Tissue-specific differentiation of phosphorus transporters coordinated with their regulators confers phosphorus homeostasis, regulating nodule development.
Abstract
Maintaining phosphorus (Pi) homeostasis in nodules is the key to nodule development and nitrogen fixation, an important source of nitrogen for agriculture and ecosystems. PHOSPHATE-TRANSPORTER1 (PHT1) and its regulator PHOSPHATE-STARVATION-RESPONSE1 (PHR1), which constitute the PHR1-PHT1 module, play important roles in maintaining Pi homeostasis in different organs. However, the PHR1-PHT1 module and its functions in nodules remain unknown. We identified one PHT1 (GmPHT1;11) and four PHR1 (GmPHR1) homologs in soybean (Glycine max) plants, which displayed specific expression patterns in different tissues in nodules, similar to previously reported GmPHT1;1 and GmPHT1;4. Through the integration of different approaches, GmPHR-GmPHT1 modules were confirmed. Combining our results and previous reports, we established multiple GmPHR-GmPHT1 modules acting in the infected or noninfected tissues in nodules. A single GmPHR had more than one GmPHT1 target, and vice versa. Therefore, overlapping and cross-talking modules monitored the wave of available Pi to maintain Pi homeostasis in nodules, which sequentially regulated nodule initiation and development. High levels of GmPHT1;11 enhanced Pi accumulation in nodules, increased nodule size, but decreased nodule number. Nitrogenase activity was also enhanced by GmPHT1;11. Our findings uncover GmPHR-GmPHT1 modules in nodules, which expands our understanding of the mechanism of maintaining Pi homeostasis in soybean plants.
Legume nodules provide agriculture and ecosystems with nitrogen from the ammonium ion (NH4+) through N2 fixation. Phosphorus (Pi) is utilized as a macronutrient in plants and affects nodule initiation, development, and N2 fixation (Tang et al., 2001; Valentine et al., 2017). Pi deficiency directly impairs many cellular functions not only by reducing the available ATP necessary for enzymatic activity, membrane transport, utilization of inorganic N sources, and photosynthesis but also by limiting the biosynthesis of macromolecules (proteins, lipids, and nucleic acids; Bosse and Kock, 1998). In root nodules, the role of Pi is vital in the metabolic energy processes driving symbiotic N2 fixation into NH3, and nodules act as strong sinks for P even when the supply of P is adequate (Israel, 1987; Drevon and Hartwig, 1997). Therefore, the availability of Pi is pivotal to nodule morphogenesis and the efficiency of N2 fixation (Drevon and Hartwig, 1997; Tang et al., 2001; Sulieman et al., 2013). The effect of Pi on nodule initiation is nodule-specific and independent of plant growth, indicating that Pi has dual functions as a critical mineral element in cells and as a signal when Pi is deficient in nodules (Gentili and Huss-Danell, 2003).
A phosphate transporter is responsible for intercellular and intracellular Pi transport (Bardin et al., 1996; Qin et al., 2012a). The PHOSPHATE TRANSPORTER1 (PHT1) genes encode a family of plasma membrane phosphate transporters (Nussaume et al., 2011). In the soybean (Glycine max) genome, there are at least 14 members (GmPHT1;1–GmPHT1;14) in this family with different names in different labs (Qin et al., 2012a; Tamura et al., 2012; Fan et al., 2013). These GmPHT1s have different affinities to Pi (Fan et al., 2013). Among them, only GmPHT1;1 (Chen et al., 2019) and GmPHT1;4 (Qin et al., 2012b) are expressed in nonfixing regions of nodules, and they also play a role in nodulation and Pi homeostasis in nodules. GmPHT1;11, GmPHT1;12, and GmPHT1;13 are arbuscular mycorrhiza-inducible phosphate transporter genes of soybean (Tamura et al., 2012), and GmPHT1;11, an early divergent gene from other GmPHT1s (Fan et al., 2013), is related to arbuscular development and leaf senescence (Inoue et al., 2014).
PHT1 is regulated by many genes at different levels (Puga et al., 2017). When Pi is deficient, PHOSPHATE STARVATION RESPONSE1 (PHR1), a constitutively expressed MYB-domain transcription factor (Rubio et al., 2001; Puga et al., 2017), promotes Pi uptake by directly inducing the expression of PHT1. PHR1 not only functions in controlling Pi transport but also plays a role in directly repressing plant defenses (Castrillo et al., 2017) by activating jasmonate biosynthesis (Khan et al., 2016). PHR1 proteins bind to specific cis-elements (P1BS, GnATATnC) of Pi starvation-inducible genes, including PHT1, phosphatase, and RNase genes (Rubio et al., 2001; Schünmann et al., 2004; Nilsson et al., 2007; Guo et al., 2015). Therefore, the PHR-PHT1 module, a regulatory module conserved across phylogenetically distant plant species, plays an important role in maintaining Pi homeostasis (Puga et al., 2017). However, there are no reports elucidating the functions of PHR-PHT1 modules in soybean nodules.
In this study, we identified PHR and PHT1 genes in soybean nodules and constructed multiple PHR-PHT1 modules. These modules formed a network and contributed to maintaining Pi homeostasis in nodules, which controls nodule development and functions in soybean. Our results show that in nodules, one GmPHR has several target GmPHT1s, and one GmPHT1 is under the control of several GmPHRs. Therefore, the divergence of GmPHR-GmPHT1 modules in a tissue-specific mode endows nodules with special characteristics during Pi signaling in soybean.
RESULTS
Soybean Nodules Harbor Multiple PHR and PHT1 Homologs
The PHT gene family is responsible for transport of Pi, and PHT1 is a subgroup of the PHT family, which mainly functions in cellular phosphate transport. In the soybean genome, 14 PHT1 homologous genes (GmPHT1;1–GmPHT1;14) have been identified in our laboratory and other laboratories, and at least four PHT1s (GmPHT1;1, GmPHT1;4, GmPHT1;11, and GmPHT1;14) are expressed in roots (Qin et al., 2012a; Fan et al., 2013). Among these four GmPHT1s, GmPHT1;1 and GmPHT1;4 were reported to function in nodules (Qin et al., 2012b; Chen et al., 2019). It has been reported that GmPHT1;11 functions in the absorption of fungus-derived phosphate and in arbuscular development (Inoue et al., 2014). We postulated that if a gene was related to nodule function, it may be expressed in roots. Therefore, GmPHT1;1, GmPHT1;11, and GmPHT1;14, working together with GmPHT1;4, may synergistically function in nodules. These GmPHT1 proteins were all found to localize on the cytoplasmic membrane (Supplemental Fig. S1), as described in a previous report (Fan et al., 2013).
One of the direct regulators of PHT1 is PHR1, a MYB transcription factor that can bind to a cis-element with an imperfect palindromic sequence (GnATATnC; [P1BS]) in its target genes and regulate its expression (Rubio et al., 2001; Bari et al., 2006; Bustos et al., 2010). Hence, the PHR1-PHT1 module plays pivotal roles in regulating Pi signaling and maintaining Pi homeostasis. In the soybean genome, it has been reported that there are 35 PHR1 genes encoding potential transcriptional factors (Xue et al., 2017), but no detailed functions have been revealed. Using the protein sequence of Arabidopsis (Arabidopsis thaliana) PHR1 as a query, we obtained four highly conserved PHR1 homologous genes in the soybean genome, named GmPHR1 to GmPHR4 here (Supplemental Fig. S2). The GmPHR1 to GmPHR4 proteins share two conserved functional domains, the Myb coiled-coil motif (a potential dimerization motif) and the Myb DNA-binding motif (Supplemental Fig. S2), indicating potential binding activity for their target genes (Rubio et al., 2001).
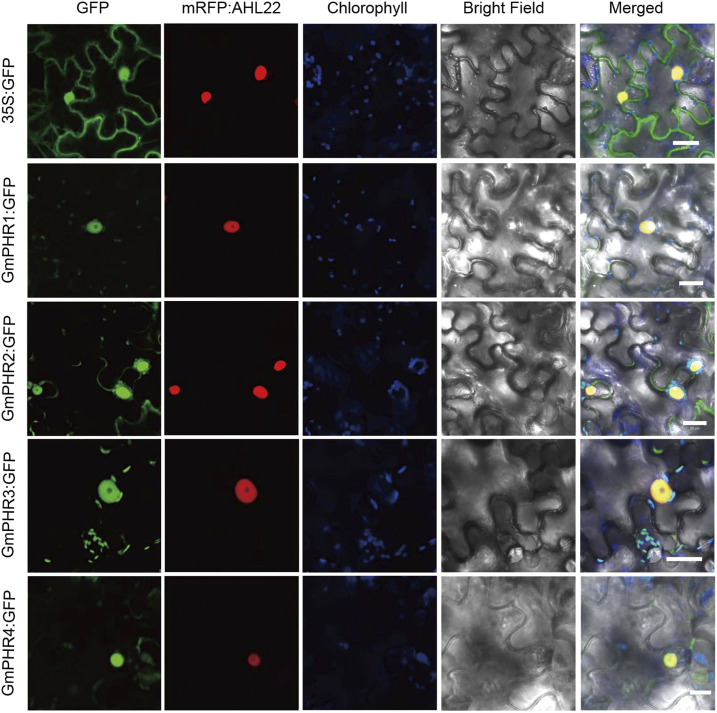
To investigate the subcellular localization of GmPHR1 to GmPHR4 proteins, we amplified their coding sequences from soybean ‘Tainlong1’ and constructed the fusion genes 35S:GmPHR1:GFP, 35S:GmPHR2:GFP, 35S:GmPHR3:GFP, and 35S:GmPHR4:GFP. Then, the corresponding vectors were infiltrated into Nicotiana benthamiana leaves mediated by Agrobacterium tumefaciens. Similar to PHR proteins in other plants (Rubio et al., 2001; Ren et al., 2012; Li et al., 2014), GmPHR1:GFP, GmPHR2:GFP, GmPHR3:GFP, and GmPHR4:GFP proteins mainly target the nucleus (Fig. 1).
Figure 1.
Localization of GmPHR proteins. GmPHR1:GFP, GmPHR2:GFP, GmPHR3:GFP, and GmPHR4:GFP fusion genes were coinfiltrated with the mRFP:AHL22 nuclear marker in N. benthamiana leaves. All GmPHR:GFP proteins mainly localized in the nuclei. All experiments were performed with at least three independent biological repeats, and all gave similar results. A single representative result is shown. Bars = 20 μm.
Tissue-Specific Divergence of the PHR-PHT1 Modules
The soybean genome underwent at least three rounds of duplication during its evolutionary process, which resulted in multiple copies of a given gene (Schmutz et al., 2010). To survive natural selection, individual copies experienced various divergences, including divergence in tissue-specific expression (Blanc and Wolfe, 2004; Wang et al., 2012; Roulin et al., 2013). We focused on the GmPHR1, GmPHR2, GmPHR3, GmPHR4, GmPHT1;1, GmPHT1;4, GmPHT1;11, and GmPHT1;14 genes in our study, due to their expression in roots (Qin et al., 2012a, 2012; Fan et al., 2013; Chen et al., 2019), which indicates that they may potentially function in nodules.
To elucidate the expression specificity of PHR-PHT1 modules in nodules, we cloned the promoter sequence approximately 2.5 kb upstream of the translational start site of these genes and individually fused them to the GUS reporter gene. The resulting vectors were introduced into soybean hairy roots by Agrobacterium rhizogenes. The transgenic hairy roots were then inoculated with rhizobia (Sinorhizobium fredii HH103) to allow nodule development. After 2 weeks, transgenic nodules and their transverse sections were stained by 5-bromo-4-chloro-3-indoxyl-β-d-glucuronide (X-gluc) to visualize the tissue-specific expression of the promoters. Regardless of Pi conditions, GmPHR2:GUS lines showed faint signals in nodules, whereas there were no signals detectable in GmPHR3:GUS nodules (Supplemental Fig. S3).
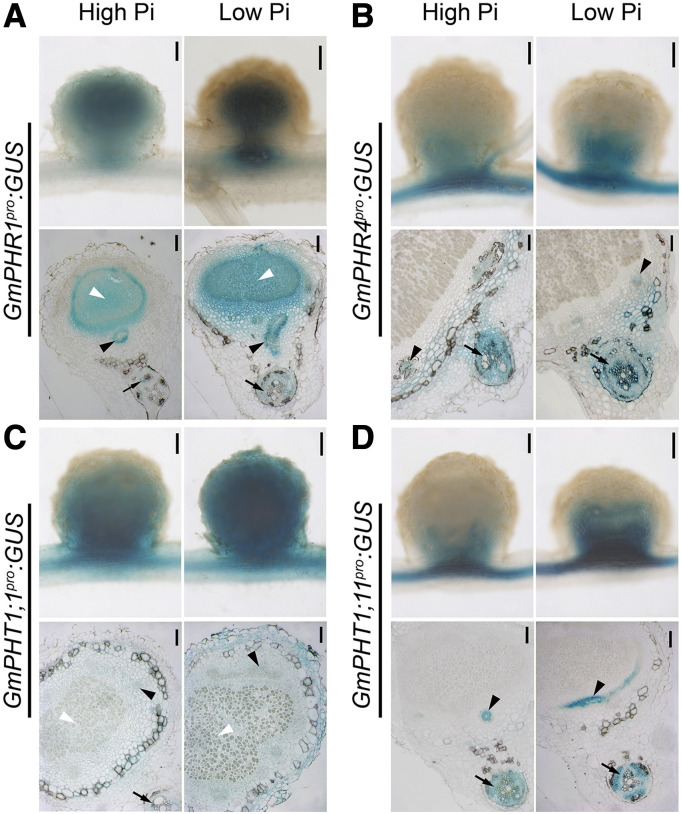
Interestingly, GmPHR1 was expressed in the entire tissue of nodules, while GmPHR4 expression was limited to non-N2-fixation regions (Fig. 2, A and B). Such patterns were independent of the concentration of available Pi in the medium, even though low Pi levels induced higher GmPHR gene expression. Similar to GmPHR, the phosphate transporter GmPHT1s also exhibited tissue-specific differentiation of gene expression. GmPHT1;1 was distributed throughout an entire nodule, consistent with a previous report (Chen et al., 2019), whereas GmPHT1;11 was only expressed in non-N2-fixing regions (Fig. 2, C and D), indicating that GmPHT1;11 was not expressed in the infected tissues. Although GmPHT1;14 was expressed in the root vascular tissue, no GUS signal was detected in the nodule tissue (Supplemental Fig. S3). A previous report showed that GmPHT1;4 is expressed in nonfixing regions of nodules (Qin et al., 2012b).
Figure 2.
Tissue-specific expression of GmPHR-GmPHT1 modules in soybean nodules. GmPHR1 (A), GmPHR4 (B), GmPHT1;1 (C), and GmPHT1;11 (D) promoters were individually fused to a GUS reporter gene, and the resulting vectors were transferred into soybean hairy roots that were cultured under high (left images) or low (right images) Pi concentrations. Nodules were stained with X-Gluc, and whole nodule tissue (top images) or semithin sections (bottom images) were observed with a microscope. Photographs were taken from sections of different nodules produced from at least two independent biological experiments. A single representative result is shown. Black arrows indicate nodule vascular bundles, white arrowheads indicate nodule infected tissues, and black arrowheads indicate root vascular tissues. Bars = 200 μm (top images) and 100 μm (bottom images).
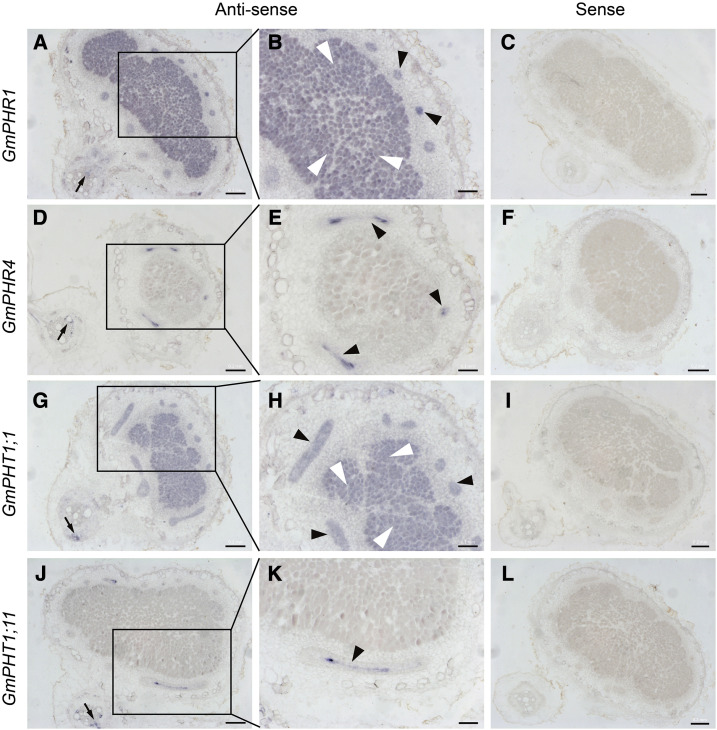
To further confirm the gene expression patterns above, we carried out RNA in situ hybridization analysis in nodules from roots of soybean plants grown in low-phosphate conditions. As Figure 3 indicated, there were no signals found on all sections hybridized by sense probes, suggesting the specificity of antisense probes. Not surprisingly, the transcript locations of GmPHR1, GmPHR4, GmPHT1;1, and GmPHT1;11 in soybean nodules (Fig. 3) matched well with the activity sites of their promoters revealed using the GUS marker (Fig. 2). Together, these results suggest that the expression specificity of different GmPHR or GmPHT1 homologs diverged in different nodule tissues over the evolutionary process.
Figure 3.
RNA in situ hybridization of GmPHR-GmPHT1 modules in soybean nodules. Nodules on soybean roots at 14 d after infection were used for RNA in situ hybridization. Digoxigenin-labeled antisense (A, B, D, E, G, H, J, and K) or sense (C, F, I, and L) probes were used for detection of the transcripts of GmPHR1 (A–C), GmPHR4 (D–F), GmPHT1;1 (G–I), and GmPHT1;11 (J–L). Images B, E, H, and K show magnifications of images A, D, G, and J, respectively. GmPHR1 and GmPHT1;1 were strongly expressed in nodule vascular bundles and infected tissues (A, B, G, and H), while GmPHR4 and GmPHT1;11 were expressed in nodule vascular bundles (D, E, J, and K). GmPHR1, GmPHR4, GmPHT1;1, and GmPHT1;11 were expressed in root vascular tissues (A, D, G, and J). No signals were found for sense probes. Three to five nodules were sectioned for each gene. Black arrows indicate nodule vascular bundles, white arrowheads indicate nodule infected tissues, and black arrowheads indicate root vascular tissues. Section thickness was 7 μm. Bars = 200 μm (A, C, D, F, G, I, J, and L) and 100 μm (B, E, H, and K).
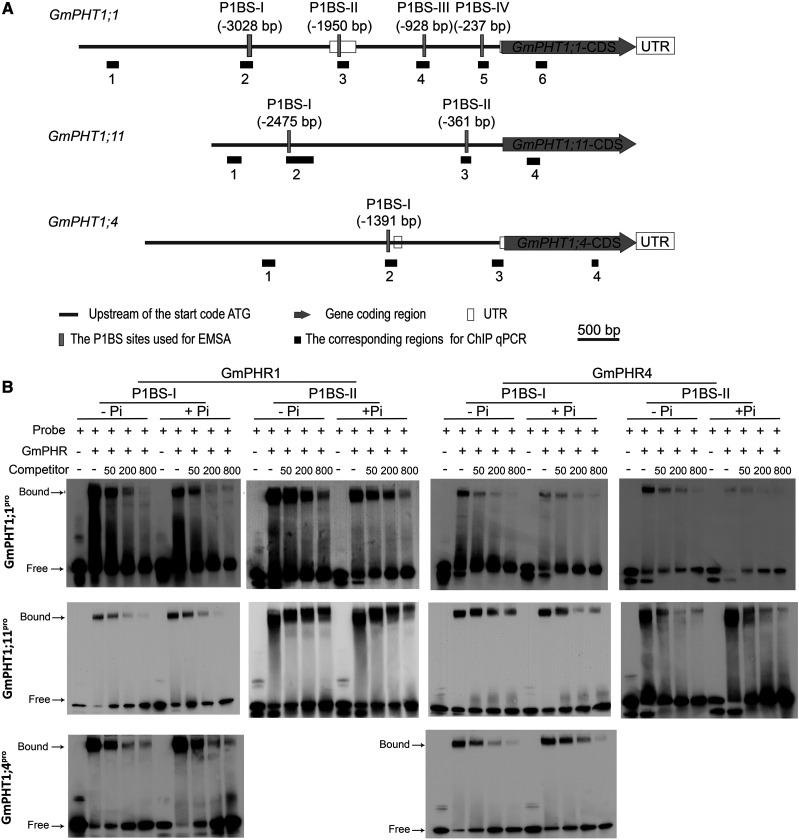
As different GmPHRs or GmPHT1s have highly conserved sequences within their family, it is of interest to clarify the relationship between individual GmPHRs and GmPHT1s to determine one-to-one correspondence relationships in soybean nodules. First, we performed an electrophoretic mobility shift assay (EMSA) to verify if these GmPHR proteins could bind to GmPHT1 promoters. As expected, both GmPHR1 and GmPHR4 directly bound to cis-element P1BS (GnATATnC) in the promoters of GmPHT1;1, GmPHT1;11, and GmPHT1;4 (Fig. 4) in vitro. There are four potential P1BSs in the GmPHT1;1 promoter (Fig. 4A); however, only two of them could be the targets of GmPHR1 and GmPHR4 proteins (Fig. 4B; Supplemental Fig. S4), suggesting that the flanking sequence of P1BS or other factors affected the physical interaction between PHR proteins and P1BS elements. Furthermore, the availability of Pi did not affect such physical interactions in vitro because there was no difference among binding assays in the presence or absence of Pi in the reaction buffers.
Figure 4.
GmPHR1 and GmPHR4 proteins bind directly to P1BS cis-elements in the promoters of GmPHT1 in vitro. A, Schematic structures of genomic genes GmPHT1;1, GmPHT1;11, and GmPHT1;4. The positions of the P1BS sites are indicated in parentheses (upstream of the start codon ATG), vertical gray bars represent the corresponding fragments for EMSA experiments (in B), and horizontal black bars display the corresponding regions for ChIP-qPCR detection (in Fig. 6). Gray bars represent the region upstream of the start codon ATG, gray arrow bars represent the gene-coding region, empty bars represent untranslated regions (UTR), vertical gray bars represent the PIBS sites used for EMSA, and black boxes represent the corresponding sites for ChIP qPCR in Figure 6. Scale bars (500 bp) are indicated above the genes. B, EMSA tests show that both GmPHR1 and GmPHR4 proteins bind to P1BS-containing fragments of GmPHT1;1 (top row), GmPHT1;11 (middle row), and GmPHT1;4 (bottom row) promoters independent of Pi concentration (+P, 1 mm KH2PO4; −P, 1 mm KCl in EMSA buffer). The fragments of P1BS-I and P1BS-II are indicated in A. Probe indicates labeled P1BS-containing fragments, while Competitor represents unlabeled P1BS-containing fragments. Numbers indicate fold of the concentration of Competitor compared with Probe. All experiments were performed with at least three biological repeats, which showed similar results. A single representative result is shown.
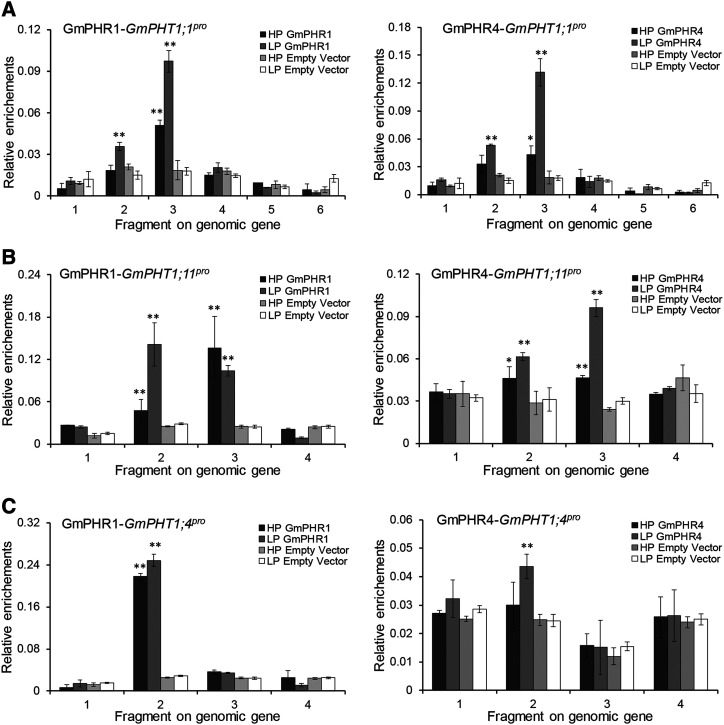
To confirm the interactions above, the chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) approach was employed to analyze transgenic hairy roots harboring the 35S:GmPHR1:GFP or 35S:GmPHR4:GFP fusion gene. The results indicate that either GmPHR1 or GmPHR4 protein could enrich in vivo special fragments of GmPHT1;1 (Fig. 5A), GmPHT1;11 (Fig. 5B), and GmPHT1;4 (Fig. 5C). However, the binding affinity was nonidentical for a given GmPHR with different P1BS-containing fragments, such as GmPHR1/GmPHR4 and GmPHT1;1 (Fig. 5A), supporting the importance of the flanking sequences of P1BS as shown above and being consistent with the general characteristics of transcription factors (Rajkumar et al., 2013). Additionally, it was observed that the concentration of available Pi did have an impact on GmPHR binding in vivo to GmPHT1. Generally, low-Pi conditions were favorable for GmPHR1 and GmPHR4 binding to their targets in vivo, indicating that the interactions between GmPHR1/4 and GmPHT1;1/4/11 were Pi stress inducible or enhanced.
Figure 5.
GmPHR1 and GmPHR4 proteins enrich P1BS cis-element-containing fragments in the promoters of GmPHT1 in hairy roots. ChIP-qPCR investigation demonstrates that both GmPHR1 (right graphs) and GmPHR4 (left graphs) enrich fragment 3 in the GmPHT1;1 promoter (A), fragments 2 and 3 in the GmPHT1;11 promoter (B), and fragment 2 in the GmPHT1;4 promoter (C) in vivo, regardless of the presence or absence of Pi in the medium, and GmPHR1 has higher affinity than GmPHR4 or GmPHT1;1. The positions of the fragments on the genomic genes are labeled as in Figure 3A. All statistical data show means ± sd from at least three biological replicates. Statistical analysis was performed using Student’s t test (*P < 0.05 and **P < 0.01).
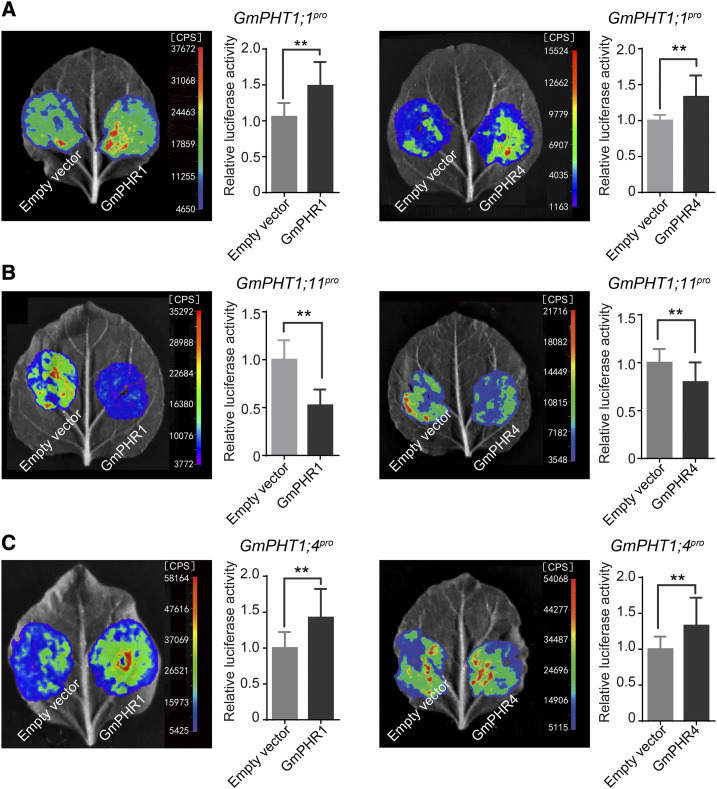
To determine the effect of GmPHR regulation on GmPHT1 transcription activity, we carried out transcriptional activation experiments using N. benthamiana leaves. We constructed the promoter-luciferase (LUC) fusion genes GmPHT1;1pro:LUC, GmPHT1;4pro:LUC, and GmPHT1;11pro:LUC, which were cotransformed with 35S:GmPHR1:GFP or 35S:GmPHR4:GFP (we used 35S:GFP as a control) into N. benthamiana leaves, mediated by A. tumefaciens. Then, the transcription activity of the GmPHT1 promoter was monitored through the fluorescence intensity produced by LUC. The results show that both GmPHR1 and GmPHR4 exerted significant effects on the intensity of LUC signals in different vectors. Both GmPHR1 and GmPHR4 enhanced the promoter activity of GmPHT1;1 (Fig. 6A) and GmPHT1;4 (Fig. 6C) but unexpectedly inhibited the promoter activity of GmPHT1;11 (Fig. 6B).
Figure 6.
GmPHR1 and GmPHR4 genes regulate the activity of GmPHT1 promoters. Both GmPHR1 and GmPHR4 genes significantly increase the activity of the GmPHT1;1 (A) and GmPHT1;4 (C) promoters but repress the activity of the GmPHT1;11 promoter (B) in N. benthamiana leaves. All statistical data are presented as means ± sd from at least three biological replicates. Statistical analysis was performed using Student’s t test (**P < 0.01).
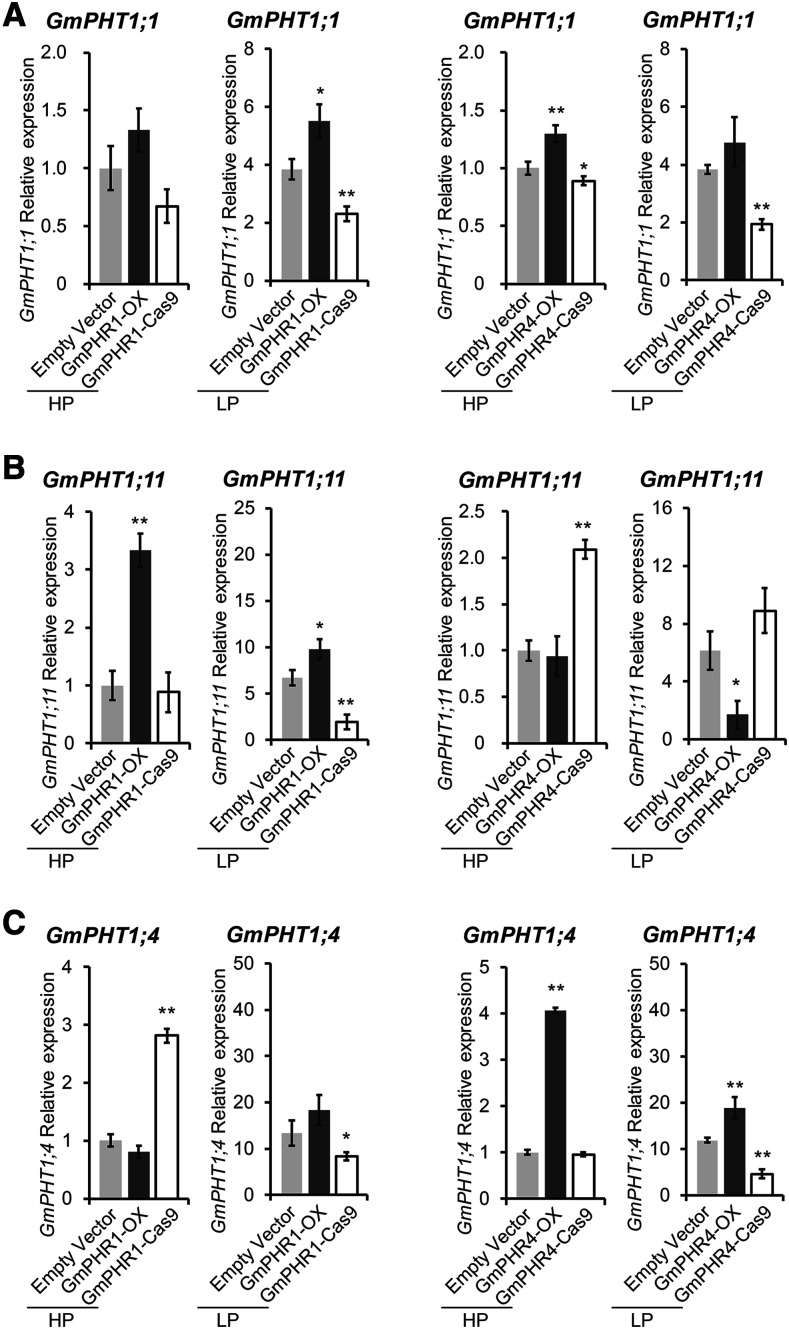
Next, we investigated the effect of changes in GmPHR expression on GmPHT1 gene expression in vivo. We took approaches of overexpression of GmPHR genes or mutating these genes by CRISPR/Cas9 editing. First, we generated hairy roots overexpressing GmPHR1 or GmPHR4 (from the 35S promoter), whose expression levels were higher than that in wild-type roots, as confirmed by reverse transcription quantitative PCR (RT-qPCR) analysis (Supplemental Fig. S5, A and C). Then, we constructed transgenic hairy roots with mutated GmPHR1 or GmPHR4 (Supplemental Figs. S6–S8) gene using CRISPR/Cas9 editing. These transgenic lines could not produce normal proteins because the coding region of their mutated genes had premature stop codons resulting in early termination, even though they had lower expression of the GmPHR1 or GmPHR4 gene, respectively (Supplemental Fig. S5, A and C). Such expression changes were independent of the availability of Pi (Supplemental Fig. S5, A and C). With these transgenic lines, RT-qPCR analysis was carried out to evaluate the variation in PHT1 expression levels affected by the change of the GmPHR1 or GmPHR4 gene under different Pi conditions. As expected, a positive effect of both GmPHR1 and GmPHR4 on GmPHT1;1 expression (Fig. 7A) was observed, consistent with the results of the promoter activity assay (Fig. 6A).
Figure 7.
GmPHR1 and GmPHR4 genes regulate GmPHT1 gene expression in soybean nodules. A, GmPHR1 gene enhances GmPHT1;1 expression under low-Pi conditions (LP), while GmPHR4 gene enhances GmPHT1;1 expression independent of the availability of Pi. B, GmPHR1 enhances while GmPHR4 represses GmPHT1;11 expression in soybean nodules independent of the availability of Pi. Evidence from overexpression of GmPHR1 or GmPHR4 was consistent with data from knocking out these genes using the CRISPR/Cas9 approach. C, Both GmPHR1 and GmPHR4 genes regulate GmPHT1;4 expression in soybean nodules depending on the availability of Pi. All statistical data show means ± sd from at least three biological replicates with six individual plants. Statistical analysis was performed using Student’s t test (*P < 0.05 and **P < 0.01). Normalization of the data was performed using the expression of the empty vector control under high-Pi conditions (HP) as a reference.
Overexpressing GmPHR4 repressed GmPHT1;11 expression, regardless of the Pi state (Fig. 7B), which also coincided with the results of the transcription activity analysis (Fig. 6B). However, as for GmPHR1 and GmPHT1;11, the results (Fig. 7B) were entirely opposite to the results of the transcription activity analysis (Fig. 6B). That is, overexpressing GmPHR1 increased GmPHT1;11 expression under both low- and high-Pi conditions in nodules (Fig. 7B). Such contradictory results suggest that there is species or tissue specificity of GmPHR1 function on GmPHT1;11. In N. benthamiana, GmPHR1 inhibits GmPHT1;11 expression, while in soybean nodules, GmPHR1 enhances GmPHT1;11 expression. In other words, the effect of GmPHR1 on GmPHT1;11 expression was dependent on another unknown factor.
The expression of GmPHT1;4 was dependent on Pi availability. When the amount of Pi was relatively low, the GmPHR4 gene enhanced GmPHT1;4 expression (Fig. 7C). However, when the amount of Pi was relatively high, overexpressing GmPHR1 repressed GmPHT1;4 transcription, while overexpression of GmPHR4 promoted GmPHT1;4 expression, and knocking out GmPHR4 did not change GmPHT1;4 expression (Fig. 7C). Because the entire nodule was used for RT-qPCR analysis here, it is reasonable to speculate that GmPHRs regulate the expression of GmPHT1s in a tissue-specific mode in nodules.
PHR-PHT1 Modules Contribute to Pi Homeostasis in Nodules
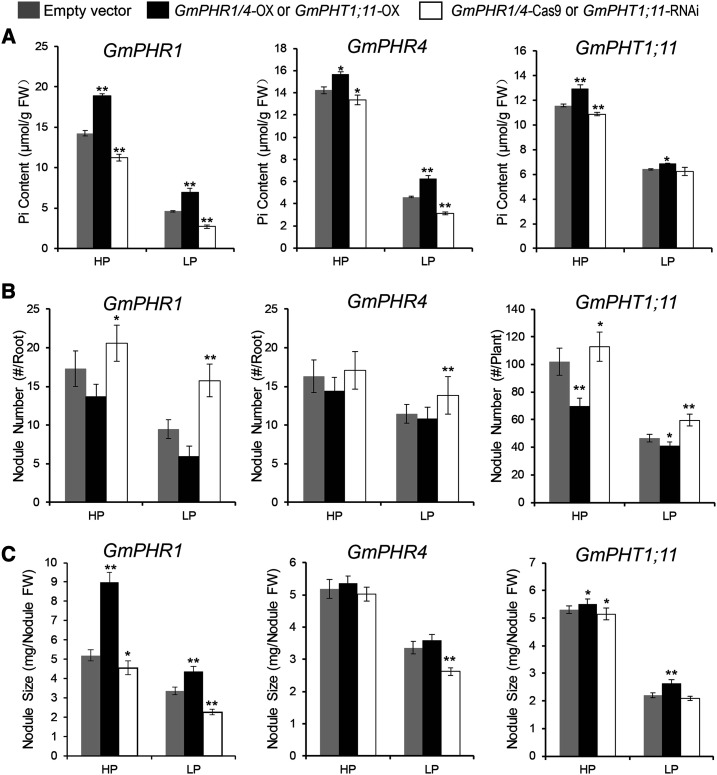
To investigate the function of GmPHR-GmPHT1 in Pi homeostasis in nodules, we determined the Pi content in different transgenic nodules using similar transgenic lines. As expected, overexpressing either GmPHR1 or GmPHR4 increased the accumulation of Pi in nodules in a Pi-independent manner (Fig. 8A). Higher levels of GmPHT1;11 also increased Pi accumulation in nodules (Fig. 8A). These results indicate that GmPHR1, GmPHR4, and GmPHT1;11 all participate in maintaining Pi homeostasis in nodules.
Figure 8.
GmPHR-GmPHT1 modules play important roles in maintaining Pi homeostasis in nodules and in regulating nodule initiation and development. For GmPHR1 and GmPHR4 genes, nodules from transgenic hairy roots were sampled; for GmPHT1;11, nodules from roots of stable transgenic plants were harvested. A, Pi homeostasis in nodules. GmPHR1, GmPHR4, and GmPHT1;11 enhance Pi accumulation in nodules. B and C, The effects of GmPHR1, GmPHR4, and GmPHT1;11 on nodule initiation (number; B) and development (size; C). All statistical data show means ± sd from at least three biological replicates. Statistical analysis was performed using Student’s t test (*P < 0.05 and **P < 0.01). FW, Fresh weight; HP, high-Pi conditions; LP, low-Pi conditions.
PHR-PHT1;11 Modules Regulate Nodule Growth and Function
Next, we investigated the function of PHR-PHT1 modules in nodulation through statistical analysis of the nodule number, size, and fresh weight of transgenic nodules as in Figure 7. Our results show that both GmPHRs and GmPHT1s exhibit functions governing the number of nodules (Fig. 8B). GmPHR1 inhibited nodule initiation, and lower GmPHR1 levels resulted in an increased number of nodules at either low or high Pi concentrations. Similarly, lower expression of GmPHR4 clearly increased the number of nodules in the low-Pi state. Additionally, it was significant that the inhibitory effect of GmPHT1;11 on nodule formation was independent of Pi concentration.
Regarding nodule size, the results were generally opposite to the number of nodules in response to Pi signaling (Fig. 8C). GmPHR1, GmPHR4, and GmPHT1;11 enhanced the growth of nodules, and GmPHR1 exerted stronger effects on nodule growth than GmPHR4 in the case of nodule size analysis. In addition, GmPHR1, GmPHR4, and GmPHT1;11 did not significantly affect the fresh weight of nodules (Supplemental Fig. S9).
To further confirm that GmPHT1;11 functions in nodulation, we constructed GmPHT1;11-Cas9-edited hairy roots (Supplemental Figs. S8, E and F, and S10). The nodulation assay indicated that mutation of GmPHT1;11 increased the number of nodules per root and nodule fresh weight but decreased nodule size. These results are consistent with the results of the stable transgenic soybean lines above (Fig. 8, B and C).
Interestingly, interactions were observed between GmPHR1 and GmPHR4 or between GmPHT1;4 and GmPHT1;11. An increase in GmPHR1 expression enhanced GmPHR4 expression under both low- and high-Pi conditions, and vice versa (Supplemental Fig. S5, B and D). However, overexpression of GmPHT1;11 decreased GmPHT1;4 expression, whereas silencing of GmPHT1;11 had no significant effect on the expression of GmPHT1;4 (Supplemental Fig. S5F). The effect of GmPHR-GmPHT1 modules on nodule growth was a comprehensive process, which relied on the interaction between different GmPHR-GmPHT1 modules, tissue specificity, and availability of Pi. These results suggest that there exists a potential compensatory mechanism among GmPHR-GmPHT1 modules, similar to previous reports (Kumar et al., 2008; Laguerre et al., 2012).
GmPHT1;11 Regulates Nitrogenase Activity in Nodules
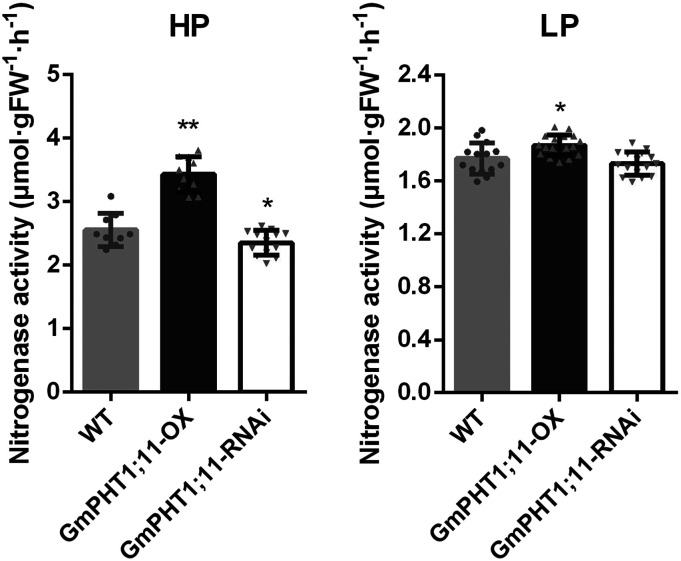
Previous reports showed that the expression level of GmPHT1;1 and GmPHT1;4 was positively related to both nitrogenase activity and the accumulation of N in soybean plants (Qin et al., 2012b; Chen et al., 2019). We also determined the nitrogenase activity in nodules of GmPHT1;11 transgenic plants according to an approach described previously (Buendia-Claveria et al., 1989). As Figure 9 shows, knockdown of the GmPHT1;11 gene inhibited the activity of nitrogenase, whereas overexpression of GmPHT1;11 enhanced nitrogenase activity.
Figure 9.
GmPHT1;11 expression contributes to the function of nodules in soybean. Nitrogenase activity is shown in lines of overexpressing (OX) and RNA interference (RNAi)-silenced GmPHT1;11 compared with their parents grown under high-Pi (HP; left) and low-Pi (LP; right) conditions. Gray dots, upward-pointing triangles, and downward-pointing triangles on the top of the columns indicate the different samples for the wild type (WT), GmPHT1;11-OX, and GmPHT1;11-RNAi, respectively. Significance was based on Student’s t test with six plants (*P < 0.05 and **P < 0.01). Experiments were repeated three times with similar results. A single representative replicate is shown here, while the other two replicates are shown in Supplemental Table S2. FW, Fresh weight.
DISCUSSION
Many studies demonstrate that Pi plays important roles in rhizobium-legume symbiosis (Israel, 1993; Tang et al., 2001; Gentili and Huss-Danell, 2003; Schulze et al., 2006; Sulieman et al., 2013; Thuynsma et al., 2014). Phosphorus not only controls the energy costs of N2 fixation (Schulze et al., 1999) but also regulates nodule formation and development (Israel, 1987) through specific effects and not a general stimulation via a plant growth effect (Gentili and Huss-Danell, 2003). N2-fixing legumes can enhance Pi utilization within the nodules to tolerate Pi deficiency (Araujo et al., 2008), and therefore, the Pi concentration in nodules does not differ significantly under high- or low-Pi conditions (Vardien et al., 2014), suggesting that Pi homeostasis in nodules is subtly controlled. The PHR-PHT1 module is one of the main regulators for Pi transport in plant cells (Bardin et al., 1996; Rubio et al., 2001; Qin et al., 2012a; Puga et al., 2017). In our study, we elucidated the role of PHR-PHT1 modules in maintaining Pi homeostasis in soybean nodules.
GmPHR Directly Controls GmPHT1 Expression in Soybean Nodules
Our study identified GmPHR1/4 and their target GmPHT1;1/4/11 transporter genes in soybean nodules. GmPHR1/4 proteins directly bound to P1BS elements on GmPHT1;1/4/11 promoters and subsequently regulated their expression. Even though their interactions were independent of Pi availability in vitro, a lower level of available Pi induced/enhanced such interactions in vivo, suggesting that GmPHR-GmPHT1 signaling in nodules was a response to phosphorus stress. Both GmPHR1 and GmPHR4 increased the expression of GmPHT1;1 and GmPHT1;4 but suppressed GmPHT1;11 expression in N. benthamiana cells. Interestingly, GmPHR1 enhanced GmPHT1;11 expression in soybean, indicating species or tissue specificity of the interaction between GmPHR1 proteins and the GmPHT1;11 promoter. We postulated that there are other unknown regulators controlling GmPHR1 activity that may be specific to soybean nodules.
There are a few reports showing that SPX (SYG1/Pho81/XPR1) proteins inhibit PHR binding to its targets in Arabidopsis (Puga et al., 2014) and rice (Oryza sativa; Wang et al., 2014). There is another possibility that the tissue specificity of gene interactions may play an important role in the regulation of GmPHR functions, because our data indicate that different GmPHRs and GmPHT1s possessed tissue-specific expression patterns. Therefore, the mechanism of GmPHRs regulating GmPHT1 expression is much more complex in soybean nodules. In almost all previous studies, PHRs were reported as enhancers of PHT1 expression (Rubio et al., 2001; Bari et al., 2006; Zhou et al., 2008; Bustos et al., 2010). However, our results demonstrate that GmPHR could be an inhibitor of PHT1 expression under special conditions, dependent on its target genes, tissue specificities, or the availability of Pi.
GmPHR-GmPHT1 Modules Diverge in a Spatial Pattern in Nodules
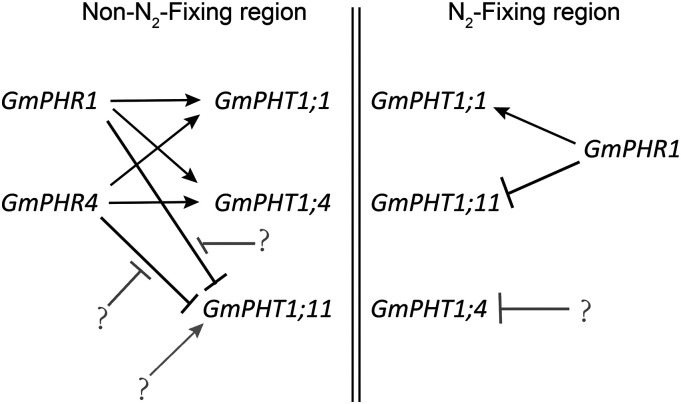
Specific expression in nodules was obvious for different GmPHR or GmPHT1 genes. Both GmPHR1 and GmPHT1;1 were expressed in whole nodule tissues, while GmPHR4 and GmPHT1;11 were expressed in noninfected tissues in nodules. It was previously reported that the expression of GmPHT1;4 was also limited in noninfected tissues in nodules (Qin et al., 2012b) and that GmPHT1;1 was expressed in whole nodules (Chen et al., 2019). Based on our observation of expression patterns and interactions between PHRs and PHT1s, in addition to previous reports (Qin et al., 2012b; Chen et al., 2019), we developed an interaction model for GmPHR-GmPHT1 modules in nodule tissues (Fig. 10). In N2-fixing regions, GmPHR1 not only enhanced GmPHT1;1 expression but also repressed GmPHT1;11. However, our results show that GmPHR1 increases GmPHT1;4 expression in N. benthamiana cells, and no GUS signal was detected for the PHT1;4 promoter in infected regions of nodules, even though the activity of the GmPHR1 promoter was detected in these regions. Therefore, the transcriptional repression of GmPHT1;4 in infected regions may be contributed by transcription factors other than PHR1.
Figure 10.
A working model of GmPHR-GmPHT1 modules in soybean nodules. Double vertical lines separate the non-N2-fixing region (left) from the N2-fixing region (right). The arrows indicate enhanced effects, and dashes indicate repressed effects. Black lines indicate the results in our study, while gray lines denote unidentified factors and their effects in our study. Question marks indicate unknown factors (see the text for details).
In non-N2-fixing regions, both GmPHR1 and GmPHR4 enhanced the expression of GmPHT1;1 and GmPHT1;4. Due to the repression effect of GmPHR1 and GmPHR4 on GmPHT1;11 in N. benthamiana cells, other unidentified factors could be involved in the activation of GmPHT1;11 transcription in non-N2-fixing regions. It is clear in our model that there were several GmPHR-GmPHT1 modules in nodules that had the following identities. (1) The effect of GmPHRs on GmPHT1s could be enhanced or repressed, depending on their targets (GmPHT1s) but independent of the availability of Pi concentration. (2) The same module exhibited different characteristics in different tissues. For example, GmPHR1 repressed GmPHT1;11 in N2-fixing regions but may not inhibit GmPHT1;11 expression in non-N2-fixing tissues. A previous study showed the importance of temporal induction for the potato (Solanum tuberosum) phosphate transporter StPT3 expression in root cells harboring arbuscular mycorrhizal structures (Karandashov et al., 2004). Our results show that the spatial issue may be also important for PHT1 expression. (3) These modules could affect each other because altering the expression of one PHR affected the expression of other PHRs, and a similar situation existed for PHT1s. For example, the change in GmPHT1;11 expression altered GmPHT1;4 expression. (4) One GmPHR had several GmPHT1 targets, and conversely, one GmPHT1 had several regulating GmPHRs, which built up the network with cross talk between GmPHR and GmPHT1. If protein movement of transcriptional factors among cells happens in nodules, the regulatory network may be more complicated. Such divergence in expression contributes to the establishment of a regulatory network of Pi homeostasis in nodules.
GmPHR-GmPHT1 Modules Participate in the Regulation of Nodule Growth and Nitrogenase Activity
Nodules are exquisite legume organs in which every process is strictly controlled. Nodules show high nitrogen fixation efficiency even under Pi-deficient conditions (Sulieman et al., 2013; Cabeza et al., 2014; Vardien et al., 2014; Valentine et al., 2017). The PHR-PHT1 module network described above may maintain Pi homeostasis in soybean nodules. Different genes in the module network displayed different effects on nodule growth and function. Therefore, abnormal expression of one of the elements in this network could disturb Pi homeostasis in nodules, resulting in abnormalities in Pi accumulation and nodule initiation, development, and N2-fixing efficiency.
Our results also suggest that nodule initiation and growth may be two dissociable processes controlled by PHR-PHT1 modules; that is, the factors favorable to nodule initiation could inhibit nodule growth, and vice versa. For example, silencing GmPHT1;11 increased nodule number but decreased nodule size. Also, overexpression of GmPHR1 or GmPHR4 reduced the number of nodules, but the nodules greatly increased in size when exposed to high levels of GmPHR transcripts.
GmPHT1 regulates N2 fixation because GmPHT1;1 enhances nitrogenase activity in soybean plants (Chen et al., 2019). In our study, we showed that the expression level of GmPHT1;11 was positively related to the activity of nitrogenase. Further investigation of the effect of interactions among GmPHRs and GmPHT1s on nodules will allow us to elucidate the mechanism underlying nitrogen fixation in soybean.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All genetically modified lines were constructed with soybean (Glycine max ‘Tianlong1’) mediated by Agrobacterium tumefaciens for stable transformation or Agrobacterium rhizogenes for hairy root transformation. All soybean plants were grown in a growth room under long-day conditions (16 h of light/8 h of dark) at 25°C with light illumination. Nicotiana benthamiana plants were grown in a growth room at 22°C with a 16-h-light/8-h-dark cycle. Light was provided by GreenPower light-emitting diode (LED) toplighting (Philips Horticulture LED) with light intensity ranging from 500 to 800 μmol m−2 s−1 for soybean or 225 to 450 μmol m−2 s−1 for N. benthamiana, depending on the distance between the plants and the LED light source.
Vector Construction and Soybean Transformation
Using the Arabidopsis (Arabidopsis thaliana) PHR1 (AT4G28610) protein sequence as a query to search its homologs in the soybean genome in Phytozome (http://www.phytozome.net), four GmPHR members (Glyma.19G167500, Glyma.03G166400, Glyma.13G126200, and Glyma.10G039700) with high similarity were identified in soybean and named as GmPHR1 to GmPHR4, sequentially. Identities between AT4G28610 and GmPHR score from 323.2 to 286.2, with e values from 1.2E−104 to 2.4E−90. Similar to AtPHR1, GmPHR1, GmPHR2, GmPHR3, and GmPHR4 proteins also contain two conserved domains: Myb and coiled-coil domains. Alignment of two domains of amino acid sequences was carried out using ClustalW programs. All GmPHT1;1, GmPHT1;4, GmPHT1;11, and GmPHT1;14 genes were identified in our previous study (Fan et al., 2013).
For the promoter analysis, fragments upstream of the transcription start sites of GmPHR1 (GmPHR1pro; 2,542 bp), GmPHR2 (GmPHR2pro; 2,453 bp), GmPHR3 (GmPHR3pro; 2,258 bp), and GmPHR4 (GmPHR4pro; 2,525 bp) were cloned into the vector Fu76 (Wang et al., 2013) with SacI and PstI cloning sites. Then, the promoter fragments were inserted between HindIII and NcoI sites to replace the Cauliflower mosaic virus (CaMV) 35S promoter in a modified pCAMBIA3301 to generate the GmPHRpro:GUS-expressing vectors. Similarly, fragments upstream of the translation start sites (ATG) of GmPHT1;1 (GmPHT1;1pro; 5,240 bp), GmPHT1;11 (GmPHT1;11pro; 2,923 bp), and GmPHT1;14 (GmPHT1;14pro; 4,988 bp) were cloned into the Fu76 vector with SacI and FspI sites. The promoter segments were then cloned into a modified pCAMBIA3301 vector with SacI and SmaI sites to generate GmPHT1pro:GUS binary vectors. For overexpressing genes, the coding regions of GmPHR1 and GmPHR4 were introduced into the entry vector Fu28 (Wang et al., 2013) with SpeI and FspI and fused to the GFP gene in the vector, then these entry clones were combined with a modified pHairyRED (Lin et al., 2011; added attR1/2 recombination sites and a CaMV 35S promoter, in our lab) to generate 35S:GmPHR:GFP-expressing constructs by LR reaction (Invitrogen). All resulting expression vectors above were introduced into A. rhizogenes strain K599 for soybean hairy root transformation (Kereszt et al., 2007). For GmPHT1;11 overexpression, the coding region of GmPHT1;11 was introduced into entry vector Fu48 (Wang et al., 2013) with XbaI and FspI enzymes to make a 3×Myc fused gene. Then, the fusion gene was cloned into pFGC5941 vector (GenBank accession no. AY310901) with AscI and AvrII. For the knockdown (GmPHT1;11-RNAi) construct, a 334-bp (bp 937–1,270) fragment of the GmPHT1;11 coding sequence was amplified and inserted into the pB7GWIWG2(I) vector (Karimi et al., 2002) in both sense and antisense orientations. Both overexpressing and silencing vectors of GmPHT1;11 were introduced into A. tumefaciens strain EHA105 for soybean genetic transformation. All sequences were confirmed by sequencing and are identical to the corresponding genes in Phytozome.
Gene Editing, Screening, and Phenotyping
Vector Construction and Testing
For genome editing in soybean, we constructed two entry vectors to individually express Cas9 and single-guide RNAs (sgRNAs). A Cas9 gene (Sun et al., 2015) driven by the CaMV 35S promoter was cloned into vector Fu76 (containing attL3/4 recombination sites) with StuI and MfeI. The GmU6 promoter (Du et al., 2016), two BsaI sites, and the sgRNA scaffold were cloned into vector Fu79 (containing attL1/2 recombination sites) with SphI and HindIII. The resulting vector was used to construct vector Fu79-sgRNA expressing two sgRNAs by cloning another set of the GmU6 promoter, two BfuAI sites, and the sgRNA scaffold with KpnI and SphI. To design sgRNA of target genes, we use the Web-based tool CRISPR-P (Lei et al., 2014; http://crispr.hzau.edu.cn/cgi-bin/CRISPR2/CRISPR). Two target sequences were synthesized and cloned into Fu79-sgRNA with BsaI and BfuAI, respectively. Finally, Fu76-Cas9 and Fu79-sgRNA were combined into a modified pHairyRED483 (with added attR1/2/3/4 recombination sites, in our lab) to generate binary vectors for gene editing. The gene-editing vectors were introduced into A. rhizogenes K599 for hairy root transformation, and then the positive hairy roots with DsRed2 fluorescence (Lin et al., 2011) were selected for further functional analysis. We also used a method similar to a previous report (Xing et al., 2014) to confirm that the CRISPR/Cas9 system worked in hairy roots, in which fragments surrounding the target sites of GmPHR1, GmPHR4, and GmPHT1;11 were amplified by PCR with three pairs of gene-specific Cas9 test primers (Supplemental Table S1). In this study, two target sites for each gene were confirmed to work well (Supplemental Figs. S6, S7, and S10).
Screening Procedure and Genotyping of Edited Nodules
A detailed procedure for screening and phenotyping CRISPR/Cas9-edited nodules is presented in Supplemental Figure S8 and includes the following steps. Step I, Remove primary roots of the seedling for inoculation of A. rhizogenes. Step II, Allow hairy roots to grow. Step III. Remove negative roots and keep positive ones after screening with the RFP marker (Lin et al., 2011). Then inoculate Sinorhizobium fredii HH103. Step IV, Allow nodule development. Step V, Name and phenotype nodules on roots individually and keep the nodules named after individual roots. Step VI, Genotype individual roots and discard nonedited roots and nodules. Step VII, Based on the genotyping data, combine phenotyping data (step V) from positive roots (edited roots) for statistical analysis of nodule number and size. Step VIII, Combine all positive edited nodules. Step IX, Determine the content of Pi in combined positive nodules. Step X, Analyze gene expression in combined positive nodules. All edited genes resulted in premature and predicted nonfunctional proteins. Around 10 to 14 of the nearly 30 clones showed a single cut, with the others having double cuts in the genomic genes. Thus, the edited efficiencies are 71.9%, 76.7%, and 76.5% for GmPHR1-Cas9, GmPHR4-Cas9, and GmPHT1;11-Cas9, respectively. However, we did not use unedited nodules for phenotyping and gene expression. Instead, all nodules used were edited.
Soybean Transformation
Soybean ‘Tianlong1’ was used as the starting material and as the wild-type control. Soybean stable genetic transformation, mediated by A. tumefaciens strain EH105, was carried out according to a cotyledon node transformation approach (Wang, 2010) to obtain stable genetically transformed plants overexpressing or RNAi silencing GmPHT1;11. Soybean root hairy transformation, mediated by A. rhizogenes strain K599, was performed following previous methods (Kereszt et al., 2007; Lin et al., 2011), using DsRed2 fluorescence as a positive selection marker and an empty vector pHairyRED (Lin et al., 2011) as a negative control.
Soybean Nodulation
A nodulation assay was performed according to a previous report (Broughton and Dilworth, 1971). Typically, the transgenic soybean lines and wild-type plants were inoculated with a suspension of S. fredii HH103 (109 bacteria, in 10 mm MgSO4; Weidner et al., 2012), then transferred to pots (23 × 15 × 23 cm) containing vermiculite. Plants were fed with the low-N nutrient solution (1 mm KNO3, 1 mm CaCl2, 10 μm iron citrate, 0.25 μm MgSO4, 0.25 μm K2SO4, 1 μm MnSO4, 2 μm H3BO3, 0.5 μm ZnSO4, 0.2 μm CuSO4, 0.1 μm CoSO4, and 0.1 μm Na2MoO4, plus 5 μm [low P] or 500 μm [high P] KH2PO4, pH 5.8). At 28 d after inoculation, soybean nodules were harvested for determining nodule number, nodule fresh weight, and soluble Pi content. Nodule size was calculated as the average fresh weight of a single nodule. Pi content was analyzed as described (Chiou et al., 2006).
Nitrogenase Activity Assays
Assay of nitrogenase activity of nodules was performed following a previous report (Buendia-Claveria et al., 1989). Cleaned roots with nodules were placed in a gas reaction bottle, and acetylene gas (10 mL each) was injected into the bottle. The reaction was carried out for 2 h in 28°C. Then, ethylene was measured by gas chromatography (7820A GC system; Agilent Technologies). The standard curve for ethylene was developed according to the standard peak area of ethylene to calculate the molar content of ethylene. Nodule fresh weight was recorded to calculate the nitrogenase activity of nodules as ethylene produced (μmol g−1 fresh weight h−1).
Total RNA Isolation and RT-qPCR
Nodules at 28 d after inoculation were collected for preparation of total RNA using Trizol reagent (Takara). The extracted RNA was treated with DNase I to remove genomic DNA and applied to synthesize first-strand cDNA using the PrimeScript RT Reagent Kit (Takara). The cDNA was diluted to 100 μL with distilled, deionized water in a 1:5 ratio, and 2 μL of the diluted cDNA was used for RT-qPCR with the SYBR Premix Ex Taq (Takara) on the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific; 120 s at 95°C, 45 cycles of 10 s at 95°C and 1 min at 60°C). Expression was determined relative to GmUKN1 (Achard and Genschik, 2009). The data were analyzed by StepOne software v2.0. All primers or oligonucleotides are listed in Supplemental Table S1. Each experiment was repeated at least three times for statistical analysis.
For sampling nodules edited by Cas9, we first selected positive transgenic hairy roots produced by A. rhizogenes harboring a pHairyRED binary vector with an RFP fluorescence marker (Lin et al., 2011). Positive hairy roots were kept and negative ones were removed. The composed seedlings with positive roots were transplanted to soil and inoculated with S. fredii HH103 for nodule development. After 4 weeks, phenotypes of nodules were individually analyzed. Genotyping of edited hairy roots was carried out through PCR with a pair of primers covering two CRISPR/Cas9 target sites. The PCR products were separated on an agarose gel, and the PCR products with at least two bands (Supplemental Fig. S8) were mixed and subcloned into the pGWC vector for transforming into Escherichia coli DH5α. Finally, 30 independent E. coli clones were sequenced to confirm the editing efficiency (Supplemental Fig. S8). cDNA prepared from the corresponding edited nodules was mixed and used for gene expression analysis (Fig. 7), whereas the nodule phenotypes of individually corresponding edited nodules were combined for analysis of nodule number and size (Fig. 8).
Subcellular Localization
The coding regions of GmPHR1, GmPHR2, GmPHR3, and GmPHR4 were introduced into the entry vector pGWC (Chen et al., 2006). The resulting entry clones were combined into pGWB5 (Nakagawa et al., 2007) to generate 35S:GmPHR:GFP binary vectors by LR reaction (Invitrogen). The expression constructs were mobilized into A. tumefaciens strain EHA105, which was infiltrated into the abaxial side of leaves of 2- to 4-week-old N. benthamiana plants. After 48 h, the fluorescence signal was visualized using a Zeiss LSM700 confocal laser scanning microscope. 35S:GFP in pGWB6 (Nakagawa et al., 2007) was used as a control and 35S:mRFP:AHL22 as a nuclear marker (Xiao et al., 2009). Analysis of GmPHT1 protein localization in onion (Allium cepa) epidermal cells was carried out as in our previous report (Fan et al., 2013).
GUS Staining
Histochemical analysis of GUS expression was performed mainly according to a previous report (Jefferson et al., 1987) with minor modifications. The transgenic hairy roots inoculated with S. fredii HH103 were grown for 15 d. The roots and nodules (at least six independent lines) were harvested and fixed in precooled 90% (v/v) acetone, rinsed with cold water, vacuum infiltrated with precooled GUS staining solution [50 mm sodium phosphate buffer, 0.2% (v/v) Triton X-100, 5 mm K4Fe(CN)6, 5 mm K3Fe(CN)6, and 1–2 mm X-gluc] and incubated at 37°C for 12 h. Then, samples were transferred through an ethanol series (20%, 30%, 50%, and 70% [v/v] ethanol) and examined with a microscope. After GUS staining, about 5-mm-long root fragments with nodules were sampled and sectioned transversely for paraffin slides (10 μm thickness) and semithin sections (4 μm thickness). Sections were detected with a light microscope (Zeiss; Discovery2.0).
RNA in Situ Hybridization
RNA in situ hybridization was carried out following a previous procedure (Bustos-Sanmamed et al., 2013). After infection with S. fredii HH103, soybean plants grew in low-phosphorus conditions for 14 d. Roots and nodules were harvested, fixed in formaldehyde-acetic acid solution (50% [v/v] ethanol, 5% [v/v] glacial acetic acid, and 3.7% [v/v] formaldehyde) for 24 h at 4°C, and embedded in paraffin. Sections (7 μm in thickness) were prepared with a microtome (Leica; RM2235). Digoxigenin-labeled antisense and sense RNA probes (Supplemental Table S1) were synthesized by in vitro transcription using the DIG RNA Labeling Kit (Roche). The hybridization signals were detected by enzyme immunoassay and enzyme-catalyzed color reaction with 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (Roche; catalog no. 11175041910). Slides were examined with a light microscope (Zeiss; Discovery2.0).
EMSA
The full-length coding sequences of GmPHR1 and GmPHR4 genes were cloned into the vector pET28a (Novagen) using BamHI and XhoI sites, and the resulting vectors were transformed into E. coli strain BL21. The recombinant proteins were purified using Ni-NTA beads (Qiagen). The biotin-labeled probes of the P1BS-containing fragments were generated by annealing the biotin-labeled oligonucleotide and its non-biotin-labeled complementary oligonucleotide. EMSA was performed according to the instructions of the Light-Shift Chemiluminescent EMSA Kit (Thermo Scientific; no. 829-07910). Components of EMSA binding reactions included 1× binding buffer, 2.5% (v/v) glycerol, 5 mm MgCl2, 50 ng μL−1 poly(dI·dC), 0.05% (v/v) Nonidet P-40, 1 unit of protein extract, 20 fmol of biotin-labeled target DNA, and 0 to 16 pmol of unlabeled target DNA. A final concentration of 1 mm Pi (KH2PO4) was added to binding reactions for +P treatment, and 1 mm KCl was replaced for −P treatment. All oligonucleotides are listed in Supplemental Table S1.
ChIP-qPCR
ChIP-qPCR assays were performed essentially as previously described (Malapeira and Mas, 2014). Chromatin was isolated from the transgenic hairy roots overexpressing GmPHR1:GFP or GmPHR4:GFP. The isolated chromatin was cross-linked with 1% (v/v) formaldehyde, sonicated, and precipitated by Anti-GFP mAb-Magnetic Beads (MBL; code no. D153-11). The precipitated DNA fragments were released by 215 mm NaCl and subjected to qPCR analysis. The primers used are listed in Supplemental Table S1.
Transcriptional Activity Assays
The regions upstream of the ATG sites of GmPHT1;1 (GmPHT1;1pro; 5,240 bp), GmPHT1;4 (GmPHT1;4pro; 1,855 bp), and GmPHT1;11 (GmPHT1;11pro; 2,923 bp) were introduced into the entry vector pGWC (Chen et al., 2006). Then, the promoters were fused to the LUC reporter gene on the pGWB35 vector (Nakagawa et al., 2007) by LR reaction (Invitrogen). The expression vectors of 35S:GmPHR1:GFP and 35S:GmPHR4:GFP were used for expressing activators, while GmPHT1;1/4/11:GUS were reporters. A. tumefaciens bacteria containing the indicated constructs were transiently transformed into the abaxial side of leaves of 2- to 4-week-old N. benthamiana plants. After 2 d of inoculation, the infiltrated leaves were harvested and sprayed with 3 mm d-luciferin (Proven and Published; no. LUCK-1G), and the fluorescence was detected using a plant imaging system (LB 985 NightSHADE; Berthold Technologies). The images were analyzed by indiGO software (Berthold Technologies). pGWB6 (35S:GFP; Nakagawa et al., 2007) was employed as a control. There were at least three biological replicates for each assay. The primers used to amplify coding regions of GmPHRs and promoter regions of GmPHT1s are listed in Supplemental Data S1.
Statistical Analyses
All experiments in our study were carried out with at least three biological replicates and at least three independent times, all of which showed similar results. For presentation, the figures show only representative results out of at least five individual experiments. Data in all bar graphs represent means ± sd from three independent experiments. All statistical analyses were determined using the SPSS software package (https://www.ibm.com/analytics/spss-statistics-software). Asterisks indicate significant differences according to Student’s t test (**, P < 0.01 and *, P < 0.05).
Accession Numbers
Sequence data for this article can be found in Phytozome (https://phytozome.jgi.doe.gov/) with the following gene identities: GmPHR1 (Glyma.19G167500), GmPHR2 (Glyma.03G166400), GmPHR3 (Glyma.13G126200), GmPHR4 (Glyma.10G039700), GmPHT1;1 (Glyma.10G186500), GmPHT1;4 (Glyma.10G036800), GmPHT1;11 (Glyma.14G188000), GmPHT1;14 (Glyma.02G005800), and GmUKN1 (Glyma.12G020500).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Subcellular localization of GmPHT1;11, GmPHT1;1, GmPHT1;4, and GmPHT1;14 proteins.
Supplemental Figure S2. Soybean PHR proteins shared high conservation with their homologs in Arabidopsis and rice.
Supplemental Figure S3. Tissue-specific expression of the promoters of GmPHR2, GmPHR3, and GmPHT1;14.
Supplemental Figure S4. GmPHR1 and GmPHR4 proteins did not bind to two P1BS-containing fragments of the PHT1;1 gene promoter.
Supplemental Figure S5. The gene interaction among GmPHR-GmPHT1 modules by RT-qPCR analysis of gene expression.
Supplemental Figure S6. Characterization of mutation of GmPHR1 Gene by CRISPR/Cas9 editing in soybean hairy roots.
Supplemental Figure S7. Knockout of GmPHT1;11 by CRISPR/Cas9 increased the number but decreased the size of nodules in soybean hair roots.
Supplemental Figure S8. A protocol for genotyping and phenotyping CRISPR/Cas9-edited hairy roots.
Supplemental Figure S9. The statistical analysis of fresh weight of nodules from gene editing or transgenic roots.
Supplemental Figure S10. Knockout of GmPHT1;11 by CRISPR/Cas9 increased the number but decreased the size of nodules in soybean hair roots.
Supplemental Table S1. Oligonucleotides used in our study.
Supplemental Table S2. Nitrogenase activity assay.
Supplemental Data Set S1. The sequences of the promoters and the coding regions of genes.
Acknowledgments
We thank Drs. Wenfeng Chen and Yanwei Sun for help with nitrogenase analysis, all friends and colleagues who helped us in any way but are not included in the author list, and LetPub (www.letpub.com) for its linguistic assistance during the preparation of the article.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant nos. 2016YFD0101005 and 2006YFD0101900), Transgenic Project (grant no. 2014ZX0800930B), and the Chinese Academy of Agricultural Sciences-Innovation Team Project and the Basal Research Fund (grant no. Y2017CG25).
References
- Achard P, Genschik P(2009) Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J Exp Bot 60: 1085–1092 [DOI] [PubMed] [Google Scholar]
- Araujo AP, Plassard C, Drevon JJ(2008) Phosphatase and phytase activities in nodules of common bean genotypes at different levels of phosphorus supply. Plant Soil 312: 129–138 [Google Scholar]
- Bardin S, Dan S, Osteras M, Finan TM(1996) A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J Bacteriol 178: 4540–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR(2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH(2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse D, Kock M(1998) Influence of phosphate starvation on phosphohydrolases during development of tomato seedlings. Plant Cell Environ 21: 325–332 [Google Scholar]
- Broughton WJ, Dilworth MJ(1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia-Claveria AM, Chamber M, Ruiz-Sainz JE(1989) A comparative study of the physiological characteristics, plasmid content and symbiotic properties of different Rhizobium fredii strains. Syst Appl Microbiol 12: 203–209 [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J(2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos-Sanmamed P, Laffont C, Frugier F, Lelandais-Brière C, Crespi M(2013) Analyzing small and long RNAs in plant development using non-radioactive in situ hybridization. Methods Mol Biol 959: 303–316 [DOI] [PubMed] [Google Scholar]
- Cabeza RA, Liese R, Lingner A, von Stieglitz I, Neumann J, Salinas-Riester G, Pommerenke C, Dittert K, Schulze J(2014) RNA-seq transcriptome profiling reveals that Medicago truncatula nodules acclimate N2 fixation before emerging P deficiency reaches the nodules. J Exp Bot 65: 6035–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo G, Teixeira PJ, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD, et al. (2017) Root microbiota drive direct integration of phosphate stress and immunity. Nature 543: 513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Qin L, Zhou L, Li X, Chen Z, Sun L, Wang W, Lin Z, Zhao J, Yamaji N, et al. (2019) A nodule-localized phosphate transporter GmPT7 plays an important role in enhancing symbiotic N2 fixation and yield in soybean. New Phytol 221: 2013–2025 [DOI] [PubMed] [Google Scholar]
- Chen QJ, Zhou HM, Chen J, Wang XC(2006) Using a modified TA cloning method to create entry clones. Anal Biochem 358: 120–125 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL(2006) Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevon JJ, Hartwig UA(1997) Phosphorus deficiency increases the argon-induced decline of nodule nitrogenase activity in soybean and alfalfa. Planta 201: 463–469 [Google Scholar]
- Du H, Zeng X, Zhao M, Cui X, Wang Q, Yang H, Cheng H, Yu D(2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol 217: 90–97 [DOI] [PubMed] [Google Scholar]
- Fan C, Wang X, Hu R, Wang Y, Xiao C, Jiang Y, Zhang X, Zheng C, Fu YF(2013) The pattern of Phosphate transporter 1 genes evolutionary divergence in Glycine max L. BMC Plant Biol 13: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili F, Huss-Danell K(2003) Local and systemic effects of phosphorus and nitrogen on nodulation and nodule function in Alnus incana. J Exp Bot 54: 2757–2767 [DOI] [PubMed] [Google Scholar]
- Guo M, Ruan W, Li C, Huang F, Zeng M, Liu Y, Yu Y, Ding X, Wu Y, Wu Z, et al. (2015) Integrative comparison of the role of the PHOSPHATE RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiol 168: 1762–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Kobae Y, Omoto E, Tanaka A, Banba M, Takai S, Tamura Y, Hirose A, Komatsu K, Otagaki S, et al. (2014) The soybean mycorrhiza-inducible phosphate transporter gene, GmPT7, also shows localized expression at the tips of vein endings of senescent leaves. Plant Cell Physiol 55: 2102–2111 [DOI] [PubMed] [Google Scholar]
- Israel DW.(1987) Investigation of the role of phosphorus in symbiotic dinitrogen fixation. Plant Physiol 84: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel DW.(1993) Symbiotic dinitrogen fixation and host‐plant growth during development of and recovery from phosphorus deficiency. Physiol Plant 88: 294–300 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW(1987) GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karandashov V, Nagy R, Wegmüller S, Amrhein N, Bucher M(2004) Evolutionary conservation of a phosphate transporter in the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 101: 6285–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A(2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kereszt A, Li D, Indrasumunar A, Nguyen CD, Nontachaiyapoom S, Kinkema M, Gresshoff PM(2007) Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat Protoc 2: 948–952 [DOI] [PubMed] [Google Scholar]
- Khan GA, Vogiatzaki E, Glauser G, Poirier Y(2016) Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol 171: 632–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Singh S, Nandwal AS, Waldia RS, Sharma MK(2008) Genotypic differences in water status, membrane integrity, ionic content, N2-fixing efficiency and dry matter of mungbean nodules under saline irrigation. Physiol Mol Biol Plants 14: 363–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguerre G, Heulin-Gotty K, Brunel B, Klonowska A, Le Quéré A, Tillard P, Prin Y, Cleyet-Marel JC, Lepetit M(2012) Local and systemic N signaling are involved in Medicago truncatula preference for the most efficient Sinorhizobium symbiotic partners. New Phytol 195: 437–449 [DOI] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL(2014) CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7: 1494–1496 [DOI] [PubMed] [Google Scholar]
- Li XH, Wang YJ, Wu B, Kong YB, Li WL, Chang WS, Zhang CY(2014) GmPHR1, a novel homolog of the AtPHR1 transcription factor, plays a role in plant tolerance to phosphate starvation. J Integr Agric 13: 2584–2593 [Google Scholar]
- Lin MH, Gresshoff PM, Indrasumunar A, Ferguson BJ(2011) pHairyRed: A novel binary vector containing the DsRed2 reporter gene for visual selection of transgenic hairy roots. Mol Plant 4: 537–545 [DOI] [PubMed] [Google Scholar]
- Malapeira J, Mas P(2014) ChIP-seq analysis of histone modifications at the core of the Arabidopsis circadian clock. Methods Mol Biol 1158: 57–69 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T(2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH(2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC(2011) Phosphate import in plants: Focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, et al. (2014) SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA 111: 14947–14952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Rojas-Triana M, de Lorenzo L, Leyva A, Rubio V, Paz-Ares J(2017) Novel signals in the regulation of Pi starvation responses in plants: Facts and promises. Curr Opin Plant Biol 39: 40–49 [DOI] [PubMed] [Google Scholar]
- Qin L, Guo Y, Chen L, Liang R, Gu M, Xu G, Zhao J, Walk T, Liao H(2012a) Functional characterization of 14 Pht1 family genes in yeast and their expressions in response to nutrient starvation in soybean. PLoS ONE 7: e47726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Zhao J, Tian J, Chen L, Sun Z, Guo Y, Lu X, Gu M, Xu G, Liao H(2012b) The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol 159: 1634–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar AS, Dénervaud N, Maerkl SJ(2013) Mapping the fine structure of a eukaryotic promoter input-output function. Nat Genet 45: 1207–1215 [DOI] [PubMed] [Google Scholar]
- Ren F, Guo QQ, Chang LL, Chen L, Zhao CZ, Zhong H, Li XB(2012) Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS ONE 7: e44005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin A, Auer PL, Libault M, Schlueter J, Farmer A, May G, Stacey G, Doerge RW, Jackson SA(2013) The fate of duplicated genes in a polyploid plant genome. Plant J 73: 143–153 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J(2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schulze J, Adgo E, Merbach W(1999) Carbon costs associated with N2 fixation in Vicia faba L. and Pisum sativum L. over a 14-day period. Plant Biol (Stuttg) 1: 625–631 [Google Scholar]
- Schulze J, Temple G, Temple SJ, Beschow H, Vance CP(2006) Nitrogen fixation by white lupin under phosphorus deficiency. Ann Bot 98: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schünmann PH, Richardson AE, Vickers CE, Delhaize E(2004) Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol 136: 4205–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulieman S, Ha CV, Schulze J, Tran LS(2013) Growth and nodulation of symbiotic Medicago truncatula at different levels of phosphorus availability. J Exp Bot 64: 2701–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y(2015) Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep 5: 10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Kobae Y, Mizuno T, Hata S(2012) Identification and expression analysis of arbuscular mycorrhiza-inducible phosphate transporter genes of soybean. Biosci Biotechnol Biochem 76: 309–313 [DOI] [PubMed] [Google Scholar]
- Tang C, Hinsinger P, Drevon JJ, Jaillard B(2001) Phosphorus deficiency impairs early nodule functioning and enhances proton release in roots of Medicago truncatula L. Ann Bot (Lond) 88: 131–138 [Google Scholar]
- Thuynsma R, Valentine A, Kleinert A(2014) Phosphorus deficiency affects the allocation of below-ground resources to combined cluster roots and nodules in Lupinus albus. J Plant Physiol 171: 285–291 [DOI] [PubMed] [Google Scholar]
- Valentine AJ, Kleinert A, Benedito VA(2017) Adaptive strategies for nitrogen metabolism in phosphate deficient legume nodules. Plant Sci 256: 46–52 [DOI] [PubMed] [Google Scholar]
- Vardien W, Mesjasz-Przybylowicz J, Przybylowicz WJ, Wang Y, Steenkamp ET, Valentine AJ(2014) Nodules from Fynbos legume Virgilia divaricata have high functional plasticity under variable P supply levels. J Plant Physiol 171: 1732–1739 [DOI] [PubMed] [Google Scholar]
- Wang K.(2010) Agrobacterium-mediated transformation of soybean and recovery of transgenic soybean plants. Iowa State University, Ames, IA: Available at: http://agron-www.agron.iastate.edu/ptf/service/Agrosoy.aspx [Google Scholar]
- Wang X, Fan C, Zhang X, Zhu J, Fu YF(2013) BioVector, a flexible system for gene specific-expression in plants. BMC Plant Biol 13: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang X, Paterson AH(2012) Genome and gene duplications and gene expression divergence: A view from plants. Ann N Y Acad Sci 1256: 1–14 [DOI] [PubMed] [Google Scholar]
- Wang Z, Ruan W, Shi J, Zhang L, Xiang D, Yang C, Li C, Wu Z, Liu Y, Yu Y, et al. (2014) Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci USA 111: 14953–14958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner S, Becker A, Bonilla I, Jaenicke S, Lloret J, Margaret I, Pühler A, Ruiz-Sainz JE, Schneiker-Bekel S, Szczepanowski R, et al. (2012) Genome sequence of the soybean symbiont Sinorhizobium fredii HH103. J Bacteriol 194: 1617–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Chen F, Yu X, Lin C, Fu YF(2009) Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol Biol 71: 39–50 [DOI] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ(2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue YB, Xiao BX, Zhu SN, Mo XH, Liang CY, Tian J, Liao H, Miriam G(2017) GmPHR25, a GmPHR member up-regulated by phosphate starvation, controls phosphate homeostasis in soybean. J Exp Bot 68: 4951–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P(2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]