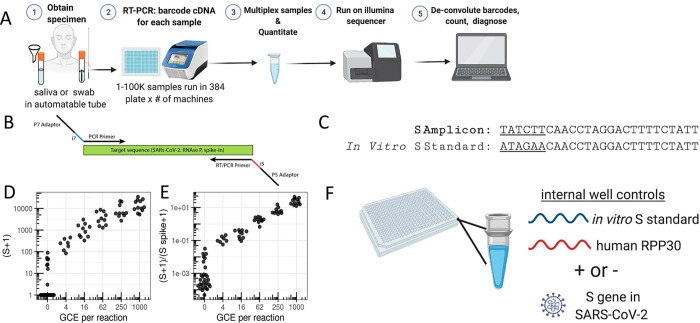
Figure 1. SwabSeq Diagnostic Testing Platform for COVID19.
A) The workflow for SwabSeq is a five step process that takes approximately 12 hours from start to finish. B) In each well, we perform RT-PCR on clinical samples. Each well has two sets of indexed primers that generate cDNA and amplicons for SARS-CoV-2 S gene and the human RPP30 gene. Each primer is synthesized with the P5 and P7 adaptors for Illumina sequencing, a unique i7 and i5 molecular barcodes, and the unique primer pair. Importantly, every well has a synthetic in vitro S standard that is key to allowing the method to work at scale. C) The in vitro S standard (abbreviated as S-Spike) differs from the virus S gene by 6 base pairs that are complemented (underlined). (D) Read count at various viral concentrations (E) Ratiometric normalization allow for in-well normalization for each amplicon (F) Every well has two internal well controls for amplification, the in vitro S standard and the human RPP30. The RPP30 amplicon serves as a control for specimen collection. The in vitro S standard is critical to SwabSeq’s ability to distinguish true negatives.