Abstract
The invasive nature and the pain caused to patients inhibit the routine use of tissue biopsy-based procedures for cancer diagnosis and surveillance. The analysis of extracellular vesicles (EVs) from biofluids has recently gained significant traction in the liquid biopsy field. EVs offer an essential “snapshot” of their precursor cells in real time and contain an information-rich collection of nucleic acids, proteins, lipids, and so on. The analysis of protein phosphorylation, as a direct marker of cellular signaling and disease progression could be an important stepping stone to successful liquid biopsy applications. Here we introduce a rapid EV isolation method based on chemical affinity called EVtrap (extracellular vesicle total recovery and purification) for the EV phosphoproteomics analysis of human plasma. By incorporating EVtrap with high-performance mass spectrometry (MS), we were able to identify over 16 000 unique peptides representing 2238 unique EV proteins from just 5 μL of plasma sample, including most known EV markers, with substantially higher recovery levels compared with ultracentrifugation. Most importantly, more than 5500 unique phosphopeptides representing almost 1600 phosphoproteins in EVs were identified using only 1 mL of plasma. Finally, we carried out a quantitative EV phosphoproteomics analysis of plasma samples from patients diagnosed with chronic kidney disease or kidney cancer, identifying dozens of phosphoproteins capable of distinguishing disease states from healthy controls. The study demonstrates the potential feasibility of our robust analytical pipeline for cancer signaling monitoring by tracking plasma EV phosphorylation.
Keywords: extracellular vesicles, exosomes, proteomics, phosphoproteomics, EVtrap, kidney cancer, renal cell carcinoma, LC–MS
Graphical Abstract
INTRODUCTION
Currently, the most widespread method for clinical cancer profiling and disease diagnosis involves a tumor biopsy, an invasive and painful procedure and one that certainly is impractical for early-stage detection. As certain cancers become more chronic diseases that require active monitoring over longer periods of time, tissue biopsies on a continuous basis are no longer a realistic scenario. As a result, “liquid biopsies”, the analysis of biofluids such as plasma, serum, and urine, have gained much attention as a potentially useful source of diagnostic biomarkers.1–4 Liquid biopsies offer numerous advantages for clinical analysis, including noninvasive collection, suitable sample collection for longitudinal disease monitoring, a better screenshot of tumor heterogeneity, higher stability and sample volumes, faster processing times, lower rejection rates, and lower cost. However, there are technical challenges and shortcomings with the most common focus of liquid biopsy, circulating tumor cells (CTCs) and cell-free DNA (cfDNA), including their heterogeneity, extreme rarity, and high fragmentation levels.5–9
In addition, current analyses overwhelmingly focus on genetic information, usually gene mutations. Whereas genetic testing is valuable, it could greatly benefit from an additional layer of biological information. The ability to detect the genome output, active proteins, can provide useful real-time information about the organism’s physiological functions and disease progression, in particular, in cancer.10–13 Oncologists understand the value of protein testing and immunoassays and can easily interpret the results. Compared with gene panel testing, immunoassays, once developed, are relatively inexpensive and are easier to adopt in clinical settings.
To help overcome some of the challenges associated with the current liquid biopsy methods, a new field has generated much interest over the past few years: profiling of cell-secreted extracellular vesicles (EVs, which include microvesicles and exosomes).14,15 EVs provide an effective and ubiquitous method for intercellular communication, stimulation of the immune system, and removal of harmful materials and serve many more functions.16–18 Because EVs are shed into every biological fluid and embody a good representation of their parent cell, analysis of the EV cargo has great promise for biomarker discovery and disease diagnosis.19,20 Previous studies also revealed that EV-based disease markers could be identified before the onset of symptoms or the physiological detection of a tumor, making them promising candidates for early-stage disease detection.21,22
Despite the significant recent excitement and efforts, a standardized method for collecting and processing EVs has not yet been developed.23 Differential centrifugation with ultracentrifugation (UC) as the final step is generally considered the “gold standard” for EV isolation (in particular, for exosomes). However, this approach is highly time-consuming (typically 6–22 h), requires dedicated equipment, and is low-throughput and thus is not suitable in a clinical setting due to its poor reproducibility.24–27 In addition, multiple studies have shown that the exosome recovery rate after ultracentrifugation is only 5–25%.27–29 Several other groups have published and commercialized new methods for EV isolation, which include polymer-induced precipitation,30,31 antibody-based capture,32,33 affinity filtration,34 size-exclusion chromatography,29,35 and so on. However, each one has its own limitations, including low recovery rate (usually similar to or slightly worse than ultracentrifugation) and high contamination levels.25–27,29,34,36–40 Whereas these can certainly be used as a faster alternative to UC, at 5–25% published yields, their efficiency of isolation still leaves much room for improvement. As a result, there is a critical need for a fast, reproducible, and inexpensive approach for EV isolation that would allow a much more complete capture and extraction of pure EVs for research and clinical purposes. This prerequisite is particularly essential for the analysis of active and tumor-specific proteins and phosphoproteins.
We have recently introduced a novel magnetic bead approach based on chemical affinity for urinary EV capture and purification.41 The technology, named EVtrap (extracellular vesicle total recovery and purification), enables the complete capture of EVs onto beads modified with a combination of hydrophilic and aromatic lipophilic groups that have high affinity toward lipid-coated EVs. These modified amphiphilic beads are capable of capturing EVs with high purity, and we found that the method enables a >95% recovery yield of all EVs present in the urine sample, a significant improvement compared with ultracentrifugation and commercial exosome isolation methods. We applied LC–MS analyses on proteins from EVs isolated by EVtrap, and all known detectable exosome markers were significantly enriched after EVtrap compared with UC. We also identified the largest number of phosphoproteins present in urinary EVs (>1000 phosphoproteins in 10 mL of urine).
Whereas the EVtrap approach showed a dramatic improvement in urinary EV analysis, blood is a much more complex milieu with many more challenges in sample preparation and analysis. The scale of free proteins and other molecules present in plasma or serum that affect EV analysis is orders of magnitude higher than that in urine. Nonetheless, blood contacts most cells and organs and is considered to be the most important biofluid for liquid biopsy. A majority of EVs from different types of cells are likely secreted into the blood. Therefore, to enable a blood-based liquid biopsy for disease diagnosis, in this study, we attempted to achieve EV isolation from plasma based on the EVtrap approach, with specific focus on EV phosphoproteomics. We carried out a comparison among EVtrap, ultracentrifugation, and other commonly used exosome isolation methods. Using western blotting, silver staining, and LC–MS analyses, we found that EVtrap captured EV markers from plasma at significantly higher levels and improved EV capture purity compared with UC and other standard EV isolation techniques.
We further utilized the EVtrap approach for EV phosphoproteome analysis from plasma. Protein phosphorylation is a key control mechanism for cellular regulatory pathways and one often targeted by drug developers to create inhibitors that block signaling pathways involved in cancer and other diseases. However, as far as liquid biopsy is concerned, until our recent study, phosphoproteins have been virtually ignored due to their perceived instability and low abundance level in biofluids.42 Phosphoproteins are typically present at substoichiometric levels, and thus the recovery rate during the sample preparation steps must be efficient to achieve their detection. To carry out the EV plasma phosphoproteome analysis, we utilized the EVtrap approach to capture EVs from 1 mL of a plasma sample and compared this method with standard ultracentrifugation. We demonstrated an efficient analysis of the plasma phosphoproteome with the EVtrap isolation method and its feasible application for disease diagnosis.
Finally, we utilized our EVtrap–LC–MS approach to examine EV proteome and phosphoproteome differences between healthy plasma controls and plasma samples from patients with chronic kidney disease (CKD) and kidney cancer. These efforts resulted in the discovery of several highly promising potential biomarkers for the noninvasive detection of renal cell carcinoma (RCC) from plasma, which we will further validate in upcoming studies.
EXPERIMENTAL SECTION
Experimental details of materials, ultracentrifugation, EV isolation by other methods, western blotting, silver staining, LC–MS analysis, and data processing are included in the Supporting Information.
Sample Collection
Plasma samples were collected under approval from the Purdue University Human Research Protection Program and the Indiana University Human Subjects Office Institutional Review Boards, and all patients properly consented before samples were collected. All frozen samples were thawed and centrifuged at 2500g for 10 min to remove platelets, apoptotic bodies, and other large particles and aggregates.
Extracellular Vesicle Isolation by EVtrap
For EVtrap characterization experiments, we used plasma from healthy individuals. For the kidney cancer analysis part of the project, we used 1 mL each of plasma from (a) five healthy individuals (no known kidney-related disease), (b) five patients with diagnosed CKD, some of whom were also diagnosed with kidney stones, and (c) five patients diagnosed with the most common form of kidney cancer, RCC. EVtrap beads were provided by Tymora Analytical as a suspension in water and were used as previously described in more detail.41 The plasma samples were diluted 20 times in the diluent buffer, the EVtrap beads were added to the samples in a 1:2 v/ v ratio, and the samples incubated by end-over-end rotation for 30 min according to the manufacturer’s instructions. After supernatant removal using a magnetic separator rack, the beads were washed with PBS, and the EVs were eluted by a 10 min incubation with 200 mM triethylamine (TEA, Millipore-Sigma). The samples were fully dried in a vacuum centrifuge.
LC–MS Sample Preparation
The isolated and dried EV samples were lysed to extract proteins using the phase-transfer-surfactant (PTS)-aided procedure.43 First, EVs were solubilized in the lysis solution containing 12 mM sodium deoxycholate, 12 mM sodium lauroyl sarcosinate, 10 mM TCEP, 40 mM CAA, and phosphatase inhibitor cocktail (Millipore-Sigma) in 50 mM Tris HCl, pH 8.5 by incubating for 10 min at 95 °C. This step also denatured, reduced, and alkylated the proteins. The samples were diluted five-fold with 50 mM triethylammonium bicarbonate and digested with Lys-C (Wako) in a 1:100 (w/w) enzyme-to-protein ratio for 3 h at 37 °C. Trypsin was added to a final 1:50 (w/w) enzyme-to-protein ratio for overnight digestion at 37 °C. The samples were acidified with trifluoroacetic acid (TFA) to a final concentration of 1% TFA. Ethyl acetate solution was added in a 1:1 ratio to the samples. The mixture was vortexed for 2 min and then centrifuged at 20 000g for 2 min to obtain aqueous and organic phases. The organic phase (top layer) was removed, and the aqueous phase was collected, dried down to <10% original volume in a vacuum centrifuge, and desalted using Top-Tip C18 tips (Glygen) according to the manufacturer’s instructions. Each sample was split into 99 and 1% aliquots for phosphoproteomic and proteomic experiments, respectively. The samples were completely dried in a vacuum centrifuge and stored at −80 °C. For the phosphoproteome analysis, the 99% portion of each sample was subjected to phosphopeptide enrichment using a PolyMAC phosphopeptide enrichment kit (Tymora Analytical) according to the manufacturer’s instructions, and the eluted phosphopeptides were dried completely in a vacuum centrifuge. For the phosphoproteomics analysis, the whole enriched sample was used, whereas for proteomics, only 50% of the sample was loaded onto LC–MS.
Data Availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE44 partner repository with the data set identifier PXD017994.
RESULTS
Isolation of Plasma EVs by EVtrap
Here we adapted the EVtrap approach for the purification of EVs from plasma samples. The collected and frozen plasma samples were thawed, and platelets and other large particles were removed by centrifugation at 2500g for 10 min. The precleared plasma samples were diluted 20-fold in PBS and incubated with EVtrap beads for 30 min.41 The captured EVs were eluted with 10 min of incubation in 200 mM TEA, and the resulting EV samples were dried in a vacuum centrifuge. The transmission electron microscopy (TEM) analysis showed that the standard cup-shaped exosomes/microvesicles recovered (Figure 1A,B). Examinations of the post-EVtrap eluted EVs by nanoparticle tracking analysis (NTA) and tunable resistive pulse sensing (TRPS) both demonstrated similar ranges of the isolated EVs, with the majority being in the 100–200 nm range (Figure 1C,D). The mode particle diameters were 137 and 134 nm, whereas the d50 values were 154 and 143 nm for NTA and TRPS, respectively. Whereas both methods showed pretty similar data, we found that TRPS produced more consistent and accurate results due to its nanopore-based single-particle analysis capabilities.
Figure 1.
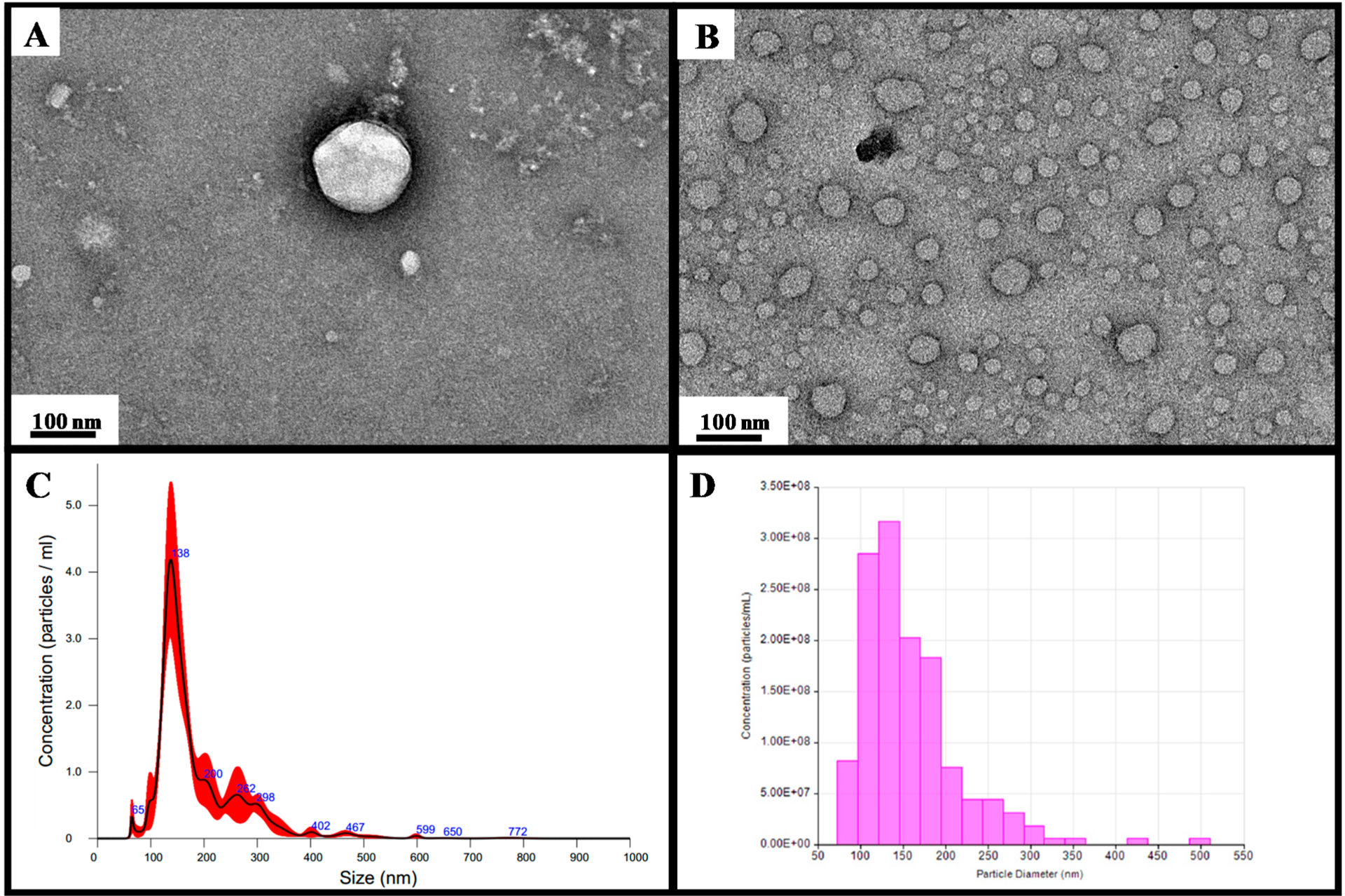
Transmission electron microscopy (TEM) images of (A) single EV and (B) multiple EVs captured from the plasma by EVtrap. TEM imaging of EVs was carried out on a Hitachi H-8100 electron microscope (Hitachi, Tokyo, Japan) with an accelerating applied potential of 200 kV. (C) Nanoparticle analysis using NTA after elution off EVtrap beads. (D) Nanoparticle analysis using TRPS after the elution of EVtrap beads.
For the initial recovery yield comparison and method validation, we used a 100 K × g ultracentrifugation “gold standard” as the control method (with and without PBS wash). An equivalent of 5 μL of plasma of the EV pellet from this ultracentrifugation (labeled as 100 K UC) was loaded on the gel for western blot analysis. The supernatant after 100 K UC was also further incubated with EVtrap beads to check if any residual exosome population was left in the supernatant after UC. Likewise, 5 μL of direct plasma was used to capture EVs by EVtrap. After 30 min of incubation, the beads were washed, and EVs were eluted and dried. Then, the internal cargo was extracted by boiling in the LDS (lithium dodecyl sulfate) sample buffer and loaded on the gel. All samples were loaded on the same gel and detected by western blotting using a primary antibody against CD9 (a common exosome marker). The experiment was carried out three separate times for quantitation, with each isolation using a plasma from the same source. (The complete blots for all three experiments are available in Supplementary Figure S1.) A representative western blot is shown in Figure 2A, and the quantitative values for each CD9 band signal are listed in the bar graph in Figure 2B. For comparison, we also loaded 0.05 μL of plasma (~4 μg protein; 1% of the amount used by other methods), which, as expected, was too low to produce any detectable CD9 signal. As the results show, the ultracentrifugation step indeed captured only a portion of the exosomes (some were likely lost during the 10 000g pretreatment), with EVtrap being able to isolate and produce greater than seven times more CD9 signal. If the EVtrap method is considered to capture the majority of exosomes, then it can be calculated than UC captures ~14% of EVs on average, a recovery rate similar to that of other studies27,28 and equivalent to what we found for urine EVs. The detection of EVs by EVtrap from the UC supernatant further confirmed the incomplete isolation by UC, showing a large percentage of EVs remaining in the supernatant (Figure 2). We note that the high-yield capture by EVtrap was achieved after only a 45 min procedure, in comparison with the >5 h needed for the UC protocol.
Figure 2.
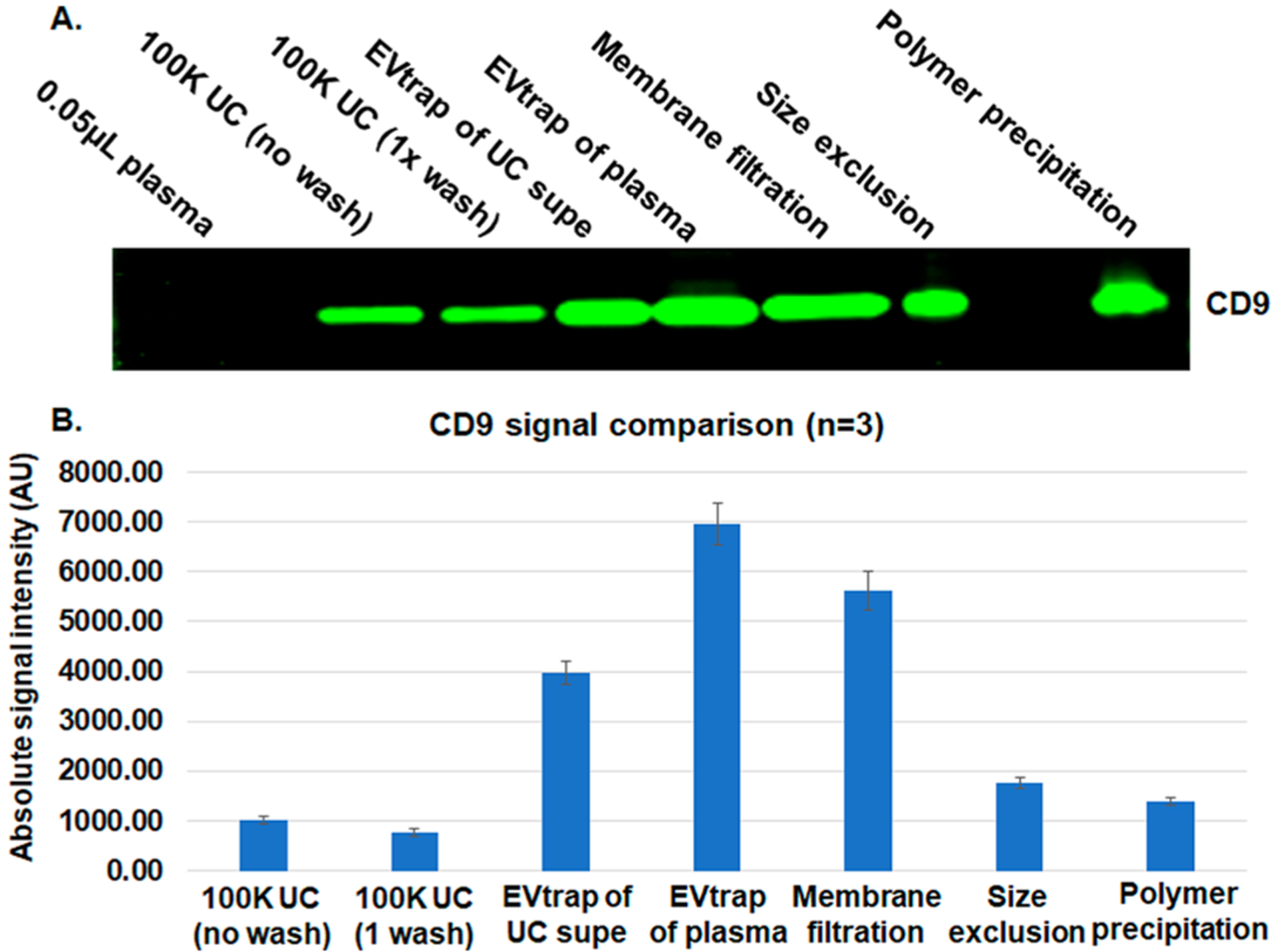
Comparison among ultracentrifugation (UC), EVtrap, and three commercial methods for exosome capture. (A) Detection of the CD9 exosome marker using western blot. The lanes were loaded as follows: 0.05 μL of plasma loaded directly, EVs isolated from plasma by UC with no additional wash steps, EVs isolated from plasma by UC with one wash step, EVs captured by EVtrap from the UC supernatant sample (leftover after UC), EVs directly captured by EVtrap from plasma, EVs captured from plasma by the membrane filtration method, EVs captured from plasma by the size exclusion method, and EVs captured from plasma by the polymer precipitation method. All loaded EV samples were from 5 μL of plasma. (B) Quantitation of WB data in panel A (n = 3).
Besides ultracentrifugation, we also sought to compare the EVtrap method with other common EV isolation approaches. We tested three common EV isolation kits: one based on the membrane affinity spin method,34 a size-based filtration tube, and a polymer-based exosome precipitation kit.30 100 μL of plasma was used in each case, and 5 μL of equivalent was loaded on the same gel as the previous samples. As the results in Figure 2 demonstrate, the alternative methods produced a somewhat similar or better exosome recovery signal compared with 100 K ultracentrifugation, matching the previously published results for these methods.27,29,34,37,38 However, the low purity of these methods is a known disadvantage (as will also be further shown in this study). Therefore, it is difficult to implement them for subsequent protein analysis due to significant contamination from free plasma proteins. Nonetheless, the EVtrap still produced the highest exosome recovery yield compared with any other approach (Figure 2A,B). All CD9 band signals from the three replicate blots were quantified, and the mean signal intensities are plotted on the bar graph in Figure 2B to show a better comparison between the methods. The standard deviation error bars among EVtrap experiments are higher than we expected, but this is likely because the three replicates were carried out on separate gels and the western blotting analyses were also performed on separate membranes.
Our overall goal was to show how much of the EV amount can be obtained from the same volume of plasma. In many cases, the volume of clinical samples is very limited; therefore, the ability to enrich a high amount of EVs from a small volume would be very advantageous. This is the reason we compared the EV recovery in each method using the same starting volume. Nonetheless, it is common to assess the EV marker signal in relation to the total protein amount. We carried out an additional experiment of CD9 western blot comparing the signal after EVtrap enrichment and 100 K ultracentrifugation. Here the recovered EVs were resuspended and lysed, and the concentration of each sample was determined by Nanodrop. Then, 5 μg of each protein amount was loaded on a gel and analyzed with anti-CD9 antibody. As the data in Supplementary Figure S2 demonstrate, the EVtrap again demonstrated a significant increase in the CD9 signal when the loading sample amount was normalized by concentration.
Assessment of Plasma EV Purity
Besides EV capture yield analysis, another important feature is the purity of the isolated EVs. This is particularly important for plasma samples because the free proteins in plasma are present at 70–80 mg/mL, levels that can significantly impede biomarker analysis. To examine the amount of contamination by free plasma proteins present after each EV capture method, we used 5 μL of plasma for each experimental treatment and detected the resulting EV sample with silver staining for total protein analysis. The samples were processed in a manner identical to those in the previous EV recovery experiments and loaded on the gel in the same sequence. As expected, the 100 K UC sample had a lower level of contamination when compared with the loading of 0.05 μL (1%) of direct plasma and much less contamination after the PBS wash (Figure 3A). The three commercial kits tested (the same as those used in the first experiment) resulted in a very high amount of contamination and therefore a low purity of EV isolation. When compared with the 1% plasma sample loaded, all three commercial methods had >1% free protein contaminants remaining. As a result, these methods would be very difficult to use for downstream proteome and phosphoproteome analyses. Perhaps this is the primary reason why most researchers use them for DNA/RNA detection only. By comparison, EVtrap isolation showed the cleanest sample compared with all three commercial products and a similar contamination level compared to ultracentrifugation.
Figure 3.
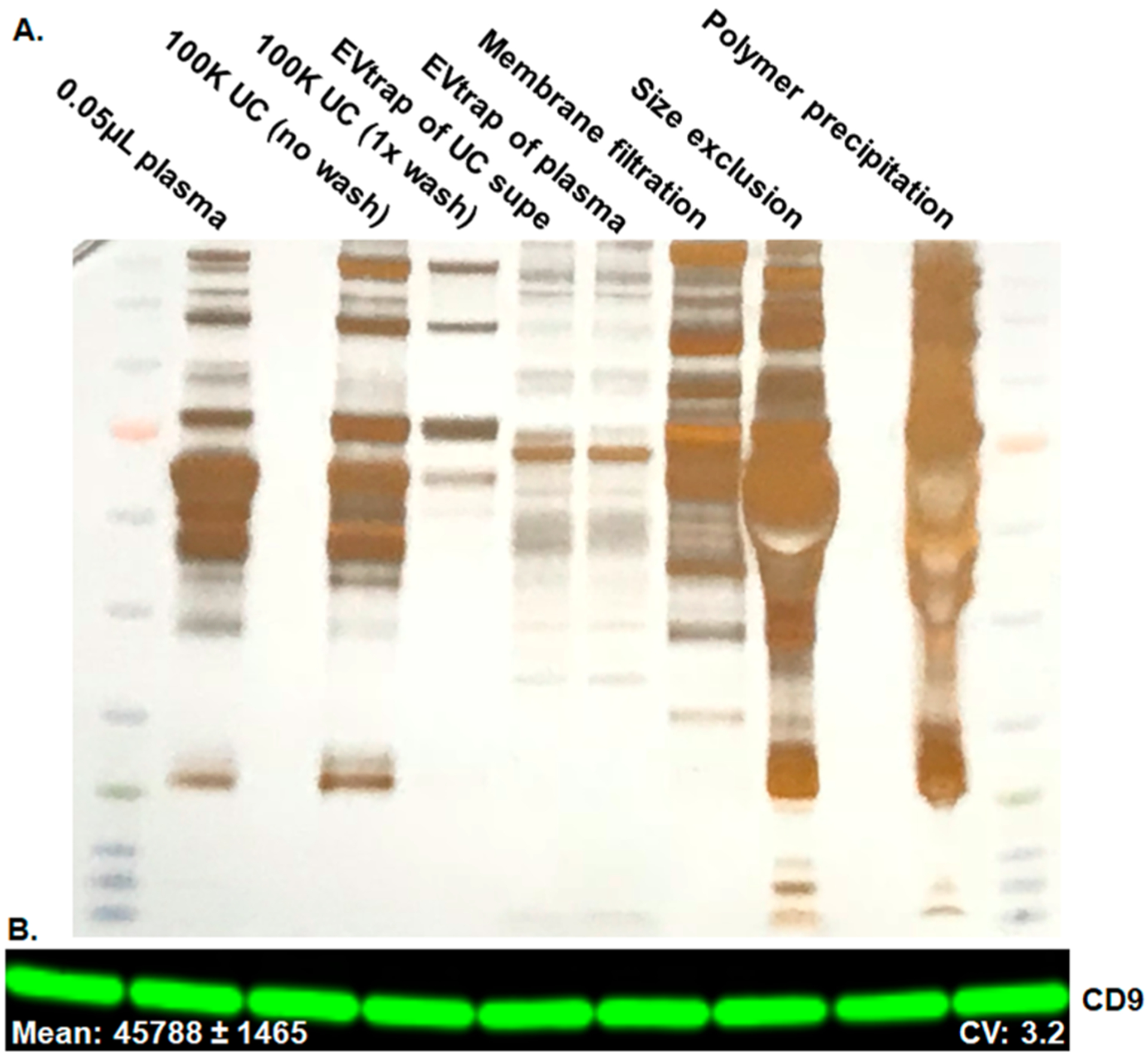
(A) Silver stain total protein detection to access the plasma protein contamination. Comparison among ultracentrifugation (UC), EVtrap, and three commercial methods for exosome capture. The lanes were loaded as follows: 0.05 μL of plasma loaded directly, EVs isolated from plasma by UC with no additional wash steps, EVs isolated from plasma by UC with one wash step, EVs captured by EVtrap from the UC supernatant sample (leftover after UC), EVs directly captured by EVtrap from plasma, EVs captured from plasma by the membrane filtration method, EVs captured from plasma by the size exclusion method, and EVs captured from plasma by the polymer precipitation method. All loaded EV samples were from 5 μL of plasma. (B) Test of EVtrap procedure reproducibility carried out using nine separate plasma samples from the same source detection of CD9 signal.
Examination of EVtrap Reproducibility
To analyze clinical samples in a high-throughput manner, a method must be able to demonstrate low experimental variability. Thus we also tested the EVtrap sample-to-sample reproducibility. The EVtrap reproducibility experiments were carried out using nine 10 μL aliquots of plasma, and the captured EV samples were then analyzed on the same blot using CD9 marker detection. The results shown in Figure 3B demonstrate outstanding sample-to-sample reproducibility with a 3.2% coefficient of variation (CV). The expanded blot and the quantitative CD9 band values are shown in Supplementary Figure S3.
LC–MS Analysis of Plasma EV Proteome and Phosphoproteome
For our preliminary phosphoproteome analyses, we used 1 mL of plasma for EVtrap capture and for ultracentrifugation as the control (100 K UC; carried out after the 10 000g centrifugation step). Our 100 K ultracentrifugation method (total UC time ~5 h, including one wash step) produced 321 unique phosphopeptides from 177 unique phosphoproteins (Supplementary Table S1). However, when EVtrap was used for the 30 min capture, we saw a significant increase in phosphoproteome identification levels. We identified 5570 unique phosphopeptides from 1593 unique phosphoproteins using only 1 mL of plasma and a single 60 min LC–MS run (Supplementary Table S2). This confirms that most phosphoproteins were simply not detectable by MS after ultracentrifugation.
To complement our phosphoproteome data, we also carried out a direct proteomics analysis on plasma EVs. For these samples, only a 5 μL equivalent of plasma was used. The 100 K UC procedure produced 3282 peptides and 406 total proteins (Supplementary Table S3). The EVtrap capture approach, on the contrary, allowed the identification of 16 266 peptides from 2238 unique proteins (Supplementary Table S4).
We further carried out the label-free quantitation of EV proteins isolated by EVtrap and by UC. Figure 4 shows the total signal increase in the identified proteins and phosphoproteins compared with UC (Supplementary Tables S5 and S6). In our experiment, we have identified 88 out of 100 common EV markers and proteins published in ExoCarta45–47 (marked EV protein data in Supplementary Table S7). Overall, the average signal increase in EV markers and the total EV proteome level is 78- and 69-fold, respectively, for EVtrap compared with UC. This is noteworthy because many other studies have shown that different methods enrich different exosome populations with various success rates.38,48 With EVtrap, it appears that the complete EV profile is recovered. Markedly, the overall increase in the phosphoproteome signal is ~85-fold. The significant increase in the phosphoproteome signal is due to most of the phosphopeptides being undetectable by LC–MS after ultracentrifugation, thus resulting in a relative abundance ratio of 100 (maximum set fold-change in Proteome Discoverer). This difference in intensities is substantial. Whereas the internal RNA/DNA molecules and many proteins can still be detected even after low EV recovery, phosphoproteins are already present at very low concentrations. After poor sample preparation, they would be essentially undetectable on a discovery instrument like a mass spectrometer. These data confirmed our hypothesis that to achieve the efficient identification and quantification of phosphoproteome in EVs, a method like EVtrap is necessary to enable simple and highly efficient EV isolation and recovery.
Figure 4.
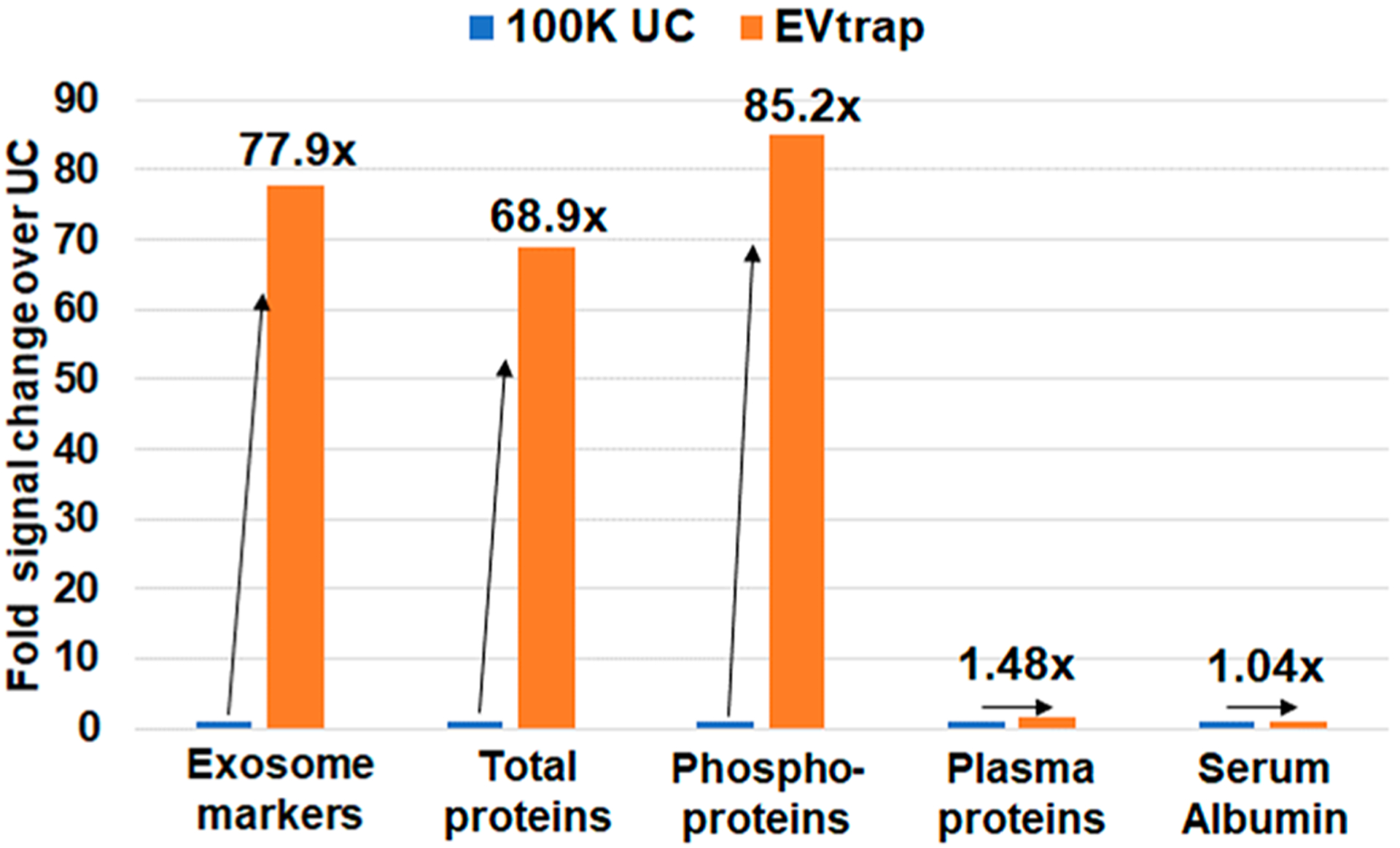
LC–MS proteome and phosphoproteome analyses of 100 K UC and EVtrap samples. The fold change in overall signal between 100 K UC and EVtrap experiments was quantified and plotted in a bar graph for: 88 identified common exosome markers, all proteins identified (excluding known contaminants), all phosphoproteins identified, serum albumin, and other high-abundant free plasma proteins (usually treated as contaminants).
Finally, for purity comparison purposes, we also quantified the signal of serum albumin and other common highly abundant plasma proteins. The quantitative LC–MS data showed a similar intensity (ratio of 1.05) of serum albumin in the EV sample isolated by EVtrap compared with UC (Figure 4). When analyzing other highly abundant plasma proteins, the average increase in signal in the EVtrap sample was 1.48-fold (marked plasma protein data in Supplementary Table S7). Whereas there is a small increase in the contaminants level of ~1.48-fold after EVtrap capture, this increase is not significant compared with the 78-fold increase in EV markers and 69-fold increase in total EV protein amount. Therefore, it is apparent from these results that EVtrap indeed significantly improves protein and phosphoprotein detection from plasma EVs without significantly increasing the level of contamination compared with the standard isolation methods like UC.
The data demonstrate that EVtrap can be applied to directly process plasma in only 30 min, which would be highly useful for routine clinical analysis. With 1 mL of plasma being sufficient to identify over 1000 phosphoproteins and 5 μL of plasma being enough to identify over 2000 unique proteins, the volume is also convenient for routine analysis.
EVtrap-based Plasma EV Phosphoproteome Analysis of Kidney Cancer and CKD Patient Samples
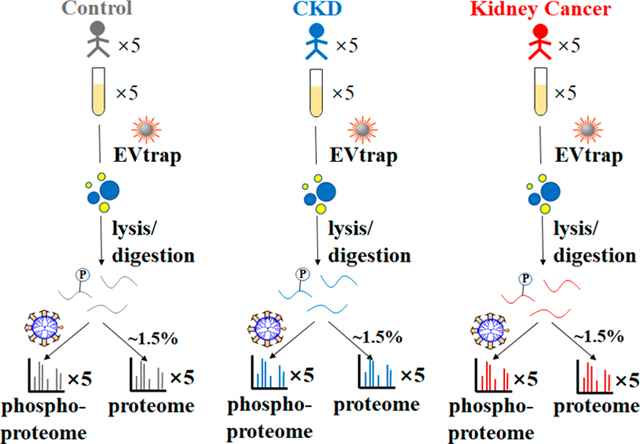
Protein phosphorylation is a determining regulatory mechanism for pathological pathways directly linked to cancer and other diseases. With one previous study from our group unveiling the possibility of using plasma EV phosphoproteins as candidate biomarkers for breast cancer,42 we believe our novel benchmarked EVtrap technique allows a more efficient and robust way for phosphoprotein biomarker discovery using plasma EVs. Here we focused on diseases related to the kidney, including kidney cancer (specifically, RCC), which accounts for ~3% of human malignancies.49 The current most common methods of diagnosis include a computerized tomography (CT) or computerized axial tomography (CAT) scan followed by an invasive tumor biopsy, which is far from optimal.50,51 In this study, we carried out preliminary efforts to detect potential biomarkers of kidney cancer using plasma EV samples to enable a noninvasive diagnosis. The general workflow for the whole process of plasma EV proteome and phosphoproteome analysis is illustrated in Figure 5 to accurately distinguish kidney cancer from noncancerous conditions; CKD patients’ plasma samples were included as an additional control in our study.52
Figure 5.
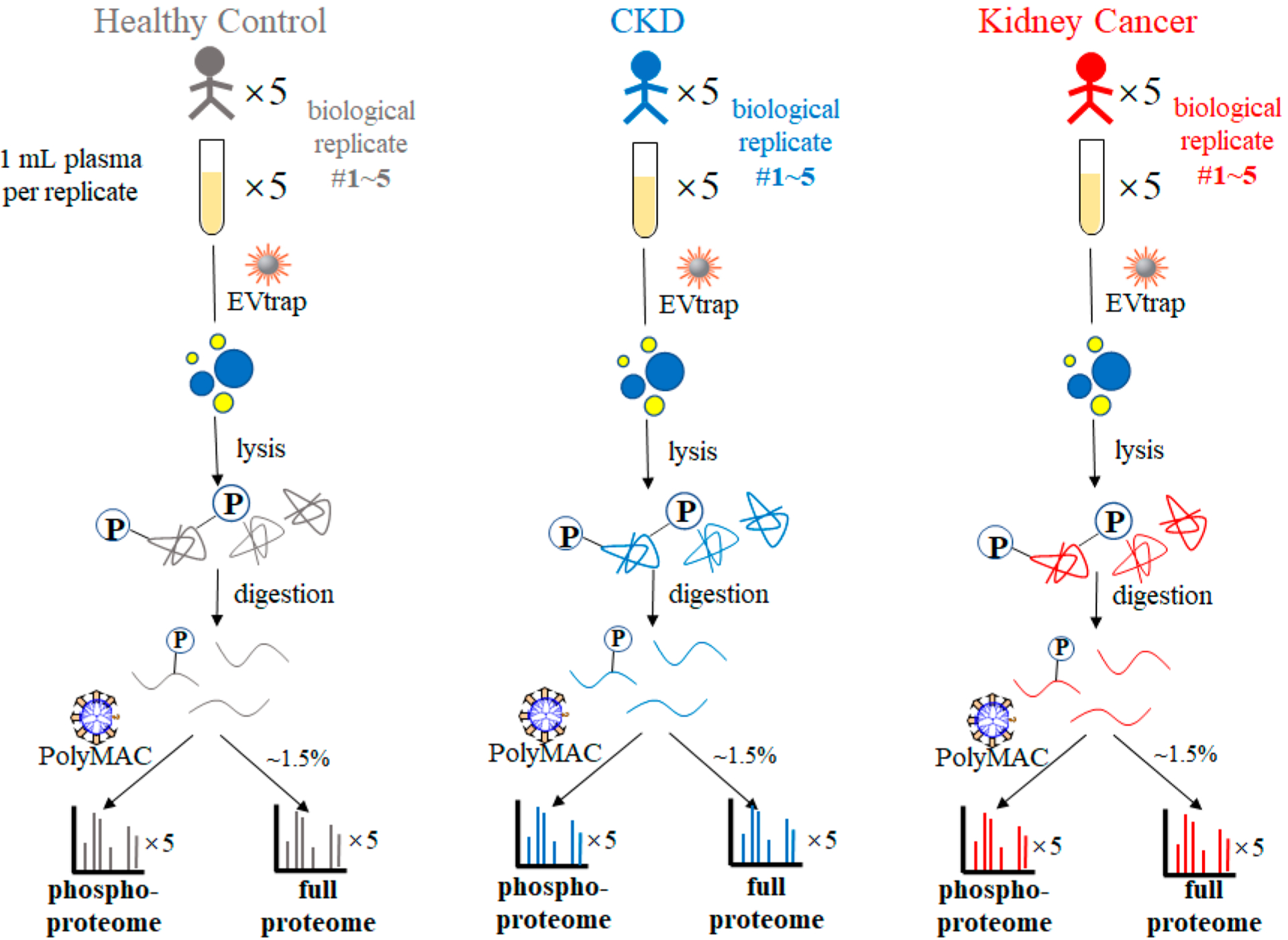
Workflow of EV proteomics and phosphoproteomics analyses of plasma samples from healthy controls and patients diagnosed with CKD and kidney cancer.
EVs were isolated from human plasma samples (n = 5 biological replicates/group × 3 groups = 15), as described in Figure 5 using our EVtrap approach. EV proteins were extracted and digested by trypsin with the aid of PTSs.43 After detergent removal and desalting, 1.5% (~1 μg) of each peptide sample was stored for direct proteome analysis. The remainder of each peptide sample was used for phosphopeptide enrichment with an in-house-developed PolyMAC dendrimer-based phosphopeptide enrichment method53 prior to LC–MS analysis. Both proteome and phosphoproteome fractions were analyzed on an Ultimate 3000 nanoLC apparatus coupled to a Q Exactive HF-X mass spectrometer.54 Additionally, an indexed Retention Time Standard containing 11 artificial synthetic peptides was spiked into each sample and utilized as an internal standard to reduce run-to-run variations and assist with peptide quantitation.55
This streamlined procedure resulted in the identification of 146 significantly changing phosphoproteins and 28 significantly changing proteins in kidney cancer samples compared with the control. Likewise, we identified 156 significantly changing phosphoproteins and 16 significantly changing proteins in CKD samples compared with the control. The comparison between the RCC and CKD samples revealed 44 phosphoproteins and 10 proteins that are significantly different between these two groups. Overall quantitative proteomics and phosphoproteomics data are available in Supplementary Tables S8 and S9. Furthermore, an in-depth data analysis was employed to obtain statistical results and generate visualized hierarchical clustering groups (heatmaps) and volcano plots (Figure 6). The proteins and phosphoproteins included in the two heatmaps (Figure 6A,B) are listed in Supplementary Table S10.
Figure 6.
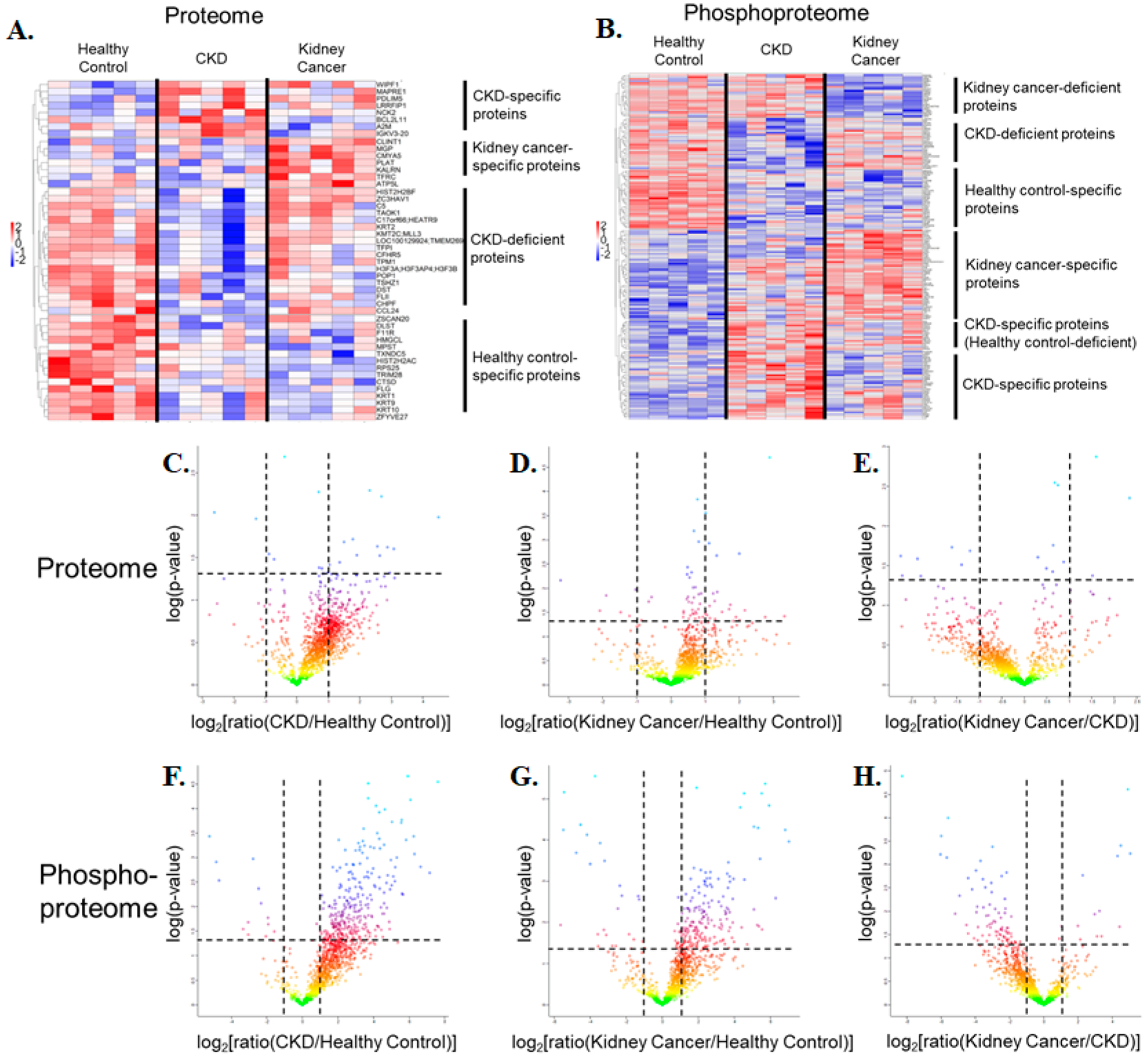
Hierarchical clustering analyses on quantitative (A) proteomics and (B) phosphoproteomics data. The gene names included in the two heatmaps (A) and (B) are listed in Supplementary Table S10. (C–H) Volcano plots representing the quantitative comparison of the plasma EV proteomes (top) and phosphoproteomes (bottom). (C) CKD versus healthy control in the full proteome. (D) Kidney cancer versus healthy control in the full proteome. (E) Kidney cancer versus CKD in the full proteome. (F) CKD versus healthy control in the phosphoproteome. (G) Kidney cancer versus healthy control in the phosphoproteome. (H) Kidney cancer versus CKD in the phosphoproteome.
Hierarchical clustering analyses on quantitative proteomics and phosphoproteomics were performed on all individual biological replicates with the threshold of the p value at 0.05. Clusters of proteins or phosphoproteins with consistent significantly changing abundance levels among categories were highlighted and annotated on the right. For a more global visualization of the quantitation results, the volcano plots with a basis of t-test statistics featured the quantitative comparison analyses of the plasma EV proteomes (top) and phosphoproteomes (bottom) between every two sample categories out of three. Proteins and phosphoproteins with considerable up- and down-regulation in diseases were exposed through a t-test permutation-based false discovery rate (FDR) (FDR = 0.05, the horizontal dashed line at −log(p value) =1.30) and a difference in fold-change (fold-change = 2, the vertical dashed line at log2(ratio) = ±1), with all five biological replicates in each group being counted. Notably, a greater number of phosphoproteins was up-regulated in disease states compared with control samples, a promising outcome for further clinical validation.
We focused on several significantly changing proteins and phosphoproteins and identified the four most promising biomarker representatives, with their relative abundances in three categories from DDA-mode quantitation sketched in linear box-and-whiskers plots (Figure 7). Indeed, prior studies substantiated the scientific rationale for these targets as possible disease biomarkers. The anchoring function of cardiomyopathy-associated protein 5 (CMYA5) is corroborated to mediate the subcellular compartmentation of protein kinase A (PKA) by binding to PRKAR2A implicated in the STRING network analysis,56,57 although no straightforward evidence currently exists underlining the cancer-correlated mechanism behind the abundance elevation (Figure 7A). Likewise, in reference to the KEGG database, the enhanced signal of the Crk-like protein (CRKL) in the phosphorylated form was supported by its critical role in the PI3K-Akt signaling pathway in cancer formation (Figure 7B).58,59 With regard to phosphoprotein LYRIC (MTDH) as a potential marker, it appears promising due to its interaction with AKT1 and HRAS, both of which serve as key components in the kidney carcinoma pathway. Moreover, the networks between MTDH and MMP9, MYC, and ZBTB16 are also essential players in cancer progression (Figure 7C).59 Beyond cancer-relevant roles, a previous finding of apolipoprotein A-IV (APOA4) as a concentration-increased protein in chronic renal disease underlines its feasibility as a kidney stone/inflammation-specific protein, which was mainly attributed to kidney metabolism (Figure 7D).60 Additional linear box-and-whiskers plots for other potential candidates are shown in Supplementary Figures S4–S6.
Figure 7.
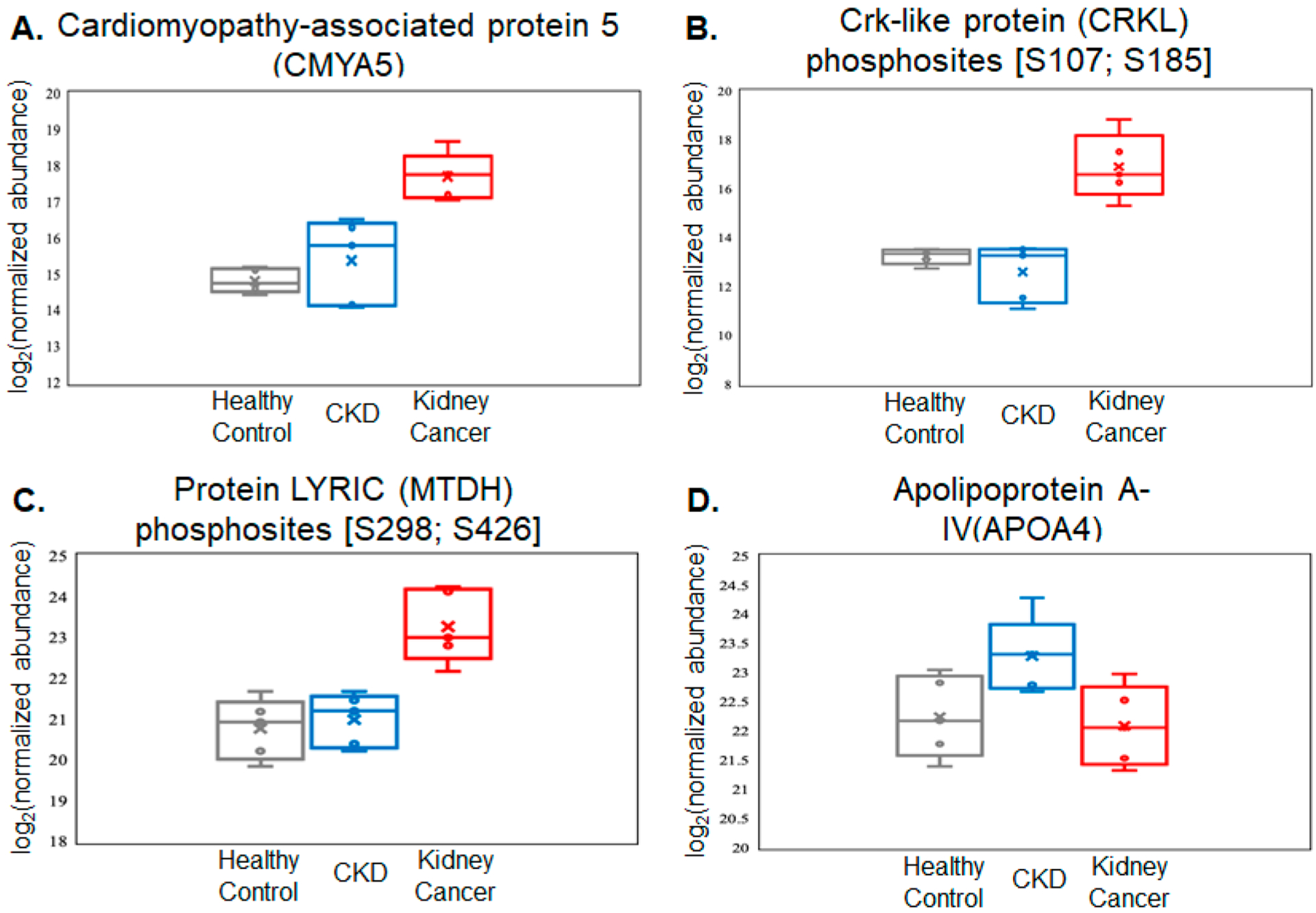
Relative abundance data of the selected four proteins and phosphoproteins as potential markers to differentiate the relevant categories. All four targets demonstrate significant up-regulation (P < 0.05) in patients diagnosed with kidney cancer or CKD compared with the other two categories. (A) Kidney cancer-specific protein marker. (B,C) Kidney-cancer-specific phosphoprotein markers. (D) CKD-specific protein marker. All values are log2 conversions of protein- or phosphosite-normalized abundance levels, as determined by LC–MS.
DISCUSSION
Along with emerging research in EVs and exosomes, total EVs and exosome capture and purification have been the focus of many recent studies that attempt to develop a simple and efficient EV isolation protocol. Here we present a novel chemical-affinity-based capture method for effective EV isolation from plasma. The EVtrap method enables the capture of the complete EV profile based on the lipid bilayer structure of these vesicles and the unique combination of the hydrophilic and aromatic lipophilic groups on the synthesized beads. The binding of these combinatorial amphiphilic groups appears to be, in part, through the shift from hydrophilicity to amphiphilicity of these groups, electrostatic interactions, and lipid affinity, and thus the EVs can be released with the increase in pH using TEA. The specificity of the binding to the lipid bilayer membrane of EVs as opposed to free lipids and lipoproteins, as well as their aggregates, is further enhanced by the additives in the diluent buffer.
We were able to directly isolate hundreds of phosphoproteins from plasma and revealed its future applications in clinical settings. Whereas our previous publication on plasma EV phosphoproteome analysis produced strong data and demonstrated outstanding potential in this field,42 the EVtrap approach would be more suitable for routine EV analysis and biomarker discovery from clinically relevant samples. The initial demonstration of this was implemented in this study of kidney cancer plasma EV phosphoproteomics. We strongly believe that the future of disease detection will depend on the robust and reproducible analysis of low-abundant signaling proteins and phosphoproteins, especially for early cancer diagnosis and monitoring. This will supplement the current assays and offer an additional layer of important information not typically available from genetic tests.
Multiple studies have shown that there is a notable discordance between genomic information and the actual proteome/phosphoproteome profile, with phosphoprotein information being much more relevant for cancer detection and molecular subtyping.10–12,61,62 A recent study published on breast cancer profiling indicated the utility of phosphoproteomic data to help clarify the highly complex genomic features.63 For example, genomic data have not been successful in treatment decisions with MTOR and PI3K inhibitors due to a myriad of mutations in multiple pathways, most of which are not actionable.64 Here the researchers were able to more confidently predict the drug response based on the phosphorylation of downstream signaling targets. In another example, close to half of HER2-positive breast cancer patients do not respond well to Herceptin.65 Likewise, a significant number of HER2-negative women do respond to Herceptin.66 In both cases, a downstream phosphorylation analysis can predict these response differences. These examples demonstrate the need to examine the more complete signaling pathways for better cancer subtyping in addition to the presence of mutations or a receptor. Being able to do this in a less invasive manner using plasma EV analysis would have an enormous public health benefit. Therefore, the successful development of methods like EVtrap will be a substantial step forward in this objective.
CONCLUSIONS
We have carried out a comprehensive assessment and comparison of the EVtrap method against several EV isolation methods for plasma proteome and phosphoproteome studies. The EVtrap method enables high recovery levels of exosomes with a low contamination level and <5% CV. All detectable exosome markers were captured at higher levels than by the common UC approach. Whereas most data in this study were generated using western blotting and LC–MS, the EVtrap-isolated vesicles can be used for different types of follow-up analyses. We expect that the captured EV cargo can also be processed for DNA/RNA examination. Because the proposed approach is magnetic-bead-based, it can be easily automated for a high-throughput screening or diagnostics assay in a hands-off manner. Researchers equipped with EVtrap will be able to uncover more low-abundant plasma biomarkers, such as those with post-translational modifications (PTMs). We hypothesize that low-abundant plasma EV proteins and phosphoproteins will one day be used for early-stage disease detection, for longitudinal monitoring, and as companion diagnostics for targeted cancer treatments, in particular, those involving kinase inhibitors.
Supplementary Material
ACKNOWLEDGMENTS
We thank the IU Health ECRO Biorepository for help in obtaining specimens used in this study. All samples were collected under IRB approved protocol no. 1011004282, Development of a Biorepository for IU Health Enterprise Clinical Research Operations. We also thank Thermo Fisher Scientific for providing our team with the Ultimate 3000 LC coupled to a Q-Exactive HF-X high-resolution MS instrument, which enabled this research. Finally, we thank Dr. Anoop Pal and Izon Science for performing the TRPS analysis of the samples. This project has been funded in part by NIH grants R41CA210772, R44CA239845, and R43AG063589 (to A.I. and W.A.T.) and by the Walther Oncology Physical Sciences & Engineering Research Embedding Program (to W.A.T. and R.S.B.).
Footnotes
The authors declare the following competing financial interest(s): A.I. and W.A.T. are principals at Tymora Analytical Operations, which developed the EVtrap beads and commercialized PolyMAC enrichment kit.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00151.
Supporting Experimental Section. Figure S1. Triplicate western blot results of comparison between UC, EVtrap, and three commercial methods for exosome capture as in Figure 2A. Figure S2. Concentration-based comparison between UC and EVtrap. Figure S3. Expanded Figure 3B. Figure S4. Linear box-and-whiskers plots for select up-regulated kidney cancer-specific phosphoproteins. Figure S5. Linear box-and-whiskers plots for select up-regulated CKD-specific phosphoproteins. Figure S6. Linear box-and-whiskers plots for select up-regulated CKD-specific proteins (PDF)
Table S1. LC–MS phosphoproteome analysis data for the 100 K UC sample. Table S2. LC–MS phosphoproteome analysis data for the EVtrap sample. Table S3. LC–MS proteome analysis data for the 100 K UC sample. Table S4. LC–MS proteome analysis data for the EVtrap sample. Table S5. Quantitative proteomics results of LC–MS analyses for the 100 K UC and EVtrap samples. Table S6. Quantitative phosphoproteomics results of LC–MS analyses for the 100 K UC and EVtrap samples. Table S7. Marked EV proteins and abundant plasma proteins from quantitative proteomics results of LC–MS analyses for the 100 K UC and EVtrap samples (proteins only). Table S8. Quantitative phosphoproteomics results of LC–MS analyses for the normal control, chronic kidney disease, and kidney cancer urine EV samples. Table S9. Quantitative proteomics results of LC–MS analyses for normal control chronic kidney disease and kidney cancer urine EV samples. Supplementary Table S10. Lists of proteins and phosphoproteins included in the two heatmaps in Figure 6A,B (ZIP)
Contributor Information
Anton Iliuk, Department of Biochemistry, Purdue University, West Lafayette, Indiana 47907, United States;; Tymora Analytical Operations, West Lafayette, Indiana 47906, United States;
Xiaofeng Wu, Department of Chemistry, Purdue University, West Lafayette, Indiana 47907, United States.
Li Li, Tymora Analytical Operations, West Lafayette, Indiana 47906, United States.
Jie Sun, College of Biological Science and Medical Engineering, Southeast University, Nanjing 210096, China.
Marco Hadisurya, Department of Biochemistry, Purdue University, West Lafayette, Indiana 47907, United States.
Ronald S. Boris, Department of Urology, Indiana University School of Medicine, Indianapolis, Indiana 46202, United States
W. Andy Tao, Department of Biochemistry, Department of Chemistry, and Purdue Center for Cancer Research, Purdue University, West Lafayette, Indiana 47907, United States;; Tymora Analytical Operations, West Lafayette, Indiana 47906, United States; College of Biological Science and Medical Engineering, Southeast University, Nanjing 210096, China
REFERENCES
- (1).Fernandez-Lazaro D; Garcia Hernandez JL; Garcia AC; Cordova Martinez A; Mielgo-Ayuso J; Cruz-Hernandez JJ Liquid Biopsy as Novel Tool in Precision Medicine: Origins, Properties, Identification and Clinical Perspective of Cancer’s Biomarkers. Diagnostics 2020, 10 (4), 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Mattox AK; Bettegowda C; Zhou S; Papadopoulos N; Kinzler KW; Vogelstein B Applications of liquid biopsies for cancer. Sci. Transl. Med 2019, 11 (507), eaay1984. [DOI] [PubMed] [Google Scholar]
- (3).Snow A; Chen D; Lang JE The current status of the clinical utility of liquid biopsies in cancer. Expert Rev. Mol. Diagn 2019, 19 (11), 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cervena K; Vodicka P; Vymetalkova V Diagnostic and prognostic impact of cell-free DNA in human cancers: Systematic review. Mutat. Res., Rev. Mutat. Res 2019, 781, 100–129. [DOI] [PubMed] [Google Scholar]
- (5).Kilgour E; Rothwell DG; Brady G; Dive C Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell 2020, 37 (4), 485–495. [DOI] [PubMed] [Google Scholar]
- (6).Nagrath S; Sequist LV; Maheswaran S; Bell DW; Irimia D; Ulkus L; Smith MR; Kwak EL; Digumarthy S; Muzikansky A; Ryan P; Balis UJ; Tompkins RG; Haber DA; Toner M Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450 (7173), 1235–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cohen JD; Li L; Wang Y; Thoburn C; Afsari B; Danilova L; Douville C; Javed AA; Wong F; Mattox A; Hruban RH; Wolfgang CL; Goggins MG; Dal Molin M; Wang TL; Roden R; Klein AP; Ptak J; Dobbyn L; Schaefer J; Silliman N; Popoli M; Vogelstein JT; Browne JD; Schoen RE; Brand RE; Tie J; Gibbs P; Wong HL; Mansfield AS; Jen J; Hanash SM; Falconi M; Allen PJ; Zhou S; Bettegowda C; Diaz LA Jr.; Tomasetti C; Kinzler KW; Vogelstein B; Lennon AM; Papadopoulos N Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359 (6378), 926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Geeurickx E; Hendrix A Targets, pitfalls and reference materials for liquid biopsy tests in cancer diagnostics. Mol. Aspects Med 2020, 72, 100828. [DOI] [PubMed] [Google Scholar]
- (9).Yan WT; Cui X; Chen Q; Li YF; Cui YH; Wang Y; Jiang J Circulating tumor cell status monitors the treatment responses in breast cancer patients: a meta-analysis. Sci. Rep 2017, 7, 43464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Seviour EG; Sehgal V; Lu Y; Luo Z; Moss T; Zhang F; Hill SM; Liu W; Maiti SN; Cooper L; Azencot R; Lopez-Berestein G; Rodriguez-Aguayo C; Roopaimoole R; Pecot CV; Sood AK; Mukherjee S; Gray JW; Mills GB; Ram PT Functional proteomics identifies miRNAs to target a p27/Myc/phospho-Rb signature in breast and ovarian cancer. Oncogene 2016, 35 (6), 801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wulfkuhle JD; Berg D; Wolff C; Langer R; Tran K; Illi J; Espina V; Pierobon M; Deng J; DeMichele A; Walch A; Bronger H; Becker I; Waldhor C; Hofler H; Esserman L; Liotta LA; Becker KF; Petricoin EF 3rd. Molecular analysis of HER2 signaling in human breast cancer by functional protein pathway activation mapping. Clin. Cancer Res 2012, 18 (23), 6426–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Li J; Zhao W; Akbani R; Liu W; Ju Z; Ling S; Vellano CP; Roebuck P; Yu Q; Eterovic AK; Byers LA; Davies MA; Deng W; Gopal YN; Chen G; von Euw EM; Slamon D; Conklin D; Heymach JV; Gazdar AF; Minna JD; Myers JN; Lu Y; Mills GB; Liang H Characterization of Human Cancer Cell Lines by Reverse-phase Protein Arrays. Cancer Cell 2017, 31 (2), 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Pishvaian MJ; Bender RJ; Matrisian LM; Rahib L; Hendifar A; Hoos WA; Mikhail S; Chung V; Picozzi V; Heartwell C; Mason K; Varieur K; Aberra M; Madhavan S; Petricoin E; Brody JR A pilot study evaluating concordance between blood-based and patient-matched tumor molecular testing within pancreatic cancer patients participating in the Know Your Tumor (KYT) initiative. Oncotarget 2017, 8, 83446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Harel M; Oren-Giladi P; Kaidar-Person O; Shaked Y; Geiger T Proteomics of microparticles with SILAC Quantification (PROMIS-Quan): a novel proteomic method for plasma biomarker quantification. Mol. Cell. Proteomics 2015, 14 (4), 1127–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Cocucci E; Meldolesi J Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015, 25 (6), 364–372. [DOI] [PubMed] [Google Scholar]
- (16).Milane L; Singh A; Mattheolabakis G; Suresh M; Amiji MM Exosome mediated communication within the tumor micro-environment. J. Controlled Release 2015, 219, 278–294. [DOI] [PubMed] [Google Scholar]
- (17).Vader P; Breakefield XO; Wood MJ Extracellular vesicles: emerging targets for cancer therapy. Trends Mol. Med 2014, 20 (7), 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Lee TH; D’Asti E; Magnus N; Al-Nedawi K; Meehan B; Rak J Microvesicles as mediators of intercellular communication in cancer–the emerging science of cellular ‘debris’. Semin. Immunopathol 2011, 33 (5), 455–467. [DOI] [PubMed] [Google Scholar]
- (19).Lin J; Li J; Huang B; Liu J; Chen X; Chen XM; Xu YM; Huang LF; Wang XZ Exosomes: novel biomarkers for clinical diagnosis. Sci. World J 2015, 2015, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Xu R; Greening DW; Zhu HJ; Takahashi N; Simpson RJ Extracellular vesicle isolation and characterization: toward clinical application. J. Clin. Invest 2016, 126 (4), 1152–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Verma M; Lam TK; Hebert E; Divi RL Extracellular vesicles: potential applications in cancer diagnosis, prognosis, and epidemiology. BMC Clin. Pathol 2015, 15, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Yang KS; Im H; Hong S; Pergolini I; Del Castillo AF; Wang R; Clardy S; Huang CH; Pille C; Ferrone S; Yang R; Castro CM; Lee H; Del Castillo CF; Weissleder R Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci. Transl. Med 2017, 9 (391), No. eaal3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lotvall J; Hill AF; Hochberg F; Buzas EI; Di Vizio D; Gardiner C; Gho YS; Kurochkin IV; Mathivanan S; Quesenberry P; Sahoo S; Tahara H; Wauben MH; Witwer KW; Thery C Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).An T; Qin S; Xu Y; Tang Y; Huang Y; Situ B; Inal JM; Zheng L Exosomes serve as tumour markers for personalized diagnostics owing to their important role in cancer metastasis. J. Extracell. Vesicles 2015, 4, 27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- s(25).Witwer KW; Buzas EI; Bemis LT; Bora A; Lasser C; Lotvall J; Nolte-’t Hoen EN; Piper MG; Sivaraman S; Skog J; Thery C; Wauben MH; Hochberg F Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Taylor DD; Shah S Methods of isolating extracellular vesicles impact down-stream analyses of their cargoes. Methods 2015, 87, 3–10. [DOI] [PubMed] [Google Scholar]
- (27).Nakai W; Yoshida T; Diez D; Miyatake Y; Nishibu T; Imawaka N; Naruse K; Sadamura Y; Hanayama R A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci. Rep 2016, 6, 33935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lamparski HG; Metha-Damani A; Yao JY; Patel S; Hsu DH; Ruegg C; Le Pecq JB Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods 2002, 270 (2), 211–226. [DOI] [PubMed] [Google Scholar]
- (29).Stranska R; Gysbrechts L; Wouters J; Vermeersch P; Bloch K; Dierickx D; Andrei G; Snoeck R Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med 2018, 16 (1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zeringer E; Li M; Barta T; Schageman J; Pedersen KW; Neurauter A; Magdaleno S; Setterquist R; Vlassov AV Methods for the extraction and RNA profiling of exosomes. World J. Methodol 2013, 3 (1), 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Niu Z; Pang RTK; Liu W; Li Q; Cheng R; Yeung WSB Polymer-based precipitation preserves biological activities of extracellular vesicles from an endometrial cell line. PLoS One 2017, 12 (10), No. e0186534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Mathivanan S; Lim JW; Tauro BJ; Ji H; Moritz RL; Simpson RJ Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol. Cell. Proteomics 2010, 9 (2), 197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Yoo CE; Kim G; Kim M; Park D; Kang HJ; Lee M; Huh N A direct extraction method for microRNAs from exosomes captured by immunoaffinity beads. Anal. Biochem 2012, 431 (2), 96–98. [DOI] [PubMed] [Google Scholar]
- (34).Enderle D; Spiel A; Coticchia CM; Berghoff E; Mueller R; Schlumpberger M; Sprenger-Haussels M; Shaffer JM; Lader E; Skog J; Noerholm M Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS One 2015, 10 (8), No. e0136133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Welton JL; Webber JP; Botos LA; Jones M; Clayton A Ready-made chromatography columns for extracellular vesicle isolation from plasma. J. Extracell. Vesicles 2015, 4, 27269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Tang YT; Huang YY; Zheng L; Qin SH; Xu XP; An TX; Xu Y; Wu YS; Hu XM; Ping BH; Wang Q Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int. J. Mol. Med 2017, 40 (3), 834–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Bijnsdorp IV; Maxouri O; Kardar A; Schelfhorst T; Piersma SR; Pham TV; Vis A; van Moorselaar RJ; Jimenez CR Feasibility of urinary extracellular vesicle proteome profiling using a robust and simple, clinically applicable isolation method. J. Extracell. Vesicles 2017, 6 (1), 1313091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Royo F; Zuniga-Garcia P; Sanchez-Mosquera P; Egia A; Perez A; Loizaga A; Arceo R; Lacasa I; Rabade A; Arrieta E; Bilbao R; Unda M; Carracedo A; Falcon-Perez JM Different EV enrichment methods suitable for clinical settings yield different subpopulations of urinary extracellular vesicles from human samples. J. Extracell. Vesicles 2016, 5, 29497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Liang LG; Sheng YF; Zhou S; Inci F; Li L; Demirci U; Wang S An Integrated Double-Filtration Microfluidic Device for Detection of Extracellular Vesicles from Urine for Bladder Cancer Diagnosis. Methods Mol. Biol 2017, 1660, 355–364. [DOI] [PubMed] [Google Scholar]
- (40).Van Deun J; Mestdagh P; Sormunen R; Cocquyt V; Vermaelen K; Vandesompele J; Bracke M; De Wever O; Hendrix A The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wu X; Li L; Iliuk A; Tao WA Highly Efficient Phosphoproteome Capture and Analysis from Urinary Extracellular Vesicles. J. Proteome Res 2018, 17 (9), 3308–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Chen IH; Xue L; Hsu CC; Paez JS; Pan L; Andaluz H; Wendt MK; Iliuk AB; Zhu JK; Tao WA Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. U. S. A 2017, 114 (12), 3175–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Masuda T; Tomita M; Ishihama Y Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res 2008, 7 (2), 731–740. [DOI] [PubMed] [Google Scholar]
- (44).Perez-Riverol Y; Csordas A; Bai J; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Perez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaino JA The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Keerthikumar S; Chisanga D; Ariyaratne D; Al Saffar H; Anand S; Zhao K; Samuel M; Pathan M; Jois M; Chilamkurti N; Gangoda L; Mathivanan S ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol 2016, 428 (4), 688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Mathivanan S; Fahner CJ; Reid GE; Simpson RJ ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40 (Databaseissue), D1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Simpson RJ; Kalra H; Mathivanan S ExoCarta as a resource for exosomal research. J. Extracell. Vesicles 2012, 1, 18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wang T; Anderson KW; Turko IV Assessment of Extracellular Vesicles Purity Using Proteomic Standards. Anal. Chem 2017, 89 (20), 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Siegel RL; Miller KD; Jemal A Cancer statistics, 2019. Ca-Cancer J. Clin 2019, 69 (1), 7–34. [DOI] [PubMed] [Google Scholar]
- (50).Ng CS; Wood CG; Silverman PM; Tannir NM; Tamboli P; Sandler CM Renal cell carcinoma: diagnosis, staging, and surveillance. AJR, Am. J. Roentgenol 2008, 191 (4), 1220–1232. [DOI] [PubMed] [Google Scholar]
- (51).Diaz de Leon A; Pedrosa I Imaging and Screening of Kidney Cancer. Radiol Clin North Am. 2017, 55 (6), 1235–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Rule AD; Bergstralh EJ; Melton LJ 3rd; Li X; Weaver AL; Lieske JC Kidney stones and the risk for chronic kidney disease. Clin. J. Am. Soc. Nephrol 2009, 4 (4), 804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Iliuk AB; Martin VA; Alicie BM; Geahlen RL; Tao WA In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol. Cell. Proteomics 2010, 9 (10), 2162–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Kelstrup CD; Bekker-Jensen DB; Arrey TN; Hogrebe A; Harder A; Olsen JV Performance Evaluation of the Q Exactive HF-X for Shotgun Proteomics. J. Proteome Res 2018, 17 (1), 727–738. [DOI] [PubMed] [Google Scholar]
- (55).Collins BC; Hunter CL; Liu Y; Schilling B; Rosenberger G; Bader SL; Chan DW; Gibson BW; Gingras AC; Held JM; Hirayama-Kurogi M; Hou G; Krisp C; Larsen B; Lin L; Liu S; Molloy MP; Moritz RL; Ohtsuki S; Schlapbach R; Selevsek N; Thomas SN; Tzeng SC; Zhang H; Aebersold R Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun 2017, 8 (1), 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Chursa U; Nunez-Duran E; Cansby E; Amrutkar M; Sutt S; Stahlman M; Olsson BM; Boren J; Johansson ME; Backhed F; Johansson BR; Sihlbom C; Mahlapuu M Overexpression of protein kinase STK25 in mice exacerbates ectopic lipid accumulation, mitochondrial dysfunction and insulin resistance in skeletal muscle. Diabetologia 2017, 60 (3), 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Snel B; Lehmann G; Bork P; Huynen MA STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28 (18), 3442–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Tsuda M; Tanaka S Roles for crk in cancer metastasis and invasion. Genes Cancer 2012, 3 (5–6), 334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Kanehisa M; Goto S; Furumichi M; Tanabe M; Hirakawa M KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38 (suppl_1), D355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Lingenhel A; Lhotta K; Neyer U; Heid IM; Rantner B; Kronenberg MF; Konig P; von Eckardstein A; Schober M; Dieplinger H; Kronenberg F Role of the kidney in the metabolism of apolipoprotein A-IV: influence of the type of proteinuria. J. Lipid Res 2006, 47 (9), 2071–2079. [DOI] [PubMed] [Google Scholar]
- (61).Chen F; Zhang Y; Parra E; Rodriguez J; Behrens C; Akbani R; Lu Y; Kurie JM; Gibbons DL; Mills GB; Wistuba II; Creighton CJ Multiplatform-based molecular subtypes of non-small-cell lung cancer. Oncogene 2017, 36 (10), 1384–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Zagorac I; Fernandez-Gaitero S; Penning R; Post H; Bueno MJ; Mouron S; Manso L; Morente MM; Alonso S; Serra V; Munoz J; Gomez-Lopez G; Lopez-Acosta JF; Jimenez-Renard V; Gris-Oliver A; Al-Shahrour F; Pineiro-Yanez E; Montoya-Suarez JL; Apala JV; Moreno-Torres A; Colomer R; Dopazo A; Heck AJR; Altelaar M; Quintela-Fandino M In vivo phosphoproteomics reveals kinase activity profiles that predict treatment outcome in triple-negative breast cancer. Nat. Commun 2018, 9 (1), 3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Huang KL; Li S; Mertins P; Cao S; Gunawardena HP; Ruggles KV; Mani DR; Clauser KR; Tanioka M; Usary J; Kavuri SM; Xie L; Yoon C; Qiao JW; Wrobel J; Wyczalkowski MA; Erdmann-Gilmore P; Snider JE; Hoog J; Singh P; Niu B; Guo Z; Sun SQ; Sanati S; Kawaler E; Wang X; Scott A; Ye K; McLellan MD; Wendl MC; Malovannaya A; Held JM; Gillette MA; Fenyo D; Kinsinger CR; Mesri M; Rodriguez H; Davies SR; Perou CM; Ma C; Reid Townsend R; Chen X; Carr SA; Ellis MJ; Ding L Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat. Commun 2017, 8, 14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Mayer IA; Abramson VG; Isakoff SJ; Forero A; Balko JM; Kuba MG; Sanders ME; Yap JT; Van den Abbeele AD; Li Y; Cantley LC; Winer E; Arteaga CL Stand up to cancer phase Ib study of pan-phosphoinositide-3-kinase inhibitor buparlisib with letrozole in estrogen receptor-positive/human epidermal growth factor receptor 2-negative metastatic breast cancer. J. Clin. Oncol 2014, 32 (12), 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Vogel CL; Cobleigh MA; Tripathy D; Gutheil JC; Harris LN; Fehrenbacher L; Slamon DJ; Murphy M; Novotny WF; Burchmore M; Shak S; Stewart SJ; Press M Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J. Clin. Oncol 2002, 20 (3), 719–726. [DOI] [PubMed] [Google Scholar]
- (66).Pogue-Geile KL; Kim C; Jeong JH; Tanaka N; Bandos H; Gavin PG; Fumagalli D; Goldstein LC; Sneige N; Burandt E; Taniyama Y; Bohn OL; Lee A; Kim SI; Reilly ML; Remillard MY; Blackmon NL; Kim SR; Horne ZD; Rastogi P; Fehrenbacher L; Romond EH; Swain SM; Mamounas EP; Wickerham DL; Geyer CE; Costantino JP; Wolmark N; Paik S Predicting Degree of Benefit From Adjuvant Trastuzumab in NSABP Trial B-31. Jnci-J. Natl. Cancer I 2013, 105 (23), 1782–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE44 partner repository with the data set identifier PXD017994.


