Abstract
Background
Radiomics analyses has been proposed to interrogate the biology of tumour as well as to predict/assess response to therapy in vivo. The objective of this work was to assess the sensitivity of radiomics features to noise, resolution, and tumour volume in the context of a co-clinical trial.
Methods
Triple negative breast cancer (TNBC) patients were recruited into an ongoing co-clinical imaging trial. Sub-typed matched TNBC patient-derived tumour xenografts (PDX) were generated to investigate optimal co-clinical MR radiomic features. The MR imaging protocol included T1-weighed and T2-weighted imaging. To test the sensitivity of radiomics to resolution, PDX were imaged at three different resolutions. Multiple sets of images with varying signal-to-noise ratio (SNR) were generated, and an image independent patch-based method was implemented to measure the noise levels. Forty-eight radiomic features were extracted from manually segmented 2D and 3D segmented tumours and normal tissues of T1- and T2- weighted co-clinical MR images.
Findings
Sixteen radiomics features were identified as volume dependent and corrected for volume-dependency following normalization. Features from grey-level run-length matrix (GLRLM), grey-level size zone matrix (GLSZM) were identified as most sensitive to noise. Radiomic features Kurtosis and Run-length variance (RLV) from GLSZM were most sensitive to changes in resolution in both T1w and T2w MRI. In general, 3D radiomic features were more robust compared to 2D (single slice) measures, although the former exhibited higher variability between subjects.
Interpretation
Tumour volume, noise characteristics, and image resolution significantly impact radiomic analysis in co-clinical studies.
Keywords: Radiomics, Breast cancer, Co-clinical, MRI, Sensitivity, TNBC, Quantitative imaging
Research in context.
Evidence before this study: Magnetic resonance imaging (MRI) radiomic analyses pipelines are widely implemented to predict and assess response to therapy as well as aid in stratification of patients. Numerous studies previously characterized the test-retest performance of radiomic features through phantom studies and clinical imaging studies.
Added value of this study: Beyond test-retest performance, a critical consideration in implementation of radiomic pipelines is site-to-site variability in image acquisition protocols such as image resolution and image characteristics such as signal-to-noise. This is especially applicable to preclinical imaging protocols where unlike clinical imaging, there are no established guidelines for best practices and there is significant variability in image acquisition parameters. In addition, a critical consideration in implementation of radiomic analysis is the sensitivity of radiomic features to target tissue volume. This study investigates the sensitivity of radiomic features to noise, resolution, and tumour volume, with emphasis on co-clinical (preclinical and clinical) harmonization of radiomic features to support translational therapeutic studies with imaging as an endpoint.
Implications of all the available evidence: Our evidence indicates that tumour volume, noise, and image resolution significantly impact radiomic features in co-clinical imaging. We propose a volume-dependency correction scheme and identify a set of robust radiomic features for co-clinical imaging studies. This work highlights the need to harmonize preclinical and clinical image acquisition protocols and radiomic analyses pipelines to enable multi-site translational studies with radiomic analysis as an endpoint.
Alt-text: Unlabelled box
1. Introduction
Co-clinical trials recently emerged as an area of investigation in which a clinical trial is coupled with a corresponding preclinical trial to inform the corresponding clinical trial [1], [2], [3], [4], [5], [6], [7], [8]. The preclinical arm of the co-clinical trial generally uses genetically engineered mouse models (GEMMs) of human cancer or patient-derived tumour xenografts (PDXs) to aid in assessing therapeutic efficacy, patient stratification, and to design optimal treatment strategies [2,9]. The emergence of GEMMs and PDXs as a co-clinical paradigm is largely motivated by the realization that established cell-lines do not recapitulate the heterogeneity of human tumours and the diversity of tumour phenotypes [10] and that better oncology models are needed to support high-impact translational cancer research. To that end, the National Cancer Institute (NCI) and numerous European entities have launched wide-ranging programs to support co-clinical trials (e.g. Co-Clinical Imaging Research Resource Program, (https://nciphub.org/groups/cirphub; EuroPDX, https://www.europdx.eu). The use of PDX, in particular, offers numerous advantages in translational imaging research; chief among them is retention of human tumour heterogeneity, which can be exploited to develop and validate image metrics of heterogeneity and response to therapy. To that end, radiomics analyses of tumours, broadly defined as the process of extracting image features from medical images, has been proposed to interrogate tumour heterogeneity in vivo [11], [12], [13].
Indeed, radiomic analyses pipelines are widely applied to predict and assess response to therapy as well as aid in stratification of patients in clinical trials [14], [15], [16], [17], [18], [19], [20]. Importantly, as preclinical imaging is a critical component in the translational drug discovery pipeline, a bi-directional translation and implementation of quantitative imaging methodologies is needed to support translational studies in assessing the efficacy of novel therapeutics and their validation in the clinical setting. However, numerous considerations are warranted in designing co-clinical imaging studies with radiomic endpoints, in particular if the objective is for preclinical imaging standards to meet or exceed clinical imaging standards. Differences in scale (human vs. mouse) yield smaller voxel sizes and reduced signal-to-noise (SNR) in preclinical imaging. Since image features are a function of voxel density, a measure of resolution-to-scale is needed to harmonize co-clinical radiomic analysis. Finally, radiomic features may be dependent on tumour volume, and ideally volume-independent features should be used in extraction of tumour features as to not bias image metrics longitudinally. These considerations have broad implications in designing co-clinical trials with radiomic endpoints. In light of these considerations, in this work we characterize the sensitivity of radiomics features to noise, image resolution, and tumour volume in context of a prospective co-clinical trial.
The sensitivity of forty-eight radiomics features, broadly categorized as first order features, second order statistics, and higher-order features, were extracted from tumour and muscle regions of pre-treatment MR images (preclinical). Measures of image noise such signal-to-noise (SNR) ratio and root mean square error (RMSE) were determined. In addition, an algorithm was implemented to calibrate noise characteristics between preclinical and clinical images. To address the impact of image resolution (vis-vis voxel size) on radiomic features, we extracted quantitative image features across three resolutions. In addition, we correlate the dependency of radiomics features on tumour volume to identify volume-independent radiomic features. The workflow is performed in the context of a triple negative breast cancer (TNBC) co-clinical trial with the primary objective of identifying robust T1w and T2w radiomic features for both preclinical and clinical imaging pipelines. We demonstrate that tumour volume, image noise, and image resolution significantly impact radiomic features, suggesting that these factors are a major consideration in designing co-clinical trials with radiomic endpoints.
2. Methods
2.1. Overview of harmonization and analytic scheme
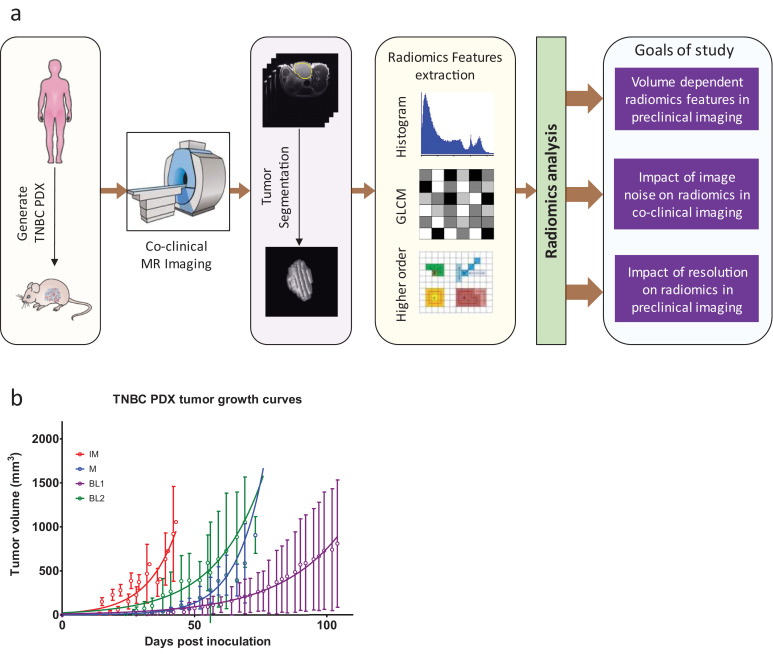
The analytic pipeline to characterize the sensitivity of image features is displayed in Fig. 1a. The co-clinical raw images were processed to annotate regions of interest (ROI), such as tumour and muscle regions slice by slice to build 3D ROIs. Histogram and texture features were extracted and analysed to investigate the effects of noise on co-clinical MR image radiomic features. The noise level was estimated from low noise (SNR>25) “baseline” images. The sensitivity to image resolution was investigated through multiple acquisitions of the same PDX during the same imaging session. Using the same data, the dependency of radiomic features on tumour volume was established.
Fig. 1.
(a) Overview of analytic methods. Patient-derived tumour xenografts (PDX) were generated from human tumour biopsies from TNBC patients. Human imaging was performed on the Siemens mMR PET/MR scanner. PDX were imaged on Agilent 4.7T or MR Solutions 7T MR scanner. Radiomics analysis, including histogram (1st order), grey level co-occurrence matrix (GLCM) or second order and higher order features, was performed on 2D and 3D segmented tumours to assess volume dependency, sensitivity to noise, and sensitivity of resolution. (b). Growth profile of TNBC PDX used in this study based on calliper measurements.
2.2. TNBC PDX model and growth characterization
Gene expression analyses of 93 TNBC patients samples (29657 unique genes/probes) was performed to identify six TNBC subtypes including 2 basal-like (BL1 and BL2), an immunomodulatory (IM), a mesenchymal (M), a mesenchymal stem–like (MSL), and a luminal androgen receptor (LAR) subtype as described previously [21,22]. IM, M, BL1, and BL2 TNBC PDX sub-types served as a preclinical platform to assess the sensitivity of radiomics features. Tumours were implanted in the 4th mammary fat pad of NSG mice. Mice were examined regularly for palpable tumours. Tumour volumes were calculated from calliper measurement using the formula 1/6*π*L*W2 with the goal of recruiting PDX with tumour volumes of ~250mm3. The variability in timing and rate of tumour growth both between and within PDX proved somewhat challenging in maintaining consistent recruitment schedule, and resulted in wide range of tumour volumes (See Fig. 1b). Details regarding animals, surgeries, and tumour xenografts were reported previously [22,23]. All animal experiments were conducted in compliance with the Guidelines for the Care and Use of Research Animals established by Washington University's Animal Studies Committee.
2.3. MR image acquisition
Clinical MR Imaging. The study protocol was approved by the institutional review board and written informed consents were obtained from all prospective patients. Twenty-five stage II or III TNBC patients were recruited into an ongoing co-clinical imaging trial (ClinicalTrial.gov ID # NCT02124902). Clinical MR imaging protocol was implemented on the Siemens mMR Simultaneous PET/MR. The MR protocol included: T1W (2D TSE multi slice, turbo factor 3, echo time (TE) 11 ms, repetition time (TR) FOV 320 × 320, spatial resolution 1.0mm × 1.0mm X 3.0mm), and T2W (2D TSE multi slice with an inversion pulse, TI=170 ms, turbo factor 11, TE = 58 ms, TR= 7040 ms, FOV 448 X448, spatial resolution 0.759mm X 0.759mm X 3.0mm).
Preclinical MR Imaging. TNBC PDX displayed highly variable growth patterns both within and between PDX. Calliper-based measurements of tumour volumes, which are known to overestimate tumour volumes, are depicted in Fig. 1b. Corresponding MR-based tumour volumes were in the range of 100 – 650 mm3. MR imaging was performed on the MR Solutions simultaneous 7T MR/PET scanner (MR Solutions, Guildford, UK). The multi-parametric (MP) MR protocol included: T1W (2D FSE multi slice, echo train length 4, echo spacing 11 ms, effective TE =11 ms, respiratory gated with effective TR = 1s, respiration rate kept around 60 breaths/min, spatial resolution 0.25mmX0.25mmX1.0mm), and T2W (2D FSE multi slice, echo train length 7, echo spacing 15 ms, effective TE = 45 ms, respiratory gated with TR = 5 s, respiration kept around 60 breaths/min. PDX mice were anesthetized with 1-2% isoflurane throughout imaging sessions. Given the lag (upwards of 6 months) in developing PDX, three cohorts of PDX were used for different experiments:
-
1)
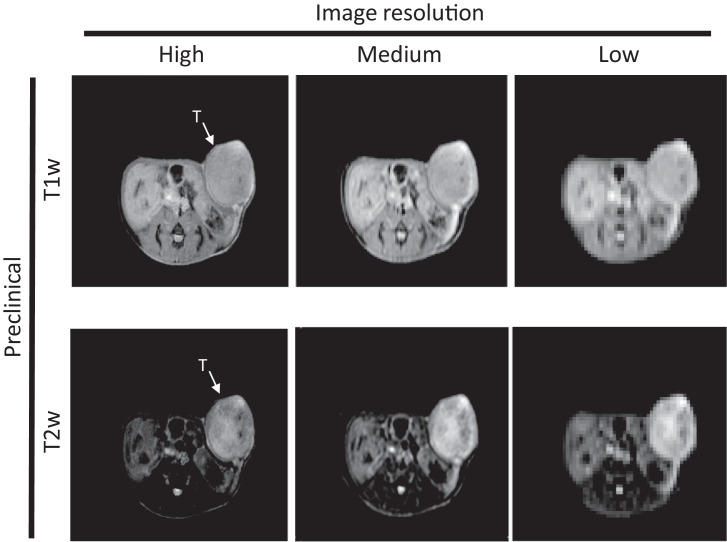
Preclinical imaging to assess sensitivity to resolution. Six PDX were imaged at three different in plane resolutions 0.125mm × 0.125mm × 1mm, 0.25mm × 0.25mm × 1mm, and 0.5mm × 0.5mm × 1mm, referred to henceforth as high, medium, and low resolution, respectively, while keeping same FOV (Fig. 2).
-
2)
Preclinical imaging to assess sensitivity to noise: Additional PDX mice were used to investigate the sensitivity to noise—5 at high resolution and 19 at medium resolution per above resolution.
-
3)
Preclinical imaging to assess dependency of radiomic features on tumour volume. The dependency of radiomic features on tumour volume was characterized using 31 image datasets from 12 PDX at different time points volumes for a total of 31 datasets. Theses scans were acquired on an Agilent 4.7T scanner with in-plane spatial resolution of 0.195 × 0.195 × 1 mm3, as previously described [24].
Fig. 2.
Single slice for each of three different resolutions. PDX were imaged at three different in plane resolutions 0.125mm×0.125mm×1mm, 0.25mm×0.25mm×1mm, and 0.5mm×0.5mm×1mm, referred to henceforth as high, medium, and low resolution, receptively, while keeping a constant FOV. High resolution left; medium resolution, middle; high resolution, right. Top row depicts T1w images, while bottom row depicts T2w MR images.
Table 1A tabulates descriptive data for clinical and preclinical studies to asses sensitivity to noise and resolution, whereas Fig. 1b tabulates the PDX descriptive data to assess dependency of radiomic features on tumour volume.
Table 1A.
Co-clinical T1w and T2w image descriptors for sensitivity to noise and resolution.
| MR Seq. | Metric | SNR (in dB) | Estimated noise level | 3D tumour |
2D slice |
|||
|---|---|---|---|---|---|---|---|---|
| Tumour volume (mm3) | Number of voxels | Largest cross section (mm3) | Number of voxels in largest cross section area | |||||
| Clinical* | T1w (N=25) | Range | 18.9– 45.4 | 0.20 – 0.68 | 1,182 – 43,309 | 268 – 2,837 | 366 – 4,600 | 93 – 1,394 |
| Avg±SD | 30.6 ± 07.6 | 0.37 ± 0.13 | 6,773 ± 8155 | 1,259 ± 760 | 1,229 ± 834 | 305 ± 254 | ||
| Median | 29.6 | 0.37 | 4725 | 984 | 1129 | 244 | ||
| 95% CL | 03.2 | 0.05 | 3366 | 321 | 344 | 105 | ||
| T2w (N=25) | Range | 21.3– 74.0 | 0.25 – 0.93 | 1,873 – 4,4691 | 832 – 23,513 | 518 – 4,567 | 230 – 2,403 | |
| Avg±SD | 35.7 ± 11.7 | 0.57 ± 0.13 | 7,603 ± 8360 | 3,592 ± 4,424 | 1,387 ± 820 | 643 ± 435 | ||
| Median | 33.1 | 0.57 | 5305 | 2244 | 1226 | 527 | ||
| 95% CL | 4.9 | 0.05 | 3451 | 1826 | 339 | 179 | ||
| Preclinical⁎⁎ | ||||||||
| High Res | T1w (N=11) | Range | 17.0– 64.4 | 0.69 – 2.56 | 174 – 467 | 11,362 – 29,896 | 33 – 83 | 2,157 – 5,287 |
| Avg±SD | 33.1 ±12.7 | 1.38 ± 0.57 | 317 ± 109 | 20,525 ± 7142 | 49 ± 15 | 3,184 ± 970 | ||
| Median | 33.1 | 1.18 | 350 | 22885 | 42.4 | 2771 | ||
| 95% CL | 8.5 | 0.38 | 73 | 4798 | 10 | 652 | ||
| T2w (N=11) | Range | 19.4– 65.4 | 0.98 – 2.46 | 174 – 456 | 11,362 – 29,201 | 33 – 80 | 2,157 – 5,119 | |
| Avg±SD | 33.8 ± 13.3 | 1.5 ± 0.43 | 309 ± 114 | 20,037 ± 7,483 | 48 ± 15 | 3,138 ± 978 | ||
| Median | 33.0 | 1.35 | 350 | 22885 | 42 | 2771 | ||
| 95% CL | 8.1 | 0.29 | 77 | 5027 | 10 | 657 | ||
| Med Res | T1w (N=25) | Range | 17.3– 63.7 | 0.45 – 1.95 | 75 – 438 | 1,447 – 7,175 | 20 – 61 | 372 – 992 |
| Avg±SD | 33.5± 11.6 | 0.96 ± 0.33 | 245 ± 123 | 4,157 ± 1901 | 38 ± 13 | 651 ± 189 | ||
| Median | 30.9 | 0.96 | 198 | 3357 | 35 | 621 | ||
| 95% CL | 4.8 | 0.13 | 51 | 785 | 5 | 78 | ||
| T2w (N=25) | Range | 18.0– 52.2 | 0.51 – 1.59 | 64 – 436 | 1,234 – 7,141 | 17 – 61 | 320 – 992 | |
| Avg±SD | 32.78± 7.8 | 0.97 ± 0.22 | 241 ± 125 | 4,091 ± 1,931 | 37 ± 13 | 639 ± 189 | ||
| Median | 30.9 | 0.94 | 190 | 3518 | 34 | 591 | ||
| 95% CL | 3.3 | 0.09 | 51 | 797 | 5 | 78 | ||
| Low Res | T1w (N=6) | Range | 20.7– 65.5 | 0.47 – 1.69 | 172 – 442 | 706 – 1,809 | 37 – 72 | 150 – 294 |
| Avg±SD | 36.6± 16.7 | 0.79 ± 0.46 | 357 ± 96 | 1,464 ± 3393 | 55 ± 13 | 225 ± 54 | ||
| Median | 31.7 | 0.66 | 386 | 1583 | 59 | 243 | ||
| 95% CL | 17.6 | 0.48 | 101 | 412 | 14 | 57 | ||
| T2w (N=6) | Range | 19.6–63.9 | 0.36 – 1.21 | 172 – 442 | 706 – 1,809 | 37 – 72 | 150 – 294 | |
| Avg±SD | 35.2± 15.5 | 0.73 ± 0.29 | 358 ± 97 | 1,467 ± 395 | 55 ± 13 | 226 ± 54 | ||
| Median | 32.2 | 0.70 | 386 | 1583 | 60 | 246 | ||
| 95% CL | 16.3 | 0.31 | 101 | 415 | 14 | 57 | ||
Clinical image voxel size (x,y,z): T1 (1.0mm, 1.0mm, 3mm); T2w (0.76mm, 0.76mm, 3mm).
Preclinical image voxel size (x,y,z): High Res T1w and T2w (0.125mm, 0.125mm, 1mm); Med Res T1w and T2w (0.25mm, 0.25mm, 1mm); Low Res T1w and T2w (0.5mm, 0.5mm, 1mm).
2.4. Image segmentation
Digital Imaging and Communications in Medicine (DICOM) and Flexible Data Format (FDF) MRI data were transferred into MATLAB (2019a) prior to texture analysis and successive analysys were executed in MATLAB. T1w and T2w MR images were analysed separately with ROIs drawn manually on every slice. Tumour volumes were calculated by determining the number of voxels in the volume of interest multiplied by the voxel dimension (mm3) for each MR sequence. In segmenting tumours at the three resolutions, ROIs were defined on high-resolution images and were applied to lower resolution images as to not bias results potentially due to ROI variability.
2.5. Extraction of radiomic features
Radiomic features were extracted per ISBI guidelines [25] using formulations from previously published work [26,27]. Forty-eight radiomics features were extracted from 2D (single slice with largest cross section) and 3D (whole tumour) segmented tumours and muscle as normal tissue ROIs. Single slice was chosen based on the largest cross section area of the tumour. MR image data were transferred into image matrix in MATLAB and features were extracted from segmented 3D/2D matrix. MATLAB R2019a was used for the radiomic feature extraction and analysis. Equal-probability quantization algorithms to quantize raw data into grey level (Ng) was implemented using histeq MATLAB functions. MR data normalization suggested by Collewet [28] was implemented. Resampling to isotropic voxel size was applied to reduce the effect of resolutions for GLCM, GLRLM, GLSZM and NGTDM matrix generation. Supplement Table S1 summarizes the features used within. The features are broadly categorized as: first order features extracted from intensity histogram, second order statistics, and higher-order features. First order global features were extracted directly from intensity histogram of raw data. Mean, variance, standard deviation, skewness, and kurtosis were extracted from first order statistics. Sixty-four fixed bin number grey level quantization was used for histogram analysis of variance, skewness, and kurtosis of first order global features [26]. Co-occurrence matrix (GLCM) features Energy, Contrast, Correlation, Homogeneity, Variance, Sum Average, and Entropy were computed with fixed bin number grey level quantization number 64. Lastly, thirty-one higher-order textures [26,[29], [30], [31]] were extracted from Grey-level run-length matrix (GLRLM), Grey-level size zone matrix (GLSZM) and Neighborhood grey-tone difference matrix (NGTDM) with fixed number grey level quantization number 64. GLCM, GLRLM, GLSZM and NGTDM were generated from 3D ROI with 26 voxels connectivity (8 connectivity in 2D).
2.6. Evaluation of volume-independent radiomic features
Radiomic features were extracted from 31 T2-weighted preclinical MR images. Voxel intensities within a region of interest (ROI) were resampled into a limited number of discrete values or bin sizes before calculating feature values. Radiomic features were extracted as described above. The magnitude of radiomic features was regressed against their corresponding tumour volume using linear and non-linear models: linear, power, log, exponential or 2nd order polynomial (see Statistical method below). In an attempt to correct for volume dependencies, the volume-dependent features were normalized by volume for subsequent analysis. Features that had a significant linear correlation with volume, were normalized by dividing by tumour volume; otherwise, if inversely proportional to tumour volume, features were normalized by multiplying by tumour volume. Nonlinear correlated radiomic features were normalized by as per the fitted model.
2.7. Evaluation of noise effects on radiomics features
Addition of External Noise. To examine the sensitivity of the selected radiomics features to SNR, the baseline images were normalized to 0-255 and then degraded by the addition of six different levels of Gaussian noise. We considered Gaussian noise in this study as Rician distributions are nearly Gaussian for SNR>2 as described previously [32]. Gaussian noise with zero mean and standard deviation (σ) proportional to σ=α×255 where α denotes the noise magnitude constant (i.e., G (0, α × 255)) was added to images. Six different levels of α (0.001, 0.005, 0.010, 0.015, 0.025, and 0.05) were added resulting in SNR range of 45 dB to 6 dB corresponding with σi=(0.255, 1.27, 2.55, 3.825, 6.735, 12.75).
Noise level estimation (Ψ). A patch based method from single noisy 2D image was used to estimate noise levels [33]. The images first normalized to 0-255 intensity scale to remove intensity dependencies. The noise level estimated (Ψ) of 3D image was calculated by taking average of noise from all multi-slice images. The minimum variance was calculated using principal component analysis (PCA) from the covariance matrix. Images were decomposed into patch sizes of 3×3, 5×5, and 7×7 depending on the test image dimension. The minimum eigenvalue of the covariance (<0.5) matrix was considered as weak texture patches. Therefore, the noise was easily measured from the selection of weakly textured patches. The richer textures were larger maximum eigenvalues. Thus, the quantitative level of the texture strength of the image patches were measures from eigenvalue of the gradient covariance matrix. Then weak patches (Wp) were selected from the gradient covariance matrix with the maximum eigenvalue less than some threshold (Th). The threshold was calculated from the inverse gamma cumulative distribution function where the significance level can also be changed manually. In practice, low significance levels indicate enhanced sensitivity to noise. An iterative framework was used to ensure the convergence of noise level σn. Manual iteration reduces the computational time compared to automatic iteration until convergence.
The signal to noise ratio (SNR) and root mean square error (RMSE) [34] were also implemented to quantify noise level of original and noisy image. SNR is represented by the ratio of the average signal intensity of tumour region over the standard deviation of the noise, given in decibel (dB). The RMSE between two images I(x,y) and I*(x,y) is given by:
where, I(x,y) is image before noise added and I*(x,y) is the image after noise added externally.
2.8. Statistical analysis
Volume-dependent radiomic features: The Pearson correlation coefficient was determined to assess the correlation between radiomic feature and the tumour volume. Correlations with P-value < 0.05 were considered significant (Table 2). We did not perform multiple hypothesis testing as to not undercount the number of potential significant correlations. Linear or nonlinear least squares regression was used to fit all significant volume-dependent radiomic features. The Akaike Information Criterion (AIC) and Bayesian Information Criterion (BIC) were calculated for each model, and the appropriate model was selected based on the minimum value of AIC and BIC. STATA 12 (StataCorp, College Station, TX) was used for regression and statistics.
Table 2.
Volume-dependent radiomic texture features.
| Features name | P-Value | Stats | Linear | Exponential | Log | Power | Polynomial |
|---|---|---|---|---|---|---|---|
| GLCM.Contrast | 0.71×10-3 | AIC | 261.3 | 261.1 | 259.6 | 260.0 | 262.2 |
| BIC | 276.1 | 275.9 | 274.4 | 274.8 | 281.9 | ||
| GLCM.Correlation | 0.37×10-3 | AIC | -144.3 | -141.9 | -147.3 | -146.5 | -144.6 |
| BIC | -129.5 | -127.1 | -132.5 | -131.7 | -124.8 | ||
| GLRLM.SRE | 0.31×10-9 | AIC | -19.6 | -6.2 | -9.9 | -19.0 | -18.1 |
| BIC | -4.8 | 8.6 | 4.9 | -4.2 | 1.6 | ||
| GLRLM.RLN | 0.73×10-6 | AIC | -297.6 | -297.2 | -297.4 | -298.7 | -287.3 |
| BIC | -282.8 | -282.4 | -282.6 | -282.9 | -267.6 | ||
| GLRLM.RP | 0.32×10-6 | AIC | -337.5 | -338.4 | -334.3 | -340.0 | -338.2 |
| BIC | -322.7 | -323.6 | -319.5 | -325.2 | -318.4 | ||
| GLSZM.SZE | 0.34×10-3 | AIC | -251.4 | -257.6 | -256.8 | -257.1 | -250.4 |
| BIC | -236.6 | -242.8 | -242.0 | -242.3 | -230.7 | ||
| GLSZM.LZE | 0.22×10-13 | AIC | -48.3 | -2.2 | -38.9 | -47.3 | -47.1 |
| BIC | -33.5 | 12.6 | -24.1 | -32.5 | -27.4 | ||
| GLSZM.ZSN | 0.32×10-3 | AIC | -206.2 | -208.8 | -215.6 | -216.4 | -211.3 |
| BIC | -191.4 | -194.0 | -200.8 | -201.6 | -191.6 | ||
| GLSZM.ZP | 0.74×10-6 | AIC | -202.6 | -203.0 | -204.1 | -203.7 | -205.9 |
| BIC | -187.8 | -188.2 | -189.3 | -188.9 | -192.2 | ||
| GLSZM.LZLGE | 0.12×10-3 | AIC | -85.1 | -84.8 | -80.3 | -79.9 | -84.7 |
| BIC | -71.0 | -70.3 | -65.5 | -65.1 | -65.0 | ||
| GLSZM.LZHGE | 0.68×10-9 | AIC | 482.9 | 493.7 | 492.7 | 483.8 | 484.1 |
| BIC | 497.7 | 508.5 | 507.5 | 498.6 | 503.8 | ||
| GLSZM.GLV | 0.14×10-5 | AIC | -381.5 | -387.7 | -375.8 | -389.7 | -383.1 |
| BIC | -366.7 | -372.9 | -361.0 | -376.0 | -363.4 | ||
| GLSZM.ZSV | 0.81×10-7 | AIC | -773.9 | -834.2 | -809.9 | -837.2 | -791.3 |
| BIC | -759.1 | -819.4 | -795.1 | -822.4 | -771.6 | ||
| NGTDM.Coarseness | 0.99×10-10 | AIC | -490.9 | -500.6 | -380.5 | -533.8 | -495.2 |
| BIC | -476.1 | -485.8 | -365.7 | -519.0 | -475.5 | ||
| NGTDM.Busyness | 0.28×10-10 | AIC | -126.3 | -115.5 | -107.4 | -120.2 | -124.1 |
| BIC | -121.5 | -120.7 | -92.6 | -105.4 | -114.4 | ||
| NGTDM.Strength | 0.52×10-8 | AIC | -31.4 | -44.7 | -49.8 | -50.0 | -46.8 |
| BIC | -16.6 | -29.9 | -35.0 | -35.2 | -27.1 |
*Minimum values of AIC and BIC were highlighted with bold
Sensitivity of resolution on radiomic features: Radiomic features were extracted from low, medium, and high-resolution images (as described above). The ratio of the radiomic feature extracted from low (L:H) and medium (M:H) resolution images was normalized to those from the corresponding high resolution image for each PDX tumour. A ratio of unity denotes agreement between the different resolutions tested. Subsequently, the averages, μL: H and μM: H and corresponding SDs and coefficient of variation (CV = SD/µ) were determined for the set of L:H and M:H ratios for each feature. The magnitude of the sensitivity is defined by the bias from unity, i.e., bL:H=1- μL: H and bM:H=1- μM: H. Tables S2 and S3 tabulates the statistics for the ratios for each radiomic feature.
Cross-correlation between radiomic features and identification of independent signatures: The cross-correlation between the 48 features was calculated using Pearson correlation. A P-value<0.05 was considered significant (Supplemental Table S4). Additionally, principle component analyses (PCA) was performed on the 48 features on the volume data.
Relative percent change (RPC) and average percentage changes (APC) noise sensitivity: Relative change in image feature was calculated from the ratio between the change in image feature (before addition of noise and following addition of noise) relative to baseline (before addition of noise). Relative percentage change (RPC) is the relative change calculated as a percentage. The average percent change (APC) was calculated by taking average of RPCs.
2.9. Role of the funding source
Funders had no role in the study design, data collection, data analysis, interpretation, and writing of the manuscript.
3. Results
Table 1A tabulates the range of estimated noise (Ψ), tumour volumes, and corresponding number of voxels (2D and 3D) in the co-clinical study. Since image features are dependent on the information density embedded in the image (and hence, the number of voxels defining an image) a consideration of resolution-to-scale is needed when harmonizing preclinical and clinical image acquisition pipelines. For the range of tumour volumes in the clinical study, the corresponding number of voxels defining a T1w and T2w imaged was on average 1,259±744 and 3,592±4,335, respectively. Of the three preclinical image resolutions tested, the medium resolution image acquisition protocol yielded comparable statistics to the clinical images in terms of estimated noise and number of voxels defining a single tumour slice (2D slice) or whole segmented tumour (3D). Table 1B summarizes descriptive data for correlation of volume to radiomic features.
Table 1B.
Co-clinical image descriptive data for volume dependent radiomic features.
| 3D Tumour Volume (mm3) | 3D tumour number of voxels | |
|---|---|---|
| Range (N=31) | 63.82-418.70 | 1673-10,976 |
| Avg ± SD | 177.1±83.2 | 4,641.6± 2,180.3 |
| Median | 172.8 | 4,530.5 |
| 95% CL | 31.1 | 814.2 |
*Image voxel size (x,y,z) :0.195 × 0.195 × 1 mm3.
3.1. Volume-dependent radiomics features
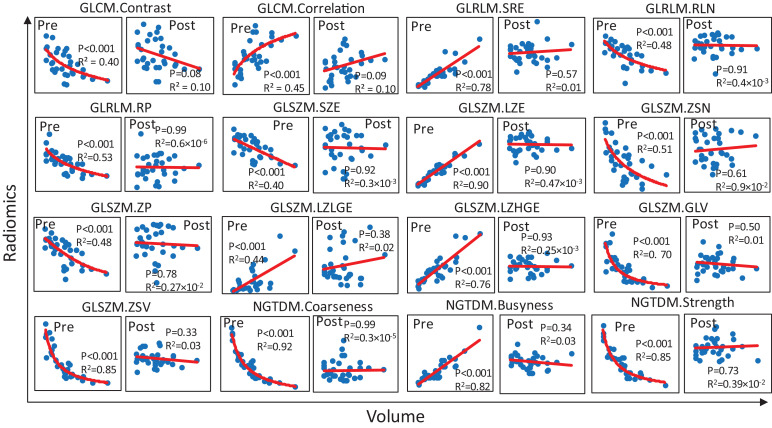
In all, 16 T2w 3D radiomic features were found to be volume dependent as tabulated in Table 2 along with P-values indicating the significance of the correlation to volume (assuming a linear model) and AIC and BIC scores for the considered models. Tumour volume has greater influence on higher order features extracted from GLSZM (8 of 16 features). In addition, tumour volume influenced select features derived from GLCM, GLRLM and NGDTM. The voxels adjacent to a given position with same grey value may have an indirect influence on volume-dependent features. To remove the dependency on volume, we attempted to normalize the volume-dependent features as described in Methods section. Following correction, volume-dependent features were no longer volume-dependent. Pre- and post- volume dependency correction regression plots is depicted in Fig. 3.
Fig. 3.
Volume-dependent radiomic features. Regression plots of 48 T2w 3D radiomic features as function of volume for sixteen volume-dependent features that were converted to volume independent. The radiomic feature is depicted before and after normalization marked with Pre and Post in subplots. The corresponding P-value and R2 values are displayed in the corresponding subplots. P-values indicate significance of linear correlation.
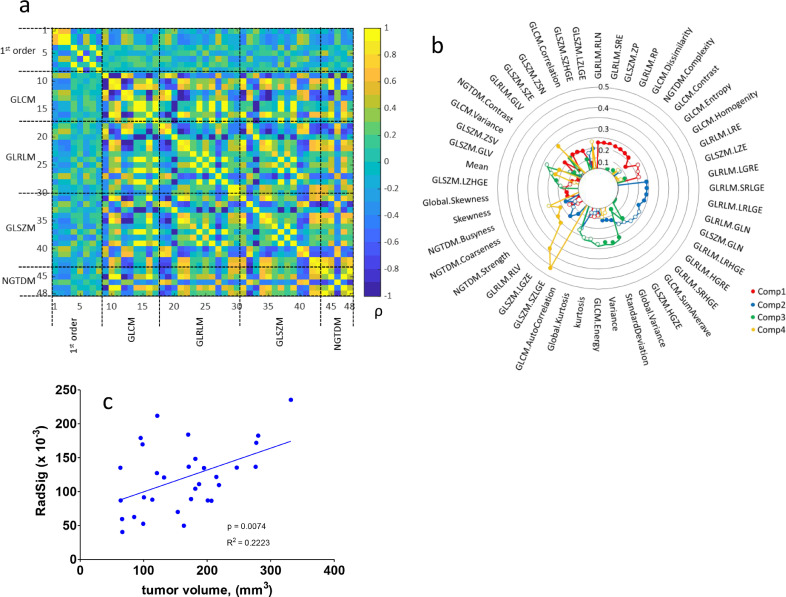
3.2. Cross-correlation of radiomic features and dimension reduction of radiomic signature
We additionally evaluated the cross-correlation of the radiomic features with the goal of identifying a panel of radiomic signatures which may be independent of tumour volume. The cross-correlation of radiomic features is depicted in heatmap of Fig. 4a. The number of within-group correlations is high in both GLCM (50%) and NGTDM (40%) groups, while the 1st order features (17.9%), the GLRLM (17.9%) and SLSZM (20%) groups have fewer within group correlations. In general, we observe that 1st order features are mildly correlated to higher order features and higher order features are highly correlated to each other. The application of PCA to derive a panel of independent radiomic signatures is depicted in the radar plot of Fig. 4b. The radar plot depicts the loading of each feature from top four principal components. To further delineate volume dependency, we tested the correlation between PCA-derived radiomic features (with loading greater to 0.1) to volume (Fig. 4c). The correlation remained significant suggesting that dimension reduction does not correct volume dependency.
Fig. 4.
Identification of independent radiomic signatures. (a) Pearson (ρ) cross-correlation between 48 T2w 3D radiomic feature. (b) Radar plot following PCA to identify independent radiomic signatures (RadSig). (c) Correlation between independent PCA-derived radiomic signature to volume. P-value indicate significance of linear regression.
3.3. Sensitivity of radiomic features to noise
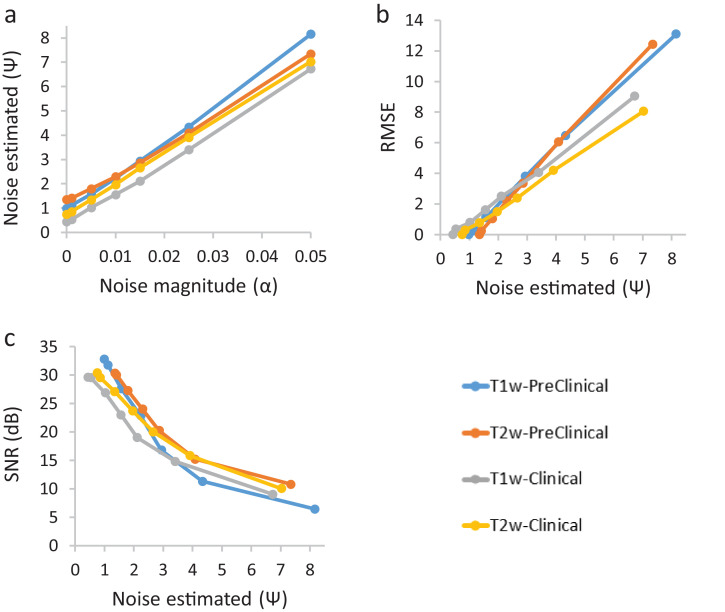
Representative co-clinical T1w and T2w MR images are displayed in Fig. 5 after adding varying magnitudes of Gaussian noise. The performance of the noise estimation algorithm was compared to RMSE and SNR. The correlation between externally added noise vs. noise measured, and the corresponding SNR and RMSE with noise measured is depicted in Fig. 6. Fig. 6a suggests a linear correlation (R2≥0.99) between added noise and estimated noise. In the absence of added noise, there are differences in the levels of noise measured between preclinical and clinical images. Fig. 6b and Fig. 6c depict the correlation between estimated noise and RMSE and SNR, respectively. With no noise added, estimated noise is greater than zero (and RMSE is zero) due to inherent noise in the baseline (prior to the addition of noise) image. RMSE increases linearly (R2≥0.99) with added Gaussian noise. There is a divergence in preclinical and clinical correlation between RMSE and noise measured starting at Ψ =3. The linear relationship (R2≥0.99) between noise estimation and RMSE (Fig. 6b) and exponential (R2≥0.95) decrease of SNR with the increase of noise (Fig. 6c) highlights the robustness of the texture-based noise estimation method. This texture-based method can be used to estimate noise on an image-by-image basis and thus define customized retrospective assessment of the diagnostic study in co-clinical study design.
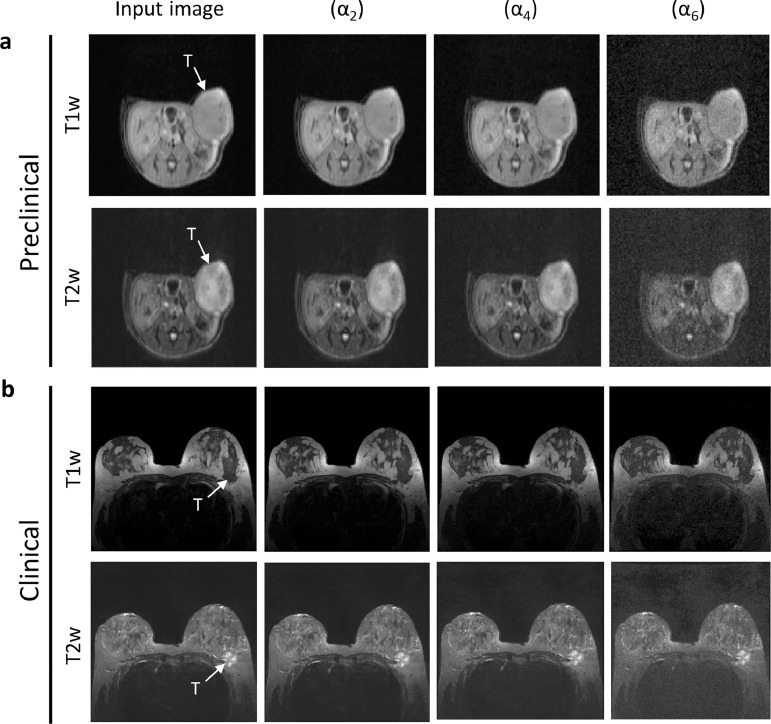
Fig. 5.
The output after added varying magnitude of (α2, α4, and α6) Gaussian noise on input co-clinical T1w and T2w breast MR image is shown in (a) and (b). SNR corresponding to α were (α1 ≈ 33dB, α2≈32dB, α3≈28dB, α4≈23dB, α5≈17dB, and α6≈11dB).
Fig. 6.
Plot of noise added of magnitude (α) into 3D MR vs noise estimated from 3D MR co-clinical image. (a) Noise added of magnitude (α) vs noise estimated using texture-based method; (b) noise estimated vs RMSE measured after added noise; (c) noise estimated vs SNR measured after added noise.
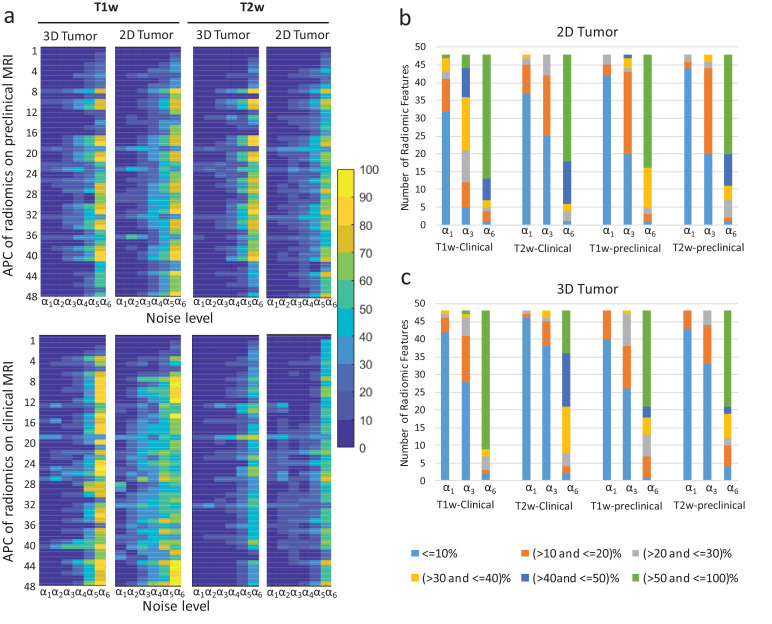
Fig. 7a displays sensitivity of radiomics features to noise (via APCs) at varying noise magnitudes. Four major cases are depicted: features extracted from single 2D slice and whole-tumour (3D) tumour of T1w and Tw2 images with addition of Gaussian noise. Features varying less than 10% (approx. SNR of 26 in T1w and 28 in T2w images) were considered as less sensitive to noise in co-clinical images. Of the 48 features examined, 38 features exhibited APC < 10% for preclinical T1w; and 42 with APC < 10% for preclinical T2w up to estimated noise Ψ ≈ 1.5 (corresponding to SNR ≈ 28); 37 features exhibited variability APC<10% for clinical T1w with estimated noise Ψ ≈ 0.9 (corresponding to SNR ≈ 30); and 42 features were variability < 10% for clinical T2w on estimated noise Ψ ≈ 1.5. Mean feature exhibited <10% variability across all realistic noise levels for T1w and T2w. In all, thirty-one features in preclinical and clinical image features exhibited APC<10% for each T1w and T2w. Fig.s 7b (2D tumour) and 7c (3D tumour) highlight the proportion of radiomic features at a given bias for a given noise level. Overall, T2w radiomic features are more robust to noise than T1w, with 3D analyses being more robust than 2D analyses.
Fig. 7.
Sensitivity of radiomic features to noise. (a) Average percent change (APC) of radiomic features as a function of noise. The X- axis corresponds to added noise level [α1 ≈ 33(SNR); α2 ≈32(SNR); α3 ≈28(SNR); α4 ≈23(SNR); α5 ≈17(SNR); α6 ≈11(SNR)], and Y-axis corresponds to the radiomic features (1- 48)(see Supplemental table S#) . (b) and (c) depict the proportion of radiomic features at different bias with increasing noise for 2D and 3D analyses, respectively. The bias is denoted by the colour-scale legend below panel C.
3.4. Sensitivity of radiomic features to resolution
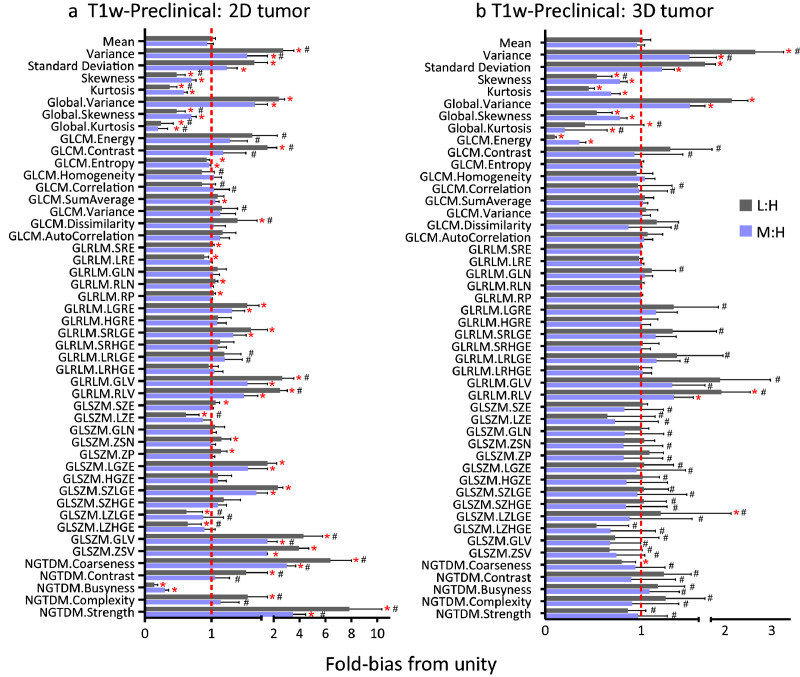
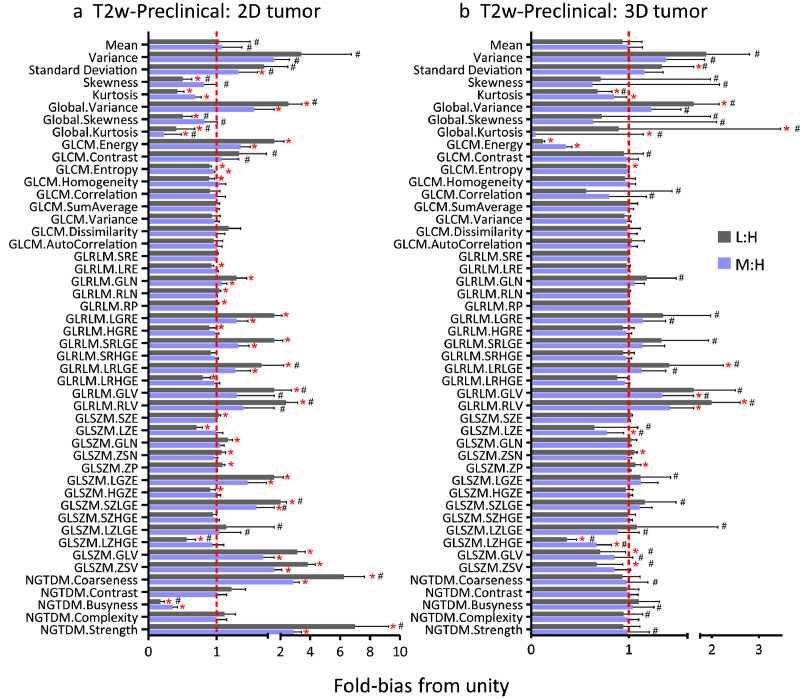
Fig. 8, Fig. 9 depict the sensitivity of T1w and T2w radiomic features to resolution, respectively. The ratio of low and medium to high (L:M and M:H, respectively) image features was used to assess the sensitivity of radiomic features to resolution. The mean and standard deviations of ratios are calculated for each feature while the coefficient of variation (CV) is provided as supplement table (Supplemental Table S2 and Table S3). To ascertain the sensitivity of the features, we determined the fold-bias of the ratio from unity. Generally, the number of significant features different from unity was higher in L:H compared to M:H. In addition, 2D radiomic features exhibited higher prevalence of bias compared to 3D image features. Many first order and GLSZM features were more sensitive to resolution than other higher order features, and generally, single slice (2D tumour) radiomics features were more sensitive to resolution than 3D based image features. The variability of the average fold-bias, as provided by the CV, is also a critical consideration for each feature. While several of the features did not exhibit significant bias (i.e., were not found to be significantly different from unity), they exhibited high CV as depicted in Figs. 8,9 and summarized in Supplemental Table S2 and Table S3. Thus, there is an added constraint in minimizing both the bias and the CV. Given the impact of reduced resolution on feature metrics, we tested whether isotropic up-sampling of resolution from low and medium to high resolution would alleviate the bias in resolution. As summarized in Supplemental Table S5, there is significant bias in image features following up-sampling, suggesting that up-sampling cannot correct for resolution bias in image features.
Fig. 8.
Sensitivity of T1w radiomics features to resolution. Ratio of low-to-high (L:H) and medium-to-high (M:H) radiomic features using either (a) a single slice (2D) or (b) the whole tumour (3D) for the analysis. Line at unity indicates perfect agreement between lower resolution and high-resolution images. *denotes radiomic features whose mean ratio is significantly different from unity using a Student's t-test; #denotes radiomic features with CV > 20%.
Fig. 9.
Sensitivity of T2w radiomics features to resolution. Ratio of low-to-high (L:H) and medium-to-high (M:H) radiomic features using either (a) a single slice (2D) or (b) the whole (3D) tumour for the analysis. Line at unity indicates perfect agreement between lower resolution and high-resolution images. *denotes radiomic features whose mean ratio is significantly different from unity using a Student's t-test; #denotes radiomic features with CV > 20%.
4. Discussion
Radiomic analysis has been proposed as a step towards realization of precision medicine by providing means to interrogate the spatial complexity of tumours in vivo [11,35]. A recent publication by the Imaging Biomarker Standardization Initiative (ISBI) identified 174 radiomic features to that end [25]. The radiomic features presented within are only a subset of features available to define phenotypes of an image as not all features are applicable for co-clinical imaging trials. Numerous studies previously characterized the robustness and test-retest performance of radiomic features [36,37] through phantom studies and clinical imaging studies. Beyond robustness of features, a consideration in implementation of radiomic pipelines is site-to-site variability in image acquisition parameters such as image resolution and image characteristics such as signal-to-noise. In addition, a critical consideration in implementation of radiomic analysis in oncologic imaging is the sensitivity of radiomic features to tumour volume. This is particularly relevant in longitudinal studies during which tumour volumes will change with the course of the disease or following therapy.
Our results indicate that numerous radiomic features are dependent on tumour volume suggesting that a normalization scheme is needed to account for the influence of tumour volume on radiomic features. Of the radiomic features tested, 16 were found to be volume dependent. We subsequently attempted to correct for volume dependencies using both linear and non-linear models and were successful in removing all volume dependencies based on statistical consideration, although for select features correlations remain. We also performed PCA in an effort to identify a radiomic signature that is volume independent. However, the panel of radiomic features following dimension reduction still correlated to volume, suggesting that dependency on volume is a major consideration in designing imaging studies with radiomic analysis as an endpoint. Importantly, the utility of the volume-dependency correction schemes will need to be tested in the context of therapeutic studies. Of consideration, in MR imaging there are trade-offs in acquisition time, voxel size (resolution), and signal-to-noise (SNR). Cattell et al. and Baessler et al. performed phantom studies to characterize the robustness of select radiomic features [36,37]. Recently, Shafiq-Ul-Hassan et al [38] characterized the dependencies of clinical computed-tomography (CT) radiomic features on voxel size. Sensitivity of radiomic features due to motion and noise in CT were also investigated using both simulated and clinical data [39]. Bagher-Ebadian et al., characterized the impact of smoothing and noise on robustness of CT radiomic features in context of head and neck cancer [40]. The sensitivity of image reconstruction in positron emission tomography (PET) has also been extensively investigated using both physical phantoms and patient data [41]. To our knowledge, however, no study has investigated the sensitivity of radiomic pipelines in the context of co-clinical MR imaging trials (preclinical and clinical).
As it relates to sensitivity of features to noise, our data suggests that performance of radiomic features can be improved significantly if SNR > 28 in particular for rich texture images. In an effort to harmonize preclinical and clinical noise levels or SNR, we developed an unbiased algorithm to harmonize noise levels and identified 31 common co-clinical radiomic features exhibiting less than 10% variability in the presence of noise ranging from SNR 28 to 33. Overall, features extracted directly from histogram were less sensitive to noise compared to second order and higher order features except skewness and kurtosis. In addition, image features derived from T1w images tended to be more susceptible to noise than T2w images. In general, features extracted from 3D exhibited less sensitivity than features extracted from 2D in both clinical and preclinical images. The implications of the latter are significant since some radiomic analysis pipelines use 2D images to characterize the heterogeneity of images, and interslice differences may produce bias in radiomic analysis. Importantly, if using 2D slices for radiomic analysis, interslice variability may further bias radiomic analysis in particular in longitudinal studies. Finally, the sensitivity of radiomic features to resolution cannot be overstated. Clinical image acquisition pipelines are well standardized within an institution and between institutions, partly since they cater to patient standard of care, but also owing to numerous initiatives such the quantitative imaging network (QIN) and the quantitative imaging biomarker alliance (QIBA). In contrast to clinical imaging, preclinical imaging acquisition pipelines are not well optimized nor standardized between institutions, and in some cases even within an institution highlighting the need for robust radiomic features to support co-clinical imaging trials.
As a way of summary, Table 3 tabulates robust radiomics features characterized in this study. A feature was considered robust if it displayed less than 10% bias in noise and resolution, in 2D and 3D analyses schemes, or is volume independent. A challenge in all radiomic analyses schemes is the lack of reproducibility within and between institutions, as published studies have been difficult to reproduce and validate [42], [43], [44], [45]. This has been partly attributed to differences in software implementations of radiomics features and the image processing schemes required to compute features [42,44,46,47]. Another source of variability, as highlighted within, is differences in image acquisition. Ideally, in designing co-clinical imaging studies, a feature should be selected that's robust in both preclinical and clinical image acquisition protocols. The implications of the work further highlight the need for consistency in imaging acquisition protocols, both within a single site, as well as in multi-site studies to enable integration of data across sites. Importantly, our data highlight the need to develop best practices in preclinical imaging to enable the translation imaging studies in support of precision medicine.
Table 3.
Summary of robust radiomic features.
 |
Dark grey shading indicates robust feature. Light green shading indicates corrected volume dependency; Light blue shading indicates bias variability in resolution with CV>20%
Declaration of Competing Interests
Dr. Roy has nothing to disclose. Dr. Whitehead has nothing to disclose. Dr. Quirk has nothing to disclose. Dr. Salter has nothing to disclose. Dr. Ademuyiwa reports personal fees from Abbvie, personal fees and other from Pfizer, personal fees and other from Immunomedics, personal fees from Astra Zeneca, personal fees from Cardinal Health, personal fees from Best Doctors, personal fees from Advance Medical, personal fees from QED, other from Seattle Genetics, other from Neoimmnune Tech, other from Astellas , personal fees from Eisai, other from RNA Diagnostics, outside the submitted work. Dr. Li reports grants from NIH U54CA224083, grants from NIH U24CA209837 during the conduct of the study. Dr. An has nothing to disclose. Dr. Shoghi reports grants from NIH U24CA209837.
Data availability
All the co-clinical data will be available for download through the Washington University School of Medicine Co-Clinical Imaging Research Resource web portal at https://c2ir2.wustl.edu/ via the co-clinical database (CCDB).
Acknowledgements
The authors acknowledge the staff of the Preclinical Imaging Facility and the Small Animal MR Facility at Mallinckrodt Institute of Radiology (MIR), Washington University School of Medicine for performing imaging studies.
Footnotes
Funding: This work was supported by NCI grants U24CA209837, U54CA224083; U2CCA233303; Siteman Cancer Center (SCC) Support Grant P30CA091842; and Internal funds provided by Mallinckrodt Institute of Radiology.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102963.
Appendix. Supplementary materials
References
- 1.Chen Z, Akbay E, Mikse O, Tupper T, Cheng K, Wang Y. Co-clinical trials demonstrate superiority of crizotinib to chemotherapy in ALK-rearranged non-small cell lung cancer and predict strategies to overcome resistance. Clin Cancer Res. 2014;20(5):1204–1211. doi: 10.1158/1078-0432.CCR-13-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clohessy JG, Pandolfi PP. Mouse hospital and co-clinical trial project–from bench to bedside. Nat Rev Clin Oncol. 2015;12(8):491–498. doi: 10.1038/nrclinonc.2015.62. [DOI] [PubMed] [Google Scholar]
- 3.Kim HR, Kang HN, Shim HS, Kim EY, Kim J, Kim DJ. Co-clinical trials demonstrate predictive biomarkers for dovitinib, an FGFR inhibitor, in lung squamous cell carcinoma. Ann Oncol. 2017;28(6):1250–1259. doi: 10.1093/annonc/mdx098. [DOI] [PubMed] [Google Scholar]
- 4.Nishino M, Sacher AG, Gandhi L, Chen Z, Akbay E, Fedorov A. Co-clinical quantitative tumor volume imaging in ALK-rearranged NSCLC treated with crizotinib. Eur J Radiol. 2017;88:15–20. doi: 10.1016/j.ejrad.2016.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owonikoko TK, Zhang G, Kim HS, Stinson RM, Bechara R, Zhang C. Patient-derived xenografts faithfully replicated clinical outcome in a phase II co-clinical trial of arsenic trioxide in relapsed small cell lung cancer. J Transl Med. 2016;14(1):111. doi: 10.1186/s12967-016-0861-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sia D, Moeini A, Labgaa I, Villanueva A. The future of patient-derived tumor xenografts in cancer treatment. Pharmacogenomics. 2015;16(14):1671–1683. doi: 10.2217/pgs.15.102. [DOI] [PubMed] [Google Scholar]
- 7.Kwong LN, Boland GM, Frederick DT, Helms TL, Akid AT, Miller JP. Co-clinical assessment identifies patterns of BRAF inhibitor resistance in melanoma. J Clin Invest. 2015;125(4):1459–1470. doi: 10.1172/JCI78954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lunardi A, Ala U, Epping MT, Salmena L, Clohessy JG, Webster KA. A co-clinical approach identifies mechanisms and potential therapies for androgen deprivation resistance in prostate cancer. Nature Genet. 2013;45(7):747–755. doi: 10.1038/ng.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho SY, Kang W, Han JY, Min S, Kang J, Lee A. An integrative approach to precision cancer medicine using patient-derived xenografts. Molecule Cells. 2016;39(2):77–86. doi: 10.14348/molcells.2016.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sulaiman A, Wang L. Bridging the divide: preclinical research discrepancies between triple-negative breast cancer cell lines and patient tumors. Oncotarget. 2017;8(68):113269–113281. doi: 10.18632/oncotarget.22916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parekh V, Jacobs MA. Radiomics: a new application from established techniques. Expert Rev Precis Med Drug Dev. 2016;1(2):207–226. doi: 10.1080/23808993.2016.1164013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avanzo M, Stancanello J, El Naqa I. Beyond imaging: The promise of radiomics. Phys Med. 2017;38:122–139. doi: 10.1016/j.ejmp.2017.05.071. [DOI] [PubMed] [Google Scholar]
- 14.Yue Y, Osipov A, Fraass B, Sandler H, Zhang X, Nissen N. Identifying prognostic intratumor heterogeneity using pre- and post-radiotherapy 18F-FDG PET images for pancreatic cancer patients. J Gastrointest Oncol. 2017;8(1):127–138. doi: 10.21037/jgo.2016.12.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kickingereder P, Gotz M, Muschelli J, Wick A, Neuberger U, Shinohara RT. Large-scale radiomic profiling of recurrent glioblastoma identifies an imaging predictor for stratifying anti-angiogenic treatment response. Clin Cancer Res. 2016;22(23):5765–5771. doi: 10.1158/1078-0432.CCR-16-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hyare H, Thust S, Rees J. Advanced MRI techniques in the monitoring of treatment of gliomas. Curr Treat Options Neurol. 2017;19(3):11. doi: 10.1007/s11940-017-0445-6. [DOI] [PubMed] [Google Scholar]
- 17.Grossmann P, Narayan V, Chang K, Rahman R, Abrey L, Reardon DA. Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro Oncol. 2017;19(12):1688–1697. doi: 10.1093/neuonc/nox092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grossmann P, Gutman DA, Dunn WD, Jr., Holder CA, Aerts HJ. Imaging-genomics reveals driving pathways of MRI derived volumetric tumor phenotype features in Glioblastoma. BMC Cancer. 2016;16:611. doi: 10.1186/s12885-016-2659-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19(1):57. doi: 10.1186/s13058-017-0846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee G, Lee HY, Park H, Schiebler ML, van Beek EJR, Ohno Y. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: State of the art. Eur J Radiol. 2017;86:297–307. doi: 10.1016/j.ejrad.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savaikar MA, Whitehead T, Roy S, Strong L, Fettig N, Prmeau T. SUV25 and microPERCIST: precision imaging of response to therapy in co-clinical FDG-PET imaging of triple negative breast cancer (TNBC) patient-derived tumor xenografts (PDX) J Nucl Med. 2019 doi: 10.2967/jnumed.119.234286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4(6):1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge X, Quirk JD, Engelbach JA, Bretthorst GL, Li S, Shoghi KI. Test-retest performance of a 1-hour multiparametric MR image acquisition pipeline with orthotopic triple-negative breast cancer patient-derived tumor xenografts. Tomography. 2019;5(3):320–331. doi: 10.18383/j.tom.2019.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zwanenburg A, Vallieres M, Abdalah MA, Aerts H, Andrearczyk V, Apte A. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020 doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vallieres M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60(14):5471–5496. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]
- 27.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cyb. 1973;Smc3(6):610–621. [Google Scholar]
- 28.Collewet G, Strzelecki M, Mariette F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging. 2004;22(1):81–91. doi: 10.1016/j.mri.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Galloway MM. Texture analysis using gray level run lengths. Comput Graphics and Image Process. 1975;4(2):8. [Google Scholar]
- 30.Dasarathy BV, Holder EB. Image characterizations based on joint gray level run length distributions. Pattern Recogn Lett. 1991;12(8):497–502. [Google Scholar]
- 31.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Tans Syst Man Cyb. 1989;19(5):1264–1274. [Google Scholar]
- 32.Gudbjartsson H, Patz S. The Rician distribution of noisy MRI data. Magn Reson Med. 1995;34(6):910–914. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xinhao Liu MT, Okutomi Masatoshi. Proceedings of the ICIP. 2012. Noise level estimation using weak textured patches of a single noisy image; pp. 665–669. 2013, February Orlando, FL, USA: IEEE. [Google Scholar]
- 34.Moraru L, Moldovanu S, Dragos Obreja C. A survey over image quality analysis techniques for brain MR images. Int. J. Radiol. 2015;2(1):29–37. [Google Scholar]
- 35.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30(9):1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cattell R, Chen S, Huang C. Robustness of radiomic features in magnetic resonance imaging: review and a phantom study. Visual Comput. Ind. Biomed. Art. 2019;2(1):19. doi: 10.1186/s42492-019-0025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baessler B, Weiss K, Pinto Dos Santos D. Robustness and reproducibility of radiomics in magnetic resonance imaging: a phantom study. Invest Radiol. 2019;54(4):221–228. doi: 10.1097/RLI.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 38.Shafiq-Ul-Hassan M, Zhang GG, Latifi K, Ullah G, Hunt DC, Balagurunathan Y. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44(3):1050–1062. doi: 10.1002/mp.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lafata K, Cai J, Wang C, Hong JC, Kelsey C, Yin FF. Sensitivity of radiomic features to acquisition noise and respiratory motion. Int J Radiat Oncol. 2017;99(2):S93–SS4. [Google Scholar]
- 40.Bagher-Ebadian H, Siddiqui F, Liu C, Movsas B, Chetty IJ. On the impact of smoothing and noise on robustness of CT and CBCT radiomics features for patients with head and neck cancers. Med Phys. 2017;44(5):1755–1770. doi: 10.1002/mp.12188. [DOI] [PubMed] [Google Scholar]
- 41.Presotto L, Bettinardi V, De Bernardi E, Belli ML, Cattaneo GM, Broggi S. PET textural features stability and pattern discrimination power for radiomics analysis: An "ad-hoc" phantoms study. Phys Med. 2018;50:66–74. doi: 10.1016/j.ejmp.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 42.Welch ML, McIntosh C, Haibe-Kains B, Milosevic MF, Wee L, Dekker A. Vulnerabilities of radiomic signature development: The need for safeguards. Radiother Oncol. 2019;130:2–9. doi: 10.1016/j.radonc.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 43.Meyer M, Ronald J, Vernuccio F, Nelson RC, Ramirez-Giraldo JC, Solomon J. Reproducibility of CT radiomic features within the same patient: influence of radiation dose and CT reconstruction settings. Radiology. 2019;293(3):583–591. doi: 10.1148/radiol.2019190928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalpathy-Cramer J, Mamomov A, Zhao B, Lu L, Cherezov D, Napel S. Radiomics of lung nodules: a multi-institutional study of robustness and agreement of quantitative imaging features. Tomography. 2016;2(4):430–437. doi: 10.18383/j.tom.2016.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berenguer R, Pastor-Juan MDR, Canales-Vazquez J, Castro-Garcia M, Villas MV, Mansilla Legorburo F. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288(2):407–415. doi: 10.1148/radiol.2018172361. [DOI] [PubMed] [Google Scholar]
- 46.Foy JJ, Robinson KR, Li H, Giger ML, Al-Hallaq H, Armato SG., 3rd Variation in algorithm implementation across radiomics software. J Med Imaging (Bellingham) 2018;5(4) doi: 10.1117/1.JMI.5.4.044505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bogowicz M, Leijenaar RTH, Tanadini-Lang S, Riesterer O, Pruschy M, Studer G. Post-radiochemotherapy PET radiomics in head and neck cancer - The influence of radiomics implementation on the reproducibility of local control tumor models. Radiother Oncol. 2017;125(3):385–391. doi: 10.1016/j.radonc.2017.10.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the co-clinical data will be available for download through the Washington University School of Medicine Co-Clinical Imaging Research Resource web portal at https://c2ir2.wustl.edu/ via the co-clinical database (CCDB).