Abstract
Background/Aims
Whereas colorectal cancer (CRC) is the third most common cancer worldwide, klotho gene has been reported as a tumor suppressor gene. Therefore, the aim of this study was to investigate the association between klotho (rs1207568 and rs564481) variants and CRC in Egyptian patients.
Materials and Methods
A case-control study comprising 100 patients with CRC and 100 age- and sex-matched healthy controls was conducted. Genotyping of klotho was performed by polymerase chain reaction with confronting two-pair primers.
Results
The frequencies of the A allele of rs1207568 and the AC haplotype were significantly higher in patients with CRC than in the controls (p=0.019 and p=0.005, respectively).
Conclusion
We propose that klotho (rs1207568 and rs564481) variants play a significant role in colorectal carcinogenesis and that the klotho protein could be a target for oncotherapy.
Keywords: Klotho, colorectal cancer, gene variation, klotho rs1207568, klotho rs564481
INTRODUCTION
Colorectal cancer (CRC) is one of the most prevalent cancers, ranking as the third most common cancer worldwide (1). The incidence rate of CRC has been increasing steadily; thus, CRC is of great international concern for researchers (2).
Although CRC ranks second in terms of cancer-related deaths (1), it is considered a preventable cancer because genetic changes precede the onset of the disease by approximately 10 years (3). Thus, the development of CRC can be prevented, and numerous research groups are currently studying these genetic changes.
Screening for precancerous colorectal lesions results in improved outcomes and is important for disease prevention. Although considerable advances have been made in diagnostic procedures for CRC, many patients with CRC are still diagnosed in late stages (4). Thus, additional studies are needed to improve our understanding of the molecular alterations involved in CRC to identify diagnostic, prognostic, and predictive markers that can enhance prevention, early detection, and treatment (5).
In 1997, the klotho protein was identified as an anti-aging protein by studies showing that mice with a klotho protein deficiency were more susceptible to aging diseases (6,7). Previous scientific research indicates an anticancer role for klotho protein in addition to its anti-aging properties, and this anticancer role has been studied in many cancers, including lung cancer, breast cancer, colon cancer, and pancreatic cancer. The anticancer activity of klotho is thought to be attributed to the dysregulation of tumor cell proliferation and apoptosis (8–11). Further research on klotho is anticipated to provide new insight into targeted cancer therapy (12).
The aim of this study was to investigate the association between two single nucleotide variants (SNVs) in the klotho gene (rs1207568, in the promoter region and rs564481, located in exon 4) and CRC in Egyptian patients.
Klotho rs1207568 (c.-395G>A) is an SNV, located in the promoter region of the klotho gene, in which guanine nucleotide is replaced by adenine (13), whereas klotho rs564481(c.1767C>T, p.His589=) is a synonymous variant, located in exon 4, where cytosine nucleotide is substituted by thymine at codon 1767 resulting in no amino acid (Histidine) change (14).
Despite the abundance of previous studies suggesting a significant role of klotho in tumorigenesis and cancer progression, data and published studies regarding the role of variations in the klotho gene (rs1207568 and rs564481) and their association with CRC are scarce. Therefore, studies in numerous populations are needed to clarify this association.
MATERIALS AND METHODS
This study included 100 histopathologically confirmed CRC cases and 100 healthy controls. Clinical data, including age; sex; and tumor localization, staging, and grading, were collected from these patients. The characteristics of the patients with CRC and controls are summarized in Table 1.
Table 1.
Descriptive data of the study subjects.
| Demographics | Patients with CRC (n=100) | Healthy Controls (n=100) | p |
|---|---|---|---|
| Age (years) | 46.6±13.9 | 43.4±9.9 | 0.061 |
| Male Sex, % | 46.0 | 37.0 | 0.169 |
| Site, % | |||
| Colon | 53.0 | — | — |
| Rectum | 47.0 | — | — |
| Differentiation Degree, % | |||
| Low | 17.0 | — | — |
| Moderate and High | 83.0 | — | — |
| Histological type, % | |||
| Mucinous | 29.0 | — | — |
| Adenocarcinoma | 71.0 | — | — |
| Tumor Stage, % | |||
| I + II | 30.0 | — | — |
| III + IV | 70.0 | — | — |
| Polyp, % | 22.0 | — | — |
| CEA (ng/mL) | 2.2 (1.2–19.4) | — | — |
| CA 19-9 (IU/mL) | 14.2 (3.7–49.0) | — | — |
Normally distributed variables are presented as mean ± standard deviation. Skewed variables are presented as median (interquartile range).
CRC: colorectal carcinoma; CEA: carcinoembryonic antigen; CA 19-9: cancer antigen 19-9.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Genotyping of SNVs in the klotho gene
Blood samples of 2 ml were collected from each study participant into ethylenediaminetetraacetic acid–containing tubes. Genomic DNA was extracted using a TINAamp genomic DNA extraction kit (Tiangen Biotech, Beijing, China). Genotyping of the rs1207568 and rs564481 variants in the klotho gene was performed using polymerase chain reaction (PCR) with confronting two-pair primers (15) according to the protocol proposed by Shimoyama et al. (16). In this assay, confronting pairs of primers (a total of four primers) were used (Operon Biotechnologies, Germany).
The following primers were used to amplify the klotho rs1207568—Forward primer 1, 5′-GTTTCGTGGACGCTCAGGTTCATTCTC-3′; Reverse primer 1, 5′-GATCCCGCCCCCAAGTCGGGA-3′; Forward primer 2, 5′-GAGAAAAGGCGCCGACCAACTTTC-3′; and Reverse primer 2, 5′-GTCCCTCTAGGATTTCGGCCAG-3′.
The PCR conditions were as follows: an initial denaturation step at 95 °C for 10 min; 35 cycles at 95 °C for 1 min, annealing at 65 °C for 1 min, and 72 °C for 1 min; and a final elongation step at 72 °C for 5 min. PCR products were evaluated using 2% agarose gels stained with ethidium bromide. The GG genotype appeared as two bands of 252 and 175 bp; the GA genotype appeared as three bands of 252, 175, and 121 bp; and the AA genotype appeared as two bands of 252 and 121 bp (Figure 1).
Figure 1.
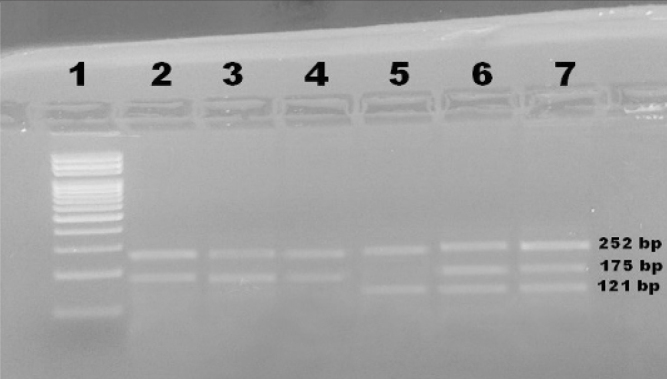
Agarose gel showing PCR-CTPP analysis of the klotho c.-395G>A variant. Lane 1 shows 100 bp DNA ladder, lanes 2, 3 & 4 show the homozygous GG genotype, lane 5 shows the homozygous AA genotype, lanes 6 and 7 show the heterozygous GA genotype.
The following primers were used to amplify the klotho rs564481—Forward primer 1, 5′-CTCAGTTTACCGACCTGAATGTTTACCTG-3′; Reverse primer 1, 5′-GTCCAGGGAGAAGCGAAAATGTGTAACA-3′; Forward primer 2, 5′-CAGATCGCTTTACTCCAGGAAATGCAC-3′; and Reverse primer 2, 5′-GAGCTCTTGAAAGCACAGTCGGGC-3′.
The PCR conditions were as follows: an initial denaturation step at 95 °C for 10 min; 30 cycles at 95 °C for 1 min, annealing at 69 °C for 1 min, and 72 °C for 1 min; and a final elongation step at 72 °C for 5 min. PCR products were evaluated using 2% agarose gels stained with ethidium bromide. The CC genotype appeared as two bands of 416 and 291 bp; the CT genotype appeared as three bands of 416, 291, and 179 bp; and the TT genotype appeared as two bands of 416 and 179 bp (Figure 2).
Figure 2.
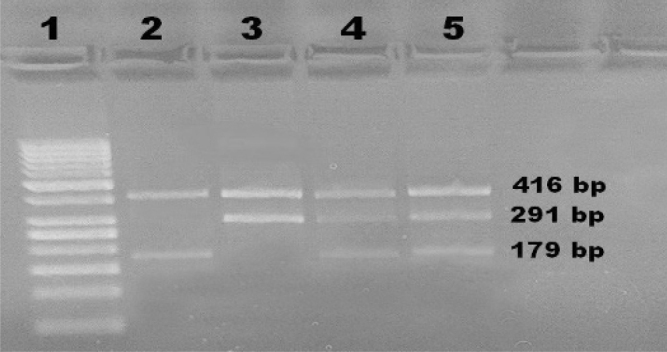
Agarose gel showing PCR-CTPP analysis of the klotho c.1767C>T variant. Lane 1 shows 50 bp DNA ladder, lane 2 shows the homozygous TT genotype, lane 3 shows the homozygous CC genotype, lanes 4 and 5 show the heterozygous CT genotype.
Statistical Analysis
The Statistical Package for Social Science, version 16.0 (SPSS Inc.; Chicago, IL, USA), was used for statistical analysis in this study. In the controls, each polymorphism was tested for Hardy–Weinberg equilibrium (HWE). The haplotype frequencies were analyzed using the Web-based calculator SNPstats (17). The chi-square test and Fisher’s exact test were used to analyze the association between klotho gene variants and CRC. The odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to determine the strength of the association.
RESULTS
The descriptive data of the patients with CRC and healthy control subjects are summarized in Table 1. The patients with CRC and healthy controls were age- and sex-matched.
In the control group, both the klotho rs1207568 and rs564481 SNVs were in HWE. The minor allele frequencies of the klotho rs1207568 and rs564481 SNVs were 0.29 and 0.22, respectively, in patients with CRC.
Table 2 shows the klotho genotypes and allele frequencies in the patients with CRC and healthy control volunteers. The frequencies of both the A allele and the combined (G/A–A/A) genotype of the klotho rs1207568 variant were significantly higher in the patients with CRC than in the controls (p=0.019 and p=0.007, respectively). However, no significant difference was found in the distribution of the klotho rs564481 variant or the allele frequencies between the patients with CRC and the healthy controls (Table 2).
Table 2.
The genotype and allele frequencies of klotho genetic variations in the CRC and healthy control groups.
| Variant | Healthy Controls (n=100) | Patients with CRC (n=100) | OR (95% CI) | p |
|---|---|---|---|---|
| rs1207568, % | ||||
| G/G | 63.0 | 44.0 | 1.00 | — |
| G/A | 36.0 | 54.0 | 2.15 (1.21–3.80) | 0.008 |
| A/A | 1.0 | 2.0 | 2.86 (0.25–32.56) | 0.570 |
| G/G | 63.0 | 44.0 | 1.00 | |
| G/A–A/A | 37.0 | 56.0 | 2.2 (1.2–3.8) | 0.007 |
| G allele | 81.0 | 71.0 | 1.00 | |
| A allele | 19.0 | 29.0 | 1.7 (1.1–2.8) | 0.019 |
| rs564481, % | ||||
| C/C | 62.0 | 57.0 | 1.00 | — |
| C/T | 34.0 | 41.0 | 1.31 (0.73–2.34) | 0.843 |
| T/T | 4.0 | 2.0 | 0.54 (0.10–3.08) | 0.683 |
| C/C | 62.0 | 57.0 | 1.00 | |
| C/T–T/T | 38.0 | 43.0 | 1.2 (0.7–2.2) | 0.470 |
| C allele | 79.0 | 78.0 | 1.00 | |
| T allele | 21.0 | 22.0 | 1.1 (0.7–1.8) | 0.716 |
CI: confidence interval; CRC: colorectal cancer; OR: odds ratio.
Regarding the klotho haplotype distribution, the AC haplotype was significantly associated with an increased risk of CRC development (OR=2.66; 95% CI 1.35–5.25; p=0.005; Table 3).
Table 3.
Analysis of klotho haplotype frequencies with the risk of CRC.
| Haplotype (rs1207568 and rs564481) | Total Frequency | Control Frequency | CRC Frequency | OR (95% CI) | p |
|---|---|---|---|---|---|
| GC | 0.5797 | 0.6401 | 0.5071 | 1.00 | — |
| AC | 0.2028 | 0.1499 | 0.2679 | 2.66 (1.35–5.25) | 0.005 |
| GT | 0.1803 | 0.1699 | 0.2029 | 1.49 (0.77–2.87) | 0.24 |
| AT | 0.0372 | 0.0401 | 0.0221 | 0.88 (0.09–8.25) | 0.91 |
CI: confidence interval; CRC: colorectal cancer; OR: odds ratio.
We further analyzed the association between klotho genotypes and clinical manifestations of CRC. However, no significant associations were found between klotho variants and any of the clinical characteristics of CRC.
DISCUSSION
In this study, we analyzed the relationship between klotho gene variants (rs1207568 and rs564481) and CRC in Egyptian patients to better understand the ways in which these genetic alterations influence CRC. To our knowledge, this study is the first to investigate the relationship between klotho variants (rs1207568 and rs564481) and CRC in this population.
The A allele of klotho rs1207568 was found to be significantly associated with an increased risk of CRC development. On the other hand, no significant association was found between the klotho rs564481 variant and CRC. However, when both the rs1207568 and rs564481 variants were considered, the frequency of the AC haplotype was significantly higher in the patients with CRC than in the healthy controls, indicating that the rs564481 variant influences CRC development, potentially through synergistic action with the rs1207568 variant.
We further studied the association of the klotho rs1207568 and rs564481 genotypes with different demographic characteristics, histological tumor types, tumor stages, and CRC tumor markers, but no significant relationships were found.
In humans, the klotho gene is located at chromosome region 13q13.1 and consists of six exons (18). Humans express two types of the klotho protein: membrane-bound klotho, which is a coreceptor of fibroblast growth factor (FGF) 23 and regulates phosphate and vitamin D homeostasis (19, 20), and secreted klotho, which acts as a humoral factor that mediates anti-aging effects through the regulation of oxidative stress, receptors of multiple growth factors, and ion channels (21, 22).
Previous research has reported a link between a low klotho protein level in the blood and an increased risk of cancers, for example, pancreatic cancer and hepatocellular cancer. The suggested mechanism acts through epigenetic modulation, including promoter methylation and histone deacetylation. In addition, klotho downregulation was found to enhance cancer cell proliferation and decrease apoptosis through possible mechanisms involving the FGF signaling pathway, the insulin-like growth factor 1 receptor pathway, or the wnt/β-catenin signaling pathway (12).
Approximately 10 variations have been detected in the klotho gene (16). In this study, we focused on only two, rs1207568 (c.-395G>A), located in the promoter region, and rs564481 (p.His589=), located in exon 4, and their relation to CRC. We found that the rs1207568 variant and the AC haplotype were significantly associated with increased CRC risk. Our results agree with those of Liu et al. (23) in Chinese patients.
Study of the functional mechanism of the klotho rs1207568 variant revealed a correlation between the A allele and a low level of klotho protein expression in human vascular tissue (24). This finding is consistent with those of previous studies, which speculated that the A allele of this genetic variant reduces transcription factor binding SP1 and hence klotho expression levels (25). However, Kawano et al. (25) failed to find such an association between the G and A alleles and klotho protein expression levels in cultured human cells.
The potential effect of the rs1207568 variant on klotho gene function remains speculative. The variation in the results of different studies could be due to the existence of regulatory sites other than those related to the rs1207568 variant that also regulate the expression and function of klotho. In addition, rs1207568 is in strong linkage disequilibrium with other functional genetic variations (such as c.110G>C, p.His589= and c.2298C>T) and could act as a surrogate for these variants (26).
Based on these studies and our results, we hypothesized that the A allele of the klotho rs1207568 variant could reduce the level or activity of klotho in humans, thereby increasing the risk of CRC.
Although rs564481, located in exon 4, is a synonymous variant where a nucleotide transition from C to T resulted in no amino acid change, previous studies have demonstrated the possibility that this type of variations affects protein function, potentially through effects on mRNA stability and processing, translation kinetics, and protein folding (27).
Aberrant expression of klotho has been observed in several cancers. Klotho acts as a tumor suppressor in most cancers, as klotho overexpression results in the suppression of cancer cell proliferation. Because klotho expression is downregulated in most cancers, knowledge about the klotho protein will likely provide new insight into therapy for cancers, including CRC (28–31).
In conclusion, our study suggests that klotho (rs1207568 and rs564481) variants play a significant role in colorectal carcinogenesis. However, further investigations are required to elucidate the detailed mechanisms of klotho genetic variations in cancers in general and in CRC specifically.
MAIN POINTS.
Colorectal cancer (CRC) is ranking as the third most common cancer worldwide and ranks second in terms of cancer-related deaths.
klotho protein has both antiaging and anticancer properties.
klotho downregulation was found to enhance cancer cell proliferation and decrease apoptosis.
The A allele of klotho rs1207568 variant was associated with increased risk of CRC in Egyptian patients.
klotho (rs1207568 and rs564481) variants play a significant role in colorectal carcinogenesis.
Footnotes
Ethics Committee Approval: Ethics committee approval was received from the Ethics Committee of Faculty of medicine, Cairo University.
Informed Consent: Informed consent was obtained from the patients who participated in this study.
Peer-review: Externally peer-reviewed.
Author Contributions: Concept – A.K., I.S., M.S.; Design – A.K.; Supervision – I.S.; Resource – A.K., I.S., A.M., M.S., Amr Kamal; Materials – A.K., M.S.; Data Collection and/or Processing – M.S., A.M., Amr Kamal; I.S.; Analysis and/or Interpretation – A.K.; Literature Search – A.K., A.M.; Writing – A.K., A.M.; Critical Reviews – I.S.
Conflict of Interest: The authors have no conflict of interest to declare.
Financial Disclosure: The authors declared that this study has received no financial support.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Pox CP. Controversies in colorectal cancer screening. Digestion. 2014;89:274–81. doi: 10.1159/000363287. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez-Pons M, Cruz-Correa M. Colorectal cancer biomarkers: where are we now? BioMed Res Int. 2015;2015 doi: 10.1155/2015/149014. 149014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zauber AG, Winawer SJ, O’Brien MJ, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687–96. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamal A, Darwish RK, Saad S, Salama M, El-Baradie TS, Mahmoud HG, Elshiwy Y. Association of osteopontin gene polymorphisms with colorectal cancer. Cancer Invest. 2017;35:71–7. doi: 10.1080/07357907.2016.1247454. [DOI] [PubMed] [Google Scholar]
- 6.Kuro-o M, Matsumura Y, Aizawa H, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 7.Kurosu H, Yamamoto M, Clark JD, et al. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–33. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf I, Levanon-Cohen S, Bose S, et al. Klotho: a tumor suppressor and a modulator of the IGF-1 and FGF pathways in human breast cancer. Oncogene. 2008;27:7094. doi: 10.1038/onc.2008.292. [DOI] [PubMed] [Google Scholar]
- 9.Pan J, Zhong J, Gan LH, Chen SJ, Jin HC, Wang X, Wang LJ. Klotho, an anti-senescence related gene, is frequently inactivated through promoter hypermethylation in colorectal cancer. Tumor Biol. 2011;32:729–35. doi: 10.1007/s13277-011-0174-5. [DOI] [PubMed] [Google Scholar]
- 10.Usuda J, Ichinose S, Ishizumi T, et al. Klotho predicts good clinical outcome in patients with limited-disease small cell lung cancer who received surgery. Lung Cancer. 2011;74:332–7. doi: 10.1016/j.lungcan.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Kuro-o M. Klotho in health and disease. Nat Rev Nephrol. 2019;15:27–44. doi: 10.1038/s41581-018-0078-3. [DOI] [PubMed] [Google Scholar]
- 12.Zhou X, Wang X. Klotho: a novel biomarker for cancer. J Cancer Res Clin Oncol. 2015;141:961–9. doi: 10.1007/s00432-014-1788-y. [DOI] [PubMed] [Google Scholar]
- 13.National Library of Medicine (US) Genetics Home Reference. Bethesda (MD): The Library; 2013. Sep 16, [reviewed 2019 July; cited 2019 Aug 11]. Available from: https://www.ncbi.nlm.nih.gov/snp/rs1207568. [Google Scholar]
- 14.National Library of Medicine (US) Genetics Home Reference. Bethesda (MD): The Library; 2013. Sep 16, [reviewed 2019 July; cited 2019 Aug 11]. Available from: https://www.ncbi.nlm.nih.gov/snp/rs564481. [Google Scholar]
- 15.Hamajima N, Saito T, Matsuo K, Kozaki KI, Takahashi T, Tajima K. Polymerase chain reaction with confronting two-pair primers for polymorphism genotyping. Cancer Sci. 2000;91:865–8. doi: 10.1111/j.1349-7006.2000.tb01026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimoyama Y, Taki K, Mitsuda Y, Tsuruta Y, Hamajima N, Niwa T. KLOTHO gene polymorphisms G-395A and C1818T are associated with low-density lipoprotein cholesterol and uric acid in Japanese hemodialysis patients. Am J Nephrol. 2009;30:383–8. doi: 10.1159/000235686. [DOI] [PubMed] [Google Scholar]
- 17.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22:1928–9. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 18.Taguchi K, Yasui T, Milliner DS, Hoppe B, Chi T. Genetic risk factors for idiopathic urolithiasis: a systematic review of the literature and causal network analysis. Euro Urol Focus. 2017;3:72–81. doi: 10.1016/j.euf.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Urakawa I, Yamazaki Y, Shimada T, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature. 2006;444:770–4. doi: 10.1038/nature05315. [DOI] [PubMed] [Google Scholar]
- 20.Kuro-o M. Klotho. Pflugers Arch. 2010;459:333–43. doi: 10.1007/s00424-009-0722-7. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M, Clark JD, Pastor JV, et al. Regulation of oxidative stress by the anti-aging hormone klotho. J Biol Chem. 2005;280:38029–34. doi: 10.1074/jbc.M509039200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuro-o M. Klotho and aging. BBA. 2009;1790:1049–58. doi: 10.1016/j.bbagen.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Cui W, Wang L, et al. Klotho gene polymorphisms are related to colorectal cancer susceptibility. Int J Clin Exp Pathol. 2015;8:7446–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Donate-Correa J, Martín-Núñez E, Martínez-Sanz R, et al. Influence of Klotho gene polymorphisms on vascular gene expression and its relationship to cardiovascular disease. J Cell Mol Med. 2016;20:128–33. doi: 10.1111/jcmm.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawano KI, Ogata N, Chiano M, et al. Klotho gene polymorphisms associated with bone density of aged postmenopausal women. JBMR. 2002;17:1744–51. doi: 10.1359/jbmr.2002.17.10.1744. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F, Zhai G, Kato BS, Hart DJ, Hunter D, Spector TD, Ahmadi KR. Association between KLOTHO gene and hand osteoarthritis in a female Caucasian population. Osteoarthritis Cartilage. 2007;15:624–9. doi: 10.1016/j.joca.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Bali V, Bebok Z. Decoding mechanisms by which silent codon changes influence protein biogenesis and function. Int J Biochem Cell Biol. 2015;64:58–74. doi: 10.1016/j.biocel.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang B, Kim J, Jeong D, et al. Klotho inhibits the capacity of cell migration and invasion in cervical cancer. Oncology Rep. 2012;28:1022–8. doi: 10.3892/or.2012.1865. [DOI] [PubMed] [Google Scholar]
- 29.Chen B, Ma X, Liu S, Zhao W, Wu J. Inhibition of lung cancer cells growth, motility and induction of apoptosis by Klotho, a novel secreted Wnt antagonist, in a dose-dependent manner. Cancer Biol Ther. 2012;13:1221–8. doi: 10.4161/cbt.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shu G, Xie B, Ren F, et al. Restoration of klotho expression induces apoptosis and autophagy in hepatocellular carcinoma cells. Cell Oncol (Dordr) 2013;36:121–9. doi: 10.1007/s13402-012-0118-0. [DOI] [PubMed] [Google Scholar]
- 31.Xie B, Zhou J, Shu G, et al. Restoration of klotho gene expression induces apoptosis and autophagy in gastric cancer cells: tumor suppressive role of klotho in gastric cancer. Cancer Cell Int. 2013;13:18. doi: 10.1186/1475-2867-13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]


