Abstract
Background
Diagnoses of Skin diseases are frequently delayed in China due to lack of dermatologists. A deep learning-based diagnosis supporting system can facilitate pre-screening patients to prioritize dermatologists’ efforts. We aimed to evaluate the classification sensitivity and specificity of deep learning models to classify skin tumors and psoriasis for Chinese population with a modest number of dermoscopic images.
Methods
We developed a convolutional neural network (CNN) based on two datasets from a consecutive series of patients who underwent the dermoscopy in the clinic of the Department of Dermatology, Peking Union Medical College Hospital, between 2016 and 2018, prospectively. In order to evaluate the feasibility of the algorithm, we used two datasets. Dataset I consisted of 7192 dermoscopic images for a multi-class model to differentiate three most common skin tumors and other diseases. Dataset II consisted of 3115 dermoscopic images for a two-class model to classify psoriasis from other inflammatory diseases. We compared the performance of CNN with 164 dermatologists in a reader study with 130 dermoscopic images. The experts’ consensus was used as the reference standard except for the cases of basal cell carcinoma (BCC), which were all confirmed by histopathology.
Results
The accuracies of multi-class and two-class models were 81.49% ± 0.88% and 77.02% ± 1.81%, respectively. In the reader study, for the multi-class tasks, the diagnosis sensitivity and specificity of 164 dermatologists were 0.770 and 0.962 for BCC, 0.807 and 0.897 for melanocytic nevus, 0.624 and 0.976 for seborrheic keratosis, 0.939 and 0.875 for the “others” group, respectively; the diagnosis sensitivity and specificity of multi-class CNN were 0.800 and 1.000 for BCC, 0.800 and 0.840 for melanocytic nevus, 0.850 and 0.940 for seborrheic keratosis, 0.750 and 0.940 for the “others” group, respectively. For the two-class tasks, the sensitivity and specificity of dermatologists and CNN for classifying psoriasis were 0.872 and 0.838, 1.000 and 0.605, respectively. Both the dermatologists and CNN achieved at least moderate consistency with the reference standard, and there was no significant difference in Kappa coefficients between them (P > 0.05).
Conclusions
The performance of CNN developed with relatively modest number of dermoscopic images of skin tumors and psoriasis for Chinese population is comparable with 164 dermatologists. These two models could be used for screening in patients suspected with skin tumors and psoriasis respectively in primary care hospital.
Keywords: Artificial intelligence, Convolutional neural network, Skin tumor, Psoriasis, Dermoscopy
Introduction
In China, the dermatologist to patient ratio is as low as 1:60,000. Also, most well-trained and experienced dermatologists are located in large cities, increasing scarcity of dermatologists in rural China.[1] Furthermore, a considerable number of Chinese dermatologists and general physicians, especially those in the remote areas, due to limited clinical experience and learning opportunities, have low diagnostic accuracy for skin diseases. Therefore, a lot of patients with skin diseases could not see a professional dermatologist in time or at all. So their diseases are often misdiagnosed or the treatments are delayed. There is an urgent need to improve the diagnosis accuracy of skin diseases in China.
As the most common malignant and benign skin tumors in the Chinese population, basal cell carcinoma (BCC), melanocytic nevus (MN), and seborrheic keratosis (SK) usually share similar clinical features, and are easily misdiagnosed. However, the preferred treatments for these tumors are quite different and a misdiagnosis could result in improper healthcare.[2–6] So it is of great significance to correctly diagnose BCC, MN, and SK.
As a major skin inflammatory disease, psoriasis has a prevalence of 2% to 4% worldwide.[7,8] It can lead to psychological problems for young people and decrease quality of life. For psoriasis diagnosis, it is crucial to rule out other conditions that result in delay of appropriate therapy, such as pityriasis rosea, lichen planus, eczema, and so on. Additionally, when scalp scaling occurs, it is necessary to rule out seborrheic dermatitis.[9] Therefore, the accurate diagnosis of psoriasis is very important.
The problem of classifying skin lesions has been a focus for the machine learning. Automated image classification can support dermatologists in their daily clinical practice and create faster and less expensive diagnosis assistance. Deep learning, a branch of machine learning, attempts to extract high-level information from data.[10] The convolutional neural network (CNN) can perform feature extraction implicitly and learn sophisticated representations of the image.[10,11] One advantage of CNN is that it is not necessary to pre-process the images for classification, which is a key step of traditional machine learning.[12] CNN systems become more accurate as the data volume increases. However, one of the largest barrier to using deep learning effectively in the medical field is the lack of datasets.[13]
Our current study was designed to investigate whether a CNN-based artificial intelligence (AI) model can be used to develop an efficient skin tumor or psoriasis classification system with a modest number of dermoscopic images. We chose dermoscopic images rather than clinical pictures for the following reasons: (1) most skin diseases have specific characteristics under dermoscopy, which is useful for improving the diagnostic accuracy of cutaneous diseases[14–17]; and (2) the dermoscopic image is a magnified image of the lesion with a simple background, few interference factors, and obvious structures, suitable for input into the computer.
In order to evaluate the feasibility of the algorithm, we used two datasets. First, BCC, MN, and SK were selected as the three main target diseases in dataset I to build a multiclass model; psoriasis, acne vulgaris, dermatofibroma, and cutaneous haemangioma, which are common types of tumors or skin inflammatory diseases, were collectively called “others.” The “others” group was used to represent the images that did not belong to the three target categories to increase the model's screening specificity. Then, we collected dataset II to include psoriasis and other inflammatory cutaneous diseases for developing a two-class model; other inflammatory diseases included seborrheic dermatitis, eczema, lichen planus, and pityriasis rosea. Furthermore, the two models’ performances were compared with the diagnoses of 164 board-certified Chinese dermatologists. This tool could potentially be used as a pre-screening tool to assist general physicians to make recommendations or for dermatologists to prioritize their efforts and lay the foundation for precision medicine and remote consultation.
Methods
Ethical approval
All procedures involving humans were carried out in accordance with the ethical standards of the 2013 Declaration of Helsinki and were approved by the Medical Ethics Committee of Peking Union Medical College Hospital. Informed consent was obtained from all participants.
Datasets
All images were consecutively collected from the clinic of the Department of Dermatology, Peking Union Medical College Hospital, between 2016 and 2018 prospectively. The patients were all Asian with the type IV of Fitzpatrick skin types. The images were all obtained by MoleMax HD 1.0 dermoscope (Derma Medical Systems, Vienna, Austria). All BCCs were confirmed by histopathology. The diagnosis of other disease was made according to the criteria in the expert consensus or guidelines published [18–21] and the annotation process is seen in Figure 1. The annotation process in details is as follows: (1) two experts who were with more than 5 years of work experience blinded with each other provided a consistent interpretation of the dermoscopic image combining the clinical image and medical histories; (2) If there is a disagreement, a third expert would be the tie breaker; (3) If the three experts still could not get an agreement, the confusing cases would be excluded. The images which have poor focus, include multiple lesions or show interference factors such as clothing fiber, written notes or hair (except for scalp psoriasis and scalp seborrheic dermatitis), and the lesions located in the nail or mucosa were also excluded. The datasets used and patient demographic characteristics are in Table 1. The category of SK did not include solar lentigo and lichen planus-like keratosis. In most cases, multiple dermoscopic images of a single lesion, including different angles or close-ups, were photographed. Some example pictures of dataset I and dataset II are shown in Figure 2.
Figure 1.
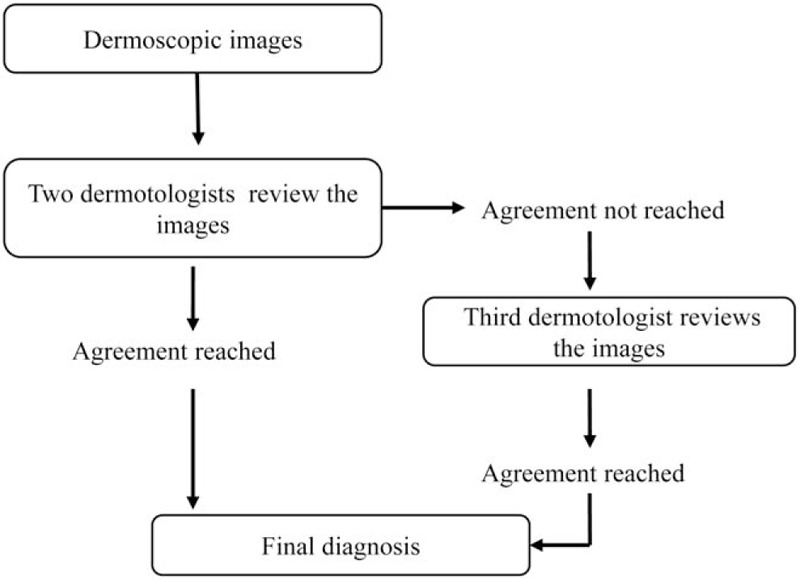
The annotation process of dermoscopic images.
Table 1.
Characteristics of dataset I and II for the multi- and two-class model respectively.

Figure 2.
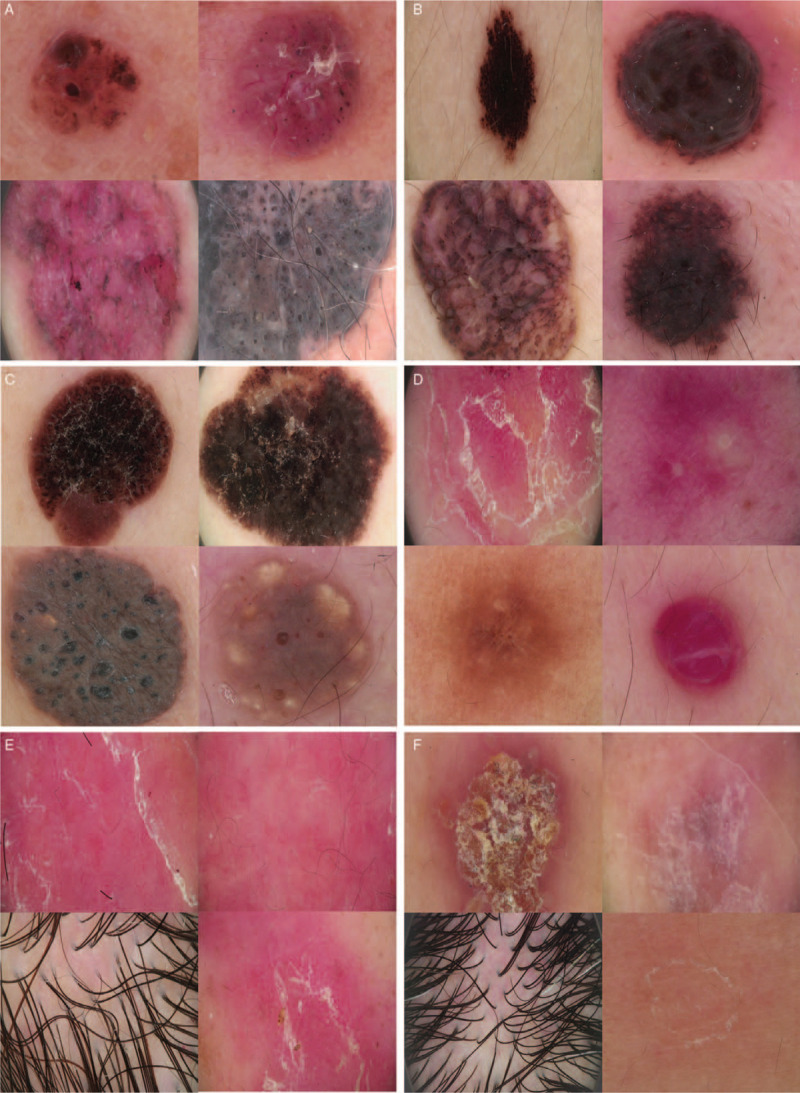
Example dermoscopic images: basal cell carcinoma (A), melanocytic nevus (B), seborrheic keratosis (C), “others” (D) including psoriasis (upper left), acne vulgaris (upper right), dermatofibroma (bottom left), and cutaneous haemangioma (bottom right). These images highlight the difficulty of classification: Psoriasis (E), other inflammatory diseases (F) involving eczema (upper left), lichen planus (upper right), seborrheic dermatitis (bottom left), and pityriasis rosea (bottom right).
CNN and deep learning algorithm
Both datasets were divided into training, validation, and testing sets in an 8:1:1 ratio.[22] Ten-fold cross-validation was performed to verify the robustness of the model. The validation set was separated from the training set to avoid overfitting.[23] The images were divided by patient to prevent images of the same patient being used in both training/validating and testing. In order to verify the methodology published by the Stanford University[24] and to test the applicability for the cases of Chinese hospitals, we developed our algorithm based on the pre-trained CNN parameters from GoogLeNet Inception v3 using the ImageNet dataset.[25] ImageNet contains over 1.28 million images for over 1000 normal life objects.[22] We re-trained the final layer with our images as input, using “ReLU” as our activation function and “Gradient Descent Optimizer” as our optimizer with a learning rate of “0.01.” “Cross entropy mean” was used to minimize the loss function.
The format of the input vectors in the input layer is a numeric matrix, where each element in the matrix is one pixel in the input image. For example, if an image has 864 pixels, multiplied by red, green, and blue layers, the number of elements in the matrix would be 2592. If there are 1000 input images, the input matrix will have the dimension of 2592 × 1000. A feature map was obtained by applying a linear filter and a non-linear function to the input matrix. For example, the hidden layer Ak, where k is the kth feature map. The filters consisted of weight Wk and bias bk. The feature map Ak was calculated using Formula 1. Then, the feature map calculated from extraction was classified. Each hidden layer was composed of multiple feature maps. There were a total of 28 layers in our model, including input and output layers. The simplified framework of our CNN model structure is shown in Figure 3. More detailed code regarding the retraining process publicly released on Github can be found at: https://github.com/tensorflow/hub/blob/master/examples/image_retraining/retrain.py.
Figure 3.
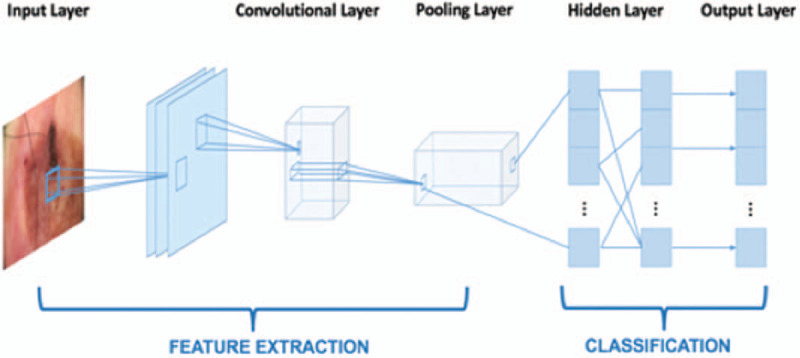
The simplified framework of convolutional neural network.
Visualization of internal features
T-distributed Stochastic Neighbor Embedding (t-SNE) plot, a method for visualizing high-dimensional data, was employed to show the internal features learned by the CNN.[24] Each point represents a skin lesion image projected from the output of the CNN's last hidden layer into two dimensions.[24]
Evaluation
Performance of CNN for the multi and two-class models
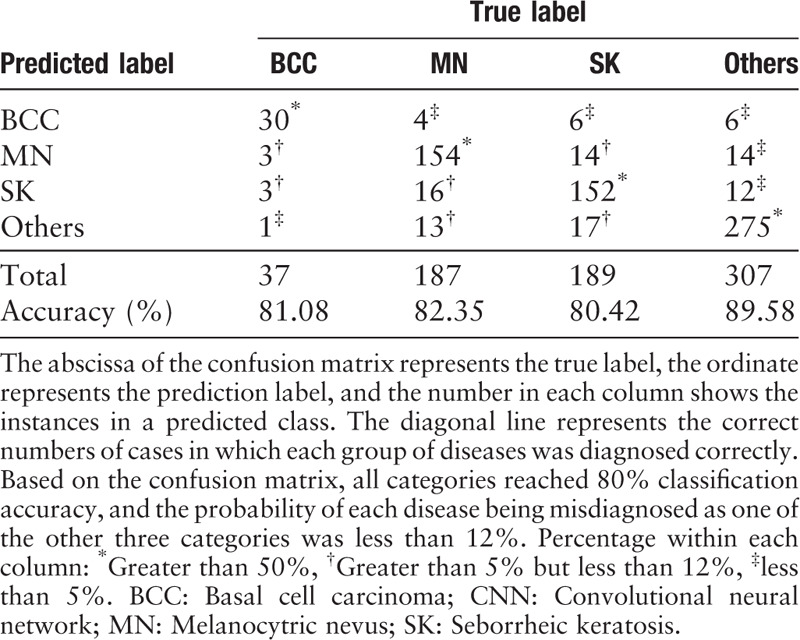
Each test image was given a probability for each of the two or four disease categories, summing to 1. The highest probability was regarded as the classification category. We calculated the accuracy, specificity, sensitivity, and area under the receiver operating characteristic (ROC) curve (AUC) for each category. The confusion matrix[26] was also employed to evaluate the multiclass models. It is a specific table layout that visualizes the performance of the algorithm. The abscissa represents the true label, the ordinate represents the prediction label, and the number in each column shows the instances in a predicted class.
Comparison between CNN and dermatologists
The classification sensitivity and specificity of our CNN-based classifier was compared against Chinese board-certified dermatologists in a reader study. In the experiment, 164 dermatologists who finished more than 10 h of systematic dermoscopic knowledge training and our algorithm classified the same 130 images; the images were selected on a patient-by-patient basis and there was no overlap of patients between the 130 images and the training, validation, and testing sets.
Each dermatologist was given a questionnaire that comprised two parts of images (70 images for the multiclass model and 60 images for the two-class model) through screen sharing. All participating dermatologists were required to answer this questionnaire in the same room at the same time. The corresponding clinical pictures were provided in a small size and no other information was offered. We should note that the clinical picture was provided to dermatologists but not to the AI. The reason is that the dermatologists will definitely look at the clinical picture firstly in the real clinical practice, they rarely directly gave a diagnosis according to the dermoscopic picture only. On the other hand, according to our experience, training AI with clinical pictures and dermoscopic images will reduce the accuracy. Therefore, when training and testing AI, solely dermoscopic images were provided. The gold standard was created by biopsy (BCC) and expert consensus (all others).
Statistical analysis
In the reader study, sensitivity, specialty, and their 95% confidence interval (CI) were calculated. Kappa coefficients were used to assess the consistency between dermatologists (or CNN) and the reference standard on the classification of each disease. Kappa coefficient >0.75 indicates good consistency, 0.40 to 0.75 indicates moderate consistency, and <0.40 indicates poor consistency. Adjusted Z-tests were used to assess differences in Kappa coefficients between dermatologists and CNN. Results were considered statistically significant at the P < 0.05 level. All analyses were carried out using SAS 9.4 (SAS Institute Inc., Cary, NC, USA).
Results
Performance of CNN for the multiclass model
The average accuracy ± standard deviation was 81.49% ± 0.88%. The fluctuation of the accuracy of classification was only ± 0.88%, demonstrating robustness of the CNN. The diagnosis sensitivity and specificity were 0.825 ± 0.060 and 0.960 ± 0.007 for BCC, 0.789 ± 0.015 and 0.923 ± 0.008 for MN, 0.749 ± 0.029 and 0.920 ± 0.008 for SK, 0.869 ± 0.017 and 0.936 ± 0.010 for the “others” group. The AUC scores of diagnosing BCC, MN, SK, and “others” were 0.972 ± 0.011, 0.952 ± 0.014, 0.933 ± 0.014, and 0.965 ± 0.005, respectively. The ROC graphs and the corresponding confusion matrix for each category from one experiment are shown in Figure 4A–4D and Table 2.
Figure 4.
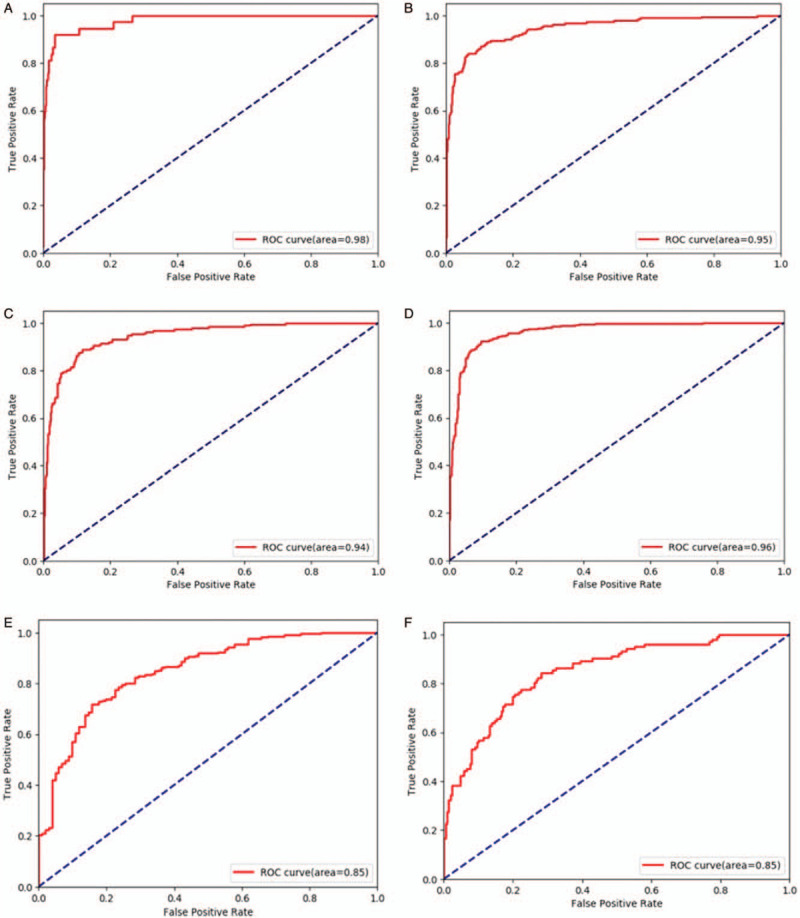
Receiver operating characteristic (ROC) graphs for basal cell carcinoma (A), melanocytic nevus (B), seborrheic keratosis (C), and other groups (D) for one test of the ten-fold cross-validation, which comes from the same test as Table 4. ROC graphs for psoriasis (E) vs. other inflammation conditions (F) for one test of the ten-fold cross-validation.
Table 2.
Confusion matrix of CNN in one test of ten-fold cross-validation.
Performance of CNN for the two-class model
The average accuracy of the “psoriasis vs. other inflammatory diseases” group was 77.02% ± 1.81%. The two-class model achieved 0.797 ± 0.040 sensitivity and 0.719 ± 0.078 specificity for psoriasis. The ROC graphs for each category from one experiment are shown in Figure 4E and 4F. And the average AUC for this binary classification model is 0.840 ± 0.022.
Comparison between CNN and dermatologists
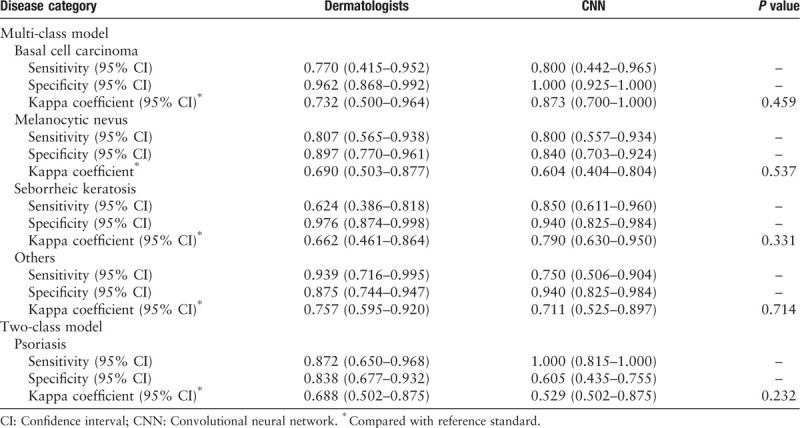
The general information of the 164 board-certified dermatologists is shown in Table 3. As Table 4 shows, in the multi-class tasks, the diagnosis sensitivity and specificity of 164 dermatologists were 0.770 (95% CI 0.415–0.952) and 0.962 (95% CI 0.868–0.992) for BCC, 0.807 (95% CI 0.565–0.938) and 0.897 (95% CI 0.770–0.961) for MN, 0.624 (95% CI 0.386–0.818) and 0.976 (95% CI 0.874–0.998) for SK, 0.939 (95% CI 0.716–0.995) and 0.875 (95% CI 0.744–0.947) for the “others” group, respectively; the diagnosis sensitivity and specificity of multi-class CNN were 0.800 (95% CI 0.442–0.965) and 1.000 (95% CI 0.925–1.000)] for BCC, 0.800 (95% CI 0.557–0.934) and 0.840 (95% CI 0.703–0.924) for MN, 0.850 (95% CI 0.611–0.960) and 0.940 (95% CI 0.825–0.984) for SK, 0.750 (95% CI 0.506–0.904) and 0.940 (95% CI 0.825–0.984) for the “others” group, respectively. In the two-class tasks, the sensitivity and specificity of dermatologists and CNN for classifying psoriasis were 0.872 (95% CI 0.650–0.968) and 0.838 (95% CI 0.677–0.932), 1.000 (95% CI 0.815–1.000) and 0.605 (95% CI 0.435–0.755), respectively. Both the dermatologists and CNN achieved at least moderate evaluation consistency with the reference standard, and there was no significant difference in Kappa coefficients between them (P > 0.05).
Table 3.
Demographics of 164 board-certified Chinese dermatologists.
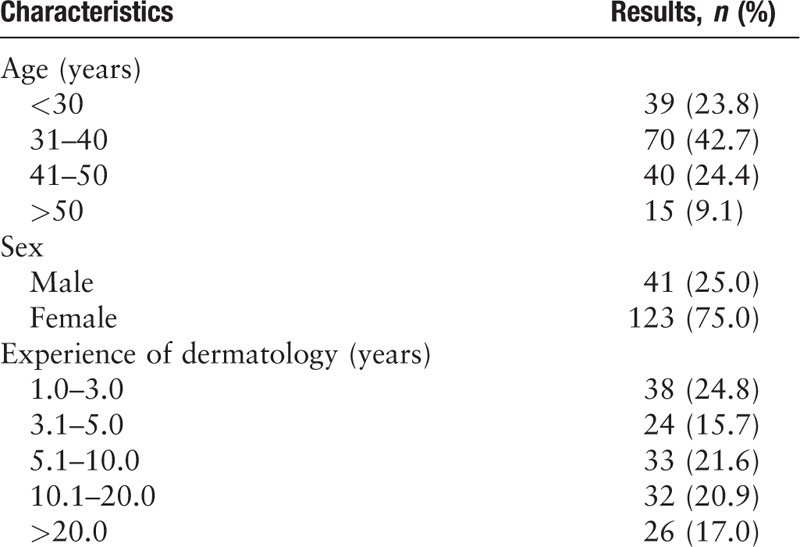
Table 4.
The classification sensitivity, specificity, and Kappa coefficient compared with reference standard of 164 board-certified dermatologists and the CNN.
Visualization of internal features
Each point in the t-SNE plot represents a dermoscopic image. Similar images are more likely to be clustered together. We used the test set from one experiment for the multiclass model in Figure 5A. All images in the experiment for the multiclass model are plotted in Figure 5B.
Figure 5.

The Stochastic Neighbor Embedding (t-SNE) plot of the multi-class model. (A) The red cluster below represents the BCC cluster. The blue cluster has several subgroups, corresponding to multiple disease types in the “others” group. The patterns for MN and SK classes are not revealed. (B) When using all the images, the pattern of MN is seen to distribute mostly in the bottom right corner. The clusters for SK are split crossed. The pattern for the “others” and BCC groups is similar to Figure (A). BCC: Basal cell carcinoma; CNN: Convolutional neural network; MN: Melanocytic nevus; SK: Seborrheic keratosis.
Discussion
In recent years, several studies have reported the application of deep learning for the classification of skin diseases, especially skin tumors.[27–37] Most of them used the pre-trained ResNet50 CNN network,[33–37] using images from the International Skin Imaging Collaboration. To verify the consistency of results published by the Stanford University[24] for Chinese hospital use cases, we developed the algorithm based on the pre-trained GoogLeNet Inception v3 CNN network for our own dataset. As for the disease spectrum, they were mostly focused on distinguishing melanoma, nevi or skin cancer images. Images of inflammatory skin conditions, including psoriasis and acne vulgaris, were innovatively added to the multiclass training dataset for skin tumor classification in this study to verify the CNN's ability to learn dermoscopic features of inflammatory skin diseases, and to determine whether CNN can distinguish skin tumors from inflammatory skin conditions. This is the first study, to the best of our knowledge, focusing on using deep-learning-based approach to automatically classify dermoscopic images for most common skin diseases in Chinese population. The study includes the most common skin tumors and inflammatory diseases in China. Melanoma is excluded from the classification list because the incidence of malignant melanoma in the Chinese population is relatively low (less than 1/100,000[38]) and the number of pictures is limited. From 2014 to 2018, we collected about 130,000 dermoscopic images that include about 14,000 cases in total. However, it contains only 25 cases of malignant melanoma (188 dermoscopic images); and 89/188 images are further excluded because they are located in the nail or with notes. Moreover, the most common type is the acral malignant melanoma (41.8%),[38] and the superficial type is relatively rare. It is relatively easy to diagnosis acral malignant melanoma as the associated features are usually easy to be distinguished from other tumors. Therefore, the skin tumors in this study do not contain malignant melanoma, but focus on three most common skin tumors (BCC, melanocytic nuvus, and SK).
In previous studies, Esteva et al[24] used 129,450 images and demonstrated that a CNN could achieve dermatologist-level classification of skin cancer, including keratinocytic and melanocytic tumors. In the study of Fujisawa et al,[22] CNNs were trained on approximately 5% (4867 images) that number of images; for 14 skin tumors, the overall classification accuracy of the CNN was 76.5%, which was a statistically higher accuracy than the board-certified dermatologists. Similar to Fujisawa's study, satisfactory overall classification accuracies (81.49% and 77.02%) were achieved using modest datasets (7192 and 3115 images) in our study, which showed that, as long as the images were correctly labeled, a well-performed CNN classifier can be developed using a much smaller training dataset.
Our previous study, which tested over 1000 dermoscopic images for the model of four classes, had an average accuracy of 87.25% ± 2.24%.[28] The robustness of our algorithm improved; however, the accuracy decreased from 87.25% to 81.49%. A possible reason could be the images in the “others” group, which included inflammatory skin conditions. It might be difficult for CNN to generate key features for classification of many different skin diseases within the same category. The results of the “Performance of CNN for the multiclass model” showed that the SK group had the worst performance, while the accuracy of SK in the reader study was the best. It demonstrated that the differences in the level of difficulty inherent to images of the test set will directly impact the diagnostic performance of the algorithms.
To serve as a reference comparator to the automated algorithm, it is of the utmost importance to include a large group of dermatologists.[39] This study compared CNN diagnostic performance to 164 Chinese dermatologists with more than 10 h of systematic dermoscopy training. To the best of our knowledge, this was the largest group of dermatologists involved for such studies. In Fujisawa's study, the accuracy of 13 board-certified dermatologists and nine dermatology trainees was compared to the accuracy of their CNN.[22] Han et al[29] compared the accuracy of a computer classifier to 16 dermatologist board members. The publication of Haenssle et al[27] included 58 dermatologists in the comparison with CNN's diagnostic performance. Our previous study included 95 experienced dermatologists who had received dermoscopy training in the differential diagnosis of pigmented nevus and SK. We concluded that performance of CNN automatic classification model is similar to that of experienced dermatologists in the two classification of pigmented nevus and SK.[40] And recently, it has been reported that deep learning outperformed 112 dermatologists or 136 of 157 dermatologists in a dermoscopic melanoma image classification task or multiclass skin cancer image classification.[33,36] This demonstrates that our models are as accurate as large-scale trained dermatologists, and may potentially be used as a pre-screening tool to assist general physicians to make recommendations or for dermatologists to prioritize their efforts, especially since a considerable number of Chinese dermatologists do not have systematic training in dermoscopy.
Furthermore, we developed a two-class model for psoriasis and other inflammatory diseases as a pilot test and achieved an average accuracy of 77.02%. The prevalence of psoriasis is about 0.59% in China[41] and the involved population reached up to 8.26 million. It is a cosmetically debilitating and chronic disease which occurs both in developing and developed countries. In many hospitals in China, misdiagnosis and missed diagnosis of psoriasis occur often. To the best of our knowledge, there is currently no differential diagnosis model for dermoscopic images of psoriasis and other inflammatory diseases for Chinese population. This model achieved 100% sensitivity during the competition of 164 dermatologists vs. computer algorithm which has the potential to help dermatologists improve the early diagnosis and treatment of psoriasis significantly. It shows that our model prefers to classify the image under the category of “psoriasis,” and all the real psoriasis images are classified correctly. We might need to adjust the weights of two classes during the training process. Since the size of our second testing set is relatively small, 22 images for psoriasis group and 38 images for the other group, the accuracy, sensitivity, and specificity results from ten-fold cross validation are more valued. And these results also indicate that our model was not over-fitting. Unfortunately, other inflammatory disease groups here included only seborrheic dermatitis, eczema, lichen planus, and pityriasis rosea. Larger datasets including data on more psoriasis-related diseases are needed to develop a more efficient classification model for psoriasis in the future.
There are several limitations to our study. First, our dataset was created using only the database of the dermatology department of the Peking Union Medical College Hospital; due to the lack of diversity, it is possible that the sensitivity and specificity will be different when a different dataset is used. Second, although CNN achieved equal accuracy to the dermatologists in our study, this classification was based on the information obtained from only dermoscopic images (for CNN) or dermoscopic and clinical images (for dermatologists). If additional information, such as location, tactility, clinical course, subjective symptoms, age, and sex of the patients, were provided, the classification accuracy may be different.[42] Further studies are needed to clarify this hypothesis. Third, the dataset covered only 11 diseases and lacked the full spectrum of skin lesions encountered in clinical practice. To overcome this issue, future research can focus on ensuring the diagnostic sensitivity and specificity of the CNN model for each disease while increasing the categories of diseases.
In conclusion, we used a CNN trained with modest numbers of dermoscopic images from the Chinese population to develop two efficient skin disease classifiers. Based on a single dermoscopic image, the performance of this CNN was comparable to 164 board-certified dermatologists with more than 10 h of dermoscopy systematic training in the classification of skin tumors and psoriasis. Next, we will apply this system to a professional medical network platform open to dermatologists in China (https://ai.dxy.cn) to find more ways to improve the models’ classification performance.
Acknowledgements
The authors would like to thank Yangzhige He of the Department of Medical Research Center, Peking Union Medical College Hospital, Chinese Academy of Medical Science & Peking Union Medical College, for her suggestions about the statistical analysis.
Funding
This work was supported by grants from the Beijing Natural Science Foundation (No. 7182127), the National Natural Science Foundation of China (No. 61871011), the National Key Research and Development Program of China (No. 2016YFC0901500), the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2019XK320024), and the CAMS Innovation Fund for Medical Sciences (No. 2017-I2M-3-020).
Conflicts of interest
None.
Footnotes
How to cite this article: Wang SQ, Zhang XY, Liu J, Tao C, Zhu CY, Shu C, Xu T, Jin HZ. Deep learning-based, computer-aided classifier developed with dermoscopic images shows comparable performance to 164 dermatologists in cutaneous disease diagnosis in the Chinese population. Chin Med J 2020;133:2027–2036. doi: 10.1097/CM9.0000000000001023
Shi-Qi Wang and Xin-Yuan Zhang contributed equally to this work.
References
- 1.Zhao S, Xie B, Li Y, Zhao X, Kuang Y, Su J, et al. Smart identification of psoriasis by images using convolutional neural networks: a case study in China. J Eur Acad Dermatol Venereol 2020; 34:518–524.. doi: 10.1111/jdv.15965. [DOI] [PubMed] [Google Scholar]
- 2.Bobadilla F, Wortsman X, Munoz C, Segovia L, Espinoza M, Jemec GB. Pre-surgical high resolution ultrasound of facial basal cell carcinoma: correlation with histology. Cancer Imaging 2008; 8:163–172.. doi: 10.1102/1470-7330.2008.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christenson LJ, Borrowman TA, Vachon CM, Tollefson MM, Otley CC, Weaver AL, et al. Incidence of basal cell and squamous cell carcinomas in a population younger than 40 years. JAMA 2005; 294:681–690.. doi: 10.1001/jama.294.6.681. [DOI] [PubMed] [Google Scholar]
- 4.Hauschild A, Egberts F, Garbe C, Bauer J, Grabbe S, Hamm H, et al. Melanocytic nevi. J Dtsch Dermatol Ges 2011; 9:723–734.. doi: 10.1111/j.1610-0387.2011.07741.x. [DOI] [PubMed] [Google Scholar]
- 5.Minagawa A. Dermoscopy-pathology relationship in seborrheic keratosis. J Dermatol 2017; 44:518–524.. doi: 10.1111/1346-8138.13657. [DOI] [PubMed] [Google Scholar]
- 6.Jackson JM, Alexis A, Berman B, Berson DS, Taylor S, Weiss JS. Current understanding of seborrheic keratosis: prevalence, etiology, clinical presentation, diagnosis, and management. J Drugs Dermatol 2015; 14:1119–1125.. [PubMed] [Google Scholar]
- 7.Armstrong AW. Psoriasis. JAMA Dermatol 2017; 153:956.doi: 10.1001/jamadermatol.2017.2103. [DOI] [PubMed] [Google Scholar]
- 8.Daniyal M, Akram M, Zainab R, Munir N, Shah SMA, Liu B, et al. Progress and prospects in the management of Psoriasis and developments in Phyto-therapeutic modalities. Dermatol Ther 2019; 32:e12866.doi: 10.1111/dth.12866. [DOI] [PubMed] [Google Scholar]
- 9.Napolitano M, Caso F, Scarpa R, Megna M, Patrì A, Balato N, et al. Psoriatic arthritis and psoriasis: differential diagnosis. Clin Rheumatol 2016; 35:1893–1901.. doi: 10.1007/s10067-016-3295-9. [DOI] [PubMed] [Google Scholar]
- 10.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521:436–444.. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 11.Kumar A, Kim J, Lyndon D, Fulham M, Feng D. An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J Biomed Health Inform 2017; 21:31–40.. doi: 10.1109/JBHI.2016.2635663. [DOI] [PubMed] [Google Scholar]
- 12.Mai SM, Sheha MA, Sharawy A. Automatic detection of melanoma skin cancer using texture analysis. Int J Comput Appl 2012; 42:22–26.. doi: 10.5120/5817-8129. [Google Scholar]
- 13.Han SS, Park GH, Lim W, Kim MS, Na JI, Park I, et al. Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: automatic construction of onychomycosis datasets by region-based convolutional deep neural network. PLoS One 2018; 13:e0191493.doi: 10.1371/journal.pone.0191493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vestergaard ME, Macaskill P, Holt PE, Menzies SW. Dermoscopy compared with naked eye examination for the diagnosis of primary melanoma: a meta-analysis of studies performed in a clinical setting. Br J Dermatol 2008; 159:669–676.. doi: 10.1111/j.1365-2133.2008.08713.x. [DOI] [PubMed] [Google Scholar]
- 15.Rosendahl C, Tschandl P, Cameron A, Kittler H. Diagnostic accuracy of dermatoscopy for melanocytic and nonmelanocytic pigmented lesions. J Am Acad Dermatol 2011; 64:1068–1073.. doi: 10.1016/j.jaad.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 16.Braun RP, Ludwig S, Marghoob AA. Differential diagnosis of seborrheic keratosis: clinical and dermoscopic features. J Drugs Dermatol 2017; 16:835–842.. [PubMed] [Google Scholar]
- 17.Errichetti E, Stinco G. Dermoscopy in general dermatology: a practical overview. Dermatol Ther (Heidelb) 2016; 6:471–507.. doi: 10.1007/s13555-016-0141-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skin Imaging Group, Combination of Tradtional and Western Medicine Dermatology; Skin Imaging Group, China International Exchange and Promotion Association for Medical and Healthcare; Skin Imaging Group, Chinese Society of Dermatology, Subcommittee on Dermatologic Surgery, China Dermatologist Association; Skin Imaging Equipment Group, Committee on Skin Disease and Cosmetic Dermatology, China Association of Medical Equipment. Dermoscopic characteristics of basal cell carcinoma: a Chinese expert consensus statement (2019) (in Chinese). Chin J Dermatol 2019; 52:371–377.. doi: 10.3760/cma.j.issn.0412-4030.2019.06.001. [Google Scholar]
- 19.National Clinical Research Center for Skin and Immune Diseases; Huaxia Shin Image and Artificial Intelligence Cooperation, China International Exchange and Promotion Association for Medical and Healthcare; Skin Imaging Group, Dermatology Branch of China International Exchange and Promotion Association for Medical and Healthcare; Skin Imaging Group, Combination of Traditional and Western Medicine Dermatology; Subcommittee on Dermatologic Surgery, China Dermatologist Association; Skin Imaging Group (Preparing), Chinese Society of Dermatology, et al. Dermoscopic features of solar lentigo, seborrheic keratosis and lichen planus-like keratosis: an expert consensus statement (in Chinese). Chin J Dermatol 2019; 52:878–883.. doi: 10.35541/cjd.20190480.20. [Google Scholar]
- 20.Qiao JJ, Zou XB, Dong HT, Xin LL. The diagnosis of dermoscopy in pigmented nevus (in Chinese). Chin J Leprosy Skin Dis 2017; 33:65–69.. [Google Scholar]
- 21.Lallas A, Kyrgidis A, Tzellos TG, Apalla Z, Karakyriou E, Karatolias A, et al. Accuracy of dermoscopic criteria for the diagnosis of psoriasis, dermatitis, lichen planus and pityriasis rosea. Br J Dermatol 2012; 166:1198–1205.. doi: 10.1111/j.1365-2133.2012.10868.x. [DOI] [PubMed] [Google Scholar]
- 22.Fujisawa Y, Otomo Y, Ogata Y, Nakamura Y, Fujita R, Ishitsuka Y, et al. Deep learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumor diagnosis. Br J Dermatol 2018; 179:373–381.. doi: 10.1111/bjd.16924. [DOI] [PubMed] [Google Scholar]
- 23.Panchal G, Ganatra A, Kosta YP, Panchal D. Behaviour analysis of multilayer perceptrons with multiple hidden neurons and hidden layers. Int J Comput Theory Eng 2011; 3:332–337.. doi: 10.7763/IJCTE.2011.V3.328. [Google Scholar]
- 24.Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542:115–118.. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016; 35:1285–1298.. doi: 10.1109/tmi.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the Inception Architecture for Computer Vision. IEEE Conference on Computer Vision and Pattern Recognition. Las Vegas, USA; 2016. p. 2818–2826. [Google Scholar]
- 27.Haenssle HA, Fink C, Schneiderbauer R, Toberer F, Buhl T, Blum A, et al. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol 2018; 29:1836–1842.. doi: 10.1093/annonc/mdy166. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Wang S, Liu J, Tao C. Towards improving diagnosis of skin diseases by combining deep neural network and human knowledge. BMC Med Inform Decis Mak 2018; 18: Suppl 2: 59.doi: 10.1186/s12911-018-0631-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol 2018; 138:1529–1538.. doi: 10.1016/j.jid.2018.01.028. [DOI] [PubMed] [Google Scholar]
- 30.Tschandl P, Argenziano G, Razmara M, Yap J. Diagnostic accuracy of content-based dermatoscopic image retrieval with deep classification features. Br J Dermatol 2018; 181:155–165.. doi: 10.1111/bjd.17189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brinker TJ, Hekler A, Utikal JS, Grabe N, Schadendorf D, Klode J, et al. Skin cancer classification using convolutional neural networks: systematic review. J Med Internet Res 2018; 20:e11936.doi: 10.2196/11936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarrete-Dechent C, Dusza SW, Liopyris K, Marghoob AA, Halpern AC, Marchetti MA. Automated dermatological diagnosis: hype or reality? J Invest Dermatol 2018; 138:2277–2279.. doi: 10.1016/j.jid.2018.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maron RC, Weichenthal M, Utikal JS, Hekler A, Berking C, Hauschild A, et al. Systematic outperformance of 112 dermatologists in multiclass skin cancer image classification by convolutional neural networks. Eur J Cancer 2019; 119:57–65.. doi: 10.1016/j.jaad.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 34.Hekler A, Utikal JS, Enk AH, Hauschild A, Weichenthal M, Maron RC, et al. Superior skin cancer classification by the combination of human and artificial intelligence. Eur J Cancer 2019; 120:114–121.. doi: 10.1016/j.ejca.2019.07.019. [DOI] [PubMed] [Google Scholar]
- 35.Brinker TJ, Hekler A, Enk AH, Berking C, Haferkamp S, Hauschild A, et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur J Cancer 2019; 119:11–17.. doi: 10.1016/j.ejca.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 36.Brinker TJ, Hekler A, Enk AH, Klode J, Hauschild A, Berking C, et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur J Cancer 2019; 113:47–54.. doi: 10.1016/j.ejca.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Brinker TJ, Hekler A, Enk AH, Klode J, Hauschild A, Berking C, et al. A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur J Cancer 2019; 111:148–154.. doi: 10.1016/j.ejca.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Jun G, Shukui Q, Jun L, Tongyu L, Lu S, Xiaohong C, et al. Chinese guidelines on the diagnosis and treatment of melanoma (2015 edition) (in Chinese). Chin Clin Oncol 2016; 5:57.doi: 10.21037/cco.2015.12.02. [DOI] [PubMed] [Google Scholar]
- 39.Marchetti MA, Codella NCF, Dusza SW, Gutman DA, Helba B, Kalloo A, et al. Results of the 2016 international skin imaging collaboration international symposium on biomedical imaging challenge: comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J Am Acad Dermatol 2018; 78:270–277.. doi: 10.1016/j.jaad.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Liu J, Zhu C, Shu C, Zhou H, Xie F, et al. Comparison of diagnostic performance of dermatologists versus deep convolutional neural network for dermoscopic images of pigmented nevus and seborrheic keratosis (in Chinese). Chin J Dermatol 2018; 51:486–489.. doi: 10.3760/cma.j.issn.0412-4030.2018.07.002. [Google Scholar]
- 41.Ding X, Wang T, Shen Y, Wang X, Zhou C, Tian S, et al. Prevalence of psoriasis in China: a population-based study in six cities. Eur J Dermatol 2012; 22:663–667.. doi: 10.1684/ejd.2012.1802. [DOI] [PubMed] [Google Scholar]
- 42.Li CX, Shen CB, Xue K, Shen X, Jing Y, Wang ZY, et al. Artificial intelligence in dermatology: past, present, and future. Chin Med J 2019; 132:2017–2020.. doi: 10.1097/CM9.0000000000000372. [DOI] [PMC free article] [PubMed] [Google Scholar]


