An intrinsically disordered protein interacts with HYPONASTIC LEAVES1 and ARGONAUTE1 in a post-miRNA processing complex to promote ARGONAUTE1 stability and miRNA activity.
Abstract
In plants, small RNAs are loaded into ARGONAUTE (AGO) proteins to fulfill their regulatory functions. MicroRNAs (miRNAs), one of the most abundant classes of endogenous small RNAs, are preferentially loaded into AGO1. Such loading, long believed to happen exclusively in the cytoplasm, was recently proposed to also occur in the nucleus. Here, we identified CONSTITUTIVE ALTERATIONS IN THE SMALL RNAS PATHWAYS9 (CARP9), a nuclear-localized, intrinsically disordered protein, as a factor promoting miRNA activity in Arabidopsis (Arabidopsis thaliana). Mutations in the CARP9-encoding gene led to a mild reduction of miRNAs levels, impaired gene silencing, and characteristic morphological defects, including young leaf serration and altered flowering time. Intriguingly, we found that CARP9 was able to interact with HYPONASTIC LEAVES1 (HYL1), but not with other proteins of the miRNA biogenesis machinery. In the same way, CARP9 appeared to interact with mature miRNA, but not with primary miRNA, positioning it after miRNA processing in the miRNA pathway. CARP9 was also able to interact with AGO1, promoting its interaction with HYL1 to facilitate miRNA loading in AGO1. Plants deficient in CARP9 displayed reduced levels of AGO1-loaded miRNAs, partial retention of miRNA in the nucleus, and reduced levels of AGO1. Collectively, our data suggest that CARP9 might modulate HYL1-AGO1 cross talk, acting as a scaffold for the formation of a nuclear post-primary miRNA-processing complex that includes at least HYL1, AGO1, and HEAT SHOCK PROTEIN 90. In such a complex, CARP9 stabilizes AGO1 and mature miRNAs, allowing the proper loading of miRNAs in the effector complex.
Posttranscriptional regulation of gene expression mediated by microRNAs (miRNAs) controls numerous processes during plant development and response to the environment (Li et al., 2017). In Arabidopsis (Arabidopsis thaliana), RNA polymerase II transcribes MIRNA loci to primary miRNAs (pri-miRNAs), which are processed into mature ∼21 nucleotides miRNA duplexes by multiple and sequential cuts (Bologna et al., 2013; Zhu et al., 2013; Moro et al., 2018). The RNase-III endonuclease DICER-LIKE1 (DCL1), together with the RNA binding protein HYPONASTIC LEAVES1 (HYL1) and the zinc finger protein SERRATE (SE), recognizes and processes pri-miRNAs into mature miRNA duplexes (Kurihara and Watanabe, 2004; Dong et al., 2008). In addition, numerous accessory proteins were identified lately to regulate the miRNA biogenesis at different stages (Rogers and Chen, 2012; Achkar et al., 2016; Manavella et al., 2019). HYL1 was proposed to remain bound to the mature miRNA duplexes and interact with HUA ENHANCER1 (HEN1), acting as a scaffold to ensure miRNA methylation (Li et al., 2005a; Yang et al., 2010; Baranauskė et al., 2015). Mature miRNAs, potentially still bound by HYL1, are then loaded into an ARGONAUTE (AGO) protein, with AGO1 as the main effector of the miRNA pathway in plants, to fulfill their functions (Fang and Qi, 2016). Loading of miRNA duplexes into AGO1 and the assembly of the RNA-induced silencing complex were initially thought to be cytoplasmic-exclusive processes (Bologna and Voinnet, 2014). This was inferred from metazoan cells where miRNA loading into the RNA-induced silencing complex occurs in the cytosol, and the fact that the homolog of the human EXPORTIN5 of Arabidopsis, known as HASTY (HST), showed impaired miRNA accumulation (Park et al., 2005). A recent report demonstrated that AGO1 is at least partially loaded with miRNA duplexes in the nucleus and then exported to the cytosol as an AGO1:miRNA complex (Bologna et al., 2018). It is not clear if the nuclear AGO1 loading is dependent on HYL1. However, some evidence suggested that this is a certain possibility: (1) AGO1 colocalizes with HYL1 in nuclear speckles (Fang and Spector, 2007); (2) HYL1 is required for the proper miRNA loading and strand selection by AGO1 (Eamens et al., 2009; Manavella et al., 2012); and (3) HYL1 remains bound to mature miRNAs after processing (Yang et al., 2010). ENHANCED MIRNA ACTIVITY1 (EMA1) and TRANSPORTIN1 (TRN1) were also shown to interact with AGO1 and modulate miRNAs loading into AGO1 (Wang et al., 2011; Cui et al., 2016). The finding of the nuclear loading of AGO1 not only created a new model for miRNAs export but also rationalized the recently identified nuclear functions of AGO1 (Dolata et al., 2016; Schalk et al., 2017; Liu et al., 2018).
Using a forward genetic screening, we identified CONSTITUTIVE ALTERATIONS IN THE SMALL RNAS PATHWAYS9 (CARP9), a predicted intrinsically disordered protein (IDP), as a new partner of the miRNA pathway. Mutations in CARP9 produced morphological alterations, a mild reduction in the miRNA accumulation, and impaired gene silencing. We found that CARP9 interacts with HYL1 in discrete nuclear speckles promoting HYL1-AGO1 interaction. Our data suggest that CARP9 did not participate in pri-miRNA processing, but instead it is associated with HYL1 and mature miRNAs in a post-miRNA-processing complex. In such a complex, we found that CARP9 also interacts with AGO1 and HEAT SHOCK PROTEIN90 (HSP90). CARP9 mutants presented low levels of AGO1-associated miRNAs, a reduction of AGO1 levels, and an apparent depletion of miRNAs in the cytoplasm. All this evidence allows us to suggest that CARP9 could be acting as a scaffold protein, connecting HYL1 to AGO1 in a post-pri-miRNAs processing complex, ensuring AGO1 stability and thus leading to the proper loading of the AGO1:miRNA complexes, likely before its export to the cytosol.
RESULTS
Identification of CARP9 from a miRNA Activity Screen
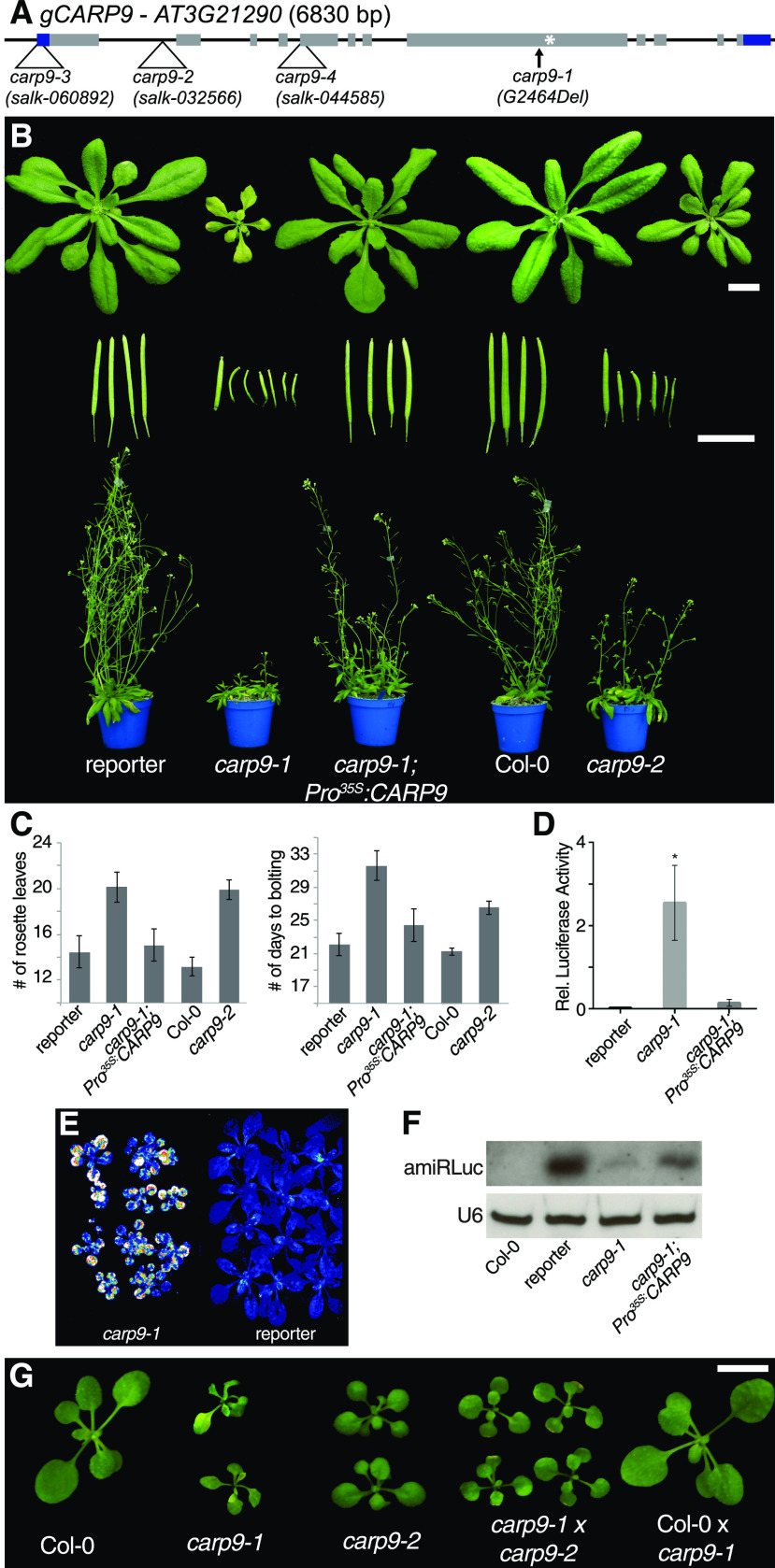
During the last years, we characterized several miRNA-deficient mutants isolated from a genetic screening based on the silencing of a luciferase reporter by an artificial miRNA (amiRLUC; Manavella et al., 2012; Francisco-Mangilet et al., 2015; Karlsson et al., 2015; Ré et al., 2020). Here, using mapping by sequencing (Sun and Schneeberger, 2015), we localized the causal mutation in one of the isolated plants, named constitutive alterations in the small RNAs pathways 9 (CARP9), to a small region of chromosome 3 (Supplemental Fig. S1A). Within this region, we detected a single-nucleotide deletion (G2464Del, AT3G21290, Chr3:7486786, The Arabidopsis Information Resource 10) resulting in a premature stop codon and an aberrant C-terminal region of 34 amino acids (Fig. 1A). Compared to the reporter plants, this mutant allele (named carp9-1) showed a reduced stature, irregular young leaf edges, pale green color, shorter and twisted siliques, a delay in flowering time, and observable defects in the architecture of flowers, including shorter stamens (Fig. 1, B and C; Supplemental Fig. S1, B and C). Transformation of the mutant with a wild-type copy of CARP9 cDNA, either expressed under the 35S promoter or its native regulatory region, fully reverted the morphological phenotype of the mutant (Fig. 1, B and C; Supplemental Fig. S1, B and C). In the overexpressing lines, we selected lines with low expression levels for the analyses. Additionally, the carp9-1 mutant showed elevated luciferase activity with a severe reduction of the amiRLUC levels (Fig. 1, D–F). All morphological defects of carp9-1 were also observed, to a lesser degree, in a mutant with a transfer DNA (T-DNA) insertion within the first intron of this gene named carp9-2 (Fig. 1, A–C; Supplemental Fig. S1B). To further confirm that the mutation in AT3G21290 was causing the observed phenotype, we crossed carp9-1 and carp9-2 mutants and analyzed the progeny. The compound heterozygote offspring showed similar phenotypes to the parental lines, confirming that the mutation in CARP9 was the cause of the observed phenotypes (Fig. 1G). In the case of a third allele, carp9-3 (SALK_060892), bearing a T-DNA insertion in the 5′ untranslated region (UTR) region of CARP9 approximately 100 bp upstream the ATG codon (Fig. 1A), we were not able to recover any homozygous line, suggesting that plants are not viable when this mutation is homozygous. Interestingly, we did not find signs of embryonic abortion or impaired germination/growth in carp9-3+/− progeny (Supplemental Fig. S1, D and E). Nevertheless, all genotyped seedlings were either wild type or heterozygous for carp9-3 mutation, suggesting a problem with the male gametes. Supporting this scenario, we were not able to detect the carp9-3 insertion in the offspring of wild-type plants fertilized with pollen of carp9-3 heterozygous plants. Similarly, we failed to detect the homozygous insertion in the SALK_044585 allele (carp9-4), which is annotated to have an insertion in the CARP9 fifth exon (Fig. 1A). All together, the analysis of these alleles suggests that CARP9 is essential for the plant development and points to carp9-1 and carp9-2 as hypomorphic alleles. In this sense, carp9-1 may still produce a truncated but active protein, while carp9-2 partially reduces CARP9 transcription as indicated by reverse transcription-quantitative PCR (RT-qPCR) analysis (Supplemental Fig. S1F).
Figure 1.
Characterization of carp9 mutants. A, Gene structure of CARP9 showing single nucleotide deletion of the carp9-1 allele and T-DNA insertion sites in carp9-2, carp9-3, carp9-4, and carp9-5. Black boxes and lines represent exons and introns, respectively; gray boxes represent coding sequence, while blue boxed shows 5′ and 3′ UTR regions. A white asterisk marks the position of the stop codon that results from the frameshift caused by the carp9-1 single nucleotide deletion (G2464Del). B, Phenotypic characterization of carp9 mutants, control lines (reporter and Col-0), and carp9-1 mutants complemented by the overexpression of the CARP9 cDNA (carp9-1; Pro35S:CARP9). Twenty-one-day-old plants, fully expanded siliques, and 40-d-old plants are displayed. Bars = 1 cm. Plants were imaged individually, digitally extracted, and mounted on a single black background panel to facilitate comparison and observation. C, Analysis of the flowering time of control, carp9 mutant, and complemented lines grown in long-day photoperiod as measured by the number of rosette leaves or the number of days to bolting. Error bars show means ± se (n ≥ 15). D, Bioluminescence activity, quantified by a luminometer, of 12 leave discs belonging to 20-d-old carp9-1 mutants and reporter plants. Error bars show means ± se (n ≥ 12). Asterisks indicate significance by two-tailed, unpaired t test Holm-Sidak corrected (*P < 0.05). E, Bioluminescence activity as measured with a CCD camera, in 20-d-old carp9-1 mutants and reporter plants. Luminescence intensity is color scaled from low (blue) to high (white). Two pots containing 12 plants each were imaged individually, digitally extracted, mounted in a single black background panel, and displayed in the figure. F, RNA blots for detecting amiRLuc. U6 was used as a loading control. G, Phenotypic features of 18-d-old carp9 mutants, control lines, and carp9-1/carp9-2 compound heterozygous mutants. Bar = 1 cm. Plants were imaged individually, digitally extracted, and mounted in a single black background panel to facilitate comparison and observation.
CARP9 Mutants Displayed Impaired Activity of the miRNA Pathway
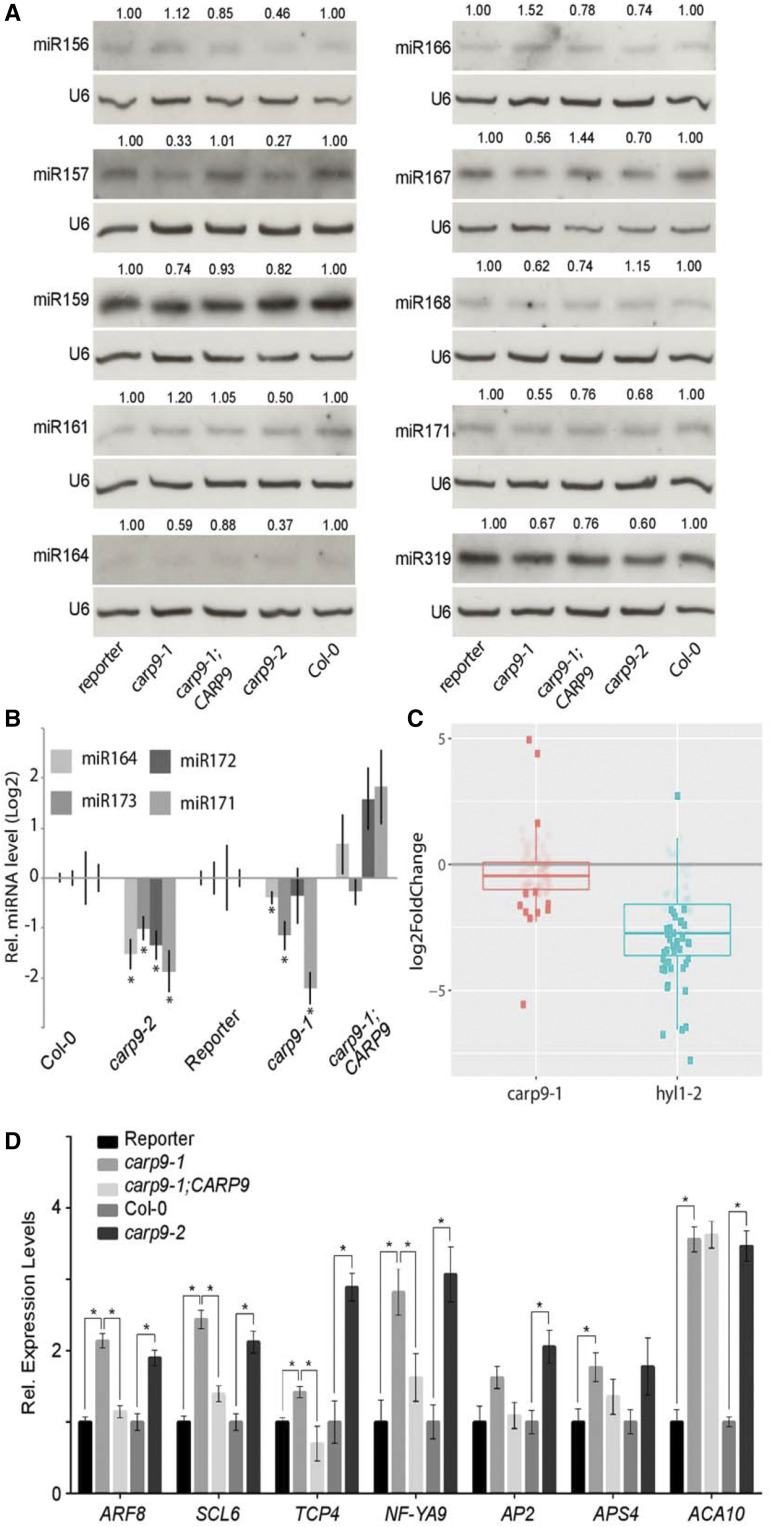
As carp9-1 was isolated as a miRNA-deficient mutant, we explored the endogenous levels of several miRNAs by RNA blots and RT-qPCR. The quantification of miRNAs revealed a mild reduction of several tested miRNAs in the single mutants, as well as in the carp9-1/carp9-2 compound heterozygous plants (Fig. 2, A and B; Supplemental Fig. S2A). As expected, the overexpression of CARP9 in carp9-1 restored miRNA levels (Fig. 2, A and B). Aiming to explore the genome-wide profile of miRNA accumulation in the carp9 mutant, we performed Illumina small RNA sequencing of carp9-1 mutants. The results were consistent across replicates (Supplemental Fig. S2B) and supported our northern blot results. We observed an overall reduction of miRNAs and miRNAs passenger strand accumulation, although to a lesser degree than hyl1-2 plants, used as controls of impaired miRNA production (Fig. 2C; Supplemental Fig. S2C; Supplemental Table S1). Only the nuclear-acting miR845 (Borges et al., 2018), miR391, and miR827 appeared overaccumulated in carp9-1 (Supplemental Table S1).
Figure 2.
CARP9 mutants present impaired miRNA activity. A, RNA blots for detecting endogenous miRNAs. U6 was used as a loading control. The relative abundance of each miRNA, indicated above each band, was calculated by measuring the band intensity using ImageJ and relativized to the corresponding control plant (reporter for carp9-1 and complemented plants, and Col-0 for carp9-2). B, miRNA levels, as measured by RT-qPCR, in mutants and control lines. Error bars show means ± 2× se (n ≥ 4). Asterisks indicate significance by two-tailed, unpaired FDR-corrected t test (*P < 0.05) were considered significant. C, Mean expression levels of individual miRNAs in carp9-1 and hyl1-2 plants relative to Col-0 plants. Horizontal segments indicate the median of the expression levels. Each dot corresponds to a single miRNA or collapsed miRNA family. Dark and light dots show differentially (false discovery rate adjusted P-value < 0.05) and not differentially accumulated miRNAs, respectively. D, Expression of miRNA targets in control and mutant plants as measured by RT-qPCR. Error bars show means ± 2× se (n ≥ 4). Asterisks indicate significance by two-tailed, unpaired t test Holm-Sidak corrected (*P < 0.05).
Coincidently with impaired activity of the miRNA pathway in the mutants, we observed an overaccumulation of several miRNA-targeted mRNAs in the single mutants and carp9-1/carp9-2 compound heterozygous plants (Fig. 2D; Supplemental Fig. S2, D and E). As expected, the 35S:CARP9 plants showed reduced miRNA target transcripts to wild-type levels (Fig. 2D; Supplemental Fig. S2D).
CARP9 Encodes an IDP of Unknown Function Conserved among Land Plants
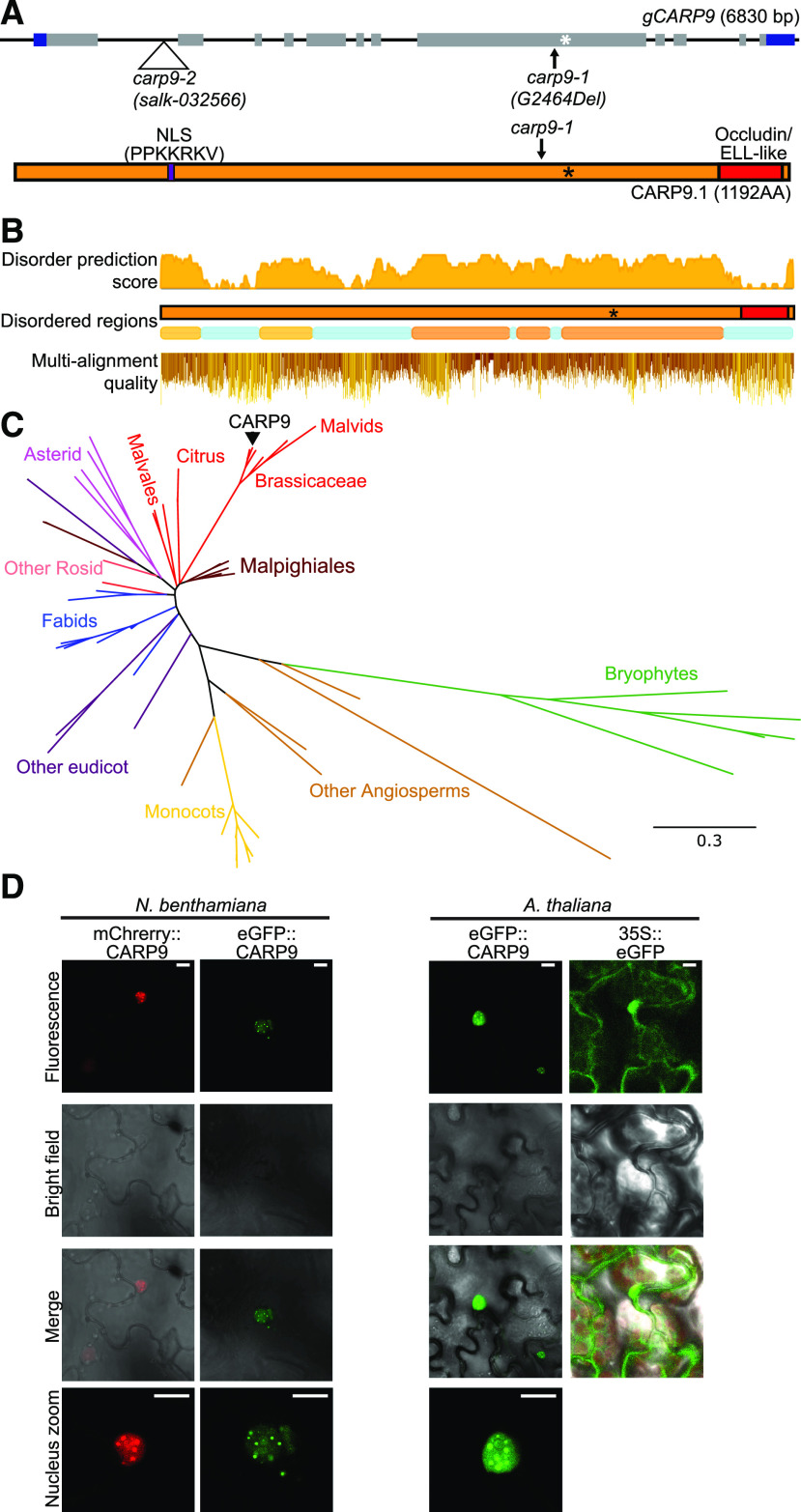
CARP9 encodes a 1,192-amino acid-long protein of unknown functions that includes a predicted nuclear localization signal (NLS) at position 238 to 245 (Fig. 3A). A single isoform is reported in the The Arabidopsis Information Resource 10 genome version (Arabidopsis Genome Initiative, 2000), while a second splicing variant is annotated in the Araport11 release (Cheng et al., 2017), but we were not able to detect it by RT-PCR and represent less than the 2% of splicing junctions in RNA sequencing experiments (Supplemental Fig. S3A). An analysis of the protein sequence searching for conserved homology domains revealed only the presence of an Occludin/elongation factor (ELL)-like domain (pfam E-value score, 8.71e-27; El-Gebali et al., 2019) in the C-terminal region, covering 8.4% of the total protein length (amino acids 1088–1187; Fig. 3A). Most of the eukaryotic RNA polymerase II ELLs contain this kind of domain and are thought to mediate protein interactions (Li et al., 2005b; Van Itallie and Anderson, 2018), but they are mostly unstudied in plants yet (Shilatifard et al., 1996). Besides this domain, a structure prediction indicated that CARP9 is an IDP with 61.1% of disordered regions (Fig. 3B; Potenza et al., 2015).
Figure 3.
Conservation analysis of CARP9 across the plant kingdom. A, Top, gene structure of CARP9 as shown in Figure 1A. Bottom, the CARP9 protein structure; in purple is marked a putative NSL signal and its amino acid sequence. The occluding/ELL-like domain is marked in red. B, Top and middle, disordered score, and regions in AtCARP9 amino acid sequence according to MobiDB (Piovesan et al., 2018). Bottom, amino acid alignment quality of CARP9-like genes using Jalview software (Waterhouse et al., 2009); positions are based on AtCARP9 full sequence. C, Phylogeny of CARP9-like genes in embryophytes. The unrooted consensus tree was generated using the maximum likelihood method. Colors represent different lineages of plant species, referenced in the figure. AtCARP9 is highlighted with a black arrow. Supplemental Figure S3B shows the fully annotated tree. D, Confocal microscopy images showing the nuclear localization of eGFP- and mCherry-tagged versions of CARP9 in Nicotiana benthamiana transiently transformed leaves (left) and stably transformed Arabidopsis plants (right). Scale bars = 5 μm.
Interestingly, proteins with long disordered regions followed by an occluding domain appeared to be exclusive of plant species, according to the InterPro database (Mitchell et al., 2019). In this sense, we performed a BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi) search of CARP9 in plant genomes. This search yielded that this gene is well conserved among embryophytes species (Fig. 3C). We could also track CARP9 orthologs in all embryophytes, including bryophytes, while they are absent in algae species, suggesting that CARP9 has evolved within land plants. Using the Maximum Likelihood method, we performed a phylogenetic analysis with these proteins (Fig. 3C; Supplemental Fig. S3B). The obtained phylogenetic tree is entirely consistent with the evolution of plant species, and we did not identify conserved duplication events for this gene. In most of the species, CARP9 orthologs correspond to a single-copy gene. Such a single copy of the gene may explain why some of the studied alleles are not viable in homozygosis, as redundancy may be absent for this gene. However, we cannot exclude partial functional redundancy with other IDPs. Protein alignment showed that the conserved regions include both the Occludin/ELL domain and the disordered region (Fig. 3B). Moreover, CARP9 paralogs are also predicted as disordered in all species. This suggests that both parts of the protein, the Occludin/ELL domain and the disordered region, might be necessary for their molecular function. IDPs are important for molecular recognition (Tompa et al., 2015) and work as a scaffold for many molecular interactions (Cortese et al., 2008). They are also particularly abundant in cellular membrane-less organelles such as nuclear bodies (Uversky et al., 2015). Furthermore, intrinsically disordered regions are essential in protein-RNA interactions, mediating both specific and nonspecific interactions (Varadi et al., 2015; Järvelin et al., 2016). Recently, Arabidopsis FLL2, also a disordered protein, was reported to be located in nuclear bodies promoting liquid-liquid phase separation (Fang et al., 2019). The fusion of CARP9 with the fluorescent proteins eGFP or mCherry, followed by confocal microscopy, revealed that the protein localizes in the nucleus, and particularly in nuclear bodies of unknown nature (Fig. 3D). Such nuclear entities could perfectly reflect specific liquid organelles or the well-known dicing bodies, subnuclear speckles of miRNA processing (Fang and Spector, 2007).
CARP9 Interacts with HYL1 but Does Not Affect miRNA Processing
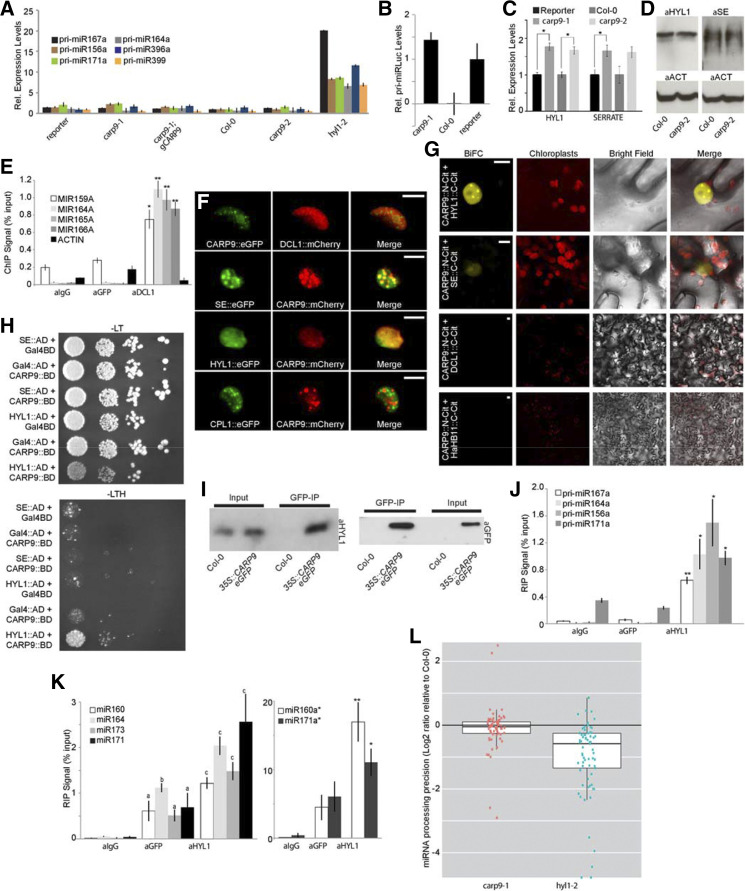
The reduced miRNA levels in carp9 mutants and the presence of an Occluddin/ELL domain in CARP9 led us to propose a putative role as an ELL controlling the transcription of genes encoding miRNA biogenesis factors or even of MIRNA genes. To test this hypothesis, we first quantified by RT-qPCR transcript levels of pri-miRNAs and genes encoding core components of the miRNA biogenesis machinery. In contrast to hyl1-2 mutants, used as a positive control for defective processing where pri-miRNAs overaccumulate, carp9-1 and carp9-2 presented normal levels of the miRNA precursors as well as pri-articfical miRLUC (Fig. 4, A and B). Among all tested genes encoding miRNA-related proteins, we observed a subtle but significant increase in HYL1 and SE transcript levels in both CARP9 mutant alleles (Fig. 4C). This result is opposed to our hypothesis of CARP9 acting as an elongator factor but compatible with a feedback response to the impaired miRNA activity in the mutants. However, immunoblot analysis of HYL1 and SE showed that these protein levels remained stable in the mutants (Fig. 4D). It has also been shown that DCL1 is recruited to MIRNA genes by the action of ELLs (Fang et al., 2015). Thus, it is possible that CARP9 does not affect MIRNA gene transcription itself but allows the recruitment of the processing machinery to the loci. We tested the capacity of CARP9 to interact with MIRNA loci by chromatin immunoprecipitation (ChIP)-qPCR using an eGFP-tagged version of the protein. Unlike DCL1, used as a positive control, CARP9 did not appear associated with the tested MIRNA loci (Fig. 4E). All together, these results suggested that CARP9 is not acting as an elongator factor to control miRNA activity. Alternatively, it is also possible that CARP9 is directly implicated in the processing of pri-miRNA, based on its particular subnuclear localization in speckles similar to dicing bodies (Fig. 3, D and E). Interestingly, transient expression of fluorescent-tagged CARP9 followed by confocal microscopy showed colocalization with SE and HYL1 in the same nuclear speckles (Fig. 4F). However, it did not colocalize, or only partially colocalized, with DCL1 and CTD-PHOSPHATASE-LIKE1 (CPL1), which dephosphorylates HYL1 to enhance pri-miRNA processing (Fig. 4F; Manavella et al., 2012). Coincidently, bimolecular fluorescence complementation (BiFC) assays showed fluorescent reconstitution when CARP9:N-CITRINE was confronted with HYL1 and SE fused to the C-CITRINE, but not with DCL1 (Fig. 4G; Supplemental Fig. S4). We used the nuclear transcription factor HaHB11 (Cabello et al., 2016) as a negative control for the assay. Since colocalization and BiFC assays indicate protein proximity but not necessarily interactions, we performed a yeast two-hybrid assay to evaluate direct protein-protein interaction among CARP9, HYL1, and SE. Only HYL1 among the tested proteins was able to interact with CARP9 (Fig. 4H). This interaction was further confirmed by a coimmunoprecipitation (co-IP) experiment using eGFP tagged CARP9 and an antibody against the endogenous HYL1 (Fig. 4I). The fact that HYL1 is known to interact with SE (Lobbes et al., 2006) may explain the positive signal detected between SE and CARP9 in BiFC assays. RNA immunoprecipitation (RIP)-qPCR assays using a eGFP-tagged CARP9 showed no association of this protein with pri-miRNAs, which appeared associated to HYL1 (Fig. 4J). Conversely, we detected abundant mature miRNAs associated with CARP9 in the IP samples (Fig. 4K). This association of CARP9 with mature miRNAs is probably through its interaction with HYL1, which is known to interact with miRNAs (Yang et al., 2010). The fact that HYL1 binds both pri-miRNA and mature miRNAs (Fig. 4, J and K; Yang et al., 2010) but CARP9 only appeared to interact with mature miRNA suggests that CARP9-HYL1 interaction occurs after pri-miRNA processing. Supporting this scenario, miRNA-processing precision, a feature impaired in hyl1-2 mutants and associated to its function as DCL1 accessory protein, was not affected in carp9-1 plants (Fig. 4L). MiRNA-processing precision was calculated by scoring the ratio of total miRNA-matched small RNA to the pool of imprecisely processed small RNAs, defined as those only partially matching the mature miRNA sequence. A late action of CARP9 in the pathway is also in line with the lack of interaction of CARP9 with DCL1 and CPL1, which are expected to act on early stages of the miRNA processing. This is also in agreement with the unchanged levels of pri-miRNAs in the mutant compared to wild type (Fig. 4A), which tend to accumulate in mutants impaired in pri-miRNA processing (Ben Chaabane et al., 2013), suggesting that CARP9 is not active during miRNA biogenesis.
Figure 4.
CARP9 interacts with HYL1 and mature miRNAs, but not with the miRNA processing machinery. A to C, Expression of pri-miRNA, pri-artificial miRLUC, HYL1, and SE in control and mutant plants as measured by RT-qPCR. D, HYL1 and SE quantification by immunoblot in samples extracted from carp9-2 and reporter plants. The detection of ACTIN was used as a loading control. E, ChIP experiment using either anti-GFP, anti-DCL1, or anti-IgG antibodies in plants that express a GFP-tagged version of CARP9 to detect MIRNAs loci associated with the proteins. Primers used for the amplification are listed in the Supplemental Table S3 and based on a previous report (Fang et al., 2015). ACTIN gene was used as control not targeted by CARP9 nor DCL1. F, Confocal microscopy images simultaneously showing the localization of CARP9 with DCL1, SE, HYL1, and CPL1 in transiently transformed N. benthamiana leaves. Scale bars = 5 μm. G, BiFC assay in N. benthamiana cells showing CARP9 interaction with HYL1 and se. Negative interactions are displayed in a wider magnification to show the negative interactions better. Positive interactions in a wider magnification are shown in Supplemental Figure S4. Scale bars = 5 μm. H, Interaction of CARP9 with HYL1, but not with se, as detected by yeast two-hybrid assays. GAL4 activation domain (AD); GAL4 DNA binding domain (BD); −LT, medium without Leu and Trp; −LTH, selective medium without Leu, Trp, and His. Each column shows a 1:10 serial dilution. I, CARP9-HYL1 interaction detected by co-IP assays. Leaves of Arabidopsis plants transformed with Pro-35S::CARP9-eGFP were immunoprecipitated using an anti-GFP antibody. Interacting HYL1 was identified using an antibody targeting the endogenous protein. J and K, pri-miRNAs (J) and mature miRNAs or miRNAs passender strands (miRNA*s; K) associated with CARP9, or HYL1, as quantified by RIP-RT-qPCR in samples extracted from plants expressing a GFP-tagged version of CARP9 and immunoprecipitated with either an anti-GFP, anti-HYL1, or anti-IgG antibodies. Values are given as a percentage to the qPCR signal detected in the input samples. L, Precisely processed miRNA reads at all highly expressed MIRNA loci. Each dot represents an individual miRNA; horizontal black bars indicate medians. miRNA levels in all samples are expressed as a ratio to the precisely processed miRNAs in Col-0 plants grown at 23°C in long-day (LD) photoperiod. In A to C, E, and J to K, error bars show means ± 2× se (n ≥ 4). Asterisks indicate significance by two-tailed, unpaired t test (*P < 0.05 and **P < 0.01).
CARP9 Interacts with AGO1; Affects Its Stability and miRNA Loading
The potential post-miRNA-processing interaction of CARP9 with HYL1 and its nuclear localization suggested that this protein might affect one of the nuclear steps of the miRNA pathway downstream of biogenesis. This could be the case of the recently reported nuclear loading of some miRNAs into AGO1 (Bologna et al., 2018). To explore this possibility, we first tested the capacity of CARP9 to interact with AGO1 by BiFC and co-IP assays. Both experiments showed that CARP9 can interact with AGO1, and this interaction occurs in the nucleus, as observed in the microscopy assays (Fig. 5. A and B; Supplemental Fig. S5, A and B). The nuclear transcription factors TCP15 and PIF4 (Ferrero et al., 2019) were used as negative controls for the assay. Interestingly, yeast two-hybrid assays showed that CARP9 also interacts with the known AGO1-partner HSP90, which can locate in the nucleus (Fig. 5C; Bologna et al., 2018). Contrarily, BiFC assays revealed that CARP9 do not interact with SQUINT (SQN), which, according to our data, interacts with AGO1 exclusively in the cytoplasm (Supplemental Fig. S5C). As CARP9 interacts with HYL1 and this protein was shown to interact with AGO1 in the nucleus (Fang and Spector, 2007), it is possible that CARP9, as many IDPs, acts as a scaffold for AGO1-HYL1 interaction. To test this hypothesis, we performed AGO1-HYL1 co-IP experiments in wild type and carp9-2 mutants treated with the proteasome inhibitor MG132. MG132 treatments equalize AGO1 levels, which are altered in carp9 mutants (Fig. 6A), allowing us a correct interpretation of the results. The experiment showed that AGO1-HYL1 interaction is partially impaired in the mutant background, suggesting that CARP9 facilitates the formation of a postprocessing miRNA complex containing AGO1, HYL1, and likely HSP90 (Fig. 5D; Supplemental Fig. S5D).
Figure 5.
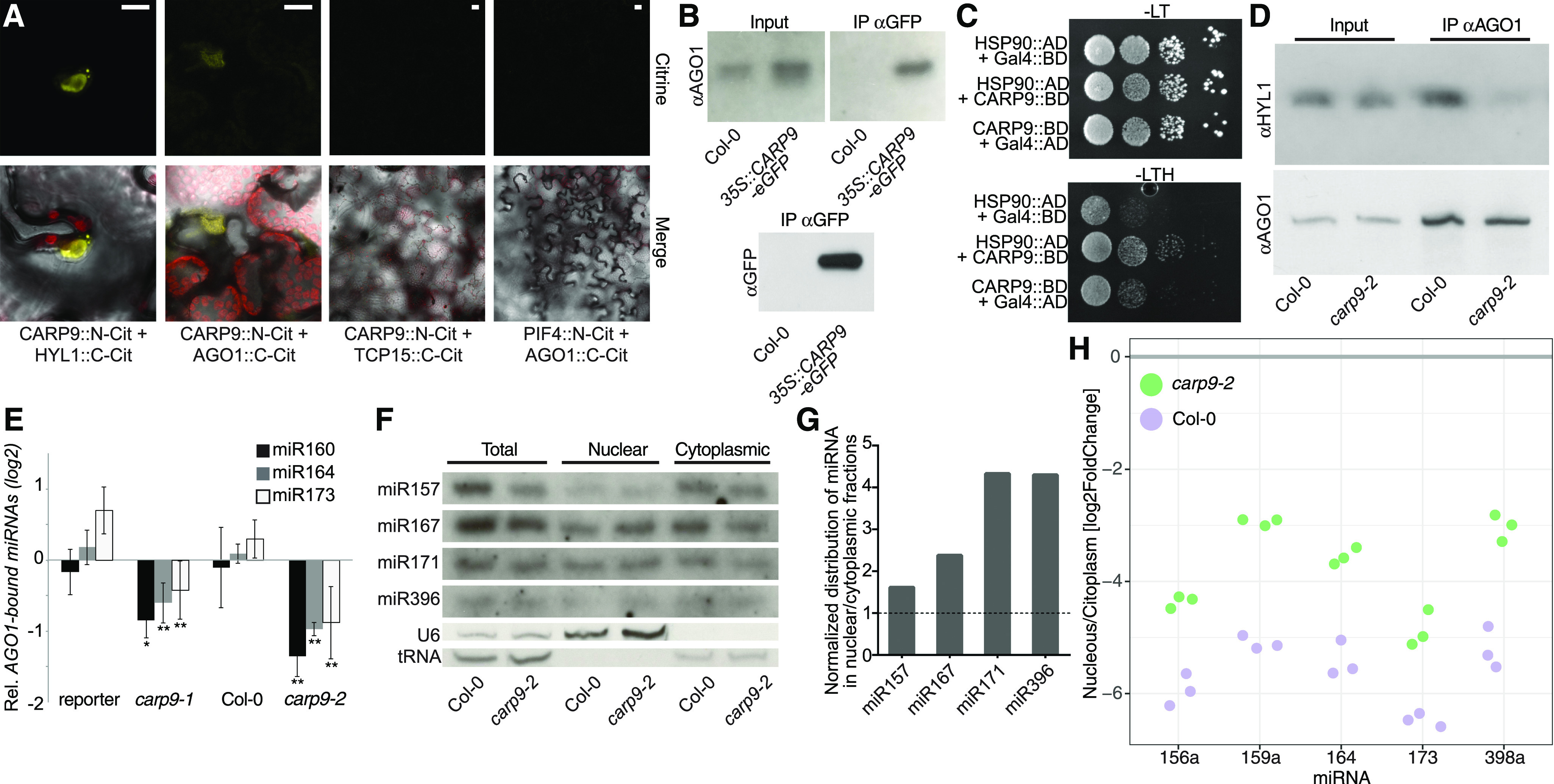
CARP9 interacts with AGO1 to modulate its nuclear miRNA loading. A, BiFC assay in N. benthamiana cells showing CAP9 interaction with AGO1 and with HYL1 as controls. The nuclear transcription factors PIF4 and TCP15 were used as negative controls and displayed in a wider magnification. Scale bars = 5 μm. B, Co-IP assays. Protein samples extracted from Col-0 wild-type plants or plants transformed with a 35S::CARP9-eGFP were immunoprecipitated using an anti-GFP antibody, AGO1-CARP9 interaction was then detected using an anti-AGO1 antibody. C, Interaction of CARP9 with HSP90 as detected by yeast two-hybrid assays. GAL4 activation domain (AD); GAL4 DNA binding domain (BD); −LT, medium without Leu and Trp; −LTH, selective medium without Leu, Trp, and His. Each column shows a 1:10 serial dilution. D, Co-IP assays. Protein samples extracted from Col-0 wild-type or carp9-2 plants were immunoprecipitated using an anti-AGO1 antibody, AGO1-HYL1 interaction was then detected using an anti-HYL1 antibody. AGO1 was detected to test IP efficiency. E, Relative amount of mature miRNA bound to AGO1 as measured by stem-loop RT-qPCR of samples immunoprecipitated using an anti-AGO1 antibody. Co-IPed miRNAs were normalized to the levels of the same miRNA in the input samples. For both mutants, the miRNA levels were then expressed as relative to their corresponding control. Error bars show means ± 2× se (n = 4). Asterisks indicate significance by two-tailed, unpaired t test (*P < 0.05 and **P < 0.01). No-antibody samples and ago1-36 mutant plants were used as negative controls for the IP experiment, not showing detectable signal in the assayed conditions. F, RNA blots for detecting miRNAs in different cell fractions. Quantification of U6 and tRNAs were used as a loading control and to monitor the purity of the nuclear/cytoplasmic fractions. G, Quantification of the miRNA distribution measured in F. Band intensity was quantified by ImageJ and normalized by the corresponding loading control. Distributions of miRNAs in the nuclear/cytoplasmic fractions were then expressed as relative to Col-0 (marked as a dashed line). H, Quantification of the miRNA distribution in nucleus versus cytoplasm fractions as measured by RT-qPCR. Each dot represents an independent replicate. Significant differences were tested with an ANOVA test: between miRNAs (P = 0.00484 and between genotypes, P= 0.00013).
Figure 6.
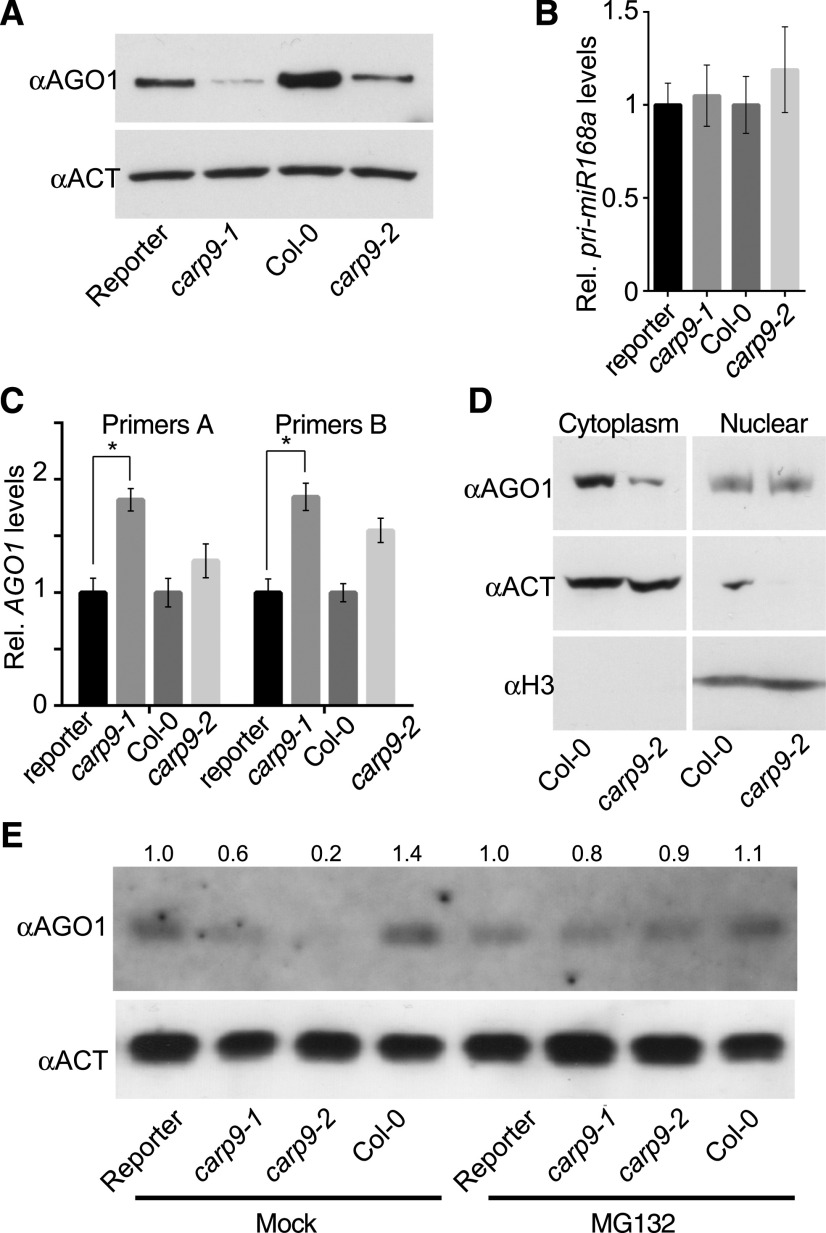
AGO1 stability is compromised in CARP9 mutants. A, AGO1 levels quantified by immunoblot in samples extracted from carp9 mutants and control plants. Levels of ACTIN were measured as a loading control. B and C, Expression of pri-miRNA168 (B) and AGO1 (C) as measured by RT-qPCR. AGO1 transcript levels were measured using two sets of primers; a pair amplifying the 3′ end of the transcript (Primers A) and a pair flanking the miR168 recognition site in the AGO1 mRNA (Primers B). Error bars show means ± 2× se (n = 4. Asterisks indicate significance by two-tailed, unpaired t test (*P < 0.05). D, AGO1 levels, as measured by immunoblots, in cytoplasmic or nuclear cell fractionated samples. ACTIN and Histone 3 (H3) were used to verify the purity of the fractions. E, Immunoblot quantification of AGO1 levels in mutant and control plants treated with the proteasome inhibitor MG132.
All together, these data suggest that CARP9 may act by stabilizing HYL1-AGO1 interaction and perhaps facilitating the loading of miRNAs into AGO1. To test whether AGO1 loading is affected in carp9 mutants, we immunoprecipitated AGO1 in wild-type and mutant plants treated with MG132 and scored the levels of associated miRNAs by RT-qPCR (RIP-qPCR). We observed a significant depletion of AGO1-associated miRNAs in the mutants when compared to wild-type plants, except for miR822, which is canonically loaded in the cytoplasm (Fig. 5E; Supplemental Fig. S5, E and F; Rajagopalan et al., 2006), suggesting that CARP9 is participating in the nuclear loading of miRNAs into AGO1 or at least stabilizing AGO1-miRNA association. This observation may explain the mild reduction in miRNA accumulation, as most likely carp9 mutation only affects the portion of miRNAs loaded in the nucleus but not the cytoplasmic loading. The impaired loading of miRNAs into AGO1 in carp9 mutants was also evident when we measured mature miRNA accumulation in nuclear and cytoplasmic fractions. RNA blot and RT-qPCR assays revealed that the nuclear portion of mature miRNAs is enriched in carp9-2, compatible with deficient AGO1 nuclear loading, and subsequent cytoplasm exportation of this miRNA fraction (Fig. 5, F–H; Supplemental Fig. S5G).
Intriguingly, we detected increased levels of AGO1 in the plants overexpressing eGFP:CARP9 construct (Fig. 5B; Supplemental Fig. S5B) and a reduction in carp9-1 and carp9-2 mutants when compared to their respective controls (Fig. 6A). Such changes in AGO1 levels cannot be attributed to the known regulation of AGO1 by miR168 (Vaucheret et al., 2004), as we did not observe an increment of miR168 or MIR168 transcript levels (Figs. 1E amd 6B; Supplemental Table S1). In contrast to AGO1 protein, AGO1 transcript levels, measured by RT-qPCR with primers designed toward the 3′ end of the mRNA (primer set A) or flanking the miR168 target site (primer set B), were higher in the mutants than in control plants, probably as a feedback response to the protein reduction or as a consequence of the lower miR168 activity (Fig. 6C; Supplemental Table S1). Nuclear/cytoplasm protein fractionation of wild-type and carp9-2 plants showed that the nuclear fraction of AGO1 is not reduced in the mutant, which is consistent with a possible cytoplasmic proteolytic degradation of AGO1 (Fig. 6D). Treatment of the mutant plants with the proteasome inhibitor MG132 partially reverted AGO1 reduction, confirming that this protein is being degraded in the mutants (Fig. 6E; Supplemental Fig. S6). Such reduced levels of AGO1 can be the cause, but also the consequence, of the deficient miRNA loading observed in the mutants, as unloaded AGO1 could become unstable and get degraded. Nevertheless, the most parsimonious explanation to our observations is that CARP9 is acting as a scaffold protein, promoting the formation of a nuclear postprocessing miRNA complex containing at least AGO1, HYL1, and HSP90. In the absence of CARP9, this hypothetical complex would be disrupted, thus altering AGO1 stability, the proper miRNA loading, and therefore the stability of miRNA duplex itself.
DISCUSSION
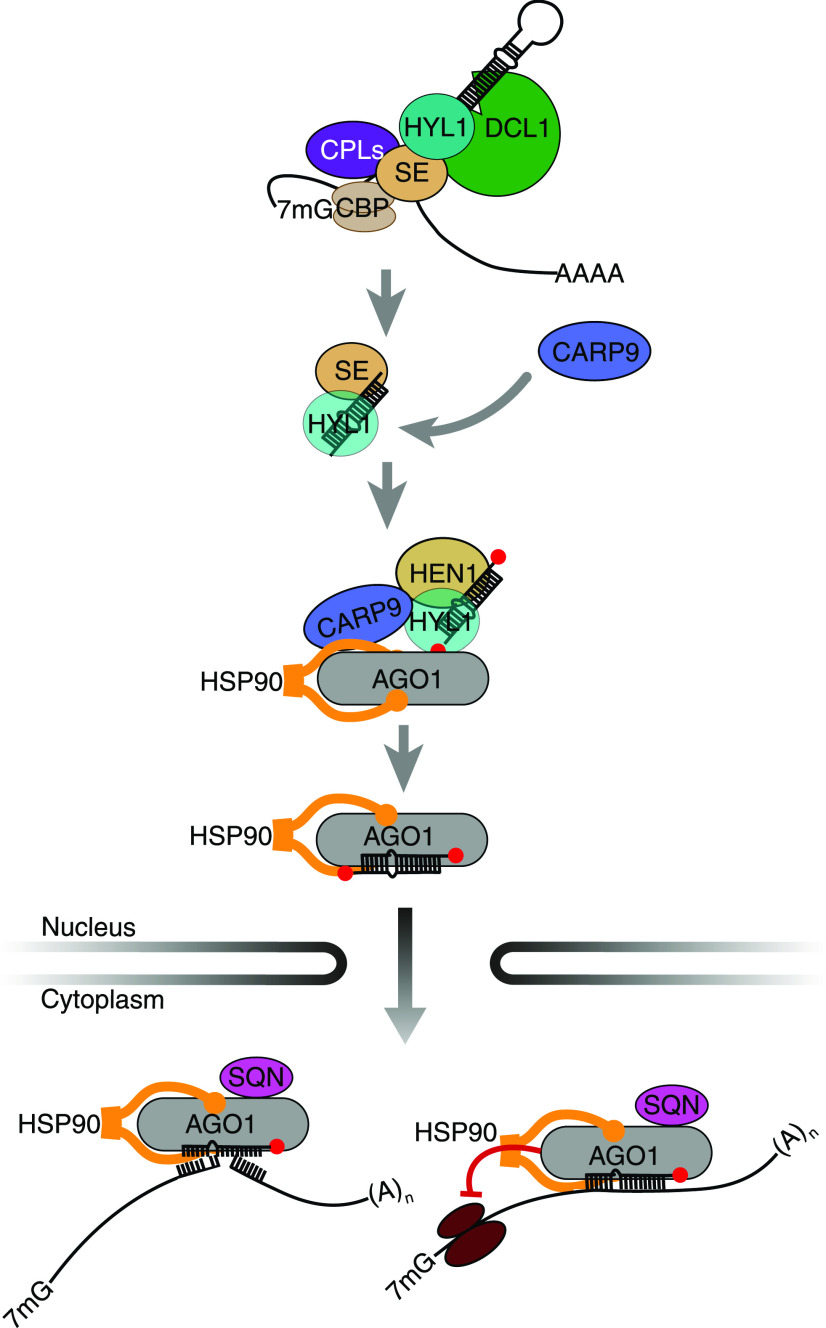
During the last years, our knowledge of the miRNA pathway in plants has grown exponentially. New functions were assigned to well-known proteins of the pathway, such as the chromatin association of AGO1 and DCL1 (Fang et al., 2015; Dolata et al., 2016; Liu et al., 2018), the role of these same proteins in DNA damage repair (Wei et al., 2012; Schalk et al., 2017), the transcriptional and epigenetic regulation of genes by SE (Ma et al., 2018; Speth et al., 2018), and the independence of some pri-miRNA of HYL1 at low temperatures (Ré et al., 2019), among other examples. A recent report demonstrated that a portion of miRNAs can be directly loaded into AGO1 inside the nucleus and exported as a complex (Bologna et al., 2018). This evidence contrasts the previous conception that miRNAs were exported to the cytoplasm by HASTY in order to be loaded into AGO1. The functional relevance of AGO1 nuclear loading is becoming important since AGO1 involvement in several nuclear processes were recently described (Dolata et al., 2016; Schalk et al., 2017; Liu et al., 2018). However, it remains unknown whether this nuclear loading of AGO1 is a passive process or if components of the processing machinery assist it. It has been previously shown that proper loading of AGO1, particularly the miRNA strand selection, is impaired in HYL1 mutants, suggesting that this processing factor might participate actively during AGO1 loading (Eamens et al., 2009; Manavella et al., 2012). Interestingly HEN1, which 2′-O-methylates mature miRNA, interacts with HYL1, but not with the processing factor SE (Baranauskė et al., 2015). This potentially also places HEN1 in a hypothetical postprocessing complex together with AGO1 and HYL1. The capacity of HYL1 to efficiently bind mature miRNAs in vitro (Yang et al., 2010) suggests that, after processing, mature miRNA might remain bound to HYL1 until loaded into AGO1. In this context, CARP9 appeared to act as a nexus among these proteins, facilitating the proper function of such a postprocessing complex (Fig. 7).
Figure 7.
A model for the role of CARP9 as a scaffold in a post-pri-miRNA processing and nuclear AGO1 loading complex. After the processing of pri-miRNAs by DCL1, the mature duplex remains associated with HYL1 and SE, which later is replaced by HEN1. CARP9 is recruited to this postprocessing complex by its binding to HYL1. Nuclear AGO1 is then associated with the complex and HSP90 interacting with CARP9, a process leading to the enhanced AGO1 stability and loading of AGO1 with the mature miRNAs. Loaded AGO1 is then exported to the cytosol to silence their target genes.
In this study, we describe CARP9, an IDP conserved among land plants, as a nuclear protein participating in miRNA activity. Interestingly, we found that CARP9 interacts with HYL1 and mature miRNAs, but not with the miRNA processing machinery or miRNA precursors, suggesting that this protein functions after the pri-miRNA-processing steps. This idea is strongly supported by a miRNA-processing precision analysis that showed that carp9-1 mutants, opposite to hyl1-2, displayed a normal processing activity (Fig. 4L).This discovery also supports previous reports proposing that HYL1 remains bound to the mature miRNA duplexes once they are produced (Fig. 5; Baranauskė et al., 2015). In this context, it is possible that after pri-miRNA processing, HYL1 transfers the mature miRNA duplexes to AGO1 in the nuclear speckles where they colocalize, a scenario compatible with the AGO1-loading defects in hyl1 mutants (Fang and Spector, 2007; Eamens et al., 2009; Manavella et al., 2012). Furthermore, HYL1 may facilitate AGO1 loading in the cytoplasm, where it is also located without reported functions (Cho et al., 2014; Achkar et al., 2018). However, and to the best of our knowledge, there is no evidence of direct interaction between HYL1 and AGO1 in the cytoplasm. Besides its interaction with HYL1, we found that CARP9 also interacts with AGO1, contributing to the miRNA loading process (Fig. 5). It is common that IDPs, such as CARP9, act as chaperones assisting the folding of other proteins, allowing interactions and even preventing their aggregation (Tompa and Kovacs, 2010). In such a scenario, CARP9 could act as a scaffold promoting AGO1-HYL1 interaction, providing stability to AGO1 and to the mature miRNA duplexes and consequently promoting loading of miRNA into AGO1. As HYL1 is also found in the cell cytoplasm, where it undergoes protein degradation during the night (Cho et al., 2014; Achkar et al., 2018), it is possible that this RNA binding protein escorts AGO1 during miRNA nuclear export, a process that would exclude CARP9 as we found it to be exclusively nuclear.
It was interesting to notice that carp9 mutant alleles displayed reduced AGO1-bound miRNAs, miRNA levels, and morphological defects compatible with miRNA-deficient mutants. However, these defects are milder than those observed in mutants of core proteins of the pathway, such as HYL1 and AGO1. Redundancy with proteins with high sequence similarity is unlikely to explain this observation, as CARP9 appeared as a single gene in most of the genomes of plants considered here, including Arabidopsis. Furthermore, plants homozygous for carp9 null alleles appeared not to be viable. It is more likely that a reduced activity of CARP9 in carp9-1 and carp9-2 alleles explains the observations. The premature stop codon in carp9-1 would allow a large portion of the protein to be translated, and the intronic T-DNA insertion of carp9-2 possibly represents a knockdown allele, as indicated by the reduced CARP9 transcripts in this mutant (Supplemental Fig. S3). Additionally, as CARP9 is likely to only affect the nuclear loading of miRNAs, the large pool of cytoplasmic loaded miRNAs are not expected to be affected, thus restricting the effect of the protein to only a subpopulation of miRNAs.
The high degree of conservation of CARP9 among land plants is notable, suggesting a crucial role of this protein for plant homeostasis. The extraordinary level of amino acid conservation in the intrinsically disordered regions, which commonly tend to diverge rapidly during evolution, reinforces the idea of an evolutionary pressure to conserve this protein. Such inference might explain why we failed to identify homozygous T-DNA mutants in the coding sequence of this gene. The identification of strong but viable loss-of-function alleles of CARP9 could help to dimension the importance of this protein in plant development and miRNA activity in the future.
CARP9 represents a new component of the miRNA pathway that links the postprocessing machinery with the miRNA effector complex. Several questions remain open; for example, HEN1 was shown to interact with HYL1 post-processing (Baranauskė et al., 2015), opening the question of whether AGO1 is loaded with already methylated miRNA or if such process occurs after loading for the fraction of nuclear-loaded miRNAs. EMA1 and TRN1 were also shown to interact with AGO1, modulating miRNA loading (Wang et al., 2011; Cui et al., 2016). However, it remains to be addressed whether these proteins also participate in the nuclear AGO1-loading process interacting with HYL1 and CARP9 or have a different role. Similarly, we showed that CARP9 interacts with AGO1 and HYL1 to facilitate loading, but the biochemical activity of CARP9 during this process is unclear. Perhaps the most intriguing question lays in the observed destabilization of AGO1 in carp9 mutants. How this process is triggered and accomplished are questions we still need to answer. In particular, it will be essential to understand how AGO1 degradation, CARP9-HYL1 interaction, and miRNA loading are interconnected.
We found three significant effects of carp9 mutations: a mild reduction in miRNA levels, a reduction in AGO1 loading, and a reduction in AGO1 protein levels. Interestingly, each of these observations can be explained in light of the other ones. In this sense, miRNA reduction and impaired AGO1 loading can be the consequence of AGO1 destabilization. It is also possible that impaired AGO1 loading destabilizes AGO1 and miRNAs. Finally, if CARP9 is directly involved in stabilizing the miRNA duplex after processing, we could also expect to see impaired AGO1 loading and stability. Perhaps the first scenario is the least likely, as AGO1 loading is impaired in carp9 even when MG132 treatments block protein degradation. Nevertheless, we cannot exclude any of these possibilities, and further studies will be necessary to discriminate these alternatives and to understand the role of CARP9 in HYL1-AGO1 cross talk precisely.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) reporter and mutant plants were grown at 23°C on soil in long-day photoperiod (16 h of light/8 h of dark). Alternatively, plants were grown on plates containing 2.2 g L−1 of Murashige and Skoog (MS) medium (pH 5.7) and 0.6% (w/v) agar in long-day conditions. Seeds were disinfected with 10% (v/v) bleach and 0.1% (w/v) SDS and stratified in 0.1% (w/v) agar for 3 d at 4°C before sowing. Mutant lines carp9-2 (SALK_032566), carp9-3 (SALK_060892), carp9-4 (SALK_044585), carp9-5 (WiscDsLox358B04), and hyl1-2 (SALK_064863) were obtained from the Arabidopsis Biological Resource Center stock center. Col-0 miRNA activity reporter lines (reporter) used for ethyl methanesulfonate mutagenesis, and thus as carp9-1 control, were previously described (Manavella et al., 2012). Transgenic lines were grown on MS plates with 50 mg mL−1 kanamycin.
DNA Constructs and Plant Transformation
The CARP9, HSP90, and SQN coding region sequences, with or without stop codon, were amplified and cloned into pEntr/D-TOPO or pCR8GW-TOPO entry vector (Thermo Fisher Scientific). The CARP9 promoter region (2558 bp upstream of the transcription start site) was amplified by PCR, fused to CARP9 cDNA fragment by PCR, and cloned into pEntr/D-TOPO entry vector. eGFP and mCherry fusion constructs were obtained by recombination of the entry clones into modified pGREEN vectors under the control of the Cauliflower mosaic virus 35S promoter. An untagged cDNA copy of CARP9 under a 35S promoter was used to rescue carp9-1 mutants. Yeast two-hybrid constructs were obtained by cloning the specific cDNAs into pEntr/D-TOPO, followed by recombination into the pDEST32 or pDEST22 vectors (Life Technologies). Refer to Supplemental Table S2 for a detailed list of constructs used in this work. Arabidopsis transgenic seedlings were selected using 50 mg mL−1 kanamycin on plates. At least 15 independent pooled T1 seedlings were used for quantitative measurement of transgenic lines. Transient infiltration of Nicotiana benthamiana leaves was performed as described previously (de Felippes and Weigel, 2010). We were unable to directly transform carp9 mutants by floral dip. Thus, all carp9 transgenic plants were obtained by crossing the mutants with transgenic Col-0 plants and then recovering the homozygosis on the mutant alleles. In the case of the experiments performed with overexpression lines, we only used plants with low expression levels of CARP9 and pooled lines to minimize variability.
RNA Analysis
Total RNA was extracted from 15-d-old plants using TRIzol reagent (Life Technologies). For RNA blots, 1 to 5 µg of total RNA were resolved in 17% (v/v) polyacrylamide gels under denaturing conditions (7 m urea) and then transferred to HyBond-N+ charged nylon membranes (Amersham) by semidry electroblotting (Tomassi et al., 2017). RNA was covalently fixed to membranes in a UV crosslinker. Membranes were hybridized overnight with DNA oligonucleotide probes labeled with a second-generation DIG oligonucleotide 3′ end labeling kit (Roche); the signal was detected using CSPD ready-to-use solution (Roche), by exposure to Amersham hyperfilm ECL (GE Healthcare Life Sciences). ImageJ was used to analyze the band intensity of small RNA blots as integrated pixel density, using the intensity of U6 bands to normalize sample loading. Reverse transcriptase reactions were performed using 1 µg of DNaseI-treated total RNA (Thermo Fisher Scientific) using the RevertAid RT reverse transcription kit (Thermo Fisher Scientific). RT-qPCRs were performed using three independent biological replicates of pooled seedlings, and ACTIN2/8 (At3g18780/At1g49240) were used as a housekeeping loading control. Stem-loop RT-qPCRs for miRNA quantification were performed as previously described (Kramer, 2011). Averages from biological triplicates and SE were calculated from 2−ΔΔCt values, and the error displayed as two times SE. Each replicate was treated as independent samples for statistical analysis. Statistical differences between samples were determined by an unpaired, two-tailed t-test analysis and corrected with Holm-Sidak method for multiple pair comparisons. All quantifications were repeated twice in independent experiments. See Supplemental Table S3 for oligonucleotide primer and probe details.
Small RNA Sequencing
Small RNA libraries were prepared as indicated by the TruSeq small RNA library prep kit (Illumina) using biological triplicates of the reporter lines and hyl1-2 mutants and duplicates of carp9-1 mutants. Fifty nanograms of small RNAs purified with the ZR small-RNA PAGE recovery kit (Zymo Research) were used as input for the library preparation. Small RNA libraries size selections were performed using the BluePippin system (SAGE Science). Single-end Illumina sequencing was performed with a HiSeq3000 apparatus. Small RNA reads were first processed to remove 3′ adapters using cutadapt version 1.9.1 (https://github.com/marcelm/cutadapt/) and then mapped using bowtie version 1.1.2 (http://bowtie-bio.sourceforge.net/). Reproducibility was tested by computing the Spearman correlation of the miRNA counts per million between all samples, converting this correlation to a distance (1-Spearman Rho) and performing a hierarchical clustering of the samples, with the “complete” agglomeration method, to show the degree of similarity between them. The references used were the databases for hairpin and mature miRNAs for Arabidopsis from miRBase (release 21), in the latter mature miRNAs with identical sequences were collapsed into single miRNAs. Additionally, reads were mapped to the Arabidopsis genome using the same software. For the differential expression analysis of the miRNAs, only reads mapping to the full-length mature miRNAs were considered, and primary alignments of reads mapping to the sense strand were counted (filtering with “samtools view-F 272”). Counts per miRNA were used as input for baySeq version 2.8.0 (https://www.bioconductor.org/packages/2.8/bioc/html/baySeq.html) to perform the differential expression analysis. For this, miRNAs with low expression levels (less than 10 counts in all samples) were discarded, and size factors were set according to the total number of reads mapping to the genome for each sample. Graphics and statistical analyses were performed in the R statistical programming environment (R Core Team, https://cran.r-project.org/) with the ggplot package. miRNA processing precision was calculated by scoring the ratio of total miRNA-matched small RNA to the pool of imprecisely processed small RNAs, defined as those only partially matching the mature miRNA sequence. All data reported in this paper is available at the European Nucleotide Archive (ENA), PRJEB37499.
Protein Analysis
For immunoblot analysis, proteins were extracted from 15-d-old pooled plants (n = 5) with 100 µL extraction buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1 mm EDTA, 10% [v/v] glycerol, 1 mm dithiothreitol, and one tablet complete protease inhibitor cocktail [Roche]) per 100 mg of grind tissue. Proteins were resolved in 8% (w/v) SDS-PAGE gels (running buffer, 25 mm Tris-base; 192 mm glycin; 0.1% [w/v] SDS) and transferred using a standard wet tank blotting (blotting buffer, 2.5 mm Tris-base; 19.2 mm Gly; 10% [v/v] methanol) to polyvinylidene difluoride membrane (Amersham). Thermo Scientific PageRuler prestained protein ladder was used to determine the Mr of the bands and to confirm transfer efficiency. AGO1, HYL1, and SE were detected using a polyclonal antibody targeting the endogenous Arabidopsis protein (Agrisera AS09527, AS06136, and AS09532; dilution 1:10,000 each). ACTIN8 (Agrisera AS132640; dilution 1:10,000), HISTONE3 (Agrisera AS10710; dilution 1:10,000), or Coomassie Blue staining were used as loading controls in different experiments. horseradish peroxidase-conjugated polyclonal antirabbit IgG (Agrisera AS09602; dilution 1:20,000) was used to detect primary antibodies. Signal was detected using enhanced chemiluminescence plus western blotting substrate (Thermo Fisher Scientific). Experiments were repeated at least twice. For co-IP assays, eGFP-CARP9 was immunoprecipitated with an anti-GFP antibody (Abcam ab290, dilution 1:1000) from samples extracted from transgenic Arabidopsis flowers transformed with 35S::eGFP:CARP9, including wild-type Col-0 flowers as a negative control, and AGO1 was immunoprecipitated with an anti-AGO1 antibody (Agrisera, dilution 1:1000) from samples extracted from pooled (n = 5) 15-d-old carp9-1, carp9-2, reporter, and wild-type Col-0 plants, using Sure Beads Protein-A magnetic beads (Bio-Rad) following the manufacturer’s instruction. eGFP:CARP was then detected in the input and IP fraction by immunoblot using an anti-GFP antibody (Abcam ab290, dilution 1:10,000). HYL1 and AGO1 were detected in the input and IP fraction using the antibodies previously described. Yeast two-hybrid assays were performed with the ProQuest two-hybrid system (Thermo Fisher Scientific). Selection medium containing 2.5 mm of 3-amino-1,2,4-triazole was used to detect interactions reducing autoactivation. CARP9 fusions to the N-terminal and C-terminal fragments of citrine, eGFP, or mCherry were used for BiFC assays and protein localization in transiently transformed N. benthamiana leaves. Fluorescent protein visualization and imaging were performed using a Leica TCS SP8 confocal microscope. The excitation wavelengths were 488, 514, and 552 nm, and emission was collected at 500 to 530 nm, 525 to 560 nm, and 600 to 630 nm for eGFP, mCitrine, and mCherry, respectively. For luciferase activity assays, 12 5-mm discs cut from mature leaves were individually embedded in 100 mm d-Luciferin potassium salt solution and bioluminescence quantified using a Fluoroskan Ascent FL plate luminometer (Thermo Scientific). Means and se were calculated (n = 12) and P values of less than 0.05 in a two-tailed, unpaired t test, Holm-Sidak-corrected, were considered significant. Alternatively, luciferase activity was detected in plants sprayed with 100 mm d-Luciferin potassium salt solution using an Orca 2-BT cooled charge-coupled device camera with 5 min integration time (Hamamatsu Photonics).
Nuclear-Cytoplasmic Fractionation
Nuclear-cytoplasmic fractionation was performed following a protocol previously described (Wang et al., 2011). In brief, samples of pooled 15-d-old plants (2.5 g) were ground on ice with 2 mL g−1 of lysis buffer (20 mm Tris-HCl pH 7.5, 20 mm KCl, 2 mm EDTA, 2.5 mm MgCl2, 25% [v/v] glycerol, 250 mm Suc, and 5 mm dithiothreitol) supplemented with 200 µm phenylmethylsulfonyl fluoride (only for proteins fractionations). The homogenate was filtered through a layer of Miracloth, and the flow-through was centrifuged at 1500g for 10 min. The supernatant (cytoplasmic fraction) was centrifuged at 10,000g and 4°C for 10 min and collected. The pellet was washed three to five times with 5 mL of NRBT buffer (20 mm Tris-HCl, pH 7.5, 25% [v/v] glycerol, 2.5 mm MgCl2, and 0.2% [v/v] Triton X-100) and then resuspended with 500 µL of NRB2 (20 mm Tris-HCl, pH 7.5, 0.25 m Suc, 10 mm MgCl2, 0.5% [v/v] Triton X-100, and 5 mm β-mercaptoethanol) supplemented with complete protease inhibitor cocktail (Roche, only for protein fractionations). The resuspension was carefully pipetted on top of 700 µL of NRB3 buffer (20 mm Tris-HCl, pH 7.5, 1.7 m Suc, 10 mm MgCl2, 0.5% [v/v] Triton X-100, and 5 mm β-mercaptoethanol, supplemented with complete protease inhibitor cocktail). The obtained Suc gradient was centrifuged at 16,000g and 4°C for 1 to 3 min. For protein extraction, the pellet was resuspended in 100 µL lysis buffer and sonicated in a Bioruptor Pico water bath (10 cycles of 30-s on/30-s off pulses at high intensity; Diagenode). After centrifugation at 16,000g for 10 min at 4°C, the supernatant was collected as the nuclear fraction. For RNA extraction, the pellet was resuspended in 200 µL lysis buffer, and then 1 mL of TRIzol reagent (Life Technologies) was added, as to the cytoplasmic fraction, followed then by a standard protocol for RNA extraction. As a quality and loading control for the fractionation, ACTIN8 and a tRNA probe (for RNA quantifications) were used as cytoplasmic markers, while histone H3 and U6 RNA (for RNA quantifications) were used as nuclear markers.
RIP and ChIP Assays
RIP assays to detect mature miRNA or pri-miRNAs bound to eGFP::CARP9, HYL1, and AGO1 were performed using four independent biological replicates following a reported protocol with a few modifications and scaled down to 50% of the volumes (Carbonell, 2017). Immunoprecipitation was performed to 4 g of tissue (flowers) using anti-GFP (AS152987, dilution 1:250), anti-HYL1 (AS06136, dilution 1:500), and anti-AGO1 (AS09527, dilution 1:500). In the case of AGO1 RIP experiments, 15-d-old UV-cross-linked plants, grown on MS agar plates and treated with MG132 for 24 h, were used as starting material. In all cases, Sure Beads Protein-A magnetic beads (Bio-Rad) were used for the immunoprecipitation. RNA was finally extracted from the IP fraction by a regular TRIzol extraction. Associated RNA quantifications were performed as described in the “RNA Analysis” section of the “Materials and Methods.” ChIP assays of eGFP::CARP9- and DCL1-associated loci were performed using anti-GFP (AS152987, dilution 1:250) and anti-DCL1 (AS122102, dilution 1:200). We first performed nuclei enriched of samples obtained from 3 g of seedlings following the same protocol described above for the cell fractioning assay. Extracted nuclei were then resuspended in 500 µL nuclei lysis buffer (50 mm Tris-HCl, pH 8, 10 mm EDTA, 1% [w/v] SDS, 1 mm phenylmethylsulfonyl fluoride, 1% [w/v] complete protease inhibitors [Roche]). Chromatin was sheared using a Bioruptor Pico (Diagenode; 10 cycles 30” ON, 30” OFF). After fragmentation, nuclear debris was pelleted and the supernatant diluted 10-fold with ChIP dilution buffer (1.1% [v/v] Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8, 167 mm NaCl). Immunoprecipitation was carried out at 4°C overnight with 100 μL of washed Sure Beads Protein-A magnetic beads (Bio-Rad) and the appropriate amount of antibody. After washing five times, the beads were resuspended in 100 μL of TE, and 1 μL of 20 mg mL−1 Proteinase K was added. Samples were incubated at 43°C for 1 h, and the protease was inactivated by incubation at 95°C for 10 min. Samples were centrifuged 1 min at maximum speed, and 1.5 μL was used for qPCR reactions. Negative controls were performed with samples in which specific antibodies were not included. Values were expressed as a % of the input signal for the same measured miRNA.
Phylogenetic Analysis
CARP9-like protein sequences of representative plant species were retrieved from Phytozome (Goodstein et al., 2012) using AtCARP9 full amino acid sequence as a query for BLASTP search with default parameters. Protein sequences with low sequence similarity were discarded (query coverage < 30%, E-value > 1.1-10). The resulting sequences were aligned using the MAFFT G-INS-1 iterative method (Katoh and Standley, 2013) and automatically trimmed using the TrimAI webserver (Capella-Gutiérrez et al., 2009) with 0.9 gap threshold fraction. Splicing variants were manually removed. Phylogenetic analysis was performed using the maximum likelihood method with IQTree default parameters (Trifinopoulos et al., 2016) using a Shimodaira-Hasegawalike approximate likelihood ratio test. The consensus tree was obtained with all compatible groups and visualized using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
CARP9 Conservation Analysis
To test conservation of CARP9 in algae species, protein sequences of AtCARP9 and Marchantia polymorpha CARP9 were used as a query for BLASTP search against NCBI nonredundant database. Domain architecture searches were made using InterProScan (Mitchell et al., 2019). Amino acid disorder score and regions were obtained from MobiDB (Piovesan et al., 2018) using full protein sequences. Alignment quality was computed with Jalview (Waterhouse et al., 2009) using the same alignment obtained for phylogenetic analysis. NLS was predicted using the cNLS Mapper (http://nls-mapper.iab.keio.ac.jp) with 7.0 as the cutoff score.
Accession Numbers
Sequence data from this article can be found in the GenBank data libraries under accession numbers AT3G21290 (CARP9); AT1G09700 (HYL1); AT1G48410 (AGO1); AT2G15790 (SQN); AT2G27100 (SE); AT1G01040 (DCL1); AT4G21670 (CPL1); AT1G69690 (TCP15); AT2G43010 (PIF4); AT5G56010 (HSP90).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotype of carp9 mutants and rescued plants.
Supplemental Figure S2. Impact of CARP9 mutation on the miRNA pathway.
Supplemental Figure S3. Conservation of CARP9 among plants.
Supplemental Figure S4. CARP9 interacts with HYL1.
Supplemental Figure S5. CARP9 interacts with AGO1.
Supplemental Figure S6. CARP9 mutations impair AGO1 stability.
Supplemental Table S1. Differentially accumulated miRNAs as shown by small RNA sequencing data analysis of carp9-1 mutants.
Supplemental Table S2. Transgenes and plasmids.
Supplemental Table S3. DNA oligonucleotide primers and probes.
Acknowledgments
We would like to thank Raquel L. Chan and Virginia Miguel for sharing the HaHB11::C-Citrine construct used as a negative control for BiFC assays. CARP9 was named in honor of Club Atlético River Plate from Argentina and Enzo Francescoli. D.A.R., J.E.M., D.A.C., A.L.A., and P.A.M. are members of CONICET; A.H.T., L.G., and F.R. are fellows of the same institution.
Footnotes
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica, Argentina (grant no. PICT 2014–1293/2017–0861), the Human Frontier Science Program, the Max Planck Society, and the International Centre for Genetic Engineering and Biotechnology.
References
- Achkar NP, Cambiagno DA, Manavella PA(2016) miRNA biogenesis: A dynamic pathway. Trends Plant Sci 21: 1034–1044 [DOI] [PubMed] [Google Scholar]
- Achkar NP, Cho SK, Poulsen C, Arce AL, Re DA, Giudicatti AJ, Karayekov E, Ryu MY, Choi SW, Harholt J, et al. (2018) A quick HYL1-dependent reactivation of microRNA production is required for a proper developmental response after extended periods of light deprivation. Dev Cell 46: 236–247.e6 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Baranauskė S, Mickutė M, Plotnikova A, Finke A, Venclovas Č, Klimašauskas S, Vilkaitis G(2015) Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic Acids Res 43: 2802–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Chaabane S, Liu R, Chinnusamy V, Kwon Y, Park JH, Kim SY, Zhu JK, Yang SW, Lee BH(2013) STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res 41: 1984–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Iselin R, Abriata LA, Sarazin A, Pumplin N, Jay F, Grentzinger T, Dal Peraro M, Voinnet O(2018) Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant microRNA pathway. Mol Cell 69: 709–719.e5 [DOI] [PubMed] [Google Scholar]
- Bologna NG, Schapire AL, Zhai J, Chorostecki U, Boisbouvier J, Meyers BC, Palatnik JF(2013) Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res 23: 1675–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bologna NG, Voinnet O(2014) The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol 65: 473–503 [DOI] [PubMed] [Google Scholar]
- Borges F, Parent JS, van Ex F, Wolff P, Martínez G, Köhler C, Martienssen RA(2018) Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat Genet 50: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello JV, Giacomelli JI, Piattoni CV, Iglesias AA, Chan RL(2016) The sunflower transcription factor HaHB11 improves yield, biomass and tolerance to flooding in transgenic Arabidopsis plants. J Biotechnol 222: 73–83 [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T(2009) trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A.(2017) Immunoprecipitation and high-throughput sequencing of ARGONAUTE-bound target RNAs from plants. Methods Mol Biol 1640: 93–112 [DOI] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD(2017) Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Cho SK, Ben Chaabane S, Shah P, Poulsen CP, Yang SW(2014) COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat Commun 5: 5867. [DOI] [PubMed] [Google Scholar]
- Cortese MS, Uversky VN, Dunker AK(2008) Intrinsic disorder in scaffold proteins: Getting more from less. Prog Biophys Mol Biol 98: 85–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Fang X, Qi Y(2016) TRANSPORTIN1 promotes the association of microRNA with ARGONAUTE1 in Arabidopsis. Plant Cell 28: 2576–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes FF, Weigel D(2010) Transient assays for the analysis of miRNA processing and function. Methods Mol Biol 592: 255–264 [DOI] [PubMed] [Google Scholar]
- Dolata J, Bajczyk M, Bielewicz D, Niedojadlo K, Niedojadlo J, Pietrykowska H, Walczak W, Szweykowska-Kulinska Z, Jarmolowski A(2016) Salt stress reveals a new role for ARGONAUTE1 in miRNA biogenesis at the transcriptional and posttranscriptional levels. Plant Physiol 172: 297–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N(2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA 105: 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens AL, Smith NA, Curtin SJ, Wang MB, Waterhouse PM(2009) The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15: 2219–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. (2019) The Pfam protein families database in 2019. Nucleic Acids Res 47(D1): D427–D432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Cui Y, Li Y, Qi Y(2015) Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat Plants 1: 15075. [DOI] [PubMed] [Google Scholar]
- Fang X, Qi Y(2016) RNAi in plants: An Argonaute-centered view. Plant Cell 28: 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Wang L, Ishikawa R, Li Y, Fiedler M, Liu F, Calder G, Rowan B, Weigel D, Li P, et al. (2019) Arabidopsis FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. Nature 569: 265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL(2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17: 818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero LV, Viola IL, Ariel FD, Gonzalez DH(2019) Class I TCP transcription factors target the gibberellin biosynthesis gene GA20ox1 and the growth-promoting genes HBI1 and PRE6 during thermomorphogenic growth in Arabidopsis. Plant Cell Physiol 60: 1633–1645 [DOI] [PubMed] [Google Scholar]
- Francisco-Mangilet AG, Karlsson P, Kim MH, Eo HJ, Oh SA, Kim JH, Kulcheski FR, Park SK, Manavella PA(2015) THO2, a core member of the THO/TREX complex, is required for microRNA production in Arabidopsis. Plant J 82: 1018–1029 [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvelin AI, Noerenberg M, Davis I, Castello A(2016) The new (dis)order in RNA regulation. Cell Commun Signal 14: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson P, Christie MD, Seymour DK, Wang H, Wang X, Hagmann J, Kulcheski F, Manavella PA(2015) KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proc Natl Acad Sci USA 112: 14096–14101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM(2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MF.(2011) Stem-loop RT-qPCR for miRNAs. Curr Protoc Mol Biol 95: 15.10.1-15.10.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y(2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X(2005a) Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Castillo-González C, Yu B, Zhang X(2017) The functions of plant small RNAs in development and in stress responses. Plant J 90: 654–670 [DOI] [PubMed] [Google Scholar]
- Li Y, Fanning AS, Anderson JM, Lavie A(2005b) Structure of the conserved cytoplasmic C-terminal domain of occludin: identification of the ZO-1 binding surface. J Mol Biol 352: 151–164 [DOI] [PubMed] [Google Scholar]
- Liu C, Xin Y, Xu L, Cai Z, Xue Y, Liu Y, Xie D, Liu Y, Qi Y(2018) Arabidopsis ARGONAUTE 1 binds chromatin to promote gene transcription in response to hormones and stresses. Dev Cell 44: 348–361.e7 [DOI] [PubMed] [Google Scholar]
- Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J(2006) SERRATE: A new player on the plant microRNA scene. EMBO Rep 7: 1052–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Castillo-Gonzalez C, Wang Z, Sun D, Hu X, Shen X, Potok ME, Zhang X(2018) Arabidopsis serrate coordinates histone methyltransferases ATXR5/6 and RNA processing factor RDR6 to regulate transposon expression. Dev Cell 45: 769–784 [DOI] [PubMed] [Google Scholar]
- Manavella PA, Hagmann J, Ott F, Laubinger S, Franz M, Macek B, Weigel D(2012) Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell 151: 859–870 [DOI] [PubMed] [Google Scholar]
- Manavella PA, Yang SW, Palatnik J(2019) Keep calm and carry on: miRNA biogenesis under stress. Plant J 99: 832–843 [DOI] [PubMed] [Google Scholar]
- Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, et al. (2019) InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47(D1): D351–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro B, Chorostecki U, Arikit S, Suarez IP, Höbartner C, Rasia RM, Meyers BC, Palatnik JF(2018) Efficiency and precision of microRNA biogenesis modes in plants. Nucleic Acids Res 46: 10709–10723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS(2005) Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA 102: 3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan D, Tabaro F, Paladin L, Necci M, Micetic I, Camilloni C, Davey N, Dosztányi Z, Mészáros B, Monzon AM, et al. (2018) MobiDB 3.0: More annotations for intrinsic disorder, conformational diversity and interactions in proteins. Nucleic Acids Res 46(D1): D471–D476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potenza E, Di Domenico T, Walsh I, Tosatto SC(2015) MobiDB 2.0: An improved database of intrinsically disordered and mobile proteins. Nucleic Acids Res 43: D315–D320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP(2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20: 3407–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ré DA, Cambiagno DA, Arce AL, Tomassi AH, Giustozzi M, Casati P, Ariel FD, Manavella PA(2020) CURLY LEAF regulates microRNA activity by controlling ARGONAUTE 1 degradation in plants. Mol Plant 13: 72–87 [DOI] [PubMed] [Google Scholar]
- Ré DA, Lang PLM, Yones C, Arce AL, Stegmayer G, Milone D, Manavella PA(2019) Alternative use of miRNA-biogenesis co-factors in plants at low temperatures. Development 146: dev172932. [DOI] [PubMed] [Google Scholar]
- Rogers K, Chen X(2012) microRNA biogenesis and turnover in plants. Cold Spring Harb Symp Quant Biol 77: 183–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk C, Cognat V, Graindorge S, Vincent T, Voinnet O, Molinier J(2017) Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc Natl Acad Sci USA 114: E2965–E2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW(1996) An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271: 1873–1876 [DOI] [PubMed] [Google Scholar]
- Speth C, Szabo EX, Martinho C, Collani S, Zur Oven-Krockhaus S, Richter S, Droste-Borel I, Macek B, Stierhof YD, Schmid M, et al. (2018) Arabidopsis RNA processing factor SERRATE regulates the transcription of intronless genes. eLife 7: e37078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Schneeberger K(2015) SHOREmap v3.0: Fast and accurate identification of causal mutations from forward genetic screens. Methods Mol Biol 1284: 381–395 [DOI] [PubMed] [Google Scholar]
- Tomassi AH, Gagliardi D, Cambiagno DA, Manavella PA(2017) Nonradioactive detection of small RNAs using digoxigenin-labeled probes. Methods Mol Biol 1640: 199–210 [DOI] [PubMed] [Google Scholar]
- Tompa P, Kovacs D(2010) Intrinsically disordered chaperones in plants and animals. Biochem Cell Biol 88: 167–174 [DOI] [PubMed] [Google Scholar]
- Tompa P, Schad E, Tantos A, Kalmar L(2015) Intrinsically disordered proteins: Emerging interaction specialists. Curr Opin Struct Biol 35: 49–59 [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ(2016) W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44(W1): W232–W235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Kuznetsova IM, Turoverov KK, Zaslavsky B(2015) Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett 589: 15–22 [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM(2018) Phosphorylation of tight junction transmembrane proteins: Many sites, much to do. Tissue Barriers 6: e1382671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M, Zsolyomi F, Guharoy M, Tompa P(2015) Functional advantages of conserved intrinsic disorder in RNA-binding proteins. PLoS One 10: e0139731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP(2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ye R, Xin Y, Fang X, Li C, Shi H, Zhou X, Qi Y(2011) An importin β protein negatively regulates microRNA activity in Arabidopsis. Plant Cell 23: 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ(2009) Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y(2012) A role for small RNAs in DNA double-strand break repair. Cell 149: 101–112 [DOI] [PubMed] [Google Scholar]
- Yang SW, Chen HY, Yang J, Machida S, Chua NH, Yuan YA(2010) Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 18: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zhou Y, Castillo-González C, Lu A, Ge C, Zhao YT, Duan L, Li Z, Axtell MJ, Wang XJ, et al. (2013) Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat Struct Mol Biol 20: 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]