Abstract
Adeno-associated virus (AAV) is one of the most important gene delivery vehicles for in vivo gene therapy. Intramuscular (i.m.) and intravascular (i.v.) injection are commonly used for AAV gene transfer. Unfortunately, the fate of AAV vectors following administration remains unclear at the histological level. Taking advantage of RNAscope, a recently developed in situ hybridization technique that can reveal high-resolution viral DNA localization information, in this study, we evaluated body-wide distribution of an AAV9 vector in the context of the cell and tissue microenvironments. We observed distinctive kinetics of cell and nuclear entry of the AAV DNA in striated muscle and liver following i.m. and i.v. injection. We also found characteristic distribution patterns of the AAV DNA in various histological structures in internal organs, including gonads and lymph nodes, following i.v. injection. Finally, we showed significantly body-wide spreading of the AAV DNA following i.m. injection. These results add a new dimension to our understanding of AAV transduction biology and provide a basis for assessing the full impact of AAV gene therapy.
Keywords: in situ hybridization, RNAscope, AAV, histology, intramuscular, intravenous
Graphical Abstract
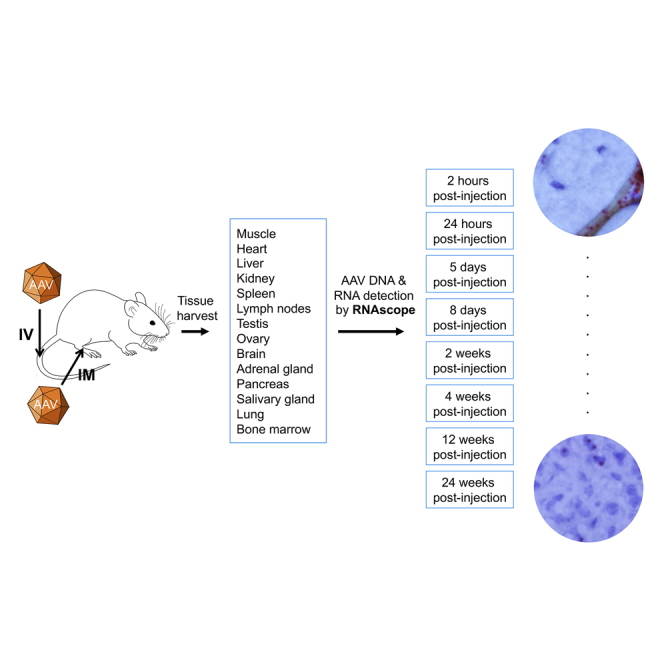
Using RNAscope, for the first time, the high-resolution body-wide organ, tissue, and cell biodistribution profile of the AAV9 vector was delineated following intravenous and intramuscular injection in a mouse model of Duchenne muscular dystrophy. These findings add a new dimension to our understanding of AAV transduction biology in mammals.
Introduction
Adeno-associated virus (AAV) was first identified as a byproduct of adenovirus characterization 50 years ago.1 Since then, accumulated efforts in elucidating AAV biology, identifying and engineering novel capsids, improving manufacturing, and characterizing safety and the immune response have transformed AAV from a nonpathogenic virus to one of the most powerful gene delivery vehicles for in vivo gene therapy. The success of AAV as a gene transfer tool is further exemplified by the approval of AAV-based medicines in the European Union and United States, and by an increasing number of ongoing clinical trials with AAVs.1,2
To date, AAV clinical trials are mainly targeted to diseases affecting striated muscles, liver, and the central nervous system (CNS). In these trials, AAV vectors are delivered via intravenous (i.v.) injection or local injections such as intramuscular (i.m.), intrathecal, subretinal, intraparenchymal, and intraventricular injection. Ideally, the AAV vector should only deliver the therapeutic gene into the nuclei of the target cells in the affected tissues. However, i.m. and i.v. injection often result in off-target distribution. i.v. injection is a prevailing regimen to transport AAV to target organs through the circulation,3 and it will unavoidably lead to off-target distribution. Surprisingly, local i.m. injection has also been suggested to reach non-muscle tissues such as the liver.4, 5, 6, 7, 8 Conventional methods to examine AAV distribution consist of detecting the viral DNA and transcript by quantitative PCR (qPCR), Southern/northern blot, and deep sequencing. Alternatively, the transgene product (protein) is examined by immunostaining, western blot, and enzymatic assays. While these techniques allow investigators to gain a general sense of the biodistribution and transduction efficiency of the AAV vector, they lack sufficient spatial and contextual resolution to delineate cellular and histological features of AAV transduction. As a consequence, we lack a thorough understanding of AAV gene transfer in vivo, especially regarding off-target distribution from i.m. or i.v. administration.
In recent clinical trials, severe adverse reactions, such as acute kidney injury and death, have been observed within days or weeks following AAV administration.1,2,9, 10, 11, 12 To elucidate underlying mechanisms of these toxic responses, we have to have an in-depth understanding of AAV distribution (especially at the early time points after AAV injection) in the context of cell communities and tissue organizations. Given the structural and cellular heterogeneity in a tissue or an organ, it is necessary to use a method that can resolve the spatial and contextual information of AAV distribution in various cell types and histological structures.
The recently developed RNAscope technique, an in situ hybridization (ISH) assay, has the potential to detect nucleic acids at single molecule resolution in an anatomically intact tissue microenvironment.13, 14, 15, 16, 17, 18, 19, 20, 21, 22 Investigators have begun to apply this powerful technique to study the infection of pathogenic viruses such as hepatitis B virus and human immunodeficiency virus.14,19,23,24 In this study, we optimized RNAscope for AAV DNA and RNA detection and systemically investigated an AAV serotype 9 vector (AAV9, referred to as “AAV” in subsequent text) transduction following i.v. and i.m. injection. We revealed the kinetics of cell and nuclear entry of the AAV genome in skeletal muscle, heart, and liver. We also characterized the distribution of the AAV genome in various histological structures in internal organs including the lung, kidney, spleen, brain, pancreas, testis, ovary, lymph node, bone marrow, adrenal gland, and salivary gland. Our study adds a new dimension to the understanding of AAV transduction biology. It also sets a foundation for comprehensively assessing the full impact of AAV gene therapy.
Results
Optimization of RNAscope for AAV DNA and RNA Detection in Muscle Tissue In Situ
The RNAscope technique has never been used for studying the AAV genome (DNA) and transcript (RNA) in muscle. To validate the specificity of RNAscope in detecting AAV DNA and RNA, we included two separate probes (Figure S1). The anti-sense probe (referred to as “GFP probe” herein) targets the sense strand. Hence, it can detect the plus strand version of the single-stranded AAV genome, the converted double-stranded AAV genome, and the RNA transcript. The sense probe (referred to as “GFP-S probe” herein) targets the anti-sense strand. This probe can detect the minus strand version of the single-stranded AAV genome and the converted double-stranded AAV genome. However, it cannot recognize the RNA transcript.
To examine the performance of the GFP and GFP-S probe, we injected 3 × 1011 viral genomes (vg) of AAV.R20-24.GFP into the tibialis anterior (TA) muscle of adult dystrophin-deficient mdx mice, the most commonly used model for Duchenne muscular dystrophy (Figures S1 and S2).25 The TA muscle was harvested at 4 weeks after AAV injection and examined by RNAscope. We first validated the RNAscope technique with three controls including a no-probe control, a positive probe control, and a negative probe control (Figure S2A). The positive probe labels the endogenous mouse peptidylprolyl isomerase B (PPIB) mRNA. The negative probe recognizes the bacterial dapB mRNA. The phosphate-buffered saline (PBS) only is the no probe control. A strong signal was detected with the PPIB probe. No signal was detected with the dapB probe. The PBS control also showed no signal (Figure S2A).
Next, we tested the GFP and GFP-S probe in AAV-injected muscle. In the absence of DNase and RNase treatment, the GFP probe revealed stronger signals in the cytoplasm, especially in the perinuclear region, while the signal detected by the GFP-S probe was mostly limited to the nuclei (Figure S2B). DNase treatment completely removed the GFP-S probe signal while the GFP probe signal was minimally affected. RNase treatment removed most (but not all) cytoplasmic signals detected by the GFP probe (the remaining cytoplasmic signal represents the AAV DNA trapped in the cytosol or cytosolic organelles). Importantly, RNase treatment has no impact on the nuclear signal detected by the GFP and GFP-S probe. These results confirmed the specificity of the GFP and GFP-S probe. In particular, the GFP-S probe only detects the AAV DNA while the GFP probe detects both AAV DNA and RNA. Furthermore, the AAV RNA can be identified by the GFP probe following DNase treatment.
Spatial and Cellular Distribution of the AAV DNA in Striated Muscle following i.v. Injection
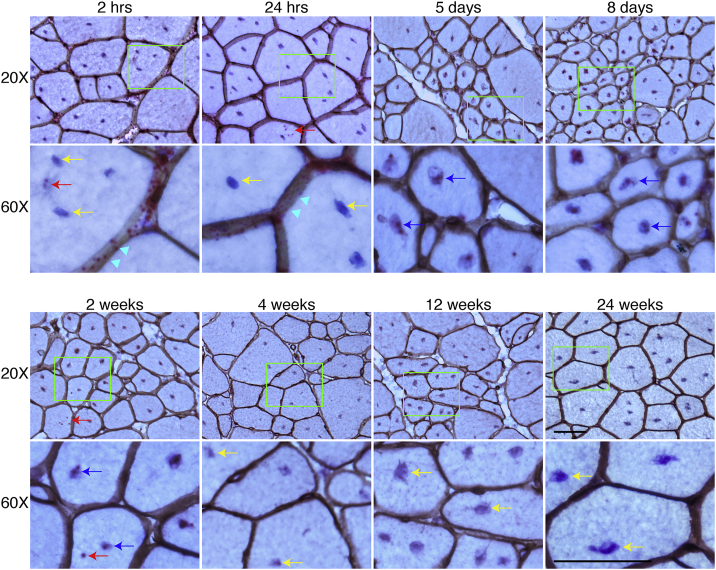
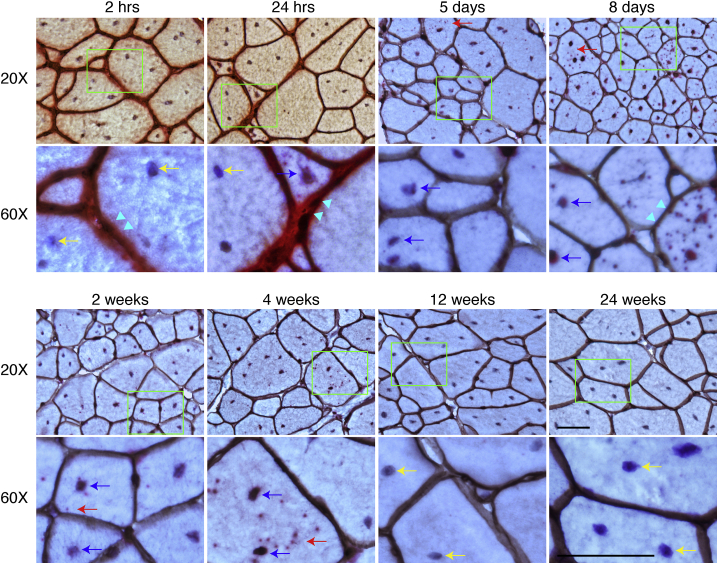
To study the kinetic distribution of the AAV DNA following i.v. injection, we delivered 6 × 1012 vg/mouse AAV.R20-24.GFP to adult mdx mice via the tail vein. Tissues were collected at 2 and 24 h, 5 and 8 days, and 2, 4, 12, and 24 weeks after injection. We marked the myonuclei with hematoxylin and the sarcolemma by laminin immunohistochemical staining. The AAV DNA was detected with the GFP-S probe. In skeletal muscle, the AAV DNA was primarily localized in the interstitial tissue between myofibers at 2 h after injection. Occasionally, the AAV DNA was found in the cytoplasm of few myofibers (Figure 1). By 24 h after injection, most of the AAV DNA remained in the interstitial space. However, there was a clear trend of the attachment of the AAV DNA on the sarcolemma. Cytosolic localization of the AAV DNA also became more obvious at this time. From day 5 to 2 weeks after injection, most of the AAV DNA had accumulated in the myonuclei although some was still visible in the cytoplasm. Starting from 4 weeks after injection, the AAV DNA became barely visible. At 24 weeks after injection, the AAV DNA became completely undetectable (Figure 1).
Figure 1.
AAV DNA Distribution in the TA Muscle at Different Time Points after i.v. Injection
AAV DNA distribution in the TA muscle at 2 and 24 h, 5 and 8 days, and 2, 4, 12, and 24 weeks after i.v. injection. Cyan arrowhead points to sarcolemma; red arrow points to cytoplasmic AAV DNA; yellow arrow points to nuclei without AAV DNA; blue arrow points to nuclei with AAV DNA. Bottom rows are the high-magnification images of the green boxed areas in the corresponding top row images. Scale bars, 50 μm (for both ×20 and ×60 original magnification).
To determine whether the lack of the AAV DNA signal was due to a complete loss of the AAV genome, we performed RNAscope with the GFP probe following DNase treatment to detect the AAV RNA (Figure S3). The AAV RNA transcript was readily visible at 2 weeks after injection and the signal persisted until 24 weeks after injection (the last time point of the study). The presence of the AAV RNA signal suggests active transcription from the AAV DNA. Hence, failure to detect AAV DNA by the GFP-S probe at 24 weeks after injection is not due to the loss of the AAV DNA in muscle cells. To further validate the continuous presence of AAV DNA at and 4 weeks after injection, we performed qPCR (Figure S4). We detected ∼120, ∼15, and ∼10 copies/diploid genome of the AAV genome at 4, 12, and 24 weeks after injection (Figure S4A).
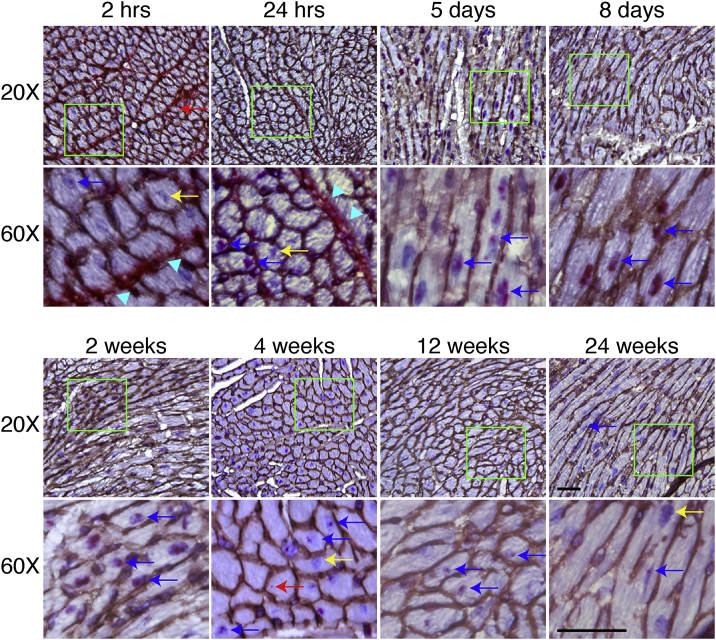
A similar but distinctive AAV DNA trafficking profile was observed in the heart (Figure 2). Consistent with what we observed in skeletal muscle, the AAV DNA was mainly localized in the interstitial tissue at 2- and 24-h time points. However, the AAV DNA was rarely seen in the cytoplasm. Rather, the AAV DNA appeared to have rapidly migrated into the nuclei after AAV entered the cardiomyocytes. Similar to our observation in skeletal muscle, the interstitial AAV DNA was completely cleared by 5 days after AAV injection. Starting from this time point, the AAV DNA was only detected in the nuclei. The nuclear AAV DNA signal appeared more intense at 8 days and 2 weeks but reduced starting from 4 weeks (Figure 2). In skeletal muscle, the definitive AAV DNA signal lasted for 2 weeks only. In contrast, the AAV DNA signal was clearly visible even at 24 weeks after injection, although only in a few cardiomyocytes (Figure 2). As expected, a robust AAV RNA signal was detected at 12 and 24 weeks after injection in cardiomyocytes (Figure S3). On qPCR, we detected ∼180, ∼100, and ∼100 copies/diploid genome at 4, 12, and 24 weeks after injection (Figure S4B).
Figure 2.
AAV DNA Distribution in the Heart at Different Time Points after i.v. Injection
AAV DNA distribution in the heart at 2 and 24 h, 5 and 8 days, and 2, 4, 12, and 24 weeks after i.v. injection. Cyan arrowhead points to sarcolemma; red arrow points to cytoplasmic AAV DNA; yellow arrow points to nuclei without AAV DNA; blue arrow points to nuclei with AAV DNA. Bottom rows are the high-magnification images of the green boxed areas in the corresponding top row images. Scale bars, 50 μm (for both ×20 and ×60 original magnification).
Spatial and Cellular Distribution of the AAV DNA in the Liver following i.v. Injection
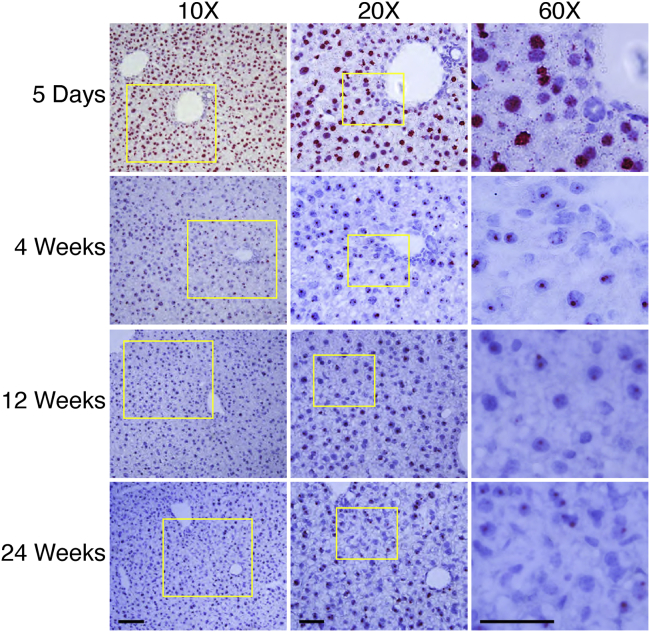
The liver is also one of the most frequently targeted organs in gene therapy. Similar to what we observed in striated muscle, the AAV DNA was mainly located outside the nuclei of the hepatocytes, either in the cytoplasm or the extracellular matrix between hepatocytes at 2 h after injection (Figure S5). At day 5 after injection, we observed uniform AAV DNA distribution throughout the entire liver, and the AAV DNA became primarily concentrated in the nuclei of the hepatocytes (Figure 3; Figure S6). At 4 weeks after injection, the cytoplasmic AAV DNA disappeared completely. At this time point, the AAV DNA signal in the nuclei was also greatly reduced compared to that of the 5-day time point. Interestingly, the number of the AAV DNA-positive nuclei became stable thereafter. There was no apparent decline from 4 weeks to the 24 weeks. Similarly, the AAV genome copy number was maintained between ∼1,100 and ∼1,400 copies/diploid genome from 4 to 24 weeks after injection on qPCR (Figure S4C).
Figure 3.
AAV DNA Distribution in the Liver at Different Time Points after i.v. Injection
At 5 days after i.v. injection, a massive amount of the AAV DNA accumulated in the nuclei, with spotty signals in the cytoplasm of hepatocytes. From 4 to 24 weeks after i.v. injection, the AAV DNA exclusively resided in the nuclei. Scale bars, 100 μm (for ×10 original magnification), 50 μm (for both ×20 and ×60 original magnification).
Spatial and Cellular Distribution of AAV in Other Tissues following i.v. Injection
Next, we evaluated AAV DNA distribution in the brain, lung, kidney, spleen, pancreas, testis, ovary, lymph node, adrenal gland, salivary gland, and bone marrow (Table 1; Figures 4, 5, 6, and 7; Figures S7–S10). Homogeneous spread of the AAV DNA was observed in the brain, lung, salivary gland, and bone marrow but not in the remaining organs we examined (Table 1; Figure S7).
Table 1.
Distribution Pattern of AAV after i.v. Injection
| Organs | Distribution Pattern |
|---|---|
| Kidney | highly accumulated in the glomeruli; scattered in the medulla; no viral DNA is visible in the Bowman’s capsule |
| Spleen | accumulated in the B cell-dominant areas of the white pulp; scattered in the red pulp |
| Lymph node | accumulated in the B cell-enriched cortical lymphoid nodules; scattered in the interfollicular zone |
| Testis | confined in the interstitium, and Leydig cells transduced; no viral DNA in seminiferous tubules |
| Ovary | scattered in central medulla and corpus luteum; thecal cells transduced; no viral DNA in the layer of zona granulosa and the oocytes |
| Adrenal glands | highly concentrated in the zona fasciculata and scattered in the zona glomerulosa and the medulla |
| Pancreas | accumulated in the β cells of the islets |
| Bone marrow and bone | robust localization in the bone marrow, with a few positive signals in the cartilage bone |
| Lung | lung alveolar cells transduced by AAV |
| Salivary glands | acinar cells transduced by AAV |
| Brain | relatively homogeneous distribution is seen in the brain, with viral DNA localized in endothelial cells, neurons, microglia, and astrocytes |
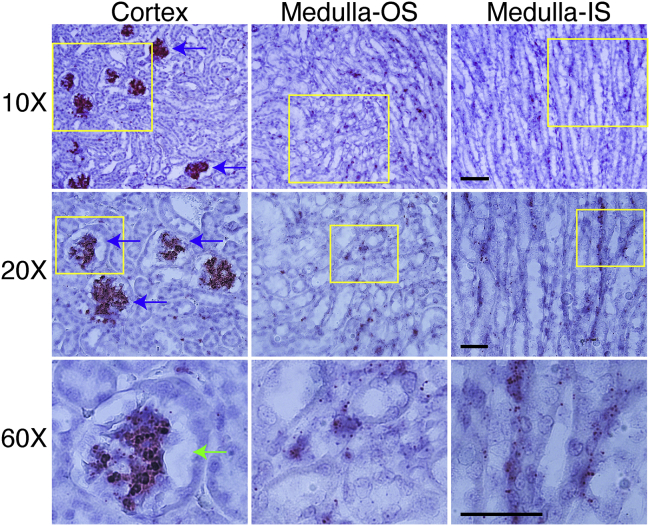
Figure 4.
AAV DNA Distribution in the Kidney at 5 Days after i.v. Injection
The distribution features of AAV are presented for both the cortex and medulla of the kidney. Clearly, the AAV DNA accumulated in the cortical glomerulus (blue arrow). Almost all glomeruli in the cortex contain a massive amount of the AAV DNA. However, no AAV DNA was observed in the Bowman’s capsule (green arrow). The AAV DNA was scattered in the medulla of the kidney, with spotty signals in the epithelium of proximal and distal convoluted tubules. OS, outer stripe; IS, inner stripe. The yellow box indicates the region amplified in higher magnification in the row below. Scale bars, 100 μm (for ×10 original magnification), 50 μm (for both ×20 and ×60 original magnification).
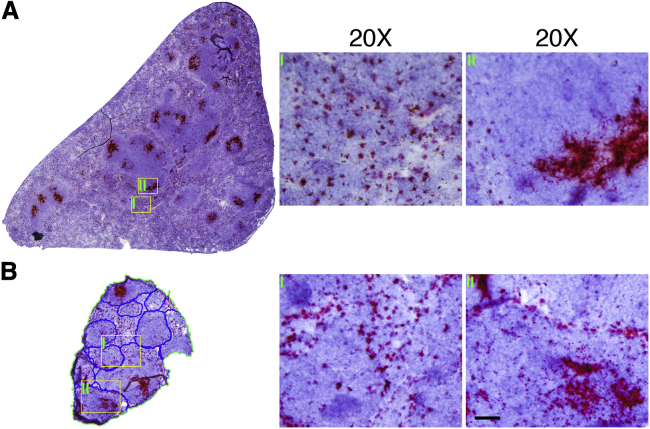
Figure 5.
AAV DNA Distribution in the Lymphoid Tissues at 5 Days after i.v. Injection
(A) AAV DNA in the spleen. Massive distribution in the spleen was observed, accompanied by highly concentrated localization in the white pulp (region II) and scattered cluster distribution in the red pulp (region I). (B) AAV DNA in the lymph node (LN). The accessory mandibular LN, which is located near the submandibular salivary glands, was harvested and the AAV DNA was examined by RNAscope. Substantial quantities of the AAV DNA were found in the LN, with distinguished accumulation in the germinal centers of the lymphoid nodules on the periphery of the LN (region II) and scattered cluster distribution in the interfollicular zone (region I). The border of lymphoid nodules, which contains germinal centers (mainly B cells), is marked with blue lines. Scale bar, 50 μm (for ×20 original magnification).
Figure 6.
AAV DNA Distribution in the Teste at 5 Days and 4 Weeks after i.v. Injection
The seminiferous tubules (S) are separated by interstitium, where Leydig cells and blood vessels are located. At 5 days after injection, AAV DNA was confined to interstitium and there was no AAV DNA inside the seminiferous tubules, which are lined with Sertoli cells and germ cells. The same distribution pattern was seen at 4 weeks after i.v. injection. Green arrow points to AAV DNA in interstitial cells; red arrow points to no AAV DNA in Sertoli and germ cells. Scale bars, 100 μm (for ×10 original magnification), 50 μm (for both ×20 and ×60 original magnification).
Figure 7.
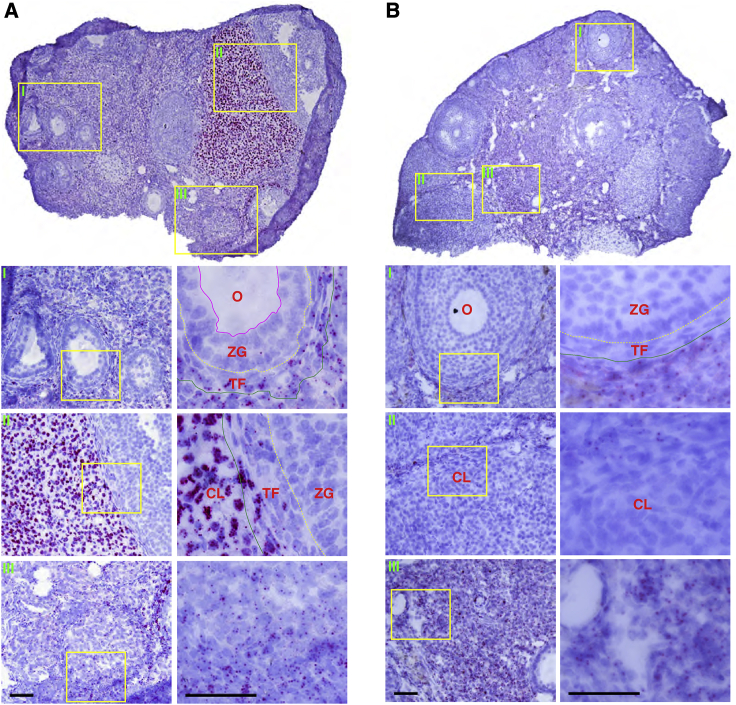
AAV DNA Distribution in the Ovary at 5 Days and 4 Weeks after i.v. Injection
(A) AAV DNA distribution in the ovary at 5 days after i.v. injection. The AAV DNA was scattered in the medulla but highly accumulated in the corpus luteum. No AAV DNA was seen in follicles. The AAV DNA was found in lutein cells and thecal cells but not in granulosa cells and oocytes. (B) AAV DNA distribution in the ovary at 4 weeks after i.v. injection. The AAV DNA in the corpus luteum and thecal cells disappeared, but the AAV DNA in the medulla persisted. No AAV was seen inside the follicles. CL, corpus luteum; O, oocyte; ZG, zona granulosa; TF, theca folliculi. Scale bars, 50 μm (for both ×20 and ×60 original magnification).
In the kidney, the AAV DNA was highly enriched in the cortical glomerulus at 5 days after injection (Figure 4). Scattered AAV DNA was also found in the medulla. Interestingly, we failed to detect the AAV DNA in the Bowman’s capsule (Figure 4). The AAV DNA persisted for 24 weeks in the glomerulus although the signal intensity was greatly reduced at the 24-week time point (Figure S8).
In the spleen, the AAV DNA was concentrated as foci in regions enriched with B cells in the white pulp (Figure 5A; Figure S9). However, in the red pulp of the spleen, the AAV DNA was evenly dispersed throughout the macrophage-enriched region (Figure 5A; Figure S9). In the lymph node, the AAV DNA mainly accumulated in the germinal center, the region dominated by B cells (Figure 5B). In the medulla of the lymph node, the AAV DNA appeared uniformly distributed in immune cells (Figure 5B).
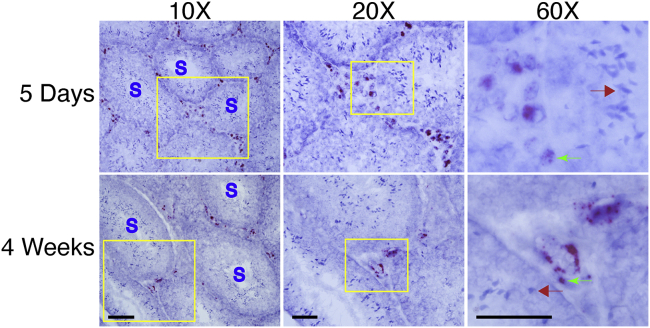
In the testis, the AAV DNA was found in Leydig cells in the interstitium (Figure 6). In the ovary, the AAV DNA was enriched in the corpus luteum and scattered in the central medulla (Figure 7). We failed to detect the AAV DNA in sperms and oocytes (Figures 6 and 7).
In the pancreas, the AAV DNA was observed in a subset of insulin-secreting β cells in the islet but barely detected in the secretory acini and interlobular duct (Figure S10A). In the adrenal gland, the AAV DNA was primarily seen in the cortex, especially in the zona fasciculata (Figure S10B).
Spatial and Cellular Distribution of AAV following i.m. Injection
i.m. injection is primarily used to deliver secreted proteins in clinical gene therapy studies (such as anti-trypsin I and factor IX), although this route is also used in preclinical studies to test gene therapy for muscle diseases. To study the kinetics of AAV distribution following i.m. injection, we injected 3 × 1011 vg/mouse of AAV.R20-24.GFP to the TA muscle of dystrophin null mice. At 2 and 24 h after injection, we observed prominent accumulation of the AAV DNA along the membrane of the muscle cells, with the sporadic deposition in the cytoplasm and nucleus at 24 h after injection (Figure 8). From 5 days to 4 weeks after injection, the AAV DNA became undetectable in the interstitial tissue. During this period, the AAV DNA was mainly found inside muscle cells with noticeable cytoplasmic distribution at 5 and 8 days and pronounced nuclear distribution at 2 and 4 weeks after injection. The AAV DNA was not detected in the TA muscle at 12 and 24 weeks after i.m. injection (Figure 8).
Figure 8.
AAV DNA Distribution in the TA Muscle at Different Time Points after i.m. Injection
At 2 and 24 h after i.m. injection, a considerable amount of the AAV DNA was seen along the muscle membrane, with occasional cytoplasmic and nuclear localization at 24 h after injection. From 5 days to four weeks after i.m. injection, most of the AAV DNA resided in the nuclei (blue arrow), with isolated cytoplasmic distribution (red arrow). From 12 to 24 weeks, no viral DNA was seen intercellularly or intracellularly. Arrowhead points to AAV DNA at the membrane; red arrow points to AAV DNA in the cytoplasm; yellow arrow points to nuclei without AAV DNA; blue arrow points to nuclei with AAV DNA. Scale bars, 50 μm (for both ×20 and ×60 original magnification).
Next, we examined the AAV DNA in neighboring muscle, including the extensor digitorum longus (EDL), gastrocnemius, and quadriceps muscles (Figure S11). Abundant AAV DNA was found in the nuclei of the immediate neighboring muscles such as the EDL and gastrocnemius. Some AAV DNA was also detected in the cytosol in these muscles. For the distantly located quadriceps, the AAV DNA was only observed in the nuclei of a small proportion of muscle cells.
To determine whether local muscle injection can lead to body-wide AAV spreading, we examined the AAV DNA in non-muscle tissues. Previous studies have shown that i.m. injection can result in AAV distribution in the liver.4, 5, 6 At 5 days after bilateral TA muscle injection, we detected substantial distribution of the viral DNA in the liver (mainly in the nuclei) (Figure 9A). To further test whether muscle injection-associated systemic delivery correlated with the AAV loading, we performed unilateral TA muscle injection (Figure 9B). The AAV DNA was easily observed in the nuclei of the hepatocytes, although to a lesser extent than what we saw following bilateral TA injection (Figure 9B). We also detected pronounced AAV DNA distribution in the kidney, spleen, tibia bone marrow, and distant lymph node (Figure 10). The AAV DNA signal was much reduced, although not absent, in the heart, pancreas, lung, brain, testis, ovary, adrenal gland, and salivary gland (Figures 10 and 11; Figure S12).
Figure 9.
AAV DNA Distribution in the Liver at 5 Days after Bilateral and Unilateral i.m. Injections
(A) Distribution of the AAV DNA in the liver after bilateral tibialis anterior (TA) injection. A significant amount of the AAV DNA was found in the nuclei of hepatocytes. (B) The AAV DNA in the liver after unilateral TA injection. Pronounced amount of the AAV DNA was detected in the liver, although to a lesser extent than that of bilateral TA injection. Scale bars, 100 μm (for ×10 original magnification), 50 μm (for both ×20 and ×60 original magnification).
Figure 10.
AAV DNA Distribution in the Kidney, LN, Spleen, Tibia Bone Marrow, and Testis at 5 Days after Bilateral TA Injections
AAV DNA distribution in the kidney, accessory mandibular LN, spleen, tibia bone marrow, and testis at 5 days after bilateral TA injections. Scale bars, 100 μm (for ×10 original magnification), 50 μm (for both ×20 and ×60 original magnification).
Figure 11.
AAV DNA Distribution in the Ovary at 5 days after Bilateral TA Injections
Distribution of the AAV DNA in the ovary at 5 days after bilateral TA injection. CL, corpus luteum; O, oocyte; ZG, zona granulosa; TF, theca folliculi. Scale bars, 50 μm (for both ×20 and ×60 original magnification).
Discussion
Current methods of examining AAV distribution rely on measuring the protein, RNA transcript, and vector genome from the lysate of all cells in an organ or a tissue. These measurements cannot provide the important spatial and contextual information of AAV distribution in different cells within a tissue. In light of the complexity of the cellular composition and structural organization of different tissues/organs, there is an urgent need to study the spatial and contextual features of AAV distribution following administration. Such knowledge is crucial to our understanding of AAV transduction and the host response.
Traditional ISH assays, such as fluorescence ISH (FISH), provide a way to visualize AAV nucleic acid in situ and have been used to study the AAV DNA in the liver,26 testis,8 and neuron tissues.27 However, sensitivity and specificity of traditional ISH assays remain unsatisfactory.13,14 RNAscope is a highly sensitive ISH assay, which can detect nucleic acids in situ at single-molecule resolution.13,14 When combined with histological and immunological staining, RNAscope provides important novel insights in regard to cellular and histological localization of nucleic acids. A few groups have studied AAV DNA and RNA in neuronal tissues using RNAscope or similar single-molecule ISHs.20,21,28,29 However, a comprehensive examination of body-wide AAV transduction has never been performed with RNAscope. To address this important knowledge gap, in this study, we optimized the RNAscope protocol for AAV DNA and RNA detection (Figures S1 and S2). We then studied the spatial and cellular distribution of the AAV DNA in various tissues/organs after i.v. and i.m. injection.
Immediately following i.v. injection (2 h), we found that most of the AAV DNA was restricted to interstitial spaces in skeletal muscle, heart, and liver (Figures 1, 2, and 3; Figure S5). Even at 24 h after injection, there was still a minimum of AAV DNA inside muscle cells in skeletal muscle and heart (Figures 1 and 2). This is in sharp contrast to what was reported in cell culture studies.30, 31, 32, 33 In these in vitro studies, AAV particles are taken up by cells rapidly (usually within 30 min) and enter the nucleus within 2–4 h. Our results suggest that the extracellular matrix may represent an important barrier for systemic AAV delivery.
We arbitrarily selected day 5 as our next time point. By this time point, most of the AAV DNA already reached the nucleus of the tissues we examined (Figures 1, 2, 3, 6, 7, 8, 9, 10, and 11). This experimental design prevented us from comprehensively evaluating potential differences in cytoplasmic trafficking in different tissues. Nevertheless, the day 5 data from skeletal muscle, heart, and liver suggest that there might be a difference in the kinetics of cytosolic trafficking in these three tissues. The AAV DNA was barely detected in the cytoplasm of cardiomyocytes, but it was found in the cytoplasm of some skeletal myofibers and many hepatocytes (Figures 1, 2, and 3). These results suggest that AAV9 can rapidly enter the nucleus of cardiomyocytes but cytosolic trafficking may take a longer time in skeletal muscle cells and liver cells. In other words, strategies that can improve cytosolic trafficking may enhance AAV delivery in skeletal muscle and liver following i.v. injection.
Another interesting finding is the clear reduction of the AAV DNA signal at later time points. The AAV DNA became barely detectable from 4 weeks onward in skeletal muscle, but it was clearly visible at 4 weeks in the heart, although less strong than that at the 2-week time point. The AAV DNA persisted in the heart until 24 weeks (Figures 1 and 2). In the liver, the AAV DNA signal dropped substantially at 4 weeks after injection but remained in the nuclei of many hepatocytes at 24 weeks (Figure 3). After entering the nucleus, the single-stranded AAV genome is converted to the double-stranded transcription-competent genome, which then forms high-molecular-weight concatemers with complexed structures.34, 35, 36 Hence, in the infected tissues, there is a mix of different forms of AAV genomes, including the single-stranded genomes, double-stranded linear monomers and multimers with different orientations, and double-stranded circular monomers and multimers with different orientations.37 While the AAV DNA was detectable in the heart and liver after 4 weeks, surprisingly, the signal was lost in skeletal muscle after 4 weeks (Figures 1, 2, and 3). Since we observed robust AAV transcription even at 24 weeks (Figure S3), we reasoned that the AAV DNA was not lost in skeletal muscle. Quantification of the AAV vector genome copy number at 4, 12, and 24 weeks after injection validated our reasoning (Figure S4). We currently do not have an explanation on why we failed to detect the AAV DNA by RNAscope at these time points. Future studies are needed to clarify the detection limit of RNAscope in the context of AAV transduction in vivo.
The body-wide AAV DNA distribution was examined at 5 days after i.v. injection (Figures 1, 2, 3, 4, 5, 6, and 7). The AAV DNA was relatively evenly distributed in skeletal muscle, heart, liver, brain, lung, salivary gland, and bone marrow (Table 1; Figures 1, 2, and 3; Figure S7). Interestingly, we found selective accumulation of the AAV DNA in certain histological structures in the kidney, spleen, lymph node, gonads, pancreas, and adrenal glands (Table 1; Figures 4, 5, 6, and 7; Figures S7–S10). In the kidney, the AAV DNA was highly concentrated in the glomerulus. This is consistent with the protein expression pattern reported by others.38,39 Taken together, these findings suggest that AAV may represent an ideal candidate vector for treating glomerular diseases.
In the spleen and lymph node, the AAV DNA was highly enriched in regions dominated by B cells (Figure 5; Figure S9). Others have shown that B cells are the predominant cell types transduced by AAV in the spleen,40 and, furthermore, AAV capsids accumulate in splenic germinal centers, B cell-dominant areas.41 Our results correlated well with these publications. Taken together, these observations suggest that B cells may possess yet unidentified mechanisms that promote AAV uptake and/or retention. This may have important implications in understanding the AAV-induced immune response.42
Germline transmission has been an ethical concern for systemic gene therapy.7 A number of groups have evaluated the risk of germline transmission in male animals following i.v. injection.7,8,43, 44, 45, 46 These studies found the AAV DNA in semen and the testis. However, it is unclear whether germ cells are transduced by AAV. It is also unclear whether AAV can reach the ovary in females. To address these important questions, we performed RNAscope on gonads from both male and female animals. In the testis, we detected the AAV DNA in interstitial Leydig cells but not in Sertoli cells and germ cells, including spermatogonia, spermatocytes, spermatids, and spermatozoa (Figure 6). In the ovary, the AAV DNA was readily detected in the corpus luteum and theca folliculi but not in oocytes (Figure 7). Our results suggest that AAV9 cannot transduce germ cells.
The pancreas and adrenal gland are the other two organs that showed disproportional transduction in different histological domains (Figure S10). In the pancreas, the AAV DNA was primarily located in the Langerhans islets in a subset of insulin-secreting β cells although not in glucagon-secreting α cells (Figures S10A). We also did not detect the AAV DNA in exocrine tissues. In the adrenal gland, the AAV DNA was enriched in the zona fasciculata, the domain responsible for secreting glucocorticoids. Interestingly, barely any AAV DNA was detected in the adrenal medulla, the region responsible for secreting adrenaline. These unique distribution patterns should be considered when designing gene therapy for treating diseases related to the pancreas and adrenal gland.
In this study, we also evaluated the kinetic distribution of the AAV DNA following i.m. injection. In the first 24 h, a pattern similar to what we saw in i.v. injection was observed following i.m. injection (Figures 1 and 8). Specifically, the AAV DNA was primarily located in the interstitial tissue with minimal uptake by muscle cells. The AAV DNA was found mainly inside muscle cells between 5 days and 2 weeks after injection (Figure 8). Interestingly, there is a striking difference in the AAV DNA distribution pattern between i.v. and i.m. injection during this period. The AAV DNA was rarely detected in the sarcoplasm following i.v. injection but was abundant in the cytosol following i.m. injection. It is unclear whether the difference is due to local AAV concentration or there may exist a fundamental difference in AAV endocytosis in muscle between two delivery routes. Future studies are needed to clarify the differences.
For i.m. injection in the TA muscle, we expect direct spreading of the AAV particles from the targeted muscle to its neighboring muscles such as the EDL and gastrocnemius. We indeed observed direct spreading (Figure S11). Interestingly, we also found the AAV DNA in the distant quadriceps, which cannot be easily explained by direct spreading (Figure S11). A more likely scenario is spreading through the circulation. In support of this notion, we found the AAV DNA in several internal organs, including the liver, kidney, spleen, heart, lung, brain, pancreas, adrenal gland, salivary gland, gonads, tibia bone marrow, and distant lymph node (Figures 9, 10, and 11; Figures S11 and S12). Additional studies suggest that the distant spreading is dose-dependent (Figure 9). Previous studies suggest that i.m. injection can result in AAV spreading to the liver, lymph node, and gonads.4, 5, 6, 7, 8 Our results suggest that systemic spreading following local injection is much broader than what has been reported. Systemic toxicity should be an important consideration when conducting local gene therapy.
We also point out that the AAV DNA distribution pattern not only holds biological implications when deciding the target cells to treat in gene therapy, but it also has important ramifications for assessing potential toxicity. Recent studies suggest that the AAV DNA sequence per se contributes to tissue toxicity.47,48 Given the relatively low therapeutic index of AAV systemic gene therapy,49 a high dose of AAV gene delivery is required. However, a high dose of AAV is associated with severe adverse effects.9,10,21 Detailed information of AAV distribution, especially in the context of cellular communities and tissue organizations, will help develop novel AAV vectors with the improved therapeutic index.
As the first study to comprehensively evaluate the spatial and cellular distributions of AAV in situ following i.v. and i.m. injection, our studies have some limitations. First, our probe cannot distinguish the single-stranded versus double-stranded AAV genome, nor can it discriminate the episomal form versus the integrated AAV genome. Further optimization of the RNAscope technique or combined use of RNAscope and other molecular techniques will be essential to address these issues. Second, we only focused on AAV9 in this study because it is currently the only AAV serotype approved by the regulatory agency for systemic gene therapy. As more AAV serotypes are being tested in clinical trials, and there is a need to study the spatial and cellular distributions of other AAV serotypes. Third, we only studied the conventional single-stranded AAV vector. Since self-complementary AAV does not require second-strand synthesis for transgene expression and has been used in some clinical trials (such as hemophilia B gene therapy), it will be interesting to compare the histological distribution patterns between the single-stranded and self-complementary AAV vectors. Fourth, we only considered eight different time points in this study. Adding additional time points (such as between 24 h and 5 days) and longer time points (such as 1, 2, and 3 years) will reveal a more complete picture of the histological features of AAV transduction. Last, considering the limit of the rodent study, there is a critical need to use RNAscope to investigate AAV transduction in large mammals.
In summary, we have cataloged the spatial and cellular information of AAV distribution following i.m. and i.v. injection. This systematic interrogation of AAV distribution throughout the body tissues has generated a detailed spatial and cellular map of AAV nucleic acids and their relationships with tissue organizations. Our findings have added a new dimension of information to AAV transduction biology.
Materials and Methods
Animal Care and Experiments
All animal experiments were approved by the Animal Care and Use Committee (ACUC) of the University of Missouri. All animal procedures were performed in accordance with NIH guidelines. Dystrophin null mdx mice (stock no. 001801) were purchased from The Jackson Laboratory. Both male and female mice were used in this study. All the mice are maintained in a specific pathogen-free animal care facility with access to food and water ad libitum.
AAV Production and Injection
In this study, a previously engineered AAV construct is used to examine AAV distribution within the whole-body tissues.50 This AAV construct carries the expression cassette of dystrophin spectrin-like repeats 20–24.GFP (R20–24.GFP), in which GFP is fused in-frame to the C terminus of dystrophin R20–24. The expression of dystrophin R20–24.GFP is driven by the cytomegalovirus (CMV) promoter and the SV40 polyadenylation (SV40 poly(A)) signal (Figure S1A).
AAV9 viruses were produced by triple-plasmid transfection in human embryonic kidney (HEK) 293 cells and three rounds of CsCl ultracentrifugation purification.51,52 The AAV titer was quantitated by real-time PCR using a Fast SYBR Green master mix kit (Applied Biosystems, Foster City, CA, USA) with a pair of primers that amplify a fragment in the CMV promoter. The primers were as follows: forward, 5′-TTACGGTAAACTGCCCACTTG-3′; reverse, 5′-CATAAGGTCATGTACTGGGCATAA-3′.
For i.m. injection, either unilateral or bilateral TA muscles of adult dystrophin null mice (3–9 months old) were injected with AAV according to our established method.50, 51, 52 Each TA muscle received a total of 3 × 1011 vg particles of AAV (in a volume of 50 μL). For i.v. injection, a total of 6 × 1012 vg particles (equivalent to 1.7–2.5 × 1011 vg/g) of AAV (in a volume of 500 μL) were injected into adult dystrophin null mice (3–9 months old) through the tail vein.
Tissue Sample Harvesting and Processing
The whole-body tissues from injected mice were harvested at different time points following i.m. and i.v. injections. The time points include 2 h, 24 h, 5 days, 8 days, 2 weeks, 4 weeks, 12 weeks, and 24 weeks after AAV injections (n = 2/sex/each time point). Except for the tibia bones, all tissues were fixed in 10% neutral buffered formalin (NBF) at 4°C overnight. Then, the fixed tissues were dehydrated in 30% sucrose solution for 3 h, embedded in Tissue-Tek OCT (Sakura Finetek), and snap-frozen in 2-methylbutane with liquid nitrogen.
For the processing of tibia bones, bilateral tibia bones, including knee joints and a small portion of distant femur bones, were harvested from injected mice, and fixed in 10% NBF at 4°C overnight. After rinsing samples with PBS, the bones were decalcified in 14% EDTA for 5 days. Then, the decalcified bones were dehydrated in 30% sucrose for 3 h, embedded in OCT, and snap-frozen in 2-methylbutane with liquid nitrogen.
RNAscope
The RNAscope kit (RNAscope 2.5 high-definition [HD] reagent kit-RED, catalog no. 322350) was purchased from Advanced Cell Diagnostics (ACD). RNAscope was performed on 8- to 10-μm tissue sections according the manufacturer’s instructions. Briefly, tissue sections were blocked with hydrogen peroxide at room temperature for 10 min and boiled in the target retrieval solution for 5 min. Then, tissue slides were covered with protease plus solution and incubated in the hybridization oven at 40°C for 30 min. After rinsing the slides with deionized water, the tissues sections were hybridized with the target probes at 40°C for 2 h. Then, sequential hybridization procedures were performed with pre-amplifier, amplifier, and label probes. The label probes were conjugated with alkaline phosphatase, which reacts with the Fast Red chromogen substrate. The red punctate signal can be viewed under a standard bright-field microscope. Lastly, the tissue sections were counterstained with hematoxylin.
In this study, two target probes were used (Figure S1). AAV DNA was detected with a GFP-S probe (ACD, catalog no. 409971) and the above-described procedure. To detect AAV RNA, the aforementioned protocol was adapted by using a GFP probe (ACD, catalog no. 400281) following DNase I treatment (1:50; Sigma, catalog no. D5319-500UG) right after the target retrieval step. The standard RNAscope protocol was also modified for different purposes. To detect AAV DNA in different tissues and organs, the duration of hydrogen peroxide blocking and/or target retrieval was adjusted according to the recommendation of the manufacturer. To specifically detect viral DNA with the GFP probe, viral RNA was eliminated by treating tissue sections with RNase A (5 mg/mL in PBS; QIAGEN, catalog no. 19101) right before adding the target probe.
Immunohistochemistry
Immunohistochemistry (IHC) was carried out either on serial sections or simultaneously with RNAscope ISH. A Vectastain ABC kit (Vector Laboratories) was used for IHC staining. Laminin IHC was co-stained with RNAscope ISH for muscle sections. After completing the whole ISH procedure, muscle sections were blocked with 1.5% goat serum for 20–30 min. A rabbit anti-laminin antibody (1:1,000; Sigma, L9393) was diluted in 1.5% goat serum. The sections with anti-laminin antibody were incubated at 4°C overnight. After rinsing tissue sections with PBS, secondary antibody was added and incubated with muscle sections at room temperature for 1 h. Then, the sections were covered with ABC mixture and the 3,3′-diaminobenzidine (DAB) chromogen substrate. At last, hematoxylin was added for counterstaining.
For serial section IHC staining, macrophages were revealed with a rat anti-mouse F4/80 antibody (1:100; Invitrogen, RM2920; clone no. CI:A3-1), pan-T cells were identified with a rat anti-mouse CD3 antibody (1:100; BD Biosciences, 555273; clone no. 17A2), pan-B cells were labeled with a rat anti-mouse CD45R/B220 antibody (1:50; BD Biosciences, 553084; clone no. RA3-6B2),53 β cells were confirmed with a mouse anti-insulin and proinsulin antibody (1:2,000; Abcam, ab8304; clone no. D6C4), and α cells were examined with a mouse anti-glucagon antibody (1:5,000; Novus Biologicals, NBP2-21803; clone no. 09). The IHC procedure is the same as the one described above for laminin staining.
AAV Genome Copy Number Quantification
AAV genome copy number in the TA muscle, heart, and liver from 4 to 24 weeks after i.v. injection was quantified as the previously published protocol.54 In brief, genomic DNA was extracted from OCT-embedded tissue samples. Quantitative TaqMan PCR assays were carried out using PrimeTime qPCR probe assays (Integrated DNA Technologies) to detect the SV40 poly(A) region. The forward primer is 5′-CCAGACATGATAAGATACATTGATGAGTT-3′, the reverse primer is 5′-AGCAATAGCATCACAAATTTCACAA-3′, and the probe is 5′-GACAAACCACAACTAGAATGCAGTGAAAAAAATGCT-3′. The data are presented as the viral genome copy number per diploid genome.
Author Contributions
Y.L. and D.D. designed the experiments. J.Z., A.P., L.W., J.F.K., K.Z., Y.Y., and Y.L. performed the experiments. D.D. and Y.L. interpreted the data and wrote the manuscript. All authors have read and approved the submission.
Conflicts of Interest
D.D. is a member of the scientific advisory board and equity holder of Solid Biosciences, LLC. D.D. has received research funding unrelated to this project from Solid Biosciences, LLC in last three years. D.D. and Y. L. have received research funding unrelated to this project from Edgewise Therapeutics in last three years. The remaining authors declare no competing interests.
Acknowledgments
We thank Drs. Stefan G. Sarafianos, Maritza N. Puray-Chavez, and Huatao Guo for assistance with techniques. We are grateful for Drs. Adam G. Schrum and Kimberly Laffey for providing the antibodies against CD3 and B220. We also thank Dr. Douglas C. Miller for the help of reviewing the histology of mouse brain tissues. This work was supported by the Duchenne Parent Project, the Netherlands (to Y.L.), National Institute of Health grants NS-90634 and AR-70517 (to D.D.), and by the Jackson Freel DMD Research Fund (to D.D.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.08.006.
Contributor Information
Dongsheng Duan, Email: duand@health.missouri.edu.
Yi Lai, Email: laiy@health.missouri.edu.
Supplemental Information
References
- 1.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 3.Duan D. Systemic delivery of adeno-associated viral vectors. Curr. Opin. Virol. 2016;21:16–25. doi: 10.1016/j.coviro.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greig J.A., Peng H., Ohlstein J., Medina-Jaszek C.A., Ahonkhai O., Mentzinger A., Grant R.L., Roy S., Chen S.J., Bell P. Intramuscular injection of AAV8 in mice and macaques is associated with substantial hepatic targeting and transgene expression. PLoS ONE. 2014;9:e112268. doi: 10.1371/journal.pone.0112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favre D., Provost N., Blouin V., Blancho G., Chérel Y., Salvetti A., Moullier P. Immediate and long-term safety of recombinant adeno-associated virus injection into the nonhuman primate muscle. Mol. Ther. 2001;4:559–566. doi: 10.1006/mthe.2001.0494. [DOI] [PubMed] [Google Scholar]
- 6.Herzog R.W., Yang E.Y., Couto L.B., Hagstrom J.N., Elwell D., Fields P.A., Burton M., Bellinger D.A., Read M.S., Brinkhous K.M. Long-term correction of canine hemophilia B by gene transfer of blood coagulation factor IX mediated by adeno-associated viral vector. Nat. Med. 1999;5:56–63. doi: 10.1038/4743. [DOI] [PubMed] [Google Scholar]
- 7.Gene therapy and the germline. Nat. Med. 1999;5:245. doi: 10.1038/6429. [DOI] [PubMed] [Google Scholar]
- 8.Arruda V.R., Fields P.A., Milner R., Wainwright L., De Miguel M.P., Donovan P.J., Herzog R.W., Nichols T.C., Biegel J.A., Razavi M. Lack of germline transmission of vector sequences following systemic administration of recombinant AAV-2 vector in males. Mol. Ther. 2001;4:586–592. doi: 10.1006/mthe.2001.0491. [DOI] [PubMed] [Google Scholar]
- 9.Srivastava A. AAV vectors: are they safe? Hum. Gene Ther. 2020;31:697–699. doi: 10.1089/hum.2020.187. [DOI] [PubMed] [Google Scholar]
- 10.Wilson J.M., Flotte T.R. Moving forward after two deaths in a gene therapy trial of myotubular myopathy. Hum. Gene Ther. 2020;31:695–696. doi: 10.1089/hum.2020.182. [DOI] [PubMed] [Google Scholar]
- 11.Duan D. Micro-dystrophin gene therapy goes systemic in Duchenne muscular dystrophy patients. Hum. Gene Ther. 2018;29:733–736. doi: 10.1089/hum.2018.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan D. Systemic AAV micro-dystrophin gene therapy for Duchenne muscular dystrophy. Mol. Ther. 2018;26:2337–2356. doi: 10.1016/j.ymthe.2018.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang F., Flanagan J., Su N., Wang L.C., Bui S., Nielson A., Wu X., Vo H.T., Ma X.J., Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deleage C., Wietgrefe S.W., Del Prete G., Morcock D.R., Hao X.P., Piatak M., Jr., Bess J., Anderson J.L., Perkey K.E., Reilly C. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog. Immun. 2016;1:68–106. doi: 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu D.C., Sunyakumthorn P., Wegner M., Schuetz A., Silsorn D., Estes J.D., Deleage C., Tomusange K., Lakhashe S.K., Ruprecht R.M. Central nervous system inflammation and infection during early, nonaccelerated simian-human immunodeficiency virus infection in rhesus macaques. J. Virol. 2018;92 doi: 10.1128/JVI.00222-18. e00222-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeler A.M., Sapp E., Chase K., Sottosanti E., Danielson E., Pfister E., Stoica L., DiFiglia M., Aronin N., Sena-Esteves M. Cellular analysis of silencing the Huntington’s disease gene using AAV9 mediated delivery of artificial micro RNA into the striatum of Q140/Q140 mice. J. Huntingtons Dis. 2016;5:239–248. doi: 10.3233/JHD-160215. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z., Portier B.P., Gruver A.M., Bui S., Wang H., Su N., Vo H.T., Ma X.J., Luo Y., Budd G.T., Tubbs R.R. Automated quantitative RNA in situ hybridization for resolution of equivocal and heterogeneous ERBB2 (HER2) status in invasive breast carcinoma. J. Mol. Diagn. 2013;15:210–219. doi: 10.1016/j.jmoldx.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Amini Chermahini G., Rashnonejad A., Harper S.Q. RNAscope in situ hybridization-based method for detecting DUX4 RNA expression in vitro. RNA. 2019;25:1211–1217. doi: 10.1261/rna.070177.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deleage C., Chan C.N., Busman-Sahay K., Estes J.D. Next-generation in situ hybridization approaches to define and quantify HIV and SIV reservoirs in tissue microenvironments. Retrovirology. 2018;15:4. doi: 10.1186/s12977-017-0387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabinski T.M., Kneynsberg A., Manfredsson F.P., Kanaan N.M. A method for combining RNAscope in situ hybridization with immunohistochemistry in thick free-floating brain sections and primary neuronal cultures. PLoS ONE. 2015;10:e0120120. doi: 10.1371/journal.pone.0120120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinderer C., Katz N., Buza E.L., Dyer C., Goode T., Bell P., Richman L., Wilson J.M. Severe toxicity in nonhuman primates and piglets following high-dose intravenous administration of an adeno-associated virus vector expressing human SMN. Hum. Gene Ther. 2018;29:285–298. doi: 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collaud F., Bortolussi G., Guianvarc’h L., Aronson S.J., Bordet T., Veron P., Charles S., Vidal P., Sola M.S., Rundwasser S. Preclinical development of an AAV8-hUGT1A1 vector for the treatment of Crigler-Najjar syndrome. Mol. Ther. Methods Clin. Dev. 2018;12:157–174. doi: 10.1016/j.omtm.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X., Lu W., Zheng Y., Wang W., Bai L., Chen L., Feng Y., Zhang Z., Yuan Z. In situ analysis of intrahepatic virological events in chronic hepatitis B virus infection. J. Clin. Invest. 2016;126:1079–1092. doi: 10.1172/JCI83339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puray-Chavez M., Tedbury P.R., Huber A.D., Ukah O.B., Yapo V., Liu D., Ji J., Wolf J.J., Engelman A.N., Sarafianos S.G. Multiplex single-cell visualization of nucleic acids and protein during HIV infection. Nat. Commun. 2017;8:1882. doi: 10.1038/s41467-017-01693-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGreevy J.W., Hakim C.H., McIntosh M.A., Duan D. Animal models of Duchenne muscular dystrophy: from basic mechanisms to gene therapy. Dis. Model. Mech. 2015;8:195–213. doi: 10.1242/dmm.018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao C.H., Nakai H., Thompson A.R., Storm T.A., Chiu W., Snyder R.O., Kay M.A. Nonrandom transduction of recombinant adeno-associated virus vectors in mouse hepatocytes in vivo: cell cycling does not influence hepatocyte transduction. J. Virol. 2000;74:3793–3803. doi: 10.1128/jvi.74.8.3793-3803.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter J.E., Gurda B.L., Yoon S.Y., Castle M.J., Wolfe J.H. In situ hybridization for detection of AAV-mediated gene expression. Methods Mol. Biol. 2019;1950:107–122. doi: 10.1007/978-1-4939-9139-6_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polinski N.K., Manfredsson F.P., Benskey M.J., Fischer D.L., Kemp C.J., Steece-Collier K., Sandoval I.M., Paumier K.L., Sortwell C.E. Impact of age and vector construct on striatal and nigral transgene expression. Mol. Ther. Methods Clin. Dev. 2016;3:16082. doi: 10.1038/mtm.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Polinski N.K., Gombash S.E., Manfredsson F.P., Lipton J.W., Kemp C.J., Cole-Strauss A., Kanaan N.M., Steece-Collier K., Kuhn N.C., Wohlgenant S.L., Sortwell C.E. Recombinant adenoassociated virus 2/5-mediated gene transfer is reduced in the aged rat midbrain. Neurobiol. Aging. 2015;36:1110–1120. doi: 10.1016/j.neurobiolaging.2014.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao P.J., Samulski R.J. Cytoplasmic trafficking, endosomal escape, and peri-nuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J. Virol. 2012;86:10462–10473. doi: 10.1128/JVI.00935-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartlett J.S., Wilcher R., Samulski R.J. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 2000;74:2777–2785. doi: 10.1128/jvi.74.6.2777-2785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lux K., Goerlitz N., Schlemminger S., Perabo L., Goldnau D., Endell J., Leike K., Kofler D.M., Finke S., Hallek M., Büning H. Green fluorescent protein-tagged adeno-associated virus particles allow the study of cytosolic and nuclear trafficking. J. Virol. 2005;79:11776–11787. doi: 10.1128/JVI.79.18.11776-11787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding W., Zhang L., Yan Z., Engelhardt J.F. Intracellular trafficking of adeno-associated viral vectors. Gene Ther. 2005;12:873–880. doi: 10.1038/sj.gt.3302527. [DOI] [PubMed] [Google Scholar]
- 34.Penaud-Budloo M., Le Guiner C., Nowrouzi A., Toromanoff A., Chérel Y., Chenuaud P., Schmidt M., von Kalle C., Rolling F., Moullier P., Snyder R.O. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J. Virol. 2008;82:7875–7885. doi: 10.1128/JVI.00649-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duan D., Sharma P., Yang J., Yue Y., Dudus L., Zhang Y., Fisher K.J., Engelhardt J.F. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent-Lacaze N., Snyder R.O., Gluzman R., Bohl D., Lagarde C., Danos O. Structure of adeno-associated virus vector DNA following transduction of the skeletal muscle. J. Virol. 1999;73:1949–1955. doi: 10.1128/jvi.73.3.1949-1955.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun X., Lu Y., Bish L.T., Calcedo R., Wilson J.M., Gao G. Molecular analysis of vector genome structures following liver transduction by conventional and self-complementary adeno-associated viral serotype vectors in murine and nonhuman primate models. Hum. Gene Ther. 2010;21:750–761. doi: 10.1089/hum.2009.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubin J.D., Nguyen T.V., Allen K.L., Ayasoufi K., Barry M.A. Comparison of gene delivery to the kidney by adenovirus, adeno-associated virus, and lentiviral vectors after intravenous and direct kidney injections. Hum. Gene Ther. 2019;30:1559–1571. doi: 10.1089/hum.2019.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hillestad M.L., Guenzel A.J., Nath K.A., Barry M.A. A vector-host system to fingerprint virus tropism. Hum. Gene Ther. 2012;23:1116–1126. doi: 10.1089/hum.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang J.F., Toulmin S.A., Brida K.L., Eisenlohr L.C., Davidson B.L. Standard screening methods underreport AAV-mediated transduction and gene editing. Nat. Commun. 2019;10:3415. doi: 10.1038/s41467-019-11321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., Bell P., Somanathan S., Wang Q., He Z., Yu H., McMenamin D., Goode T., Calcedo R., Wilson J.M. Comparative study of liver gene transfer with AAV vectors based on natural and engineered AAV capsids. Mol. Ther. 2015;23:1877–1887. doi: 10.1038/mt.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao W., Chirmule N., Schnell M.A., Tazelaar J., Hughes J.V., Wilson J.M. Route of administration determines induction of T-cell-independent humoral responses to adeno-associated virus vectors. Mol. Ther. 2000;1:323–329. doi: 10.1006/mthe.2000.0045. [DOI] [PubMed] [Google Scholar]
- 43.Favaro P., Downey H.D., Zhou J.S., Wright J.F., Hauck B., Mingozzi F., High K.A., Arruda V.R. Host and vector-dependent effects on the risk of germline transmission of AAV vectors. Mol. Ther. 2009;17:1022–1030. doi: 10.1038/mt.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Favaro P., Finn J.D., Siner J.I., Wright J.F., High K.A., Arruda V.R. Safety of liver gene transfer following peripheral intravascular delivery of adeno-associated virus (AAV)-5 and AAV-6 in a large animal model. Hum. Gene Ther. 2011;22:843–852. doi: 10.1089/hum.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuettrumpf J., Liu J.H., Couto L.B., Addya K., Leonard D.G., Zhen Z., Sommer J., Arruda V.R., Arruda V.R. Inadvertent germline transmission of AAV2 vector: findings in a rabbit model correlate with those in a human clinical trial. Mol. Ther. 2006;13:1064–1073. doi: 10.1016/j.ymthe.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z., Zhu T., Qiao C., Zhou L., Wang B., Zhang J., Chen C., Li J., Xiao X. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat. Biotechnol. 2005;23:321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- 47.Xiong W., Wu D.M., Xue Y., Wang S.K., Chung M.J., Ji X., Rana P., Zhao S.R., Mai S., Cepko C.L. AAV cis-regulatory sequences are correlated with ocular toxicity. Proc. Natl. Acad. Sci. USA. 2019;116:5785–5794. doi: 10.1073/pnas.1821000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faust S.M., Bell P., Cutler B.J., Ashley S.N., Zhu Y., Rabinowitz J.E., Wilson J.M. CpG-depleted adeno-associated virus vectors evade immune detection. J. Clin. Invest. 2013;123:2994–3001. doi: 10.1172/JCI68205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mével M., Bouzelha M., Leray A., Pacouret S., Guilbaud M., Penaud-Budloo M., Alvarez-Dorta D., Dubreil L., Gouin S.G., Combal J.P. Chemical modification of the adeno-associated virus capsid to improve gene delivery. Chem. Sci. (Camb.) 2020;11:1122–1131. doi: 10.1039/c9sc04189c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao J., Kodippili K., Yue Y., Hakim C.H., Wasala L., Pan X., Zhang K., Yang N.N., Duan D., Lai Y. Dystrophin contains multiple independent membrane-binding domains. Hum. Mol. Genet. 2016;25:3647–3653. doi: 10.1093/hmg/ddw210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel A., Zhao J., Yue Y., Zhang K., Duan D., Lai Y. Dystrophin R16/17-syntrophin PDZ fusion protein restores sarcolemmal nNOSμ. Skelet. Muscle. 2018;8:36. doi: 10.1186/s13395-018-0182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lai Y., Zhao J., Yue Y., Duan D. α2 and α3 helices of dystrophin R16 and R17 frame a microdomain in the α1 helix of dystrophin R17 for neuronal NOS binding. Proc. Natl. Acad. Sci. USA. 2013;110:525–530. doi: 10.1073/pnas.1211431109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kunder S., Calzada-Wack J., Hölzlwimmer G., Müller J., Kloss C., Howat W., Schmidt J., Höfler H., Warren M., Quintanilla-Martinez L. A comprehensive antibody panel for immunohistochemical analysis of formalin-fixed, paraffin-embedded hematopoietic neoplasms of mice: analysis of mouse specific and human antibodies cross-reactive with murine tissue. Toxicol. Pathol. 2007;35:366–375. doi: 10.1080/01926230701230296. [DOI] [PubMed] [Google Scholar]
- 54.Hakim C.H., Wasala N.B., Pan X., Kodippili K., Yue Y., Zhang K., Yao G., Haffner B., Duan S.X., Ramos J. A five-repeat micro-dystrophin gene ameliorated dystrophic phenotype in the severe DBA/2J-mdx model of Duchenne muscular dystrophy. Mol. Ther. Methods Clin. Dev. 2017;6:216–230. doi: 10.1016/j.omtm.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.