The OsABCI7 transporter interacts with OsHCF222 at thylakoid membranes to regulate reactive oxygen species homeostasis, thereby maintaining thylakoid membrane stability.
Abstract
The thylakoid membrane is a highly complex membrane system in plants and plays crucial roles in the biogenesis of the photosynthetic apparatus and plant development. However, the genetic factors involved in chloroplast development and its relationship with intracellular metabolites are largely unknown. Here, a rice (Oryza sativa) chlorotic and necrotic leaf1 (cnl1) mutant was identified and map-based cloning revealed that a single base substitution followed by a 6-bp deletion in the ATP-binding cassette transporter I family member7 (OsABCI7) resulted in chlorotic and necrotic leaves with thylakoid membrane degradation, chlorophyll breakdown, photosynthesis impairment, and cell death in cnl1. Furthermore, the expression of OsABCI7 was inducible under lower temperatures, which severely affected cnl1 chloroplast development, and etiolated cnl1 seedlings were unable to recover to a normal green state under light conditions. Functional complementation and overexpression showed that OsABCI7 could rescue the cnl1 chlorotic and necrotic phenotype. OsABCI7 interacted with HIGH CHLOROPHYLL FLUORESCENCE222 (OsHCF222) to regulate cellular reactive oxygen species (ROS) homeostasis for thylakoid membrane stability. OsABCI7 localized to thylakoid membranes, while OsHCF222 targeted to endoplasmic reticulum and chloroplasts. Exogenous application of ascorbic acid eased the yellowish leaf phenotype by increasing chlorophyll content and alleviating ROS stress in cnl1. Unlike cnl1, the CRISPR/Cas9-mediated OsHCF222 knockout lines showed chlorotic leaves but were seedling lethal. Our results provide insight into the functions of ABC transporters in rice, especially within the relationship between ROS homeostasis and stability of thylakoid membranes.
In plant leaves, mature chloroplasts develop from proplastids, a type of colorless and small precursor, with surrounded inner and outer envelopes and rudimentary inner membranes. These rudimentary membranes elaborate into an inerratic thylakoid membrane system during leaf development. As the photosynthetic machinery, the thylakoid membrane complex is composed of four major protein complexes: PSII, cytochrome b6f (Cytb6f), PSI, and ATP synthase. Thylakoid membrane biogenesis is involved in a variety of processes, such as gene transcriptional regulation, posttranslational regulation and modification, and protein complex assembly (Lyska et al., 2013). In chloroplasts, the thylakoid membrane is regularly stacked to increase the membrane area for a higher photosynthetic efficiency, and a well-developed thylakoid membrane system is also closely associated with chloroplast development (Kobayashi et al., 2007). To date, many genes exhibiting crucial regulatory roles in chloroplast development under low temperatures have been identified (Wang et al., 2016; Sun et al., 2017; Liu et al., 2018; Cui et al., 2019), while genetic factors affecting the relationship between chloroplast development and intracellular metabolites are largely unknown.
Endogenous reactive oxygen species (ROS) are mainly produced at the PSI and PSII reaction centers in thylakoids. Usually, an oxygen molecule obtains an electron and is reduced to a superoxide anion (O2−), and its disproportionation by a superoxide dismutase (SOD) produces hydrogen peroxide (H2O2; Asada et al., 1974). To protect chloroplasts from the direct damage by H2O2, the newly produced thylakoidal H2O2 is promptly scavenged; hence, H2O2 is not accumulated in intact chloroplasts (Asada, 2006). In this process, H2O2 is reduced into water by ascorbate acid (AsA) catalyzed by the plant- and algae-specific H2O2-scavenging enzyme, ascorbate peroxidase (APX; Asada, 1992). Additionally, catalase (CAT) can directly reduce H2O2 into water. Once the balance of ROS production and scavenging is broken, oxidative damage induced by ROS preferentially occurs in chloroplast thylakoids, leading to chlorophyll degradation as well as DNA damage (Ahmad et al., 2010; Gill and Tuteja, 2010; Soares et al., 2016). Besides biotic and/or abiotic stresses, mutations of relevant genes are important causes for inducing ROS bursts, resulting in various morphological variations, such as premature senescence, yellowing, and spotted leaves (Huang et al., 2016; Xu et al., 2018; Sathe et al., 2019; Wang et al., 2019).
The ATP-binding cassette (ABC) transporter is an ancient and large protein superfamily consisting of eight major subgroups (ABCA–ABCH). In plants, the members of the ABCA to ABCG subgroups are widely distributed, while the ABCH members are absent (Garcia et al., 2004). ABC proteins, with six conserved typical motifs, including ABC signature, Walker-A, Walker-B, d-loop, H-loop, and Q-loop sequence, are well known for their functions in intracellular/extracellular substrate transporting (Huang et al., 2009). Not only limited to the transport of ions, lipids, monosaccharides, hormone secondary metabolites, and xenobiotics, recent studies have demonstrated that ABC proteins are also involved in plant defense response, pathogen resistance, and male reproduction (Matsuda et al., 2012; Zhao et al., 2015; Garroum et al., 2016). Plants share the specific bacterial-type ABC transporters with prokaryotes, designed as ABCI subgroup. In Arabidopsis (Arabidopsis thaliana), both chloroplast-localized AtABCI10 and AtABCI11 (AtNAP14) proteins participate in the regulation of chloroplast metal homeostasis, and their corresponding transfer DNA mutants are totally devoid of chlorophylls with severely deformed leaf structures, aberrant chloroplasts, and defective photosynthetic capacity (Shimoni-Shor et al., 2010; Voith von Voithenberg et al., 2019). In rice (Oryza sativa), OsABCI8 is indispensable for chloroplast development by engaging iron transportation and homeostasis (Zeng et al., 2017). OsABCI12 (STAR2) interacts with STAR1 to form a complex responsible for the transport of UDP-Glc and detoxification of aluminum (Huang et al., 2009).
The rice high-chlorophyll-fluorescence (hcf) mutants are unable to utilize absorbed light efficiently (Meurer et al., 1996). Due to lack of photosynthetic autotrophy, most hcf mutants are seedling lethal yet able to survive on Suc-supplemented medium (Schult et al., 2007; Link et al., 2012; Hartings et al., 2017). Here, we isolated an ABC transporter I family member7 (OsABCI7), which interacts with HIGH CHLOROPHYLL FLUORESCENCE222 (OsHCF222) to maintain thylakoid membrane stability via regulating ROS homeostasis in rice.
RESULTS
cnl1 Is Defective in Plant Growth and Development
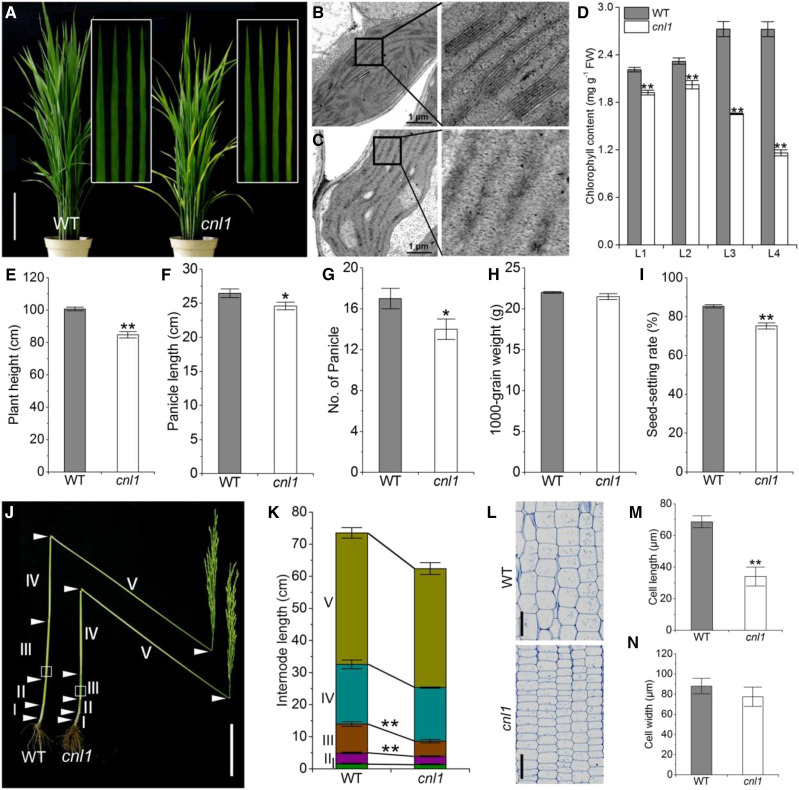
The chlorotic and necrotic leaf1 (cnl1) mutant was isolated from an ethyl methylsulfonate-induced indica rice var. Zhongjian 100 (wild type) mutant bank. Under natural field conditions, cnl1 exhibited chlorotic and necrotic phenotypes on older leaves with the vestigial thylakoid membrane ultrastructure and lower levels of chlorophyll contents ∼40 d after sowing (Fig. 1, A–D; Supplemental Fig. S1, A–C). Due to abnormal chlorophyll degradation, cnl1 showed significantly compromised net photosynthesis rate, higher intercellular CO2 concentration, and transpiration rate compared with wild type (Supplemental Fig. S1, D–G).
Figure 1.
Phenotypic characterization of cnl1. A, Wild-type (WT) and cnl1 at the tillering stage (45 d after field transplanting). Insets show magnified views of the four leaves (L1–L4) from top to bottom of wild type and cnl1. Bar = 20 cm. B and C, Chloroplast ultrastructure in mesophyll cells of L4 in wild type (B) and cnl1 (C) in A. D, Chlorophyll contents in different leaves of wild type and cnl1. L1 to L4 represent the four leaves shown in A (from left to right), respectively. E to I, Major agronomic traits of wild type and cnl1. J, Comparison of main stems from wild type and cnl1. Arrows indicate the node positions. Bar = 10 cm. K, Internode lengths of main stems in wild type and cnl1 at the mature stage. L, Longitudinal sections of the corresponding parts of internode III from wild type and cnl1 as marked by the white boxes in J. Bars = 100 μm. M and N, Comparison of cell length (M) and cell width (N) of internode III in wild type and cnl1. Data are means ± sd (n = 5). Asterisks indicate significant difference by Student’s t test (**P < 0.01 and *P < 0.05).
In addition, cnl1 also exhibited a poorer performance in major agronomic traits compared with wild type (Fig. 1, E–I). The II and III internode lengths were remarkably decreased in cnl1 compared to wild type at the heading stage (Fig. 1, J and K), and microscope observation directly revealed that the shorter stature of cnl1 resulted from the reduced length of cells in the stems (Fig. 1, L–N). These results indicated that the cnl1 mutation resulted in multiple deficiencies in plant growth and development.
CNL1 Encodes Rice ABC Transporter I Family Member7 (OsABCI7)
To determine the genetic control of the cnl1 phenotype, we crossed cnl1 with two wild type japonica rice cultivars (cv ReYan 1 and cv ORO, respectively). As shown in Table 1, the wild-type F1 plants and expected mendelian 3:1 (wild type:mutant) ratios in F2 populations indicated monogenic and recessive genetic control of the mutant trait.
Table 1. Genetic control of the cnl1 phenotype.
| Cross | F1 | No. F2 Individuals | χ2 (3:1) | |
|---|---|---|---|---|
| Wild Type | Mutant Type | |||
| cnl1 × wild type | Wild type | 127 | 34 | 1.29 |
| cnl1 × cv ReYan 1 | Wild type | 923 | 283 | 1.51 |
| cnl1 × cv ORO | Wild type | 399 | 112 | 2.59 |
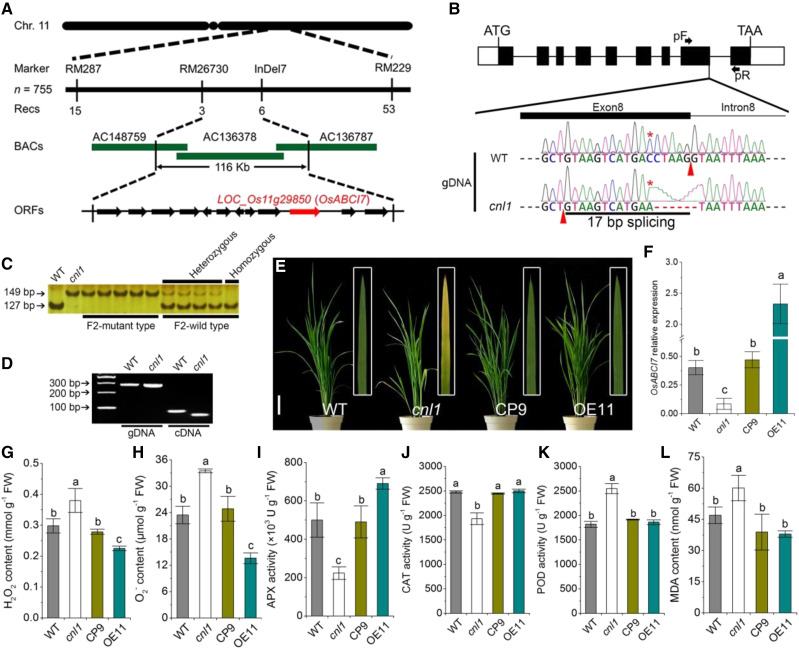
We selected 755 F2 mutant-type individuals derived from the cross cnl1 × cv ReYan 1 to map the mutation. The OsABCI7 locus preliminarily mapped to the long arm of chromosome 11 between the markers RM287 and RM229, and we narrowed the location to a 116-kb region containing 12 open reading frames (ORFs; Fig. 2A; Supplemental Table S1). Sequencing of these 12 ORFs revealed a single nucleotide substitution (C to A) followed by a 6-bp deletion at the junction of exon 8 and intron 8 of LOC_Os11g29850, resulting in a 17-bp alternative splicing deletion at the end of exon 8, which finally led to a frame shift and premature stop codon (Fig. 2B; Supplemental Figs. S2 and S3A). According to the Rice Annotation Project Database (https://rapdb.dna.affrc.go.jp/), LOC_Os11g29850 belongs to the ABC transport superfamily ABCI subgroup member 7 (referred to OsABCI7). The structural changes of OsABCI7 were intuitively visible by modeling the three-dimensional structures of wild-type (OsABCI7) and mutant (ΔOsABCI7) proteins (Supplemental Fig. S3B). Additionally, the derived cleaved amplified polymorphic sequence analysis was performed to confirm the mutation in the genomic DNA, and the agarose electrophoresis verified the presence of the 17-bp deletion in the cDNA of ΔOsABCI7 (Fig. 2, C and D). OsABCI7 encodes an ABC transport protein and contains all typical conserved motifs of ABC transporters, namely, the Walker-A, Q-loop, ABC signature, Walker-B, d-loop, and H-loop motifs (Supplemental Fig. S2).
Figure 2.
Map-based cloning of OsABCI7 and ROS detection in cnl1. A, Mapping of the OsABCI7 locus. The molecular markers and recombinant numbers are displayed. The candidate gene OsABCI7/LOC_Os11g29850 is depicted in red. B, OsABCI7 allele structure in wild type (WT) and cnl1. The white boxes, black boxes, and black lines represent untranslated regions, exons, and introns, respectively. The red stars indicate the nucleotide substitution, and red arrowheads indicate the splicing site of intron 8 in wild type and cnl1. pF and pR indicate the forward and reverse primer sites in D. C, Derived cleaved amplified polymorphic sequence confirmation of the mutation. PCR products were digested by SalI from wild type but not from cnl1 and detected by 6% polyacrylamide gel. D, PCR analysis showing the 17-bp splicing deletion of the eighth exon. The smaller band of cnl1 cDNA resulted from the 17-bp alternative upstream splicing deletion as indicated in B. gDNA, Genomic DNA. Agarose gel concentration was 3.5%. E, Phenotypes of complementary (CP) and overexpression (OE) lines at 40 d after sowing. Insets show magnified views of the bottom second leaves. Bar = 10 cm. F, Relative expression of OsABCI7 in wild-type, cnl1, and transgenic plants. Rice UBIQUITIN was used as an internal control. Data are means ± sd (n = 3). G to K, ROS levels, ROS scavenging enzyme activities and MDA contents in flag leaves of wild-type, cnl1, and transgenic plants at the heading stage. H2O2 content (G), O2− content (H), APX activity (I), CAT activity (J), peroxidase (POD) activity (K), and MDA content (L). Data are means ± sd (n = 3). Different letters indicate significant differences by one-way ANOVA and Duncan’s test (P < 0.05).
To confirm whether the mutation was responsible for the cnl1 phenotype, we performed a complementation test by introducing 5.9-kb genomic DNA containing the promoter and the full genomic coding region of OsABCI7 into cnl1-derived calli. All 15 independent positive T0 complementary lines showed the normal green phenotype, mimicking the wild type (Fig. 2E). We also overexpressed the coding sequence (CDS) of OsABCI7 in cnl1 using the vector pCAMBIA1300-UBI (Zhang et al., 2018), which confers maize (Zea mays) Ubiquitin1 promoter and the coding region for an N-terminal hemagglutinin (HA) epitope tag to the CDS (Supplemental Fig. S4, A and B), and the mutant phenotype was rescued in all 24 T0 positive overexpression lines (Fig. 2E). These results demonstrated that ΔOsABCI7 was responsible for the cnl1 phenotype, and the CDS of OsABCI7 encoded a functional protein. Compared with wild type, the expression of OsABCI7 was significantly downregulated in cnl1 but recovered to wild-type levels in complementary plants, and the expression level of OsABCI7 in overexpression plants was upregulated by about 6-fold in wild type (Fig. 2F). To investigate whether the low expression of OsABCI7 would lead to the cnl1 phenotype, we also performed RNA interference analysis of OsABCI7 and obtained 21 T0 positive lines that exhibited significantly down-regulated OsABCI7 expression with normal wild-type phenotype (Supplemental Fig. S4, D–H). We therefore concluded that the low OsABCI7 expression was unable to cause cnl1 phenotype, and the rescues of cnl1 by complementation and overexpression were due to the recovered OsABCI7 function and not the recovered OsABCI7 expression. Furthermore, similar expression levels of OsABCI7 were detected from the tip to the base of the yellowing flag leaves, indicating low relevance between OsABCI7 expression and natural leaf yellowing (Supplemental Fig. S4C).
Leaf yellowing and senescence are largely accompanied with ROS accumulation (Buchanan-Wollaston et al., 2003). Thus, we first measured the levels of ROS, including H2O2 and O2−, and found that ROS were higher in cnl1 compared with wild type (Fig. 2, G and H). We then detected the activities of ROS scavenging enzymes and the contents of malondialdehyde (MDA) and soluble proteins. The activities of APX and CAT, two key H2O2 scavenging enzymes, as well as soluble protein contents, were significantly lower in cnl1 than in the wild type (Fig. 2, I and J; Supplemental Fig. S5A). In contrast, the level of MDA, an indicator of ROS-induced lipid peroxidation, was significantly higher in cnl1 than in the wild type (Fig. 2L). Peroxidase activity was higher in cnl1 compared with wild type, while SOD activity was similar between the two genotypes (Fig. 2K; Supplemental Fig. S5B). These results suggested that ΔOsABCI7 disrupted the balance between ROS production and scavenging. Moreover, ROS accumulation in cnl1 was also confirmed by nitrotetrazolium blue chloride and 3,3′-diaminobenzidine staining (Supplemental Fig. S6C). As expected, the ROS contents and ROS scavenging enzyme activities in the complementary and overexpression line recovered to wild-type levels (Fig. 2, G–L). In addition, chlorophyll fluorescence and chlorophyll levels in the complementary and overexpression lines also recovered to the wild-type levels (Supplemental Fig. S6, A and B; Supplemental Table S2). Taken together, our results demonstrated that both the normal expression and overexpression of OsABCI7 in the cnl1 background could fully rescue the physiological and biochemical defects of cnl1. Notably, in contrast to wild type, the overexpression plants exhibited prominently decreased H2O2 and O2− contents and increased APX activities (Fig. 2, G–I). We speculated that OsABCI7 was involved in chloroplast-specific ROS scavenging since the H2O2 scavenging largely relies on APX in chloroplasts (Asada, 2006).
DNA Damage and DNA Damage Response Are Induced in cnl1
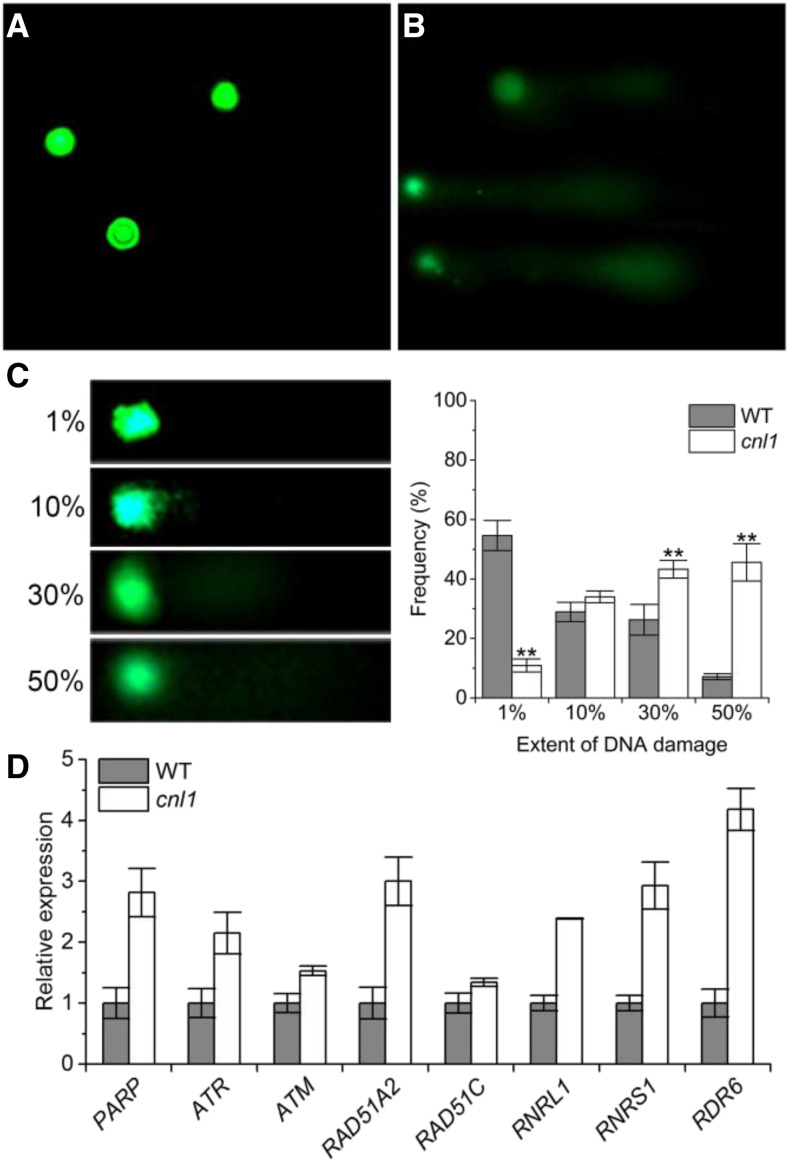
The programmed cell death (PCD) phenotype was detected in cnl1 by trypan blue staining (Supplemental Fig. S6C). Since PCD associates with DNA breakdown, we conducted the terminal deoxyribonucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay to determine the nuclear DNA fragmentation in the flag leaves of wild type and cnl1. Expectedly, few TUNEL-positive signals were detected in wild type, yet numerous nuclei exhibited TUNEL-positive signals in cnl1 (Supplemental Fig. S7). Furthermore, we performed comet assay (single cell gel electrophoresis assay), a simple and effective method for evaluating DNA damage in cells (Speit and Hartmann, 2006), to test DNA damage in wild type and cnl1. Consistent with the TUNEL assay, significantly increased DNA damage was found in cnl1 compared with wild type (Fig. 3, A–C).
Figure 3.
DNA damage detection and transcriptional level analysis of DNA damage repair- and replication-associated genes. A and B, Typical comets for DNA damage in nuclei of flag leaves from wild type (WT; A) and cnl1 (B) at the heading stage. C, Four types of DNA damage extents in each nucleus are indicated by the units 1%, 10%, 30%, or 50%. An increased unit associated with a larger comet tail. Frequency distribution of four types of DNA damage extents in wild type and cnl1. Data are means ± sd (n = 3). Asterisks indicate significant difference by Student’s t test (**P < 0.01). D, Transcriptional level analysis of DNA repair- and replication-associated genes by RT-qPCR. Rice Ubiquitin was used as an internal control. Data are means ± sd (n = 3).
DNA repair and DNA replication occur in response to DNA damage. Indeed, eight up-regulated marker genes associated with DNA repair and replication were found in cnl1 (Fig. 3D). Poly (ADP-ribose) polymerase (PARP), ataxia telangiectasia mutated (ATM), and ATM-rad3-related (ATR) are classified to the ATM/ATR-mediated signaling response (Zhou and Elledge, 2000; Culligan et al., 2006). DNA repair protein Rad51 (RAD51A2 and RAD51C) belong to the RecA/Rad1 protein family, which is required for the homologous recombination repair pathway (Durrant et al., 2007). RNRL1 and RNRS1 encode the large and small subunits of ribonucleotide reductase (RNR), respectively, regulating the deoxyribonucleotide production rate for DNA synthesis and repair (Yoo et al., 2009). RNA-dependent RNA polymerase6 (RDR6) acts in the biogenesis of multiple small RNAs (Song et al., 2012). In conclusion, ΔOsABCI7 induced PCD and severe DNA damage and up-regulated expression of marker genes associated with DNA repair and replication.
OsABCI7 Is Constitutively Expressed and OsABCI7 Localizes to the Thylakoid Membrane
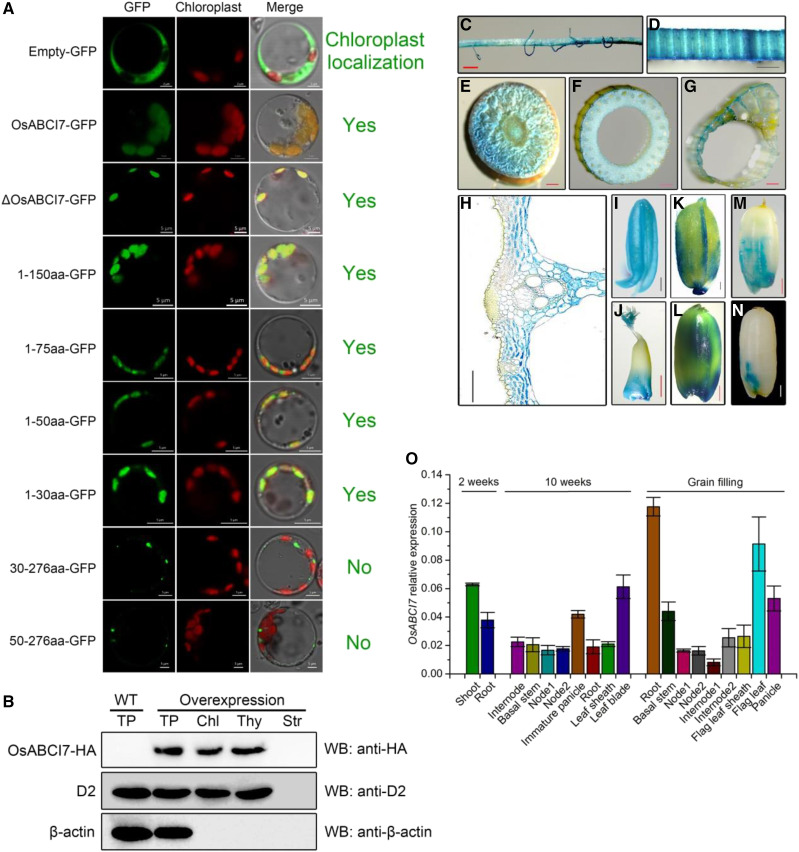
To validate if OsABCI7 localized to chloroplasts as predicted by TargetP 1.1 Server (http://www.cbs.dtu.dk/services/TargetP/), we fused the full CDS of OsABCI7 to the N terminus of GFP driven by the CaMV35S promoter and transiently expressed the construct in rice protoplasts. The OsABCI7-GFP fusion protein indeed localized to chloroplasts (Fig. 4A). Notably, the ΔOsABCI7-GFP fusion protein also localized to chloroplasts, indicating the mutation of OsABCI7 did not change its chloroplast localization (Fig. 4A). Additionally, we truncated constructs to analyze the chloroplast transit peptide, and nearly all fusion constructs with varying lengths of OsABCI7, such as OsABCI71–30aa, apparently localized to the chloroplast except for OsABCI730–276aa and OsABCI750–276aa (Fig. 4A). These demonstrated that the OsABCI71–30aa may contain a chloroplast transit peptide indispensable for OsABCI7 chloroplast targeting. To detect the precise localization of OsABCI7, we performed immunoblot analysis using total proteins, chloroplasts, thylakoid fractions, and stromal fractions extracted from OsABCI7 overexpression plants. A 32-kD protein was absent in stromal fractions while present in total proteins, chloroplasts, and thylakoid membrane fractions, confirming the thylakoid membrane localization of OsABCI7 (Fig. 4B).
Figure 4.
Subcellular localization and expression pattern of OsABCI7. A, GFP signals of different constructs in rice protoplasts. The full OsABCI7 protein contains 276 amino acids. ΔOsABCI7 represents the mutant OsABCI7 protein. Bars = 5 μm. B, Immunoblot analysis of OsABCI7. Protein samples were from OsABCI7 (fused HA-tag) overexpression seedlings. Antiserum against HA-tag (anti-HA), D2 (thylakoid marker), and β-actin were used in blotting. TP, Total protein; Chl, chloroplast; Thy, thylakoid membrane fraction; Str, stroma fraction; WT, wild type. C to N, GUS staining of OsABCI7Pro::GUS transgenic plants. Root (C), leaf blade (D), node V (E), culm (F), leaf sheath (G), leaf blade transverse slice (H), stamen (I), and seeds at different stages of development (J–N). Bars = 1 mm (B–F and H–M) and 100 μm (G). O, Relative expression of OsABCI7 in various organs at different growth stages. Data are means ± sd (n = 3).
We then detected OsABCI7 expression levels in different organs using reverse transcription quantitative PCR (RT-qPCR). OsABCI7 was expressed in all organs at different growth stages (Fig. 4O). Consistent with the RT-qPCR results, GUS expression was detected in all organs of OsABCI7Pro::GUS transgenic plants (Fig. 4, C–N), further supporting the constitutive expression of OsABCI7.
OsABCI7 Is Required for Chloroplast Development under Cold Stress and Light-Dependent Chlorophyll Synthesis
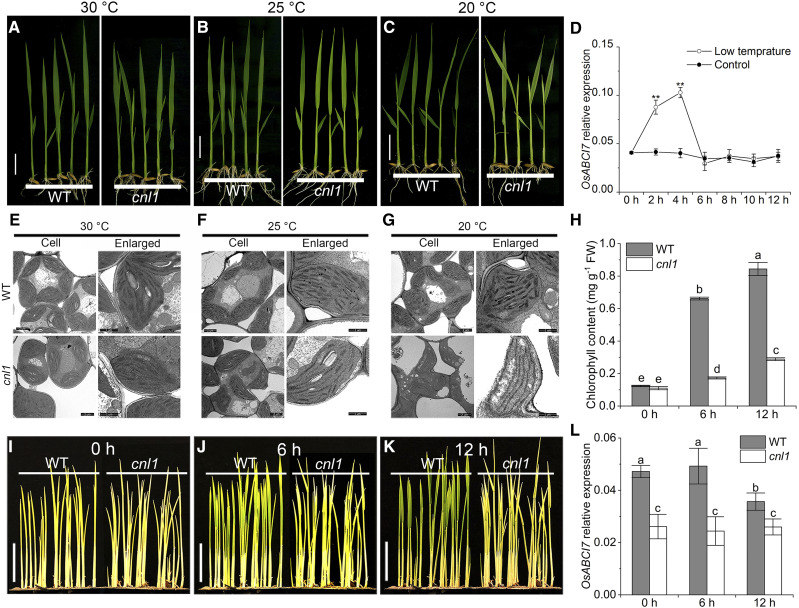
Chloroplast development and chlorophyll synthesis are affected by light and temperature in rice (Wang et al., 2016; Sun et al., 2017; Wang et al., 2017). Wild type and cnl1 were planted in a growth chamber under different temperature conditions. At 30°C, cnl1 plants displayed a wild-type-like phenotype, though the chlorophyll content of cnl1 was significantly lower than that of the wild type (Fig. 5A; Supplemental Fig. S8A). In contrast, cnl1 showed pale green leaves at 25°C, and the pale green phenotype aggravated at 20°C with rapid decrease of chlorophyll contents compared with wild type (Fig. 5, B and C; Supplemental Fig. S8A). We then explored the kinetic expression of OsABCI7 in wild type and cnl1 under low-temperature treatment at 10°C, and OsABCI7 expression increased rapidly by 2-fold after 2 h, by 2.5-fold after 4 h treatment, and recovered to the control level after 6 h of treatment (Fig. 5D), while OsABCI7 expression increased 2- to 3-fold from 4 to 10 h treatment in cnl1 (Supplemental Fig. S8B). The results indicated that OsABCI7 transcription was induced by low temperature and likely acted as a signal factor responding to cold stress.
Figure 5.
Phenotypic characterization of wild type (WT) and cnl1 at different temperatures and light-dependent chlorophyll synthesis of wild type and cnl1. A to C, Phenotypic comparison of wild-type and cnl1 seedlings at different temperatures. The wild type and cnl1 were grown in growth chambers after germination for 10 d at 30°C (A) and 25°C (B) and 16 d at 20°C (C) with constant temperatures and 14 h light/10 h dark cycles (400 μmol m−2 s−1). Bars = 2 cm. D, Kinetic analysis of OsABCI7 expression in wild type at 10°C. Data are means ± sd (n = 3). Asterisks indicate significant difference by Student’s t test (**P < 0.01). E to G, Ultrastructure of chloroplasts in mesophyll cells of wild type and cnl1 at different temperatures. All the cells were from the fully emerged third leaves of wild type and cnl1 seedlings at the three-leaf stage. Bars = 2 μm (in cell view) and 1 μm (in enlarged view). H to L, Chlorophyll contents (H), greening speed (I–K), and OsABCI7 expression (L) of etiolated wild-type and cnl1 seedlings during the greening process. Seven-day-old etiolated seedlings were exposed to light (250 μmol m−2 s−1) for 12 h. Bars = 2 cm. Data are means ± sd (n = 3). Different letters indicate significant differences by one-way ANOVA and Duncan’s test (P < 0.05).
We also investigated the ultrastructure of chloroplasts in mesophyll cells of wild type and cnl1 under different temperatures using transmission electron microscopy (TEM). At 30°C, intact chloroplast structure and well-formed grana thylakoids were observed in wild type and cnl1 (Fig. 5E). In contrast, distinctly degraded chloroplasts and reduced grana thylakoids were found in cnl1 at 25°C and 20°C compared with wild type (Fig. 5, F and G). These suggested that the developmental defects associated with the pale green phenotype in cnl1 resulted from chloroplast breakdown induced by low temperatures. Furthermore, OsABCI7 was essential for light-dependent chlorophyll synthesis during the greening of etiolated seedlings (Fig. 5, H–K), while OsABCI7 transcription in wild type and cnl1 was not relevant to the greening process (Fig. 5L). Collectively, the results suggested that OsABCI7 was involved in light-dependent chlorophyll synthesis and required for chloroplast development under low temperature.
Thylakoid Membrane Complexes Are Impaired in cnl1
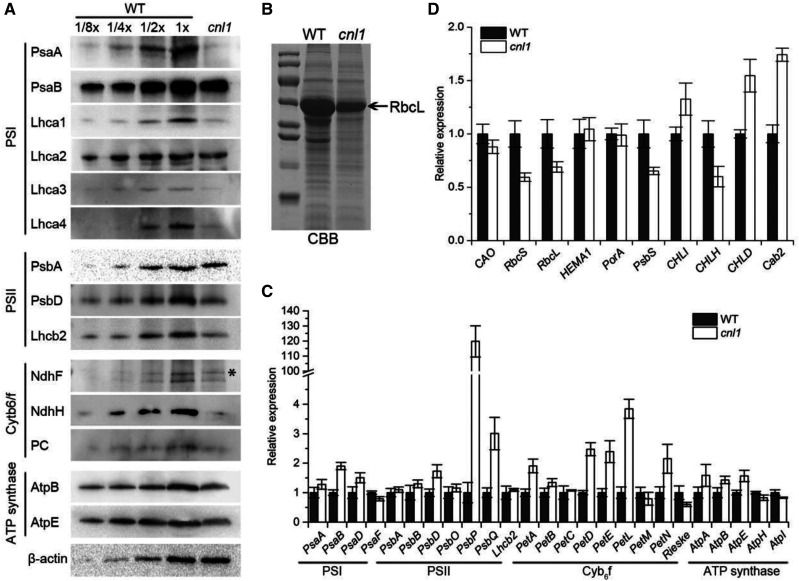
Thylakoid membrane biogenesis is regulated and assisted by many plastome- and nucleus-encoded genes, and thylakoid membrane protein complexes are composed of PSII, PSI, Cytb6f complex, and ATP synthase (Hartings et al., 2017). Upon the low photosynthetic capacity in cnl1 (Supplemental Fig. S1, D–G; Supplemental Table S2), the accumulation of major components of thylakoid membrane protein complexes in wild type and cnl1 were determined by immunoblot to test whether thylakoid membrane complexes were impaired in cnl1. We tested the core subunits of PSII and PSI complexes (plastid encoded, PsaA/B and PsbA/D), nucleus-encoded light-harvesting antenna of PSI (LHCI) chlorophyll a/b-binding proteins Lhca1 to Lhca4, and nucleus-encoded LHCII type II chlorophyll a/b-binding protein Lhcb2. A pronounced reduction was observed for the levels of all these subunits except for PsaB, Lhca2, and PsbA (Fig. 6A). Additionally, the plastome-encoded NAD(P)H dehydrogenase subunit5 (NdhF) and NAD(P)H-quinone oxidoreductase subunit H (NdhH), as well as nucleus-encoded plastocyanin, β-subunit (AtpB), and epsilon-subunit (AtpE) of ATP synthase, showed notably decreased levels in cnl1 compared with wild type (Fig. 6A). The observed reduction of almost all examined photosynthetic proteins in cnl1 might be caused by a generally impaired rate of chloroplast protein synthesis in the mutant. Besides the thylakoid membrane proteins, soluble chloroplast proteins also should be affected by this defect. Hence, we further performed polyacrylamide SDS-PAGE to analyze the total proteins extracted from wild-type and cnl1 flag leaves. As a soluble representative of total proteins, the level of large subunit of Rubisco (RbcL) was much lower in cnl1 than in the wild type (Fig. 6B). Decreased accumulation of the abundant RbcL protein indicated that the reduction of different thylakoid membrane proteins in cnl1 is the consequence of a generally defective synthesis rate of chloroplast proteins. Taken together, the results demonstrated that the thylakoid membrane protein complexes were impaired in cnl1.
Figure 6.
Levels of thylakoid membrane proteins and expression of photosynthetic genes. A, Levels of thylakoid membrane proteins detected in total proteins from flag leaves of wild type (WT) and cnl1 at the heading stage. The wild-type dilution series contained 30 μg of proteins (1×) or the indicated dilutions, and the cnl1 contained 30 μg proteins. Antibodies used for detection are indicated on the left; the asterisk indicates an unspecific signal detected by the NdhF antibody. B, Total proteins of flag leaves from wild type and cnl1 mutant separated by SDS-PAGE and indicated by Coomassie Brilliant Blue (CBB) staining. C, Levels of plastid- and nucleus-encoded mRNAs related to thylakoid membrane proteins. D, Relative expression analysis of photosynthetic genes in wild-type and cnl1. Data are means ± sd (n = 3).
We also detected the plastid- and nucleus-encoded transcripts of thylakoid membrane proteins by RT-qPCR, and 15 out of 25 thylakoid membrane protein-encoding genes were significantly up-regulated, while PsaF, Rieske, AtpH, and AtpI were significantly downregulated in cnl1 compared to wild type (Fig. 6C). Intriguingly, the immunoblot analysis showed that nearly all detected thylakoid membrane proteins were reduced in cnl1, which was not consistent with the RT-qPCR results, as most genes were upregulated. We speculated that the impaired thylakoid membrane protein accumulation was likely caused by a posttranslational defect in cnl1. The abnormal mRNA levels of photosynthesis-related genes might also imply a feedback mechanism upon the posttranslational down-regulation of thylakoid membrane proteins (Fig. 6D).
OsABCI7 Interacts with OsHCF222 in Chloroplasts
To determine the functional pathway of OsABCI7, we performed a yeast two-hybrid (Y2H) assay to screen for proteins that interact with OsABCI7. A total of 132 colonies were found positive by expressing the β-galactosidase (lacZ) reporter gene. Among 67 positive colonies sequenced, nine corresponded to rice LOC_Os03g30092. In the rice database (http://rice.plantbiology.msu.edu/), we found an orthologous gene of LOC_Os03g30092 in Arabidopsis named HIGH CHLOROPHYLL FLUORESCENCE222 (AtHCF222); hence, we referred to rice LOC_Os03g30092 as OsHCF222. To verify the localization of OsHCF222, GFP fused to the C terminus of OsHCF222 was transformed into rice protoplasts. Confocal microscopy showed GFP signals of OsHCF222-GFP fusion proteins colocalized both with the red autofluorescent signals of chloroplasts and mCherry-HDEL signals (Supplemental Fig. S9), revealing that OsHCF222 dually targeted to the chloroplasts and endoplasmic reticulum (ER), consistent with the previous localization of AtHCF222 (Hartings et al., 2017).
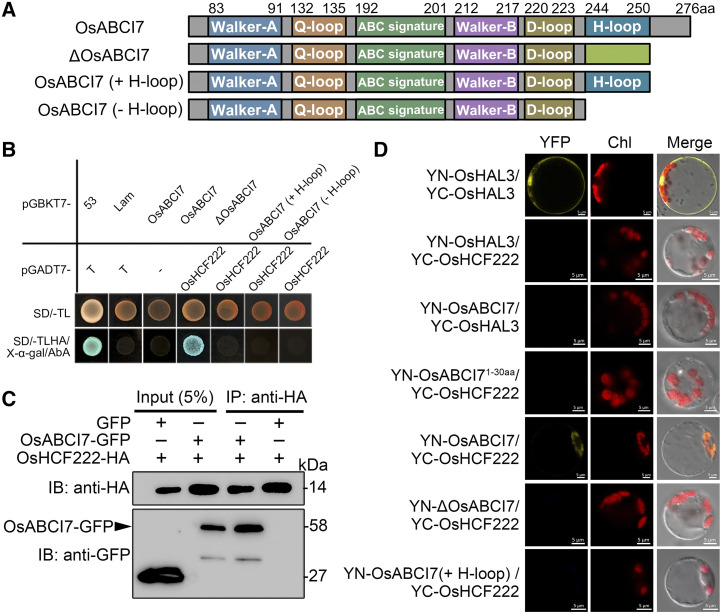
The conserved H-loop motif was deleted in ΔOsABCI7 (Fig. 7A; Supplemental Fig. S2). In yeast (Saccharomyces cerevisiae) cells and a bimolecular fluorescence complementation (BiFC) assay, we observed the interaction between OsABCI7 and OsHCF222, but no interaction between ΔOsABCI7 and OsHCF222 (Fig. 7, B and D; Supplemental Fig. S10). We therefore speculated that the H-loop motif of OsABCI7 was required for the interaction. Using Y2H and BiFC assay, we also examined the interaction between OsABCI7 with/without H-loop motif and OsHCF222. The results exhibited that both OsABCI7 (+H-loop) and OsABCI7 (−H-loop) could not interact with OsHCF222 (Fig. 7, B and D; Supplemental Fig. S10). These suggested that the C terminus of OsABCI7 was essential for the interaction between OsABCI7 and OsHCF222. Additionally, this interaction was furtherly confirmed by coimmunoprecipitation (co-IP) and BiFC assay in vivo, indicating that OsABCI7 interacts with OsHCF222 in rice chloroplasts and Nicotiana benthamiana cells (Fig. 7, C and D; Supplemental Fig. S10). All these demonstrated the interactions of OsABCI7 and OsHCF222 in vivo and vitro.
Figure 7.
Interaction of OsABCI7 and OsHCF222. A, Diagrams of the wild-type OsABCI7, mutant OsABCI7 (ΔOsABCI7), OsABCI7 with H-loop motif (OsABCI7 [+H-loop]), and OsABCI7 without H-loop motif (OsABCI7 [−H-loop]). The number of amino acids is indicated at the top. B, Y2H assay for the interaction of OsABCI7 and OsHCF222. The pGBKT7-53 and pGADT7-T pair is the positive control, the pGBKT7-Lam and pGADT7-T pair is the negative control, and the pGBKT7-OsABCI7 and pGADT7 pair is the self-activation test. SD/−TL, Synthetic dropout medium without Trp and Leu; SD/−TLHA, synthetic dropout medium without Trp, Leu, His, and Adenosine; X-α-gal, 5-bromo-4-chloro-3-indoxyl-α-d-galactopyranoside; AbA, aureobasidin A. C, Co-IP assay confirmation of OsABCI7 and OsHCF222 interaction. D, BiFC assay for OsABCI7 and OsHCF222 interaction in rice protoplasts. YN-OsHAL3/YC-OsHAL3 is the positive control, YN-OsHAL3/YC-OsHCF222 and YN-ABCI7/YC-OsHAL3 are the negative controls, YN-OsABCI71–30aa/YC-OsHCF222 is a negative control to ensure that the YFP signals of YN-OsABCI7/YC-OsHCF222 are caused by the interaction between OsABCI7 and OsHCF222 instead of the chloroplast colocalization of OsABCI7 and OsHCF222. Bars = 5 μm.
OsHCF222 Is Required for Biogenesis of the Thylakoid Membrane
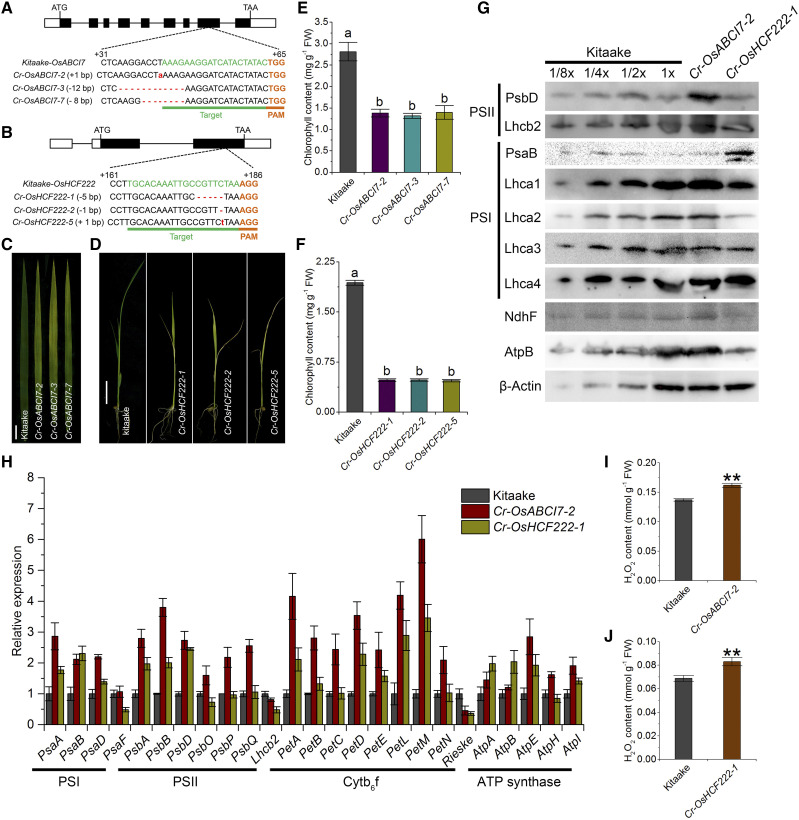
To confirm whether both OsABCI7 and OsHCF222 are essential for chloroplast development, we used CRISPR/Cas9 editing to generate Cr-OsABCI7 and Cr-OsHCF222 mutants in the cultivar Kitaake background and identified 13 Cr-OsABCI7 and 18 Cr-OsHCF222 knockout plants (Fig. 8, A and B). Expectedly, all Cr-OsABCI7 knockout plants showed similar phenotypes to cnl1 with degraded chlorophyll contents (Fig. 8, C and E; Supplemental Fig. S11), while all Cr-OsHCF222 knockout plants exhibited severe growth retardation and much paler leaf color compared with Kitaake, and, strikingly, homozygous Cr-OsHCF222 plants were lethal approximately 12 d after germination under conditions of soil cultivation (Fig. 8, D and F). Thus, all progenies of Cr-OsHCF222 homozygous plants were obtained from Cr-OsHCF222 heterozygous plants. Among 16 and 12 T1 plants randomly chosen from two independent heterozygous Cr-OsHCF222 lines, four and three homozygous plants were found, respectively, showing a mendelian 3:1 (wild type:mutant) ratio that indicated a single gene recessive nature of OsHCF222. On Suc-supplemented one-half strength Murashige and Skoog (MS) culture medium, the growth of Cr-OsHCF222 plants was slightly retarded and the leaves were paler than those of Kitaake, whereas on Suc-free one-half strength MS medium, the Cr-OsHCF222 plants showed more severe phenotypes than in Suc-supplemented one-half strength MS medium (Supplemental Fig. S12A). These indicated that the CRISPR/Cas9-mediated mutations of OsHCF222 resulted in nonautotrophic seedling-lethal mutants.
Figure 8.
Characterization of CRISPR/Cas9-edited OsABCI7 and OsHCF222 mutations in the Kitaake background. A and B, CRISPR/Cas9-mediated mutations at the target sites of OsABCI7 and OsHCF222 in representative knockout lines. The sgRNA target sequence is underlined in green, and the PAM motif is indicated in orange. C, Phenotypes of flag leaves in Kitaake and Cr-OsABCI7 knockout lines at 60 d after transplanting. D, Seedlings of T1 Cr-OsHCF222 knockout lines at 12 d after germination. Bars = 2 cm. E and F, Chlorophyll contents of Cr-OsABCI7 and Cr-OsHCF222 knockout lines in C and D. Different letters indicate significant differences by one-way ANOVA and Duncan’s test (P < 0.05). Data are means ± sd (n = 3). G, Levels of thylakoid membrane proteins tested in total proteins extracted from Kitaake, Cr-OsABCI7, and Cr-OsHCF222 knockout lines at 8 d after germination. H, Detection of mRNA levels of plastid- and nucleus-encoded genes related to thylakoid membrane proteins in Cr-OsABCI7-2 and Cr-OsHCF222-1 lines at 8 DAG. I and J, Measurements of H2O2 contents in Cr-OsABCI7-2 and Cr-OsHCF222-1 at 8 DAG. Data are means ± sd (n = 3). Asterisks indicate significant difference by Student’s t test (**P < 0.01). FW, Fresh weight.
The levels of thylakoid proteins in the representative Cr-OsABCI7-2 and Cr-OsHCF222-1 lines were compared by immunoblot analysis. In both CRISPR/Cas9 mutants, the levels of Lhcb2, Lhca3, and Lhca4 were similar to those of Kitaake, while pronounced protein reductions were found for Lhca1, Lhca2, NdhF, and AtpB in Cr-OsHCF222-1 compared with Kitaake and Cr-OsABCI7-2 (Fig. 8G). In contrast, PsbD and PsaB were highly accumulated in Cr-OsABCI7-2 and Cr-OsHCF222-1, respectively (Fig. 8G). Compared to Kitaake, the expression of photosynthesis-related genes varied in Cr-OsABCI7-2 but were all downregulated in Cr-OsHCF222-1 (Supplemental Fig. S12B). Moreover, we also analyzed the expression levels of 25 thylakoid membrane genes in both mutants, and 22 genes except PsaF, Lhcb2, and Rieske were up-regulated in Cr-OsABCI7-2 compared with Kitaake, and a similar upregulated expression pattern was detected in Cr-OsHCF222-1 compared with Kitaake (Fig. 8H). This suggested that the higher transcriptional levels of thylakoid membrane genes in Cr-OsABCI7-2 and Cr-OsHCF222-1 did not contribute to the synthesis of thylakoid proteins. Considering that OsABCI7/OsHCF222 interaction occurs in chloroplasts, the main ROS-generating organelle, we measured the H2O2 contents and found both Cr-OsABCI7-2 and Cr-OsHCF222-1 had significantly increased H2O2 levels compared with Kitaake (Fig. 8, I and J).
Exogenous Antioxidants Alleviate the cnl1 Phenotype
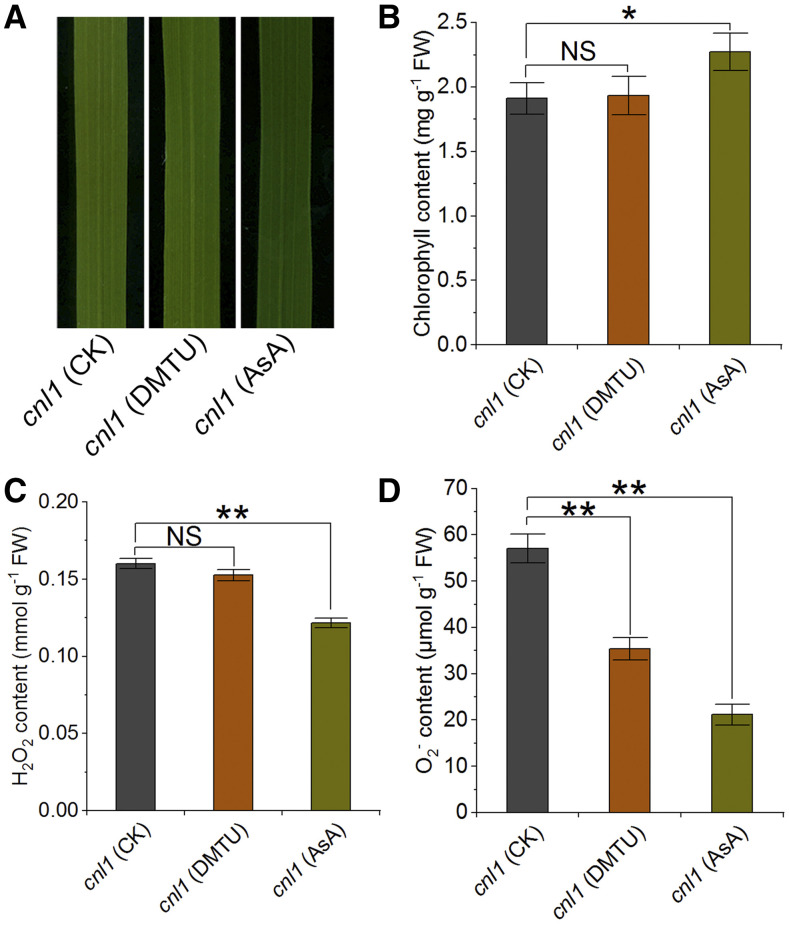
Considering the significant reduction of ROS levels in OsABCI7 overexpression plants and significant accumulation in Cr-OsHCF222 knockout plants (Fig. 2, G, H, and J), we hypothesized that OsABCI7 may interact with OsHCF222 to regulate the balance of ROS production and scavenging in chloroplasts. To demonstrate this, cnl1 seedlings were treated with the exogenous antioxidants dimethythiourea (DMTU) and AsA (Asada, 2006; Wang et al., 2018). Under normal hydroponic conditions, cnl1 exhibited chlorophyll degradation and H2O2 accumulation compared to wild type (Supplemental Fig. S13), while the cnl1 seedlings treated with 1 mm exogenous AsA displayed an eased yellowish phenotype with significantly alleviated chlorophyll degradation (Fig. 9, A and B). We then measured the contents of H2O2 and O2− in cnl1 under different antioxidant treatments. Higher levels of H2O2 and O2− were both detected in untreated cnl1 seedlings compared with cnl1 seedlings treated with 1 mm AsA (Fig. 9, C and D). As for DMTU treatment, only O2− but not H2O2 quenching was observed in treated cnl1 seedlings (Fig. 9). Upon these results, we inferred that excessive ROS was responsible for the cnl1 phenotype, and the exogenous AsA alleviated the yellowish phenotype of cnl1 by relieving the ROS stress.
Figure 9.
Responses of cnl1 to exogenous antioxidant treatment. A, Phenotypes of fully expanded top-second leaves of cnl1 under 1 mm DMTU and 1 mm AsA treatments. cnl1 was hydroponically cultured for 4 d after germination and then treated with 1 mm DMTU or 1 mm AsA for 8 d. CK, Control. B to D, Chlorophyll (B), H2O2 (C), and O2− contents (D) of cnl1 in A. Data are means ± sd (n = 3). Asterisks indicate significant difference by Student’s t test (*P < 0.05 and **P < 0.01). NS, No significance.
DISCUSSION
OsABCI7 Functions in Chloroplast Development but Its Function May Differ between Monocots and Dicots
Many leaf pigmentation mutants are usually associated with chloroplast development in rice (Wang et al., 2016; Lv et al., 2017; Sun et al., 2017; Liu et al., 2018; Cui et al., 2019). In this study, we isolated the constitutively expressed OsABCI7, which encodes a chloroplast-localized ABC transporter protein (Fig. 4). Due to the mutation of OsABCI7, cnl1 plants mainly showed chlorotic and necrotic leaves with chloroplast structure disintegration, chlorophyll degradation, and photosynthesis impairment, finally leading to the deteriorated yield performance (Fig. 1, A–I; Supplemental Fig. S1). The chloroplast development of temperature-sensitive mutants, like tsv, wsl5, and dua1, is easily hindered by low temperatures in rice (Sun et al., 2017; Liu et al., 2018; Cui et al., 2019). For cnl1, we also observed yellowish leaves with significantly decreased chlorophyll levels and degradation of chloroplast ultrastructure at 20°C and altered mRNA levels of OsABCI7 in response to the low-temperature stress (Fig. 5, A–G; Supplemental Fig. S8). Unlike most temperature-sensitive rice mutants, ΔOsABCI7 leads to disabled light-dependent chlorophyll synthesis in cnl1 during the greening process of etiolated seedlings (Fig. 5, I–K). Taken together, we conclude that OsABCI7 plays a crucial role in chloroplast biogenesis, especially under low-temperature stress and in the greening process.
The homolog of rice OsABCI7, Arabidopsis AtABCI11 (AtNAP14), participates in the regulation of chloroplast metal homeostasis and attaches to the inner envelope membrane of the chloroplast; compared to the wild type (Col-0), metal elemental contents including iron (Fe), zinc (Zn), molybdenum (Mo), and copper (Cu) are higher, while manganese (Mn) is lower in Atabci11 mutant shoot tissues, whereas Fe and Zn contents in Atabci11 were lower than those of Col-0 in root tissues (Shimoni-Shor et al., 2010; Voith von Voithenberg et al., 2019). Hence, we determined the contents of metal elements, including magnesium (Mg), potassium (K), Fe, Cu, and Zn in the shoot and root tissues of wild type and cnl1. Inconsistent with the metal distribution of Atabci11 mutants, only significant differences in Mn (1.2-fold in shoot and 2.2-fold in root) and Cu (0.8-fold in shoot and root) were detected (Supplemental Fig. S14). Mn is integral to photosynthetic electron transport, redox processes, and ROS scavenging in plants, while excessive Mn is toxic to the photosynthetic apparatus and inhibits chloroplast protein and chlorophyll synthesis (Millaleo et al., 2013; Eisenhut et al., 2018). Cu is an important minor element found in chloroplasts as a cofactor with Cu/Zn-SOD, and Cu/Zn-SOD contributes to the reduction of oxidative stress (Bowler et al., 1994; Cohu et al., 2009). The observed Mn overload and lower Cu concentration in cnl1 may explain the chlorotic and necrotic leaf phenotypes, which are likely due to the increased ROS stress generated by the overaccumulation of Mn and decreased levels of Cu/Zn-SOD. As a transporter, OsABCI7 plays an important role in regulating chloroplast metal homeostasis (Supplemental Fig. S14). Based on this, we proposed that OsABCI7 could be a component of the chloroplast transporter complex. In addition, unlike the chloroplast inner envelope membrane-localized AtABCI11, OsABCI7 targets to the thylakoid membrane in chloroplasts (Fig. 4B). Considering the pea (Pisum sativum) samples used for the in vivo localization detection of AtABCI11 (Voith von Voithenberg et al., 2019), further studies need to be conducted in Arabidopsis to confirm the true in vivo localization of AtABCI11. In conclusion, these deviations of OsABCI7 and AtABCI11 might suggest different regulatory manners between monocots and dicots.
OsABCI7 Is Required for ROS-Mediated Thylakoid Membrane Stability
During the photosynthesis process at the thylakoid membrane, O2 molecules are reduced to O2− by trapping electrons derived from water in PSI, and O2− is disproportionated to H2O2, catalyzed by chloroplastic SOD (Asada, 2006). Both overaccumulated ROS and a disrupted ROS scavenging system were detected in cnl1, indicating the impairment of ROS homeostasis (Fig. 2, G–K). As the consequence of overaccumulation of ROS, DNA fragmentation and DNA damage repair response activation were observed in cnl1, which exhibited an enhanced PCD manifested by trypan blue staining, TUNEL assay, and comet assay (Fig. 3; Supplemental Figs. S6C and S7). Accumulating evidence also demonstrates that ROS and cell death usually occur together (Roy et al., 2012; Luo et al., 2013; He et al., 2018a; Qiu et al., 2019; Zafar et al., 2020).
ROS are extremely active and able to interact with many kinds of target molecules and metabolites, like DNA, proteins, lipids, pigments, and other cellular molecules, resulting in a number of destructive cell processes (Ashraf, 2009). An accurate control of steady-state H2O2 concentration is critical to cell homeostasis, and H2O2 is directly scavenged by the antioxidant AsA with the catalysis of APX in healthy chloroplasts (Zhen et al., 2011; Ni et al., 2018). Notably, endogenous ROS contents of OsABCI7 overexpression plants were significantly lower than those of wild type, which was well explained by the higher level of APX activity in overexpression plants compared to wild type (Fig. 2, G–I). We therefore speculated that OsABCI7 is closely associated with the APX-related ROS scavenging pathway. As a consequence of OsABCI7 disruption, newly produced ROS could not be eliminated effectively in APX pathway, leading to overaccumulation of ROS in chloroplasts. Conversely, overaccumulated ROS directly gave rise to thylakoid membrane damage, chlorophyll breakdown, and photosynthetic protein degradation, which may be the reason for the initiation of the cnl1 phenotype (Figs. 1, A–D, and 6, A and B). The above hypothesis was further confirmed by the easing of the cnl1 phenotype under the treatment of exogenous antioxidant AsA (Fig. 9). In conclusion, these findings suggested that OsABCI7 is essential for ROS-mediated thylakoid membrane stability.
OsABCI7 Interacts with OsHCF222 at the Thylakoid Membrane to Regulate ROS Homeostasis
HCF proteins are required for the formation of thylakoid membrane complexes in plants (Schult et al., 2007; Link et al., 2012; Hartings et al., 2017). The Arabidopsis AtHCF222 encodes a small protein homologous to the zinc finger domain of Escherichia coli chaperone DnaJ-like protein (Hartings et al., 2017). OsHCF222 shares 59% protein identity to AtHCF222 and is also characterized by eight conserved Cys-rich (CR) domains arranged in four CxxC motif clusters (Supplemental Fig. S15). Between the Cys residues of four CxxC motifs, two Zn(II) ions are coordinated to form a topology with two C4-type zinc finger structures (Shi et al., 2005). As members of the heat shock protein family, DnaJ proteins serve as cochaperones and involve in different substrate bindings, playing important roles in multiple biotic and abiotic stress responses (Liu and Whitham, 2013; Pulido and Leister, 2018).
Our results revealed physical interaction between OsABCI7 and OsHCF222 in vitro and vivo (Fig. 7, B–D; Supplemental Fig. S10). AtHCF222 is a dually ER- and chloroplast-localized protein and has been detected in leaf whole-cell membrane fractions (Hartings et al., 2017). In this study, OsHCF222 dually localized to the ER and chloroplasts, and OsABCI7 was detected in thylakoid membrane fractions by immunoblot (Fig. 4B). Upon these findings, we speculated that OsABCI7 interacts with OsHCF222 at thylakoid membranes to regulate ROS homeostasis for maintaining thylakoid integrity. To test this assumption, CRISPR/Cas9 editing was performed to obtain the corresponding knockout mutants of OsABCI7 and OsHCF222. Unlike the Cr-OsABCI7 plants, Cr-OsHCF222 lines displayed chlorotic leaves and seedling lethality (Fig. 8, C and D). In spite of the major differences in seedling lethality and photosynthetic protein accumulation between the two types of knockout lines, ROS overaccumulation occurred in both Cr-OsABCI7 and Cr-OsHCF222 mutants (Fig. 8, G, I, and J). Rice DnaJ proteins are involved in chloroplast development and H2O2-induced DNA damage (Yamamoto et al., 2005; Zhu et al., 2015). Thus, we conclude that both OsABCI7 and OsHCF222 are essential for the thylakoid membrane development, and the OsABCI7/OsHCF222 complex plays a vital role in ROS homeostasis regulation at thylakoids. This explained that thylakoid degradation of cnl1 was closely associated with the disruption of OsABCI7/OsHCF222 complex formation in cnl1 (Fig. 7, A and B).
In summary, our data support the idea that OsABCI7 interacts with OsHCF222 to form a complex functioning at the thylakoid membranes in chloroplasts. We propose that the interaction is crucial to maintain thylakoid stability by regulating ROS homeostasis in chloroplasts. In cnl1, the disrupted interaction causes a rapid ROS burst, resulting in thylakoid instability, chloroplast breakdown, and cell death, which might in turn reinforce ROS accumulation but in a controllable manner, as cnl1 itself and the knockout lines are viable. On the other hand, OsHCF222 knockout lines are unviable, probably indicating that OsHCF222 is not only required for thylakoid stability but also for thylakoid membrane complex formation, as demonstrated in Arabidopsis (Schult et al., 2007; Link et al., 2012; Hartings et al., 2017).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The cnl1 mutant was obtained from an ethyl methylsulfonate-induced indica rice (Oryza sativa) Zhongjian100 (wild type) mutant bank. cnl1 mutants were crossed to the japonica rice ‘ReYan 1,’ ‘ORO,’ and wild type, respectively. All the parents, F1 plants, and F2 individuals were grown in the paddy field at Zhejiang province for genetic analysis and/or gene mapping. For the exogenous antioxidant treatments, plants were hydroponically cultured with Yoshida rice nutrient salt mixture (Coolaber, NSP1040) after germination in a growth chamber at 30°C, 14 h light/26°C, 10 h dark cycle. The cnl1 seedlings without antioxidant treatment were used as the control (CK). The nutrient solution containing either 1mm DMTU or 1 mm AsA was used for antioxidant treatments.
Chlorophyll Measurement, TEM, and Paraffin Sectioning
Chlorophyll content was determined by measuring the A652 using a SpectraMax i3x multi-mode microplate reader (Molecular Devices) as described previously (Kim et al., 2006). Leaves were collected to perform TEM according to a previous report (He et al., 2018a). The third internodes of the main stem from wild type and cnl1 at the mature stage were used for paraffin sections as described previously (He et al., 2017).
MDA, ROS Content, and Antioxidant Activity Measurement
The contents of MDA, as well as the activities of ROS scavenging enzymes, including peroxidase, SOD, and CAT, were determined using the kits (A003-1-2, A084-1-1, A001-1-1, and A007-1-1) following the manufacturer’s instructions (Nanjing Jiancheng Bioengineering Institute). H2O2 content was determined using the H2O2 assay kit (Nanjing Jiancheng Bioengineering Institute), and the level of O2− was quantified using a kit (R30343) following the manufacturer’s instructions (Shanghai Yuanye Bio-Technology).
Map-Based Cloning
The mutant F2 individuals from the cnl1/cv ReYan 1 cross were selected for mapping the OsABCI7 locus. Bulked segregant analysis was adopted to rapidly locate the mutation as follows: equal amounts of leaf blades from each of 10 wild-type plants, and 10 cnl1 plants were sampled for DNA extraction to form a wild-type DNA pool and a mutant DNA pool, respectively. The parents and the two DNA pools were subjected to preliminary linkage analysis of the mutation by genotyping 362 polymorphic simple sequence repeat markers covering 12 chromosomes. Subsequently, 755 cnl1 individual plants from the F2 population were genotyped to determine the physical location of OsABCI7. Primers used for mapping are listed in Supplemental Table S3.
RNA Extraction and RT-qPCR
Total RNA was extracted using a NucleoZOL reagent kit (MACHEREY-NAGEL) according to the manufacturer’s instructions. One microgram total RNA was reverse-transcribed using the ReverTra Ace RT-qPCR RT master mix with genomic DNA remover kit (Toyobo). Rice Ubiquitin (LOC_Os03g13170) was used as an internal control. Primers used for RT-qPCR are listed in Supplemental Table S3.
Generation of Transgenic Plants
For the complementation of cnl1, the wild-type allele containing the 1788-bp promoter region, entire genomic DNA region (3384 bp), and 711 bp downstream of OsABCI7 was cloned into the vector pCAMBIA1300. To generate the overexpression lines, the 831-bp CDS of OsABCI7 was cloned into the vector pCAMBIA1300-UBI under the control of the maize (Zea mays) Ubiquitin1 promoter. For the promoter activity assay of OsABCI7, the OsABCI7 promoter was inserted into the vector pCAMBIA1381Z containing the GUS gene. The CRISPR/Cas9 constructs for OsABCI7 and OsHCF222 were generated according to a previous report (Ma et al., 2015). The genetic transformations were conducted using rice embryogenic calli through Agrobacterium tumefaciens-mediated transformation (Hiei and Komari, 2008). Primers used for vector constructions are listed in Supplemental Table S3.
Subcellular Localization and Chloroplast-Localized Signal Peptide Analysis
To determine the subcellular localization of OsABCI7 and OsHCF222, the CDS of OsABCI7 and OsHCF222 amplified from wild type were fused to the N terminus of GFP in the pAN580 (GFP) vector, respectively. To determine the chloroplast-localized signal peptide region, different truncated CDS of OsABCI7 were also fused to the N terminus of GFP in the pAN580 vector. All these constructs were separately introduced into rice protoplasts for transient expression, and the GFP fluorescence signals were detected using a Zeiss lsm710 confocal laser scanning microscope (Carl Zeiss).
OsABCI7Pro::GUS Staining Assay
Different tissues from OsABCI7-promoter-GUS transgenic plants were collected at different growth stages and stained using a GUS staining kit (Coolaber, SL7160), following the manufacturer’s instructions. Images were photographed using a stereomicroscope (Leica MC120 HD).
Comet Assay
The alkaline comet assay was performed according to Wang and Liu (2006) using the Comet assay kit from Trevigen. The SYBR green I nucleic acid gel stain (S9430) purchased from Sigma-Aldrich was used for staining. Thirty to 70 comets were counted in each slide, and DNA damage was classified into four degrees as previously reported (Yuan et al., 2010).
Y2H Assay
The cDNA library constructed from Nipponbare embryogenic calli and the screening kit (Clontech) were used for Y2H assays. All protocols were carried out strictly according to the manufacturer’s user manual. The full CDS of OsABCI7 and OsHCF222 amplified from wild-type cDNA and the mutant full CDS amplified from cnl1 were cloned into the pGBKT7 or pGADT7 vector, respectively. The truncated CDS of OsABCI7 with/without specific domains from the wild type were also cloned into the pGBKT7 vector. The yeast (Saccharomyces cerevisiae) strain Y2HGold was used in the Y2H assay. Primers used are listed in Supplemental Table S3.
BiFC Assay
For BiFC assays, the entire CDS of OsABCI7, 90 bp of the N-terminal OsABCI7 CDS, the CDS of ΔOsABCI7, and OsABCI7 with the H-loop motif were cloned into the 1300-YN vector. The entire CDS of OsHCF222 was cloned into the 2300-YC vector. The new constructs were electroporated into A. tumefaciens strain GV3101 for transient expression in Nicotiana benthamiana leaves and were also introduced into rice protoplasts for transient expression. The YFP fluorescent signals in the N. benthamiana leaves were detected ∼48 h after transfection, and the YFP signals in rice protoplasts were observed about 36 h after expression.
Protein Extraction and Immunoblot Analysis
Total proteins were extracted as previously described (He et al., 2018b) and quantified into the same concentration using the BCA protein assay kit (Coolaber, SK1070). For intact chloroplast isolation, 1 g fresh leaf tissue from OsABCI7 overexpression seedlings was collected for chloroplast isolation using the chloroplast isolation kit (BestBio, BB-3622) following the manufacturer’s instructions. To directly isolate the thylakoid membranes, 1 g fresh leaf tissue was homogenized on ice with 5 mL extraction buffer (0.4 m Suc, 10 mm NaCl, 2 mm MgCl2, and 50 mm HEPES), then filtered through two layers of 40 μm nylon membrane and centrifuged for 10 min at 4°C, 5000g. After discarding the supernatant, the sediment was rinsed with 1 mL extraction buffer, then centrifuged for 10 min at 4°C, 5000g, repeating this step twice. Finally, the sediment was resuspended with the extraction buffer and stored at −80°C. Antibodies used for immunoblot analysis of thylakoid membrane proteins were purchased from Agrisera, and the products and catalog numbers are as follows: anti-PsaA (AS06172), anti-PsaB (AS10695), anti-Lhca1 (AS01005), anti-Lhca2 (AS01006), anti-Lhca3 (AS01007), anti-Lhca4 (AS01008), anti-PsbA (AS132669), anti-PsbD (AS06146), anti-Lhcb1 (AS01004), anti-Lhcb2 (AS01003), anti-NdhH (AS132712), anti-NdhF (AS132711), anti-plastocyanin (AS06141), anti-AtpB (AS05085), and anti-AtpE (AS101586). The protein levels were detected using a super ECL western blotting substrate (Coolaber, SL1350).
Co-IP Assay
For the Co-IP assay, the full CDS of OsABCI7 was cloned into pYBA1132-GFP, and the CDS of OsHCF222 was cloned into pYBA1132-HA, respectively. The constructs were transiently expressed in N. benthamiana leaves via A. tumefaciens-mediated transfection. About 60 h after transfection, fresh N. benthamiana leaves were harvested for total protein extraction using the buffer (0.4 mm Tris-HCl, pH 7.5, 5 mm NaCl, 6.25 µm MgCl2, 10 µm EDTA, and 1% Triton X-100) with proteinase inhibitor cocktail (Roche, 04693159001). The supernatant of total proteins was collected after centrifugation at 12,000g for 20 min at 4°C. The Pierce HA-tag magnetic IP/co-IP kit (88838X) was used for the Co-IP assay following the manufacturer’s manual. Anti-GFP (Abmart, M200045) and anti-HA (CWBIO, CW0092M) antibodies were used for detection in immunoblot analysis.
Accession Numbers
Sequence data from this article for the cDNA and genomic DNA of OsABCI7 and OsHCF222 can be found in the GenBank/EMBL/Gramene data libraries under accession numbers LOC_Os11g29850 and LOC_Os03g30092, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic and photosynthetic analysis of wild type and cnl1.
Supplemental Figure S2. CDS of OsABCI7 and amino acid sequence of OsABCI7.
Supplemental Figure S3. Mutation analysis of OsABCI7.
Supplemental Figure S4. Overexpression of OsABCI7 and analysis of OsABCI7-RNA-interference lines.
Supplemental Figure S5. Soluble protein contents and SOD activities in wild type, cnl1, complementary, and overexpression lines.
Supplemental Figure S6. Chlorophyll contents and histochemical analysis.
Supplemental Figure S7. TUNEL assay of wild type and cnl1.
Supplemental Figure S8. Chlorophyll contents and OsABCI7 expression analysis.
Supplemental Figure S9. Subcellular localization of OsHCF222.
Supplemental Figure S10. BiFC for the interaction of OsABCI7 and OsHCF222 in N. benthamiana.
Supplemental Figure S11. Knockout analysis of OsABCI7 by CRISPR/Cas9 technology.
Supplemental Figure S12. Phenotypes of OsHCF222 knockout line and expression of photosynthesis-related genes in OsABCI7 and OsHCF222 knockout lines.
Supplemental Figure S13. Chlorophyll and H2O2 contents in wild-type and cnl1 seedlings.
Supplemental Figure S14. Levels of metal elements in shoots and roots of wild type and cnl1.
Supplemental Figure S15. Alignment of HCF222 orthologs from different plant species.
Supplemental Table S1. List of ORFs in the 116-kb target region.
Supplemental Table S2. Chlorophyll fluorescence parameters measured in flag leaves of wild-type, cnl1, complementary, and overexpression lines.
Supplemental Table S3. Primers used in this study.
Footnotes
This work was supported by the Ministry of Science and Technology of China (grant no. 2016YFD0101104) and the State Key Laboratory of Rice Biology.
Articles can be viewed without a subscription.
References
- Ahmad P, Jaleel CA, Salem MA, Nabi G, Sharma S(2010) Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit Rev Biotechnol 30: 161–175 [DOI] [PubMed] [Google Scholar]
- Asada K.(1992) Ascorbate peroxidase—a hydrogen peroxide-scavenging enzyme in plants. Physiol Plant 85: 235–241 [Google Scholar]
- Asada K.(2006) Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol 141: 391–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K, Kiso K, Yoshikawa K(1974) Univalent reduction of molecular oxygen by spinach chloroplasts on illumination. J Biol Chem 249: 2175–2181 [PubMed] [Google Scholar]
- Ashraf M.(2009) Biotechnological approach of improving plant salt tolerance using antioxidants as markers. Biotechnol Adv 27: 84–93 [DOI] [PubMed] [Google Scholar]
- Bowler C, Van Camp W, Van Montagu M, Inze D, Asada K(1994) Superoxide dismutase in plants. Crit Rev Plant Sci 13: 199–218 [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D(2003) The molecular analysis of leaf senescence—a genomics approach. Plant Biotechnol J 1: 3–22 [DOI] [PubMed] [Google Scholar]
- Cohu CM, Abdel-Ghany SE, Gogolin Reynolds KA, Onofrio AM, Bodecker JR, Kimbrel JA, Niyogi KK, Pilon M(2009) Copper delivery by the copper chaperone for chloroplast and cytosolic copper/zinc-superoxide dismutases: Egulation and unexpected phenotypes in an Arabidopsis mutant. Mol Plant 2: 1336–1350 [DOI] [PubMed] [Google Scholar]
- Cui X, Wang Y, Wu J, Han X, Gu X, Lu T, Zhang Z(2019) The RNA editing factor DUA1 is crucial to chloroplast development at low temperature in rice. New Phytol 221: 834–849 [DOI] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB(2006) ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J 48: 947–961 [DOI] [PubMed] [Google Scholar]
- Durrant WE, Wang S, Dong X(2007) Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc Natl Acad Sci USA 104: 4223–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Hoecker N, Schmidt SB, Basgaran RM, Flachbart S, Jahns P, Eser T, Geimer S, Husted S, Weber APM, et al. (2018) The plastid envelope CHLOROPLAST MANGANESE TRANSPORTER1 is essential for manganese homeostasis in Arabidopsis. Mol Plant 11: 955–969 [DOI] [PubMed] [Google Scholar]
- Garcia O, Bouige P, Forestier C, Dassa E(2004) Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette (ABC) systems. J Mol Biol 343: 249–265 [DOI] [PubMed] [Google Scholar]
- Garroum I, Bidzinski P, Daraspe J, Mucciolo A, Humbel BM, Morel JB, Nawrath C(2016) Cuticular defects in Oryza sativa ATP-binding cassette transporter G31 mutant plants cause dwarfism, elevated defense responses and pathogen resistance. Plant Cell Physiol 57: 1179–1188 [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N(2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48: 909–930 [DOI] [PubMed] [Google Scholar]
- Hartings S, Paradies S, Karnuth B, Eisfeld S, Mehsing J, Wolff C, Levey T, Westhoff P, Meierhoff K(2017) The DnaJ-like zinc-finger protein HCF222 is required for thylakoid membrane biogenesis in plants. Plant Physiol 174: 1807–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Li L, Zhang Z, Wu JL(2018a) Identification and comparative analysis of premature senescence leaf mutants in rice (Oryza sativa L.). Int J Mol Sci 19: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Shi YF, Zhang XB, Wang HM, Xu X, Wu JL(2017) Identification of a gravitropism-deficient mutant in rice. Rice Sci 24: 109–118 [Google Scholar]
- He Y, Zhang Z, Li L, Tang S, Wu JL(2018b) Genetic and physio-biochemical characterization of a novel premature senescence leaf mutant in rice (Oryza sativa L.). Int J Mol Sci 19: 2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Komari T(2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc 3: 824–834 [DOI] [PubMed] [Google Scholar]
- Huang CF, Yamaji N, Mitani N, Yano M, Nagamura Y, Ma JF(2009) A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 21: 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang QN, Shi YF, Zhang XB, Song LX, Feng BH, Wang HM, Xu X, Li XH, Guo D, Wu JL(2016) Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J Integr Plant Biol 58: 12–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I(2006) Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc Natl Acad Sci USA 103: 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kondo M, Fukuda H, Nishimura M, Ohta H(2007) Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc Natl Acad Sci USA 104: 17216–17221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link S, Engelmann K, Meierhoff K, Westhoff P(2012) The atypical short-chain dehydrogenases HCF173 and HCF244 are jointly involved in translational initiation of the psbA mRNA of Arabidopsis. Plant Physiol 160: 2202–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Whitham SA(2013) Overexpression of a soybean nuclear localized type-III DnaJ domain-containing HSP40 reveals its roles in cell death and disease resistance. Plant J 74: 110–121 [DOI] [PubMed] [Google Scholar]
- Liu X, Lan J, Huang YS, Cao PF, Zhou CL, Ren YK, He NQ, Liu SJ, Tian YL, Nguyen T, et al. (2018) WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress. J Exp Bot 69: 3949–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Yang A, Schulte BA, Wargovich MJ, Wang GY(2013) Resveratrol induces premature senescence in lung cancer cells via ROS-mediated DNA damage. PLoS One 8: e60065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y, Shao G, Qiu J, Jiao G, Sheng Z, Xie L, Wu Y, Tang S, Wei X, Hu P(2017) White Leaf and Panicle 2, encoding a PEP-associated protein, is required for chloroplast biogenesis under heat stress in rice. J Exp Bot 68: 5147–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyska D, Meierhoff K, Westhoff P(2013) How to build functional thylakoid membranes: From plastid transcription to protein complex assembly. Planta 237: 413–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Matsuda S, Funabiki A, Furukawa K, Komori N, Koike M, Tokuji Y, Takamure I, Kato K(2012) Genome-wide analysis and expression profiling of half-size ABC protein subgroup G in rice in response to abiotic stress and phytohormone treatments. Mol Genet Genomics 287: 819–835 [DOI] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P(1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Millaleo R, Reyes-Díaz M, Alberdi M, Ivanov AG, Krol M, Hüner NP(2013) Excess manganese differentially inhibits photosystem I versus II in Arabidopsis thaliana. J Exp Bot 64: 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y, Liu H, Dai D, Mu X, Xu J, Shao S(2018) Chromogenic, fluorescent, and redox sensors for multichannel imaging and detection of hydrogen peroxide in living cell systems. Anal Chem 90: 10152–10158 [DOI] [PubMed] [Google Scholar]
- Pulido P, Leister D(2018) Novel DNAJ-related proteins in Arabidopsis thaliana. New Phytol 217: 480–490 [DOI] [PubMed] [Google Scholar]
- Qiu Z, Zhu L, He L, Chen D, Zeng D, Chen G, Hu J, Zhang G, Ren D, Dong G, et al. (2019) DNA damage and reactive oxygen species cause cell death in the rice local lesions 1 mutant under high light and high temperature. New Phytol 222: 349–365 [DOI] [PubMed] [Google Scholar]
- Roy N, Bagchi S, Raychaudhuri P(2012) Damaged DNA binding protein 2 in reactive oxygen species (ROS) regulation and premature senescence. Int J Mol Sci 13: 11012–11026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe AP, Su X, Chen Z, Chen T, Wei X, Tang S, Zhang XB, Wu JL(2019) Identification and characterization of a spotted-leaf mutant spl40 with enhanced bacterial blight resistance in rice. Rice (N Y) 12: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schult K, Meierhoff K, Paradies S, Töller T, Wolff P, Westhoff P(2007) The nuclear-encoded factor HCF173 is involved in the initiation of translation of the psbA mRNA in Arabidopsis thaliana. Plant Cell 19: 1329–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YY, Tang W, Hao SF, Wang CC(2005) Contributions of cysteine residues in Zn2 to zinc fingers and thiol-disulfide oxidoreductase activities of chaperone DnaJ. Biochemistry 44: 1683–1689 [DOI] [PubMed] [Google Scholar]
- Shimoni-Shor E, Hassidim M, Yuval-Naeh N, Keren N(2010) Disruption of Nap14, a plastid-localized non-intrinsic ABC protein in Arabidopsis thaliana results in the over-accumulation of transition metals and in aberrant chloroplast structures. Plant Cell Environ 33: 1029–1038 [DOI] [PubMed] [Google Scholar]
- Soares C, Sousa AD, Pinto A, Azenha M, Teixeira J, Azevedo RA, Fidalgo F(2016) Effect of 24-epibrassinolide on ROS content, antioxidant system, lipid peroxidation and Ni uptake in Solanum nigrum L. under Ni stress. Environ Exp Bot 122: 115–125 [Google Scholar]
- Song X, Wang D, Ma L, Chen Z, Li P, Cui X, Liu C, Cao S, Chu C, Tao Y, et al. (2012) Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J 71: 378–389 [DOI] [PubMed] [Google Scholar]
- Speit G, Hartmann A(2006) The comet assay: a sensitive genotoxicity test for the detection of DNA damage and repair. Methods Mol Biol 314: 275–286 [DOI] [PubMed] [Google Scholar]
- Sun J, Zheng T, Yu J, Wu T, Wang X, Chen G, Tian Y, Zhang H, Wang Y, Terzaghi W, et al. (2017) TSV, a putative plastidic oxidoreductase, protects rice chloroplasts from cold stress during development by interacting with plastidic thioredoxin Z. New Phytol 215: 240–255 [DOI] [PubMed] [Google Scholar]
- Voith von Voithenberg L, Park J, Stübe R, Lux C, Lee Y, Philippar K(2019) A novel prokaryote-type ECF/ABC transporter module in chloroplast metal homeostasis. Front Plant Sci 10: 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Zhang Y, Bi Z, Liu Q, Xu T, Yu N, Cao Y, Zhu A, Wu W, Zhan X, et al. (2019) Impaired function of the calcium-dependent protein kinase, OsCPK12, leads to early senescence in rice (Oryza sativa L.). Front Plant Sci 10: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu Z(2006) Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18: 350–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Dong X, Feng Y, Liu X, Wang J, Zhang Z, Li J, Zhao Y, Shi S, Tu P(2018) H2O2 and NADPH oxidases involve in regulation of 2-(2-phenylethyl)chromones accumulation during salt stress in Aquilaria sinensis calli. Plant Sci 269: 1–11 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang J, Shi X, Peng Y, Li P, Lin D, Dong Y, Teng S(2016) Temperature-sensitive albino gene TCD5, encoding a monooxygenase, affects chloroplast development at low temperatures. J Exp Bot 67: 5187–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hong X, Hu K, Wang Y, Wang X, Du S, Li Y, Hu D, Cheng K, An B, et al. (2017) Impaired magnesium protoporphyrin IX methyltransferase (ChlM) impedes chlorophyll synthesis and plant growth in rice. Front Plant Sci 8: 1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen Z, Shi YF, Wang HM, He Y, Shi L, Chen T, Wu JL, Zhang XB(2018) Functional inactivation of OsGCNT induces enhanced disease resistance to Xanthomonas oryzae pv. oryzae in rice. BMC Plant Biol 18: 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Mori Y, Ishibashi T, Uchiyama Y, Ueda T, Ando T, Hashimoto J, Kimura S, Sakaguchi K(2005) Interaction between proliferating cell nuclear antigen (PCNA) and a DnaJ induced by DNA damage. J Plant Res 118: 91–97 [DOI] [PubMed] [Google Scholar]
- Yoo SC, Cho SH, Sugimoto H, Li J, Kusumi K, Koh HJ, Iba K, Paek NC(2009) Rice virescent3 and stripe1 encoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiol 150: 388–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Luo D, Li G, Yao X, Wang H, Zeng M, Huang H, Cui X(2010) Characterization of the AE7 gene in Arabidopsis suggests that normal cell proliferation is essential for leaf polarity establishment. Plant J 64: 331–342 [DOI] [PubMed] [Google Scholar]
- Zafar SA, Patil SB, Uzair M, Fang J, Zhao J, Guo T, Yuan S, Uzair M, Luo Q, Shi J, et al. (2020) DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) encodes a cystathionine β-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytol 225: 356–375 [DOI] [PubMed] [Google Scholar]
- Zeng X, Tang R, Guo H, Ke S, Teng B, Hung YH, Xu Z, Xie XM, Hsieh TF, Zhang XQ(2017) A naturally occurring conditional albino mutant in rice caused by defects in the plastid-localized OsABCI8 transporter. Plant Mol Biol 94: 137–148 [DOI] [PubMed] [Google Scholar]
- Zhang XB, Feng BH, Wang HM, Xu X, Shi YF, He Y, Chen Z, Sathe AP, Shi L, Wu JL(2018) A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J Integr Plant Biol 60: 160–172 [DOI] [PubMed] [Google Scholar]
- Zhao G, Shi J, Liang W, Xue F, Luo Q, Zhu L, Qu G, Chen M, Schreiber L, Zhang D(2015) Two ATP binding cassette G transporters, rice ATP binding cassette G26 and ATP binding cassette G15, collaboratively regulate rice male reproduction. Plant Physiol 169: 2064–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen A, Bie Z, Huang Y, Liu Z, Lei B(2011) Effects of salt-tolerant rootstock grafting on ultrastructure, photosynthetic capacity, and H2O2-scavenging system in chloroplasts of cucumber seedlings under NaCl stress. Acta Physiol Plant 33: 2311–2319 [Google Scholar]
- Zhou BB, Elledge SJ(2000) The DNA damage response: Putting checkpoints in perspective. Nature 408: 433–439 [DOI] [PubMed] [Google Scholar]
- Zhu X, Liang S, Yin J, Yuan C, Wang J, Li W, He M, Wang J, Chen W, Ma B, et al. (2015) The DnaJ OsDjA7/8 is essential for chloroplast development in rice (Oryza sativa). Gene 574: 11–19 [DOI] [PubMed] [Google Scholar]