Abstract
Introduction
The association of Vitamin D and children with asthma is known and there are several individual studies on Vitamin D polymorphisms. However, systematic reviews on all vitamin D associated gene polymorphisms have not been done in children with asthma.
Objective
To investigate the association of Vitamin D associated gene polymorphisms and asthma in children (0–18 years) by systematic review and meta-analytic approach.
Methods
Our search included 20 full text articles of which 15 were case control studies and 5 used family based linkage disequilibrium method. Total of 2491cases and 3682 controls were included in case control studies, with mean age of 9.58 years and 10.16 years respectively. Quantitative and qualitative analysis were done.
Results
Quantitative analysis revealed significant association with protective effect of Apa1 polymorphism in allele (OR 0.81 (0.71,0.91) and homozygous major form (OR 0.83 (0.70,0.98) and Taq 1 minor allele in homozygous form (OR 0.73 (0.58,0.92) in children with asthma. However, the minor allele of Apa1(OR 1.21 (1.07,1.37), Bsm 1 in heterozygous (OR 1.35 (1.07,1.71) and homozygous minor form (OR 1.95 (1.59,2.39), major allele of Fok1(OR1.34 (1.17,1.52) and Taq1 (OR 1.22 (1.08,1.38) were found to be increasing the odds of asthma. Ethnic variations were noted in subgroup analysis. Qualitative analysis of the polymorphisms of the Vitamin D associated metabolic genes also showed significant associations.
Conclusion
Our review shows significant associations with VDR polymorphisms - Apa1, Bsm1, Fok 1, Taq 1, polymorphisms of Vitamin D metabolic genes – CYP27A1, CYP 2R1, CYP 24A1, GC and genes related to Vitamin D response element (VDRE) in children with asthma. Conducting large studies involving various ethnic regions will strengthen our knowledge on the association and aid in targeted interventions for control of asthma in children.
Keywords: Biotechnology, Genetics, Asthma in children, Vitamin D associated gene polymorphisms, Single nucleotide polymorphisms
Biotechnology; Genetics; Asthma in children; Vitamin D associated gene polymorphisms; Single nucleotide polymorphisms
1. Introduction
With 325 million people suffering worldwide, asthma is the most common non communicable disease in children with increasing prevalence of 0.02 and 0.06% respectively in the age group of 13–14 years and 6–7years respectively [1, 2]. Although it is well known that positive atopic status, exposure and sensitization to environmental allergens, and/or familial allergic diseases are significant risk factors associated with the development of asthma, recent evidence suggest that vitamin D deficiency may predispose to allergic phenotype [3, 4]. Vitamin D through its action on the immune system regulation seems to play a pivotal role in allergic diseases [5].
Genes associated with Vitamin D in metabolic pathway, CYP27B1, which increases the expression of 1,25(OH)2 D3 in cell types, CYP27A1 and CYP2R1 which influence levels of active Vitamin D, GC transport gene and Vitamin D receptor (VDR), one of the super family of nuclear receptors found in respiratory epithelial and bronchial smooth muscle that increases the expression of various cytokines and interleukins all contribute to inflammatory role of Vitamin D [6].
Number of studies have looked into the association of vitamin D levels, genotypic polymorphisms in Vitamin D associated genes with asthma as well as spirometric parameters. Based on all the studies, 5 systematic reviews have also been done. The reviews have looked into the association of VDR polymorphism with asthma in all age groups. Among them only one meta analysis done by Zhao et al had looked specifically for association in children [7, 8, 9, 10, 11]. Age can be a significant factor in analysing the association of the single nucleotide polymorphism (SNP) when all parameters are similar as shown by studies done by Poon et al and Raby et al. [12, 13]. Apart from these studies, several others have looked into the association of polymorphism of Vitamin D metabolic genes with asthma and have found them also to be contributing to the disease [14, 15, 16, 17]. Hence, there is a need to look into the strength of the association of all the genes associated with Vitamin D pathway and asthma in children independently. Our systematic review is an attempt to look into the association of all the Vitamin D related genes in children.
2. Methods
2.1. Protocol and registration
We registered the systematic review protocol with PROSPERO prior to starting the review process (CRD42017058266) [18].
2.2. Eligibility criteria for considering studies for this review
The eligibility criteria of the studies included were - [1] study should have been an original study in human population [2] case control design or family based analysis [3] Study population should comprise of children less than 18 years [4] study should have been done on association of Vitamin D associated genes and their polymorphism with asthma in children. Only studies reported in English language were included and there was no cut off date of publication or geographical barrier for exclusion.
2.3. Search strategy
We designed the search strategy based on the key words included in the study. The main search strategy consisted of searching the following electronic databases from inception till January 2020. The search combined terms for asthma, vitamin D associated gene polymorphism and pediatrics. The search terms included were (asthma OR reactive airway OR wheeze OR Asthma OR Respiratory Hypersensitivity OR Bronchial Hyperreactivity), (child OR pediatrics OR pediatric OR infan$ OR toddler$ OR boy$ OR girl$ OR kid$ OR school$ OR juvenile$ OR teen$ OR pubescen$ OR adolescen$), (“Genetic Polymorphism” OR “Genetic polymorphisms” OR “single nucleotide polymorphism” OR SNPs OR “Single nucleotide polymorphisms”), (Vitamin D, hydrocholecalciferols, 25, hydroxyvitamin D2, cholecalciferol, Vitamin D2 or Vitamin D3).
The planning of the study and protocol development was done in accordance with Cochrane handbook for systematic reviews of interventions v.5.1.0 (updated 2011) [19]. The reporting format of the study was as per preferred reporting items for systematic review and meta analysis (PRISMA) statement [20].
2.4. Search methodology
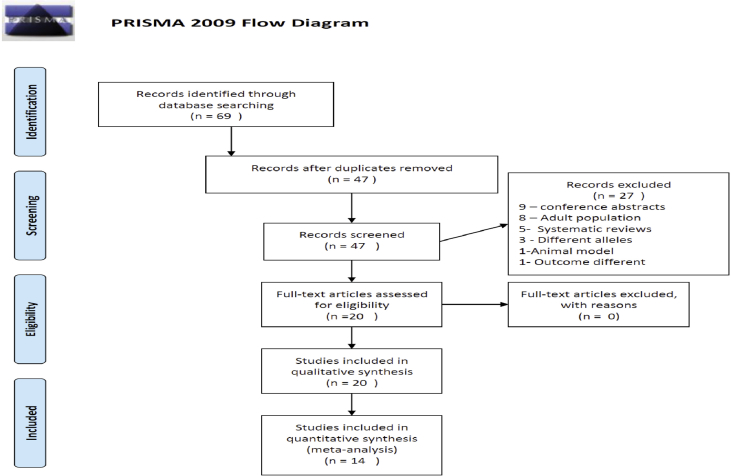
We conducted a literature search in the National Library of Medicine (NLM) MEDLINE, Google Scholar and EMBASE database for studies using the search strategy. The search filters were applied to restrict studies done in human subjects and those in English language. The search was done by two authors independently. Since, there was a time lag between the search date and completion of the review we did an initial search till March 2017 and have updated the search again in various databases until January 2020. The conflict, if any, between the authors was resolved by discussion. A total of 69 studies were obtained by searching all the listed database by each author independently. After removing duplicates there were 47 references identified and screened for eligibility criteria for inclusions. After exclusion of 27 studies due to reasons cited in diagram, 20 studies were taken for final analysis (Figure 1).
Figure 1.
PRISMA diagram of the search done [20].
2.5. Data collection and extraction
Data was extracted using a pre-tested standard data extraction form. Details of cases, controls, ethnicity, race, sex, genotypic methods were extracted for each study. Quality Scores were assessed using a modified scoring based on the quality Scoring used by Srivastava and Srivastava 2001 [21, 22].
2.6. Statistical analysis
Meta analysis was performed using the Odds ratio (OR) and the 95 % confidence intervals (CI) calculated by pooling the results from each individual study when there were similarity across the reported SNPs. Random effect model with inverse variance method of pooling data was used in consideration with the diverse nature of the studies included in this review. All data analysis was done using STATA 14 [23]. Heterogeneity in each analysis was assessed based on the I2 statistics following the guidance provided in the Cochrane handbook for classifying the heterogeneity as moderate (I2: 30–50 %), substantial (I2: 50–75 %) and considerable (I2: 75–100 %) [24]. Subgroup analyses were conducted to assess the association based on the ethnicity with major or minor allele for the SNPs as well as for individual alleles. Forest plots were employed to evaluate the weighted average of the study results. If the number of studies was not adequate enough to calculate the forest plot the results were qualitatively analysed. Publication bias was assessed based on the symmetry observed in the funnel plot.
3. Results
3.1. Demographic details of the study included
A total of 20 studies were taken for analysis. 15 studies used case control method to analyse the significance of polymorphisms and the rest had used family based linkage disequilibrium method of analysis. Among the 15 studies, study done by Munkhbayalak et al had included Taiwan and Mongolese population individually and hence they have been included separately making it as data available from 16 studies. A total of 2491 individual cases were included apart from those involved in family-based studies with a mean age of 9.58 years. The total number of controls included in the study were 3682, with a mean age of 10.16 years. One of the studies did not have age matched controls and in another age of the controls was not mentioned. Hardy Weinberg equilibrium had been calculated in almost all studies. The following table gives detail demographic characters of the studies involved (Table 1).
Table 1.
Demographic details extracted from the studies included in the analysis.
| S No | Studies | Year of pub | Country & Ethnicity | Population | Type of study | Method | Unit analyzed | HWE tested | No of cases | Mean age | No of controls | Mean age of controls | Matching bet case &Controls |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Einisman et al [25] | 2015 | Chile American |
Hospital based | Case control | PCR | Genotype | Not specified | 75 | 9.1yr (6 -15y) | 227 | 10.3 (2–12 yr) | Yes |
| 2 | Fawzy et al [26] | 2018 | Egypt Egyptian |
Hospital | Case control | RT PCR analysis | Genotype | Yes | 96 | 10 | 96 | 7–14 yrs | yes |
| 3 | Hou et al [27] | 2017 | China Asians |
Hospital | Case control | PCR- RFLP | SNP | Not specified | 70 | 8.84 +/- 3.21 | 70 | 8.04+/- 3.01 | yes |
| 4 | Hutchinson et al [28] | 2018 | Ireland European |
Hospital | Case control | RFLP | Genotype | Yes | 44 | 8.7 | 55 | Mean age not mentioned | no |
| 5 | Iordanidou et al [29] | 2014 | Greece European |
Hospital | Case control | PCR | Gene | Yes | 127 | 8.4+/- 0.2 | 91 | 9.6+/- 1.04 | Yes |
| 6 | Ismail et al [30] | 2013 | Egypt Egyptian |
Hospital Based | Case control | PCR RFLP | SNP | No | 51 | 8.6 | 33 | 7.8 | Yes |
| 7 | Leung et al [17] | 2015 | Hongkong Asians |
Hospital | Case control | Taqmann (PCR amplification) | SNP | Yes | 914 | 11+/- 0.4 | 1231 | 13.7 +/- 4.5 | Yes |
| 8 | Kilic et al [31] | 2019 | Turkish European |
Hospital | Case control | RFLP | SNP | YES | 100 | 9.5+/- 2.8 | 80 | 9.5+/-2.5 | YES |
| 9 | Maalmi et al [32] | 2013 | Tunisia African |
Population | Case control | PCR RFLP | Genotype | Yes | 155 | 9.1 (4 16yrs) | 225 | 9.5 (2–16yrs) | Yes |
| 10 | Munkhbayarlakh et al - Taiwan [33] | 2019 | Taiwan Asian |
Hospital | Case control | PCR amplification (MassARRAY) | SNP | Yes | 94 | 7.77+/- 4.09 | 129 | 28.11+/- 9.67 | no |
| 11 | Munkhbayarlakh et al - Mongolian [33] | 2019 | Mongolian Asian |
Hospital | Case control | PCR Amplification (MassARRAY) | SNP | Yes | 115 | 11.94+/- 10.28 | 256 | 27.75+/- 13.16 | No |
| 12 | Nabih et al [34] | 2014 | Egypt Egyptian |
Community | Case control | PCR | Genotype | Yes | 180 | 7.98+/-3.22 | 180 | 8.5+/- 2.22 | YES |
| 13 | NavasNazario et al [35] | 2015 | Yale European |
Community | Case control | PCR | Genotype | Yes | 81 | 5.8 | 247 | 5.6 | Yes |
| 14 | Papadopoulou et al [36] | 2015 | Cypriot European |
Community | Case control | PCR | Genotype allele | Yes | 190 | 16.9 (16–18year) | 671 | 17 (16–18yrs) | yes |
| 15 | Pillai et al [37] | 2011 | America African American |
Hospital | Case control | PCR | SNP | Yes | 139 | 11.2 (6-17y) | 74 | 11.8 (6-17y) | Yes |
| 16 |
Santos et al [38] |
2018 |
Brazil |
Hospital |
Observational, Case control |
PCR |
Promoter region |
Not specified |
60 |
8.7+/-2.5 |
17 |
10.8 = /- 2.2 |
yes |
| Studies based on family based analysis of transmission | |||||||||||||
| 1 | Bosse et al [15] | 2009 | Canada Canadian |
Family Based | FBAT | PCR | SNP | Yes | 1064 probands 210 | 17.6 | Not specified | 48.3 | No |
| 2 | Poon et al [13] | 2004 | Quebec Canadian |
Community | FBAT | FBAT | Genes &SNP | Yes | 570 subjects 223 probands | 16 (2–46yrs) | 569 | 48 (3-96y) | Not matched |
| 3 | Raby et al [12] | 2004 | America Americans |
Community | FBAT | FBAT | Genes | Yes | 1041 children 582 complete family | Girls 8.2 y boys 8.1 y | Not mentioned | 30 -55y | NO |
| 4 | Wjst et al [16] | 2006 | Germany European |
Population | Family based | PCR RFLP | Genotype | Yes | 210 families | 13.6 | 131 | Among families | No |
| 5 | Volmert et al [39] | 2004 | Germany European |
Community | FBAT | PCR | SNP | Not specified | 176 families with 352 parents 411 children | 10.7+/- 3.8 | 11.5+/- 4.3 | Yes | |
(Abbreviation foot note - HWE – Hardy-Weinberg Equilibrium, PCR – Polymerase chain reaction, RFLP – Restriction Fragment Length Polymorphism, FBAT – Family Based Analysis of transmission, SNP- Single nucleotide polymorphism, RT – Reverse transcriptase).
3.2. Quality score and assessment
Criteria was selected based on the reference by Srivastava and Srivastava [21]. Scores were categorized as good quality if score was > eight, fair > five and poor if score was < five. HWE had been analyzed in most of the studies except in four of the studies. Three of the studies were of good quality and three had score < five and fourteen of the studies had scoring between five to eight (Table 2).
Table 2.
Quality Score of the studies included.
| Criteria/Studies | Representativeness of cases | Source of controls | Ascertainment of asthma | Sample size | Quality control of genotyping methods | Hardy-Weinberg equilibrium (HWE) | Total score |
|---|---|---|---|---|---|---|---|
| Einisman et al [25] | 1 | 0.5 | 1 | 1 | 0 | 0 | 3.5 |
| Fawzy et al [26] | 1 | 1.5 | 2 | 0 | 1 | 1 | 6.5 |
| Hou et al [27] | 1 | 1.5 | 2 | 0 | 0 | 0 | 3.5 |
| Hutchinson et al [28] | 1 | 1.5 | 2 | 0 | 0 | 1 | 5.5 |
| Iordanidou et al [29] | 1 | 1.5 | 2 | 1 | 0 | 1 | 7 |
| Ismail et al [30] | 1 | 1 | 2 | 0 | 1 | 0 | 5 |
| Leung et al [17] | 1 | 1.5 | 2 | 2 | 0 | 1 | 7.5 |
| Kilic et al [31] | 1 | 1.5 | 2 | 0 | 0 | 1 | 5.5 |
| Maalmi et al [32] | 1 | 1.5 | 2 | 1 | 0 | 1 | 6.5 |
| Munkhbayarlakh et al [33] | 1 | 1.5 | 2 | 1 | 0 | 1 | 6.5 |
| Nabih et al [34] | 1 | 2 | 2 | 0 | 0.5 | 1 | 6.5 |
| NavasNazario et al [35] | 1 | 2 | 1 | 1 | 0 | 1 | 6 |
| Papadopoulou et al [36] | 2 | 2 | 2 | 1 | 0.5 | 1 | 8.5 |
| Pillai et al [37] | 1 | 1.5 | 2 | 0 | 0.5 | 1 | 6 |
| Santos et al [38] |
1 |
1.5 |
2 |
0 |
0 |
0 |
4.5 |
| Studies based on family based analysis of transmission | |||||||
| Bosse et al [15] | 2 | 2 | 2 | 2 | 1 | 1 | 9 |
| Poon et al [13] | 2 | 0 | 2 | 2 | 0.5 | 1 | 7.5 |
| Raby et al [12] | 2 | 2 | 2 | 2 | 1 | 1 | 10 |
| Wjst et al [16] | 1 | 2 | 2 | 1 | 0.5 | 1 | 7.5 |
| Vollmert et al [39] | 2 | 2 | 2 | 1 | 1 | 0.5 | 8 |
3.3. Funnel plot analysis for publication bias
The studies included for Taq 1 and Fok 1 were within the 95% confidence limit and there was no evidence of publication bias. However, the studies included in Bsm 1 and Apa 1 had number of studies outside confidence limits and hence may have had publication bias. Heterogeneity was observed for all polymorphisms in all genetic models and results tabulated (Figure 2).
Figure 2.
Funnel plot analysis for publication bias.
3.4. Analysis of the polymorphisms and asthma
The included studies were analysed quantitatively and qualitatively for association with Vitamin D related genes and asthma in children. Significance of association was found using forest plot for 15 of the case control studies for each of the polymorphisms of individual alleles and genotype association in homozygous and heterozygous state. The studies which had found significance of association by family based analysis technique (FBAT) or linkage disequilibrium (LD) were included for qualitative analysis.
3.5. Association of SNPs associated with VDR gene
The genetic variants of the VDR located in chromosome 12q13.11 have been implicated in asthmatic populations. The most studied single nucleotide polymorphisms (SNP) associated with this gene are located in last intron Apa1 (rs7975232), Bsm 1 (rs1554410) and last exon Taq1 (rs73126) near the 3' end of the gene. The Fok1 (rs2228570) located in exon 2 sequence has also been implicated in number of studies [29, 36].
3.6. Association of Apa 1 (rs7975232) polymorphism with asthma
Apa 1 polymorphism association with asthma in children was analysed in 11 case control studies and by 2 studies using FBAT techniques. A total of 2150 cases and 1878 controls were analysed in case control studies and the polymorphic major allele “A” was analysed in 7 studies. It was found to be significantly associated with protective effect in asthmatic children with (OR 0.81 (0.71,0.91) irrespective of ethnicity. The minor allele was noticed to be significantly associated increasing the odds of disease (OR 1.21 (1.07.1.37) and this significance was noticed in European population in subgroup analysis. 11 studies looked at association of homozygous form of A allele and found to be significantly associated with asthmatic children (OR 0.83 (0.70, 0.98)) but was not replicated when individual ethnic groups were analysed. No significant association was noticed in heterozygous and homozygous form of the reference allele. There was slight variation in subgroup analysis where in Asian population homozygous form of minor allele was found to be protective in asthmatic children (OR 0.61 (0.44,0.83).
In family based analysis, minor allele was noticed to be under transmitted to affected offspring in a study done by Raby et al [12]. In contrast, in the sister study done using same parameters by Poon et al [13] noticed over transmission of minor allele in affected cases. As both had similar ethnic population the difference in age group of the population could be attributed as the reason for this variation. Interestingly study done by Wjst et al done in Germany [16] had found no association in family transmission [12, 25, 27, 28, 29, 31, 32, 33, 36, 37] (Figure 3, Table 3).
Figure 3.
Forest Plot analysis of Apa 1 polymorphism.
Table 3.
Results of the tests of association for the VDR gene polymorphisms.
| SNP | TYPE | NO. OF STUDIES | TEST OF ASSOCIATION |
TEST OF HETEROGENEITY |
|
|---|---|---|---|---|---|
| OR (95% CI) | P | I2 | |||
| Apa (overall) | Allele Major | 7 | 0.81 (0.71,0.91) | 0.381 | 6.1% |
| Allele minor | 7 | 1.21 (1.07,1,37) | 0.350 | 10.3% | |
| Homozygous major | 11 | 0.83 (0.70,0.98) | 0.011 | 56.2% | |
| Heterozygous | 11 | 1.08 (0.93,1.25) | 0.000 | 88.3% | |
| Homozygous minor | 11 | 0.91 (0.77,1.09) | 0.000 | 91.4% | |
| Apa (European) | Allele Major | 4 | 0.74 (0.60,0.92) | 0.174 | 39.6% |
| Allele minor | 4 | 1.34 (1.08,1.65) | 0.159 | 42.1% | |
| Homozygous major | 4 | 0.79 (0.57,1.09) | 0.031 | 56.1% | |
| Heterozygous | 4 | 1.36 (1.01,1.82) | 0.754 | 0.0% | |
| Homozygous minor | 4 | 0.84 (0.57,1.22) | 0.008 | 74.4% | |
| Apa (American) | Allele Major | 2 | 0.82 (0.70,0.96) | 0.0% | 0.863 |
| Allele minor | 2 | 1.18 (1.00,1.39) | 0.0% | 0.989 | |
| Homozygous major | 3 | 0.81 (0.64,1.02) | 0.0% | 0.968 | |
| Heterozygous | 3 | 0.92 (0.74,1.14) | 0.0% | 0.652 | |
| Homozygous minor | 3 | 1.37 (1.04,1.80) | 0.0% | 0.902 | |
| Apa (Africans) | Allele Major | 1 | 0.95 (0.67,1.34) | ||
| Allele minor | 1 | 1.06 (0.74,1.50) | |||
| Homozygous major | 1 | 0.85 (0.56,1.30) | |||
| Heterozygous | 1 | 1.29 (0.84,1.98) | |||
| Homozygous minor | 1 | 0.46 (0.17,1.25) | |||
| Apa (Asians) | Allele Major | 0 | |||
| Allele minor | 0 | ||||
| Homozygous major | 3 | 1.17 (0.59,2.33) | 0.001 | 85.1% | |
| Heterozygous | 3 | 1.07 (0.76,1.51) | 0.000 | 97.5% | |
| Homozygous minor | 3 | 0.61 (0.44,0.83) | 0.000 | 97.7% | |
| Bsm1 (overall) | Allele (major) | 3 | 1.10 (0.90.1.34) | 0.177 | 42.3% |
| Allele (minor) | 3 | 0.91 (0.75,1,11) | 0.177 | 42.3% | |
| Homozygous major | 6 | 1.25 (0.87,1.74) | 0.038 | 60.7% | |
| Heterozygous | 6 | 1.35 (1.07,1.71) | 0.000 | 84% | |
| Homozygous minor | 3 | 1.35 (1.07,1.71) | 0.000 | ||
| Bsm1 (European) | Allele (major) | 2 | 0.93 (0.71,1.22) | 0.656 | 0.0% |
| Allele (minor) | 2 | 1.07 (0.82,1.40) | 0.656 | 0.0% | |
| Homozygous major | 2 | 0.77 (0.48,1.26) | 0.732 | 0.0% | |
| Heterozygous | 2 | 1.18 (0.81,1.72) | 0.251 | 24.2% | |
| Homozygous minor | 2 | 0.98 (0.06,1.47) | 0.338 | 0.0% | |
| Bsm1 (African) | Allele (major) | 1 | 1.34 (1.00,1.80) | ||
| Allele (minor) | 1 | 0.74 (0.55,1.00) | |||
| Homozygous major | 1 | 2.15 (1.23,3.76) | |||
| Heterozygous | 1 | 0.77 (0.51,1.16) | |||
| Homozygous minor | 1 | 0.84 (0.54,1.29) | |||
| Bsm1 (Asians) | Allele (major) | 0 | |||
| Allele (minor) | 0 | ||||
| Homozygous major | 3 | 2.07 (0.46,9.21) | 0.038 | 60.7% | |
| Heterozygous | 3 | 3.36 (2.12,5.32) | 0.003 | 83.3% | |
| Homozygous minor | 3 | 4.40 (3.18,6.09) | 0.000 | 98% | |
| Taq 1 (overall) | Allele (major) | 7 | 1.22 (1.08,1.38) | 0.000 | 76.1% |
| Allele (minor) | 7 | 1.13 (1.01,1.27) | 0.001 | 72.7% | |
| Homozygous major | 10 | 0.99 (0.86,1.14) | 0.000 | 95% | |
| Heterozygous | 10 | 1.08 (0.93,1.26) | 0.000 | 75.1% | |
| Homozygous minor | 10 | 0.73 (0.58,0.92) | 0.001 | 70.1% | |
| Taq 1 (European) | Allele (major) | 4 | 0.99 (0.80,1.23) | 0.031 | 66.2% |
| Allele (minor) | 4 | 1.01 (0.82,1.25) | 0.031 | 66.2% | |
| Homozygous major | 4 | 0.92 (0.68,1.24) | 0.060 | 59.4% | |
| Heterozygous | 4 | 1.17 (0.87,1.56) | 0.918 | 0.0% | |
| Homozygous minor | 4 | 0.87 (0.58,1.31) | 0.001 | 53% | |
| Taq 1 (American) | Allele (major) | 2 | 1.35 (1.14,1.59) | 0.001 | 90.8% |
| Allele (minor) | 2 | 1.35 (1.15,1.59) | 0.666 | 0.0% | |
| Homozygous major | 3 | 1.22 (0.98,1.52) | 0.002 | 83.4% | |
| Heterozygous | 3 | 0.98 (0.79,1.22) | 0.0 | 0.918 | |
| Homozygous minor | 3 | 0.77 (0.58,1.06) | 0.292 | 18.9% | |
| Taq 1 (Africans) | Allele (major) | 1 | 1.32 (0.98,1.78) | ||
| Allele (minor) | 1 | 0.78 (0.56,1.02) | |||
| Homozygous major | 1 | 1.14 (0.74,1.74) | |||
| Heterozygous | 1 | 1.349 (0.89,2.03) | |||
| Homozygous minor | 1 | 0.43 (0.23,0.80) | |||
| Taq 1 (Asians) | Allele (major) | 0 | |||
| Allele (minor) | 0 | ||||
| Homozygous major | 2 | 0.68 (0.51,0.92) | 0.000 | 99.4% | |
| Heterozygous | 2 | 1.08 (0.57,2.03) | 0.000 | 93.8% | |
| Homozygous minor | 2 | EXCLUDED | |||
| Fok 1 (overall) | Allele (major) | 6 | 1.34 (1.17,1.52) | 0.000 | 92% |
| Allele (minor) | 6 | 0.89 (0.78,1.02) | 0.052 | 54.3% | |
| Homozygous major | 10 | 0.96 (0.80,1.16) | 0.000 | 90.7% | |
| Heterozygous | 10 | 0.99 (0.85,1.14) | 0.000 | 90.2% | |
| Homozygous minor | 10 | 1.07 (0.90,1.27) | 0.000 | 86.3% | |
| Fok 1 (European) | Allele (major) | 2 | 1.32 (0.96,1.81) | 0.547 | 0.0% |
| Allele (minor) | 2 | 0.76 (0.55,1.04) | 0.547 | 0.0% | |
| Homozygous major | 2 | 1.61 (0.98,2.64) | 0.782 | 0.0% | |
| Heterozygous | 2 | 0.82 (0.55,1.23) | 0.650 | 0.0% | |
| Homozygous minor | 2 | 0.93 (0.55,1.54) | 0.233 | 29.6% | |
| Fok 1 (American) | Allele (major) | 2 | 0.94 (0.80,1.11) | 0.540 | 0.0% |
| Allele (minor) | 2 | 0.93 (0.79,1.10) | 0.237 | 28.3% | |
| Homozygous major | 3 | 0.94 (0.67,1.32) | 0.622 | 0.0% | |
| Heterozygous | 3 | 0.95 (0.76,1.18) | 0.280 | 21.5% | |
| Homozygous minor | 3 | 0.89 (0.70,1.12) | 0.888 | 0.0% | |
| Fok1 (Africans) | Allele (major) | 2 | 3.54 (2.66,4.72) | 0.379 | 0.0% |
| Allele (minor) | 2 | 0.89 (0.66,1.20) | |||
| Homozygous major | 2 | 2.79 (1.90,4.09) | 0.648 | 0.0% | |
| Heterozygous | 2 | 1.46 (0.98,2.16) | 0.388 | 0.0% | |
| Homozygous minor | 2 | 0.67 (0.32,1.42) | |||
| Fok1 (Asians) | Allele (major) | 0 | |||
| Allele (minor) | 0 | ||||
| Homozygous major | 2 | 0.44 (0.26,0.75) | 0.000 | 96% | |
| Heterozygous | 2 | 0.61 (0.42,0.88) | 0.000 | 98.6% | |
| Homozygous minor | 2 | 1.45 (0.94,2,23) | 0.000 | 97.2% | |
| Fok1 (Egyptian) | Allele (major) | 0 | |||
| Allele (minor) | 0 | ||||
| Homozygous major | 1 | 0.28 (0.17,0.44) | |||
| Heterozygous | 1 | 1.75 (1.14,2.68) | |||
| Homozygous minor | 1 | 2.32 (1.40,3.83) |
3.7. Association of Bsm1 (rs1544410) with asthma in children
6 studies including 757 cases and 2022 controls analysed Bsm1 allele. There was no significant association noticed in 3 of the studies which analysed individual alleles and in 6 studies involving homozygous major form. However, significant association was noticed for heterozygous (OR 1.35 (1.07, 1.71) and homozygous minor (OR 1.95 (1.59,2.39) forms. Subgroup analysis showed significant association in Asian population in heterozygous form (OR 3.36 (2.12,5.32) and homozygous minor form (OR 4.40 (3.18,6.09) not noticed in other ethnic groups [27, 29, 32, 33, 36].
In the family based analysis, Raby et al and Wjst el did not find significant transmission of Bsm 1 allele in contrast to Poon et al who had noticed significant transmission among asthmatics probably due to the difference in age group analysed. Bosse et al used two gene model of IL10 and minor allele of rs1544410 and found them to be a predictor of risk of asthma in contrast to the finding in case control studies [12, 13, 15, 16]. (Figure 4,Table3).
Figure 4.
Forest analysis of Bsm 1 polymorphism.
3.8. Association of Taq1 (rs731236) with asthma in children
Taq1 polymorphism was analysed in 10 studies which included 2080 cases and 1808 controls. The major allele, analysed in 7 studies was found to be significantly associated (OR 1.22 (1.08,1.38) which was also noticed in subgroup analysis in African and American children, with OR 1.32 (0.98,1.78) and 1.35 (1.14,1.59) respectively. No association was noticed for the homozygous major allele but the homozygous minor form was found to be associated protectively with asthmatic children (0R 0.73 (0.58–0.92). Difference was noticed in the subgroup analysis with Asian asthmatic children showing significant association in homozygous major form (OR 0.68 (0.51,0.92) and African children with homozygous minor allele (OR 0.43 (0.23,0.80). Thus, it can be inferred that Taq 1 polymorphism had varying association depending on ethnicity with asthmatic children [15, 25, 28, 29, 31, 32, 33, 36, 37].
In the family based analysis Wjst etal and Raby et al had not found significant transmission of the polymorphism. However, Poon et al had noticed significant over transmission of the major allele in the population again attributed to the difference in the age group analysed [12, 13, 16] (Figure 5, Table3).
Figure 5.
Forest plot analysis of Taq 1 polymorphism.
3.9. Association of Fok 1(rs2228570) with asthma in children
Nine case control studies with 2077 cases had analysed the association of Fok1 polymorphism with asthma. The major allele analyzed in 6 studies was found to have significant association (OR 1.34 (1.17,1.52). Subgroup analysis showed significant association of major allele in both European and African population which was not seen in American population. Minor allele did not exhibit any association. Similarly no association was noticed with alleles in homozygous or heterozygous form. In subgroup analysis, the major allele in homozygous form (OR 2.79 (1.90,4.09) and in heterozygous form (OR 1.46 (0.98,2.16) was found to increase the odds of the disease in African population. In contrast, the Egyptian population showed significant protective association of major allele in homozygous form with asthmatics (OR 0.28 (0.17,0.44). There was also significant association of minor allele in homozygous with OR 2.32 (1.40, 3.83) and in heterozygous form with OR 1.75 (1.14, 2.68) [12, 25, 29, 30, 31, 32, 33, 34, 37].
In family based analysis, Wjst et al in German population analysed Fok 1 allele but no transmission was noticed in families with asthma though significant association was found with IgE levels. Similarily none of the family based studies done by Vollmert et al in Germany. Poon et al in Canada, Nazario et al in Egypt showed association with Fok 1 allele irrespective of ethnicity [13, 16, 35, 39] (Figure 6, Table3).
Figure 6.
Forest plot analysis of Fok 1 polymorphism.
3.10. Association of other VDR polymorphisms in family based studies
There were several other VDR polymorphisms analysed by several studies. Study done by Santos et al in the promoter region gene CDX2 and in Exons 2 and 3 showed significant association with asthmatic children [38]. Poon et al. in Quebec, Canada analysed 12 SNPs in Canadian population with mean age of 16 years and found significant over transmission of six alleles (rs3782905C, rs1540339A, rs2239185C, Bsm1G, Taq1T (p < 0.05) to asthmatic children with substantial linkage disequilibrium among the haplotypes. Strong association was found between genetic variants of VDR locus and asthma [13].
However, no significant transmission of the same alleles was noticed by Raby et al in North America in slightly younger age group of children. [12], Pillai et al (rs3782905 (OR 1.44 (0.77–2.70), rs1540339 (OR 0.79 (0.42–1.49), rs2239185 (OR 0.94 (0.4–1.81) [37]. and by Wjst et al in his family based analysis in Germany of 96 SNPs. However, the study showed significant transmission of SNPs Apa 1 (rs7975232) and rs2239186 [16]. Leung et al, did not find any association of VDR polymorphisms of SNPs Apa 1 (rs7975232) or rs 2239185 [17] similar to Vollmert et al who had studied the polymorphism 117(C/T) associated with VDR gene [39]. In conclusion, there is variation in association of other polymorphisms associated with VDR.
3.11. Results of the studies that looked into SNPS associated with other vitamin D associated genes
Eight studies had analysed association of polymorphism of other Vitamin D associated genes out of which 5 of the studies had used family based transmission and linkage disequilibrium analysis to study the significance of transmission.
3.12. Review of association of CYP27A1gene polymorphisms with asthma in children
2 studies had looked into association of SNPs associated with CYP27A1gene however, the SNPs analysed by them were different. Leung et al had analysed 4 SNP and Bosse et al had analysed several SNPs and, except for one SNP rs645163 which showed significant transmission in dominant and additive model, none of the other SNPs showed any association with asthma in children [15, 17] (Table 4).
Table 4.
SNPs analyzed in various studies in Vitamin D associated genes that had significant association with children with asthma.
| Gene analysed | No of studies | Name of the study | SNP analysed | Type of analysis | Significance (p value) |
|---|---|---|---|---|---|
| CYP27A1 | 2 | Leung et al [17] | rs645163 | Logistic regression Additive Dominant |
Significant (p-0.018) Significant (0.022) |
| Bosse et al [15] | rs4674338 | FBAT | No association | ||
| CYP2R1 | 5 | Bosse et al [15] | rs11023774 | FBAT | Significant (p -0.017) |
| Pillai et al [37] | rs10766197 | Homozygous minor | Significant (p-0.044) | ||
| Wjst et al [16] | Several SNPs | Case control | Transmission present | ||
| Leung et al [17] | Several SNP | Case control | No association | ||
| Nazario et al [35] | Several SNP | Details not known | No association | ||
| CYP27B1 | 3 | Leung et al [17] | rs1048691 rs464638 rs4646536 |
Logistic regression & additive model | Significant (p-0.041) No significant association |
| Wjst et al [16] | rs238532, rs2072052, rs1048691, rs4646537, rs4646536, rs703842 |
FBAT | No association | ||
| Bosse et al [15] | FBAT | No association | |||
| CYP24A1 | 3 | Bosse et al [15] | rs8124792 rs2248359 rs927650 rs912505 |
FBAT | Significant (p - 0.030) Significant (p -0.032) No association |
| Wjst et al [16] | rs2244719 rs2296241 rs2248137 rs2762943 rs2248359 rs2426496 |
Transmission significance | No significance No significance p-0.0046 p-0.0256 p -0.0158 no significance |
||
| Pillai et al [37] |
rs2296241 rs2248137 rs2248359 rs17219315 |
Case control | Associated with decreased asthma control | ||
| GC | Ismail et al [30] | rs282679 | Case control Allele GG/TT GT/TT |
Significant OR 2.68 (1.36,5.28) Significant (p -0.04) Significant (p -0.08) |
|
| Leung et al [17] | rs2282679 rs4588 rs7041 rs222020 rs1155563 rs2298849 |
Case control | Not significant Not significant Significant Not significant Not significant Not significant |
||
| Wjst et al [16] | rs222040 rs7041 rs221999 |
FBAT Transmission approach |
0,0163 0,0249 |
||
| Nazario et al [35] | rs 7041 rs4588 |
Wild type allele Protein variant |
Significant association (p < 0.001 | ||
| Fawzy et al [26] | rs 7041 rs 4588 |
Allelic Codominant recessive Dominant Allelic Codominant Recessive Dominant |
3.08 (2.03,4.69) p < 0.0001 11.4 (4.26,30.8) p < 0.0001 10.7 (4.26,26.9) p < 0.0001 2.30 (1.25,4.23) p < 0.0001 0.44 (0.27,0.72) p < 0.001 0.39 (0.21, 0.75) p -0.0047 0.42 (0.14,1.27) p – 0.120 0.37 (0.20,0.68) p – 0.041 |
||
| Bosse et al [15] | 2 SNP | Was not included in analysis |
Abbreviation – SNP – Single nucleotide polymorphism, FBAT – Family Based analysis of transmission. The polymorphisms given in bold has been analysed & documented by several authors whereas those not in bold have been analysed only by that author.
3.13. Review of association of CYP2R1gene polymorphisms with asthma in children
Association of CYP2R1 was analysed by 5 studies. Bosse et al had analysed SNP rs11023374 and had found significant transmission among asthmatic children. However, study done by Pillai et al in SNP rs10766197 did not find association directly with asthma but found that the particular SNP was associated with decreased baseline lung functions. Similarly, no association was noticed by Wjst et al, in his analysis of 3 SNPs of CYP2R1including rs10776197. Nazario et al, in analysis of SNP rs10741657 and by Leung et al in Hongkong Chinese children who had analysed 8 SNPs including rs10776197 [15, 16, 17, 35, 37] (Table 4).
3.14. Review of association of CYP27B1gene polymorphisms with asthma in children
CYP27B1 was analysed by 3 studies done by Boss et al, Wjst et al who had analysed 6 SNPs and Leung et al who analysed 3 SNPs out of which 2 were similar to that of Wjst et al had not found any association except for Leung et al who had found significant association in only on SNP rs104861 in the additive models. Hence it can be concluded that there is no significant association of the SNPs of this gene with asthma. [15, 16, 17] (Table 4).
3.15. Review of association of CYP24A1gene polymorphisms with asthma in children
CYP24A1 was analysed in 3 studies. Boss et al had analysed 4 SNPs and found association of rs8124792 and rs2248359 with asthma and atopy. Wjst et al had analysed 6 SNPs and Pillai et al analsyed 4 SNPS. SNP rs2248359 was analysed by all the studies and found to be associated with asthma except in the study by Pillai et al where it was associated with decreased asthma control rather than directly with asthma diagnosis. SNP rs2296241 was noticed have significant transmission among asthmatic families by Wjst etal and was associated with lower lung functions by Pillai et al. Hence, there has been association of some polymorphisms of this gene with asthma [15, 16, 37] (Table 4).
3.16. Review of association of GC gene polymorphisms with asthma in children
GC gene was analysed in 6 studies. Bosse et al studied 2 SNPs associated with GC gene but as they failed the assay design, they could not be analysed. Wjst et al analysed 6 SNPs rs705120, rs222040, rs7041, rs4752, rs222011, rs221999. Among this rs 222040 and rs7041was found to be associated in families with asthma. Leung et al analysed 6 SNPs. Only one SNP was common rs7041. The SNP was found to be associated with asthma but did not survive Bonferroni correction. However, the rest of the 5 SNPs rs 2282679, rs4588, rs222020, rs1155563, rs2298849 did not have any association. Ismail et al in Egypt analysed the rs2282679 and found that children carrying the SNP have 2.22 times more likely to develop asthma which was not noticed by Leung et al. Even the genotypes were significantly associated with asthmatic children. Navas- Nazario et al in a study done in Canada, analysed the SNPs rs4588 and rs7041 and found to be significant determinants of asthma in Hispanic children. They analysed the haplotype variants combining this SNP and found that the allelic variant was protective and wild type alleles of the SNPs were associated with increased risk of asthma. Fawzy et al in Egypt also analysed the same variants rs4588 and rs7041 with both SNPs showing significant association in allelic, dominant models, homozygous and heterozygous condition and the relationship persisting even in linkage disequilibrium analysis.
Thus, all studies which had analysed rs7041 showed good strength of association. The other SNP rs228679 also shows association in one study and in the other no association [15, 16, 17, 26, 30, 35] (Table 4).
3.17. Review of association of vitamin D dependent genes associated with vitamin D dependent response element (VDRE) – IL10, CD28, IL8, IL1R1, CD86 to asthma in children
There have been 2 studies that has analysed the association of the genes that are associated with vitamin D response element (VDRE) in the nucleus of vitamin D target cells. Bosse et al has studied the SNPs associated. 5 SNPs associated with IL10 – rs1800871, rs1800872, rs1800896, rs3024490, rs4844553 were found to be associated significantly with asthma. Similarily 5 SNPs associated with IL1R1 rs950880, rs1420089, rs1946131, rs1921622, rs1861245 were analysed and 3 of them were found to be associated with asthma. One SNP rs 6435203 associated with CD28 was significantly associated with asthma. However, SNPs associated with IL8 and CD 86 did not show any association.
Wjst et al studied the transmission significance of these SNPs in German population. SNPs associated with IL10 – rs 3024498, rs151811, rs1000000076, 11 10–57 ICA, rs 1800895, rs1800894 were analysed and found 2 SNPs were associated with asthma in the families. These SNPs were different from that of Bosse et al and hence cannot be compared. Thus, SNPs associated with these genes have shown association with asthma in children [15, 16] (Table 4).
4. Discussion
Childhood asthma has several contributing factors and attempts have been made to analyse the factors especially Vitamin D, that leads to asthma and exacerbation of the clinical features. Children with asthma have been found to have increased severity of disease with lower concentrations of Vitamin D [40]. COPSAC 2000 birth cohort study had analysed that levels of Vitamin D levels in the cord blood and found that it was associated with increased risk of recurrent troublesome lung symptoms which included asthma till age of 7 years [41]. However, WAO guideline panel did not find any evidence to recommend use of Vitamin D to prevent allergic diseases [42]. This contradiction in the reports can only be sorted if there are adequate genetic studies to prove the association of Vitamin D genes and asthma. An attempt to find the significance of genetic studies was done by 5 meta-analysis but was concentrated only on VDR gene. Zhao et al, in 2017 in children had included nine studies in children among which studies from Chinese literature was also included [10]. The other four meta analysis also on VDR polymorphism, was done by Makoui et al, Tizaoui et al, Han et al and Zhang et al but had included studies in adult population [8, 9, 11, 43]. However, we know that Vitamin D has several genes associated in its pathway and each of the genes may influence the inflammatory changes in asthma and some studies had been done in those SNPs. Hence, our study was to analyse the SNPs associated with all the genes associated with Vitamin D pathway in children.
Our findings showed significant association of all polymorphisms associated with VDR. Our forest plot analysis revealed significant protective association of Apa1 polymorphism both in allele and homozygous major form and Taq 1 minor allele in homozygous form in children with asthma. The minor allele of Apa1, minor allele of Bsm 1 in heterozygous and homozygous form, major allele of Fok1 and Taq1 was found to be associated and increasing the odds of asthma.
Interesting observation in subgroup ethnicity analysis revealed, the homozygous major allele of Apa1 to have association with asthma in Asian population in contrast to all other groups. The same observation of Apa1 polymorphism was noticed by Zhao et al where the Asians had 1.5 times association with asthma not seen in Caucasians and Africans – Americans. However, Makoui et al did not notice any association of this allele. The age group analysed and hence the studies included could explain the results [9, 10].
We noticed the same ethnic difference in Fok 1 polymorphism of the major allele in homozygous form where it is noticed to be associated with asthma in Asian population and Egyptian population in contrast to other ethnicity. This ethnic difference has also been observed by Makoui et al in his study in Asian population and by Zhao et al who also observed Bsm1 and Fok1 to have opposite effects in different ethnic groups [7, 9, 10]. They had further noticed Fok 1 polymorphism to increase the asthma risk in American population, which is not shown by our study. Raby et al and Poon et al studied similar SNPs and found significantly varying type of transmission and attributed it to the age group studied.
In conclusion, it is clear that polymorphisms associated with VDR are significantly associated with asthma in children. It is essential that ethnicity and age to be taken into consideration while interpretation.
When we analysed the associations of SNPs associated with other Vitamin D related genes in the metabolic pathway - CYP27A1, CYP2R1, CYP27B1, CYP24A1, GC and the five genes related to Vitamin D response element (VDRE) - IL10, IL1RL1, CD28, CD86, IL8 we had varying results. Studies done on SNPs of IL10, IL1RL1, CD28, GC gene polymorphisms rs7041 and rs4588 showed association with asthma. Overall, the studies indicate that genes associated with Vitamin D are associated with asthma in children.
SNPs associated with CYP27A1, CYP27B1, IL8 and CD86 did not show any association in the studies done. SNPs associated with CYP2R1 showed conflicting results between 2 studies done probably due to ethnic difference.
More studies involving different population involving these genes are required to confirm this significance of association. As other meta analysis had not analysed the genes we could not get a comparative result. The influence of vitamin D related genes in the metabolic pathway is because the endogenous serum metabolite of Vitamin D (calcitriol 1.25(OH) D3) is considered a true steroid hormone contributing to the anti-inflammatory effect and hence may explain the association. However, large scale population studies done on the SNPs in Vitamin D metabolic pathway are essential to draw inferences for a genotypic correlation [14].
Our study has been the first to do a systematic review on all the genes associated with Vitamin D including the metabolic pathway which brings out the need for more studies including the SNPs. The previous meta analysis in children by Zhao et al had included eight studies [10]. Makoui et al had included 17 case control studies albeit done in adults [9]. We have included 15 case control studies with addition of 5 family-based studies that has also added significance to the analysis of the results. Vitamin D deficiency in children has been shown to be more in children with asthma [44]. Our study shows that apart from SNPs in VDR gene, SNPs associated in metabolic pathway can also contribute to this apparent association and also significant variations exist between ethnic populations. The studies have been concentrated in certain geographical regions and done several times in the same population. However, there is apparent lack of data from certain countries like India, especially where there is significant deficiency of Vitamin D in children [45]. Our study brings out the need to conduct studies on SNPs in such population to establish the association. As vitamin D is a modifiable factor it can greatly change the management of such children.
Our study had several limitations. First, we could not include all studies in the forest plot as they were not case control studies. Second, we could not include studies from other languages as only published articles in the sources cited were included. Hence, there will be an element of language bias. Thirdly, each of the genotypes had been given various representation and analysed differently in the studies which made the integration of the data difficult. Finally, a trial sequential analysis (TSA) would have given us added confirmation of the results but because of the variations in the polymorphism analysed and differences in the type of studies to analyse the significance of transmission we did not do the same.
5. Conclusions
In conclusion, our study has been able to establish relationship of vitamin D receptor polymorphisms with childhood asthma, however with significant ethnicity variations. It also shows the importance of Vitamin D associated metabolic gene polymorphisms. As vitamin D is a modifiable factor, this relationship will help us in intervention and modification of certain factors in childhood asthma. This analysis also shows the need to do large scale studies in various ethnic population preferably with similar polymorphisms to bring up guidelines on interventions with regard to Vitamin D levels and supplementation in childhood asthma.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
1. Dr. D. J. Christopher, Professor & Head, Department of Pulmonology, Christian Medical College & Hospital, Vellore gave critical inputs.
2. Dr. Somasundaram, Pediatrician, Sabari Child Care Centre helped in Data extraction.
3. Dr. Nedunchezian, Professor and Head, Department of Pediatrics, Mehta Children's Hospital, Chennai gave critical inputs.
4. Dr. Niraimathi K, Director, Fenivi Research Solutions gave inputs on statistics.
5. VIT University, Vellore for providing facilities.
References
- 1.Asthma. https://www.who.int/news-room/fact-sheets/detail/asthma [Internet]. [cited 2020 May 5]. Available from:
- 2.Pal R., Dahal S., Pal S. Prevalence of bronchial asthma in Indian children. Indian J. Community Med. 2009 Oct 1;34(4):310. doi: 10.4103/0970-0218.58389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fares M.M., Alkhaled L.H., Mroueh S.M., Akl E.A. Vitamin D supplementation in children with asthma: a systematic review and meta-analysis. BMC Res. Notes. 2015;8(1):23. doi: 10.1186/s13104-014-0961-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bener A., Bener A., Ehlayel M.S., Tulic M.K., Hamid Q. Vitamin D deficiency as a strong predictor of asthma in children. IAA. 2012;157(2):168–175. doi: 10.1159/000323941. [DOI] [PubMed] [Google Scholar]
- 5.Huang H., Porpodis K., Zarogoulidis P., Domvri K., Giouleka P., Papaiwannou A. Vitamin D in asthma and future perspectives. Drug Des. Dev. Ther. 2013 Sep 23;7:1003–1013. doi: 10.2147/DDDT.S50599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeffer P.E., Mann E.H., Hornsby E., Chambers E.S., Chen Y.-H., Rice L. Vitamin D influences asthmatic pathology through its action on diverse immunological pathways. Annals ATS. 2014 Dec;11(Supplement 5):S314–S321. doi: 10.1513/AnnalsATS.201405-204AW. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Zhang S., He C., Wang X. VDR gene polymorphisms and allergic diseases: evidence from a meta-analysis. Immunol. Invest. 2020 Feb 17;49(1–2):166–177. doi: 10.1080/08820139.2019.1674325. [DOI] [PubMed] [Google Scholar]
- 8.Tizaoui K., Berraies A., Hamdi B., Kaabachi W., Hamzaoui K., Hamzaoui A. Association of vitamin D receptor gene polymorphisms with asthma risk: systematic review and updated meta-analysis of case–control studies. Lung. 2014 Dec;192(6):955–965. doi: 10.1007/s00408-014-9648-8. [DOI] [PubMed] [Google Scholar]
- 9.Makoui M.H., Imani D., Motallebnezhad M., Azimi M., Razi B. Vitamin D receptor gene polymorphism and susceptibility to asthma: meta-analysis based on 17 case-control studies. Ann. Allergy Asthma Immunol. 2020 Jan 1;124(1):57–69. doi: 10.1016/j.anai.2019.10.014. [DOI] [PubMed] [Google Scholar]
- 10.Zhao D.-D., Yu D.-D., Ren Q.-Q., Dong B., Zhao F., Sun Y.-H. Association of vitamin D receptor gene polymorphisms with susceptibility to childhood asthma: a meta-analysis. Pediatr. Pulmonol. 2017;52(4):423–429. doi: 10.1002/ppul.23548. [DOI] [PubMed] [Google Scholar]
- 11.Han J.-C., Du J., Zhang Y.-J., Qi G.-B., Li H.-B., Zhang Y.J. Vitamin D receptor polymorphisms may contribute to asthma risk. J. Asthma. 2016 Sep 13;53(8):790–800. doi: 10.3109/02770903.2016.1158267. [DOI] [PubMed] [Google Scholar]
- 12.Raby B.A., Lazarus R., Silverman E.K., Lake S., Lange C., Wjst M. Association of vitamin D receptor gene polymorphisms with childhood and adult asthma. Am. J. Respir. Crit. Care Med. 2004 Nov 15;170(10):1057–1065. doi: 10.1164/rccm.200404-447OC. [DOI] [PubMed] [Google Scholar]
- 13.Poon A.H., Laprise C., Lemire M., Montpetit A., Sinnett D., Schurr E. Association of vitamin D receptor genetic variants with susceptibility to asthma and atopy. Am. J. Respir. Crit. Care Med. 2004 Nov;170(9):967–973. doi: 10.1164/rccm.200403-412OC. [DOI] [PubMed] [Google Scholar]
- 14.Rajaram M., Selvarajan S., Neelamegan R., Kamalanathan S., Gunaseelan V., Xavier A.S. Effects of genetic polymorphisms in Vitamin D metabolic pathway on Vitamin D level and asthma control in South Indian patients with bronchial asthma. Lung India. 2019 Nov 1;36(6):483. doi: 10.4103/lungindia.lungindia_23_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bossé Y., Lemire M., Poon A.H., Daley D., He J.-Q., Sandford A. Asthma and genes encoding components of the vitamin D pathway. Respir. Res. 2009 Oct 24;10(1):98. doi: 10.1186/1465-9921-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wjst M., Altmüller J., Faus-Kessler T., Braig C., Bahnweg M., André E. Asthma families show transmission disequilibrium of gene variants in the vitamin D metabolism and signalling pathway. Respir. Res. 2006 Apr 6;7(1):60. doi: 10.1186/1465-9921-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung T.F., Wang S.S., Tang M.F., Kong A.P., Sy H.Y., Hon K.L. Childhood asthma and spirometric indices are associated with polymorphic markers of two vitamin D 25-hydroxylase genes. Pediatr. Allergy Immunol. 2015;26(4):375–382. doi: 10.1111/pai.12392. [DOI] [PubMed] [Google Scholar]
- 18.Narmada Seshadri, Richard Kirubakaran, Radha Saraswathy. Association of vitamin D gene polymorphisms in children with asthma – a systematic review. PROSPERO 2017 CRD42017058266. Available from: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42017058266. [DOI] [PMC free article] [PubMed]
- 19.Cochrane handbook for systematic reviews of interventions. http://handbook [Internet]. [cited 2020 May 5]. Available from:
- 20.PRISMA. http://www.prisma-statement.org/ [Internet]. [cited 2020 May 5]. Available from:
- 21.Srivastava K., Srivastava A. Comprehensive review of genetic association studies and meta-analyses on miRNA polymorphisms and cancer risk. PloS One. 2012 Nov 30;7(11) doi: 10.1371/journal.pone.0050966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo J., Jin M., Zhang M., Chen K. A genetic variant in miR-196a2 increased digestive system cancer risks: a meta-analysis of 15 case-control studies. PloS One. 2012 Jan 24;7(1) doi: 10.1371/journal.pone.0030585. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3265498/ [Internet] [cited 2020 May 5] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stata: software for statistics and data science. https://www.stata.com/ [Internet]. [cited 2020 May 19]. Available from:
- 24.Cochrane handbook for systematic reviews of interventions | Cochrane training. https://training.cochrane.org/handbook [Internet]. [cited 2019 Sep 3]. Available from:
- 25.Einisman H., Reyes M.L., Angulo J., Cerda J., López-Lastra M., Castro-Rodriguez J.A. Vitamin D levels and vitamin D receptor gene polymorphisms in asthmatic children: a case–control study. Pediatr. Allergy Immunol. 2015;26(6):545–550. doi: 10.1111/pai.12409. [DOI] [PubMed] [Google Scholar]
- 26.Fawzy M.S., Elgazzaz M.G., Ibrahim A., Hussein M.H., Khashana M.S., Toraih E.A. Association of group-specific component exon 11 polymorphisms with bronchial asthma in children and adolescents. Scand. J. Immunol. 2019;89(3) doi: 10.1111/sji.12740. [DOI] [PubMed] [Google Scholar]
- 27.Hou C., Zhu X., Chang X. Correlation of vitamin D receptor with bronchial asthma in children. Exp. Ther. Med. 2018 Mar 1;15(3):2773–2776. doi: 10.3892/etm.2018.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutchinson K., Kerley C.P., Faul J., Greally P., Coghlan D., Louw M. Vitamin D receptor variants and uncontrolled asthma. Eur. Ann. Aller. Clin. Immunol. 2018 Jan;50(3):59. doi: 10.23822/EurAnnACI.1764-1489.46. [DOI] [PubMed] [Google Scholar]
- 29.Iordanidou M., Paraskakis E., Giannakopoulou E., Tavridou A., Gentile G., Borro M. Vitamin D receptor ApaI a allele is associated with better childhood asthma control and improvement in ability for daily activities. OMICS A J. Integr. Biol. 2014 Oct 29;18(11):673–681. doi: 10.1089/omi.2014.0023. [DOI] [PubMed] [Google Scholar]
- 30.Ismail M.F., Elnady H.G., Fouda E.M. Genetic variants in vitamin D pathway in Egyptian asthmatic children: a pilot study. Hum. Immunol. 2013 Dec 1;74(12):1659–1664. doi: 10.1016/j.humimm.2013.08.284. [DOI] [PubMed] [Google Scholar]
- 31.Kilic M., Ecin S., Taskin E., Sen A., Kara M. The vitamin D receptor gene polymorphisms in asthmatic children: a case-control study. Pediatr. Allergy Immunol. Pulmonol. 2019 Jun 1;32(2):63–69. doi: 10.1089/ped.2018.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maalmi H., Sassi F.H., Berraies A., Ammar J., Hamzaoui K., Hamzaoui A. Association of vitamin D receptor gene polymorphisms with susceptibility to asthma in Tunisian children: a case control study. Hum. Immunol. 2013 Feb 1;74(2):234–240. doi: 10.1016/j.humimm.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Munkhbayarlakh S., Kao H.-F., Hou Y.-I., Tuvshintur N., Bayar-Ulzii B., Narantsetseg L. Vitamin D plasma concentration and vitamin D receptor genetic variants confer risk of asthma: a comparison study of Taiwanese and Mongolian populations. World Allergy Organ. J. 2019 Nov;12(11):100076. doi: 10.1016/j.waojou.2019.100076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nabih E.S., Kamel T.B. Association between vitamin D receptor gene FokI polymorphism and atopic childhood bronchial asthma. Egypt. J. Chest Dis. Tuberc. 2014 Jul 1;63(3):547–552. [Google Scholar]
- 35.Navas-Nazario A., Li F.Y., Shabanova V., Weiss P., Cole D.E.C., Carpenter T.O. Effect of vitamin D–binding protein genotype on the development of asthma in children. Ann. Allergy Asthma Immunol. 2014 Jun 1;112(6):519–524. doi: 10.1016/j.anai.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papadopoulou A., Kouis P., Middleton N., Kolokotroni O., Karpathios T., Nicolaidou P. Association of vitamin D receptor gene polymorphisms and vitamin D levels with asthma and atopy in Cypriot adolescents: a case–control study. Multidiscip. Resp. Med. 2015 Sep 4;10(1):26. doi: 10.1186/s40248-015-0025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pillai D.K., Iqbal S.F., Benton A.S., Lerner J., Wiles A., Foerster M. Associations between genetic variants in vitamin D metabolism and asthma characteristics in young African Americans: a pilot study. J. Invest. Med. 2011 Aug;59(6):938–946. doi: 10.231/JIM.0b013e318220df41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos H.L.B.S., Silva S de, de Paula E., Pereira-Ferrari L., Mikami L., Riedi C.A. Vitamin D receptor gene mutations and vitamin D serum levels in asthmatic children. Rev. Paul. Pediatr. 2018;36(3):269–274. doi: 10.1590/1984-0462/;2018;36;3;00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vollmert C., Illig T., Altmüller J., Klugbauer S., Loesgen S., Dumitrescu L. Single nucleotide polymorphism screening and association analysis--exclusion of integrin beta 7 and vitamin D receptor (chromosome 12q) as candidate genes for asthma. Clin. Exp. Allergy. 2004 Dec;34(12):1841–1850. doi: 10.1111/j.1365-2222.2004.02047.x. [DOI] [PubMed] [Google Scholar]
- 40.Riverin B.D., Maguire J.L., Li P. Vitamin D supplementation for childhood asthma: a systematic review and meta-analysis. PloS One. 2015 Aug 31;10(8) doi: 10.1371/journal.pone.0136841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cord blood 25(OH)-Vitamin D deficiency and childhood asthma, allergy and eczema: the COPSAC2000 birth cohort study. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0099856 [Internet]. [cited 2020 May 8]. Available from: [DOI] [PMC free article] [PubMed]
- 42.Yepes-Nuñez J.J., Fiocchi A., Pawankar R., Cuello-Garcia C.A., Zhang Y., Morgano G.P. World allergy organization-McMaster university guidelines for allergic disease prevention (GLAD-P): vitamin D. World Allergy Organ. J. 2016 May 17;9(1):17. doi: 10.1186/s40413-016-0108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VDR Gene Polymorphisms and Allergic Diseases Evidence from a meta-analysis: immunological investigations: vol 49, No 1-2. https://www.tandfonline.com/doi/abs/10.1080/08820139.2019.1674325?journalCode=iimm20 [Internet]. [cited 2020 May 5]. Available from: [DOI] [PubMed]
- 44.Jat K.R., Khairwa A. Vitamin D and asthma in children: a systematic review and meta-analysis of observational studies. Lung India. 2017 Jul 1;34(4):355. doi: 10.4103/0970-2113.209227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aparna P., Muthathal S., Nongkynrih B., Gupta S.K. Vitamin D deficiency in India. J. Fam. Med. Prim. Care. 2018 Apr;7(2):324. doi: 10.4103/jfmpc.jfmpc_78_18. [DOI] [PMC free article] [PubMed] [Google Scholar]