Abstract
Genetic and pharmacological intervention studies have identified evolutionarily conserved and functionally interconnected networks of cellular energy homeostasis, nutrient-sensing, and genome damage response signaling pathways, as prominent regulators of longevity and health span in various species. Mitochondria are the primary sites of ATP production and are key players in several other important cellular processes. Mitochondrial dysfunction diminishes tissue and organ functional performance and is a commonly considered feature of the aging process. Here we review the evidence that through reciprocal and multilevel functional interactions, mitochondria are implicated in the lifespan modulation function of these pathways, which altogether constitute a highly dynamic and complex system that controls the aging process. An important characteristic of these pathways is their extensive crosstalk and apparent malleability to modification by non-invasive pharmacological, dietary, and lifestyle interventions, with promising effects on lifespan and health span in animal models and potentially also in humans.
Keywords: Aging, Mitochondria, DNA repair, Energy, Lifespan, Metabolism
1. Introduction
Human life expectancy has been increasing worldwide since the nineteenth century. This is an impressive achievement, but the increasing elderly population entails major health and socioeconomic challenges in the years ahead. Meeting these challenges requires multidisciplinary approaches and increased knowledge about the underlying biochemical mechanisms of aging.
Aging is commonly described as a progressive decline in organismal function over time associated with an increasing risk of disease and death. Aging is complex, involves multiple and interconnected processes, is arguably universal, and shows similarities but also variation both in rate and in physiological traits across and within species (Jones et al., 2014; Lopez-Otin et al., 2013). In humans, longevity is influenced by heredity, as well as, diverse environmental factors such as nutrients, lifestyle, pollution, medical care, childhood stress, and socioeconomic status (Blackburn et al., 2015; Ridout et al., 2017).
At the molecular level, aging is thought to be a consequence of a lifelong accumulation of stochastic damage to tissues and cellular components such as DNA, proteins, and lipids (Kirkwood, 2005). Genetic studies in model organisms have identified signaling pathways that can influence the rate of aging and lifespan across species, with prominent features of these pathways being evolutionarily conserved and having extensive functional interactions. These pathways likely evolved early in the evolutionary history of living cells as part of an adaptation mechanism to integrate key biological processes in response to the availability of the intra- and extracellular nutrients and energy substrates. However, a key question that has long interested biologists is how evolutionary forces could have acted upon these pathways to favor an aging process that is obviously not maximally beneficial to the fitness of the individual.
Although it has sometimes been suggested that aging is directly programmed, e.g. as a mechanism to sacrifice the individual in favor of kin, careful analysis shows such arguments to be generally untenable (Kowald and Kirkwood, 2016). Instead, the evolution of aging is explained through the fact that natural selection is relatively weak in its control of gene actions in later life and by the principle of optimization of allocation of resources between, on the one hand, growth and reproduction, and on the other hand, maintenance and repair (Kirkwood and Austad, 2000). In particular, the “disposable soma theory” of aging recognizes that under pressure of natural selection in the wild, where individuals mostly die relatively young, the optimal allocation of nutrients and energy would have placed a limited priority on the long-term maintenance of soma when balanced against the more urgent priorities of growth and reproduction. Accordingly, longer-lived species, which generally have adaptations (wings, shells etc.) enabling better survival in the wild, allocate more resources on maintenance than short-lived species. The significance of this theory is that it provides a direct connection between evolutionary understanding of why aging occurs with the extensive evidence for how aging is regulated through the signaling pathways that sense nutrient and cellular energy availability to control cell metabolism and energy production accordingly. In line with this view, the lifespan extension effects of dietary restriction and of pharmacological treatments including central regulators of metabolism in organisms as diverse as yeast and mice often converge on these signaling pathways one way or another.
If aging is driven by damage, the key challenges are to identify which cellular components are most likely to be implicated among the primary mechanisms underlying functional decline, and how the highly networked pathways involving damage and repair can best be dissected and understood. Mitochondria are essential for normal cellular and organismal function, and defects in the pathways that control mitochondrial DNA (mtDNA) maintenance and mitochondrial function are pathogenic (DeBalsi et al., 2017). Proper mitochondrial function and maintenance require the action of multiple mechanisms (Box 1). Subtle alterations and irregularities in these processes impair mitochondrial homeostasis and function with age (Kwong and Sohal, 2000), and are linked to age-associated functional declines such as sarcopenia (Ibebunjo et al., 2013; Marzetti et al., 2013), insulin resistance (Petersen et al., 2003), and brain aging (Mattson and Arumugam, 2018).
Box 1. The intricate mechanisms of maintaining mitochondrial function and homeostasis.
Mitochondria are the primary sites for the production of cellular ATP, and also play a central role in several other key cellular processes such as apoptosis (Gahl et al., 2016), cytoplasmic calcium buffering (Drago et al., 2012), and reactive oxygen species (ROS) mediated signaling pathways (Shadel and Horvath, 2015). The dual location of mitochondrial genes, the dynamic nature of mitochondria, the bidirectional mitochondria-nucleus signaling, together with the mechanisms for the degradation of dysfunctional mitochondria, form an intricate and interconnected network important for normal mitochondrial and cellular functions.
Each mitochondrion contains multiple copies of ˜16.6 kb circular mtDNA molecules. mtDNA contains 37 genes that code for two ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and 13 proteins. mtDNA is crucial for organismal development, normal function, and survival.
The mechanism of mtDNA replication is complex, and several models have been proposed (Holt and Reyes, 2012). The enzymes critical for mtDNA replication (mtDNA replisome) in vitro, are the heterodimer mtDNA polymerase γ, DNA helicase Twinkle, mitochondrial single-strand DNA-binding protein (mtSSB), mitochondrial RNA polymerase (POLRMT), topoisomerase 3α and DNA ligase III (Holt and Reyes, 2012; Nicholls et al., 2018).
mtDNA is organized into structures called nucleoids in close association with the mitochondrial inner membrane (Brown et al., 2011). Because the mitochondrial inner membrane is a major site of mitochondrial ROS production, (Cadenas and Davies, 2000) mtDNA is expected to constantly undergo oxidative damage. DNA base excision repair (BER) is the prominent DNA repair pathway for repair of oxidative DNA damage, and likely to be the most active DNA repair pathway in human mitochondria (Sykora et al., 2012). Other DNA repair activities in mammalian mitochondria have been reported, including mismatch repair (de Souza-Pinto et al., 2009), and double-strand break repair (Bacman et al., 2009; Tadi et al., 2016).
mtDNA repair and replication require a steady supply of nucleotides that are recycling inside the mitochondria, as well as those imported from the cytoplasm. Defects in genes that regulate the supply of mitochondrial nucleotides cause mtDNA depletion syndromes (MDS) characterized by severe reduction in mtDNA content and impaired mitochondrial function that usually affect one or several organs (El-Hattab and Scaglia, 2013).
Mitochondria form interconnected networks, and adapt cell-type specific morphology and undergo fusion and fission (Giedt et al., 2016). The major factors in the fusion process include mitofusin 1(MFN1), mitofusin 2 (MFN2), and optic atrophy type 1 (OPA1). The central fission proteins include the dynamin-related protein 1 (DRP1), mitochondrial fission factor (MFF), mitochondrial fission 1 protein (Fis1), and mitochondrial dynamic proteins MiD49, and MiD51 (Chan, 2012; Loson et al., 2013). Mutations in MFN2 and OPA1 cause neurological disorders, Charcot-Marie-Tooth disease (Zuchner et al., 2004), and Autosomal Dominant Hereditary Atrophy (ADOA) (Delettre et al., 2000), respectively. Diminished expression of OPA1 has been reported in cells from ataxia with oculomotor apraxia type 1 (AOA1) patients (Zheng et al., 2019).
Autophagy is the mechanism for targeting cytoplasmic material from endogenous and exogenous sources to lysosomes for degradation, such as protein aggregates and organelles (Galluzzi et al., 2017). Selective autophagic elimination of mitochondria is called mitophagy. Mitophagy is an important control system for the removal of damaged mitochondria (Rodger et al., 2018), during development and differentiation (Schweers et al., 2007), and other conditions (Al Rawi et al., 2011; Fang et al., 2019). PTEN-induced putative kinase 1 (PINK1) and Parkin, a cytosolic E3 ligase, are key factors in mediating mitophagy. Mutations in PINK1 and Parkin are linked to the hereditary form of Parkinson disease (Kitada et al., 1998; Valente et al., 2004). A number of factors and receptors for the initiation and formation of mitophagy cargo (mitophagosome) have been identified (Rodger et al., 2018).
The processes of fusion, fission, and mitophagy are tightly connected and collectively determine mitochondrial morphology and dynamics (Sebastian et al., 2017).
In this review, we discuss the highly dynamic processes that connect mitochondrial homeostasis to DNA metabolism and the conserved signaling pathways that control lifespan. We also discuss some frequently reported non-invasive life style changes and pharmacological interventions that extend lifespan and health span in a range of organisms by acting on those processes.
2. The apparent dichotomy of mitochondrial stress and ROS in disease and longevity
For mitochondria to respond properly to local energy need and because of their other key functions, signal transduction lines of communication have evolved between mitochondria and the nucleus called anterograde (nucleus to mitochondria) and retrograde (mitochondria to nucleus). Such signaling can be enhanced in response to nuclear DNA damage (Fang et al., 2016b), mtDNA damage (Chae et al., 2013), and mitochondrial stress, such as accumulation of misfolded proteins within mitochondria (Zhao et al., 2002). The key mediators of the mitochondrial retrograde signaling pathways include ROS, Ca2+, and the cellular AMP/ATP ratio.
High levels of ROS are generated in abnormal mitochondria, but also ROS produced under regular physiological conditions can generate oxidative damage to cells and tissues, and have long been considered a significant driving force in the aging process, a concept known as the “free radical theory” of aging (Harman, 1956). Emerging evidence, however, has resulted in a more nuanced view and ROS are now recognized as important signaling molecules in a number of key biological processes (Sena and Chandel, 2012), including longevity (Schulz et al., 2007). For example, while severe defects in mitochondrial function are the primary cause of a number of human diseases (Schapira, 2012), in C. elegans mild mitochondrial stress and modest increase in mitochondrial ROS levels seem to increase lifespan (Feng et al., 2001; Lee et al., 2003b). This is reminiscent of hormesis, the concept based on observations that exposure to low doses of a toxin might have beneficial effects for the organisms, for instance, by activating cellular defense systems against stress (Mattson, 2008). Accordingly, uncritical use of antioxidant supplementation to lower ROS levels seems to have adverse effects on health and may even be linked to increased mortality (Bjelakovic et al., 2014; Ristow et al., 2009). On the other hand, in longer living animal models, multiple studies have shown that enhanced mitochondrial antioxidant capacity can extend lifespan and health span and may even explain the exceptional longevity of the naked mole rat (Munro et al., 2019; Schriner et al., 2005; Shabalina et al., 2017). These results illustrate the challenging work of understanding the complex nature of the antioxidant system and mitochondrial ROS-mediated signaling and their role in health and aging, which are further complicated by the study design, type of model system (i.e. intact cells vs. isolated mitochondria, cell lines vs. animals, short-lived vs. long-lived animals, type of tissue studied), and the limitations of the available techniques (Kalyanaraman et al., 2012; Munro et al., 2019; Sanz, 2016).
3. Key nutrient and energy sensing signaling pathways that control longevity in laboratory animal models
3.1. The somatotropic axis
Genomic and intervention studies in humans and other animals have identified the evolutionarily conserved somatotropic axis consisting of growth hormone (GH), insulin and insulin-like-growth factor 1(IGF-1) signaling (IIS), and their receptors and downstream effectors, as a key signaling pathway in delaying aging and in promoting longevity across species (Brown-Borg, 2015; Kenyon et al., 1993; Morris et al., 1996; Tatar et al., 2001) (Fig. 1).
Fig. 1.
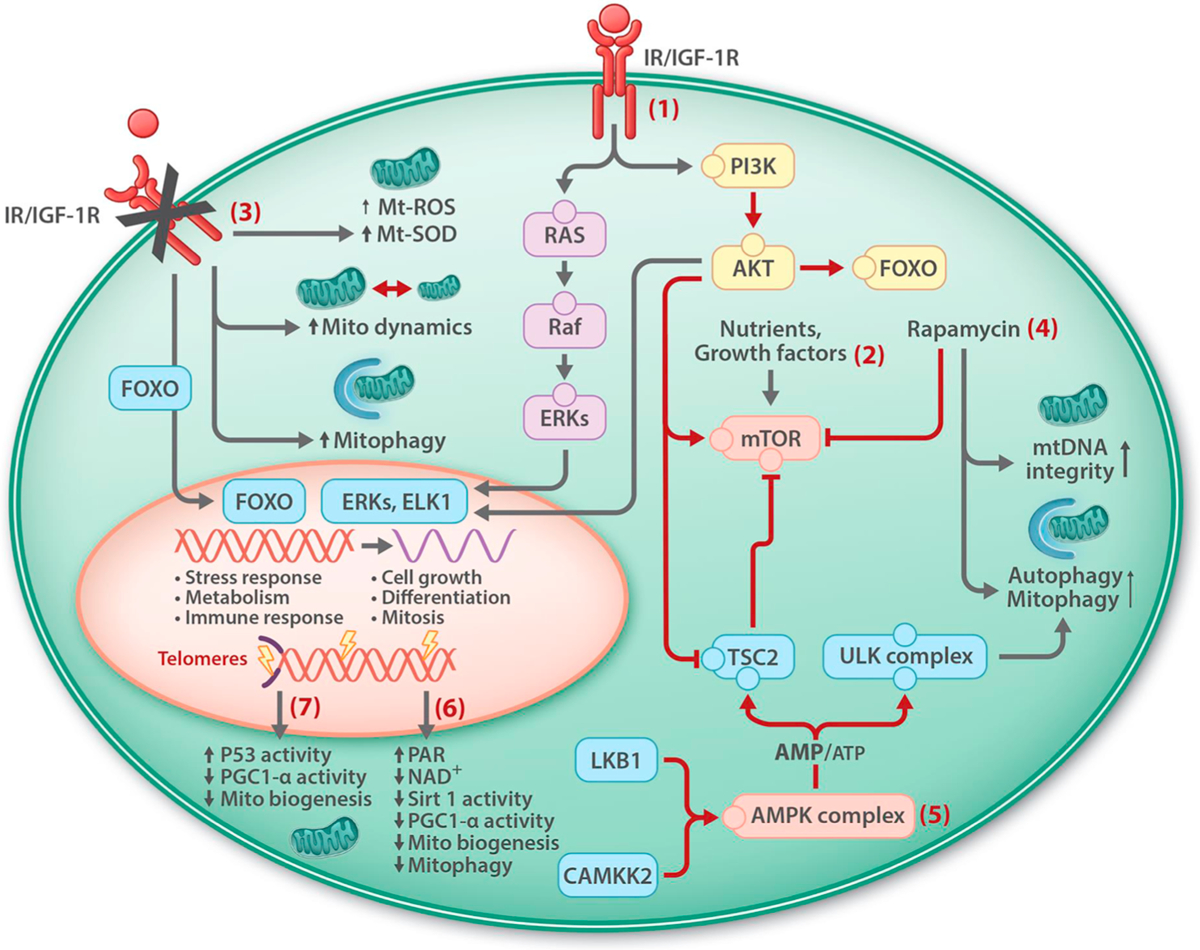
Mitochondrial homeostasis and signaling pathways that regulate longevity and health span. Signaling pathways that are activated by insulin and insulin-like growth factor 1 receptors (IR/IGF-1R) includes PI3K-Akt and Ras-Raf-ERK-MAPK pathways leading to the retention of FOXO in cytoplasm, cell growth and proliferation (1), and stimulation of mTOR, and inhibition of autophagy/mitophagy (2). Inhibition of IR/IGF-1R stimulates pathways that regulate mitochondrial maintenance and leads to the translocation of FOXO into the nucleus where it controls the expression of a number of genes including those involved in stress response and immunity (3). Rapamycin (4) and activated AMPK, following increased AMP/ATP ratio (5), inhibit mTOR and stimulate mitochondrial maintenance. NAD+ over-consumption following DNA damage, and increased PARylation, reduces SIRT1 activity and mitochondrial biogenesis and maintenance (6). Damage to telomeres may impair mitochondrial biogenesis through activation of p53 and reduced PGC1-a activity (7).
GH, also called somatotropin, is a peptide hormone produced and secreted by somatotrope cells in the anterior lobe of the pituitary gland. A key function of GH is the stimulation of IGF-1 production by the hepatic cells. Circulating IGF-1 is mainly produced by liver cells, but is also produced locally in other tissues acting in paracrine and autocrine manners. IGF-1 inhibits the release of GH from the pituitary gland through a negative-feedback (Bartke et al., 2013). The amount of circulating GH and IGF-1 decline with age (Junnila et al., 2013). Individuals carrying loss of function mutation in the GH receptor are characterized by a high level of circulating GH, dwarfism and obesity, known as Laron dwarfism (Godowski et al., 1989).
Binding of insulin and IGF-1 to their receptors results in the activation of the phosphoinositide 3-kinase (PI3K), serine threonine Akt kinase, and phosphorylation of the forkhead box protein O (FOXO) transcription factor (Fernandez and Torres-Aleman, 2012) resulting in its retention in the cytoplasm. Inhibition of IGF-1 signaling results in the nuclear localization of FOXO and activation of the expression of its target genes that are involved in various cellular processes including autophagy, apoptosis, and stress responses (Lee et al., 2003a; Murphy et al., 2003). Activated Akt suppresses autophagy by stimulating mTOR and inhibiting tuberous sclerosis complex 2 (TSC2). The Ras-Raf-ERKs-MAPK pathway is another central pathway activated by insulin and IGF-1 that promotes cell growth and proliferation (Fernandez and Torres-Aleman, 2012) (Fig. 1).
3.1.1. IIS pathway in aging
The influence of IIS signaling on longevity was first demonstrated in long-lived daf-2 and age-1 C. elegans mutants (Friedman and Johnson, 1988; Kenyon et al., 1993). The effect of daf-2 mutation on aging was suppressed in worms by an additional mutation in the daf-16 gene, revealing that DAF-2 and DAF-16 function in the same pathway (Dorman et al., 1995; Kenyon et al., 1993). Later works showed that DAF-2 is a homologue of the human insulin and IGF-1trans-membrane tyrosine kinase receptors (Kimura et al., 1997), and that AGE-1 is a worm homolog of human PI3K, and DAF-16 a human FOXO homolog acting downstream to DAF-2 and AGE-1 (Lin et al., 1997; Ogg et al., 1997). DAF-16 regulates the expression of a number genes involved in the stress-response and antioxidant defense systems, metabolism, and antimicrobial system, a sort of immune response, (Honda and Honda, 1999; Murphy et al., 2003). C. elegans and D. melanogaster have a single FOXO homolog gene, but mammals have four; FOXO1, FOXO3, FOXO4, and FOXO6 (Martins et al., 2016).
Two closely related types of mice, the Ames dwarf mice and the Snell dwarf mice carry mutations in Prop1 (Prophet of Pit-1), a transcription factor that regulates the expression of PIT-1 that drives the differentiation of the anterior pituitary gland (Sornson et al., 1996). These mice are deficient in GH, prolactin, and thyroid-stimulating hormone (TSH), have reduced levels of IGF-1 and insulin, and increased life-expectancy.
The lifespan extension effect of diminished IIS signaling has been demonstrated in yeast (Fabrizio et al., 2001), D. melanogaster (Clancy et al., 2001), and mice (Brown-Borg, 2015; Holzenberger et al., 2003; Mao et al., 2018), indicating that IIS is as an evolutionarily conserved pathway integral to the aging process.
A key question is at what time-point in life targeting IIS signaling can have beneficial effects on health and lifespan. Treating 18 months old mice with an antibody against the IGF-1 receptor improved health span and increased median lifespan by 9% in female mice (Mao et al., 2018), suggesting that inhibition of IIS signaling can delay aging even when applied in advanced age, at least for females.
The role of IIS signaling in human aging is complex and not fully understood. There is, however, some evidence suggesting that reduced IIS signaling extends lifespan and health in humans as well. The GH secretion rate was found to be lower and tightly controlled in the offspring of long-lived families (van der Spoel et al., 2016). Moreover, genetic variants of the IIS signaling pathway have been identified in long-lived individuals maybe linking the IIS pathway to longevity or to a healthy life span (Milman et al., 2014; Suh et al., 2008).
3.1.2. Mitochondria in IIS signaling
Biochemical and cell biological studies in animal models have identified an intricate interplay between mitochondrial physiology and IIS signaling. Inhibition of the C. elegans DAF-2 leads to the nuclear translocation of the DAF-16 transcription factor and the expression of the DAF-16 target genes including those involved in the regulation of cellular stress-responses and metabolism (Honda and Honda, 1999; Murphy et al., 2003). Extensive overlap was identified in the gene expression profile of daf-2 mutants and some long-lived mitochondrial mutant strains of C. elegans (Senchuk et al., 2018), suggesting the convergence of daf-2 and mitochondria retrograde signaling on the same lifespan controlling pathway(s). Protein synthesis is a highly energy consuming process in the cell. DAF-2 deficiency was accompanied by an overall slower protein turnover rate, which was particularly more significant for mitochondrial proteins (Dhondt et al., 2016). In a separate study, several mitochondrial parameters were markedly different in daf-2 worms such as increased abundance of OXPHOS proteins, higher reserve respiration capacity, and higher membrane potential (Brys et al., 2010). Thus, in C. elegans, IIS inhibition appears to influence mitochondrial bioenergetics and biogenesis by reducing the half-life of mitochondrial proteins and by enhancing the expression of the nuclear encoded mitochondrial genes.
Mitochondrial respiration progressively declines in adult C. elegans, but the rate of decline was slower in daf-2 mutant worms, concomitant with more mitochondrial ROS production although without detectable adverse effect on mtDNA integrity and protein oxidative damage (Brys et al., 2010). Acute inhibition of IIS in adult C. elegans transiently increased ROS levels, which in turn induced the expression of ROS neutralizing enzymes superoxide dismutase (SOD) and catalase followed with reduced ROS levels (Zarse et al., 2012). These results suggest that the regulation and improvement of mitochondrial function together with ROS-mediated adaptive response contribute to the life extension effect of IIS signaling impairment in C. elegans.
Mitochondrial shape, size and network are largely determined by the rates of fission and fusion, and alter in response to internal and external cues such as metabolic demand, and stress. Neuronal mitochondria in C. elegans undergo changes in density, morphology and in axonal transport frequency and distance with age (Morsci et al., 2016). In daf-2 mutants, the age-associated decline in mitochondrial morphology and density was significantly delayed and the axonal trafficking rate of mitochondria was maintained during adulthood (Morsci et al., 2016). This study demonstrated that the status of mitochondrial dynamics and distribution are closely linked to the IIS signaling and the aging process in worms.
Mitophagy is a central process in maintaining the cellular content of functional mitochondria by removing malfunctioning mitochondria. Mitophagy was markedly enhanced in daf-2 deficient C. elegans (Palikaras et al., 2015), and exposure to mitophagy enhancers increased lifespan in worms (Fang et al., 2017b; Ryu et al., 2016). Dct-1 is a nematode ortholog of the mammalian mitophagy receptors NIX/BNIP3L (Palikaras et al., 2015). DCT-1 impairment markedly shortened lifespan in daf-2 mutants, demonstrating a functional connection between mitophagy and the observed lifespan extension following IIS impairment in C. elegans (Palikaras et al., 2015).
In long-living Ames dwarf and GHR-KO mice with defects in GH/IGF-1 signaling, there was a metabolic shift to β-oxidation and significantly higher oxygen consumption rate, lower respiratory quotient (RQ), and a higher level of PGC1-α expression and more abundant OXPHOS proteins (Brown-Borg et al., 2012), connecting IIS signaling impairment in mammals to a significant shift in mitochondrial metabolism and more efficient mitochondrial biogenesis and function (Bartke and Westbrook, 2012).
Insulin signaling in adipose tissue regulates lipid and glucose metabolism. Fat-specific insulin receptor knockout mice (FIRKO) were lean and had increased lifespan (Bluher et al., 2003). The expression of genes involved in glycolysis, TCA cycle, β-oxidation, and OXPHOS proteins, were all higher in FIRKO mice with age, while they declined in control mice (Katic et al., 2007), suggesting the involvement of mitochondrial metabolism in the life extension effect of insulin signaling deprivation in the fat tissue. Disruption of the insulin receptor in adipose and muscle tissue in adult mice, however, failed to increase life expectancy (Merry et al., 2017), indicating the complex role of the mammalian IIS signaling on lifespan.
Data on mitochondrial gene expression profiles in long-lived humans with genetic variants in GH/IGF-1 pathway (Milman et al., 2014; Suh et al., 2008), and in the offspring of long-living families with reduced circulating GH hormone (van der Spoel et al., 2016), are not available and this warrants further investigation.
3.2. Mechanistic target of rapamycin (mTOR) signaling pathway
mTOR signaling is an evolutionarily conserved pathway that controls cell growth and metabolism in response to nutrients, growth factors, and cellular energy levels. mTOR is present in two functionally distinct complexes; mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) with distinct and overlapping interacting partners, and inputs and output signaling. Central to both complexes is the serine-threonine kinase mTOR, a phosphoinositide 3-kinase related protein kinase (Saxton and Sabatini, 2017).
The activity of mTORC1 is regulated in response to the availability of nutrients (amino acids, lipids, cholesterol, and glucose), growth factors (for instance insulin and insulin-like growth factors), and also the cellular energy status, i.e. AMP/ATP ratio. Activated mTORC1 promotes cell growth (accumulation of cell mass) by promoting protein, lipid and nucleotide synthesis and suppressing autophagy (Ben-Sahra et al., 2013; Ganley et al., 2009; Hosokawa et al., 2009; Peterson et al., 2011). mTORC2 controls cell growth, proliferation and cell survival by regulating lipogenesis, glucose metabolism, the actin cytoskeleton, and apoptosis (Hagiwara et al., 2012; Jacinto et al., 2004). mTOR and IIS signaling pathways functionally interact. mTORC2 phosphorylates and activates Akt (Sarbassov et al., 2005), a key protein in IIS-PI3K-Akt signaling pathway that promotes cell survival and growth in part through phosphorylation and cytoplasmic retention and inhibition of FOXO transcription factors (Fig. 1).
3.2.1. mTOR in aging
Genetic and pharmacological studies have consistently shown that inhibition of mTOR increases lifespan in yeast (Kaeberlein et al., 2005), C. elegans (Vellai et al., 2003), D. melanogaster (Kapahi et al., 2004), and in mice (Harrison et al., 2009; Wu et al., 2013b), indicating that the lifespan expansion function of the mTOR pathway is a highly conserved mechanism.
Rapamycin is an antibiotic macrolide with immunosuppressive, antifungal, anticancer, and cell proliferation inhibitory properties (Yoo et al., 2017). Rapamycin binds to its intracellular receptor FK506-binding protein 12 (FKBP12). The rapamycin-FKBP12 complex inhibits mTOR by directly binding to the FRB (FKBP12-rapamycin-binding) domain of mTOR inhibiting raptor binding mTORC1 (Brown et al., 1994; Sabatini et al., 1994; Shimobayashi and Hall, 2014; Zoncu et al., 2011). Rapamycin also inhibits mTORC2 following prolonged exposure (Sarbassov et al., 2006), which may account for the observed toxicity of chronic rapamycin treatment such as glucose intolerance and insulin resistance (Lamming et al., 2012). Rapamycin has been consistently shown to prolong lifespan in various species ranging from worm to mouse (Bjedov et al., 2010; Harrison et al., 2009; Miller et al., 2011; Robida-Stubbs et al., 2012).
Key mechanisms for the increased longevity effects of mTORC1 inhibition are thought to include an overall reduction of mRNA translation and protein synthesis, and enhanced autophagy flux (Beretta et al., 1996; Kim et al., 2011b; Saxton and Sabatini, 2017; Selman et al., 2009). The serine-threonine kinase ULK1 controls the formation of autophagosoms and autophagic flux. mTORC1 actively suppresses autophagy by phosphorylating ULK1 (Kim et al., 2011b). mTORC1 also regulates autophagy in part through phosphorylation and cytoplasmic retention of transcription factor EB (TFEB), that regulates the expression of lysosomal and autophagy genes (Martina et al., 2012; Settembre et al., 2011).
mTOR signaling controls many cellular processes and functions in a highly age and tissue-specific manner. For example, mice with adipose-specific depletion of raptor had significantly less adipose tissue, did not develop obesity and showed improved insulin sensitivity and increased mitochondrial respiration in adipose tissue (Polak et al., 2008). Disruption of raptor in skeletal muscle, on the other hand, resulted in muscle dystrophy (Bentzinger et al., 2008). In a separate study, uncontrolled mTORC1 activity in skeletal muscle resulted in severe muscle atrophy because of impaired autophagy (Castets et al., 2013). These results indicate the importance of tight regulation of the mTORC1 pathway and careful implementation of mTORC1 inhibitors in longevity studies (Arriola Apelo et al., 2016; Kraig et al., 2018).
3.2.2. Mitochondria in the mTOR pathway
Cell growth and proliferation are energy consuming processes, so, as could be expected, mTOR has been found to control mitochondrial function at several levels (Bao et al., 2015; Bentzinger et al., 2008; Betz et al., 2013; Cunningham et al., 2007; Khan et al., 2017; Lu et al., 2015; Morita et al., 2013; Polak et al., 2008; Ramanathan and Schreiber, 2009).
Subcellular localization of mTOR complexes is tightly linked to their function (Betz and Hall, 2013). Mitochondria-associated endoplasmic reticulum (ER) membranes (MAM) are part of ER that physically connects ER to mitochondria and are important for the movement of lipids and calcium between these organelles (Rizzuto et al., 1998). Growth factors stimulate localization of mTORC2 to MAM where it phosphorylates Akt that in turn phosphorylates MAM associated proteins IP3R and hexokinase II, thereby controlling mitochondrial physiology (Betz et al., 2013).
An important downstream function of mTORC1 signaling is the suppression of autophagy.
For example, sustained mTORC1 activity in skeletal muscle resulted in severe muscle atrophy probably as a result of impaired autophagy (Castets et al., 2013). A general increase in autophagy following mTORC1 inhibition could also be expected to enhance selective removal of damaged mitochondria by mitophagy. This is supported by several recent reports. TSC2 is a negative regulator of mTORC1 (Fig. 1). TSC2−/− cells showed impaired autophagy and PINK1-mediated mitophagy (Bartolome et al., 2017). Neurons from TSC2−/− mice showed impaired mitochondrial dynamics and were depleted of axonal and presynaptic mitochondria. Blocking mTORC1 restored mitochondrial homeostasis and subcellular distribution of mitochondria (Ebrahimi-Fakhari et al., 2016). In a separate study, inhibition of mTORC1 induced mitophagy in cytoplasmic hybrid (cybrid) cell lines carrying severe mtDNA defects (Gilkerson et al., 2012). Long term rapamycin treatment significantly reduced the frequency of mtDNA carrying deletion mutations in aging mice probably through stimulation of mitophagy (Bielas et al., 2018).
Leigh syndrome is a primary mitochondrial disease caused by mtDNA deletion. Treatment of a mouse model of Leigh syndrome with rapamycin improved mitochondrial function and alleviated symptoms of the disease (Johnson et al., 2013). Moreover, low dose rapamycin treatment extended lifespan in a mouse model of human mtDNA depletion syndrome probably through systemic changes in metabolism (Siegmund et al., 2017). mTOR inhibition may therefore represent a potential therapeutic target in diseases caused by defect in mtDNA maintenance systems. Collectively, these studies demonstrate that mitochondrial physiology and homeostasis are affected by the mTOR signaling pathway. We speculate that mitochondrial homeostasis is a critical mediator of the longevity effects of mTOR inhibition. Investigating this link is important, because of the key roles of mTOR signaling and mitochondrial metabolism in aging, and their malleability to regulation by non-invasive, dietary, and pharmacological interventions.
3.3. Adenosine monophosphate kinase (AMPK)
ATP is the principal energy substrate of the cell. The free energy released from the hydrolysis of ATP to ADP is harvested to drive almost all energetically unfavorable reactions in the cell. Therefore, elaborate systems have evolved to maintain sufficient and continual supply of cellular ATP. AMPK has been identified as the primary sensor of cellular ATP levels and a key regulator of cellular energy homeostasis and metabolism (Hardie et al., 2016; Herzig and Shaw, 2018). AMPK is a heterotrimer, evolutionarily conserved protein kinase consisting of αβγ subunits (Ross et al., 2016). Phosphorylation of Thr172 within the kinase domain of the α-subunit enhances the protein kinase activity of AMPK by several orders of magnitude. The tumor suppressor liver kinase B1 (LKB1) and the calcium-sensitive kinase (CAMKK2) are two major upstream kinases for the Thr172 phosphorylation (Hardie et al., 2016; Herzig and Shaw, 2018) (Fig. 1). AMPK senses the amount of available ATP in the form of high AMP/ATP or ADP/ATP ratios that increases Thr172 dephosphorylation and AMPK activation (Gowans et al., 2013). Activated AMPK regulates a number of metabolic processes (Hardie et al., 2016; Herzig and Shaw, 2018). In general, activated AMPK alters metabolism towards increased catabolic processes such as autophagy and down-regulates energy consuming anabolic, and biosynthetic processes including lipid and protein synthesis, to stimulate ATP production and to restore cellular energy homeostasis (Hardie et al., 2016; Herzig and Shaw, 2018).
3.3.1. AMPK in aging
AMPK may influence aging and lifespan through different mechanisms (Burkewitz et al., 2014). The lifespan extension effects of pharmacological and dietary interventions in model organisms often implicate autophagy. AMPK activates autophagy by phosphorylating ULK1, a critical kinase for autophagy initiation (Egan et al., 2011), and by suppressing mTORC1 via activation of its negative regulator TSC2 (Inoki et al., 2003), and phosphorylation and inhibition of the mTORC1 subunit, raptor (Gwinn et al., 2008) (Fig. 1).
AMPK also modulates the signaling pathways that promote longevity such as those involving FOXOs (Greer et al., 2007) and SIRT1 (Canto et al., 2009).
In addition to rapamycin, multiple studies have shown that compounds such as resveratrol and metformin also increase lifespan in various organisms. AMPK is involved in mediating the life extension effects of these molecules (Burkewitz et al., 2014).
Dietary restriction (DR) is the severe reduction of food intake. DR is one of the most consistent interventions to delay the onset of age-related diseases and to increase life expectancy in organisms ranging from yeast to mammals. Long-term and acute DR activates AMPK (Burkewitz et al., 2014). Like rapamycin, a key outcome of both DR and activated AMPK is the inhibition and suppression of mTOR signaling, suggesting a DR mediated lifespan regulatory function of AMPK via inhibition of mTOR signaling pathway.
In mammals, the three AMPK subunits exists in several isoforms encoded by different genes (Ross et al., 2016). Changes in the phosphorylation and the expression of different isoforms seem to be affected by age (Salminen et al., 2016), resulting in a decline in the capacity of AMPK signaling to respond to different stimuli.
AMPK activity is suppressed by IIS via Akt (Kovacic et al., 2003), and activated AMPK inhibits IIS by phosphorylating insulin receptor substrate 1 (IRS) (Tzatsos and Tsichlis, 2007), and mTOR signaling pathways through phosphorylation of TSC2 and raptor (Gwinn et al., 2008). mTOR in turn activates IIS and is stimulated by IIS (Sarbassov et al., 2005) (Fig. 1). These examples illustrate the intricate network of interactions that regulate the longevity pathways.
3.3.2. AMPK and mitochondria
The central role of AMPK in the regulation of cellular energy metabolism implies close links between AMPK activity and mitochondrial biology. Consistent with this, different mouse models of AMPK deficiency have demonstrated that AMPK influences various aspects of mitochondrial biology (Garcia-Roves et al., 2008; Lantier et al., 2014; O’Neill et al., 2011). AMPK regulates mitochondrial biogenesis by controlling the expression of mitochondrial genes. Early experiments in mice showed that a reduced ATP/AMP ratio in skeletal muscle induced the expression of PGC1-α and activated the mitochondrial biogenesis (Zong et al., 2002). This response was abrogated in mice expressing a dominant negative mutant of AMPK (Zong et al., 2002). Later work showed that phosphorylation of PGC1-α by activated AMPK enhances PGC1-α action and the induction of mitochondrial genes (Jager et al., 2007).
The dynamic changes in mitochondrial morphology and network formation by fission and fusion are integral in adaptive cell metabolism, and in the maintenance of a functional mitochondria population by mitophagy (Sebastian et al., 2017). Recent studies have implicated AMPK in an intricate signaling network that controls mitochondrial quality. An important role of AMPK in mitochondrial homeostasis is probably linked to its key function in initiating autophagy and mitophagic turnover of dysfunctional mitochondria by phosphorylation and activation of ULK1, as has been demonstrated in human cell lines and in C. elegans (Egan et al., 2011). Mitochondrial stress promoted mitochondrial fragmentation through phosphorylation of mitochondrial fission factor, MFF, by AMPK to facilitate elimination of damaged mitochondria by mitophagy in human and mouse embryonic fibroblasts (MEF) cells (Toyama et al., 2016). In C. elegans, inhibition of mitochondrial dynamics blocked AMPK- and DR-mediated increased longevity, illustrating the close interplay between mitochondrial dynamics and AMPK in regulating longevity (Weir et al., 2017). It now seems evident that mitochondrial function declines with age (Gonzalez-Freire et al., 2018). Aging-related reductions in AMPK activity and signaling may in part contribute to the reduced mitochondrial function and biogenesis over time (Reznick et al., 2007).
Altogether, evidence indicates that AMPK plays a central role in different aspects of mitochondrial homeostasis as part of an intricate signaling network with significant implications on cell metabolism and longevity.
4. Genome instability
Evidence indicates that DNA damage and mutation accumulate with age in multiple human and animal tissues, and loss of genome integrity, i.e. nuclear and mitochondrial DNA, is a hallmark of aging across species (Bua et al., 2006; Chow and Herrup, 2015; Fakouri et al., 2018; Fayet et al., 2002; Kalfalah et al., 2015; Li et al., 2017, 2016; Lopez-Otin et al., 2013; Mattson and Arumugam, 2018; Maynard et al., 2015; Vaidya et al., 2014; Zhang and Vijg, 2018; Zhang et al., 2015).
DNA is constantly damaged at high frequency by spontaneous hydrolytic decay and by various endogenous and environmental causes (De Bont and van Larebeke, 2004; Lindahl, 1993). DNA damage refers to any changes to the chemistry of DNA and misincorporated but chemically normal nucleotides, e.g. during DNA replication. Structural DNA damage can give rise to mutation during replication and thus change the original nucleotide sequence of DNA. While DNA damage can be repaired by the DNA repair machinery, mutations cannot, and will become fixed in the genome of the daughter cells (Akbari and Krokan, 2008). In the case of germ cells, mutations can potentially be passed on to the progeny, unless the cell dies or the carrier does not reproduce or dies at a pre-reproductive age (Crow, 1997).
Our genome is organized within the nucleus and mitochondria. The nuclear genome in a typical haploid cell contains approximately three billion base pairs and an estimated over 20 000 protein coding genes. By comparison, mtDNA is a very small ˜16.6 kb circular DNA, and contains only 13 protein coding genes. Despite its small size, however, mtDNA is crucial for development, normal cell function, and survival (El-Hattab and Scaglia, 2013). The transformation of mtDNA damage to mutation seems to occur at a much lower frequency than that of nuclear DNA (Valente et al., 2016). Thus, mutational load in nuclear and mtDNA may have distinct origins, and replication error is commonly considered the major source of mtDNA mutation (DeBalsi et al., 2017).
The preservation of the chemical structure and the original nucleotide sequence of the genome are essential for life. Thus, mechanisms have evolved in all living organisms to repair DNA lesions and to maintain genome stability and integrity (Iyama and Wilson, 2013; Sykora et al., 2012). Structure distorting lesions, such as UV-light, generate thymidine dimers that are repaired by the nucleotide excision repair (NER) pathway (Marteijn et al., 2014). DNA double strand breaks (DSBs) are repaired by non-homologous end-joining (NHEJ) (Kragelund et al., 2016), and recombination repair (San Filippo et al., 2008) pathways. Mismatch repair (MMS) is coupled with DNA replication and corrects wrongly inserted nucleotides during replication (Jiricny, 2006). DNA base excision repair (BER) pathway repairs a number of different damaged bases and non-coding baseless sites (AP sites) (Krokan and Bjørås, 2013). Genotoxic insults that generate breaks in one of the two DNA strands, called single-strand DNA breaks, often result in chemically modified 3´- and 5´-termini and are repaired by single-strand break repair (SSBR) pathway. BER and SSBR are enzymatically similar pathways and share proteins.
The expression of DNA repair proteins is up-regulated in long-lived specifies compared with animals with shorter lifespan (MacRae et al., 2015), and genetic variation in DNA repair can influence longevity (Debrabant et al., 2014; Keane et al., 2015; Kim et al., 2011a). These examples demonstrate the principle of the optimization of allocation of resources between maintenance and reproduction in the aging process.
Segmental progeroid syndromes, or premature aging syndromes, are a group of hereditary diseases that are characterized by some but not all clinical features of normal aging progressing at an accelerated rate (Carrero et al., 2016). Most cases of progeroid syndromes are causally linked to defects in DNA repair (Carrero et al., 2016; Kipling et al., 2004). Although defect in DNA repair does not seem to be a major cause of age-associated genome instability, premature aging diseases illustrate a key role of DNA repair in the aging process. Moreover, evidence suggests that the catalytic activity of DNA repair proteins alters or declines with age (Atamna et al., 2000; Cabelof et al., 2002; Xu et al., 2015), which may diminish the capacity of cells to cope with the burden of DNA damage resulting in the accumulation DNA lesions and genome instability over time. Although a number of studies on human materials and in laboratory animals have identified DNA repair as a key determinant of the rate of aging and lifespan, evidence showing positive effects of stimulation or increased DNA repair capacity on lifespan is limited and more work of this sort needs to be conducted in animal models (Qian et al., 2018).
A less investigated but potentially important source of DNA damage is the process of DNA repair itself. Alterations in the expression of DNA repair proteins during aging have been frequently reported for almost all DNA repair pathways (Gorbunova et al., 2007). Most DNA repair pathways are multi-enzyme and multistep processes and changes in the level of proteins can lead to loss of coordination in the repair process (Fu et al., 2012; Kidane et al., 2014; Senejani et al., 2012). For example, BER corrects various types of DNA lesions that occur frequently and in large numbers, which if left unrepaired can be mutagenic and cytotoxic (Krokan and Bjørås, 2013). However, uncontrolled BER activity, particularly under conditions of enhanced DNA lesions, can also lead to the generation of DNA repair intermediates, such as AP-sites and single-strand nicks, that are often highly mutagenic and cytotoxic (Akbari et al., 2009), leading to the activation DNA damage response (DDR) and cell death (Ebrahimkhani et al., 2014; Parrish et al., 2018). Thus the control and balance of all the individual steps in the DNA repair process are central to its function and changes that up-regulate or down-regulate the DNA repair processes can render the system dysfunctional.
Addition of a methyl group to cytosine (5mC) and its removal constitute epigenetic DNA modification processes that are important for the regulation of expression of a number of genes. DNA demethylation is controlled by the coordinated actions of Ten-eleven translocation (TET) proteins and BER. TET proteins oxidize 5mC to 5-hydroxymethylcytosine (5hmC) and further to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). These bases are excised and repaired by BER resulting in the conversion of 5mC to C (Wu and Zhang, 2017). DNA methylation/demethylation processes are dynamically regulated in brain cells and continually change in response to physiological neuronal activity (Guo et al., 2011). Aberrant and incomplete BER (Weissman et al., 2007) can result in genome instability at these sites following cycles of DNA methylation/demethylation (Mahfoudhi et al., 2016), thus contributing to age-related somatic genome instability and pathological alteration of gene expression and neurodegeneration (Leandro et al., 2015).
DNA damage contributes to aging in various ways. DNA lesions can persist over a long period of time and increase with age (Siddiqui et al., 2015). Patients with childhood cancer who were treated with DNA damaging chemotherapeutic agents show signs of accelerated aging later in life (Ness et al., 2013). DNA lesions can impede transcription, which is especially highly relevant in long genes (Vermeij et al., 2016). DNA damage can affect the large number of enhancers that are located throughout the human genome perturbing normal gene expression (Andersson et al., 2014; Vijg et al., 2017). Increased amounts of DNA lesions can result in aberrant cell cycle re-entry in post-mitotic neurons leading to apoptosis or senescence (Fielder et al., 2017), and cause stem cell exhaustion (Nijnik et al., 2007; Rossi et al., 2007). DNA lesions such as bulky adducts, hydrolytic deamination of bases and AP-sites can also become converted to mutations by various mechanisms (Helleday et al., 2014). Thus, accumulation of DNA lesions combined with alterations and decline in DNA repair capacity, contribute to genome instability and constitute a critical driving force in the aging process.
5. The mitochondrial genome in aging
Progressive alterations in the primary structure of the genomic DNA (nuclear and mitochondrial DNA) with age can compromise mitochondrial function in various ways. Mitochondria contain ˜1200 proteins (Calvo et al., 2016), only 13 of which are encoded by mtDNA and all are components of the OXPHOS system, and are essential for mitochondrial function (DeBalsi et al., 2017).
A number of pathogenic mtDNA rearrangements and single nucleotide mutations have been reported over the years (Elliott et al., 2008; http://www.mitomap.org/). A mixture of normal and mutated mtDNA molecules in a cell is known as heteroplasmy. The proportion of mutant to normal mtDNA affects mitochondrial function and determines the severity and the progression of the disease and tissue dysfunction (Schon et al., 2012). mtDNA is exclusively maternally inherited, and diseases caused by mtDNA mutation display a maternal mode of inheritance (Schon et al., 2012).
The amount of mtDNA mutations increases in multiple tissues with age (Bua et al., 2006; Corral-Debrinski et al., 1992; Fayet et al., 2002; Greaves et al., 2014; Kauppila et al., 2017; Kennedy et al., 2013; Kraytsberg et al., 2006), mostly originating from clonal expansion of mtDNA replication errors during development and spontaneous deamination of cytosine and adenine (Fayet et al., 2002; Kennedy et al., 2013). A direct link between mtDNA instability with aging phenotypes was demonstrated in mice engineered to express a proof-reading deficient variant of mitochondrial DNA polymerase γ (Kujoth et al., 2005; Trifunovic et al., 2004). These mice accumulate mtDNA mutations at large numbers and show aging features such as weight loss, alopecia, osteoporosis, and kyphosis (Kujoth et al., 2005; Trifunovic et al., 2004).
mtDNA is organized into structures called nucleoids in close association with mitochondrial inner membrane (Brown et al., 2011). Because the mitochondrial inner membrane is a major site of mitochondrial ROS production, oxidative lesions are expected to occur frequently in mtDNA (Cadenas and Davies, 2000). Current data, however, does not support a substantial contribution of oxidative mtDNA damage to age-related increase in somatic mtDNA mutations (Halsne et al., 2012; Kauppila et al., 2018; Kennedy et al., 2013).
As discussed in Box1, of several DNA repair activities that have been reported in mammalian mitochondria, BER is likely the most active DNA repair pathway in human mitochondria (Sykora et al., 2012). Alterations in mitochondrial BER activity and protein levels with age have been identified in different tissues and organisms (Garreau-Balandier et al., 2014; Gredilla et al., 2010; Szczesny et al., 2003). These changes may lower mtDNA repair capacity resulting in persistent mtDNA damage (Canugovi et al., 2014), and create DNA repair imbalance leading to the generation of genotoxic DNA repair intermediates and compromise mtDNA integrity (Harrison et al., 2005).
A somewhat overlooked consequence of mtDNA damage on aging is the effect of DNA lesions on mtDNA transcription fidelity. In addition to oxidative damage to mtDNA from ROS, environmental chemicals and endogenously produced aldehydes can generate different forms of mtDNA crosslinks and bulky adducts (Cline, 2012; Valente et al., 2016). Such DNA lesions can stop the progression of mitochondrial RNA polymerase (POLRMT) resulting in premature termination of the transcript, or the damage can be bypassed by the polymerase but result in the production of mutated transcripts (Cline, 2012; Cline et al., 2010; Sultana et al., 2017). In the nucleus, defects in transcription-coupled DNA damage repair cause Cockayne syndrome, a disease characterized by severe neurological deficiencies and accelerated aging phenotypes (Karikkineth et al., 2017), and impaired mitochondrial homeostasis (Scheibye-Knudsen et al., 2012). Thus, mtDNA transcription failure in the form of mutations and truncations can, in principle, contribute to mitochondrial dysfunction and age-associated decline in tissue function, and warrants investigation.
Damaged deoxyribonucleotide triphosphates (dNTPs) are another important source of DNA damage and mutation. Spontaneous deamination of dCTP generates dUTP that can be readily incorporated into DNA opposite adenine. Although U:A is not mutagenic, high levels of uracil incorporation followed by removal of uracil from DNA by uracil-DNA glycosylase (UNG) generate potentially mutagenic and cytotoxic AP-sites (Krokan and Bjørås, 2013). Deoxyuridine triphosphate nucleotidhydrolase (dUTPase) hydrolyses dUTP to dUMP and pyrophosphate. dUTPase is present in both nucleus and mitochondria (Ladner and Caradonna, 1997).
MTH1 protein hydrolyzes oxidized purine nucleotides such as 8-oxo-dGTP and 8-oxo-dATP to monophosphates. MTH1 is located in the cytoplasm, mitochondria and the nucleus (Kang et al., 1995). A very low amount of oxidized guanine was able to reduce the fidelity of the mitochondrial DNA polymerase γ resulting in the pre-mutagenic A:8-oxoG mismatches in DNA (Pursell et al., 2008). Moreover, primary neurons from mice deficient for Mth1 and DNA glycosylase Ogg1 (removes the oxidative base damage 8-oxoG from DNA) contained high levels of 8-oxoG in mtDNA, exhibited poor neurite outgrowth, and mitochondrial dysfunction (Leon et al., 2016). Thus, mitochondrial nucleotide pool sanitation enzymes play key roles in the preservation of mtDNA integrity, and exploring their role in the age-associated increased mtDNA damage has not been adequately addressed and merits investigation.
Ribonucleotides are frequently mis-incorporated into genomic DNA (Williams et al., 2016). Ribonucleotides in nuclear DNA can cause DNA replication stress and small and large scale genome instability (Williams et al., 2016). The presence of ribonucleotides in mtDNA has long been known (Grossman et al., 1973). The mitochondrial DNA polymerase γ incorporates ribonucleotide into DNA with varying efficiency depending on the incoming nucleotide and the presence of a 3´-terminal ribonucleotide on a primer (Kasiviswanathan and Copeland, 2011). The frequency of ribonucleotide incorporation into mtDNA is influenced by the availability of nucleotides, and is elevated in cells from individuals with mutation in genes that regulate mitochondrial nucleotide pool, suggesting high levels of ribonucleotides in mtDNA might be pathogenic (Berglund et al., 2017). The biological consequence of ribonucleotides in mtDNA is largely unknown, but seems to decrease the speed of mtDNA replication (Forslund et al., 2018). In the context of aging, it will be interesting to investigate the rate of ribonucleotide incorporation and the level of ribonucleotides in mtDNA in different tissues with age (Pawar et al., 2018).
5.1. MtDNA variants
mtDNA may influence human biology and aging in subtle ways. mtDNA has a higher mutation and evolution rate than nuclear DNA, and mtDNA shows large within-population sequence variability (Elliott et al., 2008; Wallace, 2015). Non-deleterious, beneficial mutations that improved adaptation to environmental changes, such as climate and food, and infectious diseases, were fixed in mtDNA giving rise to different lineages or haplotypes. Further nucleotide sequence changes in the haplotypes generated clusters of related mtDNA within populations known as haplogroups (Wallace, 2015). Haplogroups may provide protection or influence the severity and susceptibility to various diseases (Carelli et al., 2006; Wallace, 2018). There are, however, also reports that do not support such associations (Goncalves et al., 2018; Hudson et al., 2012).
The maternal inheritance of mtDNA is predicted to result in a selection asymmetry and increased mtDNA mutation load and reduced fitness in males (Wolff and Gemmell, 2013). Analysis of data from a female founder lineage over several generations revealed that an mtDNA variant had a negative effect on male fitness. Thus, the pre-dominant maternal transmission of mtDNA has probably contributed to the reduction of male lifespan (Milot et al., 2017).
Experiments on laboratory models and domestic animals have shown associations between specific mtDNA variants and various phenotypic traits, as well as, functional interactions with the nuclear genome (Aw et al., 2018; St John and Tsai, 2018; Vivian et al., 2017). A transgenic mouse harboring nuclear DNA from one strain and homoplasmic mtDNA haplotype from another, revealed substantial influence of mtDNA haplotype on longevity and health span (Latorre-Pellicer et al., 2016).
These results suggest that mtDNA variants may have subtle effects on health and aging. Further, experimental studies using different animal models may provide better insight into functional interactions between mtDNA haplotypes and the longevity signaling pathways.
5.2. mtDNA and inflammation
The induction of acute inflammation is an integral part of the innate immune response to defend body against pathogens, bacterial and viral infections, tissue damage and the following healing process. On the other hand, non-infectious, low-grade, chronic inflammation is a major risk factor for the development of many age-associated diseases and frailty (Franceschi et al., 2017; Lopez-Otin et al., 2013; Mattson and Arumugam, 2018).
The inflammatory response is triggered by the release of mediators such as cytokines and chemokines by tissue-resident leukocytes and other cells (Medzhitov, 2008). Non-infectious endogenous sources of induction of inflammation includes cellular DNA molecules outside mitochondria and the nucleus, e.g. in the cytoplasm. Such DNA could be sensed as pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs), by specific pattern recognition receptors (PRRs) and activation of immune response (Medzhitov, 2008; Netea et al., 2017).
The cyclic guanosine monophosphate-adenosine monophosphate synthase (cGAS) and stimulator of interferon genes (STING) signaling has been identified as a key sensor of cytoplasmic DNA (Ishikawa and Barber, 2008; Li and Chen, 2018; Wu et al., 2013a). cGAS binds to double-stranded DNA, independent of nucleotide sequence, as a result, cytosolic self-DNA (e.g. DNA released into the cytosol following nuclear DNA damage) and foreign DNA activate cGAS-STING signaling that results in the production of different inflammatory interferon and cytokines (Li and Chen, 2018; Wu et al., 2013a). Defects in the proteins that prevent the accumulation of cytosolic host DNA have been linked to Aicardi-Goutières syndrome (AGS) (Coquel et al., 2018; Crow et al., 2006; Rice et al., 2015; Yang et al., 2007), a rare systemic inflammatory autoimmune disease clinically characterized by early-onset encephalopathy, basal ganglia calcification, cerebral white matter abnormalities, and elevated level of type I interferon in serum and cerebral spinal fluid (Goutieres et al., 1998).
The release of mtDNA into cytoplasm, e.g. as a result of mitochondrial stress and damage, can activate cGAS-STING inflammatory response (West et al., 2015; White et al., 2014). Importantly, removal of damaged or stressed mitochondria by mitophagy inhibited this response (Nakahira et al., 2011; Sliter et al., 2018). Mitochondria play various pivotal roles in the regulation of innate and adaptive immune responses (Weinberg et al., 2015). Thus, the health beneficial effects of life style and compounds that stimulate mitophagy and maintain mitochondrial quality (Fang et al., 2019, 2016a; Fang et al., 2017a) are likely in part also to control chronic inflammation, a major source of age-related frailty and morbidity.
6. From nuclear DNA damage response to mitochondria dysfunction and aging
As we discussed in section 4, damage to DNA can cause a number of unpredictable adverse effects on protein coding genes and transcriptional regulatory regions. However, recent findings that DNA lesions (nuclear and telomeric) generate signaling responses that influence mitochondrial function (Fakouri et al., 2018; Fang et al., 2016b; Sahin et al., 2011), have identified a novel consequence of an overall effect of stochastic genomic damage on cell function and aging, thus connecting genome instability to mitochondrial dysfunction, two hallmarks of aging (Lopez-Otin et al., 2013; Mattson and Arumugam, 2018).
Poly (ADP-ribose) polymerases 1 and 2 (PARP1/2) are activated following binding to DNA damage to signal the site of damage to DNA repair proteins by adding ADP-ribose polymers (PAR) to themselves and to the nearby proteins while consuming nicotinamide adenine dinucleotide (NAD+) in the process (Pascal and Ellenberger, 2015).
NAD+ is an important redox coenzyme in metabolic pathways such as the citric acid cycle, and glycolysis, and is also used as co-substrate by three classes of enzymes; 1- poly (ADP-ribose) polymerases (PARP1/2), 2- sirtuins, and 3- the cyclic ADP-ribose (cADPR) synthases (CD38, CD157) (Canto et al., 2015). These enzymes compete for the available cellular NAD+ pool. The level of NAD+ declines with age (Braidy et al., 2011; Zhu et al., 2015), and in animal models of human DNA repair deficiency (Fang et al., 2016a, 2014), while NAD+ supplementation improves health span and extends lifespan in different animal models (Mouchiroud et al., 2013; Zhang et al., 2016).
Sirtuins are evolutionarily conserved protein deacetylases/deacylases implicated in the regulation of aging and longevity in various organisms (Imai and Guarente, 2016). Seven sirtuins (1–7) have been identified in human cells that localize in the nucleus (SIRT1, 6 and 7), cytoplasm (SIRT2) and in mitochondria (SIRT 3–5) (Canto et al., 2015). The nuclear SIRT1 controls and regulates transcription by deacetylating transcription factors, and modulates the catalytic activity of enzymes (Canto et al., 2015).
Several recent studies have identified a link between nuclear DNA damage and cell metabolism and mitochondrial homeostasis through NAD+ consumption. For example, increased PARP activity, as a result of prolonged genotoxic exposure, defects in DNA repair, and during the aging process, limits the level of NAD+ available for sirtuins consequently affecting the function of their downstream targets such as PGC1-α, an important regulator of mitochondrial biogenesis and proteins involved in mitophagy (Bai et al., 2011; Fang et al., 2019). Accordingly, NAD+ supplementation and PARP inhibition increases cellular NAD+ levels and improves mitochondria homeostasis in various animal models of human diseases (Fang et al., 2019, 2016a; Fang et al., 2017a) (Fig. 2).The effect of DNA damage-mediated PARP activation on mitochondrial bioenergetics and cell metabolism is complex, probably involving alternative pathways (Fakouri et al., 2018; Fang et al., 2017a; Fouquerel et al., 2014), and requires more investigation.
Fig. 2.
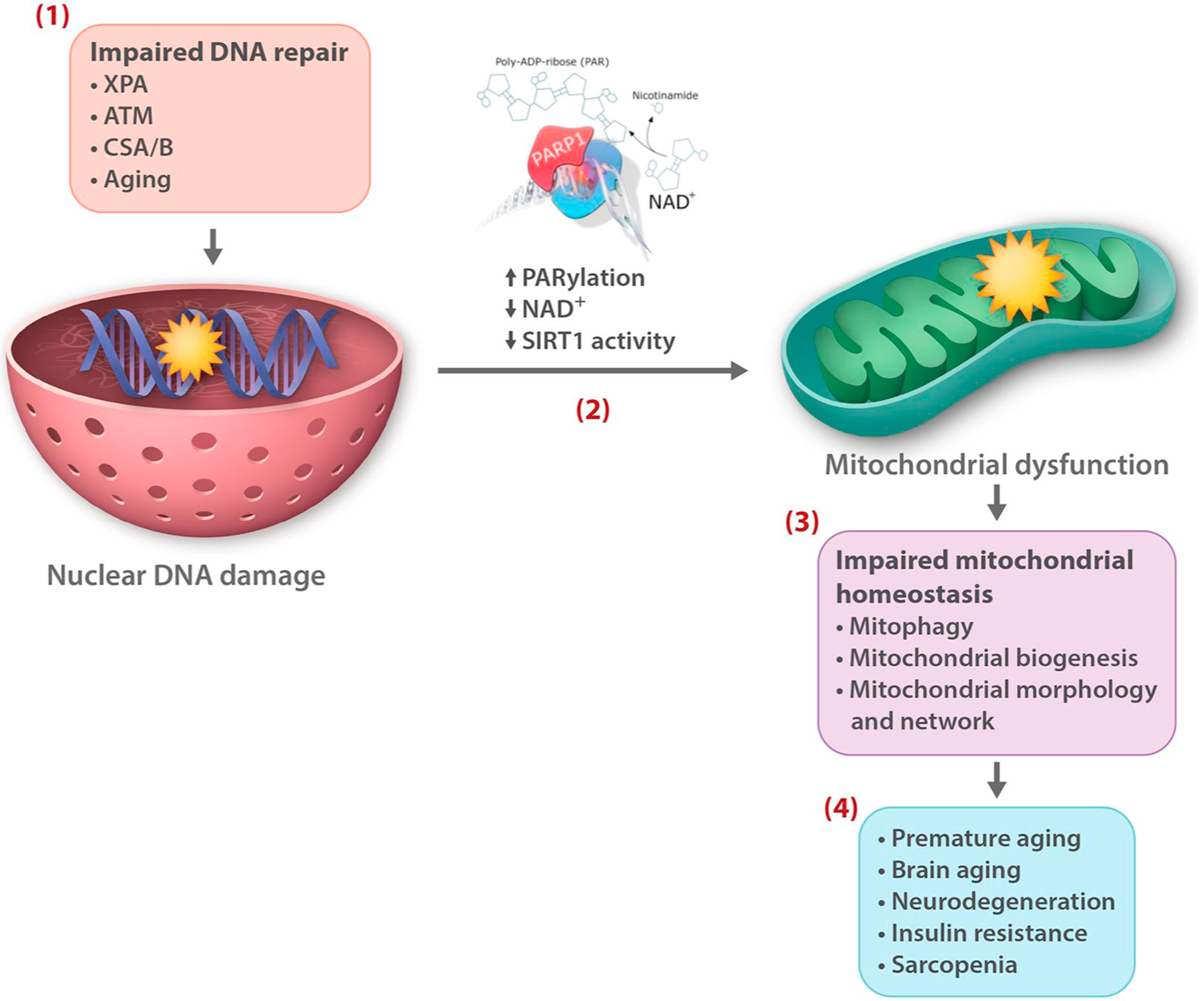
NAD+ over-consumption following persistent DNA lesions is a central link between nuclear DNA instability and mitochondrial dysfunction, two hallmarks of aging. Defects in DNA repair proteins such as in the premature aging disorders xeroderma pigmentosum group A (XPA), ataxia telangiectasia mutated (ATM), and Cockayne syndrome A and B (CSA/B), as well as age-related DNA repair imbalance and dysregulation (Aging), result in the accumulation of DNA damage and genome instability (1), leading to prolonged PARP activation (PARylation), increased NAD+ consumption, and reduced NAD+ levels and SIRT 1 activity (2). A consequence of this chain of events is impaired mitochondrial homeostasis, e.g, because of reduced autophagic turnover of mitochondria by mitophagy and mitochondrial biogenesis (3), that in DNA repair deficient disorders contribute to premature aging phenotypes, and in normal aging process leads to diminished tissue and organ function and disease (4).
7. Telomeres
The regular DNA replication machinery is not able to synthesize the ends of the chromosomes. The maintenance and the protection of the genes at the end of the chromosomes is achieved by telomeres, the tandem repeats of TTAGGG nucleotide sequences, and telomerase, a specialized reverse transcriptase, that replicates and maintains telomeres. Telomeres are partially protected from damage by a protein complex called shelterin that consists of TRF1, TRF2, POT1, RAP1, TIN2, and TPP1 subunits (de Lange, 2005).
In many human cell types, telomeres shorten throughout the life-span; however, a critical role of telomere shortening in the usual human aging process is not fully established. Most studies connecting telomere attrition to aging phenotypes have been conducted in transgenic animal models with defects in telomere maintenance genes, in which telomere shortening during normal aging is insignificant (Blackburn et al., 2015).
Cell senescence is probably a key link between telomere attrition and aging. Many cell types express telomerase at a very low level. When these cells are cultured, they undergo progressive telomere shortening during successive rounds of cell division and eventually enter a permanent cell cycle arrest known as replicative senescence. Telomeres are particularly vulnerable to oxidative damage that can result in telomere shortening and dysfunction and persistent DDR (Barnes et al., 2019; Hewitt et al., 2012). It has been speculated that the higher sensitivity of telomeres to oxidative damage was to help telomere-driven replicative senescence to block the growth of the most badly stressed cells that were at high risk of accumulating DNA damage and mutations (von Zglinicki, 2002).
Damage to telomeres can induce senescence independently of telomere size by activating DDR (Anderson et al., 2019; Hewitt et al., 2012).
7.1. Mitochondria and telomeres
Reciprocal signaling between telomere damage and mitochondrial function has been reported and appears to occur through different mechanisms. ROS emitted from dysfunctional mitochondria was identified as a major determinant of telomere damage and telomere dependent senescence, which was proposed to account for cell-to-cell variation in replicative capacity (Passos et al., 2007).
In telomerase deficient mice, persistent telomere damage and attrition was shown to activate the transcription factor p53, which in turn suppressed the expression of PGC1-α and PGC-1β genes that are considered key regulators of mitochondrial biogenesis, thus compromising mitochondria homeostasis (Sahin et al., 2011). Telomere dysfunction and p53 activation also suppressed the expression of sirtuins including the mitochondrial sirtuins 3–5, which was corrected by p53 deletion (Amano et al., 2019). Administration of the NAD+ precursor NMN, lowered the level of p53 acetylation (which can negatively affect p53 transcriptional activity), maintained telomere length, and reduced DDR. In addition, NMN treatment improved several mitochondrial parameters in transgenic mice (Amano et al., 2019). The results of this study identify a novel pathway linking impaired sirtuin activity following telomere dysfunction and DDR, to mitochondrial dysfunction, and present NAD+ supplementation as a potential therapeutic approach for telomere-related aging phenotypes and diseases.
It should, however, be noted that patients with dyskeratosis congenita, a telomere related disease caused, in part, by mutation in the components of telomerase complex, do not seem to show clear clinical features of mitochondrial disease, (Armanios and Blackburn, 2012; Scheibye-Knudsen et al., 2013), but this needs further investigation.
Other forms of apparent interplay between mitochondria and telomeres involve the dual localization of TERT, the catalytic subunit of telomerase, and the shelterin protein TIN2 in mitochondria and telomeres.
A number of studies have demonstrated the presence of TERT in mitochondria both in proliferating and post-mitotic brain neurons. However, whereas initial studies showed that localization of TERT in mitochondria compromised mitochondrial function and mtDNA integrity (Santos et al., 2004, 2006), later works demonstrated improved mitochondrial parameters and mtDNA stability (Ahmed et al., 2008; Haendeler et al., 2009).
Compared to TERT, limited data is available on the biological significance of mitochondrial localization of TIN2. Overexpression of TIN2 had a profound effect on mitochondrial morphology and network. Furthermore, shRNA mediated TIN2 knockdown improved several parameters of mitochondrial bioenergetics, (Chen et al., 2012), suggesting a link between TIN2 and mitochondrial morphology and metabolism.
The RECQL4 helicase functions in DNA repair and replication and is a central enzyme in the maintenance of genome stability (Croteau et al., 2014). RECQL4 localizes into mitochondria and contributes to telomeric DNA maintenance (Croteau et al., 2014, 2012; Ghosh et al., 2012). Individuals with defect in RECQL4 develop premature aging syndromes which may be in part related to its role in telomere integrity and mtDNA metabolism.
In addition, a number of DNA repair proteins have functions in the mitochondria and in telomeres (Akbari et al., 2014; Batenburg et al., 2012; Duxin et al., 2009; Safdar et al., 2016; Sahin et al., 2011; Aamann et al., 2010). It is not clear what drives the distribution of these proteins between mitochondria and telomeres, and this presents as an interesting avenue of future research.
Taken together, there appears to be some crosstalk between mitochondria and telomeres, and elucidating functional links between these compartments merits more investigation.
8. Mitochondria in cellular senescence
Cellular senescence has long been recognized as a distinct and central feature of aging tissues (Dimri et al., 1995). It is characterized by a state of irreversible and permanent proliferation arrest in somatic cells, together with a complex and distinct senescence-associated secretory phenotype (SASP) consisting of upregulation of pro-inflammatory cytokines, proteases, and growth and angiogenesis factors that can alter tissue homeostasis (Freund et al., 2010; Wiley and Campisi, 2016). Senescence can be induced by various stress factors including mitogenic stress, and persistent DNA damage including telomeric DNA, and activation of DDR, and is commonly thought to have been developed to counter oncogenic transformation of the affected cell (Wiley and Campisi, 2016). Over time, the number of senescent cells increase, and this results in diminished tissue function, altered tissue structure, and decline in tissue repair and regenerative capacity, collectively promoting aging and age-related phenotypes (Chapman et al., 2019). Accordingly, genetic and pharmacological clearance of senescent cells delays the onset of age-related pathologies and extends health span in mice (Baker et al., 2011; Xu et al., 2018).
Evidence suggests a key role for mitochondria in the development of cellular senescence and in the modulation of the SASP signaling. A range of mitochondrial alterations have been reported in senescent cells including mitochondrial morphology and network, mass, mitochondrial ROS production, metabolites, and overall function (Chapman et al., 2019; Kaplon et al., 2013; Moiseeva et al., 2009; Wiley and Campisi, 2016; Ziegler et al., 2015).
Controlled disruption of mitochondrial function and depletion of mitochondria in cells, have provided some clues about the molecular processes that connect mitochondrial metabolism to senescence.
Cells treated with various DDR activators and senescence inducers display increased mitochondrial mass prior to senescent cell cycle arrest (Moiseeva et al., 2009). This is via transcriptional activation of PGC1-β resulting in increased mitochondrial ROS production and DNA damage. Persistent DNA damage, gives rise to a positive feedback loop through ATM, Akt, and mTORC1 phosphorylation activation followed by enhanced PGC-1β dependent mitochondrial biogenesis, sustained DDR, cell cycle arrest and cellular senescence (Correia-Melo et al., 2016).
Mitochondrial dysfunction-associated senescence (MiDAS) is characterized by distinct SASP, and reduced NAD+/NADH ratio and AMPK activation. Activated AMPK was shown to drive growth arrest and senescence though phosphorylation of p53 that increased expression of its target senescence markers p21WAF1 and p16INK4a (Wiley et al., 2016). Senescence and MiDAS phenotypes were also detected in tissues from transgenic mice that accumulate mtDNA mutations, and dysfunctional mitochondria, and show accelerated aging phenotypes (Kujoth et al., 2005; Trifunovic et al., 2004), demonstrating MiDAS in vivo (Wiley et al., 2016).
Thus, cellular senescence might be an important consequence of age-related decline in mitochondrial homeostasis. Because of the key roles of senescence and mitochondrial dysfunction in aging and age-related tissue dysfunction and disease, more in vivo studies in animal models and in human tissues are needed.
Collectively, current data suggests that cellular senescence integrates several regulators of longevity and health span, i.e. genome instability and DDR, mitochondrial metabolism, AMPK activation, and mTOR pathway function, which are largely druggable targets, including senescent cells and SASP (Arriola Apelo et al., 2016; Bai et al., 2011; Fang et al., 2016a, a; Kirkland et al., 2017; Kraig et al., 2018; Lin and Hardie, 2018; Zhang et al., 2016), which can also be modified by lifestyle changes (Di Francesco et al., 2018; Mattson et al., 2018).
9. Dynamic modeling of modulators of longevity and health span
A central objective in aging research is to develop intervention strategies to reduce age-associated illness and frailty, and to improve quality of life at old age. The action of the longevity signaling pathways is influenced by highly dynamic and complex system of interactions and feedback loops, and is controlled at several levels including gene expression, subcellular localization, and post-translational modification, and importantly by external factors like lifestyle and various compounds (Figs. 1–3).
Fig. 3.
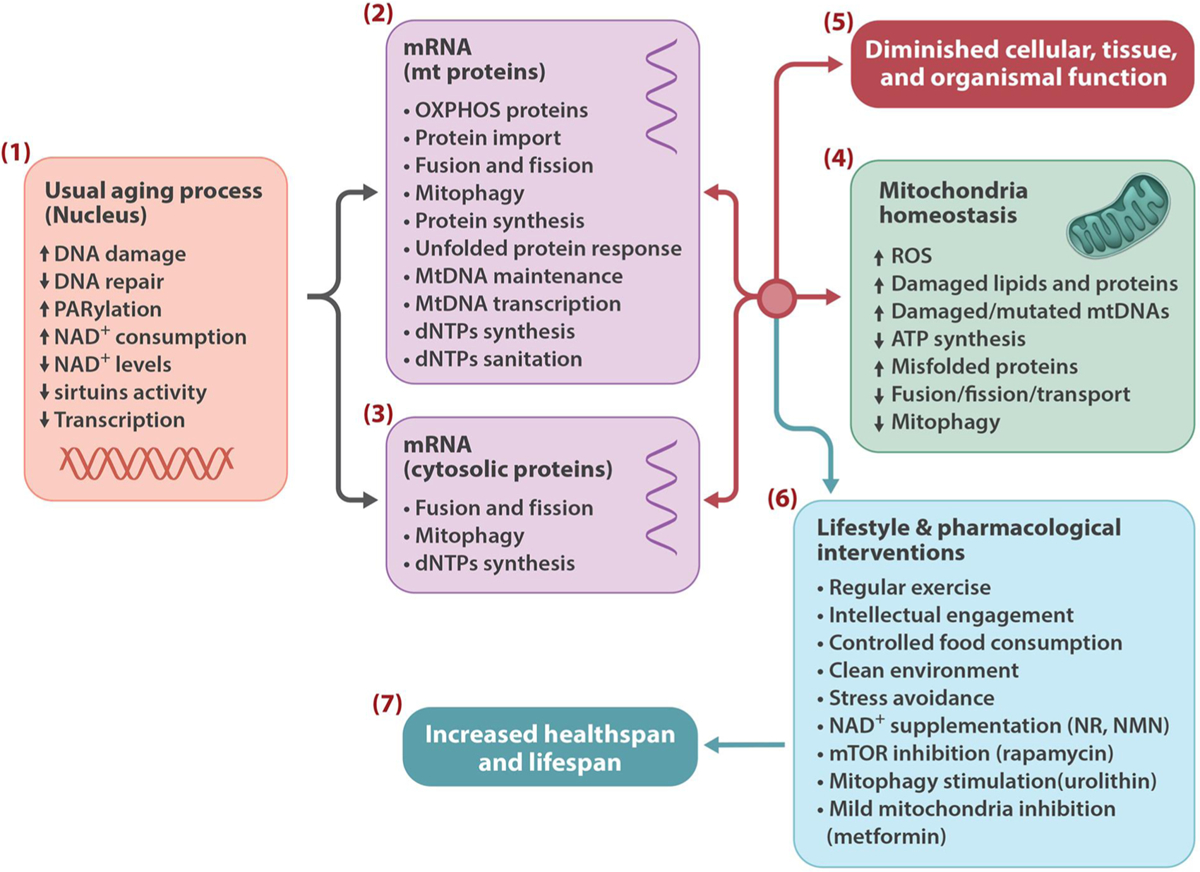
Multiple processes may link nuclear DNA instability to mitochondrial dysfunction and aging, several of which are malleable to modification by lifestyle and drug interventions. Persistent DNA damage and DNA damage response (PARylation), reduces cellular NAD+ content resulting in aberrant activity of sirtuins and their downstream targets (1). These events can affect mitochondrial proteins (2) and cytosolic proteins that control mitochondrial quality and mtDNA maintenance (3), leading to mitochondrial stress and loss of mitochondrial homeostasis (4), which ultimately result in tissue and organismal dysfunction (5). Life style and pharmacological interventions (6) may improve health span and increase lifespan by correcting these events (7).
Computer modeling has been developed to handle complex biological systems (Dalle Pezze et al., 2014; Guimera et al., 2019; Mc Auley et al., 2017). Integrated dynamic computer modeling represents a powerful tool to bring together the actions and responses determined experimentally in order to quantify and predict the outcomes of various drug interventions on the longevity regulating pathways and mitochondrial function, and to develop testable models for intervention strategies.
10. Conclusions
The biology of the aging process is complex, shows large inter-individual variations, and is largely driven by the accumulation of stochastic damage to tissues and cellular components over time.
Impaired mitochondrial function and homeostasis is now commonly recognized to constitute a central connection to multiple aspects of the aging process and age-onset frailty and diseases (Lopez-Otin et al., 2013; Mattson and Arumugam, 2018). This is largely because of the central position of mitochondria in key cellular processes, as well as, the control of mitochondrial homeostasis by numerous processes taking place within and outside the mitochondria. Interestingly, mitochondrial dysfunction has emerged as a malleable therapeutic target (Fang et al., 2019, 2016a; Ryu et al., 2016). This seems to be a research area of great opportunity going forward.
Single cell and multicellular organisms have evolved interconnected signaling mechanisms to sense and respond to genome damage, and to the environmental cues and nutrients to regulate their internal needs for growth, proliferation and maintenance. Research has consistently shown that the manipulation of some of these pathways markedly affects lifespan and health span in various model organisms. A remarkable outcome of these studies is the identification of lifestyle and pharmacological interventions that act on these pathways and can potentially delay or reduce age-associated chronic illnesses and extend lifespan (Fang et al., 2017a, b; Mattson et al., 2018) (Fig. 3). Some of the drugs that promote health span and extend lifespan in animal models are already in clinical use for other purposes, and some seem safe to be tested in human clinical trials (Dellinger et al., 2017; Martens et al., 2018; Newman et al., 2016).
With the advancing knowledge and the growing insight into the genetic and biochemistry of aging, together with continual improvement in biomarkers of biological aging, safer and target specific intervention strategies may be developed to improve mitochondrial biology and to modulate the longevity signaling pathways to provide a healthier aging for the growing elderly populations worldwide.
Acknowledgments
This work was supported by NORDEA Foundation, Denmark (02-2013-0220); EU Joint Program-Neurodegenerative Disease Research (JPND); Innovation Fund Denmark (5188-00001); Olav Thon foundation Norway (531811-710131); Novo Nordisk foundation Denmark (NNF17OC0027812); Intramural Research Program of the NIH, National Institute on Aging, USA (AG000733). We thank Marc Raley at National Institute on Aging, NIH, Baltimore, USA, for the figures.
Nomenclature
- AMPK
Adenosine monophosphate kinase
- ATM
Ataxia telangiectasia mutated
- ATP
Adenosine 5´-triphosphate
- BER DNA
base excision repair
- C. elegans
Caenorhabditis elegans
- DDR
DNA damage response
- DR
Dietary restriction
- D. melanogaster
Drosophila melanogaster
- ETC
Mitochondrial electron transport chain
- ELK
ETS-Like 1 protein
- ERK1/2
Extracellular signal-regulated kinase 1/2
- FOXO
Forkhead box O
- GH
Growth hormone
- IIS
Insulin and insulin-like-growth factor (IGF-1) signaling
- MAPK
Mitogen-activated protein kinase
- mtDNA
Mitochondrial DNA
- mTOR
Mechanistic target of rapamycin
- NAD+
Nicotinamide adenine dinucleotide (oxidized)
- NMN
Nicotinamide mononucleotide
- OXPHOS
Oxidative phosphorylation
- PGC1-α
Peroxisome proliferator-activated receptor-gamma co-activator-1α
- ROS
Reactive oxygen species
- SOD
Superoxide dismutase
- TCA cycle
Tricaboxylic acid cycle
Footnotes
Declaration of Competing Interest
None.
References
- Aamann MD, Sorensen MM, Hvitby C, Berquist BR, Muftuoglu M, Tian J, de Souza-Pinto NC, Scheibye-Knudsen M, Wilson DM 3rd, Stevnsner T, Bohr VA, 2010. Cockayne syndrome group B protein promotes mitochondrial DNA stability by supporting the DNA repair association with the mitochondrial membrane. FASEB J. 24, 2334–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Passos JF, Birket MJ, Beckmann T, Brings S, Peters H, Birch-Machin MA, von Zglinicki T, Saretzki G, 2008. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J. Cell. Sci 121, 1046–1053. [DOI] [PubMed] [Google Scholar]
- Akbari M, Krokan HE, 2008. Cytotoxicity and mutagenicity of endogenous DNA base lesions as potential cause of human aging. Mech. Ageing Dev 129, 353–365. [DOI] [PubMed] [Google Scholar]
- Akbari M, Pena-Diaz J, Andersen S, Liabakk NB, Otterlei M, Krokan HE, 2009. Extracts of proliferating and non-proliferating human cells display different base excision pathways and repair fidelity. DNA Repair (Amst.) 8, 834–843. [DOI] [PubMed] [Google Scholar]
- Akbari M, Keijzers G, Maynard S, Scheibye-Knudsen M, Desler C, Hickson ID, Bohr VA, 2014. Overexpression of DNA ligase III in mitochondria protects cells against oxidative stress and improves mitochondrial DNA base excision repair. DNA Repair (Amst.) 16, 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V, 2011. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334, 1144–1147. [DOI] [PubMed] [Google Scholar]
- Amano H, Chaudhury A, Rodriguez-Aguayo C, Lu L, Akhanov V, Catic A, Popov YV, Verdin E, Johnson H, Stossi F, Sinclair DA, Nakamaru-Ogiso E, Lopez-Berestein G, Chang JT, Neilson JR, Meeker A, Finegold M, Baur JA, Sahin E, 2019. Telomere dysfunction induces sirtuin repression that drives telomere-dependent disease. Cell Metab. 29, 1274–1290 e1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R, Lagnado A, Maggiorani D, Walaszczyk A, Dookun E, Chapman J, Birch J, Salmonowicz H, Ogrodnik M, Jurk D, Proctor C, Correia-Melo C, Victorelli S, Fielder E, Berlinguer-Palmini R, Owens A, Greaves LC, Kolsky KL, Parini A, Douin-Echinard V, LeBrasseur NK, Arthur HM, Tual-Chalot S, Schafer MJ, Roos CM, Miller JD, Robertson N, Mann J, Adams PD, Tchkonia T, Kirkland JL, Mialet-Perez J, Richardson GD, Passos JF, 2019. Length-independent telomere damage drives post-mitotic cardiomyocyte senescence. EMBO J. 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jorgensen M, Andersen PR, Bertin N, Rackham O, Burroughs AM, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume DA, Jensen TH, Suzuki H, Hayashizaki Y, Muller F, Consortium F, Forrest AR, Carninci P, Rehli M, Sandelin A, 2014. An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Blackburn EH, 2012. The telomere syndromes. Nature reviews. Genetics 13, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriola Apelo SI, Pumper CP, Baar EL, Cummings NE, Lamming DW, 2016. Intermittent administration of rapamycin extends the life span of female C57BL/6J mice. J. Gerontol. A Biol. Sci. Med. Sci 71, 876–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamna H, Cheung I, Ames BN, 2000. A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc. Natl. Acad. Sci. U.S.A 97, 686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aw WC, Towarnicki SG, Melvin RG, Youngson NA, Garvin MR, Hu Y, Nielsen S, Thomas T, Pickford R, Bustamante S, Vila-Sanjurjo A, Smyth GK, Ballard JWO, 2018. Genotype to phenotype: diet-by-mitochondrial DNA haplotype interactions drive metabolic flexibility and organismal fitness. PLoS Genet. 14, e1007735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Moraes CT, 2009. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 37, 4218–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, Schoonjans K, Schreiber V, Sauve AA, Menissierde Murcia J, Auwerx J, 2011. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 13, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM, 2011. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Ledderose C, Graf AF, Brix B, Birsak T, Lee A, Zhang J, Junger WG, 2015. mTOR and differential activation of mitochondria orchestrate neutrophil chemotaxis. J. Cell Biol 210, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RP, Fouquerel E, Opresko PL, 2019. The impact of oxidative DNA damage and stress on telomere homeostasis. Mech. Ageing Dev 177, 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A, Westbrook R, 2012. Metabolic characteristics of long-lived mice. Front. Genet 3, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A, Sun LY, Longo V, 2013. Somatotropic signaling: trade-offs between growth, reproductive development, and longevity. Physiol. Rev 93, 571–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome A, Garcia-Aguilar A, Asahara SI, Kido Y, Guillen C, Pajvani UB, Benito M, 2017. MTORC1 regulates both general autophagy and mitophagy induction after oxidative phosphorylation uncoupling. Mol. Cell. Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batenburg NL, Mitchell TR, Leach DM, Rainbow AJ, Zhu XD, 2012. Cockayne Syndrome group B protein interacts with TRF2 and regulates telomere length and stability. Nucleic Acids Res. 40, 9661–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra I, Howell JJ, Asara JM, Manning BD, 2013. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 339, 1323–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, Zorzato F, Hall MN, Ruegg MA, 2008. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 8, 411–424. [DOI] [PubMed] [Google Scholar]
- Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N, 1996. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 15, 658–664. [PMC free article] [PubMed] [Google Scholar]
- Berglund AK, Navarrete C, Engqvist MK, Hoberg E, Szilagyi Z, Taylor RW, Gustafsson CM, Falkenberg M, Clausen AR, 2017. Nucleotide pools dictate the identity and frequency of ribonucleotide incorporation in mitochondrial DNA. PLoS Genet. 13, e1006628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Hall MN, 2013. Where is mTOR and what is it doing there? J. Cell Biol 203, 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Stracka D, Prescianotto-Baschong C, Frieden M, Demaurex N, Hall MN, 2013. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl. Acad. Sci. U.S.A 110, 12526–12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielas J, Herbst A, Widjaja K, Hui J, Aiken JM, McKenzie D, Miller RA, Brooks SV, Wanagat J, 2018. Long term rapamycin treatment improves mitochondrial DNA quality in aging mice. Exp. Gerontol 106, 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L, 2010. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11, 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelakovic G, Nikolova D, Gluud C, 2014. Antioxidant supplements and mortality. Curr. Opin. Clin. Nutr. Metab. Care 17, 40–44. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Epel ES, Lin J, 2015. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198. [DOI] [PubMed] [Google Scholar]
- Bluher M, Kahn BB, Kahn CR, 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299, 572–574. [DOI] [PubMed] [Google Scholar]
- Braidy N, Guillemin GJ, Mansour H, Chan-Ling T, Poljak A, Grant R, 2011. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS One 6, e19194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL, 1994. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369, 756–758. [DOI] [PubMed] [Google Scholar]
- Brown TA, Tkachuk AN, Shtengel G, Kopek BG, Bogenhagen DF, Hess HF, Clayton DA, 2011. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell. Biol 31, 4994–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg HM, 2015. The somatotropic axis and longevity in mice. American journal of physiology. Endocrinol. Metab 309, E503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg HM, Johnson WT, Rakoczy SG, 2012. Expression of oxidative phosphorylation components in mitochondria of long-living Ames dwarf mice. Age 34, 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brys K, Castelein N, Matthijssens F, Vanfleteren JR, Braeckman BP, 2010. Disruption of insulin signalling preserves bioenergetic competence of mitochondria in ageing Caenorhabditis elegans. BMC Biol. 8, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bua E, Johnson J, Herbst A, Delong B, McKenzie D, Salamat S, Aiken JM, 2006. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet 79, 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkewitz K, Zhang Y, Mair WB, 2014. AMPK at the nexus of energetics and aging. Cell Metab. 20, 10–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelof DC, Raffoul JJ, Yanamadala S, Ganir C, Guo Z, Heydari AR, 2002. Attenuation of DNA polymerase beta-dependent base excision repair and increased DMS-induced mutagenicity in aged mice. Mutat. Res 500, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas E, Davies KJ, 2000. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic. Biol. Med 29, 222–230. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK, 2016. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J, 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Menzies KJ, Auwerx J, 2015. NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canugovi C, Shamanna RA, Croteau DL, Bohr VA, 2014. Base excision DNA repair levels in mitochondrial lysates of Alzheimer’s disease. Neurobiol. Aging 35, 1293–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli V, Achilli A, Valentino ML, Rengo C, Semino O, Pala M, Olivieri A, Mattiazzi M, Pallotti F, Carrara F, Zeviani M, Leuzzi V, Carducci C, Valle G, Simionati B, Mendieta L, Salomao S, Belfort R Jr., Sadun AA, Torroni A, 2006. Haplogroup effects and recombination of mitochondrial DNA: novel clues from the analysis of Leber hereditary optic neuropathy pedigrees. Am. J. Hum. Genet 78, 564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero D, Soria-Valles C, Lopez-Otin C, 2016. Hallmarks of progeroid syndromes: lessons from mice and reprogrammed cells. Dis. Model. Mech 9, 719–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castets P, Lin S, Rion N, Di Fulvio S, Romanino K, Guridi M, Frank S, Tintignac LA, Sinnreich M, Ruegg MA, 2013. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab. 17, 731–744. [DOI] [PubMed] [Google Scholar]
- Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, Hwang D, Park KS, 2013. A systems approach for decoding mitochondrial retrograde signaling pathways. Sci. Signal 6, rs4. [DOI] [PubMed] [Google Scholar]
- Chan DC, 2012. Fusion and fission: interlinked processes critical for mitochondrial health. Annu. Rev. Genet 46, 265–287. [DOI] [PubMed] [Google Scholar]
- Chapman J, Fielder E, Passos JF, 2019. Mitochondrial dysfunction and cell senescence: deciphering a complex relationship. FEBS Lett. [DOI] [PubMed] [Google Scholar]
- Chen LY, Zhang Y, Zhang Q, Li H, Luo Z, Fang H, Kim SH, Qin L, Yotnda P, Xu J, Tu BP, Bai Y, Songyang Z, 2012. Mitochondrial localization of telomeric protein TIN2 links telomere regulation to metabolic control. Mol. Cell 47, 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow HM, Herrup K, 2015. Genomic integrity and the ageing brain. Nature reviews. Neuroscience 16, 672–684. [DOI] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L, 2001. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science 292, 104–106. [DOI] [PubMed] [Google Scholar]
- Cline SD, 2012. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochim. Biophys. Acta 1819, 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline SD, Lodeiro MF, Marnett LJ, Cameron CE, Arnold JJ, 2010. Arrest of human mitochondrial RNA polymerase transcription by the biological aldehyde adduct of DNA, M1dG. Nucleic Acids Res. 38, 7546–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquel F, Silva MJ, Techer H, Zadorozhny K, Sharma S, Nieminuszczy J, Mettling C, Dardillac E, Barthe A, Schmitz AL, Promonet A, Cribier A, Sarrazin A, Niedzwiedz W, Lopez B, Costanzo V, Krejci L, Chabes A, Benkirane M, Lin YL, Pasero P, 2018. SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557, 57–61. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC, 1992. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat. Genet 2, 324–329. [DOI] [PubMed] [Google Scholar]
- Correia-Melo C, Marques FD, Anderson R, Hewitt G, Hewitt R, Cole J, Carroll BM, Miwa S, Birch J, Merz A, Rushton MD, Charles M, Jurk D, Tait SW, Czapiewski R, Greaves L, Nelson G, Bohlooly YM, Rodriguez-Cuenca S, Vidal-Puig A, Mann D, Saretzki G, Quarato G, Green DR, Adams PD, von Zglinicki T, Korolchuk VI, Passos JF, 2016. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 35, 724–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, Rossi ML, Canugovi C, Tian J, Sykora P, Ramamoorthy M, Wang ZM, Singh DK, Akbari M, Kasiviswanathan R, Copeland WC, Bohr VA, 2012. RECQL4 localizes to mitochondria and preserves mitochondrial DNA integrity. Aging Cell 11, 456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, Popuri V, Opresko PL, Bohr VA, 2014. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem 83, 519–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF, 1997. The high spontaneous mutation rate: is it a health risk? Proc. Natl. Acad. Sci. U.S.A 94, 8380–8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, Baumann C, Baxter P, Bertini E, Chandler KE, Chitayat D, Cau D, Dery C, Fazzi E, Goizet C, King MD, Klepper J, Lacombe D, Lanzi G, Lyall H, Martinez-Frias ML, Mathieu M, McKeown C, Monier A, Oade Y, Quarrell OW, Rittey CD, Rogers RC, Sanchis A, Stephenson JB, Tacke U, Till M, Tolmie JL, Tomlin P, Voit T, Weschke B, Woods CG, Lebon P, Bonthron DT, Ponting CP, Jackson AP, 2006. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat. Genet 38, 910–916. [DOI] [PubMed] [Google Scholar]
- Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P, 2007. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450, 736–740. [DOI] [PubMed] [Google Scholar]
- Dalle Pezze P, Nelson G, Otten EG, Korolchuk VI, Kirkwood TB, von Zglinicki T, Shanley DP, 2014. Dynamic modelling of pathways to cellular senescence reveals strategies for targeted interventions. PLoS Comput. Biol 10, e1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N, 2004. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 19, 169–185. [DOI] [PubMed] [Google Scholar]
- de Lange T, 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110. [DOI] [PubMed] [Google Scholar]
- de Souza-Pinto NC, Mason PA, Hashiguchi K, Weissman L, Tian J, Guay D, Lebel M, Stevnsner TV, Rasmussen LJ, Bohr VA, 2009. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair (Amst.) 8, 704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBalsi KL, Hoff KE, Copeland WC, 2017. Role of the mitochondrial DNA replication machinery in mitochondrial DNA mutagenesis, aging and age-related diseases. Ageing Res. Rev 33, 89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrabant B, Soerensen M, Flachsbart F, Dato S, Mengel-From J, Stevnsner T, Bohr VA, Kruse TA, Schreiber S, Nebel A, Christensen K, Tan Q, Christiansen L, 2014. Human longevity and variation in DNA damage response and repair: study of the contribution of sub-processes using competitive gene-set analysis. Eur. J. Hum. Genet 22, 1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, Pelloquin L, Grosgeorge J, Turc-Carel C, Perret E, Astarie-Dequeker C, Lasquellec L, Arnaud B, Ducommun B, Kaplan J, Hamel CP, 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet 26, 207–210. [DOI] [PubMed] [Google Scholar]
- Dellinger RW, Santos SR, Morris M, Evans M, Alminana D, Guarente L, Marcotulli E, 2017. Repeat dose NRPT (nicotinamide riboside and pterostilbene) increases NAD(+) levels in humans safely and sustainably: a randomized, double-blind, placebo-controlled study. NPJ Aging Mech. Dis 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt I, Petyuk VA, Cai H, Vandemeulebroucke L, Vierstraete A, Smith RD, Depuydt G, Braeckman BP, 2016. FOXO/DAF-16 activation slows down turnover of the majority of proteins in C. Elegans . Cell Rep 16, 3028–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Francesco A, Di Germanio C, Bernier M, de Cabo R, 2018. A time to fast. Science 362, 770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O, et al. , 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U.S.A 92, 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman JB, Albinder B, Shroyer T, Kenyon C, 1995. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics 141, 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drago I, De Stefani D, Rizzuto R, Pozzan T, 2012. Mitochondrial Ca2+ uptake contributes to buffering cytoplasmic Ca2+ peaks in cardiomyocytes. Proc. Natl. Acad. Sci. U.S.A 109, 12986–12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA, 2009. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell. Biol 29, 4274–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi-Fakhari D, Saffari A, Wahlster L, Di Nardo A, Turner D, Lewis TL Jr., Conrad C, Rothberg JM, Lipton JO, Kolker S, Hoffmann GF, Han MJ, Polleux F, Sahin M, 2016. Impaired mitochondrial dynamics and mitophagy in neuronal models of tuberous sclerosis complex. Cell Rep. 17, 1053–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimkhani MR, Daneshmand A, Mazumder A, Allocca M, Calvo JA, Abolhassani N, Jhun I, Muthupalani S, Ayata C, Samson LD, 2014. Aag-initiated base excision repair promotes ischemia reperfusion injury in liver, brain, and kidney. Proc. Natl. Acad. Sci. U.S.A 111, E4878–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ, 2011. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331, 456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hattab AW, Scaglia F, 2013. Mitochondrial DNA depletion syndromes: review and updates of genetic basis, manifestations, and therapeutic options. Neurotherapeutics 10, 186–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF, 2008. Pathogenic mitochondrial DNA mutations are common in the general population. Am. J. Hum. Genet 83, 254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD, 2001. Regulation of longevity and stress resistance by Sch9 in yeast. Science 292, 288–290. [DOI] [PubMed] [Google Scholar]
- Fakouri NB, Hou Y, Demarest TG, Christiansen LS, Okur MN, Mohanty JG, Croteau DL, Bohr VA, 2018. Toward understanding genomic instability, mitochondrial dysfunction and aging. FEBS J. [DOI] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA, 2014. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 157, 882–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Kassahun H, Croteau DL, Scheibye-Knudsen M, Marosi K, Lu H, Shamanna RA, Kalyanasundaram S, Bollineni RC, Wilson MA, Iser WB, Wollman BN, Morevati M, Li J, Kerr JS, Lu Q, Waltz TB, Tian J, Sinclair DA, Mattson MP, Nilsen H, Bohr VA, 2016a. NAD(+) replenishment improves lifespan and Healthspan in Ataxia telangiectasia models via Mitophagy and DNA repair. Cell Metab. 24, 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Chua KF, Mattson MP, Croteau DL, Bohr VA, 2016b. Nuclear DNA damage signalling to mitochondria in ageing. Nature reviews. Molecular cell biology 17, 308–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Lautrup S, Hou Y, Demarest TG, Croteau DL, Mattson MP, Bohr VA, 2017a. NAD(+) in aging: molecular mechanisms and translational implications. Trends Mol. Med 23, 899–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Waltz TB, Kassahun H, Lu Q, Kerr JS, Morevati M, Fivenson EM, Wollman BN, Marosi K, Wilson MA, Iser WB, Eckley DM, Zhang Y, Lehrmann E, Goldberg IG, Scheibye-Knudsen M, Mattson MP, Nilsen H, Bohr VA, Becker KG, 2017b. Tomatidine enhances lifespan and healthspan in C. Elegans through mitophagy induction via the SKN-1/Nrf2 pathway. Sci. Rep 7, 46208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Hou Y, Palikaras K, Adriaanse BA, Kerr JS, Yang B, Lautrup S, Hasan-Olive MM, Caponio D, Dan X, Rocktaschel P, Croteau DL, Akbari M, Greig NH, Fladby T, Nilsen H, Cader MZ, Mattson MP, Tavernarakis N, Bohr VA, 2019. Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci 22, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet G, Jansson M, Sternberg D, Moslemi AR, Blondy P, Lombes A, Fardeau M, Oldfors A, 2002. Ageing muscle: clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function. Neuromuscul. Disord 12, 484–493. [DOI] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S, 2001. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1, 633–644. [DOI] [PubMed] [Google Scholar]
- Fernandez AM, Torres-Aleman I, 2012. The many faces of insulin-like peptide signalling in the brain. Nature reviews. Neuroscience 13, 225–239. [DOI] [PubMed] [Google Scholar]
- Fielder E, von Zglinicki T, Jurk D, 2017. The DNA Damage Response in Neurons: Die by Apoptosis or Survive in a Senescence-Like State? J. Alzheimers Dis [DOI] [PubMed] [Google Scholar]
- Forslund JME, Pfeiffer A, Stojkovic G, Wanrooij PH, Wanrooij S, 2018. The presence of rNTPs decreases the speed of mitochondrial DNA replication. PLoS Genet. 14, e1007315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquerel E, Goellner EM, Yu Z, Gagne JP, Barbi de Moura M, Feinstein T, Wheeler D, Redpath P, Li J, Romero G, Migaud M, Van Houten B, Poirier GG, Sobol RW, 2014. ARTD1/PARP1 negatively regulates glycolysis by inhibiting hexokinase 1 independent of NAD+ depletion. Cell Rep. 8, 1819–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C, Garagnani P, Vitale G, Capri M, Salvioli S, 2017. Inflammaging and’ Garb-aging’ Trends Endocrinol. Metab. 28, 199–212. [DOI] [PubMed] [Google Scholar]
- Freund A, Orjalo AV, Desprez PY, Campisi J, 2010. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol. Med 16, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE, 1988. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics 118, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Calvo JA, Samson LD, 2012. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nature reviews. Cancer 12, 104–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahl RF, Dwivedi P, Tjandra N, 2016. Bcl-2 proteins bid and bax form a network to permeabilize the mitochondria at the onset of apoptosis. Cell Death Dis. 7, e2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jaattela M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Munz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Kroemer G, 2017. Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X, 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem 284, 12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Roves PM, Osler ME, Holmstrom MH, Zierath JR, 2008. Gain-of-function R225Q mutation in AMP-activated protein kinase gamma3 subunit increases mitochondrial biogenesis in glycolytic skeletal muscle. J. Biol. Chem 283, 35724–35734. [DOI] [PubMed] [Google Scholar]
- Garreau-Balandier I, Lefebvre M, Jacquard S, Caillat S, Cruz-Rodriguez L, Ishak L, Agier V, Morel F, Lachaume P, Dubessay P, Sauvaigo S, Alziari S, Vernet P, 2014. A comprehensive approach to determining BER capacities and their change with aging in Drosophila melanogaster mitochondria by oligonucleotide microarray. FEBS Lett. 588, 1673–1679. [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Rossi ML, Singh DK, Dunn C, Ramamoorthy M, Croteau DL, Liu Y, Bohr VA, 2012. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. J. Biol. Chem 287, 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedt RJ, Fumene Feruglio P, Pathania D, Yang KS, Kilcoyne A, Vinegoni C, Mitchison TJ, Weissleder R, 2016. Computational imaging reveals mitochondrial morphology as a biomarker of cancer phenotype and drug response. Sci. Rep 6, 32985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson RW, De Vries RL, Lebot P, Wikstrom JD, Torgyekes E, Shirihai OS, Przedborski S, Schon EA, 2012. Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Hum. Mol. Genet 21, 978–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godowski PJ, Leung DW, Meacham LR, Galgani JP, Hellmiss R, Keret R, Rotwein PS, Parks JS, Laron Z, Wood WI, 1989. Characterization of the human growth hormone receptor gene and demonstration of a partial gene deletion in two patients with Laron-type dwarfism. Proc. Natl. Acad. Sci. U.S.A 86, 8083–8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves VF, Giamberardino SN, Crowley JJ, Vawter MP, Saxena R, Bulik CM, Yilmaz Z, Hultman CM, Sklar P, Kennedy JL, Sullivan PF, Knight J, 2018. Examining the role of common and rare mitochondrial variants in schizophrenia. PLoS One 13, e0191153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Freire M, Scalzo P, D’Agostino J, Moore ZA, Diaz-Ruiz A, Fabbri E, Zane A, Chen B, Becker KG, Lehrmann E, Zukley L, Chia CW, Tanaka T, Coen PM, Bernier M, de Cabo R, Ferrucci L, 2018. Skeletal muscle ex vivo mitochondrial respiration parallels decline in vivo oxidative capacity, cardiorespiratory fitness, and muscle strength: the Baltimore Longitudinal Study of Aging. Aging Cell 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A, Mao Z, Hine C, 2007. Changes in DNA repair during aging. Nucleic Acids Res. 35, 7466–7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goutieres F, Aicardi J, Barth PG, Lebon P, 1998. Aicardi-Goutieres syndrome: an update and results of interferon-alpha studies. Ann. Neurol 44, 900–907. [DOI] [PubMed] [Google Scholar]
- Gowans GJ, Hawley SA, Ross FA, Hardie DG, 2013. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 18, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves LC, Nooteboom M, Elson JL, Tuppen HA, Taylor GA, Commane DM, Arasaradnam RP, Khrapko K, Taylor RW, Kirkwood TB, Mathers JC, Turnbull DM, 2014. Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing. PLoS Genet. 10, e1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredilla R, Bohr VA, Stevnsner T, 2010. Mitochondrial DNA repair and association with aging-an update. Exp. Gerontol 45, 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A, 2007. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem 282, 30107–30119. [DOI] [PubMed] [Google Scholar]
- Grossman LI, Watson R, Vinograd J, 1973. The presence of ribonucleotides in mature closed-circular mitochondrial DNA. Proc. Natl. Acad. Sci. U.S.A 70, 3339–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera AM, Shanley DP, Proctor CJ, 2019. Modelling the role of redox-related mechanisms in musculoskeletal ageing. Free Radic. Biol. Med 132, 11–18. [DOI] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H, 2011. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci 14, 1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ, 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendeler J, Drose S, Buchner N, Jakob S, Altschmied J, Goy C, Spyridopoulos I, Zeiher AM, Brandt U, Dimmeler S, 2009. Mitochondrial telomerase reverse transcriptase binds to and protects mitochondrial DNA and function from damage. Arterioscler. Thromb. Vasc. Biol 29, 929–935. [DOI] [PubMed] [Google Scholar]
- Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, Hall MN, 2012. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab. 15, 725–738. [DOI] [PubMed] [Google Scholar]
- Halsne R, Esbensen Y, Wang W, Scheffler K, Suganthan R, Bjoras M, Eide L, 2012. Lack of the DNA glycosylases MYH and OGG1 in the cancer prone double mutant mouse does not increase mitochondrial DNA mutagenesis. DNA Repair (Amst.) 11, 278–285. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Schaffer BE, Brunet A, 2016. AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 26, 190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D, 1956. Aging: a theory based on free radical and radiation chemistry. J. Gerontol 11, 298–300. [DOI] [PubMed] [Google Scholar]
- Harrison JF, Hollensworth SB, Spitz DR, Copeland WC, Wilson GL, LeDoux SP, 2005. Oxidative stress-induced apoptosis in neurons correlates with mitochondrial DNA base excision repair pathway imbalance. Nucleic Acids Res. 33, 4660–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA, 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T, Eshtad S, Nik-Zainal S, 2014. Mechanisms underlying mutational signatures in human cancers. Nat. Rev. Genet 15, 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Shaw RJ, 2018. AMPK: guardian of metabolism and mitochondrial homeostasis. Nature reviews. Molecular cell biology 19, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt G, Jurk D, Marques FD, Correia-Melo C, Hardy T, Gackowska A, Anderson R, Taschuk M, Mann J, Passos JF, 2012. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Commun 3, 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, Reyes A, 2012. Human mitochondrial DNA replication. Cold Spring Harb. Perspect. Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y, 2003. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421, 182–187. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S, 1999. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 13, 1385–1393. [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N, 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991. http://www.mitomap.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G, Sims R, Harold D, Chapman J, Hollingworth P, Gerrish A, Russo G, Hamshere M, Moskvina V, Jones N, Thomas C, Stretton A, Holmans PA, O’Donovan MC, Owen MJ, Williams J, Chinnery PF, Consortium G, 2012. No consistent evidence for association between mtDNA variants and Alzheimer disease. Neurology 78, 1038–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibebunjo C, Chick JM, Kendall T, Eash JK, Li C, Zhang Y, Vickers C, Wu Z, Clarke BA, Shi J, Cruz J, Fournier B, Brachat S, Gutzwiller S, Ma Q, Markovits J, Broome M, Steinkrauss M, Skuba E, Galarneau JR, Gygi SP, Glass DJ, 2013. Genomic and proteomic profiling reveals reduced mitochondrial function and disruption of the neuromuscular junction driving rat sarcopenia. Mol. Cell. Biol 33, 194–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai SI, Guarente L, 2016. It takes two to tango: NAD(+) and sirtuins in aging/longevity control. NPJ Aging Mech. Dis 2, 16017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Zhu T, Guan KL, 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN, 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyama T, Wilson DM 3rd, 2013. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst.) 12, 620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN, 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol 6, 1122–1128. [DOI] [PubMed] [Google Scholar]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM, 2007. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc. Natl. Acad. Sci. U.S.A 104, 12017–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiricny J, 2006. The multifaceted mismatch-repair system. Nature reviews. Molecular cell biology 7, 335–346. [DOI] [PubMed] [Google Scholar]
- Johnson SC, Yanos ME, Kayser EB, Quintana A, Sangesland M, Castanza A, Uhde L, Hui J, Wall VZ, Gagnidze A, Oh K, Wasko BM, Ramos FJ, Palmiter RD, Rabinovitch PS, Morgan PG, Sedensky MM, Kaeberlein M, 2013. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones OR, Scheuerlein A, Salguero-Gomez R, Camarda CG, Schaible R, Casper BB, Dahlgren JP, Ehrlen J, Garcia MB, Menges ES, Quintana-Ascencio PF, Caswell H, Baudisch A, Vaupel JW, 2014. Diversity of ageing across the tree of life. Nature 505, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junnila RK, List EO, Berryman DE, Murrey JW, Kopchick JJ, 2013. The GH/IGF-1 axis in ageing and longevity. Nature reviews. Endocrinology 9, 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK, 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kalfalah F, Seggewiss S, Walter R, Tigges J, Moreno-Villanueva M, Burkle A, Ohse S, Busch H, Boerries M, Hildebrandt B, Royer-Pokora B, Boege F, 2015. Structural chromosome abnormalities, increased DNA strand breaks and DNA strand break repair deficiency in dermal fibroblasts from old female human donors. Aging 7, 110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyanaraman B, Darley-Usmar V, Davies KJ, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ 2nd, Ischiropoulos H, 2012. Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med 52, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Nishida J, Iyama A, Nakabeppu Y, Furuichi M, Fujiwara T, Sekiguchi M, Takeshige K, 1995. Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J. Biol. Chem 270, 14659–14665. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S, 2004. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol 14, 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EM, Cascante M, Shlomi T, Gottlieb E, Peeper DS, 2013. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498, 109–112. [DOI] [PubMed] [Google Scholar]
- Karikkineth AC, Scheibye-Knudsen M, Fivenson E, Croteau DL, Bohr VA, 2017. Cockayne syndrome: clinical features, model systems and pathways. Ageing Res. Rev 33, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasiviswanathan R, Copeland WC, 2011. Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J. Biol. Chem 286, 31490–31500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katic M, Kennedy AR, Leykin I, Norris A, McGettrick A, Gesta S, Russell SJ, Bluher M, Maratos-Flier E, Kahn CR, 2007. Mitochondrial gene expression and increased oxidative metabolism: role in increased lifespan of fat-specific insulin receptor knock-out mice. Aging Cell 6, 827–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppila TE, Kauppila JH, Larsson NG, 2017. Mammalian mitochondria and aging: an update. Cell Metab. 25, 57–71. [DOI] [PubMed] [Google Scholar]
- Kauppila JHK, Bonekamp NA, Mourier A, Isokallio MA, Just A, Kauppila TES, Stewart JB, Larsson NG, 2018. Base-excision repair deficiency alone or combined with increased oxidative stress does not increase mtDNA point mutations in mice. Nucleic Acids Res. 46, 6642–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane M, Semeiks J, Webb AE, Li YI, Quesada V, Craig T, Madsen LB, van Dam S, Brawand D, Marques PI, Michalak P, Kang L, Bhak J, Yim HS, Grishin NV, Nielsen NH, Heide-Jorgensen MP, Oziolor EM, Matson CW, Church GM, Stuart GW, Patton JC, George JC, Suydam R, Larsen K, Lopez-Otin C, O’Connell MJ, Bickham JW, Thomsen B, de Magalhaes JP, 2015. Insights into the evolution of longevity from the bowhead whale genome. Cell Rep. 10, 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SR, Salk JJ, Schmitt MW, Loeb LA, 2013. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 9, e1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R, 1993. A C. Elegans mutant that lives twice as long as wild type. Nature 366, 461–464. [DOI] [PubMed] [Google Scholar]
- Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang L, Pradhan S, Kivela R, Pessia A, Velagapudi V, Suomalainen A, 2017. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab. 26, 419–428 e415. [DOI] [PubMed] [Google Scholar]
- Kidane D, Murphy DL, Sweasy JB, 2014. Accumulation of abasic sites induces genomic instability in normal human gastric epithelial cells during Helicobacter pylori infection. Oncogenesis 3, e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, Marino SM, Sun X, Turanov AA, Yang P, Yim SH, Zhao X, Kasaikina MV, Stoletzki N, Peng C, Polak P, Xiong Z, Kiezun A, Zhu Y, Chen Y, Kryukov GV, Zhang Q, Peshkin L, Yang L, Bronson RT, Buffenstein R, Wang B, Han C, Li Q, Chen L, Zhao W, Sunyaev SR, Park TJ, Zhang G, Wang J, Gladyshev VN, 2011a. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature 479, 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL, 2011b. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol 13, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G, 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942–946. [DOI] [PubMed] [Google Scholar]
- Kipling D, Davis T, Ostler EL, Faragher RG, 2004. What can progeroid syndromes tell us about human aging? Science 305, 1426–1431. [DOI] [PubMed] [Google Scholar]
- Kirkland JL, Tchkonia T, Zhu Y, Niedernhofer LJ, Robbins PD, 2017. The clinical potential of senolytic drugs. J. Am. Geriatr. Soc 65, 2297–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TB, 2005. Understanding the odd science of aging. Cell 120, 437–447. [DOI] [PubMed] [Google Scholar]
- Kirkwood TB, Austad SN, 2000. Why do we age? Nature 408, 233–238. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N, 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608. [DOI] [PubMed] [Google Scholar]
- Kovacic S, Soltys CL, Barr AJ, Shiojima I, Walsh K, Dyck JR, 2003. Akt activity negatively regulates phosphorylation of AMP-activated protein kinase in the heart. J. Biol. Chem 278, 39422–39427. [DOI] [PubMed] [Google Scholar]
- Kowald A, Kirkwood TB, 2016. Can aging be programmed? A critical literature review. Aging Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragelund BB, Weterings E, Hartmann-Petersen R, Keijzers G, 2016. The Ku70/80 ring in Non-Homologous End-Joining: easy to slip on, hard to remove. Front. Biosci 21, 514–527. [DOI] [PubMed] [Google Scholar]
- Kraig E, Linehan LA, Liang H, Romo TQ, Liu Q, Wu Y, Benavides AD, Curiel TJ, Javors MA, Musi N, Chiodo L, Koek W, Gelfond JAL, Kellogg DL Jr., 2018. A randomized control trial to establish the feasibility and safety of rapamycin treatment in an older human cohort: Immunological, physical performance, and cognitive effects. Exp. Gerontol 105, 53–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K, 2006. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet 38, 518–520. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Bjørås M, 2013. Base excision repair. Cold Spring Harb. Perspect. Biol 5, a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, Hofer T, Seo AY, Sullivan R, Jobling WA, Morrow JD, Van Remmen H, Sedivy JM, Yamasoba T, Tanokura M, Weindruch R, Leeuwenburgh C, Prolla TA, 2005. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science 309, 481–484. [DOI] [PubMed] [Google Scholar]
- Kwong LK, Sohal RS, 2000. Age-related changes in activities of mitochondrial electron transport complexes in various tissues of the mouse. Arch. Biochem. Biophys 373, 16–22. [DOI] [PubMed] [Google Scholar]
- Ladner RD, Caradonna SJ, 1997. The human dUTPase gene encodes both nuclear and mitochondrial isoforms. Differential expression of the isoforms and characterization of a cDNA encoding the mitochondrial species. J. Biol. Chem 272, 19072–19080. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, Guertin DA, Sabatini DM, Baur JA, 2012. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantier L, Fentz J, Mounier R, Leclerc J, Treebak JT, Pehmoller C, Sanz N, Sakakibara I, Saint-Amand E, Rimbaud S, Maire P, Marette A, Ventura-Clapier R, Ferry A, Wojtaszewski JF, Foretz M, Viollet B, 2014. AMPK controls exercise endurance, mitochondrial oxidative capacity, and skeletal muscle integrity. FASEB J. 28, 3211–3224. [DOI] [PubMed] [Google Scholar]
- Latorre-Pellicer A, Moreno-Loshuertos R, Lechuga-Vieco AV, Sanchez-Cabo F, Torroja C, Acin-Perez R, Calvo E, Aix E, Gonzalez-Guerra A, Logan A, Bernad-Miana ML, Romanos E, Cruz R, Cogliati S, Sobrino B, Carracedo A, Perez-Martos A, Fernandez-Silva P, Ruiz-Cabello J, Murphy MP, Flores I, Vazquez J, Enriquez JA, 2016. Mitochondrial and nuclear DNA matching shapes metabolism and healthy ageing. Nature 535, 561–565. [DOI] [PubMed] [Google Scholar]
- Leandro GS, Sykora P, Bohr VA, 2015. The impact of base excision DNA repair in age-related neurodegenerative diseases. Mutat. Res 776, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G, 2003a. DAF-16 target genes that control C. Elegans life-span and metabolism. Science 300, 644–647. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G, 2003b. A systematic RNAi screen identifies a critical role for mitochondria in C. Elegans longevity. Nat. Genet 33, 40–48. [DOI] [PubMed] [Google Scholar]
- Leon J, Sakumi K, Castillo E, Sheng Z, Oka S, Nakabeppu Y, 2016. 8-Oxoguanine accumulation in mitochondrial DNA causes mitochondrial dysfunction and impairs neuritogenesis in cultured adult mouse cortical neurons under oxidative conditions. Sci. Rep 6, 22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chen ZJ, 2018. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med 215, 1287–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang W, Chen Y, Guo W, Zhang J, Tang H, Xu Z, Zhang H, Tao Y, Wang F, Jiang Y, Sun FL, Mao Z, 2016. Impaired DNA double-strand break repair contributes to the age-associated rise of genomic instability in humans. Cell Death Differ. 23, 1765–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Shen L, Hu P, Huang R, Cao Y, Deng J, Yuan W, Liu D, Yang J, Gu H, Bai Y, 2017. Aging-associated mitochondrial DNA mutations alter oxidative phosphorylation machinery and cause mitochondrial dysfunctions. Biochim. Biophys. Acta [DOI] [PubMed] [Google Scholar]
- Lin SC, Hardie DG, 2018. AMPK: sensing glucose as well as cellular energy status. Cell Metab. 27, 299–313. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C, 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278, 1319–1322. [DOI] [PubMed] [Google Scholar]
- Lindahl T, 1993. Instability and decay of the primary structure of DNA. Nature 362, 709–715. [DOI] [PubMed] [Google Scholar]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G, 2013. The hallmarks of aging. Cell 153, 1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loson OC, Song Z, Chen H, Chan DC, 2013. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CL, Qin L, Liu HC, Candas D, Fan M, Li JJ, 2015. Tumor cells switch to mitochondrial oxidative phosphorylation under radiation via mTOR-mediated hexokinase II inhibition-a Warburg-reversing effect. PLoS One 10, e0121046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae SL, Croken MM, Calder RB, Aliper A, Milholland B, White RR, Zhavoronkov A, Gladyshev VN, Seluanov A, Gorbunova V, Zhang ZD, Vijg J, 2015. DNA repair in species with extreme lifespan differences. Aging 7, 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfoudhi E, Talhaoui I, Cabagnols X, Della Valle V, Secardin L, Rameau P, Bernard OA, Ishchenko AA, Abbes S, Vainchenker W, Saparbaev M, Plo I, 2016. TET2-mediated 5-hydroxymethylcytosine induces genetic instability and mutagenesis. DNA Repair (Amst.) 43, 78–88. [DOI] [PubMed] [Google Scholar]
- Mao K, Quipildor GF, Tabrizian T, Novaj A, Guan F, Walters RO, Delahaye F, Hubbard GB, Ikeno Y, Ejima K, Li P, Allison DB, Salimi-Moosavi H, Beltran PJ, Cohen P, Barzilai N, Huffman DM, 2018. Late-life targeting of the IGF-1 receptor improves healthspan and lifespan in female mice. Nat. Commun 9, 2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH, 2014. Understanding nucleotide excision repair and its roles in cancer and ageing. Nature reviews. Molecular cell biology 15, 465–481. [DOI] [PubMed] [Google Scholar]
- Martens CR, Denman BA, Mazzo MR, Armstrong ML, Reisdorph N, McQueen MB, Chonchol M, Seals DR, 2018. Chronic nicotinamide riboside supplementation is well-tolerated and elevates NAD(+) in healthy middle-aged and older adults. Nat. Commun 9, 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina JA, Chen Y, Gucek M, Puertollano R, 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins R, Lithgow GJ, Link W, 2016. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell 15, 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzetti E, Calvani R, Cesari M, Buford TW, Lorenzi M, Behnke BJ, Leeuwenburgh C, 2013. Mitochondrial dysfunction and sarcopenia of aging: from signaling pathways to clinical trials. Int. J. Biochem. Cell Biol 45, 2288–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, 2008. Hormesis defined. Ageing Res. Rev 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Arumugam TV, 2018. Hallmarks of brain aging: adaptive and pathological modification by metabolic states. Cell Metab. 27, 1176–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Moehl K, Ghena N, Schmaedick M, Cheng A, 2018. Intermittent metabolic switching, neuroplasticity and brain health. Nature reviews. Neuroscience 19, 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard S, Fang EF, Scheibye-Knudsen M, Croteau DL, Bohr VA, 2015. DNA damage, DNA repair, aging, and neurodegeneration. Cold Spring Harb. Perspect. Med 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Auley MT, Guimera AM, Hodgson D, McDonald N, Mooney KM, Morgan AE, Proctor CJ, 2017. Modelling the molecular mechanisms of aging. Biosci. Rep 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, 2008. Origin and physiological roles of inflammation. Nature 454, 428–435. [DOI] [PubMed] [Google Scholar]
- Merry TL, Kuhlow D, Laube B, Pohlmann D, Pfeiffer AFH, Kahn CR, Ristow M, Zarse K, 2017. Impairment of insulin signalling in peripheral tissue fails to extend murine lifespan. Aging Cell 16, 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, Orihuela CJ, Pletcher S, Sharp ZD, Sinclair D, Starnes JW, Wilkinson JE, Nadon NL, Strong R, 2011. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J. Gerontol. A Biol. Sci. Med. Sci 66, 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman S, Atzmon G, Huffman DM, Wan J, Crandall JP, Cohen P, Barzilai N, 2014. Low insulin-like growth factor-1 level predicts survival in humans with exceptional longevity. Aging Cell 13, 769–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milot E, Moreau C, Gagnon A, Cohen AA, Brais B, Labuda D, 2017. Mother’s curse neutralizes natural selection against a human genetic disease over three centuries. Nat. Ecol. Evol 1, 1400–1406. [DOI] [PubMed] [Google Scholar]
- Moiseeva O, Bourdeau V, Roux A, Deschenes-Simard X, Ferbeyre G, 2009. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol. Cell. Biol 29, 4495–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, McLaughlan S, Nouet Y, Pause A, Pollak M, Gottlieb E, Larsson O, St-Pierre J, Topisirovic I, Sonenberg N, 2013. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 18, 698–711. [DOI] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G, 1996. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature 382, 536–539. [DOI] [PubMed] [Google Scholar]
- Morsci NS, Hall DH, Driscoll M, Sheng ZH, 2016. Age-related phasic patterns of mitochondrial maintenance in adult Caenorhabditis elegans neurons. J. Neurosci 36, 1373–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, Guarente L, Auwerx J, 2013. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154, 430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro D, Baldy C, Pamenter ME, Treberg JR, 2019. The exceptional longevity of the naked mole-rat may be explained by mitochondrial antioxidant defenses. Aging Cell 18, e12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C, 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277–283. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM, 2011. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol 12, 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness KK, Krull KR, Jones KE, Mulrooney DA, Armstrong GT, Green DM, Chemaitilly W, Smith WA, Wilson CL, Sklar CA, Shelton K, Srivastava DK, Ali S, Robison LL, Hudson MM, 2013. Physiologic frailty as a sign of accelerated aging among adult survivors of childhood cancer: a report from the St Jude Lifetime cohort study. J. Clin. Oncol 31, 4496–4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Balkwill F, Chonchol M, Cominelli F, Donath MY, Giamarellos-Bourboulis EJ, Golenbock D, Gresnigt MS, Heneka MT, Hoffman HM, Hotchkiss R, Joosten LAB, Kastner DL, Korte M, Latz E, Libby P, Mandrup-Poulsen T, Mantovani A, Mills KHG, Nowak KL, O’Neill LA, Pickkers P, van der Poll T, Ridker PM, Schalkwijk J, Schwartz DA, Siegmund B, Steer CJ, Tilg H, van der Meer JWM, van de Veerdonk FL, Dinarello CA, 2017. A guiding map for inflammation. Nat. Immunol 18, 826–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Milman S, Hashmi SK, Austad SN, Kirkland JL, Halter JB, Barzilai N, 2016. Strategies and challenges in clinical trials targeting human aging. J. Gerontol. A Biol. Sci. Med. Sci 71, 1424–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls TJ, Nadalutti CA, Motori E, Sommerville EW, Gorman GS, Basu S, Hoberg E, Turnbull DM, Chinnery PF, Larsson NG, Larsson E, Falkenberg M, Taylor RW, Griffith JD, Gustafsson CM, 2018. Topoisomerase 3alpha is required for decatenation and segregation of human mtDNA. Mol. Cell 69, 9–23 e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, Enver T, Bell JI, Slijepcevic P, Goodnow CC, Jeggo PA, Cornall RJ, 2007. DNA repair is limiting for haematopoietic stem cells during ageing. Nature 447, 686–690. [DOI] [PubMed] [Google Scholar]
- O’Neill HM, Maarbjerg SJ, Crane JD, Jeppesen J, Jorgensen SB, Schertzer JD, Shyroka O, Kiens B, van Denderen BJ, Tarnopolsky MA, Kemp BE, Richter EA, Steinberg GR, 2011. AMP-activated protein kinase (AMPK) beta1beta2 muscle null mice reveal an essential role for AMPK in maintaining mitochondrial content and glucose uptake during exercise. Proc. Natl. Acad. Sci. U.S.A 108, 16092–16097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G, 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. Elegans. Nature 389, 994–999. [DOI] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N, 2015. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. Elegans. Nature 521, 525–528. [DOI] [PubMed] [Google Scholar]
- Parrish MC, Chaim IA, Nagel ZD, Tannenbaum SR, Samson LD, Engelward BP, 2018. Nitric oxide induced S-nitrosation causes base excision repair imbalance. DNA Repair (Amst.) 68, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal JM, Ellenberger T, 2015. The rise and fall of poly(ADP-ribose): an enzymatic perspective. DNA Repair (Amst.) 32, 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos JF, Saretzki G, Ahmed S, Nelson G, Richter T, Peters H, Wappler I, Birket MJ, Harold G, Schaeuble K, Birch-Machin MA, Kirkwood TB, von Zglinicki T, 2007. Mitochondrial dysfunction accounts for the stochastic heterogeneity in telomere-dependent senescence. PLoS Biol. 5, e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar T, Bjoras M, Klungland A, Eide L, 2018. Metabolism and DNA repair shape a specific modification pattern in mitochondrial DNA. Mitochondrion 40, 16–28. [DOI] [PubMed] [Google Scholar]
- Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI, 2003. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300, 1140–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM, 2011. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak P, Cybulski N, Feige JN, Auwerx J, Ruegg MA, Hall MN, 2008. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab. 8, 399–410. [DOI] [PubMed] [Google Scholar]
- Pursell ZF, McDonald JT, Mathews CK, Kunkel TA, 2008. Trace amounts of 8-oxo-dGTP in mitochondrial dNTP pools reduce DNA polymerase gamma replication fidelity. Nucleic Acids Res. 36, 2174–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M, Liu Z, Peng L, Tang X, Meng F, Ao Y, Zhou M, Wang M, Cao X, Qin B, Wang Z, Zhou Z, Wang G, Gao Z, Xu J, Liu B, 2018. Boosting ATM activity alleviates aging and extends lifespan in a mouse model of progeria. eLife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Schreiber SL, 2009. Direct control of mitochondrial function by mTOR. Proc. Natl. Acad. Sci. U.S.A 106, 22229–22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick RM, Zong H, Li J, Morino K, Moore IK, Yu HJ, Liu ZX, Dong J, Mustard KJ, Hawley SA, Befroy D, Pypaert M, Hardie DG, Young LH, Shulman GI, 2007. Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell Metab. 5, 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Rodero MP, Crow YJ, 2015. Human disease phenotypes associated with mutations in TREX1. J. Clin. Immunol 35, 235–243. [DOI] [PubMed] [Google Scholar]
- Ridout KK, Levandowski M, Ridout SJ, Gantz L, Goonan K, Palermo D, Price LH, Tyrka AR, 2017. Early life adversity and telomere length: a meta-analysis. Mol. Psychiatry [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M, Zarse K, Oberbach A, Kloting N, Birringer M, Kiehntopf M, Stumvoll M, Kahn CR, Bluher M, 2009. Antioxidants prevent health-promoting effects of physical exercise in humans. Proc. Natl. Acad. Sci. U.S.A 106, 8665–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T, 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280, 1763–1766. [DOI] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK, 2012. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab. 15, 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodger CE, McWilliams TG, Ganley IG, 2018. Mammalian mitophagy - from in vitro molecules to in vivo models. FEBS J. 285, 1185–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross FA, MacKintosh C, Hardie DG, 2016. AMP-activated protein kinase: a cellular energy sensor that comes in 12 flavours. FEBS J. 283, 2987–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL, 2007. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447, 725–729. [DOI] [PubMed] [Google Scholar]
- Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Felix AA, Williams EG, Jha P, Lo Sasso G, Huzard D, Aebischer P, Sandi C, Rinsch C, Auwerx J, 2016. Urolithin A induces mitophagy and prolongs lifespan in C. Elegans and increases muscle function in rodents. Nat. Med 22, 879–888. [DOI] [PubMed] [Google Scholar]
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH, 1994. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78, 35–43. [DOI] [PubMed] [Google Scholar]
- Safdar A, Khrapko K, Flynn JM, Saleem A, De Lisio M, Johnston AP, Kratysberg Y, Samjoo IA, Kitaoka Y, Ogborn DI, Little JP, Raha S, Parise G, Akhtar M, Hettinga BP, Rowe GC, Arany Z, Prolla TA, Tarnopolsky MA, 2016. Exercise-induced mitochondrial p53 repairs mtDNA mutations in mutator mice. Skelet. Muscle 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, Maser RS, Tonon G, Foerster F, Xiong R, Wang YA, Shukla SA, Jaskelioff M, Martin ES, Heffernan TP, Protopopov A, Ivanova E, Mahoney JE, Kost-Alimova M, Perry SR, Bronson R, Liao R, Mulligan R, Shirihai OS, Chin L, DePinho RA, 2011. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K, Kauppinen A, 2016. Age-related changes in AMPK activation: role for AMPK phosphatases and inhibitory phosphorylation by upstream signaling pathways. Ageing Res. Rev 28, 15–26. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H, 2008. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem 77, 229–257. [DOI] [PubMed] [Google Scholar]
- Santos JH, Meyer JN, Skorvaga M, Annab LA, Van Houten B, 2004. Mitochondrial hTERT exacerbates free-radical-mediated mtDNA damage. Aging Cell 3, 399–411. [DOI] [PubMed] [Google Scholar]
- Santos JH, Meyer JN, Van Houten B, 2006. Mitochondrial localization of telomerase as a determinant for hydrogen peroxide-induced mitochondrial DNA damage and apoptosis. Hum. Mol. Genet 15, 1757–1768. [DOI] [PubMed] [Google Scholar]
- Sanz A, 2016. Mitochondrial reactive oxygen species: Do they extend or shorten animal lifespan? Biochim. Biophys. Acta 1857, 1116–1126. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM, 2005. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM, 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22, 159–168. [DOI] [PubMed] [Google Scholar]
- Saxton RA, Sabatini DM, 2017. mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371. [DOI] [PubMed] [Google Scholar]
- Schapira AH, 2012. Mitochondrial diseases. Lancet 379, 1825–1834. [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, Wilson DM 3rd, Cooper M, Spencer R, de Cabo R, Croteau DL, Bohr VA, 2012. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J. Exp. Med 209, 855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Scheibye-Alsing K, Canugovi C, Croteau DL, Bohr VA, 2013. A novel diagnostic tool reveals mitochondrial pathology in human diseases and aging. Aging 5, 192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, DiMauro S, Hirano M, 2012. Human mitochondrial DNA: roles of inherited and somatic mutations. Nature reviews. Genetics 13, 878–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, Wallace DC, Rabinovitch PS, 2005. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science 308, 1909–1911. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M, 2007. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 6, 280–293. [DOI] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, Ney PA, 2007. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. U.S.A 104, 19500–19505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian D, Palacin M, Zorzano A, 2017. Mitochondrial dynamics: coupling mitochondrial fitness with healthy aging. Trends Mol. Med 23, 201–215. [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, Woods A, Robinson IC, Schuster E, Batterham RL, Kozma SC, Thomas G, Carling D, Okkenhaug K, Thornton JM, Partridge L, Gems D, Withers DJ, 2009. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326, 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Chandel NS, 2012. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell 48, 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senchuk MM, Dues DJ, Schaar CE, Johnson BK, Madaj ZB, Bowman MJ, Winn ME, Van Raamsdonk JM, 2018. Activation of DAF-16/FOXO by reactive oxygen species contributes to longevity in long-lived mitochondrial mutants in Caenorhabditis elegans. PLoS Genet. 14, e1007268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senejani AG, Dalal S, Liu Y, Nottoli TP, McGrath JM, Clairmont CS, Sweasy JB, 2012. Y265C DNA polymerase beta knockin mice survive past birth and accumulate base excision repair intermediate substrates. Proc. Natl. Acad. Sci. U.S.A 109, 6632–6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A, 2011. TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina IG, Vyssokikh MY, Gibanova N, Csikasz RI, Edgar D, Hallden-Waldemarson A, Rozhdestvenskaya Z, Bakeeva LE, Vays VB, Pustovidko AV, Skulachev MV, Cannon B, Skulachev VP, Nedergaard J, 2017. Improved health-span and lifespan in mtDNA mutator mice treated with the mitochondrially targeted antioxidant SkQ1. Aging 9, 315–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Horvath TL, 2015. Mitochondrial ROS signaling in organismal homeostasis. Cell 163, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimobayashi M, Hall MN, 2014. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nature reviews. Molecular cell biology 15, 155–162. [DOI] [PubMed] [Google Scholar]
- Siddiqui MS, Francois M, Fenech MF, Leifert WR, 2015. Persistent gammaH2AX: a promising molecular marker of DNA damage and aging. Mutat. Res. Rev. Mutat. Res 766, 1–19. [DOI] [PubMed] [Google Scholar]
- Siegmund SE, Yang H, Sharma R, Javors M, Skinner O, Mootha V, Hirano M, Schon EA, 2017. Low-dose rapamycin extends lifespan in a mouse model of mtDNA depletion syndrome. Hum. Mol. Genet 26, 4588–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliter DA, Martinez J, Hao L, Chen X, Sun N, Fischer TD, Burman JL, Li Y, Zhang Z, Narendra DP, Cai H, Borsche M, Klein C, Youle RJ, 2018. Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sornson MW, Wu W, Dasen JS, Flynn SE, Norman DJ, O’Connell SM, Gukovsky I, Carriere C, Ryan AK, Miller AP, Zuo L, Gleiberman AS, Andersen B, Beamer WG, Rosenfeld MG, 1996. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature 384, 327–333. [DOI] [PubMed] [Google Scholar]
- St John JC, Tsai TS, 2018. The association of mitochondrial DNA haplotypes and phenotypic traits in pigs. BMC Genet. 19, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P, 2008. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. U.S.A 105, 3438–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana S, Solotchi M, Ramachandran A, Patel SS, 2017. Transcriptional fidelities of human mitochondrial POLRMT, yeast mitochondrial Rpo41, and phage T7 single-subunit RNA polymerases. J. Biol. Chem 292, 18145–18160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora P, Wilson DM 3rd, Bohr VA, 2012. Repair of persistent strand breaks in the mitochondrial genome. Mech. Ageing Dev 133, 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Hazra TK, Papaconstantinou J, Mitra S, Boldogh I, 2003. Age-dependent deficiency in import of mitochondrial DNA glycosylases required for repair of oxidatively damaged bases. Proc. Natl. Acad. Sci. U.S.A 100, 10670–10675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadi SK, Sebastian R, Dahal S, Babu RK, Choudhary B, Raghavan SC, 2016. Microhomology-mediated end joining is the principal mediator of double-strand break repair during mitochondrial DNA lesions. Mol. Biol. Cell 27, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS, 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292, 107–110. [DOI] [PubMed] [Google Scholar]
- Toyama EQ, Herzig S, Courchet J, Lewis TL Jr., Loson OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, Shaw RJ, 2016. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 351, 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG, 2004. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423. [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Tsichlis PN, 2007. Energy depletion inhibits phosphatidylinositol 3-kinase/Akt signaling and induces apoptosis via AMP-activated protein kinase-dependent phosphorylation of IRS-1 at Ser-794. J. Biol. Chem 282, 18069–18082. [DOI] [PubMed] [Google Scholar]
- Vaidya A, Mao Z, Tian X, Spencer B, Seluanov A, Gorbunova V, 2014. Knock-in reporter mice demonstrate that DNA repair by non-homologous end joining declines with age. PLoS Genet. 10, e1004511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, Gonzalez-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW, 2004. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160. [DOI] [PubMed] [Google Scholar]
- Valente WJ, Ericson NG, Long AS, White PA, Marchetti F, Bielas JH, 2016. Mitochondrial DNA exhibits resistance to induced point and deletion mutations. Nucleic Acids Res. 44, 8513–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Spoel E, Jansen SW, Akintola AA, Ballieux BE, Cobbaert CM, Slagboom PE, Blauw GJ, Westendorp RG, Pijl H, Roelfsema F, van Heemst D, 2016. Growth hormone secretion is diminished and tightly controlled in humans enriched for familial longevity. Aging Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F, 2003. Genetics: influence of TOR kinase on lifespan in C. Elegans. Nature 426, 620. [DOI] [PubMed] [Google Scholar]
- Vermeij WP, Dolle ME, Reiling E, Jaarsma D, Payan-Gomez C, Bombardieri CR, Wu H, Roks AJ, Botter SM, van der Eerden BC, Youssef SA, Kuiper RV, Nagarajah B, van Oostrom CT, Brandt RM, Barnhoorn S, Imholz S, Pennings JL, de Bruin A, Gyenis A, Pothof J, Vijg J, van Steeg H, Hoeijmakers JH, 2016. Restricted diet delays accelerated ageing and genomic stress in DNA-repair-deficient mice. Nature 537, 427–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijg J, Dong X, Milholland B, Zhang L, 2017. Genome instability: a conserved mechanism of ageing? Essays Biochem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian CJ, Brinker AE, Graw S, Koestler DC, Legendre C, Gooden GC, Salhia B, Welch DR, 2017. Mitochondrial genomic backgrounds affect nuclear DNA methylation and gene expression. Cancer Res. 77, 6202–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Zglinicki T, 2002. Oxidative stress shortens telomeres. Trends Biochem. Sci 27, 339–344. [DOI] [PubMed] [Google Scholar]
- Wallace DC, 2015. Mitochondrial DNA variation in human radiation and disease. Cell 163, 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, 2018. Mitochondrial genetic medicine. Nat. Genet 50, 1642–1649. [DOI] [PubMed] [Google Scholar]
- Weinberg SE, Sena LA, Chandel NS, 2015. Mitochondria in the regulation of innate and adaptive immunity. Immunity 42, 406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir HJ, Yao P, Huynh FK, Escoubas CC, Goncalves RL, Burkewitz K, Laboy R, Hirschey MD, Mair WB, 2017. Dietary restriction and AMPK increase lifespan via mitochondrial network and peroxisome remodeling. Cell Metab. 26, 884–896 e885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman L, Jo DG, Sorensen MM, de Souza-Pinto NC, Markesbery WR, Mattson MP, Bohr VA, 2007. Defective DNA base excision repair in brain from individuals with Alzheimer’s disease and amnestic mild cognitive impairment. Nucleic Acids Res. 35, 5545–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS, 2015. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, McArthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, van Delft MF, Bedoui S, Lessene G, Ritchie ME, Huang DC, Kile BT, 2014. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159, 1549–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley CD, Campisi J, 2016. From ancient pathways to aging cells-connecting metabolism and cellular senescence. Cell Metab. 23, 1013–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley CD, Velarde MC, Lecot P, Liu S, Sarnoski EA, Freund A, Shirakawa K, Lim HW, Davis SS, Ramanathan A, Gerencser AA, Verdin E, Campisi J, 2016. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 23, 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Lujan SA, Kunkel TA, 2016. Processing ribonucleotides incorporated during eukaryotic DNA replication. Nature reviews. Molecular cell biology 17, 350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff JN, Gemmell NJ, 2013. Mitochondria, maternal inheritance, and asymmetric fitness: why males die younger. Bioessays 35, 93–99. [DOI] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ, 2013a. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Liu J, Chen EB, Wang JJ, Cao L, Narayan N, Fergusson MM, Rovira II, Allen M, Springer DA, Lago CU, Zhang S, DuBois W, Ward T, deCabo R, Gavrilova O, Mock B, Finkel T, 2013b. Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 4, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang Y, 2017. TET-mediated active DNA demethylation: mechanism, function and beyond. Nature reviews. Genetics 18, 517–534. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhang L, Zhang W, Meng D, Zhang H, Jiang Y, Xu X, Van Meter M, Seluanov A, Gorbunova V, Mao Z, 2015. SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle 14, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Pirtskhalava T, Farr JN, Weigand BM, Palmer AK, Weivoda MM, Inman CL, Ogrodnik MB, Hachfeld CM, Fraser DG, Onken JL, Johnson KO, Verzosa GC, Langhi LGP, Weigl M, Giorgadze N, LeBrasseur NK, Miller JD, Jurk D, Singh RJ, Allison DB, Ejima K, Hubbard GB, Ikeno Y, Cubro H, Garovic VD, Hou X, Weroha SJ, Robbins PD, Niedernhofer LJ, Khosla S, Tchkonia T, Kirkland JL, 2018. Senolytics improve physical function and increase lifespan in old age. Nat. Med 24, 1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE, 2007. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 131, 873–886. [DOI] [PubMed] [Google Scholar]
- Yoo YJ, Kim H, Park SR, Yoon YJ, 2017. An overview of rapamycin: from discovery to future perspectives. J. Ind. Microbiol. Biotechnol 44, 537–553. [DOI] [PubMed] [Google Scholar]
- Zarse K, Schmeisser S, Groth M, Priebe S, Beuster G, Kuhlow D, Guthke R, Platzer M, Kahn CR, Ristow M, 2012. Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 15, 451–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Vijg J, 2018. Somatic mutagenesis in mammals and its implications for human disease and aging. Annu. Rev. Genet 52, 397–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A, Yuan T, Yang J, Li Y, Shi L, Guan D, Pan H, Duan S, Ding Z, Li M, Yi F, Bai R, Wang Y, Chen C, Yang F, Li X, Wang Z, Aizawa E, Goebl A, Soligalla RD, Reddy P, Esteban CR, Tang F, Liu GH, Belmonte JC, 2015. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 348, 1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P, D’Amico D, Ropelle ER, Lutolf MP, Aebersold R, Schoonjans K, Menzies KJ, Auwerx J, 2016. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352, 1436–1443. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ, 2002. A mitochondrial specific stress response in mammalian cells. EMBO J. 21, 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Croteau DL, Bohr VA, Akbari M, 2019. Diminished OPA1 expression and impaired mitochondrial morphology and homeostasis in Aprataxin-deficient cells. Nucleic Acids Res. 47 (8), 4086–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XH, Lu M, Lee BY, Ugurbil K, Chen W, 2015. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc. Natl. Acad. Sci. U.S.A 112, 2876–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler DV, Wiley CD, Velarde MC, 2015. Mitochondrial effectors of cellular senescence: beyond the free radical theory of aging. Aging Cell 14, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM, 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nature reviews. Molecular cell biology 12, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI, 2002. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc. Natl. Acad. Sci. U.S.A 99, 15983–15987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL,Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schroder JM, Vance JM, 2004. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet 36, 449–451. [DOI] [PubMed] [Google Scholar]


