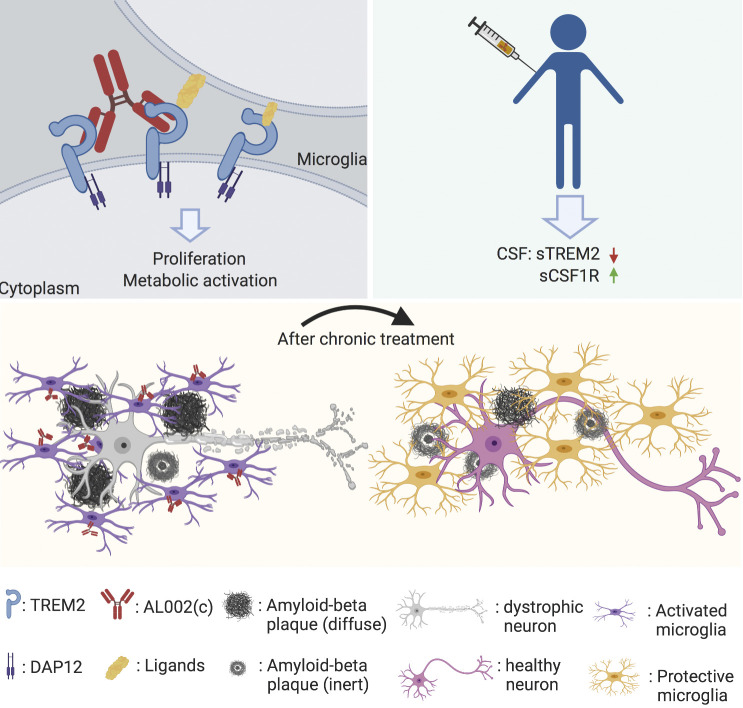
Wang et al. demonstrate that a mAb specific for the human microglial receptor TREM2 induces microglia proliferation, ameliorates Aβ-induced pathology in a mouse model of Alzheimer’s disease, and can be given safely in a first-in-human phase I clinical trial.
Abstract
TREM2 is a receptor for lipids expressed in microglia. The R47H variant of human TREM2 impairs ligand binding and increases Alzheimer’s disease (AD) risk. In mouse models of amyloid β (Aβ) accumulation, defective TREM2 function affects microglial response to Aβ plaques, exacerbating tissue damage, whereas TREM2 overexpression attenuates pathology. Thus, AD may benefit from TREM2 activation. Here, we examined the impact of an anti-human TREM2 agonistic mAb, AL002c, in a mouse AD model expressing either the common variant (CV) or the R47H variant of TREM2. Single-cell RNA-seq of microglia after acute systemic administration of AL002c showed induction of proliferation in both CV- and R47H-transgenic mice. Prolonged administration of AL002c reduced filamentous plaques and neurite dystrophy, impacted behavior, and tempered microglial inflammatory response. We further showed that a variant of AL002c is safe and well tolerated in a first-in-human phase I clinical trial and engages TREM2 based on cerebrospinal fluid biomarkers. We conclude that AL002 is a promising candidate for AD therapy.
Graphical Abstract
Introduction
Alzheimer’s disease (AD) is the most common form of senile dementia. It affects one in eight Americans over the age of 65 yr and is the sixth leading cause of death in the United States (https://www.alz.org/alzheimers-dementia/facts-figures). AD is characterized by memory and executive function deficits, followed by progressive, global cognitive decline (Long and Holtzman, 2019; Sarlus and Heneka, 2017). Brain AD pathology consists of extracellular aggregates of amyloid β (Aβ) oligomers and large insoluble plaques, intraneuronal tau hyperphosphorylation, synaptic dysfunction, and neuronal cell death (Long and Holtzman, 2019; Sarlus and Heneka, 2017). AD lesions trigger a secondary expansion of reactive microglia, which cluster around Aβ plaques, limiting their spreading (Long and Holtzman, 2019; Sarlus and Heneka, 2017). Profiling of microglia transcriptome in mouse models of Aβ accumulation has revealed that this increase in microglia numbers is associated with a robust transcriptional activation signature on a per-microglia basis, which has been referred to as disease-associated microglia (DAM), which is quite distinct from that of homeostatic microglia (Keren-Shaul et al., 2017). Recently, the analysis of the human microglial transcriptome in AD by single-nucleus RNA sequencing (RNA-seq) has revealed a microglial transcriptional response that in part recapitulates the mouse DAM signature (Mathys et al., 2019; Zhou et al., 2020).
Studies of genetic risk for sporadic AD have suggested that microglia not only respond to disease but also modulate disease course (Karch and Goate, 2015; Lambert et al., 2013). Most notably, a hypomorphic missense mutation in the microglia receptor TREM2, R47H, increases the risk of AD severalfold, as do other TREM2 variants, such as R62H, although with reduced penetrance (Jonsson et al., 2013; Guerreiro et al., 2013). TREM2 is a lipid receptor expressed in microglia and other tissue macrophages, which promotes their survival and proliferation by transmitting intracellular activating signals through the adaptor DAP12. Impaired TREM2 function in the 5XFAD mouse model of Aβ pathology restricts the ability of microglia to proliferate and accumulate around Aβ plaques to limit their pathogenic potential (Wang et al., 2015). TREM2-deficient microglia can acquire an incomplete DAM profile, or stage 1 DAM, but fail to develop a completely activated profile, or stage 2 DAM (Keren-Shaul et al., 2017). This defective microglial response leads to greater neuritic dystrophy adjacent to Aβ plaques (Yuan et al., 2016; Wang et al., 2016). The beneficial role of TREM2-dependent microglial activation has been further supported by an in vivo study showing that 5XFAD mice develop less Aβ pathology when crossed to transgenic (Tg) mice overexpressing human TREM2 (hTREM2; Lee et al., 2018). Moreover, a recent study showed that myeloid cells with potentially beneficial effects on neurodegeneration can be generated in vitro with an agonist TREM2 antibody (Cheng et al., 2018). Taken together, these findings suggest that TREM2-dependent microglial activation can delay AD onset and/or progression.
In this study, we examined the potential therapeutic impact of a mouse anti-hTREM2 agonistic mAb named AL002c, which is a variant of a mAb, called AL002, that has recently been studied in a phase I clinical trial (https://clinicaltrials.gov/ct2/show/NCT03635047). The antibody was tested in Tg mice that express either the common variant (CV) or the R47H variant of hTREM2, but lack the endogenous Trem2 gene (referred to as CV knockout [CV-KO] and R47H-KO, respectively; Song et al., 2018). We had previously shown that 5XFAD mice crossed to CV-KO (CV-KO-5XFAD) show more microglia activation and plaque coverage than 5XFAD mice crossed to R47H-KO mice (R47H-KO-5XFAD). We found that a single injection of AL002c expanded unique subpopulations of metabolically active and proliferating microglia in both CV-KO-5XFAD and R47H-KO-5XFAD mice, as assessed by single-cell RNA-seq (scRNA-seq). Moreover, prolonged treatment of both mouse models with AL002c curtailed the formation of filamentous neurotoxic plaques, as well as the extent of neurite dystrophy, and improved behavior. Attenuated pathology was associated with reduced activation of microglia in response to Aβ plaques, as indicated by diminished clustering around plaques and reduced expression of the inflammation marker SPP1. Finally, we showed that the AL002 version of AL002c can be given safely in a first-in-human phase I clinical trial and identified soluble TREM2 (sTREM2) and soluble CSF-1R (sCSF-1R) as biomarkers indicative of TREM2 engagement in humans. We conclude that antibody-mediated activation of TREM2 in microglia can provide a potential avenue of therapeutic intervention capable of attenuating the neurotoxic impact of Aβ pathology.
Results
AL002c activates hTREM2 in vitro and in vivo
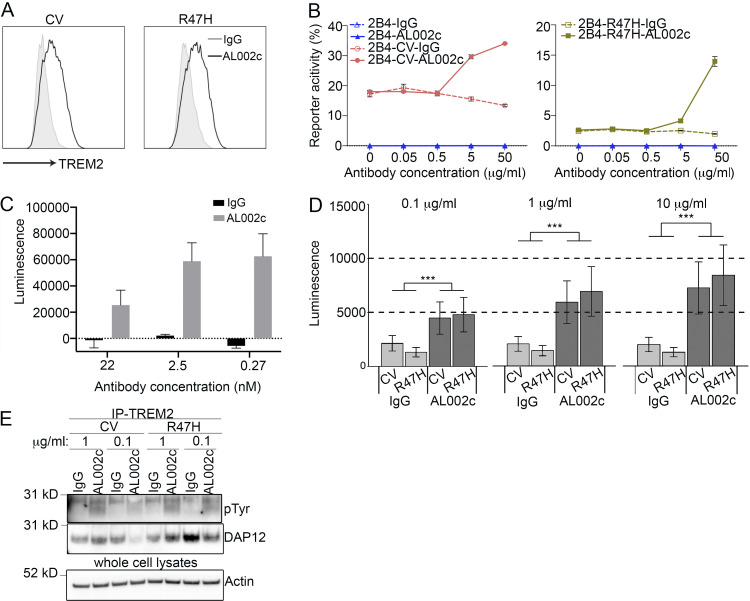
AL002c is a mouse IgG1 anti-hTREM2 mAb that was generated using the recombinant hTREM2 extracellular domain as an immunogen in a hybridoma approach, followed by humanization and affinity maturation by yeast display. In most experiments, we used AL002c with the intact Fc region, as mouse IgG1 has poor effector functions (Bruhns and Jönsson, 2015) and therefore is unlikely to markedly impact the behavior of the antibody. To validate the specificity of AL002c, we stained bone marrow–derived macrophages (BMMs) prepared from CV- and R47H-KO mice. We corroborated that AL002c binds hTREM2 and demonstrated that binding is not affected by the R47H mutation (Fig. 1 A). We further established by peptide epitope mapping that AL002c binds a region of the extracellular domain of TREM2 spanning amino acids 112–174 (not depicted).
Figure 1.
AL002c is a TREM2 agonist. (A) CV- and R47H-derived BMM were cultured for 7 d, harvested, and stained with AL002c (black solid line histograms) or isotype control (gray histograms). This experiment is representative of three independent experiments. (B) 2B4 reporter cell lines expressing hTREM2 CV or hTREM2 R47H were stimulated with grading doses of control IgG or AL002c in solution. EGFP expression was measured by flow cytometry. Data are representative of two independent experiments as mean ± SEM. (C) Human macrophages were cultured for 7 d and stimulated for 48 h with grading doses of IgG or AL002c in solution. Viability was measured by CellTiter Glow. Data are representative of 10 independent experiments as mean ± SD. (D) BMM derived from CV-KO and R47H-KO were cultured for 7 d and stimulated for 96 h with grading doses of IgG or AL002c in solution. Viability was measured by CellTiter Glow. ***, P < 0.001 by two-way ANOVA. Data are presented as mean ± SEM. (E) BMM derived from CV-KO and R47H-KO were stimulated for 15 min with two different concentrations of control IgG or AL002c. Cells were then lysed, immunoprecipitated (IP) with a distinct anti-TREM2 antibody, blotted with anti-phosphotyrosine antibody, stripped, and blotted with anti-DAP12 antibody. For control loading, whole-cell lysates were blotted with anti-actin antibody.
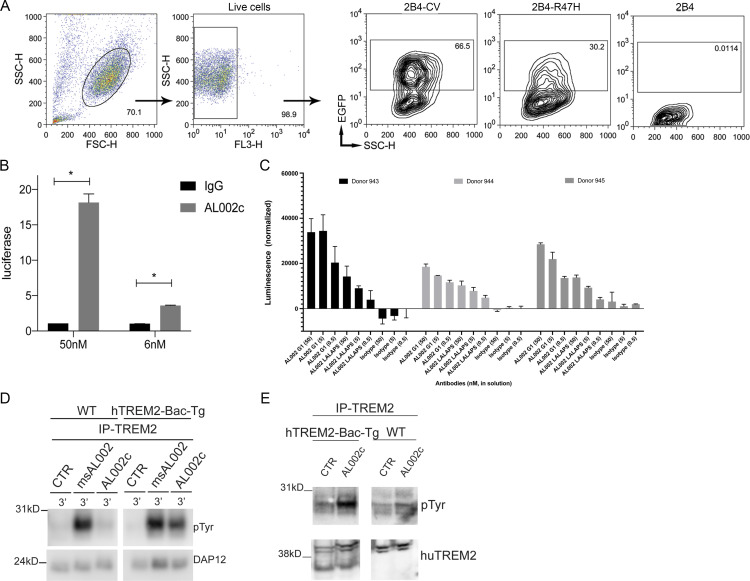
To see whether AL002c activates hTREM2, we next tested the Ca2+-driven reporter cell line 2B4 stably transfected with either hTREM2 CV or R47H together with DAP12 (Fig. S1 A; Song et al., 2017). In these reporter cells, engagement of hTREM2 promotes Ca2+ signals that lead to nuclear translocation of NFAT and NFAT-driven synthesis of enhanced GFP (EGFP), which is detected by flow cytometry. AL002c induced EGFP in both CV and R47H reporter cells, while a control IgG did not, suggesting that AL002c acts as an hTREM2 agonist (Fig. 1 B). Similar results were obtained with another CV-expressing Ca2+-driven reporter cell in which Ca2+ signals induce luciferase rather than EGFP (Fig. S1 B).
Figure S1.
AL002c activates hTREM2 signaling and function in vitro. (A) Parental, hTREM2-CV, and -R47H 2B4 reporter cell lines were stimulated with plate-bound phosphatidylcholine. Live cells were gated and tested for EGFP expression. Stimulation of hTREM2-CV, and to a minor extent hTREM2-R47H, induced EGFP. SSC, side scatter; FSC, forward scatter. (B) BW5.147G14 cells expressing hTREM2 (CV) were stimulated with two concentrations of AL002c or control IgG in medium for 6 h, and luciferase expression was measured by adding OneGlo reagent. *, P < 0.05 by unpaired two-tailed t-test. Data are presented as mean ± SEM. (C) Cell viability assay of hTREM2+ macrophages treated with different concentrations of soluble AL002c with intact Fc and mutated Fc (AL002 LALAPS). Data are presented as mean ± SD. (D) WT and hTREM2-Bac-Tg BMM were stimulated for 3 min with control IgG (CTR), anti-mouse TREM2 mAb msAL002, or anti-hTREM2 mAb AL002c, followed by cross-linking with a secondary antibody. Cells were then lysed and immunoprecipitated (IP) with a distinct anti-TREM2 antibody and blotted with anti-phosphotyrosine antibody. For control loading, the membrane was stripped and probed with anti-DAP12 antibody. (E) WT or hTREM2-Bac-Tg mice were injected at day 0 with 3% thioglycolate. At day 3, mice were injected with the AL002c or CTR antibodies. 24 h later, peritoneal cells were collected, immunoprecipitated with anti-TREM2 antibody, and blotted with anti-phosphotyrosine antibody. For control loading, whole-cell lysates were blotted with anti-hTREM2 antibody.
Previously, we demonstrated that TREM2 cooperates with CSF-1R, sustaining in vitro survival of BMMs after CSF-1 withdrawal (Otero et al., 2009; Wang et al., 2015; Ulland et al., 2017). We also showed that hTREM2 CV sustains macrophage viability more effectively than hTREM2 R47H (Song et al., 2018). Thus, we tested whether AL002c impacts in vitro survival of macrophages after CSF-1 withdrawal. Human monocyte-derived macrophages treated with AL002c were more viable than those treated with control IgG after 48 h of CSF-1 withdrawal, as measured by an ATP level–based viability assay (Fig. 1 C). Interestingly, mutations of the Fc region of AL002c that block binding to Fc receptors and complement (also known as LALAPS) only slightly quelled its prosurvival effect on myeloid cells (Fig. S1 C), suggesting that AL002c-mediated activation of hTREM2 function is largely independent of mAb-mediated cross-linking of Fc receptors. Finally, AL002c but not control IgG sustained cell viability of BMM derived from CV-KO and R47H-KO mice (Fig. 1 D), demonstrating that AL002c activates both hTREM2 variants.
To further corroborate that AL002c is an agonistic antibody, we assessed its capacity to activate DAP12 signaling in macrophages. In a first set of experiments, we generated BMM from CV-KO and R47H-KO mice and stimulated them in vitro with two different doses of AL002c or a control IgG, followed by immunoprecipitation with a distinct anti-TREM2 antibody. Analysis of the immunoprecipitates with an anti-phosphotyrosine antibody showed that AL002c induced phosphorylation of a protein with a molecular weight corresponding to DAP12, whereas the control IgG did not (Fig. 1 E). Anti-DAP12 immunoblot confirmed that TREM2 associates with DAP12 regardless of phosphorylation.
We conducted two additional sets of experiments using another Tg mouse that expresses both hTREM2 CV and endogenous mouse Trem2 (referred to as hTREM2-Bac-Tg). First, we demonstrated that AL002c induced DAP12 phosphorylation in BMM prepared from hTREM2-Bac-Tg but not in BMM from WT non-Tg B6 mice (Fig. S1 D), corroborating that AL002c selectively activates hTREM2 but not mouse Trem2 signaling. Second, we investigated whether AL002c activates DAP12 signaling in vivo in peritoneal macrophages elicited by i.p. injection of thioglycolate. After induction of peritoneal macrophages in hTREM2-Bac-Tg and WT mice, AL002c or a control IgG were directly injected in the peritoneal cavity to stimulate macrophages in vivo. Peritoneal cells were recovered, lysed, and analyzed by immunoprecipitation with anti-TREM2 and immunoblotting with anti-phosphotyrosine. AL002c but not control IgG induced DAP12 phosphorylation in peritoneal macrophages from hTREM2-Bac-Tg mice (Fig. S1 E). Altogether, these results demonstrate that AL002c functions as a TREM2 agonist for both hTREM2 CV and R47H in vitro and in vivo.
Systemically administered AL002 can cross the blood–brain barrier and attain concentrations that activate TREM2
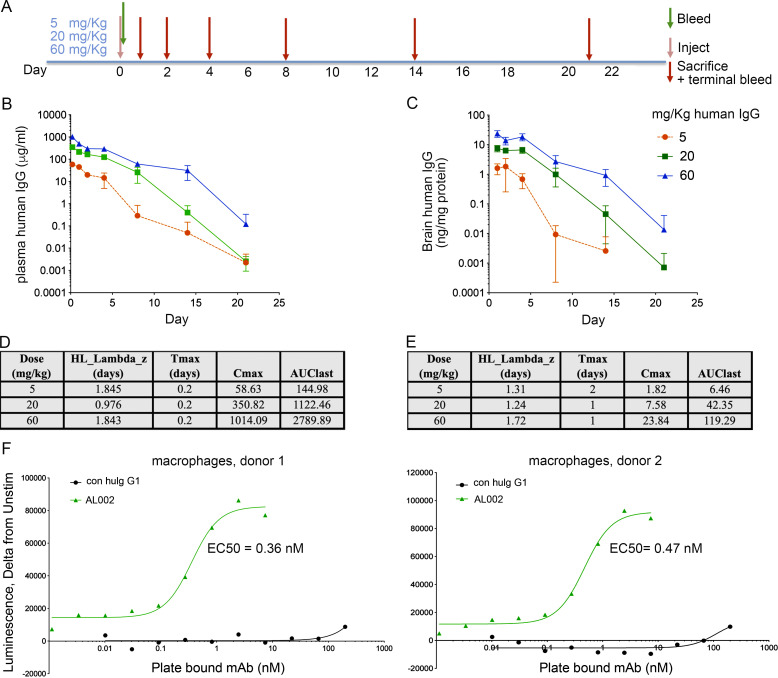
To design a dosing regimen that ensures therapeutic efficacy in the 5XFAD model, we performed a pharmacokinetic study. This determination was important, because peripherally administered mAbs cross the blood–brain barrier poorly, and hence mAb concentration is expected to be much lower in the brain than in plasma. For this experiment, we used the clinical variant of AL002c, called AL002, which is on a human IgG1 backbone, facilitating detection in mouse brain lysates and plasma using anti-human IgG. We chose i.p. injection as a route of administration. A single dose of AL002 was administered i.p. at 5, 20, and 60 mg/kg into hTREM2-Bac-Tg mice on day 0 (Fig. S2 A). Plasma and brain homogenate samples were collected for measurement of antibody levels from cohorts of mice on days 1, 2, 4, 8, 14, and 21. The concentration of human IgG in the brain was ∼0.11% of that in plasma at all time points in all mice (Fig. S2, B and C). The terminal half-life in hTREM2-Bac-Tg mice was relatively short, ranging from 1 to 1.8 d (Fig. S2, D and E). Given that hTREM2 is overexpressed in these mice, the short half-life may be due to enhanced target-mediated disposition in this model.
Figure S2.
AL002 pharmacokinetics. (A) Timeline of pharmacokinetics (PK) study of AL002 in TREM2-Bac-Tg mice. A single dose of AL002 was administered on day 0 at 5, 20, and 60 mg/kg. Different mouse cohorts were sacrificed on days 1, 2, 4, 8, 14, and 21, and plasma and brain samples were collected for measurement of antibody levels. (B) AL002 (human IgG) levels in plasma. (C) AL002 (human IgG) brain concentration in ng/ml was converted to ng/mg protein assuming a brain volume of 0.5 ml and a protein content of 38 mg. Using this calculation, we observed that ∼0.11% of AL002 penetrated in brain across all time points and animals. The terminal half-life in TREM2-Bac-Tg mice ranged from 1 to 1.8 d. Data are presented as mean ± SD. (D and E) PK parameters for plasma and brain were calculated using WinNonlin. n = 4 for each time point. AUClast, area under the curve concentration; Cmax, maximum plasma concentration; HL_Lambda_z (days), half-life; Tmax (days), time to maximum plasma concentration. (F) Functional EC50 of AL002. Viability of primary hTREM2+ macrophages from two distinct donors was measured by ATP assay after exposure of cells to different concentrations of AL002. The EC50 value of AL002 spans 0.36–0.47 nM (∼60 ng/ml).
Using the viability assay described in Fig. 1 C, we also determined the concentration of AL002 that confers half-maximal viability (EC50) of monocyte-derived macrophages in vitro (Fig. S2 F). By this approach, we found that the EC50 for AL002 is between 0.36 and 0.47 nM. Furthermore, comparison of AL002 concentrations in the brain with the calculated EC50 indicated that the concentration of AL002 in the brain is above the EC50 at all three doses on day 2 after administration. Therefore, we chose a dose of 30 mg/kg for subsequent in vivo experiments.
AL002c drives metabolic activation and proliferation of microglia in vivo
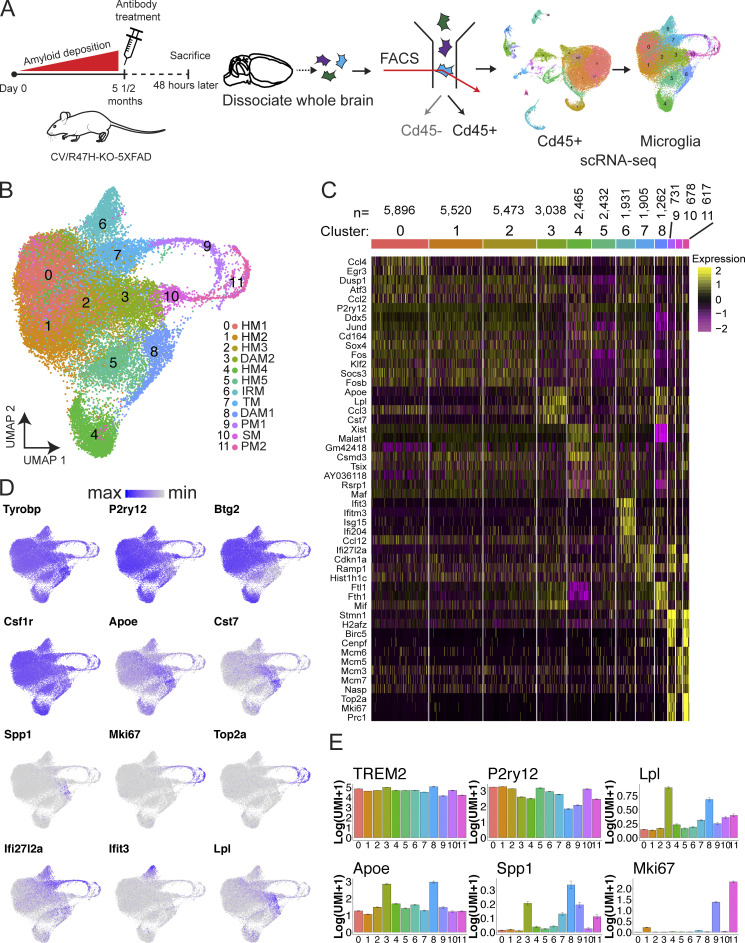
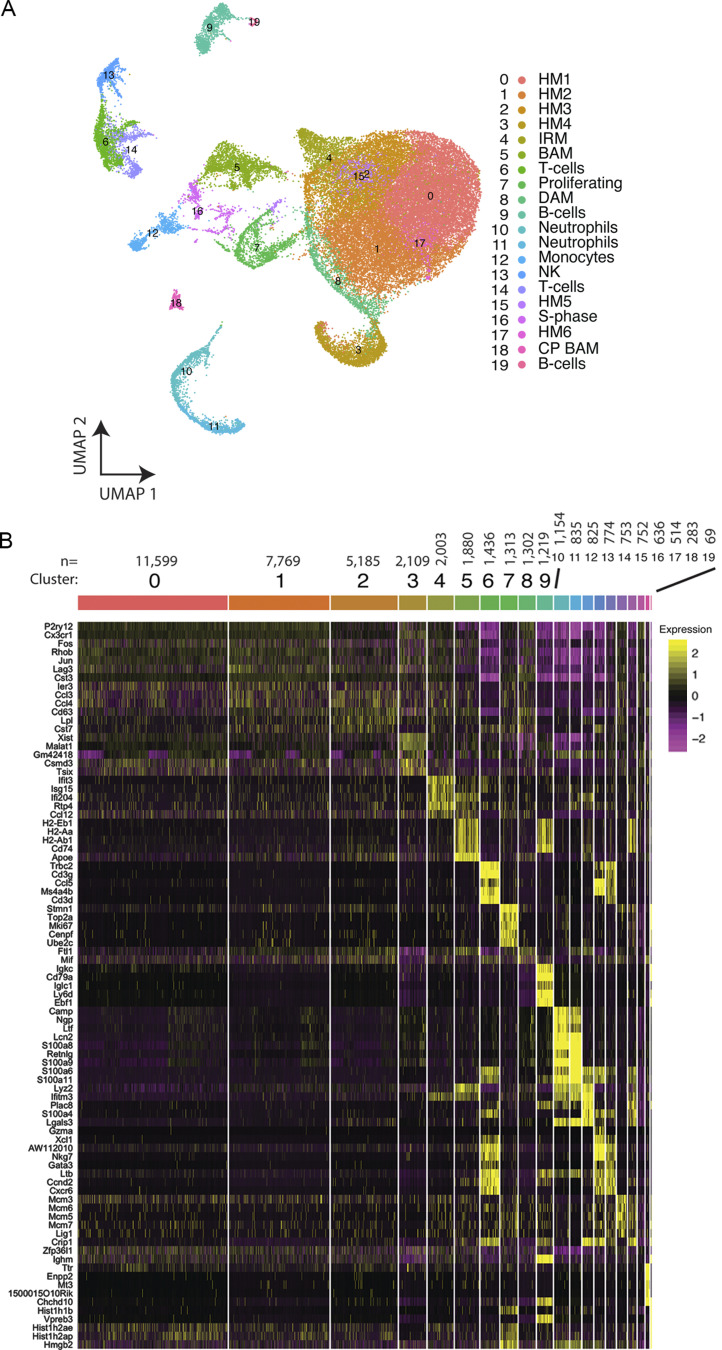
To characterize the impact of AL002c on microglial activation in vivo, we performed scRNA-seq. CV-KO-5XFAD and R47H-KO-5XFAD mice were injected with either AL002c or isotype control. After 48 h, Cd45+ cells were isolated from whole brains, and RNA transcripts from single cells were barcoded, reverse transcribed, and sequenced using the 10X Genomics platform (Fig. 2 A). Most cells had similar numbers of unique molecular identifiers (UMIs), or distinct RNA molecules (not depicted). A total of 42,409 single cells were plotted on uniform manifold approximation projection (UMAP) dimensions for visualization. Unsupervised clustering revealed a total of 12 broad cell types comprising 20 distinct clusters across all mice (Fig. S3 A). These clusters were annotated based on differential expression of known cell type–specific marker genes (Fig. S3 B). The preponderance of cells captured by our scRNA-seq dataset were microglia (clusters 0, 1, 2, 3, 4, 8, 15, and 17; ∼75% of all Cd45+ cells), with significantly lower numbers of border-associated macrophages (BAMs; clusters 5 and 18), T cells (clusters 6 and 14), B cells (clusters 9 and 19), neutrophils (clusters 10 and 11), natural killer cells (cluster 13), monocytes (cluster 12), S-phase cells (cluster 16), and proliferating G2-/M-phase cells (cluster 7; Fig. S3 B). Compared with control IgG, AL002c did not have a significant impact on any of the clusters representing nonmicroglial cell types.
Figure 2.
scRNA-seq analysis of microglia in treated mice. (A) Schematic of the scRNA-seq experiment. (B) UMAP of microglia subclustered from all Cd45+ cells. HM1-5, homeostatic microglia; TM, transitioning microglia; IRM, interferon-response microglia; PM1-2, proliferating microglia; SM, S-phase microglia. (C) Gene expression heatmap for clusters defined as microglia. Genes shown are the top gene markers for each cluster defined by Wilcoxon rank-sum test. The number of cells per cluster is denoted above the cluster label. (D) Mean normalized expression of key signature genes across clusters. Error bars represent SEM from all cells within the cluster. (E) Normalized expression of key signature genes on UMAP.
Figure S3.
scRNA-seq of Cd45+ cells from brains of AL002c- and IgG-treated mice. (A) UMAP of all Cd45+ cells. Clusters were annotated using previously identified gene markers: HM, homeostatic microglia; IRM, interferon-response microglia; NK, natural killer cells; CP BAM, choroid plexus BAMs. (B) Gene expression heatmap for all Cd45+ clusters. Genes shown are the top gene markers for each cluster defined by a Wilcoxon rank-sum test. The number of cells per cluster is denoted above the cluster label.
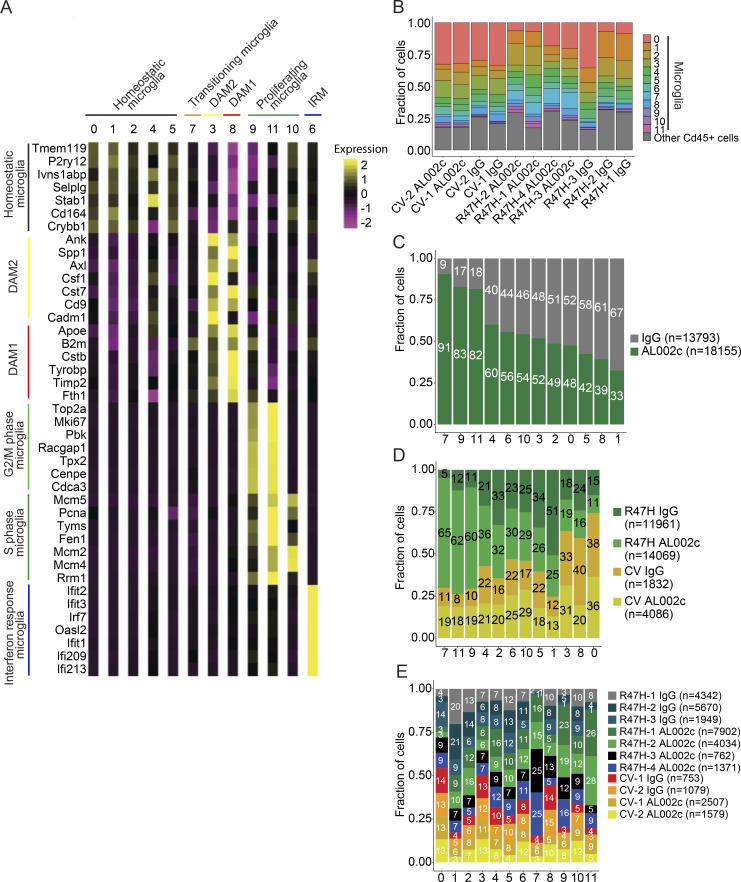
Taking advantage of the single-cell resolution of our data, we reclustered 31,948 microglia cells into 12 clusters (Fig. 2, B and C). Based on expression of published microglia subtype-specific marker genes (Sala Frigerio et al., 2019; Keren-Shaul et al., 2017; Mathys et al., 2017), we identified homeostatic microglia (clusters 0, 1, 2, 4, and 5; HM1-5), stage 1 DAM (cluster 8; DAM1), stage 2 DAM (cluster 3; DAM2), and microglia with intermediate features between homeostatic and DAM, which we define as transitioning microglia (cluster 7; Fig. 2, D and E; and Fig. 3 A). We further identified three clusters of proliferating microglia in the G2/M phase (clusters 9 and 11; PM1-2) and S-phase (cluster 10; SM), as well as one cluster expressing IFN-induced genes, which we define as IFN-response microglia (cluster 6; Fig. 2, D and E; and Fig. 3 A). There was a residual cluster of nonmicroglial cells carried over in this analysis, which was removed and no longer considered in subsequent analyses.
Figure 3.
Impact of antibody treatment and TREM2 variants on microglia clusters. (A) Comparison of genes’ mean cluster z-scores from microglial clusters (x axis) to gene signatures (y axis) taken from Sala Frigerio et al. (2019) and Keren-Shaul et al. (2017). (B) Fraction of all cells belonging to the microglia clusters or other Cd45+ cells. (C) Fraction of cells in each cluster for each antibody treatment (adjusted for number of cells belonging to each mouse). (D) Identical to C except for combined genotype and antibody categories. (E) Identical to C except for mice.
We next compared the microglia clusters obtained from mice in each experimental category (Fig. 3, B–E). Microglia represented >70% of the Cd45+ cells from all mice analyzed compared with other myeloid and lymphocytic cells (Fig. 3 B). More microglia in clusters 7 (transitioning) and 9 (G2/M phase) were present from mice injected with AL002c than from mice injected with the control antibody (Fig. 3, C–E), whereas fewer microglia were present in cluster 1 (homeostatic). These results suggested that injection of AL002c expands microglia transitioning from homeostatic to DAM and proliferating microglia, at the expense of homeostatic microglia.
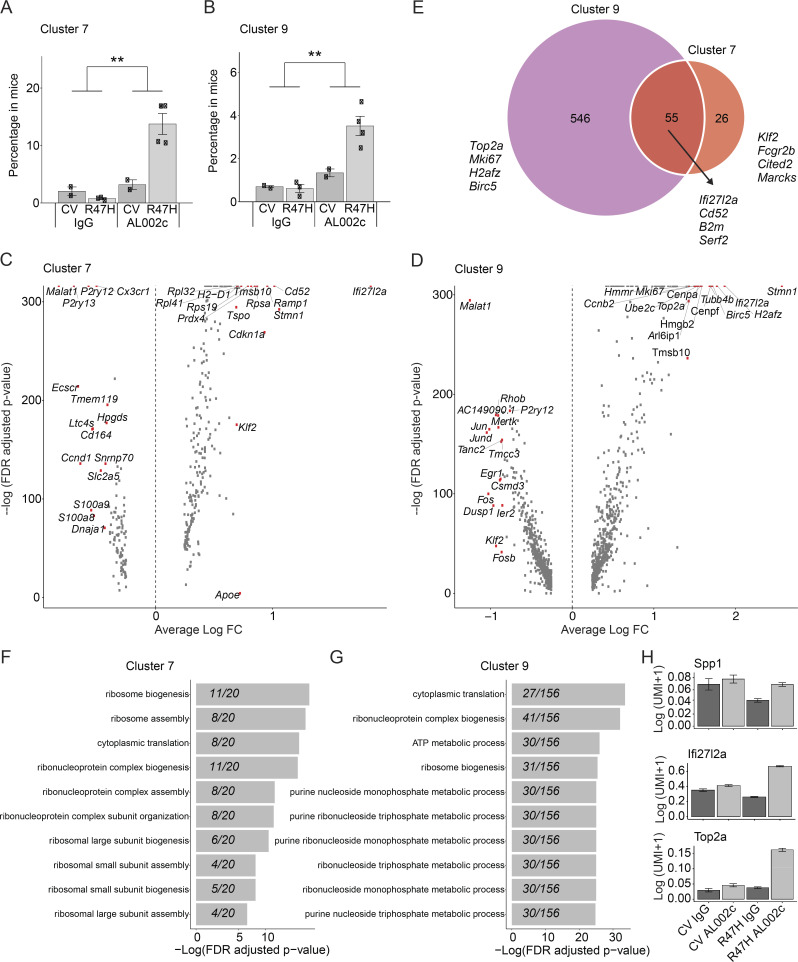
Further examination of clusters 7 and 9 corroborated that they were significantly enriched in AL002c-treated mice (Fig. 4, A and B). Interestingly, microglia were more activated and proliferative in response to AL002c in R47H mice than CV mice. We speculate that microglia expressing CV are already activated by endogenous ligands in the steady state and hence are less responsive to AL002c. Analysis of differentially expressed genes (DEGs) in clusters 7 and 1 (homeostatic microglia) showed that cluster 7 was enriched for genes encoding ribosomal proteins, indicating metabolic activation (Fig. 4 C). Similar comparison of cluster 9 versus all other clusters revealed that cluster 9 was enriched in cell cycle genes, indicating a high proliferation rate (Fig. 4 D). Clusters 7 and 9 shared 55 up-regulated DEGs (Fig. 4 E), many related to cell activation, whereas homeostatic microglia genes, such as P2ry12, were down-regulated in both clusters. Gene ontology (GO) term enrichment of biological processes supported these observations (Fig. 4, F and G). Microglia from AL002c-treated mice showed overall up-regulation of markers of activation and proliferation (Top2a, Ifi27l2a, and Spp1) across all clusters compared with microglia of control-treated mice (Fig. 4 H). We conclude that acute engagement of hTREM2 on microglia by AL002c promotes a signature of metabolic activation and proliferation distinct from that of DAM.
Figure 4.
AL002c expands metabolically active and proliferating microglia. (A and B) Bar plot of the percentage of cells in cluster 7 (**, P = 6.4 × 10−3; A) and cluster 9 (**, P = 3.4 × 10−3; B) on a per-mouse basis. Data are presented as mean ± SEM. (C) Wilcoxon rank-sum tests comparing cluster 7 to cluster 1 (homeostatic microglia). Genes above the −log(FDR adjusted P values) cutoff of 100 are annotated. (D) Identical to C except comparing cluster 9 to all other clusters. FC, fold-change. (E) Venn diagram showing the number of shared DEGs found with Wilcoxon rank-sum test for cluster 7 and cluster 9. Cluster 7’s DEGs were computed by comparing cluster 7 to cluster 1. Cluster 9’s DEGs were computed by comparing cluster 9 to all other clusters. (F and G) GO term analysis for positive fold-change DEGs in cluster 7 and cluster 9. (H) Mean normalized gene expression grouped by TREM2 variant and antibody treatment in all microglia.
Sustained AL002c treatment has a negligible impact on Aβ load in 5XFAD mice expressing hTREM2
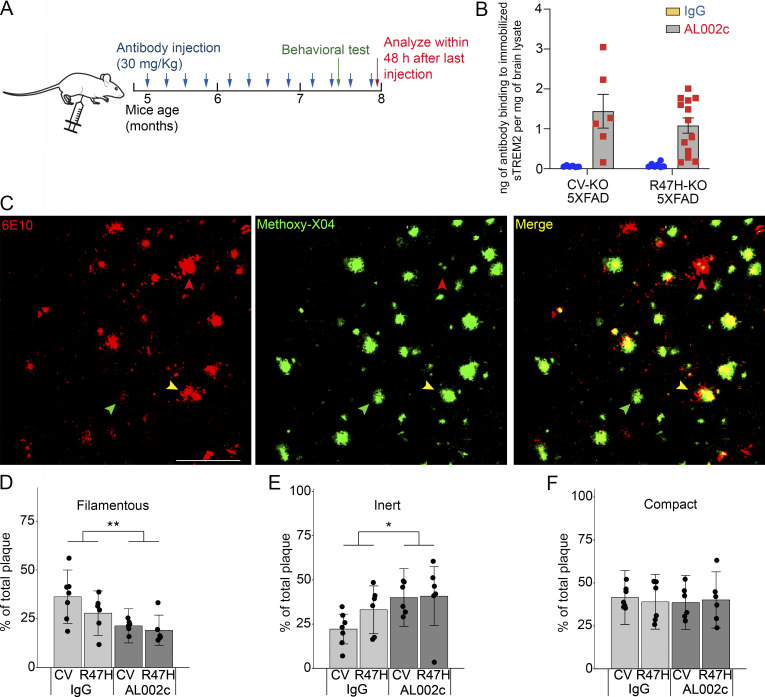
We next investigated the potential therapeutic impact of AL002c on Aβ load in the 5XFAD model of Aβ aggregation. In this model, plaques become clearly detectable at 3–4 mo of age and continue to grow for several months (Oakley et al., 2006). We crossed 5XFAD mice with CV-KO and R47H-KO to generate CV-KO-5XFAD and R47H-KO-5XFAD mice, respectively. These mice were treated with weekly i.p. injections of AL002c or a control IgG (30 mg/kg). To mimic a therapeutic treatment, we initiated injections at 5 mo of age, when mice have already developed Aβ aggregation, and continued until 8 mo of age, when Aβ plaques are still growing (Fig. 5 A). The presence of effective AL002c concentrations was verified in the cerebellum at the end of the experiment (Fig. 5 B).
Figure 5.
AL002c reduces soluble Aβ load and modifies the conformation of Aβ plaques. (A) Timeline of AL002c treatment in CV-KO-5XFAD or R47H-KO-5XFAD mice. 5-mo-old mice were injected i.p. with AL002c at 30 mg/kg weekly for 12 wk. Littermates were administered the same concentration of IgG as a control group. The behavioral test was performed after the 10th injection. Brains were harvested within 48 h after the last antibody injection. (B) AL002c concentrations in the cerebellum at the end of chronic treatment. To capture AL002c, brain lysates from AL002c- or IgG-treated mice were incubated with sTREM2 immobilized on plastic, followed by a secondary antibody. Only brain lysates containing AL002c but not control IgG test positive in this assay. (C) Representative confocal images from the cortex of CV-KO-5XFAD mice treated with AL002c stained with 6E10 (red) and methoxy-X04 (green), displaying three different forms of Aβ plaques. Red, green, and yellow arrowheads indicate filamentous, inert, and compact plaques, respectively. Bar = 100 µm. (D–F) Percentages of filamentous, inert, and compact plaques in each treatment group of mice. A total of 1,604 plaques were analyzed. CV-KO-5XFAD, AL002c, n = 6; CV-KO-5XFAD, IgG, n = 7; R47H-KO-5XFAD, AL002c, n = 6; R47H-KO-5XFAD, IgG, n = 6. Each symbol represents data of one mouse. *, P < 0.05; **, P < 0.01 by two-way ANOVA. Although the overall effect of AL002c was significant across the CV and R47H genotypes in D and E, the AL002c effect was greater in the CV than the R47H genotype. Accordingly, in pairwise comparisons, AL002c treatment was significant in the CV but not the R47H genotype (P = 0.0066 and 0.1002 for D; P = 0.00257 and 0.3275 for E; from pairwise comparison of the fitted least square mean from the linear model). No significant interaction between antibody treatment and genotype was found. Data are presented as mean ± SEM and depict one experiment representative of three independent experiments.
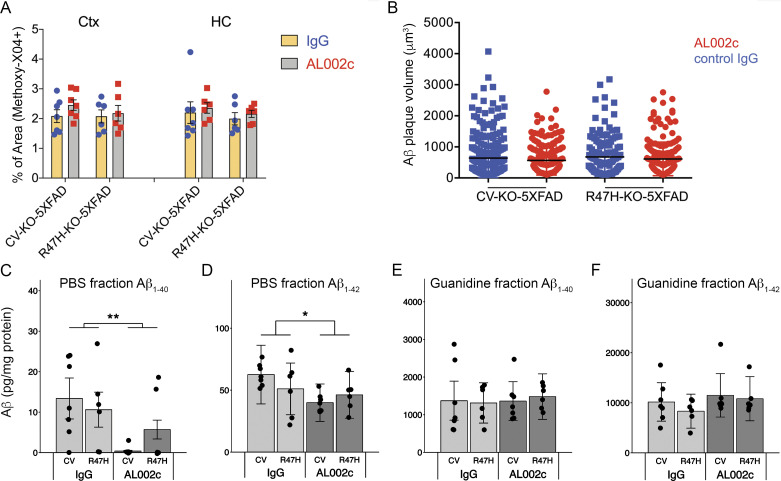
To determine the impact of AL002c on total Aβ plaque load, we stained matched coronal brain sections with methoxy-X04 and measured total fibrillar plaque area in the cortices and hippocampi by confocal fluorescence microscopy. We found no differences in the total brain area covered by methoxy-X04 between AL002c- and control IgG-treated CV-KO-5XFAD and R47H-KO-5XFAD mice (Fig. S4 A). Additionally, the overall distribution of plaques size was similar whether mice were treated with AL002c or control IgG (Fig. S4 B).
Figure S4.
Impact of AL002c treatment on total Aβ load. (A) Quantification of methoxy-X04+ area coverage in the cortex (Ctx) and hippocampus (HC) of treated mice. Each CV-KO-5XFAD treatment group, n = 7; each R47H-KO-5XFAD treatment group, n = 6. (B) Volume of individual methoxy-X04+ Aβ plaques in the cortex of indicated treatment groups. The volume of ∼1,000 plaques per condition was determined by Imaris. (C–F) PBS-soluble and PBS-insoluble guanidine-soluble fractions of hippocampus were assessed for Aβ1–40 (C and E) and Aβ1–42 (D and F) by MSD. Values for PBS-soluble Aβ1–40 were below the limit of detection in some samples. Each CV-KO-5XFAD treatment group, n = 7; each R47H-KO-5XFAD treatment group, n = 6. No differences were detected for Aβ1–40 or Aβ1–42 in PBS-insoluble guanidine-soluble fractions of cortices among treatment groups (not depicted). PBS-soluble fractions of cortices showed no detectable Aβ1–40 or Aβ1–42 by MSD. *, P < 0.05; **, P < 0.01 by two-way ANOVA. Data are presented as mean ± SEM.
We next determined levels of Aβ1–40 and Aβ1–42 in the soluble and insoluble fractions of hippocampus. Flash-frozen tissue was homogenized sequentially in PBS and guanidine solutions to obtain PBS-soluble and -insoluble fractions. Aβ1–40 and Aβ1–42 levels were measured in both fractions by the Meso Scale Discovery (MSD) approach. Hippocampal regions of AL002c-treated mice had slightly lower levels of soluble Aβ1–40 and Aβ1–42 compared with control IgG-treated mice, whereas levels of insoluble Aβ1–40 and Aβ1–42 were similar (Fig. S4, C–F). Cortices of AL002c- and control-treated mice had similar levels of insoluble Aβ1–40 and Aβ1–42 and no detectable soluble Aβ1–40 and Aβ1–42 by MSD (not depicted). We conclude that the impact of AL002c on soluble and insoluble Aβ load is small or negligible.
AL002c treatment reduces filamentous plaques and neurite dystrophy
Because TREM2 sustains the formation of a microglia barrier that promotes amyloid compaction and insulation and prevents neurite dystrophy (Yuan et al., 2016; Wang et al., 2016), we examined the impact of AL002c on amyloid compaction and neurite dystrophy. Plaques were detected by staining brain sections with mAb 6E10, which identifies Aβ aggregates whether they have β-sheet or non–β-sheet conformations. Aβ aggregates with a β-sheet conformation were further identified by staining with methoxy-X04. Using this immunofluorescence approach, we distinguished three types of plaques (Fig. 5 C). Filamentous plaques consist of Aβ aggregates with little β-sheet conformation (6E10+, methoxy-X04lo/−) and branched amyloid fibrils that protrude into the brain parenchyma, causing severe axonal swelling. Inert plaques comprise Aβ aggregates that form a compact β-sheet core with no Aβ radiating into the surrounding parenchyma (6E10−, compact methoxy-X04+ core). Compact plaques also have a compact β-sheet core but exhibit non–β-sheet protrusions (6E10+, compact methoxy-X04+ core), which are indicative of moderate neurotoxicity. In comparison to control treatment, administration of AL002c induced an overall significant reduction of filamentous plaques across the CV and R47H genotypes, which was paralleled by an increase of inert plaques, whereas the number of compact plaques was similar in the two groups (Fig. 5, D–F). In pairwise comparisons, the effect of AL002c on filamentous and inert plaques was more significant in the CV than the R47H genotype, suggesting that the ability of TREM2 to bind ligands, which is impaired in the R47H variant, may impact the effectiveness of AL002c treatment.
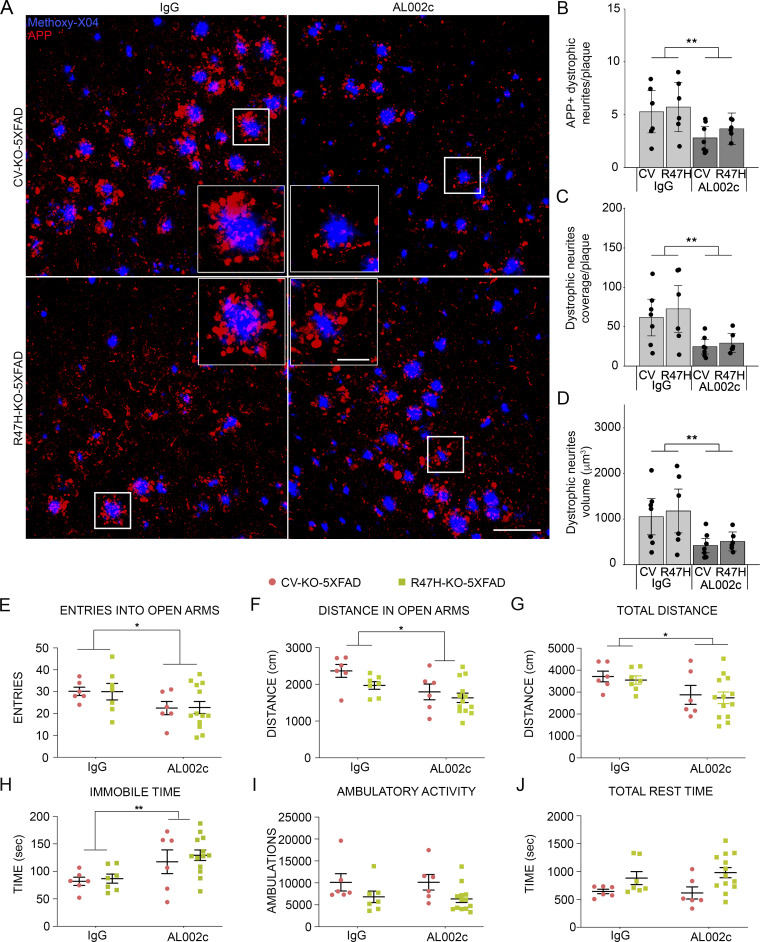
Given the differential neurotoxicity of filamentous, inert and compact plaques, we next examined whether changes in plaque profiles induced by AL002c impacted neuronal damage. We stained brain sections for the N terminus of amyloid precursor protein (APP), which accumulates in dystrophic neurites, and measured N-terminal APP+ neuronal processes within 0–30-µm spherical shells surrounding Aβ plaques (Fig. 6 A). We found that the number, volume, and coverage of dystrophic neurites per plaque were significantly decreased in mice treated with AL002c versus control IgG across the CV and R47H genotypes (Fig. 6, B–D). In pairwise comparisons, the effect of AL002c on volume and coverage of dystrophic neurites was also significant for each genotype. Thus, AL002c reduces the formation of “neuritic” plaques and the associated induction of swollen, degenerating axons and dendrites.
Figure 6.
AL002c ameliorates neuronal damage and impacts anxiety-like traits. (A) Representative confocal images from the cortex of indicated groups of mice, showing methoxy-X04-labeled Aβ plaques (blue) and surrounding N terminus-APP+ neurites (red). Bars = 100 µm; insets, 25 µm. (B–D) Quantification of number (B), coverage (C), and volume (D) of dystrophic neurites per plaque in the indicated groups of mice. Each CV-KO-5XFAD treatment group, n = 7; each R47H-KO-5XFAD treatment group, n = 6. The effect of AL002c was significant across CV and R47H genotypes: **, P < 0.01 by two-way ANOVA. In C and D, the effect of AL002c was also independently significant in the CV and R47H genotypes in pairwise comparisons (P = 0.0293 and 0.019 for C; P = 0.0371 and 0.0409 for D; from pairwise comparison of the fitted least square mean from the linear model). (E–H) Anxiety-like traits in mice behavior were evaluated by EPM and are presented as total entries into open arms (E), distance in open arms (F), total distance (G), and total immobile time (H). (I and J) AL002c treatment does not affect general locomotor activities. Total ambulations and time at rest were quantified over a 60-min period in transparent enclosures. CV-KO-5XFAD, AL002c, n = 6; CV-KO-5XFAD, IgG, n = 6; R47H-KO-5XFAD, AL002c, n = 13; R47H-KO-5XFAD, IgG, n = 7. Each symbol represents data of one mouse. *, P < 0.05; **, P < 0.01 by two-way ANOVA. No significant interaction between antibody treatment and genotype was found. Data are presented as mean ± SEM.
Mice treated with AL002c exhibit changes in exploratory drive and anxiety-like traits
We next examined whether the reduction of Aβ pathology induced by AL002c treatment was associated with behavioral changes. It has been established that 5XFAD mice have an abnormal behavior in the Elevated Plus Maze (EPM) test compared with WT mice (Braun and Feinstein, 2019; Hüttenrauch et al., 2017). EPM is designed to assess risk-taking behavior, exploratory drive, and anxiety-like traits by giving mice free access to a maze that contains both open unprotected arms and enclosed protected arms, all elevated ∼1 m from the floor. In previous studies, 5XFAD mice were shown to explore the open anxiogenic areas more than WT mice, demonstrating that Aβ pathology is associated with increased risk-taking behavior and exploratory drive, whereas anxiety-like behavior is subdued (Braun and Feinstein, 2019; Hüttenrauch et al., 2017). Therefore, we chose the EPM to explore the behavioral impact of AL002c.
When CV-KO-5XFAD and R47H-KO-5XFAD mice were tested in EPM, the number of entries and the distance traveled in open arms were significantly reduced in AL002c-treated mice compared with control IgG-treated mice (Fig. 6, E–G). In contrast, the time of immobility was increased (Fig. 6 H), indicating decreased exploration of the maze. In control tests of locomotor activity, antibody treatment did not affect general exploratory behavior or locomotion, suggesting that the observed differences were specific to the EPM (Fig. 6, I and J). We conclude that AL002c treatment restores fear of heights and open spaces and mitigates risk-taking behavior and exploratory drive in CV-KO-5XFAD and R47H-KO-5XFAD mice.
Treatment with AL002c impacts microglia response to Aβ
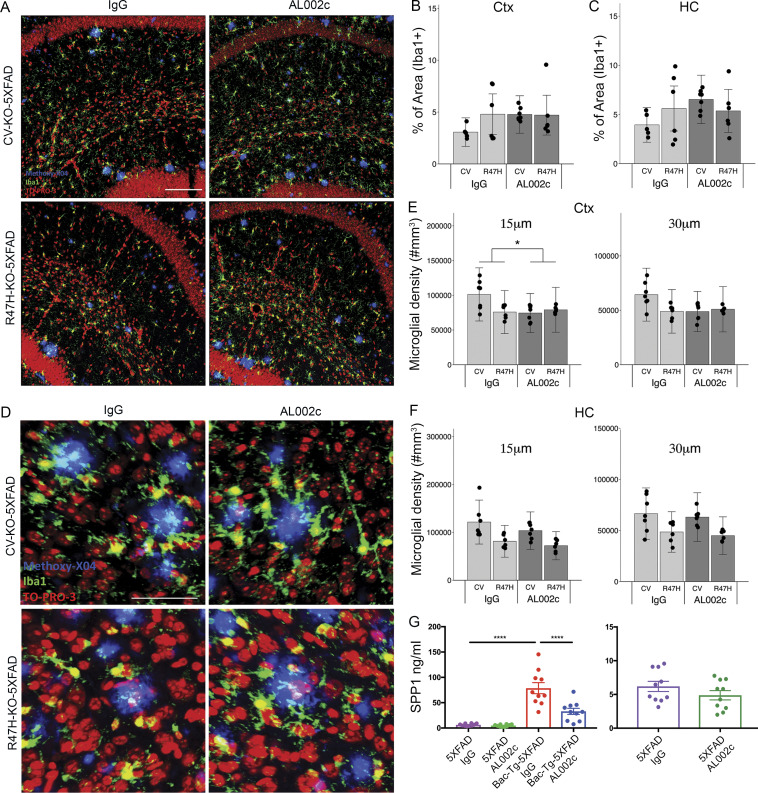
We next directly examined the impact of AL002c on microglia. We first performed immunofluorescence analysis of cortices and hippocampi by staining for the microglial marker Iba1. Quantification of total Iba1+ area throughout the brains of CV-KO-5XFAD mice and R47H-KO-5XFAD mice revealed no significant difference whether mice were treated with AL002c or control IgG (Fig. S5, A–C). We next measured the microglia clusters within 0–15-µm and 0–30-µm spherical shells surrounding Aβ plaques by positioning Iba1+ microglia and methoxy-X04+ plaques in a semiautomated workflow in Imaris and Matlab. As we reported previously (Song et al., 2018), CV-KO-5XFAD showed more microglia coverage of Aβ plaques than R47H-KO-5XFAD mice (Fig. S5, D–F). Coverage was not increased by AL002c in the hippocampi of both strains, and was in fact slightly reduced in the cortex of CV-KO-5XFAD mice (Fig. S5, D–F).
Figure S5.
Impact of AL002c treatment on microglia clustering around plaques and Spp1 production (A) Representative confocal images from the hippocampus of indicated groups of mice, showing Iba1+ microglia (green), TO-PRO3-stained nuclei (red), and methoxy-X04 labeled Aβ plaques (blue). Bar, 100 µm. (B and C) Quantification of Iba1+ area coverage in the cortex (Ctx) and hippocampus (HC) of treated mice (CV-KO-5XFAD, AL002c, n = 7; CV-KO-5XFAD, IgG, n = 5; R47H-KO-5XFAD, AL002c, n = 6; R47H-KO-5XFAD, IgG, n = 6). Data are presented as mean ± SEM. (D) Representative confocal images from the cortex of the indicated groups of mice, showing Iba1+ (green) microglia, TO-PRO3-stained nuclei (red), and methoxy-X04 labeled Aβ plaques (blue). Bar = 50 µm. CV-KO-5XFAD treatment group, n = 7; R47H-KO-5XFAD treatment group, n = 6. (E and F) Quantification of the density of microglia within a 15- or 30-µm shell around plaque surfaces within cortex or hippocampus. The figure depicts one experiment representative of two biological replicates. *, P < 0.05 by two-way ANOVA, showing significant difference for interaction between antibody treatment and genotype. Data are presented as mean ± SEM. (G) 5-mo-old 5XFAD and hTREM2-Bac-Tg × 5XFAD mice were injected i.p. with AL002c or IgG as control (n = 10) at 60 mg/kg weekly for 12 wk. Littermates were administered the same concentration of IgG as a control group. SPP1 was measured by ELISA in brain lysates of treated mice. Each symbol represents data of one mouse. ****, P < 0.0001 by one-way ANOVA. Data are presented as mean ± SEM.
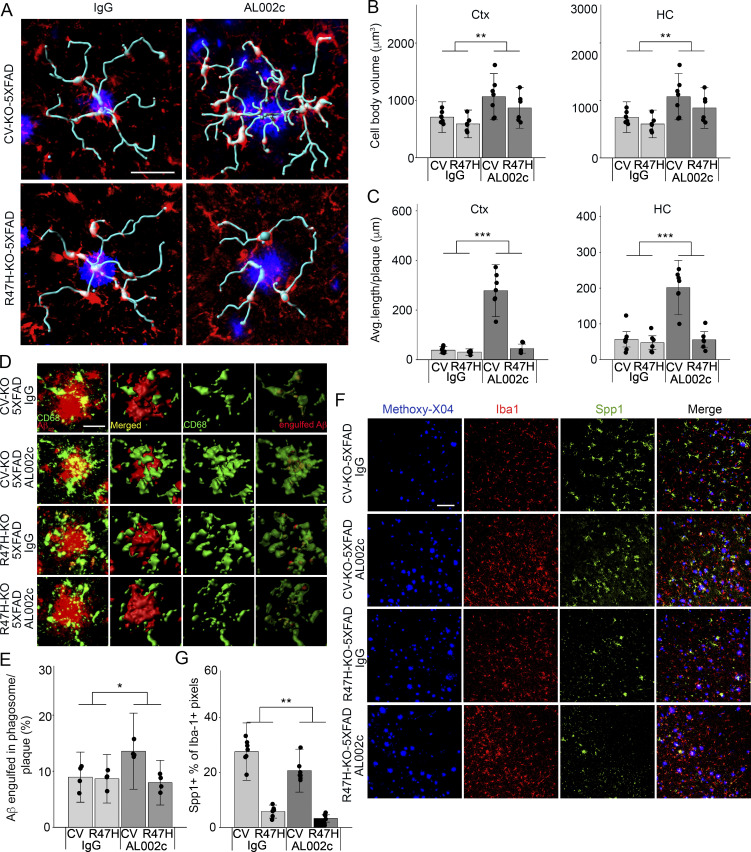
To further examine the impact of AL002c on microglia, we measured the cell body size and the total length of processes by which microglia monitor their surroundings (Davalos et al., 2005; Nimmerjahn et al., 2005). Interestingly, both parameters were increased in the cortices and hippocampi of AL002c-treated mice compared with control IgG-treated mice, suggesting increased surveillance activity (Fig. 7, A–C). Moreover, we stained microglia for CD68, a marker of activation and phagocytosis, as well as Aβ42. Three-dimensional reconstruction of Aβ42 and CD68 immunofluorescence demonstrated that AL002c treatment increased the ratio of Aβ42+CD68+ microglial phagosomes over total Aβ42, suggesting that anti-TREM2 promotes microglial activation and phagocytosis of Aβ (Fig. 7, D and E). Together, these data suggest that AL002c increases microglial surveillance and phagocytic activities, although no obvious increase in microglia clustering around plaques is detectable. Statistical analysis revealed an interaction between AL002c treatment and mouse genotypes, such that AL002c had a greater impact on the morphology and phagocytic capacity of CV-KO-5XFAD microglia than on R47H-KO-5XFAD microglia. As discussed above, the disparate impact of AL002c treatment may depend on the different ability of TREM2 variants to bind ligands.
Figure 7.
Impact of AL002c on microglial responses to Aβ. (A) Morphology of microglia associated with plaques (blue) highlighting the shape of cell bodies (red) and primary processes (cyan) in the cortex (Ctx) of indicated groups of mice. Bar = 20 µm. (B and C) Cell body volume (B) and average length of primary processes (C) of plaque-associated microglia were measured by Imaris using z-stack confocal images. Images of total 21 or 18 plaques with >180 microglia per treatment group were analyzed. Each CV-KO-5XFAD treatment group, n = 7; each R47H-KO-5XFAD treatment group, n = 6. **, P < 0.01; ***, P < 0.001 by two-way ANOVA. HC, hippocampus. (D) Representative confocal images by three-dimensional reconstruction of the cortex of indicated groups of mice, depicting engulfed Aβ (red) by CD68+ phagosome (green). Bar = 25 µm. (E) Quantification of the ratio of Aβ colocalized with CD68 over total Aβ volume. n = 4 for each treatment group. Each symbol represents data of one mouse. *, P < 0.05 by two-way ANOVA. A significant interaction was detected between antibody treatment and genotype in C and E, such that the effect of the antibody treatment is greater in the CV-KO-5XFAD than the R47H-KO-5XFAD genotype. Data are presented as mean ± SEM. (F) Representative confocal images of the cortex of the indicated groups of mice, depicting Aβ plaques (methoxy-X04, blue), Iba1 (red), and Spp1 (green). Bar = 100 µm. (G) Quantification of the percentage of Spp1+ pixels within Iba1+ pixels in the indicated genotypes of mice (CV-KO-5XFAD, AL002c, n = 7; CV-KO-5XFAD, IgG, n = 7; R47H-KO-5XFAD, AL002c, n = 6; R47H-KO-5XFAD, IgG, n = 6). Each symbol represents data of one mouse. **, P < 0.01 by two-way ANOVA. No interaction between antibody treatment and genotype was detected. Data are presented as mean ± SEM.
Finally, we stained brain sections for Iba1 and the inflammation marker Spp1. As we reported previously (Song et al., 2018), CV-KO-5XFAD showed more microglial expression of Spp1 than R47H-KO-5XFAD mice. Treatment with AL002c was associated with reduced expression of Spp1 within Iba1+ cells compared with control IgG treatment in both strains (Fig. 7, F and G). To corroborate this result, we measured soluble Spp1 in the brain lysates of hTREM2-Bac-Tg crossed with 5XFAD mice that had been chronically treated with AL002c or control IgG. AL002c reduced soluble Spp1 compared with control IgG (Fig. S5 G). These results suggest that AL002c-induced increase of microglial phagocytosis may be associated with reduced inflammatory activity.
AL002 is well tolerated in a first-in-human phase I clinical trial
Finally, we report the results from a first-in-human phase I trial of AL002, the clinical variant of AL002c. The main objectives of this phase I study (NCT03635047) were to assess the safety, tolerability, pharmacokinetics and pharmacodynamic effects of AL002. In the single ascending dose (SAD) stage of the phase I study, 56 healthy adult participants were sequentially enrolled in 10 cohorts (A–I and K) and received a single i.v. dose of AL002 ranging from 0.003 to 60 mg/kg. SAD cohorts A–C contained one participant on active drug per cohort, cohorts D–H consisted of eight participants per cohort (six active, two placebo), and cohort I included seven participants (six active, one placebo); in one open-label cohort (K), six participants were treated with active drug at 45 mg/kg. For cohorts F–K, lumbar punctures were performed before and 2 d after doses to obtain cerebrospinal fluid samples to measure sTREM2 and soluble CSF-1R (sCSF-1R) levels. All subjects were followed out to 12 wk after dosing. AL002 was generally safe and well tolerated, with no drug-related serious adverse events or dose-limiting toxicities up to the highest dose (Table 1).
Table 1. Adverse events in healthy volunteer cohorts A–K and I in the phase I clinical trial AL002-1.
| Adverse event | Single ascending dose | |||||||
|---|---|---|---|---|---|---|---|---|
| Cohorts A, B, C (0.003–0.2 mg/kg; n = 3) | Cohort D (0.6 mg/kg; n = 8) | Cohort E (2 mg/kg; n = 8) | Cohort F (6 mg/kg; n = 8) | Cohort G (15 mg/kg; n = 8) | Cohort H (30 mg/kg; n = 8) | Cohort K (45 mg/kg; n = 6) | Cohort I (60 mg/kg; n = 7) | |
| Any TEAE | 2 (66.7) | 4 (50.0) | 4 (50.0) | 7 (87.5) | 6 (75.0) | 6 (75.0) | 6 (100.0) | 6 (85.7) |
| Any treatment-related TEAE | 2 (66.7) | 2 (25.0) | 4 (50.0) | 4 (50.0) | 2 (25.0) | 5 (62.5) | 5 (83.3) | 6 (85.7) |
| Any SAE | 0 | 0 | 0 | 1 (12.5)a | 0 | 0 | 0 | 0 |
| Any treatment-related SAE | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Any TEAE leading to early disclosure | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Data are n (%). SAE, serious adverse event; TEAE, treatment-emergent adverse event.
Traumatic injury (unrelated).
sTREM2 and sCSF-1R are biomarkers of AL002 treatment
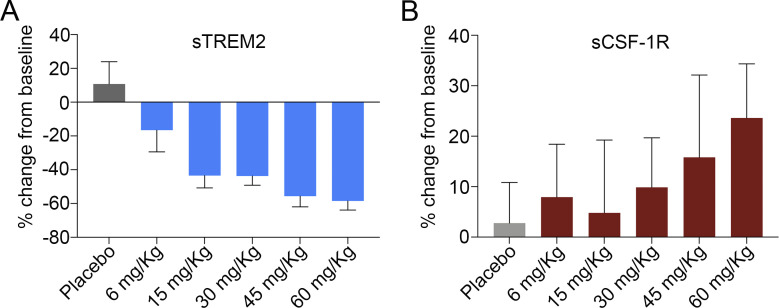
As part of the phase I study, we assessed the effect of AL002 on cerebrospinal fluid biomarkers. Administration of a single dose of AL002 caused a dose-dependent decrease in sTREM2 from baseline within 2 d that was paralleled by an increase in sCSF-1R (Fig. 8). sTREM2 is the product of proteolytic cleavage of cell-surface TREM2 (Piccio et al., 2008; Wunderlich et al., 2013; Schlepckow et al., 2017; Feuerbach et al., 2017). A decrease in cerebrospinal fluid sTREM2 may indicate that AL002 interferes with proteolytic shedding, by blocking the cleavage site; however, it may also induce the internalization of TREM2, thereby reducing surface TREM2 available for cleavage. Whether a reduction of sTREM2 in the brain contributes to the beneficial effect of the antibody remains to be determined. sCSF-1R is the cleavage product of transmembrane CSF-1R, which is only expressed by microglia in the brain. The increase of sCSF-1R may indirectly reflect the increased proliferation of microglia documented in our mouse model by scRNA-seq. Whether sCSF-1R acts a decoy receptor that regulates microglia expansion during AL002 treatment or has another function remains to be established.
Figure 8.
Biomarkers in first-in-human phase I clinical trial. (A and B) Administration of a single dose of AL002 caused a dose-dependent decrease in cerebrospinal fluid of sTREM2 (A) from baseline within 2 d that was paralleled by an increase in sCSF-1R concentration (B). P = 0.0001 for 6 mg/kg and P < 0.0001 for all other doses vs. pooled placebo control (A) and P = 0.026 at 60 mg/kg vs. pooled placebo (B) by mixed-model repeated measures analysis adjusted for multiplicities using Dunnett's test. Data are presented as mean ± SD; cohort n = 6 (placebo, 6 mg/kg, 15 mg/kg, 30 mg/kg) and 5 (45 mg/kg, 60 mg/kg).
Discussion
In this study, we found that a single injection of the hTREM2 agonistic mAb AL002c in the 5XFAD model of AD expressing the hTREM2 transgene acutely expanded microglial subsets that are metabolically active and proliferate. Moreover, prolonged systemic administration of AL002c reduced filamentous Aβ plaques and neurite damage. Subtle but significant changes in risk-taking and anxiety-like traits paralleled the reduced pathology. Immunofluorescence analyses of microglia showed signs of increased surveillance activity and Aβ phagocytosis, as well as reduced expression of the inflammatory marker Spp1. We conclude that mAb-mediated engagement of hTREM2 enhances neuroprotective microglia, restraining neurotoxic Aβ plaques and neuronal damage.
It has been thought that TREM2-dependent clustering of microglia around plaques is essential for containing Aβ plaques and associated neuropathology. In our study, mice treated with AL002c evinced neither a reduction of total Aβ load nor an increase in microglia clustering around Aβ plaques. AL002c-mediated activation of microglia may be insufficient to cope with the rapid accumulation of Aβ characteristic of the 5XFAD model. Nevertheless, AL002c was sufficient to reduce Aβ plaque toxicity and attenuate neuritic dystrophy, which is one of the important pathological features associated with AD and age-dependent neuronal dysfunction (Lombardo et al., 2003; Serrano-Pozo et al., 2010; Mucke and Selkoe, 2012). Microglia repeatedly activated by AL002c may act in part by removing oligomeric Aβ and Aβ seeds rather than plaques, as well as by clearing dystrophic neurons and producing neuroprotective factors. Attenuation of Aβ-induced pathology may also explain the observed diminution of microglial clustering and inflammation, which may be beneficial in this context. Consistent with our results, overexpression of hTREM2 in a mouse model of AD had beneficial effects on the disease in conjunction with diminished microglial clustering (Lee et al., 2018).
A very recent paper showed that injection of an agonistic anti-mouse TREM2 mAb at 3-d intervals for 10 d reduced the Aβ plaque load in the APP-NL-G-F knock-in model of AD (Schlepckow et al., 2020). Whereas we observed a clear effect of the anti-hTREM2 mAb AL002c on microglial proliferation, neurite dystrophy, and behavior, we did not detect any significant change in plaque load. Numerous factors could contribute to this discrepancy: different mAbs were administered according to disparate administration regimens, distinct mouse models of Aβ accumulation were used, and mice were analyzed at different ages. Clearly, more studies are warranted to compare various therapeutic regimens that entail different doses and/or frequencies of mAb administration, as well as disparate mouse models of AD pathology.
Our scRNA-seq analysis of microglia in AL002c-treated mice broadens our knowledge of microglia heterogeneity in the 5XFAD model by delineating a spectrum of microglial subsets that includes not only previously identified homeostatic, DAM, and IFN-responsive microglia, but also two novel subsets with distinct metabolic and proliferation profiles. These two clusters expanded after a single injection of anti-TREM2, whereas a homeostatic cluster tended to contract in parallel, suggesting that metabolically active and proliferating microglia derive from homeostatic microglia. scRNA-seq analysis at later time points of treatment will be necessary to see whether subsets that expand early after AL002c administration persist as such, convert into DAM in response to Aβ-induced tissue damage, or return into a homeostatic state when mAb activation terminates or Aβ pathology is attenuated.
In this study, we evaluated AL002c on both the CV and R47H variants of hTREM2 in vivo. Acute TREM2 stimulation had a more impressive impact on microglia proliferation in R47H than in CV mice; conversely, prolonged TREM2 stimulation had a more beneficial impact on Aβ-induced pathology in CV mice than in their R47H counterparts. Because CV is continuously bound to lipids, Aβ, and other ligands in vivo, whereas R47H is not, we speculate that R47H may be more responsive to mAb-mediated cross-linking than CV. In the long term, however, heightened responsiveness of R47H may lead to excessive activation and exhaustion. It is also possible that the long-term beneficial effects of mAb-activated microglia on brain pathology depend on the ability of TREM2 to effectively bind lipids, apoptotic cells, and Aβ.
Can anti-hTREM2 mAbs provide a valid therapeutic approach for AD? Our results show that AL002c treatment of 5XFAD mice was beneficial when Aβ aggregation was already ongoing and Aβ plaques formed, suggesting that anti-TREM2 antibodies may be helpful even if introduced at relatively late stages of disease. Future studies should further define the broad impact of AL002c treatment on memory and learning behaviors other than the subtle but significant impact on risk-taking and fear-conditioning reported here. Importantly, we reported preliminary data from the first-in-human phase I clinical trial with AL002, which indicates that AL002 administration was generally safe and well tolerated,and that its activity can be traced in the cerebrospinal fluid through two biomarkers, sTREM2 and sCSF-1R, which corroborate specific target engagement. Thus, results presented here overall support that anti-TREM2 antibodies, such as AL002, are promising candidates for AD therapy. This approach may have broader neuroprotective effects than plaque removal and may be a viable alternative to mAb treatments that remove plaques without inducing neuroprotection.
Materials and methods
Antibody generation
TREM2tm1(KOMP)Vlcg mice were immunized by ImmunoPrecise with hTREM2-Fc recombinant protein using standard procedures. Bleed titers were evaluated in in vitro assays, such as ELISA or FACS. Animals with a good immune response to the antigen were selected for fusion and given a final i.v. boost of antigen without adjuvant. Lymphocytes were isolated from the immunized animals and fused with mouse myeloma cells using polyethylene glycol (PEG 1500; Roche, 10783641001) according to the manufacturer’s instructions. Fused cells were plated into semisolid methylcellulose-based medium containing hypoxanthine, aminopterin, and thymidine for 10–12 d, allowing for single-step cloning and hybridoma selection. Single colonies were picked and transferred to 96-well plates containing culture medium with hypoxanthine-thymidine and grown for 4–5 d until mid-log-phase growth was reached. Supernatants were screened by ELISA for IgG production, isotype, and antigen specificity and by FACS for binding to native antigen on cells. Positive hybridoma clones were subcloned using a single-step cloning system to ensure monoclonality, and the subclone supernatants were rescreened by FACS to confirm specificity. Final subclones were expanded in culture, and the supernatants were purified by Protein-A affinity chromatography. Purified antibodies were tested in vitro for specificity, functional activity, binding affinity, and epitope binning. The parental clone of AL002c was selected for its ability to activate hTREM2.
Adimab yeast platform was used for humanization and affinity maturation. Briefly, for humanization of the lead antibody 9F5, Adimab generated 94 humanized versions of 9F5 by combining 21 huIgG1 versions of VH with 6 huIgG1 versions of VK, each containing 0–11 framework residue mutations. Antibodies were tested for binding to hTREM2 and agonistic activity, and the top two humanized versions were affinity matured. For affinity maturation, key amino acid residues in the heavy or light chain genes of the humanized parental clones were selectively mutagenized, and mutants that improved binding were selected through additional rounds of screening. Antibodies with improved affinity, based on Forte Bio and MSD measurements, were further screened for improvement in cell-based binding affinity and agonistic activity in a luciferase reporter assay. Finally, liability amino acids were replaced.
Mice
CV-KO-5XFAD and R47H-KO-5XFAD mice were generated as previously described (Song et al., 2018). Mice were housed in the animal facilities of Washington University in St. Louis. All animal experiments were conducted in compliance with institutional regulations, under authorized protocol #20160220 approved by the Institutional Animal Care and Use Committee. hTREM2-Bac-Tg mice were generated by using the bacterial artificial chromosome (BAC) clone CTD-3222A20 (Invitrogen/Life Technologies/Thermo Fisher Scientific). This BAC covered a contiguous region from nucleotide 41104901 to 41292419 (based on hg38 build of UCSC) spanning 187,519 nucleotides and harboring the complete sequences of the hTREM2, TREML2, TREM1, TREML1, and TREML4 genes. The purified BAC DNA was injected into mouse C57BL6/j zygotes by standard pronuclear injection techniques. Zygotes were returned to females, and the resulting pups were genotyped for the presence of the transgene. Founder animals harboring the transgene were then bred to nontransgenic animals, and progeny were screened for expression of the transgene using standard techniques.
Tissue preparation
BM was collected from femurs and tibias as described before (Ulland et al., 2017). To prepare BMM for antibody binding, survival, and immunoprecipitation assays, BM cells were counted and plated at 10 × 106 cells/100-mm Petri dish in RPMI supplemented with Glutamax, penicillin/streptomycin, nonessential amino acids, pyruvate, 10% heat-inactivated fetal bovine serum (complete RPMI) and 50 ng/ml CSF-1 (Peprotech). Cells were cultured for 7 d before use.
To perform scRNA-seq of the immune cells in brain, whole brains of 22-wk-old (5.5 mo) mice from AL002c- or control IgG-treated groups were dissected after transcardial perfusion with ice-cold PBS and dissociated as previously described (Song et al., 2018). Immune cells were recovered after Percoll (GE) separation, as described (Mildner et al., 2007).
To perform the biochemical analysis of brain after AL002c chronic treatment, brains were removed after transcardial perfusion with ice-cold PBS. Cortex and hippocampus were dissected out from left brain hemispheres and flash frozen; right brain hemispheres were fixed in 4% PFA for 2 d at 4°C and rinsed in PBS before incubation for 2 d at 4°C in 30% sucrose. Dehydrated brain tissues were frozen in a 2:1 mixture of 30% sucrose and optimal cutting temperature compound for obtaining serial 40-µm cryosections.
Antibody binding assay
BMM from CV-KO and R47H-KO were harvested at day 7 and stained in FACS buffer (2% FCS in PBS) with anti-hTREM2 AL002c-dylight 650 and isotype CTR-dylight 650. Human Fc-block (14-9161-73, eBioscience) was added to the mix. Dead cells were stained with live/dead staining kit (Invitrogen).
Survival assay
Human macrophages or BMM were harvested at day 7 of culture and plated in 96-well flat-bottom plates at 5 × 105/well in complete RPMI without CSF-1. Survival, measured as ATP content, was detected after 5-d culture with CellTiter-Glo luminescent viability assay (Promega).
EGFP reporter assay
2B4 NFAT:EGFP reporter cells expressing hTREM2-CV and 2B4-hTREM2-R47H have been described (Song et al., 2017). Phosphatidylcholine (Avanti, 840051P) was reconstituted in methanol at 100 ng/µl, and 50 µl was added to each well as a positive control. Plates were allowed to dry by evaporation at room temperature, leaving phosphatidylcholine coated on the well bottom. Next, stock solutions of AL002c or control IgG were diluted to the indicated concentrations in medium, and 50 µl of the resulting solution was added to distinct wells within the same plate coated with phosphatidylcholine. Each condition was performed in triplicate. After preparation of the plate, 50,000 cells in 50 µl of complete RPMI with 5% fetal bovine serum were added to the corresponding wells. After 24 h, the cells were transferred to FACS tubes and read on a FACSCalibur for EGFP expression.
Luciferase reporter assay
BW5.147G14 cells (ATCC) were infected with a retrovirus encoding hTREM2 and DAP12, as well as an NFAT:luciferase carrying virus (Qiagen). Cells were stimulated with two concentrations of AL002c or control IgG in solution for 6 h, and luciferase expression was measured by adding OneGlo reagent (Promega) according to manufacturer’s instructions. Luminescence was measured on the Biotek Synergy H1 plate reader.
Immunoprecipitation in vitro
Before stimulation, BMM were starved for 4 h in 1% serum RPMI. 10 × 106 cells were incubated on ice for 15 min with 1 µg of antibody per million of cells. Cells were then washed and incubated at 37°C for the indicated period of time in the presence of goat anti-mouse IgG (1.5 µg for 106 cells). After stimulation, cells were lysed with lysis buffer (1% n-dodecyl-β-d-maltoside, 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1.5 mM MgCl2, and 10% glycerol, plus protease and phosphatase inhibitors) and immunoprecipitated with anti-TREM2 (clone 237920, R&D Systems) or isotype control. Precipitated proteins were fractionated by SDS-PAGE in nonreducing conditions, transferred to PVDF membranes, and probed with anti-phosphotyrosine antibody (4G10, Millipore). TREM2 is not detected in nonreducing conditions. To confirm that each sample contained equal amounts of proteins, lysates were fractionated by SDS-PAGE in reducing conditions. Immunoblots were then stained with an anti-actin antibody (sc-47778, Santa Cruz) to allow protein loading quantification.
Immunoprecipitation in vivo
6–8-wk-old C57BL/6 mice or hTrem2-Bac-Tg mice were injected i.p. with 3 ml of 3% thioglycolate. After 3 d, when the peritoneal cavity was enriched with peritoneal macrophages (CD11b+F4/80+) expressing TREM2, mice were injected with control or AL002c (40 mg/kg) for 24 h. Peritoneal cells were recovered and immediately lysed in lysis buffer (n-dodecyl-β-d-maltoside 1%, 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1.5 mM MgCl2, and 10% glycerol, plus protease and phosphatase inhibitors) and immunoprecipitated with rat anti-human/mouse TREM2 (R&D Systems, clone 237920). Precipitated proteins were fractionated by SDS-PAGE (nonreducing conditions), transferred to nitrocellulose membranes, and probed with anti-phosphotyrosine antibody (4G10, Millipore). To confirm that all substrates were adequately immunoprecipitated, whole-cell lysates from each sample were separately fractionated by SDS-PAGE in reducing conditions and immunoblotted with goat anti-hTREM2 (R&D Systems).
scRNA-seq and computational analysis
Mice were reared for 5.5 mo. They were injected i.p. with AL002c or IgG at 30 mg/kg 48 h before sacrifice. Whole brains were perfused, dissociated, and sorted using FACS for Cd45+ cells. For AL002c treatment, we analyzed two CV-KO-5XFAD and four R47H-KO-5XFAD mice. For IgG treatment, we analyzed two CV-KO-5XFAD and three R47H-KO-5XFAD mice. scRNA-seq information has been deposited in the Gene Expression Omnibus database under accession number GSE150358.
scRNA-seq and read alignment
All Cd45+ cells were prepared via 10X Genomics v3 3′ Gene Expression Kit and sequenced on NovaSeq flow cells to achieve a read depth of 50,000 reads per cell and ∼2,000–8,000 cells per sample. Demultiplexed FASTQs were aligned to the mouse genome (mm10 build) using 10X Genomics’ CellRanger v3.0.2 with the command cellranger count with default parameters. Mouse-level matrices were concatenated to produce the raw input UMI count matrix. Cells were filtered out to remove low-viability and poorly sequenced cells. We removed cells that had <200 genes expressed and removed genes that were expressed in <3 cells, that were outside the boundary defined by the median ± 4 × absolute SD of either the number of UMIs or the number of genes detected, and that had >10% reads coming from mitochondrial genes. 42,409 Cd45+ cells remained after this filtering, with a median 7,363 UMIs per sample and average 3,855 cells per mouse. To obtain hTREM2 expression levels, FASTQs were aligned to the human genome (hg38 build). The resulting UMI counts were normalized and scaled using the mouse UMI matrix.
Normalization, dimensional reduction, and clustering
The Seurat R package (Butler et al., 2018) was used for all scRNA-seq analysis and processing. All steps were run using default parameters unless otherwise noted. Log normalization was performed on the UMI count matrix. The number of UMIs per cell and percentage of mitochondrial reads per cell were treated as undesired covariates and regressed out of the normalized expression values. The normalized expression values were scaled and centered for input into subsequent dimensional reductions of principal component analysis (PCA) and UMAP. PCA was performed using the top 2,000 variable genes identified using variance-stabilizing transformation. Cells were segmented based on their neighbors (FindNeighbors Seurat function) using the top 13 principal components (PCs) from PCA and using only the top 2,000 variable genes. The number of PCs used was based on the screen plot (i.e., a plot of the PC eigenvalues in decreasing order). Other parameters of FindNeighbors were k.param of 12. To find cell clusters (FindClusters Seurat function), a resolution of 0.4 was used. UMAP was computed using the top 13 PCs, and cells were plotted onto the first two UMAP dimensions.
Cell-type annotation
Cell types were manually classified based on well-known cell-type gene markers. To classify cell types that were not T cells, B cells, natural killer cells, or neutrophils, gene markers of distinct brain BAM subpopulations (Van Hove et al., 2019) were used. To correctly classify all Cd45+ clusters, Seurat’s AddModuleScore function was used to score each cell on each of the BAM gene signatures (Van Hove et al., 2019). Cluster 18 was identified as BAMs. Cluster 5 had a bimodal distribution of MHC class II genes that separated the population of cells into MHC class II high and low, but the granularity of clustering was not fine enough to detect these subclusters (not depicted). Cluster 12 was defined as monocytes, but we note that this cluster additionally exhibited a high dural BAM score. Proliferating cells had high expression of G2, M, and S phase genes (e.g., Top2a, Mki67, and Mcm5).
Subsetting of microglia
Based on the clustering of all Cd45+ cells, clusters 0, 1, 2, 3, 4, 7, 8, 15, and 17 were kept for finer analysis. These clusters were selected based on high microglial gene expression and low mitochondrial read percentage. Moreover, including these two clusters in further analysis did not yield any different conclusions. These clusters contained 31,948 cells and comprised the majority of Cd45+ cells (∼75%). Scaling and PCA were repeated as stated above. Cells were clustered again (k parameter 12, top 2,000 variable genes, and resolution 0.45). UMAP dimensional reduction was repeated with the top 25 PCs to help further visually separate clusters. Upon clustering this new subset of microglia, a small cluster of cells was defined by myeloid and lymphoid markers, indicating T cell and monocyte-like cells. These cells were removed for subsequent analysis.
Microglia classification and gene signature scoring
For homeostatic microglia, cycling/proliferating microglia, and IFN-response microglia gene signatures, we reran the analysis of the data from Chen et al. (2019) using only the AppNL-G-F versus C57BL/6J mice data to recover the same microglial states the authors did. We took the homeostatic microglia, cycling/proliferating microglia, and IFN-response microglia gene markers and scored them in our microglial cells to create a score for each cell. Scoring used the z-scores of that gene’s expression in all cells of a cluster. DAM1 and DAM2 genes were taken from the supplemental figures of Keren-Shaul et al. (2017). S-phase genes were taken from Tirosh et al. (2016).
Differential expression
Differential expression was performed using functions in the Seurat R package, namely FindAllMarkers and FindMarkers for cluster-defining gene markers and specific cluster comparisons, respectively. To calculate P values and average log fold-changes, the Wilcoxon rank-sum test was used in conjunction with false discovery rate (FDR) correction to adjust P values for multiple hypothesis testing. To identify cluster-defining gene markers, we compared one cluster to all other clusters. Genes that were expressed in 25% of the cells of a cluster and had absolute value >0.25 log fold-change were defined to be DEGs for that group. Cluster-defining genes were defined using this differential expression.
GO enrichment analysis
After deriving lists of DEGs for each cluster, we sought to discover which biological process these genes implicate. Using only positive expression fold-change DEGs that defined clusters, the clusterProfiler function in R was used to run hypergeometric tests to obtain probability significance levels. These P values were adjusted using FDR. We pulled out enriched genes and biological processes ontologies in which they were found. For plotting purposes, P values were transformed by applying −log base 10 to produce adjusted P values. Only the top 10 biological process hits were plotted, and the ratios of genes belonging to the GO term from the full list of DEGs were labeled.
Normalizing group-level proportions
Because of varying levels of the number of cells in each experimental group, a correction was implemented to adjust for the number of cells in each group when comparing group compositions within clusters. Group-level proportions within clusters were adjusted to account for the raw number of observations of each category in the group. For each group (e.g., cluster), we divided the observation size of each category (e.g., mouse #3) by fraction of cells in that category among all single cells. This gives a value representing the n-corrected observation size. We divided the n-corrected observation sizes by the n-corrected observation sizes summed across all categories. This is known as the n-corrected category percentage, shown in Fig. 3.
Immunofluorescence
Brain sections were blocked with PBS + 3% BSA and permeabilized with 0.25% Triton X-100 in blocking solution. Primary antibodies were added overnight at a dilution of 1:5,000 for Iba1 (rabbit polyclonal, Wako), 1:1,000 for CD68 (rat monoclonal, BioLegend), and 1:500 for Aβ42 (rabbit recombinant monoclonal, Thermo Fisher Scientific) or 1:1,000 for Aβ1–16 (6E10, BioLegend), 1:500 for Spp1 (goat polyclonal, R&D Systems), and 1:1,000 for APP (22C11 mouse mAb, Millipore) at 4°C. Secondary antibodies were added as follows: anti-goat IgG Alexa Fluor 488 (donkey polyclonal, 1:2,000; Abcam), anti-rabbit IgG Alexa Fluor 555 (donkey polyclonal, 1:1,000; Abcam), anti-rat IgG Alexa Fluor 555 (goat polyclonal, 1:1,000, Invitrogen), anti-goat IgG Alexa Fluor 647 (donkey polyclonal, 1:1,000; Abcam), anti-rabbit IgG Alexa Fluor 647 (goat recombinant polyclonal, 1:1,000; Invitrogen), anti-mouse IgG Alexa Fluor 647 (goat recombinant polyclonal, 1:2,000; Invitrogen) for 1.5 h at RT. Nuclei were counterstained with TO-PRO-3 iodide (300 nM; Thermo Fisher Scientific), and methoxy-X04 (3 µg/ml; Tocris) was used to label Aβ plaques. The confocal pictures were taken on a Nikon A1Rsi+ confocal laser-scanning microscope using a 20× 0.95-NA objective. z-Stacks with 1.1-µm steps in the z direction, 1,024 × 1,024-pixel resolution, were recorded. Three-dimensional reconstruction of microglia, plaques, and Aβ and dystrophic neurons and extraction of parameters were performed in Imaris v8.3 software (Bitplane), and further processing was analyzed using automated scripts in Matlab (MathWorks).
Image analyses
For measurement of distinct Aβ plaque morphology, 6E10, methoxy-X04, and merged z-stack images were exported from Imaris. The conformation of individual plaque in each treatment group of mice was determined as follows: filamentous, strong 6E10 staining without or less methoxy-X04 single; inert, only methoxy-X04+ or concomitant with little 6E10 staining; and compact, methoxy-X04+6E10+. The proportion of distinct plaque morphology was calculated by normalizing the total number of plaques from each z-stack image.
For measurement of methoxy-X04 area coverage, a similar region encompassing portions of cortex or hippocampus was drawn in ImageJ (National Institutes of Health) from individual z-stack image. By setting up a threshold in which a fixed intensity was much greater than the mean for the selected region, the percentage of area coverage was calculated automatically by batch processing in ImageJ. A similar procedure was performed for quantification of Iba1 coverage in the cortex and hippocampus, with different threshold settings. The Surfaces function of Imaris on methoxy-X04 images was used to calculate the volume of individual plaques.
For measurement of neurite dystrophy around plaques, the volumes of dystrophic neurites and plaque, as well as their centers of mass, were determined using the Surfaces function of Imaris on methoxy-X04 and APP image data. The number of dystrophic neurites within 30 µm of plaque surfaces was determined in Matlab as established before (Song et al., 2018). The density of microglia around plaques was calculated by a similar approach, in which the Spots function of Imaris was used to identify microglia within the Iba1 and ToPro-3 colocalized channel. The volume of microglial cell bodies was measured using the Surfaces function. Lengths of the microglia processes were determined by using the Filament function of Imaris as described before (Wang et al., 2015).
For visualization of Aβ engulfed by microglial CD68+ phagosome, the Surfaces function on Aβ+, CD68+ and Aβ+CD68+ image data was used to generate three dimensional reconstruction images. The ratio of engulfed Aβ by phagosome was calculated as the total Aβ+CD68+ volume divided by the total Aβ+ volume.
For measurement of Spp1+ pixels within Iba1+ pixels, the Spots function on z-stack image data was used to conduct the Iba1+ channel. The thresholds for Spp1+ within Iba1+ pixels were determined by visual inspection and used for all images. For each z-stack image, the percentages of Spp1+ pixels within Iba1+ pixels were calculated, and the ratio was calculated.
Aβ and Spp1 quantification
Cortical tissue and hippocampus were homogenized in PBS containing protease inhibitor (Roche) at 0.5× concentration, and remaining insoluble material was pelleted. Supernatant was collected as the PBS-soluble fraction, and the pellet was resuspended in 5.5 M guanidine and 50 mM Tris, pH 8.0, buffer, further homogenized, and incubated on a rotator for 3 h at room temperature. The samples were centrifuged again to pellet insoluble material, and the supernatant was collected as the PBS-insoluble guanidine fraction. Aβ1–40 and Aβ1–42 were measured by MSD. Spp1 was measured by ELISA (R&D Systems).
Behavioral tests
The behavioral performance of CV-KO-5XFAD and R47H-KO-5XFAD with control IgG or AL002c treatment was assessed in all mice at 8 mo of age. Locomotor activity was used to measure differences in general activity or emotionality between treatment groups. Anxiety-related behavior was assessed in the EPM similar to previously published methods (Wozniak et al., 2013; Yuede et al., 2009). All behavioral testing was conducted during the light cycle by a female experimenter blinded to experimental group. All equipment was cleaned with 2% chlorhexidine diacetate or 70% ethanol between animals. The order of tests conducted was locomotor activity and then EPM.
Open field activity/exploratory behavior
General activity levels and exploratory behavior were quantified over a 60-min period in transparent enclosures (47.6 × 25.4 × 20.6 cm) constructed of Plexiglas and surrounded by computerized photobeam instrumentation (Kinder Scientific). General activity variables (total ambulations, rearings, and time at rest) along with measures of emotionality, including time spent, distance traveled, and entries made into the central zone, were analyzed.
EPM
EPM was conducted 1 d after locomotor activity assessment by methods similar to those previously described (Wozniak et al., 2013). Briefly, animals were placed on a dimly lit black acrylic surface measuring 5 × 5 cm and elevated 63 cm above the floor, equipped with photo beam pairs. Four arms (35 cm long, 5 cm wide; two open and two with 15-cm-high walls) extended from a central area. MotorMonitor software (Kinder Scientific) quantified movement as duration, distance traveled, entries, and time at rest in each zone (closed arms, open arms, and center area).
Statistical analyses
All statistical analyses were performed using GraphPad Prism software (v8). Before analyses, all data were screened for missing values, fit between distributions and assumptions of univariate analysis, and homogeneity of variance. ANOVA, including repeated measures, was used to analyze the behavioral data. With a statistically significant interaction between main factors, simple main effects were calculated to provide clarification of statistically significant differences with regard to treatment. Where appropriate, the Greenhouse–Geisser correction was used to protect against violations of sphericity in repeated-measures analyses, and Bonferroni correction was applied to multiple pairwise comparisons and post hoc tests. Probability value for all analyses was P < 0.05, unless otherwise stated.
SAD part of the phase I study
56 healthy adult participants sequentially enrolled in 10 cohorts (A–I, K) received a single i.v. dose of AL002 ranging from 0.003 to 60 mg/kg. SAD cohorts A–C contained one participant on active drug per cohort; cohorts D–H consisted of eight participants per cohort (six active, two placebo); and cohort I included seven participants (six active, one placebo). In one open-label cohort (K), six participants were treated with active drug at 45 mg/kg. For cohorts F–K, lumbar punctures were performed before and 2 d after doses to obtain cerebrospinal fluid samples. All subjects were followed to 12 wk after dosing.
sTREM2 assay in human cerebrospinal fluid
An immunoassay method was used for the determination of sTREM2 in human cerebrospinal fluid based on an electrochemiluminescent methodology. A first anti-hTREM2 antibody was diluted in coating buffer and immobilized onto a 96-well microtiter sample plate. After blocking and washing of the plate, endogenous quality control and study samples were diluted with assay buffer, dispensed onto the sample plate, and incubated. A second anti-hTREM2 antibody binding to a different bin from the first was added as capture antibody. The plate was washed, and Sulfo-Tag streptavidin was added and incubated, followed by addition of MSD Read Buffer T. Concentrations were determined on a standard curve obtained by relative light units versus concentration. The calibration curve was generated using a 4-parameter curve fit with 1/y2 weighting. The qualified range for this method in human cerebrospinal fluid is from 0.400 to 50.0 ng/ml.
sCSF-1R assay in human cerebrospinal fluid
A commercial ELISA assay by R&D Systems was used for the determination of sCSF-1R in human cerebrospinal fluid. Human M-CSF R capture antibody was diluted in coating buffer and immobilized onto a 96-well microtiter sample plate. After blocking and washing of the plate, endogenous quality control and study samples were diluted, dispensed onto the sample plate, and incubated. Human M-CSF R detection antibody was added and incubated. The plate was washed, and Streptavidin-HRP A was subsequently added followed by a working substrate solution. The plate was incubated at ambient temperature, stopped with the addition of Sulfuric Acid Stop Solution, and read on a plate reader using two filters, 450 nm for detection and 570 nm for background. Concentrations were determined on a standard curve obtained by plotting optical density versus concentration. The calibration curve was generated using a 4-parameter curve fit with 1/y2 weighting. The qualified range for this method in human cerebrospinal fluid is from 125 to 4,000 pg/ml in 100% cerebrospinal fluid.
Statistical analysis
Two-way ANOVA was used to compare the independent effects of both antibody treatment and TREM2 variant (genotype), individually, on quantifications. The aov function in R was used with the format (data$y ~ data$antibody + data$genotype), and P values were taken from the antibody variable unless otherwise noted. scRNA-seq commonly uses n = 2 experimental design. We performed scRNA-seq on more than n = 2 per condition to increase the statistical power of our results even further, as n = 2 is common for scRNA-seq experiments.
Online supplemental material
Fig. S1 shows that AL002c activates hTREM2 signaling and function. Fig. S2 displays the pharmacokinetics of AL002c. Fig. S3 shows scRNA-seq analysis of Cd45+ brain cells from antibody-treated mice. Fig. S4 presents the impact of chronic AL002c treatment on Aβ load. Fig. S5 shows the impact of chronic AL002c treatment on microglia clustering around plaques and Spp1 secretion.
Acknowledgments
We would like to thank Jingqin Luo for help in statistical analyses. This work was supported by the National Institutes of Health (RF1AG05148501, R21 AG059176, and RF1 AG059082) and the Cure Alzheimer’s Fund.
Author contributions: S. Wang, M. Mustafa, S.V. Salazar, A. Ibrahim, H. Rhinn, T. Schwabe, and I. Tassi conducted experiments; C.M. Yuede performed behavioral tests; S. Gilfillan prepared all mouse groups; S. Wang, A. Rosenthal, I. Tassi, T. Schwabe, and M. Colonna designed experiments; P. Kong, H. Long, M. Ward, O. Siddiqui, and R. Paul analyzed human phase I clinical trial data. M. Colonna wrote the manuscript with input from S. Wang, M. Mustafa, S.V. Salazar, A. Ibrahim, H. Rhinn, I. Tassi, C.M. Yuede, A. Rosenthal, and T. Schwabe.
References
- Braun D., and Feinstein D.L.. 2019. The locus coeruleus neuroprotective drug vindeburnol normalizes behavior in the 5xFAD transgenic mouse model of Alzheimer’s disease. Brain Res. 1702:29–37. 10.1016/j.brainres.2017.12.028 [DOI] [PubMed] [Google Scholar]
- Bruhns P., and Jönsson F.. 2015. Mouse and human FcR effector functions. Immunol. Rev. 268:25–51. 10.1111/imr.12350 [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.-T., Lu A., Craessaerts K., Pavie B., Frigerio C.S., Mancuso R., Qian X., Lalakova J., Kühnemund M., Voytyuk I., et al. 2019. Spatial and temporal transcriptomics reveal microglia-astroglia crosstalk in the amyloid-β plaque cell niche of Alzheimer’s disease. bioRxiv 10.1101/719930 (Preprint posted August 12, 2019). [DOI]
- Cheng Q., Danao J., Talreja S., Wen P., Yin J., Sun N., Li C.M., Chui D., Tran D., Koirala S., et al. 2018. TREM2-activating antibodies abrogate the negative pleiotropic effects of the Alzheimer’s disease variant Trem2R47H on murine myeloid cell function. J. Biol. Chem. 293:12620–12633. 10.1074/jbc.RA118.001848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos D., Grutzendler J., Yang G., Kim J.V., Zuo Y., Jung S., Littman D.R., Dustin M.L., and Gan W.B.. 2005. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8:752–758. 10.1038/nn1472 [DOI] [PubMed] [Google Scholar]
- Feuerbach D., Schindler P., Barske C., Joller S., Beng-Louka E., Worringer K.A., Kommineni S., Kaykas A., Ho D.J., Ye C., et al. 2017. ADAM17 is the main sheddase for the generation of human triggering receptor expressed in myeloid cells (hTREM2) ectodomain and cleaves TREM2 after Histidine 157. Neurosci. Lett. 660:109–114. 10.1016/j.neulet.2017.09.034 [DOI] [PubMed] [Google Scholar]
- Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J.S.K., Younkin S., et al. ; Alzheimer Genetic Analysis Group . 2013. TREM2 variants in Alzheimer’s disease. N. Engl. J. Med. 368:117–127. 10.1056/NEJMoa1211851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenrauch M., Walter S., Kaufmann M., Weggen S., and Wirths O.. 2017. Limited Effects of Prolonged Environmental Enrichment on the Pathology of 5XFAD Mice. Mol. Neurobiol. 54:6542–6555. 10.1007/s12035-016-0167-x [DOI] [PubMed] [Google Scholar]
- Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P.V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A.I., Lah J.J., et al. 2013. Variant of TREM2 associated with the risk of Alzheimer’s disease. N. Engl. J. Med. 368:107–116. 10.1056/NEJMoa1211103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch C.M., and Goate A.M.. 2015. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry. 77:43–51. 10.1016/j.biopsych.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. 2017. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 169:1276–1290.e17. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Lambert J.C., Ibrahim-Verbaas C.A., Harold D., Naj A.C., Sims R., Bellenguez C., DeStafano A.L., Bis J.C., Beecham G.W., Grenier-Boley B., et al. ; Cohorts for Heart and Aging Research in Genomic Epidemiology . 2013. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45:1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.Y.D., Daggett A., Gu X., Jiang L.L., Langfelder P., Li X., Wang N., Zhao Y., Park C.S., Cooper Y., et al. 2018. Elevated TREM2 Gene Dosage Reprograms Microglia Responsivity and Ameliorates Pathological Phenotypes in Alzheimer’s Disease Models. Neuron. 97:1032–1048.e5. 10.1016/j.neuron.2018.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo J.A., Stern E.A., McLellan M.E., Kajdasz S.T., Hickey G.A., Bacskai B.J., and Hyman B.T.. 2003. Amyloid-β antibody treatment leads to rapid normalization of plaque-induced neuritic alterations. J. Neurosci. 23:10879–10883. 10.1523/JNEUROSCI.23-34-10879.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.M., and Holtzman D.M.. 2019. Alzheimer Disease: An Update on Pathobiology and Treatment Strategies. Cell. 179:312–339. 10.1016/j.cell.2019.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H., Adaikkan C., Gao F., Young J.Z., Manet E., Hemberg M., De Jager P.L., Ransohoff R.M., Regev A., and Tsai L.H.. 2017. Temporal Tracking of Microglia Activation in Neurodegeneration at Single-Cell Resolution. Cell Rep. 21:366–380. 10.1016/j.celrep.2017.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J.Z., Menon M., He L., Abdurrob F., Jiang X., et al. 2019. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 570:332–337. 10.1038/s41586-019-1195-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildner A., Schmidt H., Nitsche M., Merkler D., Hanisch U.K., Mack M., Heikenwalder M., Brück W., Priller J., and Prinz M.. 2007. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat. Neurosci. 10:1544–1553. 10.1038/nn2015 [DOI] [PubMed] [Google Scholar]
- Mucke L., and Selkoe D.J.. 2012. Neurotoxicity of amyloid β-protein: synaptic and network dysfunction. Cold Spring Harb. Perspect. Med. 2 a006338 10.1101/cshperspect.a006338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A., Kirchhoff F., and Helmchen F.. 2005. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 308:1314–1318. 10.1126/science.1110647 [DOI] [PubMed] [Google Scholar]
- Oakley H., Cole S.L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., et al. 2006. Intraneuronal β-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: potential factors in amyloid plaque formation. J. Neurosci. 26:10129–10140. 10.1523/JNEUROSCI.1202-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero K., Turnbull I.R., Poliani P.L., Vermi W., Cerutti E., Aoshi T., Tassi I., Takai T., Stanley S.L., Miller M., et al. 2009. Macrophage colony-stimulating factor induces the proliferation and survival of macrophages via a pathway involving DAP12 and β-catenin. Nat. Immunol. 10:734–743. 10.1038/ni.1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccio L., Buonsanti C., Cella M., Tassi I., Schmidt R.E., Fenoglio C., Rinker J. II, Naismith R.T., Panina-Bordignon P., Passini N., et al. 2008. Identification of soluble TREM-2 in the cerebrospinal fluid and its association with multiple sclerosis and CNS inflammation. Brain. 131:3081–3091. 10.1093/brain/awn217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala Frigerio C., Wolfs L., Fattorelli N., Thrupp N., Voytyuk I., Schmidt I., Mancuso R., Chen W.T., Woodbury M.E., Srivastava G., et al. 2019. The Major Risk Factors for Alzheimer’s Disease: Age, Sex, and Genes Modulate the Microglia Response to Aβ Plaques. Cell Rep. 27:1293–1306.e6. 10.1016/j.celrep.2019.03.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarlus H., and Heneka M.T.. 2017. Microglia in Alzheimer’s disease. J. Clin. Invest. 127:3240–3249. 10.1172/JCI90606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlepckow K., Kleinberger G., Fukumori A., Feederle R., Lichtenthaler S.F., Steiner H., and Haass C.. 2017. An Alzheimer-associated TREM2 variant occurs at the ADAM cleavage site and affects shedding and phagocytic function. EMBO Mol. Med. 9:1356–1365. 10.15252/emmm.201707672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlepckow K., Monroe K.M., Kleinberger G., Cantuti-Castelvetri L., Parhizkar S., Xia D., Willem M., Werner G., Pettkus N., Brunner B., et al. 2020. Enhancing protective microglial activities with a dual function TREM2 antibody to the stalk region. EMBO Mol. Med. 12 e11227 10.15252/emmm.201911227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Pozo A., William C.M., Ferrer I., Uro-Coste E., Delisle M.B., Maurage C.A., Hock C., Nitsch R.M., Masliah E., Growdon J.H., et al. 2010. Beneficial effect of human anti-amyloid-β active immunization on neurite morphology and tau pathology. Brain. 133:1312–1327. 10.1093/brain/awq056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Hooli B., Mullin K., Jin S.C., Cella M., Ulland T.K., Wang Y., Tanzi R.E., and Colonna M.. 2017. Alzheimer’s disease-associated TREM2 variants exhibit either decreased or increased ligand-dependent activation. Alzheimers Dement. 13:381–387. 10.1016/j.jalz.2016.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.M., Joshita S., Zhou Y., Ulland T.K., Gilfillan S., and Colonna M.. 2018. Humanized TREM2 mice reveal microglia-intrinsic and -extrinsic effects of R47H polymorphism. J. Exp. Med. 215:745–760. 10.1084/jem.20171529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H. II, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 352:189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulland T.K., Song W.M., Huang S.C.C., Ulrich J.D., Sergushichev A., Beatty W.L., Loboda A.A., Zhou Y., Cairns N.J., Kambal A., et al. 2017. TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell. 170:649–663.e13. 10.1016/j.cell.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hove H., Martens L., Scheyltjens I., De Vlaminck K., Pombo Antunes A.R., De Prijck S., Vandamme N., De Schepper S., Van Isterdael G., Scott C.L., et al. 2019. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci. 22:1021–1035. 10.1038/s41593-019-0393-4 [DOI] [PubMed] [Google Scholar]
- Wang Y., Cella M., Mallinson K., Ulrich J.D., Young K.L., Robinette M.L., Gilfillan S., Krishnan G.M., Sudhakar S., Zinselmeyer B.H., et al. 2015. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 160:1061–1071. 10.1016/j.cell.2015.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Ulland T.K., Ulrich J.D., Song W., Tzaferis J.A., Hole J.T., Yuan P., Mahan T.E., Shi Y., Gilfillan S., et al. 2016. TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213:667–675. 10.1084/jem.20151948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak D.F., Diggs-Andrews K.A., Conyers S., Yuede C.M., Dearborn J.T., Brown J.A., Tokuda K., Izumi Y., Zorumski C.F., and Gutmann D.H.. 2013. Motivational disturbances and effects of L-dopa administration in neurofibromatosis-1 model mice. PLoS One. 8 e66024 10.1371/journal.pone.0066024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich P., Glebov K., Kemmerling N., Tien N.T., Neumann H., and Walter J.. 2013. Sequential proteolytic processing of the triggering receptor expressed on myeloid cells-2 (TREM2) protein by ectodomain shedding and γ-secretase-dependent intramembranous cleavage. J. Biol. Chem. 288:33027–33036. 10.1074/jbc.M113.517540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Condello C., Keene C.D., Wang Y., Bird T.D., Paul S.M., Luo W., Colonna M., Baddeley D., and Grutzendler J.. 2016. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron. 90:724–739. 10.1016/j.neuron.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuede C.M., Zimmerman S.D., Dong H., Kling M.J., Bero A.W., Holtzman D.M., Timson B.F., and Csernansky J.G.. 2009. Effects of voluntary and forced exercise on plaque deposition, hippocampal volume, and behavior in the Tg2576 mouse model of Alzheimer’s disease. Neurobiol. Dis. 35:426–432. 10.1016/j.nbd.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Song W.M., Andhey P.S., Swain A., Levy T., Miller K.R., Poliani P.L., Cominelli M., Grover S., Gilfillan S., et al. 2020. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26:131–142. 10.1038/s41591-019-0695-9 [DOI] [PMC free article] [PubMed] [Google Scholar]