Grond et al. show that GRASPs or GORASPs do not have a role in stacking of Golgi cisternae in vivo but instead are required for linking cisternal rims to likely control flux of transport through the Golgi apparatus.
Abstract
In vitro experiments have shown that GRASP65 (GORASP1) and GRASP55 (GORASP2) proteins function in stacking Golgi cisternae. However, in vivo depletion of GORASPs in metazoans has given equivocal results. We have generated a mouse lacking both GORASPs and find that Golgi cisternae remained stacked. However, the stacks are disconnected laterally from each other, and the cisternal cross-sectional diameters are significantly reduced compared with their normal counterparts. These data support earlier findings on the role of GORASPs in linking stacks, and we suggest that unlinking of stacks likely affects dynamic control of COPI budding and vesicle fusion at the rims. The net result is that cisternal cores remain stacked, but cisternal diameter is reduced by rim consumption.
Introduction
Most eukaryotic cells contain stacked Golgi cisternae, but what holds the cisternae together, and what is the function of cisternal stacking? Identification of stacking factors could help address these questions.
In mammalian cells, pericentriolar stacks of Golgi cisternae are separated from each other late in G2, and by metaphase, these membranes are converted into tubules and small vesicles. Cisternal stacks are visible in telophase and after cytokinesis, when they return to the pericentriolar region of the newly formed cells (Lucocq and Warren, 1987; Pecot and Malhotra, 2006). Warren and colleagues reconstituted the fragmentation of isolated Golgi stacks using mitotic cytosol (Misteli and Warren, 1995) and the reassembly of Golgi stacks from mitotic Golgi fragments (Rabouille et al., 1995). Treatment of the disassembled Golgi membranes with an alkylating agent (N-ethyl maleimide) inhibited stack reassembly (Rabouille et al., 1995). This led to identification of a Golgi reassembly stacking protein 65 (GRASP65 [now called GORASP1]; Barr et al., 1997) that is N-terminally myristoylated. This modification, along with tight binding to the coiled-coil Golgi protein GM130 (Nakamura et al., 1997), is required for attachment of GORASP1 to cis- and medial-Golgi cisternae (Bachert and Linstedt, 2010). Remarkably, adding antibodies targeted against GORASP1, or a recombinant GORASP1 to disassembled Golgi membranes, prevented assembly of cisternae into stacks (Barr et al., 1997; Wang et al., 2003). GRASP55 (GORASP2), a protein highly homologous to the N-terminal half of GORASP1, localizes to more distal Golgi cisternae and was also subsequently shown to be required for stacking in vitro (Shorter et al., 1999). Moreover, simultaneous depletion of both GORASPs by siRNA in HeLa cells resulted in unstacked Golgi profiles, consistent with small single cisternal and Golgi fragments (Xiang and Wang, 2010; Wang et al., 2005). These data led to the conclusion that GORASPs are essential to stack Golgi cisternae (Vinke et al., 2011; Rabouille and Linstedt, 2016).
Deletion of the single GORASP gene expressed in Pichia pastoris (Levi et al., 2010) and Drosophila melanogaster cells (Kondylis et al., 2005) is largely inconsequential with respect to Golgi cisternal stacking. siRNA-based depletion of GORASP1 in HeLa cells did not affect the overall Golgi stacking, but the numbers of cisternae per stack were reduced. Interestingly, these cells enter mitosis but do not proceed beyond mitosis because of defects in spindle dynamics (Sütterlin et al., 2005). In a transgenic murine system, deletion of GORASP1 did not affect animal viability or the stacking of cisternae, but the stacks were laterally separated from each other (Veenendaal et al., 2014). This phenocopies earlier reports on the functional significance of GORASP1 and GORASP2 in connecting cisternae laterally without a requirement in the cisternal stacking (Puthenveedu et al., 2006; Feinstein and Linstedt, 2008; Jarvela and Linstedt, 2014). However, these studies addressed the effect of depleting either only one of the two GORASP orthologues.
In the mouse knockout of GORASP1 that failed to detect an obvious effect on Golgi stacking (Veenendaal et al., 2014), the levels of GORASP2 were unaffected, which raises the possibility of a functional redundancy. To address redundancy, Wang and colleagues used CRISPR–Cas9 to completely deplete GORASPs in HeLa and HEK293 cells, but these alterations led to a significant reduction (up to 75–80% of the control levels) of GM130 and golgin-45 proteins (Bekier et al., 2017). Fluorescence microscopy–based analysis with antibodies to GM130 and the late Golgi transmembrane protein TGN46 revealed that these membrane components were dispersed throughout the cytoplasm instead of localized at the Golgi as in control cells. Not clear, therefore, was whether this was due to loss of GORASPs, loss of 75–80% of GM130, golgin-45, or perhaps loss of another cellular component that might be depleted by these procedures (see Fig. 4 in Bekier et al., 2017). Also, in the electron micrographs shown in the paper, the Golgi membranes although vacuolated are clearly stacked (see Fig. 5 D in Bekier et al., 2017). In sum, there is a general agreement that loss of a single GORASP protein (by siRNA, CRISPR, or gene knockout) in mammalian cells or its orthologous gene in flies or P. pastoris has no effect on cisternal stacking. The results from the double depletion of GORASPs in HeLa and HEK293 cells are far less clear.
To address the apparent discrepancy regarding the role of GORASPs in Golgi cisternae stacking, we generated a mouse in which both GORASPs are knocked out. GORASP1 mouse knockout has been described before (Veenendaal et al., 2014). A GORASP2 knockout by similar procedure produced a mouse that appeared to have defects linked to male fertility but was otherwise apparently normal. This qualitative observation fits well with an earlier report of defective spermatogenesis in GORASP2−/− mice (Cartier-Michaud et al., 2017). The standard procedure of generating a double knockout (DKO) by crossing GORASP1- and GORASP2-lacking mice, respectively, resulted in embryonic lethality. We therefore used another procedure in which GORASP2 was conditionally deleted from GORASP1-deleted mice to generate a mouse lacking both genes. The general physiology of this mouse is under investigation, but development is delayed compared with the control, and mice are therefore smaller and lighter in weight. Also, analyses of the small intestine during early postnatal development as well as intestinal organoids reveal that GORASPs are not required to stack Golgi cisternae. The Golgi stacks are separated from each other as reported previously (Feinstein and Linstedt, 2008; Veenendaal et al., 2014), but more striking is the finding that the cisternal diameter is significantly reduced in diameter compared with control Golgi cisternae.
We discuss our findings to help resolve debates on the role of GORASPs in stacking of Golgi cisternae.
Results and discussion
Quantitation of GORASP levels in the mouse DKO
Standard procedures to obtain a double GORASP1 and GORASP2 knockout failed because of embryonic lethality. We therefore employed an alternative, albeit routine, procedure to obtain a mouse lacking GORASP1 and GORASP2.
A GORASP1 mouse knockout (Veenendaal et al., 2014) was crossed with conditional knockout (cFlox) mice expressing endogenous GORASP2, with exon 5 flanked by LoxP sites (tm1c allele; Fig. 1 A). Crossed with Rosa Cre ERT2, tamoxifen (TAM) induces Cre recombinase, eliminating GORASP2 exon 5 expression and resulting in a mouse lacking both functional GORASP1 and GORASP2 (Fig. 1, B and C). Briefly, this procedure resulted in a conditional mouse line that lacks GORASP1 (cDKO), which upon TAM application causes loss of GORASP2, thereby producing a DKO lacking both GORASP1 and GORASP2.
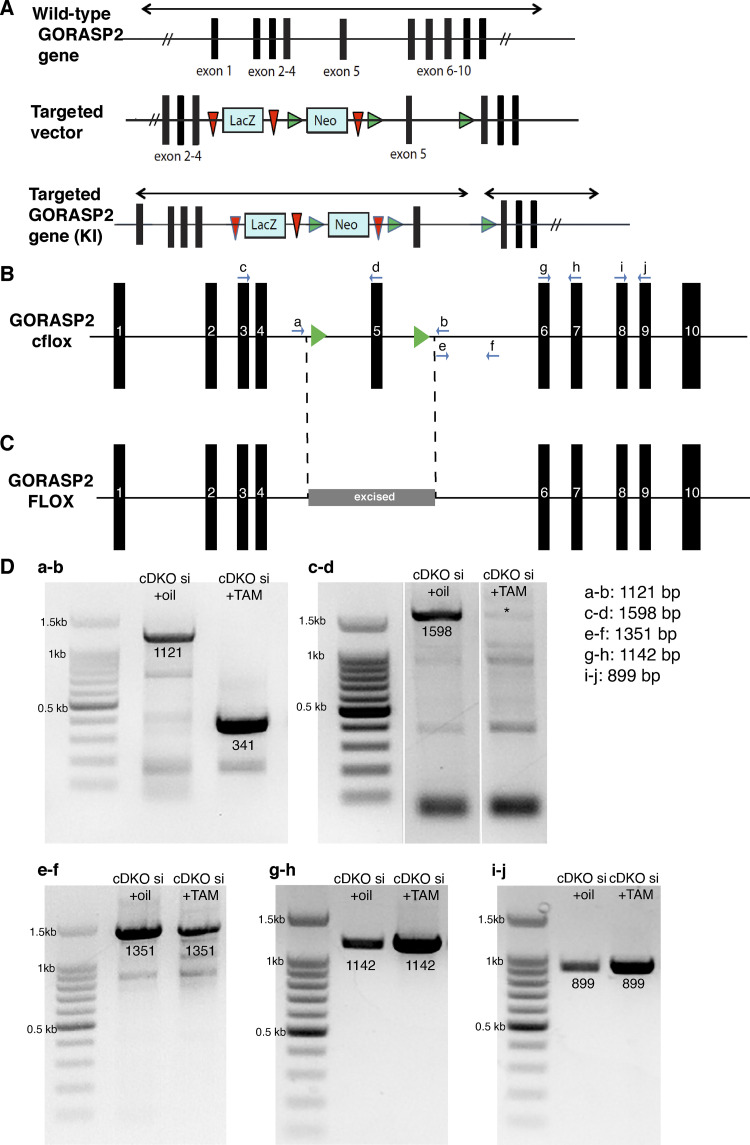
Figure 1.
Genomic GORASP2 locus in the cDKO small intestine (si) in control and TAM-treated tissue. (A) Schematic of the GORASP2 KI allele with the LacZ/Neo cassette between exons 4 and 5. (B) Schematic of the GORASP2 cFlox allele upon crossing GORASPKI with Flp-recombinase expressing mice. The exon 5 is now flanked by two LoxP sites. (C) Schematic of the GORASP2 FLOX allele upon TAM injection (after crossing with a Rosa Cre ERT2 mouse). Exon 5 is excised. (D) A series of PCRs using different combinations of primers showing that exon 5 is deleted (a–d), a part of intron 5 is still present (e and f), and exons 6 and 7 (g and h) and exons 8 and 9 (i and j) are still present in genomic DNA.
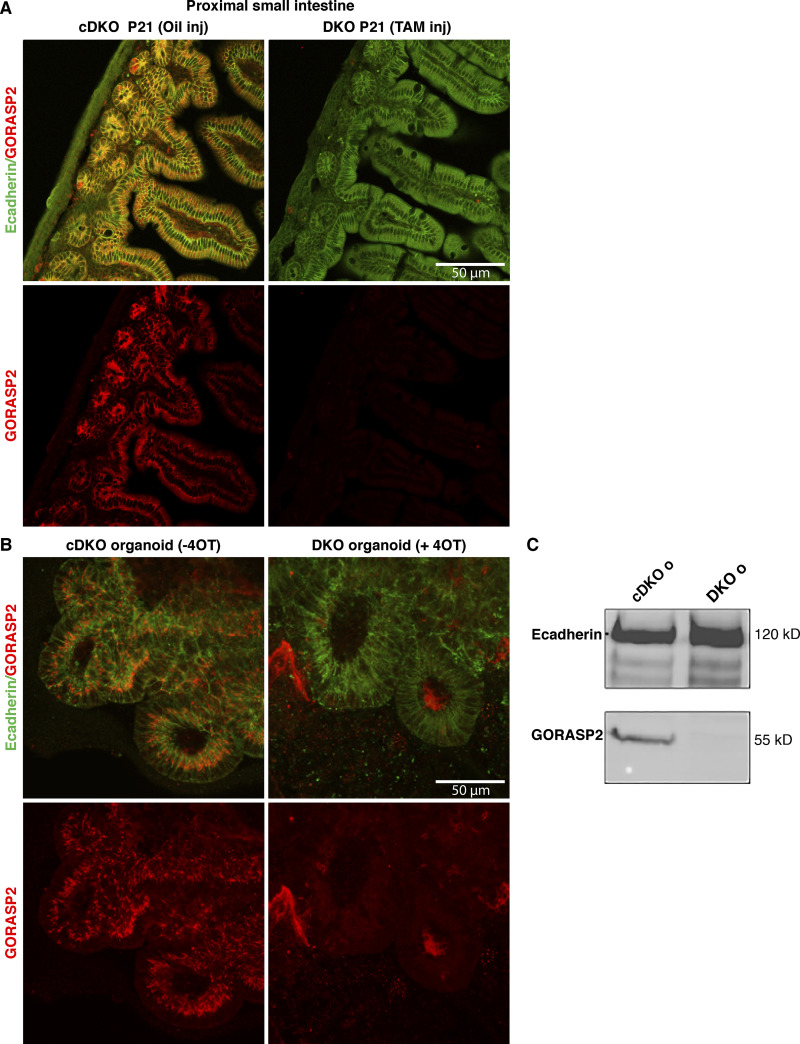
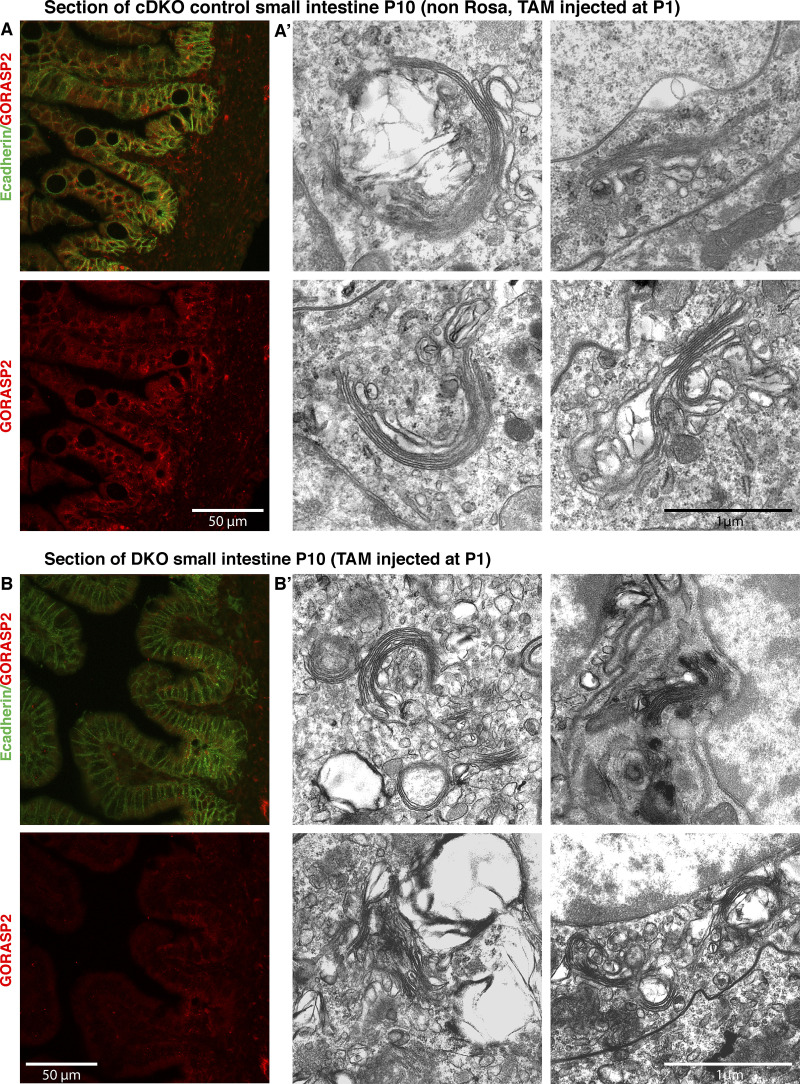
Following injection with TAM or control injection (oil) at postnatal day 1 (P1), the intestine was isolated for further analysis. In parallel, from uninjected mice (cDKO), we generated small intestine budding organoids that were then incubated with 4-hydroxy TAM (4OT) to deplete GORASP2 (DKO; see Materials and methods). These tissues were first analyzed for the levels of GORASP1 and GORASP2. As expected, these tissues were completely depleted of GORASP1. Our procedure for depletion of GORASP2 resulted in excision of exon 5 from the genomic locus (Fig. 1, C and D), whereas exons 1–4 and 6–10 are still present. This yields an exon 5–lacking mRNA (truncated by 131 bp) that is expressed at 20–30% of the levels seen in the control (Fig. S1, A–C). We checked whether this mRNA produces a protein product. The small intestine of TAM- and oil-injected mice was dissected at P10, P21, and P42 and processed for immunofluorescence (IF) using a GORASP2 antibody raised against its C-terminus (see Materials and methods). We failed to detect GORASP2 protein in any of the TAM-injected mice (Fig. 2 A for P21, Fig. 4 A for P10, and Fig. S2 for P10), while it was abundant in the control animals. We also processed the treated organoids and found a complete loss of full-length GORASP2 protein expression as shown by IF (Fig. 2 B) and Western blot (Fig. 2 C). The data suggest that deletion of exon 5 either destabilizes GORASP2 mRNA or results in a base shift, yielding a protein with an aberrant C-terminal sequence. The level of this truncated mRNA measured by RT-PCR is reduced to 20–30% when compared with nontreated controls (Fig. S1 C) and appears to be largely degraded. Furthermore, if it were to harbor a base shift, it would be translated into a protein harboring PDZ1, half of PDZ2 and a very short C-terminus due to the presence of stop codons in exon 6. Although the mRNA abundance is tested by RT-PCR only and not the more quantitative qPCR, the fact that we could not detect a polypeptide corresponding to GORASPs is a good measure of the quantitative loss of these proteins by our experimental procedure.
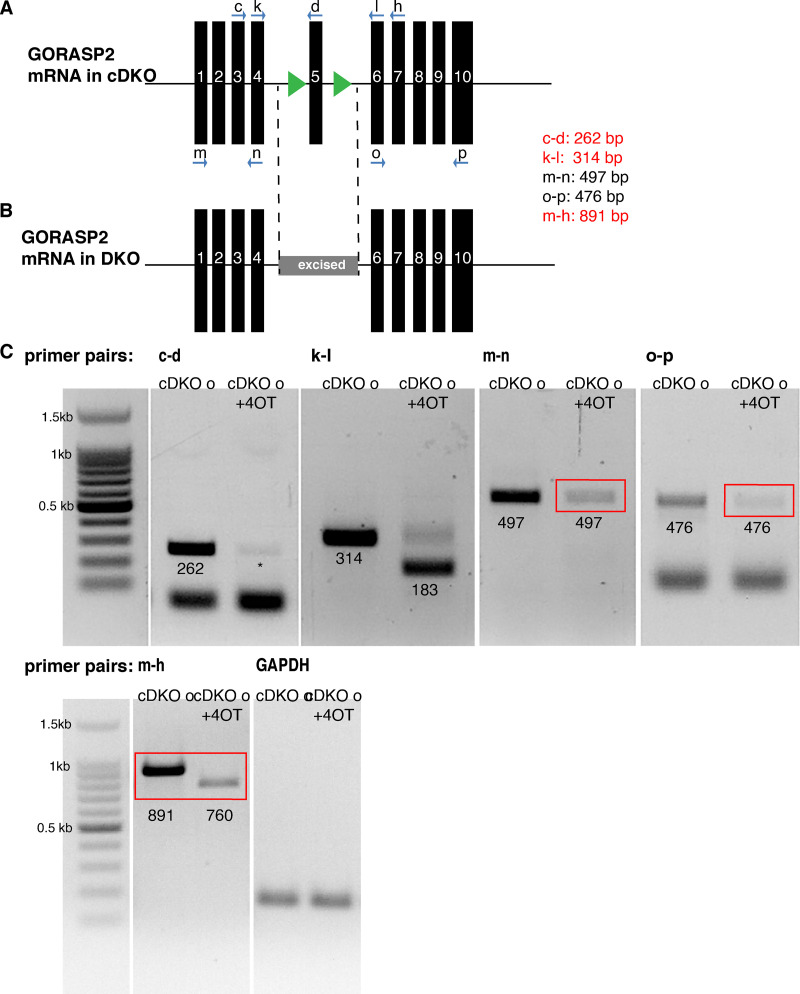
Figure S1.
GORASP2 mRNA in cDKO and DKO organoids (o). (A) Schematic of the GORASP2 cFlox mRNA. Exon 5 is now flanked by two LoxP sites. (B) Schematic of the GORASP2 FLOX allele upon 4OT treatment. Exon 5 is excised. (C) A series of PCRs using different combinations of primers showing that exon 5 is excised upon 4OT inhibition (c and d) and that the sequence between exons 4 and 6 is shorter (from 314 bp to 183 bp, k and l), exons 1–4 are produced at lower amounts than control (red box, m and n), and exons 6–10 are also expressed at lower amounts than control (red box, o and p). Exons 1–7 are produced at lower amounts and shorter sequences compared with the control.
Figure 2.
GORASP2 protein is not expressed in mouse small intestine and organoids upon TAM/4OT exposure. (A) Visualization of GORASP2 (red) and E-cadherin (green) in a section of P21 proximal small intestine. Note that GORASP2 staining is no longer visible upon TAM injection at P1. (B) Visualization of GORASP2 (red) and E-cadherin (green) in small intestine budding organoids (confocal section). Note that GORASP2 staining is no longer visible upon 4OT incubation for 7 d. Background staining can be observed in 4OT; however, these areas are clearly either outside the organoid or in the lumen of the organoid and not in the cells of the organoids. (C) Western blot showing that GORASP2 is no longer expressed in organoids (o) treated with 4OT as in B. E-cadherin is the loading control.
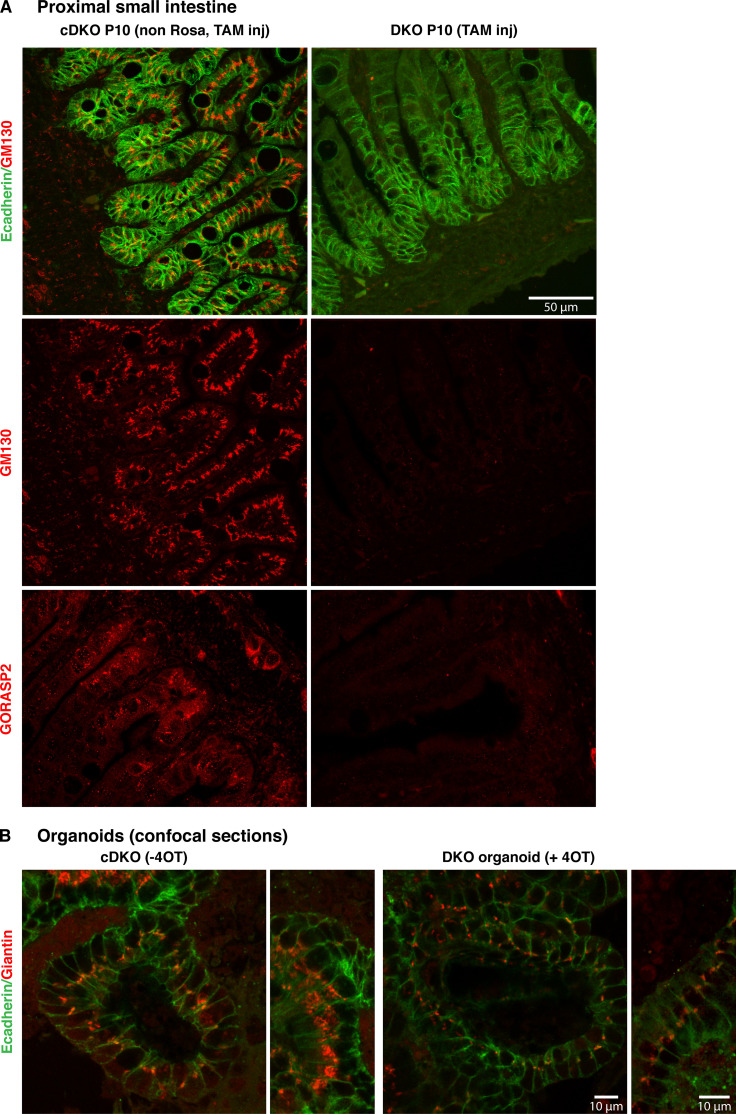
Figure 4.
GM130 is degraded in GORASPs knockout cells and does not affect cisternal stacking. (A) Visualization of GM130 (red) and E-cadherin (green) in section of P10 proximal small intestine in mice injected with either control (non Rosa, TAM injected, functions as control) or TAM at P1. Note that GM130 staining is no longer visible upon TAM injection at P1. GORASP2 visualization on sections from the same tissue confirm the loss of GORASP2 in TAM-injected tissue. (B) Visualization of Giantin (red) and E-cadherin (green) in budding organoids treated or not with 4OT (confocal section). Note that Giantin staining is still prominent in every cell after 4OT treatment, although the intensity of staining is decreased, possibly reflecting the Golgi ribbon unlinking and dispersion of the smaller stacks in the cell cytoplasm.
Figure S2.
DKO of GORASP 1 and 2 does not lead to cisternal unstacking in P10 mouse small intestine. (A and A’) Visualization of GORASP2 (red) and E-cadherin (green) in section of P10 proximal small intestine of control mice (non ROSA Cre ERT2) injected with TAM (A) and corresponding Golgi stack profiles (A’). Note that the stacks are large and contain two to four cisternae. (B and B’) Visualization of GORASP2 (red) and E-cadherin (green) in a section of P10 proximal small intestine of DKO mice injected with TAM at P1 (B) and corresponding Golgi stack profiles (B’). Note that GORASP2 staining is no longer visible upon TAM injection, yet the Golgi stacks are clearly visible. They are shorter but contain the same number of cisternae. Quantification is shown in Fig. 3 C.
Taken together, we conclude that TAM/4OT treatment of cDKO mice/cDKO organoids results in the generation of a DKO GORASP1 and GORASP2 (DKO mice/DKO organoids).
No visible loss of Golgi stacking in DKO tissue
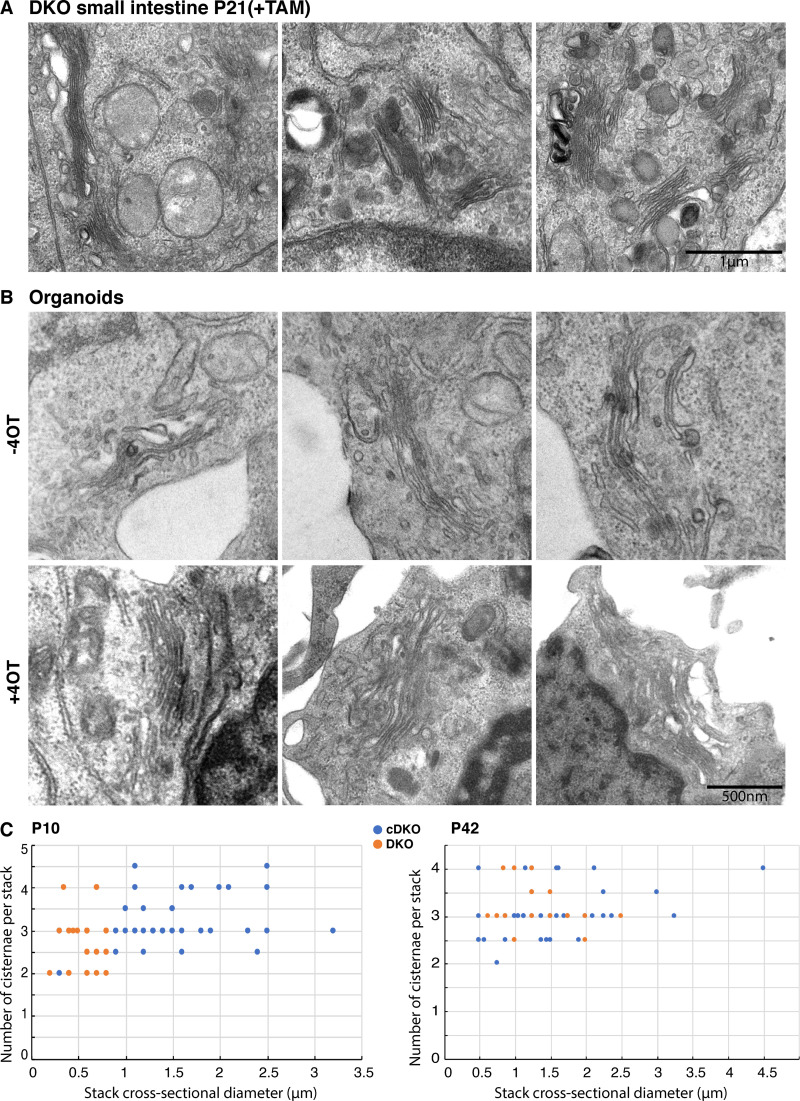
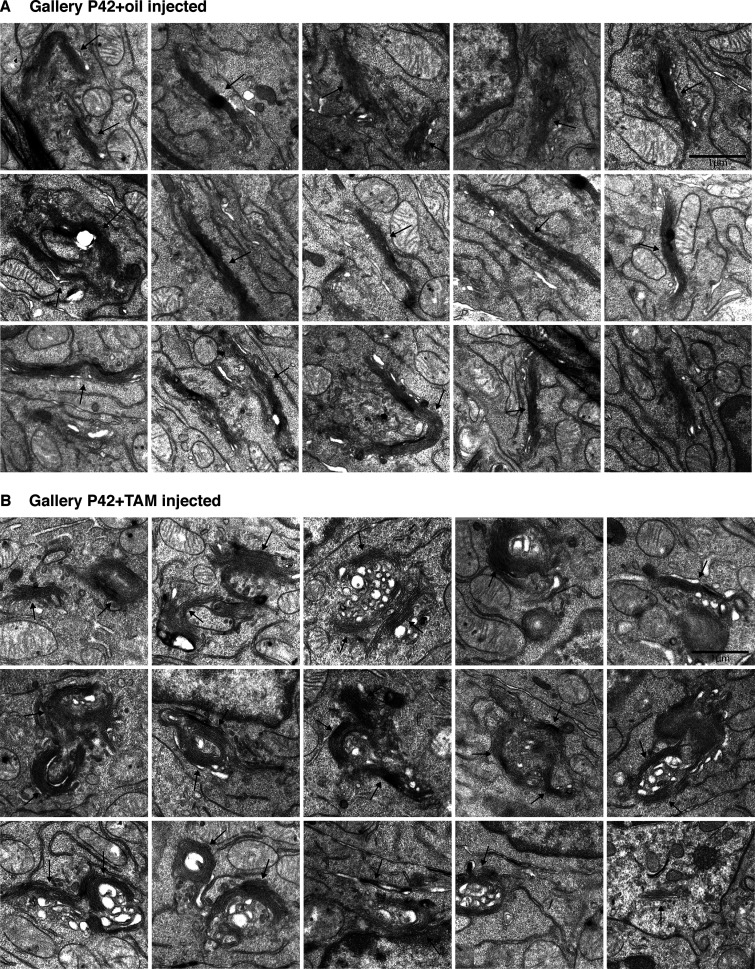
Mouse small intestine and organoids lacking GORASP1 (cDKO) and DKO (GORASP 1 and GORASP 2) were processed for conventional electron microscopy to compare and detect potential effects on the organization of the Golgi apparatus. At P10 (Fig. S2, A and B; and Fig. 3 C), P21 (Fig. 3 A), and P42 (Fig. S3, A and B; and Fig. 3 C), the Golgi stacks in the DKO small intestines were prominently visible at each time point. The number of cisternae per stack was largely unchanged, averaging approximately three cisternae per stack (mean ± SD: P10 cDKO: 3.24 ± 0.59/DKO: 2.71 ± 0.61; P42 cDKO: 3.09 ± 0.52/DKO 3.15 ± 0.46). Although not quantitated, the organoid data show no obvious change in the number of cisternae per stack (Fig. 3 B). However, we noticed a quantitative change in the dimensions of the stacked cisternae. The cross-sectional diameters of TAM-treated mice (DKO) were shorter by a range of an eighth to two thirds (mean diameter ± SD: P10 DKO, 0.58 ± 0.27 µm; P42 DKO, 1.32 ± 0.50 µm) compared with the cDKO (mean diameter ± SD: P10 cDKO, 1.52 ± 0.63 µm; P42 cDKO, 1.50 ± 0.80 µm). Of note, cells present in the stroma also exhibited Golgi stacks in both the cDKO and DKO mice. Overall, these results show that GORASP1 and GORASP2 are not required to stack the core of the Golgi cisternae in mouse small intestine, but they show clear functions at peripheral regions of the cisternae, which are shorter than their control counterpart. We have noticed that rims of the Golgi cisternae are vacuolated in the DKO. We cannot explain the reason for this morphological change, but one possibility is that lateral connections between cisternae mechanically keep them stretched. Loss of GORASPs and therefore the lateral connections affects this mechanical property causing vacuolation of the rims. This is an important issue for further analysis.
Figure 3.
DKO of GORASP1 and GORASP2 does not lead to Golgi unstacking. (A) Electron micrograph of Golgi stacks in P21 small intestine from mouse treated with TAM at P1. Note that the Golgi are still stacked. (B) Electron micrograph of Golgi stacks in small intestine organoids treated with 4OT or without 4OT treatment for 7 d. Note that the Golgi are still stacked. (C) Quantification of the stack morphometry plotting the stack cross-sectional diameter versus the number of cisternae per stack in the small intestine of mice (±TAM at P1) at P10 (n = 35 and 29) and P42 (n = 40 and 20). Blue is the cDKO and orange the DKO. Note that the cross-sectional diameter is reduced upon TAM treatment but that the number of cisternae per stack is largely unchanged.
The rims of Golgi cisternae are highly fenestrated, and conditions that exaggerate COPI vesicle formation, such as treatment with GTPγS or incubation of isolated Golgi membranes with mitotic cytosol, are reported to convert cisternal rims into small vesicles without severely affecting the core that remains stacked (Warren, 1985). This suggests that rims of the cisternae are functionally and perhaps biochemically different from the core. This also suggests the possibility that the core is stacked by a mechanism different from that linking the rims in a Golgi stack. GORASPs have the property of linking surfaces and bind tethers, golgins, and SNAREs, components needed for COPI vesicle fusion (Wei and Seemann, 2017; Ahat et al., 2019). Perhaps, this linking controls vesicle budding/fusion at the rims. Unlinking of cisternae laterally by GORASP removal would expose more cisternal membrane to COPI vesicle budding, more than enough to compensate for the diminished fusion since the needed components are no longer presented properly. This would create a new steady state with more local COPI vesicles driving increased flux. The increased local vesicles would “contain” the rim membrane and explain the decreased cisternal diameter. In other words, GORASPs control the linking of cisternal rims laterally into a Golgi ribbon, whereas another hitherto uncharacterized set of components additionally cement the core of Golgi cisternae.
GM130 does not compensate for the loss of GORASPs
The cis-Golgi–associated peripheral protein GM130 is diffusely dispersed around the Golgi in GORASP DKO CRISPR HeLa cells (see Fig. 5 A, Bekier et al., 2017). This could potentially compromise the Golgi stacking caused by the GORASP DKO. In other words, the stacking we observe in the GORASP1 and GORASP2 DKO small intestine and organoids could be due to the presence of GM130, or even its up-regulation to compensate for the loss of GORASPs (Lee et al., 2014). Therefore, we stained the same tissue sections as above for GM130 and found that while GM130 was abundant in the cDKO material, it was completely absent in the DKO tissue (Fig. 4 A). This strongly indicates that GM130 is degraded in GORASP1- and GORASP2-lacking cells. Interestingly, it has been shown earlier that depletion of GORASPs by CRISPR resulted in reduction of GM130 by 75% (Bekier et al., 2017). The severity of the effect on GM130 levels might depend on the levels of GORASPs and merits further investigation.
In contrast to GM130, another Golgi-associated protein, Giantin, was readily detected in in every cell of the organoids after 4OT treatment. However, the intensity of Giantin staining is lower compared with untreated organoids. This is likely due to dispersal of Golgi ribbon into smaller stacks, thereby giving the impression of a reduced fluorescence signal (Fig. 4 B). This needs to be quantitated by other approaches. Overall, these findings highlight the need to test the effect of GORASP loss on other Golgi-associated proteins, as it might reveal an important physiologically relevant connection of proteins that could function in concert with GORASPs to control overall features of Golgi organization. It is also important to note that we have tested the involvement of GORASPs in Golgi stacking only in intestine and intestinal organoids. It is difficult to envision a tissue-specific function of GORASPs in Golgi stacking, but we cannot formally rule out this possibility.
Altogether, our data provide a unifying framework for GORASP function. We suggest that a Golgi cisternae is composed of at least two discrete functional domains: a core that can stack in a GORASP-independent manner, and rims that are scaffolded and linked laterally by GORASPs (Ladinsky et al., 1999; Rabouille and Linstedt, 2016). Subcisternal localization of Golgi components indicate specificity in location of Golgi components associated with the core or the periphery of a cisternae (Kweon et al., 2004; Tie et al., 2018). GORASP proteins are concentrated at the rim of a cisternae (Tie et al., 2018), which fits well with their function at the rims. Depletion of GORASPs alters the physical and functional properties of cisternal rims and promotes hypervesiculation, producing a smaller, albeit stacked Golgi. Compounds like Ilimaquinone, on the other hand, control membrane cutting and not surprisingly completely vesiculate Golgi stacks (Takizawa et al., 1993). These data indicate that there are boundaries in a cisterna that separate rims from the core. GORASPs function at the rim, whereas Ilimaquinone starts cutting at the rims and continues to cut across the boundary until the entire cisterna is vesiculated.
Genetic data show that deletion of the single GORASP gene in Dictyostelium discoideum and yeast inhibits starvation-specific secretion of signal sequence lacking ACBA/Acb1 (Kinseth et al., 2007; Duran et al., 2008; Manjithaya et al., 2010). In Drosophila, depletion of the single GRASP orthologue prevents a Golgi-bypassed delivery of the transmembrane protein Integrin to the plasma membrane (Schotman et al., 2008). Clearly, genetics tells us that GORASPs have a role in unconventional secretion. This conclusion is now supported by a large number of reports showing the involvement of GORASPs in other systems, including mammalian cells (Feinstein and Linstedt, 2008; Wu et al., 2020; van Ziel et al., 2019; Noh et al., 2018). Although transport by COPII and COPI is not required for unconventional secretion, the essential role of GORASPs in this process suggests a link to the Golgi membranes. One possibility is that the rims of Golgi cisternae are a necessary source of membranes or a surface for proteins required for unconventional secretion. Loss of rim linking upon depletion of GORASPs therefore affects both processes. The rims of Golgi cisternae are essential for the flux of COPI vesicles during conventional secretion and could be the source of COPI-independent vesicles to transport specific components for Compartment for Unconventional Secretion (CUPS) biogenesis during unconventional protein secretion. The latter, we suggest, is functionally analogous to the export of Atg9 by small vesicles from late Golgi to create an autophagosome.
Materials and methods
Mouse genetics
The generation and characterization of GORASP1 knockin (KI; that act as a knockout) is described elsewhere (Veenendaal et al., 2014). Briefly, embryonic stem cells were transfected with a targeting construct (containing GORASP1tm1a(KOMP) Wtsi allele, neomycin resistance, Lacz, and two LoxP sites) by electroporation. Clones showing homologous recombination were injected into blastocysts from C57BL/6 mice and implanted in pseudopregnant mice. Through backcrossing, a mouse line homogenous for the recombination of the GORASP1 genomic locus to the targeting vector was obtained (GORASP1KI).
GORASP2 KI/KO mice were generated at McLaughlin Research Institute (Great Falls, MT) using the EUCOMM/KOMP knockout first conditional-ready targeted embryonic stem cell resource (http://www.mousephenotype.org/about-ikmc/eucomm-proram/eucomm-targeting-strategies; targeted trap tm1a allele). In short, Gorasp2tm1a(EUCOMM)Hmgu embryonic stem cells were injected into C57BL/6J-Tyr<c-2J> blastocysts to generate chimeras that were bred to C57BL/6J-Tyr<c-2J> mice. Offspring were subsequently brother–sister mated for several generations.
We first attempted to generate a double KI by crossing the mice to homozygosity, but we were unable to recover pups of the right genotype, suggesting that the loss of both GORASPs is critical for embryonic development. We therefore resorted to use the GORASP2 cFlox allele of the GORASP2 KI mouse: GORASP2 cFlox mouse. Conditional knockout (cFlox) mice were generated by breeding the tm1a allele carrying mice with Flp-recombinase–expressing mice, removing the LacZ and neomycin cassettes and resulting in mice expressing endogenous GORASP2, with exon 5 being flanked by LoxP sites (tm1c allele; Fig. 1 B). Primers 5′-CCGATCCTTTCCTCAGCCGTCAGC-3′, 5′-CCTCAAGGCCCTACCTTAGC-3′, and 5′-TGAACTGATGGCGAGCTCAGACC-3′ were used to genotype the offspring using genomic DNA extracted from ear punches (MouseDirect PCR kit; Biotools).
We then crossed the GORASP2 cFlox mouse and the GORASP1 KI mouse to a Rosa Cre-ERT2 mouse and through back-crossing generated a GORASP1 KI homologous (HOM); GORASP2 cFlox HOM; Rosa Cre ERT2 HOM mouse that we called the “cDKO” line. Upon injection of 20 µl of 5 mg/ml TAM (T5648; Sigma) 1 d after birth (P1), GORASP2 exon 5 is excised (GORASP2 FLOX; Fig. 1, C and D) and the full-length protein is no longer expressed (see Results). Mice sacrificed at P42 were reinjected at P21 and P39 with 170 µl of 5 mg/ml TAM. Controls were performed by injected oil to a parallel litter of the same genotype. Note that exons 6–10 are still present in the genomic locus. This yields a GORASP2 mRNA that misses exon 5 and a complete loss of GORASP2 protein (IF and Western blot; Fig. S1, A–C).
cDKO organoids
We generated small intestine organoids using the published procedure (Beumer et al., 2018) using a 6-wk-old female GORASP1 KI HOM; GORASP2 cFlox HOM; Rosa Cre ERT2 HOM mouse. We used Cultrex Reduced Growth Factor Basement Membrane Extract, type II (3533–005-02) instead of Matrigel. To deplete GORASP2 gene, 100 nM 4OT (H7904; Sigma) was added on budding large organoids for 7 d.
Monitoring exon 5 excision
For DNA extraction, 20 mg proximal gut tissue (flushed, washed, and snap frozen) was used as described in the DNeasy Blood & Tissue Kit (69504; Qiagen) to recover 30 ng/µl DNA.
RNA was extracted from 650–700 organoids harvested with Corning Cell Recovery Solution (354253; Corning) removing BME gel. After washing with PBS, the RNA was extracted as recommended by the RNeasy Mini Kit (Qiagen, 74104). This yielded ∼20 ng/µl of RNA. Of all samples, a total of 75 ng cDNA in 20 µl was synthesized by GoScript Reverse transcription (A5000; Promega; Table 1).
Table 1. Primers for PCR shown in Figs. 1 and S1.
| Name | Sequence (5′ to 3′) | Annealing | |
|---|---|---|---|
| a | F-GORASP2 ex4 | CCGATCCTTTCCTCAGCCGTCAGC | 62.0°C |
| b | R-GORASP2 ex4LoxP | TGTGGAGTCCTTCCTTGCTC | |
| c | F-GORASP2 ex3 | GGGTTAGCATTCGTTTCTGC | 53.4°C |
| d | R-GORASP2 ex5 | CATGCAGAGTTTGGTGTGATG | |
| e | F-GORASP2 int5 | GAGCAAGGAAGGACTCCACA | 56.8°C |
| f | R-GORASP2 int5 | CAAGAGGCCGAGATCCTGAG | |
| g | F-GORASP2 ex6 | CGAATACCTACACGCCCCTT | 57.2°C |
| h | R-GORASP2 ex7 | TGCCGAGCTAATGGAGAGTC | |
| i | F-GORASP2 ex8 | ACTGTACCATTGCCGCC | 58.4°C |
| j | R-GORASP2 ex9 | GAGCTCTGGCAAACCGACC | |
| k | F-GORASP2 ex4 | AATCCAATTCCCCAGCG | 57.7°C |
| l | R-GORASP2 ex6 | GGGGGTACCAGTCATTTGTCC | |
| m | F-GORASP2 ex1 | CAAGTGCCACGTCCCAAGAG | 58.5°C |
| n | R-GORASP2 ex4 | CTGTGAGGTCTCAATCCTGCC | |
| o | F-GORASP2 ex6 | CGAATACCTACACGCCCCTT | 57.2°C |
| p | R-GORASP2 ex10 | ATGCCAGGTAAGTTTCGGGG | |
| F-GAPDH ex3–4 | TGTCGTGGAGTCTACTGGTG | 53.1°C | |
| R-GAPDH ex3–4 | ACACCCATCACAAACATGG |
List of primers used in this study. F, Forward; R, Reverse; int, intron; ex, exon.
Western blot
1,500 organoids treated or not with 100 nM 4OT for 7 d were harvested using 1 ml Corning Cell Recovery Solution (354253; Corning) per ∼60 µl BME gel. After centrifugation, they were lysed in 100 µl RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 5 mM EDTA, 50 mM NaF, and 1 mM PMSF supplemented with Roche 1X Halt Protease inhibitor cocktail), incubated for 1 h at 4°C, and sonicated using a Bioruptor PLUS (Diagenode) for 30 cycles of 30 s spaced by 30 s of cooling down. Total protein concentration in lysates was determined using Pierce BCA Protein Assay Kit (23227; Thermo Scientific). The protein concentration in the lysates was 1.74 µg/µl. Proteins were fractionated in 10% polyacrylamide gel and blotted on polyvinylidene difluoride membrane. Milk was used as a blocking agent.
Membranes were incubated with a Rabbit anti GORASP2 (Rich, against aa 232–454), at dilution 1/1,000; a gift from M. Bekier (University of Michigan, Ann Arbor, MI; Xiang and Wang, 2010); and a mouse monoclonal antibody to E-cadherin (610182 at 1/5,000; BD Biosciences).
The visualization was performed after incubation with secondary antibodies coupled to HRP (NA931 and NA934; GE Healthcare) and Clarity Western ECL Substrate (Bio-Rad) using ImageQuant LAS4000 ECL (GE Healthcare).
IF microscopy
The small intestine of oil- and TAM-injected cDKO mice were dissected at P10, P21, and P42 and then flushed and fixed in formalin (Merck) overnight. It was then processed and embedded in paraffin.
Sections were cut on a microtome, rehydrated, boiled in citrate, blocked with 1% BSA, and stained with primary antibodies followed by fluorescent secondary antibodies as described previously (Beumer et al., 2018). Slides were observed under an SPE Leica microscope with HCX PL APO 63x 1.40NA objective and antifade mounting media (Vectashield, H1200). Images were acquired with Leica acquisition software.
Budding organoids treated with and without 100nM 4OT for 7 d and were fixed directly in the gel leading them to stick tightly to the glass coverslip that is set underneath the gel drop. Coverslips were simply removed and processed for IF after permeabilization using 0.5% Triton X-100 for 30 min. SPE Leica microscope with HCX PL APO 63x 1.40NA objective and antifade mounting media (Prolong, Invitrogen P36935) was used to capture images.
We used a rabbit anti GORASP2 antibody (Rich, against aa 232–454) from M. Bekier (Xiang and Wang, 2010) at dilution 1/200, a rabbit anti GM130 antibody (MLO8, at dilution 1/100), gift from M. Lowe (University of Manchester, Manchester, UK), a rabbit anti Giantin antibody (TA10, at dilution 1/100), gift from F. Perez (Institut Curie, Paris, France) and a mouse anti E-cadherin antibody (610182 at 1/1,000; BD Biosciences). Primary antibodies were coupled to donkey anti-rabbit Alexa Fluor 568 (A10042; Invitrogen) or goat anti-mouse Alexa Fluor 488 (A11001; Invitrogen).
Electron microscopy
A small part of the intestine that is close to the part processed for IF was fixed in Karnovsky (2% glutaraldehyde, 2% PFA, 0.25 mM CaCl2, and 0.5 mM MgCl2 in phosphate buffer [0.1 M at pH 7.4]) overnight at room temperature followed by 1% PFA at 4°C. After processing, they were embedded in epon resin. Ultrathin plastic sections were cut and examined under an FEI Tecnai T12 Spirit microscope with an Olympus Veleta camera at magnification 18,500× and acquisition software TIA V4.7 SP3 (FEI), SerialEM (University of Colorado; Slot and Geuze, 2007). Images of crypts and villi were acquired and stitched together (software IMOD 4.9; Etomo; University of Colorado) to allow a view of the small intestine section both at low and a high magnification.
Quantification
The cross-sectional diameter of each stack that is visible in the intestinal section was measured using a ruler on high magnification pictures (20,000×). Using the same pictures, the number of cisternae per stack was counted and plotted against the cross-sectional diameter.
Each recognizable Golgi area was accounted for, whether a stack is present or not. 0 in the graph means that a Golgi area was identified but that no stacks were present.
Online supplemental material
Fig. S1 shows GORASP2 mRNA in cDKO and DKO organoids. Fig. S2 shows that DKO of GORASP1 and GORASP2 does not lead to Golgi unstacking in P10 mouse small intestine. Fig. S3 shows that DKO of GORASP1 and GORASP2 does not lead to Golgi unstacking in P42 mouse small intestine.
Figure S3.
DKO of GORASP1 and GORASP2 does not lead to Golgi unstacking in P42 mouse small intestine. (A) A gallery of Golgi stack profile in sections of small intestine control mouse at P42 injected with oil at P1. (B) A gallery of Golgi stack profile in sections of small intestine mouse at P42 injected with TAM at P1. Note that as in P10, the stacks are shorter in cross-sectional diameter but that the number of cisternae per stack is unchanged upon TAM injection. Quantification is shown in Fig. 3 C. Arrows indicate Golgi structures.
Acknowledgments
We thank Graham Warren for helping us present a balanced discussion. We thank the members of the Clevers, Malhotra, and Rabouille groups for their thoughts and input on this project, specifically Laura Zeinstra for her help with creating the organoids. A special thanks to the Animal Facility at the Hubrecht Institute, Anko de Graaff, the Hubrecht Imaging Centre, and Jeroen Korving and Harry Begthel of the Histology Department. All mouse experiments were conducted under a project license granted by the Central Committee Animal Experimentation of the Dutch government and approved by the KNAW-Hubrecht Institute Animal Welfare Body.
V. Malhotra is an Institució Catalana de Recerca i Estudis Avançats professor at the Centre for Genomic Regulation, and work in his laboratory is funded by the Ministerio de Economía y Competitividad (grants SEV-2012-0208, BFU2013-44188-P, and CSD2009-00016). I. Raote acknowledges funding from the Spanish Ministry of Science and Innovation (IJCI-2017-34751).
The authors declare no competing financial interests.
Author contributions: Conceptualization, V. Malhotra and C. Rabouille; Data curation, V. Malhotra and C. Rabouille; Formal analysis, V. Malhotra and C. Rabouille; Funding acquisition, V. Malhotra and C. Rabouille; Investigation, R. Grond, T. Veenendaal, S. Corstjens, L. Delfgou, and B. El Haddouti; Methodology, J. Duran, J.H. van Es, V. Malhotra, and C. Rabouille; Project administration, V. Malhotra and C. Rabouille; Resources, J.H. van Es, V. Malhotra, and C. Rabouille; Supervision, V. Malhotra and C. Rabouille; Validation, R. Grond, V. Malhotra, and C. Rabouille; Visualization, R. Grond and C. Rabouille; Writing – original draft, V. Malhotra and C. Rabouille; Writing – review and editing, R. Grond, I. Raote, and V. Malhotra.
References
- Ahat E., Li J., and Wang Y.. 2019. New Insights Into the Golgi Stacking Proteins. Front. Cell Dev. Biol. 7:131 10.3389/fcell.2019.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachert C., and Linstedt A.D.. 2010. Dual anchoring of the GRASP membrane tether promotes trans pairing. J. Biol. Chem. 285:16294–16301. 10.1074/jbc.M110.116129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F.A., Puype M., Vandekerckhove J., and Warren G.. 1997. GRASP65, a protein involved in the stacking of Golgi cisternae. Cell. 91:253–262. 10.1016/S0092-8674(00)80407-9 [DOI] [PubMed] [Google Scholar]
- Bekier M.E. II, Wang L., Li J., Huang H., Tang D., Zhang X., and Wang Y.. 2017. Knockout of the Golgi stacking proteins GRASP55 and GRASP65 impairs Golgi structure and function. Mol. Biol. Cell. 28:2833–2842. 10.1091/mbc.e17-02-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer J., Artegiani B., Post Y., Reimann F., Gribble F., Nguyen T.N., Zeng H., Van den Born M., Van Es J.H., and Clevers H.. 2018. Enteroendocrine cells switch hormone expression along the crypt-to-villus BMP signalling gradient. Nat. Cell Biol. 20:909–916. 10.1038/s41556-018-0143-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartier-Michaud A., Bailly A.L., Betzi S., Shi X., Lissitzky J.C., Zarubica A., Sergé A., Roche P., Lugari A., Hamon V., et al. . 2017. Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis. PLoS Genet. 13 e1006803 10.1371/journal.pgen.1006803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran J.M., Kinseth M., Bossard C., Rose D.W., Polishchuk R., Wu C.C., Yates J., Zimmerman T., and Malhotra V.. 2008. The role of GRASP55 in Golgi fragmentation and entry of cells into mitosis. Mol. Biol. Cell. 19:2579–2587. 10.1091/mbc.e07-10-0998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein T.N., and Linstedt A.D.. 2008. GRASP55 regulates Golgi ribbon formation. Mol. Biol. Cell. 19:2696–2707. 10.1091/mbc.e07-11-1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvela T., and Linstedt A.D.. 2014. Isoform-specific tethering links the Golgi ribbon to maintain compartmentalization. Mol. Biol. Cell. 25:133–144. 10.1091/mbc.e13-07-0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinseth M.A., Anjard C., Fuller D., Guizzunti G., Loomis W.F., and Malhotra V.. 2007. The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell. 130:524–534. 10.1016/j.cell.2007.06.029 [DOI] [PubMed] [Google Scholar]
- Kondylis V., Spoorendonk K.M., and Rabouille C.. 2005. dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol. Biol. Cell. 16:4061–4072. 10.1091/mbc.e04-10-0938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon H.S., Beznoussenko G.V., Micaroni M., Polishchuk R.S., Trucco A., Martella O., Di Giandomenico D., Marra P., Fusella A., Di Pentima A., et al. . 2004. Golgi enzymes are enriched in perforated zones of golgi cisternae but are depleted in COPI vesicles. Mol. Biol. Cell. 15:4710–4724. 10.1091/mbc.e03-12-0881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky M.S., Mastronarde D.N., McIntosh J.R., Howell K.E., and Staehelin L.A.. 1999. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J. Cell Biol. 144:1135–1149. 10.1083/jcb.144.6.1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Tiwari N., Dunlop M.H., Graham M., Liu X., and Rothman J.E.. 2014. Membrane adhesion dictates Golgi stacking and cisternal morphology. Proc. Natl. Acad. Sci. USA. 111:1849–1854. 10.1073/pnas.1323895111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi S.K., Bhattacharyya D., Strack R.L., Austin J.R. II, and Glick B.S.. 2010. The yeast GRASP Grh1 colocalizes with COPII and is dispensable for organizing the secretory pathway. Traffic. 11:1168–1179. 10.1111/j.1600-0854.2010.01089.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq J.M., and Warren G.. 1987. Fragmentation and partitioning of the Golgi apparatus during mitosis in HeLa cells. EMBO J. 6:3239–3246. 10.1002/j.1460-2075.1987.tb02641.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R., Anjard C., Loomis W.F., and Subramani S.. 2010. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol. 188:537–546. 10.1083/jcb.200911149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T., and Warren G.. 1995. A role for tubular networks and a COP I-independent pathway in the mitotic fragmentation of Golgi stacks in a cell-free system. J. Cell Biol. 130:1027–1039. 10.1083/jcb.130.5.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Lowe M., Levine T.P., Rabouille C., and Warren G.. 1997. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 89:445–455. 10.1016/S0092-8674(00)80225-1 [DOI] [PubMed] [Google Scholar]
- Noh S.H., Gee H.Y., Kim Y., Piao H., Kim J., Kang C.M., Lee G., Mook-Jung I., Lee Y., Cho J.W., et al. . 2018. Specific autophagy and ESCRT components participate in the unconventional secretion of CFTR. Autophagy. 14:1761–1778. 10.1080/15548627.2018.1489479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot M.Y., and Malhotra V.. 2006. The Golgi apparatus maintains its organization independent of the endoplasmic reticulum. Mol. Biol. Cell. 17:5372–5380. 10.1091/mbc.e06-06-0565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M.A., Bachert C., Puri S., Lanni F., and Linstedt A.D.. 2006. GM130 and GRASP65-dependent lateral cisternal fusion allows uniform Golgi-enzyme distribution. Nat. Cell Biol. 8:238–248. 10.1038/ncb1366 [DOI] [PubMed] [Google Scholar]
- Rabouille C., and Linstedt A.D.. 2016. GRASP: A Multitasking Tether. Front. Cell Dev. Biol. 4:1 10.3389/fcell.2016.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C., Misteli T., Watson R., and Warren G.. 1995. Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system. J. Cell Biol. 129:605–618. 10.1083/jcb.129.3.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotman H., Karhinen L., and Rabouille C.. 2008. dGRASP-mediated noncanonical integrin secretion is required for Drosophila epithelial remodeling. Dev. Cell. 14:171–182. 10.1016/j.devcel.2007.12.006 [DOI] [PubMed] [Google Scholar]
- Shorter J., Watson R., Giannakou M.E., Clarke M., Warren G., and Barr F.A.. 1999. GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18:4949–4960. 10.1093/emboj/18.18.4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot J.W., and Geuze H.J.. 2007. Cryosectioning and immunolabeling. Nat. Protoc. 2:2480–2491. 10.1038/nprot.2007.365 [DOI] [PubMed] [Google Scholar]
- Sütterlin C., Polishchuk R., Pecot M., and Malhotra V.. 2005. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol. Biol. Cell. 16:3211–3222. 10.1091/mbc.e04-12-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P.A., Yucel J.K., Veit B., Faulkner D.J., Deerinck T., Soto G., Ellisman M., and Malhotra V.. 1993. Complete vesiculation of Golgi membranes and inhibition of protein transport by a novel sea sponge metabolite, ilimaquinone. Cell. 73:1079–1090. 10.1016/0092-8674(93)90638-7 [DOI] [PubMed] [Google Scholar]
- Tie H.C., Ludwig A., Sandin S., and Lu L.. 2018. The spatial separation of processing and transport functions to the interior and periphery of the Golgi stack. eLife. 7 e41301 10.7554/eLife.41301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ziel A.M., Largo-Barrientos P., Wolzak K., Verhage M., and Scheper W.. 2019. Unconventional secretion factor GRASP55 is increased by pharmacological unfolded protein response inducers in neurons. Sci. Rep. 9:1567 10.1038/s41598-018-38146-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenendaal T., Jarvela T., Grieve A.G., van Es J.H., Linstedt A.D., and Rabouille C.. 2014. GRASP65 controls the cis Golgi integrity in vivo. Biol. Open. 3:431–443. 10.1242/bio.20147757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinke F.P., Grieve A.G., and Rabouille C.. 2011. The multiple facets of the Golgi reassembly stacking proteins. Biochem. J. 433:423–433. 10.1042/BJ20101540 [DOI] [PubMed] [Google Scholar]
- Wang Y., Seemann J., Pypaert M., Shorter J., and Warren G.. 2003. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. EMBO J. 22:3279–3290. 10.1093/emboj/cdg317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Satoh A., and Warren G.. 2005. Mapping the functional domains of the Golgi stacking factor GRASP65. J. Biol. Chem. 280:4921–4928. 10.1074/jbc.M412407200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. 1985. Membrane traffic and organelle division. Trends Biochem. Sci. 10:439–443. 10.1016/0968-0004(85)90027-1 [DOI] [Google Scholar]
- Wei J.H., and Seemann J.. 2017. Golgi ribbon disassembly during mitosis, differentiation and disease progression. Curr. Opin. Cell Biol. 47:43–51. 10.1016/j.ceb.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Li T., and Zhao J.. 2020. GRASP55: A Multifunctional Protein. Curr. Protein Pept. Sci. 10.2174/1389203721666200218105302 [DOI] [PubMed] [Google Scholar]
- Xiang Y., and Wang Y.. 2010. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J. Cell Biol. 188:237–251. 10.1083/jcb.200907132 [DOI] [PMC free article] [PubMed] [Google Scholar]