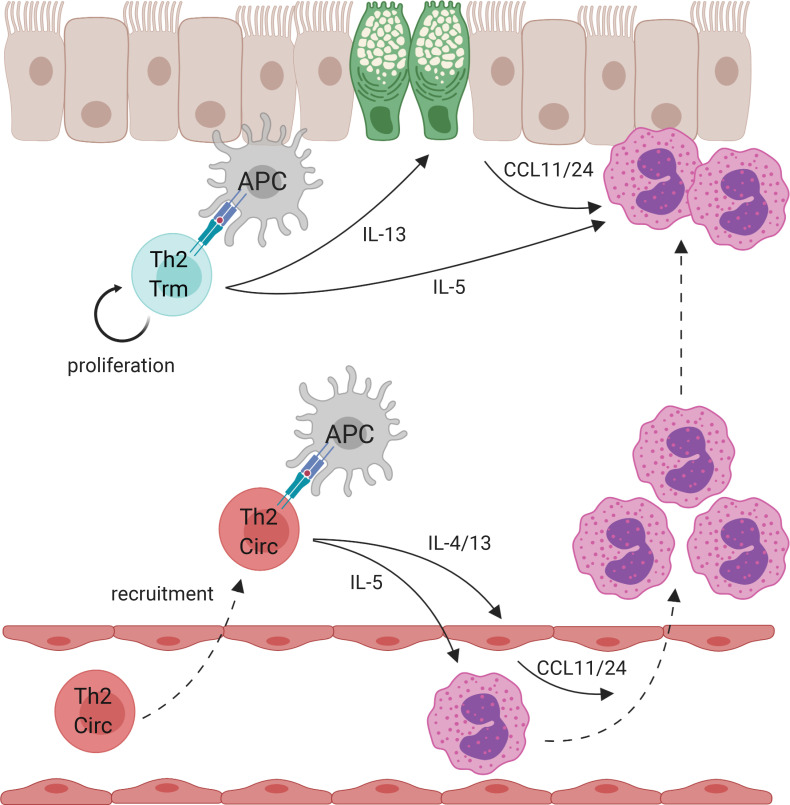
How memory Th2 cell subsets orchestrate allergic airway inflammation remains unclear. Rahimi et al. demonstrate that tissue-resident and circulating memory Th2 cells are functionally and transcriptionally distinct subsets with unique roles in promoting allergic airway disease.
Abstract
Memory CD4+ T helper type 2 (Th2) cells drive allergic asthma, yet the mechanisms whereby tissue-resident memory Th2 (Th2 Trm) cells and circulating memory Th2 cells collaborate in vivo remain unclear. Using a house dust mite (HDM) model of allergic asthma and parabiosis, we demonstrate that Th2 Trm cells and circulating memory Th2 cells perform nonredundant functions. Upon HDM rechallenge, circulating memory Th2 cells trafficked into the lung parenchyma and ignited perivascular inflammation to promote eosinophil and CD4+ T cell recruitment. In contrast, Th2 Trm cells proliferated near airways and induced mucus metaplasia, airway hyperresponsiveness, and airway eosinophil activation. Transcriptional analysis revealed that Th2 Trm cells and circulating memory Th2 cells share a core Th2 gene signature but also exhibit distinct transcriptional profiles. Th2 Trm cells express a tissue-adaptation signature, including genes involved in regulating and interacting with extracellular matrix. Our findings demonstrate that Th2 Trm cells and circulating memory Th2 cells are functionally and transcriptionally distinct subsets with unique roles in promoting allergic airway disease.
Graphical Abstract
Introduction
Asthma is an inflammatory airway disorder affecting more than 350 million individuals worldwide (Soriano et al., 2017). Although asthma is a heterogeneous syndrome, allergic airway inflammation drives asthma pathogenesis in the majority of children and half of adults (Arbes et al., 2007; Lambrecht and Hammad, 2015; Woodruff et al., 2009). The development of CD4+ T helper type 2 (Th2) cells that recognize airborne allergens is a key feature of allergic asthma (Lambrecht et al., 2019; Walker and McKenzie, 2018). Allergen-specific Th2 cells orchestrate allergic airway inflammation by producing type 2 cytokines, including IL-4, IL-5, and IL-13, which drive eosinophilic inflammation, mucus metaplasia, and airway hyperresponsiveness (Lambrecht et al., 2019; Walker and McKenzie, 2018). In addition, Th2 cells can give rise to long-lived memory Th2 cells that maintain allergen-specific immunity (Hondowicz et al., 2016; Onodera et al., 2018). Consequently, memory Th2 cells represent an attractive therapeutic target in allergic asthma, but our knowledge of memory Th2 biology in vivo remains limited.
Over the last 20 years, distinct subsets of memory T cells have been characterized that exhibit unique trafficking patterns and functions in vivo (Jameson and Masopust, 2018). Tissue-resident memory T (Trm) cells persist in previously inflamed nonlymphoid tissue (NLT), providing enhanced local immune memory (Carbone, 2015; Schenkel and Masopust, 2014). In contrast, circulating memory T cells provide global host defense (Jameson and Masopust, 2018). Most of our knowledge of Trm cell biology stems from the CD8+ T cell field, and less is known about CD4+ Trm cells. Parabiosis experiments have demonstrated that CD4+ T helper type 1 (Th1 Trm) cells are the dominant memory Th1 cell subset surveying NLT and initiating local recall responses (Beura et al., 2019). Both Th1 Trm cells and a “second wave” of recruited Th1 cells are required for optimal pathogen control in vivo (Stary et al., 2015; Iijima and Iwasaki, 2014; Glennie et al., 2015). Studies using the house dust mite (HDM) model of allergic asthma have shown that Th2 Trm cells persist in the lung in inducible bronchus-associated lymphoid tissue (iBALT) structures (Hondowicz et al., 2016; Shinoda et al., 2016; Turner et al., 2018). Interestingly, Th2 Trm cells can promote airway hyperresponsiveness and inflammatory cell recruitment even after depletion of circulating T cells, suggesting Th2 Trm cells are an important cell population orchestrating local type 2 immunity (Hondowicz et al., 2016; Turner et al., 2018). However, adoptively transferring circulating memory Th2 cells into naive mice and administering repetitive antigen challenge leads to allergic airway inflammation (Endo et al., 2011, 2015). As a result, the mechanisms whereby Th2 Trm cells and circulating memory Th2 cells collaborate in an endogenous recall response are unknown, a gap in knowledge that limits therapeutic targeting of pathogenic memory Th2 cells in allergic airway disease.
Here, we use a HDM model of allergic asthma and parabiosis to define the functions of endogenous tissue-resident and circulating memory Th2 cells. Unexpectedly, we found Th2 Trm cells and circulating memory Th2 cells performed distinct functions in vivo. Upon HDM rechallenge, circulating memory Th2 cells trafficked into the lung parenchyma and ignited perivascular inflammation to promote eosinophil and CD4+ T cell recruitment. In contrast, Th2 Trm cells proliferated near airways and promoted mucus metaplasia, airway hyperresponsiveness, and airway eosinophil activation. Transcriptional analysis revealed that Th2 Trm cells and circulating memory Th2 cells share a core Th2 gene signature but also exhibit distinct transcriptional profiles. Specifically, Th2 Trm cells express a tissue-adaptation signature, including genes involved in regulating and interacting with extracellular matrix. Our findings demonstrate that Th2 Trm cells and circulating memory Th2 cells are functionally and transcriptionally distinct subsets with unique roles in vivo, with the establishment of Th2 Trm cells being critical for the full manifestation of allergic airway disease. We propose a novel model for memory Th2 responses in the airways with implications for developing disease-modifying therapies for individuals with allergic asthma.
Results and discussion
Memory Th2 cells orchestrate the recall response to HDM in an allergen-specific manner
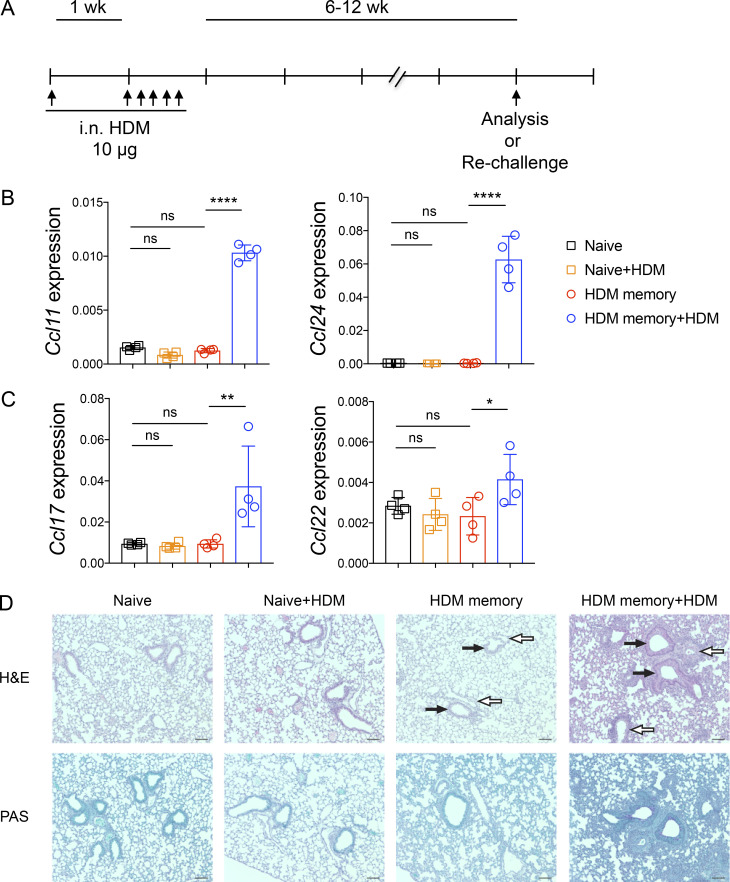
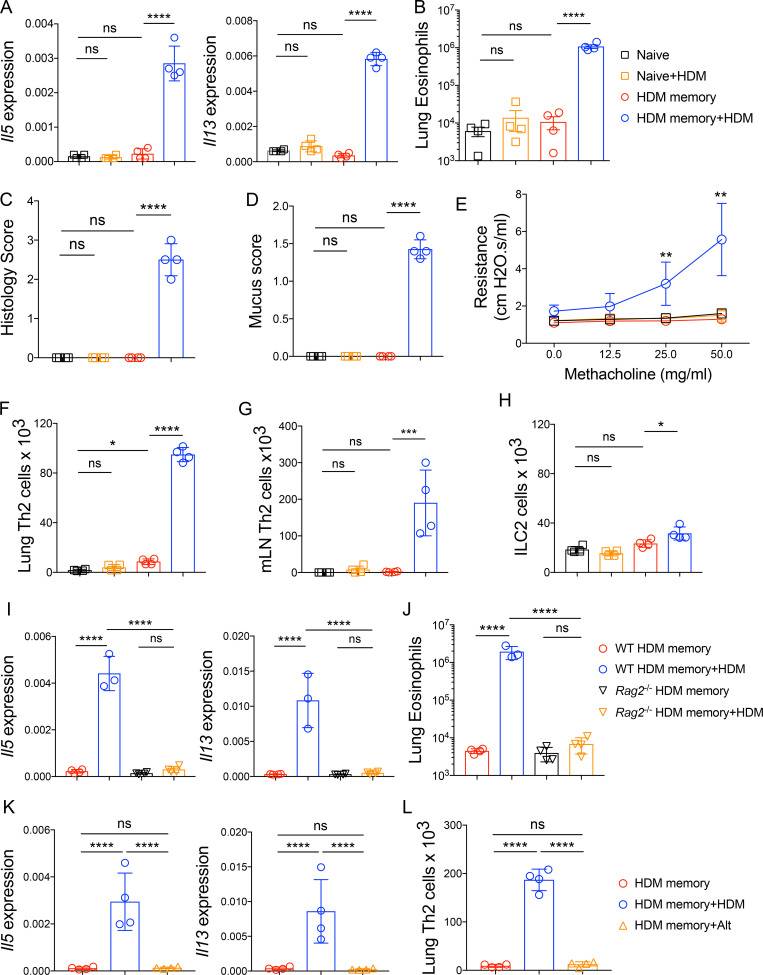
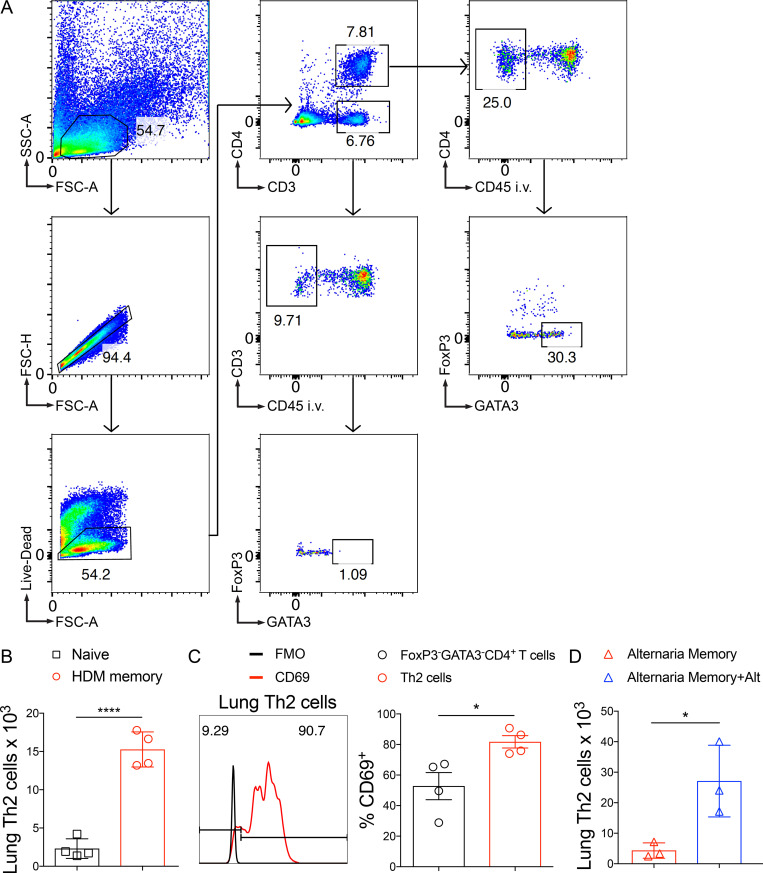
To define the function of endogenous memory Th2 cell subsets in vivo, we used a well-established murine model of allergic asthma via administration of i.n. HDM. HDM sensitization and repetitive challenge induces robust allergic airway inflammation that is dependent on Th2 cells and independent of IgE (Li et al., 2016; Hondowicz et al., 2016; McKnight et al., 2017). We sensitized and challenged mice with HDM followed by a 6–12-wk rest period to generate “HDM-memory” mice (Fig. S1 A). First, we assessed the ability of HDM-memory mice to exhibit a recall response in vivo by rechallenging with a single i.n. dose of HDM. Compared with naive mice treated with a single dose of HDM, HDM-memory mice exhibited robust induction of type 2 cytokines and chemokines within the lung as well as the cardinal features of allergic inflammation, including lung eosinophilia, mucus metaplasia, and airway hyperresponsiveness (Fig. 1, A–E; and Fig. S1, B–D). The observation that naive mice do not exhibit a significant innate type 2 response after one dose of HDM suggests that memory Th2 cells are driving the recall response in HDM-memory mice. However, previous studies have demonstrated that allergen exposure can lead to both memory Th2 cells as well as memory-like group 2 innate lymphoid cells (ILC2) with enhanced responsiveness to allergens (Martinez-Gonzalez et al., 2016, 2017). We sought to determine the relative contribution of memory Th2 cells and memory-like ILC2 to the recall response in HDM-memory mice. To do so, we initially assessed the proliferative response of memory Th2 cells and ILC2 to HDM rechallenge in HDM-memory mice. To examine Th2 cells in the lung parenchyma, we performed i.v. injection with fluorophore-labeled anti-CD45 antibody to label intravascular leukocytes before lung harvest as previously described (Fig. S2 A; Anderson et al., 2014; Galkina et al., 2005). We performed intranuclear staining for FoxP3 and GATA3, allowing us to identify Th2 cells as FoxP3−GATA3+CD4+ T cells (Fig. S2 A). Consistent with previous reports, compared with naive controls, we observed an increase in the number of Th2 cells persisting in the lung parenchyma of HDM-memory mice (Fig. 1 F and Fig. S2 B; Hondowicz et al., 2016; Turner et al., 2018). Memory Th2 cells persisting in the lung parenchyma of HDM-memory mice expressed high levels of CD69, consistent with being Trm cells (Fig. S2 C). Upon HDM treatment, naive mice did not develop a significant increase in lung Th2 cells, whereas HDM-memory mice exhibited a robust expansion of Th2 cells within the lung parenchyma and mediastinal LN (mLN; Fig. 1, F and G). In contrast, we did not observe a difference in the number of ILC2 between naive and HDM-memory mice during homeostasis (Fig. 1 H). In addition, ILC2 from HDM-memory mice minimally expanded upon HDM rechallenge (Fig. 1 H).
Figure S1.
Allergen rechallenge promotes type 2 chemokine expression, lung inflammation, and mucus metaplasia. C57BL/6 mice were sensitized with 10 µg i.n. HDM followed by 10 µg daily challenges of i.n. HDM on days 7–11. After 6–12 wk of rest, HDM-memory mice were left untreated or rechallenged with a single dose of i.n. HDM followed by tissue harvest 72 h later. (A) Schematic of i.n. HDM treatment. (B) Lung Ccl11 and Ccl24 relative RNA levels assessed via qPCR. (C) Lung Ccl17 and Ccl22 relative RNA levels assessed via qPCR. (D) Representative images of lung H&E and PAS staining in indicated groups. White arrows indicate blood vessels, and black arrows indicate airways. Scale bars represent 100 µm. Representative data show individual mice with mean ± SEM from one of two independent experiments with three or four mice per group. For statistical analysis, one-way ANOVA analysis with Holm-Sidak’s testing was used for statistical analysis of multiple groups. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Figure 1.
Memory Th2 cells orchestrate the recall response to HDM in an allergen-specific manner. (A–H) C57BL/6 mice were sensitized and challenged with i.n. HDM, rested for 6–12 wk, and left untreated or rechallenged with a single dose of i.n. HDM followed by tissue harvest 72 h later. (A) Lung Il5 and Il13 relative RNA levels assessed via qPCR. (B) Lung parenchymal (anti-CD45 i.v. unlabeled) eosinophils were quantitated via flow cytometry. (C) Lung histology scores. (D) Mucus scores. (E) Airway resistance was measured in indicated groups after increasing doses of methacholine. (F and G) Th2 cells (FoxP3−GATA3+CD4+ T cells) were quantitated by flow cytometry in the (F) lung parenchyma (anti-CD45 i.v. unlabeled) and (G) mLNs in indicated groups. (H) Quantitation of lung parenchymal ILC2 (anti-CD45 i.v. unlabeled, lineage−CD4−Thy1.2+CD127+ST2+ cells). (I and J) C57BL/6 and Rag2−/− mice were sensitized and challenged with i.n. HDM, rested for 6–12 wk, and left untreated or rechallenged with a single dose of i.n. HDM followed by tissue harvest 72 h later. (I) Lung Il5 and Il13 relative RNA levels assessed via qPCR. (J) Lung parenchymal (anti-CD45 i.v. unlabeled) eosinophils were quantitated via flow cytometry. (K and L) C57BL/6 mice were sensitized and challenged with i.n. HDM, rested for 6–12 wk, and left untreated or rechallenged with a single dose of i.n. HDM or i.n. A. alternata followed by tissue harvest 72 h later. (K) Lung Il5 and Il13 relative RNA levels assessed via qPCR. (L) Th2 cells (FoxP3−GATA3+CD4+ T cells) were quantitated by flow cytometry in the lung parenchyma (anti-CD45 i.v. unlabeled). Representative data show individual mice with mean ± SEM from one of three independent experiments with four mice per group (A–H) or mean ± SEM from one of two independent experiments with three or four mice per group (I–L). One-way ANOVA analysis with Holm-Sidak’s testing was used for statistical analysis of multiple groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. ns, not significant.
Figure S2.
Identification of Th2 Trm cells via flow cytometry. C57BL/6 mice were sensitized and challenged with i.n. HDM and then rested for 6–12 wk followed by lung tissue harvest. (A) Representative flow cytometry on lung cells in HDM-memory mice to identify Th2 cells (anti-CD45 i.v. unlabeled, FoxP3−GATA3+CD4+ T cells). (B) Quantitation of lung Th2 cells (anti-CD45 i.v. unlabeled, FoxP3−GATA3+CD4+ T cells) in naive and HDM-memory mice via flow cytometry. (C) Representative histogram demonstrating CD69 staining (and fluorescence minus one control staining) of lung Th2 cells from HDM-memory mice (left panel) with quantitation of percent CD69+ cells from lung Th2 cells and FoxP3−GATA3−CD4+ T cell populations (right panel). (D) C57BL/6 mice were sensitized and challenged with i.n. A. alternata, rested for 6–8 wk, and left untreated or rechallenged with a single dose of i.n. A. alternata. Quantitation of lung Th2 cells (anti-CD45 i.v. unlabeled, FoxP3−GATA3+CD4+ T cells) in indicated groups. Representative data show individual mice with mean ± SEM from one of two independent experiments with three or four mice per group. For statistical analysis, a two-tailed t test was performed for parametric data, and a two-tailed Mann-Whitney U test was performed for nonparametric data. *, P < 0.05; ****, P < 0.0001. FSC-A, forward scatter area; FSC-H, forward scatter height; SSC-A, side scatter area.
To further investigate the potential role of memory-like ILC2 to the HDM recall response, we took advantage of the observation that allergen-experienced ILC2 can acquire memory-like properties independently of T cells (Martinez-Gonzalez et al., 2016). We generated WT and Rag2−/− HDM-memory mice and left them untreated or rechallenged with HDM. We found Rag2−/− HDM-memory mice failed to induce significant expression of type 2 cytokines or eosinophilia within the lung (Fig. 1, I and J). These results indicate that HDM treatment promotes memory Th2 cell development but does not efficiently induce memory-like ILC2. These findings are consistent with previous studies demonstrating that Th2 cells are the functionally dominant type 2 lymphocyte in the primary response to HDM (Li et al., 2016; Hondowicz et al., 2016). Finally, given that Th2 cells are capable of responding to innate signals, including IL-33, in an antigen-independent manner, we investigated whether the memory Th2 cell recall response was allergen-specific (Guo et al., 2009, 2015; Minutti et al., 2017). To do so, we used Alternaria alternata fungal extract, which is well characterized to promote IL-33 release and allergic airway inflammation (Snelgrove et al., 2014; Causton et al., 2018). Specifically, we administered a single 10-µg dose of either HDM or Alternaria alternata extract to HDM-memory mice. While HDM rechallenge induced robust type 2 cytokine expression and Th2 cell expansion, we did not observe increased expression of type 2 cytokines or Th2 cell expansion with A. alternata treatment (Fig. 1, K and L). As a control, we generated A. alternata–memory mice and left them untreated or rechallenged with a single dose of A. alternata, which promoted robust Th2 cell expansion within the lung (Fig. S2 D). In summary, memory Th2 cells proliferate and orchestrate allergic inflammation in the HDM model of allergic airway disease in an allergen-specific manner.
Th2 Trm cells and circulating memory Th2 cells collaborate to promote Th2 cell expansion and type 2 cytokine production within the lung
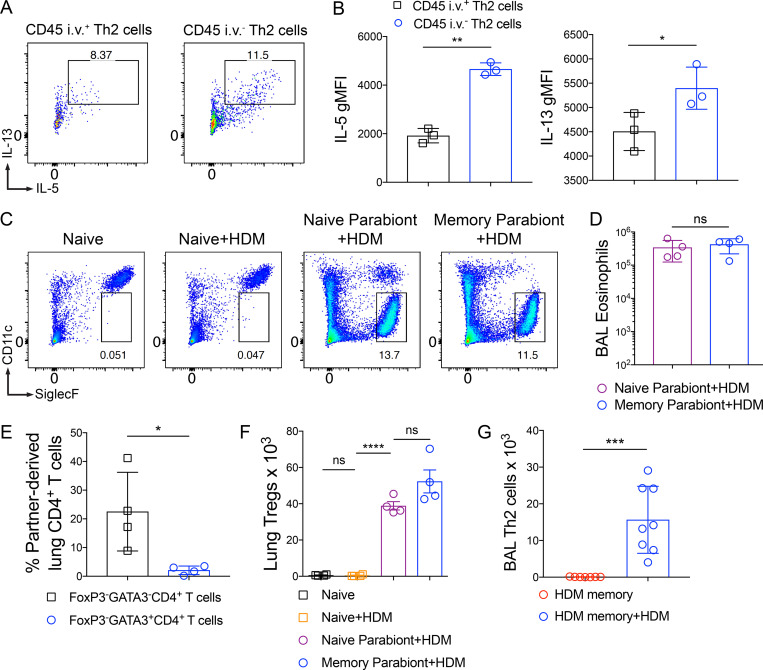
Next, we sought to define the functions of Th2 Trm cells and circulating memory Th2 cells. Studies on Trm cells in other experimental systems have suggested that Trm cells are more potent cytokine producers than their circulating counterparts (Strutt et al., 2018; Smolders et al., 2018; Hombrink et al., 2016; Oja et al., 2018). To assess the ability of Th2 Trm cells and circulating memory Th2 cells to produce type 2 cytokines upon reactivation, we isolated lung CD4+ T cells from HDM-memory mice after anti-CD45 i.v. injection, performed ex vivo treatment with anti-CD3 and anti-CD28, and assessed the ability of intraparenchymal Th2 Trm (anti-CD45 i.v. negative) and intravascular memory Th2 cells (anti-CD45 i.v. positive) to produce type 2 cytokines (Fig. S3, A and B). While both memory Th2 cell subsets are capable of producing IL-5 and IL-13, we found that Th2 Trm cells within the lung parenchyma produced a greater amount of type 2 cytokines on a per-cell basis (Fig. S3, A and B).
Figure S3.
Function and trafficking of tissue-resident and circulating memory Th2 cells in HDM-memory mice. (A and B) C57BL/6 mice were sensitized and challenged with i.n. HDM and rested for 6–12 wk. (A) Lung cells from HDM-memory mice underwent CD4+ T cell negative selection followed by treatment with anti-CD3 and anti-CD28 for 12–16 h. Representative flow cytometry of anti-CD45 i.v. unlabeled (intraparenchymal) and labeled (intravascular) Th2 cells after intracellular cytokine staining for IL-5 and IL-13. (B) Geometric mean fluorescence intensity (gMFI) for IL-5 and IL-13 expression in indicated groups. (C and D) CD45.2 HDM-memory mice were surgically conjoined to CD45.1 naive mice. After 3–4 wk, both parabionts received a single dose of i.n. HDM with harvest of mLN and lung after 72 h. (C) Representative flow cytometry of lung parenchymal eosinophils (anti-CD45 i.v. unlabeled, CD11cloSiglec-F+ cells) from indicated groups. (D) Quantitation of BAL eosinophils from indicated groups. (E) CD45.2 and CD45.1 HDM-memory mice were surgically conjoined. After 3–4 wk, lungs from both parabionts were harvested. Quantitation of percent partner-derived lung memory CD4+ T cells (anti-CD45 i.v. unlabeled, FoxP3−GATA3−CD4+ T cells and FoxP3−GATA3+CD4+ T cells) assessed via flow cytometry. (F) CD45.2 HDM-memory mice were surgically conjoined to CD45.1 naive mice. After 3–4 wk, both parabionts received a single dose of i.n. HDM with harvest of lungs after 72 h. Lung FoxP3+CD4+ T cells (T regs) were quantitated via flow cytometry. (G) Individual HDM-memory mice were left untreated or received a single dose of i.n. HDM with BAL after 72 h. BAL Th2 cells were quantitated via flow cytometry. Representative data show individual mice with mean ± SEM from one of two independent experiments with three or four mice per group (A and B), or one of three independent experiments with three or four mice per group (C, D, and F) or cumulative data from two independent experiments (E and G). For statistical analysis, a two-tailed t test was performed for parametric data (B and G). One-way ANOVA analysis with Holm-Sidak’s testing was used for statistical analysis of multiple groups with paired two-tailed t tests for comparison of parabionts (D and F). A two-tailed Mann-Whitney U test was performed for nonparametric data (E). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
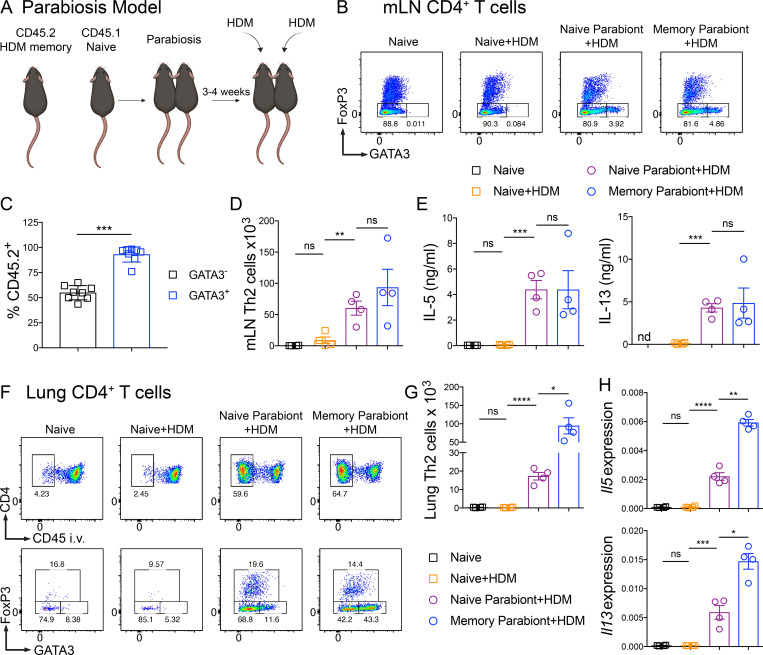
To investigate the mechanisms whereby Th2 Trm cells enhance the HDM recall response in vivo, we used a parabiosis system in which we surgically conjoined congenic HDM-memory mice (CD45.2 memory parabiont) and naive mice (CD45.1 naive parabiont). Parabiosis leads to a shared vasculature such that circulating memory Th2 cells equilibrate between both parabionts, but only the CD45.2 memory parabiont have Th2 Trm cells. After 3–4 wk of parabiosis, we administered a single dose of i.n. HDM to both parabionts (Fig. 2 A). First, we assessed whether the parabiosis system had effectively led to chimerism of the circulating memory T cell compartment. While CD45.1 naive, nonparabiotic mice administered a single dose of HDM did not induce an increase in Th2 cells within the mLN after 72 h, naive and memory parabionts exhibited a robust population of Th2 cells in the mLN (Fig. 2 B). To confirm that the Th2 cells within the mLN of both parabionts were derived from memory Th2 cells, we assessed the expression of CD45.2 within the GATA3− and GATA3+CD4+ T cell populations. GATA3+CD4+ T cells from the mLN were overwhelming derived from the CD45.2 memory parabiont, whereas GATA3−CD4+ T cells showed ∼50:50 chimerism (Fig. 2 C). In addition, quantitation of Th2 cells from the mLN revealed similar numbers of Th2 cells in the two parabionts (Fig. 2 D). Finally, total mLN cells from the naive and memory parabionts restimulated ex vivo with HDM produced similar levels of IL-5 and IL-13 (Fig. 2 E). In summary, these results demonstrate that parabiosis effectively transferred endogenous, circulating memory Th2 cells in vivo.
Figure 2.
Th2 Trm cells and circulating memory Th2 cells collaborate to promote Th2 cell expansion and type 2 cytokine production within the lung. (A–H) CD45.2 HDM-memory mice were surgically conjoined to CD45.1 naive mice. After 3–4 wk, both parabionts received a single dose of i.n. HDM with harvest of mLN and lung after 72 h. (A) Schematic of the parabiosis experiment. (B) Representative flow cytometry of mLN CD4+ T cells isolated from indicated groups demonstrating FoxP3 and GATA3 expression. (C) Percentage of CD45.2+ cells (from memory parabiont) of FoxP3−GATA3− and FoxP3−GATA3+ CD4+ T cells from the mLN of naive and memory parabionts. (D) Quantitation of Th2 cells from mLNs from indicated groups. (E) Total mLN cells were restimulated ex vivo with 25 µg/ml HDM for 72 h and levels of IL-5 and IL-13 protein in the supernatant was measured via ELISA. (F) Representative flow cytometry of lung CD4+ T cells from indicated groups demonstrating FoxP3 and GATA3 expression. (G) Lung parenchymal Th2 cells (anti-CD45 i.v. unlabeled, FoxP3−GATA3+CD4+ T cells) were quantitated via flow cytometry. (H) Lung Il5 and Il13 relative RNA levels assessed via qPCR. Representative data show individual mice with mean ± SEM from one of three independent experiments with three or four mice per group. For statistical analysis, one-way ANOVA analysis with Holm-Sidak’s testing was used for statistical analysis of multiple groups with paired two-tailed t tests for comparison of naive and memory parabiont groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Next, we turned to the HDM recall response within the lung. Within the lung parenchyma (anti-CD45 i.v. negative), CD45.1 naive, nonparabiotic mice had few CD4+ T cells and after a single dose of HDM there was no increase (Fig. 2, F and G). In contrast, naive parabionts possessing circulating memory Th2 cells exhibited a robust accumulation of CD4+ T cells within the lung parenchyma (Fig. 2, F and G). However, memory parabionts had significantly more Th2 cells (defined by being FoxP3−GATA3+CD4+ cells) within the lung (Fig. 2, F and G). In support of Th2 Trm cells driving an enhanced type 2 response, expression of type 2 cytokines within the lung was higher in memory parabionts than naive parabionts following HDM challenge (Fig. 2 H). Of note, naive parabionts did induce greater type 2 cytokine expression compared with naive, nonparabiotic mice treated with HDM. Consequently, we found that Th2 Trm and circulating memory Th2 cells collaborate to promote Th2 cell expansion and type 2 cytokine production within the lung upon HDM rechallenge.
Th2 Trm and circulating memory Th2 cells perform nonredundant functions upon HDM rechallenge
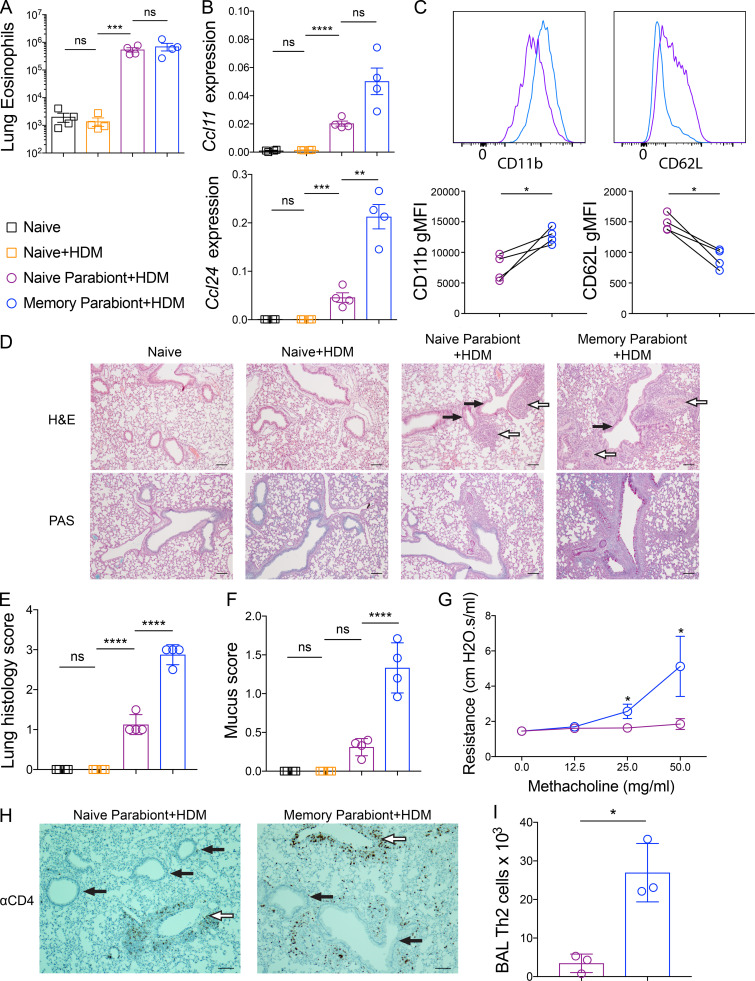
To define the roles of Th2 Trm cells and circulating memory Th2 cells in promoting the cardinal features of allergic airway inflammation within the lung, we first enumerated the number of eosinophils within the lung parenchyma of naive and memory parabionts as well as naive controls after HDM challenge. Unexpectedly, we found comparable numbers of eosinophils within the lung parenchyma and bronchoalveolar lavage (BAL) of both parabionts, suggesting that circulating memory Th2 cells were necessary and sufficient to promote eosinophil mobilization and recruitment into the lung and airways (Fig. 3 A and Fig. S3, C and D). The eosinophil-attracting chemokines CCL11 (eotaxin-1) and CCL24 (eotaxin-2) are induced by IL-4 and IL-13 and are well characterized to promote eosinophil recruitment into the lung parenchyma via CCR3 (Griffith et al., 2014). Consequently, we compared Ccl11 and Ccl24 expression within the lung of naive and memory parabionts as well as naive controls after HDM challenge. Compared with naive controls, naive parabionts acquired the ability to induce expression of Ccl11 and Ccl24 but to levels less than memory parabionts (Fig. 3 B). Consequently, it remained unclear why the enhanced CCR3 ligand expression within the lungs of the memory parabionts did not result in greater eosinophil recruitment into the lung. Since CCL11 and CCL24, as well as IL-5, have been shown to induce recruitment as well as activation of eosinophils, we investigated the expression of several well-characterized eosinophil activation markers, including CD11b and CD62L, which are up-regulated and down-regulated, respectively, upon eosinophil activation (Johansson, 2014; Lukacs, 2001). We found significantly higher expression of CD11b and lower expression of CD62L on eosinophils present in the BAL of memory parabionts, consistent with enhanced activation (Fig. 3 C). These findings demonstrate that Th2 Trm cells and circulating memory Th2 cells both contribute to Ccl11 and Ccl24 expression within the lung upon HDM rechallenge. In addition, our results suggest that Th2 Trm cell–driven CCL11 and CCL24 production does not significantly enhance the recruitment of eosinophils from the blood into the lung parenchyma; instead, Trm cells in the memory parabiont enhanced eosinophil activation within the airways.
Figure 3.
Th2 Trm cells and circulating memory Th2 cells perform nonredundant functions upon HDM rechallenge. (A–I) CD45.2 HDM-memory mice were surgically conjoined to CD45.1 naive mice. After 3–4 wk, both parabionts received a single dose of i.n. HDM with harvest of lung or BAL after 72 h. (A) Quantitation of eosinophils (anti-CD45 i.v. unlabeled, CD11cloSiglec-F+ cells) from the lung parenchyma of indicated groups. (B) Lung Ccl11 and Ccl24 relative RNA levels assessed via qPCR. (C) Representative histograms demonstrating BAL eosinophil cell surface expression of CD11b and CD62L (top panel) and geometric mean fluorescence intensity (gMFI; bottom panel) from naive parabionts (purple circles) and memory parabionts (blue circles) as determined by flow cytometry. (D) H&E-stained (top panel) and PAS-stained (bottom panel) lung sections from indicated groups. White arrows indicate blood vessels, and black arrows indicate airways. Scale bars represent 100 µm. (E) Lung histology scores. (F) Mucus scores. (G) Airway resistance was measured in indicated group after increasing doses of methacholine. (H) Immunohistochemistry for lung CD4+ T cells in naive and memory parabionts 72 h after HDM challenged. White arrows indicate blood vessels, and black arrows indicate airways. Scale bars represent 100 µm. (I) BAL Th2 cells (FoxP3−GATA3+CD4+ T cells) were quantitated in indicated groups via flow cytometry. Representative data show individual mice with mean ± SEM from one of three independent experiments with three to four mice per group (A–F and H–I) or mean ± SEM from two cumulative experiments with four mice per group (G). For statistical analysis, one-way ANOVA analysis with Holm-Sidak’s testing was used for statistical analysis of multiple groups with paired two-tailed t tests for comparison on naive and memory parabiont groups. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We then assessed the pattern and severity of inflammation within the lung parenchyma from naive and memory parabionts as well as naive controls after HDM treatment. The lung inflammation induced by circulating memory Th2 cells in the naive parabiont mice was mainly perivascular, consistent with our data that recruited Th2 cells are sufficient for eosinophil recruitment (Fig. 3 D). In contrast, memory parabionts with Th2 Trm cells exhibited more peribronchial inflammation than naive parabionts (Fig. 3 D). Memory parabionts had higher lung inflammation scores, predominantly due to airway inflammation (Fig. 3 E). In support of the observation that Th2 Trm cells were required for efficient peribronchial inflammation, we found greater mucus metaplasia in memory parabionts (Fig. 3, D and F). In murine models of allergic asthma, allergen treatment and administration of methacholine increases airway resistance, which requires airway mucus production (Evans et al., 2015). To assess the role of Th2 Trm cells and circulating memory Th2 cells in airway hyperresponsiveness, we measured airway resistance in naive and memory parabionts after HDM treatment and increasing doses of methacholine. In support of our histological findings, increased airway resistance only occurred in memory parabionts rechallenged with HDM (Fig. 3 G). We also found that CD4+ T cells in naive parabionts treated with HDM localized around blood vessels and did not accumulate near airways, whereas memory parabionts treated with HDM exhibited accumulations of CD4+ T cells around both blood vessels and airways (Fig. 3 H). Finally, we performed BAL in naive and memory parabionts challenged with HDM followed by flow cytometry for FoxP3−GATA3+CD4+ T cells and found markedly greater numbers of Th2 cells in the airways of memory parabionts (Fig. 3 I). These results demonstrate that Th2 Trm cells and circulating memory Th2 cells both contribute to the recall response to HDM and perform nonredundant function in vivo. Circulating memory Th2 cells are sufficient to promote perivascular inflammation and eosinophil recruitment, while Th2 Trm cells promote peribronchial inflammation, including mucus metaplasia, airway hyperresponsiveness, and airway eosinophil activation.
Th2 Trm cells drive the airway Th2 response to HDM via in situ proliferation but are dispensable for CD4+ T cell recruitment
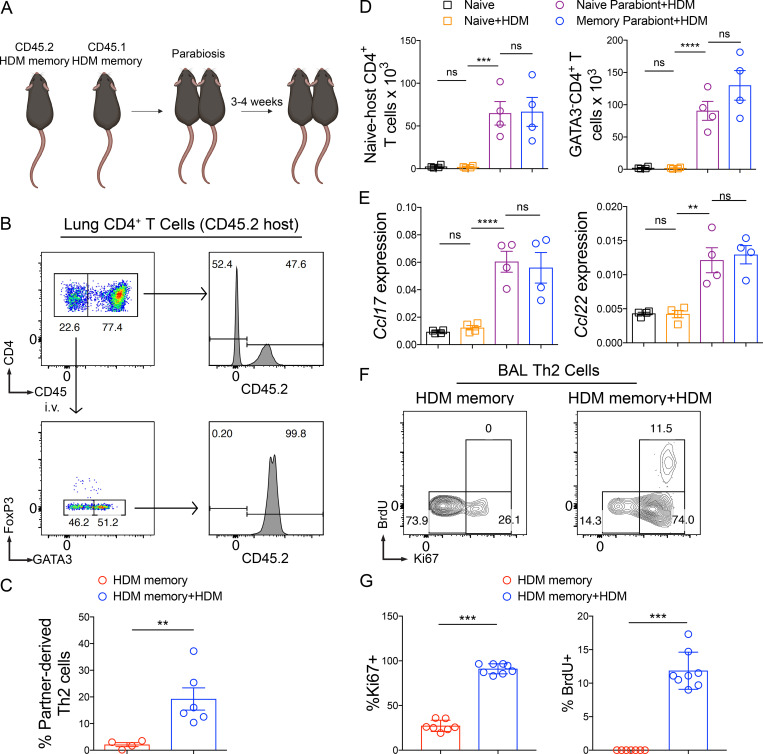
We subsequently turned to the mechanisms regulating the trafficking of Th2 Trm cells and circulating memory Th2 cells during homeostasis and HDM rechallenge. Although Trm cells are the dominant memory T cell population in NLT during homeostasis, a population of recirculating (nonresident) memory T cells can dynamically survey NLT (Bromley et al., 2013; Gerlach et al., 2016). In addition, in a murine model of influenza infection, lung CD8+ Trm cells wane with time due to apoptosis, with a half-life of 5 d, and require circulating memory T cells to “reseed” the Trm cell compartment during homeostasis (Slütter et al., 2017). To investigate the extent to which circulating memory Th2 cells recirculate or reseed the Th2 Trm cell pool within the lung parenchyma during homeostasis, we surgically conjoined congenic (CD45.2 and CD45.1) HDM-memory mice (Fig. 4 A). After 3–4 wk, we found that memory Th2 cells within the lung parenchyma were overwhelming derived from the host, indicating the absence of significant memory Th2 cell recirculation or circulating memory Th2 cells reseeding the Th2 Trm cell pool in the HDM model, at least during this time frame (Fig. 4, B and C). In contrast, FoxP3−GATA3−CD4+ T cells (non-Th2 cells) within the lung parenchyma were a mix of tissue-resident and recirculating memory CD4+ T cells (Fig. S3 E). To investigate Th2 cell trafficking during HDM rechallenge, we administered a single dose of HDM to both memory parabionts and found a significant influx of partner-derived Th2 cells (Fig. 4 C). Consequently, Th2 Trm cells do not require reseeding from circulating memory Th2 cells, and circulating memory Th2 cells only traffic into the lung parenchyma upon HDM rechallenge.
Figure 4.
Th2 Trm cells drive the airway Th2 response to HDM via in situ proliferation but are dispensable for CD4+ T cell recruitment. (A and B) CD45.2 and CD45.1 HDM-memory mice were surgically conjoined. After 3–4 wk, both HDM-memory parabionts were left untreated or received a single dose of i.n. HDM with harvest of lungs after 72 h. (A) Schematic of parabiosis experiment. (B) Representative flow cytometry of lung CD4+ T cells from untreated CD45.2 memory parabiont demonstrating CD45.2 expression of anti-CD45 labeled CD4+ T cells. In addition, FoxP3 and GATA3 expression of anti-CD45 unlabeled CD4+ T cells as well as CD45.2 expression of FoxP3−GATA3+CD4+ T cells. (C) Quantitation of percentage of partner-derived lung Th2 cells (anti-CD45 i.v. unlabeled, FoxP3−GATA3+CD4+ T cells) assessed via flow cytometry from untreated and HDM-rechallenged memory parabionts. (D and E) CD45.2 HDM-memory mice were surgically conjoined to CD45.1 naive mice. After 3–4 wk, both parabionts received a single dose of i.n. HDM with harvest of lung after 72 h. (D) Lung naive-host–derived CD4+ T cells (anti-CD45 i.v. unlabeled, FoxP3−CD45.2−CD4+ T cells) and anti-CD45 i.v. unlabeled FoxP3−GATA3−CD4+ T cells were quantitated via flow cytometry. (E) Lung Ccl17 and Ccl22 relative expression assessed via qPCR. (F and G) Individual HDM-memory mice were left untreated or received a single dose of i.n. HDM with BAL after 72 h. BrdU was injected i.p. 2 h before BAL. (F) Representative flow cytometry of BAL Th2 cells (FoxP3−GATA3+CD4+ T cells) from HDM-memory mice without and with HDM rechallenge demonstrating Ki67 and BrdU expression. (G) Quantitation of the percentage of Ki67+ and BrdU+ BAL Th2 cells in indicated groups. Representative data show individual mice with mean ± SEM from two cumulative experiments with four to six mice per group (A–C), mean ± SEM from one of three independent experiments with three or four mice per group (D and E), or mean ± SEM from two cumulative experiments with eight mice per group (F and G). For statistical analysis, two-tailed Mann-Whitney U testing was performed for nonparametric data. One-way ANOVA analysis with Holm-Sidak’s testing was used for statistical analysis of multiple groups with paired two-tailed t tests for comparison on naive and memory parabiont groups. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Next, we investigated the mechanism whereby tissue-resident memory to HDM enhances Th2 cell expansion (as demonstrated in Fig. 2, F and G). Trm cells within NLT have been demonstrated to enhance local expansion of T cells in two ways. First, in models of type 1 immunity, CD8+ and Th1 Trm cells have been shown to enhance the recruitment of circulating T cells (Schenkel et al., 2013; Glennie et al., 2015). Second, while Trm cells have long been assumed to be terminally differentiated and proliferate poorly, recent studies have shown that CD8+ Trm cells can proliferate in situ (Park et al., 2018; Beura et al., 2018). To determine whether Th2 Trm cells promote enhanced CD4+ T cell recruitment from the circulation, we returned to our parabiosis system involving HDM-memory and naive mice. Specifically, to measure CD4+ T cell recruitment, we investigated the number of conventional CD4+ T cells derived from the naive mouse that trafficked into the lung parenchyma upon HDM challenge. CD45.1-naive, nonparabiotic mice treated with HDM did not exhibit CD4+ T cell recruitment, suggesting innate signals induced by a single dose of HDM are insufficient to promote CD4+ T cell recruitment (Fig. 4 D). In contrast, both naive and memory parabionts exhibited similar recruitment of naive-host–derived CD45.2−CD4+ T cells into the lung parenchyma (Fig. 4 D). In addition, the number of FoxP3−GATA3−CD4+ T cells (non-Th2 cells) and regulatory T (T reg) cells in the lung parenchyma was similar in the two parabionts, suggesting that tissue-resident memory is dispensable for CD4+ T cell recruitment (Fig. 4 D and Fig. S3 F). The chemokine receptor CCR4 is well described to promote CD4+ T cell recruitment into the lung during type 2 immune responses via expression of the CCR4 ligands CCL17 and CCL22, which are induced by IL-4 and IL-13 (Mikhak et al., 2009, 2013; Perros et al., 2009; Faustino et al., 2013; Afshar et al., 2013). We examined the expression of Ccl17 and Ccl22 after HDM administration in naive and memory parabionts as well as naive controls. CD45.1-naive, nonparabiotic mice treated with HDM did not exhibit increased expression of either chemokine in the lung after HDM administration. In contrast, naive parabionts treated with HDM induced Ccl17 and Ccl22 expression in the lung similar to those of memory parabionts, consistent with our observations of similar T reg cell and CD4+ T cell recruitment (Fig. 4 E). Consequently, in agreement with circulating memory Th2 cells being sufficient to promote perivascular inflammation and lung eosinophilia, we found circulating memory Th2 cells were sufficient to promote CD4+ T cell recruitment into the lung parenchyma upon HDM treatment.
The observations that Th2 Trm cells enhance the number of Th2 cells within the lung and BAL without increasing CD4+ T cell recruitment suggested that Th2 Trm cells enhance the Th2 response within the airways primarily by proliferating in situ. To confirm this, we took advantage of our observation that Th2 Trm cells are required for Th2 cell expansion in the BAL (demonstrated in Fig. 3 I). We left individual HDM-memory mice untreated or rechallenged with a single dose of HDM. 2 h before harvest, we administered an i.p. injection of BrdU and performed BAL to assess Ki67 expression and BrdU uptake in BAL Th2 cells. During homeostasis, the majority of BAL Th2 cells exhibited low expression of Ki67 and did not uptake BrdU, consistent with being in the G0 phase of the cell cycle (Fig. 4, F and G). Upon HDM rechallenge, airway Th2 cells dramatically increased in number and increased Ki67 expression, with a subset acquiring BrdU (Fig. 4, F and G; andFig. S3 G). Our various experimental approaches demonstrate that Th2 Trm cells are the dominant memory Th2 cell subset surveying the lung parenchyma during homeostasis and respond to HDM rechallenge by robustly proliferating in situ near airways but are dispensable for CD4+ T cell recruitment into the lung parenchyma.
Shared and distinct transcriptional profiles of Th2 Trm cells and circulating memory Th2 cells
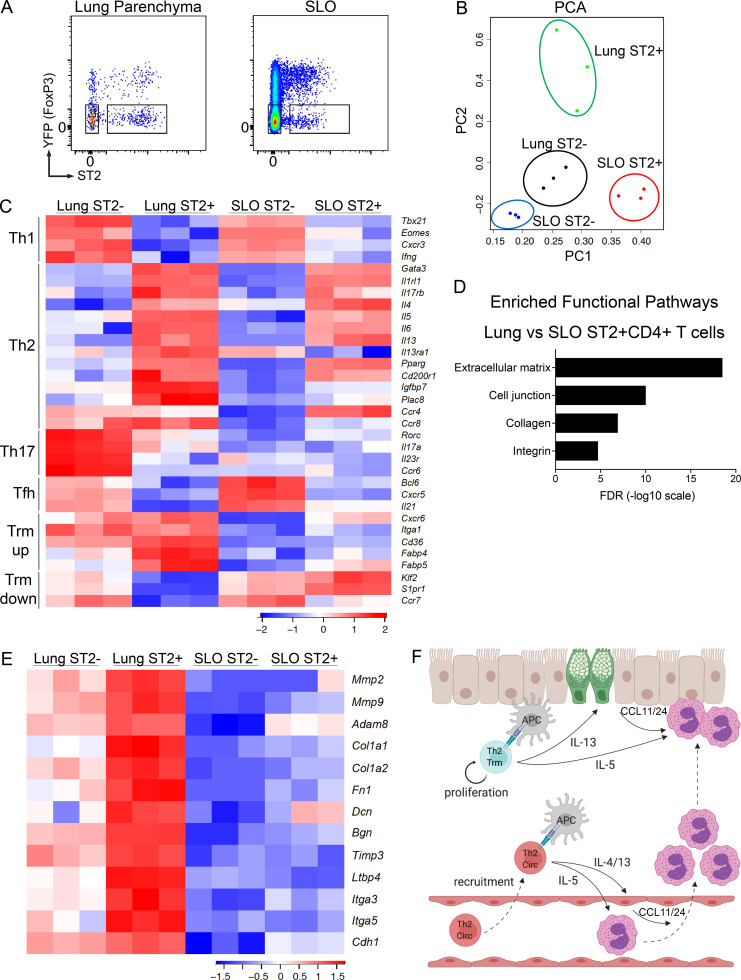
Studies on CD8+ Trm and Th1 Trm cells have demonstrated that these cells exhibit a transcriptional profile distinct from their circulating counterparts (Mackay et al., 2013; Wakim et al., 2012; Pan et al., 2017; Mackay et al., 2016; Strutt et al., 2018; Beura et al., 2019). Given the distinct trafficking patterns and functions of Th2 Trm cells and circulating memory Th2 cells that we have elicited here, we sought to define the transcriptional profiles of these two memory Th2 cell subsets via RNA sequencing (RNA-seq) analysis. We used FoxP3YFPCre reporter mice to generate HDM-memory mice and then sorted YFP−ST2+CD4+ T cells from the lung parenchyma and secondary lymphoid organs (SLO; Fig. 5 A). We took this approach to avoid fixation and permeabilization, which is required for intranuclear staining for FoxP3 (to exclude T reg cells) and GATA3 (to identify Th2 cells). Rather, we used YFP fluorescence to exclude T reg cells and the IL-33 receptor ST2 to identify Th2 cells, the latter marker having been shown to be a reliable surface marker for Th2 cells in the murine HDM model (Tibbitt et al., 2019). Along with the above memory Th2 cell populations, we also sorted YFP−ST2−CD4+ T cells from the lung and YFP−ST2−CD44+CD4+ T cells from SLO as control populations of non-Th2 cells. We excluded CD44−CD4+ T cells from the SLO to avoid a mixed population of naive and memory CD4+ T cells in our control population. Principal-component analysis of differentially expressed genes demonstrated that lung ST2+CD4+ T cells and SLO ST2+CD4+ T cells were transcriptionally distinct subsets (Fig. 5 B). We next compared canonical T helper gene signatures in our various memory CD4+ T cell populations. As expected, Th1, Th17, and Tfh gene signatures were enriched in the memory ST2−CD4+ T cells from the lung and SLO (Fig. 5 C). Lung and SLO memory ST2+CD4+ T cells exhibited similar expression of many canonical Th2 genes, including Gata3, Il1rl1 (ST2), Il17rb1 (IL-25R), Il4, Il5, and Il3 (Fig. 5 C). Lung memory ST2+CD4+ T cells expressed higher levels of the IL-13 receptor Il13ra1 compared with SLO memory ST2+CD4+ T cells, indicating that Th2 Trm cells exhibit enhanced responsiveness to IL-13 within the lung parenchyma (Fig. 5 C). A recent study performing single-cell RNA-seq analysis on CD4+ T cells from the primary HDM model demonstrated that effector Th2 cells within the lung are a predominant source of IL-6 and also identified several Th2-specific transcripts, including Pparg, Cd200r1, Igfbp7, and Plac8 (Tibbitt et al., 2019). PPARγ has been shown to be required for effective induction of IL-5 and IL-13 in effector Th2 cells (Angela et al., 2016; Chen et al., 2017; Nobs et al., 2017; Tibbitt et al., 2019). However, whether these Th2 cell–specific genes are expressed in the memory phase and the relative expression within Th2 Trm cells and circulating memory Th2 cells is not known. We found lung and SLO memory ST2+CD4+ T cells express similar levels of Il6, Pparg, and Cd200r1 (Fig. 5 C). In contrast, lung memory ST2+CD4+ T cells expressed higher levels of Igfbp7 and Plac8 than SLO memory ST2+CD4+ T cells (Fig. 5 C). While the function of Igfbp7 and Plac8 in Th2 cell biology remains unclear, our results suggest these genes may play an important role in Th2 Trm function. Lastly, while the chemokine receptors CCR4 and CCR8 have been shown to be expressed by Th2 cells (Griffith et al., 2014), we found differential expression in lung and SLO memory ST2+CD4+ T cells, with Ccr4 being preferentially expressed in SLO memory ST2+CD4+ T cells and Ccr8 being preferentially expressed in lung memory ST2+CD4+ T cells (Fig. 5 C), indicating these two memory Th2 cell subsets differentially respond to type 2 chemokines in vivo.
Figure 5.
Shared and distinct transcriptional profiles of Th2 Trm and circulating memory Th2 cells. (A–E) FoxP3YFPCre mice were sensitized and challenged with intranasal HDM and rested for 6 wk followed by lung and LN/spleen (SLO) harvest. (A) Representative flow cytometry of lung and SLO CD4+ T cells indicating YFP and ST2 expression as well as sorting gates for YFP−ST2− and YFP−ST2+ populations. (B) Principal component analysis (PCA) plot based on differentially expressed genes from RNA-seq data of indicated memory CD4+ T cell populations. (C) Heatmap of expression values of T cell subset genes, including Th1, Th2, Th17, Tfh, and Trm up-regulated and down-regulated genes. (D) Bar graph of statistical significance (false discovery rate [FDR] by the DAVID tool) of pathway enrichment among genes overexpressed in lung YFP−ST2+CD4+ T cells relative to SLO YFP−ST2+CD4+ T cells. (E) Heatmap of a selected subset of genes known to be involved in ECM biology and cell adhesion. Data are from three independent experiments with five HDM-memory mice pooled for each experiment (cell sort and RNA isolation). (F) Model of memory Th2 cell subsets during allergen rechallenge. Transcriptionally distinct memory Th2 cell subsets include Th2 Trm cells persisting in the lung parenchyma (indicated in blue) and circulating memory Th2 cells in the blood (Th2 Circ; indicated in red). After allergen rechallenge, circulating memory Th2 cells traffic into the lung parenchyma and produce type 2 cytokines, promoting perivascular inflammation, CCL11 and CCL24 production, and eosinophil recruitment. Activation of Th2 Trm cells leads to in situ proliferation and production of type 2 cytokines near the airways, which promotes mucus metaplasia (green epithelial cells), CCL11 and CCL24 production, and eosinophil activation. CCL11 and CCL24 are likely produced from multiple structural and immune cell types following allergen rechallenge. Created with BioRender.com.
Next, we assessed the expression of genes known to play a role in the maintenance and/or function of Trm in vivo. CD8+ Trm cells express the chemokine receptor CXCR6 and CD49a, which constitutes the α-subunit of the α1β1 integrin and binds to collagen (Szabo et al., 2019). CD8+ Trm cells within the lung require CXCR6 to localize to airways, and α1β1 is required for effective CD8+ T cell immunity within the lung (Ray et al., 2004; Richter and Topham, 2007; Wein et al., 2019). We found lung ST2+CD4+ T cells express higher levels of Cxcr6 as well as CD49a (Itga1) than circulating memory ST2+CD4+ T cells (Fig. 5 C). In addition, CD8+ Trm cells have been shown to require exogenous lipid uptake and metabolism, including fatty acid–binding proteins, for survival in NLT (Pan et al., 2017). Similar to CD8+ Trm, Th2 Trm cells express higher levels of the lipid scavenging molecule Cd36 as well as fatty acid–binding proteins (Fabp4 and Fabp5) than circulating memory Th2 cells (Fig. 5 C; Pan et al., 2017). Lastly, the establishment of CD8+ Trm cells, as well as tissue-resident natural killer T cells and natural killer cells, requires the transcription factors Blimp1 and Hobit, which directly promote the down-regulation of pathways involved in egress from NLTs, including the genes Klf2, S1pr1, and Ccr7 (Mackay et al., 2016). We found that Th2 Trm cells down-regulated tissue egress genes relative to circulating memory Th2 cells (Fig. 5 C). However, we did not find that Th2 Trm cells preferentially express Prdm1 (Blimp1) or Znf683 (Hobit) or the CD8+ Trm cell–promoting transcription factor Runx3 (Milner et al., 2017; Mackay et al., 2016). Transcription factors critical for Trm cell differentiation, such as Runx3, can be expressed at similar levels in Trm cells and circulating memory T cells, indicating that transcription factor function, rather than relative expression, drives Trm cell development (Milner et al., 2017).
To identify additional unique pathways differentially expressed between lung and SLO memory ST2+CD4+ T cells, we performed pathway enrichment analysis using the DAVID tool (Huang et al., 2009). This analysis of lung and SLO memory ST2+CD4+ T cells in our HDM model revealed the enrichment of ECM-regulating and interacting genes among differentially expressed genes (Fig. 5 D). Compared with SLO memory ST2+CD4+ T cells, lung memory ST2+CD4+ T cells expressed higher transcript levels of metalloproteases, such as Mmp2, Mmp9, and Adam8; ECM components, such as collagen (col1a1 and col1a2), fibronectin (fn1), Decorin (Dcn), and Byglycan (Bgn); and regulators of ECM, including tissue inhibitor of metalloproteases Timp3 and the TGF-β binding protein Ltbp4 (Fig. 5 E). In addition, genes involved in cell-to-ECM and cell-to-cell interaction, including Itga3, Itga5, and Cdh1, were enriched in lung memory ST2+CD4+ T cells compared with SLO memory ST2+CD4+ T cells (Fig. 5 E). Our findings indicate that Th2 Trm cells and circulating memory Th2 cells share a core Th2 gene signature, while Th2 Trm cells uniquely express a tissue-adaptation program, which includes genes involved in lipid metabolism and ECM biology. Notably, metalloproteases, ECM components, and specific integrins have also been shown to be enriched in CD8+ and Th1 Trm cells in other experimental systems, suggesting that regulation and interaction with the ECM may play an important role in the development, maintenance, and function of Trm cells in various tissues (Mackay et al., 2013; Wakim et al., 2012; Pan et al., 2017; Beura et al., 2019). Lastly, a recent study using Aspergillus fumigatus fungal extract to promote airway inflammation in mice found that effector CD4+ T cells expressing CD69 are enriched for genes involved in ECM biology and fibrosis, indicating that effector CD4+ T cells are also capable of directly regulating the ECM within the lung during active inflammation (Ichikawa et al., 2019).
Previous studies investigating the function of Th2 Trm cells have relied on strategies to decrease the numbers of circulating T cells, including treatment with the S1P1 agonist FTY720 or anti-Thy1 antibody (Hondowicz et al., 2016; Turner et al., 2018; Ichikawa et al., 2019). Notably, decreasing circulating T cells during allergen rechallenge did not significantly reduce the number of allergen-specific CD4+ T cells within the lung or the allergen recall response, suggesting circulating memory Th2 cells do not play a significant role (Hondowicz et al., 2016; Turner et al., 2018; Ichikawa et al., 2019). While FTY720 efficiently sequesters naive T cells in LNs, it is less efficient at causing lymphopenia of memory T cells (Hofmann et al., 2006). In addition, FTY720 treatment has been shown to alter the biology of multiple cell types beyond conventional T cells, including increasing endothelial permeability (Shea et al., 2010; Oo et al., 2011), potentially altering inflammatory cell recruitment, and potently inhibiting T reg cell function (Wolf et al., 2009). Likewise, anti-Thy1 antibody likely targets multiple cell types, including T reg cells. Parabiosis allowed us to define the function of Th2 Trm cells and circulating memory Th2 cells in an unmanipulated rechallenge response, demonstrating that both Th2 cell subsets contribute to the recall response and perform distinct functions. While our parabiosis model allows us to define the role of memory Th2 cell subsets in vivo, one limitation of our study is that we have not defined the additional lung-resident changes in HDM-memory mice that promote the recall response to HDM. Specifically, the niches maintaining Th2 Trm cells within the lung remain poorly delineated. Notably, there appear to be differences in the tissue environment supporting distinct types of Trm cells. Lung CD8+ Trm cells have been shown to persist in “repair-associated memory depots” independently of conventional iBALT (Takamura et al., 2016). Similarly, Th1 Trm cells in the female reproductive tract have also been shown to persist in “macrophage lymphocyte clusters” independently of tertiary lymphoid structures (Iijima and Iwasaki, 2014). In contrast, memory Th2 cells persisting in the lung of mice after allergic inflammation have been shown to localize to iBALT-like structures, supported by IL-7 from lymphatic endothelial cells (Shinoda et al., 2016). Future studies will be needed to define the unique features of the peribronchial niche supporting the development, maintenance, and function of Th2 Trm cells in vivo.
In summary, we have used a HDM model of allergic airway disease, parabiosis, and transcriptional analysis to define the functions and transcriptional profiles of tissue-resident and circulating memory Th2 cells. We demonstrate that Th2 Trm cells within the lung and circulating memory Th2 cells are transcriptionally distinct subsets that exhibit nonoverlapping trafficking patterns during homeostasis. Upon allergen rechallenge, Th2 Trm cells and circulating memory Th2 cells both contribute to the increased numbers of Th2 cells and type 2 cytokine expression in the lung and perform nonredundant functions in vivo (Fig. 5 F). Consequently, Th2 Trm cells represent a unique Th2 cell subset with a distinct tissue-adaptation gene signature that are required for the full manifestation of allergic airway disease in vivo. Defining the unique mechanisms supporting the development and maintenance of Th2 Trm cells will be critical to developing novel therapeutic approaches for allergic asthma.
Materials and methods
Mice
C57BL/6J (WT) and CD45.1 CRL mice were obtained from Charles River Laboratories. Rag2-deficient and C57BL6/J control mice were obtained from Jackson Laboratories. FoxP3YFPCre mice were obtained from Jackson Laboratories and bred and maintained within our facility. All mice analyzed were sex and aged matched (6–12 wk old). All mice were bred and maintained in specific pathogen–free conditions at the animal facility of the Massachusetts General Hospital and used under a study protocol approved by Massachusetts General Hospital Subcommittee on Research Animal Care.
Mouse treatments
To induce allergic airway inflammation, mice were anesthetized with intramuscular injection of ketamine/xylazine (Patterson Veterinary) and sensitized via i.n. administration with 10 µg HDM (Dermatophagoides pteronyssinus extracts; Greer Laboratories) in 40 µl sterile PBS on day 0. On day 7, mice were challenged with 10 µg HDM via i.n. route daily for 5 d. 6–12 wk later, mice were either left untreated or rechallenged with 10 µg HDM via i.n. route once or 10 µg of A. alternata extract (Greer Laboratories). For Th2 proliferation studies, 1–2 mg BrdU (Thermo Fisher Scientific) was injected i.p., and tissue was harvested 2 h later.
Parabiosis surgery
CD45.2 C57BL/6J mice were sensitized and challenged with HDM as outlined and rested for 3–4 wk to generate memory Th2 cells in vivo. CD45.2 HDM-memory C57BL/6J mice and naive CD45.1 B6 mice were surgically conjoined to achieve parabiosis. Specifically, mice underwent hair removal along opposite lateral flanks with the use of hair clippers and depilatory cream. Skin was then wiped clean of fur and sterilized with topical betadine solution. Mirrored incisions were then made on the lateral aspects of both mice from forelimb to hindlimb. 4.0 sutures were placed around the olecranon and knee joints to secure the upper and lower extremities of the mice. Dorsal and ventral skin was approximated with a running 4.0 suture and surgical staples. At the end of the surgery, mice received subcutaneous enrofloxacin antibiotic as well as buprenorphine and flunixin for pain control. Enrofloxacin antibiotic was subsequently administered via drinking water for 2 wk. s.c. buprenorphine and flunixin was administered as needed every 12 h for 48 h. Parabionts were both challenged with 10 µg HDM i.n., and tissues were collected at 72 h.
Tissue harvest and leukocyte preparation
Prior to tissue harvest, mice were anesthetized with ketamine/xylazine (Patterson Veterinary) and injected i.v. with 3 µg fluorophore-labeled anti-CD45 (30-F11; BioLegend) mAb through the retro-orbital sinus to label intravascular leukocytes. 3 min after anti-CD45 antibody injection, tissues were harvested. For BAL, the trachea was exposed, cannulated, and infused with 3 ml cold PBS with 0.5 mM EDTA. Lung lobes and mLNs were removed, minced with scissors, and digested at 37°C for 20–30 min in digestion buffer (0.52 U/ml Liberase TM [Roche] and 60 U/ml DNase I [Roche] in RPMI 1640 [Cellgro] with 5% FBS). Digested tissue was strained through a 70-µm filter to generate a single-cell suspension.
Flow cytometry
Single cells were incubated with anti-mouse CD16/32 (93, TruStain fcX; BioLegend) to block Fc receptors. Staining was performed with Fixable Viability Dye eF780 or eF506 (eBioscience), to identify dead cells.The following fluorochrome-conjugated anti-mouse mAbs were used: CD3e-AF488 (145-2C11), CD4-PeCy7 (GK1.5), Foxp3-APC (FJK-16s), GATA3-PE (L50-823), CD45-APC-eFluor780 (30-F11), CD11c-BV605 (N418), Siglec-F-PE (E50-2440), CD11b-BUV737 (M1/70), MHCII-PeCy7 (M5/114.15.2), CD45.2-BUV395 (104), CD62L-FITC (MEL14), BrdU-AF647 (3D4), ST2-BV421 (U29-93), CD127-PeCy7 (A7R34), and Thy1.2-PerCP-eFluor710 (30-H12). Flow cytometric analysis was performed using a LSR Fortessa X-20 flow cytometer (BD Biosciences) and FlowJo software (Tree Star). Intracellular staining was performed using eBioscience Fixation Permeabilization buffers. BrdU staining was performed following the BD Biosciences manufacture’s protocol.
Ex vivo T cell restimulation
mLNs were harvested and single-cell suspensions were obtained. 0.25 × 106 total LN cells were cultured in 250 μl of RPMI 1640 media (Cellgro) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS, Sigma), 50 µM 2-mercaptoethanol, 2 mM Glutamax (Gibco), 5 mM Hepes, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, and 100 U/ml penicillin-streptomycin (all from Lonza) and stimulated with 25 µg/ml HDM (Greer Laboratories) for 72 h at 37°C, 5% CO2. Supernatant culture cytokine levels for IL-5 (BioLegend) and IL-13 (eBioscience) were measured by ELISA using BioLegend and eBioscience kits, respectively, and according to the manufacturer’s instructions.
qPCR RNA levels
RNA was extracted from lung tissue in Trizol using the RNeasy Mini kit (Qiagen), and cDNA was reverse transcribed using SuperScript III First Strand (Invitrogen) following manufacturer’s guidelines. Quantitative PCR (qPCR) reactions were performed on a LightCylcer 96 Instrument (Roche) using FastStart Essential DNA Green Master (Roche) and normalized to B2m or Gapdh using the following primers: Il5, 5′-CTCTGTTGACAAGCAATGAGACG-3′ (forward) and 5′-TCTTCAGTATGTCTAGCCCCTG-3′ (reverse); Il13, 5′-CCTGGCTCTTGCTTGCCTT-3′ (forward) and 5′-GGTCTTGTGTGATGTTGCTCA-3′ (reverse); Ccl11, 5′-TCCACAGCGCTTCTATTCCTG-3′ (forward) and 5′-GGAGCCTGGGTGAGCCA-3′ (reverse); Ccl24, 5′-ATT CTGTGACCATCCCCTCAT-3′ (forward) and 5′-TGTATGTGCCTCTGAACCCAC-3′ (reverse); Ccl17, 5′-CAG GGATGCCATCGTGTTTC-3′ (forward) and 5′-CACCAATCTGATGGCCTTCTT-3′ (reverse); Ccl22, 5′-TAC ATCCGTCACCCTCTGCC-3′ (forward) and 5′-CGGTTATCAAAACAACGCCAG-3′ (reverse); B2m, 5′-CCCGTTCTTCAGCATTTGGA-3′ (forward) and 5′-CCGAACATACTGAACTGCTACGTAA-3′ (reverse); Gapdh, 5′-GGCAAATTCAACGGCACAGT-3′ (forward) and 5′-AGATGGTGATGGGCTTCCC-3′ (reverse). Results indicated as relative expression indicate copies per B2m or Gapdh.
Histology
Lung samples were fixed in buffered 10% formalin solution. Paraffin-embedded sections were cut (5 mm) and stained with H&E or periodic acid–Schiff (PAS). Lung H&E histology scoring was performed by a pathologist from the Harvard Pathology Core who was blinded to the groups. Goblet cells were enumerated from PAS-stained sections using a numerical scoring system (0, <5% goblet cells; 1, 5–25%; 2, 25–50%; 3, 50–75%; 4, >75%). 20–50 airways per mouse were evaluated, and the sum of goblet cell scoring from each mouse was divided by the number of airways and presented as a mucus score as previously described (Patel et al., 2019). All images are shown at 10× magnification.
Measurement of airway resistance
Airway resistance was measured using the SCIREQ flexiVent system. Mice were anesthetized with ketamine/xylazine and intubated via tracheotomy. The mice were placed on a ventilator and paralyzed with pancuronium (1 mg/kg). After 5 min, the mice were first challenged with PBS and then progressively challenged with methacholine (12.5, 25, and 50 mg/ml), and airway resistance was measured using the flexiVent software system.
RNA-seq analysis (accession code GSE149779)
Sequencing was performed on Illumina HiSeq 2500 instrument, resulting in ∼30 million of 50-bp reads per sample. Sequencing reads were mapped in a splice-aware fashion to the mouse reference transcriptome (mm9 assembly) using STAR (Dobin et al., 2013). Read counts over transcripts were calculated using HTSeq based on the Ensembl annotation for GRCm37/mm9 assembly (Anders et al., 2015). For differential expression analysis, we used the EdgeR method and classified genes as differentially expressed based on the cutoffs of twofold change in expression value and false discovery rates <0.05 (Robinson et al., 2010).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 7 software. Statistical parameters are reported in the figure legends, but generally, P values were determined for multiple comparisons by one-way ANOVA analysis with Holm-Sidak’s testing. An unpaired or paired two-tailed Student’s t test was used for two-group comparisons for normally distributed data. Mann-Whitney U testing was used for two-group comparisons for nonnormally distributed data. In all figures, statistical significance is indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). No statistical method was used to predetermine sample size. The number of mice used in each experiment to reach statistical significance was determined on the basis of preliminary data. No animals were excluded from the analyses. Blinding was used for lung H&E history scoring. Data met assumptions of statistical methods used, and variance was similar between groups that were statistically compared.
Online supplemental material
Fig. S1 shows that HDM rechallenge promotes type 2 chemokine expression within the lung as well as lung inflammation and mucus metaplasia. Fig. S2 shows representative gating strategy and quantitation of Th2 Trm cells within the lung. Fig. S3 shows additional data demonstrating the functions and trafficking of Th2 Trm cells and circulating memory Th2 cells.
Acknowledgments
We thank Dr. Hui Zheng from the Massachusetts General Hospital (MGH) Biostatistics Center for thoughtful input on statistical analyses. We thank Dr. Roderick Bronson from the Rodent Pathology Core at Harvard Medical School for scoring histology samples. We thank members of the MGH Next Generation Sequencing Core facility and Flow Cytometry Core facility for technical assistance.
This work was supported by the National Institutes of Health (grant R01 AI040618 to A.D. Luster, Training Program in Pulmonary Immunology and Allergy grant T32 HL116275 to A.D. Luster, grant K08 HL140173 to R.A. Rahimi, and grant P30 DK040561 to R.I. Sadreyev). Cytometric findings reported here were performed in the MGH Department of Pathology Flow and Image Cytometry Research Core, which obtained support from the National Institutes of Health Shared Instrumentation program (grants 1S10OD012027-01A1, 1S10OD016372-01, 1S10RR020936-01, and 1S10RR023440-01A1).
Author contributions: R.A. Rahimi designed, performed, analyzed, and interpreted experiments and wrote the manuscript. K. Nepal performed experiments and collected and assembled data. M. Cetinbas and R.I. Sadreyev assisted with RNA-seq data analysis and presentation as well as manuscript preparation. A.D. Luster conceptualized and designed the study, analyzed and interpreted data, and contributed to manuscript preparation.
References
- Afshar R., Strassner J.P., Seung E., Causton B., Cho J.L., Harris R.S., Hamilos D.L., Medoff B.D., and Luster A.D.. 2013. Compartmentalized chemokine-dependent regulatory T-cell inhibition of allergic pulmonary inflammation. J. Allergy Clin. Immunol. 131:1644–1652. 10.1016/j.jaci.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.G., Mayer-Barber K., Sung H., Beura L., James B.R., Taylor J.J., Qunaj L., Griffith T.S., Vezys V., Barber D.L., et al. 2014. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9:209–222. 10.1038/nprot.2014.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angela M., Endo Y., Asou H.K., Yamamoto T., Tumes D.J., Tokuyama H., Yokote K., and Nakayama T.. 2016. Fatty acid metabolic reprogramming via mTOR-mediated inductions of PPARγ directs early activation of T cells. Nat. Commun. 7:13683 10.1038/ncomms13683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbes S.J. Jr., Gergen P.J., Vaughn B., and Zeldin D.C.. 2007. Asthma cases attributable to atopy: results from the Third National Health and Nutrition Examination Survey. J. Allergy Clin. Immunol. 120:1139–1145. 10.1016/j.jaci.2007.07.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Mitchell J.S., Thompson E.A., Schenkel J.M., Mohammed J., Wijeyesinghe S., Fonseca R., Burbach B.J., Hickman H.D., Vezys V., et al. 2018. Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19:173–182. 10.1038/s41590-017-0029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Fares-Frederickson N.J., Steinert E.M., Scott M.C., Thompson E.A., Fraser K.A., Schenkel J.M., Vezys V., and Masopust D.. 2019. CD4+ resident memory T cells dominate immunosurveillance and orchestrate local recall responses. J. Exp. Med. 216:1214–1229. 10.1084/jem.20181365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley S.K., Yan S., Tomura M., Kanagawa O., and Luster A.D.. 2013. Recirculating memory T cells are a unique subset of CD4+ T cells with a distinct phenotype and migratory pattern. J. Immunol. 190:970–976. 10.4049/jimmunol.1202805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone F.R. 2015. Tissue-Resident Memory T Cells and Fixed Immune Surveillance in Nonlymphoid Organs. J. Immunol. 195:17–22. 10.4049/jimmunol.1500515 [DOI] [PubMed] [Google Scholar]
- Causton B., Pardo-Saganta A., Gillis J., Discipio K., Kooistra T., Rajagopal J., Xavier R.J., Cho J.L., and Medoff B.D.. 2018. CARMA3 mediates allergic lung inflammation in response to alternaria alternata. Am. J. Respir. Cell Mol. Biol. 59:684–694. 10.1165/rcmb.2017-0181OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Tibbitt C.A., Feng X., Stark J.M., Rohrbeck L., Rausch L., Sedimbi S.K., Karlsson M.C.I., Lambrecht B.N., Karlsson Hedestam G.B., et al. 2017. PPAR-γ promotes type 2 immune responses in allergy and nematode infection. Sci. Immunol. 2 eaal5196 10.1126/sciimmunol.aal5196 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Iwamura C., Kuwahara M., Suzuki A., Sugaya K., Tumes D.J., Tokoyoda K., Hosokawa H., Yamashita M., and Nakayama T.. 2011. Eomesodermin controls interleukin-5 production in memory T helper 2 cells through inhibition of activity of the transcription factor GATA3. Immunity. 35:733–745. 10.1016/j.immuni.2011.08.017 [DOI] [PubMed] [Google Scholar]
- Endo Y., Hirahara K., Iinuma T., Shinoda K., Tumes D.J., Asou H.K., Matsugae N., Obata-Ninomiya K., Yamamoto H., Motohashi S., et al. 2015. The interleukin-33-p38 kinase axis confers memory T helper 2 cell pathogenicity in the airway. Immunity. 42:294–308. 10.1016/j.immuni.2015.01.016 [DOI] [PubMed] [Google Scholar]
- Evans C.M., Raclawska D.S., Ttofali F., Liptzin D.R., Fletcher A.A., Harper D.N., McGing M.A., McElwee M.M., Williams O.W., Sanchez E., et al. 2015. The polymeric mucin Muc5ac is required for allergic airway hyperreactivity. Nat. Commun. 6:6281 10.1038/ncomms7281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino L., da Fonseca D.M., Takenaka M.C., Mirotti L., Florsheim E.B., Guereschi M.G., Silva J.S., Basso A.S., and Russo M.. 2013. Regulatory T cells migrate to airways via CCR4 and attenuate the severity of airway allergic inflammation. J. Immunol. 190:2614–2621. 10.4049/jimmunol.1202354 [DOI] [PubMed] [Google Scholar]
- Galkina E., Thatte J., Dabak V., Williams M.B., Ley K., and Braciale T.J.. 2005. Preferential migration of effector CD8+ T cells into the interstitium of the normal lung. J. Clin. Invest. 115:3473–3483. 10.1172/JCI24482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach C., Moseman E.A., Loughhead S.M., Alvarez D., Zwijnenburg A.J., Waanders L., Garg R., de la Torre J.C., and von Andrian U.H.. 2016. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity. 45:1270–1284. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glennie N.D., Yeramilli V.A., Beiting D.P., Volk S.W., Weaver C.T., and Scott P.. 2015. Skin-resident memory CD4+ T cells enhance protection against Leishmania major infection. J. Exp. Med. 212:1405–1414. 10.1084/jem.20142101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J.W., Sokol C.L., and Luster A.D.. 2014. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu. Rev. Immunol. 32:659–702. 10.1146/annurev-immunol-032713-120145 [DOI] [PubMed] [Google Scholar]
- Guo L., Wei G., Zhu J., Liao W., Leonard W.J., Zhao K., and Paul W.. 2009. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. Proc. Natl. Acad. Sci. USA. 106:13463–13468. 10.1073/pnas.0906988106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Huang Y., Chen X., Hu-Li J., Urban J.F. Jr., and Paul W.E.. 2015. Innate immunological function of TH2 cells in vivo. Nat. Immunol. 16:1051–1059. 10.1038/ni.3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Brinkmann V., and Zerwes H.G.. 2006. FTY720 preferentially depletes naive T cells from peripheral and lymphoid organs. Int. Immunopharmacol. 6:1902–1910. 10.1016/j.intimp.2006.07.030 [DOI] [PubMed] [Google Scholar]
- Hombrink P., Helbig C., Backer R.A., Piet B., Oja A.E., Stark R., Brasser G., Jongejan A., Jonkers R.E., Nota B., et al. 2016. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 17:1467–1478. 10.1038/ni.3589 [DOI] [PubMed] [Google Scholar]
- Hondowicz B.D., An D., Schenkel J.M., Kim K.S., Steach H.R., Krishnamurty A.T., Keitany G.J., Garza E.N., Fraser K.A., Moon J.J., et al. 2016. Interleukin-2-Dependent Allergen-Specific Tissue-Resident Memory Cells Drive Asthma. Immunity. 44:155–166. 10.1016/j.immuni.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Ichikawa T., Hirahara K., Kokubo K., Kiuchi M., Aoki A., Morimoto Y., Kumagai J., Onodera A., Mato N., Tumes D.J., et al. 2019. CD103hi Treg cells constrain lung fibrosis induced by CD103lo tissue-resident pathogenic CD4 T cells. Nat. Immunol. 20:1469–1480. 10.1038/s41590-019-0494-y [DOI] [PubMed] [Google Scholar]
- Iijima N., and Iwasaki A.. 2014. T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science. 346:93–98. 10.1126/science.1257530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson S.C., and Masopust D.. 2018. Understanding Subset Diversity in T Cell Memory. Immunity. 48:214–226. 10.1016/j.immuni.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.W. 2014. Activation states of blood eosinophils in asthma. Clin. Exp. Allergy. 44:482–498. 10.1111/cea.12292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht B.N., and Hammad H.. 2015. The immunology of asthma. Nat. Immunol. 16:45–56. 10.1038/ni.3049 [DOI] [PubMed] [Google Scholar]
- Lambrecht B.N., Hammad H., and Fahy J.V.. 2019. The Cytokines of Asthma. Immunity. 50:975–991. 10.1016/j.immuni.2019.03.018 [DOI] [PubMed] [Google Scholar]
- Li B.W.S., de Bruijn M.J.W., Tindemans I., Lukkes M., KleinJan A., Hoogsteden H.C., and Hendriks R.W.. 2016. T cells are necessary for ILC2 activation in house dust mite-induced allergic airway inflammation in mice. Eur. J. Immunol. 46:1392–1403. 10.1002/eji.201546119 [DOI] [PubMed] [Google Scholar]
- Lukacs N.W. 2001. Role of chemokines in the pathogenesis of asthma. Nat. Rev. Immunol. 1:108–116. 10.1038/35100503 [DOI] [PubMed] [Google Scholar]
- Mackay L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. 2013. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14:1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- Mackay L.K., Minnich M., Kragten N.A.M., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352:459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- Martinez-Gonzalez I., Mathä L., Steer C.A., Ghaedi M., Poon G.F.T., and Takei F.. 2016. Allergen-Experienced Group 2 Innate Lymphoid Cells Acquire Memory-like Properties and Enhance Allergic Lung Inflammation. Immunity. 45:198–208. 10.1016/j.immuni.2016.06.017 [DOI] [PubMed] [Google Scholar]
- Martinez-Gonzalez I., Mathä L., Steer C.A., and Takei F.. 2017. Immunological Memory of Group 2 Innate Lymphoid Cells. Trends Immunol. 38:423–431. 10.1016/j.it.2017.03.005 [DOI] [PubMed] [Google Scholar]
- McKnight C.G., Jude J.A., Zhu Z., Panettieri R.A. Jr., and Finkelman F.D.. 2017. House dust mite-induced allergic airway disease is independent of IgE and FcεRIα. Am. J. Respir. Cell Mol. Biol. 57:674–682. 10.1165/rcmb.2016-0356OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhak Z., Fukui M., Farsidjani A., Medoff B.D., Tager A.M., and Luster A.D.. 2009. Contribution of CCR4 and CCR8 to antigen-specific T(H)2 cell trafficking in allergic pulmonary inflammation. J. Allergy Clin. Immunol. 123:67–73.e3. 10.1016/j.jaci.2008.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhak Z., Strassner J.P., and Luster A.D.. 2013. Lung dendritic cells imprint T cell lung homing and promote lung immunity through the chemokine receptor CCR4. J. Exp. Med. 210:1855–1869. 10.1084/jem.20130091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner J.J., Toma C., Yu B., Zhang K., Omilusik K., Phan A.T., Wang D., Getzler A.J., Nguyen T., Crotty S., et al. 2017. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 552:253–257. 10.1038/nature24993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minutti C.M., Drube S., Blair N., Schwartz C., McCrae J.C., McKenzie A.N., Kamradt T., Mokry M., Coffer P.J., Sibilia M., et al. 2017. Epidermal Growth Factor Receptor Expression Licenses Type-2 Helper T Cells to Function in a T Cell Receptor-Independent Fashion. Immunity. 47:710–722.e6. 10.1016/j.immuni.2017.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobs S.P., Natali S., Pohlmeier L., Okreglicka K., Schneider C., Kurrer M., Sallusto F., and Kopf M.. 2017. PPARγ in dendritic cells and T cells drives pathogenic type-2 effector responses in lung inflammation. J. Exp. Med. 214:3015–3035. 10.1084/jem.20162069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oja A.E., Piet B., Helbig C., Stark R., van der Zwan D., Blaauwgeers H., Remmerswaal E.B.M., Amsen D., Jonkers R.E., Moerland P.D., et al. 2018. Trigger-happy resident memory CD4+ T cells inhabit the human lungs. Mucosal Immunol. 11:654–667. 10.1038/mi.2017.94 [DOI] [PubMed] [Google Scholar]
- Onodera A., Kokubo K., and Nakayama T.. 2018. Epigenetic and Transcriptional Regulation in the Induction, Maintenance, Heterogeneity, and Recall-Response of Effector and Memory Th2 Cells. Front. Immunol. 9:2929 10.3389/fimmu.2018.02929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oo M.L., Han D.K., Hla T., Oo M.L., Chang S., Thangada S., Wu M., and Rezaul K.. 2011. Engagement of S1P 1 -degradative mechanisms leads to vascular leak in mice Find the latest version : Engagement of S1P 1 -degradative mechanisms leads to vascular leak in mice. J. Clin. Invest. 121:2290–2300. 10.1172/JCI45403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Tian T., Park C.O., Lofftus S.Y., Mei S., Liu X., Luo C., O’Malley J.T., Gehad A., Teague J.E., et al. 2017. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature. 543:252–256. 10.1038/nature21379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.L., Zaid A., Hor J.L., Christo S.N., Prier J.E., Davies B., Alexandre Y.O., Gregory J.L., Russell T.A., Gebhardt T., et al. 2018. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol. 19:183–191. 10.1038/s41590-017-0027-5 [DOI] [PubMed] [Google Scholar]
- Patel D.F., Gaggar A., Blalock J.E., Gregory L.G., Lloyd C.M., and Snelgrove R.J.. 2019. Response to Comment on “An extracellular matrix fragment drives epithelial remodeling and airway hyperresponsiveness”. Sci. Transl. Med. 11 eaaw0462 10.1126/scitranslmed.aaw0462 [DOI] [PubMed] [Google Scholar]
- Perros F., Hoogsteden H.C., Coyle A.J., Lambrecht B.N., and Hammad H.. 2009. Blockade of CCR4 in a humanized model of asthma reveals a critical role for DC-derived CCL17 and CCL22 in attracting Th2 cells and inducing airway inflammation. Allergy. 64:995–1002. 10.1111/j.1398-9995.2009.02095.x [DOI] [PubMed] [Google Scholar]
- Ray S.J., Franki S.N., Pierce R.H., Dimitrova S., Koteliansky V., Sprague A.G., Doherty P.C., de Fougerolles A.R., and Topham D.J.. 2004. The collagen binding alpha1beta1 integrin VLA-1 regulates CD8 T cell-mediated immune protection against heterologous influenza infection. Immunity. 20:167–179. 10.1016/s1074-7613(04)00021-4 [DOI] [PubMed] [Google Scholar]
- Richter M.V., and Topham D.J.. 2007. The alpha1beta1 integrin and TNF receptor II protect airway CD8+ effector T cells from apoptosis during influenza infection. J. Immunol. 179:5054–5063. 10.4049/jimmunol.179.8.5054 [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., and Masopust D.. 2014. Tissue-resident memory T cells. Immunity. 41:886–897. 10.1016/j.immuni.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., Fraser K.A., Vezys V., and Masopust D.. 2013. Sensing and alarm function of resident memory CD8+ T cells. Nat. Immunol. 14:509–513. 10.1038/ni.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea B.S., Brooks S.F., Fontaine B.A., Chun J., Luster A.D., and Tager A.M.. 2010. Prolonged exposure to sphingosine 1-phosphate receptor-1 agonists exacerbates vascular leak, fibrosis, and mortality after lung injury. Am. J. Respir. Cell Mol. Biol. 43:662–673. 10.1165/rcmb.2009-0345OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda K., Hirahara K., Iinuma T., Ichikawa T., Suzuki A.S., Sugaya K., Tumes D.J., Yamamoto H., Hara T., Tani-Ichi S., et al. 2016. Thy1+IL-7+ lymphatic endothelial cells in iBALT provide a survival niche for memory T-helper cells in allergic airway inflammation. Proc. Natl. Acad. Sci. USA. 113:E2842–E2851. 10.1073/pnas.1512600113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slütter B., Van Braeckel-Budimir N., Abboud G., Varga S.M., Salek-Ardakani S., and Harty J.T.. 2017. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2 eaag2031 10.1126/sciimmunol.aag2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolders J., Heutinck K.M., Fransen N.L., Remmerswaal E.B.M., Hombrink P., Ten Berge I.J.M., van Lier R.A.W., Huitinga I., and Hamann J.. 2018. Tissue-resident memory T cells populate the human brain. Nat. Commun. 9:4593 10.1038/s41467-018-07053-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelgrove R.J., Gregory L.G., Peiró T., Akthar S., Campbell G.A., Walker S.A., and Lloyd C.M.. 2014. Alternaria-derived serine protease activity drives IL-33-mediated asthma exacerbations. J. Allergy Clin. Immunol. 134:583–592.e6. 10.1016/j.jaci.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano J.B., Abajobir A.A., Abate K.H., Abera S.F., Agrawal A., Ahmed M.B., Aichour A.N., Aichour I., Eddine Aichour M.T., Alam K., et al. ; GBD 2015 Chronic Respiratory Disease Collaborators . 2017. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir. Med. 5:691–706. 10.1016/S2213-2600(17)30293-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary G., Olive A., Radovic-Moreno A.F., Gondek D., Alvarez D., Basto P.A., Perro M., Vrbanac V.D., Tager A.M., Shi J., et al. 2015. VACCINES. A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells. Science. 348 aaa8205 10.1126/science.aaa8205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt T.M., Dhume K., Finn C.M., Hwang J.H., Castonguay C., Swain S.L., and McKinstry K.K.. 2018. IL-15 supports the generation of protective lung-resident memory CD4 T cells. Mucosal Immunol. 11:668–680. 10.1038/mi.2017.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.A., Miron M., and Farber D.L.. 2019. Location, location, location: Tissue resident memory T cells in mice and humans. Sci. Immunol. 4:1–12. 10.1126/sciimmunol.aas9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura S., Yagi H., Hakata Y., Motozono C., McMaster S.R., Masumoto T., Fujisawa M., Chikaishi T., Komeda J., Itoh J., et al. 2016. Specific niches for lung-resident memory CD8+ T cells at the site of tissue regeneration enable CD69-independent maintenance. J. Exp. Med. 213:3057–3073. 10.1084/jem.20160938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbitt C.A., Stark J.M., Martens L., Ma J., Mold J.E., Deswarte K., Oliynyk G., Feng X., Lambrecht B.N., De Bleser P., et al. 2019. Single-Cell RNA Sequencing of the T Helper Cell Response to House Dust Mites Defines a Distinct Gene Expression Signature in Airway Th2 Cells. Immunity. 51:169–184.e5. 10.1016/j.immuni.2019.05.014 [DOI] [PubMed] [Google Scholar]
- Turner D.L., Goldklang M., Cvetkovski F., Paik D., Trischler J., Barahona J., Cao M., Dave R., Tanna N., D’Armiento J.M., et al. 2018. Biased Generation and In Situ Activation of Lung Tissue-Resident Memory CD4 T Cells in the Pathogenesis of Allergic Asthma. J. Immunol. 200:1561–1569. 10.4049/jimmunol.1700257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim L.M., Woodward-Davis A., Liu R., Hu Y., Villadangos J., Smyth G., and Bevan M.J.. 2012. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J. Immunol. 189:3462–3471. 10.4049/jimmunol.1201305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.A., and McKenzie A.N.J.. 2018. TH2 cell development and function. Nat. Rev. Immunol. 18:121–133. 10.1038/nri.2017.118 [DOI] [PubMed] [Google Scholar]
- Wein A.N., McMaster S.R., Takamura S., Dunbar P.R., Cartwright E.K., Hayward S.L., McManus D.T., Shimaoka T., Ueha S., Tsukui T., et al. 2019. CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J. Exp. Med. 216:2748–2762. 10.1084/jem.20181308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A.M., Eller K., Zeiser R., Dürr C., Gerlach U.V., Sixt M., Markut L., Gastl G., Rosenkranz A.R., and Wolf D.. 2009. The sphingosine 1-phosphate receptor agonist FTY720 potently inhibits regulatory T cell proliferation in vitro and in vivo. J. Immunol. 183:3751–3760. 10.4049/jimmunol.0901011 [DOI] [PubMed] [Google Scholar]
- Woodruff P.G., Modrek B., Choy D.F., Jia G., Abbas A.R., Ellwanger A., Koth L.L., Arron J.R., and Fahy J.V.. 2009. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am. J. Respir. Crit. Care Med. 180:388–395. 10.1164/rccm.200903-0392OC [DOI] [PMC free article] [PubMed] [Google Scholar]