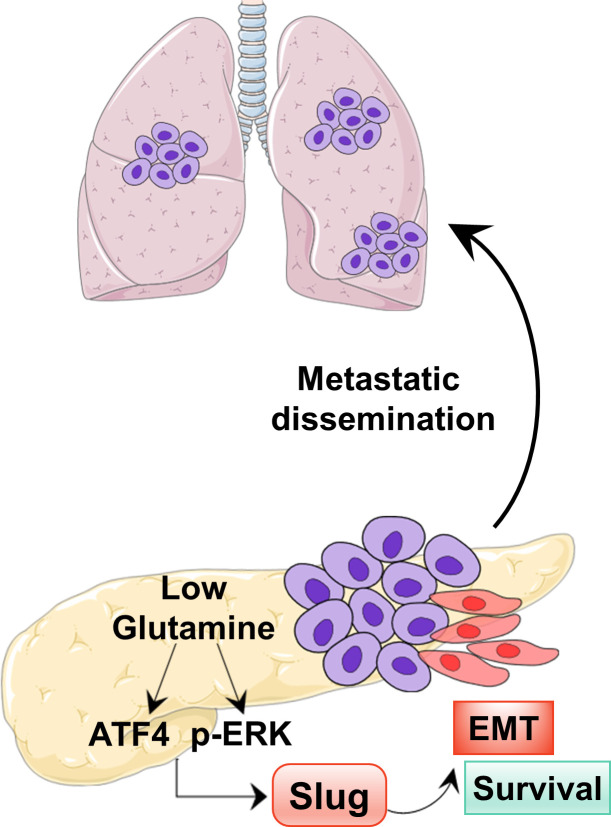
Pancreatic ductal adenocarcinoma (PDAC) cells rely heavily on glutamine metabolism. Recouvreux et al. demonstrate that nutrient stress caused by glutamine deprivation drives epithelial-mesenchymal transition (EMT) and aggressive behaviors in PDAC cells through up-regulation of the EMT master regulator Slug.
Abstract
Tumor cells rely on glutamine to fulfill their metabolic demands and sustain proliferation. The elevated consumption of glutamine can lead to intratumoral nutrient depletion, causing metabolic stress that has the potential to impact tumor progression. Here, we show that nutrient stress caused by glutamine deprivation leads to the induction of epithelial–mesenchymal transition (EMT) in pancreatic ductal adenocarcinoma (PDAC) cells. Mechanistically, we demonstrate that glutamine deficiency regulates EMT through the up-regulation of the EMT master regulator Slug, a process that is dependent on both MEK/ERK signaling and ATF4. We find that Slug is required in PDAC cells for glutamine deprivation–induced EMT, cell motility, and nutrient stress survival. Importantly, we decipher that Slug is associated with nutrient stress in PDAC tumors and is required for metastasis. These results delineate a novel role for Slug in the nutrient stress response and provide insight into how nutrient depletion might influence PDAC progression.
Graphical Abstract
Introduction
Glutamine is a critical nutrient in cancer that contributes to a wide array of biosynthetic and metabolic processes such as the synthesis of proteins, lipids, and other amino acids; de novo nucleotide production; hexosamine biosynthesis; the urea cycle; and glutathione production (Cluntun et al., 2017). Pancreatic ductal adenocarcinoma (PDAC) cells rely heavily on glutamine utilization to fulfill their metabolic and biosynthetic requirements, and therefore, it is not surprising that they are exquisitely sensitive to glutamine withdrawal (Biancur et al., 2017; Son et al., 2013). Highlighting the importance of glutamine in PDAC tumors, glutamine contributes the most to TCA cycle metabolites relative to other nutrient sources (Hui et al., 2017). PDAC tumors are poorly vascularized and often encounter a paucity of nutrients. Indeed, glutamine is the most depleted amino acid in human PDAC (Kamphorst et al., 2015), and regional glutamine deficiencies within PDAC tumors can modulate adaptation mechanisms through signal transduction (Lee et al., 2019). However, little is known about how glutamine deficiency in the tumor microenvironment might affect PDAC progression.
A key step in PDAC progression is epithelial–mesenchymal transition (EMT). EMT is considered a critical process for the initiation of the metastatic cascade, as it allows cancer cells to exhibit increased cell motility and acquire invasive features (Lu and Kang, 2019; Nieto et al., 2016). Lineage tracing of epithelial cells in a genetically engineered mouse model of PDAC (KPC model, Pdx1-Cre; LSL-KRasG12D/+; LSL-Tp53R172H/+) demonstrated that EMT and the dissemination of tumor cells occur early in PDAC development (Rhim et al., 2012). However, the role of EMT in PDAC has been a focus of controversy in recent years, particularly regarding the contribution of the different EMT transcription factors (EMT-TFs), Snail, Twist, and Zeb1, to PDAC metastasis (Aiello et al., 2017; Krebs et al., 2017; Zheng et al., 2015). For instance, genetic depletion of Snail or Twist in the KPC model has no effect on the development of metastasis (Zheng et al., 2015), but genetic ablation of Zeb1 profoundly reduced the metastatic capacity of PDAC tumors (Krebs et al., 2017). While it is established that pancreatic cancer cells can escape their epithelial properties in different ways, leading to distinct modes of invasion (Aiello et al., 2018), how nutrient stress contributes to EMT, metastasis, and the regulation of EMT-TFs in PDAC has not been explored.
Here, we examined global transcriptional changes to identify pathways that were modulated by glutamine starvation in both murine and human PDAC cells. We show that glutamine depletion leads to the activation of the EMT transcriptional program and to properties associated with enhanced tumor aggressiveness. In this study, we identify a novel mechanism of EMT induction in PDAC that is orchestrated by the EMT-TF Snai2 or Slug (from here on, Slug) upon glutamine starvation. We demonstrate that glutamine depletion–induced Slug is a major driver of EMT, motility, and survival in glutamine-deprived conditions. Moreover, suppression of Slug in vivo profoundly abrogated the metastatic capacity of PDAC cells. These results shed light on the specific contributions of EMT-TFs in regulating metastasis in PDAC and point to the induction of Slug-driven EMT as a novel adaptive response to nutrient stress.
Results
Glutamine deprivation induces EMT and promotes migration and invasion in PDAC cells
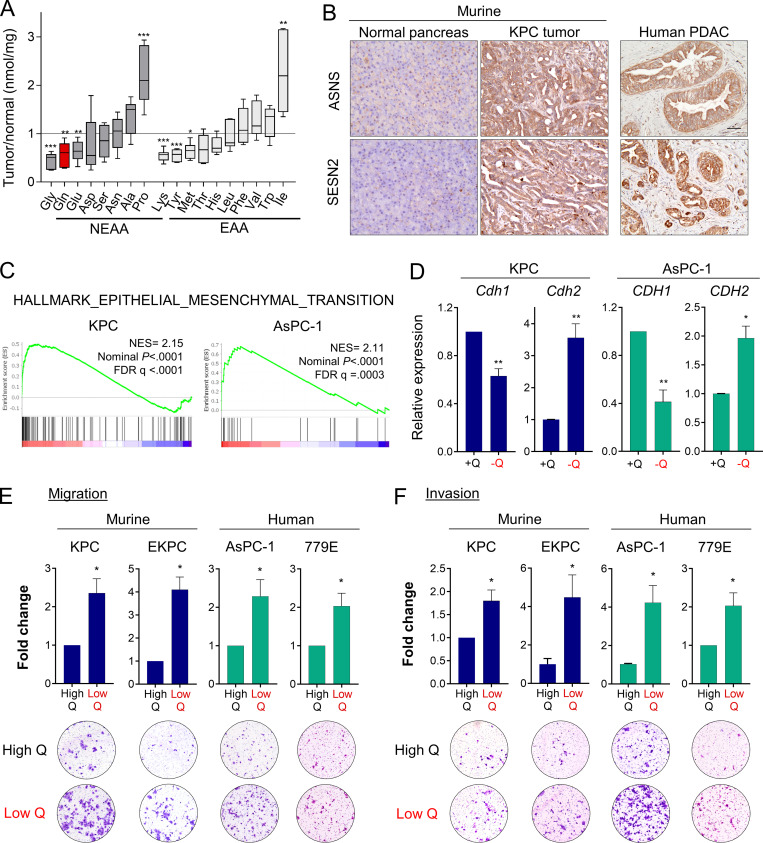
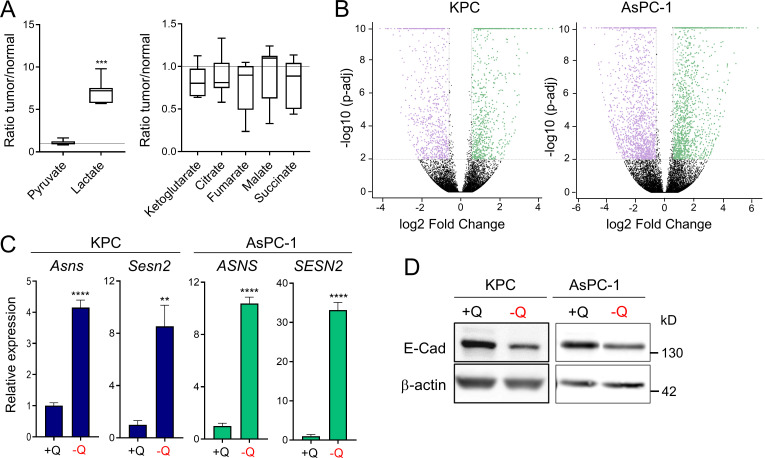
We previously reported that glutamine is the most depleted amino acid in human PDAC tumors relative to adjacent nonneoplastic tissue (Kamphorst et al., 2015). To further study nutrient scarcity in PDAC, we used an orthotopic syngeneic mouse model of PDAC that recapitulates the histopathological complexity of the human disease (Bayne et al., 2012). In this model, murine PDAC cells originating from animals of the genotype Pdx1-Cre; LSL-KRasG12D/+; LSL-Tp53R172H/+ (KPC) were surgically implanted into the tail of the pancreas (Hingorani et al., 2005). When orthotopic tumors were palpable, we quantified polar metabolites within the tumors using gas chromatography–mass spectrometry. Relative to normal age- and sex-matched pancreas tissue, murine PDAC tumors exhibited significantly lower amounts of many amino acids, including nonessential amino acids such as glycine, glutamine, and glutamate, as well as essential amino acids such as lysine, tyrosine, and methionine (Fig. 1 A). Lactate was also increased in PDAC tumors, whereas tricarboxylic acid cycle intermediates were unchanged (Fig. S1 A). As we previously observed in human PDAC (Kamphorst et al., 2015), glutamine was among the most depleted amino acids in the murine orthotopic tumors. Glutamine metabolism is particularly relevant to PDAC, since PDAC cancer cells uniquely rely on glutamine utilization as a major carbon and nitrogen source to sustain cell proliferation and tumor growth (Hosios et al., 2016; Hui et al., 2017; Son et al., 2013). Consistent with PDAC tumors displaying a paucity of nutrients, we found that murine and human PDAC tumors express asparagine synthetase (ASNS) and Sestrin2 (SESN2), both markers of metabolic stress that are highly induced upon glutamine deprivation (Fig. 1 B and Fig. S1 C; Tajan et al., 2018; Ye et al., 2010). Altogether, these results indicate that both murine and human PDAC tumors are depleted of nutrients, with the vital amino acid glutamine being among the most deficient metabolites.
Figure 1.
Glutamine deprivation induces EMT and promotes aggressive behaviors in PDAC cells. (A) Quantification of amino acids in orthotopic KPC tumors relative to normal murine pancreatic tissue. NEAA, nonessential amino acids; EAA, essential amino acids. Data are presented as box and whiskers plots. Vertical lines extend to the minimum and maximum values. (B) Immunohistochemical staining using antibodies specific to ASNS and SESN2 in normal murine pancreas, KPC orthotopic pancreatic tumor, and human PDAC specimen. Scale bar, 50 µm. (C) GSEA plots derived from RNA-seq data of glutamine-deprived versus control (glutamine-replete) KPC or AsPC-1 cells. NES, normalized enrichment score; FDR, false discovery rate. (D) Relative mRNA expression of the indicated genes measured by qPCR in KPC or AsPC-1 in glutamine-replete or glutamine-free medium for 24 h. Results are expressed relative to control cells in glutamine-replete conditions (+Q). Data are expressed as mean ± SEM; n = 5 (KPC) and n = 4 (AsPC-1) independent experiments. (E and F) Migration (E) and invasion (F) capacity of the indicated PDAC cells cultured in glutamine-replete medium (High Q, 4 mM) or glutamine starvation conditions (Low Q, 0 or 0.2 mM). Quantification is shown relative to the High Q condition and expressed as mean ± SEM; n = 3–5 independent experiments, each performed in triplicate. Representative images of migrating/invading cells stained with crystal violet are shown. Statistical significance was determined using unpaired two-tailed Student’s t test. The quantification of amino acids in A was generated from one experiment with n = 6 mice per cohort. The GSEA plots in C were generated from a single RNA-seq experiment that included n = 5 replicates per condition. Results are representative of two independent experiments in B and a pool of three to five independent experiments in D, E, and F. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure S1.
Glutamine deprivation induces metabolic stress and EMT in PDAC cells. (A) Quantification of metabolites in orthotopic KPC tumors relative to murine normal pancreas tissue. Data are presented as box and whiskers plots. Vertical lines extend to the minimum and maximum values. n = 6. (B) Volcano plots showing differentially expressed genes identified by RNA-seq in KPC or AsPC-1 cells grown in glutamine-replete versus glutamine-free medium for 24 h (fold-change ≥2 and less than or equal to −2; P < 0.05). n = 5 samples per group. (C) Relative gene expression of metabolic stress markers Asns and Sesn2 from RNA-seq analysis of KPC or AsPC-1 cells grown in glutamine-replete versus glutamine-free medium for 24 h. Results are expressed relative to the control cells (+Q). Data are expressed as mean ± SEM; n = 5 replicates. (D) Immunoblots assessing E-cadherin (E-Cad) protein expression in KPC or AsPC-1 cells in glutamine-replete or glutamine-free medium for 24 h. β-actin was used as a loading control. Statistical significance was determined using unpaired two-tailed Student’s t test. Data were generated from one experiment with n = 6 tumors (A) or n = 5 samples (C) or are representative of three independent experiments (D). **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To broadly explore pathways that are modulated by glutamine depletion, we analyzed global transcriptional changes by RNA sequencing (RNA-seq) in murine KPC and human AsPC-1 cells cultured in glutamine-free medium for 24 h. We found 1,387 and 3,005 genes differentially expressed upon glutamine deprivation (fold-change ≥1.5 and less than or equal to −1.5; P < 0.01) in KPC and AsPC-1 cells, respectively (Fig. S1 B). Gene set enrichment analysis (GSEA) identified the EMT signature among the most significantly enriched pathways up-regulated in the glutamine-starved cells (Fig. 1 C). EMT is a cellular process by which cancer cells lose their epithelial characteristics and acquire a mesenchymal phenotype that allows them to increase cell motility and adopt invasive behaviors (Nieto et al., 2016). Consistent with the induction of EMT, expression of the epithelial marker E-cadherin (CDH1) was decreased, whereas expression of N-cadherin (CDH2), a mesenchymal marker, was enhanced in response to glutamine depletion (Fig. 1 D and Fig S1 D). We next tested whether the induction of this nutrient stress–driven EMT signature was linked to migration and/or invasion. Importantly, we found that limitation of glutamine enhanced both the migration and invasion capacities of murine (KPC, EKPC) and human (AsPC-1, 779E) PDAC cells (Fig. 1, E and F). These findings demonstrate that nutrient stress driven by glutamine starvation can induce EMT and lead to increased aggressive behaviors in PDAC.
The EMT-TF Slug is induced by glutamine deprivation via ERK signaling and ATF4 activation
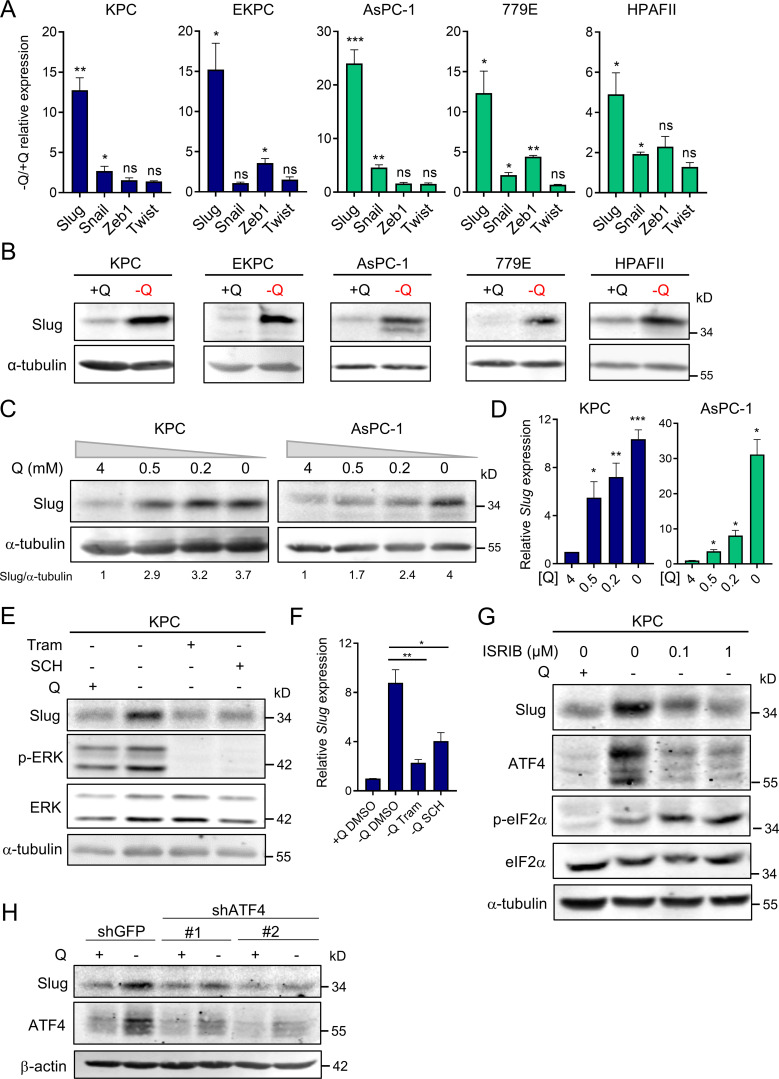
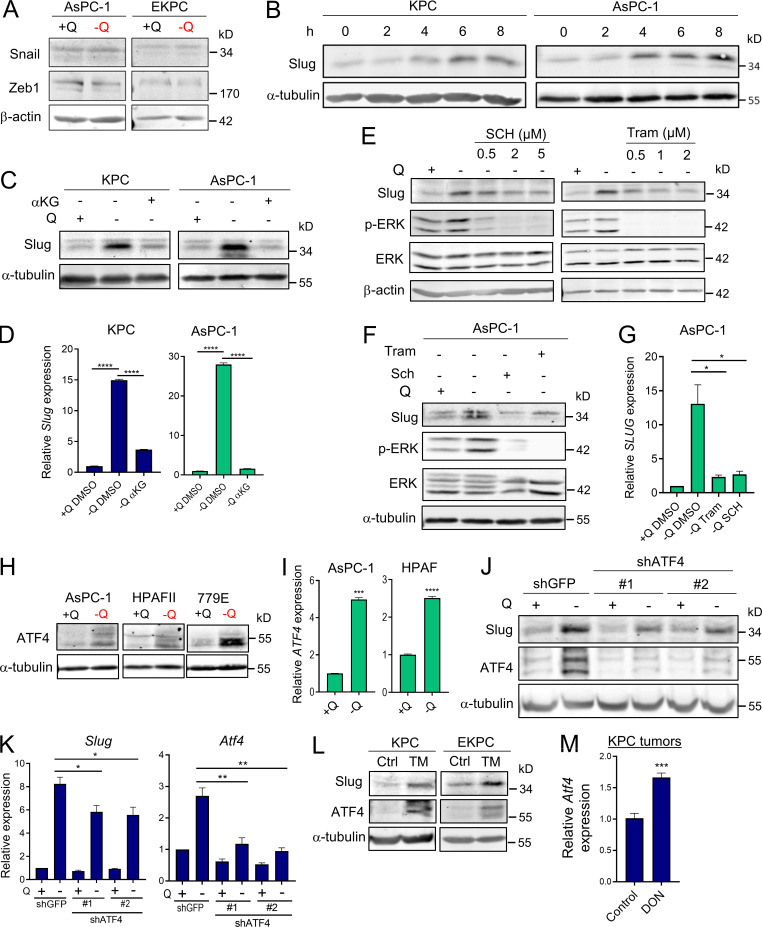
EMT is a complex biological process that involves the loss of cell polarity and epithelial cell–cell contact, reorganization of the actin cytoskeleton, and an increased ability to degrade extracellular matrix. These intricate processes are orchestrated by a set of EMT-TFs, including Snail, Slug, Twist, and Zeb1, that act as master regulators of EMT (De Craene and Berx, 2013; Lamouille et al., 2014). The relevance of each of these EMT-TFs can vary depending on context and the EMT-initiating stimulus; therefore, we examined the extent of induction for each EMT-TF upon glutamine starvation. Strikingly, we found that only Slug expression was robustly and uniformly increased, at both the transcript and protein levels, in all the PDAC cell lines tested (Fig. 2, A and B). In a few cases, Snail and Zeb1 transcripts were slightly induced by glutamine starvation, but the protein levels were unaffected (Fig. 2 A and Fig. S2 A). Notably, up-regulation of Slug by glutamine deprivation was rapid and sensitive to glutamine levels. Time-course experiments revealed a prompt induction of Slug protein starting at 4 h of glutamine starvation (Fig. S2 B), and dose–response assays showed the gradual enhancement of Slug protein and transcript levels as glutamine concentrations were decreased (Fig. 2, C and D). Importantly, Slug expression was heightened at both subphysiological (0.2 mM) and physiological (0.5 mM) concentrations of glutamine. To ascertain whether glutaminolysis has a role in Slug regulation, we cultured PDAC cells in glutamine-free medium supplemented with cell-permeable α-ketoglutarate (αKG), a downstream metabolite of glutamine catabolism. Addition of αKG was sufficient to rescue glutamine depletion–induced Slug expression at both the protein and mRNA levels in murine and human PDAC cells (Fig. S2, C and D).
Figure 2.
Slug is induced by glutamine deprivation via ERK signaling and ATF4. (A) Relative expression of Slug, Snail, Zeb1, and Twist transcript levels measured by qPCR in the indicated PDAC cells cultured in glutamine-replete or glutamine-free medium for 24 h. Results are expressed relative to control cells in glutamine-replete conditions (+Q). Data are expressed as mean ± SEM; n = 3–5 independent experiments. (B) Immunoblots of Slug expression in the indicated PDAC cells cultured in glutamine-replete or glutamine-free medium for 24 h. α-tubulin was used as a loading control. (C) Immunoblots assessing Slug expression in response to the indicated glutamine concentrations in KPC and AsPC-1 cells. α-tubulin was used as a loading control. Slug/α-tubulin ratios relative to 4 mM Q are shown. (D) Relative Slug mRNA expression by qPCR at the indicated glutamine concentrations in KPC and AsPC-1 cells. Results are expressed relative to the 4-mM Q concentration. Data are expressed as mean ± SEM; n = 3 independent experiments. (E) Immunoblots assessing protein expression of Slug, phospho-ERK (p-ERK), and ERK in KPC cells treated with vehicle (DMSO), 2 µM Tram or 2 µM SCH772984 (SCH) in glutamine-free medium for 5 h. α-tubulin was used as loading control. The immunoblots shown are representative of three independent experiments. (F) Relative Slug mRNA expression measured by qPCR in KPC cells under the same conditions described in E. Data are expressed as mean ± SEM; n = 3 independent experiments. (G) Immunoblots showing protein expression of Slug, ATF4, p-eIF2α (Ser51), and eIF2α in KPC cells treated with vehicle (DMSO) or ISRIB at the indicated concentrations in glutamine-free medium. α-tubulin was used as loading control. (H) Immunoblots assessing Slug and ATF4 protein expression in KPC cells stably expressing a nontargeting shRNA control (shGFP) or two independent short hairpins targeting ATF4 (shATF4#1 and shATF4#2) in glutamine-replete or glutamine-free medium for 24 h. β-actin was used as a loading control. Statistical significance was determined using unpaired two-tailed Student’s t test. Results are representative of at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Figure S2.
Slug expression is induced by glutamine deprivation via ERK and ATF4. (A) Immunoblots assessing Snail and Zeb1 protein expression in AsPC-1 or EKPC cells cultured in glutamine-replete or glutamine-free medium for 24 h. β-actin was used as a loading control. (B) Immunoblots showing Slug protein expression upon glutamine starvation for 0–8 h in KPC and AsPC-1 cells. α-tubulin was used as a loading control. Slug/α-tubulin ratios are shown relative to t = 0. (C) Immunoblots assessing Slug protein expression in glutamine-replete or glutamine-free medium supplemented with cell-permeable αKG, 7 mM) for 24 h in KPC and AsPC-1 cells. α-tubulin was used as a loading control. Blots are representative of at least three independent experiments. (D) Relative Slug mRNA expression determined by qPCR in KPC and AsPC-1 cells under the same conditions described in C. Data are expressed as mean ± SEM. (E) Immunoblots assessing protein expression of Slug, phospho-ERK (p-ERK), and ERK in KPC cells treated with vehicle (DMSO) or the indicated concentrations of Tram or SCH772984 (SCH) in glutamine-free medium for 5 h. β-actin was used as loading control. The immunoblots shown are representative of three independent experiments. (F) Immunoblots assessing protein expression of Slug, phospho-ERK (p-ERK), and ERK in AsPC-1 cells treated with vehicle (DMSO), 2 µM Tram, or 2 µM SCH in glutamine-free medium for 5 h. α-tubulin was used as loading control. The immunoblots shown are representative of three independent experiments. (G) Relative SLUG mRNA expression measured by qPCR in AsPC-1 cells under the same conditions described in F. Data are expressed as mean ± SEM; n = 3 independent experiments. (H) Immunoblots assessing protein expression of ATF4 in the indicated human PDAC cell lines in glutamine-replete or glutamine-free medium for 24 h. α-tubulin was used as loading control. (I) Relative ATF4 mRNA expression measured by qPCR in AsPC-1 and HPAFII cells under the same conditions described in H. Data are expressed as mean ± SEM. (J) Immunoblots assessing Slug and ATF4 protein expression in EKPC shGFP, shATF4#1, or shATF4#2 cells incubated in glutamine-replete or glutamine-free medium for 24 h. α-tubulin was used as loading control. (K) Relative gene expression of Slug and ATF4 evaluated by qPCR in KPC shGFP, shATF4#1, or shATF4#2 cells incubated in glutamine-replete or glutamine-free medium for 24 h. Results are expressed relative to shGFP/+Q condition. Data are shown as mean ± SEM; n = 3 independent experiments. (L) Immunoblotting of Slug and ATF4 in KPC and EKPC cells treated with vehicle (DMSO) or 1 µg/ml Tunicamycin (TM) for 24 h in glutamine-replete conditions. α-tubulin was used as a loading control. (M) Relative ATF4 mRNA expression measured by qPCR in KPC subcutaneous tumors from mice treated with vehicle-only control (Ctrl) or DON (10 mg/kg). Results are expressed relative to Ctrl group. Data are expressed as mean ± SEM; n = 6. Statistical significance was determined using unpaired two-tailed Student’s t test. Data in D were generated from one experiment with n = 3 replicates. Data in M were generated from one experiment with n = 6 mice per cohort. Data are representative of two (B and I) or three (A, C, E–H, and J–L) independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We recently reported that glutamine deprivation in human PDAC cells activates ERK signaling (Lee et al., 2019); therefore, we next examined whether Slug up-regulation by glutamine restriction was dependent on MEK/ERK signal transduction. We found that suppression of MEK/ERK signaling, as confirmed by the extent of ERK phosphorylation (p-ERK), via administration of either ERK or MEK inhibitors, abrogated glutamine deprivation–induced Slug expression in both murine and human PDAC cells (Fig. 2 E and Fig. S2, E and F). This observed suppression of Slug expression by ERK inhibition occurs at the level of transcription (Fig. 2 F and Fig. S2 G). Our results indicate that ERK activation mediates Slug induction upon glutamine starvation. It was previously reported that MEK/ERK signaling is required for activation of the integrated stress response (ISR) through phosphorylated eukaryotic initiation factor 2α (p-eIF2α)/ATF4 in the context of nutrient starvation (Thiaville et al., 2008). ATF4 is thought to regulate the cellular response to a variety of different stresses, including amino acid starvation. Amino acid starvation, through the accumulation of uncharged transfer RNAs, triggers a GCN2-dependent activating phosphorylation of eIF2α, which in turn inhibits global cap-dependent translation and favors the differential translation of transcripts containing an upstream open reading frame, such as ATF4 (Pakos-Zebrucka et al., 2016; Ye et al., 2010). Because of the established link between MEK/ERK signaling and p-eIF2α/ATF4 (Thiaville et al., 2008), we evaluated whether this pathway was responsible for Slug up-regulation in glutamine-starved cells. We observed an increase in both p-eIF2α and ATF4 upon glutamine starvation, indicative of the activation of ISR in KPC cells (Fig. 2 G). Similarly, glutamine deprivation induced ATF4 expression at both the protein and mRNA levels in human PDAC cell lines (Fig. S2, H and I). Inhibition of ATF4 by treatment with the ISR inhibitor ISRIB, which prevents the inhibition of translation downstream of p-eIF2α (Sidrauski et al., 2013), resulted in a reduction of glutamine starvation–induced Slug expression (Fig. 2 G). Consistent with previous reports, ISRIB did not block eIF2α phosphorylation and in fact further increased the level of eIF2α phosphorylation (Sidrauski et al., 2013). Similarly, attenuation of ATF4 expression by shRNA-mediated knockdown was also able to rescue the glutamine depletion–driven Slug induction in KPC and EKPC cells (Fig. 2 H and Fig. S2, J and K). ATF4 can be robustly activated by ER stress; indeed, treatment with the ER-stress inducer tunicamycin in glutamine-replete medium enhanced both ATF4 and Slug expression in KPC and EKPC PDAC cells (Fig. S2 L). Collectively, these data demonstrate that glutamine depletion–driven Slug induction is orchestrated by MEK/ERK signaling and by activation of ISR through p-eIF2α and ATF4.
Slug expression is enhanced by glutamine deprivation in vivo and coincides with metabolic stress markers in PDAC tumors
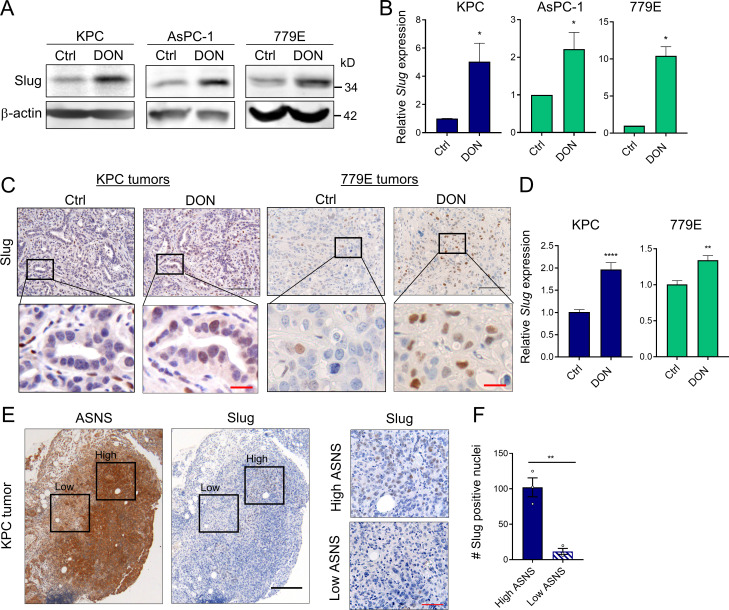
To determine whether glutamine deficiency induces Slug expression in vivo, we used the glutamine analogue 6-diazo-5-oxo-l-norleucine (DON) that broadly inhibits glutamine-using enzymes, hence mimicking glutamine deprivation (Lemberg et al., 2018). Consistent with our previous results, glutamine inhibition by DON treatment in vitro substantially enhanced Slug protein and transcript levels in murine KPC and human AsPC-1 and 779E cell lines (Fig. 3, A and B). To evaluate the in vivo effects of DON treatment on Slug expression, we administered DON for 5 d to C57BL/6J or athymic animals bearing heterotopic tumors derived from either KPC or 779E cells, respectively. We found that DON treatment significantly increased Slug expression in the tumors, as assessed by immunohistochemistry and quantitative real-time PCR (qPCR; Fig. 3, C and D). Interestingly, DON treatment induced ATF4 mRNA expression in KPC heterotopic tumors, suggesting that ATF4 may play a role in Slug regulation in vivo as well (Fig. S2 M). We conclude from these data that pharmacological blockade of glutamine metabolism has the capacity to enhance Slug expression in vivo.
Figure 3.
Slug expression is enhanced by a glutamine antagonist in vivo and coincides with metabolic stress in PDAC tumors. (A) Immunoblots of Slug protein expression in the indicated PDAC cells treated for 24 h with DON compared with vehicle-only control (Ctrl) cells. β-actin was used as loading control. Results are representative of three independent experiments. (B) Relative Slug mRNA expression assessed by qPCR in the indicated cell lines in the same conditions described in A. Data are expressed as mean ± SEM; n = 3 independent experiments. (C) Immunohistochemical staining of Slug protein in KPC or 779E heterotopic tumors from mice treated with vehicle-only control (Ctrl) or DON (10 mg/kg). The inset boxes indicate the areas further magnified below. Black scale bar, 100 µm; red scale bar, 20 µm. (D) Relative Slug mRNA expression assessed by qPCR in KPC or 779E subcutaneous tumors from mice treated with vehicle-only control (Ctrl) or DON (10 mg/kg). Results are expressed relative to Ctrl group. Data are expressed as mean ± SEM; n = 10 KPC and n = 5 779E tumors. (E) Immunohistochemical staining of ASNS and Slug protein in KPC orthotopic tumors. The inset boxes indicate the areas of high and low ASNS and Slug expression in the further magnified images to the right. The images shown are representative of three different tumors. Black scale bar, 200 µm; red scale bar, 50 µm. (F) Quantification of Slug-positive nuclei in areas of high or low ASNS expression in the KPC orthotopic tumors shown in E. Results are expressed as the mean ± SEM; n = 3 tumors. Four to five images of high or low ASNS areas per tumor were quantified at 40× magnification. Statistical significance was determined using unpaired two-tailed Student’s t test. Data are representative of three independent experiments (A and B), two independent experiments (C and D), or one experiment with n = 3 mice per cohort (E and F). *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
We and others have demonstrated that tumors display amino acid deficiencies that lead to regional metabolic stress (Lee et al., 2019; Pan et al., 2016); therefore, we assessed whether intratumoral Slug expression coincides with an established metabolic stress marker. We immunostained serial tumor sections originating from KPC-derived orthotopic tumors with antibodies specific to Slug and to the stress marker ASNS. Notably, we observed that tumor regions with high levels of Slug coincided with the highest expression levels of ASNS, whereas tumor regions where ASNS expression was lower were often associated with a decreased number of Slug-positive cells (Fig. 3, E and F). Taken together, these results demonstrate that Slug is up-regulated in response to nutrient stress in PDAC tumors.
Slug mediates EMT and nutrient stress survival in PDAC cells
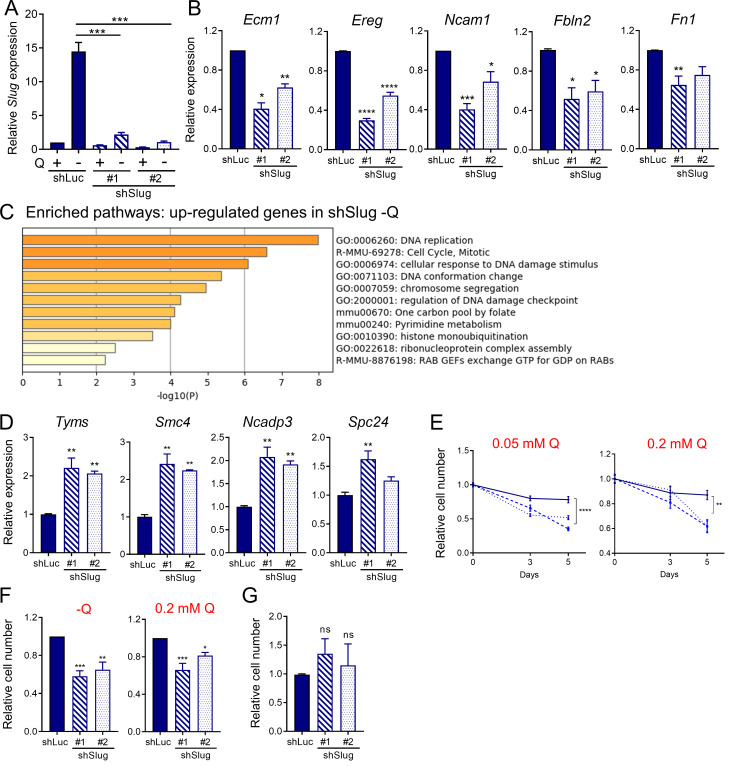
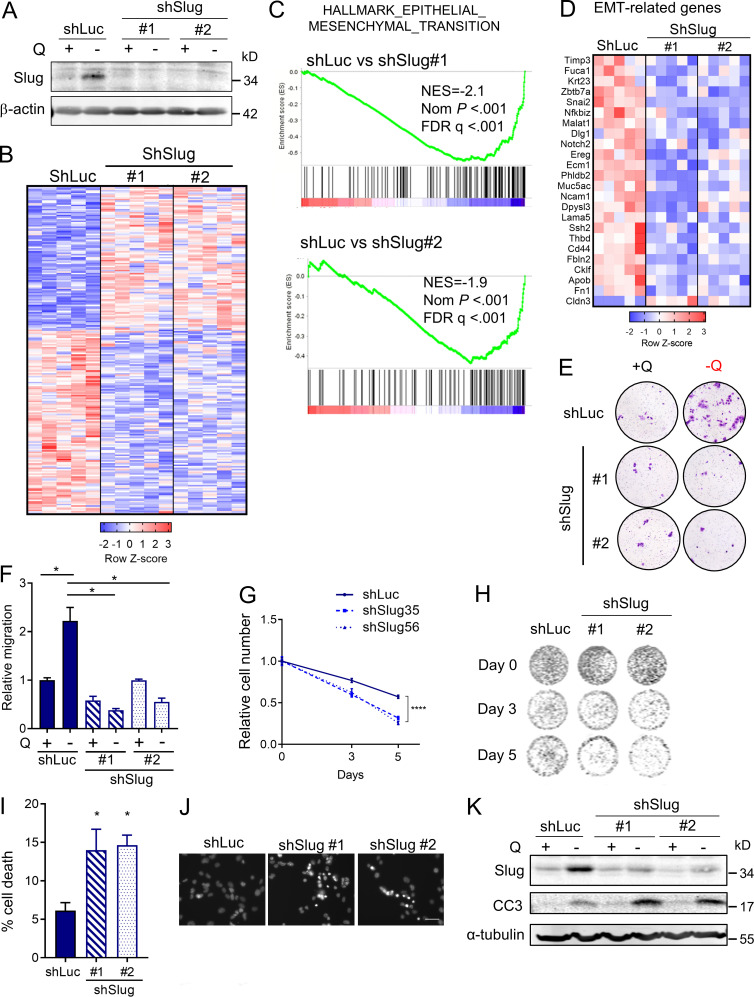
Having established a unique link between EMT and glutamine deprivation in PDAC, we sought to elucidate whether Slug was regulating EMT, and possibly other biological functions, in glutamine-starved cells. To this end, we generated stable Slug knockdown cell lines via lentiviral transduction with two different Slug-targeting shRNAs and a luciferase-targeting shRNA hairpin as a negative control (shSlug#1, shSlug#2, and shLuciferase [shLuc]). Slug knockdown in KPC cells was confirmed by Western blot and qPCR (Fig. 4 A and Fig. S3 A). To examine the broad impact of Slug depletion in the context of glutamine deprivation, we performed RNA-seq analysis on shSlug and shLuc cells that were deprived of glutamine for 24 h. We identified 226 genes that were differentially expressed in both Slug-targeting shRNAs (fold-change ≥1.5 and less than or equal to −1.5; P < 0.05; Fig. 4 B). GSEA analysis confirmed the requirement of Slug for the induction of a transcriptional EMT program under glutamine deprivation, as demonstrated by the down-regulation of the EMT gene signature and other EMT associated pathways in shSlug cells (Fig. 4 C and Table S1). Several of the EMT-related genes inhibited in the Slug-depleted cells were validated by qPCR (Fig. 4 D and Fig. S3 B). Consistent with a role for Slug in EMT caused by glutamine withdrawal, suppression of Slug abolished the increased cell motility observed under glutamine starvation (Fig. 4, E and F). Together, these results establish a clear causal link between Slug induction and nutrient stress–driven EMT.
Figure S3.
Slug regulates EMT and survival upon glutamine starvation. (A) Relative Slug mRNA expression by qPCR in KPC Slug knockdown cells compared with control (shLuc) in glutamine-replete or glutamine-free medium for 24 h. Results are expressed relative to shLuc/+Q condition. Data are shown as mean ± SEM; n = 5 independent experiments. (B) Relative expression of the indicated EMT-related genes in KPC Slug knockdown compared with shLuc in glutamine-free medium for 24 h. Results are expressed relative to shLuc. Data are shown as mean ± SEM; n = 3 independent experiments. (C) Chart showing enriched pathways among the genes up-regulated in Slug-depleted KPC cells compared with control (shLuc) in glutamine-free conditions. The graph was generated with Metascape. (D) Relative expression of the indicated genes in KPC Slug knockdown compared with shLuc in glutamine-free medium for 24 h. Results are expressed relative to shLuc. The results are representative of three independent experiments. Data are presented as the mean ± SEM of three replicates. (E) Relative cell number assessed by Syto60 staining in KPC shLuc, shSlug#1, or shSlug#2 cells incubated in medium containing 0.05 or 0.2 mM glutamine for 3 and 5 d. Representative data are shown for n = 3 independent experiments with three to five replicates each. (F) Relative cell number assessed by crystal violet staining in KPC shLuc, shSlug#1, or shSlug#2 cells incubated in glutamine-free medium or medium containing 0.2 mM glutamine for 5 or 6 d. Data are normalized to day 0 for each cell line and are shown relative to the shLuc control. Results are expressed as mean ± SEM; n = 5 independent experiments. (G) Relative cell number assessed by crystal violet staining in KPC shLuc, shSlug#1, or shSlug#2 incubated in complete medium for 48 h. Results are expressed as mean ± SEM; n = 4 independent experiments. ns, not significant. Statistical significance was determined using unpaired Student’s t test in A and one-way ANOVA followed by Dunnett's test for multiple comparisons in B–G. All data are representative of at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001.
Figure 4.
Slug mediates EMT and nutrient stress survival in PDAC cells. (A) Immunoblot assessing Slug expression in KPC cells stably expressing a nontargeting shRNA control (shLuc) or two independent short hairpins targeting Slug (shSlug#1 and shSlug#2) in glutamine-replete or glutamine-free medium. β-actin was used as a loading control. (B) Heatmap of RNA-seq data depicting differentially expressed genes in KPC shLuc, shSlug#1, and shSlug#2 under glutamine deprivation for 24 h (fold-change ≥1.5 and less than or equal to −1.5; P < 0.05). n = 5 samples per group. (C) GSEA plots of RNA-seq data from KPC shLuc versus shSlug#1 or shSlug#2 under glutamine deprivation for 24 h showing inhibition of the EMT gene signature in Slug knockdown cells compared with shLuc. NES, normalized enrichment score; FDR, false discovery rate. (D) Heatmap showing EMT-related genes that are differentially expressed in KPC shLuc versus shSlug#1 and shSlug#2 after glutamine starvation for 24 h. (E) Representative images of a Transwell migration assay comparing KPC shLuc, shSlug#1, or shSlug#2 incubated in glutamine-replete or glutamine-free medium for 72 h. Cells were stained with crystal violet. (F) Quantification of the migration capacity of KPC shLuc, shSlug#1, or shSlug#2 cells from E. Results are presented relative to the shLuc/+Q condition and are representative of n = 3 independent experiments. Data are expressed as the mean ± SEM of triplicate wells. (G) Relative cell number as assessed by Syto60 fluorescent staining of KPC shLuc, shSlug#1, and shSlug#2 cells cultured in glutamine-free medium for 5 d. Results are presented relative to shLuc/day 0 and are representative data of five independent experiments, each performed with five replicates per group. (H) Representative images for Syto60 staining under the conditions described in G. (I) Quantification of cell death in KPC shLuc, shSlug#1, and shSlug#2 cells under glutamine deprivation for 48 h as evaluated by Hoechst staining. Data are expressed as mean ± SEM; n = 3 independent experiments. (J) Representative images of Hoechst-stained KPC shLuc, shSlug#1, and shSlug#2 cells under the conditions described in I. Scale bar, 50 µm. (K) Immunoblot assessing Slug and cleaved caspase 3 (CC3) protein in KPC shLuc, shSlug#1, and shSlug#2 cells cultured in glutamine-replete or glutamine-free medium for 72 h. α-tubulin was used as a loading control. The results are representative of three independent experiments. Statistical significance was determined using unpaired two-tailed Student’s t test in F and one-way ANOVA followed by Dunnett's test in G and I. Data in B, C, and D were generated from a single RNA-seq experiment with n = 5 replicates per condition. Data are representative of at least three independent experiments (A and E–K). *, P < 0.05; ****, P < 0.001.
Interestingly, GSEA and Metascape (Zhou et al., 2019b) analyses of the up-regulated genes in glutamine-starved Slug knockdown cells revealed a pronounced enrichment in proliferation/survival pathways, such as DNA replication, DNA damage, and pyrimidine metabolism, among others (Fig. S3 C and Table S2). We validated the RNA-seq results for a subset of these Slug targets, including Tyms, Smc4, Spc24, and Ncapd3, by qPCR (Fig. S3 D). These findings suggested that when glutamine is scarce, Slug normally functions as a repressor of these proliferation pathways, possibly aiding to decrease metabolic demands under nutrient stress conditions. Glutamine contributes to nucleotide biosynthesis, and restriction of glutamine supply can cause cell cycle arrest, DNA damage, and/or apoptosis in different cellular contexts (Gaglio et al., 2009; Son et al., 2013; Tajan et al., 2018; Zhang et al., 2014). It has also been well established that EMT confers resistance to cell death induced by various stimuli, including DNA damage and chemotherapeutic agents (De Craene and Berx, 2013; Vega et al., 2004; Zheng et al., 2015). In particular, Slug has a protective role against DNA damage and exerts anti-apoptotic effects by antagonizing p53 (Kurrey et al., 2009; Wu et al., 2005). Based on these paradigms, we reasoned that Slug induction during glutamine starvation could be part of an adaptive response that serves to confer a survival advantage to PDAC cells. To test this notion, we assessed survival capacity of Slug knockdown cells relative to control cells in glutamine-free medium. We found that Slug suppression significantly diminished cell viability upon glutamine starvation, as measured by Syto60 staining and by crystal violet staining (Fig. 4, G and H; and Fig. S3 F). This observed decrease in survival was also evident when cells were cultured in nonzero subphysiological glutamine concentrations (Fig. S3, E and F). Importantly, Slug suppression had no effect on the proliferative capacity of KPC cells in glutamine-replete medium (Fig. S3 G), suggesting that the survival role of Slug might be most relevant in the setting of nutrient stress.
We next evaluated whether the effects of Slug suppression on nutrient stress survival are due to an increase in cell death upon glutamine restriction. Nuclear condensation, as visualized by Hoechst 33342 staining, can be used to distinguish apoptotic cells from healthy cells (Crowley et al., 2016). Hoechst 33342 staining showed a significant increase in the percentage of apoptotic cells in Slug-knockdown cells that were deprived of glutamine relative to control cells (Fig. 4, I and J). Consistent with apoptotic enhancement, we observed a strong induction of cleaved caspase-3 protein in Slug-deficient cells starved of glutamine for 72 h (Fig. 4 K). Collectively, these results indicate that Slug mediates PDAC cell survival under glutamine deprivation by suppressing apoptosis.
Slug regulates metastasis in PDAC
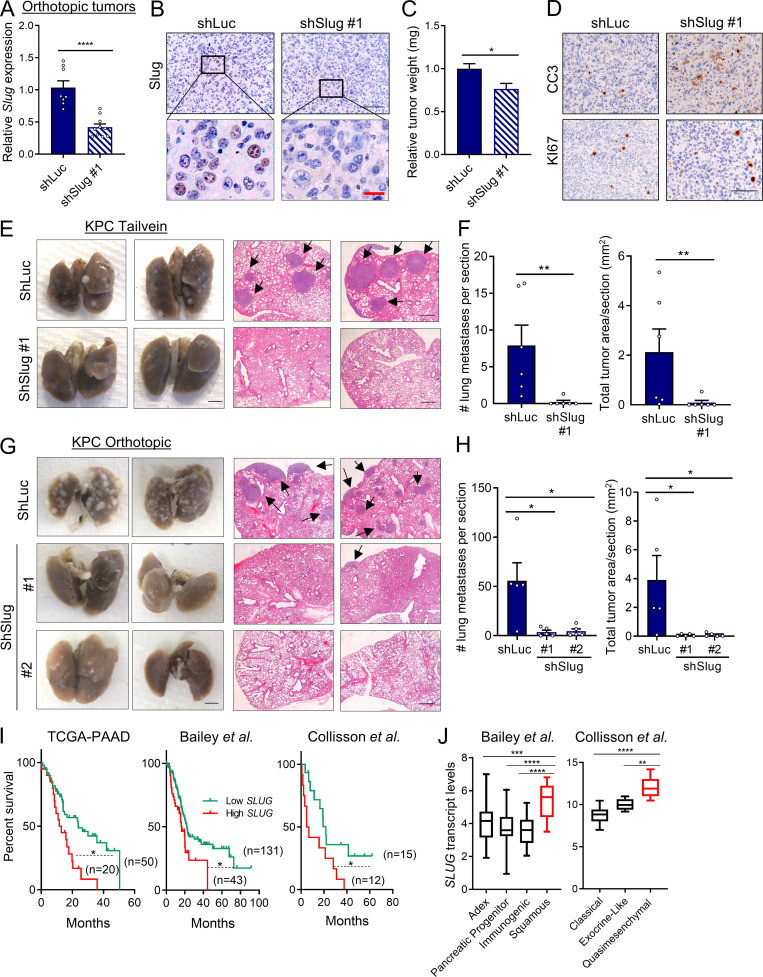
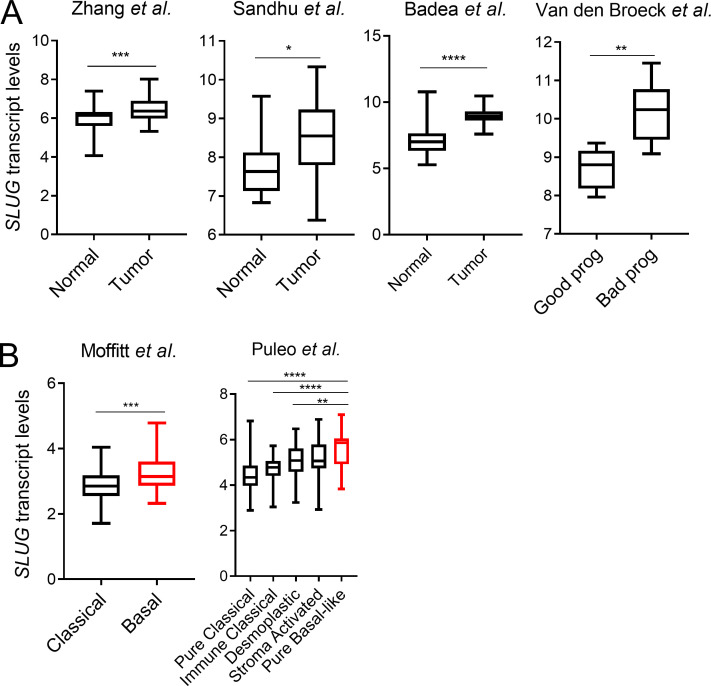
Activation of the EMT program is considered a critical step in tumor progression and metastatic disease (Lu and Kang, 2019; Nieto et al., 2016). Previous studies in PDAC models have examined the EMT-TFs Snail, Twist, and Zeb1. None of these EMT-TFs seem to be required for PDAC tumor initiation or growth; however, in terms of metastatic capacity, only Zeb1 has been clearly shown to regulate metastasis in PDAC (Aiello et al., 2017; Krebs et al., 2017; Zheng et al., 2015). These data suggest that the EMT-TFs might differentially regulate metastatic potential in PDAC depending on context. Notably, very little is known about the contribution of Slug to tumor growth and metastasis in PDAC. To examine any broad effects of Slug depletion on primary tumors in vivo, we used an orthotopic syngeneic mouse model of PDAC using EKPC cells engineered to stably knockdown Slug. Depletion of Slug expression in the orthotopic tumors was confirmed by qPCR and immunostaining (Fig. 5, A and B). Although Slug knockdown did not affect tumor initiation, there was an ∼25% reduction in tumor burden (Fig. 5 C). This observed suppression in tumor growth was likely due to increased apoptosis as opposed to decreased proliferation, as Slug-knockdown tumors displayed normal levels of KI-67 but enhanced staining of cleaved caspase-3 (Fig. 5 D). These data suggest that Slug may exert a moderate but significant prosurvival effect in these nutrient-poor primary tumors by suppressing apoptosis. Next, we evaluated the role of Slug in metastasis using two mouse models. As a first approach, we used an experimental model of lung metastases, in which tumor cells are i.v. injected into systemic circulation (Khanna and Hunter, 2005). KPC shSlug or shLuc control cells were i.v. injected into the tail veins of C57BL/6J mice; 25 d after injection, lung tissue was harvested and examined for macro- and micrometastases. Although shLuc control cells efficiently colonized the lung, giving rise to large tumors, Slug knockdown strikingly abolished the ability of KPC cells to colonize the lung, as evidenced by the reduction of both the number and total area of lung foci (Fig. 5, E and F). As a second approach, we used the KPC orthotopic syngeneic model to analyze the development of spontaneous metastasis. In this setting, Slug knockdown also abrogated the ability of KPC cells to form distant metastases from the primary tumor, as demonstrated by the significantly decreased number of metastatic foci and total tumor area in the lungs (Fig. 5, G and H). These results establish that Slug is required for metastatic dissemination in murine PDAC models. Lastly, linking Slug to human PDAC, our analyses of SLUG expression in publicly available datasets revealed that SLUG is more highly expressed in PDAC tumors compared with normal tissue (Fig. S4 A), and that increased SLUG transcript levels are associated with poor survival of PDAC patients (Fig. 5 I). Additionally, SLUG expression is enhanced in PDAC subtypes that are associated with the worst outcome (Fig. 5 J and Fig. S4 B; Badea et al., 2008; Collisson et al., 2011; Van den Broeck et al., 2012; Moffitt et al., 2015; Sandhu et al., 2015; Bailey et al., 2016; Puleo et al., 2018). These observations suggest that Slug may play a role in the metastatic progression of PDAC tumors in patients.
Figure 5.
Slug regulates metastasis in PDAC. (A) Relative Slug mRNA expression evaluated by qPCR in EKPC shLuc or EKPC shSlug#1 orthotopic tumors. Results are expressed relative to shLuc. Data are expressed as mean ± SEM; n = 8 shLuc tumors; n = 10 shSlug#1 tumors. (B) Immunohistochemical staining of Slug protein in EKPC shLuc or EKPC-shSlug#1 orthotopic tumors. The inset boxes indicate the areas further magnified below. Black scale bar, 100 µm; red scale bar, 20 µm. (C) Relative tumor weight of EKPC shLuc or EKPC shSlug#1 orthotopic tumors. Data are expressed as mean ± SEM; n = 8 shLuc tumors; n = 10 shSlug#1 tumors. (D) Immunohistochemical staining of cleaved caspase 3 (CC3) and KI67 in EKPC shLuc or EKPC shSlug#1 orthotopic tumors. Images are representative of n = 3 tumors. Scale bar, 100 µm. (E) Lung colonization capacity of KPC shLuc and KPC shSlug#1 cells injected into the tail vein of syngeneic mice. Representative images of whole lungs and H&E-stained lung sections are shown. The arrows indicate lung metastatic foci. Images are representative of n = 6 mice per group. Scale bars: left panel, 2.5 mm; right panel, 500 µm. (F) Quantification of lung colonies (number and area per section) after tail vein injection as described in E. Data are presented as mean ± SEM; n = 6 mice per group. (G) Representative images of macroscopic and H&E-stained lungs of mice bearing KPC shLuc, KPC shSlug#1, or KPC shSlug#2 orthotopic tumors. The arrows indicate lung metastatic foci. Scale bars: left panel, 2.5 mm; right panel, 500 µm. (H) Quantification of lung metastatic colonies (number and area per section) originated from KPC shLuc, KPC shSlug#1, or KPC shSlug#2 orthotopic tumors as described in G. Data are presented as the mean ± SEM for n = 5 mice per group. (I) Kaplan–Meier survival analysis of the indicated datasets comparing low and high SLUG expression in PDAC patients. (J) Box plots representing SLUG transcript levels across different PDAC subtypes from the indicated public datasets. Data are presented as box and whiskers plots. Vertical lines extend to the minimum and maximum values. ADEX, aberrantly differentiated endocrine exocrine. Statistical significance was determined using unpaired two-tailed Student’s t test in A and C; Mann–Whitney nonparametric U test in F and H; log-rank (Mantel–Cox) test in I; and one-way ANOVA followed by Tukey test for multiple comparisons in J. Data were pooled from two cohorts (A–D, G, and H) and are representative of two independent experiments (E and F). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S4.
Slug is more highly expressed in human PDAC and is associated with poor prognosis. (A) Box plots showing SLUG mRNA expression in the indicated datasets comparing normal pancreas versus PDAC tumors and good prognosis versus bad prognosis. (B) Box plots showing SLUG mRNA expression in the indicated datasets comparing different PDAC subtypes. Data in A and B are presented as box and whiskers plots. Vertical lines extend to the minimum and maximum values. Statistical significance was determined using unpaired two-tailed Student’s t test in A and one-way ANOVA followed by Tukey multiple comparisons in B. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Discussion
Our work demonstrates a previously unappreciated link between glutamine depletion–induced nutrient stress and the induction of EMT and aggressive behaviors in PDAC cells. We decipher a novel mechanism by which glutamine scarcity selectively up-regulates the expression of the EMT master regulator Slug to drive EMT and enhance invasiveness. Moreover, Slug depletion leads to the suppression of metastasis, establishing an important role for this EMT-TF in PDAC aggressiveness.
The role of Slug in metastatic capacity of PDAC has not previously been reported, despite the high Slug expression in human PDAC tumors (Hotz et al., 2007) and its association with poor survival. Our findings contribute to the understanding of the specific roles of the different EMT-TFs in PDAC by positioning Slug as an important driver of metastasis in response to nutrient stress in PDAC tumors. Besides the role of Slug in the induction of EMT and aggressiveness in response to glutamine starvation, we also show that Slug functions to promote cell survival under nutrient limitation. The protective role of Slug in glutamine-deprived conditions is associated with the prevention of apoptosis, possibly related to the repression of genes involved in cell proliferation. Our work also points to a pivotal role of glutamine availability in the induction of Slug-dependent functions in PDAC cells. We delineated that the induction of Slug upon glutamine depletion requires activation of the MEK/ERK signaling pathway, as well as activation of the ISR through p-eIF2α/ATF4. These signaling pathways are mutually dependent to modulate survival upon amino acid deprivation (Thiaville et al., 2008). Indeed, a recent study demonstrated the therapeutic benefit of combining MEK inhibition with asparagine deprivation by treatment with l-asparaginase in both melanoma and PDAC tumors (Pathria et al., 2019). Hence, our findings integrate the observed activation of MEK/ERK signaling in response to glutamine starvation through epidermal growth factor receptor (EGFR) in PDAC (Lee et al., 2019) and the role of ERK signaling in the induction of Slug expression in other tumor types (Chen et al., 2009; Liu et al., 2016; Zhou et al., 2019a). Additionally, our data demonstrating that the p-eIF2α/ATF4 pathway regulates Slug expression suggest that Slug might represent a novel and important mediator of ISR that contributes to cell survival by preventing apoptosis.
We demonstrate that in PDAC tumors, Slug expression is coincident with the metabolic stress marker ASNS, which is highly induced in response to glutamine starvation. These results are in line with previous observations in melanoma tumors that revealed that tumor core regions with low glutamine levels were associated with increased histone methylation and elevated expression of genes involved in dedifferentiation (Pan et al., 2016). ASNS is a well-described ATF4 target gene (Gwinn et al., 2018). Hence, its coexpression with Slug in specific intratumoral regions suggests that either ATF4 activity or expression is higher in these areas. We recently demonstrated that, similar to melanoma tumors, PDAC tumor cores are deficient of glutamine and other nonessential amino acids (Lee et al., 2019). EGFR activation was enhanced in these tumor cores, and since p-ERK is a readout of EGFR signaling, it would not be surprising that p-ERK levels were enhanced in regions of Slug expression or in DON-treated tumors. Although glutamine depletion-induced EMT is exhibited by both PDAC and melanoma cells, the specific EMT-TFs used might be different, since Slug expression is actually down-regulated in glutamine-deprived melanoma cells (Falletta et al., 2017). Along the same lines, breast cancer cells that were selected to grow independently of glutamine show increased metastatic capacity due to the overexpression of cyclooxygenase 2 (COX-2 or PTGS2; Singh et al., 2012). Interestingly, in our RNA-seq analyses, we observed a prominent induction of COX-2 upon glutamine depletion in PDAC cells, although its up-regulation was Slug independent. It might be interesting to evaluate whether COX-2 mediates EMT and metastasis in PDAC, and whether it plays a role in Slug induction, as was previously reported in bladder cancer cells (Adhim et al., 2011). Irrespective of the mechanism used, the regulation of EMT by nutrient availability points to the EMT program as an important aspect of the metabolic reprogramming that occurs in tumors (García-Jiménez and Goding, 2019; Sciacovelli and Frezza, 2017), and it is worth further examination to evaluate whether this adaptation can be targeted therapeutically. Based on our results, one possibility is that the suppression of both ATF4 and Slug expression via p-eIF2α inhibition might represent an attractive therapeutic strategy in PDAC to prevent or diminish metastasis in the adjuvant setting.
Materials and methods
Cells and cell culture conditions
AsPC-1, HPAFII, and HEK 293T cells were obtained from the American Type Culture Collection; 779E are epithelial cells established from a moderate- to poorly differentiated patient-derived tumor (Villarino et al., 2017). KPC cells originated from Pdx1-Cre; LSL-KRasG12D/+; LSL-Tp53R172H/+ mice were kindly provided by Dr. Robert H. Vonderheide (University of Pennsylvania, Philadelphia, PA; Bayne et al., 2012) and subcloned in our laboratory to give rise to the KPC#65 clone used in this study. EKPC cells originate from Pdx1-Cre; LSL-EYFP/+ LSL-KRasG12D/+; LSL-Tp53R172H/+ mice. All cell lines were maintained in 100 units/ml penicillin/streptomycin under 5% CO2 at 37°C and were routinely tested for mycoplasma contamination by ABM’s PCR Mycoplasma detection kit. AsPC-1 cells were cultured in RPMI 1640 (Corning) supplemented with 10% FBS and 1 mM sodium pyruvate. HPAFII, KPC, EKPC, and 779E cells were cultured in DMEM supplemented with 10% FBS. Glutamine deprivation experiments were performed in glutamine-free RPMI 1640 (Corning) or glutamine-free DMEM (Corning) supplemented with 10% dialyzed FBS for 24 h unless otherwise indicated. For glutamine dose–response experiments, glutamine-free RPMI or DMEM was supplemented with the indicated glutamine concentrations. The following inhibitors were used in this study at the indicated durations and concentrations: DON (Sigma-Aldrich), ISRIB (Sigma-Aldrich), Tunicamycin (MP Biomedicals), Sch772984 (CHEMIETEK), and Trametinib (Tram; Selleckchem).
Generation of stable cell lines by lentiviral transduction
Lentiviral vectors (pLKO.1) containing shRNA constructs were obtained from the TRC Lentiviral shRNA Library (Dharmacon). The following shRNA sequences were used: shSlug#1, 5′-ACCCTATACCTGTCATACCAA-3′; shSlug#2, 5′-CCCATATCTCTATGAAAGTTA-3′; shATF4 #1, 5′-ATAGTCATCTAAGAGACCTAG-3′; and shATF4 #2, 5′-ACTCGAAGGTATCTTTGTCCG-3′. shLuc, 5′-CGCTGAGTACTTCGAAATGTC-3′, and shGFP, 5′-GTCGAGCTGGACGGCGACGTA-3′, were used as controls. HEK 293T cells were cotransfected with 12 µg of each shRNA containing pLKO.1 plasmid, 8 µg of psPAX2 packaging plasmid, and 4 µg of pMD2.G envelope plasmid using Lipofectamine 2000 (Invitrogen) in 10-cm plates. After 16 h, medium was replaced with fresh 10% FBS-DMEM without antibiotics, and 48 h later, the viral supernatant was collected and filtered through a 0.45-µm filter. KPC and EKPC cells were transduced with viral supernatant and 8 µg/ml polybrene, and stable cells were selected with 7 µg/ml puromycin.
In vivo studies
C57BL/6J female mice and NU/J athymic nude female mice ages 5–6 wk were purchased from the Jackson Laboratory. All animals were housed in sterile caging and maintained under pathogen-free conditions. All experimental procedures in mice were approved by the Institutional Animal Care and Committee of Sanford Burnham Prebys (SBP) Medical Discovery Institute (approval number 17-092).
Heterotopic tumor model
500,000 KPC or 779E cells suspended in 100 µl of 1:1 PBS:Matrigel were subcutaneously injected in both flanks of syngeneic C57BL/6J mice or immunodeficient nude mice, respectively, under isoflurane anesthesia. Tumor growth was monitored by volume measurement with calipers. When tumors reached a volume of 250–300 mm3, the mice were divided into two groups and received sterile water (vehicle; control group) or 10 mg/kg DON (DON-treated group) via i.p. administration. Mice were injected every other day for 5 d. On the last day, mice were euthanized 4–5 h after the last injection. Tumor tissue was collected in 10% formalin for immunostaining, and a portion of the tissue was snap-frozen in liquid nitrogen for qPCR analysis.
Orthotopic tumor model
12,500 murine PDAC cells were injected (50 µl, 1:1 PBS:Matrigel) in the tail of the pancreas via a small abdominal incision in the left flank of anesthetized C57BL/6J female mice. Tumor growth was monitored by palpation under isoflurane anesthesia. For metabolite quantification, KPC tumor-bearing mice were euthanized 37 d after cell injection, and tumor pieces were snap-frozen in liquid nitrogen. To evaluate the effects of Slug knockdown in primary PDAC tumors, EKPC-shLuc or EKPC-shSlug#1 tumor-bearing mice were euthanized in paired cohorts 15–29 d after implantation. Tumors were weighed and tissue was collected in 10% formalin for histological analysis or snap-frozen in liquid nitrogen for qPCR analysis. To evaluate the effects of Slug knockdown on lung metastases, KPC-shLuc, KPC-shSlug#1, or KPC-shSlug#2 tumor-bearing mice were euthanized 30–40 d after implantation. The lungs were excised and collected in 10% formalin for histological analysis and metastasis quantification.
Experimental lung metastasis model
200,000 KPC cells suspended in PBS were injected i.v. into the tail vein of syngeneic C57BL/6J mice. After 25 d, lungs were collected in 10% formalin for histological analysis.
Lung metastasis quantification
Fixed lungs were paraffin embedded, sectioned, and stained with H&E at the SBP histology core following standard procedures. Lung metastasis number and tumor area quantification was performed in images obtained at 10× magnification from three H&E-stained lung sections per sample, separated by 50 µm each. Metastasis area was measured using ImageJ (National Institutes of Health).
Immunohistochemistry
Normal pancreas or PDAC tumor tissue was collected and fixed in 10% formalin. Fixed tissue was embedded in paraffin and sectioned by the histology core at SBP. Human primary pancreatic cancer tissues were fixed in 10% formalin and paraffin embedded at the Moores Cancer Center at the University of California, San Diego according to standard protocols. All studies with human tissues were approved by the Institutional Review Board for human subjects research of the University of California, San Diego (Human Research Protection Program protocol 090401). Tissue sections were deparaffinized and rehydrated, and antigen retrieval was performed by microwave-heating the slides in 10 mM sodium citrate (pH 6). Endogenous peroxidases were quenched in 3% hydrogen peroxide. For Slug staining, tissue sections were permeabilized with 0.3% Triton X-100 for 20 min at room temperature. Sections were blocked in 2% BSA and 10% goat serum in PBS for 1 h at room temperature and incubated with primary antibodies diluted in 2% BSA/PBS overnight at 4°C. After washing, sections were incubated with biotinylated goat anti-rabbit secondary antibody for 1.5 h at room temperature followed by incubation with the Vectastain Elite ABC HRP Kit (Vector Labs) and the DAB HRP Substrate Kit (Vector Labs) for signal amplification and detection. Nuclear counterstaining was performed by hematoxylin staining. Images were captured with a bright-field Olympus CX-31 microscope coupled with Infinity camera and Infinity capture software (Lumenera). The following primary antibodies and dilutions were used: ASNS (1:500, ProteinTech, 14681-1-AP), SESN2 (1:500, ProteinTech, 10795-1-AP), Slug (1:100, Cell Signaling Technologies, 9585), cleaved caspase 3 (1:1,000, Cell Signaling Technologies, 9664), and KI-67 (1:400, Thermo Fisher Scientific, MA5-14520).
Slug and ASNS were immunostained in consecutive tumor sections, and quantification of Slug positive nuclei was performed at 40× magnification in areas of high or low ASNS expression. Positive nuclei were manually counted with the cell counter function of ImageJ in four to five images corresponding to either low or high ASNS staining in three different EKPC orthotopic tumors.
Polar metabolite extraction and quantification
KPC orthotopic tumor and normal pancreas tissue from sex- and age-matched C57BL/6J mice were collected and flash-frozen in liquid nitrogen. 20–30 mg of tissue sample was transferred to 2-ml round-bottom tubes (Qiagen) with the addition of 5-mm stainless steel beads (Qiagen) and ice-cold 50% methanol/20 mM l-norvaline in a volume of 450 ml. Tubes were shaken at the maximum speed for 2 min using the Tissuelyser (Qiagen), vortexed, and placed on dry ice for 30 min. After thawing on ice, samples were centrifuged at 15,000 g for 10 min at 4°C. The supernatant was then transferred to a new tube, mixed (vortexed) with chloroform (220 ml), and centrifuged at 15,000 g for 15 min at 4°C. The top layer was dried using a Speedvac, derivatized, and analyzed using gas chromatography–mass spectrometry to quantify small polar metabolites as previously described (Ratnikov et al., 2015).
RNA extraction and qPCR
Total RNA was extracted from cells or tumor tissue with the PureLink RNA Mini Kit (Thermo Fisher Scientific). For qPCR, cDNA was synthesized from 1,000 ng of total RNA using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). cDNA samples were diluted 1:10 in RNase-free water before being subjected to qPCR. qPCR was performed in triplicate with the SYBR Premix Taq II master mix (Takara) and specific primers on the LightCycler 96 (Roche). Relative target gene expression was determined by comparing average threshold cycles with that of housekeeping genes (18s or Rpl13) by the ΔΔCT method. Specific primers are listed in Table 1.
Table 1. List of specific primers.
| Gene | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|
| mCdh1 | ATCCTCGCCCTGCTGATT | ACATTGTCCCGGGTATCATC |
| mCdh2 | GCCATCATCGCTATCCTTCT | CCGTTTCATCCATACCACAAA |
| mSlug | CATTGCCTTGTGTCTGCAAG | AGAAAGGCTTTTCCCCAGTG |
| mSnail | CTTGTGTCTGCACGACCTGT | CAGGAGAATGGCTTCTCACC |
| mZeb1 | TGGAGTTCAAAGGTTGTCGTT | TTGCCACATCAACACTGGTC |
| mTwist | CTGCCCTCGGACAAGCTGAG | CTAGTGGGACGCGGACATGG |
| mAtf4 | CGGGTGTCCCTTTCCTCTTC | TGAAGAGCGCCATGGCTTAG |
| mEcm1 | TGGCCCACTTTCCTAAACCC | CAGGCGGGTTTTCCTTCTGT |
| mEreg | TGCTTTGTCTAGGTTCCCACC | GGCGGTACAGTTATCCTCGG |
| mNcam1 | TGCCAAAGAAGCCAACATGG | CGGACTGGCTGTGTCTTGAA |
| mFbln2 | CTGCCCAGGTATCTCCCAAC | CACTCATTGATGTCTTCACAGGAC |
| mFn1 | CGAAGAGCCCTTACAGTTCCA | ATCTGTAGGCTGGTTCAGGC |
| mTyms | TCCGTTATGCTGGTGGTTGG | AATTCATCTCTCAGGCTGTATCGT |
| mSmc4 | CACAGCGACGAAGAGATGGA | TCACCTGGAGGCTTTGCG |
| mSpc24 | AGGAGCTCAGGGAGATGGAG | GTGGTGGAACTGCAGAGGG |
| mNcadp3 | GGATGAGGTGCCTTTCCCAT | TCGCCCACGTCATATCGTTC |
| 18s | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
| hCDH1 | GGTCTGTCATGGAAGGTGCT | GATGGCGGCATTGTAGGT |
| hCDH2 | CTCCATGTGCCGGATAGC | CGATTTCACCAGAAGCCTCTAC |
| hSLUG | CCATGCCTGTCATACCACAA | ACAGTGATGGGGCTGTATGC |
| hSNAIL | AGGATCTCCAGGCTCGAAAG | TCGGATGTGCATCTTGAGG |
| hZEB1 | CGAAACGCGAGGTTTTGTA | CTAGACAGGAAATCCCACACAA |
| hTWIST | GGCATCACTATGGACTTTCTCTATT | GGCCAGTTTGATCCCAGTATT |
| hRPL13 | GTTCGGTACCACACGAAGGT | TGGGGAAGAGGATGAGTTTG |
RNA-seq analysis
Libraries were prepared from isolated total RNA using the QIAseq UPX 3′ Transcriptome from Qiagen. Unique molecular identifiers and sample barcodes were introduced during first-strand synthesis, and all samples were pooled before second-strand synthesis and library preparation. The pooled libraries were paired end sequenced (124 × 28) on the NextSeq 500 using the Mid output V2.5 kit (Illumina). Read data were processed using the Qiagen GeneGlobe UPX 3′ Transcriptome primary and Secondary analysis tools. Read data were aligned to the human hg19 or mouse mm10 genome, and EdgeR (Robinson et al., 2010) outputs were used to call differentially expressed genes. Volcano plots were made using DEseq2 (Love et al., 2014) in Rosalind (Onramp Bio). GSEA was performed using the Broad Institute platform (v4.0.3; Subramanian et al., 2005). Gene sets of the hallmark gene sets, C2 curated gene sets, and C5 GO gene sets from the Molecular Signature Database collection were used for analysis with default settings. All RNA-seq data are available at Gene Expression Omnibus accession number GSE150875.
Analysis of human PDAC online datasets
SLUG gene expression data from human PDAC public datasets were used for survival analysis, tumor versus normal comparison, and PDAC subtypes comparison. Data were obtained from The Cancer Genome Atlas, EMBL-EBI Array Express, and the Gene Expression Omnibus (accession numbers E-MTAB-6134, GSE36924, GSE17891, GSE28735, GSE60978, GSE15471, GSE42952, GSE17891, and GSE71729).
Migration and invasion assays
PDAC cells were cultured in glutamine-deprivation conditions as follows: 72 h in glutamine-free medium (–Q) for AsPC-1 and 779E cells; 6 d in 0.2 mM glutamine for KPC cells; and 12 d in 0.2 mM glutamine for EKPC cells. KPC shSlug cells were incubated for 72 h in –Q. Culture medium was replaced every 2–3 d. After glutamine starvation, cells were trypsinized and seeded in serum-free medium in 8-µm-pore Transwell control inserts (BD Biocoat) for migration assays or in Matrigel-coated inserts (BD Biocoat Matrigel Invasion Chamber) for invasion assays in 24-well plates. Culture medium containing 10% FBS was used as chemoattractant in the wells containing the inserts. After 24 h, cells that migrated/invaded through the insert membrane pores were fixed in 70% ethanol and stained with 0.05% crystal violet. Nonmigrating cells in the upper surface of the membrane were removed with a cotton swab. The stained inserts were dried overnight, and the membranes were removed with a scalpel and mounted on glass slides to take images at 10× magnification with an Olympus CX-31 microscope coupled with an Infinity camera. Each experiment was performed in triplicate wells for at least three independent experiments. The number of migrating or invading cells was counted in four fields within each Transwell. Results are expressed relative to the control group in complete medium. Control wells were seeded in parallel to assess proliferation for the duration of the assays. Cells were seeded at the same density and medium conditions as in the Transwell inserts, and after 24 h, cells were stained with crystal violet and quantified. We did not observe any effects on proliferation in any of the cells during this 24-h period.
Immunoblotting
Cells were lysed in radioimmunoprecipitation assay buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, and 1% Triton X-100) with protease and phosphatase inhibitors (Roche). Protein concentrations were measured using the DC Protein Assay Kit (Bio-Rad). 15–20-µg protein samples were run in SDS-PAGE followed by protein transfer using Mini Gel Tank and Mini Blot Module (Life Technologies). Immunoblotting was detected using near-infrared fluorescence (LI-COR) and the Odyssey CLx imager (LI-COR). The following primary antibodies were used: E-cadherin (1:1,000, Cell Signaling Technologies, 3195), Slug (1:500, Cell Signaling Technologies, 9585), Snail (1:500, Cell Signaling Technologies, 3879), Zeb1 (1:500, Sigma-Aldrich, HPA027524), ATF4 (1:1,000, Cell Signaling Technologies, 11815), p-ERK (T202/Y204; 1:2,000, Cell Signaling Technologies, 4370), ERK (1:1,000; Cell Signaling Technologies, 4695), p-eIF2α (1:500, Cell Signaling Technologies, 9721), eIF2α (1:1,000, Cell Signaling Technologies, 9722), cleaved caspase 3 (1:1,000, Cell Signaling Technologies, 9664), α-tubulin (1:10,000, Sigma-Aldrich, T6074), and β-actin (1:20,000, Sigma-Aldrich, A1978). To analyze phosphorylation of proteins, membranes were probed with phospho-specific antibodies first, stripped with NewBlot IR Stripping Buffer (LI-COR), and then reprobed with pan-antibodies. The band fluorescence intensities were quantified with ImageStudio Lite software (LI-COR).
Cell number quantification by Syto60 and crystal violet
Cells were seeded in complete culture medium at a density of 5,000 cells per well in a 96-well plate. 24–48 h after seeding, cells were rinsed with PBS and incubated in glutamine-free DMEM supplemented with 10% dialyzed FBS. At the indicated time points, cells were rinsed in PBS, fixed with 4% paraformaldehyde, and stained with Syto60 (Molecular Probes, S11342), a red fluorescent nucleic acid stain. Plates were then scanned with an Odyssey Imager (LI-COR) at 700 nm, and relative cell number was determined by the intensity of the fluorescent dye using ImageStudio Lite software (LI-COR). The plates were then stained with 0.5% crystal violet, washed, and dried overnight. The crystal violet plates were scanned, and cell number was calculated as relative stained area determined using ImageJ. Three to five replicates per condition were used in each experiment.
Cell death assay by Hoechst staining
KPC shLuc, shSlug#1, and shSlug#2 cells were seeded in 24-well plates in complete medium. 24 h after seeding the cells, the medium was replaced with glutamine-free medium supplemented with 10% dialyzed FBS. After 48 h, the cells were fixed with 4% paraformaldehyde and stained with 0.5 µg/ml Hoechst 33342 and 0.1% Triton X-100. Stained cells were imaged at 20× magnification using the EVOS FL Cell Imaging System (Thermo Fisher Scientific). Apoptotic cells showing nuclear condensation (bright staining intensity) were quantified in five different fields of triplicate wells of three independent experiments.
Statistical analysis
All graphs and statistical analysis were done using Prism software v8 (GraphPad). Results are shown as means ± SEM. Statistical significance was determined by the unpaired two-tailed Student’s t test with Welch’s correction, when appropriate, or by Mann–Whitney nonparametric U test. Comparison of more than two groups was done by one-way ANOVA followed by Tukey or Dunnett's test for multiple comparisons. Survival curves were calculated using log-rank (Mantel–Cox) test. P values <0.05 were considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Online supplemental material
Fig. S1 relates to Fig. 1 and shows additional data for the metabolite analyses, volcano plots for RNA-seq data, and Western blot for E-cadherin expression in +Q and –Q. Fig. S2 relates to Fig. 2 and shows additional data on the regulation of Slug by glutamine deprivation through Erk signaling and ATF4. Fig. S3 relates to Fig. 4 and shows qPCR validation of Slug-regulated genes identified by RNA-seq and additional data on the effect of Slug knockdown on cell growth in glutamine-deprived conditions. Fig. S4 relates to Fig. 5 and shows additional analyses of Slug transcript levels in PDAC public datasets. Table S1 and Table S2 relate to Fig. 4 and show the gene sets related to EMT and proliferation pathways that are differentially enriched in Slug knockdown KPC cells.
Supplementary Material
shows EMT-related gene sets down in shSlug -Q (vs. shLuc -Q).
shows GSEA enrichment in shSlug -Q (vs. shLuc -Q)
Acknowledgments
We are grateful to members of the Commisso laboratory for their helpful comments and discussions. We thank Dr. Robert Vonderheide for providing KPC cells. We thank Drs. Olga Zagnitko and David Scott at the SBP Cancer Metabolism Core for performing the metabolite quantification experiments. We thank Buddy Charbono for performing the tail vein injections. We thank Guillermina Garcia and Monica Sevilla at the SBP Histology Core for their assistance with sample processing.
This work was supported by National Institutes of Health grant R01CA207189 to C. Commisso. SBP core services are supported by National Cancer Institute Cancer Center support grant P30CA030199.
Author contributions: M.V. Recouvreux and C. Commisso conceived the study and wrote the manuscript. M.V. Recouvreux performed the majority of the experiments. M.R. Moldenhauer, K.M.O. Galenkamp, M. Jung, and Y. Zhang performed and assisted with experiments; B. James performed the transcriptomic analysis; A. Lowy provided human specimens and 779E cells; and A. Bagchi provided EKPC cells.
References
- Adhim Z., Matsuoka T., Bito T., Shigemura K., Lee K.M., Kawabata M., Fujisawa M., Nibu K., and Shirakawa T.. 2011. In vitro and in vivo inhibitory effect of three Cox-2 inhibitors and epithelial-to-mesenchymal transition in human bladder cancer cell lines. Br. J. Cancer. 105:393–402. 10.1038/bjc.2011.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello N.M., Brabletz T., Kang Y., Nieto M.A., Weinberg R.A., and Stanger B.Z.. 2017. Upholding a role for EMT in pancreatic cancer metastasis. Nature. 547:E7–E8. 10.1038/nature22963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello N.M., Maddipati R., Norgard R.J., Balli D., Li J., Yuan S., Yamazoe T., Black T., Sahmoud A., Furth E.E., et al. . 2018. EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev. Cell. 45:681–695.e4. 10.1016/j.devcel.2018.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea L., Herlea V., Dima S.O., Dumitrascu T., Popescu I., et al. . 2008. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 55:2016–2027. [PubMed] [Google Scholar]
- Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., Miller D.K., Christ A.N., Bruxner T.J., Quinn M.C., et al. ; Australian Pancreatic Cancer Genome Initiative . 2016. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 531:47–52. 10.1038/nature16965 [DOI] [PubMed] [Google Scholar]
- Bayne L.J., Beatty G.L., Jhala N., Clark C.E., Rhim A.D., Stanger B.Z., and Vonderheide R.H.. 2012. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 21:822–835. 10.1016/j.ccr.2012.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biancur D.E., Paulo J.A., Małachowska B., Quiles Del Rey M., Sousa C.M., Wang X., Sohn A.S.W., Chu G.C., Gygi S.P., Harper J.W., et al. . 2017. Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat. Commun. 8:15965 10.1038/ncomms15965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhu G., Li Y., Padia R.N., Dong Z., Pan Z.K., Liu K., and Huang S.. 2009. Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 69:9228–9235. 10.1158/0008-5472.CAN-09-1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluntun A.A., Lukey M.J., Cerione R.A., and Locasale J.W.. 2017. Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer. 3:169–180. 10.1016/j.trecan.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., Cooc J., Weinkle J., Kim G.E., Jakkula L., et al. . 2011. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17:500–503. 10.1038/nm.2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley L.C., Marfell B.J., and Waterhouse N.J.. 2016. Analyzing Cell Death by Nuclear Staining with Hoechst 33342. Cold Spring Harb. Protoc. 2016 pdb.prot087205 10.1101/pdb.prot087205 [DOI] [PubMed] [Google Scholar]
- De Craene B., and Berx G.. 2013. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 13:97–110. 10.1038/nrc3447 [DOI] [PubMed] [Google Scholar]
- Falletta P., Sanchez-Del-Campo L., Chauhan J., Effern M., Kenyon A., Kershaw C.J., Siddaway R., Lisle R., Freter R., Daniels M.J., et al. . 2017. Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev. 31:18–33. 10.1101/gad.290940.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio D., Soldati C., Vanoni M., Alberghina L., and Chiaradonna F.. 2009. Glutamine deprivation induces abortive s-phase rescued by deoxyribonucleotides in k-ras transformed fibroblasts. PLoS One. 4 e4715 10.1371/journal.pone.0004715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Jiménez C., and Goding C.R.. 2019. Starvation and Pseudo-Starvation as Drivers of Cancer Metastasis through Translation Reprogramming. Cell Metab. 29:254–267. 10.1016/j.cmet.2018.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn D.M., Lee A.G., Briones-Martin-Del-Campo M., Conn C.S., Simpson D.R., Scott A.I., Le A., Cowan T.M., Ruggero D., and Sweet-Cordero E.A.. 2018. Oncogenic KRAS Regulates Amino Acid Homeostasis and Asparagine Biosynthesis via ATF4 and Alters Sensitivity to L-Asparaginase. Cancer Cell. 33:91–107.e6. 10.1016/j.ccell.2017.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani S.R., Wang L., Multani A.S., Combs C., Deramaudt T.B., Hruban R.H., Rustgi A.K., Chang S., and Tuveson D.A.. 2005. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 7:469–483. 10.1016/j.ccr.2005.04.023 [DOI] [PubMed] [Google Scholar]
- Hosios A.M., Hecht V.C., Danai L.V., Johnson M.O., Rathmell J.C., Steinhauser M.L., Manalis S.R., and Vander Heiden M.G.. 2016. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev. Cell. 36:540–549. 10.1016/j.devcel.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz B., Arndt M., Dullat S., Bhargava S., Buhr H.J., and Hotz H.G.. 2007. Epithelial to mesenchymal transition: expression of the regulators snail, slug, and twist in pancreatic cancer. Clin. Cancer Res. 13:4769–4776. 10.1158/1078-0432.CCR-06-2926 [DOI] [PubMed] [Google Scholar]
- Hui S., Ghergurovich J.M., Morscher R.J., Jang C., Teng X., Lu W., Esparza L.A., Reya T., Le Zhan J., Yanxiang Guo J., et al. . 2017. Glucose feeds the TCA cycle via circulating lactate. Nature. 551:115–118. 10.1038/nature24057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst J.J., Nofal M., Commisso C., Hackett S.R., Lu W., Grabocka E., Vander Heiden M.G., Miller G., Drebin J.A., Bar-Sagi D., et al. . 2015. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75:544–553. 10.1158/0008-5472.CAN-14-2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna C., and Hunter K.. 2005. Modeling metastasis in vivo. Carcinogenesis. 26:513–523. 10.1093/carcin/bgh261 [DOI] [PubMed] [Google Scholar]
- Krebs A.M., Mitschke J., Lasierra Losada M., Schmalhofer O., Boerries M., Busch H., Boettcher M., Mougiakakos D., Reichardt W., Bronsert P., et al. . 2017. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 19:518–529. 10.1038/ncb3513 [DOI] [PubMed] [Google Scholar]
- Kurrey N.K., Jalgaonkar S.P., Joglekar A.V., Ghanate A.D., Chaskar P.D., Doiphode R.Y., and Bapat S.A.. 2009. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 27:2059–2068. 10.1002/stem.154 [DOI] [PubMed] [Google Scholar]
- Lamouille S., Xu J., and Derynck R.. 2014. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15:178–196. 10.1038/nrm3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.W., Zhang Y., Jung M., Cruz N., Alas B., and Commisso C.. 2019. EGFR-Pak Signaling Selectively Regulates Glutamine Deprivation-Induced Macropinocytosis. Dev. Cell. 50:381–392.e5. 10.1016/j.devcel.2019.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberg K.M., Vornov J.J., Rais R., and Slusher B.S.. 2018. We’re Not “DON” Yet: Optimal Dosing and Prodrug Delivery of 6-Diazo-5-oxo-L-norleucine. Mol. Cancer Ther. 17:1824–1832. 10.1158/1535-7163.MCT-17-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Ye D., Xu D., Liao Y., Zhang L., Liu L., Yu W., Wang Y., He Y., Hu J., et al. . 2016. Autocrine epiregulin activates EGFR pathway for lung metastasis via EMT in salivary adenoid cystic carcinoma. Oncotarget. 7:25251–25263. 10.18632/oncotarget.7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., and Kang Y.. 2019. Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Dev. Cell. 49:361–374. 10.1016/j.devcel.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt R.A., Marayati R., Flate E.L., Volmar K.E., Loeza S.G., Hoadley K.A., Rashid N.U., Williams L.A., Eaton S.C., Chung A.H., et al. . 2015. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 47:1168–1178. 10.1038/ng.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M.A., Huang R.Y., Jackson R.A., and Thiery J.P.. 2016. Emt: 2016. Cell. 166:21–45. 10.1016/j.cell.2016.06.028 [DOI] [PubMed] [Google Scholar]
- Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., and Gorman A.M.. 2016. The integrated stress response. EMBO Rep. 17:1374–1395. 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M., Reid M.A., Lowman X.H., Kulkarni R.P., Tran T.Q., Liu X., Yang Y., Hernandez-Davies J.E., Rosales K.K., Li H., et al. . 2016. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat. Cell Biol. 18:1090–1101. 10.1038/ncb3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathria G., Lee J.S., Hasnis E., Tandoc K., Scott D.A., Verma S., Feng Y., Larue L., Sahu A.D., Topisirovic I., et al. . 2019. Translational reprogramming marks adaptation to asparagine restriction in cancer. Nat. Cell Biol. 21:1590–1603. 10.1038/s41556-019-0415-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleo F., Nicolle R., Blum Y., Cros J., Marisa L., Demetter P., Quertinmont E., Svrcek M., Elarouci N., Iovanna J., et al. . 2018. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology. 155:1999–2013.e3. 10.1053/j.gastro.2018.08.033 [DOI] [PubMed] [Google Scholar]
- Ratnikov B., Aza-Blanc P., Ronai Z.A., Smith J.W., Osterman A.L., and Scott D.A.. 2015. Glutamate and asparagine cataplerosis underlie glutamine addiction in melanoma. Oncotarget. 6:7379–7389. 10.18632/oncotarget.3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim A.D., Mirek E.T., Aiello N.M., Maitra A., Bailey J.M., McAllister F., Reichert M., Beatty G.L., Rustgi A.K., Vonderheide R.H., et al. . 2012. EMT and dissemination precede pancreatic tumor formation. Cell. 148:349–361. 10.1016/j.cell.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu V., Bowitz Lothe I.M., Labori K.J., Lingjærde O.C., Buanes T., Dalsgaard A.M., Skrede M.L., Hamfjord J., Haaland T., Eide T.J., et al. . 2015. Molecular signatures of mRNAs and miRNAs as prognostic biomarkers in pancreatobiliary and intestinal types of periampullary adenocarcinomas. Mol. Oncol. 9(4):758–771. 10.1016/j.molonc.2014.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciacovelli M., and Frezza C.. 2017. Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. FEBS J. 284:3132–3144. 10.1111/febs.14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C., Acosta-Alvear D., Khoutorsky A., Vedantham P., Hearn B.R., Li H., Gamache K., Gallagher C.M., Ang K.K., Wilson C., et al. . 2013. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife. 2 e00498 10.7554/eLife.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Tai K., Madan S., Raythatha M.R., Cady A.M., Braunlin M., Irving L.R., Bajaj A., and Lucci A.. 2012. Selection of metastatic breast cancer cells based on adaptability of their metabolic state. PLoS One. 7 e36510 10.1371/journal.pone.0036510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., Perera R.M., Ferrone C.R., Mullarky E., Shyh-Chang N., et al. . 2013. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 496:101–105. 10.1038/nature12040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. . 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajan M., Hock A.K., Blagih J., Robertson N.A., Labuschagne C.F., Kruiswijk F., Humpton T.J., Adams P.D., and Vousden K.H.. 2018. A Role for p53 in the Adaptation to Glutamine Starvation through the Expression of SLC1A3. Cell Metab. 28:721–736.e6. 10.1016/j.cmet.2018.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville M.M., Pan Y.X., Gjymishka A., Zhong C., Kaufman R.J., and Kilberg M.S.. 2008. MEK signaling is required for phosphorylation of eIF2alpha following amino acid limitation of HepG2 human hepatoma cells. J. Biol. Chem. 283:10848–10857. 10.1074/jbc.M708320200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broeck A., Vankelecom H., Van Eijsden R., Govaere O., and Topal B.. 2012. Molecular markers associated with outcome and metastasis in human pancreatic cancer. J. Exp. Clin. Cancer Res. 31:68 10.1186/1756-9966-31-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega S., Morales A.V., Ocaña O.H., Valdés F., Fabregat I., and Nieto M.A.. 2004. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev. 18:1131–1143. 10.1101/gad.294104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino N., Signaevskaia L., van Niekerk J., Medal R., Kim H., Lahmy R., Scully K., Pinkerton A., Kim S., Lowy A., et al. . 2017. A screen for inducers of bHLH activity identifies pitavastatin as a regulator of p21, Rb phosphorylation and E2F target gene expression in pancreatic cancer. Oncotarget. 8:53154–53167. 10.18632/oncotarget.18587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W.S., Heinrichs S., Xu D., Garrison S.P., Zambetti G.P., Adams J.M., and Look A.T.. 2005. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 123:641–653. 10.1016/j.cell.2005.09.029 [DOI] [PubMed] [Google Scholar]
- Ye J., Kumanova M., Hart L.S., Sloane K., Zhang H., De Panis D.N., Bobrovnikova-Marjon E., Diehl J.A., Ron D., and Koumenis C.. 2010. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 29:2082–2096. 10.1038/emboj.2010.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fan J., Venneti S., Cross J.R., Takagi T., Bhinder B., Djaballah H., Kanai M., Cheng E.H., Judkins A.R., et al. . 2014. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol. Cell. 56:205–218. 10.1016/j.molcel.2014.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C.C., LeBleu V.S., and Kalluri R.. 2015. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 527:525–530. 10.1038/nature16064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Gross K.M., and Kuperwasser C.. 2019a Molecular regulation of Snai2 in development and disease. J. Cell Sci. 132:jcs235127. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., and Chanda S.K.. 2019b Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10:1523 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows EMT-related gene sets down in shSlug -Q (vs. shLuc -Q).
shows GSEA enrichment in shSlug -Q (vs. shLuc -Q)