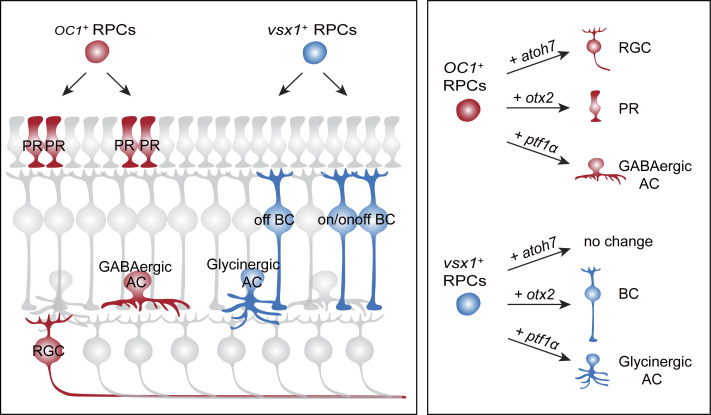
Combining in vivo lineage tracing and single-cell RNA- and ATAC-seq, Wang et al. identified a repertoire of stereotyped neurogenic lineages in the developing zebrafish retina, revealing the lineage logic of neuron type/subtype specification and respecification in a vertebrate CNS structure.
Abstract
How astounding neuronal diversity arises from variable cell lineages in vertebrates remains mostly elusive. By in vivo lineage tracing of ∼1,000 single zebrafish retinal progenitors, we identified a repertoire of subtype-specific stereotyped neurogenic lineages. Remarkably, within these stereotyped lineages, GABAergic amacrine cells were born with photoreceptor cells, whereas glycinergic amacrine cells were born with OFF bipolar cells. More interestingly, post-mitotic differentiation blockage of GABAergic and glycinergic amacrine cells resulted in their respecification into photoreceptor and bipolar cells, respectively, suggesting lineage constraint in cell subtype specification. Using single-cell RNA-seq and ATAC-seq analyses, we further identified lineage-specific progenitors, each defined by specific transcription factors that exhibited characteristic chromatin accessibility dynamics. Finally, single pro-neural factors could specify different neuron types/subtypes in a lineage-dependent manner. Our findings reveal the importance of lineage context in defining neuronal subtypes and provide a demonstration of in vivo lineage-dependent induction of unique retinal neuron subtypes for treatment purposes.
Graphical Abstract
Introduction
In invertebrate species, such as Caenorhabditis elegans and Drosophila, diverse neuron types derive from predetermined stereotyped cell lineages (Brody and Odenwald, 2000; Isshiki et al., 2001; Pearson and Doe, 2004; Sulston, 1976; Sulston et al., 1983; Udolph et al., 1995). In the vertebrates, neuronal types are, however, generated within highly variable cell lineages (He et al., 2012; Holt et al., 1988; Pearson and Doe, 2004; Turner et al., 1990). It has raised a big challenge for the past 30 years to resolve the question as to how vertebrate neuronal diversity arises from these highly variable cell lineages.
Increasing evidence suggests the functional relevance of lineage-related neurons. In the developing mouse cortex, lineage-related neurons are preferentially connected (Li et al., 2012). However, the fate specification of lineage-related neurons is still mostly unknown. On the other hand, current studies on neuron fate specification are limited mainly to identifying molecules essential for specifying different neuronal types/subtypes (Brown et al., 2001; Cepko, 2014; Hatakeyama et al., 2001). Unfortunately, single molecules, in most cases, are insufficient to specify multipotent neural stem/progenitor cells into specific neuron types/subtypes, which are highly demanding for regenerative medicine (Brzezinski et al., 2012; Hatakeyama et al., 2001; Powell and Jarman, 2008; Salie et al., 2005). It is urgent to clarify the influence of lineage context that neural stem/progenitor cells intrinsically provide on their neuronal production.
The vertebrate retina is a unique central nervous system (CNS) structure that has long been used to study vertebrate neuronal type/subtype diversification due to its well-characterized neuron types/subtypes and laminar organization (Connaughton et al., 2004; Masland, 2001). The retina comprises six major cell types, including retinal ganglion cells (RGCs), amacrine cells (ACs), bipolar cells (BCs), horizontal cells (HCs), photoreceptor cells (PRs), and Müller glial cells (MCs), with their cell bodies and processes located in specific layers (Fig. 1 A). This laminar location and cell morphology allow unambiguous identification of all major types. Moreover, the retina exhibits a vast diversity of cell subtypes (>60 classes; Masland, 2012). For instance, glutamatergic bipolar cells can be subdivided into ON and OFF subtypes, which initiate the light-ON and light-OFF responses, respectively (Connaughton, 2011; Shekhar et al., 2016). According to neurotransmitter phenotypes, amacrine cells are mainly classified as Gamma Aminobutyric Acid (GABA)-ergic and glycinergic (Balasubramanian and Gan, 2014; Cherry et al., 2009; Marc and Cameron, 2001; Menger et al., 1998).
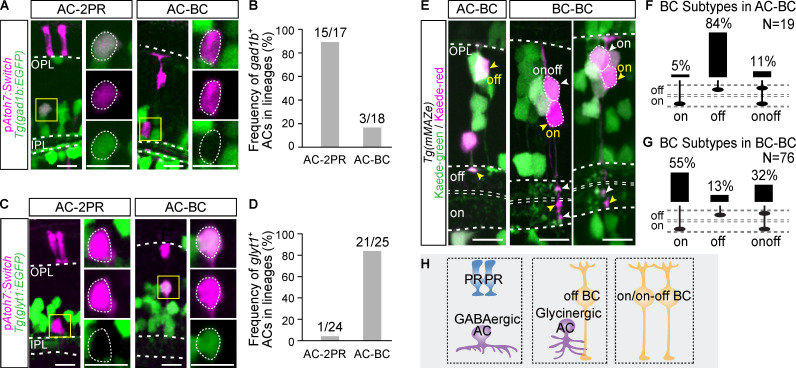
Figure 1.
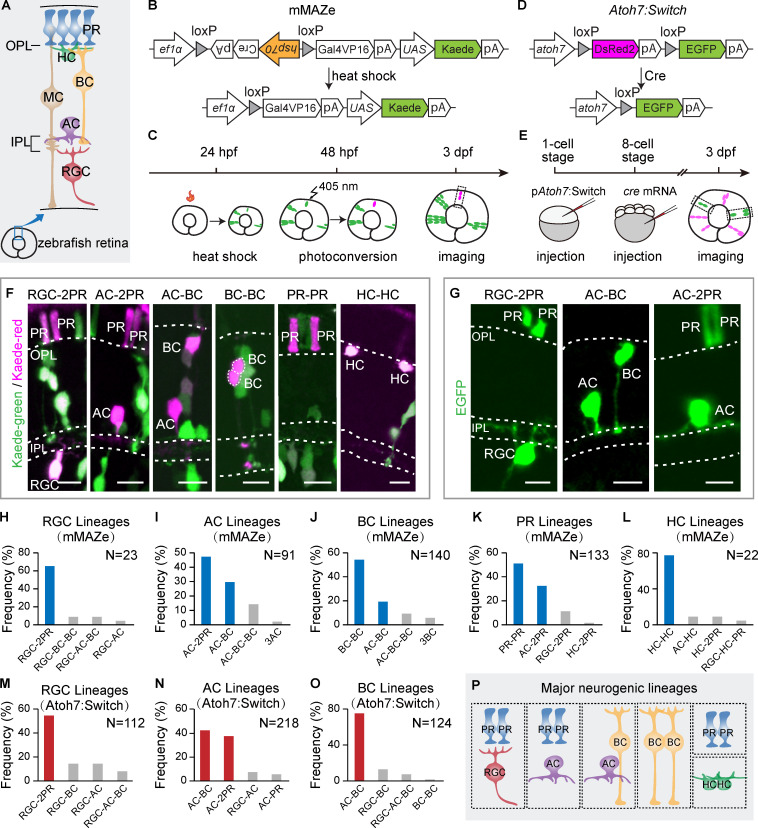
Major neurogenic lineages in the zebrafish retina. (A) Schematic of zebrafish retina structure. OPL, outer plexiform layer. IPL, inner plexiform layer. (B) Schematic of the mMAZe construct. (C) The working flow of lineage analysis of 48-hpf RPCs using mMAZe. (D) Schematic of the atoh7:Switch plasmid (pAtoh7:Switch). (E) The working flow of lineage analysis of atoh7+ PRCs using atoh7:Switch. (F) Representatives of major neurogenic lineages traced by mMAZe. (G) Representatives of major neurogenic lineages traced by atoh7:Switch. (H–L) The top four neurogenic lineages, which were analyzed using mMAZe and produce RGCs (H), ACs (I), BCs (J), PRs (K), or HCs (L) are listed. Major neurogenic lineages of each neuron type (frequency >15%) are highlighted in blue. (M–O) The top four lineages, which were analyzed using atoh7:Switch and produce RGCs (M), ACs (N), or BCs (O) are listed. Major neurogenic lineages of each neuron type (frequency >15%) are highlighted in red. (P) Summary graph of six major neurogenic lineages. Scale bars, 10 µm.
Early retinal progenitor cells (RPCs) can produce all retinal cell types (Turner and Cepko, 1987; Turner et al., 1990). As development progresses, late RPCs become neurogenic and produce lineages with biased types. In the retina, there appears to be substantial fate bias at the terminal division of RPCs (Cepko, 2014). Early studies showed that cone photoreceptors and horizontal cells are generated in homotypic pairs by dedicated precursors (Godinho et al., 2007; Rompani and Cepko, 2008; Suzuki et al., 2013). More interestingly, retinal major cell types can also be produced by heterotypic terminal lineages consisting of two different retinal types (Hafler et al., 2012; He et al., 2012; Turner et al., 1990). Consistently, earlier live imaging showed that single atoh7-expressing RPCs frequently produced one RGC and one progenitor that often produce a pair of PRs (He et al., 2012; Poggi et al., 2005). These findings leave the questions as to whether different retinal neuron subtypes also derive from specific lineages, and if so, whether these specific lineages are the results of dedicated progenitors.
Specific types of neurons are direct products of either asymmetric neurogenic divisions, giving rise to a progenitor and a differentiating neuron (PD division), or symmetric neurogenic divisions producing two differentiating neurons (DD division). Neurogenic RPCs are those undergoing PD or DD divisions, thereby producing neurogenic lineages. A rapid development of the zebrafish retina (∼60 h; Glass and Dahm, 2004) makes it possible for a systematic in vivo analysis of neurogenic lineages. Combining in vivo lineage tracing, single-cell RNA sequencing (scRNA-seq), and single-cell ATAC (Assay for Transposase Accessible Chromatin) sequencing (scATAC-seq), we found that retinal neuron types/subtypes were largely derived from stereotypic neurogenic lineages and revealed the underlying molecular mechanisms.
Results
Identification of six major neurogenic lineages for all retinal neuron types
For studying neurogenic lineages in the zebrafish retina, we developed two lineage tracing methods, termed “modified mosaic analysis in zebrafish” (mMAZe; Collins et al., 2010) and “atoh7:Switch,” and analyzed ∼1,000 lineages derived from single RPCs in vivo (Table S1 and Table S2).
In the mMAZe method, heat shock at 24 h postfertilization (hpf) allowed the Cre-dependent mosaic induction of kaede, a photoconvertible protein. Kaede was photoconverted from kaede-green to kaede-red to precisely label single RPCs at 48 hpf (Fig. 1, B and C), at which stage RPCs are mostly undergoing the final cell divisions. The resulting lineages were analyzed at 3 d postfertilization (dpf; Fig. 1 C), when embryonic development of the retina is mostly completed. Retinal cell types within lineages were then identified according to their cell body location and morphology (Fig. 1 A). We collected 511 lineages, among which 58% were two-cell or three-cell lineages (Fig. S1 B). We focused on two-cell and three-cell lineages because an earlier time-lapse study showed that 97% of neurogenic RPCs produce two or three neurons (He et al., 2012).
Figure S1.
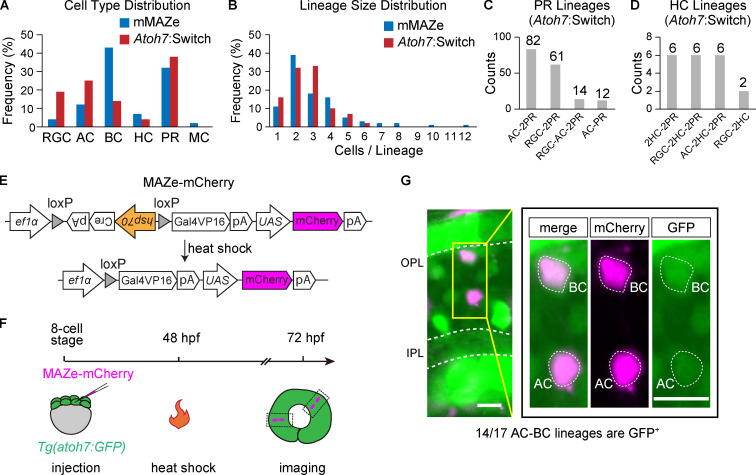
Characteristics of lineages traced by mMAZe and atoh7:Switch. (A) Cell type distribution of lineages traced by mMAZe (1,595 cells in total) and atoh7:Switch (1,302 cells in total). (B) Size (the number of cells per lineage) distribution of lineages traced by mMAZe (n = 511) and atoh7:Switch (n = 484). (C) Top4 lineages traced by atoh7:Switch which produce PR(s). (D) Top4 lineages traced by atoh7:Switch which produce HCs. (E–G) Atoh7 expression in AC-BC lineages. (E) Schematics of MAZe-mCherry. (F) Workflow to analyze atoh7 expression in AC-BC. Sparse mCherry expression was achieved by heat shock at 48 hpf. Spatially isolated AC-BC lineages were analyzed at 72 hpf. (G) Atoh7 expression (labeled by GFP) in AC-BC lineages. Image in yellow rectangle was zoomed in on the right panel. All images presented are in Z-stack. Scale bar, 10 µm.
Consistent with previous studies, PRs and HCs were mainly generated as homotypic pairs (PR-PR and HC-HC; Fig. 1, F, K, and L; He et al., 2012; Rompani and Cepko, 2008; Suzuki et al., 2013). Interestingly, we found that RGCs, ACs, and BCs were mainly derived from four major neurogenic lineages with stereotyped cell type composition (Fig. 1, F and H–J). RGC-2PR accounted for 65% of RGC-containing lineages (n = 15/23; Fig. 1, F and H). ACs, the major interneurons in retina, were predominantly born together with excitatory neurons. Specifically, AC-2PR and AC-BC lineages made up 47% (n = 43/91) and 30% (n = 27/91) of AC-containing lineages, respectively (Fig. 1, F and I). BC-BC and AC-BC lineages contributed to 54% (n = 76/140) and 19% (n = 27/140) of BC-containing lineages, respectively (Fig. 1, F and J). We also found a few cases of BC-MC lineages (n = 7; Table S1).
To better characterize neurogenic lineages producing early-born retinal types, including RGCs and ACs, we also developed atoh7:Switch. Atoh7, a basic helix-loop-helix (bHLH) transcription factor (TF), is among the first set of pro-neural TFs, and most RGCs and ACs are derived from atoh7-expressing RPCs (atoh7+ RPCs; Jusuf et al., 2011; Masai et al., 2000; Poggi et al., 2005). EGFP could sparsely mark individual atoh7-expressing RPCs through Cre-dependent stochastic removal of the loxP-DsRed2-loxP cassette, and the resulting lineages were analyzed at 3 dpf (Fig. 1, D and E). We analyzed in total 484 lineages derived from atoh7+ RPCs (atoh7+ lineages), among which 135 were two-cell lineages, and 181 were three-cell lineages (Fig. S1 B). In all lineages, no MC was detected (Fig. S1 A), consistent with the fact that MCs are atoh7− (Vitorino et al., 2009). In line with the results from mMAZe, RGC-2PR was the major neurogenic lineage of RGC production, while AC-BC and AC-2PR gave rise to the majority of ACs (Fig. 1, G, M, and N). Interestingly, atoh7+ RPCs were mostly biased toward AC-BC and produced few BC-BC (Fig. 1, G and O). Meanwhile, independent analysis showed that ∼80% AC-BC lineages were atoh7+ (Fig. S1, E–G). PRs and HCs were mainly born as homotypic pairs as sub-lineages of atoh7+ RPC (Fig. S1, C and D). Together, RGC-2PR, AC-2PR, AC-BC, BC-BC, HC-HC, and PR-PR were characterized as the major neurogenic lineages that produce all retinal neuronal types (Fig. 1 P).
AC and BC subtypes arise from distinct neurogenic lineages
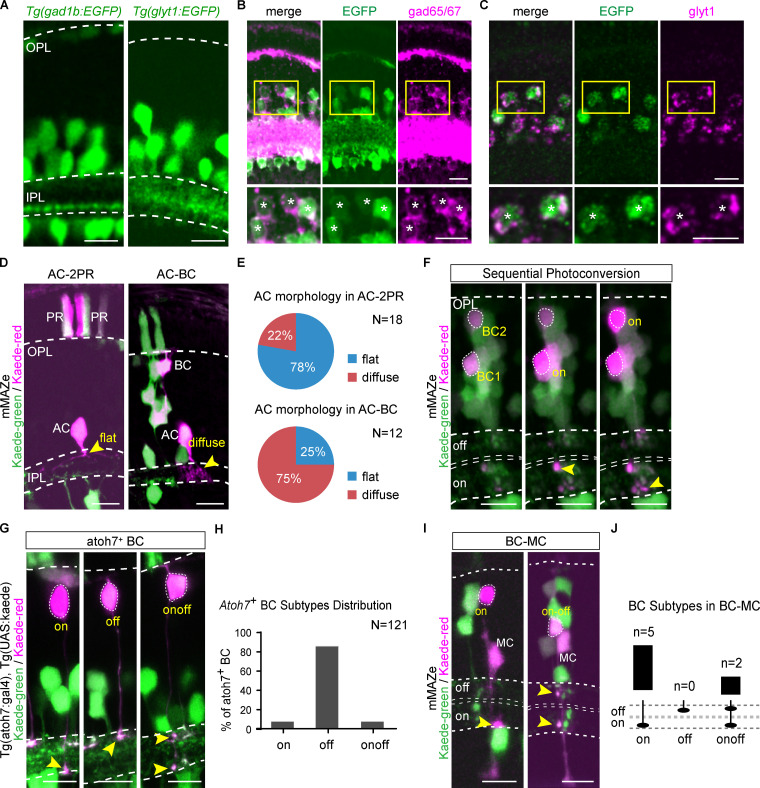
More than one major neurogenic lineages described above could produce the same retinal type, such as ACs from both AC-2PR and AC-BC lineages, and BCs from BC-BC and AC-BC lineages. This interesting observation raised the question as to whether the same neuron type produced by different lineages belongs to different subtypes. It is known that in the inner plexiform layer (IPL), GABAergic ACs are mainly mono-stratified, whereas most glycinergic ACs have diffuse terminals (Fig. S2 A; Menger et al., 1998). We found that in the clones derived from 48-hpf RPCs using mMAZe, 14/18 ACs produced in the AC-2PR lineage were mono-stratified, whereas 9/12 ACs within the AC-BC lineage had diffuse terminals (Fig. S2, D and E), suggesting that ACs in AC-BC and AC-2PR lineages were biased to be glycinergic and GABAergic, respectively. To identify AC subtypes directly, we generated two zebrafish lines, Tg(gad1b:EGFP) and Tg(glyT1:EGFP), which specifically label GABAergic and glycinergic ACs, respectively (Fig. S2, A–C). Next, we analyzed the atoh7+ lineages in both transgenic lines. We found that ACs within the AC-2PR lineage were predominantly GABAergic (15/17; Fig. 2, A–D). In contrast, ACs within the AC-BC lineages were mostly glycinergic (21/25; Fig. 2, A–D). Together, our results showed that ACs generated by AC-2PR and AC-BC lineages bias to two different neurotransmitter-specific subtypes (Fig. 2 H).
Figure S2.
AC and BC subtypes. (A) Signal of Tg(gad1b:EGFP) and Tg(glyt1:EGFP). (B) Verify Tg(gad1b:EGFP) by double immunostaining of EGFP and gad65/67. 142/144 EGFP+ ACs were gad65/67+. Lower panels show the zoom-in result, and the colabeled cells are indicated by asterisks. (C) Verify Tg(glyt1:EGFP) by RNA in situ hybridization of glyt1 with immunostaining of EGFP. 130/132 EGFP+ ACs were glyt1+. Lower panels show the zoom-in result, and the colabeled cells are indicated by asterisks. (D and E) AC morphology in AC-2PR and AC-BC lineages, which were traced by mMAZe (D) and distribution summary is shown in E. (F) An example illustrates sequential photoconversion. BC 1 was determined as ON-subtype after the first photoconversion, and then BC 2 was determined as ON-subtype after the second photoconversion. (G and H) Atoh7+ BC subtypes were determined by photoconverting kaede-green to kaede-red (G). The distribution summary is shown in H. (I and J) All BCs from BC-MC lineages were ON- or ONOFF-subtype (I), and the statistical results are shown in J. All images are presented in Z-stacks. Cell terminals are indicated by arrowheads. Scale bar, 10 µm.
Figure 2.
AC and BC subtypes in major neurogenic lineages. (A and B) Analysis of GABAergic ACs in AC-2PR and AC-BC lineages. Representative images are shown in A. Merged channels and separated channels of ACs in yellow rectangles are shown in the right panels. The statistical results are summarized in B. (C and D) Analysis of glycinergic ACs in AC-2PR and AC-BC lineages. Representative images are shown in C. Merged channels and separated channels of ACs in yellow rectangles are shown in the right panels. The statistic results are summarized in D. (E–G) Analysis of BC subtypes in AC-BC and BC-BC lineages traced by mMAZe. Representative images of BC subtypes in E, statistical results of BC subtypes in AC-BC (F), and BC-BC (G). (H) Summary graphic of AC and BC subtypes in different lineages. Scale bar, 10 µm.
We also examined BC subtypes (ON, OFF, and ONOFF) within the BC-BC and AC-BC lineages based on the location of axon terminals within IPL with the help of sequential photoconversion (Fig. S2 F). We found that, in the lineages derived from 48-hpf RPCs using mMAZe, 84% (n = 16/19) of BCs from AC-BC lineages were OFF subtype (Fig. 2, E and F), whereas 87% (n = 66/76) BCs from BC-BC lineages were ON or ONOFF subtype (Fig. 2, E and G). As expected, >80% of atoh7+ BCs were OFF subtype (Fig. S2, G and H), which was consistent with the result that most AC-BC lineages were atoh7+ (Fig. S1, E–G). Interestingly, all seven BCs from BC-MC lineages belong to ON or ONOFF subtype (Fig. S2, I and J). Thus, distinct BC subtypes were also derived from different lineages (Fig. 2 H).
Fate respecification of ACs is lineage-dependent after ptf1α knockout
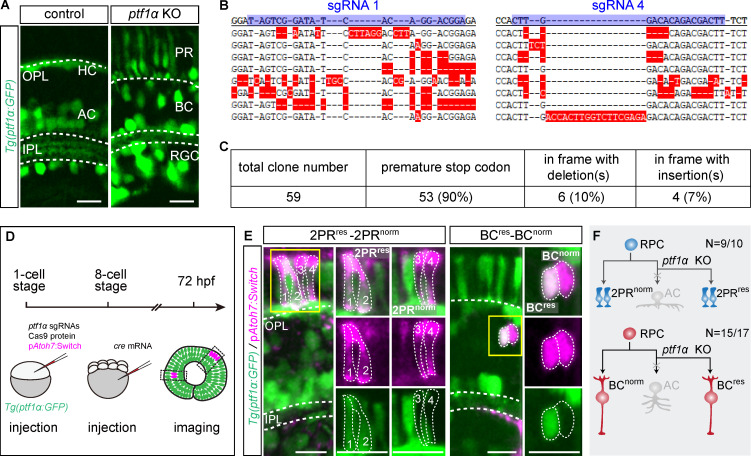
Pancreas-specific transcription factor 1α (ptf1α), a bHLH family member, is expressed exclusively in postmitotic ACs and HC precursors (Fujitani et al., 2006; Godinho et al., 2007). ACs and HCs are known to be respecified into glutamatergic neurons following ptf1α knockdown (Jusuf et al., 2011). We validated this finding by knocking out ptf1α through injecting a set of four CRISPR/Cas9 ribonucleoprotein complexes in the Tg(ptf1α:GFP) fish line, which destroyed ptf1α highly efficiently in G0 zebrafish (Fig. 3, B and C; Wu et al., 2018), and observed EGFP+ RGCs, BCs, and PRs, indicating the respecification of ACs or HCs into glutamatergic neurons (Fig. 3 A). To further determine how ACs are respecified within the AC-2PR and AC-BC lineages, we analyzed these two lineages in Tg(ptf1α:GFP) after ptf1α knockout (Fig. 3 D). Surprisingly, we found that the vast majority of ACs in the AC-2PR lineages were respecified into two PRs (n = 9/10), resulting in new lineages containing two normal PRs and two respecified PRs (Fig. 3, E and F). In contrast, ACs in the AC-BC lineages were predominantly respecified into BCs (n = 15/17), resulting in new lineages consisting of one normal BC and one respecified BC (Fig. 3, E and F). These results demonstrated that ACs of AC-2PR and AC-BC lineages were respecified into their sister cell types when ptf1α was depleted, suggesting that fates of sister neurons are intrinsically constrained in mother cells before the last division, pointing to the existence of lineage-specific neurogenic RPCs.
Figure 3.
AC respecification in AC-2PR and AC-BC lineages after ptf1α knockout (KO). (A) Signal of Tg(ptf1α:GFP) in control and after knocking out ptf1α. (B and C) Verification of ptf1α knockout efficiency through injecting four CRISPR/Cas9 ribonucleoprotein complexes in G0 zebrafish. (B) Examples of eight alleles around targeted sites of sgRNA 1 and sgRNA 4. The first line is the WT sequence, sgRNA targeted sites are highlighted in blue, and mismatches are highlighted in red. (C) Summary of characteristics of 59 ptf1α alleles analyzed. (D) The workflow to analyze AC respecification. (E) 9/10 ACs in AC-2PR lineages were respecified as two PRs, and the resulting lineages were 2PRres-2PRnorm (left panel). 15/17 ACs in AC-BC lineages were respecified as single BC, and the resulting lineages were BCres-BCnorm (right panel). Zoom-in images within yellow rectangles are shown in the right panels. (F) The summary graphic illustrates the AC respecification in AC-2PR and AC-BC lineages after ptf1α knockout. res, respecified; norm, normal. Scale bar, 10 µm.
Defining RPC heterogeneity by single-cell RNA sequencing
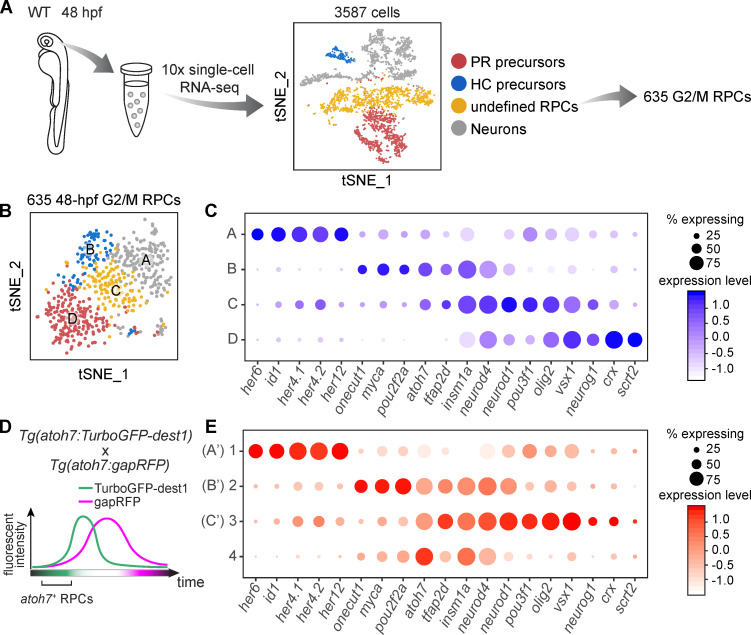
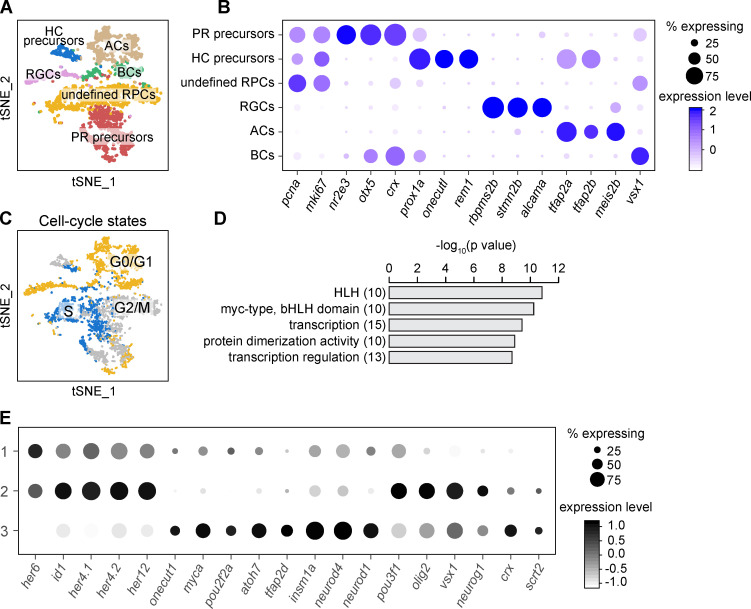
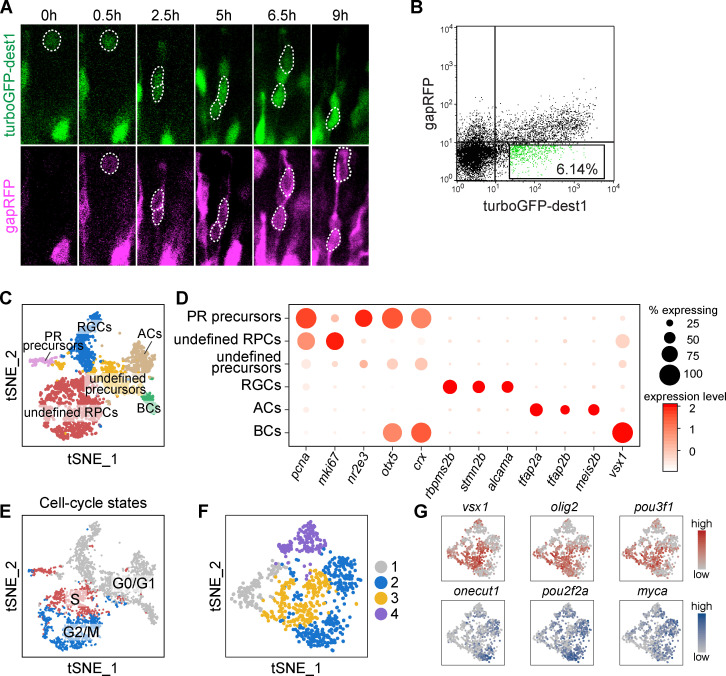
To search for lineage-specific RPCs, we analyzed the data of scRNA-seq of 48-hpf RPCs (Xu et al., 2020; Fig. 4 A). Besides the identification of precursors of PRs and HCs according their putative markers (Fig. 4 A; and Fig. S3, A and B), we further analyzed the rest of the progenitors (termed as “undefined RPCs”) by clustering (Fig. 4 A; and Fig. S3, A and B). To minimize cell cycle influence (Fig. S3 C and Fig. S4 E), we analyzed undefined RPCs in G2/M phase, which were aggregated into four clusters (Clusters A–D; Fig. 4 B). GO analysis of their top featured genes showed enrichment of TFs (Fig. S3 D). Cluster A expressed TFs specific to early RPCs at proliferative stage, such as her6, id1, and her12 (Fig. 4 C; Bai et al., 2007; Ohtsuka et al., 2001; Scholpp et al., 2009). The other three clusters (Clusters B, C, and D) expressed TFs related to neurogenesis. RPCs of Cluster D were atoh7− with the high expression of crx and scrt2, while Clusters B and C were atoh7+ (Fig. 4 C). Cluster B cells specifically expressed onecut1, myca, and pou2f2a, whereas Cluster C cells shared the featured TFs with Cluster B (atoh7, tfap2d) and Cluster D (pou3f1, olig2, vsx1, neurog1; Fig. 4 C). Together, we identified five clusters of neurogenic RPCs (PR precursors, HC precursors, and Clusters B, C, and D).
Figure 4.
Defining RPC heterogeneity by scRNA-seq. (A) The workflow of scRNA-seq of 48-hpf retina and primary data analysis. (B) Four clusters of undefined RPCs in G2/M phase in A. (C) Top feature TFs of Clusters A–D in B. The size of each circle is the percentage of cells expressing the marker in each cluster, and its intensity represents the scaled expression level. (D) A schematic showing the experimental design to enrich atoh7+ RPCs. (E) Gene expression pattern of atoh7+ RPCs. Genes listed are featured TFs of 48-hpf RPCs in C. Clusters 1–3 show similar expression patterns as Clusters A–C of 48-hpf RPCs and are labeled as A’, B’, and C’.
Figure S3.
Single-cell RNA sequencing of 48-hpf retina. (A) The t-SNE plot of the 3,587 qualified cells of zebrafish retina at 48 hpf and the annotation. (B) Featured genes of each cell cluster in A. (C) Cell-cycle distribution of cells in A. Cells during the same cell-cycle phase tend to cluster together. (D) GO analysis of top featured genes of 48-hpf RPCs showed the enrichment of transcription factors. HLH, helix-loop-helix. (E) Gene expression pattern of the three populations of 48-hpf early RPCs. Genes listed are the featured TFs of 48-hpf RPCs in (Fig. 4 C).
Figure S4.
Single-cell RNA sequencing of atoh7+ RPCs. (A) Time-lapse result of Tg(atoh7:turboGFP-dest1::atoh7:gapRFP) retina. TurboGFP-dest1 signal appears and disappears earlier than that of gapRFP. (B) FACS analysis of retinal cells from the Tg(atoh7:turboGFP-dest1::atoh7:gapRFP) embryos. The GFP+RFP− cells for scRNA-seq are highlighted. (C) The t-SNE plot showing the clustering result of GFP+RFP− cells in B and the annotation. (D) Featured genes of each cell cluster in C. (E) Cell-cycle distribution of cells in C. Cells during the same cell cycle phase tend to cluster together. (F) Four clusters of atoh7+ G2/M RPCs. (G) Expression patterns of cluster 2–specific (onecut1, pou2f2a, myca) and cluster 3–specific (vsx1, olig2, pou3f1) TFs of cells in F.
In addition, we performed further analysis on early RPCs (Cluster A; Fig. 4 C). They could be divided into three clusters (Fig. S3 E). Clusters 1 and 2 highly expressed early markers (e.g., her6 and her12). Interestingly, they also weakly expressed different neurogenesis-related TFs, suggesting that different early RPCs might generate distinct neurogenic RPCs. On the other hand, we also observed that some large-size lineages (more than three cells per lineage) occurred frequently in the lineages traced by atoh7:Switch and mMAZe, such as 4BCs, RGC-AC-BC-2PR, and RGC-2AC-BC-2PR (Table S1 and Table S2), suggesting that lineage-specific progenitors might exist in earlier RPCs. The link between early RPCs and neurogenic lineages needs future investigation.
We also performed scRNA-seq analysis of enriched atoh7+ RPCs. To efficiently label and isolate atoh7+ RPCs, the conventional transgenic reporter line Tg(atoh7:gapRFP) is limited due to the stability of fluorescent protein, in which most labeled cells are neurons rather than progenitor cells. We therefore constructed Tg(atoh7:turboGFP-dest1), in which turboGFP-dest1 exhibits the fast protein maturation and degradation (Fig. 4 D and Fig. S4 A; Evdokimov et al., 2006; Li et al., 1998). In the Tg(atoh7:TurboGFP-dest1::atoh7:gapRFP) retina, GFP+RFP− cells, which are enriched with atoh7+ RPCs, were collected for scRNA-seq (Fig. S4 B). Besides PR precursors (Fig. S4, C and D), atoh7+ RPCs were aggregated into four clusters (Clusters 1–4; Fig. S4 F). Consistently, Clusters 1–3 showed very similar featured TFs as Clusters A–C of 48-hpf RPCs (Fig. 4, C and E). Cells in Cluster 4 had no specific highly expressed genes while weakly expressing the featured TFs of Clusters 2 and 3 (Fig. 4 E and Fig. S4, F and G), suggesting Cluster 4 may represent a transitional state. Thus, the independent scRNA-seq analysis validated two clusters of atoh7+ neurogenic RPCs.
TF-defined RPCs generate distinct neurogenic lineages
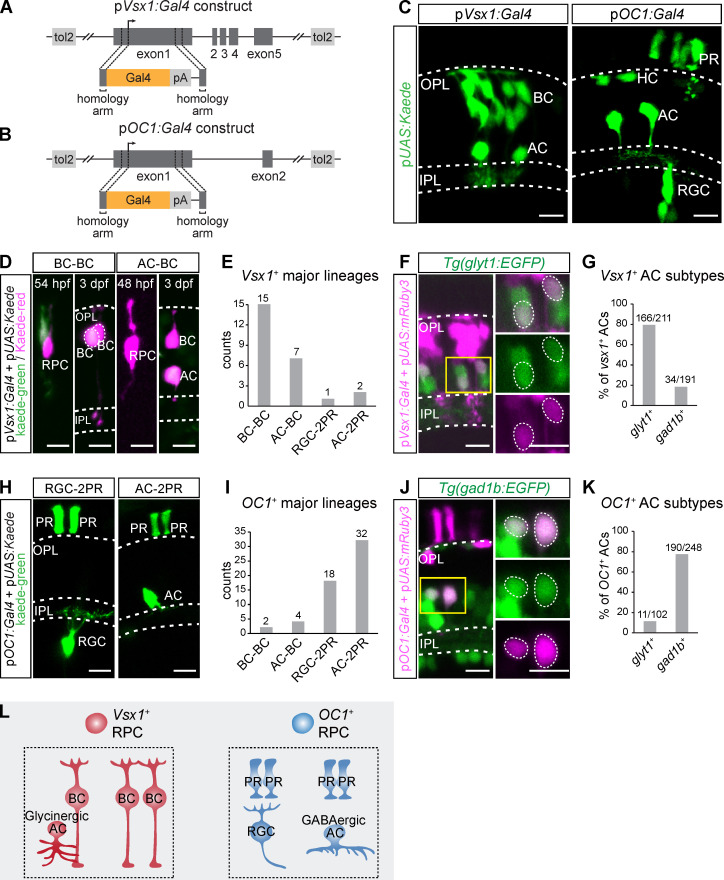
Since we have known that HCs and PRs are generated by dedicated progenitor cells, we next examined whether the major neurogenic lineages of RGC-2PR, AC-2PR, AC-BC, and BC-BC were produced by intrinsically different RPCs. We traced the lineages derived from vsx1-expressing RPCs (Clusters C and D of 48-hpf RPCs) and onecut1 (OC1)-expressing RPCs (Cluster B of 48-hpf RPCs; Fig. 4 B). We created Bacteria Artificial Chromosome (BAC) plasmids of vsx1:Gal4 and OC1:Gal4 (Fig. 5, A and B), which allowed the labeling of vsx1- or OC1-expressing RPCs with kaede (UAS:kaede).
Figure 5.
Lineage tracing of vsx1+ and OC1+ RPCs. (A and B) Schematics of vsx1:Gal4 (A) and OC1:Gal4 (B) BAC constructs. (C) Vsx1+ cells and OC1+ cells in the retina at 3 dpf. (D and E) Lineage tracing of vsx1+ RPCs by the photoconversion of kaede-green to kaede-red at 48–54 hpf. Representative images are shown in D. The statistical result is shown in E. (F and G) Representative images (F) and the statistical result (G) show vsx1+ AC subtypes. (H and I) Lineage tracing of OC1+ RPCs by collecting spatially isolated lineages. Representative images are shown in H. The statistical result is shown in I. (J and K) Representative images (J) and the statistical result (K) show OC1+ AC subtypes. (L) A summary graphic illustrates lineage bias of vsx1+ and OC1+ PRCs. Scale bar, 10 µm.
We found that vsx1-expressing RPCs predominantly generate ACs and BCs (Fig. 5 C). The photoconversion of individual vsx1-expressing RPCs around 48 hpf enabled the single-cell lineage tracing (Fig. 5 D), and found vsx1-expressing RPCs produced far more BC-BC (n = 15) and AC-BC (n = 7) lineages than RGC-2PR (n = 1) and AC-2PR (n = 2) lineages (Fig. 5, D and E). Moreover, nearly 80% of vsx1+ ACs were glycinergic (Fig. 5, F and G), consistent with the earlier result that ACs in AC-BC were glycinergic-biased (Fig. 2, A–D). Note that vsx1-expressing RPCs included atoh7− (Cluster D of 48-hpf RPCs) and atoh7+ clusters (Cluster C of 48-hpf RPCs; Fig. 4 B). We have shown that BC-BC lineages were atoh7− lineages while AC-BC lineages were atoh7+ (Fig. 1, J and O). Thus, BC-BC lineages were likely derived from vsx1+atoh7− RPCs, while AC-BC lineages were derived from vsx1+atoh7+ RPCs.
However, OC1-expressing RPCs preferentially produce RGCs, ACs, HCs, and PRs (Fig. 5 C). We performed the lineage tracing of OC1+ RPCs by analyzing spatially well-isolated clones (Fig. 5 H), and found AC-2PR (n = 32) and RGC-2PR (n = 18) lineages were much more abundant than AC-BC (n = 4) and BC-BC (n = 2) lineages (Fig. 5, H and I). Consistently, >80% of OC1+ ACs were GABAergic (Fig. 5, J and K).
In conclusion, transcript-defined RPCs produced distinct sets of major neurogenic lineages, that is, vsx1+ RPCs bias toward BC-BC and AC-BC lineages, whereas OC1+ RPCs bias toward RGC-2PR and AC-2PR lineages (Fig. 5 L).
A developmental landscape of chromatin accessibility of lineage-specific TFs
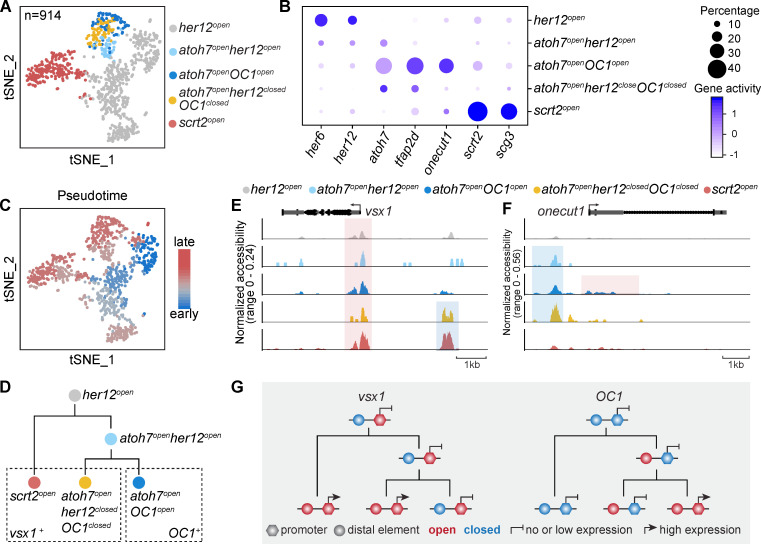
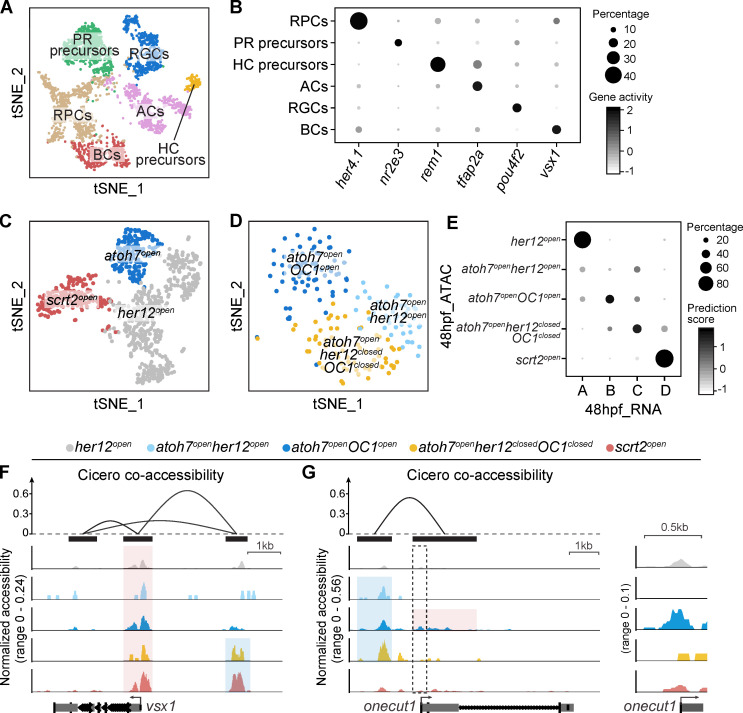
To gain more insights into the developmental establishment of lineage-specific neurogenic RPCs at the chromatin level, we obtained chromatin accessibility profiles of 4,058 qualified 48-hpf retinal cells by scATAC-seq. We performed clustering analysis on these cells and quantified gene activities in each cell by assessing chromatin accessibility (Stuart et al., 2019; see Materials and methods). Among these clusters, we identified five RPC populations, including her12open, scrt2open, atoh7openher12open, atoh7openOC1open, and atoh7openher12closedOC1closed (Fig. 6 A). Interestingly, their gene activity pattern was consistent with the gene expression pattern obtained by scRNA-seq (Fig. 4 C and Fig. 6 B). Further integration analysis also showed the strong correlation among these five RPC populations of scATAC-seq data and Clusters A–D of 48-hpf RPCs in the scRNA-seq data (Fig. S5 E). Note that scrt2open and atoh7openher12closedOC1closed represented vsx1+ RPCs producing AC-BC and BC-BC lineages, while atoh7openOC1open represented OC1+ RPCs producing RGC-2PR and AC-2PR lineages (Fig. 5). The pseudo-time analysis further showed the developmental trajectory of these five RPC populations (Fig. 6 C). Specifically, her12open were RPCs at the earliest stage and gave rise to scrt2open and atoh7openher12open, which in turn developed into atoh7openOC1open and atoh7openher12closedOC1closed (Fig. 6 D).
Figure 6.
Different chromatin accessibility dynamics of vsx1 and OC1 in 48-hpf RPCs. (A) A t-SNE plot of 48-hpf RPCs based on their chromatin accessibility. Clusters are annotated by their specific chromatin accessibility. (B) A dot plot of gene activity of the clusters in A. (C) A t-SNE plot of 48-hpf RPCs based on their chromatin accessibility. Cells are colored by their pseudo-time. (D) A schematic illustrates the developmental trajectory and lineage markers of 48-hpf RPCs. (E and F) Coverage plots of proximal (highlighted in red) and distal (highlighted in blue) elements of vsx1 (E) and OC1 (F) in different clusters of 48-hpf RPCs. (G) A summary schematic shows the different chromatin accessibility dynamics of vsx1 and OC1 in 48-hpf RPCs.
Figure S5.
ScATAC-seq of 48-hpf retina. (A and B) The clustering result of 48-hpf retinal cells. Clusters are shown in a t-SNE plot (A) and annotated by their specific gene activities. Gene activity patterns of all clusters are shown in a dot plot (B). (C and D) Clustering results of all RPCs (C) and atoh7open RPCs (D). (E) Correlation among the five RPC populations of 48-hpf scATAC-seq data (Fig. 6 A) and Clusters A–D of 48-hpf scRNA-seq data (Fig. 4 B) with the minimal prediction scores of 0.3. (F and G) Cicero co-accessibility among elements surrounding vsx1 (F, top) and OC1 (G, top). Peak positions are marked by black bars. The proximal opening (highlighted in red) and distal opening (highlighted in blue) in the 48-hpf RPCs are shown in the coverage plots. The proximal element of OC1 (marked by the dashed box) is magnified (G, right).
Next, we examined chromatin accessibility of lineage-specific TFs, including vsx1 and OC1, in these five ATAC-based RPC populations. Notably, vsx1 and OC1 had different characteristic dynamics of their chromatin accessibility. The promoter region (around transcription start site) of vsx1 was open in all RPCs, while its distal element exhibited specific accessibility in vsx1+ RPCs (atoh7openher12closedOC1closed and scrt2open RPCs; Fig. 6 E), suggesting that chromatin accessibility of the distal element rather than the promoter region accounts for lineage-specific vsx1 expression. On the contrary, the promoter region of OC1 became accessible specifically in OC1+ RPCs (Atoh7openOC1open), while its distal elements of OC1 were accessible in all three atoh7openRPCs (Fig. 6 F and Fig. S5 G). These results suggest that the distal regulatory element of OC1 is primed earlier than the lineage-specific opening of its promoters. It is exciting that the role of this priming of distal elements may be clarified in the future.
Together, we demonstrated the characteristics of chromatin accessibility dynamics of lineage-specific TFs (Fig. 6 G).
Single TFs specify different retinal neuron types/subtypes in a stereotyped lineage-dependent manner
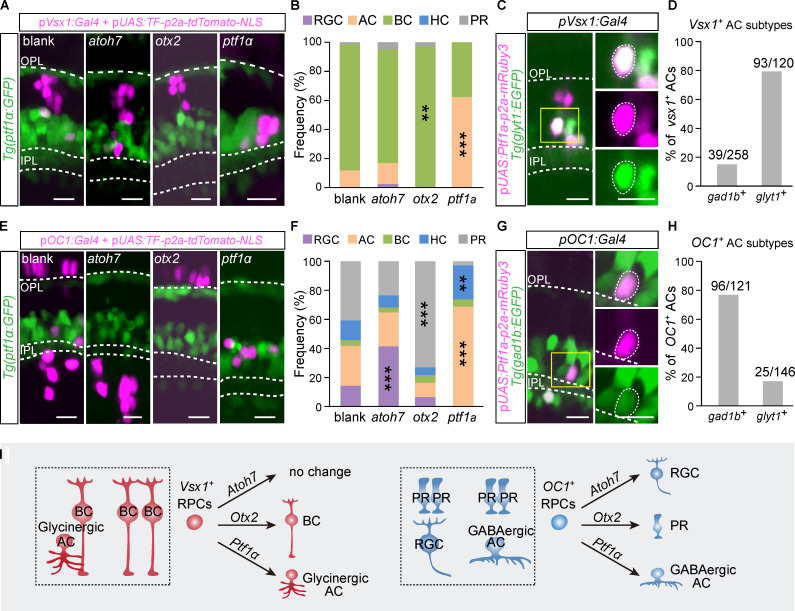
Previous studies have shown the essential roles of TFs in neuronal fate specification. For instance, atoh7 is required for specifying RGCs (Kay et al., 2001; Wang et al., 2001), whereas otx2 and ptf1α are necessary for specifying BCs/PRs (Koike et al., 2007; Li et al., 2015; Nishida et al., 2003) and ACs/HCs (Fujitani et al., 2006; Jusuf and Harris, 2009; Nakhai et al., 2007), respectively. However, these TFs are not sufficient to commit all RPCs into single neuron types, which is likely due to the RPC heterogeneity. Since lineage-specific RPCs represent relatively homogenous progenitors, we then overexpressed individual TFs in these lineage-specific RPCs and examined their fate outputs. Using the Gal4/Upstream Activation Sequence (UAS) system, we overexpressed individual TFs in vsx1+ or OC1+ RPCs using vsx1 or OC1 promoters and reported by nucleus-localized tdTomato (tdTomato-NLS). We performed the overexpression in the Tg(ptf1a: GFP) transgenic line to facilitate the identification of retinal cell types according to their relative positions related to ACs and HCs.
The overexpression of atoh7 in OC1+ RPCs led to a significant increase in RGC production (Fig. 7, E and F), whereas the overexpression in vsx1+ RPCs did not affect RGC production (Fig. 7, A and B). Thus, overexpression of a common TF could promote the generation of a neuronal type in one lineage but not in another. Furthermore, otx2 overexpression led to the induction of different neuronal types in vsx1+ and OC1+ RPCs. Specifically, vsx1+ RPCs mainly (∼97%) produced BCs (Fig. 7, A and B), whereas OC1+ RPCs mostly (∼73%) generated PRs (Fig. 7, E and F). It indicates that the overexpression of a common TF could promote the production of distinct neuronal types in different lineages. Finally, when we overexpressed ptf1α, vsx1+ RPCs mostly produced glycinergic ACs (79%; Fig. 7, A–D), whereas OC1+ RPCs mainly generated GABAergic ACs (78%; Fig. 7, E–H). Thus, the overexpression of a common TF could promote the production of different neuronal subtypes in different lineages.
Figure 7.
Overexpression of atoh7, otx2, and ptf1α in vsx1+ and OC1+ RPCs. (A) Representative images of vsx1+ cells (labeled by tdTomato-NLS) after overexpressing atoh7, otx2, and ptf1α in vsx1+ RPCs with no TF overexpression as the blank. Tg(ptf1α:GFP) was used to indicate cell body location. (B) Cell type distribution of vsx1+ cells in A. The significant increase of each neuron type compared with blank is shown. (C and D) Vsx1+ AC subtypes after ptf1α overexpression. Representative image of glyT1+ ACs is shown in C. Zoom-in images in the yellow rectangle are shown on the right side. Statistic distribution of vsx1+ AC subtypes is shown in D. (E) Representative images of OC1+ cells (labeled by tdTomato-NLS) after overexpressing atoh7, otx2, and ptf1α in OC1+ RPCs with no TF overexpression in the blank. (F) Cell type distribution of OC1+ cells in E. The significant increase of each neuron type compared with blank is shown. (G and H) OC1+ AC subtypes after ptf1α overexpression. The image of a gad1b+ AC is shown in G. Zoom-in images in the yellow rectangle are shown on the right side. Statistical distribution of OC1+ AC subtypes is shown in H. (I) A summary graph. Scale bar, 10 µm. **, P < 0.001; ***, P < 0.0001 (Fisher's exact test).
Taken together, the overexpression of single TFs could reprogram transcript-defined RPCs into single neuronal types or subtypes in a manner that depends on their lineage context (Fig. 7 I).
Discussion
Over the past three decades, increasingly sophisticated clonal analyses allowed us to appreciate the high variability of cell lineages derived from single RPCs during early neurogenic stages (Holt et al., 1988; Turner et al., 1990). In this study, we focused on a specific population of RPCs at a later stage that will undergo PD or DD division to give rise to neurons directly. Using newly developed in vivo single-cell lineage tracing methods, we discovered six major neurogenic lineages, each generating stereotypic retinal cell types and subtypes. Then we performed scRNA-seq analysis of neurogenic RPCs and revealed RPC heterogeneity. More importantly, different transcript-defined RPCs bias to generate distinct major neurogenic lineages, indicating the existence of lineage-specific neurogenic RPCs in the developing retina. Interestingly, three well-known pro-neurogenic TFs, when overexpressed in lineage-specific RPCs, could markedly bias the production of neuronal types or subtypes in a lineage-dependent manner.
Developmental emergence of heterogeneity in RPCs
Our study showed that neurogenic RPCs undergoing PD and DD divisions predominantly gave rise to small clones (mainly two- or three-cell clones), which exhibited an obvious bias in neuron types (Fig. 1 and Fig. 2). Combining scRNA-seq, in vivo lineage tracing, and genetic manipulations, we identified transcript-defined RPCs (such as atoh7, OC1, and vsx1) that are destined to generate biased neurogenic lineages. In addition, clustering analysis of 48-hpf early RPCs showed their heterogeneity in terms of the expression of pro-neural TFs, suggesting that the emergence of lineage-specific RPCs might be earlier (Fig. S3 E).
Our findings raised the interesting question as to how such transcript-defined RPCs arise. Previous studies found that a stochastic model incorporating stochastic expression of TFs could well explain clonal cell-type variability in zebrafish (Boije et al., 2015). Stochastic fate choices can act through either intrinsic or environmental mechanisms. Gene oscillation is one mechanism to create stochastic gene expression. For instance, many genes express in an oscillation manner in neural stem cells, such as hes1, ascl1, olig2, Ngn2, and Dll1 (Imayoshi et al., 2015; Shimojo et al., 2008). At any given point, cells within a population are staying at different expression phases and respond differently even to the same stimulus (Kobayashi et al., 2009). Alternatively, RPCs follow stochastic migration trajectories via interkinetic nuclear migration in the retinal neuro-epithelium, where an apical-basal gradient of Delta, a ligand for the Notch activation, is present (Del Bene et al., 2008). As a result, different levels of Notch exposure may result in stochastic fate choices of individual RPCs. Future mechanistic study on the stochastic transcription of TFs might yield insight into the emergence of transcript-defined RPCs and provide an integrated picture by bridging the gap between stochastic and deterministic cell fate regulation in the vertebrate CNS.
Lineage-specific RPCs generate distinct retinal neuron types
Using a systematic in vivo lineage analysis, we identified a set of six major neurogenic lineages that are responsible for the production of all retinal cell types (Fig. 1). Interestingly, different subtypes of ACs and BCs were generated by different RPCs, as GABAergic ACs and glycinergic ACs derived from OC1+ and vsx1+ RPCs, respectively, while OFF and ON BCs were segregated in atoh7+ and atoh7− lineages (Fig. 2). Although we have identified the lineage-dependent generation of AC and BC subtypes, we did not obtain direct evidence for more finely subdivided subtypes, such as different subtypes of OFF-subtype BCs (s1-s3). Local environmental cues and neuron–neuron interaction during the formation of IPL may be important players. Single-cell sequencing and lineage analysis of more defined RPC subpopulations may reveal to what extent lineage-dependent and -independent mechanisms contribute to neuronal subtype specification, and a more complete picture of the specification of diverse neuronal diversity in the zebrafish retina.
However, we have not excluded the existence of additional major neurogenic lineages, such as AC-AC, which has been shown to be one of the major lineages for AC production (He et al., 2012), but was absent in our study using the atoh7:Switch labeling method. This discrepancy could be due to the low transcriptional activity of the atoh7 promoter in the RPCs that give rise to AC-AC. Future studies are needed to examine the subtype of ACs in the AC-AC lineages.
Whether similar lineage-specific progenitors exist in developing mammalian cortex is still controversial. Through the lineage tracing using MADM labeling, cortical progenitors marked at the early neurogenic stages were found to produce both deep and superficial layer neurons but were seldom restricted to specific neuron types (Gao et al., 2014). Cux2+ cortical progenitors can be intrinsically specified into only upper-layer neurons (Franco et al., 2012), suggesting the presence of lineage-specific cortical progenitors. On the other hand, contradictory results were also reported (Guo et al., 2013; Eckler et al., 2015). Recent studies, however, favor the possibility of the coexistence of progenitors with or without lineage restriction (Garcia-Moreno and Molnar, 2015; Llorca et al., 2019). Using combined approaches, cortical progenitors marked at the onset of the neurogenesis can generate translaminar (∼80%), deep layer–restricted (∼10%), and superficial layer–restricted (∼10%) lineages in the developing mouse neocortex (Llorca et al., 2019). In the future, more systematic single-cell transcriptome and lineage analyses of neural progenitors are needed for a better characterization of lineage-specific progenitors in the developing mammalian cortex.
Temporal generation of different neuron types
In the developing vertebrate retina, distinct retinal types occur in a temporally sequential but overlapping manner (Cepko, 2014). Our analyses revealed the lineage-specific progenitors, providing new insights into this temporal generation of different neurons. Interestingly, our data showed that OC1-expressing RPCs produced RGCs, GABAergic ACs, which are early-born. On the contrary, vsx1-expressing RPCs gave rise to glycinergic ACs and BCs, which are the late-born neurons. One possibility is the earlier occurrence of OC1-expressing RPCs than vsx1-expressing RPCs. Alternatively, onecut1-expressing RPCs exhibit shorter cell-cycle lengths than vsx1-expressing RPCs. To distinguish the two possibilities, future analysis is required. Meanwhile, within three-cell lineages (RGC-2PR and AC2-PR), we observed the sequential generation of RGC/AC and PRs. Thus, within the stereotyped lineages derived from lineage-specific RPCs, the generation of distinct retinal cell types conforms to the conserved temporal order we observed at the population level.
Functional significance of clonally related neurons within stereotyped lineage
To what extent the lineage origin of neurons contributes to the neural circuit assembly remains an outstanding question under dispute in recent years. In the developing mouse cortex, clonally related cortical neurons are preferentially connected (Yu et al., 2009). Even long-range connections among clonally related neurons were found in frontal and sensory cortices (Ren et al., 2019). However, no preference for the connection among clonally related hippocampus neurons was found (Xu et al., 2014). In the retina, light signals are channeled into ON and OFF parallel circuits, which begin with ON and OFF BCs, respectively (Demb and Singer, 2015; Popova, 2014). We found distinct major neurogenic lineages gave rise to ON and OFF BCs, which suggests a potential lineage basis of the development of ON and OFF circuits. Are glycinergic ACs connected with OFF BCs within AC-BC? If so, how do they integrate with other neurons to assemble a complete OFF circuit? Further experiments addressing these questions will provide insights into the relationship between the lineage origins of neurons and the formation of retina microcircuits.
Lineage-dependent respecification of retinal neurons and its implication
Reprogramming of neural progenitor cells into specific neuron types represents a new approach in regenerative medicine. A significant challenge in neuronal reprogramming is to efficiently generate the unique cell type that is needed. One reason that leads to the challenge is only a small part of progenitors respond to the TF overexpression. For instance, atoh7 promoted the generation of RGC in OC1+ RPCs but did not affect vsx1+ RPCs. Another reason is that an individual TF could specify more than one neuron type, such as ptf1α for GABAergic ACs, glycinergic ACs and HCs, as well as otx2 for PRs and BCs (Fujitani et al., 2006; Jusuf et al., 2011; Koike et al., 2007; Nishida et al., 2003). With the identification of lineage-specific RPCs, we were able to get unique retinal types efficiently through in vivo cell reprogramming by the overexpression of single TFs. Thus, we have demonstrated the possibility of lineage context–dependent production of specific neuron types. It would be of interest to determine whether the lineage-dependent cell reprogramming could also occur in the mammalian CNS as well as neural progenitor cells derived from human stem cells, thus providing the cell resource of high purity for cell therapy.
Materials and methods
Zebrafish husbandry
Zebrafish lines were maintained and bred at 27°C on 14-h-light/10-h-dark cycles. Zebrafish embryos were obtained from natural spawning of fish lines and raised in embryo medium (NaCl 5.03 mM, KCl 0.17 mM, CaCl2 • 2H2O 0.33 mM, MgSO4 • 7H2O 0.33 mM, and methylene blue 0.0002% [wt/vol]) at 28.5°C. The embryos were staged by hpf before 72 hpf as previously described (Kimmel et al., 1995) and by dpf after that. Embryos used for imaging were treated with 0.003% phenylthiourea (Sigma-Aldrich, P7629) from 12 hpf to avoid pigmentation. Zebrafish sex cannot be determined until 25 dpf (Takahashi, 1977), so the sex of the experimental animals was unknown. All animal procedures performed in this study were approved by the Animal Use Committee of the Institute of Neuroscience, Chinese Academy of Sciences (NA-045-2019).
Zebrafish transgenic lines
The following published transgenic lines were used: Tg(UAS:kaede) (Scott and Baier, 2009), Tg(ptf1α:GFP) (Godinho et al., 2005), Tg(atoh7:gal4) (Maddison et al., 2009), and Tg(atoh7:GFP) (Masai et al., 2003). The following transgenic lines were generated in this study: Tg(gad1b:EGFP), Tg(glyt1:EGFP), Tg(mMAZe), and Tg(atoh7:turboGFP-dest1). They were created by co-injecting 10 ng/µl plasmids with 50 ng/µl tol2 mRNA into WT embryos at the one-cell stage. Injected embryos were screened at 3–5 dpf for positive founder. Founders with germline transmission were crossed with WT fish to generate stable transgenic lines.
Plasmid construction
BAC plasmids
atoh7:Switch, gad1b:EGFP, glyt1:EGFP, vsx1:gal4, and OC1:gal4 were generated according to the previous protocol (Suster et al., 2011). BAC plasmids containing genes of interest were ordered from commercial companies (Table 1).
Table 1. Original BAC plasmids information.
| Gene | BAC ID | Company |
|---|---|---|
| atoh7 | DKEY-51A16 | Source BioScience |
| gad1b | DKEY-251E16 | |
| slc6a9(glyt1) | CH211-261P7 | BACPAC Resources Center |
| vsx1 | CH211-67N1 | |
| OC1 | CH211-277D2 |
These original BAC plasmids were first electroporated into the SW105 bacteria strain. The iTol2 cassette with 50-bp homologies on each end targeting the BAC backbone was amplified by PCR and inserted into the BAC plasmids via recombineering in SW105. Next, the cassettes including reporter genes (loxP-DsRed2-loxP-EGFP for atoh7:Switch, EGFP for gad1b:EGFP and glyt1:EGFP, Gal4 for vsx1:gal4 and OC1:gal4) and an kanamycin-resistance gene (neo) flanked by FRT sites (FRT-neo-FRT) were inserted into the start codons of genes of interest via recombineering. The FRT-neo-FRT cassettes were then excised by the induction of L-arabinose (Sigma-Aldrich, A3256). The final BAC plasmids were extracted using the commercial kit NucleoBond BAC 100 (MACHEREY-NAGEL, 740579) following the manufacturer’s protocol.
mMAZe and MAZe-mCherry
The plasmid of mMAZe was created using the gateway cloning technology as described (Kwan et al., 2007). Entry clones, p5E-MAZe and pME-Kaede, were generated by performing BP clonase reactions (Gateway BP Clonase II Enzyme Mix, Invitrogen, 11789020) of an 8-kb MAZe fragment and a Kaede fragment with the pDONR P4-P1R and pDONR 221 vectors, respectively. Next, p5E-MAZe, pME-Kaede, p3E-polyA, and the destination vector pDestTol2pA2 were assembled in an LR reaction (Gateway LRClonase II Enzyme Mix, Invitrogen, 11791020) to generate the final plasmid of mMAZe. The vectors of pDONR P4-P1R,pDONR 221, p3E-polyA, and pDestTol2pA2 were kind gifts from the National Institute of Genetics (Mishima, Japan). For the construction of MAZe-mCherry, kaede of mMAZe was replaced by mCherry.
Conventional plasmids
For the plasmids of UAS:kaede, UAS:mRuby3, UAS:atoh7-p2a-tdTomato-NLS, UAS:otx2-p2a-tdTomato-NLS, UAS:ptf1α-p2a-tdTomato-NLS, UAS:ptf1α-p2a-mRuby3, and atoh7:turboGFP-dest1, DNA fragments of the corresponding cassettes for each plasmid were inserted into the pDestTol2pA2 vector (Kwan et al., 2007) through homologue recombination using the ClonExpressMultiS One Step Cloning Kit (Vazyme, C113-02). The coding fragments of Atoh7, Otx2, and Ptf1α were amplified from the cDNA library of 48-hpf zebrafish. For atoh7:turboGFP-dest1, the atoh7 promoter ranges from 7.8 kb upstream of the atoh7 coding sequence.
Confocal imaging and kaede photoconversion
Embryos at the desired stage were anesthetized by 0.04% tricaine mesylate (MS-222; Sigma-Aldrich, A5040) and embedded in 1% low-melting agarose (Sigma-Aldrich, A0701). Retinas were imaged using the inverted laser-scanning confocal microscope (Olympus, FV1200) under 30× (oil, NA = 1.05) or 60× (water, NA = 1.20) objectives. If further experiment was needed, embryos were immediately released from the agarose and were allowed to develop in the embryo medium. For kaede photoconversion, pulses of 405-nm laser were applied to desired single cells with kaede-green signals until the kaede-green proteins were converted to kaede-red.
Lineage tracing
Lineage tracing using mMAZe
To efficiently label single RPCs, Tg(mMAZe::UAS:kaede) embryos at 24 hpf were treated with heat shock at 39°C for 15 min. After that, kaede photoconversion in single progenitors was performed at 48 hpf. Typically, two to three progenitors were photoconverted in each embryo. The embryos were then raised in embryo medium at 28.5°C separately. Lineages with kaede-red signals were obtained at 3 dpf.
Lineage tracing using atoh7:Switch
WT embryos were injected with the atoh7:Switch BAC plasmid (10 ng/µl) together with tol2 mRNA (50 ng/μl) at the one-cell stage, followed by cre mRNA (10 ng/µl) injection into yolk at the 8- or 16-cell stage. For the systematic analysis (Fig. 1), spatially isolated atoh7+ lineages (EGFP signals of atoh7:Switch) with less than seven cells were obtained at 72 hpf by confocal imaging (Olympus, FV1200). For the analysis of AC subtypes in WT and AC respecification after ptf1α knockout, sparsely distributed atoh7+ lineages (DsRed2 signals of atoh7:Switch) were collected at the stage depending on the experiments.
Lineage tracing of vsx1+ RPCs and OC1+ RPCs
vsx1:gal4 or OC1:gal4 BAC plasmids were injected with UAS:kaede plasmid. At 48–54 hpf, individual vsx1+ kaede+ retinal cells were photoconverted, and then the lineages were analyzed at 3 dpf. However, the OC1 promoter was too weak to trigger a strong enough kaede signal for photoconversion, so we collected spatially isolated OC1+ cells (containing two to five cells) as lineages derived from single RPCs.
Analysis of cell subtypes in major neurogenic lineages
AC subtypes
To analyze the morphology of ACs traced using mMAZe (Fig. S2, D and E), only ACs whose stratification could be identified clearly were taken into account. ACs that terminated flatly in one or two layers in IPL were assigned as “flat,” and ACs that terminated diffusely were assigned as “diffuse.” To analyze the AC subtypes in lineages of AC-BC and AC-2PR (Fig. 2, A–D), lineage tracing using atoh7:Switch was performed in Tg(gad1b:EGFP) or Tg(glyt1:EGFP). At 4 dpf, embryos with EGFP (signal of Tg(gad1b:EGFP) or Tg(glyT1:EGFP)) and DsRed2 (signal of atoh7:Switch) signals were selected. DsRed2 and EGFP signals were amplified by whole-mount immunostaining before imaging.
BC subtypes
BC subtypes were characterized by the locations of their axon terminals in the inner plexiform layer at 4–5 dpf. In AC-BC and BC-MC lineages traced by mMAZe, axon terminals of BCs were easily identified. Kaede-red signals could be enhanced by rephotoconversion if needed. In BC-BC lineages obtained by mMAZe, BC subtypes were determined by sequential photoconversion (Fig. S2 F). Only those BCs whose subtype could be assigned unambiguously were taken into account.
Ptf1α knockout
Four sgRNAs targeting the coding sequence of ptf1α were designed using the CRISPRScan online tool (https://www.crisprscan.org/; Moreno-Mateos et al., 2015). To generate the template for in vitro transcription of the sgRNAs, a DNA fragment of standard sgRNA scaffold (Gagnon et al., 2014) was amplified with a mixture of four forward primers and one reverse primer. Forward primers consist of three elements: the T7 promoter sequence, sgRNA-targeting sequence, scaffold-targeting sequence. The primers used are listed in Table 2.
Table 2. Primers used to synthesize ptf1α sgRNA.
| Primer | Sequence (5′ to 3′) |
|---|---|
| Forward primer 1 | TGTAATACGACTCACTATAGGTAGTCGGATATCACAGGAGTTTTAGAGCTAGAAAT |
| Forward primer 2 | TGTAATACGACTCACTATAGGGGCTTACTGAAGAGGCGGGTTTTAGAGCTAGAAAT |
| Forward primer 3 | TGTAATACGACTCACTATAGGCAGTCGGACATGCCAATTGTTTTAGAGCTAGAAAT |
| Forward primer 4 | TGTAATACGACTCACTATAGGAAGTCGTCTGTGTCCAAGGTTTTAGAGCTAGAAAT |
| Reverse primer | AAAAAAAGCACCGACTCGGTGCCAC |
The sgRNAs were then in vitro transcribed and purified using the LiCl precipitation approach (MEGAscript T7 Transcription Kit, Invitrogen, AM1334). To knock out ptf1α, the mixture of the four sgRNAs (in total 200 ng/µl) and Cas9 protein (400 ng/µl; Novoprotein, E365-01A) were coinjected into the yolk of Tg(ptf1α: GFP) embryos at the one-cell stage.
To quantify ptf1α sgRNA efficiency, five Tg(ptf1α:EGFP) embryos with ectopic BCs, PRs, and RGCs after injecting ptf1α sgRNA and cas9 protein were mixed and treated with NaOH to extract the genomic DNA. Ptf1α DNA fragments were amplified with primers flanking the sgRNA target sites. The primers’ sequences are ptf1α-sgtest-F (5′-3′) ATGGACACTGTGTTGGATCC, and ptf1α-sgtest-R (5′-3′) CATACTTGTTCCTCGGTGGC. These amplified products were inserted into Blunt Zero Vector (TransGen Biotech, CB501-01) and then sequenced with M13F primer. Tg(ptf1α:GFP) embryos had no injection treatment in control groups. The conclusion was drawn based on three independent replicates.
Quantification of the respecified lineages of AC-BC and AC-2PR after Ptf1α knockout
Lineages consisting of two normal PRs (DsRed2+GFP−) and one or more respecified neurons (DsRed2+GFP+) after ptf1α knockout were taken as candidates of respecified lineages derived from AC-2PR. In total, 10 candidates were collected: 9 2PRres-2PRnorm (Fig. 3, E and F) and 1 RGCres-2PRnorm. However, from the lineage tracing results using atoh7:Switch in WT embryos (Table S2), these candidates could also come from 2HC-2PR (n = 6) and AC-2HC-2PR (n = 6). Compared with the AC-2PR lineages (n = 82), these two lineages only took up ∼12.8% (12/94) of the contribution. Similarly, lineages consisting of one normal BC (DsRed2+GFP−) and one or more respecified neurons (DsRed2+GFP+) were taken as candidates derived from AC-BC. Of all 17 candidates, 15 were BCres-BCnorm (Fig. 3, E and F), and 2 were RGCres-BCnorm. These candidates could only come from AC-BC since BC-HC(s) lineage was absent in atoh7+ lineages.
Whole-mount immunostaining
Embryos were fixed in 10% formalin (Sigma-Aldrich, ht5011-1CS) at 4°C overnight, washed with 1× PBT (1× PBS with 0.25% Triton X-100), incubated in 0.05% trypsin-EDTA on ice for 45 min, followed by two quick washes and a 5-min wash with 1× PBT. Non-specific binding was blocked by incubating the samples with the blocking buffer (2% sheep serum, 1% DMSO [Sigma-Aldrich, D2438], diluted with 1× PBT), and then embryos were incubated with the primary antibodies (diluted by the blocking buffer) at 4°C overnight. The primary antibodies were rabbit polyclonal anti-DsRed2 (1:500; Takara Bio, 632496) and chicken monoclonal anti-GFP (1:2,000; Abcam, ab13970). Embryos were washed by 1× PBT and then incubated with the secondary antibodies (diluted by 1× PBT with 2% BSA) at room temperature for 4 h. The secondary antibodies were Alexa Fluor 594 goat anti-rabbit (1:1,000; Yeasen, 33112ES60), and Alexa Fluor 488 goat anti-chicken (1:1,000; Yeasen, 34606ES60). All procedures were performed in darkness from this step onward. Embryos were then washed by 1× PBT and finally kept in 1× PBS at 4°C before imaging.
Slice immunostaining
Tg(gad1b:EGFP) embryos at 4 dpf were fixed by 4% PFA and cryo-sectioned with 20 µm width. The slices were sequentially treated with 1× PBS, Antigen Retrieval Solution (Beyotime, P0090), and 5% BSA. The slices were then incubated with chicken-anti-GFP (Abcam, ab13970, 1:5,000 in 5% BSA/PBS) and rabbit-anti-gad65/67 (Abcam, ab11070; 1:500 in 5% BSA/PBS) at 4°C for >16 h, washed with 1× PBS, incubated with Alexa Fluor 594 goat anti-rabbit and Alexa Fluor 488 donkey anti-chicken for 3 h at room temperature. The slices were washed with 1× PBS, air dried, and mounted with coverslip. The signals were checked on a confocal microscope (Olympus, FV1200) under 60× (water, NA = 1.20) objective. Rabbit anti-gad65/67 was omitted in the control group. The conclusion was drawn based on two independent replicates.
RNA probe synthesis
To synthesize glyt1 antisense RNA probe, partial glyt1 coding region was amplified from cDNA library with primers: glyt1-F/T7-glyt1-R. For sense probe (control), the template was amplified with primers: T7-glyt1-F/glyt1-R. Then RNA probes were synthesized through T7 RNA polymerase (Promega, P2077) according to the manual (Table 3).
Table 3. Primers used to synthesize RNA probes.
| Primer name | Sequence (5′-3′) |
|---|---|
| glyt1-F | AGCAGTTCCAGGAGAGCCAG |
| T7-glyt1-R | TAATACGACTCACTATAGGGGCCAGACAACCCAGGATAGG |
| T7-glyt1-F | TAATACGACTCACTATAGGGAGCAGTTCCAGGAGAGCCAG |
| Glyt1-R | GCCAGACAACCCAGGATAGG |
RNA in situ hybridization with immunostaining
This experiment needs 3 d in total. On the first day, Tg(gad1b:EGFP) embryos at 4 dpf were fixed and cryo-sectioned with 20 µm width. Slices were fixed again with 4% PFA for 10 min and then washed with 1× PBS. Endogenous peroxidases were blocked by 0.1% H2O2 treatment for 30 min and slices were then washed with 1× PBS. Slices were treated with 10 µg/ml proteinase K (Sigma-Aldrich, V900887) in Tris-EDTA (TE) buffer at 37°C for 10 min, fixed again with 4% PFA, washed with 1× PBS, treated with 0.2 M HCl for 10 min, and washed with 1× PBS. Slices were treated with 0.1 M triethanol amine-HCl (pH 8.0) for 1 min. 1/400 volume acetic anhydrate was added to the solution, and incubation was continued for 10 min. The slices were washed with 1× PBS and then dehydrated sequentially with 60%, 80%, 95%, 100%, and 100% ethanol for 90 s. The slices were air-dried and incubated with hybridization buffer (50% formamide, 10 mM Tris-HCl, pH 8.0, 200 µg/ml yeast tRNA, 10% dextran sulfate, 1× Denhardt’s solution, 600 mM NaCl, 0.25% SDS, and 1 mM EDTA, pH 8.0) containing 1 µg/ml antisense or sense RNA probe at 60°C for >16 h. On the second day, slices were washed sequentially with 2× Standard Saline Citrate (SSC)/50% formamide at 60°C for 30 min, TNE (10 mM Tris-HCl, pH 7.5, 0.5 M NaCl, and 1 mM EDTA) at 37°C for 10 min, 20 μg/ml RNaseA in TNE at 37°C for 10 min, 2× SSC at 60°C for 20 min, 0.2× SSC at 60°C for 20 min, 0.1× SSC for 20 min, TN buffer (100 mM Tris-HCl, pH 7.5, and 0.15 M NaCl) for 5 min, and TNB buffer (TN buffer with 0.5% Blocking reagent) for 30 min. Finally, the slices were incubated with anti-dig-peroxidase (Roche, 11207733910, 1:500) and rabbit-anti-GFP tag (Proteintech, 50430–2-AP, 1:500) at 4°C for >16 h. On the third day, TSA detection was performed with cy3 labeling (PerkinElmer, NEL753001KT) according to the manufacturer’s instructions. Slices were incubated with 488-goat-anti-rabbit for 3 h and washed with 1× PBS. Coverslips were mounted, and the signal was checked on a confocal microscope (Olympus, FV1200) under 60× (water, NA = 1.20) objective. The conclusion was drawn based on two independent replicates.
Single-cell RNA-seq library construction of atoh7+ RPCs
Retinas of Tg(atoh7:gapRFP::atoh7:turboGFP-dest1) embryos at around 44 hpf were dissected in Ca2+ free medium (116.6 mM NaCl, 0.67 mM KCl, 4.62 mM Tris base, and 0.4 mM EDTA, pH 7.8) and digested by 100 µl 0.25% trypsin (Biosharp, BS051C)–EDTA at 37°C for 8 min. The digestion was then terminated by 100 µl 6% BSA (Sigma-Aldrich, B2064). Following that, the single-cell suspension was filtered with a 40-µm cell strainer. TurboGFP+ cells were obtained with fluorescence-activated cell sorting (MoFloXDP, Beckman Coulter) and centrifuged at 500 g, 4°C, for 5 min. Then the cells were washed once with PBS-0.04% BSA and resuspended in 30 µl PBS–0.04% BSA for cell loading. About 7,000 TurboGFP+ cells were loaded, and the single-cell RNA-seq library was prepared using the Chromium Single Cell 3′ Library & Gel Bead Kit v2 (10x Genomics, PN-120237) following the manufacturer’s protocol. After that, the library was sequenced on the Illumina NovaSeq 6000 (paired-end: 150 bp for both reads, Novogene).
Single-cell ATAC-seq library construction
About 10 zebrafish retinas at 48 hpf were dissected in DMEM/F12 medium and digested by 100 µl papain (Worthington Biochemical Corporation, LS003126) solution at 37°C for 17 min. During the digestion, we pipetted the tissues about four times. The digestion was then terminated by 400 µl washing buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, and 1% BSA). Subsequently, the single-cell suspension was filtered with a 40-µm cell strainer and centrifuged at 500 g, 4°C, for 5 min. The cell pellet was resuspended by 50 µl PBS–0.04% BSA and centrifuged at 300 g, 4°C, for 5 min. Next, 45 µl supernatant was removed and replaced by 45 µl chilled lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, 0.1% NP-40, 0.01% Digitonin [Invitrogen, BN2006], and 1% BSA). The sample was then gently pipetted three times and incubated on ice for 3.5 min. After that, 50 µl chilled wash buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% Tween-20, and 1% BSA) was added to the tube without pipetting. Immediately, the sample was centrifuged at 500 g, 4°C, for 5 min. 95 µl supernatant was removed, and 45 µl chilled diluted nuclei buffer (10x Genomics) was added without pipetting. Again, the sample was centrifuged at 500 g, 4°C, for 5 min. After removing all supernatant, the nuclei were resuspended with 10 µl chilled diluted nuclei buffer. About 12,000 single nuclei were loaded, and the single-cell ATAC-seq library was prepared using the Chromium Single Cell ATAC Library & Gel Bead Kit (10x Genomics, PN-1000111) following the manufacturer’s protocol. After that, the library was sequenced on the Illumina NovaSeq 6000 (paired-end: 50 bp for both reads, Novogene).
scRNA-seq data analysis
Raw data were aligned (zebrafish genome, GRCz10), filtered, and counted using Cell Ranger software (v2.1.0, 10x Genomics). On average, 51,235 and 181,837 reads per cell were obtained, and 3,621 and 3,340 cells were recovered for whole retina at 48 hpf and atoh7 samples, respectively. The following analyses were performed using the R package “Seurat” (v2.3.4; Butler et al., 2018). We first filtered out low-abundance genes (detected in fewer than three cells), poor libraries (with <200 genes or >5% of transcripts coming from mitochondrial genes), and cell doublets (with >4,000 genes). The filtered data (15,667 genes × 3,587 cells for whole retina at 48 hpf and 14,907 genes × 3,298 cells for atoh7 sample) were then log normalized and scaled with the default parameters. Highly variable genes (with the mean expression between 0.0125 and 3 and standard deviation of at least 0.5) were selected and used for principal components analysis. After that, the graph-based clustering was performed with the PCs defined by the PCElbowPlot function. We found marker genes for each cluster using the FindAllMarkers function (with genes detected at a minimum percentage of 25%) and identified the clusters with known marker genes. In both samples, clusters expressing proliferating markers, such as pcna or mki67 (Gerdes et al., 1984; Miyachi et al., 1978), were defined as RPCs. Among them, PR precursors were characterized due to their specific expression of nr2e3, otx5, and crx (Peng et al., 2005; Sauka-Spengler et al., 2001); one cluster in a 48-hpf sample with the specific expression of prox1a was defined as HC precursors (Godinho et al., 2007); the remaining clusters were named as undefined RPCs. We identified neurons (RGCs, ACs, and BCs) for their expression of rbpms2b, tfap2a, and vsx1, separately (Diekmann and Stuermer, 2009; Jin et al., 2015; Passini et al., 1997). The clustering result was visualized using t-distributed stochastic neighbor embedding (t-SNE) with the cells grouped by their identities (Fig. S3 A). Gene expression patterns of marker genes for each group of cells were shown using the DotPlot function (Fig. S3 B).
Next, we analyzed the undefined RPCs in detail. Using the CellCycleScoring function, we assigned each cell with the cell-cycle state of G0/G1 phase, S phase, or G2/M phase. However, we can see that cell cycle played important roles in clustering in both samples. To minimize the cell cycle influence on clustering, we extracted undefined RPCs at G2/M phase (635 cells for 48-hpf sample and 849 cells for atoh7 sample). Similar to the above processing, the expression data were normalized, scaled, and further clustered. The 48-hpf sample was clustered into four clusters. After the same analysis for differential gene expression, we performed GO analysis of the top 15 featured genes for all clusters using the DAVID online tool (https://david.ncifcrf.gov/; Huang et al., 2009) and found an enrichment of TFs (Fig. S3 D). The atoh7 sample was clustered into six clusters. Among them, four clusters (merged into two groups, groups 2 and 3) had very similar TF expression patterns to the two atoh7+ clusters in the 48-hpf sample (Fig. 4, C and E). Actually, the only difference between the two clusters in each group was the expression of cell cycle genes. Finally, we extracted the 48-hpf early RPCs (Cluster A; Fig. 4, B and C) and performed the normalization, dimensionality reduction, and clustering analysis as previously described.
scATAC-seq data analysis
The raw datasets were aligned (zebrafish genome, GRCz10), filtered, and counted using the Cell Ranger ATAC software (v1.1, 10x Genomics). On average, 3,798 fragments per cell were obtained, and 4,200 cells were recovered.
The clustering analyses were performed using the R package “Signac” (v0.1.6; Stuart et al., 2019). We first computed quality control (QC) metrics and removed the outliers. Specifically, we filtered out low-quality cells (with <15% peak-region fragments or >10 folds of mono-nucleosome to nucleosome-free fragments) and doublets (with >10,000 peak-region fragments). We then normalized the filtered data (107,585 peaks × 4,058 cells) by the RunTFIDF function and ran a singular value decomposition using “LSI” with all peaks. Next, we performed graph-based clustering by FindNeighbors and FindClusters functions using the first 20 dimensions of reduction as an input. In total, we got 21 clusters at a resolution of 1.4. We quantified gene activities for each gene using the CreateGeneActivityMatrix function through counting the number of fragments that located at the gene body and 500 bp upstream of transcription start site, and normalized the data using the NormalizeData function. The identity of each cluster was then assigned according to the activities of known marker genes of specific cell types (Fig. S5 B). Clusters with high activity of nr2e3 and rem1 were assigned as PR precursors (clusters 2, 4, and 21) and HC precursors (cluster 17), respectively. ACs (clusters 9, 13, 14, 15, and 16), RGCs (clusters 10, 11, 12, and 19), and BCs (clusters 3 and 6) were assigned for the high activities of tfap2a, pou4f2, and vsx1, separately. Clusters (clusters 5, 7, 8, 18, and 20) that had high her4.1 activity but low activities for the neuronal marker genes were characterized as RPCs. Cluster 1 had a very low fraction of peak-region fragments, so we removed this cluster of cells as it may represent low-quality cells or technical artifacts.
To further study the chromatin characteristics of RPCs, we extracted 914 48-hpf RPCs and performed the normalization, dimensionality reduction, and clustering steps as previously described. We obtained five clusters at a resolution of 0.6. Clusters 1, 4, and 5 had high activities for her12 and her6 (combined as group her12open). Cluster 3 had high atoh7 and tfap2d activities (termed as atoh7open). Cluster 2 had specific scrt2 and scg3 activities (termed as scrt2open; Fig. S5 C). Atoh7open RPCs were further subclustered into three clusters (atoh7openher12open, atoh7openOC1open, and atoh7openher12closedOC1closed) at a resolution of 0.8 (Fig. S5 D). Next, we performed the integration analysis of 48-hpf scATAC-seq and scRNA-seq data using the FindTransferAnchors and TransferData functions of “Signac.” Specifically, 914 48-hpf RPCs with ATAC-based gene activities were anchored to the 635 48-hpf RPCs at G2/M phase with gene expression levels. As a result, each cell in the scATAC-seq data was assigned with four prediction scores for the Clusters A–D of scRNA-seq data, which were shown in a dotplot with a cutoff of 0.3 (Fig. S5 E). The read coverage of regions near vsx1 and OC1 in each group were plotted by the CoveragePlot function (Fig. 6, E and F).
We performed the pseudo-time analysis of the 914 RPCs by “Monocle3” (v0.2.0; Cao et al., 2019). We first processed the data using “LSI” and then continued with the standard dimensionality reduction using “UMAP.” Next, we transferred the clustering result by “Signac” to this dataset. By the default learn graph and order cells functions, we obtained the pseudo-time of each cell. Then we transferred the pseudo-time information back to the “Signac” dataset and visualized them in the t-SNE plot (Fig. 6 C).
For the coaccessibility analysis to predict the enhancer-promoter connections, we ran the standard process of “Cicero” (v1.3.4.3; Pliner et al., 2018) for the 914 RPCs. After that, we obtained a “Cicero coaccessibility” score between −1 and 1 (a higher number indicates higher coaccessibility) between each pair of peaks. Among the peaks, those overlapped with gene transcription start sites were defined as promoters, and the others were distal elements. The “Cicero coaccessibility” scores between the promoter and its related distal elements of vsx1 or OC1 were shown (Fig. S5, F and G).
Single TFs overexpression
To overexpress single TFs (atoh7, otx2, or ptf1α) in vsx1+ RPCs (or OC1+ RPCs), plasmids of vsx1:Gal4 (or OC1:Gal4; 15 ng/µl) and UAS:TF-p2a-tdTomatoNLS (UAS:tdTomato-NLS in blank group; 15 ng/µl), together with tol2 mRNA (50 ng/µl) were injected into the cells of Tg(ptf1α:EGFP) embryos at the one-cell stage. At 3 dpf, cells with tdTomato-NLS signals in each group were collected unbiasedly. Cell identity was finally characterized by their cell body location.
For the statistical analysis of cell-composition difference between control groups and TF overexpressed groups (Fig. 7), we obtained in total 170–446 cells (including RGCs, ACs, BCs, HCs, and PRs) from >10 embryos for each group. Fisher’s exact test was used for the statistical test. To test the difference of a specific cell type, for instance, the difference of OC1+ RGCs between the blank group and the atoh7 overexpression group, 446 cells were collected in the blank group (pOC1:Gal4 + pUAS:tdTomato-NLS), including 64 RGCs and 382 non-RGCs (123 ACs, 17 BCs, 61 HCs, and 181 PRs; first bar of Fig. 7 F). In the experimental group (pOC1:Gal4 + pUAS:Atoh7-p2a-tdTomato-NLS), 330 cells were collected, including 137 RGCs and 193 non-RGCs (77 ACs, 11 BCs, 28 HCs, and 77 PRs; second bar of Fig. 7 F). The input for Fisher’s exact test is shown in Table 4.
Table 4. The representative contigency table for Fisher’s exact test.
| Control | Atoh7 misexpression | |
|---|---|---|
| RGC | 64 | 137 |
| Non-RGC | 382 | 193 |
The result (P < 2.2e-16) showed that RGC ratio in the atoh7 misexpression group was significantly increased compared with the blank group. Similar analyses were performed for all the cell types between blank groups and TF-misexpressed groups. The significant P values are shown in Fig. 6: *, P < 0.01; **, P < 0.001; and ***, P < 0.0001.
Image processing
Image analysis was performed with FV10-ASW 4.0 (Olympus), Imaris 7.6.5 (Bitplane), and ImageJ (National Institutes of Health). Cell types and cell numbers were determined manually. For presentation, maximal intensity projection of several z-sections was used, and brightness and contrast of each channel (GFP or RFP) were adjusted separately. All manipulations were applied for the whole pictures, and then the regions of interest were selected and cropped from the whole picture for presentation.
Data availability
Atoh7 scRNA-seq and 48-hpf scATAC-seq datasets generated in this study have been deposited in GEO under accession no. GSE150839. 48-hpf scRNA-seq data is from GSE122680 (Xu et al., 2020).
Online supplemental material
Fig. S1 shows characteristics of lineages traced by mMAZe and atoh7:Switch. Fig. S2 displays AC and BC subtypes. Fig. S3 shows scRNA-seq of 48-hpf retina. In Fig. S4 shows scRNA-seq of atoh7+ RPCs. Fig. S5 shows scATAC-seq of 48-hpf retina. Table S1 lists lineages traced by mMAZe. Table S2 lists lineages traced by atoh7:Switch.
Supplementary Material
lists lineages traced by mMAZe.
lists lineages traced by atoh7:Switch.
Acknowledgments
We thank Dr. Muming Poo, Dr. Patricia Jusuf, and Dr. William A. Harris for discussion and manuscript editing. We thank Dr. Min Zhang and Zhenning Zhou from the Molecular and Cellular Biology Core Facility of the Institute of Neuroscience (ION) for the library construction of scRNA-seq and scATAC-seq. We thank Haiyan Wu, Songlin Qian, and Lijuan Quan from the FACS Facility of ION.
This research was funded by grants from the Shanghai Municipal Science and Technology Major Project (grant no. 2018SHZDZX05), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB32000000), the State Key Laboratory of Neuroscience, Shanghai Basic Research Field Project (grant no. 18JC1410100), and the National Natural Science Foundation of China (grant no. 31871035).
The authors declare no competing financial interests.
Author contributions: M. Wang and L. Du: conceptualization, data curation, formal analysis, methodology, validation, visualization, writing of original draft, and review and editing. A.C. Lee: conceptualization, data curation, formal analysis, methodology, validation, writing-original draft. H. Qin and Y. Li: methodology. J. He: conceptualization, writing of original draft, review and editing, funding acquisition, and supervision.
References
- Bai G., Sheng N., Xie Z., Bian W., Yokota Y., Benezra R., Kageyama R., Guillemot F., and Jing N.. 2007. Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev. Cell. 13:283–297. 10.1016/j.devcel.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Balasubramanian R., and Gan L.. 2014. Development of Retinal Amacrine Cells and Their Dendritic Stratification. Curr. Ophthalmol. Rep. 2:100–106. 10.1007/s40135-014-0048-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boije H., Rulands S., Dudczig S., Simons B.D., and Harris W.A.. 2015. The Independent Probabilistic Firing of Transcription Factors: A Paradigm for Clonal Variability in the Zebrafish Retina. Dev. Cell. 34:532–543. [DOI] [PMC free article] [PubMed]
- Brody T., and Odenwald W.F.. 2000. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev. Biol. 226:34–44. 10.1006/dbio.2000.9829 [DOI] [PubMed] [Google Scholar]
- Brown N.L., Patel S., Brzezinski J., and Glaser T.. 2001. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 128:2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski J.A. IV, Prasov L., and Glaser T.. 2012. Math5 defines the ganglion cell competence state in a subpopulation of retinal progenitor cells exiting the cell cycle. Dev. Biol. 365:395–413. 10.1016/j.ydbio.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. . 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 566:496–502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko C. 2014. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat. Rev. Neurosci. 15:615–627. 10.1038/nrn3767 [DOI] [PubMed] [Google Scholar]
- Cherry T.J., Trimarchi J.M., Stadler M.B., and Cepko C.L.. 2009. Development and diversification of retinal amacrine interneurons at single cell resolution. Proc. Natl. Acad. Sci. USA. 106:9495–9500. 10.1073/pnas.0903264106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R.T., Linker C., and Lewis J.. 2010. MAZe: a tool for mosaic analysis of gene function in zebrafish. Nat. Methods. 7:219–223. 10.1038/nmeth.1423 [DOI] [PubMed] [Google Scholar]
- Connaughton V.P. 2011. Bipolar cells in the zebrafish retina. Vis. Neurosci. 28:77–93. 10.1017/S0952523810000295 [DOI] [PubMed] [Google Scholar]
- Connaughton V.P., Graham D., and Nelson R.. 2004. Identification and morphological classification of horizontal, bipolar, and amacrine cells within the zebrafish retina. J. Comp. Neurol. 477:371–385. 10.1002/cne.20261 [DOI] [PubMed] [Google Scholar]
- Del Bene F., Wehman A.M., Link B.A., and Baier H.. 2008. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cll. 134:1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demb J.B., and Singer J.H.. 2015. Functional Circuitry of the Retina. Annu. Rev. Vis. Sci. 1:263–289. 10.1146/annurev-vision-082114-035334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann H., and Stuermer C.A.. 2009. Zebrafish neurolin-a and -b, orthologs of ALCAM, are involved in retinal ganglion cell differentiation and retinal axon pathfinding. J. Comp. Neurol. 513:38–50. 10.1002/cne.21928 [DOI] [PubMed] [Google Scholar]
- Eckler M.J., Nguyen T.D., McKenna W.L., Fastow B.L., Guo C., Rubenstein J.L.R., and Chen B.. 2015. Cux2-positive radial glial cells generate diverse subtypes of neocortical projection neurons and macroglia. Neuron. 86(4):1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov A.G., Pokross M.E., Egorov N.S., Zaraisky A.G., Yampolsky I.V., Merzlyak E.M., Shkoporov A.N., Sander I., Lukyanov K.A., and Chudakov D.M.. 2006. Structural basis for the fast maturation of Arthropoda green fluorescent protein. EMBO Rep. 7:1006–1012. 10.1038/sj.embor.7400787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S.J., Gil-Sanz C., Martinez-Garay I., Espinosa A., Harkins-Perry S.R., Ramos C., and Müller U.. 2012. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 337:746–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y., Fujitani S., Luo H., Qiu F., Burlison J., Long Q., Kawaguchi Y., Edlund H., MacDonald R.J., Furukawa T., et al. . 2006. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 133:4439–4450. 10.1242/dev.02598 [DOI] [PubMed] [Google Scholar]
- Gagnon J.A., Valen E., Thyme S.B., Huang P., Akhmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., and Schier A.F.. 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 9 e98186 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P., Postiglione M.P., Krieger T.G., Hernandez L., Wang C., Han Z., Streicher C., Papusheva E., Insolera R., Chugh K., et al. . 2014. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 159(4):775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno F., and Molnar Z.. 2015. Subset of early radial glial progenitors that contribute to the development of callosal neurons is absent from avian brain. Proc. Natl. Acad. Sci. USA. 112(36). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J., Lemke H., Baisch H., Wacker H.H., Schwab U., and Stein H.. 1984. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 133:1710–1715. [PubMed] [Google Scholar]
- Glass A.S., and Dahm R.. 2004. The zebrafish as a model organism for eye development. Ophthalmic Res. 36:4–24. 10.1159/000076105 [DOI] [PubMed] [Google Scholar]
- Godinho L., Mumm J.S., Williams P.R., Schroeter E.H., Koerber A., Park S.W., Leach S.D., and Wong R.O.. 2005. Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development. 132:5069–5079. 10.1242/dev.02075 [DOI] [PubMed] [Google Scholar]
- Godinho L., Williams P.R., Claassen Y., Provost E., Leach S.D., Kamermans M., and Wong R.O.. 2007. Nonapical symmetric divisions underlie horizontal cell layer formation in the developing retina in vivo. Neuron. 56:597–603. 10.1016/j.neuron.2007.09.036 [DOI] [PubMed] [Google Scholar]
- Guo C., Eckler M.J., McKenna W.L., McKinsey G.L., Rubenstein J.L., and Chen B.. 2013. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 80(5):1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafler B.P., Surzenko N., Beier K.T., Punzo C., Trimarchi J.M., Kong J.H., and Cepko C.L.. 2012. Transcription factor Olig2 defines subpopulations of retinal progenitor cells biased toward specific cell fates. Proc. Natl. Acad. Sci. USA. 109:7882–7887. 10.1073/pnas.1203138109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J., Tomita K., Inoue T., and Kageyama R.. 2001. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 128:1313–1322. [DOI] [PubMed] [Google Scholar]
- He J., Zhang G., Almeida A.D., Cayouette M., Simons B.D., and Harris W.A.. 2012. How variable clones build an invariant retina. Neuron. 75:786–798. 10.1016/j.neuron.2012.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C.E., Bertsch T.W., Ellis H.M., and Harris W.A.. 1988. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron. 1:15–26. 10.1016/0896-6273(88)90205-X [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Imayoshi I., Ishidate F., and Kageyama R.. 2015. Real-time imaging of bHLH transcription factors reveals their dynamic control in the multipotency and fate choice of neural stem cells. Front. Cell. Neurosci. 9:288 10.3389/fncel.2015.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T., Pearson B., Holbrook S., and Doe C.Q.. 2001. Drosophila Neuroblasts Sequentially Express Transcription Factors which Specify the Temporal Identity of Their Neuronal Progeny. Cll. 106:511–521. [DOI] [PubMed] [Google Scholar]
- Jin K., Jiang H., Xiao D., Zou M., Zhu J., and Xiang M.. 2015. Tfap2a and 2b act downstream of Ptf1a to promote amacrine cell differentiation during retinogenesis. Mol. Brain. 8:28 10.1186/s13041-015-0118-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusuf P.R., and Harris W.A.. 2009. Ptf1a is expressed transiently in all types of amacrine cells in the embryonic zebrafish retina. Neural Dev. 4:34 10.1186/1749-8104-4-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusuf P.R., Almeida A.D., Randlett O., Joubin K., Poggi L., and Harris W.A.. 2011. Origin and determination of inhibitory cell lineages in the vertebrate retina. J. Neurosci. 31:2549–2562. 10.1523/JNEUROSCI.4713-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay J.N., Finger-Baier K.C., Roeser T., Staub W., and Baier H.. 2001. Retinal ganglion cell genesis requires lakritz, a Zebrafish atonal Homolog. Neuron. 30:725–736. 10.1016/S0896-6273(01)00312-9 [DOI] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., and Schilling T.F.. 1995. Stages of embryonic development of the zebrafish. Dev. Dyn. 203:253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Mizuno H., Imayoshi I., Furusawa C., Shirahige K., and Kageyama R.. 2009. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 23:1870–1875. 10.1101/gad.1823109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike C., Nishida A., Ueno S., Saito H., Sanuki R., Sato S., Furukawa A., Aizawa S., Matsuo I., Suzuki N., et al. . 2007. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol. Cell. Biol. 27:8318–8329. 10.1128/MCB.01209-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., and Chien C.B.. 2007. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236:3088–3099. 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C.-C., and Kain S.R.. 1998. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 273:34970–34975. 10.1074/jbc.273.52.34970 [DOI] [PubMed] [Google Scholar]
- Li Y., Lu H., Cheng P.L., Ge S., Xu H., Shi S.H., and Dan Y.. 2012. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature. 486:118–121. 10.1038/nature11110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.H., Zhou L., Li Z., Wang Y., Shi J.T., Yang Y.J., and Gui J.F.. 2015. Zebrafish Lbh-like Is Required for Otx2-mediated Photoreceptor Differentiation. Int. J. Biol. Sci. 11:688–700. 10.7150/ijbs.11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorca A., Ciceri G., Beattie R., Wong F.K., Diana G., Serafeimidou-Pouliou E., Fernandez-Otero M., Streicher C., Arnold S.J., Meyer M., et al. . 2019. A stochastic framework of neurogenesis underlies the assembly of neocortical cytoarchitecture. Elife. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison L.A., Lu J., Victoroff T., Scott E., Baier H., and Chen W.. 2009. A gain-of-function screen in zebrafish identifies a guanylate cyclase with a role in neuronal degeneration. Mol. Genet. Genomics. 281:551–563. 10.1007/s00438-009-0428-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc R.E., and Cameron D.. 2001. A molecular phenotype atlas of the zebrafish retina. J. Neurocytol. 30:593–654. 10.1023/A:1016516818393 [DOI] [PubMed] [Google Scholar]
- Masai I., Stemple D.L., Okamoto H., and Wilson S.W.. 2000. Midline signals regulate retinal neurogenesis in zebrafish. Neuron. 27:251–263. 10.1016/S0896-6273(00)00034-9 [DOI] [PubMed] [Google Scholar]
- Masai I., Lele Z., Yamaguchi M., Komori A., Nakata A., Nishiwaki Y., Wada H., Tanaka H., Nojima Y., Hammerschmidt M., et al. . 2003. N-cadherin mediates retinal lamination, maintenance of forebrain compartments and patterning of retinal neurites. Development. 130:2479–2494. 10.1242/dev.00465 [DOI] [PubMed] [Google Scholar]
- Masland R.H. 2001. Neuronal diversity in the retina. Curr. Opin. Neurobiol. 11:431–436. 10.1016/S0959-4388(00)00230-0 [DOI] [PubMed] [Google Scholar]
- Masland R.H. 2012. The neuronal organization of the retina. Neuron. 76:266–280. 10.1016/j.neuron.2012.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menger N., Pow D.V., and Wassle H.. 1998. Glycinergic amacrine cells of the rat retina. J. Comp. Neurol. 401:34–46. [DOI] [PubMed] [Google Scholar]
- Miyachi K., Fritzler M.J., and Tan E.M.. 1978. Autoantibody to a nuclear antigen in proliferating cells. J. Immunol. 121:2228–2234. [PubMed] [Google Scholar]
- Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., and Giraldez A.J.. 2015. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 12:982–988. 10.1038/nmeth.3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhai H., Sel S., Favor J., Mendoza-Torres L., Paulsen F., Duncker G.I., and Schmid R.M.. 2007. Ptf1a is essential for the differentiation of GABAergic and glycinergic amacrine cells and horizontal cells in the mouse retina. Development. 134:1151–1160. 10.1242/dev.02781 [DOI] [PubMed] [Google Scholar]
- Nishida A., Furukawa A., Koike C., Tano Y., Aizawa S., Matsuo I., and Furukawa T.. 2003. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 6:1255–1263. 10.1038/nn1155 [DOI] [PubMed] [Google Scholar]
- Ohtsuka T., Sakamoto M., Guillemot F., and Kageyama R.. 2001. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J. Biol. Chem. 276:30467–30474. 10.1074/jbc.M102420200 [DOI] [PubMed] [Google Scholar]
- Passini M.A., Levine E.M., Canger A.K., Raymond P.A., and Schechter N.. 1997. Vsx-1 and Vsx-2: differential expression of two paired-like homeobox genes during zebrafish and goldfish retinogenesis. J. Comp. Neurol. 388:495–505. [DOI] [PubMed] [Google Scholar]
- Pearson B.J., and Doe C.Q.. 2004. Specification of temporal identity in the developing nervous system. Annu. Rev. Cell Dev. Biol. 20:619–647. 10.1146/annurev.cellbio.19.111301.115142 [DOI] [PubMed] [Google Scholar]
- Peng G.H., Ahmad O., Ahmad F., Liu J., and Chen S.. 2005. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum. Mol. Genet. 14:747–764. 10.1093/hmg/ddi070 [DOI] [PubMed] [Google Scholar]
- Pliner H.A., Packer J.S., McFaline-Figueroa J.L., Cusanovich D.A., Daza R.M., Aghamirzaie D., Srivatsan S., Qiu X., Jackson D., Minkina A., et al. . 2018. Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol. Cell. 71:858–871.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggi L., Vitorino M., Masai I., and Harris W.A.. 2005. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J. Cell Biol. 171:991–999. 10.1083/jcb.200509098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova E. 2014. ON-OFF Interactions in the Retina: Role of Glycine and GABA. Curr. Neuropharmacol. 12:509–526. 10.2174/1570159X13999150122165018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell L.M., and Jarman A.P.. 2008. Context dependence of proneural bHLH proteins. Curr. Opin. Genet. Dev. 18:411–417. 10.1016/j.gde.2008.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S.Q., Li Z., Lin S., Bergami M., and Shi S.H.. 2019. Precise Long-Range Microcircuit-to-Microcircuit Communication Connects the Frontal and Sensory Cortices in the Mammalian Brain. Neuron. 104:385–401.e3. 10.1016/j.neuron.2019.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompani S.B., and Cepko C.L.. 2008. Retinal progenitor cells can produce restricted subsets of horizontal cells. Proc. Natl. Acad. Sci. USA. 105:192–197. 10.1073/pnas.0709979104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salie R., Niederkofler V., and Arber S.. 2005. Patterning molecules; multitasking in the nervous system. Neuron. 45:189–192. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T., Baratte B., Shi L., and Mazan S.. 2001. Structure and expression of an Otx5-related gene in the dogfish Scyliorhinus canicula: evidence for a conserved role of Otx5 and Crxgenes in the specification of photoreceptors. Dev. Genes Evol. 211:533–544. 10.1007/s00427-001-0191-2 [DOI] [PubMed] [Google Scholar]
- Scholpp S., Delogu A., Gilthorpe J., Peukert D., Schindler S., and Lumsden A.. 2009. Her6 regulates the neurogenetic gradient and neuronal identity in the thalamus. Proc. Natl. Acad. Sci. USA. 106:19895–19900. 10.1073/pnas.0910894106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E.K., and Baier H.. 2009. The cellular architecture of the larval zebrafish tectum, as revealed by gal4 enhancer trap lines. Front. Neural Circuits. 3:13 10.3389/neuro.04.013.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar K., Lapan S.W., Whitney I.E., Tran N.M., Macosko E.Z., Kowalczyk M., Adiconis X., Levin J.Z., Nemesh J., Goldman M., et al. . 2016. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 166:1308–1323.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H., Ohtsuka T., and Kageyama R.. 2008. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 58:52–64. 10.1016/j.neuron.2008.02.014 [DOI] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive Integration of Single-Cell Data. Cell. 177:1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J.E. 1976. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 275:287–297. 10.1098/rstb.1976.0084 [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., and Thomson J.N.. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100:64–119. 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Suster M.L., Abe G., Schouw A., and Kawakami K.. 2011. Transposon-mediated BAC transgenesis in zebrafish. Nat. Protoc. 6:1998–2021. 10.1038/nprot.2011.416 [DOI] [PubMed] [Google Scholar]
- Suzuki S.C., Bleckert A., Williams P.R., Takechi M., Kawamura S., and Wong R.O.. 2013. Cone photoreceptor types in zebrafish are generated by symmetric terminal divisions of dedicated precursors. Proc. Natl. Acad. Sci. USA. 110:15109–15114. 10.1073/pnas.1303551110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H. 1977. Juvenile hermaphroditism in the zebrafish,Brachydanio rerio. Bull. Fac. Fish. Hokkaido Univ. 28:57–65. [Google Scholar]
- Turner D.L., and Cepko C.L.. 1987. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 328:131–136. 10.1038/328131a0 [DOI] [PubMed] [Google Scholar]
- Turner D.L., Snyder E.Y., and Cepko C.L.. 1990. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 4:833–845. 10.1016/0896-6273(90)90136-4 [DOI] [PubMed] [Google Scholar]
- Udolph G., Lüer K., Bossing T., and Technau G.M.. 1995. Commitment of CNS progenitors along the dorsoventral axis of Drosophila neuroectoderm. Science. 269:1278–1281. 10.1126/science.7652576 [DOI] [PubMed] [Google Scholar]
- Vitorino M., Jusuf P.R., Maurus D., Kimura Y., Higashijima S., and Harris W.A.. 2009. Vsx2 in the zebrafish retina: restricted lineages through derepression. Neural Dev. 4:14 10.1186/1749-8104-4-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.W., Kim B.S., Ding K., Wang H., Sun D., Johnson R.L., Klein W.H., and Gan L.. 2001. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 15:24–29. 10.1101/gad.855301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R.S., Lam I.I., Clay H., Duong D.N., Deo R.C., and Coughlin S.R.. 2018. A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev. Cell. 46:112–125.e4. [DOI] [PubMed] [Google Scholar]
- Xu H.T., Han Z., Gao P., He S., Li Z., Shi W., Kodish O., Shao W., Brown K.N., Huang K., et al. . 2014. Distinct lineage-dependent structural and functional organization of the hippocampus. Cell. 157:1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Tang X., Jin M., Zhang H., Du L., Yu S., and He J.. 2020. Unifying developmental programs for embryonic and postembryonic neurogenesis in the zebrafish retina. Development. 147 dev185660 10.1242/dev.185660 [DOI] [PubMed] [Google Scholar]
- Yu Y.C., Bultje R.S., Wang X., and Shi S.H.. 2009. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 458:501–504. 10.1038/nature07722 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists lineages traced by mMAZe.
lists lineages traced by atoh7:Switch.
Data Availability Statement
Atoh7 scRNA-seq and 48-hpf scATAC-seq datasets generated in this study have been deposited in GEO under accession no. GSE150839. 48-hpf scRNA-seq data is from GSE122680 (Xu et al., 2020).