Abstract
Background:
To investigate the correlation between growth arrest-specific transcript 5 (GAS5) gene polymorphism and the risk and prognosis of prostate cancer in Chinese Han population.
Methods:
Sanger sequencing was used to analyze genotypes at the rs17359906 and rs1951625 loci of the GAS5 gene in 218 prostate cancer patients and 220 healthy controls. The follow-up period was from August 2016 to August 2019, and the relationships between GAS5 gene polymorphisms at the rs17359906 and rs1951625 loci and the recurrence-free survival rate of prostate cancer patients were analyzed.
Results:
GAS5 A-allele carriers at the rs17359906 locus were 3.44 times more likely to develop prostate cancer than G-allele carriers (95% confidence interval (CI): 2.38–4.96, P < .001). Carriers of the GAS5 A allele at the rs1951625 locus had a 1.40-fold higher risk of prostate cancer than carriers of the G allele (95% CI: 1.05–1.86, P = .027). Plasma prostate-specific antigen (PSA), body mass index (BMI), and rs17359906 and rs1951625 loci were independent risk factors for prostate cancer. GAS5 AA genotype and A-allele carriers (GA + AA) at the rs1951625 locus were significantly correlated with Gleason scores ≤7 (P < .05). GAS5 genes rs17359906 G > A and rs1951625 G > A were associated with high plasma PSA levels. The recurrence-free survival rate of patients with prostate cancer with AA genotype at the rs17359906 locus of GAS5 (66.67%) was significantly lower than that of the GA genotype (76.47%), whereas the GG genotype was the highest (91.96%), and the difference was statistically significant (P = .002). The recurrence-free survival rate of patients with prostate cancer with the AA genotype at the rs1951625 locus of GAS5 (75.00%) was significantly lower than that of the GA genotype (81.82%), whereas the GG genotype was the highest (87.76%) with a statistically significant difference (P = .025).
Conclusion:
GAS5 rs17359906 G > A and rs1951625 G > A are significantly associated with an increased risk of prostate cancer and a reduction in three-year relapse-free survival.
Keywords: gene polymorphisms, growth arrest-specific transcript 5, prostate cancer, relapse-free survival
1. Introduction
Prostate cancer is a common solid malignant tumor in men. In recent years, the incidence of prostate cancer worldwide has increased.[1–3] With the increasing aging trend of the Chinese population and changes in diet, the incidence of prostate cancer in China has also increased.[4,5] Radical prostatectomy is one of the most effective treatments for early and middle stage prostate cancer, however about 20% of patients will experience biochemical recurrence within 5 years after surgery.[6] Most patients experiencing postoperative biochemical recurrence have reduced life expectancies. A large proportion of patients experience local tumor recurrence or distant metastasis after surgery.[7] Biochemical recurrence occurs in 20% to 50% of patients after radical radiotherapy.[8] Therefore, further research on the pathogenesis of prostate cancer is of great significance in the prevention and treatment of the disease.
The incidence of prostate cancer is strongly associated with family history,[9,10] suggesting that there may be genetic susceptibility to the pathogenesis of prostate cancer. With the development of the Human Genome Project and Whole Genome Association Analysis (GWAS),[11] there is increasing evidence that the susceptibility of prostate cancer is related to single nucleotide polymorphisms (SNPs). Through genome-wide association studies, more than 50 SNP sites related to prostate cancer risk have been identified, and this dataset has grown with the dissemination and development of gene sequencing technology.[12] A genome-wide association study of prostate cancer patients in the Chinese population found that only 2 SNPs, 9q31.2 (rs817826) and 19q13.4 (rs103294), were associated with prostate cancer in the Chinese population, which also confirms that there is a difference in the genetic susceptibility to prostate cancer between the Chinese, European, and American populations.[12] We therefore have reason to believe that in the future as more genome-wide association studies are performed in patients with prostate cancer, SNPs will be more widely used to predict prostate cancer risk in the Chinese population.
The GAS5 gene is located on human chromosome 1q25 and contains 12 exons. This gene does not encode a protein.[13] Studies have shown that long non-coding RNA (lncRNA) GAS5 expression is different in a variety of human tumor tissues and in normal tissues. Although we cannot rule out the possibility that this differential expression level is a secondary change caused by tumors, studies have also confirmed that GAS5 can play an important inhibitory role in the malignant transformation of cells by regulating a variety of cell signaling pathways.[14–16] Pickard et al[17,18] found that the expression level of GAS5 in metastatic prostate cancer cell lines LNCaP and PC3 was lower than that in normal prostate cell lines and non-metastatic prostate cancer cell lines, and overexpression of GAS5 not only increases the basal apoptosis rate of prostate cancer cell lines 22Rv1 and PC3, but also enhances the role of apoptosis stimulating factors. It is therefore speculated that GAS5 is a potentially important target for prostate cancer treatment.
The association between GAS5 polymorphism and tumor risk has been widely studied. For example, Lin et al[19] found that GAS5 rs145204276 polymorphism is associated with a risk of lymph node metastasis in patients with prostate cancer. Wang et al[20] found that polymorphism at rs55829688 in the promoter region of GAS5 is related to the risk of colorectal cancer and the down-regulation of GAS5 expression levels.
In this study, we selected GAS5 rs17359906 and rs1951625 from the lncRNASNP2 database (http://bioinfo.life.hust.edu.cn/lncRNASNP#!/). The selection of these 2 SNP sites was based on Genome-wide association analyses with family (GWAF) >0.05, and SNP-affected lncRNA and microRNA binding and lncRNA structure, and minor allele frequency (MAF) >0.05. The purpose of this study was to investigate the correlation between GAS5 polymorphisms and the occurrence and prognosis of prostate cancer, and to provide evidence for the prevention and treatment of prostate cancer.
2. Materials and methods
2.1. Patients
In this study, we enrolled 218 male prostate cancer patients (case groups), who were treated in our hospital from January 2015 to August 2016, aged 45 to 89 years, with an average age of (70.54 ± 12.04) years. Prostate cancer was confirmed in all patients by pathological examination. We recruited 220 healthy men, aged 45 to 87 years, with an average age of (70.64 ± 11.19) years, to form the control group. Patients were included in the control group provided that they had not been diagnosed with cancer before entering the study and had PSA test scores in the normal range. Patients with a history of tumors such as bladder cancer, lung cancer, liver cancer, prostate transitional cell carcinoma, squamous cell carcinoma, fibroadenomas, basal cell carcinoma, undifferentiated tumors, or sarcomatoid carcinoma, and patients with vascular diseases, severe endocrine diseases, liver diseases, or severe nutritional metabolic diseases were excluded. We limited the population to members of the Chinese Han population to reduce the possibility of population stratification. Demographic data, collected for all patients, included: age, body mass index (BMI), smoking status, drinking status, and plasma PSA levels. We also collected clinical pathological data such as Clinical T stage and Gleason score in patients with prostate cancer. This study was performed with the approval of the Medical Ethics Committee of Zhuji People's Hospital of Zhejiang Province, and all subjects signed written informed consent forms prior to the commencement of the study.
2.2. GAS5 gene polymorphism analysis
In this study, we analyzed the polymorphisms at the rs17359906 and rs1951625 loci of GAS5. We used the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) to extract genomic DNA from peripheral venous blood of the patients, and used the extracted genomic DNA as a template to perform a PCR reaction to synthesize DNA fragments containing SNP sites. The primer information was: rs17359906: 5′-ATC TGC ACC CAG CAC CAT AC-3′ (Fw); 5′-TGG TAT GTT ACC TGC ATC ATT GG-3’ (Rv). rs1951625: 5’-ACT GCA CCC GCA GTT AAG AA-3′ (Fw); 5′-CTT AAG TGC CTG CAT TCC GC-3′ (Rv). The PCR reaction system contained about 50 ng of genomic DNA, 10× buffer 2.0 μL, 2 mM dNTP 0.4 μL, 25 mM Mg2 + 0.8 μL, 10 mM forward and reverse primers 0.4 μL each, 1 U/μL Taq enzyme 0.15 μL, ddH2O 14.85 μl, and a total volume of 20 μL. Conditions for PCR amplification: 95°C for 5 minutes, then 95°C for 30 seconds; 58°C for 30 seconds; 72°C for 30 seconds, for a total of 35 cycles, and then 72°C for 5 minutes. The PCR products were sequenced by Sanger, and the sequencing work was completed by Suzhou Jinweizhi Biotechnology Co., Ltd. Five percent of the samples were randomly selected for repeated verification, and the results of the 2 verifications were found to be consistent.
2.3. Follow-up
The follow-up of 218 patients with prostate cancer in this study was performed by outpatient and telephone follow-up. The deadline for follow-up was August 2019, and no patients were lost to follow-up. The follow-up time ranged from 6 to 36 months.
2.4. Statistical analysis
Statistical analysis of the data in this study was performed using SPSS 20.0 software (SPSS Inc., Chicago). The goodness-of-fit χ2-test was used to assess whether the genotype of the SNP loci conformed to Hardy-Weinberg equilibrium. Continuous variables were expressed as (mean ± SD), and statistical analysis was performed by t-test. The categorical variables were expressed as [n (%)], and the statistical analysis was performed using the χ2-test. Logistic regression was used to analyze the correlation between GAS5 rs17359906 and rs1951625 loci and the risk of prostate cancer, calculated odds ratio (OR) and 95% confidence interval (CI), and adjusted age, BMI, smoking status, and drinking status. Kaplan-Meier Curve in Graphpad prism 7 (GraphPad Software, Inc., San Diego, CA, USA) was used to analyze the three-year relapse-free survival of patients with different genotypes of GAS5 at the rs17359906 and rs1951625 loci.
3. Results
3.1. Demographic and clinical data
Table 1 compares the demographic and clinical data of prostate cancer patients and the control group in this study. There were no differences in the age, smoking status, drinking status, and control group of prostate cancer patients (P > .05). The BMI and plasma PSA levels of prostate cancer patients were significantly higher than those of the control group (P < .05). Clinical T stages of prostate cancer patients were as follows: 48 were T1, 46 were T2, 55 were T3, and 69 were T4. Among patients with prostate cancer, 129 had a Gleason score ≤7, and 89 had a Gleason score >7.
Table 1.
Comparison of demographic and clinical data of prostate cancer patients and control groups.
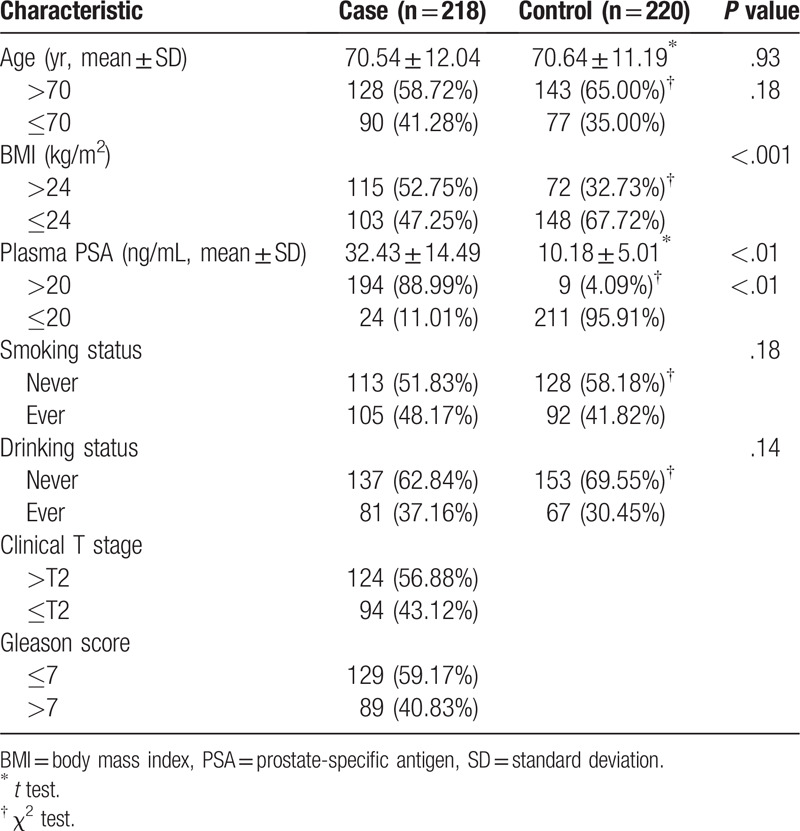
3.2. GAS5 gene polymorphism locus genotype and allele frequency
The rs17359906 and rs1951625 loci of GAS5 in the case and control groups were all in Hardy-Weinberg equilibrium (P > .05) (Table 2). Taking the GAS5 rs17359906 locus GG genotype as a reference, after adjusting for age, BMI, smoking status, and drinking status, GA and AA genotype patients had a significantly greater risk of prostate cancer. The risk in the GA genotype was 3.65 times (95% CI: 2.32–5.74, P < .001) and in the AA genotype the risk was 6.68 times (95% CI: 2.45–18.21, P < .001) greater than in the GG genotype. Carriers of the A allele were 3.44 times more likely to have prostate cancer than those of the G allele (95% CI: 2.38–4.96, P < .001). Taking the GAS5 rs1951625 locus GG genotype as a reference, after adjusting for age, BMI, smoking status, and drinking status, there was no significant correlation between GA and AA genotypes and the risk of prostate cancer (P > .05). However, carriers of the A allele had a 1.40-fold higher risk of prostate cancer than carriers of the G allele (95% CI: 1.05–1.86, P = .027) (Table 2).
Table 2.
Association of SNPs at rs17359906 and rs1951625 loci of GAS5 with prostate cancer risk.
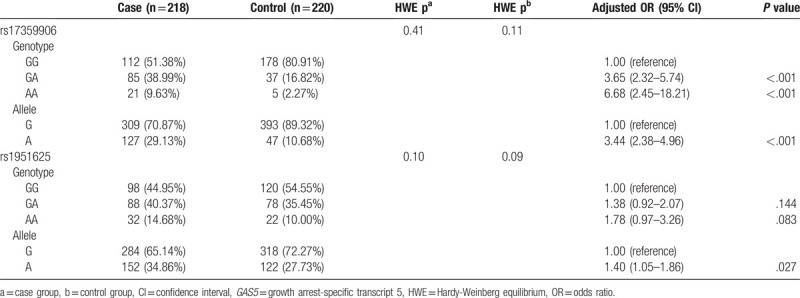
3.3. Different genetic patterns of GAS5 SNP loci and risk of prostate cancer
GAS5 rs17359906 locus dominant model (adjusted OR = 4.01, 95% CI: 2.61–6.16, P < .001), recessive model (adjusted OR = 4.58, 95% CI: 1.70–12.39, P = .002), and additive model (adjusted OR = 1.58, 95% CI: 1.17–2.13, P = .004) were all significantly associated with an increased risk of prostate cancer (Table 3). The rs1951625 locus dominant model, recessive model, and additive model all showed no significant correlation with prostate cancer risk (P > .05, Table 3).
Table 3.
Correlation between GAS5 SNP loci and the risk of prostate cancer in different genetic models.
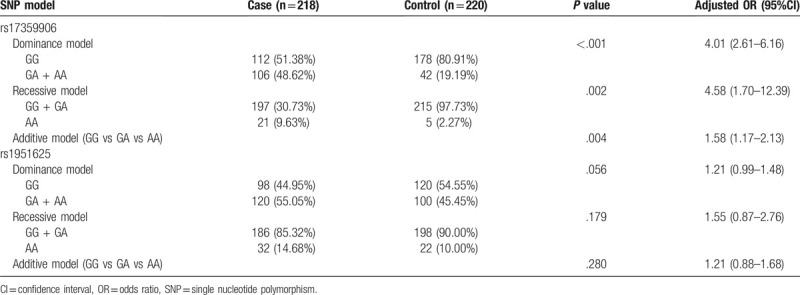
3.4. Hierarchical analysis
Stratified analysis of general patient data showed that the stratification of age, BMI, smoking status, and drinking status did not affect the association between the SNP at the GAS5 rs17359906 site and the risk of prostate cancer. The populations of GA and AA genotypes at the rs17359906 locus had a greater risk of developing prostate cancer than those of GG genotypes (P < .05, Table 4).
Table 4.
Hierarchical analysis of the association between SNPs at the rs17359906 locus of GAS5 and prostate cancer risk.

BMI and drinking status affected the association between SNPs at the GAS5 rs1951625 locus and prostate cancer risk. Among patients with BMI ≤ 24 kg/m2 and those who never drink, the risk of prostate cancer is high for GA and AA genotypes when they carry the GAS5 at the rs1951625 locus. However, there was no significant increase in the risk of prostate cancer in men with a BMI > 24 kg/m2 and those who had never had a drink when they carried GA and AA genotypes of GAS5 at the rs1951625 locus (P > .05, Table 5). The risk of prostate cancer in patients with GA and AA genotypes of the GAS5 gene at the rs1951625 locus and who were <70 or ≥70 years of age, never smoked, or ever smoked, and never drank or ever drank, compared favorably with the risk of patients with GG genotypes. There was no significant difference in the risk of developing prostate cancer in these men (P > .05, Table 5).
Table 5.
Hierarchical analysis of the association between the SNP at the rs1951625 locus of the GAS5 and the risk of prostate cancer.
3.5. Multivariate analysis of the risk of prostate cancer
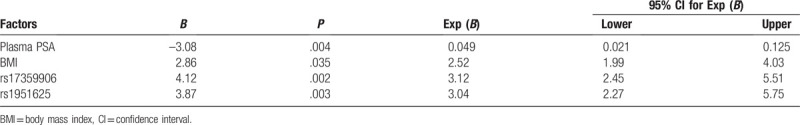
Plasma PSA, BMI, rs17359906, rs1951625, and other variables were included in the logistic regression model for multivariate correlation regression analysis. The results showed that plasma PSA, BMI, rs17359906, and rs1951625 were independent risk factors for prostate cancer (Table 6).
Table 6.
Multivariate analysis of the risk of prostate cancer.
3.6. Correlation between the SNPs of the rs17359906 and rs1951625 loci of GAS5 and clinical parameters
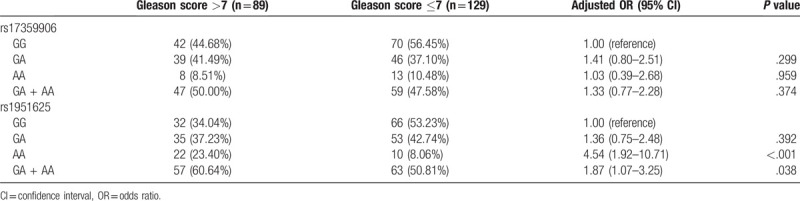
The distribution of GAS5 genotypes at the rs17359906 and rs1951625 loci between Clinical T stage >T2 and Clinical T stage ≤T2 is shown in Table 7. The results indicate that there was no significant correlation between SNPs at the GAS5 rs17359906 and rs1951625 loci and Clinical T stage (P > .05, Table 7). The distribution of GAS5 genotypes at the rs17359906 and rs1951625 loci for patients with Gleason score ≤7 and Gleason score >7 are shown in Table 8. The results indicate that there was no significant correlation between SNPs at the rs17359906 locus of GAS5 and Gleason score (P > .05), and that the AA genotype at the rs1951625 locus of GAS5 and the carrier of the A allele (GA + AA) were significantly associated with a Gleason score >7 (P < .05, Table 8).
Table 7.
Correlation between SNPs at the GAS5 rs17359906 and rs1951625 loci and clinical T stage.
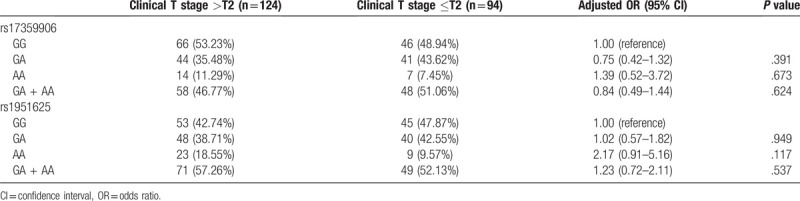
Table 8.
Correlation between SNPs at the rs17359906 and rs1951625 loci of GAS5 and Gleason score.
3.7. Correlation between SNPs at rs17359906 and rs1951625 of GAS5 and plasma PSA levels
Correlation between SNPs at GAS5 rs17359906 and rs1951625 loci and plasma PSA levels in the case and control groups was analyzed. The results of univariate analysis of variance showed that there was a statistically significant difference in plasma PSA levels between the case and control groups with different genotypes at the rs17359906 and rs1951625 loci of GAS5 (P < .05). However, in subjects with plasma PSA levels exceeding 20 ng/mL, there was no correlation between the frequency of different genotypes at rs17359906 and the risk of prostate cancer (P > .05) (Table 9). In subjects with plasma PSA levels below 20 ng/mL, the GA genotype at rs17359906 was associated with an increased risk of prostate cancer (P < .05) (Table 9). In subjects with plasma PSA levels exceeding 20 ng/mL, the GA genotype frequency at rs1951625 was associated with an increased risk of prostate cancer (P < .05) (Table 9). In subjects with plasma PSA levels below 20 ng/mL, the genotype frequency at rs1951625 was not associated with prostate cancer risk (P > .05) (Table 9). The mean plasma PSA level of the allele carriers of point A was significantly higher than that of the G-allele carriers (Fig. 1), suggesting that the GAS5 gene rs17359906 G > A and rs1951625 G > A mutations are associated with elevated plasma PSA levels.
Table 9.
Correlation between GAS5 rs17359906 and rs1951625 SNP and prostate cancer risk in subjects with different plasma PSA levels.
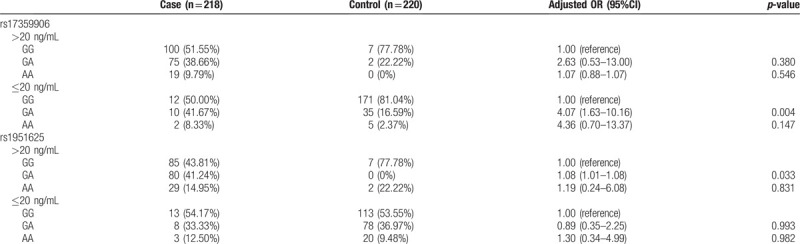
Figure 1.
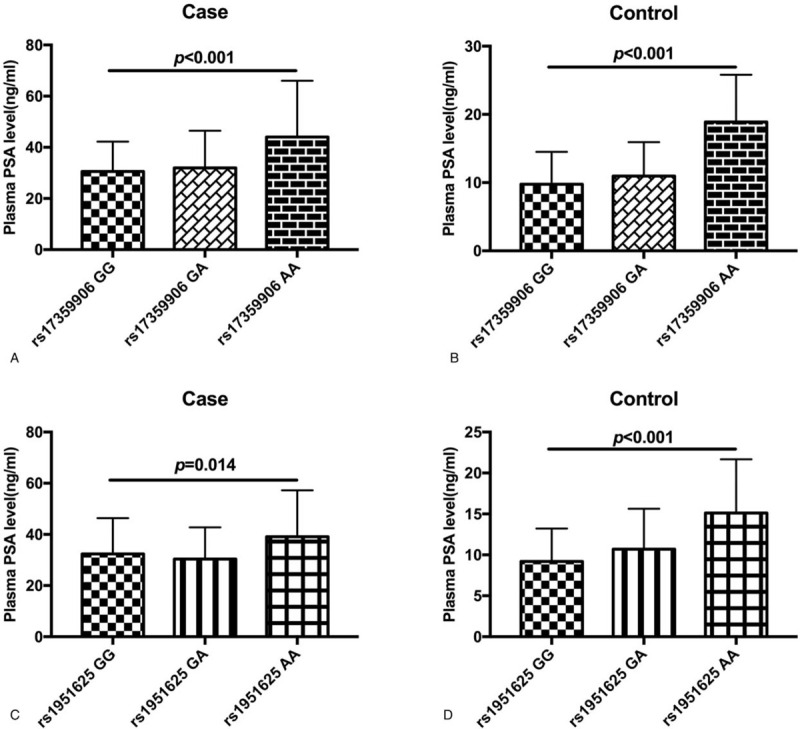
Comparison of plasma PSA levels in patients with different genotypes at the rs17359906 and rs1951625 loci of GAS5. (A) Comparison of plasma PSA levels in patients with prostate cancer with different genotypes at the GAS5 rs17359906 locus. (B) Comparison of plasma PSA levels in the control group with different genotypes at the GAS5 rs17359906 locus. (C) Comparison of plasma PSA levels in patients with prostate cancer with different genotypes at the GAS5 rs1951625 locus. (D) Comparison of plasma PSA levels in the control group with different genotypes at the GAS5 rs1951625 locus.
3.8. GAS5 SNPs at the rs17359906 and rs1951625 loci and recurrence-free survival in patients with prostate cancer
In this study, we followed the progress of disease in 218 patients with prostate cancer using outpatient and telephone follow-up methods. As of August 2019, no patients were lost to follow-up. The follow-up time ranged from 6 months to 36 months. Of the 218 patients, 36 patients relapsed during follow-up, and the 36-month recurrence-free survival rate was 83.49%. The recurrence-free survival rate of patients with prostate cancer of the AA genotype at the GAS5 rs17359906 locus (66.67%) was significantly lower than that of the GA genotype (76.47%) at the same locus, and the recurrence-free survival rate of patients with the GG genotype at this locus was the highest (91.96%). (P = .002, Fig. 2A). The recurrence-free survival rate of patients with prostate cancer of the AA genotype at the GAS5 rs1951625 locus (75.00%) was significantly lower than that of the GA genotype (81.82%) at the same locus. The recurrence-free survival rate of patients with the GG genotype at this locus was the highest (87.76%). (P = .025, Fig. 2B).
Figure 2.
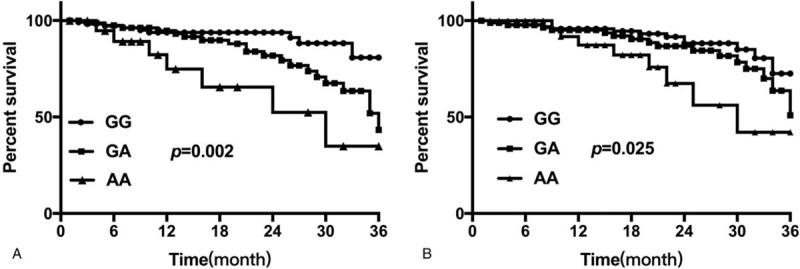
Kaplan-Meier curve analysis of 36-month recurrence-free survival rate of patients with prostate cancer. (A) Comparison of 36-month recurrence-free survival rate of patients with prostate cancer with different genotypes at the rs17359906 locus of GAS5. (B) Comparison of the 36-month relapse-free survival rate of patients with prostate cancer with different genotypes at the rs1951625 locus of GAS5.
4. Discussion
In this study, we examined 2 SNP loci on the GAS gene associated with the risk of prostate cancer, namely rs17359906 and rs1951625. Our results indicate that GAS5 rs17359906 G > A and rs1951625 G > A mutations are significantly associated with an increased risk of prostate cancer and a reduction in three-year relapse-free survival rates in patients with prostate cancer.
Prostate cancer has surpassed lung cancer in the United States and is now first among malignant tumors that endanger men's health.[21,22] Although the incidence of prostate cancer in Asia is lower than in Europe and the United States, it has increased significantly in recent years. Prostate cancer usually progresses slowly, and tumor-bearing patients often have good long-term survival rates,[23,24] however some patients’ tumors grow rapidly, and may metastasize and transform into castration-resistant prostate cancer in a short time. In such cases traditional endocrine therapy is no longer effective and the patient's condition deteriorates rapidly.[25,26] The cause of this heterogeneity in prostate cancer is currently unknown. Therefore, it is of great significance to investigate the pathogenesis of prostate cancer, to find new pathogenic factors and prognostic evaluation factors, which will facilitate individualized treatment to patients according to the unique nature of their disease.
Initially, non-coding RNA was thought to be “transcription noise” and have no biological function. However, as research into epigenetics and genomics intensified, researchers discovered that these RNAs can regulate gene expression at epigenetic, transcription, and post-transcription levels.[27–29] Likewise, it was found that lncRNA plays an important role in various biological processes such as cell differentiation, proliferation, apoptosis, migration, and infiltration.[30–32] The GAS5 plays an important role in the occurrence and development of tumors and is abnormally expressed in various tumors, such as lung cancer,[33] gastric cancer,[34] and triple negative breast cancer (TNBC).[35] Studies have shown that lncRNA GAS5 plays an important role in the occurrence of prostate cancer. For example, Xue et al[36] showed that lncRNA GAS5 mediates the AKT/mTOR signaling pathway by targeting miR-103 in prostate cancer. In addition, Pickard et al[17] found that lncRNA GAS5 promotes the apoptosis of prostate cancer cells, and abnormally low expression of lncRNA GAS5 may significantly reduce the effectiveness of chemotherapy drugs.
In this study, we used bioinformatics techniques to find 2 structures that may affect GAS5 binding to microRNA and lncRNA GAS5. The MAFs of these 2 SNP sites is >0.05. The MAF of the rs17359906 locus in the 1000 genomes database is 0.081 and the MAF of the rs1951625 locus in the 1000 genomes database is 0.2714, which is not significantly different from the findings in this research paper, indicating that the selected population is representative.
Case-control studies found that the GAS5 rs17359906 locus A allele and the GAS5 rs1951625 locus A allele were significantly associated with an increased risk of prostate cancer. The GAS5 rs17359906 locus dominant model, recessive model, and additive model were also significantly associated with increased prostate cancer risk, but no correlation was found between the GAS5 rs1951625 locus dominant model, recessive model, additive model, and prostate cancer risk. This indicates that the rs17359906 locus and the rs1951625 locus are susceptible factors for the development of prostate cancer. Further research shows that rs17359906 and rs1951625 are similar independent risk factors for prostate cancer to plasma PSA and BMI. To our knowledge, there have as yet been no research studies conducted on the correlation between rs17359906 and rs1951625 loci and prostate cancer risk, hence the association between these loci and the risk of prostate cancer has not yet been fully elucidated. This study is the first to focus on these 2 SNP loci.
We analyzed the correlation between the GAS5 rs17359906 and rs1951625 loci and Gleason scores and Clinical T stages to determine the relationship between GAS5 polymorphisms and disease progression. The results showed that the GAS5 rs1951625 locus A allele was associated with a Gleason score >7, indicating that carriers of the GAS5 rs1951625 locus A allele were likely to have highly malignant disease with a poor prognosis. The results of follow-up analysis also confirmed that the carriers of GAS5 rs17359906 locus A allele and rs1951625 locus A allele had poor prognoses. Although we did not find a significant correlation between the rs17359906 locus SNP and the Gleason score, we speculate that this may be related to our relatively small sample size.
At present, PSA is the most widely used indicator for prostate cancer screening. It is used clinically to guide biopsy, evaluate the degree of malignancy of prostate cancer, and detect recurrence.[37–39] Several studies have shown that PSA levels are positively related to an increase in postoperative Gleason score. The higher the PSA level after surgery, the higher the Gleason score. In this study, we found that GAS5 rs17359906 G > A and rs1951625 G > A mutations were associated with elevated plasma PSA levels, which further confirmed that GAS5 rs17359906 G > A and rs1951625 G > A were associated with poor prognosis of prostate cancer. Interestingly, from hierarchical analysis of plasma PSA levels, we found that in subjects with plasma PSA levels exceeding 20 ng/mL, there was no correlation between the frequency of different genotypes at rs17359906 and the risk of prostate cancer. (P > 0.05). In subjects with plasma PSA levels below 20 ng/mL, the GA genotype at rs17359906 was associated with an increased risk of prostate cancer (P < .05). In subjects with plasma PSA levels exceeding 20 ng/mL, the GA genotype frequency at rs1951625 was associated with an increased risk of prostate cancer (P < .05). In subjects with plasma PSA levels below 20 ng/mL, the genotype frequency of rs1951625 was not associated with prostate cancer risk (P > .05). The reason for the analysis may be related to the small sample size. In the stratified study of plasma PSA levels, the sample size of some genotype subjects is small, which affects the objectivity of statistical analysis. It is necessary to further expand the sample size for the study.
There are some limitations to this study. First, we did not analyze the correlation between GAS5 rs17359906 and rs1951625 loci SNP and lncRNA GAS5 expression levels and RNA structure. Therefore, there is a lack of mechanism research to support the findings of this study. Secondly, the correlation of research results in this study needs further demonstration in a study with a larger sample size. In addition, there is a lack of evidence from in vitro and in vivo studies to support the findings of this study. These are topics for future research.
5. Conclusion
The results of this study indicate that GAS5 rs17359906 G > A and rs1951625 G > A were significantly associated with increased risk of prostate cancer and reduced three-year relapse-free survival. Furthermore, the GAS5 rs17359906 and rs1951625 loci are independent risk factors for prostate cancer.
Author contributions
Study design: Lisha Zhao, Weihong Zheng, Chen Li.
Data collection: Lisha Zhao, Weihong Zheng.
Data analysis: Lisha Zhao, Weihong Zheng, Chen Li.
Interpretation of data: Lisha Zhao, Weihong Zheng, Chen Li.
Draft manuscript: Lisha Zhao.
Review manuscript: Chen Li.
Footnotes
Abbreviations: BMI = body mass index, CI = confidence interval, GAS5 = growth arrest-specific transcript 5, GWAF = genome-wide association analyses with family, GWAS = Whole Genome Association Analysis, lncRNA = long non-coding RNA, MAF = minor allele frequency, OR = odds ratio, SNPs = single nucleotide polymorphisms, TNBC = triple negative breast cancer.
How to cite this article: Zhao L, Zheng W, Li C. Association of long-chain non-coding RNA GAS5 gene polymorphisms with prostate cancer risk and prognosis in Chinese Han population. Medicine. 2020;99:36(e21790).
This work was supported by the grant Laboratory Work Research Association Project of Zhejiang University (YB201908).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
- [1].Zhai Z, Zheng Y, Li N, et al. Incidence and disease burden of prostate cancer from 1990 to 2017: results from the Global Burden of Disease Study 2017. Cancer 2020;126:1969–78.. doi: 10.1002/cncr.32733. [DOI] [PubMed] [Google Scholar]
- [2].Sharma R. The burden of prostate cancer is associated with human development index: evidence from 87 countries, 1990–2016. EPMA J 2019;10:137–52.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Teoh JYC, et al. Global incidence of prostate cancer in developing and developed countries with changing age structures. PLoS One 2019;14:e0221775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ye D, Zhu Y. Epidemiology of prostate cancer in China: an overview and clinical implication. Zhonghua Wai Ke Za Zhi 2015;53:249–52.. [PubMed] [Google Scholar]
- [5].Zhao F, et al. Trends in treatment for prostate cancer in China: preliminary patterns of care study in a single institution. J Cancer 2018;9:1797–803.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Freedland SJ, Moul JW. Prostate specific antigen recurrence after definitive therapy. J Urol 2007;177:1985–91.. [DOI] [PubMed] [Google Scholar]
- [7].Ginzburg S, et al. Prostate cancer biochemical recurrence rates after robotic-assisted laparoscopic radical prostatectomy. JSLS 2012;16:443–50.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Pollack A, et al. Prostate cancer radiotherapy dose response: an update of the fox chase experience. J Urol 2004;171:1132–6.. [DOI] [PubMed] [Google Scholar]
- [9].Brazel AJ, Vernimmen D. The complexity of epigenetic diseases. J Pathol 2016;238:333–44.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ishak MB, Giri VN. A systematic review of replication studies of prostate cancer susceptibility genetic variants in high-risk men originally identified from genome-wide association studies. Cancer Epidemiol Biomarkers Prev 2011;20:1599–610.. [DOI] [PubMed] [Google Scholar]
- [11].Stegeman S, et al. A large-scale analysis of genetic variants within putative miRNA binding sites in prostate cancer. Cancer Discov 2015;5:368–79.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Xu J, et al. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet 2012;44:1231–5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen X, et al. Long non-coding RNA GAS5 and ZFAS1 are prognostic markers involved in translation targeted by miR-940 in prostate cancer. Oncotarget 2018;9:1048–62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Huang W, et al. LncRNA GAS5-AS1 inhibits glioma proliferation, migration, and invasion via miR-106b-5p/TUSC2 axis. Hum Cell 2020;33:416–26.. [DOI] [PubMed] [Google Scholar]
- [15].Li J, et al. LncRNA GAS5 inhibits Th17 differentiation and alleviates immune thrombocytopenia via promoting the ubiquitination of STAT3. Int Immunopharmacol 2020;80:106127. [DOI] [PubMed] [Google Scholar]
- [16].Li G, et al. LncRNA GAS5 regulates the proliferation, migration, invasion and apoptosis of brain glioma cells through targeting GSTM3 expression. The effect of LncRNA GAS5 on glioma cells. J Neurooncol 2019;143:525–36.. [DOI] [PubMed] [Google Scholar]
- [17].Pickard MR, Mourtada-Maarabouni M, Williams GT. Long non-coding RNA GAS5 regulates apoptosis in prostate cancer cell lines. Biochim Biophys Acta 2013;1832:1613–23.. [DOI] [PubMed] [Google Scholar]
- [18].Mourtada-Maarabouni M, et al. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009;28:195–208.. [DOI] [PubMed] [Google Scholar]
- [19].Lin CY, et al. Impact of GAS5 genetic polymorphism on prostate cancer susceptibility and clinicopathologic characteristics. Int J Med Sci 2019;16:1424–9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang Y, Wu S, Yang X, et al. Association between polymorphism in the promoter region of lncRNA GAS5 and the risk of colorectal cancer [published correction appears in Biosci Rep. 2019 May 21;39(5):]. Biosci Rep 2019;39(4):BSR20190091.Published 2019 Apr 16. doi:10.1042/BSR20190091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Boring CC, Squires TS, Health CW., Jr Cancer statistics for African Americans. CA Cancer J Clin 1992;42:7–17.. [DOI] [PubMed] [Google Scholar]
- [22].DeSantis CE, et al. Cancer statistics for African Americans, 2016: progress and opportunities in reducing racial disparities. CA Cancer J Clin 2016;66:290–308.. [DOI] [PubMed] [Google Scholar]
- [23].Boorjian SA, et al. A critical analysis of the long-term impact of radical prostatectomy on cancer control and function outcomes. Eur Urol 2012;61:664–75.. [DOI] [PubMed] [Google Scholar]
- [24].Thompson I, et al. Guideline for the management of clinically localized prostate cancer: 2007 update. J Urol 2007;177:2106–31.. [DOI] [PubMed] [Google Scholar]
- [25].D’Amico AV, et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA 1998;280:969–74.. [DOI] [PubMed] [Google Scholar]
- [26].Bader DA, McGuire SE. Tumour metabolism and its unique properties in prostate adenocarcinoma. Nat Rev Urol 2020;17:214–31.. [DOI] [PubMed] [Google Scholar]
- [27].Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol 2010;7:582–5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Birgani MT, et al. Long non-coding RNA SNHG6 as a potential biomarker for hepatocellular carcinoma. Pathol Oncol Res 2018;24:329–37.. [DOI] [PubMed] [Google Scholar]
- [29].Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett 2018;418:41–50.. [DOI] [PubMed] [Google Scholar]
- [30].Wang Y, et al. LncRNA AB073614 regulates proliferation and metastasis of colorectal cancer cells via the PI3K/AKT signaling pathway. Biomed Pharmacother 2017;93:1230–7.. [DOI] [PubMed] [Google Scholar]
- [31].Lian Y, et al. Knockdown of pseudogene derived from lncRNA DUXAP10 inhibits cell proliferation, migration, invasion, and promotes apoptosis in pancreatic cancer. J Cell Biochem 2018;119:3671–82.. [DOI] [PubMed] [Google Scholar]
- [32].Chen S, et al. Macrophage infiltration promotes invasiveness of breast cancer cells via activating long non-coding RNA UCA1. Int J Clin Exp Pathol 2015;8:9052–61.. [PMC free article] [PubMed] [Google Scholar]
- [33].Dong L, et al. Upregulation of long noncoding RNA GAS5 inhibits lung cancer cell proliferation and metastasis via miR-205/PTEN axis. Med Sci Monit 2019;25:2311–9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li Y, Gu J, Lu H. The GAS5/miR-222 axis regulates proliferation of gastric cancer cells through the PTEN/Akt/mTOR pathway. Dig Dis Sci 2017;62:3426–37.. [DOI] [PubMed] [Google Scholar]
- [35].Li S, et al. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed Pharmacother 2018;104:451–7.. [DOI] [PubMed] [Google Scholar]
- [36].Xue D, Zhou C, Lu H, et al. LncRNA GAS5 inhibits proliferation and progression of prostate cancer by targeting miR-103 through AKT/mTOR signaling pathway [published online ahead of print, 2016 Oct 14]. Tumour Biol 2016;doi: 10.1007/s13277-016-5429-8. [DOI] [PubMed] [Google Scholar]
- [37].Mao Z, et al. Diagnostic performance of PCA3 and hK2 in combination with serum PSA for prostate cancer. Medicine (Baltimore) 2018;97:e12806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen B, Macapinlac HA, Lu Y. Superscan 18F-fluciclovine PET/CT of PSA-negative prostate cancer bone metastases. Clin Nucl Med 2019;44:337–8.. [DOI] [PubMed] [Google Scholar]
- [39].Armstrong AJ, et al. A phase 2 multimodality trial of docetaxel/prednisone with sunitinib followed by salvage radiation therapy in men with PSA recurrent prostate cancer after radical prostatectomy. Prostate Cancer Prostatic Dis 2016;19:100–6.. [DOI] [PubMed] [Google Scholar]


