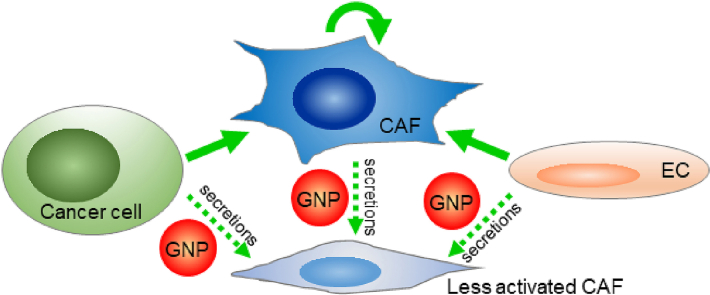
Abstract
Cancer-associated fibroblasts (CAFs) are a major constituent of the tumor microenvironment (TME) and an important contributor to cancer progression and therapeutic resistance. Regulation of CAF activation is a promising strategy to influence cancer outcomes. Here, we report that ovarian cancer cells (OCs) and TME cells promote the activation of ovarian CAFs, whereas gold nanoparticles (GNPs) of 20 nm in diameter inhibit the activation, as demonstrated by the changes in cell morphology, migration, and molecular markers. GNPs exert the effect by altering the levels of multiple fibroblast activation or inactivation proteins, such as TGF-β1, PDGF, uPA and TSP1, secreted by OCs and TME cells. Thus, GNPs represent a potential tool to help understand multicellular communications existing in the TME as well as devise strategies to disrupt the communication.
Keywords: Gold nanoparticle (GNP), Cancer-associated fibroblast (CAF), Tumor microenvironment (TME), Fibroblast activation, Morphology, Migration
Graphical abstract
Highlights
-
•
OCs and TME cells promote the activation of ovarian CAFs.
-
•
GNPs of 20 nm in diameter inhibit ovarian CAFs activation induced by OCs and TME cells.
-
•
GNPs alter the levels of fibroblast activation or inactivation proteins secreted by OCs and TME cells.
1. Introduction
Cancer-associated fibroblasts (CAFs) are a major component of the tumor microenvironment (TME) and regulate cancer metabolism, angiogenesis, immunity, growth, migration, invasion, metastasis and therapeutic resistance [[1], [2], [3]]. The crosstalk between CAFs and cancer cells or TME cells, such as endothelial cells (ECs), maintains the mutual activation state of the cells and promotes tumor cell viability and survival [4]. Therapeutic strategies to disrupt the link between CAFs and cancer cells have great promise and include: eliminating or reprogramming CAFs; eliciting immune responses to CAF antigens; and blocking CAF-secreted factors and cancer cell receptors by small molecule inhibitors, vaccines, antibodies or cells [1,5]. Understanding the molecular machineries responsible for multicellular communication is essential for devising effective therapeutic strategies. Due to their self-therapeutic nature [6], gold nanoparticles (GNPs) have a critical role to play in elucidating the multicellular communications that exist in TME.
Based on their tunable optical, chemical and biological characteristics, GNPs are under investigation in many preclinical settings [7]. Upon interacting with a biological system, either in vivo or in vitro, nanoparticle adsorbs various molecules to rapidly form a protein corona on the surface; this protein corona significantly impacts the biological properties of the particle [8]. The composition and structure of the corona is dynamic and dependent on both specific and non-specific interactions between that particle and proteins. Previously, we reported the physicochemical parameters responsible for the evolution of the dynamic protein corona around 20-nm diameter GNPs [9,10]. We have also demonstrated the anti-angiogenic properties of GNPs; GNPs inhibited several heparin-binding growth factors, and thereby, suppressed the growth of both ovarian and pancreatic tumors [[9], [10], [11]]. We further reported the ability of GNPs to transform activated pancreatic CAFs to quiescence as well as to inhibit EC angiogenic phenotype in vitro by disrupting cellular communications from cancer cells and CAFs [12,13]. In the current study, we examine the effects of ovarian cancer cells (OCs), ECs and CAFs themselves on CAF activation in vitro, and whether and how GNPs can reverse the process.
2. Materials and methods
2.1. Preparation of 20 nm GNPs
GNPs of 20 nm in diameter were prepared as described previously [12,13]. Briefly, 5 ml of 10 mM tetrachloroauric acid trihydrate (HAuCl4·3H2O, 520918, Sigma-Aldrich, St. Louis, MO) in 185 ml of endotoxin-free water (786671, G Biosciences, St. Louis, MO) was boiled in a 500 ml flask. Fifteen milliliters of 1% sodium citrate (1613859, Sigma-Aldrich) was preheated to 70 °C and added to the flask 3 min later. The solution was boiled for 15 min with stirring till the color changed to dark purple. The solution was moved to room temperature (RT) and stirred for 16 h. The GNPs were concentrated by centrifugation at 10000 rpm at 10 °C for 25 min before use. To determine GNP concentration, the original solution was concentrated 16 times, and OD522 and OD800 of both original and concentrated solutions were measured. Concentration of the original preparation was calculated as 30.018 × Initial Volume × (OD520-OD800) of As Concentrated/[As Synthesized Volume × (OD520-OD800) of As Synthesized].
2.2. GNP characterization
The GNPs were initially characterized with UV–visible spectroscopy (Spectrostar Nano, BMG Labtech, Cary, NC), dynamic light scattering (Zetasizer Nano ZS, Malvern Panalytical, Malvern, UK) and zeta potential measurements (Zetasizer Nano ZS). The Energy-dispersive X-ray spectroscopy (EDS) elemental analysis was performed using Kevex Quantum 10 mm2 X-ray detector. Powder X-ray diffraction (XRD) analyses were performed in the School of Sciences at the University of Oklahoma using a Rigaku Ultima IV diffractometer. Cu–K-alpha radiation (40 kV, 44 mA) was used in focusing geometry with a curved graphite monochromator and scintillation detector. Data analysis was completed using MDI Jade Pro with the ICDD (International Center for Diffraction Data) PDF4+ database. Transmission Electron Micrographs (TEM) were acquired using a 200-kV LaB6 JEOL 2000FX (JEOL, USA) equipped with a Gatan (1k x 1k) digital camera (Gatan, USA).
2.3. Cell culture
Human epithelial ovarian cancer cell lines: A2780-CP20 (CP20) was a kind gift from Dr. Anil K. Sood (MD Anderson Cancer Center, Houston, TX); OV90 was purchased from American Type Culture Collection (Manassas, VA); and OVCAR4 was purchased from the National Cancer Institute (Bethesda, MD). All the cells were grown in RPMI 1640 (10-040-CV, Corning) supplemented with 10% Fetal Bovine Serum (FBS, 16000–044, Life technologies Carlsbad, CA) and 1% Penn-Strep (15140–122, Life Technologies). Ovarian cancer associated fibroblasts (CAFs) were isolated and identified in this lab [13], and cultured in DMEM:F12 (10–090, Corning, NY) with 15% FBS and 1% Penn-Strep. The CAFs TAF18 and TAF19 were used up to passage 8 in this study. Human umbilical vein endothelial cells (HUVEC, Lonza, Walkersville, MD) and human microvascular endothelial cells (HMEC, a kind gift from Professor Xin Zhang, OUHSC Stephenson Cancer Center, OKC, OK) [14] were grown in Endothelial Cell Growth Medium-2 BulletKit (EGM, CC-3162) and used up to passage 8. All cells were kept at 37 °C in humidified 95% air and 5% CO2.
2.4. Preparation of cell supernatants
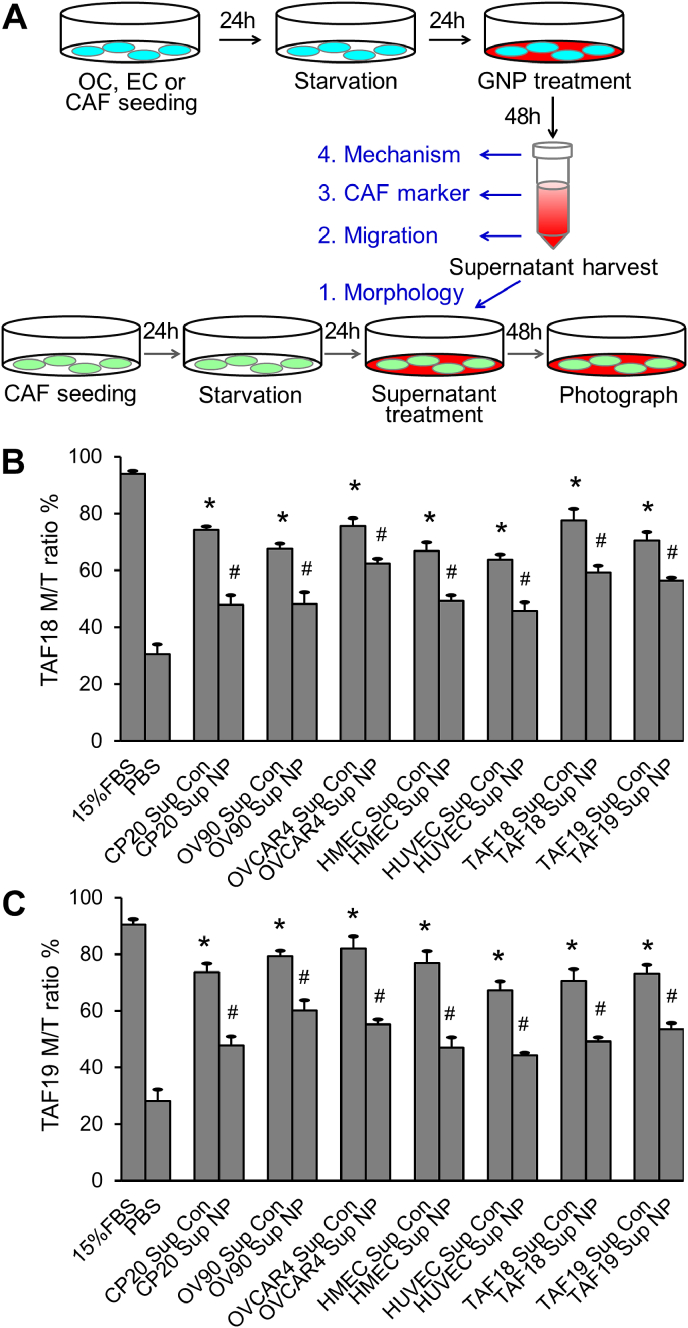
Supernatants from untreated or GNP-treated cancer cells, CAFs, or ECs were prepared as described previously [11,13] and illustrated in Fig. 2A. Briefly, 0.7 million cells were seeded onto culture dishes (T1110, Thomas Scientific) of 10 cm in diameter. Media were replaced with serum-free RPMI 1640 for cancer cells, DMEM:F12 for CAFs, or Endothelial Cell Basal Medium-2 (EBM, CC-3156, Lonza) for ECs. The media were replaced again 24 h later with fresh serum-free media (SFM) with or without 40 μg/ml 20-nm GNPs for 48 h. The media were collected, centrifuged at 1200 rpm for 6 min to remove cell debris, then centrifuged at 10000 rpm at 10 °C for 25 min to remove the remaining GNPs, and used immediately or stored at −80 °C. The supernatants were diluted with an equal volume of SFM before functional assays or used undiluted for mechanistic studies.
Fig. 2.
Supernatants of OCs, CAFs or ECs treated with GNPs change CAF morphology. (A) Schematic view of cell supernatant preparation and morphology assay. (B) TAF18, (C) TAF19: radio of multi-spindled (M) cell number to "multi-spindled + single-spindled (total)" cell number (M/T ratio). Experiments were performed in triplicate and repeated 3 times. Sup: supernatant, Con: control, NP: GNP. *, p < 0.05, compared to PBS, #, p < 0.05, compared to corresponding Con.
2.5. Morphology assay
TAF18 or TAF19 cells were seeded 10000 cells/well to 24-well plates. After starvation with serum-free DMEM:F12 medium for 24 h, cells were treated with supernatants collected from different cells treated with or without GNP (40 μg/ml) for 48 h. The supernatants were diluted with equal volumes of fresh DMEM:F12 before use. Cells were photographed using phase contrast microscope (Fig. 2A). Multi-spindled fibroblasts and single-spindled fibroblasts were counted with ImageJ. Ratios of multi-spindled (M) to “multi-spindled + single-spindled (total)” (M/T ratio) were calculated. 15% FBS in DMEM:F12 medium or PBS diluted with an equal volume of DMEM:F12 were respectively used as positive or non-treatment controls. Experiments were performed in triplicate and repeated 3 times.
2.6. Migration assay
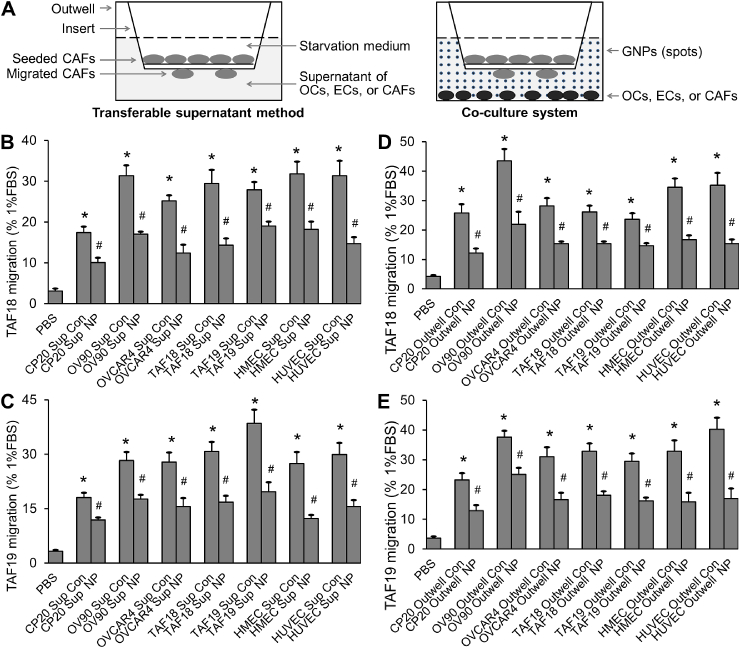
CAFs (TAF18 or TAF19) were starved in DMEM:F12 for 24 h, trypsinized, and seeded onto each transwell (3422, Corning) at 40,000 cells in 100 μl. CAF migration was induced by supernatants diluted with DMEM:F12 of equal volume for 16 h. In the co-culture system, cancer cells, CAFs, or ECs were seeded to outwells at 50,000 cells/well. The cells were starved in SFM for 24 h and then incubated with or without 40 μg/ml GNPs in 700 μl SFM for 36 h. Half the volume of the media was replaced by fresh SFM immediately before placing the migration inserts with starved CAFs to the plates for migration for 16 h (Fig. 3A). At the termination of the experiments, cells were fixed and stained with 0.2% crystal violet (C0775, Sigma-Aldrich). Cells inside the inserts were removed using cotton swabs. Cells that had migrated across the membrane were photographed and counted with ImageJ. DMEM: F12 containing 1% FBS or PBS diluted with an equal volume of DMEM:F12 were respectively used as the positive or nontreatment controls. For quantification, migration by 1% FBS was set as 100%. Migration fold increase was calculated as (Control supernatant or co-culture − PBS)/PBS; decrease percentage as (Control − GNPs)/Control × 100. Experiments were performed in duplicate and repeated 3 times.
Fig. 3.
Supernatants of OCs, CAFs or ECs treated with GNPs, or co-cultured OCs, CAFs or ECs treated with GNP, inhibit CAF migration. (A) Schematic view of migration assays. (B, C) Migration induced by transferable supernatants. TAF18 or TAF19 migration quantification, expressed as percentage of migration induced by 1% FBS medium (positive control). (D, E) Migration induced by co-cultured cells. TAF18 or TAF19 migration quantification, expressed as percentage of migration induced by 1% FBS medium. Experiments were performed in duplicate and repeated 3 times. Sup: supernatant, Outwell: CAF co-cultured cell, Con: control, NP: GNP. *, p < 0.05, compared to PBS, #, p < 0.05, compared to corresponding Con.
2.7. Western blotting
TAF18 and TAF19 cells were starved for 24 h and then incubated with supernatants from different cells incubated with or without 40 μg/ml GNPs for 48 h. Cells were collected after washing with cold PBS three times, and lysed with RIPA buffer (BP-115, Boston BioProducts, Ashland, MA) containing Protease Inhibitor Cocktail (78440, ThermoFisher). Lysate protein concentration was determined using BCA Protein Assay Kit (23227, Thermo-Fisher). Lysates (5 μg proteins/lane) were separated on 10% or 6% SDS-PAGE gels and transferred to PVDF membranes (1620177, Bio-Rad, Hercules, CA). The membranes were blocked in 4% BSA/PBST (0.1% Tween-20, P1379, Sigma-Aldrich, in PBS) at RT for 1 h, and then incubated with primary antibodies at 4 °C for 16 h. HRP-coupled secondary antibodies were applied after washing for 1 h at RT. Signals were visualized with Clarity Western ECL Substrates (1705061, Bio-Rad). Blots were imaged with Color LaserJet Pro MFO M477fdn (HP, Palo Alto, CA). Primary antibodies used in this study were as follows: mouse anti-fibronectin (1:1000, #610077, BD Biosciences, San Jose, CA), rabbit anti-α-SMA (1:1000, ab5694, Abcam, Burlingame, CA), and mouse anti-Tubulin (1:5000, #66031, Proteintech, Rosemont, IL). Secondary antibodies were as follows: goat anti-rabbit IgG (1:10000, A6154, Sigma) and goat anti-mouse IgG (1:10000, A4416, Sigma).
2.8. Antibody array assay
Supernatants (1 ml each) from CP20 or TAF18 cells treated with or without 40 μg/ml GNPs for 48 h were incubated with a cocktail of biotinylated detection antibodies from the Proteome Profiler Human Angiogenesis Antibody Array kit (# ARY007, R&D Systems, Minneapolis, MN) at RT for 1 h. The mixtures were then incubated with antibody array membranes at 4 °C for 16 h. Captured proteins were visualized by streptavidin-HRP and chemiluminescent detection reagents. Films were developed for three different exposure times. Blot images were scanned and the pixel density of the spots was analyzed using ImageJ. Ratio of GNP-treated to control was calculated. Images of the 3 exposure times were analyzed, and overexposed or underexposed spots were excluded.
2.9. Statistics
Data were expressed as mean ± SEM. Statistics was performed with one-way ANOVA Tukey's post-test. P ≤ 0.05 was considered significant statistically.
3. Results and discussion
3.1. Characterizations of GNPs
In our previous work we compared GNPs of 5, 10, 20, 50 or 100 nm diameter and showed that those of 20 nm inhibited the proliferation of a variety of cell types most effectively [9,10]. We thus prepared 20 nm GNPs as detailed in the Materials and Methods section. The hydrodynamic diameter (Fig. 1A) and charge (Fig. 1B) of these GNPs were about 24.5 nm and −42.1 mV as determined by dynamic light scattering (DLS) and Zeta potential measurements, respectively. The UV–visible spectrum of as synthesized GNPs showed a surface plasmon resonance (SPR) band at around 520 nm (Fig. 1C), indicating the formation of spherical GNPs. The EDS spectrum (Fig. 1D) verified that the nanoparticles are made from gold. Powder XRD analysis (Fig. 1E) confirmed the crystalline structure of the GNPs with no other phases or impurities present. Observed diffraction peaks are appropriately broadened as expected for nanocrystals. The intensities of the dark color correspond to reported relative diffraction peak intensities. Diffraction peaks are indexed to corresponding diffraction planes (111), (200), and (220) according to gold metal with face centered cubic structure. Lattice constant determined from XRD pattern is a = 4.078 Å, which is in good agreement with standard diffraction pattern of cubic gold metal (ICDD Pattern No. 4–784). TEM (Fig. 1F) confirmed the size and spherical shape of the GNPs. Importantly, lattice fringes (bright and dark stripes corresponding to the lattice spacing of the gold crystal lattice) can be clearly seen in the HRTEM micrograph (highlighted by white arrows) and confirm the crystalline structure of gold nanoparticles. As for the diffraction pattern (Fig. 1G) of the GNPs obtained using TEM, the diffraction rings for planes (111), (200), (220), and (311) are clearly visible and further approve crystallinity of the gold nanoparticles. These results identified the synthesized particles as gold nanoparticles with desired size, shape, structure and purity.
Fig. 1.
Characterizations of GNPs. The size (A), zeta potential (B) and UV–visible spectrum (C) of synthesized GNPs (D) Energy-dispersive X-ray spectroscopy (EDS) of the GNPs. The observed Cu signal is due to the copper grid used as substrate in TEM. (E) Power XRD analysis of the GNPs. Reference diffraction pattern is shown as a ribbon along the bottom of the figure. (F) TEM of GNPs. Low resolution (left) and high resolution (middle and right) TEM micrographs. Fringes of the gold crystal lattice are highlighted by white arrows. (G) Diffraction pattern of GNPs obtained using TEM. The diffraction rings correspond to (111), (200), (220) and (311) planes.
3.2. Supernatants of OCs or TME cells treated with GNPs change CAF morphology
CAFs are multi-spindled, migratory, highly secretory, and proliferative; in contrast, resting or quiescent fibroblasts are single-spindled and less active [1,2]. To assess if cancer cells or TME cells activate CAFs, and whether GNPs disrupt the process, we treated primary ovarian CAFs (TAF18 and TAF19 cells) with the supernatants of OCs (CP20, OV90 and OVCAR4), ECs (HMEC, HUVEC), or CAFs themselves, and determined changes in morphology. To prepare the supernatants (Fig. 2A), cells were seeded at 0.7 million/10 cm dish in complete serum containing media. After 24 h, media were replaced with serum-free media (SFM). After a further 24 h media were replaced with fresh SFM with or without 40 μg/ml GNPs and cells were incubated for 48 h. The media were then collected, centrifuged to remove cell debris and GNPs, and used to treat CAFs [11,13]. To assess CAF morphology (Fig. 2A), TAF18 or TAF19 cells were seeded at 10,000 cells/well in 24-well plates in complete media. After 24 h, cells were starved with serum free DMEM: F12 media for 24 h, and then treated for 48 h with the collected supernatants diluted with an equal volume of fresh serum-free DMEM:F12. These diluted supernatants provided adequate nutrition for the cells and cells remained in an observably healthy state. The positive control was DMEM: F12 containing 15% FBS, and PBS diluted with an equal volume of DMEM: F12 without serum was the non-treatment control. Following the specified treatments, cells were photographed using phase contrast microscope (Fig. S1). Multi-spindled fibroblasts and single-spindled fibroblasts were then counted using ImageJ, and the ratio of multi-spindled (M) to “multi-spindled + single-spindled (total, T)” was calculated (Fig. 2B and C); this M/T ratio is a measure of CAF activation. Treatment of both CAF cell lines with any supernatant resulted in relatively more multi-spindled fibroblasts than did PBS; GNPs inhibited the effect in all cases. The M/T ratio of CAFs treated with control supernatants increased in a range from 2.1 (for TAF18 treated with HUVEC control supernatant) to 2.9 (for TAF19 treated with OVCAR4 control supernatant) fold compared to PBS (Fig. 2B and C). However, treatment with supernatants from GNP-treated cells inhibited the morphology changes; the GNPs caused from a 17.5% (TAF18 treated with OVCAR4 GNP supernatant) to a 38.8% (TAF19 treated with HMEC GNP supernatant) decrease in the M/T ratio compared to control supernatant. These results suggest that OCs and TME cells (here the ECs and CAFs themselves) activate CAFs, and that GNPs inhibit this activation.
3.3. GNPs decrease CAF migration induced by OCs or TME cells
To support our findings, we examined the effects of OCs and TME cells as well as GNPs on CAF migration. Migration is another important characteristic of activated CAFs and we assessed it using both transferable supernatant and co-culture system (Fig. 3A) [13]. In the transferable supernatant method (Fig. S2A, 3A-3C), TAF18 and TAF19 cells were starved in serum-free DMEM:F12 for 24 h before trypsinization and seeding (40,000 cells/insert) to the inserts in Boyden chamber system. Cell migration was induced for 16 h by supernatants from CP20, OV90, OVCAR4, HMEC, HUVEC, TAF18 or TAF19 cells treated with or without 40 μg/ml GNPs. In the co-culture system (Fig. S2B, 3A, 3D, 3E), the migration-inducing cells (cancer cells, CAFs, or ECs) were seeded to 24-well plates at 50,000 cells per well – this served as the outwell in the Boyden chamber system. Cells were starved in SFM for 24 h starting on the following day, then cultured with or without 40 μg/ml GNPs in SFM for 36 h, and finally co-cultured for 16 h with TAF18 or TAF19 cells which were seeded to the inserts. For both methods, DMEM: F12 containing 1% FBS was used as positive control, while PBS diluted with equal volume of DMEM: F12 was used as non-treatment control. Migrating cells were stained, photographed and counted. Both supernatants and co-cultured cells stimulated CAF migration, while GNPs inhibited the effect. Control supernatants caused from 5.5 (TAF19 treated with CP20 control supernatant) to 11.7 (TAF19 treated with TAF19 control supernatant) fold increases in CAF migration compared to PBS (Fig. 3B and C). In contrast, supernatants from GNPs-treated cells decreased CAF migration by between 31.9% (TAF18 treated with TAF19 GNP supernatant) and 55.3% (TAF19 treated with HMEC GNP supernatant) compared to the corresponding controls. Similarly, nontreated co-cultured cells increased the migration of CAFs by between 5.6 (TAF18 co-cultured with TAF19) and 10.9 (TAF19 co-cultured with HUVEC) fold compared to PBS (Fig. 3D and E); GNP-treated co-cultured cells decreased the migration by 33.2% (TAF19 co-cultured with GNP-treated OV90) to 57.7% (TAF19 co-cultured with GNP-treated HUVEC) compared to the corresponding controls. These results confirm that OCs and TME cells activate CAFs, while GNPs reverse this activation.
3.4. GNPs inhibit the expression of CAF activation markers
We proceeded to examine changes in the molecular phenotype of CAFs that occur in response to GNP exposure. We assessed expression of αSMA and fibronectin as markers of CAF activation [2,15]. TAF18 cells were starved for 24 h and then incubated with supernatants from OCs and TME cells incubated with or without 40 μg/ml GNPs. Expression of αSMA and fibronectin was then determined by Western blot of cell lysates (5 μg). Both proteins were down-regulated by most supernatants from cells treated with GNPs, compared to the corresponding controls (Fig. 4). These data support our cellular phenotype changes as well as the effectiveness of GNPs for blunting the ability of OCs and TME cells to activate CAFs.
Fig. 4.
Expression of CAF activation markers in TAF18 incubated with supernatants from cells treated with or without GNP. Tubulin was used as loading control.
3.5. GNPs alter the secretions from OCs and TME cells
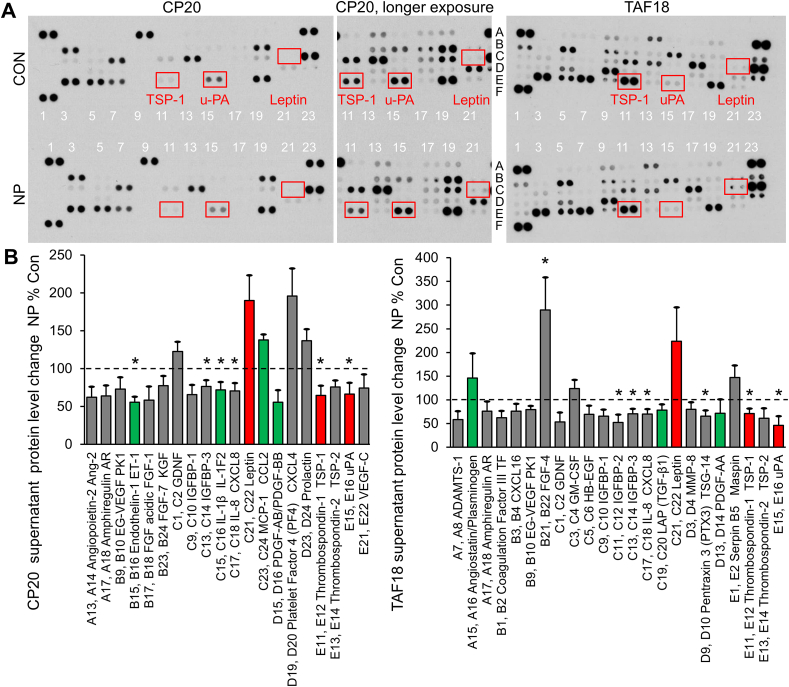
To investigate the underlying molecular mechanisms by which GNPs inhibit OCs- and TME-induced CAF activation, we focused on alterations in cytokines, growth factors, enzymes and extracellular matrix components in the supernatants of OCs and CAFs. It is known that some cytokines and growth factors, such as FGF2, HGF, IL6, PDGF, SDF1 and TGFβ induce CAF activation [16]. We performed an antibody array assay which could detect expression of 55 proteins associated with CAF activation, angiogenesis and tumor progression. Supernatants from CP20 or TAF18 cells treated with or without 40 μg/ml GNPs for 48 h were incubated with the antibody array. The intensity and size of the resulted dot blots reflect the expression of the detected proteins in the supernatants (Fig. 5A). To quantify the expression, pixel density of each spot was analyzed using ImageJ, and the ratios of GNP-treated supernatant to control were calculated. The levels of 20 proteins in CP20 supernatants were changed by at least 20% by GNP treatment (Fig. 5B). Among these proteins, Endothelin-1 [17], IL-1β [18], PDGF-AB/BB [16], CXCL4 [19] and uPA (urokinase-type plasminogen activator) [[20], [21], [22]] were shown to activate CAFs or fibroblasts, whereas leptin [23,24] and TSP1 [25,26] were shown to be context-dependent activators or inhibitors of fibroblasts. Endothelin-1, IL-1β, PDGF, TSP1 and uPA were found to be downregulated, whereas CXCL4 and leptin were upregulated, by GNPs [Fig. 5, colored]. Similarly, the levels of 23 proteins in TAF18 supernatants were changed by at least 20% by GNPs (Fig. 5B). TGF-β1 [16], PDGF-AA [16] and uPA were activators of CAFs or fibroblasts, plasminogen [22] was an inhibitor of fibroblasts, and leptin and TSP1 were context-dependent. TGF-β1, PDGF-AA, TSP1 and uPA were downregulated, whereas leptin was upregulated, by GNPs [Fig. 5, colored]. Noticeably, expression changes of uPA, leptin and TSP-1 were similar between CP20 and TAF18 supernatants [Fig. 5, red]. These results suggested that GNPs alter the levels of multiple proteins secreted by OCs and TME cells, and that the combination of the changes suppress the activation of CAFs.
Fig. 5.
Identification of GNP-altered CAF activation-related proteins. (A) Typical images of the antibody arrays incubated with CP20 or TAF18 supernatants. (B) Pixel density ratio of GNP to control spots. Images of 3 exposure times were analyzed, and overexposed or underexposed spots were excluded. Shown are those proteins with at least 20% level changes. Con: control, NP: GNP. *, p < 0.05.
4. Conclusions
We demonstrated that OCs and TME cells activate ovarian CAFs. GNPs of 20 nm in diameter inhibit the activation, as illustrated by the changes in cell morphology, migration, and molecular markers. Mechanistically, GNPs alter the levels of multiple fibroblast activation or inactivation proteins, and the combination of the changes disrupt the signaling to CAFs and depress their activation. This work provides evidence that GNPs can serve as an important tool to decipher multicellular communications within the TME and could help devise strategies to disrupt such communication.
CRediT authorship contribution statement
Yushan Zhang: Conceptualization, Data curation, Investigation, Methodology, Writing - review & editing. Chandra Kumar Elechalawar: Data curation, Investigation, Methodology, Validation. Md Nazir Hossen: Data curation, Investigation, Methodology, Validation. Emmy R. Francek: Data curation, Investigation, Methodology, Validation. Anindya Dey: Data curation, Investigation, Methodology, Validation. Stefan Wilhelm: Conceptualization, Funding acquisition, Supervision, Writing - review & editing. Resham Bhattacharya: Conceptualization, Funding acquisition, Supervision, Writing - review & editing. Priyabrata Mukherjee: Conceptualization, Funding acquisition, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This study was supported by National Institutes of Health Grants CA220237, CA136494, CA213278 (to P.M.) and HL120585 (to both P.M. and R.B.). We thank the Peggy and Charles Stephenson Cancer Center at the University of Oklahoma Health Sciences Center for a seed grant and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the NIH under grant number P20 GM103639 for the use of Histology and Immunohistochemistry Core. Research reported in this publication was also supported in part by the NCI Cancer Center Support Grant P30CA225520 awarded to the University of Oklahoma Stephenson Cancer Center and used the Office of Cancer Research. The authors thank Drs. Andy E. Madden, Preston Larson and Julian Sabisch for technical assistance with EDS, XRD and TEM. The authors thank Daniel Morton for editorial assistance.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2020.08.009.
Abbreviations
- αSMA
α‑smooth muscle actin
- CAF
cancer-associated fibroblast
- EC
endothelial cell
- GNP
gold nanoparticle
- OC
ovarian cancer cell
- SFM
serum-free media
- TME
tumor microenvironment
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Valkenburg K.C., de Groot A.E., Pienta K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018;15(6):366–381. doi: 10.1038/s41571-018-0007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalluri R. The biology and function of fibroblasts in cancer. Nat. Rev. Canc. 2016;16(9):582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 3.Koliaraki V., Henriques A., Prados A., Kollias G. Unfolding innate mechanisms in the cancer microenvironment: the emerging role of the mesenchyme. J. Exp. Med. 2020;217(4) doi: 10.1084/jem.20190457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsuyada A., Chow A., Wu J., Somlo G., Chu P., Loera S., Luu T., Li A.X., Wu X., Ye W., Chen S., Zhou W., Yu Y., Wang Y.Z., Ren X., Li H., Scherle P., Kuroki Y., Wang S.E. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Canc. Res. 2012;72(11):2768–2779. doi: 10.1158/0008-5472.CAN-11-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi H., Enomoto A., Woods S.L., Burt A.D., Takahashi M., Worthley D.L. Cancer-associated fibroblasts in gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol. 2019;16(5):282–295. doi: 10.1038/s41575-019-0115-0. [DOI] [PubMed] [Google Scholar]
- 6.Arvizo R.R., Bhattacharyya S., Kudgus R.A., Giri K., Bhattacharya R., Mukherjee P. Intrinsic therapeutic applications of noble metal nanoparticles: past, present and future. Chem. Soc. Rev. 2012;41(7):2943–2970. doi: 10.1039/c2cs15355f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabuzu D., Cirja A., Puiu R., Grumezescu A.M. Biomedical applications of gold nanoparticles. Curr. Top. Med. Chem. 2015;15(16):1605–1613. doi: 10.2174/1568026615666150414144750. [DOI] [PubMed] [Google Scholar]
- 8.Mahmoudi M., Bertrand N., Zope H., Farokhzad O.C. Emerging understanding of the protein corona at the nano-bio interfaces. Nano Today. 2006;11:817–832. [Google Scholar]
- 9.Arvizo R.R., Rana S., Miranda O.R., Bhattacharya R., Rotello V.M., Mukherjee P. Mechanism of antiangiogenic property of gold nanoparticles: role of nanoparticle size and surface charge. Nanomedicine. 2011;7:580–587. doi: 10.1016/j.nano.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arvizo R.R., Saha S., Wang E., Robertson J.D., Bhattacharya R., Mukherjee P. Inhibition of tumor growth and metastasis by a self-therapeutic nanoparticle. Proc. Natl. Acad. Sci. U. S. A. 2013;110(17):6700–6705. doi: 10.1073/pnas.1214547110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saha S., Xiong X., Chakraborty P.K., Shameer K., Arvizo R.R., Kudgus R.A., Dwivedi S.K., Hossen M.N., Gillies E.M., Robertson J.D., Dudley J.T., Urrutia R.A., Postier R.G., Bhattacharya R., Mukherjee P. Gold nanoparticle reprograms pancreatic tumor microenvironment and inhibits tumor growth. ACS Nano. 2016;10(12):10636–10651. doi: 10.1021/acsnano.6b02231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hossen M.N., Rao G., Dey A., Robertson J.D., Bhattacharya R., Mukherjee P. Gold nanoparticle transforms activated cancer-associated fibroblasts to quiescence. ACS Appl. Mater. Interfaces. 2019;11(29):26060–26068. doi: 10.1021/acsami.9b03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Xiong X., Huai Y., Dey A., Hossen M.N., Roy R.V., Elechalawar C.K., Rao G., Bhattacharya R., Mukherjee P. Gold nanoparticles disrupt tumor microenvironment - endothelial cell cross talk to inhibit angiogenic phenotypes in vitro. Bioconjugate Chem. 2019;30(6):1724–1733. doi: 10.1021/acs.bioconjchem.9b00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C., Fu C., Wren J.D., Wang X., Zhang F., Zhang Y.H., Connel S.A., Chen T., Zhang X.A. Tetraspanin-enriched microdomains regulate digitation junctions. Cell. Mol. Life Sci. 2018;75(18):3423–3439. doi: 10.1007/s00018-018-2803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nurmik M., Ullmann P., Rodriguez F., Haan S., Letellier E. In search of definitions: cancer-associated fibroblasts and their markers. Int. J. Canc. 2020;146(4):895–905. doi: 10.1002/ijc.32193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuzet S.E., Gaggioli C. Fibroblast activation in cancer: when seed fertilizes soil. Cell Tissue Res. 2016;365(3):607–619. doi: 10.1007/s00441-016-2467-x. [DOI] [PubMed] [Google Scholar]
- 17.Leask A. Potential therapeutic targets for cardiac fibrosis: TGFbeta, angiotensin, endothelin, CCN2, and PDGF, partners in fibroblast activation. Circ. Res. 2010;106(11):1675–1680. doi: 10.1161/CIRCRESAHA.110.217737. [DOI] [PubMed] [Google Scholar]
- 18.Dakin S.G., Buckley C.D., Al-Mossawi M.H., Hedley R., Martinez F.O., Wheway K., Watkins B., Carr A.J. Persistent stromal fibroblast activation is present in chronic tendinopathy. Arthritis Res. Ther. 2017;19(1):16. doi: 10.1186/s13075-016-1218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Kroef M., Carvalheiro T., Rossato M., de Wit F., Cossu M., Chouri E., Wichers C.G.K., Bekker C.P.J., Beretta L., Vazirpanah N., Trombetta E., Radstake T., Angiolilli C. CXCL4 triggers monocytes and macrophages to produce PDGF-BB, culminating in fibroblast activation: Implications for systemic sclerosis. J. Autoimmun. 2020;111 doi: 10.1016/j.jaut.2020.102444. [DOI] [PubMed] [Google Scholar]
- 20.Ciavarella S., Laurenzana A., De Summa S., Pilato B., Chilla A., Lacalamita R., Minoia C., Margheri F., Iacobazzi A., Rana A., Merchionne F., Fibbi G., Del Rosso M., Guarini A., Tommasi S., Serrati S. u-PAR expression in cancer associated fibroblast: new acquisitions in multiple myeloma progression. BMC Canc. 2017;17(1):215. doi: 10.1186/s12885-017-3183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marudamuthu A.S., Shetty S.K., Bhandary Y.P., Karandashova S., Thompson M., Sathish V., Florova G., Hogan T.B., Pabelick C.M., Prakash Y.S., Tsukasaki Y., Fu J., Ikebe M., Idell S., Shetty S. Plasminogen activator inhibitor-1 suppresses profibrotic responses in fibroblasts from fibrotic lungs. J. Biol. Chem. 2015;290(15):9428–9441. doi: 10.1074/jbc.M114.601815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosh A.K., Vaughan D.E. PAI-1 in tissue fibrosis. J. Cell. Physiol. 2012;227(2):493–507. doi: 10.1002/jcp.22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen P., Wu R., Zhu W., Jiang Z., Xu Y., Chen H., Zhang Z., Chen H., Zhang L., Yu H., Wang J., Hu X. Hypoxia preconditioned mesenchymal stem cells prevent cardiac fibroblast activation and collagen production via leptin. PloS One. 2014;9(8) doi: 10.1371/journal.pone.0103587. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Martinez-Martinez E., Jurado-Lopez R., Valero-Munoz M., Bartolome M.V., Ballesteros S., Luaces M., Briones A.M., Lopez-Andres N., Miana M., Cachofeiro V. Leptin induces cardiac fibrosis through galectin-3, mTOR and oxidative stress: potential role in obesity. J. Hypertens. 2014;32(5):1104–1114. doi: 10.1097/HJH.0000000000000149. discussion 1114. [DOI] [PubMed] [Google Scholar]
- 25.Bao Q., Zhang B., Suo Y., Liu C., Yang Q., Zhang K., Yuan M., Yuan M., Zhang Y., Li G. Intermittent hypoxia mediated by TSP1 dependent on STAT3 induces cardiac fibroblast activation and cardiac fibrosis. Elife. 2020;9 doi: 10.7554/eLife.49923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu M.P., Young M.J., Tzeng C.C., Tzeng C.R., Huang K.F., Wu L.W., Chou C.Y. A novel role of thrombospondin-1 in cervical carcinogenesis: inhibit stroma reaction by inhibiting activated fibroblasts from invading cancer. Carcinogenesis. 2008;29(6):1115–1123. doi: 10.1093/carcin/bgn077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.