Abstract
Nearly all chronic human infections are associated with alterations in the memory B cell (MBC) compartment, including a large expansion of CD19hiT-bethi MBC in the peripheral blood of HIV-infected individuals with chronic viremia. Despite their prevalence, it is unclear how these B cells arise and whether they contribute to the inefficiency of antibody-mediated immunity in chronic infectious diseases. We addressed these questions by characterizing T-bet-expressing B cells in lymph nodes (LN) and identifying a strong T-bet signature among HIV-specific MBC associated with poor immunologic outcome. Confocal microscopy and quantitative imaging revealed that T-bethi B cells in LN of HIV-infected chronically viremic individuals distinctly accumulated outside germinal centers (GC), which are critical for optimal antibody responses. In single-cell analyses, LN T-bethi B cells of HIV-infected individuals were almost exclusively found among CD19hi MBC and expressed reduced GC-homing receptors. Furthermore, HIV-specific B cells of infected individuals were enriched among LN CD19hiT-bethi MBC and displayed a distinct transcriptome, with features similar to CD19hiT-bethi MBC in blood and LN GC B cells (GCBC). LN CD19hiT-bethi MBC were also related to GCBC by B cell receptor (BCR)–based phylogenetic linkage but had lower BCR mutation frequencies and reduced HIV-neutralizing capacity, consistent with diminished participation in GC-mediated affinity selection. Thus, in the setting of chronic immune activation associated with HIV viremia, failure of HIV-specific B cells to enter or remain in GC may help explain the rarity of high-affinity protective antibodies.
INTRODUCTION
Naïve B cells respond to foreign antigens by proliferating and differentiating into two major populations, antibody-secreting plasma cells and memory B cells (MBC), which serve as sentinels for rapid recall responses (1-3). Effective, sustained immunologic memory responses to T cell-dependent pathogens are mediated by antibody affinity maturation in self-resolving germinal centers (GC). The specialized structure of GC within secondary lymphoid tissues allows antigen-specific B cells to cycle between the light zone where those with higher affinity are selected by T follicular helper (TFH) cells and the dark zone where expansion, immunoglobulin (Ig) class-switching and somatic hypermutation occur (4). When pathogens or other stimuli persist and cause chronic immune activation and inflammation, lymphoid tissues undergo hyperplastic alterations, typically manifested by expanded GC that merge into large poorly defined anatomic structures (5). In addition to loss of structural integrity, chronic inflammatory conditions also alter processes that affect immune responses. In chronic viral infections, such as those caused by HIV and lymphocytic choriomeningitis virus, where proinflammatory conditions persist, multiple inhibitory and regulatory events are triggered to counter the hyperactivation and protect tissues (6). These events have been associated with poor outcomes as a result of the emergence of dysfunctional or exhausted lymphocyte populations (7, 8), in addition to dysregulation of populations involved in generating immunity (9).
Repetitive or persistent cellular stimulation in vivo has been associated with the development of unique cellular populations, including B cells that express the transcription factor T-bet. T-bet+ B cells have been described in mouse models involving repetitive stimulation and in humans involving infectious and non-infectious chronic inflammatory processes and cytokine dysregulation (1, 10-13). T-bet is best known for its critical role as a transcriptional regulator of several immune lineages, including interferon-γ (IFN-γ)–secreting T helper type 1 (TH1) cells (14). In B cells, T-bet induces mouse Ig isotype switching to IgG2a (15) and has been shown in a number of murine models to be required for clearance of virus (16-18). However, in humans, a similar role has yet to be established, and certain conditions that regulate B cell T-bet expression in mice, namely Toll-like receptor (TLR) engagement and certain cytokine milieus (19, 20), have also been associated with B cell–associated autoimmune pathologies (21-23). Thus, it remains unclear, especially in humans, whether and under what circumstances does expression of T-bet in B cells provide immunologic benefit. In addition, very little is known regarding the genesis of T-bet+ B cells in human lymphoid tissues, although extrafollicular (EF) monocytoid T-bet+ B cells have been described in various lymphadenopathies (24-26) and suspected in systemic lupus erythematous (SLE) (22). There is also uncertainty as to how T-bet+ B cells that reside in lymphoid tissues relate to those that have been described in the peripheral blood in various conditions (22, 23, 27-30). Here, we provide insight into these issues by establishing relationships between T-bet–expressing B cells and HIV-specific counterparts in the peripheral blood and lymph node (LN) with an approach that combined quantitative and positional imaging with functional, transcriptomic, and computational B cell receptor (BCR) gene analyses.
RESULTS
Enrichment of non-GC T-bet+ B cells in LN of HIV-infected individuals
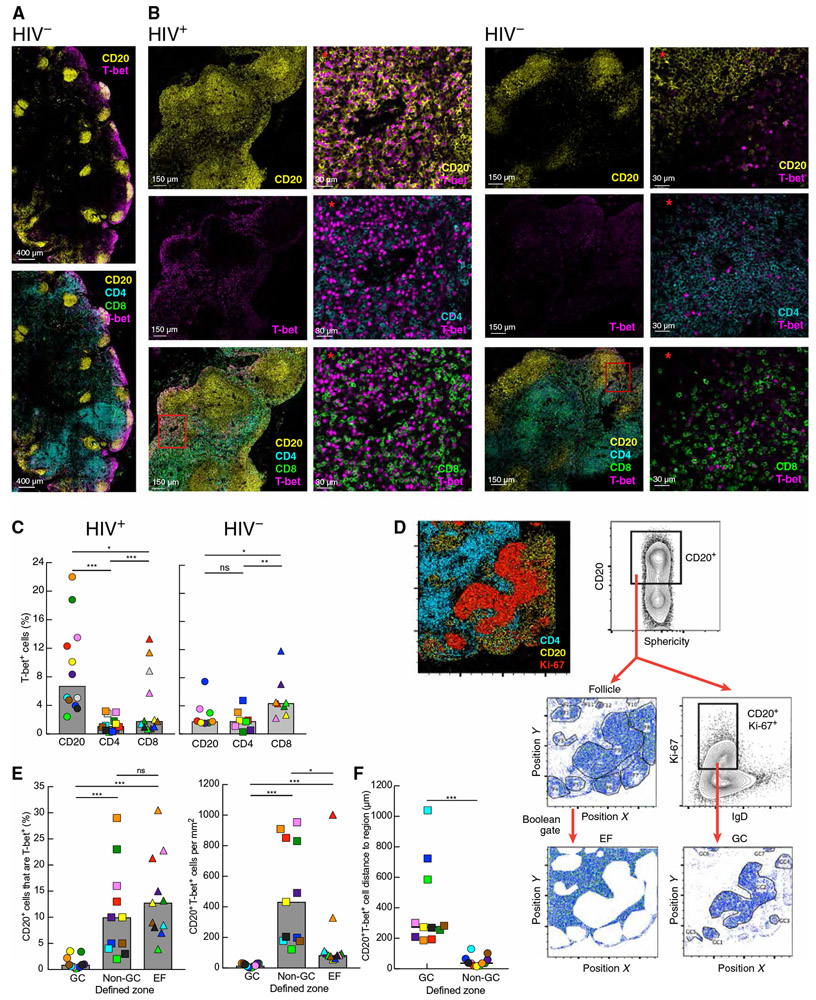
To investigate T-bet–expressing B cells in reactive (enlarged due to benign hyperplasia) and unreactive LN, we performed multiplexed confocal microscopy in combination with quantitative imaging analysis (histocytometry) on tissue sections obtained from HIV-infected and HIV-uninfected individuals (Table 1). All individuals in the HIV group were viremic at time of study. In HIV-uninfected individuals, B cell follicles were small and unreactive, and T-bet expression tended to be concentrated near the subcapsular sinus (Fig. 1, A and B). In contrast, LN sections of HIV-infected individuals contained large reactive follicles with prominent GC and diffused expression of T-bet throughout the tissue (Fig. 1B). There were significantly more reactive follicles in the LN of HIV-infected compared to HIV-uninfected individuals (P < 0.0001; fig. S1A). Frequencies of T-bet expression were significantly higher in CD20+ B cells than in CD4+ (P = 0.0005) and CD8+ (P = 0.0122) T cells in HIV-infected individuals, whereas they were highest in CD8+ T cells and not significantly different between CD20+ B cells and CD4+ T cells in HIV-uninfected individuals (Fig. 1C). When data points were compared cross-sectionally by HIV status, frequencies of T-bet in B cells, but not T cells, were significantly higher in HIV-infected than in HIV-uninfected individuals (P = 0.0012).
Table 1. Study participants.
Participants are described individually for HIV+ group (top) and by group (bottom). Colors match with those in Figs. 1 (C, E, and F), 2 (B and C), 3A, 4 (A to C and E), and 6 (B and C) and fig. S1B and S2 (A and E). Excision sites were inguinal, except cervical for HIV13 and axillary for HIV2, HIV6, HIV10 and two of the HIV− group. Numbers in the last two rows are medians (range) or geometric mean (HIV RNA); bold indicates significant difference between the two groups. N/A, not applicable or not available. HIV neutralize, reported as geometric mean of titer.
| Participants HIV+ group |
Age (years) |
Gender (M/F) |
CD4+ T cell % |
CD4+ T cell (count/μl) |
CD8+ T cell (count/μl) |
CD19+ B cell (count/μl) |
HIV RNA (copies/ml) |
Years infected |
HIV neutralize |
|---|---|---|---|---|---|---|---|---|---|
| HIV1 | 23 | M | 25 | 532 | 1193 | 149 | 5924 | 1–5 | 22 |
| HIV2 | 26 | F | 19 | 304 | 1120 | 80 | 1804 | 1–5 | <20 |
| HIV3 | 26 | M | 23 | 336 | 746 | 161 | 847,180 | <1 | <20 |
| HIV4 | 29 | M | 16 | 253 | 823 | 142 | 33,691 | 1–5 | <20 |
| HIV5 | 25 | M | 45 | 714 | 524 | 175 | 1486 | <1 | <20 |
| HIV6 | 21 | M | 28 | 758 | 1055 | 568 | 35,576 | 1–5 | N/A |
| HIV7 | 53 | M | 43 | 404 | 376 | 94 | 20,594 | >20 | 79 |
| HIV8 | 34 | M | 27 | 583 | 1123 | 194 | 148 | 10–20 | 29 |
| HIV9 | 36 | F | 36 | 1206 | 823 | 268 | 94 | 1–5 | 25 |
| HIV10 | 44 | M | 20 | 494 | 1407 | 444 | 159 | >20 | 28 |
| HIV11 | 47 | M | 40 | 579 | 449 | 290 | 7199 | <1 | <20 |
| HIV12 | 39 | F | 27 | 551 | 1061 | 306 | 267 | 10–20 | 72 |
| HIV13 | 26 | M | 35 | 995 | 967 | 398 | 272 | 1–5 | N/A |
| HIV | 29 (21–53) |
10/3 |
27 (16–45) |
551 (253–1206) |
967 (376–1407) |
194 (80—568) |
2689 | ||
| HIV− | 40 (19–57) |
12/6 |
48 (37–66) |
632 (322–1401) |
286 (180–593) |
171 (95 – 275) |
N/A |
Fig. 1. Accumulation of T-bet+ B cells in non-GC areas of LN in HIV infection.
(A and B) Images of cells expressing T-bet alone or in combination with staining for CD20+ B cells, CD4+, or CD8+ T cells in LN sections isolated from HIV-uninfected (A and B, right) and HIV-infected (B, left) individuals. Red asterisks indicate enlarged images from the red boxed inset illustrating differences in T-bet expression patterns between cell populations. (C) Frequencies of T-bet+ cells among B and T cells were quantified by histocytometry in LN sections from HIV-infected (n = 12) and HIV-uninfected (n = 8) individuals. (D) Histocytometry plots and gating strategy for defining areas within LN sections. Ki-67 expression was used to define GC (Ki-67+IgD−) and non-GC (Ki-67−) zones within CD20+ follicles, and the extrafollicular (EF) zone was defined by Boolean gate subtraction. (E) Frequencies of CD20+ cells expressing T-bet (left graph) and numbers of CD20+T-bet+ (right graph) cells in each zone (n = 11 HIV-infected) were quantified by histocytometry. (F) Distance of CD20+T-bet+ cells to GC and non-GC zones (n = 11 HIV-infected). In (C), (E), and (F), each individual is color-coded per Table 1, and shaded bars represent medians. *P < 0.05, **P < 0.01, ***P < 0.001 by Wilcoxon matched-pairs signed rank test after obtaining significance by Friedman analysis of variance (ANOVA) test on full set; ns, not significant.
To evaluate where T-bet+ B cells were located in the LN, we defined three distinct zones: GC, non-GC, and EF, based on expression of CD20, IgD, and Ki-67 (Fig. 1D). In HIV-infected individuals, the percentage of T-bet–expressing B cells was significantly higher in non-GC and EF compared to the GC zone (both P = 0.001; Fig. 1E). When the number of B cells per zone was considered (fig. S1B), there were significantly more T-bet–expressing B cells in non-GC than in GC (P = 0.001) and EF (P = 0.0137) zones (Fig. 1E). A similar pattern was observed for the HIV-negative group (fig. S1C). To further investigate the positioning of T-bet–expressing B cells within follicles, distance analyses to defined GC and non-GC zones were performed (fig. S1D). When the number of CD20+T-bet+ cells was plotted as a function of incremental minimum distance to each zone, the vast majority of T-bet–expressing B cells were located within non-GC zones (distance, 0; illustrated in fig. S1D) and significantly more distant to the GC zone in both HIV-infected (P = 0.001; Fig. 1F) and HIV-uninfected (P = 0.0078; fig. S1E) individuals. Collectively, these data demonstrate that T-bet+ B cells primarily accumulate in non-GC areas of follicles in LN of both HIV-infected and HIV-uninfected individuals, although to a lesser extent in the latter group.
Distinct phenotype of non-GC CD19hiT-bethi LN MBC
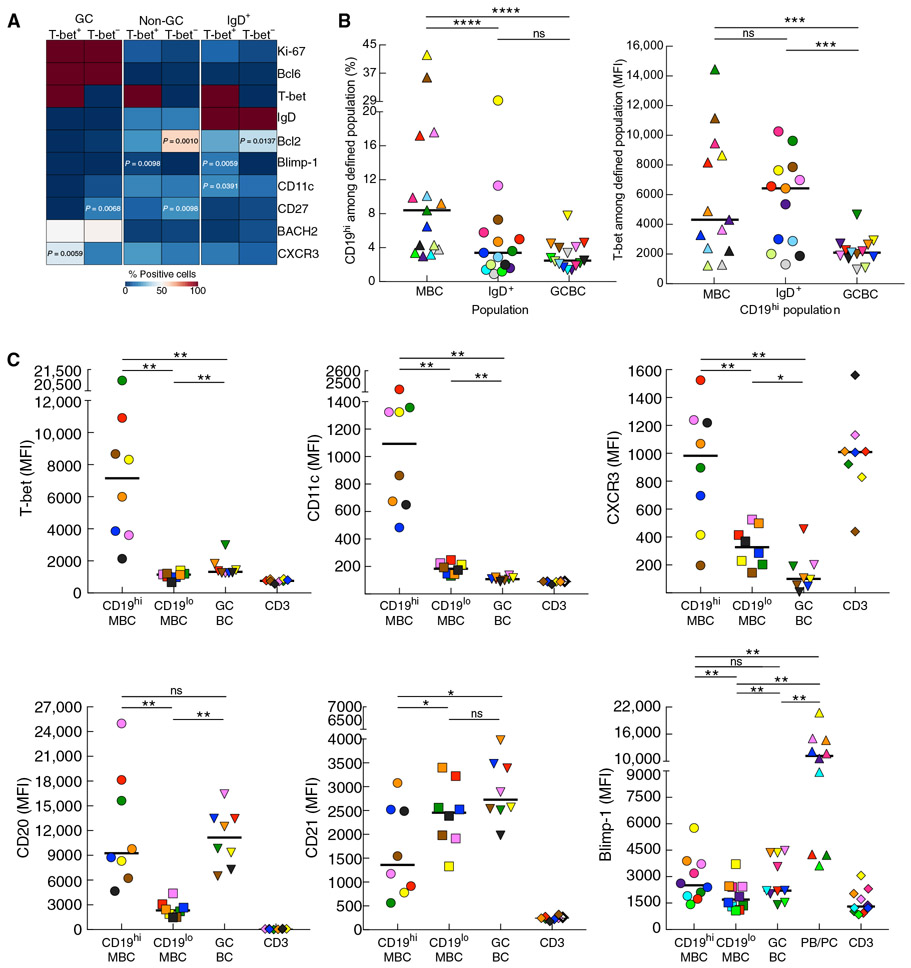
To further characterize T-bet+ B cells in LN of HIV-infected individuals, we performed additional phenotyping on LN sections and used a heatmap approach to depict expression of various markers relative to T-bet among B cells (CD20+) located in GC (Ki-67+ or Bcl6+) and non-GC areas of follicles (Fig. 2A). Relative to T-bet−, T-bet+ B cells in GC and non-GC, as well as IgD+ B cells of non-GC, tended to express higher Blimp-1, CD11c, and CXCR3 and lower CD27 and Bcl2, consistent with previous observations (22). However, differences reached significance for relatively few pair-wise comparisons (Fig. 2A), likely reflecting limits in sensitivity of this approach combined with a relative paucity of T-bet+ B cells, especially in the GC (fig. S2A). To overcome these limitations and extend phenotypic analyses, we performed conventional flow cytometry on single LN cell preparations using the surface marker CD19 to identify all B cells and CD38, CD27, and IgD to delineate the major LN B cell populations in humans (31). When compared to HIV-uninfected individuals, LN of HIV-infected individuals were significantly enriched in GC B cells (GCBC; IgD−CD38+) and plasmablasts/cells (PB/PC; P < 0.0001 each; fig. S2B), consistent with differences in frequencies of reactive follicles (fig. S1A) and HIV-induced lymph-adenopathy. Furthermore, a population of CD19hi cells with a profile enriched for CD21loCD27+/− was present among non-GC LN B cells of HIV-infected individuals (fig. S2C) and similar to peripheral blood MBC that we have previously described in the context of HIV-associated activation and exhaustion (32).
Fig. 2. T-bet expression and signature profile among CD19hi LN MBC of HIV-infected individuals.
(A) Heatmap depicting expression of several markers based on T-bet expression in defined populations from histocytometric analyses. (B) Frequencies of CD19hi (left) and T-bet intensities (right) among defined populations measured by conventional flow cytometry on LN mononuclear cells isolated from HIV-infected individuals (n = 15 and 13, respectively). (C) Quantitative analyses of mean fluorescence intensity (MFI) for each signature-defining marker by B cell population or CD3+ cells performed by conventional flow cytometry on LN mononuclear cells (n = 8 to 10). In (B) and (C), each individual is color-coded per Table 1, and black horizontal bars represent medians. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Wilcoxon matched-pairs signed rank test after obtaining significance by Friedman ANOVA test on B cell subpopulations.
Upon further evaluation of non-GC LN B cells, the CD19hi population was significantly enriched among MBC (IgD−CD38−) when compared to IgD+ B cells and GCBC (P < 0.0001 each; Fig. 2B). When T-bet expression was considered among each of the three CD19hi populations, intensities were significantly lower in GCBC compared to MBC and IgD+ B cells (P = 0.0002 each; Fig. 2B), consistent with the histocytometric analyses. There was no difference in T-bet expression between CD19hi MBC and CD19hiIgD+ B cells (Fig. 2B). However, this may be a reflection of enrichment of class-unswitched memory populations, including class-unswitched CD21loCD27− MBC, among the latter as opposed to changes in naïve B cells (CD21hiCD27−; fig. S2D). Given the heterogeneity within the LN IgD+ population, we focused further phenotyping on class-switched LN B cells. The intensities of T-bet, CD11c, CXCR3, and CD20 were significantly higher, and CD21 was significantly lower in and on CD19hi compared to CD19lo MBC (P = 0.0078 each for higher and P = 0.0234 for lower expression; Fig. 2C). Similar differences were observed between CD19hi MBC and GCBC (P = 0.0078 each for higher and P = 0.0156 for lower expression), except for the intensity of CD20, which was equally high on CD19hi MBC and GCBC (Fig. 2C), consistent with known high expression of CD20 on GCBC (2). Although intensities of T-bet in GCBC were much lower than in CD19hi MBC and nearly undetectable by confocal microscopy, they were nonetheless significantly higher than in CD19lo MBC (P = 0.0078; Fig. 2C). Expression of Blimp-1 was also similar in intensity between GCBC and CD19hi MBC, and both intensities were significantly higher than in CD19lo MBC (P = 0.006 each), yet all were significantly lower than intensities in PB/PC (P = 0.006 each; Fig. 2C). We conclude that LN-derived CD19hi MBC are T-bethiCD20hiCD21lo CD11c+CXCR3+ (Fig. 2C), similar to the distinct signature profile associated with tissue-like MBC in HIV and nonconventional B cells that have been described in other inflammatory disease settings (1, 10-13). The increased expression of Blimp-1 in CD19hi MBC and the higher Blimp-1/lower CD27 of T-bet+ relative to T-bet− in non-GCBC of LN sections are also consistent with double-negative (IgD−/CD27−) B cells recently described in SLE (22), although differences in expression of CD27 by flow cytometry did not reach significance (fig. S2E).
Enrichment of HIV-specific B cells among CD19hi LN MBC with a distinct transcriptome
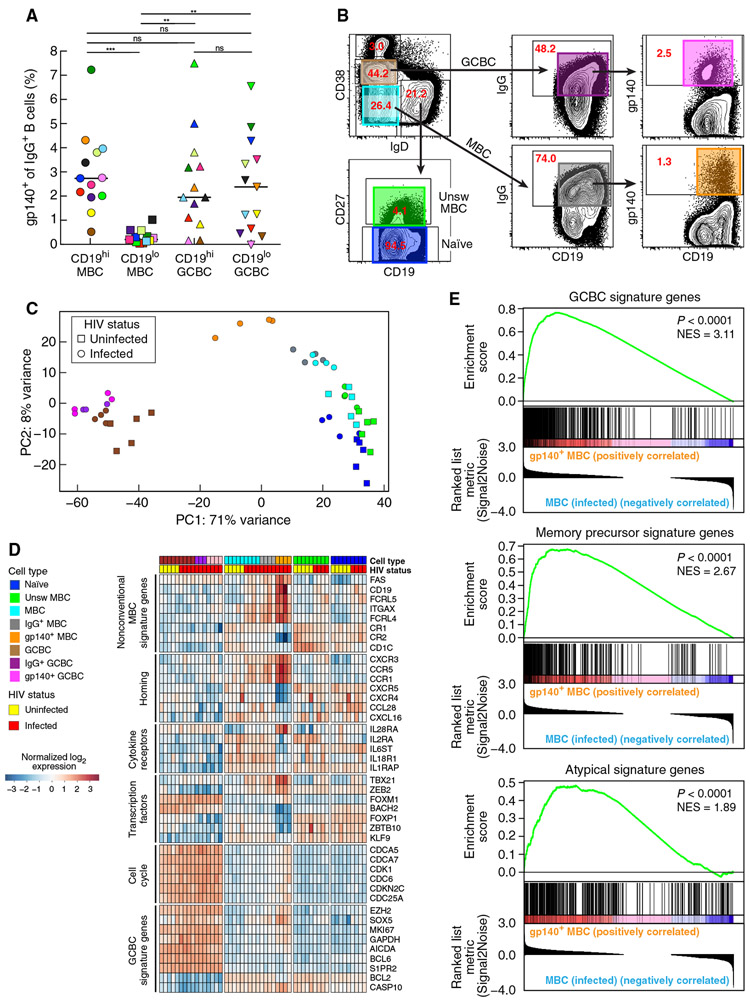
Given previous observations that HIV envelope–specific IgG+ B cells in the peripheral blood of infected individuals were enriched among MBC with a CD20hiCD21loCD11c+CXCR3+ profile (32), we performed similar analyses on LN B cells of HIV-infected individuals. Frequencies of B cells with an IgG-BCR specific for the HIV envelope, as evaluated by the binding of gp140 protein to IgG+ B cells, were significantly enriched among CD19hi compared to CD19lo MBC (P = 0.0006 each), whereas frequencies between CD19hi and CD19lo GCBC were not significantly different (Fig. 3A). This observation prompted us to perform gene expression analysis by RNA sequencing (RNA-seq) on LN-derived HIV-specific IgG+ MBC and GCBC. We also compared their profiles to those of parental (non-antigen and non-IgG selected) counterparts and other sorted B cell populations isolated from the LN of HIV-infected and HIV-uninfected individuals (Fig. 3B). Unsupervised principal component analysis (PCA) of expressed genes revealed a clear separation along PC1 of all GCBC populations from all other populations studied (Fig. 3C). Along PC2, the three class-switched MBC populations from the HIV-infected individuals were largely separate from the remaining populations. A certain degree of intermixing between the other B cell populations was observed, namely naïve and both MBC populations of HIV-uninfected and naïve and class-unswitched MBC of HIV-infected individuals. Hierarchical clustering using PC1 and PC2 revealed that, among the cluster of class-switched MBC from HIV-infected individuals, those that were HIV specific formed a separate branch (fig. S3A, magenta). Each additional PC (beyond PC1 and PC2) accounted for less than 5% of the variance; there was no visually identifiable clustering by PC3 and PC4 (fig. S3B).
Fig. 3. Unique transcriptional profile for HIV-specific MBC in LN of infected individuals.
(A) Frequencies of HIV gp140+ B cells among CD19hi and CD19lo MBC and GCBC measured by conventional flow cytometry on LN mononuclear cells isolated from HIV-infected individuals (n = 13). Each individual is color-coded per Table 1, and black horizontal bars represent medians. **P < 0.01, ***P < 0.001 by Wilcoxon matched-pairs signed rank test after obtaining significance by Friedman ANOVA test on full set. (B) Sorting strategy used to isolate multiple LN B cell populations used for RNA-seq (each population is indicated by color-coded boxes). The numbers refer to the percentage of cells within the gated population relative to the total number of cells shown in the dot plot. (C) Unsupervised principal components analysis of sorted populations using all expressed genes, performed on HIV-infected participants HIV1, HIV3, HIV5, and HIV7 (Table 1) and HIV-uninfected participants (n = 5). (D) Heatmap depicting scaled, normalized log2 expression values of selected differentially expressed genes. (E) Gene set enrichment analysis of GCBC (top), memory precursor (middle), and atypical B cell signature genes (bottom) ranked by differential expression between HIV gp140+ and total MBC in HIV-infected individuals; NES, normalized enrichment score.
A heatmap of selected genes, arranged by LN B cell population (Fig. 3D), revealed that HIV-specific MBC carried many features observed for nonconventional circulating MBC markers (22, 23, 29, 32, 33). Among transcription factors, TBX21 (T-bet) was highly expressed in HIV-specific MBC, along with increased expression of CD19, ITGAX (CD11c), and CXCR3 and decreased expression of CR2 (CD21). This mirrors the flow cytometric profile of CD19hi LN MBC, except for PRMD1 (Blimp-1), which, along with the other B cell terminal differentiation factor IRF4, was not differentially expressed among HIV-specific MBC when compared to other sorted MBC populations (fig. S3C and table S1). Other markers associated with nonconventional MBC, namely, putative inhibitory receptors FCRL4 and FCRL5 (32, 34-36), were also highly expressed among HIV-specific LN MBC (Fig. 3D). Among this subset of genes that were unique to HIV-specific LN MBC, only TBX21 was differentially expressed by HIV status; its expression was higher in all parental populations, including naïve B cells, of HIV-infected versus HIV-uninfected individuals (table S1).
In PCA, although HIV-specific MBC did not form a cluster distinct from its parental populations (fig. S3A), its positioning along PC1 was closer to that of GCBC than any of the other non-GCBC populations (Fig. 3C). FAS (CD95), a GCBC-defining marker associated with high susceptibility to CD95-mediated apoptosis (37) and also overexpressed on FCRL4+ tonsillar MBC (38), was strongly expressed in HIV-specific MBC (Fig. 3D). Other GCBC-signature genes, including several proteins involved in cell cycling of GCBC, were weakly expressed in all non-GCBC populations, except HIV-specific MBC (Fig. 3D). However, LN GCBC and HIV-specific MBC transcriptional profiles differed by the expression of transcription factors associated with B cell development and differentiation and of homing receptors. The transcription factor BACH2, a regulator of GCBC differentiation to MBC (39), was strongly expressed in all GCBC and naïve B cells, yet expression was low in all MBC populations, the lowest of which was in HIV-specific MBC (Fig. 3D). Homing receptors CXCR3, CXCR4, and CXCR5 were also differentially expressed between LN GCBC and HIV-specific MBC (Fig. 3D). In addition, expression of S1PR2, a member of the sphingosine-1-phosphate receptor family essential for GC positioning of B cells in follicles (40) and shown to be down-regulated in memory precursor cells along with BACH2 (41), a regulator of S1PR2 (42), was also lower in LN-derived HIV-specific IgG+ MBC compared to GCBC (Fig. 3D).
Despite the aforementioned differences, gene set enrichment analysis (GSEA) revealed that GCBC-signature genes were positively and significantly overrepresented among genes expressed by HIV-specific MBC compared to its parental MBC (P < 0.0001; Fig. 3E). GSEA also revealed that HIV-specific MBC shared characteristics with memory precursors (P < 0.0001; Fig. 3E) (41). When the analysis was performed against MBC-signature genes associated with T-bet–expressing B cells that are expanded in individuals chronically exposed to malaria (27), a similar pattern was observed (P < 0.0001; Fig. 3E). Together, these observations suggest that LN-derived HIV-specific MBC share features of GCBC, memory precursors, and circulating T-bet+ MBC signatures.
Transcriptional features in LN CD19hiT-bet+ MBC and GCBC extend to phenotype
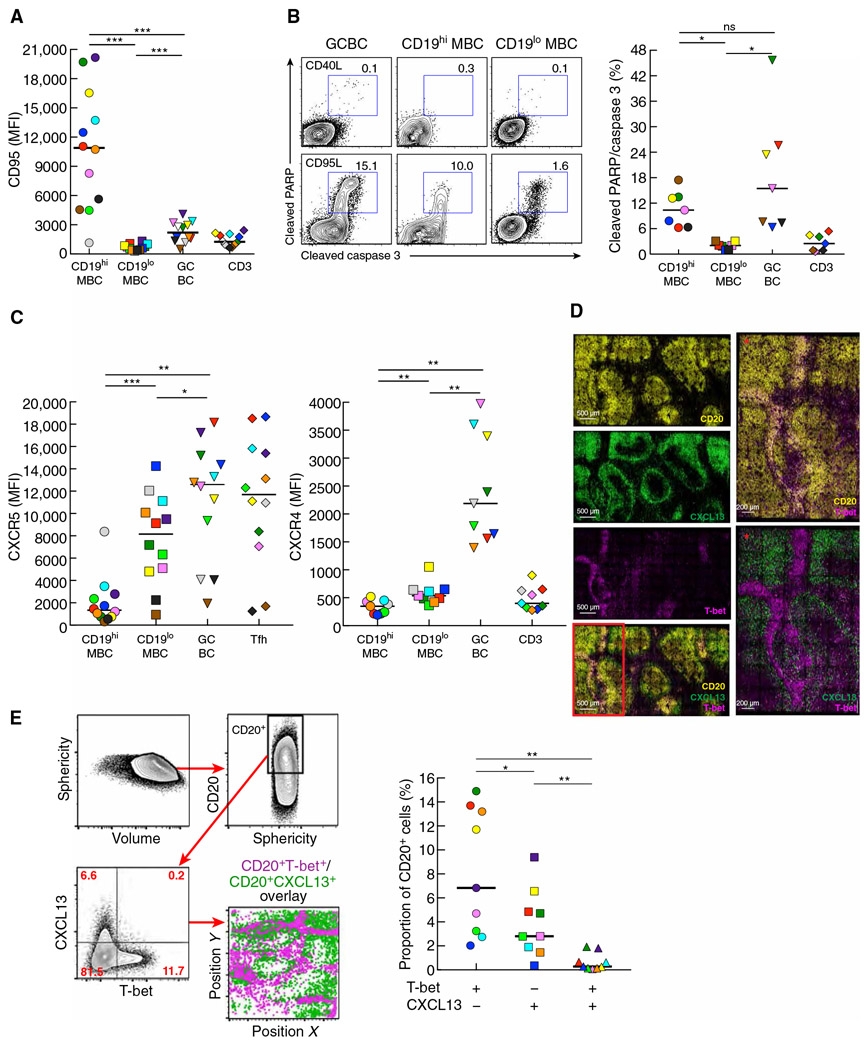
Given the uniquely high expression of FAS in HIV-specific MBC, we investigated cell surface expression of CD95 and susceptibility to apoptosis among various LN cell populations of HIV-infected individuals. Consistent with the enrichment of HIV-specific B cells among the parental CD19hi LN MBC, CD95 expression was significantly higher on this population when compared to CD19lo LN MBC (P = 0.0005) and GCBC (P = 0.001; Fig. 4A). CD95 expression on GCBC was also significantly higher compared to CD19lo LN MBC (P = 0.0005; Fig. 4A). Susceptibility to CD95-mediated apoptosis, expected to be the highest for CD19hi LN MBC, was instead similar to that of GCBC, although as expected, frequencies of apoptosis were significantly higher in these two populations when compared to CD19lo LN MBC (P = 0.0156 each; Fig. 4B).
Fig. 4. Profile of T-bet+ MBC in LN of HIV-infected individuals similar to GCBC but consistent with non-GC location.
(A) Expression of CD95 evaluated by conventional flow cytometry performed on LN mononuclear cells isolated from HIV-infected individuals (n = 12). (B) Apoptosis depicted and quantified by conventional flow cytometry performed on LN mononuclear cells isolated from HIV-infected individuals (n = 7), after incubation for 30 min with CD95L or CD40L (control). (C) Expression of CXCR5 and CXCR4 evaluated by conventional flow cytometry performed on LN mononuclear cells isolated from HIV-infected individuals (n = 9 to 12). (D) Images obtained by confocal microscopy depicting expression patterns of CD20, CXCL13, and T-bet in an LN section from an HIV-infected individual. Red asterisks are enlarged images from the red boxed inset illustrating distinct areas of expression for T-bet+ and CXCL13+ cells. (E) Imaged data from (D) were converted to flow data with gating scheme shown for delineation of CD20+ cells and depiction of CXCL13 by T-bet expression in CD20+ cells in dot plot and location. Quantification of frequencies of single or double CXCL13 and T-bet expression performed by histo-cytometry on LN sections from HIV-infected individuals (n = 9). In (A) to (C) and (E), each individual is color-coded per Table 1, and black horizontal bars represent medians. *P < 0.05, **P < 0.01, ***P < 0.001 by Wilcoxon matched-pairs signed rank test after obtaining significance by Friedman ANOVA test on B cell subpopulations.
Analyses by conventional flow cytometry revealed that intensities of CXCR5 and CXCR4 were significantly lower on CD19hi LN MBC when compared to GCBC (P = 0.0024 and 0.0039, respectively) and CD19lo LN MBC (P = 0.0005 and 0.0039, respectively; Fig. 4C), consistent with transcriptional profiles. CXCL13, a ligand for CXCR5 and a marker of GC-derived B cells (43), was evaluated by confocal microscopy of LN tissue of HIV-infected individuals. The non-GC location of T-bet+ cells contrasted with the GC location of cells expressing CXCL13 (Fig. 4D). The mutually exclusive nature of this expression pattern among B cells was confirmed by histocytometry of CD20-gated cells, where the positioning of CD20+T-bet+ and CD20+CXCL13+ cells was distinctly non-overlapping, and the percent of single B cells expressing either T-bet or CXCL13 was significantly higher than B cells expressing both T-bet and CXCL13 (P = 0.0039 each; Fig. 4E). Together, these data demonstrate that HIV-specific LN MBC are enriched among CD19hi LN MBC, and both share features with GCBC, yet both also have strong T-bet expression profiles that are associated with positioning outside the GC.
TH1 conditions favor accumulation of B cells outside GC
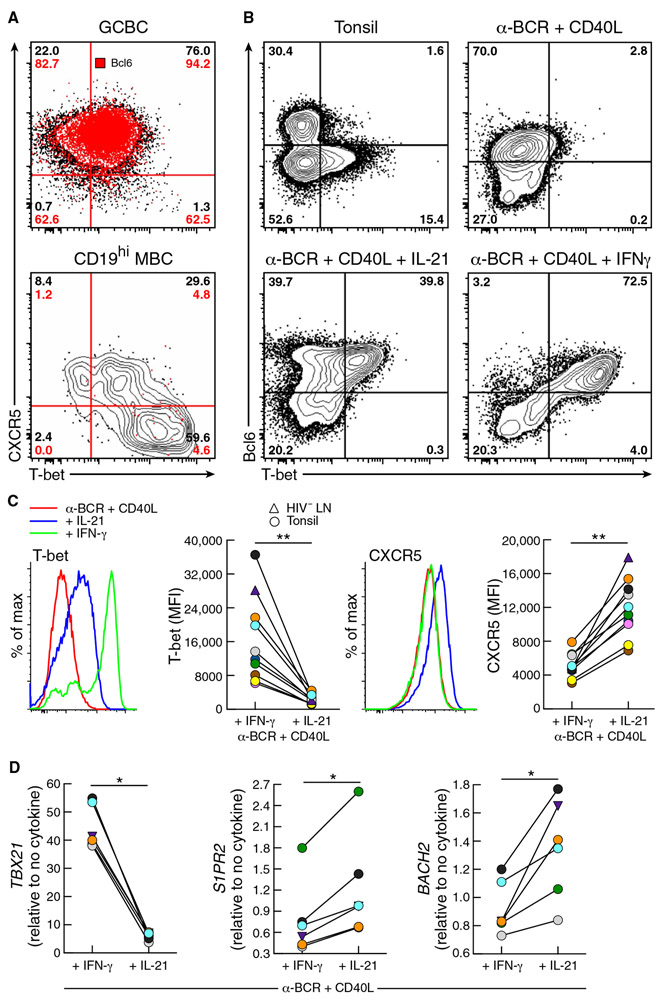
To identify factors that may be contributing to the positioning of T-bet+ LN B cells outside GC, we first examined patterns of T-bet expression among LN B cell populations. In GCBC but not CD19hi MBC, Bcl6 was coexpressed with T-bet (Fig. 5A), similar to profiles of TFH cells during early GC reactions (44). In LN B cells of HIV-infected individuals, T-bet+/Bcl6+ GCBC expressed high intensities of CXCR5, whereas increased intensities of T-bet in CD19hi MBC were accompanied by diminished expression of CXCR5 (Fig. 5A), consistent with patterns for T-bet and CXCR5 by LN B cell population shown respectively in Figs. 2 and 4. Given that CXCR5-deficient B cells are excluded from wild-type GC in chimeric mice (45) and that follicular positioning of LN T-bet+ B cells shown in Fig. 1B for HIV-infected individuals was similar to that of anergic B cells displaying reduced CXCR5 expression (46), we considered T-bet–related factors associated with chronic HIV viremia that could modulate expression of CXCR5 on B cells. In chronic simian immunodeficiency virus (SIV) infection, high expression of T-bet has been reported in the context of increased frequencies of TH1-polarized TFH cells (47). Accordingly, we considered whether such conditions could explain the positioning and accumulation of T-bet+ B cells outside GC in HIV-infected individuals. We used an in vitro system to stimulate naïve-enriched (IgD+) B cells isolated from LN or tonsil of HIV-uninfected individuals with anti-BCR and CD40L with or without polarizing cytokines. At baseline and in the absence of cytokines, few tonsil cells expressed T-bet and Bcl6, whereas T-bet expression increased in the presence of the TFH cytokine interleukin-21 (IL-21) and more intensely with the TH1 cytokine IFN-γ (Fig. 5B). In the presence of IFN-γ, intensities of T-bet were significantly higher (P = 0.002), and those of CXCR5 were significantly lower (P = 0.002) compared to IL-21 (Fig. 5C). Furthermore, we evaluated the effect of these conditions on the expression of S1PR2 and BACH2. In vitro, both S1PR2 and BACH2 were modestly yet consistently and significantly reduced (P = 0.0312 each), whereas TBX21 was significantly increased (P = 0.0312) in the presence of IFN-γ compared to IL-21 (Fig. 5D). Collectively, these findings demonstrate that under the TH1-polarizing conditions that prevail in chronic HIV viremia, expression of T-bet in B cells is high, whereas expression of GC-positioning receptors CXCR5 and S1PR2 is low, consistent with the profile of CD19hi MBC and their accumulation outside GC.
Fig. 5. TH1 and TFH cytokines modulate expression of T-bet and migration receptors.
(A) Conventional flow cytometry performed on LN GCBC and CD19hi MBC depicting the expression of Bcl6, T-bet, and CXCR5 expression of an HIV-infected individual. Red pattern and numbers identify Bcl6 frequencies within each quadrant. (B) Conventional flow cytometry performed on tonsillar B cells isolated from HIV-negative donors depicting Bcl6 and T-bet expression before and after 48 hours of stimulation of IgD+ cells with anti-human BCR and CD40L with or without IFN-γ or IL-21. (C) Histograms and comparison (n = 10) of T-bet and CXCR5 protein expression under conditions in (B). (D) Comparison (n = 6) of gene expression for TBX21, S1PR2, and BACH2 under conditions in (B). *P < 0.05, **P < 0.01 by Wilcoxon matched-pairs signed ranked test.
CD19hi MBC display low mutation frequencies and HIV-neutralizing capacity
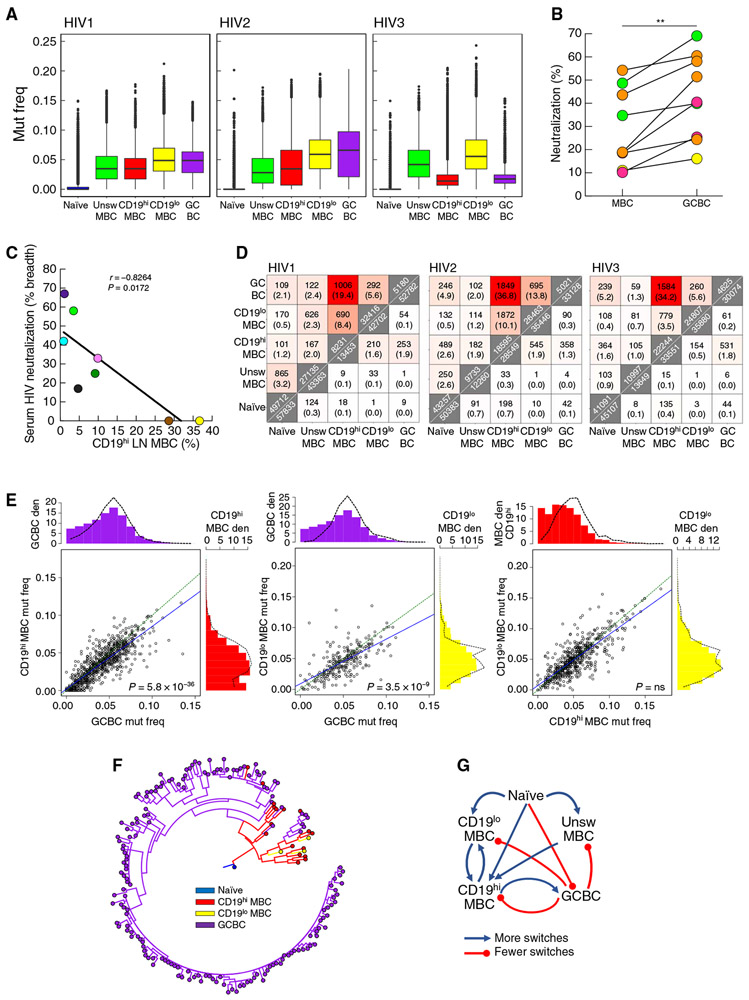
We then considered the effect of the non-GC location of CD19hiT-bet+ LN MBC on their ability to undergo affinity maturation by evaluating mutation frequencies in the BCR. Accordingly, LN B cell populations of four HIV-infected individuals were bulk-sorted (fig. S4A), and deep sequencing was performed on the Ig heavy-chain variable region (IGHV). As expected, mutation frequencies were lowest in naïve B cells and were consistently higher in CD19lo compared to CD19hi MBC (Fig. 6A and fig. S4B). In addition, we single-cell–sorted HIV-specific LN B cells of four HIV-infected individuals and, although CD19 intensity was not included in the sorting strategy due to the paucity of CD19lo HIV-specific MBC, mutation frequencies were significantly higher among HIV-specific GCBC versus MBC (P = 0.0047 for participant HIV7 and P < 0.0001 for the remainder; fig. S4B). The low mutation frequencies among HIV-specific MBC also extended to significantly lower neutralization capacity of HIV-specific monoclonal antibodies (mAbs) that were cloned and reconstituted from this compartment, when compared to GCBC-derived counterparts (P = 0.0039; Fig. 6B). The screening of serum obtained from the HIV-infected individuals at time of LN biopsy (Table 1) for HIV-neutralizing activity using standard HIV pseudotype and indicator cell line assay (48) revealed relatively weak potencies overall (Table 1). Nonetheless, among the HIV-infected individuals who were known to have been infected for at least 1 year (Table 1), the frequency of CD19hi LN MBC was inversely correlated with serologic HIV-neutralizing breadth (Fig. 6C). Thus, the serum and mAb analyses suggest that CD19hiT-bet+ LN MBC are linked to poor HIV-specific antibody responses and reflect the low mutation frequencies of the BCR from CD19hi and HIV-specific LN MBC (both of which have high expression of T-bet).
Fig. 6. BCR of CD19hi MBC related to GCBC but with decreased mutation frequency and neutralization capacity.
(A) Determination of IGHV mutation frequencies (mut freq) among B cell populations sorted from LN of three HIV-infected individuals (described in Table 1). Data are presented as box-and-whiskers plots showing the two inner quartiles (boxes) with median (black lines), 1.5 times the inner quartile range (whiskers) and outliers (black circles). (B) HIV pseudotype neutralization by clonally related or epitope target-related mAbs reconstituted from HIV-specific MBC and GCBC of four HIV-infected individuals (related clones/targets are color-coded by individual per table S1). Each value is % neutralization of one clone or median neutralization of 2 to 11 related clones from each source. **P < 0.01 by Wilcoxon matched-pairs signed rank test. (C) Breadth of serologic HIV pseudotype neutralization activity correlated with frequency of CD19hi LN MBC from HIV-infected individuals (n = 9, each individual is color-coded per Table 1). (D) Heatmaps indicating the numbers of shared clonal families (top triangle) and sequences (bottom triangle) from three HIV-infected individuals (identified in Table 1). Along the diagonal are total numbers of clones (top) or unique sequences (bottom). Percentages shown in parentheses were calculated by dividing the number of shared clones or unique sequences by the total in the smaller of the two samples. (E) Scatter-plots show the median IGHV mutation frequency of the two indicated populations for each shared clone from HIV-infected participant HIV1 (Table 1). Green equivalence and blue linear regression lines are depicted. The density (den) histograms show the mutation frequency of all sequences (bars) or sequences within shared clones between the two populations (dashed lines). P values by Wilcoxon signed rank test followed by Bonferroni correction. (F) Example of a multipopulation lineage tree with predicted internal node types. (G) Summary of predicted switches within lineage trees. Blue lines indicate more observed than expected switches; red lines indicate fewer observed than expected switches.
Next, BCR clonal relationships within and between the five populations shown in fig. S4A were determined using a clustering-based approach with a fixed distance threshold (fig. S4C) (49). Heatmaps of BCR clone and sequence overlap between B cell populations were generated for three individuals with sufficient sequencing depth in all five populations for meaningful analysis. Clonal diversity was highest for naïve B cells and lowest for GCBC, as reflected by the generalized Hill diversity index (fig. S4D). In terms of BCR relationships between populations, the overlap of clonal families was consistently highest between GCBC and CD19hi MBC and lowest between naïve and CD19lo MBC (Fig. 6D). Overlap of clonal families was intermediate between the two MBC (CD19hi and CD19lo) populations and between CD19lo MBC and GCBC (Fig. 6D). A similar pattern was observed for sequence overlap (Fig. 6D), although not as consistently observed as for clones.
Last, given the critical role of the GC reaction in affinity maturation and immunologic memory (1-4), we further analyzed the overlap between CD19hi MBC, CD19lo MBC, and GCBC. First, we considered mutation frequencies and phylogenies of clonal families that had members in at least two of these populations (shared clones). Within shared clones, median GCBC mutation frequencies were significantly higher (P < 0.0001 for all comparisons) than those of either CD19hi (all three HIV-infected individuals) or CD19lo MBC (individuals HIV1 and HIV2), whereas mutation frequencies between CD19hi and CD19lo MBC were not different (Fig. 6E and fig. S5A). Second, we used a maximum parsimony approach (50) to predict unobserved intermediate cell types in lineage trees spanning multiple cell types (Fig. 6F and fig. S5B). Within these lineages, differences between expected (calculated as described in Materials and Methods) and observed switches from one population to another were determined. In all three individuals examined, there were significantly more switches between CD19hi MBC and CD19lo MBC (P < 0.001 for CD19hi to CD19lo MBC and P = 0.00274 for CD19lo to CD19hi MBC) than expected, indicating that these populations were more closely located to each other in lineage trees than expected (shown schematically in Fig. 6G and values in fig. S5C). Furthermore, there was a consistent and significant overrepresentation of switches from CD19hi MBC to GCBC (P < 0.001), which contrasted with an underrepresentation of switches from GCBC to CD19hi MBC (P < 0.001; values in fig. S5C). This indicated that CD19hi MBC sequences were located closer to the root of lineage trees relative to GCBC sequences than expected, based on their relative sequence abundance.
DISCUSSION
In this study, we identified a unique LN population of human MBC associated with HIV viremia and characterized by strong T-bet expression and poor affinity maturation. High T-bet expression was concentrated in B cells located in non-GC areas of LN, with enrichment among CD19hi MBC and, more importantly, HIV-specific MBC. These LN T-bethi B cells displayed signature genes and phenotypes that have been reported for nonconventional MBC in the peripheral blood under immune-activating and inflammatory conditions of both infectious and non-infectious nature (1, 10-13) and may thus represent their lymphoid tissue source or counterpart. Furthermore, despite being enriched in non-GC areas/populations and having low BCR mutation frequencies, T-bethi B cell populations (CD19hi and HIV-specific MBC) nonetheless were closely related to GCBC, as revealed by GSEA and BCR lineage analyses. Collectively, these findings suggest that CD19hiT-bethi MBC are either recruited into the GC and exit early or remain outside the GC but originate from a common precursor with GCBC.
The transcriptional analyses we performed on HIV-specific LN MBC demonstrated similarities with T-bet–expressing B cells in the peripheral blood of individuals with chronic malaria exposure (27) and in individuals with active SLE (22). In both those studies, although the cells studied were not derived from lymphoid tissues, the data presented led the authors to suggest a distinct EF differentiation program, driven in the case of SLE by chronic TLR-7 stimulation. However, although parallels between these studies are clear, a chronic stimulus with non-GC expansion, consistent with decreased CXCR5 expression (22), other features appear to be unique to each condition. The GSEA analyses of HIV-specific LN MBC, as well as the BCR sequence and phylogenetic analyses of CD19hi MBC that revealed a relationship with LN GCBC, did not find evidence of signatures associated with PB/PC differentiation and TLR signaling that have been reported for SLE (22). Although T-bet expression was linked to increased Blimp-1 and decreased CD27, as observed for SLE (22), neither was differentially regulated in HIV-specific MBC by RNA-seq analysis. Furthermore, IRF4 (interferon regulatory factor), another transcription factor required for PC differentiation (2), was not up-regulated in HIV-specific MBC, suggesting that in HIV infection, Blimp-1 may be linked to increased T-bet expression but not to terminal differentiation. It is tempting to speculate that cytokine polarization (IL-21 in SLE and IFN-γ in HIV) and B cell stimulus (TLR in SLE and HIV-specific BCR in HIV) may account for differences in T-bet programs that were associated with GCBC in HIV and EF plasmablasts in SLE (22, 23). There was also evidence from the comparison of transcriptional profiles by population that T-bet was increased in HIV-infected compared to HIV-uninfected individuals. In this regard, the IFN-γ conditions in chronic HIV/SIV infection that have been associated with expansion of TH1-like TFH cells and loss of TFH cells with favorable B cell helper function (47, 51, 52) were also found to promote high expression of T-bet and reduced expression of GC-positioning receptors CXCR5 and S1PR2 in vitro. These conditions recapitulated the T-bethiCXCR5loS1PR2lo phenotype observed among HIV-specific MBC, despite an otherwise significant GCBC signature.
Despite the evidence from overall transcriptional and BCR sequence data of commonalities between the LN T-bet–expressing MBC populations and GCBC, the imaging data demonstrated that T-bet+ B cells were almost exclusively located outside the GC. BCR sequence analyses also revealed lower BCR mutation frequencies for the two T-bet+ populations investigated: bulk sorted CD19hi MBC when compared to corresponding GCBC and CD19lo MBC or the single-cell sorted HIV-specific MBC (enriched in CD19hi cells) when compared to corresponding HIV-specific GCBC. These findings are consistent with the non-GC location of the T-bet+ B cells, as observed in intact tissue sections, suggesting restricted affinity maturation from restricted or altered TFH cell help (9). The low BCR mutation frequencies among CD19hi MBC are also consistent with their positioning at the base of lineage trees occurring more often than that of a GCBC and the multipopulation prediction algorithm showing significantly more switches occurring from CD19hi MBC to GCBC than any other pair. These observations suggest either that CD19hi MBC are the source of GCBC or that CD19hi MBC arise from cells in the GC but exit without further or with slower accumulation of mutations in the EF zone. These events may occur more frequently during persistent viremia when conditions favor the establishment of CD19hiT-bethi MBC. When considering the two LN MBC populations, although CD19hi cells had lower BCR mutation frequencies compared to CD19lo cells when all sequences were considered, the difference was not significant when clones among the GCBC and two MBC populations were considered, suggesting a degree of intermixing between MBC populations, as previously shown for MBC in the peripheral blood (53). However, independent interconversion events of the two MBC populations from GCBC cannot be excluded.
Several key factors are likely responsible for the accumulation of CD19hiT-bethi MBC outside GC: decreased expression of GC homing and positioning receptors CXCR4, CXCR5, and S1PR2, as well as the near absence of CXCL13, a marker associated with GC activity (43). Furthermore, the discordance between much higher expression of CD95 on CD19hi MBC compared to GCBC and similar susceptibilities to apoptosis suggested that other factors involved in apoptosis were likely different between CD19hi MBC and GCBC. Whereas T-bet expression was associated with decreased expression of Bcl2, consistent with other findings (22), expression of Bcl2 in MBC populations was nonetheless higher than in GCBC. In addition, the transcriptional program of HIV-specific CD19hiT-bethi MBC contained several features similar to those reported for precursor MBC relative to their GC source: down-regulation of S1PR2, BACH2, and BCL6, as well as up-regulation of CXCR3, BCL2, and ZEB2 (41). Thus, despite the close association between HIV-specific (enriched in T-bethiCD19hi) LN MBC and GCBC based on GSEA, certain features help explain the non-GC location of the vast majority of T-bet–expressing LN B cells. TH1-polarizing conditions, which we show to decrease expression of GC homing and positioning receptors, may also explain the early exit from or lack of entry into GC of CD19hiT-bethi MBC.
Although T-bet–expressing B cells have an uncertain role in humans, they are required in mice for effective antiviral responses (16, 17, 54). What may be important in dictating immunologic benefit is the degree of T-bet expression that some T-bet is needed to resolve infection and maintain immunologic memory, but overexpression during chronic activation and inflammation leads to diminished functionality and plasticity (17, 55). T-bet overexpression in the context of deficient antiviral responses is also associated with increased expression of inhibitory receptors (29). Both in the peripheral blood and now shown in LN, we and others have found that the HIV-specific response is enriched among MBC that overexpress T-bet and multiple inhibitory receptors (28, 32, 56, 57). These features likely contributed to the limited functionality of HIV-specific LN MBC, both in terms of affinity maturation and HIV-neutralizing capacity of derived antibodies, given that the overexpression of inhibitory receptors has been shown to directly restrict B cell responses in HIV-infected individuals (56). In addition, the combined expression of the inhibitory receptor FCRL4 and T-bet in monocytoid B cells residing in LN outside GC has been associated with restricted affinity maturation (58). Similar observations of overexpression of T-bet and multiple immune-regulating receptors have been made in other conditions involving persisting pathogens, reviewed in (12), suggesting a common underlying mechanism of immune-mediated pathogenesis.
It is important to note some limitations of this study. Quantitative imaging, although a tremendous resource, lacks the sensitivity of conventional flow cytometry to distinguish certain populations, such as CD19hi versus CD19lo MBC, and is unable to identify HIV-specific B cells. Differences in BACH2 were also more muted in the imaging analyses than in RNA-seq, although it may be that the lowest expression in HIV-specific MBC is unique to chronic antigen-stimulated MBC. Furthermore, although T-bet was the key to all observations, it presented technical challenges in that its detection required intracellular staining, which was incompatible with the recovery of RNA suitable for RNA-seq and BCR analyses, and leads to high background when staining with the gp140 probe to identify HIV-specific B cells. As a result of these limitations, few of the observations could apply to all B cells that expressed T-bet or were HIV specific. BCR repertoire analysis was performed on a limited number of individuals, and it is important to note that the phylogenetic analyses performed did not account for uncertainty in tree topology or potential biases in sampling of cell types. Last, although it would have been ideal to extend the BCR mutational analyses to B cell populations in LN of HIV-uninfected individuals, the near absence of the most critical population, CD19h MBC, in these individuals precluded any meaningful comparisons.
Despite these limitations, collectively, our findings suggest that certain settings of chronic immune activation that favor T-bethi expression are detrimental to the generation of effective humoral immunity, both as a result of accumulation outside GC and induction of immune-regulating mechanisms. It is tempting to speculate that these same conditions that have a limiting effect on affinity maturation may enable the pathogen to persist but protect the host from damage associated with chronic immune activation. It is also possible that these conditions are regulated differently in HIV-infected individuals who develop broadly HIV-neutralizing antibodies. A better understanding of such regulators and the LN architecture where these antibodies develop will provide further insight into adaptive immunity in chronic infectious diseases and how responses to pathogens may be improved by therapeutic intervention.
MATERIALS AND METHODS
Study design
Our objectives were to locate, characterize, and compare LN T-bet expression in B cells of HIV-infected and HIV-uninfected individuals. Biopsies of palpable inguinal, cervical, or axillary LN and research phlebotomy were performed at the National Institutes of Health (NIH) Clinical Research Center in Bethesda, MD under protocols approved by the National Institute of Allergy and Infectious Diseases (NIAID) Institutional Review Board (ClinicalTrials.gov identifiers: NCT00039689, NCT00001316, and NCT00001281). All participants provided written informed consent. Participants included 13 HIV-infected viremic individuals and 18 HIV-uninfected individuals with a palpable LN who volunteered as healthy controls (Table 1). A portion of the tissue was fixed in formalin, and LN mononuclear cells (LNMC) were isolated by mechanical disruption and filtered with a 70-μm cell strainer. Tonsils were obtained from District of Columbia Center for AIDS Research Basic Science Core and Children’s National Medical Center from patients undergoing tonsillectomies. The use of these tonsils for this study was determined to be exempt from review by the NIH Institutional Review Board in accordance with the guidelines issued by the Office of Human Research Protections. Primary data are reported in data file S1.
Apoptosis detection by flow cytometry
LNMC were cultured with either FasL (500 ng/ml; Enzo) or CD40L (500 ng/ml; Enzo) for 30 min at 37°C, stained for surface markers, fixed with Lysing Solution (BD Biosciences), permeabilized with Permeabilizing Solution 2 (BD Biosciences), then stained with cleaved caspase-3 Alexa Fluor 647 (clone D3E9), and cleaved poly (adenosine diphosphate-ribose) polymerase (PARP)–phycoerythrin (clone D64E10; Cell Signaling Technology). Fluorescence-activated cell sorting (FACS) analyses were performed on a FACS Canto II flow cytometer or LSR Fortessa (BD Biosciences), with data analyses performed using FlowJo software v9.9.6 (TreeStar Inc.). Phenotypic analyses are described in Supplementary Materials and Methods.
Multiplex imaging analyses
Formalin-fixed, paraffin-embedded LN tissue sections were stained with titrated amounts of antibodies and imaged using a SP5 X-WLL confocal microscope (Leica). After correction for fluorochrome spillover, multiparameter images were further analyzed by histocytometry, as described in Supplementary Materials and Methods.
RNA-seq library preparation and analysis
Naïve, class-unswitched MBC, GCBC, and MBC from HIV-uninfected individuals, as well as naïve, class-unswitched MBC, GCBC, IgG+ GCBC, HIV-specific IgG+ GCBC, MBC, IgG+ MBC, and HIV-specific IgG+ MBC from HIV-infected individuals were sorted, as shown in Fig. 3B, using a FACSAria II (BD Biosciences) into TRIzol LS (Sigma). Total RNA from sorted subsets was extracted and concentrated using the RNA Clean and Concentrator kit (Zymo Research). Libraries were prepared using the Ovation SoLo RNA-Seq System (NuGEN), according to the manufacturer’s instructions. Sequencing was conducted on a NextSeq 550 (Illumina) with an average of 31 million reads per sample. Reads were mapped to the human genome (GRCh37/hg19) using STAR (59). DESeq2 was used to calculate differential gene expression from uniquely mapped read counts (60). GSEA was performed with 10,000 gene set permutations using the GSEA software (61). Genes previously identified as being significantly overexpressed in atypical B cells compared to conventional MBC or memory precursors compared to memory cells (Benjamini-Hochberg adjusted P ≤ 0.01; fold change, ≥1.5) were defined as atypical signature genes (27) or memory precursor genes (41), respectively. GCBC signature genes were derived by comparing differentially expressed genes from GCBC and MBC from healthy HIV-uninfected individuals (Benjamini-Hochberg adjusted P ≤ 0.001; fold change, ≥1.5).
In vitro stimulation of tissue-derived IgD+ B cells
Cell suspensions were prepared from tonsil tissue, as described for LNMC. IgD+ B cells, enriched in naïve B cells, were isolated by magnetic bead-based fractionation with biotinylated anti-IgD (Miltenyi Biotec), followed by anti-biotin microbeads (Miltenyi Biotec), according to the manufacturer’s specifications. IgD+ B cells (1 × 106) were seeded per well in a 48-well flat bottom plate in RPMI 1640/10% fetal bovine serum/1% penicillin-streptomycin with goat F(ab′)2 anti-human IgG/A/M (10 μg/ml; Jackson ImmunoResearch Laboratories) and CD40L (500 ng/ml; Enzo), with or without IFN-γ or IL-21 (50 ng/ml; R&D Systems). After stimulation, cells were collected for phenotyping or RNA extraction.
BCR sequencing library preparation and analysis
Naïve, class-unswitched MBC, CD19hi MBC, CD19lo MBC, and GCBC were sorted into TRIzol LS, and total RNA was isolated as in RNA-seq analyses. BCR library preparation was adapted from Schanz et al. (62). Briefly, complementary DNA synthesis and the first round of polymerase chain reaction (PCR) were performed as described. The second PCR was adjusted to the following: 98°C for 2 min; 5 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 50 s; 3 cycles of 95°C for 30 s, 68°C for 30 s, 72°C for 50 s, and 72°C for 10 min. Amplicons were visualized on agarose gel, purified by using the QIAquick Gel Extraction Kit (Qiagen), and sequenced on a MiSeq (Illumina).
Isotypes were assigned to sequences as previously described (62). Poor-quality reads were removed, and duplicate reads were collapsed into unique sequences using pRESTO v0.5.8 (63). Only sequences with at least two duplicate reads were processed further. Initial assignment of V(D)J germline gene annotations was performed using IgBLAST v1.7.0 (64) against the International Immunogenetics Information System (IMGT) database (65) and subsequently processed using Change-O v0.4.1 (66). Individual genotypes were computationally inferred using TIgGER v0.2.11 (67) and used to finalize V(D)J annotations. Productively rearranged heavy-chain sequences were clustered into clonally related lineages, and full-length clonal-consensus germline sequences were reconstructed for each clone. Using Alakazam v0.2.10 (66), duplicate IMGT-gapped V(D)J sequences within each clone were collapsed, with the exception of duplicates derived from different cell subsets or isotypes. For each processed sequence, mutation frequency was calculated using SHazaM v0.1.9 (66). Phylogenetic trees were constructed using the Fitch parsimony algorithm (68) and the “pratchet” optimization function from phangorn v2.2.0 (69). Internal nodes of each lineage tree were labeled using a maximum parsimony algorithm. A detailed description can be found in Supplemental Materials and Methods.
Evaluation of HIV neutralization
Neutralization of cloned mAbs was determined by using a single-round infectivity assay (48), performed as previously described (53). Briefly, HIV luciferase pseudovirus typed with a JRFL or HXB2 envelope was incubated with a fourfold dilution series of purified antibody for 2 hours before being added to TZM-bl cells for 48 hours. The median of percent neutralization at maximum mAb concentration (40 μg/ml) was calculated. Pairing of MBC-GCBC mAbs by individual was matched by HIV pseudotype and either by clonal family or by recognition of the same defined epitope within gp140 (70). Neutralization activity of serum was determined with a six-member panel using fivefold dilutions of sera and applying a 5-parameter equation to calculate the 50% reciprocal inhibitory dilution (ID50). Breadth was determined on serum samples of individuals infected for at least 1 year by calculating the percent of HIV-1 Env-pseudoviruses of the six-member panel of isolates that achieved an ID50. The six-member panel consisted of the following strains with clade indicated in parentheses: Q259.17 (A), Q461.e2 (AD), TH976.17 (AE), YU2.DG (B), CH070.1 (BC), and 0013095-2.11 (C).
Statistical methods
Statistical analyses were performed using GraphPad Prism software (GraphPad) and R (71). Mann-Whitney U test (for unpaired variables) and Wilcoxon signed rank test (for paired variables) were performed. For testing of multiple linked parameters, the data were corrected by the Bonferroni (Fig. 6E and fig. S5A) or the Benjamini-Hochberg (Fig. 3D) method or as described in Supplemental Materials and Methods. P < 0.05 was considered statistically significant.
Supplementary Material
Fig. S1. Additional histocytometric analyses of HIV-infected and HIV-uninfected LN sections.
Fig. S2. Additional analyses of LN B cell populations.
Fig. S3. Additional RNA-seq analyses.
Fig. S4. Resident LN B cell populations have differences in BCR mutation frequency and diversity.
Fig. S5. Characterization of intraclonal relationships.
Table S1. Differential gene expression analysis for selected genes and populations.
Table S2. Antibodies used in imaging analyses.
Table S3. HIV-infected participants for additional phenotyping.
Data file S1. Primary data.
Acknowledgments:
We thank the participants for their willingness to take part in this study. We thank M. Hand, S. Jones, and C. Rehm for clinic-related support; R. Jiang for helpful discussion on raw sequence processing; O. Pybus for helpful discussions of phylogenetic methods; and A. S. Fauci for critical reading of the paper. We thank M. Proschan for help with statistical analyses. We thank the Yale Center for Research Computing for use of the research computing infrastructure.
Funding: This work was funded by the Intramural Research Program of the NIAID of the NIH and NIH extramural grants R01AI104739 and AI102766. J.Q.Z. was supported by a Gruber Science Fellowship. K.B.H. was supported by a PhRMA Foundation Postdoctoral Fellowship in Informatics.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. RNA-seq data have been deposited to the Genome Expression Omnibus under accession number GSE119234. Processed BCR sequencing data have been deposited at DDBJ/ENA/GenBank under the accessions: KCIZ00000000, KCJA00000000, KCJB00000000, KCJC00000000, KCJD00000000, KCJE00000000, KCJF00000000, KCJG00000000, KCJH00000000, KCJI00000000, KCJJ00000000, KCJK00000000, KCJL00000000, KCJM00000000, KCJN00000000, KCJO00000000, KCJP00000000, KCJQ00000000, KCJR00000000, and KCJS00000000. Raw BCR sequencing data are available from the Sequence Read Archive under accession SRP159625. Code has been deposited to Zenodo (doi:10.5281/zenodo.3405436).
REFERENCES AND NOTES
- 1.Good-Jacobson KL, Strength in diversity: Phenotypic, functional, and molecular heterogeneity within the memory B cell repertoire. Immunol. Rev 284, 67–78 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Seifert M, Kuppers R, Human memory B cells. Leukemia 30, 2283–2292 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Kurosaki T, Kometani K, Ise W, Memory B cells. Nat. Rev. Immunol 15, 149–159 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Victora GD, Nussenzweig MC, Germinal centers. Annu. Rev. Immunol 30, 429–457 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Dimopoulos Y, Moysi E, Petrovas C, The lymph node in HIV pathogenesis. Curr. HIV/AIDS Rep 14, 133–140 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Beyer M, Abdullah Z, Chemnitz JM, Maisel D, Sander J, Lehmann C, Thabet Y, Shinde PV, Schmidleithner L, Kohne M, Trebicka J, Schierwagen R, Hofmann A, Popov A, Lang KS, Oxenius A, Buch T, Kurts C, Heikenwalder M, Fatkenheuer G, Lang PA, Hartmann P, Knolle PA, Schultze JL, Tumor-necrosis factor impairs CD4+ T cell–mediated immunological control in chronic viral infection. Nat. Immunol 17, 593–603 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Wherry EJ, Kurachi M, Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moir S, Fauci AS, B-cell exhaustion in HIV infection: The role of immune activation. Curr. Opin. HIV AIDS 9, 472–477 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Crotty S, T follicular helper cell differentiation, function, and roles in disease. Immunity 41 , 529–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karnell JL, Kumar V, Wang J, Wang S, Voynova E, Ettinger R, Role of CD11c+ T-bet+ B cells in human health and disease. Cell. Immunol 321, 40–45 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Knox JJ, Myles A, Cancro MP, T-bet+ memory B cells: Generation, function, and fate. Immunol. Rev 288, 149–160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Portugal S, Obeng-Adjei N, Moir S, Crompton PD, Pierce SK, Atypical memory B cells in human chronic infectious diseases: An interim report. Cell. Immunol 321, 18–25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winslow GM, Papillion AM, Kenderes KJ, Levack RC, CD11c+ T-bet+ memory B cells: Immune maintenance during chronic infection and inflammation? Cell. Immunol 321, 8–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazarevic V, Glimcher LH, Lord GM, T-bet: A bridge between innate and adaptive immunity. Nat. Rev. Immunol 13, 777–789 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng SL, Szabo SJ, Glimcher LH, T-bet regulates IgG class switching and pathogenic autoantibody production. Proc. Natl. Acad. Sci. U.S.A 99, 5545–5550 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnett BE, Staupe RP, Odorizzi PM, Palko O, Tomov VT, Mahan AE, Gunn B, Chen D, Paley MA, Alter G, Reiner SL, Lauer GM, Teijaro JR, Wherry EJ, Cutting edge: B cell-intrinsic T-bet expression is required to control chronic viral infection. J. Immunol 197, 1017–1022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piovesan D, Tempany J, Di Pietro A, Baas I, Yiannis C, O'Donnell K, Chen Y, Peperzak V, Belz GT, Mackay CR, Smyth GK, Groom JR, Tarlinton DM, Good-Jacobson KL, c-Myb regulates the T-bet-dependent differentiation program in B cells to coordinate antibody responses. Cell Rep. 19, 461–470 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Cooper L, Hailes L, Sheikh A, Zaph C, Belz GT, Groom JR, Good-Jacobson KL, Assessing the role of the T-box transcription factor Eomes in B cell differentiation during either Th1 or Th2 cell-biased responses. PLOS ONE 13, e0208343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu N, Ohnishi N, Ni L, Akira S, Bacon KB, CpG directly induces T-bet expression and inhibits IgG1 and IgE switching in B cells. Nat. Immunol 4, 687–693 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Naradikian MS, Myles A, Beiting DP, Roberts KJ, Dawson L, Herati RS, Bengsch B, Linderman SL, Stelekati E, Spolski R, Wherry EJ, Hunter C, Hensley SE, Leonard WJ, Cancro MP, Cutting edge: IL-4, IL-21, and IFN-γ interact to govern T-bet and CD11c expression in TLR-activated B cells. J. Immunol 197, 1023–1028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naradikian MS, Hao Y, Cancro MP, Age-associated B cells: Key mediators of both protective and autoreactive humoral responses. Immunol. Rev 269, 118–129 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Jenks SA, Cashman KS, Zumaquero E, Marigorta UM, Patel AV, Wang X, Tomar D, Woodruff MC, Simon Z, Bugrovsky R, Blalock EL, Scharer CD, Tipton CM, Wei C, Lim SS, Petri M, Niewold TB, Anolik JH, Gibson G, Lee FE, Boss JM, Lund FE, Sanz I, Distinct effector B cells induced by unregulated toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity 49, 725–739.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S, Wang J, Kumar V, Karnell JL, Naiman B, Gross PS, Rahman S, Zerrouki K, Hanna R, Morehouse C, Holoweckyj N, Liu H, Autoimmunity Molecular Medicine T, Manna Z, Goldbach-Mansky R, Hasni S, Siegel R, Sanjuan M, Streicher K, Cancro MP, Kolbeck R, Ettinger R, IL-21 drives expansion and plasma cell differentiation of autoreactive CD11chiT-bet+ B cells in SLE. Nat. Commun 9, 1758 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bob R, Falini B, Marafioti T, Paterson JC, Pileri S, Stein H, Nodal reactive and neoplastic proliferation of monocytoid and marginal zone B cells: An immunoarchitectural and molecular study highlighting the relevance of IRTA1 and T-bet as positive markers. Histopathology 63, 482–498 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Falini B, Tiacci E, Pucciarini A, Bigerna B, Kurth J, Hatzivassiliou G, Droetto S, Galletti AV, Gambacorta M, Orazi A, Pasqualucci L, Miller I, Kuppers R, Dalla-Favera R, Cattoretti G, Expression of the IRTA1 receptor identifies intraepithelial and subepithelial marginal zone B cells of the mucosa-associated lymphoid tissue (MALT). Blood 102, 3684–3692 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Johrens K, Shimizu Y, Anagnostopoulos I, Schiffmann S, Tiacci E, Falini B, Stein H, T-bet-positive and IRTA1-positive monocytoid B cells differ from marginal zone B cells and epithelial-associated B cells in their antigen profile and topographical distribution. Haematologica 90, 1070–1077 (2005). [PubMed] [Google Scholar]
- 27.Obeng-Adjei N, Portugal S, Holla P, Li S, Sohn H, Ambegaonkar A, Skinner J, Bowyer G, Doumbo OK, Traore B, Pierce SK, Crompton PD, Malaria-induced interferon-γ drives the expansion of Tbethi atypical memory B cells. PLOS Pathog. 13, e1006576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knox JJ, Buggert M, Kardava L, Seaton KE, Eller MA, Canaday DH, Robb ML, Ostrowski MA, Deeks SG, Slifka MK, Tomaras GD, Moir S, Moody MA, Betts MR, T-bet+ B cells are induced by human viral infections and dominate the HIV gp140 response. JCI Insight 2, e92943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burton AR, Pallett LJ, McCoy LE, Suveizdyte K, Amin OE, Swadling L, Alberts E, Davidson BR, Kennedy PT, Gill US, Mauri C, Blair PA, Pelletier N, Maini MK, Circulating and intrahepatic antiviral B cells are defective in hepatitis B. J. Clin. Invest 128, 4588–4603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frasca D, Diaz A, Romero M, D'Eramo F, Blomberg BB, Aging effects on T-bet expression in human B cell subsets. Cell. Immunol 321, 68–73 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Moysi E, Pallikkuth S, De Armas LR, Gonzalez LE, Ambrozak D, George V, Huddleston D, Pahwa R, Koup RA, Petrovas C, Pahwa S, Altered immune cell follicular dynamics in HIV infection following influenza vaccination. J. Clin. Invest 128, 3171–3185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moir S, Ho J, Malaspina A, Wang W, DiPoto AC, O'Shea MA, Roby G, Kottilil S, Arthos J, Proschan MA, Chun TW, Fauci AS, Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J. Exp. Med. 205, 1797–1805 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss GE, Crompton PD, Li S, Walsh LA, Moir S, Traore B, Kayentao K, Ongoiba A, Doumbo OK, Pierce SK, Atypical memory B cells are greatly expanded in individuals living in a malaria-endemic area. J. Immunol 183, 2176–2182 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portugal S, Tipton CM, Sohn H, Kone Y, Wang J, Li S, Skinner J, Virtaneva K, Sturdevant DE, Porcella SF, Doumbo OK, Doumbo S, Kayentao K, Ongoiba A, Traore B, Sanz I, Pierce SK, Crompton PD, Malaria-associated atypical memory B cells exhibit markedly reduced B cell receptor signaling and effector function. ELife 4, 10.7554/eLife.07218, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan RT, Kim CC, Fontana MF, Feeney ME, Jagannathan P, Boyle MJ, Drakeley BJ, Ssewanyana I, Nankya F, Mayanja-Kizza H, Dorsey G, Greenhouse B, FCRL5 delineates functionally impaired memory B Cells associated with plasmodium falciparum exposure. PLOS Pathog. 11, e1004894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehrhardt GR, Hsu JT, Gartland L, Leu CM, Zhang S, Davis RS, Cooper MD, Expression of the immunoregulatory molecule FcRH4 defines a distinctive tissue-based population of memory B cells. J. Exp. Med 202, 783–791 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu YJ, Joshua DE, Williams GT, Smith CA, Gordon J, MacLennan IC, Mechanism of antigen-driven selection in germinal centres. Nature 342, 929–931 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Ehrhardt GR, Hijikata A, Kitamura H, Ohara O, Wang JY, Cooper MD, Discriminating gene expression profiles of memory B cell subpopulations. J. Exp. Med 205, 1807–1817 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinnakasu R, Inoue T, Kometani K, Moriyama S, Adachi Y, Nakayama M, Takahashi Y, Fukuyama G, Okada T, Kurosaki T, Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol 17, 861–869 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Green JA, Suzuki K, Cho B, Willison LD, Palmer D, Allen CD, Schmidt TH, Xu Y, Proia RL, Coughlin SR, Cyster JG, The sphingosine 1-phosphate receptor S1P2 maintains the homeostasis of germinal center B cells and promotes niche confinement. Nat. Immunol 12, 672–680 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laidlaw BJ, Schmidt TH, Green JA, Allen CD, Okada T, Cyster JG, The Eph-related tyrosine kinase ligand Ephrin-B1 marks germinal center and memory precursor B cells. J. Exp. Med 214, 639–649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hipp N, Symington H, Pastoret C, Caron G, Monvoisin C, Tarte K, Fest T, Delaloy C, IL-2 imprints human naive B cell fate towards plasma cell through ERK/ELK1-mediated BACH2 repression. Nat. Commun 8, 1443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Havenar-Daughton C, Lindqvist M, Heit A, Wu JE, Reiss SM, Kendric K, Belanger S, Kasturi SP, Landais E, Akondy RS, McGuire HM, Bothwell M, Vagefi PA, Scully E, Investigators IPCP, Tomaras GD, Davis MM, Poignard P, Ahmed R, Walker BD, Pulendran B, McElrath MJ, Kaufmann DE, Crotty S, CXCL13 is a plasma biomarker of germinal center activity. Proc. Natl. Acad. Sci. U. S. A 113, 2702–2707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, Schwartzberg PL, Hager GL, O'Shea JJ, Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35, 919–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen CD, Ansel KM, Low C, Lesley R, Tamamura H, Fujii N, Cyster JG, Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol 5, 943–952 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Ekland EH, Forster R, Lipp M, Cyster JG, Requirements for follicular exclusion and competitive elimination of autoantigen-binding B cells. J. Immunol 172, 4700–4708 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Moukambi F, Rabezanahary H, Rodrigues V, Racine G, Robitaille L, Krust B, Andreani G, Soundaramourty C, Silvestre R, Laforge M, Estaquier J, Early loss of splenic Tfh Cells in SIV-infected rhesus macaques. PLOS Pathog. 11, e1005287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarzotti-Kelsoe M, Bailer RT, Turk E, Lin CL, Bilska M, Greene KM, Gao H, Todd CA, Ozaki DA, Seaman MS, Mascola JR, Montefiori DC, Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 409, 131–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gupta NT, Adams KD, Briggs AW, Timberlake SC, Vigneault F, Kleinstein SH, Hierarchical clustering can identify B cell clones with high confidence in Ig repertoire sequencing data. J. Immunol 198, 2489–2499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano T, Lu L, Liu P, Pybus OG, Viral gene sequences reveal the variable history of hepatitis C virus infection among countries. J Infect Dis 190, 1098–1108 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Boswell KL, Paris R, Boritz E, Ambrozak D, Yamamoto T, Darko S, Wloka K, Wheatley A, Narpala S, McDermott A, Roederer M, Haubrich R, Connors M, Ake J, Douek DC, Kim J, Petrovas C, Koup RA, Loss of circulating CD4 T cells with B cell helper function during chronic HIV infection. PLOS Pathog. 10, e1003853 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Velu V, Mylvaganam G, Ibegbu C, Amara RR, Tfh1 cells in germinal centers during chronic HIV/SIV infection. Front. Immunol 9, 1272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meffre E, Louie A, Bannock J, Kim LJ, Ho J, Frear CC, Kardava L, Wang W, Buckner CM, Wang Y, Fankuchen OR, Gittens KR, Chun TW, Li Y, Fauci AS, Moir S, Maturational characteristics of HIV-specific antibodies in viremic individuals. JCI Insight 1, e84610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rubtsova K, Rubtsov AV, van Dyk LF, Kappler JW, Marrack P, T-box transcription factor T-bet, a key player in a unique type of B-cell activation essential for effective viral clearance. Proc. Natl. Acad. Sci. U. S. A 110, E3216–E3224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dominguez CX, Amezquita RA, Guan T, Marshall HD, Joshi NS, Kleinstein SH, Kaech SM, The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med 212, 2041–2056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kardava L, Moir S, Wang W, Ho J, Buckner CM, Posada JG, O'Shea MA, Roby G, Chen J, Sohn HW, Chun TW, Pierce SK, Fauci AS, Attenuation of HIV-associated human B cell exhaustion by siRNA downregulation of inhibitory receptors. J. Clin. Invest 121, 2614–2624 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kardava L, Sohn H, Youn C, Austin JW, Wang W, Buckner CM, Justement JS, Melson VA, Roth GE, Hand MA, Gittens KR, Kwan RW, Sneller MC, Li Y, Chun TW, Sun PD, Pierce SK, Moir S, IgG3 regulates tissue-like memory B cells in HIV-infected individuals. Nat. Immunol 19, 1001–1012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lazzi S, Bellan C, Tiacci E, Palummo N, Vatti R, Oggioni M, Amato T, Schuerfeld K, Tonini T, Tosi P, Falini B, Leoncini L, IRTA1+ monocytoid B cells in reactive lymphadenitis show a unique topographic distribution and immunophenotype and a peculiar usage and mutational pattern of IgVH genes. J. Pathol 209, 56–66 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schanz M, Liechti T, Zagordi O, Miho E, Reddy ST, Gunthard HF, Trkola A, Huber M, High-throughput sequencing of human immunoglobulin variable regions with subtype identification. PLOS ONE 9, e111726 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vander Heiden JA, Yaari G, Uduman M, Stern JN, O'Connor KC, Hafler DA, Vigneault F, Kleinstein SH, pRESTO: A toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics 30, 1930–1932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye J, Ma N, Madden TL, Ostell JM, IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Giudicelli V, Chaume D, Lefranc MP, IMGT/GENE-DB: A comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 33, D256–D261 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, Kleinstein SH, Change-O: A toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gadala-Maria D, Yaari G, Uduman M, Kleinstein SH, Automated analysis of high-throughput B-cell sequencing data reveals a high frequency of novel immunoglobulin V gene segment alleles. Proc. Natl. Acad. Sci. U. S. A 112, E862–E870 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fitch WM, Toward defining course of evolution: Minimum change for a specific tree topology. Syst. Zool. 20, 406–416 (1971). [Google Scholar]
- 69.Schliep KP, Phangorn: Phylogenetic analysis in R. Bioinformatics 27, 592–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kardava L, Moir S, Shah N, Wang W, Wilson R, Buckner CM, Santich BH, Kim LJ, Spurlin EE, Nelson AK, Wheatley AK, Harvey CJ, McDermott AB, Wucherpfennig KW, Chun TW, Tsang JS, Li Y, Fauci AS, Abnormal B cell memory subsets dominate HIV-specific responses in infected individuals. J. Clin. Invest 124, 3252–3262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R. Core Team, A language environment for statistical computing (R Foundation for Statistical Computing, 2018). [Google Scholar]
- 72.Gerner MY, Kastenmuller W, Ifrim I, Kabat J, Germain RN, Histo-cytometry: A method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petrovas C, Ferrando-Martinez S, Gerner MY, Casazza JP, Pegu A, Deleage C, Cooper A, Hataye J, Andrews S, Ambrozak D, Del Rio Estrada PM, Boritz E, Paris R, Moysi E, Boswell KL, Ruiz-Mateos E, Vagios I, Leal M, Ablanedo-Terrazas Y, Rivero A, Gonzalez-Hernandez LA, McDermott AB, Moir S, Reyes-Teran G, Docobo F, Pantaleo G, Douek DC, Betts MR, Estes JD, Germain RN, Mascola JR, Koup RA, Follicular CD8 T cells accumulate in HIV infection and can kill infected cells in vitro via bispecific antibodies. Sci. Transl. Med 9, eaag2285 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edgar RC, Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 75.Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G, IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol 27, 55–77 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Hill MO, Diversity and evenness: A unifying notation and its consequences. Ecology 54, 427–432 (1973). [Google Scholar]
- 77.Chao A, Hsieh TC, Chazdon RL, Colwell RK, Gotelli NJ, Unveiling the species-rank abundance distribution by generalizing the Good-Turing sample coverage theory. Ecology 96, 1189–1201 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Sankoff D, Minimal mutation trees of sequences. SIAM J. Appl. Math 28, 35–42 (1975). [Google Scholar]
- 79.El-Kebir M, Satas G, Raphael BJ, Inferring parsimonious migration histories for metastatic cancers. Nat. Genet 50, 718–726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoehn KB, Lunter G, Pybus OG, A phylogenetic codon substitution model for antibody lineages. Genetics 206, 417–427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Additional histocytometric analyses of HIV-infected and HIV-uninfected LN sections.
Fig. S2. Additional analyses of LN B cell populations.
Fig. S3. Additional RNA-seq analyses.
Fig. S4. Resident LN B cell populations have differences in BCR mutation frequency and diversity.
Fig. S5. Characterization of intraclonal relationships.
Table S1. Differential gene expression analysis for selected genes and populations.
Table S2. Antibodies used in imaging analyses.
Table S3. HIV-infected participants for additional phenotyping.
Data file S1. Primary data.