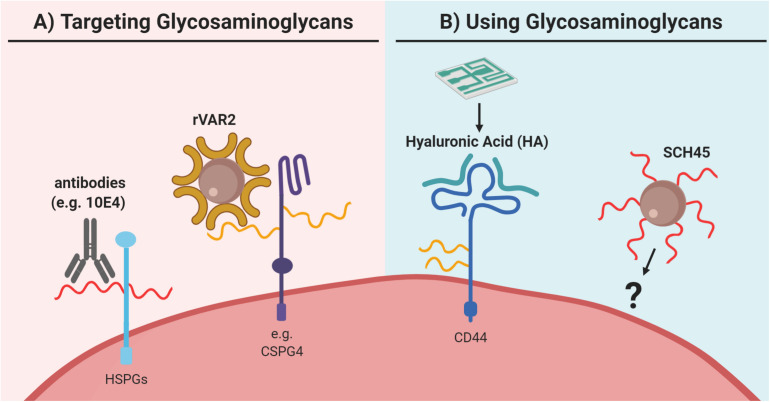
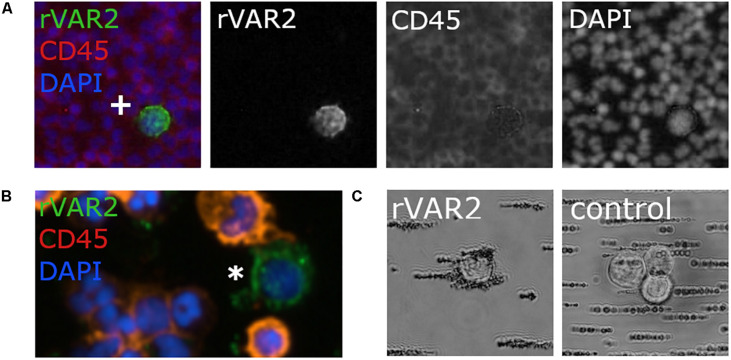
Abstract
Circulating tumor cells (CTCs) are accessible by liquid biopsies via an easy blood draw. They represent not only the primary tumor site, but also potential metastatic lesions, and could thus be an attractive supplement for cancer diagnostics. However, the analysis of rare CTCs in billions of normal blood cells is still technically challenging and novel specific CTC markers are needed. The formation of metastasis is a complex process supported by numerous molecular alterations, and thus novel CTC markers might be found by focusing on this process. One example of this is specific changes in the cancer cell glycocalyx, which is a network on the cell surface composed of carbohydrate structures. Proteoglycans are important glycocalyx components and consist of a protein core and covalently attached long glycosaminoglycan chains. A few CTC assays have already utilized proteoglycans for both enrichment and analysis of CTCs. Nonetheless, the biological function of proteoglycans on clinical CTCs has not been studied in detail so far. Therefore, the present review describes proteoglycan functions during the metastatic cascade to highlight their importance to CTCs. We also outline current approaches for CTC assays based on targeting proteoglycans by their protein cores or their glycosaminoglycan chains. Lastly, we briefly discuss important technical aspects, which should be considered for studying proteoglycans.
Keywords: cancer, circulating tumor cells, diagnostic, glycosaminoglycan, liquid biopsy, metastasis, proteoglycan, VAR2CSA
Introduction
During cancer progression, metastatic spread occurs when cancer cells disseminate from the primary tumor and travel to a distant site to form a metastasis (Micalizzi et al., 2017). This can emerge through three major routes: the blood circulation, the lymphatic system, or via serosal or mucosal surfaces (Fidler, 1978). However, cancer cell dissemination through the blood is thought to be the main route of metastasis (Lambert et al., 2017), and the subset of cancer cells that have entered the blood circulation is named circulating tumor cells (CTCs). Intravasation of CTCs into the blood stream is believed to be one of the rate-limiting steps for metastasis formation and can occur through either an active invasion or passive shedding of cells from the tumor (Cavallaro and Christofori, 2001; Bockhorn et al., 2007; van Zijl et al., 2011). Only a minority of cancer cells reaching the blood circulation manages to survive shear stress, escape immune surveillance, avoid detachment-induced cell death, extravasate at the distant site, and finally establish a metastasis (Massague and Obenauf, 2016). Thus, the process of metastasis is both a complex and inefficient process (van Zijl et al., 2011; Reymond et al., 2013).
In addition to representing the primary tumor, CTCs have also been shown to exit from metastatic lesions (Kim et al., 2009). Such cells have the potential to travel back to the primary tumor site (called tumor self-seeding) or create another metastasis (Mentis et al., 2020). Therefore, CTCs could represent both the primary tumor and potential metastatic lesions (Kim et al., 2009), making CTC analyses highly relevant even years after surgical removal of the primary tumor. Hence, CTC analyses could provide important information about disease progression and relapse. Furthermore, molecular analyses of CTCs including mutational profiling, could provide the basis for personalized therapies in the future (Greene et al., 2012). Thus, CTCs are currently evaluated for clinical diagnostics. However, CTC analysis remains technically challenging, not only due to the rarity of CTCs among billions of normal blood cells, but also due to their inherent high degree of cellular plasticity complicating the choice of detection markers (Alix-Panabieres et al., 2017). Accordingly, CTC enrichment and detection strategies must be based on highly specific biomarkers to achieve the necessary assay specificity and sensitivity. Moreover, targeting a broader CTC population would be beneficial to ensure that the liquid biopsy better reflects the heterogenic cancer cell population.
As a part of discovering novel CTC targets, many strategies focus on proteins known to play an active role in metastatic seeding. Although solid tumors differ in their metastatic patterns, they share certain mechanistic similarities for metastasis formation, which are summarized as the metastatic cascade (Figure 1) (de Groot et al., 2017; Lambert et al., 2017; Riggi et al., 2018). Notably, the majority of steps in this process concerns the interaction between the cancer cells and the surrounding extracellular matrix (ECM). Therefore, novel clinically relevant CTC targets may be discovered within the pericellular layer called the glycocalyx.
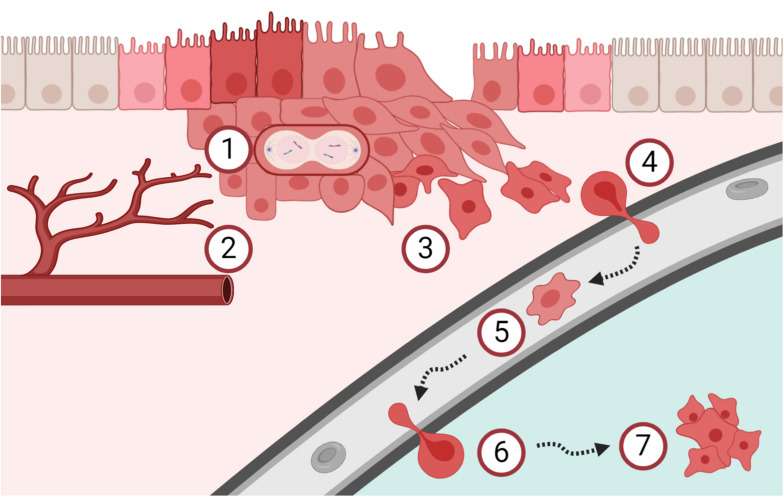
FIGURE 1.
Schematic overview of the metastatic cascade. (1) Cancer cells start to proliferate uncontrolled and (2) tumor angiogenesis is mandatory to support continued tumor growth, already early during carcinogenesis. (3) The process of epithelial-mesenchymal transition increases the migration and invasion capacity of cancer cells. (4) Cancer cells intravasate into the blood circulation and (5) are then called circulating tumor cells (CTCs). CTCs are easily accessible by liquid biopsies and are currently investigated as tool for cancer diagnostics and surveillance. (6) A subpopulation of these CTCs has the potential to extravasate and (7) form metastasis in secondary organs. Clearly, the metastatic process is very complex and many of these steps are interconnected. Please refer to the main text for details and references.
The glycocalyx is a thick network of carbohydrates bound to glycoproteins, glycolipids, and proteoglycans (Figure 2A; reused from Okada et al., 2017). It is present on cell surfaces throughout the entire human body and constitutes a physical barrier between the cell and the surrounding microenvironment (Tarbell and Cancel, 2016). The glycocalyx plays a crucial role for receptor–ligand interactions of cancer cells and their surroundings, enabling migration as well as intra- and extravasation. Furthermore, the composition of the glycocalyx is thought to influence the transportation and survival of CTCs in the bloodstream (Mitchell and King, 2014). However, very few studies have investigated the glycocalyx of CTCs. At present, it is best studied in endothelial cells, where it serves as a physical and electrostatic barrier as well as a mechanotransducer toward other cells, the extracellular matrix (ECM), or shear forces of the blood (Reitsma et al., 2007; Butler and Bhatnagar, 2019). To convey signaling, growth factors, chemokines, and other interaction partners have to navigate through this dense structure. The glycocalyx, which extends beyond the length of most surface receptors, has a dual role in signaling by creating a physical hindrance for ligand receptor interactions or by promoting binding once interaction partners are in close proximity to each other (Kuo et al., 2018). Moreover, certain glycocalyx components are involved in chemokine storage and oligomerization, which strongly modulates their signaling strength (Salanga and Handel, 2011). Therefore, glycocalyx changes can have various effects on cellular behavior and, not surprisingly, cancer cells show specific alterations in their glycocalyx.
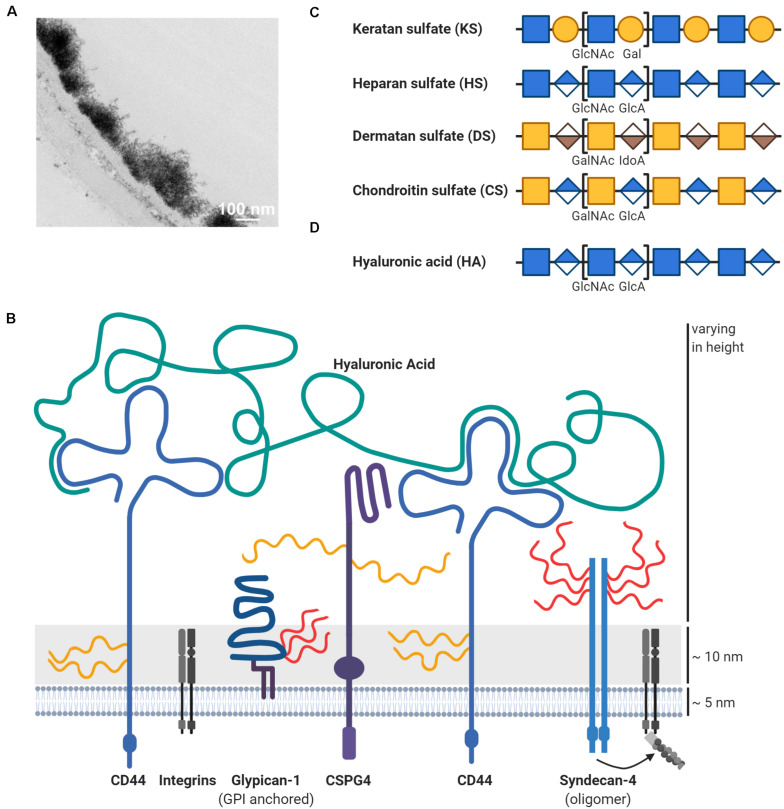
FIGURE 2.
Schematic overview of proteoglycans in the glycocalyx. (A) The glycocalyx is a dense network of carbohydrates on the cell surface, here exemplified on top of the endothelium. This transmission electron microscopic image with lanthanum nitrate staining was reused from Okada et al. (2017) under CC BY 4.0 license. (B) The thick carbohydrate layer on the cell surface extends beyond the length of membrane proteins like integrins. One important glycocalyx component are proteoglycans, which consist of a protein core (blue) and covalently attached glycosaminoglycans such as heparan sulfate (HS; in red) or chondroitin sulfate (CS; in yellow). Depicted are some proteoglycans, which are mentioned and discussed throughout the review like chondroitin sulfate proteoglycan 4 (CSPG4). Hyaluronic acid (HA; in green) is another important glycosaminoglycan component in the glycocalyx, but is distinct through the lack of a protein core. Hyaluronic acid is attached to the cell surface via interactions with its receptors like CD44, which is itself a proteoglycan. Other glycocalyx components like glycoproteins are not shown due to abstractification. Depiction of the disaccharides units for (C) glycosaminoglycans attached to proteoglycans (keratan sulfate/KS, heparan sulfate/HS, dermatan sulfate/DS, and chondroitin sulfate/CS) and (D) of hyaluronic acid (HA), which is non-covalently attached to its receptors. Glycosaminoglycans can be subjected to further modifications, such as sulfation or epimerization, which is not shown for simplicity. Please refer to the main text for details and references.
The cancer cell glycocalyx is a highly dynamic structure from which most of the components have been linked to the acquisition of oncogenic phenotypes (Daniotti et al., 2015; Buffone and Weaver, 2020). For instance, aberrant glycosylations including hypersialylation support immune evasion mechanisms (Pearce and Läubli, 2016). Moreover, increased expression of bulky glycoproteins like mucin-1 has been linked to aggressive cancers (Paszek et al., 2014; Barnes et al., 2018) and associated with poor survival outcome in patients (Kufe, 2009). This might be explained by the bulkiness of the mucin-1 ectodomain shaped by numerous glycosylations, which facilitates integrin clustering, cell signaling, and cell proliferation (Paszek et al., 2014; Woods et al., 2017; Kuo et al., 2018). Naturally, mucin-1 is of interest as a target for therapy (Pillai et al., 2015), due to its high involvement in cancer. Likewise, CTCs were found to express high mucin-1 levels (Paszek et al., 2014), and mucin-1 has also been explored for CTC capture and detection (Muller et al., 2012; Strati et al., 2013; Schehr et al., 2016).
Another important component of the glycocalyx are proteoglycans (Figure 2B) with multiple implications in metastatic dissemination of cancer cells and tumor cell growth (Iozzo and Sanderson, 2011; Vitale et al., 2019). Proteoglycans can be secreted into the ECM or located intracellularly as well as on the cell surface either directly embedded in the plasma membrane or anchored by a glycosylphosphatidylinositol (GPI)-linker (Iozzo and Schaefer, 2015). Proteoglycans consist of two functional units: protein core and glycosaminoglycan (GAG) chains (Walimbe and Panitch, 2019). Most commonly, the assembly of GAG chains occur from a tetrasaccharide linker region covalently attached to serine residues within the protein core (Esko and Zhang, 1996). The GAG family is classified by their chemical composition and includes chondroitin sulfate (CS), dermatan sulfate (DS), keratan sulfate (KS), heparan sulfate (HS), and hyaluronic acid (HA) (Figures 2C,D) (Toole, 2004; Bulow and Hobert, 2006). In general, GAGs consist of long linear repeats of disaccharide units consisting of hexuronic acids and hexosamines. The hexuronic acid epimers comprise D-glucuronic acid (GlcA) for CS/HA and L-iduronic acid (IdoA) for DS, whereas the hexosamine units consist of an N-acetyl-D-glucosamine (GlcNAc) for HS/KS/HA, and an N-acetyl-D-galactosamine (GalNAc) for CS/DS (Schaefer and Schaefer, 2010; Ghiselli and Maccarana, 2016; Pomin and Mulloy, 2018). The structures of GAGs are extremely diverse, as their synthesis in the Golgi apparatus is not based on a precise template, but on a redundant network of enzymes that seems to be regulated based on tissue and cell types (Dick et al., 2012; Mikami and Kitagawa, 2013; Chen Y. H. et al., 2018).
Besides the variation in the monosaccharide composition, the molecular diversity of GAGs also results from varying polymer lengths and extensive post-translational modifications such as sulfations and epimerizations along the chain (Bulow and Hobert, 2006). GAG sulfation patterns often determine their biological function and serve as specific recognition motifs for a wide variety of growth factors, cytokines, chemokines, and pathogens (Xu and Esko, 2014; Mizumoto et al., 2015; Pinho and Reis, 2015). Therefore, alterations in the GAG composition of proteoglycans in cancers have received a lot of interest (Sweet et al., 1976; Chandrasekaran and Davidson, 1979; De Klerk et al., 1984). A well-studied example is the change in sulfation patterns of GAGs, which likely depends on the specific cancer type. Some studies have reported high expression of CS 4-O-sulfotransferases in both ovarian and breast cancers, while a study on cancerous lung tissues found elevated 6-O-sulfated CS, compared to nonmalignant tissue (Cooney et al., 2011; Oliveira-Ferrer et al., 2015; Li et al., 2017). Similarly, various HS sulfotransferases have been found upregulated in different cancers. These include 6-O-sulfotransferases in ovarian and colorectal cancer; 3-O-sulfotransferases in breast and pancreatic cancer; along with N-deacetylase and N-sulfotransferases in hepatocellular carcinomas (Tatrai et al., 2010; Song et al., 2011; Hatabe et al., 2013; Cole et al., 2014; Vijaya Kumar et al., 2014). Moreover, several studies have reported increases in CS quantity or in expression of CS polymerization genes in malignant tissues, suggesting that CS polymers are pro-tumorigenic (Momose et al., 2016; Li et al., 2017; Hou et al., 2019).
Interestingly, some proteoglycans may be modified with different GAG types simultaneously, as seen for syndecans, which can carry both CS/DS and HS, dependent on the structure of the ectodomain (Kokenyesi and Bernfield, 1994; Iozzo and Schaefer, 2015). Similarly, the HA-binding proteoglycan, versican, undergoes alternative splicing of exons encoding the GAG-attachment region resulting in altered GAG display. Notably, expression of distinct versican isotypes was shown to facilitate cancer progression in multiple cancer types (Dours-Zimmermann and Zimmermann, 1994; Theocharis et al., 2015; Zhangyuan et al., 2020). The protein cores of proteoglycans are, however, not just scaffolds for GAG extension, since they also directly bind ligands and mediate intracellular signaling in GAG-independent manners.
In contrast to the rest of the GAGs, HA stands out by not being covalently attached to a protein core (Figures 2B,D). Instead, HA is synthesized as an unmodified polysaccharide at the plasma membrane, where it is extruded from the cell surface and cleaved off into the ECM (Weigel and DeAngelis, 2007; Itano, 2008). In most cells, HA is an abundant structural component of the glycocalyx, where it interacts with receptors and surface proteoglycans via their hyaluronan-binding motifs and regulates the viscosity of the glycocalyx by its ability to retain water (Toole, 2001, 2004). Upon binding, HA triggers activation of a range of signaling pathways involved in cell proliferation, differentiation, motility, and adhesion, thereby influencing processes such as development, tissue homeostasis, and carcinogenesis (Turley et al., 2002; Toole, 2004; Liu et al., 2019). Although HA is extensively involved in cancer, it will not be discussed in detail throughout this review, as it is not considered a proteoglycan due to its lack of a protein core.
Altogether, proteoglycans compose a highly heterogeneous group of proteins that diverge by structural alterations of the protein core as well as by differences in their GAGs with regard to chain number, type, length, and post-translational modifications. Notably, proteoglycans are important integrators for cell signaling events with direct implications for carcinogenesis and cancer progression (Iozzo and Sanderson, 2011; Pinho and Reis, 2015; Nikitovic et al., 2018). In spite of this, the functions of proteoglycans and their GAGs in relation to CTCs are currently understudied. Therefore, this review will highlight examples of proteoglycans involved in the metastatic cascade with potential links to CTC biology. More specifically, we will discuss how proteoglycans play active roles in cancer cell proliferation, migration, survival, plasticity, and invasion with a dedicated focus on the function of both the protein core and the GAG chains. Finally, we provide an overview of proteoglycans that are currently evaluated for CTC technologies and briefly highlight some of the technical aspects to consider when studying proteoglycans.
Proteoglycans in the Metastatic Cascade
Cancer Cell Proliferation
Cancers show deregulation of their cell proliferation by various mechanisms. Proteoglycans can influence cell growth by interacting with growth factors, either via their core proteins or through their GAG chains, as observed for HS chains of heparan sulfate proteoglycans (HSPGs) (Knelson et al., 2014). Enzymes modifying GAGs may hence influence tumor growth (Morla, 2019) as demonstrated by sulfatases interfering with growth factor signaling through HS desulfation (Ai et al., 2003; Peterson et al., 2010; Vicente et al., 2015).
However, proteoglycans also influence tumor growth by GAG–independent mechanisms. The transmembrane chondroitin sulfate proteoglycan 4 (CSPG4) has been shown to positively regulate cancer cell proliferation in various cancer entities (Wang et al., 2011; Jamil et al., 2016; Hsu et al., 2018) and is currently under investigation for CTC capture and identification, as described in detail later. Studies have found that CSPG4 is involved in growth signaling by interaction through both its cytoplasmic domain and ectodomain (Yang et al., 2009; Stallcup, 2017). Through the extracellular part, CSPG4 potentiates the mitogen-activated protein kinase (MAPK) cascade by high-affinity, largely GAG-independent binding of growth factors, which are thus likely presented to their cognate receptors by CSPG4 (Nishiyama et al., 1996; Goretzki et al., 1999; Stallcup, 2002; Price et al., 2011). In glioma cell models, phosphorylation of the cytoplasmic CSPG4 domain induced proliferation, which was mediated by interaction with integrins (Makagiansar et al., 2007; Stallcup, 2017). Furthermore, CSPG4-mediated activation of one of the same integrins induced chemoresistance and survival in tumor cells (Chekenya et al., 2008). Thus, CSPG4 is an example of a proteoglycan positively regulating growth and survival via its protein domain.
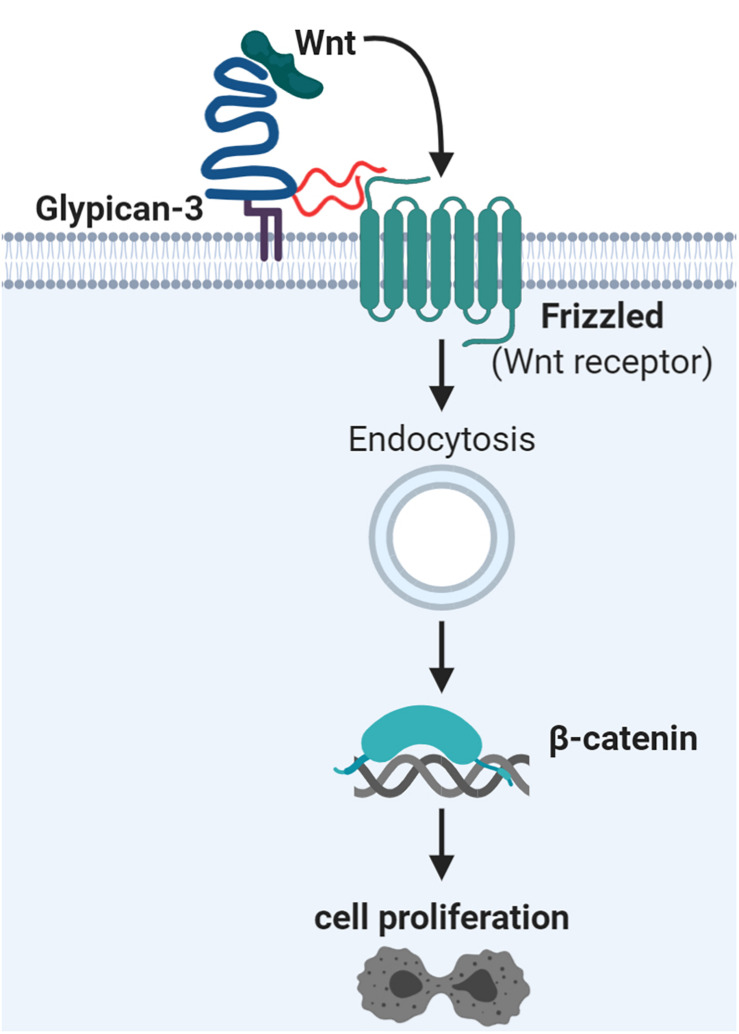
The proteoglycan glypican-3 (GPC3) has also been shown to increase cell proliferation. GPC3 influence several central signaling pathways in hepatocellular carcinoma (Kolluri and Ho, 2019) and is also evaluated for CTC capture as described later. GPC3 and other glypicans are GPI-anchored and known to carry HS chains (Filmus et al., 2008; Yoneda et al., 2012), but were also demonstrated to carry CS chains (Chen and Lander, 2001; Toledo et al., 2020). Their GAG chains are located close to the cell membrane due to their proximity to the C-terminus, which is thought to be critical for their interaction with surface receptors (Filmus et al., 2008). GPC3 overexpression increased cell proliferation in vitro and in vivo for liver cancer cells by enhancing Wnt signaling (Figure 3) (Capurro et al., 2005). Mutagenesis of the GAG attachment site in GPC3 revealed that the HS GAGs were not essential for binding of Wnt ligands (Capurro et al., 2005). Supporting this, the Wnt binding site on GPC3 has recently been located to a hydrophobic groove, which works independently of GAG chains (Li et al., 2019). However, the GAG chains of GPC3 are essential for direct interaction with the Wnt receptors, the Frizzled proteins (Capurro et al., 2014). Upon Wnt stimulation a ternary complex is formed and endocytosed (Capurro et al., 2014). Generally, endocytosis of Wnt signaling complexes seems to be important for canonical Wnt signaling with final stabilization and nuclear accumulation of β-catenin and subsequent gene expression changes (Brunt and Scholpp, 2018). In addition, this signaling axis could be a potential therapeutic target for hepatocellular carcinoma based on a monoclonal antibody recognizing the HS chains of GPC3 (Gao et al., 2014). Overall, it was suggested that GPC3 works as a bridging protein between Wnt and its receptor thereby inducing cell proliferation (Li et al., 2019). The exact interaction dependencies could rely on the expression levels of all three partners (Wnt ligands, Wnt receptors, and GPC3) (Li et al., 2019).
FIGURE 3.
Glypican-3 signaling supports Wnt signaling and hepatocellular proliferation. Glypican-3 (shown in blue) can carry two glycosaminoglycan chains of heparan sulfate (HS; in red) or chondroitin sulfate (CS; not shown). It has been determined that these glycosaminoglycan chains are essential for interaction with Frizzled proteins, the Wnt receptors, but not for Wnt ligand binding. The ternary complex of glypican-3, Frizzled, and Wnt ligand becomes endocytosed as part of canonical Wnt signaling. This leads to nuclear accumulation of β-catenin and subsequent gene expression changes, stimulating cell proliferation. Details and references are given in the main text.
However, proteoglycans can also act as negative regulators of cancer biology. One example for this is decorin, which is modified with a single CS or DS side chain. Decorin can act as an inhibitor of cell proliferation by hampering growth signaling. This repression is thought to occur through growth factor sequestering as well as receptor internalization and degradation, mediated by binding to the decorin core protein (Jarvinen and Prince, 2015). For example, de novo expression of decorin in breast cancer cell lines suppressed proliferation and anchorage-independent growth (Santra et al., 2000). Consistently, 30% of decorin-knockout mice formed spontaneous intestinal tumors (Bi et al., 2008), highlighting its potential role as tumor suppressor.
To sum up, proteoglycans appear to have a multi-facetted and important role in cancer cell proliferation by diverse mechanisms, which can vary across different cancer types. When CTCs reach the metastatic site, they often go into an inactive dormancy state (Sosa et al., 2014). Reactivation of cell proliferation is therefore an important factor for establishment of clinically relevant metastatic lesions, in which proteoglycans are actively involved (Elgundi et al., 2019) and which will also be discussed later in more detail.
Angiogenesis in Cancer
Oxygen supply is essential for cells and their metabolism. Ex vivo measurement on xenografts revealed that oxygen perfuses only to around 100 μm deep into the tumor tissue (Olive et al., 1992). Therefore, cancer cells must secure sustained blood supply at an early stage, which can happen by different mechanisms (Xu et al., 2016; Lugano et al., 2020). Several proteoglycans are involved in the complex process of tumor angiogenesis (Iozzo and Sanderson, 2011; Chiodelli et al., 2015). Interestingly, increased vascularization could be observed already in premalignant lesions (Menakuru et al., 2008), possibly explaining how CTCs can be shed already from early stage cancers (Husemann et al., 2008; Stott et al., 2010; Rhim et al., 2012; Zhang et al., 2014; Tsai et al., 2016; Murlidhar et al., 2017). Studies on early cancer cell dissemination are of high clinical importance as it enables the use of CTCs in screening programs for early cancer detection.
One central molecule for angiogenesis is the vascular endothelial growth factor (VEGF) (Ferrara et al., 2003), which has been linked to different proteoglycans as for example biglycan. Cancer cells have been shown to overexpress biglycan (Zhu et al., 2013; Hu et al., 2014; Andrlova et al., 2017; Jacobsen et al., 2017), which has two potential GAG attachment sites carrying either CS or DS chains (Valiyaveettil et al., 2004). Interestingly, biglycan is a homolog of decorin (Fisher et al., 1989), but seems to have tumor promoting capacities by angiogenesis induction in contrast to decorin (Schaefer et al., 2017). Indeed, elevated biglycan levels induced higher density of blood vessels and increased tumor growth in vivo of colorectal cancer xenografts via induction of VEGF expression (Hu et al., 2016). In endothelial cells, biglycan binds to Toll-like receptor 2 (TLR2) and TLR4 with activation of the transcription factor family nuclear factor-κB (NFκB). This subsequently leads to increased levels of hypoxia-inducible factor 1-alpha (HIF1α), which drives VEGF expression (Hu et al., 2016) and could finally lead to tumor angiogenesis. VEGF can potentially regulate expression of another proteoglycan linked to cancer and angiogenesis, namely endocan (Grigoriu et al., 2006; Rennel et al., 2007; Roudnicky et al., 2013). Indeed, endocan was detected in the tumor vasculature (Maurage et al., 2009; Roudnicky et al., 2013) as well as in cancer cells (Rennel et al., 2007; Maurage et al., 2009; Xu et al., 2019). Interestingly, it was suggested by Rocha et al. (2014) that endocan binding replaced VEGF from fibronectin in the ECM, creating a positive feedback loop. In head and neck cancer, endocan was strongly co-expressed with angiopoietin-2 (Xu et al., 2019), which can regulate vascular permeability during intra- and extravasation processes (Garcia-Roman and Zentella-Dehesa, 2013) and potentially affect the dissemination of CTCs during metastasis. Importantly, endocan expression is associated with poor survival rate and might also be used as serum biomarker in cancer patients (Grigoriu et al., 2006; Roudnicky et al., 2013; Kim et al., 2018). Biglycan and endocan are important proteoglycans in angiogenesis and thus tumor progression. However, they might not be ideal candidates for CTC technologies as secreted proteoglycans might not be stable targets for cell analysis.
The neuropilins is another proteoglycan family involved in angiogenesis (Ellis, 2006; Niland and Eble, 2019). Neuropilin-1 helps to bind VEGF to the cell surface and forms a trimeric complex together with VEGF receptor 2 (VEGFR2), which was suggested to act as a potential bridge between cancer cells and endothelial cells (Soker et al., 1998, 2002). Indeed, neuropilin-1 was detected in tumor cells of different cancer entities as well as in endothelial cells of the tumor vasculature (Jubb et al., 2012). Overexpression of neuropilin-1 increased xenograft growth (Miao et al., 2000; Hu et al., 2007). Neuropilin-1 carries a single GAG chain of HS or CS, dependent on the cell type (Shintani et al., 2006; Frankel et al., 2008). However, the exact role of the GAG chain is not fully understood. Mutagenesis of the GAG attachment site in neuropilin-1 increased glioma cell invasion (Frankel et al., 2008). Interestingly, global removal of CS by chondroitinase ABC enzyme treatment led to decreased invasion in the same cells. Neuropilin-1 is also physiologically expressed as a GAG-deficient splice variant (namely NRP1-Δ7), which attributes to 10–30% of total neuropilin-1 transcripts depending on cell type or tissue (Hendricks et al., 2016). Importantly, NRP1-Δ7 acted anti-tumorigenic and diminished tumor vascularization in prostate cancer xenografts in vivo (Hendricks et al., 2016). Moreover, soluble neuropilin-1 isoforms with anti-tumorigenic functions have been described, which block VEGFR signaling (Gagnon et al., 2000; Cackowski et al., 2004). Overexpression of soluble neuropilin-1 in cancer cells led to disturbed tumor vascularization and cancer cell apoptosis in xenografts (Gagnon et al., 2000). Overall, proteoglycans are connected to tumor angiogenesis and to VEGF signaling with various effects.
Epithelial-to-Mesenchymal Transition, Migration, and Intravasation
Another important milestone for cancer cells is to gain migratory capacities to leave the primary tumor and invade the surrounding tissue. During the gastrulation phase of embryogenesis, epithelial-to-mesenchymal transition (EMT) causes stationary epithelial cells to undergo major changes into motile mesenchymal-like cells in order to form new germ layers. Molecular changes in transcription factor networks and gene expression facilitate the loss of cell polarity and cytoskeletal reorganization, resulting in an increased migratory capacity (Lim and Thiery, 2012; Lamouille et al., 2014). Cancer cells imitate this developmental EMT program and several studies suggest that proteoglycans are actively involved in this part of cancer progression, thus supporting the relevance of proteoglycans as targets for CTC capture. Situated in the glycocalyx of cancer cells, proteoglycans provide a contact link between the cell membrane and the surrounding ECM, thereby playing a central role in regulating cancer cell adhesion and migration. Some proteoglycans are downregulated in order to enable detachment from the basement membrane facilitating invasion, others are shed from the surface as a different mode of regulation, and some maintain their function throughout the invasive phase. Importantly, the current standard for CTC isolation is based on antibodies against epithelial cell adhesion molecule (EpCAM), which is often downregulated during EMT (Gorges et al., 2012; Hyun et al., 2016). Thus, understanding the process of EMT in terms of proteoglycan regulation is important for their evaluation as alternative CTC-target candidates.
One important modulator of EMT processes is transforming growth factor β (TGFβ), which is known to drive progression of late state malignancies by promoting invasion (Akhurst and Derynck, 2001; Xu et al., 2009). Indeed, TGFβ regulates a multitude of genes with potential cancer-specific effects (Ranganathan et al., 2007; Kowli et al., 2013). Several proteoglycans are connected to TGFβ-signaling. An example of this is the expression of HS-carrying syndecan-4, which was positively regulated by TGFβ in lung cancer A549 cells (Toba-Ichihashi et al., 2016). Expression of this proteoglycan further induced upregulation of the EMT transcription factor zinc finger protein SNAI1 (sometimes referred to as snail) (Toba-Ichihashi et al., 2016), thereby fueling the migratory behavior. This is somewhat surprising, since syndecan-4 plays a well-established role in focal adhesion sites together with integrins, thereby promoting the adhesive phenotype of cancer cells (Echtermeyer et al., 1999; Saoncella et al., 1999).
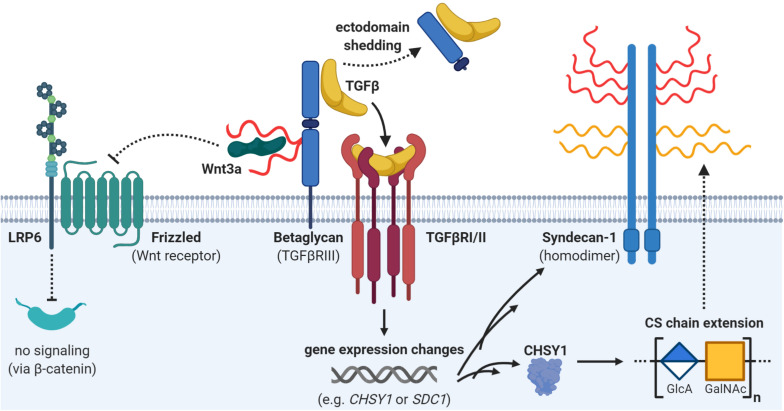
Syndecan-1 can also be affected by TGFβ and was suggested as a poor prognostic factor in breast cancer (Hayashida et al., 2006; Nikitovic et al., 2014). Incubation of mouse mammary epithelial cells with TGFβ changed the GAG composition of syndecan-1 from being mainly HS modified to carry nearly equal amounts of HS and CS (Rapraeger, 1989). Notably, increased CS display was not only mediated by attachment of more GAG chains, but also by increased length of individual CS chains (Figure 4). This is in line with a later study showing that TGFβ induced expression of CS synthase 1, a key enzyme involved in the elongation of CS and DS GAG chains (Hu et al., 2015). In addition, other GAG polymer-modifying enzymes have been shown to be transcriptionally affected by TGFβ treatment (Tiedemann et al., 2005; Mohamed et al., 2019), suggesting a role of specific GAG modifications on proteoglycans in regulating the cellular response toward this cytokine.
FIGURE 4.
Interplay of proteoglycans and transforming growth factor beta (TGFβ) signaling. The proteoglycan betaglycan is a co-receptor for TGFβ and brings it to the TGFβ receptor (heterodimer of TGFβRI and TGFβRII). However, ectodomain shedding of betaglycan might attenuate TGFβ signaling. Furthermore, betaglycan also dampens Wnt signaling by sequestering Wnt ligands with its heparan sulfate (HS) side chains (in red). Active TGFβ signaling can affect the expression of many different genes such as SDC1 or CHSY1. Upregulation of the enzyme chondroitin sulfate synthase 1 (CHSY1) can potentially lead to elongation of chondroitin sulfate (CS) chains on proteoglycans. Indeed, TGFβ can also upregulate one potential CHSY1 targets, namely syndecan-1, which can carry both chondroitin sulfate (CS; in yellow) or heparan sulfate (HS; in red). Details and references are provided in the main text.
TGFβ signaling is mediated through heteromeric complex formation of type I and type II receptors (Weiss and Attisano, 2013). However, co-receptors like type III TGFβ receptor, also known as betaglycan, can modulate ligand presentation to the type II receptor (Figure 4). Betaglycan is a cell membrane proteoglycan which may carry both CS and HS GAG chains (Cheifetz et al., 1988). Sulfated HS-modifications on betaglycan have been proven to sequester the Wnt3a ligand and thereby inhibiting proliferation by dampening Wnt signaling (Jenkins et al., 2016). In contrast, TGFβ binding is mediated by the protein core of the proteoglycan, and is therefore insensitive to point mutations disrupting the GAG-attachment sites (Lopez-Casillas et al., 1994). As any other membrane proteoglycan, betaglycan can undergo ectodomain shedding (Weiss and Attisano, 2013). A soluble form of the receptor was shown to result in reduced ligand availability due to its high-affinity interaction with TGFβ and thus decreased TGFβ signaling (Elderbroom et al., 2014). In line with this, it was shown that increased betaglycan expression decreased the invasive behavior of breast cancer cells in vitro in response to TGFβ stimulation and that this effect was abrogated when betaglycan was expressed in a shedding-impaired mutant form (Elderbroom et al., 2014). Importantly, metastatic lesions showed lower betaglycan expression compared to matched primary tumors (Hempel et al., 2007). In ovarian cancer cell lines this seems to be mediated through epigenetic silencing, as expression was restored upon epigenetic-acting drugs (Hempel et al., 2007). This indicates that betaglycan might be involved in the dissemination processes and thus the investigation of its biological function in CTCs and metastasis formation would be interesting.
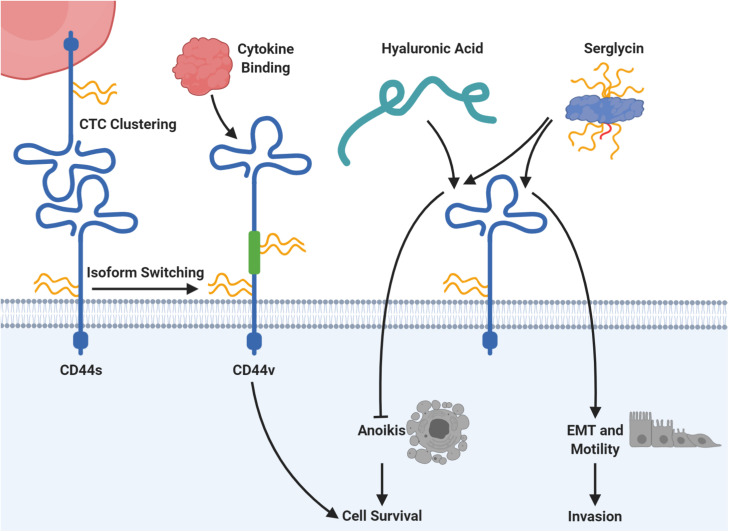
In addition, proteoglycans might affect cancer cell migration independently of TGFβ signaling. For example, a number of studies have demonstrated a role of serglycin in malignant transformation as described below. Serglycin can carry up to eight CS or HS chains (Kolset and Tveit, 2008) and is widely expressed by hematopoietic cells as well as embryonic stem cells, where it serves functions in storage of intracellular granules and secretion of inflammatory mediators (Toyama-Sorimachi et al., 1995; Schick et al., 2001; Abrink et al., 2004). Elevated serglycin expression was reported for cancer cells in patient tissues and has been linked to aggressive cancer cell phenotypes in vitro (Korpetinou et al., 2015). Further, serglycin was identified as an unfavorable prognostic factor in patients suffering from a range of cancers, including glioblastoma (Roy et al., 2017), liver (He et al., 2013), lung (Guo et al., 2017), and nasopharynx (Li et al., 2011). Secreted serglycin from cancer cells was shown to be primarily CS-modified, and transgenic expression of serglycin lacking the GAG attachment site led to decreased migratory capacity of breast cancer cells in vitro (Korpetinou et al., 2013). This observed GAG-dependency was further supported by another study focused on lung cancer cells (Guo et al., 2017). Here it was demonstrated that serglycin exerts its functional role during migration by binding to the cluster of differentiation 44 antigen (CD44) with downstream activation of EMT (Figure 5). Blocking of the post-translational CS modification on serglycin abrogated the effect on motility (Guo et al., 2020). CD44 itself is a transmembrane proteoglycan expressed by various cell types (Goodison et al., 1999; Gronthos et al., 2001; Domev et al., 2012) and is normally involved in hematopoiesis, inflammation, and wound healing (Johnson et al., 2000; Dimitroff et al., 2001b; Cichy and Puré, 2003). CD44 is also involved in several important steps during metastasis formation and has been explored as a CTC target in several studies as described later.
FIGURE 5.
CD44 is connected to several mechanisms of circulating tumor cell (CTC) survival and plasticity. The protein core of CD44 can carry chondroitin sulfate (CS; in yellow), heparan sulfate (HS; in red), or dermatan sulfate (DS; not shown). Homophilic CD44 interactions allow clustering of CTCs, which supports seeding and colonization at the metastatic site in mice. Moreover, expression of the standard CD44 protein form (CD44s) can be switched to expression of CD44 variants (CD44v), which include additional exons via alternative splicing (depicted in green). Those CD44v forms may carry additional glycosaminoglycan chains in dependency of the included exons and promote binding of cancer-related cytokines with downstream signaling for cell survival. The proteoglycan serglycin can carry eight glycosaminoglycan chains of heparan sulfate (HS; in red) or chondroitin sulfate (CS; in yellow) and binds to CD44. Serglycin binding to CD44 induces epithelial-mesenchymal transition (EMT) as well as cell motility and has been proven to be dependent on its CS chains. Moreover, the serglycin-CD44 interaction prevents the induction of anoikis, a specialized form of apoptosis. For this, serglycin competes with hyaluronic acid, which has the same survival promoting effect. Please refer to the specific subsections for details and references.
When cells gain migratory and invasive capacity, this is often associated with increased remodeling and breakdown of the ECM, which finally enables breaching of the endothelial basement membrane and intravasation into the blood circulation (Eccles, 1999). One contributing factor is the secretion of different proteases by cancer cells, including for example matrix metalloproteinases (MMPs) (Lynch and Matrisian, 2002). MMPs are produced in a catalytically inactive form, which requires proteolytic cleavage for activation. Interestingly, CSPG4 has been shown to facilitate assembly of a ternary complex consisting of pro-MMP2, MMP-cleaving enzyme (MT3-MMP), and the proteoglycan itself at the cell surface of melanoma cells, leading to cleavage and thus activation of MMP2 (Iida et al., 2007). While the interaction with MT3-MMP was shown to be mediated through the core protein of CSPG4, the association with pro-MMP2 was depending on the CS-modification (Iida et al., 2007).
Another enzyme playing an important role in accessing the blood circulation is heparanase, which cleaves HS polysaccharides located in the basement membrane and on the cancer cell surface, leading to increased invasive behavior of cancers (Sanderson et al., 2017; Masola et al., 2018; Elgundi et al., 2019). Several studies have provided evidence that heparanase plays a major role in progression of a variety of cancers including liver cancer (Koliopanos et al., 2001), sarcomas (Cassinelli and Lanzi, 2020), ovarian cancer (Zhang et al., 2013), breast cancer (Maxhimer et al., 2005), and colon cancer (Nobuhisa et al., 2005). In addition to this, it has recently been shown that overexpression of heparanase promotes the formation of cell clusters in MDA-MB-231 breast cancer cells, most likely by modulating the level of intercellular adhesion molecule 1 (ICAM-1) and phosphorylation status of downstream kinases (Wei et al., 2018). Furthermore, the enhanced ability to form clusters correlated with increased number of metastatic foci in the lungs upon tail-vein injection into mice (Wei et al., 2018). Together this data strongly suggests that invasion and intravasation are mediated through HS degradation in the ECM and possibly at the cell surface, leaving behind HS-associated core proteins without this modification.
Surviving the Journey Through Circulation
For normal cells, detachment from the extracellular matrix leads to cell death through a mechanism of detachment-induced apoptosis, called anoikis (Strilic and Offermanns, 2017). Therefore, CTCs must overcome this major challenge in order to survive in circulation. Studies have pointed toward several cellular strategies to circumvent apoptotic signaling, some involving proteoglycans as important players.
Increased syndecan-4 and heparanase expression have been reported in anoikis-resistant rat endothelial cells (Carneiro et al., 2014). Studying these cell lines, Carneiro et al. (2014) also detected increased level of HS in the culture medium, whereas cell lysates contained increased levels of CS. As another example of proteoglycan involvement in anoikis resistance, overexpression of serglycin in a lung cancer cell line led to increased survival in an anchorage-independent growth assay (Guo et al., 2017). This effect was dependent on CD44 expression. In line with this, increased CD44 expression caused by EMT induction also led to anoikis resistance in immortalized human mammary epithelial cells (Cieply et al., 2015). However, in this study the ability of anchorage-independent growth relied on the hyaluronan-binding capacity of CD44. Interestingly, an early study indicated that serglycin and hyaluronan were competing for the binding to CD44 (Figure 5) (Toyama-Sorimachi et al., 1995). The prominent role of CD44 in escaping anoikis was further strengthened by a study linking the CD44-expressing subsets of two hepatocellular carcinoma cell lines to anoikis resistance (Okabe et al., 2014). Importantly, this study also observed increased CD44 expression in CTCs compared to patient-matched primary tumor biopsies, again highlighting a potential role of CD44 in CTC analysis.
The ability of the cancer cells to establish an immunosuppressive microenvironment, and thereby escape elimination by the immune system, is considered one of the hallmarks of cancer (Hanahan and Weinberg, 2011). Leaving the primary tumor site and entering the hostile environment of the blood circulation further elevate the requirement for immune cell evasion. One specific strategy for suppressing an immune attack is upregulation of CD47, which constitutes an anti-phagocytotic “do-not-eat-me”-signal on the surface of cancer cells (Jaiswal et al., 2009). CD47 is a proteoglycan carrying both CS and HS, and is widely expressed on white blood cells, where it functions as a receptor for thrombospondin-1 in a GAG-dependent manner (Kaur et al., 2011). Interestingly, CD47 expression was upregulated in colorectal CTCs compared to corresponding primary tumor tissue (Steinert et al., 2014). In addition, Baccelli et al. (2013) characterized the metastasis-initiating subpopulation of breast cancer CTCs as positive for EpCAM, tyrosine-protein kinase Met (cMet), CD44, and CD47 (Baccelli et al., 2013). Another study found that blocking of CD47 on 4T1 mouse breast cancer cells prior to tail vein injection significantly reduced the number of lung metastases in mice (Lian et al., 2019).
Another proteoglycan-based mechanism was shown to provide a strategy to avoid the secretion of lytic granules from NK cells, which would be lethal to the cancer cells. Baccelli et al. (2013) demonstrated that expression of telomeric repeat-binding factor 2 (TRF2) controlled a cell-extrinsic pathway, involving upregulation of HS glucosamine 3-O-sulfotransferase 4 (HS3ST4), thereby dampening immune surveillance by NK cells (Biroccio et al., 2013). Further, it was revealed that TRF2 overexpression led to upregulated expression of two HS-carrying proteoglycans, glypican-6 and versican, both of which were shown to decrease NK cell degranulation (Cherfils-Vicini et al., 2019). However, whether TRF2 and associated changes in HSPGs play a role for CTC survival in the circulation still needs to be shown.
Extravasation and Colonization at the Metastatic Site
To form metastatic lesions, CTCs must extravasate and enter the distal tissue. This crucial step in the metastatic cascade is highly inefficient, as the vast majority of CTCs undergo apoptosis, and only a small fraction of the surviving cells succeed in forming metastatic colonies (Massague and Obenauf, 2016; Rejniak, 2016). During extravasation, CTCs adhere to and cross the vascular endothelium in the process of transendothelial migration (TEM) (Reymond et al., 2013). Indeed, multiple factors influence cancer cell extravasation. For instance, capillaries lined with fenestrated endothelial cells and a discontinuous basal membrane in the liver and bone marrow facilitate CTC invasion (Aird, 2007; Strilic and Offermanns, 2017) and contribute to the high incidence of bone and liver metastases (Nguyen et al., 2009; Budczies et al., 2015).
There is ample evidence that CTCs exert what has become known as “leukocyte mimicry,” since many of the adhesion and TEM mechanisms are shared with leukocytes (Strell and Entschladen, 2008). Especially, the selectin family of adhesion molecules, important for hematopoietic progenitor cell homing to the bone marrow, have been implicated in cancer cell extravasation (Dimitroff et al., 2001a, 2004). This is mainly mediated by sialofucosylated carbohydrate ligands, particularly the sialyl Lewis (sLeX) structures, which are primarily found on leukocytes (Fukuda et al., 1999) as well as on various cancer cells (Majuri et al., 1995; Renkonen et al., 1997). A particular sialofucosylated glycoform of the proteoglycan CD44 termed hematopoietic cell E-/L-selectin ligand (HCELL) mediates selectin tethering (Sackstein, 2004). E-selectin is highly expressed on endothelial cells in the bone marrow (Burdick et al., 2012). In cooperation with carcinoembryonic antigen, HCELL facilitates cancer cell rolling through binding to E-selectin (Hanley et al., 2006; Thomas et al., 2008), strongly supporting the hypothesis of HCELL-mediated CTC arrest in the vasculature, a crucial step in CTC extravasation (Lee et al., 2014). Interestingly, studies have investigated the presence of over-sulfated GAGs as alternative ligands for selectins in cancer extravasation mechanisms (Martinez et al., 2013), highlighting the importance of altered display of GAGs.
Besides robust adhesion of CTCs to the endothelial wall, CTC clusters also seem to be important for metastatic seeding and outgrowth. In the circulation, CTCs can exist as single cancer cells or as clusters of cancer cells. The prevalence of polyclonal CTC clusters correlates with poor prognosis in patients, and is believed to be an important component for metastatic success (Aceto et al., 2014; Gundem et al., 2015; Cheung et al., 2016). In a recent study, CD44 was identified as a key component in clustering of cancer cells both in patient-derived xenograft (PDX) models in mice and in metastatic breast cancer patients (Liu et al., 2019). Mechanistically, CD44 formed homophilic interactions independent of HA on the cancer cell surface (Figure 5), which in turn triggered activation of a serine/threonine-protein kinase 2 (PAK2) and focal adhesion kinase (FAK) dependent signaling cascade. Knockout of CD44 resulted in loss of CTC cluster formation and reduced lung colonization and metastasis in PDX models.
CD44 is a multi-functional proteoglycan for colonization and priming of the metastatic niche (Zoller, 2011). The standard CD44 (CD44s) comprises exons 1–5 and 16–20, while the splice variants (CD44v) also include various combinations of exons 7–15, whereas exon 6 is missing in humans (Naor et al., 2002). In several cancers isoform switching via alternative splicing of CD44 is frequently observed (Johnson et al., 2000). For example, CD44v3, CD44v6, and CD44v10 have been implicated in cancer and are the only CD44 isoforms that contain binding sites for cancer-related cytokines and chemokines (Chen C. et al., 2018; Wang Z. et al., 2018). In colorectal cancer, CD44v6 positive cells are able to form metastatic lesions in the liver and lung through interaction with osteopontin (Huang et al., 2012). Importantly, the CD44 protein core can carry HS, CS, KS, or DS, but the GAG content is highly dependent on the isoform and exons involved (Bennett et al., 1995; Greenfield et al., 1999; Clark et al., 2004). Furthermore, cytokines secreted in the tumor microenvironment (e.g., hepatocyte growth factor and stromal-derived factor 1a), increased CD44v6 expression, and assisted colorectal cancer stem cells in colonization and survival through activation of the phosphatidylinositol 3-kinase-protein kinase B (PI3K-AKT) pathway (Todaro et al., 2014).
Once the CTCs have managed to extravasate into the tissue, the nature of the ECM at the secondary site dictates whether the disseminated cancer cells will proliferate into overt metastases, enter a dormant state, or undergo apoptosis (Ghajar et al., 2013; Sosa et al., 2014; Peinado et al., 2017; Goddard et al., 2018). One way in which cancers prime the pre-metastatic niche is through exosome secretion, which facilitates organ-specific engraftment of cancer cells (Simons and Raposo, 2009; Peinado et al., 2012; Hoshino et al., 2015). Interestingly, HS has been shown to play a role in the syndecan-1 mediated formation of the syntenin-ALG2-interacting protein X (ALIX) complex (Baietti et al., 2012; Thompson et al., 2013; Roucourt et al., 2015). Following vesicular release, HSPGs also take part in exosome docking and delivery of vesicular cargo to the recipient cell. This dual role of HSPGs in exosome-mediated crosstalk between cells is fostered by fibronectin that interacts with HS displayed on the surface of exosomes and functions as a heparan sulfate/HS-binding ligand on target cells (Purushothaman et al., 2016; Colombo et al., 2019). Another study reported a correlation between the expression of glypican-1 on the exosomal surface and the tumor burden in pancreatic cancer patients (Melo et al., 2015), supporting a prognostic value of proteoglycans associated with exosomes in carcinogenesis.
At the metastatic site, proteoglycans also contribute by promoting cancer cell engraftment and colonization (Fares et al., 2020). The potential role of serglycin in metastatic dissemination has been investigated in a mouse model of breast cancer, where knockout of serglycin resulted in CTCs unable to establish metastatic tumors although not affecting primary tumor formation (Roy et al., 2016). Correspondingly, increased serglycin expression was shown to facilitate liver colonization by cancer cells in a patient-derived xenograft model of non-small-cell lung cancer (NSCLC) (Guo et al., 2017) as well as to promote hepatocellular carcinoma metastasis to the bone (He et al., 2014).
In summary, proteoglycans are connected to all steps of the metastatic cascade. Notably, some proteoglycans appear to play active roles in several aspects of cancer progression, highlighting these as potential key players of the cancer cell surface. One such proteoglycan is CD44, which is highly involved in EMT, helps to prevent anoikis due to its HA-receptor function, and furthermore takes actions in generating CTC clusters and extravasation, thereby enabling a successful arrival at the metastatic site (Figure 5). Another key proteoglycan seems to be CSPG4, with important roles for the regulation of cancer cell growth and invasion. Furthermore, the studies on CSPG4 presented here demonstrate how transmembrane proteoglycans possess multiple modes of action by engaging with other receptors or signaling molecules through either their cytoplasmic domain, ectodomains, or their GAG chains. With this central role in metastasis and CTC biology, proteoglycans could be an interesting target for CTC technologies. Indeed, proteoglycans are already studied and partly utilized for CTC identification and capture by different technologies. The following section will hence provide a detailed overview on proteoglycans as CTC targets.
Proteoglycans in Circulating Tumor Cell Diagnostics
Circulating tumor cell detection assays have spurred increasing clinical interest since the prognostic value in progression-free and overall survival was established in patients with metastatic colorectal (Cohen et al., 2009), breast (Cristofanilli et al., 2004), prostate (de Bono et al., 2008), and lung (Krebs et al., 2011) cancer. CTC enumeration from patient blood samples has also demonstrated clinical relevance for several other cancer types such as pancreatic cancer (Kurihara et al., 2008; Bidard et al., 2013; Effenberger et al., 2018) or hepatocellular carcinoma (Schulze et al., 2013; Qi et al., 2018). The presence of detectable levels of CTCs in the peripheral blood is associated with the metastatic capacity of the disease (Allard et al., 2004; Cristofanilli et al., 2004). However, low levels of CTCs have been reported in non-metastatic disease for several cancer indications before and after surgery (Thorsteinsson et al., 2011; Franken et al., 2012; Gazzaniga et al., 2013). Additional studies suggest that CTCs are even shed from premalignant lesions and this opens the possibility for using CTC detection for early diagnosis of cancer (Husemann et al., 2008; Stott et al., 2010; Rhim et al., 2012; Zhang et al., 2014; Tsai et al., 2016; Murlidhar et al., 2017). CTC assays might also have potential as a tool for predicting treatment efficacy and monitoring disease (Schochter et al., 2019; Yang et al., 2019), thereby providing real-time, non-invasive information about the disease by liquid biopsies. Furthermore, many CTC assays do not only enable enumeration but also downstream analyses such as genomic, transcriptomic, proteomic, or phenotypic characterization of cancer cells. Therefore, studying CTCs can also bring novel insight into aspects of metastasis formation, which are still not fully understood (Chaffer and Weinberg, 2011). Despite the interest and potential in analyzing CTCs, the methods are rarely implemented in the clinical setting, as CTC identification requires highly specific markers and an extreme assay sensitivity. Many CTC methods struggle to reach the needed sensitivity, as it is a technical challenge to detect few cancer cells in billions of normal blood cells.
Several CTC enrichment technologies ranging in complexity have been developed (Kowalik et al., 2017; Dianat-Moghadam et al., 2020). From whole blood, CTCs can be enriched along with leukocytes by density fractionation or a simple lysis of the erythrocytes. The crude cell enrichment can be analyzed by direct antibody staining and examined by, e.g., microscopy (Hillig et al., 2015; Werner et al., 2015) or flow cytometry (Hristozova et al., 2012; Watanabe et al., 2014; Lopresti et al., 2019). Because of the rarity of the CTCs, an additional cancer cell-specific enrichment step is, however, often preferred. CTCs can be enriched from the leukocytes based on distinct biophysical properties such as size, density, deformability, or charge (Liu et al., 2015; Mitchell et al., 2015; Shaw Bagnall et al., 2015). Following CTC enrichment, the detection of CTCs will still rely on a staining step, distinguishing for example cytokeratin (CK)-positive CTCs from the remaining CD45-positive leukocytes. Other systems for CTC isolation use cancer- or tissue-specific antibodies to enrich for CTCs (Coumans and Terstappen, 2015) or even leukocyte cell surface proteins like CD45 to deplete for leukocytes (Ozkumur et al., 2013; Karabacak et al., 2014). The positive selection of CTCs is evidently very dependent on highly specific cancer or tissue markers. In order to demonstrate high potential for clinical application, extensive validation of CTC capture platforms must reveal robust clinical sensitivity and specificity (Parkinson et al., 2012). Most pilot studies do not present large-scale clinical data and should hence be interpreted with caution. Inclusion of healthy controls becomes crucial to demonstrate the specificity of the capture and/or detection strategy. Alternatively, some studies apply downstream molecular analyses to verify the tumor origin of detected CTCs, e.g., by mutation detection, which supports the reliability of the CTC assay (Gasch et al., 2013; Muller et al., 2014). From a more technical perspective, pre-analytical conditions such as blood tubes, storage time, and temperature as well as choice of antibody clones can have a huge effect on assay performance (Qin et al., 2014; Ilie et al., 2018; Wu et al., 2020), making comparisons across studies difficult. Furthermore, in the light of exploring proteoglycans as potential CTC targets, the consideration of technical assay parameters become crucial for, e.g., sustaining the GAG stability.
The current clinical standard for CTC enumeration is the CellSearch® platform, which is approved by the American Food and Drug Administration (FDA) for monitoring patients with metastatic breast, colorectal, and prostate cancer. CellSearch® relies on cell enrichment using anti-EpCAM antibody-coated ferrofluid and CTC detection via fluorescent anti-CK antibody labeling (Liberti et al., 2001; Allard et al., 2004; Coumans and Terstappen, 2015). EpCAM-based capture approaches are, however, rarely efficient for epithelial cancers with downregulated EpCAM expression, likely due to EMT, or cancers of mesenchymal origin. Therefore, several studies have been focusing on finding novel markers, which can distinguish EpCAM-low or -negative CTCs from normal blood cells with high specificity and sensitivity to broaden the spectrum of detectable CTC subpopulations (Lampignano et al., 2017; Nicolazzo et al., 2019). As a part of this, multiple strategies using proteoglycans for CTC enrichment or identification are currently under investigation. See Table 1 for an overview on the most used proteoglycans and their applications. Some of them are highlighted in the following.
TABLE 1.
Proteoglycans used for isolation or identification of clinical circulating tumor cells.
| Indication | Used for | Finding | References | |
| Carbonic anhydrase 9 (CA9) | RCC | Immunomagnetic capture | Combined approach with CD147, captured CTCs in 94% of patients. CTC amount correlated with disease stage. | Liu et al. (2016) |
| CTC detection (On−chip Sort®) | CTCs were detected in 46% of patients (combined CA9 and EpCAM staining). | Naoe et al. (2019) | ||
| CD44 | BC/TNBC | CTC detection (flow cytometry) | NACT caused significant changes in the quantity of the CTC subsets present in patient blood samples. | Kaigorodova et al. (2018) |
| CTC detection (ns) | CTC clusters were associated with poor OS. CTC clusters showed higher CD44 expression. | Liu et al. (2019) | ||
| CRC | CTC detection (flow cytometry | CD133+CD54+CD44+ CTC subset was significantly associated with liver metastasis and had a prognostic value in CRC patients. | Fang et al. (2017) | |
| Gastric cancer | CTC detection (FACS) | The amount of EpCAM+CD44+ CTCs, but not EpCAM+CD44− CTCs, correlated with disease progression and venous invasion. | Watanabe et al. (2017) | |
| Negative immunomagnetic enrichment (anti-CD45) | CD44 was a marker for tumorigenic CTCs. | Toyoshima et al. (2015) | ||
| NSCLC | Immunomagnetic capture | CD44+ CTCs were associated with lymphatic invasion and tumor size. CD44+ CTCs were more sensitive to TRAIL-induced apoptosis. | Yan-Bin et al. (2020) | |
| OSCC | Immunomagnetic capture | CD44+ CTCs showed increased self-renewal capability and chemotherapy-resistance. Clinical correlation between increased CD44v6 and loco-regional aggressiveness and recurrence. | Patel et al. (2016) | |
| Chondroitin sulfate proteoglycan 4 (CSPG4) | Melanoma | Immunomagnetic capture | Analysis of RNA suggested that CSPG4+ CTCs were distinct from CTCs enriched by another melanoma marker, ABCB5. | Aya-Bonilla et al. (2019) |
| Immunomagnetic capture | Significant difference between CTC numbers in healthy controls, stage I/II and stage III/IV, using multiple markers. Decrease in CTC numbers during treatments was associated with longer OS and shorter response to treatment. | Freeman et al. (2012), Klinac et al. (2014) | ||
| Immunomagnetic capture (with subsequent depletion of CD45+ cells) | 1–20 CTCs found per 5 mL blood and BRAF genetic heterogeneity was detected among CTCs. | Sakaizawa et al. (2012) | ||
| Immunomagnetic capture | ≥2 CTCs correlated with OS for stage III and IV patients. CTC-positivity correlated with stage. | Ulmer et al. (2004) | ||
| Immunomagnetic capture and IF | MCAM/CSPG4-positive CTCs and RAS/RAF mutational status were associated. | Gorges et al. (2019) | ||
| CellSearch® Melanoma Kit | CTC positivity in early-stage disease correlated with OS at 24 months. | Anand et al. (2019) | ||
| Strong correlation between CTC positivity and PFS and OS; and between CTC numbers and ctDNA-levels. | Bidard et al. (2014) | |||
| Prospective study of stage IV patients. Baseline CTC-positivity independently predicted poorer PFS after 180 days. | Hall et al. (2018) | |||
| ≥2 CTCs at baseline was an independent prognostic factor for poor OS. | Khoja et al. (2013) | |||
| CellSearch® Melanoma Kit | Prospective study of 243 stage III patients. CTC-positivity at baseline independently predicted poorer relapse-free survival within 6 and 54 months. | Lucci et al. (2020) | ||
| Retrospective study; baseline ≥ 2 CTCs correlated with OS in stage III/IV patients. 95% of healthy subjects had no CTCs. | Rao et al. (2011) | |||
| Difference in CTC positivity for stage I/II vs. IV. Only 2.9% of healthy subject had CTCs. | Roland et al. (2015) | |||
| CTC detection (flow cytometry) | Early-stage CTCs expressed mainly one marker, late-stage CTCs expressed more. 42% of CTCs expressed CSPG4. | Gray et al. (2015) | ||
| CTC detection (flow cytometry) | CTCs were found in 14/22 patients. | Liu et al. (2011) | ||
| CTC detection (IF) | 5% of patients had ≥100 CTCs/mL. In these, unique clonal populations were identified. | Ruiz et al. (2015) | ||
| Characterization by Surface-enhanced Raman spectroscopy with αCSPG4 | CTC surface marker levels, including CSPG4, were altered during treatment. | Tsao et al. (2018) | ||
| C-X-C chemokine receptor type 4 (CXCR4) | HCC | CTC detection (RISH) | CTCs were detected in 89.9% of all patients and CXCR4 expression was associated with different CTC subsets. | Bai et al. (2020) |
| NSCLC | CTC detection (flow cytometry) | CXCR4 expression was increased on EpCAM- CTCs. | Yin et al. (2015) | |
| CTC detection (flow cytometry) | CXCR4+ CTCs showed potential as a predictive marker for OS in NSCLC patients. | Reckamp et al. (2009) | ||
| Glypican-3 (GPC3) | HCC | Immunomagnetic capture and CTC detection (flow cytometry) | High GPC3+ CTC amount correlated with shortened disease-free survival in non-metastatic HCC patients. | Hamaoka et al. (2019) |
| Immunomagnetic capture | Capture cocktail (together with anti-ASGPR, anti-EpCAM) found higher CTC numbers than each antibody alone. | Court et al. (2018) | ||
| CTC detection (ImageStream®) | 12.5% of all found CTCs were GPC3+. | Ogle et al. (2016) | ||
| HCC/CCA | CTC detection (IHC) | 1 out of 14 patients with CTCs had GPC3+ CTCs. | Nam et al. (2016) | |
| Syndican-1 (SDC1/CD138) | MM | Immunomagnetic capture | CD138+ CTCs strongly correlated with disease burden and treatment response. | Wang et al. (2019) |
| MM | Immunomagnetic capture | 20–184 CD138+ CTCs detected in patient blood/mL (2–5/mL healthy blood). Patients in remission had fewer CTCs than other patients. | Qasaimeh et al. (2017) |
+, positive; ABCB5, ATP-binding cassette sub-family B member 5; ASGPR, asialoglycoprotein receptor; BC, breast cancer; CA9, carbonic anhydrase 9; CCA, cholangiocarcinoma; CD, cluster of differentiation; CK, cytokeratin; CRC, colorectal cancer; CSPG4, chondroitin sulfate proteoglycan 4; CTC, circulating tumor cell; ctDNA, circulating tumor DNA; CXCR4, C-X-C chemokine receptor type 4; EpCAM, epithelial cell adhesion molecule; FACS, fluorescence-activated cell sorting; GPC3, glypican-3; HCC, hepatocellular carcinoma; IF, immunofluorescence; IHC, immunohistochemistry; ISET, isolation by size of epithelial tumor cells; MCAM, melanoma cell adhesion molecule; MM, multiple myeloma; NACT, neoadjuvant chemotherapy; ns, not specified; OSCC, oral squamous cell carcinoma; OS, overall survival; PFS, progression-free survival; RBC, red blood cell; RCC, renal cell carcinoma; RISH: RNA in situ hybridization; SDC1, syndican-1; TNBC, triple-negative breast cancer; TRAIL, TNF-related apoptosis-inducing ligand.
A well-known example is the CellSearch® Circulating Endothelial Cell Kit, which can be used for the enrichment of circulating melanoma cells that are EpCAM-negative by nature (Rao et al., 2011; Khoja et al., 2013). After capture using ferrofluid coupled with antibodies against melanoma cell adhesion molecule (MCAM), circulating melanoma cells are identified by staining with antibodies against high molecular weight melanoma-associated antigen (HMW-MAA), also known as CSPG4.
As described earlier, CSPG4 has been linked to many aspects of the metastatic cascade, including proliferation, migration, as well as ECM-remodeling and is expressed across many cancer types (Ilieva et al., 2018). Moreover, CSPG4 is expressed in a majority of melanoma lesions (Real et al., 1985) and is a well-characterized surface marker for melanoma (Ilieva et al., 2018). Multiple retrospective studies using the MCAM/CSPG4 CellSearch® Kit have found that CTC levels detected at baseline correlates with overall survival in late-stage melanoma (Rao et al., 2011; Khoja et al., 2013; Bidard et al., 2014) (Table 1). Recently, two large prospective studies also evaluated the prognostic significance of MCAM/CSPG4-positive CTCs in cutaneous melanoma. In a study of 93 stage IV patients, Hall et al. (2018) found that presence of CTCs at baseline was associated with shorter progression-free survival after 6 months compared to CTC-negative patients. Later, the same research group showed that CTC-positivity at baseline for stage III patients (n = 243) was an independent predictor of relapse-free survival within 6 and 54 months (Lucci et al., 2020). The CTC levels in these studies were not associated with primary tumor characteristics, such as ulcerations, tumor thickness, and mutational status. Therefore, MCAM/CSPG4-positive CTC numbers may add additional information on top of clinicopathological characteristics for clinicians to foresee the disease course in the future.
Interestingly, CTC-negative melanoma patients have been found to have better progression-free or relapse-free survival compared to CTC-positive patients (Hall et al., 2018; Lucci et al., 2020). However, a significant proportion of late-stage melanoma patients still appear to have no CTCs detectable by CellSearch® (Rao et al., 2011; Roland et al., 2015; Hall et al., 2018). This has also been reported for other CSPG4-based methods (Ulmer et al., 2004; Ruiz et al., 2015) as well as for CSPG4-independent isolation methods (Khoja et al., 2014; Aya-Bonilla et al., 2019). This may simply be due to the rare nature of CTCs. However, there could be CSPG4-negative CTC subpopulations, which are not captured by CSPG4-dependent strategies. In fact, one study found that of 31 melanoma patients with CTCs detectable by other markers, only 42% had CSPG4-positive CTCs (Gray et al., 2015), suggesting a need for multi-marker approaches.
During the past decade, multiple other studies investigated the potential of CSPG4 for CTC capture and/or identification in melanoma (Table 1). Up to 4 CTCs per mL blood was found using CSPG4 immunomagnetic capture (Sakaizawa et al., 2012), which is similar to the reported CTC numbers using the MCAM/CSPG4 CellSearch® kit (Khoja et al., 2013). However, a study by Ruiz et al. (2015) using a CSPG4-based immunofluorescent microscopy approach without prior enrichment step identified a mean of 14.9 CTCs per 1 mL blood samples from melanoma patients (n = 40), without potential CTC hits in healthy control samples (n = 10). These variations in CSPG4-positive CTCs can be explained to some extent by the use of different CTC enrichment strategies, varying markers for CTC identification or other technical differences in the assays.
Commonly mutated genes in melanoma, such as BRAF and NRAS (Colombino et al., 2012), are upstream activators of ERK signaling (Savoia et al., 2019). As CSPG4 expression has been connected to ERK signaling (Ampofo et al., 2017), these mutations might be particularly important in CSPG4-positive CTCs. Indeed, a recent study found that CTCs enriched by CSPG4-based method presented more RAS/RAF mutated cells compared to CTCs isolated only by physical properties (Gorges et al., 2019). Since some therapeutic approaches target the serine/threonine-protein kinase B-raf (BRAF) (Holderfield et al., 2014), it is possible that the CSPG4 expression might also decrease in response to this form of treatment, which could affect the prospect of using CSPG4 alone to monitor CTC numbers. Indeed, initial longitudinal study of CTC heterogeneity in 10 stage IV melanoma patients suggested that expression of CSPG4 on CTCs may be downregulated in response to BRAF and mitogen-activated protein kinase kinase (MEK)-inhibiting therapy (Tsao et al., 2018). However, to our knowledge, none of the major studies on CSPG4-positive CTCs in cutaneous melanoma have yet found any correlation between CTC levels and BRAF-mutational status or adjuvant therapy.
Another recent study revealed that the transcriptomic profile of CSPG4-enriched CTC populations from six patients was dominated by up-regulation of tumor necrosis factor alpha (TNFα)/nuclear factor kappa B (NF-κB) as well as signal transducer and activator of transcription (STAT) pathways (Aya-Bonilla et al., 2019). Both signaling pathways have central roles for cell proliferation as well as cell survival (Wu and Zhou, 2010; Igelmann et al., 2019). Furthermore, in silico analysis found other genes upregulated in the CSPG4-enriched population to be connected to metastasis, tumor growth, and melanoma biology (Aya-Bonilla et al., 2019), which indicates an interesting biological role of CSPG4-positive CTCs for melanoma progression.
Overall, CSPG4 is a relevant surface marker for melanoma CTCs and is hence evaluated in many studies. Although little is still known about the biological role of CSPG4-positive CTCs, they might represent a specific subpopulation. This potential bias should be considered when using only CSPG4 for CTC capture or CTC identification.
CD44 has also been explored for CTC analysis. CD44 is widely expressed (Goodison et al., 1999), and as previously described it acts as a receptor for a variety of ligands. Particularly well-described is the interaction with HA, which constitutes a major part of the glycocalyx and ECM (Banerjee et al., 2016). Upregulation of CD44 confers tumorigenicity, metastatic capacity, and drug resistance to primary tumor cells as well as CTCs (Naor et al., 2002; Fitzgerald and McCubrey, 2014). The abnormal expression of CD44 splice variants is associated with treatment refractoriness, recurrence, and prognosis (Katoh et al., 2015), and overexpression of both CD44s and variants serves a long list of biological functions across many cancer types (Chen C. et al., 2018). Since isoform switching introduces new cancer-related antigens, development of both anti-CD44s and anti-CD44v antibodies has attracted much interest.
As a CTC isolation tool, anti-CD44 antibodies have been used to capture CTCs from cancer patient blood (see Table 1). Yan-Bin et al. (2020) investigated the CD44-positive CTC abundance in NSCLC patients by immunomagnetic enrichment and evaluated the correlation to clinical characteristics. None or very few CD44-positive cells were detected in the 30 controls in contrast to frequent CTC observations in the 128 patient samples. Detected CTCs associated negatively with serum TNF-related apoptosis-inducing ligand (TRAIL) levels, suggesting that CD44-positive CTCs could be more vulnerable to TRAIL-induced apoptosis through death receptor 4 and 5 signaling (Yan-Bin et al., 2020). A small study on gastric cancer patients (n = 26) and healthy controls (n = 10) associated increased prevalence of EpCAM- and CD44-positive CTCs in patients with tumor depth, disease progression, and venous invasion (Watanabe et al., 2017). Consequently, CD44-based CTC detection was suggested to reflect the malignant potential of the tumor. The authors, however, disregard EpCAM-positive cells found in all healthy controls and the few double positive cells found in 2 healthy controls as either non-specific immunological reactions or contaminating skin cells. Again, this discrepancy highlights the demand for CTC validation, as for example via genomics. Another study analyzed CD44-positive CTCs isolated by immunomagnetic enrichment from 30 oral squamous cell carcinoma (OSCC) patients and 15 healthy controls (Patel et al., 2016). Self-renewal and proliferation capability of the CD44-positive cells were observed by increased sphere-forming capacity unlike the CD44-negative sorted population. Moreover, cisplatin resistance assays confirmed a drug-resistant phenotype associated with the CD44-positive population. This was specifically associated with high transcript levels of CD44v6, as opposed to CD44s, as well as elevated levels of the stemness marker NANOG. Furthermore, the different expression levels strongly correlated with the primary tumor profile and, importantly, clinicopathological parameters such as late-stage, loco-regional aggressiveness, and relapse. The findings suggest that detection of CD44v6-positive CTCs could be used to predict disease progression, therapy outcome, and recurrence.
In addition, CD44 is being evaluated for novel therapeutic approaches against CTCs. For instance, in vivo homophilic CD44-mediated CTC clustering of metastatic breast cancer cells in mice was largely inhibited by the administration of anti-CD44 neutralizing antibody, leading to decreased metastatic capacity (Liu et al., 2019). In summary, numerous studies of applications to target CD44-positive CTCs underline its potential in therapy and as a valuable marker for prognosis and treatment response.
Another interesting proteoglycan for clinical purposes is glypican-3 (GPC3), which is upregulated amongst several cancer entities with highest positive case rates in hepatocellular carcinoma (HCC). Importantly, GPC3 has been reported to discriminate between HCC and non-malignant lesions (Zhu et al., 2001; Wang et al., 2008) or other liver-associated cancers like cholangiocarcinoma (CCA) (Man et al., 2005). Nowadays GPC3 is included in a diagnostic HCC panel together with glutamine synthetase and heat shock protein 70, according to guidelines of the European Society for Medical Oncology (ESMO) (Vogel et al., 2019) and the American Association for the Study of Liver Diseases (AASLD) (Marrero et al., 2018). Furthermore, GPC3 might also be used as a serum biomarker (Capurro et al., 2003; Hippo et al., 2004) and is exploited for different targeted cancer therapy approaches (Sawada et al., 2012; Feng et al., 2013).
Several studies have been utilizing GPC3 for analysis of CTCs in HCC patients (Table 1). Anti-GPC3 antibodies have been used for positive immunomagnetic enrichment of CTCs (Court et al., 2018; Hamaoka et al., 2019). Hamaoka et al. (2019) found in a prospective, single-institution study that most of the 85 examined HCC patients had GPC3-positive CTCs with a median of 3 CTCs in 8 mL blood samples, whereas negative controls (in total n = 27) such as healthy individuals (n = 12) or individuals with inflammatory diseases (n = 4), only had a median of 1 GPC3-positive cell in the blood samples (Hamaoka et al., 2019). Moreover, patients with 5 or more CTCs showed shorter disease-free survival compared to patients with fewer GPC3-positive CTCs. Another study by Court et al. (2018) analyzed GPC3 in a capture cocktail together with antibodies against asialoglycoprotein receptor (ASGPR) and EpCAM. Importantly, the combined capture approach with all three targets, isolated higher CTC numbers in patients than each antibody alone. This approach detected CTCs in 96.7% of all HCC patients (n = 61) with a median of 6 CTCs per 4 mL blood. In contrast, in healthy controls (n = 8) maximum one potential CTC hit was found. Moreover, CTC numbers were increased in more advanced stages compared to early stages. This effect was even more pronounced for the subfraction of vimentin-positive CTCs, which presumably are generated by EMT processes. This highlights the importance of CTC capture strategies independent of potential EMT target proteins like EpCAM, which are often downregulated during EMT. Indeed, varying or low EpCAM expression has already been reported for CTCs originating from other cancer entities (Hyun et al., 2016; de Wit et al., 2018) and should be taken into consideration when designing or interpreting CTC capture assays.
In summary, GPC3 is currently evaluated as a therapeutic target, serum biomarker, and importantly for CTC analyses, where it has been used both for capture (Court et al., 2018; Hamaoka et al., 2019) and identification (Ogle et al., 2016) of CTCs in HCC. Since only few studies have been performed, further studies are needed to prove the feasibility of GPC3 for clinical CTC analyses. Although GPC3 is a well-established diagnostic marker for HCC, further characterization or validation of the potential GPC3-positive CTC hits, for example via molecular analyses, is to our knowledge missing so far.
As described, there is a great diversity among the proteoglycans associated with different cancer types. The different proteoglycans facilitate distinctive processes in the metastatic cascade and their universal expression suggests that proteoglycans are an essential feature for all cancers. The complexity is further expanded when considering the GAG composition. An increasing number of studies indicate that at least some of the functions of proteoglycans are exerted through specific GAG chains. However, only a few studies focused on targeting the GAG part or GAG composition of proteoglycans when isolating CTCs.
A wide variety of qualitative and quantitative methods has been developed for studying glycocalyx components. As the biosynthesis of glycans is non-template driven and complex, their analysis may often be challenging. Several approaches take advantage of the large repertoire of glycan-binding proteins and antibodies to distinguish between different glycan classes. For large screenings, glycan microarrays have been developed that may probe for different glycan classes or subclasses. This approach has been used to screen breast CTCs for glycan markers, which identified a specific O-glycan epitope as a potential target (Wang et al., 2015). Microarrays and cell-based libraries have also been developed to screen for GAG-binding proteins (Rogers and Hsieh-Wilson, 2012) and these may be useful for identifying GAG-based CTC targeting reagents. However, microarrays for detection of cell-surface GAGs, which could be useful for identifying GAG biomarkers on CTCs, have not been constructed. CS and HS-specific antibodies, such as CS56 and 10E4, and GAG-binding proteins, like fibroblast growth factor, are also commonly used in flow cytometry and microscopy-based assays (Figure 6A). These may assess the relative levels of GAGs, however, they do not convey specific structural information due to their low specificity or affinity toward their targets (Yamagata et al., 1987; Smetsers et al., 2004; ten Dam et al., 2007). For this, GAGs will have to be isolated and analyzed, often by chromatography, mass spectrometry, or nuclear magnetic resonance. The structural characterization of GAGs is challenging due to heterogeneity of the polymers. Hence, analysis is often limited to disaccharide analysis, which does not allow for sequencing of intact GAG chains. This is even more technical challenging for CTCs because of the limited input material due to their low abundance. Similarly, while different proteoglycan core proteins can be probed with antibodies, fine structural analysis of their GAG attachment sites is only achieved by glycoproteomic methods. While these analyses may be laborious, they are highly descriptive and may provide novel insight into structural alterations on cancer cells, both on the protein and GAG level. For example, one study found that several major ECM proteoglycans had elevated levels of N-glycosylation in pancreatic cancer tissues (Pan et al., 2014). In addition, another study identified novel CS linkage region modifications in CS glycopeptides from the inter-α-trypsin inhibitor complex, which is abundant in plasma from cancer patients (Gomez Toledo et al., 2015). To our knowledge, glycoproteomics has not been used for analysis of CTCs, and could potentially help identify novel targets.
FIGURE 6.
Utilization of glycosaminoglycans for capture of circulating tumor cells (CTCs). (A) Glycosaminoglycans can be directly targeted as for example via antibodies like 10E4, which binds to heparan sulfate (HS; in red) of heparan sulfate proteoglycans (HSPGs). However, to our knowledge, this approach has not been explored for CTC capture. Furthermore, for CTC capture the recombinant protein VAR2CSA (rVAR2) can be used, which binds to oncofetal chondroitin sulfate (in yellow) as for example identified on chondroitin sulfate proteoglycan 4 (CSPG4). (B) Glycosaminoglycans have been applied in the reversed approach as capture agent for CTC enrichment. Here, glycosaminoglycan-based probes were used to capture CTCs. For example, a microfluidic chip has been coated with hyaluronic acid (HA; in green) to capture CTCs via its interaction with the HA-receptor CD44. Similarly, the heparan sulfate-based probe SCH45 has been coupled to magnetic beads to capture CTCs in hepatocellular carcinoma in a microfluidic setup, but the exact cellular target of SCH45 in these CTCs remains unknown. Generally, both strategies are relatively new for CTC capture and clearly further extensive validation is needed. Please refer to the main text for details and references.
If succeeding in finding specific binding moieties, the GAG chains would be an alternative novel approach for CTC enrichment or detection. We have previously shown the use of the recombinant VAR2CSA malaria protein (rVAR2) as a novel CTC-targeting reagent (Figures 6A, 7) (Agerbaek et al., 2018; Bang-Christensen et al., 2019; Sand et al., 2020). rVAR2 binds to a distinct type of CS, termed oncofetal CS, expressed by placental as well as cancer cells (Salanti et al., 2015). The native VAR2CSA binds to a specific CS oligosaccharide motif in the placenta during normal physiological conditions (Gowda, 2006; Ayres Pereira et al., 2016; Toledo et al., 2020). A study using a library of cells with knockouts of GAG biosynthesis genes, indicated that 4-O-sulfated CS is essential for rVAR2 binding (Chen Y. H. et al., 2018). The specific oncofetal CS-carrying proteoglycans have been examined by screening of rVAR2 binding to more than 3500 cell surface proteins (Salanti et al., 2015) as well as by rVAR2-affinity chromatography coupled to glycoproteomics, using tumor and placenta samples (Toledo et al., 2020). These studies showed that the distinct oncofetal CS is displayed on multiple proteoglycans such as CSPG4 or CD44 in cancer cells, indicating an important function of oncofetal CS in the disease development. Moreover, rVAR2 binds to cancer cells independently of tumor origin and oncofetal CS is expressed both in primary and metastatic lesions (Salanti et al., 2015). This has also been shown in a metastatic murine model, where rVAR2 binding furthermore inhibited integrin signaling and seeding of CTCs (Clausen et al., 2016). As studies have also indicated that rVAR2 binds to cancer cells independent of EMT processes (Agerbaek et al., 2018; Bang-Christensen et al., 2019), oncofetal CS could be an advantageous target for CTC enrichment. In line with this, rVAR2-coated paramagnetic beads have been used to capture CTCs from blood samples of different carcinoma patients (n = 44) and glioma patients (n = 10) in small proof-of concept studies (Agerbaek et al., 2018; Bang-Christensen et al., 2019). Therefore, the rVAR2-based approach offers an alternative capture approach, demonstrating how GAG-targeting can allow the capture of CTCs independently of single target proteins, like EpCAM.
FIGURE 7.
Recombinant VAR2CSA (rVAR2) can be used for staining and capture of potential circulating tumor cells (CTCs). (A) Immunofluorescence staining of the colorectal cancer cells COLO205 (marked with cross) with rVAR2 (green), anti-CD45 (in red) to mark normal blood cells, and DAPI (in blue) to mark cell nuclei. (B) One potential CTC hit (green by rVAR2 stain; marked with white asterisk) in a blood sample from a colorectal cancer patient with the same staining as described in panel A. (C) Magnetic beads coated with rVAR2 bind specifically to COLO205 cells, compared to non-rVAR2 control beads. Pictures were kindly provided by Mette Ø. Agerbæk, Amalie M. Jørgensen, and Nicolai T. Sand.
Actually, the reversed approach can also be utilized for CTC capture (Figure 6B). For example, Gopinathan et al. (2020) coated a synthetic HS-based octasaccharide probe (SCH45) onto magnetic beads, that were used in combination with a microfluidic chip to isolate CTCs from 65 advanced or metastatic cholangiocarcinoma patients. Single CTCs or CTC clusters were detected in all samples with ≥ 1 CTCs per 1 mL blood, even in patients with no distant metastases. Previous comparable CellSearch®-based studies found only 17% of CCA patients positive for ≥2 CTCs per 7.5 mL blood (Yang et al., 2016). However, only three healthy controls were included in the HS-based study. Moreover, the authors reported that studies evaluating whether this approach could be employed to capture EpCAM-negative CTCs are currently ongoing (Gopinathan et al., 2020). In addition it would be interesting to identify the binding target of the SCH45-coated beads in order to characterize the captured CTC population.
Another approach exploited the GAG-receptor function of CD44 in order to capture CTCs (Wang M. et al., 2018). Purified HA, the ligand of CD44, was coated to a microfluidic chip and showed 91% retrieval of CD44-overexpressing A549 cells spiked into blood. Also other cancer cell lines from different cancer entities were captured with comparable efficiencies. Although also lacking healthy controls, the study found between 1–18 putative CTCs per 1 mL blood from 9 of 10 NSCLC and 5 of 5 breast cancer patients as detected through CK- and DAPI-staining.
The utilization of GAGs for CTC technologies is a relatively new approach. Most studies have been limited to smaller pilot studies so far and further molecular characterization of the putative CTC hits is needed to prove their cancer-origin and thus the reliability of the CTC assay. Clearly, the establishment of specific GAGs as biomarkers for clinical CTC diagnostics needs extensive validation in large-scale studies in the future. However, GAGs have the potential to capture or identify broader and more heterogenous CTC populations as they are often independent of a single protein and thus might be less prone to gene expression changes associated with different or transient cancer cell phenotypes.
Concluding Remarks
Circulating tumor cell analyses have the potential to allow prognostic and predictive insights by convenient liquid biopsies. However, novel biomarkers are needed to enable the necessary assay sensitivity and specificity to detect CTCs. Another unsolved problem is that most CTC assays introduce biases in regards to which CTC subpopulations can be captured as they are often based on single biomarkers. Therefore, a CTC capture approach based on a combination of several biomarkers could be beneficial. Another solution for this problem could be the targeting of cancer-specific changes in the GAGs (the “GAGome”), or other known glycocalyx components, which should in principle, allow the capture of more heterogenous CTC populations. Studies on clinical CTCs and their proteoglycans, GAGs, or general glycocalyx structure are still not strongly represented, probably due to associated technical challenges of glycocalyx characterizations. However, structural insights would be beneficial for improving or defining novel CTC capturing strategies based on proteoglycans or their GAGs and to explore whether these strategies then better reflect the heterogenic cancer cell population.
Author Contributions
TA, SB-C, AJ, CL, CS, NS, and MA conceived and drafted the manuscript. TA designed the figures with scientific input from co-authors. All authors critically revised the manuscript.
Conflict of Interest
TC, AS, and MA are shareholders in VAR2 Pharmaceuticals holding the intellectual property rights to use rVAR2 for binding oncofetal chondroitin sulfate and to capture circulating tumor cells. VarCT Diagnostics is a subsidiary to VAR2 Pharmaceuticals.
The remaining authors declare that the research was conducted in the absence of any other commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to anyone whose work was not presented in this review due to space requirements. Schematic figures were created with BioRender.com. Figure 2A was reused from Okada et al. (2017), with slight modifications to remove the original panel labeling, under CC BY 4.0 license (https://creativecommons.org/licenses/by/4.0/).
Footnotes
Funding. This research was funded by Independent Research Fund Denmark (Grant Number 8020-00446A), Innovation Fund Denmark (Grant Numbers 7038-00212B and 9090-00024B), HARBOEFONDEN, and Svend Andersen Fonden. TA was supported by the Bridge Translational Excellence Programme, funded by the Novo Nordisk Foundation (Grant Number NNF18SA0034956).
References
- Abrink M., Grujic M., Pejler G. (2004). Serglycin is essential for maturation of mast cell secretory granule. J. Biol. Chem. 279 40897–40905. 10.1074/jbc.M405856200 [DOI] [PubMed] [Google Scholar]
- Aceto N., Bardia A., Miyamoto D. T., Donaldson M. C., Wittner B. S., Spencer J. A., et al. (2014). Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158 1110–1122. 10.1016/j.cell.2014.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agerbaek M. O., Bang-Christensen S. R., Yang M. H., Clausen T. M., Pereira M. A., Sharma S., et al. (2018). The VAR2CSA malaria protein efficiently retrieves circulating tumor cells in an EpCAM-independent manner. Nat. Commun. 9:3279. 10.1038/s41467-018-05793-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai X., Do A. T., Lozynska O., Kusche-Gullberg M., Lindahl U., Emerson C. P. (2003). QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J. Cell Biol. 162 341–351. 10.1083/jcb.200212083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aird W. C. (2007). Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ. Res. 100 158–173. 10.1161/01.RES.0000255691.76142.4a [DOI] [PubMed] [Google Scholar]
- Akhurst R. J., Derynck R. (2001). TGF-beta signaling in cancer–a double-edged sword. Trends Cell Biol. 11 S44–S51. 10.1016/s0962-8924(01)02130-4 [DOI] [PubMed] [Google Scholar]
- Alix-Panabieres C., Mader S., Pantel K. (2017). Epithelial-mesenchymal plasticity in circulating tumor cells. J. Mol. Med. 95 133–142. 10.1007/978-3-030-35805-1_2 [DOI] [PubMed] [Google Scholar]
- Allard W. J., Matera J., Miller M. C., Repollet M., Connelly M. C., Rao C., et al. (2004). Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 10 6897–6904. 10.1158/1078-0432.CCR-04-0378 [DOI] [PubMed] [Google Scholar]
- Ampofo E., Schmitt B. M., Menger M. D., Laschke M. W. (2017). The regulatory mechanisms of NG2/CSPG4 expression. Cell. Mol. Biol. Lett. 22:4. 10.1186/s11658-017-0035-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K., Roszik J., Gombos D., Upshaw J., Sarli V., Meas S., et al. (2019). Pilot study of circulating tumor cells in early-stage and metastatic Uveal Melanoma. Cancers 11:856. 10.3390/cancers11060856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrlova H., Mastroianni J., Madl J., Kern J. S., Melchinger W., Dierbach H., et al. (2017). Biglycan expression in the melanoma microenvironment promotes invasiveness via increased tissue stiffness inducing integrin-beta1 expression. Oncotarget 8 42901–42916. 10.18632/oncotarget.17160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya-Bonilla C., Gray E. S., Manikandan J., Freeman J. B., Zaenker P., Reid A. L., et al. (2019). Immunomagnetic-Enriched Subpopulations of Melanoma Circulating Tumour Cells (CTCs) exhibit distinct transcriptome profiles. Cancers 11:157. 10.3390/cancers11020157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres Pereira M., Mandel Clausen T., Pehrson C., Mao Y., Resende M., Daugaard M., et al. (2016). Placental sequestration of plasmodium falciparum malaria parasites is mediated by the interaction between VAR2CSA and chondroitin sulfate A on Syndecan-1. PLoS Pathog. 12:e1005831. 10.1371/journal.ppat.1005831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccelli I., Schneeweiss A., Riethdorf S., Stenzinger A., Schillert A., Vogel V., et al. (2013). Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 31 539–544. 10.1038/nbt.2576 [DOI] [PubMed] [Google Scholar]
- Bai T., Mai R. Y., Ye J. Z., Chen J., Qi L. N., Tang J., et al. (2020). Circulating tumor cells and CXCR4 in the prognosis of hepatocellular carcinoma. Transl. Cancer Res. 9 1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti M. F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., et al. (2012). Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14 677–685. 10.1038/ncb2502 [DOI] [PubMed] [Google Scholar]
- Banerjee S., Modi S., Mcginn O., Zhao X., Dudeja V., Ramakrishnan S., et al. (2016). Impaired synthesis of stromal components in response to minnelide improves vascular function, drug delivery, and survival in pancreatic cancer. Clin. Cancer Res. 22 415–425. 10.1158/1078-0432.CCR-15-1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang-Christensen S. R., Pedersen R. S., Pereira M. A., Clausen T. M., Loppke C., Sand N. T., et al. (2019). Capture and detection of circulating glioma cells using the recombinant VAR2CSA malaria protein. Cells 8:998. 10.3390/cells8090998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J. M., Kaushik S., Bainer R. O., Sa J. K., Woods E. C., Kai F., et al. (2018). A tension-mediated glycocalyx-integrin feedback loop promotes mesenchymal-like glioblastoma. Nat. Cell Biol. 20 1203–1214. 10.1038/s41556-018-0183-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett K. L., Jackson D. G., Simon J. C., Tanczos E., Peach R., Modrell B., et al. (1995). CD44 isoforms containing exon V3 are responsible for the presentation of heparin-binding growth factor. J. Cell Biol. 128 687–698. 10.1083/jcb.128.4.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Tong C., Dockendorff A., Bancroft L., Gallagher L., Guzman G., et al. (2008). Genetic deficiency of decorin causes intestinal tumor formation through disruption of intestinal cell maturation. Carcinogenesis 29 1435–1440. 10.1093/carcin/bgn141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidard F. C., Huguet F., Louvet C., Mineur L., Bouche O., Chibaudel B., et al. (2013). Circulating tumor cells in locally advanced pancreatic adenocarcinoma: the ancillary CirCe 07 study to the LAP 07 trial. Ann. Oncol. 24 2057–2061. 10.1093/annonc/mdt176 [DOI] [PubMed] [Google Scholar]
- Bidard F. C., Madic J., Mariani P., Piperno-Neumann S., Rampanou A., Servois V., et al. (2014). Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int. J. Cancer 134 1207–1213. 10.1002/ijc.28436 [DOI] [PubMed] [Google Scholar]
- Biroccio A., Cherfils-Vicini J., Augereau A., Pinte S., Bauwens S., Ye J., et al. (2013). TRF2 inhibits a cell-extrinsic pathway through which natural killer cells eliminate cancer cells. Nat. Cell Biol. 15 818–828. 10.1038/ncb2774 [DOI] [PubMed] [Google Scholar]
- Bockhorn M., Jain R. K., Munn L. L. (2007). Active versus passive mechanisms in metastasis: do cancer cells crawl into vessels, or are they pushed? Lancet Oncol. 8 444–448. 10.1016/S1470-2045(07)70140-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunt L., Scholpp S. (2018). The function of endocytosis in Wnt signaling. Cell. Mol. Life Sci. 75 785–795. 10.1007/s00018-017-2654-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budczies J., Von Winterfeld M., Klauschen F., Bockmayr M., Lennerz J. K., Denkert C., et al. (2015). The landscape of metastatic progression patterns across major human cancers. Oncotarget 6 570–583. 10.18632/oncotarget.2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffone A., Weaver V. M. (2020). Don’t sugarcoat it: how glycocalyx composition influences cancer progression. J. Cell Biol. 219:e201910070. 10.1083/jcb.201910070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulow H. E., Hobert O. (2006). The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 22 375–407. 10.1146/annurev.cellbio.22.010605.093433 [DOI] [PubMed] [Google Scholar]
- Burdick M. M., Henson K. A., Delgadillo L. F., Choi Y. E., Goetz D. J., Tees D. F. J., et al. (2012). Expression of E-selectin ligands on circulating tumor cells: Cross-regulation with cancer stem cell regulatory pathways? Front. Oncol. 2:103. 10.3389/fonc.2012.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler P. J., Bhatnagar A. (2019). Mechanobiology of the abluminal glycocalyx. Biorheology 56 101–112. [DOI] [PubMed] [Google Scholar]
- Cackowski F. C., Xu L., Hu B., Cheng S. Y. (2004). Identification of two novel alternatively spliced Neuropilin-1 isoforms. Genomics 84 82–94. 10.1016/j.ygeno.2004.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurro M., Martin T., Shi W., Filmus J. (2014). Glypican-3 binds to Frizzled and plays a direct role in the stimulation of canonical Wnt signaling. J. Cell Sci. 127 1565–1575. 10.1242/jcs.140871 [DOI] [PubMed] [Google Scholar]
- Capurro M., Wanless I. R., Sherman M., Deboer G., Shi W., Miyoshi E., et al. (2003). Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology 125 89–97. 10.1016/s0016-5085(03)00689-9 [DOI] [PubMed] [Google Scholar]
- Capurro M. I., Xiang Y. Y., Lobe C., Filmus J. (2005). Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 65 6245–6254. 10.1158/0008-5472.CAN-04-4244 [DOI] [PubMed] [Google Scholar]
- Carneiro B. R., Pernambuco Filho P. C., Mesquita A. P., Da Silva D. S., Pinhal M. A., Nader H. B., et al. (2014). Acquisition of anoikis resistance up-regulates syndecan-4 expression in endothelial cells. PLoS One 9:e116001. 10.1371/journal.pone.0116001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassinelli G., Lanzi C. (2020). Heparanase: a potential therapeutic target in sarcomas. Adv. Exp. Med. Biol. 1221 405–431. 10.1007/978-3-030-34521-1_15 [DOI] [PubMed] [Google Scholar]
- Cavallaro U., Christofori G. (2001). Cell adhesion in tumor invasion and metastasis: loss of the glue is not enough. Biochim. Biophys. Acta 1552 39–45. 10.1016/s0304-419x(01)00038-5 [DOI] [PubMed] [Google Scholar]
- Chaffer C. L., Weinberg R. A. (2011). A perspective on cancer cell metastasis. Science 331 1559–1564. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran E. V., Davidson E. A. (1979). Glycosaminoglycans of normal and malignant cultured human mammary cells. Cancer Res. 39 870–880. [PubMed] [Google Scholar]
- Cheifetz S., Andres J. L., Massague J. (1988). The transforming growth factor-beta receptor type III is a membrane proteoglycan. Domain structure of the receptor. J. Biol. Chem. 263 16984–16991. [PubMed] [Google Scholar]
- Chekenya M., Krakstad C., Svendsen A., Netland I. A., Staalesen V., Tysnes B. B., et al. (2008). The progenitor cell marker NG2/MPG promotes chemoresistance by activation of integrin-dependent PI3K/Akt signaling. Oncogene 27 5182–5194. 10.1038/onc.2008.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Zhao S., Karnad A., Freeman J. W. (2018). The biology and role of CD44 in cancer progression: therapeutic implications. J. Hematol. Oncol. 11:64. 10.1186/s13045-018-0605-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. L., Lander A. D. (2001). Mechanisms underlying preferential assembly of heparan sulfate on glypican-1. J. Biol. Chem. 276 7507–7517. 10.1074/jbc.M008283200 [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Narimatsu Y., Clausen T. M., Gomes C., Karlsson R., Steentoft C., et al. (2018). The GAGOme: a cell-based library of displayed glycosaminoglycans. Nat. Methods 15 881–888. 10.1038/s41592-018-0086-z [DOI] [PubMed] [Google Scholar]
- Cherfils-Vicini J., Iltis C., Cervera L., Pisano S., Croce O., Sadouni N., et al. (2019). Cancer cells induce immune escape via glycocalyx changes controlled by the telomeric protein TRF2. EMBO J. 38:e100012. 10.15252/embj.2018100012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K. J., Padmanaban V., Silvestri V., Schipper K., Cohen J. D., Fairchild A. N., et al. (2016). Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. U.S.A. 113 E854–E863. 10.1073/pnas.1508541113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodelli P., Bugatti A., Urbinati C., Rusnati M. (2015). Heparin/Heparan sulfate proteoglycans glycomic interactome in angiogenesis: biological implications and therapeutical use. Molecules 20 6342–6388. 10.3390/molecules20046342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichy J., Puré E. (2003). The liberation of CD44. J. Cell Biol. 161 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B., Koontz C., Frisch S. M. (2015). CD44S-hyaluronan interactions protect cells resulting from EMT against anoikis. Matrix Biol. 48 55–65. 10.1016/j.matbio.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. A. F., Lin F., Greiling D., An J., Couchman J. R. (2004). Fibroblast invasive migration into fibronectin/fibrin gels requires a previously uncharacterized dermatan sulfate-CD44 proteoglycan. J. Invest. Dermatol. 122 266–277. 10.1046/j.0022-202X.2004.22205.x [DOI] [PubMed] [Google Scholar]
- Clausen T. M., Pereira M. A., Al Nakouzi N., Oo H. Z., Agerbaek M. O., Lee S., et al. (2016). Oncofetal chondroitin sulfate glycosaminoglycans are key players in integrin signaling and tumor cell motility. Mol. Cancer Res. 14 1288–1299. 10.1158/1541-7786.MCR-16-0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. J., Punt C. J., Iannotti N., Saidman B. H., Sabbath K. D., Gabrail N. Y., et al. (2009). Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol. 20 1223–1229. 10.1016/j.yexmp.2018.12.006 [DOI] [PubMed] [Google Scholar]
- Cole C. L., Rushton G., Jayson G. C., Avizienyte E. (2014). Ovarian cancer cell heparan sulfate 6-O-sulfotransferases regulate an angiogenic program induced by heparin-binding epidermal growth factor (EGF)-like growth factor/EGF receptor signaling. J. Biol. Chem. 289 10488–10501. 10.1074/jbc.M113.534263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombino M., Capone M., Lissia A., Cossu A., Rubino C., De Giorgi V., et al. (2012). BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J. Clin. Oncol. 30 2522–2529. 10.1200/JCO.2011.41.2452 [DOI] [PubMed] [Google Scholar]
- Colombo M., Giannandrea D., Lesma E., Basile A., Chiaramonte R. (2019). Extracellular vesicles enhance multiple myeloma metastatic dissemination. Int. J. Mol. Sci. 20:3236. 10.3390/ijms20133236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney C. A., Jousheghany F., Yao-Borengasser A., Phanavanh B., Gomes T., Kieber-Emmons A. M., et al. (2011). Chondroitin sulfates play a major role in breast cancer metastasis: a role for CSPG4 and CHST11 gene expression in forming surface P-selectin ligands in aggressive breast cancer cells. Breast Cancer Res. 13:R58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumans F., Terstappen L. (2015). Detection and characterization of circulating tumor cells by the CellSearch approach. Methods Mol. Biol. 1347 263–278. 10.1007/978-1-4939-2990-0_18 [DOI] [PubMed] [Google Scholar]
- Court C. M., Hou S., Winograd P., Segel N. H., Li Q. W., Zhu Y., et al. (2018). A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transpl. 24 946–960. 10.1002/lt.25062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofanilli M., Budd G. T., Ellis M. J., Stopeck A., Matera J., Miller M. C., et al. (2004). Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 351 781–791. 10.1053/j.seminoncol.2006.03.016 [DOI] [PubMed] [Google Scholar]
- Daniotti J. L., Lardone R. D., Vilcaes A. A. (2015). Dysregulated expression of glycolipids in tumor cells: from negative modulator of anti-tumor immunity to promising targets for developing therapeutic agents. Front. Oncol. 5:300. 10.3389/fonc.2015.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bono J. S., Scher H. I., Montgomery R. B., Parker C., Miller M. C., Tissing H., et al. (2008). Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 14 6302–6309. 10.1158/1078-0432.CCR-08-0872 [DOI] [PubMed] [Google Scholar]
- de Groot A. E., Roy S., Brown J. S., Pienta K. J., Amend S. R. (2017). Revisiting seed and soil: examining the primary tumor and cancer cell foraging in metastasis. Mol. Cancer Res. 15 361–370. 10.1158/1541-7786.MCR-16-0436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Klerk D. P., Lee D. V., Human H. J. (1984). Glycosaminoglycans of human prostatic cancer. J. Urol. 131 1008–1012. [DOI] [PubMed] [Google Scholar]
- de Wit S., Manicone M., Rossi E., Lampignano R., Yang L., Zill B., et al. (2018). EpCAM(high) and EpCAM(low) circulating tumor cells in metastatic prostate and breast cancer patients. Oncotarget 9 35705–35716. 10.18632/oncotarget.26298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianat-Moghadam H., Azizi M., Eslami S. Z., Cortes-Hernandez L. E., Heidarifard M., Nouri M., et al. (2020). The role of circulating tumor cells in the metastatic cascade: biology, technical challenges, and clinical relevance. Cancers 12:867. 10.3390/cancers12040867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick G., Akslen-Hoel L. K., Grondahl F., Kjos I., Prydz K. (2012). Proteoglycan synthesis and Golgi organization in polarized epithelial cells. J. Histochem. Cytochem. 60 926–935. 10.1369/0022155412461256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitroff C. J., Lechpammer M., Long-Woodward D., Kutok J. L. (2004). Rolling of human bone-metastatic prostate tumor cells on human bone marrow endothelium under shear flow is mediated by E-selectin. Cancer Res. 64 5261–5269. [DOI] [PubMed] [Google Scholar]
- Dimitroff C. J., Lee J. Y., Rafii S., Fuhlbrigge R. C., Sackstein R. (2001a). Cd44 is a Major E-Selectin Ligand on human hematopoietic progenitor cells. J. Cell Biol. 153 1277–1286. 10.1083/jcb.153.6.1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitroff C. J., Lee J. Y., Schor K. S., Sandmaier B. M., Sackstein R. (2001b). Differential L-Selectin Binding Activities of Human Hematopoietic Cell L-Selectin Ligands, HCELL and PSGL-1. J. Biol. Chem. 276 47623–47631. 10.1074/jbc.M105997200 [DOI] [PubMed] [Google Scholar]
- Domev H., Amit M., Laevsky I., Dar A., Itskovitz-Eldor J. (2012). Efficient engineering of vascularized ectopic bone from human embryonic stem cell–derived mesenchymal stem cells. Tissue Eng. Part A 18 2290–2302. 10.1089/ten.TEA.2011.0371 [DOI] [PubMed] [Google Scholar]
- Dours-Zimmermann M. T., Zimmermann D. R. (1994). A novel glycosaminoglycan attachment domain identified in two alternative splice variants of human versican. J. Biol. Chem. 269 32992–32998. [PubMed] [Google Scholar]
- Eccles S. A. (1999). Heparanase: breaking down barriers in tumors. Nat. Med. 5 735–736. 10.1038/10455 [DOI] [PubMed] [Google Scholar]
- Echtermeyer F., Baciu P. C., Saoncella S., Ge Y., Goetinck P. F. (1999). Syndecan-4 core protein is sufficient for the assembly of focal adhesions and actin stress fibers. J. Cell Sci. 112(Pt 20), 3433–3441. [DOI] [PubMed] [Google Scholar]
- Effenberger K. E., Schroeder C., Hanssen A., Wolter S., Eulenburg C., Tachezy M., et al. (2018). Improved risk stratification by circulating tumor cell counts in pancreatic cancer. Clin. Cancer Res. 24 2844–2850. 10.1158/1078-0432.CCR-18-0120 [DOI] [PubMed] [Google Scholar]
- Elderbroom J. L., Huang J. J., Gatza C. E., Chen J., How T., Starr M., et al. (2014). Ectodomain shedding of TbetaRIII is required for TbetaRIII-mediated suppression of TGF-beta signaling and breast cancer migration and invasion. Mol. Biol. Cell 25 2320–2332. 10.1091/mbc.E13-09-0524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgundi Z., Papanicolaou M., Major G., Cox T. R., Melrose J., Whitelock J. M., et al. (2019). Cancer metastasis: the role of the extracellular matrix and the heparan sulfate proteoglycan perlecan. Front. Oncol. 9:1482. 10.3389/fonc.2019.01482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis L. M. (2006). The role of neuropilins in cancer. Mol. Cancer Ther. 5 1099–1107. [DOI] [PubMed] [Google Scholar]
- Esko J. D., Zhang L. (1996). Influence of core protein sequence on glycosaminoglycan assembly. Curr. Opin. Struct. Biol. 6 663–670. 10.1016/s0959-440x(96)80034-0 [DOI] [PubMed] [Google Scholar]
- Fang C., Fan C., Wang C., Huang Q., Meng W., Yu Y., et al. (2017). Prognostic value of CD133(+) CD54(+) CD44(+) circulating tumor cells in colorectal cancer with liver metastasis. Cancer Med. 6 2850–2857. 10.1002/cam4.1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares J., Fares M. Y., Khachfe H. H., Salhab H. A., Fares Y. (2020). Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct. Target. Ther. 5:28. 10.1038/s41392-020-0134-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M., Gao W., Wang R., Chen W., Man Y. G., Figg W. D., et al. (2013). Therapeutically targeting glypican-3 via a conformation-specific single-domain antibody in hepatocellular carcinoma. Proc. Natl. Acad. Sci. U.S.A. 110 E1083–E1091. 10.1073/pnas.1217868110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N., Gerber H.-P., LeCouter J. (2003). The biology of VEGF and its receptors. 9, 669–676. 10.1038/nm0603-669 [DOI] [PubMed] [Google Scholar]
- Fidler I. J. (1978). Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Res. 38 2651–2660. 10.1158/0008-5472.CAN-16-1330 [DOI] [PubMed] [Google Scholar]
- Filmus J., Capurro M., Rast J. (2008). Glypicans. Genome Biol. 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher L. W., Termine J. D., Young M. F. (1989). Deduced protein sequence of bone small proteoglycan I (biglycan) shows homology with proteoglycan II (decorin) and several nonconnective tissue proteins in a variety of species. J. Biol. Chem. 264 4571–4576. [PubMed] [Google Scholar]
- Fitzgerald T. L., McCubrey J. A. (2014). Pancreatic cancer stem cells: association with cell surface markers, prognosis, resistance, metastasis and treatment. Adv. Biol. Regul. 56 45–50. 10.1016/j.jbior.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Frankel P., Pellet-Many C., Lehtolainen P., D’abaco G. M., Tickner M. L., Cheng L., et al. (2008). Chondroitin sulphate-modified neuropilin 1 is expressed in human tumour cells and modulates 3D invasion in the U87MG human glioblastoma cell line through a p130Cas-mediated pathway. EMBO Rep. 9 983–989. 10.1038/embor.2008.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken B., De Groot M. R., Mastboom W. J., Vermes I., Van Der Palen J., Tibbe A. G., et al. (2012). Circulating tumor cells, disease recurrence and survival in newly diagnosed breast cancer. Breast Cancer Res. 14:R133. 10.1186/bcr3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman J. B., Gray E. S., Millward M., Pearce R., Ziman M. (2012). Evaluation of a multi-marker immunomagnetic enrichment assay for the quantification of circulating melanoma cells. J. Transl. Med. 10:192. 10.1186/1479-5876-10-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M., Hiraoka N., Yeh J.-C. (1999). C-Type Lectins and Sialyl Lewis X oligosaccharides: versatile roles in cell–cell interaction. J. Cell Biol. 147 467–470. 10.1083/jcb.147.3.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon M. L., Bielenberg D. R., Gechtman Z., Miao H. Q., Takashima S., Soker S., et al. (2000). Identification of a natural soluble neuropilin-1 that binds vascular endothelial growth factor: in vivo expression and antitumor activity. Proc. Natl. Acad. Sci. U.S.A. 97 2573–2578. 10.1073/pnas.040337597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Kim H., Feng M., Phung Y., Xavier C. P., Rubin J. S., et al. (2014). Inactivation of Wnt signaling by a human antibody that recognizes the heparan sulfate chains of glypican-3 for liver cancer therapy. Hepatology 60 576–587. 10.1002/hep.26996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Roman J., Zentella-Dehesa A. (2013). Vascular permeability changes involved in tumor metastasis. Cancer Lett. 335 259–269. 10.1016/j.canlet.2013.03.005 [DOI] [PubMed] [Google Scholar]
- Gasch C., Bauernhofer T., Pichler M., Langer-Freitag S., Reeh M., Seifert A. M., et al. (2013). Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin. Chem. 59 252–260. 10.1373/clinchem.2012.188557 [DOI] [PubMed] [Google Scholar]
- Gazzaniga P., Gianni W., Raimondi C., Gradilone A., Lo Russo G., Longo F., et al. (2013). Circulating tumor cells in high-risk nonmetastatic colorectal cancer. Tumour Biol. 34 2507–2509. 10.1007/s13277-013-0752-9 [DOI] [PubMed] [Google Scholar]
- Ghajar C. M., Peinado H., Mori H., Matei I. R., Evason K. J., Brazier H., et al. (2013). The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol. 15 807–817. 10.1038/ncb2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli G., Maccarana M. (2016). Drugs affecting glycosaminoglycan metabolism. Drug Discov. Today 21 1162–1169. [DOI] [PubMed] [Google Scholar]
- Goddard E. T., Bozic I., Riddell S. R., Ghajar C. M. (2018). Dormant tumour cells, their niches and the influence of immunity. Nat. Cell Biol. 20 1240–1249. 10.1038/s41556-018-0214-0 [DOI] [PubMed] [Google Scholar]
- Gomez Toledo A., Nilsson J., Noborn F., Sihlbom C., Larson G. (2015). Positive mode LC-MS/MS analysis of chondroitin sulfate modified glycopeptides derived from light and heavy chains of the human Inter-alpha-Trypsin inhibitor complex. Mol. Cell. Proteomics 14 3118–3131. 10.1074/mcp.M115.051136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodison S., Urquidi V., Tarin D. (1999). CD44 cell adhesion molecules. Mol. Pathol. 52 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinathan P., Chiang N.-J., Bandaru A., Sinha A., Huang W.-Y., Hung S.-C., et al. (2020). Exploring circulating tumor cells in cholangiocarcinoma using a novel glycosaminoglycan probe on a microfluidic platform. Adv. Healthc. Mater. 9:1901875. 10.1002/adhm.201901875 [DOI] [PubMed] [Google Scholar]
- Goretzki L., Burg M. A., Grako K. A., Stallcup W. B. (1999). High-affinity binding of basic fibroblast growth factor and platelet-derived growth factor-AA to the core protein of the NG2 proteoglycan. J. Biol. Chem. 274 16831–16837. 10.1074/jbc.274.24.16831 [DOI] [PubMed] [Google Scholar]
- Gorges K., Wiltfang L., Gorges T. M., Sartori A., Hildebrandt L., Keller L., et al. (2019). Intra-patient heterogeneity of circulating tumor cells and circulating tumor DNA in blood of melanoma patients. Cancers 11:1685. 10.3390/cancers11111685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorges T. M., Tinhofer I., Drosch M., Rose L., Zollner T. M., Krahn T., et al. (2012). Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 12:178. 10.1186/1471-2407-12-178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda D. C. (2006). Role of chondroitin-4-sulfate in pregnancy-associated malaria. Adv. Pharmacol. 53 375–400. 10.1016/S1054-3589(05)53018-7 [DOI] [PubMed] [Google Scholar]
- Gray E. S., Reid A. L., Bowyer S., Calapre L., Siew K., Pearce R., et al. (2015). Circulating melanoma cell subpopulations: their heterogeneity and differential responses to treatment. J. Invest. Dermatol. 135 2040–2048. 10.1038/jid.2015.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene B. T., Hughes A. D., King M. R. (2012). Circulating tumor cells: the substrate of personalized medicine? Front. Oncol. 2:69. 10.3389/fonc.2012.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield B., Wang W.-C., Marquardt H., Piepkorn M., Wolff E. A., Aruffo A., et al. (1999). Characterization of the heparan sulfate and chondroitin sulfate assembly sites in CD44. J. Biol. Chem. 274 2511–2517. 10.1074/jbc.274.4.2511 [DOI] [PubMed] [Google Scholar]
- Grigoriu B. D., Depontieu F., Scherpereel A., Gourcerol D., Devos P., Ouatas T., et al. (2006). Endocan expression and relationship with survival in human non-small cell lung cancer. Clin. Cancer Res. 12 4575–4582. 10.1158/1078-0432.CCR-06-0185 [DOI] [PubMed] [Google Scholar]
- Gronthos S., Franklin D. M., Leddy H. A., Robey P. G., Storms R. W., Gimble J. M. (2001). Surface protein characterization of human adipose tissue-derived stromal cells. J. Cell. Physiol. 189 54–63. 10.1002/jcp.1138 [DOI] [PubMed] [Google Scholar]
- Gundem G., Van Loo P., Kremeyer B., Alexandrov L. B., Tubio J. M. C., Papaemmanuil E., et al. (2015). The evolutionary history of lethal metastatic prostate cancer. Nature 520 353–357. 10.1016/j.urolonc.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. Y., Chiu C. H., Wang M. J., Li F. A., Chen J. Y. (2020). Proteoglycan serglycin promotes non-small cell lung cancer cell migration through the interaction of its glycosaminoglycans with CD44. J. Biomed. Sci. 27:2. 10.1186/s12929-019-0600-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. Y., Hsu H. S., Tyan S. W., Li F. Y., Shew J. Y., Lee W. H., et al. (2017). Serglycin in tumor microenvironment promotes non-small cell lung cancer aggressiveness in a CD44-dependent manner. Oncogene 36 2457–2471. 10.1038/onc.2016.404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C. S., Ross M., Bowman Bauldry J. B., Upshaw J., Karhade M. G., Royal R., et al. (2018). Circulating tumor cells in stage IV melanoma patients. J. Am. Coll. Surg. 227 116–124. 10.1016/j.jamcollsurg.2018.04.026 [DOI] [PubMed] [Google Scholar]
- Hamaoka M., Kobayashi T., Tanaka Y., Mashima H., Ohdan H. (2019). Clinical significance of glypican-3-positive circulating tumor cells of hepatocellular carcinoma patients: a prospective study. PLoS One 14:e0217586. 10.1371/journal.pone.0217586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2011). Hallmarks of cancer: the next generation. Cell 144 646–674. [DOI] [PubMed] [Google Scholar]
- Hanley W. D., Napier S. L., Burdick M. M., Schnaar R. L., Sackstein R., Konstantopoulos K. (2006). Variant isoforms of CD44 are P- and L-selectin ligands on colon carcinoma cells. FASEB J. 20 337–339. [DOI] [PubMed] [Google Scholar]
- Hatabe S., Kimura H., Arao T., Kato H., Hayashi H., Nagai T., et al. (2013). Overexpression of heparan sulfate 6-O-sulfotransferase-2 in colorectal cancer. Mol. Clin. Oncol. 1 845–850. 10.3892/mco.2013.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida K., Johnston D. R., Goldberger O., Park P. W. (2006). Syndecan-1 expression in epithelial cells is induced by transforming growth factor beta through a PKA-dependent pathway. J. Biol. Chem. 281 24365–24374. 10.1074/jbc.M509320200 [DOI] [PubMed] [Google Scholar]
- He J., Zeng Z. C., Xiang Z. L., Yang P. (2014). Mass spectrometry-based serum peptide profiling in hepatocellular carcinoma with bone metastasis. World J. Gastroenterol. 20 3025–3032. 10.3748/wjg.v20.i11.3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Zhou X., Qu C., Tang Y., Zhang Q., Hong J. (2013). Serglycin (SRGN) overexpression predicts poor prognosis in hepatocellular carcinoma patients. Med. Oncol. 30:707. 10.1007/s12032-013-0707-4 [DOI] [PubMed] [Google Scholar]
- Hempel N., How T., Dong M., Murphy S. K., Fields T. A., Blobe G. C. (2007). Loss of betaglycan expression in ovarian cancer: role in motility and invasion. Cancer Res. 67 5231–5238. 10.1158/0008-5472.CAN-07-0035 [DOI] [PubMed] [Google Scholar]
- Hendricks C., Dubail J., Brohee L., Delforge Y., Colige A., Deroanne C. (2016). A novel physiological glycosaminoglycan-deficient splice variant of neuropilin-1 is anti-tumorigenic in vitro and in vivo. PLoS One 11:e0165153. 10.1371/journal.pone.0165153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillig T., Horn P., Nygaard A. B., Haugaard A. S., Nejlund S., Brandslund I., et al. (2015). In vitro detection of circulating tumor cells compared by the CytoTrack and CellSearch methods. Tumour Biol. 36 4597–4601. 10.1007/s13277-015-3105-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippo Y., Watanabe K., Watanabe A., Midorikawa Y., Yamamoto S., Ihara S., et al. (2004). Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Res. 64 2418–2423. 10.1158/0008-5472.can-03-2191 [DOI] [PubMed] [Google Scholar]
- Holderfield M., Deuker M. M., Mccormick F., Mcmahon M. (2014). Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer 14 455–467. 10.1038/nrc3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A., Costa-Silva B., Shen T. L., Rodrigues G., Hashimoto A., Tesic Mark M., et al. (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527 329–335. 10.1038/nature15756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X. M., Zhang T., Da Z., Wu X. A. (2019). CHPF promotes lung adenocarcinoma proliferation and anti-apoptosis via the MAPK pathway. Pathol. Res. Pract. 215 988–994. 10.1016/j.prp.2019.02.005 [DOI] [PubMed] [Google Scholar]
- Hristozova T., Konschak R., Budach V., Tinhofer I. (2012). A simple multicolor flow cytometry protocol for detection and molecular characterization of circulating tumor cells in epithelial cancers. Cytometry A 81 489–495. 10.1002/cyto.a.22041 [DOI] [PubMed] [Google Scholar]
- Hsu S.-H. C., Nadesan P., Puviindran V., Stallcup W. B., Kirsch D. G., Alman B. A. (2018). Effects of chondroitin sulfate proteoglycan 4 (NG2/CSPG4) on soft-tissue sarcoma growth depend on tumor developmental stage. J. Biol. Chem. 293 2466–2475. 10.1074/jbc.M117.805051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Guo P., Bar-Joseph I., Imanishi Y., Jarzynka M. J., Bogler O., et al. (2007). Neuropilin-1 promotes human glioma progression through potentiating the activity of the HGF/SF autocrine pathway. Oncogene 26 5577–5586. 10.1038/sj.onc.1210348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Shi C., Tian Y., Zhang Y., Xu C., Chen H., et al. (2015). TGF-beta induces up-regulation of chondroitin sulfate synthase 1 (CHSY1) in nucleus pulposus cells through MAPK signaling. Cell. Physiol. Biochem. 37 793–804. 10.1159/000430396 [DOI] [PubMed] [Google Scholar]
- Hu L., Duan Y. T., Li J. F., Su L. P., Yan M., Zhu Z. G., et al. (2014). Biglycan enhances gastric cancer invasion by activating FAK signaling pathway. Oncotarget 5 1885–1896. 10.18632/oncotarget.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Zang M. D., Wang H. X., Li J. F., Su L. P., Yan M., et al. (2016). Biglycan stimulates VEGF expression in endothelial cells by activating the TLR signaling pathway. Mol. Oncol. 10 1473–1484. 10.1016/j.molonc.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Pan C., Hu H., Zheng S., Ding L. (2012). Osteopontin-enhanced hepatic metastasis of colorectal cancer cells. PLoS One 7:e47901. 10.1371/journal.pone.0047901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husemann Y., Geigl J. B., Schubert F., Musiani P., Meyer M., Burghart E., et al. (2008). Systemic spread is an early step in breast cancer. Cancer Cell 13 58–68. 10.1016/j.ccr.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Hyun K. A., Koo G. B., Han H., Sohn J., Choi W., Kim S. I., et al. (2016). Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 7 24677–24687. 10.18632/oncotarget.8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igelmann S., Neubauer H. A., Ferbeyre G. (2019). STAT3 and STAT5 activation in solid cancers. Cancers 11:1428. 10.3390/cancers11101428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida J., Wilhelmson K. L., Ng J., Lee P., Morrison C., Tam E., et al. (2007). Cell surface chondroitin sulfate glycosaminoglycan in melanoma: role in the activation of pro-MMP-2 (pro-gelatinase A). Biochem. J. 403 553–563. 10.1042/BJ20061176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilie M., Hofman V., Leroy S., Cohen C., Heeke S., Cattet F., et al. (2018). Use of circulating tumor cells in prospective clinical trials for NSCLC patients - standardization of the pre-analytical conditions. Clin. Chem. Lab. Med. 56 980–989. 10.1515/cclm-2017-0764 [DOI] [PubMed] [Google Scholar]
- Ilieva K. M., Cheung A., Mele S., Chiaruttini G., Crescioli S., Griffin M., et al. (2018). Chondroitin sulfate proteoglycan 4 and its potential as an antibody immunotherapy target across different tumor types. Front. Immunol. 8:1911. 10.3389/fimmu.2017.01911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo R. V., Sanderson R. D. (2011). Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J. Cell. Mol. Med. 15 1013–1031. 10.1111/j.1582-4934.2010.01236.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo R. V., Schaefer L. (2015). Proteoglycan form and function: a comprehensive nomenclature of proteoglycans. Matrix Biol. 42 11–55. 10.1016/j.matbio.2015.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itano N. (2008). Simple primary structure, complex turnover regulation and multiple roles of hyaluronan. J. Biochem. 144 131–137. 10.1093/jb/mvn046 [DOI] [PubMed] [Google Scholar]
- Jacobsen F., Kraft J., Schroeder C., Hube-Magg C., Kluth M., Lang D. S., et al. (2017). Up-regulation of biglycan is associated with poor prognosis and PTEN deletion in patients with prostate cancer. Neoplasia 19 707–715. 10.1016/j.neo.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S., Jamieson C. H., Pang W. W., Park C. Y., Chao M. P., Majeti R., et al. (2009). CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 138 271–285. 10.1016/j.cell.2009.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil N. S. M., Azfer A., Worrell H., Salter D. M. (2016). Functional roles of CSPG4/NG2 in chondrosarcoma. Int. J. Exp. Pathol. 97 178–186. 10.1111/iep.12189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvinen T. A., Prince S. (2015). Decorin: a growth factor antagonist for tumor growth inhibition. Biomed Res. Int. 2015:654765. 10.1155/2015/654765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins L. M., Singh P., Varadaraj A., Lee N. Y., Shah S., Flores H. V., et al. (2016). Altering the proteoglycan state of transforming growth factor beta type III receptor (TbetaRIII)/Betaglycan modulates canonical wnt/beta-catenin signaling. J. Biol. Chem. 291 25716–25728. 10.1074/jbc.M116.748624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P., Maiti A., Brown K. L., Li R. (2000). A role for the cell adhesion molecule CD44 and sulfation in leukocyte–endothelial cell adhesion during an inflammatory response? Biochem. Pharmacol. 59 455–465. 10.1016/s0006-2952(99)00266-x [DOI] [PubMed] [Google Scholar]
- Jubb A. M., Strickland L. A., Liu S. D., Mak J., Schmidt M., Koeppen H. (2012). Neuropilin-1 expression in cancer and development. J. Pathol. 226 50–60. [DOI] [PubMed] [Google Scholar]
- Kaigorodova E. V., Savelieva O. E., Tashireva L. A., Tarabanovskaya N. A., Simolina E. I., Denisov E. V., et al. (2018). Heterogeneity of circulating tumor cells in neoadjuvant chemotherapy of breast cancer. Molecules 23:727. 10.3390/molecules23040727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabacak N. M., Spuhler P. S., Fachin F., Lim E. J., Pai V., Ozkumur E., et al. (2014). Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protoc. 9 694–710. 10.1038/nprot.2014.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh S., Goi T., Naruse T., Ueda Y., Kurebayashi H., Nakazawa T., et al. (2015). Cancer stem cell marker in circulating tumor cells: expression of CD44 Variant Exon 9 is strongly correlated to treatment refractoriness, recurrence and prognosis of human colorectal cancer. Anticancer Res. 35 239–244. [PubMed] [Google Scholar]
- Kaur S., Kuznetsova S. A., Pendrak M. L., Sipes J. M., Romeo M. J., Li Z., et al. (2011). Heparan sulfate modification of the transmembrane receptor CD47 is necessary for inhibition of T cell receptor signaling by thrombospondin-1. J. Biol. Chem. 286 14991–15002. 10.1074/jbc.M110.179663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoja L., Lorigan P., Zhou C., Lancashire M., Booth J., Cummings J., et al. (2013). Biomarker utility of circulating tumor cells in metastatic cutaneous melanoma. J. Invest. Dermatol. 133 1582–1590. 10.1038/jid.2012.468 [DOI] [PubMed] [Google Scholar]
- Khoja L., Shenjere P., Hodgson C., Hodgetts J., Clack G., Hughes A., et al. (2014). Prevalence and heterogeneity of circulating tumour cells in metastatic cutaneous melanoma. Melanoma Res. 24 40–46. 10.1097/CMR.0000000000000025 [DOI] [PubMed] [Google Scholar]
- Kim K. H., Lee H. H., Yoon Y. E., Na J. C., Kim S. Y., Cho Y. I., et al. (2018). Clinical validation of serum endocan (ESM-1) as a potential biomarker in patients with renal cell carcinoma. Oncotarget 9 662–667. 10.18632/oncotarget.23087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. Y., Oskarsson T., Acharyya S., Nguyen D. X., Zhang X. H., Norton L., et al. (2009). Tumor self-seeding by circulating cancer cells. Cell 139 1315–1326. 10.1016/j.cell.2009.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinac D., Gray E. S., Freeman J. B., Reid A., Bowyer S., Millward M., et al. (2014). Monitoring changes in circulating tumour cells as a prognostic indicator of overall survival and treatment response in patients with metastatic melanoma. BMC Cancer 14:423. 10.1186/1471-2407-14-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knelson E. H., Nee J. C., Blobe G. C. (2014). Heparan sulfate signaling in cancer. Trends Biochem. Sci. 39 277–288. 10.1016/j.tibs.2014.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokenyesi R., Bernfield M. (1994). Core protein structure and sequence determine the site and presence of heparan sulfate and chondroitin sulfate on syndecan-1. J. Biol. Chem. 269 12304–12309. [PubMed] [Google Scholar]
- Koliopanos A., Friess H., Kleeff J., Shi X., Liao Q., Pecker I., et al. (2001). Heparanase expression in primary and metastatic pancreatic cancer. Cancer Res. 61 4655–4659. [PubMed] [Google Scholar]
- Kolluri A., Ho M. (2019). The role of glypican-3 in regulating Wnt, YAP, and hedgehog in liver cancer. Front. Oncol. 9:708. 10.3389/fonc.2019.00708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolset S. O., Tveit H. (2008). Serglycin–structure and biology. Cell. Mol. Life Sci. 65 1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpetinou A., Papachristou D. J., Lampropoulou A., Bouris P., Labropoulou V. T., Noulas A., et al. (2015). Increased expression of serglycin in specific carcinomas and aggressive cancer cell lines. Biomed Res. Int. 2015:690721. 10.1155/2015/690721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpetinou A., Skandalis S. S., Moustakas A., Happonen K. E., Tveit H., Prydz K., et al. (2013). Serglycin is implicated in the promotion of aggressive phenotype of breast cancer cells. PLoS One 8:e78157. 10.1371/journal.pone.0078157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalik A., Kowalewska M., Gozdz S. (2017). Current approaches for avoiding the limitations of circulating tumor cells detection methods-implications for diagnosis and treatment of patients with solid tumors. Transl. Res. 185 58–84.e15. 10.1016/j.trsl.2017.04.002 [DOI] [PubMed] [Google Scholar]
- Kowli S., Velidandla R., Creek K. E., Pirisi L. (2013). TGF-beta regulation of gene expression at early and late stages of HPV16-mediated transformation of human keratinocytes. Virology 447 63–73. 10.1016/j.virol.2013.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. G., Sloane R., Priest L., Lancashire L., Hou J. M., Greystoke A., et al. (2011). Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 29 1556–1563. 10.1200/JCO.2010.28.7045 [DOI] [PubMed] [Google Scholar]
- Kufe D. W. (2009). Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer 9 874–885. 10.1038/nrc2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J. C.-H., Gandhi J. G., Zia R. N., Paszek M. J. (2018). Physical biology of the cancer cell glycocalyx. Nat. Phys. 14 658–669. 10.1038/s41567-018-0186-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara T., Itoi T., Sofuni A., Itokawa F., Tsuchiya T., Tsuji S., et al. (2008). Detection of circulating tumor cells in patients with pancreatic cancer: a preliminary result. J. Hepatobiliary Pancreat. Surg. 15 189–195. 10.1007/s00534-007-1250-5 [DOI] [PubMed] [Google Scholar]
- Lambert A. W., Pattabiraman D. R., Weinberg R. A. (2017). Emerging biological principles of metastasis. Cell 168 670–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S., Xu J., Derynck R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampignano R., Schneck H., Neumann M., Fehm T., Neubauer H. (2017). Enrichment, isolation and molecular characterization of EpCAM-negative circulating tumor cells. Adv. Exp. Med. Biol. 994 181–203. 10.1007/978-3-319-55947-6_10 [DOI] [PubMed] [Google Scholar]
- Lee N., Barthel S. R., Schatton T. (2014). Melanoma stem cells and metastasis: mimicking hematopoietic cell trafficking? Lab. Invest. 94 13–30. 10.1038/labinvest.2013.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Li L., Joo E. J., Son J. W., Kim Y. J., Kang J. K., et al. (2017). Glycosaminoglycans and glycolipids as potential biomarkers in lung cancer. Glycoconj. J. 34 661–669. 10.1007/s10719-017-9790-7 [DOI] [PubMed] [Google Scholar]
- Li N., Wei L., Liu X., Bai H., Ye Y., Li D., et al. (2019). A frizzled-like cysteine-rich domain in glypican-3 mediates Wnt binding and regulates hepatocellular carcinoma tumor growth in mice. Hepatology 70 1231–1245. 10.1002/hep.30646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-J., Ong C. K., Cao Y., Xiang Y.-Q., Shao J.-Y., Ooi A., et al. (2011). Serglycin is a theranostic target in nasopharyngeal carcinoma that promotes metastasis. Cancer Res. 71 3162–3172. 10.1158/0008-5472.CAN-10-3557 [DOI] [PubMed] [Google Scholar]
- Lian S., Xie R., Ye Y., Lu Y., Cheng Y., Xie X., et al. (2019). Dual blockage of both PD-L1 and CD47 enhances immunotherapy against circulating tumor cells. Sci. Rep. 9:4532. 10.1038/s41598-019-40241-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti P. A., Rao C. G., Terstappen L. W. M. M. (2001). Optimization of ferrofluids and protocols for the enrichment of breast tumor cells in blood. J. Magn. Magn. Mater. 225 301–307. [Google Scholar]
- Lim J., Thiery J. P. (2012). Epithelial-mesenchymal transitions: insights from development. Development 139 3471–3486. 10.1242/dev.071209 [DOI] [PubMed] [Google Scholar]
- Liu H. Y., Qian H. H., Zhang X. F., Li J., Yang X., Sun B., et al. (2015). Improved method increases sensitivity for circulating hepatocellular carcinoma cells. World J. Gastroenterol. 21 2918–2925. 10.3748/wjg.v21.i10.2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Tian Z., Zhang L., Hou S., Hu S., Wu J., et al. (2016). Combined cell surface carbonic anhydrase 9 and CD147 antigens enable high-efficiency capture of circulating tumor cells in clear cell renal cell carcinoma patients. Oncotarget 7 59877–59891. 10.18632/oncotarget.10979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Taftaf R., Kawaguchi M., Chang Y. F., Chen W., Entenberg D., et al. (2019). Homophilic CD44 interactions mediate tumor cell aggregation and polyclonal metastasis in patient-derived breast cancer models. Cancer Discov. 9 96–113. 10.1158/2159-8290.CD-18-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Fusi A., Klopocki E., Schmittel A., Tinhofer I., Nonnenmacher A., et al. (2011). Negative enrichment by immunomagnetic nanobeads for unbiased characterization of CTCs from peripheral blood of cancer patients. J. Transl. Med. 9:70. 10.1186/1479-5876-9-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Casillas F., Payne H. M., Andres J. L., Massague J. (1994). Betaglycan can act as a dual modulator of TGF-beta access to signaling receptors: mapping of ligand binding and GAG attachment sites. J. Cell Biol. 124 557–568. 10.1083/jcb.124.4.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopresti A., Malergue F., Bertucci F., Liberatoscioli M. L., Garnier S., Dacosta Q., et al. (2019). Sensitive and easy screening for circulating tumor cells by flow cytometry. JCI Insight 5:e128180. 10.1172/jci.insight.128180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucci A., Hall C., Patel S. P., Narendran B., Bauldry J. B., Royal R., et al. (2020). Circulating tumor cells and early relapse in node-positive melanoma. Clin. Cancer Res. 26, 1886–1895. 10.1158/1078-0432.CCR-19-2670 [DOI] [PubMed] [Google Scholar]
- Lugano R., Ramachandran M., Dimberg A. (2020). Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 77 1745–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch C. C., Matrisian L. M. (2002). Matrix metalloproteinases in tumor-host cell communication. Differentiation 70 561–573. 10.1046/j.1432-0436.2002.700909.x [DOI] [PubMed] [Google Scholar]
- Majuri M. L., Niemela R., Tiisala S., Renkonen O., Renkonen R. (1995). Expression and function of alpha 2,3-sialyl- and alpha 1,3/1,4-fucosyltransferases in colon adenocarcinoma cell lines: role in synthesis of E-selectin counter-receptors. Int. J. Cancer 63 551–559. 10.1002/ijc.2910630416 [DOI] [PubMed] [Google Scholar]
- Makagiansar I. T., Williams S., Mustelin T., Stallcup W. B. (2007). Differential phosphorylation of NG2 proteoglycan by ERK and PKCα helps balance cell proliferation and migration. J. Cell Biol. 178 155–165. 10.1083/jcb.200612084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man X. B., Tang L., Zhang B. H., Li S. J., Qiu X. H., Wu M. C., et al. (2005). Upregulation of Glypican-3 expression in hepatocellular carcinoma but downregulation in cholangiocarcinoma indicates its differential diagnosis value in primary liver cancers. Liver Int. 25 962–966. 10.1111/j.1478-3231.2005.01100.x [DOI] [PubMed] [Google Scholar]
- Marrero J. A., Kulik L. M., Sirlin C. B., Zhu A. X., Finn R. S., Abecassis M. M., et al. (2018). Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American association for the study of liver diseases. Hepatology 68 723–750. 10.1002/hep.29913 [DOI] [PubMed] [Google Scholar]
- Martinez P., Vergoten G., Colomb F., Bobowski M., Steenackers A., Carpentier M., et al. (2013). Over-sulfated glycosaminoglycans are alternative selectin ligands: insights into molecular interactions and possible role in breast cancer metastasis. Clin. Exp. Metastasis 30 919–931. 10.1007/s10585-013-9592-7 [DOI] [PubMed] [Google Scholar]
- Masola V., Bellin G., Gambaro G., Onisto M. (2018). Heparanase: a multitasking protein involved in extracellular matrix (ECM) remodeling and intracellular events. Cells 7:236. 10.3390/cells7120236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J., Obenauf A. C. (2016). Metastatic colonization by circulating tumour cells. Nature 529 298–306. 10.1038/nature17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurage C. A., Adam E., Mineo J. F., Sarrazin S., Debunne M., Siminski R. M., et al. (2009). Endocan expression and localization in human glioblastomas. J. Neuropathol. Exp. Neurol. 68 633–641. 10.1097/NEN.0b013e3181a52a7f [DOI] [PubMed] [Google Scholar]
- Maxhimer J. B., Pesce C. E., Stewart R. A., Gattuso P., Prinz R. A., Xu X. (2005). Ductal carcinoma in situ of the breast and heparanase-1 expression: a molecular explanation for more aggressive subtypes. J. Am. Coll. Surg. 200 328–335. 10.1016/j.jamcollsurg.2004.10.034 [DOI] [PubMed] [Google Scholar]
- Melo S. A., Luecke L. B., Kahlert C., Fernandez A. F., Gammon S. T., Kaye J., et al. (2015). Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. 523, 177–182. 10.1038/nature14581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menakuru S. R., Brown N. J., Staton C. A., Reed M. W. (2008). Angiogenesis in pre-malignant conditions. Br. J. Cancer 99 1961–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentis A. A., Grivas P. D., Dardiotis E., Romas N. A., Papavassiliou A. G. (2020). Circulating tumor cells as Trojan Horse for understanding, preventing, and treating cancer: a critical appraisal. Cell. Mol. Life Sci. 10.1007/s00018-020-03529-4[Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H. Q., Lee P., Lin H., Soker S., Klagsbrun M. (2000). Neuropilin-1 expression by tumor cells promotes tumor angiogenesis and progression. FASEB J. 14 2532–2539. 10.1096/fj.00-0250com [DOI] [PubMed] [Google Scholar]
- Micalizzi D. S., Maheswaran S., Haber D. A. (2017). A conduit to metastasis: circulating tumor cell biology. Genes Dev. 31 1827–1840. 10.1101/gad.305805.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami T., Kitagawa H. (2013). Biosynthesis and function of chondroitin sulfate. Biochim. Biophys. Acta 1830 4719–4733. [DOI] [PubMed] [Google Scholar]
- Mitchell M. J., Castellanos C. A., King M. R. (2015). Surfactant functionalization induces robust, differential adhesion of tumor cells and blood cells to charged nanotube-coated biomaterials under flow. Biomaterials 56 179–186. 10.1016/j.biomaterials.2015.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell M. J., King M. R. (2014). Physical biology in cancer. 3. The role of cell glycocalyx in vascular transport of circulating tumor cells. Am. J. Physiol. Cell Physiol. 306 C89–C97. 10.1152/ajpcell.00285.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto S., Yamada S., Sugahara K. (2015). Molecular interactions between chondroitin-dermatan sulfate and growth factors/receptors/matrix proteins. Curr. Opin. Struct. Biol. 34 35–42. 10.1016/j.sbi.2015.06.004 [DOI] [PubMed] [Google Scholar]
- Mohamed R., Dayati P., Mehr R. N., Kamato D., Seif F., Babaahmadi-Rezaei H., et al. (2019). Transforming growth factor-beta1 mediated CHST11 and CHSY1 mRNA expression is ROS dependent in vascular smooth muscle cells. J. Cell Commun. Signal. 13 225–233. 10.1007/s12079-018-0495-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose T., Yoshimura Y., Harumiya S., Isobe K., Kito M., Fukushima M., et al. (2016). Chondroitin sulfate synthase 1 expression is associated with malignant potential of soft tissue sarcomas with myxoid substance. Hum. Pathol. 50 15–23. 10.1016/j.humpath.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Morla S. (2019). Glycosaminoglycans and Glycosaminoglycan Mimetics in Cancer and Inflammation. Int. J. Mol. Sci. 20:1963. 10.3390/ijms20081963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller C., Holtschmidt J., Auer M., Heitzer E., Lamszus K., Schulte A., et al. (2014). Hematogenous dissemination of glioblastoma multiforme. Sci. Transl. Med. 6:247ra101. [DOI] [PubMed] [Google Scholar]
- Muller V., Riethdorf S., Rack B., Janni W., Fasching P. A., Solomayer E., et al. (2012). Prognostic impact of circulating tumor cells assessed with the CellSearch System and AdnaTest Breast in metastatic breast cancer patients: the DETECT study. Breast Cancer Res. 14:R118. 10.1186/bcr3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murlidhar V., Reddy R. M., Fouladdel S., Zhao L., Ishikawa M. K., Grabauskiene S., et al. (2017). Poor prognosis indicated by venous circulating tumor cell clusters in early-stage lung cancers. Cancer Res. 77 5194–5206. 10.1158/0008-5472.CAN-16-2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S. J., Yeo H. Y., Chang H. J., Kim B. H., Hong E. K., Park J. W. (2016). a new cell block method for multiple immunohistochemical analysis of circulating tumor cells in patients with liver cancer. Cancer Res. Treat. 48 1229–1242. 10.4143/crt.2015.500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoe M., Kusaka C., Ohta M., Hasebe Y., Unoki T., Shimoyama H., et al. (2019). Development of a highly sensitive technique for capturing renal cell cancer circulating tumor cells. Diagnostics 9:96. 10.3390/diagnostics9030096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naor D., Nedvetzki S., Golan I., Melnik L., Faitelson Y. (2002). CD44 in Cancer. Crit. Rev. Clin. Lab. Sci. 39 527–579. [DOI] [PubMed] [Google Scholar]
- Nguyen D. X., Bos P. D., Massague J. (2009). Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer 9 274–284. 10.1038/nrc2622 [DOI] [PubMed] [Google Scholar]
- Nicolazzo C., Gradilone A., Loreni F., Raimondi C., Gazzaniga P. (2019). EpCAM(low) circulating tumor cells: gold in the waste. Dis. Markers 2019:1718920. 10.1155/2019/1718920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitovic D., Berdiaki A., Spyridaki I., Krasanakis T., Tsatsakis A., Tzanakakis G. N. (2018). Proteoglycans-biomarkers and targets in cancer therapy. Front. Endocrinol. 9:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitovic D., Kouvidi K., Voudouri K., Berdiaki A., Karousou E., Passi A., et al. (2014). The motile breast cancer phenotype roles of proteoglycans/glycosaminoglycans. Biomed Res. Int. 2014:124321. 10.1155/2014/124321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niland S., Eble J. A. (2019). Neuropilins in the context of tumor vasculature. Int. J. Mol. Sci. 20:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A., Lin X. H., Giese N., Heldin C. H., Stallcup W. B. (1996). Interaction between NG2 proteoglycan and PDGF α-receptor on O2A progenitor cells is required for optimal response to PDGF. J. Neurosci. Res. 43 315–330. [DOI] [PubMed] [Google Scholar]
- Nobuhisa T., Naomoto Y., Ohkawa T., Takaoka M., Ono R., Murata T., et al. (2005). Heparanase expression correlates with malignant potential in human colon cancer. J. Cancer Res. Clin. Oncol. 131 229–237. 10.1007/s00432-004-0644-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle L. F., Orr J. G., Willoughby C. E., Hutton C., Mcpherson S., Plummer R., et al. (2016). Imagestream detection and characterisation of circulating tumour cells - A liquid biopsy for hepatocellular carcinoma? J. Hepatol. 65 305–313. 10.1016/j.jhep.2016.04.014 [DOI] [PubMed] [Google Scholar]
- Okabe H., Ishimoto T., Mima K., Nakagawa S., Hayashi H., Kuroki H., et al. (2014). CD44s signals the acquisition of the mesenchymal phenotype required for anchorage-independent cell survival in hepatocellular carcinoma. Br. J. Cancer 110 958–966. 10.1038/bjc.2013.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Takemura G., Suzuki K., Oda K., Takada C., Hotta Y., et al. (2017). Three-dimensional ultrastructure of capillary endothelial glycocalyx under normal and experimental endotoxemic conditions. Crit. Care 21:261. 10.1186/s13054-017-1841-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive P. L., Vikse C., Trotter M. J. (1992). Measurement of oxygen diffusion distance in tumor cubes using a fluorescent hypoxia probe. Int. J. Radiat. Oncol. Biol. Phys. 22 397–402. 10.1016/0360-3016(92)90840-e [DOI] [PubMed] [Google Scholar]
- Oliveira-Ferrer L., Hessling A., Trillsch F., Mahner S., Milde-Langosch K. (2015). Prognostic impact of chondroitin-4-sulfotransferase CHST11 in ovarian cancer. Tumour Biol. 36 9023–9030. 10.1007/s13277-015-3652-3 [DOI] [PubMed] [Google Scholar]
- Ozkumur E., Shah A. M., Ciciliano J. C., Emmink B. L., Miyamoto D. T., Brachtel E., et al. (2013). Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 5:179ra147. 10.1126/scitranslmed.3005616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan S., Chen R., Tamura Y., Crispin D. A., Lai L. A., May D. H., et al. (2014). Quantitative glycoproteomics analysis reveals changes in N-glycosylation level associated with pancreatic ductal adenocarcinoma. J. Proteome Res. 13 1293–1306. 10.1021/pr4010184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson D. R., Dracopoli N., Petty B. G., Compton C., Cristofanilli M., Deisseroth A., et al. (2012). Considerations in the development of circulating tumor cell technology for clinical use. J. Transl. Med. 10:138. 10.1186/1479-5876-10-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek M. J., Dufort C. C., Rossier O., Bainer R., Mouw J. K., Godula K., et al. (2014). The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 511 319–325. 10.1038/nature13535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Shah K., Mirza S., Shah K., Rawal R. (2016). Circulating tumor stem like cells in oral squamous cell carcinoma: an unresolved paradox. Oral Oncol. 62 139–146. 10.1016/j.oraloncology.2016.10.019 [DOI] [PubMed] [Google Scholar]
- Pearce O. M. T., Läubli H. (2016). Sialic acids in cancer biology and immunity. Glycobiology 26 111–128. 10.1093/glycob/cwv097 [DOI] [PubMed] [Google Scholar]
- Peinado H., Aleckovic M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., et al. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 18 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H., Zhang H., Matei I. R., Costa-Silva B., Hoshino A., Rodrigues G., et al. (2017). Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer 17 302–317. 10.1038/nrc.2017.6 [DOI] [PubMed] [Google Scholar]
- Peterson S. M., Iskenderian A., Cook L., Romashko A., Tobin K., Jones M., et al. (2010). Human Sulfatase 2 inhibits in vivo tumor growth of MDA-MB-231 human breast cancer xenografts. BMC Cancer 10:427. 10.1186/1471-2407-10-427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai H. K., Pourgholami C. M., Chua L. T., Morris L. D. (2015). MUC1 as a potential target in anticancer therapies. Am. J. Clin. Oncol. 38 108–118. 10.1097/COC.0b013e31828f5a07 [DOI] [PubMed] [Google Scholar]
- Pinho S. S., Reis C. A. (2015). Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15 540–555. 10.1038/nrc3982 [DOI] [PubMed] [Google Scholar]
- Pomin V. H., Mulloy B. (2018). Glycosaminoglycans and proteoglycans. Pharmaceuticals 11:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M. A., Colvin Wanshura L. E., Yang J., Carlson J., Xiang B., Li G., et al. (2011). CSPG4, a potential therapeutic target, facilitates malignant progression of melanoma. Pigment Cell Melanoma Res. 24 1148–1157. 10.1111/j.1755-148X.2011.00929.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman A., Bandari S. K., Liu J., Mobley J. A., Brown E. E., Sanderson R. D. (2016). Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J. Biol. Chem. 291 1652–1663. 10.1074/jbc.M115.686295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qasaimeh M., Wu Y., Bose S., Menachery A., Talluri S., Gonzalez G., et al. (2017). Isolation of circulating plasma cells in multiple myeloma using CD138 antibody-based capture in a microfluidic device. Sci. Rep. 7:45681. 10.1038/srep45681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L. N., Xiang B. D., Wu F. X., Ye J. Z., Zhong J. H., Wang Y. Y., et al. (2018). Circulating tumor cells undergoing EMT provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res. 78 4731–4744. 10.1158/0008-5472.CAN-17-2459 [DOI] [PubMed] [Google Scholar]
- Qin J., Alt J. R., Hunsley B. A., Williams T. L., Fernando M. R. (2014). Stabilization of circulating tumor cells in blood using a collection device with a preservative reagent. Cancer Cell Int. 14:23. 10.1186/1475-2867-14-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan P., Agrawal A., Bhushan R., Chavalmane A. K., Kalathur R. K., Takahashi T., et al. (2007). Expression profiling of genes regulated by TGF-beta: differential regulation in normal and tumour cells. BMC Genomics 8:98. 10.1186/1471-2164-8-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C., Bui T., Connelly M., Doyle G., Karydis I., Middleton M. R., et al. (2011). Circulating melanoma cells and survival in metastatic melanoma. Int. J. Oncol. 38 755–760. 10.3892/ijo.2011.896 [DOI] [PubMed] [Google Scholar]
- Rapraeger A. (1989). Transforming growth factor (type beta) promotes the addition of chondroitin sulfate chains to the cell surface proteoglycan (syndecan) of mouse mammary epithelia. J. Cell Biol. 109 2509–2518. 10.1083/jcb.109.5.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real F. X., Houghton A. N., Albino A. P., Cordon-Cardo C., Melamed M. R., Oettgen H. F., et al. (1985). Surface antigens of melanomas and melanocytes defined by mouse monoclonal antibodies: specificity analysis and comparison of antigen expression in cultured cells and tissues. Cancer Res. 45 4401–4411. [PubMed] [Google Scholar]
- Reckamp K., Figlin R., Burdick M., Dubinett S., Elashoff R., Strieter R. (2009). CXCR4 expression on circulating pan-cytokeratin positive cells is associated with survival in patients with advanced non-small cell lung cancer. BMC Cancer 9:213. 10.1186/1471-2407-9-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitsma S., Slaaf D. W., Vink H., Van Zandvoort M. A. M. J., Oude Egbrink M. G. A. (2007). The endothelial glycocalyx: composition, functions, and visualization. Pflugers Arch. 454 345–359. 10.1007/s00424-007-0212-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejniak K. A. (2016). Circulating tumor cells: when a solid tumor meets a fluid microenvironment. Adv. Exp. Med. Biol. 936 93–106. 10.1007/978-3-319-42023-3_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkonen J., Paavonen T., Renkonen R. (1997). Endothelial and epithelial expression of sialyl Lewis(x) and sialyl Lewis(a) in lesions of breast carcinoma. Int. J. Cancer 74 296–300. [DOI] [PubMed] [Google Scholar]
- Rennel E., Mellberg S., Dimberg A., Petersson L., Botling J., Ameur A., et al. (2007). Endocan is a VEGF-A and PI3K regulated gene with increased expression in human renal cancer. Exp. Cell Res. 313 1285–1294. 10.1016/j.yexcr.2007.01.021 [DOI] [PubMed] [Google Scholar]
- Reymond N., D’agua B. B., Ridley A. J. (2013). Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer 13 858–870. [DOI] [PubMed] [Google Scholar]
- Rhim A. D., Mirek E. T., Aiello N. M., Maitra A., Bailey J. M., Mcallister F., et al. (2012). EMT and dissemination precede pancreatic tumor formation. Cell 148 349–361. 10.1016/j.cell.2011.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggi N., Aguet M., Stamenkovic I. (2018). Cancer metastasis: a reappraisal of its underlying mechanisms and their relevance to treatment. Annu. Rev. Pathol. Mech. Dis. 13 117–140. 10.1146/annurev-pathol-020117-044127 [DOI] [PubMed] [Google Scholar]
- Rocha S. F., Schiller M., Jing D., Li H., Butz S., Vestweber D., et al. (2014). Esm1 modulates endothelial tip cell behavior and vascular permeability by enhancing VEGF bioavailability. Circ. Res. 115 581–590. 10.1161/CIRCRESAHA.115.304718 [DOI] [PubMed] [Google Scholar]
- Rogers C. J., Hsieh-Wilson L. C. (2012). Microarray method for the rapid detection of glycosaminoglycan-protein interactions. Methods Mol. Biol. 808 321–336. 10.1007/978-1-61779-373-8_22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland C. L., Ross M. I., Hall C. S., Laubacher B., Upshaw J., Anderson A. E., et al. (2015). Detection of circulating melanoma cells in the blood of melanoma patients: a preliminary study. Melanoma Res. 25 335–341. 10.1097/CMR.0000000000000168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roucourt B., Meeussen S., Bao J., Zimmermann P., David G. (2015). Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 25 412–428. 10.1038/cr.2015.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudnicky F., Poyet C., Wild P., Krampitz S., Negrini F., Huggenberger R., et al. (2013). Endocan is upregulated on tumor vessels in invasive bladder cancer where it mediates VEGF-A-induced angiogenesis. Cancer Res. 73 1097–1106. 10.1158/0008-5472.CAN-12-1855 [DOI] [PubMed] [Google Scholar]
- Roy A., Attarha S., Weishaupt H., Edqvist P. H., Swartling F. J., Bergqvist M., et al. (2017). Serglycin as a potential biomarker for glioma: association of serglycin expression, extent of mast cell recruitment and glioblastoma progression. Oncotarget 8 24815–24827. 10.18632/oncotarget.15820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Femel J., Huijbers E. J., Spillmann D., Larsson E., Ringvall M., et al. (2016). Targeting serglycin prevents metastasis in murine mammary carcinoma. PLoS One 11:e0156151. 10.1371/journal.pone.0156151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz C., Li J., Luttgen M. S., Kolatkar A., Kendall J. T., Flores E., et al. (2015). Limited genomic heterogeneity of circulating melanoma cells in advanced stage patients. Phys. Biol. 12:016008 10.1088/1478-3975/12/1/016008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackstein R. (2004). The bone marrow is akin to skin: HCELL and the biology of hematopoietic stem cell homing. J. Invest. Dermatol. 122 1061–1069. 10.1111/j.0022-202X.2004.09301.x [DOI] [PubMed] [Google Scholar]
- Sakaizawa K., Goto Y., Kiniwa Y., Uchiyama A., Harada K., Shimada S., et al. (2012). Mutation analysis of BRAF and KIT in circulating melanoma cells at the single cell level. Br. J. Cancer 106 939–946. 10.1038/bjc.2012.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanga C. L., Handel T. M. (2011). Chemokine oligomerization and interactions with receptors and glycosaminoglycans: the role of structural dynamics in function. Exp. Cell Res. 317 590–601. 10.1016/j.yexcr.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanti A., Clausen T. M., Agerbaek M. O., Al Nakouzi N., Dahlback M., Oo H. Z., et al. (2015). Targeting human cancer by a glycosaminoglycan binding malaria protein. Cancer Cell 28 500–514. 10.1016/j.ccell.2015.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sand N. T., Petersen T. B., Bang-Christensen S. R., Ahrens T. D., Loppke C., Jorgensen A. M., et al. (2020). Optimization of rVAR2-based isolation of cancer cells in blood for building a robust assay for clinical detection of circulating tumor cells. Int. J. Mol. Sci. 21:2401. 10.3390/ijms21072401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson R. D., Elkin M., Rapraeger A. C., Ilan N., Vlodavsky I. (2017). Heparanase regulation of cancer, autophagy and inflammation: new mechanisms and targets for therapy. FEBS J. 284 42–55. 10.1111/febs.13932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra M., Eichstetter I., Iozzo R. V. (2000). An anti-oncogenic role for decorin: down-regulation of ErbB2 leads to growth suppression and cytodifferentiation of mammary carcinoma cells. J. Biol. Chem. 275 35153–35161. 10.1074/jbc.M006821200 [DOI] [PubMed] [Google Scholar]
- Saoncella S., Echtermeyer F., Denhez F., Nowlen J. K., Mosher D. F., Robinson S. D., et al. (1999). Syndecan-4 signals cooperatively with integrins in a Rho-dependent manner in the assembly of focal adhesions and actin stress fibers. Proc. Natl. Acad. Sci. U.S.A. 96 2805–2810. 10.1073/pnas.96.6.2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoia P., Fava P., Casoni F., Cremona O. (2019). Targeting the ERK signaling pathway in melanoma. Int. J. Mol. Sci. 20:1483. 10.3390/ijms20061483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada Y., Yoshikawa T., Nobuoka D., Shirakawa H., Kuronuma T., Motomura Y., et al. (2012). Phase I trial of a glypican-3-derived peptide vaccine for advanced hepatocellular carcinoma: immunologic evidence and potential for improving overall survival. Clin. Cancer Res. 18 3686–3696. 10.1158/1078-0432.CCR-11-3044 [DOI] [PubMed] [Google Scholar]
- Schaefer L., Schaefer R. M. (2010). Proteoglycans: from structural compounds to signaling molecules. Cell Tissue Res. 339 237–246. 10.1007/s00441-009-0821-y [DOI] [PubMed] [Google Scholar]
- Schaefer L., Tredup C., Gubbiotti M. A., Iozzo R. V. (2017). Proteoglycan neofunctions: regulation of inflammation and autophagy in cancer biology. FEBS J. 284 10–26. 10.1111/febs.13963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schehr J. L., Schultz Z. D., Warrick J. W., Guckenberger D. J., Pezzi H. M., Sperger J. M., et al. (2016). High specificity in circulating tumor cell identification is required for accurate evaluation of programmed death-ligand 1. PLoS One 11:e0159397. 10.1371/journal.pone.0159397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick B. P., Gradowski J. F., San Antonio J. D. (2001). Synthesis, secretion, and subcellular localization of serglycin proteoglycan in human endothelial cells. Blood 97 449–458. 10.1182/blood.v97.2.449 [DOI] [PubMed] [Google Scholar]
- Schochter F., Friedl T. W. P., Degregorio A., Krause S., Huober J., Rack B., et al. (2019). Are Circulating Tumor Cells (CTCs) ready for clinical use in breast cancer? An overview of completed and ongoing trials using CTCs for clinical treatment decisions. Cells 8:1412. 10.3390/cells8111412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze K., Gasch C., Staufer K., Nashan B., Lohse A. W., Pantel K., et al. (2013). Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int. J. Cancer 133 2165–2171. 10.1002/ijc.28230 [DOI] [PubMed] [Google Scholar]
- Shaw Bagnall J., Byun S., Begum S., Miyamoto D. T., Hecht V. C., Maheswaran S., et al. (2015). Deformability of tumor cells versus blood cells. Sci. Rep. 5:18542. 10.1038/srep18542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani Y., Takashima S., Asano Y., Kato H., Liao Y., Yamazaki S., et al. (2006). Glycosaminoglycan modification of neuropilin-1 modulates VEGFR2 signaling. EMBO J. 25 3045–3055. 10.1038/sj.emboj.7601188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M., Raposo G. (2009). Exosomes–vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21 575–581. 10.1016/j.ceb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- Smetsers T. F., Van De Westerlo E. M., Ten Dam G. B., Overes I. M., Schalkwijk J., Van Muijen G. N., et al. (2004). Human single-chain antibodies reactive with native chondroitin sulfate detect chondroitin sulfate alterations in melanoma and psoriasis. J. Invest. Dermatol. 122 707–716. 10.1111/j.0022-202X.2004.22316.x [DOI] [PubMed] [Google Scholar]
- Soker S., Miao H. Q., Nomi M., Takashima S., Klagsbrun M. (2002). VEGF165 mediates formation of complexes containing VEGFR-2 and neuropilin-1 that enhance VEGF165-receptor binding. J. Cell. Biochem. 85 357–368. 10.1002/jcb.10140 [DOI] [PubMed] [Google Scholar]
- Soker S., Takashima S., Miao H. Q., Neufeld G., Klagsbrun M. (1998). Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 92 735–745. 10.1016/s0092-8674(00)81402-6 [DOI] [PubMed] [Google Scholar]
- Song K., Li Q., Jiang Z. Z., Guo C. W., Li P. (2011). Heparan sulfate D-glucosaminyl 3-O-sulfotransferase-3B1, a novel epithelial-mesenchymal transition inducer in pancreatic cancer. Cancer Biol. Ther. 12 388–398. 10.4161/cbt.12.5.15957 [DOI] [PubMed] [Google Scholar]
- Sosa M. S., Bragado P., Aguirre-Ghiso J. A. (2014). Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat. Rev. Cancer 14 611–622. 10.1038/nrc3793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallcup W. B. (2002). The NG2 proteoglycan: past insights and future prospects. J. Neurocytol. 31 423–435. 10.1023/a:1025731428581 [DOI] [PubMed] [Google Scholar]
- Stallcup W. B. (2017). NG2 proteoglycan enhances brain tumor progression by promoting beta-1 integrin activation in both Cis and trans orientations. Cancers 9:31. 10.3390/cancers9040031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert G., Scholch S., Niemietz T., Iwata N., Garcia S. A., Behrens B., et al. (2014). Immune escape and survival mechanisms in circulating tumor cells of colorectal cancer. Cancer Res. 74 1694–1704. 10.1158/0008-5472.CAN-13-1885 [DOI] [PubMed] [Google Scholar]
- Stott S. L., Lee R. J., Nagrath S., Yu M., Miyamoto D. T., Ulkus L., et al. (2010). Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci. Transl. Med. 2:25ra23. 10.1126/scitranslmed.3000403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati A., Kasimir-Bauer S., Markou A., Parisi C., Lianidou E. S. (2013). Comparison of three molecular assays for the detection and molecular characterization of circulating tumor cells in breast cancer. Breast Cancer Res. 15:R20. 10.1186/bcr3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strell C., Entschladen F. (2008). Extravasation of leukocytes in comparison to tumor cells. Cell Commun. Signal. 6:10. 10.1186/1478-811X-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strilic B., Offermanns S. (2017). Intravascular survival and extravasation of tumor cells. Cancer Cell 32 282–293. 10.1016/j.ccell.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Sweet M. B., Thonar E. M., Immelman A. R. (1976). Glycosaminoglycans and proteoglycans of human chondrosarcoma. Biochim. Biophys. Acta 437 71–86. 10.1016/0304-4165(76)90348-2 [DOI] [PubMed] [Google Scholar]
- Tarbell J. M., Cancel L. M. (2016). The glycocalyx and its significance in human medicine. J. Intern. Med. 280 97–113. [DOI] [PubMed] [Google Scholar]
- Tatrai P., Egedi K., Somoracz A., Van Kuppevelt T. H., Ten Dam G., Lyon M., et al. (2010). Quantitative and qualitative alterations of heparan sulfate in fibrogenic liver diseases and hepatocellular cancer. J. Histochem. Cytochem. 58 429–441. 10.1369/jhc.2010.955161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam G. B., Van De Westerlo E. M., Purushothaman A., Stan R. V., Bulten J., Sweep F. C., et al. (2007). Antibody GD3G7 selected against embryonic glycosaminoglycans defines chondroitin sulfate-E domains highly up-regulated in ovarian cancer and involved in vascular endothelial growth factor binding. Am. J. Pathol. 171 1324–1333. 10.2353/ajpath.2007.070111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theocharis A. D., Skandalis S. S., Neill T., Multhaupt H. A. B., Hubo M., Frey H., et al. (2015). Insights into the key roles of proteoglycans in breast cancer biology and translational medicine. Biochim. Biophys. Acta 1855 276–300. 10.1016/j.bbcan.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S. N., Zhu F., Schnaar R. L., Alves C. S., Konstantopoulos K. (2008). Carcinoembryonic antigen and CD44 variant isoforms cooperate to mediate colon carcinoma cell adhesion to E- and L-selectin in shear flow. J. Biol. Chem. 283 15647–15655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. A., Purushothaman A., Ramani V. C., Vlodavsky I., Sanderson R. D. (2013). Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J. Biol. Chem. 288 10093–10099. 10.1074/jbc.C112.444562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsson M., Soletormos G., Jess P. (2011). Low number of detectable circulating tumor cells in non-metastatic colon cancer. Anticancer Res. 31 613–617. [PubMed] [Google Scholar]
- Tiedemann K., Olander B., Eklund E., Todorova L., Bengtsson M., Maccarana M., et al. (2005). Regulation of the chondroitin/dermatan fine structure by transforming growth factor-beta1 through effects on polymer-modifying enzymes. Glycobiology 15 1277–1285. 10.1093/glycob/cwj027 [DOI] [PubMed] [Google Scholar]
- Toba-Ichihashi Y., Yamaoka T., Ohmori T., Ohba M. (2016). Up-regulation of Syndecan-4 contributes to TGF-beta1-induced epithelial to mesenchymal transition in lung adenocarcinoma A549 cells. Biochem. Biophys. Rep. 5 1–7. 10.1016/j.bbrep.2015.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro M., Gaggianesi M., Catalano V., Benfante A., Iovino F., Biffoni M., et al. (2014). CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell 14 342–356. 10.1016/j.stem.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Toledo A. G., Pihl J., Spliid C. B., Persson A., Nilsson J., Pereira M. A., et al. (2020). An Affinity-Chromatography and Glycoproteomics Workflow to Profile the Chondroitin Sulfate Proteoglycans that interact with malarial VAR2CSA in the Placenta and in Cancer. Glycobiology. 10.1093/glycob/cwaa039[Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toole B. P. (2001). Hyaluronan in morphogenesis. Semin. Cell Dev. Biol. 12 79–87. [DOI] [PubMed] [Google Scholar]
- Toole B. P. (2004). Hyaluronan: from extracellular glue to pericellular cue. Nat. Rev. Cancer 4 528–539. 10.1038/nrc1391 [DOI] [PubMed] [Google Scholar]
- Toyama-Sorimachi N., Sorimachi H., Tobita Y., Kitamura F., Yagita H., Suzuki K., et al. (1995). A novel ligand for CD44 is serglycin, a hematopoietic cell lineage-specific proteoglycan. Possible involvement in lymphoid cell adherence and activation. J. Biol. Chem. 270 7437–7444. 10.1074/jbc.270.13.7437 [DOI] [PubMed] [Google Scholar]
- Toyoshima K., Hayashi A., Kashiwagi M., Hayashi N., Iwatsuki M., Ishimoto T., et al. (2015). Analysis of circulating tumor cells derived from advanced gastric cancer. Int. J. Cancer 137 991–998. 10.1002/ijc.29455 [DOI] [PubMed] [Google Scholar]
- Tsai W. S., Chen J. S., Shao H. J., Wu J. C., Lai J. M., Lu S. H., et al. (2016). Circulating tumor cell count correlates with colorectal neoplasm progression and is a prognostic marker for distant metastasis in non-metastatic patients. Sci. Rep. 6:24517. 10.1038/srep24517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao S. C.-H., Wang J., Wang Y., Behren A., Cebon J., Trau M. (2018). Characterising the phenotypic evolution of circulating tumour cells during treatment. Nat. Commun. 9:1482. 10.1038/s41467-018-03725-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley E. A., Noble P. W., Bourguignon L. Y. (2002). Signaling properties of hyaluronan receptors. J. Biol. Chem. 277 4589–4592. [DOI] [PubMed] [Google Scholar]
- Ulmer A., Schmidt-Kittler O., Fischer J., Ellwanger U., Rassner G., Riethmüller G., et al. (2004). Immunomagnetic enrichment, genomic characterization, and prognostic impact of circulating melanoma cells. Clin. Cancer Res. 10 531–537. 10.1158/1078-0432.ccr-0424-03 [DOI] [PubMed] [Google Scholar]
- Valiyaveettil M., Achur R. N., Muthusamy A., Gowda D. C. (2004). Characterization of chondroitin sulfate and dermatan sulfate proteoglycans of extracellular matrices of human umbilical cord blood vessels and Wharton’s jelly. Glycoconj. J. 21 361–375. 10.1023/B:GLYC.0000046276.77147.b2 [DOI] [PubMed] [Google Scholar]
- van Zijl F., Krupitza G., Mikulits W. (2011). Initial steps of metastasis: cell invasion and endothelial transmigration. Mutat. Res. 728 23–34. 10.1016/j.mrrev.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente C. M., Lima M. A., Yates E. A., Yates H. B., Toma L. (2015). Enhanced tumorigenic potential of colorectal cancer cells by extracellular sulfatases. Mol. Cancer Res. 13 510–523. 10.1158/1541-7786.MCR-14-0372 [DOI] [PubMed] [Google Scholar]
- Vijaya Kumar A., Salem Gassar E., Spillmann D., Stock C., Sen Y. P., Zhang T., et al. (2014). HS3ST2 modulates breast cancer cell invasiveness via MAP kinase- and Tcf4 (Tcf7l2)-dependent regulation of protease and cadherin expression. Int. J. Cancer 135 2579–2592. 10.1002/ijc.28921 [DOI] [PubMed] [Google Scholar]
- Vitale D., Kumar Katakam S., Greve B., Jang B., Oh E. S., Alaniz L., et al. (2019). Proteoglycans and glycosaminoglycans as regulators of cancer stem cell function and therapeutic resistance. FEBS J. 286 2870–2882. 10.1111/febs.14967 [DOI] [PubMed] [Google Scholar]
- Vogel A., Cervantes A., Chau I., Daniele B., Llovet J. M., Meyer T., et al. (2019). Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 30 871–873. [DOI] [PubMed] [Google Scholar]
- Walimbe T., Panitch A. (2019). Proteoglycans in biomedicine: resurgence of an underexploited class of ECM molecules. Front. Pharmacol. 10:1661. 10.3389/fphar.2019.01661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Liu X., Hsieh B., Bruce R., Somlo G., Huang J., et al. (2015). Exploring glycan markers for immunotyping and precision-targeting of breast circulating tumor cells. Arch. Med. Res. 46 642–650. 10.1016/j.arcmed.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. L., Anatelli F., Zhai Q. J., Adley B., Chuang S. T., Yang X. J. (2008). Glypican-3 as a useful diagnostic marker that distinguishes hepatocellular carcinoma from benign hepatocellular mass lesions. Arch. Pathol. Lab. Med. 132 1723–1728. 10.1043/1543-2165-132.11.1723 [DOI] [PubMed] [Google Scholar]
- Wang J., Svendsen A., Kmiecik J., Immervoll H., Skaftnesmo K. O., Planagumà J., et al. (2011). Targeting the NG2/CSPG4 proteoglycan retards tumour growth and angiogenesis in preclinical models of GBM and melanoma. PLoS One 6:e23062. 10.1371/journal.pone.0023062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Xiao Y., Lin L., Zhu X., Du L., Shi X. (2018). A microfluidic chip integrated with hyaluronic acid-functionalized electrospun chitosan nanofibers for specific capture and nondestructive release of CD44-overexpressing circulating tumor cells. Bioconj. Chem. 29 1081–1090. 10.1021/acs.bioconjchem.7b00747 [DOI] [PubMed] [Google Scholar]
- Wang N., Tesfaluul N., Li J., Gao X., Liu S., Yue B. (2019). Enrichment of circulating myeloma cells by immunomagnetic beads combined with flow cytometry for monitoring minimal residual disease and relapse in patients with multiple myeloma. Ann. Hematol. 98 2769–2780. 10.1007/s00277-019-03833-5 [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhao K., Hackert T., Zoller M. (2018). CD44/CD44v6 a reliable companion in cancer-initiating cell maintenance and tumor progression. Front. Cell Dev. Biol. 6:97. 10.3389/fcell.2018.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Uehara Y., Yamashita N., Fujimura Y., Nishio K., Sawada T., et al. (2014). Multicolor detection of rare tumor cells in blood using a novel flow cytometry-based system. Cytometry A 85 206–213. 10.1002/cyto.a.22422 [DOI] [PubMed] [Google Scholar]
- Watanabe T., Okumura T., Hirano K., Yamaguchi T., Sekine S., Nagata T., et al. (2017). Circulating tumor cells expressing cancer stem cell marker CD44 as a diagnostic biomarker in patients with gastric cancer. Oncol. Lett. 13 281–288. 10.3892/ol.2016.5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R. R., Sun D. N., Yang H., Yan J., Zhang X., Zheng X. L., et al. (2018). CTC clusters induced by heparanase enhance breast cancer metastasis. Acta Pharmacol. Sin. 39 1326–1337. 10.1038/aps.2017.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel P. H., DeAngelis P. L. (2007). Hyaluronan synthases: a decade-plus of novel glycosyltransferases. J. Biol. Chem. 282 36777–36781. 10.1074/jbc.R700036200 [DOI] [PubMed] [Google Scholar]
- Weiss A., Attisano L. (2013). The TGFbeta superfamily signaling pathway. Wiley Interdiscip. Rev. Dev. Biol. 2 47–63. [DOI] [PubMed] [Google Scholar]
- Werner S. L., Graf R. P., Landers M., Valenta D. T., Schroeder M., Greene S. B., et al. (2015). Analytical validation and capabilities of the epic CTC platform: enrichment-free circulating tumour cell detection and characterization. J. Circ. Biomark. 4:3. 10.5772/60725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods E. C., Kai F., Barnes J. M., Pedram K., Pickup M. W., Hollander M. J., et al. (2017). A bulky glycocalyx fosters metastasis formation by promoting G1 cell cycle progression. eLife 6:e25752. 10.7554/eLife.25752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Raba K., Guglielmi R., Behrens B., Van Dalum G., Flugen G., et al. (2020). Magnetic-based enrichment of rare cells from high concentrated blood samples. Cancers 12:E933. 10.3390/cancers12040933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhou B. P. (2010). TNF-α/NF-κB/Snail pathway in cancer cell migration and invasion. Br. J. Cancer 102 639–644. 10.1038/sj.bjc.6605530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Esko J. D. (2014). Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 83 129–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Chen X., Huang Z. (2019). Identification of ESM1 overexpressed in head and neck squamous cell carcinoma. Cancer Cell Int. 19:118. 10.1186/s12935-019-0833-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Lamouille S., Derynck R. (2009). TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 19 156–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Vilanova G., Gomez H. (2016). A mathematical model coupling tumor growth and angiogenesis. PLoS One 11:e0149422. 10.1371/journal.pone.0149422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M., Kimata K., Oike Y., Tani K., Maeda N., Yoshida K., et al. (1987). A monoclonal antibody that specifically recognizes a glucuronic acid 2-sulfate-containing determinant in intact chondroitin sulfate chain. J. Biol. Chem. 262 4146–4152. [PubMed] [Google Scholar]
- Yan-Bin S., Guang-Hao S., Shun X., Jing-Jing X. (2020). Tumor-suppressive activity of sTRAIL on circulating CD44+ cells in patients with non-small cell lung cancer. Biol. Chem. 401 417–422. 10.1515/hsz-2019-0339 [DOI] [PubMed] [Google Scholar]
- Yang C., Xia B. R., Jin W. L., Lou G. (2019). Circulating tumor cells in precision oncology: clinical applications in liquid biopsy and 3D organoid model. Cancer Cell Int. 19:341. 10.1186/s12935-019-1067-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Price M. A., Li G. Y., Bar-Eli M., Salgia R., Jagedeeswaran R., et al. (2009). Melanoma proteoglycan modifies gene expression to stimulate tumor cell motility, growth, and epithelial-to-mesenchymal transition. Cancer Res. 69 7538–7547. 10.1158/0008-5472.CAN-08-4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. D., Campion M. B., Liu M. C., Chaiteerakij R., Giama N. H., Ahmed Mohammed H., et al. (2016). Circulating tumor cells are associated with poor overall survival in patients with cholangiocarcinoma. Hepatology 63 148–158. 10.1002/hep.27944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Wang Y., Yin H., Chen W., Jin G., Ma H., et al. (2015). Circulating tumor cells enriched by the depletion of leukocytes with Bi-antibodies in non-small cell lung cancer: potential clinical application. PLoS One 10:e0137076. 10.1371/journal.pone.0137076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A., Lendorf M. E., Couchman J. R., Multhaupt H. A. (2012). Breast and ovarian cancers: a survey and possible roles for the cell surface heparan sulfate proteoglycans. J. Histochem. Cytochem. 60 9–21. 10.1369/0022155411428469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Chan H., Wei L., Pan Z., Zhang J., Li L. (2013). Overexpression of heparanase in ovarian cancer and its clinical significance. Oncol. Rep. 30 2279–2287. 10.3892/or.2013.2701 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Shiratsuchi H., Lin J., Chen G., Reddy R. M., Azizi E., et al. (2014). Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget 5 12383–12397. 10.18632/oncotarget.2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhangyuan G., Wang F., Zhang H., Jiang R., Tao X., Yu D., et al. (2020). VersicanV1 promotes proliferation and metastasis of hepatocellular carcinoma through the activation of EGFR–PI3K–AKT pathway. Oncogene 39 1213–1230. 10.1038/s41388-019-1095-9 [DOI] [PubMed] [Google Scholar]
- Zhu Y. H., Yang F., Zhang S. S., Zeng T. T., Xie X., Guan X. Y. (2013). High expression of biglycan is associated with poor prognosis in patients with esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 6 2497–2505. [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. W., Friess H., Wang L., Abou-Shady M., Zimmermann A., Lander A. D., et al. (2001). Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut 48 558–564. 10.1136/gut.48.4.558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. (2011). CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 11 254–267. 10.1038/nrc3023 [DOI] [PubMed] [Google Scholar]