The tumor suppressor PTEN is essential for epithelial morphogenesis. Qi et al. identify Abi1, a core adaptor protein in the WAVE regulatory complex, as a new PTEN substrate. PTEN dephosphorylates Abi1 and causes Abi1 degradation through calpains and thus down-regulates the WAVE regulatory complex to induce epithelial differentiation and polarization.
Abstract
The tumor suppressor PTEN is essential for early development. Its lipid phosphatase activity converts PIP3 to PIP2 and antagonizes the PI3K–Akt pathway. In this study, we demonstrate that PTEN’s protein phosphatase activity is required for epiblast epithelial differentiation and polarization. This is accomplished by reconstitution of PTEN-null embryoid bodies with PTEN mutants that lack only PTEN’s lipid phosphatase activity or both PTEN’s lipid and protein phosphatase activities. Phosphotyrosine antibody immunoprecipitation and mass spectrometry were used to identify Abi1, a core component of the WASP-family verprolin homologous protein (WAVE) regulatory complex (WRC), as a new PTEN substrate. We demonstrate that PTEN dephosphorylation of Abi1 at Y213 and S216 results in Abi1 degradation through the calpain pathway. This leads to down-regulation of the WRC and reorganization of the actin cytoskeleton. The latter is critical to the transformation of nonpolar pluripotent stem cells into the polarized epiblast epithelium. Our findings establish a link between PTEN and WAVE-Arp2/3–regulated actin cytoskeletal dynamics in epithelial morphogenesis.
Introduction
Phosphatase and tensin homolog (PTEN) is one of the most frequently mutated tumor suppressor genes in human cancers. PTEN germline mutations cause tumor predisposition syndromes such as Cowden disease (Sansal and Sellers, 2004). In mice, global deletion of the Pten gene arrests embryonic development between embryonic day 6.5 (E6.5) and E9.5, depending upon domains disrupted and genetic backgrounds. This effect delineates the essential role PTEN plays in embryogenesis (Di Cristofano et al., 1998; Podsypanina et al., 1999; Stambolic et al., 1998). Conditional ablation of PTEN results in tumor formation in most tissues examined (Kishimoto et al., 2003; Knobbe et al., 2008). We and others have shown that PTEN-null embryonic stem (ES) cells fail both to form a polarized epiblast epithelium during embryoid body (EB) morphogenesis and to differentiate into derivatives of three germ layers when transplanted into syngeneic mice (Di Cristofano et al., 1998; Qi et al., 2015). However, it remains unknown which differentiation pathway is dysregulated and how epiblast polarity is disrupted upon PTEN loss (Di Cristofano and Pandolfi, 2000; Frank and Miranti, 2013).
PTEN functions as both an inositol phospholipid phosphatase and a protein phosphatase capable of acting on phosphothreonine, phosphoserine, and phosphotyrosine. Its well-documented lipid phosphatase activity converts phosphatidylinositol (3,4,5)-trisphosphate (PIP3) to phosphatidylinositol 4,5-bisphosphate (PIP2) and antagonizes the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)–Akt pathway, thereby inhibiting cell proliferation, survival and migration (Carracedo and Pandolfi, 2008). There is increasing evidence of important PI3K-independent activities contributing to PTEN’s tumor-suppressive functions (Bassi et al., 2013; Davidson et al., 2010; Dey et al., 2008; Leslie et al., 2007, 2009; Poon et al., 2010; Raftopoulou et al., 2004; Song et al., 2011; Tang and Eng, 2006; Zhang et al., 2011). One of the most challenging questions to be addressed is whether additional PTEN substrates exist (Di Cristofano and Pandolfi, 2000; Song et al., 2012). Finding such substrates will clarify the extent to which the PTEN and the PI3K–Akt pathway overlap and provide new insights into the function of PTEN in embryonic development and tumor suppression. Several phosphoproteins, including focal adhesion kinase, have been proposed as candidate substrates of PTEN’s protein phosphatase activity (Gu et al., 1999; Tamura et al., 1998). However, none of these have been confirmed with much confidence, and the identification of specific sites of dephosphorylation and other mechanistic details are absent (Leslie et al., 2009). In addition, the relative contributions of PTEN’s lipid and protein phosphatase activities as well as its phosphatase-independent activities to its role in embryonic development and tumor suppression are not well understood (Di Cristofano et al., 1998; Fournier et al., 2009; Martin-Belmonte et al., 2007; Podsypanina et al., 1999; Suzuki et al., 1998).
The WAVE (WASP-family verprolin homologous protein) regulatory complex (WRC) is a pentameric complex consisting of Abl-interactor 1 (Abi1 or its paralogue, Abi2 or Abi3), Nck-associated protein 1 (Nckap1 or Nckap1L), WAVE2 (or WAVE1 and WAVE3), cytoplasmic FMR1-interacting protein 1 (Cyfip1 or Cyfip2), and hematopoietic stem progenitor cell 300 (also termed Brick1; Mendoza, 2013). WAVE proteins possess the VCA (verprolin-homology, central, acid regions) motif at their C-terminus. The VCA motif binds to an actin monomer and the Arp2/3 complex, stimulating Arp2/3 complex–mediated nucleation of actin polymerization (Pollard and Borisy, 2003). Structural analysis showed that WRC formation inhibits WAVE activation via Cyfip1 binding to the WAVE VCA motif (Chen et al., 2010). Rac1-GTP binding to Cyfip1, acidic phospholipids (especially PIP3), and kinase-mediated phosphorylation of WAVE2 and other WRC subunits can coordinately activate WAVE by releasing the VCA motif from its inhibitory interaction with Cyfip1 (Lebensohn and Kirschner, 2009; Mendoza, 2013; Steffen et al., 2004). Within the WRC, Abi1 acts as a core scaffold protein to mediate membrane recruitment and stabilization of WRC subunits (Kunda et al., 2003; Leng et al., 2005). shRNA-mediated knockdown or genetic ablation of Abi1 leads to destabilization and degradation of the WRC (Dubielecka et al., 2011; Innocenti et al., 2005). However, it remained unknown how WRC stability is regulated by extracellular cues and intracellular signaling pathways.
In this study, we performed phosphotyrosine immunoprecipitation using PTEN-null EBs reconstituted with wild-type PTEN as well as the mutant forms that lack either PTEN’s lipid phosphatase activity (PTEN G129E) or both its lipid and protein phosphatase activity (PTEN C124S; Liliental et al., 2000; Maier et al., 1999; Myers et al., 1998). Mass spectrometry has identified Abi1 as a new substrate for PTEN and established a novel link between PTEN and the WRC. PTEN dephosphorylates Abi1 and causes its degradation through the calpain pathway, thereby promoting epithelial differentiation and polarization.
Results
PTEN’s protein phosphatase activity is required for epiblast differentiation and polarization
ES cell–derived EBs recapitulate key developmental processes of peri-implantation embryogenesis. These include endoderm differentiation, basement membrane assembly, epiblast polarization, and proamniotic cavity formation (Fig. S1; Li et al., 2003). Previously, we have shown that PTEN is essential for epiblast polarization and cavitation during EB morphogenesis (Qi et al., 2015). This is confirmed by a recent study on E5.5 Pten−/− embryos generated by crossing Ptenfl/fl mice with Sox2-Cre mice (Meng et al., 2017). In rescue experiments using PTEN mutants G129E or C124S, we demonstrated that PTEN’s lipid phosphatase activity is required for epiblast elongation while its protein phosphatase activity is essential for apical polarization (Qi et al., 2015). Because Pten−/− ES cells cannot form the epiblast epithelium during EB morphogenesis or contribute to the chimeric organism when introduced into wild-type blastocysts, we reasoned that they may have differentiation defects (Di Cristofano et al., 1998). To test this possibility, we analyzed the expression of the pluripotency and epithelial apical markers in wild-type– and PTEN mutant–rescued Pten−/− EBs.
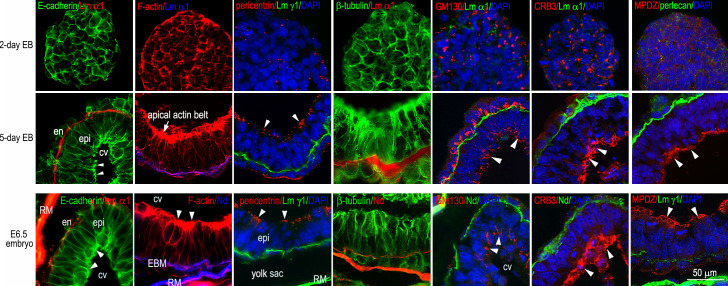
Figure S1.
EB differentiation recapitulates the process of epiblast polarization during mouse peri-implantation development. 2- and 5-d EBs as well as E6.5 mouse embryos were immunostained for E-cadherin, pericentrin, β-tubulin, GM130, CRB3, and MUPP1. The basement membrane was identified by immunofluorescence of nidogen (Nd), perlecan, or the laminin α1 or γ1 chain. F-actin was visualized with rhodamine-phalloidin. The nucleus was counterstained with DAPI. As EBs begin to form the epiblast epithelium, the adherens junction receptor E-cadherin gradually concentrates on the apical side of the epiblast (arrowheads). Cortical F-actin is reorganized to form an apical actin belt (the arrow and arrowheads) and longitudinal filaments. Pericentrin, a component of the microtubule organization center (MTOC), also moves to the apex, where it marks the minus ends of microtubules. Microtubules, labeled with β-tubulin antibody, form a longitudinal array parallel to the lateral plasma membranes. Meanwhile, nuclei are repositioned from the center to the basal side, while the Golgi protein GM130 is localized to the apical side of the epiblast (arrowheads). The polarity markers CRB3 and MPDZ are translocated from a vesicular compartment and the cell cortex, respectively, to the apical surface of the epiblast (arrowheads). cv, central cavity; EBM, embryonic basement membrane; en, endoderm; epi, epiblast; RM, Reichert’s membrane.
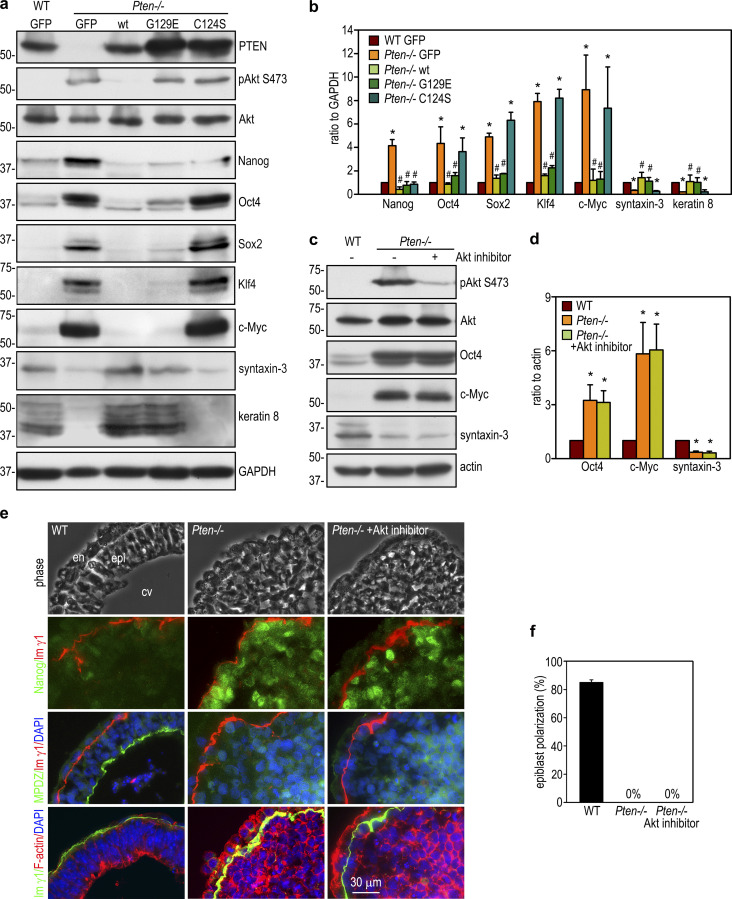
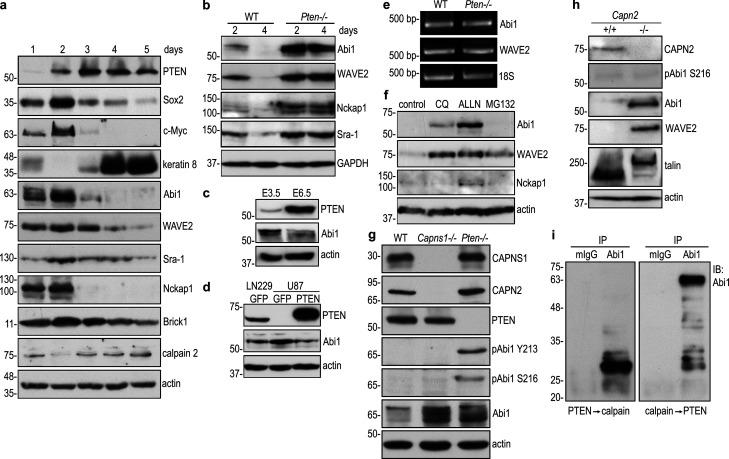
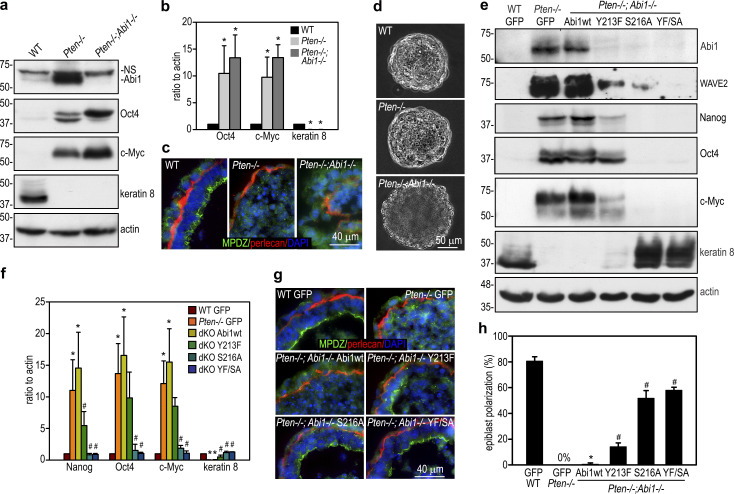
PTEN’s lipid phosphatase activity converts PIP3 to PIP2 and antagonizes the effect of PI3K. In the absence of PTEN, PIP3 accumulates in cell membranes and both recruits and activates Akt. As expected, Akt phosphorylation at S473 was higher in 5-d Pten−/− EBs than in wild-type controls, and this was suppressed by stable transfection with wild-type PTEN, but not the G129E or C124S mutant (Fig. 1 a). In comparison with wild-type EBs, Pten−/− EBs expressed increased levels of the pluripotency transcription factors Nanog, Oct4, Sox2, and Klf4 as well as the oncoprotein c-Myc; the latter promotes stem cell proliferation and metabolic flux (Cliff et al., 2017; Ralston and Rossant, 2010; Fig. 1, a and b). On the other hand, Pten−/− EBs expressed lower levels of the epithelial apical markers syntaxin-3 and keratin 8. This differentiation defect could be rescued by stable reconstitution of Pten−/− EBs with wild-type PTEN or the G129E mutant, deficient in PTEN’s lipid phosphatase activity. In contrast, reconstitution with PTEN C124S, deficient in both PTEN’s lipid and protein phosphatase activities, largely failed to rescue differentiation, despite Nanog down-regulation. These results suggest that the protein phosphatase activity of PTEN is required for differentiation of pluripotent stem cells into the epiblast epithelium.
Figure 1.
PTEN’s protein phosphatase activity regulates epiblast epithelial differentiation and polarization. (a) 5-d wild-type GFP EBs and Pten−/− EBs stably transfected with GFP, wild-type PTEN (wt), mutant PTEN G129E, or C124S were analyzed by immunoblotting. (b) The blots were analyzed by densitometry and the intensity of markers for pluripotency and differentiation was plotted as a ratio relative to GAPDH. Mean ± SD. *, P < 0.01 versus wild-type GFP; #, P < 0.05 versus Pten−/− GFP. (c) Wild-type and Pten−/− EBs were cultured for 24 h in suspension and then either treated with the Akt inhibitor or the vehicle control for 4 d. EB lysates were analyzed by immunoblotting. (d) The blots were analyzed by densitometry, and the ratio of Oct4, c-Myc, and syntaxin relative to actin was plotted. Mean ± SD. *, P < 0.001 versus wild type. (e) 5-d EBs were immunostained for Nanog, MPDZ, and the basement membrane laminin γ1 chain (lm γ1). The apical actin belt of polarized epiblast was visualized by rhodamine-phalloidin. Nuclei were counterstained with DAPI. cv, central cavity; en, endoderm; epi, epiblast. (f) EBs with the polarized epiblast epithelium were counted by phase-contrast microscopy and plotted as a percentage of total EBs examined. Mean ± SD. n = 335–398 for each group.
To further rule out the role of PTEN’s lipid phosphatase activity in epiblast differentiation, we treated Pten−/− EBs with Akt inhibitor. Akt inhibition reduced Akt phosphorylation at S473 but did not alter the expression of pluripotency and differentiation markers (Fig. 1, c and d). Similarly, it failed to induce epiblast polarization. This is evidenced by the lack of apical accumulation of polarity protein MPDZ (multiple PDZ domain crumbs polarity complex component) and the failure in formation of an apical actin belt, a hallmark of epithelial apical polarization (Fig. 1, e and f). These results suggest Akt overactivation is not the mechanism by which PTEN ablation causes defects in epiblast differentiation and polarization.
Identification of Abi1 as a new substrate of PTEN
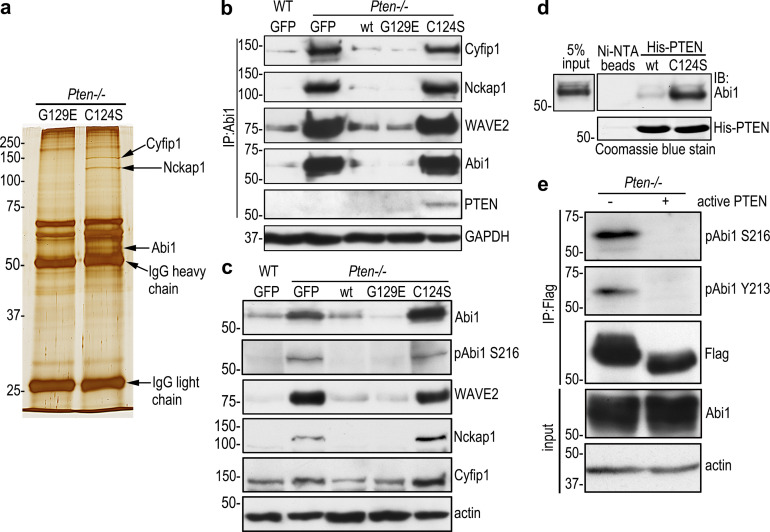
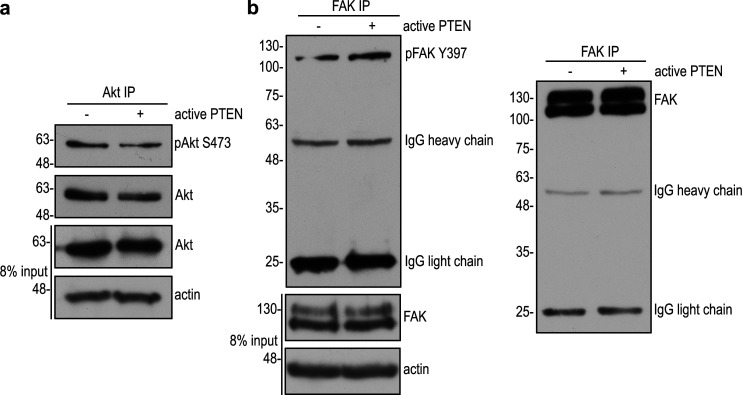
To identify substrates for PTEN’s protein phosphatase activity, we performed coimmunoprecipitation of Pten−/− EBs rescued with G129E or C124S using anti-phosphotyrosine antibody conjugated to agarose beads under a stringent condition (1% NP-40, 0.25% deoxycholate, 700 mM NaCl, and 25 mM Tris, pH 7.4). Silver staining of SDS-polyacrylamide gels demonstrated that proteins of 145 kD, 125 kD, and 60 kD were selectively immunoprecipitated from C124S-reconstituted EBs (Fig. 2 a). Mass spectrometry identified them as Cyfip1, Nckap1, and Abi1, respectively. Notably, Abi1 was found to be phosphorylated at Y213 and S216 by mass spectrometry. Given that Cyfip1, Nckap1, and Abi1 are WRC components, we tested whether these proteins would form a complex in differentiating EBs. Indeed, Cyfip1 and Nckap1 were detected in the Abi1 immunoprecipitates together with WAVE2 (Fig. 2 b). Interestingly, Abi1 specifically coimmunoprecipitated with substrate-trapping mutant PTEN C124S, which binds to PTEN substrates but cannot dephosphorylate them (Myers et al., 1998; Shinde and Maddika, 2016). Complex formation seems to depend on the level of the WRC proteins, which were markedly increased together with pAbi1 S216 in 5-d Pten−/− EBs expressing GFP or C124S, but not in those rescued with wild-type PTEN or G129E (Fig. 2 c). This suggests that PTEN requires its protein phosphatase activity to dephosphorylate Abi1 and down-regulate the WRC. Next, we performed an in vitro substrate-trapping assay by incubating either recombinant His-tagged wild-type PTEN or the C124S mutant with Pten−/− EB lysates. Affinity pull-down experiments using nickel-nitrilotriacetic acid agarose beads revealed that the affinity binding of PTEN C124S to Abi1 was much greater than that of wild-type PTEN (Fig. 2 d). To further confirm Abi1 as a new substrate of PTEN, we expressed Flag-tagged Abi1 in Pten−/− EBs and performed immunoprecipitation with Flag-tag antibody conjugated to agarose beads. Treatment of the immunoprecipitates with recombinant active PTEN eliminated its tyrosine phosphorylation at Y213 and serine phosphorylation at S216 (Fig. 2 e). In contrast, recombinant PTEN was unable to dephosphorylate pAkt S473 and pFAK Y397, even with a longer incubation time (Fig. S2). This confirmed that the phosphatase activity of recombinant PTEN toward Abi1 is specific. Taken together, these results demonstrate that Abi1 is a new substrate of PTEN and likely involved in epiblast epithelial morphogenesis.
Figure 2.
Abi1 is a new PTEN substrate. (a) 5-d Pten−/− EBs stably transfected with PTEN G129E or C124S were immunoprecipitated with anti-phosphotyrosine antibody-agarose beads. Analysis of the immunoprecipitates by SDS-PAGE and silver staining followed by mass spectrometry identified that the WAVE regulatory complex (WRC) subunits Cyfip1, Nckap1, and Abi1 were selectively present in the C124S immunoprecipitates. (b) 5-d WT GFP EBs and Pten−/− EBs stably transfected with GFP, wild-type PTEN, PTEN G129E, or C124S were immunoprecipitated (IP) with anti-Abi1 antibody. The WRC subunits Cyfip1, Nckap1, and WAVE2 as well as PTEN were detected by immunoblotting. (c) Direct immunoblot analysis of the above EBs (IP input controls). (d) Substrate-trapping assay. 5-d Pten−/− EB lysates were incubated with N-terminally His-tagged wild-type or mutant PTEN bound to NTA agarose beads with NTA agarose beads only as a negative control. Affinity pull-down products were analyzed by immunoblotting for Abi1. (e) Lysates of 5-d EBs stably transfected with N-terminally Flag-tagged Abi1 were immunoprecipitated with Flag antibody-agarose beads. The immunoprecipitates were incubated either with or without 0.1 µg/µl active PTEN. Immunoblotting was performed using Abi1 phospho-specific antibodies to pY213 and pS216.
Figure S2.
Active recombinant PTEN does not dephosphorylate phospho-Akt S473 or phospho-FAK Y397. (a) AML12 mouse hepatocytes were treated with 10 nM insulin for 15 min and immunoprecipitated with anti-Akt antibody. The immunoprecipitates were incubated either with or without 0.1 µg/µl active recombinant PTEN (SignalChem; catalog number P23-20G-10) for 30 min in the reaction buffer containing 20 mM Tris, pH 7.5, 150 mM NaCl, 50 µM MgCl2, 0.05% Tween-20, and 0.2% 2-mercaptolethanol. After incubation, the samples were analyzed by immunoblotting for phospho-Akt S473 (pAkt S473) and Akt. (b) Human fibroblasts were cultured on gelatin-coated dishes overnight and immunoprecipitated with anti-focal adhesion kinase (FAK) antibody. The immunoprecipitates were treated either with or without active recombinant PTEN as described above. The treated samples were then analyzed by immunoblotting for phospho-FAK Y397 (pFAK Y397) and FAK. IP, immunoprecipitation.
PTEN dephosphorylation of Abi1 causes its degradation through the calpain and lysosomal pathways
During EB differentiation, clusters of loosely attached ES cells undergo compaction, a process in which cells are packed tightly to form a solid ball (Li et al., 2003). Cells on the surface of this ball cannot be distinguished from one another. Compaction occurs around day 2 of EB morphogenesis and is followed by endoderm differentiation on day 3 and epiblast polarization on day 4. PTEN was expressed at low levels on day 1, gradually up-regulated to reach its peak on day 3, and remained high through day 5 (Fig. 3 a). All of the WRC subunits were observed to be up-regulated on day 2 and quickly down-regulated on day 3. The expression level of WRC proteins was positively correlated with that of pluripotency markers and negatively correlated with that of PTEN, calpain 2, and the epithelial differentiation marker keratin 8 (Qi et al., 2015). To determine the fate of Abi1 after dephosphorylation by PTEN, we analyzed Abi1 in wild-type and Pten−/− EBs by immunoblotting. Abi1 and other WRC components were detected at low levels in 2-d wild-type EBs and were found to be down-regulated in 4-d EBs (Fig. 3 b; Qi et al., 2015). During early mouse embryonic development, up-regulation of PTEN was associated with down-regulation of Abi1 (Fig. 3 c). In the absence of PTEN, the protein levels of Abi1 and other WRC components were markedly elevated in both 2-and 4-d EBs. Similarly, the Abi1 protein level was higher in PTEN-deficient glioblastoma cells. Reintroduction of PTEN caused a reduction of Abi1 (Fig. 3 d). However, mRNA levels of Abi1 and WAVE2 in 4-d wild-type and Pten−/− EBs were similar, which indicates that in Pten−/− EBs, increased Abi1 and WAVE2 proteins are not due to enhanced transcription (Fig. 3 e). These results suggest that PTEN dephosphorylation of Abi1 induces WRC degradation.
Figure 3.
PTEN-mediated dephosphorylation of Abi1 leads to Abi1 degradation by calpains. (a) Wild-type EBs were cultured for 1–5 d and analyzed by immunoblotting. (b) 2- and 4-d wild-type and Pten−/− EBs were analyzed by immunoblotting. (c) E3.5 and E6.5 embryos were isolated from timed wild-type pregnant mice and analyzed by immunoblotting. (d) The protein level of Abi1 was higher in PTEN-negative (U87) human glioblastoma cells when compared with those that were PTEN positive (LN229). Subsequent restoration of PTEN expression in U87 cells down-regulated Abi1. (e) RT-PCR analysis for Abi1 and WAVE2 in 4-d wild-type and Pten−/− EBs. (f) 4-d EBs were treated with the lysosomal inhibitor chloroquine (CQ), the calpain inhibitor ALLN, or the proteasome inhibitor MG-132 for 16 h. Immunoblotting was performed to detect the WAVE complex subunits. (g) Wild-type, calpain small subunit 1 (Capns1)–, and Pten-null (−/−) EBs were cultured for 5 d and analyzed by immunoblotting. (h) Immunoblot analysis of 5-d wild-type and calpain 2 (Capn2)-null EBs showed that Abi1 and WAVE2 were significantly increased in Capn2−/− EBs. Intact talin was detected in Capn2−/− EBs, but not in wild-type EBs. Cleaved talin (the rod domain) was mainly detected in wild-type EBs. (i) Abi1 was immunoprecipitated from 5-d Pten−/− EBs with anti-Abi1 monoclonal antibody. Preimmune mouse IgG (mIgG) served as a control. The immunoprecipitates were incubated with active PTEN for 30 min, washed with PBS, and then treated with active porcine calpain 1 for an additional 30 min. Enzymatic digestion in the reverse order of above was also performed with mouse IgG immunoprecipitates as control. Digestion products were analyzed by immunoblotting using rabbit anti-Abi1 antibody.
To determine the degradation pathway of Abi1, we incubated 4-d wild-type EBs with the lysosomal inhibitor chloroquine, the calpain inhibitor N-acetyl-leucyl-leucyl-norleucinal (ALLN), or the proteasome inhibitor MG-132 for 16 h. In vehicle control, Abi1 is barely detectable. Treatment with chloroquine slightly increased Abi1 protein levels over control (Fig. 3 f). Notably, treatment with ALLN caused a marked increase of Abi1, WAVE2, and Nckap1, whereas MG-132 had little effect. These findings suggest that Abi1 is mainly degraded by calpain and to a lesser extent via the lysosomal pathway. Protein sequence analysis revealed that Abi1 contains multiple PEST sequences (rich in proline [P], glutamic acid [E], serine [S], and threonine [T]), which are often found in proteins with short half-lives and act as a signal for degradation (Rogers et al., 1986; Shumway et al., 1999). Computation prediction indicates that S216 in the first PEST sequence is a calpain cleavage site (CaMPBD; calpain.org), which suggests that calpain may mediate Abi1 degradation.
Calpains are a family of calcium-dependent cysteine proteases (Ono and Sorimachi, 2012). The conventional, ubiquitously expressed calpain-1 (μ-calpain) and calpain-2 (m-calpain) isoforms are heterodimers consisting of large catalytic subunits of calpain 1 (CAPN1) or 2 (CAPN2), respectively, and are heterodimerized with the common small regulatory subunit 1 (CAPNS1). CAPNS1 is required for the stability and activity of both calpain catalytic subunits (Arthur et al., 2000). Targeted deletion of the Capns1 or Capn2 gene in mice leads to embryonic lethality (Arthur et al., 2000; Dutt et al., 2006; Zimmerman et al., 2000). To test if the conventional calpains mediate Abi1 degradation, we examined Abi1 expression in 5-d EBs derived from wild-type and Capns1−/− ES cells (Arthur et al., 2000). Ablation of Capns1 eliminated CAPN2 expression and markedly increased Abi1 protein abundance (Fig. 3 g). The phosphorylation level of Abi1 at S216 and Y213 was very low in wild-type EBs and comparable to that found in Capns1−/− EBs. In contrast, phospho-Abi1 Y213 and S216 were readily detected in Pten−/− EBs. This result suggests that the Abi1 protein accumulation in Capns1 EBs is mostly dephosphorylated by PTEN. Ablation of Capn2, which is known to cleave the cytoskeletal protein talin, was associated with accumulation of full-length talin and greatly reduced levels of talin fragments (Fig. 3 h; Calle et al., 2006). Capn2 ablation also led to an increase in Abi1 and WAVE2 protein levels but did not increase Abi1 phosphorylation at S216 (Fig. 3 h). To further test whether Abi1 is dephosphorylated by PTEN and subsequently degraded by calpains, we immunoprecipitated Abi1 from 5-d Pten−/− EBs and treated the immunoprecipitates with recombinant PTEN. This was followed by treatment with active calpain 1. Immunoblot analysis of the treated immunoprecipitates primarily detected an ∼29-kD Abi1 fragment (Fig. 3 i). However, when the reverse was done (i.e., the immunoprecipitates were first incubated with calpain 1 followed by PTEN), intact Abi1 was the main band detected by immunoblotting. These results suggest that Abi1 is mainly degraded by conventional calpains following dephosphorylation by PTEN during epiblast morphogenesis. Following Abi1 degradation, other WRC subunits are degraded likely due to reduced stability of the complex (Dubielecka et al., 2011; Innocenti et al., 2004).
PTEN-mediated down-regulation of Abi1 promotes epiblast differentiation and polarization
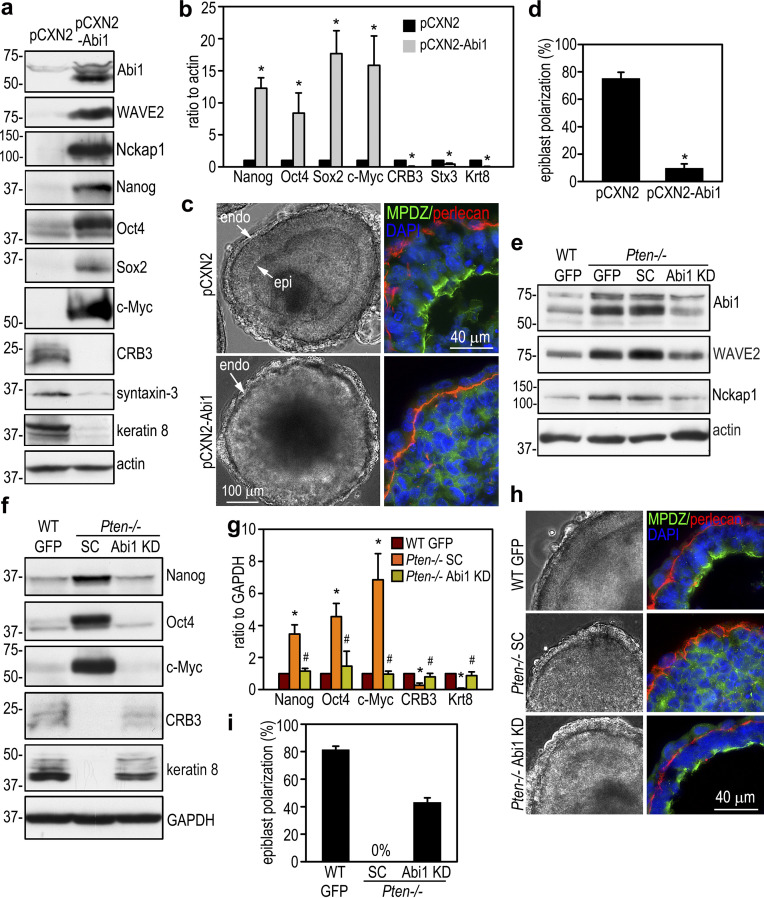
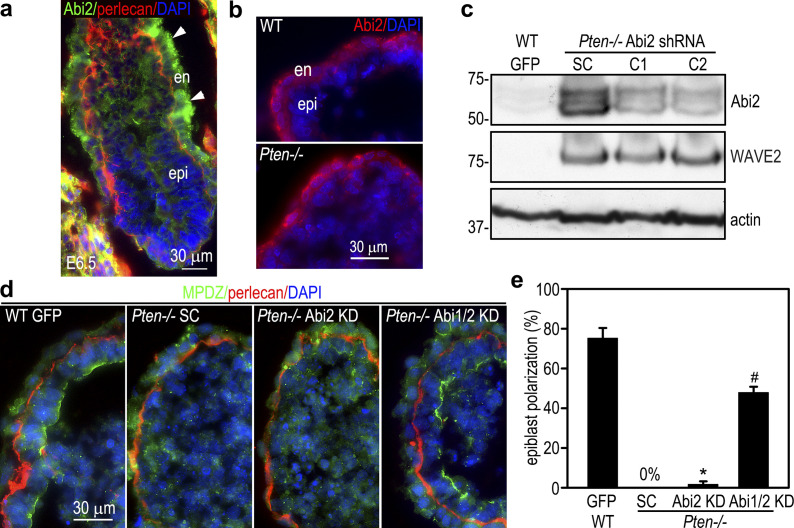
To examine the effect of Abi1 elevation on epiblast differentiation and polarization, we stably transfected wild-type ES cells with human Abi1. As shown in Fig. 4, a and b, overexpression of Abi1 in 5-d EBs inhibited epiblast epithelial differentiation. This was evidenced by high-level expression of the pluripotency markers, low-level expression of the epithelial differentiation marker keratin 8, as well as low-level expression of apical polarity proteins CRB3 and syntaxin-3. Only a small percentage (9.7 ± 3.2%) of the EBs contained the polarized epiblast epithelium and a central cavity (Fig. 4, c and d). By contrast, 75.2 ± 4.5% of the EBs transfected with the vector alone displayed epiblast polarization, although endoderm differentiation was similar in the two groups. Given that Abi1 is markedly up-regulated after PTEN ablation, we next tested if knockdown of Abi1 in Pten−/− EBs rescues epiblast differentiation and polarity. To this end, Pten−/− ES cells were stably transfected with vectors encoding shRNAs targeting Abi1, scrambled shRNAs, or GFP. Abi1 knockdown clones with Abi1 protein levels depleted to that of the wild-type GFP EBs were chosen for further analysis (Fig. 4 e). Immunoblots show that knockdown of Abi1 decreased WAVE2 and Nckap1, likely caused by disassembly of the WRC and subsequent degradation of these subunits. Abi1 depletion was associated with reduced expression of pluripotency transcription factors and increased expression of markers for epithelial differentiation (Fig. 4, f and g). Quantification of the formation of the columnar epiblast epithelium and cavitation in 5-d EBs revealed >50% rescue of epiblast polarity in comparison with the wild-type EBs (Fig. 4, h and i). However, shRNA-mediated stable knockdown of Abi2, a paralogue of Abi1 mainly expressed in the endoderm, did not rescue epiblast polarity in Pten−/− EBs (Fig. S3). Knockdown of both Abi1 and Abi2 did not further increase the rescue efficiency as compared with Abi1 depletion alone. These results suggest that PTEN-induced down-regulation of Abi1 promotes epiblast differentiation and polarization.
Figure 4.
PTEN regulates epiblast differentiation and polarization via down-regulation of Abi1. (a) 5-d EBs stably transfected with either Abi1 (pCXN2-Abi1) or vector alone (pCXN2) were analyzed by immunoblotting. (b) The blots were quantified by densitometry, and the ratio of pluripotency and differentiation markers relative to actin was plotted. Mean ± SD. *, P < 0.01 versus the pCXN2 group. Krt8, keratin 8; Stx3, syntaxin-3. (c and d) 5-d EBs were analyzed by live phase-contrast microscopy and immunostaining. endo, endoderm; epi, epiblast. EBs with the polarized epiblast epithelium were counted and plotted as a percentage of the total EBs examined. n = 967–1,021 for each group. *, P < 0.01 versus pCXN2. (e) Pten−/− ES cells were stably transfected with Abi1 shRNA (Abi1 KD), the scrambled control (SC), or GFP. 5-d EBs were analyzed by immunoblotting. Wild-type EBs stably expressing GFP served as controls. (f) Immunoblots of 5-d EBs show that knockdown of Abi1 down-regulated Nanog, Oct4, and c-Myc and up-regulated CRB3 and keratin 8. (g) The blots were quantified by densitometry, and the data were plotted. Mean ± SD. *, P < 0.001 versus WT GFP; #, P < 0.01 versus Pten−/− scrambled control. (h and i) Live phase-contrast and immunofluorescence micrographs show epiblast polarization in Abi1-knockdown EBs. EBs with the polarized epiblast epithelium were counted and plotted as a percentage of the total EBs examined. n = 1,025–1,186 for each group. Mean ± SD.
Figure S3.
Abi2 is mainly expressed in endoderm and not involved in epiblast polarization. (a) E6.5 mouse embryos were immunostained for Abi2 and basement membrane perlecan. Abi2 was mainly localized to the visceral endoderm (en; arrowheads). epi, epiblast. (b) Immunostaining of 5-d wild-type (WT) and Pten−/− EBs showed localization of Abi2 at the outer endoderm layer. (c) Immunoblots show that shRNA-mediated knockdown of Abi2 in Pten−/− EBs (clone 1 and 2, C1 and C2) did not alter the expression of WAVE2, although both Abi2 and WAVE2 were up-regulated in Pten−/− EBs. (d and e) 5-d EBs were immunostained for the apical polarity marker MPDZ and basement membrane perlecan. Nuclei were stained with DAPI. Quantification of epiblast epithelial formation by live phase microscopy revealed that knockdown of Abi2 failed to rescue epiblast polarity in Pten−/− EBs. Knockdown of both Abi1 and Abi2 did not show any synergistic effect. n = 374–414 for each group. Mean ± SD. *, P < 0.01 versus WT GFP; #, P < 0.01 versus Abi2 KD. SC, the scrambled shRNA.
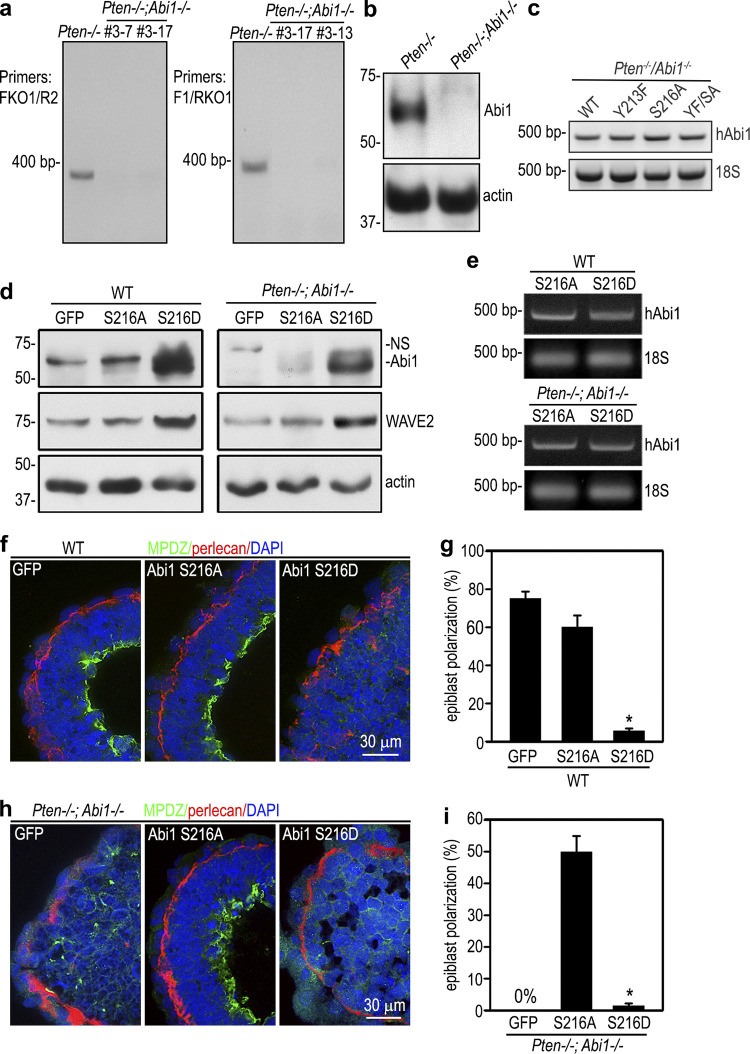
To further determine the effect of PTEN dephosphorylation of Abi1 on epiblast differentiation and polarization, we disrupted the Abi1 gene in Pten−/− ES cells using CRISPR-Cas9 technology (Fig. S4, a and b). Analysis of three independent ES cell clones showed that ablation of Abi1 in Pten−/− EBs failed to rescue epiblast differentiation and polarization (Fig. 5, a–c). However, we observed less compaction of 2-d Pten−/−;Abi1−/− EBs compared with wild-type and Pten−/− EBs, which suggests that Abi1 plays a role in compaction (Fig. 5 d). The Pten−/−;Abi1−/− ES cells were then reconstituted with wild-type or mutant Abi1 containing PTEN dephosphorylation site mutations (Y213F, S216A, or Y213F/S216A; Fig. 5 e). Stable ES cell clones were selected based on GFP fluorescence and similar expression levels of the Abi1 mRNA (Fig. S4 c). Immunoblot analysis revealed that the Abi1 mutants were largely degraded, with degradation accompanied by reduced WAVE2, Nanog, Oct4, and c-Myc and increased keratin 8 in S216A and Y213F/S216A EBs (Fig. 5, e and f). These changes were not seen in wild-type Abi1-reconstituted double-knockout EBs. Further, these changes correlate with a significant rescue of epiblast polarity (Fig. 5, g and h). In the Abi1 Y213F-reconstituted EBs, decrease of Abi1 was associated with only a moderate reduction of pluripotency markers and a slight increase of keratin 8. The rescue efficiency of epiblast polarity was also lower. We also stably transfected wild-type and Pten−/−;Abi1−/− EBs with the phosphomimetic mutant of Abi1 (S216D). The protein level of S216D was much higher in both EBs, although the mRNA level was comparable (Fig. S4, d and e). Expression of Abi1 S216D inhibited EB morphogenesis in wild-type EBs and failed to rescue epiblast differentiation in Pten−/−;Abi1−/− EBs (Fig. S4, f–i). Taken together, these results suggest that PTEN-mediated dephosphorylation and down-regulation of Abi1 promote epiblast differentiation and polarization.
Figure S4.
Pten/Abi1 double-knockout EBs. (a) PCR genotyping was performed with two sets of primers spanning the sgRNA targeting site using DNA extracted from 5-d Pten−/− and Pten−/−;Abi1−/− EBs (clones 3-7, 3-17, and 3-13). FKO1, forward knockout primer 1; R2, reverse primer 2; F1, forward primer 1; RKO1, reverve knockout primer 1. (b) Immunoblots show loss of Abi1 expression in 5-d Pten−/−;Abi1−/− EBs (clone 3-13). (c) RT-PCR analysis of 5-d Pten−/−;Abi1−/− EBs stably transfected with wild-type (WT) human Abi1 (hAbi1) and the mutants Y213F, S216A, and Y213F/S216A (YF/SA) all showed similar expression levels of human Abi1 mRNAs. (d) Wild-type and Pten−/−;Abi1−/− EBs stably transfected with Abi1 S216A, S216D, or GFP were cultured for 5 d and analyzed by immunoblotting. Actin served as a loading control. (e) The mRNA level of transfected human Abi1 (hAbi1) in 5-d EBs were analyzed by RT-PCR. 18S RNA served as an internal control. (f and g) 5-d wild-type EBs stably transfected with Abi1 S216A, S216D, or GFP were stained for MPDZ. Basement membrane was stained for perlecan. The formation of a polarized epiblast layer was quantified by live phase-contrast microscopy and plotted as a percentage of total EBs examined. n = 651–707 for each group. Mean ± SD. *, P < 0.05 S216D versus S216A. (h and i) 5-d Pten−/−;Abi1−/− EBs stably transfected with Abi1 S216A, S216D, or GFP were stained for MPDZ. Basement membrane was stained for perlecan. Epiblast polarization was quantified and plotted as a percentage of total EBs examined. Mean ± SD. *, P < 0.01 versus the S216A group. n = 544–590 for each group.
Figure 5.
Rescue Pten and Abi1 double-knockout EBs with PTEN dephosphorylation mutant Abi1. (a) 5-d wild-type, Pten−/−, and Pten−/−;Abi1−/− EBs were analyzed by immunoblotting. A nonspecific (NS) band appeared in the Abi1 blot using 1% SDS lysis buffer. (b) The blots were analyzed by densitometry, and the ratio of Oct4, c-Myc, and keratin 8 to actin was plotted. Mean ± SD. *, P < 0.01 versus WT. (c) 5-d EBs were coimmunostained for MPDZ and perlecan. (d) Live phase-contrast micrographs show less compaction of 2-d Pten−/−;Abi1−/− EBs. (e) Pten−/−;Abi1−/− EBs were stably reconstituted with wild-type (wt) Abi1, Abi1 Y213F, S216A, or Y213F/S216A (YF/SA) with wild-type GFP and Pten−/− GFP EBs serving as controls. 5-d EBs were subjected to immunoblot analysis. (f) The blots were quantified by densitometry, and the ratio of Nanog, Oct4, c-Myc, and keratin 8 to actin was plotted. Mean ± SD. *, P < 0.01 versus WT GFP; #,P < 0.01 versus Pten−/−;Abi1−/− (double knockout [dKO]) Abi1wt. (g and h) 5-d EBs were coimmunostained for MPDZ and perlecan. EBs with the polarized epiblast epithelium were counted by live phase microscopy and plotted as a percentage of total EBs examined. n = 662–701 for each group. Mean ± SD. *, P < 0.01 versus WT GFP; #, P < 0.01 versus double-knockout Abi1 wild type.
PTEN regulates epiblast polarization mainly through down-regulation of the WRC
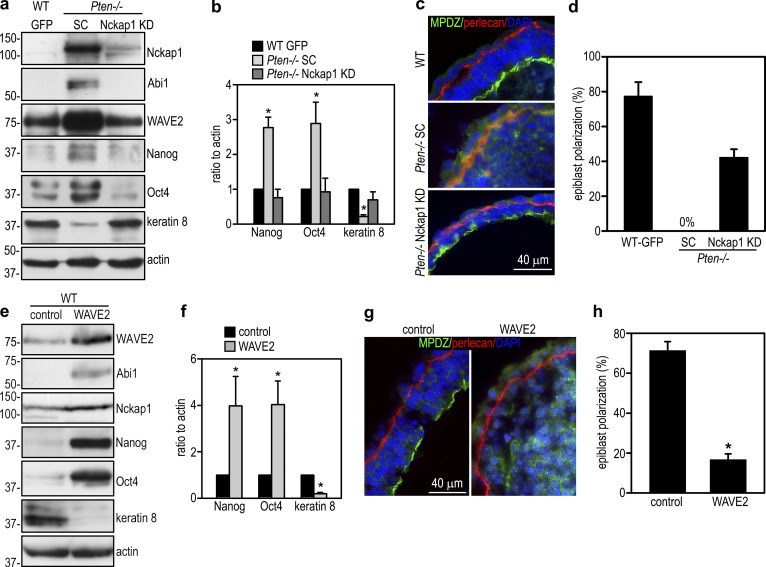
In addition to being a core subunit of the WRC, Abi1 has been shown to interact with syntaxin-1, mDia1, Abl kinases, the p85 subunit of PI3K, N-WASP, mDia2, and Eps8 (Echarri et al., 2004; Innocenti et al., 2003; Ryu et al., 2009). To test whether rescue of the Pten−/− epiblast phenotype by Abi1 depletion is mediated by down-regulation of the WRC, we depleted Nckap1 in Pten−/− ES cells by stable transfection of Nckap1 shRNAs. Our microarray data revealed that the other family member Nckap1L is not expressed in EBs (Data S1, microarray spreadsheet). Therefore, Nckap1 is a unique subunit shared by all WRCs in EBs. shRNA-mediated knockdown of Nckap1 led to a reduction in Abi1 and WAVE2 protein levels in 5-d Pten−/− EBs (Fig. 6 a). This is accompanied by decreased expression of Nanog and Oct4 and increased expression of keratin 8 (Fig. 6, a and b). We quantified epiblast epithelial formation by live phase-contrast microscopy and immunostaining for apical polarity marker MPDZ. This revealed that over ∼40% of Pten−/− EBs with Nckap1 knockdown formed a polarized epiblast layer encompassing a central cavity and achieved similar rescue efficiency (>50%) as shRNA-mediated Abi1 depletion (Fig. 6, c and d). To further address the consequence of WRC elevation, we stably transfected wild-type ES cells with WAVE2. Overexpression of WAVE2 increased Abi1, Nckap1, Nanog, and Oct4 and decreased keratin 8 in 5-d EBs (Fig. 6, e and f). These changes led to impaired epiblast polarization (Fig. 6, g and h). Taken together, these results suggest that PTEN promotes epiblast epithelial differentiation and polarization mainly through down-regulation of the WRC.
Figure 6.
PTEN promotes epiblast differentiation and polarization via down-regulation of the WRC. (a and b) 5-d wild-type GFP and Pten−−/− EBs stably transfected with Nckap1 shRNA or the scrambled control (SC) were analyzed by immunoblotting. Blots were quantified by densitometry, and the ratio of Nanog, Oct4, and keratin 8 to actin was plotted. Mean ± SD. *, P < 0.05 versus WT GFP and Pten−/− Nckap1 knockdown (KD). (c) 5-day EBs were coimmunostained for MPDZ and perlecan. (d) EBs with polarized epiblast epithelium were counted and plotted as a percentage of total EBs examined. n = 1,090–1,166 for each group. Mean ± SD. (e and f) 5-d normal EBs stably transfected either with pCXN2-WAVE2 or only the vector were subjected to immunoblot analysis. Blots were quantified and the ratio of Nanog, Oct4, and keratin 8 to actin was plotted. Mean ± SD. *, P < 0.05 versus the vector control. (g) 5-d EBs were immunostained for MPDZ and perlecan. (h) Formation of the polarized epiblast epithelium in 5-d EBs were quantified by phase-contrast microscopy and plotted as a percentage of total EBs examined. n = 724–750 for each group. Mean ± SD. *, P < 0.01 versus control.
Down-regulation of Abi1 is required for the contribution of Pten−/− ES cells to the polarized epiblast epithelium
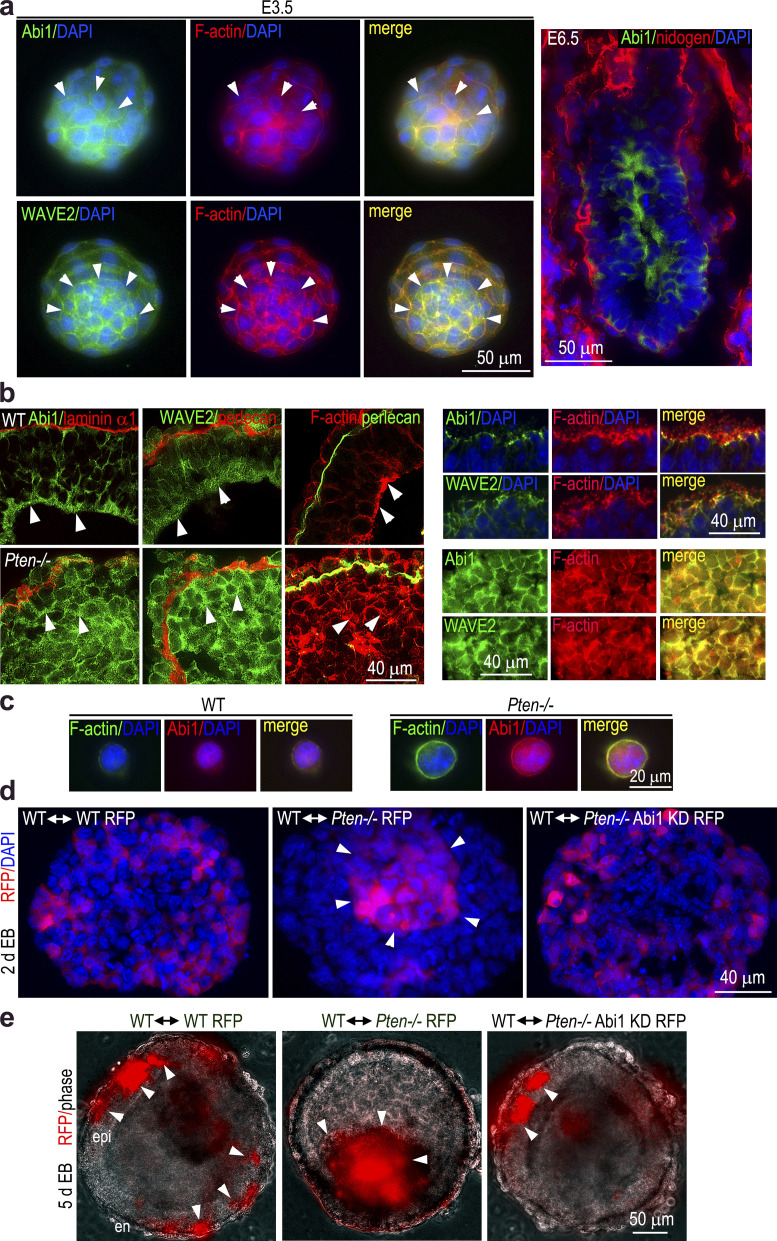
Immunofluorescence microscopy showed that in the inner cell mass of mouse blastocysts, Abi1 and WAVE2 were enriched at the cell cortex where they colocalized with F-actin (Fig. 7 a). In E6.5 embryos, Abi1 was mainly detected on the apical side of polarized epiblast. Similarly, in normal 5-d EBs, Abi1 and WAVE2 were mainly localized on the apical side of the epiblast epithelium and associated with the apical actin belt (Fig. 7 b). In contrast, these were enriched at the cell cortex of nonpolar cells in the absence of PTEN. The increased accumulation of WRC proteins at the cell cortex was accompanied by strong F-actin staining in these cells. To further examine the association of Abi1 with the cortical actin cytoskeleton, we isolated epiblast cells from 5-d EBs by removing the outer endoderm layer with a hypotonic buffer. Isolated epiblast cells were plated on gelatin-coated glass coverslips for 2 h. In wild-type epiblast cells, low levels of Abi1 were mainly detected intracellularly. This was accompanied by weak F-actin staining at the cell cortex (Fig. 7 c). In contrast, Abi1 accumulated at the cell cortex at much higher levels and colocalized with strong cortical F-actin in Pten−/− epiblast cells. These results suggest that Abi1 and its binding partner WAVE2 are mainly involved in actin nucleation associated with the apical surface in polarized epiblast cells. In the absence of PTEN, increased accumulation and activation of the WRC may enhance actin nucleation at the cell cortex, thereby inhibiting actin cytoskeleton reorganization associated with epiblast polarization. This may explain the failure of Pten−/− ES cells to contribute to the differentiated tissues when injected into wild-type mouse blastocysts (Di Cristofano et al., 1998).
Figure 7.
Immunofluorescence and chimeric EB analysis. (a) E3.5 embryos were stained for Abi1, WAVE2, and F-actin. Arrowheads define the inner cell mass. E6.5 embryos were stained for Abi1 and basement membrane nidogen. (b) 5-d EBs were immunostained for Abi1 and WAVE2. F-actin was stained with rhodamine-phalloidin. The basement membrane was immunostained for the laminin α1 chain or perlecan. Arrowheads point to the apical surface of polarized epiblast in wild-type EBs and the cell cortex in Pten−/− EBs. The upper right panels show colocalization of Abi1 and WAVE2 with F-actin at the apex of the polarized epiblast in wild-type EBs. The lower right panels show colocalization of Abi1 and WAVE2 with F-actin at the cell cortex in Pten−/− EBs. (c) Isolated epiblast cells were plated on gelatin-coated glass coverslips for 2 h and immunostained for Abi1. F-actin was stained with FITC-phalloidin. (d) Unlabeled wild-type ES cells were mixed with RFP-labeled wild-type, Pten−/−, or Pten−/− with Abi1 knockdown (KD) ES cells in a ratio of 2:1 and cultured in hanging drops for 24 h. The EBs were cultured in suspension for additional 2 d, and cryosections were stained with DAPI to reveal the nucleus. Arrowheads point to an aggregate of Pten−/− RFP cells. (e) The EBs formed in hanging drops were further cultured for 4 d and examined by live phase-contrast and fluorescence microscopy. Both wild-type RFP and Pten−/− Abi1 KD RFP ES cells contributed to the polarized epiblast epithelium (epi), whereas Pten−/− RFP cells formed an aggregate inside the cavity.
To test this hypothesis, we performed chimeric analysis by intermixing wild-type ES cells with RFP–labeled wild-type or Pten−/− ES cells in hanging drops for EB formation. Analysis of 3-d chimeric EBs showed that RFP-labeled wild-type cells could intermix with unlabeled cells, whereas RFP Pten−/− cell formed an aggregate in the EB center (Fig. 7 d). After 5 d of differentiation, both wild-type and Pten−/− cells contributed to endoderm formation (Fig. 7 e). However, only wild-type ES cells contributed to the polarized epiblast epithelium. A cluster of Pten−/− cells was observed in the central cavity. In contrast to the wild-type↔Pten−/− RFP chimeric EBs, Pten−/− ES cells with Abi1 knockdown behaved like wild-type RFP cells and could intermix with unlabeled wild-type cells in 3-d EBs and contribute to both the endoderm and the polarized epiblast epithelium in 5-d EBs. Given that tissues with high surface tension are sorted to the center and those with low surface tension go to the periphery, these results suggest that the increased Abi1 and actin nucleation at the cell cortex can impair symmetry-breaking polarization of epiblast cells (Foty and Steinberg, 2005). Further, symmetry-breaking apical polarization may require relaxation of the cortical actin cytoskeleton (Lomakin et al., 2015).
Discussion
Establishment and maintenance of epithelial polarity is essential for embryonic development and adult physiology. Loss of polarity is a hallmark of cancer. During the formation of epithelial tissues from a solid mass of undifferentiated cells such as the inner cell mass of the blastocyst, the morphogenetic transformation involves epithelial differentiation, polarization, and cavitation. Despite the fact that PTEN plays a fundamental role in these processes, there is limited understanding of the molecular mechanisms whereby PTEN regulates epithelial morphogenesis. In this study, we identified Abi1 as a new PTEN substrate that links PTEN’s protein phosphatase activity to epiblast epithelial differentiation and polarization. PTEN dephosphorylates Abi1 and causes Abi1 degradation through the calpain pathway and thus down-regulates the WRC and induces reorganization of the actin cytoskeleton and apical polarization.
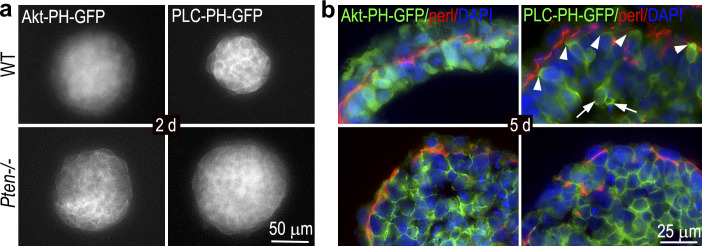
The lipid phosphatase activity of PTEN was shown to be important for apical polarization and lumen formation in the three-dimensional MDCK cell culture system (Martin-Belmonte et al., 2007). In that system, PTEN at the apical plasma membrane mediates the enrichment of PIP2 by converting PIP3 to PIP2. The phospholipid-binding protein Annexin 2 binds to PIP2 and recruits Cdc42 to the apical surface. Cdc42 in turn recruits atypical protein kinase C to induce apical polarization and lumen formation. However, we have shown that in Pten−/− EBs, PTEN G129E, deficient in the lipid phosphatase activity, largely rescues apical polarity and cavitation, although it inhibits epiblast elongation (Qi et al., 2015). Furthermore, before epiblast polarization, PIP2, visualized with PLC-PH-GFP, is localized uniformly to the plasma membrane in wild-type and Pten−/− EBs (Fig. S5). In differentiated normal EBs, PIP2 is mainly detected at the basal rather than apical plasma membrane of the epiblast epithelium. PIP2 is also enriched at the plasma membrane of the centrally located cells in both wild-type and Pten−/− EBs. In contrast, PIP3, detected with Akt-PH-GFP, is mainly intracellular in wild-type EBs and at the plasma membrane in Pten−/− EBs, indicating that PTEN is responsible for the removal of PIP3 from the plasma membrane (Fig. S5). Lastly, treatment of Pten−/− EBs with PI3K or Akt inhibitors fail to rescue the epiblast phenotype. Taken together, the results from Pten−/− EBs suggest that PTEN’s lipid phosphatase activity is not required for apical polarization or cavitation during epiblast morphogenesis. The discrepancy in PTEN mechanisms between the MDCK and the EB system is unknown. During EB morphogenesis, epiblast polarization is coupled with differentiation of pluripotent stem cells into epithelial cells. Nevertheless, these studies suggest that the underlying molecular mechanisms whereby PTEN regulates epithelial polarity may vary depending on the epithelial tissues/cells analyzed.
Figure S5.
Localization of PIP3 and PIP2 in EBs. (a) Wild-type (WT) and Pten−/− ES cells were stably transfected either with the Akt PH domain-GFP (Akt-PH-GFP) or the phospholipase C (PLC) PH domain-GFP (PLC-PH-GFP) vector with expressed fusion proteins specifically bind to PIP3 and PIP2, respectively. Fluorescent micrographs of 2-d live EBs show that PIP3 was mainly intracellular in wild-type EBs and was localized to the plasma membrane in Pten−/− EBs. PIP2 was mainly detected at the plasma membrane in both wild-type and Pten−/− EBs. (b) Cryosections of 5-d EBs were immunostained for basement membrane perlecan (perl). PIP3 was localized intracellularly in both endoderm and polarized epiblast in wild-type EBs, whereas it was found at the plasma membrane in Pten−/− EBs. PIP2 was detected mainly on the basal side of polarized epiblast cells (arrowheads) and the centrally located cells undergoing apoptosis (arrows) in wild-type EBs. In the absence of PTEN, PIP2 was detected at the plasma membrane of nonpolar epiblast cells.
During ES cell differentiation into the columnar epiblast epithelium, the nonpolar ES cells must first exit from the pluripotent ground state. PTEN regulates pluripotency exit likely through dephosphorylation and down-regulation of Abi1. This is supported by the finding that shRNA-mediated down-regulation of Abi1 in Pten−/− EBs suppressed the expression of the core pluripotency transcription factors Nanog and Oct4. Reintroduction of PTEN dephosphorylation mutant Abi1 into Pten−/−; Abi1−/− EBs also reduced expression of these transcription factors. In contrast, overexpression of Abi1 in wild-type EBs sustained pluripotency of EBs in the 5-d experimental period. The regulation of pluripotency by Abi1 is likely to be mediated by the WRC, because manipulation of the WRC subunits Nckap1 and WAVE2 yield similar results. The mechanisms by which the WRC regulates ES cell pluripotency and epithelial differentiation remains unknown. The WRC-mediated assembly of cell cortical actin cytoskeleton may play a role. The cell cortex is a thin F-actin meshwork beneath the plasma membrane (Chugh and Paluch, 2018). WRC-Arp2/3 and mDia are the main drivers of actin nucleation for the assembly of the cortical actin cytoskeleton (Bovellan et al., 2014). The primary function of cortical actin is to control cell shape required for cell migration, division, polarization, and tissue morphogenesis (Chalut and Paluch, 2016; Coravos et al., 2017; Tsankova et al., 2017). The cortical actin cytoskeleton undergoes drastic reorganization during developmental transitions. As ES cells exit from pluripotency and begin to differentiate, actin changes from cortical organization to stress fibers (Chalut and Paluch, 2016). Similarly, culturing cells on a soft substrate, which increases the cortical actin, helps to maintain ES cell pluripotency and promote the reprogramming of somatic cells to stem cells (David et al., 2019; Guo et al., 2014; Higuchi et al., 2014). It would be interesting to determine whether the WRC regulates pluripotency exit by affecting cortical actin assembly.
Our microarray analysis revealed that Abi3, Nckap1L, and WAVE3 are not expressed in EBs while the level of WAVE1 and Cyfip2 is low (Data S1). In addition, immunostaining revealed that Abi2 is predominantly expressed in the endoderm in both E6.5 mouse embryos and EBs (Fig. S3, a and b). Therefore, in the epiblast Abi1 mainly interacts with Nckap1, WAVE2, Sra-1, and Brick1 to form the WRC, where Abi1 serves as a core adaptor protein to hold the complex together (Gautreau et al., 2004; Innocenti et al., 2005; Kunda et al., 2003). In normal mouse blastocysts, Abi1 and WAVE2 are enriched at the cell cortex in the inner cell mass and colocalize with F-actin. This suggests that the WRC is activated and mediates actin polymerization at this location. During epiblast polarization, Abi1 and WAVE2 are significantly reduced at the basolateral membrane and relocated to the apical side, where they colocalize with the apical actin belt. Lomakin et al. reported that in epithelial cells, actin competes between two distinct networks, one for the formation of cortical actin networks and the other for polarized dendritic growth (Lomakin et al., 2015). Relaxation of the actomyosin cytoskeleton using a myosin II inhibitor promotes cell polarization. Similarly, reduction of the cortical actin network may be required for the formation of the apical actin belt during the transition of the compact inner cell mass to polarized epiblast. In Pten−/− EBs, increased amounts of Abi1 and excessive WRC activation may lock actin in the cell cortex and thus prevent epiblast differentiation and polarization. Furthermore, in the Caenorhabditis elegans intestinal epithelium, similar relocation of the WRC is suggested to facilitate the lateral movement of E-cadherin to the apical side and promote the assembly of apical adherens junctions (Sasidharan et al., 2018). Taken together, our data support the following hypothesis: during epiblast morphogenesis, PTEN dephosphorylates Abi1 and down-regulates the WRC at the cell cortex. This causes reorganization of the actin cytoskeleton to facilitate the formation of the apical actin belt and adherens junctions. Loss of PTEN leads to accumulation of PIP3 (Fig. S5) and phosphorylated Abi1 at the cell cortex, prevents actin cytoskeleton reorganization, and inhibits epithelial differentiation (Dubielecka et al., 2010; Mendoza et al., 2011; Xiong et al., 2008).
After dephosphorylation by PTEN, Abi1 is degraded by calpains. Abi1 S216 is a candidate site for calpain cleavage. It has been reported that phosphorylation of calpain cleavage sites or nearby residues inhibits calpain cleavage of its substrates (Falet et al., 1998; Li et al., 2016; Zamorano et al., 2005). During epiblast polarization, the Abi1 protein is down-regulated in parallel with up-regulation of PTEN and calpain 2 (Qi et al., 2015). In Pten−/− EBs, the Abi1 protein is markedly elevated accompanied by increased phosphorylation at S216. There is no parallel elevation of mRNA. By contrast, disruption of the Capns1 or Capn2 gene increases the level of the Abi1 protein, but not phosphorylation, which is likely due to accumulation of nonphosphorylated Abi1. We have also shown that incubation of phosphorylated Abi1, first with PTEN and then with calpain, causes Abi1 degradation. However, reversing that enzymatic digestion order yields largely intact Abi1. Furthermore, the PTEN-dephosphorylation mimetic mutants S216A, Y213F, and S216A/Y213F are degraded after introduced into Pten−/−; Abi1−/− EBs. On the contrary, wild-type Abi1 or the phosphomimetic mutant S216D are not. PTEN dephosphorylation and calpain degradation of Abi1 can lead to the disassembly of the WRC and subsequent degradation of WRC components. This overall reduction of the WRC may be responsible for reduced F-actin at the cell cortex during epiblast morphogenesis.
We detected Abi1 phosphorylation in Pten−/− EBs rescued with the C124S mutant, but not with the G129E mutant. Gain- and loss-of-function analysis suggests that PTEN dephosphorylation of Abi1 is important for epiblast differentiation and polarization. However, it must be noted that the G129E mutation is associated with Cowden disease, a condition that increases the likelihood of several different cancers in patients (Liaw et al., 1997; Myers et al., 1998). The protein phosphatase activity reported here is unlikely to be involved in the pathogenesis of Cowden disease and cancers with the PTEN G129E mutation. Whether PTEN dephosphorylation of Abi1 contributes to its tumor suppressor function warrants further investigation.
Materials and methods
Cell culture and EB differentiation
The ES cell lines used for this study were wild-type R1, Pten−/−, Capns1−/−, and Capn2+/− ES cells (Arthur et al., 2000; Di Cristofano et al., 1998; Dutt et al., 2006). Capn2−/− ES cell clones were generated by screening Capn2+/− ES cells at 2 mg/ml G418. All the ES cells were cultured on mitomycin C–treated Sandos inbred mouse-derived 6-thioguanine- and ouabain-resistant (STO) cells and supplemented with 103 U/ml leukemia-inhibiting factor. EB differentiation was initiated from ES cell aggregates in suspension culture as described previously (Li et al., 2002). Briefly, ES cells cultured for 3–4 d were disassociated to small clusters of ∼20 cells with 0.05% trypsin-0.53 mM EDTA. ES cell clumps were allowed to pellet by gravity and collected for culturing on Nunc Petri dishes in DMEM containing 15% FBS, 1× nonessential amino acids (Thermo Fisher Scientific), 2 mM L-glutamine, 1 mM sodium pyruvate, and 0.1 mM 2-mercaptoethanol. FBS used for EB culture was selected from various sources and batches for maximal EB differentiation. U87 human malignant glioma cells were purchased from ATCC and cultured in Eagle’s minimum essential medium supplemented with 10% FBS. LN229 human glioblastoma cells were also purchased from ATCC and cultured in DMEM supplemented with 5% FBS.
Mouse embryos
E3.5 and E6.5 embryos were isolated from timed pregnant C57BL/6J mice. For immunostaining, E6.5 embryos were fixed in 3% paraformaldehyde for 10 min and mounted in optimal-cutting-temperature compound for the preparation of cryosections. E3.5 embryos were also fixed for 10 min for whole-mount immunofluorescence. For immunoblotting, freshly isolated embryos were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 0.25% sodium deoxycholate), which also contained protease and phosphatase inhibitor cocktails (MilliporeSigma). The animal studies were conducted in accordance with regulations of the Institutional Animal Care and Use Committee at Rutgers University, New Brunswick, New Jersey. The experimental protocol was approved by the Rutgers Institutional Animal Care and Use Committee.
Antibodies and cDNA constructs
Rabbit polyclonal antibodies (pAbs) to pAkt (Ser473, catalog number 9271), Akt (catalog number 9272, Research Resource Identifier [RRID]:AB_329827), Klf4 (catalog number 4038, RRID:AB_2265207), and calpain 2 (catalog number 2539, RRID:AB_2069843); rabbit mAb to WAVE2 (catalog number 3659, RRID:AB_2216981), PTEN (cat. #9188, RRID:AB_2253290), and c-Myc (catalog number 13987, RRID:AB_2631168); and mouse mAb to Sox2 (catalog number 4900, RRID:AB_1056051) were obtained from Cell Signaling. Mouse Abi1 mAb (catalog number D147-3, RRID:AB_592744) was obtained from MBL. Rabbit anti-Sra-1 pAb (catalog number 07-531, RRID:AB_390148) and rat anti-laminin γ1 mAb (catalog number 05-206, RRID:AB_309655) were obtained from Upstate Biotechnology. pAbs to Oct4 (catalog number sc-9081, RRID:AB_2167703) and Sos1/2 (catalog number sc-259, RRID:AB_2270724) and perlecan mAb (catalog number sc-33707, RRID:AB_627714) were obtained from Santa Cruz Biotechnology. Abi1 (catalog number A5106, RRID:AB_2220843) and Nckap1 (catalog number N3788, RRID:AB_1841025) pAbs; Flag-tag (catalog number F3165, RRID:AB_259529), β-actin (catalog number A5441, RRID:AB_476744), β-tubulin (catalog number T4026, RRID:AB_477577) mAbs; and Flag-tag mAb-agarose beads (catalog number A2220, RRID:AB_10063035) were obtained from MilliporeSigma. Nanog and Syntaxin-3 pAb (catalog number ab4113, RRID:AB_2198665) were obtained from Abcam. Cytokeratin-8 mAb (catalog number TROMA-I, RRID:AB_531826) was obtained from Developmental Studies Hybridoma Bank. Rabbit anti-calpain small subunit 1 pAb (catalog number PA5-82266, RRID:AB_ 2789426) was purchased from ThermoFisher Scientific. Phospho-Abi1 S216 was provided by Dr. Zonghan Dai (Texas Tech University Health Science Center, Amarillo, TX). Phospho-Abi1 Y213 was obtained from Dr. Leszek Kotula (New York State Institute for Basic Research in Developmental Disabilities, New York, NY). Abi2 pAb was kindly provided by Dr. Ann Marie Pendergast (Duke University, Durham, NC). MPDZ and mDia2 pAbs were provided by Dr. Makoto Adachi and Dr. Shuh Narumiya (Kyoto University, Kyoto, Japan), respectively. CRB3 pAb was from Dr. Ben Margolis (University of Michigan, Ann Arbor, MI). Cy3-, Cy5-, and horseradish peroxidase–conjugated secondary antibodies were obtained from Jackson ImmunoResearch. Alexa Fluor 488–conjugated secondary antibodies were obtained from Molecular Probes.
The shRNA vectors targeting specifically mouse Abi1, Abi2, Nckap1, and the control scrambled sequences were obtained from OriGene. The effective sequences selected by immunoblot analysis of stably transfected ES cell lines are 5′-TCAGAGACCAAGGACGCATTAGTGGAAGT-3′ (Abi1), 5′-ACTGCGAGAATAACTACATCCAGTCACCA-3′ (Abi2), and 5′-ACGCCTTGGTCAGATGATTGTGGACTATG-3′ (Nckap1). The PTEN wild-type, C124S, and G129E vectors were obtained from Addgene and subcloned to pCXN2-IRES-GFP and pET-45b (Qi et al., 2015; Takahashi et al., 2006; Tamura et al., 1998). Wild-type and Y213F Abi1 cDNAs were kindly provided by Dr. Leszek Kotula (New York State Institute for Basic Research in Developmental Disabilities, New York, NY) and subcloned to pCXN2-IRES-GFP (Xiong et al., 2008). Abi1 S216A and Y213F/S216A mutations were generated using the Agilent QuikChange II site-directed mutagenesis kit. Mouse WAVE2 was amplified by RT-PCR from wild-type mouse ES cells and cloned into pCXN2-IRES-GFP. All the constructs were confirmed by DNA sequencing.
Chemical and protein reagents
Akt1/2 kinase inhibitor (catalog number A6730) and chloroquine (Chemical Abstracts Service number 50–63-50) were purchased from MilliporeSigma and used at final concentrations of 0.5 µM and 100 µM, respectively. ALLN was purchased from Calbiochem (Chemical Abstracts Service number 110044–82-1) and used at a final concentration of 0.5 µM. MG-132 was purchased from Selleckchem (catalog number S2619) and used at a final concentration of 0.1 µM. Active PTEN (catalog number P23-20G-10) was purchased from SignalChem. Immunoprecipitates were digested for 30 min with 0.1 µg/µl active PTEN in PTEN dilution buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 50 µM MgCl2, 0.05% Tween-20, and 0.2% 2-mercaptoethanal) provided by the manufacturer. Active porcine calpain 1 (1 U/µg protein, catalog number 1138) was purchased from BioVision. Digestion was performed for 30 min in 50 mM Tris buffer, pH 7.5, containing 150 mM NaCl, 1 mM EDTA, 1mM DTT, 2 mM CaCl2, and 0.5 U/µl active calpain 1.
Stable transfection of ES cells
For stable expression of Abi1 or WAVE2, wild-type ES cells were transfected with the corresponding vectors using Lipofectamine 3000 reagent (Invitrogen). Stable ES cell clones were selected with 500 µg/ml G418. GFP-positive colonies were cloned and expanded on Sandos inbred mouse-derived 6-thioguanine- and ouabain-resistant (STO) feeders. Reconstitution of Pten−/− ES cells with wild-type PTEN, G129E, or C124S was reported previously (Qi et al., 2015). For reconstitution of G418-resistant Pten−/−;Abi1−/− ES cells with either wild-type or mutant Abi1, selection of stable clones was based on GFP fluorescence and verified by RT-PCR detection of human Abi1 mRNAs. For knockdown of Abi1 or Nckap1, specific shRNAs and scrambled controls in the pRFP-C-RS vector were introduced into Pten−/− ES cells by transfection using Lipofectamine 3000 reagent. The cells were selected with 1 µg/ml puromycin, and RFP-positive clones were expanded and grown in medium containing puromycin. Abi2 knockdown was achieved by stable transfection of Pten−/− ES cells with Abi2 shRNAs in the pGFP-C-RS vector. For Abi1 and Abi2 double knockdown, Pten−/− ES cells with Abi1 knockdown were transfected with the effective Abi2 shRNA, and GFP/RFP double-positive cells were selected and confirmed by immunoblotting.
Generation of Pten−/−; Abi1−/− ES cell lines
Pten−/−; Abi1−/− ES cell lines were generated by disrupting the Abi1 gene in Pten−/− ES cells using the CRISPR-Cas9 approach as described previously (Ran et al., 2013). Briefly, the oligonucleotide pair encoding mouse Abi1-targeting sgRNA (top: 5′-CACCGACTACATGACTAGTCCTGCG-3′; bottom: 5′-AAACCGCAGGACTAGTCATGTAGTC-3′) was designed with the online CRISPR Design Tool (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) and cloned into the pSpCas9(BB)-2A-GFP vector (Addgene; 48138) to generate pSpCas9(BB)-Abi1-sgRNA. Pten−/− ES cells were transfected with pSpCas9(BB)-Abi1-sgRNA using Lipofectamine 3000. 48 h after transfection, the brightest 1% GFP-positive cells were sorted by FACS into 96-well plates (one cell per well). Abi1 knockout clones were screened by PCR genotyping (primer sequences are Forward 1, 5′-TGCCATCGTGCCTAGTTTGTAT-3′; Reverse knockout 1, 5′-CAATGACTACATGACTAGTCCTGC-3′; Forward knockout 1, 5′-GCTTCCAAGCCTCGCAGG-3′, Reverse 2 5′-TGCACAATACTCCTCTTCCCTG-3′) and further confirmed by DNA sequencing and immunoblotting.
Immunofluorescence
EB processing and immunostaining were performed as described previously (Li and Yurchenco, 2006). Briefly, EBs were fixed with 3% paraformaldehyde for 10 min at room temperature and washed three times with PBS. Fixed EBs were incubated in 7.5% sucrose-PBS at room temperature for 3 h and then in 15% sucrose-PBS at 4°C overnight. EBs were imbedded in Tissue-Tek O.C.T. Compound and sectioned on a Leica cryostat. For immunostaining, EB sections were incubated with primary antibodies at room temperature for 60 min followed by washing with PBS three times. Secondary antibody incubation time was 30 min. Nuclei were counterstained with 1 µg/ml DAPI in PBS for 10 min. Slides were viewed with a Nikon inverted fluorescence microscope (Eclipse TE2000) using a Nikon Plan Fluor 40×/1.30 oil objective at 20°C, and digital images were acquired with a Hamamatsu CCD (charge-coupled device) camera controlled by IP Lab software (Scanalytics). The fluorochromes used were Alexa Fluor 488, Cy3, and Cy5 (in case GFP was expressed). Three-color RGB (red-green-blue) images were constructed from single-channel images using IP Lab. Immunofluorescence intensity was analyzed using ImageJ software (National Institutes of Health) with six biological replicates. EBs containing the polarized epiblast epithelium were counted by live phase microscopy with a Nikon 10× phase objective. The data were collected in a blinded fashion. The percentage of epiblast polarization was calculated as EBs with polarized epiblast/total EBs counted × 100%.
Immunoprecipitation and immunoblotting
EBs were collected by allowing them to settle through gravity’s action and washed once in PBS. For immunoprecipitation and analysis of only cytoplasmic proteins, EBs were lysed in RIPA buffer containing protease and phosphatase inhibitor cocktails. For analysis of nuclear proteins, SDS lysis buffer containing 1% SDS and 50 mM Tris, pH 7.4, was used. Immunoprecipitation and immunoblotting were performed as described previously (Liu et al., 2009). Briefly, protein concentrations of cell lysates were determined using Pierce BCA protein assay reagents (Thermo Fisher Scientific). Protein samples (0.5–1.0 mg protein per sample) were precleared with 20 µl of 50% protein A/G Plus-agarose bead slurry (Santa Cruz Biotechnology; catalog number sc-2003). Precleared samples were incubated with antibody at 4°C for 2 h and then precipitated with 30 µl protein A/G-agarose beads overnight under gentle rotation. Immunoprecipitates were analyzed by SDS-polyacrylamide gel electrophoresis. The specificity of antibodies was confirmed using preimmune IgG as a control. For immunoblotting, proteins were transferred onto polyvinylidene fluoride membranes, which were probed with appropriate dilutions of primary antibody. Enhanced chemiluminescence reagent was used for signal development. To quantify immunoblots, the blots were scanned into computer at 600 dpi in TIFF format using an Epson dual lens transparency scanner. Band intensity (mean gray value) was measured using ImageJ software (National Institutes of Health) with the background subtracted.
Mass spectrometry
EB lysates (1.5 mg protein for each sample) were immunoprecipitated with anti-phosphotyrosine antibody–agarose beads (MilliporeSigma; catalog number 16–101). The immunoprecipitates were washed three times with Tris-buffered saline containing 0.4% Tween-20 and then analyzed in 4–10% SDS-polyacrylamide gradient gels. The experiments were repeated three times. Bands of interest were cut out of silver-stained gels and used for in-gel trypsin digests (specificity, carboxyl side of K and R). Samples were analyzed in duplicates by liquid chromatography–tandem mass spectometry using an LTQ Orbitrap Velos mass spectrometer (ThermoFisher) at Rutgers University Biological Mass Spectrometry facility as described previously (Xie et al., 2015). Peak lists were generated using Proteome Discover version 1.4 (Orbitrap Velos) with a minimum signal/noise of 1.5. The Global Proteome Machine version 2.2.1 (GPM) Cyclone XE (Beavis Informatics) was used to search mass spectrometry data against a unique mouse genome assembly based on GRCh38.79 (January 2012; 41,214 total genes, 22,592 protein-coding genes).
Substrate-trapping assay
Wild-type PTEN and C124S mutant cDNAs were subcloned into a pQE-31 vector (Qiagen) with an N-terminal His tag. The constructs were verified by DNA sequencing. Recombinant proteins were expressed in BL21 bacterial cells by IPTG induction and purified by affinity chromatography using nickel-nitrilotriacetic acid (NTA) agarose beads according to the manufacturer’s instructions (Qiagen). PTEN proteins bound to the agarose beads were quantified by densitometry analysis of Coomassie blue–stained gels; bovine serum albumin was used as a standard. A substrate-trapping assay was performed as described previously (Mercan and Bennett, 2010). Briefly, 5-d Pten−/− EBs were treated with 1 mM pervanadate and 100 nM calyculin A for 30 min to allow for accumulation of tyrosine and serine/threonine phosphorylation on proteins. The EBs were lysed in RIPA buffer. EB lysates were incubated with 10 mM DTT for 10 min on ice to inactivate phosphatase inhibitors. 1.5 mg EB lysates was then incubated with 10 µg His-tagged wild-type or mutant PTEN bound to NTA agarose beads at 4°C for 3 h. NTA agarose beads only served as a negative control. Affinity pull-down products were analyzed by immunoblotting for Abi1.
RT-PCR
Total RNA was isolated with TRIzol reagent (ThermoFisher Scientific) and reverse transcribed to cDNA using a SuperScript VILO cDNA synthesis kit as per manufacturer’s instructions (ThermoFisher Scientific). PCR reactions were performed using Bio-Rad MyCycler thermocycler and Taq DNA polymerase with ThermoPol buffer (New England Biolabs). The PCR primers for mouse Abi1 are 5′-GAGACCAAGGACGCATAGTG-3′ (forward) and 5′-GAGTTGGGCTATCAGCAATG-3′ (reverse). The primers for mouse WAVE2 are 5′-GCCGTTAGTAACCAGGAACATC-3′ (forward) and 5′-GGTTCACATTCCCTCTATTTGG-3′ (reverse). The primers for human Abi1 are 5′-GCTAGGCTTGGAAGTCAGCA-3′ (forward) and 5′-TCAACTGAGGCATAGGGGGA-3′ (reverse). The primers for 18S RNA are 5′-TCAAGAACGAAAGTCGGAGG-3′ (forward) and 5′-GGACATCTAAGGGCATCACA-3′ (reverse). PCR products were electrophoretically resolved on 2% agarose gels.
Statistical analysis
All the immunoblots shown are representative results of three or four independent experiments. Quantitative data are presented as mean ± SD. Statistical differences between multiple groups were evaluated by one-way ANOVA. Statistical analysis between two groups was performed using unpaired Student’s t test.
Online supplemental material
Fig. S1 shows that EB differentiation recapitulates the process of epiblast polarization during mouse peri-implantation development. Fig. S2 shows that active recombinant PTEN does not dephosphorylate phospho-Akt S473 or phospho-FAK Y397. Fig. S3 shows that Abi2 is mainly expressed in endoderm and not involved in epiblast polarization. Fig. S4 shows Pten/Abi1 double-knockout EBs. Fig. S5 shows localization of PIP3 and PIP2 in EBs. Data S1 shows microarray analysis of 2-d EBs. Total RNA was isolated from 2-d normal EBs with TRIZOL reagent and reverse transcribed to cRNA. Fragmented cRNAs were hybridized to mouse genome 430 2.0 microarray chips (Affymetrix). Hybridization data were analyzed by MAS software (v5.0; Affymetrix).
Supplementary Material
shows microarray analysis of 2-d EBs.
Acknowledgments
Pten-null ES cells were kindly provided by Dr. Pier Paolo Pandolfi (Harvard Medical School, Boston, MA).
The authors acknowledge support from the National Cancer Institute to S. Li (grant R21CA201715).
The authors declare no competing financial interests.
Author contributions: Conceptualization, S. Li and Y. Qi; Methodology, Y. Qi, J. Liu, and S. Li; Formal analysis, S. Li; Investigation, Y. Qi, J. Liu, J. Chao, and S. Li; Resources, P.A. Greer; Data curation, S. Li and Y. Qi; Writing of original draft, S. Li and Y. Qi; Writing – review and editing, S. Li, Y. Qi, J. Chao, and P.A. Greer; Supervision, S. Li; Funding acquisition, S. Li.
References
- Arthur J.S., Elce J.S., Hegadorn C., Williams K., and Greer P.A.. 2000. Disruption of the murine calpain small subunit gene, Capn4: calpain is essential for embryonic development but not for cell growth and division. Mol. Cell. Biol. 20:4474–4481. 10.1128/MCB.20.12.4474-4481.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi C., Ho J., Srikumar T., Dowling R.J., Gorrini C., Miller S.J., Mak T.W., Neel B.G., Raught B., and Stambolic V.. 2013. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 341:395–399. 10.1126/science.1236188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovellan M., Romeo Y., Biro M., Boden A., Chugh P., Yonis A., Vaghela M., Fritzsche M., Moulding D., Thorogate R., et al. . 2014. Cellular control of cortical actin nucleation. Curr. Biol. 24:1628–1635. 10.1016/j.cub.2014.05.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calle Y., Carragher N.O., Thrasher A.J., and Jones G.E.. 2006. Inhibition of calpain stabilises podosomes and impairs dendritic cell motility. J. Cell Sci. 119:2375–2385. 10.1242/jcs.02939 [DOI] [PubMed] [Google Scholar]
- Carracedo A., and Pandolfi P.P.. 2008. The PTEN-PI3K pathway: of feedbacks and cross-talks. Oncogene. 27:5527–5541. 10.1038/onc.2008.247 [DOI] [PubMed] [Google Scholar]
- Chalut K.J., and Paluch E.K.. 2016. The Actin Cortex: A Bridge between Cell Shape and Function. Dev. Cell. 38:571–573. 10.1016/j.devcel.2016.09.011 [DOI] [PubMed] [Google Scholar]
- Chen Z., Borek D., Padrick S.B., Gomez T.S., Metlagel Z., Ismail A.M., Umetani J., Billadeau D.D., Otwinowski Z., and Rosen M.K.. 2010. Structure and control of the actin regulatory WAVE complex. Nature. 468:533–538. 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugh P., and Paluch E.K.. 2018. The actin cortex at a glance. J. Cell Sci. 131:jcs186254. 10.1242/jcs.186254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff T.S., Wu T., Boward B.R., Yin A., Yin H., Glushka J.N., Prestegaard J.H., and Dalton S.. 2017. MYC Controls Human Pluripotent Stem Cell Fate Decisions through Regulation of Metabolic Flux. Cell Stem Cell. 21:502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coravos J.S., Mason F.M., and Martin A.C.. 2017. Actomyosin Pulsing in Tissue Integrity Maintenance during Morphogenesis. Trends Cell Biol. 27:276–283. 10.1016/j.tcb.2016.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David B.G., Fujita H., Yasuda K., Okamoto K., Panina Y., Ichinose J., Sato O., Horie M., Ichimura T., Okada Y., et al. . 2019. Linking substrate and nucleus via actin cytoskeleton in pluripotency maintenance of mouse embryonic stem cells. Stem Cell Res. (Amst.). 41 101614 10.1016/j.scr.2019.101614 [DOI] [PubMed] [Google Scholar]
- Davidson L., Maccario H., Perera N.M., Yang X., Spinelli L., Tibarewal P., Glancy B., Gray A., Weijer C.J., Downes C.P., et al. . 2010. Suppression of cellular proliferation and invasion by the concerted lipid and protein phosphatase activities of PTEN. Oncogene. 29:687–697. 10.1038/onc.2009.384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey N., Crosswell H.E., De P., Parsons R., Peng Q., Su J.D., and Durden D.L.. 2008. The protein phosphatase activity of PTEN regulates SRC family kinases and controls glioma migration. Cancer Res. 68:1862–1871. 10.1158/0008-5472.CAN-07-1182 [DOI] [PubMed] [Google Scholar]
- Di Cristofano A., and Pandolfi P.P.. 2000. The multiple roles of PTEN in tumor suppression. Cell. 100:387–390. 10.1016/S0092-8674(00)80674-1 [DOI] [PubMed] [Google Scholar]
- Di Cristofano A., Pesce B., Cordon-Cardo C., and Pandolfi P.P.. 1998. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19:348–355. 10.1038/1235 [DOI] [PubMed] [Google Scholar]
- Dubielecka P.M., Machida K., Xiong X., Hossain S., Ogiue-Ikeda M., Carrera A.C., Mayer B.J., and Kotula L.. 2010. Abi1/Hssh3bp1 pY213 links Abl kinase signaling to p85 regulatory subunit of PI-3 kinase in regulation of macropinocytosis in LNCaP cells. FEBS Lett. 584:3279–3286. 10.1016/j.febslet.2010.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubielecka P.M., Ladwein K.I., Xiong X., Migeotte I., Chorzalska A., Anderson K.V., Sawicki J.A., Rottner K., Stradal T.E., and Kotula L.. 2011. Essential role for Abi1 in embryonic survival and WAVE2 complex integrity. Proc. Natl. Acad. Sci. USA. 108:7022–7027. 10.1073/pnas.1016811108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutt P., Croall D.E., Arthur J.S., Veyra T.D., Williams K., Elce J.S., and Greer P.A.. 2006. m-Calpain is required for preimplantation embryonic development in mice. BMC Dev. Biol. 6:3 10.1186/1471-213X-6-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echarri A., Lai M.J., Robinson M.R., and Pendergast A.M.. 2004. Abl interactor 1 (Abi-1) wave-binding and SNARE domains regulate its nucleocytoplasmic shuttling, lamellipodium localization, and wave-1 levels. Mol. Cell. Biol. 24:4979–4993. 10.1128/MCB.24.11.4979-4993.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falet H., Pain S., and Rendu F.. 1998. Tyrosine unphosphorylated platelet SHP-1 is a substrate for calpain. Biochem. Biophys. Res. Commun. 252:51–55. 10.1006/bbrc.1998.9593 [DOI] [PubMed] [Google Scholar]
- Foty R.A., and Steinberg M.S.. 2005. The differential adhesion hypothesis: a direct evaluation. Dev. Biol. 278:255–263. 10.1016/j.ydbio.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Fournier M.V., Fata J.E., Martin K.J., Yaswen P., and Bissell M.J.. 2009. Interaction of E-cadherin and PTEN regulates morphogenesis and growth arrest in human mammary epithelial cells. Cancer Res. 69:4545–4552. 10.1158/0008-5472.CAN-08-1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S.B., and Miranti C.K.. 2013. Disruption of prostate epithelial differentiation pathways and prostate cancer development. Front. Oncol. 3:273 10.3389/fonc.2013.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A., Ho H.Y., Li J., Steen H., Gygi S.P., and Kirschner M.W.. 2004. Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci. USA. 101:4379–4383. 10.1073/pnas.0400628101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Tamura M., Pankov R., Danen E.H., Takino T., Matsumoto K., and Yamada K.M.. 1999. Shc and FAK differentially regulate cell motility and directionality modulated by PTEN. J. Cell Biol. 146:389–403. 10.1083/jcb.146.2.389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Wang Y., Sachs F., and Meng F.. 2014. Actin stress in cell reprogramming. Proc. Natl. Acad. Sci. USA. 111:E5252–E5261. 10.1073/pnas.1411683111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi S., Watanabe T.M., Kawauchi K., Ichimura T., and Fujita H.. 2014. Culturing of mouse and human cells on soft substrates promote the expression of stem cell markers. J. Biosci. Bioeng. 117:749–755. 10.1016/j.jbiosc.2013.11.011 [DOI] [PubMed] [Google Scholar]
- Innocenti M., Frittoli E., Ponzanelli I., Falck J.R., Brachmann S.M., Di Fiore P.P., and Scita G.. 2003. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 160:17–23. 10.1083/jcb.200206079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L.B., Steffen A., Stradal T.E., Di Fiore P.P., Carlier M.F., and Scita G.. 2004. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 6:319–327. 10.1038/ncb1105 [DOI] [PubMed] [Google Scholar]
- Innocenti M., Gerboth S., Rottner K., Lai F.P., Hertzog M., Stradal T.E., Frittoli E., Didry D., Polo S., Disanza A., et al. . 2005. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol. 7:969–976. 10.1038/ncb1304 [DOI] [PubMed] [Google Scholar]
- Kishimoto H., Hamada K., Saunders M., Backman S., Sasaki T., Nakano T., Mak T.W., and Suzuki A.. 2003. Physiological functions of Pten in mouse tissues. Cell Struct. Funct. 28:11–21. 10.1247/csf.28.11 [DOI] [PubMed] [Google Scholar]
- Knobbe C.B., Lapin V., Suzuki A., and Mak T.W.. 2008. The roles of PTEN in development, physiology and tumorigenesis in mouse models: a tissue-by-tissue survey. Oncogene. 27:5398–5415. 10.1038/onc.2008.238 [DOI] [PubMed] [Google Scholar]
- Kunda P., Craig G., Dominguez V., and Baum B.. 2003. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 13:1867–1875. 10.1016/j.cub.2003.10.005 [DOI] [PubMed] [Google Scholar]
- Lebensohn A.M., and Kirschner M.W.. 2009. Activation of the WAVE complex by coincident signals controls actin assembly. Mol. Cell. 36:512–524. 10.1016/j.molcel.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y., Zhang J., Badour K., Arpaia E., Freeman S., Cheung P., Siu M., and Siminovitch K.. 2005. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc. Natl. Acad. Sci. USA. 102:1098–1103. 10.1073/pnas.0409120102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie N.R., Yang X., Downes C.P., and Weijer C.J.. 2007. PtdIns(3,4,5)P(3)-dependent and -independent roles for PTEN in the control of cell migration. Curr. Biol. 17:115–125. 10.1016/j.cub.2006.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie N.R., Maccario H., Spinelli L., and Davidson L.. 2009. The significance of PTEN’s protein phosphatase activity. Adv. Enzyme Regul. 49:190–196. 10.1016/j.advenzreg.2008.12.002 [DOI] [PubMed] [Google Scholar]
- Li S., and Yurchenco P.D.. 2006. Matrix assembly, cell polarization, and cell survival: analysis of peri-implantation development with cultured embryonic stem cells. Methods Mol. Biol. 329:113–125. [DOI] [PubMed] [Google Scholar]
- Li S., Harrison D., Carbonetto S., Fassler R., Smyth N., Edgar D., and Yurchenco P.D.. 2002. Matrix assembly, regulation, and survival functions of laminin and its receptors in embryonic stem cell differentiation. J. Cell Biol. 157:1279–1290. 10.1083/jcb.200203073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Edgar D., Fässler R., Wadsworth W., and Yurchenco P.D.. 2003. The role of laminin in embryonic cell polarization and tissue organization. Dev. Cell. 4:613–624. 10.1016/S1534-5807(03)00128-X [DOI] [PubMed] [Google Scholar]
- Li Y., Luo X., Sun Y., Cui Z., Liu Y., Liu R., and Guo X.. 2016. High Stoichiometry Phosphorylation of Talin at T144/T150 or S446 Produces Contrasting Effects on Calpain-mediated Talin Cleavage and Cell Migration. J. Cancer. 7:1645–1652. 10.7150/jca.14192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw D., Marsh D.J., Li J., Dahia P.L., Wang S.I., Zheng Z., Bose S., Call K.M., Tsou H.C., Peacocke M., et al. . 1997. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 16:64–67. 10.1038/ng0597-64 [DOI] [PubMed] [Google Scholar]
- Liliental J., Moon S.Y., Lesche R., Mamillapalli R., Li D., Zheng Y., Sun H., and Wu H.. 2000. Genetic deletion of the Pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr. Biol. 10:401–404. 10.1016/S0960-9822(00)00417-6 [DOI] [PubMed] [Google Scholar]
- Liu J., He X., Corbett S.A., Lowry S.F., Graham A.M., Fässler R., and Li S.. 2009. Integrins are required for the differentiation of visceral endoderm. J. Cell Sci. 122:233–242. 10.1242/jcs.037663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin A.J., Lee K.C., Han S.J., Bui D.A., Davidson M., Mogilner A., and Danuser G.. 2015. Competition for actin between two distinct F-actin networks defines a bistable switch for cell polarization. Nat. Cell Biol. 17:1435–1445. 10.1038/ncb3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D., Jones G., Li X., Schönthal A.H., Gratzl O., Van Meir E.G., and Merlo A.. 1999. The PTEN lipid phosphatase domain is not required to inhibit invasion of glioma cells. Cancer Res. 59:5479–5482. [PubMed] [Google Scholar]
- Martin-Belmonte F., Gassama A., Datta A., Yu W., Rescher U., Gerke V., and Mostov K.. 2007. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell. 128:383–397. 10.1016/j.cell.2006.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M.C. 2013. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin. Cell Dev. Biol. 24:272–279. 10.1016/j.semcdb.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M.C., Er E.E., Zhang W., Ballif B.A., Elliott H.L., Danuser G., and Blenis J.. 2011. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol. Cell. 41:661–671. 10.1016/j.molcel.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Cai K.Q., Moore R., Tao W., Tse J.D., Smith E.R., and Xu X.X.. 2017. Pten facilitates epiblast epithelial polarization and proamniotic lumen formation in early mouse embryos. Dev. Dyn. 246:517–530. 10.1002/dvdy.24503 [DOI] [PubMed] [Google Scholar]
- Mercan F., and Bennett A.M.. 2010. Analysis of protein tyrosine phosphatases and substrates. Curr Protoc Mol Biol. Chapter 18:Unit 18 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M.P., Pass I., Batty I.H., Van der Kaay J., Stolarov J.P., Hemmings B.A., Wigler M.H., Downes C.P., and Tonks N.K.. 1998. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc. Natl. Acad. Sci. USA. 95:13513–13518. 10.1073/pnas.95.23.13513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y., and Sorimachi H.. 2012. Calpains: an elaborate proteolytic system. Biochim. Biophys. Acta. 1824:224–236. 10.1016/j.bbapap.2011.08.005 [DOI] [PubMed] [Google Scholar]
- Podsypanina K., Ellenson L.H., Nemes A., Gu J., Tamura M., Yamada K.M., Cordon-Cardo C., Catoretti G., Fisher P.E., and Parsons R.. 1999. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc. Natl. Acad. Sci. USA. 96:1563–1568. 10.1073/pnas.96.4.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., and Borisy G.G.. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- Poon J.S., Eves R., and Mak A.S.. 2010. Both lipid- and protein-phosphatase activities of PTEN contribute to the p53-PTEN anti-invasion pathway. Cell Cycle. 9:4450–4454. 10.4161/cc.9.22.13936 [DOI] [PubMed] [Google Scholar]
- Qi Y., Liu J., Saadat S., Tian X., Han Y., Fong G.H., Pandolfi P.P., Lee L.Y., and Li S.. 2015. PTEN induces apoptosis and cavitation via HIF-2-dependent Bnip3 upregulation during epithelial lumen formation. Cell Death Differ. 22:875–884. 10.1038/cdd.2014.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou M., Etienne-Manneville S., Self A., Nicholls S., and Hall A.. 2004. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 303:1179–1181. 10.1126/science.1092089 [DOI] [PubMed] [Google Scholar]
- Ralston A., and Rossant J.. 2010. The genetics of induced pluripotency. Reproduction. 139:35–44. 10.1530/REP-09-0024 [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S., Wells R., and Rechsteiner M.. 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 234:364–368. 10.1126/science.2876518 [DOI] [PubMed] [Google Scholar]
- Ryu J.R., Echarri A., Li R., and Pendergast A.M.. 2009. Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol. Cell. Biol. 29:1735–1748. 10.1128/MCB.01483-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansal I., and Sellers W.R.. 2004. The biology and clinical relevance of the PTEN tumor suppressor pathway. J. Clin. Oncol. 22:2954–2963. 10.1200/JCO.2004.02.141 [DOI] [PubMed] [Google Scholar]
- Sasidharan S., Borinskaya S., Patel F., Bernadskaya Y., Mandalapu S., Agapito M., and Soto M.C.. 2018. WAVE regulates Cadherin junction assembly and turnover during epithelial polarization. Dev. Biol. 434:133–148. 10.1016/j.ydbio.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde S.R., and Maddika S.. 2016. PTEN modulates EGFR late endocytic trafficking and degradation by dephosphorylating Rab7. Nat. Commun. 7:10689 10.1038/ncomms10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumway S.D., Maki M., and Miyamoto S.. 1999. The PEST domain of IkappaBalpha is necessary and sufficient for in vitro degradation by mu-calpain. J. Biol. Chem. 274:30874–30881. 10.1074/jbc.274.43.30874 [DOI] [PubMed] [Google Scholar]
- Song M.S., Carracedo A., Salmena L., Song S.J., Egia A., Malumbres M., and Pandolfi P.P.. 2011. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 144:187–199. 10.1016/j.cell.2010.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.S., Salmena L., and Pandolfi P.P.. 2012. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13:283–296. 10.1038/nrm3330 [DOI] [PubMed] [Google Scholar]
- Stambolic V., Suzuki A., de la Pompa J.L., Brothers G.M., Mirtsos C., Sasaki T., Ruland J., Penninger J.M., Siderovski D.P., and Mak T.W.. 1998. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 95:29–39. 10.1016/S0092-8674(00)81780-8 [DOI] [PubMed] [Google Scholar]
- Steffen A., Rottner K., Ehinger J., Innocenti M., Scita G., Wehland J., and Stradal T.E.. 2004. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23:749–759. 10.1038/sj.emboj.7600084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., de la Pompa J.L., Stambolic V., Elia A.J., Sasaki T., del Barco Barrantes I., Ho A., Wakeham A., Itie A., Khoo W., et al. . 1998. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr. Biol. 8:1169–1178. 10.1016/S0960-9822(07)00488-5 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Morales F.C., Kreimann E.L., and Georgescu M.M.. 2006. PTEN tumor suppressor associates with NHERF proteins to attenuate PDGF receptor signaling. EMBO J. 25:910–920. 10.1038/sj.emboj.7600979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura M., Gu J., Matsumoto K., Aota S., Parsons R., and Yamada K.M.. 1998. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 280:1614–1617. 10.1126/science.280.5369.1614 [DOI] [PubMed] [Google Scholar]
- Tang Y., and Eng C.. 2006. PTEN autoregulates its expression by stabilization of p53 in a phosphatase-independent manner. Cancer Res. 66:736–742. 10.1158/0008-5472.CAN-05-1557 [DOI] [PubMed] [Google Scholar]
- Tsankova A., Pham T.T., Garcia D.S., Otte F., and Cabernard C.. 2017. Cell Polarity Regulates Biased Myosin Activity and Dynamics during Asymmetric Cell Division via Drosophila Rho Kinase and Protein Kinase N. Dev. Cell. 42:143–155. [DOI] [PubMed] [Google Scholar]
- Xie X.J., Hsu F.N., Gao X., Xu W., Ni J.Q., Xing Y., Huang L., Hsiao H.C., Zheng H., Wang C., et al. . 2015. CDK8-Cyclin C Mediates Nutritional Regulation of Developmental Transitions through the Ecdysone Receptor in Drosophila. PLoS Biol. 13 e1002207 10.1371/journal.pbio.1002207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X., Cui P., Hossain S., Xu R., Warner B., Guo X., An X., Debnath A.K., Cowburn D., and Kotula L.. 2008. Allosteric inhibition of the nonMyristoylated c-Abl tyrosine kinase by phosphopeptides derived from Abi1/Hssh3bp1. Biochim. Biophys. Acta. 1783:737–747. 10.1016/j.bbamcr.2008.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamorano J., Rivas M.D., Setien F., and Perez-G M.. 2005. Proteolytic regulation of activated STAT6 by calpains. J. Immunol. 174:2843–2848. 10.4049/jimmunol.174.5.2843 [DOI] [PubMed] [Google Scholar]
- Zhang S., Huang W.C., Li P., Guo H., Poh S.B., Brady S.W., Xiong Y., Tseng L.M., Li S.H., Ding Z., et al. . 2011. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat. Med. 17:461–469. 10.1038/nm.2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman U.J., Boring L., Pak J.H., Mukerjee N., and Wang K.K.. 2000. The calpain small subunit gene is essential: its inactivation results in embryonic lethality. IUBMB Life. 50:63–68. 10.1080/15216540050176610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows microarray analysis of 2-d EBs.