Super-resolution imaging shows that G1/S transcription factors in budding yeast are spatially organized in small clusters that increase in number, not molecular content, as the transcription factor copy number increases with cell growth in G1. This spatial and temporal organization of the G1/S regulon might help coordinate the Start transition.
Abstract
In budding yeast, the transcription factors SBF and MBF activate a large program of gene expression in late G1 phase that underlies commitment to cell division, termed Start. SBF/MBF are limiting with respect to target promoters in small G1 phase cells and accumulate as cells grow, raising the questions of how SBF/MBF are dynamically distributed across the G1/S regulon and how this impacts the Start transition. Super-resolution Photo-Activatable Localization Microscopy (PALM) mapping of the static positions of SBF/MBF subunits in fixed cells revealed each transcription factor was organized into discrete clusters containing approximately eight copies regardless of cell size and that the total number of clusters increased as cells grew through G1 phase. Stochastic modeling using reasonable biophysical parameters recapitulated growth-dependent SBF/MBF clustering and predicted TF dynamics that were confirmed in live cell PALM experiments. This spatio-temporal organization of SBF/MBF may help coordinate activation of G1/S regulon and the Start transition.
Introduction
Budding yeast cells must achieve a characteristic critical size before the commitment to division in late growth (G1) phase, an event termed Start (Hartwell et al., 1974; Johnston et al., 1977). Start depends on activation of an extensive G1/S transcriptional regulon comprised of ∼200 genes that function in macromolecular biosynthesis, bud emergence, DNA replication, spindle pole body duplication, and other critical processes (Jorgensen and Tyers, 2004). The G1/S transcriptional program is controlled by two master transcription factor (TF) complexes, SCB-binding factor (SBF) and MCB Binding Factor (MBF), each made up of a DNA binding subunit, Swi4 and Mbp1, respectively, and a common activator subunit, Swi6 (Koch et al., 1993). SBF and MBF recognize specific sites in G1/S promoter regions, called SCB (Swi4/6 cell cycle box) and MCB (MluI cell cycle box) sites, with some degree of overlapping specificity (Koch et al., 1993; Iyer et al., 2001; Bean et al., 2005). ChipSeq experiments have delineated >450 binding sites for Swi4, Mbp1, and Swi6 in the genome (Iyer et al., 2001; Park et al., 2013; Simon et al., 2001; Lee et al., 2002), although the agreement between these various studies is only partial (Ferrezuelo et al., 2010).
Prior to Start, a transcriptional repressor called Whi5 binds to and inhibits SBF. At Start, this inhibition is alleviated by phosphorylation of Whi5 and SBF by the G1 cyclin (Cln)-Cdc28 protein kinase complexes, which disrupts the SBF-Whi5 interaction and drives nuclear export of Whi5 (Costanzo et al., 2004; de Bruin et al., 2004). The upstream G1 cyclin, Cln3, is thought to initiate a positive feedback loop in which SBF-dependent expression of CLN1/2 further amplifies (Cln)-Cdc28 activity and thus SBF activation (Skotheim et al., 2008). The growth-dependent trigger for Start remains unclear but likely depends on a combination of factors including the accumulation of transcriptional activators, nutrient signaling, and metabolic flux (Jorgensen et al., 2004; Schmoller et al., 2015; Talarek et al., 2017; Dorsey et al., 2018; Litsios et al., 2019). Absolute measurements of the concentrations of the G1/S TFs in single G1 phase cells demonstrated that the SBF/MBF copy numbers are sub-saturating with respect to the target promoters in small cells, and that TF levels increase as cells grow, suggesting titration of the G1/S promoters (Dorsey et al., 2018).
Based on recent Swi6 ChipSeq data, bioinformatics approaches have been used to map the Swi6 target sites onto a 3D model of the budding yeast G1 phase genome (Park et al., 2013; Capurso et al., 2016; Duan et al., 2010). This model predicted functional 3D hotspots for Swi6 binding, in particular the MSB2 and ERG11 genes. A combination of ChipSeq and chromatin capture data suggests many transcription factors in budding yeast, including Swi4 and Swi6, have targets sites that cluster in space (Ben-Elazar et al., 2013; Duan et al., 2010). Swi4 and Swi6 have been shown to be associated with highly transcriptionally active gene clusters (Tsochatzidou et al., 2017). While these domains seem to separate regions of similarly timed replication origins, their relation to the timing of the Start transition has not been characterized. Despite the strong inference of TF clustering from these studies, the spatial and temporal organization of the G1/S TFs and their target sites has not been directly observed.
Here, we have used a super-resolution method, Photo-Activatable Localization Microscopy (PALM; Betzig et al., 2006; Rust et al., 2006), to map the static and dynamic positions of fusions of Swi4, Mbp1, and Swi6 with the photoactivatable protein mEos3.2 (Zhang et al., 2012) expressed from their natural loci in fixed and live budding yeast cells. PALM images of fixed cells provided 2D projections of the 3D organization of these proteins in the nucleus. We found that the TFs organize into clusters that increase in number from ∼5 clusters in small G1 phase cells to ∼30 clusters in large G1 phase cells, while the number of molecules per cluster, approximately eight monomers (four dimers), was independent of cell size. Given that throughout most of G1 phase, SBF/MBF copy numbers are limiting with respect to the ∼200 G1/S promoters (Dorsey et al., 2018), the observed SBF/MBF clustering suggests close spatial proximity of several promoter sites within each cluster. A mathematical model and Monte Carlo computer simulations based on simple biophysical assumptions predicted TF clustering if the target promoters were clustered. The simulations also indicated that protein interactions mediated by Swi6 enhance the clustering of the SBF/MBF DNA binding subunits, Swi4 and Mbp1. Finally, the simulations predicted that SBF/MBF subunits alternate between a highly confined state, in which they are trapped within G1/S promoter clusters, and a highly dynamic state, in which they hop rapidly between clusters. Live cell single particle tracking (spt)-PALM verified the prediction of distinct slow sub-diffusive and fast diffusive dynamic modes for these factors. Overall, these results suggest that the promoters of the G1/S regulon are spatially organized into discrete clusters that are successively titrated by increasing TF copy number as cells grow.
Results
The G1/S transcription factors are clustered in yeast nuclei
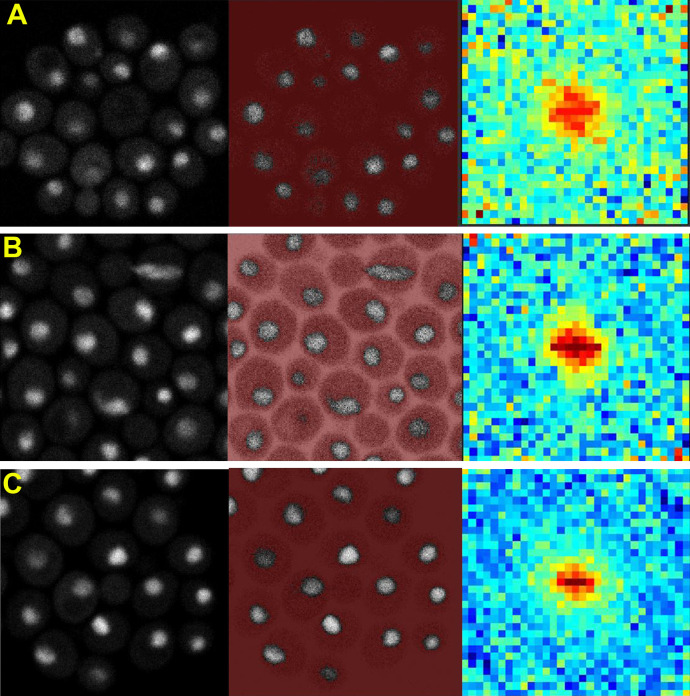
Super-resolution PALM imaging of mEos3.2 fusions of Swi4, Mbp1, and Swi6 in single nuclei from fixed cells grown on rich (synthetic complete [SC] + 2% glucose) medium revealed nonhomogeneous distributions for each protein (Fig. 1). No size phenotype was observed for these strains, as with our prior studies on strains expressing analogous GFP fusions (Dorsey et al., 2018), indicating that these crucial TFs retain their function when fused to fluorescent proteins. The super-resolution detection images with a resolution of ∼25 nm (Fig. 1) resulted from the super-position of all detections of all Swi4-, Mbp1-, and Swi6-mEos3.2 molecules, respectively, over the ∼30,000–40,000 frames acquired for a given field of view (FOV). However, since each individual TF molecule was cross-linked by fixation and thus immobile, and was detected multiple times in successive frames (up to 100), we averaged its mean position over multiple detections to obtain a molecular image (Betzig et al., 2006) at a 6-nm resolution (Fig. 1, lower panels for each TF as indicated). It was apparent from both the detection and the molecular images that in all G1 phase cells (small, medium, and large), most nuclear Swi4, Mbp1, and Swi6 molecules were organized in discrete clusters of a few molecules. Clusters of Swi6, Mbp1, and Swi4 were also observed in cells grown on SC+2% glycerol, a poor carbon source (Fig. 2). The SBF repressor Whi5 (Costanzo et al., 2004) also formed clusters (Fig. 2).
Figure 1.
Super-resolution PALM imaging reveals clustering of Swi4-, Mbp1-, and Swi6-mEos3.2 in fixed budding yeast cells. (A–C) PALM imaging of from Swi4- (A), Mbp1- (B), and Swi6-mEos3.2 (C). For all panels: top panels in A–C: Composite images of the phase contrast and PALM detection images with, left to right, the full FOV and zoomed images of a small, medium, and large cell (indicated by the yellow circles in the full FOV image). Scale bar for the full FOV is 5 µm for Swi4-mEos3.2 and Swi6-mEos3.2 and 10 µm for Mbp1-mEos3.2. Scale bars for the zoomed composite images are 1 µm. Detection image outputs (pink dots; one for each detection) were obtained with the Thunderstorm plugin from ImageJ and were not filtered for blinking. High-intensity purple in the image corresponds to out-of-focus beads. Lower panels in A–C: Zoomed molecular images of the small, medium, and large cells in the composite images in the top panels of A–C. Molecular images were created as described in the text and corrected for blinking. Scale bars in panels, left to right, are 0.15 µm, 0.24 µm, and 0.3 µm for Swi4-mEos3.2, 0.21 µm, 0.21 µm, and 0.21 µm for Mbp1-mEos3.2 and 0.15 µm, 0.15 µm, and 0.27 µm for Swi6-mEos3.2.
Figure 2.
Super-resolution PALM imaging of Whi5-mEos3.2 in SC + 2% glucose, Swi4-mEos3.2, Mbp1-mEos3.2, and Swi6-mEos3.2 in SC + 2% glycerol and NLS-mEos3.2-NLS in SC + 2% glucose. (A) PALM imaging of Whi5-mEos3.2. Top panels: Composite phase contrast image and the detection image (red dots; one for each detection) output produced with Thunderstorm. The detection image is not filtered for blinking. Saturated red intensity in the image corresponds to out-of-focus beads or compromised cells. Left to right are the full FOV and zoomed cells 1 (small), 2 (medium), and 3 (large) described below are indicated by yellow circles and numbers. Cells were grown in SC + 2% glucose. Scale bar is 5 µm for full FOV and 1 µm for each of the zoomed images. Bottom panels: Molecular images of the nuclei of indicated cells 1–3, respectively, left to right (from yellow squares in top panels). Molecular images were created as described in the text and corrected for blinking. Scale bars, 0.3 µm. (B) Molecular images of cells in SC + 2% glycerol medium expressing, left to right, Swi4-mEos3.2, Mbp1-mEos3.2, and Swi6-mEos3.2, respectively. Scale bars are 0.18, 0.24, and 0.3 µm, left to right, respectively. (C) PALM imaging of the NLS-mEos3.2-NLS control. Top panels: Composite phase contrast images and the detection images (red dots; one for each detection) output was produced with Thunderstorm. The detection image is not filtered for blinking. Images from left to right correspond to the full FOV, and cells 1–3, respectively. Scale bar in full FOV is 5 µm, and 1 µm for the zoomed cells 1–3. Bottom panels: Molecular images of the nuclei of indicated cells 1–3, respectively, left to right (from yellow squares in top panels). Molecular images were created as described in the text and corrected for blinking. Scale bars, 0.3 µm.
The number of clusters of SBF/MBF increases with cell size while copy number per cluster remains constant
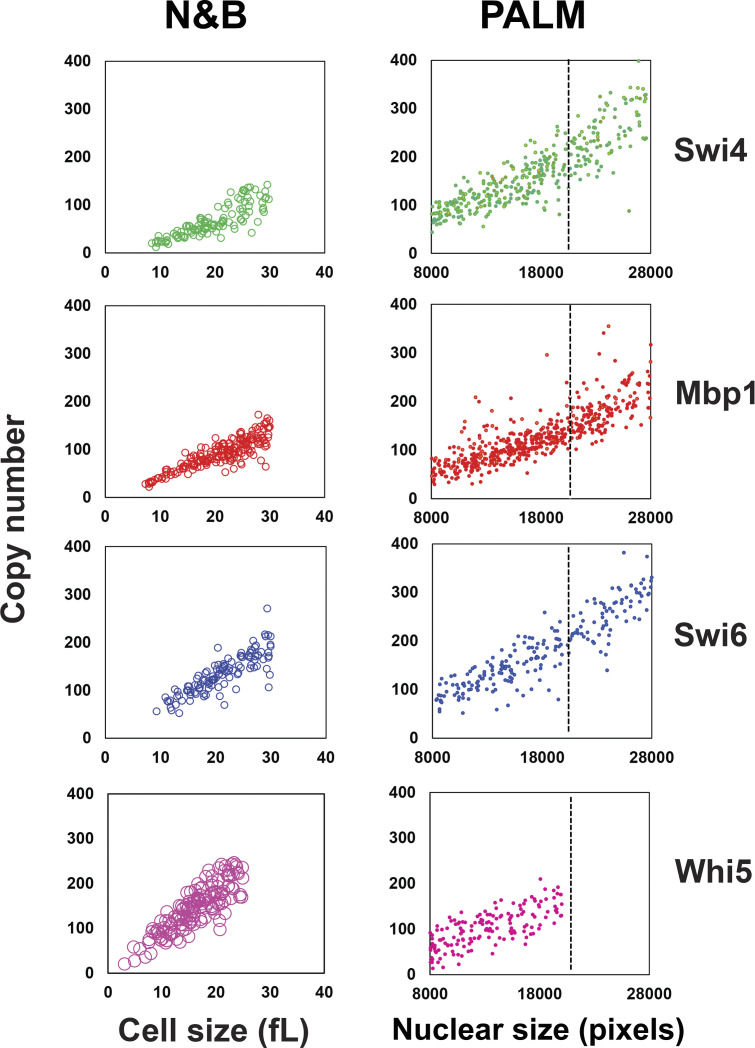
The number of molecules detected in each nucleus was obtained from analysis of the blinking-corrected molecular images (Fig. 1, lower panels for each TF as indicated). The nuclear copy numbers of Swi4, Mbp1, Swi6, and Whi5 increased with cell size (Fig. 3, A–D), consistent with our previously reported size-dependent increase in G1/S copy number determined by Number and Brightness (N&B) fluctuation microscopy (Dorsey et al., 2018). Given their nuclear size, most cells to the left of the black dashed lines in Fig. 3 were expected to be in G1 phase (see Materials and methods, Fig. 4 A). Consistently, preStart cells showing nuclear Whi5 were on the left of the black dashed lines (Fig. 3, D, H, and L). The average number of proteins per nucleus was in reasonably good agreement with values determined by N&B of 50–100 copies in small G1 phase cells and 100–200 copies in large G1 phase cells (Fig. S1), although we detected somewhat fewer molecules in the PALM experiments, especially in large cells. This difference is most likely due to the limited depth of field in the PALM experiments (see Materials and methods). We note that PALM microscopy is not as reliable as N&B for particle counting due to blinking of mEos3.2, imperfect correction thereof (Lee et al., 2012), incomplete activation of mEos3.2, and exclusion of out-of-focus particles.
Figure 3.
Copy numbers and the number of clusters per cell for Swi4-mEos3.2, Mbp1-mEos3.2, Swi6-mEos3.2, and Whi5-mEos3.2 increase with cell size. (A–D) Total number of Swi4-, Mbp1-, Swi6-, and Whi5-mEos3.2 molecules, respectively, in each nucleus as a function of nuclear size, a proxy for cell size. Each point represents an individual nucleus. (E–H) Number of Swi4-, Mbp1-, Swi6-, and Whi5-mEos3.2 clusters per nucleus as a function of cell size. (I–L) Average number of Swi4-, Mbp1-, Swi6-, and Whi5-mEos3.2 molecules per cluster for each nucleus as a function of cell size. Moving from left to right, panels correspond to Swi4-mEos3.2 (green, 370 cells), Mbp1-mEos3.2 (red, 536 cells), Swi6-mEos3.2 (blue, 222 cells) and Whi5-mEos3.2 (pink, 155 cells). Nuclear size is given in pixels. To the left of the dashed lines at 20,000 pixels, the majority of cells are in G1 phase as determined by the presence of Whi5-mEos3.2 in the nucleus (D, H, and L).
Figure 4.
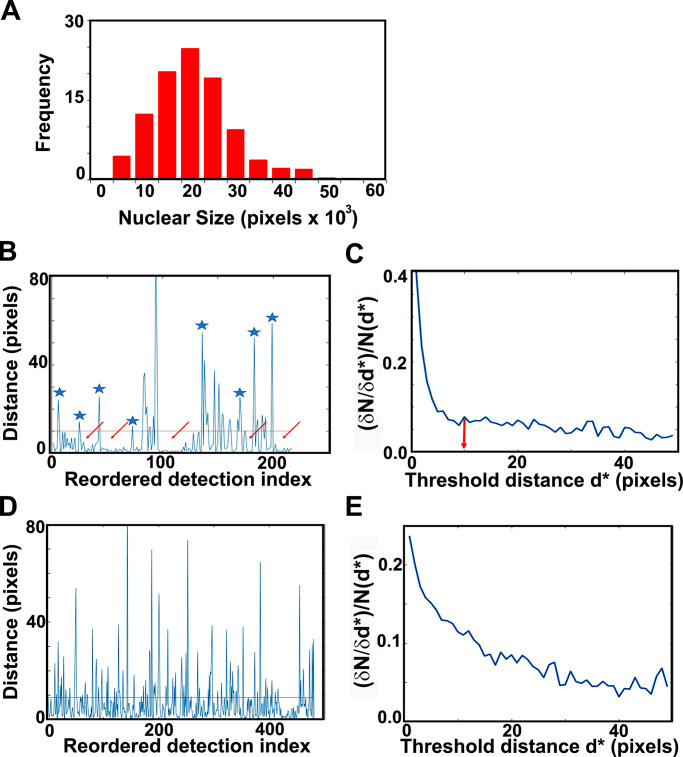
G1/S TF cluster statistics. (A) Nuclear size distribution, as determined by our masking algorithm. Distribution of nuclear sizes (in pixel units; 1 pixel = 3 nm) obtained from our entire dataset of Mbp1-mEos3.2 cells grown in SC + 2% glucose medium. This distribution resembles the cell size distribution of asynchronously growing cultures; therefore, we used it as a proxy for cell size and in particular the size of G1/S cells (mode size, 20,000 pixels). Assuming spherical cells and nuclei, and the 1/7 karyoplasmic to cytoplasmic ratio (Jorgensen et al., 2007), this corresponds to a cell size of 30 fl. The Mbp1-mEos3.2 strain was chosen for this purpose, because unlike Swi4 and Swi6, Mbp1 nuclear signal shows no cell cycle dependence, yielding a (masked) nuclear size distribution that mirrors the entire cell size distribution. (B and C) Cluster quantification algorithm. Example calculations in B and C are for Swi4. (B) Distance to the next particle (vertical axis, in high resolution pixel units, 1 pixel = 3 nm) as a function of the position of the particle in the nearest-neighbor ranked list (see Materials and methods and supplemental figures for details). Clusters appear as valleys (arrows), wherein inter-particle distances fall below the chosen cluster detection threshold (10 pixels = 30 nm, red line), and are separated by larger distances (blue asterisks). (C) Relative variation of the number of clusters (vertical axis, cluster count change upon 3 nm threshold increment over initial cluster count ratio, (δN/δd*)/N(d*) as a function of the specific choice of the cluster detection threshold, d*, (horizontal axis, in high-resolution pixel unit, 1 pixel = 3 nm). (D and E) Quantification of NLS-mEos3.2-NLS PALM data reveals no significant clustering. (D) Distance to the next particle (vertical axis, in high-resolution pixel units, 1 pix = 3 nm) as a function of the position of the particle in the nearest-neighbor ranked list (see Materials and methods section for details). The absence of large valleys in this plot (as compared with B) is indicative of the absence of NLS-mEos3.2-NLS clustering. (E) Relative variation of the number of detected putative clustsers (vertical axis, cluster count change upon 3 nm threshold increment over initial cluster count ratio) as a function of the specific choice of the cluster detection threshold (horizontal axis, in high-resolution pixel unit, 1 pixel = 3 nm). The absence of obvious threshold value beyond which the relative cluster counts does not change is indicative of an even distribution of NLS-mEos3.2-NLS molecules, and of a lack of actual cluster formation.
Figure S1.
Comparison of the copy numbers of Swi4-GFP, Mbp1-GFP, Swi6-GFP, and Whi5-GFP determined by scanning number and brightness and PALM. Left panels: Copy number vs. cell size (in fl) for Swi4, Mbp1, Swi6, and Whi5 as indicated, determined by scanning Number and Brightness (sN&B; Dorsey et al., 2018). Only G1 cells are shown. Right panels: Copy number vs. nuclear size (in pixels) determined by PALM for Swi4-mEos3.2, Mbp1-mEos3.2, Swi6-mEos3.2, and Whi5-mEos3.2 as indicated. A dotted line has been placed at the critical cell size below which the majority of cells are in G1 phase. As cells become larger, the efficiency of PALM detection decreases due to out-of-focus and hence undetected molecules. This effect is particularly clear for Mbp1 and Swi6.
To quantify the number of clusters in each nucleus and the number of molecules in each cluster, we developed a custom cluster identification algorithm based on a simplification of the OPTICS ranking algorithm (Kriegel et al., 2011). Briefly, our algorithm iteratively reorders the list of all molecular detections within each individual nucleus such that each molecule is listed adjacent to its nearest neighbor in real space (see Materials and methods). We then compute the list of distances between each molecule and the next on the list (see Fig. 4 B, for an example). To define and distinguish clusters, a distance threshold (i.e., threshold spike amplitude separating two different clusters) of 10 high-resolution pixels, corresponding to 10 pixels × 3 nm/pixel = 30 nm, was chosen (Fig. 4 B, red horizontal line). This threshold value was chosen because the relative change in cluster number with respect to the threshold did not change significantly beyond this critical distance of 30 nm (Fig. 4 C). Finally, since Mbp1, Swi4 and Swi6 predominantly occur as dimers (Dorsey et al., 2018), we defined a cluster as a group of molecules larger than at least 2 dimers (i.e., 4 molecules). Unlike previously published cluster detection algorithms (Mazouchi and Milstein, 2015), our algorithm detected the small clusters observed for these proteins, even for the sparse clusters found in small G1 cells. The mEos3.2 protein has been characterized biophysically and is essentially monomeric (Wang et al., 2014), making it unlikely to be responsible for the clustering of Swi4, Mbp1, and Swi6. As a control we constructed a strain expressing from a plasmid mEos3.2 molecules fused at both their N and C termini to nuclear localization signal (NLS) peptides to ensure nuclear localization. As expected, the images of the cells producing the NLS-mEos3.2-NLS construct did not show significant cluster formation (Fig. 2, bottom panels). Moreover, unlike the G1/S TFs, cluster analysis of the NLS-mEos3.2-NLS images showed very few wide valleys (corresponding to clusters) in the reordered distance plots (Fig. 4 D). In addition, the threshold distance dependence of cluster counts exhibited no obvious cutoff value (Fig. 4 E), as expected for evenly distributed molecules, compared with the sharp decrease to a constant level observed for the G1/S TFs (Fig. 4 C). These results demonstrated that the clustering observed for the G1/S TFs was not due to optical or physical-chemical properties of the mEos3.2 tag.
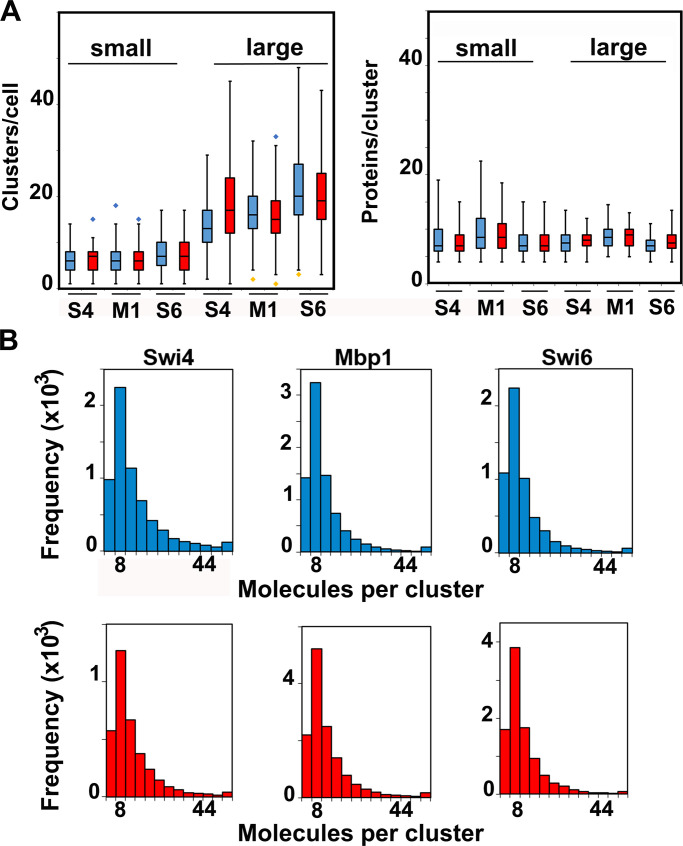
For each TF, the number of clusters increased with cell size (Fig. 3, E–G; and Fig. 5 A) in both glucose and glycerol medium. In contrast, most clusters contained a cell size–independent number of approximately eight molecules (four dimers), although some were significantly larger (Fig. 3, I–K; and Fig. 5, B and C). Regardless of cell size, 85% of all molecules were located in clusters, whose lateral extension was in the 30–80 nm range (Fig. 1). These results suggested that as TF copy number increased with cell growth, TFs formed new clusters rather than associating with existing ones. The number of clusters for each TF in the largest cells reached ∼20–30, much lower than the ∼200 G1/S promoters and the ∼600 target sites across all G1/S promoters (Iyer et al., 2001; Ferrezuelo et al., 2010). Interestingly, Swi6 clusters had about the same average number of molecules as Swi4 or Mbp1, such that the higher copy number of Swi6 molecules (with respect to Swi4 or Mbp1) was reflected by a larger number of Swi6-containing clusters. However, as the total number of Swi6 clusters for any cell size was significantly smaller than the sum of Swi4 and Mbp1 clusters, we inferred that most clusters contained both MBF and SBF. The number of Whi5 clusters at Start was lower than the number of target Swi6 clusters (compare Fig. 3 H to Fig. 3 G), consistent with the molecular images (Fig. 2, E–G, top panels), while the number of Whi5 molecules per cluster was slightly higher (Fig. 3 L).
Figure 5.
Cluster statistics are largely independent of carbon source. (A) Number of clusters (left panel) and number of TF protein molecules per cluster (right panel) for Swi4-mEos3.2 (S4), Swi6-mEos3.2 (S6), and Mbp1-mEos3.2 (M1) in small cells (<20,000 pixels, left of each panel) and large cells (>20,000 pixels, right of each panel). Cells grown in glucose (blue) and glycerol (red). The box and whisker plots represent the distribution of values across three to five experiments, each encompassing >100 cells for each condition, and blue and yellow diamonds represent distribution outliers. The difference in cluster counts between small and large cells was statistically significant in each condition (P values <1e-40). There were significantly more Swi6-mEos3.2 clusters than Swi4-mEos3.2 or Mbp1-mEos3.2 clusters, in both large and small cells and in glucose and glycerol (P values ranging from 10e-23 to 0.0197). (B) Histograms showing molecular content for all clusters of Swi4-mEos3.2, Mbp1-mEos3.2, and Swi6-mEos3.2 as indicated in all nuclei for cells grown in SC + 2% glucose (blue) and SC + 2% glycerol (red). The frequency unit (vertical axis) is actual cluster counts within each bin across all datasets (3–5 experiments, >100 cells each) for each condition.
Given that Swi4 and Mbp1 copy numbers are only in slight excess with respect to the number of target G1/S promoters in large cells at the end of G1 phase (Dorsey et al., 2018), the organization of the TFs into ∼30 clusters (Fig. 3, E–H) indicated that G1/S promoters might also be clustered to facilitate synchronous expression of the G1/S regulon. In this view, most G1/S promoters would be spatially organized into ∼30 clusters (7–10 promoters each) that are progressively titrated by G1/S TFs as cells grow. In contrast, the limiting number of Swi4-Mbp1 dimers might have been expected to partially populate such hypothetical promoter clusters even in small cells (Dorsey et al., 2018), and newly synthesized molecules would also randomly distribute across all clusters of target sites. This alternative scenario would result in a constant number of clusters that increase in TF copy number per cluster as cells grow. Our data unequivocally support the former model in which most G1/S promoters are spatially organized into clusters that are successively titrated by the increasing copy number of G1/S TFs as cells grow.
A quantitative model couples G1/S DNA promoter clusters to TF clusters
We developed a simple mathematical model and used Monte Carlo computer simulations to explore the biophysical parameters that might explain the observed spatial patterns of the G1/S TFs (Fig. 6). The goal was to identify the minimum number of parameters with physically reasonable values that could reproduce the observed experimental clustering, and in particular the counter-intuitive result that SBF/MBF form clusters even in small G1 cells where the TF copy numbers barely exceed the ∼30 G1/S promoter clusters revealed by our data in large cells. The dynamic Monte Carlo model was based on the SBF/MBF binding module of our previously published Start model (Dorsey et al., 2018), with an additional constraint that G1/S promoters first were organized into preformed clusters (see Materials and methods). We assumed no higher order interactions between SBF and MBF complexes. The SBF/MBF binding module encompasses mass action–driven binding of Swi4 and Mbp1 dimers to Swi6 dimers, their binding to DNA, and the converse dissociation reactions. This equilibrium mathematical model was first converted to mass action–like ordinary differential equations and then into stochastic simulations using the Gillespie algorithm (Gillespie, 1976). To explicitly account for diffusion, we employed a well-established method in which the cell is discretized onto a 3D spatial mesh (Jose et al., 2013; Bernstein, 2005; Earnest et al., 2018). Unless otherwise specified, we used a nuclear diffusion coefficient of 2 µm2/s (Thattikota et al., 2018). Model equations, assumptions, and parameters are detailed in the Materials and methods section.
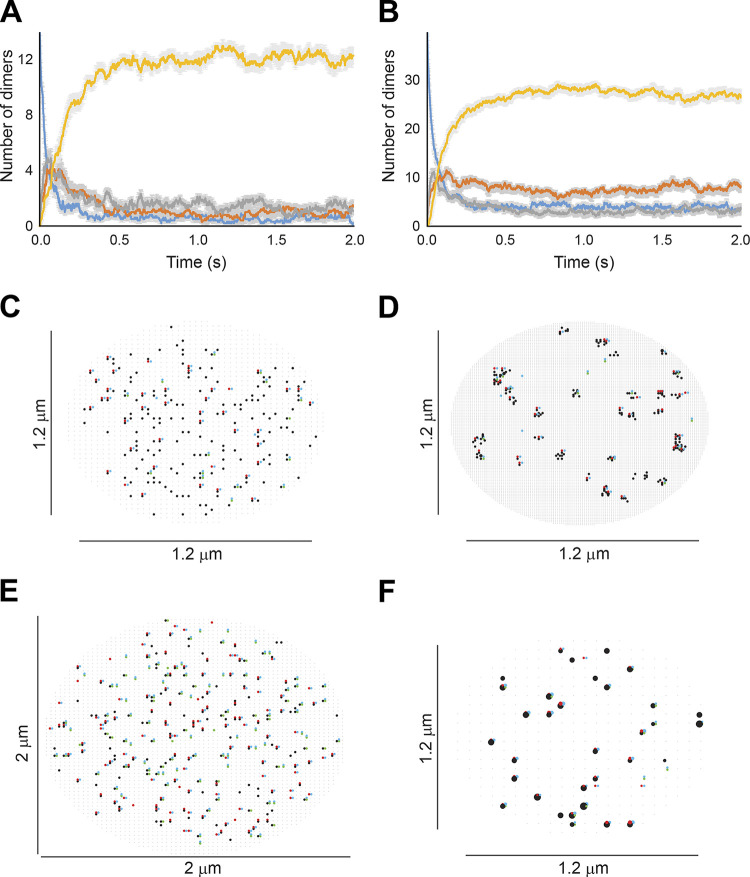
Figure 6.
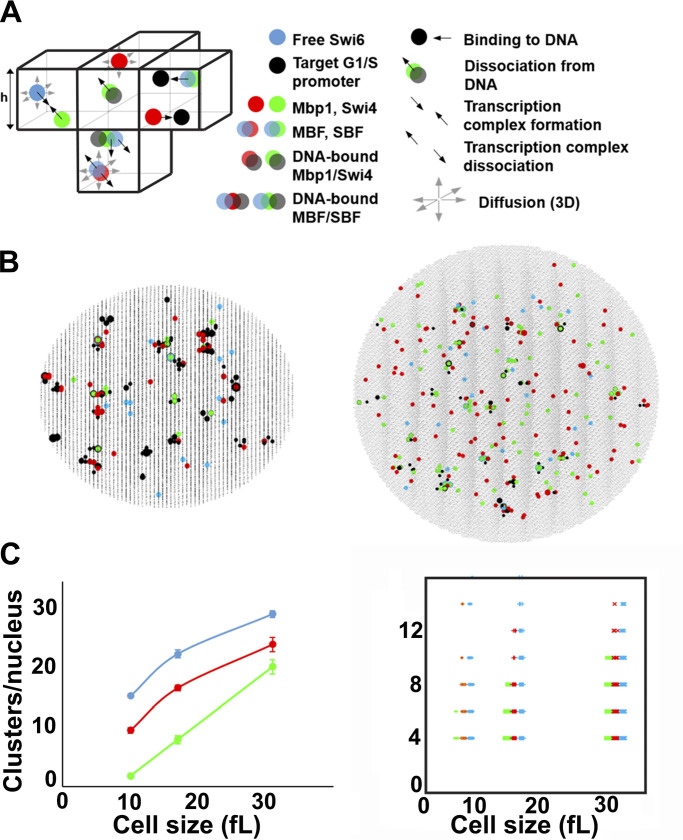
Stochastic modeling predicts Swi4, Mbp1, and Swi6 clustering. (A) Schematic of SBF/MBF binding model (left), encompassing mobile Swi4 dimers (green dots), Mbp1 dimers (red dots), Swi6 dimers (blue dots), and immobile G1/S DNA promoters (black dots), moving and interacting in the nucleus discretized in infinitesimal volume elements (cubes separated by thin black lines; only a few of them are shown here). Swi4/Mbp1 can associate with Swi6 to form mobile SBF/MBF, and/or bind (immobile) promoter DNA. For illustrative purposes, the leftmost element shows SBF formation from Swi4 and Swi6 (convergent thick black arrows). The bottommost element shows MBF dissociation into Mbp1 and Swi6 (divergent thick black arrows) and diffusion (thin gray arrows). Also shown is SBF dissociation from DNA. The rightmost element shows Mbp1 and SBF association with two promoters within a cluster. All interactions (i.e., promoter DNA binding and dissociation, complex formation and dissociation, diffusion) accounted for in the model are indicated (right). The corresponding propensities (in s-1) were derived from Bernstein (2005) and are listed below for all five reaction types respectively: , *, , *, and , where h is the mesh size, and the reaction “on” rates (and the equilibrium constants such that ) are defined in Materials and methods. (B) 2D projection of the 3D output of a typical simulation showing clusters of Swi4 dimers (green dots), Mbp1 dimers (red), Swi6 dimers (blue), and G1/S DNA promoters (black dots) in small (10 fl, left) and large (31.5 fl, right) cells. (C) Left: Number of Swi4 (green), Mbp1 (red), and Swi6 (cyan) clusters per nucleus (vertical axis) as a function of the size of simulated cells (horizontal axis). Data points indicate the cluster number averaged over 10 independent simulations for each cell size, while error bars indicate the standard error of the mean. Right: Scatter plot showing the number of Swi4 (green), Mbp1 (red), and Swi6 (cyan) molecules per cluster (vertical axis) as a function of the size of simulated cells (horizontal axis). Data points represent individual clusters and were gathered from 10 independent simulations for each cell size.
SBF/MBF dimer copy number values as a function of cell size were taken from previous determinations by N&B microscopy (Dorsey et al., 2018). The average concentrations of Mbp1 and Swi6 of 110 and 150 nM in G1 cells of all sizes (Dorsey et al., 2018) were converted to dimeric copy numbers of 42 and 57, respectively, in small cells, and increased approximately threefold to 131 and 178 in large cells. Swi4 concentration was much lower in small cells, 50 nM (i.e., dimeric copy number 15), and doubled as cells grew in G1, leading to a dimeric copy number of 109 in large cells (Dorsey et al., 2018). These parameters ensured stable and predominant formation of DNA-bound SBF and MBF complexes, confirming that the equilibrium regimen previously predicted (Dorsey et al., 2018) is reached kinetically when molecular noise is accounted for (Fig. S2, A and B). In this minimal model, the ∼200 G1/S promoters were preorganized and randomly distributed across 35 clusters, themselves randomly distributed within the cell nucleus, yielding typically 4–12 promoters per cluster. Pre-clustering the promoters in this manner in the model successfully predicted the formation of TF clusters (Fig. 6 B). Importantly, spontaneous TF cluster formation was not observed in simulations when G1/S promoters were not preclustered, despite using identical simulation parameters (Fig. S2, C and E). Nor did the particular choice of mesh size affect cluster formation (Fig. S2, D and F).
Figure S2.
TF formation kinetics and effects of target promoter clustering and mesh size. (A) Typical time evolution of the number of all Swi4-containing transcription factor complexes in small (10 fl) cells, showing DNA-free Swi4 (dimer; blue), DNA-free SBF (gray), DNA-bound Swi4d (dimer; orange), and DNA-bound SBF (yellow). (B) Typical time evolution of the number of all Mbp1-containing transcription factor complexes: DNA-free Mbp1d (dimer; blue), DNA-free MBF (gray), DNA-bound Mbp1d (dimer; orange), and DNA-bound MBF (yellow). Plots are for averages over 10 independent simulations. Gray-shaded regions indicate the standard error of the mean at each time point for each species. (C and E) Effect of promoter clustering on G1/S TF clustering in simulations. 2D projections of the 3D output of a typical simulation in the absence of G1/S promoters clustering, showing Swi4 dimers (green dots), Mbp1 dimers (red), Swi6 dimers (blue), and G1/S DNA promoters (black dots) in a small (10 fl, panel C) and a large (31.5 fl, panel E) cell. (D and F) Simulation mesh size does not affect G1/S TF clustering. (D) Mesh size, 10 nm. (F) Mesh size, 60 nm. Both graphs show Swi4 dimers (green dots), Mbp1 dimers (red), Swi6 dimers (blue), and G1/S DNA promoters (black dots) in a typical small cell (10 fl). In C–F, not all molecules/sites are shown at some pixels where Swi4, Mbp1, Swi6, and G1/S promoters dots overlap.
In our simulations, and in agreement with our experimental observations, a substantial fraction of promoter clusters was free from binding of TFs in small cells (Fig. 6 B, left; and Fig. S2, D and F, black dots). In contrast, in larger cells, close to the critical size at the end of G1 phase, most if not all clusters and promoters were bound with either fully formed SBF/MBF or Swi4 or Mbp1 dimers (Fig. 6 B, right). We computed cluster statistics by counting the number of particles of each type within each promoter cluster (retaining clusters with four or more molecules, i.e., two dimers, to compare with our experiments) across 10 independent simulations for each cell size. The number of Swi4, Mbp1, and Swi6 clusters increased from ∼5–10 in small cells to ∼15–20 for Swi4 and Mbp1 and ∼25–30 for Swi6 in large cells (Fig. 6 C, left). The number of molecules per cluster in the model was between 4 and 12, with no particular size dependence (Fig. 6 C, right), in reasonable agreement with our experimental observations (Fig. 3). This distribution mirrored the multinomial distribution of promoter cluster size that arose from the random distribution of G1/S promoters across clusters.
Scaling arguments provide a plausible explanation for why DNA-binding TFs (i.e., Swi4 and Mbp1 dimers) would be trapped in promoter clusters. TF motion in live cells on the PALM timescale (see below) and quantitative analysis of diffusion using Arbitrary region Raster Scanning Image Correlation Spectroscopy (ARICS; Brown et al., 2008; Hendrix et al., 2016; Fig. S3) indicated binding and dissociation from DNA in the 10–20 ms scale, corresponding to typical off-rates koff∼67s−1, and with Kd = 0.02 μM (Dorsey et al., 2018), to on-rates ∼3,350 s−1 μM−1. For each TF dimer, the time spent diffusing without binding DNA (mean free time) in a nuclear region where the available binding site density is d (μM) is then . During to, the distance traveled by the TF (diffusional jump) is with Dnuc = 2 μm2s−1. If binding sites form clusters, must be compared with the cluster’s geometrical size L, L = 30 nm here. If , on average TFs diffuse within a single cluster between two binding events provided binding sites are available. In contrast, if , TFs escape their original cluster before rebinding. The critical binding site density above which diffusing TFs are more likely to be trapped in clusters corresponds to Ljump = L, i.e., μM. In a cluster of lateral extension L = 30 nm with eight sites, d = 492 μM , hence trapping of TFs within clusters. In support of this view, simulations with 100-fold slower dynamics showed slightly reduced cluster number and/or size for all proteins (Table 1, compare rows 1 and 2). Molecules approaching nearly saturated clusters have a higher propensity to diffuse away because Ljump increases due to a lack of free sites. This explains why the number of particles trapped within each DNA cluster does not significantly increase with cell size even though SBF/MBF particle counts increase.
Figure S3.
ARICS analysis of diffusion of G1/S TF-GFP fusion constructs. (A–C) Strains expressing GFPmut3 fusion constructs of Swi4, Mbp1, and Swi6 from their natural loci were as previously described (Dorsey et al., 2018). ARICS analysis (Hendrix et al., 2016) allows RICS analysis of arbitrary regions of an FOV. ROI were selected based on intensity threshold values that selected only pixels in the nuclei. Spatio-temporal correlation of the intensity values was performed as described previously (Digman et al., 2005). Left panels correspond to grayscale intensity images. Middle panels correspond to arbitrary thresholding in which only gray pixels are analyzed and red pixels are eliminated. Right panels show the RICS image. (A) Swi4-GFPmut3, (B) Mbp1-GFPmut3, and (C) Swi6-GFPmut3. Cells were grown in SC + 2% glucose medium. Note that RICS analysis is limited to timescales faster than ∼50 ms, with any slower dynamics appearing to be immobile on the RICS timescale. Thus, these spatio-temporal correlation patterns are a signature of TF motion at shorter timescales.
Table 1. Sensitivity analysis of TF clustering simulations in very small cells.
| Parameters | Reaction/result | Number of molecules/cluster | Number of clusters/nucleus | ||||
|---|---|---|---|---|---|---|---|
| Swi4 | Mbp1 | Swi6 | Swi4 | Mbp1 | Swi6 | ||
| kon1,3 s/m = 3,500 kon2,4 s/m = 35,000 | Default parameters | 4.27 | 5.15 | 5.38 | 2.2 | 10.1 | 15.2 |
| 100-fold lower kon1,2,3,4 s/m | All dynamics slow down | 4.09 | 5.26 | 5.38 | 2.1 | 9.7 | 13.9 |
| 20-fold larger Ks/m1 | With kon1 constant, this is an increased rate for Swi4d/Mbp1d dissociating from DNA relative to SBF/MBF | 4.27 | 4.90 | 5.39 | 2.2 | 11.8 | 17.0 |
| 20-fold larger Ks/m2 | With kon2 constant, this is an increased rate for Swi6 dissociation from Swi4/Mbp1 | 4.09 | 4.97 | 4.33 | 2.3 | 11.5 | 0.6 |
kon1s/m, on rate for Swi4/Mbp1 binding to DNA; kon2s/m, on rate for Swi6 binding to Swi4 or Mbp1; kon3s/m, on rate for SBF/MBF binding to DNA; kon4s/m, on rate for Swi6 binding to Swi4/Mbp1 DNA complexes; Ks/m1, dissociation constant for monomer Swi4/Mbp1 binding to DNA; Ks/m2, dissociation constant for monomer Swi6 binding monomer Swi4 and Mbp1. Simulated cell size was 10 fl.
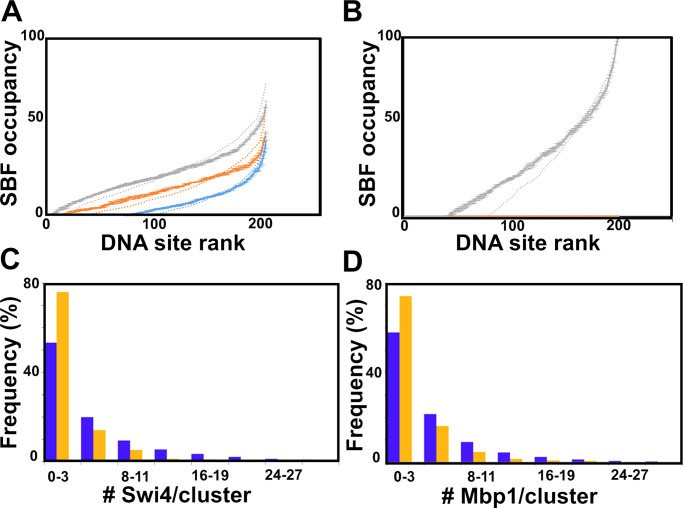
We then asked how clustering might affect the coordination of the G1/S transition. The residence time of the TFs on promoters, when averaged over all promoters and simulation times, was unaffected by clustering because it depended only on the on/off rates. However, ranking individual G1/S promoters according to their average occupancy by SBF (SBF site occupancy) revealed that clustering homogenized SBF occupancy across promoters (Fig. 7 A). This effect was enhanced in short test periods (e.g., 1 s) in large cells close to the G1/S transition (Fig. 7 B). In this situation, clustering reduced by approximately twofold the number of G1/S promoters that were never bound by SBF during the test time (occupancy = 0). If the G1/S transition was triggered in this time window, the expression of all SBF-bound genes would therefore be more correlated. This modeling result suggested that clustering might facilitate the synchronous expression of the G1/S regulon.
Figure 7.
Clustering improves DNA simulated site occupancy and synchrony, while Swi6 contributes to experimental Swi4 and Mbp1 clustering. (A) Clustering homogenizes DNA occupancy. Promoters are ranked by SBF occupancy. Promoter occupancy by SBF is represented as fraction of time in the steady-state (vertical axis, percent of 5 s test time) during which each individual G1/S promoter (represented by individual dots) is occupied by SBF in small (blue; cell volume = 10 fl), medium size (orange; cell volume = 17 fl), and large (gray; cell volume = 31.5 fl) cells. To facilitate data visualization, promoters were ranked according to increasing occupancy, which was averaged at each ranking position (rather than each individual promoter) over five simulations conducted in the presence of G1/S promoter clusters. Error bars represent the standard error of the mean. Promoter occupancy is defined similarly but in the absence of G1/S promoter clustering is shown as dotted lines for comparison. (B) Clustering improves Start synchrony. Promoter SBF occupancy represented as fractions of time in the steady-state (vertical axis, percent of short 1 s test time) as in A in large cells (i.e., close to the G1/S transition). As in A, promoters were ranked according to increasing SBF occupancy, which was averaged at each ranking position over five simulations conducted in the presence of G1/S promoter clusters. Error bars represent the standard error of the mean. Promoter occupancy defined similarly but in the absence of G1/S promoter clustering (and thus, TF clustering; see Fig. S2, C and E) is shown as a dotted line for comparison. The number of promoters that are never SBF-bound during this critical time where the cell is susceptible to trigger Start at any time is twofold larger in the absence of clustering, indicating that clustering may improve Start synchrony. (C and D) Histograms of the number of (C) Swi4 molecules per cluster and (D) the number of Mbp1 molecules per cluster in WT (purple) and swi6Δ (orange) cells.
Swi6 contributes to Swi4 and Mbp1 clustering
One of the surprising observations in both the experiments and the modeling was that TF clusters were observed in small cells in which the TF copy numbers are much lower than the number of target sites. Thus, a small number of DNA clusters were populated by the TFs, while many other clusters remained unoccupied. These observations implied that the TFs do not distribute randomly to all promoters, even though our model assumed the same affinity. To address this question, we focused on Swi6 clustering. Because Swi6 does not directly bind to DNA, its clustering is dependent on interaction with Swi4 and Mbp1. Given the model parameters and the TF concentrations at equilibrium, the system evolved in a regimen where most Swi4/Mbp1 molecules were Swi6-bound, and hence promoters were mostly occupied by SBF/MBF (Fig. S2, A and B). The dissociation of Swi6 from Swi4/Mbp1-bound DNA, which occurred regularly during the simulations given the koff values, would create a Swi6-enriched region around a partially populated cluster. Swi4 and Mbp1 dimers diffusing into the vicinity of such a cluster have a probability to interact with the local Swi6 to form new SBF/MBF complexes, in addition to their probability of interacting with free promoters, or diffusing out of the cluster. Thus, the presence of Swi6 increases the probability of localizing Mbp1 and Swi4 to clusters. This effect would not occur around empty clusters, favoring the formation and DNA-binding of new SBF/MBF complexes within partially populated clusters.
We computationally tested this hypothesis by modulating SBF/MBF affinity for DNA and Swi6 affinity for Mbp1/Swi4. In small cells, strengthening SBF/MBF DNA binding enhanced Swi6 clustering by increasing the number of molecules per cluster (Table 1, compare rows 1 and 3 for Swi6, last column). In contrast, decreasing the Swi6 affinity for Swi4 and Mbp1 markedly reduced the number and size of Swi6 clusters (Table 1, compare rows 1 and 4 for Swi6). Both of these results directly follow from the fact that Swi6 forms clusters via binding to Swi4/Mbp1. Interestingly, decreasing the Swi6 affinity for Swi4 and Mbp1 also slightly reduced the molecular content of Swi4/Mbp1 clusters, while slightly increasing the number of clusters (Table 1, compare rows 1 and 4 for Swi4 and Mbp1), in agreement with a scenario where local Swi6 enrichment contributes to Swi4/Mbp1 accumulation at already populated promoter clusters. We experimentally tested this model prediction by determining the statistics of Swi4-mEOS3.2 and Mbp1-mEos3.2 clustering in swi6Δ cells. We found that deletion of SWI6 decreased the fraction of clusters (defined as containing at least four dimers), for both Swi4 and Mbp1 (Fig. 7, C and D, respectively). These experimental observations support the model conclusions based on the sensitivity testing of Swi6 interaction affinity, and suggest that SBF and MBF complexation statistically enhances their clustering.
Collectively, these local Swi6 concentration effects offer a rationale for the observation of clusters of all three proteins even at low copy number in small cells. The number of molecules per cluster is set by the trade-off between two phenomena. The first is the saturation of target sites within a promoter cluster; the higher the cluster saturation, the higher the probability that the TFs will escape. The second is the local Swi6-enrichment around partially populated clusters that improves the likelihood of binding new Swi4/Mbp1 molecules (as SBF/MBF complexes) to the promoters in these clusters. We note that allosteric effects of Swi6 binding on Mbp1 and Swi4 affinity for their promoters could also contribute to SBF/MBF clustering effects. Moreover, other factors such as chromatin states, other protein partners, or different promoter affinities not accounted for in our model could also contribute to the observed clustering in small cells.
Single particle tracking PALM in live cell nuclei reveals multiple modes of TF mobility
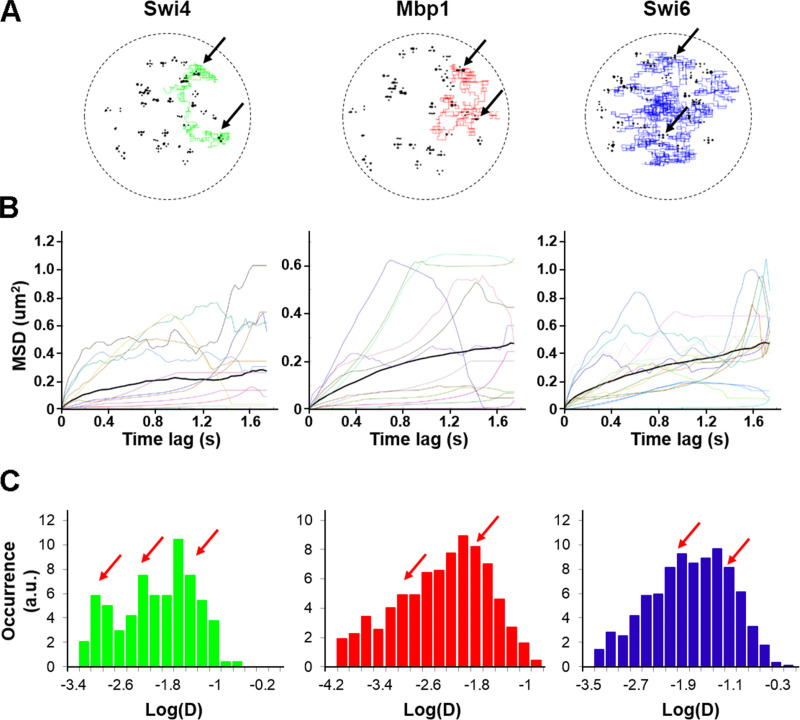
A key prediction of our mathematical model was that the G1/S TFs should display very different kinds of motion ranging from slow, confined diffusion within the neighborhood of promoter clusters to faster diffusion between clusters (Fig. 8). In our model, this combination of slow and fast motion modes along the same single molecule trajectories (Fig. 8 A), with relative weights that mirror the fractions of time spent within and between clusters, yielded downward curvature of the mean squared displacement (MSD) curves (Fig. 8 B). This behavior, characteristic of anomalous subdiffusion, was reflected by short-term effective diffusion coefficients ranging over orders of magnitude (Fig. 8 C).
Figure 8.
Simulated dynamics of Swi4, Mbp1, and Swi6 are multimodal. (A) Example trajectories of individual Swi4 (left, green), Mbp1 (middle, red), and Swi6 (right, blue) dimers from the simulations in 3D, showing for each dimer particle alternations between anomalous sub-diffusive motion confined within cluster and fast, freely diffusive motion between clusters. Black circles represent DNA G1/S promoter target sites. Black arrows indicate positions of multiple binding/unbinding events within given promoter clusters. (B) Example of MSD versus time lag curves corresponding to individual Swi4 (left), Mbp1 (middle), and Swi6 (right) dimer trajectories (color curves) of one small cell simulation. The thick black curves represent the MSD averaged over all the trajectories for each protein. Computation of MSD curves were restricted to the steady-state section of each trajectory, corresponding to simulated times beyond 0.6 s from the onset of the simulation. (C) Histograms of single-trajectory diffusion coefficients extracted from the slope of linear fits of the first four points of individual MSD curves from B. Data were gathered from five to seven independent simulations. Red arrows indicate the approximate positions of main peaks underlying the distribution. We note that simulations predicted a significant fraction of quasi-immobile molecules with an effective diffusion coefficient lower than 0.001. We did not observe this fraction in experiments, possibly due to instrument jitter, which sets a lower bound to the slowness of quantifiable motions.
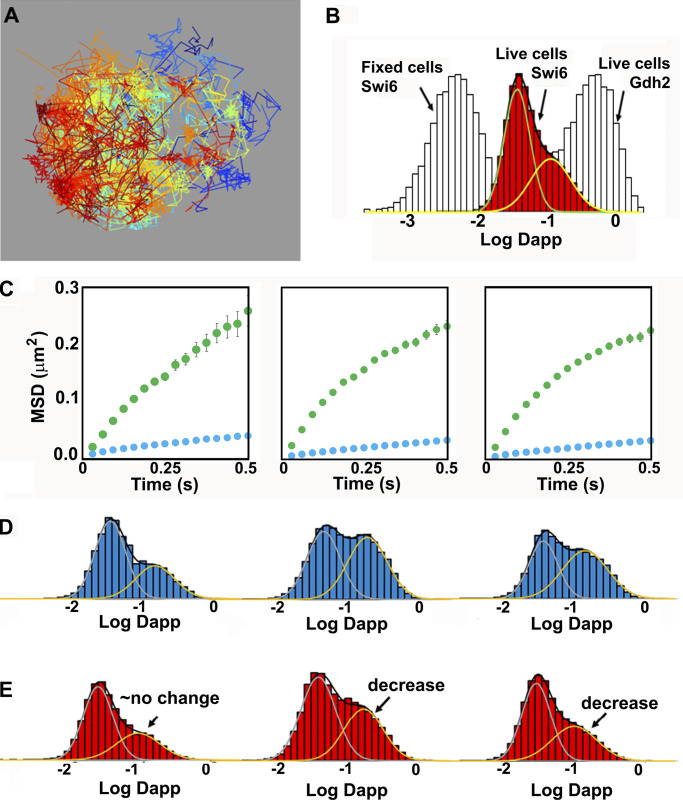
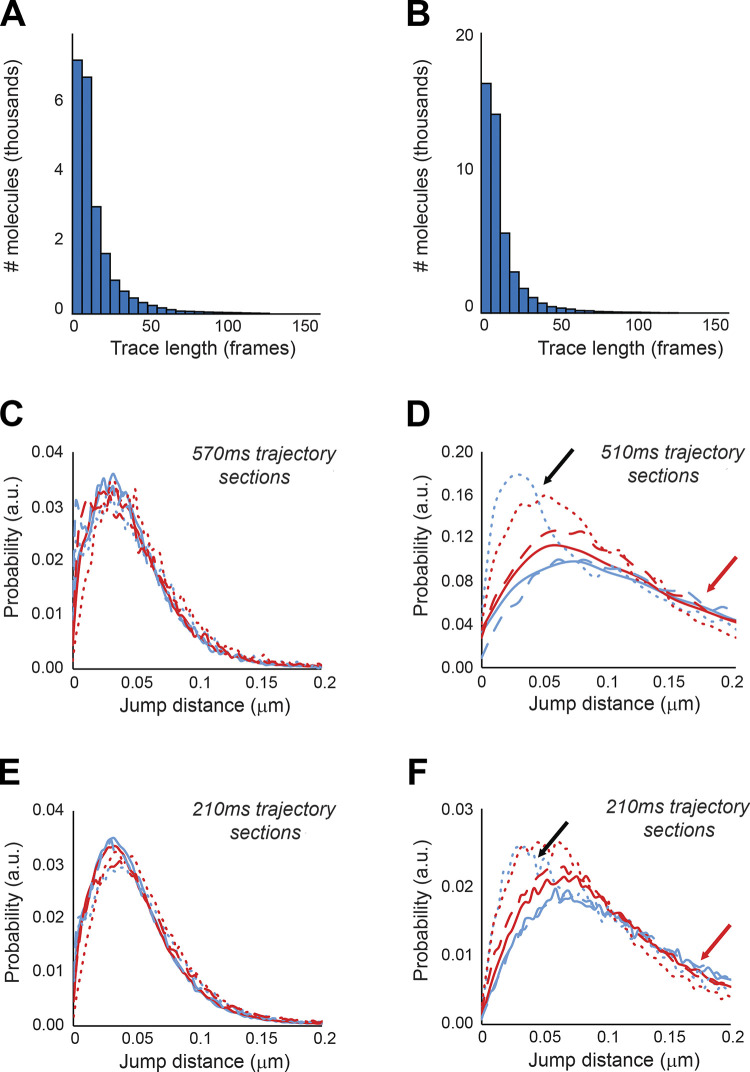
To test these predictions, we measured the dynamics of Swi4, Swi6 and Mbp1 using single particle tracking sptPALM in live cells (Videos 1, 2, and 3). After analysis of the trajectories (see Materials and methods), images of overlaid individual trajectories for each nucleus were produced (e.g., Fig. 9 A). We note the similarity of the image in Fig. 9 A, in terms of the space mapped out by the overlaid trajectories with previous models of the yeast nucleus (Duan et al., 2010; Wong et al., 2012). The trajectories of individual molecules were similar to those from our simulations (compare Fig. 8 B and Fig. 9 C), corresponding to a mixture of smaller and larger MSDs. Analysis of the individual experimental MSD in terms of apparent diffusion coefficients in the linear regimen at short timescales yielded effective single molecule diffusion coefficients whose distribution could be fitted reasonably well with super-positions of two Gaussian modes, in both glucose and glycerol medium (Fig. 9, B, D, and E). Experimental trajectory length distributions were similar in both carbon sources tested, and the majority of trajectories were shorter than 25 frames (Fig. S4 A).
Video 1.
Example trajectory of a Swi4-mEos3.2 molecule in a live yeast cell nucleus. Video was made using the MTT software (Sergé et al., 2008) on one of the live cell PALM FOV acquisitions. Frametime was 30 ms. Pixel size was 120 nm. Frames are colored from blue to red as time increases. Both free and confined diffusion are apparent in this trajectory.
Video 2.
Example trajectory of Mbp1-mEos3.2 in a live yeast cell nucleus. Frametime is 30 ms. Video was made using the MTT software (Sergé et al., 2008) on one of the live cell PALM FOV acquisitions. Pixel size was 120 nm. Frames are colored from blue to red as time increases. Both free and confined diffusion are apparent in this trajectory.
Video 3.
Example trajectory of Swi6-mEos3.2 in a live yeast cell nucleus. Frametime is 30 ms. Video was made using the MTT software (Sergé et al., 2008) on one of the live cell PALM FOV acquisitions. Pixel size was 120 nm. Frames are colored from blue to red as time increases. Both free and confined diffusion are apparent in this trajectory.
Figure 9.
Experimental sptPALM reveals bimodal dynamics of G1/S TFs. (A) Experimental individual molecule trajectories of Swi6-mEos3.2 in the nucleus of a live cell grown in SC + 2% glucose medium. Each individual trajectory is represented by a different color. (B) Histogram of the log of apparent diffusion coefficients, Dapp, calculated from all individual trajectories of Swi6-mEos3.2 (red) for all cells in three FOV (∼5,000 trajectories per FOV). The distribution was fit to two Gaussians. The distribution for Swi6 Log Dapp obtained from spt tracking in fixed cells (light gray, immobile, except for instrument jitter) and the distribution of Log Dapp of mEos3.2-Gdh2 (glutamate dehydrogenase; also light gray) diffusing freely in the cytoplasm are plotted for comparison and indicated by the arrows. (C) Plots of the average mean squared displacements for all trajectories of mEos3.2-tagged Swi4-mEos3.2, Mbp1-mEos3.2, and Swi6-mEos3.2 are shown for SC + 2% glucose medium. Two classes of motion are apparent in all cases, one fast (green) and one slow (blue). (D and E) Distributions of diffusion coefficients from two component analysis of complete individual molecule trajectories for mEos3.2-labeled Swi4-mEos3.2, Mbp1-mEos3.2, and Swi6-mEos3.2 (left, middle, and right) in SC + 2% glucose (D) and SC + 2% glycerol (E) medium. A decrease in the fraction of the fast component is apparent for Mbp1 and Swi6 in the poor carbon source. The average reduced normalized root sum square (RSS) value for a two Gaussian distribution fitting model was 6.4, whereas a single distribution model yielded a reduced RSS value of 27.8, demonstrating that the dynamics involved have at least two modes of diffusion.
Figure S4.
G1/S single particle trajectories length and jump size distributions. (A and B) Trajectory length distributions for sptPALM on (A) Swi4-mEos3.2 and (B) Mbp1-mEos3.2 in live cells. Data are shown for Swi4-mEos3.2 and Mbp1-mEos3.2 in cells grown in SC + 2% glucose. Results were identical for cells grown in SC + 2% glycerol. JDDs for Swi4 (dotted lines), Mbp1 (filled lines), and Swi6 (dashed lines) single molecules in fixed cells (C and E) as a control for instrument jitter and (D and F) in live cells. Colors in all JDD distributions correspond to cells grown in SC + 2% glucose medium (blue) and SC + 2% glycerol medium (red). (C and D) Distributions of jump distances over large times (570 ms and 510 ms, respectively, for fixed and live cells with 20 and 18 points, respectively). (E and F) distributions of jump distances over shorter times (210 ms, eight-point trajectories for both fixed and live cells). Red arrows indicate occurrences of large jumps in live cells distributions that exceed confined diffusion (black arrows).
The average apparent fast diffusion coefficients for each TF were ∼0.1 µm2/s in both simulations and experiments, ∼10–20-fold slower than that observed for glutamate dehydrogenase (mEos3.2-Gdh2), which diffuses freely in the cytoplasm (Fig. 9 B). This value was also considerably slower than the diffusion coefficient of free nuclear proteins evaluated by Raster Scanning Image Correlation Spectroscopy (RICS; Thattikota et al., 2018). The lower mobility of this component, with respect to free protein diffusion, was consistent with the fixed cell data showing that 10–15% of the molecules were outside of the clusters and were likely undergoing a combination of free diffusion and nonspecific DNA binding. The second, much slower apparent diffusion coefficient, 0.01–0.03 µm2/s, was consistent with observed localization of ∼85% of the TFs in clusters. This value was 10-fold faster than the apparent diffusion of immobile molecules in fixed cells, which arose from instrument jitter and localization precision (Fig. 9 B). The slow TF diffusion component of 0.01–0.03 µm2/s constituted a signature of actual TF motion on the scale of the PALM frame time (30 ms), supporting the values of <30 ms for the off-rates of DNA-TF complexes (koff) used in our modeling. We note that the apparent diffusion coefficient of this pool of slowly moving G1/S TFs is also at least one order of magnitude faster than the diffusion coefficient previously reported for chromosomal motion, estimated at ∼0.0005 µm2/s (Marshall et al., 1997). More recent single particle tracking experiments on the two copies of the yeast GAL1 promoter revealed motion timescales of 0.1–30 s (Backlund et al., 2014), although faster chromosomal motion on the PALM frame time could conceivably contribute to the observed TF motion. ARICS analysis (Hendrix et al., 2016) of Swi4-, Mbp1- and Swi6-GFP fluorescence fluctuations for nuclear pixels yielded diffusion coefficients of 0.015, 0.05, and 0.11, respectively, for the three proteins (Fig. S3), also in reasonable agreement with the experimental sptPALM results and simulations.
Since individual molecules sampled both fast and slow dynamic modes in a single trajectory, these apparent diffusion coefficients conflate the two modes of motion, which results in the apparent anomalous nature of TF diffusion. To avoid mixing different modes of motion over single trajectories, we analyzed the dynamics of individual molecules using a Jump-Distance Distribution (JDD) approach (Menssen and Mani, 2018; Tollis, 2015). JDDs for Swi4, Swi6, and Mbp1 in glucose and glycerol-grown cells all showed a main peak (black arrows) corresponding to a jump distance ranging from 30 to 50 nm (for Swi4) to 60 to 80 nm (for Swi6 and Mbp1), for a duration of 7 × 30 ms = 0.21 s over eight trajectory points, in agreement with apparent cluster sizes on PALM images (Fig. S4 B, bottom panels). Furthermore, the position of this peak was barely altered when plotting the JDD on larger timescales (18 trajectory points, 17 × 30ms = 0.51s, Fig. S4 B, top panels), emphasizing that this peak corresponded to confined molecules, for which the volume of diffusion (and hence the jump distance) did not depend on the sampling time. In comparison, free nuclear diffusion in our quasi-2D excitation volume would yield a typical jump of ∼1 µm, emphasizing how strongly the molecular motion is restricted. Comparison of JDDs acquired in live (Fig. S4 B, right panels) and fixed (Fig. S4 B, left panels) cells revealed that jump distances are significantly larger in live cells, indicating that the peak at small jump distance in this case was not due to instrument jitter but represented actual motion. The long tail in the JDD in live cells (Fig. S4 B, right panels, red arrows) indicated that a small fraction of the particles displays fast motion, in agreement with the MSD analysis and model predictions.
The JDDs could not be well fitted using simple models such as free diffusion, anomalous diffusion, anisotropic motion along and around linear tracks, or a mixed two-mode model of free diffusion. Rather, our live cell data were best characterized by a superposition of an anomalous diffusion component (low JDD main peak, 65–80% of the molecules; see Table 2, columns 3 and 4) with a faster anisotropic motion (20–35% of the molecules) characterized in our fitting by a unidirectional motion along a linear track (Table 2, column 5) with variance around the track (Table 2, columns 6 and 7). The latter component may correspond in part to anisotropic diffusion along DNA (von Hippel and Berg, 1989).
Table 2. JDD parameters from live cell sptPALM experiments.
| Fraction of anomalously diffusing molecules (%) | Anomalous diffusion coefficient (μm2/sa) | Anomalous power (a) | Velocity along tracks (μm/s) | Variance around tracks | Diffusion coefficient around tracks (μm2/s) | |
|---|---|---|---|---|---|---|
| Mbp1, glu | 66.26 | 0.0091 | 0.73 | 0.1198 | 0.007833 | 0.1263 |
| Mbp1, gly | 68.80 | 0.0080 | 0.74 | 0.1043 | 0.00760 | 0.1226 |
| Swi4, glu | 80.80 | 0.0054 | <0.4833 | 0.1577 | 0.006205 | 0.1001 |
| Swi4, gly | 81.64 | 0.0044 | 0.6038 | 0.5251 | 0.004683 | 0.0755 |
| Swi6, glu | 65.46 | 0.0089 | 0.8263 | 0.0976 | 0.007743 | 0.1249 |
| Swi6, gly | 75.56 | 0.0065 | 0.6767 | 0.1066 | 0.006898 | 0.1113 |
These results were also in good quantitative agreement with the cluster analysis in fixed cells that yielded ∼15% and ∼85% of TFs outside and inside clusters, respectively, at the time of fixation. Thus, at the short timescale of JDD computation, TFs exhibited anomalous motion, confirming that DNA-binding/unbinding dynamics were faster than the ∼100 ms regimen. We note that both JDD and MSD analyses were performed across time intervals that exceeded 100 ms, a time interval during which fast diffusing molecules (D ∼2–3 µm2/s) travel an average of 400–500 nm. This linear distance is comparable to the depth of our microscope field in the z-direction, and therefore a small fraction of such fast diffusing molecules might have escaped the microscope field and not be accounted for in our analysis. Thus, it is possible that we slightly underestimated the fraction of fast-diffusing molecules. Analysis using either the JDD or the two component MSD of complete trajectories revealed a lower mobile fraction for Swi4 compared with either Swi6 or Mbp1 (Fig. S4 B), perhaps due to higher affinity (i.e., lower off-rates) of Swi4 for its target sites on DNA. This was also the case for the ARICS analysis above. Moreover, Mbp1 and Swi6 mobility decreased in glycerol medium compared with glucose (Fig. 9, D and E). This result is consistent with stronger specific binding of Mbp1 in the poor carbon source and is in agreement with the conditional large cell size phenotype of the Δmbp1 mutant reported previously (Dorsey et al., 2018). Collectively, these results suggest a mechanism whereby G1/S TFs populate discrete clusters and can also jump between clusters.
Discussion
Super-resolution spatial mapping of Swi4, Mbp1, and Swi6 molecules in fixed cells revealed that these TFs do not distribute randomly in yeast nuclei, but are organized into discrete clusters of approximately eight molecules, even in small cells in which they are limiting with respect to their promoter targets. The results also demonstrate that the number of clusters increases as cells grow, while the number of molecules in each cluster remains constant. In our stochastic model, based on reasonable biophysical parameters, clustering the ∼200 G1/S promoters was sufficient for clustering of the G1/S TFs. The distribution of G1/S TFs cluster sizes (in number of molecules) that we observed strongly resembles the multinomial distribution of promoter cluster sizes that one would expect by randomly distributing ∼200 promoters across 30–35 clusters (i.e., typically 4–12 promoters per cluster), although we cannot exclude that larger clusters are formed in vivo via biologically active processes. Although our results do not rule out explicitly the converse possibility that G1/S TFs might spontaneously assemble into clusters, any such mechanism would need to counteract free diffusion.
Importantly, clusters are observed in small cells where the TFs are severely limiting with respect to promoter target sites, and the number of molecules per cluster does not change with increasing copy number or size. Our simulations demonstrate that the balance between the effects of Swi6 local concentration on the one hand and target site saturation versus diffusion propensity on the other, is sufficient to explain the existence of clusters in small cells and the observed successive titration of new clusters as cells grow. The simulations suggest that the occupation of some sites within a cluster by SBF or MBF tends to sequester Mbp1 and Swi4 molecules via transient interactions with the Swi6 activator already present in the cluster. However, as target sites within any given cluster are bound by the increasing TF copy numbers as cells grow, the decreased number of unbound target sites available decreases the Swi4 or Mbp1 binding propensity. Diffusion out of the cluster eventually becomes statistically more probable than DNA rebinding. This interpretation is consistent with the bimodal dynamics of the G1/S TF we observe by sptPALM in live cells. Overall, these results indicate that cluster size and the distribution of TFs across clusters can be tuned by the promoter content of preformed clusters and by relative affinities of TF subunits for each other and for target sites on DNA. These modeling results, based on known concentrations and reasonable affinity and diffusion constants, account well for the observation of clusters even in small cells, but do not rule out other mechanisms of cluster formation.
Both general and specific transcription factors have been observed to form clusters. For example, in budding yeast, the transcriptional repressor Mig1 forms clusters of similar size as we observe for Swi4, Mbp1, and Swi6 (Wollman et al., 2017). Like the G1/S TFs, these clusters also exhibit mixed dynamic properties, and interestingly, the number and copy number content of Mig1 clusters increases upon glucose repression. In mammalian cells, RNA polymerase II and its associated Mediator complex are colocalized in much larger stable clusters (>300 nm, ∼300 molecules) that exhibit properties of phase separated condensates (Cho et al., 2018). Even larger clusters have been observed using super-resolution imaging for the transcription factor STAT3 (Gao et al., 2017). These examples suggest that TF clustering is a common phenomenon, but that different TFs can exhibit distinct clustering behavior. The small size of the clusters observed here is inconsistent with phase separation.
While our observations can be accounted for by simple biochemical phenomena and captured in a mathematical model based on minimal assumptions, the observed clustering of the G1/S TFs must be coupled at some level to the global organization of the yeast genome (Duan et al., 2010; Lazar-Stefanita et al., 2017; Taddei and Gasser, 2012; Wong et al., 2012; Zimmer and Fabre, 2011). Thus, our results lead to several open questions. The nature of the preformed G1/S promoter clusters inferred from our model is unknown at this juncture. It is possible that condensin- and/or cohesin-mediated chromosome looping could play a role in organizing the G1/S promoters, perhaps in conjunction with other factors that bind specific promoter regions. Condensin and cohesin are primarily implicated in replication, division, and silencing of RNA genes (Lazar-Stefanita et al., 2017; Machín et al., 2004), but have also been associated with retrotransposons and transcription of highly active genes, such as Pol II housekeeping genes and Pol III targets near centromeres (Noma, 2017; Kim et al., 2016; Hsieh et al., 2015). Cohesin appears to form smaller domains (84 kb) than does condensin (∼350 kb), both of which are smaller than the Topologically Associated Domains (TADs) typically found in mammalian cells (∼1,000 kb; Dixon et al., 2012; Nora et al., 2012; Sexton et al., 2012; Bintu et al., 2018; Szabo et al., 2018; Cattoni et al., 2017).
Expression of the G1/S regulon is not completely coherent, and early CLN1/2 expression is important to activate commitment before transcribing all 200 genes in the regulon (Eser et al., 2011). Furthermore, TADs and chromosomal location do not strongly influence the timing of CLN1 expression in budding yeast (Eser et al., 2017), although these results do not rule out a contribution of G1/S promoter clustering to Start synchrony. Recent Micro-C experiments in yeast have revealed much smaller chromosomally interacting domains at the scale of nucleosomes (Hsieh et al., 2016, 2015), a much higher resolution than prior Hi-C experiments. Micro-C experiments in mammalian cells established transcription-linked contacts within and between TADs (Hsieh et al., 2020; Krietenstein et al., 2020). It is possible that the clusters observed here correspond to similar promoter–promoter contacts within cohesin-mediated chromosomally interacting domains.
It remains to be determined if the promoter clusters deduced from our observations of TF clustering in budding yeast are populated in a discrete order as cells grow in G1 phase or whether clusters have defined or random promoter compositions. It is also unclear whether clusters can exchange promoters over time as opposed to being of fixed composition. Regardless of the underlying static or dynamic mechanisms, the localization of G1/S promoters within discrete clusters may help coordinate the G1/S transcriptional program once Start is triggered. Higher-level organization of the genome may contribute to the efficiency of other cell state transitions that depend on complex gene regulons.
Materials and methods
Strains and sample preparation
The Saccharomyces cerevisiae strains used in this study (Table 3) were constructed in the S288C (BY4741) background by PCR-based homologous recombination integration of a mEOS3.2-HisMX cassette (pDM810) at the 3′ end of each reading frame at each endogenous locus. Relevant strain genotypes are provided in Table 3.
Table 3. Key resources.
| Reagent or resource | Source | Identifier |
|---|---|---|
| Experimental models: yeast strains | ||
| BY4741 | S288C-derived strain, parental strain for the MATa haploid gene deletion collection | |
| MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | ||
| Swi4-mEos3.2 | This study | MT5032 |
| swi4::SWI4-mEOS3.2-HisMX | ||
| Mbp1-mEos3.2 | This study | MT5031 |
| mbp1::MBP1-mEOS3.2-HisMX | ||
| Swi6-mEos3.2 | This study | MT5033 |
| swi6::SWI6-mEOS3.2-HisMX | ||
| Gdh2-mEOS3.2 | This study | MT5062 |
| gdh2::GDH2-mEOS3.2-HisMX | ||
| NLS-mEos3.2-NLS | This study, by transformation of BY4741 with pMT 4649 | |
| <pGAL1-NLS-mEOS3.2-NLS LEU2 CEN> | ||
| swi6Δ Swi4-mEos3.2 | This study | MT5064 |
| swi6::KanMX swi4::SWI4-mEOS3.2-HisMX | ||
| swi6Δ Mbp1-mEos3.2 | This study | MT5063 |
| swi6::KanMX mbp1::MBP1-mEOS3.2 | ||
| Software and algorithms | ||
| MTT SPT analysis | Available online from http://ciml-e12.univ-mrs.fr/App.Net/mtt/ | |
| sptPALM_CBS single particle trajectory analysis | Available from nollmann@cbs.cnrs.fr upon request. | |
| Molecular detection and cluster analysis routine—MATLAB | This study, available from royerc@rpi.edu upon request. | |
| Mathematical modeling routines—MATLAB | This study, available from royerc@rpi.edu upon request. |
The pGAL1-NLS-mEOS3.2-NLS strain was generated by transformation of the BY4741 parental strain with plasmid pMT4649, which was constructed by Gibson assembly of an SV40 NLS domain, the mEOS3.2 sequence amplified from an mEOS3.2-HisMX cassette (pDM810), and a second SV40 NLS domain into a pGAL1 CEN plasmid. All plasmid constructions were verified by sequence analysis, and all strain constructions were verified by PCR of the integrated locus and sequence analysis.
The TF-mEos3.2 fusion strains were grown on SC–His + 2% glucose plates for 3 d. Fresh colonies were picked and grown in SC–His media supplemented with a rich (2% glucose) or poor (2% glycerol) carbon nutrient source until stationary phase. Prior to imaging, strains grown on rich or poor carbon sources were diluted to 0.3 OD and allowed to grow until OD 0.7 in fresh SC-His + 2% glucose or SC-His + 2% glycerol medium. A 1-ml sample of OD 0.7 culture was pelleted at 3,000 rpm and washed with fresh media. The sample was concentrated 10× by removing 900 ml of media, and the cells were resuspended. A 5-µl sample of the cells was placed on a Con A–coated no. 1 coverslip with 100 nm Tetraspec fluorescent beads and allowed to adhere to the surface for 4 min. The coverslip was then placed on a 2% agar SC – His + carbon source pad as previously described (Dorsey et al., 2018) and immediately imaged. For fixed cells, a 1-ml sample of cells was washed with PBS buffer, the buffer was removed, and then 500 µl of a 4% paraformaldehyde solution was added and allowed to react for 20 min. The sample was washed extensively with PBS and placed on a no. 1 coverslip coated with Con A and 100 nm Tetraspec fluorescent beads similar to live cells. Fixed cell samples were treated the same as live cells from this point on.
PALM microscope
Imaging was performed with a custom-built PALM/STORM system based on a Nikon inverted Ti-U Eclipse microscope and similar to that described in (Fiche et al., 2013). A CFI Plan Apo Lambda 100×/1.40 NA oil objective was used, and images were collected on an Andor iXon Ultra 897 EMCCD camera. A 561-nm laser (Sapphire 561–150 CW CDRH) at 0.2 kW/cm2 and a 405-nm laser (OBIS 405 nm LX 50 mW) at 0.3 W/cm2 were directed into the microscope objective in a Koehler illumination configuration with the aid of a pair of beam-expanding lenses (150/30 mm, Thor Labs) and a quad-band (405/488/561/640 nm, Chroma) filter for mEos3.2 excitation and activation, respectively. Emission was collected with a 600/50 nm BP filter (Chroma) mounted in an automated filter wheel (Thor Labs). The image was magnified with a set of lenses (150/250 mm) to an effective pixel size of 120 nm, creating a 61.44 × 61.44 µm2 (512 × 512 pixel) imaging area. Excitation and activation power were controlled using an acousto-optical tunable filter (AOTFnC400.650-TN, AA Optoelectrionics).
Super-resolution image acquisition
Samples were placed in a cylindrical sample chamber and mounted on the microscope at 21°C. After locking in the optimum imaging plane of the sample, a bright field reference image was acquired. For each FOV, 30,000–40,000 frames (exposure time 30 ms) were collected within a data acquisition. The 561 nm CW laser at a fixed power was used for excitation of mEos3.2 molecules in the red-shifted state. Photoactivation was achieved by continually increasing the power of a 405 nm CW laser diode until mEos3.2 photoswitching was no longer observed (Lee et al., 2012). For each FOV, 30,000–40,000 frames were collected for each acquisition. The x–y drift was fit to a smoothing function with the frame number as the independent variable (Fig. S5, A and B). Active correction for drift in the z-plane was performed as previously described (Fiche et al., 2013). The imaging focal plane was locked in position (within 10 nm) using an autofocus program by tracking the reflection of a 785 nm IR source (OBIS 785 nm LX 100 mW) from the sample coverslip with a Thor Labs CCD camera and continuous incremental adjustments with a Fast PIFOC Piezo Nanofocusing Z-Drive (PI; Fig. S5 C). Software controls and data acquisition for the microscope stage, laser excitation and activation power, autofocus, and camera were written in Labview 2015 (National Instruments). The super-resolution images corresponded to 2D representations of 3D objects, since the microscope depth of field (∼500 nm) is larger than macromolecular structures. Hence, some degree of clustering could in principle arise from super-position of molecules in different z-planes. Nonetheless, the extensive degree of clustering observed exceeded what may be expected from 2D super-position of randomly distributed molecules in 3D (see simulations in Fig. S2, C and E).
Figure S5.
PALM Acquisition and analysis. (A–C) Positional drift correction in x, y, and z axes respectively, as described in the Materials and methods section and Supplemental materials. (D) Detection localization uncertainty histogram for all detections in a typical FOV. (E) Standard error of the mean of the molecular localizations for a typical FOV. (F) Blinking time constant determination. (G) Example of the results of the nuclear masking algorithm.
PALM analysis
PALM image stacks were analyzed and x–y drift—corrected using the Thunderstorm ImageJ plugin. If images drifted more than 2 pixels (120 nm effective pixel size) in either x or y directions, then the data were discarded. For acquisitions within the threshold drift range, molecular detection positions were drift-corrected using the image-correlation drift algorithm from Thunderstorm that cross-correlated the signal from 100-nm fluorescent beads (Tetraspek, Anaspec) used as fiducial markers. The average frame number for an individual molecule was then used to compute its total associated drift, which was added to its averaged x–y position.
The average resolution for each individual localization was 25 nm (Fig. S5 D), which allowed an optimal super-resolution image pixel size of 12 nm. The MATLAB script MTT (Sergé et al., 2008) was used to connect molecular trajectories (i.e., positions of the same molecule from one frame to the next), and the sptPALM_CBS (Fiche et al., 2013) script was used to analyze and filter the trajectories. For fixed cell analysis, nuclei were masked with the MATLAB gaussfit function (Fig. S5 G) using our custom SuperResolution MATLAB script. This script also corrected for over-counting due to mEos3.2 blinking (Fig. S5 F; Lee et al., 2012), with spatial and temporal cutoffs of 10 nm and 2 s. Since most molecules were found within the nucleus, the nuclear mask sets boundaries within which the number of molecules per nucleus, number of clusters per nucleus, and number of molecules per cluster were determined. This procedure yielded the overlaid corrected detection images (see Fig. 1, A–C). The distributions of masked nuclear sizes resembled the cell size distribution of asynchronous cultures, and therefore we used the mode of this distribution (∼20,000 pixels) to characterize the upper limit size of the G1 cells nuclei. Of note, in a spherical model for both cells and nuclei with a karyoplasmic ratio of ∼1/7 (Jorgensen et al., 2007), this size is equivalent to ∼30 fl, a good estimate of the G1/S critical size of WT cells in SC + 2% glucose.
The drift-corrected positions of each individual molecule detected in multiple (5–100) successive frames were averaged to yield the molecular images (see Fig. 1, lower panels for each TF as indicated). The pixel size in these images was chosen as 3 nm, half the value of the standard error of the mean for all molecular positions within a dataset (6 nm; Fig. S5 E). The number of molecules per nucleus and the cell size in pixels were also generated by this script. Live cell sptPALM trajectories were analyzed as MSD as a function of time shift along each trajectory using the sptPALM_CBS MATLAB script (Fiche et al., 2013). To disentangle distinct molecular motion modes along individual trajectories, trajectories were also analyzed using the JDD approach (Menssen and Mani, 2018; Tollis, 2015) in which trajectories are subdivided into short sections (8 points, 210-ms time bins) and analyzed collectively. Separate experiments were performed on six different days for Mbp1, and three different days for Swi4 and Swi6, for a total of 28 FOV for fixed cells and 14 for live cells for Mbp1, 10 FOV for fixed cells and 6 FOV for live cells for Swi4, and 15 FOV for fixed cells and 8 FOV for live cells for Swi6. Despite the inherent limitations of PALM for particle counting, the super-resolution images of Swi4, Mbp1, and Swi6 correspond to a reasonable representation of their actual distribution in the nucleus for several reasons. First, nonactivated (and hence unobserved) mEos3.2 molecules should be randomly distributed and hence unlikely to bias the fraction of molecules detected in clusters vs. nonclustered molecules. Second, our low-intensity, continuous switching illumination parameters allow for relatively efficient photoactivation. Moreover, because the three TFs diffuse as dimers (Dorsey et al., 2018), the majority of dimers are detected via at least one of their constituent monomers. Finally, yeast chromosomes bearing the G1/S TF target sites occupy a limited volume of the nucleus, which limits problems of detection due to depth of field. Thus, the number of clusters per cell and the cell size invariance of cluster size are reasonably well determined.
Fixed cell cluster analysis
We used the nuclear super-resolution molecular images to develop custom cluster analysis scripts in the MATLAB environment. For each nucleus, we ordered the particles in a list such that each particle was next to its nearest neighbor in the nucleus using an algorithm based on the OPTICS ranking algorithm (Kriegel et al., 2011). This list is referred to as the nearest-neighbor ranked particle list. We computed the list of distances between each particle and the next across this list (see Fig. 4 B). Plotting this list revealed two characteristic features: valleys (red arrows), wherein the distance between consecutive particles is small, separated by spikes corresponding to large distances (blue asterisks). Valleys represent particle clusters, wherein the interparticle distance is small, whereas spikes are characteristic of the distance between clusters. These plots provided a tool to count the number of clusters in the nucleus, through the definition of a distance threshold (red line in Fig. 4 B), such that particles separated by a distance lower than the threshold were assigned to the same cluster. We computed the relative variation of the total number of clusters (across our entire dataset for each protein and each FOV, Swi4, Mbp1, and Swi6) as a function of threshold (see Fig. 4 C). We found that the number of detected clusters is threshold-dependent for values <10 super-resolution pixels (30 nm), whereas for values >30 nm, there is little dependence on the threshold. This indicates that most clustered particles were within a 30-nm neighborhood of their closest neighbor, a distance that thus represented a logical cluster size detection threshold. The nearest-neighbor ranked list of particles provided a simple means to count clusters within each nucleus, to count the number of particles within each cluster, and to correlate these data with nuclear size.
Live cell MSD and JDD analyses
To analyze single particle tracking data and gain insight on the dynamic motion features at the molecular scale, we used two methods, each with complementary advantages and disadvantages. First, we selected entire individual trajectories (Fig. 9 A and Videos 1, 2, and 3), and computed the MSD as a function of time shift along each trajectory (Fig. 9 C) using the sptPALM_CBS MATLAB script (Fiche et al., 2013). Although this analysis revealed a sub-diffusion type motion with downward curvature at large times, below 200 ms the MSD curves could be fit reasonably well with a linear function. The slope provided the trajectory-averaged instantaneous diffusion coefficient (as shown in Fig. 9, B, D, and E). We processed simulated in silico individual trajectories in the same manner (Fig. 8).
Although MSD analysis is a powerful method to reveal particle confinement, because dynamics are averaged over entire trajectories (and further averaged across different trajectories), this approach only provided a global view of particle dynamics on the seconds timescale. To disentangle distinct molecular motion modes along individual trajectories, trajectories were also analyzed using the JDD (Fig. S4 B) approach introduced in Tollis (2015) and further developed in Menssen and Mani (2018). Trajectories were subdivided into short sections (eight points, 210-ms time bins) and analyzed collectively. From these sub-trajectories, a JDD (i.e., the distance covered along any given sub-trajectory within the 210 ms of its duration) was computed for all data from a given experiment. This approach increases the likelihood of observing a unique mode of molecular motion along a shorter fraction of single molecule trajectories. We fitted the experimental JDDs with molecular motion models, including free (Brownian) diffusion, anomalous diffusion, directed transport along linear tracks, or more complex models that incorporate two of the classical motion models discussed above (i.e., where the population of sub-trajectories includes two subpopulations with different underlying transport modes). To select among these competing models, we used a Bayesian model selection procedure (see Tollis, 2015; Menssen and Mani, 2018), which outputs the probability of each underlying motion model in a manner that balances the fitting quality of a given model to its complexity.
Mathematical modeling and Monte Carlo simulations
To model SBF/MBF formation and binding to target promoters, we used our previously published Start model (Dorsey et al., 2018). The model comprises a mass-action kinetics-based SBF/MBF binding module, resolution of which yields the concentrations of DNA-bound and DNA-free SBF/MBF complexes, and of Swi6-free Swi4/Mbp1 dimers bound to DNA in the cell nucleus as a function of cell size. From previous fluctuation microscopy-based measurements (Dorsey et al., 2018), we used the size-independent nuclear Mbp1 and Swi6 concentrations (respectively, 110 nM and 150 nM) and the Swi4 concentration that doubles linearly between early G1 (50 nM in 14 fl cells) and late G1 phase (100 nM in 35 fl cells). We assumed that dissociation constant (Kd) values for interactions of the DNA binding proteins (Swi4 or Mbp1 = DBP) to DNA were unaffected by Swi6 binding, and vice versa. In addition, our previous brightness data revealed that all measured Start proteins were predominantly dimeric (Dorsey et al., 2018). Thus, we reduced the model complexity by neglecting the equilibrium concentrations of protein complexes formed with monomer DNA-binding protein (DBP, Swi4, or Mbp1) and/or Swi6. As a result, in the steady-state, the SBF/MBF binding module reduces to eight reactions (with effective dissociation constants derived in (Dorsey et al., 2018):
| (Eq. 1) |
where dim stands for dimer and can be either Swi4 or Mbp1, and represent a DBP-free and DBP dimer–bound target promoter, respectively, and are fully formed DNA-free and DNA-bound DBP dimer–Swi6 dimer SBF and MBF complexes, and the microscopic dissociation constants respectively characterize monomer DBP-DNA, monomer DBP-Swi6, and dimer DBP-DNA binding, with s and m lowercase subscripts standing for Swi4 and Mbp1, respectively. It is noteworthy that the dissociation constant of is not the microscopic DBP/monomeric-Swi6 constant but an effective dimer-DBP/dimer-Swi6 dissociation constant that involves multiple interactions. Unless otherwise specified, we used the following default values: Ks1 = Km1 = 100 nM, Ks3 = Km3 = 20 nM, and Ks2 = 20 nM < Km2 = 50 nM (Dorsey et al., 2018).
The mass action-like ordinary differential equations corresponding to this equilibrium relate the variation of the concentrations of the transcription complexes (right-hand side of Eq. 1) to their formation rates ( *[interacting species], left-hand side of Eq. 1) and their dissociation rates ( x [TF complex]):
Similar equations are used for the transcription complexes components, DNA, Swi6dim and DBPdim, expressed with the same rates. Default kinetic on-off rates are given in main text Table 1. These Ordinary Differential Equations (ODEs) were converted to stochastic simulations using the Gillespie algorithm modified to account for diffusion (Jose et al., 2013; Bernstein, 2005). The Gillespie algorithm is a particular class of Monte Carlo simulation algorithm originally developed to stochastically simulate biochemical systems with molecules binding to and dissociating from each other in a homogeneous, well-mixed solution (Gillespie, 1976). For any given state of the system at a given time, the algorithm associates each type of species (individual molecule or complex) a propensity (in s−1) to convert into another (i.e., a reaction). The sum of all propensities is then used to randomly determine the time to the next reaction, and another random number is generated to determine the type of the next reaction such that reactions with high propensities are more likely to be chosen than reactions with low propensities. Propensities are next reevaluated at the new time, and successive iterations of this algorithm simulate the stochastic behavior of the chemical system as a function of time. The Gillespie framework has been successfully used by us and others to address various cell biological questions including, for instance, RNA secondary structure folding kinetics (Clote and Bayegan, 2018), stochastic gene expression (Ferguson et al., 2012), and yeast polarity establishment (Jose et al., 2013).
The inclusion of diffusion in this framework is straightforward. It requires partition of the reaction-diffusion volume into small elements. It considers identical molecules in different elements as different diffusing-reacting species. Finally, it extends the list of possible reactions between species, such that regular reactions (converting species A into B) are only possible between molecules within the same volume-element i (A,i -> B,i), and that molecule A diffusing from one element i to a neighbor j is a reaction that converts the species A,i into the species A,j. We divided the nuclear volume into infinitesimal volume elements (in 3D Cartesian coordinates, with xyz mesh-size = 30 nm, the maximal size that still provides sub-cluster resolution). We defined the following for each diffusible particle (i.e., free Swi4, Swi6, Mbp1 dimers, and DNA-free SBF and MBF): (1) a propensity to isotropically diffuse to a neighboring element (in six directions, see gray arrows), (2) bind to another particle (black arrows pointing toward each other), or (3) bind to DNA (black arrows pointing toward a DNA site), as defined by the model Eq. 1 (see Fig. 6 A). In contrast, DNA-bound species are not assumed to diffuse at all, since chromosomal motion is expected to be negligible on the short time scale of our measurements and simulations (<2 s and 10 s, respectively; Marshall et al., 1997). However, DNA-bound species have propensities to dissociate from each other or from DNA (black arrows pointing away from DNA sites) according to the model Eq. 1. Importantly, for a given set of on rates and microscopic Kd’s (which define the effective Kd’s for dimer interactions; see Dorsey et al., 2018), propensities depend on the mesh size (Fig. 6 A). This follows from the fact that propensities are defined for individual particles or pairs of particles for complex formation. In the latter case, for any individual particles of type A and B, the rate of A–B complex formation depends linearly on the apparent concentration of particle B, which is proportional to the inverse of the element volume (). This term is absent in complex dissociation events (Bernstein, 2005). The presence of the term in the propensity of diffusion events follows from the second-order spatial derivative in the diffusion equation. One critical condition for the use of the Gillespie algorithm is that the reaction-diffusion volumes are well mixed. This holds as long as the simulated number of diffusion events is significantly larger than the number of biochemical reaction events. This condition is fulfilled in our simulations since diffusion events typically exceed reactions by two to three orders of magnitude.
This algorithm was implemented numerically in MATLAB, for small, medium, and large cells with nuclear radii of 0.67, 0.8, and 0.98 µm (corresponding to cell volumes, respectively, of 10, 17.1, and 31.5 fl), using a cell size–independent karyoplasmic ratio (Jorgensen et al., 2007) on a h = 0.03 µm 3D mesh and 200 DNA promoters randomly distributed across 35 clusters, themselves randomly distributed within the nucleus for each simulation. Specifically, each promoter assigned to a cluster was positioned either within the element containing the cluster center, or in an immediate neighbor element (a total of seven possible positions). Given the mesh size of h = 30 nm, this procedure ensured that DNA promoters belonging to the same cluster are within a 30–60-nm distance from each other, in agreement with observed cluster sizes (Fig. 1). We recorded the simulation data every millisecond. Unless otherwise specified, we used a nuclear diffusion coefficient of 2 µm2/s, concentrations of 110 nM for Mbp1 and 150 nM for Swi6 in cells of all sizes, and 50 and 100 nM for Swi4 in small and large cells, respectively, previously determined by N&B-based absolute measurements (Dorsey et al., 2018). All species were assumed dimeric to yield the total number of Mbp1, Swi4, and Swi6 dimers in small (42, 15, 57), medium (71, 37, 97), and large (131, 109, 178) simulated cells.
Online supplemental material
Fig. S1 shows comparison of the copy numbers of GFP fusions of Swi4, Mbp1, Swi6, and Whi5 determined by scanning number and brightness and copy numbers of their mEos3.2 fusions by PALM. Fig. S2 shows TF formation kinetics and effects of target promoter clustering and mesh size. Fig. S3 shows ARICS analysis of diffusion of G1/S TF-GFP fusion constructs. Fig. S4 shows G1/S single particle trajectories length and jump size distributions. Fig. S5 shows PALM acquisition and analysis. Video 1 shows an example of Swi4-mEos3.2 trajectory. Video 2 shows an example of Mbp1-mEos3.2 trajectory. Video 3shows an example of Swi6-mEos3.2 trajectory.
Acknowledgments
We thank Derek McCusker (Centre National pour la Recherche Scientifique, Paris, France) for providing the mEOS3.2 plasmid.
This work was supported by the National Science Foundation (PHY 1806638 to C.A. Royer), the Canadian Institutes of Health Research (FDN-167277 to M. Tyers) and a Canada Research Chair in Systems and Synthetic Biology (to M. Tyers).
The authors declare no competing financial interests.
Author contributions: Conceptualization: L. Black, S. Tollis, M. Tyers, and C.A. Royer; Methodology: S. Tollis, M. Tyers, C.A. Royer, J-B. Fiche, and M. Nollmann; Software: S. Tollis, M. Nollmann, J-B. Fiche, S. Notley, B. Crevier, and J. Bigness; Formal analysis: S. Tollis, C.A. Royer, L. Black, and G. Fu; Investigation: L. Black, S. Tollis, G. Fu, and S. Dorsey; Resources: S. Tollis, J. Cheng, and G. Ghazal; Writing – original draft preparation: L. Black, S. Tollis, M. Tyers, and C.A. Royer; Writing – review and editing: S. Tollis, M. Tyers, and C.A. Royer; Supervision: S. Tollis, M. Tyers, and C.A. Royer; Project administration: S. Tollis, M. Tyers, and C.A. Royer; Funding acquisition: M. Tyers and C.A. Royer.
References
- Backlund M.P., Joyner R., Weis K., and Moerner W.E.. 2014. Correlations of three-dimensional motion of chromosomal loci in yeast revealed by the double-helix point spread function microscope. Mol. Biol. Cell. 25:3619–3629. 10.1091/mbc.e14-06-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean J.M., Siggia E.D., and Cross F.R.. 2005. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics. 171:49–61. 10.1534/genetics.105.044560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Elazar S., Yakhini Z., and Yanai I.. 2013. Spatial localization of co-regulated genes exceeds genomic gene clustering in the Saccharomyces cerevisiae genome. Nucleic Acids Res. 41:2191–2201. 10.1093/nar/gks1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein D. 2005. Simulating mesoscopic reaction-diffusion systems using the Gillespie algorithm. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 71 041103 10.1103/PhysRevE.71.041103 [DOI] [PubMed] [Google Scholar]
- Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., and Hess H.F.. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 313:1642–1645. 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Bintu B., Mateo L.J., Su J.H., Sinnott-Armstrong N.A., Parker M., Kinrot S., Yamaya K., Boettiger A.N., and Zhuang X.. 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 362 eaau1783 10.1126/science.aau1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.M., Dalal R.B., Hebert B., Digman M.A., Horwitz A.R., and Gratton E.. 2008. Raster image correlation spectroscopy (RICS) for measuring fast protein dynamics and concentrations with a commercial laser scanning confocal microscope. J. Microsc. 229:78–91. 10.1111/j.1365-2818.2007.01871.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capurso D., Bengtsson H., and Segal M.R.. 2016. Discovering hotspots in functional genomic data superposed on 3D chromatin configuration reconstructions. Nucleic Acids Res. 44:2028–2035. 10.1093/nar/gkw070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoni D.I., Cardozo Gizzi A.M., Georgieva M., Di Stefano M., Valeri A., Chamousset D., Houbron C., Déjardin S., Fiche J.B., González I., et al. . 2017. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nat. Commun. 8:1753 10.1038/s41467-017-01962-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W., Spille J., Hecht M., Lee C., and Li C.. 2018. Mediator and RNA polymerase II clusters associate in transcription- dependent condensates. Science. 415:412–415. 10.1126/science.aar4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clote P., and Bayegan A.H.. 2018. RNA folding kinetics using Monte Carlo and Gillespie algorithms. J. Math. Biol. 76:1195–1227. 10.1007/s00285-017-1169-7 [DOI] [PubMed] [Google Scholar]
- Costanzo M., Nishikawa J.L., Tang X., Millman J.S., Schub O., Breitkreuz K., Dewar D., Rupes I., Andrews B., and Tyers M.. 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 117:899–913. 10.1016/j.cell.2004.05.024 [DOI] [PubMed] [Google Scholar]
- de Bruin R.A., McDonald W.H., Kalashnikova T.I., Yates J. III, and Wittenberg C.. 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 117:887–898. 10.1016/j.cell.2004.05.025 [DOI] [PubMed] [Google Scholar]
- Digman Michelle A., Brown Claire M., Sengupta Parijat, Wiseman Paul W., Horwitz Alan R., and Gratton Enrico. 2005. Measuring Fast Dynamics in Solutions and Cells with a Laser Scanning Microscope. Biophysical Journal. 89(2):1317–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., and Ren B.. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 485:376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey S., Tollis S., Cheng J., Black L., Notley S., Tyers M., and Royer C.A.. 2018. G1/S Transcription Factor Copy Number Is a Growth-Dependent Determinant of Cell Cycle Commitment in Yeast. Cell Syst. 6:539–554.e11. 10.1016/j.cels.2018.04.012 [DOI] [PubMed] [Google Scholar]
- Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y.J., Lee C., Shendure J., Fields S., Blau C.A., Noble W.S., et al. . 2010. A three-dimensional model of the yeast genome. Nature. 465:363–367. 10.1038/nature08973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest T.M., Cole J.A., and Luthey-Schulten Z.. 2018. Simulating biological processes: stochastic physics from whole cells to colonies. Rep. Prog. Phys. 81 052601 10.1088/1361-6633/aaae2c [DOI] [PubMed] [Google Scholar]
- Eser U., Falleur-Fettig M., Johnson A., and Skotheim J.M.. 2011. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol. Cell. 43:515–527. 10.1016/j.molcel.2011.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eser U., Chandler-Brown D., Ay F., Straight A.F., Duan Z., Noble W.S., and Skotheim J.M.. 2017. Form and function of topologically associating genomic domains in budding yeast. Proc. Natl. Acad. Sci. USA. 114:E3061–E3070. 10.1073/pnas.1612256114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M.L., Le Coq D., Jules M., Aymerich S., Radulescu O., Declerck N., and Royer C.A.. 2012. Reconciling molecular regulatory mechanisms with noise patterns of bacterial metabolic promoters in induced and repressed states. Proc. Natl. Acad. Sci. USA. 109:155–160. 10.1073/pnas.1110541108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F., Colomina N., Futcher B., and Aldea M.. 2010. The transcriptional network activated by Cln3 cyclin at the G1-to-S transition of the yeast cell cycle. Genome Biol. 11:R67 10.1186/gb-2010-11-6-r67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiche J.B., Cattoni D.I., Diekmann N., Langerak J.M., Clerte C., Royer C.A., Margeat E., Doan T., and Nöllmann M.. 2013. Recruitment, assembly, and molecular architecture of the SpoIIIE DNA pump revealed by superresolution microscopy. PLoS Biol. 11 e1001557 10.1371/journal.pbio.1001557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Chen J., Cai M., Xu H., Jiang J., Tong T., and Wang H.. 2017. Clustered localization of STAT3 during the cell cycle detected by super-resolution fluorescence microscopy. Methods Appl. Fluoresc. 5 024004 10.1088/2050-6120/aa6ab5 [DOI] [PubMed] [Google Scholar]
- Gillespie D. 1976. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 22:403–464. 10.1016/0021-9991(76)90041-3 [DOI] [Google Scholar]
- Hartwell L.H., Culotti J., Pringle J.R., and Reid B.J.. 1974. Genetic control of the cell division cycle in yeast. Science. 183:46–51. 10.1126/science.183.4120.46 [DOI] [PubMed] [Google Scholar]
- Hendrix J., Dekens T., Schrimpf W., and Lamb D.C.. 2016. Arbitrary-Region Raster Image Correlation Spectroscopy. Biophys. J. 111:1785–1796. 10.1016/j.bpj.2016.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T.H.S., Weiner A., Lajoie B., Dekker J., Friedman N., and Rando O.J.. 2015. Mapping Nucleosome Resolution Chromosome Folding in Yeast by Micro-C. Cell. 162:108–119. 10.1016/j.cell.2015.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T.S., Fudenberg G., Goloborodko A., and Rando O.J.. 2016. Micro-C XL: assaying chromosome conformation from the nucleosome to the entire genome. Nat. Methods. 13:1009–1011. 10.1038/nmeth.4025 [DOI] [PubMed] [Google Scholar]
- Hsieh T.S., Cattoglio C., Slobodyanyuk E., Hansen A.S., Rando O.J., Tjian R., and Darzacq X.. 2020. Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Mol. Cell. 78:539–553.e8. 10.1016/j.molcel.2020.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V.R., Horak C.E., Scafe C.S., Botstein D., Snyder M., and Brown P.O.. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 409:533–538. 10.1038/35054095 [DOI] [PubMed] [Google Scholar]
- Johnston G.C., Pringle J.R., and Hartwell L.H.. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105:79–98. 10.1016/0014-4827(77)90154-9 [DOI] [PubMed] [Google Scholar]
- Jorgensen P., and Tyers M.. 2004. How cells coordinate growth and division. Curr. Biol. 14:R1014–R1027. 10.1016/j.cub.2004.11.027 [DOI] [PubMed] [Google Scholar]
- Jorgensen P., Rupeš I., Sharom J.R., Schneper L., Broach J.R., and Tyers M.. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491–2505. 10.1101/gad.1228804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Edgington N.P., Schneider B.L., Rupes I., Tyers M., and Futcher B.. 2007. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell. 18:3523–3532. 10.1091/mbc.e06-10-0973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose M., Tollis S., Nair D., Sibarita J.B., and McCusker D.. 2013. Robust polarity establishment occurs via an endocytosis-based cortical corralling mechanism. J. Cell Biol. 200:407–418. 10.1083/jcb.201206081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.D., Tanizawa H., Iwasaki O., and Noma K.. 2016. Transcription factors mediate condensin recruitment and global chromosomal organization in fission yeast. Nat. Genet. 48:1242–1252. 10.1038/ng.3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C., Moll T., Neuberg M., Ahorn H., and Nasmyth K.. 1993. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 261:1551–1557. 10.1126/science.8372350 [DOI] [PubMed] [Google Scholar]
- Kriegel H.P., Kröger P., Sander J., and Zimek A.. 2011. Density-based clustering. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 1:231–240. 10.1002/widm.30 [DOI] [Google Scholar]
- Krietenstein N., Abraham S., Venev S.V., Abdennur N., Gibcus J., Hsieh T.S., Parsi K.M., Yang L., Maehr R., Mirny L.A., et al. . 2020. Ultrastructural Details of Mammalian Chromosome Architecture. Mol. Cell. 78:554–565.e7. 10.1016/j.molcel.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar-Stefanita L., Scolari V.F., Mercy G., Muller H., Guérin T.M., Thierry A., Mozziconacci J., and Koszul R.. 2017. Cohesins and condensins orchestrate the 4D dynamics of yeast chromosomes during the cell cycle. EMBO J. 36:2684–2697. 10.15252/embj.201797342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., et al. . 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 298:799–804. 10.1126/science.1075090 [DOI] [PubMed] [Google Scholar]
- Lee S.-H., Shin J.Y., Lee A., and Bustamante C.. 2012. Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM). Proc. Natl. Acad. Sci. USA. 109:17436–17441. 10.1073/pnas.1215175109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsios A., Huberts D.H.E.W., Terpstra H.M., Guerra P., Schmidt A., Buczak K., Papagiannakis A., Rovetta M., Hekelaar J., Hubmann G., et al. . 2019. Differential scaling between G1 protein production and cell size dynamics promotes commitment to the cell division cycle in budding yeast. Nat. Cell Biol. 21:1382–1392. 10.1038/s41556-019-0413-3 [DOI] [PubMed] [Google Scholar]
- Machín F., Paschos K., Jarmuz A., Torres-Rosell J., Pade C., and Aragón L.. 2004. Condensin regulates rDNA silencing by modulating nucleolar Sir2p. Curr. Biol. 14:125–130. 10.1016/S0960-9822(04)00002-8 [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Straight A., Marko J.F., Swedlow J., Dernburg A., Belmont A., Murray A.W., Agard D.A., and Sedat J.W.. 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7:930–939. 10.1016/S0960-9822(06)00412-X [DOI] [PubMed] [Google Scholar]
- Mazouchi A., and Milstein J.N.. 2015. Fast Optimized Cluster Algorithm for Localizations (FOCAL): a spatial cluster analysis for super-resolved microscopy. Bioinformatics. 32:747–754. 10.1093/bioinformatics/btv630 [DOI] [PubMed] [Google Scholar]
- Menssen R., and Mani M.. 2018. A jump distance based parameter inference scheme for particulate trajectories in biological settings. https://www.biorxiv.org/content/early/2018/12/10/238238.
- Noma K.I. 2017. The Yeast Genomes in Three Dimensions: Mechanisms and Functions. Annu. Rev. Genet. 51:23–44. 10.1146/annurev-genet-120116-023438 [DOI] [PubMed] [Google Scholar]
- Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J., et al. . 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 485:381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Lee Y., Bhupindersingh G., and Iyer V.R.. 2013. Widespread misinterpretable ChIP-seq bias in yeast. PLoS One. 8 e83506 10.1371/journal.pone.0083506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.J., Bates M., and Zhuang X.. 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods. 3:793–795. 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoller K.M., Turner J.J., Kõivomägi M., and Skotheim J.M.. 2015. Dilution of the cell cycle inhibitor Whi5 controls budding-yeast cell size. Nature. 526:268–272. 10.1038/nature14908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergé A., Bertaux N., Rigneault H., and Marguet D.. 2008. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods. 5:687–694. 10.1038/nmeth.1233 [DOI] [PubMed] [Google Scholar]
- Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., and Cavalli G.. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 148:458–472. 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Simon I., Barnett J., Hannett N., Harbison C.T., Rinaldi N.J., Volkert T.L., Wyrick J.J., Zeitlinger J., Gifford D.K., Jaakkola T.S., et al. . 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 106:697–708. 10.1016/S0092-8674(01)00494-9 [DOI] [PubMed] [Google Scholar]
- Skotheim J.M., Di Talia S., Siggia E.D., and Cross F.R.. 2008. Positive feedback of G1 cyclins ensures coherent cell cycle entry. Nature. 454:291–296. 10.1038/nature07118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q., Jost D., Chang J.M., Cattoni D.I., Papadopoulos G.L., Bonev B., Sexton T., Gurgo J., Jacquier C., Nollmann M., et al. . 2018. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci. Adv. 4 eaar8082 10.1126/sciadv.aar8082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A., and Gasser S.M.. 2012. Structure and function in the budding yeast nucleus. Genetics. 192:107–129. 10.1534/genetics.112.140608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talarek N., Gueydon E., and Schwob E.. 2017. Homeostatic control of START through negative feedback between Cln3-Cdk1 and Rim15/Greatwall kinase in budding yeast. eLife. 6 e26233 10.7554/eLife.26233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattikota Y., Tollis S., Palou R., Vinet J., Tyers M., and D’Amours D.. 2018. Cdc48/VCP Promotes Chromosome Morphogenesis by Releasing Condensin from Self-Entrapment in Chromatin. Mol. Cell. 69:664–676.e5. 10.1016/j.molcel.2018.01.030 [DOI] [PubMed] [Google Scholar]
- Tollis S. 2015. A Jump Distance-based Bayesian analysis method to unveil fine single molecule transport features. http://arxiv.org/abs/1506.01112.
- Tsochatzidou M., Malliarou M., Papanikolaou N., Roca J., and Nikolaou C.. 2017. Genome urbanization: clusters of topologically co-regulated genes delineate functional compartments in the genome of Saccharomyces cerevisiae. Nucleic Acids Res. 45:5818–5828. 10.1093/nar/gkx198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P.H., and Berg O.G.. 1989. Facilitated target location in biological systems. J. Biol. Chem. 264:675–678. 10.1074/jbc.RA117.000130 [DOI] [PubMed] [Google Scholar]
- Wang S., Moffitt J.R., Dempsey G.T., Xie X.S., and Zhuang X.. 2014. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc. Natl. Acad. Sci. USA. 111:8452–8457. 10.1073/pnas.1406593111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman A.J.M., Shashkova S., Hedlund E.G., Friemann R., Hohmann S., and Leake M.C.. 2017. Transcription factor clusters regulate genes in eukaryotic cells. eLife. 6 e27451 10.7554/eLife.27451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H., Marie-Nelly H., Herbert S., Carrivain P., Blanc H., Koszul R., Fabre E., and Zimmer C.. 2012. A predictive computational model of the dynamic 3D interphase yeast nucleus. Curr. Biol. 22:1881–1890. 10.1016/j.cub.2012.07.069 [DOI] [PubMed] [Google Scholar]
- Zhang M., Chang H., Zhang Y., Yu J., Wu L., Ji W., Chen J., Liu B., Lu J., Liu Y., et al. . 2012. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods. 9:727–729. 10.1038/nmeth.2021 [DOI] [PubMed] [Google Scholar]
- Zimmer C., and Fabre E.. 2011. Principles of chromosomal organization: lessons from yeast. J. Cell Biol. 192:723–733. 10.1083/jcb.201010058 [DOI] [PMC free article] [PubMed] [Google Scholar]