The tomato DELLA protein PROCERA promotes abscisic acid-induced stomatal closure and gene expression by upregulating expression of the ABA transporter ABA-IMPORTING TRANSPORTER1 in guard cells.
Abstract
Plants reduce transpiration through stomatal closure to avoid drought stress. While abscisic acid (ABA) has a central role in the regulation of stomatal closure under water-deficit conditions, we demonstrated in tomato (Solanum lycopersicum) that a gibberellin response inhibitor, the DELLA protein PROCERA (PRO), promotes ABA-induced stomatal closure and gene transcription in guard cells. To study how PRO affects stomatal closure, we performed RNA-sequencing analysis of isolated guard cells and identified the ABA transporters ABA-IMPORTING TRANSPORTER1.1 (AIT1.1) and AIT1.2, also called NITRATE TRANSPORTER1/PTR TRANSPORTER FAMILY4.6 in Arabidopsis (Arabidopsis thaliana), as being upregulated by PRO. Tomato has four AIT1 genes, but only AIT1.1 and AIT1.2 were upregulated by PRO, and only AIT1.1 exhibited high expression in guard cells. Functional analysis of AIT1.1 in yeast (Saccharomyces cerevisiae) confirmed its activity as an ABA transporter, possibly an importer. A clustered regularly interspaced short palindromic repeats-Cas9–derived ait1.1 mutant exhibited an increased transpiration, a larger stomatal aperture, and a reduced stomatal response to ABA. Moreover, ait1.1 suppressed the promoting effects of PRO on ABA-induced stomatal closure and gene expression in guard cells, suggesting that the effects of PRO on stomatal aperture and transpiration are AIT1.1-dependent. Previous studies suggest a negative crosstalk between gibberellin and ABA that is mediated by changes in hormone biosynthesis and signaling. The results of this study suggest this crosstalk is also mediated by changes in hormone transport.
The growth-promoting hormone GA regulates central developmental processes throughout the plant life cycle, from germination to stem elongation, leaf expansion, flowering, and fruit development (Yamaguchi, 2008). GA also affects plant response to abiotic stresses, such as salinity and drought (Achard et al., 2006; Colebrook et al., 2014, Nir et al., 2017). The output of GA activity on plant development and response to the environment depends on complex interactions with other hormones (Weiss and Ori, 2007). The negative interaction between GA and the stress hormone abscisic acid (ABA) has been studied for many years in numerous plant species. These studies suggest that GA and ABA negatively affect each other’s biosynthesis and signaling (Shu et al., 2018).
The nuclear DELLA proteins suppress almost all GA responses by interacting with various transcription factors (Hauvermale et al., 2012; Locascio et al., 2013). When GA binds to its receptor GIBBERELLIN-INSENSITIVE DWARF1 (GID1), it increases the affinity of the latter to DELLA. The generation of GID1-GA-DELLA complex leads to DELLA degradation via the ubiquitin-proteasome pathway, which is mediated by the F-box protein SLEEPY1 (Sasaki et al., 2003; Dill et al., 2004; Griffiths et al., 2006; Harberd et al., 2009; Hauvermale et al., 2012). DELLA destruction in the proteasome leads to transcriptional reprogramming and activation of GA responses. The ability of DELLA to interact with numerous transcriptional regulators is a key factor in the crosstalk between GA and other hormones. For example, DELLA interaction with JASMONATE ZIM DOMAIN proteins mediates the effect of GA on jasmonic acid (JA) activity (Hou et al., 2010), and its interaction with BRASSINAZOLE-RESISTANT1 mediates the crosstalk with brassinosteroids (Li et al., 2012).
The N-terminal region of DELLA (the DELLA domain) is important for the interaction with GID1 and therefore, mutations in this region interfere with the interaction (Harberd et al., 2009). These dominant, gain-of-function mutations stabilize DELLA, leading to constitutive inhibition of GA responses. The C-terminal region of DELLA (the GRAS domain) plays a major role in repressing GA responses by interacting with numerous transcription factors (Yoshida et al., 2014). Mutations in the C-terminal region are recessive, and exhibit constitutive GA responses (Sun and Gubler, 2004; Harberd et al., 2009). Tomato (Solanum lycopersicum) has one DELLA protein, called PROCERA (PRO; Jasinski et al., 2008; Livne et al., 2015). The tomato loss-of-function mutant pro is tall and exhibits increased GA responses (Van Tuinen et al., 1999; Bassel et al., 2008; Fleishon et al., 2011), whereas the gain-of-function proGF mutant is dwarf due to constitutive inhibition of GA responses (Zhu et al., 2019).
The crosstalk between GA and ABA has been studied for many years, mainly in seeds (Piskurewicz et al., 2008; Liu and Hu, 2018; Shu et al., 2018). The balance between the two hormones regulates dormancy versus germination; high ABA to GA ratio promotes dormancy, whereas the opposite promotes germination (Razem et al., 2006). The transcription factor ABSCISIC ACID-INSENSITIVE4 (ABI4) promotes seed dormancy in Arabidopsis (Arabidopsis thaliana) by the suppression of GA accumulation and the promotion of ABA biosynthesis (Shu et al., 2013). In Sorghum bicolor, SbABI4 promotes the transcription of the GA deactivating gene SbGA2ox3 (Cantoro et al., 2013). Moreover, the transcription factor GERMINATION INSENSITIVE TO ABA MUTANT2 promotes GA biosynthesis while reducing ABA production (Xiong et al., 2018). In Arabidopsis seeds, DELLA promotes the expression of the RING ubiquitin E3 ligase XERICO that is involved in ABA accumulation. It also increases the expression of the transcription factor ABI5 that inhibits seed germination, and interacts with the ABA signaling components ABI3 (Lim et al., 2013). ABA, in turn, stabilizes the Arabidopsis DELLA protein REPRESSOR OF GA1-3 LIKE-2 and inhibits GA signaling (Piskurewicz et al., 2008). In tomato, the lack of DELLA activity in seeds suppresses desiccation tolerance due to inhibition of ABA-induced gene expression (Livne et al., 2015). Taken together, these studies suggest that GA and ABA negatively interact at the hormone biosynthesis and signaling levels (Shu et al., 2018).
Previously we suggested a crosstalk between GA/DELLA and ABA in the regulation of stomatal movement in tomato. Overexpressing the Arabidopsis GA METHYLTRANSFERASE1 gene in tomato reduces GA levels and whole-plant transpiration (Nir et al., 2014). Transgenic tomato plants overexpressing the Arabidopsis or the tomato stable DELLA mutant proteins rgaΔ17 or proΔ17, respectively, exhibit lower GA activity and reduced stomatal aperture and transpiration compared with wild-type controls. Overexpressing pro∆17 specifically in guard cells was sufficient to reduce stomatal aperture. On the other hand, pro loss-of-function mutant plants exhibit increased transpiration rate, faster water loss under water-deficit conditions, and larger stomatal pore area (Nir et al., 2017). The effects of pro∆17 on stomatal closure and water loss were suppressed in the ABA-deficient sitiens mutant, indicating that these effects of DELLA are ABA-dependent (Nir et al., 2017). We found that DELLA promotes ABA responses, including ABA-induced stomatal closure and reactive oxygen species accumulation in guard cells after ABA application. Because DELLA is a transcription regulator, it is yet unclear how PRO affects ABA-induced stomatal closure. PRO did not affect ABA accumulation in leaves, thus we speculated that it affects ABA signaling or uptake into guard cells via transcriptional regulation of ABA signaling component or transporter genes (Nir et al., 2017).
Several ABA transporters have been identified and characterized in Arabidopsis, including the ATP-BINDING CASSETTE (ABC) transporters ABCG25 and ABCG40, and ABA-IMPORTING TRANSPORTER1 (AIT1), also called NITRATE TRANSPORTER1.2 (NRT1.2), or NRT1/PTR TRANSPORTER FAMILY4.6 (NPF4.6). ABCG25 is expressed in vascular tissues and functions as an ABA exporter (Kuromori et al., 2010). ABCG40 is an ABA importer that was localized to the guard-cell plasma membrane (Kang et al., 2010). AIT1 is expressed in the vascular tissues of inflorescence stems and the ait1 mutant exhibited increased water loss due to open stomata (Kanno et al., 2012). Loss of ABCG25 increases the sensitivity to ABA whereas the loss of ABCG40 and AIT1 reduce the sensitivity to the hormone (Kuromori et al., 2018).
Here we studied the mechanism by which the tomato DELLA protein PRO increases ABA responses in guard cells. RNA-sequencing (RNA-seq) analysis of isolated guard cells identified the ABA transporter AIT1.1 as upregulated by PRO. The loss of AIT1.1 suppressed the effect of PRO on guard-cell ABA responses.
RESULTS
PRO Promoted ABA Responses in Guard Cells
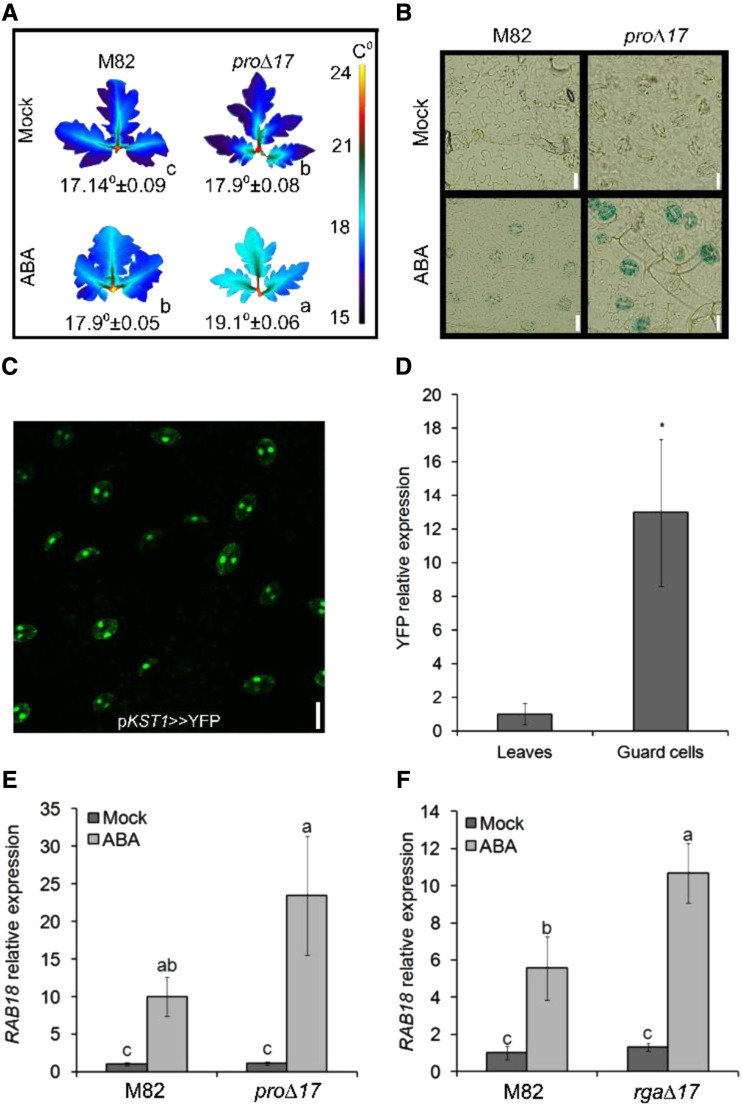
To support our previous suggestion that DELLA promotes ABA responses in guard cells (Nir et al., 2017), we tested the effect of PRO on ABA-inhibition of transpiration and ABA-induced gene expression in guard cells. Thermal imaging of M82 and transgenic plants overexpressing the stable DELLA protein pro∆17 (35S:pro∆17; Nir et al., 2017) showed higher leaf-surface temperature in the transgenic line, after the application of ABA, indicating lower transpiration rate (Fig. 1A). To examine the effect of DELLA on ABA-induced transcription, we have generated transgenic M82 plants expressing the GUS reporter gene under the regulation of the Arabidopsis ABA-induced promoter MAPKKK18 (Okamoto et al., 2013). The transgene was then introgressed into 35S:pro∆17 plants by crosses. The GUS signal in ABA-treated leaves was observed in guard cells and was stronger in 35S:pro∆17 compared to M82 (Fig. 1B). These results suggest that PRO promotes ABA physiological and transcriptional responses in guard cells.
Figure 1.
PRO promotes ABA responses in guard cells. A, Thermal imaging of leaves (leaf no. 4 below the apex) taken from M82 and 35S:pro∆17 treated or not (Mock) with 10 μm of ABA. Leaves were digitally extracted for comparison. Number below leaves are the calculated leaf-surface temperature and the values are means of three plants, measured 20 times ± se. Small letters above the numbers represent significant differences between respective treatments (Tukey–Kramer HSD test, P < 0.05). B, Representative images of GUS staining of epidermal peels treated or not (Mock) with 10 μm of ABA. Peels were taken from leaf no. 4 below the apex of M82 and 35S:pro∆17 expressing the reporter GUS under the regulation of the MAPKKK18 promoter. C, YFP signal in guard cells of pKST1>>YFP transactivated epidermal peel. Scale bars = 20 μm. D, YFP expression in whole leaf tissue and guard-cell–enriched samples. Values are means of four biological replicates ± se. Stars above the columns represents significant differences between respective treatments by Student’s t test (P < 0.05). E and F, RT-qPCR analysis of RAB18 expression in guard-cell–enriched samples isolated from leaves no. 3 and 4 below the apex of M82 and 35S:pro∆17 (E) or 35S:rga∆17 (F). Values in E and F are means of four biological replicates ± se. Small letters above the columns represent significant differences between respective treatments by Tukey–Kramer HSD (P < 0.05). The value for leaves in D was set to 1 and the value for M82 Mock in E and F was set to 1.
Because DELLA is a transcription regulator, we hypothesized that PRO affects transpiration and stomatal movement by regulating the expression of ABA/stomatal-related genes in guard cells. To study the interaction between DELLA and ABA in the regulation of gene expression, we first developed a rapid and efficient guard-cell isolation protocol to minimize the effect of the isolation on gene expression (see “Materials and Methods”). To validate the procedure, we have used plants expressing the YELLOW FLUORESCENT PROTEIN (YFP) under the guard-cell–specific promoter KST1 (Fig. 1C; Nir et al., 2017). Quantitative reverse-transcription PCR (RT-qPCR) analysis of RNA extracted from whole leaf tissue or isolated guard cells showed ∼13-fold higher YFP expression in the guard-cells enriched samples (Fig. 1D). We then used this procedure to test the response of the ABA-induced gene RAB18 (Nir et al., 2017) to ABA in guard-cell enriched samples taken from M82, 35S:pro∆17, and 35S:rga∆17. The expression of RAB18 after ABA treatment was higher in the transgenic lines (Fig. 1, E and F).
Global Expression Response to PRO in Guard Cells
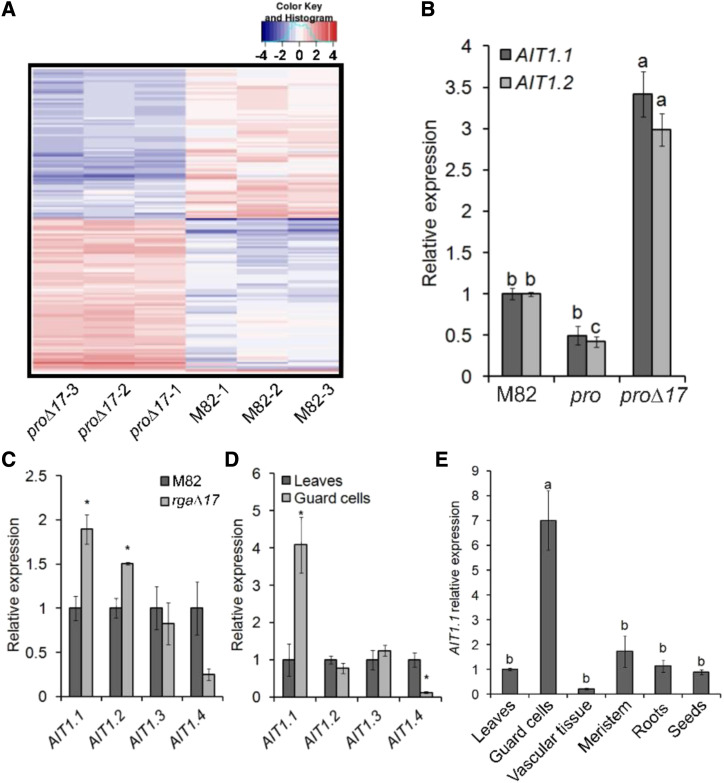
We next explored the mechanism by which DELLA promotes ABA-induced stomatal closure. To this end, we examined the global effect of PRO on guard-cell–transcriptional activity by performing RNA-seq analysis to guard-cell–enriched samples taken from M82, 35S:pro∆17, and pro. Using a twofold increase or decrease cutoff (adjusted P value for multiple comparisons ≤ 0.05), we identified 162 PRO-regulated genes (81 upregulated and 81 downregulated; Fig. 2A; Supplemental Dataset S1; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143999). We then searched for differentially expressed genes related to stomatal closure and/or ABA. Among these genes (Supplemental Table S1), we identified three putative ABA transporters: the tomato homologs of the Arabidopsis ABCG40 and two AIT1 genes, also called in Arabidopsis NPF4.6 or NRT1.2 (Supplemental Fig. S1; Kanno et al., 2012). We named the tomato proteins AIT1.1 and AIT1.2. The expression of ABCG40 and AIT1.2 in the RNA-seq analysis was extremely low (Supplemental Table S1). It is worth noting that although we reported previously that two ABA receptors PYRABACTIN RESISTANCE1(PYR1) and PYR1-like8-1 are upregulated in 35S:pro∆17 (Nir et al., 2017), in the RNA-seq analysis we did not find them among the differentially expressed genes.
Figure 2.
RNA-seq analysis identified the ABA transporter AIT1.1 as upregulated by PRO in guard cells. A, Clustered heatmap of PRO-regulated genes (proΔ17 versus M82, three samples each) generated from RNA-seq analysis shows 81 PRO upregulated and 81 downregulated genes. Genes were grouped based on their pattern of expression. Coloring of the genes is according to the color bar on the upper-right side (Log2 fold change). The complete list of PRO-regulated genes is provided in Supplemental Dataset 1. B, RT-qPCR analysis of AIT1.1 and AIT1.2 expression in M82, pro, and 35S:proΔ17 (proΔ17) guard cells isolated from leaves no. 3 and 4 below the apex. Values are means of three biological replicates ± se. Lowercase letters represent significant differences between lines by Tukey–Kramer HSD (P < 0.05). C, Expression of all tomato AIT1 genes in M82 and 35S:rgaΔ17 (rgaΔ17) isolated guard cells. D, Expression of all tomato AIT1 genes in leaves and isolated guard cells. E, Expression of AIT1.1 in different tissues: leaves (leaf no. 4 below the apex), guard cells, vascular tissue (isolated from leaf no. 4 below the apex), meristems (apices including leaf primordia), young roots, and imbibed seeds. Values in C, D and E are means of four replicates ± se. Stars (C and D) and lowercase letters (E) above the columns represent significant differences between respective treatments by Student’s t test (P < 0.05). The values for M82 in B and C were set to 1 and the values for leaves in D and E were set to 1.
We first validated the results of the RNA-seq for the effect of PRO on the expression of ABCG40, AIT1.1, and AIT1.2 in guard-cell–enriched samples by RT-qPCR. This analysis did not confirm the effect of PRO on ABCG40 (Supplemental Fig. S2), but it did confirm PRO’s effect on AIT1.1 and AIT1.2. These genes were up- and downregulated in 35S:pro∆17 and pro, respectively (Fig. 2B). Tomato has four AIT1 homologs that we named AIT1.1 to AIT1.4 (Supplemental Fig. S1). We analyzed the expression of all four AIT1 homologs in M82 and 35S:rga∆17 guard cells and only AIT1.1 and AIT1.2 were upregulated by DELLA in guard cells (Fig. 2C). We then analyzed the expression of all AIT1s in M82 guard-cell–enriched samples compared to whole leaf tissue, and only AIT1.1 exhibited significantly higher expression in guard cells (Fig. 2D). Kanno et al. (2012) found that the Arabidopsis AIT1 is expressed in the vascular tissue, using the reporter line. We used the same approach and generated transgenic M82 plants expressing the GUS reporter under the regulation of the AIT1.1 promoter (∼1,400 bp upstream of the start codon). The tomato AIT1.1 also showed high GUS activity in vascular tissues (Supplemental Fig. S3), but not in guard cells. We therefore analyzed AIT1.1 expression in various tissues of M82 plants by RT-qPCR (mature leaves, shoot apices, vascular tissue, guard cells, roots, and imbibed seeds), and found the highest expression in guard cells (Fig. 2E). These results suggest that the promoter used to express GUS did not provide the authentic spatial expression pattern.
PRO Promoted ABA Responses via the ABA Transporter AIT1.1
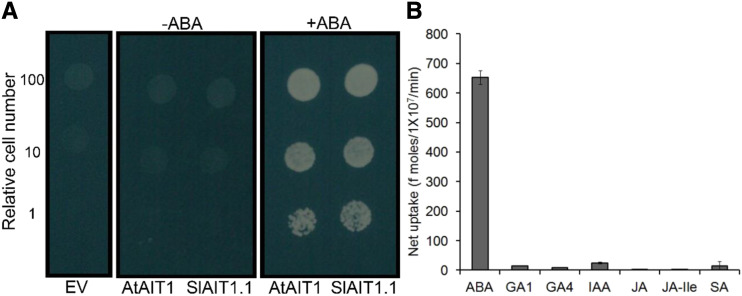
Functional analysis of the Arabidopsis AIT1 protein in yeast cells suggests that it operates as an ABA importer (Kanno et al., 2012). When expressed in yeasts, (Saccharmyces cerevisiae) the tomato AIT1.1 induced the interactions between the Arabidopsis ABA receptor PYR1 and the protein phosphatase 2C ABI1 under a relatively low ABA concentration (0.5 μm) in the growth media, as the Arabidopsis AIT1 protein did, while the interactions were not observed in the absence of ABA (Fig. 3A). We further confirmed that AIT1.1 mediated cellular ABA uptake by directly quantifying the molecules taken into the yeast cells by liquid chromatography tandem mass-spectrometry (LC-MS/MS; Fig. 3B). We also examined the substrate selectivity of AIT1.1 against several other hormones. It appeared that AIT1.1 transported GA1, GA4, and indole-3-acetic acid (IAA) to some extent; however, the selectivity was much lower compared to ABA (Fig. 3B).
Figure 3.
AIT1.1 mediates ABA uptake into yeast cells. A, Effects of AIT1.1 on the interactions between the ABA receptor and protein phosphatase 2C. Tomato AIT1.1 (SlAIT1.1) or Arabidopsis AIT1 (AtAIT1) was expressed in yeast containing a yeast two-hybrid system with the Arabidopsis PYR1 ABA receptor fused to the GAL4 DNA binding domain and the ABI1 protein phosphatase fused to the GAL4 activation domain, and the cells were inoculated on selection media (SD, -Leu, -Trip, -Ura, and -His) containing 0.5 μm of ABA (+ABA) or without ABA (−ABA). An empty vector (EV) was transformed as a negative control. Photos were taken 3 d after inoculation. B, Hormone transport activities of AIT1.1. Yeast cells expressing tomato AIT1.1 were incubated with solutions containing 10 μm of ABA, GA1, GA4, IAA, JA, JA-Ile, or salicylic acid (SA), and the amounts of compounds taken into the cells were quantified with LC-MS/MS.
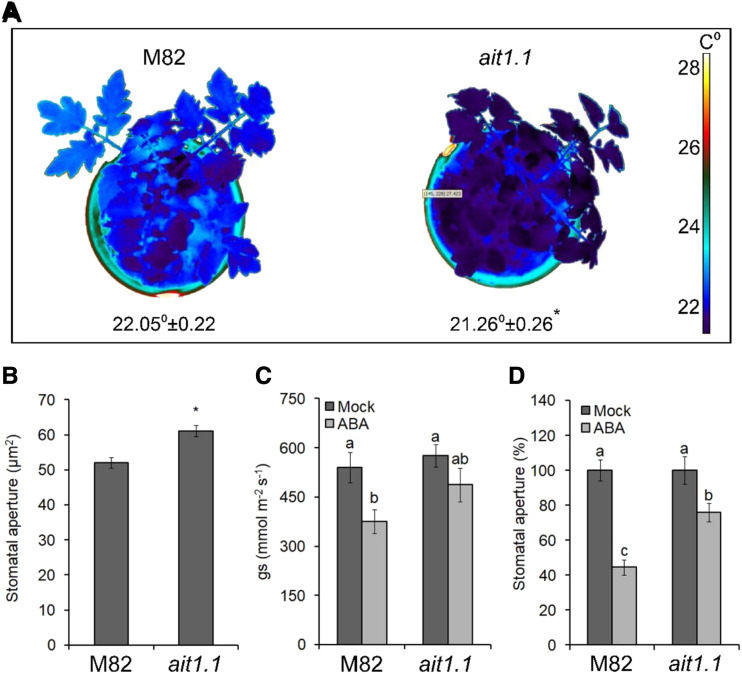
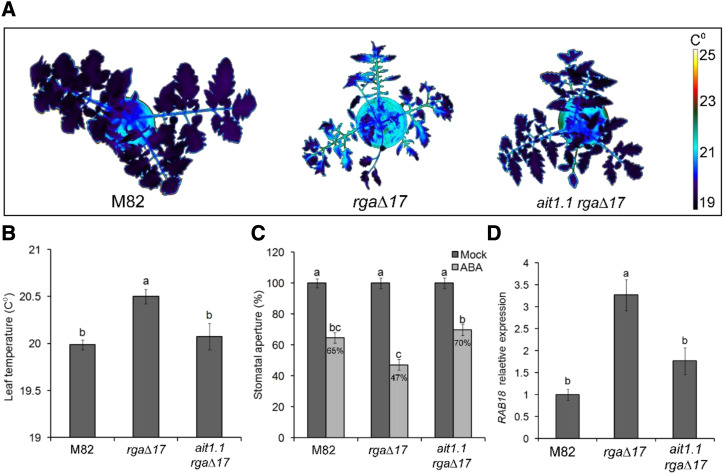
Although AIT1.2 was upregulated by PRO, it showed very low expression levels (∼20-fold lower than AIT1.1; Supplemental Table S1) and its expression in guard cells was similar to whole leaf tissue. We therefore focused further on AIT1.1. We generated clustered regularly interspaced short palindromic repeats-Cas9 (CRISPR-Cas9)–derived ait1.1 mutant and obtained two independent alleles. Both mutations caused a frame shift and premature stop codon (Supplemental Fig. S4). Homozygous plants of the two lines exhibited mild growth suppression (Supplemental Fig. S5, A and B). Thermal imaging showed lower leaf-surface temperature in ait1.1, suggesting that transpiration in ait1.1 is higher than in M82 (Fig. 4A; Supplemental Fig. S5C). The ait1.1 plants exhibited larger stomatal aperture (Fig. 4B; Supplemental Fig. S5D), higher stomatal conductance (measured as gs) after ABA treatment (Fig. 4C), and partial inhibition of stomatal closure in response to application of ABA to epidermal peels (Fig. 4D).
Figure 4.
Loss of the ABA transporter AIT1.1 increased transpiration and inhibited ABA responses in guard cells. A, Thermal imaging of M82 and CRISPR-Cas9–derived ait1.1 mutant. Images were digitally extracted for comparison. Numbers below plants are the leaf-surface temperature, and the values are means of three replicates, each measured 20 times ± se. Star above the number represents significant differences between lines by Student’s t test (P < 0.05). B, Stomatal aperture measured on imprints of abaxial epidermis taken at 11:00 am from leaves no. 3 and 4 below the apex. Values are means of four replicates, each with ∼100 measurements (stomata) ± se. Star above the column represents significant difference between respective treatments (Student’s t test, P < 0.05). C, Stomatal conductance (gs) in the fourth leaf below the apex in M82 and ait1.1 plants, 1 h after treatment with 10 μm of ABA (or Mock). Values are means of six measurements taken from three different plants ± se. D, Stomatal aperture in M82 and ait1.1 epidermal peels (taken from leaves no. 3 and 4 below the apex) treated or not treated (Mock) with 10 μm of ABA. One hour after the ABA treatment, stomatal aperture was measured. Values are mean percentage of mock of four replicates, each with ∼100 measurements (stomata) ± se. Different letters above the columns in C and D represent significant differences between lines and treatments by Tukey–Kramer HSD test (P < 0.05).
We then introgressed the 35S:rga∆17 transgene into the ait1.1 background by crosses and generated homozygous ait1.1 plants overexpressing rga∆17. We confirmed the presence of the transgene (35S:rga∆17) by the phenotype (shorter stem and smaller, darker, and more serrated leaves; Supplemental Fig. S6), and the ait1.1 mutation by sequencing. Stable DELLA (35S:rga∆17) inhibited germination, but the loss of AIT1.1 suppressed this effect and promoted germination (Supplemental Fig. S7), suggesting that the inhibition effect of DELLA on germination in tomato (Zhu et al., 2019) is AIT1.1-dependent. We next tested the effect of the mutation on DELLA activity in guard cells. The loss of AIT1.1 suppressed the effect of stable DELLA overexpression on transpiration as indicated by the lower leaf-surface temperature in 35S:rga∆17 ait1.1 plants compared to 35S:rga∆17 (Fig. 5, A and B; Supplemental Fig. S8). In addition, stomatal response to ABA in epidermal peels, and the expression of the ABA-induced gene RAB18 in isolated guard cells, were suppressed in 35S:rga∆17 ait1.1 compared to 35S:rga∆17 (Fig. 5, C and D). These results suggest that AIT1.1 is required for DELLA to promote ABA responses in guard cells.
Figure 5.
ait1.1 suppressed the effect of PRO on ABA responses in guard cells. A, Thermal imaging of M82, 35S:rgaΔ17 (rgaΔ17), and rgaΔ17 ait1.1. Images were digitally extracted for comparison. B, Leaf-surface (leaves no. 3 and 4 below the apex) temperature of M82, rgaΔ17, and rgaΔ17 ait1.1. plants. Values are means of three replicates measured 20 times ± se. C, Stomatal aperture in M82, rgaΔ17, and rgaΔ17 ait1.1 epidermal peels (from leaves no. 3 and 4 below the apex) treated or not treated (Mock) with 10 μm of ABA. One hour after the ABA treatment, stomatal aperture was measured. Values are mean percentage of Mock of four replicates, each with ∼100 measurements (stomata) ± se. D, RT-qPCR analysis of RAB18 expression in M82, rgaΔ17, and rgaΔ17 ait1.1 guard cells, isolated from leaves no. 3 and 4 below the apex. Values are means of four biological replicates ± se. Different letters above the columns in B and D represent significant differences between lines by Student’s t test (P < 0.05). Different letters above the columns in C represent significant differences between lines and treatments by Tukey–Kramer HSD test (P < 0.05). The values for M82 were set to 1.
DISCUSSION
Drought avoidance is a major plant-adaptation strategy to survive transient water-deficit conditions (Kooyers, 2015). To avoid dehydration, plants close their stomata to reduce transpiration and can use the available water in the soil more slowly and for a longer time before the next rain comes (Martin-St Paul et al., 2017; Gupta et al., 2020). While ABA has a major role in the regulation of transpiration under water-deficit conditions, accumulating evidence suggests that GA antagonizes these ABA responses. Increased GA levels or activity promote stomatal opening in Commelina benghalensis, Vicia faba, Arabidopsis, and tomato (Santakumari and Fletcher, 1987; Göring et al., 1990; Nir et al., 2017; Sukiran et al., 2020). Reduced GA activity and DELLA accumulation suppress canopy expansion and xylem hydraulic conductivity and promotes stomatal closure, all leading to lower transpiration (Nir et al., 2014, 2017; Illouz-Eliaz et al., 2020). It was suggested that water deficiency inhibits GA accumulation to promote adaptation to drought (Colebrook et al., 2014).
Because ABA-induced stomatal closure is mediated by the phosphorylation of ion channels and not by activation of gene transcription (Cutler et al., 2010; Kim et al., 2010; Munemasa et al., 2015), it was unclear how the transcriptional regulator DELLA (PRO) affects ABA-induced stomatal closure. We hypothesized that PRO affects the transcription of either the ABA signaling component or the ABA transporter (Nir et al., 2017). RNA-seq analysis of isolated guard cells (M82, 35S:pro∆17, and pro) identified the ABA transporter AIT1.1 as upregulated by PRO. The Arabidopsis AIT1, also called NRT1.2 or NPF4.6, is an ABA importer (Kanno et al., 2012). This gene is also upregulated by DELLA in the Arabidopsis shoot apical meristems (Serrano-Mislata et al., 2017). The Arabidopsis ait1 mutant exhibited increased water loss due to open stomata (Kanno et al., 2012); however, expression analysis (based on reporter line) suggests that AIT1 is active in the vascular tissue of inflorescence stems, but not in stomata. Although the promoter:GUS line suggested that the tomato AIT1.1 gene is also active in the vascular tissue, RT-qPCR analysis showed that the highest expression levels of this gene is in guard cells. This, together with the open-stomatal phenotype of the ait1.1 mutant, suggest that protein function in tomato to be an ABA transporter, possibly as an importer, in guard cells. The source for ABA that stimulates stomatal closure under water-deficit conditions is still not clear. Several studies suggest that ABA is produced in the phloem companion cells and/or in guard cells (Bauer et al., 2013; Kuromori et al., 2014; Merilo et al., 2018). Other studies suggest that the leaf mesophyll cells are the major site of ABA production under water-deficit conditions (McAdam and Brodribb, 2018). The localization of AIT1.1 in guard cells and its possible activity as an importer, regulating ABA influx into guard cells, supports the hypothesis that at least part of the ABA that stimulates stomatal closure in response to drought comes from other cells—either leaf mesophyll or companion cells.
Because ait1.1 mutant exhibited only partial inhibition of stomatal closure in response to exogenous ABA, AIT1.1 is probably not the only ABA transporter in tomato guard cells. Still, among the AIT1 group of transporters, AIT1.1 is the most dominant one, based on its expression level. In Arabidopsis, an importer from another group, the ABC transporter ABCG40, is active in guard cells (Kang et al., 2010; Kuromori et al., 2014). We identified the ABCG40 homolog in the RNA-seq analysis of isolated guard cells as PRO-upregulated. However, this was not confirmed by RT-qPCR analysis. Thus, ABCG40 may contribute to ABA uptake into tomato guard cells, but probably does not mediate the effect of PRO on guard-cell ABA responses.
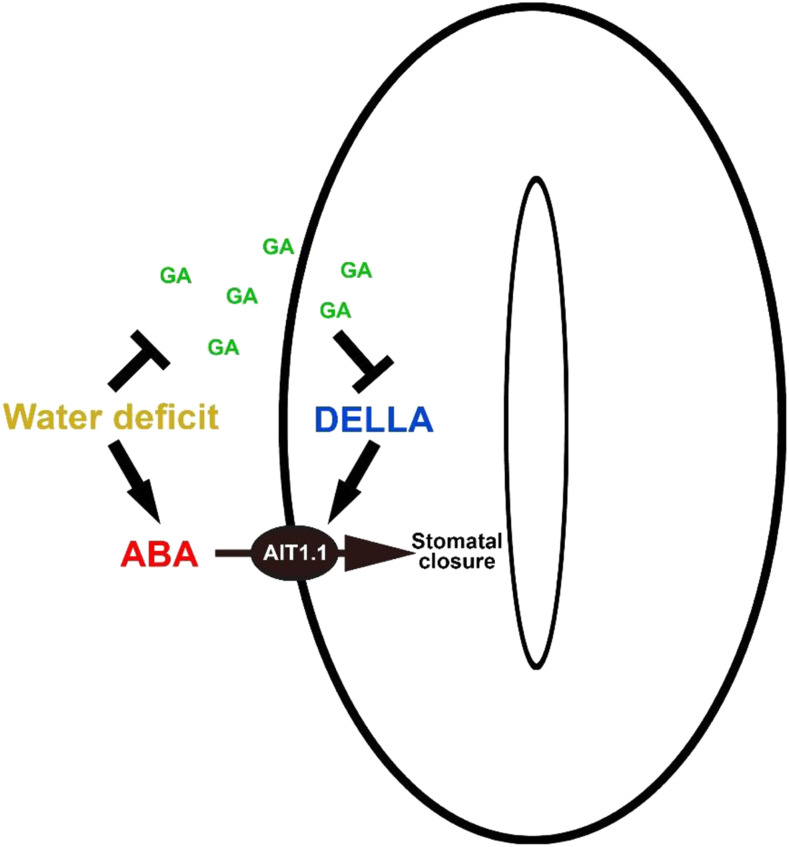
The up- and downregulation of AIT1.1 by stable PRO and pro loss of function, respectively, and the suppression of PRO-promoted ABA responses in ait1.1 guard cells, suggests that AIT1.1 mediates the effect of PRO on ABA-induced stomatal closure. It is possible that water deficiency reduces the levels of active GAs (Colebrook et al., 2014), leading to PRO accumulation in guard cells (Nir et al., 2017). PRO promotes the expression of the ABA importer AIT1.1, facilitating ABA uptake into guard cells, leading to faster stomatal closure (Fig. 6).
Figure 6.
Suggested model of the crosstalk between GA and ABA in guard cells. Water-deficit conditions suppress GA accumulation (Colebrook et al., 2014), leading to DELLA (PRO) stabilization. In guard cells, PRO promotes the expression of the ABA importer AIT1.1, facilitating ABA uptake into guard cells, and stomatal closure.
The crosstalk between GA and ABA has been investigated for many years (Weiss and Ori, 2007; Shu et al., 2018). This crosstalk is largely dependent on DELLA; DELLA promotes ABA synthesis and signaling, and ABA promotes DELLA stability—and therefore inhibits GA signaling (Achard et al., 2006; Piskurewicz et al., 2008; Lim et al., 2013; Liu and Hou, 2018). Here we bring evidence that PRO (DELLA) promotes ABA responses in guard cells via the upregulation of the ABA transporter AIT1.1, suggesting that the crosstalk between GA and ABA is mediated at multiple levels through hormone biosynthesis, signaling, and transport.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Hormone Treatments
Tomato (Solanum lycopersicum) plants in M82 background (sp/sp) were used throughout this study. The pro mutant was in the M82 background (Fleishon et al., 2011). The CRISPR-derived ait1.1 mutant and the transgenic lines 35S:rga∆17 (Livne et al., 2015), 35S:pro∆17, pKST1:LhG4, OP:YFP (Nir et al., 2017), and pMAPKKK18:GUS were generated in the M82 background. Plants were grown in a growth room set to a photoperiod of 12/12-h night/day, light intensity of 200 μmol m−2 s−1 and 25°C, and irrigated to saturation. In other experiments, plants were grown in a greenhouse under natural day-length conditions, light intensity of 700 to 1,000 μmol m−2 s−1, and temperature of 18°C to 30°C. The seeds were harvested from ripe fruits and treated with 1% (v/v) sodium hypochlorite followed by 1% (w/v) Na3PO4 and 12 water, and incubated with 10% (w/v) Suc overnight in 37°C. Seeds were stored dry at room temperature. (±)-ABA dissolved in DMSO (Sigma-Aldrich) was applied to plants by spraying.
CRISPR/Cas9 Mutagenesis, Tomato Transformation, and Selection of Mutant Alleles
Four single-guide RNAs (sgRNAs; Supplemental Table S2) were designed to target the AIT1.1 gene, using the CRISPR-P tool (http://cbi.hzau.edu.cn/crispr). Vectors were assembled using the Golden Gate cloning system, as described by Weber et al. (2011). Final binary vectors, pAGM4723, were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. The constructs were transferred into M82 cotyledons using transformation and regeneration methods described by McCormick (1991). Kanamycin-resistant T0 plants were grown and independent transgenic lines were selected and self-pollinated to generate homozygous transgenic lines. The genomic DNA of each plant was extracted, and genotyped by PCR for the presence of the Cas9 construct. The CRISPR-Cas9–positive lines were further genotyped for mutations using a forward primer to the upstream sequence of the sgRNA1 target and a reverse primer downstream of the sgRNA2 target sequence. The target genes in all mutant lines were sequenced. Several homozygous and heterozygous lines were identified, and independent mutant lines for each gene were selected for further analysis. The Cas9 construct was segregated out by crosses to M82.
Molecular Cloning/Construct and Transactivation
To generate the ABA-reporter transgenic plants, the plasmid containing the AtMAPKKK18 promoter were kindly supplied by Assaf Mosquna (Okamoto et al., 2013). To generate pMAPKKK18:GUS the MAPKKK18 promoter was inserted into the pRITA vector downstream to the GUS start codon into the KpnI and BamHI sites (Steiner et al., 2012). Primers sequences for cloning are presented in Supplemental Table S3. This construct was then cloned into the pART27 binary vector and introduced to the A. tumefaciens strain GV3101 by electroporation to generate transgenic M82 tomato plants (as described above). To specifically express YFP in guard cells, the LhG4 transactivation system (Moore et al., 1998) with the KST1 promoter was used, with pKST1:LhG4 as the driver line and OP:YFP as the responder line. The cross between these lines generated the transactivated line pKST1>>YFP.
pAIT1.1:GUS Molecular Cloning
The 1,380 bp upstream to the AIT1.1 start codon were amplified by PCR (−49 to −1,429). pAIT1.1 was inserted into the KpnI and BamHI sites in the pRITA vector downstream to the GUS start. Primers sequences for cloning are presented in Supplemental Table S3. This construct was then cloned into the pART27 binary vector and introduced to A. tumefaciens strain GV3101 by electroporation to generate transgenic M82 tomato plants (as described above).
Library Constructions and Sequencing
Total RNA was extracted from isolated guard cells using an RNeasy Plant Mini Kit (Qiagen). Libraries were prepared at the Crown Institute for Genomics (The Nancy & Stephen Grand Israel National Center for Personalized Medicine [G-INCPM], Weizmann Institute of Science). Five-hundred nanograms of total RNA for each sample were processed using the in-house poly-A–based RNA-seq protocol. Libraries were evaluated with the tools Qubit and TapeStation (the INCPM Unit, Weizmann Institute of Science). Sequencing libraries were constructed with barcodes to allow multiplexing of nine samples on one lane of a HiSeq 2500 machine (Illumina), using the Single-Read 60 protocol (v4), yielding a median of 29.4 million reads per sample.
Sequence Data Analysis
Stretches of Poly-A/T and Illumina adapters were trimmed from the reads using the tool Cutadapt (Martin, 2011); resulting reads shorter than 30 bp were discarded. Reads were mapped to the S. lycopersicum reference genome (release SL3.00) using the program STAR (with “End-To-End” option and “out Filter Mismatch Nover Lmax = 0.04”; Dobin et al., 2013). The annotation file was downloaded from the International Tomato Genome Sequencing Project (3.2; Sol Genomics). Expression levels for each gene were quantified with the program htseq-count (Anders et al., 2015), using the gtf file given above (in “Union” mode). Differential expression gene analysis was performed using the program DESeq2 (Love et al., 2014) with the “betaPrior,” “cooksCutoff,” and independent filtering parameters set to “False.” Raw P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg (1995). The RNA-seq data discussed in this publication have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through Gene Expression Omnibus Series accession number GSE:143999 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE143999).
RNA Extraction and cDNA Synthesis
Total RNA was extracted with an RNeasy Plant Mini Kit (Qiagen). For synthesis of cDNA, SuperScript II reverse transcriptase (18064014; Invitrogen) and 3 mg of total RNA were used according to the manufacturer’s instructions.
RT-qPCR Analysis
RT-qPCR analysis was performed using an Absolute Blue qPCR SYBR Green ROX Mix kit (AB-4162/B; Thermo Fisher Scientific). Reactions were performed using a Rotor-Gene 6000 Cycler (Corbett Research). A standard curve was obtained using dilutions of the cDNA sample. The expression was quantified using the Rotor-Gene’s software (Corbett Research). Three independent technical repeats were performed for each sample. Relative expression was calculated by dividing the expression level of the examined gene by that of SlACTIN. The target-gene-to-ACTIN ratio was then averaged. All primer sequences are presented in Supplemental Table S3.
Isolation of Guard Cells
Fully expanded leaves (leaf no. 3 or 4 below the apex) without the central veins were ground twice, for 60 s each time, in a blender containing 100 mL of cold distilled water. The blended mixture was poured onto a 100-μm nylon mesh (Sefar) and the remaining epidermal peels were rinsed thoroughly with 0.5 L of cold deionized water. The peels were then transferred into 2-mL tubes and frozen in liquid nitrogen.
Microscopy and Confocal Imaging Analysis
Imaging was done using a confocal laser scanning microscope model SP8 (Leica Microsystems) with an HCX PL APO CS 20×/0.70 dry objective (Leica Microsystems) for YFP in epidermal strips. YFP was excited with the 514-nm laser line, and the 520- to 560-nm filter was used for emission. Images were later analyzed using the “Fit-Ellipse” tool from the software ImageJ (http://rsb.info.nih.gov/ij/).
Thermal Imaging
Thermal images were obtained using a model no. A655sc FOV 15 camera (FLIR Systems). The camera was mounted vertically about 50 cm above the plants. Mean temperature of leaflets from leaves no. 3 and 4 below the apex were calculated using the customized region-of-interest tool, according to the manufacturer’s instructions.
GUS Staining
Histochemical detection of GUS activity was performed using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide as described in Donnelly et al. (1999). Samples were put on glass coverslips and photographed under a model no. ICC50 W bright-field inverted microscope (Leica Microsystems). Images were later analyzed using the software ImageJ. A microscopic ruler (Olympus) was used for size calibration.
Transport Assays
Yeast two-hybrid assays were performed using the ProQuest Two-Hybrid System with Gateway Technology (Thermo Fisher Scientific). Arabidopsis (Arabidopsis thaliana) ABI1 and PYR1 cDNAs cloned in pENTR-d-TOPO were introduced into pDEST22 and pDEST32, respectively, by LR recombination reactions. Tomato AIT1.1 cloned in pENTR-d-TOPO was introduced into pYES-DEST52 in which the GAL1 promoter had been replaced with the AHD promoter (Kanno et al., 2012). Arabidopsis AIT1 was cloned in pYES-DEST52 as in a previous study (Kanno et al., 2012). The pDEST22, pDES32, and pYES-DEST52 constructs were transformed into a yeast (Saccharmyces cerevisiae) strain BY20249. Several (∼10) independent colonies were mixed and precultured in media (synthetic defined [SD], -Leu, -Trp, and -Ura) overnight at 30°C and then inoculated on selection media (SD, -Leu, -Trp, -Ura, -His, and 1 mm of 3AT) with (+) or without (−) ABA.
For direct measurements of transport activities by LC-MS/MS, AIT1.1 was cloned into the standard pYES-DEST52 vector and transformed into a yeast strain INVSc1. Assays were performed as described in Kanno et al. (2016). Hormones were extracted from yeast cells with 1 mL of acetone containing 1% (v/v) acetic acid, and the supernatants after centrifugation were dried with N2 gas. The extracts were dissolved in 1 mL of water containing 1% (v/v) acetic acid and loaded onto a cartridge column (1-cc Oasis Wax; Waters) that had been pretreated with 0.5 mL of acetonitrile and then with water containing 1% (v/v) acetic acid. After washing with 1 mL of water containing 1% (v/v) and then with 80% (v/v) acetonitrile, ABA, GA1, GA4, IAA, JA, and JA-Ile were eluted with 80% (v/v) acetonitrile containing 1% (v/v) acetic acid. Salicylic acid was then eluted with 2 mL of acetonitrile containing 5% (v/v) formic acid. The eluents containing hormones were dried with N2 gas and then dissolved in 50 μL of water containing 1% (v/v) acetic acid to be analyzed by LC-MS/MS. LC-MS/MS analysis was performed as described in Kanno et al. (2016).
Stomatal Aperture Measurements
Stomatal aperture was determined using the rapid imprinting technique described by Geisler et al. (2000). Light-bodied vinylpolysiloxane dental resin (eliteHD+; Zhermack Clinical) was attached to the abaxial side of the terminal leaflet (leaf no. 4 below the apex) and then removed as soon as it dried (minutes). The resin epidermal imprints were covered with transparent nail polish, which was removed after it dried, and served as a mirror image of the resin imprint. The nail-polish imprints were put on glass coverslips and photographed under a light microscope, as described above.
Stomatal Response to ABA
Abaxial epidermal peels taken from leaf no. 4 below the apex were incubated in stomatal opening buffer, containing 20 mm of potassium chloride and 5 mm of MES at pH 6.15 (Wigoda et al., 2006) for 2 h under light conditions (400 μmol m−2 s−1). The peels were then transferred to a fresh stomatal opening buffer with or without 10 μm of ABA, under the same light conditions. After 60 min, the peels were put on glass coverslips and photographed under the bright-field inverted microscope as described above. Stomatal images were later analyzed using the software ImageJ for stomatal aperture measurements as described above.
Germination Assay
Fresh seeds were germinated in petri dishes on Murashige & Skoog medium in a growth room set to a photoperiod of 12/12-h night/day, with a light intensity of 200 μmol m−2 s−1, and at a temperature of 25°C. Germination was scored when the radicle pierced the seed coat.
Statistical Analyses
All assays were conducted with three or more biological replicates and analyzed using the software JMP (SAS Institute). Means comparison was conducted using ANOVA with post hoc Tukey–Kramer Honestly Significant Difference (HSD; for two comparisons) and Student’s t tests (for one comparison; P < 0.05). Summary of the statistical parameters, and significant differences, are presented in Supplemental Table S4.
Accession Numbers
Sequence data from this article can be found in the Sol Genomics Network (https://solgenomics.net/) under the following accession numbers: ACTIN, Solyc11g005330; RAB18, Solyc05g053600 ABCG40, Solyc02g084850; AIT1.1, Solyc05g006990; AIT1.2, Solyc05g007000; AIT1.3, Solyc04g005790; and AIT1.4, Solyc03g113250.
Supplemental Data
The following supplemental data are available.
Supplemental Figure S1. Molecular phylogenetic analysis of AIT1 protein sequences in tomato and Arabidopsis.
Supplemental Figure S2. PRO activity has no effect on the expression of the tomato ABCG40 homolog.
Supplemental Figure S3. AIT1.1 promoter drives expression in the vascular tissue.
Supplemental Figure S4. Sequence analyses of ait1.1 no. 1 and ait1.1 no. 7 CRISPR mutant alleles.
Supplemental Figure S5. Loss of AIT1.1 suppresses growth, and increases transpiration and stomatal aperture.
Supplemental Figure S6. Plant and leaf phenotypes of 35S:rga∆17 ait1.1.
Supplemental Figure S7. Loss of AIT1.1 promotes germination of 35S:rga∆17 seeds.
Supplemental Figure S8. Thermal imaging of M82, 35S:rga∆17, and 35S:rga∆17 ait1.1 leaves.
Supplemental Table S1. ABA- and guard-cell–related PRO-regulated genes (fold change > ±2).
Supplemental Table S2. Guide RNAs used in this study.
Supplemental Table S3. Primers used in this study.
Supplemental Table S4. Summary of statistical parameters and significant differences.
Supplemental Dataset S1. Results of the RNA-seq analysis.
Acknowledgments
We thank Naomi Ori, Menachem Moshelion (The Hebrew University of Jerusalem), and Yuval Eshed (Weizmann Institute of Science) for their valuable discussions and suggestions. We thank Dr. Assaf Mosquna (The Hebrew University of Jerusalem) for providing the MAPKKK18 promoter. We thank Drs. Tali Shalit and Gilgi Friedlander (INCPM unit, Weizmann Institute of Science) for their help with the bioinformatic analysis.
Footnotes
This work was supported in part by the Israel Science Foundation (grant no. 779/15 to D.W.).
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, van der Straeten D, Peng J, Harberd NP(2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W(2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel GW, Mullen RT, Bewley JD(2008) Procera is a putative DELLA mutant in tomato (Solanum lycopersicum): Effects on the seed and vegetative plant. J Exp Bot 59: 585–593 [DOI] [PubMed] [Google Scholar]
- Bauer H, Ache P, Lautner S, Fromm J, Hartung W, Al-Rasheid KA, Sonnewald S, Sonnewald U, Kneitz S, Lachmann N, et al. (2013) The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr Biol 23: 53–57 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y(1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc 57: 289–300 [Google Scholar]
- Cantoro R, Crocco CD, Benech-Arnold RL, Rodríguez MV(2013) In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: Possible role of this interaction in the expression of seed dormancy. J Exp Bot 64: 5721–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebrook EH, Thomas SG, Phillips AL, Hedden P(2014) The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol 217: 67–75 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR(2010) Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun T-P(2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR(2013) STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG(1999) Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol 215: 407–419 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE(2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishon S, Shani E, Ori N, Weiss D(2011) Negative reciprocal interactions between gibberellin and cytokinin in tomato. New Phytol 190: 609–617 [DOI] [PubMed] [Google Scholar]
- Geisler M, Nadeau J, Sack FD(2000) Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by the too many mouths mutation. Plant Cell 12: 2075–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring H, Koshuchowa S, Deckert C(1990) Influence of gibberellic acid on stomatal movement. Biochem Physiol Pflanz 186: 367–374 [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang Z-L, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Rico-Medina A, Caño-Delgado AI(2020) The physiology of plant responses to drought. Science 368: 266–269 [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y(2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM(2012) Gibberellin signaling: A theme and variations on DELLA repression. Plant Physiol 160: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Lee LY, Xia K, Yan Y, Yu H(2010) DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894 [DOI] [PubMed] [Google Scholar]
- Illouz-Eliaz N, Nissan I, Nir I, Ramon U, Shohat H, Weiss D(2020) Mutations in the tomato gibberellin receptors suppress xylem proliferation and reduce water loss under water-deficit conditions. J Exp Bot 71: 3603–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Tattersall A, Piazza P, Hay A, Martinez-Garcia JF, Schmitz G, Theres K, McCormick S, Tsiantis M(2008) PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato. Plant J 56: 603–612 [DOI] [PubMed] [Google Scholar]
- Kang J, Hwang J-U, Lee M, Kim Y-Y, Assmann SM, Martinoia E, Lee Y(2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc Natl Acad Sci USA 107: 2355–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Hanada A, Chiba Y, Ichikawa T, Nakazawa M, Matsui M, Koshiba T, Kamiya Y, Seo M(2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci USA 109: 9653–9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Oikawa T, Chiba Y, Ishimaru Y, Shimizu T, Sano N, Koshiba T, Kamiya Y, Ueda M, Seo M(2016) AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun 7: 13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI(2010) Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooyers NJ.(2015) The evolution of drought escape and avoidance in natural herbaceous populations. Plant Sci 234: 155–162 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Miyaji T, Yabuuchi H, Shimizu H, Sugimoto E, Kamiya A, Moriyama Y, Shinozaki K(2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc Natl Acad Sci USA 107: 2361–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Seo M, Shinozaki K(2018) ABA transport and plant water stress responses. Trends Plant Sci 23: 513–522 [DOI] [PubMed] [Google Scholar]
- Kuromori T, Sugimoto E, Shinozaki K(2014) Intertissue signal transfer of abscisic acid from vascular cells to guard cells. Plant Physiol 164: 1587–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q-F, Wang C, Jiang L, Li S, Sun SS, He J-X(2012) An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci Signal 5: ra72. [DOI] [PubMed] [Google Scholar]
- Lim S, Park J, Lee N, Jeong J, Toh S, Watanabe A, Kim J, Kang H, Kim DH, Kawakami N, et al. (2013) ABA-insensitive3, ABA-insensitive5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25: 4863–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hou X(2018) Antagonistic regulation of ABA and GA in metabolism and signaling pathways. Front Plant Sci 9: 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livne S, Lor VS, Nir I, Eliaz N, Aharoni A, Olszewski NE, Eshed Y, Weiss D(2015) Uncovering DELLA-independent gibberellin responses by characterizing new tomato PROCERA mutants. Plant Cell 27: 1579–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Blázquez MA, Alabadí D(2013) Genomic analysis of DELLA protein activity. Plant Cell Physiol 54: 1229–1237 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S(2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.(2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 1–10 [Google Scholar]
- Martin-St Paul N, Delzon S, Cochard H(2017) Plant resistance to drought depends on timely stomatal closure. Ecol Lett 20: 1437–1447 [DOI] [PubMed] [Google Scholar]
- McAdam SAM, Brodribb TJ(2018) Mesophyll cells are the main site of abscisic acid biosynthesis in water-stressed leaves. Plant Physiol 177: 911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S.(1991) Transformation of tomato with Agrobacterium tumefaciens In Linclsey H, ed, Plant Tissue Culture Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–9 [Google Scholar]
- Merilo E, Yarmolinsky D, Jalakas P, Parik H, Tulva I, Rasulov B, Kilk K, Kollist H(2018) Stomatal VPD response: There is more to the story than ABA. Plant Physiol 176: 851–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore I, Gälweiler L, Grosskopf D, Schell J, Palme K(1998) A transcription activation system for regulated gene expression in transgenic plants. Proc Natl Acad Sci USA 95: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI(2015) Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 28: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir I, Moshelion M, Weiss D(2014) The Arabidopsis gibberellin methyl transferase 1 suppresses gibberellin activity, reduces whole-plant transpiration and promotes drought tolerance in transgenic tomato. Plant Cell Environ 37: 113–123 [DOI] [PubMed] [Google Scholar]
- Nir I, Shohat H, Panizel I, Olszewski N, Aharoni A, Weiss D(2017) The tomato DELLA protein PROCERA acts in guard cells to promote stomatal closure. Plant Cell 29: 3186–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Peterson FC, Defries A, Park SY, Endo A, Nambara E, Volkman BF, Cutler SR(2013) Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc Natl Acad Sci USA 110: 12132–12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L(2008) The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razem FA, Baron K, Hill RD(2006) Turning on gibberellin and abscisic acid signaling. Curr Opin Plant Biol 9: 454–459 [DOI] [PubMed] [Google Scholar]
- Santakumari M, Fletcher RA(1987) Reversal of triazole-induced stomatal closure by gibberellic acid and cytokinins in Commelina benghalensis. Physiol Plant 71: 95–99 [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong D-H, An G, Kitano H, Ashikari M, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Serrano-Mislata A, Bencivenga S, Bush M, Schiessl K, Boden S, Sablowski R(2017) DELLA genes restrict inflorescence meristem function independently of plant height. Nat Plants 3: 749–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q(2013) ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet 9: e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Zhou W, Chen F, Luo X, Yang W(2018) Abscisic acid and gibberellins antagonistically mediate plant development and abiotic stress responses. Front Plant Sci 9: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D(2012) The Arabidopsis o-linked n-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24: 96–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukiran AN, Steel PG, Knight MR(2020) Basal stomatal aperture is regulated by GA-DELLAs in Arabidopsis. J Plant Physiol 250: 153182. [DOI] [PubMed] [Google Scholar]
- Sun T-P, Gubler F(2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55: 197–223 [DOI] [PubMed] [Google Scholar]
- Van Tuinen A, Peters AHLJ, Kendrick RE, Zeevaart JAD, Koornneef M(1999) Characterization of the procera mutant of tomato and the interaction of gibberellins with end-of-day far-red light treatments. Physiol Plant 106: 121–128 [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S(2011) A modular cloning system for standardized assembly of multigene constructs. PLoS One 6: e16765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss D, Ori N(2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144: 1240–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigoda N, Ben-Nissan G, Granot D, Schwartz A, Weiss D(2006) The gibberellin-induced, cysteine-rich protein GIP2 from Petunia hybrida exhibits in planta antioxidant activity. Plant J 48: 796–805 [DOI] [PubMed] [Google Scholar]
- Xiong W, Ye T, Yao X, Liu X, Ma S, Chen X, Chen M-L, Feng Y-Q, Wu Y(2018) The dioxygenase GIM2 functions in seed germination by altering gibberellin production in Arabidopsis. J Integr Plant Biol 60: 276–291 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S.(2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yoshida H, Hirano K, Sato T, Mitsuda N, Nomoto M, Maeo K, Koketsu E, Mitani R, Kawamura M, Ishiguro S, et al. (2014) DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc Natl Acad Sci USA 111: 7861–7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Kang X, Lor VS, Weiss D, Olszewski N(2019) Characterization of a semidominant dwarfing PROCERA allele identified in a screen for CRISPR/Cas9-induced suppressors of loss-of-function alleles. Plant Biotechnol J 17: 319–321 [DOI] [PMC free article] [PubMed] [Google Scholar]