Cytoplasmic cilia, which are found in human and Drosophila sperm, are unique in that the axoneme is exposed to the cytoplasm. Fingerhut and Yamashita show that localization of a novel RNP granule containing axonemal dynein mRNAs facilitates incorporation of these axonemal proteins, promoting cytoplasmic cilia formation.
Abstract
Cytoplasmic cilia, a specialized type of cilia in which the axoneme resides within the cytoplasm rather than within the ciliary compartment, are proposed to allow for the efficient assembly of very long cilia. Despite being found diversely in male gametes (e.g., Plasmodium falciparum microgametocytes and human and Drosophila melanogaster sperm), very little is known about cytoplasmic cilia assembly. Here, we show that a novel RNP granule containing the mRNAs for axonemal dynein motor proteins becomes highly polarized to the distal end of the cilia during cytoplasmic ciliogenesis in Drosophila sperm. This allows for the incorporation of these axonemal dyneins into the axoneme directly from the cytoplasm, possibly by localizing translation. We found that this RNP granule contains the proteins Reptin and Pontin, loss of which perturbs granule formation and prevents incorporation of the axonemal dyneins, leading to sterility. We propose that cytoplasmic cilia assembly requires the precise localization of mRNAs encoding key axonemal constituents, allowing these proteins to incorporate efficiently into the axoneme.
Introduction
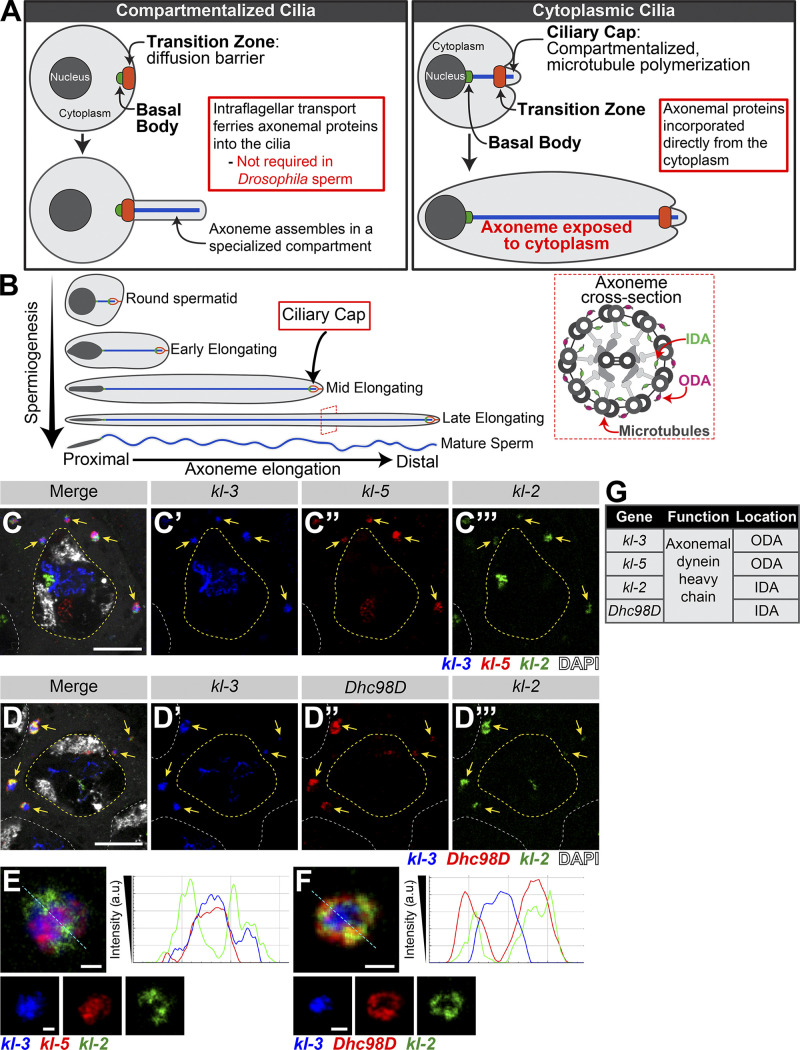
Cilia are microtubule-based structures present on the surface of many cells. These specialized cellular compartments can be nonmotile primary cilia that largely function in signaling or motile cilia that can either move extracellular materials (e.g., lung multiciliated cells) or allow for cell motility (e.g., Chlamydomonas reinhardtii flagellum, sperm in many species; Ishikawa and Marshall, 2011). It is well established that most cilia are separated from the bulk cytoplasm (Fig. 1 A), which serves to concentrate signaling molecules for rapid response to extracellular signals received by the cilia, and that the ciliary gate at the base of the cilia forms a diffusion barrier through which molecules must be selectively transported (Reiter et al., 2012; Wheway et al., 2018). However, recent studies identified an additional type of cilia, called cytoplasmic cilia, in which the axoneme (the microtubule-based core of the cilia) is exposed to the cytoplasm (Fig. 1 A; Avidor-Reiss et al., 2017; Avidor-Reiss and Leroux, 2015; Dawson and House, 2010; Fawcett et al., 1970; Sinden et al., 1976; Tates, 1971; Tokuyasu, 1975). Cytoplasmic cilia are found in human and Drosophila melanogaster sperm as well as in Plasmodium falciparum and Giardia intestinalis. There are two proposed advantages to cytoplasmic cilia: (1) faster assembly, as the cell does not need to rely on ciliary transport mechanisms, allowing for the assembly of longer cilia; and (2) proximity to mitochondria for energy (Avidor-Reiss and Leroux, 2015; Sinden et al., 2010). Despite being found across diverse taxa, very little is known about how cytoplasmic cilia are assembled and whether their assembly bears similarity to that of traditional compartmentalized cilia (Desai et al., 2018).
Figure 1.
Axonemal dynein heavy chain mRNAs colocalize in an RNP granule in SCs. (A) Diagram comparing and contrasting traditional compartmentalized cilia and cytoplasmic cilia. Shown are the nucleus (dark gray), cytoplasm (light gray), basal body (green), transition zone (orange), and axoneme (blue). (B) Diagram of Drosophila spermiogenesis with stages of spermatid elongation. Shown are the nucleus (dark gray), cytoplasm (light gray), transition zone (green), ciliary cap (orange), and axoneme (blue). Axoneme cross-section image showing location of axonemal dynein arms. Shown are the microtubules and other structural components (gray), IDA (green), and ODA (magenta). (C and D) smFISH against axonemal dynein heavy chain transcripts in SCs showing kl-3, kl-5, and kl-2 mRNAs (C) or kl-3, kl-2, and Dhc98D mRNAs (D) in kl-granules. Shown are kl-3 (blue), kl-2 (green), kl-5 (red, C), Dhc98D (red, D), DAPI (white), kl-granules (yellow arrows), SC nuclei (yellow dashed line), neighboring SC nuclei (white dashed line). Scale bars: 10 µm. (E and F) smFISH against kl-3, kl-5, and kl-2 (E) or kl-3, kl-2, and Dhc98D (F) showing a single kl-granule. Shown are kl-3 (blue), kl-2 (green), kl-5 (red; E), Dhc98D (red; F). Intensity plots are shown for the regions marked by the cyan dashed line. Scale bars: 1 µm. (G) Table listing the genes focused on in this study, their function, and their localization within the axoneme.
Cytoplasmic ciliogenesis has been proposed to occur in two stages (Fig. 1 A; Avidor-Reiss and Leroux, 2015). In the first stage, microtubules are polymerized in a small compartmentalized region, which is similar to canonical compartmentalized cilia, at the most distal end of the cilia (Gottardo et al., 2013; Tokuyasu, 1975). This region is gated by a transition zone (Caudron and Barral, 2009; Kwitny et al., 2010; Vieillard et al., 2016). This entire compartmentalized region, called the ciliary cap or the growing end, migrates away from the basal body, which is docked at the nuclear membrane (Basiri et al., 2014; Fawcett et al., 1970). The ciliary cap does not change in size as the cilia elongates. The continued polymerization of microtubules inside the ciliary cap displaces recently synthesized microtubules out of the compartmentalized region, exposing them to the cytoplasm (Fig. 1 A). The second stage is axoneme maturation, in which additional axonemal proteins (e.g., axonemal dyneins, the motor proteins that confer motility by allowing axonemal microtubules to slide against each other; Fig. 1 B), are added to the bare microtubule structure after it emerges from the ciliary cap (Tates, 1971; Tokuyasu, 1975). Axoneme maturation was inferred to occur in the cytoplasm based on the dispensability of ciliary transport mechanisms and the inefficiency of relaying on diffusion through the transition zone (Avidor-Reiss and Leroux, 2015; Avidor-Reiss et al., 2004; Breslow et al., 2013; Briggs et al., 2004; Han et al., 2003; Hoeng et al., 2008; Kee et al., 2012; Lin et al., 2013; Sarpal et al., 2003). However, how this maturation process occurs in the cytoplasmic compartment to allow for cytoplasmic cilia formation remains unknown.
Drosophila spermatogenesis provides an excellent model for the study of cytoplasmic ciliogenesis (Fig. 1 B), owing to rich cytological knowledge of spermatogenesis and the conservation of almost all known ciliary proteins (Zur Lage et al., 2019). Developing spermatids elongate from 15 µm to 1,900 µm (1.9 mm; Tates, 1971; Tokuyasu, 1975). Within mature sperm, the cytoplasmic cilia are 1,800 µm, and the ciliary caps (the compartmentalized region) are only ∼2 µm. Ciliogenesis starts in premeiotic spermatocytes (SCs), which assemble short primary (compartmentalized) cilia (Fabian and Brill, 2012; Gottardo et al., 2013; Riparbelli et al., 2012; Tates, 1971). Prior to axoneme elongation, these primary cilia, which were docked at the plasma membrane in SCs, invaginate, forming the ciliary cap. During axoneme elongation, the majority of the length of the cilia will be exposed to the cytoplasm, as described above. Accordingly, axoneme assembly in Drosophila does not require intraflagellar transport (IFT; Han et al., 2003; Sarpal et al., 2003), the process used by traditional compartmentalized cilia to ferry axonemal proteins from the cytoplasm into the ciliary compartment for incorporation (Rosenbaum and Witman, 2002). Other cytoplasmic cilia have been found not to require IFT for their assembly (Avidor-Reiss and Leroux, 2015; Briggs et al., 2004; Hoeng et al., 2008), leading to the appreciation of a distinct type of cilia; based on the dispensability of IFT, it was postulated that axoneme maturation must occur in the cytoplasm, hence the term cytoplasmic cilia.
It has long been known that SCs transcribe almost all genes whose protein products are needed postmeiotically and that these mRNAs may not be translated until days later when proteins are needed (Barckmann et al., 2013; Olivieri and Olivieri, 1965). We previously showed that the Y-linked testis-specific axonemal dynein heavy chain genes kl-3 and kl-5, as well as the testis-specific axonemal dynein intermediate chain Dic61B, are transcribed in SCs (Fingerhut et al., 2019). However, axoneme elongation does not begin until after meiosis, suggesting that these mRNAs may not be translated until later in development. Intriguingly, we previously showed that kl-3 and kl-5 mRNAs localize to cytoplasmic granules in SCs. RNP granules (e.g., stress granules, P granules, and germ granules) are known to play critical roles in mRNA regulation, such as mediating the subcellular localization of mRNAs and controlling the timing of translation (Anderson and Kedersha, 2009; Buchan, 2014; Medioni et al., 2012). Therefore, we decided to investigate the role of this novel RNA granule in the mRNA regulatory mechanisms that ensure proper axoneme assembly and found that it plays an essential role in mediating the incorporation of axonemal proteins, providing the first insights into the molecular mechanism of cytoplasmic cilia maturation. We show four axonemal dynein heavy chain mRNAs, including kl-3 and kl-5, colocalize in these novel granules in late SCs along with the AAA+ (ATPases associated with diverse cellular activities) proteins Reptin (Rept) and Pontin (Pont). These RNP granules are segregated during the meiotic divisions and localize to the distal end of the cytoplasmic compartment as the axoneme elongates during spermiogenesis. We further show that Rept and Pont are required for RNP granule formation and that RNP granule formation is necessary for accumulation and incorporation of the axonemal dynein proteins into the axoneme. Our data suggest that cytoplasmic cilia maturation relies on the local translation of axonemal components such that they can be incorporated into the bare microtubule structure as it emerges from the ciliary cap.
Results
Axonemal dynein heavy chain mRNAs colocalize in RNP granules in SCs
In our previous study, we analyzed the expression of the Y-linked axonemal dynein genes kl-3 and kl-5 and showed that these two mRNAs localized to cytoplasmic granules in late SCs (Fingerhut et al., 2019). Using single-molecule RNA FISH (smFISH), we found that mRNAs for four testis-specific axonemal dynein heavy chain genes (the Y-chromosome genes kl-2, kl-3, and kl-5, as well as the autosomal gene Dhc98D; Carvalho et al., 2000; Goldstein et al., 1982; Hardy et al., 1981; Zur Lage et al., 2019) colocalize in RNP granules in the cytoplasm of late SCs, with each SC containing several of these cytoplasmic granules (Fig. 1, C and D). We termed these granules “kl-granules” after the three Y-linked constituent mRNAs. It should be noted that robust transcription of these genes is still ongoing in SC nuclei (visible as bright nuclear signal; Fig. 1, C and D) but these are nascent transcripts that still contain intronic RNA, whereas the kl-granules in the cytoplasm do not contain intronic RNA, as we showed previously (Fingerhut et al., 2019). The present study focuses on the fate of these cytoplasmic RNPs. mRNAs within a kl-granule appear spatially suborganized: kl-3 and kl-5 mRNAs, which encode outer dynein arm (ODA) dynein heavy chain proteins, cluster together in the core of the kl-granule while kl-2 and Dhc98D mRNAs, which encode inner dynein arm (IDA) dynein heavy chain proteins, localize peripherally (Fig. 1, E–G). This is similar to the subcompartmentalization observed in other RNP granules, including the germ granules in the Drosophila ovary, stress granules, P granules, nucleoli, and the dynein axonemal particles in human and Xenopus multiciliated cells (Boisvert et al., 2007; Jain et al., 2016; Lee et al., 2020a Preprint; Little et al., 2015; Trcek et al., 2015; Wang et al., 2014). We noted that kl-granule formation is unlikely to be dependent upon any one mRNA constituent, as RNAi-mediated knockdown of kl-3, kl-5, kl-2, or Dhc98D (bam-gal4>UAS-kl-3TRiP.HMC03546 or bam-gal4>UAS-kl-5TRiP.HMC03747 or bam-gal4>UAS-kl-2GC8807 or bam-gal4>UAS-Dhc98DTRiP.HMC06494) did not perturb granule formation despite efficient knockdown (Fig. S1).
Figure S1.
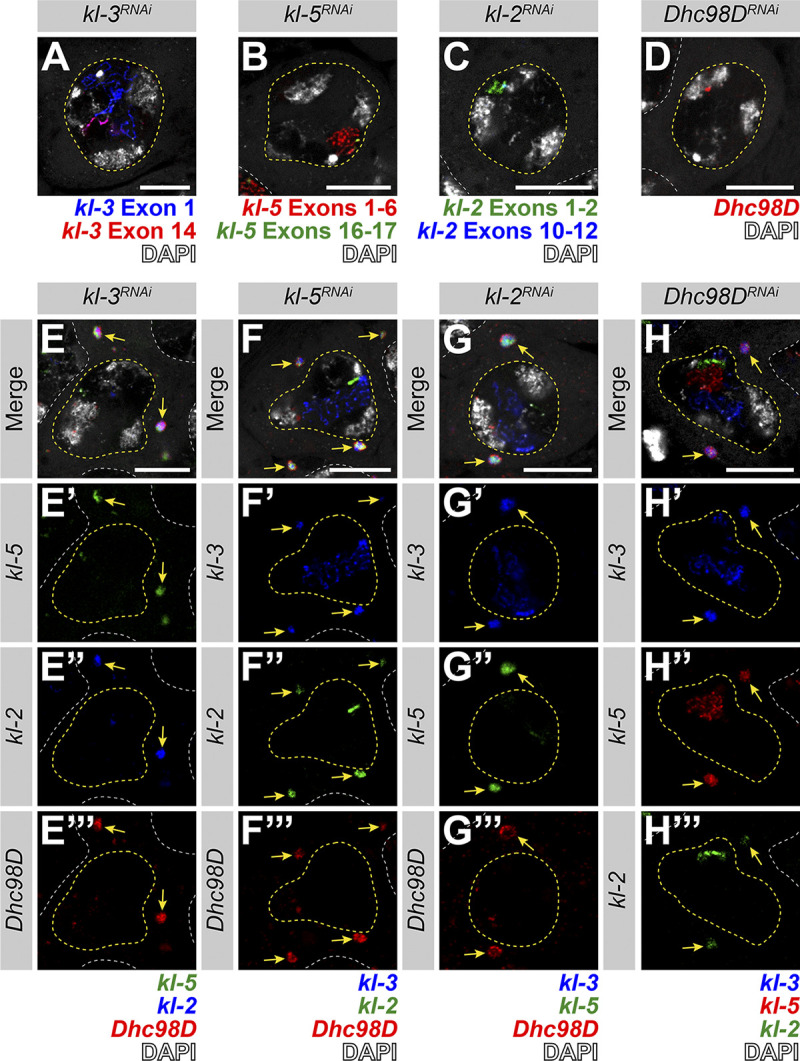
kl-granule formation is not dependent upon any one mRNA constituent. (A–D) smFISH against each known kl-granule mRNA constituent following RNAi of that constituent shows successful knockdown (no remaining cytoplasmic signal). Note that we use multiple smFISH probe sets for some mRNAs targeted against different regions of the transcript (see Table S1). (A) kl-3 exon 1 (blue), kl-3 exon 14 (red) and DAPI (white). (B) kl-5 exons 1–6 (red), kl-5 exons 16–17 (green), and DAPI (white). (C) kl-2 exons 1–2 (green), kl-2 exons 10–12 (blue), and DAPI (white). (D) Dhc98D (red) and DAPI (white). For all, SC nuclei, yellow dashed line; neighboring SC nuclei, white dashed line. Scale bars: 10 µm. (E–H) smFISH against the other three constituent mRNAs after RNAi of the fourth mRNA. Note that the color used to represent each smFISH probe corresponds to the probe sets in A–D. (E) kl-5 (green), kl-2 (blue), Dhc98D (red) and DAPI (white). (F) kl-3 (blue), kl-5 (green), Dhc98D (red), and DAPI (white). (G) kl-3 (blue), kl-5 (green), Dhc98D (red) and DAPI (white). (H) kl-3 (blue), kl-5 (red), kl-2 (green), and DAPI (white). For all, SC nuclei, yellow dashed line; neighboring SC nuclei, white dashed line; kl-granules, yellow arrows. Scale bars: 10 µm.
We conclude that mRNAs for the testis-specific axonemal dynein heavy chains localize to novel RNP granules, which we termed kl-granules, in late SCs.
kl-granules segregate during meiotic divisions and localize to the distal end of elongating spermatids
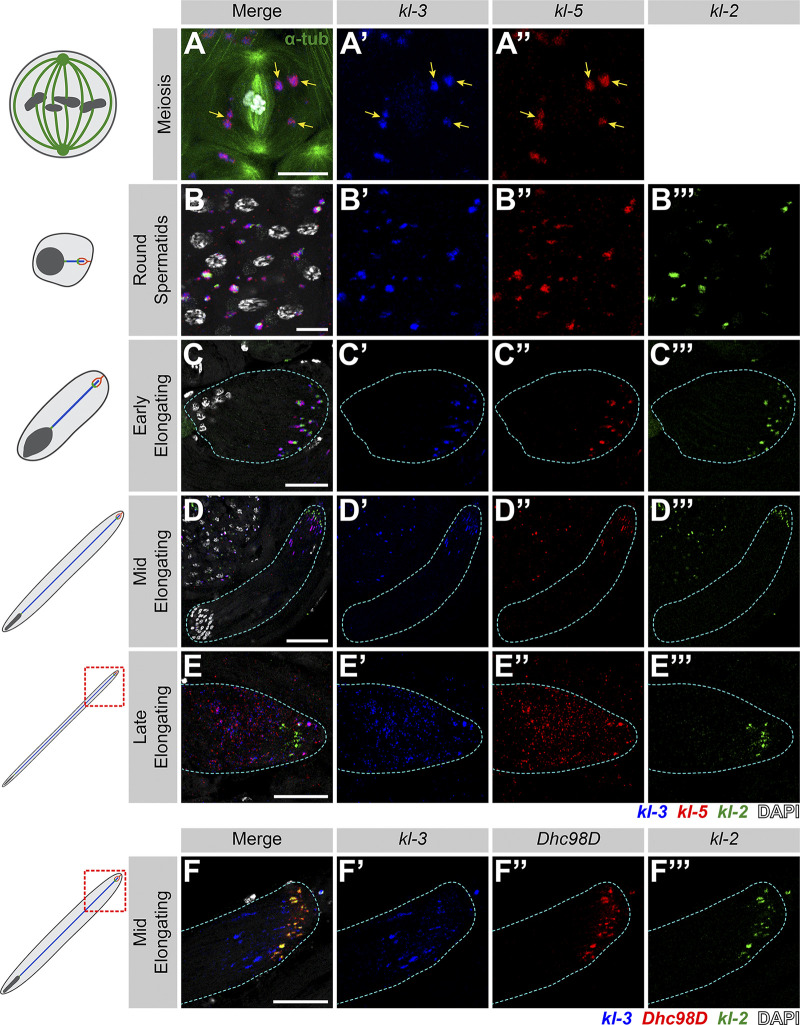
As kl-granules contain mRNAs for axonemal proteins that are only necessary for spermiogenesis, we next followed the fate of the kl-granules through meiosis and into spermiogenesis. kl-granules segregate through the two sequential meiotic divisions (Fig. 2 A) such that each resulting haploid spermatid receives a relatively equal amount of kl-granule (Fig. 2 B). Upon completion of meiosis, the resultant spermatids are interconnected due to incomplete cytokinesis during the four mitotic divisions that occur early in germ cell development and the two meiotic divisions, forming a cyst of 64 spermatids (Fuller, 1993; Hime et al., 1996). As the axoneme starts to elongate within each spermatid, the nuclei cluster to the proximal end of the cyst while the axoneme elongates unidirectionally away from the nuclei with the ciliary caps clustered at the distal end of the cyst (Fig. 1 B; Fabian and Brill, 2012). Strikingly, we found that kl-granules become localized to the distal end of elongating spermatid cysts (Fig. 2 C). This polarized localization remains as the axoneme continues to elongate (Fig. 2, D and E). At later stages of elongation, kl-granules begin to dissociate and mRNAs become more diffusely localized at the distal end (Fig. 2, D and E). Interestingly, kl-3 and kl-5 mRNAs (encoding ODA proteins) separate from kl-2 and Dhc98D mRNAs (encoding IDA proteins; Fig. 2, D and F). It is of note that this separation pattern correlates with the subgranule localization of constituent mRNAs described above (kl-3 and kl-5 localize to the core of the kl-granules [Fig. 1 E] whereas kl-2 and Dhc98D localize to the periphery of the kl-granules [Fig. 1 F]) and was observed in 100% of elongating spermatid cysts (n = 269).
Figure 2.
kl-granules segregate during the meiotic divisions and localize to the distal end of elongating spermatids. (A) smFISH against kl-3 and kl-5 during meiosis. Shown are kl-3 (blue), kl-5 (red), α-tubulin-GFP (green), DAPI (white), and kl-granules (yellow arrows). Scale bar: 10 µm. (B–E) smFISH against kl-3, kl-5, and kl-2 during spermiogenesis. The round spermatid (B), early elongating spermatid (C), mid elongating spermatid (D), and late elongating spermatid (E) stages are shown. Shown are kl-3 (blue), kl-5 (red), kl-2 (green), DAPI (white), and a spermatid cyst (cyan dashed line). Scale bars: 10 µm (B), 25 µm (C and E), or 50 µm (D). (F) smFISH against kl-3, Dhc98D, and kl-2 in mid elongating spermatids. kl-3 (blue), Dhc98D (red), kl-2 (green), DAPI (white), spermatid cyst (cyan dashed line). Scale bar: 25 µm.
These results show that kl-granules exhibit stereotypical localization to the growing end of spermatids after being segregated during meiosis, implying that programmed positioning of kl-granules may play an important role during spermatid elongation and axoneme maturation.
The AAA+ proteins Rept and Pont colocalize with kl-granules
To further understand how kl-granules form and their potential function, we sought to identify a protein that localizes to kl-granules. In our previous study, we screened for proteins involved in the expression of the Y-linked axonemal dynein genes (Fingerhut et al., 2019). Rept and Pont, two AAA+ proteins (Puchades et al., 2020), were included in this screen because of their high expression in the testis and their involvement in RNP complex formation in other systems (Mao and Houry, 2017; Robinson et al., 2013). Also, studies in Drosophila, mouse, zebrafish, Chlamydomonas, and Xenopus have specifically implicated Rept and Pont in axoneme/motile cilia assembly and/or sperm motility, although the underlying mechanism remains unknown (Dafinger et al., 2018; Huizar et al., 2018; Li et al., 2017; Stolc et al., 2005; Tammana and Tammana, 2017; Zhao et al., 2013; Zur Lage et al., 2018).
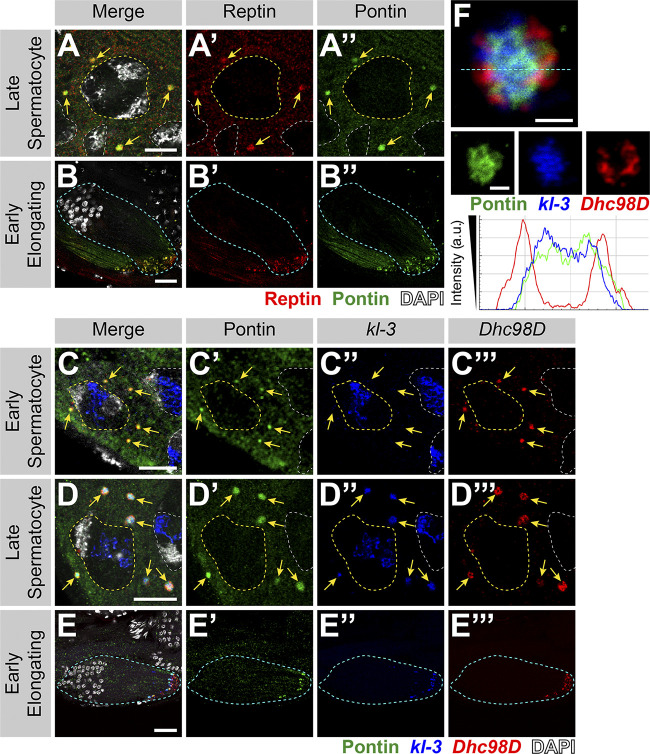
We found that Rept and Pont colocalize in cytoplasmic granules from the SC stage through elongating spermatids (Fig. 3, A and B). Immunofluorescent staining combined with smFISH showed that Rept and Pont colocalize with kl-granules. Pont first colocalizes with Dhc98D mRNA in early SCs (Fig. 3 C) and with all other kl-granule constituent mRNAs in later SCs (Fig. 3 D) and throughout spermatid elongation (Fig. 3 E). Close examination of kl-granules in late SCs revealed that Pont is not evenly distributed within a kl-granule and rather concentrates near the core with kl-3 and kl-5 mRNAs (Figs. 1 E and 3 F). In contrast, kl-2 and Dhc98D mRNAs occupy the periphery of the kl-granule (Fig. 1 F), where Pont is less concentrated.
Figure 3.
Rept and Pont colocalize with kl-granules. (A and B) Rept and Pont colocalization in SCs (A) and early elongating spermatids (B). Shown are Rept (red), Pont (green), DAPI (white), SC nuclei (yellow dashed line, A), neighboring SC nuclei (white dashed line,; A), kl-granules (yellow arrows; C), and a spermatid cyst (cyan dashed line; B). Scale bars: 10 µm (A) or 25 µm (B). (C–E) Immunofluorescent staining combined with smFISH for Pont protein and kl-3 and Dhc98D mRNAs in early SCs (C), late SCs (D), and early elongating spermatids (E). Shown are Pont (green), kl-3 (blue), Dhc98D (red), DAPI (white), SC nuclei (yellow dashed line; C and D), neighboring SC nuclei (white dashed line; C and D), kl-granules (yellow arrows; C and D), and a spermatid cyst (cyan dashed line; E). Bar: 10 µm (C and D) or 25 µm (E). (F) Immunofluorescent staining combined with smFISH for Pont protein and kl-3 and Dhc98D mRNAs in a single kl-granule. Pont (green), kl-3 (blue), and Dhc98D (red). Intensity plot is shown for the region marked by the cyan dashed line. Scale bar: 1 µm.
We conclude that Rept and Pont localize to kl-granules together with axonemal dynein heavy chain mRNAs. It is interesting to note that previous studies have proposed that Rept and Pont function as chaperones in the assembly of axonemal dynein motors (complexes containing a combination of dynein heavy, intermediate, and light chains; Huizar et al., 2018; Li et al., 2017; Zur Lage et al., 2018). However, the role of mRNA in the previously reported Rept- and Pont-containing chaperon complexes remains unexplored. Interestingly, a recent study reported the presence of RNA in this chaperon complex (although the identity of the RNAs remains unknown), raising the possibility that mRNA localization may be a universal mechanism (Drew et al., 2020 Preprint; see Discussion).
Rept and Pont are required for kl-granule assembly
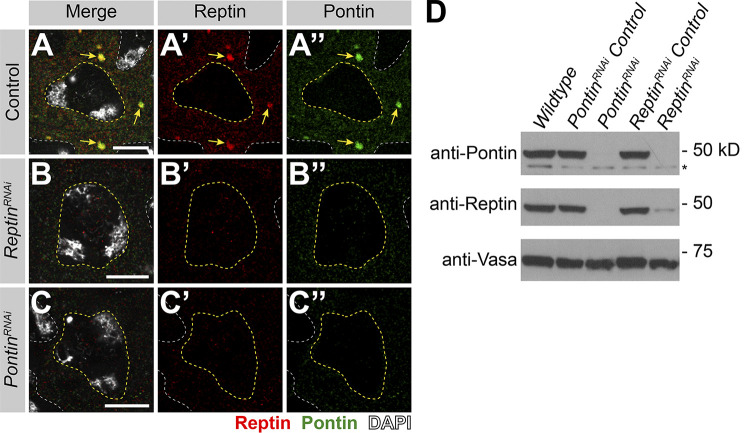
To explore the function of Rept and Pont in kl-granule formation, we performed RNAi-mediated knockdown of either rept or pont (bam-gal4>UAS-reptKK105732 or bam-gal4>UAS-pontKK101103). In addition to eliminating the targeted protein, depletion of rept resulted in loss of Pont and vice versa, reminiscent of findings from previous studies, likely because these proteins stabilize each other as components of the same complex (Fig. S2; Gorynia et al., 2011; Li et al., 2017; Rivera-Calzada et al., 2017; Venteicher et al., 2008).
Figure S2.
RNAi of rept or pont results in loss of both proteins. (A–C) Rept and Pont protein expression in SCs in control (A), rept RNAi (bam-gal4>UAS-reptKK105732; B), or pont RNAi (bam-gal4>UAS-pontKK101103; C). Shown are Rept (red), Pont (green), DAPI (white), SC nuclei (yellow dashed line), neighboring SC nuclei (white dashed line), and kl-granules (yellow arrow). Scale bars: 10 µm. (D) Western blot for Pont and Rept in the indicated genotypes. Asterisk indicates nonspecific band.
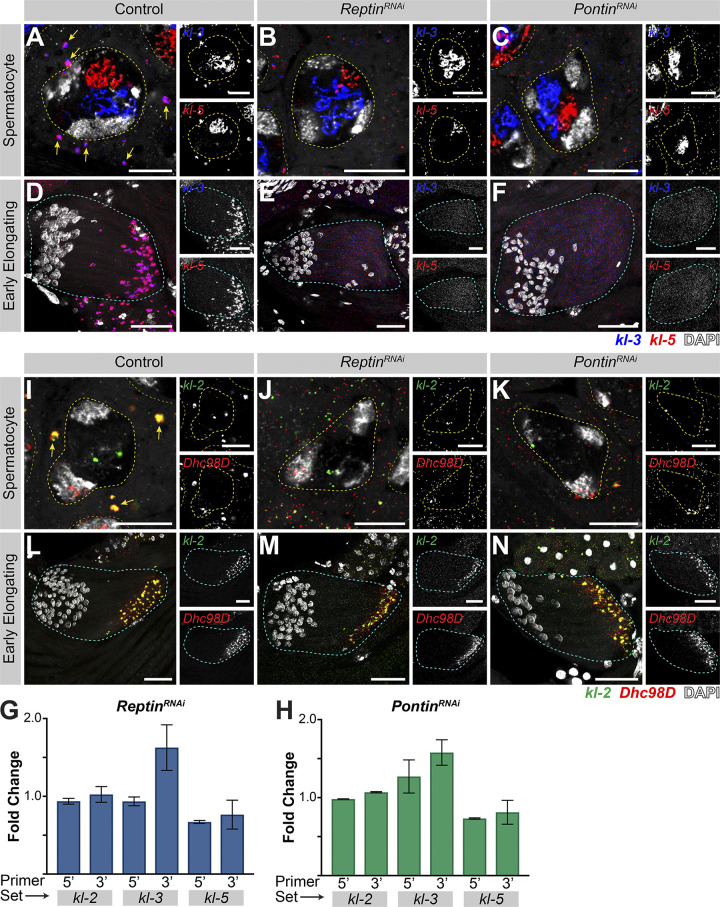
We next determined whether Rept and Pont are needed for kl-granule assembly. Indeed, knockdown of rept or pont resulted in disruption of kl-granules. smFISH clearly detected the presence of dispersed kl-3 and kl-5 mRNAs in late SCs, suggesting that rept and pont are required for kl-granule formation (Fig. 4, A–C; note that nuclear signal was oversaturated in order to focus on the dispersed cytoplasmic signal). This effect was more pronounced in elongating spermatids where kl-3 and kl-5 mRNAs were diffuse throughout the entire cyst in the RNAi conditions (Fig. 4, D–F). Quantitative RT-PCR further demonstrated that mRNA levels were not reduced compared with cross-sibling controls (Fig. 4, G and H), demonstrating that kl-granule formation is not required for mRNA stability. This is in accordance with observations in other systems, which suggest that RNA granule formation is not required for mRNA stability and may be more important for mRNA localization or translation (Bley et al., 2015; Lee et al., 2020b).
Figure 4.
Rept and Pont are required for kl-granule assembly. (A–F) smFISH against kl-3 and kl-5 in control (A and D), rept RNAi (bam-gal4>UAS-reptKK105732; B and E), or pont RNAi (bam-gal4>UAS-pontKK101103; C and F) SCs (A–C, single z plane) and early elongating spermatids (D–F, z-projection). Shown are kl-3 (blue), kl-5 (red), DAPI (white), SC nuclei (yellow dashed lines), neighboring SC nuclei (narrow yellow dashed lines), SC kl-granules (yellow arrows), and a spermatid cyst (cyan dashed line). Scale bars: 10 µm (A–C) or 25 µm (D–F). (G and H) Quantitative RT-PCR in rept RNAi (bam-gal4>UAS-reptKK105732, G) or pont RNAi (bam-gal4>UAS-pontKK101103, H) for kl-3, kl-5, and kl-2 using two primer sets per gene, as indicated (see Table S1). Data were normalized to GAPDH and sibling controls and represent at least two biological replicates, each reaction performed in technical triplicate. Error bars represent SD. (I–N) smFISH against kl-2 and Dhc98D in control (I and L), rept RNAi (bam-gal4>UAS-reptKK105732, J and M), or pont RNAi (bam-gal4>UAS-pontKK101103; K and N) SCs (I–K, single z plane) and early elongating spermatids (L–N, z-projection). Shown are kl-2 (green), Dhc98D (red), DAPI (white), SC nuclei (yellow dashed lines), neighboring SC nuclei (narrow yellow dashed lines), SC kl-granules (yellow arrows), and a spermatid cyst (cyan dashed line). Scale bars: 10 µm (I–K) or 25 µm (L–N).
Interestingly, knockdown of rept or pont had a somewhat different effect on kl-2 and Dhc98D mRNAs. smFISH for kl-2 and Dhc98D following RNAi of either rept or pont showed loss of kl-granule localization in late SCs similar to that seen for kl-3 and kl-5 (Fig. 4, I–K). However, in elongating spermatids, kl-2 and Dhc98D mRNAs appeared to localize properly at the distal end of the cyst (Fig. 4, L–N). Considering that Pont primarily colocalized with kl-3 and kl-5 mRNAs (Fig. 3 F), this may suggest that other proteins play a role in kl-granule formation or that there may be a differential requirement for these proteins between different mRNAs and over developmental time.
In conclusion, Rept and Pont are critical for assembling kl-granules.
kl-granule assembly is required for efficient Kl-3 translation and sperm motility
Previous studies in Drosophila and mouse demonstrated that Rept and Pont are required for male fertility (Li et al., 2017; Zur Lage et al., 2018). We confirmed that seminal vesicles, where mature motile sperm are stored after exiting the testis, were empty in rept or pont RNAi testes (Fig. 5, A–C), as was observed for kl-3, kl-5, kl-2, or Dhc98D RNAi testes (Fig. S3; Fingerhut et al., 2019; Zur Lage et al., 2018).
Figure 5.
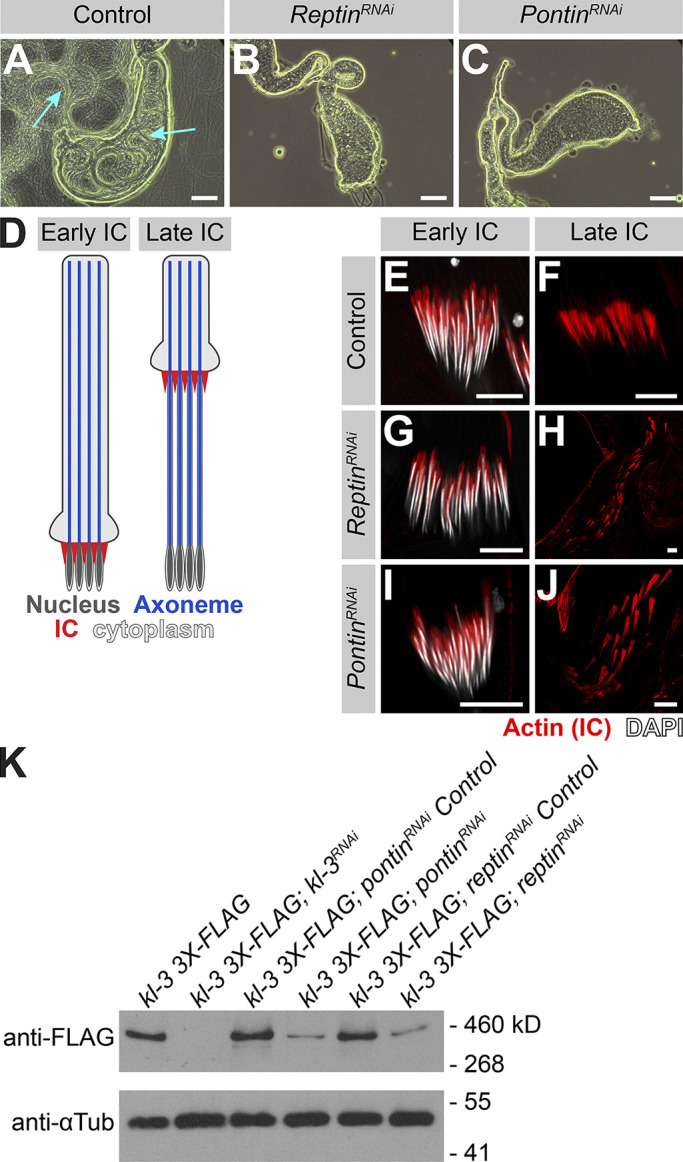
kl-granule assembly is required for efficient Kl-3 translation and sperm motility. (A–C) Phase-contrast images of seminal vesicles in control (A), rept RNAi (bam-gal4>UAS-reptKK105732; B), and pont RNAi (bam-gal4>UAS-pontKK101103; C). Cyan arrows point to mature sperm. Scale bars: 100 µm. (D) Schematic of IC progression during individualization. Shown are the nucleus (dark gray), axoneme (blue), ICs (red), and cytoplasm (light gray). (E–J) Phalloidin staining of early and late ICs in the indicated genotypes. Shown are phalloidin (actin, red) and DAPI (white). Scale bars: 10 µm. (K) Western blot for Kl-3-3X FLAG in the indicated genotypes.
Figure S3.
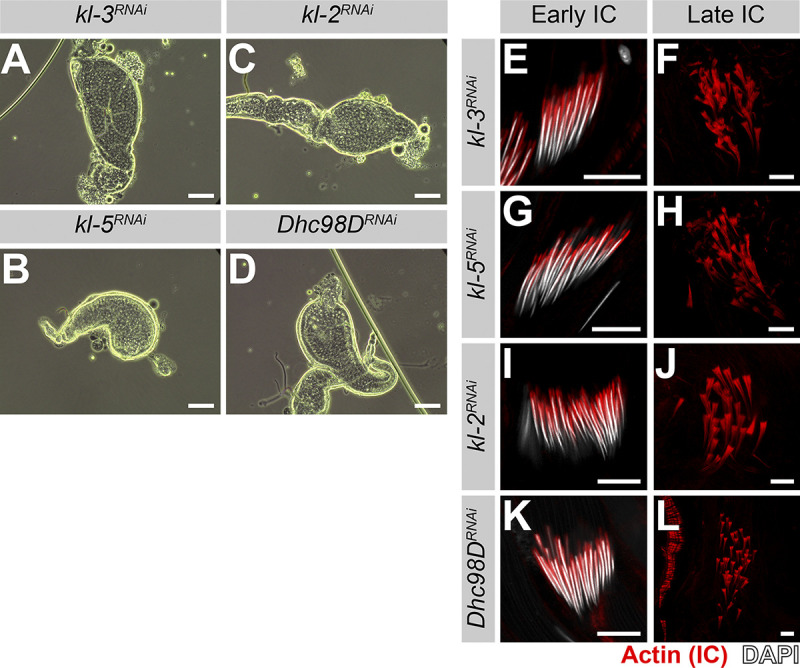
RNAi of kl-3, kl-5, kl-2 or Dhc98D results in the same sterility phenotype seen in rept or pont RNAi testes. (A–D) Phase-contrast images of seminal vesicles in kl-3 RNAi (bam-gal4>UAS-kl-3TRiP.HMC03546; A), kl-5 RNAi (bam-gal4>UAS-kl-5TRiP.HMC03747; B), kl-2 RNAi (bam-gal4>UAS-kl-2GC8807; C) and Dhc98D RNAi (bam-gal4>UAS-Dhc98DTRiP.HMC06494; D). Bar: 100 µm. (E–L) Phalloidin staining of early and late ICs in the indicated genotypes. Phalloidin (Actin, red) and DAPI (white). Bar: 10 µm.
We further characterized the sterility phenotype of rept and pont RNAi testes and found that spermiogenesis fails during individualization. As sperm develop as cysts, the process of individualization removes excess cytoplasm from the spermatids and separates the cyst into individual sperm via actin-rich individualization complexes (ICs; Fabian and Brill, 2012). The ICs form around the nuclei at the proximal end of the cyst and progress evenly toward the distal end (Fig. 5 D). It is well established that defects in axoneme assembly, including loss of axonemal dynein motor proteins, perturb IC progression (Fatima, 2011; Fingerhut et al., 2019; Wang et al., 2019). We found that RNAi-mediated knockdown of rept or pont does not affect IC assembly but does result in disorganized IC progression (Fig. 5, E–J), as is observed following knockdown of kl-3, kl-5, kl-2, or Dhc98D (Fig. S3; Fingerhut et al., 2019).
As previous studies have implicated Rept and Pont in male fertility and axonemal dynein motor assembly and the observed individualization defects are characteristic of axonemal defects, we analyzed Kl-3 protein levels following rept or pont RNAi. Western blotting using total testis extracts revealed that Kl-3 protein levels are drastically reduced following knockdown of rept or pont (Fig. 5 K). Taken together, our results demonstrate that Rept and Pont are required for mRNAs to congress in the kl-granule, which in turn is required for efficient translation. This defect in axonemal dynein expression is likely the cause of sterility in rept and pont RNAi testes, but it should be noted that due to the myriad of functions attributed to Rept and Pont (Mao and Houry, 2017; Robinson et al., 2013), it is possible that Rept and Pont play additional roles in the testis that are also important for fertility.
kl-granule formation and localization are required for cytoplasmic cilia maturation
Precise mRNA localization is a widely used mechanism to ensure that proteins are concentrated where they are needed (Medioni et al., 2012). As described above, kl-granules localize to the distal end of elongating spermatids (Fig. 2), where bare axonemal microtubules are first exposed to the cytoplasm after being displaced from the ciliary cap as new microtubules are polymerized. We therefore postulated that the kl-granule may function in cytoplasmic cilia maturation. We first determined whether kl-granules localize within the ciliary cap or within the cytoplasmic compartment. By using Unc-GFP to mark the ring centriole, a structure at the base of the ciliary cap near the transition zone at the boundary between the cytoplasmic and compartmentalized regions (Baker et al., 2004; Phillips, 1970), we found that kl-granules are located within the cytoplasmic compartment, immediately proximal to the ciliary cap (Fig. 6 A). Similarly, we found that FLAG-tagged Kl-3 protein (expressed from the endogenous locus; see Materials and methods) occupies the same region proximal to the ciliary cap as the kl-granules and that Kl-3 protein is restricted to the cytoplasmic compartment while the microtubules extend into the compartmentalized region (i.e., the ciliary cap; Fig. 6 B). These results indicate that while the axonemal microtubules are polymerized within the ciliary cap, axoneme maturation (the incorporation of axonemal dyneins and other axonemal proteins) may occur within the cytoplasmic compartment, as has been proposed (Avidor-Reiss and Leroux, 2015).
Figure 6.
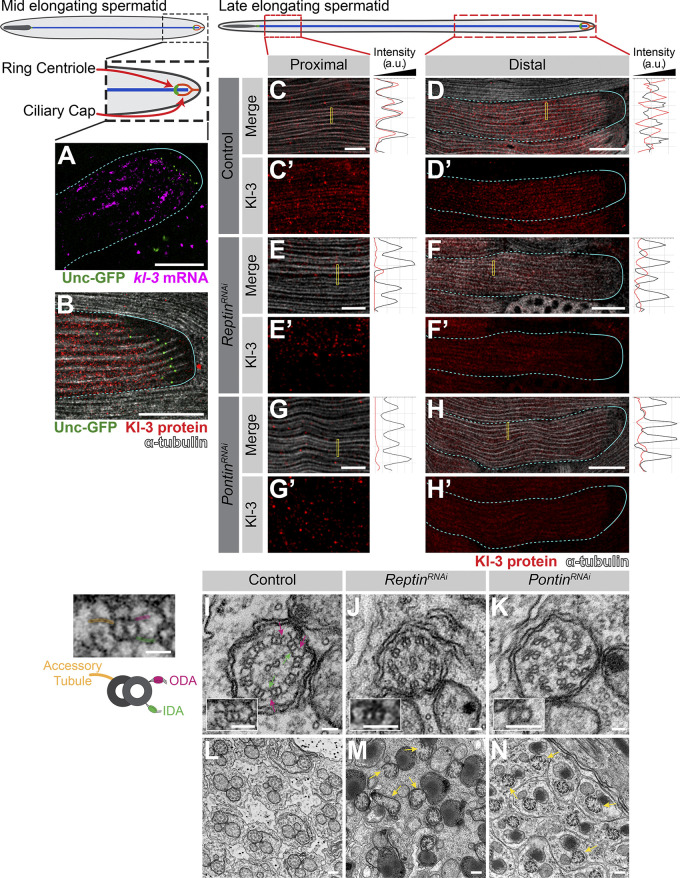
kl-granule formation and localization are required for cytoplasmic cilia maturation. (A) smFISH against kl-3 in flies expressing Unc-GFP. Shown are kl-3 (magenta), Unc-GFP (ring centriole, green), and a spermatid cyst (cyan, dashed line: cytoplasmic region; solid line: compartmentalized region). Scale bar: 20 µm. (B) Kl-3-3X FLAG protein in flies expressing Unc-GFP. Shown are Kl-3 (red), Unc-GFP (ring centriole, green), α-tubulin (white), and spermatid cyst (cyan, dashed line: cytoplasmic region, solid line: compartmentalized region). Scale bar: 20 µm. (C–H) Kl-3-3X FLAG protein expression in control (C and D), rept RNAi (bam-gal4>UAS-reptKK105732; E and F), and pont RNAi (bam-gal4>UAS-pontKK101103; G and H) proximal (C, E, and G) and distal (D, F, and H) regions of late elongating spermatids. Shown are Kl-3 (red), α-tubulin-GFP (white), and a spermatid cyst (cyan, dashed line: cytoplasmic region; solid line: compartmentalized region). Intensity plots are shown for the regions within the yellow rectangles. Scale bars: 5 µm (C, E, and G) or 25 µm (D, F, and H). (I–N) Transmission EM images of control (I and L), rept RNAi (bam-gal4>UAS-reptKK105732; J and M), and pont RNAi (bam-gal4>UAS-pontKK101103; K and N) axonemes. Magenta arrows, ODA; green arrows, IDA; yellow arrows, broken axonemes. The control single doublet enlarged image is duplicated to the left of the figure and colored to match the diagram. Scale bars: 50 nm (I–K), 200 nm (L–N), or 25 nm (diagram left of I).
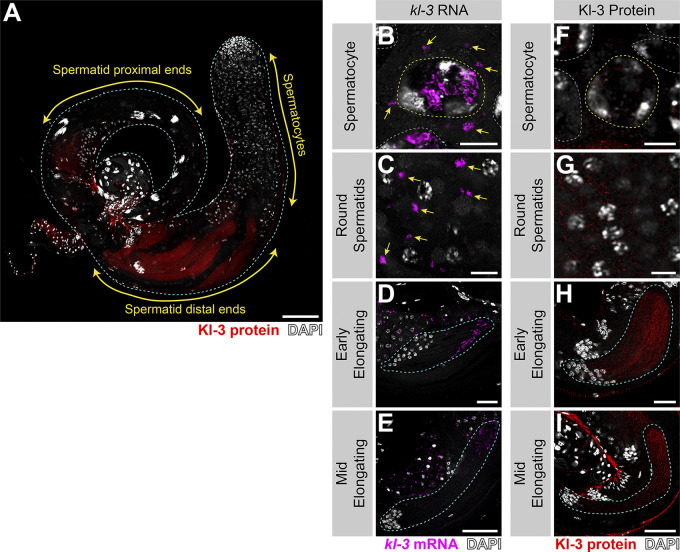
Detailed examination of Kl-3 protein within elongating spermatid cysts provided insights into where Kl-3 protein may be translated and incorporated into the growing axoneme (Fig. 6, C and D; and Fig. S4). Kl-3 protein is not present in SCs and becomes weakly detectable in round spermatids, but it does not robustly accumulate until elongating spermatids, where it becomes strongly enriched at the distal end, correlating with kl-granule localization (Figs. S4 and 2). At the distal end of the cyst, Kl-3 protein was predominantly observed in the cytoplasm while being excluded from the axonemal microtubules (Fig. 6 D; see the right panel for intensity plot showing mutually exclusive localization of microtubules and Kl-3). This suggests that Kl-3 protein at the distal end may represent the pool of newly translated Kl-3 before it is incorporated into the axoneme, which is also consistent with the presence of kl-granules at this location (Fig. S4). In contrast to the distal end, Kl-3 protein was observed to colocalize with axonemal microtubules at the proximal end (Fig. 6 D; see the right panel for intensity plot showing colocalization of microtubules and Kl-3), suggesting that Kl-3 protein has been successfully incorporated into the axoneme. These results suggest that Kl-3 protein may be translated at the distal end, where the kl-granules localize, and that the diffuse cytoplasmic Kl-3 protein may be the newly synthesized pool, which is subsequently incorporated into the axoneme.
Figure S4.
Kl-3 translation correlates with kl-granule dissociation and is enriched at the distal end. (A) Kl-3 3X FLAG protein expression in a wild-type testis. Shown are Kl-3 (red), DAPI (white), and testis outline (cyan dashed line). Scale bar: 100 µm. (B–E) smFISH for kl-3 in the indicated developmental stages. Shown are kl-3 (magenta), DAPI (white), SC nuclei (yellow dashed line), neighboring SC nuclei (white dashed line), a spermatid cyst (cyan dashed line), and kl-granules (yellow arrows). (F–I) Kl-3 3X FLAG protein in the indicated developmental stages. Shown are Kl-3 (red), DAPI (white), SC nuclei (yellow dashed line), neighboring SC nuclei (white dashed line), and a spermatid cyst (cyan dashed line). Scale bars: 10 µm (B, C, F, and G), 25 µm (D and H), and 50 µm (E and I).
Following RNAi-mediated knockdown of rept or pont, which prevents kl-granule formation (Fig. 4) and drastically reduces Kl-3 protein levels (Fig. 5 K), we still observed Kl-3 protein in the cytoplasm at the distal end (Fig. 6, F and H), although at a much reduced level. However, Kl-3 protein was never observed to colocalize with the axonemal microtubules at the proximal end upon rept or pont RNAi (Fig. 6, E and G), suggesting that Rept and Pont are required for incorporation of Kl-3 into the axoneme.
Consistent with this notion, transmission EM revealed that the ODAs and IDAs are largely absent from the axonemes following rept or pont RNAi (Fig. 6, I–K). Additional gross axonemal defects (e.g., broken axonemes) were present in the RNAi conditions (Fig. 6, L–N), suggesting additional impairments to axoneme assembly. These results suggest that mRNA localization via formation of kl-granules is required for axonemal dynein motor proteins to incorporate into the axoneme.
Discussion
Cytoplasmic cilia have been found in organisms as diverse as Plasmodium and humans (Avidor-Reiss et al., 2017; Avidor-Reiss and Leroux, 2015; Dawson and House, 2010; Fawcett et al., 1970; Sinden et al., 1976; Tates, 1971; Tokuyasu, 1975). While it has been proposed that axoneme maturation proceeds through the direct incorporation of axonemal proteins from the cytoplasm, this model remained untested (Avidor-Reiss and Leroux, 2015). Our study provides the first insights into the mechanism of cytoplasmic cilia formation. Our results show that localization of axonemal dynein mRNAs facilitates the maturation of cytoplasmic cilia by concentrating axonemal dynein proteins, likely through localized translation, allowing for the efficient incorporation of axonemal dynein proteins into bare axonemal microtubules directly from the cytoplasm.
Mechanism for cytoplasmic cilia maturation
It has been proposed that cytoplasmic cilia assemble in two steps (Avidor-Reiss and Leroux, 2015): first, microtubules are polymerized within a small compartmentalized region of the cilia; then, as the bare microtubules are displaced from this region, axonemal proteins are incorporated directly from the cytoplasm during the maturation step. Previous studies that have shown that IFT, the process used by traditional compartmentalized cilia to ferry axonemal proteins into the ciliary compartment, is dispensable for Drosophila spermiogenesis, and that the genomes of some other organisms known to form cytoplasmic cilia (e.g., Plasmodium) do not encode IFT and/or transition zone proteins (Avidor-Reiss and Leroux, 2015; Avidor-Reiss et al., 2004; Breslow et al., 2013; Briggs et al., 2004; Han et al., 2003; Hoeng et al., 2008; Kee et al., 2012; Lin et al., 2013; Sarpal et al., 2003). These studies led to the notion that maturation of cytoplasmic cilia ought to happen in the cytoplasm, although direct evidence has been lacking.
Our study, which identified a novel RNP granule, the kl-granule, composed of axonemal dynein heavy chain mRNAs and the proteins Rept and Pont, provides the first molecular insights into cytoplasmic cilia maturation. Our results show that axonemal dynein heavy chain mRNAs (kl-3, kl-5, kl-2, and Dhc98D) congress into kl-granules in SCs. We further show that Rept and Pont are required for kl-granule assembly and the robust translation of axonemal dynein mRNAs. We demonstrate that the polarized localization of kl-granule mRNAs and their protein products within the cytoplasmic compartment allows for the incorporation of axonemal dyneins into the axoneme, facilitating the maturation step in cytoplasmic cilia assembly (Fig. 7). Our results refine the proposed two-step model for cytoplasmic cilia assembly by demonstrating that concentrating axonemal proteins within distal regions of the cytoplasm is critical for maturation. Our data suggest that kl-granule mRNAs are locally translated at the distal end as kl-3 mRNA and Kl-3 protein are both polarized at this end (Figs. S4 and 6), although we cannot exclude other alternative possibilities. Thus, axoneme maturation proceeds in a stepwise fashion, allowing for the efficient assembly of this very long cilia. This model implies that the proximal region of the axoneme is more mature than the distal region, as axonemal proteins are still cytoplasmic at the distal end while they have incorporated at the proximal end, a notion that is supported by previous studies that looked at axoneme ultrastructure and tubulin dynamics within the axoneme (Noguchi et al., 2011; Sinden et al., 2010; Tokuyasu, 1975).
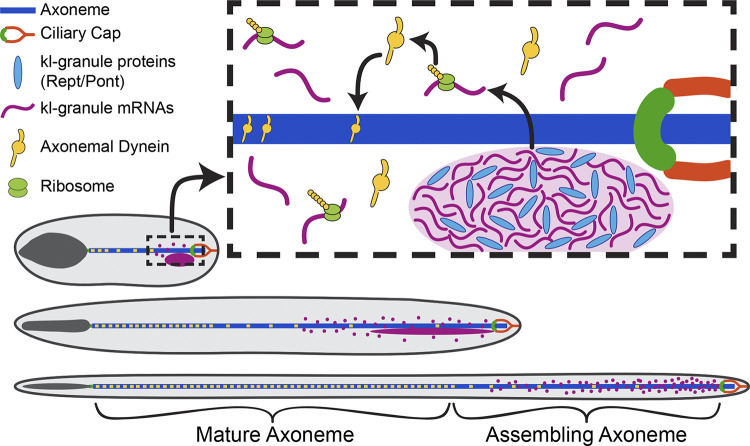
Figure 7.
Model for cytoplasmic cilia maturation. The kl-granule (light purple) localizes immediately proximal to the ciliary cap (orange) and transition zone (green) within the cytoplasmic compartment. Constituent mRNAs (purple) are likely locally translated (ribosomes, lime green), and their proteins (axonemal dyneins, yellow) are incorporated into the axoneme (blue) as the microtubules are displaced from the ciliary cap. In this way, cytoplasmic cilia maturation is progressive, with axonemal proteins being added to the bare microtubules as elongation proceeds.
Function of Rept and Pont in dynein assembly
A wide range of functions have been assigned to Rept- and Pont-containing complexes, including roles in chromatin remodeling, transcription regulation, DNA repair, and ribosome assembly (Mao and Houry, 2017). They can act alone, together, or as part of larger complexes (Huen et al., 2010; Kakihara and Saeki, 2014). Among these, previous studies have proposed that Rept and Pont are dynein arm preassembly factors, chaperones that take individual dynein motor subunits (i.e., the heavy, intermediate, and light chain proteins) and stabilize and assemble them into a motor unit in the cytoplasm that is then ferried into the cilia for incorporation (Desai et al., 2018; Fabczak and Osinka, 2019; Fowkes and Mitchell, 1998). These assembly factors include R2TP and R2TP-like complexes (which include Rept [RUVBL2] and Pont [RUVBL1]) in association with dynein axonemal assembly factors (Fabczak and Osinka, 2019). Our data suggest that Rept and Pont are involved in RNP complex assembly. While previous studies have demonstrated that Rept, Pont, R2TP, and dynein axonemal assembly factors are needed for axonemal dynein incorporation (Huizar et al., 2018; Li et al., 2017; Liu et al., 2019; Yamaguchi et al., 2018; Zhao et al., 2013; Zur Lage et al., 2018), our study is the first to demonstrate the importance of axonemal dynein mRNAs with these complexes, showing that Rept and Pont are required for axonemal dynein mRNAs to localize to kl-granules. We cannot however exclude the possibility that Rept and Pont may also be playing more traditional roles in protein stability (Kl-3 protein may be translated in the rept and pont RNAi conditions but degraded due to lack of chaperone activity) or complex assembly (similarly, Kl-3 protein may be translated but degraded if it cannot complex with dynein intermediate and light chains). Interestingly, a recent study reported the presence of RNA in dynein assembly complexes in Xenopus multiciliated cells (Drew et al., 2020 Preprint), and while the identity of these RNAs is unknown, it is intriguing to postulate that similarities may exist between kl-granules and these previously identified dynein assembly particles. However, important differences exist as well. First, while kl-3 mRNA is present in kl-granules, no puncta are observed for Kl-3 protein (Figs. 6 and S4), indicating that Kl-3 protein does not concentrate within kl-granules (or another granule) as dyneins do in the dynein preassembly complexes reported in other systems (Dafinger et al., 2018; Huizar et al., 2018). Additionally, dynein preassembly complexes were found to contain proteins (e.g., Wdr78 [Huizar et al., 2018]), where the Drosophila homologue (Dic61B) mRNAs are not constituents of the kl-granules (Fig. S5; see below). Therefore, the kl-granule may be a novel adaptation of a Rept- and Pont-containing dynein arm assembly complex specifically found in cytoplasmic cilia and is distinct from its role as a dynein preassembly factor in other systems.
Figure S5.
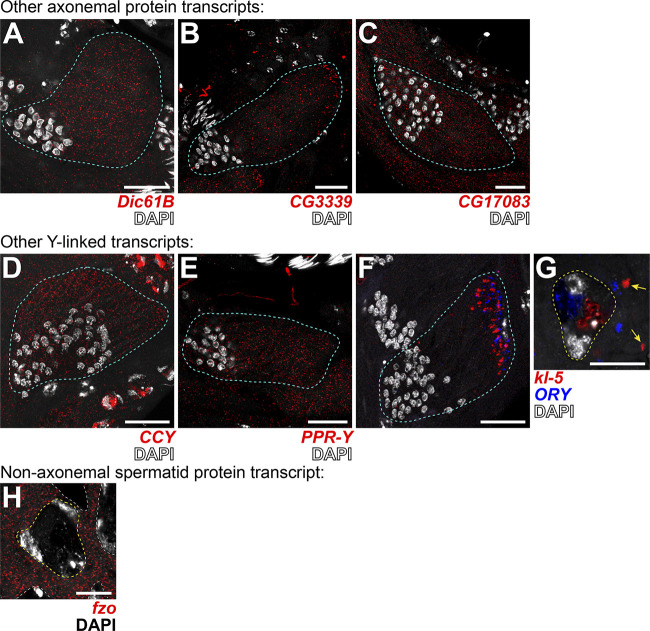
Transcripts for other axonemal, Y-linked, and spermatid proteins do not localize to kl-granules. (A–G) smFISH against other axonemal (A–C), Y-linked (D–G), or spermatid-essential (H) transcripts in wild type. (A) Dic61B (dynein intermediate chain, red) and DAPI (white). (B) CG3339 (axonemal dynein heavy chain, red) and DAPI (white). (C) CG17083 (ODA docking complex, red) and DAPI (white). (D) CCY (red) and DAPI (white). (E) PPR-Y (red) and DAPI (white). (F and G) kl-5 (red), ORY (blue), DAPI (white), and kl-granules (yellow arrows). (G) fzo (red) and DAPI (white). For all, spermatid cyst, cyan; SC nuclei, yellow dashed line; neighboring SC nuclei, white dashed line. Note that because Fzo is translated early in spermiogenesis, an SC image was used. Scale bars: 25 µm (A–F) and 10 µm (G and H).
It is likely that additional protein components of the kl-granule remain to be discovered. Structural analyses in previous studies have identified mechanisms by which other proteins interact with Rept and Pont (Rivera-Calzada et al., 2017); however, Rept and Pont do not have any RNA-binding domains (Mao and Houry, 2017). Therefore, it is likely that additional proteins, not Rept and Pont themselves, physically interact with constituent mRNAs for kl-granule formation. Our data also support the existence of additional proteins governing kl-granule dynamics. For example, as spermatids elongate, the ODA and IDA mRNAs separate slightly from each other while remaining polarized at the distal end (Fig. 2). Moreover, in the absence of Rept and Pont, the IDA mRNAs are still able to congress at the distal end of the elongating spermatid cyst, after failing to form kl-granules in SCs. In contrast, localization of the ODA mRNAs entirely depends on Rept and Pont, as ODA mRNAs remain diffuse throughout spermatogenesis following rept or pont RNAi. Finally, Pont more strongly colocalizes with the ODA mRNAs within the kl-granule (Fig. 3 F), which altogether suggests that there are additional proteins that can sort and specify the fate of these kl-granule mRNAs both alongside or in the absence of Rept and Pont. The identity of these additional proteins is the subject of further study. Alternatively, there could be a differential requirement for Rept and Pont and/or their associated proteins between ODA and IDA mRNAs and over developmental time. Determining the involvement of the other dynein arm preassembly factors is of particular interest, especially considering the existence of multiple dynein arm assembly complexes that have been shown to differentially regulate IDA and ODA assembly (Fabczak and Osinka, 2019; Yamaguchi et al., 2018). It is also appealing to posit the existence of testis-specific factors that may help to distinguish the role of Rept and Pont in cytoplasmic cilia formation from their role in the assembly of other cellular bodies.
Purpose of mRNA localization to kl-granules
Interestingly, we found that not all mRNAs for axonemal/spermiogenesis proteins localize to kl-granules (Fig. S5). mRNAs for other axonemal proteins (the dynein intermediate chain Dic61B, the dynein heavy chain CG3339, and the ODA docking complex component CG17083; Zur Lage et al., 2019), as well as mRNAs for other Y-linked transcripts (CCY and PPR-Y; Carvalho et al., 2001) and a nonaxonemal spermatid protein (fzo; Hales and Fuller, 1997), did not localize to kl-granules. Instead they remain evenly distributed throughout the cytoplasm, despite also being important for sperm maturation (Fig. S5). Additionally, we previously reported that mRNAs for the Y-liked gene ORY also gather in cytoplasmic RNA granules in late SCs (Fingerhut et al., 2019); however, these RNA granules are distinct from the kl-granule (Fig. S5, F and G).
In particular, it is intriguing that Dic61B mRNA, an IDA intermediate chain that needs to bind to the IDA heavy chains Kl-2 and Dhc98D, is located differently (diffusely) within the spermatid cyst. Dynein preassembly is believed to be important for dynein protein stability and a prerequisite for axonemal incorporation (Fabczak and Osinka, 2019; Fowkes and Mitchell, 1998). An intriguing possibility is that temporal/spatial regulation of dynein mRNAs plays a role in helping the ordered assembly of dynein complexes. It will be of future interest to determine when and where during spermiogenesis dynein complexes are formed in the cytoplasmic cilia as well as what factors are necessary for their formation. A comprehensive understanding of kl-granule mRNAs and proteins would allow for further study into this temporal/spatial regulatory mechanism and a more thorough understanding of how kl-granules function in the maturation of cytoplasmic cilia.
In summary, our study provides the first insights into the mechanism of cytoplasmic cilia maturation: mRNAs for axonemal dynein motor proteins are localized at the distal end of the axoneme within the cytoplasmic compartment, which allows for efficient maturation of cytoplasmic cilia.
Materials and methods
Fly husbandry
All fly stocks were raised on standard Bloomington medium at 25°C, and young flies (1- to 5-d-old adults) were used for all experiments. Flies used for wild-type experiments were the standard laboratory wild-type strain yw (y1w1). The following fly stocks were used: bam-GAL4:VP16 (Bloomington Drosophila Stock Center [BDSC]: 80579), UAS-kl-3TRiP.HMC03546 (BDSC: 53317), UAS-kl-5TRiP.HMC03747 (BDSC:55609), UAS-Dhc98DTRiP.HMC06494 (BDSC: 77181), and C(1)RM/C(1;Y)6, y1w1f1/0 (BDSC: 9460) were obtained from the BDSC. UAS-kl-2GC8807 (Vienna Drosophila Resource Center [VDRC]: v19181), UAS-reptKK105732 (VDRC: v103483), and UAS-pontKK101103 (VDRC: v105408) were obtained from the VDRC. unc-GFP (GFP-tagged unc expressed by the endogenous promoter) and Ub-α-tubulin84B-GFP were a gift from Cayentano Gonzalez (IRB Barcelona, Barcelona, Spain; Baker et al., 2004; Rebollo et al., 2004), and bam-gal4 was a gift from Dennis McKearin (Howard Hughes Medical Institute, Chevy Chase, MD; Chen and McKearin, 2003). The kl-3-FLAG strain was constructed using CRISPR-mediated knock-in of a 3X-FLAG tag at the C-terminus of kl-3 as previously described (Fingerhut et al., 2019).
smFISH
All solutions used for RNA FISH were RNase-free. Testes from 2- to 3-d-old flies were dissected in 1X PBS and fixed in 4% formaldehyde in 1X PBS for 30 min. Then testes were washed briefly in 1X PBS and permeabilized in 70% ethanol overnight at 4°C. Testes were briefly rinsed with wash buffer (2X saline-sodium citrate, 10% formamide) and then hybridized overnight at 37°C in hybridization buffer containing 2X saline-sodium citrate, 10% dextran sulfate (Sigma-Aldrich; D8906), 1 mg/ml Escherichia coli tRNA (Sigma-Aldrich; R8759), 2 mM Vanadyl Ribonucleoside complex (NEB; S142), 0.5% BSA (Ambion; AM2618), and 10% formamide. Following hybridization, samples were washed three times in wash buffer for 20 min each at 37°C and mounted in VECTASHIELD with DAPI (Vector Labs). Images were acquired using an upright Leica TCS SP8 confocal microscope with a 63× oil immersion objective lens (NA 1.4) and processed using Adobe Photoshop and ImageJ software.
Fluorescently labeled probes were added to the hybridization buffer to a final concentration of 100 nM. Probes against kl-3, kl-5, kl-2, Dhc98D, CG3339, Dic61B, CG17083, CCY, PPR-Y, ORY, and fzo mRNAs were designed using the Stellaris RNA FISH Probe Designer (Biosearch Technologies; available online at www.biosearchtech.com/stellarisdesigner). Each set of custom Stellaris RNA FISH probes was labeled with Quasar 670, Quasar 570, or Fluorescein (Table S1).
For strains expressing GFP (e.g., unc-GFP or Ub-α-tubulin84B-GFP), the overnight permeabilization in 70% ethanol was omitted.
Immunofluorescence staining
Testes were dissected in 1X PBS, transferred to 4% formaldehyde in 1X PBS, and fixed for 30 min. Testes were then washed in 1X PBST (PBS containing 0.1% Triton X-100) for at least 60 min followed by incubation with primary antibodies diluted in 1X PBST with 3% BSA at 4°C overnight. Samples were washed for at least 1 h in 1X PBST, incubated with secondary antibody in 1X PBST with 3% BSA at 4°C overnight, washed as above, and mounted in VECTASHIELD with DAPI (Vector Labs). Images were acquired using an upright Leica TCS SP8 confocal microscope with a 63× oil immersion objective lens (NA 1.4) and processed using Adobe Photoshop and ImageJ software.
The following primary antibodies were used: anti–α-tubulin (1:100, mouse; Sigma-Aldrich; T6199), anti-FLAG (1:500, rabbit; Invitrogen; PA1-984B), anti-Rept (1:200, rabbit, gift of Andrew Saurin, Institut de Biologie du Développement de Marseille, Marseille, France [Diop et al., 2008], anti-Pont (1:200; guinea pig, this study), Phalloidin-Alexa Fluor 546 or 488 (1:200; Thermo Fisher Scientific; A22283 or A12379). The Pont antibody was generated by injecting a peptide (CKVNGRNQISKDDIEDVH, targeting 18 aa from the C-terminal end of Pont) in guinea pigs (Covance). Alexa Fluor–conjugated secondary antibodies (Life Technologies) were used at a dilution of 1:200.
A modified version of Stefanini’s fixative (4% formaldehyde, 0.18% wt/vol picric acid [Ricca Chemical 5860], 0.3 M Pipes, pH 7.5 [Alfa Aesar; J63617], and 0.05% Tween-20) was used in order to detect Kl-3 (Müller, 2008). No signal was detectable using traditional formaldehyde fixation.
Immunofluorescence staining with smFISH
To combine immunofluorescent staining with smFISH, testes from 2- to 3 d-old-flies were dissected in 1X PBS and fixed in 4% formaldehyde in 1X PBS for 30 min. Then testes were washed briefly in PBS and permeabilized in 70% ethanol overnight at 4°C (unless from a strain expressing GFP, in which case this step was omitted). Testes were then washed with 1X PBS and blocked for 30 min at 37°C in blocking buffer (1X PBS, 0.05% BSA, 50 µg/ml E. coli tRNA, 10 mM Vanadyl Ribonucleoside complex, and 0.2% Tween-20). Primary antibodies were diluted in blocking buffer and incubated at 4°C overnight. The testes were washed with 1X PBS containing 0.2% Tween-20, reblocked for 5 min at 37°C in blocking buffer, and incubated 4°C overnight in blocking buffer containing secondary antibodies. Then, testes were washed with 1X PBS containing 0.2% Tween-20 and refixed for 10 min before continuing the smFISH, starting from the brief rinse with wash buffer.
Quantitative RT-PCR
Total RNA from testes (50 pairs/sample) was extracted using TRIzol (Invitrogen) according to the manufacturer’s instructions. 1 µg total RNA was reverse transcribed using SuperScript III Reverse transcription (Invitrogen) followed by qPCR using Power SYBR Green reagent (Applied Biosystems) on a QuantStudio 6 Real-Time PCR system (Applied Biosystems). Primers for quantitative PCR were designed to amplify only mRNA. The genes analyzed by qPCR are all predicted to contain megabase sized introns, and primers were designed to span these large introns such that a product would be detect only if the intron had been spliced out (Fingerhut et al., 2019). Two primer sets were used for each gene, one near the 5′ end and another closer to the 3′ end. Relative expression levels were normalized to GAPDH and cross-sibling controls. All reactions were done in technical triplicates with at least two biological replicates. Graphical representation is inclusive of all replicates. Primers used are listed in Table S1.
Western blot
Testes (40 pairs/sample) were dissected in Schneider’s media at room temperature within 30 min, the media was removed, and the samples were frozen at −80°C until use. After thawing, testes were then lysed in 200 μl of 2X Laemmli Sample Buffer + βME (Bio-Rad; 161–0737). For Kl-3, samples were separated on a NuPAGE Tris-Acetate gel (Invitrogen; 3–8%, 1.5 mm), and for Rept and Pont, samples were separated on a Novex Tris-Glycine gel (Invitrogen; 10%, 1 mm) with the appropriate running buffer in a Xcell SureLock mini-cell electrophoresis system (Invitrogen). For Kl-3, proteins were transferred using the XCell II blot module (Invitrogen) onto a polyvinylidene fluoride membrane (Immobilon-P, Millipore) using NuPAGE transfer buffer (Invitrogen) without added methanol. For Rept and Pont, transfer buffer contained 20% methanol. Membranes were blocked in 1X TBST (0.1% Tween-20) containing 5% nonfat milk, followed by incubation with primary antibodies diluted in 1X TBST containing 5% nonfat milk. Membranes were washed with 1X TBST, followed by incubation with secondary antibodies diluted in 1X TBST containing 5% nonfat milk. After washing with 1X TBST, detection was performed using the Pierce ECL Western Blotting Substrate enhanced chemiluminescence system (Thermo Fisher Scientific). Primary antibodies used were anti–α-tubulin (1:2,000, mouse; Sigma-Aldrich; T6199), anti-FLAG (1:2,500, mouse; Sigma-Aldrich; F1804), anti-Rept (1:2,000, rabbit; gift of Andrew Saurin), anti-Pont (1:2,000, guinea pig; this study), and anti-Vasa (1:3,000, rabbit; Santa Cruz Biotechnology; D-260). The secondary antibodies were HRP-conjugated goat anti-mouse IgG, anti-rabbit IgG, or anti-guinea pig IgG (1:10,000; Abcam).
Phase contrast microscopy
Seminal vesicles were dissected in 1X PBS and transferred to slides for live observation by phase contrast on a Leica DM5000B microscope with a 40× objective (NA 0.75) and imaged with a QImaging Retiga 2000R Fast 1394 Mono Cooled camera. Images were adjusted in Adobe Photoshop.
Transmission EM
Testes were fixed for 1 h or overnight (at 4°C) with 2.5% glutaraldehyde in 0.1M Sorensen’s buffer, pH 7.4. Samples were rinsed twice for 5 min each with 0.1 M Sorensen’s buffer and postfixed for 1 h in 1% osmium tetroxide in 0.1 M Sorensen’s buffer. Next, testes were rinsed twice in double distilled water for 5 min each and en bloc stained with 2% uranyl acetate in double distilled water for 1 h. The samples were then dehydrated in increasing concentrations of ethanol, rinsed with acetone, and embedded in epon epoxy resin. Thin sections were mounted on Formvar/carbon-coated slotted grids and poststained with uranyl acetate and lead citrate. Samples were examined on a JEOL1400 transmission electron microscope and images captured using a sCMOS XR401 custom engineered optic camera by AMT (Advanced Microscopy Techniques).
Online supplemental material
Fig. S1 shows efficiency of RNAi knockdown of kl-3, kl-5, kl-2, and Dhc98D by smFISH and lack of dependence upon a one of those transcripts for kl-granule formation (related to Fig. 1). Fig. S2 shows that RNAi of rept or pont results in efficient knockdown of both products (related to Fig. 4). Fig. S3 shows the sterility phenotype of kl-3, kl-5, kl-2, and Dhc98D RNAi flies (related to Fig. 5). Fig. S4 shows Kl-3 protein distribution and stage-matched kl-3 mRNA (related to Fig. 6). Fig. S5 shows smFISH for other axonemal, Y-linked, and spermatid-essential transcripts (related to Discussion).
Supplementary Material
showssmFISH probes and quantitative RT-PCR primers used in this study.
Acknowledgments
We thank Drs. Dennis McKearin, Cayentano Gonzalez, and Andrew Saurin, the BDSC, and the VDRC for reagents. We thank Sasha Meshinchi and the University of Michigan Microscopy Core for help with EM experiments. We thank the Yamashita laboratory and Drs. Sue Hammoud and Joshua Bembenek for discussion and comments on the manuscript, Drs. Tomer Avidor-Reiss and Tony Mahowald for helpful suggestions, and Dr. Jiandie Lin (University of Michigan, Ann Arbor, MI) for sharing equipment.
This work was supported by the Howard Hughes Medical Institute (Y.M. Yamashita) and National Institutes of Health cellular and molecular biology training grant T32-GM007315 (J.M. Fingerhut).
The authors declare no competing financial interests.
Author contributions: J.M. Fingerhut conceived the project and conducted experiments. J.M. Fingerhut and Y.M. Yamashita designed experiments, analyzed the data, and wrote the manuscript.
References
- Anderson P., and Kedersha N.. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10:430–436. 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T., and Leroux M.R.. 2015. Shared and Distinct Mechanisms of Compartmentalized and Cytosolic Ciliogenesis. Curr. Biol. 25:R1143–R1150. 10.1016/j.cub.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T., Maer A.M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., and Zuker C.S.. 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 117:527–539. 10.1016/S0092-8674(04)00412-X [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T., Ha A., and Basiri M.L.. 2017. Transition Zone Migration: A Mechanism for Cytoplasmic Ciliogenesis and Postaxonemal Centriole Elongation. Cold Spring Harb. Perspect. Biol. 9:a028142. 10.1101/cshperspect.a028142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker J.D., Adhikarakunnathu S., and Kernan M.J.. 2004. Mechanosensory-defective, male-sterile unc mutants identify a novel basal body protein required for ciliogenesis in Drosophila. Development. 131:3411–3422. 10.1242/dev.01229 [DOI] [PubMed] [Google Scholar]
- Barckmann B., Chen X., Kaiser S., Jayaramaiah-Raja S., Rathke C., Dottermusch-Heidel C., Fuller M.T., and Renkawitz-Pohl R.. 2013. Three levels of regulation lead to protamine and Mst77F expression in Drosophila. Dev. Biol. 377:33–45. 10.1016/j.ydbio.2013.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basiri M.L., Ha A., Chadha A., Clark N.M., Polyanovsky A., Cook B., and Avidor-Reiss T.. 2014. A migrating ciliary gate compartmentalizes the site of axoneme assembly in Drosophila spermatids. Curr. Biol. 24:2622–2631. 10.1016/j.cub.2014.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley N., Lederer M., Pfalz B., Reinke C., Fuchs T., Glaß M., Möller B., and Hüttelmaier S.. 2015. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res. 43 e26 10.1093/nar/gku1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F.M., van Koningsbruggen S., Navascués J., and Lamond A.I.. 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8:574–585. 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- Breslow D.K., Koslover E.F., Seydel F., Spakowitz A.J., and Nachury M.V.. 2013. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. J. Cell Biol. 203:129–147. 10.1083/jcb.201212024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs L.J., Davidge J.A., Wickstead B., Ginger M.L., and Gull K.. 2004. More than one way to build a flagellum: comparative genomics of parasitic protozoa. Curr. Biol. 14:R611–R612. 10.1016/j.cub.2004.07.041 [DOI] [PubMed] [Google Scholar]
- Buchan J.R. 2014. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 11:1019–1030. 10.4161/15476286.2014.972208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A.B., Lazzaro B.P., and Clark A.G.. 2000. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc. Natl. Acad. Sci. USA. 97:13239–13244. 10.1073/pnas.230438397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A.B., Dobo B.A., Vibranovski M.D., and Clark A.G.. 2001. Identification of five new genes on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 98:13225–13230. 10.1073/pnas.231484998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron F., and Barral Y.. 2009. Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell. 16:493–506. 10.1016/j.devcel.2009.04.003 [DOI] [PubMed] [Google Scholar]
- Chen D., and McKearin D.M.. 2003. A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development. 130:1159–1170. 10.1242/dev.00325 [DOI] [PubMed] [Google Scholar]
- Dafinger C., Rinschen M.M., Borgal L., Ehrenberg C., Basten S.G., Franke M., Höhne M., Rauh M., Göbel H., Bloch W., et al. . 2018. Targeted deletion of the AAA-ATPase Ruvbl1 in mice disrupts ciliary integrity and causes renal disease and hydrocephalus. Exp. Mol. Med. 50:1–17. 10.1038/s12276-018-0108-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S.C., and House S.A.. 2010. Life with eight flagella: flagellar assembly and division in Giardia. Curr. Opin. Microbiol. 13:480–490. 10.1016/j.mib.2010.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai P.B., Dean A.B., and Mitchell D.R.. 2018. Cytoplasmic preassembly and trafficking of axonemal dyneins. Dyneins. 1:140–161. [Google Scholar]
- Diop S.B., Bertaux K., Vasanthi D., Sarkeshik A., Goirand B., Aragnol D., Tolwinski N.S., Cole M.D., Pradel J., Yates J.R. III, et al. . 2008. Reptin and Pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep. 9:260–266. 10.1038/embor.2008.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew K., Lee C., Cox R.M., Dang V., Devitt C.C., Papoulas O., Huizar R.L., Marcotte E.M., and Wallingford J.B.. 2020. A systematic, label-free method for identifying RNA-associated proteins in vivo provides insights into vertebrate ciliary beating. bioRxiv. doi: 10.1101/2020.02.26.966754 Preprint posted March 2, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabczak H., and Osinka A.. 2019. Role of the Novel Hsp90 Co-Chaperones in Dynein Arms’ Preassembly. Int. J. Mol. Sci. 20:6174 10.3390/ijms20246174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian L., and Brill J.A.. 2012. Drosophila spermiogenesis: Big things come from little packages. Spermatogenesis. 2:197–212. 10.4161/spmg.21798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima R. 2011. Drosophila Dynein intermediate chain gene, Dic61B, is required for spermatogenesis. PLoS One. 6 e27822 10.1371/journal.pone.0027822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett D.W., Eddy E.M., and Phillips D.M.. 1970. Observations on the fine structure and relationships of the chromatoid body in mammalian spermatogenesis. Biol. Reprod. 2:129–153. 10.1095/biolreprod2.1.129 [DOI] [PubMed] [Google Scholar]
- Fingerhut J.M., Moran J.V., and Yamashita Y.M.. 2019. Satellite DNA-containing gigantic introns in a unique gene expression program during Drosophila spermatogenesis. PLoS Genet. 15 e1008028 10.1371/journal.pgen.1008028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowkes M.E., and Mitchell D.R.. 1998. The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol. Biol. Cell. 9:2337–2347. 10.1091/mbc.9.9.2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller M.T. 1993. Spermatogenesis In The Development of Drosophila Melanogaster. Vol. Vol. 1 Bate M., and Arias A.M., editors. Cold Spring Harbor Laboratory Press, New York: pp. 71–148. [Google Scholar]
- Goldstein L.S., Hardy R.W., and Lindsley D.L.. 1982. Structural genes on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 79:7405–7409. 10.1073/pnas.79.23.7405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorynia S., Bandeiras T.M., Pinho F.G., McVey C.E., Vonrhein C., Round A., Svergun D.I., Donner P., Matias P.M., and Carrondo M.A.. 2011. Structural and functional insights into a dodecameric molecular machine - the RuvBL1/RuvBL2 complex. J. Struct. Biol. 176:279–291. 10.1016/j.jsb.2011.09.001 [DOI] [PubMed] [Google Scholar]
- Gottardo M., Callaini G., and Riparbelli M.G.. 2013. The cilium-like region of the Drosophila spermatocyte: an emerging flagellum? J. Cell Sci. 126:5441–5452. 10.1242/jcs.136523 [DOI] [PubMed] [Google Scholar]
- Hales K.G., and Fuller M.T.. 1997. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 90:121–129. 10.1016/S0092-8674(00)80319-0 [DOI] [PubMed] [Google Scholar]
- Han Y.G., Kwok B.H., and Kernan M.J.. 2003. Intraflagellar transport is required in Drosophila to differentiate sensory cilia but not sperm. Curr. Biol. 13:1679–1686. 10.1016/j.cub.2003.08.034 [DOI] [PubMed] [Google Scholar]
- Hardy R.W., Tokuyasu K.T., and Lindsley D.L.. 1981. Analysis of spermatogenesis in Drosophila melanogaster bearing deletions for Y-chromosome fertility genes. Chromosoma. 83:593–617. 10.1007/BF00328522 [DOI] [PubMed] [Google Scholar]
- Hime G.R., Brill J.A., and Fuller M.T.. 1996. Assembly of ring canals in the male germ line from structural components of the contractile ring. J. Cell Sci. 109:2779–2788. [DOI] [PubMed] [Google Scholar]
- Hoeng J.C., Dawson S.C., House S.A., Sagolla M.S., Pham J.K., Mancuso J.J., Löwe J., and Cande W.Z.. 2008. High-resolution crystal structure and in vivo function of a kinesin-2 homologue in Giardia intestinalis. Mol. Biol. Cell. 19:3124–3137. 10.1091/mbc.e07-11-1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen J., Kakihara Y., Ugwu F., Cheung K.L., Ortega J., and Houry W.A.. 2010. Rvb1-Rvb2: essential ATP-dependent helicases for critical complexes. Biochem. Cell Biol. 88:29–40. 10.1139/O09-122 [DOI] [PubMed] [Google Scholar]
- Huizar R.L., Lee C., Boulgakov A.A., Horani A., Tu F., Marcotte E.M., Brody S.L., and Wallingford J.B.. 2018. A liquid-like organelle at the root of motile ciliopathy. eLife. 7:e38497. 10.7554/eLife.38497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., and Marshall W.F.. 2011. Ciliogenesis: building the cell’s antenna. Nat. Rev. Mol. Cell Biol. 12:222–234. 10.1038/nrm3085 [DOI] [PubMed] [Google Scholar]
- Jain S., Wheeler J.R., Walters R.W., Agrawal A., Barsic A., and Parker R.. 2016. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell. 164:487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakihara Y., and Saeki M.. 2014. The R2TP chaperone complex: its involvement in snoRNP assembly and tumorigenesis. Biomol. Concepts. 5:513–520. 10.1515/bmc-2014-0028 [DOI] [PubMed] [Google Scholar]
- Kee H.L., Dishinger J.F., Blasius T.L., Liu C.J., Margolis B., and Verhey K.J.. 2012. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat. Cell Biol. 14:431–437. 10.1038/ncb2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwitny S., Klaus A.V., and Hunnicutt G.R.. 2010. The annulus of the mouse sperm tail is required to establish a membrane diffusion barrier that is engaged during the late steps of spermiogenesis. Biol. Reprod. 82:669–678. 10.1095/biolreprod.109.079566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Cox R.M., Papoulas O., Horani A., Drew K., Devitt C.C., Brody S.L., Marcotte E.M., and Wallingford J.B.. 2020a Functional partitioning of a liquid-like organelle during assembly of axonemal dyneins. bioRxiv. doi: 10.1101/2020.04.21.052837 (Preprint posted April 21, 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.S., Putnam A., Lu T., He S., Ouyang J.P.T., and Seydoux G.. 2020b Recruitment of mRNAs to P granules by condensation with intrinsically-disordered proteins. eLife. 9:e52896. 10.7554/eLife.52896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhao L., Yuan S., Zhang J., and Sun Z.. 2017. Axonemal dynein assembly requires the R2TP complex component Pontin. Development. 144:4684–4693. 10.1242/dev.152314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C., Niewiadomski P., Lin B., Nakamura H., Phua S.C., Jiao J., Levchenko A., Inoue T., Rohatgi R., and Inoue T.. 2013. Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nat. Chem. Biol. 9:437–443. 10.1038/nchembio.1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S.C., Sinsimer K.S., Lee J.J., Wieschaus E.F., and Gavis E.R.. 2015. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat. Cell Biol. 17:558–568. 10.1038/ncb3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Wang L., and Pan J.. 2019. Chlamydomonas WDR92 in association with R2TP-like complex and multiple DNAAFs to regulate ciliary dynein preassembly. J. Mol. Cell Biol. 11:770–780. 10.1093/jmcb/mjy067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.Q., and Houry W.A.. 2017. The Role of Pontin and Reptin in Cellular Physiology and Cancer Etiology. Front. Mol. Biosci. 4:58 10.3389/fmolb.2017.00058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medioni C., Mowry K., and Besse F.. 2012. Principles and roles of mRNA localization in animal development. Development. 139:3263–3276. 10.1242/dev.078626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller H.A. 2008. Immunolabeling of embryos. Methods Mol. Biol. 420:207–218. 10.1007/978-1-59745-583-1_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T., Koizumi M., and Hayashi S.. 2011. Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr. Biol. 21:805–814. 10.1016/j.cub.2011.04.016 [DOI] [PubMed] [Google Scholar]
- Olivieri G., and Olivieri A.. 1965. Autoradiographic study of nucleic acid synthesis during spermatogenesis in Drosophila melanogaster. Mutat. Res. 2:366–380. 10.1016/0027-5107(65)90072-2 [DOI] [PubMed] [Google Scholar]
- Phillips D.M. 1970. Insect sperm: their structure and morphogenesis. J. Cell Biol. 44:243–277. 10.1083/jcb.44.2.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchades C., Sandate C.R., and Lander G.C.. 2020. The molecular principles governing the activity and functional diversity of AAA+ proteins. Nat. Rev. Mol. Cell Biol. 21:43–58. 10.1038/s41580-019-0183-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebollo E., Llamazares S., Reina J., and Gonzalez C.. 2004. Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol. 2 E8 10.1371/journal.pbio.0020008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J.F., Blacque O.E., and Leroux M.R.. 2012. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 13:608–618. 10.1038/embor.2012.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli M.G., Callaini G., and Megraw T.L.. 2012. Assembly and persistence of primary cilia in dividing Drosophila spermatocytes. Dev. Cell. 23:425–432. 10.1016/j.devcel.2012.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Calzada A., Pal M., Munoz-Hernandez H., Luque-Ortega J.R., Gil-Carton D., Degliesposti G., Skehel J.M., Prodromou C., Pearl L.H., and Llorca O.. 2017. The Structure of the R2TP Complex Defines a Platform for Recruiting Diverse Client Proteins to the HSP90 Molecular Chaperone System. Structure. 25:1145–1152.e1144. [DOI] [PMC free article] [PubMed]
- Robinson S.W., Herzyk P., Dow J.A., and Leader D.P.. 2013. FlyAtlas: database of gene expression in the tissues of Drosophila melanogaster. Nucleic Acids Res. 41(D1):D744–D750. 10.1093/nar/gks1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J.L., and Witman G.B.. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813–825. 10.1038/nrm952 [DOI] [PubMed] [Google Scholar]
- Sarpal R., Todi S.V., Sivan-Loukianova E., Shirolikar S., Subramanian N., Raff E.C., Erickson J.W., Ray K., and Eberl D.F.. 2003. Drosophila KAP interacts with the kinesin II motor subunit KLP64D to assemble chordotonal sensory cilia, but not sperm tails. Curr. Biol. 13:1687–1696. 10.1016/j.cub.2003.09.025 [DOI] [PubMed] [Google Scholar]
- Sinden R.E., Canning E.U., and Spain B.. 1976. Gametogenesis and fertilization in Plasmodium yoelii nigeriensis: a transmission electron microscope study. Proc. R. Soc. Lond. B Biol. Sci. 193:55–76. 10.1098/rspb.1976.0031 [DOI] [PubMed] [Google Scholar]
- Sinden R.E., Talman A., Marques S.R., Wass M.N., and Sternberg M.J.. 2010. The flagellum in malarial parasites. Curr. Opin. Microbiol. 13:491–500. 10.1016/j.mib.2010.05.016 [DOI] [PubMed] [Google Scholar]
- Stolc V., Samanta M.P., Tongprasit W., and Marshall W.F.. 2005. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. USA. 102:3703–3707. 10.1073/pnas.0408358102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammana D., and Tammana T.V.S.. 2017. Human DNA helicase, RuvBL1 and its Chlamydomonas homologue, CrRuvBL1 plays an important role in ciliogenesis. Cytoskeleton (Hoboken). 74:251–259. 10.1002/cm.21377 [DOI] [PubMed] [Google Scholar]
- Tates A.D. 1971. Cytodifferentiation during Spermatogenesis in Drosophila Melanogaster: An Electon Microsope Study. Vol. Ph.D. Rijksuniversiteit, Leiden. [Google Scholar]
- Tokuyasu K.T. 1975. Dynamics of spermiogenesis in Drosophila melanogaster. VI. Significance of “onion” nebenkern formation. J. Ultrastruct. Res. 53:93–112. 10.1016/S0022-5320(75)80089-X [DOI] [PubMed] [Google Scholar]
- Trcek T., Grosch M., York A., Shroff H., Lionnet T., and Lehmann R.. 2015. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat. Commun. 6:7962 10.1038/ncomms8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher A.S., Meng Z., Mason P.J., Veenstra T.D., and Artandi S.E.. 2008. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 132:945–957. 10.1016/j.cell.2008.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieillard J., Paschaki M., Duteyrat J.L., Augière C., Cortier E., Lapart J.A., Thomas J., and Durand B.. 2016. Transition zone assembly and its contribution to axoneme formation in Drosophila male germ cells. J. Cell Biol. 214:875–889. 10.1083/jcb.201603086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.T., Smith J., Chen B.C., Schmidt H., Rasoloson D., Paix A., Lambrus B.G., Calidas D., Betzig E., and Seydoux G.. 2014. Regulation of RNA granule dynamics by phosphorylation of serine-rich, intrinsically disordered proteins in C. elegans. eLife. 3 e04591 10.7554/eLife.04591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu R., Cheng Y., Cao H., Wang Z., Zhu T., Jiang J., Zhang H., Wang C., Qi L., et al. . 2019. RSBP15 interacts with and stabilizes dRSPH3 during sperm axoneme assembly in Drosophila. J. Genet. Genomics. 46:281–290. 10.1016/j.jgg.2019.05.001 [DOI] [PubMed] [Google Scholar]
- Wheway G., Nazlamova L., and Hancock J.T.. 2018. Signaling through the Primary Cilium. Front. Cell Dev. Biol. 6:8 10.3389/fcell.2018.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Oda T., Kikkawa M., and Takeda H.. 2018. Systematic studies of all PIH proteins in zebrafish reveal their distinct roles in axonemal dynein assembly. eLife. 7:e36979. 10.7554/eLife.36979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Yuan S., Cao Y., Kallakuri S., Li Y., Kishimoto N., DiBella L., and Sun Z.. 2013. Reptin/Ruvbl2 is a Lrrc6/Seahorse interactor essential for cilia motility. Proc. Natl. Acad. Sci. USA. 110:12697–12702. 10.1073/pnas.1300968110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur Lage P., Stefanopoulou P., Styczynska-Soczka K., Quinn N., Mali G., von Kriegsheim A., Mill P., and Jarman A.P.. 2018. Ciliary dynein motor preassembly is regulated by Wdr92 in association with HSP90 co-chaperone, R2TP. J. Cell Biol. 217:2583–2598. 10.1083/jcb.201709026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur Lage P., Newton F.G., and Jarman A.P.. 2019. Survey of the Ciliary Motility Machinery of Drosophila Sperm and Ciliated Mechanosensory Neurons Reveals Unexpected Cell-Type Specific Variations: A Model for Motile Ciliopathies. Front. Genet. 10:24 10.3389/fgene.2019.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
showssmFISH probes and quantitative RT-PCR primers used in this study.