Lee et al. show that circulating NS1 protein does not contribute to dengue pathogenesis in mice infected with highly virulent DENV2 strains. Instead, structural prME region is found to play a critical role in the in vivo fitness and virulence of these strains.
Abstract
Dengue is a major public health concern in the tropical and subtropical world, with no effective treatment. The controversial live attenuated virus vaccine Dengvaxia has boosted the pursuit of subunit vaccine approaches, and nonstructural protein 1 (NS1) has recently emerged as a promising candidate. However, we found that NS1 immunization or passive transfer of NS1 antibodies failed to confer protection in symptomatic dengue mouse models using two non–mouse-adapted DENV2 strains that are highly virulent. Exogenous administration of purified NS1 also failed to worsen in vivo vascular leakage in sublethally infected mice. Neither method of NS1 immune neutralization changed the disease outcome of a chimeric strain expressing a vascular leak-potent NS1. Instead, virus chimerization involving the prME structural region indicated that these proteins play a critical role in driving in vivo fitness and virulence of the virus, through induction of key proinflammatory cytokines. This work highlights that the pathogenic role of NS1 is DENV strain dependent, which warrants reevaluation of NS1 as a universal dengue vaccine candidate.
Graphical Abstract
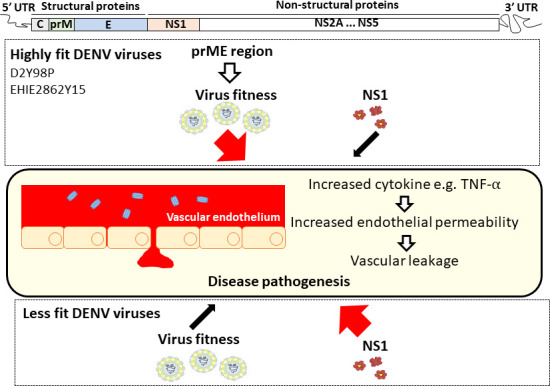
Introduction
Dengue virus (DENV) is a mosquito-borne virus responsible for an estimated 390 million annual infections in the tropical and subtropical world (Bhatt et al., 2013). The virus exists as four antigenically distinct serotypes (DENV1–4). Infection with DENV results in a wide spectrum of disease manifestations ranging from mild to life-threatening conditions, the latter being characterized by vascular leakage with or without hemorrhage development (World Health Organization, 2009; Kyle and Harris, 2008). DENV serotype cross-reactive immunity has been proposed, and to a certain extent demonstrated, to represent a risk for the development of severe dengue. Specifically, preexisting antibodies raised during a previous heterotypic DENV infection that recognize the structural components of the virion could enhance DENV uptake and facilitate its replication within FcγR-bearing cells, leading to increased disease severity, a phenomenon termed antibody-dependent enhancement (ADE; Halstead et al., 2002; Halstead, 2003). There is currently no effective therapeutic or vaccine against dengue, which mainly stems from our lack of understanding of dengue pathogenesis associated with limited availability of relevant symptomatic animal models (Yauch and Shresta, 2008; Chan et al., 2015). The only licensed vaccine, Dengvaxia, which consists of chimeric live attenuated tetravalent DENV, not only offers limited protective efficacy, particularly toward DENV2 strains, but also was found to predispose immunologically dengue-naive individuals to an increased risk of developing severe disease (Capeding et al., 2014; Villar et al., 2015; Aguiar et al., 2016; Halstead, 2017). This latter observation led to the suspension of dengue vaccination programs in the Philippines (Fatima and Syed, 2018). Thus, safer and more effective second-generation vaccines are urgently needed. In the wake of the Dengvaxia setback and controversy, subunit vaccine candidates have regained some traction (Lam et al., 2016). While the envelope protein (E protein) of DENV has been the most popular subunit vaccine candidate, a number of studies have also proposed DENV nonstructural protein 1 (NS1) as a potential candidate (Schlesinger et al., 1987; Henchal et al., 1988; Falgout et al., 1990; Beatty et al., 2015; Gonçalves et al., 2015; Wan et al., 2014, 2017; Lai et al., 2017). NS1 is a glycosylated nonstructural protein (46–55 kD) that homodimerizes after posttranslational modification, becomes membrane associated, and participates in RNA replication within membrane-bound replication complexes. Soluble hexameric NS1 (sNS1) is made of three dimers and a central lipid cargo, and is secreted into the extracellular milieu (Gutsche et al., 2011; Muller and Young, 2013; Watterson et al., 2016). Cell surface–associated NS1 dimers and sNS1 hexamers are highly immunogenic (Muller and Young, 2013). sNS1 is detected in the blood at concentrations ≤50 µg/ml and typically follows viremia in dengue patients (Young et al., 2000; Libraty et al., 2002; Alcon et al., 2002), and thus is widely used as an early diagnostic marker (Peeling et al., 2010; Hang et al., 2009; Chuansumrit et al., 2011). Furthermore, recent studies have reported a role for sNS1 in dengue pathogenesis, specifically in causing vascular leakage, a key clinical manifestation of severe dengue (Beatty et al., 2015; Modhiran et al., 2015). As such, immunity raised against NS1, through either direct immunization with purified sNS1 or passive transfer of anti-NS1 immune serum, was found to afford protection against lethal DENV infection in a mouse model (Beatty et al., 2015). These findings thus support that NS1 represents a promising subunit vaccine candidate and a safer alternative to avoid the risk of ADE, which is a major roadblock in the development of vaccines that rely on protective immunity against the structural components of DENV. However, the role of NS1 in dengue pathogenesis and the in vivo protective efficacy of NS1 immunity had been demonstrated in mouse models that either produce mild symptoms such as local skin hemorrhage and prolonged bleeding time (Wan et al., 2014, 2017; Lai et al., 2017) or use mouse-adapted DENV2 strains (Schlesinger et al., 1987; Henchal et al., 1988; Falgout et al., 1990; Costa et al., 2006; Beatty et al., 2015).
Here, the role of NS1 in dengue pathogenesis as well as the protective potential of NS1 immunity were investigated in symptomatic dengue mouse models established with a non–mouse-adapted DENV2 strain, namely D2Y98P (Tan et al., 2010; Ng et al., 2014; Martínez Gómez et al., 2016). In these models, neutralization of circulating sNS1 through antibody transfer or active NS1 immunization did not confer protection against D2Y98P-induced disease, thus suggesting a limited role of sNS1 in the pathogenesis of D2Y98P. Similar observations were made with a DENV2 clinical isolate that currently circulates in Singapore and Malaysia. Instead, we showed that the precursor membrane prME region drives the in vivo virulence of D2Y98P virus through induction of key proinflammatory cytokines.
Results
No protective efficacy of NS1 immunity in A129 and AG129 ADE mouse models
The protective efficacy of NS1 was evaluated in symptomatic mouse models of dengue that have been established with the non–mouse-adapted DENV2 D2Y98P strain (Tan et al., 2010; Ng et al., 2014; Martínez Gómez et al., 2016). D2Y98P derives from a 1998 clinical isolate from the DENV2 cosmopolitan genotype and has been exclusively passaged for ∼20 rounds in C6/36 mosquito cells (Tan et al., 2010). E protein nucleotide sequence or the entire coding sequence of D2Y98P genome was compared with that of a DENV2 prototype strain (NGC strain; Añez et al., 2016) and six other DENV2 Singapore isolates that were collected between 2007 and 2010 (Lee et al., 2012). The results indicated 94% identity with NGC and 98% identity with the six Singapore isolates, thus supporting that D2Y98P can be considered a representative of DENV2 strains that circulate in Singapore (Table S1).
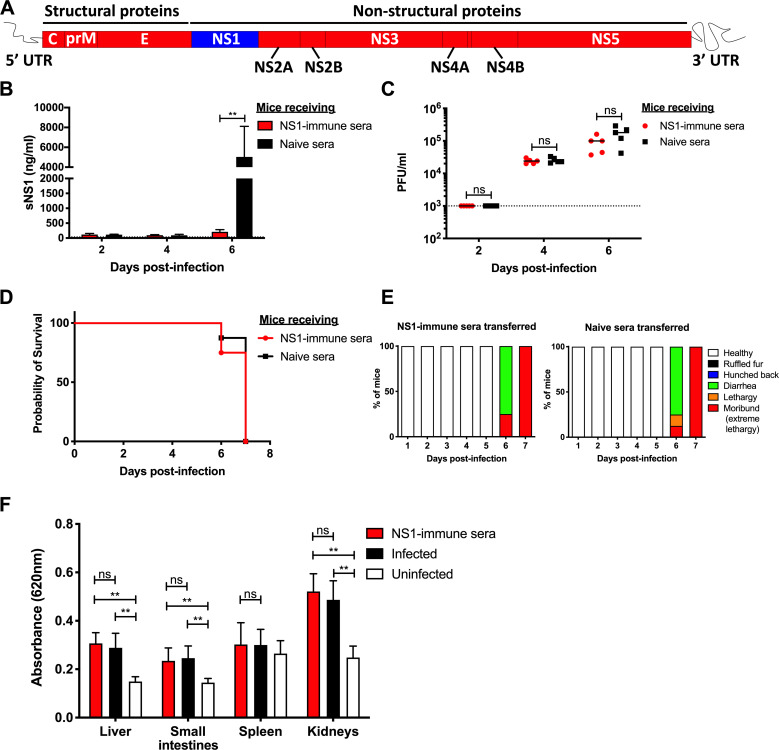
Adult A129 mice (deficient in type I IFN pathway) were administered commercially available purified hexameric NS1 from DENV2 strain 16681 three times according to a previously published immunization protocol (Beatty et al., 2015). High titers of NS1-specific IgG antibodies were measured in the serum of the immunized mice (Fig. S1, A and B), with the majority of the IgG1 subclass (Fig. S1 C). A control group consisting of mice administered OVA according to the same immunization regimen was included. OVA- and NS1-immunized mice were then administered DENV1-immune serum 1 d before infection with D2Y98P strain to produce lethal infection in an ADE setting (Fig. 1 A).
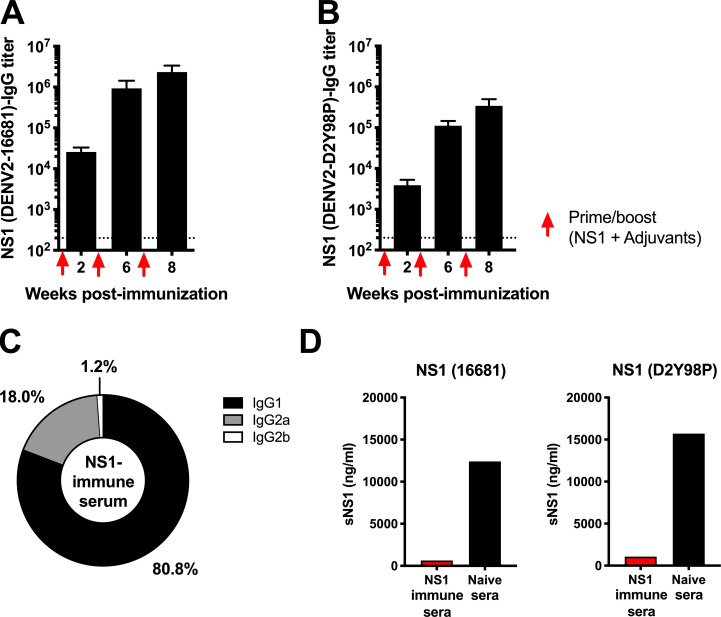
Figure S1.
NS1-specific IgG titers in NS1 immune serum. A129 mice were immunized i.p. three times with 20 µg NS1(16681) or OVA with MPLA and Addavax as adjuvants at wks 2, 6, and 8. (A and B) NS1-specific IgG antibody titers were measured by ELISA using NS1(16681) (A) or NS1(D2Y98P) (B) as coating antigen. Titers were calculated as the reciprocal of the highest serum dilution with absorbance corresponding to 3× the absorbance of blank wells. The dotted line indicates the limit of detection. (C) The percentages of specific IgG subclasses were determined. (D) 5×-diluted NS1 immune serum was coincubated with 20 µg/ml NS1(16681) or NS1(D2Y98P), and the amount of free NS1 was measured by sandwich ELISA.
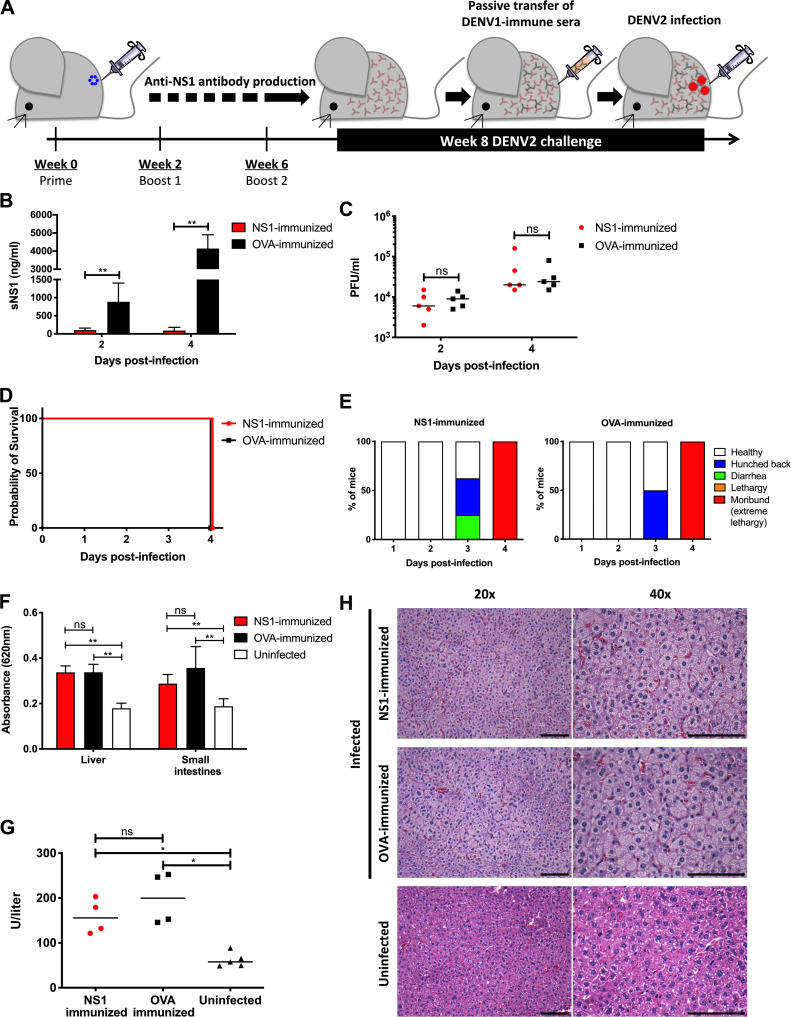
Figure 1.
NS1 immunization in A129 ADE infection model. (A) A129 mice were immunized i.p. three times with 20 µg NS1(16681) or OVA with MPLA and Addavax as adjuvants. 8 wk after immunization, mice were passively transferred with DENV1-immune serum 1 d before i.v. challenge with 106 PFU DENV2 (D2Y98P). (B and C) Circulating sNS1 levels (B) and viremia titers (C) were measured (n = 5) at days 2 and 4 p.i. (D and E) Mice (n = 8) were monitored daily upon challenge. Kaplan–Meier survival curve and clinical scores are shown. (F) Vascular leakage was quantified by Evans blue dye extravasation assay in liver and small intestines at day 4 p.i. (n = 5). (G and H) Systemic aspartate aminotransferase levels were measured (n = 4–5; G) and histological analysis of liver was performed at day 4 p.i. (n = 4–5; H). Images were taken at 20× and 40× magnification. Representative sections are shown. Scale bar, 10 µm. Data are representative of at least two independent experiments. Data were analyzed by nonparametric Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ns, not significant. Results were expressed as averages ± SD (B and F).
NS1(16681) and NS1(D2Y98P) share 97% amino acid identity (Table S2), and the ability of immune serum raised against NS1(16681) to bind to and neutralize NS1(D2Y98P) was verified by ELISA and sandwich ELISA, respectively (Fig. S1, B and D). Consistently, significant reduction in circulating sNS1 was observed in NS1-immunized mice compared with OVA-immunized animals after viral challenge (Fig. 1 B), indicating that NS1(16681) immunization successfully neutralized circulating sNS1 produced during infection with D2Y98P virus. However, comparable viremia titers were measured between NS1-immunized mice and OVA controls (Fig. 1 C). Furthermore, NS1- and OVA-immunized mice displayed similar disease manifestations and progression, including hunched back, diarrhea, and lethargy, and eventually all the animals from both groups were moribund by day 4 post-infection (p.i.; Fig. 1, D and E). Vascular leakage in the liver and small intestines from NS1-immunized animals was as extensive as that measured in the OVA control group (Fig. 1 F). Systemic levels of the liver enzyme aspartate aminotransferase were also comparable between NS1- and OVA-immunized groups (Fig. 1 G), and massive cytoplasmic vacuolation was apparent in the hepatocytes from both infected groups (Fig. 1 H), indicating that reduction in sNS1 levels brought about by NS1 immunization did not protect the mice from DENV-induced severe liver damage (Martínez Gómez et al., 2016). Therefore, despite effective neutralization of circulating sNS1, active immunization with purified NS1 failed to provide protection against lethal DENV2 challenge in an A129 ADE model.
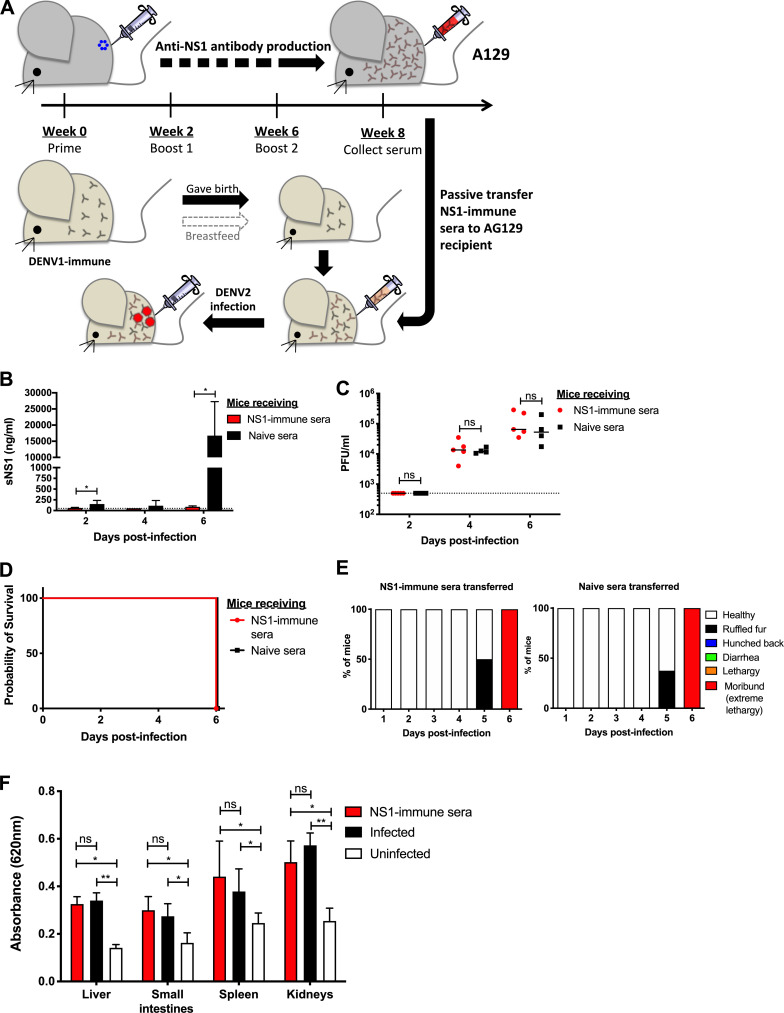
The protective potential of NS1 immunity was further explored in another lethal ADE model using AG129 mice (deficient in types I and II IFN pathways), a model in which mice born to DENV1-immune dams exhibit extensive vascular leakage upon DENV2 infection (Ng et al., 2014). Because AG129 mice lack a functional IFN-γ signaling pathway, direct immunization with purified NS1 may result in suboptimal immune responses that may affect the protective potential of NS1 immunity (van den Broek et al., 1995). To address this possibility, an NS1 immune serum was instead generated in A129 mice according to the same immunization protocol described above. 1 d before DENV2 infection, AG129 mice born to DENV1-immune dams were then passively transferred with the NS1 immune serum (Fig. 2 A), which led to effective neutralization of circulating sNS1 after D2Y98P challenge (Fig. 2 B). However, comparable viremia titers (Fig. 2 C), survival rate (Fig. 2 D), clinical symptoms (Fig. 2 E), and vascular leakage (Fig. 2 F) were observed between mice administered with NS1 immune serum and the control group transferred with naive serum.
Figure 2.
Passive NS1 immunity in AG129 ADE infection model. (A) AG129 mice born to DENV1-immune mothers were passively transferred with NS1 immune serum collected from NS1(16681)-immunized A129 mice (Fig. 1 A) or with naive serum. 1 d after transfer, the mice were challenged s.c. with 103 PFU D2Y98P. (B and C) Systemic sNS1 level) and viremia titers (C) were measured at days 2, 4, and 6 p.i. (n = 4–5). (D and E) Mice (n = 8) were monitored daily upon challenge. Kaplan–Meier survival curve and clinical score are shown. (F) Vascular leakage was assessed in indicated tissues at day 6 p.i. (n = 4–5). Data are representative of at least two independent experiments. Data were analyzed by nonparametric Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ns, not significant. Results were expressed as averages ± SD (B and F).
Together, our data indicate that neither active NS1 immunization nor passive transfer of NS1 immune serum conferred significant protection in both A129 and AG129 ADE models, despite effective neutralization of circulating sNS1. These observations imply that in these ADE infection models, sNS1 does not play a critical role in dengue pathogenesis.
Lack of protective NS1 immunity is independent of mouse background
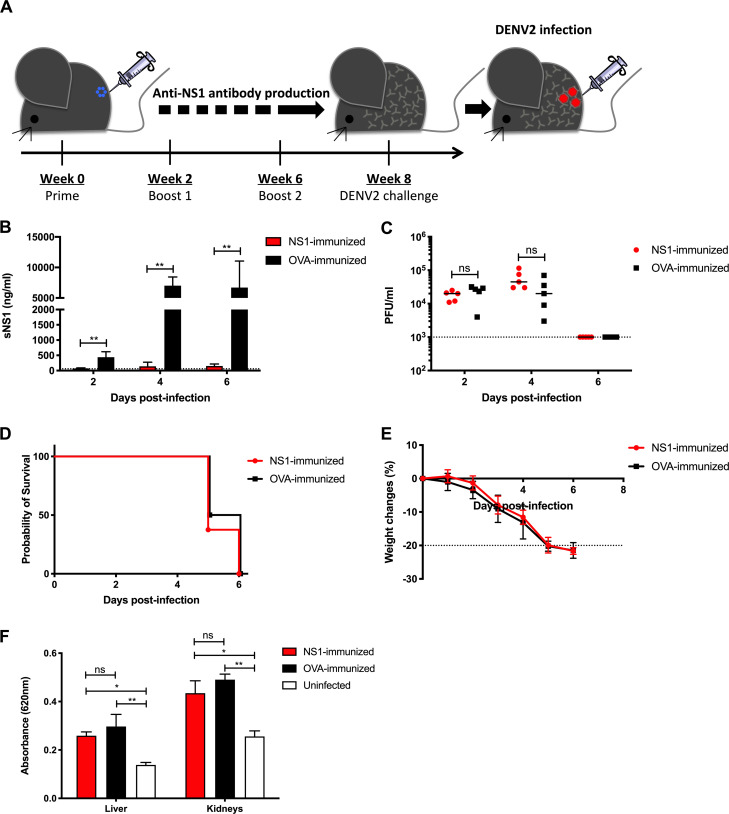
Previous studies that reported NS1-mediated protection were conducted in mouse strains such as IFNAR−/−; Beatty et al., 2015) and STAT1−/− (Wan et al., 2017; Lai et al., 2017), both on the C57BL/6 background. We thus questioned whether the discrepancy between our observations and the earlier studies could be explained by a difference in mouse background. Hence, IFNAR−/− mice of C56BL/6 background were immunized with NS1 or OVA according to the same immunization regimen above, followed by lethal challenge with D2Y98P (Fig. 3 A). NS1-immunized IFNAR−/− mice had significantly reduced sNS1 levels after D2Y98P challenge compared with OVA-immunized controls, indicating effective neutralization of sNS1 in circulation (Fig. 3 B). However, comparable viremia titers (Fig. 3 C), survival rates (Fig. 3 D), weight loss profiles (Fig. 3 E), and vascular leakage (Fig. 3 F) were measured between NS1-immunized and OVA control groups. These data indicate that the lack of protection mediated by NS1 immunity is likely independent of mouse background.
Figure 3.
NS1 immunization in IFNAR−/− primary infection model. (A) IFNAR−/− mice were immunized with NS1(16681) or OVA as described in the legend of Fig. 1. 8 wk after immunization, mice were challenged s.c. with 105 PFU D2Y98P. (B and C) Systemic sNS1 levels (B) and viremia titers (C) were measured at days 2, 4, and 6 p.i. (n = 5). (D and E) Mice (n = 6–8) were monitored daily upon challenge. Kaplan–Meier survival curve and weight loss profile are shown. (F) Vascular leakage was assessed in liver and kidneys at day 4 p.i. (n = 4–5). Data are representative of at least two independent experiments. Data were analyzed by nonparametric Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ns, not significant. Results were expressed as averages ± SD (B, E, and F).
Lack of NS1-mediated protective efficacy was also observed with another circulating DENV2 Singapore clinical isolate
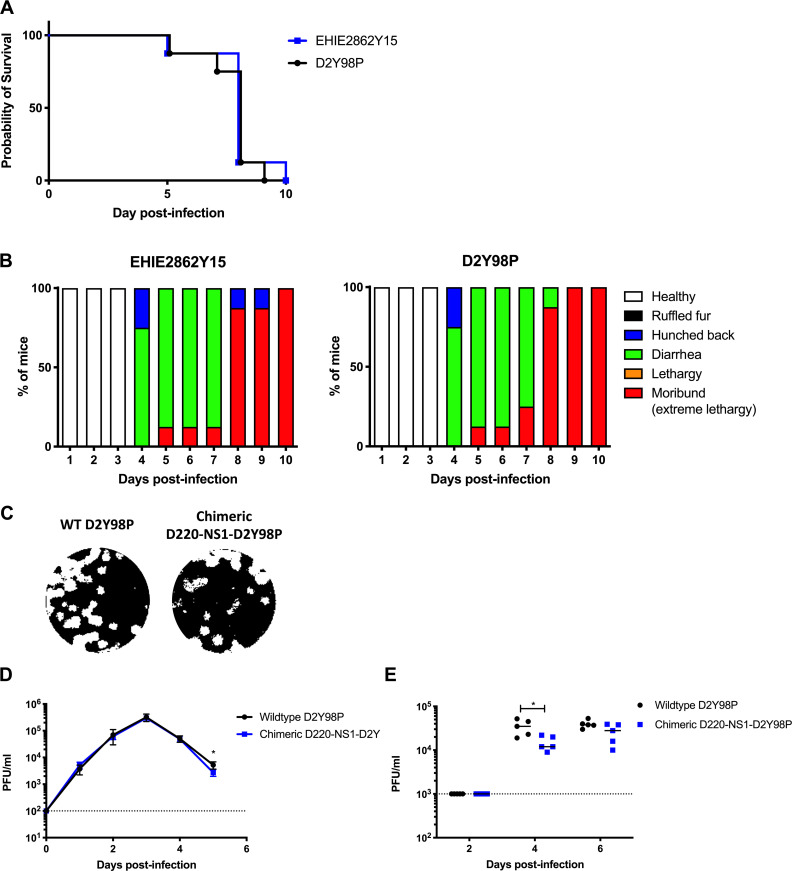
Although D2Y98P is not mouse adapted, the virus had been passaged in the C6/36 cell line for several rounds, during which some mutations in the viral genome may have modified the virus fitness. In particular, the presence of a phenylalanine (F) at position 52 in NS4B was found to play an important role in D2Y98P in vivo and in vitro fitness (Grant et al., 2011; Tan et al., 2010). As none of the other DENV2 strains whose genome sequence is available in the public database harbors an F at this position, we proposed that this particular amino acid was likely acquired during in vitro passages. To examine whether the findings made with D2Y98P can be extended to other DENV2 strains, we used another DENV2 clinical isolate, EHIE2862Y15, which has been passaged in the C6/36 cell line for no more than four rounds. This virus belongs to the cosmopolitan genotype clade Ib that has been circulating in Singapore and Malaysia since 2013 and has been associated with a high fatality rate (Ng et al., 2015). EHIE2862Y15 shares 98.6% nucleotide identity and 99.6% amino acid identity with D2Y98P (Table S3). Interestingly, although EHIE2862Y15 does not harbor an F at position 52 in NS4B, infection with EHIE2862Y15 gave rise to symptomatic infection in AG129 mice, with a disease kinetic profile similar to that of D2Y98P (Fig. S2, A and B).
Figure S2.
In vivo and in vitro fitness of EHIE2862Y15 strain and chimeric D220-NS1-D2Y98P in AG129 mice. (A and B) In vivo fitness of EHIE2862Y15 strain. Mice(n = 8) were challenged s.c. with 106 PFU of EHIE2862Y15 or D2Y98P virus. Mice were monitored daily upon challenge. Kaplan–Meier survival curve (A) and clinical scores (B) are shown. (C–E) In vitro and in vivo fitness of D220-NS1-D2Y98P chimeric strain. Plaque morphology (C) and in vitro growth profile (D) in BHK cells; viremia titers (E) in AG219 mice (n = 5) s.c. infected with 103 PFU of WT D2Y98P or chimeric D220-NS1-D2Y98P. Dotted line denotes the limit of detection. Data were analyzed by nonparametric Mann–Whitney U test. *, P < 0.05. Results were expressed as averages ± SD (D).
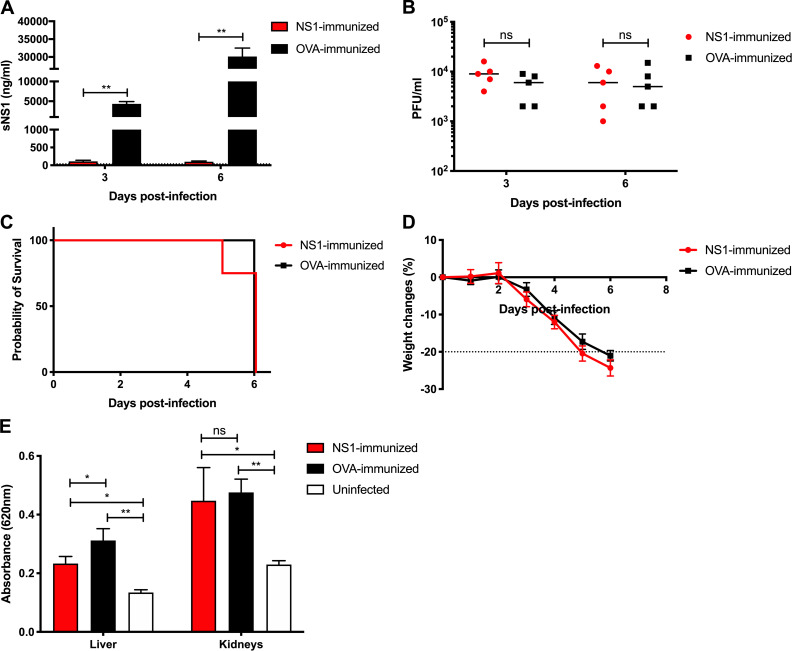
To evaluate the role of NS1 in the context of EHIE2862Y15 infection, IFNAR−/− mice were immunized with purified NS1(16681) followed by lethal viral challenge. Despite the effective neutralization of circulating sNS1 (Fig. 4 A), mice were not protected from lethal EHIE2862Y15 challenge, as evidenced by comparable viremia titers (Fig. 4 B), survival rates (Fig. 4 C), and weight loss profiles (Fig. 4 D). Furthermore, significant vascular leakage was measured in the liver and kidneys from NS1- and OVA-immunized mice compared with uninfected controls, although the extent of vascular leakage in the liver of NS1-immunized mice was slightly but significantly lower than in OVA-immunized animals (Fig. 4 E). Overall, these data support that the lack of NS1 immunity-mediated protection is not restricted to the D2Y98P strain but also applies to a clinically relevant DENV2 isolate that is circulating in Singapore.
Figure 4.
Protective efficacy of NS1 immunity against a low-passaged DENV2 clinical isolate. IFNAR−/− mice were immunized with NS1(16681) or OVA as described in the legend of Fig. 1. 8 wk after immunization, mice were s.c. challenged with 105 PFU EHIE2862Y15. (A and B) Systemic sNS1 levels (A) and viremia titers (B) were measured at days 3 and 6 p.i. (n = 5). (C and D) Mice (n = 8) were monitored daily upon challenge. Kaplan–Meier survival curve and weight loss profile are shown. (E) Vascular leakage was assessed in the liver and kidneys at day 4 p.i. (n = 4–5). Data are representative of at least two independent experiments. Data were analyzed by nonparametric Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ns, not significant. Results were expressed as averages ± SD (A, D, and E).
Exogenous administration of sNS1 did not worsen dengue disease severity
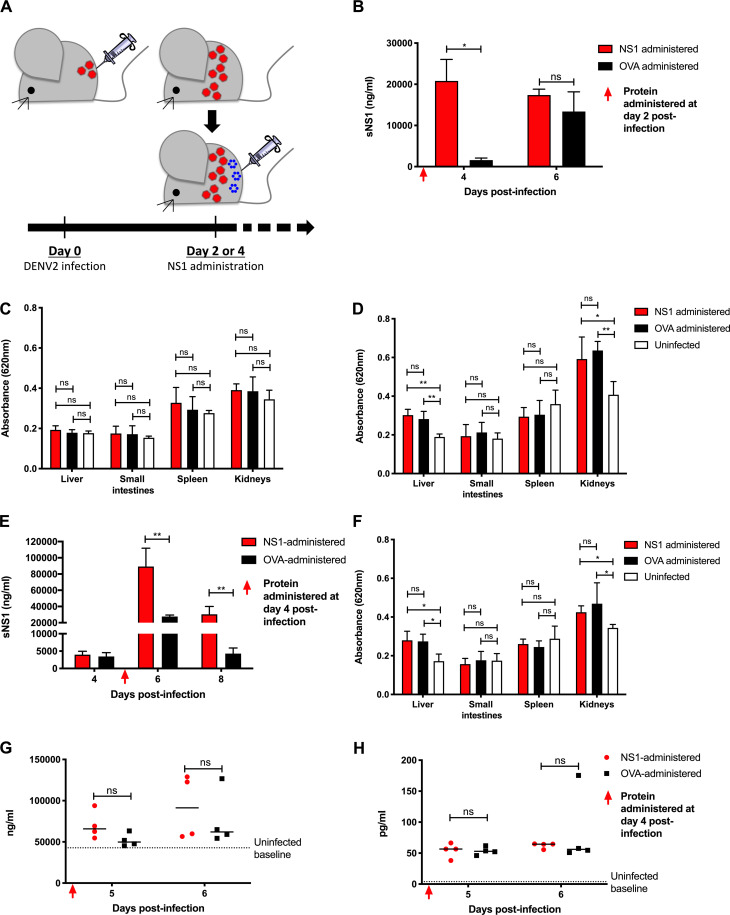
To assess the pathogenic role of sNS1 during D2Y98P infection, we tested whether the exogenous administration of purified sNS1(D2Y98P) could aggravate disease severity in AG129 mice infected with a sublethal dose of D2Y98P (Fig. 5 A). Administration of purified sNS1(D2Y98P) at day 2 p.i. led to significantly higher sNS1 levels measured at day 4 p.i. compared with mice administered OVA (Fig. 5 B). No significant vascular leakage was detected, however, in any of the infected mice from both groups at this time point (Fig. 5 C). Vascular leakage was also measured at day 6 p.i., when comparable levels of sNS1 were measured in both groups (Fig. 5 B). It is likely that at this later time point, the sNS1 level mainly results from virus replication, and that exogenously added sNS1 had been cleared from the circulation and/or deposited onto the endothelial layer and internalized by endothelial cells. Although no significant vascular leakage was observed in the small intestine and spleen from both infected groups, liver and kidneys did display significant vascular leak compared with uninfected controls (Fig. 5 D). However, the extent of vascular leakage was not greater in mice administered with exogenous sNS1 compared with the OVA control group (Fig. 5 D).
Figure 5.
Administration of NS1(D2Y98P) to AG129 mice infected with a sublethal dose of D2Y98P. (A) AG129 mice were challenged s.c. with 103 PFU D2Y98P. (B–H) At day 2 p.i. (B–F) or day 4 p.i. (E–H), infected mice were i.v. administered 10 mg/kg NS1(D2Y98P) or OVA. (B and E) Systemic levels of sNS1 were measured at various time points p.i. (n = 4–5). (C, D, and F) Vascular leakage was evaluated in various tissues at day 4 (C) or day 6 (D and F) p.i. (n = 4–5). (G and H) Plasma heparan sulfate (n = 4; G) and TNF-α levels (n = 4; H) were measured at day 6 p.i. Data are representative of at least two independent experiments. Data were analyzed by nonparametric Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ns, not significant. Results were expressed as averages ± SD (B–F).
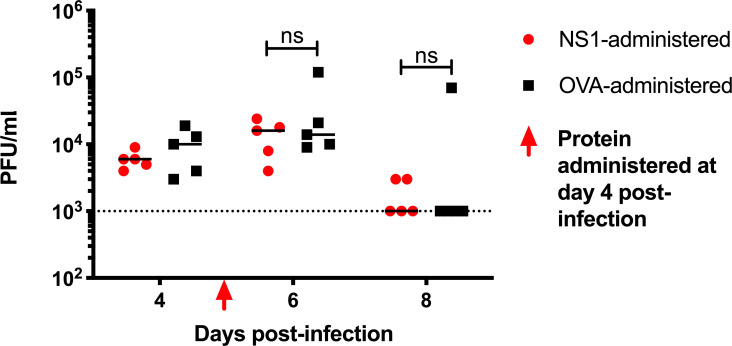
Because vascular leakage was more prominent at day 6 p.i. in this sublethal AG129 model, we reasoned that delaying the exogenous administration of sNS1 from day 2 p.i. to day 4 p.i. may have a greater impact on vascular leakage. Significantly higher circulating sNS1 levels were measured at day 6 p.i. in mice administered with purified sNS1 (Fig. 5 E). However, these mice failed to exhibit greater vascular leakage than the OVA control group (Fig. 5 F). Viremia titers were also not enhanced with sNS1 administration (Fig. S3).
Figure S3.
Viremia titers in D2Y98P-infected AG129 mice administered NS1. AG129 mice were challenged s.c. with 103 PFU D2Y98P. At day 4 p.i., D2Y98P-infected mice were i.v. administered 10 mg/kg NS1(16681) or OVA. Dotted line denotes the limit of detection. Data were analyzed by nonparametric Mann–Whitney U test. ns, not significant.
Previous studies have reported the ability of sNS1 to activate TLR4 on immune cells leading to the production of vasoactive cytokines TNF-α (Modhiran et al., 2015) and causing endothelial glycocalyx disruption (Puerta-Guardo et al., 2016). However, D2Y98P-infected mice administered with exogenous sNS1 did not display significantly higher levels of TNF-α compared with OVA controls (Fig. 5 G). Likewise, the levels of heparan sulfate in circulation, indicative of glycocalyx degradation, were not enhanced with NS1 administration (Fig. 5 H). These findings are consistent with the observation that exogenous administration of sNS1 did not increase vascular leakage in D2Y98P-infected mice.
D2Y98P virulence is independent of the intrinsic ability of NS1 to increase vascular permeability.
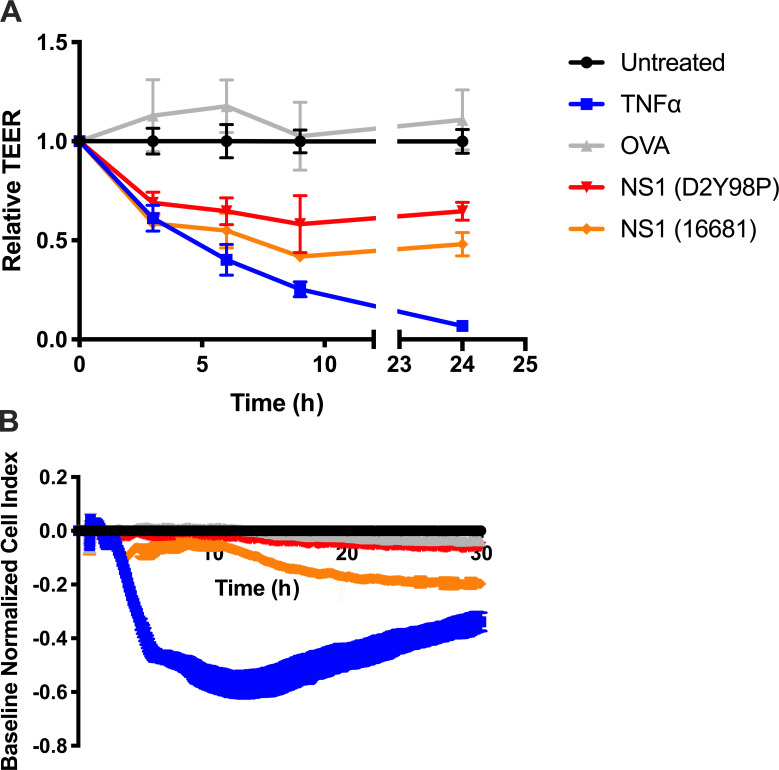
Absence of NS1-mediated protective immunity and the lack of increased vascular leakage observed upon exogenous administration of NS1(D2Y98P) to sublethally infected mice prompted us to test the ability of purified NS1(D2Y98P) to interact and disrupt the endothelial integrity compared with NS1 from DENV2 strain 16681 recently reported to be vascular leak potent (Puerta-Guardo et al., 2019). Two different in vitro permeability assays were conducted, namely transendothelial electrical resistance (TEER) assay and real-time cell analysis (RTCA), as previously described (Chen et al., 2016). TEER results showed that NS1(D2Y98P) was able to increase vascular permeability as effectively as NS1(16681) (Fig. S4 A). In contrast, RTCA indicated that NS1(D2Y98P) did not significantly disturb the endothelial monolayer, unlike NS1(16681) (Fig. S4 B). Therefore, these contradicting findings did not allow us to establish whether NS1(D2Y98P) had a defect in interacting with and/or disrupt the vascular endothelium.
Figure S4.
Permeability assays. (A) TEER assay: Confluent HMVEC monolayers cultured on Transwell inserts were incubated with 10 µg/ml of purified sNS1 from 16681 or D2Y98P strain, 10 µg/ml OVA (negative control), or 5 ng/ml TNF-α (positive control). Measurements were taken at the indicated time points using an EVOM resistance meter. (B) RTCA: HMVEC monolayers cultured on E-plate were incubated with 10 µg/ml of purified sNS1 from 16681 or D2Y98P strain, 10 µg/ml OVA (negative control), or 5 ng/ml TNF-α (positive control). Measurements were taken in real time every 15 s using an xCELLigence RTCA MP instrument. Data are representative of two independent experiments. Results were expressed as averages ± SD.
To determine whether the dispensable role of NS1 in D2Y98P-induced pathology is attributed to weaker ability of the protein to interact and disrupt the endothelium, we generated a chimeric virus consisting of D2Y98P expressing a vascular leak potent NS1 (Fig. 6 A). NS1 from D220 strain was selected, since this DENV2 strain was previously shown to depend on NS1 for its virulence and ability to cause vascular leak in vivo (Beatty et al., 2015). The in vitro fitness of D220-NS1-D2Y98P was comparable to WT D2Y98P strain, as evidenced by similar plaque sizes (Fig. S2 C) and growth kinetic profiles in BHK cells (Fig. S2 D). Viremia titers in AG129 mice infected with a sublethal dose of the virus strains (103 PFU/mouse) were also comparable, although a slight but significantly lower virus titer was measured in D220-NS1-D2Y98P–infected mice at day 4 p.i. (Fig. S2 E). These data thus indicate that swapping NS1 from D220 virus into the backbone of D2Y98P did not drastically affect the in vitro and in vivo fitness of the virus. Next, the pathogenic role of NS1(D220) in the context of D2Y98P infection was evaluated. Passive administration of NS1 immune serum effectively neutralized circulating sNS1 (Fig. 6 B) but did not protect against D220-NS1-D2Y98P challenge. Mice receiving NS1 immune serum displayed similar viremia titers (Fig. 6 C), disease manifestations (Fig. 6, D and E), and vascular leakage (Fig. 6 F) compared with the control group administered naive serum. These results indicate that expression of a vascular leak potent NS1 into the backbone of D2Y98P virus did not allow this protein to play a critical role in disease pathogenesis. Together, the data suggest that the in vivo fitness and virulence of D2Y98P virus is independent of the intrinsic ability of NS1 to increase vascular permeability and support that other viral determinants are likely involved.
Figure 6.
Passive NS1 immunity in an ADE model using D220-NS1-D2Y98P chimeric virus. (A) D220-NS1-D2Y98P chimeric virus consists of NS1 coding region from D220 strain (blue) in the backbone of D2Y98P strain (red). AG129 mice born to DENV1-immune mothers were passively transferred with NS1 immune serum as described in Fig. 2 A. 1 d after transfer, the mice were challenged s.c. with 103 PFU of chimeric D220-NS1-D2Y98P. (B and C) Systemic levels of sNS1 (B) and viremia titers (C) were measured at days 2, 4, and 6 p.i. (n = 5). (D and E) Mice (n = 8) were monitored daily upon challenge. Kaplan–Meier survival curve and clinical score are shown. (F) Vascular leakage was assessed in various tissues at day 6 p.i. (n = 5). Data are representative of at least two independent experiments. Data were analyzed by nonparametric Mann–Whitney U test. **, P < 0.01; ns, not significant. Results were expressed as averages ± SD (B and F).
In vivo virulence of D2Y98P is driven by structural prME region
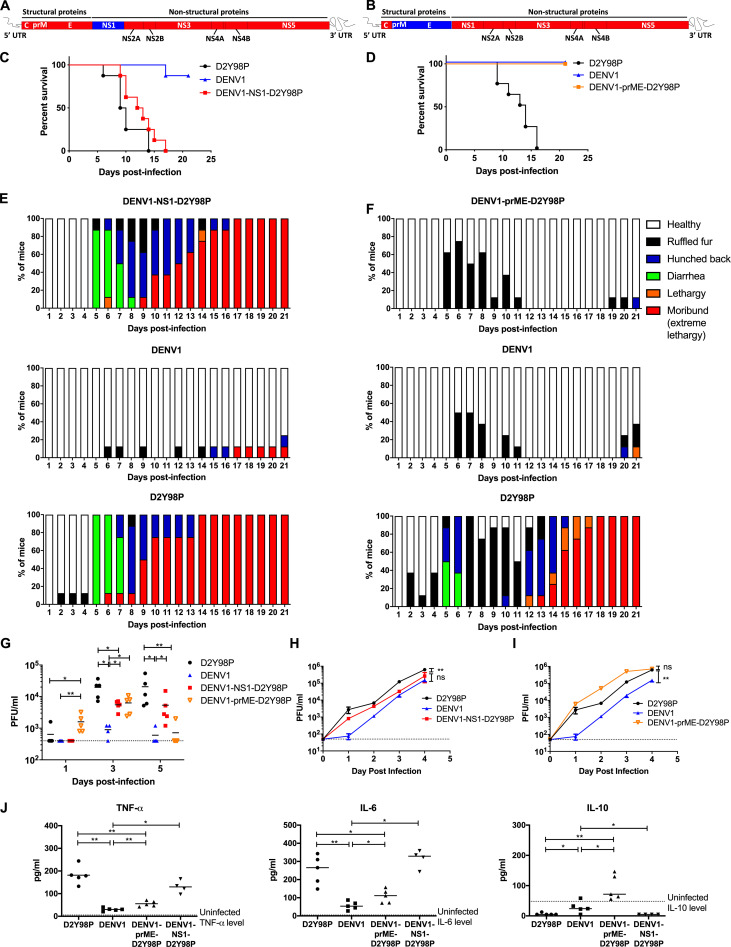
The in vivo fitness of DENV was reported to be linked to its organ and cell tropism, which is mainly mediated by the prME structural region that harbors the receptor-binding site (Modis et al., 2004; Prestwood et al., 2008). To explore the mechanisms involved in D2Y98P fitness and virulence, we generated a chimeric virus in which the prME region from D2Y98P was swapped with that of a DENV1 strain, 05K3903DK1, which typically gives rise to asymptomatic transient viremia in AG129 mice (Fig. 7 G). We also engineered another chimeric virus in which NS1 from D2Y98P was replaced by that of DENV1 (Fig. 7, A and B).
Figure 7.
In vivo and in vitro characterization of DENV1-NS1-D2Y98P and DENV1-prME-D2Y98P chimeric viruses. (A and B) DENV1-NS1-D2Y98P (A) or DENV1-prME-D2Y98P (B) chimeric strains consist of NS1 (A) or (B) coding region from DENV1 strain (blue) cloned into the backbone of D2Y98P strain (red). AG129 mice were infected s.c. with 106 PFU of D2Y98P, DENV1, DENV1-NS1-D2Y98P, or DENV1-prME-D2Y98P. Mice (n = 8) were monitored daily upon challenge. (C–F) Kaplan–Meier survival curves (C and D) and clinical scores (E and F) are shown. (G) Viremia titers were measured at days 1, 3, and 5 p.i. (n = 4). (H and I) In vitro virus titer in Vero cells infected with respective viruses. The viral kinetic curves were compared and analyzed by linear regression, which takes into consideration the slope and Y-intercept. (J) TNF-α, IL-6, and IL-10 systemic levels were measured at day 5 p.i. (n = 4–5). Data are representative of at least two independent experiments. Data between WT and chimeric viruses were compared and analyzed by nonparametric Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ns, not significant. Results were expressed as averages ± SD (H and I).
Mice infected with DENV1-NS1-D2Y98P displayed survival profiles, clinical manifestations, and disease progression that were comparable to those infected with WT D2Y98P virus (Fig. 7, C and E). On the other hand, mice infected with DENV1-prME-D2Y98P remained asymptomatic throughout the experiment, similar to WT DENV1 (Fig. 7, D and F). Interestingly, the peak viremia titer (day 3 p.i.) in mice infected with DENV1-prME-D2Y98P was similar to that measured in mice infected with DENV1-NS1-D2Y98P (Fig. 7 G), although the infection outcome induced by the two viruses differed drastically. Together, these observations support that prME, but not NS1, drives D2Y98P virulence in mice. Swapping either NS1 or prME from DENV1 into D2Y98P did significantly impact the viremia titers (Fig. 7 G), suggesting that these proteins play a role in viral replication. The role of NS1 in DENV replication has indeed been well established (Fan et al., 2014). Furthermore, previous work reported the existence of specific interactions between NS1 and prM/E that facilitate membrane budding or conformational changes in the structural proteins necessary for formation of nucleocapsids (Scaturro et al., 2015). This suggests that some compatibility between the two proteins is required for optimal viral replication. Consistently, when comparing the kinetic profile in Vero cells of D2Y98P with that of DENV1-NS1-D2Y98P, a mild but significant defect could be observed with the chimeric strain (Fig. 7 H). In contrast, the in vitro kinetic profile of DENV1-prME-D2Y98P was superimposable to that of WT D2Y98P (Fig. 7 I). This latter observation implied that the lower viremia titers measured in mice infected with DENV1-prME-D2Y98P cannot be explained by an intrinsic defect in viral replication.
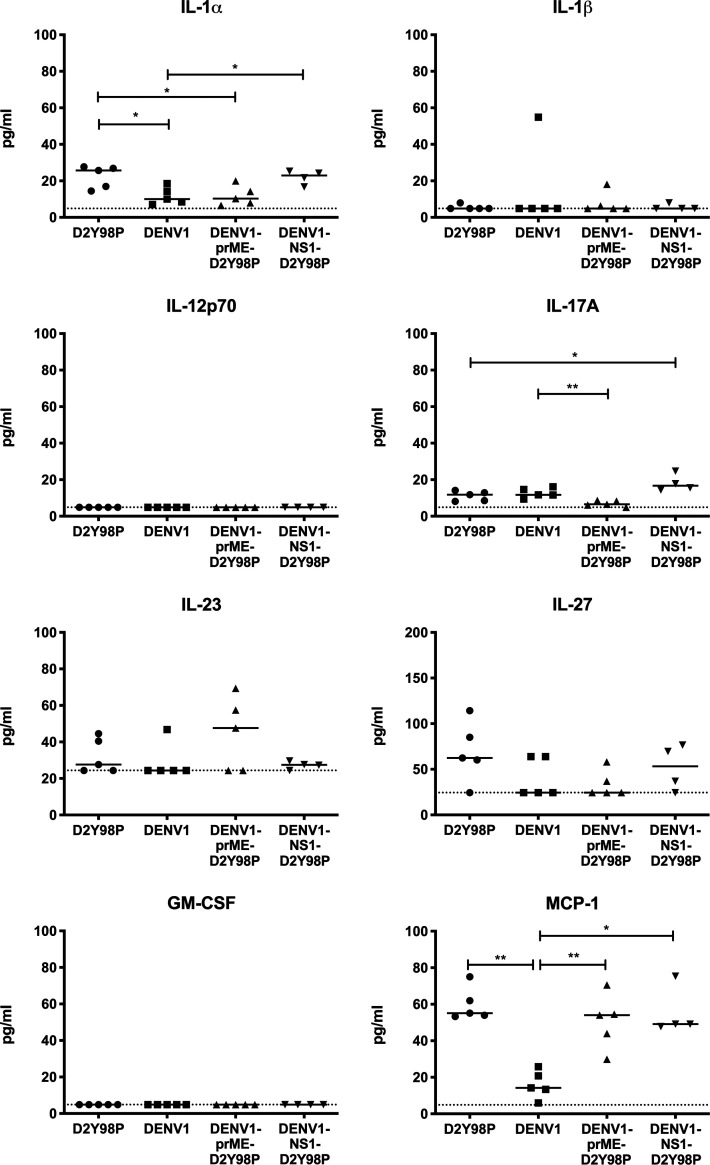
To further understand the basis of the prME-driven virulence of D2Y98P, we examined the cytokine profile in mice infected with the chimeric and WT strains (Fig. 7 J and Fig. S5). Comparable high levels of key proinflammatory cytokines TNF-α and IL-6 were measured in mice infected with DENV1-NS1-D2Y98P and WT D2Y98P (Fig. 7 J). On the other hand, mice infected with DENV1-prME-D2Y98P displayed significantly reduced levels of these proinflammatory cytokines compared with D2Y98P-infected mice, which were nonetheless higher than the levels measured in DENV1-infected animals (Fig. 7 J). Furthermore, DENV1-prME-D2Y98P was unable to suppress the production of antiinflammatory IL-10 (Fig. 7 J). Therefore, our data suggest that the in vivo virulence of D2Y98P virus is mainly driven by its structural component, prME, through its ability to induce the production of key proinflammatory cytokines.
Figure S5.
Cytokine profile in AG129 mice infected with chimeric and parental viruses. AG129 mice (n = 4–5) were infected s.c. with 106 PFU of D2Y98P, DENV1, DENV1-prME-D2Y98P, or DENV1-NS1-D2Y98P virus. At day 5 p.i., the systemic levels of various cytokines was measured. Data were analyzed by nonparametric Mann–Whitney U test. *, P < 0.05; **, P < 0.05.
Discussion
The mechanisms that drive the fitness and virulence of DENV remain largely unknown and are likely to be multifactorial. They include the ability to infect and replicate effectively in various host cells as well as the ability to evade or counteract the host defenses (Muñoz-Jordan et al., 2003; Mukhopadhyay et al., 2005; Muñoz-Jordán et al., 2005; Hsieh et al., 2011, 2014).
Beyond its role in DENV intracellular replication, recent literature has reported a critical role of NS1 in dengue pathogenesis by interacting with the endothelium and inducing vascular leakage, a clinical feature of severe dengue (Beatty et al., 2015; Modhiran et al., 2015). Consequently, NS1 immunity was found to protect against dengue disease (Beatty et al., 2015; Costa et al., 2006; Lai et al., 2017; Wan et al., 2014, 2017). These in vivo studies were conducted with DENV clinical isolates or mouse-adapted strains which either required very high dosages to induce lethality (DENV2-454009A, D220, and DENV2-3295) or induced only mild clinical symptoms (DENV2-16681, DENV1-8700828, DENV3-8700829, DENV4-59201818, and DENV2-RJ; Orozco et al., 2012; Beatty et al., 2015; Costa et al., 2006; Lai et al., 2017; Wan et al., 2014, 2017; Chan et al., 2019), thus implying that the in vivo fitness of these DENV strains is rather poor.
Here, we used a non–mouse-adapted DENV2 strain (D2Y98P) that is highly virulent in mice (Tan et al., 2010; Ng et al., 2014; Martínez Gómez et al., 2016). In that context, and using several symptomatic models in different mouse backgrounds, we did not observe a role for circulating sNS1 in dengue pathogenesis, neither could we see a protective role for NS1 immunity. Similar observations were made with another DENV2 clinical isolate that currently circulates in Singapore and Malaysia, and that is equally virulent in mice.
Impaired ability to interact with and disrupt the vascular endothelium may explain the dispensability of NS1 in D2Y98P-induced disease. The in vitro permeability assays conducted (TEER and RTCA) led to different outcomes and did not allow us to conclude whether NS1 produced by D2Y98P virus is less potent than NS1(16681) in increasing endothelial permeability. Regardless, upon introducing vascular leak potent NS1(D220) into the backbone of D2Y98P, we still did not observe a role for NS1 in disease pathogenesis. This latter approach therefore supported that the virulence and fitness of D2Y98P virus is independent of NS1 intrinsic ability to induce vascular permeability and is driven by other viral determinants. We propose that the role of NS1 in disease pathogenesis may be limited to strains of relatively poor fitness and upon mouse adaptation.
Consistently, the role of NS1 in human dengue disease has remained controversial. A hospital-based study in dengue patients reported a positive association between circulating sNS1 levels and dengue disease severity and proposed that levels >600 ng/ml are predictive of severe dengue (Libraty et al., 2002). However, a closer examination of the data revealed that the correlation was not absolute: a proportion of patients with self-limiting dengue fever displayed levels of sNS1 >600 ng/ml, and conversely, a subset of patients who developed severe dengue had sNS1 levels below the detectable range or considerably lower than 600 ng/ml (Libraty et al., 2002). Furthermore, a more recent prospective study conducted in Vietnam found no association between sNS1 level and severe dengue (Fox et al., 2011). All these observations therefore suggest that the correlation between sNS1 levels and disease severity is not absolute and that sNS1 levels may be strain dependent. A separate prospective study indeed reported that the level of circulating sNS1 produced in infected individuals differs greatly across DENV serotypes and even among strains of the same DENV2 serotype (Duyen et al., 2011).
Currently, little is known about the viral determinants that influence DENV virulence in humans. A number of studies have reported that some lineages seem to be more virulent than others, based on their ability to replicate in the host and give high viremia titers (Rico-Hesse et al., 1997; Leitmeyer et al., 1999; Armstrong and Rico-Hesse, 2001; Cologna et al., 2005). DENV strains with greater replicative ability in their host are thought to spread more rapidly and successfully than those with lower fitness (Guzman and Harris, 2015; Cologna et al., 2005). As an NS protein that plays a critical role in viral replication, NS1 has been naturally proposed to be a key determinant in driving the fitness of DENV, and studies have shown that single amino acid substitutions within NS1 could affect viral production and consequently disease severity (Rodriguez-Roche et al., 2005, 2011; Chan et al., 2019). Other mechanisms have also been proposed and include the ability to evade, deceive, or interfere with the host defenses, through production of subgenomic viral RNA (Finol and Ooi, 2019), immature viral particles (Rodenhuis-Zybert et al., 2010), or specific interactions between viral and host proteins (Muñoz-Jordan et al., 2003; Muñoz-Jordán et al., 2005).
Our data support that prME drives the in vivo virulence of D2Y98P. As prM and E form the outer protein shell of the virion, these proteins have been found to play a critical role in receptor binding and entry into the host cell, thereby driving virus tropism (Mukhopadhyay et al., 2005; Hsieh et al., 2011, 2014). Specifically, the two N-linked glycans on E protein at positions 67 and 153 have been shown to be crucial for binding to DC-SIGN (dendritic cell–specific intercellular adhesion molecule-3–grabbing nonintegrin) and mannose receptors (Pokidysheva et al., 2006; Tassaneetrithep et al., 2003; Miller et al., 2008). Although both N-glycosylation sites are conserved among DENV strains, the sugar composition and branching structure present at each site differ among serotypes and even among strains within the same serotype (Yap et al., 2017). The sugar composition at the surface of the viral particles is likely to impact binding efficacy of the virus to its lectin-type host receptors, thereby influencing cell and organ tropism. In addition to receptor binding, glycans on E protein may play an important role in shaping the host immune responses, through modulation of the lectin pathway of complement activation, and induction of proinflammatory cytokines through C-type lectin domain family 5 member A (CLEC5A) receptor activation (Chen et al., 2008). Using a chimerization approach, we showed that swapping the prME region from DENV1 into D2Y98P virus did not modify the in vitro viral replication rate but significantly impaired the induction of proinflammatory cytokines in vivo, which resulted in asymptomatic disease. We have indeed previously shown that proinflammatory cytokines, in particular TNFα, play a critical role in D2Y98P-induced disease, whereby administration of anti-TNFα antibodies partially or completely protected mice from succumbing to the infection (Ng et al., 2014; Martínez Gómez et al., 2016). Whether the glycan structures on (DENV1)E protein differ from those found on (D2Y98P)E and are responsible for the phenotype observed remains to be determined experimentally and is beyond the scope of this study. It is interesting to note, however, that the prME region from DENV2 Singapore clinical isolate, EHIE2862Y15, shares 100% amino acid identity with the prME of D2Y98P.
In conclusion, our work supports that the pathogenic role of sNS1 and consequently the protective efficacy of NS1 immunity are not seen in the context of infection with virulent DENV2 strains such as D2Y98P and EHIE2862Y15, whose virulence relies on their structural components. Hence, we propose that the pathogenic role of sNS1 is likely to be DENV strain dependent. Given the highly diverse and vast number of DENV strains circulating in the population (Kyle and Harris, 2008; Lee et al., 2012; Hapuarachchi et al., 2016), disease pathology in infected individuals could be driven by different key viral determinants, with sNS1 playing a major or minor role. Hence, it is critical to reevaluate the protective potential of NS1 vaccination against a variety of circulating DENV strains to avoid repeating the Dengvaxia scenario.
Materials and methods
Ethics statement
All the animal experiments were carried out in accordance with the guidelines of the National Advisory Committee for Laboratory Animal Research. Animal facilities are licensed by the regulatory body Agri-Food and Veterinary Authority of Singapore. The described animal experiments were approved by the Institutional Animal Care and Use Committee from National University of Singapore (NUS) under protocol numbers R14-0992 and R16-0422.
Cell lines and viruses
C6/36 Aedes albopictus cell line (ATCC; CRL-1660) was maintained in Leibotvitz’s L-15 medium (Gibco) supplemented with 10% FBS (Gibco) at 28°C. Baby hamster kidney-21 (BHK-21; ATCC; CCL-10) cell line was maintained in RPMI 1640 (Gibco) supplemented with 10% FBS and cultured at 37°C with 5% CO2. African green monkey kidney epithelial (Vero; ATCC; CCL-81) cell line was maintained in DMEM (Gibco) supplemented with 10% FBS and cultured at 37°C with 5% CO2. Human dermal blood microvascular endothelial cells (HMVECs; Lonza; CC-2811) were maintained in EGM-2MV BulletKit medium (Lonza) at 37°C with 5% CO2. DENV1 (Dengue 1 05K3903DK; GenBank accession no. EU081242) was isolated during the 2005 dengue outbreak in Singapore. DENV2 (Dengue D2Y98P; GenBank accession no. JF327392) derives from a clinical strain isolated in Singapore in 1998 that had been exclusively passaged in C6/36 cells and plaque purified twice in BHK-21 cells. EHIE2862Y15 (GenBank accession no. MK513444) was a DENV2 clinical strain isolated in Singapore in 2015. All DENV stocks were propagated in C6/36 cell line maintained in Leibovitz’s L-15 medium supplemented with 2% FBS as previously described (Ng et al., 2014). Harvested culture supernatants containing the virus particles were stored at −80°C. Virus titers were determined by plaque assay in BHK-21 cells as described below.
Virus quantification
Virus titer was quantified by plaque assay in BHK-21 cells as previously described (Ng et al., 2014). Briefly, 40,000 cells/well were seeded in 24-well plates (Nunc) 1 d before plaque assay. Cell monolayers were then infected with 10-fold serially diluted viral suspensions in RPMI 1640 supplemented with 2% FBS. After 1-h incubation at 37°C with CO2, overlay medium (RPMI 1640 containing 1% [wt/vol] carboxymethyl cellulose and 2% FBS) was added to each well. After incubation for 4 d (DENV2) or 5 d (DENV1) at 37°C with CO2, cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) and stained with 0.05% crystal violet (Sigma-Aldrich). Plaques were counted, adjusted by dilution, and expressed as the number of plaque forming units per milliliter.
Chimeric virus construction and production
Fragments from D2Y98P, D220 (GenBank accession no. HQ541799), and DENV1 (05K3903DK1) genomes were obtained by PCR amplification or de novo synthesis. Primer sequences used for amplification are listed in Table S4. PCR was performed using Q5 Hot Start high-fidelity DNA polymerase (New England Biolabs) according to the manufacturer’s instructions. All PCR products were purified from agarose gel using QIAquick gel extraction kit (Qiagen) following the manufacturer’s instructions. D220-NS1-D2Y98P, DENV1-NS1-D2Y98P, and DENV1-prME-D2Y98P chimeric viruses were constructed using DNA assembly of PCR fragments by Gibson assembly technique as previously described (Siridechadilok et al., 2013). The pACYC177 plasmid containing CMV promoter, hepatitis delta virus ribozyme, and SV40 poly(A) signal was a kind gift from Ooi Eng Eong (Duke-NUS). The vector and viral DNA fragments were assembled using Gibson Assembly Master Mix (New England Biolabs) at 50°C for 2 h. The assembly mix (15 µl) was then directly transfected into BHK-21 cells using Lipofectamine-2000 (Thermo Fisher Scientific) in Opti-MEM medium (Thermo Fisher Scientific) as described in the manufacturer’s protocol. After incubation for 4 h at 37°C with CO2, the medium was changed to MEM medium (Gibco) supplemented with 10% FBS. Culture supernatants containing the virus were collected daily from day 1 to day 5 after transfection, and the presence of virus was verified by plaque assay. The chimeric viruses were eventually propagated in C6/36 cell line for two passages.
NS1 immunization and mouse infection
5–6-wk-old A129 mice (129/Sv mice deficient in IFN-α/β receptors) were immunized i.p. three times (weeks 0, 2, and 6) with 20 µg of hexameric NS1 from DENV2 16681 strain (The Native Antigen Company) or OVA protein (InvivoGen) adjuvanted with 1 µg of monophosphoryl lipid A (MPLA; InvivoGen) and 1:1 volume AddaVax (InvivoGen) as described previously (Beatty et al., 2015). 2 wk after the last administration, A129 mice were challenged with 106 PFU DENV2 (D2Y98P) i.v. 1 d after i.v. administration of DENV1-immune serum to trigger ADE.
Following the same NS1(16681) immunization regimen in A129 mice, NS1 immune serum was collected 2 wk after the third immunization and heat inactivated at 56°C for 30 min before storage at −80°C. Antibody titers of immune serum were quantified by ELISA. The stored immune sera were used for passive transfer experiment into AG129 mice (129/Sv mice deficient in IFN-α/β and IFN-γ receptors) born to DENV1-immune mothers. 1 d after administration of NS1 immune serum (150 µl per mouse), these AG129 mice were s.c. challenged with 103 PFU of DENV2 (D2Y98P or chimeric D220-NS1-D2Y98P).
5–7-wk-old IFNAR−/− mice (C57BL/6 deficient in IFN-α/β receptors) were subjected to the same immunization regimen with NS1(16681) or OVA protein as described above, and were challenged s.c. with 105 PFU DENV2 (D2Y98P or EHIE2862Y15) 2 wk after the third immunization.
Administration of NS1 protein
5–6-wk-old AG129 mice were infected s.c. with 103 PFU (sublethal dose) of D2Y98P. 2 or 4 d p.i., 10 mg/kg of purified NS1(D2Y98P) (The Native Antigen Company) or OVA protein (InvivoGen) was administered i.v. to the infected mice.
Primary infection with chimeric viruses
5–6-wk-old AG129 mice were infected s.c. with 106 PFU of parental (D2Y98P or DENV1) or chimeric (DENV1-NS1-D2Y98P or DENV1-prME-D2Y98P) viruses.
Quantification of vascular leakage
Vascular leakage was assessed by Evans blue dye extravasation as previously described (Ng et al., 2014). Briefly, mice were i.v. administered 0.5% (wt/vol) Evans blue dye (Sigma-Aldrich) in PBS adjusted to the weight of the mouse (10 µl/g body weight). 2 h after administration, mice were euthanized and extensively perfused with PBS. Organs (liver, small intestines, spleen, and kidneys) were harvested and weighed. Evans blue dye was extracted from the organs by the addition of N,N-dimethylformamide (Sigma-Aldrich; adjusted to 4 ml/g wet tissue) and incubated overnight at 37°C. Extracted dye was read at 620 nm, and data were expressed as absolute absorbance.
Histology analysis
Mice were sacrificed at the indicated time points, and the liver was harvested and fixed immediately with 4% paraformaldehyde in PBS. Fixed tissues were processed for embedding, sectioning and staining with H&E (Department of Pathology, NUS). Slides were viewed under a microscope (Leica), and images were captured.
ELISAs
Levels of systemic IgG antibodies specific to NS1 protein were quantified via indirect ELISA. Purified NS1 protein from D2Y98P or 16681 (10 ng/well) diluted in PBS was coated onto 96-well enzyme immunoassay plates (Corning Costar) overnight at 4°C. Plates were washed three times with wash buffer (0.05% Tween 20 in PBS) and blocked with reagent diluent (2% BSA in wash buffer) for 1 h at 37°C. Serially diluted serum samples were added to the wells and incubated at 37°C. Plates were washed three times before the addition of HRP-conjugated anti-mouse IgG (H+L; Bio-Rad; 170-6516) at 1:3,000 and anti-mouse IgG1, IgG2a, and IgG2b (Abcam ab97240, ab97245, and ab97250) at 1:10,000. Plates were incubated for 1 h at 37°C. After the final three washes, detection was performed by the addition of o-phenylene-diamine dihydrochloride substrate SigmaFast (Sigma-Aldrich) and incubated for 30 min at room temperature. The reaction was stopped upon the addition of 2 N H2SO4. Absorbance was read at 490 nm, and antibody titer was determined by nonlinear regression as the reciprocal of the highest serum dilution with absorbance corresponding to three times the absorbance of blank wells.
Levels of systemic sNS1 in mice during the course of infection or NS1 in in vitro experiments were quantified via sandwich ELISA as described previously (Watanabe et al., 2012). Mouse anti-NS1 Mab62.1 (a kind gift from Subhash Vasudevan, Duke-NUS, and Christiane Ruedl, Nanyang Technological University; 0.1 µg/well) diluted in PBS was coated onto 96-well enzyme immunoassay plates overnight at 4°C. After washing and blocking as described above, diluted serum samples (from 1:100 to 1:2,500) were added to the wells and incubated for 2.5 h at 37°C. A standard curve was established by twofold serial dilution of recombinant NS1 (D2Y98P or 16681) from 25 to 0.39 ng/ml. Plates were washed five times before the addition of HRP-conjugated mouse anti-NS1 Mab56.2 (a kind gift from Subhash Vasudevan and Christiane Ruedl; 25 ng/well), made by conjugating HRP to Mab56.2 using NH2 peroxidase labeling kit (Abnova) and incubated for 1.5 h at room temperature. After the final washes, detection was performed with the addition of tetramethylbenzidine (R&D Systems) for 30 min at room temperature. The reaction was stopped with 2 N H2SO4. Absorbance was read at 450 nm. The concentration of NS1 was calculated based on the standard curve and expressed as the concentration of NS1 in nanograms per milliliter.
Levels of circulating TNF-α in infected mice were measured using the Mouse TNF-α Immunoassay Quantikine ELISA kit (R&D Systems) according to the manufacturer’s instructions. Levels of heparan sulfate in circulation were quantified using Mouse Heparan Sulfate ELISA kit (G-Biosciences) according to the manufacturer’s instructions.
Multiplex cytokine and soluble mediator detection
Levels of cytokines and soluble mediators present in plasma of infected mice were quantified using LEGENDplex Mouse Inflammatory Panel kit (BioLegend) according to the manufacturer’s protocol. Samples were run using Attune Nxt flow cytometer and analyzed using FlowJo software.
In vitro permeability assays
To test the ability of NS1 to induce endothelial permeability in vitro, a TEER assay was conducted as described previously (Beatty et al., 2015). HMVECs (50,000 cells/well) were seeded onto 24-well Transwell insert membranes (PET, 0.4-µm pore size; Millipore Millicell) coated with 50 µg/ml collagen (Corning). Lower and upper chambers were filled with EGM-2MV BulletKit medium and cultured for 2 d at 37°C with 5% CO2. 10 µg/ml of purified sNS1 from D2Y98P or 16681 strain diluted with culture medium was added onto the upper chamber of the Transwell, while the lower chamber was replaced with fresh culture medium. TEER was measured at 3-h intervals using an EVOM2 Epithelial Volt/Ohm meter (World Precision Instruments). 10 µg/ml of OVA protein and 5 ng/ml of TNF-α (R&D Systems) were used as negative and positive controls, respectively. Untreated Transwell inserts seeded with HMVECs and inserts containing medium only were also included. Relative TEER was expressed as follows: [resistance (treatment) − resistance (medium only)]/[resistance (untreated cells) − resistance (medium only)].
The xCELLigence RTCA system (ACEA, Biosciences) was also used to measure the ability of NS1 to disrupt endothelial monolayers. HMVECs (50,000 cells/well) were seeded onto 96-well E-plates (ACEA, Biosciences). Seeded cells were placed into the RTCA station for real-time measurement of electrical impedance. When a confluent monolayer formed, cells were treated with 10 µg/ml of purified sNS1 from D2Y98P or 16681 strain. OVA (10 µg/ml) and TNFα (5 ng/ml) were included as negative and positive controls, respectively. Electrical impedance reflecting the permeability of the endothelial monolayer was monitored in real time using the RTCA MP system, with readings taken every 15 s. Readings were expressed as baseline normalized cell index by normalizing the cell index measured at the time of treatment and setting untreated control as the baseline.
In vitro DENV infection
Vero cells were infected at a multiplicity of infection of 0.1 with D2Y98P, DENV1, DENV1-prME-D2Y98P, and DENV1-NS1-D2Y98P. Plates were incubated at 37°C for 1 h with rocking every 15 min for viral adsorption. Each well was rinsed twice with PBS before addition of 200 µl of DMEM containing 2% FBS. The plates were incubated for 4 d, and the culture supernatant was collected at indicated time points p.i. Viral quantification was performed by plaque assay.
Statistical analysis
Data analyses were performed using Graphpad Prism 6.0. Statistical comparison was conducted using nonparametric Mann–Whitney U test or linear regression. Comparison of survival rates was performed using log-rank (Mantel–Cox) test. Differences were considered significant (*) at P < 0.05.
Online supplemental material
Fig. S1 shows the characterization of the NS1 polyclonal immune serum generated upon immunization with purified NS1 protein. Fig. S2 shows the in vivo and in vitro fitness of EHIE2862Y15 strain and chimeric D220-NS1-D2Y98P in AG129 mice. Fig. S3 displays the viremia titers in D2Y98P-infected AG129 mice administered NS1. Fig. S4 shows the results of NS1-induced permeability in vitro assays. Fig. S5 presents the cytokine profile measured in AG129 mice infected with D2Y98P, DENV1, DENV1-prME-D2Y98P, or DENV1-NS1-D2Y98P virus. Table S1 shows the comparison of D2Y98P genome against DENV2 NGC strain and Singapore DENV2 clinical isolates. Table S2 compares NS1 amino acid sequence between D2Y98P, D220, and 16681 strains. Table S3 highlights the amino acid differences between EHIE2862Y15 and D2Y98P strains. Table S4 compiles the primer sequences used for PCR amplification.
Supplementary Material
shows a comparison of D2Y98P genome against DENV2 NGC strain and Singapore DENV2 clinical isolates.
shows an NS1 amino acid sequence comparison between D2Y98P, D220, and 16681 strains.
shows an amino acid sequence comparison between EHIE2862Y15 and D2Y98P strains.
lists primers used for PCR amplification.
Acknowledgments
We thank Subhash Vasudevan (Duke-NUS) and Christiane Ruedl (Nanyang Technological University) for the kind gift of mouse monoclonal anti-NS1 antibodies for ELISA. We also thank Ooi Eng Eong and Antonio Bertoletti (Duke-NUS) for sharing pACYC177 plasmid and the use of xCELLigence equipment for RTCA experiments, respectively.
This work was funded by the Ministry of Health - Singapore (CBRG13nv005 and MOHIAFCAT2/3/011 to S. Alonso).
Author contributions: P.X. Lee, D.H.R. Ting, C.P.H. Boey, E.T.X. Tan, J.Z.H. Chia, F. Idris, Y. Oo, L.C. Ong, Y.L. Chua, and C. Hapuarachchi performed the experiments; P.X. Lee, D.H.R. Ting, and S. Alonso designed the experiments and wrote the manuscript; P.X. Lee, D.H.R. Ting, C. Hapuarachchi, L.C. Ng, and S. Alonso analyzed the data.
References
- Aguiar M., Stollenwerk N., and Halstead S.B.. 2016. The risks behind Dengvaxia recommendation. Lancet Infect. Dis. 16:882–883. 10.1016/S1473-3099(16)30168-2 [DOI] [PubMed] [Google Scholar]
- Alcon S., Talarmin A., Debruyne M., Falconar A., Deubel V., and Flamand M.. 2002. Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J. Clin. Microbiol. 40:376–381. 10.1128/JCM.40.02.376-381.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Añez G., Heisey D.A., Volkova E., and Rios M.. 2016. Complete Genome Sequences of Dengue Virus Type 1 to 4 Strains Used for the Development of CBER/FDA RNA Reference Reagents and WHO International Standard Candidates for Nucleic Acid Testing. Genome Announc. 4 e01583-15 10.1128/genomeA.01583-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong P.M., and Rico-Hesse R.. 2001. Differential susceptibility of Aedes aegypti to infection by the American and Southeast Asian genotypes of dengue type 2 virus. Vector Borne Zoonotic Dis. 1:159–168. 10.1089/153036601316977769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty P.R., Puerta-Guardo H., Killingbeck S.S., Glasner D.R., Hopkins K., and Harris E.. 2015. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci. Transl. Med. 7 304ra141 10.1126/scitranslmed.aaa3787 [DOI] [PubMed] [Google Scholar]
- Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O., et al. 2013. The global distribution and burden of dengue. Nature. 496:504–507. 10.1038/nature12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capeding M.R., Tran N.H., Hadinegoro S.R., Ismail H.I., Chotpitayasunondh T., Chua M.N., Luong C.Q., Rusmil K., Wirawan D.N., Nallusamy R., et al. ; CYD14 Study Group . 2014. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet. 384:1358–1365. 10.1016/S0140-6736(14)61060-6 [DOI] [PubMed] [Google Scholar]
- Chan K.W., Watanabe S., Kavishna R., Alonso S., and Vasudevan S.G.. 2015. Animal models for studying dengue pathogenesis and therapy. Antiviral Res. 123:5–14. 10.1016/j.antiviral.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Chan K.W.K., Watanabe S., Jin J.Y., Pompon J., Teng D., Alonso S., Vijaykrishna D., Halstead S.B., Marzinek J.K., Bond P.J., et al. 2019. A T164S mutation in the dengue virus NS1 protein is associated with greater disease severity in mice. Sci. Transl. Med. 11 eaat7726 10.1126/scitranslmed.aat7726 [DOI] [PubMed] [Google Scholar]
- Chen H.R., Chuang Y.C., Lin Y.S., Liu H.S., Liu C.C., Perng G.C., and Yeh T.M.. 2016. Dengue Virus Nonstructural Protein 1 Induces Vascular Leakage through Macrophage Migration Inhibitory Factor and Autophagy. PLoS Negl. Trop. Dis. 10 e0004828 10.1371/journal.pntd.0004828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.T., Lin Y.L., Huang M.T., Wu M.F., Cheng S.C., Lei H.Y., Lee C.K., Chiou T.W., Wong C.H., and Hsieh S.L.. 2008. CLEC5A is critical for dengue-virus-induced lethal disease. Nature. 453:672–676. 10.1038/nature07013 [DOI] [PubMed] [Google Scholar]
- Chuansumrit A., Chaiyaratana W., Tangnararatchakit K., Yoksan S., Flamand M., and Sakuntabhai A.. 2011. Dengue nonstructural protein 1 antigen in the urine as a rapid and convenient diagnostic test during the febrile stage in patients with dengue infection. Diagn. Microbiol. Infect. Dis. 71:467–469. 10.1016/j.diagmicrobio.2011.08.020 [DOI] [PubMed] [Google Scholar]
- Cologna R., Armstrong P.M., and Rico-Hesse R.. 2005. Selection for virulent dengue viruses occurs in humans and mosquitoes. J. Virol. 79:853–859. 10.1128/JVI.79.2.853-859.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S.M., Paes M.V., Barreto D.F., Pinhão A.T., Barth O.M., Queiroz J.L., Armôa G.R., Freire M.S., and Alves A.M.. 2006. Protection against dengue type 2 virus induced in mice immunized with a DNA plasmid encoding the non-structural 1 (NS1) gene fused to the tissue plasminogen activator signal sequence. Vaccine. 24:195–205. 10.1016/j.vaccine.2005.07.059 [DOI] [PubMed] [Google Scholar]
- Duyen H.T., Ngoc T.V., Ha T., Hang V.T., Kieu N.T., Young P.R., Farrar J.J., Simmons C.P., Wolbers M., and Wills B.A.. 2011. Kinetics of plasma viremia and soluble nonstructural protein 1 concentrations in dengue: differential effects according to serotype and immune status. J. Infect. Dis. 203:1292–1300. 10.1093/infdis/jir014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Bray M., Schlesinger J.J., and Lai C.J.. 1990. Immunization of mice with recombinant vaccinia virus expressing authentic dengue virus nonstructural protein NS1 protects against lethal dengue virus encephalitis. J. Virol. 64:4356–4363. 10.1128/JVI.64.9.4356-4363.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Liu Y., and Yuan Z.. 2014. Critical role of Dengue Virus NS1 protein in viral replication. Virol. Sin. 29:162–169. 10.1007/s12250-014-3459-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima K., and Syed N.I.. 2018. Dengvaxia controversy: impact on vaccine hesitancy. J. Glob. Health. 8 010312 10.7189/jogh.08.020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finol E., and Ooi E.E.. 2019. Evolution of Subgenomic RNA Shapes Dengue Virus Adaptation and Epidemiological Fitness. iScience. 16:94–105. 10.1016/j.isci.2019.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox A., Le N.M., Simmons C.P., Wolbers M., Wertheim H.F., Pham T.K., Tran T.H., Trinh T.M., Nguyen T.L., Nguyen V.T., et al. 2011. Immunological and viral determinants of dengue severity in hospitalized adults in Ha Noi, Viet Nam. PLoS Negl. Trop. Dis. 5 e967 10.1371/journal.pntd.0000967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves A.J., Oliveira E.R., Costa S.M., Paes M.V., Silva J.F., Azevedo A.S., Mantuano-Barradas M., Nogueira A.C., Almeida C.J., and Alves A.M.. 2015. Cooperation between CD4+ T Cells and Humoral Immunity Is Critical for Protection against Dengue Using a DNA Vaccine Based on the NS1 Antigen. PLoS Negl. Trop. Dis. 9 e0004277 10.1371/journal.pntd.0004277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant D., Tan G.K., Qing M., Ng J.K., Yip A., Zou G., Xie X., Yuan Z., Schreiber M.J., Schul W., et al. 2011. A single amino acid in nonstructural protein NS4B confers virulence to dengue virus in AG129 mice through enhancement of viral RNA synthesis. J. Virol. 85:7775–7787. 10.1128/JVI.00665-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsche I., Coulibaly F., Voss J.E., Salmon J., d’Alayer J., Ermonval M., Larquet E., Charneau P., Krey T., Mégret F., et al. 2011. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc. Natl. Acad. Sci. USA. 108:8003–8008. 10.1073/pnas.1017338108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman M.G., and Harris E.. 2015. Dengue. Lancet. 385:453–465. 10.1016/S0140-6736(14)60572-9 [DOI] [PubMed] [Google Scholar]
- Halstead S.B. 2003. Neutralization and antibody-dependent enhancement of dengue viruses. Adv. Virus Res. 60:421–467. 10.1016/S0065-3527(03)60011-4 [DOI] [PubMed] [Google Scholar]
- Halstead S.B. 2017. Dengvaxia sensitizes seronegatives to vaccine enhanced disease regardless of age. Vaccine. 35:6355–6358. 10.1016/j.vaccine.2017.09.089 [DOI] [PubMed] [Google Scholar]
- Halstead S.B., Lan N.T., Myint T.T., Shwe T.N., Nisalak A., Kalyanarooj S., Nimmannitya S., Soegijanto S., Vaughn D.W., and Endy T.P.. 2002. Dengue hemorrhagic fever in infants: research opportunities ignored. Emerg. Infect. Dis. 8:1474–1479. 10.3201/eid0812.020170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang V.T., Nguyet N.M., Trung D.T., Tricou V., Yoksan S., Dung N.M., Van Ngoc T., Hien T.T., Farrar J., Wills B., et al. 2009. Diagnostic accuracy of NS1 ELISA and lateral flow rapid tests for dengue sensitivity, specificity and relationship to viraemia and antibody responses. PLoS Negl. Trop. Dis. 3 e360 10.1371/journal.pntd.0000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapuarachchi H.C., Koo C., Rajarethinam J., Chong C.S., Lin C., Yap G., Liu L., Lai Y.L., Ooi P.L., Cutter J., et al. 2016. Epidemic resurgence of dengue fever in Singapore in 2013-2014: A virological and entomological perspective. BMC Infect. Dis. 16:300 10.1186/s12879-016-1606-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henchal E.A., Henchal L.S., and Schlesinger J.J.. 1988. Synergistic interactions of anti-NS1 monoclonal antibodies protect passively immunized mice from lethal challenge with dengue 2 virus. J. Gen. Virol. 69:2101–2107. 10.1099/0022-1317-69-8-2101 [DOI] [PubMed] [Google Scholar]
- Hsieh S.C., Wu Y.C., Zou G., Nerurkar V.R., Shi P.Y., and Wang W.K.. 2014. Highly conserved residues in the helical domain of dengue virus type 1 precursor membrane protein are involved in assembly, precursor membrane (prM) protein cleavage, and entry. J. Biol. Chem. 289:33149–33160. 10.1074/jbc.M114.610428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh S.C., Zou G., Tsai W.Y., Qing M., Chang G.J., Shi P.Y., and Wang W.K.. 2011. The C-terminal helical domain of dengue virus precursor membrane protein is involved in virus assembly and entry. Virology. 410:170–180. 10.1016/j.virol.2010.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle J.L., and Harris E.. 2008. Global spread and persistence of dengue. Annu. Rev. Microbiol. 62:71–92. 10.1146/annurev.micro.62.081307.163005 [DOI] [PubMed] [Google Scholar]
- Lai Y.C., Chuang Y.C., Liu C.C., Ho T.S., Lin Y.S., Anderson R., and Yeh T.M.. 2017. Antibodies Against Modified NS1 Wing Domain Peptide Protect Against Dengue Virus Infection. Sci. Rep. 7:6975 10.1038/s41598-017-07308-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J.H., Ong L.C., and Alonso S.. 2016. Key concepts, strategies, and challenges in dengue vaccine development: an opportunity for sub-unit candidates? Expert Rev. Vaccines. 15:483–495. 10.1586/14760584.2016.1106318 [DOI] [PubMed] [Google Scholar]
- Lee K.S., Lo S., Tan S.S., Chua R., Tan L.K., Xu H., and Ng L.C.. 2012. Dengue virus surveillance in Singapore reveals high viral diversity through multiple introductions and in situ evolution. Infect. Genet. Evol. 12:77–85. 10.1016/j.meegid.2011.10.012 [DOI] [PubMed] [Google Scholar]
- Leitmeyer K.C., Vaughn D.W., Watts D.M., Salas R., Villalobos I., de Chacon C., Ramos C., and Rico-Hesse R.. 1999. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 73:4738–4747. 10.1128/JVI.73.6.4738-4747.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libraty D.H., Young P.R., Pickering D., Endy T.P., Kalayanarooj S., Green S., Vaughn D.W., Nisalak A., Ennis F.A., and Rothman A.L.. 2002. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis. 186:1165–1168. 10.1086/343813 [DOI] [PubMed] [Google Scholar]
- Martínez Gómez J.M., Ong L.C., Lam J.H., Binte Aman S.A., Libau E.A., Lee P.X., St John A.L., and Alonso S.. 2016. Maternal Antibody-Mediated Disease Enhancement in Type I Interferon-Deficient Mice Leads to Lethal Disease Associated with Liver Damage. PLoS Negl. Trop. Dis. 10 e0004536 10.1371/journal.pntd.0004536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.L., de Wet B.J., Martinez-Pomares L., Radcliffe C.M., Dwek R.A., Rudd P.M., and Gordon S.. 2008. The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog. 4 e17 10.1371/journal.ppat.0040017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modhiran N., Watterson D., Muller D.A., Panetta A.K., Sester D.P., Liu L., Hume D.A., Stacey K.J., and Young P.R.. 2015. Dengue virus NS1 protein activates cells via Toll-like receptor 4 and disrupts endothelial cell monolayer integrity. Sci. Transl. Med. 7 304ra142 10.1126/scitranslmed.aaa3863 [DOI] [PubMed] [Google Scholar]
- Modis Y., Ogata S., Clements D., and Harrison S.C.. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature. 427:313–319. 10.1038/nature02165 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Kuhn R.J., and Rossmann M.G.. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13–22. 10.1038/nrmicro1067 [DOI] [PubMed] [Google Scholar]
- Muller D.A., and Young P.R.. 2013. The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Res. 98:192–208. 10.1016/j.antiviral.2013.03.008 [DOI] [PubMed] [Google Scholar]
- Muñoz-Jordan J.L., Sánchez-Burgos G.G., Laurent-Rolle M., and García-Sastre A.. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA. 100:14333–14338. 10.1073/pnas.2335168100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Jordán J.L., Laurent-Rolle M., Ashour J., Martínez-Sobrido L., Ashok M., Lipkin W.I., and García-Sastre A.. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004–8013. 10.1128/JVI.79.13.8004-8013.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J.K., Zhang S.L., Tan H.C., Yan B., Martinez J.M., Tan W.Y., Lam J.H., Tan G.K., Ooi E.E., and Alonso S.. 2014. First experimental in vivo model of enhanced dengue disease severity through maternally acquired heterotypic dengue antibodies. PLoS Pathog. 10 e1004031 10.1371/journal.ppat.1004031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L.C., Chem Y.K., Koo C., Mudin R.N.B., Amin F.M., Lee K.S., and Kheong C.C.. 2015. 2013 dengue outbreaks in Singapore and Malaysia caused by different viral strains. Am. J. Trop. Med. Hyg. 92:1150–1155. 10.4269/ajtmh.14-0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco S., Schmid M.A., Parameswaran P., Lachica R., Henn M.R., Beatty R., and Harris E.. 2012. Characterization of a model of lethal dengue virus 2 infection in C57BL/6 mice deficient in the alpha/beta interferon receptor. J. Gen. Virol. 93:2152–2157. 10.1099/vir.0.045088-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeling R.W., Artsob H., Pelegrino J.L., Buchy P., Cardosa M.J., Devi S., Enria D.A., Farrar J., Gubler D.J., Guzman M.G., et al. 2010. Evaluation of diagnostic tests: dengue. Nat. Rev. Microbiol. 8(12, Suppl):S30–S38. 10.1038/nrmicro2459 [DOI] [PubMed] [Google Scholar]
- Pokidysheva E., Zhang Y., Battisti A.J., Bator-Kelly C.M., Chipman P.R., Xiao C., Gregorio G.G., Hendrickson W.A., Kuhn R.J., and Rossmann M.G.. 2006. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell. 124:485–493. 10.1016/j.cell.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Prestwood T.R., Prigozhin D.M., Sharar K.L., Zellweger R.M., and Shresta S.. 2008. A mouse-passaged dengue virus strain with reduced affinity for heparan sulfate causes severe disease in mice by establishing increased systemic viral loads. J. Virol. 82:8411–8421. 10.1128/JVI.00611-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerta-Guardo H., Glasner D.R., and Harris E.. 2016. Dengue Virus NS1 Disrupts the Endothelial Glycocalyx, Leading to Hyperpermeability. PLoS Pathog. 12 e1005738 10.1371/journal.ppat.1005738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerta-Guardo H., Glasner D.R., Espinosa D.A., Biering S.B., Patana M., Ratnasiri K., Wang C., Beatty P.R., and Harris E.. 2019. Flavivirus NS1 Triggers Tissue-Specific Vascular Endothelial Dysfunction Reflecting Disease Tropism. Cell Rep. 26:1598–1613.e8. 10.1016/j.celrep.2019.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Hesse R., Harrison L.M., Salas R.A., Tovar D., Nisalak A., Ramos C., Boshell J., de Mesa M.T., Nogueira R.M., and da Rosa A.T.. 1997. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology. 230:244–251. 10.1006/viro.1997.8504 [DOI] [PubMed] [Google Scholar]
- Rodenhuis-Zybert I.A., van der Schaar H.M., da Silva Voorham J.M., van der Ende-Metselaar H., Lei H.Y., Wilschut J., and Smit J.M.. 2010. Immature dengue virus: a veiled pathogen? PLoS Pathog. 6 e1000718 10.1371/journal.ppat.1000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Roche R., Alvarez M., Gritsun T., Halstead S., Kouri G., Gould E.A., and Guzman M.G.. 2005. Virus evolution during a severe dengue epidemic in Cuba, 1997. Virology. 334:154–159. 10.1016/j.virol.2005.01.037 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Roche R., Sanchez L., Burgher Y., Rosario D., Alvarez M., Kouri G., Halstead S.B., Gould E.A., and Guzman M.G.. 2011. Virus role during intraepidemic increase in dengue disease severity. Vector Borne Zoonotic Dis. 11:675–681. 10.1089/vbz.2010.0177 [DOI] [PubMed] [Google Scholar]
- Scaturro P., Cortese M., Chatel-Chaix L., Fischl W., and Bartenschlager R.. 2015. Dengue Virus Non-structural Protein 1 Modulates Infectious Particle Production via Interaction with the Structural Proteins. PLoS Pathog. 11 e1005277 10.1371/journal.ppat.1005277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger J.J., Brandriss M.W., and Walsh E.E.. 1987. Protection of mice against dengue 2 virus encephalitis by immunization with the dengue 2 virus non-structural glycoprotein NS1. J. Gen. Virol. 68:853–857. 10.1099/0022-1317-68-3-853 [DOI] [PubMed] [Google Scholar]
- Siridechadilok B., Gomutsukhavadee M., Sawaengpol T., Sangiambut S., Puttikhunt C., Chin-inmanu K., Suriyaphol P., Malasit P., Screaton G., and Mongkolsapaya J.. 2013. A simplified positive-sense-RNA virus construction approach that enhances analysis throughput. J. Virol. 87:12667–12674. 10.1128/JVI.02261-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan G.K., Ng J.K., Trasti S.L., Schul W., Yip G., and Alonso S.. 2010. A non mouse-adapted dengue virus strain as a new model of severe dengue infection in AG129 mice. PLoS Negl. Trop. Dis. 4 e672 10.1371/journal.pntd.0000672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassaneetrithep B., Burgess T.H., Granelli-Piperno A., Trumpfheller C., Finke J., Sun W., Eller M.A., Pattanapanyasat K., Sarasombath S., Birx D.L., et al. 2003. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 197:823–829. 10.1084/jem.20021840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Broek M.F., Müller U., Huang S., Aguet M., and Zinkernagel R.M.. 1995. Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J. Virol. 69:4792–4796. 10.1128/JVI.69.8.4792-4796.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar L., Dayan G.H., Arredondo-García J.L., Rivera D.M., Cunha R., Deseda C., Reynales H., Costa M.S., Morales-Ramírez J.O., Carrasquilla G., et al. ; CYD15 Study Group . 2015. Efficacy of a tetravalent dengue vaccine in children in Latin America. N. Engl. J. Med. 372:113–123. 10.1056/NEJMoa1411037 [DOI] [PubMed] [Google Scholar]
- Wan S.W., Chen P.W., Chen C.Y., Lai Y.C., Chu Y.T., Hung C.Y., Lee H., Wu H.F., Chuang Y.C., Lin J., et al. 2017. Therapeutic Effects of Monoclonal Antibody against Dengue Virus NS1 in a STAT1 Knockout Mouse Model of Dengue Infection. J. Immunol. 199:2834–2844. 10.4049/jimmunol.1601523 [DOI] [PubMed] [Google Scholar]
- Wan S.W., Lu Y.T., Huang C.H., Lin C.F., Anderson R., Liu H.S., Yeh T.M., Yen Y.T., Wu-Hsieh B.A., and Lin Y.S.. 2014. Protection against dengue virus infection in mice by administration of antibodies against modified nonstructural protein 1. PLoS One. 9 e92495 10.1371/journal.pone.0092495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Tan K.H., Rathore A.P., Rozen-Gagnon K., Shuai W., Ruedl C., and Vasudevan S.G.. 2012. The magnitude of dengue virus NS1 protein secretion is strain dependent and does not correlate with severe pathologies in the mouse infection model. J. Virol. 86:5508–5514. 10.1128/JVI.07081-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson D., Modhiran N., and Young P.R.. 2016. The many faces of the flavivirus NS1 protein offer a multitude of options for inhibitor design. Antiviral Res. 130:7–18. 10.1016/j.antiviral.2016.02.014 [DOI] [PubMed] [Google Scholar]
- World Health Organization 2009. Dengue: guidelines for diagnosis, treatment, prevention and control. http://apps.who.int/iris/bitstream/10665/44188/1/9789241547871_eng.pdf?ua=1 (accessed June 16, 2020) [PubMed]
- Yap S.S.L., Nguyen-Khuong T., Rudd P.M., and Alonso S.. 2017. Dengue Virus Glycosylation: What Do We Know? Front. Microbiol. 8:1415 10.3389/fmicb.2017.01415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch L.E., and Shresta S.. 2008. Mouse models of dengue virus infection and disease. Antiviral Res. 80:87–93. 10.1016/j.antiviral.2008.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P.R., Hilditch P.A., Bletchly C., and Halloran W.. 2000. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 38:1053–1057. 10.1128/JCM.38.3.1053-1057.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows a comparison of D2Y98P genome against DENV2 NGC strain and Singapore DENV2 clinical isolates.
shows an NS1 amino acid sequence comparison between D2Y98P, D220, and 16681 strains.
shows an amino acid sequence comparison between EHIE2862Y15 and D2Y98P strains.
lists primers used for PCR amplification.