Arabidopsis glucosyl sphingolipid-modulated plasmodesmal gating involves modulation of the glycosylphosphatidylinositol-anchored PdBG2 protein secretion machinery and a change in callose accumulation.
Abstract
Plasma membranes encapsulated in the symplasmic nanochannels of plasmodesmata (PD) contain abundant lipid rafts, which are enriched with sphingolipids (SLs) and sterols. Reduction of sterols has highlighted the role played by lipid raft integrity in the intercellular trafficking of glycosylphosphatidylinositol (GPI)-anchored PD proteins, particularly in affecting callose enhancement. The presence of callose at PD is strongly attributed to the regulation of callose accumulation and callose degradation by callose synthases and β-1,3-glucanases (BGs), respectively. SLs are implicated in signaling and membrane protein trafficking; however, the underlying processes linking SL composition to the control of symplasmic apertures remain unknown. The wide variety of SLs in plants prompted us to investigate which SL molecules are important for regulating symplasmic apertures in Arabidopsis (Arabidopsis thaliana). We introduced several potential SL pathway inhibitors and genetically modified SL contents using two independent SL pathway mutants. We were able to modulate callose deposition to control symplasmic connectivity through perturbations of SL metabolism. Alteration in glucosylhydroxyceramides or related SL composition particularly disturbed the secretory machinery for the GPI-anchored PdBG2 protein, resulting in an overaccumulation of callose. Moreover, our results revealed that SL-enriched lipid rafts link symplasmic channeling to PD callose homeostasis by controlling the targeting of GPI-anchored PdBG2. This study elevates our understanding of the molecular linkage underlying intracellular trafficking and precise targeting of GPI-anchored PD proteins incorporating glucosyl SLs.
Plasmodesmata (PD), plant-specific symplasmic channels, cross the plant cell wall and physically connect the cytoplasm and endoplasmic reticulum (ER) of contiguous cells. These intercellular channels play important roles in multicellular events during plant development by allowing the molecular exchange of signaling molecules such as transcription factors, RNAs, and growth regulators (Zambryski and Crawford, 2000; Maule, 2008; Wu et al., 2018). The PD plasma membrane (PM) is distinct from the general cellular PM and is characterized by the enrichment of sterols and sphingolipid (SL) species (Grison et al., 2015; Iswanto and Kim, 2017; Mamode Cassim et al., 2019). The domains enriched with those special lipids are referred to as the membrane microdomain compartments, also called lipid rafts (Mongrand et al., 2010).
The dynamic regulation of the size exclusion limit of PD particularly involves the modulation of callose deposition; callose is synthesized by callose synthases and degraded by β-1,3-glucanases (BGs; Verma and Hong, 2001; Levy et al., 2007a; Chen and Kim, 2009). The importance of sterols in the function of PD has well been reported; the perturbation of sterol biosynthesis pathways affects symplasmic intercellular connectivity by changing the subcellular localization of glycosylphosphatidylinositol (GPI)-anchored PD proteins, namely, plasmodesmata callose binding-1 (PDCB1) and plasmodesmata-associated β-1,3-glucanase (PdBG2), which is followed by callose accumulation (Grison et al., 2015). Recently, by characterizing PHLOEM UNLOADING MODULATOR encoding a putative enzyme required for the biosynthesis of SLs with very-long-chain fatty acid, SLs were shown to modulate plasmodesmal ultrastructure and phloem unloading (Yan et al., 2019). In addition, an increase in ceramide content resulted in a reduced callose deposition in Arabidopsis (Arabidopsis thaliana) cells (Bi et al., 2014). However, the involvement of SLs in callose-mediated PD regulation has not yet been established.
GPI lipid anchoring refers to the posttranslational modification of many cell surface proteins with a glycolipid. Lipid rafts provide platforms for GPI-anchored proteins, enabling them to stay at the cellular membrane (Mongrand et al., 2010). Modulations in lipid raft components have been implicated in the secretory machinery affecting certain PM proteins, in which the ABCB19 and PIN2 translocations are perturbed along with the depletion of sterols and SLs (Yang et al., 2013; Wattelet-Boyer et al., 2016). GPI-anchored PD proteins contain a signal peptide at their N-terminal domain, and their GPI modification occurs in the ER. Attachment of the GPI moiety to the C terminus (ω-site) of the polypeptide occurs after proteolytic cleavage of a C-terminal propeptide from the proprotein (Udenfriend and Kodukula, 1995). Moreover, the GPI anchor undergoes remodeling either in the glucan chain or in the lipid moiety by the addition of a saturated fatty acid chain. GPI remodeling is crucial for the attachment events of GPI-anchored proteins to the membrane lipid raft during translocation (Muñiz and Zurzolo, 2014; Kinoshita, 2015).
The SL synthetic pathway is initiated by the activation of Ser palmitoyl transferase, which is involved in the conversion of Ser and palmitoyl-CoA to 3-ketosphinganine; 3-ketosphinganine then proceeds to form ceramides as intermediate products in the metabolism of SL (Michaelson et al., 2016). Ceramide undergoes modification to produce glycosphingolipids, and glycosylinositolphosphorylceramides (GIPCs) and glucosylceramides (GlcCers) are two major classes of glycosphingolipids found in plants (Warnecke and Heinz, 2003; Kim et al., 2013). Approximately 64% of all SLs are GIPCs, and several studies have shown that critical roles played by GlcCers and GIPCs in intracellular trafficking and plant developmental events (Markham and Jaworski, 2007; Msanne et al., 2015; Fang et al., 2016). Furthermore, GlcCers can be divided into six groups with distinct molecular compositions depending upon the existence and attributes of types of long-chain bases (LCBs), double bonds, and hydroxy fatty acids (Hamanaka et al., 2002).
Here, using pharmacological and genetic approaches, we show that the perturbation of SL metabolic pathways results in the alteration of symplasmic connectivity in a manner that is linked to the PD callose level. In addition, we found that the attenuation of Glucosylhydroxyceramides (GlcHCers) alters the subcellular localization of GPI-anchored PdBG2 and PDCB1 proteins but not for non-GPI-anchored PDLP1 protein. We also report that there are at least two independent secretory pathways for GPI-anchored PD protein and non-GPI-anchored PD protein. In summary, our studies reveal that GlcHCers or related SL composition are important for the regulation of symplasmic channeling through the modulation of PD targeting of GPI-anchored PdBG2 protein and the callose level.
RESULTS
Perturbation in SL Metabolism Alters PD Permeability in Arabidopsis Hypocotyl and Root Tip
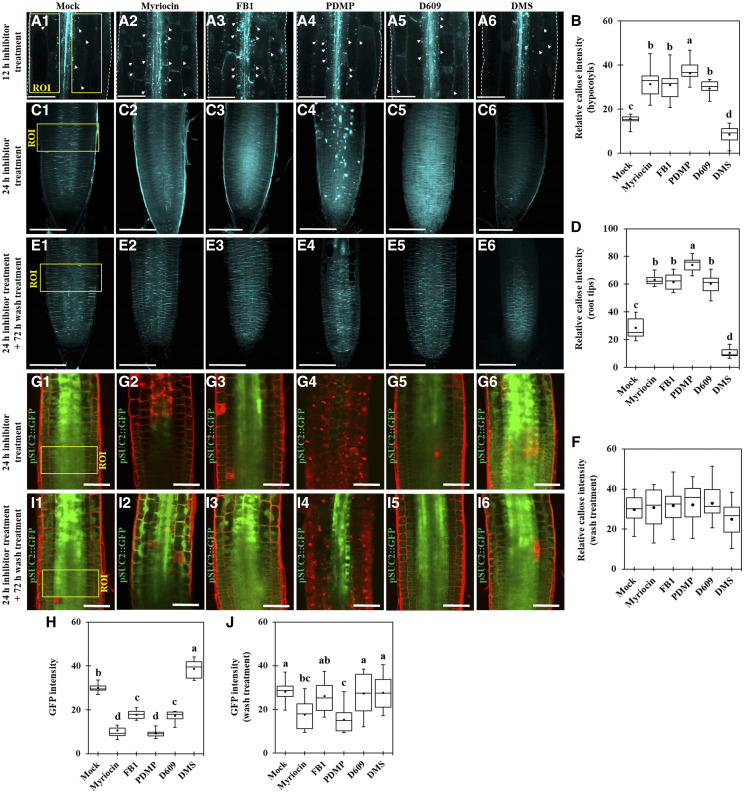
To test if SLs are important in PD regulation, we used several potential inhibitors of SL metabolism (Supplemental Fig. S1; Edsall et al., 1998; Singh et al., 2000; Coursol et al., 2003; Sperling and Heinz, 2003; Wright et al., 2003; Falcone et al., 2004; Chen et al., 2006, 2014; Delgado et al., 2006; Kang et al., 2008; Worrall et al., 2008; Melser et al., 2010; Markham et al., 2011; Su et al., 2011; Adibhatla et al., 2012; Michaelson et al., 2016). We performed PD permeability assays using 8-hydroxypyrene-1,3,6-trisulfonic acid (HPTS; Han et al., 2014) and Aniline Blue callose staining assays with etiolated Arabidopsis hypocotyls after treatment with SL synthetic pathway inhibitors. PD permeability, as shown by HPTS movement, was reduced in seedlings treated with myriocin, fumonisin B1 (FB1), DL-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol hydrochloride (PDMP), and tricyclodecan-9-yl-xanthogenate (D609), but seedlings treated with D-erythro-N,N-dimethylsphingosine (DMS) displayed enhanced PD permeability (Supplemental Fig. S2). Accordingly, PD permeability was inversely associated with callose deposition; higher PD permeability was found at a lower level of callose deposition (Fig. 1, A and B). Furthermore, we also investigated the action of SL synthetic pathway inhibitors in PD regulation at the Arabidopsis root tip. At 24 h after the treatment of SL inhibitors such as myriocin, FB1, PDMP, and D609, the callose levels were significantly increased as compared with the mock condition, whereas DMS treatment reduced the callose level (Fig. 1, C and D).
Figure 1.
Perturbation in the SL pathway alters PD permeability in Arabidopsis hypocotyls and root tips. A1 to A6, Confocal images show Aniline Blue-stained callose in Arabidopsis etiolated hypocotyls after SL inhibitor treatment (12 h). White arrowheads show Aniline Blue-stained callose at PD. Bars = 100 μm. B, Quantitative data show the relative callose intensity of Arabidopsis etiolated hypocotyls after SL inhibitor treatment (A1–A6; n = 15). C1 to C6, Confocal images show Aniline Blue-stained callose in Arabidopsis root tips after SL inhibitor treatment (24 h). Bars = 100 μm. D, Quantitative data show the relative callose intensity of Arabidopsis root tips after SL inhibitor treatment (C1–C6; n = 15). E1 to E6, Confocal images show Aniline Blue-stained callose in Arabidopsis root tips. Images were captured right after wash treatment (replaced to the normal agar plates, 72 h) from SL inhibitor treatment (24 h). Bars = 100 μm. F, Quantitative data show the relative callose intensity of Arabidopsis root tips after wash treatment (E1–E6; n = 9). G1 to G6, Confocal images of root tips of Arabidopsis transgenic plants expressing pSUC2::GFP. Images were captured right after SL inhibitor treatment (24 h). Bars = 50 μm. H, Quantitative data show the GFP fluorescence intensity of Arabidopsis root tips expressing pSUC2::GFP after SL inhibitor treatment (G1–G6; n = 12). I1 to I6, Confocal images of root tips of Arabidopsis transgenic plants expressing pSUC2::GFP. Images were captured right after wash treatment from SL inhibitor treatment (24 h). Bars = 50 μm. J, Quantitative data show the GFP fluorescence intensity of Arabidopsis root tips expressing pSUC2::GFP after wash treatment (I1–I6; n = 12). Yellow boxes indicate regions of interest (ROI) for callose quantification (A1–A6 and C1–C6) and GFP fluorescence intensity quantification (G1–G6 and I1–I6). Three independent biological replicates were performed for each experiment, and statistical significance was determined by one-way ANOVA with the Tukey-Kramer test. Boxes in box plots shows 25th to 75th percentiles. Black dots and lines in boxes indicate mean and median, respectively. Significant differences are indicated by the presence of different lowercase letters above each box plot (P < 0.05).
Next, we also used a transgenic plant expressing GFP under the control of a companion cell-specific AtSUC2 promoter (pSUC2::GFP; Benitez-Alfonso et al., 2013) to test if the symplasmic diffusion of GFP can be affected by SL pathway inhibitor-altered callose accumulation. The GFP fluorescence intensity at the root tip was observed 24 h after mock or inhibitor treatment. The GFP fluorescence intensities in seedlings treated with myriocin, FB1, PDMP, and D609 were significantly reduced compared with the GFP fluorescence intensity in seedlings subjected to the mock treatment. In contrast, DMS treatment resulted in enhanced PD permeability, as shown by the higher intensity of the GFP fluorescence (Fig. 1, G and H). Although we cannot exclude the possibility that some SL pathway inhibitors also altered pSUC2-mediated gene expression or protein synthesis, in general, GFP fluorescence intensities in the root meristems were inversely associated with callose deposition. We next tested whether the effects of SL inhibitor treatment on callose level and GFP movement were reversible. When SL inhibitor-treated seedlings were placed back into Murashige and Skoog (MS) control medium for 72 h, we observed no obvious differences between callose levels (Fig. 1, E and F) as well as GFP fluorescence intensities (Fig. 1, I and J) in mock condition and SL inhibitor-treated seedlings. These results strongly suggest that perturbation of SL metabolic pathways alters PD permeability by modulating callose accumulation.
GlcHCers Are Important for GPI-Anchored PD Protein Localization
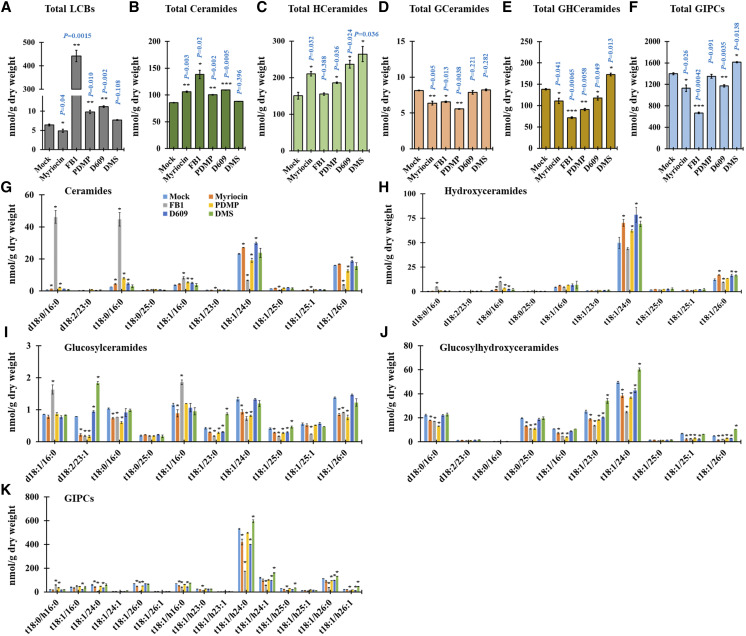
To explore the role of SL metabolites in the regulation of PD permeability, we performed SL profile analyses in 14-d-old Arabidopsis seedlings after a 24-h inhibitor treatment. After myriocin treatment, the total contents of SLs and individual contents of several SL molecules such as LCBs, GlcCers, GlcHCers, and GIPCs were strongly reduced (Fig. 2, A, D–F, and I–K). FB1-treated seedlings displayed reductions in the total contents of SLs and several SL molecules such as GlcCers, GlcHCers, and GIPCs (Fig. 2, D–F and I–K), but the total contents of LCBs and ceramides were significantly increased (Fig. 2, A and B). However, several ceramide molecules with very-long-chain fatty acids were significantly reduced; conversely, ceramide molecules with long-chain fatty acids such as d18:0/16:0, t18:1/16:0, and t18:1/16:0 were highly elevated (Fig. 2G). PDMP-treated seedlings resulted in the attenuation of total contents of GlcCers, GlcHCers, and GIPCs (Fig. 2, D–F), but the total contents of SLs and several SL molecules such as LCBs, ceramides, and hydroxyceramides were elevated (Fig. 2, A–C, G, and H). Moreover, after D609 treatment, the total contents of GIPCs and GlcHCers were significantly reduced (Fig. 2, E and F), but the total contents and several SL molecules of LCBs, ceramides, and hydroxyceramides were significantly elevated (Fig. 2, A–C, G, and H). Next, DMS-treated seedlings resulted in the enhancement of the total contents of hydroxyceramides, GlcHCers, and GIPCs without disturbing the other SL molecule levels (Fig. 2, C, E, and F). Based on the callose phenotype, four SL inhibitor treatments (myriocin, FB1, PDMP, and D609) showed callose accumulation enhancement, whereas DMS treatment showed the reverse result. When we observed the SL results, we found that myriocin-, FB1-, PDMP-, or D609-treated seedlings similarly resulted in the attenuation of GlcHCer level; conversely, the level of GlcHCers was elevated in the DMS treatment. Altogether, our results suggested that the GlcHCer level is important for the alteration of callose-mediated PD permeability.
Figure 2.
SL inhibitor treatment alters SL content. A to F, Measurement of SLs from wild-type plants after SL inhibitor treatment (24 h) included total LCBs (A), total ceramides (B), total hydroxyceramides (C), total GlcCer (D), total GlcHCer (E), and total GIPC (F). G to K, SL species characterized by LCB (d18:0, d18:1, d18:2, t18:0, and t18:1) and fatty acids (16:0–26:1) from wild-type plants after SL inhibitor treatment (24 h) included ceramides (G), hydroxyceramides (H), GlcCer (I), GlcHCer (J), and GIPC (K). Measurements are averages of four independent biological experiments (n = 200). Data are means ± sd. Statistical significance was determined by two-tailed Student’s t test (*P < 0.05, **P < 0.01, and ***P < 0.001).
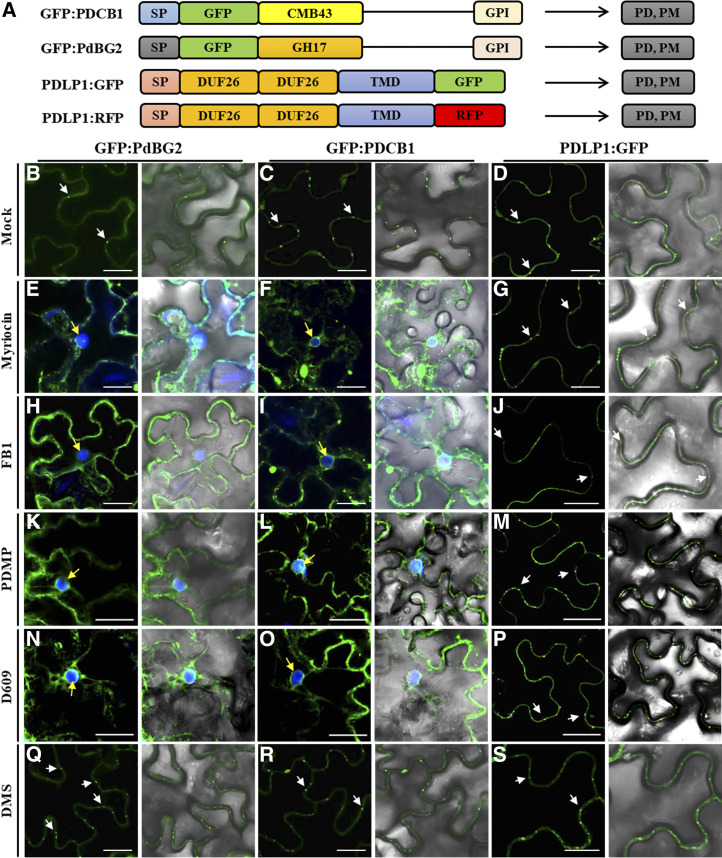
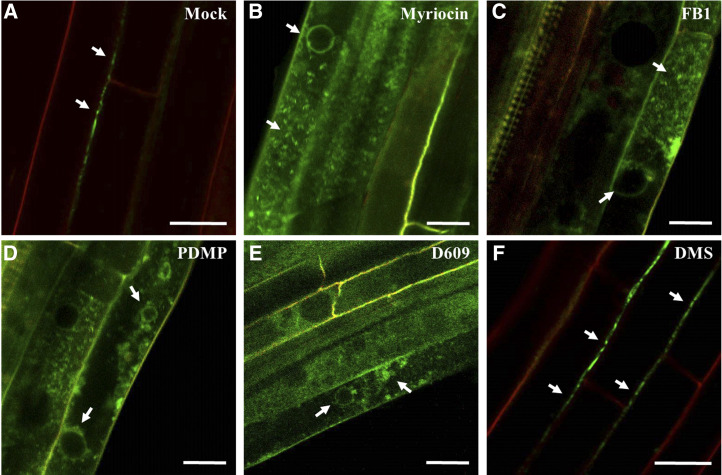
It was reported that GPI-anchored PdBG2 and PDCB1 proteins are mislocalized in the presence of fenpropimorph (Fen), a potential inhibitor of sterol metabolism that affects lipid raft organization (Grison et al., 2015). Thus, we investigated the subcellular localization of GPI-anchored PD proteins (PdBG2 and PDCB1) after SL inhibitor treatment in Nicotiana benthamiana leaves. We constructed several PD reporters for this study (Fig. 3A). First, we observed subcellular localization of GFP:PdBG2 and GFP:PDCB1 with PDLP1:RFP, and we found that sterol depletion due to Fen treatment changed the subcellular localization of GFP:PdBG2 and GFP:PDCB1 but not that of PDLP1:RFP (Supplemental Fig. S3). Next, we tested the localization of those PD reporters after SL inhibitor treatment. GPI-anchored PdBG2 and PDCB1 were mislocalized in the presence of myriocin, FB1, PDMP, and D609, whereas the localization of PDLP1 was not altered (Fig. 3). Interestingly, DMS treatment did not change the subcellular localization of any PD reporters (Fig. 3, Q–S). The β-1,3 glucanase PdBG2 has been shown to regulate callose accumulation; thus, next we asked whether the subcellular localization alteration of this GPI-anchored PdBG2 protein affected the callose level in the SL inhibitor-treated N. benthamiana leaves. Our callose analysis showed that the leaves treated with myriocin, FB1, PDMP, or D609 showed significantly increased callose accumulation; by contrast, DMS-treated leaves exhibited the attenuation of callose accumulation in comparison with mock treatment (Supplemental Fig. S4). In addition, we also generated transgenic Arabidopsis plants overexpressing GFP:PdBG2 and observed its subcellular localization after SL inhibitor treatment. Arabidopsis seedlings subjected to the mock or DMS treatment did not change GFP:PdBG2 subcellular localization; the GFP fluorescence pattern was punctated at PD-PM. Consistently, as shown in the transient expression of GFP:PdBG2, myriocin-, FB1-, PDMP-, or D609-treated seedlings showed mislocalization of GFP:PdBG2, the punctate PD-PM fluorescence pattern was abolished, and fluorescence was strongly detected at cytoplasm (Fig. 4). Our results suggest that the alteration of GPI-anchored PdBG2 localization is a possible reason for the reduced PD callose level after the treatment of a subset of SL pathway inhibitors, resulting in GlcHCer reduction.
Figure 3.
GPI-anchored PD proteins are mislocalized in the presence of SL inhibitors. A, Structural organization of GFP:PDCB1, GFP:PdBG2, and PDLP1:GFP. Chimeric constructs consist of the signal peptides (SP) of PDCB1 and PdBG2, followed by the coding sequence of GFP fused to the callose-binding domain (CBM-43) for PDCB1, GH17 for PdBG2, and C-terminal GPI anchor signals. For PDLP1, the chimeric construct consists of the signal peptide of PDLP1, followed by double DUF26 domain, single transmembrane domain (TMD), and the coding sequence of GFP. B to S, Confocal images of leaf epidermal cells of N. benthamiana expressing fluorescent fusion proteins of PdBG2, PDCB1, and PDLP1 after SL inhibitor treatment, as follows: mock (B–D), 0.1 μm myriocin (E–G), 5 μm FB1 (H–J), 50 μm PDMP (K–M), 40 μm D609 (N–P), and 30 μm DMS (Q–S). PD localization is indicated by white arrows. 4′,6-Diamino-phenylindole (DAPI) staining was used to determine nuclear localization (yellow arrows). Bars = 10 μm.
Figure 4.
Subcellular localization of PdBG2 after SL inhibitor treatment in Arabidopsis hypocotyls. Confocal images show Arabidopsis hypocotyls expressing fluorescent fusion protein of PdBG2 after SL inhibitor treatment (24 h) as follows: mock (A), 0.1 μm myriocin (B), 5 μm FB1 (C), 50 μm PDMP (D), 40 μm D609 (E), and 30 μm DMS (F). White arrows indicate GFP:PdBG2 localizations. Red signals show propidium iodide staining of the cell wall. Bars = 20 μm.
GlcHCers Are Associated with PD Permeability
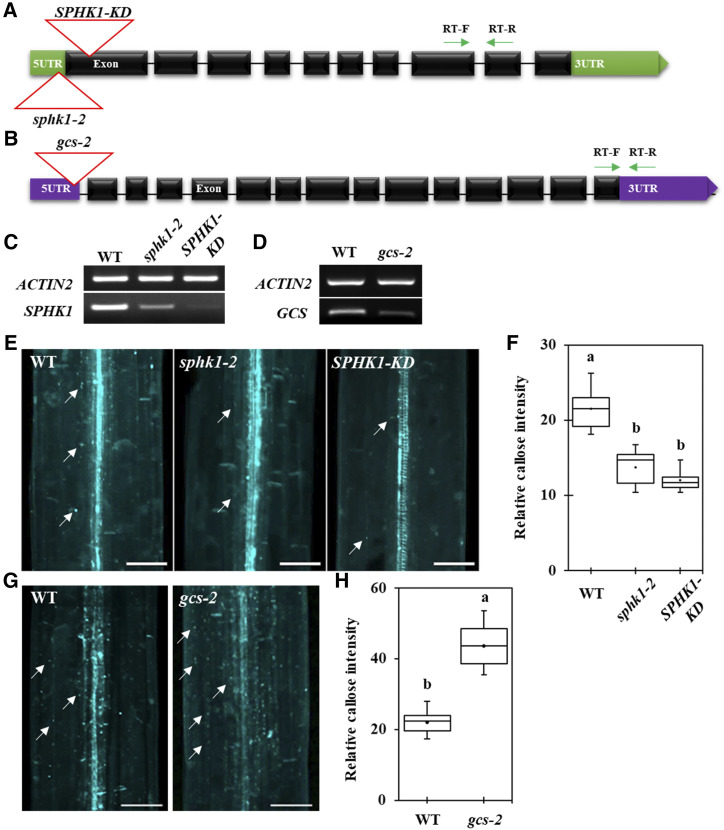
In plants, DMS inhibits SPHKs and PDMP inhibits GCS. In Arabidopsis, SPHK1 was reported to catalyze the formation of sphingosine-1-phosphate (S1P) through the phosphorylation of sphingosine, and GCS is responsible for the formation of GlcCers (Sperling and Heinz, 2003; Silva et al., 2007; Kajiwara et al., 2008; Raffaele et al., 2009; Loizides-Mangold et al., 2012). Based on the SL profiling results after SL inhibitor treatment, GlcHCers were significantly suppressed in the PDMP-treated seedling condition; however, DMS-treated seedlings firmly showed high GlcHCers in comparison with seedlings subjected to the mock condition. It has been reported that SPHK1 overexpression plants showed excess callose deposition at Arabidopsis rosette leaves, and it was restricted in the SPHK1-KD mutant plant upon the FB1-triggered cell death condition (Qin et al., 2017). Therefore, we employed sphk1 (sphk1-2 and SPHK1-KD; Fig. 5, A and C) and gcs mutants in the next experiment to test if the two mutants phenocopy the PD permeability and GlcHCer profiles of the plants treated with DMS and PDMP. The null gcs mutant in Arabidopsis showed seedling lethality, but the partial suppression of GCS with RNA interference resulted in viable and fertile plants (Msanne et al., 2015). We selected another weak mutant allele of the GCS gene, gcs-2, for which the T-DNA insertion site was predicted to be located in the promoter region, resulting in a slightly reduced transcription level (Fig. 5, B and D).
Figure 5.
Callose level is altered in the sphk1-2, SPHK1-KD, and gcs-2 mutants. A, Gene organization of SPHK1 and its T-DNA insertion alleles. Green arrows indicate RT primer locations for C. B, Gene organization of GCS and its T-DNA insertion allele. Green arrows indicate RT primer locations for D. C, SPHK1 transcript expression with alleles containing the T-DNA insertion. D, GCS transcript expression with alleles containing the T-DNA insertion. E, Confocal images show Aniline Blue-stained callose in Arabidopsis etiolated hypocotyls of different genotypes (the wild type [WT], sphk1-2, and SPHK1-KD). F, Quantitative data show the relative callose intensity of E. G, Confocal images show Aniline Blue-stained callose in Arabidopsis etiolated hypocotyls of different genotypes (the wild type and gcs-2). H, Quantitative data show the relative callose intensity of G. In E and G, white arrows show Aniline Blue-stained callose at PD. Bars = 80 μm. In F and H, statistical significance was determined by one-way ANOVA with the Tukey-Kramer test (n = 15, three independent biological experiments). Boxes shows 25th to 75th percentiles. Black dots and lines in boxes indicate the mean and median, respectively. Significant differences are indicated by the presence of different lowercase letters above each box plot (P < 0.05).
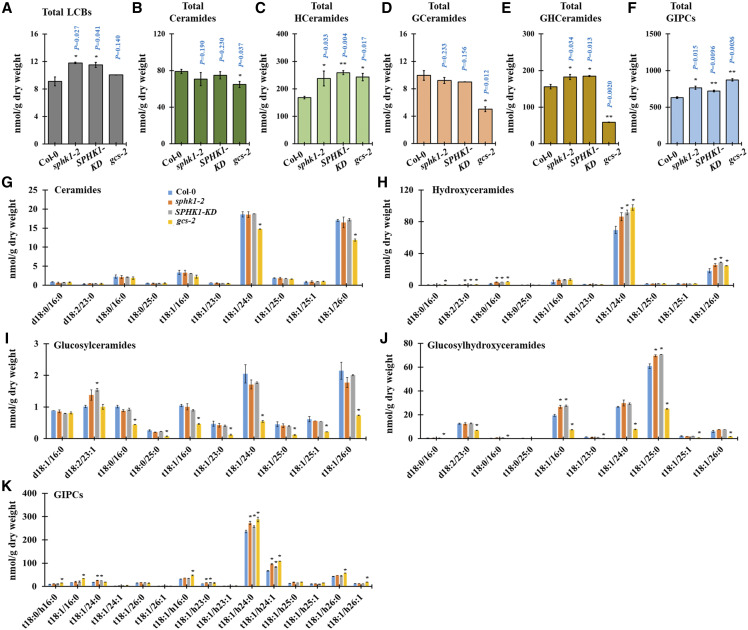
To confirm these hypotheses, we tested PD permeability in the sphk1-2, SPHK1-KD, and gcs-2 mutants by performing HPTS and Aniline Blue staining at Arabidopsis hypocotyls. The SPHK1-KD and sphk1-2 mutants displayed enhanced PD permeability, depicted by the strong HPTS movement and the reduction in callose deposition (Fig. 5, E and F; Supplemental Fig. S5). Conversely, the gcs-2 mutant displayed reduced PD permeability, depicted by the retardation of HPTS movement and the overaccumulation of callose deposition (Fig. 5, G and H; Supplemental Fig. S5). We next conducted SL profile analyses for the sphk1-2, SPHK1-KD, and gcs-2 mutants. The total contents and several molecules of LCBs, hydroxyceramides, GlcCers, GlcHCers, and GIPCs were up-regulated in the sphk1-2 and SPHK1-KD mutants (Fig. 6). On the other hand, the gcs-2 mutant showed no significant difference in the LCB level, but the proportions of hydroxyceramides and GIPCs were significantly enriched in comparison with the ecotype Colombia-0 (Col-0) plant. Then, we found a greater attenuation in the proportion of numerous molecules and total contents of GlcCers and GlcHCers (Fig. 6), indicating that the gcs-2 plant also impairs GlcHCer production. Our data showed that the callose deposition phenotype and the proportion of GlcHCers at PDMP- or DMS-treated seedlings were phenocopied by gcs-2 or sphk1 (sphk1-2 and SPHK1-KD) mutants, respectively. These data indicate that GlcHCers play the critical role in PD gating via the modulation of callose deposition. We also examined if a perturbation in SL pathways disrupts the gene expression of PdBG2. Myriocin-, FB1-, PDMP-, or D609-treated seedlings did not change the PdBG2 expression level as compared with mock treatment, whereas up-regulation of the PdBG2 transcript level was observed in the DMS-treated seedlings and sphk1 mutants (Supplemental Fig. S6). This suggests that DMS treatment or the sphk1 mutation might trigger another signaling pathway in addition to the GlcHCer-mediated one.
Figure 6.
SL homeostasis is altered in the sphk1-2, SPHK1-KD, and gcs-2 mutants. A to F, Measurement of SLs from sphk1-2, SPHK1-KD, and gcs-2 mutant plants included total LCBs (A), total ceramides (B), total hydroxyceramides (C), total GlcCer (D), total GlcHCer (E), and total GIPC (F). G to K, SL species characterized by LCB (d18:0, d18:1, d18:2, t18:0, and t18:1) and fatty acids (16:0–26:1) from sphk1-2, SPHK1-KD, and gcs-2 mutant plants included ceramides (G), hydroxyceramides (H), GlcCer (I), GlcHCer (J), and GIPC (K). Measurements are averages of four independent biological experiments (n = 200). Data are means ± sd. Statistical significance was determined by two-tailed Student’s t test (*P < 0.05 and **P < 0.01).
GlcHCers or Related SL Composition Are Required for the Targeting of GPI-Anchored PD Proteins
Since myriocin-, FB1-, PDMP-, and D609-suppressed GlcHCer levels only disrupted the subcellular localization of the GPI-anchored PD proteins and not that of PDLP1 protein, we hypothesized that the GPI-anchored PdBG2 and PDLP1 proteins might use different cargo machinery in a GlcHCer-enriched vesicle-dependent manner. The secretory pathways for GPI-anchored proteins and non-GPI-anchored transmembrane or secretory proteins are distinct in yeast (Saccharomyces cerevisiae) and mammalian cells (Funato and Riezman, 2001; Muñiz et al., 2001; Watanabe et al., 2008; Castillon et al., 2009; Rivier et al., 2010; Muñiz and Zurzolo, 2014; Paladino et al., 2014; Muñiz and Riezman, 2016). During protein secretion in yeast, GPI-anchored proteins are segregated from other proteins and delivered to their final destination by SL-enriched vesicles (Silva et al., 2007; Kajiwara et al., 2008; Loizides-Mangold et al., 2012). So far, there is no clear evidence that GPI-anchored proteins are sorted differently from other transmembrane proteins in plants.
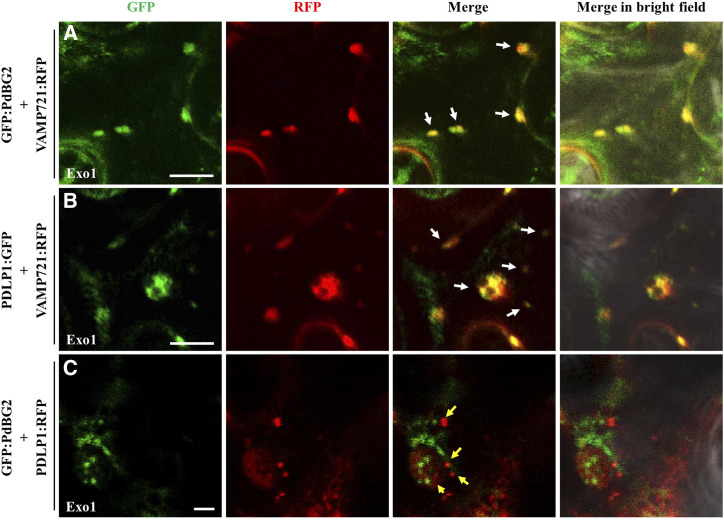
To address the question of whether PdBG2 and PDLP1 proteins are delivered by different pools of secretory vesicles, we used 2-(4-fluorobenzoylamino)-benzoic acid methyl ester (Exo1) to block membrane trafficking between the ER and the Golgi apparatus (Feng et al., 2003; Mishev et al., 2013). First, we examined the subcellular localization of GFP-PdBG2 and PDLP1-GFP after Exo1 treatment in N. benthamiana leaves. Cytosolic GFP signals surrounding the nucleus were detected after Exo1 treatment for all proteins (Supplemental Fig. S7), and the depicted GFP signals seem to be retained in the vesicle compartments. To confirm these results, we next generated a fusion protein of a well-known vesicle marker involved in the secretory pathway of the plant system: VAMP721 protein was selected (Caillaud et al., 2014; Karnahl et al., 2017). We first determined the subcellular localization of VAMP721 subjected to mock or Exo1 treatment in N. benthamiana leaves. We observed no obvious differences between VAMP721 subcellular localizations in Exo1-treated and control plants (Supplemental Fig. S8). To obtain further insight into the mechanisms controlling the secretion of PDLP1 and GPI-anchored PdBG2 proteins, we performed colocalization assays with three different combinations, GFP:PdBG2-VAMP72:RFP, PDLP1:GFP-VAMP721:RFP, and GFP:PdBG2-PDLP1:RFP, in the presence of Exo1. Interestingly, the cytosolic GFP signals from PdBG2 and PDLP1 were colocalized with VAMP721:RFP (Fig. 7, A and B), indicating that PDLP1 or PdBG2 is located at VAMP721-enriched vesicles. However, the cytosolic GFP signal from PdBG2 was not colocalized with RFP signal from PDLP1 (Fig. 7C), suggesting that VAMP721/PDLP1 and VAMP721/PdBG2 use different secretary vesicles, which presumably is attributed to the lipid raft and nonraft vesicles. Taken together, our results suggest that there are at least two different vesicles involved in PD protein delivery, and GPI-anchored PdBG2 may require a lipid raft-enriched vesicle that is associated with the GlcHCer composition. (Supplemental Fig. S9). Unfortunately, we could not test this hypothesis because we could not identify any published lipid marker protein localized in trafficking vesicles.
Figure 7.
PdBG2 and PDLP1 proteins are secreted with different cargo machinery. A, Confocal images show colocalization between GFP:PdBG2 and VAMP721:RFP after Exo1 (10 μm) treatment (12 h). B, Confocal images show colocalization between PDLP1:GFP and VAMP721:RFP after Exo1 (10 μm) treatment (12 h). In A and B, white arrows indicate that GFP and RFP signals were perfectly merged. C, Confocal images show different localization between GFP:PdBG2 and PDLP1:RFP after Exo1 (10 μm) treatment (12 h). Yellow arrows indicate that GFP and RFP signals were not colocalized. The fusion proteins were transiently expressed in N. benthamiana leaf epidermal cells. Bars = 10 µm (A and B) and 2 µm (C)
DISCUSSION
SL Pathways Control the Cellular Localization and Transcriptional Regulation of PdBG2
BGs are key players of the callose degradation event. There are 50 BG-related genes that have been identified in Arabidopsis (Doxey et al., 2007; Kumar et al., 2015). The biological functions of some BGs are tightly connected to the functions of the PD; loss of function in PD-localized BGs results in the reduction of the PD size exclusion limit caused by excessive callose accumulation in the PD (Doxey et al., 2007; Levy et al., 2007a, 2007b; Benitez-Alfonso et al., 2013). The most prominent and well studied of Arabidopsis BGs are AtBG_papp, PdBG1, and PdBG2. Among them, AtBG_papp and PdBG2 are GPI-anchored PD proteins that are attributed to the specialized domains called lipid rafts/microdomains at PD-PM (Levy et al., 2007b; Grison et al., 2015). The lipid raft is a unique platform composed by phospholipids, sterols, and SLs that is highly enriched in the PD-PM. Moreover, it was shown that PdBG2 protein, which attributes with lipid rafts at PD, is mislocalized and induced callose deposition upon sterol depletion (Mongrand et al., 2004; Borner et al., 2005; Grison et al., 2015; Iswanto and Kim, 2017), and this subcellular localization alteration was also observed in the N. benthamiana leaves transiently expressing GFP:PdBG2 after Fen treatment (Supplemental Fig. S3). Thus, our callose deposition data indicate that perturbation in the glucosyl SL composition, including GlcHCer level, is determined by the subcellular localization of PdBG2 protein. Although we could not detect any subcellular localization defect of DMS-treated seedlings, probably there is another molecular mechanism that contributes to the SL-mediated callose deposition.
To address this question, first we investigated the transcriptional activity of PdBG2 after SL inhibitor treatment and in the sphk1 (SPHK1-KD and sphk1-2) as well as gcs-2 mutants. Intriguingly, reverse transcription (RT)-PCR showed that myriocin-, FB1-, PDMP-, or D609-treated seedlings did not change the PdBG2 expression level as compared with mock treatment, whereas the gcs-2 mutant displayed a slight reduction of PdBG2 expression in comparison with the Col-0 plant. Moreover, up-regulation of the PdBG2 transcript level was observed in the DMS-treated seedlings and sphk1 mutants (Supplemental Fig. S6). The latter result implies that posttranslational cellular activity of PdBG2 protein is not solely responsible for regulating callose turnover upon GlcHCer fluctuation. Second, it is known that SPHK1 is employed to produce S1P (Supplemental Fig. S1). In plants, the SL metabolite S1P is involved in guard cell signal transduction (Ng et al., 2001; Coursol et al., 2003, 2005; Worrall et al., 2008; Puli et al., 2016). S1P in guard cells triggers alterations in nitric oxide, cytoplasmic calcium, and cytoplasmic pH (Ng et al., 2001; Puli et al., 2016). Reactive oxygen species and cytoplasmic calcium are well-known triggers of callose accumulation, probably through posttranslational regulation of callose synthases (Xu et al., 2017; Wu et al., 2018). Thus, a reduction of S1P by DMS treatment or in sphk1 mutants may accompany a diminution of callose synthase activity by posttranslational modification.
Interestingly, recent reports showed that PLASMODESMATA-LOCATED PROTEIN5 (PDLP5), a member of the plasmodesmal receptor-like protein family found in Arabidopsis, specifically binds to phytosphinganine (t18:0). PDLP5 is highly accumulated in the leaf epidermal cells of the sld1 sld2 double mutant, where trihydroxy LCB or phytosphinganine t18:0 is elevated. Similarly, PDLP5 overexpression plants and the sld1 sld2 double mutant show decreased PD permeability through callose accumulation (Chen et al., 2012; Wang et al., 2013; Liu et al., 2020; Vu et al., 2020). However, GlcCer proportions are strongly attenuated in the sld1 sld2 double mutant (Chen et al., 2012), which led us to speculate that GlcHCers might also be impaired. Consistent with our results, reductions in the GlcCer and GlcHCer compositions decrease PD permeability due to an enhancement in callose accumulation that is particularly caused by misregulation of the PdBG2 protein.
Secretory Machinery of PdBG2 Is Dependent on the GlcHCer Levels or Related SL Composition
This study demonstrates that there are different secretory mechanisms for GPI-anchored PdBG2 protein and non-GPI-anchored PDLP1 protein in plants (Fig. 7). The PD proteins harboring the GPI anchor in their C-terminal domain, such as PdBG2 and PDCB1, undergo lipid remodeling and GPI modification during protein sorting. The SLs are the LCBs, ceramides, hydroxyceramides, GlcCers, GlcHCers, and GIPCs, and along with sterols, the SLs serve as the basic materials for the formation of lipid rafts or microdomains. We wondered how two different types of PD proteins are differentially delivered to the secretory cargo machineries. In this study, we suggest that lipid raft-enriched vesicles are assumed to be recruited during the sorting of GPI-anchored PD proteins, but not for non-GPI-anchored PD proteins. Moreover, the attenuation of the more complex SLs such as GlcHCers caused by SL inhibitors (myriocin, FB1, PDMP, and D609) or the gcs mutation impaired the subcellular localization of GPI-anchored PdBG2 protein. This observation indicates that the sorting of GPI-anchored PdBG2 protein is dependent on GlcHCer levels or related SL composition.
Since GPI-anchored PD protein targeting is dependent on SL-enriched lipid rafts, the PDs might be hot spots in which GPI-anchored proteins are enriched. Indeed, many PD proteins have been revealed to be GPI anchored, including several BGs, the PDCB family members, REMORIN, PD-localized GRAIN SETTING DEFECT1, and LYSIN MOTIF DOMAIN-CONTAINING GLYCOSYL-PHOSPHATIDYL-INOSITOL-ANCHORED PROTEIN2 (Raffaele et al., 2009; Faulkner et al., 2013; Perraki et al., 2014). PDCB1, another GPI-anchored protein, physically binds to callose and is localized in the PDs. Plants overexpressing PDCB1 display enhanced callose accumulation and reduced PD permeability (Simpson et al., 2009). Not all GPI-anchored proteins are PD proteins. How GPI-anchored PD proteins and GPI-anchored non-PD proteins are differentially sorted remains to be solved. Our study demonstrated that GlcHCers or related SL composition play key roles in the formation of the lipid rafts that are attributed to the vesicle property and that are responsible for the PD targeting of GPI-anchored PdBG2 protein.
CONCLUSION
Overall, our study shows the role of SL molecules in the symplasmic continuity framework. One important finding is that GlcHCers are keys to the intercellular trafficking machinery of GPI-anchored PD proteins for regulating callose deposition and PD permeability. Our results, demonstrate that the subcellular localization of GlcHCer-modulated GPI-anchored PD proteins is dependent on lipid raft integrity. The appropriate function of Exo1 to block the secretory cargo transportation from ER to Golgi enabled us to show that there are at least two different cargo machineries between GPI-anchored PD proteins and non-GPI-anchored PD proteins, and it seems that lipid raft-enriched secretion vesicles are required for the transportation of GPI-anchored PD proteins but not for the transportation of non-GPI-anchored PD proteins (Supplemental Fig. S9).
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Col-0 and the following mutants were used: SPHK1-KD (Sail_794_B01; Worrall et al., 2008; Qin et al., 2017), sphk1-2 (Salk_042166), and gcs-2 (SK31705). The following constructs for the PD reporter were previously described: p35S::SP:GFP:PdBG2 (Simpson et al., 2009; Grison et al., 2015), p35S::SP:GFP:PDCB1 (Simpson et al., 2009; Grison et al., 2015), and p35S::PDLP1:GFP (Caillaud et al., 2014). For Gateway cloning, the genes of interest were amplified with Gateway-compatible primers (Supplemental Table S1). The PCR fragments were cloned into a donor vector (pDONR207) to generate entry clones. The resultant entry clones were subsequently transformed into Gateway binary vectors (pMDC83 and pH7RWG2.0). For plasmids cloned by flanking PCR, the genes of interest were amplified with gene-specific primers (Supplemental Table S1), then PCR fragments were cloned into donor vector pDONR207 and a destination vector (pEarlyGate 100) using a standard Gateway cloning system. The pSUC2::GFP construct was transformed into the Col-0 background. Seeds were grown on MS (Duchefa Biochemie) agar medium plates (pH 5.9 with KOH, left at 4°C for 2 d in darkness) and then grown in a 16-h-light/8-h-dark cycle (long-day conditions). Nicotiana benthamiana plants were grown at 24°C in long-day conditions. Plants were grown for 4 weeks before Agrobacterium tumefaciens-mediated transient transformation was performed. To generate stable expression in Arabidopsis, a floral dipping method (Clough and Bent, 1998) was used.
Inhibitor Treatments
For the PD permeability analyses (callose deposition and HPTS movement) using Arabidopsis hypocotyls, Col-0 seedlings were grown vertically on normal (without additional SL inhibitors) MS plates for 3 d in darkness. After 3 d in the dark condition, the seedlings were transferred to MS plates containing the following SL inhibitors: 0.1 µm myriocin (Santa Cruz), 5 µm FB1 (Santa Cruz), 50 µm PDMP (Santa Cruz), 100 µm D609 (Santa Cruz), and 30 µm DMS (Santa Cruz); they were left in the dark condition for 12 h prior to measurement of callose deposition and fluorescence signal intensity. For the PD permeability analyses (callose deposition and GFP fluorescence intensity) using Arabidopsis root tips, Col-0 and Col-0 overexpressing pSUC2::GFP seedlings were grown vertically on normal MS plates for 7 d in the light condition. After 7 d, the seedlings were transferred to MS plates containing SL inhibitors for 24 h prior to measurement of callose deposition and GFP signal intensity. Washout experiments were conducted by placing 24-h-treated seedlings onto MS agar plates subjected to the mock condition for another 72 h before confocal microscopy observation.
Transient Expression Analysis
N. benthamiana plants were grown at 24°C under long-day conditions. The plants were grown for 4 weeks before A. tumefaciens-mediated transient transformation was performed. Mature 4-week-old leaves were then transformed with the A. tumefaciens strain GV3101 containing the appropriate plasmids, and signals were observed 48 h after the transformation. Plasmolysis was performed by incubating the leaf samples in 1 m mannitol solution for 15 min. The PM was stained with 5 µm FM4-64 solution (Invitrogen Thermo Fisher Scientific) prior to plasmolysis. For the SL inhibitor treatments, 0.1 µm myriocin, 5 µm FB1, 50 µm PDMP, 100 µm D609, or 20 µm DMS were applied at 36 h after the A. tumefaciens transformation. For the nuclear staining, samples were stained with 8 µg mL−1 DAPI (Sigma) solution for 10 min.
PD Permeability Assessments
HPTS loading assays and Aniline Blue staining (Han et al., 2014; Kumar et al., 2016) were used to assess the permeability of the PD. GFP fluorescence intensity and callose signal intensity were measured using ImageJ software (https://imagej.nih.gov/ij/). For additional information on the quantification of callose using Aniline Blue staining, see Zavaliev et al. (2011, 2013). Data were analyzed by one-way ANOVA with the Tukey-Kramer test for statistical significance (Supplemental Table S2).
RNA Extraction, RT, and RT-PCR
Total RNA from Arabidopsis seedlings (Col-0, sphk1-2, SPHK1-KD, and gcs-2) was extracted using the RNeasy Plant Mini Kit (Qiagen), according to the manufacturer’s instructions. Aliquots of RNA (1 µg) from each sample were reverse transcribed to cDNA using the QuantiTect Reverse Transcription Kit (Qiagen), according to the manufacturer’s instructions. The first-strand cDNA was used as a template for quantitative PCR amplification. RT-PCR analyses were conducted using the gene-specific primer pairs for PdBG2 and ACTIN2 (Supplemental Table S1) under the following conditions: 95°C for 10 min followed by 40 cycles of 95°C for 10 s, 55°C for 30 s, and 72°C for 30 s. The expression levels of the different genes were normalized to that of ACTIN2. All experiments were repeated independently three times.
Confocal Microscopy
Confocal fluorescence microscopy was performed with an Olympus FV1000-LDPSU inverted confocal microscope using a 20×/0.8 oil-immersion objective or a 40×/1.3 oil-immersion objective. GFP was excited with a laser using a 488-nm beam splitter. RFP and FM4-64 were excited with a laser using a 543-nm beam splitter. Aniline Blue and DAPI were excited with a laser using a 405-nm beam splitter. Signal intensities from GFP (pSUC2:GFP) and Aniline Blue (callose detection) were quantified with ImageJ software for statistical data.
SL Extraction and SL Analysis
We identified and quantified a total of 115 SL molecular species based on their characteristic tandem mass spectrometry fragmentation patterns, including three LCBs, 23 ceramides, 30 hydroxyceramides, 30 GlcCers, and 29 GIPCs, from the Arabidopsis seedlings using liquid chromatography-tandem mass spectrometry (Supplemental Table S3). For plant sample preparation for SL inhibitor treatments, seeds (Col-0) were grown on normal MS medium for 14 d. At 10 d old, seedlings were transferred into MS liquid medium containing mock (dimethyl sulfoxide), 0.1 µm myriocin, 5 µm FB1, 50 µm PDMP, 100 µm D609, or 30 µm DMS and were grown for 24 h. After 24 h in SL inhibitor treatments, seedlings were immediately frozen in liquid nitrogen and ground to a fine powder (n = 200, four independent biological experiments). For plant sample preparation for mutant backgrounds (Col-0, sphk1-2, SPHK1-KD, and gcs-2), seeds were grown on normal MS medium for 14 d. Then, 14-d-old seedlings were frozen in liquid nitrogen and ground to a fine powder (n = 200, four independent biological experiments). For plant SL analysis, the total lipids were extracted from 3 mg of lyophilized Arabidopsis seedlings using the combined upper phase (220 μL) and lower phase (110 μL) of methyl-tert-butyl ether:methanol:water (100:30:35 [v/v/v]) described previously (Chen et al., 2013). Extracts were reconstituted in 100 μL of chloroform:methanol (1:9 [v/v]).
SL profiling was performed using a Nexera2 LC system (Shimadzu) connected to a triple quadrupole mass spectrometer (LC-MS 8040; Shimadzu) with a reverse-phase Kinetex C18 column (100 × 2.1 mm, 2.6 μm; Phenomenex) for chromatographic separations of lipids. Mobile phase A consisted of water:methanol (1:9, v/v) containing 10 mm ammonium acetate, and mobile phase B consisted of isopropanol:methanol (5:5, v/v) containing 10 mm ammonium acetate. To achieve chromatographic separation, a gradient elution program was optimized as follows: 0 min, 30% B; 0 to 15 min, 95% B; 15 to 20 min, 95% B; and 20 to 25 min, 30% B. The flow rate was 200 μL min−1. Sample volumes of 5 μL were injected for each run. To achieve SL quantifications, the calculated ratio of analyte and internal standard is multiplied by the concentration of the internal standard to obtain the concentration for each lipid species (Xia and Jemal, 2009; Buré et al., 2013; Lee et al., 2017; Im et al., 2019). Since there is no commercial internal standard for the quantification of GIPC molecular species, ganglioside GM1 is often used as an alternative to it (Markham and Jaworski, 2007; Tellier et al., 2014). We performed quantitative analysis of SLs using one-point calibrations of each target SL species (dihydrosphingosine d17:0/LCBs, non-hydroxy-phytoceramide [t18:0/8:0]/ceramide, α-hydroxy-phytoceramide [t18:0/h6:0]/hydroxyceramide, glucosylceramide [d18:1/12:0]/GlcCer or GlcHCer, and GM1 [d18:1/18:0]/GIPC with known concentration). Non-hydroxy-phytoceramide [(t18:0/8:0), molecular weight = 443.6] and α-hydroxy-phytoceramide [(t18:0/h6:0), molecular weight = 431.6] were synthesized by Kyungpook University, and other internal standards were purchased from Matreya or Avanti Polar Lipids. The distribution or absolute amount of SLs is very dynamic depending on the tissues or plant growth stages (Supplemental Table S4; Markham et al., 2006; Markham and Jaworski, 2007; Chao et al., 2011; Tellier et al., 2014; Msanne et al., 2015; Wu et al., 2015; Xie et al., 2015).
Accession Numbers
Sequence data for genes characterized in this article can be found at The Arabidopsis Information Resource (https://www.arabidopsis.org/) under accession numbers: PdBG2 (AT2G01630), PDCB1 (AT5G61130), PDLP1 (AT5G43980), SPHK1 (AT4G21540), and GCS (AT2G19880).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. SL biosynthesis pathways, including their potential inhibitors.
Supplemental Figure S2. HPTS movement analysis in etiolated Arabidopsis hypocotyls.
Supplemental Figure S3. GPI-anchored PdBG2 and PDCB1 proteins are mislocalized after Fen treatment.
Supplemental Figure S4. SL inhibitor treatment alters callose level in N. benthamiana leaves.
Supplemental Figure S5. HPTS movement analysis in the sphk1-2, SPHK1-KD, and gcs-2 mutants.
Supplemental Figure S6. Transcriptomic analysis of PdBG2 in the alteration of SL compositions.
Supplemental Figure S7. Exo1 inhibits secretory machinery of PdBG2 and PDLP1 proteins.
Supplemental Figure S8. Exo1 treatment does not change the cellular localization of VAMP721.
Supplemental Figure S9. Schematic model of the role of lipid rafts in the regulation of the PD.
Supplemental Table S1. Primers used in this study
Supplemental Table S2. Summary of statistical tests.
Supplemental Table S3. Retention times and selected reaction monitoring transitions of the Arabidopsis SLs identified.
Supplemental Table S4. Distribution of sphingolipids from various tissues of Arabidopsis.
Footnotes
This work was supported by the National Research Foundation of Korea (grant no. NRF–2018R1A2A1A05077295 to J.-Y.K.) and the Rural Development Administration, Republic of Korea (Systems and Synthetic Agrobiotech Center grant no. PJ01137901 to J.-Y.K. and the Program for New Plant Breeding Techniques grant no. PJ01478401).
References
- Adibhatla RM, Hatcher JF, Gusain A(2012) Tricyclodecan-9-yl-xanthogenate (D609) mechanism of actions: A mini-review of literature. Neurochem Res 37: 671–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Faulkner C, Pendle A, Miyashima S, Helariutta Y, Maule A(2013) Symplastic intercellular connectivity regulates lateral root patterning. Dev Cell 26: 136–147 [DOI] [PubMed] [Google Scholar]
- Bi FC, Liu Z, Wu JX, Liang H, Xi XL, Fang C, Sun TJ, Yin J, Dai GY, Rong C, et al. (2014) Loss of ceramide kinase in Arabidopsis impairs defenses and promotes ceramide accumulation and mitochondrial H2O2 bursts. Plant Cell 26: 3449–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, Napier JA, Beale MH, Lilley KS, Dupree P(2005) Analysis of detergent-resistant membranes in Arabidopsis: Evidence for plasma membrane lipid rafts. Plant Physiol 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buré C, Ayciriex S, Testet E, Schmitter JM(2013) A single run LC-MS/MS method for phospholipidomics. Anal Bioanal Chem 405: 203–213 [DOI] [PubMed] [Google Scholar]
- Caillaud MC, Wirthmueller L, Sklenar J, Findlay K, Piquerez SJ, Jones AM, Robatzek S, Jones JD, Faulkner C(2014) The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog 10: e1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon GA, Watanabe R, Taylor M, Schwabe TM, Riezman H(2009) Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic 10: 186–200 [DOI] [PubMed] [Google Scholar]
- Chao DY, Gable K, Chen M, Baxter I, Dietrich CR, Cahoon EB, Guerinot ML, Lahner B, Lü S, Markham JE, et al. (2011) Sphingolipids in the root play an important role in regulating the leaf ionome in Arabidopsis thaliana. Plant Cell 23: 1061–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Pan Q, Gao Y, Yang X, Wang S, Peppelenbosch MP, Kong X(2014) DMS triggers apoptosis associated with the inhibition of SPHK1/NF-κB activation and increase in intracellular Ca2+ concentration in human cancer cells. Int J Mol Med 33: 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Han G, Dietrich CR, Dunn TM, Cahoon EB(2006) The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18: 3576–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Markham JE, Cahoon EB(2012) Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J 69: 769–781 [DOI] [PubMed] [Google Scholar]
- Chen S, Hoene M, Li J, Li Y, Zhao X, Häring HU, Schleicher ED, Weigert C, Xu G, Lehmann R(2013) Simultaneous extraction of metabolome and lipidome with methyl tert-butyl ether from a single small tissue sample for ultra-high performance liquid chromatography/mass spectrometry. J Chromatogr A 1298: 9–16 [DOI] [PubMed] [Google Scholar]
- Chen XY, Kim JY(2009) Callose synthesis in higher plants. Plant Signal Behav 4: 489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coursol S, Fan LM, Le Stunff H, Spiegel S, Gilroy S, Assmann SM(2003) Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654 [DOI] [PubMed] [Google Scholar]
- Coursol S, Le Stunff H, Lynch DV, Gilroy S, Assmann SM, Spiegel S(2005) Arabidopsis sphingosine kinase and the effects of phytosphingosine-1-phosphate on stomatal aperture. Plant Physiol 137: 724–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado A, Casas J, Llebaria A, Abad JL, Fabrias G(2006) Inhibitors of sphingolipid metabolism enzymes. Biochim Biophys Acta 1758: 1957–1977 [DOI] [PubMed] [Google Scholar]
- Doxey AC, Yaish MW, Moffatt BA, Griffith M, McConkey BJ(2007) Functional divergence in the Arabidopsis beta-1,3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol Biol Evol 24: 1045–1055 [DOI] [PubMed] [Google Scholar]
- Edsall LC, Van Brocklyn JR, Cuvillier O, Kleuser B, Spiegel S(1998) N,N-Dimethylsphingosine is a potent competitive inhibitor of sphingosine kinase but not of protein kinase C: Modulation of cellular levels of sphingosine 1-phosphate and ceramide. Biochemistry 37: 12892–12898 [DOI] [PubMed] [Google Scholar]
- Falcone S, Perrotta C, De Palma C, Pisconti A, Sciorati C, Capobianco A, Rovere-Querini P, Manfredi AA, Clementi E(2004) Activation of acid sphingomyelinase and its inhibition by the nitric oxide/cyclic guanosine 3′,5′-monophosphate pathway: Key events in Escherichia coli-elicited apoptosis of dendritic cells. J Immunol 173: 4452–4463 [DOI] [PubMed] [Google Scholar]
- Fang L, Ishikawa T, Rennie EA, Murawska GM, Lao J, Yan J, Tsai AY, Baidoo EE, Xu J, Keasling JD, et al. (2016) Loss of inositol phosphorylceramide sphingolipid mannosylation induces plant immune responses and reduces cellulose content in Arabidopsis. Plant Cell 28: 2991–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ(2013) LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 110: 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Yu S, Lasell TK, Jadhav AP, Macia E, Chardin P, Melancon P, Roth M, Mitchison T, Kirchhausen T(2003) Exo1: A new chemical inhibitor of the exocytic pathway. Proc Natl Acad Sci USA 100: 6469–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K, Riezman H(2001) Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J Cell Biol 155: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grison MS, Brocard L, Fouillen L, Nicolas W, Wewer V, Dörmann P, Nacir H, Benitez-Alfonso Y, Claverol S, Germain V, et al. (2015) Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell 27: 1228–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka S, Hara M, Nishio H, Otsuka F, Suzuki A, Uchida Y(2002) Human epidermal glucosylceramides are major precursors of stratum corneum ceramides. J Invest Dermatol 119: 416–423 [DOI] [PubMed] [Google Scholar]
- Han X, Hyun TK, Zhang M, Kumar R, Koh EJ, Kang BH, Lucas WJ, Kim JY(2014) Auxin-callose-mediated plasmodesmal gating is essential for tropic auxin gradient formation and signaling. Dev Cell 28: 132–146 [DOI] [PubMed] [Google Scholar]
- Im SS, Park HY, Shon JC, Chung IS, Cho HC, Liu KH, Song DK(2019) Plasma sphingomyelins increase in pre-diabetic Korean men with abdominal obesity. PLoS ONE 14: e0213285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iswanto AB, Kim JY(2017) Lipid raft, regulator of plasmodesmal callose homeostasis. Plants (Basel) 6: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara K, Watanabe R, Pichler H, Ihara K, Murakami S, Riezman H, Funato K(2008) Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol Biol Cell 19: 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MS, Jung SY, Jung KM, Kim SK, Ahn KH, Kim DK(2008) D609, an inhibitor of phosphatidylcholine-specific phospholipase C, inhibits group IV cytosolic phospholipase A2. Mol Cells 26: 481–485 [PubMed] [Google Scholar]
- Karnahl M, Park M, Mayer U, Hiller U, Jürgens G(2017) ER assembly of SNARE complexes mediating formation of partitioning membrane in Arabidopsis cytokinesis. eLife 6: e25327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jung JH, Lee SB, Go YS, Kim HJ, Cahoon R, Markham JE, Cahoon EB, Suh MC(2013) Arabidopsis 3-ketoacyl-coenzyme A synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol 162: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T.(2015) Structural changes of GPI anchor after its attachment to proteins: Functional significance. Adv Exp Med Biol 842: 17–25 [DOI] [PubMed] [Google Scholar]
- Kumar R, Kumar D, Hyun TK, Kim JY(2015) Players at plasmodesmal nano-channels. J Plant Biol 58: 75–86 [Google Scholar]
- Kumar R, Wu SW, Iswanto AB, Kumar D, Han X, Kim JY(2016) A strategy to validate the role of callose-mediated plasmodesmal gating in the tropic response. J Vis Exp 53513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Mok HJ, Lee DY, Park SC, Kim GS, Lee SE, Lee YS, Kim KP, Kim HD(2017) UPLC-QqQ/MS-based lipidomics approach to characterize lipid alterations in inflammatory macrophages. J Proteome Res 16: 1460–1469 [DOI] [PubMed] [Google Scholar]
- Levy A, Erlanger M, Rosenthal M, Epel BL(2007a) A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J 49: 669–682 [DOI] [PubMed] [Google Scholar]
- Levy A, Guenoune-Gelbart D, Epel BL(2007b) Beta-1,3-glucanases: Plasmodesmal gate keepers for intercellular communication. Plant Signal Behav 2: 404–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu NJ, Zhang T, Liu ZH, Chen X, Guo HS, Ju BH, Zhang YY, Li GZ, Zhou QH, Qin YM, et al. (2020) Phytosphinganine affects plasmodesmata permeability via facilitating PDLP5-stimulated callose accumulation in Arabidopsis. Mol Plant 13: 128–143 [DOI] [PubMed] [Google Scholar]
- Loizides-Mangold U, David FP, Nesatyy VJ, Kinoshita T, Riezman H(2012) Glycosylphosphatidylinositol anchors regulate glycosphingolipid levels. J Lipid Res 53: 1522–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamode Cassim A, Gouguet P, Gronnier J, Laurent N, Germain V, Grison M, Boutté Y, Gerbeau-Pissot P, Simon-Plas F, Mongrand S(2019) Plant lipids: Key players of plasma membrane organization and function. Prog Lipid Res 73: 1–27 [DOI] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG(2007) Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 21: 1304–1314 [DOI] [PubMed] [Google Scholar]
- Markham JE, Li J, Cahoon EB, Jaworski JG(2006) Separation and identification of major plant sphingolipid classes from leaves. J Biol Chem 281: 22684–22694 [DOI] [PubMed] [Google Scholar]
- Markham JE, Molino D, Gissot L, Bellec Y, Hématy K, Marion J, Belcram K, Palauqui JC, Satiat-Jeunemaître B, Faure JD(2011) Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23: 2362–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maule AJ.(2008) Plasmodesmata: Structure, function and biogenesis. Curr Opin Plant Biol 11: 680–686 [DOI] [PubMed] [Google Scholar]
- Melser S, Batailler B, Peypelut M, Poujol C, Bellec Y, Wattelet-Boyer V, Maneta-Peyret L, Faure JD, Moreau P(2010) Glucosylceramide biosynthesis is involved in Golgi morphology and protein secretion in plant cells. Traffic 11: 479–490 [DOI] [PubMed] [Google Scholar]
- Michaelson LV, Napier JA, Molino D, Faure JD(2016) Plant sphingolipids: Their importance in cellular organization and adaption. Biochim Biophys Acta 1861: 1329–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishev K, Dejonghe W, Russinova E(2013) Small molecules for dissecting endomembrane trafficking: A cross-systems view. Chem Biol 20: 475–486 [DOI] [PubMed] [Google Scholar]
- Mongrand S, Morel J, Laroche J, Claverol S, Carde JP, Hartmann MA, Bonneu M, Simon-Plas F, Lessire R, Bessoule JJ(2004) Lipid rafts in higher plant cells: Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J Biol Chem 279: 36277–36286 [DOI] [PubMed] [Google Scholar]
- Mongrand S, Stanislas T, Bayer EM, Lherminier J, Simon-Plas F(2010) Membrane rafts in plant cells. Trends Plant Sci 15: 656–663 [DOI] [PubMed] [Google Scholar]
- Msanne J, Chen M, Luttgeharm KD, Bradley AM, Mays ES, Paper JM, Boyle DL, Cahoon RE, Schrick K, Cahoon EB(2015) Glucosylceramides are critical for cell-type differentiation and organogenesis, but not for cell viability in Arabidopsis. Plant J 84: 188–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz M, Morsomme P, Riezman H(2001) Protein sorting upon exit from the endoplasmic reticulum. Cell 104: 313–320 [DOI] [PubMed] [Google Scholar]
- Muñiz M, Riezman H(2016) Trafficking of glycosylphosphatidylinositol anchored proteins from the endoplasmic reticulum to the cell surface. J Lipid Res 57: 352–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñiz M, Zurzolo C(2014) Sorting of GPI-anchored proteins from yeast to mammals: Common pathways at different sites? J Cell Sci 127: 2793–2801 [DOI] [PubMed] [Google Scholar]
- Ng CK, Carr K, McAinsh MR, Powell B, Hetherington AM(2001) Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 410: 596–599 [DOI] [PubMed] [Google Scholar]
- Paladino S, Lebreton S, Tivodar S, Formiggini F, Ossato G, Gratton E, Tramier M, Coppey-Moisan M, Zurzolo C(2014) Golgi sorting regulates organization and activity of GPI proteins at apical membranes. Nat Chem Biol 10: 350–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraki A, Binaghi M, Mecchia MA, Gronnier J, German-Retana S, Mongrand S, Bayer E, Zelada AM, Germain V(2014) StRemorin1.3 hampers Potato virus X TGBp1 ability to increase plasmodesmata permeability, but does not interfere with its silencing suppressor activity. FEBS Lett 588: 1699–1705 [DOI] [PubMed] [Google Scholar]
- Puli MR, Rajsheel P, Aswani V, Agurla S, Kuchitsu K, Raghavendra AS(2016) Stomatal closure induced by phytosphingosine-1-phosphate and sphingosine-1-phosphate depends on nitric oxide and pH of guard cells in Pisum sativum. Planta 244: 831–841 [DOI] [PubMed] [Google Scholar]
- Qin X, Zhang RX, Ge S, Zhou T, Liang YK(2017) Sphingosine kinase AtSPHK1 functions in fumonisin B1-triggered cell death in Arabidopsis. Plant Physiol Biochem 119: 70–80 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Bayer E, Lafarge D, Cluzet S, German Retana S, Boubekeur T, Leborgne-Castel N, Carde JP, Lherminier J, Noirot E, et al. (2009) Remorin, a Solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier AS, Castillon GA, Michon L, Fukasawa M, Romanova-Michaelides M, Jaensch N, Hanada K, Watanabe R(2010) Exit of GPI-anchored proteins from the ER differs in yeast and mammalian cells. Traffic 11: 1017–1033 [DOI] [PubMed] [Google Scholar]
- Silva LC, de Almeida RF, Castro BM, Fedorov A, Prieto M(2007) Ceramide-domain formation and collapse in lipid rafts: Membrane reorganization by an apoptotic lipid. Biophys J 92: 502–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson C, Thomas C, Findlay K, Bayer E, Maule AJ(2009) An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AT, Radeff JM, Kunnel JG, Stern PH(2000) Phosphatidylcholine-specific phospholipase C inhibitor, tricyclodecan-9-yl xanthogenate (D609), increases phospholipase D-mediated phosphatidylcholine hydrolysis in UMR-106 osteoblastic osteosarcoma cells. Biochim Biophys Acta 1487: 201–208 [DOI] [PubMed] [Google Scholar]
- Sperling P, Heinz E(2003) Plant sphingolipids: Structural diversity, biosynthesis, first genes and functions. Biochim Biophys Acta 1632: 1–15 [DOI] [PubMed] [Google Scholar]
- Su HC, Ma CT, Lin CF, Wu HT, Chuang YH, Chen LJ, Tsao CW(2011) The acid sphingomyelinase inhibitors block interferon-α-induced serotonin uptake via a COX-2/Akt/ERK/STAT-dependent pathway in T cells. Int Immunopharmacol 11: 1823–1831 [DOI] [PubMed] [Google Scholar]
- Tellier F, Maia-Grondard A, Schmitz-Afonso I, Faure JD(2014) Comparative plant sphingolipidomic reveals specific lipids in seeds and oil. Phytochemistry 103: 50–58 [DOI] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K(1995) How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem 64: 563–591 [DOI] [PubMed] [Google Scholar]
- Verma DP, Hong Z(2001) Plant callose synthase complexes. Plant Mol Biol 47: 693–701 [DOI] [PubMed] [Google Scholar]
- Vu MH, Iswanto ABB, Lee J, Kim JY(2020) The role of plasmodesmata-associated receptor in plant development and environmental response. Plants (Basel) 9: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sager R, Cui W, Zhang C, Lu H, Lee JY(2013) Salicylic acid regulates plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell 25: 2315–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke D, Heinz E(2003) Recently discovered functions of glucosylceramides in plants and fungi. Cell Mol Life Sci 60: 919–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R, Castillon GA, Meury A, Riezman H(2008) The presence of an ER exit signal determines the protein sorting upon ER exit in yeast. Biochem J 414: 237–245 [DOI] [PubMed] [Google Scholar]
- Wattelet-Boyer V, Brocard L, Jonsson K, Esnay N, Joubès J, Domergue F, Mongrand S, Raikhel N, Bhalerao RP, Moreau P, et al. (2016) Enrichment of hydroxylated C24- and C26-acyl-chain sphingolipids mediates PIN2 apical sorting at trans-Golgi network subdomains. Nat Commun 7: 12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall D, Liang YK, Alvarez S, Holroyd GH, Spiegel S, Panagopulos M, Gray JE, Hetherington AM(2008) Involvement of sphingosine kinase in plant cell signalling. Plant J 56: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BS, Snow JW, O’Brien TC, Lynch DV(2003) Synthesis of 4-hydroxysphinganine and characterization of sphinganine hydroxylase activity in corn. Arch Biochem Biophys 415: 184–192 [DOI] [PubMed] [Google Scholar]
- Wu JX, Wu JL, Yin J, Zheng P, Yao N(2015) Ethylene modulates sphingolipid synthesis in Arabidopsis. Front Plant Sci 6: 1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SW, Kumar R, Iswanto ABB, Kim JY(2018) Callose balancing at plasmodesmata. J Exp Bot 69: 5325–5339 [DOI] [PubMed] [Google Scholar]
- Xia YQ, Jemal M(2009) Phospholipids in liquid chromatography/mass spectrometry bioanalysis: Comparison of three tandem mass spectrometric techniques for monitoring plasma phospholipids, the effect of mobile phase composition on phospholipids elution and the association of phospholipids with matrix effects. Rapid Commun Mass Spectrom 23: 2125–2138 [DOI] [PubMed] [Google Scholar]
- Xie LJ, Chen QF, Chen MX, Yu LJ, Huang L, Chen L, Wang FZ, Xia FN, Zhu TR, Wu JX, et al. (2015) Unsaturation of very-long-chain ceramides protects plant from hypoxia-induced damages by modulating ethylene signaling in Arabidopsis. PLoS Genet 11: e1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Cheval C, Laohavisit A, Hocking B, Chiasson D, Olsson TSG, Shirasu K, Faulkner C, Gilliham M(2017) A calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol 215: 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Yadav SR, Paterlini A, Nicolas WJ, Petit JD, Brocard L, Belevich I, Grison MS, Vaten A, Karami L, et al. (2019) Sphingolipid biosynthesis modulates plasmodesmal ultrastructure and phloem unloading. Nat Plants 5: 604–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Richter GL, Wang X, Młodzińska E, Carraro N, Ma G, Jenness M, Chao DY, Peer WA, Murphy AS(2013) Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J 74: 37–47 [DOI] [PubMed] [Google Scholar]
- Zambryski P, Crawford K(2000) Plasmodesmata: Gatekeepers for cell-to-cell transport of developmental signals in plants. Annu Rev Cell Dev Biol 16: 393–421 [DOI] [PubMed] [Google Scholar]
- Zavaliev R, Levy A, Gera A, Epel BL(2013) Subcellular dynamics and role of Arabidopsis β-1,3-glucanases in cell-to-cell movement of tobamoviruses. Mol Plant Microbe Interact 26: 1016–1030 [DOI] [PubMed] [Google Scholar]
- Zavaliev R, Ueki S, Epel BL, Citovsky V(2011) Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma 248: 117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]