Summary
Two opposing viewpoints are held regarding the need for understanding a drug's molecular target and mechanism of action. One extreme viewpoint is that it is unnecessary, because, after all, there are many beneficial drugs in use for which the target and mechanism of action remain unknown. A second extreme viewpoint is that target identification and mechanism of action should be elucidated very early in the drug discovery process due to the tangible benefits provided by this knowledge. I offer an intermediate perspective that considers the complexity of the disease of interest, the existence of a standard-of-care treatment, and the resources available to the investigator.
Subject Areas: Health Sciences, Pharmaceutical Science
Health Sciences; Pharmaceutical Science
Introduction
Drug discovery has evolved from the identification of active substances in traditional medicines to the direct search for new medicines using high-throughput screening campaigns, fragment-based screening, virtual screening, and other approaches (Leveridge et al., 2018). However, under debate are questions regarding the importance of revealing the specific molecular target and mechanism of action (MoA) for a new drug, and when during the process of drug discovery should such information should be obtained.
Some authors have issued strong or categorical statements that target identification (TID) of a new drug and elucidating the MoA is essential early in the drug discovery process (MedChemComm Editorial, 2014), and certainly before initiating human clinical trials (NatMed Editorial, 2010; Moffat et al., 2017). From the author's own experience, the timeline for obtaining such data has been accelerated by reviewers of grants and manuscripts to very early phases in the timeline, even before showing efficacy of a compound in an animal model for the disease of interest. The value of TID and elucidating MoA has been extensively debated with widely varying opinions. Several questions need to be posed and answered first before delving into the broader issue of whether and when TID/MoA is required to forward a new drug to human trials: (1) Is TID/MoA required for US Food and Drug Administration (FDA) approval of a new drug? (2) What value does knowledge of TID/MoA bring to the process of drug discovery and potential future use in humans? (3) When and under what circumstances should drug discovery researchers dedicate time and resources to elucidating TID/MoA? (4) Should the peer review process require TID/MoA in grant applications and submitted manuscripts?
The term “target” is used most often in the scientific literature to describe the specific molecular target (protein, RNA molecule, etc.) that a drug interacts with to initiate a biological response. MoA is often used synonymously with “target,” although some investigators reserve this term to describe the drug's action at a higher level of biological complexity, referring to a cell signaling system or processes that are impacted by the drug through its interaction with a specific molecular target. The simplest examples of molecular targets include inhibitors of enzymes, agonists, or antagonists of cell surface receptors and blockers of plasma membrane transporters. For instance, the most widely used drug in the world, acetylsalicylic acid or aspirin, is classed as a non-selective cyclooxygenase inhibitor (Vane and Botting, 2003). Interestingly, various preparations or decoctions containing salicylate were used for centuries for the treatment of fever or pain before the discovery that acetylsalicylic acid inhibited cyclooxygenase activity in 1971. The drug memantine, used for managing Alzheimer disease, exhibits antagonist activity to the N-methyl-D-aspartate receptor (Rogawski and Wenk, 2003). Fluoxetine, an antidepressant classed as a selective serotonin reuptake inhibitor, blocks the activity of serotonin plasma membrane transporters and the cellular import of this neurotransmitter (Owens et al., 2001). For this discussion, TID/MoA are used interchangeably as they go hand in hand. Knowing the specific molecular target immediately commands the question of how a drug's interaction with that target alters processes at the cellular or tissue level to effect a phenotypic change. TID/MoA is brought to the forefront when considering two general approaches used to assay and screen compounds while searching for new drugs during the preclinical phase of drug discovery: target-based screens and phenotypic screens. A brief discussion of these two general approaches for drug discovery is necessary because the priority a researcher assigns to TID/MoA often aligns with their preferred approach.
Target and Phenotypic Screens
Target-based screens offer a reductionist approach to drug discovery, generally employing in vitro biochemical assays to search through a library of small molecules. They are based on developing assays to detect compounds that interact with a specific molecular entity, most often a protein, which is known or hypothesized from basic research to be involved in processes impaired in a disease of interest. Phenotypic screens, in contrast, employ a holistic approach most often at the level of the cell, although tissues, organs, or even whole animals can be employed for obtaining the desired readouts (Aulner et al., 2019; Clatworthy et al., 2018). They are based on assays that test whether small molecules exert a desirable phenotypic change in the biological material that is utilized (Clatworthy et al., 2018; Zheng et al., 2013). Perhaps the most popular type of phenotypic assay is the high-content assay, which employs imaging technologies often with cell-based readouts to follow the effect of compounds on cultured cells (Varkuti et al., 2020). Ironically, phenotypic screens formed the foundation for drug discovery before target-based screens became popular in the 1980s (Zheng et al., 2013).
Target-based strategies offer numerous advantages for drug discovery (Zheng et al., 2013). The strategy is efficient, cost effective, and offers extremely high throughput given that they often feature an endpoint assay for the activity of a molecule on a per-well basis using microtiter plates with 384 or 1,536 wells. The analog development phase of drug discovery (Hughes et al., 2011) used to dial in desirable properties and dial out undesirable ones is accelerated because the initial screens are designed with prior knowledge of a specific molecular target hypothesized or known to be relevant to a disease of interest. For instance, the lead compound in developing the drug imatinib, which inhibits a chimeric Abl protein-tyrosine kinase that causes chronic myelogenous leukemia, was a compound that inhibited the protein-serine/threonine kinase, protein kinase C (Roskoski, 2015). Analog development from this lead compound led to derivatives that inhibited protein-tyrosine kinases and abolished the activity against protein kinase C. Thus, an effective medicine was developed by chemically migrating the activity of the lead compound away from the initial target to a related target, all because the specific molecular target was known in advance. Furthermore, with an effective drug in hand and knowledge of the molecular target, drug developers can design new generations of drugs from the original with increased efficacy and reduced side effects (Zheng et al., 2013). The value of TID to personalized medicine is powerfully illustrated by the drug trastuzumab and its derivatives. Trastuzumab was a first-in-class immunotherapeutic targeting the HER2 tyrosine kinase receptor and is used to treat patients with HER2-overexpressing breast tumors (Barginear et al., 2013; Lewis Phillips et al., 2008). Obviously, such an important advance was impossible without prior knowledge about HER2 expression levels in some types of breast cancer cells. The combination therapy that eliminated HIV-1 infection from causing death within a year after infection to providing a near normal lifespan highlights the importance of TID. The most recent guidelines for HIV-1 treatment recommend a two- or three-part combination of nucleoside analogs to inhibit the virally encoded reverse transcriptase enzyme and a small molecule that inhibits the viral integrase protein (Saag et al., 2018; Guidelines, 2019). These advances depended entirely on knowledge about the specific proteins required for the replication of the viral genome and its integration into the host's genome gained from basic science research.
A major disadvantage of the target-based approach is that an in-depth understanding of the cause of the disease is required for success, as illustrated by the breast cancer and HIV-1 examples described earlier. In both cases, there existed essential prior knowledge that provided a rational TID/MoA for drug development. One major reason that drugs discovered from target-based strategies fail in clinical trials has been incomplete preclinical target validation, e.g., obtaining crystal clear evidence that the target chosen is intimately related to the disease of interest and should provide therapeutic value (Gashaw et al., 2014; Zheng et al., 2013). Furthermore, there are many complex diseases, especially when considering brain disorders, for which few reasonable molecular targets are known. Extensive efforts have been made to develop therapeutics for Alzheimer disease based on the convincing biological evidence that Aβ oligomers/amyloid drives the onset of genetic forms of the disease, but to date, the costly clinical trials for these have failed (Aisen, 2019). There are several possible reasons for this outcome, but included among them is that sporadic forms of the disease may be spurred by a different mechanism. In addition, key molecular targets have been difficult to find even for the bacterial disorder tuberculosis, due to its impenetrable cell wall and its persistence in several different microenvironments within the host (Kumar et al., 2017). Given these difficulties, one extreme opinion could be that drug development for some indications should wait until basic science provides a clear path forward. However, this defeatist attitude sidelines the millions of individuals suffering from such complex conditions. A final issue that exists from target-based strategies and the assumption that the drug's interaction with the original target is that science has a way of throwing in surprises. A recent study found that CRISPR-based genomic knockouts of 6 different protein targets for 10 different anti-cancer drugs failed to block the drugs' killing effects on cancer cells (Lin et al., 2019), indicating that the original targets were imposters (see also Settleman et al., 2018; Giuliano et al., 2018). So even if one believes during the process of drug development that a certain target should offer therapeutic value, in the end, it may prove to be a false target.
Phenotypic-based drug discovery offers distinct advantages over target-based drug discovery. As phenotypic screens are performed with cells, tissues, organs, or whole animals, they are performed in a biological context rather than in an in vitro, cell-free assay. Thus, they may mimic the biological conditions required for therapeutic efficacy, reducing the possibility of failure at subsequent steps, and providing an enrichment in compounds with increased potential for treating complex diseases and effective translation into the clinic. They also cast a broader net. The phenotypic change observed with a putative drug is at the level of the cell or higher, and because of this, the drug could be interacting with multiple targets that produce the desired effect. Indeed, as any drug might interact with multiple targets that differ across cell types and tissues, one could argue that it is best to aim as high in the complexity hierarchy as possible, because the ultimate effect on the organism is a balance between all the positive and negative effects of a drug. Recent research in human genetics offers a good analogy. Candidate gene approaches focus on a single or small number of genes that are thought to be involved in a disease of interest, analogous to the pre-selection of molecular targets for target-based screens. In contrast, genome-wide screens search the genome for DNA sequence polymorphisms that could identify many genes involved in the disease. Thus, phenotypic screens usually provide more hits as starting points for drug discovery. Furthermore, the strategy is agnostic to the molecular target; it is not contaminated by preconceived ideas of which molecular targets are most relevant. A strong argument for adopting a phenotypic screen is that human biology is complicated and the clues available to develop effective therapeutics especially for complex indications are limited. Phenotypic screens also offer the potential for providing alternative pathways for developing therapeutics by uncovering the hidden biology in human disease.
An increased emphasis in phenotypic screening occurred from a frequently cited study (Swinney and Anthony, 2011) reporting that this strategy offered a superior approach over target-based strategies in finding first-in-class drugs (see also Moffat et al., 2017; Swinney and Xia, 2014). However, this conclusion has been challenged using an alternative definition of what constitutes a phenotypic screen and considering data across a longer time frame (Eder et al., 2014). This debate appears to have polarized the drug discovery community into two camps, segregated, in part, due to the personal biases that influence perspective and decision-making processes. For instance, biochemists and molecular biologists may generally be more enamored with target-based strategies because of their focus at the level of the molecule, whereas neuroscientists, immunologists, and cardiovascular researchers, as examples, may be partial to phenotypic strategies due to the complexity of the organ systems of their interest. These biases, along with perhaps instincts of territorial protection, need to be recognized and discarded as much as humanly possible in the interest of providing the world's population with new medicines. Both strategies should be accepted as valuable for drug discovery and employed strategically. Target-based strategies would be the priority when the basic science of a disease has uncovered attractive and validated targets; phenotypic strategies would the priority for complex indications that remain in search of the cause.
Phenotypic screens do have their downside. They are generally less efficient, more expensive, and offer a lower throughput than target-based screens. Most importantly, phenotypic approaches are not initiated with a specific molecular target in mind (Brown and Wobst, 2020; Moffat et al., 2017) and so the target remains unknown until that research avenue is broached. This could reduce the probability of dialing out potential side effects, as the target(s) and off-target(s) are hidden. In addition, although more hits are generally recovered from phenotypic screens as mentioned earlier, having multiple chemical entities in hand may offer difficulties in choosing the best lead to optimize (Leveridge et al., 2018). Moreover, TID is a complex and resource-intensive process that can take years or decades to solve (MedChemComm Editorial, 2014; Lederman, 2014). Lithium offers a prime example. This drug forms the first line of pharma for long-term management of bipolar disorder with its use dating back to the nineteenth century (Won and Kim, 2017). Although there is growing evidence to indicate that it may have multiple MoAs, including direct inhibition of glycogen synthase kinase, potentiation of the cell's mechanisms for protection against oxidation, inhibition of inositol monophosphatase, and enhancement of the actions of the cAMP response element-binding protein; a clear understanding of how lithium stabilizes mood remains unknown. This is despite research reported in over 4,000 articles in PubMed across the last 10 years identified using the keywords “lithium mechanism of action.” Given this downside, those that favor phenotypic screens emphasize their advantages described above over TID during early stages of the drug discovery process.
When Is Target Identification Needed?
This brings us back to the three unanswered questions posed initially that stem from the dichotomy of target versus phenotypic strategies. (1) Is MoA absolutely required for FDA approval of a new drug? (2) When and under what circumstances should drug discovery researchers dedicate time and resources to TID/MoA? (3) Should the peer review process require TID/MoA in grant applications and submitted manuscripts? The answers to these questions are a matter of perspective and the weight one assigns to the advantages and disadvantages of the two approaches. Nevertheless, a fine-grained consideration of the constraints and circumstances of different drug discovery projects offers perhaps an intermediate perspective to the polarized ones that have been expressed to date.
First, understanding the MoA for a new drug is not required for FDA approval (Brown and Wobst, 2020; Moffat et al., 2017). Between 10% and 20% of currently approved drugs have no known target or clear MoA (Moffat et al., 2017). In addition, other regulatory agencies in the world beyond the FDA will approve a new drug as long as it is safe and effective in disease treatment (Zheng et al., 2013). And as indicated with the examples of aspirin and lithium earlier, some drugs with an unknown or uncertain MoA have been used for decades or longer. The fact that there exist today efficacious drugs that have no target or MoA argues convincingly that the absence of TID/MoA should not hold a drug back from reaching those whose lives would be enhanced with its availability.
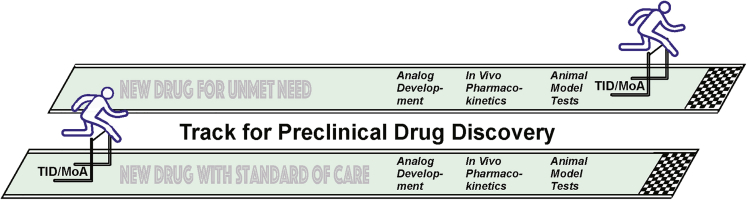
Second, when during a phenotypic drug discovery effort should TID/MoA be prioritized? The advantages of TID for a new drug are clear as discussed earlier. However, are there factors or conditions that should influence when across the stages of drug development that TID/MoA research should be pursued? Two huge factors that should be considered are the need for the new therapeutic and the complexity of the indication. For drug discovery aimed at developing second- or third-generation therapeutics, one could reasonably argue that TID/MoA should be obtained early in the process to add value, as existing treatments are available to those in need (Figure 1). For others, especially the neurological and psychiatric diseases for which TID/MoA is much more elusive due to the complexity of the central nervous system and how it functions, it would be prudent to postpone TID/MoA studies until after analog development, in vivo pharmacokinetics, and in vivo efficacy in an animal model are demonstrated (Figure 1). It makes little sense to set aside large resources for TID/MoA research if the new drug is fated to fail the threshold for in vivo efficacy. The drug discovery process is risky, and some academic investigators have argued that MoA should be pursued early to help discover new biology even if the drug discovery efforts end up failing. This path allows the investigator to provide a return for the investment that NIH or other agencies have made. The strong counterarguments to this perspective are that it dilutes an investigator's effort from the main goal of finding an effective medication, and the drug discovery goal could be completely derailed by the consumption of resources from TID/MoA studies.
Figure 1.
Paths for Preclinical Drug Discovery
The perspective expressed in the previous paragraph aligns with the general viewpoints of small biotech firms (Haasen et al., 2017; Moffat et al., 2017). Small companies are generally prepared to forward new drugs even into clinical trials without TID/MoA, as it diverts limited resources away from the central goal of delivering a new medicine. Drug discovery investigators with limited resources in academic positions would fall into the same camp. Big pharma, in contrast, views TID/MoA as crucial for moving compounds forward due to the expenses involved in late-phase clinical trials and the potential for toxic side effects (Weaver and Valentin, 2018). Obviously, the process and timeline for developing each new drug is unique, and this demands a flexible viewpoint rather than dogmatism.
Third, should the peer review process require investigators to include TID/MoA pursuits in grant applications and submitted manuscripts that focus on phenotypic assays and screens? The answer to this question is not a simple “yes” or “no,” but like all complex issues, “it depends.” However, any reasonable answer starts with having reviewers and editors making judgments based on the strength of the arguments that the author presents. After that, all the circumstances surrounding the proposed grant project or scientific report need to be considered. Some of these are rather obvious, such as the disease being targeted in drug discovery research, the resources available to the investigator, and the anticipated difficulty and timeline in obtaining hard data on TID/MoA, and so forth. As mentioned earlier in the article, this author holds the opinion that reviewers should not require TID/MoA data before in vivo efficacy data for complex indications of unmet need. Each situation will have its own unique set of circumstances beyond those listed that needs to be taken into account.
In summary, I argue that the prioritization of when to pursue TID/MoA studies depends on the nature of the indication and whether there exists an unmet need. There is no debate whether TID is important. The multiple arguments for knowing the target have been discussed in the article and are compelling. The debate concerns “when” during the preclinical phases of a phenotypic drug discovery project should TID/MoA become actionable. This should generally be early in the process for indications that have a standard-of-care treatment. However, for indications with an unmet need, efforts made for TID/MoA should occur at a time after showing efficacy of a new drug in an animal model, due to the extensive demands on resources that the effort may take.
Acknowledgments
I would like to thank my colleagues, Ted Kamenecka, Louis Scampavia, and Timothy Spicer and two anonymous reviewers for their comments on the manuscript. Funding for drug discovery research to the author has been provided by NIH grant 5R01MH109957.
References
- Aisen P.S. Editorial: failure after failure. What next in AD drug development? J. Prev. Alzheimers Dis. 2019;6:150. doi: 10.14283/jpad.2019.23. [DOI] [PubMed] [Google Scholar]
- Aulner N., Danckaert A., Ihm J., Shum D., Shorte S.L. Next-generation phenotypic screening in early drug discovery for infectious diseases. Trends Parasitol. 2019;35:559–570. doi: 10.1016/j.pt.2019.05.004. [DOI] [PubMed] [Google Scholar]
- Barginear M.F., John V., Budman D.R. Trastuzumab-DM1: a clinical update of the novel antibody-drug conjugate for HER2-overexpressing breast cancer. Mol. Med. 2013;18:1473–1479. doi: 10.2119/molmed.2012.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.G., Wobst H.J. Opportunities and challenges in phenotypic screening for neurodegenerative disease research. J. Med. Chem. 2020;63:1823–1840. doi: 10.1021/acs.jmedchem.9b00797. [DOI] [PubMed] [Google Scholar]
- Clatworthy A.E., Romano K.P., Hung D.T. Whole-organism phenotypic screening for anti-infectives promoting host health. Nat. Chem. Biol. 2018;14:331–341. doi: 10.1038/s41589-018-0018-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder J., Sedrani R., Wiesmann C. The discovery of first-in-class drugs: origins and evolution. Nat. Rev. Drug Discov. 2014;13:577–587. doi: 10.1038/nrd4336. [DOI] [PubMed] [Google Scholar]
- Gashaw I., Ziegelbauer K., Asadullah K., Bechem M. Molecular target validation in preclinical drug discovery. Drug Target Rev. 2014;1 https://www.drugtargetreview.com/article/821/molecular-target-validation-in-preclinical-drug-discovery/ [Google Scholar]
- Giuliano C.J., Lin A., Smith J.C., Palladino A.C., Sheltzer J.M. MELK expression correlates with tumor mitotic activity but is not required for cancer growth. Elife. 2018 doi: 10.7554/eLife.32838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidelines . Department of Health and Human Services. DDHS Panel on Antiretroviral Guidelines for Adults and Adolescents; 2019. Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents Living with HIV.https://aidsinfo.nih.gov/contentfiles/lvguidelines/adultandadolescentgl.pdf [Google Scholar]
- Haasen D., Schopfer U., Antczak C., Guy C., Fuchs F., Selzer P. How phenotypic screening influenced drug discovery: lessons from five years of practice. Assay Drug Dev. Technol. 2017;15:239–246. doi: 10.1089/adt.2017.796. [DOI] [PubMed] [Google Scholar]
- Hughes J.P., Rees S., Kalindjian S.B., Philpott K.L. Principles of early drug discovery. Br. J. Pharm. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Chettiar S., Parish Tanya. Current challenges in drug discovery for tuberculosis. Expert Opin. Drug Discov. 2017;12:1–4. doi: 10.1080/17460441.2017.1255604. [DOI] [PubMed] [Google Scholar]
- Lederman S. Life Science Leader Magazine; 2014. The Evolving Role of Drug Mechanism of Action in Drug Discovery and Development.https://www.lifescienceleader.com/doc/the-evolving-role-of-drug-mechanism-of-action-in-drug-discovery-and-development-0001 [Google Scholar]
- Leveridge M., Chung C.-W., Gross J.W., Phelps C.B., Green D. Integration of lead discovery tactics and the evolution of the lead discovery toolbox. SLAS Discov. 2018;23:881–897. doi: 10.1177/2472555218778503. [DOI] [PubMed] [Google Scholar]
- Lewis Phillips G.D., Li G., Dugger D.L., Crockers L.M., Parsons K.L., Mai E., Blattler W.A., Lambert J.M., Chair R.V.J., Lutz R.J. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008;68:9280–9290. doi: 10.1158/0008-5472.CAN-08-1776. [DOI] [PubMed] [Google Scholar]
- Lin A., Giuliano C.J., Palladino A., John K.M., Abramowicz C., Yuan M.L., Sausville E.L., Lukow D.A., Liu L., Chait A.R. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 2019 doi: 10.1126/scitranslmed.aaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MedChemComm Editorial Chemical biology for target identification and validation. Med. Chem. Commun. 2014;5:244–246. [Google Scholar]
- Moffat J.G., Vincent F., Lee J.A., Eder J., Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- NatMed Editorial Mechanism matters. Nat. Med. 2010;16:347. doi: 10.1038/nm0410-347. [DOI] [PubMed] [Google Scholar]
- Owens M.J., Knight D.L., Nemeroff C.B. Second-generation SSRIs: human monoamine transporter binding profile of escitalopram and R-fluoxetine. Biol. Psychiatry. 2001;50:345–350. doi: 10.1016/s0006-3223(01)01145-3. [DOI] [PubMed] [Google Scholar]
- Rogawski M.A., Wenk G.L. The neuropharmacological basis for the use of memantine in the treatment of Alzheimer's disease. CNS Drug Rev. 2003;9:275–308. doi: 10.1111/j.1527-3458.2003.tb00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskoski R., Jr. A historical overview of protein kinases and their targeted small molecule inhibitors. Pharmacol. Res. 2015;100:1–23. doi: 10.1016/j.phrs.2015.07.010. [DOI] [PubMed] [Google Scholar]
- Saag M.S., Benson C.A., Gandhi R.T., Hoy J.F., Landovitz R.J., Mugavero M.J., Sax P.E., Smith D.M., Thompson M.A., Buchbinder S.P. Antiretroviral drugs for treatment and prevention of HIV infection in adults: recommendations of the International antiviral Society-USA panel. JAMA. 2018;320:379–396. doi: 10.1001/jama.2018.8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settleman J., Sawyers C.L., Hunter T. Challenges in validating candidate therapeutic targets in cancer. Elife. 2018;7 doi: 10.7554/eLife.32402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinney D.C., Anthony J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011;10:507–519. doi: 10.1038/nrd3480. [DOI] [PubMed] [Google Scholar]
- Swinney D.C., Xia S. The discovery of medicines for rare diseases. Future Med. Chem. 2014;6:987–1002. doi: 10.4155/fmc.14.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vane J.R., Botting R.M. The mechanism of action of aspirin. Thromb. Res. 2003;110:225–228. doi: 10.1016/s0049-3848(03)00379-7. [DOI] [PubMed] [Google Scholar]
- Varkuti B.H., Kepiro M., Liu Z., Vick K., Avchalumov Y., Pacfico R., MacMullen C.M., Kamenecka T.M., Puthanveettil S.V., Davis R.L. Neuron-based high-content assay and screen for CNS active mitotherapeutics. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aaw8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R.J., Valentin J.-P. Today’s challenges to de-risk and predict drug safety in human “mind-the-gap. Soc. Toxicol. 2018;167:307–321. doi: 10.1093/toxsci/kfy270. [DOI] [PubMed] [Google Scholar]
- Won E., Kim Y.K. An oldie but goodie: lithium in the treatment of bipolar disorder through neuroprotective and neurotrophic mechanisms. Int. J. Mol. Sci. 2017;18:2679. doi: 10.3390/ijms18122679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W., Thorne N., McKew J.C. Phenotypic screens as a renewed approach for drug discovery. Drug Discov. Today. 2013;18:1067-1073. doi: 10.1016/j.drudis.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]