Abstract
Maintenance of genomic integrity depends on the spatiotemporal recruitment and regulation of DNA damage response and repair proteins at DNA damage sites. These highly dynamic processes have been widely studied using laser microirradiation coupled with fluorescence microscopy. Laser microirradiation has provided a powerful methodology to identify and determine mechanisms of DNA damage response pathways. Here we describe methods used to analyze protein recruitment dynamics of fluorescently tagged or endogenous proteins to laser-induced DNA damage sites using laser scanning and fluorescence microscopy. We further describe multiple applications employing these techniques to study additional processes at DNA damage sites including transcription. Together, we aim to provide robust visualization methods employing laser-microirradiation to detect and determine protein behavior, functions and dynamics in response to DNA damage in mammalian cells.
Keywords: Laser microirradiation, DNA repair, DNA damage, Transcription, Fluorescence microscopy, Live-cell imaging
1. Introduction
Genome integrity is constantly challenged by endogenous, as well as exogenous sources of DNA damage including replication stress, reactive cellular metabolites, UV, ionizing radiation, and other mutagens including various chemicals. These threats generate a multitude of different DNA lesions including various base damages and single as well as double-strand breaks in the DNA. These lesions must be identified and faithfully repaired in order to avoid mutations that can result in genomic instability [1]. Defects in the ability to maintain genome stability are known to contribute to several human diseases and conditions including neurodegeneration, aging, and cancer.
To protect the genome, organisms employ highly orchestrated surveillance and repair mechanisms that are collectively known as DNA damage response (DDR) pathways [2, 3]. These pathways consist of DNA damage sensor proteins that detect damage, which then activate signaling pathways that promote checkpoint responses and DNA repair reactions. The spatiotemporal regulation of these processes is vital for coordinating these activities within chromatin to ensure genome integrity and cell survival [4, 5]. Following DNA damage, a multitude of proteins are localized to DNA damage sites in an ordered manner [4, 6]. For example, the MRE11-RAD50-NBS1 (MRN) complex and PARP detect DNA double-strand break (DSBs) and DNA single-strand breaks respectively [7]. For DSBs, these sensor proteins promote the phosphorylation of histone H2AX on serine 139 residue (γH2AX) via activation and recruitment of DDR kinases including ATM [8, 9]. The mediator of DNA damage checkpoint 1 (MDC1) binds to phosphorylated histone H2AX amplifying downstream DNA damage signaling. The ATM kinase also phosphorylates MDC1 and this modification is recognized by the RNF8 E3 ubiquitin ligase. RNF8 initiates an ubiquitin signaling cascade at the DNA damage site for recruitment of the DNA repair factors 53BP1 and BRCA1 [10–12].
Significant progress has been made to identify and characterize key proteins involved in these DNA damage response pathways. A key method that has been integral to our identification of these proteins has been the use of laser microirradiation. This technique allows localized tracks of DNA damage to be generated, including DSBs, in cells using lasers coupled to fluorescent microscopes. Upon DNA damage instigation, proteins can be analyzed in live or fixed cells to study the spatial, temporal and coordinated dynamics of DDR proteins and the pathways that they regulate at DNA damage sites.
In addition to studying DNA damage response factor recruitment using laser microirradiation, this technique can also be used to study other nuclear processes at DNA damage sites and protein dynamics within cells [13, 14]. For example, various types of DNA lesions result in local inhibition of transcription, which is thought to be critical for limiting the production of anomalous transcripts and to avoid conflicts between transcription and DDR machinery [15]. Several groups have shown that cells coordinate transcriptional responses with the DDR. For example, the ATM kinase promotes transcriptional silencing through DNA damage responsive histone ubiquitylations by RNF8 and RNF168 [16]. In another example, KDM5A, a H3K4 lysine demethylase, is recruited to laser-induced DNA damage sites where it acts to demethylate chromatin to facilitate recruitment of the ZMYND8-NuRD chromatin-remodeling complex [17–19]. Together, these factors enforce transcriptional repression at DNA damage sites, which promotes homologous recombination repair of DSBs, potentially by the removal of transcriptional conflicts. In addition to recruitment dynamics of DDR proteins, laser microirradiation has provided key experimental evidence for understanding transcriptional responses that occur within damaged regions of chromatin, an important process that is as yet not well defined.
In this chapter, we describe detailed methods for employing laser-induced DNA damage in living cells to study recruitment dynamics to DNA lesions of DDR factors. We also provide an assay to study nascent transcription at laser-induced DNA damage sites. These methods can be utilized for detecting, quantifying, and analyzing protein recruitment and transcriptional dynamics at DNA damage sites. These methods also provide insights into the spatiotemporal dynamics of DDR and transcription proteins that occur at DNA damage sites, processes that require coordinated responses of these pathways to ensure genome and epigenome stability.
2. Materials
2.1. Equipment
Microscope equipped with a 405 nm 50 mW laser and CO2/humidity chamber for live imaging (see Note 1).
A 37 °C incubator with 5% CO2 for growing human cells.
2.2. Cell Culture
Glass bottomed dishes with glass well size 12 mm (WillCo Wells, GWST-3512) or 35 mm high grid-500 (ibidi, 81168).
5-Bromo-2′-deoxyuridine (BrdU).
Hoechst 33342.
Adherent human cell line (e.g., U2OS, HeLa, RPE).
Cells are cultured in DMEM medium supplemented with 10% FBS and penicillin–streptomycin–glutamine.
Opti-MEM for transfection.
0.25% trypsin.
Phosphate-buffered saline.
Transfection reagent (e.g., Fugene HD [Promega] or other lipid-based reagent).
Expression vector for fluorescence-tagged gene of interest.
2.3. Immunofluorescence
Fixation buffer: 4% formaldehyde in PBS.
2% Formaldehyde: dilute 4% formaldehyde to 2% with 1× PBS.
0.5% Triton X-100 in PBS.
Microscope coverslips.
CSK (cytoskeleton) buffer: 10 mM PIPES, pH 6.8; 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.5% (v/v) Triton X-100.
Blocking and antibody solution (1× PBS + 3% BSA).
Anti-fade mounting medium.
4′,6-Diamidino-2-phenylindole (DAPI).
Diamond tipped pen.
2.4. Antibodies
Anti-γH2AX (Millipore, 05–636).
Anti-ZMYND8 (Bethyl, A302–089A).
Alexa Fluor 488 goat anti-rabbit IgG (Invitrogen, A11034).
Alexa Fluor 594 goat anti-mouse IgG (Invitrogen, A11032).
2.5. Nascent Transcription Analysis
3. Methods
3.1. Recruitment of Fluorescently Tagged Proteins to DNA Damage Sites Using Laser Microirradiation and Live-Cell Imaging
Day 1: Seeding cells. Seed 5–7 × 104 cells/1.5 mL in a 12 mm glass bottomed dishes. These experiments were optimized for human U2OS cancer cells.
Incubate at 37 °C in 5% CO2 for 24 h.
Day 2: Transfection of fluorescently tagged gene of interest. Transfect vector containing fluorescence tagged target gene for protein expression (as explained below).
Remove medium from target cells.
Add 1.5 mL regular medium without Penicillin/Streptomycin.
Prepare transfection mixture: add 1.5 μg of expression construct to 150 μL of Opti-MEM and add 4.5 μL of Fugene HD transfection reagent. Mix gently by pipetting and incubate tubes for 15 min at room temperature (see Note 2).
Add the transfection mixture dropwise onto the target cells, mix by gently moving the plate forward and backward, side to side. Once mixed, the plate is placed back into the incubator.
-
Day 3: Sensitization of transfected cells for laser microirradiation (see Note 3). Cells are incubated with photo sensitizer containing media before laser microirradiation. This treatment allows for the generation of DNA damage using lower laser powers, which helps to reduce phototoxicity to the cell. For example, it has been calculated that six to ten times higher laser power is required to induce DNA damage using various lasers without presensitization [14]. This can be done using BrdU or Hoechst 33342.
BrdU sensitization: BrdU is a synthetic nucleoside that is an analog of thymidine and is incorporate into newly synthesized DNA. For BrdU, the CH3 group of thymidine is substituted with a bromine atom, which makes BrdU incorporated cells are more sensitive to DNA damage. Incubate cells with 10 μM BrdU containing media 24 h prior to laser microirradiation to allow all of the cells to complete one S-phase and therefore all cells will have incorporated BrdU and be presensitized to laser damage.
Hoechst 33342 sensitization: This compound binds to the minor groove of DNA, which results in sensitizing to laser damage. Incubate cells with 10 μg/mL Hoechst 33342 containing media 10 min prior to laser microirradiation.
Day 4: Laser Microirradiation: Laser induced DNA damage. Turn on the CO2/humidity/temperature-controlled chamber and the confocal laser scanning microscope. Once the chamber has reached the desired parameters (e.g., 5% CO2 and 37 °C), place the dish containing the cells into the chamber (see Note 4).
Visualize the fluorescent protein using the appropriate filters/lasers to locate and focus on the cells that will be damaged (see Notes 5 and 6).
Adjust laser power, HV (current voltage to PMT), gain (amplification of the signal from the PMT), and offset (cut off the higher or lower signal) control. Normally, 0.1% laser power and 500 HV setting are a good starting point for scanning. Adjustment of these parameters can be used to get a better quality image after starting the image scan.
Once the image settings are set, laser DNA damage can be induced using the stimulation settings for the 405 nm laser. A defined region of interest (ROI) is drawn within the cell of interest (Fig. 1a). ROIs can include a line, circle, or box. For the laser, set the scan speed to 20.0 μs/pixel first. Activate the 405 laser using 150 frames at 60% laser power (see Note 7). This is a general setting that will change depending on the application and set up of the microscope including objectives. These settings will need to be empirically optimized by the user based on experimental design and goals.
After putting in the settings and defining the ROI, the laser damage is delivered and imaging and quantification of the fluorescence intensity of the sample is performed, including an identical ROI in an undamaged region of the same cell (Fig. 1b–e, see Notes 8 and 9).
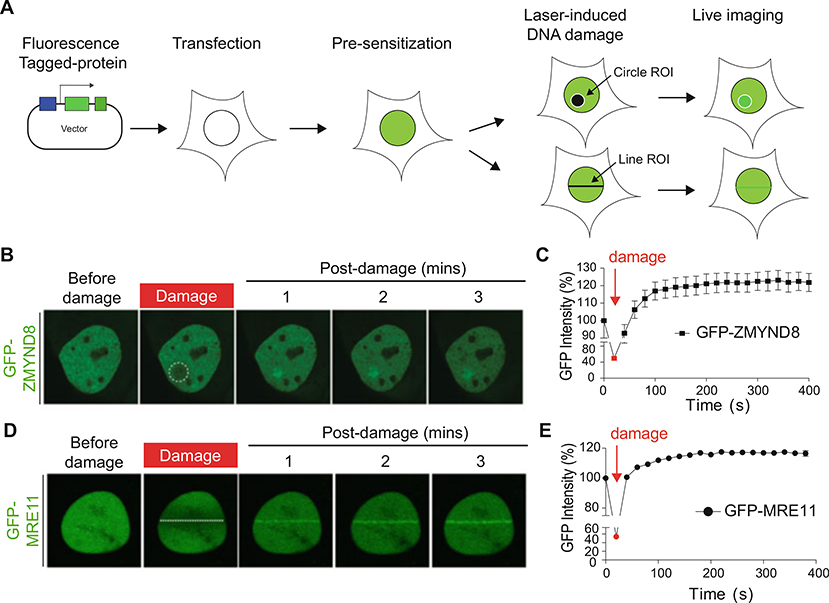
Fig. 1.
Laser microirradiation and live imaging of fluorescently tagged repair protein translocation to DNA damage sites. (a) Schematic illustration of live imaging for protein recruitment to DNA damage sites. (b) Laser damage-induced recruitment of GFP-tagged DDR protein ZMYND8. The white circle indicates the laser path defined by the ROI. (c) Quantification of the GFP intensity using laser microirradiation. The fluorescence intensity of GFP-ZMYND8 in b of damaged versus undamaged regions are plotted as the relative GFP intensity at each time point. Error bars indicate SEM; n > 10. (d) Laser damage-induced recruitment of GFP-tagged DDR protein MRE11. The white line indicates the laser path defined by the ROI. (e) Quantification of d as in c
3.2. Data Analysis of Subheading 3.1
At least ten cells are needs for quantification of kinetics for repair protein recruitment to DNA damage site.
Fluorescence intensities values that are measured by FluoView FV3000 software at DNA damage site and an undamaged region are compared. Intensity of damaged region (ROID)/intensity of undamaged region (ROIU). These are normalized to fluorescence intensity at time zero of the experiment predamage.
Relative fluorescence quantified for each time points are normalized with relative fluorescence predamage (Fig. 1c, e). After DNA damage (ROID/ROIu)/before DNA damage (ROID/ROIu) × 100.
Fluorescence intensities values measured by FluoView FV3000 software and relative fluorescence are calculated using Microsoft Excel and Prism software to create graphs showing the fluorescence intensity as a measure of time post-damage (Fig. 1c, e).
3.3. Endogenous Protein Recruitment to DNA Damage Sites Using Laser Microirradiation
Day 1: Seed Cells. Cells are counted and 5–7 × 104 cells are seeded/dish in glass bottom dishes. For endogenous protein recruitment, it is useful to laser damage cells that are in a defined region. For this a diamond pen is used to etch a cross in the glass (Fig. 2a) or identifiable grids can be used. This makes it easier to locate the damaged cells for imaging once processing of the samples has been performed (see Note 10).
Day 2: Presensitize cells as in Subheading 3.1, step 8. Cells are then microirradiated using the same procedures as in Subheading 3.1 (Fig. 2b, see Note 6).
-
Day 3: Perform laser damage followed by immunofluorescence. After laser damage, cells are treated with CSK buffer or Triton X-100 to permeabilize the cells followed by fixation for preparation for antibody staining (see Note 11).
CSK extraction:
Wash cells three times with 1 mL of 1× PBS, 5 min each wash.
Place dish with cells on ice. Add 500 μL cold CSK buffer and incubate for 5 min. (This is the only step that is required to be performed on ice; other steps are done at room temperature [RT].)
Remove the CSK buffer following three times wash with 1 mL of 1× PBS, 5 min each wash.
Add 500 μL of fixation buffer at RT for 15 min to fix the cells.
Remove the fixation buffer followed by washing in 1 mL of PBS three times, 5 min each wash.
Add 500 μL of 3% BSA blocking solution for 15 min at RT.
Triton X-100 permeabilization:
Wash cells with 1 mL of PBS three times, 5 min each wash.
Add 500 μL of fixation buffer for 15 min to fix cells at RT.
Remove fixation buffer and wash in 1 mL of PBS three times, 5 min each wash.
Add 0.5% Triton X-100 for 15 min at RT.
Remove Triton X-100 buffer and wash in 1 mL of PBS three times, 5 min each wash.
Add 500 μL of 3% BSA blocking solution for 15 min at RT.
During the blocking step, dilute primary antibodies with 3% BSA (in 1× PBS) solution. If dual staining with two antibodies is required, ensure the two antibodies are derived from different hosts to avoid cross-contamination. For example, to stain with ZMYND8 and γH2AX antibodies at the same time, use mouse anti-γH2AX and rabbit anti-ZMYND8.
After blocking, add 100 μL of antibody-BSA mix into the small well of the glass bottom dish.
Incubate with primary antibodies for 1 h at RT. Certain antibodies may need incubation at 4 ºC overnight (see Notes 12 and 13).
Wash cells with 1 mL of PBS three times, 5 min each wash.
Incubate with secondary antibodies for 1 h at RT. Since secondary antibodies are light sensitive, this step should be done in reduced light conditions, which may be accomplished by putting the reaction container in the dark or by covering the reaction with aluminum foil.
Wash cells with 1 mL of PBS three times, 5 min each wash.
Incubate with 0.1–1 μg/mL DAPI for 5 min.
Wash cells 1 mL of PBS three times, 5 min each wash.
Add 10 μL of mounting solution to the small well of the glass bottom dish and cover with a 10 mm coverslip.
Nail polish can be used to seal the coverslip at the bottom of the dish. If the samples will be analyzed immediately, this step is not necessary and dishes can be stored in a humidified box at 4 ºC for up to a week.
Analyze the samples by fluorescence microscopy to detect the desired fluorophores used for the experiment (Fig. 2d).
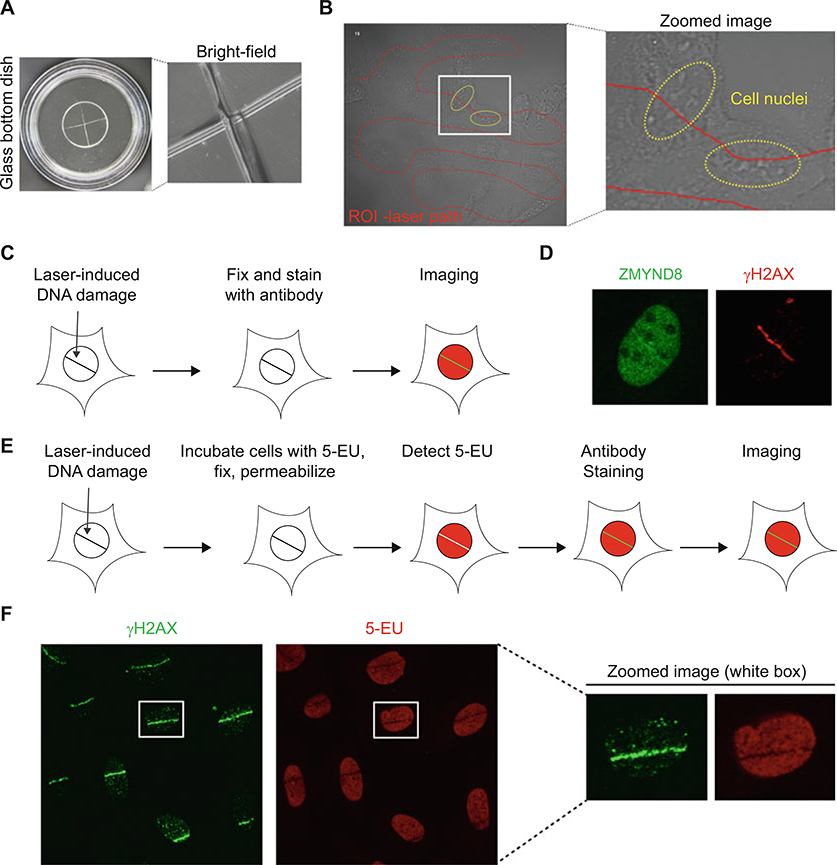
Fig. 2.
Immunofluorescence analysis of endogenous protein recruitment and transcription in the DNA damage site. (a) Experimental setup for laser microirradiation of fixed samples. Left: etched crossed line on glass bottom dish. Right: zoomed image using 60× oil immersion objective and bright-field microscopy. (b) Free drawn ROI line on U2OS human cancer cells within glass bottomed dishes viewed by bright-field microscopy. Red line indicates the ROI and yellow circle indicates the cell nuclei. (c) Schematic illustration for analysis of endogenous protein recruitment and transcription in DNA damage site. (d) Endogenous ZMYND8 translocation to DNA damage site. DNA damage was induced by laser microirradiation and stain with ZMYND8 and γH2AX antibodies. (e) Schematic of nascent transcription and laser microirradiation technique. (f) Transcription analysis in the DNA damage site using 5-EU staining. γH2AX is marker for the DNA damage region
3.4. Data Analysis of Subheading 3.3
Immunofluorescence intensity in the DNA damage site can be measured by ImageJ software.
The number of cells displaying the labeled protein of interest that is colocalized with a DNA damage marker, for example γH2AX, is determined and compared to the total number of damaged cells. At least 100 damaged cells should be analyzed.
3.5. Analysis of Transcription at Laser-Induced DNA Damage Sites (Fig. 2e, f)
Day 1: Seed cells. Cells are split, counted, and seeded as in Subheading 3.1, step 1.
Day 2: Laser damage and transcriptional analysis. After 5 min following DNA damage induction, remove medium and add 100 μL of medium with EU (1 mM) to the glass well. 100 μL is enough to cover the 12 mm glass well of the dish; use more volume if a large dish is used to ensure that the cells are covered by the medium.
Incubate the cells for 1 h, remove the medium and wash cells three times with 1 mL of PBS, 5 min each wash. Fix cells with 500 μL of 2% formaldehyde in PBS at room temperature for 10 min.
After fixation, remove the fixative and wash three times with 1 mL of PBS, 5 min each wash. Add 500 μL of 0.5% Triton X-100 in PBS to each well to permeabilize the cells. Incubate 15 min at room temperature.
During the permeabilization step, prepare the Click-iT reaction cocktail mix following the manufacturer’s instructions.
After permeabilization, wash each well three times with 1 mL of PBS, 5 min each wash. Add 100 μL of Click-iT cocktail mix to the 12 mm glass dish well and incubate 30 min. After this step, all procedures should be performed to reduce light exposure.
After incubation, remove the Click-iT cocktail mix. Rinse each well with 500 μL of Click-iT reaction rinse buffer.
Perform immunofluorescence staining procedures with desired antibodies as described in Subheading 3.3 (see Notes 11–13).
Image and analyze samples by fluorescence microscopy (Fig. 2e, f) as described in Subheading 3.3. For example, γH2AX staining can be used to verify and detect damaged DNA.
Acknowledgments
The K.M.M. laboratory is supported by the NIH National Cancer Institute (R01CA198279 and RO1CA201268) and the American Cancer Society (RSG-16–042-01-DMC).
Footnotes
These protocols and settings for laser damage have been optimized and conducted using a FLUOVIEW FV1000 and FV3000 Olympus confocal laser scanning microscope system with a 405 nm laser. LSM systems with additional lasers can also be used to generate laser microirradiation. We refer users to these protocols, resources and additional information [20–24].
This protocol is optimized for U2OS cells. Transfection efficiency will be different depending on the cell type and transfection reagent. The transfection method should be modified or used with different transfection reagents that are determined empirically for each cell type. For laser microirradiation in difficult to transfect cell types, nucleofector™ kit (Lonza) or other electroporation techniques can be used [25, 26].
Experiments can also be performed without presensitization to ensure that the laser damage is dependent on this procedure, which reduces the laser power required and therefore the potential cellular toxicity. Cell cycle analysis of damage recruitment, including RPA and BRCA1, which should only occur in S/G2 phases of the cell cycle, can also be used to monitor damage strength and biological effects of laser damage [27]. For recruitment of endogenous proteins, DNA damage markers including γH2AX or 53BP1 are used to control for DSB formation. Other markers can be used for other types of DNA damage [6].
Allow heat chamber to stabilize for 5–10 min after setting required temperature and CO2. Maintain humidity of chamber by adding and replenishing water as needed. The laser source should be turned on for 10–15 min before use, which should be consistent for experiments to ensure that the laser is stable when used.
Choosing which cells to analyze by microirradiation is critical for obtaining consistent results for both live and fixed cell imaging and analysis. It is important to select cells with good expression of the protein of interest as well as cells that have normal morphology. Cells with very high expression of ectopically expressed tagged protein or abnormal cell shape or size should be avoided to reduce potential artifacts due to overexpression. However, it is always a good idea to check recruitment between different expression levels as well as compare with tagged and endogenously expressed proteins.
A very important detail for this experiment is to ensure that cells are in focus before damaging the cells. For fluorescence experiments, this can be done while imaging the tagged protein of interest. For recruitment studies of endogenous proteins, a focused cell should have the nucleus distinguished clearly using bright-field microscopy; while the nucleoli will appear bright and white in color (Fig. 2b). Having properly focused cells ensures that the laser path is concentrated within the center of the cell, which will provide better laser stripes of DNA damage for analysis.
Intensity of 405 nm lasers can vary due to laser power and microscope design. It is important to calibrate the optimum laser intensity required to induce DNA damage stripes or marks. To maintain the same laser power between experiments, a laser power meter can be used to measure 405 nm laser intensity.
Phenol red in culture medium is a chromophore and can increase the level of background fluorescence. This can be avoided by using phenol red free cell culture medium such as FluoroBrite DMEM (Thermo Scientific, #A1896701).
Many proteins have been shown to be recruited to the sites of DNA damage. These can act as positive controls to test the settings and conditions for these experiments. Good positive controls for DNA double-strand breaks include 53BP1, RNF168, KAP1, BRCA1, RAD51 and others, which can be found in the literature.
Before seeding cells, use a diamond tipped pen to carefully etch a cross on the glass bottom, which can help to track cells that have been damaged for downstream analyses including immunofluorescence and imaging. Alternatively, use a grid glass bottom dish, which contains grids with 500 μm repeat distance on the bottom glass. It can help to locate damaged cells without the need to use a diamond pen but these dishes are larger which requires more antibodies than the 12mm dishes. Make a cross gently, as the glass bottom is very fragile. Applying excessive pressure while etching the glass could damage glass bottom resulting in breakage and leakage.
Proper fixation and permeabilization are important for successful immunofluorescence staining. Choice of fixative and permeabilization reagent and incubation time should be optimized based on cell type, antibody and protein of interest. For example, CSK removes all soluble proteins so this condition works well for chromatin associated proteins. Antibody datasheets may provide information about suitable reagents and incubation times.
Concentration of primary antibody, blocking solution and incubation time should be optimized based on the antibody. For anti-γH2AX, a dilution of 1:500 in 3% BSA/1× PBS is used for an incubation time of 1 h at room temperature.
If the imaging cannot be performed immediately, the dishes can be stored at 4 ºC in humidified chamber in the dark for at least a week. Waiting for longer times could result in dried out samples and/or a loss of fluorescence signal.
Prepare stock solutions following the instructions provided in the kit.
References
- 1.Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411 (6835):366–374. 10.1038/35077232 [DOI] [PubMed] [Google Scholar]
- 2.Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40(2):179–204. 10.1016/j.molcel.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461(7267):1071–1078. 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polo SE, Jackson SP (2011) Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 25 (5):409–433. 10.1101/gad.2021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sulli G, Di Micco R, d’Adda di Fagagna F (2012) Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat Rev Cancer 12(10):709–720. 10.1038/nrc3344 [DOI] [PubMed] [Google Scholar]
- 6.Aleksandrov R, Dotchev A, Poser I, Krastev D, Georgiev G, Panova G, Babukov Y, Danovski G, Dyankova T, Hubatsch L, Ivanova A, Atemin A, Nedelcheva-Veleva MN, Hasse S, Sarov M, Buchholz F, Hyman AA, Grill SW, Stoynov SS (2018) Protein dynamics in complex DNA lesions. Mol Cell 69 (6):1046–1061.e1045. 10.1016/j.molcel.2018.02.016 [DOI] [PubMed] [Google Scholar]
- 7.Petrini JH, Stracker TH (2003) The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol 13(9):458–462 [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, Paull TT (2005) ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308 (5721):551–554. 10.1126/science.1108297 [DOI] [PubMed] [Google Scholar]
- 9.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273 (10):5858–5868 [DOI] [PubMed] [Google Scholar]
- 10.Jackson SP, Durocher D (2013) Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 49(5):795–807. 10.1016/j.molcel.2013.01.017 [DOI] [PubMed] [Google Scholar]
- 11.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318 (5856):1637–1640. 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Elledge SJ (2007) Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A 104(52):20759–20763. 10.1073/pnas.0710061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gassman NR, Wilson SH (2015) Microirradiation tools to visualize base excision repair and single-strand break repair. DNA Repair (Amst) 31:52–63. 10.1016/j.dnarep.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mistrik M, Vesela E, Furst T, Hanzlikova H, Frydrych I, Gursky J, Majera D, Bartek J (2016) Cells and stripes: a novel quantitative photo-manipulation technique. Sci Rep 6:19567 10.1038/srep19567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svejstrup JQ (2010) The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem Sci 35 (6):333–338. 10.1016/j.tibs.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 16.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA (2010) ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141(6):970–981. 10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong F, Chiu LY, Cox B, Aymard F, Clouaire T, Leung JW, Cammarata M, Perez M, Agarwal P, Brodbelt JS, Legube G, Miller KM (2015) Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev 29(2):197–211. 10.1101/gad.252189.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong F, Clouaire T, Aguirrebengoa M, Legube G, Miller KM (2017) Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J Cell Biol 216 (7):1959–1974. 10.1083/jcb.201611135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong F, Miller KM (2018) Double duty: ZMYND8 in the DNA damage response and cancer. Cell Cycle 17(4):414–420. 10.1080/15384101.2017.1376150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adam S, Dabin J, Chevallier O, Leroy O, Baldeyron C, Corpet A, Lomonte P, Renaud O, Almouzni G, Polo SE (2016) Real-time tracking of parental histones reveals their contribution to chromatin integrity following DNA damage. Mol Cell 64(1):65–78. 10.1016/j.molcel.2016.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Britton S, Coates J, Jackson SP (2013) A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol 202 (3):579–595. 10.1083/jcb.201303073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong X, Cruz GMS, Silva BA, Wakida NM, Khatibzadeh N, Berns MW, Yokomori K (2018) Laser microirradiation to study in vivo cellular responses to simple and complex DNA damage. J Vis Exp (131). 10.3791/56213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukas C, Bartek J, Lukas J (2005) Imaging of protein movement induced by chromosomal breakage: tiny ‘local’ lesions pose great ‘global’ challenges. Chromosoma 114(3):146–154. 10.1007/s00412-005-0011-y [DOI] [PubMed] [Google Scholar]
- 24.Xie S, Mortusewicz O, Ma HT, Herr P, Poon RY, Helleday T, Qian C (2015) Timeless interacts with PARP-1 to promote homologous recombination repair. Mol Cell 60 (1):163–176. 10.1016/j.molcel.2015.07.031 [DOI] [PubMed] [Google Scholar]
- 25.Dobbin MM, Madabhushi R, Pan L, Chen Y, Kim D, Gao J, Ahanonu B, Pao PC, Qiu Y, Zhao Y, Tsai LH (2013) SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat Neurosci 16 (8):1008–1015. 10.1038/nn.3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang WY, Pan L, Su SC, Quinn EJ, Sasaki M, Jimenez JC, Mackenzie IR, Huang EJ, Tsai LH (2013) Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat Neurosci 16(10):1383–1391. 10.1038/nn.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hustedt N, Durocher D (2016) The control of DNA repair by the cell cycle. Nat Cell Biol 19 (1):1–9. 10.1038/ncb3452 [DOI] [PubMed] [Google Scholar]