Abstract
A growing awareness of the diversity and ubiquity of microbes (eukaryotes, prokaryotes, and viruses) associated with larger ‘host’ organisms has led to the realisation that many diseases thought to be caused by one primary agent are the result of interactions between multiple taxa and the host. Even where a primary agent can be identified, its effect is often moderated by other symbionts. Therefore, the one pathogen–one disease paradigm is shifting towards the pathobiome concept, integrating the interaction of multiple symbionts, host, and environment in a new understanding of disease aetiology. Taxonomically, pathobiomes are variable across host species, ecology, tissue type, and time. Therefore, a more functionally driven understanding of pathobiotic systems is necessary, based on gene expression, metabolic interactions, and ecological processes.
Keywords: pathobiome, pathobiotic, disease, symbiome, microbiome, symbiont
Highlights
Animal and plant diseases are increasingly recognised to result from interactions between host-associated bacteria, eukaryotes, and viruses, their host, and the environment.
The diversity and function of host-associated organisms are diverse and incompletely understood.
Multidisciplinary studies, including high-throughput sequencing ‘omics, can be used to reveal both the structure and function of pathobiomes, which may not be discernible from taxonomic analyses alone.
Both ‘normal’ and ‘disease’ pathobiomes vary over time and between host tissues and organs.
Understanding pathobiotic systems presents not only challenges to current disease diagnostic practices and legislation associated with this, but also diverse new opportunities for mitigating disease and optimising on-farm growing conditions.
Disease in a Microbe-Dominated World
The pathobiome (see Glossary) concept arose from human studies in which disruption of a health-promoting and ecologically stable gut microbiome resulted in dysbiosis: a microbiome community of low-diversity and modified metabolic state, exposing the gut to invasion by, and proliferation of, pathogenic agents 1, 2. Dysbiotic communities can subvert the immune system and lead to further deleterious effects [3]. This concept is being adopted for research into the pathology of other animals and plants because attempts to explain syndromic conditions by identifying a single pathogenic agent are often incomplete (i.e., the one pathogen–one disease paradigm is often insufficient to explain many diseases 4, 5, 6).
Pathobiomes differ from those assemblages representing healthy or ‘normal’ states. What is ‘normal’ likely encompasses a range of assemblages that need to be understood before a pathobiome can be reliably distinguished from them. There is a lack of consistency in defining ‘pathobiome’ in the literature, ranging from a single pathogenic agent interacting with its biotic and abiotic environments (e.g., [5]) to the effects of interacting communities of microbes on host health [7]. Our synthesis (Box 1) is based on the effects of multiple symbionts, across all domains of life, on host health. The term ‘microbiome’ generally excludes eukaryotes; therefore, in this review, we use the term ‘symbiome’ to describe the whole assemblage of associated organisms excluding the host, and ‘symbiont’ for individual taxa within that assemblage. This definition is concordant with an inclusive scheme of symbiosis acknowledged in [9], which ranges from neutralism (neutral effect on both partners) to mutual beneficial effects and mutual antagonistic effects, and all other possible combinations of neutral, beneficial, and antagonistic effects. The duration of the association need not necessarily be long-term, as interactions can be effective on even short timescales; great variability in duration of association is both possible and likely. This inclusive definition is not inconsistent with some previous usages of the term, and is required by the large diversity of associations that are being revealed by ongoing research, most of which are not currently recognised, and an even greater proportion of which are not characterised.
Box 1. What Is a Pathobiome?
We use the term ‘pathobiome’ to refer to the set of host-associated organisms (crucially encompassing prokaryotes, eukaryotes, and viruses) associated with reduced (or potentially reduced) health status, as a result of interactions between members of that set and the host. These interactions are inevitably moderated by the environment within the host and immediately surrounding it (Figure I). A pathobiome comprises a host with anything from two non-host lineages up to a complex symbiont community. However, a full understanding of pathobiomes cannot rely on inventories of the organisms involved alone, but should incorporate the nature of their interactions and relationship with their host in the context of all relevant factors (ecological, chemical, physical, genetic, immunological, etc.). Therefore, we prefer the use of the term ‘pathobiotic’ to capture this bigger picture in addition to the solely organismal set inferred by the suffix ‘-biome’.
Pathobiomes should be considered as host compartment specific, and are also likely to be context dependent (e.g., moderated by host condition and response, environmental conditions, etc.) 8, 10 (see Figure 1 in main text). The pathobiome may be temporally structured, differing according to the stage and severity of disease, from subclinical to late clinical stages and death (see Figure 2 in main text). Therefore, a range of host–microbe scenarios can legitimately be considered ‘pathobiomes’, even within a single host species. Furthermore, because most hosts, especially aquatic organisms, live in a ‘microbial soup,’ predicting the risk of pathobiont infection will depend on understanding the composition and activity of environmental microbiota. Ectoparasites and attached epibionts (including opportunistic or environmentally driven biofouling; e.g., [11]) can also usefully be considered members of a pathobiotic community.
To fully understand pathobiome-mediated disease aetiology, it is important to integrate other factors, as for single pathogen models. In 1974, Snieszko outlined a fundamental model of the interactions between pathogen, host, and environment, in which disease is an outcome of their interaction [12]. This was recently updated to include the microbiota of hosts and their environment, and anthropogenic factors acting on those, (i.e., animal husbandry including antibiotic application, climate change, pollution, and species introductions) [4]. However, the presence of more than one pathogen was not required in either of these models.
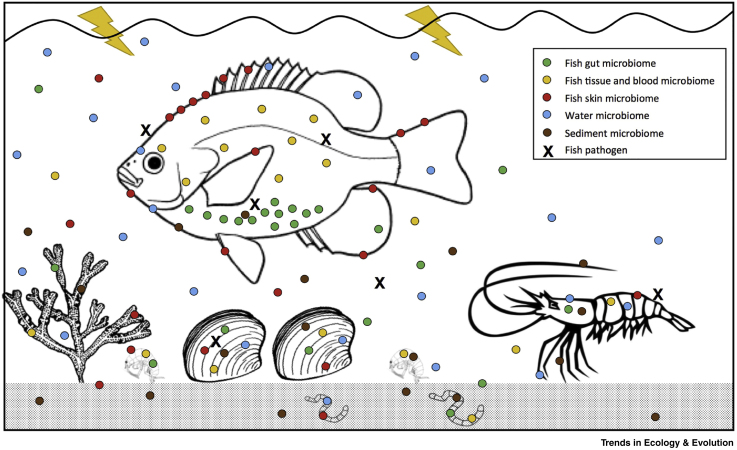
Figure I.
Microbial Complexity in a Typical Host–Symbiont–Environment System.
Symbionts (including viruses, bacteria, fungi, and eukaryotes) in the fish gut, other tissues and organs, blood, and from the skin surface can be released to the water column and may appear in samples of sediment and within other hosts residing in the same environment. Similarly, microbes that originate within the water column and sediment may be detected on the skin surface and within the gut and tissues of the fish. Extension of this principle to consider exchange of symbionts between other hosts from the same system (in this case crustaceans, molluscs, annelids, and macroalgae) underlines the intricacy of potential symbiont relations, even within simple systems. Disruption to the microbial profile within different compartments of the fish (i.e., gut, tissue, blood, or skin) caused by infection with pathogens, starvation, overcrowding, in-breeding, or via a range of environmental perturbations that may also affect other hosts within the system, is consistent with the development of a pathobiome (see Figure 1 in main text). In the diseased state, the detection of specific pathogens (here denoted by ‘X’) may occur alongside an altered microbial profile, the combined effects of which may elicit the clinical signs detected in the host.
Alt-text: Box 1
What Are Pathogens?
Microbial diversity is now known to be more diverse than previously thought in both environmental and host-associated habitats. Examples include high levels of protistan 13, 14, 15, 16, 17, bacterial [18], and viral [19] diversity revealed by environmental (e)D/RNA-style studies of host-associated (e.g. tissue, gut contents, and skin epibionts) and actual environmental samples (e.g., water, sediment, and soils) 20, 21. This diversity includes many lineages that are (or may become) host associated and can form part of pathobiomes. These include ‘cryptic’ or ‘emerging’ pathogens, which fall into several categories: (i) new lineages that are more or less closely related to known pathogens; (ii) previously unknown pathogens related to free-living lineages; and (iii) lineages that act transiently or opportunistically as pathogens, including commensals switching to pathogenic mode [22]. Our appreciation of what constitutes a ‘pathogen’ has diversified, and will probably continue to do so as we better understand the complexity of the pathogenic process.
Context-Dependent Pathogenesis and Cryptic Infections
Opportunistic and transient pathogenic modes depend on context. For example, most ciliates, abundant and diverse protists in most environments, are generally free-living heterotrophs, but they may take advantage of larger hosts for nutrition, sometimes triggered by a wounded or stressed host, with pathogenic effects resulting from aggressive feeding [22]. Such opportunism may involve multiple agents, amplifying the pathogenic affect. Vibrio spp. are frequently detected opportunistic bacterial pathogens in aquatic environments, associated with a range of hosts [23]. Photosynthetic algae, both bacterial and eukaryotic, can also infect a range of vertebrate hosts, causing dermatological and systemic pathologies. Examples of algal infections include Chlorochytrium in cichlid fish, Chlorella in sheep and other vertebrates 24, 25, and cyanobacteria associated with black band disease of corals [26]. Cryptic infections include pathogens with dormant life stages that may asymptomatically infect a diversity of hosts or tissues, and be triggered to cause negative effects in response to environmental changes [9].
Pathogenic Transitions
Perturbation of a balanced, ‘healthy’ microbiome into a pathobiome is a high-level example of a pathogenic transition [6]. However, pathogenic transitions have many triggers, including temperature, toxicity, physical site in host, the surrounding microbiota, shifts in symbiont community structure, host resistance, nutritional, and reproductive status, and environmental stressors [27].
Many potential pathogens can be common members of a symbiome in the absence of disease, but an accumulation or change in the relative abundance of recognised pathogens in a host can result in greater disease incidence or severity. Ryan reports that it is ‘normal’ to detect a range of enteropathogens in infants and young children in resource-limited settings, and suggests that elevated levels of one or more of these contributes to a tipping point between non-diarrhoeal and diarrhoeal states [28]. This ‘pathogen excess’ context for ascribing particular pathogens to an outbreak is a clear pathobiome scenario.
Interactions within the Symbiome: Mechanisms and Dynamics of Pathobiomes
Interactions between Low Numbers of Symbionts
Concurrent infections are common, and co-infection can result in antagonism or synergism among infecting agents 29, 30, 31. Even apparently simple interactions can produce a diversity of outcomes. For example, interactions between strains of Aeromonas resulted in enhanced virulence compared with the effects of individual strains [32]. These enhanced effects were only seen in pairs of strains belonging to the same species. Enhanced deleterious effects can also result from pathobiotic relationships between symbionts from different domains of life. For example, increased mortality was observed in channel catfish, Ictalurus punctatus, when infected by both the trematode Bolbophorus damnificus and the enteric septicaemia-causing bacterium Edwardsiella ictaluri [33]. Contrastingly, the virulence of one microbe may be reduced by co-infection with another, as has been shown for the bacterium Acinetobacter baumannii, which inhibits virulence factors of the yeast Candida albicans during infection of Caenorhabditis elegans [34]. More complex relationships can occur, for example when some symbionts are parasites of others: plant-attacking wireworms (Coleoptera) are more susceptible to the ascomycete fungus Metarhizium brunneum, often highly prevalent as latent infections, in the absence of certain bacteria in the microbiome of the wireworms, which may mediate the resistance of wireworms to the fungus [35].
Pathobiotic Mechanisms
Mechanisms underlying symbiont interactions include quorum sensing (QS); for example, signal sharing between the non-pathogenic bacterium Erwinia toletana and pathogenic Pseudomonas savastanoi pv. savastanoi in olive knot disease of Olea europaea [36]. These two symbionts co-localise in the knot, likely facilitating signal sharing and metabolic interaction. The latter could also include sharing of metabolites; in silico recreation of their metabolic pathways indicates complementing metabolic pathways. In response to QS, quorum quenching enables some bacteria to outcompete their rivals by inhibiting QS pathways, another means by which bacteria can interact in a pathobiome context [37]. Specialisms, including those associated with pathogenicity, can evolve in bacteria by mutualistic sharing of gene products and associated loss of genes by inefficient producers, leading to an interdependency between taxa (Black Queen Hypothesis [38]).
Bleaching disease in the red alga Delisea pulchra is caused by host stress and invasion by secondary bacterial pathogens from several families (Rhodobacteraceae, Saprospiraceae, and Flavobacteriaceae), but no single bacterial lineage has all bleaching-enriched bacterial genes [39]. A related concept is the distribution between co-infecting lineages of bacteria of ‘community-dependent essential genes’ required for infection [40]. Such situations may be a host-associated parallel of that recently determined in freshwater Actinomycetes, which have highly streamlined genomes but are interdependent: they need to coexist with strains of complementary capacities to flourish [41]. This genomic streamlining strategy has balancing benefits in speed of growth and metabolic lability, which are likely advantageous in limiting and/or challenging environments.
Virulence capacity can be enhanced in a pathobiotic system by horizontal gene transfer, which could explain the emergence of some opportunistic pathogens [37]. Bacteria may become (more) pathogenic by acquiring and utilising DNA from other (e.g., co-occurring) bacteria 42, 43, via plasmids, genomic islands, transposons, insertion sequence elements, and bacteriophages [44]. The importance of microorganisms as vectors of other microbes or viruses into the holobiome should not be overlooked, given, for example, the role of the protistan root parasite Polymyxa in transmitting a range of pathogenic viruses into crop plants [45]. The pathogenic and functional consequences of hyperparasitic associations have not received much attention (but see, for example, [46]), compounded by the fact that the extent of microbial hyperparasitism is likely underestimated (e.g., 47, 48).
More Diverse Communities of Symbionts
Symbiont community shifts and dysbioses analogous to those studied in humans have recently been reported in other animals, plants, and macroalgae, such as impoverished and less stable bacterial community networks associated with diseased Acropora corals relative to healthy individuals [7]. When diversity and community connectivity are reduced, invasion by pathogens may be more likely, such as the reduced bacterial diversity of asymptomatic bark tissue of horse chestnut (Aesculus hippocastanum) with disease caused by Pseudomonas syringae pv aesculi [49]. In this case, symptomatic compared with asymptomatic tissue harboured elevated levels of plant disease agents and opportunistic pathogens, such as Serratia and Brenneria, suggesting enhanced opportunity for further infection by other pathogens.
Although symbiont community shifts are often strongly associated with host health shifts, describing these does not in itself reveal the mechanisms by which they operate. Cause and effect can rarely be determined from the association alone (Figure 1). Symbiome-wide association studies often result in spurious associations between specific microbes and host disease state [50]. Whether a symbiont community shift occurs before, and causes a change in, health status, or is a consequence of a change in health from a different cause, is a key question for understanding pathobiotic dynamics, and is increasingly being addressed in experimental studies (e.g., 51, 52).
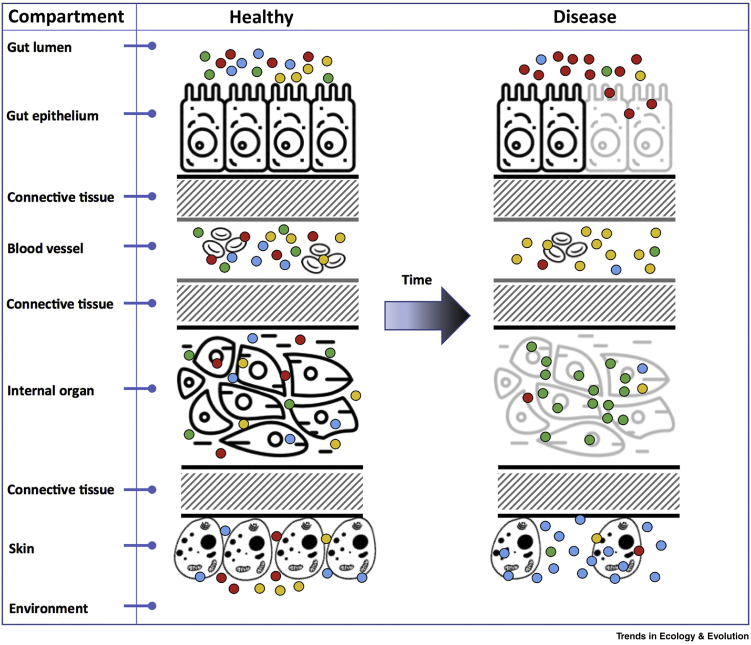
Figure 1.
Different Pathobiomes in Discrete Compartments of the Host.
Defining the pathobiome necessitates the contextualisation of data pertaining to the symbiont profile with observations consistent with definition of a disease state (e.g., clinical signs). Since disease is often associated with specific host compartments (e.g., gut lumen, skin, epithelia of internal organs, etc.) it is relevant to consider the pathobiome also operating within these compartments. Furthermore, given that many diseases involve a progressive departure from the healthy state, the pathobiome associated with this progression may also be expected to change, leading to specific symbiont profiles (pathobiomes) in early, established, and late stages of disease (see Figure 2). The recognition of such spatial and temporal changes in the symbiont profile of a given host (and its compartments) has the potential to complicate definition of a single ‘pathobiome’ associated with a specific disease state. In the model shown here, the composition of a simple microbiome (containing four taxa, defined by dots of different colours) may differ spatially in one or more compartments and, in time, in a host undergoing progression from healthy to diseased states (diseased tissue in grey). We propose that changes in profile occurring within specific compartments may also differ (i.e., the pathobiome associated with the skin will not necessarily be the same as that observed in the blood or gut). We do not predict whether changes in profile drive the appearance of disease within specific compartments or, conversely, that diseased tissue precedes changes in the profile. Furthermore, it is not assumed that a pathobiome is less diverse than that observed during health. As such, the pathobiome associated with diseased tissue should be defined relative to the symbiont profile occurring in the healthy state.
In the horse chestnut example described earlier, further studies using either temporal sampling or experimental manipulation of the symbiome composition should determine whether the reduced bacterial diversity of asymptomatic tissue of diseased trees is an a priori state that predisposed the tree to disease, or whether the compromised health state of the trees itself drove changes in the bacterial community. Parallel work from a tomato model system suggests that the phyllosphere microbiome is directly linked to reduced P. syringae colonization, and that the degree of protection is dependent on microbiome diversity [53]. Recent work on Sea Star Wasting Disease, adopting a temporal sampling approach, showed that microbiome shifts towards imbalance and colonisation by both pathogenic and opportunistic bacteria were consistently found at disease onset [54].
Some taxonomic characteristics of pathobiomes are frequently encountered; for example, certain bacterial taxa (notably Vibrio and Photobacterium) are consistently elevated in diseased animals 55, 56, 57, 58, 59. Patterns such as these can be uncovered through microbiome-wide association studies, where microbial consortia can be linked to disease states [60]. However, it is also clear that microbiomes associated with both health and disease differ between host species, and are additionally influenced by the ecological and geographical provenance of their hosts 59, 60, 61, 62. Geographically disparate Cladocora corals harbour a common core microbiome, suggested as key players in the coral holobiont, but microbial assemblages associated with diseased corals were more influenced by sampling location [61]. The inference that pathobiomes may be particularly influenced by geographical distance and local ecological conditions is consistent with the ‘Anna Karenina’ principle, which posits that dysbiotic individuals vary more in microbial community composition compared with healthy individuals [63]. This may occur via ‘priority effects’ [64] mediated by the local conditions. Also referred to as the primary symbiont hypothesis, these effects have been reported in plants: bottlenecks in endophyte diversity associated with individual plant seeds are proposed to impact subsequent plant microbiome diversity [65].
Pathobiotic communities are not always less diverse than healthy ones 61, 66. In the Cladocera example described earlier [61], there was no significant difference in symbiont diversity between healthy and diseased individuals at the most pristine of the sites sampled. Similarly, a study of white band and white patch diseases of coralline algae showed that, although diseased tissue in both cases had compositionally distinct bacterial communities dominated by different operational taxonomic units (OTUs) compared with healthy tissue, the bacterial diversity of band disease was comparable to (and in some cases higher than) healthy tissue, whereas patch disease communities were significantly less diverse [66]. A similar association has been observed between human cervical intraepithelial neoplasia disease and increased vaginal microbiome diversity [67].
Primary Causative Agents in a Pathobiome Context
Some diseases thought to be caused by a primary pathogen in fact have different aetiologies (the ‘moving target hypothesis’ [68]). For example, white pox disease (WPX) of the coral Acropora, originally ascribed to Serratia marcescens, is now thought to have multiple aetiologic agents, with elevated Vibrio abundance consistently associated with WPX relative to healthy samples [55]. Vibrio spp. may act within, or emerge from, nonspecific heterotrophic blooms rather than acting as primary pathogens.
Even cases where a primary agent is recognised can be elucidated by investigations into the associated pathobiome, because the presence of other pathogens or particular symbiont profiles may increase their chances of infection and/or subsequent virulence. Acute hepatopancreatic necrosis disease (AHPND) of shrimp is specifically associated with toxin genes expressed from the plasmid of Vibrio parahaemolyticus. An increase in copy number of the AHPND toxin-producing plasmid has been associated with a shift of the microbiome to domination by Vibrio and Candidatus bacilloplasma, and decreases in complexity and species connectivity. A co-occurrence pattern was predicted between V. parahaemolyticus and several resident and transit members within Candidatus bacilloplasma and cyanobacteria. Conversely, the key hub taxon in healthy shrimp was Lactobacillus, which appeared to inhibit the growth of Vibrio, the pathogen that becomes the main hub in diseased shrimp [29]. Therefore, the one pathogen–one disease paradigm should be reassessed even in cases where a primary pathogen has been considered the sole pathogenic agent.
Ecological Processes and Interaction Networks
Bacterial communities are shaped by a balance of facilitative and antagonistic processes, including competition for shared resources, interactions via metabolic and molecular signalling, and production of other molecules that negatively or positively affect other members of the community 69, 70, 71. Pathobiomes can be modelled as outcomes of networks of interacting symbionts. This has been shown in the pathobiome associated with the causal agent of oak powdery mildew (the ascomycete fungus Erysiphe alphitoides) in oak (Quercus robur) phylloplane communities [72]. The type (facilitative–antagonistic) and strength of pairwise bacterial interactions can reliably predict the outcome of invasions in more complex communities, and antagonistic communities can directly inhibit invading microbes through both antagonistic interactions and resource competition [73].
Structural equation modelling has been used to show that the significant differences between gut bacterial communities of cohabiting normal, overgrown, and retarded shrimp were strongly related to digestive activities, in turn affecting shrimp growth rate [74]. Gut communities of the normally grown shrimp showed higher taxonomic diversity and evenness than either abnormal category, hypothesised to result as a legacy of priority effects during larval gut colonisation ([64] and see earlier).
Interactions with the Host
Commensal bacteria are critical for the maturation of host innate and adaptive immunity responses. Protection against pathogens is mediated by the immunity response and host microbiome [37]. For example, host stress, diet, and antibiotic use can have consequences for the composition of its microbiome, potentially leading to dysbiosis. In turn, dysbiosis can lead to enhanced horizontal gene transfer, promoting further pathogen evolution by transferring and amplifying virulence and antimicrobial resistance. Therefore, interaction with the host is an essential feature of pathobiome mechanisms, and pathobiomes can promote their own evolutionary trajectory to further pathogenicity.
Symbiomes Associated with Resistance to Disease
Some symbiont communities protect from disease. Amphibian skin microbiota can be significantly different from that of the surrounding environment, the dermal habitat selecting and promoting rare lineages [75]. Frogs resistant to the chytrid pathogen Batrachochytrium dendrobatidis (Bd) had different skin microbiota depending on whether Bd was present at the sampling site. Where it was absent, non-susceptible frogs had similar skin bacterial communities dominated by Pseudomonas and Acinetobacter. However, where Bd was present, the skin microbiomes were less diverse due to increased relative abundance of bacteria that inhibit the pathogen 76, 77. An agricultural parallel is provided by disease-suppressive soils, which variously have the properties of suppressing a range of plant pathogens. Multifactorial analyses, combined with the recent ability to identify a range of taxa in such complex environments via molecular techniques, is starting to reveal a variety of mechanisms by which suppression operates 78, 79.
Considerations of time are important in symbiont–host relationships (Figures 1 and 2), and are probably often overlooked. For example, Knutie et al. showed that bacterial diversity in tadpoles was negatively correlated with establishment of an ascarid gut worm in the corresponding adult frogs, while bacterial diversity during parasite exposure as adults was not correlated with parasite establishment [80].
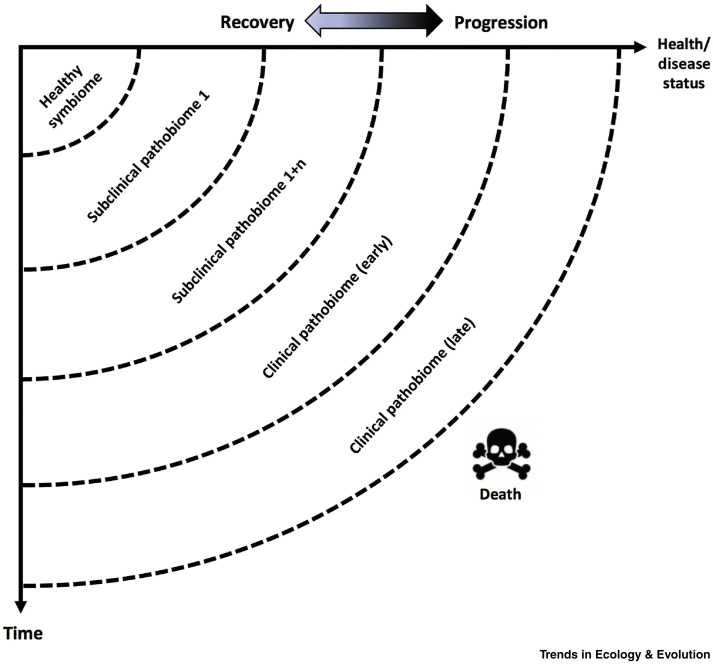
Figure 2.
Temporal Shifts in the Pathobiome Relating to Host Health and Disease Status over Time.
The healthy symbiome (top left of figure, and associated with a particular body compartment; see Figure 1) is altered (to a pathobiome) when the host enters a progressive disease state. During early (‘subclinical’) stages of disease, the altered symbiome may precede detection of specific symptoms (in humans) and signs (in humans, animals, and plants). During later stages of disease, where symptoms and clinical signs are apparent, the pathobiome may be further altered in specific host compartments, although not necessarily as a direct cause or effect of those symptoms and signs detected. Where recovery of the host occurs (e.g., where infection by a specific agent is cleared, a treatment is provided, or an immune response occurs), reversion to a healthy symbiome may occur. In other cases, where disease progression leads to death, the disease-associated pathobiome gives way to microbial decay of host tissues. In the latter, the consortium of microbes associated with the decaying host and its compartments should not be classified as a ‘pathobiome’.
Experimental Insights into Pathobiomes
Experimental approaches are key for determining the most important factors in pathobiome operation, distinguishing between cause and effect, and avoiding the danger of making spurious associations between symbiomes and host health status 51, 52. Existing frameworks, such as Koch’s Postulates, will require adaptation away from single agents to pathobiomes [81]. Most existing experimental studies investigate the functional complexities of pathobiomes, from genomic, metabolomics, and microbial ecology perspectives, or combinations thereof.
Multi-omics, combining large molecular data sets such as metagenomes, metatranscriptomes, and metaproteomes, are becoming increasingly feasible as technology costs decrease and analytical capacity increases. Broberg et al. used these techniques to characterise host–microbiota relationships associated with Acute Oak Decline in both community and functional terms [82], for example by functional annotation of phytopathogenic virulence genes [83]. They revealed polymicrobial–host interactions and measured the relative lesion activity of several bacteria, including some not previously known to be associated with this disease. Multiple ‘omic’ data can be used to integrate taxonomic information, microbial networks, and bacterial defensive function and metabolism, to enable intervention and mitigation of diseases, such as chytridiomycosis of amphibians [76].
Given that microbiome community structures are often highly variable, functional traits, such as gene expression patterns at different levels (e.g., core and non-core functions), and metabolic interactions, may provide complementary perspectives with different patterns of variability, from which to understand pathobiotic mechanisms ([84] and references therein).
Somewhat contrastingly, reductionist approaches can be used to disentangle the complex interactions of microbiota and host, including experiments using synthetic microbial communities to complement environmental studies. Thus, the effects of modifying microbial, host, and environmental parameters can be measured within a testable system [85]. Using specific pathogen-free (SPF) animals could offer a ‘blank canvas’ on which host–symbiont–environment experiments can be conducted [86].
Hypothesis-free integrative systems biology approaches [87] may be developed to generate novel hypotheses that were previously undefinable due to the complexity of pathobiotic systems. Well-designed network analyses can reveal a lot about how symbiont interactions operate, for example the phylogenetic and spatial distribution of different kinds of interaction (e.g., competitive, cooperative, or exploitative). A study of polymicrobial urinary tract infections found that competitive and cooperative interactions were more strongly represented than exploitative ones, and that simple pairwise interaction models may be insufficient to predict higher-order interactions; different interaction dynamics were ascribed to different bacterial taxa [88].
Applied Pathobiotics
Knowledge about pathobiotic systems can facilitate their manipulation, either from within the host or via the surrounding environment. De Bruijn et al. reviewed a range of ways to achieve this, including emerging practices of introducing or augmenting beneficial microbes, and taking into account different host life stages, nutrition, genotype, and so on [89]. There is potential for monitoring and manipulating the environmental microbiome as a tool for managing and monitoring host health, particularly in aquaculture settings [90], but significant research is required before this becomes a reliable reality. Extending the (human) microbial dysbiosis concept to ecosystems should enable definitions of disease and pathogenesis in those systems to be reconsidered [91] from a holobiont perspective 6, 92, 93, 94.
An improved understanding of the spatial and temporal changes as a host symbiome transitions to a pathobiome state will undoubtedly provide a new perception of what we mean by ‘disease’ (e.g., [54] and Figure 1, Figure 2). In pathobiotic settings, detection of individual ‘known’ agents (of which the diagnostician has pre-existing knowledge and established protocols to detect) will inevitably underestimate the role of other elements of the symbiome before the observation of clinical signs or symptoms. In extreme cases, it may be that application of specific diagnostics towards single known agents has the potential to incorrectly assign causality to that agent, disregarding the contributions of other symbionts. Where data sets relating to the symbiome are available for large numbers of hosts exhibiting certain disease states, or where data relating to predisease (healthy) hosts are available, early-stage interventions to avoid development of a pathobiome state could be envisaged [95]. In such cases, temporal analyses that reveal the potential for shifts in the symbiome to equate to specific symptoms or clinical signs (i.e., progression towards the disease state) will be required for specific host–disease models (Figure 1, Figure 2).
We predict that understanding the symbiome and formation of the pathobiome will become a crucial element of mitigating disease in global food production systems 96, 97. In most cases, disease outbreaks are considered departures from the normal health status of the host associated with infection by a specific agent. Accordingly, bodies such as the World Organisation for Animal Health (OIE) define a list of emerging and notifiable diseases associated with infection by specific agents, against which diagnostic tests and management strategies can be designed. Due to the focus on specific pathogens, the elimination of the pathogen threat forms the basis of most biosecurity strategies. Global standard setting provided by such bodies is important for controlling the of spread of animal pathogens via cross-border trade [98]. However, such legislation does not take into account broader scale symbiont profiling of hosts and, therefore, the role that the symbiome may have in agent-associated disease outbreaks. Furthermore, it does not consider the effect of specific pathogen exclusion on susceptibility of hosts to remaining components of the symbiome or to the emergence of new diseases associated with these symbionts.
The increasing propensity to farm hosts with an artificially altered symbiome (e.g., in the case of SPF or even specific-pathogen resistant, SPR, hosts) may require more careful analysis in the light of the pathobiome concept. Exclusion of specific pathogens from host systems via these means has potential to either create space for (previously excluded) perfect competitors [99] or to alter in some way the remaining symbiome to the detriment of the host [100]. Although successful strategies for prevention of specific pathogens exist within certain farming systems, the role of pathogen exclusion in driving alternative pathogen emergence (possibly via a disrupted symbiome) requires further consideration.
Concluding Remarks
The essence of the pathobiome concept is to provide a framework within which the combined effect of more than one symbiont on host health can be understood, taking into account environmental influences and host response and/or condition. We have shown that pathobiotic systems are diverse in terms of the types and numbers of symbiont involved, and the processes by which their influence manifests. What constitutes a pathogen is strongly context dependent. Similarly, pathobiotic effects vary from the activity of a primary pathogen being modified by other symbionts to truly synergistic activity of multiple symbionts. Much work is required to identify consistent and predictive aspects of pathobiotic systems (see Outstanding Questions). These must be temporally dynamic, and take into account different compartments within the host, as well as the external environment. Describing pathobiotic systems in terms of functional traits, rather than, or in addition to, taxonomic profiles may be key to this understanding. The pathobiome concept may eventually be extended beyond the individual, leading to models in which pathobiotic elements shared between individuals form part of ecological networks that determine health and persistence at the host population level.
Outstanding Questions.
Given the increasing recognition that host-associated microbiomes have a role in shaping the outcome of infection and disease, how and when should we draw a line between a pathogen and a pathobiome?
Just as disease has classically been modelled as an interaction between the host and pathogen genotype (a G × G interaction), how can we usefully move towards a predictive framework when the outcome is the result of interactions among many genotypes?
What factors determine the variation within ‘normal’ microbiomes and pathobiomes, and can they always be reliably distinguished from each other?
Is the best basis for distinguishing pathobiomes their taxonomic profiles or functional characteristics?
Are low diversity microbiomes associated with disease generally a result of pre-existing pathogen infections, or do they have a wider range of causes and predispose hosts to infection?
Can a framework, such as Koch’s Postulates, be updated in the light of knowledge of pathobiotic systems, to identify causative relationships between pathobiomes and diseases?
What factors enable hosts to maintain ‘healthy’ microbiomes under conditions where conspecifics develop dysbioses and/or pathobiomes?
In terms of treatment options for pathobiome-related diseases, can we devise combination treatments to reduce the likelihood of resistance evolution and increase the likelihood of success?
Can the likelihood of disease emergence be predicted by sampling and undertaking molecular analyses of the environments in which hosts live (e.g., aquaculture ponds or agricultural soils)?
How might existing ecological and evolutionary theory (e.g., community ecology, network analyses, or coevolution) provide insights into the pathobiome and be leveraged for the management or treatment of diseases?
Alt-text: Outstanding Questions
Acknowledgements
D.B. and G.D.S. were supported by Defra Research Project C7277C (FC1214) and BBSRC/Newton Fund project BB/N00504X/1, which also supported C.R.T. H.C.W. was supported by the Ministry of Science and Technology, Taiwan, project 108-2314-B-006-096-MY3. B.K. was supported by NSF award IOS-1838299. This work was also partly supported by the EU H2020 program, no. 678589; VIVALDI.
Glossary
- Dysbiosis
a microbiome community of low-diversity and modified metabolic state, exposing the gut to invasion by, and proliferation of, pathogenic agents.
- Holobiont
a unit of biological organisation comprising a host and its associated bacteria, Archaea, viruses, and eukaryotes.
- Microbiome
a term used variably, from (implicitly) including only bacteria and Archaea, to a more inclusive usage incorporating bacteria, Archaea, viruses, protists, and fungi. However, due to this ambiguity, and because it does not generally include larger symbionts, such as symbiotic metazoans and multicellular algae and/or plants, we prefer the use of the term ‘symbiome’ to include the full range of host-associated organisms, and use ‘microbiome’ in the original more (but not exclusively) prokaryote-oriented sense, where investigated as such by studies cited in this review, and when referring to environmental (i.e., not necessarily host-associated) microbial diversity.
- Pathobiome
the set of host-associated organisms (encompassing prokaryotes, eukaryotes, and viruses) associated with reduced (or potentially reduced) health status, as a result of interactions between members of that set and the host.
- Pathobiotic
adjective describing a compromised health status associated with a symbiont community interacting with its host in the context of all relevant factors (ecological, chemical, physical, genetic, immunological, etc.), where such factors are known. It may simply refer to a pathobiome or a syndromic condition if no other information is available.
- Symbiome
the full range of host-associated organisms and viruses, but excluding the host.
- Symbiont
a member of the symbiome.
- Symbiosis
interactions or associations between different species that may be mutually beneficial, antagonistic, or neutral, or any combination of those. The interactions or associations may persist on short or long timescales.
References
- 1.Defazio J. The opposing forces of the intestinal microbiome and the emerging pathobiome. Surg. Clin. North Am. 2014;94:1151–1161. doi: 10.1016/j.suc.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krezalek M.A. The shift of an intestinal ‘microbiome’ to a ‘pathobiome’ governs the course and outcome of sepsis following surgical injury. Shock. 2016;45:475–482. doi: 10.1097/SHK.0000000000000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stecher B. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat. Rev. Microbiol. 2013;11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 4.Vayssier-Taussat M. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front. Cell. Infect. Microbiol. 2014;4:29. doi: 10.3389/fcimb.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vayssier-Taussat M. Emerging horizons for tick-borne pathogens: from the ‘one pathogen-one disease’ vision to the pathobiome paradigm. Fut. Microbiol. 2015;10:2033–2043. doi: 10.2217/fmb.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitlik S.D., Koren O. How holobionts get sick–toward a unifying scheme of disease. Microbiome. 2017;5:64. doi: 10.1186/s40168-017-0281-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sweet M.J., Bulling M.T. On the importance of the microbiome and pathobiome in coral health and disease. Front. Mar. Sci. 2017;4:9. [Google Scholar]
- 8.Okamura B. Hidden infections and changing environments. Integr. Comp. Biol. 2016;56:620–629. doi: 10.1093/icb/icw008. [DOI] [PubMed] [Google Scholar]
- 9.Martin B.D., Schwab E. Current usage of symbiosis and associated terminology. Int. J. Biol. 2013;5:32–45. [Google Scholar]
- 10.Schwartzman J.A., Ruby E.G. Stress as a normal cue in the symbiotic environment. Trends Microbiol. 2016;24:414–424. doi: 10.1016/j.tim.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burris Z.P., Dam H.G. Deleterious effects of the ciliate epibiont Zoothamnium sp. on fitness of the copepod Acartia tonsa. J. Plankt. Res. 2014;36:788–799. [Google Scholar]
- 12.Snieszko S.F. The effects of environmental stress on outbreaks of infectious diseases of fish. J. Fish Biol. 1974;6:197–208. [Google Scholar]
- 13.Hartikainen H. Mikrocytids are a broadly distributed and divergent radiation of parasites in aquatic invertebrates. Curr. Biol. 2014;24:807–812. doi: 10.1016/j.cub.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 14.Busby P.E. Fungal endophytes: modifiers of plant disease. Plant Mol. Biol. 2016;90:645–655. doi: 10.1007/s11103-015-0412-0. [DOI] [PubMed] [Google Scholar]
- 15.Lynch S.V., Pedersen O. The human intestinal microbiome in health and disease. New Engl. J. Med. 2016;375:2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 16.Ward G.M. A new phylogeny and eDNA insight into paramyxids: an increasingly important but enigmatic clade of protistan parasites of marine invertebrates. Int. J. Parasitol. 2016;46:605–619. doi: 10.1016/j.ijpara.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Ward G.M. Environmental sequencing fills the gap between parasitic haplosporidians and free-living giant amoebae. J. Eukaryot. Microbiol. 2018;65:574–586. doi: 10.1111/jeu.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koskella B. The microbiome beyond the horizon of ecological and evolutionary theory. Nat. Ecol. Evol. 2017;1:1606–1615. doi: 10.1038/s41559-017-0340-2. [DOI] [PubMed] [Google Scholar]
- 19.Shi M. Redefining the invertebrate RNA virosphere. Nature. 2016;540:539–543. doi: 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 20.de Vargas C. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 21.Bass D. Applications of environmental DNA methods in parasitology. Trends Parasitol. 2015;31:499–513. doi: 10.1016/j.pt.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 22.Overstreet R.M., Lotz J.M. Host-symbiont relationships: understanding the change from guest to pest. Adv. Env. Microbiol. 2016;3:27–64. [Google Scholar]
- 23.Chatterjee S., Haldar S. Vibrio related diseases in aquaculture and development of rapid and accurate identification methods. J. Marine Sci. Res. Dev. 2012;S1:002. [Google Scholar]
- 24.Philbey A.W. Algal infection in sheep grazing irrigated pasture. Aust. Vet. J. 2001;79:212–214. doi: 10.1111/j.1751-0813.2001.tb14583.x. [DOI] [PubMed] [Google Scholar]
- 25.Yanong R.P.E. Algal dermatitis in cichlids. J. Am. Vet. Med. Assoc. 2002;220:1353–1358. doi: 10.2460/javma.2002.220.1353. [DOI] [PubMed] [Google Scholar]
- 26.Seveso D. The cellular stress response of the scleractinian coral Goniopora columna during the progression of the black band disease. Cell Stress Chap. 2017;22:225–236. doi: 10.1007/s12192-016-0756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai H.C. Pathogenesis of acute hepatopancreatic necrosis disease (AHPND) in shrimp. Fish Shellfish Immunol. 2015;47:1006–1014. doi: 10.1016/j.fsi.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Ryan E.T. The intestinal pathobiome: its reality and consequences among infants and young children in resource-limited settings. J. Infect. Dis. 2013;208:1732–1733. doi: 10.1093/infdis/jit509. [DOI] [PubMed] [Google Scholar]
- 29.Chen W.Y. Microbiome dynamics in a shrimp grow-out pond with possible outbreak of acute hepatopancreatic necrosis disease. Sci. Rep. 2017;7:9395. doi: 10.1038/s41598-017-09923-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otta S.K. Association of dual viral infection with mortality of Pacific white shrimp (Litopenaeus vannamei) in culture ponds in India. Virus Dis. 2014;25:63–68. doi: 10.1007/s13337-013-0180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McArdle A.J. When do co-infections matter? Curr. Opin. Infect. Dis. 2018;31:209–215. doi: 10.1097/QCO.0000000000000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mosser T. Exposure to pairs of Aeromonas strains enhances virulence in the Caenorhabditis elegans infection model. Front. Microbiol. 2015;6:1218. doi: 10.3389/fmicb.2015.01218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labrie L. Effect of sublethal exposure to the trematode Bolbophorus spp. on the severity of enteric septicemia of catfish in channel catfish fingerlings. J. Aquat. Anim. Health. 2004;14:231–237. [Google Scholar]
- 34.Peleg A.Y. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14585–14590. doi: 10.1073/pnas.0805048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabaluk T. Metarhizium brunneum – an enzootic wireworm disease and evidence for its suppression by bacteria symbionts. J. Invertebr. Pathol. 2017;150:82–87. doi: 10.1016/j.jip.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 36.Passos da Silva D. Bacterial multispecies studies and microbiome analysis of a plant disease. Microbiology. 2014;160:556–566. doi: 10.1099/mic.0.074468-0. [DOI] [PubMed] [Google Scholar]
- 37.Derome N. Bacterial opportunistic pathogens of fish, and fungal secondary invaders of fish. Adv. Env. Microbiol. 2016;3:81–108. [Google Scholar]
- 38.Morris J.J. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio. 2012;3 doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zozaya-Valdés E. Microbial community function in the bleaching disease of the marine macroalgae Delisea pulchra. Environ. Microbiol. 2017;19:3012–3024. doi: 10.1111/1462-2920.13758. [DOI] [PubMed] [Google Scholar]
- 40.Ibberson C.B. Co-infecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat. Microbiol. 2017;2:17079. doi: 10.1038/nmicrobiol.2017.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuenschwander S.M. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J. 2018;12:185–198. doi: 10.1038/ismej.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diene S.M. The rhizome of the multidrug-resistant Enterobacter aerogenes genome reveals how new ‘killer bugs’ are created because of a sympatric lifestyle. Mol. Biol. Evol. 2013;30:369–383. doi: 10.1093/molbev/mss236. [DOI] [PubMed] [Google Scholar]
- 43.Georgiades K., Raoult D. Defining pathogenic bacterial species in the genomic era. Front. Microbiol. 2010;1:151. doi: 10.3389/fmicb.2010.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurst M.R.H. Non-spore-forming bacterial entomopathogens: their toxins, hosts and the environment: why be a pathogen. Adv. Env. Microbiol. 2016;3:169–220. [Google Scholar]
- 45.Smith M.J. Ribosomal DNA analyses reveal greater sequence variation in Polymyxa species than previously thought and indicate the possibility of new ribotype-host-virus associations. Environ. Microbiol. Rep. 2013;5:143–150. doi: 10.1111/1758-2229.12026. [DOI] [PubMed] [Google Scholar]
- 46.Parratt S.R. Local adaptation at higher trophic levels: contrasting hyperparasite-pathogen infection dynamics in the field and laboratory. Mol. Ecol. 2017;26:1964–1979. doi: 10.1111/mec.13928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dheilly N.M. Parasite microbiome project: systematic investigation of microbiome dynamics within and across parasite-host interactions. mSystems. 2017;2 doi: 10.1128/mSystems.00050-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stentiford G.D. Hyperspora aquatica n. gn., n. sp. (Microsporidia), hyperparasitic in Marteilia cochillia (Paramyxida), is closely related to crustacean-infecting microspordian taxa. Parasitology. 2017;144:186–199. doi: 10.1017/S0031182016001633. [DOI] [PubMed] [Google Scholar]
- 49.Koskella B. A signature of tree health? Shifts in the microbiome and the ecological drivers of horse chestnut bleeding canker disease. New Phytol. 2017;215:737–746. doi: 10.1111/nph.14560. [DOI] [PubMed] [Google Scholar]
- 50.Menon R. Interactions between species introduce spurious associations in microbiome studies. PLoS Comp. Biol. 2018;14 doi: 10.1371/journal.pcbi.1005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogel C. The Arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol. 2016;212:192–207. doi: 10.1111/nph.14036. [DOI] [PubMed] [Google Scholar]
- 52.Aguirre de Cárcer D. Infection with diverse immune–modulating poxviruses elicits different compositional shifts in the mouse gut microbiome. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berg M., Koskella B. Nutrient-and dose-dependent microbiome-mediated protection against a plant pathogen. Curr. Biol. 2018;28:2487–2492. doi: 10.1016/j.cub.2018.05.085. [DOI] [PubMed] [Google Scholar]
- 54.Lloyd M.M., Pespeni M.H. Microbiome shifts with onset and progression of Sea Star Wasting Disease revealed through time course sampling. Sci. Rep. 2018;8:16476. doi: 10.1038/s41598-018-34697-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kemp K.M. Abundance and multilocus sequence analysis of Vibrio bacteria associated with diseased elkhorn coral (Acropora palmata) of the Florida Keys. Appl. Environ. Microbiol. 2017;84 doi: 10.1128/AEM.01035-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chauhan A. Metagenomic assessment of the eastern oyster-associated microbiota. Genome Announc. 2014;2 doi: 10.1128/genomeA.01083-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Z., Wang L. Metagenomic 16S rRNA sequencing analysis of Pacific oyster (Crassostrea gigas) microbiota from the Puget Sound region in the United States. Genome Announc. 2017;5 doi: 10.1128/genomeA.00468-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.King W.L. Characterisation of the Pacific oyster microbiome during a summer mortality event. Microb. Ecol. 2018;77:502–512. doi: 10.1007/s00248-018-1226-9. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen T.V. Omics approaches to investigate host–pathogen interactions in mass mortality outbreaks of Crassostrea gigas. Rev. Aquacult. 2018 doi: 10.1111/raq.12294. Published online October 12, 2018. [DOI] [Google Scholar]
- 60.Gilbert J.A. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016;535:94. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- 61.Rubio-Portillo E. Biogeographic differences in the microbiome and pathobiome of the coral Cladocora caespitosa in the western Mediterranean Sea. Front. Microbiol. 2018;9:22. doi: 10.3389/fmicb.2018.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aires T. Host and environmental specificity in bacterial communities associated to two highly invasive marine species (Genus Asparagopsis) Front. Microbiol. 2016;7:559. doi: 10.3389/fmicb.2016.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zanefeld J.R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat. Microbiol. 2017;2:17121. doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- 64.Sprockett D. Role of priority effects in the early-life assembly of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2018;15:197–205. doi: 10.1038/nrgastro.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Newcombe G. A hypothetical bottleneck in the plant microbiome. Front. Microbiol. 2018;9:1645. doi: 10.3389/fmicb.2018.01645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meistertzheim A.L. Pathobiomes differ between two diseases affecting reef building coralline algae. Front. Microbiol. 2017;8:1686. doi: 10.3389/fmicb.2017.01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitra A. Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep. 2015;5:16865. doi: 10.1038/srep16865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sutherland K.P. Shifting white pox aetiologies affecting Acropora palmata in the Florida Keys, 1994–2014. Philos. Trans. R. Soc. Lond. B. 2016;371:20150205. doi: 10.1098/rstb.2015.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conn V.M. Endophytic Actinobacteria induce defense pathways in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2008;21:208–218. doi: 10.1094/MPMI-21-2-0208. [DOI] [PubMed] [Google Scholar]
- 70.Gond S.K. Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol. Res. 2014;172:79–87. doi: 10.1016/j.micres.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 71.Kaltenpoth M., Engl T. Defensive microbial symbionts in Hymenoptera. Funct. Ecol. 2014;28:315–327. [Google Scholar]
- 72.Jakuschkin B. Deciphering the pathobiome: intra- and interkingdom interactions involving the pathogen Erysiphe alphitoides. Microb. Ecol. 2016;72:870–880. doi: 10.1007/s00248-016-0777-x. [DOI] [PubMed] [Google Scholar]
- 73.Li M. Facilitation promotes invasions in plant-associated microbial communities. Ecol. Lett. 2019;22:149–158. doi: 10.1111/ele.13177. [DOI] [PubMed] [Google Scholar]
- 74.Xiong J. The underlying ecological processes of gut microbiota among cohabitating retarded, overgrown and normal shrimp. Microb. Ecol. 2017;73:988–999. doi: 10.1007/s00248-016-0910-x. [DOI] [PubMed] [Google Scholar]
- 75.Walke J.B. Amphibian skin may select for rare environmental microbes. ISME J. 2014;8:2207–2217. doi: 10.1038/ismej.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rebollar E.A. Using ‘omics’ and integrated multi-omics approaches to guide probiotic selection to mitigate chytridiomycosis and other emerging infectious diseases. Front. Microbiol. 2016;7:68. doi: 10.3389/fmicb.2016.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walke J.B. Dominance-function relationships in the amphibian skin microbiome. Environ. Microbiol. 2017;19:3387–3397. doi: 10.1111/1462-2920.13850. [DOI] [PubMed] [Google Scholar]
- 78.Schlatter D. Disease suppressive soils: new insights from the soil microbiome. Phytopathology. 2017;107:1284–1297. doi: 10.1094/PHYTO-03-17-0111-RVW. [DOI] [PubMed] [Google Scholar]
- 79.Durán P. Microbial community composition in take-all suppressive soils. Front. Microbiol. 2018;9:2198. doi: 10.3389/fmicb.2018.02198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Knutie S.A. Early-life disruption of amphibian microbiota decreases later-life resistance to parasites. Nat. Commun. 2017;8:86. doi: 10.1038/s41467-017-00119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh V.P. Koch’s postulates, microbial dysbiosis and inflammatory bowel disease. Clin. Microbiol. Infect. 2016;22:594–599. doi: 10.1016/j.cmi.2016.04.018. [DOI] [PubMed] [Google Scholar]
- 82.Broberg M. Integrated multi-omic analysis of host–microbiota interactions in acute oak decline. Microbiome. 2018;6:21. doi: 10.1186/s40168-018-0408-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Doonan J. Genomic analysis of bacteria in the Acute Oak Decline pathobiome. Microb. Genet. 2019;5 doi: 10.1099/mgen.0.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Louca S. Probing the metabolism of microorganisms. Science. 2017;358:1264–1265. doi: 10.1126/science.aar2000. [DOI] [PubMed] [Google Scholar]
- 85.Vorholt J.A. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe. 2017;22:142–155. doi: 10.1016/j.chom.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 86.Le Roux F. Oysters and Vibrios as a model for disease dynamics in wild animals. Trends Microbiol. 2016;24:568–580. doi: 10.1016/j.tim.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 87.Robinson S.W. Current advances in systems and integrative biology. Comput. Struct. Biotechnol. J. 2014;11:35–46. doi: 10.1016/j.csbj.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Vos M.G.J. Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections. Proc. Natl. Acad. Sci. U. S. A. 2017;114:10666–10671. doi: 10.1073/pnas.1713372114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Bruijn I. Exploring fish microbial communities to mitigate emerging diseases in aquaculture. FEMS Microbiol. Ecol. 2018;94 doi: 10.1093/femsec/fix161. [DOI] [PubMed] [Google Scholar]
- 90.Bentzon-Tilia M. Monitoring and managing microbes in aquaculture - towards a sustainable industry. Microb. Biotechnol. 2016;9:576–584. doi: 10.1111/1751-7915.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Egan S., Gardiner M. Microbial dysbiosis: rethinking disease in marine ecosystems. Front. Microbiol. 2016;7:991. doi: 10.3389/fmicb.2016.00991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bosch T.C.G., McFall-Ngai M.J. Metaorganisms as the new frontier. Zoology. 2011;114:185–190. doi: 10.1016/j.zool.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Skillings D. Holobionts and the ecology of organisms: multi-species communities or integrated individuals? Biol. Philos. 2016;31:875–892. [Google Scholar]
- 94.Faure D. Holobiont: a conceptual framework to explore the eco-evolutionary and functional implications of host-microbiota interactions in all ecosystems. New Phytol. 2018;218:1321–1324. doi: 10.1111/nph.15199. [DOI] [PubMed] [Google Scholar]
- 95.Bakken J.S. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011;9:1044–1049. doi: 10.1016/j.cgh.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stentiford G.D. New paradigms to solve the global aquaculture disease crisis. PLoS Pathol. 2017;13 doi: 10.1371/journal.ppat.1006160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ab Rahman S.F.S. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018;267:102–111. doi: 10.1016/j.plantsci.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 98.Lightner D.V. Global transboundry disease politics: the OIE perspective. J. Invertebr. Pathol. 2012;110:184–187. doi: 10.1016/j.jip.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 99.Leggett H.C. Generalism and the evolution of parasite virulence. Trends Ecol. Evol. 2013;28:592–596. doi: 10.1016/j.tree.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 100.Eswarappa S.M. Within-host dynamics of multi-species infections: facilitation, competition and virulence. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038730. [DOI] [PMC free article] [PubMed] [Google Scholar]