Abstract
E2F transcription factors (E2Fs) were found to be related with cell activities and disease progression among a variety of different tumors, including regulating cell division and cell proliferation. In the analysis, it aimed to focus on transcriptional and survival information of E2Fs in gastric cancer (GC) from Gene Expression Profiling Interactive Analysis (GEPIA), Kaplan-Meier plotter, cBioPortal, Database for Annotation, Visualization and Integrated Discovery (DAVID), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and Oncomine databases. It was found that the expression of E2F1/2/3/5/7/8 in GC tissues was obviously higher than the normal. Of interest, none of the E2Fs was related with pathological stages. Nevertheless, high expression of E2F2/3/5/7/8 was related with better survival data, except E2F6 regarding shorter first-progression (FP) survival. High expression levels of E2F2/5/7/8 have significant correlations with overall survival (OS) in patients with intestinal and diffuse GC, and this prognostic value is not affected by gender. Oppositely, the lower level of E2F1/4 illustrated superior survival data. Moreover, increased expression of E2F1 in GC tissues might play an important role in the development of GC. Collectively, E2F1 could be a potential therapeutic target for patients with GC. E2F1/2/3/5/7/8 might be original prognostic predictors of GC.
Keywords: gastric cancer, E2F transcription factors, predictive factors, gene targets, prognosis, bioinformatics analysis
Graphical Abstract
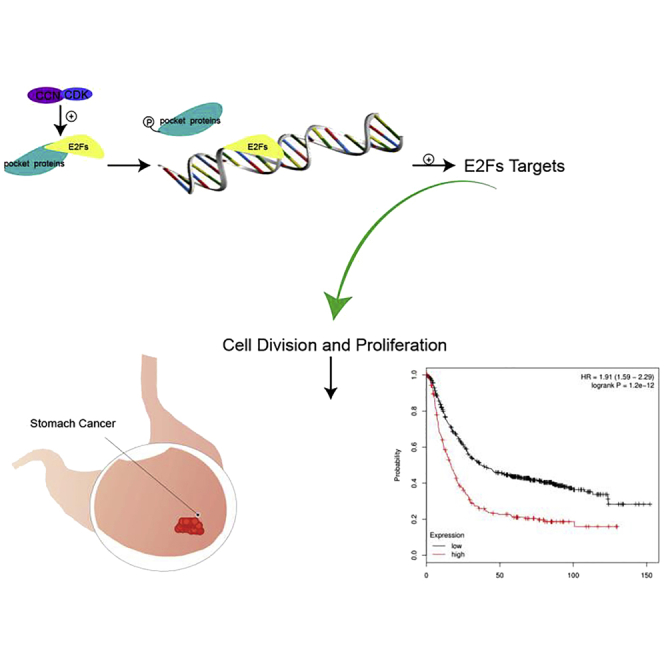
E2F transcription factors (E2Fs) play a great part in cell cycle and proliferation, regulating tumor development, and progression. By studying data from online analysis databases, expression of E2Fs in gastric cancer is distinct from the normal ones. E2F1/2/3/5/7/8 could be original prognostic factors in gastric cancer, and E2F1 might be a therapeutic target.
Introduction
Gastric cancer (GC) has been being a severe and social health problem among Eastern-Asia regions, especially in developing and less-developed countries, where the cancer survival rates were poor relatively.1, 2, 3 According to a recent analysis, GC was the fifth most common incident of cancer overall, and the third most common cause of cancer deaths for men at the same time. Besides, GC incidence increased by 25% between 2007 and 2017 globally.4
E2F transcription factors (E2Fs) are a family of genes that are considered to play a significant role in cell cycle, necrosis, and regulating DNA duplication in cells of mammal.5, 6, 7 Moreover, E2Fs regulates not only transcription of cellular genes that are essential for cell division but also many genes in chromatin assembly and condensation, chromosome separation, DNA repairing, and checkpoint control.8, 9, 10 The transcription factors are classified into two opposite groups: transcriptional activators (E2F1/2/3a) and suppressors (E2F3b/4–8).8,11 E2F family components were found to be related with cell activities and disease progression among a variety of different tumors, such as bladder cancer, lung cancer, breast cancer, prostate cancer, and ovarian cancer.12, 13, 14, 15 E2F1–6 has highly conserved DNA binding and dimerization domains and binds DNA target sequences to DNA polymerase-1(DP1) or DNA polymerase-2(DP2) as heterodimers. Overexpression of E2F7 and E2F8 can significantly delay proliferation of mouse embryonic fibroblasts, due to unique structural characteristics of the two members.8
A study explained the upregulated expression of E2F1/3/4 was associated with poor prognosis in GC, whereas high expression of E2F2/5/6/7 indicted a good prognosis under the condition of human epidermal growth factor receptor 2 (HER-2) negative conversely. In addition, cancer treatment of E2F family members with a 5-fluorouracil (5-FU)-based adjuvant showed a significantly poor prognosis.16 However, the study only analyzed E2F1–7, except E2F8, and the potential targets of precision therapy have not been described yet. Meanwhile, finding new factors for the accurate prediction of prognosis and therapeutic targets of GC is of great significance. In the analysis, we studied the prognostic indicators and potential target values of E2Fs (E2F1/2/3/4/5/6/7/8) in patients with GC.
Results
Transcriptional Levels of E2Fs in GC
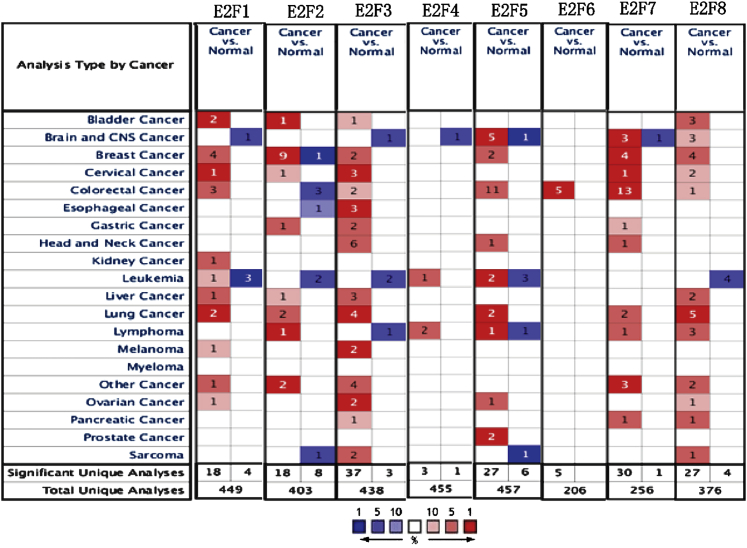
All of the E2F family members were studied in human multiple tumor tissues. The expression levels of E2Fs in multiple human cancers were compared with those in normal organ tissues from the Oncomine database (www.oncomine.org) (Figure 1). In summary, E2Fs are generally upregulated in a variety of tumors.
Figure 1.
The Transcription Levels of E2Fs in Different Types of Cancers
Correlations between Expression Levels of E2Fs and Clinical Characteristics in GC
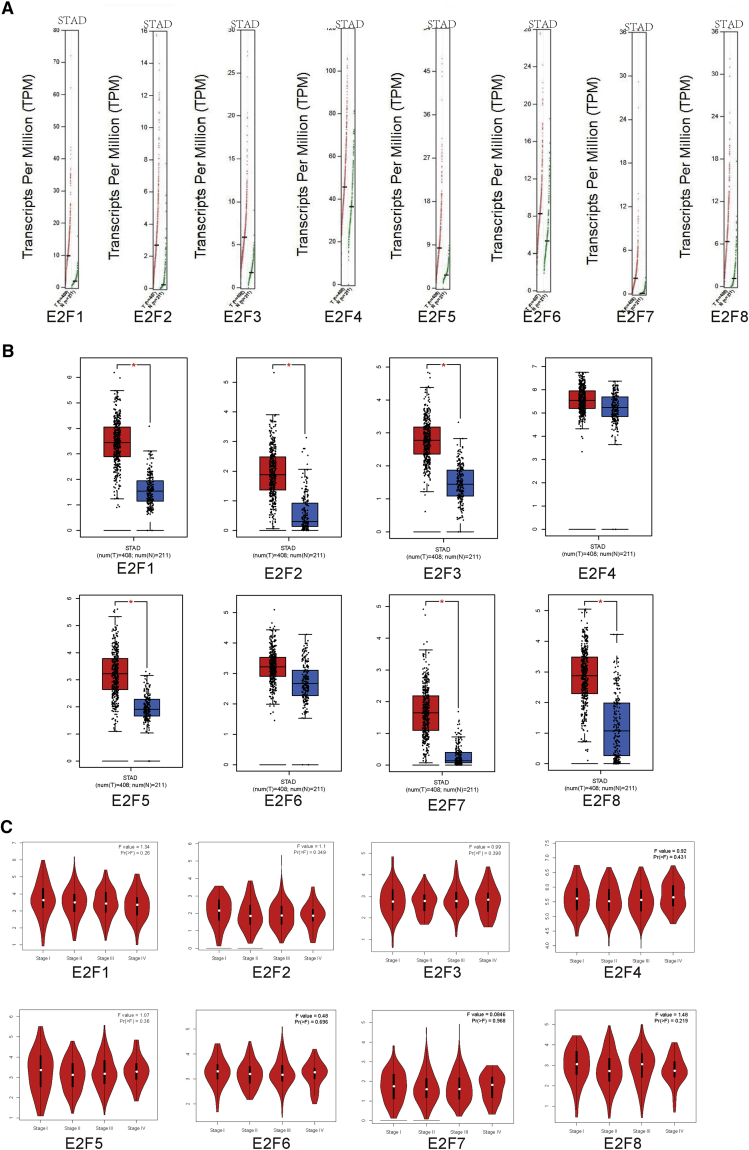
We made comparisons between E2F expression with normal gastric tissues from the GEPIA2 (Gene Expression Profiling Interactive Analysis) database (http://gepia.cancer-pku.cn/). Through analysis of dot plots and boxplots, the transcriptional levels of E2F1/2/3/5/7/8 obviously increased compared with normal samples, whereas E2F4/6 implied no apparent difference in expression (Figures 2A and 2B). Of interest, no significant difference existed between the expression of E2Fs and pathological stages of stomach adenocarcinoma through violin plots (Figure 2C).
Figure 2.
The Expression of E2Fs and Relations between E2Fs and Tumor Stages in GC
(A) Gene Expression Profile (dot plots ). (B) Expression on Box Plots. (C) Pathological Stage Plots (violin plots).
Association of E2F Expression with Prognosis in GC
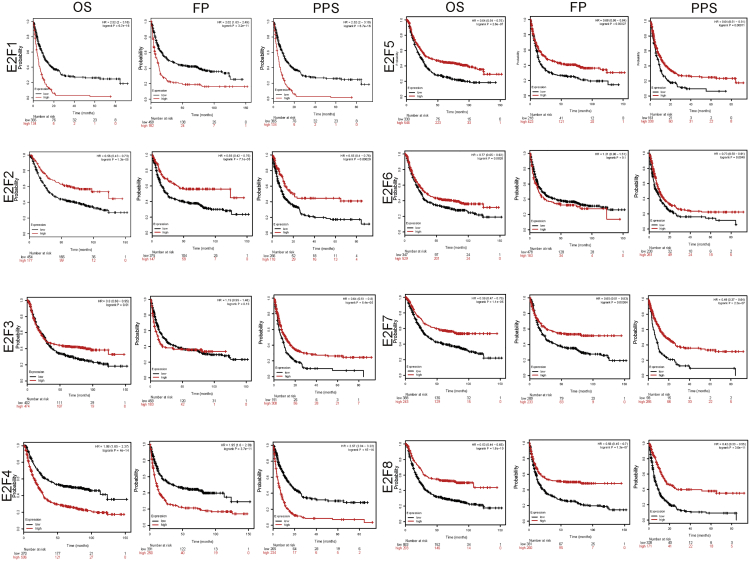
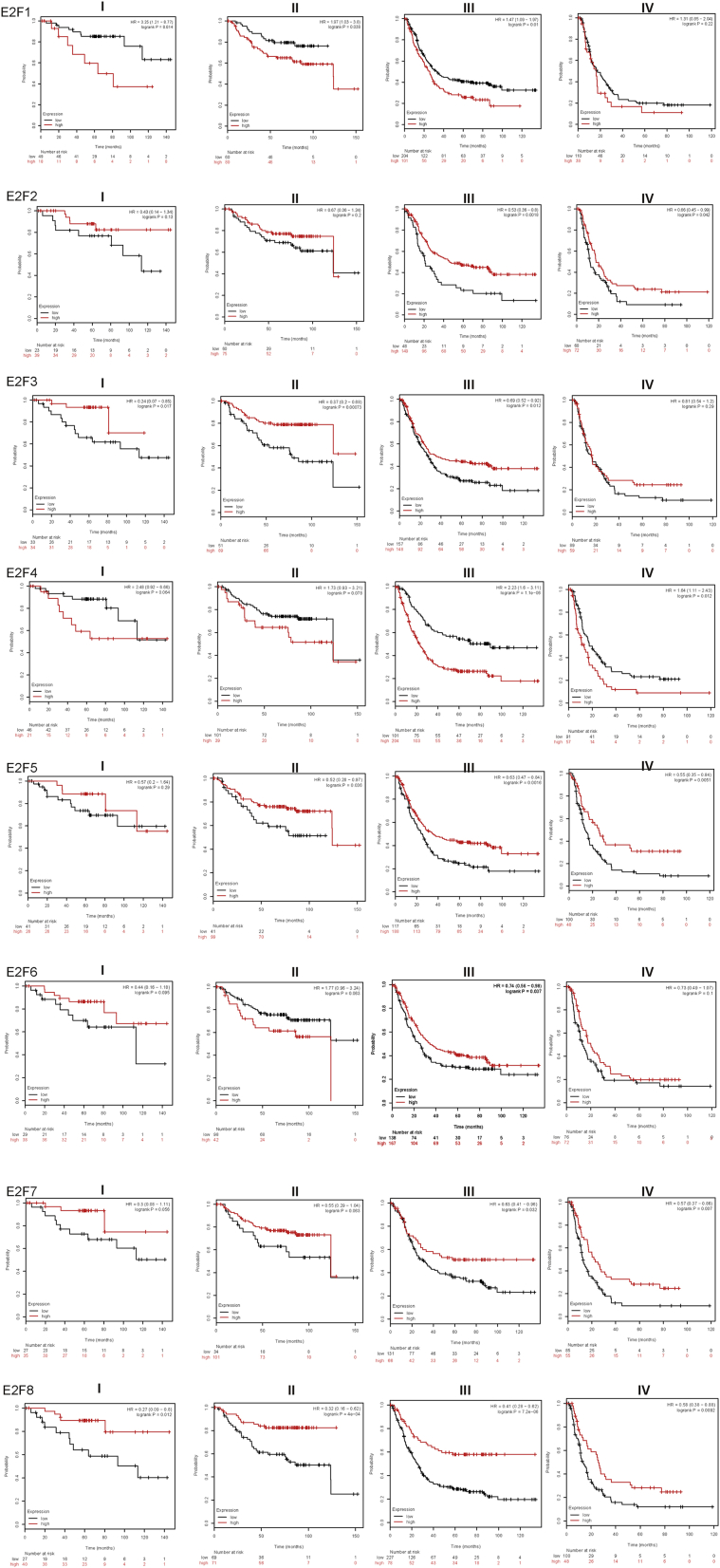
We analyzed the relationship with expression level of E2F members and prognostic value in patients with GC by using Kaplan-Meier Plotter (2019 version; http://kmplot.com/analysis/index.php?p=service&cancer=gastric). The number of patients evaluated was 1,056, and the Jetset best probe set was chosen in the Kaplan-Meier plotter database. Results concluded from the curve were that overexpression levels of E2F2/3/5/6/7/8 were related with long overall survival (OS) and post-progression survival (PPS) and that E2F family elements showed a positive effect on first-progression (FP) survival except E2F6. Conversely, the high levels of E2F1 and E2F4 indicated poor OS, FP, and PPS in all GC patients (Figure 3). The relationships among transcriptional levels of E2Fs, Her-2 status, tumor stages, and OS were also analyzed in all GC patients (Figures 4 and 5). We found that the expression levels of E2F1/3/6 showed no statistical differences in OS among all patients with stage IV GC.
Figure 3.
The Prognostic Value of E2Fs in Gastric Cancer
Figure 4.
The Overall Survival Prognostic Value of E2Fs in Gastric Cancer under Different Conditions of Her-2 (A) Her-2 positive status. (B) Her-2 negative status.
Figure 5.
The Overall Survival Prognostic Value of E2Fs in Four Different Tumor Stages of Gastric Cancer
Additionally, E2F2/4/5/7 transcriptional levels appeared on statistical significance for OS in patients with stage I GC, and there was no statistical difference of E2F2/4/6/7 expression linked with OS among patients with stage II GC.
E2F3/6/7/8 and the positive condition of Her-2 might influence OS together through analyzing the survival curves downloaded from the Kaplan-Meier plotter. Moreover, the prognostic value of E2Fs was not influenced by the negative condition of Her-2 in all patients with GC.
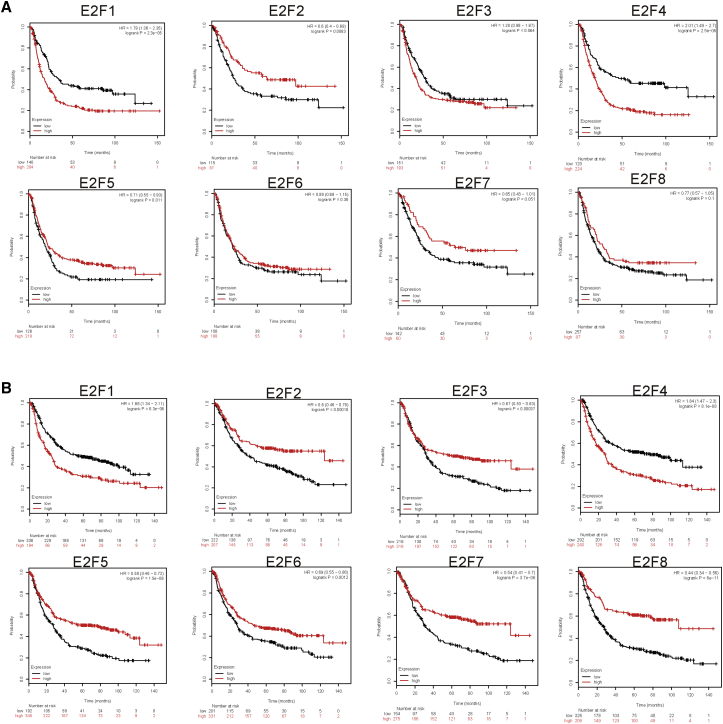
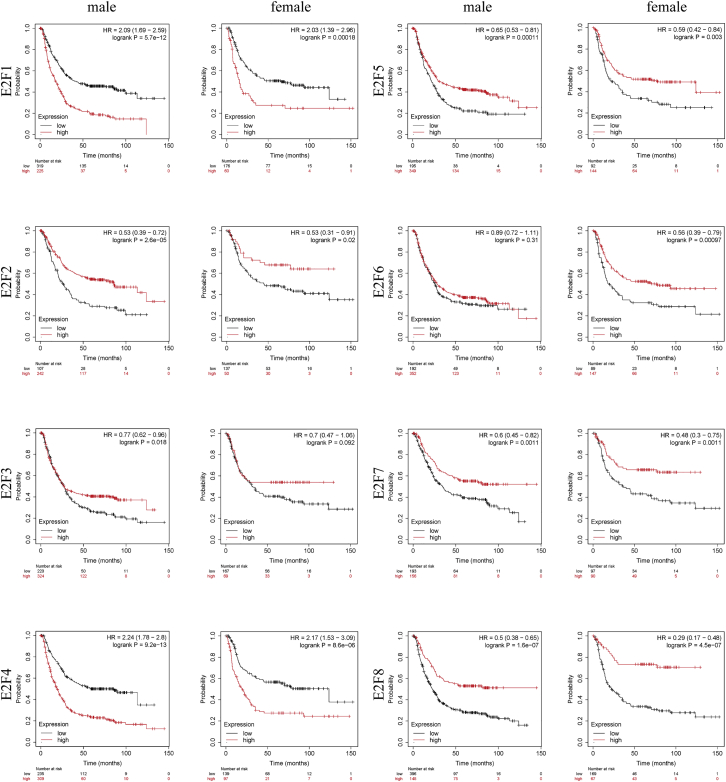
We analyzed whether E2F expression exerted distinct effects on OS in different genders. It was found that except for E2F3 in females and E2F6 in males, the expression levels of E2Fs were significantly correlated with OS of patients, which revealed no relation to genders (Figure 6).
Figure 6.
The Overall Survival Prognostic Value of E2Fs in Different Genders in Patients with Gastric Cancer
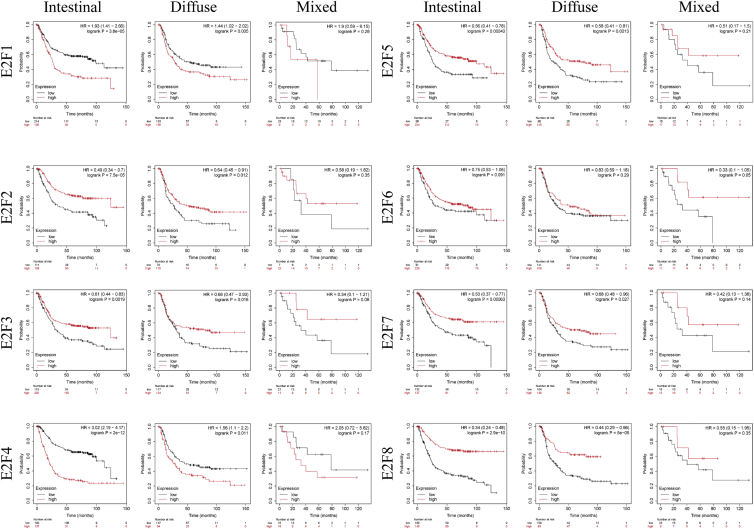
Apart from genders, the associations between pathological types by Lauren classification and OS were also investigated (Figure 7). High expression of E2F2/3/5/7/8 was correlated with superior OS in both intestinal and diffuse GC, whereas high expression of E2F1 and E2F4 was significantly correlated with adverse OS (p < 0.05). There was no significant correlation between E2F expression and OS in all mixed GC (p > 0.05).
Figure 7.
The Overall Survival Prognostic Value of E2Fs in Different Pathologic Types by Lauren Classification in Gastric Cancer
The Coexpression and Interactions of E2Fs
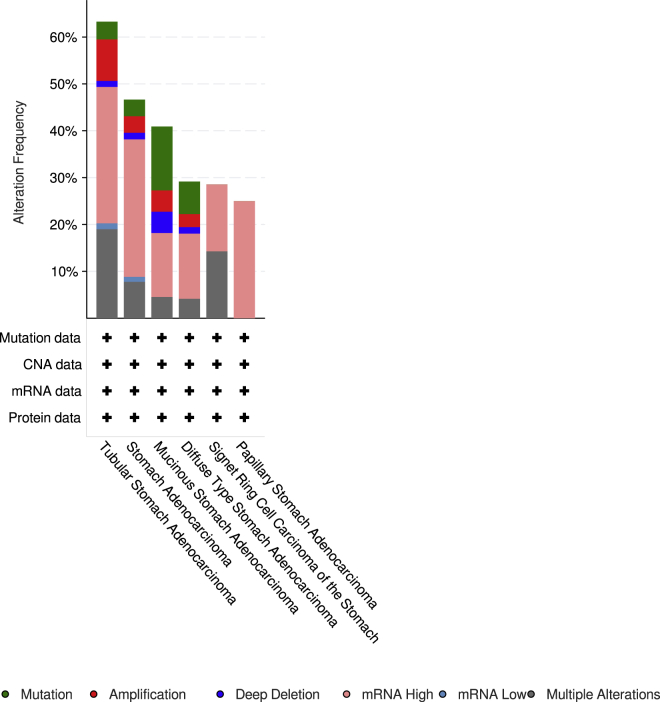
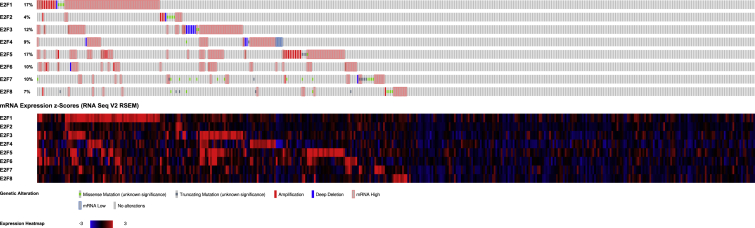
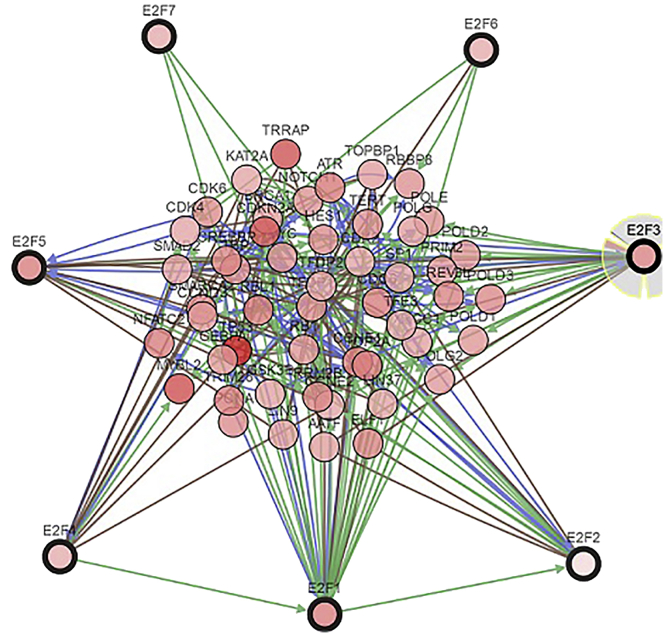
The E2F alterations, correlations, and networks were studied among patients with stomach adenocarcinoma from the cBioPortal dataset (The Cancer Genome Atlas [TCGA], Firehose Legacy). E2F genes were altered in 214 (52%) of queried 415 patients/samples. The analysis showed that alterations of E2Fs were detected in 6 different pathological types (tubular stomach adenocarcinoma, stomach adenocarcinoma, mucinous stomach adenocarcinoma, signet ring cell carcinoma of the stomach, diffuse-type stomach adenocarcinoma, and papillary stomach adenocarcinoma). The highest ratio of E2F factor changes among the 6 sample groups was over 60% in the figure. Moreover, high mRNA expression of E2Fs accounts for most of the length in the chart (Figures 8 and 9). In addition, we calculated the Pearson’s correlation coefficient between E2Fs and the network by selecting mRNA expression Z scores (RNA Seq V2 RSEM) from the cBioPortal dataset (TCGA, Firehose Legacy). The table delivered positive but nondistinctive relations among E2F members (Figure 10). E2Fs and the 50 most-frequently altered neighbor genes were in the network, which included cell-division genes, such as CCNE1, CCNE2, CDC6, CDK2, CDK4, CDK6, etc. (Figure 11).
Figure 8.
E2Fs Genomic Alteration Types in Gastric Cancer Detailed Types
Figure 9.
E2Fs Expression Analysis in Gastric Cancer
Figure 10.
E2Fs Coexpression Analysis in Gastric Cancer
Figure 11.
E2F Interaction Analysis in Gastric Cancer
The Functions and Signaling Pathways of E2Fs
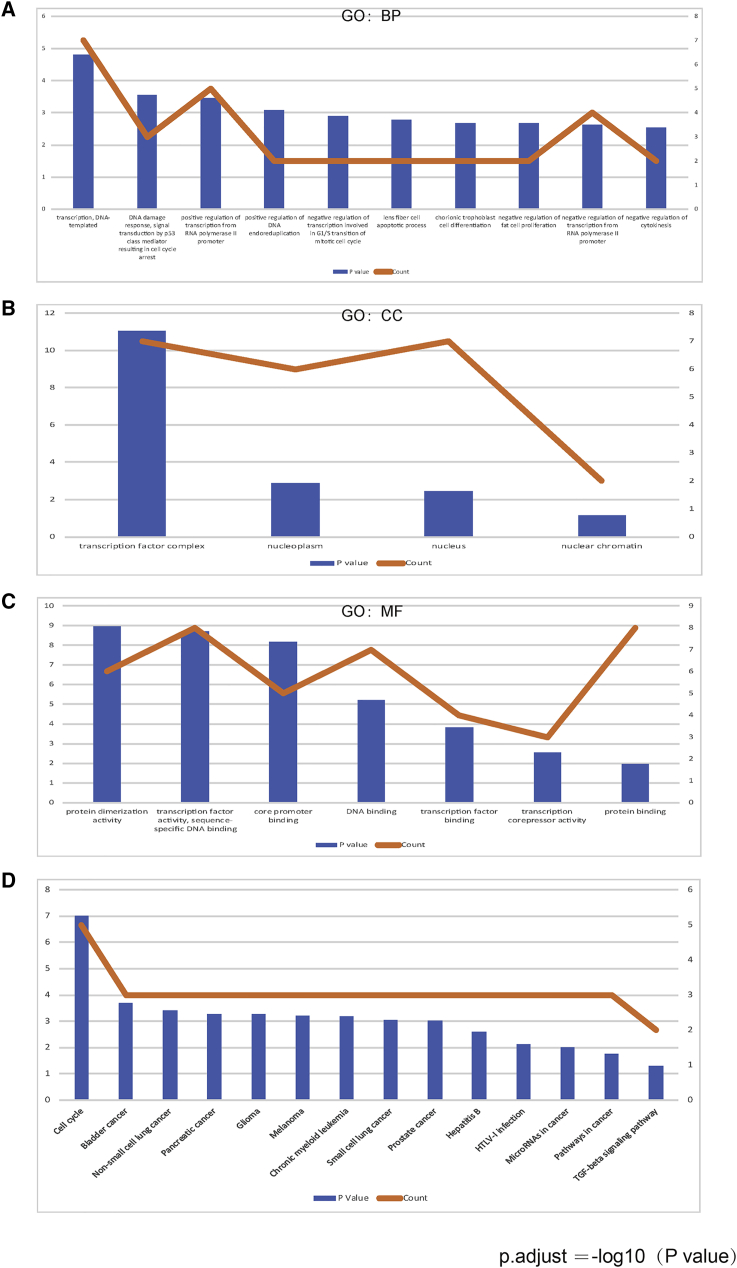
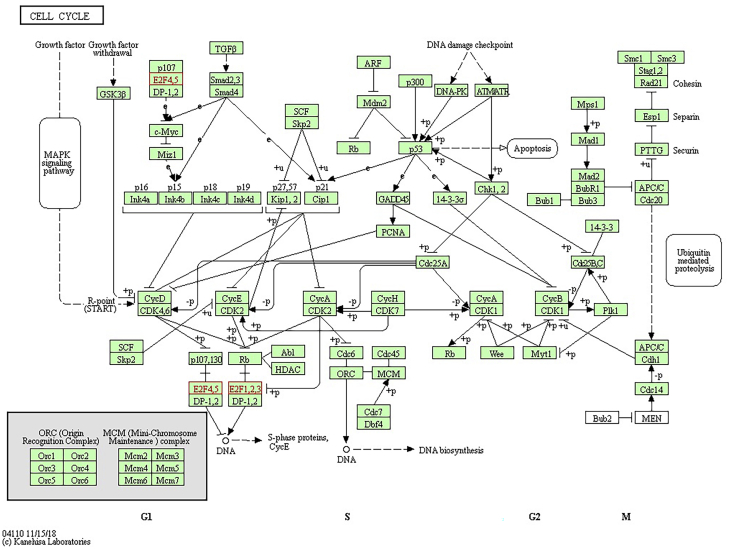
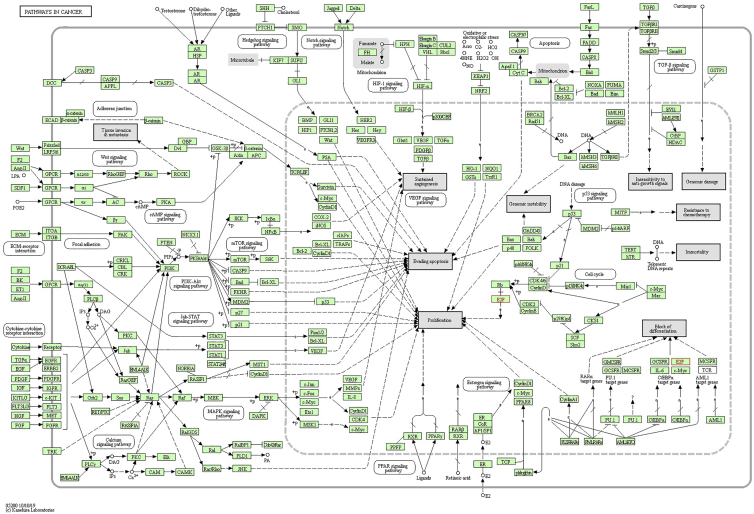
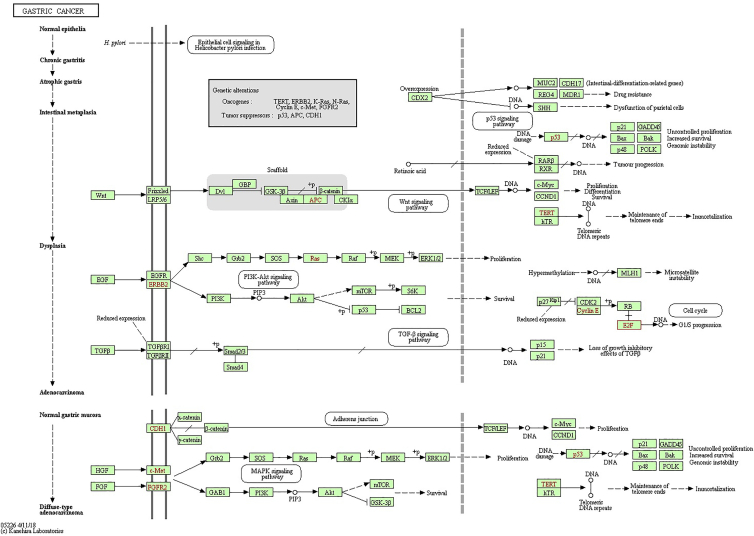
The functional patterns of E2Fs and other genes related with E2F changes were presented through making use of Database for Annotation, Visualization and Integrated Discovery (DAVID). More specially, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were studied in DAVID. The functional value of E2Fs was mainly classified into 3 types: biological processes (BPs), cellular components (CCs), and molecular functions (MFs). GO functional enrichment analysis suggested that GO: 0006351 (transcription, DNA templated) and GO: 0006977 (DNA damage response, signal transduction by p53 class mediator resulting in cell-cycle arrest) were significantly regulated in BPs (Figure 12A). Meanwhile, GO: 0046983 (protein dimerization activity) and GO: 0003700 (transcription factor activity, sequence-specific DNA binding) were also strongly interlinked with E2Fs in MFs (Figure 12B). GO: 0005667 (transcription factor complex) was the most closely related with E2F changes in CCs (Figure 12C). As for KEGG analysis, 14 different aspects were associated with E2F expression and alterations, which implied multiple pathways between E2Fs and some other related genes (Figure 12D). Among the pathways, hsa04110 (cell cycle), hsa05200 (pathways in cancer), and hsa04350 (transforming growth factor beta [TGF-β] signaling pathway) were linked with E2Fs in the process of gastric tumor evolution. TGF-β and E2Fs were related with transformation from normal epithelial cells into GCs. E2Fs regulated cell proliferation and apoptosis through the mitogen-activated protein kinase (MAPK) signaling pathway, p53 signaling pathway, and phosphatidylinositol 3-kinase (PI3K)-protein kinase B (AKT)-mammalian target of rapamycin (mTOR) signaling pathway, which were involved in the development and metastasis of tumors. E2F1/2/3/4/5 played a significant role in cell cycle and took part in the GC occurrence and development by analyzing the KEGG pathway database (https://www.kegg.jp/kegg/pathway.html) (Figures 13, 14, and 15).
Figure 12.
The Functions and KEGG Enrichment Analysis of E2Fs
(A–C) GO enrichment analysis predicted the functional roles of host target genes based on three aspects, including biological processes (A), cellular components (B), and molecular functions (C). (D) The KEGG enrichment analysis of E2Fs.
Figure 13.
Signaling Pathways of E2Fs in Gastric Cancer (Cell Cycle)
Figure 14.
Signaling Pathways of E2Fs in Gastric Cancer (Pathways in Cancer)
Figure 15.
Signaling Pathways of E2Fs in Gastric Cancer (TGF-β Signaling Pathway)
Discussion
E2F family components were partially proven to be associated with a variety of different tumors, such as bladder cancer, lung cancer, breast cancer, prostate cancer, and ovarian cancer.12, 13, 14, 15 As for GC, survival data concerning E2F1/2/3/4/5/6/7 linked with GC was illustrated,16 whereas more details and correlations between E2Fs and GC were not described in previous documents. Herein, our analysis is the first to investigate the prognostic value of E2Fs with GC, along with the influence of Her-2 conditions, genders, pathological classifications, and tumor stages on survival. Moreover, the role of E2Fs as potential therapeutic targets for future treatment strategies is also stated in this paper. We aim to unfold deeper exposure to E2Fs to make contributions to the development of clinical medicine, optimizing treatment therapy and prolonging lifetimes for the sick with GC.
High expression of E2F1 appeared in the early stage of GC, whereas low expression appeared in the late stage.17 Another study has shown that LSINCT5 was a direct transcriptional target of E2F1. LSINCT5 was significantly overexpressed in metastatic GC tissues and played an important role in epithelial mesenchymal transformation, promoting cell migration and invasion.18 E2F1-mediated activation of LSINCT5, as a regulator of cell migration and invasion, constitutes the mechanism between the E2F1-mediated pathway and long non-coding RNA (lncRNA) regulating cell migration and invasion.19
In addition, E2F1 partially promoted GC cell growth by inducing terminal differentiation-induced non-coding RNA (TINCR) transcription. TINCR could bind to STAU1 (staufen1) protein and affected the stability and expression of CDKN2B mRNA, thereby regulating the proliferation of GC cells and accelerating the progression of GC. Data showed that E2F1 could be involved in regulating the stemness of tumor cells, which meant that reducing the level of E2F1 protein equaled inhibiting the stemness of cancer cells.20 That upregulation of E2F1 enhanced the S-phase arrest of cell cycle might account for the possible mechanism of E2F1 in GC resistance in vivo and in vitro. High expression of E2F1enhanced the development of multidrug resistance (MDR) in GC.21
High expression of E2F1 appeared in the early stage of GC, whereas low expression appeared in the advanced stage. E2F1 was found highly expressed in patients of early clinical stage and was relevant to a short survival time. In stages III–IV GC patients, there was no significant difference in the prognosis between E2F1 positive and negative expression.17 However, it was found in our study that the E2F1 expression level was not statistically significant for OS in phase IV GC. In addition, the high level of microRNA (miR)-135a in GC was associated with a shorter survival time. The mechanism of miR-135a acting on GC appeared to be the inhibition of E2F1 expression and Sp1/DAPK2 pathway signaling.22 In contrast, the high expression of miR-34a could enhance the anti-tumor immune effect of dendritic cells (DCs) in GC, and miR-34a targeted DAPK2 and Sp1 to participate in the inactivation of E2F1. The data revealed the mediating mechanism of E2F1 controlling DC anti-tumor immunity through miR-34a-dependent downregulation of E2F1 expression and suggested its contribution to GC immunotherapy. In terms of immunity: E2F1, a negative regulatory gene, could lead to immature DCs when it is in an active state, thus stopping the production of antigen and suppressing the immune response.23 Overexpression of E2F1 accelerated the apoptosis of GC cells, significantly inhibited the growth and proliferation of GC cells, and blocked the cell cycle from stepping into the S phase. Additionally, E2F1 also reduced the movement and invasion ability of GC cells.24 On the other hand, CDK inhibitor p16(INK4a) also regulated transcription and apoptosis by controlling the expression of two major transcriptional regulators, AUF1 and E2F1. Overexpression of E2F1 increased Bax expression and inhibited bcl-2, cyclin D1, Skp2, and c-MYC expression in tumor tissues. E2F1 inhibited the growth of cancer cells by regulating various signaling pathways, which might play an important role in targeting therapy of GC.25 Another study showed that in all GC patients, the high mRNA expression of three members of E2Fs (E2F1, E2F3, E2F4) was significantly correlated with poor OS. However, increased expression of E2F2, E2F5, E2F6, and E2F7 was significantly associated with favorable OS, especially in the advanced clinical stages of GC patients. Except for E2F1/3/4, the rest of E2Fs were of significant value for prognosis. High expression of E2F2/5/6/7 had better OS data in late-stage GC, and these factors indicated good prognosis when Her-2 was negative, whereas high expression of E2F2/5 indicated superior prognosis when Her-2 was positive.16 In our analysis, expression level of E2F1 in GC was significantly higher than the normal tissues in GEPIA. We found the low expression of E2F1 indicated better OS, PPS, and FP data by using the Kaplan-Meier plotter online tool.
E2F2 had high expression in GC tissues, and it could be targeted and inhibited directly by miR-31, playing an important part in tumor suppression.26 Moreover, miR-26a was stated to improve the sensitivity of GC cells to cisplatin chemotherapy drugs by targeting E2F2 so that the efficacy of chemotherapy was enhanced.27 In our research, the expression level of E2F2 was higher than in normal samples from the GEPIA dataset. High expression of E2F2 indicated a better OS, PPS, and FP by using the Kaplan-Meier plotter. Of note, the predictive value of E2F2 was mainly reflected in an advanced stage of GC, with no significance in stage I and stage II.
E2F3, a direct target gene of miR-449a, is an important transcription factor involved in accelerating tumor cell proliferation and metastasis. Overexpression of E2F3 rivaled the inhibitory effect of miR-449a mimics on proliferation and apoptosis of GC cells.28 As a tumor suppressor, overexpression of miR-203a could prevent the proliferation of GC cells. One possible mechanism speculated was that miR-203a might play a role in anti-tumor effects in GC by targeting E2F3.29 The expression of E2F3 was affected by miR-577, an important regulatory factor, and the change of miR-577 expression led to excessive proliferation of GC cells.30 In our study, the expression level of E2F3 was several-fold higher than that in normal samples by analyzing the GEPIA dataset. Additionally, the high expression level of E2F3 was significantly linked with a satisfied OS, FP, and PPS in all of the patients with GC by analyzing the Kaplan-Meier plotter. There was no statistical difference in OS of the E2F3 level among that of all patients with stage IV of GC.
Notably, the serine (AGC)13 repeat mutation in E2F4 was found only in squamous cell carcinoma but not in adenocarcinoma, which suggested that E2F4 might be involved in the transformation from adenocarcinoma into squamous cell carcinoma.31 Some scholars have pointed out that TGFβRII, IGFIIR, BAX, and E2F4 gene repeat coding regions were the targets of microsatellite instability (MSI). These four genes played a large part in the development of microsatellite instable GC.32 Besides, E2F4 and hMSH3 were mutated in all tumor types.33 However, the change of E2F4 was always an integral multiple of three nucleotides lost or acquired in the tumor of microsatellite instability-high (MSI-H), without causing code shift mutation and within the normal polymorphism range. GC patients from North America with MSH-H and MSH-L (n = 127) had a median survival of 541 and 587 days, respectively, whereas patients with microsatellite-stable (MSS) had a median survival of 265 days.34 The increased binding of E2F4 with the RAD51 promoter seemed to enhance the biological function of pci-24781 and cisplatin (CDDP) combined therapy, which significantly reduced the growth of GC cells in vivo, promoted apoptosis, and inhibited cloning of cancer cells.35 Another research illustrated that pRb2 /p130 was a key tumor-suppressor gene and that its tumor-suppressor activity was mainly through the interaction with E2F4 and E2F5 transcription factors to regulate the cell cycle negatively.36 Enrichment analysis revealed that cyclins interacted with transcription factors, such as FOXM1, SIN3A, NFYA, and E2F4, in GC and that high expression of these cyclins was associated with a poor prognosis.37 In our analysis, the expression level of E2F4 showed no statistical difference compared with normal samples from the GEPIA dataset, but we found that the low expression of E2F4 indicated better OS, PPS, and FP data by the Kaplan-Meier plotter. There was a significant statistical difference for OS between stages III and IV of E2F4 in all GC patients. When Her-2 was negative, there was still statistical difference for OS in patients with GC.
E2F5 was found highly expressed in a variety of tumors, such as glioblastoma and prostate cancer.38,39 However, E2F5 expression and prognostic effect in GC have not been reported. It has been reported that miR-106b could accelerate the cell cycle of GC cells and promote the progression of GC by regulating the expression of E2F5.40 miRNA-34a enhanced the therapeutic effect of paclitaxel-sensitive GC cells by targeting E2F5. Therefore, the miRNA-34a/E2F5 axis may be a promising therapeutic target for overcoming GC chemotherapy resistance.41 In the present analysis, we demonstrated that the expression level of E2F5 showed a significant statistical difference compared with normal samples from the GEPIA dataset. With the use of the Kaplan-Meier plotter, we found that high expression of E2F5 indicated better OS, PPS, and FP data. There was a significant statistical difference, which was not influenced by the Her-2 condition, for OS among stages II/III/IV of E2F5 in all GC patients.
E2F6 is a member of the E2Fs with the functions of controlling cell cycle and regulating tumor progression.42, 43, 44 Several studies have clarified the role of E2F6, a component of the Wnt signaling pathway, and SMUG1, a component of the BER signaling pathway, in gastric adenocarcinoma. E2F6/SMUG1 was more common in poorly differentiated tumors and could be considered as an invasive phenotype of GC.45 Interestingly, targets of miR-31 include E2F6 and SMUG1. Induction of miR-31 expression in MKN-45 resulted in a significant decrease in E2F6 and SMUG1 genes. Induction of miR-31 expression could increase drug sensitivity and reduce tumor cell migration and GC cell invasion.46 Additionally, E2F6 regulated the proliferation, invasion, and apoptosis of GC cells by inhibiting the expression of CASC2, suggesting that the E2F6/CASC2 axis was another regulator of GC progression.43 Other scholars have pointed out that E2F6 regulated hypoxia-induced apoptosis of tumor cells by regulating E2F1.44 In our findings, the expression level of E2F6 revealed no significantly statistical difference compared with normal samples from the GEPIA dataset. However, with the use of the Kaplan-Meier plotter, it was found that high expression of E2F6 indicated better OS and PPS data, except for FP. There was significant statistical difference for OS among stages I/II/III of E2F6 in all GC patients.
E2F7 has been reported in a variety of tumors.47,48 Zhang et al.49 have analyzed the relationships between E2Fs and the prognosis of GC. They believed that the mRNA level of E2F7 was related to GC cell invasion and tumor differentiation.49 We found the expression level of E2F7 was higher than that in normal samples from the GEPIA dataset. High expression of E2F7 indicated a better OS, PPS, and FP by using the Kaplan-Meier plotter. Of note, the predictive value of E2F7 was mainly reflected in middle- and advanced-stage GC, with no significance in stage I and stage II.
E2F8 was also expressed higher in other types of tumors and was closely related to tumor prognosis in preceding documents.50, 51, 52 A study has shown that E2F8 was targeted by miR-223-5p, inhibiting tumor progression in non-small cell lung cancer (NSCLC).53 Although the prognostic value of E2F8 has been studied in GC,49 there was no definite and relevant study on the cellular mechanism of E2F8 on GC. In our analysis, we explored that the expression level of E2F8 appeared with a significant statistical difference compared with the normal tissue. High expression of E2F8 indicated a better OS, PPS, and FP by using the Kaplan-Meier plotter. We also demonstrated that there was no correlation between E2F8 expression and tumor stage in all patients with GC.
Conclusions
In this study, the expression and prognostic value of E2Fs in GC were comprehensively analyzed for the first time. Our results suggested that E2F1 could be a potential target of precise treatment, and E2F1/2/3/5/7/8 could be deemed as potential prognostic factors for patients with GC. Although subsequent clinical trials and data are still required for further verification, our findings could be a promising start for the discovery of novel prognostic predictors and the development of new drugs for the treatment of GC.
Materials and Methods
Ethics Statement
This study was approved by the Academic Committee of Central South University and conducted according to the principles expressed in the Declaration of Helsinki. All of the datasets were retrieved from the publishing literature, so it was confirmed that all written, informed consent was obtained.
Oncomine
Oncomine (https://www.oncomine.org/resource/login.html) is a cancer microarray database and web-based data-mining platform54 that is currently the world’s largest oncogene database and integrated data-mining platform. It integrates GEO and TCGA RNA and RNA sequencing (RNA-seq) data and has the most comprehensive cancer mutation spectrum, gene expression data, and related clinical information. Besides, it contains 715 datasets and 86,733 sample information. Based on this database, researchers can conduct gene differential expression analysis and other studies.
GEPIA2
The GEPIA (http://gepia.cancer-pku.cn/) server has been running for 2 years and processed ∼280,000 analysis requests for ∼110,000 users from 42 countries. Present GEPIA2, an updated and enhanced version of GEPIA with 198,619 subtypes and 84 cancer subtypes, has extended the quantification of gene expression from gene level to transcriptional level and could support the analysis and comparison of specific cancer subtypes.55 Correlations between expression level of E2Fs and clinical characteristics in GC were assessed according to the GEPIA2 dataset.
The cBioPortal for Cancer Genomics
The cBioPortal for Cancer Genomics (http://www.cbioportal.org/) was established for exploration of visualization, analysis, and downloading of large-scale cancer genomics datasets.56,57 The stomach adenocarcinoma (TCGA, Firehose Legacy) data, including 415 patients with mRNA data (RNA-seq v.2), was selected for analysis of E2Fs. Coexpression and the network of E2Fs were estimated by using the cBioPortal online tool.
The Kaplan-Meier Plotter
The prognostic value of E2Fs was estimated by using the online dataset, Kaplan-Meier plotter (https://kmplot.com/analysis/), including correlations of gene mRNA expression and survival analysis of breast, lung, and ovarian cancer and GC.58 In the dataset, transcriptomic data of 1,065 patients with GC were evaluated.59 To assess the correlations of survival data and E2Fs, along with tumor stages and Her-2 conditions, genders, and pathological classifications, only the Jetset best probe was chosen and recommended by the website. Association of mRNA expression of E2Fs with the prognosis in GC was analyzed from the Kaplan-Meier plotter, including OS, PPS, and FP survival.
Functional Enrichment Analysis
In this study, GO annotation analysis and the KEGG pathway (https://www.kegg.jp/kegg/pathway.html) enrichment analysis were performed to predict pathways and BPs of E2Fs by using DAVID (https://david.ncifcrf.gov/). By modulating the signaling pathways, E2Fs were able to participate in the development of GC, which was achieved by downloading diagrams from the KEGG pathway website.
Author Contributions
C.H. conducted the conception and design of the study and revised the final version to be published. X.L. collected and analyzed all of the data and drafted the article.
Conflicts of Interest
The authors declare no competing interests.
References
- 1.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Fitzmaurice C., Abate D., Abbasi N., Abbastabar H., Abd-Allah F., Abdel-Rahman O., Abdelalim A., Abdoli A., Abdollahpour I., Abdulle A.S.M., Global Burden of Disease Cancer Collaboration Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019 doi: 10.1001/jamaoncol.2019.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attwooll C., Lazzerini Denchi E., Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004;23:4709–4716. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H.Z., Tsai S.Y., Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trimarchi J.M., Lees J.A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 8.Maiti B., Li J., de Bruin A., Gordon F., Timmers C., Opavsky R., Patil K., Tuttle J., Cleghorn W., Leone G. Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 2005;280:18211–18220. doi: 10.1074/jbc.M501410200. [DOI] [PubMed] [Google Scholar]
- 9.Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R.A., Dynlacht B.D. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nevins J.R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 11.Sozzani R., Maggio C., Varotto S., Canova S., Bergounioux C., Albani D., Cella R. Interplay between Arabidopsis activating factors E2Fb and E2Fa in cell cycle progression and development. Plant Physiol. 2006;140:1355–1366. doi: 10.1104/pp.106.077990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos M., Martínez-Fernández M., Dueñas M., García-Escudero R., Alfaya B., Villacampa F., Saiz-Ladera C., Costa C., Oteo M., Duarte J. In vivo disruption of an Rb-E2F-Ezh2 signaling loop causes bladder cancer. Cancer Res. 2014;74:6565–6577. doi: 10.1158/0008-5472.CAN-14-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rennhack J., Andrechek E. Conserved E2F mediated metastasis in mouse models of breast cancer and HER2 positive patients. Oncoscience. 2015;2:867–871. doi: 10.18632/oncoscience.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shackney S.E., Chowdhury S.A., Schwartz R. A Novel Subset of Human Tumors That Simultaneously Overexpress Multiple E2F-responsive Genes Found in Breast, Ovarian, and Prostate Cancers. Cancer Inform. 2014;13(Suppl 5):89–100. doi: 10.4137/CIN.S14062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C.L., Liu D., Nakano J., Yokomise H., Ueno M., Kadota K., Wada H. E2F1 overexpression correlates with thymidylate synthase and survivin gene expressions and tumor proliferation in non small-cell lung cancer. Clin. Cancer Res. 2007;13:6938–6946. doi: 10.1158/1078-0432.CCR-07-1539. [DOI] [PubMed] [Google Scholar]
- 16.Manicum T., Ni F., Ye Y., Fan X., Chen B.C. Prognostic values of E2F mRNA expression in human gastric cancer. Biosci. Rep. 2018;38 doi: 10.1042/BSR20181264. BSR20181264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J.G., Yu J.W., Wu H.B., Zheng L.H., Ni X.C., Li X.Q., Du G.Y., Jiang B.J. Expressions and clinical significances of c-MET, p-MET and E2f-1 in human gastric carcinoma. BMC Res. Notes. 2014;7:6. doi: 10.1186/1756-0500-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi P., Lin W.R., Zhang M., Huang D., Ni S.J., Zhu X.L., Bai Q.M., Sheng W.Q., Du X., Zhou X.Y. E2F1 induces LSINCT5 transcriptional activity and promotes gastric cancer progression by affecting the epithelial-mesenchymal transition. Cancer Manag. Res. 2018;10:2563–2571. doi: 10.2147/CMAR.S171652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu T.P., Wang Y.F., Xiong W.L., Ma P., Wang W.Y., Chen W.M., Huang M.D., Xia R., Wang R., Zhang E.B. E2F1 induces TINCR transcriptional activity and accelerates gastric cancer progression via activation of TINCR/STAU1/CDKN2B signaling axis. Cell Death Dis. 2017;8:e2837. doi: 10.1038/cddis.2017.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enjoji S., Yabe R., Tsuji S., Yoshimura K., Kawasaki H., Sakurai M., Sakai Y., Takenouchi H., Yoshino S., Hazama S. Stemness Is Enhanced in Gastric Cancer by a SET/PP2A/E2F1 Axis. Mol. Cancer Res. 2018;16:554–563. doi: 10.1158/1541-7786.MCR-17-0393. [DOI] [PubMed] [Google Scholar]
- 21.Yan L.H., Wei W.Y., Cao W.L., Zhang X.S., Xie Y.B., Xiao Q. Overexpression of E2F1 in human gastric carcinoma is involved in anti-cancer drug resistance. BMC Cancer. 2014;14:904. doi: 10.1186/1471-2407-14-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan L.H., Chen Z.N., Li-Li, Chen J., Wei W.E., Mo X.W., Qin Y.Z., Lin Y., Chen J.S. miR-135a promotes gastric cancer progression and resistance to oxaliplatin. Oncotarget. 2016;7:70699–70714. doi: 10.18632/oncotarget.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan L.H., Chen Z.N., Li L., Chen J., Mo X.W., Qin Y.Z., Wei W.E., Qin H.Q., Lin Y., Chen J.S. E2F-1 promotes DAPK2-induced anti-tumor immunity of gastric cancer cells by targeting miR-34a. Tumour Biol. 2016 doi: 10.1007/s13277-016-5446-7. Published online October 4, 2016. [DOI] [PubMed] [Google Scholar]
- 24.Xie Y., Wang C., Li L., Ma Y., Yin Y., Xiao Q. Overexpression of E2F-1 inhibits progression of gastric cancer in vitro. Cell Biol. Int. 2009;33:640–649. doi: 10.1016/j.cellbi.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Wei W.Y., Yan L.H., Wang X.T., Li L., Cao W.L., Zhang X.S., Zhan Z.X., Yu H., Xie Y.B., Xiao Q. E2F-1 overexpression inhibits human gastric cancer MGC-803 cell growth in vivo. World J. Gastroenterol. 2015;21:491–501. doi: 10.3748/wjg.v21.i2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H., Zhang X., Liu Y., Ni Z., Lin Y., Duan Z., Shi Y., Wang G., Li F. Downregulated miR-31 level associates with poor prognosis of gastric cancer and its restoration suppresses tumor cell malignant phenotypes by inhibiting E2F2. Oncotarget. 2016;7:36577–36589. doi: 10.18632/oncotarget.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen L., Cheng F., Zhou Y., Yin C. MiR-26a enhances the sensitivity of gastric cancer cells to cisplatin by targeting NRAS and E2F2. Saudi J. Gastroenterol. 2015;21:313–319. doi: 10.4103/1319-3767.166206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Li H., Zhang R., Liu J., Liu J. MicroRNA-449a inhibits proliferation and induces apoptosis by directly repressing E2F3 in gastric cancer. Cell. Physiol. Biochem. 2015;35:2033–2042. doi: 10.1159/000374010. [DOI] [PubMed] [Google Scholar]
- 29.Yang H., Wang L., Tang X., Bai W. miR-203a suppresses cell proliferation by targeting E2F transcription factor 3 in human gastric cancer. Oncol. Lett. 2017;14:7687–7690. doi: 10.3892/ol.2017.7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Z., Zhang W., Deng F. MicroRNA-577 inhibits gastric cancer growth by targeting E2F transcription factor 3. Oncol. Lett. 2015;10:1447–1452. doi: 10.3892/ol.2015.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woo D.K., Lee W.A., Kim Y.I., Kim W.H. Microsatellite instability and alteration of E2F-4 gene in adenosquamous and squamous cell carcinomas of the stomach. Pathol. Int. 2000;50:690–695. doi: 10.1046/j.1440-1827.2000.01105.x. [DOI] [PubMed] [Google Scholar]
- 32.Chen G.T., Zhu Z.G., Yin H.R., Liu B.Y., Ji J., Zhang J., Lin Y.Z. [The relationship between frameshift mutations of transforming growth factor-beta type II receptor, insulin growth factor II receptor, bcl-2 associated X protein, E2F4 and microsatellite instability in gastric carcinoma] Zhonghua Wai Ke Za Zhi. 2006;44:344–348. [PubMed] [Google Scholar]
- 33.Semba S., Ouyang H., Han S.Y., Kato Y., Horii A. Analysis of the candidate target genes for mutation in microsatellite instability-positive cancers of the colorectum, stomach, and endometrium. Int. J. Oncol. 2000;16:731–737. doi: 10.3892/ijo.16.4.731. [DOI] [PubMed] [Google Scholar]
- 34.Schneider B.G., Bravo J.C., Roa J.C., Roa I., Kim M.C., Lee K.M., Plaisance K.T., Jr., McBride C.M., Mera R. Microsatellite instability, prognosis and metastasis in gastric cancers from a low-risk population. Int. J. Cancer. 2000;89:444–452. doi: 10.1002/1097-0215(20000920)89:5<444::aid-ijc8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 35.He W.L., Li Y.H., Hou W.J., Ke Z.F., Chen X.L., Lu L.Y., Cai S.R., Song W., Zhang C.H., He Y.L. RAD51 potentiates synergistic effects of chemotherapy with PCI-24781 and cis-diamminedichloroplatinum on gastric cancer. World J. Gastroenterol. 2014;20:10094–10107. doi: 10.3748/wjg.v20.i29.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cito L., Indovina P., Forte I.M., Pentimalli F., Di Marzo D., Somma P., Barone D., Penon A., Penon D., Ceccherini E. pRb2/p130 localizes to the cytoplasm in diffuse gastric cancer. J. Cell. Physiol. 2015;230:802–805. doi: 10.1002/jcp.24805. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H.P., Li S.Y., Wang J.P., Lin J. Clinical significance and biological roles of cyclins in gastric cancer. OncoTargets Ther. 2018;11:6673–6685. doi: 10.2147/OTT.S171716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang D.Z., Wang Y.P., Liu J., Hui X.B., Wang X.D., Chen X., Liu D. MicroRNA-129-3p suppresses tumor growth by targeting E2F5 in glioblastoma. Eur. Rev. Med. Pharmacol. Sci. 2018;22:1044–1050. doi: 10.26355/eurrev_201802_14387. [DOI] [PubMed] [Google Scholar]
- 39.Li S.L., Sui Y., Sun J., Jiang T.Q., Dong G. Identification of tumor suppressive role of microRNA-132 and its target gene in tumorigenesis of prostate cancer. Int. J. Mol. Med. 2018;41:2429–2433. doi: 10.3892/ijmm.2018.3421. [DOI] [PubMed] [Google Scholar]
- 40.Yao Y.L., Wu X.Y., Wu J.H., Gu T., Chen L., Gu J.H., Liu Y., Zhang Q.H. Effects of microRNA-106 on proliferation of gastric cancer cell through regulating p21 and E2F5. Asian Pac. J. Cancer Prev. 2013;14:2839–2843. doi: 10.7314/apjcp.2013.14.5.2839. [DOI] [PubMed] [Google Scholar]
- 41.Li L., Wu C., Zhao Y. miRNA-34a enhances the sensitivity of gastric cancer cells to treatment with paclitaxel by targeting E2F5. Oncol. Lett. 2017;13:4837–4842. doi: 10.3892/ol.2017.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kherrouche Z., De Launoit Y., Monté D. Human E2F6 is alternatively spliced to generate multiple protein isoforms. Biochem. Biophys. Res. Commun. 2004;317:749–760. doi: 10.1016/j.bbrc.2004.03.099. [DOI] [PubMed] [Google Scholar]
- 43.Li Y., Jiang L., Lv S., Xu H., Fan Z., He Y., Wen H. E2F6-mediated lncRNA CASC2 down-regulation predicts poor prognosis and promotes progression in gastric carcinoma. Life Sci. 2019;232:116649. doi: 10.1016/j.lfs.2019.116649. [DOI] [PubMed] [Google Scholar]
- 44.Yang W.W., Shu B., Zhu Y., Yang H.T. E2F6 inhibits cobalt chloride-mimetic hypoxia-induced apoptosis through E2F1. Mol. Biol. Cell. 2008;19:3691–3700. doi: 10.1091/mbc.E08-02-0171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Korourian A., Roudi R., Shariftabrizi A., Kalantari E., Sotoodeh K., Madjd Z. Differential role of Wnt signaling and base excision repair pathways in gastric adenocarcinoma aggressiveness. Clin. Exp. Med. 2017;17:505–517. doi: 10.1007/s10238-016-0443-0. [DOI] [PubMed] [Google Scholar]
- 46.Korourian A., Madjd Z., Roudi R., Shariftabrizi A., Soleimani M. Induction of miR-31 causes increased sensitivity to 5-FU and decreased migration and cell invasion in gastric adenocarcinoma. Bratisl. Lek Listy. 2019;120:35–39. doi: 10.4149/BLL_2019_005. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z.L., Bi X.W., Liu P.P., Lei D.X., Wang Y., Li Z.M., Jiang W.Q., Xia Y. Expressions and prognostic values of the E2F transcription factors in human breast carcinoma. Cancer Manag. Res. 2018;10:3521–3532. doi: 10.2147/CMAR.S172332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo H., Zhang L. MicroRNA-30a suppresses papillary thyroid cancer cell proliferation, migration and invasion by directly targeting E2F7. Exp. Ther. Med. 2019;18:209–215. doi: 10.3892/etm.2019.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X., Ni Z., Duan Z., Xin Z., Wang H., Tan J., Wang G., Li F. Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA co-regulatory network analysis. PLoS ONE. 2015;10:e0116979. doi: 10.1371/journal.pone.0116979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang A.P., Liu L.G., Chen M.M., Liu F., You H., Liu L., Yang H., Xun Y., Liu J., Wang R.X. Integrated analysis of 10 lymphoma datasets identifies E2F8 as a key regulator in Burkitt’s lymphoma and mantle cell lymphoma. Am. J. Transl. Res. 2019;11:4382–4396. [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Y.L., Ning G., Chen L.B., Lian Y.F., Gu Y.R., Wang J.L., Chen D.M., Wei H., Huang Y.H. Promising diagnostic and prognostic value of E2Fs in human hepatocellular carcinoma. Cancer Manag. Res. 2019;11:1725–1740. doi: 10.2147/CMAR.S182001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morrell B.C., Zhang L., Schütz L.F., Perego M.C., Maylem E.R.S., Spicer L.J. Regulation of the transcription factor E2F8 gene expression in bovine ovarian cells. Mol. Cell. Endocrinol. 2019;498:110572. doi: 10.1016/j.mce.2019.110572. [DOI] [PubMed] [Google Scholar]
- 53.Dou L., Han K., Xiao M., Lv F. miR-223-5p Suppresses Tumor Growth and Metastasis in Non-Small Cell Lung Cancer by Targeting E2F8. Oncol. Res. 2019;27:261–268. doi: 10.3727/096504018X15219188894056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pandey A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du P., Zhao J., Wang J., Liu Y., Ren H., Patel R., Hu C., Zhang W., Huang G. Sine Oculis Homeobox Homolog 1 Regulates Mitochondrial Apoptosis Pathway Via Caspase-7 In Gastric Cancer Cells. J. Cancer. 2017;8:636–645. doi: 10.7150/jca.16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szász A.M., Lánczky A., Nagy Á., Förster S., Hark K., Green J.E., Boussioutas A., Busuttil R., Szabó A., Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]