Abstract
Over the past few years, different Computer-Aided Diagnosis (CAD) systems have been proposed to tackle skin lesion analysis. Most of these systems work only for dermoscopy images since there is a strong lack of public clinical images archive available to evaluate the aforementioned CAD systems. To fill this gap, we release a skin lesion benchmark composed of clinical images collected from smartphone devices and a set of patient clinical data containing up to 21 features. The dataset consists of 1373 patients, 1641 skin lesions, and 2298 images for six different diagnostics: three skin diseases and three skin cancers. In total, 58.4% of the skin lesions are biopsy-proven, including 100% of the skin cancers. By releasing this benchmark, we aim to support future research and the development of new tools to assist clinicians to detect skin cancer.
Keywords: Skin cancer, Skin lesion, Clinical data, Cancer research, Computer-Aided Diagnosis (CAD)
Specifications Table
| Subject | Cancer Research and Computer Vision and Pattern Recognition |
| Specific subject area | Automated Skin Cancer detection |
| Type of data | Images and Metadata |
| How data were acquired | All data were collected through smartphone devices using an application developed specifically to this work |
| Data format | Portable Network Graphics (PNG) and Comma Separated Values (CSV) file formats |
| Parameters for data collection | All data are collected during the patient appointment |
| Description of data collection | Each sample in this dataset consists of an clinical image and a set of patient clinical data that contains up to 21 features |
| Data source location | Institution: Federal University of Espírito Santo (UFES) Region: Espírito Santo Country: Brazil |
| Data accessibility | Dataset is available on https://data.mendeley.com/datasets/zr7vgbcyr2/1 |
| Related research article | Andre G. C. Pacheco and Renato A. Krohling, ”The impact of patient clinical information on automated skin cancer detection.” Computers in biology and medicine 116 (2020): 103545. https://doi.org/10.1016/j.compbiomed.2019.103545 |
Value of the Data
-
•
This dataset is useful to support future research and the development of new tools to detect skin cancer without using dermoscopy images.
-
•
Automated skin cancer detection using dermoscopy images is an important and promising task. However, in emerging countries and in remote/rural areas there is a strong lack of medical tools and experts to assist the population. Thus, this dataset is an effort to assist researchers to develop tools, in particular, to assist skin cancer detection in these types of areas.
-
•
This dataset may be used as a training data to develop Computer-Aided Diagnosis (CAD) systems to deal with skin cancer.
-
•
Also, this data may be helpful for educational purposes, i.e., to train medical students to identify skin cancer.
-
•
Beyond the clinical images collected from smartphones, this dataset also contains the patient clinical data related to each image. It may help researchers to understand the relationship between these two types of data and how they can be used to improve skin cancer detection.
1. Data
1.1. Overview
Over the past few years, different skin lesion datasets composed of dermoscopy images have been fomenting the development of CAD systems for skin cancer analysis [1]. The Atlas of Dermoscopy [2] was the first well-known dataset containing over one thousand skin lesion images. In 2018, Tschandl, Rosendahl, and Kittler [3] released the HAM10000 dataset, a large collection of multi-source dermoscopy images of common pigmented skin lesions containing over 10 thousand samples. One year later, Combalia et al. [4] presented the BCN20000 dataset, which contains around twenty-thousand images of skin cancer, including lesions found in hard-to-diagnose locations (nails and mucosa). Together, both HAM10000 and BCN20000 made up the majority part of the International Skin Imaging Collaboration (ISIC) archive1, a public repository that plays an important role for both the purposes of clinical training and for supporting technical research toward automated algorithmic analysis. Since 2016, this organization hosts the ISIC challenge, an open competition that has been boosting automated skin cancer detection algorithms. Recently, the ISIC 2020 challenge2 was released using 33,126 dermoscopy training images of unique benign and malignant skin lesions from over 2000 patients from the archive.
Developing CAD systems to detect skin cancer using dermoscopy images is an important and promising task [5]. However, in emerging countries [6] and in remote/rural areas [7] there is a strong lack of medical tools and experts to assess such type of data. In these places, CAD systems embedded on smartphones may be a low-cost solution, in particular, to assist non-expert clinicians to detect skin cancer [8]. Alves et al. [9] attached a special type of dermatoscope to smartphones to collect a skin lesion dataset using different levels of focus. However, this dataset is not publicly available nor annotated by experts, and it still depends on the dermatoscope, which is costly and may not be available in emerging countries. Side-stepping the dependence on dermatoscope, Udrea et al. [10] used the SkinVision3 database to gather a large skin lesion dataset collected from smartphone cameras (clinical images). However, this is a private dataset. In addition, the skin lesion diagnostic is obtained only from the raw images, there are no biopsy-proven samples, and the dataset does not contain patient demographics.
In order to support future research and the development of new tools to detect skin cancer, we present the PAD-UFES-20, a dataset composed of clinical images of skin lesions and patient clinical data related to each skin lesion collected from different smartphone devices. The dataset contains 2298 samples of six different types of skin lesions, three cancers and three skin diseases. In addition, each image has up to 21 patient clinical features including the patient’s age, skin lesion location, Fitzpatrick skin type, and skin lesion diameter. The dataset was collected from 2018 to 2019 and, to the best of our knowledge, is the first skin lesion archive that: (1) it is publicly available, (2) contains clinical images collected from smartphones, (3) includes patient demographics.
1.2. Description
All samples within PAD-UFES-20 represents a skin lesion of a patient that is composed of an image and a set of metadata. A patient may have one or more skin lesions and a skin lesion may have one or more images. In total, there are 1373 patients, 1641 skin lesions, and 2298 images present in the dataset. Although the number of total samples is not as high as the HAM10000 or BCN20000 datasets, we would like to highlight that this is the first step towards building a public dataset of this type. For comparative purposes, the ISIC archive started in 2016 [11] containing only 1279 samples, and today it is over 40 thousand samples. Moreover, we aim to update the dataset every two years by including more samples and more skin lesions. All data records are publicly available at Mendeley Data [12].
1.2.1. Images
Since the images in this dataset are collected using different smartphone devices they present different resolutions, sizes, and lighting conditions. Essentially, an application to detect skin cancer using clinical images needs to deal with such variability. Thus, it aims to simulate the real world. All images are available in Portable Network Graphics (PNG) file format. In addition, all images are raw, i.e., we do not apply any image processing to enhance visualization.
1.2.2. Meta-data
The metadata associated with each skin lesion is composed of 26 attributes: 21 patient clinical features, four identifying features (patient ID, lesion ID, Image ID, and if the sample is biopsy-proven), and a diagnostic label. All attributes are available in a Comma Separated Values (CSV) file format, where each line represents a skin lesion and each column a metadata attribute. We describe all attributes in Table 1.
Table 1.
Description of each attribute present in the metadata CSV file.
| Attribute | Description |
|---|---|
| patient_id | a string representing the patient ID – example: PAT_1234 |
| lesion_id | a string representing the lesion ID – example: 123 |
| img_id | a string representing the image ID, which is a composition of the patient ID, lesion ID, and a random number – example: PAT_1234_123_000 |
| smoke | a boolean to map if the patient smokes cigarettes |
| drink | a boolean to map if the patient consumes alcoholic beverages |
| background_father and background_mother | a string representing the country in which the patient’s father and mother descends. Note: many patients descend from Pomerania, a region between Poland and Germany. Although it is not a country, we decided to keep the nomenclature, since they identify themselves as Pomeranians descendants. |
| age | an integer representing the patient’s age |
| pesticide | a boolean to map if the patient uses pesticides |
| gender | a string representing the patient’s gender |
| skin_cancer_history | a boolean to map if the patient or someone in their family has had skin cancer in the past |
| cancer_history | a boolean to map if the patient or someone in their family has had any type of cancer in the past |
| has_piped_water | a boolean to map if the patient has access to piped water in their home |
| has_sewage_system | a boolean to map if the patient has access to a sewage system in their home |
| fitspatrick | a integer representing the Fitspatrick skin type [13] |
| region | a string representing one of the 15 macro-regions previously described |
| diameter_1 and diameter_2 | a float representing the skin lesions’ horizontal and vertical diameters |
| diagnostic | a string representing the skin lesion diagnostic – BCC, SCC, ACK, SEK, MEL, or NEV |
| itch | a boolean to map if the skin lesion itches |
| grew | a boolean to map if the skin lesion has recently grown |
| hurt | a boolean to map if the skin lesion hurts |
| changed | a boolean to map if the skin lesion has recently changed |
| bleed | a boolean to map if the skin lesion has bled |
| elevation | a boolean to map if the skin lesion has an elevation |
| biopsed | a boolean to map if the diagnostic comes from clinical consensus or biopsy |
It is important to note that some attributes may be missing for some skin lesions. In brief, patient_id, lesion_id, img_id, age, region, and biopsed are always present. The remaining attributes depend on the patient’s answers during the appointment. Missing values are left blank in the CSV file. When a patient does not know the answer for some question – for example, he/she does not know their father’s background – we fill the feature as UNK (unknown).
2. Materials and methods
The Dermatological and Surgical Assistance Program (in Portuguese: Programa de AssistȬncia Dermatolgica e Cirurgica - PAD) at the Federal University of Espírito Santo (UFES) is a nonprofit program that provides free skin lesion treatment, in particular, to low-income people who cannot afford private treatment. For historical reasons, the Espírito Santo state has received thousands of immigrants from Europe throughout the 19th century. As Brazil is a tropical country, most of these immigrants and their descendants were/are not adapted to this climate. As a result, there is a high incidence of skin lesions/cancer in this state and the PAD plays a fundamental role to assist these people [14].
In late 2017, the Nature Inspired Computing Laboratory (LABCIN-UFES) and the PAD started a partnership that resulted in the creation of a web-based platform and a multi-platform smartphone application to collect and store patient clinical data and skin lesion images. In this section, we present a brief description of this system and the data collection.
2.1. Software description
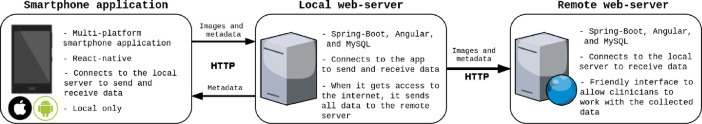
The PAD provides full skin lesion treatment, from the screening to the surgical process (if needed), in 11 different countryside cities in Espírito Santo state. Most of these places are rural areas without Internet access, which is an important requisite to take into account. In this context, the software infrastructure is composed of three parts: a local web-server, a remote web-server, and a multi-platform smartphone application. Basically, all data is collected using the smartphone application, which locally connects to the local web-server to store data. After the data collection is done, as soon as the local web-server gets access to the internet, it synchronizes all data with the remote one. This software structure is illustrated in Fig. 1.
Fig. 1.
An illustration of the software structure that we developed to collect data at the PAD.
Regarding technologies applied to develop the software, the smartphone application was developed using React-Native4, which is an open-source library, based on Javascript5, for building user interfaces for both Android and iOS. Both local and remote web-servers were developed using two main frameworks: Angular6 and Spring-Boot7. Angular is a open-source framework, based on Typescript8, for developing efficient and sophisticated single-page applications. Spring-Boot is also an open-source framework, based on Java9, that is used to create a micro service on the server side. The database is managed using MySQL10.
Beyond storing all data in an organized and structured way, the remote web-server offers a friendly interface to clinicians to access the collected data. This is important for three reasons: (1) it is used to train medical students to identify the lesions; (2) it is important to keep tracking patient lesions since evolution is an important feature to pay attention to detect skin cancer [15]; (3) it helps clinicians with statistics about skin lesions and patients, which is relevant to understand the behavior of the disease over Espírito Santo state. This software is also open-sourced and the code is publicly available on Github11.
2.2. Data collection
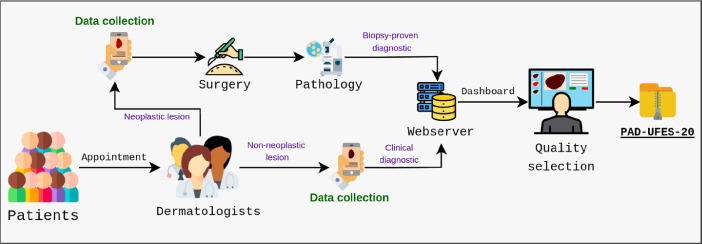
Fig. 2 summarizes our data collection workflow. First of all, the patients have an appointment with a group of up to three senior dermatologists (at least 15 years of experience) that assesses the skin lesion. If the group identifies a neoplasm, the skin lesion is removed through surgical procedure – performed by medical students under the supervision of two senior plastic surgeons of the PAD – and sent to the Pathological Anatomy Unit of the University Hospital Cassiano Antȳnio Moraes (HUCAM) at the UFES to perform histopathology examination. On the other hand, if the group has a consensus that there is no neoplasm, they do not request a biopsy. In both cases, we collect images and clinical data. Later, when the biopsy result is available, it is filled for those lesions in which it was requested. All data is stored in a web-server and the final step is a quality selection to review every single sample that was collected in the previous steps.
Fig. 2.
Data collection workflow of the PAD-UFES-20 dataset.
The goal of the quality selection step is to review the patient clinical data and remove poor quality images. All data from the appointment are filled by senior medical students and the images are collected using different types of smartphone devices. In addition, the smartphone application allows the user to crop the image in order to select only the region of interest. As a result, images in different conditions are upload to the server. Thereby, during the quality selection, we delete those images according to the following rules:
-
•
The image resolution is very poor and it is not possible to identify the lesion
-
•
The patient may be identified because of a tattoo, for example
-
•
The lesion is completely occluded by hair or ink marking
Regarding the patient clinical data, we review all samples to correct typos and information that are clearly wrong, for example, birth dates before 1900 or skin lesions’ diameter that are much bigger than it looks on the image according to visual inspection. For these cases, we re-checked the physical files in order to fix the information. If it is not possible to fix it, we remove the wrong information from the clinical data and it becomes missing data. Lastly, we translate all clinical data from Brazilian-Portuguese to English.
2.3. Data selection
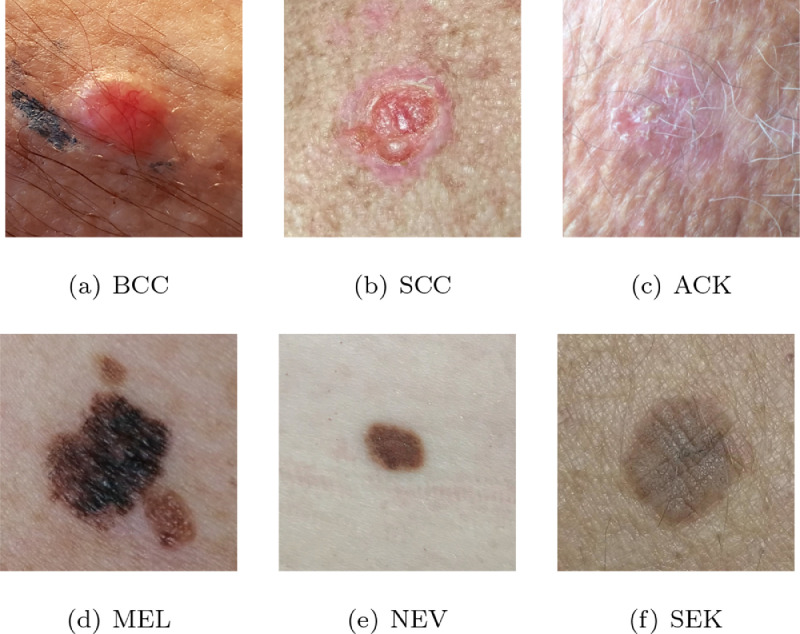
The dataset was collected during 2018 and 2019. In total, there are over 50 types of skin lesions that were collected during this period. However, most of them are rare and contain only a few samples. For this reason, we selected the seven most common skin lesions diagnosed at PAD, which are: (1) Basal Cell Carcinoma (BCC), (2) Squamous Cell Carcinoma (SCC), (3) Actinic Keratosis (ACK), (4) Seborrheic Keratosis (SEK), (5) Bowens disease (BOD), (6) Melanoma (MEL), and (7 Nevus (NEV). As the Bowens disease is considered SCC in situ [13], we clustered them together, which results in six skin lesions in the dataset, three skin cancers (BCC, MEL, and SCC) and three skin disease (ACK, NEV, and SEK). An image sample and the quantity of each of these six skin lesions present in PAD-UFES-20 dataset are presented in Fig. 3 and in Table 2, respectively. It is important to note that all samples diagnosed as skin cancer are proved by biopsy. For those cases in which the pathology yielded a biopsy that is inconclusive, we removed the sample from the dataset. In Table 2 is also described the percentage of samples diagnosed as ACK, NEV, and SEK that are also proved by biopsy. As presented in Table 1, we provide this information in the metadata file through the biopsy attribute.
Fig. 3.
A sample of each type of skin lesion present in PAD-UFES-20 dataset.
Table 2.
The number of samples and the % of biopsy-proven for each type of skin lesion present in PAD-UFES-20 dataset.
| Diagnostic | Næ of samples | % biopsied |
|---|---|---|
| Actinic Keratosis (ACK) | 730 | 24.4% |
| Basal Cell Carcinoma of skin (BCC) | 845 | 100% |
| Malignant Melanoma (MEL) | 52 | 100% |
| Melanocytic Nevus of Skin (NEV) | 244 | 24.6% |
| Squamous Cell Carcinoma (SCC) | 192 | 100% |
| Seborrheic Keratosis (SEK) | 235 | 6.4% |
| Total | 2298 | 58.4% |
To conclude, there are approximately 120 different anatomical regions used by the PAD’s dermatologists and pathologists. We clustered these regions in 15 macro-regions that are more frequent and have more potential to raise a skin lesion, such as follows: face, scalp, nose, lips, ears, neck, chest, abdomen, back, arm, forearm, hand, thigh, shin, and foot. As skin lesions have preferences for some regions of the body [13], [15], it is an important feature to consider.
2.4. Technical validation
The histopathology procedure involves the following steps: (1) the collection of a skin fragment, (2) tissue fixation in formaldehyde (at a concentration of 10%), (3) macroscopic analysis of the skin fragment, (4) histological processing, (5) producing the microscope slides, and (6) a microscopic study with diagnosis’ formulation and interpretation [16]. As described in Table 2, 58.4% of the skin lesions in PAD-UFES-20 dataset are biopsy-proven. This number is compatible with other skin lesion datasets described in literature, for instance, the HAM10000 dataset [3] has 53.3% of biopsy-proven samples.
Regarding the metadata features, they were collected according to the anamnesis of a patient, which beyond the skin lesion screening, dermatologists also consider the anatomical region, diameter, ulceration, itching, bleeding, among others characteristics of the skin lesion [13], [15]. In addition, risk factors are also taken into accounts such as exposure to chemicals, cancer history, and the type of the skin [17]. Combining patient clinical data and skin lesion images has been shown to improve the efficacy of CAD systems for skin cancer detection [5], [18].
2.5. Additional notes
As we previously stated, there are two main characteristics that differ the PAD-UFES-20 dataset from other skin lesion datasets available on literature: the clinical images collected from smartphone devices and the set of patient clinical data. Beyond educational purposes, this dataset aims to support the development of CAD systems embedded in smartphones, in particular, to assist clinicians/non-experts to handle skin lesions in remote places. In addition, the dataset contains samples of six of the most commonly known skin lesions [13], including pigmented and non-pigmented ones.
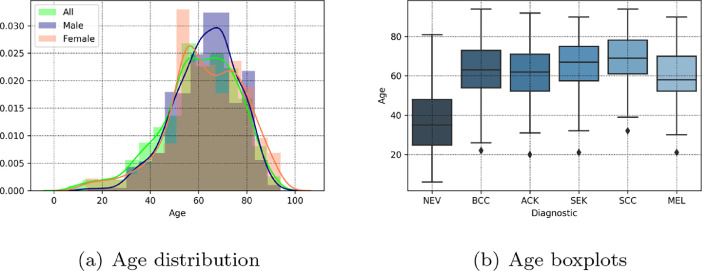
All data are collected in 11 different cities in Espírito Santo state, Brazil. Most of the patients are European immigrants descendants and are or have been farm workers with many hours of sun exposure per day. In addition, the patient average age is approximately 60 years old, but it may vary according to the diagnostic – see Fig. 4. Thus, it is important to note this data represents a specific population in a particular region in Brazil.
Fig. 4.
The patients age distribution according to gender and the age boxplots for each diagnostic.
Another important aspect is the raw image. We do not apply any image processing algorithm into the collected images. A suggestion to enhance the images would be using color constancy algorithms, which have been shown to be helpful for automated skin cancer detection [5], [19]. Also, as we may note in Table 2, the dataset is imbalanced, in particular, for melanoma, the deadliest case of skin cancer. Unfortunately, imbalanced datasets are quite common for skin lesion datasets. For instance, in HAM10000 [3] and BCN20000 [4] datasets, we may find an imbalance among the diagnostic labels of approximately 58:1 and 40:1, respectively. For automated skin cancer detection, common algorithms to deal with this issue are oversampling and weighted loss functions [5].
Lastly, in supplementary materials, we provide the medical questionnaire that results in the patient’s clinical data and a CSV file in which we link all medical terms used in this paper with the SNOMED CT concepts12.
Ethics statement
The dataset was collected along with the Dermatological and Surgical Assistance Program (PAD) of the Federal University of Espírito Santo. The program is managed by the Department of Specialized Medicine and was approved by the university ethics committee (næ 500002/478) and the Brazilian government through Plataforma Brasil (næ 4.007.097), the Brazilian agency responsible for research involving human beings. In addition, all data is collected under patient consent and the patients privacy is completely preserved.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgments
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001; the Conselho Nacional de Desenvolvimento Científico e Tecnólogico (CNPq) – grant n. 309729/2018-1 – and the Fundação de Amparo a Pesquisa e Inovação do Espírito Santo (FAPES) – grant n. 575/2018. We also thank the support of the Secretary of Health of the Espírito Santo state (SESA), the municipal administration of the 11 cities in which PAD takes place, and the Lutheran church of the Espírito Santo state. Lastly, we acknowledge the work of Prof. Carlos Cley and Prof. Luiz F. S. de Barros who founded the PAD in 1987.
Footnotes
https://challenge2020.isic-archive.com
Supplementary material associated with this article can be found, in the online version, at 10.1016/j.dib.2020.106221
Contributor Information
Andre G.C. Pacheco, Email: agcpacheco@inf.ufes.br.
Patricia H.L. Frasson, Email: patricia.frasson@ebserh.gov.br.
Renato A. Krohling, Email: rkrohling@inf.ufes.br.
Appendix A. Supplementary materials
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
References
- 1.Celebi M.E., Codella N., Halpern A. Dermoscopy image analysis: overview and future directions. IEEE J. Biomed. Health Inform. 2019;23(2):474–478. doi: 10.1109/JBHI.2019.2895803. [DOI] [PubMed] [Google Scholar]
- 2.Argenziano G., Soyer H., De Giorgi V., Piccolo D., Carli P., Delfino M. Interactive Atlas of Dermoscopy. EDRA Medical Publishing & New Media; Milan, Italy: 2000. [Google Scholar]
- 3.Tschandl P., Rosendahl C., Kittler H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data. 2018;5:180161. doi: 10.1038/sdata.2018.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. Combalia, N.C. Codella, V. Rotemberg, B. Helba, V. Vilaplana, O. Reiter, A.C. Halpern, S. Puig, J. Malvehy, BCN20000: dermoscopic lesions in the wild, arXiv preprint arXiv:1908.02288(2019).
- 5.Pacheco A.G.C., Krohling R.A. The impact of patient clinical information on automated skin cancer detection. Comput. Biol. Med. 2020;116:103545. doi: 10.1016/j.compbiomed.2019.103545. [DOI] [PubMed] [Google Scholar]
- 6.Scheffler R.M., Liu J.X., Kinfu Y., Dal Poz M.R. Forecasting the global shortage of physicians: an economic-and needs-based approach. Bull. World Health Org. 2008;86:516–523B. doi: 10.2471/BLT.07.046474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng H., Berk-Krauss J., Feng P.W., Stein J.A. Comparison of dermatologist density between urban and rural counties in the United States. JAMA Dermatol. 2018;154(11):1265–1271. doi: 10.1001/jamadermatol.2018.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castro P.B., Krohling B.A., Pacheco A.G.C., Krohling R.A. Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN) IEEE; 2020. An app to detect melanoma using deep learning: an approach to handle imbalanced data based on evolutionary algorithms; pp. 1–8. [Google Scholar]
- 9.Alves J., Moreira D., Alves P., Rosado L., Vasconcelos M.J.M. Automatic focus assessment on dermoscopic images acquired with smartphones. Sensors. 2019;19(22):4957. doi: 10.3390/s19224957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Udrea A., Mitra G., Costea D., Noels E., Wakkee M., Siegel D., de Carvalho T., Nijsten T. Accuracy of a smartphone application for triage of skin lesions based on machine learning algorithms. J. Eur. Acad.Dermatol. Venereol. 2020;34(3):648–655. doi: 10.1111/jdv.15935. [DOI] [PubMed] [Google Scholar]
- 11.Marchetti M.A., Codella N.C., Dusza S.W., Gutman D.A., Helba B., Kalloo A., Mishra N., Carrera C., Celebi M.E., DeFazio J.L. Results of the 2016 international skin imaging collaboration international symposium on biomedical imaging challenge: comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J. Am. Acad. Dermatol. 2018;78(2):270–277. doi: 10.1016/j.jaad.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.A.G.C. Pacheco, G.R. Lima, A.S. Salomão, B.A. Krohling, I.P. Biral, G.G. de Angelo, F.C.R. Alves, et al., PAD-UFES-20: a skin lesion dataset composed of patient data and clinical images collected from smartphones, 2020, (Mendeley Data, v1). Http://dx.doi.org/10.17632/zr7vgbcyr2.1. [DOI] [PMC free article] [PubMed]
- 13.Wolff K., Johnson R.A., Saavedra A.P., Roh E.K. Fitzpatrick’s Color Atlas and Synopsis of Clinical Dermatology. 8. McGraw-Hill Education; New York, USA: 2017. [Google Scholar]
- 14.Frasson P.H.L., Duque D.S., Pinto E.B., Dalvi G.C., Madalon S.Z., Nunes T.A., De-Vargas P.R. Profile of skin cancer in Pomeranian communities of the state of Espírito Santo. Rev. Colégio Bras. Cirurg. 2017;44(2):187–193. doi: 10.1590/0100-69912017002013. [DOI] [PubMed] [Google Scholar]
- 15.Azulay R.D. Dermatologia. 7. Guanabara Koogan; Rio de Janeiro, Brazil: 2017. [Google Scholar]
- 16.Werner B. Biópsia de pele e seu estudo histológico: por quê? para quê? como? Ana. Brasil. Dermatol. 2009;84(5):507–513. doi: 10.1590/s0365-05962009000500010. [DOI] [PubMed] [Google Scholar]
- 17.Duarte A.F., Sousa-Pinto B., Haneke E., Correia O. Risk factors for development of new skin neoplasms in patients with past history of skin cancer: a survival analysis. Sci. Rep. 2018;8(1):1–6. doi: 10.1038/s41598-018-33763-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kharazmi P., Kalia S., Lui H., Wang Z., Lee T. A feature fusion system for basal cell carcinoma detection through data-driven feature learning and patient profile. Skin Res. Technol. 2018;24(2):256–264. doi: 10.1111/srt.12422. [DOI] [PubMed] [Google Scholar]
- 19.Barata C., Celebi M.E., Marques J.S. Improving dermoscopy image classification using color constancy. IEEE J. Biomed. Health Inform. 2014;19(3):1146–1152. doi: 10.1109/JBHI.2014.2336473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/
Supplementary Raw Research Data. This is open data under the CC BY license http://creativecommons.org/licenses/by/4.0/