Chan et al. show that natural killer (NK) cells can be reprogrammed by breast cancer cells to promote metastasis. Reprogramming can be blocked by targeting NK cell inhibitory receptors TIGIT or KLRG1 or inhibiting DNA methyltransferases, which suggests new approaches to prevent or treat metastasis.
Abstract
Natural killer (NK) cells have potent antitumor and antimetastatic activity. It is incompletely understood how cancer cells escape NK cell surveillance. Using ex vivo and in vivo models of metastasis, we establish that keratin-14+ breast cancer cells are vulnerable to NK cells. We then discovered that exposure to cancer cells causes NK cells to lose their cytotoxic ability and promote metastatic outgrowth. Gene expression comparisons revealed that healthy NK cells have an active NK cell molecular phenotype, whereas tumor-exposed (teNK) cells resemble resting NK cells. Receptor–ligand analysis between teNK cells and tumor cells revealed multiple potential targets. We next showed that treatment with antibodies targeting TIGIT, antibodies targeting KLRG1, or small-molecule inhibitors of DNA methyltransferases (DMNT) each reduced colony formation. Combinations of DNMT inhibitors with anti-TIGIT or anti-KLRG1 antibodies further reduced metastatic potential. We propose that NK-directed therapies targeting these pathways would be effective in the adjuvant setting to prevent metastatic recurrence.
Introduction
Metastatic disease is the major driver of breast cancer mortality (Siegel et al., 2017). Adjuvant chemotherapy is used after locoregional control to prevent metastatic recurrence but is not sufficiently effective, because we do not fully understand how metastases form. Although the loss of immunosurveillance is critical to breast cancer metastasis, immune checkpoint blockade has not been as effective in treating metastatic breast cancer as in melanoma or lung cancer (Adams et al., 2019). This clinical observation suggests that the tumor microenvironment in metastatic breast cancer is complex and that inhibiting programmed cell death protein 1 and ligand (PD-1/PD-L1) signaling is not sufficient to restore a robust antitumor immune response.
Natural killer (NK) cells are key components of the innate immune system and have potent antitumor and antimetastatic activity (López-Soto et al., 2017). Accordingly, breast cancer cells must overcome NK cell surveillance to form distant metastases, yet we currently have a limited understanding of how metastatic cancer cells escape NK cell regulation. Others have shown that breast cancer cells, through a dormant state, down-regulate activating receptors to evade NK cells (Malladi et al., 2016). However, we do not fully understand how breast cancer cells escape NK cell–mediated immunosurveillance during transit through the circulation and initial seeding of distant organs. Mechanistic studies have also been limited by the availability of appropriate models to study NK cell–cancer cell interactions in physiologically realistic 3D settings.
Breast tumors exhibit significant molecular heterogeneity, potentially explaining observed differences in metastatic potential and treatment response (Janiszewska et al., 2019; Marusyk et al., 2012). We previously demonstrated that keratin-14 (K14) defines a subpopulation of breast cancer cells that lead collective invasion, systemic dissemination, and colonization of distant organs (Cheung et al., 2013, 2016; Cheung and Ewald, 2016). In the present study, we used novel ex vivo cocultures and in vivo metastasis models to understand the cellular interactions between NK cells and K14+ cancer cells and to elucidate the molecular mechanisms by which breast cancer cells escape NK cell immunosurveillance to establish distant metastases.
Results and discussion
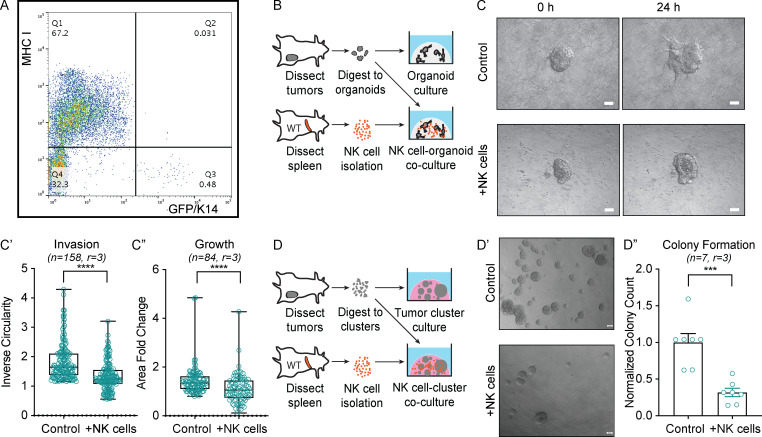
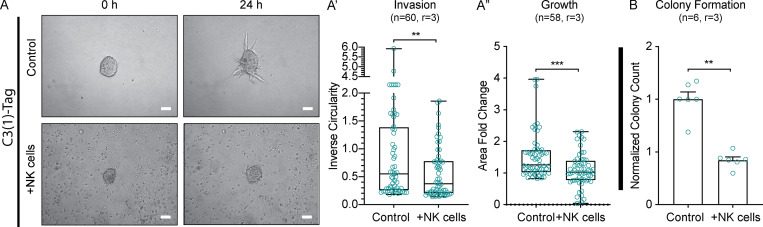
To determine how K14+ cells evade immunosurveillance, we isolated K14− and K14+ cells by FACS from MMTV-PyMT (Guy et al., 1992) tumors with a genetically encoded K14 fluorescent reporter, then stained for major histocompatibility complex (MHC) class I molecules, which are are key inhibitors of NK cell activity (Morvan and Lanier 2016). We observed a striking inverse relationship between K14 status and MHC class I expression, suggesting that K14+ cancer cells are susceptible to NK cell–mediated cytotoxicity (Fig. 1 A). Motivated by this observation, we developed a novel NK cell–tumor organoid ex vivo coculture system (Fig. 1; and Videos 1, 2, 3, and 4). Briefly, freshly isolated NK cells from the spleens of healthy congenic mice were activated with IL-2 or IL-15 and then embedded with organoids derived from mammary tumors in 3D collagen I gels. We used time-lapse differential interface contrast (DIC) microscopy to determine the impact of healthy NK (hNK) cells on invasion in both the MMTV-PyMT and C3(1)-Tag (Maroulakou et al., 1994) models of murine mammary tumors (Figs. 1 C and S1 A). Coculture with hNK cells reduced tumor organoid invasion by ∼40% in PyMT organoids and ∼50% in C3(1)-Tag organoids within the first 24 h of culture (Fig. 1, C and C′; and Fig. S1, A and A′). Organoid growth was decreased 20–30% in both models, suggesting that hNK cells preferentially target cancer cells involved in collective invasion (Figs. 1 C″ and S1 A″). We next sought to model the effect of hNK cells on distant organ seeding, using a colony-forming assay in which two to three cell clusters are embedded in Matrigel (Fig. 1 D; Cheung et al., 2016; Padmanaban et al., 2019). Coculture with hNK cells reduced colony formation by ∼70% in clusters derived from MMTV-PyMT mice and 60% in clusters derived from C3(1)-Tag mice (Fig. 1, D′ and D″; and Fig. S1 B).
Figure 1.
NK cells limit early stages of metastasis in ex vivo models of breast cancer. (A) Dot plot of GFP+ K14+ and K14− tumor cells stained for MHC class I expression. (B) Schema of hNK cell-tumor organoid coculture. Tumor organoids were isolated from dissected MMTV-PyMT mammary tumors, and hNK cells were isolated from the spleens of FVB/n mice. Tumor organoids were cultured alone or in coculture with hNK cells in collagen I gels. (C) Representative DIC images of MMTV-PyMT tumor organoids alone (top) or in coculture with hNK cells (bottom) at 0 and 24 h. Scale bar, 50 µm. (C′ and C″) Boxplots of inverse circularity (C′) and area fold change (C″) of MMTV-PyMT tumor organoids alone or in coculture with hNK cells. Error bars represent 5th to 95th percentile. ****, P < 0.0001 by Mann–Whitney U test. (D) Schema of hNK cell-tumor cluster coculture. Tumor clusters were isolated from dissected MMTV-PyMT mammary tumors, and hNK cells were isolated from the spleens of WT mice. Tumor clusters were cultured alone or in coculture with hNK cells in Matrigel. (D′) Representative DIC images of MMTV-PyMT tumor colonies alone (top) or in coculture with hNK cells (bottom) at 24 h. Scale bar, 50 µm. (D″) Quantification of normalized colony formation count from MMTV-PyMT tumor clusters cultured alone or in coculture with hNK cells. Colony count was normalized to control. Mean is represented; ***, P < 0.001 by Mann–Whitney U test.
Video 1.
Control MMTV-PyMT organoid in monoculture. DIC time-lapse video of a control MMTV-PyMT organoid invading in a collagen I gel. Video frame rate is 10 frames per second.
Video 2.
Coculture of MMTV-PyMT organoid with hNK cells limits invasion. DIC time-lapse video of a MMTV-PyMT organoid in coculture with hNK cells, within a collagen I gel. The hNK cells initially suppress invasion completely, but at later time points, the organoid begins to invade. Video frame rate is 10 frames per second.
Video 3.
Control C3(1)-Tag organoid in monoculture. DIC time-lapse video of a control C3(1)-Tag organoid invading in a collagen I gel. Video frame rate is 10 frames per second.
Video 4.
Coculture of C3(1)-Tag organoid with hNK cells limits invasion. DIC time-lapse video of a C3(1)-Tag organoid in coculture with hNK cells, within a collagen I gel. The hNK cells initially suppress invasion completely, but at later time points, the organoid begins to invade. Video frame rate is 10 frames per second.
Figure S1.
hNK cells limit invasion, growth, and colony formation in the C3(1)-Tag mouse model of breast cancer. (A) Representative DIC images of tumor organoids alone (top) or in coculture with hNK cells (bottom) at 0 and 24 h. Scale bar, 50 µm. (A′ and A″) Boxplot of inverse circularity of tumor organoids alone or in coculture with hNK cells (A′) and area fold change of tumor organoids alone or in coculture with hNK cells (A″). Error bars represent 5th to 95th percentile. **, P < 0.01; ***, P < 0.001 by Mann–Whitney U test. (B) Quantification of normalized colony counts from tumor clusters cultured alone or in coculture with hNK cells. **, P < 0.01 by Mann–Whitney U test.
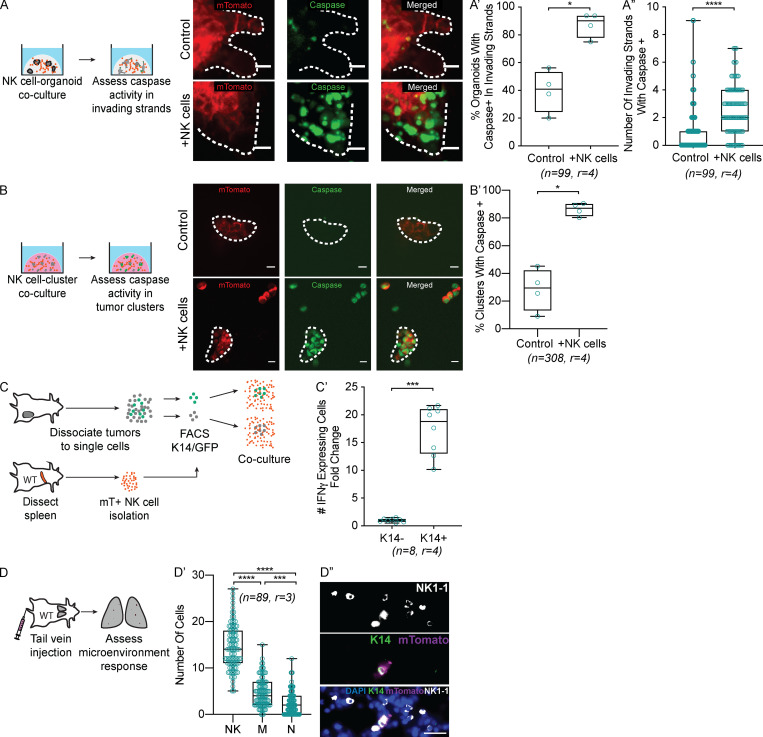
We then sought to identify the cellular mechanism by which hNK cells limit invasion and colony formation, with the hypothesis of direct cytotoxicity. A caspase-3/7 biosensor enabled real-time analysis of apoptosis dynamics during interactions between hNK cells and tumor organoids. We observed caspase 3/7 activity within invasive cancer cells when cocultured with hNK cells during the first 12–24 h (Video 5). In the organoid invasion assays, ∼43% of monoculture organoids and ∼85% of hNK cell cocultured organoids exhibited caspase activity, with multiple caspase biosensor+ invasion strands observed per organoid (Fig. 2, A–A″). Consistent with their MHC class I–negative status, caspase activity localized to K14+ cancer cells within the invasive strand (Fig. S2 A). We next examined apoptosis at 6–8 h in the colony-forming assay and observed caspase+ cells in ∼25% of monoculture and ∼85% of coculture clusters (Fig. 2, B and B′). As before, apoptosis was enriched in K14+ cells within the clusters (Fig. S2 B). Together, these data suggest that hNK cells limit invasion and colony formation through direct cytotoxicity of K14+ cells. As molecular validation of this concept, we cultured hNK cells with either FACS-sorted K14+ or K14− cancer cells and assayed for IFNγ expression (Fig. 2 C). We observed a >17-fold increase of IFNγ-producing cells among hNK cells cocultured with K14+ cells, relative to K14− coculture (Fig. 2 C′). This result provides direct evidence that hNK cells preferentially respond to K14+ cells.
Video 5.
hNK cells activate caspase in invading tumor cells. Live confocal imaging of hNK cells (red) interacting with invading tumor cells (blue, circled), leading to activation of caspase (green) in tumor cells. Scale bar, 20 µm. Video frame rate is 10 frames per second.
Figure 2.
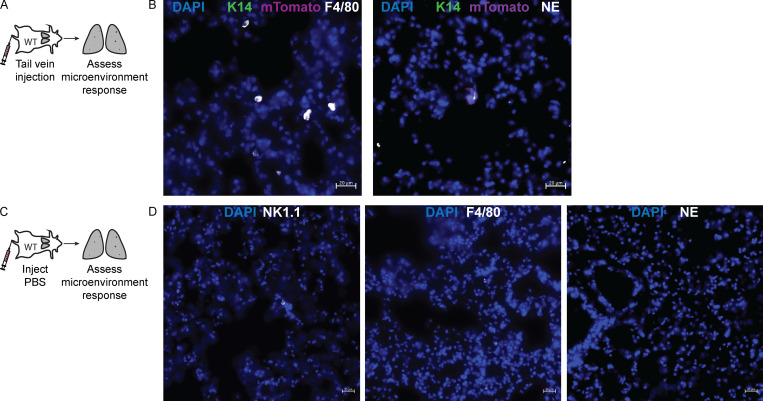
hNK cells induce apoptosis in K14+ invasive breast cancer cells. (A) Representative confocal images of the invading strands of tumor organoids (mTomato+) and caspase activity (green) cultured alone (top) or in coculture with hNK cells (bottom). Scale bar, 10 µm. (A′ and A″) Boxplot of the percentage of organoids with caspase activity in invading strands per biological replicate (A′) and the total number of invading strands with caspase activity (A″) cultured alone or in coculture with hNK cells. *, P < 0.05; ****, P < 0.0001 by Mann–Whitney U test. (B) Representative confocal images of tumor clusters organoids (mTomato+) and caspase activity (green) cultured alone (top) or in coculture with hNK cells (bottom). Scale bar, 10 µm. (B′) Boxplot of the percentage of tumor clusters per biological replicate with caspase activity in tumor clusters cultured alone or in coculture with hNK cells. *, P < 0.05 by Mann–Whitney U test. (C) Schema for assessing IFNγ activity in hNK cells in response to coculture with K14+ or K14− tumor cells. hNK cells were taken from ROSAmT/mG mice and in coculture with K14+ or K14− cells from K14-actin-GFP;MMTV-PyMT mice. In this experiment, hNK cells were fluorescently labeled with mTomato and K14+ cells were labeled with GFP. (C′) Boxplot of IFNγ expression among hNK cells after coculture with K14+ and K14− cells and normalized to K14− cells. Error bars represent 5th to 95th percentile. ***, P < 0.001 by Mann–Whitney U test. (D) Schema for assessment of the innate immune response to an initial metastatic seed. Tumor clusters from the mammary tumors of K14-actin-GFP;MMTV-PyMT;ROSAmT/mG mice were injected into the tail veins of immunocompetent mice, and the lung microenvironment was assessed after 6 h. (D′) Boxplot of the number of NK cells, macrophages, and neutrophils around a metastatic seed. Error bars represent 5th to 95th percentile. ***, P < 0.001; ****, P < 0.0001 by Kruskal–Wallis test. (D″) Representative slide scanned images of lung tissue field of view containing a K14+ (green), metastatic seed (magenta), surrounded by NK cells (NK1.1, white). Scale bar, 20 µm.
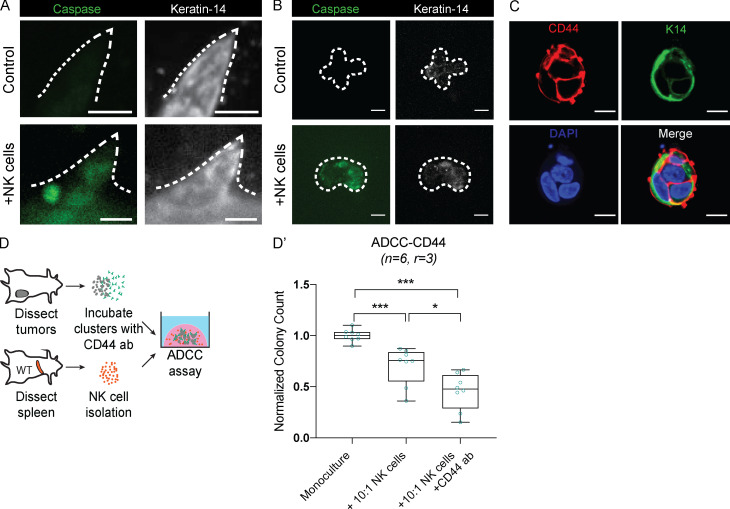
Figure S2.
hNK cells induce caspase activity in K14+ invasive cells, and hNK cell cytotoxicity can be increased by using a CD44 antibody specific to K14+ cells. (A and B) Representative confocal images of tumor organoids (A) and tumor clusters (B) stained for caspase activity (green) and K14 (white) among tumor organoids cultured alone (top) or in coculture with hNK cells (bottom). Scale bar, 10 µm. (C) Representative confocal images of staining tumor cell clusters for CD44 and K14. Scale bar, 10 µm. (D) Schema for the ADCC assay. Tumor clusters were isolated from MMTV-PyMT mammary tumors and incubated with a CD44 antibody before being in coculture with hNK cells at a reduced ratio of 10 NK cells to 1 tumor cell. (D′) Boxplot of the normalized colony count. Error bars represent 5th to 95th percentile. ns, not significant; *, P < 0.05; ***, P < 0.001 by Mann–Whitney U test.
The specificity of NK cells against their target cells can be increased through antibody-dependent cell-mediated cytotoxicity (ADCC), in which NK cell receptors bind to antibodies against specific tumor antigens (Clynes et al., 2000). We therefore hypothesized that hNK cell targeting of K14+ cells could be enhanced with ADCC. Prior RNA-sequencing (RNA-seq) analysis of K14+ cells revealed high expression of the cell surface receptor CD44 relative to K14− cells, and immunofluorescence revealed double-positive (K14+ CD44+) cancer cell clusters (Fig. S2 C). We therefore modified our coculture assay to pretreat clusters with anti-CD44 antibody before coculture with an intermediate concentration of hNK cells (Fig. S2 D). The combination of CD44 pretreatment and hNK cell coculture further decreased colony formation relative to NK cell coculture alone (Fig. S2 D′). Taken together, our results provide functional evidence that hNK cells are selectively targeting K14+ cells and that their efficacy can be increased by ADCC-based strategies.
Having demonstrated that hNK cells can reduce colony formation ex vivo by apoptotic targeting of K14+ cells, we next characterized the acute response of hNK cells to metastatic seeds in vivo. We injected K14-GFP+ mTomato+ cancer cell clusters into immunocompetent mice and assayed NK cell, neutrophil, and macrophage abundance at 6 h (Fig. 2 D). The most frequent responders to K14+ tumor cell cluster were NK cells, whereas few were observed in PBS-treated lungs (Fig. 2, D′ and D″; and Fig. S3, A–D). NK cells are therefore ideally positioned to mediate the early response to the arrival of metastatic cancer cells in distant organs.
Figure S3.
Quantification of macrophage and neutrophil response to early metastatic seeds in the lungs. (A) Schema for assessment of the innate immune response to an initial metastatic seed. Tumor clusters from the mammary tumors of K14-actin-GFP;MMTV-PyMT;ROSAmT/mG mice were injected into the tail veins of immunocompetent mice, and the lung microenvironment was assessed for macrophages and neutrophils after 6 h. (B) Representative slide scanned images of early metastatic seeds staining for F4/80 (macrophages, white) and neutrophil-elastase (neutrophils, white) around K14+ (green) metastatic seeds (magenta). Scale bar, 20 µm. (C) Schema for the control experiment in which PBS was injected into immunocompetent host mice, and the lung microenvironment was assessed for macrophages and neutrophils after 6 h. (D) Representative slide scanned images of staining for NK1.1 (NK cells, white), F4/80 (macrophages, white), and neutrophil-elastase (neutrophils, white) around tumor clusters. Scale bar, 20 µm.
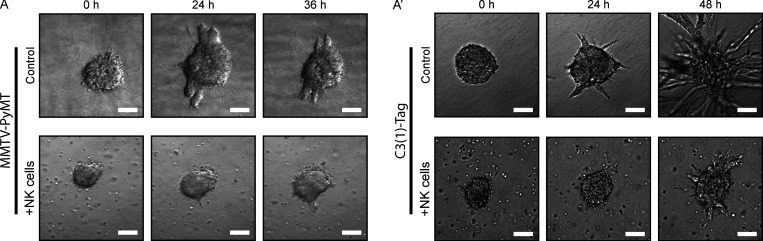
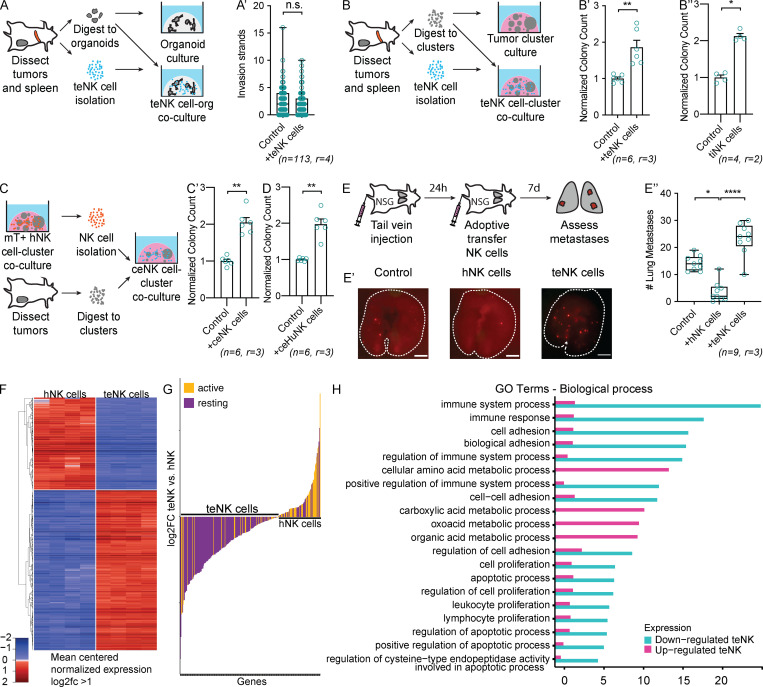
Because MMTV-PyMT and C(3)1-Tag are both highly metastatic (Guy et al., 1992; Maroulakou et al., 1994), their cancer cells must have developed strategies to overcome NK surveillance. Consistent with this concept, we observed that both invading organoids and growing colonies were able to partially overcome hNK cell cytotoxicity by 36–48 h of culture (Fig. S4, A and A′; and Videos 2 and 4). We hypothesized that the cancer cells were reprogramming NK cell differentiation. To test this hypothesis, we isolated NK cells from the spleens of MMTV-PyMT tumor-bearing mice and placed them in coculture with tumor organoids (Fig. 3 A). Tumor-exposed NK (teNK) cells did not limit organoid invasion (Fig. 3 A′). We next placed teNK cells in coculture with tumor cell clusters (Fig. 3 B). Surprisingly, they promoted colony formation by almost twofold (Fig. 3 B′). We used teNKs from the spleen because they can readily be isolated in large numbers. To validate that their behavior was consistent with that of NK cells in contact with tumor cells in vivo, we isolated tumor-infiltrating NK (tiNK) cells from primary tumors and generated cocultures with a similar schema. We observed that tiNK cells promoted colony formation to a similar degree as teNK cells (Fig. 3 B″). These data suggest that cancer cells can reprogram NK cells to support metastatic progression. We next sought to model tumor education of NK cells ex vivo through exposure to tumor clusters. We first cocultured fluorescently labeled hNK cells from control FVB/n mice with cell clusters derived from MMTV-PyMT tumors for 48 h, processed to single cells, used FACS to isolate culture-educated NK (ceNK) cells, then cocultured these ceNKs with new tumor cell clusters (Fig. 3 C). Strikingly, coculture with ceNK increased colony formation by twofold, similar to the effect of freshly isolated teNK cells (Fig. 3 C′). To functionally validate our murine findings in human models and assess whether human NK cells could be educated, we used a parallel schema to Fig. 3 C by culture-educating human NK-92 cells with MCF-7 cell clusters. We found that culture-educated human NK (ceHuNK) cells can also promote colony formation (Fig. 3 D). To test whether teNK cells promote colony formation in vivo, we performed a tail vein assay with fluorescently labeled MMTV-PyMT tumor clusters, followed by adoptive transfer of teNK cells or hNK cells (Fig. 3 E). Consistent with the results of our ex vivo colony-forming assay, hNK cells significantly reduced the number of macrometastases, and teNK cells significantly increased the number of macrometastases relative to hNK cells (Figs. 3, E′ and E″). These results demonstrate that hNK cells can limit metastasis formation and that tumor education can coopt NK cells to promote metastasis.
Figure S4.
Breast cancer organoids are able to overcome hNK cell cytotoxicity over time in 3D culture. (A and A′) Representative tumor organoids isolated from MMTV-PyMT (A) and C3(1)-Tag (A′) mice placed in 3D collagen I alone (top) or in coculture with hNK cells from FBV/n mice (bottom). Although hNK cells are initially able to limit tumor organoid invasion in both models at 24 h, by 36–48 h, tumor organoids are able to invade despite hNK cell activity. Scale bar, 50 µm.
Figure 3.
teNK cells promote colony formation. (A) Schema for teNK cell-tumor organoid coculture. (A′) Boxplot of tumor organoid invasion strands of tumor organoids cultured alone or in coculture with teNK cells. Error bars represent 5th to 95th percentile. ns, not significant by Mann–Whitney U test. (B) Schema for teNK cell-tumor cluster coculture. (B′) Normalized colony count of tumor clusters cultured alone or in coculture with teNK cells. Mean with SEM is represented. **, P < 0.01 by Mann–Whitney U test. (B″) Normalized colony count of tumor clusters cultured alone or in coculture with tiNK cells. Mean with SEM is represented. *, P < 0.05 by Mann–Whitney U test. (C) Schema for generating ceNK cells. (C′) Normalized colony count of MMTV-PyMT tumor clusters cultured alone or in coculture with ceNK cells. Mean with SEM is represented. **, P < 0.01 by Mann–Whitney U test. (D) Normalized colony count of MCF-7 cell clusters cultured alone or in coculture with ceHuNK cells. Mean with SEM is represented. **, P < 0.01 by Mann–Whitney U test. (E) Schema of the adoptive transfer of NK cells following a tail vein injection of cancer cells. (E′) Representative whole-lung images. Macrometastases were identified based on their mTomato expression. Scale bar, 4 mm. (E″) Boxplot of the number of lung macrometastases. Error bars represent 5th to 95th percentile. *, P < 0.05; ****, P < 0.0001 by Kruskal–Wallis test. (F) Heat map displaying z-scores for the variance-stabilized transform of gene expression for differentially expressed genes with absolute value of log2(fold change) >1 between hNK cells and teNK cells. Hierarchical clustering was used to order the genes. (G) Waterfall plot of genes associated with an active and resting NK cell phenotype, expressed by teNK cells and hNK cells. (H) Gene ontology enrichment analysis in “biological process” category for differentially expressed genes up- and down-regulated by teNK cells. Four categories with the lowest P value related to the immune system, metabolic processes, apoptosis, and proliferation are displayed.
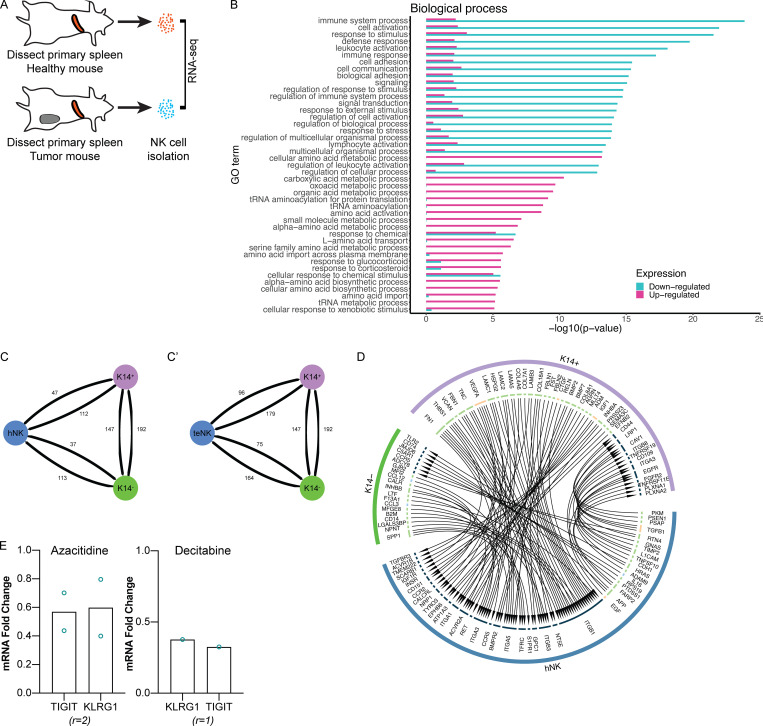
To identify the molecular mechanisms underlying tumor education of NK cells, we performed RNA-seq analysis of hNK cells and teNK cells isolated from FVB/n and MMTV-PyMT mice, respectively (Fig. S5 A). Thresholds of twofold up- or down-regulated and false discovery rate <5% yielded 2,604 differentially expressed genes between teNK cells and hNK cells (Fig. 3 F), with 1,574 genes increased and 1,030 decreased in teNK cells relative to hNK cells (Table S1). We next used the ImmuCC gene signatures (Chen et al., 2018) and deconvoluted the data using CIBERSORT (Newman et al., 2015), a method of resolving relative fractions of cell types from complex mixtures. Using a subset of the gene signatures comprising NK cells, dendritic cells, and neutrophils, >95% of the relative fraction of cells were identified as NK cells (Table S2). Compared with other innate immune cell types, the differential genes expressed by the teNK population corresponded to a resting NK cell phenotype (Fig. 3 G and Table S3). Genes up-regulated by teNK cells had gene ontology annotations relating to the negative regulation of extrinsic apoptosis, and genes down-regulated by teNK cells included those associated with the proliferation, adhesion, and activation of immune response to tumor cells and the positive regulation of apoptotic processes (Figs. 3 H and S5 B).
Figure S5.
RNA-seq analysis of hNK cells and teNK cells reveals differences in identity and biological processes. Receptor–ligand analysis of hNK cells and K14+ or K14− cells reveals interactions between NK cells and cancer cells. Treatment with DNMT inhibitors alter gene expression of inhibitory receptors. (A) Schema for RNA-seq analysis of hNK cells and teNK cells. (B) Gene ontology enrichment analysis in “biological process” category for genes differentially expressed between hNK and teNK cells. 30 categories with the lowest P value associated with up- or down-regulated teNK cells are displayed. (C and C′) Network representation of total receptor–ligand pairs between hNK cells (C) or teNK cells (C′) and K14+ or K14− cells as identified by the databases included in the iTalk algorithm. (D) Relationship map of receptor–ligand pairs of hNK cells and K14+ or K14− cells as identified by the databases included in the iTalk algorithm. (E) Treatment of teNK cells with azacitidine or decitabine alters gene expression of TIGIT and KLRG1 by qPCR. Because of logistical constraints during the COVID-19 pandemic, the assays were able to be conducted only twice and once, respectively.
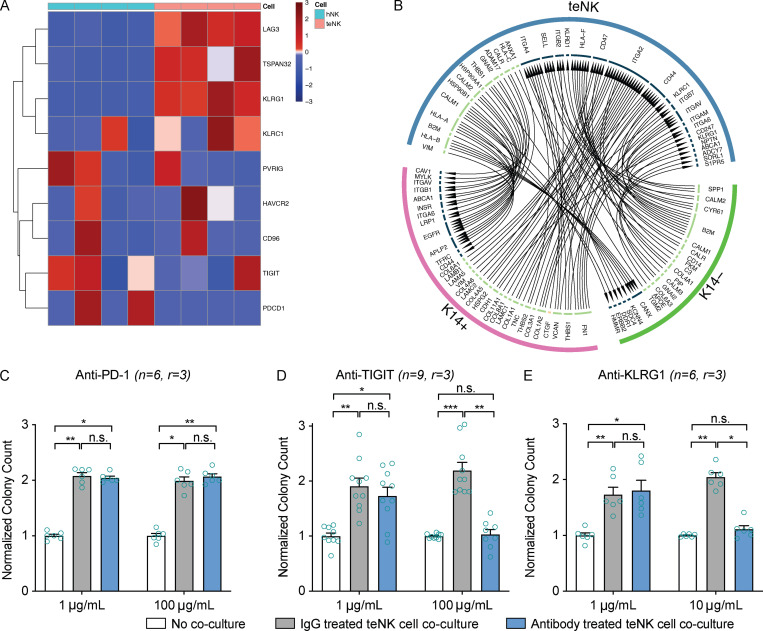
We next sought to identify molecular strategies to reverse the metastasis-promoting effect of teNK cells. Analysis of RNA-seq data revealed increased expression of inactivating receptors in teNK cells compared with hNK cells (Fig. 4 A). We computationally identified ligand–receptor pairs potentially responsible for signaling between NK cells and tumor cells. Applying the R package iTalk (Wang et al., 2019), which includes 2,648 known receptor–ligand pairs, we found ligand–receptor pairings between highly transcribed genes on K14+ and K14− cells and the most significantly expressed genes on teNK cells or hNK cells. Overall, there were slightly more pairings between both types of NK cells and K14+ cells than with K14− cells (Fig. S5, C and C′). Given our prior result showing that K14+ cells increase IFNγ production in hNK cells, we identified ligand–receptor interactions that could lead to IFNγ production on NK cells and apoptosis in K14+ cells. Ligand–receptor pairings that fit these criteria were TNF-related apoptosis-inducing ligand and receptor 2 (TRAILR2–TRAIL), Caveolin-1–HRAS (Harvey rat sarcoma), and TNF receptor associated factor 2–TNF superfamily member 4 on K14+ cells and hNK cells, respectively (Fig. S5 D). Among ligand–receptor pairings on tumor cells and teNK cells, Cdh1 on K14+ cells were suggested to bind to Klrg1 on teNK cells (Fig. 4 B).
Figure 4.
The teNK cell phenotype can be reversed. (A) Heat map of z-scores of gene expression by hNK cells and teNK cells of genes related to NK cell inhibitory signaling. Hierarchical clustering was used to order the genes. (B) Relationship map of receptor-ligand pairs between teNK cells and K14+ or K14− cells as identified by the iTalk algorithm. (C–E) Normalized colony count from antibody-treated control assays and teNK cell–MMTV-PyMT tumor cluster coculture assays. Mean with SEM is represented. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Kruskal–Wallis test.
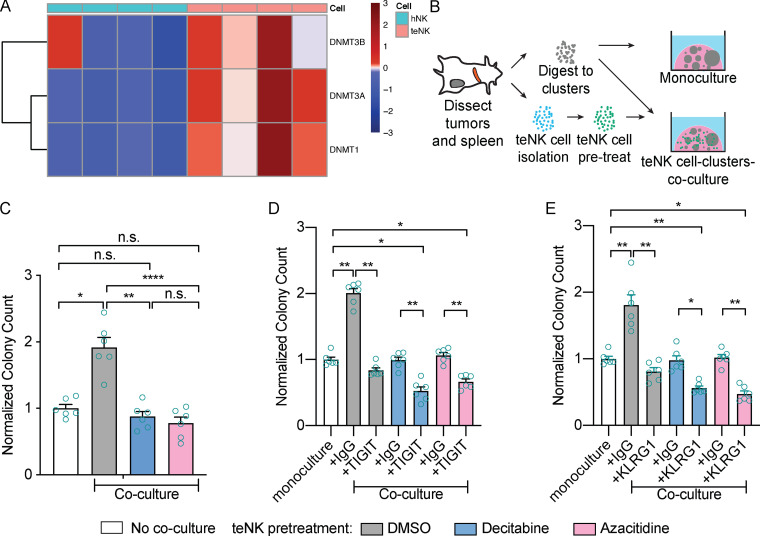
We next investigated the capacity of candidate immunotherapies to reverse the effect of teNK cells on colony formation. Anti-PD-1 therapy has been reported to restore the function of anergic NK cells (Hsu et al., 2018). However, consistent with their limited clinical efficacy in breast cancer (Dirix et al., 2018), treatment with anti-PD-1 antibodies did not limit the colony-promoting effect of teNK cells (Fig. 4 C). We next targeted receptors identified in our RNA expression analysis, specifically T cell immunoreceptor with Ig and ITIM domains (TIGIT) and killer cell leptin-like receptor G1 (KLRG1). TIGIT is an emerging immune checkpoint on NK cells and T cells, and anti-TIGIT therapies are in clinical trials (Zhang et al., 2018). KLRG1, a marker of senescent NK cells and T cells and a recently reported inhibitor of NK cell function (Müller-Durovic et al., 2016), was highly expressed by teNK cells. Strikingly, treatment with either anti-TIGIT or anti-KLRG1 antibodies neutralized the effect of teNK cells and reduced colony formation (Fig. 4, D and E). The broad gene expression differences between hNK and teNK cells motivated us to identify potential epigenetic regulators of NK cell state. We found that DNA methyltransferases (DNMT1, DNMT3a, and DNMT3b) were highly differentially expressed by teNK cells, relative to NK cells (Fig. 5 A). We then sought to validate the functional significance of these findings in our coculture models. To avoid potential tumor-intrinsic impact of DNMT inhibition, we pretreated teNK cells with Food and Drug Administration (FDA)-approved DNMT inhibitors, decitabine and azacitidine, for 24 h before coculture (Fig. 5 B). Pretreatment with DNMT inhibitors also neutralized the effect of teNK cells in coculture (Fig. 5 C) and reduced gene expression of TIGIT and KLRG1 (Fig. S5 E). There is growing evidence of synergy between epigenetic modifiers and immune checkpoint inhibitors (Topper et al., 2019). We hypothesized that the combination of FDA-approved DNAMT inhibitors and anti-TIGIT or anti-KLRG1 would prevent colonies from being formed in coculture. After pretreatment with decitabine and azacitidine, treatment of cocultures with anti-TIGIT (Fig. 5 D) or anti-KLRG1 (Fig. 5 E) blocking antibodies reduced the number of colonies below the monoculture controls.
Figure 5.
Pretreatment of teNK cells with FDA-approved DNMT inhibitors neutralizes the teNK cell phenotype. (A) Heat map of z-scores of gene expression by hNK cells and teNK cells of genes related to DNMTs. Hierarchical clustering was used to order the genes. (B) Schema of pretreatment of teNK cells before coculture with tumor clusters. (C) Normalized colony count from DMSO control or DNMT inhibitor pretreated teNK cell–MMTV-PyMT tumor cluster coculture assays versus monoculture controls. Mean with SEM is represented. n.s., not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by Mann–Whitney U test. (D and E) Normalized colony count from DMSO or DNMT inhibitor pretreated teNK cells and antibody treated monoculture control assays and teNK cell–tumor cluster coculture assays. Mean with SEM is represented. *, P < 0.05; **, P < 0.01 by Mann–Whitney U test.
In this study, we sought to understand the cellular and molecular basis of NK cell–cancer cell interactions during breast cancer metastasis. Prior work from our laboratory revealed that clusters of K14+ cancer cells pioneer collective invasion and distant metastasis across breast cancer subtypes (Cheung et al., 2013, 2016; Cheung and Ewald, 2016) and that K14 expression correlates with quantitative measures of invasion in human breast tumor samples (Padmanaban et al., 2020). Here we show that NK cells are the most abundant early responder to cancer cell clusters in the lung, that K14+ cancer cells typically lack MHC class I expression, and that hNK cells preferentially target K14+ cancer cells for cytotoxic death. Our varied 3D culture assays revealed that hNK coculture induced a mild reduction in tumor organoid growth, moderate reduction in organoid invasion, and a strong reduction in colony formation in culture and metastatic outgrowth in vivo. The effect on colony formation was increased further by treatment with antibodies targeting cell surface receptors on K14+ cells, presumably through ADCC. The relative impact of NK cells on cancer progression was most pronounced in assays modeling later stages of metastasis, similar to those seen in the adjuvant breast cancer setting. These observations support the view of NK cells as negative regulators of metastasis (López-Soto et al., 2017; Krasnova et al., 2017).
It remained unclear how metastases ever emerged from NK surveillance to reach clinical significance. Our first clue came toward the end of long time-lapse videos of NK cell–cancer cell cocultures. After an initial period of complete control, the cancer cells eventually began to grow and invade. This observation inspired us to discover that teNK cells isolated from the spleens of tumor-bearing mice were able to promote colony formation in vitro and metastasis formation in vivo. We then validated that tiNK cells isolated from the primary tumor similarly promoted colony formation in vitro and that we could replicate the NK education process in culture with either murine or human NK cells. Our demonstration that cancer cells can directly reprogram NK cells to promote metastatic colony formation adds to the concept of NK cell plasticity (Gao et al., 2017) and to prior mechanisms of evasion of NK cell surveillance (Cherfils-Vicini et al., 2019). Just as other immune cells demonstrate a range of molecular and functional phenotypes that are guided by specific environmental contexts, we have shown that exposure to metastatic seeds is sufficient to alter the transcriptomic and functional state of NK cells.
Because NK cell function is regulated at the level of inhibitory and activating cell surface receptors, we conducted bulk RNA-seq comparisons of the transcriptomes of hNK and teNK cells. We observed up-regulation of inhibitory receptors and a shift toward a resting NK state in teNK cells. We next demonstrated that antibodies directed toward either TIGIT or KLRG1 were sufficient to eliminate the metastasis-promoting effects of teNK cells and that the combination of DNMT inhibition and anti-TIGIT or anti-KLRG1 antibody treatment further reduced metastatic potential. To our knowledge, we are the first to demonstrate that cancer cells can convert NK cells to an alternative metastasis-promoting cell state and the first to demonstrate KLRG1’s role in NK cell cooption. Further, the synergistic effects of epigenetic modification with inhibitory receptor blockade suggests a viable clinical strategy to activate teNK cells to target and eliminate breast cancer metastases.
Beyond the specific conclusions of the study, our novel ex vivo models combine primary metastatic breast cancer cells with primary NK cells to allow investigators to capture the cellular and molecular dynamics of immune control and immune escape. These models should be readily adaptable to other cancer types and offer a scalable way to identify new molecular targets for NK cell–directed immunotherapies, test combination therapies, and inform their potential clinical utilization. Furthermore, our analysis of receptor–ligand pairing between teNK cells and tumor cells suggests a diverse range of additional novel drug targets for further validation. Our results suggest that antibodies targeting NK inhibitory receptors could be effective in eliminating metastatic breast cancer cells, either alone or in combination with epigenetic therapies. Combined with our observation that NK cells are abundant early responders to disseminated breast cancer cells, our data provide preclinical rationale for the concept of NK cell–directed immunotherapies in the adjuvant setting for breast cancer patients with high risk of metastatic recurrence.
Materials and methods
Mouse lines and breeding
All mouse husbandry and procedures performed were in accordance with an animal protocol approved by the Johns Hopkins School of Medicine Animal Care and Use Committee. All study mice were female and were backcrossed and maintained on the FVB/n background. FVB/N-Tg(MMTV-PyVT)634Mul/J (MMTV-PyMT) (Guy et al., 1992), FVB-Tg(C3-1-TAg)cJeg/JegJ (C3(1)-Tag) (Maroulakou et al., 1994), B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (ROSAmT/mG), and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were obtained from the Jackson Laboratory. K14-actin-GFP mice were a generous gift from Elaine Fuchs (Rockefeller University, New York, NY). For sorting of K14+ cells, MMTV-PyMT mice were crossed with K14-GFP-actin mice.
Tumor organoid and tumor cluster isolation
Primary tumor organoids were isolated from murine mammary tumors by stepwise mechanical disruption, enzymatic digestion, and differential centrifugation according to our published protocols (Nguyen-Ngoc et al., 2015). Briefly, tumors were harvested from MMTV-PyMT or C3(1)-Tag mice with 20-mm tumors, mechanically disrupted with a scalpel, and digested on a shaker for 1 h at 37°C in collagenase solution (DMEM-F12 [10565-018; Gibco Life Technologies] with 2 mg/ml collagenase [C2139; Sigma-Aldrich], 2 mg/ml trypsin [27250-018; Gibco Life Technologies], 5% [vol/vol] FBS [F0926; Sigma-Aldrich], 5 µg/ml insulin [I9278; Sigma-Aldrich], and 50 µg/ml gentamicin [15750; Gibco Life Technologies]). The suspension was centrifuged at 400 g to remove cellular debris and undigested tissue, and the pellet was treated with 2 U/µl DNase (D4263; Sigma-Aldrich). The epithelial organoids were enriched and separated from stromal cells by a series of differential centrifugations, after which organoids of ∼100–500 epithelial cells were obtained. Tumor cell clusters of two to five cells were obtained by taking organoids and further digesting them with 1× TrypLE (12604013; Thermo Fisher Scientific) for 10 min at 37°C. Cells were resuspended in PBS (−Ca2+, −Mg2+), filtered through a 40-µm filter, and resuspended again at a density of 2 × 106 cells/ml.
NK cell isolation
Primary splenocytes were isolated by mechanical dissociation from spleens of WT FVB/n, MMTV-PyMT, or C3(1)-Tag mice or from the tumors of MMTV-PyMT mice. NK cells isolated from WT FVB/n mice were defined as hNK cells, and those isolated from tumor-bearing mice were defined as teNK cells. NK cells isolated from MMTV-PyMT tumors were defined as tiNK cells. The NK cells were purified from isolated splenocytes using EasySep Mouse NK Cell Isolation Kit per the manufacturer’s protocol (19855; StemCell Technologies). The isolated cells were then positively selected by FACS by gating on CD49b+/CD3− cells and CD45+/CD49b+/CD3− for NK cells isolated from tumors. NK cells were cultured in medium at 37°C containing 200 U/ml recombinant mouse IL-2 (402-ML-100; R&D Systems), 5 ng/ml recombinant mouse IL-15 (447-ML-010; R&D Systems), or 5 ng/ml recombinant mouse IL-15:IL-15R complex (14-8152-62; Thermo Fisher Scientific) for 16 h before use in assays. Pretreatment of teNK cells was performed with decitabine and azacitidine at 1-µM doses for 24 h (NCI Development Therapeutics Program).
NK cell–organoid and NK cell–tumor cluster coculture
Organoids were embedded at a density of 1.5 organoids/µl with 30:1 or 10:1 hNK or teNK cells (NK:tumor cells) into neutralized, fibrillar rat-tail collagen I (354236; Corning) onto 24- or 96-well glass-bottomed plates over a 37°C heating block to generate a NK cell–organoid coculture. Tumor clusters were embedded at a density of 100 clusters/µl into Matrigel (354230; BD Biosciences) onto 24-well or 96-well glass bottomed plates over a 37°C heating block to generate NK cell-tumor cluster coculture. Human NK cell-tumor cluster cocultures were generated by embedding human NK-92 cells (ATCC CRL-2408) and MCF7 tumor cell clusters (ATCC HTB-22) into Matrigel. Gels were allowed to polymerize at 37°C for 1 h, after which medium containing RPMI 1690 (11875093; Gibco), 1% insulin-transferrin-selenium (51500-056; Gibco), 1% penicillin-streptomycin (P4333; Sigma-Aldrich), 2.4 nM FGF2 (F0291; Sigma-Aldrich) and supplemented with 200 U/ml recombinant mouse IL-2, 5 ng/ml recombinant mouse IL-15, or 5 ng/ml recombinant mouse IL-15:IL-15Ra complexes, was added to the wells. ceNK cells were produced by first generating a mTomato+ hNK cell–tumor cluster coculture for 48 h. ceHuNK cells were generated by first generating RFP+ human NK-92 by incubating cells with CellTracker Red (C34552; Thermo Fisher Scientific) for 24 h before coculture. Coculture gels were then digested by pipetting in cold PBS-EDTA buffer, comprising PBS and 20 mM EDTA. The solubilized matrix-NK cell mix was centrifuged for 5 min at 400 g, 4°C, and washed with PBS, and NK cells were isolated by FACS.
ADCC assay
ADCC assays were performed using tumor clusters isolated from MMTV-PyMT mice, and cocultures were formed with hNK cells at an intermediate 10:1 NK:tumor cell ratio. Tumor clusters were incubated with 5 µg/ml of CD44 antibody (14-0441-82; eBioscience) or medium alone for another 30 min at 37°C and washed twice with medium to remove unbound antibodies. Colony formation was assessed at the end of 24 h.
DIC
DIC imaging was performed using a LD Plan-Neofluar 20×/0.4 Korr Ph2 objective lens, a Zeiss AxioObserver Z1, and an AxioCam MRM camera. DIC time-lapse videos were collected at 15-min acquisition intervals for 24 h, maintaining temperature at 37°C and CO2 at 5%.
Quantification of organoid invasion, organoid growth, and colony formation
Invasion of organoids was quantified by manually tracing organoid boundaries from DIC images and measuring the circularity with ImageJ (National Institutes of Health). Circularity is defined as the ratio of the square of the perimeter to 4π times the area. A circle, which has the maximum area for a given perimeter, has a value of 1; organoids with multiple invasive strands have much higher values. Inverse circularity was graphed to provide a relationship between increased invasion and increased inverse circularity (Figs. 1 C′ and S1 A′′). For growth, paired images for each organoid were obtained at 0 and 24 h after plating, and growth was represented as a fold change in projected area in 24 h. Colony formation was performed by counting all tumor colonies in each well across multiple Z planes. Fig. 1 D′ is a representation of colonies counted.
K14+-tumor cell/NK coculture assay
Single-cell suspensions of K14+ and K14− cells were generated from MMTV-PyMT;K14-actin-GFP mammary tumor organoids and separated by FACS. Equal numbers of K14+ or K14− cells were placed in suspension with hNK cells isolated from ROSAmTmG mice at a 10:1 tumor:NK cell ratio for 4 h at 37°C. Samples were fixed and permeabilized using BD CytoFix/CytoPerm (554714; BD Biosciences) and analyzed by flow cytometry for IFNγ.
Flow cytometry and cell sorting
MMTV-PyMT; K14− actin-GFP organoids were harvested for tumor and subsequently dissociated into single-cell suspensions using TrypLE. The GFP+/K14+ cells were analyzed and sorted using either a MoFlo Legacy or XDP Cytometer (Beckman Coulter). Data acquisition and sorting were performed with Summit software. hNK cells were isolated as a CD3−/CD49b+/mTomato+ cell population from the spleens of ROSAmTmG mice. Propidium iodide or DAPI was used as a viability marker for sorting. FCS files generated from FACS experiments were reanalyzed by FlowJo software for data representation.
Tail vein assays
Mammary tumor organoids from MMTV-PyMT;ROSAmTmG were trypsinized into small clusters using TrypLE (Thermo Fisher Scientific) at 37°C for 10 min. Cells were resuspended in DMEM-F12 at a concentration of 106 cells/ml. Host FVB/n and NSG mice for these experiments were 6–10-wk-old female mice. The tail veins of these mice were dilated by exposure to a heat lamp for ∼1 min. Using a 26.5-gauge needle, 200 µl cells was injected via the tail vein of the mouse. In NSG mice, tail vein assays were followed by adoptive transfer of 500,000 NK cells in 200 µl of PBS on the next day. Cytokine support with 1 µg recombinant mouse IL-15:IL-15Ra complexes (14-8152-62; Thermo Fisher Scientific) was delivered intraperitoneally for 3 d. Lungs from mice were collected 7 d from the date of injection and examined for macrometastases. Macrometastases were counted based on expression of mTomato+ under the dissection microscope. Representative images in Fig. 3 E′ were collected using an iPhone X.
Immunofluorescence
Organoid cocultures and tumor clusters cultured in 3D collagen I were fixed using 4% PFA (15714S; Electron Microscopy Sciences), and colonies in Matrigel were fixed using 1% PFA for 10 min. Residual formaldehyde was washed two times in Dulbecco's Phosphate-Buffered Saline. The gels were permeabilized using 0.5% Triton X-100 (X100-500ML; Sigma-Aldrich) for 30 min at room temperature. Samples were blocked for 2 h with 10% FBS/1% BSA/0.2% Triton X-100/PBS at room temperature, incubated with primary antibodies diluted in 1% FBS/1% BSA/0.2% Triton X-100/PBS overnight at 4°C, then washed three times for 10 min each using PBS. Secondary antibodies were added diluted in 1% FBS/1% BSA/0.2% Triton X-100/PBS and incubated for 3 h at room temperature. Samples were washed three times with PBS and stored at 4°C until imaged. Confocal imaging was conducted on a spinning disk microscope (Solamere Technology Group) with a Prime 95B scientific complementary-symmetry metal–oxide–semiconductor camera (Photometrics). An LD Plan-Neofluar 20×/0.4 Korr Ph2 objective lens (Zeiss) was used for single and time-lapse image acquisition. For time-lapse imaging, images were acquired at 10- or 15-min intervals. For z stacks, 2-µm spacing was used. Acquisition of both time-lapse and still images was performed using μManager (Edelstein et al., 2010).
For confocal videos, hNK cells were isolated from ROSAmTmG mice, tumor cell nuclei were labeled using Sir-DNA (CY-SC007; Cytoskeleton), and caspase activity was assessed using CellEvent Caspase-3/7 Green Detection Reagent (C10423; Thermo Fisher Scientific). Images were collected every 10 min using 2-µm spacing for 24 h under standard incubation conditions. Videos were collected in parallel using one to three channels (excitation at 488, 561, and 647 nm). Imaris 8 (Bitplane) was used to analyze videos, export individual TIFFs, and adjust brightness and contrast of the images in each channel to maximize the clarity. ImageJ and Imaris were used to adjust brightness and contrast of the images in each channel to maximize clarity, place scale bars, and export as TIFFs. Image adjustments were always made across entire images.
Before lung harvest, cardiac perfusion with PBS and 1% PFA was performed. Lungs from tail vein assays were then harvested and fixed in 1% PFA for 4 h at 4°C. Fixed tumor cells and lungs were transferred into 25% sucrose/PBS overnight at 4°C, embedded into optimal cutting temperature compound (4583; TissueTek) and frozen at −80°C. Sections (20 µm thick) were cut onto Superfrost Plus Gold Microscope slides (15-188-48; Thermo Fisher Scientific) at −20°C using a cryostat. The OCT was removed from these slides by incubating with PBS for 1 h at room temperature. The sections were processed and stained identical to gel embedded cocultures, covered with #1.5 High Precision Cover Glasses (CG15KH; Thor Labs), and images were acquired using a Zeiss Axio Scan.Z1 and analyzed with Zen Imaging software.
Antibodies
Primary antibodies used include anti-NK1.1 (1:200; 553162; BD Biosciences), anti-F4/80 (1:200; 123122; BioLegend), anti-neutrophil-elastase (1:200; ab68672; Abcam), anti-K14 (1:200; PRB-155P; Covance), anti-CD49b (1:200; StemCell Technologies), anti-CD3 (1:200; StemCell Technologies), anti-CD44 (1:200; 14-0441-82; eBioscience), anti-IFN-γ (1:100; 11-7311-82; Thermo Fisher Scientific), and DAPI (1:1,000; D571; Invitrogen). All secondary antibodies used were Alexa Fluor conjugates (1:200; Invitrogen). Treatment of monocultures and cocultures described in the main text were performed with anti-TIGIT antibody (clone 1G9, BE0274; BioXCell) and anti-KLRG1 antibody (clone 2F1, 16-5893-82; Thermo Fisher Scientific).
RNA extraction and quantitative PCR (qPCR)
RNA was extracted with the RNeasy Mini Kit (74104; Qiagen) following the manufacturer’s protocol. cDNA was synthesized from 100 ng total RNA using the SuperScript IV VILO Master Mix (11766050; Thermo Fisher Scientific). Synthesized cDNA was diluted in RNAase-free water before RT-qPCR. qPCR was conducted using the SsoAdvanced Universal SYBR Green Supermix (1725271; Bio-Rad) with 500 pg cDNA and 500 nM primers per reaction. Reactions were run in triplicate on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Target gene expression values were normalized to GAPDH expression, and fold change was calculated as 2-ΔΔCt. RT-qPCR primers used were KLRG1 forward, 5′-GCTCACATCTCCTTACATTTCCG-3′, and reverse, 5′-TCCTCAAGCCGATCCAGTA-3′; and TIGIT forward, 5′-CTGCCTTCCTCGCTACAG-3′, and reverse, 5′-GTAAGATGACAGAGCCACCTTC-3′.
High-throughput RNA-seq and gene-set analysis
For each replicate, primary hNK cells and teNK cells were isolated and cultured in cytokine-supplemented medium for 12–16 h. The cells were then snap-frozen, and total RNA was extracted using RNeasy (74104; Qiagen). The sequencing library was prepared using the TruSeq Stranded mRNA Sample Kit (RS-122-2101; Illumina). Briefly, ≥400 pg of total RNA isolated per sample was converted to double-stranded cDNA, end-repaired, A-tailed, and ligated with Illumina indexed adapters. The PCR amplified library was purified using Agencourt RNAClean XP (A63987; Beckman Coulter) magnetic beads and run on an Agilent High Sensitivity DNA Chip for quality check. The library was then sequenced in a paired-end 150-bp cycle using Illumina NextSeq 500 (The Johns Hopkins School of Medicine Deep Sequencing and Microarray Core Facility).
The mouse genome was obtained in FASTA format (GRCm38) from Ensembl version 95 and gene set annotation in GTF format. The hisat2 indices were built from the genome index using hisat2-build (Kim et al., 2015) from Hisat2 version 2.1.0. Raw RNAseq paired-end reads were aligned to the genome using hisat2 and trimming the first base (using optional flag −5 1). The total reads per sample ranged from 37 to 62 million, and the alignment mapping rate was 88–90%. The DNA fragments had been labeled with unique molecular identifiers. NuDup (v2.3) was used to mark the unique molecular identifiers and keep the read pair with the highest mapping quality. HTSeq was used to count reads mapping to individual genes by processing the sorted bam files with accepted reads (Anders et al., 2015).
We next used DESeq2 (Love et al., 2014) to estimate differential gene expression between hNK and teNK cells from the counts generated by HTSeq. Using standard DESeq2 parameters genes with no reads were excluded, and those with P values set to the nominal value of 1 were removed. Additionally, we removed all genes with ≤10 total reads.
External databases
Gene ontology
Gene ontology pathway analysis was performed using the R package goseq (Young et al., 2010).
ImmuCC and CIBERSORT
The mouse-specific immune cell signatures from ImmuCC (Chen et al., 2018), based on the expression of 511 genes, were obtained and loaded them along with the list of genes differentially expressed between teNK and hNK into CIBERSORT (Newman et al., 2015). CIBERSORT uses signature expression profiles for cells to deconvolute the gene expression from a mixture of cells. The algorithm quantifies relative levels of distinct cell types.
iTALK software package
iTALK (Wang et al., 2019) is an R package based on 2,648 unique receptor–ligand pairs. Using the previously published RNA-seq data for K14+ and K14− cells (Cheung et al., 2016) with mean expression >10 and the differentially expressed genes identified in this study, the algorithm identified 648 receptor–ligand pairs between hNK and K14+ and K14− cells and 853 pairs between teNK and K14+ and K14− cells. We ordered the receptor–ligand pairs based on the product of the mean expression for each cell type.
Statistical analysis
All statistical tests were performed in Prism (GraphPad). All tests used are described in the figure legends. P values <0.05 were considered significant.
Data availability
Next-generation RNA sequencing data is available on NCBI GEO (accession no. GSE152790). Any additional information may be provided by the corresponding author upon request.
Code availability
Most bioinformatic analyses relied on publicly available tools, as specified above. The limited custom code used is available on GitHub (https://github.com/baderzone/nk_2020).
Online supplemental material
Fig. S1 demonstrates that hNK cells limit invasion, growth, and colony formation in the C3(1)-Tag mouse model of breast cancer. Fig. S2 demonstrates that NK cells limit invasion and colony formation by inducing apoptosis in K14+ invasive breast cancer cells. Further, hNK cell cytotoxicity can be increased by using a CD44 antibody specific to K14+ cells. Fig. S3 shows quantification of macrophage and neutrophil response to early metastatic seeds in the lungs. Fig. S4 demonstrates that breast cancer organoids are able to overcome hNK cell cytotoxicity over time in 3D culture. Fig. S5 shows that RNA-seq analysis of hNK cells and teNK cells reveals differences in identity and biological processes. Receptor–ligand analysis was performed between hNK cells and K14+ or K14− cells. Treatment with azacitidine reduced the gene expression of KLRG1 and TIGIT. Table S1 shows genes differentially expressed by teNK cells relative to hNK cells. Table S2 shows CIBERSORT deconvolution analysis of genes expressed by hNK cells and teNK cells using immune cell signatures from ImmuCC. Table S3 shows genes from ImmuCC used to determine active and resting NK phenotypes. The relative fraction of cells from hNK cell and teNK cell subset are consistent with gene signatures related to NK cells. Video 1 is a DIC time-lapse video of a MMTV-PyMT organoid in monoculture. Video 2 is a DIC time-lapse video of a MMTV-PyMT organoid in coculture with hNK cells. Video 3 is a DIC time-lapse video of a C3(1)-Tag organoid in monoculture. Video 4 is a DIC time-lapse video of a C3(1)-Tag organoid in coculture with hNK cells.Video 5 shows live confocal imaging of hNK cells activating caspase in invading tumor cells.
Supplementary Material
lists genes differentially expressed by teNK cells relative to hNK cells.
shows CIBERSORT deconvolution analysis of genes expressed by hNK cells and teNK cells using immune cell signatures from ImmuCC: the relative fraction of cells from hNK cell and teNK cell subset are consistent with gene signatures related to NK cells.
lists genes from ImmuCC used to determine active and resting NK phenotypes.
Acknowledgments
We thank Dr. Sudipto Ganguly (Johns Hopkins University, Baltimore, MD) for generously providing anti-TIGIT antibody for the drug experiments. We thank Haiping Hao from the Microarray and Deep Sequencing Core Facility for technical assistance.
A.J. Ewald received support for this project through grants from the Breast Cancer Research Foundation/Pink Agenda (BCRF-19-048), Twisted Pink, Hope Scarves, and the National Institutes of Health/National Cancer Institute (U01CA217846, U54CA2101732, 3P30CA006973). J.S. Bader received support for this project through a grant from the National Institutes of Health/National Cancer Institute (U01CA217846). I.S. Chan received support from National Institutes of Health/National Cancer Institute (T32CA009071) and the Conquer Cancer Foundation Young Investigator Award. Both A.J. Ewald and J.S. Bader received support from the Jayne Koskinas Ted Giovanis Foundation for Health and Policy and the Breast Cancer Research Foundation, private foundations committed to critical funding of cancer research. The opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and not necessarily those of the Jayne Koskinas Ted Giovanis Foundation for Health and Policy or the Breast Cancer Research Foundation, or their respective directors, officers, or staffs.
E.M. Jaffee receives grant funding from AduroBiotech, Bristol Myers Squibb, and Amgen and serves on advisory boards for CSTONE, Adaptive Biotech, DragonFly, and Genocea. Through a licensing agreement between JHU and AduroBiotech, both JHU and E.M. Jaffee have the potential to receive royalty payments in the future. J.S. Bader is a founder and director of Neochromosome and a member of the scientific advisory board of AI Therapeutics. A.J. Ewald’s spouse is an employee of ImmunoCore. A.J. Ewald and V. Padmanaban are listed as inventors on a patent application related to the use of antibodies as cancer therapeutics. A.J. Ewald is an inventor of a patent related to the use of K14 as a prognostic indicator for breast cancer outcomes.
Author contributions: I.S. Chan and A.J. Ewald conceptualized the project and designed the experiments. I.S. Chan, G. Ramakrishnan, V. Padmanaban, J.C. Ramirez, M. Dunworth, and H. Zhang performed the experiments, and I.S. Chan performed data analysis. I.S. Chan, E.M. Jaffee, and A.J. Ewald contributed to the interpretation of the experimental results. H. Knútsóttir, M. Warrier, and J.S. Bader performed RNA-seq analysis. I.S. Chan, H. Knútsóttir, J.S. Bader, and A.J. Ewald contributed to the interpretation of the sequencing data. I.S. Chan and A.J. Ewald wrote the manuscript with useful input from all authors.
References
- Adams S., Gatti-Mays M.E., Kalinsky K., Korde L.A., Sharon E., Amiri-Kordestani L., Bear H., McArthur H.L., Frank E., Perlmutter J., et al. . 2019. Current Landscape of Immunotherapy in Breast Cancer: A Review. JAMA Oncol. 5:1205 10.1001/jamaoncol.2018.7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., and Huber W.. 2015. HTSe—a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Quan L., Huang A., Zhao Q., Yuan Y., Yuan X., Shen Q., Shang J., Ben Y., Qin F.X., et al. . 2018. seq-ImmuCC: Cell-Centric View of Tissue Transcriptome Measuring Cellular Compositions of Immune Microenvironment From Mouse RNA-Seq Data. Front. Immunol. 9:1286 10.3389/fimmu.2018.01286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils-Vicini J., Iltis C., Cervera L., Pisano S., Croce O., Sadouni N., Győrffy B., Collet R., Renault V.M., Rey-Millet M., et al. . 2019. Cancer cells induce immune escape via glycocalyx changes controlled by the telomeric protein TRF2. EMBO J. 38 e100012 10.15252/embj.2018100012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., and Ewald A.J.. 2016. A collective route to metastasis: Seeding by tumor cell clusters. Science. 352:167–169. 10.1126/science.aaf6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Gabrielson E., Werb Z., and Ewald A.J.. 2013. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 155:1639–1651. 10.1016/j.cell.2013.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K.J., Padmanaban V., Silvestri V., Schipper K., Cohen J.D., Fairchild A.N., Gorin M.A., Verdone J.E., Pienta K.J., Bader J.S., et al. . 2016. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc. Natl. Acad. Sci. USA. 113:E854–E863. 10.1073/pnas.1508541113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes R.A., Towers T.L., Presta L.G., and Ravetch J.V.. 2000. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat. Med. 6:443–446. 10.1038/74704 [DOI] [PubMed] [Google Scholar]
- Dirix L.Y., Takacs I., Jerusalem G., Nikolinakos P., Arkenau H.T., Forero-Torres A., Boccia R., Lippman M.E., Somer R., Smakal M., et al. . 2018. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast Cancer Res. Treat. 167:671–686. 10.1007/s10549-017-4537-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N.. 2010. Computer control of microscopes using microManager. Curr. Protoc. Mol. Biol. Chapter 14, Unit 14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Souza-Fonseca-Guimaraes F., Bald T., Ng S.S., Young A., Ngiow S.F., Rautela J., Straube J., Waddell N., Blake S.J., et al. . 2017. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat. Immunol. 18:1004–1015. 10.1038/ni.3800 [DOI] [PubMed] [Google Scholar]
- Guy C.T., Cardiff R.D., and Muller W.J.. 1992. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12:954–961. 10.1128/MCB.12.3.954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J., Hodgins J.J., Marathe M., Nicolai C.J., Bourgeois-Daigneault M.C., Trevino T.N., Azimi C.S., Scheer A.K., Randolph H.E., Thompson T.W., et al. . 2018. Contribution of NK cells to immunotherapy mediated by PD-1/PD-L1 blockade. J. Clin. Invest. 128:4654–4668. 10.1172/JCI99317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiszewska M., Tabassum D.P., Castaño Z., Cristea S., Yamamoto K.N., Kingston N.L., Murphy K.C., Shu S., Harper N.W., Del Alcazar C.G., et al. . 2019. Subclonal cooperation drives metastasis by modulating local and systemic immune microenvironments. Nat. Cell Biol. 21:879–888. 10.1038/s41556-019-0346-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S.L.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12:357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnova Y., Putz E.M., Smyth M.J., and Souza-Fonseca-Guimaraes F.. 2017. Bench to bedside: NK cells and control of metastasis. Clin. Immunol. 177:50–59. 10.1016/j.clim.2015.10.001 [DOI] [PubMed] [Google Scholar]
- López-Soto A., Gonzalez S., Smyth M.J., and Galluzzi L.. 2017. Control of Metastasis by NK Cells. Cancer Cell. 32:135–154. 10.1016/j.ccell.2017.06.009 [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malladi S., Macalinao D.G., Jin X., He L., Basnet H., Zou Y., de Stanchina E., and Massagué J.. 2016. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell. 165:45–60. 10.1016/j.cell.2016.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroulakou I.G., Anver M., Garrett L., and Green J.E.. 1994. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc. Natl. Acad. Sci. USA. 91:11236–11240. 10.1073/pnas.91.23.11236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A., Almendro V., and Polyak K.. 2012. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer. 12:323–334. 10.1038/nrc3261 [DOI] [PubMed] [Google Scholar]
- Morvan M.G., and Lanier L.L.. 2016. NK cells and cancer: you can teach innate cells new tricks. Nat. Rev. Cancer. 16:7–19. 10.1038/nrc.2015.5 [DOI] [PubMed] [Google Scholar]
- Müller-Durovic B., Lanna A., Covre L.P., Mills R.S., Henson S.M., and Akbar A.N.. 2016. Killer Cell Lectin-like Receptor G1 Inhibits NK Cell Function through Activation of Adenosine 5′-Monophosphate-Activated Protein Kinase. J. Immunol. 197:2891–2899. 10.4049/jimmunol.1600590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., and Alizadeh A.A.. 2015. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12:453–457. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc K.V., Shamir E.R., Huebner R.J., Beck J.N., Cheung K.J., and Ewald A.J.. 2015. 3D culture assays of murine mammary branching morphogenesis and epithelial invasion. Methods Mol. Biol. 1189:135–162. 10.1007/978-1-4939-1164-6_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanaban V., Krol I., Suhail Y., Szczerba B.M., Aceto N., Bader J.S., and Ewald A.J.. 2019. E-cadherin is required for metastasis in multiple models of breast cancer. Nature. 573:439–444. 10.1038/s41586-019-1526-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanaban V., Tsehay Y., Cheung K.J., Ewald A.J., and Bader J.S.. 2020. Between-tumor and within-tumor heterogeneity in invasive potential. PLOS Comput. Biol. 16 e1007464 10.1371/journal.pcbi.1007464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., and Jemal A.. 2017. Cancer Statistics, 2017. CA Cancer J. Clin. 67:7–30. 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- Topper M.J., Vaz M., Marrone K.A., Brahmer J.R., and Baylin S.B.. 2019. The emerging role of epigenetic therapeutics in immuno-oncology. Nat. Rev. Clin. Oncol. 17:75–90. 10.1038/s41571-019-0266-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang R., Zhang S., Song S., Jiang C., Han G., Wang M., Ajani J., Futreal A., and Wang L.. 2019. iTALK: an R Package to Characterize and Illustrate Intercellular Communication. bioRxiv. (Preprint posted January 4, 2019). [Google Scholar]
- Young M.D., Wakefield M.J., Smyth G.K., and Oshlack A.. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14 10.1186/gb-2010-11-2-r14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bi J., Zheng X., Chen Y., Wang H., Wu W., Wang Z., Wu Q., Peng H., Wei H., et al. . 2018. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti-tumor immunity. Nat. Immunol. 19:723–732. 10.1038/s41590-018-0132-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists genes differentially expressed by teNK cells relative to hNK cells.
shows CIBERSORT deconvolution analysis of genes expressed by hNK cells and teNK cells using immune cell signatures from ImmuCC: the relative fraction of cells from hNK cell and teNK cell subset are consistent with gene signatures related to NK cells.
lists genes from ImmuCC used to determine active and resting NK phenotypes.
Data Availability Statement
Next-generation RNA sequencing data is available on NCBI GEO (accession no. GSE152790). Any additional information may be provided by the corresponding author upon request.