Abstract
Childhood obesity and its consequences are a significant public health problem worldwide. Gut microbiota has a potential role in the development of. In the current datasets, we present 16S rDNA amplicon metasequencing of the gut microbiome of adolescents with normal weight, obesity, and obesity with irritable bowel syndrome (IBS) carried out using the Illumine platform. The datasets presented in this report are partly shown in the research article named “Composition and Structure of Gut Microbiome in Adolescents with Obesity and Different Breastfeeding Duration” [1]. The amplicon metasequencing data were deposited at NCBI SRA as BioProject PRJNA604466. A total of 22 phyla, 34 classes, and 231 genera were revealed. Three groups of adolescents had 196 core amplicon sequence variant (ASV), whereas 45, 24, and 1 ASV were unique for adolescents with normal body weight, obesity, and obesity with IBS, respectively. The metagenomic data were first obtained for adolescents from Eastern Siberian, Russia. They have the potential for predictive analysis, which is crucial for understanding microbial community dynamics and their role in the development of the intestinal microbiome. Considering the recent focus on gut microbiota, new datasets are needed to determine the association between gut microbes and the weight of adolescents from previously unexplored regions such as Siberia, Russia.
Keywords: Amplicon metasequencing, Human gut microbiome, Obesity, Adolescents, Normal weight, Irritable bowel syndrome
Specifications Table
| Subject | Microbiology |
| Specific subject area | Applied Microbiology |
| Type of data | Table Figure 16S rDNA illumine sequences |
| How data were acquired | Amplicon metasequencing using Illumina HiSeq platform |
| Data format | Raw data in FASTQ format Analyzed |
| Parameters for data collection | Fecal samples were collected for microbial analysis from adolescents over 15 years old, who signed voluntary informed consent forms. The data collection was approved by the Ethics Committee of Scientific centre for Family Health and Human Reproduction Problems (protocol N6 from 21 December 2015). Adolescents were grouped according to body mass index into three groups: normal body mass (23 persons), obesity (BMI ≥ 95 percentile) (20), and obesity with irritable bowel syndrome (IBS) (10). |
| Description of data collection | Total DNA was extracted from stool samples, the V3-V4 variable regions of 16S rDNA were amplified, and paired-end sequenced according to Illumina protocol using Illumine platform. |
| Data source location | Institution: Scientific centre for Family Health and Human Reproduction Problems City: Irkutsk Country: Russia |
| Data accessibility | Raw data were deposited to NCBI Repository name: SRA Data identification number: BioProject PRJNA604466, BioSamples from SAMN13972309 to SAMN13972361. Direct URL to data: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA604466 |
| Related research article | Belkova N.L., Nemchenko U.M., Pogodina A.V., Feranchuk S.I., Romanitsa A.I., Novikova E.A., Rychkova L.V. Composition and Structure of Gut Microbiome in Adolescents with Obesity and Different Breastfeeding Duration. // Bulletin of Experimental Biology and Medicine. 2019. V. 167, No. 6. P. 759–762. Translated from Byulleten’ Eksperimental'noi Biologii i Meditsiny, Vol. 167, No. 6, pp. 717–721. DOI 10.1007/s10517–019–04617–7 |
Value of the Data
-
•
These are the first amplicon metagenomic datasets of feces collected from adolescents with normal body mass, obesity, and obesity with IBS from Eastern Siberia, Russia.
-
•
The data provides valuable information about the diversity of bacterial communities in the gut microbiome of adolescents with normal body mass, obesity, and obesity with IBS.
-
•
The data are useful for defining gut microbiome peculiarities associated with normal weight, obesity, and obesity with IBS in adolescents.
-
•
Raw sequence data can be used for various additional bioinformatics processing.
1. Data Description
Obesity is a multifactorial disease that develops as a result of a complex interaction of genetic, environmental, socioeconomic, and behavioral factors which leads to an imbalance between energy intake and energy expenditure [2]. The extra weight in childhood and adolescence carries over into adulthood and is associated with cardiometabolic risk factors such as hypertension and the disturbance of lipid, carbohydrate, or purine metabolism [3], [4], [5]. There are two current hypotheses concerning human gut microbiome and obesity. According to one, there are specific peculiarities in the gut microbiome of obese patients, which contribute to a more effective extraction of energy from foods and its deposit in the form of fats [6]. In this way, the gut microbiome stimulates the development of obesity. According to another hypothesis, geography is one of the factors affecting the gut microbiome. Eating habits and diet can alter the gut microbiota in such a way that its metabolic activity will maximize energy from absorbed food [7,8]. Many studies have recently focused on the role of the gut microbiome in the genesis of functional gastrointestinal disorders, including irritable bowel syndrome (IBS) [9]. Changes in the gut microbiota composition have been implicated in the onset and maintenance of IBS. They have also been suggested as an essential factor influencing energy metabolism and the development of obesity [10]. The gut microbiota may be the link between obesity and IBS, which may be necessary for the treatment of both diseases.
The amplicon metagenomic datasets comprise raw sequencing data acquired through the V3–V4 variable region sequencing of feces from adolescents with normal weight, obesity, and obesity with IBS from Eastern Siberia, Russia. The data files (reads in FASTQ format) were deposited at NCBI SRA as BioProject PRJNA604466. The datasets presented in this report are partly shown in the research article named “Composition and Structure of Gut Microbiome in Adolescents with Obesity and Different Breastfeeding Duration” [1]. The Illumina sequencer produced 4 209 582 paired-end reads in total. After the quality filter and contigs assembly, 3 406 651 reads with an average read length of 417 bp were left. The number of sequences in the samples varied from 51 586 to 77 290. The averaged values for the primary analysis of individual samples, as well as Shannon, Simpson, ACE and Chao1 indices, and general characteristics of the adolescents are shown in Supplement Table 1 by groups formed at the sampling stage of the research.
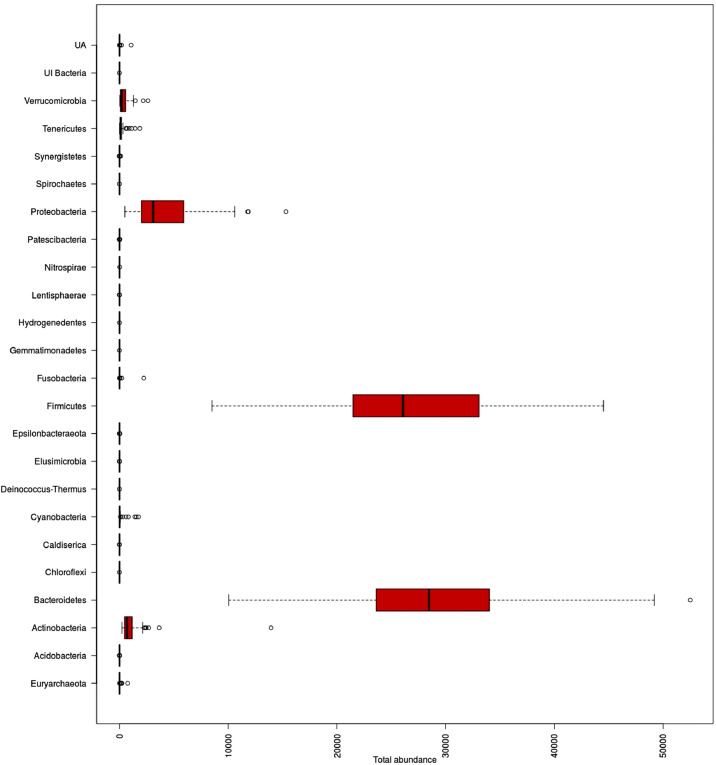
The QIIME2 analysis showed that the reads represented two microbial Kingdoms (Bacteria and Archaea). Overall, the reads represented 22 phyla, 34 classes, and 231 genera. The bacterial diversity was found to be highest, with >99.9% of the total reads, followed by Archaea with only 0.04%. Within the bacterial fractions, the phylum Bacteroidetes was dominant, with 46.1% of the total reads, followed by the phylum Firmicutes (43.1%) and Proteobacteria (7.2%) (Fig. 1). Archaea diversity was poorly represented with the single phylum Euryarchaeota.
Fig. 1.
Total representation of the phyla list and their average frequencies per sample.
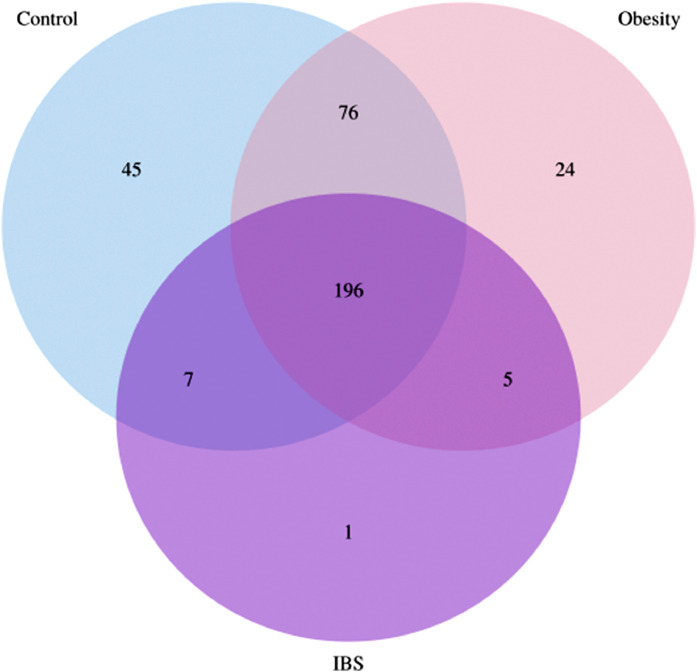
In total, 2 890 phylotypes (ASV) were determined. The core and unique ASVs for three groups of adolescents are presented in Fig. 2. Three groups of adolescents had 196 core ASV, whereas 45, 24, and 1 ASV were unique for adolescents with normal weight, obesity, and obesity with IBS, respectively.
Fig. 2.
Venn diagram of core and unique ASV in the gut microbiome of three group adolescents: normal-weight, obese, and obese with IBS.
We have presented the first datasets on the bacterial diversity of gut microbiome of adolescents with normal body mass, obesity, and obesity with IBS from Eastern Siberia, Russia, based on Illumina sequencing technology.
2. Experimental design, materials, and methods
2.1. Study cohort
The sampling was made up of adolescent patients referred to the Clinic of Scientific centre for Family Health and Human Reproduction Problems (Irkutsk).
Inclusion criteria were: 11–17 years of age, normal body mass (body mass index (BMI) < 85 percentile for relative age and sex) or obesity (BMI ≥ 95 percentile), and written informed consent from parents/primary caregivers and participants. Exclusion criteria were: treatment with pre- and/or probiotics in the last six months, antibiotic therapy during the previous three months, having acute infectious gastroenteritis in the last six months, and the presence of inflammatory bowel diseases and severe chronic diseases. A pediatric gastroenterologist made the diagnosis of IBS. All diagnosed children fulfilled Rome IV criteria for IBS. None of them were vegetarian, vegan, or followed a special diet (i.e., low-FODMAP diet). The data collection was approved by the Ethics Committee of Scientific centre for Family Health and Human Reproduction Problems (protocol N6 from 21 December 2015).
2.2. Sample collection and treatment
Fecal sampling and total DNA isolation were conducted according to the requirements of the International Human Microbiome Standards (IHMS). Standard IHMS_SOP 03 V2 includes feces collection by a patient, transportation, and further sample preparation in a laboratory setting, which takes from 4 to 24 h. DNA from fecal samples was isolated using zirconia beads (BeadBug™, 0.5 mm Zirconium beads, Sigma, USA) and the QIAamp DNA stool kit (Qiagen, Germany) according to the manufacturer's protocol. DNA quality was tested by 1% agarose gel electrophoresis and the concentration was determined using a spectrophotometer NanoDrop™ (Thermo Scientific, USA).
2.3. Library preparation and sequencing
Amplicon metagenomic analysis was conducted at the Novogene Company (China). We analysed the V3-V4 variable regions of the 16S rRNA gene, which allowed us to extract a fragment of 466 bps.
2.4. Analysis of metagenome datasets
Metagenomic data was processed using QIIME2 software (Version 2019.4, https://qiime2.org) [11]. The forward and reverse reads were independently quality filtered and grouped using the q2-dada plugin probability model. Individually grouped forward and reverse reads were then merged, and the chimeras were filtered. Taxonomic classification was performed using the QIIME2 implementation of the RDP naïve Bayesian classifier [12] and the SILVA database V132. Diversity indices Chao1, Shannon, Simpson, Gini, and Fisher alpha were calculated with the q2-diversity plugin.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgements
This work was done according to the program of the Ministry of Science and Higher Education of the Russian Federation [program number 0542–2019–0016].
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.106141.
Appendix. Supplementary materials
References
- 1.Belkova N.L., Nemchenko U.M., Pogodina A.V., Feranchuk S.I., Romanitsa A.I., Novikova E.A., Rychkova L.V. Composition and structure of gut microbiome in adolescents with obesity and different breastfeeding duration. Bull. Exp. Biol. Med. 2019;167:759–762. doi: 10.1007/s10517-019-04617-7. [DOI] [PubMed] [Google Scholar]
- 2.Styne D.M., Arslanian S.A., Connor E.L., Farooqi I.S., Murad M.H., Silverstein J.H., Yanovski J.A. Pediatric obesity-assessment, treatment, and prevention: an endocrine society clinical practice guideline. J. Clin. Endocrinol. Metab. 2017;102:709–757. doi: 10.1210/jc.2016-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh A.S., Mulder C., Twisk J.W., Van Mechelen W., Chinapaw M.J. Tracking of childhood overweight into adulthood: a systematic review of the literature. Obes. Rev. 2008;9:474–488. doi: 10.1111/j.1467-789X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 4.Pogodina A.V., Dolgikh V.V., Rychkova L.V. Uric acid and factors of cardiometabolic risk in adolescents with arterial hypertension. Kardiologiya. 2014;54:36–42. doi: 10.18565/cardio.2014.7.36-42. [DOI] [PubMed] [Google Scholar]
- 5.Pogodina A., Rychkova L., Kravtzova O., Klimkina J., Kosovtzeva A. Cardiometabolic risk factors and health-related quality of life in adolescents with obesity. Child. Obes. 2017;13:499–506. doi: 10.1089/chi.2016.0330. [DOI] [PubMed] [Google Scholar]
- 6.Ley R.E. Obesity and the human microbiome. Curr. Opin. Gastroenterol. 2010;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 7.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M., Magris M., Hidalgo G., Baldassano R.N., Anokhin A.P., Heath A.C., Warner B., Reeder J., Kuczynski J., Caporaso J.G., Lozupone C.A., Lauber C., Clemente J.C., Knights D., Knight R., Gordon J.I. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angelakis E., Bachar D., Yasir M., Musso D., Djossou F., Melenotte C., Robert C., Davoust B., Gaborit B., Azhar E.I., Bibi F., Dutour A., Raoult D. Comparison of the gut microbiota of obese individuals from different geographic origins. New Microbes New Infect. 2018;27:40–47. doi: 10.1016/j.nmni.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajilic-Stojanovic M., Biagi E., Heilig H.G., Kajander K., Kekkonen R.A., Tims S. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141:1792–1801. doi: 10.1053/j.gastro.2011.07.043. [DOI] [PubMed] [Google Scholar]
- 10.R. Tambucci P, Quitadamo, Ambrosi M., De Angelis P., Angelino G., Stagi</i> S., Verrotti A., Staiano A., Farello G. Association between obesity/overweight and functional gastrointestinal disorders in children. J. Pediatr. Gastroenterol. Nutr. 2019;68:517–520. doi: 10.1097/mpg.0000000000002208. [DOI] [PubMed] [Google Scholar]
- 11.Bolyen E., Rideout J.R., Dillon M.R. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R., Huttley G.A., Gregory Caporaso J. Optimising taxonomic classification of marker‐gene amplicon sequences with QIIME 2’s q2‐feature‐classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.