Abstract
MET amplifications, mutations, and fusions are drivers of oncogenesis. More contemporary diagnostic assays can maximize the likelihood of detecting these alterations. de novo MET alterations can serve as primary drivers of cancer growth or as secondary drivers that mediate resistance to targeted therapy for another oncogene such as sensitizing EGFR mutations. Single-agent or combination MET-directed targeted therapy is active in MET-amplified cancers. While the likelihood of benefit increases with increasing levels of MET amplification, no consensus exists on diagnostic cutoffs for MET copy number gains by fluorescence in-situ hybridization or next-generation sequencing. MET mutations include those affecting the kinase or extracellular domains, and those that result in exon 14 skipping biology. The activity of type I or II MET tyrosine kinase inhibitors (TKIs) varies by MET mutation category. Type I MET TKIs bind the kinase ATP pocket in the active state, whereas type II inhibitors bind the inactive state. The biology and response to targeted therapy of MET fusions are underexplored. While MET expression in the absence of a genomic marker of MET dependence is a poor predictor of MET-targeted therapy benefit, MET expression in the context of pathogenic MET alterations may select for response.
INTRODUCTION
Dysregulation of the c-MET tyrosine kinase (hereafter simplified as MET) is an established driver of oncogenesis1. Compared to many other proto-oncogenes, MET is unique in that three different genomic states can lead to clinically-relevant oncogenesis: amplification, mutation, and fusion. All three of these states present diagnostic challenges in the clinic. Furthermore, these can be identified in two major contexts - as primary or secondary drivers of cancer growth. Primary MET dependence is exemplified by tumors that rely solely on overactive MET signaling to fuel growth. Secondary MET dependence is characterized by reliance on another oncogenic driver (e.g. mutant EGFR) and concurrent dependence on MET. Secondary MET dependence can be de novo or acquired, following the selective pressures of inhibitors directed against the primary driver.
Identifying tumors that are oncogenically addicted to MET is crucial because multiple MET-directed therapeutics are available in the clinic. This has been hindered on a diagnostic level due to (1) the lack of standardized cutoffs and testing methodology for MET-dependent states such as MET amplification that are measured as a continuous variable, and (2) the inability of older assays to more reliably capture both MET copy number gains and the wide variety of MET mutations and MET fusions that lead to oncogenesis. While no MET-directed targeted therapy is currently approved for MET-dependent tumors, several agents have recently gained breakthrough designation from regulatory authorities. This has happened largely secondary to the adoption of more advanced diagnostic technologies that more effectively identify MET-dependent cancers, and the contemporary strategy of molecular enrichment for these tumors on prospective targeted therapy trials.
MET AMPLIFICATION
MET copy number gains can occur either through polysomy or amplification. Polysomy occurs when multiple copies of chromosome 7 that carries MET are present. This can occur through chromosomal or whole genome duplication10,11. The presence of multiple chromosomes results in an increase in the number of MET copies. With amplification, MET undergoes regional or focal copy number gains without chromosome 7 duplication12 (Figure 1). In contrast to polysomy, true amplification is more likely to lead to oncogene addiction12. These findings parallel data in breast cancer where tumors with HER2 copy number gains secondary to polysomy behave similarly to HER2-negative tumors13. Focal MET amplification can lead to elevations in MET expression, receptor activation, and ligand-independent downstream signaling in preclinical models14,15.
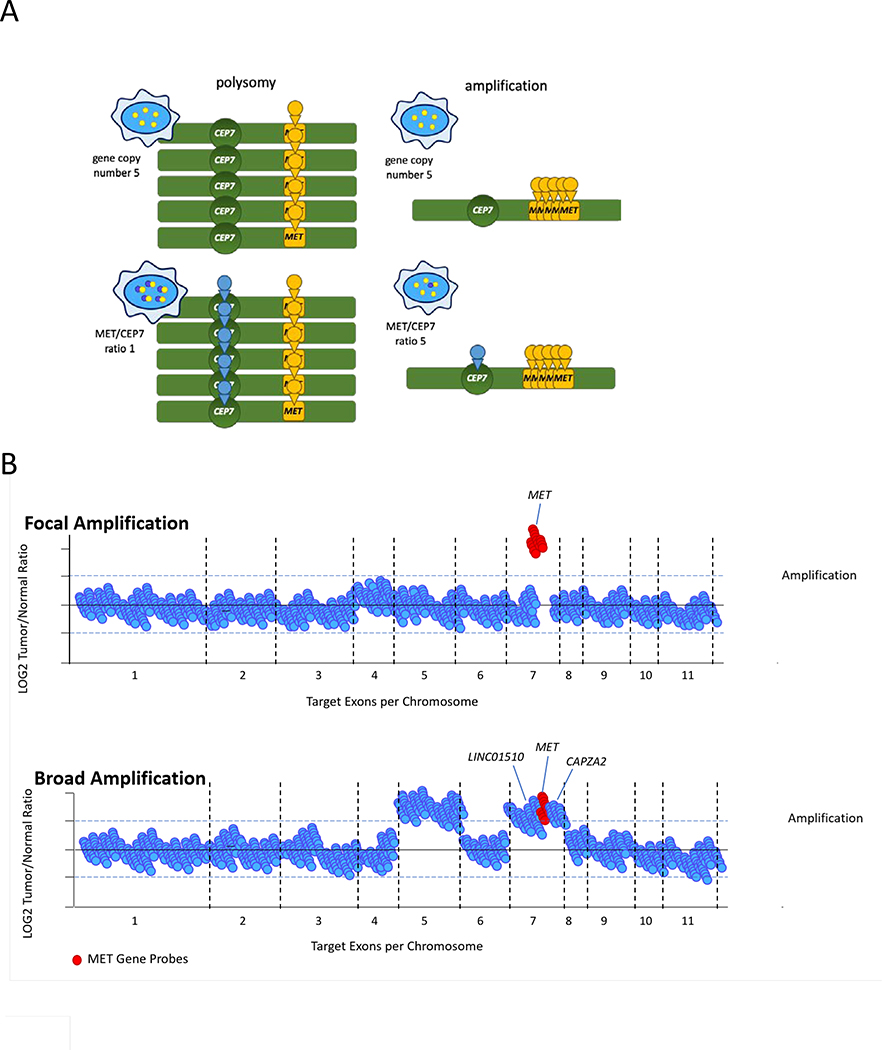
Figure 1. MET amplification diagnosis.
(A) The identification of MET gene copy number by FISH only requires a single colored probe (yellow) against MET that is counted to determine the number of copies of the gene. This strategy cannot differentiate polysomy from true focal amplification as the absolute number of chromosomes that contain MET cannot be determined. In contrast, the use of an additional probe targeting centromere 7 (CEP7, blue) allows this determination. The MET/CEP7 ratio thus helps differentiate whole genome duplication/polysomy (low MET/CEP7 ratio) from focal amplification (high MET/CEP7 ratio). (B) Using next-generation sequencing, focal MET amplification can be distinguished from broad chromosomal gains that include MET. In the latter, adjacent genes such as LINC01510 and CAPZA2 are concurrently amplified. Focal MET amplification is associated with a higher likelihood of MET-dependence for oncogenesis.
Diagnosis
Various assays can detect MET copy number changes. These include fluorescence in-situ hybridization (FISH), quantitative real-time polymerase chain reaction (qRT-PCR), and next-generation sequencing (NGS)16. The latter can be utilized for tumor or plasma circulating tumor DNA (ctDNA) testing. Unfortunately, cutoff points that define MET amplification vary within each assay.
Fluorescence in situ hybridization
FISH is a commonly used technique employing fluorophore-coupled DNA fragments to recognize and tag genomic regions of interest. One or more colored fluorophores may be used during testing. Following fluorophore treatment, the gene sequences of interest in the cell nucleus will fluoresce with one or more probe colors. The number of signals identified in a cell nucleus indicates the number of copies of the gene of interest (Figure 1). Signals from a predetermined number of cells are counted and the number of signals per cell are averaged. Because FISH is performed under a microscope, signals from malignant cells may be differentiated from those of normal cells. While challenging, this can be helpful in samples with low tumor content. However, some tumors may be prone to tissue sectioning artifacts and signals from one cell may overlap another. This reduces the number of evaluable cells and double signals may appear due to overlap.
MET amplification can be defined by FISH in two major ways. The first method relies on determining gene copy number (GCN). Using the Cappuzzo criteria, MET amplification is defined as the presence of 5 or more copies of MET per cell (MET GCN ≥5)17–19. Alternate definitions include a MET GCN of ≥620 and a MET GCN of ≥1521,22. Unfortunately, GCN is unable to distinguish true amplification from polysomy. The second method surpasses this liability by taking the ratio of MET to CEP7 (centromere 7 enumeration probe); a separate fluorophore is used for the latter. Determination of this ratio adjusts for the number of chromosomes present and more accurately identifies MET amplification in the absence of chromosomal duplication. A MET/CEP7 ratio ≥2.0 is commonly used to define amplification18,19,23–27. Others have categorized the degree of amplification into three groups using MET/CEP7 ratios: low ≥1.8 to ≤2.2; intermediate >2.2 to <5, and high ≥512.
Next-generation sequencing
Methods for determining MET copy number and cutoffs for MET amplification vary by NGS platform28–30. Similar to FISH, there is no consensus on a single definition. In general, the sequenced gene of interest is normalized against a comparator such as archived sequences from normal samples. Some platforms go one step further and incorporate normal samples from patients (i.e. peripheral blood) into the normalization in addition to using a comparator29. The subsequent bioinformatic pipelines by which GCNs are called also vary by platform and are often proprietary. Advantages to NGS include the identification of other genomic alterations, in addition to differentiating broad chromosomal gains from focal amplification as different regions on a chromosome are tiled, sequenced, and averaged over the cells sampled.
Two types of assays are used in the clinic: amplicon-based NGS and hybrid capture-based NGS. These differ by the method of tumor DNA enrichment31 and are more extensively discussed in a subsequent section of this review. Briefly, hybrid capture-based NGS can more accurately assess of copy number variations of MET and other genes. By contrast, with the amplicon-based approach (in which the amplified regions are limited solely to stretches of DNA flanked by established primers) the genomic territory covered is limited, significant sequence bias can be introduced and sequence replicates cannot be removed, and thus the true sequence coverage depth is affected.
Several technical issues should be considered when utilizing NGS. First, identifying copy number gains/losses is dependent on tumor purity and sample selection as normal cells are often admixed with malignant cells during analysis28–30. Second, poor DNA quality (e.g. from old tumor samples) can increase noise and make it difficult to accurately analyze copy number alterations. Third, large genomic alterations can present a computational challenge and decrease call accuracy32. Finally, studies of the concordance between MET amplification by NGS and FISH have not been done. This makes it difficult to interpret NGS results considering FISH has been better studied.
Other assays
NGS of plasma ctDNA has been used to detect MET amplification. Methods for calling are similar to that of NGS of tumor33,34. Although ctDNA is less invasive and has the potential to pick up more tumor heterogeneity than tumor biopsies, plasma calls depend on the sensitivity and resolution of the platform and relies on rates of tumor shedding33–36. As such, studies have shown that amplification can be missed using this method33,37,38. qRT-PCR of tumor has also been used to detect MET amplification, although the performance of this assay is not well characterized compared to FISH/NGS39–47. Again, cutoffs vary and no clear standard definition for MET amplification has been proposed.
Clinical Features
De novo MET amplification
De novo MET amplification is found across a wide variety of solid tumors. It is identified <1%−5% of NSCLC25,48–51, <1%−10% of gastric cancers19,21,23,52–54, 2–4% of colorectal cancers55,56, 13% of type I papillary renal cell carcinomas (PRCCs), 3% of type II PRCCs57, and at lower frequencies in esophageal carcinoma and hepatocellular carcinoma (HCC)18,24,58. In NSCLC, studies have not shown a strong association between MET amplification and smoking48,59. In the TCGA and cBioPortal databases, MET amplification is also found in less well-studied cancer types, including glioblastoma, melanoma, gynecologic cancers, and lymphoma60–64. In many of these cancers, MET amplification confers a poor prognosis, although these studies were conducted before effective MET-targeted therapies were widely tested17,19,21,23,25. The prognostic implications of MET amplification are unknown against the current backdrop of targeted therapeutics. It is important to recognize that the actual frequency MET amplification varies from study to study and the true frequency of MET amplification within a particular context is challenging to determine. This is unsurprising given that no consensus exists on which assay or cutoff to use, as was discussed extensively in the prior section on diagnosis.
The NSCLC literature has been instrumental in highlighting that, compared to lower-level MET amplification, high-level MET amplification is more likely to indicate oncogenic MET-dependence. In one series, NSCLCs that had high-level MET amplification by FISH (MET/CEP7 ratio ≥5) did not harbor other oncogenic drivers (such as EGFR mutations or ALK fusions), whereas low (MET/CEP7 ≥1.8 to ≤2.2) or intermediate (MET/CEP7 >2.2 to <5) levels of MET amplification were found to overlap more commonly with other oncogenes12. A recent study of crizotinib (a MET TKI) in MET-amplified NSCLC showed that response was enriched in the intermediate to high MET-amplified tumors compared to those with low-level MET amplification or polysomy (MET GCN >6 and MET/CEP7 <1.8), lending support to the use of MET/CEP7 ratio over MET GCN in evaluating MET-dependency65. The cutpoints of the intermediate and high groups were modified to intermediate (MET/CEP7 >2.2 to <4) and high (MET/CEP7 ≥4) to enrich the high MET group for potential responders66. In a separate report, focal MET amplification by NGS was thought to more likely represent a true oncogenic driver state compared to broad gains on chromosome 7 that includes MET67.
Acquired resistance
Whereas tumors with de novo high-level MET amplification are primarily dependent on the MET pathway for growth, tumors that are reliant on other oncogenes (such as mutant EGFR) can develop secondary dependence on the MET pathway as a mechanism of resistance to targeted therapy68. In NSCLC, depending on the cutpoints and assays used, acquired MET amplification is identified in 5–20% of cancers with sensitizing EGFR mutations that develop resistance to 1st, 2nd, and 3rd generation EGFR TKIs69–72. EGFR-mutant cell lines with MET amplification rely on MET-mediated bypass signaling through the PI3K signaling pathway, and that MET-mediated activation of ERBB3 is needed to initiate PI3K signaling43,68. MET amplification has also been described as a mechanism of resistance to ALK inhibitors in ALK-fusion positive NSCLC73. Acquired MET amplification has also been identified in unselected colorectal cancers treated with anti-EGFR monoclonal antibodies74 and in BRAF V600E-mutant colon cancers treated with EGFR inhibitors41,75.
When MET amplification occurs as a secondary driver of resistance, this can occur in a subclonal population. Cutoffs for selecting MET-dependency in this context will need to consider that, when employing an assay that interrogates nucleic acids derived from a mixed population of tumor cells, the existence of MET amplification in a subpopulation of cells and its absence in other cells may result in a lower apparent level of MET copy number change or the lack of a detectable change in MET compared to if single cells were sequenced or if FISH were performed76. The use of complementary assays such as FISH may thus need to be considered in situations where NGS testing fails to detect subclonal MET copy number changes.
Targeted Therapy
De novo amplification
One of the earliest studies to examine the activity of MET-directed targeted therapy by the degree of MET amplification was PROFILE 100177. This phase I trial with an expansion cohort for MET-amplified NSCLCs examined the activity of crizotinib by varying levels of amplification: low (MET/CEP7 ≥1.8 to <2.2), intermediate (MET/CEP7 ≥2.2 to <5), and high (MET/CEP7 ≥5)78. The objective response rate (ORR) was highest (50%) in the high MET amplification group compared to an ORR of 33% and 20% in the low and intermediate groups, respectively. In an update of PROFILE 1001 for which the cutoff between intermediate- and high-level MET amplification was changed to a MET/CEP7 ratio of 466, the best outcomes were consistently achieved in high-level MET-amplified NSCLCs (ORR 40%, median PFS 6.7 months) (Figure 2). A separate clinical trial studied the activity of the selective MET TKI capmatinib in MET-amplified NSCLCs79. Tumors were classified by MET GCN: GCN <4, GCN ≥4 to <6, and GCN ≥6. Similarly, the ORR was highest (47%) in tumors with a MET GCN ≥6.
Figure 2. Targeted therapy for MET amplification.
In six prospective phase I/II clinical trials, patients with cancers possessing higher levels of MET amplification or gene copy number derived increased benefit from MET-directed targeted therapy. Trials included patients with non-small cell lung cancer (NSCLC), papillary renal cell cancer (PRCC), and other solid tumors. While cutoffs for MET amplification/gene copy number varied, the cutoff of a MET/CEP7 ratio or MET gene copy number of 4 was chosen for consistency for trials that used FISH; patient-level data were reviewed to calculate response rates. For the only trial which employed next-generation sequencing (NGS), the trial cutoff of MET gene copy number ≥6 was used. The objective response rate is depicted by cancers that fall below these cutoffs (light red circles) and cancers that met or exceeded these cutoffs (red circles). The size of each circle represents the size of the subpopulation within each trial (with the smallest circle representing an n of 2), and each row represents a single trial. §MET amplification was determined by FISH. ǂMET amplification was determined by NGS.
Finally, the activity of the selective c-MET TKI savolitinib was studied in MET-amplified PRCCs80. As opposed to the two prior studies that used FISH, NGS was utilized and MET amplification was defined as ≥6 copies of MET. MET-amplified PRCCs were more likely to respond (ORR 43%) compared to PRCCs with a GCN <6 (ORR 0%). A summary of targeted therapy outcomes by MET amplification status is featured in Table 1.
Table 1.
Targeted therapy outcomes by MET copy number status
| Drug | Trial (n) | Amplification Criteria | Assay Used | All Patients | MET Amplification Status | |||
|---|---|---|---|---|---|---|---|---|
| Solid tumors | MET-Amplified | |||||||
| SAR125844193 | Phase I (n=72)* | MET GCN >4 and MET/CEP7 ≤2.0a | FISH | ORR | - | 17% (5/29) | ||
| capmatinib198 | Phase I (n=38)€ | MET/CEP7 ≥ 2.0 or MET GCN ≥5b | FISH |
MET
GCN <4 |
MET 4< GCN <6 |
MET
GCN ≥6 |
||
| ORR | 0% (0/36) | 0% (0/22) | 0% (0/6) | 0% (0/3) | ||||
| 95%CI | (0–9.3%) | - | - | - | ||||
| AMG 337199 | Phase I* (n=111) | MET/CEP7 ≥2.0 | FISH | MET/CEP7 <4 | MET/CEP7 ≥ 4 | |||
| mDOR | 6.6 mo | - | - | |||||
| ORR | 10% (11/111) | 0% (0/2) | 60% (6/10) | |||||
| 95%CI | (5–17%) | - | - | |||||
| Gastroesophageal cancers | MET-Amplified | |||||||
| AMG 337200 | Phase II (n=60)¥ | MET/CEP7 ≥2.0 | FISH | mDOR | - | 6.0 mo | ||
| 95%CI | - | (3.7–16.7) | ||||||
| ORR | - | 18% (8/45) | ||||||
| 95%CI | - | (8%–32%) | ||||||
| foretinib201 | Phase II (n=74) | MET/CEP7 ≥2.0 | FISH | mPFS | 1.7 mo | - | ||
| 95%CI | (1.6 –1.8) | - | ||||||
| ORR | 0% (0/71) | 0% (0/3) | ||||||
| Hepatocellular carcinoma | ||||||||
| capmatinib202 | Phase II (n=30) | MET/CEP7 ≥2.0 or GCN ≥5a | FISH | ORR | - | 10% (3/30) | ||
| 95%CI | - | (2.1–27%) | ||||||
| Non-small cell lung cancers | MET/CEP7 ≥1.8 and ≤2.2 | MET/CEP7 >2.2 – <4.0 | MET/CEP7 ≥4.0 | |||||
| crizotinib | Phase I66 (n=37) | MET/CEP7 ≥1.8 | FISH | mPFS | - | 1.8 mo | 1.9 mo | 6.7 mo |
| 95%CI | - | (0.8–14.0) | (1.3–5.5) | (3.4–7.4) | ||||
| ORR | - | 33% (1/3) | 14% (2/14) | 40% (8/20) | ||||
| 95%CI | - | (0.8–91%) | (2%–43%) | (19%–64%) | ||||
| Phase II203 (n=25)* | MET GCN ≥6 and IHC 2+/3+ | FISH | MET/CEP7 ≥1.8 and ≤2.2 | MET/CEP7 >2.2 –<5.0 | MET/CEP7 ≥5.0 | |||
| mPFS | 5.0 mo | - | 4.4 mo | NE | ||||
| 95%CI | (2.7–7.3) | - | (2.9–5.9) | - | ||||
| ORR | 31% (5/16) | - | 36% | 0% (0/2) | ||||
| 95%CI | (5.2–71.4%) | - | (6.0–77%) | - | ||||
| Phase II65 (n=25)* | MET GCN ≥6 and IHC 2+/3+ | FISH | mPFS | 3.2 mo | - | - | - | |
| 95%CI | (1.9–3.7) | - | - | - | ||||
| ORR | 32% (8/25) | - | - | - | ||||
| capmatinib79 | Phase I (n=44) | MET/CEP7 ≥2.0 or MET GCN ≥5c,d | FISH |
MET
GCN <4 |
MET 4< GCN <6 |
MET
GCN ≥6 |
||
| ORR | 20% (11/55) | 0% (0/17) | 17% (2/12) | 47% (7/15) | ||||
| 95%CI | - | - | (2–48%) | (21–73%) | ||||
| Non-small cell lung cancers (EGFR-mutant) | MET-amplified | |||||||
| osimertinib/ savolitinib 300 mg daily204 | Phase I (n=36) | Previously treated with 1st/2nd generation EGFR-TKI and T790M negative; MET/CEP7 ratio ≥2 or MET GCN ≥5 by FISH or MET GCN ≥5 by NGSd | FISH or NGS | mPFS | - | 9.1 mo | ||
| 95%CI | - | (5.4–12.9) | ||||||
| ORR | - | 64% | ||||||
| 95%CI | - | (46%–79%) | ||||||
| osimertinib/ savolitinib 300 or 600 mg daily204 | Phase I (n=51) | Previously treated with 1st/2nd generation EGFR-TKI and T790M negative; MET/CEP7 ratio ≥2 or MET GCN ≥5 by FISH or MET GCN ≥5 by NGSd | FISH or NGS | mPFS | - | 9.0 | ||
| 95%CI | - | (5.5–11.9) | ||||||
| ORR | - | 65% (33/51) | ||||||
| 95%CI | - | (50%–78%) | ||||||
| osimertinib/ savolitinib 300 or 600 mg daily204 | Phase I (n=18) | Previously treated with 1st/2nd generation EGFR-TKI and T790M positive MET/CEP7 ratio ≥2 or MET GCN ≥5 by FISH or MET GCN ≥5 by NGSd | FISH or NGS | mPFS | - | 11.0 mo | ||
| 95%CI | - | (4.0-NE) | ||||||
| ORR | - | 67% (12/18) | ||||||
| 95%CI | - | (41–87%) | ||||||
| osimertinib/ savolitinib 300 or 600 mg daily204 | Phase I (n=69) | Previously treated with 3rd generation EGFR-TKI; MET/CEP7 ratio ≥2 or MET GCN ≥5 by FISH or MET GCN ≥5 by NGSd | FISH or NGS | mPFS | - | 5.4 mo | ||
| 95%CI | - | (4.1–8.0) | ||||||
| ORR | - | 30% (21/69) | ||||||
| 95%CI | - | (20–43%) | ||||||
|
gefitinib/ tepotinib205 |
Phase II (n=12) | MET/CEP7 ratio ≥2 or MET GCN ≥5; after prior EGFR-TKI and T790M negative | FISH | mPFS | - | 16.6 mo | ||
| 90%CI | - | (8.3-NE) | ||||||
| ORR | - | 67% (8/12) | ||||||
| 90%CI | - | (39%–88%) | ||||||
|
gefitinib/ savolitinib206 |
Phase I (n=44) | MET/CEP7 ratio ≥2 or MET GCN ≥5; after prior EGFR-TKI | FISH | ORR | - | 25% (11/44) | ||
| gefitinib/capmatinib82 | Phase II (n=100) | GCN ≥5 or IHC 2+/3+ in >50%; then, GCN ≥5 plus IHC 2+/3+; then, GCN ≥4 or IHC 3+; MET/CEP7 ≥1.8 | FISH |
MET
GCN <4 |
MET
4 ≤ GCN <6 |
MET
GCN ≥6 |
||
| mPFS | 5.5 mo | 3.9 mo | 5.4 mo | 5.5 mo | ||||
| 95%CI | (3.8–5.6) | (3.7–5.6) | (3.7–7.5) | (4.2–7.3) | ||||
| ORR | 29% (29/100) | 12% (12/41) | 22% (4/18) | 47% (17/36) | ||||
| Papillary renal cell carcinoma | MET GCN <6 and not MET-driven§ | MET GCN ≥6 | ||||||
| savolitinib80 | Phase II (n=79)* | Focal MET GCN ≥6 | NGS | ORR | 10% (8/79) | 0% (0/32) | 43% (3/7) | |
MET positivity also defined as
immunohistochemistry (IHC) ≥50% with 2+/3+
H-score ≥150 or IHC ≥50% with 2+/3+ or H-score ≥50 for hepatocellular carcinoma and glioblastoma
H-score ≥150 or IHC ≥50% with 2+/3+
IHC ≥50 with % 3+.
Available patient level data with MET gene copy numbers or MET/CEP7 ratio and response results were used to produce these results.
Response and outcome data derived from dosing regimen #3 cohort of the study, but response rates by MET amplification status in this cohort was not available.
Only data from patients receiving a stable dose in the dose expansion cohort were used.
One patient with partial response had both a MET exon 14 alteration and MET copy number 2.3.
Only patients from cohort 1 was used.
Not MET-driven defined as no MET focal amplification, no MET kinase domain mutation, no HGF amplification, and no chromosome 7 gain. CI, confidence interval; ORR, objective response rate; mPFS, median progression-free survival; mDOR, median duration of response; GCN, gene copy number; NE, not estimable; NGS, next-generation sequencing; FISH, fluorescence in-situ hybridization; IHC, immunohistochemistry; mo, month(s); HCC, hepatocellular carcinoma; GBM, glioblastoma; PRCC, papillary renal cell carcinoma
Acquired resistance
In tumors with primary dependence on another oncogene and secondary dependence on MET, combination therapy targeting both the primary driver and MET is active. For example, the combination of an EGFR TKI and a MET TKI is active in EGFR-mutant NSCLCs that acquire MET amplification after progression on a prior EGFR TKI. In a trial of osimertinib and capmatinib, the ORR was 25%81 in EGFR-mutant NSCLCs that acquired MET amplification (GCN ≥5 or MET/CEP7 ratio ≥2 by FISH) after progression on osimertinib. In a trial of gefitinib and tepotinib, the ORR was 67% in EGFR-mutant NSCLCs that acquired MET amplification (GCN ≥5 or MET/CEP7 ratio ≥2 by FISH) after progression on a 1st generation EGFR TKI. Interestingly, the degree of activity of combination therapy also increases with increased MET copy number (Figure 1). One study of EGFR-mutant NSCLCs with MET amplification82 evaluated the activity of gefitinib and capmatinib by MET GCN: GCN <4, GCN ≤4 to <6, and GCN ≥6. The ORR was highest (47%) in patients with GCNs ≥6. Other agents such the EGFR-MET bispecific antibody JNJ-372 have shown promising activity in EGFR-mutant cancers that have become resistant to EGFR TKI therapy and the activity of these drugs should be characterized further in tumors with acquired MET amplification.
Taken together, higher levels of MET amplification can predict for increased benefit from MET-directed targeted therapy. This applies to (1) single-agent MET inhibitor therapy for MET-amplified cancers, and (2) combination therapy that includes a MET inhibitor for cancers that develop MET amplification as a mechanism of resistance to targeted therapy directed against the primary oncogenic driver. The need for standardized definitions of MET amplification thus has substantial implications not only for the diagnosis of MET-dependent cancers, but for the identification of cancers that are more oncogenically addicted to MET with a higher likelihood of benefit from MET-directed targeted therapy.
MET MUTATION
Genomic diversity
Activating mutations can occur at a diverse range of positions along MET. These include alterations involving the kinase domain, intronic splice sites that flank exon 14, and the extracellular domain.
Kinase domain mutations
MET mutations were first described in hereditary HPRCC83. These germline MET mutations include V1092I, H1094R/Y M1131T, V1188L, V1220I, M1250T, and D1228H/N/V. MET mutation increases MET kinase activity and leads to phenotypic transformation or tumor focus formation in vitro84. It also induces tumor formation in mouse models in vivo84,85. Later studies discovered that MET mutations occur in sporadic PRCCs. These are found in up to 15% of cases57,86,87, predominantly in type 1 and less commonly in type 2 PRCC57. The spectrum of MET mutations in sporadic PRCCs includes V1092I, H1094l/R/Y, N1100Y, H1106D, M1131T, V1188L, L1195V, V1220I, D1228H/N/V, Y1230H, Y1230A/C/D/H, Y1235D, and M1250T/I83,88,89 (Figure 3). Interestingly, some of these mutations are less active than others and concomitant MET amplification may be necessary to increase oncogenesis85,86,90. Activating MET mutations have been found in other cancers, including HCC and head and neck cancers91,92. In the latter, MET Y1235D mutation is found in up to 14% of cases93. In addition to being found de novo, MET kinase domain mutations can emerge as a mechanism of acquired resistance to MET TKI therapy. In MET exon 14 NSCLCs (discussed in detail below), MET Y1230C, Y1230H, D1228H, and D1228N have been found to mediate resistance to prior MET TKI therapy with crizotinib94–98 by disrupting drug binding94. MET kinase domain mutations have also emerged as resistance mechanisms in EGFR-mutant lung cancers who were treated with a combination of an EGFR TKI and a MET TKI for MET-amplification-mediated acquired resistance99.
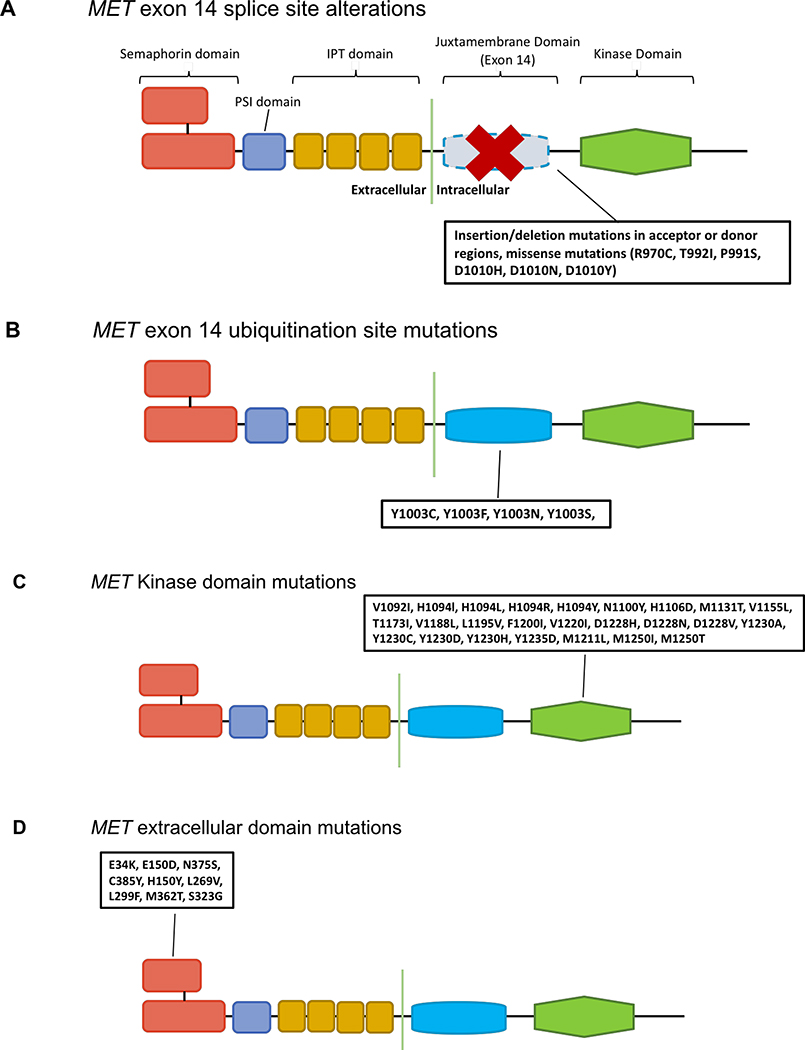
Figure 3. MET mutations.
The MET protein consists of the extracellular semaphorin (SEMA) domain in red, a plexin-semaphorin-integrin (PSI) domain in purple, four immunoglobulin-plexin-transcription (IPT) repeats in yellow, a juxtamembrane domain (encoded by exon 14) in blue, and a kinase domain in green. A) MET exon 14 splice site alterations result in exon 14 exclusion. This results in the absence of the ubiquitin binding site of the juxtamembrane domain, impaired MET degradation, and increased MET signaling. B) Missense mutations in the juxtamembrane domain prevent spliceosome binding or modify the Y1003 ubiquitination site. These ultimately recapitulate the biology of MET exon 14 splice site alterations. C) Mutations in the kinase domain lead to the increased activation of the MET kinase and can be associated with conformational changes that favor the xDFG-out state. D) Other mutations can occur in the semaphorin domain where hepatocyte growth factor, the ligand for MET, binds. The impact of these mutations on MET function are unclear.
MET exon 14-alterations
When MET is activated, the Y1003 residue of the juxtamembrane domain that is encoded by exon 14 is phosphorylated. This mediates c-Cbl E3-ligase binding and subsequent ubiquitination and degradation of the MET receptor100. MET exon 14 alterations are a heterogeneous group of MET mutations that convergently interfere with this process. While MET exon 14 alterations were first identified in small cell lung cancer (SCLC)101, later studies revealed that these mutations are more commonly found in NSCLC, with a prevalence of 3–4%102–105. These are further enriched in sarcomatoid carcinomas (occurring in 9–22% of cases), an aggressive NSCLC variant that can be highly resistant to chemotherapy106. MET exon 14 alterations occur at lower frequencies in other cancers, including gastric cancers and neuroblastomas103,107,108. Beyond being found de novo in various malignancies, MET exon 14 alterations have been shown to mediate resistance to EGFR TKIs in EGFR-mutant NSCLC109,110. For NSCLCs with MET exon 14 skipping, a higher proportion of patients have a smoking history compared to those whose cancers harbour other drivers such as ALK/ROS1/RET fusions, although several independent reports have noted that a proportion of patients with these alterations are never smokers, consistent with the GEOMETRY trial results111–113.
Most MET exon 14 alterations are mutations that interfere with RNA splicing. In the wild-type state, the intronic regions of MET pre-mRNA are spliced out before the transcript is translated into a protein. Mutations that occur in regions that flank MET exon 14 (i.e. the polypyrimidine tract, splice donor, or splice acceptor regions) effectively disrupt normal splicing and result in exon 14 being skipped16. The loss of the encoded juxtamembrane domain leads to the loss of the Y1003 ubiquitin binding site on MET. Consequently, MET degradation is decreased and MET expression increases, thus driving oncogenesis114–116. Most of these splice site mutations take the form of insertions or deletions with a wide range of sizes. In addition, missense mutations that result in D1010 substitution (i.e. D1010H/N/Y) disrupt splicing117.
MET mutations that do not directly affect splicing can recapitulate a similar biology. For instance, mutations that lead to Y1003 substitution (i.e. Y1003F/N/S) interfere with c-Cbl E3-ligase binding115. Like MET exon 14 RNA splice site mutations, Y1003 substitution transforms normal cells, leads to increased cell proliferation, and promotes tumor growth114,115. Large deletions that encompass exon 14 result in the loss of the juxtamembrane domain that carries Y1003.
Other mutations
The semaphorin (SEMA) domain of MET protein interacts with HGF (the ligand of MET) and is involved in dimerization that leads to receptor activation118. Mutations in this domain include E34K, E150D, H150Y, L269V, L299F, S323G, M362T, N375S, and C385Y (Figure 3)102,119–122. N375S is the most common of these and occurs in 3–14% of NSCLCs123. Whether SEMA domain mutations (particularly N375S) are activating is a point of contention. While N375S mutation has been shown to participate in carcinogenesis by activating downstream SRC and ERK1/2 signaling124, data have shown that some SEMA domain mutations decrease the binding affinity of HGF and are found in individuals without cancer123,125.
Diagnosis
DNA-based sequencing
As MET exon 14 alterations are highly heterogenous, it is important to select an NGS assay that is poised to capture this wide variety of mutations103. As mentioned previously, amplicon-based NGS and hybrid-based NGS are the two main types of assays used in the clinic. In amplicon-based NGS, genes of interest are sequenced using an established set of DNA primers that flank defined genomic regions126. Although this approach can offer a shorter turn-around time and improved capture of difficult regions, it is less tolerant of sequence variability and more prone to sequencing bias and allelic dropout, especially for repetitive sequences. Many MET exon 14 alterations, particularly indels that result in splicing defects, can lie outside these amplified regions and be missed127,128. Furthermore, mutations such as large insertion/deletions may involve a primer binding site and interfere with primer binding; this likewise prevents amplicon-based assays from detecting these alterations126. Consistently, studies have demonstrated that a significant fraction of MET exon 14 alterations can be missed by amplicon-based NGS127,128.
In contrast, hybrid capture-based NGS platforms avoid some of the issues associated with amplicon-based NGS128. Tumor DNA is sheared, captured by long oligonucleotide baits, and then amplified. There is overlap between these unique DNA fragments so duplicate sequences can be identified and removed and the challenges of sequencing repetitive sequences can be adjusted for with careful bait design. By tiling over appropriate intronic regions, hybrid capture-based NGS forestalls primer binding issues and can detect mutations that are further into the intron. The use of a capture based approach also corrects for some of the sequencing bias and allele drop-out issues that occur with amplicon-based NGS126. With respect to calling missense mutations, hybrid capture-based NGS also outperforms amplicon-based NGS. Although both platforms usually have high coverage depth of genes of interest to ensure accuracy, amplicon-based platforms are more commonly associated with false-positives or false-negatives consequently affecting the threshold of sensitivity that the assay can attain.
RNA-based sequencing
It is important to point out that while DNA-based sequencing detects mutations that lie within regions that are predicted to result in MET exon 14 skipping, it cannot confirm the absence of the exon that occurs at the RNA level. As such, RNA-based sequencing platforms can complement DNA-based NGS128. Given the diversity of MET exon 14 splice site alterations, it can be difficult to interpret whether some mutations identified by DNA-based NGS truly result in exon 14 skipping. Interpretations can be made based on proximity to splice donor/acceptor regions, but this becomes challenging with alterations that occur deeper into the intron. In contrast, RNA sequencing directly identifies the loss of exon 14 transcription. In addition, the challenges associated with the need to sequence large intronic regions in DNA-based NGS are avoided with RNA-based sequencing methods, given the absence of intronic regions in RNA. Furthermore, whereas the effect of a new mutation on METex14 can be difficult to interpret with DNA-based platforms, RNA sequencing directly demonstrates whether exon 14 has been transcribed.
Assays using anchored multiplex PCR (AMP, Archer Technologies) have been utilized to identify MET exon 14 alterations. Using this method, RNA is used as template to generate complementary DNA (cDNA) from tumor129. As the cDNA is sequenced and amplified, molecular barcodes are incorporated to quantify sequences and correct sequencing errors. The loss of exon 14 is then identified in cDNA. One study examined the utility of AMP RNA sequencing to identify MET exon 14 alterations in 232 NSCLCs that were “driver-negative” by DNA-based hybrid capture NGS130. Six MET exon 14 skipping alterations were identified by RNA sequencing that DNA-based NGS did not detect. An analysis of the genomic sequences identified by DNA-based NGS was then retroactively performed. In five of six cases, novel MET exon 14 splice site alterations were detected after further manual review127. In the remaining case, MET mutation was not detected, suggesting that MET exon 14 skipping occurred by another mechanism.
Molecular counting using the nCounter system (Nanostring Technologies) has also been used to identify MET exon 14 loss at the RNA level. The system uses probes to detect MET transcripts of interest, utilizing a fluorescently tagged 5’ reporter probe and biotinylated 3’ capture probes131. In contrast to AMP, it does not require the reverse transcription of RNA to cDNA and does not require amplification. Instead, target-specific color-coded probes detect the sequences of interest and the color codes are counted and tabulated using a digital analyzer for image acquisition131. Further studies of this method are needed, particularly as it has not been compared to AMP for the detection of MET exon 14 skipping132,133.
Targeted therapy
Kinase domain mutations
MET-directed targeted therapy is active against select MET kinase domain mutations. It is critical to recognize that the activity of various MET TKIs against specific mutations is variable. Specifically, while type II MET inhibitors such as cabozantinib and foretinib have preclinical activity against several kinase domain mutations (e.g. D1228N, M1250T, H1094Y/L134), type I MET inhibitors such as crizotinib do not have substantial activity against these alterations. As such, MET kinase domain mutations have emerged as mechanisms of resistance to crizotinib in MET-amplified and MET exon 14-altered cancers. The ability of these mutations to switch the conformation of the MET kinase from an active (xDFG-in) to an inactive (xDFG-out) conformation may be contributory as type I MET inhibitors preferentially bind to the former while type II MET inhibitors bind to the latter135.
Data in tumors with de novo MET kinase domain mutations is largely limited to PRCC. In a study of foretinib in patients with HPRCC with germline MET mutations, the ORR was 50%136. In cancers where MET kinase domain mutations were acquired as mechanisms of resistance to prior MET TKIs, data is limited to NSCLCs94–96,98. Patients were identified with cancers that developed MET kinase domain mutations as resistance mechanisms to the combination of an EGFR TKI and crizotinib (administered to EGFR-mutant cancers with MET amplification-mediated resistance to a prior EGFR TKI)137. TKI type switching to the type II MET inhibitor cabozantinib resulted in the re-establishment of disease control.
MET exon 14 alterations
As opposed to kinase domain mutations that can alter the conformation of the MET kinase, MET exon 14 alterations harbor a MET kinase domain that is presumably similar to the wild-type MET receptor kinase domain100. Thus, both type I and type II MET TKIs are poised to inhibit MET exon 14-altered cancers and have shown preclinical activity in these models. The clinical activity of MET inhibition in MET exon 14-altered NSCLCs was first prospectively established with crizotinib, a type Ia multikinase MET inhibitor77. The phase I PROFILE 1001 study accrued patients with advanced MET exon 14-altered NSCLCs to an expansion cohort of the trial. The ORR was 32% and the median PFS was 7.3 months (Table 2). These results supported the inclusion of crizotinib in the NCCN guidelines for this indication. It also supported the US FDA’s decision to grant crizotinib Breakthrough Therapy Designation for MET exon 14-altered NSCLCs in 2018138.
Table 2.
Targeted therapy in MET exon 14-altered lung cancers
| Inhibitor Type | Type Ia | Type Ib | ||
|---|---|---|---|---|
| Drug | Crizotinib77 | Capmatinib113 | Tepotinib140 | Savolitinib207 |
| Objective response rate (ORR) | 32% (95% CI 21–45) n=21/65 |
52% (95% CI NR) n=17/31 |
||
| ORR by treatment | ||||
| Naïve (no systemic therapy) Pre-treated (no prior c-MET therapy) |
68% (95% CI 48–84) n=19/28 41% (95% CI 29–53) n=28/69 |
44% (95% CI 22–69) n=8/18 45% (95% CI 28–64) n=15/33 |
||
| ORR by mutation type | ||||
| Insertion/deletion Point Mutation |
0% (95% CI NR) n=0/4 36% (95% CI NR) n=12/33 |
NR | NR | 43% (95% CI NR) n=6/14 59% (95% CI NR) n=10/17 |
| ORR by splice site | ||||
| Splice acceptor Splice donor |
31% (95% NR) n=5/16 32% (95% CI NR150) n=12/37 |
NR | NR | 42% (95% CI NR) n=5/12 58% (95% CI NR) n=11/19 |
| ORR by MET amplification | ||||
| Concurrently amplified Wild-type |
0% (95% CI NR) n=0/2 NR (95% CI NR) n=NR |
NR | NR | 100% (95% CI NR) n=5/5 35% (95% CI NR) n=8/23 |
| Median progression-free survival | 7.3 months (95% CI 5.4–9.1) |
12.3 months (95% CI 6.3-NR) |
NR | |
| Median progression-free survival | ||||
| Naïve (no systemic therapy) Pre-treated (no prior MET TKI) |
9.7 months (95% CI 5.5–13.9) 5.4 months (95% CI 4.1–7.0) |
|||
NR, not reported
Newer agents such as the selective type Ib MET inhibitors such as capmatinib, tepotinib, and savolitinib have since been explored in MET exon 14-altered NSCLCs113,139,140 (Table 2). Notably, selective MET inhibitors such as capmatinib are more potent compared to crizotinib117. In the GEOMETRY trial, the ORRs with capmatinib were 68% and 41% in treatment-naïve and chemotherapy pre-treated patients, respectively113. In the VISION trial, the ORR with tepotinib was 44%140. In a phase 2 trial of savolitinib, the ORR was 52%139. The durability of disease control and relative toxicity of these agents compared to crizotinib has not yet been well characterized.
Given that the response rates to MET TKIs vary widely, several investigators have attempted to identify subgroups within MET exon 14-altered NSCLCs that are less likely to respond to therapy77,139. The PROFILE 1001 study of crizotinib first noted that response did not significantly vary by MET exon 14 alteration region (splice acceptor vs donor site involvement), mutation type (insertion/deletion vs base substitution), or concurrent MET amplification77 (Table 2). This was subsequently confirmed by studies of selective MET inhibitors such as savolitinib139.
In terms of acquired resistance in patients with initial benefit, on-target mechanisms such as MET amplification and MET kinase domain mutations have been identified. While the role that HGF plays in resistance remains to be determined, HGF amplification has been found in the setting of acquired resistance97,141. While preclinical data supporting the role of the latter in MET exon14-altered NSCLCs is not available, in models of MET-amplified HCC cancer cell lines, HGF can reduce the sensitivity to MET TKIs in MET-amplified cancer cell lines and patient-derived xenografts142.
MET FUSION
Clinicopathologic features
The MET gene was originally identified as an oncogene after chemically transformed osteosarcoma cell lines were found to have the TPR-MET fusion147. MET fusions were thereafter identified in gastric cancers, thyroid carcinoma, PRCC, lung adenocarcinoma, HCC, glioma, and sarcoma. The exact frequency within these cancers is poorly defined, although these are enriched in gliomas and found in 12% of cases148,149. Beyond TPR-MET, multiple other fusions have been identified60,61,105,148,150–161 (Figure 4). MET fusion can occur through intrachromosomal (e.g. PTPRZ1-MET, CLIP2-MET, CAPZA2-MET, ST7-MET) or interchromosomal fusion155, each in about half of cases. In pediatric glioblastomas, intrachromosomal fusion appears to be more common and frequently involves PTPRZ1148. MET fusion can also result from paracentric or pericentric inversion - the latter appears to be preferred.
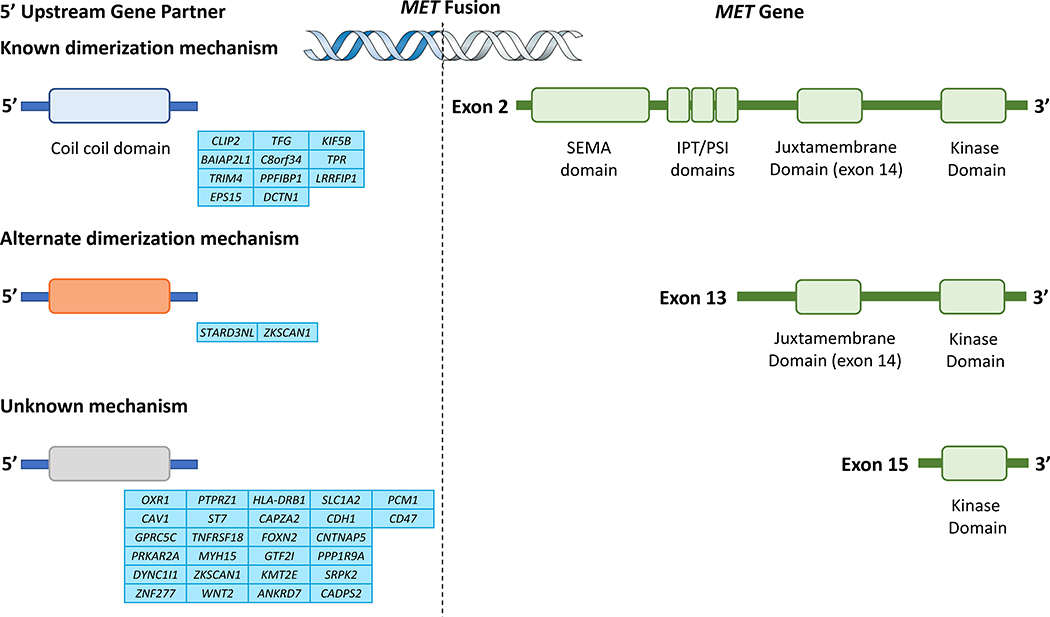
Figure 4. MET Fusion.
A wide variety of MET fusions have been identified. These fusions can result in constitutive MET activation in different ways. The 5’ upstream partners CLIP2, TFG, KIF5B, BAIAP2L1, C8orf34, TPR have coiled-coil domains that promote chimeric oncoprotein dimerization. Other domains (e.g. the MENTAL domain of STARD3NL) can mediate alternate methods of homodimerization. The 3’ MET gene typically includes the kinase domain, however, fusions that include the juxtamembrane domain, or larger regions of the gene have been identified. Interestingly, certain fusions such as TPR-MET result in the exclusion of exon 14. The biology of this fusion is thus thought to be similar to that of MET exon 14-altered cancers. The exons at which the breakpoints for MET can occur are noted in the diagram.
These fusions often include the kinase domain on exon 15148,150,151,153,154,160,162,163 and many upstream partners provide dimerization domains, resulting in ligand-independent constitutive MET activation. In addition, select fusion events (e.g. TPR-MET) have been found to exclude exon 14, enabling behavior similar to MET exon 14 alteration biology162. Interestingly, fusions that include exon 14 appear (e.g. KIF5B-MET, PTPRZ-MET) to be less oncogenic than fusions that exclude exon 14148. In PTPRZ-MET, the PTPRZ promoter is fused to the full-length MET gene that includes the MET dimerization domain on exon 2; this results in both MET overexpression and increased downstream signaling activation148,164.
Diagnosis and targeted therapy
A number of assays can detect MET fusions. These include FISH, RT-PCR, and NGS148,150,165,166. How well each of these performs has not been well-studied and complex/novel rearrangements can make FISH inadequate for detecting many MET fusions167. This section will thus focus primarily on NGS, the increasingly preferred assay in the clinic. DNA-based NGS can reliably detect a wide variety of MET fusions, although several general features of fusions make it difficult for even DNA-based hybrid capture NGS to capture all events168. First, repetitive intronic DNA sequences can occur in fusions breakpoints. Because these can also occur at other sites of the genome and hybrid capture produces short reads, using baits can result in reads that are unmappable to the reference genome30. As a result, these baits are excluded in contemporary assays, increasing the chance of missing genomic fusion breakpoints. Second, the intronic regions of select fusion partners can be prohibitively long, making it challenging and impractical to tile these sequences130. Third, DNA-based NGS is limited in its ability to detect novel gene fusion partners167,169. As discussed earlier, these issues can be avoided with RNA-based AMP NGS130,169,170. In support of this, the same study of DNA-based NGS “driver-negative” NSCLCs identified actionable gene fusions in 12% (27 of 232) of cases using RNA-based NGS130. This suggests that AMP can complement DNA-based NGS in detecting MET fusions, especially in settings where no driver is found.
The utility of MET-directed targeted therapy has been incredibly underexplored in MET fusion-positive cancers. MET TKIs induce apoptosis in TPR-MET-transformed cell lines171 and isolated clinical responses to crizotinib have been described in MET fusion-positive lung adenocarcinomas and gliomas148,155,172. In the PROFILE 1001 study, one patient was included in the MET exon 14 cohort given that his cancer harbored a MET fusion that resulted in exon 14 skipping77. A confirmed response was achieved. Multiple clinical trials are underway to evaluate the efficacy of MET TKIs in cancers including those with MET fusions (www.clinicaltrials.gov: NCT02978261, NCT03993873, NCT01639508).
MET OVEREXPRESSION
The role of MET expression in oncogenesis should be considered in a number of contexts. First, MET can be transcriptionally induced in cancer cells in the setting of hypoxia/inflammation to activate proliferation, decrease apoptosis, and promote migration. Tumors can thus rely on MET signaling even in the absence of a genomic driver such as MET amplification, mutation, or fusion. Such states could theoretically be addressed by MET-directed targeted therapy although the clinical data in this setting with monoclonal antibodies has been disappointing. Thankfully, newer strategies such as biparatopic antibodies, antibody combinations, and antibody-drug conjugates are being explored. Second, MET can be overexpressed in cancers that harbor an activating genomic signature, including those with primary/secondary MET amplification or MET exon 14 alterations.
Diagnosis
Immunohistochemistry
A number of MET IHC antibodies have been utilized. These include monoclonal antibodies [e.g. SP44 (Ventana), cMET (R&D systems), MET4 (Dako)], polyclonal antibodies (e.g. polyclonal MET AF276 (R&D systems) and antibodies to phosphorylated MET [pMET Y1349 (Epitomics)]173–177. Of these, SP44, a rabbit monoclonal anti-total MET antibody clone, is commonly used. The comparative performance of these antibodies is not known. The extent and intensity of IHC staining as assessed by a pathologist and provides a semi-quantitative measure of MET protein expression. Various scoring systems have been used to define both MET expression and overexpression by IHC48,175,178. Most commonly, staining is based on a 0–3+ scale corresponding to negative (0), weak (1+), moderate (2+), or strong (3+)175. Staining that is 1+ or greater provides evidence for MET expression. A common cutoff for MET overexpression is 2+ in at least 50% of the cells (MetMab criterion)175. Another scoring system, the H-score, multiplies the percentage of cells with 1+, 2+, or 3+ staining by the staining intensity score179. H-scores range from 0–300; ≥200 usually denotes overexpression but cutpoints vary48,180. Investigators have also used a median H-score (of the range of H-scores obtained from samples exclusively within a given study) as a cut point for overexpression; this approach makes standardization across studies difficult.
Mass spectrometry
Selected reaction monitoring mass spectrometry (SRM-MS) utilizes serial steps of ionization and fragmentation of tumor proteins to quantify the mass of the MET protein per sample (in amol/μg). SRM-MS can measure MET from formalin-fixed paraffin-embedded tissue and maintains reproducibility in samples a year old and beyond181. Compared to IHC, SRM-MS can be less dependent on interobserver bias and may detect lower levels of expression. However, SRM-MS cannot differentiate between tumor and normal tissue and MET quantification may be influenced by admixed stroma or inflammatory infiltrates. Furthermore, SRM-MS is more complicated than IHC and requires more expensive technology, making it less widely adopted. IHC is more routinely used in diagnostic pathology laboratories while SRM-MS use has largely been investigational.
Screening
The presence of MET overexpression has been investigated as a screening tool for activating MET alterations. Unfortunately, MET overexpression has not reliably detected MET amplification or MET exon 14 alterations and there is little data on MET fusion detection59. This contrasts with ALK-rearranged NSCLC where ALK overexpression by IHC strongly correlates with the presence of an ALK rearrangement by FISH182.
MET amplification
MET IHC overexpression has not been strongly correlated with MET amplification183–185. This may be secondary to the inclusion of lower levels of MET amplification that do not result in substantial protein expression, or expression may be variably modulated by post-transcriptional/translational factors. In NSCLC, MET amplification (MET/CEP7 ratio >2.2) was only detected in one of 74 patients (1%) with MET overexpression (H-score ≥ 200) in a series from the Lung Cancer Mutation Consortium (LCMC)186. In gastric cancer, a MET IHC H-score of 150 had a 75% sensitivity and 78% specificity to detect MET amplification (MET/CEP7 ratio >2.0 and GCN >4.0)181. All the above studies utilized FISH and there is little data on NGS.
MET mutation
Interestingly, while MET exon 14-altered NSCLCs are expected to overexpress MET, not all tumors are MET-positive by IHC or SRM-MS97,112,187. In 25 patients with MET exon 14-altered lung cancer, only 16 patients (64%) were MET IHC positive (2+ and 3+) and about a third of cases were not found to express MET by SRM-MS187. Similar to MET-amplified cancers, the expression of MET may be modulated by post-transcriptional/translational factors.
It is thus not surprising that MET overexpression does not highly correlate with the presence of MET exon 14 alteration. In the LCMC series discussed above, MET exon 14 alteration was only detected in two of 74 patients (3%) with MET overexpression (H-score ≥ 200)186. The sensitivity and specificity of MET IHC overexpression for MET exon 14 alterations is variable. IHC had a 90% sensitivity and 47% specificity for predicting MET exon 14 mutation in NSCLC188, and 20% sensitivity and 83% specificity in sarcomatoid lung carcinoma180. IHC has not routinely been explored as a screening tool for other MET mutations (e.g. kinase domain mutations) that do not result in exon 14 alteration.
Targeted therapy
In isolation, MET overexpression is not consistently predictive of benefit from MET-directed therapy. Reasons include the challenge of defining expression versus overexpression for a continuous variable, and overexpression may not be not equivalent to a MET-dependent state. While many cancers overexpress MET by IHC, the frequency of overexpression is variable and dependent on cutoff point used. For example, this ranges from 24–66% in NSCLCs184,188 and 28%−63% in gastric cancers23,52,189. Multiple therapeutic anti-MET antibodies (onartuzumab, emibetuzumab)190,191, anti-hepatocyte growth factor (HGF) antibodies (ficlatuzumab, rilotumumab)178,192 and TKIs (crizotinib, cabozatinib, tivantinib, SAR125844, tepotinib)193–196 have been tested in clinical trials. The overall activity of these drugs as monotherapies for MET-overexpressing cancers was low (Table 3). For example, in a phase III trial for MET-overexpressing HCCs, there was no difference (p=0.81) in the median OS between patients who received tivantinib (8.4 months) versus placebo (9.1 months). In the same population of MET-overexpressing HCCs, tepotinib resulted in a short median PFS of 2.8 months and the degree of MET IHC positivity did not select for improved activity. Given the limited therapeutic success with antibody-directed therapies, many drug development programs combined these therapies with chemotherapy, or EGFR-directed targeted therapy. The latter strategy was chosen given the preclinical synergy of this combination and the identification of secondary MET dependence following EGFR TKI resistance. Unfortunately, while select phase 2 trials seemed promising (with improvements in PFS and OS), subsequent confirmatory phase III trials did not show a benefit in MET-overexpressing cancers (Table 3) and the level of MET expression did not correlate with increased benefit. Additionally, patients with MET-overexpressing cancers by IHC were equally responsive to MET-directed therapy or placebo (Supplemental Table 1).
Table 3.
Targeted therapy outcomes by MET expression status
| MET IHC | Drug | Disease | Trial (n) | IHC (−) | IHC (+) | |||
|---|---|---|---|---|---|---|---|---|
| Polyclonal antibody | ||||||||
| AF276 | rilotumumab + chemotherapy173 | prostate cancer | phase II (n=144) | + low | + high | |||
| mPFS | - | 3.6 mo | 2.9 mo | |||||
| 80% CI | - | (2.8–4.0) | (2.7–3.5) | |||||
| Monoclonal antibodies | ||||||||
| MET4 | rilotumumab + chemotherapy174 | gastric/GEJ cancer | phase Ib/II (n=9 for Ib, n=121 for II) | mPFS | 6.8 mo | 4.4 mo | ||
| mOS | 11.1 mo | 10.6 mo | ||||||
| ORR | 6/19 (32%) | 20/40 (50%) | ||||||
| SP44 | tivantinib194 | hepatocellular carcinoma | phase II (n=71) | mOS | 3.8 mo | 9.0 mo | ||
| tivantinib + chemotherapy208 | colorectal cancer | phase II (n=117) | mPFS | 11 mo | 7.9 mo | |||
| mOS | NE | 22.3 mo | ||||||
| ORR | 11/36 (31%) | 24/54 (44%) | ||||||
| onartuzumab + chemotherapy209 | triple negative breast cancer | phase II (n=185) | + low | + high | ||||
| mPFS | 7.3 mo | 5.7 mo | 10.3 mo | |||||
| mPFS | - | 8.5 mo | 4.4 mo | |||||
| onartuzumab + erlotinib210 | EGFR-mutant non-small cell lung cancer | phase II (n=61) | 95% CI | - | (7.1–12.4) | (0.7-NE) | ||
| ORR | 35/53 (66%) | 7/8 (87%) | ||||||
| onartuzumab + erlotinib175 | non-small cell lung cancer | phase II (n=137) | 1+ | 2+ | 3+ | |||
| mPFS | 1.4 mo | 1.4 mo | 4.1 mo | 2.7 mo | ||||
| ORR | 1/32 (3%) | 3/35 (9%) | ||||||
| Unknown antibody | capmatinib | Non-small cell lung cancer | phase I (n=52) | 1+ | 2+ | 3+ | ||
| ORR | - | - | 2/12 (17%) | 9/37 (24%) | ||||
New MET antibody strategies have thus been explored. These include mixtures of antibodies directed against different epitopes on the MET protein (e,g, Sym015) that have shown preclinical activity against MET overexpressing and MET exon 14-altered cell lines. In addition, antibody drug conjugates have been explored. These drugs have the advantage of binding MET-expressing cancer cells regardless of MET signal dependence and lower levels of expression may be sufficient for payload delivery. As an example, a study of the antibody-drug conjugate telisotuzumab vedotin did not find a correlation between MET expression levels and benefit in a variety of solid tumors. It is important to keep in mind that, in the same vein, as MET can be expressed on normal lung tissues197, pulmonary toxicity can be an issue.
While MET expression is a poor predictor of benefit from MET-directed therapy in the absence of evidence of a genomic correlate for MET dependence, the predictive nature of MET expression is being recontextualized in cancers that are oncogenically addicted to MET. In one study of MET exon 14-altered NSCLCs, MET expression was surprisingly heterogeneous, where 31% (n=5/16) did not have MET protein detectable by SRM-MS97. In addition, the ORR with crizotinib was higher in cases with detectable MET expression by SRM-MS (55%, n=6/11) compared to those that did not express MET (0%, n=0/5). This suggests that, in the right context, prospectively identifying MET expression may maximize benefit with MET-directed targeted therapy. Furthermore, the role that MET expression plays in MET-amplification has not been well defined; this should be explored prospectively on ongoing trials of MET-directed therapies.
CONCLUSIONS
The process of identifying MET-dependent cancers is complex. From a diagnostic perspective, clinically-meaningful cutoffs need to be standardized for continuous variables such as the level of MET amplification or MET expression. Diagnostic migration towards more comprehensive and technically sophisticated assays will likely be required to maximize the likelihood of detection MET amplification, mutation, and fusion. Assays such as next-generation sequencing should be considered for the detection of these alterations in both tumor and plasma, and ideally in both DNA and RNA. The effective detection of MET-dependent cancers is critical given that MET-directed targeted therapy is active in many of these cancers. Importantly, the level of activity of these therapies can be modulated by the type of alteration identified and the degree of oncogenic addiction to MET pathway signaling.
Supplementary Material
Box 1 |. Pathophysiology of MET in cancer and immunotherapy.
Under physiologic conditions, hepatocyte growth factor (HGF), the ligand for MET, regulates the epithelial-to-mesenchymal transition for tissue repair and embryogenesis2. In cancer, increased MET activity promotes tumor growth by providing anti-apoptotic and pro-migratory signals3. Furthermore, the MET gene is regulated by transcriptional factors including hypoxia-inducible factor 1α (HIF-1α)4. In preclinical models, the inhibition of angiogenesis results in hypoxic stress that leads to MET-mediated local invasion and distant metastasis5,6.
MET also plays a role in regulating the immune microenvironment. During tissue repair, upregulation of HGF and increased HGF-MET autocrine signaling promotes an immunosuppressive environment by converting immunologically active macrophages (M1 phenotype) to a growth-stimulating (M2) phenotype and by inducing tolerogenic dendritic cells7,8. MET inhibition can abrogate these effects while concurrently increasing PD-L1 expression9. Thus, the combination of immunotherapy (e.g. with an immune checkpoint inhibitor) and MET-targeted therapy is currently being explored in multiple ongoing clinical trials (NCT03914300; NCT04151563; NCT02406208; NCT03866382; NCT03793166; NCT02819596; NCT03742349; NCT03484923).
Box 2 |. Type 1 and type 2 MET inhibitors.
Type I MET tyrosine kinase inhibitors (TKIs) target the ATP-binding pocket of the active form of MET143. Type Ia agents, such as the multikinase ALK, ROS1, and MET inhibitor crizotinib, interact with MET moieties such as the Y1230 residue, the hinge region, and the solvent front G1163 residue. Type Ib inhibitors tend to be more MET-selective agents that, in contrast to type Ia agents, do not interact with G1163117. Type Ib inhibitors include capmatinib, tepotinib, and savolitinib. Unsurprisingly, these have been shown in vitro to overcome solvent front substitutions (e.g. G1163E/R) that render resistance to crizotinib.
Type II MET TKIs (cabozantinib, merestinib, and glesatinib) are likewise ATP-competitive, but bind the ATP pocket in the inactive state by extending to a hydrophobic back pocket117,144,145. Binding to this configuration allows these agents to act against MET kinase domain mutations that result in resistance to type Ia and type Ib agents117,146. These include the kinase domain mutations MET D1228E/G/H/N, and MET Y1230C/D/S/H/N.
Conversely, resistance to type II MET inhibitors can occur by MET L1195 and MET F1200 mutations117,144,145. Thus, switching between type I and type II MET inhibitors might be effective in MET-dependent cancers depending on the specific resistance mutation that emerges.h
KEY POINTS.
The degree of MET amplification is a continuous variable that can be measured by FISH or NGS. No consensus exists on diagnostic cutoffs for MET amplification.
Solid tumors that harbor high-level MET amplification have a higher likelihood of benefit with single-agent or combination MET-directed therapy compared to those with lower MET copy number gains.
MET mutations are highly heterogeneous and can range from those that involve the MET kinase domain to those that result in MET exon 14 alteration biology.
The activity of type I or type II MET tyrosine kinase inhibitors can substantially differ by MET mutation type.
A wide variety of MET fusions have been identified, but the biology of these alterations and their response to targeted therapy are not well characterized.
MET overexpression in the absence of a known driver of MET dependence is a poor predictor of benefit from MET-directed targeted therapy.
Acknowledgements
We are grateful to Dr. Clare Wilhelm for his thorough review and assistance with our manuscript. Some of the results shown here are in whole or part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Disclosures
Dr. Guo reports grants from National Institutes of Health/National Cancer Institute, during the conduct of the study. Dr. Luo reports a T32-CA009207 grant from the National Institutes of Health, during the conduct of the study. Dr. Chang reports grants from National Institutes of Health/National Cancer Institute, during the conduct of the study. Dr. Rekhtman reports grants from National Institutes of Health/National Cancer Institute, during the conduct of the study. Dr. Arcila reports grants from National Institutes of Health/National Cancer Institute, during the conduct of the study; personal fees from Biocartis and Invivoscribe, outside the submitted work. Dr. Drilon reports grants from National Cancer Institute, during the conduct of the study; personal fees from Medscape, personal fees from OncLive, personal fees from PeerVoice, personal fees from Physicians Education Resources, personal fees from Targeted Oncology, personal fees from Research to Practice, personal fees and other from TP Therapeutics, personal fees and other from AstraZeneca, grants, personal fees and other from Pfizer, personal fees and other from Blueprint Medicines, personal fees and other from Ignyta/Genentech/Roche, personal fees and other from Takeda/Ariad/Millenium, personal fees and other from Helsinn, personal fees and other from Beigene, personal fees and other from BergenBio, personal fees and other from Hengrui Therapeutics, grants, personal fees and other from Exelixis, personal fees and other from Loxo/Bayer/Lilly, personal fees and other from Tyra Biosciences, personal fees and other from Verastem, personal fees and other from MORE Health, non-financial support from Merck, grants from Teva, grants from Taiho, personal fees from Wolters Kluwer, personal fees and other from Loxo, grants from GlaxoSmithKline, personal fees from Lilly, grants from PharmaMar, non-financial support from Puma, personal fees and other from Abbvie, grants from Foundation Medicine, grants and non-financial support from Ignyta/Roche, outside the submitted work.
References
- 1.Bradley CA et al. Targeting c-MET in gastrointestinal tumours: rationale, opportunities and challenges. Nat Rev Clin Oncol 14, 562–576, doi: 10.1038/nrclinonc.2017.40 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R & Weinberg RA The basics of epithelial-mesenchymal transition. J Clin Invest 119, 1420–1428, doi: 10.1172/JCI39104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sennino B, Ishiguro-Oonuma T, Schriver BJ, Christensen JG & McDonald DM Inhibition of c-Met reduces lymphatic metastasis in RIP-Tag2 transgenic mice. Cancer research 73, 3692–3703, doi: 10.1158/0008-5472.CAN-12-2160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennacchietti S et al. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer cell 3, 347–361, doi: 10.1016/s1535-6108(03)00085-0 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Paez-Ribes M et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer cell 15, 220–231, doi: 10.1016/j.ccr.2009.01.027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu KV et al. VEGF inhibits tumor cell invasion and mesenchymal transition through a MET/VEGFR2 complex. Cancer cell 22, 21–35, doi: 10.1016/j.ccr.2012.05.037 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi W, Lee J, Lee J, Lee SH & Kim S Hepatocyte Growth Factor Regulates Macrophage Transition to the M2 Phenotype and Promotes Murine Skeletal Muscle Regeneration. Front Physiol 10, 914, doi: 10.3389/fphys.2019.00914 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas SK & Mantovani A Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 11, 889–896, doi: 10.1038/ni.1937 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Li H et al. MET Inhibitors Promote Liver Tumor Evasion of the Immune Response by Stabilizing PDL1. Gastroenterology 156, 1849–1861 e1813, doi: 10.1053/j.gastro.2019.01.252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zack TI et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45, 1134–1140, doi: 10.1038/ng.2760 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reams AB & Roth JR Mechanisms of gene duplication and amplification. Cold Spring Harb Perspect Biol 7, a016592, doi: 10.1101/cshperspect.a016592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noonan SA et al. Identifying the Appropriate FISH Criteria for Defining MET Copy Number-Driven Lung Adenocarcinoma through Oncogene Overlap Analysis. J Thorac Oncol 11, 1293–1304, doi: 10.1016/j.jtho.2016.04.033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanden Bempt I et al. Polysomy 17 in breast cancer: clinicopathologic significance and impact on HER-2 testing. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 26, 4869–4874, doi: 10.1200/JCO.2007.13.4296 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Smolen GA et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci U S A 103, 2316–2321, doi: 10.1073/pnas.0508776103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutterbach B et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res 67, 2081–2088, doi: 10.1158/0008-5472.CAN-06-3495 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Drilon A, Cappuzzo F, Ou SI & Camidge DR Targeting MET in Lung Cancer: Will Expectations Finally Be MET? Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 12, 15–26, doi: 10.1016/j.jtho.2016.10.014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cappuzzo F et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol 27, 1667–1674, doi: 10.1200/JCO.2008.19.1635 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato H et al. Gene amplification of EGFR, HER2, FGFR2 and MET in esophageal squamous cell carcinoma. International journal of oncology 42, 1151–1158, doi: 10.3892/ijo.2013.1830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graziano F et al. Genetic activation of the MET pathway and prognosis of patients with high-risk, radically resected gastric cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 29, 4789–4795, doi: 10.1200/JCO.2011.36.7706 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Yin X et al. Relationships between Chromosome 7 Gain, MET Gene Copy Number Increase and MET Protein Overexpression in Chinese Papillary Renal Cell Carcinoma Patients. PloS one 10, e0143468, doi: 10.1371/journal.pone.0143468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HE et al. MET in gastric carcinomas: comparison between protein expression and gene copy number and impact on clinical outcome. Br J Cancer 107, 325–333, doi: 10.1038/bjc.2012.237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagatsuma AK et al. Expression profiles of HER2, EGFR, MET and FGFR2 in a large cohort of patients with gastric adenocarcinoma. Gastric Cancer 18, 227–238, doi: 10.1007/s10120-014-0360-4 (2015). [DOI] [PubMed] [Google Scholar]
- 23.An X et al. MET amplification is not rare and predicts unfavorable clinical outcomes in patients with recurrent/metastatic gastric cancer after chemotherapy. Cancer 120, 675–682, doi: 10.1002/cncr.28454 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Kondo S et al. Clinical impact of c-Met expression and its gene amplification in hepatocellular carcinoma. Int J Clin Oncol 18, 207–213, doi: 10.1007/s10147-011-0361-9 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Go H et al. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J Thorac Oncol 5, 305–313, doi: 10.1097/JTO.0b013e3181ce3d1d (2010). [DOI] [PubMed] [Google Scholar]
- 26.Lennerz JK et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 29, 4803–4810, doi: 10.1200/JCO.2011.35.4928 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jardim DL et al. Analysis of 1,115 patients tested for MET amplification and therapy response in the MD Anderson Phase I Clinic. Clinical cancer research : an official journal of the American Association for Cancer Research 20, 6336–6345, doi: 10.1158/1078-0432.CCR-14-1293 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Frampton GM et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31, 1023–1031, doi: 10.1038/nbt.2696 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng DT et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 17, 251–264, doi: 10.1016/j.jmoldx.2014.12.006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia EP et al. Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Archives of pathology & laboratory medicine 141, 751–758, doi: 10.5858/arpa.2016-0527-OA (2017). [DOI] [PubMed] [Google Scholar]
- 31.Hung SS et al. Assessment of Capture and Amplicon-Based Approaches for the Development of a Targeted Next-Generation Sequencing Pipeline to Personalize Lymphoma Management. J Mol Diagn 20, 203–214, doi: 10.1016/j.jmoldx.2017.11.010 (2018). [DOI] [PubMed] [Google Scholar]
- 32.McCombie WR, McPherson JD & Mardis ER Next-Generation Sequencing Technologies. Cold Spring Harb Perspect Med 9, doi: 10.1101/cshperspect.a036798 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanman RB et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PloS one 10, e0140712, doi: 10.1371/journal.pone.0140712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paweletz CP et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clinical cancer research : an official journal of the American Association for Cancer Research 22, 915–922, doi: 10.1158/1078-0432.CCR-15-1627-T (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guibert N et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 29, 1049–1055, doi: 10.1093/annonc/mdy005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leighl NB et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 25, 4691–4700, doi: 10.1158/1078-0432.CCR-19-0624 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Combaret V et al. Influence of neuroblastoma stage on serum-based detection of MYCN amplification. Pediatric blood & cancer 53, 329–331, doi: 10.1002/pbc.22009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janku F et al. Development and Validation of an Ultradeep Next-Generation Sequencing Assay for Testing of Plasma Cell-Free DNA from Patients with Advanced Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 23, 5648–5656, doi: 10.1158/1078-0432.CCR-17-0291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Tang ET & Du Z Detection of MET Gene Copy Number in Cancer Samples Using the Droplet Digital PCR Method. PloS one 11, e0146784, doi: 10.1371/journal.pone.0146784 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu CW et al. Comparison of the c-MET gene amplification between primary tumor and metastatic lymph nodes in non-small cell lung cancer. Thorac Cancer 8, 417–422, doi: 10.1111/1759-7714.12455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bardelli A et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer discovery 3, 658–673, doi: 10.1158/2159-8290.CD-12-0558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galimi F et al. Genetic and expression analysis of MET, MACC1, and HGF in metastatic colorectal cancer: response to met inhibition in patient xenografts and pathologic correlations. Clinical cancer research : an official journal of the American Association for Cancer Research 17, 3146–3156, doi: 10.1158/1078-0432.CCR-10-3377 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Bean J et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A 104, 20932–20937, doi: 10.1073/pnas.0710370104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Onozato R et al. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol 4, 5–11, doi: 10.1097/JTO.0b013e3181913e0e (2009). [DOI] [PubMed] [Google Scholar]
- 45.Chen HJ et al. Clinicopathologic and molecular features of epidermal growth factor receptor T790M mutation and c-MET amplification in tyrosine kinase inhibitor-resistant Chinese non-small cell lung cancer. Pathol Oncol Res 15, 651–658, doi: 10.1007/s12253-009-9167-8 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Onitsuka T et al. Acquired resistance to gefitinib: the contribution of mechanisms other than the T790M, MET, and HGF status. Lung cancer 68, 198–203, doi: 10.1016/j.lungcan.2009.05.022 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Voutsina A et al. Combined analysis of KRAS and PIK3CA mutations, MET and PTEN expression in primary tumors and corresponding metastases in colorectal cancer. Mod Pathol 26, 302–313, doi: 10.1038/modpathol.2012.150 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Aisner DL et al. The Impact of Smoking and TP53 Mutations in Lung Adenocarcinoma Patients with Targetable Mutations-The Lung Cancer Mutation Consortium (LCMC2). Clin Cancer Res 24, 1038–1047, doi: 10.1158/1078-0432.CCR-17-2289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schildhaus HU et al. MET amplification status in therapy-naive adeno- and squamous cell carcinomas of the lung. Clinical cancer research : an official journal of the American Association for Cancer Research 21, 907–915, doi: 10.1158/1078-0432.CCR-14-0450 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Okuda K, Sasaki H, Yukiue H, Yano M & Fujii Y Met gene copy number predicts the prognosis for completely resected non-small cell lung cancer. Cancer Sci 99, 2280–2285, doi: 10.1111/j.1349-7006.2008.00916.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okamoto I et al. Multiplex genomic profiling of non-small cell lung cancers from the LETS phase III trial of first-line S-1/carboplatin versus paclitaxel/carboplatin: results of a West Japan Oncology Group study. Oncotarget 5, 2293–2304, doi: 10.18632/oncotarget.1906 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janjigian YY et al. MET expression and amplification in patients with localized gastric cancer. Cancer Epidemiol Biomarkers Prev 20, 1021–1027, doi: 10.1158/1055-9965.EPI-10-1080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng N et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut 61, 673–684, doi: 10.1136/gutjnl-2011-301839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsugawa K et al. Amplification of the c-met, c-erbB2 and epidermal growth factor receptor gene in human gastric cancers: Correlation to clinical features. Oncology 55, 475–481, doi:Doi 10.1159/000011898 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Raghav K et al. MET amplification in metastatic colorectal cancer: an acquired response to EGFR inhibition, not a de novo phenomenon. Oncotarget 7, 54627–54631, doi: 10.18632/oncotarget.10559 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang M et al. MET amplification, expression, and exon 14 mutations in colorectal adenocarcinoma. Human pathology 77, 108–115, doi: 10.1016/j.humpath.2018.03.024 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Pal SK et al. Characterization of Clinical Cases of Advanced Papillary Renal Cell Carcinoma via Comprehensive Genomic Profiling. Eur Urol 73, 71–78, doi: 10.1016/j.eururo.2017.05.033 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Lee SJ et al. A survey of c-MET expression and amplification in 287 patients with hepatocellular carcinoma. Anticancer Res 33, 5179–5186 (2013). [PubMed] [Google Scholar]
- 59.Park S et al. MET amplification, protein expression, and mutations in pulmonary adenocarcinoma. Lung cancer 90, 381–387, doi: 10.1016/j.lungcan.2015.10.022 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Gao J et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 6, pl1, doi: 10.1126/scisignal.2004088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404, doi: 10.1158/2159-8290.CD-12-0095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brennan CW et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477, doi: 10.1016/j.cell.2013.09.034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cancer Genome Atlas, N. Genomic Classification of Cutaneous Melanoma. Cell 161, 1681–1696, doi: 10.1016/j.cell.2015.05.044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cancer Genome Atlas Research, N. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615, doi: 10.1038/nature10166 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moro-Sibilot D et al. Crizotinib in c-MET- or ROS1-positive NSCLC: results of the AcSe phase II trial. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 30, 1985–1991, doi: 10.1093/annonc/mdz407 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Camidge DR et al. Crizotinib in patients (pts) with MET-amplified non-small cell lung cancer (NSCLC): Updated safety and efficacy findings from a phase 1 trial. Journal of Clinical Oncology 36, 9062–9062, doi: 10.1200/JCO.2018.36.15_suppl.9062 (2018). [DOI] [Google Scholar]
- 67.Ali SM et al. Prospective comprehensive genomic profiling of advanced gastric carcinoma cases reveals frequent clinically relevant genomic alterations and new routes for targeted therapies. Oncologist 20, 499–507, doi: 10.1634/theoncologist.2014-0378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Engelman JA et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043, doi: 10.1126/science.1141478 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Sequist LV et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3, 75ra26, doi: 10.1126/scitranslmed.3002003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu HA et al. Concurrent Alterations in EGFR-Mutant Lung Cancers Associated with Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clinical cancer research : an official journal of the American Association for Cancer Research 24, 3108–3118, doi: 10.1158/1078-0432.CCR-17-2961 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schoenfeld AJ et al. Tissue-based molecular and histological landscape of acquired resistance to osimertinib given initially or at relapse in patients with EGFR-mutant lung cancers. Journal of Clinical Oncology 37, 9028–9028, doi: 10.1200/JCO.2019.37.15_suppl.9028 (2019). [DOI] [Google Scholar]
- 72.Oxnard GR et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 4, 1527–1534, doi: 10.1001/jamaoncol.2018.2969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gouji T, Takashi S, Mitsuhiro T & Yukito I Crizotinib can overcome acquired resistance to CH5424802: is amplification of the MET gene a key factor? J Thorac Oncol 9, e27–28, doi: 10.1097/jto.0000000000000113 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Parseghian CM, Napolitano S, Loree JM & Kopetz S Mechanisms of Innate and Acquired Resistance to Anti-EGFR therapy: A Review of Current Knowledge with a Focus on Rechallenge Therapies. Clinical cancer research : an official journal of the American Association for Cancer Research, doi: 10.1158/1078-0432.CCR-19-0823 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pietrantonio F et al. MET-Driven Resistance to Dual EGFR and BRAF Blockade May Be Overcome by Switching from EGFR to MET Inhibition in BRAF-Mutated Colorectal Cancer. Cancer Discov 6, 963–971, doi: 10.1158/2159-8290.CD-16-0297 (2016). [DOI] [PubMed] [Google Scholar]
- 76.Camidge DR & Davies KD MET Copy Number as a Secondary Driver of Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor Resistance in EGFR-Mutant Non-Small-Cell Lung Cancer. J Clin Oncol 37, 855–857, doi: 10.1200/jco.19.00033 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Drilon A et al. OA12.02 Updated Antitumor Activity of Crizotinib in Patients with MET Exon 14-Altered Advanced Non-Small Cell Lung Cancer. Journal of Thoracic Oncology 13, S348, doi: 10.1016/j.jtho.2018.08.300 (2018). [DOI] [Google Scholar]
- 78.Camidge DR et al. Efficacy and safety of crizotinib in patients with advanced c-MET-amplified non-small cell lung cancer (NSCLC). Journal of Clinical Oncology 32, 8001–8001, doi: 10.1200/jco.2014.32.15_suppl.8001 (2014). [DOI] [Google Scholar]
- 79.Schuler MH et al. Phase (Ph) I study of the safety and efficacy of the cMET inhibitor capmatinib (INC280) in patients (pts) with advanced cMET+ non-small cell lung cancer (NSCLC). Journal of Clinical Oncology 34, 9067–9067, doi: 10.1200/JCO.2016.34.15_suppl.9067 (2016). [DOI] [Google Scholar]
- 80.Frigault MM et al. Abstract 4541: MET gene copy number gains evaluated by NGS is more predictive than other methods to enrich for papillary RCC patients sensitive to savolitinib, a selective MET inhibitor. Cancer Research 78, 4541–4541, doi: 10.1158/1538-7445.am2018-4541 (2018). [DOI] [Google Scholar]
- 81.Sequist LV et al. Abstract CT033: TATTON Phase Ib expansion cohort: Osimertinib plus savolitinib for patients (pts) with EGFR-mutant, MET-amplified NSCLC after progression on prior third-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI). Cancer Research 79, CT033–CT033, doi: 10.1158/1538-7445.Am2019-ct033 (2019). [DOI] [Google Scholar]
- 82.Wu YL et al. Phase Ib/II Study of Capmatinib (INC280) Plus Gefitinib After Failure of Epidermal Growth Factor Receptor (EGFR) Inhibitor Therapy in Patients With EGFR-Mutated, MET Factor-Dysregulated Non-Small-Cell Lung Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 36, 3101–3109, doi: 10.1200/JCO.2018.77.7326 (2018). [DOI] [PubMed] [Google Scholar]
- 83.Schmidt L et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet 16, 68–73, doi: 10.1038/ng0597-68 (1997). [DOI] [PubMed] [Google Scholar]
- 84.Jeffers M et al. Activating mutations for the met tyrosine kinase receptor in human cancer. Proc Natl Acad Sci U S A 94, 11445–11450, doi: 10.1073/pnas.94.21.11445 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Graveel C et al. Activating Met mutations produce unique tumor profiles in mice with selective duplication of the mutant allele. Proceedings of the National Academy of Sciences of the United States of America 101, 17198–17203, doi: 10.1073/pnas.0407651101 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Durinck S et al. Spectrum of diverse genomic alterations define non-clear cell renal carcinoma subtypes. Nat Genet 47, 13–21, doi: 10.1038/ng.3146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Albiges L et al. MET is a potential target across all papillary renal cell carcinomas: result from a large molecular study of pRCC with CGH array and matching gene expression array. Clinical cancer research : an official journal of the American Association for Cancer Research 20, 3411–3421, doi: 10.1158/1078-0432.CCR-13-2173 (2014). [DOI] [PubMed] [Google Scholar]
- 88.Schmidt L et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene 18, 2343–2350, doi: 10.1038/sj.onc.1202547 (1999). [DOI] [PubMed] [Google Scholar]
- 89.Lorenzato A et al. Novel somatic mutations of the MET oncogene in human carcinoma metastases activating cell motility and invasion. Cancer research 62, 7025–7030 (2002). [PubMed] [Google Scholar]
- 90.Zhuang Z et al. Trisomy 7-harbouring non-random duplication of the mutant MET allele in hereditary papillary renal carcinomas. Nat Genet 20, 66–69, doi: 10.1038/1727 (1998). [DOI] [PubMed] [Google Scholar]
- 91.Park WS et al. Somatic mutations in the kinase domain of the Met/hepatocyte growth factor receptor gene in childhood hepatocellular carcinomas. Cancer research 59, 307–310 (1999). [PubMed] [Google Scholar]
- 92.Aebersold DM et al. Prevalence and clinical impact of Met Y1253D-activating point mutation in radiotherapy-treated squamous cell cancer of the oropharynx. Oncogene 22, 8519–8523, doi: 10.1038/sj.onc.1206968 (2003). [DOI] [PubMed] [Google Scholar]
- 93.Ghadjar P et al. MET Y1253D-activating point mutation and development of distant metastasis in advanced head and neck cancers. Clin Exp Metastasis 26, 809–815, doi: 10.1007/s10585-009-9280-9 (2009). [DOI] [PubMed] [Google Scholar]
- 94.Bahcall M et al. Acquired METD1228V Mutation and Resistance to MET Inhibition in Lung Cancer. Cancer Discov 6, 1334–1341, doi: 10.1158/2159-8290.CD-16-0686 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Heist RS et al. Acquired Resistance to Crizotinib in NSCLC with MET Exon 14 Skipping. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 11, 1242–1245, doi: 10.1016/j.jtho.2016.06.013 (2016). [DOI] [PubMed] [Google Scholar]
- 96.Ou SI et al. Emergence of Preexisting MET Y1230C Mutation as a Resistance Mechanism to Crizotinib in NSCLC with MET Exon 14 Skipping. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 12, 137–140, doi: 10.1016/j.jtho.2016.09.119 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Guo R et al. MET inhibitor resistance in patients with MET exon 14-altered lung cancers. Journal of Clinical Oncology 37, 9006–9006, doi: 10.1200/JCO.2019.37.15_suppl.9006 (2019). [DOI] [Google Scholar]
- 98.Li A et al. Acquired MET Y1248H and D1246N Mutations Mediate Resistance to MET Inhibitors in Non-Small Cell Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 23, 4929–4937, doi: 10.1158/1078-0432.CCR-16-3273 (2017). [DOI] [PubMed] [Google Scholar]
- 99.Liang Y, Chen Q, Lin Y, Liu H & Qiu B P3.01–60 A Novel MET D1246H Mutation After Progression of EGFR-TKI/MET Inhibitor Combined Therapy in a NSCLC Patient with Acquired MET Amplification. Journal of Thoracic Oncology 13, S890, doi: 10.1016/j.jtho.2018.08.1620 (2018). [DOI] [Google Scholar]
- 100.Peschard P, Ishiyama N, Lin T, Lipkowitz S & Park M A conserved DpYR motif in the juxtamembrane domain of the Met receptor family forms an atypical c-Cbl/Cbl-b tyrosine kinase binding domain binding site required for suppression of oncogenic activation. The Journal of biological chemistry 279, 29565–29571, doi: 10.1074/jbc.M403954200 (2004). [DOI] [PubMed] [Google Scholar]
- 101.Ma PC et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer research 63, 6272–6281 (2003). [PubMed] [Google Scholar]
- 102.Ma PC et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer research 65, 1479–1488, doi: 10.1158/0008-5472.CAN-04-2650 (2005). [DOI] [PubMed] [Google Scholar]
- 103.Frampton GM et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 5, 850–859, doi: 10.1158/2159-8290.CD-15-0285 (2015). [DOI] [PubMed] [Google Scholar]
- 104.Schrock AB et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 11, 1493–1502, doi: 10.1016/j.jtho.2016.06.004 (2016). [DOI] [PubMed] [Google Scholar]
- 105.Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550, doi: 10.1038/nature13385 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu X et al. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 34, 794–802, doi: 10.1200/JCO.2015.62.0674 (2016). [DOI] [PubMed] [Google Scholar]
- 107.Lee J et al. Gastrointestinal malignancies harbor actionable MET exon 14 deletions. Oncotarget 6, 28211–28222, doi: 10.18632/oncotarget.4721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yan B et al. Identification of MET genomic amplification, protein expression and alternative splice isoforms in neuroblastomas. J Clin Pathol 66, 985–991, doi: 10.1136/jclinpath-2012-201375 (2013). [DOI] [PubMed] [Google Scholar]
- 109.Suzawa K et al. Acquired MET Exon 14 Alteration Drives Secondary Resistance to Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor in EGFR-Mutated Lung Cancer. JCO Precis Oncol 3, doi: 10.1200/PO.19.00011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schoenfeld AJ et al. Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations As Early Resistance Mechanisms to First-line Osimertinib in EGFR-Mutant Lung Cancer. Clin Cancer Res, doi: 10.1158/1078-0432.CCR-19-3563 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tong JH et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clinical cancer research : an official journal of the American Association for Cancer Research 22, 3048–3056, doi: 10.1158/1078-0432.CCR-15-2061 (2016). [DOI] [PubMed] [Google Scholar]
- 112.Awad MM et al. MET Exon 14 Mutations in Non–Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. Journal of Clinical Oncology 34, 721–730, doi: 10.1200/JCO.2015.63.4600 (2016). [DOI] [PubMed] [Google Scholar]
- 113.Wolf J et al. Capmatinib (INC280) in METΔex14-mutated advanced non-small cell lung cancer (NSCLC): Efficacy data from the phase II GEOMETRY mono-1 study. Journal of Clinical Oncology 37, 9004–9004, doi: 10.1200/JCO.2019.37.15_suppl.9004 (2019). [DOI] [Google Scholar]
- 114.Kong-Beltran M et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res 66, 283–289, doi: 10.1158/0008-5472.CAN-05-2749 (2006). [DOI] [PubMed] [Google Scholar]
- 115.Peschard P et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell 8, 995–1004, doi: 10.1016/s1097-2765(01)00378-1 (2001). [DOI] [PubMed] [Google Scholar]
- 116.Abella JV et al. Met/Hepatocyte growth factor receptor ubiquitination suppresses transformation and is required for Hrs phosphorylation. Molecular and cellular biology 25, 9632–9645, doi: 10.1128/MCB.25.21.9632-9645.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fujino T et al. Sensitivity and Resistance of MET Exon 14 Mutations in Lung Cancer to Eight MET Tyrosine Kinase Inhibitors In Vitro. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 14, 1753–1765, doi: 10.1016/j.jtho.2019.06.023 (2019). [DOI] [PubMed] [Google Scholar]
- 118.Kong-Beltran M, Stamos J & Wickramasinghe D The Sema domain of Met is necessary for receptor dimerization and activation. Cancer cell 6, 75–84, doi: 10.1016/j.ccr.2004.06.013 (2004). [DOI] [PubMed] [Google Scholar]
- 119.Liu S et al. Functional consequence of the MET-T1010I polymorphism in breast cancer. Oncotarget 6, 2604–2614, doi: 10.18632/oncotarget.3094 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Seiwert TY et al. The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer research 69, 3021–3031, doi: 10.1158/0008-5472.CAN-08-2881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stella GM et al. MET mutations in cancers of unknown primary origin (CUPs). Human mutation 32, 44–50, doi: 10.1002/humu.21374 (2011). [DOI] [PubMed] [Google Scholar]
- 122.de Melo Gagliato D et al. Analysis of MET genetic aberrations in patients with breast cancer at MD Anderson Phase I unit. Clinical breast cancer 14, 468–474, doi: 10.1016/j.clbc.2014.06.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Krishnaswamy S et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res 15, 5714–5723, doi: 10.1158/1078-0432.CCR-09-0070 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kong LR, Binte Mohamed Salleh NA, Tan TZ, Kappei D & Goh BC P1.02–041 Characterization of MET-N375S as an Activating Mutation in Squamous Cell Carcinoma of the Lung: Topic: Driver Genes in NSCLC, Resistance, and Other. Journal of Thoracic Oncology 12, S512, doi: 10.1016/j.jtho.2016.11.624 (2017). [DOI] [Google Scholar]
- 125.Tengs T et al. A transforming MET mutation discovered in non-small cell lung cancer using microarray-based resequencing. Cancer letters 239, 227–233, doi: 10.1016/j.canlet.2005.08.007 (2006). [DOI] [PubMed] [Google Scholar]
- 126.Samorodnitsky E et al. Evaluation of Hybridization Capture Versus Amplicon-Based Methods for Whole-Exome Sequencing. Human mutation 36, 903–914, doi: 10.1002/humu.22825 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Poirot B et al. MET Exon 14 Alterations and New Resistance Mutations to Tyrosine Kinase Inhibitors: Risk of Inadequate Detection with Current Amplicon-Based NGS Panels. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 12, 1582–1587, doi: 10.1016/j.jtho.2017.07.026 (2017). [DOI] [PubMed] [Google Scholar]
- 128.Davies KD et al. DNA-Based versus RNA-Based Detection of MET Exon 14 Skipping Events in Lung Cancer. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 14, 737–741, doi: 10.1016/j.jtho.2018.12.020 (2019). [DOI] [PubMed] [Google Scholar]
- 129.Hao M et al. QTug.sau-3B is a Major Quantitative Trait Locus for Wheat Hexaploidization. G3, doi: 10.1534/g3.114.013078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Benayed R et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clinical cancer research : an official journal of the American Association for Cancer Research 25, 4712–4722, doi: 10.1158/1078-0432.CCR-19-0225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kulkarni MM Digital multiplexed gene expression analysis using the NanoString nCounter system Curr Protoc Mol Biol Chapter 25, Unit25B 10, doi: 10.1002/0471142727.mb25b10s94 (2011). [DOI] [PubMed] [Google Scholar]
- 132.Paik PK et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. [DOI] [PMC free article] [PubMed]
- 133.Sunami K et al. Multiplex Diagnosis of Oncogenic Fusion and MET Exon Skipping by Molecular Counting Using Formalin-Fixed Paraffin Embedded Lung Adenocarcinoma Tissues. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 11, 203–212, doi: 10.1016/j.jtho.2015.10.005 (2016). [DOI] [PubMed] [Google Scholar]
- 134.Berthou S et al. The Met kinase inhibitor SU11274 exhibits a selective inhibition pattern toward different receptor mutated variants. Oncogene 23, 5387–5393, doi: 10.1038/sj.onc.1207691 (2004). [DOI] [PubMed] [Google Scholar]
- 135.Hari SB, Merritt EA & Maly DJ Sequence determinants of a specific inactive protein kinase conformation. Chemistry & biology 20, 806–815, doi: 10.1016/j.chembiol.2013.05.005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Choueiri TK et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J Clin Oncol 31, 181–186, doi: 10.1200/JCO.2012.43.3383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kang J et al. Osimertinib and Cabozantinib Combinatorial Therapy in an EGFR-Mutant Lung Adenocarcinoma Patient with Multiple MET Secondary-Site Mutations after Resistance to Crizotinib. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 13, e49–e53, doi: 10.1016/j.jtho.2017.10.028 (2018). [DOI] [PubMed] [Google Scholar]
- 138.US Food and Drug Administration. Breakthrough Therapy Approvals, https://www.fda.gov/drugs/nda-and-bla-approvals/breakthrough-therapy-approvals (2019).
- 139.Lu S et al. Abstract CT031: Preliminary efficacy and safety results of savolitinib treating patients with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations. Cancer Research 79, CT031–CT031, doi: 10.1158/1538-7445.am2019-ct031 (2019). [DOI] [Google Scholar]
- 140.Paik PK et al. Phase II study of tepotinib in NSCLC patients with METex14 mutations. Journal of Clinical Oncology 37, 9005–9005, doi: 10.1200/JCO.2019.37.15_suppl.9005 (2019). [DOI] [Google Scholar]
- 141.Rotow JK et al. Co-occurring Alterations in the RAS-MAPK Pathway Limit Response to MET Inhibitor Treatment in MET Exon 14 Skipping Mutation-Positive Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 26, 439–449, doi: 10.1158/1078-0432.CCR-19-1667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pennacchietti S et al. Microenvironment-derived HGF overcomes genetically determined sensitivity to anti-MET drugs. Cancer Res 74, 6598–6609, doi: 10.1158/0008-5472.CAN-14-0761 (2014). [DOI] [PubMed] [Google Scholar]
- 143.Roskoski R Jr. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacological research 103, 26–48, doi: 10.1016/j.phrs.2015.10.021 (2016). [DOI] [PubMed] [Google Scholar]
- 144.Engstrom LD et al. Glesatinib Exhibits Antitumor Activity in Lung Cancer Models and Patients Harboring MET Exon 14 Mutations and Overcomes Mutation-mediated Resistance to Type I MET Inhibitors in Nonclinical Models. Clinical cancer research : an official journal of the American Association for Cancer Research 23, 6661–6672, doi: 10.1158/1078-0432.CCR-17-1192 (2017). [DOI] [PubMed] [Google Scholar]
- 145.Tiedt R et al. A drug resistance screen using a selective MET inhibitor reveals a spectrum of mutations that partially overlap with activating mutations found in cancer patients. Cancer research 71, 5255–5264, doi: 10.1158/0008-5472.CAN-10-4433 (2011). [DOI] [PubMed] [Google Scholar]
- 146.Yan SB et al. LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models. Investigational new drugs 31, 833–844, doi: 10.1007/s10637-012-9912-9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cooper CS et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 311, 29–33, doi: 10.1038/311029a0 (1984). [DOI] [PubMed] [Google Scholar]
- 148.International Cancer Genome Consortium PedBrain Tumor, P. Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nature medicine 22, 1314–1320, doi: 10.1038/nm.4204 (2016). [DOI] [PubMed] [Google Scholar]
- 149.Bao ZS et al. RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res 24, 1765–1773, doi: 10.1101/gr.165126.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stransky N, Cerami E, Schalm S, Kim JL & Lengauer C The landscape of kinase fusions in cancer. Nat Commun 5, 4846, doi: 10.1038/ncomms5846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Soman NR, Correa P, Ruiz BA & Wogan GN The TPR-MET oncogenic rearrangement is present and expressed in human gastric carcinoma and precursor lesions. Proc Natl Acad Sci U S A 88, 4892–4896, doi: 10.1073/pnas.88.11.4892 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim P, Jia P & Zhao Z Kinase impact assessment in the landscape of fusion genes that retain kinase domains: a pan-cancer study. Brief Bioinform 19, 450–460, doi: 10.1093/bib/bbw127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Plenker D et al. Structural Alterations of MET Trigger Response to MET Kinase Inhibition in Lung Adenocarcinoma Patients. Clinical cancer research : an official journal of the American Association for Cancer Research 24, 1337–1343, doi: 10.1158/1078-0432.CCR-17-3001 (2018). [DOI] [PubMed] [Google Scholar]
- 154.Pan Y et al. Detection of Novel NRG1, EGFR, and MET Fusions in Lung Adenocarcinomas in the Chinese Population. J Thorac Oncol 14, 2003–2008, doi: 10.1016/j.jtho.2019.07.022 (2019). [DOI] [PubMed] [Google Scholar]
- 155.Davies KD et al. Dramatic Response to Crizotinib in a Patient with Lung Cancer Positive for an HLA-DRB1-MET Gene Fusion. JCO Precis Oncol 2017, doi: 10.1200/PO.17.00117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu J, Li X & Peng J A Novel CAV1-MET Fusion in SCLC Transformation Responds to Crizotinib and Osimertinib Treatment. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 14, e126–e128, doi: 10.1016/j.jtho.2019.01.025 (2019). [DOI] [PubMed] [Google Scholar]
- 157.Subramaniam DS et al. RNA-Seq analysis of glioma tumors to reveal targetable gene fusions. Journal of Clinical Oncology 35, 2019–2019, doi: 10.1200/JCO.2017.35.15_suppl.2019 (2017). [DOI] [Google Scholar]
- 158.Kim HP et al. Novel fusion transcripts in human gastric cancer revealed by transcriptome analysis. Oncogene 33, 5434–5441, doi: 10.1038/onc.2013.490 (2014). [DOI] [PubMed] [Google Scholar]
- 159.Kudlow B et al. in Annals of Oncology (2016) 27 (6): 401–406. 10.1093/annonc/mdw380. [DOI] [Google Scholar]
- 160.Yeh I et al. Activating MET kinase rearrangements in melanoma and Spitz tumours. Nat Commun 6, 7174, doi: 10.1038/ncomms8174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70, doi: 10.1038/nature11412 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vigna E, Gramaglia D, Longati P, Bardelli A & Comoglio PM Loss of the exon encoding the juxtamembrane domain is essential for the oncogenic activation of TPR-MET. Oncogene 18, 4275–4281, doi: 10.1038/sj.onc.1202791 (1999). [DOI] [PubMed] [Google Scholar]
- 163.Rodrigues GA & Park M Dimerization mediated through a leucine zipper activates the oncogenic potential of the met receptor tyrosine kinase. Molecular and cellular biology 13, 6711–6722, doi: 10.1128/mcb.13.11.6711 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Petrini I Biology of MET: a double life between normal tissue repair and tumor progression. Ann Transl Med 3, 82, doi: 10.3978/j.issn.2305-5839.2015.03.58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Flucke U et al. TFG-MET fusion in an infantile spindle cell sarcoma with neural features. Genes, chromosomes & cancer 56, 663–667, doi: 10.1002/gcc.22470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gow CH et al. Oncogenic Function of a KIF5B-MET Fusion Variant in Non-Small Cell Lung Cancer. Neoplasia 20, 838–847, doi: 10.1016/j.neo.2018.06.007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Heyer EE et al. Diagnosis of fusion genes using targeted RNA sequencing. Nat Commun 10, 1388, doi: 10.1038/s41467-019-09374-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Treangen TJ & Salzberg SL Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 13, 36–46, doi: 10.1038/nrg3117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang Q, Xia J, Jia P, Pao W & Zhao Z Application of next generation sequencing to human gene fusion detection: computational tools, features and perspectives. Brief Bioinform 14, 506–519, doi: 10.1093/bib/bbs044 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Levin JZ et al. Targeted next-generation sequencing of a cancer transcriptome enhances detection of sequence variants and novel fusion transcripts. Genome Biol 10, R115, doi: 10.1186/gb-2009-10-10-r115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sattler M et al. A novel small molecule met inhibitor induces apoptosis in cells transformed by the oncogenic TPR-MET tyrosine kinase. Cancer research 63, 5462–5469 (2003). [PubMed] [Google Scholar]
- 172.Zhu YC et al. Identification of a novel crizotinib-sensitive MET-ATXN7L1 gene fusion variant in lung adenocarcinoma by next generation sequencing. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 29, 2392–2393, doi: 10.1093/annonc/mdy455 (2018). [DOI] [PubMed] [Google Scholar]
- 173.Ryan CJ et al. Targeted MET inhibition in castration-resistant prostate cancer: a randomized phase II study and biomarker analysis with rilotumumab plus mitoxantrone and prednisone. Clin Cancer Res 19, 215–224, doi: 10.1158/1078-0432.CCR-12-2605 (2013). [DOI] [PubMed] [Google Scholar]
- 174.Iveson T et al. Rilotumumab in combination with epirubicin, cisplatin, and capecitabine as first-line treatment for gastric or oesophagogastric junction adenocarcinoma: an open-label, dose de-escalation phase 1b study and a double-blind, randomised phase 2 study. The Lancet. Oncology 15, 1007–1018, doi: 10.1016/S1470-2045(14)70023-3 (2014). [DOI] [PubMed] [Google Scholar]
- 175.Spigel DR et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 31, 4105–4114, doi: 10.1200/JCO.2012.47.4189 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lahat G et al. The expression of c-Met pathway components in unclassified pleomorphic sarcoma/malignant fibrous histiocytoma (UPS/MFH): a tissue microarray study. Histopathology 59, 556–561, doi: 10.1111/j.1365-2559.2011.03946.x (2011). [DOI] [PubMed] [Google Scholar]
- 177.Srivastava AK et al. Pharmacodynamic Response of the MET/HGF Receptor to Small-Molecule Tyrosine Kinase Inhibitors Examined with Validated, Fit-for-Clinic Immunoassays. Clinical cancer research : an official journal of the American Association for Cancer Research 22, 3683–3694, doi: 10.1158/1078-0432.CCR-15-2323 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gordon MS et al. Safety, pharmacokinetics, and pharmacodynamics of AMG 102, a fully human hepatocyte growth factor-neutralizing monoclonal antibody, in a first-in-human study of patients with advanced solid tumors. Clin Cancer Res 16, 699–710, doi: 10.1158/1078-0432.CCR-09-1365 (2010). [DOI] [PubMed] [Google Scholar]
- 179.Stenger M Calculating H-Score, https://ascopost.com/issues/april-10-2015/calculating-h-score/ (2015).
- 180.Mignard X et al. c-MET Overexpression as a Poor Predictor of MET Amplifications or Exon 14 Mutations in Lung Sarcomatoid Carcinomas. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 13, 1962–1967, doi: 10.1016/j.jtho.2018.08.008 (2018). [DOI] [PubMed] [Google Scholar]
- 181.Catenacci DV et al. Absolute quantitation of Met using mass spectrometry for clinical application: assay precision, stability, and correlation with MET gene amplification in FFPE tumor tissue. PloS one 9, e100586, doi: 10.1371/journal.pone.0100586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sholl LM et al. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 8, 322–328, doi: 10.1097/JTO.0b013e31827db604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Watermann I et al. Improved diagnostics targeting c-MET in non-small cell lung cancer: expression, amplification and activation? Diagn Pathol 10, 130, doi: 10.1186/s13000-015-0362-5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Bubendorf L et al. Prevalence and clinical association of MET gene overexpression and amplification in patients with NSCLC: Results from the European Thoracic Oncology Platform (ETOP) Lungscape project. Lung Cancer 111, 143–149, doi: 10.1016/j.lungcan.2017.07.021 (2017). [DOI] [PubMed] [Google Scholar]
- 185.Casadevall D et al. MET expression and copy number heterogeneity in nonsquamous non-small cell lung cancer (nsNSCLC). Oncotarget 6, 16215–16226, doi: 10.18632/oncotarget.3976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Guo R et al. MET IHC Is a Poor Screen for MET Amplification or MET Exon 14 Mutations in Lung Adenocarcinomas: Data from a Tri-Institutional Cohort of the Lung Cancer Mutation Consortium. J Thorac Oncol 14, 1666–1671, doi: 10.1016/j.jtho.2019.06.009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Descarpentries C et al. Optimization of Routine Testing for MET Exon 14 Splice Site Mutations in NSCLC Patients. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 13, 1873–1883, doi: 10.1016/j.jtho.2018.08.2023 (2018). [DOI] [PubMed] [Google Scholar]
- 188.Lambros L & Uguen A MET Immunohistochemistry Should Be Avoided in Selecting Non-small-cell Lung Cancers Requiring MET Exon 14 Skipping Mutation Analysis. Clin Lung Cancer 20, e418–e420, doi: 10.1016/j.cllc.2018.12.002 (2019). [DOI] [PubMed] [Google Scholar]
- 189.Nakajima M et al. The prognostic significance of amplification and overexpression of c-met and c-erb B-2 in human gastric carcinomas. Cancer 85, 1894–1902, doi: (1999). [DOI] [PubMed] [Google Scholar]
- 190.Nishio M et al. Phase I study of the anti-MET antibody onartuzumab in patients with solid tumors and MET-positive lung cancer. Investigational new drugs 33, 632–640, doi: 10.1007/s10637-015-0227-5 (2015). [DOI] [PubMed] [Google Scholar]
- 191.Sakai D et al. A non-randomized, open-label, single-arm, Phase 2 study of emibetuzumab in Asian patients with MET diagnostic positive, advanced gastric cancer. Cancer chemotherapy and pharmacology 80, 1197–1207, doi: 10.1007/s00280-017-3445-z (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Tabernero J et al. A pharmacodynamic/pharmacokinetic study of ficlatuzumab in patients with advanced solid tumors and liver metastases. Clinical cancer research : an official journal of the American Association for Cancer Research 20, 2793–2804, doi: 10.1158/1078-0432.CCR-13-1837 (2014). [DOI] [PubMed] [Google Scholar]
- 193.Angevin E et al. A first-in-human phase I study of SAR125844, a selective MET tyrosine kinase inhibitor, in patients with advanced solid tumours with MET amplification. Eur J Cancer 87, 131–139, doi: 10.1016/j.ejca.2017.10.016 (2017). [DOI] [PubMed] [Google Scholar]
- 194.Santoro A et al. Tivantinib for second-line treatment of advanced hepatocellular carcinoma: a randomised, placebo-controlled phase 2 study. Lancet Oncol 14, 55–63, doi: 10.1016/S1470-2045(12)70490-4 (2013). [DOI] [PubMed] [Google Scholar]
- 195.Goyal L et al. A phase 2 and biomarker study of cabozantinib in patients with advanced cholangiocarcinoma. Cancer 123, 1979–1988, doi: 10.1002/cncr.30571 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li AN, Yang J-J, Zhang X & Wu Y-L Impact of different MET alterations on the efficacy of crizotinib in non-small-cell lung cancer. Journal of Clinical Oncology 34, e20622–e20622, doi: 10.1200/JCO.2016.34.15_suppl.e20622 (2016). [DOI] [Google Scholar]
- 197.Tsao MS et al. Differential expression of Met/hepatocyte growth factor receptor in subtypes of non-small cell lung cancers. Lung Cancer 20, 1–16, doi: 10.1016/s0169-5002(98)00007-5 (1998). [DOI] [PubMed] [Google Scholar]
- 198.Bang YJ et al. Phase 1 study of capmatinib in MET-positive solid tumor patients: Dose escalation and expansion of selected cohorts. Cancer Sci 111, 536–547, doi: 10.1111/cas.14254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hong DS et al. Phase I Study of AMG 337, a Highly Selective Small-molecule MET Inhibitor, in Patients with Advanced Solid Tumors. Clinical cancer research : an official journal of the American Association for Cancer Research 25, 2403–2413, doi: 10.1158/1078-0432.CCR-18-1341 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Van Cutsem E et al. A Multicenter Phase II Study of AMG 337 in Patients with MET-Amplified Gastric/Gastroesophageal Junction/Esophageal Adenocarcinoma and Other MET-Amplified Solid Tumors. Clin Cancer Res 25, 2414–2423, doi: 10.1158/1078-0432.CCR-18-1337 (2019). [DOI] [PubMed] [Google Scholar]
- 201.Shah MA et al. Phase II study evaluating 2 dosing schedules of oral foretinib (GSK1363089), cMET/VEGFR2 inhibitor, in patients with metastatic gastric cancer. PloS one 8, e54014, doi: 10.1371/journal.pone.0054014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Qin S et al. A phase II study of the efficacy and safety of the MET inhibitor capmatinib (INC280) in patients with advanced hepatocellular carcinoma. Ther Adv Med Oncol 11, 1758835919889001, doi: 10.1177/1758835919889001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Landi L et al. Crizotinib in MET-Deregulated or ROS1-Rearranged Pretreated Non-Small Cell Lung Cancer (METROS): A Phase II, Prospective, Multicenter, Two-Arms Trial. Clinical cancer research : an official journal of the American Association for Cancer Research 25, 7312–7319, doi: 10.1158/1078-0432.CCR-19-0994 (2019). [DOI] [PubMed] [Google Scholar]
- 204.Sequist LV et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol, doi: 10.1016/S1470-2045(19)30785-5 (2020). [DOI] [PubMed] [Google Scholar]
- 205.Wu Y et al. MA09.09 Long-Term Outcomes to Tepotinib Plus Gefitinib in Patients with EGFR-Mutant NSCLC and MET Dysregulation: 18-Month Follow-Up. Journal of Thoracic Oncology 14, S284, doi: 10.1016/j.jtho.2019.08.571 (2019). [DOI] [Google Scholar]
- 206.Yang J et al. OA 09.06 A Phase Ib Trial of Savolitinib plus Gefitinib for Chinese Patients with EGFR-Mutant MET-Amplified Advanced NSCLC. Journal of Thoracic Oncology 12, S1769, doi: 10.1016/j.jtho.2017.09.379 (2017). [DOI] [Google Scholar]
- 207.Lu S et al. Abstract CT031: Preliminary efficacy and safety results of savolitinib treating patients with pulmonary sarcomatoid carcinoma (PSC) and other types of non-small cell lung cancer (NSCLC) harboring MET exon 14 skipping mutations. Cancer research 79, CT031–CT031, doi: 10.1158/1538-7445.am2019-ct031 (2019). [DOI] [Google Scholar]
- 208.Eng C et al. A randomized, placebo-controlled, phase 1/2 study of tivantinib (ARQ 197) in combination with irinotecan and cetuximab in patients with metastatic colorectal cancer with wild-type KRAS who have received first-line systemic therapy. International journal of cancer. Journal international du cancer 139, 177–186, doi: 10.1002/ijc.30049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Diéras V et al. Randomized, phase II, placebo-controlled trial of onartuzumab and/or bevacizumab in combination with weekly paclitaxel in patients with metastatic triple-negative breast cancer. Annals of Oncology 26, 1904–1910, doi: 10.1093/annonc/mdv263 (2015). [DOI] [PubMed] [Google Scholar]
- 210.Kishi K et al. First-line onartuzumab plus erlotinib treatment for patients with MET-positive and EGFR mutation-positive non-small-cell lung cancer. Cancer treatment and research communications 18, 100113, doi: 10.1016/j.ctarc.2018.10.004 (2019). [DOI] [PubMed] [Google Scholar]
- 211.Catenacci DVT et al. Rilotumumab plus epirubicin, cisplatin, and capecitabine as first-line therapy in advanced MET-positive gastric or gastro-oesophageal junction cancer (RILOMET-1): a randomised, double-blind, placebo-controlled, phase 3 trial. The Lancet. Oncology 18, 1467–1482, doi: 10.1016/S1470-2045(17)30566-1 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Spigel DR et al. Results From the Phase III Randomized Trial of Onartuzumab Plus Erlotinib Versus Erlotinib in Previously Treated Stage IIIB or IV Non–Small-Cell Lung Cancer: METLung. Journal of Clinical Oncology 35, 412–420, doi: 10.1200/JCO.2016.69.2160 (2016). [DOI] [PubMed] [Google Scholar]
- 213.Bendell JC et al. A Phase II Randomized Trial (GO27827) of First-Line FOLFOX Plus Bevacizumab with or Without the MET Inhibitor Onartuzumab in Patients with Metastatic Colorectal Cancer. The oncologist 22, 264–271, doi: 10.1634/theoncologist.2016-0223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Shah MA et al. A Randomized Phase II Study of FOLFOX With or Without the MET Inhibitor Onartuzumab in Advanced Adenocarcinoma of the Stomach and Gastroesophageal Junction. The oncologist 21, 1085–1090, doi: 10.1634/theoncologist.2016-0038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hirsch FR et al. Efficacy and Safety Results From a Phase II, Placebo-Controlled Study of Onartuzumab Plus First-Line Platinum-Doublet Chemotherapy for Advanced Squamous Cell Non-Small-Cell Lung Cancer. Clin Lung Cancer 18, 43–49, doi: 10.1016/j.cllc.2016.05.011 (2017). [DOI] [PubMed] [Google Scholar]
- 216.Wakelee H et al. Efficacy and Safety of Onartuzumab in Combination With First-Line Bevacizumab- or Pemetrexed-Based Chemotherapy Regimens in Advanced Non-Squamous Non-Small-Cell Lung Cancer. Clin Lung Cancer 18, 50–59, doi: 10.1016/j.cllc.2016.09.013 (2017). [DOI] [PubMed] [Google Scholar]
- 217.Shah MA et al. Effect of Fluorouracil, Leucovorin, and Oxaliplatin With or Without Onartuzumab in HER2-Negative, MET-Positive Gastroesophageal Adenocarcinoma: The METGastric Randomized Clinical Trial. JAMA Oncol 3, 620–627, doi: 10.1001/jamaoncol.2016.5580 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Rimassa L et al. Tivantinib for second-line treatment of MET-high, advanced hepatocellular carcinoma (METIV-HCC): a final analysis of a phase 3, randomised, placebo-controlled study. Lancet Oncol 19, 682–693, doi: 10.1016/S1470-2045(18)30146-3 (2018). [DOI] [PubMed] [Google Scholar]
- 219.Scagliotti G et al. Phase III Multinational, Randomized, Double-Blind, Placebo-Controlled Study of Tivantinib (ARQ 197) Plus Erlotinib Versus Erlotinib Alone in Previously Treated Patients With Locally Advanced or Metastatic Nonsquamous Non-Small-Cell Lung Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 33, 2667–2674, doi: 10.1200/JCO.2014.60.7317 (2015). [DOI] [PubMed] [Google Scholar]
- 220.Yoshioka H et al. A randomized, double-blind, placebo-controlled, phase III trial of erlotinib with or without a c-Met inhibitor tivantinib (ARQ 197) in Asian patients with previously treated stage IIIB/IV nonsquamous nonsmall-cell lung cancer harboring wild-type epidermal growth factor receptor (ATTENTION study). Annals of Oncology 26, 2066–2072, doi: 10.1093/annonc/mdv288 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.