Cryptochrome2 interacts with transcription factors TOE1 and TOE2 in a blue-light-dependent manner, thereby regulating CO and FT activity and photoperiodic flowering.
Abstract
Cryptochromes are photolyase-like, blue-light (BL) photoreceptors found in various organisms. Arabidopsis (Arabidopsis thaliana) cryptochromes (CRYs; CRY1, and CRY2) mediate many light responses including photoperiodic floral initiation. Cryptochromes interact with COP1 and SPA1, causing the stabilization of CONSTANS (CO) and promotion of FLOWERING LOCUS T (FT) transcription and flowering. The AP2-like transcriptional factor TOE1 negatively regulates FT expression and flowering by indirectly inhibiting CO transcriptional activation activity and directly binding to FT. Here, we demonstrate that CRY1 and CRY2 physically interact with TOE1 and TOE2 in a BL-dependent manner in flowering regulation. Genetic studies showed that mutation of TOE1 and TOE2 partially suppresses the late-flowering phenotype of cry1 cry2 mutant plants. BL-triggered interactions of CRY2 with TOE1 and TOE2 promote the dissociation of TOE1 and TOE2 from CO, resulting in alleviation of their inhibition of CO transcriptional activity and enhanced transcription of FT. Furthermore, we show that CRY2 represses TOE1 binding to the regulatory element within the Block E enhancer of FT. These results reveal that TOE1 and TOE2 act as downstream components of CRY2, thus partially mediating CRY2 regulation of photoperiodic flowering through modulation of CO activity and FT transcription.
Light is one of the most important environmental cues for plants. Not only does light provide photosynthetic energy for plants, but also different light qualities, quantities, directions, and periodicity guide plant physiological responses and developmental processes, including seed germination, photomorphogenesis, phototropism, flowering time, shade avoidance, and stomatal development and closure. Plants have evolved five different types of photoreceptors that perceive various wavelengths of light, which include the red-light (RL)/far-red-light (FRL) receptors phytochromes (Quail, 2002); the blue-light receptors cryptochromes (CRYs; Cashmore et al., 1999), phototropins (Briggs and Christie, 2002), and LOV/F-box/Kelch-domain proteins (Ito et al., 2012); and the UV-B photoreceptor UVR8 (Rizzini et al., 2011; Christie et al., 2012).
CRYs are conserved flavin-containing proteins that have been identified in various organisms, including bacteria, fruit flies (Drosophila melanogaster), plants, and mammals (Partch and Sancar, 2005). The Arabidopsis (Arabidopsis thaliana) genome encodes two homologous cryptochromes, CRY1 and CRY2, which primarily mediate photomorphogenesis under blue light (BL; Ahmad and Cashmore, 1993) and photoperiodic control of flowering (Guo et al., 1998), respectively. Both CRY1 and CRY2 have been shown to participate in major physiological processes of the plant, such as the circadian clock, BL induction of stomatal opening, guard cell development, phototropic curvature, and abiotic stress response (Somers et al., 1998; Mao et al., 2005; Kang et al., 2009; Consentino et al., 2015; D’Amico-Damião and Carvalho, 2018; Zhao et al., 2019). CRYs basically comprise a N-terminal photolyase homology-related domain (CNT) and a CRY-specific C-terminal extension domain (CCE, also known as CCT; Yang et al., 2000; Sang et al., 2005; Yu et al., 2010). The CNT domain is shown to mediate the homo-dimerization of CRYs (Sang et al., 2005; Yu et al., 2007b), and it has been demonstrated recently that the homo-dimerization of CRY2 is inhibited by Blue Light Inhibitors of Cryptochromes1 and 2 in a BL-dependent manner (Wang et al., 2016). The CCE/CCT domain is responsible for mediating CRY signaling by interacting with CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) and SUPPRESSOR OF PHYA-105 1 (Yang et al., 2000, 2001; Wang et al., 2001; Yu et al., 2007b; Lian et al., 2011; Liu et al., 2011). Recently, CNT1 has been shown to regulate hypocotyl elongation under BL independent of the CRY1 C terminus (CCE1/CCT1; He et al., 2015), which is mediated through inhibition of phytohormone signaling via the direct interactions of CNT1 with Aux/IAA proteins, AUXIN RESPONSE FACTORS, and BRI1-EMS-SUPPRESSOR1 (BES1; Wang et al., 2018b; Xu et al., 2018; Mao et al., 2020), and inhibition of the transcriptional activity of HBI1 via CNT1-HBI1 interaction (Wang et al., 2018a). Arabidopsis CRY2 accumulates exclusively in the nucleus, whereas CRY1 localizes to both nucleus and cytosol (Cashmore et al., 1999; Guo et al., 1999; Kleiner et al., 1999; Wu and Spalding, 2007). CRY2 undergoes BL-dependent phosphorylation, which is catalyzed by four closely related Photoregulatory Protein Kinases and two casein kinases (Shalitin et al., 2002; Tan et al., 2013; Liu et al., 2017). CRY1 can also be phosphorylated under BL (Shalitin et al., 2003; Ozgür and Sancar, 2006). Only phosphorylated CRY2 protein is rapidly degraded by the 26S proteasome system in response to BL, whereas CRY1 is stable (Ahmad et al., 1998; Lin et al., 1998; Yu et al., 2007a).
Flowering is the process by which plants transform from vegetative development to reproductive development. Proper regulation of flowering time is crucial for reproductive success. CRY2 is a major photoreceptor that positively regulates photoperiodic flowering in Arabidopsis (Guo et al., 1998), whereas CRY1 plays a minor role in promoting floral induction (Mockler et al., 1999; Liu et al., 2008b). The B box–type zinc finger transcriptional activator CONSTANS (CO) activates transcription of the florigen gene FLOWERING LOCUS T (FT) in leaves (Kardailsky et al., 1999; Onouchi et al., 2000; Samach et al., 2000). After its translation in leaves, FT protein migrates to the shoot apex where it activates the transcription of floral meristem identity genes and induces flowering (Corbesier et al., 2007). COP1 is a major negative regulator of both photomorphogenesis and floral initiation (Deng et al., 1992; McNellis et al., 1994). COP1 interacts with CO, thus promoting its ubiquitination and degradation (Jang et al., 2008; Liu et al., 2008b), and the interactions of CRY1 and CRY2 with COP1 and SPA1 may result in the repression of the COP1/SPA complexes, thereby stabilizing CO and causing FT transcription and induction of flowering (Wang et al., 2001; Yang et al., 2001; Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011). CRY2 binds directly to the bHLH transcription factor CIB1, promoting its transcriptional activity and FT transcription and thus accelerating floral initiation (Liu et al., 2008a). CIB1 physically interacts with CO and they form a complex with CRY2 that regulates FT expression (Liu et al., 2018). It has been recently reported that CRY2 cooperates with CIB1 to regulate FT transcription by enhancing the DNA affinity and transcriptional activity of CIB1 under BL (Yang et al., 2018). Another pivotal transcription factor family, B-box proteins also play critical roles in light-mediated flowering time control at the transcriptional and post-translational levels in plants (Song et al., 2020).
Members of the APETALA2 (AP2)-like gene family, including TARGET OF EAT1 (TOE1), TOE2, TOE3, SCHLAFMÜTZE (SMZ), SCHNARCHZAPFEN (SNZ), and AP2, encode a class of transcriptional repressors. Loss-of-function mutants of these genes show an early-flowering phenotype (Aukerman and Sakai, 2003; Mathieu et al., 2009; Yant et al., 2010), indicating that they negatively regulate flowering. These transcriptional repressors are the target genes of microRNA172 (miR172), and overexpression of miR172 leads to early flowering (Aukerman and Sakai, 2003; Yant et al., 2010). TOE1 binds to the promoter and downstream region of FT, directly suppressing FT expression (Zhai et al., 2015; Zhang et al., 2015), whereas TOE1 and TOE2 physically interact with CO and COL, inhibiting their transcriptional activities and indirectly repressing FT transcription (Zhang et al., 2015). Moreover, TOE1 interacts with FLAVIN-BINDING, KELCH REPEAT, F-BOX1, thus interfering with its interaction with CO, resulting in CO degradation (Zhang et al., 2015).
Here, we report that TOE1 and TOE2 are further CRY1- and CRY2-interacting proteins functioning in the regulation of photoperiodic flowering. Through a series of protein–protein interaction assays, we found that CRY1 and CRY2 physically interact with TOE1 and TOE2 in a BL-dependent manner. Genetic interaction studies indicated that TOE1 and TOE2 act partially downstream of CRY1 and CRY2 in the regulation of flowering time under long days (LDs). We demonstrate that BL-triggered interactions of CRY2 with TOE1 and TOE2 repress their interactions with CO, resulting in attenuation of TOE1 and TOE2 inhibition of CO transcriptional activity. Furthermore, CRY2 represses TOE1 binding to the 3′ downstream regulatory region of FT in Arabidopsis. Our results reveal new layers of the molecular mechanisms through which CRY2 regulates CO activity, FT transcription, and photoperiodic flowering through their physical interactions with TOE1 and TOE2.
RESULTS
CRY1 and CRY2 Physically Interact with TOEs in Yeast Cells and In Vitro
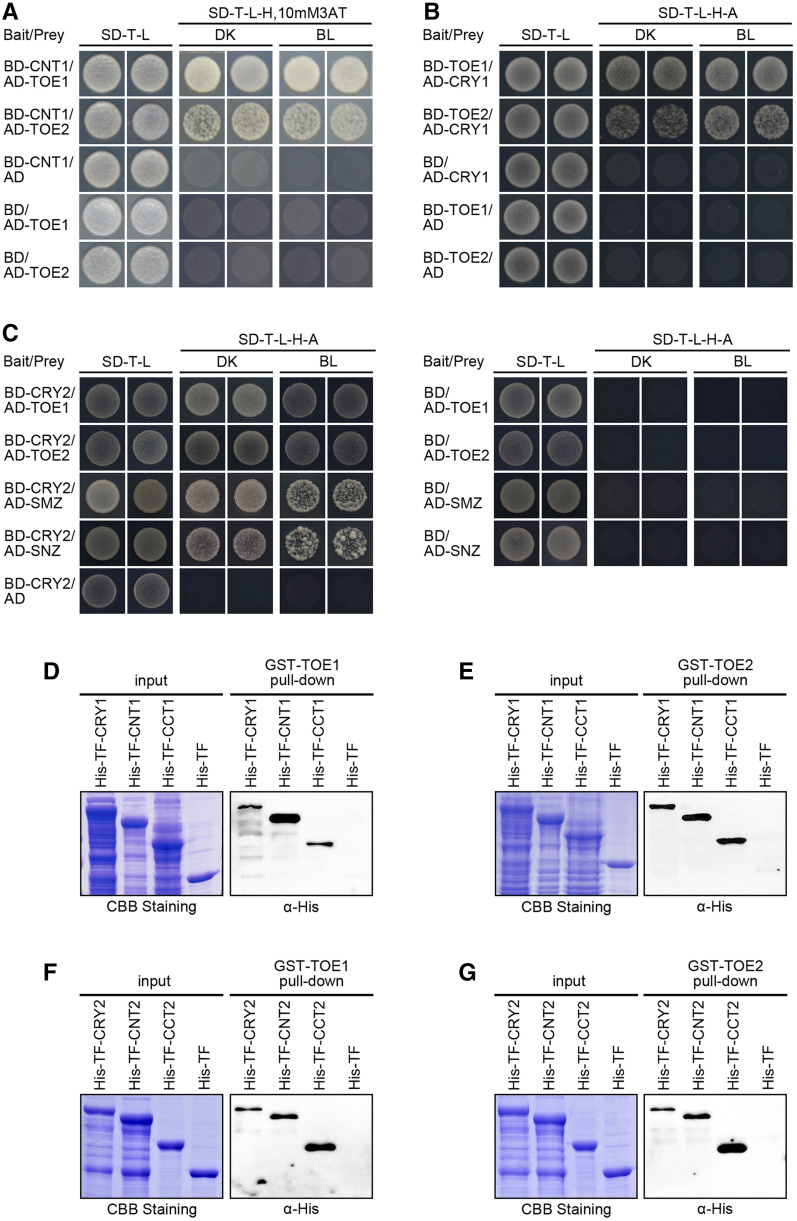
We reported previously that the N-terminal photolyase homology-related domain of CRY1 (CNT1) alone is able to mediate BL inhibition of hypocotyl elongation (He et al., 2015), and we have recently identified several CNT1-interacting proteins such as Aux/IAA proteins, BIM1 and BES1, HBI1, and AGB1 (Lian et al., 2018; Wang et al., 2018a; Wang et al., 2018b; Xu et al., 2018). Through GAL4 yeast two-hybrid screening using CNT1 as a bait, we identified TOE1 as a potential further CNT1-interacting protein (Fig. 1A). TOE1 is a member of AP2-like family of transcription factors that interacts with CO and inhibits its activation of FT to regulate flowering (Zhang et al., 2015). To determine whether the full-length CRY1 might interact with TOE1, we performed GAL4 yeast two-hybrid assays by cotransforming yeast (Saccharomyces cerevisiae) cells with a bait construct expressing the GAL4 binding domain (BD) fused to TOE1, together with a prey construct expressing the GAL4 transcriptional activation domain (AD) fused to CRY1. We found that CRY1 interacted with TOE1 in both darkness (DK) and BL (Fig. 1B). To explore whether CNT1 and CRY1 might interact with other AP2-like family members, we performed GAL4 yeast two-hybrid assays and found that both CNT1 and CRY1 interacted with TOE2 in both DK and BL in yeast cells (Fig. 1, A and B). Moreover, yeast two-hybrid assays showed that both CNT2 and CRY2 interacted with TOE1, TOE2, SMZ, and SNZ in both DK and BL (Fig. 1C; Supplemental Fig. S1).
Figure 1.
CRY1 and CRY2 physically interact with TOEs in yeast and in vitro. A, Yeast two-hybrid assays showing the interactions of CNT1 with TOE1 and TOE2. Yeast cells coexpressing the indicated combinations of constructs were grown on nonselective (SD-T-L) or selective media (SD-T-l-H) with 10 mm of 3AT in DK or BL (30 μmol m−2 s−1). “CNT1” denotes the CRY1 N terminus. B and C, Yeast two-hybrid assays showing the interactions of CRY1 with TOE1 and TOE2 (B), and CRY2 with TOE1, TOE2, SMZ, and SNZ (C). Yeast cells coexpressing the indicated combinations of constructs were grown on nonselective (SD-T-l) or selective media (SD-T-L-H-A) in DK or BL (30 μmol m−2 s−1). D to G, In vitro pull-down assays showing the interactions of CRY1, CNT1, and CCT1 with TOE1 (D) and TOE2 (E), and CRY2, CNT2, and CCT2 with TOE1 (F) and TOE2 (G). GST-TOE1 and GST-TOE2 served as bait. His-TF (as negative control), His-TF-CRY1, -CNT1, -CCT1, -CRY2, -CNT2, and -CCT2 served as preys. Input denotes Coomassie Brilliant Blue (CBB) staining. CCT1 denotes CRY1 C terminus. CNT2 and CCT2 denote CRY2 N- and C termini, respectively.
To further verify the interactions of CRY1 with TOE1 and TOE2, we performed pull-down assays using GST-tagged TOE1 or TOE2 protein as bait, and His-TF-tagged CRY1 proteins expressed in Escherichia coli as prey. As shown in Figure 1, D and E, CRY1 was pulled down by TOE1 and TOE2 proteins, whereas the control His-TF protein was not. To determine whether CNT1 or CCT1 might mediate the interactions of CRY1 with TOE1 and TOE2, we subsequently performed pull-down assays with His-TF-tagged CNT1 and CCT1 proteins expressed in E. coli. The results showed that both CNT1 and CCT1 were pulled down by TOE1 and TOE2 (Fig. 1, D and E). Consistent with these results, CRY2 and its N- and C-termini (CNT2 and CCT2) were pulled down by TOE1 and TOE2 (Fig. 1, F and G). Taken together, these results indicate that the full-length CRY1 and CRY2, via both their N- and C-termini, interact with TOE1 and TOE2 in vitro.
AP2-like family proteins contain two conserved AP2 domains with DNA-binding activity consisting of 60 to 70 amino acid residues (Weigel, 1995). To map the domains that might be required for mediating the interactions of TOE1 and TOE2 with CRY2, we made prey constructs expressing the various TOE1 and TOE2 fragments lacking either one or both AP2 domains or N- or C-terminal sequences (Supplemental Fig. S2, A and C) and performed yeast two-hybrid assays. The results showed that only the N-terminal TOE1 (amino acids 1–292) and TOE2 (amino acids 1–325) harboring both AP2 domains interacted with CRY2 (Supplemental Fig. S2, B and D), indicating that the entire N-terminal domain of TOE1 and TOE2 comprising both AP2 domains are essential for interaction of these proteins with CRY2.
CRY1 Physically Interacts with TOE1 and TOE2 in Plant Cells
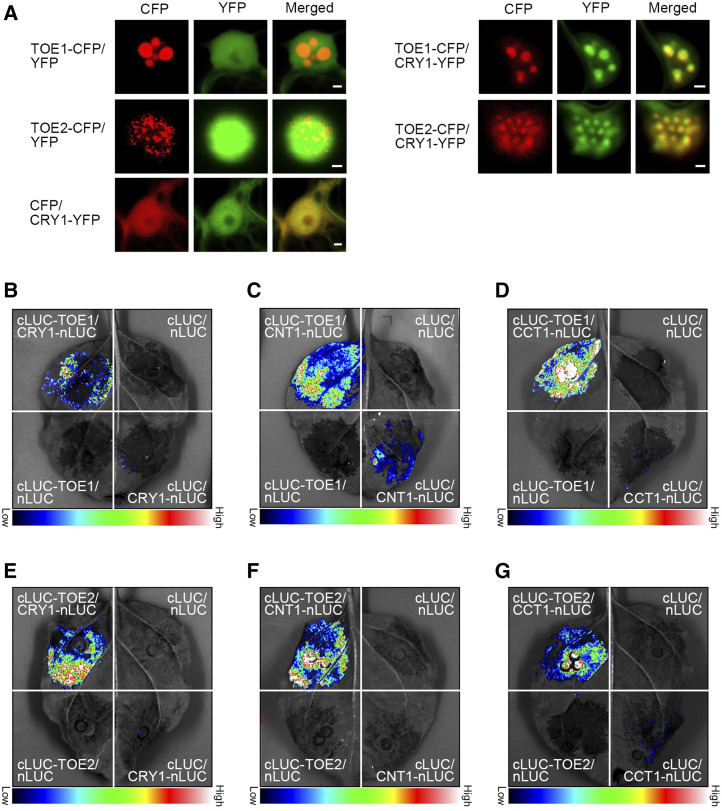
To examine whether CRY1 interacts with TOE1 and TOE2 in vivo, we first performed protein colocalization studies by transiently expressing CRY1 tagged with yellow fluorescent protein (YFP) together with TOE1 or TOE2 tagged with cyan fluorescent protein (CFP) in Nicotiana benthamiana leaves. As shown in Figure 2A, when expressed individually, TOE1-CFP and TOE2-CFP proteins were localized to the nuclear bodies (NBs) of N. benthamiana cells, whereas CRY1-YFP was not. However, when coexpressed with TOE1-CFP and TOE2-CFP, CRY1-YFP was localized to the same NBs of TOE1-CFP and TOE2-CFP (Fig. 2A), indicating interactions of CRY1 with TOE1 and TOE2 in plant cells. We then carried out bimolecular fluorescence complementation (BiFC) assays by coexpressing CRY1 or CNT1 or CCT1 tagged with the carboxyl-terminal half of YFP (cYFP; CRY1-cYFP, CNT1-cYFP, and CCT1-cYFP) and TOE1 or TOE2 tagged with the amino-terminal portion of YFP (nYFP; nYFP-TOE1 and nYFP-TOE2) in N. benthamiana cells. As shown in Supplemental Figure S3, YFP fluorescence signals were clearly observed when nYFP-TOE1 or nYFP-TOE2 was coexpressed with CRY1-cYFP or CNT1-cYFP or CCT1-cYFP, whereas no YFP signals were detected in N. benthamiana cells coexpressing nYFP-TOE1 or nYFP-TOE2 or CCT1-cYFP or CNT1-cYFP or CRY1-cYFP with the control proteins, indicating interactions of CRY1, CNT1, and CCT1 with TOE1 and TOE2 in plant cells.
Figure 2.
CRY1 physically interacts with TOE1 and TOE2 in plant cells. A, Protein colocalization assays indicating interactions of CRY1 with TOE1 and TOE2 in N. benthamiana cells. The constructs encoding TOE1 and TOE2 proteins fused to CFP and those encoding CRY1 proteins fused to YFP were cotransformed into N. benthamiana leaf epidermal cells, and TOE1 and TOE2 proteins were localized to the same NBs of CRY1 proteins. The images show overlays of fluorescence views. Scale bars = 2 μm. B to G, Split-LUC complementation imaging assays indicating the interactions of CRY1 (B), CNT1 (C), and CCT1 (D) with TOE1, and the interaction between CRY1 (E), CNT1 (F), and CCT1 (G) and TOE2 in N. benthamiana cells. Empty vectors were used as negative controls.
Next, we performed split luciferase complementation (split-LUC) assays in N. benthamiana leaves to confirm the interactions of CRY1 with TOE1 and TOE2, with CRY1 and TOE1 or TOE2 being fused to the N- and C-terminal halves of firefly luciferase (CRY1-nLUC and cLUC-TOE1 or cLUC-TOE2), respectively. As shown in Figure 2, B and E, the LUC activity was reconstituted when cLUC-TOE1 or cLUC-TOE2 and CRY1-nLUC were coexpressed in N. benthamiana leaves, whereas basically no LUC activities were detected when cLUC and CRY1-nLUC or cLUC-TOE1 and nLUC or cLUC-TOE2 and nLUC were coexpressed. These results demonstrate that CRY1 interacts with TOE1 and TOE2 in plant cells. Further split-LUC assays showed that both CNT1 and CCT1 interacted with TOE1 and TOE2 in N. benthamiana cells (Fig. 2, C–G).
CRY2 Physically Interacts with TOE1 and TOE2 in Plant Cells
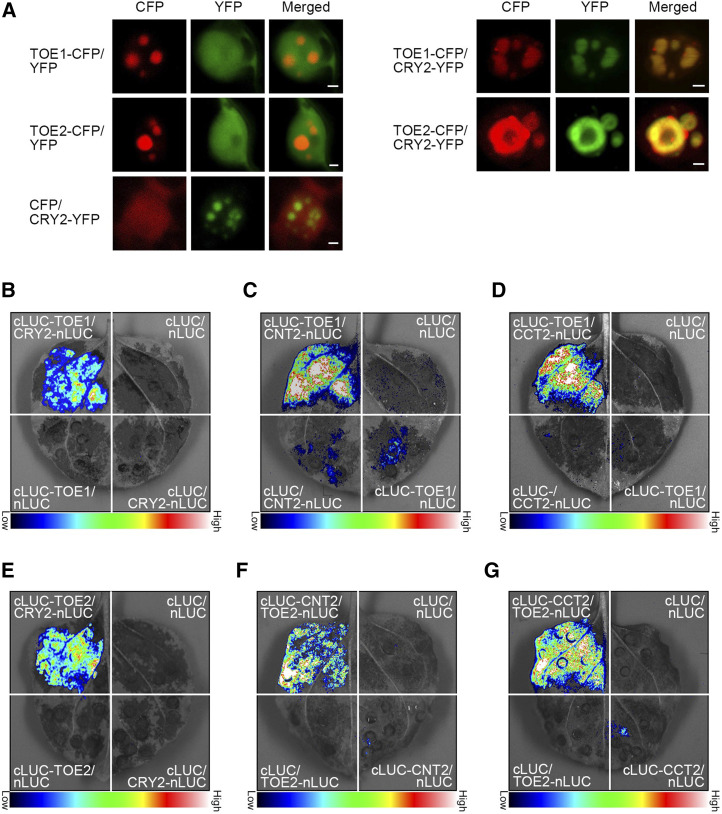
With the demonstration that CRY1 interacts with TOE1 and TOE2 in vivo, we then explored whether CRY2 also interacts with these two proteins in plant cells. First, protein colocalization assays indicated that CRY2 was colocalized with TOE1 and TOE2 in the same NBs of N. benthamiana cells (Fig. 3A). Second, BiFC assays indicated that CRY2, CNT2, and CCT2 interacted with TOE1 and TOE2 in N. benthamiana cells (Supplemental Fig. S4). Moreover, split-LUC assays demonstrated that the full-length CRY2, CNT2, and CCT2 interacted with TOE1 and TOE2 (Fig. 3, B–G). These results, in conjunction with those shown above (Figs. 1 and 2; Supplemental Fig. S3), demonstrate that both CRY1 and CRY2 interact with TOE1 and TOE2, and that both their N- and C-termini interact with TOE1 and TOE2.
Figure 3.
CRY2 physically interacts with TOE1 and TOE2 in plant cells. A, Protein colocalization assays indicating the interactions of CRY2 with TOE1 and TOE2 in N. benthamiana cells. The constructs encoding TOE1 and TOE2 proteins fused to CFP and those encoding CRY2 proteins fused to YFP were cotransformed into N. benthamiana leaf epidermal cells, and TOE1 and TOE2 proteins were localized to the same NBs of CRY2 proteins. The images show overlays of fluorescence views. Scale bars = 2 μm. B to D, Split-LUC complementation imaging assays indicating the interactions of CRY2 (B), CNT2 (C), and CCT2 (D) with TOE1 in N. benthamiana cells. Empty vectors were used as negative controls. E to G, Split-LUC complementation imaging assays indicating the interactions of CRY2 (E), CNT2 (F) and CCT2 (G) with TOE2 in N. benthamiana cells. Empty vectors were used as negative controls.
CRY1 and CRY2 Physically Interact with TOE1 and TOE2 in a BL-Dependent Manner In Planta
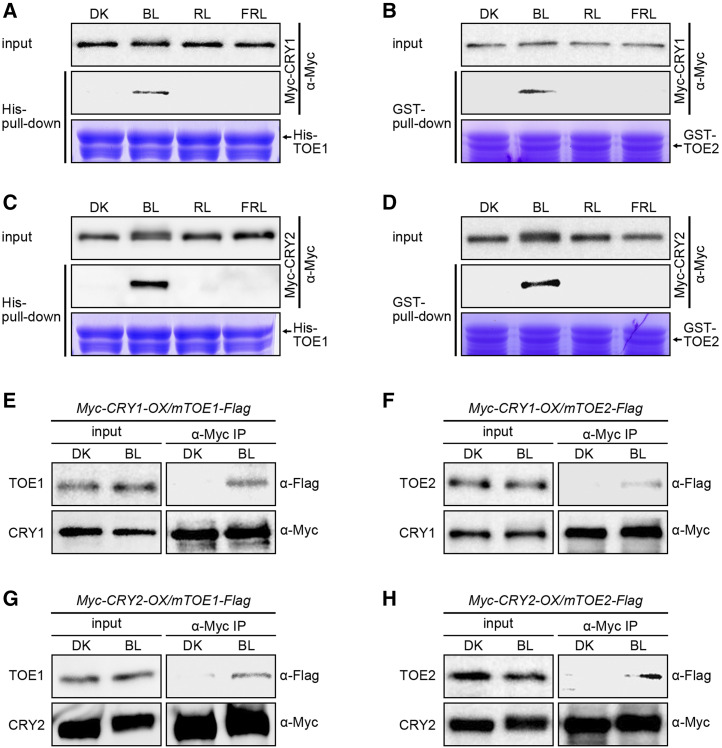
To investigate the effects of light quality on the interactions of CRY1 and CRY2 with TOE1 and TOE2, we performed semi-in vivo His-TOE1 and GST-TOE2 pull-down experiments using Myc-CRY1 and Myc-CRY2 proteins extracts from Myc-CRY1-OX and Myc-CRY2-OX seedlings adapted in DK or exposed to BL, RL, or FRL as preys. The results showed that both TOE1 and TOE2 pulled down CRY1 and CRY2 in the extracts prepared from the seedlings exposed to FL (Fig. 4, A–D), but not from those either adapted in DK or exposed to RL or FRL (Fig. 4, A–D), indicating that CRY1 and CRY2 interact with TOE1 and TOE2 in a BL-dependent manner.
Figure 4.
CRY1 and CRY2 physically interact with TOE1 and TOE2 in a BL-specific manner. A to D, Semi-in vivo pull-down assays showing BL-specific interactions of CRY1 with TOE1 (A) and TOE2 (B) and CRY2 with TOE1 (C) and TOE2 (D). His-TF-TOE1 served as bait in A and C, and GST-TOE2 served as bait in B and D. Myc-CRY1- and CRY2-containing protein extracts from 5-d-old etiolated Myc-CRY1-OX and Myc-CRY2-OX seedlings that were adapted to DK or exposed to BL (20 μmol m−2 s−1), RL (20 μmol m−2 s−1), or FRL (10 μmol m−2 s−1) for 1 h served as preys. The prey proteins pulled down by His-TF-TOE1 and GST-TOE2 were detected with anti-Myc antibody. The amount of His-TF-TOE1 and GST-TOE2 was shown in Coomassie Brilliant Blue staining gel. E to H, Co-IP assays showing BL-induced interaction of CRY1 with TOE1 (E) and TOE2 (F) and CRY2 with TOE1 (G) and TOE2 (H) in Arabidopsis. Five-day-old white-light-grown double transgenic seedlings coexpressing Myc-CRY1 and mTOE1-Flag (Myc-CRY1-OX/mTOE1-Flag) or Myc-CRY1 and mTOE2-Flag (Myc-CRY1-OX/mTOE2-Flag) or Myc-CRY2 and mTOE1-Flag (Myc-CRY2-OX/mTOE1-Flag) or Myc-CRY2 and mTOE2-Flag (Myc-CRY2-OX/mTOE2-Flag) were adapted in the dark for 3 d and then treated with 100 μm of MG132 for 4 h, and finally transferred back to DK or exposed to BL (30 μmol m−2 s−1) for 1 h. The IP (CRY1 and CRY2) and co-IP signals (TOE1 and TOE2) were detected by immunoblots probed with anti-Myc and -Flag antibodies. Transgenic Arabidopsis seedlings expressing the mutated TOE1 and TOE2 genes that comprise the mutated miR172 target sites but encode the same amino acid sequences as the wild-type TOE1 and TOE2 genes, which were fused to the DNA-fragment–encoding Flag tag under the control of the native promoters of TOE1 and TOE2 in wild-type background (mTOE1-Flag and mTOE2-Flag). We chose one line for each of the mTOE1-Flag and mTOE2-Flag transgenic Arabidopsis (mTOE1-Flag12 and mTOE2-Flag16) and genetically crossed with Myc-CRY1-OX or Myc-CRY2-OX plants to generate the double transgenic lines expressing both Myc-CRY1 or Myc-CRY2 and TOE1-Flag or TOE2-Flag (Myc-CRY1-OX/mTOE1-Flag, Myc-CRY2-OX/mTOE1-Flag, Myc-CRY1-OX/mTOE2-Flag, and Myc-CRY2-OX/mTOE2-Flag).
Next, we performed coimmunoprecipitation (co-IP) assays to further confirm whether CRY1 and CRY2 might interact with TOE1 and TOE2 in a BL-dependent manner in vivo. To do this, we first generated transgenic Arabidopsis seedlings expressing mutated TOE1 and TOE2 genes that comprise the mutated miR172 target sites but encode the same amino acid sequences as the wild-type TOE1 and TOE2 genes (Jung et al., 2007), which were fused to the DNA fragment encoding the Flag tag under the control of the native promoters of TOE1 and TOE2 in wild-type background (mTOE1-Flag and mTOE2-Flag). These plants showed very pronounced late-flowering phenotypes under LDs, and we chose one line for each of the mTOE1-Flag and mTOE2-Flag transgenes (mTOE1-Flag#12 and mTOE2-Flag#16) and genetically crossed these with Myc-CRY1-OX or Myc-CRY2-OX plants to generate the double transgenic lines expressing both Myc-CRY1 or Myc-CRY2 and TOE1-Flag or TOE2-Flag (Myc-CRY1-OX/mTOE1-Flag, Myc-CRY2-OX/mTOE1-Flag, Myc-CRY1-OX/mTOE2-Flag, and Myc-CRY2-OX/mTOE2-Flag). Co-IP assays with these double transgenic seedlings adapted in the dark or exposed to BL showed that both Myc-CRY1 and Myc-CRY2 coimmunoprecipitated with TOE1-Flag and TOE2-Flag in BL, but not in DK (Fig. 4, E–H), indicating that CRY1 and CRY2 interact with TOE1 and TOE2 in a BL-dependent manner in planta.
TOE1 and TOE2 Genetically Act Downstream of CRY1 and CRY2 in the Regulation of Flowering Time under LDs
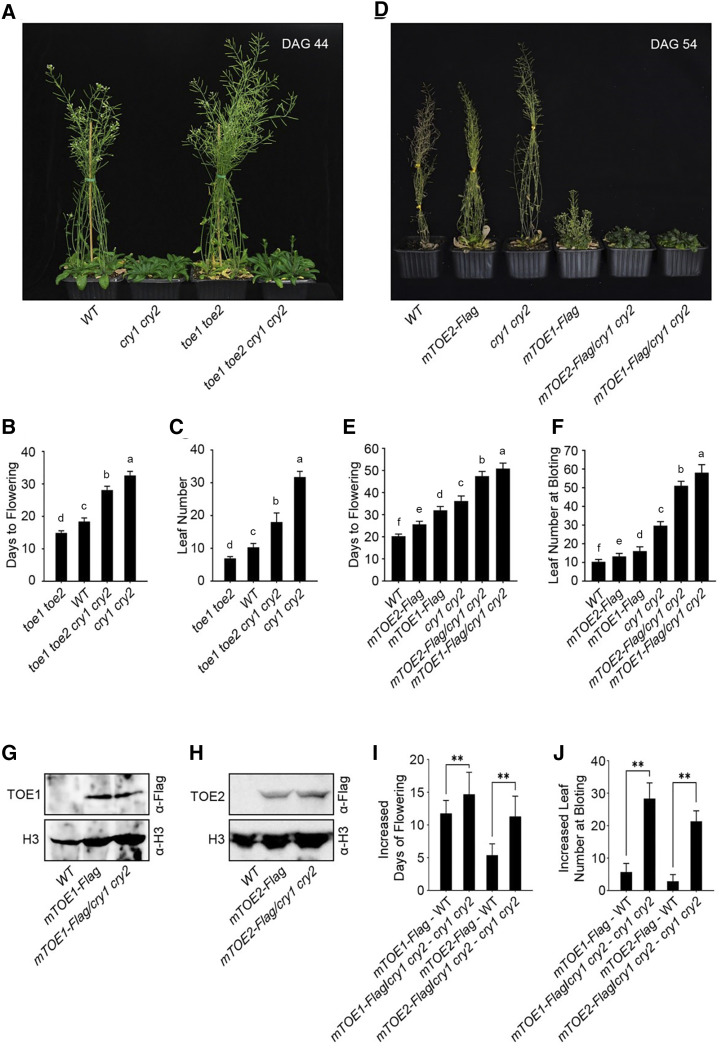
As CRY1 and CRY2 were shown to physically interact with TOE1 and TOE2, we explored whether CRY1 and CRY2 might genetically interact with TOE1 and TOE2 in the regulation of photoperiodic flowering. To do this, we generated a toe1 toe2 cry1 cry2 quadruple mutant by genetic crossing between toe1 toe2 and cry1 cry2 double mutants and analyzed the resulting flowering-time phenotype by measuring both the days to flowering and the number of rosette leaves at the time of bolting in plants grown in LDs illuminated by white light. The results showed that the toe1 toe2 cry1 cry2 quadruple mutant flowered significantly earlier than the cry1 cry2 double mutant, but significantly later than the toe1 toe2 double mutant (Fig. 5, A–C). We then performed reverse transcription quantitative PCR (RT-qPCR) assays to analyze the expression of FT and CO in 14-d-old plants at Zeitgeber time (ZT)16 under LDs. As shown in Supplemental Figure S5A, FT expression in the toe1 toe2 cry1 cry2 quadruple mutant was higher than in the cry1 cry2 double mutant, but lower than in the toe1 toe2 double mutant, which is consistent with the flowering phenotypes of these genotypes of plants. The expression of CO in wild type, the toe1 toe2 and cry1 cry2 double mutants, and the toe1 toe2 cry1 cry2 quadruple mutant was comparable (Supplemental Fig. S5B).
Figure 5.
TOE1 and TOE2 act partially downstream of CRY1 and CRY2 to regulate flowering under LDs. A, Wild type (WT), cry1 cry2, toe1 toe2, and cry1 cry2 toe1 toe2 mutant plants were photographed at 44 d after germination (DAG) under LD conditions (16-h-light/8-h-darkness). B and C, Flowering time of wild type, cry1 cry2, toe1 toe2, and cry1 cry2 toe1 toe2 plants as indicated by days to flowering under LDs (B) and the number of rosette leaves at bolting under LDs (C). Values shown are the mean ± sd of 30 individual plants. Lowercase letters indicate statistically significant differences between means for days to flowering (B) and means for the number of rosette leaves at bolting under LDs (C) of the indicated genotypes, as determined by a one-way ANOVA, followed by Fisher’s least significant difference (LSD) test (P < 0.05). D, Wild type, mTOE2-Flag, cry1 cry2, mTOE1-Flag, mTOE2-Flag/cry1 cry2, and mTOE1-Flag/cry1 cry2 plants were photographed at DAG 54 under LD conditions. E and F, Flowering time of wild type, mTOE2-Flag, cry1 cry2, mTOE1-Flag, mTOE2-Flag/cry1 cry2, and mTOE1-Flag/cry1 cry2 plants as indicated by days to flowering under LDs (E) and the number of rosette leaves at bolting under LDs (F). Values shown are the mean ± sd of 30 individual plants. Lowercase letters indicate statistically significant differences between means for days to flowering (E) and means for the number of rosette leaves at bolting under LDs (F) of the indicated genotypes, as determined by a one-way ANOVA, followed by Fisher's LSD test (P < 0.05). G and H, Western-blot analyses showing the expression level of TOE1-Flag in mTOE1-Flag and mTOE1-Flag/cry1 cry2 plants, and TOE2-Flag in mTOE2-Flag and mTOE2-Flag/cry1 cry2 plants. TOE1- and TOE2-Flag were detected by anti-Flag antibody. I, Comparison of increased days to flowering of mTOE1-Flag relative to wild type, mTOE2-Flag relative to WT, mTOE1-Flag/cry1 cry2 relative to cry1 cry2, and mTOE2-Flag/cry1 cry2 relative to cry1 cry2. These data were generated from E. J, Comparison of increased number of rosette leaves at bolting of mTOE1-Flag relative to WT, mTOE2-Flag relative to wild type, mTOE1-Flag/cry1 cry2 relative to cry1 cry2, and mTOE2-Flag/cry1 cry2 relative to cry1 cry2. These data were generated from F. Values shown are the mean ± sd of 30 individual plants. Asterisks indicate significant differences between different genotype plants by Student’s t test (**P < 0.01).
Given the result that CRYs interact with TOE1 and TOE2, and that CRYs have other downstream components such as COP1, SPAs, and CIBs (Wang et al., 2001; Liu et al., 2008a, 2008b, 2011; Lian et al., 2011; Zuo et al., 2011), these data indicate that TOE1 and TOE2 act partially downstream of CRY1 and CRY2 in the regulation of FT expression and flowering time in LDs.
Next, we genetically crossed mTOE1-Flag12 and mTOE2-Flag16 lines with the cry1 cry2 mutant to generate the plants expressing mTOE1-Flag and mTOE2-Flag in the cry1 cry2 mutant background (mTOE1-Flag/cry1 cry2 and mTOE2-Flag/cry1 cry2). Western-blot analysis indicated that TOE1-Flag and TOE2-Flag proteins were expressed at similar levels in wild-type and cry1 cry2 mutant backgrounds (Fig. 5, G and H). Flowering-time analysis in LDs showed that expression of TOE1-Flag and TOE2-Flag in cry1 cry2 mutant background led to more inhibitory effects on flowering than in wild-type background (Fig. 5, D–F). Specifically, the severity of the late-flowering phenotype measured by increases in both the days to flowering and the number of rosette leaves at bolting in mTOE1-Flag/cry1 cry2 and mTOE2-Flag/cry1 cry2 plants was significantly more pronounced than that in mTOE1-Flag and mTOE2-Flag plants (Fig. 5, I and J), indicating a role of CRY1 and CRY2 in the repression of TOE1 and TOE2 inhibition of flowering. These results further suggest that TOE1 and TOE2 function partially downstream of CRY1 and CRY2 in the regulation of flowering time under LDs.
CRY2 Promotes the Dissociation of TOE1 and TOE2 from CO
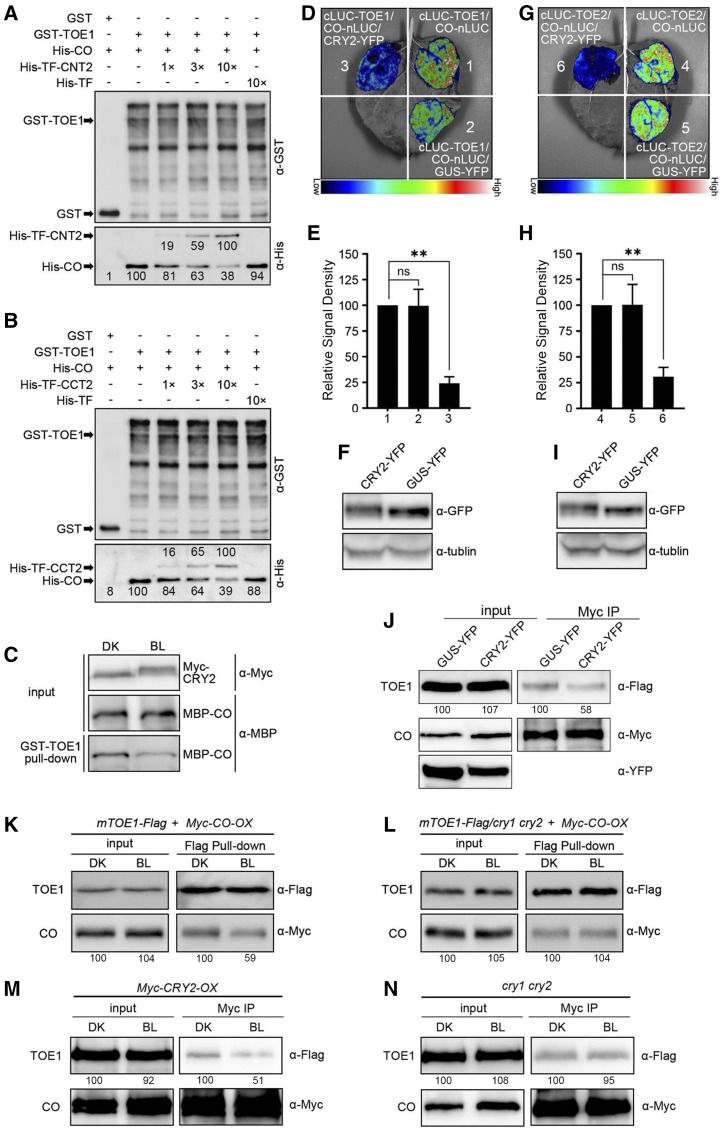
It has been reported that TOE1 interacts with CO and causes inhibited CO transcriptional activity (Zhang et al., 2015). Based on our results that CRYs interact with TOE1 and TOE2, and that TOE1 and TOE2 act partially downstream of CRY1 and CRY2 in the repression of flowering, we postulated that the interactions of CRY2 with TOE1 and TOE2 might interfere with the associations of TOE1 and TOE2 with CO, and promote CO transcriptional activity and FT expression. To test this hypothesis, we expressed and purified GST-TOE1, His-CO, His-TF-CNT2, and His-TF-CCT2 fusion proteins from E. coli and performed pull-down assays to evaluate the effects of increasing concentrations of His-TF-CNT2 or His-TF-CCT2 on the capacity of GST-TOE1 binding to His-CO. As expected, His-CO was clearly pulled down by GST-TOE1 (Fig. 6, A and B). Interestingly, the binding capacity of GST-TOE1 to His-CO was progressively reduced as the amounts of His-TF-CNT2 or His-TF-CCT2 increased, but was much less affected by the control protein His-TF (Fig. 6, A and B). As both CNT2 and CCT2 interact with TOE1 (Figs. 1F and 3, C and D; Supplemental Figs. S1 and S4), these results indicate that the interaction of the N- or C terminus of CRY2 with TOE1 inhibited the association of TOE1 with CO in vitro. We then performed pull-down assays to determine whether the TOE1-CO interaction might be affected by CRY2 using protein extracts prepared from BL-irradiated or DK-adapted Myc-CRY2-OX seedlings. As shown in Figure 6C, less CO protein was pulled down by TOE1 protein in the presence of the CRY2 protein extracts abstracted from Myc-CRY2-OX seedlings exposed to BL than from those adapted in the dark. Given the demonstration that CRY2 interacts with TOE1 in a BL-dependent manner in semi-in vivo pull-down assays (Fig. 4C), these results indicate that BL-induced interaction of CRY2 with TOE1 promotes the dissociation of TOE1 from CO.
Figure 6.
CRY2 represses the interactions of CO with TOE1 and TOE2 in vitro and in vivo. A and B, GST pull-down assays showing repression of the association of CO with TOE1 by CNT2 (A) and CCT2 (B). GST-TOE1 served as bait. His-TF served as negative control. The relative band intensity was normalized to the indicated sample for each image and shown near each band. C, Semi-in vivo pull-down assays showing repression of the association of CO with TOE1 by CRY2 in a BL-specific manner. GST-TOE1 served as bait. Myc-CRY2 protein extracted from Myc-CRY2-OX seedlings that were grown for 5 d in white light and then transferred to the dark for 3 d. Seedlings were then initially treated with 100 μm of MG132 for 4 h and finally transferred into DK or exposed to BL (20 μmol m−2 s−1) and MBP-CO served as preys. The prey proteins pulled down by GST-TOE1 were detected with anti-Myc antibody and anti-MBP antibody. D to F, Split-LUC complementation imaging assays indicating CRY2 represses the interaction of CO with TOE1 in N. benthamiana cells. Lower luminescence intensity was observed after CRY2-YFP cotransformed into N. benthamiana leaf epidermal cells compared with the control plants, which were cotransformed GUS-YFP (D). E, The luminescence intensity was quantitatively analyzed. Data are represented as the mean of biological triplicates ± sd (n = 4). Asterisks indicated significant difference by Student’s t test (**P < 0.01). F, The expression of CRY2-YFP and GUS-YFP was detected by anti-GFP antibody. G to I, Split-LUC complementation imaging assays indicating CRY2 represses the interaction of CO with TOE2 in N. benthamiana cells. Lower luminescence intensity was observed after CRY2-YFP cotransformed into N. benthamiana leaf epidermal cells compared with the control plants that were cotransformed GUS-YFP (G). H, The luminescence intensity was quantitatively analyzed. Data are represented as the mean of biological triplicates ± sd (n = 4). Asterisks indicated significant difference by Student’s t test (**P < 0.01). I, The expression of CRY2-YFP and GUS-YFP was detected by anti-GFP antibody. J, Co-IP assays showing that CRY2 represses the interaction of CO with TOE1 in N. benthamiana. TOE1-Flag, Myc-CO, and GUS-YFP or CRY2-YFP were coexpressed in N. benthamiana leaves. Myc-CO served as bait, and TOE1-Flag served as prey. GUS-YFP and CRY2-YFP were detected with anti-GFP antibody. The assays were repeated three times with similar results. K to l, Semi in vivo pull-down assays showing CRY2 inhibition of the interaction of TOE1 with CO (K). TOE1-Flag protein extracts served as bait, which were prepared from mTOE1-Flag and mTOE1-Flag/cry1 cry2 seedlings grown for 5 d in white light and then transferred to the dark for 3 d, treated with 100 μm of MG132 for 4 h, and adapted to DK or exposed to BL (30 μmol m−2 s−1). Myc-CO protein extracts from BL-adapted Myc-CO-OX seedlings served as prey. The assays were repeated three times with similar results. M and N, Co-IP assays showing CRY2 inhibition of the interaction of TOE1 with CO (M) in Arabidopsis protoplasts. TOE1-Flag and Myc-CO were coexpressed in Myc-CRY2-OX and cry1 cry2 protoplasts. The transformed protoplasts were exposed to BL (30 μmol m−2 s−1) for 16 h and then treated with 100 μm of MG132 for 1 h, and adapted to DK or exposed to BL for 2 h. TOE1-Flag served as prey and Myc-CO served as bait. The assays were repeated twice with similar results. The relative band intensity was normalized to the sample adapted to DK for each image and shown below each lane (J–N).
Next, we performed split-LUC assays to verify the capacities of the respective interactions of TOE1 and TOE2 with CO in the absence and the presence of CRY2 in N. benthamiana. As anticipated, TOE1 and TOE2 interacted with CO strongly in the absence of the third protein or in the presence of the control protein GUS (Fig. 6, D, E, G, and H; Supplemental Fig. S6, A and B). Interestingly, TOE1 and TOE2 interacted with CO to a lesser extent in the presence of CRY2 than in the presence of GUS. Western-blot analyses indicated that CRY2 and GUS proteins were expressed at similar levels in these assays (Fig. 6, F and I). Furthermore, we performed co-IP assay using the transient expression system of N. benthamiana. TOE1-Flag, Myc-CO, and GUS-YFP or CRY2-YFP proteins were coexpressed in N. benthamiana leaves. As shown in Figure 6J, CRY2-YFP significantly repressed the interaction of TOE1 with CO in N. benthamiana cells compared with the GUS-YFP control.
To further explore whether CRY2-TOE1 interaction might interfere with TOE1-CO interaction in Arabidopsis, we first performed semi-in vivo pull-down assays with the protein extracts prepared from Myc-CO-overexpressing (Myc-CO-OX) seedlings, and mTOE1-Flag and mTOE1-Flag/cry1 cry2 seedlings either BL-exposed or DK-adapted. The results showed that less CO was pulled down by TOE1 protein extracted from mTOE1-Flag BL-exposed seedlings than that from DK-adapted seedlings (Fig. 6K), whereas a similar amount of CO protein was pulled down by TOE1 protein extracts from mTOE1-Flag/cry1 cry2 BL-exposed and DK-adapted seedlings (Fig. 6L). We then performed co-IP assays with Myc-CRY2-OX and cry1 cry2 protoplasts coexpressing TOE1-Flag and Myc-CO that were DK-adapted or exposed to BL. The results showed that more TOE1-Flag was coimmunoprecipitated with Myc-CO in the Myc-CRY2-OX DK-adapted protoplasts than that from BL-exposed protoplasts (Fig. 6M), whereas a similar amount of TOE1-Flag protein was coimmunoprecipitated in cry1 cry2 protoplasts (Fig. 6N). Taken together, these results indicate that BL-induced interaction of CRY2 with TOE1 promotes the dissociation of TOE1 from CO in plants.
CRY2 Inhibits TOE1 and TOE2 Repression of CO Transcriptional Activation Activity
To examine whether CRY2 might regulate the transcriptional repression activity of TOE1 and TOE2 on CO, we performed Dual-Luciferase (Dual-LUC) assays with the reporter construct expressing LUC under the control of the 2.1-kb promoter of FT, FTpro:LUC (Supplemental Fig. S7A). The effector constructs expressing CO-YFP and/or TOE1-CFP or TOE2-CFP and/or CRY2-YFP were transiently coexpressed in N. benthamiana leaf cells with the reporter construct in different combinations. The results showed that, consistent with CO being a direct positive regulator of FT (Kardailsky et al., 1999; Onouchi et al., 2000; Samach et al., 2000), CO alone strongly activated the FTpro:LUC reporter activity (Supplemental Fig. S7, B and C). When CRY2-YFP was coexpressed with CO-YFP, FTpro activity was basically unaffected. However, when TOE1-CFP or TOE2-CFP was coexpressed with CO-YFP, FTpro activity was severely inhibited, confirming negative regulation of CO transcriptional activation activity by TOE1 and TOE2. When CRY2-YFP was coexpressed with TOE1-CFP or TOE2-CFP, the inhibition of CO transcriptional activation activity by TOE1 or TOE2 was clearly alleviated (Supplemental Fig. S7, B and C). By contrast, TOE1 or TOE2 repression of the transcriptional activation activity of CO was not affected by the control protein YFP. These results suggest that CRY2 inhibits TOE1 and TOE2 repression of CO transcriptional activation activity.
CRY2 Inhibits TOE1 Binding to the 3′ Downstream Regulatory Region of FT
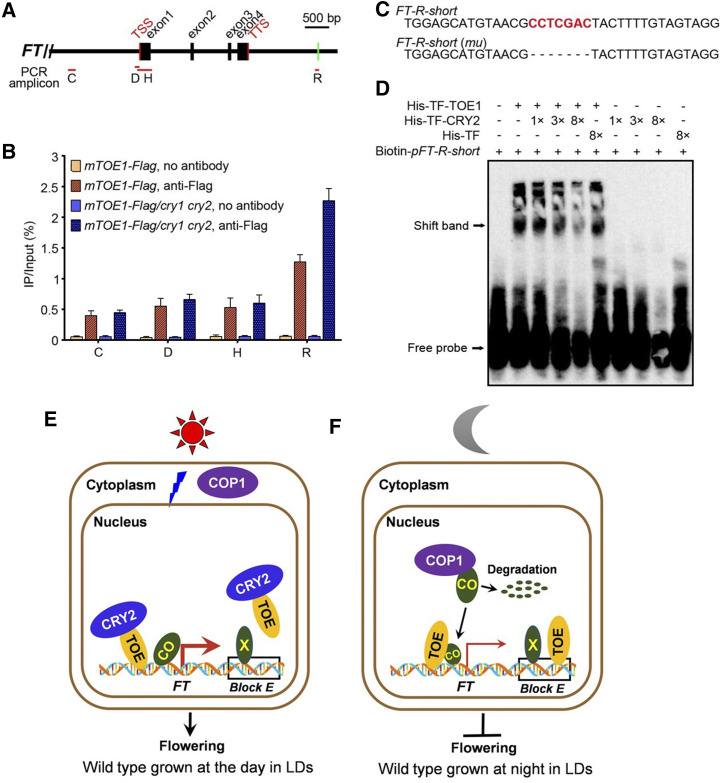
Given that TOE1 binds directly to the promoter and the 3ʹ downstream regulatory region of FT (Zhai et al., 2015; Zhang et al., 2015), we asked whether the CRY2-TOE1 interaction might affect TOE1 DNA-binding activity. To test this possibility, we first performed chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) assays using wild-type and mTOE1-Flag plants with anti-Flag antibody to confirm the TOE1 binding site at the FT genomic region. Coimmunoprecipitated DNA was quantified by RT-qPCR with the primer sets covering the upstream and downstream regulatory regions as well as the gene body of the FT locus (Fig. 7A). Consistent with previous studies (Zhai et al., 2015; Zhang et al., 2015), TOE1 was enriched at regions C, D, H, and R. However, the TOE1 enrichment level at region R was markedly higher than that at other regions (Supplemental Fig. S8A), indicating that TOE1 may preferentially bind to region R of the FT chromatin in our experimental conditions.
Figure 7.
CRY2 inhibits TOE1 binding to the 3′ downstream regulatory region of FT. A, Schematic drawing of the FT genomic region and locations of fragments amplified in ChIP experiments. Positions of the transcription start site (TSS) and transcription termination site (TTS) are indicated. Red lines denote fragments amplified in ChIP-qPCR (B; Supplemental Fig. S8A). Green bar including CCTCGAC element used as an EMSA probe (pFT-R-short) is indicated. B, ChIP-qPCR assays showing CRY2 inhibition of DNA-binding activity of TOE1 in Arabidopsis. The histogram shows the enrichment of the C, D, H, and R region of the FT chromatin. ChIP-qPCR was performed with 14-d-old mTOE1-Flag seedlings in wild-type and cry1 cry2 mutant backgrounds grown under LDs and harvested in ZT16. Genomic DNA fragments were immunoprecipitated by anti-Flag antibody and no-antibody immunoprecipitate served as control. IP/input (%) was calculated by comparison with the cycle threshold values between immunoprecipitate and input. Values are means ± sd of two independent ChIP assays. C, The core sequences of probes for binding by TOE1 in D and Supplemental Figure S8B. Probe FT-R-short is from the FT region R, containing CCTCGAC elements. FT-R-short (mu) denotes EMSA probe FT-R-short lack CCTCGAC element. D, EMSAs showing CRY2 inhibition of TOE1 DNA-binding ability. EMSA was performed with His-TF-TOE1 and His-TF proteins using Biotin-pFT-R-short as probes. The terms “1×,” “3×,” and “8×” indicate the amount of His-TF-CRY2 and His-TF relative to that of His-TF-TOE1. Free probe and TOE1-DNA–specific shift band are indicated by arrows. E, Model showing the action of CRY2, TOE, COP1, and CO in wild-type plants. During the light period in LDs, as COP1 and CO are localized in the cytoplasm and the nucleus, respectively, CO is stable and accumulates. BL-triggered CRY2-TOE interaction inhibits not only the association of TOE with CO to enhance the transcriptional activity of CO, but also the binding of TOE to the 3′ regulatory element within Block E enhancer of FT, leading to FT transcription and flowering. “X” denotes the unknown transcriptional activators binding to the element within the Block E enhancer that promotes FT transcription. The thick red arrow denotes high-level FT transcription. F, A model showing the action of TOE, COP1, and CO during the dark period in LDs. In DK, as COP1 and CO are localized in the nucleus and interact, CO undergoes ubiquitination and degradation, resulting in low-level CO accumulation and repression of FT transcription and flowering. Moreover, TOE binds to the regulatory element within Block E, leading to further inhibition of FT expression and flowering. The thin red arrow denotes low-level FT transcription.
To determine whether CRY2 regulates the DNA-binding activity of TOE1, we performed ChIP-qPCR with transgenic wild-type and cry1 cry2 mutant plants expressing mTOE1-Flag (mTOE1-Flag and mTOE1-Flag/cry1 cry2) grown under LDs and harvested at ZT16. To our surprise, we found that there was basically no difference in TOE1 enrichment at regions C, D, and H in wild-type and cry1 cry2 mutant backgrounds. However, markedly more TOE1 was enriched at region R in cry1 cry2 mutant than in wild-type background (Fig. 7B). These results indicate that CRY2 may preferentially inhibit TOE1 binding to region R lying downstream of the FT locus in planta.
Next, we performed DNA electrophoretic mobility shift assays (EMSAs) to confirm whether CRY2 might affect TOE1 binding to region R of FT containing TBS-like motif CCTCGAC in vitro. To do this, we first examined the binding of TOE1 to the TBS-like motif CCTCGAC in the absence of CRY2 by EMSA using affinity-purified His-TF-TOE1 fusion protein from E. coli and the probe, specifically the downstream regulatory region of the FT gene that contains the TBS-like sequence (Fig. 7C). As shown in Supplemental Figure S8B, the His-TF-TOE1 fusion protein bound to the probe in a protein concentration-dependent manner, and the binding capacity was reduced by the competition from cold competitor probes, whereas the control protein His-TF did not bind to this probe. Moreover, His-TF-TOE1 was not able to bind to the mutant probe lacking the TBS-like motif CCTCGAC. These results demonstrated that TOE1 bound to region R of FT (Zhai et al., 2015) and that the TBS-like motif CCTCGAC was required for TOE1 binding, which is consistent with the previous study. We then evaluated the binding capacity of TOE1 to region R of FT in the presence of His-TF-CRY2 fusion protein expressed from E. coli by EMSA. As shown in Figure 7D, the probe DNA fragments bands shifted by TOE1 protein decreased as the amounts of His-TF-CRY2 protein increased, whereas they were not affected by His-TF. These ChIP-qPCR and EMSA data, in conjunction with the above-mentioned result that CRY2 interacts with TOE1 in vitro and in vivo (Figs. 1, C and F, 3, A and B, and 4, C and G; Supplemental Fig. S4), indicate that CRY2 represses TOE1 binding to region R of FT containing the TBS-like motif CCTCGAC in vitro and in vivo.
DISCUSSION
TOEs Are Downstream Factors of CRY2 that Regulate Flowering
In the photoperiod flowering pathway, CRY2 is the primary photoreceptor that acts to induce floral initiation under LDs (Guo et al., 1998), whereas CRY1 plays a minor role in the induction of flowering (Mockler et al., 1999; Liu et al., 2008b). It has been established that CRY2 can regulate flowering through the CRY2-COP1-CO pathway, in which CRY2 interacts with COP1 and COP1 interacts with CO (Wang et al., 2001; Jang et al., 2008; Liu et al., 2008b). The interaction of COP1 with CO promotes the ubiquitination and degradation of CO, and the interaction of CRY2 with COP1 may inhibit COP1 activity and stabilize CO to promote flowering. CRY2 can also interact with SPA1 to inactivate the COP1/SPA complex and stabilize CO to induce flowering (Zuo et al., 2011). Other than through the 26S proteasome-dependent pathway, CRY2 can regulate flowering through direct regulation of the transcription factor CIB1, a transcriptional activator that directly binds to FT and promotes its transcription and thus flowering (Liu et al., 2008a). BL-triggered interaction of CRY2 with CIB1 promotes the transcriptional activity of CIB1, and FT transcription and floral initiation in Arabidopsis (Liu et al., 2008a). There are a number of CIBs acting redundantly to promote flowering (Liu et al., 2013). CIB1 interacts with CO, and CRY2, CIB1, and CO form a complex that stimulates FT transcription and floral initiation (Liu et al., 2018).
To date, the known downstream factors of CRY2 that mediate CRY2 regulation of photoperiodic flowering are COP1, CIBs, and SPAs (Wang et al., 2001; Liu et al., 2008a, 2011, 2013; Lian et al., 2011; Zuo et al., 2011). In this study, we have identified TOE1 and TOE2 as further downstream factors of CRY2 that mediate CRY2 control of flowering. Through a series of protein–protein interaction studies including yeast two-hybrid assays, in vitro pull-down assays, protein colocalization assays, BiFC, and split-LUC assays, we demonstrate that CRY1 and CRY2 physically interact with TOE1 and TOE2 (Figs. 1–3; Supplemental Figs. S3 and S4). Semi-in vivo pull-down assays and co-IP assays show that CRY1 and CRY2 physically interact with TOE1 and TOE2 in a BL-dependent manner (Fig. 4). Both the N- and C-termini of CRY1 and CRY2 are able to interact with TOE1 and TOE2 (Figs. 1–3; Supplemental Figs. S1, S3, and S4). Genetic interaction studies with the toe1 toe2 cry1 cry2 quadruple mutant and the cry1 cry2 and the toe1 toe2 double mutants demonstrate that TOE1 and TOE2 act partially downstream of CRY1 and CRY2 to mediate CRY regulation of flowering (Fig. 5, A–C; Supplemental Fig. S5). Moreover, expression of TOE1-Flag and TOE2-Flag in cry1 cry2 mutant background leads to a significantly more severe late-flowering phenotype than in wild-type background (Fig. 5, D–J). Taken together, the results in this study demonstrate that TOE1 and TOE2 act as downstream factors in CRY2-mediated regulation of flowering in Arabidopsis.
Given that a previous study showed that the spatial expression patterns of miR156-target SPLs are altered by miR156-resistant SPLs driven by their native promoters (Xu et al., 2016b), it is perhaps possible that the expression of miR172-resistant TOE1 and TOE2 driven by their native promoters in the transgenic plants generated in this study for flowering phenotype and co-IP assays may have affected the spatial expression pattern of TOE1 and TOE2 and the reliability of our co-IP assays. It was reported that TOE1 and TOE2 interact with CO in the regulation of flowering (Zhang et al., 2015), and that TOE1 and TOE2 interact with JAZ in the repression of flowering (Zhai et al., 2015), suggesting that the spatial expression pattern of TOE1 and TOE2 is similar to that of CO and FT. Moreover, like CO, FT, and CRY2 (Takada and Goto, 2003; An et al., 2004; Endo et al., 2007), TOE1 is also expressed in the vascular tissue of leaves (Jung et al., 2007). In contrast to the early-flowering phenotype of toe1 toe2 mutant (Aukerman and Sakai, 2003; Mathieu et al., 2009), the expression of miR172-resistant SMZ driven by the phloem companion cell-specific SUC2 promoter and TOE1 and TOE2 under the control of their native promoters leads to a pronounced late-flowering phenotype (Mathieu et al., 2009; Fig. 5, D–J in this study). Taken together, these results indicate that miR172-resistant TOE1 and TOE2 driven by their native promoters are likely expressed in the vascular tissue of leaves, leading to repressed floral initiation, and thus our co-IP data are of physiological significance.
The CRY2-TOE Interaction Leads to Dissociation of TOE from CO and Enhanced Transcriptional Activation Activity of CO
The following evidence suggests that CRY2 may antagonize TOE1 and TOE2, thus promoting flowering through inhibition of TOE1 and TOE2 activity: (1) The cry1 cry2 mutant shows a late-flowering phenotype in LDs, whereas the toe1 toe2 mutant exhibits an early-flowering phenotype (Fig. 5, A–C); (2) The loss of function of TOE1 and TOE2 in the cry1 cry2 mutant leads to significant suppression of the late-flowering phenotype of the cry1 cry2 mutant (Fig. 5, A–C), whereas expression of mTOE1 and mTOE2 resistant to miR172 cleavage leads to a significantly more severe late-flowering phenotype in the cry1 cry2 mutant background than in the wild-type background (Fig. 5, D–J); (3) CRY2 physically interacts with TOE1 and TOE2 in a BL-dependent manner (Fig. 3); and (4) TOE1 and TOE2 physically interact with CO, inhibiting the transcriptional activation activity of CO (Zhang et al., 2015). For these reasons, we explored whether CRY2 interacts with TOE1 and TOE2, thus inhibiting the association of TOE1 and TOE2 with CO and alleviating the inhibitory effects of TOE1 and TOE2 on CO transcriptional activation activity. We showed by pull-down assays with CNT2 and CCT2 proteins expressed in E. coli, and CRY2-containing protein extracts from Arabidopsis seedlings overexpressing CRY2, that either CNT2 or CCT2 is able to promote the disassociation of TOE1 from CO and that CRY2 interferes with the interactions of TOE1 and TOE2 with CO in a BL-dependent manner (Fig. 6, A–C). Split-LUC assays and co-IP assays showed that CRY2 represses the interactions of TOE1 with CO in N. benthamiana (Fig. 6, D–J). Further semi-in vivo pull-down and co-IP assays using transient expression in Arabidopsis protoplasts confirmed CRY2 inhibition of the interaction of TOE1 with CO in Arabidopsis (Fig. 6, K–N). These results demonstrate that CRY2 inhibits the associations of TOE1 and TOE2 with CO through CRY2-TOE1 and CRY2-TOE2 interactions. This conclusion is further supported by Dual-LUC assays using the reporter gene comprising the FT promoter as the indicator of CO transcriptional activation activity, which showed that CRY2 inhibits the repression of the CO transcriptional activation activity by TOE1 and TOE2 (Supplemental Fig. S7). A similar mechanism has been reported recently for CRY1 in the regulation of photomorphogenesis. Specifically, CRY1 interacts with the G-protein β-subunit AGB1, and AGB1 interacts with HY5 (Lian et al., 2018). The interaction of AGB1 with HY5 leads to the inhibition of HY5-DNA binding activity, and the CRY1-AGB1 interaction promotes the disassociation of AGB1 from HY5, which alleviates the inhibitory effects of AGB1 on HY5, thus promoting HY5 transcriptional activity and photomorphogenesis.
Because miR172 expression level increases as plants grow, TOE1 and TOE2 were highly and lowly expressed in young seedlings and in rosette leaves, respectively (Aukerman and Sakai, 2003; Mathieu et al., 2009; Wu et al., 2009b). A previous study showed that SMZ is highly expressed in hypocotyl, cotyledons, and 7-d-old young seedlings, but barely detectable in leaves at rosette stage and flowers (Mathieu et al., 2009). However, the TOE1/SMZ loss-of-function mutant shows a dramatically early-flowering phenotype, suggesting that TOE1/SMZ play an important role in the regulation of flowering. FT expression gradually increases as plants develop, but its expression is detected on DAG 4 and plateaus around DAG 6, proceeding floral commitment around DAG 9 and DAG 10 under LDs (Kobayashi et al., 1999). Coincidently, TOE1/SMZ expression gradually decreases after their high expression levels in hypocotyl, cotyledons, and 7-d-old seedlings. Based on these reports and our observation that toe1 toe2 mutant and mTOE1-Flag and TOE2-Flag plants don’t show obvious CRY2-related phenotypes other than flowering time, we postulate that the CRY2-TOE interactions may primarily contribute to flowering time regulation by repressing the TOE-CO interactions, thus promoting FT expression during early developmental stage starting from DAG 4.
CRY2 Represses TOE1 Binding to the Regulatory Region within the Block E Enhancer of FT
The AP2-like protein family comprises a type of transcriptional repressor that contains two conserved DNA-BD consisting of 60 to 70 amino acid-residue AP2 domains (Weigel, 1995). Through yeast two-hybrid assays, we showed that the entire N-terminal domain of TOE1 and TOE2, comprising the two AP2 domains, is essential for TOE1 and TOE2 interactions with CRY2 (Supplemental Fig. S2). It was shown that TOE1 inhibits CO transcriptional activation activity via direct TOE1-CO interaction (Zhang et al., 2015), and also represses FT expression by directly binding to the promoter and the 3′ downstream regulatory region of FT (Zhai et al., 2015; Zhang et al., 2015). We showed reproducibly by ChIP-qPCR assays that TOE1 preferentially binds to region R of the FT 3′ downstream regulatory sequence containing the TBS-like motif CCTCGAC (Supplemental Fig. S8A), and that CRY2 markedly inhibits TOE1 binding to region R of FT in Arabidopsis, but hardly affects TOE1 binding to other regions of FT (Fig. 7, A and B). Further EMSAs confirmed that CRY2 represses TOE1 binding to region R of FT containing the TBS-like motif in vitro (Fig. 7, C and D). It has been reported that CRY1 and the UV-B photoreceptor UVR8 interacts with transcription factors to directly regulate their DNA-binding activity. For examples, CRY1 and UVR8 interact directly with BES1, a pivotal transcriptional factor in the brassinosteroid signaling pathway, leading to inhibition of BES1 DNA-binding activity and BR signaling, thus promoting photomorphogenesis (Liang et al., 2018; Wang et al., 2018b). CRY1 also interacts with BZR1 and HBI1, inhibiting their DNA-binding activity and repressing hypocotyl elongation (Wang et al., 2018a; He et al., 2019).
It remains poorly understood how the binding of TOE1 to the 3ʹ downstream region of the FT locus leads to the repression of FT transcription and late flowering. A recent interesting study demonstrated that FT contains a novel enhancer element, Block E, which is 445-bp long and located 1.4 kb downstream of FT (Zicola et al., 2019). The TOE1 binding region R sequence containing the TBS-like motif resides within the Block E enhancer element (Franco-Zorrilla et al., 2014). Silencing of this enhancer in FT by inverted-repeats–mediated DNA methylation leads to dramatic downregulation of FT expression and late flowering (Zicola et al., 2019), demonstrating that this enhancer may recruit transcriptional activators to promote FT expression. Moreover, a previous study by a ChIP-chip assay has shown that SMZ, the homolog of TOE1, also binds preferentially to the regulatory element within the Block E enhancer of FT (Mathieu et al., 2009). Consistent with this, FT is considerately downregulated and flowering is significantly delayed in both the dominant mutant of SMZ, namely smz-d, and in transgenic plants overexpressing SMZ (Mathieu et al., 2009), demonstrating that this enhancer may also recruit transcriptional repressors like TOE1 and SMZ to inhibit FT expression and flowering. Recently, it was demonstrated that the binding of TOE1 to the 3′ downstream enhancer of GLABRA1, which encodes a MYB transcription factor essential for trichome initiation, leads to repression of GLABRA1 expression and inhibition of abaxial trichome initiation (Wang et al., 2019). Based on these reports and our findings, we tentatively propose that the CRY2-mediated inhibition of TOE1 binding to the regulatory element in Block E enhancer of FT may partially be involved in mediating the positive regulation of FT expression and flowering by CRY2.
A Model
Based on previous reports and the findings obtained in this study, we propose a regulatory framework for the regulation of photoperiodic flowering by CRY2, TOE, and CO (Supplemental Fig. S9), in which CRY2 positively regulates FT expression by negatively regulating TOE, thus leading to both enhanced transcriptional activation activity of CO on FT and inhibition of the direct binding of TOE to FT. To further illustrate how TOE and CO function in CRY2-regulated flowering, we propose the possible modes of actions of these proteins in the wild-type and cry1 cry2 mutant plants (Fig. 7, E and F). Of note, to clearly illustrate the significance of the CRY2-TOE interaction, we have not included in the model all the CRY2-interacting proteins such as CIBs and SPAs, which play important roles in mediating CRY2 regulation of photoperiodic flowering (Liu et al., 2008a, 2013; Zuo et al., 2011). When wild-type plants are grown under LDs, CRY2 is activated, COP1 is predominantly localized to the cytoplasm (Osterlund and Deng, 1998), CO mRNA peaks in the afternoon (Suárez-López et al., 2001; Valverde et al., 2004), and CO protein is able to accumulate (Jang et al., 2008; Liu et al., 2008b). BL-triggered CRY2-TOE interaction on the one hand interferes with the association of TOE with CO, which promotes the transcriptional activation activity of CO and FT transcription, and on the other hand prohibits the binding of TOE to the regulatory region within the Block E enhancer of FT, which promotes FT expression (Fig. 7E). When cry1 cry2 mutant plants are grown in LDs, CRY2 is absent and thus COP1 is predominantly localized to the nucleus (Osterlund and Deng, 1998) where it interacts with CO and promotes its ubiquitination and degradation (Jang et al., 2008; Liu et al., 2008b). A low level of CO protein accumulates, and its activity is suppressed by TOE, leading to repression of FT transcription. At the same time, TOE is able to bind to the regulatory element within Block E of FT and further inhibits FT expression (Fig. 7F). Given the findings showing that both the transcriptional repressors TOE1 and SMZ and the possible transcriptional activators (designated as “X”) may be recruited by the Block E enhancer of FT (Mathieu et al., 2009; Zhai et al., 2015; Zicola et al., 2019), and the emerging evidence showing that the Block E enhancer is involved in the regulation of FT expression and flowering (Zicola et al., 2019), extensive studies will be required to define the complex mechanisms by which the transcriptional regulators and this enhancer cooperate to coordinate FT expression and CRY2-regulated photoperiodic flowering.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants used were of the Columbia ecotype. The cry1 cry2 (hy4-104 cry2-1) mutant and the transgenic lines overexpressing Myc-tagged full-length CRY1 and CRY2 (Myc-CRY1-OX and Myc-CRY2-OX) were described in Mao et al. (2005) and Sang et al. (2005). Seeds of toe1 (SALK_069677) and toe2 (SALK_065370) were obtained from the Arabidopsis Biological Resource Center (https://www.arabidopsis.org). The toe1 toe2 double mutant was generated by crossing parental single mutants, and the homozygous double mutant was obtained by PCR genotyping. All the primers for genotyping are listed in Supplemental Table S1. Construction of pCambia1300-3×Flag was described in Xu et al. (2018). The mutant full-length complementary DNA (cDNA) sequences encoding TOE1 or TOE2 (mTOE1 and mTOE2) were driven by native promoters of TOE1 and TOE2, respectively, and constructed into pCambia1300-3×Flag to generate vectors expressing TOE1-Flag and TOE2-Flag (mTOE1-Flag and mTOE2-Flag). All of the constructs used were confirmed by DNA sequencing. All the primers for vector construction in this study are listed in Supplemental Table S1.
All constructs were transferred into Agrobacterium tumefaciens strain GV3101 and then transformed into wild-type Arabidopsis by the floral-dip method (Clough and Bent, 1998). Transgenic lines expressing TOE1-Flag and TOE2-Flag in wild-type background (mTOE1-Flag and mTOE2-Flag) were screened on MS plates containing 50 μg/mL of hygromycin (Roche) to generate transgenic lines of mTOE1-Flag12 and mTOE2-Flag16. The mTOE1-Flag12 and mTOE2-Flag16 in wild-type background were introgressed into cry1 cry2 mutant background by genetic crossing to generate mTOE1-Flag/cry1 cry2 and mTOE2-Flag/cry1 cry2 plants, which were confirmed by phenotypic and western-blot analyses. The transgenic line expressing both Myc-CRY1 or Myc-CRY2 and mTOE1-Flag or mTOE2-Flag (Myc-CRY1-OX/mTOE1-Flag, Myc-CRY2-OX/mTOE1-Flag, Myc-CRY1-OX/mTOE2-Flag, and Myc-CRY2-OX/mTOE2-Flag) were obtained by genetic crossing between mTOE1-Flag12 or mTOE2-Flag16 and Myc-CRY1-OX or Myc-CRY2-OX plants, which was confirmed by western-blot analysis.
Seeds were sterilized with 20% (v/v) bleach before being sown on one-half strenght Murashige and Skoog basal medium (MS; Sigma-Aldrich) with 1% (w/v) Suc and kept at 4°C for 3 d, and then exposed to white light (100 μmol m−2 s−1, produced by cool-white fluorescent lamps) for 24 h. Experiments involving BL, RL, and FRL illumination were performed as described in Jia et al. (2014) and Luo et al. (2014). Light intensity was measured with an ILT2400-A Radiometric Photometer (International Light Technologies).
Yeast Two-Hybrid Assay
The GAL4 yeast two-hybrid screening and yeast two-hybrid assays were performed according to the manufacturer’s instructions (Matchmaker user’s manual; Clontech). The Arabidopsis cDNA library cloned in the prey vector pGAD-T7 (AD) was made by the OE BioTech. The bait plasmid pGBK-T7 (BD) and the prey library DNA were cotransformed into the yeast strain AH109. Approximately 1 × 107 transformants were screened each time to select colonies that were grown under BL with 10 mm of 3-Amino-1,2,4-triazole.
Construction of BD-CNT1, BD-CRY2, BD-CNT2, and AD-CRY1 cassettes were described in Wang et al., 2018a, 2018b). For yeast two-hybrid assays, the cDNA fragments of TOE1 and TOE2 were cloned into the BD vector, and the fragments of TOE1, TOE2, SMZ, SNZ, and various TOE1 and TOE2 fragments lacking either one or both AP domains or N- or C-terminal sequences were individually cloned into the AD vector. Combinations of BD and AD vectors were cotransformed into AH109 cells via the PEG/LiAc transformation procedure. Transformed yeast cells were spread on either SD-Trp/Leu/His/Ade (SD-T-l-H-A) plates or SD-Trp/Leu/His (SD-T-l-H) plates supplemented with 1 mm, 5 mm, or 10 mm of 3-Amino-1,2,4-triazole, then one half of these plates were exposed to BL (30 μmol m−2 s−1) and the other half were kept in the dark for the interaction test (Luo et al., 2014; Xu et al., 2016a; Wang et al., 2018b).
Protein Colocalization Study
Construction of 35S::CFP, 35S::YFP, 35S::CRY1-YFP, and 35S::CRY2-YFP cassettes was described in Lian et al. (2011) and Wang et al. (2018a, 2018b). The cassettes comprising 35S::TOE1-CFP and 35S::TOE2-CFP were cloned individually into the pHB vector (Mao et al., 2005). Overnight cultures of A. tumefaciens strain GV3101 harboring the constructs expressing CFP-fusion proteins, YFP-fusion proteins, or p19 plasmid were collected by centrifugation, diluted by MS liquid medium to OD600 = 0.6 individually and incubated with 200 μm of acetosyringone and 10 mm of MES (pH 5.6) for 3 h at room temperature. Then, the mixture of A. tumefaciens harboring the constructs expressing CFP and YFP fusion proteins, and p19 plasmid with a volume ratio of 1:1:1, was introduced into Nicotiana benthamiana leaf epidermal cells by infiltration. After incubation for 2 to 3 d in dim light, protein colocalization was detected by confocal microscopy (TCS SP5II confocal laser scanning microscope; Leica).
BiFC Assay
The vectors used to make constructs for BiFC assays were pXY104 and pXY106, which carry fragments encoding the C- and N-terminal halves of YFP (cYFP and nYFP), respectively (Luo et al., 2014; Lu et al., 2015). Construction of 35S::CRY1-cYFP, 35S::CNT1-cYFP, 35S::CCT1-cYFP, and 35S::CRY2-cYFP cassettes was described in Lian et al. (2018), Wang et al. (2018b), and Xu et al. (2018). The fragments encoding CNT2 and CCT2 were fused to the fragment encoding the C terminus of YFP to generate 35S::CNT2-cYFP and 35S::CCT2-cYFP, and the fragments encoding TOE1 and TOE2 were fused to the fragment encoding the N terminus of YFP to generate 35S::nYFP-TOE1 and 35S::nYFP-TOE2, respectively. All these vectors were transformed into A. tumefaciens strain GV3101. GV3101 cultures harboring constructs expressing nYFP fusion proteins and cYFP fusion proteins were mixed at a ratio of 1:1 and introduced into N. benthamiana leaves through infiltration. After incubation in DK for 2 to 3 d in dim light, the N. benthamiana leaf samples were subjected to detection of expression of the various fluorescent proteins by confocal microscopy (TCS SP5II; Leica). Three independent experiments were performed, and one representative result is shown.
Split-LUC Assay
For the split-LUC experimental analysis of the interaction of TOE1 and TOE2 with CRY1, CNT1, CCT1, CRY2, CNT2, CCT2, and CO, the coding sequences of CRY1, CNT1, CCT1, CRY2, CNT2, CCT2, TOE2, and CO were amplified and then cloned individually into pCambia1300-nLUC vector (Chen et al., 2008). The coding sequences of TOE1, TOE2, CNT2, and CCT2 were cloned individually into pCambia1300-cLUC vector (Chen et al., 2008). All these vectors were transformed into A. tumefaciens strain GV3101. GV3101 cells transformed with the nLUC- or cLUC-fused proteins were mixed at a ratio of 1:1 and introduced into N. benthamiana leaves through infiltration. After incubation in DK for 2 to 3 d, the N. benthamiana leaves were treated with 1 mm of l-luciferin sodium salt (Yeasen) and kept in DK for 10 min, and then the LUC signal was detected by a luminescent imaging workstation (5200; Tanon).
For the split-LUC assays to determine the effect of CRY2 on the interaction of TOE1 or TOE2 with CO, 35S::cLUC-TOE1 or 35S::cLUC-TOE2 together with 35S::CO-nLUC constructs were coexpressed in the presence of 35S::CRY2-YFP or 35S::GUS-NLS-YFP (negative control) a ratio of 1:1:2. Construction of the 35S::GUS-NLS-YFP cassette was described in Wang et al. (2018a). Luminescence intensities were detected by Tanon imaging software after the treatment.
Pull-Down Assays with Proteins Expressed in Escherichia coli
Pull-down assays were performed as described previously with minor modifications (Xu et al., 2016a). Construction of pcold-TF-CNT1 and pcold-TF-CCT1 cassettes was described in Wang et al. (2018a). The fragments of CRY1, CRY2, CNT2, and CCT2 were cloned into pcold-Trigger Factor (TF; TaKaRa Bio). The fragments of TOE1 and TOE2 were cloned into pGEX-4T-1 and the fragments of CO were cloned into pET-32a vector. His-CO, His-TF-CRY1, His-TF-CNT1 (residues 1–489), His-TF-CCT1 (residues 490–681), His-TF-CRY2, His-TF-CNT2 (residues 1–485), His-TF-CCT2 (residues 486–612), His-TF, GST, GST-TOE1, and GST-TOE2 proteins were expressed in E. coli (Rosetta; CWBIO). His-TF, His-TF-CNT2, His-TF-CCT2, and His-CO were purified according to the manufacturer’s protocol (Qiagen). His-TF and GST were expressed in E. coli harboring the empty pcold-TF and pGEX-4T-1 vectors, which served as negative control. For the pull-down assays to detect the interaction of CRY1/CNT1/CCT1/CRY2/CNT2/CCT2 proteins with TOE1/TOE2, the bait proteins GST-TOE1 or GST-TOE2 were first incubated with 10-μL GST Glutathione beads (MagneGST Glutathione particles; Promega) in non-EDTA buffer (50 mm of Tris-HCl at pH 7.5, 150 mm of NaCl, 0.2% [v/v] Triton-x-100, and 10% [v/v] glycerol) at 4°C for 2 h, followed by three washes with the same buffer. The beads were then resuspended in 1 mL of non-EDTA buffer, and 50 to 100 ng of His-TF-CRY1, His-TF-CNT1, His-TF-CCT1, His-TF-CRY2, His-TF-CNT2, His-TF-CCT2, or His-TF (negative control) prey proteins were added into the solution. The mixture was incubated at 4°C for 1 h and then washed three times. Proteins were eluted into SDS loading buffer and analyzed by western blot. Prey proteins were detected by anti-His antibody (NewEast).
For the pull-down assays to determine the effect of CNT2 or CCT2 on the interaction of TOE1 with CO, bait proteins (GST and GST-TOE1) were first incubated with 10-μL GST Glutathione beads in non-EDTA buffer at 4°C for 2 h and then washed three times with the same buffer. The beads were then resuspended in 1-mL non-EDTA buffer. His-CO and His-TF-CNT2 or His-TF-CCT2 or His-TF proteins were added to the solution. The mixture was incubated for another 1 h at 4°C and washed with non-EDTA buffer three times. Proteins were eluted into SDS loading buffer and analyzed by western blot. Prey proteins were detected by anti-His antibody (NewEast) and bait proteins were visualized by anti-GST antibody (BPI; GeneTex).
Semi-In Vivo Pull-Down Assays with Arabidopsis Protein Extracts
For BL-specific CRY1/CRY2-TOE1/TOE2 interaction assays, the fragments of TOE1 were cloned into pcold-TF vector. His-TF-TOE1 or GST-TOE2 bait proteins were first incubated with 20-μL Ni-NTA agarose beads (Qiagen) or 10-μL GST Glutathione beads (MagneGST Glutathione particles; Promega) for 2 h and then washed three times with lysis buffer (50 mm of Tris-HCl at pH 7.5, 150 mm of NaCl, 0.2% [v/v] Triton-X-100, and 10% [v/v] glycerol). Myc-CRY1- and -CRY2-containing protein extracts from 5-d-old etiolated Myc-CRY1-OX and Myc-CRY2-OX seedlings that were adapted to DK or exposed to BL (20 μmol m−2 s−1), RL (20 μmol m−2 s−1), or FRL (10 μmol m−2 s−1) for 0.5 h served as preys. All the seedlings with different dark or light treatments were homogenized with non-EDTA lysis buffer plus 1 mm of Pefabloc (Sigma-Aldrich), 1× cocktail, and 50 μm of MG132. After centrifugation, the supernatant was incubated with TOE1 protein-bound Ni-NTA agarose beads or TOE2 protein-bound GST beads for 30 min and washed three to four times with 1 mL of lysis buffer, and then the precipitates were eluted into 30 μL of SDS 2× loading buffer and subjected to western-blot analysis with anti-Myc antibody (EMD Millipore).
For analysis of the effects of CRY2 on the interaction of TOE1 with CO, a fragment encoding the full-length CO was cloned into pcoldIII-MBP vector. GST-TOE1 was used as bait, whereas MBP-CO, expressed in E. coli, and Myc-CRY2 protein, that was extracted from Myc-CRY2-OX seedlings that were grown for 5 d in white light and then transferred to DK for 3 d, followed by initial treatment with 100 μm of MG132 for 4 h and then continued incubated in DK or exposure to BL (20 μmol m−2 s−1), served as preys. They were first incubated with 10-μL GST Glutathione beads (Promega) for 2 h and then washed three times. All seedlings were harvested in dim green safe light and homogenized in lysis buffer. After centrifugation, the supernatant and MBP-CO was incubated with baits for 1 h and washed three to four times with 1 mL of lysis buffer each time, and then the precipitates were eluted into 30 μL of SDS 2× loading buffer and subjected to western-blot analysis with anti-Myc (EMD Millipore) and anti-MBP antibody (New England Biolabs).
For analysis of the effects of CRY2 inhibition of the interaction of TOE1 with CO, baits were the TOE1-Flag protein extracts prepared from mTOE1-Flag and mTOE1-Flag/cry1 cry2 seedlings that were grown for 5 d in white light and then transferred to DK for 3 d, followed by initial treatment with 100 μm of MG132 for 4 h and then continued incubation in DK or exposure to BL (30 μmol m−2 s−1). Myc-CO protein extracts from BL-adapted Myc-CO-OX seedlings served as prey. Total protein concentration was determined by Bradford assay (Bio-Rad) and equal amounts of total protein in 1 mL of lysis buffer were incubated with 20 μL of anti-Flag agarose beads at 4°C for 1 h. The immunoprecipitate was washed two to three times with wash buffer. The precipitates were eluted into 30 μL of 3×Flag peptide and subjected to western-blot analysis with anti-Flag (Sigma-Aldrich) and anti-Myc (EMD Millipore) antibodies.
Co-IP Assay
Co-IP assays in Arabidopsis were performed by methods described previously with minor modifications (Lian et al., 2011). For co-IP assays of BL-dependent CRY1/CRY2-TOE1/TOE2 interaction, Myc-CRY1-OX/mTOE1-Flag or Myc-CRY1-OX/mTOE2-Flag or Myc-CRY2-OX/mTOE1-Flag or Myc-CRY2-OX/mTOE2-Flag seedlings were grown in white light (100 μmol m−2 s−1) for 5 d and then adapted in DK for 3 d. After being treated with 100 μm of MG132 for 4 h, one half of the seedlings were exposed to 30 μmol of m−2 s−1 BL for 1 h, and the other half were maintained in DK for 1 h. All the seedlings were harvested in dim green safe light and homogenized in lysis buffer (Luo et al., 2014). Total protein concentration was determined by Bradford assay (Bio-Rad) and equal amounts of total protein in 1 mL of lysis buffer were incubated with anti-Myc agarose beads (Sigma-Aldrich) at 4°C for 1.5 h. The immunoprecipitate was washed two to three times with wash buffer (Luo et al., 2014). The precipitates were eluted into 30 μL of SDS 2× loading buffer and subjected to western-blot analysis with anti-Flag (Sigma-Aldrich) and anti-Myc (EMD Millipore) antibodies.
For co-IP assays in N. benthamiana, the constructs harboring 35S::mTOE1-Flag and 35S::Myc-CO were used. The assays were performed as described in Mao et al. (2020). For co-IP assays in Arabidopsis protoplast, the TOE1-Flag and Myc-CO coding sequence were cloned into the pDT1 vector (He et al., 2016). The plasmids were extracted from the E. coli strain using a Plasmid Maxi Preparation Kit (Tiangen). Mesophyll protoplasts were isolated form 3- to 4-week–old plants using 3M Magic Tape (3M) as previously described (Wu et al., 2009a). The protoplasts transformed with the pDT1-ACT2pro::Myc-CO-UBQ10pro::TOE1-Flag construct coexpressing Myc-CO and TOE1-Flag were exposed to BL (30 μmol m−2 s−1) for 16 h, treated with 100 μm of MG132 for 1 h, and adapted to DK or exposed to BL for 2 h. These protoplasts were then collected and homogenized in lysis buffer containing 1% (v/v) Triton-X-100, and the supernatant was incubated with anti-Myc agarose (Sigma-Aldrich). The beads were then washed two to three times with lysis buffer and eluted into SDS loading buffer.
Western Blotting
Arabidopsis seeds of different genotypes were grown on MS medium for 14 d under LD conditions illuminated by white light (100 μmol m−2 s−1). Plant material (0.5 g) was fixed, ground, and used to extract nuclear proteins using ChIP assays. The protein supernatant, after quantification by Bradford assay (Bio-Rad), was subjected to western-blot analysis with the related antibody. The relative band intensity was quantified with the software ImageJ (http://rsb.info.nih.gov/ij).
Flowering Studies
Flowering studies was performed according to the methods described in Liu et al. (2008b), but with minor modifications. Seedlings were germinated on MS medium and then transferred to soil and grown in LDs illuminated by white light (100 μmol m−2 s−1). Flowering time was measured by scoring the number of rosette leaves at flowering and the number of days from germination to bolting. At least 30 plants were analyzed.
RT-qPCR
Seeds were germinated on MS plates supplemented with 2% (w/v) Suc and placed at 4°C for 3 d, and then transferred to LD (16-h-light/8-h-dark) conditions with white light (100 μmol m−2 s−1) at 22°C for 14 d and harvested at ZT16. Total RNA was extracted with RNA plant Plus Reagent following the manufacturer’s instructions (Tiangen). After the genomic DNA contamination was removed by DNase I, the first-strand cDNA was synthesized using iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad). qPCR was carried out using the CFX96 Touch Real-Time PCR System (Bio-Rad) with SYBR Premix Ex Taq II (Takara). The thermal profile for qPCR was 95°C for 1 min, followed by 45 cycles of 95°C for 5 s and 60°C for 20 s. The expression of FT and CO was normalized against the expression of the endogenous control gene PP2A. All experiments were performed with three independent biological replicates in three technical repetitions each. The RT-qPCR primers are listed in Supplemental Table S1.
Dual-LUC Assay
Transient transcription dual-LUC assays in N. benthamiana plants were carried out as described in Wang et al. (2018a) and Xu et al. (2018). An ∼2-kb fragment upstream of the start codon of FT was amplified and cloned into pGreenII0800-LUC vector (Hellens et al., 2005), which expresses Renilla LUC driven by the 35S promoter serving as an internal reference and firefly LUC driven by the FT promoter serving as a reporter. A. tumefaciens culture (OD600 = 0.6) harboring the related constructs was mixed; the lack of any effector in all groups was supplemented with liquid MS medium to the same volume. The mixture was introduced into N. benthamiana leaves by infiltration and kept in dim white light for 3 d, and the samples were then collected for dual-LUC assays with the Dual-Luciferase Reporter System (Promega) according to the manufacturer’s instructions. Renilla LUC activity for each reaction was used as an internal control.
EMSAs
EMSAs were performed using biotin-labeled probes and the Light Shift Chemiluminescent EMSA kit (Thermo). The proteins used in this experiment were expressed and purified as described above, then 0.5 μg of His-TF, His-TF-TOE1, and His-TF-CRY2 were utilized. Biotin-labeled probes and unlabeled probes were commercially synthesized and annealed (GENEWIZ). The binding reaction was carried out in 10-μL binding buffer (10× Binding buffer: 100 mm of MgCl2, 1 μg μL−1 Poly [dI·dC], 2.5% [v/v] glycerol, and 1% [v/v] Nonider P-40) with a 200-fM probe and 500 ng of His-TF-TOE1 protein (1×) according to the manufacturer’s instructions (Thermo Fisher Scientific; Zhang et al., 2014). For the cold competitor, 4 pM of unlabeled probe was added into the reaction mixture. After 20-min incubation at room temperature, the reactions were resolved by 6% native polyacrylamide gel at 4°C, and then the DNA–protein complexes were transferred to a nylon membrane. Biotin-labeled probes were detected by HRP-conjugated streptavidin and visualized with an Enhanced Chemiluminescent Detection Kit (Pierce).
ChIP-qPCR Assay
ChIP was performed according to the methods described previously with minor modifications (Zhang et al., 2014). Briefly, wild-type, mTOE1-Flag, and mTOE1-Flag/cry1 cry2 seedlings were grown in white light (100 μmol m−2 s−1) for 14 d and then were harvested at ZT16. All the seedlings harvested in dim green safe light and fixed in fixation buffer by vacuum infiltration at 4°C. Gly was added to a final concentration of 0.125 m to stop the reaction under vacuum. The sonicated protein–DNA complexes were immunoprecipitated with or without (negative control) anti-Flag antibody (Sigma-Aldrich). The resulting DNA was used for qPCR. IP/input was calculated by comparing the cycle threshold values between immunoprecipitate and input. The primers used for qPCR are listed in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AT4G08920 (CRY1), AT1G04400 (CRY2), AT2G28550 (TOE1), AT5G60120 (TOE2), AT3G54990 (SMZ), AT2G39250 (SNZ), AT5G15840 (CO), and AT1G65480 (FT).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. CNT2 physically interacts with TOE1 and TOE2 in yeast.
Supplemental Figure S2. CRY2 physically interacts with the N terminus of TOE1 and TOE2 in yeast.
Supplemental Figure S3. CRY1 physically interacts with TOE1 and TOE2 in plant cells.
Supplemental Figure S4. CRY2 physically interacts with TOE1 and TOE2 in plant cells.
Supplemental Figure S5. RT-qPCR assay showing antagonistic regulation of FT expression by CRYs and TOEs.
Supplemental Figure S6. CO physically interacts with TOE1 and TOE2 in vivo.
Supplemental Figure S7. CRY2 inhibits the repression of CO transcriptional activation activity by TOE1 and TOE2.
Supplemental Figure S8. TOE1 binds preferentially to the 3′ downstream regulatory region of FT.
Supplemental Figure S9. A regulatory framework for CRY2, TOE, and CO in the regulation of flowering.
Supplemental Table S1. The primers used in this study.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant no. 2017YFA0503802) and the National Natural Science Foundation of China (grant no. 31900609 to Z.-L.M., and grant nos. 31530085, 91217307, and 90917014 to H.-Q.Y.).
References
- Ahmad M, Cashmore AR(1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Cashmore AR(1998) Chimeric proteins between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10: 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H(2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs WR, Christie JM(2002) Phototropins 1 and 2: Versatile plant blue-light receptors. Trends Plant Sci 7: 204–210 [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, Liu D(1999) Cryptochromes: Blue light receptors for plants and animals. Science 284: 760–765 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM(2008) Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, O’Hara A, Kelly SM, Hothorn M, Smith BO, Hitomi K, et al. (2012) Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Consentino L, Lambert S, Martino C, Jourdan N, Bouchet PE, Witczak J, Castello P, El-Esawi M, Corbineau F, d’Harlingue A, et al. (2015) Blue-light dependent reactive oxygen species formation by Arabidopsis cryptochrome may define a novel evolutionarily conserved signaling mechanism. New Phytol 206: 1450–1462 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- D’Amico-Damião V, Carvalho RF(2018) Cryptochrome-related abiotic stress responses in plants. Front Plant Sci 9: 1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, Quail PH(1992) COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Endo M, Mochizuki N, Suzuki T, Nagatani A(2007) CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R(2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111: 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Duong H, Ma N, Lin C(1999) The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue-light–dependent post-transcriptional mechanism. Plant J 19: 279–287 [DOI] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C(1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- He G, Liu J, Dong H, Sun J(2019) The blue-light receptor CRY1 Interacts with BZR1 and BIN2 to Modulate the phosphorylation and nuclear function of BZR1 in repressing BR signaling in Arabidopsis. Mol Plant 12: 689–703 [DOI] [PubMed] [Google Scholar]
- He SB, Wang WX, Zhang JY, Xu F, Lian HL, Li L, Yang HQ(2015) The CNT1 domain of Arabidopsis CRY1 alone is sufficient to mediate blue light inhibition of hypocotyl elongation. Mol Plant 8: 822–825 [DOI] [PubMed] [Google Scholar]
- He Z, Liu B, Wang X, Bian M, He R, Yan J, Zhong M, Zhao X, Liu X(2016) Construction and validation of a dual-transgene vector system for stable transformation in plants. J Genet Genomics 43: 199–207 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA(2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Song YH, Imaizumi T(2012) LOV domain-containing F-box proteins: Light-dependent protein degradation modules in Arabidopsis. Mol Plant 5: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G(2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia KP, Luo Q, He SB, Lu XD, Yang HQ(2014) Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol Plant 7: 528–54024126495 [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM(2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ(2009) Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis. Plant Cell 21: 2624–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D(1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kleiner O, Kircher S, Harter K, Batschauer A(1999) Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J 19: 289–296 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T(1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Lian H, Xu P, He S, Wu J, Pan J, Wang W, Xu F, Wang S, Pan J, Huang J, et al. (2018) Photoexcited CRYPTOCHROME 1 interacts directly with G-protein β subunit AGB1 to regulate the DNA-binding activity of HY5 and photomorphogenesis in Arabidopsis. Mol Plant 11: 1248–1263 [DOI] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ(2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Mei S, Shi C, Yang Y, Peng Y, Ma L, Wang F, Li X, Huang X, Yin Y, et al. (2018) UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev Cell 44: 512–523.e5 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR(1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95: 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C(2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C(2008a) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ(2008b) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang Q, Deng W, Wang X, Piao M, Cai D, Li Y, Barshop WD, Yu X, Zhou T, et al. (2017) Molecular basis for blue-light–dependent phosphorylation of Arabidopsis cryptochrome 2. Nat Commun 8: 15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li X, Li K, Liu H, Lin C(2013) Multiple bHLH proteins form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet 9: e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Li X, Ma D, Chen Z, Wang JW, Liu H(2018) CIB1 and CO interact to mediate CRY2-dependent regulation of flowering. EMBO Rep 19: e45762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, Yang HQ(2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8: 467–478 [DOI] [PubMed] [Google Scholar]
- Luo Q, Lian HL, He SB, Li L, Jia KP, Yang HQ(2014) COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26: 2441–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ(2005) From the cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102: 12270–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, He S, Xu F, Wei X, Jiang L, Liu Y, Wang W, Li T, Xu P, Du S, et al. (2020) Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytol 225: 848–865 [DOI] [PubMed] [Google Scholar]
- Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M(2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng XW(1994) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C(1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Onouchi H, Igeño MI, Périlleux C, Graves K, Coupland G(2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Deng XW(1998) Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J 16: 201–208 [DOI] [PubMed] [Google Scholar]
- Ozgür S, Sancar A(2006) Analysis of autophosphorylating kinase activities of Arabidopsis and human cryptochromes. Biochemistry 45: 13369–13374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Sancar A(2005) Photochemistry and photobiology of cryptochrome blue-light photopigments: The search for a photocycle. Photochem Photobiol 81: 1291–1304 [DOI] [PubMed] [Google Scholar]
- Quail PH.(2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G(2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, Yang HQ(2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17: 1569–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C(2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767 [DOI] [PubMed] [Google Scholar]
- Shalitin D, Yu X, Maymon M, Mockler T, Lin C(2003) Blue-light–dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15: 2421–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA(1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Song Z, Bian Y, Liu J, Sun Y, Xu D(2020) B-box proteins: Pivotal players in light-mediated development in plants. J Integr Plant Biol [DOI] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G(2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Takada S, Goto K(2003) TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. Plant Cell 15: 2856–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan ST, Dai C, Liu HT, Xue HW(2013) Arabidopsis casein kinase1 proteins CK1.3 and CK1.4 phosphorylate cryptochrome2 to regulate blue light signaling. Plant Cell 25: 2618–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G(2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW(2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wang L, Zhou CM, Mai YX, Li LZ, Gao J, Shang GD, Lian H, Han L, Zhang TQ, Tang HB, et al. (2019) A spatiotemporally regulated transcriptional complex underlies heteroblastic development of leaf hairs in Arabidopsis thaliana. EMBO J 38: e100063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zuo Z, Wang X, Gu L, Yoshizumi T, Yang Z, Yang L, Liu Q, Liu W, Han YJ, et al. (2016) Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li L, Xu P, Lian H, Wang W, Xu F, Mao Z, Zhang T, Yang H(2018a) CRY1 interacts directly with HBI1 to regulate its transcriptional activity and photomorphogenesis in Arabidopsis. J Exp Bot 69: 3867–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu X, Li L, Lian H, Mao Z, Xu P, Guo T, Xu F, Du S, Cao X, et al. (2018b) Photoexcited CRYPTOCHROME1 interacts with dephosphorylated BES1 to regulate brassinosteroid signaling and photomorphogenesis in Arabidopsis. Plant Cell 30: 1989–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D.(1995) The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell 7: 388–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS(2009a) Tape-Arabidopsis Sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS(2009b) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Spalding EP(2007) Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc Natl Acad Sci USA 104: 18813–18818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, He S, Zhang J, Mao Z, Wang W, Li T, Hua J, Du S, Xu P, Li L, et al. (2018) Photoactivated CRY1 and phyB interact directly with AUX/IAA proteins to inhibit auxin signaling in Arabidopsis. Mol Plant 11: 523–541 [DOI] [PubMed] [Google Scholar]
- Xu F, Li T, Xu PB, Li L, Du SS, Lian HL, Yang HQ(2016a) DELLA proteins physically interact with CONSTANS to regulate flowering under long days in Arabidopsis. FEBS Lett 590: 541–549 [DOI] [PubMed] [Google Scholar]
- Xu M, Hu T, Zhao J, Park MY, Earley KW, Wu G, Yang L, Poethig RS(2016b) Developmental functions of miR156-regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes in Arabidopsis thaliana. PLoS Genet 12: e1006263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR(2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HQ, Wu YJ, Tang RH, Liu D, Liu Y, Cashmore AR(2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Yang L, Mo W, Yu X, Yao N, Zhou Z, Fan X, Zhang L, Piao M, Li S, Yang D, et al. (2018) Reconstituting Arabidopsis CRY2 signaling pathway in mammalian cells reveals regulation of transcription by direct binding of CRY2 to DNA. Cell Rep 24: 585–593 e584 [DOI] [PubMed] [Google Scholar]
- Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M(2010) Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22: 2156–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin C(2007a) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19: 3146–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C(2010) The cryptochrome blue light receptors. Arabidopsis Book 8: e0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C(2007b) Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci USA 104: 7289–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q, Li C(2015) Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 27: 2814–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao QP, Zhu JD, Li NN, Wang XN, Zhao X, Zhang X(2019) Cryptochrome-mediated hypocotyl phototropism was regulated antagonistically by gibberellic acid and sucrose in Arabidopsis. J Integr Plant Biol 62: 614–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang L, Zeng L, Zhang C, Ma H(2015) Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev 29: 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, He SB, Li L, Yang HQ(2014) Auxin inhibits stomatal development through MONOPTEROS repression of a mobile peptide gene STOMAGEN in mesophyll. Proc Natl Acad Sci USA 111: E3015–E3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zicola J, Liu L, Tänzler P, Turck F(2019) Targeted DNA methylation represses two enhancers of FLOWERING LOCUS T in Arabidopsis thaliana. Nat Plants 5: 300–307 [DOI] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C(2011) Blue-light–dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]