actin expression was used as an internal control. The expression of ciRS-7 was evaluated using the 2ΔΔCt method. The primers used for ciRS-7 and β-actin were as follows: ciRS-7, Forward primer (F: 5′–3′): TCAACTGGCTCAATATCCATGTC, Reverse primer (R: 5′–3′): ACCTTGACACAGGTGCCAT; β-actin (F: 5′–3′): GTGGCCGAGGACTTTGATTG, (R: 5′–3′): CCTGTAACAACGCATCTCATATT. All experiments were performed in triplicate.
Cell transfection
To silence ciRS-7 expression, two small interfering RNAs (siRNAs) (siRNA1 and siRNA2) targeting ciRS-7 and the negative control (siNC) were designed and synthesized by the GenePharm Company (Suzhou, China). siRNAs were then transiently transfected into cells using Lipofectamine 2000 (Invitrogen). Further assays were performed after the transfected cells were cultured for 48 h. The sequences were as follows: siRNA1: 5′-GCACCTGTGTCAAGGTCTTTT-3′; siRNA2: 5′-CTGTTCAGAGTGGATCGTTT-3′; siNC: 5′-TTCTCCGAACGTGTCACGTTT-3′.
CCK-8 assay
To assess the initial effect of ciRS-7 on cell proliferation, we conducted the CCK-8 assay. Briefly, 1500 cells in 200 μL media with 10% FBS were seeded into 96-well plates and cultured and the absorbance value at 450 nm (A450nm) was measured at different time points (0, 24, 48, 72, and 96 h). An aliquot of CCK-8 solution (10 μL) (Dojindo, Japan) was added to the plates at each time point and incubated at 37°C for 2 h, followed by the detection of the A450nm. All experiments were performed in triplicate.
Transwell invasion assays
To evaluate the influence of ciRS-7 on cell invasion, transwell invasion assays were used. The transwell chambers used in the present study were coated with matrigel (8 μm pore size; Corporation, MA, USA). Briefly, 7 × 104 cells in 150 μL media without FBS were seeded into the upper chamber and 650 μL complete media (without cells) was used in the lower chamber. The cells were incubated for 24 h and the cells that invaded the chamber were collected. The invaded cells were fixed in 4% paraformaldehyde for 10 min and the fixed cells were subsequently stained with crystal violet for 15 min. Invasive cells were calculated and photographed from five random fields at a magnification of 100 times (×100). All experiments were performed at least three times.
Western blotting
Cellular proteins were extracted using radio immunoprecipitation assay buffer, and the protein concentration was measured using a bicinchoninic acid protein assay kit (Solarbio). Proteins were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (12%). After being transferred onto nitrocellulose membranes, the membranes were blocked using 5% evaporated milk for 2 h at room temperature (RT). The membranes were then incubated with primary antibodies at 4°C overnight. The membranes were then washed with Tris-buffered saline with Tween-20 and subsequently incubated with the corresponding secondary antibodies for 1.5 h at RT. Protein bands were detected using the electrochemiluminescence reagent and captured. β-actin was used as the control.
Statistical analysis
SPSS 19.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 5.0 (GraphPad Software Co., Ltd, San Diego, CA, USA) were used for all statistical analyses. Data are present as the mean ± standard deviation. The Student's t test was applied to determine the differences between two groups. The Chi-square test was used to analyze the relationship between the expression of ciRS-7 and features of ccRCC. Two-way analysis of variance was used to analyze cell proliferation in the different groups. Progression-free survival (PFS) was determined using a Kaplan-Meier plot and the difference between the groups was determined by the log-rank test. A P < 0.05 was considered to be significant.
Results
ciRS-7 was up-regulated in ccRCC
To determine the expression of ciRS-7 in ccRCC, we initially performed qPCR analysis to detect ciRS-7 expression in 786O 769P, and HK-2 cells. ciRS-7 expression in both 786O and 769P cells was higher than that in HK-2 cells, suggesting a higher expression of ciRS-7 in ccRCC [Figure 1A]. Subsequently, ciRS-7 expression was analyzed in paired tumor and adjacent normal tissues using qPCR analysis and higher ciRS-7 expression in ccRCC tissues was identified [Figure 1B]. Collectively, these results demonstrated the up-regulation of ciRS-7 in ccRCC.
Figure 1.
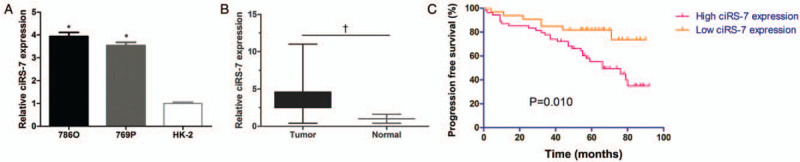
ciRS-7 was overexpressed in ccRCC. (A) The expressions of ciRS-7 in both 786O and 769P cells were higher than that observed in HK-2 cells. (B) qPCR analysis showed that ciRS-7 was up-regulated in ccRCC tissues. (C) Kaplan-Meier plot of patients in the high or low ciRS-7 expression group. ∗P < 0.01, +P < 0.001. ccRCC: Clear cell renal cell carcinoma; qPCR: Quantitative reverse transcription polymerase chain reaction.
ciRS-7 was associated with advanced pathological features in patients with ccRCC
To determine the relationship between ciRS-7 and certain pathological features of ccRCC, ciRS-7 expression in all the patients with ccRCC was categorized as high or low expression. The median expression value was then used as a threshold as previously demonstrated.[16] Statistical analyses revealed that ciRS-7 expression was significantly higher in advanced clinical stage (III + IV) patients than in low clinical stage (I + II, χ2 = 13.257, P = 0.009) patients. Furthermore, the expression of ciRS-7 was correlated with tumor diameter (χ2 = 3.829, P = 0.050) and distant metastasis (χ2 = 7.176, P = 0.007). However, ciRS-7 expression was not associated with other characteristics such as gender, age, and Fuhrman grading [Table 1]. The Kaplan-Meier plot demonstrated a shorter PFS in patients with high ciRS-7 expression compared with those with low ciRS-7 expression [Figure 1C].
ciRS-7 silencing significantly inhibited ccRCC cell proliferation
To evaluate the influence of ciRS-7 on cell proliferation, ciRS-7 was silenced using siRNAs targeting it. We first confirmed the silencing efficiency of this process [Figure 2A]. As shown in Figure 2B, the A450nm values of the cells transfected with the siRNAs were lower than those transfected with siNC, indicating that the silencing of ciRS-7 significantly suppressed the proliferative ability of the ccRCC cells.
Figure 2.
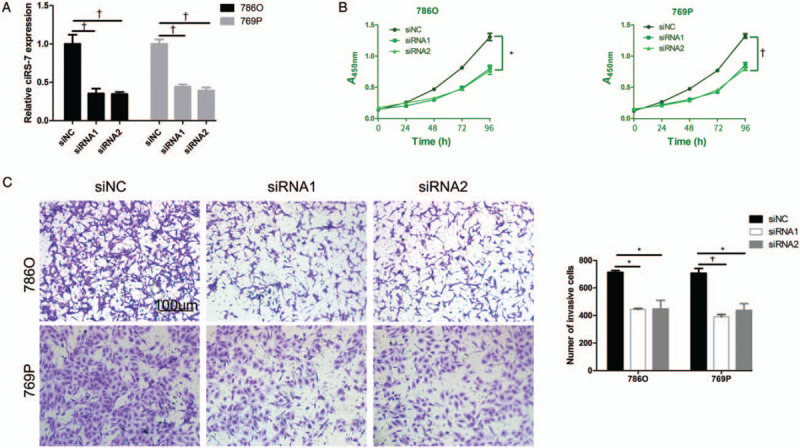
ciRS-7 suppression inhibited ccRCC cell proliferation and invasion. (A) The silencing efficiency of ciRS-7 expression in 786O and 769P cells using siRNAs was verified using qPCR. (B) CCK-8 assays demonstrated that the proliferation of 786O and 769P cells transfected with siRNAs was inhibited. (C) After transfection with the siRNAs, the numbers of invasive 786O and 769P cells were notably increased compared with control. The invaded cells were fixed in 4% paraformaldehyde for 10 min and the fixed cells were subsequently stained with crystal violet for 15 min (original magnification ×100). ∗P < 0.01, +P < 0.001. ccRCC: Clear cell renal cell carcinoma; CCK-8: Cell counting kit-8; qPCR: Quantitative reverse transcription polymerase chain reaction; siRNAs: Small interfering RNAs.
ciRS-7 silencing suppressed ccRCC cell invasion
Since high ciRS-7 expression was found to be correlated with metastasis in ccRCC, we next determined whether ciRS-7 had an effect on cell invasion. To this end, we performed transwell invasion assays with matrigel after silencing ciRS-7 in the cells. As shown in Figure 2C, the transwell invasion assays revealed that the numbers of invasive cells in the siRNA groups were less than that observed in the siNC group, indicating the promoting effect of ciRS-7 on cell invasion.
ciRS-7 silencing inactivated the epidermal growth factor receptor (EGFR)/Akt signaling pathway
Since silencing ciRS-7 significantly suppressed the proliferative and invasive abilities of cells, we next investigated the possible mechanisms by which ciRS-7 promoted ccRCC progression. It has been previously reported that ciRS-7 activates the EGFR/C-Raf proto-oncogene serine/threonine protein kinase/mitogen-activated protein kinase pathway through the regulation of miR-7 activity.[17] Therefore, we wanted to determine whether ciRS-7 was responsible for the activation of EGFR in ccRCC. Our results demonstrated that ciRS-7 silencing resulted in reduced phosphorylated-EGFR (p-EGFR) expression, whereas total EGFR expression was not altered [Figure 3]. In addition, phosphorylated-Akt (p-Akt) expression, but not total Akt expression, in cells transfected with the siRNAs was decreased compared with controls [Figure 3]. EGFR activation is responsible for the activation of Akt, which plays vital roles in cell proliferation, invasion, and other malignant behaviors. Collectively, our results suggested that ciRS-7 may promote cell proliferation and invasion through the EGFR/Akt signaling pathway.
Figure 3.
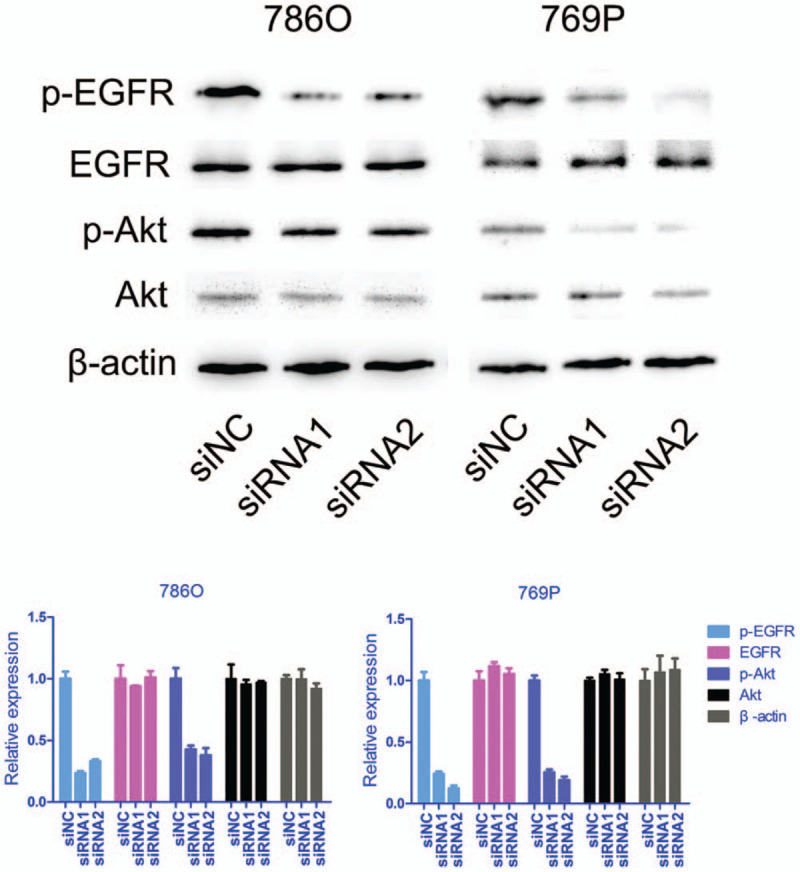
ciRS-7 suppression inhibited p-EGFR and p-Akt expression. The expressions of EGFR, p-EGFR, total Akt, and p-Akt in 786O and 769P cells transfected with siRNAs against ciRS-7 were analyzed by Western blotting. p-EGFR: Phosphorylated epidermal growth factor receptor; p-Akt: Phosphorylated serine/threonine kinase.
Discussion
In the present study, we demonstrated ciRS-7 up-regulation in ccRCC and its significant association with tumor diameter, clinical stage, and distant metastasis. Meanwhile, ciRS-7 overexpression indicated a poor PFS in patients with ccRCC. These data suggest that ciRS-7 is a useful prognostic factor for ccRCC. In addition, ciRS-7 silencing markedly inhibited ccRCC cell proliferation and invasion and contributed to inactivation of the EGFR/Akt signaling pathway. Considering the vital roles of the EGFR/Akt signaling pathway in cell proliferation and invasion, our data primarily uncovered the potential mechanism of action of ciRS-7 in ccRCC progression.
CircRNAs represents a class of non-coding RNAs. Since their first discovery in a viroid in 1976, many circRNAs have been identified in various cells and tissues.[18–21] As such, investigators have found that circRNAs exhibit different expression patterns and play diverse roles in various physiological and pathological processes. For instance, circZNF292 has been reported to play vital roles in the growth and angiogenesis of glioma.[9] Recently, circRNAs have become the focus of cancer research. Many investigations have demonstrated the abnormal expression of circRNAs and their involvement in tumor progression and prognosis. For instance, the circRNA circ_0013958 has been reported to be up-regulated in lung adenocarcinoa and be associated with advanced clinical stage and lymphatic metastasis.[22] As a circ-RNA, ciRS-7 has been reported to be abnormally expressed and play vital roles in various cancers, including RCC, neuroblastoma, and hepatocellular carcinoma.[23,24] In hepatic cellular cancer, ciRS-7 expression has been shown to be elevated and likely promotes cell proliferation and invasion by inhibiting miR-7 expression. In addition, ciRS-7 up-regulation in gastric cancer (GC) tissues is indicative of poor prognosis in patients with GC. An in vivo study demonstrated that ciRS-7 is responsible for the malignant phenotype by controlling activation of the phosphoinositide 3-kinase/ serine/threonine kinase signaling pathway.[14] These findings suggest that the role of ciRS-7 in various cancers is complex. However, the expression and role of ciRS-7 in ccRCC remain unknown. In the present study, our data suggest that ciRS-7 plays a key role in ccRCC progression. When ciRS-7 expression was silenced with siRNAs, ccRCC cell proliferation and invasion were significantly inhibited, demonstrating that ciRS-7 may promote ccRCC progression by strengthening the proliferative and invasive abilities of ccRCC cells, which was similar with previous studies. Additionally, suppressing ciRS-7 expression in ccRCC cells reduced the expressions of p-EGFR and p-Akt. A previous study demonstrated that EGFR activation activated Akt, which in turn played vital roles in cell proliferation, invasion, and other malignant behaviors in ccRCC. Therefore, our data demonstrated the vital roles of ciRS-7 in activating the EGFR/Akt signaling pathway in ccRCC. However, the exact mechanisms of ciRS-7 in activating EGFR/Akt signaling pathway still need further investigation. For instance, exploring the relationship between ciRS-7 and EGFR expression in fresh ccRCC tissues may further strengthen the theory of ciRS-7 in activating the EGFR/Akt signaling pathway. Moreover, as epidermal growth factor was a vital ligand that could activate EGFR/Akt pathway by interacting with its receptor EGFR, the relationship between ciRS-7 and EGF should also be a future research direction. Collectively, these data indicate that ciRS-7 may promote ccRCC progression through activating the EGFR/Akt signaling pathway and may serve as a ccRCC therapeutic target.
Acknowledgements
The authors thank the Department of Clinical Laboratory for technological support.
Conflicts of interest
None.
Footnotes
How to cite this article: Zhao YH, Wang Z, Zhang N, Cui T, Zhang YH. Effect of ciRS-7 expression on clear cell renal cell carcinoma progression. Chin Med J 2020;133:2084–2089. doi: 10.1097/CM9.0000000000000867
References
- 1.Zhuang J, Tu X, Cao K, Guo S, Mao X, Pan J, et al. The expression and role of tyrosine kinase ETK/BMX in renal cell carcinoma. J Exp Clin Cancer Res 2014; 33:25.doi: 10.1186/1756-9966-33-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattei J, Da SR, Sehrt D, Molina WR, Kim FJ. Targeted therapy in metastatic renal carcinoma. Cancer Lett 2014; 343:156–160.. doi: 10.1016/j.canlet.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 3.Gallou C, Joly D, Mejean A, Staroz F, Martin N, Tarlet G, et al. Mutations of the VHL gene in sporadic renal cell carcinoma: definition of a risk factor for VHL patients to develop an RCC. Hum Mutat 1999; 13:464–475.. doi: 10.1002/(SICI)1098-1004(1999)13:6<464::AID-HUMU6>3.0.CO;2-AA. [DOI] [PubMed] [Google Scholar]
- 4.Drabkin HA, Gemmill RM. Obesity, cholesterol, and clear-cell renal cell carcinoma (RCC). Adv Cancer Res 2010; 107:39–56.. doi: 10.1016/S0065-230X(10)07002-8. [DOI] [PubMed] [Google Scholar]
- 5.Turajlic S, Larkin J, Swanton C. SnapShot: renal cell carcinoma. Cell 2015; 163:1556.doi: 10.1016/j.cell.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016; 17:205–211.. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 7.Bao L, Qin S, Li C, Guo Z, Zhao L. Regulatory networks of circRNAs related to transcription factors in Populus euphratica Oliv. heteromorphic leaves. Biosci Rep 2019; 39:BSR20190540.doi: 10.1042/BSR20190540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang C, Xu D, You Z, Xu K, Tian W. Dysregulated circRNAs and ceRNA network in esophageal squamous cell carcinoma. Front Biosci (Landmark Ed) 2019; 24:277–290.. doi: 10.2741/4717. [DOI] [PubMed] [Google Scholar]
- 9.Yang P, Qiu Z, Jiang Y, Dong L, Yang W, Gu C, et al. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/beta-catenin signaling pathway. Oncotarget 2016; 7:63449–63455.. doi: 10.18632/oncotarget.11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan C, Zhang Y, Hao X, Gao J, Chen X, Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol Cancer 2019; 18:136.doi: 10.1186/s12943-019-1069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niu C, Zhao L, Guo X, Shen Y, Shao Y, Liu F. Diagnostic accuracy of circRNAs in esophageal cancer: a meta-analysis. Dis Markers 2019; 2019:9673129.doi: 10.1155/2019/9673129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu CH, et al. Circular RNA ciRS-7 promotes the proliferation and metastasis of pancreatic cancer by regulating miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat Dis Int 2019; 18:580–586.. doi: 10.1016/j.hbpd.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Su C, Han Y, Zhang H, Li Y, Yi L, Wang X, et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-kappaB signalling. J Cell Mol Med 2018; 22:3097–3107.. doi: 10.1111/jcmm.13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang Z, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on gastric cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem 2018; 119:440–446.. doi: 10.1002/jcb.26201. [DOI] [PubMed] [Google Scholar]
- 15.Peng L, Yuan XQ, Li GC. The emerging landscape of circular RNA ciRS-7 in cancer (review). Oncol Rep 2015; 33:2669–2674.. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 16.Yan B, Zhang W, Mao XW, Jiang LY. Circular RNA ciRS-7 correlates with advance disease and poor prognosis, and its down-regulation inhibits cells proliferation while induces cells apoptosis in non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2018; 22:8712–8721.. doi: 10.26355/eurrev_201812_16636. [DOI] [PubMed] [Google Scholar]
- 17.Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res 2017; 23:3918–3928.. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012; 7:e30733.doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P. Some yeast mitochondrial RNAs are circular. Cell 1980; 19:313–319.. doi: 10.1016/0092-8674(80)90505-x. [DOI] [PubMed] [Google Scholar]
- 20.Wang A, Wang J, Liu Y, Zhou Y. Mechanisms of long non-coding RNAs in the assembly and plasticity of neural circuitry. Front Neural Circuits 2017; 11:76.doi: 10.3389/fncir.2017.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol 2017; 1008:199–222.. doi: 10.1007/978-981-10-5203-3_7. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J 2017; 284:2170–2182.. doi: 10.1111/febs.14132. [DOI] [PubMed] [Google Scholar]
- 23.Yu L, Gong X, Sun L, Zhou Q, Lu B, Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One 2016; 11:e158347.doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013; 73:5609–5612.. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]


