The pssRNAit web server designs functional small interfering RNAs for precise gene silencing in plants and provides a path to effectively study gene functions and phenotypes in plants.
Abstract
We report an advanced web server, the plant-specific small noncoding RNA interference tool pssRNAit, which can be used to design a pool of small interfering RNAs (siRNAs) for highly effective, specific, and nontoxic gene silencing in plants. In developing this tool, we integrated the transcript dataset of plants, several rules governing gene silencing, and a series of computational models of the biological mechanism of the RNA interference (RNAi) pathway. The designed pool of siRNAs can be used to construct a long double-strand RNA and expressed through virus-induced gene silencing (VIGS) or synthetic transacting siRNA vectors for gene silencing. We demonstrated the performance of pssRNAit by designing and expressing the VIGS constructs to silence Phytoene desaturase (PDS) or a ribosomal protein-encoding gene, RPL10 (QM), in Nicotiana benthamiana. We analyzed the expression levels of predicted intended-target and off-target genes using reverse transcription quantitative PCR. We further conducted an RNA-sequencing-based transcriptome analysis to assess genome-wide off-target gene silencing triggered by the fragments that were designed by pssRNAit, targeting different homologous regions of the PDS gene. Our analyses confirmed the high accuracy of siRNA constructs designed using pssRNAit. The pssRNAit server, freely available at https://plantgrn.noble.org/pssRNAit/, supports the design of highly effective and specific RNAi, VIGS, or synthetic transacting siRNA constructs for high-throughput functional genomics and trait improvement in >160 plant species.
RNA interference (RNAi) is a powerful tool for silencing genes of interest for plant functional genomics and trait improvement. The two popular ways to achieve RNAi in plants are stable transformation of plants using hairpin RNAi vectors or virus-induced gene silencing (VIGS; Pandey et al., 2015). RNAi is a multistep cellular pathway of posttranscriptional gene silencing in which Dicer Like enzyme (DCL) cleaves the long double-strand RNA (dsRNA) into a series of ∼21 nucleotide duplexes of small interfering RNAs (siRNAs; Fukudome and Fukuhara, 2017), then delivers the guide strand of siRNAs to an Argonaute (AGO) protein, a catalytic engine of the RNA-inducing silencing complex (RISC), to form an active RISC that further binds to the complementary region of mRNA, resulting in targeted mRNA cleavage and degradation (Ahmed et al., 2009; Mi et al., 2008; Wilson and Doudna, 2013).
The silencing efficacy of siRNA is mainly determined by its sequence features, class of AGO protein catalytic, and binding affinity with the intended mRNA, while the silencing specificity of siRNA is mainly determined by its lack of binding affinity with the nonintended mRNAs (Ahmed et al., 2015; Jagla et al., 2005). The induction of RNAi in a plant cell is usually accomplished by expressing a 300- to 1,200-bp gene-specific sequence tag (GST) as long dsRNA (Hilson et al., 2004). The silencing specificity of GST is achieved by <70% identity with off-target genes using BLASTN. Based on sequence complementarity between siRNA and mRNA and the preferences of nucleotides in siRNA, computational tools were developed to design long-dsRNA RNAi constructs for plant gene silencing (Thareau et al., 2003; Naito et al., 2005; Xu et al., 2006; Ahmed et al., 2015). However, the designed long dsRNAs are processed at undefined positions by DCL, resulting in a pool of siRNAs with mixed and unknown features, such as noneffective, nonspecific, and toxic siRNAs, which may cause nonfunctional and off-target gene silencing (Jackson et al., 2003; Jackson et al., 2006; Burchard et al., 2009), as well as cell toxicity (Yi et al., 2005; Grimm et al., 2006; Olejniczak et al., 2010). Furthermore, a slight variation at the terminal sequence of a siRNA during cleavage by DCL can switch the loading properties from antisense to sense siRNA to RISC, creating more off-target silencing effects (Khvorova et al., 2003; Schwarz et al., 2003; Ahmed et al., 2009). These serious limitations in the designing of RNAi constructs have not been adequately addressed. Basically, available RNAi designing tools mainly focused on the rules of binding of siRNA to its targets, though rules governing other steps of the plant RNAi pathway, including processing and biogenesis of siRNAs, loading of siRNAs into the RISC complex, binding of siRNAs to nonintended targets, and toxicity associated with siRNAs, are also crucial for determining effective, specific and nontoxic gene silencing in plants. Thus, the rules governing all of these crucial steps of RNAi need to be implemented to achieve better RNAi designing tools.
Here, we report pssRNAit, a pipeline to design RNAi constructs for effective, specific, and nontoxic gene silencing in plants. We developed a series of computational models, methods, and tools that mimic and analyze the biological actions of every important step in the plant RNAi pathway to facilitate and streamline RNAi construct design in the pssRNAit back-end pipeline, including (1) designing highly effective siRNAs using a support vector machine (SVM) model; (2) estimating the accessibility of target sites by analyzing the two-dimensional structure of mRNA using the RNAup program in the Vienna Package to design siRNAs against the accessible regions of the target mRNA (Mückstein et al., 2006); (3) applying the RISC binder to select the siRNA whose antisense strand, but not sense strand, will be loaded by the RISC machinery to execute gene silencing (Ahmed et al., 2009); and (4) predicting off-target genes in species-specific transcript libraries using psRNATarget (Dai and Zhao, 2011; Dai et al., 2018). Furthermore, the back-end pipeline intelligently selects a pool of siRNAs with a minimum of off-targets, if any, mimicking a function where canonical microRNA (miRNA), along with isomeric sibling miRNAs (isomiRs) with terminal variants, bind to the same target mRNA and improve the gene silencing specificity (Ahmed and Zhao, 2011). The performance of pssRNAit was evaluated using two independent datasets of siRNA, which give a high correlation coefficient between actual and predicted efficacy. Furthermore, by doing VIGS and RNA-sequencing (RNA-seq) in Nicotiana benthamiana, we experimentally confirmed that the RNAi constructs designed by pssRNAit achieved highly effective and specific gene silencing in plants.
RESULTS
SVM Model for Effective siRNA Prediction
We developed a series of regression models using the SVMlight V5.00 package (Joachims, 1999) following a 5-fold cross-validation schema on a training dataset that consists of 2,182 siRNAs (TR2182). The developed models were further tested on an independent test dataset consisting of 249 siRNAs (TE249). Both the training and independent test datasets were obtained from the work published by Huesken et al. (2005). Under the 5-fold cross-validation schema, we achieved the best model with a correlation coefficient of 0.712 (R = 0.712, R2 = 0.504, mean absolute error [MAE] = 0.112, and root mean squared error [RMSE] = 0.141) between actual and predicted efficacy using hybrid features of siRNAs. The hybrid features for the SVM model consist of efficacy scores calculated by 12 different siRNA design models combined with the frequency of mono-, di-, and trinucleotides and a binary pattern that uses an input vector size of 180 (12 + 4 + 16 + 64 + 84). The performance of our model showed a significant increase in efficacy compared to the tool desiRm (R = 0.670, R2 = 0.448, MAE = 0.118, and RMSE = 0.148) on the same training dataset (Ahmed and Raghava, 2011). Our model was obtained on the SVM parameters with the radial-basis-function (RBF) kernel (g = 0.0001, c = 1, j = 1) and named pssRNAit2182. The performance of pssRNAit2182 was further evaluated on the independent dataset TE249 and achieved a correlation coefficient of 0.686 (R = 0.686, R2 = 0.466, MAE = 0.142, and RMSE = 0.122). Therefore, a final SVM model, which achieved a correlation coefficient of 0.709 (R = 0.709, R2 = 0.500, MAE = 0.113, and RMSE = 0.142) between actual and predicted efficacy, was developed using the whole curated dataset, TR2431 (TR2182 + TE249). Moreover, we evaluated our pssRNAit2431 with other methods on different benchmarking data consisting of 419 siRNAs (TE419) obtained from different sources (Ichihara et al., 2007; Ahmed and Raghava, 2011). Supplemental Table S1 provides detailed information about the training and testing data sources, and Supplemental Table S2 lists siRNA sequences and their silencing scores in the training and testing data. The performance of pssRNAit2431, which is based on the SVM model, is better than other well-known siRNA design algorithms on our combined training set (TR2431) and on an independent TE419 dataset (Table 1; Ichihara et al., 2007; Ahmed and Raghava, 2011 ).
Table 1. Performance of siRNA silencing efficacy of our model, pssRNAit, and five other tools on training dataset TR2431 and testing dataset TE419.
Values represent the Pearson correlation coefficient between observed and predicted siRNA efficacy. Dataset TR2431 was used to train the models, whereas dataset TE419 was used as an independent test set. The asterisk indicates the Pearson correlation coefficient of desiRm obtained for dataset TR2182.
| Algorithms | TR2431 | TE419 |
|---|---|---|
| i-Score | 0.635 | 0.557 |
| s-Biopredsi | 0.665 | 0.546 |
| ThermoComposition21 | 0.635 | 0.577 |
| DSIR | 0.687 | 0.554 |
| desiRm* | 0.670* | 0.558 |
| pssRNAit | 0.709 | 0.568 |
Implementation and Back-End Pipeline of pssRNAit
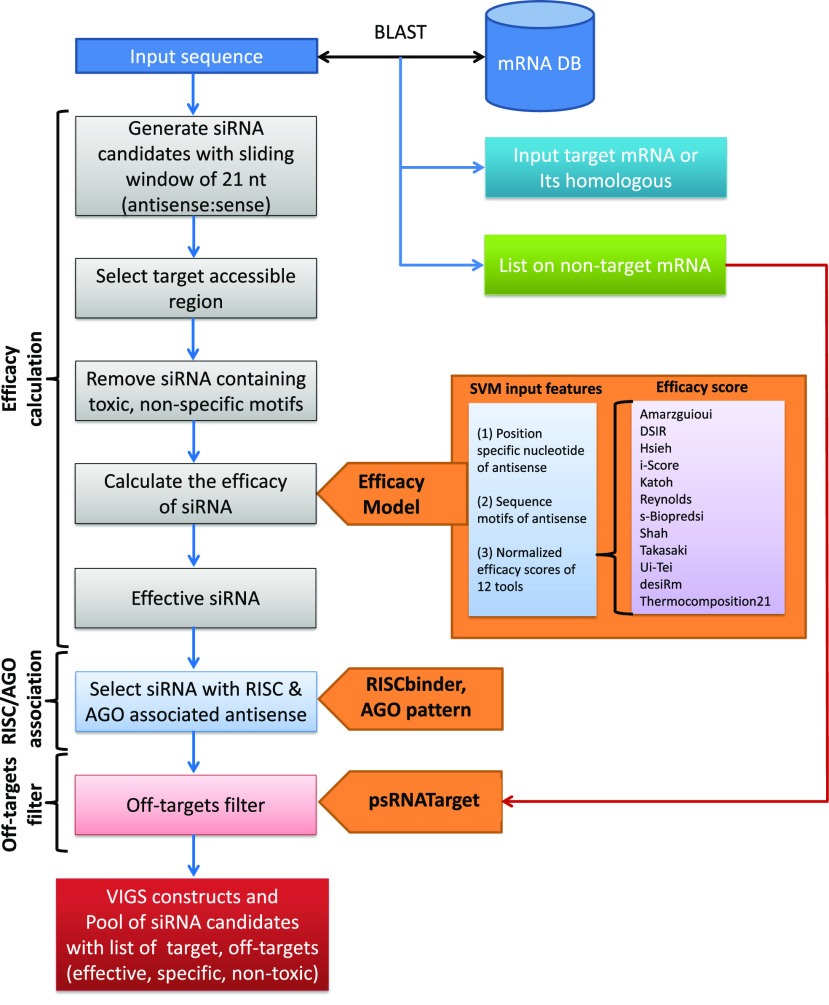
The back-end pipeline of pssRNAit consists of a series of integrated tools, along with transcript libraries, rules, and computational models, that systematically mimic the actions of the plant RNAi pathway. These integrated tools are connected logically and executed step by step to design more effective, highly specific, and nontoxic siRNAs for RNAi constructs (VIGS/synthetic transacting siRNA [syn-tasiRNA]/long dsRNA) for gene silencing in plants (Fig. 1). The back-end pipeline of pssRNAit is run on a powerful, high-performance computing platform, namely BioGrid, that distributes and runs parallel jobs spanning several hosts consisting of >1,000 central processing unit cores on a Linux cluster, which significantly enhances the speed of analysis and return of results.
Figure 1.
Workflow depicting the design of GST for VIGS/siRNAs by pssRNAit pipeline.
Principles and Back-end Pipeline of pssRNAit
The pssRNAit tool was developed by integrating six important features of the RNAi pathway as follows:
Intended and nonintended target transcripts
To clearly define the intended (target) and nonintended (off-target) transcripts, we include a comprehensive database of complementary DNA (cDNA)/transcript libraries from >160 plant species (Supplemental Table S3) in pssRNAit. When a user transcript sequence is submitted, pssRNAit conducts a BLAST search against the cDNA/transcript libraries and, depending on sequence similarity, returns the suggested target and off-target transcripts.
Removing nonspecific and toxic siRNAs
pssRNAit applies a sliding window to generate all possible 21-nucleotide-long antisense siRNA candidates along the user-submitted transcript sequence and later excludes nonspecific and toxic siRNAs using the following criteria: (1) avoid contiguous motifs of >2× CAN; (2) avoid repeats of the same nucleotide more than four times, such as GGGGG, AAAAA, CCCCC, and UUUUU; (3) avoid uninterrupted repeats of >2× CUG (Lawlor et al., 2012), >2× CCG, and 2× CGG (Krzyzosiak et al., 2012), as well as AU-rich motifs (WUAAAUW; Ahmed et al., 2011); and (4) avoid UGUGU and GUCCUUCAA in both strands and UGGC motifs in the antisense strand of siRNA.
Selecting effective siRNAs using the SVM model
After removing nonspecific and toxic siRNAs, the silencing efficiency of the antisense strand of each individual siRNA is calculated by our highly accurate SVM model. Briefly, to design highly effective siRNAs, we developed a highly accurate regression model, pssRNAit2431, using the SVMlight V5.00 package (Joachims, 1999) that achieved a correlation coefficient of 0.709 (R = 0.709, R2 = 0.500, MAE = 0.113, and RMSE = 0.142) between actual and predicted efficacy using 5-fold cross-validation on the TR2431 dataset (Huesken et al., 2005).
Selecting RISC loading siRNA
The purpose of this step is to select the antisense strand of siRNA that has the potential to load into the RISC complex. Only one strand of siRNA is loaded into the RISC complex for gene silencing, while the other is degraded (Khvorova et al., 2003; Schwarz et al., 2003). It is crucial to avoid loading the wrong (sense) strand into RISC, which leads to the silencing of off-target genes. We have developed a highly accurate SVM model, RISC binder, and have integrated it in the back-end pipeline of pssRNAit to predict the RISC loading strand of siRNA using its sequence and motif features (Krol et al., 2004; Ahmed et al., 2009). Plants have different classes of AGOs distinguished by their loading affinity with small RNAs (sRNAs) and gene-silencing features. For example, Arabidopsis (Arabidopsis thaliana) has 10 different classes of AGOs (Mi et al., 2008; Fang and Qi, 2016). Therefore, we have implemented a feature for sorting siRNAs in which the user can select the siRNAs loading to a specific AGO class based on the 5′-terminal nucleotide of the antisense strand (Mi et al., 2008). As suggested by previous findings, antisense strands with U as the 5′-terminal nucleotide would mostly sort into AGO1 and AGO10, those with A would mostly sort into AGO2, AGO4, AGO6, AGO7, and AGO9; and those with C would mostly sort into AGO5 (Mi et al., 2008; Fang and Qi, 2016).
Target site accessibility
Binding of siRNA to the targeted mRNA is a critical step in the RNAi pathway. However, the formation of secondary structures in mRNA can impair siRNA access to the target site. To evaluate target site accessibility, pssRNAit uses the RNAup program of the Vienna Package (Mückstein et al., 2006), which calculates the energy required to “open” the secondary structure around the mRNA target site. To improve performance, we only calculate the energy for a fragment of the sequence including the target region (20 bp) and the flanking 17-bp upstream and 13-bp downstream regions, as described previously (Dai and Zhao, 2011). Here, less energy required to open the secondary structure around the target site indicates a higher possibility of binding with siRNA and, consequently, more effective gene silencing.
Selecting highly specific siRNA
RNAi technology is associated with a serious problem, off-target gene silencing, due to the short length of siRNA. It is crucial to design effective siRNAs with minimum off-target gene silencing to avoid misinterpretation of experimental results. The currently available siRNA designing tools filter out off-target siRNAs using sequence similarity of nucleotides 2 to 7 in the 5′ region (seed) or the entire sequence of siRNA with unintended targets (Xu et al., 2006; Shah et al., 2007; Park et al., 2008; Fernandez-Pozo et al., 2015). However, the silencing efficiency of siRNA is determined by the type of nucleotide and its position in the antisense strand, as well as several other factors (Ahmed et al., 2015; Han et al., 2018). Therefore, sequence homology tools such as BLAST and Bowtie are not suitable to remove off-targeting siRNAs. pssRNAit employs two strategies to select highly specific siRNAs and remove off-target siRNAs.
Removing off-targeting siRNA. siRNA with near-perfect complementarity can trigger miRNA-like off-target gene silencing because miRNA and siRNA both bind with their targets according to the same basic principle. Therefore, pssRNAit was implemented with our previously developed miRNA target prediction tool, psRNATarget (Dai and Zhao, 2011; Dai et al., 2018) to find the off-targets.
Selecting a pool of siRNA against a gene. Designing siRNA without any off-targeting is not always practically possible because of the small size of siRNA. Therefore, removing off-targeting siRNA, as described in the previous paragraph, is not adequate to avoid unintended gene silencing. Consequently, pssRNAit is programmed to intelligently select a pool of siRNAs that have two features: (1) all siRNAs bind to the gene of interest with high efficacy; and (2) siRNAs bind with a minimum of off-target genes with low efficacy. These strategies, mimicking the isomiR-like function in which canonical miRNA and its terminal variant isomiRs bind to the same target to further increase the specificity and silencing efficiency of target genes while minimizing off-target effects (Ahmed and Zhao, 2011; Ahmed et al., 2014).
Web Interfaces of pssRNAit
A publicly available web interface of pssRNAit was developed using the front-end web interfaces in Groovy and back-end interfaces in Java. The pssRNAit can be freely accessed at https://plantgrn.noble.org/pssRNAit/.
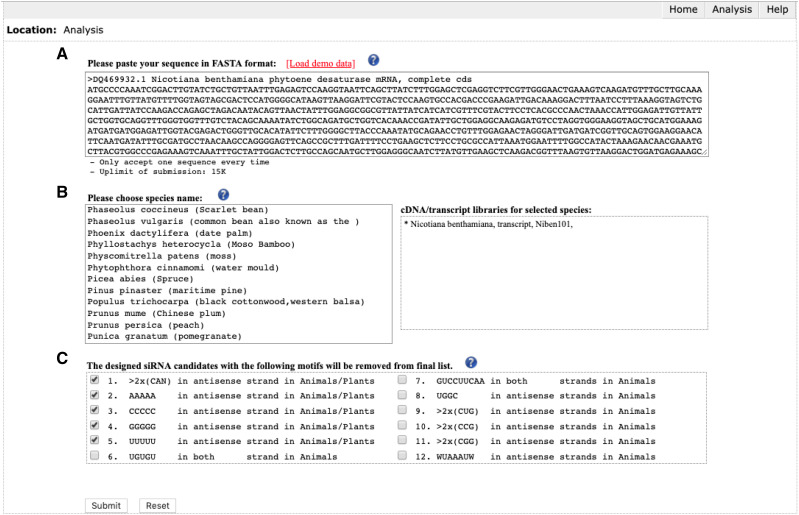
Input: pssRNAit accepts input of the mRNA/cDNA sequence in FASTA format (Fig. 2A). The user chooses a plant species in which the RNAi will be conducted, and the available cDNA/transcript libraries of the chosen plant species will be automatically loaded for genome-wide off-target assessment (Fig. 2B). pssRNAit provides options to remove siRNAs containing toxic and nonspecific sequence motifs (Fig. 2C). Upon submission, the pssRNAit will call upon its back-end pipeline to design the siRNAs and return the output results in tables. An enhanced job-queue management system gives users a session identification number to track submitted job progress and retrieve final results.
Figure 2.
Input interfaces of pssRNAit. Users can paste mRNA sequences in FASTA format (A), select plant species (B), and exclude toxic and non-specific sequence motifs (C) for siRNA design.
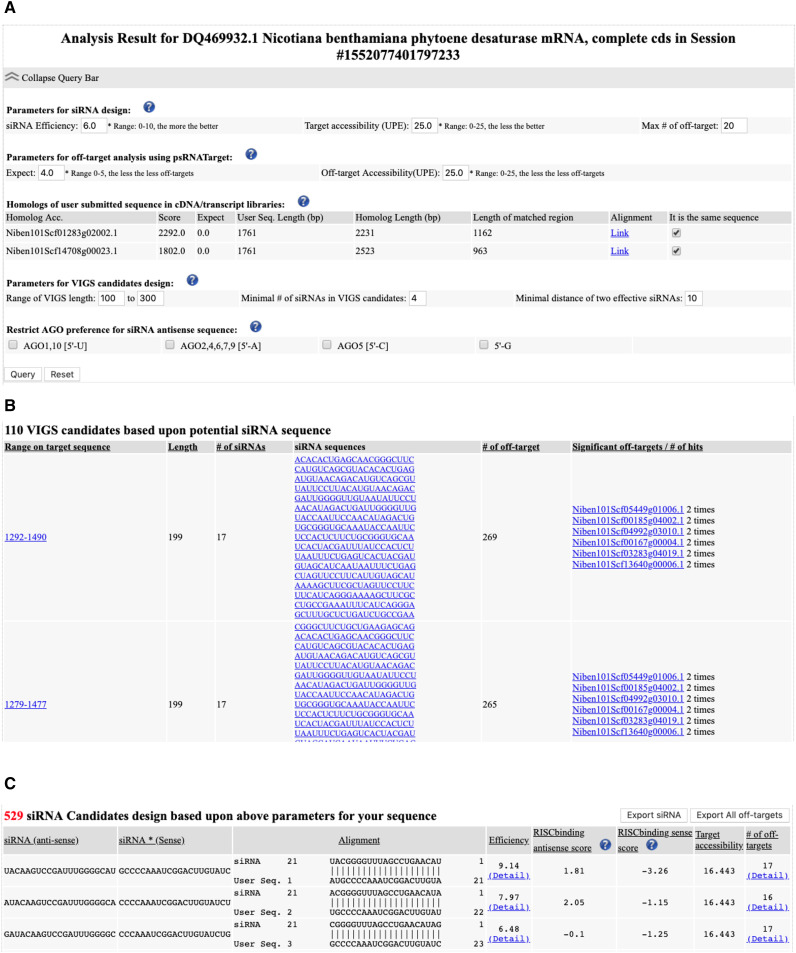
Output: The main output table (Fig. 3) consists of three sections: (1) The query bar (Fig. 3A). This displays parameters for siRNA design, off-target analysis, VIGS candidate design, and siRNA loading preference to specific AGO proteins. It also displays target or off-target transcript sequences from the reference cDNA libraries. In addition, options are available to change the siRNA designing parameters. (2) GST for VIGS (Fig. 3B). The table contains information about putative GST candidates based on potent siRNAs for VIGS analysis. In addition, detailed information about substantial off-targets and the number of hits by siRNA is provided. (3) siRNA candidates (Fig. 3C). The table contains antisense and sense sequences of siRNA, alignment of antisense siRNA binding with the target site, silencing efficiency of siRNA, loading score of antisense and sense strands into RISC, target binding accessibility, and number of off-targets. Here, users can click the provided links to retrieve detailed efficacy scores from 12 different siRNA design tools and off-target information.
Figure 3.
Screenshots of a sample output. A, The query interface for adjusting parameters to filter analysis results. B, A list of output GST candidates for VIGS. C, A list of output siRNA candidates.
pssRNAit-Designed siRNA Candidates Precisely Silenced the NbPDS Gene
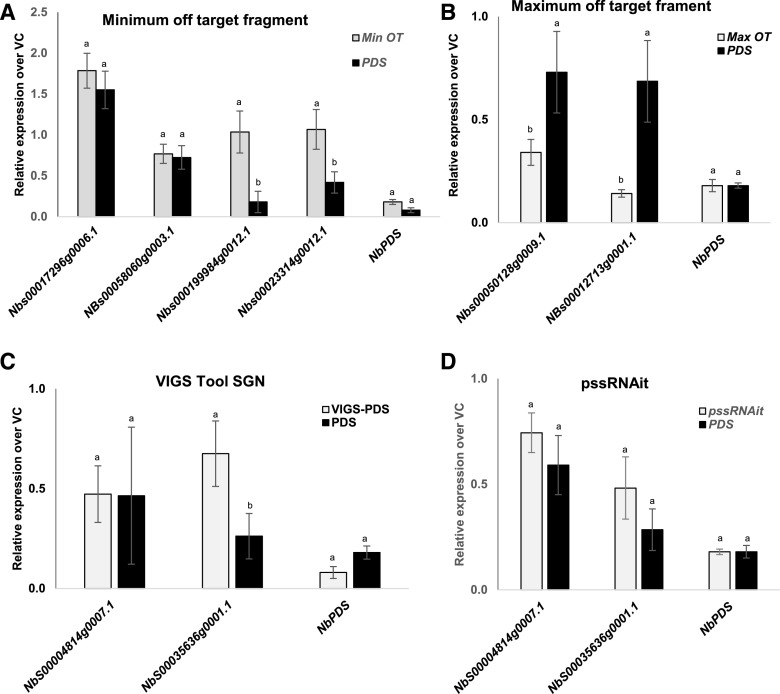
siRNAs targeting NbPDS designed by pssRNAit were cloned into the Tobacco rattle virus vector (pTRV2; TRV2::NbPDS-MinOT [minimum off-target]; TRV2::NbPDS-MaxOT [maximum off-target]; and TRV2::NbPDS) and used for VIGS in N. benthamiana (Senthil-Kumar and Mysore, 2014). All these constructs silenced the endogenous NbPDS transcripts, resulting in a photobleached leaf phenotype (Fig. 4). The widely used conventional constructs (TRV2::NbPDS and TRV2::VIGS-PDS) and pssRNAit designed for minimum (TRV2::NbPDS-MinOT) or maximum (TRV2::NbPDS-MaxOT) off-targeting (see “Materials and Methods”) all showed 80% to 90% effectiveness in silencing the NbPDS gene (Fig. 4A). TRV2::NbPDS-MinOT and TRV2::NbPDS-MaxOT showed 55% and 40% reductions in chlorophyll content, respectively, compared to the vector control (TRV2::GFP; GFP has no sequence similarity to plant genomic DNA and thus will not cause gene silencing; Fig. 4, B and C).
Figure 4.
Silencing efficiency of pssRNAit-designed RNAi fragments against the PDS gene in N. benthamiana. A, Phenotypic response of minimum (Min OT)/maximum (Max OT) off-target regions in comparison with other VIGS construct. B, Effectiveness of the silencing as assessed by the number of leaves that turned white versus the number of green leaves. C, Percent reduction in chlorophyll content compared to the GFP fragment used as a control. A minimum of five leaves from each plant and 10 plants were used to assess efficiency. Bars represent the mean ± se. Different lowercase letters on data points indicate a significant difference (P < 0.05) between the control and PDS-silenced plants determined by two-way ANOVA with Tukey's HSD mean-separation test.
Furthermore, using reverse transcription quantitative PCR (RT-qPCR), we showed that the TRV2::NbPDS-MinOT construct not only silenced minimum off-targets, but also provoked effective target gene silencing (Fig. 5A). In contrast, the TRV2::NbPDS-MaxOT construct showed a higher level of silencing of predicted off-targets, suggesting that the pssRNAit can accurately predict off-target genes (Fig. 5B). Further, the off-target genes predicted by the SGN-VIGS conventional prediction tool (http://vigs.solgenomics.net) were also predicted by pssRNAit and did not show any significant variation in gene expression (Fig. 5, C and D).
Figure 5.
Expression analysis of off-target genes in N. benthamiana plants expressing maximum off-target (MaxOT), minimum off-target (MinOT), VIGS-SGN, and pssRNAit predicted RNAi constructs in comparison with the vector control (VC) and NbPDS fragment. A and B, Expression level of off-target genes when using the minimum (A) and maximum (B) off-target construct. C and D, VIGS-SGN predicted (C) and pssRNAit–predicted (D) constructs. The off-target genes were identified using pssRNAit and the levels of expression were assessed using RT-qPCR. In each case, a minimum of three biological replicates were used for expression analysis. Bars represent the mean ± se. Different lowercase letters on data points indicate a significant difference (P < 0.05) between PDS and the respective minimum or maximum off-targets, determined by two-way ANOVA with Tukey's HSD mean-separation test.
pssRNAit-Designed VIGS Constructs That Show Accurate Genome-Wide Off-Target Prediction
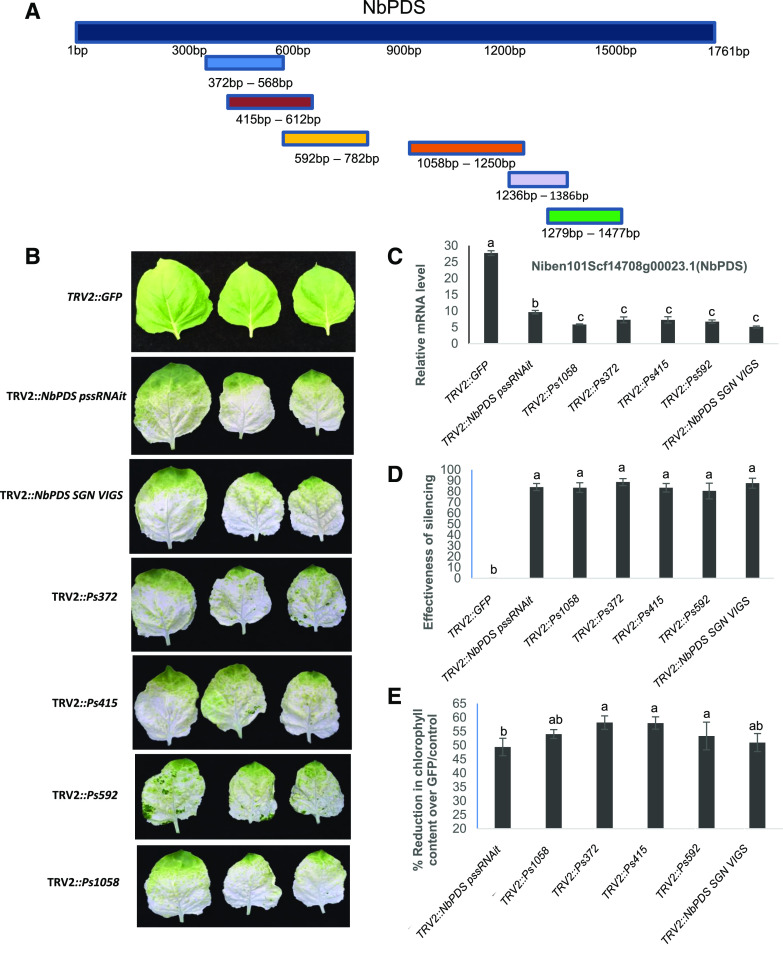
To experimentally assess the genome-wide off-target gene-silencing efficiency, four VIGS constructs that silence the same NbPDS were designed using pssRNAit (Fig. 6, A and B). Two constructs (Ps415 and Ps372) were predicted to result in greater numbers of off-targets, while two other constructs (Ps592, Ps1058) were predicted to have lower numbers of off-targets (Table 2). All these constructs were able to silence NbPDS (Fig. 6C) with ∼90% efficacy (Fig. 6D). Furthermore, the chlorophyll content due to PDS silencing was reduced by 40% to 55% in all silenced plants (Fig. 6E).
Figure 6.
VIGS constructs and their effectiveness in silencing the PDS gene in N. benthamiana. A, Schematic representation of the VIGS fragments located on the NbPDS gene, based on the webserver prediction. Ps series VIGS fragments and TRV2::NbPDS pssRNAit fragments were predicted using pssRNAit. The TRV2::NbPDS SGN VIGS fragment was designed using the VIGS SGN tool. B, Expression analysis showing silencing of NbPDS in N. benthamiana VIGS plants. C, Phenotype of the silenced plants. The top three leaves were photographed and assessed for effectiveness. D, Effectiveness of the silencing as assessed by the number of leaves that turned white versus the number of green leaves. E, Percent reduction in chlorophyll content in silenced plants compared to the GFP fragment as a control. Average values of three biological replicates were used to generate bar graphs and experiments were repeated three times with similar results. Error bars indicate the se. Different letters above the bars indicate a significant difference from two-way ANOVA at P < 0.05 with Tukey’s HSD mean-separation test (α = 0.05).
Table 2. Identity of RNAi sequences cloned into the TRV2 vector against the N. benthamina PDS gene and their off-target genes predicted by pssRNAit at default parameters.
The National Center for Biotechnology Information accession number for the full-length NbPDS sequence used in this study is DQ469932.1.
| Name of Construct | Range of Target Sequence | Length | No. of siRNAs | Construct Sequences | No. of Off-Targets | Significant Off-Targets (No. of Hits) |
|---|---|---|---|---|---|---|
| Ps1058 | 1058–1250 | 193 | 13 | TGAATGAGGATGGAAGTGTCAAATGTTTTATACTGAATAATGGCAGTACAATTAAAGGAGATGCTTTTGTGTTTGCCACTCCAGTGGATATCTTGAAGCTTCTTTTGCCTGAAGACTGGAAAGAGATCCCATATTTCCAAAAGTTGGAGAAGCTAGTGGGAGTTCCTGTGATAAATGTCCATATATGGTTTGA | 239 | Niben101Scf04220g03010.1 (2) |
| Niben101Scf01732g02009.1 (2) | ||||||
| Ps592 | 592–782 | 191 | 13 | ATGCCTAACAAGCCAGGGGAGTTCAGCCGCTTTGATTTTCCTGAAGCTCTTCCTGCGCCATTAAATGGAATTTTGGCCATACTAAAGAACAACGAAATGCTTACGTGGCCCGAGAAAGTCAAATTTGCTATTGGACTCTTGCCAGCAATGCTTGGAGGGCAATCTTATGTTGAAGCTCAAGACGGTTTAAG | 199 | Niben101Scf09867g02016.1 (2) |
| Niben101Scf06725g02017.1 (2) | ||||||
| Ps415 | 415–612 | 198 | 13 | CTGGAGGCAAGAGATGTCCTAGGTGGGAAGGTAGCTGCATGGAAAGATGATGATGGAGATTGGTACGAGACTGGGTTGCACATATTCTTTGGGGCTTACCCAAATATGCAGAACCTGTTTGGAGAACTAGGGATTGATGATCGGTTGCAGTGGAAGGAACATTCAATGATATTTGCGATGCCTAACAAGCCAGGGGAG | 203 | Niben101Scf18745g01012.1 (2) |
| Niben101Scf10385g00005.1 (2) | ||||||
| Niben101Scf17044g01001.1 (2) | ||||||
| Niben101Scf03965g00005.1 (2) | ||||||
| Niben101Scf00397g08002.1 (2) | ||||||
| Ps372 | 372–568 | 197 | 14 | TACAGCAAAATATCTGGCAGATGCTGGTCACAAACCGATATTGCTGGAGGCAAGAGATGTCCTAGGTGGGAAGGTAGCTGCATGGAAAGATGATGATGGAGATTGGTACGAGACTGGGTTGCACATATTCTTTGGGGCTTACCCAAATATGCAGAACCTGTTTGGAGAACTAGGGATTGATGATCGGTTGCAGTGGA | 233 | Niben101Scf10385g00005.1 (2) |
| Niben101Scf17044g01001.1 (2) | ||||||
| Niben101Scf03965g00005.1 (2) | ||||||
| NbPDS PssRNAit | 1279–1477 | 119 | 17 | CTGCTCTTCAGCAGAAGCCCGTTGCTCAGTGTGTACGCTGACATGTCTGTTACATGTAAGGAATATTACAACCCCAATCAGTCTATGTTGGAATTGGTATTTGCACCCGCAGAAGAGTGGATAAATCGTAGTGACTCAGAAATTATTGATGCTACAATGAAGGAACTAGCGAAGCTTTTCCCTGATGAAATTTCGGCAG | 265 | Niben101Scf05449g01006.1 (2) |
| Niben101Scf00185g04002.1 (2) | ||||||
| Niben101Scf04992g03010.1 (2) | ||||||
| Niben101Scf00167g00004.1 (2) | ||||||
| Niben101Scf03283g04019.1 (2) | ||||||
| Niben101Scf13640g00006.1 (2) |
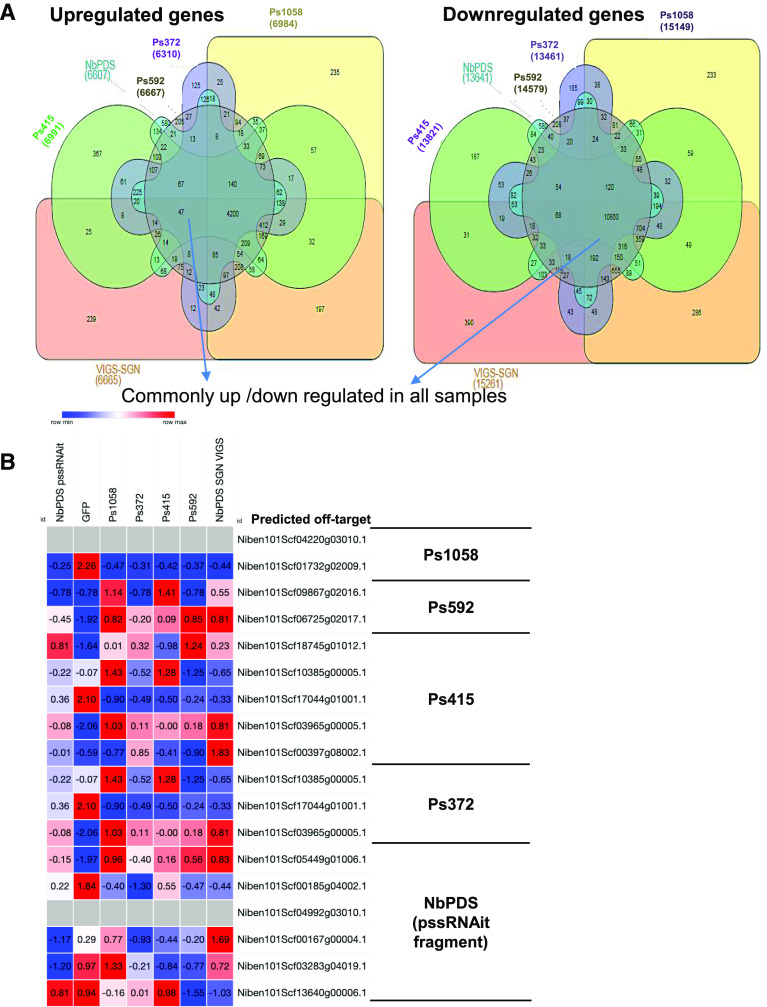
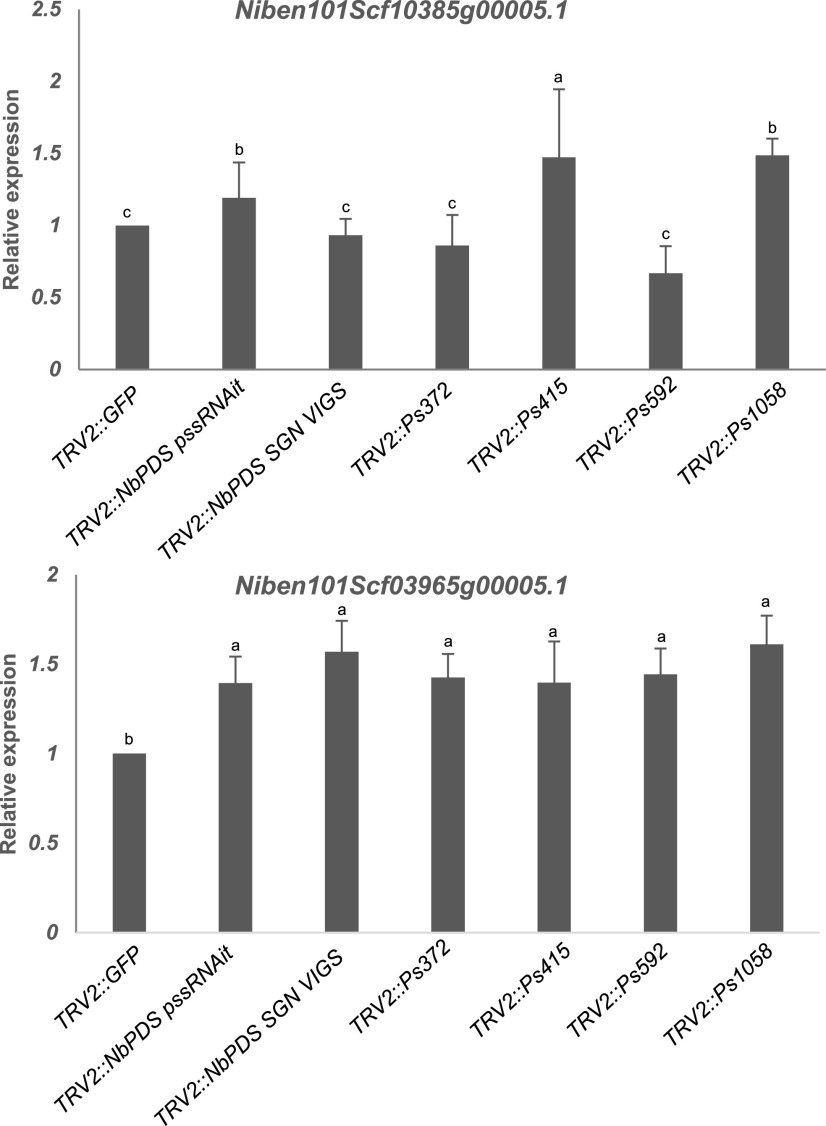
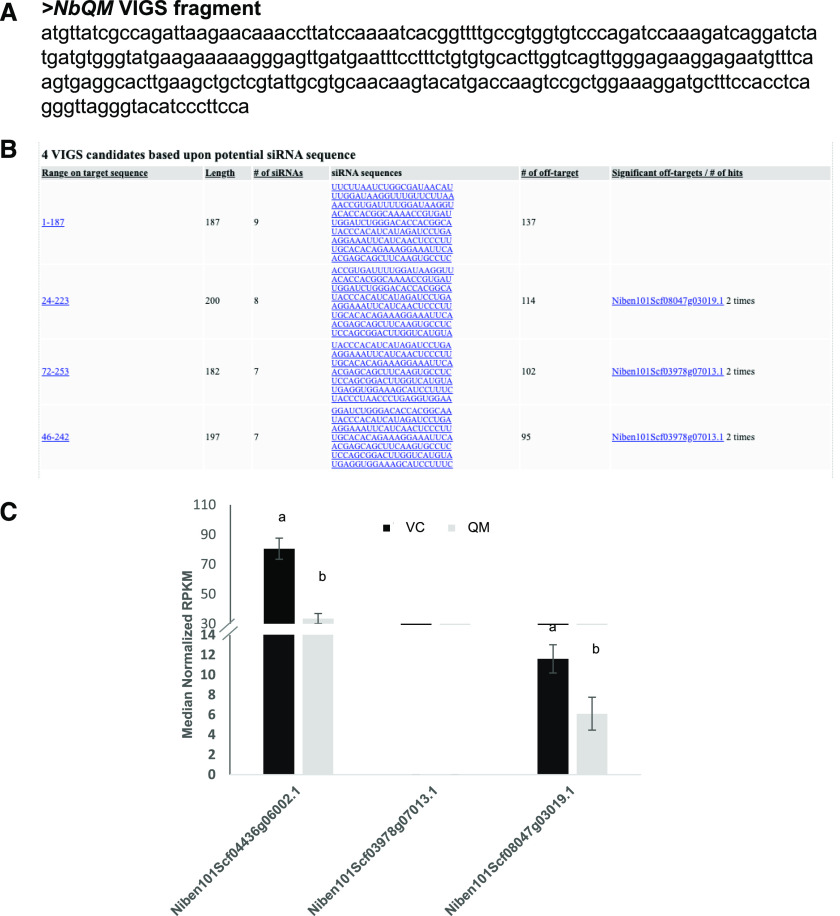
To assess the whole-genome off-targets for all of the GSTs shown in Figure 6A, we performed an RNA-seq experiment on VIGS plants. The data were analyzed, and a web interface was created for easy access to all of the off-targets and indirect effects of gene silencing (https://bioinfo.noble.org/vigs/4100/transcript/profile/5?sessionid=vigs). RNA-seq revealed that 6,991, 6,310, 6,667, 6,984, 6,665, and 6,607 genes were upregulated, while 13,821, 13,461, 14,579, 15,149, 15,261, and 13,641 genes were downregulated, in Ps415, Ps372, Ps592, Ps1058, NbPDS SGN VIGS, and NbPDS pssRNAit VIGS-construct-silenced plants, respectively, compared to TRV2::GFP-inoculated control plants. (Fig. 7A). Furthermore, 4,454 genes were commonly upregulated, and 11,092 genes were commonly downregulated in all VIGS-construct-silenced plants. This is likely due to downstream effects of NbPDS silencing. There are a few unique potential off-target genes present in addition to those predicted by pssRNAit in all of the samples (Fig. 7A). The Ps372 construct showed the highest efficiency and specificity. The down regulation of predicted off-target genes for the Ps1058 fragment was observed, however some of them were upregulated and showed a similar trend across all tested VIGS constructs (Fig. 7B). A few of the predicted off-targets common to fragments Ps372 and Ps415 showed downregulation in respective silenced plants. However, Niben101Scf10385g00005.1 was upregulated in Ps415 silenced plants. The upregulation could be due to inverse regulation between the expression of NbPDS and potential off-target genes. RNA-seq analyses showed that a few of these predicted off-targets (Niben101Scf01732g02009.1, Niben101Scf17044g01001.1, and Niben101Scf00185g04002.1) were commonly downregulated in all silencing constructs (Fig. 7B). The RT-qPCR analysis of Niben101Scf10385g00005.1 (predicted off-target for Ps415) showed downregulation of the gene in Ps415, Ps372, and Ps592 VIGS plants. The Niben101Scf03965g00005.1 predicted for Ps372 did not show downregulation in any of the other PDS silenced plants (Fig. 8). Further validation of pssRNAit-predicted off-targets has been confirmed by silencing the QM/RPL10 (Ribosomal protein L10) homolog in N. benthamiana (Rocha et al., 2008) by VIGS, revealing a 50% reduction in both target and off-target genes. Niben101Scf03978g07013 and Niben101scf08047g03019 are predicted off-targets, of which the former was not detected in either sample, whereas the latter showed a 50% reduction in expression (Fig. 9). These results suggest that pssRNAit was effective in designing VIGS constructs for efficient target gene silencing with a minimum of off-targets.
Figure 7.
Differential expression of genes from VIGS plants. A, Venn diagrams showing the number of genes that are up- or downregulated in VIGS N. benthamiana plants. B, Heat map of transcript levels of off-target genes identified in respective VIGS plants. At right are the genes predicted to be off-targets for the fragments used for VIGS. RNA-Seq analysis was carried out from the 3-week-old VIGS plants with three biological replicates. The expression values were transformed into Z-scores (across each row).
Figure 8.
Expression analysis of off-target genes of VIGS PDS fragment. Off-target genes were identified using pssRNAit, and level of expression was assessed using RT-qPCR. Average values of three biological replicates were used to generate bar graphs. Error bars indicate the se. Different letters above the bars indicate a significant difference from two-way ANOVA at P < 0.05 with Tukey’s HSD mean-separation test (α = 0.05).
Figure 9.
Target and off-target gene expression analysis from whole-genome RNA-seq analysis from the QM/RPL10 gene silenced N. benthamiana plants. A, Nucleotide sequence of the NbQM gene used for VIGS in N. benthamiana. B, Predicted off-targets using pssRNAit for the NbQM gene. C, Niben 101Scf04436g06002.1 is a target gene showing 50% downregulation in QM-silenced plants relative to the vector control (VC). Niben101Scf03978g07013 and Niben101scf08047g03019 are predicted off-targets, the former of which was not detected in either sample, whereas Niben101scf08047g03019 showed a 50% reduction in expression. As predicted, the expression of off-target sites is reduced in the QM-silenced plants, indicating that pssRNAit could identify off-targets precisely. Error bars indicate the se from three biological replicates. Different letters above the bars indicate a significant difference from two-way ANOVA at P < 0.05 determined by Tukey’s HSD mean-separation test (α = 0.05).
pssRNAit-Designed siRNA Can Be Expressed as syn-tasiRNA
tasiRNAs are a class of sRNAs that originates from tasiRNA-generating (TAS) transcripts through binding of a specific miRNA and cleavage of the TAS transcript (Felippes and Weigel, 2009). Subsequently, the cleaved TAS product is converted into dsRNA and processed by DCL4 to produce a phased array of 21-nucleotide sRNAs starting from the miRNA cleavage site (Felippes & Weigel, 2009). Generating synthetic transacting siRNA (syn-tasiRNA) constructs has been adopted to suppress multiple endogenous genes in a plant at a time (Carbonell et al., 2014). Therefore, we evaluated the performance of our tool for use as syn-tasiRNAs for efficient gene silencing in plants. For this, we collected experimental data from syn-tasiRNAs generated from expressed AtTAS1c, and their intended-target and off-target genes in Arabidopsis (Carbonell et al., 2014). Detailed data for expressed syn-tasiRNAs derived from the AtTAS1c construct and their intended target and off-target genes are given in Supplemental Table S4. Carbonell et al. (2014) cloned and generated two syn-tasiRNA sequences at a time using four different AtTAS1c-based syn-tasiRNA constructs: (1) AtTAS1c-d3&d4Trich for syn-tasiRNA-Trich and syn-tasiRNA-Trich; (2) AtTAS1c-d3&d4Ft for syn-tasiRNA-Ft and syn-tasiRNA-Ft; (3) AtTAS1c-d3Trich-d4Ft for syn-tasiRNA-Trich and syn-tasiRNA-Ft; and (4) AtTAS1c-d3Ft-d4Trich for syn-tasiRNA-Ft and syn-tasiRNA-Trich, where syn-tasiRNA-Trich targeted the gene TRIPTYCHON (TRY; AT5G53200) and syn-tasiRNA-Ft targeted the gene FLOWERING LOCUS T (FT; AT1G65480).
The AtTAS1c-d3&d4Trich and AtTAS1c-d3&d4Ft oligonucleotide sequences were selected and submitted into pssRNAit, and the predicted results were compared with experimentally verified siRNAs and their intended target and possible off-target genes. We observed that pssRNAit predicted the same siRNA (UCCCAUUCGAUACUGCUCGCC) and its target gene (TRY) and two off-target genes (ENHANCER OF TRIPTYCHON AND CAPRICE2 [ETC2; AT2G30420] and CAPRICE [CPC; AT2G46410]) as experimentally known for AtTAS1c-d3&d4Trich (Supplemental Table S5). Furthermore, FT is not an off-target gene for AtTAS1c-d3&d4Trich, as predicted by pssRNAit and confirmed experimentally (Supplemental Table S5). In addition, our tool predicted the same siRNA (UUGGUUAUAAAGGAAGAGGCC) and its target gene (FT) as experimentally known for AtTAS1c-d3&d4Ft (Supplemental Table S6). Furthermore, CPC, ETC2, and TRY are not off-target genes for AtTAS1c-d3&d4Ft, as predicted by pssRNAit and also confirmed experimentally (Supplemental Table S6).
The results clearly demonstrate that pssRNAit can accurately predict the functional siRNAs generated from AtTAS1c constructs, as well as their target and off-target genes, and therefore, our tool could also be used to design and express functional syn-tasiRNA constructs for effective and specific gene silencing in plants. Furthermore, we have evaluated our tool on experimentally known gene silencing data in Arabidopsis from articles by Xu et al. (2006) and Hilson et al. (2004). We found that pssRNAit predicted several functional siRNAs for the experimentally silenced gene and its off-targets (Supplemental Table S7).
DISCUSSION
Posttranscriptional gene silencing (PTGS) using RNAi is a powerful technique for studying gene function and improving crop traits. However, in addition to silencing of the intended target gene, unintended off-target gene silencing and cytotoxicity have been observed during RNAi (Xu et al., 2006; Grimm, 2011; Senthil-Kumar and Mysore, 2011). These limitations can obscure the correct interpretation of gene function and the proper use of RNAi technology in crop improvement. In spite of the widespread use of RNAi for PTGS in plants, none of the bioinformatics tools currently available can design highly effective, specific, and nontoxic RNAi constructs. The primary reason for this is that the findings and rules identified by RNAi studies have not been fully implemented in the currently available RNAi algorithms. For potent and precise gene silencing, siRNAs should be designed with State C, in which the antisense bind only to Real-Target, and State J, in which the antisense bind to Real-Target as well as the minimal number of off-targets, while discarding other options, as illustrated in Supplemental Figure S1. Furthermore, additional strategies could be applied in State J of Supplemental Figure S1 to increase the silencing specificity by (1) selecting the lowest number of siRNAs that bind to the same off-target mRNA and (2) selecting all siRNAs that are noneffective against off-target mRNAs.
In the past, we developed the RNAiScan server to design siRNAs and their off-target prediction in plants (Xu et al., 2006). The back-end pipeline only implemented a traditional rule developed by Ui-Tei et al. (2004) to find the efficient siRNAs and BLAST to identify off-target genes. In addition, the RNAiScan lacked continuous implementation of the up-to-date rules governing PTGS RNAi pathways. A SGN-VIGS tool (http://vigs.solgenomics.net/) was developed to design 200- to 400–nucleotide-long VIGS constructs for silencing a plant gene (Fernandez-Pozo et al., 2015), but the tool lacks information about candidate siRNAs and rules for calculating the silencing efficacy of VIGS constructs. The functional sequence motifs identified from our tool are commonly present in several coding regions of plant and animal transcripts and therefore targeting them may lead to nonspecific gene silencing (Yu et al., 2011; Lawlor et al., 2012; Zhao et al., 2014). Several studies show that some sequence motifs in siRNA cause a toxic effect in transfected animal cells in a target-independent fashion, resulting in reduced cell viability (Sledz et al., 2003; Judge et al., 2005; Armstrong et al., 2008; McAllister and Samuel, 2009). Furthermore, the SGN-VIGS tool implemented the Bowtie tool to find potential off-target genes. Bowtie was developed to find only perfect and mismatch short reads in the genome and does not implement the position and nucleotide-specific mismatch rules known to find targets of siRNAs. Another tool, P-SAMS (http://p-sams.carringtonlab.org), has been developed to design siRNAs for plant artificial miRNAs and syn-tasiRNAs (Fahlgren et al., 2016). However, P-SAMS does not implement the rule to predict the silencing efficiency of siRNA, and the off-target is identified based on a 6- to 20-nucleotide perfect match. Furthermore, P-SAMS did not recognize the 200- to 400-nucleotide-long constructs of mRNA required for VIGS. These tools have not implemented rules for (1) siRNA accessibility to target transcripts; (2) siRNA loading to the RISC; and (3) removing nonspecific and toxic siRNA. Importantly, most of these tools lack comprehensive experimental validation of the intended and off-target gene expression levels to check the accuracy of the designed RNAi constructs (Lück et al., 2019). With pssRNAit, we have addressed the limitations of the available RNAi design tools and provided experimental evidence of its efficacy. pssRNAit-predicted minimum off-targets and maximum off-targets showed >75% gene silencing efficiency. The off-targets were not downregulated in minimum off-target constructs, and downregulation was seen only in maximum off-target plants (Figs. 4 and 5). The performance of the pssRNAit SVM model was evaluated on two independent datasets of siRNAs that gave a high correlation coefficient between actual and predicted efficacy. Furthermore, we experimentally evaluated the performance of pssRNAit by using VIGS to design and express the RNAi construct of NbPDS in N. benthamiana. The predicted and shortlisted four fragments with minimum off-target and maximum off-target produced efficiently gene-silenced plants with higher chlorophyll reduction and downregulation of the targeted PDS gene (Figs. 6 and 7). We analyzed the expression level of the intended target and predicted off-target genes using RNA-seq and RT-qPCR. There are several genes commonly upregulated and downregulated in all VIGS-construct-silenced plants. This variation could be due to downstream effects of NbPDS gene silencing. Disruption of the PDS gene results in albino and dwarf phenotypes and affects transcripts of ∼20 metabolic pathway genes, including those associated with chlorophyll, carotenoid, and gibberellin biosynthesis in Arabidopsis (Qin et al., 2007). In the case of QM/RPL10 VIGS plants, many of the genes involved in translation mechanisms were similarly downregulated, which could be due to its role in the regulation of transcription, as suggested earlier (Zorzatto et al., 2015). Finally, pssRNAit predicted the functional siRNAs from experimentally known syn-tasiRNA sequences, demonstrating that pssRNAit-designed siRNAs could be expressed as syn-tasiRNAs for efficient gene silencing (Supplemental Tables S4–S6).
pssRNAit is a fast-computing pipeline for designing effective, specific, and nontoxic RNAi constructs to silence genes of interest in plants. The back-end pipeline of pssRNAit implemented a series of computational approaches integrated with comprehensive transcript libraries from >120 plant species, several rules, and computational models that mimic the biological mechanism of the RNAi pathway. Users also can request the addition of other published transcript libraries in pssRNAit. The experimental results presented in this manuscript suggest that pssRNAit is an excellent online tool for designing highly effective and specific siRNAs, facilitating the advancement of functional genomics and trait improvement in plants.
MATERIALS AND METHODS
Features Used for the SVM Model Predicting Effective siRNAs
For SVM model development, we used sequence features of the 21-nucleotide antisense strand, including frequency of mono-, di-, and trinucleotides and a position-specific binary pattern. Most importantly, we also integrated 12 different siRNA design tools in our model, which were incorporated in a cascade fashion. A previous study showed the importance of the cascade strategy for substantial improvement in the performance of the SVM model (Bhasin and Raghava, 2004). In the first layer of the cascade, the efficacy score of siRNA is predicted using 12 different tools. In the second layer, these efficacy scores become 12 input features for the SVM model. These siRNA tools include first-generation, second-generation algorithms (Amarzguioui and Prydz, 2004), DSIR (Hsieh et al., 2004; Vert et al., 2006), i-Score(Reynolds et al., 2004; Ichihara et al., 2007; Katoh and Suzuki, 2007), s-Biopredsi (Takasaki et al., 2004; Ui-Tei et al., 2004; Huesken et al., 2005; Shah et al., 2007 ), desiRm (Ahmed and Raghava, 2011), and Thermocomposition21 (Ichihara et al., 2007). The design rule is taken either from the corresponding literature or from Ichihara et al. (2007). However, the range of efficacy scores predicted by these tools varies. Therefore, we normalized them in the range 0 to 10, where 0 indicates no silencing and 10 indicates 100% silencing of mRNA. Scores from rule-based algorithms were normalized using Equation 1, while scores coming from machine-learning algorithms were multiplied or divided by 10 to get them in the approximate range of 0 to 10.
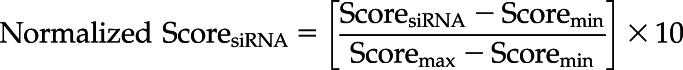 |
(1) |
where Scoremin and Scoremax are the minimum and maximum scores, respectively, achieved by an algorithm.
Plant Material and Growth Conditions
Wild-type Nicotiana benthamiana was used for all experiments. Seeds were germinated and seedlings were grown in plastic pots (20-cm diameter) with BM7 potting mixture (SUNGRO Horticulture Distribution). Two-week-old seedlings were transplanted to 10-cm-diameter round pots containing BM7, with one plant per pot. Fertilizer (N:P:K 20:10:20) with a soluble trace-element mix (ScottsMiracle-Gro) was applied with water. Greenhouse conditions were kept at 23° ± 3°C and 70% relative humidity under 16 h extended daylight with supplemental lighting at 100 mE m−2 s−1 light intensity. Three-week-old plants were used for experiments. Detailed growth conditions are described in previous publications (Senthil-Kumar et al., 2013, Senthil-Kumar and Mysore, 2014).
VIGS Constructs and Bacterial Strains
The pTRV1 and pTRV2 VIGS vectors (Senthil-Kumar and Mysore, 2014) were obtained from Dr. Dinesh-Kumar, University of CA Davis. Gateway-based cloning according to the manufacturer’s recommendations (Invitrogen) was used for preparation of constructs in this study. Primer sequences are given in Supplemental Table S8. Gene fragments to be cloned into the pTRV2 vector were amplified either from synthesized gBlock sequences or from cDNA. A fragment of the GFP gene (Senthil-Kumar and Mysore, 2014) was cloned into the pTRV2 vector and used as a control. Ten siRNAs predicted by pssRNAit software from the full-length NbPDS sequence were assembled, and artificial sequences were synthesized as gBlocks by Integrated DNA Technologies (Supplemental Table S9).
For cDNA synthesis, RNA was extracted from leaves and first-strand cDNA was synthesized using oligo 15 primers (Integrated DNA Technologies; Huesken et al., 2005). The cDNA pool was used for PCR amplification of the PDS gene sequence (Supplemental Table S9). Cloning of NbPDS derived 10 siRNAs, selected based on in silico analysis of the NbPDS sequence and predicted to cause efficient endogenous PDS gene silencing with minimal off-target gene silencing, which were assembled and custom cloned into the pTRV2 VIGS vector, resulting in the TRV2::NbPDS-MinOT construct. Similarly, the siRNA assembly that is predicted to cause maximal off-target gene silencing was cloned into the pTRV2 VIGS vector, resulting in the TRV2::NbPDS-MaxOT construct.
To test and validate the effectiveness of the pssRNAit tool on the NbPDS gene for VIGS, the minimum number of off-target genes was predicted from two fragments at 1,058 to 1,250 bp and 592 to 782 bp (Ps1058 and Ps592, respectively) and the maximum number from fragments at 415 to 612 bp and 372 to 568 bp (Ps415 and Ps372, respectively). To clone the NbPDS fragments predicted by pssRNAit and the SGN-VIGS tool, the genes were amplified and cloned into the pQUIET II TRV2 vector, yielding TRV2::NbPDS pssRNAit (the fragment designed by pssRNAit) and TRV2::NbPDS SGN VIGS (the fragment designed by the SGN-VIGS tool), respectively. The QM/RPL10 gene VIGS fragment was predicted using pssRNAit and cloned into the TRV2 vector. The pssRNAit-predicted fragments cloned into pTRV2 were designated as TRV2::Ps1058, TRV2::Ps592, TRV2::Ps415, and TRV2::Ps372 vectors, and QM/RPL10 as TRV2::NbQM vector. Details of these constructs are given in Tables 2 and 3 and Supplemental Tables S8 to S11. TRV2::GFP was used as the vector control in this study (Senthil-Kumar and Mysore, 2014). All the pTRV2 derivatives were confirmed by sequencing. Plasmids were introduced into Agrobacterium tumefaciens strain GV2260 by electroporation.
Table 3. Identity of RNAi sequences cloned into the TRV2 vector against the N. benthamina PDS gene predicted by SGN VIGS at default parameters.
The National Center for Biotechnology Information accession number for the full-length NbPDS sequence used in this study is DQ469932.1, and N. benthamiana version 0.4.4 was selected as the database.
| Name of Construct | Range on Target Sequence | Length | No. of siRNAs | Construct sequences | No. of Off-Targets | Significant Off-Targets (No. of Hits) |
|---|---|---|---|---|---|---|
| NbPDS (SGN VIGS tool fragment) | 1236–1386 | 150 | Not available | CCATATATGGTTTGACAGAAAACTGAAGAACACATCTGATAATCTGCTCTTCAGCAGAAGCCCGTTGCTCAGTGTGTACGCTGACATGTCTGTTACATGTAAGGAATATTACAACCCCAATCAGTCTATGTTGGAATTGGTATTTGCACCC | Not Available | Off-target prediction is not available in the SGN VIGS tool |
| NbPDS (conventionally used fragment) | 855–1263 | 409 | Not available | GGCACTCAACTTTATAAACCCTGACGAGCTTTCGATGCAGTGCATTTTGATTGCTTTGAACAGATTTCTTCAGGAGAAACATGGTTCAAAAATGGCCTTTTTAGATGGTAACCCTCCTGAGAGACTTTGCATGCCGATTGTGGAACATATTGAGTCAAAAGGTGGCCAAGTCAGACTAAACTCACGAATAAAAAAGATCGAGCTGAATGAGGATGGAAGTGTCAAATGTTTTATACTGAATAATGGCAGTACAATTAAAGGAGATGCTTTTGTGTTTGCCACTCCAGTGGATATCTTGAAGCTTCTTTTGCCTGAAGACTGGAAAGAGATCCCATATTTCCAAAAGTTGGAGAAGCTAGTGGGAGTTCCTGTGATAAATGTCCATATATGGTTTGACAGAAAACTGAAG | Not Available | Conventionally used fragment for VIGS |
VIGS
The A. tumefaciens strain GV2260 containing pTRV1 or pTRV2 and its derivatives were grown at 28°C in Luria-Bertani medium containing appropriate antibiotics. After 24 h, the cells were harvested and resuspended in the infiltration buffer (10 mm MES [pH 5.5] and 200 mm acetosyringone) to a final absorbance of 0.6 OD (at 600 nm) and incubated for 2 h with shaking at 28°C room temperature. For leaf infiltration, each A. tumefaciens strain containing pTRV1 and pTRV2 or its derivatives was mixed in a 1:1 ratio in MES buffer (pH 5.5) and infiltrated into the lower leaves using a 1-mL needleless syringe (Senthil-Kumar et al., 2007; Senthil-Kumar et al., 2013; Senthil-Kumar and Mysore, 2014). The infiltrated plants were maintained at a temperature range of 23°C to 25°C for effective viral infection and spread.
RT-qPCR
RT-qPCR was performed to determine the transcript levels of the endogenous NbPDS gene and other predicted off-target genes. Total RNA was extracted from silenced and mock-infiltrated plants and the first-strand cDNA was synthesized using oligo dT primers (Huesken et al., 2005). RT-qPCR was performed with PRISM 7000 (Applied Biosystems) using SYBR green (Applied Biosystems). The primers used for quantifying the relative transcripts were designed using Primer Express Software 2.0. For the relative quantification of gene transcripts between silenced and nonsilenced plants, a standard curve method was applied according to the manufacturer’s protocol (Applied Biosystems User Bulletin). As a control for silenced and mock-infiltrated plants, the parallel reaction using N. benthamiana Elongation factor1-α (EF1α) was performed, and the data obtained were used to normalize NbPDS transcripts. Each sample was run in triplicate and repeated twice from pooled samples of three independently silenced and mock-infiltrated plants. The calculations were performed as previously described (Senthil-Kumar and Mysore, 2014) and the percent reduction was determined.
Effectiveness of Gene Silencing
The effectiveness of VIGS is defined as the ability of the insert (NbPDS) to induce silencing and produce gene silencing symptoms, and it was determined by counting all the leaves (small and big), including leaves of secondary shoots that showed photobleaching or yellowing (Senthil-Kumar et al., 2007). Effectiveness of gene silencing was calculated using the formula
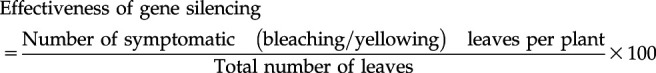 |
(2) |
Chlorophyll Estimation
The efficiency of gene silencing in this study was also assessed by quantifying the reduction in chlorophyll in the leaves, as the NbPDS gene used here regulates chlorophyll biosynthesis. A higher reduction in chlorophyll indicates a higher efficiency of gene silencing (Senthil-Kumar et al., 2007). Chlorophyll was extracted from 100 mg leaf tissue in an acetone:dimethylsulfoxide 1:1 (v/v) mix and the supernatant was made up to a known volume. The absorbance was recorded at 663 nm and 645 nm using a UV-visible spectrophotometer (model DU800, Beckman Coulter). Total chlorophyll was estimated (Hiscox and Israelstam, 1979) and expressed as the percent reduction relative to the corresponding control.
Differential Gene Expression Analysis by RNA-Seq
The leaf samples from three biological replicates from 4-week-old NbPDS- (different fragments shown in Fig. 6) or NbQM/RPL10-silenced N. benthamiana were harvested and immediately frozen in liquid nitrogen. Total RNA was extracted and sent to the Noble Research Institute Genomics Core Facility for analysis. RNA samples with high quality (RNA integrity >7.5, as assessed by the Agilent 2100 Bioanalyzer) were used for library preparation following the manufacturer's instructions (Illumina). Paired-end reads were generated from 21 libraries sequenced with Illumina 70 HiSeq 2000. After adapter sequences and low-quality reads were removed, the remaining reads were aligned to the annotated N. benthamiana reference transcriptome (Bombarely et al., 2012; https://solgenomics.net/organism/1490/view) and the gene-wise raw counts were calculated. The differential analysis was carried out using the DESeq tool (Anders and Huber, 2010). Genes with >2-fold change in expression and P < 0.05 were considered differentially expressed. Data from all of the 21 samples (seven treatments × three biological replicates) have been uploaded into our GEAUniversal gene expression atlas platform (https://bioinfo.noble.org/vigs/) for data normalization, visualization, and differential expression analysis using RNA-Seq analysis software (Li and Dewey, 2011; Love et al., 2014). The normalized expression data for all seven treatments can be directly visualized at https://bioinfo.noble.org/vigs/4100/transcript/profile/5?sessionid=vigs.
Statistical Analysis
Two-way ANOVA was carried out according to Fisher (1960). Data points with different lowercase letters indicate significant differences (P < 0.05) between samples as determined by Tukey’s honestly significant difference (HSD) mean-separation test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number PRJNA525845 (https://www.ncbi.nlm.nih.gov/sra/PRJNA525845).
Supplemental Data
The following supplemental materials are for this article.
Supplemental Figure S1. siRNA binding possibilities with intended and off-targets.
Supplemental Table S1. Sources of siRNA datasets used in pssRNAit tool development.
Supplemental Table S2. Dataset of siRNAs and their silencing scores used for developing the SVM model.
Supplemental Table S3. A list of preloaded cDNA/transcript libraries in pssRNAit.
Supplemental Table S4. Expression of AtTAS1c-based syn-tasiRNAs in Arabidopsis Col-0 T1 transgenic plants and analysis of their target gene silencing.
Supplemental Table S5. pssRNAit-designed siRNA and predicted target and off-targets of the AtTAS1c-d3&d4Trich construct in Arabidopsis at default threshold except for the expected value (3.0).
Supplemental Table S6. pssRNAit-designed siRNA and predicted target and off-targets of the AtTAS1c-d3&d4Ft construct in Arabidopsis at default threshold except for the expected value (3.0).
Supplemental Table S7. Performance of pssRNAit on experimentally known gene silencing data in Arabidopsis.
Supplemental Table S8. Details of primer sequences used in this study.
Supplemental Table S9. Details of gene and gBlock sequences used in this study.
Supplemental Table S10. siRNAs designed by pssRNAit tool against NbPDS gene with minimum off-target genes.
Supplemental Table S11. siRNAs designed by pssRNAit tool against NbPDS gene with maximum off-target genes.
Footnotes
This work was supported by the National Science Foundation, Division of Biological Infrastructure (grant nos. 0960897 and 1458597 to P.X.Z.), the Oklahoma Center for the Advancement of Science and Technology (grant no. PSB11–004 to P.X.Z.), and the Noble Research Institute.
Articles can be viewed without a subscription.
References
- Ahmed F, Ansari HR, Raghava GP(2009) Prediction of guide strand of microRNAs from its sequence and secondary structure. BMC Bioinformatics 10: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F, Benedito VA, Zhao PX(2011) Mining functional elements in messenger RNAs: Overview, challenges, and perspectives. Front Plant Sci 2: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F, Dai X, Zhao PX(2015) Bioinformatics tools for achieving better gene silencing in plants. Methods Mol Biol 1287: 43–60 [DOI] [PubMed] [Google Scholar]
- Ahmed F, Raghava GPS(2011) Designing of highly effective complementary and mismatch siRNAs for silencing a gene. PLoS One 6: e23443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F, Senthil-Kumar M, Lee S, Dai X, Mysore KS, Zhao PX(2014) Comprehensive analysis of small RNA-seq data reveals that combination of miRNA with its isomiRs increase the accuracy of target prediction in Arabidopsis thaliana. RNA Biol 11: 1414–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed F, Zhao P(2011) A comprehensive analysis of isomiRs and their targets using high-throughput sequencing data for Arabidopsis thaliana. J Nat Sci Biol Med 2: 32–33 [Google Scholar]
- Amarzguioui M, Prydz H(2004) An algorithm for selection of functional siRNA sequences. Biochem Biophys Res Commun 316: 1050–1058 [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W(2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong ME, Gantier M, Li L, Chung WY, McCann A, Baugh JA, Donnelly SC(2008) Small interfering RNAs induce macrophage migration inhibitory factor production and proliferation in breast cancer cells via a double-stranded RNA-dependent protein kinase-dependent mechanism. J Immunol 180: 7125–7133 [DOI] [PubMed] [Google Scholar]
- Bhasin M, Raghava GP(2004) Analysis and prediction of affinity of TAP binding peptides using cascade SVM. Protein Sci 13: 596–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB(2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact 25: 1523–1530 [DOI] [PubMed] [Google Scholar]
- Burchard J, Jackson AL, Malkov V, Needham RH, Tan Y, Bartz SR, Dai H, Sachs AB, Linsley PS(2009) MicroRNA-like off-target transcript regulation by siRNAs is species specific. RNA 15: 308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A, Takeda A, Fahlgren N, Johnson SC, Cuperus JT, Carrington JC(2014) New generation of artificial microRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol 165: 15–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX(2011) psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res 39: W155–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX(2018) psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res 46(W1): W49–W54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Hill ST, Carrington JC, Carbonell A(2016) P-SAMS: A web site for plant artificial microRNA and synthetic trans-acting small interfering RNA design. Bioinformatics 32: 157–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Qi Y(2016) RNAi in plants: An Argonaute-centered view. Plant Cell 28: 272–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FF, Weigel D(2009) Triggering the formation of tasiRNAs in Arabidopsis thaliana: The role of microRNA miR173. EMBO Rep 10: 264–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N, Rosli HG, Martin GB, Mueller LA(2015) The SGN VIGS tool: User-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol Plant 8: 486–488 [DOI] [PubMed] [Google Scholar]
- Fisher RA.(1960) The Design of Experiments, Ed 7 Hafner Publishing, New York [Google Scholar]
- Fukudome A, Fukuhara T(2017) Plant dicer-like proteins: Double-stranded RNA-cleaving enzymes for small RNA biogenesis. J Plant Res 130: 33–44 [DOI] [PubMed] [Google Scholar]
- Grimm D.(2011) The dose can make the poison: Lessons learned from adverse in vivo toxicities caused by RNAi overexpression. Silence 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA(2006) Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441: 537–541 [DOI] [PubMed] [Google Scholar]
- Han Y, He F, Chen Y, Liu Y, Yu H(2018) SiRNA silencing efficacy prediction based on a deep architecture. BMC Genomics 19(Suppl 7): 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al. (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res 14(10B): 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox JD, Israelstam GF(1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can J Bot 57: 1332–1334 [Google Scholar]
- Hsieh AC, Bo R, Manola J, Vazquez F, Bare O, Khvorova A, Scaringe S, Sellers WR(2004) A library of siRNA duplexes targeting the phosphoinositide 3-kinase pathway: Determinants of gene silencing for use in cell-based screens. Nucleic Acids Res 32: 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesken D, Lange J, Mickanin C, Weiler J, Asselbergs F, Warner J, Meloon B, Engel S, Rosenberg A, Cohen D, et al. (2005) Design of a genome-wide siRNA library using an artificial neural network. Nat Biotechnol 23: 995–1001 [DOI] [PubMed] [Google Scholar]
- Ichihara M, Murakumo Y, Masuda A, Matsuura T, Asai N, Jijiwa M, Ishida M, Shinmi J, Yatsuya H, Qiao S, et al. (2007) Thermodynamic instability of siRNA duplex is a prerequisite for dependable prediction of siRNA activities. Nucleic Acids Res 35: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS(2003) Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21: 635–637 [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS(2006) Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12: 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagla B, Aulner N, Kelly PD, Song D, Volchuk A, Zatorski A, Shum D, Mayer T, De Angelis DA, Ouerfelli O, et al. (2005) Sequence characteristics of functional siRNAs. RNA 11: 864–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachims T.(1999) Making large-scale support vector machine learning practical In Schölkopf B, Burges CJC, and Smola AJ, eds, Advances in Kernel Methods. MIT Press, Cambridge, MA, pp 169–184 [Google Scholar]
- Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I(2005) Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol 23: 457–462 [DOI] [PubMed] [Google Scholar]
- Katoh T, Suzuki T(2007) Specific residues at every third position of siRNA shape its efficient RNAi activity. Nucleic Acids Res 35: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD(2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ(2004) Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem 279: 42230–42239 [DOI] [PubMed] [Google Scholar]
- Krzyzosiak WJ, Sobczak K, Wojciechowska M, Fiszer A, Mykowska A, Kozlowski P(2012) Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res 40: 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor KT, O’Keefe LV, Samaraweera SE, van Eyk CL, Richards RI(2012) Ubiquitous expression of CUG or CAG trinucleotide repeat RNA causes common morphological defects in a Drosophila model of RNA-mediated pathology. PLoS One 7: e38516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN(2011) RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S(2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lück S, Kreszies T, Strickert M, Schweizer P, Kuhlmann M, Douchkov D(2019) siRNA-Finder (si-Fi) software for RNAi-target design and off-target prediction. Front Plant Sci 10: 1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister CS, Samuel CE(2009) The RNA-activated protein kinase enhances the induction of interferon-β and apoptosis mediated by cytoplasmic RNA sensors. J Biol Chem 284: 1644–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. (2008) Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133: 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mückstein U, Tafer H, Hackermüller J, Bernhart SH, Stadler PF, Hofacker IL(2006) Thermodynamics of RNA-RNA binding. Bioinformatics 22: 1177–1182 [DOI] [PubMed] [Google Scholar]
- Naito Y, Yamada T, Matsumiya T, Ui-Tei K, Saigo K, Morishita S(2005) dsCheck: Highly sensitive off-target search software for double-stranded RNA-mediated RNA interference. Nucleic Acids Res 33: W589–W591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejniczak M, Galka P, Krzyzosiak WJ(2010) Sequence-non-specific effects of RNA interference triggers and microRNA regulators. Nucleic Acids Res 38: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P, Senthil-Kumar M, Mysore KS(2015) Advances in plant gene silencing methods. Methods Mol Biol 1287: 3–23 [DOI] [PubMed] [Google Scholar]
- Park YK, Park SM, Choi YC, Lee D, Won M, Kim YJ(2008) AsiDesigner: Exon-based siRNA design server considering alternative splicing. Nucleic Acids Res 36: W97–W103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Gu H, Ma L, Peng Y, Deng XW, Chen Z, Qu LJ(2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17: 471–482 [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A(2004) Rational siRNA design for RNA interference. Nat Biotechnol 22: 326–330 [DOI] [PubMed] [Google Scholar]
- Rocha CS, Santos AA, Machado JP, Fontes EP(2008) The ribosomal protein L10/QM-like protein is a component of the NIK-mediated antiviral signaling. Virology 380: 165–169 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD(2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Hema R, Anand A, Kang L, Udayakumar M, Mysore KS(2007) A systematic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus-induced gene silencing. New Phytol 176: 782–791 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Lee H-K, Mysore KS(2013) VIGS-mediated forward genetics screening for identification of genes involved in nonhost resistance. J Vis Exp 2013: e51033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS(2011) Caveat of RNAi in plants: The off-target effect. Methods Mol Biol 744: 13–25 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS(2014) Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat Protoc 9: 1549–1562 [DOI] [PubMed] [Google Scholar]
- Shah JK, Garner HR, White MA, Shames DS, Minna JD(2007) sIR: siRNA Information Resource, a web-based tool for siRNA sequence design and analysis and an open access siRNA database. BMC Bioinformatics 8: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR(2003) Activation of the interferon system by short-interfering RNAs. Nat Cell Biol 5: 834–839 [DOI] [PubMed] [Google Scholar]
- Takasaki S, Kotani S, Konagaya A(2004) An effective method for selecting siRNA target sequences in mammalian cells. Cell Cycle 3: 790–795 [PubMed] [Google Scholar]
- Thareau V, Déhais P, Serizet C, Hilson P, Rouzé P, Aubourg S(2003) Automatic design of gene-specific sequence tags for genome-wide functional studies. Bioinformatics 19: 2191–2198 [DOI] [PubMed] [Google Scholar]
- Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K(2004) Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res 32: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert JP, Foveau N, Lajaunie C, Vandenbrouck Y(2006) An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinformatics 7: 520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA(2013) Molecular mechanisms of RNA interference. Annu Rev Biophys 42: 217–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Zhang Y, Kang L, Roossinck MJ, Mysore KS(2006) Computational estimation and experimental verification of off-target silencing during posttranscriptional gene silencing in plants. Plant Physiol 142: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR(2005) Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 11: 220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Teng X, Bonini NM(2011) Triplet repeat-derived siRNAs enhance RNA-mediated toxicity in a Drosophila model for myotonic dystrophy. PLoS Genet 7: e1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Guo C, Sutharzan S, Li P, Echt CS, Zhang J, Liang C(2014) Genome-wide analysis of tandem repeats in plants and green algae. G3 (Bethesda) 4: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorzatto C, Machado JP, Lopes KV, Nascimento KJ, Pereira WA, Brustolini OJ, Reis PA, Calil IP, Deguchi M, Sachetto-Martins G, et al. (2015) NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature 520: 679–682 [DOI] [PMC free article] [PubMed] [Google Scholar]