SAP AND MIZ1 DOMAIN-CONTAINING LIGASE1 (SIZ1) negatively regulates in vitro shoot regeneration by suppressing transcriptional activation of reprogramming regulators.
Abstract
Plants form calluses and regenerate new organs when incubated on phytohormone-containing media. While accumulating evidence suggests that these regenerative processes are governed by transcriptional networks orchestrating wound response and developmental transitions, it remains unknown if posttranslational regulatory mechanisms are involved in this process. In this study, we demonstrate that SAP AND MIZ1 DOMAIN- CONTAINING LIGASE1 (SIZ1), an E3 ligase-catalyzing attachment of the SMALL UBIQUITIN-LIKE MODIFIER (SUMO) to proteins, regulates wound-induced signal transduction and organ regeneration in Arabidopsis (Arabidopsis thaliana). We show that loss-of-function mutants for SIZ1 exhibit overproduction of shoot meristems under in vitro tissue culture conditions, while this defect is rescued in a complementation line expressing pSIZ1::SIZ1. RNA sequencing analysis revealed that siz1-2 mutants exhibit enhanced transcriptional responses to wound stress, resulting in the hyper-induction of over 400 genes immediately after wounding. Among them, we show that elevated levels of WOUND INDUCED DEDIFFERENTIATION1 (WIND1) and WIND2 contribute to the enhanced shoot regeneration observed in siz1 mutants, as expression of the dominant-negative chimeric protein WIND1-SRDX (SUPERMAN repression domain) in siz1-3 mutants partly rescues this phenotype. Although compromised SIZ1 function does not modify the transcription of genes implicated in auxin-induced callus formation and/or pluripotency acquisition, it does lead to enhanced induction of cytokinin-induced shoot meristem regulators such as WUSCHEL, promoting the formation of WUSCHEL-expressing foci in explants. This study thus suggests that SIZ1 negatively regulates shoot regeneration in part by repressing wound-induced developmental reprogramming.
Plants display a remarkable capacity for regeneration and reconstruct of both shoots and roots following severe injury. This feature is widely exploited in vitro, where regenerants are produced via de novo organogenesis induced by incubating explants on media supplemented with exogenous phytohormones (Skoog and Miller, 1957; Sugiyama, 2015). In a routinely used two-step procedure, explants are initially incubated on auxin-rich callus-inducing medium (CIM) to generate a pluripotent cell mass, called callus, and subsequently transferred to cytokinin-rich shoot-inducing medium (SIM) to induce shoot formation (Valvekens et al., 1988). Recent studies using Arabidopsis (Arabidopsis thaliana) have uncovered several key mechanisms involved in the reactivation of developmental processes in response to external stimuli that lead to induction of de novo organogenesis. For instance, upon incubation of explants on CIM, auxin triggers callus formation through the reactivation of lateral root formation (Sugimoto et al., 2010). Auxin-activated AUXIN RESPONSE FACTOR7 (ARF7) and ARF19 are known to promote expression of LATERAL ORGAN BOUNDARY DOMAIN16 (LBD16), LBD18, and LBD29, which then promote cell proliferation (Fan et al., 2012). Following the induction of callus formation, root meristem regulators WUSCHEL-RELATED HOMEOBOX5 (WOX5) and PLETHORA1 (PLT1) are broadly expressed in developing callus cells (Atta et al., 2009; Sugimoto et al., 2010). This gain of root meristem-like identity within callus cells is crucial for the acquisition of shoot regeneration competency, as illustrated by the requirement of root meristem regulators PLT3, PLT5, PLT7, and LBD16 for shoot formation (Kareem et al., 2015; Liu et al., 2018). Upon transfer to SIM, cytokinin and auxin induce reprogramming of pluripotent callus, allowing the acquisition of shoot meristem identity. A key molecular event underlying this cell fate transition is the transcriptional activation of a homeobox gene, WUSCHEL (WUS), which is induced in promeristems within the first few days after transfer to SIM (Atta et al., 2009). This is largely mediated by cytokinin signaling components ARABIDOPSIS RESPONSE REGULATOR1 (ARR1), ARR10, and ARR12, which together with homeodomain Leu zipper III transcription factors such as PHABULOSA, PHAVOLUTA, and REVOLUTA, directly induce WUS expression (Meng et al., 2017; Zhang et al., 2017). Other important regulators of shoot regeneration are the APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) transcription factors ENHANCER OF SHOOT REGENERATION1 (ESR1) and ESR2, which are up-regulated on SIM and directly activate the expression of the shoot meristem regulators CUP SHAPED COTYLEDON (CUC1) and CUC2 (Ikeda et al., 2006; Matsuo et al., 2011). CUCs are necessary for induction of the homeodomain transcriptional factor SHOOT MERISTEMLESS (STM), which is required for shoot meristem formation and in vitro regeneration alike (Aida et al., 1999; Daimon et al., 2003).
Although exogenous hormone application alone is often insufficient to induce shoot regeneration on tissue culture, wounding of tissues during excision of explants can provide the trigger necessary for cellular reprogramming and subsequent shoot regeneration (Iwase et al., 2015). A gene encoding a wound-inducible AP2/ERF transcription factor, WOUND-INDUCED DEDIFFERENTIATION1 (WIND1), and its homologs WIND2 through WIND4 promote callus formation and acquisition of regeneration competency (Iwase et al., 2011, 2015, 2017). Expression of WIND1 is enhanced upon cutting and promotes callus formation via activation of the cytokinin response (Iwase et al., 2011) and shoot regeneration via direct up-regulation of ESR1 expression (Iwase et al., 2017). Another important signaling component after wounding is jasmonic acid (JA; León et al., 2001; Koo et al., 2009), but its contribution to organ regeneration seems to be context dependent. For instance, JA promotes de novo root regeneration from leaf cuttings (Zhang et al., 2019a), but it represses callus formation in wounded hypocotyls (Ikeuchi et al., 2017). Wounding also activates other stress-associated plant hormone pathways such as those mediated by abscisic acid (ABA), ethylene, or salicylic acid (SA; León et al., 2001; Wang et al., 2002; Ogawa et al., 2010), but it is currently unclear if these pathways are involved in organ regeneration.
Wounding, as well as other abiotic stresses such as heat and drought, induces transcription-independent signals that act as important primary triggers to alter the expression of stress-responsive genes. Posttranslational modification of proteins represents a key layer of regulation that enables intricate control of protein function in many signaling pathways mediating environmental responses and developmental processes. Advantages of posttranslational modification include enabling rapid activation of protein function in response to acute stress, as well as energy efficient fine tuning of transcriptional responses appropriate for the perceived stress (Mazzucotelli et al., 2008). While several transcriptional cascades were shown to transduce stress and/or hormonal cues to activate the expression of developmental regulators, little is known about the contribution of posttranslational regulation to the control of organ regeneration. Plants utilize a combination of different protein modifications to regulate protein activity, including the extensively characterized phosphorylation and ubiquitination (Bachmair et al., 2001; Mazzucotelli et al., 2008; Guerra et al., 2015). In addition, conjugation of the SMALL UBIQUITIN-LIKE MODIFIER (SUMO) peptide has been implicated in regulating a number of important transcriptional regulators (Castro et al., 2012; Mazur and van den Burg, 2012; Augustine and Vierstra, 2018). SUMOylation is a multistep process and involves sequentially SUMO activation by E1 enzymes, SUMO conjugation by E2 enzymes, and ligation catalyzed by E3 enzymes. Unlike ubiquitination, for which over 1,400 putative E3 ligases have been found in Arabidopsis, SUMOylation is catalyzed by only two E3 ligases, the SAF-A/B, Acinus, and PIAS (SAP) and Msx-interacting-zinc finger (MIZ) DOMAIN-CONTAINING LIGASE1 (SIZ1) and METHYL METHANESULFONATE-SENSITIVE21/HIGH PLOIDY2 (MMS2/HPY2; Ishida et al., 2009, 2012; Kwak et al., 2016; Liu et al., 2016). Several environmental cues, such as heat, reactive oxygen species, cold, and drought stress, lead to accumulation of SUMOylated proteins in plants (Kurepa et al., 2003; Catala et al., 2007; Castro et al., 2012; Miller et al., 2013), and most of these stress-induced SUMOylation events are attributed to SIZ1 (Elrouby and Coupland, 2010; Miller et al., 2010; Castro et al., 2012; Rytz et al., 2018). Accumulation of SUMOylated proteins occurs within 10 min of heat stress and is thought to be transient (Kurepa et al., 2003; Saracco et al., 2007). Recent proteomic studies identified more than 1,000 putative SUMOylated proteins in plants and revealed that around 80% of these SUMOylation targets are nuclear-localized proteins, including transcription factors, chromatin remodeling enzymes, and histones (Shiio and Eisenman, 2003; Rytz et al., 2018). Targets of stress-induced SUMOylation include TOPLESS-RELATED1 (TPR1), implicated in the SA-mediated pathogen response (Niu et al., 2019), ABA-INSENSITIVE5 (ABI5), a key regulator of ABA signaling (Miura et al., 2009), and JASMONATE ZIM6 (JAZ6), a regulator of JA signaling (Srivastava et al., 2018). Additionally, several important developmental regulators were found to be SUMO targeted. For instance, a recent study demonstrated that SUMOylation of ARF7 promotes its interaction with its negative regulator INDOLEACETIC ACID-INDUCED PROTEIN3 (IAA3), consequently contributing to root branching toward water (Orosa-Puente et al., 2018).
Given that organ regeneration is the outcome of the reactivation of developmental processes upon wound stress, we sought to investigate whether SIZ1-mediated SUMOylation is involved in the regulation of de novo organogenesis. We show that SIZ1 represses in vitro shoot regeneration, as siz1 mutants exhibit overproduction of shoots in the tissue culture condition. Our gene expression analysis revealed that the transcriptional wound response following explant preparation is strongly enhanced in these mutants. Our transcriptome data further uncovered that regulators of shoot meristem formation are highly activated upon transfer of siz1 mutants to SIM. This study thus demonstrates that SIZ1 is required for the regulation of shoot regeneration by modulating the expression of key regeneration regulators.
RESULTS
SIZ1 Negatively Regulates Shoot Regeneration
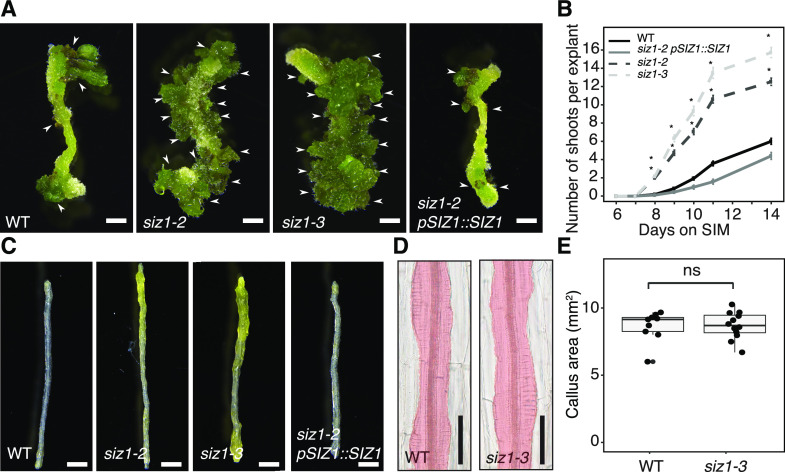
In order to investigate the role of SIZ1 in the control of de novo organogenesis, we tested whether Arabidopsis mutants for SIZ1 display altered regenerative responses using a two-step tissue culture procedure. As previously described (Valvekens et al., 1988), we cut hypocotyl segments and incubated them on CIM for 4 d before transferring them to SIM. Under our culture conditions, wild-type explants start to regenerate shoots around 9 d after transfer to SIM, leading to the formation of, on average, five visible shoots per explant at 14 d (Fig. 1, A and B). In contrast, shoot regeneration is dramatically enhanced in two SIZ1 loss-of-function mutants, siz1-2 and siz1-3, with new shoots appearing by 8 d on SIM and more than 12 shoots forming at 14 d (Fig. 1, A and B). We also found that explants of these siz1 mutants already appear green by 7 d on SIM (Fig. 1C), supporting that the initiation of shoot developmental programs is advanced by the siz1 mutation. These enhanced shoot regeneration phenotypes in the siz1-2 mutant are rescued in siz1-2 pSIZ1::SIZ1 plants expressing SIZ1 under the control of its native promoter (Fig. 1, A–C), clearly demonstrating that SIZ1 negatively regulates shoot regeneration.
Figure 1.
SIZ1 negatively regulates shoot regeneration in vitro. A, Images showing shoot regeneration phenotypes in hypocotyl explants of siz1-2, siz1-3, and siz1-2 pSIZ1::SIZ1 compared to Col-0 wild type (WT). Explants were incubated on CIM for 4 d then on SIM for 14 d. Scale bars = 1 mm. Arrows indicate regenerated shoots. B, Time series quantification of shoot regeneration for the genotypes pictured in A. Explants were incubated on CIM for 4 d followed by SIM for the indicated number of days. Sample sizes are wild type (n = 68), siz1-2 (n = 103), siz1-3 (n = 99), siz1-2 pSIZ1::SIZ1 (n = 47). Error bars represent ± SE. Asterisks indicate significant differences by one-way ANOVA and post-hoc Tukey for each genotype compared to wild type at the same time point (*P < 0.05). C, Images showing accelerated greening of hypocotyl explants in siz1-2 and siz1-3 compared to wild type and siz1-2 pSIZ1::SIZ1. Explants were incubated on CIM for 4 d followed by SIM for 7 d. Scale bars = 1 mm. D, DIC images showing similar callus production in wild-type and siz1-3 explants following 4 d of CIM incubation. Area marked in red highlights the callus-containing region. Scale bars = 50 µm. E, Quantification of projected callus area as shown in D. Sample sizes are wild type (n = 9) and siz1-3 (n = 12). ns, Not significant as evaluated by Wilcoxon rank-sum test (P = 0.917).
Since preincubation of explants on CIM is a prerequisite for shoot regeneration on SIM (Che et al., 2007), we next tested if the siz1-3 mutation causes enhanced callus formation on CIM, which could be responsible for the subsequent promotion of shoot regeneration on SIM. As shown in Figure 1, D and E, both callus size and morphology are indistinguishable between wild type and siz1-3 plants after 4 d on CIM, the time point at which the explants are transferred to SIM. It is therefore unlikely that enhanced shoot regeneration on SIM is a secondary consequence of enhanced callus formation during CIM incubation. Nevertheless, we did observe that after prolonged incubation on CIM, the siz1 mutants eventually develop substantially larger callus (Supplemental Fig. S1), suggesting that SIZ1 also negatively regulates callus formation during long-term CIM incubation.
The siz1 Mutation Causes Hyperactivation of the Wound Response
In order to explore the molecular basis underlying enhanced shoot regeneration in the siz1 mutant, we performed genome-wide transcriptomic analysis using RNA sequencing (RNA-seq). We compared gene expression in wild-type and siz1-2 plants at four time points: within 15 min after excising hypocotyls (after cutting), after 4 d of incubation on CIM and after either 4 or 6 d on SIM (Fig. 2A). Among these time points, we observed the most drastic differences in the global gene expression pattern immediately after cutting, when 1,375 genes are up-regulated and 912 genes are down-regulated in siz1-2 compared to wild type (edge-R; false discovery rate < 0.05; Fig. 2B; Supplemental Table S1). The differences in the gene expression profile are far less prominent at 4 d on CIM when only 187 genes are up-regulated and 66 genes are down-regulated (Fig. 2B). After transfer to SIM, however, the gene expression pattern is again profoundly affected by the siz1 mutation. After 4 d on SIM, we detected 593 genes up-regulated and 640 genes down-regulated, and by 6 d on SIM, the number of misexpressed genes was further increased (Fig. 2B). Interestingly, we found that misexpressed genes are largely distinct after cutting on CIM and on SIM (Fig. 2C). For instance, we detected that only 15% of the genes (211 genes out of 1,375 genes, P = 6.7e−37; hypergeometric test) up-regulated after cutting are also up-regulated on SIM. We observed a similar trend for down-regulated genes in siz1-2, as the overlap between genes down-regulated after cutting on CIM and on SIM is small (Supplemental Fig. S2A). Accordingly, Gene Ontology (GO) enrichment analysis demonstrated that genes associated with unique biological processes are up-regulated in siz1-2 at each of these time points (Fig. 2D). In contrast, we observed less distinct GO enrichment among down-regulated genes (Supplemental Fig. S2B). Using these datasets, we also found that while some genes of the SUMOylation pathway are differentially expressed in the wild type across the different time points, expression of SIZ1 is fairly stable (Supplemental Fig. S3), implying that the activity of SIZ1 is not regulated at the transcriptional level.
Figure 2.
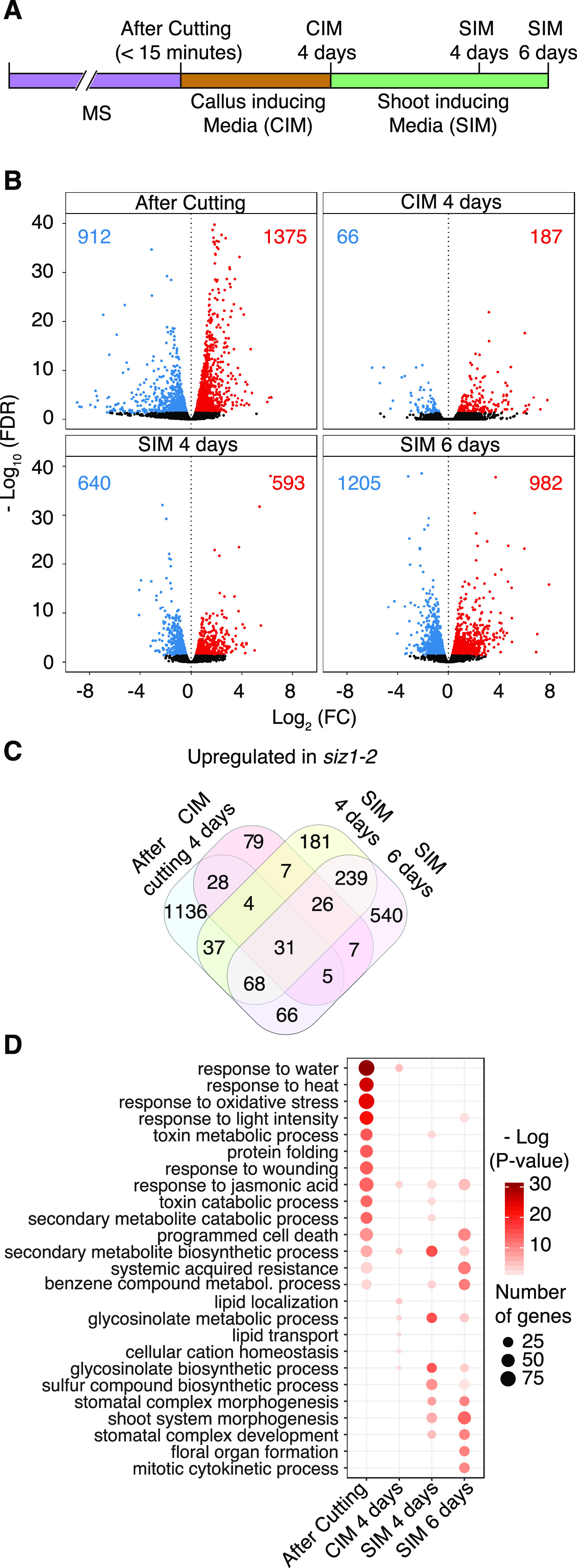
SIZ1 alters gene expression at multiple time points during in vitro shoot regeneration. A, Graphical depiction of the RNA-seq analysis experimental setup. B, Volcano plots showing differentially expressed genes (edgeR, false discovery rate [FDR] < 0.05) between siz1-2 and wild-type (WT) explants. Significantly up-regulated or down-regulated genes in siz1-2 are depicted as red or blue dots, respectively. Red or blue numbers indicate the total number of up-regulated or down-regulated genes. C, Venn diagram showing the overlap between genes up-regulated in siz1-2 at different time points. D, GO enrichment analysis for genes up-regulated in siz1-2. Displayed are the top 15 most enriched GO categories among up-regulated genes at any given time point. Highly redundant GO categories were manually removed.
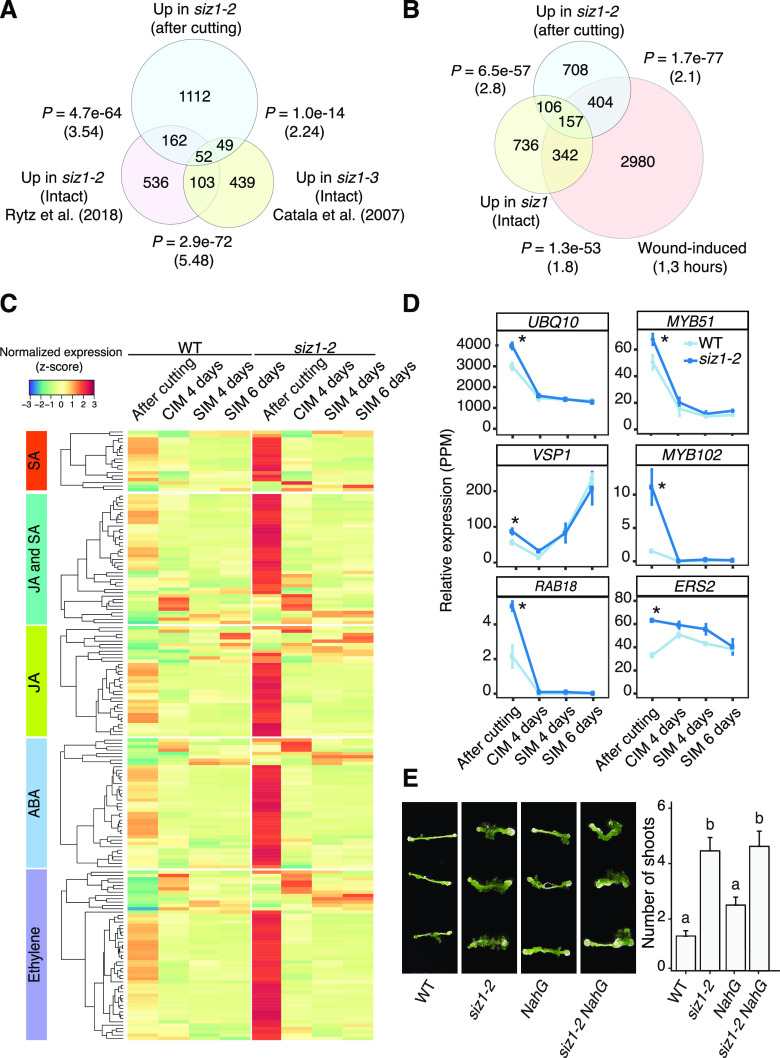
Among genes up-regulated in siz1-2 immediately after cutting, we found that those associated with stress response are strongly represented. GO categories such as “response to water” (P = 2.5e−31), “response to oxidative stress” (P = 6.7e−25), and “response to wounding” (P = 1.1e−14) are highly represented (Fig. 2D), implying that the stress response is hyperactivated in siz1-2. To test whether this apparent hyper response is caused by cutting or constitutively present in siz1 mutants even in nonstress conditions, we examined the overlap between the genes highly expressed upon cutting in our dataset, and those up-regulated in intact siz1-2 and siz1-3 plants as reported in other studies (Catala et al., 2007; Rytz et al., 2018). As shown in Figure 3A, we found that more than 80% of the genes present in our dataset (1,112 genes out of 1,375 genes) are unique to it, supporting the idea that a substantial proportion of the transcriptional up-regulation we detected in siz1 mutants is caused by cutting. We observed a significant overlap of our dataset with either of the previously published ones (Catala et al., 2007; Rytz et al., 2018; P = 4.7e−64 and P = 1.0e−14; hypergeometric test), suggesting that our dataset includes some genes constitutively activated in siz1. To further examine how many of the genes in our dataset are induced by wounding, we compared our dataset with previously published transcriptomic data for wounded hypocotyls (Ikeuchi et al., 2017). As shown in Figure 3B, more than 40% of genes (561 genes out of 1375 genes, P = 1.7e−77; hypergeometric test, representation factor = 2.1) up-regulated in siz1 are induced within 3 h following cutting of hypocotyls in wild type. Importantly, 404 genes out of these 561 genes are up-regulated only after cutting and not in intact siz1 plants (Fig. 3B), indicating that siz1 mutants display a hypersensitive response to wounding stimuli.
Figure 3.
Many genes associated with stress-related hormone signaling are hyperactivated after wounding in siz1 mutants. A, Overlap between genes up-regulated in siz1-2 compared to the wild type (WT) after cutting, based on our RNA-seq analysis, and those up-regulated in intact siz1 mutant seedlings, as determined in previous studies (Catala et al., 2007; Rytz et al., 2018). The significance of the overlap between pairs of gene sets was evaluated by a hypergeometric test. The calculated representation factor is shown in brackets. B, Overlap between genes up-regulated in siz1-2 after cutting, genes induced at 1 and/or 3 h after wounding in hypocotyls (Ikeuchi et al., 2017), and genes up-regulated in siz1 intact plants (Catala et al., 2007; Rytz et al., 2018). The significance of the overlap between pairs of gene sets was evaluated by a hypergeometric test. The calculated representation factor is shown in brackets. C, Heatmap showing the time course expression of genes hyperactivated after cutting in siz1-2 compared to wild type. Genes associated with a GO category containing one of the following terms, JA, SA, ethylene, or ABA, are included. Normalized expression was calculated for each gene across all time points, using data from both genotypes. D, Relative expression of a selection of genes which are associated with one or more of the phytohormone GO terms shown in C in siz1-2 and in wild type explants. Error bars represent ± SE. Asterisks indicate significant differences by edgeR comparative analysis between wild type and siz1-2 (*P < 0.05). E, Images (left) and quantification (right) of shoot regeneration from hypocotyl explants of wild type, siz1-2, NahG, and siz1-2 NahG plants. All explants were incubated on CIM for 4 d and then on SIM for 14 d. Values represent mean number of shoots produced per explant, and error bars represent ± SE. Sample sizes are wild type (n = 37), siz1-2 (n = 44), NahG (n = 48), and siz1-2 NahG (n = 38). Different letters indicate significant differences based on one-way ANOVA and post-hoc Tukey test (P < 0.05).
Phytohormones including SA, JA, ABA, and ethylene are known to play complex interdependent roles in regulating the response to wounding (McConn et al., 1997; Birkenmeier and Ryan, 1998; Wang et al., 2002; Yamada et al., 2004). Accordingly, GO categories such as response to JA (P = 3.89e−14) and response to SA (P = 4.24e−9) are enriched among genes up-regulated in the siz1-2 mutant (Fig. 2D; Supplemental Table S2). We indeed found that around 380 genes implicated in SA- and/or JA-mediated signaling are transiently induced by wounding in wild-type hypocotyls and that their induction is more pronounced in siz1-2 (Fig. 3C). These include SA-induced UBIQUITIN10 (UBQ10; Blanco et al., 2005) and MYB DOMAIN PROTEIN51 (MYB51), the latter of which encodes a transcription factor acting as a major regulator of stress-induced glucosinolate biosynthesis (Frerigmann and Gigolashvili, 2014). JA-induced VEGETATIVE STORAGE PROTEIN1 (VSP1; Ellis and Turner, 2002; Nemhauser et al., 2006) is also among genes highly expressed in siz1-2 after wounding (Fig. 3, C and D; Supplemental Table S3). Among genes up-regulated in siz1 mutants, those implicated in SA or JA response strongly overlap, while those associated with ABA or ethylene response are clearly more distinct (Supplemental Fig. S4).
Previous studies have shown that siz1 mutants exhibit an autoimmune response due to the accumulation of SUPPRESSOR OF NPR1-1, CONSTITUTIVE1 (SNC1) protein and consequential increase in SA levels (Gou et al., 2017; Hammoudi et al., 2018; Niu et al., 2019). Our RNA-seq data, however, showed that the expression of SNC1 and several SA-induced genes is mostly comparable after cutting between wild-type and siz1-2 explants (Supplemental Fig. S5). We did observe up-regulation of SNC1 and SA-induced genes such as PATHOGENESIS RELATED2 (PR2) and PR5 in siz1-2 explants on SIM (Supplemental Fig. S5), implying that SA signaling is enhanced on SIM. The dwarf phenotype caused by SA accumulation in siz1 mutants can be suppressed by introduction of a bacterial salicylate hydroxylase, NahG, which degrades this phytohormone (Lee et al., 2007; Miura et al., 2010). In order to investigate if hyperaccumulation of SA is responsible for the enhanced shoot regeneration phenotype in siz1 mutants, we compared the shoot regeneration phenotype in siz1-2 mutants expressing NahG (siz1-2 NahG; Lee et al., 2007) to the siz1-2 single mutant. As shown in Figure 3E, the introduction of NahG does not affect the enhanced shoot regeneration phenotype in siz1-2, indicating that this phenotype is independent of SA signaling.
Further investigation of differentially expressed genes after cutting revealed that 39 genes associated with the GO category “cellular response to abscisic acid stimulus” are strongly expressed in siz1-2 plants after cutting (Fig. 3C; Supplemental Table S3). SIZ1 is known to negatively regulate ABA signaling by SUMOylating the transcription factor ABI5, leading to the down-regulation of its direct target gene RESPONSE TO ABA18 (RAB18; Miura et al., 2009). In our dataset, the expression of RAB18 is up-regulated in siz1-2 (Fig. 3, C and D; Supplemental Table S3), implying that ABI5-mediated ABA signaling is hyperactivated in siz1-2 after wounding. Similarly, we found that 51 genes associated with GO categories “response to ethylene” or “cellular response to ethylene stimulus” are strongly expressed in siz1-2 after cutting (Fig. 3, C and D; Supplemental Table S3). These include an ethylene-induced ethylene receptor ETHYLENE RESPONSE SENSOR2 (ERS2; Wang et al., 2002; Nemhauser et al., 2006), suggesting that ethylene signaling is also affected in the siz1-2 mutant after wounding.
When Arabidopsis hypocotyls are subjected to wounding without any external hormone application, they develop calluses from wound sites (Iwase et al., 2011). Given that the siz1 mutants display enhanced transcriptional responses to wounding, we next tested if this leads to enhanced callus formation at wound sites. As previously reported (Iwase et al., 2011), we cut the top end of hypocotyls and incubated the explants on hormone-free medium. We found that callus induction is more pronounced in siz1-2 and siz1-3 mutants since nearly 75% of their hypocotyl explants produce calluses within 4 d after cutting, as opposed to 56% for the wild type (Supplemental Fig. S6). Importantly, this enhanced callus formation is not caused by SA accumulation, as introduction of NahG does not abolish this phenotype (Supplemental Fig. S6). These data together support that SIZ1 is required to prevent hyperactivation of the wound response.
Elevated WIND1 Expression Contributes to the Enhanced Shoot Regeneration Phenotype in siz1 Mutants
Wounding induces transcriptional activation of many key regulators that mediate cellular reprograming and organ regeneration (Iwase et al., 2011; Ikeuchi et al., 2017; Rymen et al., 2019). According to our RNA-seq dataset, some of these wound-induced reprogramming regulators are up-regulated in siz1-2 plants (Fig. 4A). Consistent with previous reports (Iwase et al., 2011; Ikeuchi et al., 2017; Rymen et al., 2019), the expression of both WIND1 and WIND2 is transiently elevated upon wounding in our dataset, and these genes are hyperactivated after cutting in siz1-2 plants (Fig. 4B). Our reverse transcription quantitative PCR (RT-qPCR) analysis further showed that the expression of these and several other wound-induced genes is also enhanced in siz1-2 NahG plants (Fig. 4C), strongly suggesting that this transcriptional activation is not due to the SA-dependent autoimmunity. We have previously shown that constitutive overexpression of WIND1 promotes shoot regeneration when uncut seedlings are incubated on SIM without CIM preculture (Iwase et al., 2015). Similarly, we found that the higher expression of WIND1 during CIM and SIM incubation improves shoot regeneration in LEXA-VP16-ER-(XVE-)WIND1 hypocotyl explants cultured in the presence of 17 β-estradiol (Supplemental Fig. S7). To further examine if enhanced expression of WIND1 or WIND2 is responsible for the enhanced shoot regeneration phenotype in siz1 mutants, we crossed plants expressing WIND1 fused with the SUPERMAN repression domain (SRDX), pWIND1::WIND1-SRDX (WIND1-SRDX), with the siz1-3 mutant. As shown in Figure 4D, the siz1-3 WIND1-SRDX explants regenerate a significantly reduced number of shoots compared to siz1-3 plants, demonstrating that enhanced shoot regeneration in siz1 mutants is partially dependent on WIND1. To further explore whether SIZ1 acts in the same pathway with WIND1, we compared genes up-regulated in 35S:WIND1 plants (Iwase et al., 2011) with our dataset. Around 12% of genes up-regulated in siz1-2 plants after cutting (163 of 1,375 genes) and 23% of genes up-regulated in siz1-2 at 4 d on CIM (43 of 187 genes) are also overexpressed in 35S:WIND1 seedlings (Fig. 4E; Supplemental Table S4), indicating that SIZ1- and WIND1-mediated pathways regulate a significantly overlapping set of genes. Taken together, these results strongly suggest that the hyperactive wound response in siz1 mutants contributes to their enhanced ability to regenerate shoots.
Figure 4.
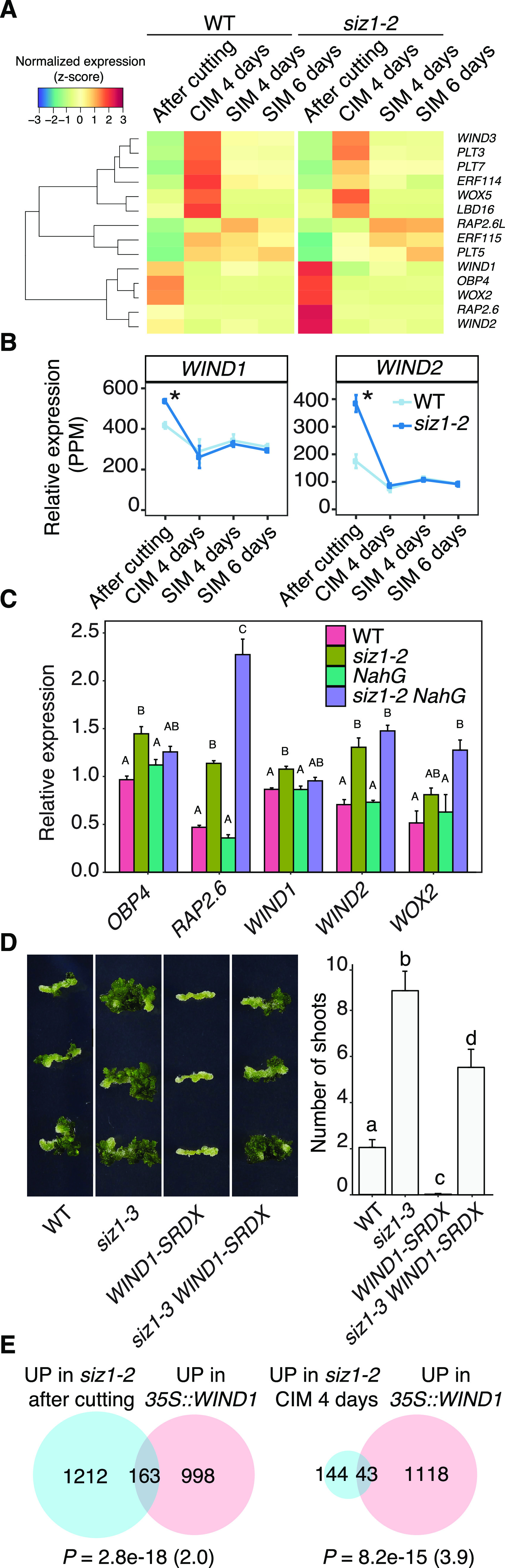
Elevated Shoot Regeneration in siz1 Mutants Is Partially Dependent on WIND1. A, Heatmap showing the relative expression, based on our RNA-seq analysis, of transcription factors identified as wound inducible and involved in cellular reprograming (Ikeuchi et al., 2017). Normalized expression was calculated for each gene across all time points, using data from both genotypes. B, WIND1 and WIND2 are up-regulated in siz1-2 after cutting. Line plots show relative expression from RNA-seq analysis. Error bars represent ± SE. Asterisks indicate significant differences by edgeR comparative analysis between wild type (WT) and siz1-2 (*P < 0.05). C, RT-qPCR analysis showing the expression of wound-induced transcription factors in wild type, siz1-2, NahG, and siz1-2 NahG hypocotyl explants after cutting. Gene expression levels are normalized to PP2A (n = 3, biological replicates). Error bars represent ± SE. Different letters indicate significant differences between genotypes for each gene based on one-way ANOVA and post-hoc Tukey test (P < 0.05). D, Images (left) and quantification (right) of shoot regeneration from hypocotyl explants of wild type, siz1-3, WIND1-SRDX (pWIND1::WIND1-SRDX), and siz1-3 WIND1-SRDX. All explants were incubated on CIM for 4 d then on SIM for 14 d. Values represent mean number of shoots produced per explant, and error bars represent ± SE. Sample sizes are wild type (n = 36), siz1-3 (n = 34), WIND1-SRDX (n = 36), and siz1-3 WIND1-SRDX (n = 38). Different letters indicate significant differences based on one-way ANOVA and posthoc Tukey test (P < 0.05). E, Overlap between genes up-regulated in siz1-2 after cutting or after 4 d on CIM and 35S:WIND1 seedlings (Iwase et al., 2011). P value was calculated by a hypergeometric test, and the representation factor is shown in brackets.
Expression of CIM-Induced Callus-Associated Genes Is Not Affected in siz1 Mutants
When performing GO analysis on the 187 genes up-regulated in siz1-2 explants compared to the wild type after a 4-d incubation on CIM, we observed a lower degree of functional enrichment compared to other time points (Fig. 2D; Supplemental Table S2). Wound stress is known to promote the production of secondary metabolites (Bodnaryk, 1992; Cheong et al., 2002). Consistent with this effect, we found that GO categories such as “secondary metabolite biosynthetic process” (P = 4.68e−5) and “glycosinolate metabolic process” (P = 4.1e−4) are significantly enriched at this time point, although this enrichment was also found at other time points (Fig. 2D; Supplemental Table S2). These data are consistent with the enhanced wound response observed in siz1 mutants.
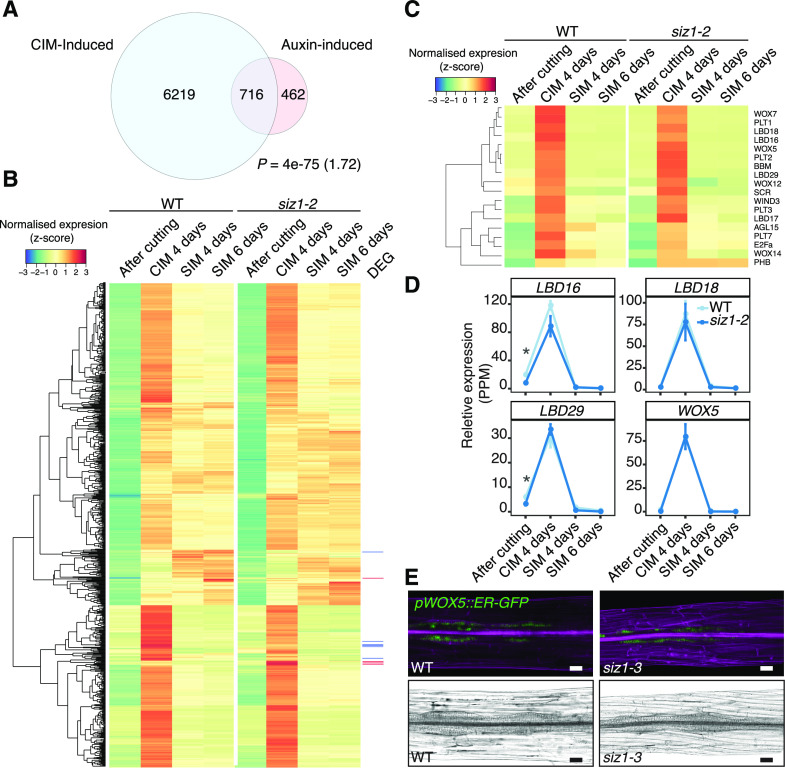
Given that auxin is primarily responsible for cell cycle re-entry and acquisition of regeneration competency during CIM incubation (Fan et al., 2012; Ikeuchi et al., 2013), we further examined whether genes that are both auxin induced, according to previous reports, and CIM-induced, according to our dataset, are differentially expressed in siz1-2. Among the 6935 genes in our dataset that are significantly induced in wild-type explants after 4 d of CIM incubation compared to after cutting, 716 genes are induced by auxin according to previously published transcriptome datasets (Fig. 5A; Supplemental Table S5; Nemhauser et al., 2006; Goda et al., 2008; Omelyanchuk et al., 2017). As shown in Figure 5B, more than 50% of these auxin-induced genes are specifically expressed on CIM, although a substantial proportion of genes also continue to be expressed after transfer to SIM. Importantly, only 10 of these genes are differentially expressed in siz1-2 plants (Fig. 5B; Supplemental Table S5), suggesting that the overall transcriptional response to auxin is not altered by the SIZ1 mutation. Most of these differentially expressed genes appeared specifically induced upon CIM incubation and are clustered in our heat map (Fig. 5B). We should note, in particular, that key regulators of callus formation, such as LBD16, LBD18, and LBD29, as well as regulators of pluripotency acquisition, like PLT1, PLT2, and WOX5, are not misexpressed in siz1-2 explants on CIM (Fig. 5, C and D; Supplemental Table S5).
Figure 5.
Expression of CIM-induced callus genes is largely unchanged in the siz1-2 mutant. A, Venn diagram showing overlap between auxin-inducible (IAA-induced) genes and CIM-inducible genes. The IAA-induced gene list was generated by combining datasets from Nemhauser et al. (2006; 430 genes), Omelyanchuk et al. (2017; 789 genes), and AtGeneExpress (ExpressionSet: 1007965859; Goda et al., 2008; 103 genes). CIM-induced genes are those up-regulated in the wild type (WT) after a 4-d incubation on CIM compared to after cutting (edgeR, false discovery rate [FDR] < 0.01). The significance of overlap between pairs of gene sets was evaluated by a hypergeometric test. The calculated representation factor is shown in brackets. B, Heatmap showing an absence of significant changes in expression between siz1-2 and wild-type plants, based on our RNA-seq analysis, for genes inducible by both IAA and CIM incubation identified in A. Normalized expression was calculated for each gene across all time points, using data from both genotypes. Differentially expressed genes (DEG) are marked in red for up-regulated genes in siz1-2 at CIM 4 d and in blue for down-regulated genes. C, Heatmap showing an absence of significant changes in expression between siz1-2 and wild-type plants for transcriptional regulators that are involved in callus formation and pluripotency acquisition according to Ikeuchi et al. (2019). Gene expression was normalized as in B. D, Relative expression of a selection of genes from the heatmap in C in siz1-2 versus wild-type plants. Error bars represent ± SE. Asterisks inidacte statistical significance of the difference is indicated based on edgeR analysis from the RNA-seq data (*P < 0.05). E, Expression of the pWOX5::ER-GFP marker (green), visualized using confocal microscopy, in siz1-3 and wild-type hypocotyl explants after 4 d on CIM. Samples were stained with propidium iodide (magenta) to stain cell walls (top). Images of the same hypocotyl explants visualized using light microscopy (bottom). Scale bars = 50 µm.
In order to further explore the expression pattern of WOX5 in siz1 mutants, we introduced the WOX5-GFP marker (pWOX5::ER-GFP; Blilou et al., 2005) into the siz1-3 mutant and examined its expression in CIM-induced callus. As shown in Figure 5E, the pattern of WOX5-GFP expression in developing callus is comparable between wild-type and siz1-3 explants after 4 d on CIM. It is thus unlikely that SIZ1 influences the efficiency of shoot regeneration by altering cellular response to auxin on CIM. We should note, however, that we did observe an increased number of GFP-positive cells in siz1-3 plants by 9 d on CIM (Supplemental Fig. S1C), suggesting that SIZ1 may modulate the auxin-triggered transcriptional response under prolonged explant incubation on CIM.
Expression of SIM-Induced Shoot Meristem Genes Is More Pronounced in siz1 Mutants
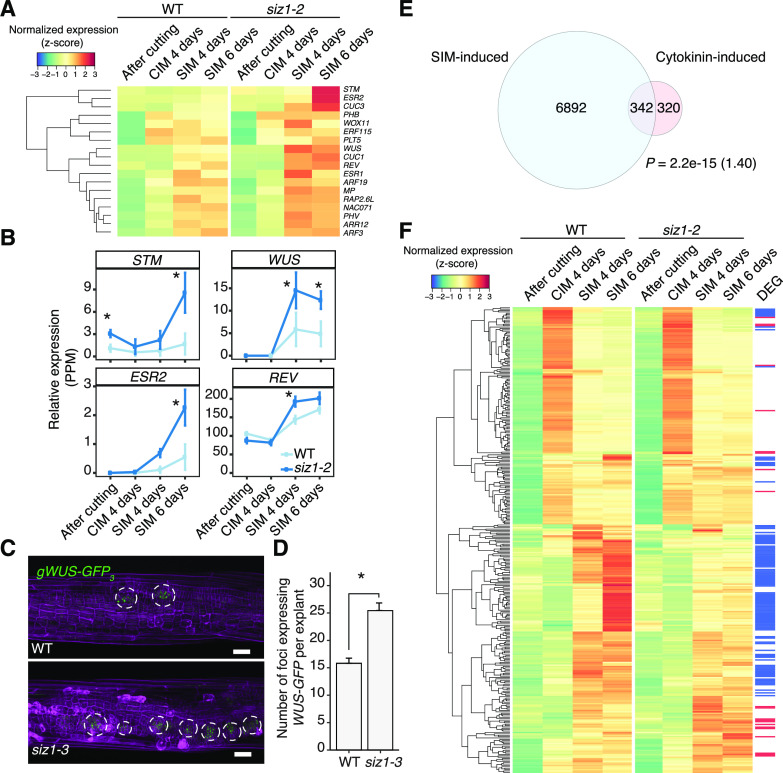
Our GO analysis showed a clear enrichment of GO categories such as “shoot system morphogenesis” (P = 1.58e−7) and “stomatal complex development” (P = 3.74e−6) among the 593 genes up-regulated in siz1-2 after 4 d of SIM incubation (Fig. 2D; Supplemental Table S2). Importantly, calluses developing in wild-type and siz1-2 explants is morphologically comparable at this time point (Supplemental Fig. S8), implying that this transcriptional activation is not the consequence of early shoot formation in siz1-2 plants. These two GO categories become even more strongly overrepresented among the 982 genes up-regulated at 6 d (Fig. 2D; Supplemental Table S2). As expected, genes that are up-regulated in siz1-2 compared to the wild type on SIM include key regulators of shoot meristem development such as REV, ESR2, STM, and WUS (Fig. 6, A and B). In order to further characterize these early transcriptional changes on SIM, we introduced the gWUS-GFP3 construct (Tucker et al., 2008) into the siz1-3 mutant and examined gWUS-GFP3 expression in explants following SIM incubation. It was previously described that WUS expression is broadly distributed in callus cells at 4 d on SIM, and it becomes spatially confined to form foci by 6 d (Zhang et al., 2017). While WUS-expressing foci were observed in hypocotyl explants in both wild-type and siz1-3 plants after a 6-d SIM incubation, the number of these foci is greatly increased in siz1-3 explants compared to the wild type (Fig. 6, C and D). This suggests that the higher level of WUS expression detected in our RNA-seq dataset is due to the higher abundance of WUS-expressing cells rather than its elevated expression in individual cells.
Figure 6.
SIZ1 affects the expression of SIM-induced shoot meristem genes. A, Heatmap showing transcriptional regulators up-regulated in siz1-2 that are highly expressed during SIM incubation and also known to be involved in regeneration according to Ikeuchi et al. (2019). Normalized expression was calculated for each gene across all time points, using data from both genotypes. B, Relative expression in siz1-2 versus wild type (WT) plants of selected shoot meristem genes from the heatmap in A. Error bars represent ± SE. Asterisks indicate significant differences by edgeR comparative analysis between wild type and siz1-2 (*P < 0.05). C, Expression of the gWUS-GFP3 reporter (green), visualized using confocal microscopy, in siz1-3 and wild-type hypocotyl explants after 4 d on CIM followed by 6 d on SIM. Samples were stained with propidium iodide (magenta) to stain cell walls. Scale bars = 50 µm. D, Quantification of the number of gWUS-GFP3-expressing foci after 6 d on SIM, observed as in C. Sample sizes are wild type (n = 33) and siz1-3 (n = 36). Significance was tested with two-tailed Welch’s t test (*P = 2.9e−7) and error bars represent ± SE. E, Venn diagram showing the overlap between cytokinin-inducible genes and SIM-inducible genes. The list of cytokinin-induced genes was created by combining data from Nemhauser et al. (2006; 332 genes), Bhargava et al. (2013; 422 genes), and AtGeneExpress (ExpressionSet: 1008031453; 60 genes). SIM-inducible genes are those up-regulated in the wild type after 4 or 6 d of incubation on SIM compared to after cutting, based on our RNA-seq data (edgeR, false discovery rate [FDR] < 0.01). The significance of overlap between pairs of gene sets was evaluated by a hypergeometric test. The calculated representation factor is shown in brackets. F, Heatmap showing the expression of genes which are both cytokinin inducible and SIM inducible, as identified in E, in siz1-2 and wild-type plants. Differentially expressed genes (DEG) are marked in red for up-regulated genes in siz1-2 at SIM 4 d or 6 d and in blue for down-regulated genes.
Given that cytokinin induces WUS expression during shoot regeneration on SIM (Zhang et al., 2017), we next investigated whether transcriptional responses to cytokinin are altered in siz1-2. Among the 7,234 genes that are induced in wild-type explants at 4 or 6 d on SIM compared to after cutting, we found 342 genes that are cytokinin inducible based on previously published transcriptomic datasets (Fig. 6E; Nemhauser et al., 2006; Goda et al., 2008; Bhargava et al., 2013). We detected 22 genes among these cytokinin-induced genes that are overinduced in siz1-2 explants on SIM, while 110 genes are down-regulated (Fig. 6F; Supplemental Table S5). Most of these differentially expressed genes are clustered together in our heat map and are induced specifically on SIM (Fig. 6F). To further examine the level of cytokinin response in the wild type and siz1 mutants, we incubated hypocotyl explants on Gamborg B5 media that contain 0 to 500 ng mL−1 6-benzylaminopurine (BA). As shown in Supplemental Figure S9, siz1 hypocotyl explants, but not wild-type explants, display increased callus growth in response to BA. These data together suggest that the cytokinin response is altered in siz1 mutants, which may contribute to the enhanced expression of shoot meristem regulators.
DISCUSSION
In this study, we demonstrate that the SUMO E3 ligase SIZ1 negatively regulates in vitro shoot regeneration in Arabidopsis, revealing a role for posttranslational modification in the control of organ regeneration. Our data suggest that the siz1 mutants exhibit an elevated response to wound stress after explant preparation and that this is partly responsible for the enhancement of shoot regeneration. SIZ1-mediated SUMOylation may also directly regulate the expression of shoot meristem genes, and identification of SUMOylated proteins under tissue culture conditions will be essential to gain further molecular insights into this control. Given that the transcript level of SIZ1 does not change markedly under our experimental conditions (Supplemental Fig. S3), SIZ1-mediated SUMOylation is likely activated by posttranscriptional mechanisms. Very little is known as to how SIZ1 is activated and how it specifies its targets under given conditions, although under some abiotic stresses such as low temperature, drought, and high salt, its activity is regulated by CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1)-dependent ubiquitination (Kim et al., 2016a; Mazur et al., 2019). Further elucidation of these molecular mechanisms will be useful for understanding how wounding and/or hormonal cues modulate plant regeneration.
Roles of SIZ1 in the Wound Response
Various biotic and/or abiotic stresses such as pathogen infection and exposure to heat are known to induce massive SUMOylation of the proteome in plants (Kurepa et al., 2003; Catala et al., 2007; Castro et al., 2012; Miller et al., 2013). Given that loss-of-function mutants of SIZ1 are often hypersensitive to these stresses, SIZ1-mediated SUMOylation is thought to limit the stress response, thereby maintaining physiological homeostasis via posttranslational regulation (Augustine and Vierstra, 2018). Although we did not establish whether wound stress promotes similar accumulation of SUMOylated proteins, our transcriptome data suggest that SIZ1-mediated SUMOylation is required to restrict several hormone signaling pathways activated by wounding (Fig. 3, A–D). SIZ1 is reported to suppress JA signaling, and accordingly we saw a significant enrichment of JA-related genes such as VSP1 (Fig. 3D; Ellis and Turner, 2002; Srivastava et al., 2018), implying that the JA-mediated stress response is enhanced in siz1 mutants. Additionally, SIZ1-mediated regulation of ABA signaling might be involved in the wounding response. As ABI5-dependent genes are strongly expressed in siz1-2 plants after cutting (Fig. 3, C and D), it is possible that SIZ1 negatively regulates this pathway upon wounding. Whether ABA plays a major role in the wound response is not established, but it is known to function in response to drought and oxidative stress (Shinozaki and Yamaguchi-Shinozaki, 1996; Finkelstein et al., 2002; Zhao et al., 2016). These stresses may also be experienced by the explant after cutting, given that genes involved in response to drought are overexpressed in siz1 explants (Fig. 2D; Supplemental Table S2). Further analysis of mutants for ABI5 or ABA-RESPONSIVE ELEMENT BINDING (AREB) transcription factors, which are major regulators of ABA signaling (Yoshida et al., 2010), could help elucidate the role of ABA in the response to wounding.
Roles of SIZ1 in Shoot Regeneration
The siz1 mutants display various developmental phenotypes including early flowering (Jin et al., 2008; Kong et al., 2017), reduced secondary cell wall thickening (Liu et al., 2019), reduced germination (Kim et al., 2016b), and reduced hypocotyl growth (Lin et al., 2016; Hammoudi et al., 2018; Zhang et al., 2019b). This study uncovered another developmental role of SIZ1 through the finding that the siz1 mutation causes enhanced shoot regeneration in tissue culture (Fig. 1, A and B). Since in vitro shoot regeneration is a multistep process that requires cutting followed by incubation on CIM and SIM, we dissected the effect of siz1 mutations at each step of the procedure by RNA-seq analysis. Interestingly, we saw the most prominent misexpression of genes previously implicated in regeneration either immediately after cutting or upon incubation on SIM (Figs. 2C; Supplemental Fig. S2A). Considering that wound stress is a key trigger for shoot regeneration (Iwase et al., 2015, 2017), an interesting hypothesis is that SIZ1 acts by suppressing the wound stress response and that the exaggerated wound response upon cutting in siz1 mutants promotes subsequent shoot regeneration. Part of the exaggerated response includes up-regulation of genes related to the stress-induced defense hormones SA, JA, and ABA (Fig. 3C). The role of SIZ1 in suppressing the SA response is well established, and some aspects of the siz1 phenotype are due to the autoimmunity caused by hyperaccumulation of SA (Lee et al., 2007; Miura et al., 2010; Gou et al., 2017; Hammoudi et al., 2018). We provide genetic evidence, however, that the wound-induced transcriptional activation as well as the enhanced shoot regeneration phenotype in siz1 plants is not due to accumulation of SA (Figs. 3E and 4C). The SA-independent developmental phenotype of siz1 mutants is also reported for both hypocotyl elongation and thermotolerance (Yoo et al., 2006; Lin et al., 2016). Exploring the functional link between these processes may give some clues to elucidate how SIZ1 functions in shoot regeneration. JA signaling is also negatively regulated by SUMOylation (Srivastava et al., 2018), and it was recently reported that pretreatment with JA before cutting explants promotes shoot regeneration, demonstrating that increased JA signaling enhances shoot regeneration (Park et al., 2019). Since this JA-mediated enhancement of shoot regeneration is dependent on COI1 (Park et al., 2019), the importance of the SUMO-dependent regulation of COI1 and its targets in shoot regeneration would be worth investigating in further studies. Whether ABA signaling participates in shoot regeneration is not well established, but it does promote shoot regeneration from embryo explants (Paulraj et al., 2014). It will thus be interesting to test whether the shoot regeneration phenotype in siz1 mutants is dependent on ABA signaling.
We have previously reported that WIND1 regulates shoot regeneration via transcriptional activation of ESR1, which is required for the induction of several shoot meristem regulators such as its paralog ESR2, WUS, and STM (Matsuo et al., 2011; Iwase et al., 2017). Indeed, the expression levels of WIND1, ESR2, WUS, and STM are elevated in siz1, although we could not detect significantly different expression of ESR1 between wild-type and siz1 plants at the time points we tested. Since ESR1 expression is generally very low and declines after several days on SIM (Iwase et al., 2017), further expression analysis at different time points may be necessary to detect a possible up-regulation of ESR1 in siz1. Genetic evidence nevertheless shows that WIND1 partly mediates the enhanced shoot regeneration phenotype observed in siz1. How SIZ1 regulates WIND1 expression is not clear at this point, but one possibility is through the ABA-mediated pathway, as SIZ1 negatively regulates ABA signaling (Miura et al., 2009), and ABA induces WIND1 expression within 30 min of application (Winter et al., 2007). Since incorporation of WIND1-SRDX does not fully suppress the shoot regeneration phenotype in siz1 mutants, we predict that SIZ1 regulates other pathways that function in parallel to the WIND1-mediated pathway. It is also possible that some factors acting downstream of WIND1 are repressed by SIZ1-dependent mechanisms, as WIND1-SRDX plants in the siz1-3 background regenerate shoots.
Our transcriptome analysis showed that many cytokinin-induced regulators of the shoot meristem are highly elevated in siz1 (Fig. 6), and consistently, we found that siz1 mutants are hypersensitive to exogenously supplied cytokinin (Supplemental Fig. S9), suggesting that the overall cytokinin response is modified. Some type-B response regulators, such as ARR1 and ARR2, that are responsible for cytokinin signaling have been identified as candidates for SUMOylation by proteomics (Rytz et al., 2018); thus, the possibility that SIZ1 directly regulates the cytokinin response should be further explored. Our data also suggested that the auxin response is only marginally affected by the siz1 mutation in calluses formed at 4 d on CIM. This is in contrast to other studies, which showed that SUMOylation regulates auxin responses in nutrient deficiency control (Miura et al., 2011) and water-induced lateral root formation (Orosa-Puente et al., 2018). These results therefore imply that the regulation of auxin signaling through SUMOylation might be context dependent. We did observe that auxin-induced callus formation is enhanced in siz1 mutants after prolonged incubation on CIM (Supplemental Fig. S1); thus, we do not exclude the possibility that the enhanced shoot regeneration phenotype in siz1 mutants is a result of undetectable enhancement in auxin response. Generation of transgenic plants that permit inducible complementation of SIZ1 function should help further reveal when SIZ1 is required during the multistep processes of in vitro shoot regeneration.
Conclusion
This study suggests that SIZ1 represses in vitro shoot regeneration by preventing the transcriptional hyperactivation of the wound response. While this repressive mechanism might be important to balance the physiological response to wound stress in natural contexts, eliminating this additional layer of regulation facilitates shoot regeneration in tissue culture conditions where maximizing the stress response does not appear to cause any unfavorable consequences. It might be that protein SUMOylation underlies regenerative recalcitrance in some plant species, and therefore, targeted inhibition of SIZ1-mediated SUMOylation, for instance by chemical inhibitors, may help improve regeneration efficiency in these plants.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) lines used in this study are in the Columbia (Col-0) ecotype, and the following genotypes were used: siz1-2 (SALK_065397), siz1-3 (SALK_034008), siz1-2 pSIZ1::SIZ1 (Miura et al., 2005; Jin et al., 2008), NahG (Delaney et al., 1994), siz1-2 nahG (Lee et al., 2007), WIND1-SRDX (Iwase et al., 2011), XVE-WIND1 (Iwase et al., 2011), pWOX5::ER-GFP (Blilou et al., 2005), and gWUS-GFP3 (Tucker et al., 2008). Double mutants were generated by crossing the siz1-3 mutant with the respective mutant lines. Genotyping of homozygous siz1-3 individuals by PCR was done according to the SIGnAL T-DNA primer design (http://signal.salk.edu/tdnaprimers.2.html; left genomic primer, 5′-TCCCTCGTAGACATCTGATGG-3′; right genomic primer, 5′-AAAGAGAGAGTGAGCGAAGGG-3′; and left t-DNA border primer, 5′-GATGCACTCGAAATCAGCCAATTTTAGAC-3′). Selection of nonsegregating lines containing the gWUS-GFP3 construct in siz1-3 gWUS-GFP3 plants was confirmed by PCR (5′-CCCTTGCGCTTTCTCTTGAGC-3′, 5′-TTGAAGTCGATGCCCT-3′) with more than 12 individual plants. Selection of homozygous lines containing the WIND1-SRDX construct in siz1-3 WIND1-SRDX plants was confirmed by PCR (5′-CAGTGGAACGAGACGTTCTCG-3′, 5′-AGCGAAACCCAAACCGAGTTCGAG-3′) with more than 12 individual plants. Selection of homozygous lines containing the construct pWOX5::ER-GFP in siz1-3 pWOX5::ER-GFP plants was confirmed by observing GFP signal in the root apical meristem. Seeds were stratified for 3 d at 4°C and then sown onto Murashige and Skoog media containing 1% (w/v) Suc and 0.6% (w/v) Gelzan (Sigma-Aldrich). Seedlings were incubated under constant fluorescent white light (∼50 µmol m−2 s−1) at 22°C.
In Vitro Tissue Culture
Seedlings were grown in the dark for 7 d to induce etiolation. Hypocotyl explants (around 10 mm long) were excised from etiolated seedlings using a razor blade and incubated on CIM (Gamborg B5 medium with 0.25% [w/v] Gelzan, 0.5 µg mL−1 2,4-dichlorophenoxyacetic acid [2,4-d] and 0.05 µg mL−1 kinetin) for 4 d under constant light. Hypocotyl explants were then transferred to SIM (Gamborg B5 medium with 0.25% (w/v) Gelzan, 0.15 µg mL−1 IAA and 0.5 µg mL−1 2-IPA) and incubated for several days to induce shoot regeneration. To test the cytokinin response, hypocotyl explants were incubated on Gamborg B5 media containing 0 to 500 ng mL−1 6-BA.
To quantify callus growth on CIM, individual calli were first dabbed on a Kimwipe to remove media and then weighed using a XS104 balance (Mettler Toledo). Projection of the callus area from calli grown for 4 d on CIM was quantified from differential interference contrast images taken with an OLYMPUS BX51 microscope and the area of visible callus was quantified using ImageJ software. For calli grown on media containing BA, images of calli were taken with a DSLR camera (Canon EOS 9000D), and the area of each callus was calculated using ImageJ software. To quantify shoot regeneration on SIM, the number of shoots visible per explant was counted. Shoots were defined as regions with viable leaves with trichomes and appearing to arise from a single meristem when visualized from the top with an OLYMPUS SZX7 microscope. The wound-induced callus assay was performed by cutting the hypocotyl of 7-d-old dark-grown seedlings about 3 mm below the shoot apical meristem, then incubating them on Murashige and Skoog media in constant light at 22°C for 4 d. The presence of callus was assessed by formation of more than 3 callus cells protruding from the cut site when visualized with a SZX7 microscope.
Transcriptomic Analysis by RNA-seq
Total RNA was extracted from ∼50 hypocotyl explants either immediately after cutting, following 4 d of incubation on CIM, or following either 4 or 6 d of incubation on SIM. Samples for different genotypes within the same biological replicate set were incubated on the same plate, and three biological replicates were used. Wild-type and siz1-2 explants were harvested simultaneously, one replicate at a time. Total RNA was isolated using the RNeasy plant mini kit (Qiagen) following the manufacturer’s instructions. Isolated RNA was subjected to library preparation with the Kapa stranded mRNA sequencing kit (KK8420, Kapa Biosystems) and Illumina-compatible FastGene adapters (NGSAD24, Nippon Genetics). Single-end sequencing was performed on an Illumina NextSeq500 platform, and mapping was carried out using Bowtie2 (Langmead and Salzberg, 2012), with over 85% of reads uniquely mapped to the TAIR10 Arabidopsis genome, resulting in 8 to 15.5 million mapped reads per sample.
Differences in expression between the wild type and mutants was calculated with the edgeR package (Robinson et al., 2010) in R using the weighted-trimmed-mean method to calculate normalization values, and the HTSFilter package (Rau et al., 2013) was used to filter lowly expressed genes. Volcano plots were made using the ggplot2 package (Wickham, 2016), and clusterProfiler (Yu et al., 2012) was used for GO analysis. The GO enrichment data were simplified using the “simplify” function, and redundant GO categories, referring to the same biological functions, were further removed manually. The heatmaps.2 function in the gplots package was used to generate heatmaps. Venn diagrams were generated using an online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/), and for weighted Venn diagrams the Vennerable package (https://r-forge.r-project.org/projects/vennerable) in R was used.
Gene Expression Analysis by RT-qPCR
Total RNA was extracted from freshly excised hypocotyls of 7-d-old dark-grown seedlings using a RNeasy plant mini kit (Qiagen). Extracted RNA was reverse transcribed using a PrimeScript RT-PCR kit with DNase I (perfect real time; Takara) in accordance with the accompanying protocol. Transcript levels were determined by qPCR using a THUNDERBIRD SYBR qPCR mix kit (Toyobo) and an Mx399P QPCR system (Agilent). The expression of the PP2A gene was used as a reference. Primer sets used in this study are listed in Supplemental Table S6.
Confocal Microscopy Analysis and Imaging
For all fluorescent marker lines, explants were mounted in either water or propidium iodide before imaging with a Leica SP5 confocal microscope with an HCXPLAPO CS 20x0.70 DRY UV lens. GFP was excited at 488 nm. A z-stack was taken through the sample, and projected images were generated using the ImageJ Bio-Formats plugin with sum of slices option.
Accession Numbers
RNA-seq data have been deposited to Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE141188. Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: SIZ1 (AT5G60410), WIND1 (AT1G78080), WIND2 (AT1G22190), WUS (AT2G17950), WOX5 (AT3G11260), RAB18 (AT5G66400), MYB51 (AT1G18570), MYB102 (AT4G21440), VSP1 (AT5G24780), STM (AT1G62360), ESR2 (AT1G24590), ARF7 (AT5G20730), ARF19 (AT1G19220), OBP4 (AT5G60850), WOX2 (AT5G59340), and RAP2.6 (AT1G43160).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. SIZ1 suppresses callus formation on CIM.
Supplemental Figure S2. Overview of down-regulated genes in the siz1-2 mutant during shoot regeneration.
Supplemental Figure S3. Expression of SIZ1 and other genes implicated in SUMOylation during shoot regeneration.
Supplemental Figure S4. Venn diagram showing the overlap of JA, SA, ABA, and ethylene signaling genes up-regulated in the siz1-2 mutant after cutting.
Supplemental Figure S5. Expression of SNC1 and other genes involved in SA-induced autoimmunity during shoot regeneration.
Supplemental Figure S6. SIZ1 negatively regulates wound-induced callus formation.
Supplemental Figure S7. Overexpression of WIND1 can promote shoot regeneration.
Supplemental Figure S8. Observation of explants after incubation on SIM for 4 and 6 d.
Supplemental Figure S9. Callus formation induced by exogenously supplied BA.
Supplemental Table S1. List of genes differentially expressed in the siz1-2 mutant at each time point.
Supplemental Table S2. List of enriched GO categories among genes differentially expressed in the siz1-2 mutant.
Supplemental Table S3. List of genes up-regulated in the siz1-2 mutant after cutting and associated with JA, SA, ABA, or ethylene-related GO categories.
Supplemental Table S4. List of genes up-regulated in both 35S::WIND1 and siz1-2 plants.
Supplemental Table S5. List of genes induced by auxin and CIM or cytokinin and SIM.
Supplemental Table S6. List of primers used in RT-qPCR analysis.
Acknowledgments
The authors are grateful to the members of Sugimoto’s lab for discussions and comments on the manuscript. They thank Mariko Mouri, Chika Ikeda, Noriko Doi, and Akiko Hanada for technical assistance.
Footnotes
This work was supported by Ministry of Education, Culture, Sports, Science, and Technology (grant no. 15H05961 to K.S.) and the Japan Society for the Promotion of Science (grant nos. 17H03704 and 20H03284 to K.S.). D.C. is supported by a fellowship from Ministry of Education, Culture, Sports, Science, and Technology, A.L. is supported by a fellowship from the University of Tokyo, and both M.I. and D.S.F. are supported by a fellowship from Japan Society for the Promotion of Science.
Articles can be viewed without a subscription.
References
- Aida M, Ishida T, Tasaka M(1999) Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563–1570 [DOI] [PubMed] [Google Scholar]
- Atta R, Laurens L, Boucheron-Dubuisson E, Guivarc’h A, Carnero E, Giraudat-Pautot V, Rech P, Chriqui D(2009) Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J 57: 626–644 [DOI] [PubMed] [Google Scholar]
- Augustine RC, Vierstra RD(2018) SUMOylation: Re-wiring the plant nucleus during stress and development. Curr Opin Plant Biol 45(Pt A): 143–154 [DOI] [PubMed] [Google Scholar]
- Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F(2001) Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci 6: 463–470 [DOI] [PubMed] [Google Scholar]
- Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang YH, Schaller GE, Loraine A, Kieber JJ(2013) Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-seq in Arabidopsis. Plant Physiol 162: 272–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier GF, Ryan CA(1998) Wound signaling in tomato plants. Evidence that aba is not a primary signal for defense gene activation. Plant Physiol 117: 687–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco F, Garretón V, Frey N, Dominguez C, Pérez-Acle T, Van der Straeten D, Jordana X, Holuigue L(2005) Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol Biol 59: 927–944 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B(2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Bodnaryk RP.(1992) Effects of wounding on glucosinolates in the cotyledons of oilseed rape and mustard. Phytochemistry 31: 2671–2677 [Google Scholar]
- Castro PH, Tavares RM, Bejarano ER, Azevedo H(2012) SUMO, a heavyweight player in plant abiotic stress responses. Cell Mol Life Sci 69: 3269–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua N-H(2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19: 2952–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P, Lall S, Howell SH(2007) Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226: 1183–1194 [DOI] [PubMed] [Google Scholar]
- Cheong YH, Chang H-S, Gupta R, Wang X, Zhu T, Luan S(2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129: 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daimon Y, Takabe K, Tasaka M(2003) The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol 44: 113–121 [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Ellis C, Turner JG(2002) A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215: 549–556 [DOI] [PubMed] [Google Scholar]
- Elrouby N, Coupland G(2010) Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA 107: 17415–17420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Xu C, Xu K, Hu Y(2012) LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res 22: 1169–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD(2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl): S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H, Gigolashvili T(2014) MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant 7: 814–828 [DOI] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Gou M, Huang Q, Qian W, Zhang Z, Jia Z, Hua J(2017) Sumoylation E3 ligase SIZ1 modulates plant immunity partly through the immune receptor gene SNC1 in arabidopsis. Mol Plant Microbe Interact 30: 334–342 [DOI] [PubMed] [Google Scholar]
- Guerra D, Crosatti C, Khoshro HH, Mastrangelo AM, Mica E, Mazzucotelli E(2015) Post-transcriptional and post-translational regulations of drought and heat response in plants: a spider’s web of mechanisms. Front Plant Sci 6: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoudi V, Fokkens L, Beerens B, Vlachakis G, Chatterjee S, Arroyo-Mateos M, Wackers PFK, Jonker MJ, van den Burg HA(2018) The Arabidopsis SUMO E3 ligase SIZ1 mediates the temperature dependent trade-off between plant immunity and growth. PLoS Genet 14: e1007157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Banno H, Niu QW, Howell SH, Chua NH(2006) The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol 47: 1443–1456 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Favero DS, Sakamoto Y, Iwase A, Coleman D, Rymen B, Sugimoto K(2019) Molecular mechanisms of plant regeneration. Annu Rev Plant Biol 70: 377–406 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Iwase A, Rymen B, Lambolez A, Kojima M, Takebayashi Y, Heyman J, Watanabe S, Seo M, De Veylder L, et al. (2017) Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol 175: 1158–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Sugimoto K, Iwase A(2013) Plant callus: Mechanisms of induction and repression. Plant Cell 25: 3159–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K(2009) SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21: 2284–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Yoshimura M, Miura K, Sugimoto K(2012) MMS21/HPY2 and SIZ1, two Arabidopsis SUMO E3 ligases, have distinct functions in development. PLoS One 7: e46897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A, Harashima H, Ikeuchi M, Rymen B, Ohnuma M, Komaki S, Morohashi K, Kurata T, Nakata M, Ohme-Takagi M, et al. (2017) WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell 29: 54–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase A, Mita K, Nonaka S, Ikeuchi M, Koizuka C, Ohnuma M, Ezura H, Imamura J, Sugimoto K(2015) WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed. J Plant Res 128: 389–397 [DOI] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al. (2011) The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol 21: 508–514 [DOI] [PubMed] [Google Scholar]
- Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, et al. (2008) The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J 53: 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareem A, Durgaprasad K, Sugimoto K, Du Y, Pulianmackal AJ, Trivedi ZB, Abhayadev PV, Pinon V, Meyerowitz EM, Scheres B, et al. (2015) PLETHORA genes control regeneration by a two-step mechanism. Curr Biol 25: 1017–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Jang IC, Seo HS(2016a) COP1 controls abiotic stress responses by modulating AtSIZ1 function through its E3 ubiquitin ligase activity. Front Plant Sci 7: 1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SI, Kwak JS, Song JT, Seo HS(2016b) The E3 SUMO ligase AtSIZ1 functions in seed germination in Arabidopsis. Physiol Plant 158: 256–271 [DOI] [PubMed] [Google Scholar]
- Kong X, Luo X, Qu GP, Liu P, Jin JB(2017) Arabidopsis SUMO protease ASP1 positively regulates flowering time partially through regulating FLC stability. J Integr Plant Biol 59: 15–29 [DOI] [PubMed] [Google Scholar]
- Koo AJK, Gao X, Jones AD, Howe GA(2009) A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J 59: 974–986 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung D-Y, Vierstra RD(2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278: 6862–6872 [DOI] [PubMed] [Google Scholar]
- Kwak JS, Son GH, Kim S-I, Song JT, Seo HS(2016) Arabidopsis HIGH PLOIDY2 sumoylates and stabilizes flowering locus C through its E3 ligase activity. Front Plant Sci 7: 530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL(2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, et al. (2007) Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J 49: 79–90 [DOI] [PubMed] [Google Scholar]
- León J, Rojo E, Sánchez-Serrano JJ(2001) Wound signalling in plants. J Exp Bot 52: 1–9 [DOI] [PubMed] [Google Scholar]
- Lin XL, Niu D, Hu ZL, Kim DH, Jin YH, Cai B, Liu P, Miura K, Yun DJ, Kim WY, et al. (2016) An Arabidopsis SUMO E3 ligase, SIZ1, negatively regulates photomorphogenesis by promoting COP1 activity. PLoS Genet 12: e1006016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yu H, Li L(2019) SUMO modification of LBD30 by SIZ1 regulates secondary cell wall formation in Arabidopsis thaliana. PLoS Genet 15: e1007928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Hu X, Qin P, Prasad K, Hu Y, Xu L(2018) The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture. Plant Cell Physiol 59: 734–743 [DOI] [PubMed] [Google Scholar]
- Liu Y, Lai J, Yu M, Wang F, Zhang J, Jiang J, Hu H, Wu Q, Lu G, Xu P, et al. (2016) The Arabidopsis SUMO E3 ligase AtMMS21 dissociates the E2Fa/DPa complex in cell cycle regulation. Plant Cell 28: 2225–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo N, Makino M, Banno H(2011) Arabidopsis ENHANCER OF SHOOT REGENERATION (ESR)1 and ESR2 regulate in vitro shoot regeneration and their expressions are differentially regulated. Plant Sci 181: 39–46 [DOI] [PubMed] [Google Scholar]
- Mazur MJ, Kwaaitaal M, Mateos MA, Maio F, Kini RK, Prins M, van den Burg HA(2019) The SUMO conjugation complex self-assembles into nuclear bodies independent of SIZ1 and COP1. Plant Physiol 179: 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur MJ, van den Burg HA(2012) Global SUMO proteome responses guide gene regulation, mRNA biogenesis, and plant stress responses. Front Plant Sci 3: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzucotelli E, Mastrangelo AM, Crosatti C, Guerra D, Stanca AM, Cattivelli L(2008) Abiotic stress response in plants: When post-transcriptional and post-translational regulations control transcription. Plant Sci 174: 420–431 [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J(1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng WJ, Cheng ZJ, Sang YL, Zhang MM, Rong XF, Wang ZW, Tang YY, Zhang XS(2017) Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD(2010) Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA 107: 16512–16517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MJ, Scalf M, Rytz TC, Hubler SL, Smith LM, Vierstra RD(2013) Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol Cell Proteomics 12: 449–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Gong Q, Ma S, Jin JB, Yoo CY, Miura T, Sato A, Bohnert HJ, Hasegawa PM(2011) SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiol 155: 1000–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM(2009) Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106: 5418–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Miura T, Hasegawa PM(2010) SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol 51: 103–113 [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. (2005) The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J(2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Niu D, Lin XL, Kong X, Qu GP, Cai B, Lee J, Jin JB(2019) SIZ1-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis. Mol Plant 12: 215–228 [DOI] [PubMed] [Google Scholar]
- Ogawa T, Ara T, Aoki K, Suzuki H, Shibata D(2010) Transient increase in salicylic acid and its glucose conjugates after wounding in Arabidopsis leaves. Plant Biotechnol 27: 205–209 [Google Scholar]
- Omelyanchuk NA, Wiebe DS, Novikova DD, Levitsky VG, Klimova N, Gorelova V, Weinholdt C, Vasiliev GV, Zemlyanskaya EV, Kolchanov NA, et al. (2017) Auxin regulates functional gene groups in a fold-change-specific manner in Arabidopsis thaliana roots. Sci Rep 7: 2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orosa-Puente B, Leftley N, von Wangenheim D, Banda J, Srivastava AK, Hill K, Truskina J, Bhosale R, Morris E, Srivastava M, et al. (2018) Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362: 1407–1410 [DOI] [PubMed] [Google Scholar]
- Park OS, Bae SH, Kim SG, Seo PJ(2019) JA-pretreated hypocotyl explants potentiate de novo shoot regeneration in Arabidopsis. Plant Signal Behav 14: 1618180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulraj S, Lopez-Villalobos A, Yeung EC(2014) Abscisic acid promotes shoot regeneration in Arabidopsis zygotic embryo explants. In Vitro Cell Dev Biol Plant 50: 627–637 [Google Scholar]
- Rau A, Gallopin M, Celeux G, Jaffrézic F(2013) Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics 29: 2146–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK(2010) edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymen B, Kawamura A, Lambolez A, Inagaki S, Takebayashi A, Iwase A, Sakamoto Y, Sako K, Favero DS, Ikeuchi M, et al. (2019) Histone acetylation orchestrates wound-induced transcriptional activation and cellular reprogramming in Arabidopsis. Commun Biol 2: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rytz TC, Miller MJ, McLoughlin F, Augustine RC, Marshall RS, Juan YT, Charng YY, Scalf M, Smith LM, Vierstra RD(2018) SUMOylome profiling reveals a diverse array of nuclear targets modified by the SUMO ligase SIZ1 during heat stress. Plant Cell 30: 1077–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD(2007) Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145: 119–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y, Eisenman RN(2003) Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA 100: 13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K(1996) Molecular responses to drought and cold stress. Curr Opin Biotech 7: 161–167 [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO(1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 11: 118–130 [PubMed] [Google Scholar]
- Srivastava AK, Orosa B, Singh P, Cummins I, Walsh C, Zhang C, Grant M, Roberts MR, Anand GS, Fitches E, et al. (2018) SUMO suppresses the activity of the jasmonic acid receptor CORONATINE INSENSITIVE1. Plant Cell 30: 2099–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Jiao Y, Meyerowitz EM(2010) Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev Cell 18: 463–471 [DOI] [PubMed] [Google Scholar]
- Sugiyama M.(2015) Historical review of research on plant cell dedifferentiation. J Plant Res 128: 349–359 [DOI] [PubMed] [Google Scholar]
- Tucker MR, Hinze A, Tucker EJ, Takada S, Jürgens G, Laux T(2008) Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development 135: 2839–2843 [DOI] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Van Lijsebettens M(1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KLC, Li H, Ecker JR(2002) Ethylene biosynthesis and signaling networks. Plant Cell 14(Suppl): S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H.(2016) ggplot2: Elegant graphics for data analysis, Ed 2 Springer, New York [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ(2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Nishimura M, Hara-Nishimura I(2004) The slow wound-response of gammaVPE is regulated by endogenous salicylic acid in Arabidopsis. Planta 218: 599–605 [DOI] [PubMed] [Google Scholar]
- Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun D-J, Bressan RA, Hasegawa PM(2006) SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol 142: 1548–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K(2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y(2012) clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16: 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Zhao F, Chen L, Pan Y, Sun L, Bao N, Zhang T, Cui CX, Qiu Z, Zhang Y, et al. (2019a) Jasmonate-mediated wound signalling promotes plant regeneration. Nat Plants 5: 491–497 [DOI] [PubMed] [Google Scholar]
- Zhang L, Han Q, Xiong J, Zheng T, Han J, Zhou H, Lin H, Yin Y, Zhang D(2019b) Sumoylation of BRI1-EMS-SUPPRESSOR 1 (BES1) by the SUMO E3 ligase SIZ1 negatively regulates brassinosteroids signaling in Arabidopsis thaliana. Plant Cell Physiol 60: 2282–2292 [DOI] [PubMed] [Google Scholar]
- Zhang T-Q, Lian H, Zhou C-M, Xu L, Jiao Y, Wang J-W(2017) A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29: 1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Chan Z, Gao J, Xing L, Cao M, Yu C, Hu Y, You J, Shi H, Zhu Y, et al. (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc Natl Acad Sci USA 113: 1949–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]