Abstract
Assessment of internal tandem duplications in FLT3 (FLT3-ITDs) and their allelic ratio (AR) is recommended by clinical guidelines for diagnostic workup of acute myeloid leukemia and traditionally performed through capillary electrophoresis (CE). Although significant progress has been made integrating FLT3-ITD detection within contemporary next-generation sequencing (NGS) panels, AR estimation is not routinely part of clinical NGS practice because of inherent biases and challenges. In this study, data from multiple NGS platforms—anchored multiplex PCR (AMP), amplicon [TruSeq Custom Amplicon (TSCA)], and hybrid-capture—were analyzed through a custom algorithm, including platform-specific measures of AR. Sensitivity and specificity of NGS for FLT3-ITD status relative to CE were 100% (42/42) and 99.4% (1076/1083), respectively, by AMP on an unselected cohort and 98.1% (53/54) and 100% (48/48), respectively, by TSCA on a selected cohort. Primer analysis identified criteria for ITDs to escape detection by TSCA, estimated to occur in approximately 9% of unselected ITDs. Allelic fractions under AMP or TSCA were highly correlated to CE, with linear regression slopes near 1 for ITDs not duplicating primers, and systematically underestimated for ITDs duplicating a primer. Bias was alleviated in AMP through simple adjustments. This article provides an approach for targeted computational FLT3-ITD analysis for NGS data from multiple platforms; AMP was found capable of near perfect sensitivity and specificity with relatively accurate estimates of ARs.
Since their discovery in 1996, internal tandem duplications (ITDs) in the fms-related tyrosine kinase 3 (FLT3) gene, or FLT3-ITDs for short, have been recognized as one of the most frequent somatic alterations in acute myeloid leukemia (AML), occurring in approximately 25% of new AML diagnoses.1,2 Clonal FLT3-ITDs are in-frame insertions that range from 3 to 300 bp in size, occur throughout the juxtamembrane domain with occasional extension into tyrosine kinase domain 1, and result in constitutive tyrosine kinase activation. Their presence confers a poor prognosis and is an indication for targeted therapy, with Food and Drug Administration approval of the multikinase inhibitor midostaurin in combination with induction chemotherapy in newly diagnosed AML and the second-generation FLT3 inhibitor gilteritinib in the setting of relapse.3,4 Studies have shown that the higher relapse rates and shorter overall survival associated with FLT3-ITDs are influenced by ITD burden, which is typically measured by the allelic ratio (AR) of mutant alleles over wild-type alleles.5, 6, 7, 8, 9, 10 Recently revised guidelines for AML risk stratification from the European Leukemia Network (ELN) define FLT3-ITD high AML as having AR ≥0.5 and categorize such cases as either intermediate or adverse risk, dependent on the presence or absence of a concurrent NPM1 mutation.11 Accordingly, FLT3-ITD testing and determination of AR have been incorporated into ELN recommendations for newly diagnosed AML. Despites these recommendations, AR data are not often part of clinical practice because of various substantial challenges.2
FLT3-ITDs are traditionally detected and quantified by PCR amplification of genomic DNA using primers flanking FLT3 exons 14 to 15 followed by fragment (sizing) analysis via capillary electrophoresis (CE).12,13 Given the increasing adoption of targeted next-generation sequencing (NGS) into routine clinical practice, molecular laboratories have also started to integrate FLT3-ITD assessment into comprehensive hematologic DNA-based NGS panels.14, 15, 16 Standard NGS pipelines typically detect ITDs only when captured as insertions during alignment, thus missing longer ITDs while systematically underestimating allelic ratios of shorter ITDs because of recognition of only a partial subset of mutant reads. Although the development of specialized FLT3-ITD algorithms has improved recognition of mutant reads, few studies have adequately addressed AR estimation and some algorithms are limited to specific NGS platforms.14,16, 17, 18, 19, 20, 21, 22, 23, 24 In particular, a trend for AR underestimation by NGS relative to CE has been consistently shown.17,21,24,25
This study explored whether a novel algorithm for FLT3-ITD detection and AR determination can function across multiple NGS platforms using different target enrichment strategies. This approach is relevant because different NGS assays and informatic methods may introduce a bias in AR and consequently result in false classification into FLT3-ITD high versus low categories. The presented computational approach harmonizes FLT3-ITD analysis across multiple NGS platforms, allows direct performance comparison of different enrichment strategies, and addresses the AR bias observed in prior NGS studies.
Materials and Methods
Sample Selection
DNA from blood or bone marrow was tested by one to two of three targeted NGS panels based on different enrichment strategies: i) anchored multiplex PCR (AMP; ArcherDx, Boulder, CO) performed clinically at Massachusetts General Hospital (MGH) on unselected samples from 2018 (MGH cohort: n = 1125), ii) amplicon based [TruSeq Custom Amplicon (TSCA); Illumina, San Diego, CA] performed clinically at Brigham and Women's Hospital (BWH) on selected samples enriched for FLT3-ITDs from 2014 to 2019 (BWH cohort: n = 102) and on separate AML samples from 2014 to 2016 (AML cohort: n = 32), and iii) hybrid-capture (HC) performed for research on the same AML cohort (n = 32), each followed by 2 × 151 bp (AMP), 2 × 150 bp (TSCA), or 2 × 101 bp (HC) paired-end Illumina sequencing.26
Fragment Analysis
CE was performed clinically (MGH cohort) and experimentally (BWH cohort) on an Applied Biosystems (Foster City, CA) 3500 Genetic Analyzer using standard FLT3-ITD primers.12 Clinical reports (MGH) provided a categorical interpretation of FLT3-ITD status (positive or negative). AR was calculated from CE data as the ratio of the area under the curve of an FLT3-ITD variant divided by the area under the curve of the FLT3 wild-type product. In each case, the FLT3-ITD with greatest AR was referred to as the primary CE ITD, whereas additional FLT3-ITDs were labeled secondary CE ITDs. CE was unavailable for the AML cohort.
Sequencing of FLT3
All NGS panels targeted FLT3 exons 14 to 15. AMP and TSCA primers are shown in Figure 1A. AMP incorporated unique molecular identifiers (UMIs) during library preparation, and its clinical pipeline used Novoalign for alignment to hg19, followed by an ensemble variant calling approach validated to detect variants at allelic frequencies >10% from DNA inputs of 200 ng. TSCA and its clinical pipeline were described previously and shown to reliably detect variants at allelic frequencies >5% from DNA inputs of 250 ng.15
Figure 1.
Anchored multiplex PCR (AMP) and TruSeq Custom Amplicon (TSCA) assays. A: Location of AMP and TSCA primers (solid arrows) near FLT3 exons 14 and 15 (rectangular boxed areas drawn with dashed lines), along with schematic 150-bp reads (dashed arrows) derived from these primers. AMP primers P1 to P7 were paired with a universal primer (not shown), yielding variable-length amplicons. Only anchored end reads are shown. An additional AMP primer P0 (not shown) targeted FLT3 further upstream. TSCA primers were paired with one another (F1-R1 and F2-R2). Genomic segments potentially duplicated in internal tandem duplications (ITDs) are depicted in light green and blue. B:FLT3-ITD within exon 14 (green). AMP primer P1 is TSCA-like because its anchored end reads reach the mutant junction (MJ). P2 is disruptive because it overlaps the ITD but cannot extend across MJ, and can only bind effectively to the second duplicated section because the MJ generates nonalignment of the end of the primer in the first half of the ITD (depicted as P2 with an X though the primer). The rest are hybrid-capture (HC)–like because MJ can only be captured within their variable nonanchored end reads (although P3 and P4 are noncontributory as their reads never capture MJ). TSCA primers F1 and R1 surround the ITD such that MJ is contained within F1-R1 amplicons and reached by reads from F1 (but not R1). F2-R2 amplicons never contain MJ. C:FLT3-ITD extending into intron 14 (blue). AMP primer P3 is TSCA-like and duplicated (denoted by P3a and P3b, which bind to the first and second halves of the duplication, respectively). P5 is TSCA-like, and the rest are HC-like (although P4 is noncontributory as its reads never capture MJ). For duplicated P3, only (anchored-end) reads from P3a capture MJ, whereas reads from P3b never capture MJ and are indistinguishable from wild type (WT). TSCA primer sites for R1 and F2 are duplicated in the ITD (denoted by R1a and R1b, and F2a and F2b). Amplicons F1-R1b, F2a-R2, and F2a-R1b (mixed product) contain and sequence MJ, whereas F1-R1a and F2b-R2 amplicons are indistinguishable from wild type. Note: these ITD examples have been chosen to illustrate terminology and primer duplication but do not appear to occur empirically; for instance, ITDs extending this far into intron 14 likely offer no competitive advantage for clonal selection to occur, as splicing probably removes the duplicated portion once enough of the region around the splice donor site is duplicated.
Custom FLT3-ITD Informatics
A novel FLT3-specific pipeline was developed to detect, characterize, assess, and quantify FLT3-ITDs from NGS data (Figure 2), and is available for download (https://github.com/ht50/FLT3_ITD_ext, last accessed March 25, 2020). A 1500-bp genomic segment of FLT3 (chromosome 13: 7607428-28608937 in hg19) containing exons 14 to 15 centrally and referred to as the FLT3 target locus was used for re-alignments at various steps of the algorithm. BWA-MEM version 0.7.17 (https://sourceforge.net/projects/bio-bwa/files, last accessed September 7, 2018) was the default aligner, unless otherwise specified.27 However, bowtie2 version 2.3.4.3 (https://sourceforge.net/projects/bowtie-bio/files/bowtie2, last accessed September 17, 2018) and Novoalign version V3.0.7.00 (Novocraft, Selangor, Malaysia) were also tested and found to be acceptable. By default, quality filters were not applied to maximize capture of mutant reads, with the view that low-quality sequences were adequate for identifying tandem duplications or insertions, with a few mismatches tolerated.
-
1.
INPUT: FASTQ formatted sequences were selectively extracted from general pipeline BAM files. Specifically, only unmapped sequences and reads with general alignments to the FLT3 target locus were extracted (eg, by the efficient command samtools view <bamfile> 13:28607428-28608937). Entire raw FASTQ files may also be used in the absence of pre-existing alignment files. Adapters were trimmed using BBDuk (version last modified August 29, 2018) if not already performed within the general pipelines.
-
2.
LOCAL RE-ALIGNMENT: Paired-end reads were locally aligned to the FLT3 target locus. Reads satisfying relatively stringent criteria (concordant alignments with insertions totaling ≤2 bp, deletions totaling ≤2 bp, soft clips totaling ≤2 bp, and edit distance ≤5 bp in each read of a pair) were categorized as wild type and removed from further analysis.
-
3.
IN SILICO EXTENSION: Reads aligning with soft clips ≥6 bp or net insertions ≥3 bp (considered as individual unpaired reads for this step) were extended in silico when possible to reach both ends of the FLT3 target locus, based on secondary alignment locations of soft-clipped ends together with primary alignment locations for non–soft-clipped ends. When a soft-clipped end did not have a corresponding secondary alignment, local re-alignment of the soft clip with relaxed settings (seed length of 6 and threshold of 9) was attempted to place the soft clip and extend the read.
-
4.
CLUSTERING: Extended sequences were ordered by frequency and clustered by sumaclust version 1.0.31 (a greedy centroid-based clustering algorithm; https://git.metabarcoding.org/obitools/sumaclust, last accessed December 26, 2018) using a score threshold of 5, with each resulting centroid sequence considered an ITD candidate.28
-
5.
ALIGNMENT-BASED ANNOTATION AND GROUPING: ITD candidates were aligned to the FLT3 target locus, and insertions or soft clips that emerged were further characterized relative to the FLT3 target locus through secondary alignments or additional iterative applications of the aligner (Figure 3). Resulting alignment data (primary, secondary, aligned soft clips, and/or aligned insertions) were merged together to provide a general structural annotation of an ITD along with mismatches relative to the general structure. ITD candidates having the same structural annotation that were suspected to differ only by sequencing error were grouped together, and thus candidates were further reduced by retaining only one candidate (of greatest frequency) per structural annotation. Near-exact duplication of a genomic segment spanning c0 to c1 with intervening nontemplated insert N had the structural annotation format c.c1_(c1 + 1)insN/c0–c1 (in contrast to standard Human Genome Variation Society nomenclature as generic insertions), thus enabling efficient identification of ITDs duplicating primer sites of TSCA or AMP, which was necessary for the allelic ratio calculation method. More generally, annotations may have the structural format a0_a1delinsN1/b0–b1/N2/c0–c1…, plus mismatches.
-
6.
EVALUATION OF ITD CANDIDATES: Paired reads not previously categorized as wild type were aligned directly to the final set of candidate ITD genomes and categorized as mutant if aligning across the left or right boundary of a mutant junction by at least 10 bp on either side and satisfying the same stringent criteria as step 1 but relative to an ITD candidate (concordant alignments with insertions totaling ≤2 bp, deletions totaling ≤2 bp, soft clips totaling ≤2 bp, and edit distance ≤5 bp in each read of a pair). Alignments were visualized using samtools tview. Total mutant read counts per ITD candidate, depth of coverage by genomic position, and breadth of coverage by genomic position (defined as the maximum sequencing radius surrounding that position within a read across all mutant reads) were used to evaluate the strength of evidence of an ITD candidate. UMI processing was performed at this stage with UMI-tools version 0.5.5 (https://github.com/CGATOxford/UMI-tools, last accessed January 30, 2019) or fgbio version 0.4.0 (Fulcrum Genomics, Phoenix, AZ) applied separately to reads of each ITD candidate and to wild-type reads.29 Of note, a targeted approach such as the above is needed to properly utilize UMIs, because most default UMI algorithms potentially lose FLT3-ITD reads because of reliance on 5′ and/or 3′ alignment positions of fragments to the reference for initial grouping of reads before further deduplication. Even if Compact Idiosyncratic Gapped Alignment Report (CIGAR) strings of paired reads are used for grouping, this may fail to separate cases with multiple ITDs sharing a boundary point, which is not uncommon.
-
7.
MANUAL RESCUE IN TSCA: To account for mutant junctions that were potentially barely reached by TSCA reads, short soft-clipped reads lacking in silico extensions in step 3 and not belonging to any mutant ITD category in step 6 were rescued for additional evaluation when a sufficient number of reads derived from the same TSCA primer (≥5% as a proxy for clonality) were soft clipped at the same genomic location with the same soft-clipped sequence. The existence of an ITD was inferred when paired reads from the opposite direction/primer reached beyond the soft-clip site without deviation from reference, generating the appearance of disagreement. This property, referred to as divergent paired alignments, was relatively specific to tandem duplications and near-exact tandem duplications, versus general insertions or deletions (Supplemental Figure S1). Sequence details of rescued ITDs were generally not characterizable, unless soft clipping also occurred in the corresponding partner read so that the duplicated sequence was approximately bounded on either side by both soft clips.
Figure 2.
Overview of the custom internal tandem duplication in FLT3 (FLT3-ITD) algorithm. 1: An input FASTQ or BAM file may contain reads (Rs; light gray boxed area) crossing the mutant junction (MJ) of an ITD with duplicated segment D (light blue boxed area) and nontemplated insert N (red boxed area). 2: These reads are locally aligned to the FLT3 target locus (A; light gray boxed area), and their soft clips (S; dark gray boxed area) due to MJ are locally aligned [S′ (black boxed area); A′ (gray boxed area)]. 3: The local alignments are used for 5′ and 3′ extension in silico. 4: Extended reads are clustered into candidate ITD representatives. 5: Candidate ITDs are characterized by alignment-based annotation (Figure 3). 6: Breadth and depth of coverage are assessed through paired reads aligning to an ITD genome and spanning MJ, with unique molecular identifier reduction at this step. Estimation of allelic ratio is platform specific and not depicted herein (see Materials and Methods and Figure 4).
Figure 3.
Alignment-based annotation. Local alignment was applied to an extended read representing a candidate internal tandem duplication (ITD) and iteratively applied to insertions and soft clips to align as many segments of the extended read as possible (D, genomic segment in light blue with boundaries c0 and c1; D1 and D2, first and second instance of D in the extended read, also both in light blue, where D1 additionally contains a point mutation in yellow versus its reference sequence in dark blue; N, nontemplated insert in red). Resulting alignments were merged together when possible, ideally resulting in the same maximal alignments independent of local aligner used. Maximal alignments were then converted to a structural annotation together with mismatches. In this schematic example, the structural annotation c.c1_(c1 + 1)insN/c0–c1 specifies an insertion of N between coordinates c1 and (c1 + 1) followed by insertion of the segment spanning coordinates c0 to c1, thus corresponding structurally to near-exact duplication of c0–c1 with intervening nontemplated insert N. The mismatch component (D1.y:A>T) further specifies a concurrent single-nucleotide variant (SNV) at the first of two positions aligning to location y in the maximal alignment. Most annotations in practice had no mismatches and were either near-exact ITDs (structural format above) or exact ITDs (c.c0_c1dup); however, the notation can describe more complex scenarios [eg, c.c1_(c1 + 1)ins/c0–c1/N/c0–c1 would correspond to a near-exact triplication with intervening nontemplated insert N between the second and third instances of c0–c1].
Allelic Ratios and Fractions by NGS
Platform-specific formulas were conceived and used to calculate either AR or equivalently allelic fraction (AF) of an ITD (possibly among multiple ITDs indexed by i), where:
| (1) |
| (2) |
All approaches utilized counts of paired reads, whereas standard pipelines sometimes considered reads separately as if derived from single-end sequencing; this single-end approach may work for single-nucleotide variants and general insertions/deletions but was not applicable to ITDs because of divergent paired alignments. The approach for AMP was based on the approaches for both TSCA and HC and will be discussed last.
In TSCA, evaluation of AR was straightforward for FLT3-ITDs confined to exon 14, or more generally surrounded by a primer pair (Figure 1B). In this case, mutant and wild-type amplicons derived from the amplicon primer pair F1-R1 were distinct from one another by NGS and could be counted separately, under the assumption of adequate read lengths for reaching the mutant junction (Supplemental Figure S1). This concept of an ITD encompassed within an amplicon exactly parallels CE, except NGS assesses the insert by sequencing instead of sizing and may use different primers and chemistry. This led to the following simple formula, where rMJ denoted number of paired read alignments to the mutant genome with coverage of the mutant junction (MJ) and rTotal denoted total number of paired reads derived from F1-R1.
| (3) |
Direct assessment of mutant and wild-type amplicons was no longer possible when primer binding sites were duplicated in an FLT3-ITD, thus enabling extra PCR products indistinguishable from wild type (Figure 1C). ITDs originating in exon 14 and extending into intron 14 or exon 15 generally contained duplicated binding sites for both F2 and R1 (denoted F2a, F2b, R1a, and R1b), yielding three possible products amplifying across the mutant junction (F1-R1b, F2a-R2, and F2a-R1b) and two possible products indistinguishable from wild type (F1-R1a and F2b-R2). The two wild-type mimics were moreover capable of co-amplification from a single mutant allele, further skewing counts. Various simple adjustments were explored to attempt to correct for wild-type mimics, including the following formulas, where D denoted the duplicated region of an FLT3-ITD, as characterized by the detection algorithm:
| (4) |
| (5) |
These formulas generally assumed an absence of false negatives, an equal likelihood of primer binding among competing sites, and equivalent PCR efficiency of corresponding products. For instance, for D containing both F2 and R1, the AF adjustment was derived by assuming an equal likelihood of F2 and R1 binding to i) F2a and R1a, ii) F2a and R1b, iii) F2b and R1a, or iv) F2b and R1b. Scenario 1 then yields an equal likelihood of sequencing MJ (F2a-R2) versus reference only (F1-R1a), and similarly for scenario 4. Scenario 2 always yields sequencing of MJ (F1-R1b, F2a-R2, or F2a-R1b), whereas scenario 3 yields sequencing of two reference-only amplicons (F1-R1a and F2b-R2) concurrently from the same DNA strand. Thus, a single ITD genome with duplicated F2 and R1 yields an average of 0.5 amplicons spanning MJ and 0.75 amplicons spanning reference sequence only (or 1.25 total amplicons), so that the observed count rMJ represents half the actual mutant population while generating an extra rMJ/2 amplicons in total, thereby rationalizing the above adjustment formula if the assumptions hold true. However, the validity of the assumptions was felt to be doubtful in cases of long ITDs, where widely differing amplicon lengths were expected to cause significant bias in the assay.
In HC, random fragmentation generated proportionally more unambiguous mutant read pairs (rMJ) from mutant alleles (based on sequencing of the unique mutant junction MJ of an ITD) than unambiguous wild-type read pairs (rWT) from wild-type alleles, because definitive assignment of wild type sequence requires sequencing across both boundary junctions J1 and J2 as well as the intervening wild-type sequence to rule out duplication (Figure 4). Reads containing either J1 or J2 and not MJ were ambiguous between mutant and wild type. The following formula was designed to assess comparable sets of mutant and wild-type reads to the extent possible, where rJ1 and rJ2 denoted read pairs (mutant, wild type, or ambiguous) containing J1 and J2, respectively:
| (6) |
Figure 4.
Read ambiguity under hybrid-capture or anchored multiplex PCR. For a given internal tandem duplication (ITD) with duplicated genomic segment depicted in light blue, identification of an unambiguous mutant read only required capture of the mutant junction (MJ), whereas identification of an unambiguous wild-type (WT) read required sequencing through the entire segment spanning both boundaries J1 and J2 to rule out duplication; indeed, once ITD size exceeded read length, unambiguous wild-type reads could no longer be identified. Rather, reads sequencing only one of J1 or J2 and not MJ were ambiguous between wild type and mutant. Such read ambiguity and the relative mismatch in criteria for unambiguous reads made direct assessment of allelic ratio infeasible in general and necessitated an alternative indirect approach. Note that the figure depicts single-end reads for simplicity.
Of note, another possible strategy was to directly calculate AR from rWT and a comparable subset of rMJ defined by requiring a buffer of size |D| around MJ and averaging over left and right buffers; however, these sets diminished proportionally as |D| increased, to the point that the strategy stopped working for |D| above the read length.
In AMP, a combination of the TSCA and HC approaches was used. For each ITD, primers were classified on the basis of their location relative to the ITD as: i) disruptive if containing the ITD or overlapping the 3′ end of the ITD relative to primer direction, ii) TSCA-like if the mutant junction was reachable from the anchored end under the employed read lengths, and iii) HC-like otherwise (Figure 1B). TSCA-like and HC-like primers were further subclassified as duplicated if contained strictly within the ITD and not duplicated otherwise (Figure 1C). Reads derived from disruptive primers were not used in AF estimation because such primers were either not capable of binding to mutant alleles or only capable of generating reference sequence, and thus prone to inflating wild-type estimates. Reads from TSCA-like primers were treated according to the TSCA approach and classified as mutant when containing MJ and otherwise as reference. Reads from HC-like primers were treated according to the HC approach and queried as to whether they contained MJ, J1, and/or J2. The base (unadjusted) allelic fraction AF was then calculated as follows:
| (7) |
Because duplicated primers may bind to two different sites within an ITD genome, where only one amplifies across the mutant junction, the following simple adjustment was incorporated for these cases, assuming equal likelihood of primer binding among competing sites and at most a single product from a primer per ITD molecule (ie, no fragmentation between duplicated primer sites):
| (8) |
Also of note, AMP employed two rounds of primers to increase specificity; however, the initial round of primers was upstream and did not need to be considered in this analysis.
Results
Sensitivity and Specificity of Clinical NGS Detection for FLT3-ITDs
First, sensitivity and specificity of clinical NGS platforms (AMP and TSCA) were evaluated for categorical FLT3-ITD status (positive or negative) under custom and standard clinical pipelines (Table 1), because categorical determination is the principal clinical need (eg, to approve targeted therapy). As gold standard, CE data were used when available (MGH and BWH cohorts) and otherwise HC data processed by the custom pipeline (AML cohort), because HC has been shown to perform well for FLT3-ITD detection and typically has outperformed other platforms in general NGS studies.17,24,30,31
Table 1.
Confusion Matrices (AMP/TSCA versus CE/HC): Comparison of FLT3-ITD Status Based on the Custom Algorithm or the Clinically Reported Calls Within Clinical NGS Panels (AMP or TSCA) Relative to CE or HC
| MGH cohort | Pos (CE) | Neg (CE) |
|---|---|---|
| Custom FLT3-ITD pipeline (AMP) | ||
| Pos | 42 | 7 |
| Neg | 0 | 1076 |
| Clinical NGS calls (AMP) | ||
| Pos | 16 | 2 |
| Neg | 26 | 1081 |
| Novoalign unfiltered (AMP) | ||
| Pos | 26 | 6 |
| Neg | 16 | 1077 |
| BWH cohort | Pos (CE) | Neg (CE) |
|---|---|---|
| Custom FLT3-ITD pipeline (TSCA) | ||
| Pos | 53 | 0 |
| Neg | 1 | 48 |
| Clinical NGS calls (TSCA) | ||
| Pos | 50 | 0 |
| Neg | 4 | 48 |
| AML cohort | Pos (HC) | Neg (HC) |
|---|---|---|
| Custom FLT3-ITD pipeline (TSCA) | ||
| Pos | 5 | 0 |
| Neg | 0 | 27 |
| Clinical NGS calls (TSCA) | ||
| Pos | 5 | 0 |
| Neg | 0 | 27 |
AML, acute myeloid leukemia; AMP, anchored multiplex PCR; BWH, Brigham and Women's Hospital; CE, capillary electrophoresis; HC, hybrid-capture; ITD, internal tandem duplication; MGH, Massachusetts General Hospital; Neg, negative; NGS, next-generation sequencing; Pos, positive; TSCA, TruSeq Custom Amplicon.
On AMP data, the custom FLT3-ITD pipeline was 100% (42/42) sensitive and 99.4% (1076/1083) specific relative to clinical CE reports. The seven false positives moreover i) represented molecular minimal residual disease in a post-treatment context with rare reads demonstrating the same ITD sequence found at initial diagnosis (two of seven cases), ii) had a small CE peak at the same ITD size as NGS but below analytical sensitivity of the CE assay (three of seven cases), or iii) both (two of seven cases), overall suggesting increased sensitivity of AMP over CE, similar to prior NGS studies.17 Detected FLT3-ITDs were supported by mutant reads derived from an average of 4.52 AMP primers, which should alleviate the rare PCR pitfall of allelic dropout (see Performance of AMP Primers for details). The standard clinical AMP pipeline based on Novoalign was not optimized for detection of FLT3-ITDs because the clinical CE assay served this purpose; thus, its sensitivity was considerably less at 38.1% (16/42). This improved to 61.9% (26/42) on re-analysis of Novoalign output, allowing for variant calls below the limit of detection of the clinical pipeline, given the tendency of Novoalign to underestimate FLT3-ITD allelic fraction because of ignored soft-clipped mutant reads.
On TSCA data, the custom FLT3-ITD pipeline was 98.1% (53/54) sensitive and 100% (48/48) specific for categorical FLT3-ITD status relative to CE (BWH cohort) and 100% (5/5) sensitive and 100% (27/27) specific relative to HC (AML cohort), where the single false negative (BWH cohort) had a relatively long ITD by CE (108 bp). The standard clinical TSCA pipeline showed comparable effectiveness, with slightly reduced sensitivity of 92.6% (50/54) in the BWH cohort. However, the BWH cohort had selection bias for shorter ITDs and the AML cohort was small; thus, TSCA performance may differ in unselected clinical populations. Indeed, analysis of TSCA primers identified escape criteria for ITDs arising from exon 14 that theoretically escaped detection under 2 × 150 bp sequencing relative to the starting coordinate (c0) and end coordinate (c1) of the duplicated region, and such ITDs appeared in approximately 9% (3/35) of FLT3-ITD–positive patients from the unselected MGH cohort (see Characterization of TSCA False Negatives for details).
Next, how often pipelines characterized ITDs of the same size as the primary CE (or HC) ITDs was evaluated, given their importance as the largest contributors to AR (Table 2). The custom FLT3-ITD pipeline successfully characterized ITDs of the same size as all 42 of 42 primary CE ITDs (21 to 198 bp) from the AMP-based MGH cohort, 48 of 54 primary CE ITDs (6 to 90 bp) from the TSCA-based BWH cohort, and 5 of 5 primary HC ITDs (24 to 195 bp) from the TSCA-based AML cohort. In comparison, the standard clinical NGS pipelines characterized ITDs of the same size as 15 of 42 primary CE ITDs (24 to 54 bp) from AMP/MGH, 34 of 54 primary CE ITDs (6 to 75 bp) from TSCA/BWH, and 3 of 5 primary HC ITDs (24 to 66 bp) from TSCA/AML. Discrepancies in AMP between custom and standard clinical pipelines were due to longer ITDs that could not be aligned as insertions by Novoalign, as well as shorter ITDs where nonrecognition of soft-clipped ITD reads resulted in underestimation of allelic fraction by Novoalign below the clinical limit of detection (see NGS-Based Determination of Allelic Fraction for details). Discrepancies in TSCA were due in part to probable misannotation of ITDs by the standard clinical TSCA pipeline, indicative of NGS annotation challenges. This included eight cases where CE and the custom algorithm agreed on number and size of ITDs, but the standard clinical pipeline characterized ITDs of different size.
Table 2.
AMP/TSCA Performance on 1° and 2° ITDs
| MGH cohort (AMP) | NGS ITDs in total | Same size as 1° CE (total 1° CE ITDs) | Same size as 2° CE (total 2° CE ITDs) | Other size | Unknown size |
|---|---|---|---|---|---|
| Custom FLT3-ITD | 80 | 42 (42) | 17 (17) | 21 | 0 |
| Clinical NGS calls | 22 | 15 (42) | 3 (17) | 4 | 0 |
| Novoalign unfiltered | 39 | 23 (42) | 8 (17) | 8 | 0 |
| BWH cohort (TSCA) | |||||
| Custom FLT3-ITD | 72 | 48 (54) | 17 (20) | 3 | 4 |
| Clinical NGS calls | 53 | 34 (54) | 3 (20) | 12 | 4 |
| AML cohort (TSCA) | Same size as 1° HC (total 1° HC ITDs) | Same size as 2° HC (total 2° HC ITDs) | |||
|---|---|---|---|---|---|
| Custom FLT3-ITD | 7 | 5 (5) | 2 (2) | 0 | 0 |
| Clinical NGS calls | 5 | 3 (5) | 2 (2) | 0 | 0 |
The sensitivity of the custom FLT3-ITD algorithm and clinical standard pipelines within clinical NGS panels (AMP and TSCA) to detect ITDs of the same size as CE or HC was the highest (100%) when applying the custom algorithm to the MGH AMP data. The ability to detect the primary ITD is particularly important because it contributes the most to an assessment of overall allelic ratio.
1°, Primary; 2°, secondary; AML, acute myeloid leukemia; AMP, anchored multiplex PCR; BWH, Brigham and Women's Hospital; CE, capillary electrophoresis; HC, hybrid-capture; ITD, internal tandem duplication; MGH, Massachusetts General Hospital; NGS, next-generation sequencing; TSCA, TruSeq Custom Amplicon.
The relatively high sequence complexity of the FLT3 target locus facilitated prediction of ITD sequences in cases of incomplete breadth of coverage, which was a common occurrence in TSCA. Application of BWA-MEM showed that every 23-bp subsequence of this locus was unique within the human genome and every 9-bp subsequence was unique within the locus itself (Supplemental Table S1). Thus, secondary alignments matching at least 9 bp determined placement in the locus, and 6 bp was sufficient in most instances. Indeed, resulting size predictions from TSCA data were invariably confirmed experimentally by CE. Three FLT3-ITDs, however, had minimal sequencing past their mutant junctions under TSCA, yielding soft clips as small as 1 bp, such that it was not possible to predict the ITD sequence (Supplemental Figure S1). Their alignments demonstrated a property relatively specific to ITDs (versus general insertions) termed divergent paired alignments, where either a read contained a soft clip or insertion while its pair overlapped the site of the soft clip or insertion but did not reach MJ and therefore did not deviate from reference or both reads of a pair contained soft clips and/or insertions but at different reference locations at opposite ends of the duplication. These NGS cases were clinically reported as positive for ITDs of indeterminate size and position; experimental CE revealed sizes of 60 to 72 bp. In contrast to TSCA, both AMP and HC consistently covered the entire duplicated regions of clonal FLT3-ITDs up to 198 bp (AMP) and 195 bp (HC) in size by mutant paired-end reads containing MJ, thus providing support for an ITD based on breadth of coverage in addition to depth.
Performance of AMP Primers
Under AMP, several FLT3 primers in both the forward direction (P0-P4) and reverse direction (P5-P7) were capable in principle of detecting ITDs within exons 14 to 15 (Figure 1). For each FLT3-ITD detected by AMP (N = 76) in the MGH cohort, an average of 4.52 primers (median, 5; range, 1 to 6) yielded paired-end reads sequencing the MJ, and cases where only one primer sequenced the MJ were always associated with extremely low ITD burden (five ITDs with two to four total UMI reads each and all below limit of detection by CE). An average of 1.77 primers (median, 2; range, 1 to 3) yielded sequencing within anchored-end reads only, although analysis of primer design suggested the possibility of rare ITDs not captured by anchored-end reads (eg, ITDs starting early within the P2 primer site and ending late within the P3 primer site). Mutant junctions of these rare ITDs are predicted to be captured within nonanchored end reads of multiple primers on the basis of the empirical data, although not guaranteed a priori. The primer P2, located centrally in exon 14, was extremely effective at detecting ITDs in the MGH cohort. All 76 of 76 (100%) ITDs had their mutant junctions sequenced by paired-end reads derived from P2, and 69 of 76 (90.8%) by anchored-end reads from P2 (Supplemental Table S2). Among FLT3 primers, P2 also generated the most paired reads containing the mutant junction for the vast majority of ITDs (65/76; 85.5%), whereas P3 located at the exon 14/intron 14 boundary generated the most such reads for the longest ITDs (7/76; 9.2%) ranging from 153 to 198 bp, and P4 never generated any reads with mutant junctions because no clonal ITDs in the cohort extended sufficiently far into exon 15 (Supplemental Figure S2). Primers P2 and P3 also appeared to demonstrate the best PCR efficiencies, as they accounted for 22.0% and 24.1% of all reads in the FLT3 exon 14 to 15 region on average, whereas the reverse primer P5 accounted for only 4.4% on average (Supplemental Figure S3).
The consistent capture and detection of ITDs by multiple AMP primers, as described above, should have the added benefit of alleviating the rare PCR pitfall of allelic dropout, which traditionally occurs because of single-nucleotide variants or small insertions/deletions within a primer binding site. Indeed, the population database Genome Aggregation Database (gnomAD) lists rare single-nucleotide variants within the genomic regions targeted by AMP primers P0 to P7 as well as by primers used in TSCA and CE (eg, nine and three single-nucleotide variants associated with the standard CE primers [chromosome 13: 28608330 to 28608352(−) and chromosome 13: 28608024 to 28608046(+)]). Although this study did not uncover any instances of allelic dropout, a prior study described a 75-bp ITD detected by CE but missed by amplicon-based NGS due to an in-cis 3-bp deletion nearby the NGS primer binding site, causing allelic dropout.16 In principle, allelic dropout may also occur from a small ITD entirely contained within a primer binding site, including, for instance, ITDs within the first 22 bp of exon 14 targeted by the forward CE primer [chromosome 13: 28608330 to 28608352(−)]. Although such small ITDs arising this early in exon 14 do not seem to appear in the literature and the juxtamembrane domain itself does not begin until base pair 10 of exon 14, at least one case has been encountered in clinical practice with a small insertion in this region, which was subclonal and deemed to have uncertain clinical significance. Overall, AMP should be considerably less susceptible in theory to false negatives associated with allelic dropout compared with CE and TSCA, which do not provide redundancy of primers. Allelic ratio estimation would likely be affected in AMP but may be corrected by filtering out all reads from the offending primer.
Characterization of TSCA False Negatives
Analysis of TSCA primers (Figure 1) allowed for identification of a subgroup of ITDs arising from exon 14 that theoretically escaped detection by 2 × 150 bp sequencing. TSCA primer pairs were noted to produce wild-type amplicons spanning FLT3 c.1705-51 to c.1837+53 (F1-R1) and c.1836 to c.1942+34 (F2-R2) by design, so that wild-type reads from F1, R1, and F2 ordinarily spanned the 150-bp segments c.1705-51 to c.1803, c.1741 to c.1837+53, and c.1836 to c.1895 (including the 90-bp intron 14), respectively (Supplemental Figure S4). The mutant junction of an ITD with starting coordinate (c0) in exon 14 and end coordinate (c1) was thus reached by F1 if c1 < c.1803, by R1 if c0 > c.1741, and by F2 if c.1837+25 ≤ c1 < c.1895 (because F2 is 27 bp). Therefore, an ITD could be missed by TSCA if the coordinates fell within the following parameters:
1. c0 < c.1741 (in e14)
AND
2. either c.1803 < c1 < c.1837+24 (in e14/i14) or c1 > c.1895 (in e15)
ITDs not satisfying the above criteria manifested at least as divergent paired alignments under manual review (Supplemental Figure S1); however, further relaxation of the criteria (eg, by 9 bp) was required for a guaranteed ability to informatically determine c0 and c1 (Supplemental Table S1). From this analysis, it was also evident that longer reads (eg, 2 × 250 bp) should in principle enable 100% sensitivity for reaching mutant junctions of arbitrary FLT3-ITDs and have been used successfully in other studies.21
These criteria defined a subgroup of relatively long ITDs, with the shortest being 63 bp in theory (c.1741_1803dup). False negatives in the BWH cohort (93 and 108 bp by CE) were suspected to satisfy the criteria; however, sequencing was not available to confirm this. The prevalence of these ITDs among general FLT3-ITDs is thought to be relatively rare (Figure 5). Of the study groups, the unselected MGH cohort likely provided the best estimate, where they were found in approximately 9% (3/35) of FLT3-ITD–positive patients as the variants c.1732-1803dup (72 bp), c.1831_1832insGGCC/1740-1831 (96 bp), and c.1834_1835insCC/1717-1834 (120 bp). In comparison, they were found in presumably approximately 4% (2/49) of FLT3-ITD–positive patients of the BWH cohort (however, this was likely an underestimate because of selection bias), and 0% (0/5) in the AML cohort, which experienced imprecision because of small sample size.
Figure 5.
TruSeq Custom Amplicon (TSCA) escape criteria relative to study cohort internal tandem duplications in FLT3 (FLT3-ITDs). Three ITDs from the Massachusetts General Hospital (MGH) cohort, detected by anchored multiplex PCR, are predicted to escape detection under TSCA relative to current assay design and 2 × 150 bp sequencing due to reads not reaching the mutant junction. The specific variants were c.1732-1803dup (72 bp), c.1831_1832insGGCC/1740-1831 (96 bp), and c.1834_1835insCC/1717-1834 (120 bp). Dotted lines indicate relative location of c1 (exon 14, intron 14, or exon 15); dashed lines indicate ITDs satisfying the TSCA escape criteria. AML, acute myeloid leukemia; BWH, Brigham and Women's Hospital. c0, start genomic coordinate of duplicated genomic region in an ITD; c1, end genomic coordinate of duplicated genomic region in an ITD; N, nontemplated insert.
Landscape of FLT3-ITDs Detected and Characterized by NGS
Overall, the custom FLT3-ITD algorithm identified 109 unique FLT3-ITDs between 15 and 198 bp in size from the study cohorts, composed of 63 exact duplications and 46 near-exact duplications with intervening nontemplated inserts between 1 and 11 bp (Supplemental Figure S5 and Supplemental Table S3). In addition, five ectopic insertions/deletions up to 27 bp in net size were found, including one variant (c.1780delinsGAAAGGTCCCGTGTCC) with a relatively high allelic fraction of 33% to 34% by CE/NGS and a blast count of 31% by flow cytometry, indicative of loss of heterozygosity. The FLT3-ITDs appeared in 86 different patients, with 75 unique ITDs appearing as primary NGS clones, 43 as secondary NGS subclones, and 9 common to more than one patient, where the most common ITD (c.1770-1793dup) appeared in 6 patients. NGS was performed clinically at multiple time points for 18 patients with persistent ITDs, and the primary NGS ITD always remained the same, except for one instance where a new clone emerged and became primary. By contrast, the set of secondary NGS ITDs never remained the same between different time points in the subset of seven patients with multiple NGS ITDs and multiple samples. Overall, multiple NGS ITDs ranging from 2 to 7 were found in 27 of 108 cases. All FLT3-ITDs originated within the juxtamembrane domain of exon 14, and most were entirely contained within exon 14, whereas minorities extended into the early portions of intron 14 and exon 15. Because ITDs extending into intron 14 duplicate its splice donor site, theoretically mutant and/or wild-type protein may be produced, depending on splice site usage. Thus, the empirical absence of ITDs extending into later portions of intron 14 may be due to a lack of competitive advantage for clonal selection to occur, under the hypothesis that wild-type protein is preferentially produced once enough of the 5′ end of intron 14 is duplicated to not disrupt use of the associated splice donor site. Intron 14 was also checked for potential occult in-frame stop codons in the setting of duplication that might preclude clonal selection, and none were found.
NGS-Based Determination of AF
Most FLT3-ITDs characterized by the custom algorithm did not contain duplicated primer sites of the NGS assay used (49/64 ITDs detected by AMP and CE; 74/74 ITDs detected by TSCA and CE; 5/7 ITDs detected by TSCA and HC). For these cases, orthogonal assays produced similar allelic fraction estimates, with mean absolute errors of 1.95% (AMP versus CE), 2.53% (TSCA versus CE), and 3.26% (TSCA versus HC) (Table 3 and Figure 6). The term error is used herein for convenience, as a true gold standard assay for AR has in many respects not yet been established.2 Methods ignoring UMIs were also evaluated, which may be relevant for laboratories using AMP technology without UMIs. AF estimates based on raw paired read counts performed slightly worse than with UMIs (mean absolute error of 2.19% versus 1.95%) but better than simple deduplication (mean absolute error of 8.99%), which experienced a greater degree of read deduplication from the more prevalent population (wild type versus mutant). Because almost all FLT3-ITDs in MGH/AMP had an AF <0.5 by CE (ie, more wild-type than mutant alleles), most cases had greater deduplication of wild-type reads, resulting in overestimation of AF (average error of 8.30% by deduplication method) (Supplemental Figure S6A). The clinical Novoalign pipeline without custom ITD informatics systematically underestimated AF because Novoalign recognized only a fraction K of mutant reads (those sufficiently spanning the entire ITD to align with an insertion), with K decreasing as ITD size increased; average K was approximately 0.51 for ITDs <40 bp in size and approximately 0.09 for ITDs >40 bp (Supplemental Figure S6B). The theoretical upper limit of detection by Novoalign is at most 75 bp because greater length duplications cannot be spanned by a 151-bp read; however, additional read support past the duplication is also necessary where the amount may depend on sequence content and aligner parameters. Indeed, the longest ITD called by Novoalign in the data was 60 bp, whereas the shortest significant ITD (AF ≥ 0.05 by CE) missed by Novoalign was 54 bp.
Table 3.
AF Performance
| AF methods | N | MAE, % | Average error, % | Range errors, % | Linear regression | R2 |
|---|---|---|---|---|---|---|
| No duplicated primer sites | ||||||
| AMP base versus CE | 49 | 1.95 | 0.95 | −7.0 to 9.0 | 1.02x + 0.005 | 0.980 |
| AMP raw versus CE | 49 | 2.19 | 1.09 | −7.2 to 9.9 | 1.04x + 0.004 | 0.978 |
| AMP deduplication versus CE | 49 | 8.99 | 8.30 | −16.3 to 23.2 | 0.94x + 0.093 | 0.859 |
| AMP standard versus CE | 34 | 13.99 | −13.61 | −76.6 to 2.8 | 0.20x + 0.025 | 0.296 |
| ITD size < 40 bp | 21 | 0.51x + 0.007 | 0.926 | |||
| ITD size 40–60 bp | 13 | 0.09x + 0.009 | 0.400 | |||
| TSCA versus CE | 74 | 2.53 | 0.83 | −13.7 to 11.1 | 1.04x − 0.001 | 0.983 |
| TSCA versus HC | 5 | 3.26 | −0.05 | −6.3% to 7.1 | 1.05x − 0.010 | 0.889 |
| ITDs with duplicated primer sites | ||||||
| AMP base versus CE | 15 | 9.73 | −9.70 | −24.9 to 0.3 | 0.59x − 0.005 | 0.843 |
| AMP adjusted versus CE | 15 | 6.21 | 1.48 | −10.3 to 16.3 | 0.99x + 0.017 | 0.801 |
AF estimates were compared under various methods within clinical next-generation sequencing panels (AMP and TSCA) versus CE or HC. Only ITDs detected by both AMP/TSCA and CE/HC were included. Not utilizing unique molecular identifiers (AMP raw) had relatively minimal effect for ITDs without duplicated primer sites. A simple AF adjustment for ITDs with duplicated primer sites improved the linear regression slope from considerably <1 (AMP base), indicative of underestimation, to near 1 (AMP adjusted); however R2 remained moderate.
AF, allelic fraction; AMP, anchored multiplex PCR; CE, capillary electrophoresis; HC, hybrid-capture; ITD, internal tandem duplication; MAE, mean absolute error; TSCA, TruSeq Custom Amplicon.
Figure 6.
Allelic fraction (AF) by next-generation sequencing (NGS) versus capillary electrophoresis (CE) for internal tandem duplications in FLT3 not duplicating a primer site. Anchored multiplex PCR (AMP; A) and TruSeq Custom Amplicon (TSCA) (B) produce highly correlated AF estimates to CE with linear regression slopes near 1 and intercepts near 0.
Fifteen ITDs in the MGH cohort contained one to two duplicated primer sites under AMP and had sizes between 54 and 198 bp. The AMP base method showed moderate correlation but consistently underestimated AF (in 14/15 or 93% of ITDs) relative to CE (Figure 7A), with mean absolute error of 9.73% and mean error of −9.70% (Table 3). The single ITD that was not underestimated had AF <0.01 by both CE and NGS (ie, CV was expected to be large), whereas all other ITDs had AF >0.05. Application of a simple adjustment to account for the duplicated primer sites yielded linear regression slopes (AMP versus CE) of 0.585 before adjustment (indicative of underestimation) and 0.992 after adjustment, with intercepts of −0.5% (before) and 1.7% (after), R2 of 84.3% (before) and 80.1% (after), and mean absolute errors of 9.7% (before) and 6.2% (after) (Figure 7B).
Figure 7.
Allelic fraction (AF) by next-generation sequencing (NGS) versus capillary electrophoresis (CE) for internal tandem duplications in FLT3 (FLT3-ITDs) containing duplicated primer sites. A: Anchored multiplex PCR (AMP) without adjustments systematically underestimates AF relative to CE with a linear regression slope considerably <1. B: A simple adjustment improves the linear regression slope; however, correlation remains moderate.
No ITDs in the BWH cohort contained duplicated primer sites under TSCA. Two ITDs in the AML cohort of sizes 180 bp (c.1749_1838dup) and 195 bp (c.1735_1839dup) each contained two duplicated primer sites (R1 and F2) under TSCA, and their AF estimates were considerably smaller under TSCA versus HC. The 180-bp ITD had AF estimates of 11.1% (base) and 23.4% (adjusted) under TSCA versus 48.0% under HC, whereas the 195-bp ITD had AF estimates of 0.4% (base) and 0.6% (adjusted) under TSCA versus 15.5% under HC. Mutant junctions of these ITDs unexpectedly did not appear in mixed F2-R1 amplicons and were found only in F1-R1 or F2-R2 amplicons with inferred insert sizes >400 bp compared with wild-type insert sizes around 230 bp. Moreover, the 195-bp ITD (c.1735_1839dup) satisfied a subset of escape criteria (c0 ≤ c.1741, and c1 ≥ c.1803) and was not detected in F1-R1 amplicons. Because HC has generally performed better than TSCA, according to studies in the literature, and has produced AR classifications consistent with CE, the large discrepancy between TSCA and HC was presumably related to significantly reduced efficiency in TSCA of the considerably larger mutant amplicons versus wild-type amplicons, a factor not accounted for in the adjusted formula; however, CE data were not available for confirmation.24,30, 31, 32 The absence of the mixed amplicon was unexplained but theoretically could have resulted if TSCA amplicons targeted opposite strands of FLT3, thereby generally precluding mixed products under the TSCA extension-ligation step.
Classification into FLT3-ITD high and low categories, defined by an AR cutoff of 0.5 per ELN risk stratification guidelines (equivalently, an AF cutoff of one-third or approximately 0.33), was highly concordant between NGS and CE (Supplemental Figure S7). For cases without ITDs duplicating primer sites, the concordance rate was 100% (33/33) between AMP and CE and 92.6% (50/54) between TSCA and CE. For AMP cases with ITDs duplicating primer sites, the concordance rate versus CE was 92.9% (13/14) before adjustment, where the discordant case was considerably underestimated under AMP (ΔAF = −25%). Informatic adjustment to account for duplicated primer sites reduced the AF underestimation from −25% to −8.7% (without changing discordant status). However, doing so resulted in three initially concordant but underestimated cases (base ΔAF = −0.5%, −9.4%, and −7.8%) to become overestimated and discordant (adjusted ΔAF = 16%, 12%, and 9.8%), for an overall decrease in concordance rate to 71.4% (10/14) on adjustment, despite improved overall performance, as described earlier, relative to mean absolute error and regression slope. One theoretical source of overestimation was due to allelic differences (wild-type versus mutant ITD) associated with noncontributory primers in the setting of primer duplication, which was not accounted for in the current adjustment model (vide infra for further details).
The definitions of FLT3-ITD high and low are not specifically described by ELN guidelines for cases with multiple ITDs. Two natural definitions arise based on AR of the dominant ITD or sum of ARs over all ITDs. These definitions generated discrepant categorizations relative to one another for 1 case under CE from the BWH cohort (of 16 cases with multiple ITDs) and 2 cases under both CE and AMP from the MGH cohort (of 7 cases with multiple ITDs). For instance, the discrepant BWH case demonstrated 2 ITDs with ARs of 0.45 and 0.40 under CE, yielding categorizations of FLT3-ITD low under definition 1 but of FLT3-ITD high using definition 2; by comparison under TSCA, the ARs were 0.51 and 0.37 for an FLT3-ITD high status using either definition. Definition 2 was adopted in the present study, which yielded slightly more concordances between CE and NGS, such as in the above BWH case. However, until a standard guideline emerges, clinicians might decide to determine categorizations on a case-by-case basis, and a molecular laboratory should ideally strive to report all individual ARs and their sum if possible.
Potential Adjustment Biases and Challenges Estimating AF Because of Primer Duplication
The current adjustment model implicitly assumes independence of primers, which may be favored as input DNA increases relative to primer abundance, whereas in practice, competition among primers will invariably occur to some extent. Such primer competition is difficult to quantify and unaccounted for in the model, which may lead to slight overestimation of AF. To illustrate this, consider the toy scenario of an ITD duplicating P2 and extending near (or disrupting) P3, such that P3 reads cannot capture the MJ but P2 and P3 likely compete in a mutually exclusive manner for DNA fragments (wild type or mutant) (Supplemental Figure S7). Supposing these are the only primers competing for A wild-type alleles and B mutant-ITD alleles with equal primer binding efficiency and an infinite pool of primers, then P2 would bind A/2 wild-type alleles, B/3 mutant alleles capturing MJ, and B/3 mutant alleles without capture of MJ (while P3 would bind the remaining alleles). According to the current model, the base AF (for TSCA like and duplicated P2) is then (B/3)/(A/2 + 2B/3) = 2B/(3A + 4B), which underestimates the true AF, whereas the adjusted AF is (2B/3)/(A/2 + 2B/3) = 4B/(3A + 4B) would overestimate true AF:
| (9) |
| (10) |
In terms of proportional errors:
| (11) |
| (12) |
| (13) |
In other words, base AF is a relatively substantial underestimate of true AF, whereas adjusted AF becomes a milder overestimate in comparison. In fact, the average of base and adjusted AF continues to underestimate true AF in the toy scenario; however, this would no longer be guaranteed under different scenarios (eg, three mutually exclusive primers). Empirically, on the level of individual ITDs duplicating primer sites in the data (15 ITDs from 14 cases), base AF underestimated CE-measured AF in 14 of 15 (93.3%) ITDs (with the single outlier at AF < 0.01), whereas adjusted AF overestimated 8 of 15 (53.3%) ITDs. It is uncertain how often assumptions of the toy scenario hold in practice, and a larger data set may be informative.
Discussion
The present study developed a novel FLT3-ITD algorithm applicable across multiple NGS platforms, with sensitivities and specificities of 100% and 99.4%, respectively, on an unselected cohort (MGH/AMP) and 98.1% and 100%, respectively, on a selected cohort (BWH/TSCA), where false positives were almost certainly due to increased sensitivity of NGS relative to CE. By contrast, the original clinical pipelines were 38.1% sensitive and 99.8% specific (MGH/AMP) and 92.6% sensitive and 100% specific (BWH/TSCA). Benefits of NGS over CE include greater genomic coverage, potentially increased sensitivity, and extraction of ITD sequence information. A recent study indicated that nucleotide composition of ITDs, particularly the presence of nontemplated nucleotide content, may impact response to FLT3 inhibition and induction chemotherapy, whereas prior studies have reported inferior outcomes associated with ITDs extending into tyrosine kinase domain 1 or located closer to the C-terminus.9,22,33,34 By using this algorithm, a case was encountered with a 30-bp ITD at initial diagnosis along with a 180-bp ITD extending into tyrosine kinase domain 1, where the 30-bp ITD subsequently disappeared on treatment while the 180-bp subclone persisted at low level. The clinical significance is uncertain; however, the case highlights the potential utility of comprehensive ITD analysis. In theory, characterization of the translated mutant protein of an ITD may also be beneficial. Although clonal insertions in exons 14 to 15 have almost universally corresponded to in-frame elongations, a recurrent out-of-frame deletion has been observed, resulting in truncation, loss of function, and dominant negative effects in vitro.35 Implications for targeted FLT3 therapy are unknown; however, loss of efficacy would be predicted. In principle, it is similarly possible for sequencing to reveal FLT3 insertions that are out of frame or give rise to premature stop codons. The FLT3-ITD c.1841-1861, reported in two cases of acute promyelocytic leukemia and notable for being contained entirely in tyrosine kinase domain 1, translates to a nonsense variant p.A620_F621ins∗(COSV54057070).36
Multiple specialized algorithms have been designed to handle ITDs or ITD-sized insertions/deletions, including ITDseek and AIH for amplicon data, F-TAFI and breakpointSearch for HC data, and Genomon ITDetector, ITD Assembler, and HeatITup for general somatic ITDs.14,16,18, 19, 20,22,24 The current algorithm leverages soft clips and secondary alignments similar to ITDetector and ITDseek but differs by using in silico extension, alignment-based annotation, UMI handling, and platform-specific AR estimates. Theoretical limitations of this approach include an inability to detect large purely nontemplated insertions, because the algorithm requires either separate alignments anchoring opposite ends of an individual read to FLT3 or a single alignment recognizing the entire insertion to proceed to the extension step. De novo assembly is more suitable for these cases and may be capable of piecing together multiple reads across such mutant segments. However, large nontemplated insertions in FLT3 have not been reported. The largest such insert was 36 bp in a recent study and 27 bp in this study.24 Large insertions in the form of internal tandem triplicates have been submitted rarely to the Catalogue of Somatic Mutations in Cancer (COSMIC, https://cancer.sanger.ac.uk/cosmic), but the current algorithm should detect them almost as well as de novo assembly, because both methods require a read sequencing across both mutant junctions to distinguish an internal tandem triplicate from an ITD unless imbalance of read coverage is somehow leveraged. Moreover, this iterative alignment-based approach to annotations should recognize such variants as triplicates. By default, the algorithm assumes and uses human genome assembly GRCh37 (hg19) for input BAM files and output VCF files; however, version 1.0 and later of the script is also compatible with GRCh38 (hg38).
The current algorithm may be adapted to other loci (eg, exon 15 of BCOR for detection of recurrent BCOR-ITDs). Because each locus has its own unique genomic complexity, separate assessment of performance relative to assay design and algorithm parameters is warranted. The algorithm may also be adapted to analyze FLT3-ITD transcripts within RNA-sequencing assays. Although allelic ratio is defined by ELN guidelines in terms of DNA fragment analysis, many RNA-based studies have shown analogous prognostic implications of FLT3-ITD mRNA level and one pediatric AML study reported greater prognostic significance through RNA-based measurements versus DNA-based measurements.37,38 An additional benefit of RNA sequencing is assessment of the actual spliced FLT3-ITD transcripts, particularly those extending into intron 14 or exon 15, where splicing can only be predicted under DNA sequencing because of duplication of splice sites.38 RNA-based assays have also been hypothesized to have greater sensitivity because of possible overexpression of mutant ITD alleles; however, comparisons of RNA and DNA fragment analysis in the above pediatric AML study did not demonstrate such overexpression.38 By contrast, specificity might be decreased because of the relative abundance of artifacts in RNA-based NGS assays, at least extrapolating from experience within clinical laboratories. Further studies of RNA-based FLT3-ITD assessment will be useful as laboratories increasingly deploy RNA-based clinical NGS assays and to explore optimal assays for minimal residual disease detection.
Accurate AR estimates were achieved by the algorithm through platform-specific methods. By contrast, prior NGS studies have consistently reported AR underestimates relative to CE, including two recent studies showing linear regression slopes of approximately 0.40 to 0.75.17,21,24,25 Multiple sources of underestimation are possible: i) unrecognized mutant reads, which are generally minimized via modern FLT3-ITD algorithms, ii) duplicated or disrupted primer sites by ITDs in PCR-based assays, and iii) mismatched mutant and wild-type populations. To address ii, informatic methods for TSCA and AMP identified problematic primer-ITD duos through alignment-based annotation and attempted to correct bias through simple adjustments. This was reasonably effective under AMP, whereas TSCA appeared to experience reduced efficiency related to significantly longer amplicons; AMP was significantly less prone in theory to size-related experimental biases because its fragment sizes were driven by random hexamer priming or enzymatic shearing of one end.32 To address iii, relevant primers were identified and restricted to by ITD in TSCA and AMP, whereas for HC, an indirect calculation of AR through AF was formulated, and mutant and reference junctions were used to define comparable populations. Conceptually, HC is more compatible with AF than AR because of ambiguous reads; AF may also be more informative in rare situations of multiple ITDs without wild-type alleles (eg, mixtures of cell lines with homozygous FLT3-ITD variants), where AF correctly quantifies relative amounts while AR would be infinite for each ITD; the asymptotic behavior of AR toward infinity may additionally yield less robust regression analyses.
The clinical NGS assays were AMP or amplicon based (TSCA), and many laboratories opt for these technologies because of simplified workflow, smaller amounts of required DNA, and generally faster turnaround times.30 Findings from the current study support the use of AMP-based NGS as a sensitive and specific test for FLT3-ITD detection with relatively accurate estimates of AR and redundant protection against allelic dropout, which has been described in a commercial TSCA assay.16 For amplicon-based NGS, this approach allowed inference of ITD annotations when there was only partial sequencing coverage because of primer locations relative to ITD boundaries. Analysis establishing escape criteria of ITDs missed by amplicon sequencing was further suggested, which, in this TSCA assay, was satisfied by approximately 9% of primary ITDs from an unselected cohort. Thus, TSCA was highly specific but slightly less sensitive than AMP, although utilization of longer read lengths or different primer sets may improve on this in the future. Finally, for HC, it is believed that adoption of the modified AR formula, in conjunction with recognition of all unambiguous mutant reads based on alignments to the ITD genome, will address trends for underestimation seen in the literature. HC is likely optimal at both FLT3-ITD detection and AR estimation and is not subject to issues of duplicated or disrupted primer sites; however, head-to-head studies will be necessary to confirm this hypothesis.
The original motivation for developing a custom FLT3-ITD algorithm was to enhance locally developed NGS pipelines. For AMP, in particular, our laboratory does not use the Archer vendor supported pipeline as our institution relies on our own library preparations, adapters, and informatics. Successful FLT3-ITD assessment using the Archer assay together with its informatics pipeline has been described in conference abstracts, including accurate detection of low AF variants, although whether there are follow-up publications providing further details is not known. Thus, although a comparison against this algorithm would be informative, it is not the intent of this study to suggest its implementation alongside the commercially available Archer pipeline, but rather to offer a flexible solution applicable to multiple platforms for laboratories that may not have an effective algorithm for detecting and quantifying FLT3-ITDs. Subsequently, the algorithm was successfully deployed with only minor adjustments in the analysis of FLT3-ITDs from yet another NGS platform using an NEBNext library preparation (New England Biolabs, Ipswich, MA). These enhancements have been made available in the latest version of the code.
In summary, the current article provides a lightweight FLT3-ITD–specific algorithm applicable to multiple NGS platforms for detection, annotation, and assessment of ITDs accompanied by platform-specific methods to quantify AR. Overall, targeted informatics should be considered for nonstandard variants of clinical importance in targeted NGS panels, and the benefits of the targeted approach to FLT3-ITD analysis, including full capture of mutant reads, annotation of ITD structure, proper use of UMIs, and accurate AR estimates, are demonstrated.
Footnotes
Supported in part by NIH grant R01 CA225655 (J.K.L.).
Disclosures: L.P.L. owns equity in ArcherDx and is on the scientific advisory board as a consultant. A.S.K. has received consulting fees from LabCorp. R.C.L. has received research funding from Jazz and MedImmune and consulting fees from Takeda.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2020.06.006.
Contributor Information
Annette S. Kim, Email: askim@bwh.harvard.edu.
Valentina Nardi, Email: vnardi@partners.org.
Supplemental Data
Supplemental Figure S1.
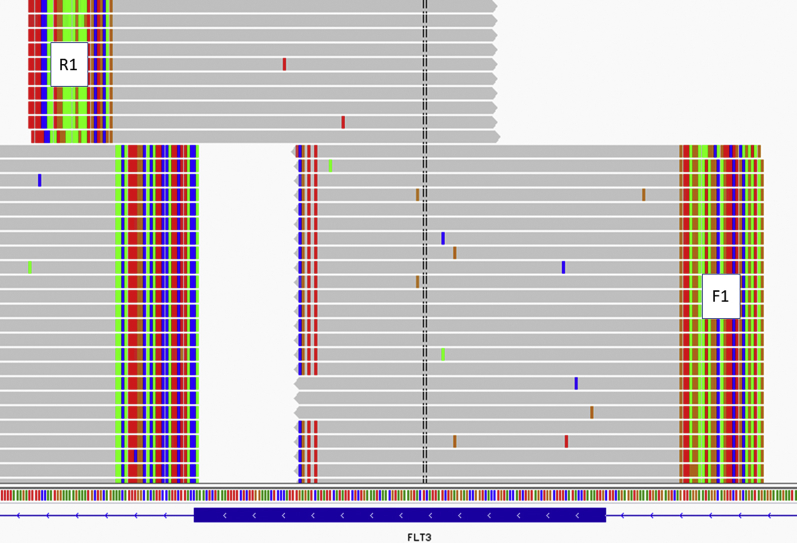
Divergent paired alignments. Mutant reads from F1 in a TSCA case with high allelic ratio just reach the mutant junction where they are soft clipped (6 bp) while their partners from R1 appear to be wild type in the area of the soft clip, as depicted in Integrative Genomics Viewer (Broad Institute, Cambridge, MA) with wild-type sequence in gray and nonreference sequence in green (A), red (T), blue (C), or brown (G). All primer sequences are soft clipped by convention.
Supplemental Figure S2.

Performance by anchored multiplex PCR (AMP) primer. Heat map of mutant reads by AMP primer for 76 internal tandem duplications in FLT3 (FLT3-ITDs) detected in the Massachusetts General Hospital (MGH) cohort. An average of 4.5 AMP primers (median, 5) generated reads capturing the mutant junction of each ITD, and always included the primer P2. This primer redundancy should alleviate situations of allelic dropout.
Supplemental Figure S3.

Relative efficiencies of anchored multiplex PCR (AMP) primers. Proportion of reads by primer across the FLT3 target locus showed consistent trends typical of PCR-based next-generation sequencing. Primers P2 and P3 were the most efficient at capturing reads, whereas P5 was the least efficient. MGH, Massachusetts General Hospital; mut, mutant; wt, wild type.
Supplemental Figure S4.
TSCA escape criteria. Given an exact internal tandem duplication (ITD) with duplicated region D (blue) consisting of nucleotides between coordinates c0 and c1 and contained entirely within exon 14 (pink), reaching the mutant junction (MJ) within reads derived from primers F1 or R1 of an FLT3-ITD amplicon depends on both ITD location and length of sequencing reads.
Supplemental Figure S5.
Landscape of detected internal tandem duplications (ITDs). A total of 123 FLT3-ITDs (109 unique) and 5 ectopic insertions of sizes between 3 and 198 bp were captured by next-generation sequencing from the study cohorts using the custom pipeline. The vast majority were patient specific (advantageous for minimal residual disease and contaminant detection), whereas c.1770-1793dup appeared in six patients. Dashed lines delineate exon boundaries relative to exact duplications. AML, acute myeloid leukemia; BWH, Brigham and Women's Hospital; MGH, Massachusetts General Hospital.
Supplemental Figure S6.
Allelic fraction (AF) error by anchored multiplex PCR (AMP) under various informatic modifications relative to capillary electrophoresis (CE). A: Simple deduplication (dedup), which is commonly applied to hybrid-capture data, resulted in systematic bias because of a greater degree of read deduplication from the more prevalent population (wild type versus mutant). B: The standard clinical pipeline based on Novoalign (or similarly other general aligners) systematically underestimated AF because of omitting soft-clipped mutant reads. Stratification by internal tandem duplication (ITD) length showed a linear correlation between error and AF under the standard pipeline because the fraction of soft-clipped mutant reads increased for longer ITDs.
Supplemental Figure S7.
Internal tandem duplication in FLT3 (FLT3-ITD) high [allelic ratio (AR) ≥ 0.5] and low (AR < 0.5) status, with total AR used for cases with multiple ITDs. Plots show allelic fraction (AF), where the threshold AR = 0.5 is equivalent to AF = AR/(1 + AR) = 1/3. A: TSCA versus capillary electrophoresis (CE). A total of 50 of 54 (93%) cases were categorized concordantly (21 high and 29 low), whereas 2 cases were high by TSCA but low by CE (ΔAF = 6.9% and 0.7%) and 2 cases were low by TSCA but high by CE (ΔAF = −4.9% and −2.7%). B: Anchored multiplex PCR (AMP) versus CE. All 33 of 33 (100%) cases without any ITDs duplicating primers were concordant (11 high and 22 low), whereas 13 of 14 (93%) cases with an ITD duplicating an AMP primer were initially concordant under base AF (1 high and 10 low) and the remaining discordant case was high by CE but low by AMP. Informatic adjustment to account for duplication of the primer sites appeared to grossly correct the underestimation bias; however, three previously underestimated low cases became overestimated and discordantly high. Adj, adjusted; NGS, next-generation sequencing.
References
- 1.Nakao M., Yokota S., Iwai T., Kaneko H., Horiike S., Kashima K., Sonoda Y., Fujimoto T., Misawa S. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10:1911–1918. [PubMed] [Google Scholar]
- 2.Daver N., Schlenk R.F., Russell N.H., Levis M.J. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia. 2019;33:299–312. doi: 10.1038/s41375-018-0357-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone R.M., Mandrekar S.J., Sanford B.L., Laumann K., Geyer S., Bloomfield C.D., Thiede C., Prior T.W., Döhner K., Marcucci G., Lo-Coco F., Klisovic R.B., Wei A., Sierra J., Sanz M.A., Brandwein J.M., de Witte T., Niederwieser D., Appelbaum F.R., Medeiros B.C., Tallman M.S., Krauter J., Schlenk R.F., Ganser A., Serve H., Ehninger G., Amadori S., Larson R.A., Döhner H. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med. 2017;377:454–464. doi: 10.1056/NEJMoa1614359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perl A.E., Altman J.K., Cortes J., Smith C., Litzow M., Baer M.R., Claxton D., Erba H.P., Gill S., Goldberg S., Jurcic J.G., Larson R.A., Liu C., Ritchie E., Schiller G., Spira A.I., Strickland S.A., Tibes R., Ustun C., Wang E.S., Stuart R., Röllig C., Neubauer A., Martinelli G., Bahceci E., Levis M. Selective inhibition of FLT3 by gilteritinib in relapsed or refractory acute myeloid leukaemia: a multicentre, first-in-human, open-label, phase 1-2 study. Lancet Oncol. 2017;18:1061–1075. doi: 10.1016/S1470-2045(17)30416-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitman S.P., Archer K.J., Feng L., Baldus C., Becknell B., Carlson B.D., Carroll A.J., Mrózek K., Vardiman J.W., George S.L., Kolitz J.E., Larson R.A., Bloomfield C.D., Caligiuri M.A. Absence of the wild-type allele predicts poor prognosis in adult de novo acute myeloid leukemia with normal cytogenetics and the internal tandem duplication of FLT3: a cancer and leukemia group B study. Cancer Res. 2001;61:7233–7239. [PubMed] [Google Scholar]
- 6.Thiede C., Steudel C., Mohr B., Schaich M., Schäkel U., Platzbecker U., Wermke M., Bornhäuser M., Ritter M., Neubauer A., Ehninger G., Illmer T. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: association with FAB subtypes and identification of subgroups with poor prognosis. Blood. 2002;99:4326–4335. doi: 10.1182/blood.v99.12.4326. [DOI] [PubMed] [Google Scholar]
- 7.Gale R.E., Green C., Allen C., Mead A.J., Burnett A.K., Hills R.K., Linch D.C. The impact of FLT3 internal tandem duplication mutant level, number, size, and interaction with NPM1 mutations in a large cohort of young adult patients with acute myeloid leukemia. Blood. 2008;111:2776–2784. doi: 10.1182/blood-2007-08-109090. [DOI] [PubMed] [Google Scholar]
- 8.Schnittger S., Bacher U., Haferlach C., Alpermann T., Kern W., Haferlach T. Diversity of the juxtamembrane and TKD1 mutations (exons 13-15) in the FLT3 gene with regards to mutant load, sequence, length, localization, and correlation with biological data. Genes Chromosomes Cancer. 2012;51:910–924. doi: 10.1002/gcc.21975. [DOI] [PubMed] [Google Scholar]
- 9.Schlenk R.F., Kayser S., Bullinger L., Kobbe G., Casper J., Ringhoffer M., Held G., Brossart P., Lübbert M., Salih H.R., Kindler T., Horst H.A., Wulf G., Nachbaur D., Götze K., Lamparter A., Paschka P., Gaidzik V.I., Teleanu V., Späth D., Benner A., Krauter J., Ganser A., Döhner H., Döhner K. Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood. 2014;124:3441–3449. doi: 10.1182/blood-2014-05-578070. [DOI] [PubMed] [Google Scholar]
- 10.Linch D.C., Hills R.K., Burnett A.K., Khwaja A., Gale R.E. Impact of FLT3(ITD) mutant allele level on relapse risk in intermediate-risk acute myeloid leukemia. Blood. 2014;124:273–276. doi: 10.1182/blood-2014-02-554667. [DOI] [PubMed] [Google Scholar]
- 11.Döhner H., Estey E., Grimwade D., Amadori S., Appelbaum F.R., Büchner T., Dombret H., Ebert B.L., Fenaux P., Larson R.A., Levine R.L., Lo-Coco F., Naoe T., Niederwieser D., Ossenkoppele G.J., Sanz M., Sierra J., Tallman M.S., Tien H.F., Wei A.H., Löwenberg B., Bloomfield C.D. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiyoi H., Naoe T., Yokota S., Nakao M., Minami S., Kuriyama K., Takeshita A., Saito K., Hasegawa S., Shimodaira S., Tamura J., Shimazaki C., Matsue K., Kobayashi H., Arima N., Suzuki R., Morishita H., Saito H., Ueda R., Ohno R., Leukemia Study Group of the Ministry of Health and Welfare (Kohseisho) Internal tandem duplication of FLT3 associated with leukocytosis in acute promyelocytic leukemia. Leukemia. 1997;11:1447–1452. doi: 10.1038/sj.leu.2400756. [DOI] [PubMed] [Google Scholar]
- 13.Murphy K.M., Levis M., Hafez M.J., Geiger T., Cooper L.C., Smith B.D., Small D., Berg K.D. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J Mol Diagn. 2003;5:96–102. doi: 10.1016/S1525-1578(10)60458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKerrell T., Moreno T., Ponstingl H., Bolli N., Dias J.M., Tischler G., Colonna V., Manasse B., Bench A., Bloxham D., Herman B., Fletcher D., Park N., Quail M.A., Manes N., Hodkinson C., Baxter J., Sierra J., Foukaneli T., Warren A.J., Chi J., Costeas P., Rad R., Huntly B., Grove C., Ning Z., Tyler-Smith C., Varela I., Scott M., Nomdedeu J., Mustonen V., Vassiliou G.S. Development and validation of a comprehensive genomic diagnostic tool for myeloid malignancies. Blood. 2016;128:e1–e9. doi: 10.1182/blood-2015-11-683334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kluk M.J., Lindsley R.C., Aster J.C., Lindeman N.I., Szeto D., Hall D., Kuo F.C. Validation and implementation of a custom next-generation sequencing clinical assay for hematologic malignancies. J Mol Diagn. 2016;18:507–515. doi: 10.1016/j.jmoldx.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Au C.H., Wa A., Ho D.N., Chan T.L., Ma E.S. Clinical evaluation of panel testing by next-generation sequencing (NGS) for gene mutations in myeloid neoplasms. Diagn Pathol. 2016;11:11. doi: 10.1186/s13000-016-0456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spencer D.H., Abel H.J., Lockwood C.M., Payton J.E., Szankasi P., Kelley T.W., Kulkarni S., Pfeifer J.D., Duncavage E.J. Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J Mol Diagn. 2013;15:81–93. doi: 10.1016/j.jmoldx.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Chiba K., Shiraishi Y., Nagata Y., Yoshida K., Imoto S., Ogawa S., Miyano S. Genomon ITDetector: a tool for somatic internal tandem duplication detection from cancer genome sequencing data. Bioinformatics. 2015;31:116–118. doi: 10.1093/bioinformatics/btu593. [DOI] [PubMed] [Google Scholar]
- 19.Kadri S., Zhen C.J., Wurst M.N., Long B.C., Jiang Z.F., Wang Y.L., Furtado L.V., Segal J.P. Amplicon indel hunter is a novel bioinformatics tool to detect large somatic insertion/deletion mutations in amplicon-based next-generation sequencing data. J Mol Diagn. 2015;17:635–643. doi: 10.1016/j.jmoldx.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Rustagi N., Hampton O.A., Li J., Xi L., Gibbs R.A., Plon S.E., Kimmel M., Wheeler D.A. ITD assembler: an algorithm for internal tandem duplication discovery from short-read sequencing data. BMC Bioinformatics. 2016;17:188. doi: 10.1186/s12859-016-1031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schranz K., Hubmann M., Harin E., Vosberg S., Herold T., Metzeler K.H., Rothenberg-Thurley M., Janke H., Bräundl K., Ksienzyk B., Batcha A.M.N., Schaaf S., Schneider S., Bohlander S.K., Görlich D., Berdel W.E., Wörmann B.J., Braess J., Krebs S., Hiddemann W., Mansmann U., Spiekermann K., Greif P.A. Clonal heterogeneity of FLT3-ITD detected by high-throughput amplicon sequencing correlates with adverse prognosis in acute myeloid leukemia. Oncotarget. 2018;9:30128–30145. doi: 10.18632/oncotarget.25729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwartz G.W., Manning B., Zhou Y., Velu P., Bigdeli A., Astles R., Lehman A.W., Morrissette J.J.D., Perl A.E., Li M., Carroll M., Faryabi R.B. Classes of ITD predict outcomes in AML patients treated with FLT3 inhibitors. Clin Cancer Res. 2019;25:573–583. doi: 10.1158/1078-0432.CCR-18-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mack E.K.M., Marquardt A., Langer D., Ross P., Ultsch A., Kiehl M.G., Mack H.I.D., Haferlach T., Neubauer A., Brendel C. Comprehensive genetic diagnosis of acute myeloid leukemia by next-generation sequencing. Haematologica. 2019;104:277–287. doi: 10.3324/haematol.2018.194258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He R., Devine D.J., Tu Z.J., Mai M., Chen D., Nguyen P.L., Oliveira J.L., Hoyer J.D., Reichard K.K., Ollila P.L., Al-Kali A., Tefferi A., Begna K.H., Patnaik M.M., Alkhateeb H., Viswanatha D.S. Hybridization capture-based next generation sequencing reliably detects FLT3 mutations and classifies FLT3-internal tandem duplication allelic ratio in acute myeloid leukemia: a comparative study to standard fragment analysis. Mod Pathol. 2020;33:334–343. doi: 10.1038/s41379-019-0359-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim B., Kim S., Lee S.T., Min Y.H., Choi J.R. FLT3 internal tandem duplication in patients with acute myeloid leukemia is readily detectable in a single next-generation sequencing assay using the pindel algorithm. Ann Lab Med. 2019;39:327–329. doi: 10.3343/alm.2019.39.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Z., Liebers M., Zhelyazkova B., Cao Y., Panditi D., Lynch K.D., Chen J., Robinson H.E., Shim H.S., Chmielecki J., Pao W., Engelman J.A., Iafrate A.J., Le L.P. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 27.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997. [Google Scholar]
- 28.Kopylova E., Navas-Molina J.A., Mercier C., Xu Z.Z., Mahé F., He Y., Zhou H.W., Rognes T., Caporaso J.G., Knight R. Open-source sequence clustering methods improve the state of the art. mSystems. 2016;1:e00003-15. doi: 10.1128/mSystems.00003-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samorodnitsky E., Jewell B.M., Hagopian R., Miya J., Wing M.R., Lyon E., Damodaran S., Bhatt D., Reeser J.W., Datta J., Roychowdhury S. Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum Mutat. 2015;36:903–914. doi: 10.1002/humu.22825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hung S.S., Meissner B., Chavez E.A., Ben-Neriah S., Ennishi D., Jones M.R., Shulha H.P., Chan F.C., Boyle M., Kridel R., Gascoyne R.D., Mungall A.J., Marra M.A., Scott D.W., Connors J.M., Steidl C. Assessment of capture and amplicon-based approaches for the development of a targeted next-generation sequencing pipeline to personalize lymphoma management. J Mol Diagn. 2018;20:203–214. doi: 10.1016/j.jmoldx.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 32.Levis M. FLT3 mutations in acute myeloid leukemia: what is the best approach in 2013? Hematology Am Soc Hematol Educ Program. 2013;2013:220–226. doi: 10.1182/asheducation-2013.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer M., Schnetzke U., Spies-Weisshart B., Walther M., Fleischmann M., Hilgendorf I., Hochhaus A., Scholl S. Impact of FLT3-ITD diversity on response to induction chemotherapy in patients with acute myeloid leukemia. Haematologica. 2017;102:e129–e131. doi: 10.3324/haematol.2016.157180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kayser S., Schlenk R.F., Londono M.C., Breitenbuecher F., Wittke K., Du J., Groner S., Späth D., Krauter J., Ganser A., Döhner H., Fischer T., Döhner K. Insertion of FLT3 internal tandem duplication in the tyrosine kinase domain-1 is associated with resistance to chemotherapy and inferior outcome. Blood. 2009;114:2386–2392. doi: 10.1182/blood-2009-03-209999. [DOI] [PubMed] [Google Scholar]
- 35.Sandhöfer N., Bauer J., Reiter K., Dufour A., Rothenberg M., Konstandin N.P., Zellmeier E., Tizazu B., Greif P.A., Metzeler K.H., Hiddemann W., Polzer H., Spiekermann K. The new and recurrent FLT3 juxtamembrane deletion mutation shows a dominant negative effect on the wild-type FLT3 receptor. Sci Rep. 2016;6:28032. doi: 10.1038/srep28032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takenokuchi M., Kawano S., Nakamachi Y., Sakota Y., Syampurnawati M., Saigo K., Tatsumi E., Kumagai S. FLT3/ITD associated with an immature immunophenotype in PML-RARα leukemia. Hematol Rep. 2012;4:e22. doi: 10.4081/hr.2012.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider F., Hoster E., Unterhalt M., Schneider S., Dufour A., Benthaus T., Mellert G., Zellmeier E., Kakadia P.M., Bohlander S.K., Feuring-Buske M., Buske C., Braess J., Heinecke A., Sauerland M.C., Berdel W.E., Büchner T., Wörmann B.J., Hiddemann W., Spiekermann K. The FLT3ITD mRNA level has a high prognostic impact in NPM1 mutated, but not in NPM1 unmutated, AML with a normal karyotype. Blood. 2012;119:4383–4386. doi: 10.1182/blood-2010-12-327072. [DOI] [PubMed] [Google Scholar]
- 38.Cucchi D.G.J., Denys B., Kaspers G.J.L., Janssen J.J.W.M., Ossenkoppele G.J., de Haas V., Zwaan C.M., van den Heuvel-Eibrink M.M., Philippé J., Csikós T., Kwidama Z., de Moerloose B., de Bont E.S.J.M., Lissenberg-Witte B.I., Zweegman S., Verwer F., Vandepoele K., Schuurhuis G.J., Sonneveld E., Cloos J. RNA-based FLT3-ITD allelic ratio is associated with outcome and ex vivo response to FLT3 inhibitors in pediatric AML. Blood. 2018;131:2485–2489. doi: 10.1182/blood-2017-12-819508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.