Chiusa et al. show that the RNA-DNA binding protein FUS plays a profibrotic role by binding to the collagen IV gene promoter and commencing its transcription. Reducing nuclear FUS inhibits collagen IV transcription, suggesting that targeting FUS offers a new antifibrotic therapy.
Abstract
Excessive accumulation of collagen leads to fibrosis. Integrin α1β1 (Itgα1β1) prevents kidney fibrosis by reducing collagen production through inhibition of the EGF receptor (EGFR) that phosphorylates cytoplasmic and nuclear proteins. To elucidate how the Itgα1β1/EGFR axis controls collagen synthesis, we analyzed the levels of nuclear tyrosine phosphorylated proteins in WT and Itgα1-null kidney cells. We show that the phosphorylation of the RNA-DNA binding protein fused in sarcoma (FUS) is higher in Itgα1-null cells. FUS contains EGFR-targeted phosphorylation sites and, in Itgα1-null cells, activated EGFR promotes FUS phosphorylation and nuclear translocation. Nuclear FUS binds to the collagen IV promoter, commencing gene transcription that is reduced by inhibiting EGFR, down-regulating FUS, or expressing FUS mutated in the EGFR-targeted phosphorylation sites. Finally, a cell-penetrating peptide that inhibits FUS nuclear translocation reduces FUS nuclear content and collagen IV transcription. Thus, EGFR-mediated FUS phosphorylation regulates FUS nuclear translocation and transcription of a major profibrotic collagen gene. Targeting FUS nuclear translocation offers a new antifibrotic therapy.
Introduction
Kidney fibrosis and other organ-specific fibrotic diseases are characterized by excessive deposition of ECM components, mainly collagens, ultimately leading to loss of organ function. Many factors control collagen homeostasis, including growth factor and matrix receptors such as integrins (Itgs; Coelho and McCulloch, 2016; Rayego-Mateos et al., 2018).
Itgs are transmembrane receptors for ECM components composed of noncovalently bound α and β subunits that heterodimerize to produce 24 different transmembrane receptors (Hynes, 2002; Pan et al., 2016). Itgα1β1 is a major collagen IV receptor that prevents injury-mediated kidney fibrosis by negatively regulating EGF receptor (EGFR) tyrosine kinase activity and the assembly of the NADPH oxidase responsible for the generation of profibrotic reactive oxygen species (ROS; Chen et al., 2004, 2010; Wang et al., 2015). A mechanism whereby Itgα1β1 negatively regulates the phosphorylation levels and activity of EGFR is recruiting and activating the tyrosine phosphatase TCPTP (Mattila et al., 2005). Accordingly, cells lacking Itgα1β1 do not recruit and activate TCPTP and thereby display increased basal levels of tyrosine phosphorylated EGFR, ROS production, and collagen expression (Chen et al., 2007).
In addition to controlling ROS levels, EGFR can exert its profibrotic action by regulating the total levels or activation of transcription factors such as FOXM1 (forkhead box M1) or STATs (Penke et al., 2018; Quesnelle et al., 2007; Su et al., 2015; Xu and Shu, 2013). We hypothesized that the Itgα1β1/EGFR axis regulates collagen production by controlling tyrosine phosphorylation of nuclear factors that interact with collagen gene regulatory elements. Therefore, we evaluated the levels of tyrosine phosphorylated nuclear proteins in WT and Itgα1knockout (Itgα1KO) kidney cells by immunoprecipitation with anti-phosphotyrosine antibody followed by mass spectrometry. We found that the RNA-DNA binding protein fused in sarcoma (FUS) was more phosphorylated in the Itgα1KO cells compared with their WT counterparts.
FUS is an RNA-DNA binding protein expressed predominantly in the nucleus of cells, where it regulates DNA repair transcription, RNA splicing, and export to the cytoplasm (Ederle and Dormann, 2017). FUS contains an uncommon nuclear localization sequence (NLS) motif called PY-NLS located at the C-terminus of the protein (Ederle and Dormann, 2017). FUS nuclear translocation is mediated by the binding of the PY-NLS motif to the nuclear import adaptor transportin/karyopherin-β2 (Lee et al., 2006).
Missense mutations of FUS have been identified as a cause of familial amyotrophic lateral sclerosis (ALS). These mutations result in subcellular mislocalization of FUS that is retained in cytoplasmic inclusions, leading to neuronal cytotoxicity (Kwiatkowski et al., 2009; Vance et al., 2009). In addition to the missense mutations, mutations within the NLS or truncation mutations that result in impaired interaction of FUS with transportin/karyopherin-β2 have been also associated with familial ALS (Bosco et al., 2010; DeJesus-Hernandez et al., 2010; Dormann et al., 2010; Kent et al., 2014).
Although mutations of FUS that prevent its nuclear translocation are a major cause of neurotoxicity in ALS, preventing FUS nuclear translocation in nonneuronal cells might be beneficial in reducing the transcription of genes implicated in fibrosis. Interestingly, patients with ALS show decreased levels of collagen in skin and serum (Ono et al., 1998; Tsukie et al., 2014); and FUS binds the collagen X promoter (Gu et al., 2014) and SP1, a transcriptional activator involved in collagen synthesis and fibrosis (Ghosh et al., 2013). Thus, it is conceivable that nuclear FUS acts as a profibrotic factor by positively regulating collagen production.
Interestingly, FUS contains several phosphorylation sites. Among them, phosphorylation of Tyr526 by the family of Src kinases reduces FUS interaction with transportin, leading to cytoplasmic accumulation of FUS (Darovic et al., 2015). We found that murine and human FUS contain two EGFR-targeted phosphorylation sites, namely Y6 and Y296 in murine and Y6 and Y304 in human FUS, suggesting that FUS can be phosphorylated by this growth factor receptor. However, the process through which EGFR controls the phosphorylation, nuclear translocation, and function of FUS is unknown.
In this study, we show that the Itgα1β1/EGFR axis controls FUS tyrosine phosphorylation and function. In the absence of Itgα1β1, increased basal EGFR phosphorylation promotes FUS tyrosine phosphorylation and FUS nuclear translocation. In the nucleus, phosphorylated FUS binds to the promoter of the collagen IV α2 chain and drives collagen IV gene transcription. We also demonstrate that inhibition of EGFR by erlotinib, siRNA-mediated down-regulation of FUS, expression of FUS mutated in the EGFR tyrosine phosphorylation sites, or treatment with a cell-penetrating peptide designed to compete with FUS binding to transportin leads to decreased FUS nuclear localization and subsequently reduced collagen IV expression. Thus, reducing FUS nuclear translocation in kidney cells offers a new approach to controlling excessive production of collagen, thereby limiting excessive matrix deposition, a hallmark of fibrosis in the kidneys and other organs.
Results
The RNA-DNA binding protein FUS is increased in the nuclei of cells lacking Itgα1β1
Itgα1β1 down-regulates collagen production by negatively controlling the tyrosine phosphorylation levels of EGFR (Chen et al., 2007). Consistent with this finding, cells lacking Itgα1β1 show increased baseline tyrosine phosphorylation levels of EGFR and increased levels of collagens I and IV (Chen et al., 2004; Wang et al., 2015), two major collagen types up-regulated in fibrotic diseases.
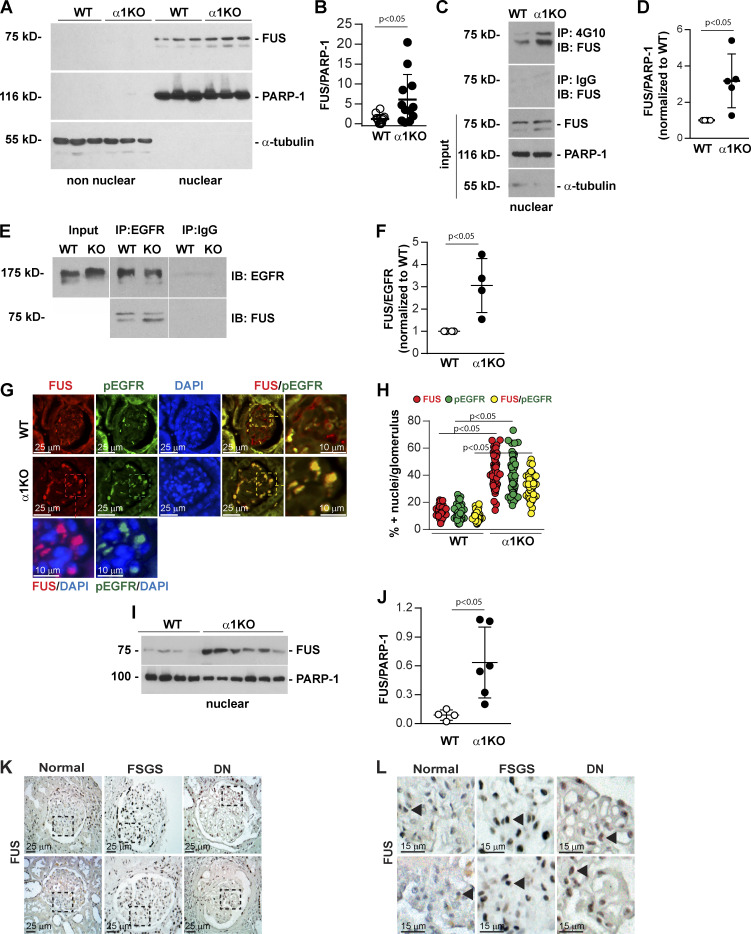
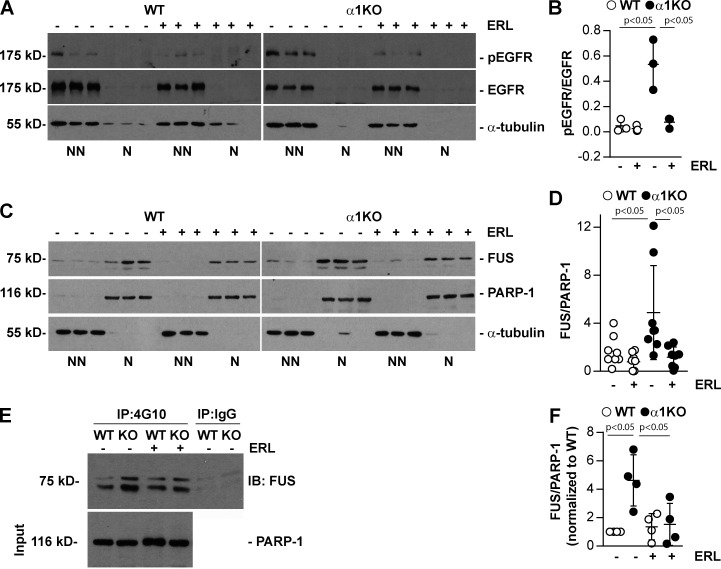
Therefore, we analyzed whether the Itgα1β1/EGFR axis can also regulate the tyrosine phosphorylation levels of transcription factors that regulate genes encoding key components of the extracellular matrix. To this end, we performed immunoprecipitation of nuclear proteins from WT and Itgα1KO mesangial cells with anti-phosphotyrosine antibody. Subsequent mass spectrometry analyses of immunoprecipitates identified FUS in the Itgα1KO cells but not in their WT counterparts, suggesting increased tyrosine phosphorylation of this nuclear factor in cells lacking Itgα1β1. The RNA-DNA binding protein FUS has the potential to regulate matrix synthesis, since patients with ALS, a disease characterized by mutations that prevents FUS translocation to the nucleus (Kwiatkowski et al., 2009; Vance et al., 2009), display overall decreased collagen levels in skin and plasma (Ono et al., 1998; Tsukie et al., 2014). Furthermore, our analysis of the murine and human bidirectional promoter of collagen IV α1 and α2 chains revealed the presence of several FUS-responsive elements (Fig. S1). Western blot analysis demonstrated that Itgα1KO mesangial cells have increased levels of nuclear FUS compared with WT cells (Fig. 1, A and B). Moreover, immunoprecipitation analysis of nuclear fractions using anti-phosphotyrosine antibody revealed that nuclear FUS was more tyrosine phosphorylated in Itgα1KO cells compared with WT cells (Fig. 1, C and D).
Figure S1.
The collagen IV gene promoter contains FUS-responsive elements. (A and B) Analysis of the murine (A) and human (B) bidirectional promoter of collagen IV α1 and/or α2 chains revealed the presence of several FUS-responsive elements (underlined in red).
Figure 1.
Increased nuclear FUS levels in Itgα1KO cells and mice. (A) Levels of FUS were analyzed by Western blotting in nonnuclear and nuclear fractions (20 µg/lane) from serum-starved WT and Itgα1KO (α1KO) mesangial cells (n = 3 samples). PARP-1 (nuclear marker) or α-tubulin (nonnuclear marker) was analyzed to evaluate fraction purity. (B) Nuclear FUS and PARP-1 bands were quantified by densitometry, and values represent the FUS/PARP-1 ratio. Circles represent individual values (n = 12 with four experiments performed) and bars represent mean ± SD. (C) Nuclear fractions (400 µg) from WT and α1KO cells were immunoprecipitated with anti-4G10 antibody or IgG control, and the immunoprecipitates were analyzed by Western blot for levels of FUS. Input represents nuclear lysates (20 µg/lane) analyzed for levels of FUS, PARP-1, and α-tubulin. (D) Immunoprecipitated FUS and PARP-1 bands were quantified by densitometry, and values represent FUS/PARP1 ratio normalized to WT cells. Circles represent single values (n = 5 experiments performed) and bars represent mean ± SD. (E) Cell lysates (200 µg) from WT and α1KO cells were immunoprecipitated (IP) with anti-EGFR antibody or IgG control, and the immunoprecipitates were analyzed by Western blot for levels of EGFR and FUS. Input represents cell lysates (20 µg/lane) analyzed for levels of EGFR. (F) Immunoprecipitated FUS and EGFR bands were quantified by densitometry, and values represent FUS/EGFR ratio normalized to WT cells. Circles represent single values (n = 4 experiments performed) and bars represent mean ± SD. (G) Paraffin kidney sections from WT and Itgα1KO mice were stained with anti-FUS (green) and anti-pEGFR (red) antibody. Increased nuclear FUS expression and colocalization of FUS with pEGFR are evident in the glomeruli of Itgα1KO mice. (H) The numbers of FUS-, pEGFR-, and FUS/pEGFR-positive nuclei were evaluated, and values are expressed as percentage of total number of nuclei per glomerulus. Circles represent single glomeruli (43 for WT and 55 for Itgα1KO mice) and bars represent mean ± SD (n = 4 WT and 5 Itgα1KO mice). (I and J) Nuclear fractions (20 µg/lane) from kidneys of four WT and six α1KO mice were analyzed by Western blot for levels of FUS and PARP-1. FUS and PARP-1 bands were quantified and analyzed as indicated above. Circles represent single mice and bars represent mean ± SD. (K and L) Paraffin kidney sections from healthy controls or subjects with focal segmental glomerulosclerosis (FSGS) or diabetic nephropathy (DN; two subjects per group) were stained with anti-FUS antibody. Nuclear FUS staining (arrowhead) was detected in both glomeruli and tubules (K). Increased FUS staining was evident in the glomeruli of kidneys from subjects with DN or FSGS compared with healthy controls (L). Two-tailed t test was used for statistical analysis.
FUS forms a complex with EGFR
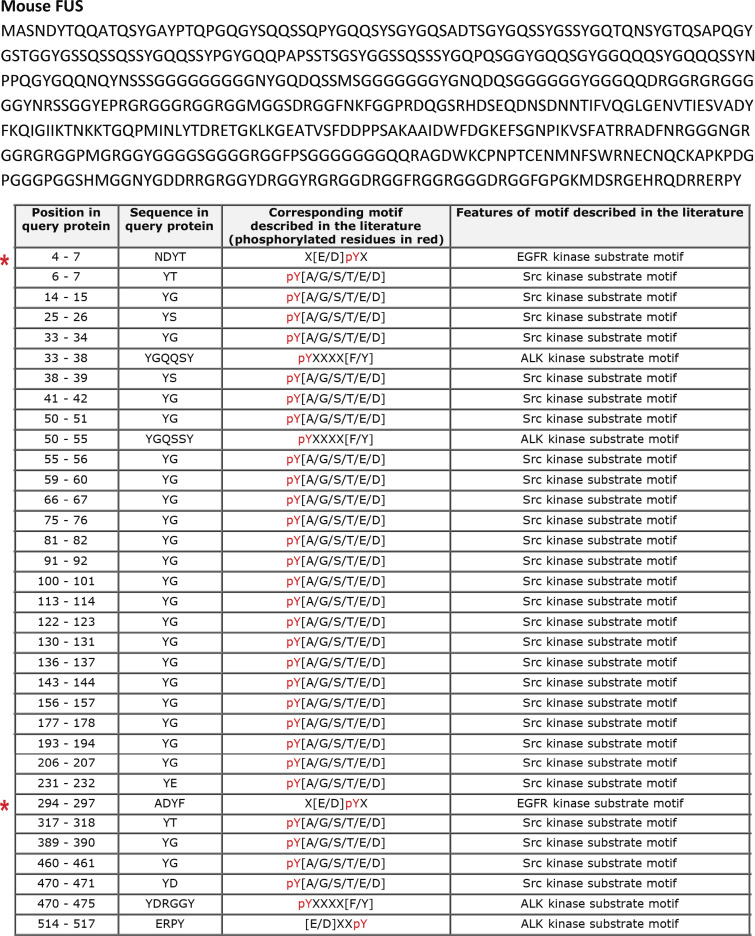
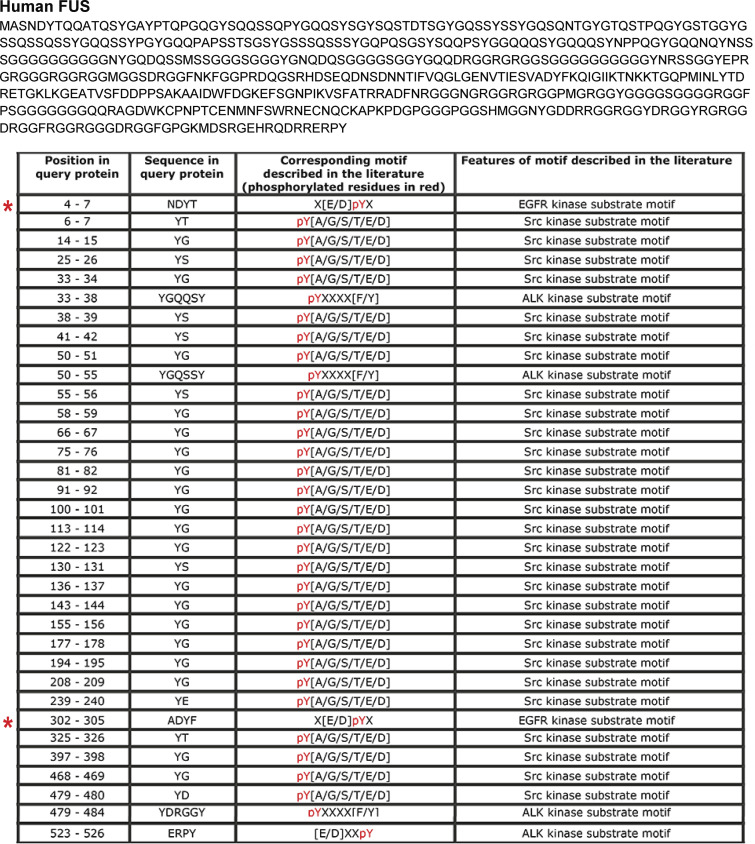
To identify kinases involved in FUS tyrosine phosphorylation, we analyzed the human and mouse FUS amino acid sequences with Phospho Motif Finder and found 34 tyrosine phosphorylation sites. Among them, we identified two (Y6 and Y296 in mouse and Y6 and Y304 in human) potential substrates for EGFR (Figs. S2 and S3). To determine whether EGFR can form a complex with FUS, we performed immunoprecipitation assays using anti-EGFR or IgG control antibody followed by Western blotting of FUS. We observed EGFR/FUS interaction in both WT and Itgα1KO mesangial cells, although it was more evident in the latter (Fig. 1, E and F). No EGFR/FUS interaction was observed in cell lysates immunoprecipitated with IgG control antibody (Fig. 1, E and F).
Figure S2.
Murine FUS contains tyrosine phosphorylation sites for EGFR kinase. Analysis of the murine FUS amino acid sequences with Phospho Motif Finder revealed Y6 and Y296 (red asterisks) as potential substrates for EGFR.
Figure S3.
Human FUS contains tyrosine phosphorylation sites for EGFR kinase. Analysis of the human FUS amino acid sequences with Phospho Motif Finder revealed Y6 and Y304 (red asterisks) as potential substrates for EGFR.
To determine the localization of FUS in healthy and diseased organs, we stained kidney sections of WT and Itgα1KO mice with anti-FUS antibody. FUS localized in the nuclei of both WT and Itgα1KO mice; however, more FUS-positive glomerular cells were detected in the latter group (Fig. 1, G and H). Staining with anti-pEGFR antibodies revealed EGFR activation primarily in the glomeruli of Itgα1KO mice, and its staining was primarily nuclear (Fig. 1 G), consistent with our previous finding that loss of Itgα1β1 leads to basal up-regulation of EGFR phosphorylation (Chen et al., 2007) and that activated EGFR can translocate to the nucleus (Lo et al., 2005). To confirm increased FUS nuclear localization in Itgα1KO mice, we evaluated FUS expression in nuclear fractions of kidney cortices. Indeed, we detected significantly increased content of FUS in kidney cortices from Itgα1KO mice compared with WT mice (Fig. 1, I and J). Moreover, we analyzed kidney sections from healthy subjects or subjects with focal segmental glomerulosclerosis or diabetic nephropathy to determine whether levels of nuclear FUS are increased in human fibrotic kidneys. Staining with anti-FUS antibody revealed low levels of nuclear FUS in the kidneys of healthy subjects. In striking contrast, nuclear FUS was evident in the glomeruli of diseased kidneys (Fig. 1, K and L). Thus, nuclear FUS expression is up-regulated in human fibrotic kidney.
Increased nuclear FUS in Dsk5 mutant mice
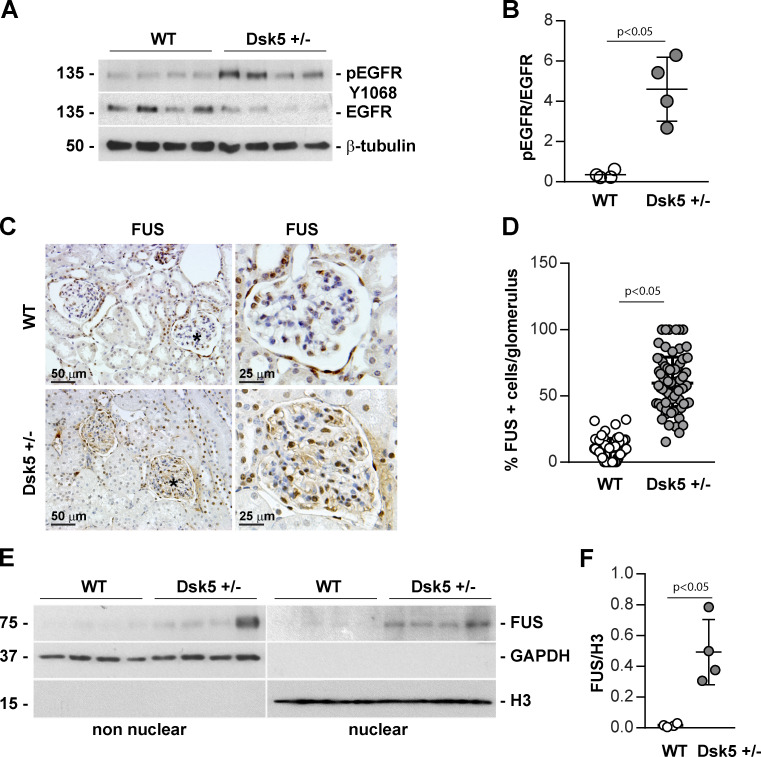
To confirm that increased EGF activation correlates with increased FUS nuclear localization, we used Dsk5 mutant mice, which carry a gain-of-function mutation in an allele of EGFR gene. These mice have increased basal EGFR kinase activity (Fig. 2, A and B) and develop progressive glomerulopathy, albuminuria, loss of podocytes, and tubulointerstitial fibrosis (Zhang et al., 2019). Paraffin kidney sections from WT and Dsk5 heterozygote mice stained with anti-FUS antibody revealed a significant increase of nuclear FUS in both glomerular and tubular cells of Dks5 heterozygote mice compared with their WT counterparts (Fig. 2, C and D). FUS expression in nuclear fractions of kidney cortices isolated from WT and Dsk5 heterozygote mice also revealed a significant increase in FUS in nuclear fractions of kidney cortices in the latter group (Fig. 2, E and F). Thus, increased EGFR activation correlates with FUS nuclear localization.
Figure 2.
Increased nuclear FUS in Dsk5 heterozygote mice expressing constitutively active EGFR kinase. (A and B) pEGFR and total EGFR in kidney tissue lysates (20 µg/lane) from four WT and four Dsk5 heterozygote (Dsk5+/−) mice were analyzed by Western blotting. pEGFR and EGFR bands were quantified by densitometry analysis, and values are expressed as pEGFR/EGFR ratio. Circles represent single mice (four WT and four Dsk5+/−) and bars represent mean ± SD. (C) Immunohistochemical detection of FUS in paraffin kidney sections from control (WT) and Dsk5+/− male mice. Increased nuclear FUS is evident in Dsk5+/− mice. (D) Glomerular FUS-positive cells as well as total cells were counted, and values are expressed as percentage of FUS-positive cells/glomerulus. Circles represent single glomerular values (57 WT and 82 Dsk5+/−) and bars represent mean ± SD (n = 3 mice). (E) Nonnuclear and nuclear fractions (20 µg/lane) of kidneys from four WT and four Dsk5+/− mice were analyzed by Western blot for levels of FUS, GAPHD (nonnuclear marker), and Histone 3 (H3, nuclear marker). (F) FUS and H3 bands were quantified by densitometry, and values are expressed as FUS/H3 ratio. Circles represent single mice (four WT and four Dsk5+/−) and bars represent mean ± SD. Two-tailed t test was used for statistical analysis.
Inhibition of EGFR phosphorylation with Erlotinib decreased FUS nuclear translocation
To further confirm that activated EGFR regulates FUS tyrosine phosphorylation levels and subsequent FUS nuclear translocation, we treated WT and Itgα1KO mesangial cells with Erlotinib, a small-molecule inhibitor of EGFR kinase. Western blot analysis of pEGFR levels in these cells revealed that the inhibitor significantly decreased baseline EGFR phosphorylation in Itgα1KO cells, with no obvious effect on WT cells, in which no basal pEGFR was detected (Fig. 3, A and B). Indeed, Erlotinib treatment significantly decreased nuclear FUS compared with the cells treated with vehicle only. This effect was more evident in Itgα1KO cells (Fig. 3, C and D). Finally, immunoprecipitation assays of nuclear fractions using anti-phosphotyrosine antibody showed a significant reduction in tyrosine phosphorylated FUS only in Itgα1KO cells treated with Erlotinib compared with Itgα1KO cells treated with vehicle (Fig. 3, E and F). No overall changes in tyrosine phosphorylated nuclear FUS levels were detected in WT cells treated with vehicle or erlotinib, consistent with the fact that there is no baseline activation of EGFR in these cells (Fig. 3, E and F). Cumulatively, our data indicate that activated EGFR regulates tyrosine phosphorylation of FUS and its nuclear localization.
Figure 3.
Inhibition of EGFR reduces FUS nuclear levels. (A and C) Serum-starved WT and Itgα1KO mesangial cells (n = 3 samples) were untreated or treated with Erlotinib (ERL, 5 µM for 18 h). Nonnuclear (NN) and nuclear (N) fractions (20 µg/lane) were analyzed by Western blot for levels of FUS, pEGFR, and total EGFR. PARP-1 or α-tubulin was analyzed to evaluate fraction purity. (B and D) Nonnuclear pEGFR and total EGFR or nuclear FUS and PARP-1 bands were quantified by densitometry, and values represent pEGFR/EGFR or FUS/PARP-1 ratio. Circles represent individual values (one representative experiment performed in triplicate is shown in B, two experiments performed in triplicate are shown in D), and bars represent mean ± SD. (E) Nuclear fractions (400 µg) from WT and Itgα1KO cells, treated as indicated above, were immunoprecipitated (IP) with anti-4G10 antibody or IgG control, and the immunoprecipitates were analyzed by Western blot for levels of FUS. Input represents nuclear lysates (20 µg/lane) analyzed for levels of PARP-1. (F) Immunoprecipitated FUS and PARP-1 bands were quantified by densitometry, and values represent FUS/PARP1 ratio normalized to WT cells. Circles represent single values (n = 4 experiments performed) and bars represent mean ± SD. One-way ANOVA and two-tailed t test were used for statistical analysis.
FUS mediates collagen IV production in mesangial cells
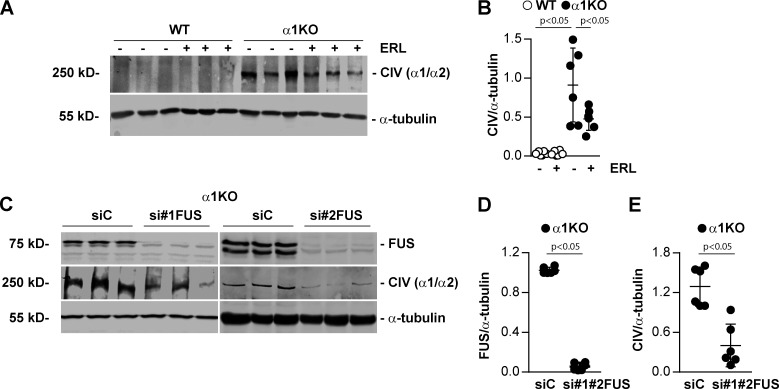
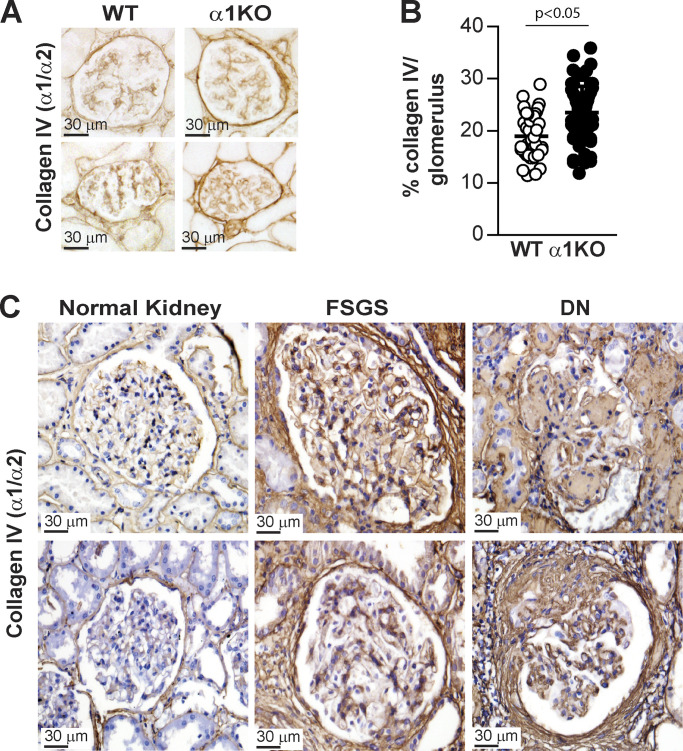
In view of our prior findings that activated EGFR signaling induces the expression of collagen IV α1 and α2 chains in kidney cells (Chen et al., 2007; Wang et al., 2015), together with the evidence that Dsk5 heterozygote mice develop progressive glomerulopathy and tubulointerstitial fibrosis (Zhang et al., 2019), we hypothesized that FUS mediates collagen production. In support of this hypothesis, studies of patients with mutations in the FUS gene that disable nuclear translocation of FUS protein demonstrated decreased collagen levels in the skin and serum (Ono et al., 1998; Tsukie et al., 2014). Therefore, we analyzed further the link between the EGFR/FUS signaling pathway and collagen production in WT and Itgα1KO mesangial cells treated with Erlotinib. Western blot analysis showed significantly increased collagen IV α1 and α2 chains (here referred to as collagen IV) levels in Itgα1KO cells treated with vehicle compared with WT cells (Fig. 4, A and B). In contrast, Erlotinib significantly decreased collagen IV levels in Itgα1KO cells, consistent with Erlotinib-mediated reduction in EGFR phosphorylation and FUS nuclear translocation. Moreover, we found that siRNA-mediated down-regulation of FUS significantly decreased collagen IV protein levels in Itgα1KO cells compared with cells treated with scrambled siRNA control (Fig. 4, C–E). To further confirm the correlation between nuclear FUS and collagen IV levels, we stained kidney sections from WT and Itgα1KO mice (Fig. 5, A and B) as well as from healthy subjects or subjects with focal segmental glomerulosclerosis or diabetic nephropathy (Fig. 5 C), with anti-collagen IV antibody. Increased collagen IV staining was evident in the glomeruli of Itgα1KO mice compared with WT mice. In addition, increased glomerular collagen IV staining was evident in human fibrotic kidneys compared with kidneys of healthy subjects. Thus, collagen IV expression in mesangial cells or glomeruli is dependent on EGFR kinase activity and the nuclear localization of FUS.
Figure 4.
Inhibition of EGFR or down-regulation of FUS reduces collagen production. (A) Cell lysates (50 µg/lane) from WT and Itgα1KO cells (n = 3 samples) untreated or treated with Erlotinib (ERL, 5 µM for 18 h) were analyzed for levels of collagen IV α1 and α2 chains (CIV α1/α2) and α-tubulin. (B) CIV and α-tubulin bands were quantified by densitometry, and values represent CIV/α-tubulin ratio. Circles represent single values (n = 6 with two experiments performed) and bars represent mean ± SD. (C) Cell lysates (20 µg/lane) from serum-starved Itgα1KO cells transfected with siRNA control (siC) or two different siRNAs against FUS (si#1FUS and si#2FUS; n = 3 samples) were analyzed by Western blot for levels of FUS, CIV α1/α2, and α-tubulin. (D and E) CIV and α-tubulin or FUS and α-tubulin bands were quantified by densitometry, and values represent CIV/α-tubulin and FUS/α-tubulin ratio. Circles represent single values (n = 6) and bars represent mean ± SD. One-way ANOVA and two-tailed t test were used for statistical analysis.
Figure 5.
Increased glomerular collagen IV deposition in Itgα1KO mice and human fibrotic kidneys. (A and C) Paraffin kidney sections from WT and Itgα1KO mice (A) as well as from healthy controls or subjects with focal segmental glomerulosclerosis (FSGS) or diabetic nephropathy (DN; B, two subjects per group) were stained with anti-collagen IV α1 and α2 chain antibody. Increased glomerular collagen IV staining is evident in the glomeruli of Itgα1KO mice and in the glomeruli of kidneys from subjects with FSGS or DN compared with healthy controls. (B) The amount of glomerular collagen IV in A was quantified using ImageJ and expressed as percentage collagen IV levels/glomerulus. Circles represent single glomeruli (44 WT and 72 Itgα1KO) and bars represent mean ± SD (n = 4 WT and 6 Itgα1KO mice). Two-tailed t test was used for statistical analysis.
FUS tyrosine phosphorylation regulates FUS nuclear translocation
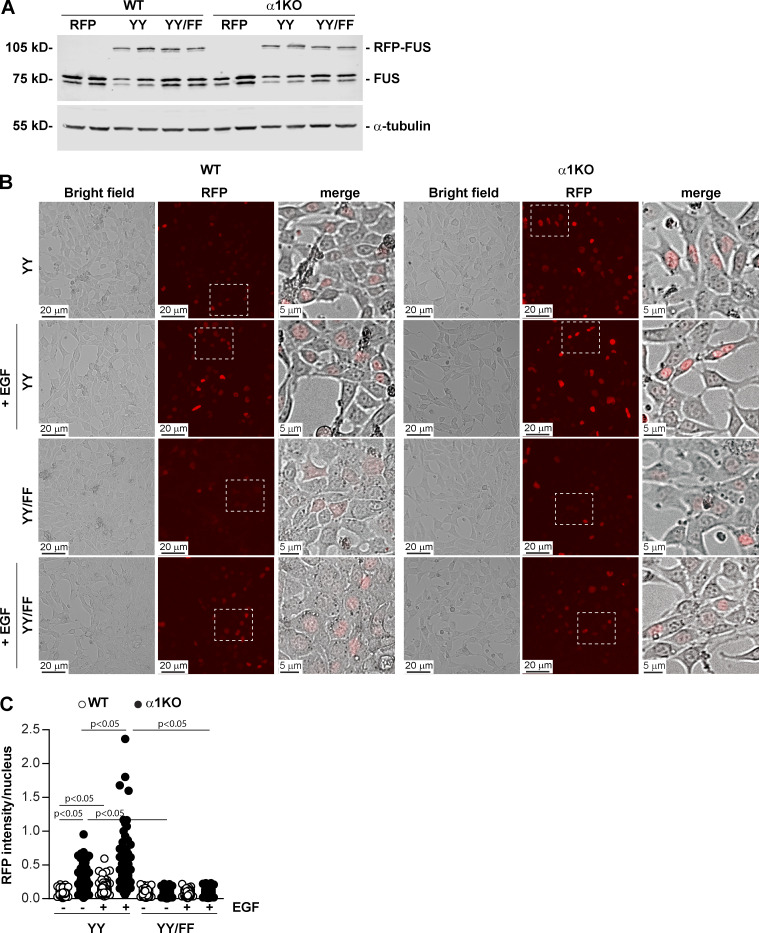
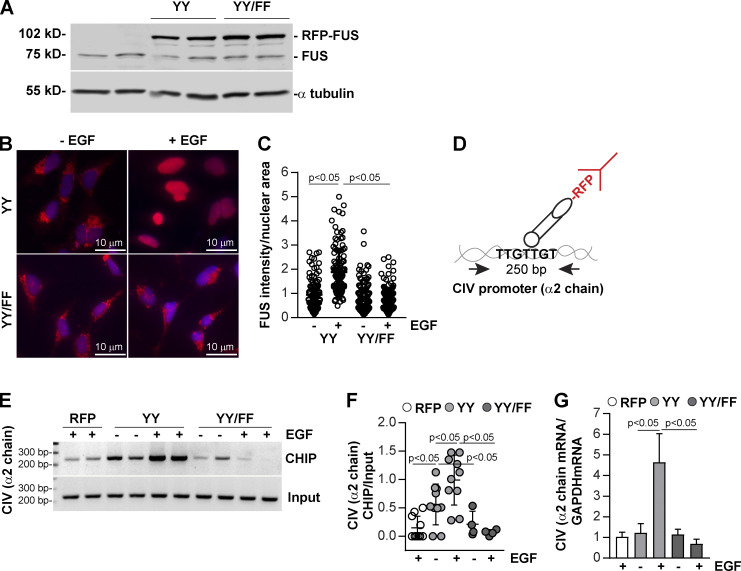
To establish whether EGFR-mediated FUS phosphorylation is a key step in controlling FUS nuclear translocation, we transiently transfected WT and Itgα1KO mesangial cells with WT murine FUS or FUS carrying Y-to-F substitutions in the putative EGFR phosphorylation sites Y6 and Y296 (FUS-Y6/296F) fused to RFP (Fig. S2). Western blot analysis showed comparable levels of RFP-FUS constructs in both WT and Itgα1KO mesangial cells, indicating that the Y-to-F mutations do not affect protein expression (Fig. 6 A).
Figure 6.
EGF-mediated FUS phosphorylation induces FUS nuclear translocation. (A) WT and Itgα1KO mesangial cells were transiently transfected with pRFP-C1 empty vector (RFP), or pRFP-C1 vector fused to WT murine FUS (YY) or FUS construct with mutated tyrosines 6 and 296 (YY/FF). After 48 h, cell lysates (20 µg/lane) were analyzed for levels of endogenous and RFP-FUS (two transfections are shown). (B and C) Serum-starved mesangial cells expressing the RFP-FUS constructs described in A were left unstimulated or stimulated with EGF for 30 min (20 ng/ml). Before and after treatment, the cells were imaged, and the nuclear RFP intensity was analyzed using ImageJ. Values in C represent RFP intensity/nuclear area. Circles represent single nuclei (n = 200–208 with two experiments performed) and bars represent mean ± SD. One-way ANOVA and two-tailed t test were used for statistical analysis.
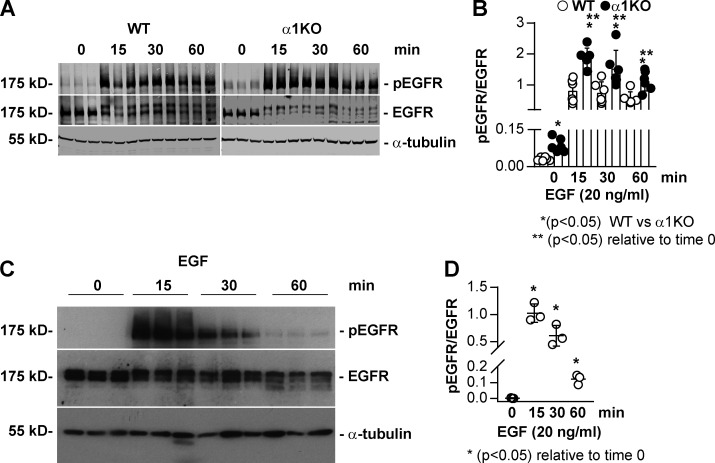
Immunofluorescence images of WT and Itgα1KO cells transfected with WT RFP-FUS revealed significantly higher RFP nuclear intensity in Itgα1KO cells compared with WT cells (Fig. 6, B and C), consistent with basal activation of EGFR being present only in the Itgα1KO cells (Fig. 3, A and B). Treatment of cells with EGF significantly increased RFP intensity in both WT and Itgα1KO cells, although it was significantly higher in the latter group (Fig. 6, B and C). To determine whether the differences in nuclear FUS between EGF-treated WT and Itgα1KO cells were due to differences in EGFR activation in these cells, we analyzed the levels of pEGFR in cells treated with EGF for different times. Both WT and Itgα1KO cells showed robust EGFR activation upon ligand treatment. However, EGFR activation was significantly higher in the Itgα1KO cells at all time points analyzed (Fig. S4 A), consistent with their loss of negative regulation of EGFR activation. In contrast to cells expressing WT RFP-FUS, both WT and Itgα1KO cells expressing the RFP-FUS-Y6/296F mutant showed RFP intensity similar to that detected in WT cells expressing WT RFP-FUS that remained unchanged after EGF treatment (Fig. 6, B and C). Thus, EGFR appears to mediate FUS nuclear translocation by phosphorylating Y6 and Y296 in FUS.
Figure S4.
Increased baseline and EGF-induced EGFR phosphorylation in Itgα1KO mesangial cells. (A and C) Serum-starved WT and Itgα1KO mesangial (A) or HEK293 (C) cells were treated with EGF (20 ng/ml) for the times indicated. Equal amounts of lysate (40 µg/lane) were analyzed by Western blot for levels of phosphorylated and total EGFR as well as α-tubulin to verify equal loading. (B and D) pEGFR and EGFR bands were quantified by densitometry. Values represent pEGFR/EGFR ratio and are mean ± SD of two experiments performed in triplicate (B) or one representative experiment performed in triplicate (D). One-way ANOVA and two-tailed t test were used for statistical analysis.
FUS binds to the collagen IV α1 and α2 chain promoter and promotes collagen transcription
To further define the role of nuclear FUS in collagen production, we used human embryonic kidney (HEK) 293 cells, since they allow a higher transfection efficiency compared with mesangial cells and we have recently used them to analyze protein/DNA interaction (Chiusa et al., 2019). Like mesangial cells, HEK293 cells express endogenous EGFR that is autophosphorylated upon EGF treatment (Fig. S4 B). We transiently transfected HEK293 cells with RFP-FUS or RFP-FUS-Y6/296F constructs (Fig. 7 A) and analyzed nuclear RFP after EGF treatment. In cells expressing WT or mutated RFP-FUS, RFP was evident in the cytoplasm and nuclei, although the intensity was higher in the cytoplasm (Fig. 7, B and C). After EGF treatment, nuclear RFP intensity significantly increased only in cells transfected with WT RFP-FUS, whereas no changes were observed in cells expressing the RFP-FUS-Y6/296F mutant (Fig. 7, B and C).
Figure 7.
FUS binds to the bidirectional promoter of collagen IV α1 and α2 chains and drives collagen IV α2 chain transcription. (A) HEK293 cells were transiently transfected with pRFP-C1 vector fused to WT FUS (YY) or FUS construct mutated in tyrosines 6 and 296 (YY/FF). After 48 h, cell lysates (20 µg/lane) were analyzed for the levels of endogenous and RFP-FUS (two transfections shown). (B) Serum-starved HEK expressing the RFP-FUS constructs described in A were left untreated or treated with EGF (20 ng/ml). After 1 h, the cells were fixed, permeabilized, and incubated with anti-RFP antibody followed by Alexa Fluor 555–conjugated secondary antibody. (C) Nuclear RFP intensity was analyzed using ImageJ, and values represent RFP intensity/nuclear area. Circles represent single nuclei (n = 115–124 with two experiments performed) and bars represent mean ± SD. (D) Overview of the ChIP assay performed in HEK cells expressing RFP empty vector (RFP), RFP-FUS (YY), or RFP-mutated FUS (YY/FF) constructs. (E) Cross-linked DNA–protein complexes from cells (n = 2 samples) untreated or treated with EGF (20 ng/ml) for 30 min were immunoprecipitated with RFP-TRAP beads. After DNA elution and purification, DNA fragments were amplified using collagen IV (CIV) α2 chain promoter primers spanning a putative FUS binding site. Input represents amplified CIV promoter in total DNA isolated from untreated or EGF-treated cells before immunoprecipitation. (F) CIV ChIP and input CIV bands were quantified by densitometry. Circles represent single values (n = 2–5 experiments performed in duplicate) and bars represent mean ± SD. (G) Serum-starved HEK expressing the constructs described in E were left untreated or treated with EGF (20 ng/ml). After 3 h, the levels of collagen IV α2 mRNA were analyzed by reverse transcription rqPCR. Bars and errors are mean ± SEM of three to six experiments performed in duplicate. One-way ANOVA and two-tailed t test were used for statistical analysis.
Based on our observation that the bidirectional promoter of human collagen IV α1 and α2 chains contains several FUS-responsive elements (Fig. S1), we next determined whether FUS forms a complex with this collagen IV promoter and whether this is dependent on EGF stimulation. We performed the chromatin immunoprecipitation (ChIP) assay on HEK293 cells expressing WT or mutated RFP-FUS treated with vehicle or EGF. To do this, nuclei were immunoprecipitated with RFP-TRAP beads, and the immunoprecipitated DNA was amplified using primers spanning a FUS binding motif at position 2,032–3,161 in the collagen IV α2 chain promoter (Fig. 7 D; and Fig. S1). ChIP analysis showed a significant increase in binding of FUS to this collagen IV promoter region in cells transfected with WT RFP-FUS compared with cells transfected with RFP vector control or RFP-FUS-Y6/296F (Fig. 7, E and F). Treatment with EGF significantly increased binding of FUS to the collagen IV promoter only in cells expressing WT, but not mutated RFP-FUS (Fig. 7, E and F).
The ChIP analysis suggested that collagen IV α2 chain might be transcriptionally controlled by FUS. To determine the functionality of our finding, we measured collagen IV α2 chain mRNA in HEK293 cells expressing WT or mutated RFP-FUS treated with vehicle or EGF. Treatment with EGF significantly enhanced collagen IV α2 chain mRNA synthesis only in cells expressing WT RFP-FUS (Fig. 7 G), consistent with the finding that EGF enhances nuclear translocation and binding to collagen IV α2 chain promoter of WT RFP-FUS, but not RFP-FUS-Y6/296 mutant.
Inhibiting FUS interaction with its nuclear import shuttle transportin 1 reduces FUS nuclear translocation and collagen production
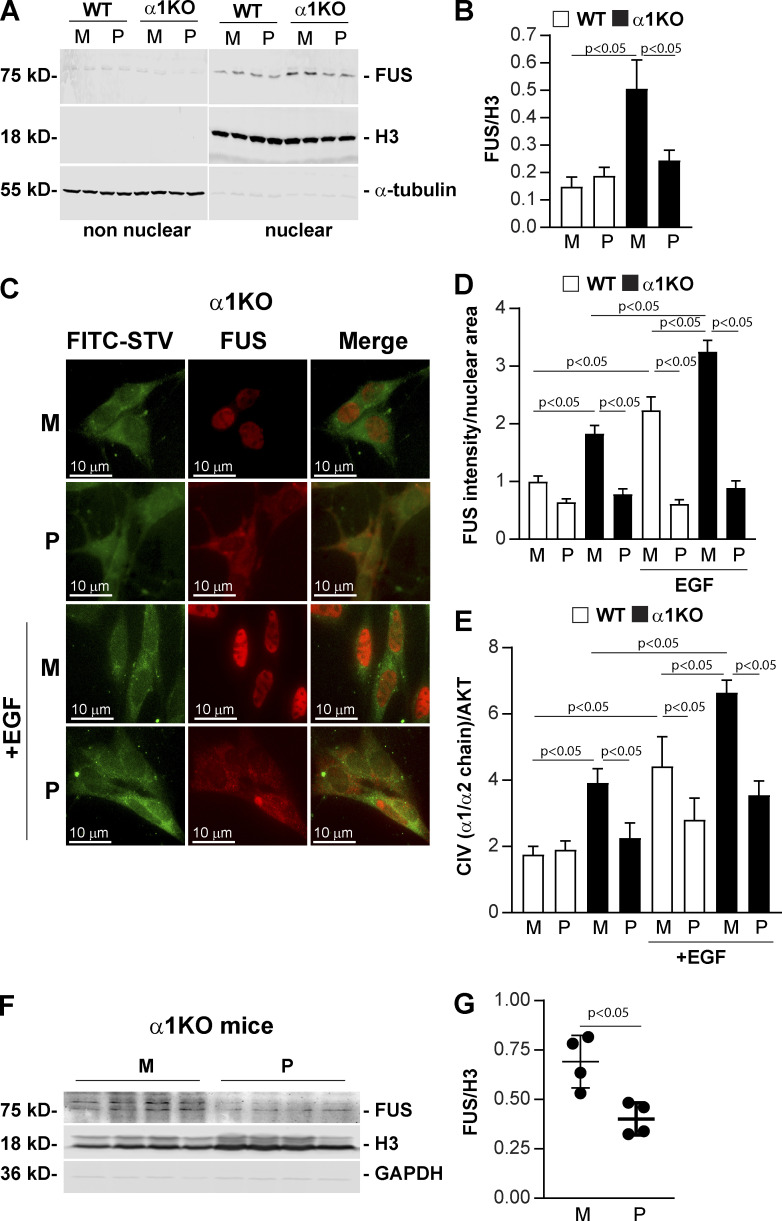
We hypothesized that a cell-penetrating peptide inhibitor of FUS nuclear translocation would reduce collagen IV production, thereby providing a new anti-fibrotic tool. To assess this concept, we designed, produced, and tested a biotin-labeled cell-penetrating chimeric peptide that inhibits FUS nuclear translocation by preventing its interaction with transportin 1 (CP-FUS-NLS), and the mutated peptide (CP-mutFUS-NLS) was designed as an inactive control. We treated WT and Itgα1KO mesangial cells with these two peptides and then analyzed the levels of FUS in nonnuclear and nuclear fractions. CP-mutFUS-NLS–treated Itgα1KO cells showed significantly higher levels of nuclear FUS compared with CP-mutFUS-NLS–treated WT cells (Fig. 8, A and B), consistent with the fact that this peptide is not able to prevent FUS/transportin-1 interaction. In contrast, treatment of cells with CP-FUS-NLS significantly reduced nuclear FUS, and this effect was primarily evident in Itgα1KO cells (Fig. 8, A and B). Consistent with this finding, baseline nuclear FUS was evident in Itgα1KO mesangial cells treated with CP-mutFUS-NLS, while highly prominent cytoplasmic staining was evident in cells treated with CP-FUS-NLS (Fig. 8, C and D). Importantly, treatment with EGF increased nuclear FUS intensity only in Itgα1KO cells treated with CP-mutFUS-NLS, but not CP-FUS-NLS (Fig. 8, C and D). To determine the physiological relevance of these findings, we analyzed collagen IV levels in WT and Itgα1KO cells treated with the two peptides with or without EGF. In the absence of EGF, Itgα1KO cells treated with CP-mutFUS-NLS produced more collagen IV than WT cells treated with CP-mutFUS-NLS, and this basal increased collagen IV production was significantly down-regulated after treatment with CP-FUS-NLS (Fig. 8 E). Treatment with EGF increased collagen IV production in both CP-mutFUS-NLS–treated WT and Itgα1KO cells, although this effect was more prominent in the latter group (Fig. 8 E). In contrast, treatment of cells with CP-FUS-NLS significantly reduced EGF-mediated collagen IV production in both WT and Itgα1KO cells (Fig. 8 E). Finally, to determine whether CP-FUS-NLS peptide can reduce FUS nuclear levels in vivo, we treated Itgα1KO mice with both inactive and active peptides and observed a significant decrease in FUS levels in kidney nuclear fractions of mice treated with CP-FUS-NLS compared with those observed in CP-mutFUS-NLS–treated mice (Fig. 8, F and G). Cumulatively, these results document that the CP-FUS-NLS peptide reduces FUS nuclear translocation and collagen IV production in mesangial cells as well as FUS nuclear levels in vivo.
Figure 8.
Cell-penetrating FUS-NLS peptides inhibit FUS nuclear translocation and collagen IV synthesis. (A) Analysis of FUS subcellular localization in WT and Itgα1KO mesangial cells (n = 2 per treatment shown) treated with biotin-labeled CP-FUS-NLS (AAVALLPAVLLALLAPK(Biot)RRGEHRQDRRERPY, P) or inactive CP-mutFUS-NLS (AAVALLPAVLLALLAPK(Biot)REGEHREDREERGA, M), both at 0.4 µM. After 1 h, nonnuclear and nuclear fractions (20 µg/lane, n = 2 samples) were analyzed by Western blot for content of FUS, H3 (nuclear marker) or α-tubulin (nonnuclear marker). (B) Nuclear FUS and H3 bands were quantified by densitometry, and values represent the FUS/H3 ratio. Values are the mean ± SEM of four experiments performed at least in duplicate. (C) Immunofluorescence tracking FUS in serum-starved WT and Itgα1KO mesangial cells. Cells were treated with biotinylated cell-penetrating WT (P) or mutated (M) FUS-NSL (both at 0.2 µM) and left unstimulated or stimulated with EGF (20 ng/ml). After 1-h treatment, cells were fixed, permeabilized, and incubated with anti-FUS antibody (red) and FITC-conjugated streptavidin (green). (D) The intensity of nuclear FUS was analyzed using ImageJ, and values represent FUS intensity/nuclear area. Bars and errors are mean ± SEM of two experiments with 112–167 cells analyzed in each group. (E) Serum-starved HEK293 cells treated with cell-penetrating biotinylated WT (P) or mutated (M) FUS-NSL peptide (both at 0.4 µM) were left untreated or treated with EGF (20 ng/ml). After 24 h, the levels of collagen IV (CIV) and AKT in total cell lysates (40 µg/lane) were analyzed by Western blot. CIV and AKT bands were quantified by densitometry, and values represent CIV/AKT ratio. Bars and errors are mean ± SEM of three experiments performed at least in duplicate. (F) Itgα1KO male mice (n = 4) received i.p. injections of cell-penetrating biotinylated WT (P) or mutated (M) FUS-NSL peptide (35.67 µg/g body weight and 33.82 µg/g body weight for WT and mutated FUS-NLS peptide, respectively) twice a day. After 3 d, kidney nuclear fractions (30 µg/lane) were analyzed by Western blot for levels of FUS, H3, and GAPDH. (G) FUS and H3 bands were quantified by densitometry, and values are expressed as FUS/H3 ratio. Bars and errors are mean ± SEM of four mice. One-way ANOVA and two-tailed t test were used for statistical analysis.
Discussion
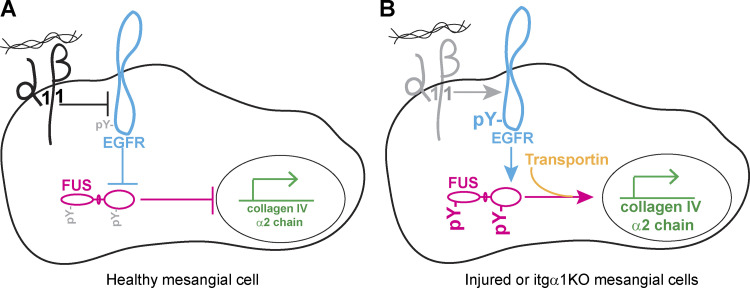
In this study, we show that the signaling axis between Itgα1β1 and EGFR regulates collagen production through the tyrosine phosphorylation and subsequent nuclear translocation of the DNA-RNA binding protein FUS. Increased basal (as observed in cells deficient in Itgα1β1) or ligand-triggered EGFR activation leads to the tyrosine phosphorylation of FUS and its nuclear translocation. In the nucleus, phosphorylated FUS binds to the collagen IV gene promoter, driving its transcription (Fig. 9).
Figure 9.
Schematic depiction of FUS regulation by the Itgα1β1/EGFR axis. (A) In healthy mesangial cells, activation of Itgα1β1 negatively regulates EGFR activation, thus reducing EGFR-mediated FUS phosphorylation, FUS nuclear translocation, and transcription of the collagen IV α2 chain. (B) In cells lacking Itgα1β1, increased EGFR activation results in enhanced FUS phosphorylation and FUS-mediated collagen transcription.
EGFR is a receptor tyrosine kinase activated by several ligands that triggers different functions such as cell proliferation, migration, and production of ECM components. EGFR is highly expressed in cells of the glomerulus, the filtering unit of the kidney, where it plays a key role in the initiation, development, and progression of glomerular injury. In the partial nephrectomy model, for example, inhibition of EGFR reduces glomerular fibrosis (Liu et al., 2012), and in mice and humans with rapidly progressive glomerulonephritis, expression of the ligand HB-EGF by podocytes promotes EGFR phosphorylation and activation, thus contributing to glomerular injury (Bollée et al., 2011). Moreover, mice lacking HB-EGF expression, specifically in endothelial cells, show decreased glomerular EGFR activation and decreased angiotensin-II–mediated glomerular injury (Zeng et al., 2016). EGFR promotes fibrosis by activating transcription factors (Quesnelle et al., 2007; Xu and Shu, 2013), mediating inflammatory response, transactivating profibrotic receptors (Rayego-Mateos et al., 2018), or promoting the assembly of the NADPH oxidase complex, thus stimulating production of ROS (Chen et al., 2007).
Herein, we provide new evidence that EGFR interacts with FUS and regulates its function. We show that EGFR can promote nuclear translocation of FUS by affecting its phosphorylation. FUS contains several tyrosine phosphorylation sites, and depending on the cell type and the FUS domains, its phosphorylation can have both protective or deleterious effects. Phosphorylation of Tyr526 at the C terminus by the family of Src kinases reduces FUS interaction with transportin, resulting in accumulation of FUS in the cytoplasm (Darovic et al., 2015). Moreover, DNA damage, which occurs in patients with frontotemporal lobar degeneration, leads to the cytoplasmic accumulation of FUS via phosphorylation of its N terminus by the DNA-dependent protein kinase (Deng et al., 2014). On the other hand, FUS is intrinsically prone to aggregate in pathological amyloid-like fibrils, and phosphorylation of FUS at its N-terminal intrinsically disordered region by DNA-dependent protein kinase reduces its aggregation, thus preventing FUS-associated cytotoxicity (Luo et al., 2018; Monahan et al., 2017). Our study indicates that EGFR-mediated FUS phosphorylation enhances nuclear FUS translocation and collagen production. These findings, together with the evidence that expression of FUS with mutated tyrosine residues targeted by EGFR kinase prevents EGFR-mediated collagen production, indicate that phosphorylated FUS is a key mediator of the EGFR-mediated profibrotic pathway.
Of interest, FUS is an RNA-DNA binding protein that has been well studied in the context of ALS or frontotemporal lobar degeneration. In these neurological disorders, mutations and/or posttranslational modifications of FUS that prevent its interaction with transportin, and subsequent nuclear translocation, are detrimental. They cause accumulation of FUS in the cytoplasm, leading to the formation of insoluble aggregates and neuronal toxicity. In addition, FUS mutations might lead to neurotoxicity by altering the translation of genes associated with mitochondrial function, resulting in reduction of mitochondrial size (Nakaya and Maragkakis, 2018). While cytoplasmic FUS leads to neuronal toxicity and death, studies from patients with ALS suggest that FUS could control the production and levels of profibrotic molecules including collagens. In patients with ALS, the serum levels of C-terminal propeptide of type I procollagen (a marker of type I collagen synthesis) are significantly lower, while the levels of C-terminal telopeptide of type I collagen (a marker of type I collagen degradation) are significantly higher than those detected in healthy controls (Ono et al., 2000). These findings, together with the observation that the skin collagen content in ALS patients is significantly lower than that observed in healthy subjects (Ono et al., 2000), indicate that the lower levels of collagen in ALS are most likely due to increased degradation. In addition to collagen I, decreased urinary collagen IV levels, which seem to correlate with decreased collagen IV immunoreactivity in skin, are observed in ALS patients, suggesting a defect in collagen IV production (Ono et al., 1999). Consistent with these findings, we provide evidence that nuclear FUS binds to the bidirectional promoter of collagen IV α1 and α2 chains, commencing its transcription. Another mechanism whereby FUS can regulate collagen transcription is by binding factors such as the nuclear paraspeckle assembly transcript 1 (Lagier-Tourenne et al., 2012; Lourenco et al., 2015). This factor promotes cell proliferation and progression of diabetic nephropathy by regulating mTOR-mediated signaling (Huang et al., 2019). Accordingly, we show that the levels of nuclear FUS are up-regulated in kidneys of subjects with fibrotic diseases, including diabetic nephropathy.
Our study, together with the finding that FUS binds the promoter of collagen X α1 chain (Gu et al., 2014), suggests that FUS might control the transcription of ECM genes involved in fibrotic responses. We analyzed the promoters of several ECM components including fibrillar collagen I, laminins, and fibronectin and found FUS-responsive elements in most of them (Table S1). In addition to controlling the synthesis of ECM components, FUS seems to play a role in inflammation. Overexpression of FUS in murine and human astrocytes leads to up-regulation of several genes implicated in inflammatory responses, including CD68, IL-6, and Csf2, to name a few (Ajmone-Cat et al., 2019). Analysis of the promoters of these reported genes revealed the presence of FUS-responsive elements in most of them (Table S1).
Although the studies highlighted above suggest that FUS plays a fibrotic and inflammatory role, published evidence suggests that this RNA-DNA binding protein could also prevent collagen production and deposition by regulating the expression of manganese superoxide dismutase gene transcription, thus reducing production of profibrotic ROS (Dhar et al., 2014). Moreover, in osteoclasts, FUS binds microphthalmia-associated transcription factor, promoting the transcription of cathepsin K involved in bone reabsorption and collagen degradation (Bronisz et al., 2014). Recently, the circular RNA circFndc3b has been shown to play a protective role in cardiac repair after myocardial infarction by enhancing neovascularization and improving left ventricular function by interacting with FUS and regulating VEGF expression (Garikipati et al., 2019). Thus, FUS can contribute to or slow down the development of fibrosis, and these effects seem to be organ and cell type dependent.
We sought to limit the availability of FUS in the nucleus by designing and testing a cell-penetrating peptide containing the NLS of FUS, thus competing with the binding of FUS to transportin. A similar approach was successfully applied by us to inhibition of the stress-responsive transcription factors’ access to the nucleus in experimental models of inflammation caused by microbial, autoimmune, and metabolic insults (Hawiger and Zienkiewicz, 2019). The noninvasive method of intracellular delivery of NLS-bearing peptides and anti-inflammatory proteins is based on the membrane-translocating hydrophobic segment. This segment allows crossing of the cell membrane phospholipid bilayer without engagement of the chirally specific receptor/transporter mechanisms while bypassing the endosomal compartment (Veach et al., 2004). Herein, we provide new evidence that cell-penetrating peptides carrying the PY-FUS-NLS sequence compete with FUS/transportin interaction, thereby reducing FUS nuclear localization in mice and kidney cells as well as FUS-mediated collagen transcription in kidney cells. These findings offer a novel approach to limiting the fibrotic process in the kidneys and possibly other organs. However, it remains to be determined whether and how the cell-penetrating peptide targeting nuclear translocation of transcription factor FUS can be selectively guided to a selective organ affected by the fibrotic process.
In conclusion, our analysis of the Itgα1β1/EGFR-triggered signaling pathways identified the RNA-DNA binding protein FUS as a profibrotic downstream target. We found that nuclear FUS regulates expression of collagen IV by directly binding to its promoter. These findings, combined with evidence that inhibition of FUS nuclear translation reduces collagen production, reveal a previously unrecognized role of FUS and open new therapeutic options for the treatment of fibrotic diseases.
Materials and methods
Cells
HEK 293 cells were obtained from ATCC and cultured in DMEM with 10% FBS. Cells were tested for mycoplasma upon purchase and were characterized and authenticated by the vendor. Immortalized mesangial cells (MCs), isolated from WT and Itgα1KO mice crossed onto the immortomouse background, were propagated at 33°C in DMEM with 10% FBS and 100 IU/ml IFNγ. Cells were cultured at 37°C without IFNγ for at least 4 d before use, as this is the optimal time for immortalized MCs to acquire a phenotype similar to that of freshly isolated primary MCs.
Nonnuclear (cytosolic) and nuclear cell fractions
Cells were harvested in 10 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, protease inhibitors (Roche Applied Science), and 5 mM NaVO3, and supernatant (nonnuclear) and pellet (nuclear) fractions were separated by centrifugation (400 g for 4 min at 4°C). Nuclear fractions were lysed in the buffer specified above containing 25% glycerol.
Kidney cortices (10 mg) were homogenized in 250 mM sucrose, 10 mM Hepes, pH 7.4, 5 mM KCl, 1.5 mM EDTA, pH 8.0, 5 mM Na3VO4, and protease inhibitors. After 15 min on ice, tissue lysates were centrifuged as described above. The nuclear fraction was resuspended in 20 mM Hepes, pH 7.4, 0.4 M NaCl, 2.5% glycerol, 1 mM EDTA, pH 8.0, 0.5 mM NaF, and protease inhibitors.
Western blotting
Cell or tissue lysates were resolved in 8% or 4–20% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated with the primary antibodies described in Table S2 followed by HRP-conjugated secondary antibodies and an ECL kit (Pierce) or IRDye fluorescent dyes secondary antibodies and a LiCor Odyssey infrared imaging system. Immunoreactive bands were quantified by densitometry analysis using ImageJ (National Institutes of Health) or Image Studio Lite.
Immunoprecipitation
Cell fractions (200–500 μg proteins) were precleared using 40 μl of protein G sepharose (GE Healthcare; SE-75184). The lysates were then incubated with 20 μl of protein G sepharose together with 1 μg of anti-phosphotyrosine (4G10) or mouse IgG control. After 18 h, the immunoprecipitates were washed with 50 mM Tris, pH 7.2, 150 mM NaCl, and 1% Triton X-100; eluted in 50 mM Tris-HCl, pH 6.8, containing 2% SDS, 10% glycerol,1% β-mercaptoethanol, 12.5 mM EDTA, and 0.02% bromophenol blue; and analyzed by Western blot as indicated above.
Immunohistochemistry and immunofluorescence
Mouse or human paraffin kidney sections were stained with mouse anti-FUS or anti-collagen IV α1 and α2 chains antibody (Table S2) followed by HRP-conjugated anti-mouse secondary antibody and Sigma Fast DAB chromogenic tablets (Sigma-Aldrich) dissolved in water. Phase-contrast images were taken using an Olympus Bx51 upright microscope (20× or 40× lens) equipped with an Olympus DP72 camera. Images were captured using the cellSens Standard software. Images were saved as Tiff files and processed using Photoshop (Adobe Systems). The levels of glomerular collagen IV–positive structures in kidneys of WT (n = 4) and Itgα1KO (n = 6) mice were evaluated in Tiff images using ImageJ.
For double immunostaining, paraffin kidney sections were stained with mouse anti-FUS and rabbit anti-pEGFR (Y1173) antibodies followed by Alexa Fluor 488 anti-mouse and Alexa Fluor 555 anti-rabbit antibodies (Alexa Fluor; A32723 and A21428) and mounted using ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific; P36931). Slides were analyzed using an LSM 710 META inverted point scanning confocal microscope with the following magnification and numerical aperture of the objective lenses: 63×/1.40 Plan-Apochromat oil, NA 1.4. Slides were treated with water-based embedding medium before analysis. Images were taken by acquisition software Zen. Images were saved as Tiff and processed using Photoshop.
Serum-starved cells expressing RFP-FUS or RFP-FUS-Y6/296F, plated in multiwell chamber slides (Millipore; PEZG S0416), were treated with vehicle (serum-free medium) or 20 ng/ml EGF for 0.5–1 h. Bright-field and fluorescence images were taken on live cells kept in serum-free medium before and after EGF treatment using an Olympus IX81 microscope with the following magnification and numerical aperture of the objective lenses: UPlanApo20XO3, NA 0.7 in a 512 × 512-pixel array, 12 bits. The microscope is equipped with a 37°C stage, a CO2 incubator, a Hamamatsu Photonics C9100-02 electron-multiplying charge-coupled device camera, and SlideBook 4.1 acquisition software. Images were saved as Tiff and processed using Photoshop.
In some experiments, cells expressing RFP-FUS or RFP-FUS-Y6/296F were fixed with 4% PFA in PBS after EGF treatment, and slides were mounted using ProLong Gold Antifade Mountant with DAPI. For double staining, cells were stained with anti-RFP antibody followed by Alexa Fluor 555–conjugated secondary antibody and FITC-conjugated streptavidin and mounted as indicated above. Confocal images were collected using the LSM710 META inverted confocal microscope described above. The intensity of nuclear RFP-positive structures was quantified in Tiff images using ImageJ.
Plasmid generation
pRFP-C1 murine FUS plasmid was obtained by replacing the GFP cDNA from pEGFR-C1 murine FUS plasmid with the RFP cDNA using NheI and SacI restriction enzymes. To generate pRFP-C1 FUS Y6/296F, we used QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technology; 200521) following the manufacturer’s instructions. The following primers were used: FUS-Y6F, 5′-GCTTCAAACGACTTTAC-3′; FUS Y296F, 5′-GTAATCAGCCACAGATTC-3′. PCR conditions were as follows: 95°C/1 min; 95°C/50 s, 60°C/50 s, 68°C/7.5 min for 18 cycles; and 68°C/7 min. Empty pRFP-C1 vector was generated by releasing FUS cDNA with SacI and BamHI restriction enzymes.
Design and development of cell-penetrating nuclear import inhibitors of FUS
A cell-penetrating chimeric peptide, an inhibitor of nuclear translocation of FUS, was designed based on analysis of published crystal structures of FUS C terminus PY NLS docking to its nuclear import adaptor protein, transportin 1 (a.k.a. karyopherin β2; PDB accession nos. 4FQ3 and 5YVG). Sequence of a cell-penetrating inactive control peptide was established upon mutagenesis of the key NLS amino acids. The WT sequence, derived from the C-terminus of human FUS, and its inactive control sequence were merged with the cell-penetrating motif derived from the signal sequence hydrophobic region (SSHR) of fibroblast growth factor 4 (Lin et al., 1995). This cell-penetrating motif attached to peptides or proteins crosses the plasma membrane through phospholipid bilayer without involving the chi rally specific receptor/transporter mechanism. Moreover, SSHR-based motif bypasses the endosomal compartment, thereby escaping the potential degradation of the attached cargo (NLS) by lysosomal proteases (Veach RA et al., 2004). Both peptides were labeled with biotin coupled to the ε-amino group of the lysine inserted between the FUS-NLS and SSHR motifs. To reduce hydrophobicity of biotin-labeled peptides, the nonbinding serine 513 was replaced with arginine. The synthesis of a WT peptide biot-CP-FUS-NLS (AAVALLPAVLLALLAPK(Biot)RRGEHRQDRRERPY; 31 aa; MW 3,763 D), and its inactive control peptide, biot-CP-mutFUS-NLS (AAVALLPAVLLALLAPK(Biot)REGEHREDREERGA; 31 aa; MW 3,578 D) was conducted on an automated peptide synthesizer FOCUS XC (AAPPTec) using standard Fmoc chemistry protocols. After chain assembly was completed, the lysine’s side chain protection group, ivDde, was selectively removed by 2% hydrazine/dimethylformamide solution (vol/vol) followed by biotin coupling. Crude peptides were removed from resin with a TFA cleavage cocktail and purified by dialysis against double-distilled water in 2-kD membrane (Spectra/Por 7; Spectrum Laboratories). Purity and structure of the final products were verified by analytical C18 RP HPLC (Beckman Coulter GOLD System) and MALDI mass spectroscopy (Voyager Elite; PerSeptive Biosystems).
Cell treatment and transfection
Serum-starved kidney mesangial cells or HEK293 cells were treated for different times with EGF (20 ng/ml; R&D Systems; 236-EG) or Erlotinib (5 μM; Millipore Sigma; CDS022564), and cell lysates or nonnuclear and nuclear fractions were analyzed by Western blot analysis.
Mesangial cells or HEK293 cells were transfected with 0.2–0.4 μg of pRFP-C1 or pRFP-C1 carrying WT or mutated FUS with Lipofectamine 2000 (Life Technology). After 48 h, cells were washed with PBS and incubated with serum-free medium. After 24 h, cells were left untreated; treated with EGF, Erlotinib, or the appropriate vehicle control (serum free medium and DMSO final concentration 0.0005%, respectively) for different times; and processed for immunofluorescence or Western blot analysis.
To down-regulate FUS expression, kidney mesangial cells derived from Itgα1KO mice were transfected with 20 nM of FUS si#1RNA, FUS si#2RNA, or nontargeting siRNA (Ambion; 4390815, 16704, and 4390843, respectively) using Lipofectamine 2000. After 48 h, cells were washed with PBS and incubated in serum-free medium. After 24 h, cells were processed for Western blot analysis.
Serum-starved mesangial cells were incubated with cell-penetrating biot-CP-FUS-NLS and its inactive control peptide biot-CP-mutFUS-NLS (0.2–0.4 μM). After 30 min, cells were treated with vehicle (serum-free medium) or EGF (20 ng/ml). After 0.5–24 h, cells were processed for subcellular fractionation, Western blot analysis, or immunofluorescence.
Analysis of gene promoters for presence of FUS-responsive elements
Promoter sequences of the various genes encoding ECM components or inflammatory factors (Table S1) were obtained from www.ensembl.org or www.switchgeargenomics.com. Sequences were analyzed for the presence of the FUS-responsive element TxxxxGT (Tan et al., 2012).
ChIP
Serum-starved HEK293 cells expressing RFP-FUS or RFP-FUS-Y6/296F were treated with vehicle (serum-free medium) or EGF (20 ng/ml). After 30 min, cells were harvested, and the nuclear FUS-DNA complex was isolated using ChIP-IT Express Enzymatic Magnetic Chromatin Immunoprecipitation Kit (Active Motif; 53009) according to the manufacturer’s instructions. Briefly, proteins and DNA were cross-linked with 1% PFA. After 15 min, the cross-link reaction was stopped by incubation with Glycine Stop-Solution. After chromatin shearing, FUS was immunoprecipitated using RFP-TRAP beads (Cromteck). DNA was eluted from the beads with Elution Buffer AM2. DNA amplification was performed using the following human collagen IV α2 promoter primers: 5′-CTTGGAGAAGGCAGCTCGT-3′ (forward) and 5′-TGCCCAAAGCAACGAAAACG-3′ (reverse).
Reverse transcription real-time quantitative PCR (rqPCR)
RNA was isolated from cells with Agilent Total RNA Isolation Kit (Agilent Technologies). cDNA synthesis was performed using 0.5 mg RNA with iScript cDNA Synthesis Kit (Bio-Rad), and reverse transcription rqPCR was performed with the SYBR green method using iQ Real-Time Sybr Green PCR Supermix Kit (Bio-Rad). Fluorescence was acquired at each cycle on a CFX96 system (Bio-Rad) using the following cycling conditions: 95°C/5 min; 95°C/45 s, 57°C/45 s, 72°C/90 s for 30 cycles; and 72°C/10 min. The quantitation cycle values were analyzed using the CFX96 system and normalized to GAPDH levels. Primers used for the human collagen IV α2 chain and GAPDH were α2 forward, 5′-ATTCCTTCCTCATGCACACGG-3′; α2 reverse, 5′-AGAAGCTGTACTTGTTGGCGT-3′; GADPH forward, 5′-TGGAGAAACCTGCCAAGTATGA-3′; and GADPH reverse, 5′-GAAGAGTGGGAGTTGCTGTTGA-3′.
In vivo treatment with cell-penetrating FUS NLS peptides
All in vivo experiments were performed according to institutional animal care guidelines and conducted in Association for Assessment and Accreditation of Laboratory Animal Care–accredited facilities. 6–8-wk-old male BALB/c WT and ItgαKO mice received biotinylated cell-penetrating FUS-NLS (35.67 μg/g) or biotinylated cell-penetrating inactive mutated FUS-NLS (33.82 μg/g) peptide (∼10−8 mol/g for both peptides) dissolved in PBS via intraperitoneal (i.p.) injections. Mice received two i.p. injections per day for 3 d. On the fourth day, mice received one i.p. injection and were sacrificed 2 h later. Kidneys were immediately harvested and used for fractionation and Western blot analysis.
Statistical analysis
Data are shown as mean ± SD or SEM. Unpaired two-tailed t test was used to evaluate statistically significant differences (P < 0.05) between two groups. GraphPad Prism software, one-way ANOVA followed by two-tailed t test when appropriate, was used to evaluate statistically significant differences (P < 0.05) among multiple groups. Data distribution was assumed to be normal, but this was not formally tested.
Online supplemental material
Fig. S1 shows that the collagen IV gene promoter contains FUS-responsive elements. Fig. S2 shows that murine FUS contains tyrosine phosphorylation sites for EGFR kinase. Fig. S3 shows that human FUS contains tyrosine phosphorylation sites for EGFR kinase. Fig. S4 shows increased baseline and EGF-induced EGFR phosphorylation in Itgα1KO mesangial cells. Table S1 lists genes containing FUS-responsive elements. Table S2 lists primary antibodies used.
Supplementary Material
is a partial list of human or murine profibrotic and inflammatory gene promoters analyzed for the presence of FUS-responsive elements (TxxxxGT).
lists the primary antibodies used for Western blot and/or immunohistochemistry.
Acknowledgments
A.Pozzi is the recipient of a Department of Veterans Affairs Senior Research Career Scientist Award. This work was supported in part by Department of Veterans Affairs Merit Reviews 1I01BX002025 (A. Pozzi), 1I01BX002196 (R. Zent), 101BX000320 (R.C. Harris), and 101BX002750 (J. Hawiger); and National Institutes of Health grants R01-DK119212 (A. Pozzi), R01-DK069921 (R. Zent), R01-DK099467 (R.M. Vanacore), R01-DK56942 (A.B. Fogo), R0-1-DK051265 (R.C. Harris and M-Z. Zhang), and P30-DK114809 (A. Pozzi, R.C. Harris, M-Z. Zhang, R. Zent, A.B. Fogo, and M.H. Wilson).
The authors declare no competing financial interests.
Author contributions: M. Chiusa: formal analysis, investigation, project administration, review, and editing; W. Hu: investigation; X. Chen: investigation, review, and editing; J.A. Bentz: investigation; M-Z. Zhang: resources, review, and editing; R.C. Harris: resources, review, and editing; R.M. Vanacore: investigation, review, and editing; G. Remuzzi: resources, review, and editing; A. Benigni: resources, review, and editing; S. Mili: resources, review, and editing; A.B. Fogo: resources, review, and editing; J. Zienkiewicz: resources, review, and editing; W. Luo: investigation, review, and editing; M.H. Wilson: methodology, review, and editing; R. Zent: resources, review, and editing; J. Hawiger: resources, review, and editing; A. Pozzi: funding acquisition, conceptualization, investigation, visualization, original draft, review, and editing.
References
- Ajmone-Cat M.A., Onori A., Toselli C., Stronati E., Morlando M., Bozzoni I., Monni E., Kokaia Z., Lupo G., Minghetti L., et al. . 2019. Increased FUS levels in astrocytes leads to astrocyte and microglia activation and neuronal death. Sci. Rep. 9:4572 10.1038/s41598-019-41040-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollée G., Flamant M., Schordan S., Fligny C., Rumpel E., Milon M., Schordan E., Sabaa N., Vandermeersch S., Galaup A., et al. . 2011. Epidermal growth factor receptor promotes glomerular injury and renal failure in rapidly progressive crescentic glomerulonephritis. Nat. Med. 17:1242–1250. 10.1038/nm.2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco D.A., Lemay N., Ko H.K., Zhou H., Burke C., Kwiatkowski T.J. Jr., Sapp P., McKenna-Yasek D., Brown R.H. Jr., and Hayward L.J.. 2010. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet. 19:4160–4175. 10.1093/hmg/ddq335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronisz A., Carey H.A., Godlewski J., Sif S., Ostrowski M.C., and Sharma S.M.. 2014. The multifunctional protein fused in sarcoma (FUS) is a coactivator of microphthalmia-associated transcription factor (MITF). J. Biol. Chem. 289:326–334. 10.1074/jbc.M113.493874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Abair T.D., Ibanez M.R., Su Y., Frey M.R., Dise R.S., Polk D.B., Singh A.B., Harris R.C., Zent R., et al. . 2007. Integrin alpha1beta1 controls reactive oxygen species synthesis by negatively regulating epidermal growth factor receptor-mediated Rac activation. Mol. Cell. Biol. 27:3313–3326. 10.1128/MCB.01476-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Moeckel G., Morrow J.D., Cosgrove D., Harris R.C., Fogo A.B., Zent R., and Pozzi A.. 2004. Lack of integrin alpha1beta1 leads to severe glomerulosclerosis after glomerular injury. Am. J. Pathol. 165:617–630. 10.1016/S0002-9440(10)63326-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Whiting C., Borza C., Hu W., Mont S., Bulus N., Zhang M.Z., Harris R.C., Zent R., and Pozzi A.. 2010. Integrin alpha1beta1 regulates epidermal growth factor receptor activation by controlling peroxisome proliferator-activated receptor gamma-dependent caveolin-1 expression. Mol. Cell. Biol. 30:3048–3058. 10.1128/MCB.00892-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiusa M., Hu W., Liao H.J., Su Y., Borza C.M., de Caestecker M.P., Skrypnyk N.I., Fogo A.B., Pedchenko V., Li X., et al. . 2019. The Extracellular Matrix Receptor Discoidin Domain Receptor 1 Regulates Collagen Transcription by Translocating to the Nucleus. J. Am. Soc. Nephrol. 30:1605–1624. 10.1681/ASN.2018111160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho N.M., and McCulloch C.A.. 2016. Contribution of collagen adhesion receptors to tissue fibrosis. Cell Tissue Res. 365:521–538. 10.1007/s00441-016-2440-8 [DOI] [PubMed] [Google Scholar]
- Darovic S., Prpar Mihevc S., Župunski V., Gunčar G., Štalekar M., Lee Y.B., Shaw C.E., and Rogelj B.. 2015. Phosphorylation of C-terminal tyrosine residue 526 in FUS impairs its nuclear import. J. Cell Sci. 128:4151–4159. 10.1242/jcs.176602 [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M., Kocerha J., Finch N., Crook R., Baker M., Desaro P., Johnston A., Rutherford N., Wojtas A., Kennelly K., et al. . 2010. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum. Mutat. 31:E1377–E1389. 10.1002/humu.21241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q., Holler C.J., Taylor G., Hudson K.F., Watkins W., Gearing M., Ito D., Murray M.E., Dickson D.W., Seyfried N.T., et al. . 2014. FUS is phosphorylated by DNA-PK and accumulates in the cytoplasm after DNA damage. J. Neurosci. 34:7802–7813. 10.1523/JNEUROSCI.0172-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar S.K., Zhang J., Gal J., Xu Y., Miao L., Lynn B.C., Zhu H., Kasarskis E.J., and St Clair D.K.. 2014. FUsed in sarcoma is a novel regulator of manganese superoxide dismutase gene transcription. Antioxid. Redox Signal. 20:1550–1566. 10.1089/ars.2012.4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D., Rodde R., Edbauer D., Bentmann E., Fischer I., Hruscha A., Than M.E., Mackenzie I.R., Capell A., Schmid B., et al. . 2010. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 29:2841–2857. 10.1038/emboj.2010.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederle H., and Dormann D.. 2017. TDP-43 and FUS en route from the nucleus to the cytoplasm. FEBS Lett. 591:1489–1507. 10.1002/1873-3468.12646 [DOI] [PubMed] [Google Scholar]
- Garikipati V.N.S., Verma S.K., Cheng Z., Liang D., Truongcao M.M., Cimini M., Yue Y., Huang G., Wang C., Benedict C., et al. . 2019. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 10:4317 10.1038/s41467-019-11777-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A.K., Quaggin S.E., and Vaughan D.E.. 2013. Molecular basis of organ fibrosis: potential therapeutic approaches. Exp. Biol. Med. (Maywood). 238:461–481. 10.1177/1535370213489441 [DOI] [PubMed] [Google Scholar]
- Gu J., Lu Y., Li F., Qiao L., Wang Q., Li N., Borgia J.A., Deng Y., Lei G., and Zheng Q.. 2014. Identification and characterization of the novel Col10a1 regulatory mechanism during chondrocyte hypertrophic differentiation. Cell Death Dis. 5 e1469 10.1038/cddis.2014.444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawiger J., and Zienkiewicz J.. 2019. Decoding inflammation, its causes, genomic responses, and emerging countermeasures. Scand. J. Immunol. 90 e12812 10.1111/sji.12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Xu Y., Ge X., Xu B., Peng W., Jiang X., Shen L., and Xia L.. 2019. Long noncoding RNA NEAT1 accelerates the proliferation and fibrosis in diabetic nephropathy through activating Akt/mTOR signaling pathway. J. Cell. Physiol. 234:11200–11207. 10.1002/jcp.27770 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: bidirectional, allosteric signaling machines. Cell. 110:673–687. 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- Kent L., Vizard T.N., Smith B.N., Topp S.D., Vance C., Gkazi A., Miller J., Shaw C.E., and Talbot K.. 2014. Autosomal dominant inheritance of rapidly progressive amyotrophic lateral sclerosis due to a truncation mutation in the fused in sarcoma (FUS) gene. Amyotroph. Lateral Scler. Frontotemporal Degener. 15:557–562. 10.3109/21678421.2014.920033 [DOI] [PubMed] [Google Scholar]
- Kwiatkowski T.J. Jr., Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. . 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 323:1205–1208. 10.1126/science.1166066 [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C., Polymenidou M., Hutt K.R., Vu A.Q., Baughn M., Huelga S.C., Clutario K.M., Ling S.C., Liang T.Y., Mazur C., et al. . 2012. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 15:1488–1497. 10.1038/nn.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.J., Cansizoglu A.E., Süel K.E., Louis T.H., Zhang Z., and Chook Y.M.. 2006. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 126:543–558. 10.1016/j.cell.2006.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.Z., Yao S.Y., Veach R.A., Torgerson T.R., and Hawiger J.. 1995. Inhibition of nuclear translocation of transcription factor NF-kappa B by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J. Biol. Chem. 270:14255–14258. 10.1074/jbc.270.24.14255 [DOI] [PubMed] [Google Scholar]
- Liu N., He S., Tolbert E., Gong R., Bayliss G., and Zhuang S.. 2012. Suramin alleviates glomerular injury and inflammation in the remnant kidney. PLoS One. 7 e36194 10.1371/journal.pone.0036194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H.W., Hsu S.C., Ali-Seyed M., Gunduz M., Xia W., Wei Y., Bartholomeusz G., Shih J.Y., and Hung M.C.. 2005. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 7:575–589. 10.1016/j.ccr.2005.05.007 [DOI] [PubMed] [Google Scholar]
- Lourenco G.F., Janitz M., Huang Y., and Halliday G.M.. 2015. Long noncoding RNAs in TDP-43 and FUS/TLS-related frontotemporal lobar degeneration (FTLD). Neurobiol. Dis. 82:445–454. 10.1016/j.nbd.2015.07.011 [DOI] [PubMed] [Google Scholar]
- Luo F., Gui X., Zhou H., Gu J., Li Y., Liu X., Zhao M., Li D., Li X., and Liu C.. 2018. Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat. Struct. Mol. Biol. 25:341–346. 10.1038/s41594-018-0050-8 [DOI] [PubMed] [Google Scholar]
- Mattila E., Pellinen T., Nevo J., Vuoriluoto K., Arjonen A., and Ivaska J.. 2005. Negative regulation of EGFR signalling through integrin-alpha1beta1-mediated activation of protein tyrosine phosphatase TCPTP. Nat. Cell Biol. 7:78–85. 10.1038/ncb1209 [DOI] [PubMed] [Google Scholar]
- Monahan Z., Ryan V.H., Janke A.M., Burke K.A., Rhoads S.N., Zerze G.H., O’Meally R., Dignon G.L., Conicella A.E., Zheng W., et al. . 2017. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36:2951–2967. 10.15252/embj.201696394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya T., and Maragkakis M.. 2018. Amyotrophic Lateral Sclerosis associated FUS mutation shortens mitochondria and induces neurotoxicity. Sci. Rep. 8:15575 10.1038/s41598-018-33964-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Imai T., Takahashi K., Jinnai K., Yamano T., Nagao K., Shimizu N., and Yamauchi M.. 1998. Decreased type IV collagen of skin and serum in patients with amyotrophic lateral sclerosis. Neurology. 51:114–120. 10.1212/WNL.51.1.114 [DOI] [PubMed] [Google Scholar]
- Ono S., Imai T., Matsubara S., Takahashi K., Jinnai K., Yamano T., and Shimizu N.. 1999. Decreased urinary concentrations of type IV collagen in amyotrophic lateral sclerosis. Acta Neurol. Scand. 100:111–116. 10.1111/j.1600-0404.1999.tb01048.x [DOI] [PubMed] [Google Scholar]
- Ono S., Imai T., Tsumura M., Takahashi K., Jinnai K., Suzuki M., Tagawa A., and Shimizu N.. 2000. Increased serum hyaluronic acid in amyotrophic lateral sclerosis: relation to its skin content. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 1:213–218. 10.1080/14660820050515214 [DOI] [PubMed] [Google Scholar]
- Pan L., Zhao Y., Yuan Z., and Qin G.. 2016. Research advances on structure and biological functions of integrins. Springerplus. 5:1094 10.1186/s40064-016-2502-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penke L.R., Speth J.M., Dommeti V.L., White E.S., Bergin I.L., and Peters-Golden M.. 2018. FOXM1 is a critical driver of lung fibroblast activation and fibrogenesis. J. Clin. Invest. 128:2389–2405. 10.1172/JCI87631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesnelle K.M., Boehm A.L., and Grandis J.R.. 2007. STAT-mediated EGFR signaling in cancer. J. Cell. Biochem. 102:311–319. 10.1002/jcb.21475 [DOI] [PubMed] [Google Scholar]
- Rayego-Mateos S., Rodrigues-Diez R., Morgado-Pascual J.L., Valentijn F., Valdivielso J.M., Goldschmeding R., and Ruiz-Ortega M.. 2018. Role of Epidermal Growth Factor Receptor (EGFR) and Its Ligands in Kidney Inflammation and Damage. Mediators Inflamm. 2018 8739473 10.1155/2018/8739473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T.H., Shiau C.W., Jao P., Liu C.H., Liu C.J., Tai W.T., Jeng Y.M., Yang H.C., Tseng T.C., Huang H.P., et al. . 2015. Sorafenib and its derivative SC-1 exhibit antifibrotic effects through signal transducer and activator of transcription 3 inhibition. Proc. Natl. Acad. Sci. USA. 112:7243–7248. 10.1073/pnas.1507499112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A.Y., Riley T.R., Coady T., Bussemaker H.J., and Manley J.L.. 2012. TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements. Proc. Natl. Acad. Sci. USA. 109:6030–6035. 10.1073/pnas.1203028109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukie T., Masaki H., Yoshida S., Fujikura M., and Ono S.. 2014. Decreased Amount of Collagen in The Skin of Amyotrophic Lateral Sclerosis in The Kii Peninsula of Japan. Acta Neurol. Taiwan. 23:82–89. [PubMed] [Google Scholar]
- Vance C., Rogelj B., Hortobágyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. . 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 323:1208–1211. 10.1126/science.1165942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veach R.A., Liu D., Yao S., Chen Y., Liu X.Y., Downs S., and Hawiger J.. 2004. Receptor/transporter-independent targeting of functional peptides across the plasma membrane. J. Biol. Chem. 279:11425–11431. 10.1074/jbc.M311089200 [DOI] [PubMed] [Google Scholar]
- Wang H., Chen X., Su Y., Paueksakon P., Hu W., Zhang M.Z., Harris R.C., Blackwell T.S., Zent R., and Pozzi A.. 2015. p47(phox) contributes to albuminuria and kidney fibrosis in mice. Kidney Int. 87:948–962. 10.1038/ki.2014.386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., and Shu H.K.. 2013. Transcription factor interactions mediate EGF-dependent COX-2 expression. Mol. Cancer Res. 11:875–886. 10.1158/1541-7786.MCR-12-0706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F., Kloepfer L.A., Finney C., Diedrich A., and Harris R.C.. 2016. Specific endothelial heparin-binding EGF-like growth factor deletion ameliorates renal injury induced by chronic angiotensin II infusion. Am. J. Physiol. Renal Physiol. 311:F695–F707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.Z., Sasaki K., Li Y., Li Z., Pan Y., Jin G.N., Wang Y., Niu A., Wang S., Fan X., et al. . 2019. The Role of the EGF Receptor in Sex Differences in Kidney Injury. J. Am. Soc. Nephrol. 30:1659–1673. 10.1681/ASN.2018121244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is a partial list of human or murine profibrotic and inflammatory gene promoters analyzed for the presence of FUS-responsive elements (TxxxxGT).
lists the primary antibodies used for Western blot and/or immunohistochemistry.