A transcriptional regulator maintains compact plant architecture and decreased leaf angle by regulating GA synthesis and auxin signaling pathways.
Abstract
Plant-specific GROWTH-REGULATING FACTORs (GRFs) participate in central developmental processes, including leaf and root development; inflorescence, flower, and seed formation; senescence; and tolerance to stresses. In rice (Oryza sativa), there are 12 GRFs, but the role of the miR396-OsGRF7 regulatory module remains unknown. Here, we report that OsGRF7 shapes plant architecture via the regulation of auxin and GA metabolism in rice. OsGRF7 is mainly expressed in lamina joints, nodes, internodes, axillary buds, and young inflorescences. Overexpression of OsGRF7 causes a semidwarf and compact plant architecture with an increased culm wall thickness and narrowed leaf angles mediated by shortened cell length, altered cell arrangement, and increased parenchymal cell layers in the culm and adaxial side of the lamina joints. Knockout and knockdown lines of OsGRF7 exhibit contrasting phenotypes with severe degradation of parenchymal cells in the culm and lamina joints at maturity. Further analysis indicated that OsGRF7 binds the ACRGDA motif in the promoters of a cytochrome P450 gene and AUXIN RESPONSE FACTOR12, which are involved in the GA synthesis and auxin signaling pathways, respectively. Correspondingly, OsGRF7 alters the contents of endogenous GAs and auxins and sensitivity to exogenous phytohormones. These findings establish OsGRF7 as a crucial component in the OsmiR396-OsGRF-plant hormone regulatory network that controls rice plant architecture.
Plant architecture is a major factor that determines grain productivity in cereal crop species (Wang and Li, 2008). Rice (Oryza sativa) is a staple food for more than half of the global population, and rice plant architecture has been continually improved by breeders to achieve high productivity. Rice plant architecture is mainly defined by plant height, the spatial pattern of leaves, and tiller and inflorescence branching patterns (Khush, 1995). Plant height and tiller branching determine the biomass and harvest index, while the spatial pattern of leaves, including leaf shape and angle, influences the photosynthesis rate and therefore the accumulation of carbohydrates (Sinclair and Sheehy, 1999; Springer, 2010; Xing and Zhang, 2010). During plant architecture determination, several factors, especially plant hormones, have been reported to affect the development of the leaf angle, plant height, and tiller number, thus modulating rice plant architecture.
Phytohormones regulate many physiological processes that largely influence growth, differentiation, and development (Luo et al., 2016). Auxins and cytokinins mainly control the size and number of plant leaves, culms, inflorescences, and grains by regulating cell size and number (Schaller et al., 2015; Lavy and Estelle, 2016). Auxins also negatively control lamina joint inclination via asymmetric adaxial-abaxial cell division (Zhang et al., 2015). Consistent with these findings, the auxin early response gain-of-function rice mutant leaf inclination1, which encodes the indole-3-acetic acid (IAA) amido synthetase OsGH3-1, showed reduced free auxin levels and enlarged leaf angles due to stimulated cell elongation on the adaxial side of the lamina joints (Zhao et al., 2013). Overexpression of AUXIN RESPONSE FACTOR19 (OsARF19) resulted in an enlarged leaf angle via an increased expression level of GH3s and decreased free IAA content (Zhang et al., 2015). GAs control plant height and tillering by regulating cell elongation and cell division (Kobayashi et al., 1988; Magome et al., 2013; Gao et al., 2016). Blocking the synthesis of GA reduces plant height and therefore increases lodging resistance (Sasaki et al., 2002; Chen et al., 2015). Thus, the integration of multiple phytohormone signaling pathways will appropriately coordinate plant architecture development.
Increasing evidence indicates that microRNAs (miRNAs) also participate in hormone synthesis or signal transduction and plant development (Tang and Chu, 2017). miR396 is one of the most conserved miRNA families in monocots and dicots (Liu et al., 2009). Studies have shown that repressing the expression of OsmiR396 in miR396 mimicry (MIM396) transgenic lines resulted in enlarged panicles (Gao et al., 2015). GROWTH-REGULATING FACTORs (GRFs), the targets of OsmiR396, are conserved plant-specific transcription factors. In rice, the GRF family comprises 12 members. All GRF genes have the conserved QLQ and WRC domains in their N-terminal regions. The QLQ domain is essential for protein-protein interaction, and the WRC domain contains a functional nuclear localization signal (Choi et al., 2004).
GRFs interact with small cofactors called GRF-INTERACTING FACTORs (GIFs) to form a functional complex to regulate plant growth and development (Kim and Kende, 2004). GIFs have been reported to participate in cell proliferation during leaf development, root meristem homeostasis, and grain size determination (Kim and Kende, 2004; Li et al., 2016; Ercoli et al., 2018). In rice, the GIF family is composed of three members: OsGIF1, OsGIF2, and OsGIF3. OsGRF6 interacts with these three OsGIFs to regulate inflorescence architecture (Liu et al., 2014; Gao et al., 2015), OsGRF10 interacts with OsGIF1 and OsGIF2 to regulate floral organogenesis (Liu et al., 2014), and OsGRF4 interacts with OsGIF1 to regulate grain size (Li et al., 2016). These findings highlight the crucial roles of the OsmiR396-OsGRFs-OsGIFs module in determining the complexity of the regulatory network of rice growth and development.
In this study, we report that OsGRF7 and OsmiR396e form a molecular node that regulates plant architecture via hormone-related genes, including OsARF12 (Wang et al., 2014; Li et al., 2020) and the cytochrome P450 gene OsCYP714B1 (Magome et al., 2013), which are involved in the auxin signaling pathway and GA synthesis, respectively. In vivo and in vitro assays indicate that these genes are directly regulated by OsGRF7. OsGRF7 also alters the contents of corresponding endogenous phytohormones and sensitivity to exogenous phytohormones. Our study demonstrates that OsGRF7 is a critical regulator of the shaping of plant architecture through GA and IAA signaling networks.
RESULTS
OsGRF7 Modulates Plant Architecture in Rice
A previous study showed that OsmiR396 regulates rice inflorescence branching and grain yield by modulating the expression of OsGRF6 (Gao et al., 2015). Here, we observed that MIM396 transgenic lines presented compact plant architecture. The leaf angle of the MIM396 transgenic lines was reduced by 26.9% at the seedling stage compared with that of the wild-type YuetaiB (YB; Supplemental Fig. S1, A and B). A similar phenotype was also observed for the second and third fully expanded leaves from the top of MIM396 plants at the tillering stage (Supplemental Fig. S1, C–E). Corresponding to the significant decrease in the abundance of OsmiR396 family members in MIM396-3 transgenic lines (Supplemental Fig. S1F), the expression levels of GRF family members, especially OsGRF7, were significantly increased (Supplemental Fig. S1G).
To elucidate the functions of OsGRF7, we performed a genetic transformation of OsGRF7 and found that seven out of 13 OsGRF7 overexpression (GRF7OE) lines and four out of six OsGRF7 RNA interference (RNAi; GRF7RNAi) lines showed altered plant architecture (Fig. 1, A and B). Compared with that of the wild type, the plant height of the GRF7OE lines was reduced by ∼20% (Fig. 1A; Supplemental Table S1), corresponding to the contraction of each internode in the GRF7OE lines (Supplemental Fig. S2, A and B).
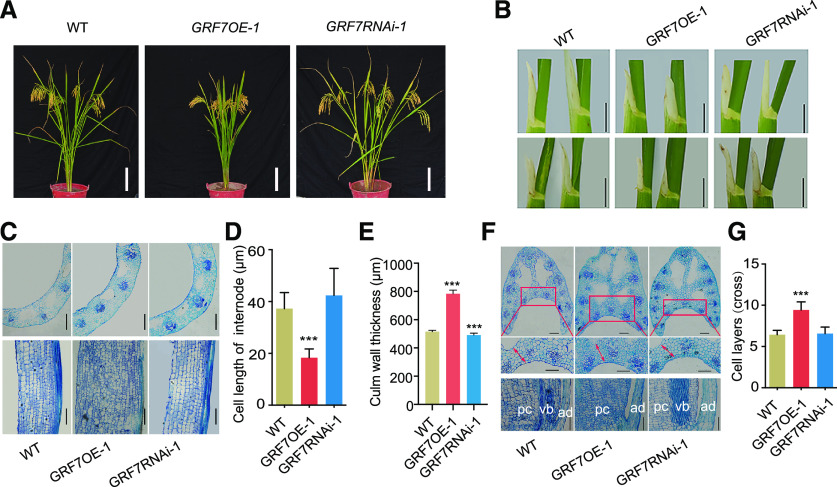
Figure 1.
Phenotypic characterization. A, Gross morphology of wild-type (WT), GRF7OE-1, and GRF7RNAi-1 plants. Bars = 15 cm. B, Comparison of the flag leaf angles (top row) and the top second leaf angles (bottom row) between OsGRF7 transgenic lines. Bars = 1 cm. C, Transverse (top row) and longitudinal (bottom row) sections of internodes in wild-type, GRF7OE-1, and GRF7RNAi-1 plants. Bars = 200 μm. D, Cell length of the internode. E, Culm wall thickness of OsGRF7 transgenic lines. F, Transverse (top row) and longitudinal (bottom row) sections of the lamina joint of flag leaves. Red boxed areas in the top row are enlarged in the middle row. Red arrows indicate adaxial side cell layers measured in G. ad, Adaxial surface of the lamina joint; pc, parenchymal cell; vb, vascular bundle. Bars = 200 μm. G, Adaxial side cell layers of the transverse sections of lamina joints. In D, E, and G, values are means ± sd of five biological replicates. Asterisks indicate significant difference by two-tailed Student’s t test (***P < 0.001).
The flag leaf angles of the wild type as well as the GRF7OE-1, GRF7OE-2, GRF7RNAi-1, and GRF7RNAi-2 transgenic lines were 5.5°, 1.9°, 1.3°, 9.5°, and 6.3°, respectively (Fig. 1B; Supplemental Table S1). Moreover, the plant height, effective panicle, leaf angle, leaf length, and leaf width were strongly correlated with the relative expression levels of OsGRF7 in the transgenic plants (Supplemental Fig. S3), meaning that OsGRF7 regulates plant architecture in a dose-dependent manner.
We further crossed GRF7RNAi-1 with MIM396-3 and found that the leaf angles were significantly decreased in the GRF7RNAi-1/MIM396-3 hybrid compared with those in GRF7RNAi-1 (Supplemental Fig. S4, A and B). The first fully expanded leaf angles from the top of the main stalk decreased from 17.4° in the GRF7RNAi-1 transgenic lines to 12.6° in the GRF7RNAi-1/MIM396-3 hybrid, and the third fully expanded leaf angles decreased from 26.5° in the GRF7RNAi-1 transgenic lines to 16.7° in the GRF7RNAi-1/MIM396-3 hybrid (Supplemental Fig. S4, C and D), suggesting that OsmiR396-OsGRF7 is involved in leaf angle determination.
To further validate the function of OsGRF7 in rice plant architecture determination, 74 independent OsGRF7 knockout (GRF7KO) lines were generated using CRISPR/Cas9 against two target sites within the second exon of OsGRF7 (Supplemental Fig. S5A). After sequence identification, we obtained four independent GRF7KO homologous lines in which OsGRF7 was prematurely terminated (Supplemental Fig. S5A; Supplemental Table S2). Consistent with the leaf angles of the GRF7RNAi transgenic lines, the angles of the second leaves of GRF7KO-2, GRF7KO-14, GRF7KO-64, and GRF7KO-66 mutants were 27.6°, 26.1°, 32.9°, and 36.1°, respectively (Supplemental Fig. S5, B–D), which are significantly larger than those of the wild-type line (14.8°). After analyzing the plant height of GRF7KO lines, we found that the plant height of GRF7KO lines was significantly increased when compared with the wild type (Supplemental Fig. S5, B and E). These results demonstrate that OsGRF7 is a critical node for plant architecture determination in rice.
GRFs are conserved and plant-specific transcription factors. Phylogenetic analysis of the GRFs from rice and Arabidopsis (Arabidopsis thaliana) showed that OsGRF7 was closely related to the Arabidopsis homologs AtGRF1 and AtGRF2 (Supplemental Fig. S6). After analyzing the mRNA expression of OsGRFs in the OsGRF7 transgenic lines, we found that when OsGRF7 was down-regulated, multiple OsGRFs showed increased expression levels, especially OsGRF4, OsGRF6, OsGRF8, and OsGRF9 (Supplemental Fig. S7). At the same time, OsGRF4, OsGRF6, OsGRF8, and OsGRF9 shared similar expression patterns with OsGRF7 (Choi et al., 2004), suggesting that these GRFs may compensate for the loss of OsGRF7 in the GRF7RNAi and GRF7KO lines.
OsGRF7 Promotes Periclinal Division in Lamina Joints and Internodes
To clarify the cellular mechanism of OsGRF7 in the control of plant architecture, we observed the microstructure of the uppermost internodes and lamina joints. The results showed that GRF7OE-1 plants had more cell layers in the uppermost internode than wild-type and GRF7RNAi-1 plants (Fig. 1C). Longitudinal sections revealed that the internode parenchymal cells of the GRF7OE-1 plants were not only shorter but also rounder than those of the wild type and GRF7RNAi-1 (Fig. 1, C and D), and the culm wall thickness of the GRF7OE-1 plants was significantly increased compared with that of the wild type (Fig. 1E), corresponding to the short, sturdy, and thick culm of the former (Fig. 1A).
The leaf angle increases after the lamina joint emerges from the prior leaf sheath. After analyzing the dynamic lamina joint growth from lamina joint initiation to the maturation of the flag leaf, we found that the GRF7OE-1 transgenic lines showed enlarged vascular bundles during the lamina joint development stage, while GRF7RNAi-1 and GRF7KO-14 showed the opposite phenotype (Supplemental Fig. S8). Additional examination of cross sections and longitudinal sections of the lamina joints revealed that, compared with the wild-type and GRF7RNAi-1 plants, the GRF7OE-1 plants showed enhanced adaxial cell proliferation and repressed cell elongation (Fig. 1, F and G), suggesting that overexpression of OsGRF7 promoted the cell periclinal division on the adaxial side of the lamina joint and thus increased cell layers on the adaxial side that caused narrow lamina joint bending (Fig. 1B) and the compact phenotype of GRF7OE-1 plants.
OsGRF7 Is Mainly Repressed by OsmiR396e
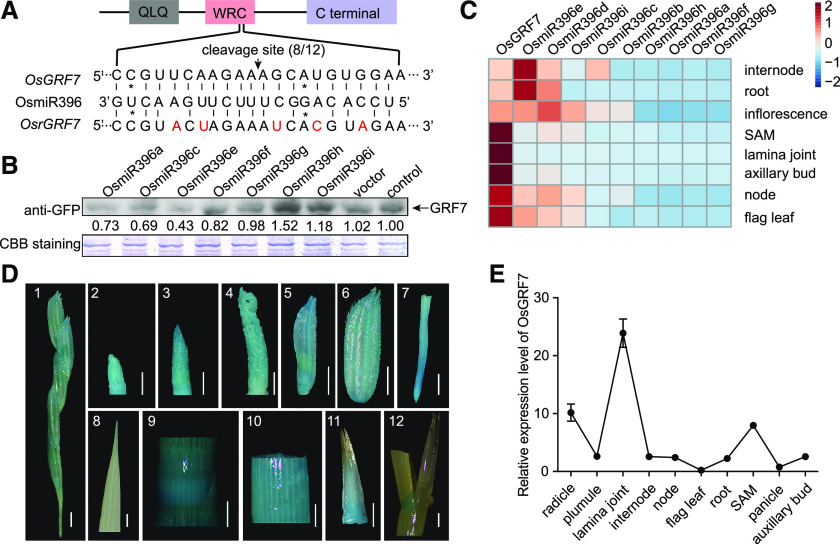
GRFs are known to be highly regulated by miR396 (Omidbakhshfard et al., 2015). Here, we found that OsmiR396 could directly cleave OsGRF7 mRNA in vivo at the site within the OsmiR396 pairing region (Fig. 2A). Moreover, we performed transient expression assays of OsGRF7 using the OsmiR396-sensitive construct 35S:OsGRF7 and the OsmiR396-resistant construct 35S:OsrGRF7 in rice protoplasts under the background of OsmiR396s (Fig. 2A). Reverse transcription quantitative PCR (RT-qPCR) analysis showed that the transcript level of OsGRF7 significantly decreased when OsmiR396-sensitive OsGRF7 was coexpressed with OsmiR396s, which contrasted with the slight decrease in OsGRF7 under the background of OsrGRF7 together with coexpression of OsmiR396 members (Supplemental Fig. S9). These results demonstrate that OsGRF7 is repressed by OsmiR396.
Figure 2.
OsGRF7 is mainly repressed by OsmiR396e and expressed in various tissues. A, The OsGRF7 cleavage site sequence complementary to OsmiR396. The position corresponding to the 5′ end of the cleaved OsGRF7 mRNA determined by 5′ RACE (rapid amplification of cDNA end) and the frequency of 5′ RACE clones corresponding to the cleavage site is shown by the arrow. 8/12 means eight of 12 clones have an OsmiR396 cleavage site. The mutated sites in OsrGRF7 are marked in red. The asterisks indicate mismatched sites between OsmiR396 and OsGRF7. The gray box indicates the QLQ domain, the pink box indicates the WRC domain, and the purple box indicates the C-terminal end of OsGRF7. B, Protein immunoblotting of OsGRF7 coexpressed with OsmiR396s in GRF7-GFP transgenic rice protoplasts. Vector and control mean with or without the empty vector, respectively. Coomassie Brilliant Blue (CBB) staining was used as a loading control. The relative protein amounts were determined by ImageJ (National Institutes of Health). C, Relative expression levels of OsGRF7 and OsmiR396s were detected by RT-qPCR in internode, root, inflorescence (10 cm), node, flag leaf, shoot apical meristem (SAM), axillary bud, and lamina joint. D, GRF7pro:GUS expression patterns in transgenic plants. Images are as follows: 1, small panicle branch; 2 to 4, inflorescence at different stages; 5 and 6, spikelet; 7, root tip; 8, flag leaf; 9, node; 10, internode; 11, axillary bud; 12, lamina joint of the 30-d seedling. Bars = 2 mm. E, Relative expression levels of OsGRF7 in different tissues analyzed with RT-qPCR. OsUBI was used as an internal reference. Values are means ± sd of three biological replicates.
In rice, there are nine miR396s that show various expression profiles (Kozomara and Griffiths-Jones, 2014), implying that each of them may have different functions. To further understand the regulatory activity of OsmiR396 members on OsGRF7, we transiently expressed Ubi:miR396s in GRF7-GFP transgenic rice protoplasts. Immunoblotting assays revealed that at the same OsmiR396 transcript level (Supplemental Fig. S10), the OsGRF7 protein content sharply decreased under the OsmiR396e background (Fig. 2B). Correspondingly, OsmiR396e exhibited the opposite expression pattern of OsGRF7 in different tissues (Fig. 2C), implying that OsmiR396e is mainly responsible for the cleavage of OsGRF7.
We further examined the spatiotemporal expression pattern of OsGRF7, and the results showed that OsGRF7 was prominently expressed in the axillary buds, lamina joints, nodes, and internodes as well as highly expressed in young inflorescences and florets (Fig. 2D), corresponding to the RT-qPCR results showing that OsGRF7 was expressed at different levels in various tissues (Fig. 2E). The multitissue expression pattern of OsGRF7 was consistent with its pleiotropic roles in controlling plant height, leaf size and angle, and tiller number.
Furthermore, transient expression of the GRF7-GFP fusion protein in rice protoplasts showed that OsGRF7 was expressed specifically in the nucleus (Supplemental Fig. S11A), which is consistent with the nuclear localization pattern in GRF7-GFP transgenic lines (Supplemental Fig. S11B), implying that OsGRF7 functions in the nucleus. This is in accordance with the transcriptional activity of the OsGRF7 C terminus (amino acids 177–430) verified in the yeast expression system (Supplemental Fig. S11C) and the interaction of the OsGRF7 N terminus with GIFs (Supplemental Fig. S11, D and E) such as AtGRF1 and AtGRF2 in Arabidopsis (Kim and Kende, 2004). These results suggest that OsGRF7 and OsGIFs work together to modulate gene expression in the nucleus.
OsGRF7 Directly Modulates the Expression of Multiple GA/IAA-Related Genes
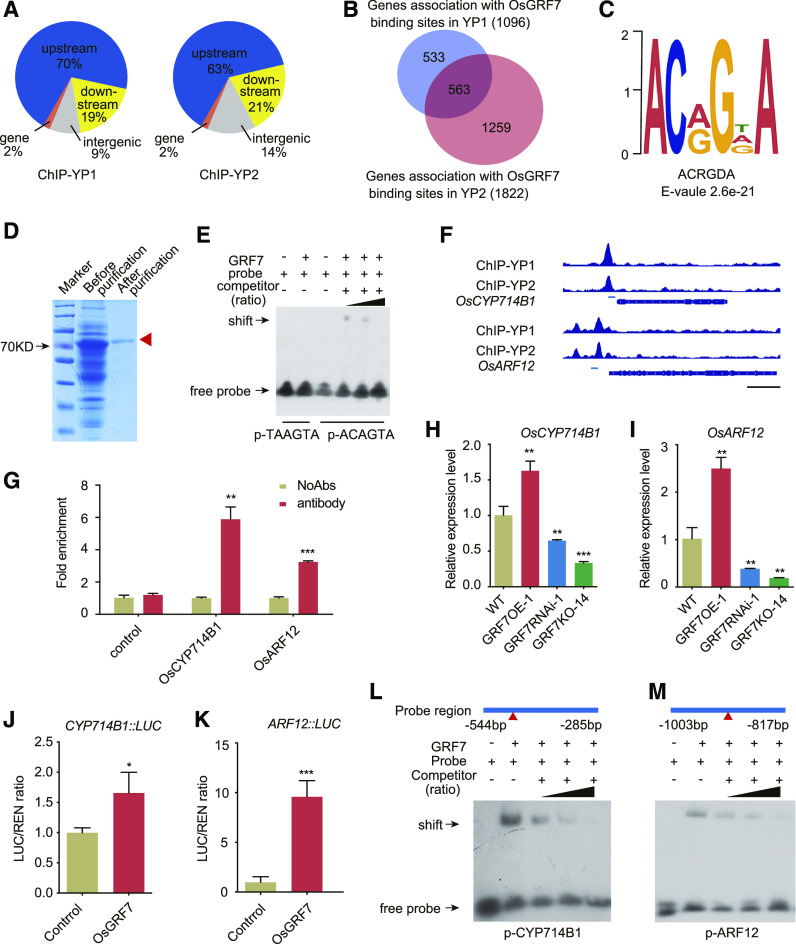
To further elucidate how OsGRF7 influences plant architecture, we performed chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) against the GFP antibodies using GRF7-GFP transgenic lines. In general, the more active metabolism of young inflorescences than of the leaves or seedlings makes the ChIP assay more practical; thus, young inflorescences of the GRF7-GFP lines were used to identify the potential targets and binding motifs of OsGRF7. By comparing the ChIP-seq data between YP1 (young inflorescences with a length of ∼0.5 cm) and YP2 (young inflorescences with a length of ∼2 cm) with the input data, we detected 1,321 and 2,290 OsGRF7-bound peaks, respectively (P < 1e−5, one-tailed Student’s t test). Of these, approximately 70% and 63% of the peaks were in the 5′ upstream regions of genes in YP1 and YP2, respectively (Fig. 3A), implying that OsGRF7 functions mainly in the regulation of gene expression, consistent with its nuclear localization (Supplemental Fig. S11, A and B). These peaks were assigned to the closed genes, therefore giving rise to 1,096 and 1,822 genes (Supplemental Data S1 and S2), respectively, and were found with 563 overlapping genes that accounted for 51.8% in YP1 and 30.9% in YP2 (Fig. 3B).
Figure 3.
ChIP analysis of the OsGRF7 target genes. A, Distribution of OsGRF7-binding peaks in the rice genome. B, Genes reproducibly associated with OsGRF7-binding sites in YP1 or YP2. C, Putative OsGRF7-binding motif predicted by DREME. D, Expression and purification of OsGRF7 protein in Escherichia coli. The red arrowhead indicates the purified OsGRF7 protein. E, EMSA validation of interaction between OsGRF7 and motif ACRGDA (ACAGTA). TAAGTA was used as the control. F, OsGRF7-binding peaks in the promoters of OsCYP714B1 and OsARF12. Blue boxes indicate probe locations. Bar = 1 kb. G, ChIP-qPCR validation of OsGRF7-binding sites in the promoters of OsCYP714B1 and OsARF12. The fold enrichment was normalized against the promoter of OsUBI. No addition of antibodies (NoAbs) served as a negative control. H and I, Relative expression levels of OsCYP714B1 and OsARF12 in the lamina joint of OsGRF7 transgenic lines were detected with RT-qPCR. OsUBI was used as an internal reference. J and K, OsGRF7 activates OsCYP714B1 and OsARF12 promoter-luciferase fusion constructs in transient transactivation assays. LUC/REN, Firefly luciferase-to-renilla luciferase ratio. In G to K, values are means ± sd of three biological replicates. Asterisks indicate significant difference by two-tailed Student’s t test (*P < 0.05; **P < 0.01; and ***P < 0.001). L and M, EMSA validation of binding between OsGRF7 and the promoters of OsCYP714B1 and OsARF12. Two-fold, 10-fold, and 100-fold unmodified probes were used as competitors. The presence (+) or absence (−) of components in the reaction mixture is indicated. Red triangles indicate the sites of ACRGDA motifs on the promoter region.
After we performed a Discriminative Regular Expression Motif Elicitation (DREME; Bailey, 2011) analysis of the filtered ChIP-seq sequences, an OsGRF7-binding motif, ACRGDA (E = 2.6e-21), was predicted (Fig. 3C), which was further validated by an electrophoretic mobility shift assay (EMSA) in vitro (Fig. 3, D and E). After analyzing these ChIP-seq-identified genes with OsGRF7-binding sites individually, we identified 31 functionally known plant architecture-related genes (Supplemental Table S3). These genes could be classified into four categories: plant hormone-related genes, transcription factors, enzymes, and others. Fifteen of them were related to plant hormones, especially OsARF12 and OsCYP714B1, which showed direct regulation by OsGRF7, as verified by ChIP-seq (Fig. 3F). ChIP-qPCR confirmed the in vivo association of OsGRF7 with ACRGDA-containing promoter fragments from OsARF12 and OsCYP714B1 (Fig. 3G). Similarly, the abundances of mRNAs encoding OsARF12 and OsCYP714B1 were relatively enhanced in GRF7OE-1 transgenic lines (Fig. 3, H and I). OsGRF7 activated the transcription of OsARF12 and OsCYP714B1 by their promoters in transactivation assays (Fig. 3, J and K). Finally, EMSA analysis demonstrated the binding of OsGRF7 to ACRGDA-containing promoter fragments from OsARF12 and OsCYP714B1 (Fig. 3, L and M). These results demonstrate that these genes are the direct targets of OsGRF7.
We further searched the promoter regions of the genes detected by ChIP-seq and found that the promoter regions of IAA-related genes (OsARF3, OsARF4, OsARF8, OsARF9, OsPIN1b, OsPIN1d, and OsPIN8) and GA-related genes (OsSLR1 and OsSLRL1) contained several ACRGDA motifs. RT-qPCR comparisons of the wild type, GRF7OE-1, GRF7RNAi-1, and GRF7KO-14 indicated that OsGRF7 up-regulated the expression of these IAA/GA-related genes (Fig. 4A). Additionally, ChIP-qPCR also confirmed the activation of OsGRF7 on these genes through the ACRGDA-binding site in vivo (Fig. 4, B–J), as OsGRF7 greatly enhanced the expression of the LUCIFERASE reporter gene driven by these promoters (Fig. 4, K and L). Taken together, these results support the conclusion that OsGRF7 directly regulates auxin and GA signaling pathway-related genes.
Figure 4.
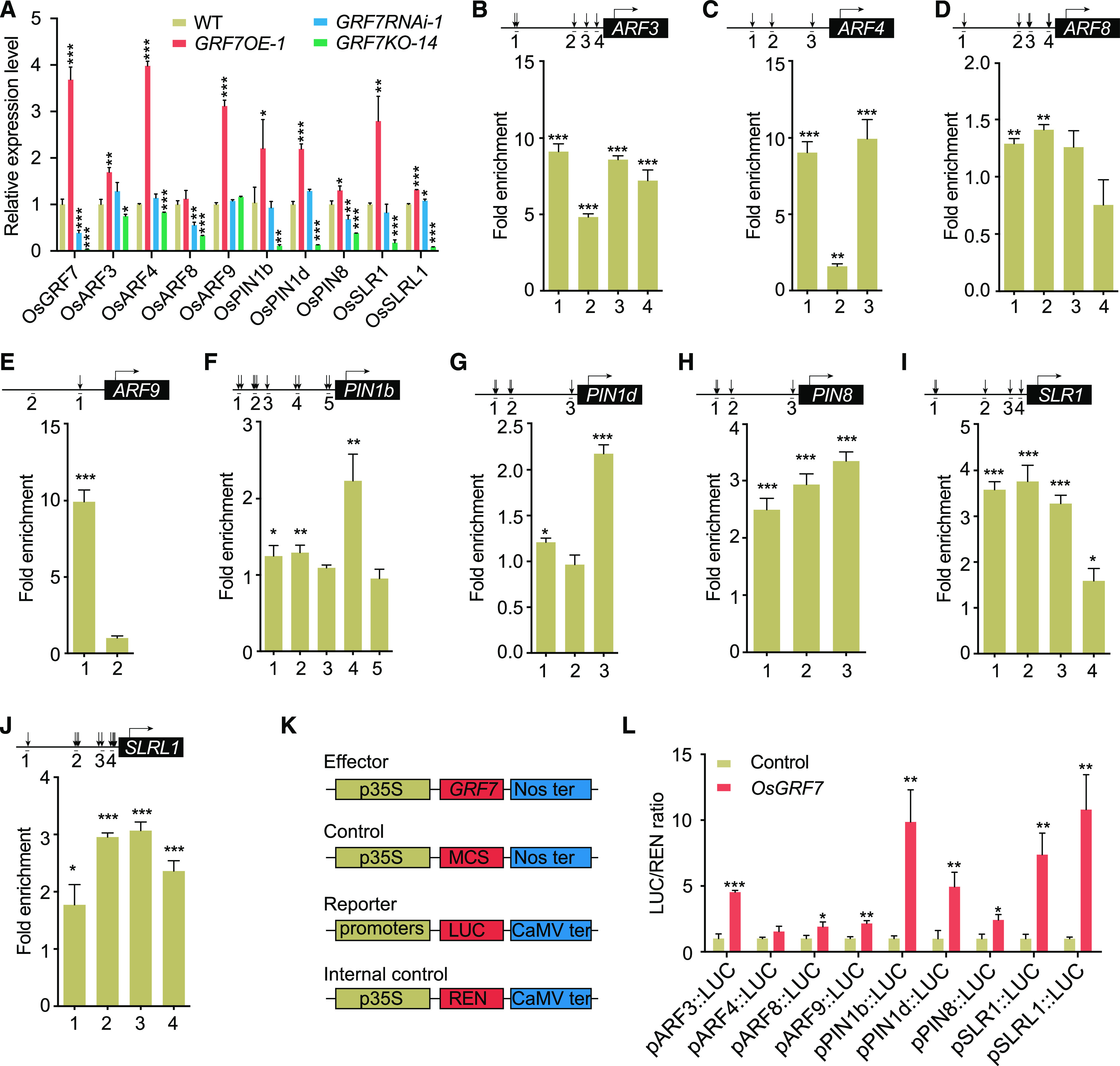
OsGRF7 regulates the expression of multiple IAA/GA-related genes. A, Relative expression levels of auxin signaling-related and GA synthesis-related genes in the lamina joint of the OsGRF7 transgenic lines. OsUBI was used as an internal reference. WT, Wild type. B to J, GRF7-GFP mediated the ChIP-qPCR enrichment (relative to the promoter region of OsUBI) of ACRGDA-containing fragments (marked with arrows) from the promoters of auxin signaling-related genes (ARF3, ARF4, ARF8, ARF9, PIN1b, PIN1d, and PIN8) and GA synthesis-related genes (SLR1 and SLRL1). The promoter region of OsUBI was used as a control. K and L, OsGRF7 activates ARF3, ARF4, ARF8, ARF9, PIN1b, PIN1d, PIN8, SLR1, and SLRL1 promoter-luciferase fusion constructs in transient transactivation assays. In B to J and L, values are means ± sd of three biological replicates. Asterisks indicate significant difference by two-tailed Student’s t test (*P < 0.05; **P < 0.01; and ***P < 0.001).
The oscyp714b1 and osarf12 Mutant Lines Are Partial Phenotypic Copies of OsGRF7 Transgenic Lines
To further validate the functions of OsGRF7-regulated genes in plant architecture determination, we collected two mutants with mutations in OsGRF7 direct target genes, oscyp714b1 and osarf12. Of these, the oscyp714b1 mutant, which has a T-DNA insertion in the promoter region of OsCYP714B1 (Supplemental Fig. S12A), presented increased expression of OsCYP714B1 (Supplemental Fig. S12B). A previous report indicated that OsCYP714B1 participates in GA synthesis and that high expression of OsCYP714B1 represses the plant height by affecting cell elongation (Magome et al., 2013). Here, we analyzed the plant height of oscyp714b1 and found that it was reduced by ∼17.9% relative to that of the wild type (Supplemental Fig. S12, C and D), consistent with the reduced plant height of the GRF7OE lines (Fig. 1A).
For the osarf12 mutant, a T-DNA insertion in its third intron prevented its transcription (Supplemental Fig. S12, E and F). It exhibited an enlarged leaf angle, with the second leaf angle from the top of the main stalk increasing by approximately 73% compared with that of the wild type (Supplemental Fig. S12, G and H), which was partially similar to that which occurred for the GRF7RNAi lines. These investigations showed that the phenotype of each mutant was partially a copy of the plant architecture of the OsGRF7 transgenic lines, demonstrating that these genes function downstream of OsGRF7 to jointly control plant architecture in rice.
OsGRF7 Regulates the Contents of Endogenous GA and IAA
We also analyzed the contents of GA and IAA in 15-d-old seedlings of GRF7OE-1, GRF7RNAi-1, and wild-type plants. The results showed that the GA1 content was increased in GRF7OE-1 (Fig. 5A). Unfortunately, we did not detect any content of GA4 at this stage (15-d-old seedlings), mainly because the GA1 was predominantly abundant at the vegetative stage, while the level of GA4 was extremely high at the reproductive stage (Kobayashi et al., 1988). In rice, both GA1 and GA4 are bioactive forms derived from GA12, and GA4 is more active than GA1. However, their contents usually exhibit an opposite pattern, and thus, an increase in GA1 leads to a relatively low GA4 content; thus, the dynamic balance between GA1 and GA4 can effectively modulate the plant height (Magome et al., 2013).
Figure 5.
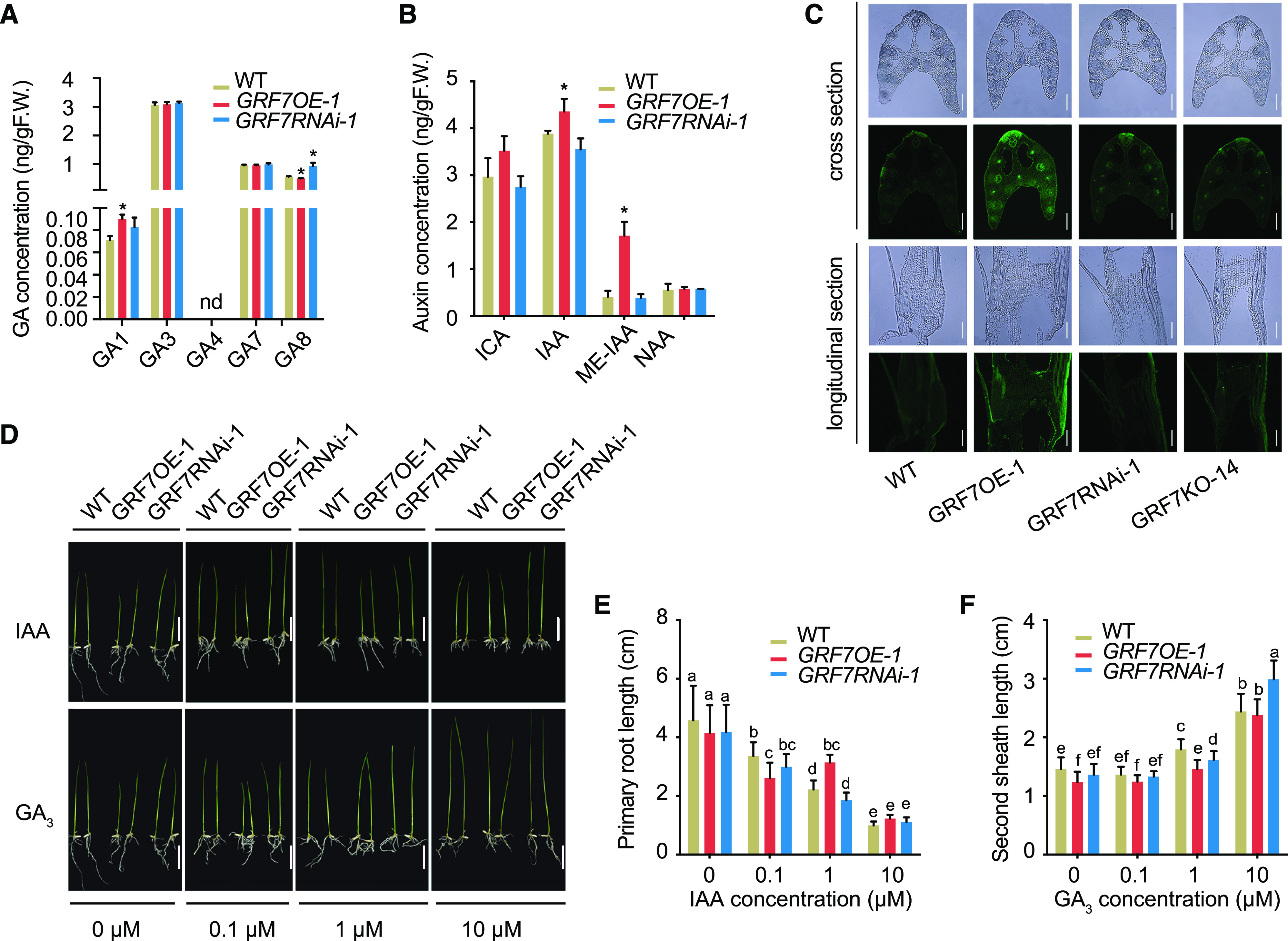
Endogenous phytohormone contents and exogenous phytohormone sensitivity test of OsGRF7 transgenic lines. A and B, Quantification of GA (A) and IAA (B) derivatives in OsGRF7 transgenic seedlings detected with liquid chromatography-tandem mass spectrometry. Values are means ± sd of three biological replicates. Asterisks indicate significant difference by two-tailed Student’s t test (*P < 0.05). F.W., Fresh weight; ICA, indole-3-carboxaldehyde; ME-IAA, methyl indole-3-acetate; NAA, 1-naphthylacetic acid; nd, not detected (below the detection limit); WT, wild type. C, Immunohistochemical observation of IAA at the lamina joint of OsGRF7 transgenic lines. Anti-IAA antibody and Alexa 488-conjugated goat anti-rabbit IgG antibody were used. Bars = 100 μm. D, Phenotypes of OsGRF7 transgenic seedlings treated by different concentrations of phytohormones. The same control was used for 0 μm IAA and GA. Bars = 2 cm. E, Effects of different concentrations of IAA on primary root length in the wild type, GRF7OE-1, and GRF7RNAi-1. Values are means ± sd (n = 15, three biological replicates and five plants per replicate). F, Effects of different concentrations of GA3 on the second sheath length in wild-type, GRF7OE-1, and GRF7RNAi-1 seedlings. Values are means ± sd (n = 15, three biological replicates and five plants per replicate). In E and F, different letters denote significant differences (P < 0.05) from Duncan’s multiple range test.
With respect to auxin, we found that the IAA and methyl-IAA contents of GRF7OE-1 significantly increased (Fig. 5B), which is consistent with the erect leaf angle of those plants (Zhang et al., 2015). Correspondingly, immunohistochemical observations revealed that the adaxial side of the GRF7OE-1 transgenic lines had more auxin than did the GRF7RNAi-1 and GRF7KO-14 lines (Fig. 5C). Auxin is critical in regulating the adaxial/abaxial cell growth of leaves, and the increased auxin content on the adaxial side of the lamina joints in the GRF7OE-1 transgenic line promoted cell division, which is consistent with the increased cell layers on the adaxial side of the GRF7OE-1 transgenic line (Fig. 1, F and G). These data reveal that OsGRF7 directly regulates the synthesis of endogenous GA and IAA in rice.
OsGRF7 Regulates the Sensitivity to Exogenous IAA and GA Treatment
We further treated OsGRF7 transgenic lines and investigated their responses to IAA and GA. After a 5-d treatment with different concentrations of IAA, we found that the primary root length of OsGRF7 transgenic lines decreased significantly under different IAA concentrations, while the primary root length of the GRF7RNAi-1 and wild-type seedlings decreased more than that of the GRF7OE-1 transgenic lines under 1 μm IAA, meaning that the GRF7OE lines are less sensitive to auxin than the wild type (Fig. 5, D and E).
GA treatment caused GRF7OE-1 seedlings to grow rapidly to the height of the wild-type and GRF7RNAi-1 plants with increasing GA content, although the second sheath length of GRF7OE-1 was shorter than that of the wild type (Fig. 5, D and F), meaning that GA could rescue the semidwarf phenotype of the GRF7OE plants. This is in agreement with the concept that high expression of OsCYP714B1 will lead to an increase in GA1 but a reduction in the content of highly active GA4 (Magome et al., 2013) because OsCYP714B1 encodes GA13 oxidase, which converts GA12 to GA1, in rice. In summary, these results demonstrate that GRF7OE lines are less sensitive to exogenous phytohormone treatments.
DISCUSSION
GRFs, the targets of OsmiR396, are members of a conserved, plant-specific gene family and are involved in many plant developmental processes, such as flowering, leaf morphogenesis, root development, and seed formation (Liu et al., 2009; Baucher et al., 2013). Increasing numbers of studies have documented that GRFs positively regulate leaf size by promoting cell proliferation and expansion in plant species such as Arabidopsis and rice (Rodriguez et al., 2010; Omidbakhshfard et al., 2015). In Arabidopsis, GRFs mainly participate in leaf and cotyledon growth, and cell division mediated by AtGRFs is important for polarized cell differentiation along the adaxial-abaxial axis during leaf morphogenesis (Wang et al., 2011). Overexpression of AtGRF1 and AtGRF2 resulted in stalks and leaves that were shorter and larger than those of the wild type (Kim et al., 2003). In contrast, a triple insertional null mutant of AtGRF1 to AtGRF3 had smaller leaves and cotyledons (Kim et al., 2003), and a quadruple mutant of AtGRF1 to AtGRF4 displayed much smaller leaves than its parental mutants (Kim and Lee, 2006), revealing that AtGRFs function mainly in plant height and leaf development. Recently, OsGRF1, OsGRF3, OsGRF4, OsGRF6, and OsGRF10 have been characterized to be involved in the regulation of plant development, especially in grain and floret development (Luo et al., 2005; Kuijt et al., 2014; Che et al., 2015; Gao et al., 2015; Tang et al., 2018), while their roles in plant architecture determination are still poorly understood. The OsmiR396-OsGRF module balanced growth and rice blast disease resistance, and overexpression of OsGRFs, including OsGRF6, OsGRF7, OsGRF8, and OsGRF9, enhanced blast resistance. However, the developmental roles of OsGRF7 have not been investigated in detail (Chandran et al., 2019).
Here, we found that OsGRF7, in combination with OsGIFs (Supplemental Fig. S11, C–E), modulated a wide range of rice plant morphological characteristics, including plant height, leaf length and width, and leaf angle (Fig. 1; Supplemental Table S1). This is in agreement with the results of the amino acid sequence-based phylogenetic analysis showing that OsGRF7 was the closest homolog of AtGRF1 and AtGRF2 (Supplemental Fig. S6). So far, 12 members of the OsGRF gene family have been identified in rice (Choi et al., 2004). Previously, OsGRF1, OsGRF4, OsGRF6, and OsGRF10 were found to regulate plant height and leaf size (Che et al., 2015; Hu et al., 2015; Li et al., 2018; Tang et al., 2018; Lu et al., 2020). We found that overexpression of OsGRF7 resulted in decreased plant height (Supplemental Fig. S3D) and altered leaf shape, suggesting that this gene promotes cell division and represses cell elongation in the culm and lamina joint (Fig. 1, C–F). Supporting this, plant height increased in GRF7KO lines; however, GRF7RNAi transgenic lines did not simply perform opposite to phenotypes of GRF7OE lines. The apparent conflict between GRF7RNAi and GRF7OE lines might be explained by compensatory changes in expression of other GRF genes when OsGRF7 is down-regulated, as evidenced by increased transcript levels of OsGRF4, OsGRF6, OsGRF8, and OsGRF9 (Supplemental Fig. S7). Alternatively, we cannot rule out the possibility that the GRF7RNAi constructs had off-target effects and/or that OsGRF7 negatively regulates other GRF genes.
Interestingly, unlike AtGRFs in Arabidopsis, OsGRF7 was expressed mainly around the vascular bundle of leaves and culms (Fig. 1; Supplemental Fig. S8), which promoted cell expansion and cell proliferation of parenchymal cells through periclinal division, especially on the adaxial side of the lamina joints, leading to reduced leaf angle and culm length in GRF7OE-1 transgenic lines, implying that OsGRF7 plays a vital role in plant architecture establishment. These findings reflect the functional diversification of structural orthologous genes in phylogenetically distant species.
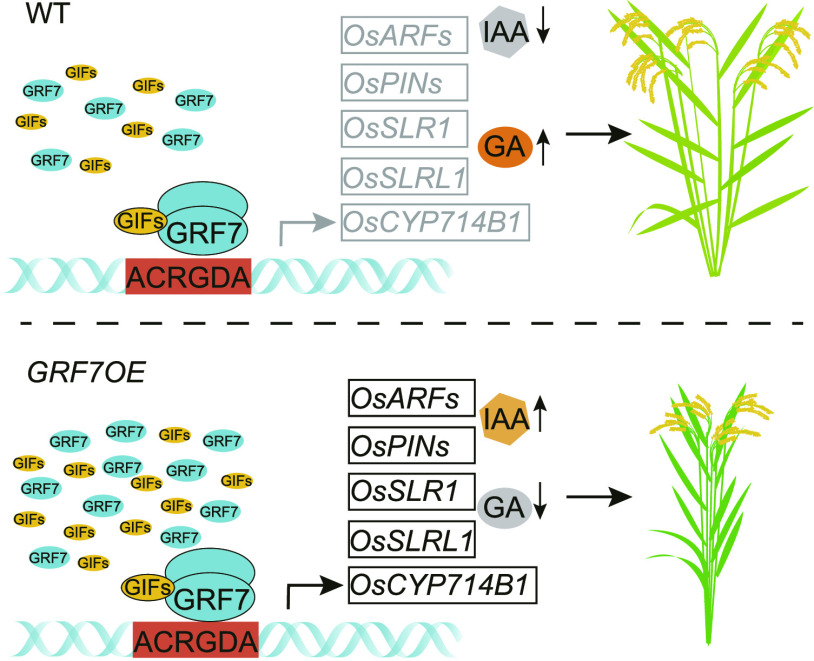
Plant hormones regulate many physiological processes that mainly influence growth, differentiation, and development (Luo et al., 2016). Recently, miRNAs have been found to modulate not only a wide range of agronomic characteristics but also many phytohormone-related biological processes (Djami-Tchatchou et al., 2017; Tang and Chu, 2017). These miRNAs function through their targets to modulate downstream genes involved in plant hormone perception and signaling, receptor proteins, or even enzyme synthesis (Tang and Chu, 2017). Here, we found that OsGRF7 can directly regulate a series of GA/IAA-related genes (Figs. 3 and 4) and can fine-tune the hormone balance of GAs and IAAs (Fig. 5, A–C). Hence, they coordinately regulate the development of internodes, leaves, and lamina joints and ultimately shape rice plant architecture (Fig. 6). A reduced auxin level causes an enhanced leaf angle due to the stimulation of cell elongation and repression of the division of parenchymal cells on the adaxial side, suggesting a negative effect of auxin on leaf inclination (Zhao et al., 2013; Zhou et al., 2017). Indeed, a high auxin level is detected at the lamina joint of GRF7OE-1 transgenic lines (Fig. 5, B and C), which is consistent with the increased expression of OsGRF7 in the lamina joints (Fig. 2, D and E). Combined with the cytological observations, our results showed that auxin is synthesized and undergoes polar transport, resulting in a high local auxin concentration on the adaxial side of the lamina joint, stimulating cell division but repressing cell elongation in GRF7OE-1 transgenic lines (Fig. 5C). These findings greatly expand our understanding of the involvement of the miR396-GRF module in hormone balance, signaling, and plant architecture determination.
Figure 6.
Proposed model for the regulation of rice plant architecture by OsGRF7. In GRF7OE plants, OsGRF7 coactivates with OsGIFs to up-regulate the expression of downstream hormone-related genes through binding to ACRGDA motifs. Then the synthesis of GA4 is inhibited, and the synthesis of IAA is promoted. Finally, the GRF7OE plants display a semidwarf and compact plant architecture. WT, Wild type.
As such, short plant stature has been the main target for the improvement of lodging resistance in rice breeding because it affords a lower risk to lodging (Ookawa et al., 2010). Here, GRF7OE lines exhibited decreased plant height and sturdy stalks resulting from decreased culm length (Supplemental Fig. S2) and increased culm wall thickness (Fig. 1, C and E). The size of the vascular bundles was significantly and negatively correlated with the lodging index, and increasing the size of vascular bundles can improve the lodging resistance in rice (Zhang et al., 2016). Cytological observations revealed that the GRF7OE lines had relatively large vascular bundles in the culms and leaves, and the parenchymal cells in the GRF7RNAi and GRF7KO leaves degraded faster than did those in the wild type and the GRF7OE lines as the leaves matured (Supplemental Fig. S8), meaning that the leaves and culms of the GRF7RNAi and GRF7KO lines senesced earlier and the parenchymal tissues became thinner as the plant matured. Consistent with this idea, auxin was concentrated mainly at the lamina joint and vascular bundles, and an apparent local auxin gradient from the cortical tissue inward was observed (Fig. 5C), reflecting that auxin modulates the leaf and culm development and senescence. An optimum level of auxin is required for cell division, differentiation, and elongation; vascular development; and organ patterning (Woodward and Bartel, 2005). Auxin can also promote vascular bundle development to regulate leaf angle (Zhang et al., 2015). These cytological characteristics in the GRF7RNAi and GRF7KO lines will cause the leaves to drop and the plants to lodge when encountering catastrophic weather with strong wind and heavy rain. In contrast, the high auxin content in the GRF7OE-1 transgenic lines increased the vascular bundle size both in the lamina joints and in the internodes (Fig. 1, C and F; Supplemental Fig. S8), resulting in erect leaf angles and sturdy stalks, which can be used in modern breeding for enhancing rice lodging resistance.
The leaf angle of rice is an important agronomic trait that constitutes the foundation of plant architecture. Increased leaf angles can expose leaf blades to more light but decreased light capture for the canopy, which is adverse for dense planting. In contrast, erect plant architecture is favored by crop breeders because it improves photosynthetic efficiency and increases plant density, thus improving the accumulation of leaf nitrogen for grain filling and increasing grain yield (Wang et al., 2018). The selection of novel genes controlling leaf angle and plant height is vital to further improve rice plant architecture. Our characterization of the OsGRF7 gene provides knowledge of the molecular basis of plant architecture control, and the utilization of OsGRF7 could be a favorable option for improving plant architecture and increasing lodging resistance.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa ssp. indica) YB was used as the transgenic receptor in this study. The osarf12 and oscyp714b1 mutants (under Dongjin, a japonica accession) were obtained from the Rice Functional Genomics Express Database of Korea (Jeong et al., 2002). MIM396-3 is the same as in our previous study (Gao et al., 2015). GRF7OE-1, GRF7OE-2, GRF7RNAi-1, and GRF7RNAi-2 transgenic lines are the same as in our previous study (Chandran et al., 2019). The T4 generation of the OsGRF7 overexpression and RNAi transgenic lines was used for the experiments. All the rice plants used in this study were grown in either Wuhan (May to October) or Hainan (December to April), China, during 2013 to 2018. To investigate the agronomic traits, 10 plants were planted in each row, and the middle five plants of each row were harvested to measure the agronomic traits. The plant-to-plant spacing was 16.5 cm within each row, and the row-to-row spacing was 26.7 cm. Field management practices essentially followed normal agricultural practices.
Phenotype Measurement
The plant height, effective panicle number, flag leaf length and width, and leaf angle were measured at the maturity stage (more than 95% of grains were matured). The plant height was measured from the lowest node to the flag leaf of the main tiller, and the effective panicle number was counted with the panicle bearing more than 10 fully developed seeds. The flag leaf of the main tiller was used to measure the flag leaf length and width, and the leaf sheath was imaged and measured with ImageJ software (https://imagej.nih.gov/ij/).
Vector Construction and Plant Transformation
The 1.3-kb full-length cDNA of OsGRF7 was amplified from the young inflorescence of YB and recombined into the overexpression vector pH7WG2D (Invitrogen) derived by the cauliflower mosaic virus 35S promoter. To generate GRF7RNAi transgenic plants, 316 bp of cDNA (661–977 bp) was inserted into a pANIC8A (Arabidopsis Biological Resource Center) vector. For the construction of GRF7-GFP, full-length cDNA of OsGRF7 was recombined into the plant expression vector pGWB5 (Invitrogen). An ∼2-kb promoter fragment of OsGRF7 was cloned into the pGWB3 vector (Invitrogen) to create the GRF7pro:GUS reporter gene construct. These vectors were introduced into the Agrobacterium tumefaciens strain EHA105 and transformed into YB. Primers used in this study are listed in Supplemental Data S3.
Generation of the OsGRF7 Knockout Mutant Using the CRISPR/Cas9 System
The CRISPR/Cas9 knockout plasmid for OsGRF7 was constructed as previously reported (Ma et al., 2015). We generated two single guide RNA (sgRNA) constructs (designed in the second exon; Supplemental Fig. S5A), in which the sgRNA was driven by the rice U6 promoter and the plant-optimized Cas9 was driven by the Ubi promoter. The integrated sgRNA expression cassettes were amplified and cloned into the CRISPR/Cas9 vector pYLCRISPR/Cas9Pubi-H (Ma et al., 2015). The construct was introduced into the wild-type variety YB. Then the 74 independent transgenic lines were subjected to PCR amplification and sequencing analysis. The T2 generation of GRF7KO lines was used for the experiments. The genotypes of GRF7KO lines are listed in Supplemental Table S2.
Histological Analysis
Plant tissues were fixed in FAA solution (3.7% [v/v] formaldehyde, 50% [v/v] ethanol, and 5% [v/v] acetic acid) and embedded in Paraplast Plus (Sigma-Aldrich). Eight-micrometer-thick sections were stained with Toluidine Blue for light microscopic analysis. Five independent sections were microscopically examined and photographed to measure the cell layers, culm thickness, and cell length; 10 independent sections were microscopically examined and photographed to measure the area of the large vascular bundles (Leica).
5′ RACE Analysis
Total RNA from YB young inflorescence was isolated with TRIzol reagent (Invitrogen), ligated to RNA adaptors, and synthesized into cDNA using SuperScript III according to the manufacturer’s instructions (Invitrogen). 5′ RACE was performed with the GeneRace kit (Invitrogen).
Transient Expression, Protein Extraction, and Immunoblotting
The coding sequence of OsrGRF7 was synthesized and ligated downstream of the 35S promoter. For OsmiR396/Os(r)GRF7 coexpression experiments, an equal amount of plasmids (3 μg) containing Ubi:miR396s and 35S:(r)GRF7 was mixed before transformation into rice protoplasts. After transformation, protoplasts were placed at 28°C for 24 h before RNA extraction. The OsmiR396 precursors were ligated downstream of the ubiquitin promoter with restriction site BamHI in the pCAMBIA1301 vector. Protoplasts isolated from etiolated rice seedlings of GRF7-GFP transgenic lines were transfected with 3 μg of Ubi:miR396s or empty vector plasmids by 40% (w/v) polyethylene glycol. After incubation for 16 h, the OsGRF7 protein was detected by western-blot analysis using an anti-GFP antibody. Total RNA was extracted to detect the transcript levels of OsmiR396s.
Total RNA Isolation and RT-qPCR
Total RNA (2 μg) was extracted and synthesized into cDNA using the Moloney murine leukemia virus reverse transcriptase (Invitrogen) according to the user’s manual. RT-qPCR was performed by Light Cycler 480 II (Roche) with SYBR Green PCR Mixture (Roche) according to the manufacturer’s instructions. The OsUBI gene was used as the internal control. Three technical replicates were performed to generate the average value, and three biological repeats were performed for each analysis.
Histochemical GUS Staining
Different tissues were stained with GUS staining buffer (100 mm sodium phosphate, 10 mm EDTA, 0.1% [v/v] Triton X-100, and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-GlcA [Sigma], pH 7) overnight at 37°C. All samples were observed with an SLR camera (Nikon SMZ-645). Two independent lines were analyzed for GUS staining.
Subcellular Localization and Bimolecular Fluorescence Complementation
For subcellular localization, the full-length OsGRF7 cDNA was cloned into pGWB5 (Invitrogen). For bimolecular fluorescence complementation, the full-length OsGRF7 cDNA was amplified with restriction sites XbaII and ClaI and cloned into pSPYNE (N-terminal end of YFP), and the full-length OsGRF7, OsGIF1, OsGIF2, and OsGIF3 cDNAs were amplified with restriction sites XbaII and ClaI and inserted into pSPYCE (C-terminal end of YFP; Walter et al., 2004). Rice protoplasts were isolated as described previously (Yoo et al., 2007), with some modifications. After overnight incubation in the darkness, the fluorescence signals and bright-field images of the protoplasts were taken with an FV1000 confocal system (Olympus). Empty vectors of bimolecular fluorescence complementation constructs were used as a negative control.
Transactivation Assay
To test the transactivation activity, the full-length or truncated OsGRF7 with restriction sites BamHI and EcoRI were amplified and cloned into pGBKT7 (Clontech). The plasmids were transformed into yeast strain AH109, then the transformants were plated on either synthetic dropout nutrient medium (SD)/-Trp or SD/-Trp-His-Leu-Ade medium.
Yeast Two-Hybrid Assay
The full-length cDNAs of OsGIF1, OsGIF2, OsGIF3, and OsGRF7 with restriction sites EcoRI and BamHI were cloned into the prey vector pGADT7 (Clontech), and the full-length or truncated cDNA of OsGRF7 with restriction sites BamHI and EcoRI was cloned into pGBKT7 (Clontech). The prey and bait plasmids were cotransformed into AH109 and plated on SD/-Leu-Trp medium for 3 d at 30°C. Interactions between bait and prey were further tested on SD/-Trp-His-Leu-Ade medium.
ChIP-Seq and Bioinformatics Analysis
The transgenic line GRF7-GFP was used for ChIP assays according to the method of Feng et al. (2012) with some modifications. Briefly, 5 g of inflorescences less than 0.5 cm (YP1) or less than 2 cm (YP2) was harvested and cross-linked with 1% (v/v) formaldehyde under vacuum for 30 min and then ground into powder in liquid nitrogen. The chromatin complexes were isolated, sonicated, and incubated with the anti-GFP antibodies (Abcam, ab290). The precipitated DNA was recovered and dissolved in 1× Tris-EDTA for later in-depth analysis.
Illumina sequencing libraries were constructed with the above-prepared DNA samples by the ThruPLEX DNA-seq Kit mainly according to the manufacturer’s instructions. Then, the library containing 270- to 330-bp fragments was purified and sequenced with the Illumina HiSeq3000 system. Clean reads were mapped to the rice genome using Bowtie (Langmead et al., 2009). MACS (Liu, 2014) was used with a P value cutoff of 10−5 for binding peaking. The reads per million around the peak summit (±5 kb) was documented with a 100-nucleotide window size to calculate the average enrichment level of the peak. To predict binding motifs, the flanking sequences of all peak summits (±50 bp) were filtered by the RepeatMasker Web Server (http://www.repeatmasker.org), and the masked sequences were subjected to DREME (Bailey, 2011).
ChIP-qPCR
The prepared DNA in ChIP was quantified using qPCR with their primers. PCR was performed in triplicate for each sample, and the values were normalized to the input sample to obtain the enrichment fold. The fold enrichment was calculated against the OsUBI promoter. No addition of antibodies served as a negative control. Three biological replicates were performed for each analysis.
Transient Transactivation Assay
Approximately 2-kb promoter regions from each of OsARF3, OsARF8, OsPIN1b, OsPIN1d, OsPIN8, and OsSLR1 with restriction sites XhoI and BamHI; OsARF9 and OsSLRL1 with restriction sites XhoI and BglII; and OsARF4, OsARF12, and OsCYP714B1 with restriction sites HindIII and BamHI were amplified from YB and then cloned into a pGreenII-0800-Luc (EK-Bioscience) vector containing the renilla luciferase reporter gene driven by the 35S promoter and the firefly luciferase reporter gene, thus generating the vectors containing specific promoters fused to firefly luciferase. Transient transactivation assays were performed using rice protoplasts, and the Dual-Luciferase Reporter Assay System (Promega) was used to detect the luciferase activity, with the renilla luciferase gene as the internal control. Three biological replicates were performed for each analysis.
Expression and Purification of His-GRF7 Fusion Proteins in Escherichia coli
The full-length cDNA fragment was amplified with restriction sites BamHI and EcoRI and fused into expression vector pCOLD. To extract recombinant protein, Rosetta (DE3) cells carrying pCOLD-GRF7 plasmid were inoculated at 37°C to an optical density. Then, isopropyl-β-d-thiogalactoside was added to a final concentration of 0.2 mm, and cultures were grown for 24 h at 15°C. Cultured cells were harvested, lysed, and centrifuged, then the supernatant was purified by His-binding-resin (GE Healthcare) using the AKTA Prime Plus protein purification system (GE Healthcare).
EMSA
Oligonucleotides were synthesized and labeled by digoxin. Promoter regions were amplified, labeled by digoxin, and purified as the probe. Then, 200 ng of DNA probe, 2 μg of protein (His-GRF7), 2 μL of 10× binding buffer (100 mm Tris, 500 mm KCl, and 10 mm DTT, pH 7.5), 1 μL of 50% (v/v) glycerol, 1 μL of 1% (v/v) Nonidet P-40, 1 μL of 1 m KCl, 1 μL of 0.5 m EDTA, 1 μL of 1 mg mL−1 poly(dI-dC), and double-distilled water were mixed to a final volume of 20 μL (Liu et al., 2014), reacted for 20 min at 25°C, electrophoresed on 3.5% (w/v) native polyacrylamide gels, and then transferred to N+ nylon membranes (Millipore) in TBE buffer (44.5 mm Tris-borate and 1 mm EDTA) at 200 mA at 4°C for 1 h. Digoxin-labeled DNA was detected by the CDP-Star easy to use kit (Roche).
Measuring Endogenous Phytohormones
Contents of IAA and GA were analyzed using a liquid chromatography-electrospray ionization-tandem mass spectrometry system. To detect endogenous GA and IAA, 15 seeds of the OsGRF7 transgenic lines were planted in a container filled with soil with 2- × 2-cm spacing in a phytotron. Fresh 15-d-old seedlings were harvested, weighed, and then immediately ground into powder in liquid nitrogen. Since tissue close to the soil may be contaminated, approximately 0.5 cm of tissue near the soil was removed with scissors. After being extracted with 1 mL of 80% (v/v) methanol at 4°C for 12 h, the extract was centrifuged at 12,000g under 4°C for 15 min. The supernatant was collected and evaporated to dryness under a nitrogen gas stream and then reconstituted in 100 mL of 95% (v/v) acetonitrile. The supernatant was collected for liquid chromatography-mass spectrometry analysis after the solution was centrifuged. Each series of experiments was performed in biological triplicates.
Immunohistochemical Observation of IAA
To cross-link IAA, excised lamina joints were prefixed for 2 h in 3% (w/v) 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (Sigma-Aldrich) at room temperature and then transferred to FAA at 4°C for 24 h. Eight-micrometer-thick sections were deparaffinized and hydrated. Antigen retrieval was carried out by rinsing the slides in 0.1 m sodium citrate buffer and microwave heated at high power for 5 min. After being washed three times with 10 mm phosphate-buffered saline (PBS) for 10 min, the slides were subsequently incubated in 10 mm PBS containing 0.1% (v/v) Tween 20, 1.5% (v/v) Gly, and 5% (w/v) bovine serum albumin (BSA) for 45 min at 22°C. Samples were then rinsed in a regular salt rinse solution (RSRS; 10 mm PBS, 0.88% [w/v] NaCl, 0.1% [v/v] Tween 20, and 0.8% [w/v] BSA) and washed briefly with 10 mm PBS containing 0.8% (w/v) BSA (PBS+BSA) solution. After the application of anti-IAA antibodies to each slide, samples were incubated overnight in a humidity chamber at 4°C. After hybridization, samples were subjected to a series of vigorous washes, twice with a high-salt rinse solution (10 mm PBS, 2.9% [w/v] NaCl, 0.1% [v/v] Tween 20, and 0.1% [w/v] BSA) for 15 min, once with RSRS for 15 min, and briefly with PBS+BSA. The Alexa 488-conjugated goat anti-mouse IgG antibodies were then placed on each slide, and these were incubated for 4 to 6 h in a humidity chamber at room temperature. After washing with RSRS twice for 15 min, samples were mounted with an antifade reagent, covered with a cover slip, and observed with a fluorescence microscope (Sakata et al., 2010).
Plant Hormone Treatment
For hormone treatment, dehulled seeds were sown on one-half strength Murashige and Skoog agar medium containing different phytohormones. After 5 d of growth, the lengths of the root and the second sheath were analyzed using ImageJ software (National Institutes of Health). Three biological replicates were performed, and five plants for each replicate were collected for data analysis.
Phylogenetic Analysis
For phylogenetic analysis of OsGRF7 homologs in rice and Arabidopsis (Arabidopsis thaliana), 12 rice GRF protein sequences and nine Arabidopsis GRF protein sequences were obtained in PLAZA 2.5 (Van Bel et al., 2012). The amino acid sequences were aligned using the ClustalX program (http://www.clustal.org/clustal2/). The phylogenetic tree was constructed using MEGA X (Kumar et al., 2018) based on the maximum likelihood method with 1,000 bootstrap replicates.
Statistical Analysis
Statistical analyses were performed using IBM SPSS statistics, version 20.0 (IBM). The two-tailed Student’s t test was used for comparing the agronomic traits of each transgenic line with the wild type. The correlation analysis was performed with GraphPad Prism 8 (https://www.graphpad.com/scientific-software/prism/) to generate the r and P values.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: OsGRF7, Os12g0484900; OsGIF1, Os03g0733600; OsGIF2, Os11g0615200; OsGIF3, Os12g0496900; OsCYP714B1, Os07g0681300; OsARF12, Os04g0671900; and OsUBI, Os03g0234200. High-throughput sequencing data created in this study have been deposited in the Gene Expression Omnibus database (GSE109802).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotypic and molecular analysis of MIM396 transgenic lines.
Supplemental Figure S2. Comparison of internode length between the wild type and OsGRF7 transgenic lines.
Supplemental Figure S3. Relationship between OsGRF7 expression levels and phenotypic traits of the OsGRF7 transgenic lines.
Supplemental Figure S4. Genetic analysis between OsmiR396 and OsGRF7.
Supplemental Figure S5. Phenotype and molecular characterization of OsGRF7 knockout lines.
Supplemental Figure S6. Phylogenetic analysis of GRFs in Arabidopsis and rice.
Supplemental Figure S7. Expression analysis of OsGRF members in the OsGRF7 transgenic lines.
Supplemental Figure S8. Cytological dynamics of lamina joint from initiation to maturation.
Supplemental Figure S9. Regulation of OsGRF7 by OsmiR396s.
Supplemental Figure S10. The transcription levels of the OsmiR396 family in GRF7-GFP transgenic rice protoplasts were analyzed with RT-PCR.
Supplemental Figure S11. Analysis of interactions between OsGRF7 and OsGIFs.
Supplemental Figure S12. Architecture and molecular identification of oscyp714b1 and osarf12 mutants.
Supplemental Table S1. Agronomic traits of OsGRF7 transgenic lines.
Supplemental Table S2. Genotype analysis of GRF7KO transgenic lines.
Supplemental Table S3. Functional categories of genes associated with OsGRF7 binding sites.
Supplemental Data Set S1. Genes associated with OsGRF7-binding sites identified by ChIP-seq in YP1 (<0.5 cm).
Supplemental Data Set S2. Genes associated with OsGRF7-binding sites identified by ChIP-seq in YP2 (<2 cm).
Supplemental Data Set S3. Primers used in this article.
Acknowledgments
We thank Dr. Kun Wang and Zhenying Shi for their critical reading and advice during the preparation of this article as well as Dr. Ai Qin for providing assistance in the bioinformatic analysis.
Footnotes
This work was supported by the National Key Research and Development Program (grant no. 2016YFD0100903), the National Transgenic Research and Development Program (grant no. 2016ZX08001004–001–002), and the National Natural Science Foundation of China (grant no. 31870322).
Articles can be viewed without a subscription.
References
- Bailey TL.(2011) DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics 27: 1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M, Moussawi J, Vandeputte OM, Monteyne D, Mol A, Pérez-Morga D, El Jaziri M(2013) A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco. Plant Biol (Stuttg) 15: 892–898 [DOI] [PubMed] [Google Scholar]
- Chandran V, Wang H, Gao F, Cao XL, Chen YP, Li GB, Zhu Y, Yang XM, Zhang LL, Zhao ZX, et al. (2019) miR396-OsGRFs module balances growth and rice blast disease-resistance. Front Plant Sci 9: 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che R, Tong H, Shi B, Liu Y, Fang S, Liu D, Xiao Y, Hu B, Liu L, Wang H, et al. (2015) Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants 2: 15195. [DOI] [PubMed] [Google Scholar]
- Chen X, Lu S, Wang Y, Zhang X, Lv B, Luo L, Xi D, Shen J, Ma H, Ming F(2015) OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. Plant J 82: 302–314 [DOI] [PubMed] [Google Scholar]
- Choi D, Kim JH, Kende H(2004) Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol 45: 897–904 [DOI] [PubMed] [Google Scholar]
- Djami-Tchatchou AT, Sanan-Mishra N, Ntushelo K, Dubery IA(2017) Functional roles of microRNAs in agronomically important plants: Potential as targets for crop improvement and protection. Front Plant Sci 8: 378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercoli MF, Ferela A, Debernardi JM, Perrone AP, Rodriguez RE, Palatnik JF(2018) GIF transcriptional coregulators control root meristem homeostasis. Plant Cell 30: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS(2012) Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Wang K, Liu Y, Chen Y, Chen P, Shi Z, Luo J, Jiang D, Fan F, Zhu Y, et al. (2015) Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat Plants 2: 15196. [DOI] [PubMed] [Google Scholar]
- Gao S, Fang J, Xu F, Wang W, Chu C(2016) Rice HOX12 regulates panicle exsertion by directly modulating the expression of ELONGATED UPPERMOST INTERNODE1. Plant Cell 28: 680–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, et al. (2015) A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant 8: 1455–1465 [DOI] [PubMed] [Google Scholar]
- Jeong DH, An S, Kang HG, Moon S, Han JJ, Park S, Lee HS, An K, An G(2002) T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol 130: 1636–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush GS.(1995) Breaking the yield frontier of rice. GeoJournal 35: 329–332 [Google Scholar]
- Kim JH, Choi D, Kende H(2003) The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J 36: 94–104 [DOI] [PubMed] [Google Scholar]
- Kim JH, Kende H(2004) A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA 101: 13374–13379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee BH(2006) GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J Plant Biol 49: 463–468 [Google Scholar]
- Kobayashi M, Yamaguchi I, Murofushi N, Ota Y, Takahashi N(1988) Fluctuation and localization of endogenous gibberellins in rice. Agric Biol Chem 52: 1189–1194 [Google Scholar]
- Kozomara A, Griffiths-Jones S(2014) miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt SJ, Greco R, Agalou A, Shao J, ’t Hoen CC, Overnäs E, Osnato M, Curiale S, Meynard D, van Gulik R, et al. (2014) Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol 164: 1952–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K(2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL(2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Estelle M(2016) Mechanisms of auxin signaling. Development 143: 3226–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Gao F, Xie K, Zeng X, Cao Y, Zeng J, He Z, Ren Y, Li W, Deng Q, et al. (2016) The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol J 14: 2134–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y, et al. (2018) Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560: 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li J, Chen Z, Wei Y, Qi Y, Wu C(2020) OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnol J doi:10.1111/pbi.13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Song Y, Chen Z, Yu D(2009) Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol Plant 136: 223–236 [DOI] [PubMed] [Google Scholar]
- Liu H, Guo S, Xu Y, Li C, Zhang Z, Zhang D, Xu S, Zhang C, Chong K(2014) OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol 165: 160–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.(2014) Use Model-based Analysis of ChIP-Seq (MACS) to analyze short reads generated by sequencing protein-DNA interactions in embryonic stem cells. Methods Mol Biol 1150: 81–95 [DOI] [PubMed] [Google Scholar]
- Lu Y, Meng Y, Zeng J, Luo Y, Feng Z, Bian L, Gao S(2020) Coordination between GROWTH-REGULATING FACTOR1 and GRF-INTERACTING FACTOR1 plays a key role in regulating leaf growth in rice. BMC Plant Biol 20: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo AD, Liu L, Tang ZS, Bai XQ, Cao SY, Chu CC(2005) Down-regulation of OsGRF1 gene in rice rhd1 mutant results in reduced heading date. J Integr Plant Biol 47: 745–752 [Google Scholar]
- Luo X, Zheng J, Huang R, Huang Y, Wang H, Jiang L, Fang X(2016) Phytohormones signaling and crosstalk regulating leaf angle in rice. Plant Cell Rep 35: 2423–2433 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Magome H, Nomura T, Hanada A, Takeda-Kamiya N, Ohnishi T, Shinma Y, Katsumata T, Kawaide H, Kamiya Y, Yamaguchi S(2013) CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc Natl Acad Sci USA 110: 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard MA, Proost S, Fujikura U, Mueller-Roeber B(2015) Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol Plant 8: 998–1010 [DOI] [PubMed] [Google Scholar]
- Ookawa T, Hobo T, Yano M, Murata K, Ando T, Miura H, Asano K, Ochiai Y, Ikeda M, Nishitani R, et al. (2010) New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat Commun 1: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF(2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata T, Oshino T, Miura S, Tomabechi M, Tsunaga Y, Higashitani N, Miyazawa Y, Takahashi H, Watanabe M, Higashitani A(2010) Auxins reverse plant male sterility caused by high temperatures. Proc Natl Acad Sci USA 107: 8569–8574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al. (2002) Green revolution: A mutant gibberellin-synthesis gene in rice. Nature 416: 701–702 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Bishopp A, Kieber JJ(2015) The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 27: 44–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair TR, Sheehy JE(1999) Erect leaves and photosynthesis in rice. Science 283: 1456–145710206873 [Google Scholar]
- Springer N.(2010) Shaping a better rice plant. Nat Genet 42: 475–476 [DOI] [PubMed] [Google Scholar]
- Tang J, Chu C(2017) MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat Plants 3: 17077. [DOI] [PubMed] [Google Scholar]
- Tang Y, Liu H, Guo S, Wang B, Li Z, Chong K, Xu Y(2018) OsmiR396d affects gibberellin and brassinosteroid signaling to regulate plant architecture in rice. Plant Physiol 176: 946–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M, Proost S, Wischnitzki E, Movahedi S, Scheerlinck C, Van de Peer Y, Vandepoele K(2012) Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol 158: 590–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang B, Smith SM, Li J(2018) Genetic regulation of shoot architecture. Annu Rev Plant Biol 69: 437–468 [DOI] [PubMed] [Google Scholar]
- Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X(2011) miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot 62: 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang S, Sun C, Xu Y, Chen Y, Yu C, Qian Q, Jiang DA, Qi Y(2014) Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol 201: 91–103 [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J(2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59: 253–279 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B(2005) Auxin: Regulation, action, and interaction. Ann Bot 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Zhang Q(2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61: 421–442 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J(2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang S, Wang S, Xu Y, Yu C, Shen C, Qian Q, Geisler M, Jiang A, Qi Y(2015) The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ 38: 638–654 [DOI] [PubMed] [Google Scholar]
- Zhang W, Wu L, Wu X, Ding Y, Li G, Li J, Weng F, Liu Z, Tang S, Ding C, et al. (2016) Lodging resistance of japonica rice (Oryza sativa L.): Morphological and anatomical traits due to top-dressing nitrogen application rates. Rice (N Y) 9: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SQ, Xiang JJ, Xue HW(2013) Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol Plant 6: 174–187 [DOI] [PubMed] [Google Scholar]
- Zhou LJ, Xiao LT, Xue HW(2017) Dynamic cytology and transcriptional regulation of rice lamina joint development. Plant Physiol 174: 1728–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]