Sicari et al. discuss the early secretory pathway in the SARS-CoV-2 infection cycle and provide a perspective on potential therapeutic implications.
Abstract
Similar to other RNA viruses, SARS-CoV-2 must (1) enter a target/host cell, (2) reprogram it to ensure its replication, (3) exit the host cell, and (4) repeat this cycle for exponential growth. During the exit step, the virus hijacks the sophisticated machineries that host cells employ to correctly fold, assemble, and transport proteins along the exocytic pathway. Therefore, secretory pathway–mediated assemblage and excretion of infective particles represent appealing targets to reduce the efficacy of virus biogenesis, if not to block it completely. Here, we analyze and discuss the contribution of the molecular machines operating in the early secretory pathway in the biogenesis of SARS-CoV-2 and their relevance for potential antiviral targeting. The fact that these molecular machines are conserved throughout evolution, together with the redundancy and tissue specificity of their components, provides opportunities in the search for unique proteins essential for SARS-CoV-2 biology that could also be targeted with therapeutic objectives. Finally, we provide an overview of recent evidence implicating proteins of the early secretory pathway as potential antiviral targets with effective therapeutic applications.
Introduction
As obligatory parasites, viruses must find a suitable host cell, deliver into it their genome (either DNA or RNA), and hijack the endogenous molecular machineries for their own replication (Su et al., 2016). Thus, some scientists exploit viruses as Trojan horses to deliver genes into target cells, whereas others search for the Achilles heels of those causing diseases. To find a suitable host, viruses bind to receptors expressed on the cell surface. Selective binding activates membrane fusion, allowing the viral genome to enter the target cell. Fusion can occur directly at the plasma membrane (e.g., human immunodeficiency virus) or within endocytic compartments (e.g., influenza). In the cytoplasm, the viral genome is replicated while enough structural and nonstructural proteins are produced. All the essential components must then be assembled and surrounded with an envelope composed of viral proteins in combination with host cell–derived membranes. Viruses exploit the ability of human cells to produce, assemble, and transport to the cell surface receptors, transporters, and other complex proteins (Su et al., 2016). For severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and other coronaviruses (CoVs), these steps take place in organelles of the early secretory pathway, the ER, the ER–Golgi intermediate compartment (ERGIC), and the Golgi apparatus. SARS-CoV-2, a member of the SARS-CoV family, is responsible for a global pandemic that started in 2019 (Lake, 2020; Myint, 1994; Andersen et al., 2020). Because SARS-CoV-2 shares 79% of the genome with SARS-CoV, one can use knowledge obtained on the viruses that caused prior pandemics, such as SARS and Middle East Respiratory Syndrome (Lake, 2020; Lim et al., 2016). CoV-2 infection starts when its spike (S) protein binds to angiotensin I–converting enzyme 2 (ACE2) receptors on the host cell membrane (Lake, 2020; Letko et al., 2020). CoV-2 does so with higher affinity than SARS-CoV (Wan et al., 2020; Wu et al., 2020). In the cytosol, the virus is uncoated, and its RNA genome is used to produce replicases and nonstructural proteins (nsps). Replicases generate subgenomic RNA capable of producing the structural proteins spike (S), nucleocapside (N), membrane (M), and envelope (E) proteins. The group of proteins helping the assembly and transport of infective viral particles are referred to as accessory proteins (ORFs). A large number of these proteins are cotranslationally translocated into the ER to be glycosylated, folded, and assembled in preparation for virus budding and release via the secretory pathway. Thus, virion spread critically depends on recruiting the most efficient secretory machineries of host cells (Su et al., 2016; Fig. 1).
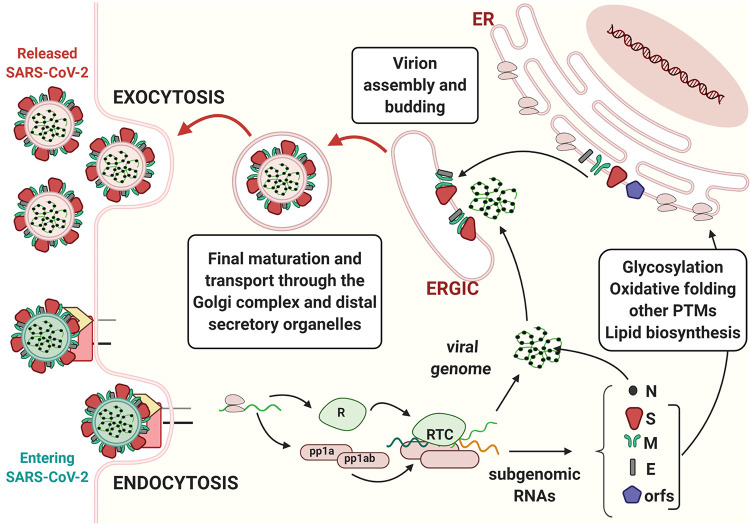
Figure 1.
The journey of SARS-CoV-2 in the host cell. Coronavirus binds to cognate receptors on target cells via the spike proteins (dark red). This drives conformational changes that promote fusion of the virus with the host cell’s plasma membrane (entry by endocytosis and green membrane–containing virion, bottom left). In the cytoplasm, viral capsids are uncoated, and the viral RNA genome is translated, producing two poly-proteins (pp1a and pp1ab). These polypeptides are then proteolytically processed by both host and viral proteases, thereby generating nonstructural proteins (nsps) and leading to the formation of the replicase–polymerase complex (RTC). The latter is responsible for the replication of the viral genome and for the production of subgenomic RNAs, which are translated into the structural proteins nucleocapsid (N), spike (S), membrane (M), and envelope (E). In addition to these genomic elements shared by other CoVs, the SARS-CoV-2 genome also contains eight open reading frames (ORFs) that drive the production of accessory proteins. S, M, and E structural proteins and some accessory proteins are co-translationally translocated into the ER, where they undergo diverse post-translational modifications, including disulfide bond formation and N-linked glycosylation. Structural proteins concentrate in the ERGIC, where they assemble around the newly formed genome–nucleocapsid complexes. Mature virions are further modified (e.g., O-glycosylated) as they proceed through the Golgi complex and later stations of the secretory pathway before being released in the extracellular milieu (release by exocytosis and pink membrane–containing virions, top left). The membrane of the virus derives from the host cell, which synthetizes it in the ER.
Proteins of the early secretory pathway bound by SARS-CoV-2
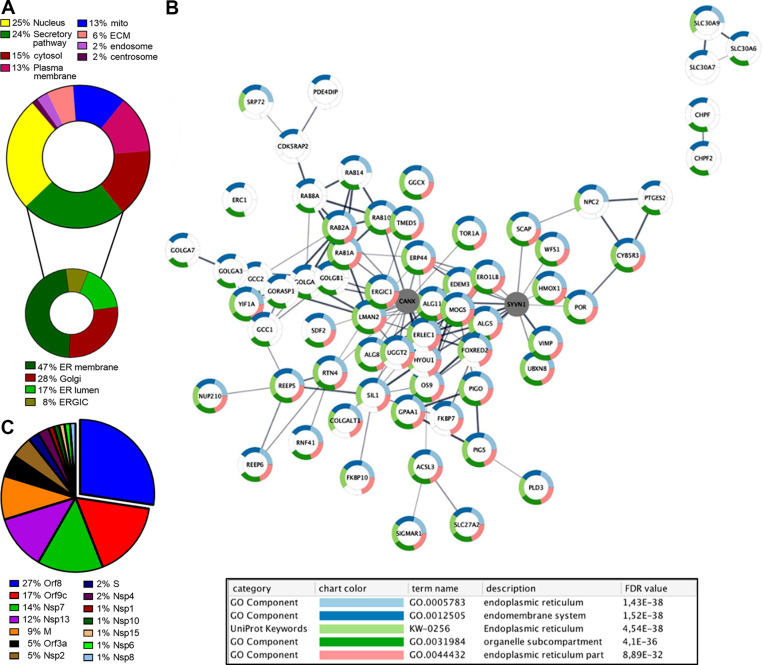
As the entire world asks for ways to stop CoV-2, many laboratories are investigating the virus’s Achilles heel(s). Because the virus is an obligatory parasite, a powerful strategy to identify endogenous molecules essential for its replication starts with the expression of suitably tagged CoV-2 proteins in human cells. The host interactors are identified by biochemical methods and grouped on the basis of their known functions. Experiments along these lines have been performed in a cooperative effort led by the Krogan laboratory in a study aiming to reposition approved drugs against the virus (Gordon et al., 2020). Taking advantage of affinity purification mass spectrometry assays, the authors, using HEK293T cells as a model, highlighted 66 putative targetable CoV-2–host protein–protein interactions among the 332 identified (Gordon et al., 2020). Many (∼24%) involved proteins from the host secretory pathway (Fig. 2, A and B). These observations are in line with those of previous analyses of other viruses but also revealed a number of proteins specific for CoV-2, the most representative being orf8 (43%), orf9c (12%), nsp7 (11%), nsp13 (10%), and M (7%; Fig. 2 C and Table 1). Hereafter, we comment on the interactions involving elements of the secretory pathway and how these could be targeted to combat CoV-2. This approach has great potential but also some limitations. Perhaps the most relevant caveat is that each viral protein was expressed independently of the other SARS-CoV-2 proteins in human cells. On the one hand, this is key to identifying specific interactors in sufficient quantities. On the other hand, however, it might yield artifacts due to the absence of other physiological partner(s) or accessories, nonstructural proteins encoded in the viral genome or produced by the host cell upon viral infection. Overexpression per se might elicit ER stress responses that do not necessarily reflect what happens in an infected cell. In addition, considering that SARS-CoV-2 can infect different human cell types, the HEK293-based interactome might miss tissue-specific factors and introduce biases. For instance, similar experiments performed in A549 cells produced slightly different results in terms of interactors (Stukalov et al., 2020), because protein–protein interaction assays depend on the gene expression patterns specific for each cell line (Table 2). Nonetheless, the interactome data obtained in HEK293 and A549 cells clearly identify commonly enriched pathways, including ER-to-cytosol trafficking, protein quality control, inflammatory response, and DNA damage (Stukalov et al., 2020).
Figure 2.
Secretory pathway in CoV-2 infection. (A) The SARS-CoV-2 interactome was subdivided on the basis of host cell compartments (upper panel) and further sorted according to subcompartments of the secretory pathway (lower panel). (B) Secretory pathway cluster of the SARS-CoV-2 interactome. (C) Percentage of SARS-COV-2–derived proteins that interact with secretory pathway components.
Table 1. Proteins of the early secretory pathway found to interact with SARS-CoV-2.
| orf8 | orf9c | nsp7 | nsp13 | M | orf3a | nsp2 | S | nsp4 | nsp1 | nsp10 | nsp15 | nsp6 | nsp8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHPF | ALG8 | ACSL3 | CDK5RAP2 | COQ8B | ALG5 | GIGYF2 | GOLGA7 | ALG11 | COLGALT1 | ERGIC1 | RNF41 | SIGMAR1 | SRP72 |
| CHPF2 | ERMP1 | CYB5R3 | ERC1 | PMPCA | CLCC1 | POR | ZDHHC5 | NUP210 | |||||
| EDEM3 | NDFIP2 | HS2ST1 | GCC1 | REEP5 | HMOX1 | RAP1GDS1 | |||||||
| ERLEC1 | PIGO | LMAN2 | GCC2 | REEP6 | TRIM59 | SLC27A2 | |||||||
| ERO1B | PIGS | MOGS | GGCX | RTN4 | |||||||||
| ERp44 | RETREG3 | PTGES2 | GOLGA2 | SLC30A7 | |||||||||
| FKBP10 | SCAP | RAB10 | GOLGA3 | SLC30A9 | |||||||||
| FKBP7 | SLC30A6 | RAB14 | GOLGB1 | YIF1A | |||||||||
| FOXRED2 | TAPT1 | RAB1A | GORASP1 | ||||||||||
| HYOU1 | TMED5 | RAB2A | PDE4DIP | ||||||||||
| NPC2 | TMEM97/SIGMAR2 | RAB8A | |||||||||||
| OS9 | UBXN8 | SELENOS | |||||||||||
| PCSK6 | WFS1 | ||||||||||||
| PLD3 | GPAA1 | ||||||||||||
| PLEKHF2 | |||||||||||||
| POFUT1 | |||||||||||||
| POGLUT2 | |||||||||||||
| POGLUT3 | |||||||||||||
| SDF2 | |||||||||||||
| SIL1 | |||||||||||||
| TM2D3 | |||||||||||||
| TOR1A | |||||||||||||
| UGGT2 |
List of host proteins (columns) related to the secretory pathway that interact with different viral proteins (first line). Proteins derived from the input signature (Gordon et al., 2020) have been divided based on the cellular compartment localization (Fig. 2 A). In Fig. 2 B, candidates related to ER and Golgi compartments have been sorted based on the interacting viral protein.
Table 2. Conserved features in SARS-CoV-2 and SARS-CoV interactomes.
| HEK293 (Gordon et al., 2020) | A549 (Stukalov et al., 2020) | ||
|---|---|---|---|
| Secretory pathway components | SARS-CoV-2 | SARS-CoV | SARS-CoV-2 |
| ACSL3 | nsp7 | - | - |
| ALG11 | nsp4 | orf7b | orf7b |
| ALG5 | orf3a | orf7b | orf7b |
| ALG8 | orf9c | orf7b | orf7b |
| CDK5RAP2 | nsp13 | - | - |
| CHPF | orf8 | - | - |
| CHPF2 | orf8 | - | - |
| CLCC1 | orf3a | orf3a | orf3 |
| COLGALT1 | nsp1 | - | - |
| COQ8B | M | M | M |
| CYB5R3 | nsp7 | - | - |
| EDEM3 | orf8 | - | - |
| ERC1 | nsp13 | - | - |
| ERGIC1 | nsp10 | - | - |
| ERLEC1 | orf8 | orf8 | orf8 |
| ERMP1 | orf9c | orf7b | orf7b |
| ERO1B | orf8 | - | - |
| ERp44 | orf8 | - | - |
| FKBP10 | orf8 | - | - |
| FKBP7 | orf8 | - | - |
| FOXRED2 | orf8 | - | - |
| GCC1 | nsp13 | - | - |
| GCC2 | nsp13 | - | - |
| GGCX | nsp13 | - | - |
| GIGYF2 | nsp2 | - | - |
| GOLGA2 | nsp13 | - | - |
| GOLGA3 | nsp13 | - | - |
| GOLGA7 | S | - | S |
| GOLGB1 | nsp13 | orf7b | orf7b |
| GORASP1 | nsp13 | - | - |
| GPAA1 | orf9c | nsp6 | nsp6 |
| HMOX1 | orf3a | - | - |
| HS2ST1 | nsp7 | - | - |
| HYOU1 | orf8 | - | - |
| LMAN2 | nsp7 | orf7b | orf7b |
| MOGS | nsp7 | - | - |
| NDFIP2 | orf9c | orf3a | orf3 |
| NPC2 | orf8 | - | - |
| NUP210 | nsp4 | - | - |
| OS9 | orf8 | orf8 | orf8 |
| PCSK6 | orf8 | - | - |
| PDE4DIP | nsp13 | N | N |
| PIGO | orf9c | orf7b | orf7b |
| PIGS | orf9c | nsp6 | nsp6 |
| PLD3 | orf8 | orf7b | orf7b |
| PLEKHF2 | orf8 | - | - |
| PMPCA | M | orf8b | - |
| POFUT1 | orf8 | - | - |
| POGLUT2 | orf8 | - | - |
| POGLUT3 | orf8 | - | - |
| POR | nsp2 | - | - |
| PTGES2 | nsp7 | - | - |
| RAB10 | nsp7 | - | - |
| RAB14 | nsp7 | orfa | orf3 |
| RAB1A | nsp7 | orf3a | orf3 |
| RAB2A | nsp7 | orf3a | orf3 |
| RAB8A | nsp7 | orf3a | orf3 |
| RAP1GDS1 | nsp2 | nsp2 | nsp2 |
| REEP5 | M | orf8 | orf7b |
| REEP6 | M | - | - |
| RETREG3 | orf9c | orf7b | orf7b |
| RNF41 | nsp15 | - | - |
| RTN4 | M | - | M |
| SCAP | orf9c | orf7b | orf7b |
| SDF2 | orf8 | orf8 | - |
| SELENOS | nsp7 | orf7b | orf7b |
| SIGMAR1 | nsp6 | - | - |
| SIL1 | orf8 | - | - |
| SLC27A2 | nsp2 | - | - |
| SLC30A6 | orf9c | - | - |
| SLC30A7 | M | - | - |
| SLC30A9 | M | - | - |
| SRP72 | nsp8 | - | - |
| TAPT1 | orf9c | orf7b | orf7b |
| TM2D3 | orf8 | - | - |
| TMED5 | orf9c | - | - |
| TMEM97/SIGMAR2 | orf9c | orf3a | orf3 |
| TOR1A | orf8 | - | orf3 |
| TRIM59 | orf3a | orf7b | orf7b |
| UBXN8 | orf9c | - | - |
| UGGT2 | orf8 | orf8 | orf8 |
| WFS1 | orf9c | nsp6 | nsp6 |
| YIF1A | M | - | - |
| ZDHHC5 | S | S | S |
Proteins listed in Table 1 were matched with human protein–protein interactions obtained in A549 cells expressing SARS-CoV or SARS-CoV-2 proteins (Stukalov et al., 2020). The comparison highlights cell type– and virus-specific differences within an overall similarity of the interactome.
The role of glycosylation and protein quality control in SARS-CoV-2 infections
Most CoVs bud at the ERGIC level (Fig. 1) and are then transported along the exocytic pathway (Klumperman et al., 1994; Stertz et al., 2007). The domains facing the ER lumen undergo N-glycosylation, disulfide bond formation, and other posttranslational modifications necessary for viral tropism and host cell specificity. Addition of N-glycans occurs cotranslationally in the ER, and their processing continues in downstream compartments (Fung and Liu, 2018; Liang et al., 2019; Oostra et al., 2006). The glycosylation profiles of the CoV-2 S protein revealed 86% conservation of the N-sites with SARS-CoV strains (Watanabe et al., 2020). It has been proposed that conservation of the N-glycans in the receptor-binding domain is important for recognition by a neutralizing antibody (CR3022) raised against SARS-CoV. Indeed, the lower affinity of CR3022 for CoV-2 than for other SARS strains might reflect the absence of a specific glycan in the N370 residue (Yuan et al., 2020). Interestingly, two O-glycosylation sites are instead present only in CoV-2 S protein. These O-glycans are thought to hinder S protein epitopes from host immune system responses and to improve ACE2 receptor binding (Watanabe et al., 2020). CoV-2 protein orf8 might support O-glycosylation by binding POGLUT2, POGLUT3, two O-glucosyltransferases, and POFUT1, an O-fucosyltransferase (Table 1). These enzymes have been shown to be pivotal for the proper glycosylation and presentation on the plasma membrane of the NOTCH1 receptor (Takeuchi et al., 2018; Yang et al., 2019; Okamura and Saga, 2008), confirming once more the importance of glycan processing in the maturation and transport of CoV-2 S and other proteins.
Accordingly, many viral accessory proteins interact with components of the host cell’s secretory pathway. We still have a limited understanding of how the assembly of viral structural proteins is coordinated. In SARS-CoV, COPI binds the KxHxx motif found in the cytosolic domain of the S protein. This motif is necessary for S to accumulate in ERGIC and interact with M and is hence crucial for assembly (McBride et al., 2007). CoV-2 S protein presents the same motif. In both HEK293 and A549 cells, it interacts with two Golgi-localized proteins, ZDHHC5 and GOLGA7 (Gordon et al., 2020; Stukalov et al., 2020), which form a complex localized to the plasma membrane (Ko and Dixon, 2018; Ohno et al., 2006). ZDHHC5 is an acyltransferase that can localize to the ER, Golgi apparatus, and plasma membrane and controls Golgi trafficking and palmitoylation of anthrax toxin (Sergeeva and van der Goot, 2019). GOLGA7 is a Golgin subfamily member that assists ZDHHC5 activity on the plasma membrane (Ko et al., 2019). The ZDHHC–GOLGA7 complex may help S protein to reach the ERGIC compartment and favor assembly.
Interestingly, ERp44, SLC30A6, and SLC30A7 were found to interact with orf8, orf9c, and M proteins, respectively. ERp44 cycles between the ER and the Golgi to retrieve select clients back to the ER (Anelli et al., 2002; Otsu et al., 2006). This process is regulated by the pH and zinc gradients between the two compartments (Vavassori et al., 2013; Sannino et al., 2014; Watanabe et al., 2017). Two Golgi-resident transporters (SLC30A6/ZnT6 and SLC30A7/ZnT7) provide sufficient zinc to activate ERp44 (Watanabe et al., 2019). Moreover, ERp44 binds the lectin ERGIC53/LMAN1, which promotes the production of infective CoVs (Klaus et al., 2013). LMAN2 (VIP36) and ERGIC1, two ERGIC53-like lectins, also interact with orf7 and nsp10, respectively (Table 1). VIP36 has been shown to control transport of high-mannose proteins (Gupta, 2012; Mages et al., 2008). Thus, it could be that CoV-2 takes advantage of ERp44- and lectin-dependent ER-to-Golgi transport processes to accumulate in the ERGIC for budding.
The formation of proper disulfide bonds is key for the folding, assembly, and recognition of ACE2 receptors by Spike (Oostra et al., 2007). CoV-2 orf8 protein binds to the ER-resident oxidase Ero1β (Table 1). orf8-mediated Ero1β interaction may be important for disulfide bond formation in protein S or for the formation of orf8 multimers. Considering that Ero1β is a powerful oxidase induced by the unfolded protein response (UPR), this interaction might also reflect a response to ER stress. CoV-2 orf8 protein was also found to bind FKBP10 and FKBP7 (Table 1), two ER-resident peptidyl-prolyl cis-trans isomerases, and UGGT2 (Table 1), a key element in the glycoprotein quality control machinery and part of a large multichaperone in the ER lumen. Other CoV-2 protein interactors are part of the sophisticated machineries that guarantee protein homeostasis within the exocytic compartment; that is, the balance between protein production, secretion, and degradation, a process generally referred to as “proteostasis” (Klaips et al., 2018). For instance, orf8 binds numerous proteins involved in ER-associated degradation (ERAD), a key pathway for limiting protein condensation and aggregation in the early secretory compartment (Sun and Brodsky, 2019). orf8 binds EDEM3 (ER degradation enhancing α-mannosidase like protein 3) and OS9 (Table 1), two ERAD-related proteins that are abundant in coatomer complex II–independent vesicles. These structures are referred to as “EDEMosomes” and are key elements in proteostasis adjustments because they normally control the levels of ERAD components (Araki and Nagata, 2011). Interestingly, during SARS-CoV infection, EDEMosomes are used for forming double-membrane vesicles (DMVs) that recruit the replication–transcription complexes (Reggiori et al., 2010). DMVs are also important to protecting the viral genome from sensors in the cytosol and from IFNs (Zinzula and Tramontano, 2013). Finally, EMC1, a subunit of the ER membrane protein complex, was found to interact with orf8 (Table 1). With EMC4 and EMC7, EMC1 is thought to engage the Rab7 GTPase in late endosomes (Bagchi et al., 2020). Bridging late endosomes and the ER, this interaction might enable efficient entry and/or excretion of viral particles. EMC proteins also associate with Sec61 (Chitwood and Hegde, 2019), a component of the translocon mediating the entry of nascent polypeptides into the ER, and have been presented by Gordon et al. (2020) as a SARS-CoV-2 modulator.
ER homeostasis
The above lines of evidence suggest that the biogenesis of effective viral proteins requires efficient assembly and transport lines in the host’s secretory protein factory. How can CoV-2 divert them to its advantage? In general, viral infection induces ER stress via the strain imposed by the massive flux of viral proteins on the cell’s folding and transport machineries (Zhang and Wang, 2012). ER stress induces activation of three ER resident receptors, inositol-requiring enzyme 1α (IRE1), protein kinase R–like ER kinase (PERK), and activating transcription factor 6 (ATF6), which in turn promote a transcriptional program named the “UPR” (Hetz et al., 2015). These receptors have all been implied in CoV infection. For instance, S protein from SARS-CoV presents a UPR-activating domain mapped in the central region of the S1 subunit (between 201 and 400 aa) that acts in an N-glycosylation–independent manner (Siu et al., 2014) and is highly conserved among CoVs. SARS-CoV orf3a activates ATF4 and CHOP promoter activities, suggesting the involvement of the PERK branch during the infection (Minakshi et al., 2009). PERK activation induces phosphorylation of the α-subunit of the eukaryotic initiation factor α (eIF2α), leading to translation suppression and activation of a downstream signaling pathway known as the “integrated stress response.” Under the integrated stress response, translation of a majority of cellular mRNAs is suppressed while mRNAs containing small upstream ORFs in their 5′UTR are translated, among them the proapoptotic transcription factor ATF4. During CoV infection, eIF2α phosphorylation, global host mRNA translation inhibition, and ATF4 translation were detected (Bechill et al., 2008). However, it remains unknown how virus mRNAs can still be translated normally when eIF2α is phosphorylated. An attractive hypothesis, based on recently published data, is that SARS-CoV-2 infection impacts protein phosphorylation profiles. For instance, LARP1, which interacts with the N viral protein, has been found to be less phosphorylated in CoV-2–infected cells. This modification increases the affinity of LARP1 for the 3′UTRs of mRNAs encoding ribosomal proteins, thus attenuating protein synthesis (Bouhaddou et al., 2020; Stukalov et al., 2020).
ER stress induces the cytosolic kinase and RNase activities of IRE1α. This leads to the nonconventional splicing of X-box binding protein 1 (XBP1) mRNA, RNA degradation via IRE1α-dependent RNA decay (RIDD), and JNK phosphorylation. XBP1 spliced (XBP1s) is a transcription factor that promotes transcription of genes encoding proteins involved in entry into the ER, folding, glycosylation, ERAD, lipid biogenesis, and vesicular trafficking (Huh et al., 2010). RIDD activity has been described as either adaptive or terminal (Maurel et al., 2014). It has been shown to degrade ER-associated mRNA to facilitate ER homeostasis (adaptive) or mRNA encoding prosurvival proteins, thereby contributing to ER stress–induced cell death (terminal). SARS-CoV infection failed to induce XBP1 splicing, RIDD, or JNK phosphorylation (DeDiego et al., 2014; Versteeg et al., 2007), suggesting that the IRE1 branch of the UPR remains inactive during SARS-CoV infection. However, CoV-2 nsp6 protein binds Sigma receptor 1. This ER-resident integral membrane protein, together with SR2 (TMEM97), which interacts with CoV-2 orf9c, modulates calcium fluxes (Table 1) and controls IRE1 activation (Mori et al., 2013; Rosen et al., 2019). Thus, CoV-2 may affect IRE1 activity through its interaction with Sigma receptor 1. Last, ER stress induces ATF6 cleavage and its ER-to-Golgi translocation and finally an ATF6 transcriptional program, whose main target genes consist of ER chaperones (i.e., GRP78/BiP or GRP94) and ERAD components (Shoulders et al., 2013). Thus, activation of the ATF6 branch enhances the ER protein–folding capacity and homeostatic balance (Adachi et al., 2008). Overexpression of the SARS-CoV S protein leads to the activation of both GRP78 and GRP94 promoters (Siu et al., 2014), suggesting ATF6 activation. Notably, the small amounts of BiP found on the cell surface have been proposed to help CoV entry and have been suggested as a target to impede SARS-CoV-2 infection (Ha et al., 2020). ATF6 cleavage and nuclear translocation of the released cytosolic fragment were observed following SARS-CoV orf8ab transfection. In addition, orf8ab has also been shown to localize to the ER and to interact with the luminal domain of ATF6 (Sung et al., 2009). These observations are consistent with the fact that many proteins that interact with CoV-2 orf8 are encoded by ATF6 target genes. Moreover, the cytosolic ATF6 fragment has been shown to be phosphorylated and activated by p38 MAPK (Luo and Lee, 2002), one of the most active kinases upon SARS-CoV-2 infection (Bouhaddou et al., 2020), supporting a role for ATF6 in CoV-2 infection. Collectively, these studies indicate the importance of evaluating the precise impact of CoV-2 proteins on the activation of ER stress receptors and induction of the UPR.
Interestingly, CoV-2 M and nsp7 proteins were found to bind molecules involved in the regulation of ER morphology. Among them, receptor expression–enhancing proteins are implicated in formation and restructuring of the ER network, as well as intracellular transport of receptors to the plasma membrane. YIF1A recycles between the ER and the Golgi with a major ERGIC localization, maintaining homeostasis of the early secretory pathway (Yoshida et al., 2008). Rab7a modulates ER morphology by controlling ER stress responses (Mateus et al., 2018). Thus, these CoV-2 interactions may be crucial to controlling ER morphology and homeostasis, limiting induction of ER stress and apoptosis. Other links between ER structure and CoV-2 infection are the orf9c–SCAP interaction and nsp13/GORASP1 or GOLGA2 binding (Table 1). In particular, SCAP regulates the ER-to-Golgi translocation and activation of sterol regulatory element–binding protein (SREBP). SREBP target genes are involved in lipid biosynthesis and metabolic homeostasis, two key processes during virus infection (Yuan et al., 2019). orf9c-mediated SCAP binding may be used to promote lipid synthesis via SREBP activation for generating the envelopes needed for CoV-2 replication. Thus, available SCAP-targeting drugs and pre-SREBP inhibitors that have shown some inhibitory effects on Middle East Respiratory Syndrome–CoV replication, together with treatment using the cholesterol metabolite 25-hydroxycholesterol, could be interesting to test (Yuan et al., 2019). nsp13 binds GORASP1 and GOLGA2 and other proteins regulating Golgi structure and vesicular transport (Table 1). Especially during ER stress, the GORASP1–GOLGA2 complex mediates the unconventional trafficking of select glycoproteins to the cell membrane. Together, these results suggest that CoV-2 may control ER–Golgi homeostasis maintenance by acting on diverse ER–Golgi-related structures and pathways.
Targeting ER homeostasis
Taken together, the above observations suggest that CoV-2 reshapes the entire secretory pathway to its own advantage. Therefore, repositioning available compounds to target the secretory pathway and UPR components could represent a strategy against CoV-2. Inhibitors for ER stress receptors are available and employed in numerous pathologies, including cancer and metabolic diseases. PERK/eIF2α inhibitors are GSK2606414, Salubrinal/Guanabenz, and integrated stress response inhibitor. The use of these drugs may lead to exaggerated protein load in the ER lumen and increased ER stress induction by preventing eIF2α phosphorylation and translation inhibition (Matsuoka and Komoike, 2015; Hetz et al., 2013). IRE1 RNase inhibition with kinase-inhibiting RNase attenuators (KIRAs; KIRA6 and KIRA8), toyocamycin, STF-083010, 4µ8C, MKC series, and B-I09 could be used to dampen XBP1 mRNA splicing and RIDD activity (Cubillos-Ruiz and Glimcher, 2015). Although no drugs directly targeting ATF6 are available, indirect inhibition was obtained using nelfinavir. This compound targets human immunodeficiency virus proteases and also inhibits the proteases responsible for ATF6 cleavage (Guan et al., 2015; Sicari et al., 2019). To slow down viral spread, it could be ideal to target the protein quality control machinery, which in turn would dampen viral assembly and maturation. For instance, compounds that target glycosylation enzymes, such as Kinefusine and iminosugar derivative 1-deoxynojirimycin, which, respectively, inhibit mannosidase and α-glucosidase, are used in the clinic to dampen the morphogenesis of enveloped viruses. In addition, these drugs alter ACE2 receptor glycosylation and transport to the cell surface, thus contributing to prevention of virus entry (Winchester, 2009). Exacerbating ER stress to induce apoptosis could be obtained by targeting ER enzymes or chaperones. Several protein disulfide isomerase inhibitors are available, including PACMA31, which specifically targets the cysteine residue present in the active site of protein disulfide isomerase proteins and blocking their capability to bind cargoes (Kaplan et al., 2015).
Finally, downstream stations of the secretory pathway could be targeted using Golgicide A (GA), an inhibitor of ER–Golgi transport vesicles. It was already shown that GA is able to inhibit protein secretion, Shiga toxin trafficking in the endosomal compartment, and replication of coxsackievirus (Alborzinia et al., 2018). In this perspective, GA could be used to block virus spreading and maturation, acting on Golgi proteins that interact with nsp13. Eeyarestatin I targets ERAD by binding p97, thus eliciting ER stress. Interestingly, this drug interferes with the trafficking of a Shiga-like toxin (Wang et al., 2010). Moreover, ER/Golgi/Sec pathway-related compounds have been proposed by Gordon et al. (2020). Among the 66 U.S. Food and Drug Administration–approved drugs, they suggested PS30613 (ER protein processing, Sec61 inhibitor); IHVR-19029 (ER protein processing); FK-506 (FKBP binder); zotatifin (EIF4E2/H, eiF4a inhibitor); rapamycin and rapalogs (LARP1, FKBP15, FKBP7/10, mammalian target of rapamycin inhibitor); diverse SIGMAR1/2 modulators, such as chloroquine, PB28, and haloperidol E-52862, PD-144418, and RS-PPCC; as well as diverse SIGMAR1/2 direct modulators, such as the antagonist PD-144418 and the agonist RS-PPCC (Gordon et al., 2020). However, owing to the huge numbers of substrates of some of the above targets, these protocols might lack the specificity needed to guarantee a reasonable therapeutic window.
Prediction of new anti–SARS-CoV-2 candidates using BioInfoMiner
To evaluate whether the select interaction of CoV-2 with components of the secretory pathway could represent an actionable strategy, we sought to identify therapeutic targets through signature-driven pharmacogenomic analysis using three distinct ontological vocabularies (MGI Mammalian Phenotype, Reactome, and Gene Ontology). To this end, the whole CoV-2 human host interactome (Gordon et al., 2020) was imported into BioInfoMiner (bioinfominer.com; Lhomond et al., 2018). This allowed mapping the viral interactome to a consensus semantic tree graph that provides a systemic functional overview. This was used to interrogate the L1000 Connectivity Map repository, statistically selecting nontrivial network perturbagens, namely compounds that, when administered in cellular models, elicit perturbation in gene subsets of the input signature (Table 3). This approach differs in the sense that it exploits the whole vector of the derived signature, which in our case integrates core regulatory proteins linking broadly perturbed modules of the host cellular physiology, in order to prioritize compounds that affect a critical subset of them, ordered according to the significance of the enrichment score, measuring their overlap with the proposed targeted gene sets of the L1000 repository.
Table 3. Potential perturbagens of the interactions between SARS-CoV-2 and proteins of the early secretory pathway.
| Rank | Cell line | Perturbagen | Dose (μm) | Duration (h) | Score | Perturbed genes |
|---|---|---|---|---|---|---|
| 1 | MCF7 | Epicatechin monogallate | 10.0 | 6 | 1,000 | GOLGB1, PDE4DIP, TOR1A |
| 2 | PC3 | s1154 | 10.0 | 6 | 0.923 | HMOX1, PDE4DIP |
| 3 | PC3 | BRD-A82197375 | 10.0 | 6 | 0.738 | TOR1A |
| 4 | A375 | ganciclovir | 10.0 | 6 | 0.600 | GOLGB1, HMOX1 |
| 5 | MCF7 | estrone | 10.0 | 6 | 0.554 | HMOX1, PDE4DIP |
| 6 | A549 | GDC-0879 | 3.33 | 24 | 0.431 | HMOX1 |
| 7 | VCAP | sertaconazole_nitrate | 10.0 | 24 | 0.323 | PDE4DIP |
| 8 | BT20 | radicicol | 1.11 | 3 | 0.292 | HMOX1, HYOU1 |
| 9 | VCAP | BRD-A16581344 | 10.0 | 24 | 0.231 | HYOU1, PDE4DIP |
| 10 | A549 | SB-216763 | 10 | 24 | 0.231 | PDE4DIP |
| 11 | VCAP | cetirizine_dihydrochloride | 10.0 | 6 | 0.231 | HYOU1, PDE4DIP |
| 12 | HCC515 | bcl-2_inhibitor | 10.0 | 24 | 0.169 | GOLGB1, HMOX1, TAPT1 |
| 13 | HA1E | betamethasone | 10.0 | 24 | 0.169 | GOLGB1, HMOX1, PDE4DIP |
| 14 | MCF7 | t0513-6584 | 10.0 | 6 | 0.154 | TOR1A |
| 15 | MCF7 | e6_berbamine | 10.0 | 6 | 0.154 | PDE4DIP |
| 16 | A549 | dabrafenib | 0.12 | 24 | 0.154 | HMOX1 |
| 17 | THP1 | BRD-K31342827 | 12.12 | 6 | 0.154 | HMOX1, HYOU1, TAPT1 |
| 18 | A375 | BRD-K19410523 | 10.0 | 6 | 0.154 | HMOX1, PDE4DIP |
| 19 | MCF10A | NVP-AUY922 | 10 | 3 | 0.138 | HMOX1, HYOU1, WFS1 |
| 20 | PC3 | mls-0435429 | 10.0 | 6 | 0.123 | HMOX1, PDE4DIP |
| 21 | BT20 | radicicol | 3.33 | 3 | 0.123 | HMOX1, HYOU1 |
| 22 | HA1E | np-010914 | 10.0 | 6 | 0.108 | ERMP1, HYOU1, PDE4DIP |
| 23 | HME1 | geldanamycin | 3.33 | 3 | 0.108 | HMOX1, HYOU1, PDE4DIP |
| 24 | MCF7 | metergoline | 10.0 | 24 | 0.108 | TOR1A |
| 25 | A549 | HY-11007 | 10 | 24 | 0.108 | TOR1A |
| 26 | MCF7 | BRD-K05593511 | 10.0 | 6 | 0.077 | GOLGB1, PDE4DIP |
The table highlights compounds, ordered according to their statistical scoring, which perturb gene subsets of the input signature (last column), when administered in the indicated cell line. The input gene signature has been derived from the BioInfoMiner analysis, using the MGIMP ontology, for the CoV-2 secretome (87 protein interactions).
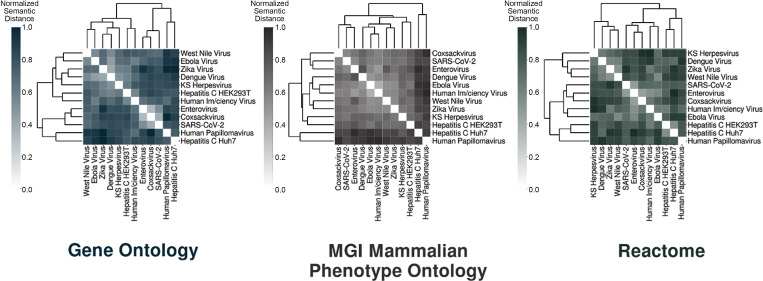
Furthermore, because BioInfoMiner enables delineation of the systemic tree snapshot of the mode of host infection, we exploited this feature to provide a detailed comparison of 12 viral interactome functional profiles. This network-aided phylogenetic analysis measured the enrichment of the vertices of this graph, estimated the functional similarities of the semantic tree graphs of the viral interactomes, and applied agglomerative clustering for the pathogens under analysis. Thus, CoV-2 is robustly found to functionally share the highest similarities with enteroviruses (rhinovirus C15 and coxsackievirus A10; Fig. 3).
Figure 3.
Network-aided phylogenetic analysis of 12 viral pathogen infection models. The graphs depict functional comparisons of 12 virus–host protein interactomes, using BioInfoMiner with the indicated vocabularies (Gene Ontology, MGI Mammalian Phenotype, and Reactome). For each graph, the comparison estimates the degree of their semantic similarities via agglomerative clustering to construct the phylogenetic tree. The similarity of two viruses was calculated by averaging the values derived from three different semantic similarity measures (Resnik, 1999; Aggregate IC [Song et al., 2014]; and XGraSM [Mazandu et al., 2016]) in conjunction with the average best matches approach (Mazandu et al., 2016).
The ontological terms obtained from the whole CoV-2 interactome with the three BioInfoMiner vocabularies (MGI Mammalian Phenotype, Reactome, and Gene Ontology) revealed many proteins related to the secretory pathway among the perturbed genes (last columns in Table 2). Starting from the idea that CoV-2 exploits host-derived secretory pathway components for assembly, budding, and spreading, we repeated the same bioinformatic analysis using the secretory pathway component list (Table 1). We observed enrichment for five host-derived virus-interacting proteins (GOLGB1, PDE4DIP, TOR1A, HMOX1, and HYOU1) involved in different processes and related to quality control and ER–Golgi homeostasis maintenance. Specifically, GOLGB1 interacts with CoV-2 nsp13, and it is key for Golgi organization and long intercisternal communication. Interestingly, it is also an antigen in chronic rheumatoid arthritis. On the basis of our analysis, GOLGB1 could be a target of diverse classes of compounds, such as catechins, ganciclovir, and cetirizine-dihydrochloride. Catechins, already proposed as potential anti–influenza virus agents, inhibit neuraminidase and specifically interact with HA (Müller and Downard, 2015). Moreover, because catechins also alter the viral membrane, they may also interfere with particle maturation. Cetirizines present anti-inflammatory effects and decrease expression of leukocyte intercellular adhesion molecule 1 membrane receptors in nasal epithelial cells (Ciprandi et al., 1995). This receptor is a major target for respiratory viruses and reasonably also for SARS-CoV-2. Finally, ganciclovir is an anti-Herpesviridae family drug (cytomegalovirus, Epstein-Barr virus, or herpes simplex virus); it blocks DNA replication, and it has already been tested in patients with CoV-2 infection in combination with two other antiviral drugs to minimize the potential for septic shock and inflammation (Yan et al., 2020). A second possible therapeutic target is PDE4DIP, which anchors components of the cAMP-dependent pathway involved in cell movement and migration to the Golgi stacks (Robinson et al., 2014; Andruska et al., 2015(Andruska et al., 2015)). nsp13 interaction with PDE4DIP may help viral particle movement during the exit step. Among PDE4DIP-targeting drugs, estrone has been employed against influenza virus infection and other respiratory diseases (Robinson et al., 2014). A third target that came up in our analysis is TOR1A, a member of the AAA+ ATPase superfamily. Because its depletion impairs herpes simplex virus 1 replication and induces improper envelope formation (Maric et al., 2014), TOR1A might facilitate envelope formation of SARS-CoV-2. HMOX1 and HYOU1 are ER residents triggered by oxygen deprivation that interact with orf8 and orf3a, respectively. Their presence in the top hits supports the involvement of hypoxia and ER stress in CoV-2 infection, possibly reflecting an antiapoptotic role. HMOX1 and HYOU1 are targets of Radicicol, an HSP90 inhibitor, which blocks vesicular stomatitis virus replication (Born et al., 2013).
Conclusions and perspectives
The SARS-CoV-2 genome encodes for nonstructural and accessory proteins, each with different specific host–protein binding patterns. Although none of them is essential for replication in other CoVs, some of them help with virus assembly and virulence, recruiting elements of the host cell’s secretory pathway compartments. Indeed, these represent a considerable part of the SARS-CoV-2 host–protein interactome, whose pathophysiological significance in the context of SARS-CoV-2 infection requires further functional studies.
Our bioinformatic analyses unveiled that some of those proteins, such as TOR1A, GOLGB1, or HMOX1, could represent attractive targets for diverse classes of compounds (Table 3). However, we still do not know how these proteins impact CoV-2 biology. In-depth studies on their structure and localization during CoV-2 infection are necessary to better characterize the related phenotypes. Synergistic effects may be obtained when canonical treatments (protease and/or RNA polymerase inhibitors) are accompanied by drugs targeting secretory pathway components, the rationale being that viral propagation depends on the efficiency of secretory organelles. Given the strong interconnections with apoptosis and innate immunity, the UPR elicited by CoV-2 may modulate host antiviral responses from different points of view. Reducing ER stress and enhancing protein folding, it may promote chaperone production and massive production of suitably glycosylated S proteins, helping CoV replication. The expansion of the ER (likely facilitated by SCAP-SREBP) may provide a source of membranes for DMV production and budding. ERAD may be important to guarantee stoichiometric virus assembly, though its role during infection remains to be understood. Accumulation in apoptotic bodies might shield viruses from immune recognition (Hay and Kannourakis, 2002). CoV-2 highlights once more the Jekyll-and-Hyde dualism of the UPR. On the one hand, the UPR attenuates global translation and activates innate immunity. On the other hand, exaggerated immune response activation is associated with tissue damage and immunopathogenesis, typical in CoV infection (Dandekar and Perlman, 2005). For all these reasons, studies on the interconnections between CoVs, inflammation, and UPR ramifications are needed. To conclude, identification of new secretory pathway–related targets for antiviral compounds may help the development of more specific anti-CoV treatments but also may help to draft a roadmap for future studies on virus biogenesis.
Acknowledgments
We thank Dr. van der Goot and Dr. Saraste for their critical comments on Fig. 1.
E. Chevet was funded by the Institut National de la Santé et de la Recherche Médicale, Agence Nationale de la Recherche (eRARE-ERAAT), Institut National du Cancer (INCa PLBIO_2017, PLBIO_2018, and PLBIO_2019), and Fondation pour la Recherche Médicale (Equipe Labelisée 2018; Frm DEQ20180339169). D. Sicari was supported by an Associazione Italiana per la Ricerca sul Cancro fellowship for Abroad. A. Chatziioannou was supported by the project “ELIXIR-GR, The Greek Research Infrastructure for Data Management and Analysis in Life Sciences” (MIS 5002780), which is co-funded by the European Commission within the research infrastructures program of Horizon 2020 Framework Programme and national funds. T. Koutsandreas was supported by Marie Skłodowska-Curie actions grant MSCA-RISE-734749 (Interfere to Suppress or Promote IRE1 [INSPIRED]). R. Sitia was funded by Associazione Italiana per la Ricerca sul Cancro.
The authors declare no competing financial interests.
Author contributions: D. Sicari, R. Sitia, and E. Chevet wrote the manuscript. A. Chatziioannou and T. Koutsandreas performed all the bioinformatic analysis and the interpretation of the results.
References
- Adachi, Y., Yamamoto K., Okada T., Yoshida H., Harada A., and Mori K.. 2008. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct. Funct. 33:75–89. 10.1247/csf.07044 [DOI] [PubMed] [Google Scholar]
- Alborzinia, H., Ignashkova T.I., Dejure F.R., Gendarme M., Theobald J., Wölfl S., Lindemann R.K., and Reiling J.H.. 2018. Golgi stress mediates redox imbalance and ferroptosis in human cells. Commun. Biol. 1:210. 10.1038/s42003-018-0212-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, K.G., Rambaut A., Lipkin W.I., Holmes E.C., and Garry R.F.. 2020. The proximal origin of SARS-CoV-2. Nat. Med. 26:450–452. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andruska, N., Zheng X., Yang X., Helferich W.G., and Shapiro D.J.. 2015. Anticipatory estrogen activation of the unfolded protein response is linked to cell proliferation and poor survival in estrogen receptor α-positive breast cancer. Oncogene. 34:3760–3769. 10.1038/onc.2014.292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anelli, T., Alessio M., Mezghrani A., Simmen T., Talamo F., Bachi A., and Sitia R.. 2002. ERp44, a novel endoplasmic reticulum folding assistant of the thioredoxin family. EMBO J. 21:835–844. 10.1093/emboj/21.4.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, K., and Nagata K.. 2011. Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol. 3. a007526. 10.1101/cshperspect.a007526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi, P., Torres M., Qi L., and Tsai B.. 2020. Selective EMC subunits act as molecular tethers of intracellular organelles exploited during viral entry. Nat. Commun. 11:1127. 10.1038/s41467-020-14967-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechill, J., Chen Z., Brewer J.W., and Baker S.C.. 2008. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J. Virol. 82:4492–4501. 10.1128/JVI.00017-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born, E.J., Hartman S.V., and Holstein S.A.. 2013. Targeting HSP90 and monoclonal protein trafficking modulates the unfolded protein response, chaperone regulation and apoptosis in myeloma cells. Blood Cancer J. 3. e167. 10.1038/bcj.2013.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhaddou, M., Memon D., Meyer B., White K.M., Veronica V., Marrero M.C., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., et al. 2020. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 10.1016/j.cell.2020.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood, P.J., and Hegde R.S.. 2019. The role of EMC during membrane protein biogenesis. Trends Cell Biol. 29:371–384. 10.1016/j.tcb.2019.01.007 [DOI] [PubMed] [Google Scholar]
- Ciprandi, G., Buscaglia S., Pesce G., Passalacqua G., Rihoux J.P., Bagnasco M., and Canonica G.W.. 1995. Cetirizine reduces inflammatory cell recruitment and ICAM-1 (or CD54) expression on conjunctival epithelium in both early- and late-phase reactions after allergen-specific challenge. J. Allergy Clin. Immunol. 95:612–621. 10.1016/S0091-6749(95)70324-1 [DOI] [PubMed] [Google Scholar]
- Cubillos-Ruiz, J.R., and Glimcher L.H.. 2015. Targeting abnormal ER stress responses in tumors: a new approach to cancer immunotherapy. Oncoimmunology. 5. e1098802. 10.1080/2162402X.2015.1098802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar, A.A., and Perlman S.. 2005. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 5:917–927. 10.1038/nri1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDiego, M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., and Enjuanes L.. 2014. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 194:124–137. 10.1016/j.virusres.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, T.S., and Liu D.X.. 2018. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 13:405–430. 10.2217/fvl-2018-0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., O’Meara M.J., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., et al. 2020. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. bioRxiv. 10.1101/2020.03.22.002386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, M., Su L., Yuan Y.-C., Li H., and Chow W.A.. 2015. Nelfinavir and nelfinavir analogs block site-2 protease cleavage to inhibit castration-resistant prostate cancer. Sci. Rep. 5:9698. 10.1038/srep09698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, G.S. 2012. Animal Lectins: Form, Function and Clinical Applications. Springer-Verlag Wien, Vienna. 1088 pp. [Google Scholar]
- Ha, D.P., Van Krieken R., Carlos A.J., and Lee A.S.. 2020. The stress-inducible molecular chaperone GRP78 as potential therapeutic target for coronavirus infection. J. Infect. 10.1016/j.jinf.2020.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, S., and Kannourakis G.. 2002. A time to kill: viral manipulation of the cell death program. J. Gen. Virol. 83:1547–1564. 10.1099/0022-1317-83-7-1547 [DOI] [PubMed] [Google Scholar]
- Hetz, C., Chevet E., and Harding H.P.. 2013. Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 12:703–719. 10.1038/nrd3976 [DOI] [PubMed] [Google Scholar]
- Hetz, C., Chevet E., and Oakes S.A.. 2015. Proteostasis control by the unfolded protein response. Nat. Cell Biol. 17:829–838. 10.1038/ncb3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh, W.J., Esen E., Geahlen J.H., Bredemeyer A.J., Lee A.H., Shi G., Konieczny S.F., Glimcher L.H., and Mills J.C.. 2010. XBP1 controls maturation of gastric zymogenic cells by induction of MIST1 and expansion of the rough endoplasmic reticulum. Gastroenterology. 139:2038–2049. 10.1053/j.gastro.2010.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, A., Gaschler M.M., Dunn D.E., Colligan R., Brown L.M., A.G. Palmer, III, Lo D.C., and Stockwell B.R.. 2015. Small molecule-induced oxidation of protein disulfide isomerase is neuroprotective. Proc. Natl. Acad. Sci. USA. 112:E2245–E2252. 10.1073/pnas.1500439112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaips, C.L., Jayaraj G.G., and Hartl F.U.. 2018. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 217:51–63. 10.1083/jcb.201709072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus, J.P., Eisenhauer P., Russo J., Mason A.B., Do D., King B., Taatjes D., Cornillez-Ty C., Boyson J.E., Thali M., et al. 2013. The intracellular cargo receptor ERGIC-53 is required for the production of infectious arenavirus, coronavirus, and filovirus particles. Cell Host Microbe. 14:522–534. 10.1016/j.chom.2013.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman, J., Locker J.K., Meijer A., Horzinek M.C., Geuze H.J., and Rottier P.J.. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523–6534. 10.1128/JVI.68.10.6523-6534.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, P.J., and Dixon S.J.. 2018. Protein palmitoylation and cancer. EMBO Rep. 19:e46666. 10.15252/embr.201846666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, P.J., Woodrow C., Dubreuil M.M., Martin B.R., Skouta R., Bassik M.C., and Dixon S.J.. 2019. A ZDHHC5-GOLGA7 protein acyltransferase complex promotes nonapoptotic cell death. Cell Chem. Biol. 26:1716–1724.e9. 10.1016/j.chembiol.2019.09.014 [DOI] [PubMed] [Google Scholar]
- Lake, M.A.. 2020. What we know so far: COVID-19 current clinical knowledge and research. Clin. Med. (Lond.). 20:124–127. 10.7861/clinmed.2019-coron [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko, M., Marzi A., and Munster V.. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5:562–569. 10.1038/s41564-020-0688-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lhomond, S., Avril T., Dejeans N., Voutetakis K., Doultsinos D., McMahon M., Pineau R., Obacz J., Papadodima O., Jouan F., et al. 2018. Dual IRE1 RNase functions dictate glioblastoma development. EMBO Mol. Med. 10:1–19. 10.15252/emmm.201707929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, J.Q., Fang S., Yuan Q., Huang M., Chen R.A., Fung T.S., and Liu D.X.. 2019. N-Linked glycosylation of the membrane protein ectodomain regulates infectious bronchitis virus-induced ER stress response, apoptosis and pathogenesis. Virology. 531:48–56. 10.1016/j.virol.2019.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, Y.X., Ng Y.L., Tam J.P., and Liu D.X.. 2016. Human coronaviruses: a review of virus–host interactions. Diseases. 4:26. 10.3390/diseases4030026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, S., and Lee A.S.. 2002. Requirement of the p38 mitogen-activated protein kinase signalling pathway for the induction of the 78 kDa glucose-regulated protein/immunoglobulin heavy-chain binding protein by azetidine stress: activating transcription factor 6 as a target for stress-induced phosphorylation. Biochem. J. 2002;366:787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mages, J., Freimüller K., Lang R., Hatzopoulos A.K., Guggemoos S., Koszinowski U.H., and Adler H.. 2008. Proteins of the secretory pathway govern virus productivity during lytic gammaherpesvirus infection. J. Cell. Mol. Med. 12:1974–1989. 10.1111/j.1582-4934.2008.00235.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric, M., Haugo A.C., Dauer W., Johnson D., and Roller R.J.. 2014. Nuclear envelope breakdown induced by herpes simplex virus type 1 involves the activity of viral fusion proteins. Virology. 460-461:128–137. 10.1016/j.virol.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus, D., Marini E.S., Progida C., and Bakke O.. 2018. Rab7a modulates ER stress and ER morphology. Biochim. Biophys. Acta Mol. Cell Res. 1865:781–793. 10.1016/j.bbamcr.2018.02.011 [DOI] [PubMed] [Google Scholar]
- Matsuoka, M., and Komoike Y.. 2015. Experimental evidence shows salubrinal, an eIF2α dephosphorylation inhibitor, reduces xenotoxicant-induced cellular damage. Int. J. Mol. Sci. 16:16275–16287. 10.3390/ijms160716275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel, M., Chevet E., Tavernier J., and Gerlo S.. 2014. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 39:245–254. 10.1016/j.tibs.2014.02.008 [DOI] [PubMed] [Google Scholar]
- Mazandu, G.K., Chimusa E.R., and Mulder N.J.. 2016. Gene Ontology semantic similarity tools: survey on features and challenges for biological knowledge discovery. Brief. Bioinform. 10.1093/bib/bbw067 [DOI] [PubMed] [Google Scholar]
- McBride, C.E., Li J., and Machamer C.E.. 2007. The cytoplasmic tail of the severe acute respiratory syndrome coronavirus spike protein contains a novel endoplasmic reticulum retrieval signal that binds COPI and promotes interaction with membrane protein. J. Virol. 81:2418–2428. 10.1128/JVI.02146-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakshi, R., Padhan K., Rani M., Khan N., Ahmad F., and Jameel S.. 2009. The SARS coronavirus 3a protein causes endoplasmic reticulum stress and induces ligand-independent downregulation of the type 1 interferon receptor. PLoS One. 4. e8342. 10.1371/journal.pone.0008342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, T., Hayashi T., Hayashi E., and Su T.P.. 2013. Sigma-1 receptor chaperone at the ER-mitochondrion interface mediates the mitochondrion-ER-nucleus signaling for cellular survival. PLoS One. 8. e76941. 10.1371/journal.pone.0076941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, P., and Downard K.M.. 2015. Catechin inhibition of influenza neuraminidase and its molecular basis with mass spectrometry. J. Pharm. Biomed. Anal. 111:222–230. 10.1016/j.jpba.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Myint, S.H.. 1994. Human coronaviruses: a brief review. Rev. Med. Virol. 4:35–46. 10.1002/rmv.1980040108 [DOI] [Google Scholar]
- Ohno, Y., Kihara A., Sano T., and Igarashi Y.. 2006. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta. 1761:474–483. 10.1016/j.bbalip.2006.03.010 [DOI] [PubMed] [Google Scholar]
- Okamura, Y., and Saga Y.. 2008. Pofut1 is required for the proper localization of the Notch receptor during mouse development. Mech. Dev. 125:663–673. 10.1016/j.mod.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Oostra, M., de Haan C.A.M., de Groot R.J., and Rottier P.J.M.. 2006. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 80:2326–2336. 10.1128/JVI.80.5.2326-2336.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostra, M., de Haan C.A.M., and Rottier P.J.M.. 2007. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 81:13876–13888. 10.1128/JVI.01631-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu, M., Bertoli G., Fagioli C., Guerini-Rocco E., Nerini-Molteni S., Ruffato E., and Sitia R.. 2006. Dynamic Retention of Ero1α and Ero1β in the Endoplasmic Reticulum by Interactions with PDI and ERp44. Antioxid. Redox Signal. 8(3–4):274–282. 10.1089/ars.2006.8.274 [DOI] [PubMed] [Google Scholar]
- Reggiori, F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A.M., and Molinari M.. 2010. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 7:500–508. 10.1016/j.chom.2010.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnik, P. 1999. Semantic Similarity in a Taxonomy: An Information-Based Measure and its Application to Problems of Ambiguity in Natural Language. J. Artif. Intell. Res. 11. 10.1613/jair.514 [DOI] [Google Scholar]
- Robinson, D.P., Hall O.J., Nilles T.L., Bream J.H., and Klein S.L.. 2014. 17β-Estradiol protects females against influenza by recruiting neutrophils and increasing virus-specific CD8 T cell responses in the lungs. J. Virol. 88:4711–4720. 10.1128/JVI.02081-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen, D.A., Seki S.M., Fernández-Castañeda A., Beiter R.M., Eccles J.D., Woodfolk J.A., and Gaultier A.. 2019. Modulation of the sigma-1 receptor-IRE1 pathway is beneficial in preclinical models of inflammation and sepsis. Sci. Transl. Med. 11. eaau5266. 10.1126/scitranslmed.aau5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannino, S., Anelli T., Cortini M., Masui S., Degano M., Fagioli C., Inaba K., and Sitia R.. 2014. Progressive quality control of secretory proteins in the early secretory compartment by ERp44. J. Cell Sci. 127:4260–4269. 10.1242/jcs.153239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva, O.A., and van der Goot F.G.. 2019. Anthrax toxin requires ZDHHC5-mediated palmitoylation of its surface-processing host enzymes. Proc. Natl. Acad. Sci. USA. 116:1279–1288. 10.1073/pnas.1812588116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoulders, M.D., Ryno L.M., Genereux J.C., Moresco J.J., Tu P.G., Wu C., J.R. Yates, III, Su A.I., Kelly J.W., and Wiseman R.L.. 2013. Stress-independent activation of XBP1s and/or ATF6 reveals three functionally diverse ER proteostasis environments. Cell Rep. 3:1279–1292. 10.1016/j.celrep.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicari, D., Fantuz M., Bellazzo A., Valentino E., Apollonio M., Pontisso I., Di Cristino F., Dal Ferro M., Bicciato S., Del Sal G., et al. 2019. Mutant p53 improves cancer cells’ resistance to endoplasmic reticulum stress by sustaining activation of the UPR regulator ATF6. Oncogene. 38:6184–6195. 10.1038/s41388-019-0878-3 [DOI] [PubMed] [Google Scholar]
- Siu, K.L., Chan C.P., Kok K.H., Woo P.C.Y., and Jin D.Y.. 2014. Comparative analysis of the activation of unfolded protein response by spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus HKU1. Cell Biosci. 4:3. 10.1186/2045-3701-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, E., Nelson B.L., and Pegden C.D.. 2014. Advanced tutorial: Input uncertainty quantification. Proceedings of the Winter Simulation Conference 2014. 10.1109/WSC.2014.7019886 [DOI] [Google Scholar]
- Stertz, S., Reichelt M., Spiegel M., Kuri T., Martínez-Sobrido L., García-Sastre A., Weber F., and Kochs G.. 2007. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 361:304–315. 10.1016/j.virol.2006.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukalov, A., Girault V., Grass V., Bergant V., Karayel O., Urban C., Haas D.A., Huang Y., Oubraham L., Wang A., et al. 2020. Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. bioRxiv. 10.1101/2020.06.17.156455 [DOI] [PubMed] [Google Scholar]
- Su, S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., and Gao G.F.. 2016. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 24:490–502. 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Z., and Brodsky J.L.. 2019. Protein quality control in the secretory pathway. J. Cell Biol. 218:3171–3187. 10.1083/jcb.201906047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S.C., Chao C.Y., Jeng K.S., Yang J.Y., and Lai M.M.C.. 2009. The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology. 387:402–413. 10.1016/j.virol.2009.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, H., Schneider M., Williamson D.B., Ito A., Takeuchi M., Handford P.A., and Haltiwanger R.S.. 2018. Two novel protein O-glucosyltransferases that modify sites distinct from POGLUT1 and affect Notch trafficking and signaling. Proc. Natl. Acad. Sci. USA. 115:E8395–E8402. 10.1073/pnas.1804005115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavassori, S., Cortini M., Masui S., Sannino S., Anelli T., Caserta I.R., Fagioli C., Mossuto M.F., Fornili A., van Anken E., et al. 2013. A pH-regulated quality control cycle for surveillance of secretory protein assembly. Mol. Cell. 50:783–792. 10.1016/j.molcel.2013.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg, G.A., van de Nes P.S., Bredenbeek P.J., and Spaan W.J.M.. 2007. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. J. Virol. 81:10981–10990. 10.1128/JVI.01033-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y., Shang J., Graham R., Baric R.S., and Li F.. 2020. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 94:e00127-20. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Shinkre B.A., Lee J.G., Weniger M.A., Liu Y., Chen W., Wiestner A., Trenkle W.C., and Ye Y.. 2010. The ERAD inhibitor Eeyarestatin I is a bifunctional compound with a membrane-binding domain and a p97/VCP inhibitory group. PLoS One. 5. e15479. 10.1371/journal.pone.0015479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S., Harayama M., Kanemura S., Sitia R., and Inaba K.. 2017. Structural basis of pH-dependent client binding by ERp44, a key regulator of protein secretion at the ER-Golgi interface. Proc. Natl. Acad. Sci. USA. 114:E3224–E3232. 10.1073/pnas.1621426114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, S., Amagai Y., Sannino S., Tempio T., Anelli T., Harayama M., Masui S., Sorrentino I., Yamada M., Sitia R., et al. 2019. Zinc regulates ERp44-dependent protein quality control in the early secretory pathway. Nat. Commun. 10:603. 10.1038/s41467-019-08429-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., Allen J.D., Wrapp D., Mclellan J.S., and Crispin M.. 2020. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchester, B.G.. 2009. Iminosugars, from botanical curiosities to licensed drugs. Tetrahedron Asymmetry. 20:645–651. 10.1016/j.tetasy.2009.02.048 [DOI] [Google Scholar]
- Wu, F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. 2020. A new coronavirus associated with human respiratory disease in China. Nature. 579:265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y., Shin W.I., Pang Y.X., Meng Y., Lai J., You C., Zhao H., Lester E., Wu T., and Pang C.H.. 2020. The first 75 days of novel coronavirus (SARS-CoV-2) outbreak: recent advances, prevention, and treatment. Int. J. Environ. Res. Public Health. 17:2323. 10.3390/ijerph17072323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Zhang D., Qin H., Liu S., and Yan Q.. 2019. poFUT1 promotes endometrial decidualization by enhancing the O-fucosylation of Notch1. EBioMedicine. 44:563–573. 10.1016/j.ebiom.2019.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, Y., Suzuki K., Yamamoto A., Sakai N., Bando M., Tanimoto K., Yamaguchi Y., Sakaguchi T., Akhter H., Fujii G., et al. 2008. YIPF5 and YIF1A recycle between the ER and the Golgi apparatus and are involved in the maintenance of the Golgi structure. Exp. Cell Res. 314:3427–3443. 10.1016/j.yexcr.2008.07.023 [DOI] [PubMed] [Google Scholar]
- Yuan, S., Chu H., Chan J.F.W., Ye Z.W., Wen L., Yan B., Lai P.M., Tee K.M., Huang J., Chen D., et al. 2019. SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat. Commun. 10:120. 10.1038/s41467-018-08015-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., and Wilson I.A.. 2020. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 368:630–633. 10.1126/science.abb7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., and Wang A.. 2012. Virus-induced ER stress and the unfolded protein response. Front Plant Sci. 3:293. 10.3389/fpls.2012.00293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzula, L., and Tramontano E.. 2013. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit. Antiviral Res. 100:615–635. 10.1016/j.antiviral.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]