Abstract
Transient modulation of genes involved in immunity, without exerting a permanent change in the DNA code, can be an effective strategy to modulate the course of many inflammatory conditions. CRISPR-Cas9 technology represents a promising platform for achieving this goal. Truncation of guide RNA (gRNA) from 5’ end, enables the application of a nuclease competent Cas9 protein for transcriptional modulation of genes, allowing multi-functionality of CRISPR. Here, we introduce an enhanced CRISPR-based transcriptional repressor to reprogram immune homeostasis in vivo. In this repressor system, two transcriptional repressors heterochromatin protein 1 (HP1a) and Krüppel associated box (KRAB) are fused to MS2 coat protein and subsequently recruited by gRNA aptamer binding to a nuclease competent CRISPR complex containing truncated gRNAs. With the enhanced repressor, we demonstrate transcriptional repression of the Myeloid differentiation primary response 88 (Myd88) gene in vitro and in vivo. We demonstrate that this strategy can efficiently downregulate Myd88 expression in lung, blood and bone marrow of Cas9 transgenic mice, which receive systemic injection of adeno-associated virus- (AAV)2/1 carrying truncated gRNAs targeting Myd88 and MS2-Hp1aKRAB cassette. This downregulation is accompanied by changes in downstream signaling elements such as TNF-α and ICAM-1. Myd88 repression leads to decrease in immunoglobulin G (IgG) production against AAV2/1 and AAV2/9 and the strategy modulates IgG response against AAV cargos. It improves the efficiency of a subsequent AAV9/CRISPR treatment for repression of Proprotein convertase subtilisin/kexin type 9 (PCSK9), a gene when repressed can lower blood cholesterol levels. We also demonstrate that CRISPR-mediated Myd88 repression can act as a prophylactic measure against septicemia in both Cas9 transgenic and C57BL/6J mice. When delivered by nanoparticles, this repressor can serve as a therapeutic modality to influence the course of septicemia. Collectively, we report that CRISPR-mediated repression of endogenous Myd88 can effectively modulate host immune response against AAV-mediated gene therapy and influence the course of septicemia. The ability to control Myd88 transcript levels using a CRISPR-based synthetic repressor can be an effective strategy for AAV-based CRISPR therapies, as this pathway serves as a key node in induction of humoral immunity against AAV serotypes.
Keywords: CRISPR/Cas9, AAV, in vivo CRISPR, immunomodulation, multifunctional Cas9, endotoxemia, LPS, transcriptional repression, Myd88, AAV antibodies, Septicemia
Introduction
Recent repurposing of the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) system for transcriptional modulation is now opening a myriad of therapeutic opportunities at the level of transcription, which was once considered as “undruggable”. Transcriptional control over genes involved in immunity can generate a universal therapeutic modality for a broad range of acute or chronic inflammatory conditions in humans as well infectious diseases at the time of pandemics. In addition, such control provides a powerful means for biological discoveries. However, despite the great potential, there have been limited studies that have translated CRISPR transcriptional tools in vivo, with far fewer that explore the utility of the system for transcriptional repression1–9. Prior CRISPR-based transcriptional repressors in vivo operated based on catalytically “dead” Cas9 protein (dCas9) fused to Kruppel-associated box domain (KRAB) domain, the current gold standard for dCas9-based repression studies10–17. But yet it is not entirely clear where a KRAB-based in vivo repressor stands in comparison with recently reported “enhanced” CRISPR repressors18.
Additionally, a useful genetic engineering platform should employ both transcriptional control and gene editing on demand to allow a high level of control at both the DNA and RNA level (e.g. to simultaneously modulate immune responses), a goal achievable through changing the length of guide RNAs (gRNAs) from the 5’ end when using Cas9 nuclease 19. Yet, it is not known if truncated gRNAs can provide effective means for synthetic repression of transcription in vivo, giving rise to physiologically relevant phenotypes.
Here, we set out to determine whether we can achieve synthetic immunomodulation in vivo using a CRISPR-based enhanced transcriptional repressor. Myeloid differentiation primary response 88 (MyD88) is a key node in innate and adaptive immune responses, acting as an essential adaptor molecule for a number of signaling pathways including Toll-like receptor (TLR), response to septicemia, and formation of adaptive immunity against viruses such as Adeno-associated virus (AAV)20–23. MYD88 activating mutations are implicated in a number of lymphoid malignancies, in particular Waldenström macroglobulinemia and activated B-cell diffuse large B-cell lymphomas24. However, it is not clear whether we can achieve control over its transcription in vivo. Given the central role of MyD88 signaling in innate and adaptive immunity21 we sought to examine synthetic transcriptional modulation over this locus in vivo.
Results
CRISPR-mediated repression with MS2-Hp1aKRAB is superior to MS2-KRAB in vitro
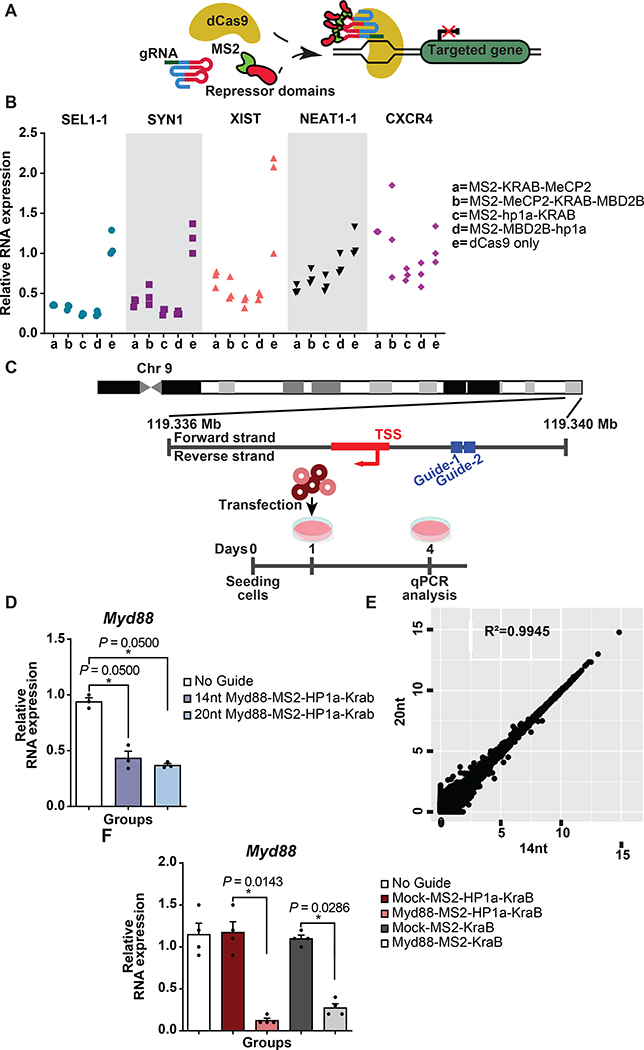
We previously reported “enhanced” CRISPR-based transcriptional repressors in vitro developed by direct fusion of a set of modulators to catalytically dead Cas9 protein (MeCP2, MBD2 or HP1a)18. We first devised an experiment to determine which transcriptional repression domain from our previously published candidates can lead to efficient transcriptional repression when fused to the MS2 coat protein (referred here to as MS2) and recruited to the CRISPR complex by gRNA aptamer binding (Fig. 1A)18. Quantitative real time polymerase chain reaction (qRT-PCR) analysis of a set of target genes in Human Embryonic Kidney 293 (HEK293FT) cells established that MS2-HP1aKRAB [heterochromatin protein 1 (HP1a)- Krüppel associated box (KRAB)] enabled efficient repression across the genes we tested (Fig. 1B).
Figure 1. Aptamer-mediated CRISPR repression in vitro.
(A) Schematic of aptamer-mediated recruitment of repressor domains to CRISPR complex.
(B) mRNA expression of targeted genes following aptamer-mediated recruitment of repressor domains to CRISPR complex in HEK293FT cells. Fold changes were quantified relative to dCas9 only control group (N=3 biologically independent samples).
(C) Top: Schematic representation of the gRNA binding sites targeted to the promoter of Myd88; Bottom: Schematic of experiment design. Mouse neuroblastoma (N2A) cells were transfected with either 14 nucleotide or 20 nucleotide Myd88 gRNA pairs together with dCas9 plasmid and MS2-HP1aKRAB cassette. Expression levels of Myd88 mRNA were analyzed using qRT-PCR three days post transfection.
(D) Fold changes of mRNA of Myd88 were quantified relative to the No Guide group (N = 3 biologically independent samples). The bars represent the mean + S.E.M.
(E) Mean expression levels of 24476 protein-coding and 16648 non-coding RNA genes following targeting Myd88 gene are shown. For visualization purposes, the values were transformed to a log2(TPM+1) scale. R denotes the Pearson correlation coefficient between two groups (N = 3 biologically independent samples). The bars represent the mean + S.E.M.
(F) qRT-PCR analysis of Myd88 mRNA expression levels post LPS treatment in N2A cells. Fold changes were quantified relative to the expression level of cells receiving non-targeting Mock gRNA (N = 4 biologically independent samples). The bars represent the mean + S.E.M. Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney U test. p value ≤ 0.05 was considered significant (*p ≤ 0.05). Statistical source data are provided in Source data fig. 1.
To translate these findings in vivo, we set out to utilize nuclease competent Streptococcus Pyogenes (Sp) -Cas9 transgenic mice as they enable us to eliminate potential confounding effects associated with delivery of Cas9. Therefore, we devised a pair of truncated gRNAs that target Cas9 nuclease and MS2-HP1aKRAB to the Myd88 promoter (Fig. 1C). This strategy allows Cas9 nuclease to be repurposed to a nuclease null protein for transcriptional repression19. We first compared the functionality of the truncated gRNA compared to the full-length gRNA in Mouse neuroblastoma (N2A) cells (Fig. 1C). RNA sequencing showed that transcriptional repression using truncated gRNA is as efficient and specific as traditional 20nt gRNA-based repression in vitro (Fig. 1D-E). Moreover, this strategy yielded similar efficiency in repressing the Myd88 locus as when a dCas9-Hp1aKRAB fusion protein is used with comparable levels of dCas9 and Hp1aKRAB (Extended Data Fig. 1A-B). Next, we set out to examine MS2-HP1aKRAB-mediated repression of endogenous mouse Myd88 levels in vitro and compared the efficiency with commonly used KRAB-based transcriptional repression. We used a previously reported non-targeting mock gRNA as a control25. qRT-PCR for Myd88 demonstrated the in vitro functionality of the gRNAs and superiority of MS2-HP1aKRAB in repression of endogenous Myd88 (Fig. 1F).
CRISPR-mediated repression of Myd88 locus can efficiently be achieved in vivo by recruitment of MS2-HP1aKRAB to gRNA
To test this repressor in vivo, we pursued delivery through packaging gRNAs and MS2-repression cassettes within Adeno-Associated Viruses (AAVs). Different AAV serotypes have been used to deliver CRISPR in vivo. The most common serotype has been AAV9, which has high affinity to parenchymal cell populations26,27. Here, we employed a hybrid AAV2/1 serotype , which is a recombinant AAV consisting of AAV2 inverted terminal repeats, and AAV1 Rep and Cap genes (here on referred to only as AAV1 for simplicity). AAV1 has been shown to be effective in transduction of components of the immune system and non-parenchymal cells such as dendritic and endothelial cells28–30. Moreover, AAV1 capsid can induce MyD88 signaling as part of the pathways of immunity against AAVs in the host31,32. Our assessment of AAV1 tissue affinity revealed the highest expression in blood, lung, and bone marrow (Extended Data Fig. 2). Subsequently, we performed systemic delivery of AAV1/Myd88 gRNA or control AAV1/Mock gRNA with MS2-HP1aKRAB or MS2-KRAB cassettes to Cas9 nuclease transgenic mice (Fig. 2A). Three weeks after injections, blood, lung, and bone marrow were harvested and Myd88 expression was assessed by qRT-PCR (Fig. 2B). Compared to uninjected controls, AAV delivery led to an increase in Myd88 across different tissues we tested. Treatment with CRISPR to repress endogenous Myd88 with HP1aKRAB led to a significant reduction in the level of Myd88 in blood (~84%), lung (~75%), and bone marrow (~63%) as compared to the mock gRNA-treated group, in agreement with high affinity of AAV1 for these tissues. Administration of the KRAB domain alone led to a less pronounced repression of Myd88 in lung (~52%), blood (~59%), and bone marrow (~34%), with slightly higher variation among the animals tested (Fig.2B).
Figure 2-. CRISPR-based targeted Myd88 repression in vitro and in vivo using MS2 repressors.
(A) Schematic of experiments demonstrating retro-orbital injection of AAV1/Myd88 or Mock repressors to Cas9 nuclease transgenic mice.
(B) qRT-PCR analysis of Myd88 expression levels in lung, blood, and bone marrow of Cas9 transgenic mice 3 weeks post retro-orbital injections of 1E+12 GC of AAV1/Myd88 or AAV1/Mock vectors carrying MS2- HP1a -KRAB or MS2-KRAB (N = 4 mice for injected groups except for the following: Mock/MS2-KRAB group N=3 mice, Myd88/MS2-HP1aKRAB in bone marrow N=7 mice, Myd88/MS2-HP1aKRAB in blood and lung N= 5 mice, and N = 2 mice for not injected group). Fold changes are relative to universal control. The bars represent the mean + S.E.M.
(C and D) Fold-change in the expression level of Icam-1 (C), and Tnfα (D) mRNA relative to the universal control. (N = 4 mice for injected groups except for the following: Mock/MS2-KRAB group N=3 mice, Myd88/MS2-HP1aKRAB in bone marrow N=7 mice, Myd88/MS2-HP1aKRAB in lung N=5 mice, and N = 2 mice for not injected group). The bars represent the mean + S.E.M.
(E) Volcano plot showing significance versus expression of differentially expressed genes between bone marrow samples collected from mice treated with Myd88-Ms2-HP1aKRAB versus Myd88-MS2-KRAB. Points above the dotted line represent genes significantly (adj. p-value <0.05) up and down regulated. Highly downregulated genes in the presence of MS2-HP1aKRAB are a family of immunoglobulin heavy and light chains (N=2 mice). Statistical analysis was performed using the two-tailed t test and Benjamini-Hochberg multiple comparisons adjustment. Universal control is a blood sample collected from an uninjected Cas9 transgenic mouse. Statistical analysis for panel A-E was performed using the non-parametric one-tailed Mann-Whitney U test. A p value ≤ 0.05 was considered significant (*p ≤ 0.05, **p ≤ 0.01). Statistical source data are provided in Source data fig. 2.
To assess the potency of repression in rewiring the downstream gene regulatory network, we evaluated the levels of tumor necrosis factor-α (TNF-α) and intercellular adhesion molecule-1 (ICAM-1), two signaling elements directly modulated by the MyD88 signaling pathway33–35. Myd88 targeting with MS2-HP1aKRAB led to a significant reduction in Icam-1 and Tnfα expression across multiple tissues, whereas targeting with MS2-KRAB did not lead to a similar consistent effect (Fig. 2C-D and Supplementary information Table1).
To perform a systematic assessment of the repression efficiency of MS2-HP1aKRAB system as compared to MS2-KRAB, we performed next generation RNA sequencing on the bone marrow of mice treated with these constructs. MS2-HP1aKRAB-treated mice expressed lower Myd88 levels compared to MS2-KRAB-treated ones (Extended Data Fig. 3A), which was accompanied with changes in downstream signaling pathways such as Il1β. Of note, GO Enrichment analysis revealed that Myd88-MS2-HP1aKRAB-treated mice had significant downregulation of signaling pathways implicated in the immune and defense response against foreign organisms and bacteria, which are pathways associated with MyD88 function (Extended Data Fig. 3B). Similarly, the Reactome database revealed the TLR pathway as one of the highly significant downregulated pathways in the presence of MS2-HP1aKRAB (Extended Data Fig. 3C). This evidence suggests that modulation of Myd88 and its downstream immune pathways is most effective with the MS2- HP1a KRAB repressor in vivo.
Interestingly, volcano plotting of differentially expressed genes revealed the constant region of heavy chain of immunoglobulin G1 and G2 (Ighg1 and Ighg2b) and other immunoglobulin-related heavy and light chain genes as most downregulated with HP1a KRAB relative to KRAB (Fig. 2E). This is interesting finding in light of the mouse genetic background (C57BL/6), which has been shown to produce high level of IgGs36.
CRISPR-mediated repression of Myd88 leads to modulation of humoral response against AAV-mediated gene therapy and the efficacy of its function
Prior studies demonstrate that viral DNA stimulates TLR (i.e. TLR9), which in turn activates MyD88 and initiates downstream signaling events leading to adaptive immunity and antibody production against AAVs8,16. In light of prior evidence and the observed repression of the immunoglobulin pathway, we asked whether there was a decrease in the AAV-specific humoral response following treatment with AAV1 carrying Myd88-targeting gRNAs and MS2-HP1aKRAB cassettes (here on referred to as AAV1/Myd88 for simplicity) as compared to control viruses carrying mock gRNAs (AAV1/Mock). Three weeks after injection, we measured immunoglobulin G (IgG) response against the AAV1 capsid. We detected a 50% decrease in plasma IgG2a levels against AAV1 in AAV1/Myd88 group compared to AAV1/Mock-treated animals. (Fig. 3A).
Figure 3-. Prophylactic administration of AAV1/MyD88 in vivo leads to modulation of humoral immunity against AAV.
(A) Top: Schematic of experiments demonstrating delivery of AAV-Myd88 or Mock repressors to Cas9 nuclease transgenic mice; Bottom: Analysis of anti-AAV1 IgG2A antibody by ELISA. Optical density values are quantified relative to the AAV1/Mock group (N=8 mice).
(B) Top: Schematic of experiments demonstrating Cas9 transgenic mice treated with AAV1/Myd88 or AAV1/Mock at day 1, followed by a second administration of AAV1/Mock on day 21; Bottom: Anti-AAV1 IgG1 and total IgG antibody measured by ELISA at different time points. Optical density values are quantified relative to a value of AAV1/Mock+AAV1/Mock group (N=4 mice).
(C) Top: Schematic of experiments demonstrating Cas9 transgenic mice treated with AAV1/Myd88 or AAV1/Mock at day 1, followed by a second administration of AAV9/SaCas9 on day 21; Bottom: Analysis of Anti-AAV9 IgG and Anti Sa-Cas9 levels in mice sera. Optical density values are quantified relative to the AAV1/Mock+AAV9/SaCas9 group (N=4 mice).
(D) Top: Schematic of experiments demonstrating Cas9 transgenic mice treated with AAV1/Myd88 or AAV1/Mock at day 1, followed by a second administration of AAV9/LacZ on day 21; Bottom: Analysis of Anti-AAV9 IgG in mice sera and LacZ mRNA levels in blood. Relative optical density values are quantified relative to the AAV1/Mock+AAV9/LacZ group (N=4 mice).
(E) Top: Schematic of the experiment. Cas9 nuclease transgenic mice were treated with AAV1/Myd88 or AAV1/Mock vectors via retro-orbital injection followed by a second and third injection of AAV9/PCSK9 vectors on day 7 and 21.
(F) Analysis of anti-AAV9 IgG and total IgG antibody measured by ELISA. Optical density values are quantified relative to the AAV1/Mock+AAV9/PCSK9+AAV9/PCSK9 group (N=4 mice).
(G and H) Plasma samples collected from treated animals were assayed for (G) PCSK9 and (H) Cholesterol at days 0, 7, 21, and 30 (N=4 mice). Panel A-H data are expressed as mean + S.E.M. (Mock= Mock-HP1aKRAB, Myd88= Myd88-HP1aKRAB, PCSK9= PCSK9-HP1aKRAB). Statistical analyses were performed using the non-parametric one-tailed Mann-Whitney U test. A p value ≤ 0.05 was considered significant (*p ≤ 0.05, and ****p ≤ 0.0001). Statistical source data are provided in Source data fig. 3.
Antibody formation against the AAV capsid is an important barrier to re-administration of AAV-based gene therapies, often leading to rapid clearance of the virus and other deleterious effects related to destruction of the virus or transduced cells by immune system. To further probe the prophylactic effect of Myd88 repression on modulating humoral immunity upon AAV1 re-administration, we asked whether pre-treatment with AAV1/Myd88 can influence IgG level against AAV1 upon re-administration of AAV1/Mock. Analysis of IgG1 and IgG2A in the plasma demonstrated lower levels after initial Myd88 repression, hinting to the potential of this strategy in modulating humoral response to AAV1 re-administration (Fig. 3B and Extended Data Fig. 4A-B).
To examine the extensibility of this strategy to modulate response against other AAV serotypes, we pre-treated mice with AAV1/Myd88 or AAV1/Mock and, 7 days later, systemically injected them with AAV9 that carried a LacZ or Staphylococcus aureus Cas9 (Sa-Cas9) cassette. In both instances, analysis of total IgG levels against AAV9 demonstrated that Myd88 repression led to a lower antibody response against AAV9. Moreover, this was accompanied by significantly lower antibodies against Sa-Cas9 and higher transcript levels of LacZ in the blood (Fig. 3C-D). These data demonstrate that modulation of immunoglobulin production through Myd88 repression can influence the humoral response against more than one AAV serotype and its cargo. Higher LacZ expression in blood in this context hints to potentially higher efficiency of the gene therapy using this approach.
To further explore this notion in the context of CRISPR therapies, we pre-treated the mice with AAV1/Myd88 or AAV1/Mock and then subjected them to two rounds of AAV9-based gene therapies 7 and 14 days apart (Fig. 3E). In this case, AAV9 carries a cassette for CRISPR-mediated repression of Proprotein convertase subtilisin/kexin type 9 (PCSK9), similar to the strategy we employed for Myd88 repression. PCSK9 is an enzyme encoded by the PCSK9 gene. This enzyme binds to the low-density lipoprotein (LDL) receptor at the surface of hepatocytes and initiates ingestion of the LDL receptor. Therefore, when PCSK9 is blocked or repressed, more LDL receptors are present to remove LDL from blood which, lowers blood LDL-cholesterol levels. This enzyme has been the target of previous in vivo CRISPR applications37–39.
Our data show that the AAV1/Myd88 pre-treated group has decreased Myd88 expression (Extended Data Fig. 4C-D) as well as lower IgG1 and total IgG levels against AAV9 compared to the control (Fig. 3F). This observation was accompanied by better PCSK9 repression and lower plasma cholesterol levels, suggesting increased efficiency of the gene therapies (Fig. 3G-H). Altogether, these data present an exciting opportunity to modulate humoral immunity against AAV, possibly through prophylactic repression of Myd88 with a tool inherently suited to perform both gene editing and epigenetic modulation (nuclease competent CRISPR).
CRISPR mediated Myd88 repression does not create visible adverse effect in long term.
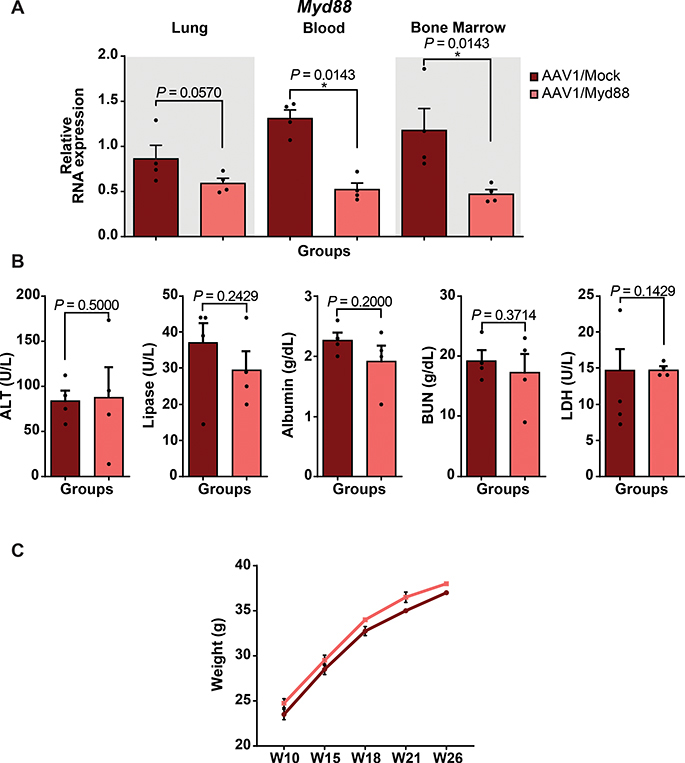
Next, we probed the long-term efficacy of AAV1/Myd88 repression in vivo to further assess its durability and possible negative consequences. Analysis of Myd88 transcripts in lung, blood, and bone marrow twenty-three weeks after injection showed Myd88 repression in the AAV1/Myd88 group (Fig. 4A). To assess possible negative consequences of long-term reduction of MyD88 level, we analyzed some key indicators of major internal organ functions including Blood Urea Nitrogen (BUN) for Kidney, Alanine Transaminase (ALT) and Albumin for liver, Lipase for pancreas, and Lactic Acid Dehydrogenase (LDH) as a marker of tissue damage. None of these markers were significantly different than mock-treated groups (Fig. 4B). Moreover, tracking the weight of the mice suggested that there were not any detectable deleterious effects on the general health and well-being as all animals demonstrated comparable weights (Fig. 4C).
Figure 4. Long-term efficacy of AAV/Myd88 repression in vivo.
(A) qRT-PCR analysis of in vivo Myd88 expression level in lung, blood, and bone marrow of Cas9 transgenic mice five months post retro-orbital injections of 1E+12 GC of AAV/Myd88 or AAV/Mock vectors (N = 4 mice). The bars represent the mean + S.E.M.
(B) Assessing the expression of a panel of general health markers in plasma samples collected from mice. About 5 months after AAV delivery, plasma samples were collected form mice and the concentration of ALT, Lipase, Albumin, BUN, and LDH were assessed in different groups. The bars represent the mean + S.E.M (N = 4 mice).
(C) Body weight measurements of mice injected with 1E+12 GC of AAV/Myd88 or AAV/Mock vectors carrying MS2-HP1a-KRAB showing similar growth condition (N = 4 mice). The bars represent the mean ± S.E.M. (Mock= Mock-HP1a-KRAB, Myd88= Myd88-HP1a-KRAB). Fold change expression levels were quantified relative to the universal control. Universal control is a blood sample collected from an unjected mouse. Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney U test. A p value ≤ 0.05 was considered significant (*P ≤ 0.05). Statistical source data are provided in Source data fig. 4.
CRISPR mediated Myd88 repression in vivo can act as a prophylactic measure against septicemia in Cas9 transgenic and C57BL/6 mice
We then asked whether this strategy could act as a prophylactic modality during septicemia, when there is an augmented systemic immune response. Septicemia is a pressing medical issue due to the emergence of antibiotic-resistance and rising longevity of patients suffering from chronic diseases40. Moreover, high mortality rates due to septicemia still remain a medical challenge following trauma in the battlefield, highlighting the need for novel prevention strategies41.
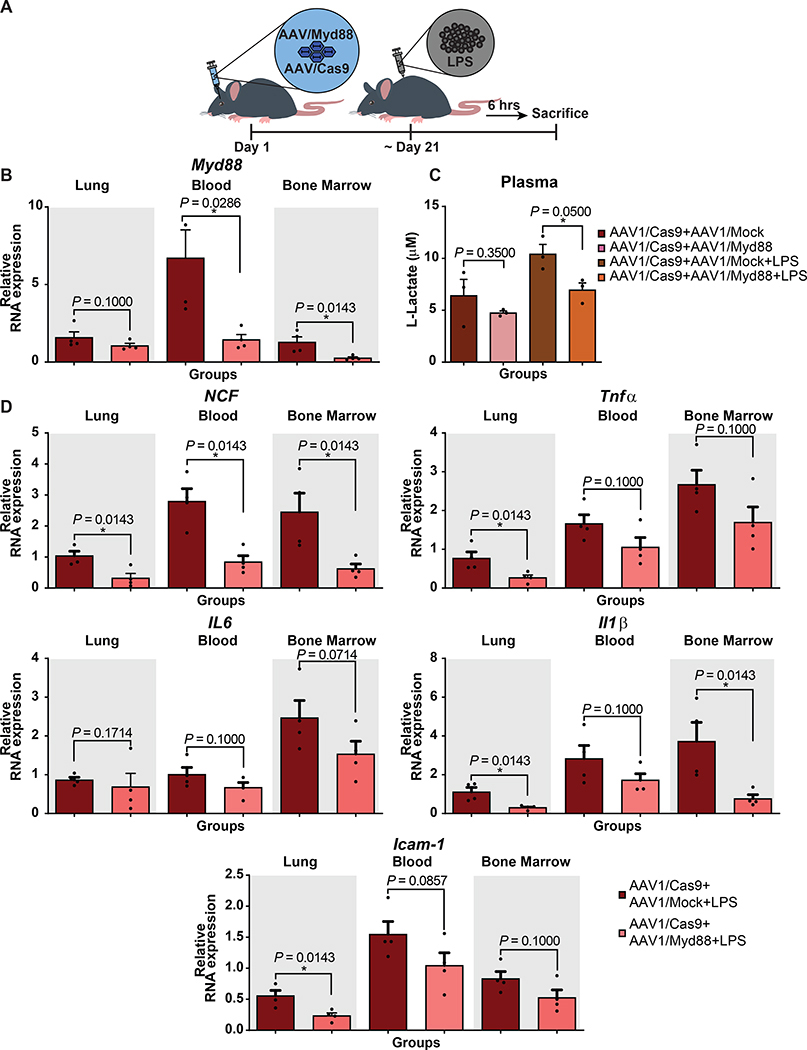
We pre-treated Cas9 mice with AAV1/Myd88 or AAV1/Mock and three weeks later subjected them to systemic lipopolysaccharides (LPS) (from Escherichia coli 0127:B8) treatment. Six hours following LPS, we harvested lung, blood, and bone marrow and assessed the transcript levels of Myd88 and major inflammatory cytokines (Fig. 5A). We observed significant repression of Myd88 in lung (61%), blood (80%), and bone marrow (76%) compared to AAV1/Mock-treated mice (Fig. 5B). In response to LPS, plasma lactate level, a systemic marker associated with septicemia and tissue damage42, was significantly lower when mice were pre-treated with the AAV1/Myd88 repression cassette before LPS exposure, indicating a reduced systemic injury (Fig. 5C). Additionally, Myd88 repression prevented upregulation of a wide range of inflammatory and immune-related cytokines that are directly or indirectly downstream of Myd88 signaling such as Icam-1, Tnfα, Ncf, Il6, Ifn-α, Ifn-β, Ifn-γ, and Stat4 (Fig. 5D and Extended Data Fig. 5-6). Analysis of plasma and lung cytokine levels using a quantitative ELISA-based chemiluminescent assay revealed lower level of cytokines in Myd88-repressed mice (Fig. 5E).
Figure 5-. CRISPR-based modulation of host inflammatory response can be a prophylactic measure against LPS-mediated septicemia in Cas9 transgenic and WT mice.
(A) Schematic of the experimental design to assess the protective effect of CRISPR-mediated MyD88 repression in septicemia. 1E+12 GC of AAV vectors were injected to Cas9-expressing mice via retro-orbital injection and approx. 3 weeks later they were treated i.p. with LPS (5mg/kg). 6 hours post LPS injection mice were sacrificed.
(B) qRT-PCR analysis of in vivo Myd88 expression relative to the universal control following LPS injection (N = 6 mice for injected groups, except Bone Marrow of Myd88 group which is N=5 mice, and N = 2 mice for Not Injected group). The bars represent the mean + S.E.M.
(C) Circulating L-lactate in plasma samples collected from mice 6 hours post LPS injection (N=3 mice). The bars represent the mean + S.E.M.
(D) qRT-PCR analysis of Icam-1, and Tnfα mRNA expression in lung, blood, and bone marrow quantified relative to the universal control following LPS injection (N = 6 mice for injected groups, except Bone Marrow of Myd88 group which is N=5 mice, and N = 2 mice for Not Injected group). The bars represent the mean + S.E.M.
(E) Measurement of a panel of inflammatory cytokines in lung and plasma using multiplex-ELISA assay; values are displayed in the heatmaps as log base 10 of the measured concentration (N=3 mice). Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney U test. A p value ≤ 0.05 was considered significant (*p ≤ 0.05, **p ≤ 0.01). Statistical source data are provided in Source data fig. 5.
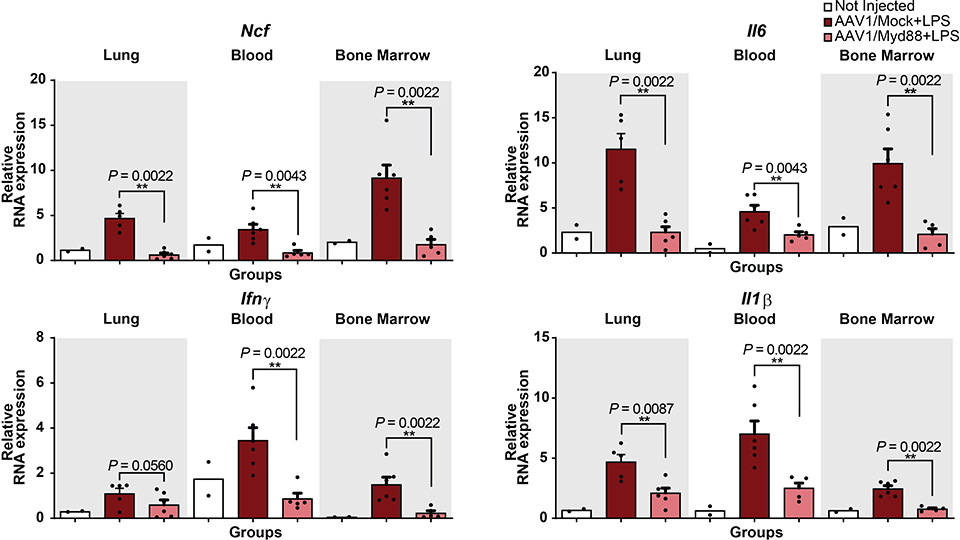
To explore whether we can achieve similar outcomes by simultaneous delivery of Cas9 and gRNA-MS2-Hp1aKRAB cassettes to wild-type animals, we examined a dual AAV1 system in which a second virus carries a Sp-Cas9 nuclease cassette (Fig. 6A). This strategy was capable of decreasing Myd88 transcripts in C57BL/6 mice, both in the presence and absence of septicemia, leading to phenotypically relevant response similar to what we observed in Cas9 transgenic animals (Fig. 6B-D and Extended Data Fig. 7).
Figure 6-. Developing protection following LPS-mediated septicemia using a dual AAV CRISPR/Cas9 strategy with AAV1/Cas9 and AAV1 carrying gRNA-MS2-HP1aKRAB.
(A) Schematic of the experiments. C57BL/6 mice received total 2E+12 GC of AAV1/Cas9 and AAV1/Myd88 or AAV1/Mock vectors via retro-orbital injection and approx. 3 weeks later they were treated i.p. with LPS (5mg/kg). 6 hours post LPS injection mice were sacrificed.
(B) qRT-PCR for Myd88 expression was performed on blood, bone marrow, and lung samples collected from mice (N=4 mice for all groups expect for blood of Mock treated group which is N=3 mice). The bars represent the mean + S.E.M.
(C) Six hours post LPS injection plasma samples were collected form mice and the concentration of L-Lactate was assessed in different groups (N=3). The bars represent the mean + S.E.M.
(D) qRT-PCR analysis of Icam-1, Tnfα, Ncf, Il6, and Il1β mRNA expression in blood, bone marrow, and lung. Fold change expression levels were quantified relative to the universal control (N = 4 mice). The bars represent the mean + S.E.M. (Mock= Mock-HP1aKRAB, Myd88= Myd88-HP1aKRAB). Universal control is a blood sample collected from an uninjected Cas9 transgenic mouse. Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney U test. A p value ≤ 0.05 was considered significant (*p ≤ 0.05). Statistical source data are provided in Source data fig. 6.
Nanoparticle mediated delivery of Myd88 targeting CRISPR super-repressors after exposure to LPS can serve as a therapeutic modality against septicemia
To examine the therapeutic potential of this approach after exposure to LPS, we sought to deliver CRISPR plasmids to C57BL/6 though a nanoparticle-based approach, as they enable faster and more feasible delivery for CRISPR-based gene modulation as compared to the AAV-based system. Given the significance of liver damage after intra-peritoneal LPS exposure and the notion that majority of the nanoparticles delivered systematically accumulate in the liver and lungs, we focused on studying the liver. We first examined whether AAV1/Myd88 can repress Myd88 expression in liver. Having difficulty repressing Myd88 in the liver with our current pair of gRNAs, we designed another pair targeting a different region of the Myd88 promoter. The new gRNAs led to Myd88 repression in the liver upon AAV mediated delivery to Cas9 transgenic mice. This was also observed following LPS injury (Extended Data Fig. 8A-B). Next, we set out to examine the therapeutic effect of this system in C57BL/6 mice 2 hours post exposure to LPS. We injected the mice with nanoparticles carrying Cas9, gRNAs, and MS2-HP1aKRAB cassettes and examined the systemic inflammatory response against LPS (Fig 7A). 72 hours post CRISPR delivery, blood, lung, liver, and bone marrow were harvested and Myd88 expression was assessed by qRT-PCR. Treatment led to a reduction in the levels of Myd88 in the blood, lung, bone marrow, and liver as compared to the Mock treated group (Fig. 7B). This Myd88 repression prevented upregulation of a wide range of inflammatory markers followed by LPS exposure (Fig. 7C). Analysis of plasma markers of tissue damage of the liver showed that we could modulate the detrimental effects of LPS injection (Fig. 7D). In particular, high density lipoproteins (HDL) have been shown to increase following LPS treatment to eliminate systemic LPS in order to protect tissues from damage43 and has been associated with MyD88 signaling44. In accordance with this, we found Myd88 repression decreased HDL, low density lipoproteins (LDL), and cholesterol as compared to Mock-treated groups. In addition, Myd88 repression decreased ALT and Aspartate Aminotransferase (AST), two markers of hepatocyte damage, which further suggests that this approach can be effective therapeutically (Fig. 7D).
Figure 7-. Therapeutic delivery of nanoparticles carrying DNA encoding Myd88-targeting CRISPR confers protection against LPS-mediated septicemia.
(A) Schematic of the experiment. C57BL/6 mice were treated i.p. with LPS(2.5mg/kg). 2 hours later, mice received Cas9 and Myd88 or Mock vectors via retro-orbital injection using nanoparticles. 72 hours post retro-orbital injection mice were sacrificed.
(B-C) Lung, blood, and bone marrow samples were collected from mice. The expression levels of Myd88 and a panel of immune-related genes were assessed by qRT-PCR.
(B) qRT-PCR analysis of Myd88 repression following LPS injection and CRISPR-mediated therapy. (N=10 mice). The bars represent the mean + S.E.M.
(C) qRT-PCR analysis of Icam-1, Tnfα, Ncf, Il6, and Il1β mRNA expression in different tissues. Fold changes were quantified relative to the universal control. (N=10 mice). The bars represent the mean + S.E.M.
(D) Plasma concentration of Cholesterol (N = 11 mice for Cas9+Mock+LPS, N = 9 mice for Cas9+Myd88+LPS), plasma concentration of HDL (N = 11 mice for Cas9+Mock+LPS, N = 8 mice for Cas9+Myd88+LPS), plasma concentration of LDL (N = 10 mice for Cas9+Mock+LPS, N = 8 mice for Cas9+Myd88+LPS), plasma concentration of ALT (N = 7 mice for Cas9+Mock+LPS, N = 4 mice for Cas9+Myd88+LPS), and plasma concentration of AST (N = 8 mice for Cas9+Mock+LPS, N = 5 mice for Cas9+Myd88+LPS). The bars represent the mean + S.E.M. (Mock=Mock-HP1aKRAB, Myd88= Myd88-HP1aKRAB). Universal control is a blood sample collected from an uninjected Cas9 transgenic mouse. Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney U test. A p value ≤ 0.05 was considered significant (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001). Statistical source data are provided in Source data fig. 7.
Discussion
In summary, we provide a potent transcriptional therapeutic modality for synthetic control of immune response in vivo using a newly developed CRISPR-based transcriptional super-repressor against endogenous Myd88. We show that this system is effective in modulating downstream immune signaling and can create a visible protective phenotype in vivo. This notion is especially attractive in the case of delivery using a less common AAV serotype (AAV2/1) known to target smaller cellular populations in vivo (e.g. non-parenchymal cells).
We demonstrate that targeting the Myd88 locus with AAV1/CRISPR generates less immunoglobulin against AAV1 and AAV9 and modulates general immunoglobulin expression patterns, consistent with prior reports on the failure of generation of an antigen-specific IgG2a response in MyD88−/− animals45. The ability to control Myd88 transcript levels using a CRISPR-based synthetic repressor is of significance in light of the common challenges involved with AAV-based clinical gene therapies, as this pathway has been shown to be a key node in induction of humoral immunity against many AAV serotypes and not just AAV2/1 in vivo. Moreover, we argue that this method can be a powerful tool to dissect biological questions at the level of Myd88 transcription. Here, we demonstrate that a prophylactic regime that represses Myd88 can be used to increase efficiency of subsequent viral-based gene delivery by preventing a surge in humoral response.
This strategy was also effective in modulating the systemic inflammatory response against (LPS)-induced endotoxemia both prophylactically and therapeutically. CRISPR-mediated endogenous repression of Myd88 prevented upregulation of a wide range of inflammatory markers and conferred a protective phenotype. Further studies are needed to address the extensibility of CRISPR super repressors to other endogenous genes of the immune system and to define target tissues and cellular players, as well as to characterize the applicability to other infectious diseases. However, the ability to modulate host immune response using this strategy is a promising step towards generating a universal yet targeted tool to prevent exaggerated inflammatory response and severe tissue damage in the context of emerging infectious diseases.
HP1a protein contains a chromodomain (CD) and a chromoshadow domain (CSD), which interacts with methylated H3K9 and H3K9-specific histone methylases, including SetDB1 and Suv39h1/2, respectively 46,47. In this study to minimize the potential nonspecific effects of ectopic HP1a expression, we used a truncated form of HP1a containing only the CSD. Several questions remain about how and if this truncated version still leads to the spread of chromatin repression marks beyond the targeted loci. HP1 proteins can undergo CSD-mediated dimerization, but such homodimerization alone cannot explain the ability of HP1a proteins to spread along chromatin48–50. Further analysis is needed to look at the genome wide effects of using truncated HP1a protein.
Taken together, we demonstrate the promise of CRISPR-based transcriptional regulation as a readily programmable tool for modulating inflammatory conditions and protecting against an infectious condition. Employment of a nuclease competent Cas9 and a truncated gRNA in this study (the step-by-step protocol can be found at the Nature Protocol Exchange [51]) opens up an opportunity for simultaneous application of CRISPR for targeted gene editing while modulating the immune response, which makes CRISPR-mediated gene repression superior to other systems such as shRNA-mediated repression.
Methods
Vector Design and Construction
MS2 Fusion constructs
To construct the MS2-fused transcriptional repressors, the specific domains of interest were amplified from vectors previously published in our group and subsequently cloned into the pcDNA3-MS2-VP64 backbone (Addgene plasmid ID: 79371). The pcDNA3-MS2-VP64 vector was digested with NotI and AgeI to remove the VP64 domain and then the amplified repressors were cloned into this backbone via the Gibson Assembly method.
U6-gRNA-MS2 plasmids
To generate these plasmids, 14bp or 20bp guide sequences were inserted into sgRNA-MS2 cloning backbone (Addgene plasmid ID: 61424) at the BbsI site via golden gate-based reaction. All the gRNA sequences are listed in the Table 2 in supplementary information.
AAV vectors
Following cloning of the gRNAs into a U6-sgRNA-MS2 backbone, the U6-gRNA encoding region was amplified from this vector and inserted within gateway entry vectors using golden gate reaction. Using the same method, the repressor or activator domain and a truncated human EF1a promoter (Gift from Dr. Noah Davidsohn, Dr. Church lab) were cloned into gateway entry vectors. Further sub-cloning of all these components into AAV backbone via gateway reaction (Invitrogen) generated final AAV vectors. Cas9 plasmids were purchased from Addgene (AAV-CMVc-Cas9 #106431 and pAAV-RSV-SpCas9 #85450).
AAV packaging and purification
AAV vectors were digested by SmaI digest to test the integrity of ITR regions before virus production. Verified AAV vectors were used to generate AAV2/1-Myd88, AAV2/1 MockgRNA and AAV2/1 GFP and AAV2/9-Pcsk9 by PackGene® Biotech, LLC. The virus titers were quantified via Real-time SYBR Green PCR at 1.5E+13 GC/ml against standard curves using linearized parental AAV vectors.
Cell culture
HEK293FT and Neuro-2a cell lines (purchased from ATCC) were maintained in Dulbecco’s modified Eagle’s medium (DMEM - Life Technologies) with 10% fetal bovine serum (FBS - Life Technologies), 2mM glutamine, 1.0 mM sodium pyruvate (Life Technologies) and 1% streptomycin– penicillin mix (Gibco) in incubators at 37 °C and 5% CO2.
Transfection of in vitro cultured cells
HEK293FT cells were seeded approximately 50,000 cells per well in 24-well plates and transfected the next day. HEK293FT cells were co-transfected with plasmids encoding gRNA (10 ng), dCas9 or dCas9-H1aKRAB (200 ng), MS2-fused repressor (100 ng), puromycin resistant gene (50 ng), and Enhanced Blue Fluorescent Protein (EBFP) as a transfection control (25 ng). Polyethylenimine (PEI) (Polysciences) was used to transfect HEK293FT cells. Transfection complexes were prepared according to manufacturer’s instructions. Cells were treated with 0.5ug/ml puromycin (Gibco-life tech) at 24 hours post-transfection. Cells were collected 72 hours post-transfection and total RNA was collected from cells using RNAeasy Plus Mini Kit (Qiagen).
Neuro-2a cells were seeded approximately 50,000 cells per well in 24-well plates and transfected the next day. Cells were co-transfected with plasmids encoding gRNA (10–100 ng), Cas9 nuclease (70 ng), dCas9 or dCas9-H1aKRAB (200 ng), and EBFP as a transfection control (25 ng), and a Puromycin resistance gene (50ng). Plasmids were delivered to Neuro-2a cells with Lipofectamine LTX. Cells were treated with 0.5ug/ml puromycin (Gibco-life tech) at 24 hours post-transfection. For the experiment shown in Figure 1F, 3 days later, cells were treated with LPS at the concentration of 10ug/ml (LPS was added to induce Myd88 expression) and after 5 hours total RNA was collected from cells using RNEasy Plus Mini Kit (Qiagen).
Quantitative RT-PCR (qRT-PCR) Analysis
Cells or tissues were lysed, and RNA was extracted using RNEasy Plus Mini Kit (Qiagen) or Trizol (Life Technologies) followed by cDNA synthesis using the High-Capacity RNA-to-cDNA Kit (Thermo Fisher). qRT-PCR was performed using SYBR Green PCR Master Mix (Thermo Fisher). All analyses were normalized to 18S rRNA and fold-changes were calculated against No gRNA control groups for in vitro transfection experiments and a universal control for in vivo experiments (2−ΔΔCt). Universal control is a blood sample collected from an uninjected Cas9 transgenic mouse, which did not receive any AAV injection and was kept as the reference throughout all analyses for comparison of values among different organs. Primer sequences for qPCR are listed in Table 3 in supplementary information.
Plasma analysis
After harvesting mice, plasma samples were aliquoted and stored at −80 °C. Plasma levels of cholesterol were measured via a colorimetric assay according to the manufacturer’s instructions (Thermo- Scientific Total Cholesterol Reagents #TR13421). Plasma Pcsk9 protein levels were quantified by ELISA according to the manufacturer’s instructions (R&D Systems #MPC900).
ELISA-based chemiluminescent assay
Lung samples were lysed using 1× cell lysis buffer (Cell Signaling) (ratio of 100 mg of tissue to 1 ml of buffer) followed by homogenization and sonication of the lysed tissue. The assay was performed using the Q-Plex™ Mouse Cytokine – Screen (16-Plex) kit (Quansys Biosciences) following the manufacturer’s protocol. Briefly, samples or calibrators were added into wells of a 96 well plate arrayed with analyte specific antibodies that capture GMCSF, IL-1α, IL-1β, IL-4, IL-5, IL-6, IL-12p70, IL-17, MCP-1, MIP-1α, RANTES, and TNFα. Plates were washed and biotinylated analyte specific antibodies were added. After washing, streptavidin-horseradish peroxidase (SHRP) was added. Following an additional wash, the amount of SHRP remaining on each location of the array was measured with the addition of a chemiluminescent substrate.
Antibody ELISA
Anti-AAV antibody assay
Fifty microliters of AAV particles diluted in 1× coating buffer (13 mM sodium carbonate, 35 mM sodium bicarbonate buffer, pH 9.2) containing 2× 109per viral particles were added to each well in a Microlon® high protein binding 96-well plate (Greiner) and incubated overnight. Wells were washed three times with 1× Tris Buffered Saline + Tween-20 (TBST, Bethyl) and blocked with 1× Tris Buffered Saline + 1% BSA (Bethyl) for 1 hour at RT. Wells were washed three times with TBST. The standard curve for Figure 2B was generated using purified mouse antibody (Mouse host IgG2a anti-AAV1 (Fitzgerald-MBS830111), Mouse IgG1 unlabeled - Southern Biotech clone 15H6, Mouse IgG2a unlabeled) in twofold dilutions in TBST + 1% BSA + 1:500 negative control mouse plasma, beginning from a concentration of 10,000 ng/ml AAV1 antibody. The standards were added to the plate followed by diluting the plasma samples (samples were diluted 1:500 for Figure 2B and no dilution for Figure 2D and 2F) and incubated for 1 hour at RT. Wells were washed four times with TBST and then goat anti-mouse HRP antibody was added at a concentration of 1:500 and incubated for 1 hour at RT. Wells were washed four times with wash buffer and TMB substrate was added to the wells. Reactions were terminated by adding 0.18 M H2SO4 after development of the standard curve (15 minutes). Finally, absorbance was measured at 450 wavelengths using a plate reader (BioTek). Absorbance results were exported and analyzed in Excel.
Anti SaCas9 antibody assay
Microplates were coated with 50 μl per well at 1 μg/ml Sacas9 protein diluted in 50mM carbonate buffer at pH 9.0. Plates were incubated overnight at 4° C. Wells were washed three times with 1× Tris Buffered Saline + Tween-20 (TBST, Bethyl). Wells were blocked with 200 μl/well of blocking buffer (PBS containing 1% BSA and 0.02% azide) and incubated overnight at 4° C. Wells were washed three times with 1× PBS+0.02% azide. Plasma samples and control were added to the wells at 50 μl/well diluted in blocking buffer and incubated 1 hour at room temperature. Antibodies may be serially diluted for determining titer or diluted to previously determined working concentration for screening assays or antigen quantitation. Wells were washed three times with PBS containing 0.05% Tween-20. Goat anti-mouse HRP antibody was added at a concentration of 1:500 and incubated for 1 hour at RT. Wells were washed. Reactions were terminated by adding 0.18 M H2SO4 after 15 minutes. Finally, absorbance was measured at 450 wavelengths using a plate reader (BioTek). Absorbance results were exported and analyzed in Excel.
Lactate assay
Blood samples were collected using EDTA coated tubes. Samples were centrifuged at 1,000 × g for 10 minutes. Plasma was collected and stored at −80°C. Lactate assay was performed following the manufacturer’s protocol (L-Lactate Assay Kit, Cayman Chemicals). Briefly, samples were deproteinated by adding 0.5 M MPA. After pelleting the protein, supernatant was added to Potassium Carbonate and centrifuged at 10,000 × g for five minutes at 4° C. Samples were diluted four-fold and added to the designated wells. Next, assay buffer cofactor mixture, Fluorometric Substrate, and Enzyme Mixture were added to each well. Plate was incubated for 20 minutes at RT and the fluorescence was measured using an excitation wavelength of 530–540 nm and an emission wavelength of 585–595 nm. Absorbance results were exported and analyzed in Excel according to the manufacturer’s protocol.
Examination of liver injury after LPS injection
Plasma samples were sent to IDEXX Laboratories to measure a panel of tissue injury markers including ALT, Cholesterol, LDL, HDL, BUN, Albumin, LDH, and Lipase.
Animals
Animal studies were conducted with adherent to the guideline for the care and use of laboratory animals of the NIH. All the experiments with animals were approved by the Institutional Animal Care and Use Committee (IACUC) at Arizona State University and have been performed according to institutional guidelines. All the experiments were performed on at least 3 mice of 6–8 weeks old per group. Both male and female were included in experiments. The sample size in each group is indicated in each figure legend.
Both male and female Rosa26-Cas9 knockin mice (JAX Stock number 026179) and male C57BL/6 mice (JAX Stock number: 000664) were used for AAV/CRISPR repression experiments.
Retro-Orbital injections
AAV particles were delivered to mice through retro-orbital injection of the venous sinus. Animals were anesthetized with 3% isoflurane and virus particles were injected to the left eye with 100 microliters of AAV solution (1E+11 to 1E+12 genome copy per mouse).
Tissue harvest
Mice were euthanized via CO2 inhalation. Tissue samples taken from liver, lung, bone marrow and blood were collected in RLT Plus buffer (Qiagen) and frozen or snap frozen for RNA analysis.
In vivo LPS Administration
Mice were given intraperitoneal (i.p.) injection of lipopolysaccharides (from Escherichia coli 0127:B8 (Sigma-Aldrich, St. Louis, MO, USA) dissolved in phosphate-buffered saline (PBS) at a concentration of 2–5 mg/ml. Mice were euthanized 6–72 hours post LPS injection (timeline is included in schematics) via CO2 inhalation.
In vivo Pepjet administration
Mice received 60 μg of DNA containing 10 μg Cas9 and 50 μg Myd88-HP1aKRAB or Mock-HP1aKRAB via retro-orbital injection. PepJet TM reagent (SignaGen Laboratories, Catalog #: SL100501) was used for in vivo transfection. DNA was mixed with PepJet at the ratio of PepJetTM (μL): DNA (μg) 2:1 and prepared according to the manufacturer’s protocol.
RNA Sequencing and Data Analysis
In vitro experiments
N2A cells were co-transfected with 10ng gRNA targeting Myd88 loci, respectively. 200ng dCas9 constructs, 100ng MS2-HP1aKRAB, 50ng puromycin resistant gene, and 25ng transfection control. Cells were treated with 0.5ug/ml puromycin (Gibco-life tech) at 24 hours post-transfection. Total RNA was extracted 72 hours post transfection using RNeasy Plus Mini Kit (Qiagen) and sent to UCLA TCGB core on dry ice. Ribosomal RNA depletion and paired end read library preparation were performed at UCLA core followed by RNA sequencing using NextSeq500. Coverage was 20 million reads per sample. FASTQ files with pair-ended 75bp reads were then aligned to the mouse GRCm38 reference genome sequence (Ensembl release 90) with STAR, and uniquely-mapped read counts (an average of 14.8 million reads per sample) were obtained with Cufflink. The read counts for each sample were then normalized for the library size to CPM (counts per million reads) with edgeR. Custom R scripts were then used to generate plots.
In vivo experiments
RNA was extracted from mice bone marrow samples using RNeasy Plus Mini Kit (Qiagen) followed by globin mRNA depletion using GLOBINclear™ Kit, mouse/rat kit (Thermofisher). None-directional library preparation was performed at Novogene Corporation Inc. followed by RNA sequencing using Illumina Nova Platform with paired-end 150 run (2×150 bases). Coverage was minimum 25 million reads per sample. FASTQ files were then aligned to mouse genome sequence using STAR software and uniquely mapped read counts were visualized with Integrative Genomics Viewer (IGV). Gene expression level was calculated by the number of mapped reads. According to all gene expression level (RPKM or FPKM) of each sample, correlation coefficient of sample between groups was calculated. Read counts obtained from Gene Expression Analysis were used for differential expression analysis and differential expression analysis of different groups was performed using the DESeq2 R package. Hierarchical clustering analysis was carried out of log10 (FPKM+1) of union differential expression genes, within all comparison groups. ClusterProfiler software was used for enrichment analysis, including GO Enrichment, DO Enrichment, KEGG Enrichment and Reactome Enrichment.
Statistical analysis and reproducibility
All in vitro experiments shown were done in triplicates with similar results obtained. All in vivo experiments were repeated in at least three biological replicates with similar results obtained. Mice were randomly allocated to control or experimental conditions. Experimenters were not blinded to conditions during data collection or analyses. Statistical analyses are included in the figure legends. Data are presented as the mean + SEM. N = number of individual transfections for in vitro experiments and N = number of animals for in vivo experiments. Statistical analyses were performed using prims 7 Software (GraphPad) using the non-parametric one-tailed Mann-Whitney U test. p value ≤ 0.05 was considered significant (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001).
Data availability
RNA–seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE152412. Source data are provided with this paper. All other data supporting the findings are available upon reasonable request. All materials are available upon completion of a material transfer agreement.
Extended Data
Extended Data Fig. 1: Evaluation of endogenous Myd88 gene expression using different CRISPR-mediated repressor circuits.
(A-B) N2A cells were transfected with Myd88 gRNA pairs along with either dCas9 plasmid fused to HP1aKRAB or dCas9 and MS2-HP1aKRAB on two separate cassettes. Expression levels of (A) Myd88, (B) dCas9, and HP1aKRAB are quantified relative to No-Guide group (N=3 independent samples) The bars represent the mean + S.E.M. Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney test. A p value ≤ 0.05 was considered significant (*P ≤ 0.05). Statistical source data are provided in Source data extended data fig. 1.
Extended Data Fig. 2: In vivo analysis of AAV1 tropism towards different tissues.
AAV1-GFP was delivered to C57BL/6 mice via retro-orbital injection. GFP expression was assessed in different tissues by qRT-PCR. Average fold change expression levels are indicated above each group and are quantified relative to not injected mice (N=3 for not injected group, N=4 for AAV-GFP group, N=5 for AAV-GFP group in spleen, and N=6 for AAV-GFP group in liver). Statistical source data are provided in Source data extended data fig. 2.
Extended Data Fig. 3: RNA-seq analyses of bone marrow samples collected from mice treated with AAV1/Myd88-MS2-HP1aKr-ab versus AAV1/Myd88-MS2-Krab.
(A) Scatter plot comparing expression of genes (Fragments Per Kilobase of transcript per Million mapped reads FPKM) in two replicates of bone marrow from Myd88-MS2-HP1aKRAB versus Myd88-MS2-KRAB. Myd88, Il1β, Icam1, Tnfa and Il6 are highlighted in red and the most downregulated genes in Myd88-Ms2-HP1a-KRAB groups as compared to MyD88-MS2-KRAB are highlighted in Cyan (N=2 mice).
(B) GO enrichment bar graph comparing bone marrow samples collected from mice treated with AAV1/Myd88-MS2-HP1aKrab versus AAV1/Myd88-MS2-Krab. The top 20 significantly enriched terms in the GO enrichment analysis are displayed. Note that pathways such as defense response to bacteria, which are associated with Myd88 signaling are mostly down regulated when HP1aKRAB was used (N=2 mice).
(C) Reactome Enrichment bar graph displaying the top 20 enriched genes in the Reactome database comparing in the BM samples of Myd88-MS2-HP1aKRAB versus Myd88-MS2-KRAB (N=2 mice). Statistical analysis was performed using the two-tailed t test and the method of multiple comparisons adjustments was Benjamini-Hochberg.
Extended Data Fig. 4: Evaluation of endogenous Myd88 gene expression following multiple AAV administration.
(A) Schematic of experiments demonstrating Cas9 transgenic mice treated with AAV1/Myd88 or AAV1/Mock at day 1, followed by a second administration of AAV1/Mock on day 21.
(B) qRT-PCR analysis of Myd88 expression level in lung, blood, and bone marrow of Cas9 transgenic mice (N = 4 mice). Fold changes are relative to universal control. The bars represent the mean + S.E.M.
(C) Schematic of the experiment. Cas9 nuclease transgenic mice were treated with AAV1-Myd88 or AAV1-Mock vectors via retro-orbital injection followed by a second and third injection of AAV9-PCSK9 vectors on day 7 and 21.
(D) qRT-PCR analysis of Myd88 expression level in lung, blood, and bone marrow of Cas9 transgenic mice (N = 4 mice). The bars represent the mean + S.E.M. (Mock= Mock-HP1aKRAB, Myd88= Myd88-HP1aKRAB, PCSK9= PCSK9-HP1aKRAB). Fold changes are relative to universal control. Universal control is a blood sample collected from an uninjected Cas9 transgenic mouse. Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney U test. A p value ≤ 0.05 was considered significant (*P ≤ 0.05). Statistical source data are provided in Source data fig. 4.
Extended Data Fig. 5: Analysis of a set of immune-related transcripts following LPS injury.
qRT-PCR analysis of Ncf, Il6, Ifnγ, and Il1β mRNA expression in lung, blood, and bone marrow quantified relative to the universal control following LPS injection (N = 6 mice for injected groups except for Blood and Bone marrow of Myd88+LPS N=5, and N = 2 mice for Not Injected group). The bars represent the mean + S.E.M. (Mock= Mock-HP1aKRAB, Myd88= Myd88-HP1aKRAB). Universal control is a blood sample collected from an uninjected Cas9 transgenic mouse. Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney U test. A p value ≤ 0.05 was considered significant (*P ≤ 0.05 and **P ≤ 0.01). Statistical source data are provided in Source data extended data fig. 5.
Extended Data Fig. 6: Assessing the level of a panel of immune related genes in lung and bone marrow following LPS injection.
qRT-PCR analysis of in vivo CD68, Infα, Infβ, CD4, Cxcl1, and Stat4 relative to the universal control following LPS injection in lung and bone marrow. (N = 6 mice for injected groups except for Bone Marrow of Myd88+LPS group, Lung of Mock+LPS, and Lung of Myd88+LPS for Cxcl1 N=5 mice, and Lung of of Mock+LPS group for Cxcl1 N=4 mice, and N = 2 mice for Not Injected group). The bars represent the mean + S.E.M. (Mock= Mock-HP1aKRAB, Myd88= Myd88-HP1aKRAB). Universal control is a blood sample collected from an uninjected Cas9 transgenic mouse. Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney U test A p value ≤ 0 05 was considered significant (*P ≤ 0 05 and **P ≤ 0 01). Statistical source data are provided in Source data extended data fig. 6.
Extended Data Fig. 7: Targeted gene silencing in wild-type mice using a dual CRISPR/Cas9 system with AAV1/Cas9 and AAV1 carrying gRNA-MS2-HP1aKRAB.
AAV1 viruses were delivered to wild-type mice via retro-orbital injection. qRT-PCR analysis was performed to assess Myd88, Icam-1, Tnfα, Ncf, Il6, and Il1β mRNA expression in blood, bone marrow, and lung. Fold change expression levels were quantified relative to the universal control (N = 4 mice). The bars represent the mean + S.E.M. (Mock= Mock-HP1aKRAB, Myd88= Myd88-HP1aKRAB). Universal control is a blood sample collected from an uninjected Cas9 transgenic mouse. Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney U test. A p value ≤ 0.05 was considered significant (*P ≤ 0.05 and **P ≤ 0.01). Statistical source data are provided in Source data extended data fig. 7.
Extended Data Fig. 8: Assessing the repression efficiency of AAV1-Myd88 targeting a different region of Myd88 in liver.
(A) qRT-PCR analysis of in vivo Myd88 expression in liver samples 3 weeks post retro-orbital injection of AAV1 in Cas9 transgenic animals. Gene expression fold-change was quantified relative to the universal control (N = 2 mice for Not Injected group, N=4 mice for Mock, N=7 mice for Myd88Guideset1, N=3 mice for Myd88Guideset2). The bars represent the mean + S.E.M.
(B) qRT-PCR analysis of in vivo Myd88 expression in liver samples 6 hours post LPS injection. Fold change expression levels were quantified relative to the universal control (N = 2 mice for Not Injected group, N=6 mice for Mock, N=4 mice for Myd88Guideset2). The bars represent the mean + S.E.M. Universal control is a blood sample collected from an uninjected Cas9 transgenic mouse. Statistical analysis was performed using the non-parametric one-tailed Mann-Whitney U test. A p value ≤ 0.05 was considered significant (*P ≤ 0.05 and **P ≤ 0.01). Statistical source data are provided in Source data extended data fig. 8.
Supplementary Material
Acknowledgments
This work was primarily supported by RO1 grant from National Institute of Biomedical Imaging and Bioengineering (R01EB024562), startup fund by the School of Biological and Health Systems Engineering of Ira. A Fulton Schools of Engineering at Arizona State University, Pittsburgh Liver Research Center, University of Pittsburgh School of Medicine (NIH/NIDDK P30DK120531), NIH 8-U01-EB029372–02 as well as DARPA Young Faculty Award (D16AP00047) to S.K. S.K. and M.R.E are also partly supported by an R01 from National Institute of Biomedical Imaging and Bioengineering (EB028532), an R01 from the National Heart, Lung, and Blood Institute (HL141805). A.C. is supported by a Career Awards for Medical Scientists from the Burroughs Welcome Fund. We would like to thank Pouya Amrollahi for helping in the figures. We thank the Molecular Epidemiology Analytics Core and Angela Bond at Arizona State University for the Multiplex-ELISA service. We thank all members of Kiani and Ebrahimkhani labs for their assistance and insightful discussions. We also thank Novogene Corporation Inc. and UCLA Genomic Core for RNA sequencing and initial analysis.
Footnotes
Competing interest
Samira Kiani is a co-founder of SafeGen Therapeutics LLC. An international patent application has been filed for this work (PCT/US19/60285).
References
- 1.Moreno AM et al. In Situ Gene Therapy via AAV-CRISPR-Cas9-Mediated Targeted Gene Regulation. Mol Ther 26, 1818–1827, doi: 10.1016/j.ymthe.2018.04.017 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thakore PI et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nat Commun 9, 1674, doi: 10.1038/s41467-018-04048-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng Y et al. CRISPR interference-based specific and efficient gene inactivation in the brain. Nat Neurosci 21, 447–454, doi: 10.1038/s41593-018-0077-5 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Zhou H et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat Neurosci 21, 440–446, doi: 10.1038/s41593-017-0060-6 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Liao HK et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 171, 1495–1507 e1415, doi: 10.1016/j.cell.2017.10.025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breinig M et al. Multiplexed orthogonal genome editing and transcriptional activation by Cas12a. Nature methods 16, 51–54 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Matharu N et al. CRISPR-mediated activation of a promoter or enhancer rescues obesity caused by haploinsufficiency. Science 363, eaau0629 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L, Zhao L, Gao Y, Xu J & Han R Empower multiplex cell and tissue-specific CRISPR-mediated gene manipulation with self-cleaving ribozymes and tRNA. Nucleic acids research 45, e28–e28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu X et al. High-fidelity CRISPR/Cas9-based gene-specific hydroxymethylation rescues gene expression and attenuates renal fibrosis. Nature communications 9, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert LA et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert LA et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearns NA et al. Functional annotation of native enhancers with a Cas9–histone demethylase fusion. Nature methods 12, 401–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thakore PI et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nature methods 12, 1143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thakore PI, Black JB, Hilton IB & Gersbach CA Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nature methods 13, 127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konermann S et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature 500, 472–476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Russa MF & Qi LS The new state of the art: Cas9 for gene activation and repression. Molecular and cellular biology 35, 3800–3809 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evers B et al. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nature biotechnology 34, 631 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Yeo NC et al. An enhanced CRISPR repressor for targeted mammalian gene regulation. Nature methods 15, 611 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiani S et al. Cas9 gRNA engineering for genome editing, activation and repression. Nat Methods 12, 1051–1054, doi: 10.1038/nmeth.3580 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X & Yang Y Targeting the TLR9-MyD88 pathway in the regulation of adaptive immune responses. Expert Opin Ther Targets 14, 787–796, doi: 10.1517/14728222.2010.501333 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janssens S & Beyaert R A universal role for MyD88 in TLR/IL-1R-mediated signaling. Trends Biochem Sci 27, 474–482 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Warner N & Nunez G MyD88: a critical adaptor protein in innate immunity signal transduction. J Immunol 190, 3–4, doi: 10.4049/jimmunol.1203103 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Plant L, Wan H & Jonsson A-B MyD88-dependent signaling affects the development of meningococcal sepsis by nonlipooligosaccharide ligands. Infection and immunity 74, 3538–3546 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu X et al. MYD88 L265P mutation in lymphoid malignancies. Cancer research 78, 2457–2462 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Liao H-K et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nature communications 6, 6413 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Castle MJ, Turunen HT, Vandenberghe LH & Wolfe JH in Gene Therapy for Neurological Disorders 133–149 (Springer, 2016). [Google Scholar]
- 27.Merkel SF et al. Trafficking of adeno‐associated virus vectors across a model of the blood–brain barrier; a comparative study of transcytosis and transduction using primary human brain endothelial cells. Journal of neurochemistry 140, 216–230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S et al. Efficient transduction of vascular endothelial cells with recombinant adeno-associated virus serotype 1 and 5 vectors. Human gene therapy 16, 235–247 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veron P et al. Major subsets of human dendritic cells are efficiently transduced by self-complementary adeno-associated virus vectors 1 and 2. J Virol 81, 5385–5394, doi: 10.1128/JVI.02516-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y & Song S Distinct immune responses to transgene products from rAAV1 and rAAV8 vectors. Proc Natl Acad Sci U S A 106, 17158–17162, doi: 10.1073/pnas.0909520106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudres M et al. MyD88 signaling in B cells regulates the production of Th1-dependent antibodies to AAV. Mol Ther 20, 1571–1581, doi: 10.1038/mt.2012.101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu J, Huang X & Yang Y The TLR9-MyD88 pathway is critical for adaptive immune responses to adeno-associated virus gene therapy vectors in mice. J Clin Invest 119, 2388–2398, doi: 10.1172/JCI37607 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin X, Kong J, Wu Q, Yang Y & Ji P Effect of TLR4/MyD88 signaling pathway on expression of IL-1beta and TNF-alpha in synovial fibroblasts from temporomandibular joint exposed to lipopolysaccharide. Mediators Inflamm 2015, 329405, doi: 10.1155/2015/329405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park GS & Kim JH LPS Up-Regulates ICAM-1 Expression in Breast Cancer Cells by Stimulating a MyD88-BLT2-ERK-Linked Cascade, Which Promotes Adhesion to Monocytes. Mol Cells 38, 821–828, doi: 10.14348/molcells.2015.0174 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu M et al. MyD88-dependent interplay between myeloid and endothelial cells in the initiation and progression of obesity-associated inflammatory diseases. J Exp Med 211, 887–907, doi: 10.1084/jem.20131314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van den Akker TW, de Glopper-van der Veer E, Radl J, & Benner R The influence of genetic factors associated with the immunoglobulin heavy chain locus on the development of benign monoclonal gammapathy in ageing IgH-congenic mice. Immunology 65, 31 (1988). [PMC free article] [PubMed] [Google Scholar]
- 37.Thakore PI et al. RNA-guided transcriptional silencing in vivo with S. aureus CRISPR-Cas9 repressors. Nature communications 9, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abifadel M et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nature genetics 34, 154 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Maxwell KN & Breslow JL Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proceedings of the National Academy of Sciences 101, 7100–7105 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang H et al. Sepsis induces hematopoietic stem cell exhaustion and myelosuppression through distinct contributions of TRIF and MYD88. Stem cell reports 6, 940–956 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma X-Y, Tian L-X & Liang H-P Early prevention of trauma-related infection/sepsis. Military Medical Research 3, 33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho S-Y & Choi J-H Biomarkers of sepsis. Infection & chemotherapy 46, 1–12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao Z et al. Blood-borne lipopolysaccharide is rapidly eliminated by liver sinusoidal endothelial cells via high-density lipoprotein. The Journal of Immunology 197, 2390–2399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dandekar A et al. Toll-like receptor (TLR) signaling interacts with CREBH to modulate high-density lipoprotein (HDL) in response to bacterial endotoxin. Journal of Biological Chemistry 291, 23149–23158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnare M et al. Toll-like receptors control activation of adaptive immune responses. Nat Immunol 2, 947–950, doi: 10.1038/ni712 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Hiragami K & Festenstein R Heterochromatin protein 1: a pervasive controlling influence. Cellular and molecular life sciences CMLS 62, 2711–2726 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultz DC, Ayyanathan K, Negorev D, Maul GG & Rauscher FJ SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes & development 16, 919–932 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canzio D et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Molecular cell 41, 67–81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meehan RR, Kao CF & Pennings S HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. The EMBO journal 22, 3164–3174 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canzio D, Larson A & Narlikar GJ Mechanisms of functional promiscuity by HP1 proteins. Trends in cell biology 24, 377–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moghadam F, et al. Synthetic immunomodulation with a CRISPR super-repressor in vivo. Protoc. Exch 10.21203/rs.3.pex-1027/v1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA–seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE152412. Source data are provided with this paper. All other data supporting the findings are available upon reasonable request. All materials are available upon completion of a material transfer agreement.