Abstract
The crumbs (crb) apical polarity genes are essential for the development and functions of epithelia. Adult zebrafish retinal neuroepithelium expresses three crb genes (crb1, crb2a, and crb2b); however, it is unknown whether and how Crb1 differs from other Crb proteins in expression, localization, and functions. Here, we show that, unlike zebrafish Crb2a and Crb2b as well as mammalian Crb1 and Crb2, zebrafish Crb1 does not localize to the subapical regions of photoreceptors and Müller glial cells; rather, it localizes to a small region of cone outer segments: the cell membranes surrounding the axonemes. Moreover, zebrafish Crb1 is not required for retinal morphogenesis and photoreceptor patterning. Interestingly, Crb1 promotes rod survival under strong white light irradiation in a previously unreported non--cell-autonomous fashion; in addition, Crb1 delays UV and blue cones' chromatin condensation caused by UV light irradiation. Finally, Crb1 plays a role in cones' responsiveness to light through an arrestin-translocation-independent mechanism. The localization of Crb1 and its functions do not differ between male and female fish. We conclude that zebrafish Crb1 has diverged from other vertebrate Crb proteins, representing a neofunctionalization in Crb biology during evolution.
SIGNIFICANCE STATEMENT Apicobasal polarity of epithelia is an important property that underlies the morphogenesis and functions of epithelial tissues. Epithelial apicobasal polarity is controlled by many polarity genes, including the crb genes. In vertebrates, multiple crb genes have been identified, but the differences in their expression patterns and functions are not fully understood. Here, we report a novel subcellular localization of zebrafish Crb1 in retinal cone photoreceptors and evidence for its new functions in photoreceptor maintenance and light responsiveness. This study expands our understanding of the biology of the crb genes in epithelia, including retinal neuroepithelium.
Keywords: apicobasal polarity, axoneme, Crumbs, light damage, photoreceptor, zebrafish
Introduction
Crumbs (Crb) apical polarity proteins are Type I single-pass transmembrane proteins. They are essential for a variety of epithelial tissues in both invertebrates and vertebrates. Their extracellular domains mediate cell-cell interactions, and their cytoplasmic C-terminal domains mediate intracellular signaling (Pocha and Knust, 2013). The prototype Crb was first discovered in Drosophila melanogaster (Tepass et al., 1990), in which Crb localizes to the subapical regions (SARs) of cell membranes, apical to the septate junctions in epithelial cells (Tepass et al., 1990). Subsequently, three classes of Crb orthologs have been identified in vertebrates: Crb1, Crb2, and Crb3, with Crb1 and Crb2 carrying large extracellular domains like the fly Crb, and Crb3 missing most part of the extracellular domain (den Hollander et al., 1999; Makarova et al., 2003; Lemmers et al., 2004; van den Hurk et al., 2005; Bulgakova and Knust, 2009). For example, the zebrafish genome contains five crb genes (crb1, crb2a, crb2b, crb3a, and crb3b) (Omori and Malicki, 2006). In vertebrate epithelial cells, Crb proteins normally localize to SARs, immediately apical to the tight junctions (Mehalow et al., 2003; Roh et al., 2003; van de Pavert et al., 2004; van den Hurk et al., 2005; Omori and Malicki, 2006; Zou et al., 2012; Pellissier et al., 2014). The multiplicity of vertebrate crb genes and their structural variations suggest diversified Crb functions.
Indeed, since the identification of Crb in the fruit fly, Crb proteins have been found to play essential roles in many biological properties and processes, such as apicobasal polarity and integrity of epithelia (Letizia et al., 2013), cell-cell adhesions and cellular patterning (Grawe et al., 1996; Tepass, 1996; Lemmers et al., 2004; Zou et al., 2012), cell proliferation and survival (Pellikka and Tepass, 2017), Hippo signaling (Chen et al., 2010; Pan, 2010; Parsons et al., 2010), protein trafficking (Pocha et al., 2011a), photoreceptor morphogenesis (Izaddoost et al., 2002; Pellikka et al., 2002; Nam and Choi, 2003), etc. Given the multiplicity of vertebrate crb genes, it is possible that this broad spectrum of Crb functions is deferentially allocated to various crb genes.
This possibility of differential allocation of functions among crb genes is supported by genetic evidence in humans. For example, ∼10% of Leber's congenital amaurosis (LCA) and retinitis pigmentosa (RP) are because of the malfunction of human CRB1 caused by >300 pathogenic mutations (den Hollander et al., 1999; Abouzeid et al., 2006; Vallespin et al., 2007; Bujakowska et al., 2012; Corton et al., 2013; Tiab et al., 2013). However, LCA and RP have not been found to be associated with human CRB2 and CRB3 (Slavotinek, 2016), although some missense mutations in CRB2 were linked to reduced visual acuity, mild optic atrophy, and irregular retinal pigmentation (van den Hurk et al., 2005; Lamont et al., 2016). The preferential association of CRB1 with RP and LCA may suggest that, in humans, CRB1 carries more retinal functions than CRB2 and CRB3 do; alternatively, mutations of CRB2 and CRB3 are embryonic lethal and their retinal phenotypes are not available. Regardless, the etiology of CRB1-related LCA or RP (Quinn et al., 2017) and how the crb genes differ from one another in vertebrate retinal functions remain elusive.
To understand the functional diversification of Crb proteins, we chose to use zebrafish retina as our model because zebrafish retina expresses three crb genes: crb1, crb2a, and crb2b (Omori and Malicki, 2006; Zou et al., 2012). Of these three genes, the cell type-specific gene expression patterns, subcellular localizations, and some functions of crb2a and crb2b have been characterized previously: Crb2a localizes to the SARs of undifferentiated retinal epithelial cells and plays an important role in establishing and maintaining the apicobasal polarity and integrity of the retinal neuroepithelium (Malicki and Driever, 1999; Omori and Malicki, 2006). In developed retina, Crb2a localizes to the SARs of the inner segments of all photoreceptors as well as the apical processes of Müller glial cells (Zou et al., 2012). By contrast, Crb2b is restrictively expressed, as Crb2b lf (long isoform) and Crb2b-sf (short isoform), in the red, green, and blue cones (RGB cones) of developed retina and localizes to the SARs of RGB cones (Zou et al., 2012; Fang et al., 2017). Both Crb2a and Crb2b are required for maintaining the mirror symmetric and pentameric (R-G-B-R-G) organization of RGB cone photoreceptors by mediating cell-cell adhesions (Zou et al., 2012). However, Crb1's cell type-specific expression pattern, protein localization, and functions in the retina remain unknown.
Thus, we here characterize zebrafish Crb1's retinal expression pattern, subcellular localization, and functions. We show that zebrafish Crb1 is not only different from zebrafish Crb2a and Crb2b, but also different from human and mouse Crb1 and Crb2, suggesting that it may represent a neofunctionalization of Crb proteins during evolution.
Materials and Methods
Zebrafish strains and animal care
Tubingen WT, Tg(−5.5opn1sws1:EGFP)kj transgenic (Takechi et al., 2003), Tg (LCRRH2-RH2-1:GFP)pt112 transgenic (Fang et al., 2013), Tg(−3.7rho:EGFP)kj2 transgenic (Hamaoka et al., 2002), and crb1sa12558 mutant (simplified as crb1 mutant hereafter; Zebrafish International Resource Center, catalog #ZL9088.07; Busch-Nentwich et al., 2012) zebrafish were maintained on a 14 h light/10 h dark cycle at 28.5°C. Zebrafish embryos were raised in an incubator at 28.5°C. Animal care and handling were in accordance with the guidelines of the University of Pittsburgh.
Phylogenetic analysis of Crb amino acid sequences
The phylogenetic relationships of the extracellular domain, intracellular domain, and full-length sequences of Crb proteins of 14 species were analyzed with the MEGA X program (http://www.megasoftware.net/). We use the neighbor-joining method to generate the unrooted phylogenetic trees. In addition, Crb amino acid sequences were compared pairwise with VectorNTI and Genedoc (Nicholas et al., 1997). The animal species and the GenBank accession numbers of these Crb proteins are as follows: fruit fly (Drosophila busckii; Crb, ALC47276.1); nematode (Caenorhabditis elegans; Crb1, CCD66913.1; Crb2a, BAA92157.1; Crb2b, BAA92158.1; Crb3, NP_001041224.1); human (Homo sapiens; Crb1, NP_957705.1; Crb2, NP_775960.4; Crb3, AAQ89047.1); rhesus monkey (Macaca mulatta; Crb1, XP_014976303.1; Crb2, XP_001083811.3; Crb3, NP_001253010.1); chimpanzee (Pan troglodytes; Crb1, XP_009438564.1; Crb2, XP_528426.4; Crb3, XP_016790312.2); house mouse (Mus musculus; Crb1, AAL65131.1; Crb2, NP_001157038.1; Crb3, AAH24462.1); Norway rat (Rattus norvegicus; Crb1, NP_001100652.1; Crb2, NP_001129233.1; Crb3, NP_001020832.1); white-footed mouse (Peromyscus leucopus; Crb1, XP_028745692.1; Crb2, XP_028743732.1; Crb3, XP_028718368.1); Chinese alligator (Alligator sinensis; Crb1, XP_006036765.1; Crb2, XP_014382035.2; Crb3, XP_025059628.1); mainland tiger snake (Notechis scutatus; Crb1, XP_026527503.1; Crb2, XP_026546320.1; Crb3, XP_026537729.1); green sea turtle (Chelonia mydas; Crb1, XP_007063718.1; Crb2, XP_007061020.1; Crb3, XP_027676516.1); fugu (Takifugu rubripes; Crb1, ENSTRUT00000002807.2; Crb2, XP_029685596.1; Crb3, XP_029692235.1); medaka (Oryzias latipes; Crb1, XP_011472343.1; Crb2, XP_023814513.1; Crb3, XP_011476413.2); and zebrafish (Danio rerio; Crb1, NP_001038408.1; Crb2a, NP_001038764.1; Crb2b-lf, DQ314737; Crb3a, AAI33838.1; Crb3b, AAI33972.1).
Immunohistochemistry
Zebrafish larvae and adult eyes were fixed with 4% PFA at room temperature for 30 min and 2 h, respectively. After washes with PBS, the fixed samples were stored in 40% sucrose (in PBS) at 4°C before immunohistochemistry using the procedure described previously (Zou et al., 2012).
Antibodies used for this study were either purchased from companies or made by the X.W. laboratory; they are listed as follows. Rabbit polyclonal antibodies: anti-Crb1aa952-1258, anti-Crb2aaa97-457, and anti-Crb2b-lfaa466-773 (1:300) (Zou et al., 2012, 2013). Mouse monoclonal antibodies: anti-ZO-1 (1:300; Invitrogen, catalog #339100; RRID:AB_87181), anti-γ-tubulin (1:3000; Sigma Millipore, catalog #T3320; RRID:AB_261655), anti-acetylated tubulin (1:3000; Sigma Millipore, catalog #T7451; RRID:AB_609894), and Zpr1 anti-arrestin3a (1:300; ZIRC, catalog #AB_174435; RRID:AB_10013803). Secondary antibodies: Cy3-conjugated goat anti-rabbit IgG (1:300; Jackson ImmunoResearch Laboratories, catalog #111-165-144; RRID:AB_2338006), Cy3-conjugated goat anti-mouse IgG (1:300; Jackson ImmunoResearch Laboratories, catalog #115-165-166; RRID:AB_2338692), and Cy5-conjugated donkey anti-rabbit IgG (1:300; Jackson ImmunoResearch Laboratories, catalog #711-175-152; RRID:AB_2340607). In addition, Cy3-conjugated peanut agglutinin (PNA) (1:100; Vector Laboratories, catalog #CL-1073; RRID:AB_2313597) was used to stain red cone outer segments. AlexaFluor-488-conjugated phalloidin (1:300; Thermo Fisher Scientific, catalog #A12379; RRID:AB_2759222) and AlexaFluor-647-conjugated phalloidin (1:300; Thermo Fisher Scientific, catalog #A22287; RRID:AB_2620155) were used to stain F-actin. DAPI (1:1000; Thermo Fisher Scientific, catalog #D1306, RRID:AB_2629482) was used to stain the nuclei. Images were taken with a Fluoview FV1000 confocal microscope (Olympus).
Genotyping crb1 mutant embryos
crb1 mutants were genotyped with the Cleaved Amplified Polymorphic Sequence method (Konieczny and Ausubel, 1993). Specifically, a forward primer (5′-GGTCACGAGTGCCAGAA-3′) and a reverse primer (5′-TGCCATTCACCATCAGATACA-3′) were used to amplify a 375 bp DNA fragment that contains the crb1sa12558 mutation site. The PCR products amplified from either embryonic or adult tail fin DNA were then digested with PstI, which cuts the PCR products of WT but not mutant DNA into two fragments of 154 and 221 bp.
Electroretinography (ERG)
To evaluate the physiological function of Crb1, the ERG of WT and crb1 mutant larvae was measured at 6 dpf using the method described previously (Nelson et al., 2019). Briefly, the isolated larval eyes were placed in an inverted lid of a 35 mm culture dish in the recording chamber and were perfused with MEM (Thermo Fisher Scientific, catalog #11090-099) at 0.07 ml/min. The MEM, equilibrated with 95% O2 and 5% CO2, contained 20 mm L-aspartate (Sigma Millipore) to block postsynaptic glutamatergic photoreceptor mechanisms. The eyes were exposed to 10 successive 7 step white light irradiance-response sequences at 0.5 log unit increments. The white light was obtained from a 150 W Xenon arc through UV compliant optics (maximal irradiance of 0.15 µW/cm2). All four zebrafish cone types, including UV cones, are stimulated to saturation by this source (Nelson and Singla, 2009). Brightness levels were preadjusted to cover the anticipated response range between the threshold and saturation. The total time from the start of the first flash to the end of the protocol was 23 min 40 s. Each intensity-response sequence required ∼2 min 22 s.
Light damage treatments
To determine whether crb genes are required for maintaining photoreceptors under damaging light irradiation, adult fish were exposed to either strong white light or UV light irradiation. For white light irradiation, 3 mpf (months post fertilization) WT fish and crb1 mutants were treated as described previously (Thomas and Thummel, 2013). Specifically, 24 crb1 mutants and 24 WT fish (12 females and 12 males for each group) were kept in two tanks (positioned side by side at a distance of 29 cm) in a dark chamber for 2 d. Then, fish were irradiated for up to 3 d with two 250 W halogen utility lamps (Designers Edge L860 Portable, 4000 Lumen output), with two lamps on each side of the two-tank unit at a distance of 37 cm. To reduce the overheating stress, fish were kept in circulating aquatic system water during the treatment. Fish were killed at 0 and 3 dpl (days post the start of light treatment) and evaluated for the effects of the loss of Crb1 on photoreceptor survival. To quantify photoreceptors, we took confocal images of DAPI-stained retinal sections under a 60× objective lens and counted all photoreceptor nuclei in radial retinal sections that span a width of 210 μm. In addition, the thicknesses of inner nuclear layer (INL) were measured with Fiji software (Schindelin et al., 2012).
For UV light irradiation, 8 mpf-old fish were first transferred to a 1 L glass beaker containing 450 ml aquatic system water and then adapted in darkness for 1.5 h. The adapted fish were then exposed to UV light for 2 h in the irradiation chamber of a Gel Doc XR+ Imaging System (Bio-Rad, catalog #1708195, equipped with Ushio G8T5E Mercury-Arc UV-B light bulbs, 7.2 W, 306 nm UV output). The treated fish were then maintained in the aquatic system water for 12, 24, and 36 h before being killed. Then their eyes were enucleated, fixed with 4% PFA, and processed for histochemistry.
The measurements of the length of cone outer segments
To determine whether the loss of Crb1 affects the length of cone outer segments, the eyes from dark-adapted 6 dpf and 3 mpf WT and crb1 mutant fish were fixed with 4% PFA for 1 h at room temperature, cryo-sectioned, and immunostained with Cy3-conjugated PNA. The lengths of cone outer segments, revealed by the PNA staining, were measured with Fiji software (Schindelin et al., 2012).
Arrestin3a translocation analysis
Melanin pigmentation of zebrafish larvae was blocked by adding 0.003% phenylthiourea starting at 24 hpf. Fish larvae were dark-adapted starting at 5 dpf, then fixed at 6 dpf with 4% PFA for 1 h or, alternatively, exposed to ambient light for either 30 min or 4 h after the dark adaptation before fixation. Each sample group contained 30 larvae. These larvae were analyzed by confocal immunohistochemistry to evaluate the effects of the loss of Crb1 on the translocation of arrestin3a during the dark-to-light transition. Specifically, confocal images of arrestin3a were collected under nonsaturating laser powers. With Fiji software (Schindelin et al., 2012), we measured and calculated the average arrestin3a intensities of a circular area of 1 μm in diameter both at the inner segments, 2-3 μm below the ellipsoid, and at the outer segments, 1–2 μm above the ellipsoid (see Fig. 10A). Then the ratios of arrestin3a signal intensities in the outer segments to that in the inner segments were calculated.
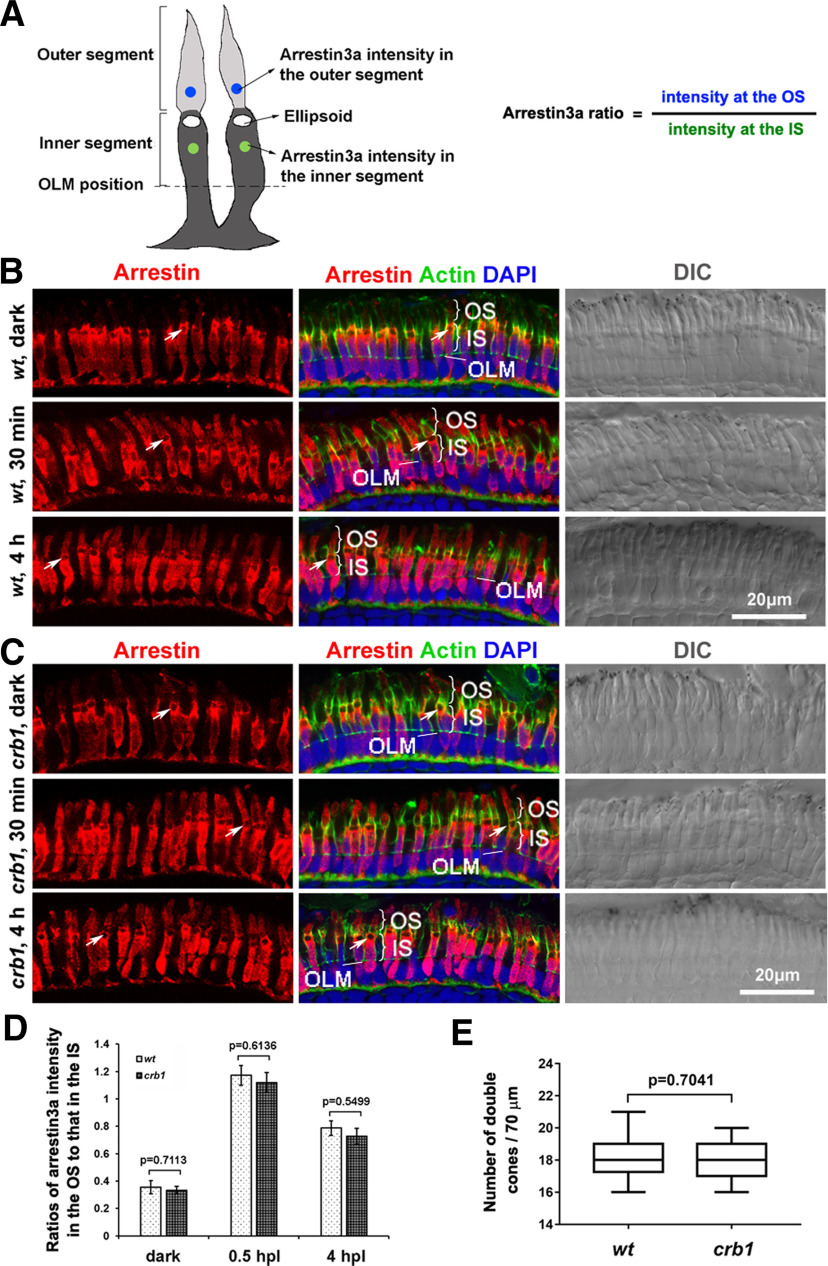
Figure 10.
The effect of the loss of Crb1 on the translocation of arrestin3a. A, Diagram represents the method of using the ratios of arrestin3a immunostaining intensities to evaluate the translocation of arrestin3a between the inner segments (IS) and the outer segments (OS). Green dots and blue dots represent the locations used to measure arrestin3a intensities, namely, at the centers of the IS and OS, respectively. B, C, Examples of arrestin3a distribution patterns between the IS and the OS of double cones in 6 dpf WT (B) and crb1 mutant retina (C) in dark, 30 min of light exposure, and 4 h of light exposure. Arrestin3a (red) was visualized with the Zpr1 antibodies, with counterstaining of actin (green) and DAPI (blue) as well as DIC images. Arrows indicate ellipsoids. Scale bar, 20 μm. D, Quantifications and comparison of arrestin3a intensity ratios between WT and crb1 mutant fish; 28 cells from 3 fish were quantified for each group. p values were generated by Student's t test, two-tailed hypothesis. E, Densities of arrestin3a-positive cones in WT and crb1 mutant retina at 6 dpf. Eighteen sections from 9 WT fish and 11 sections from 9 crb1 mutant fish were used for quantification. p values were generated by Student's t test, two-tailed hypothesis.
Experimental design and statistical analysis
This study was aimed at revealing the cellular expression pattern and subcellular localization of Crb1 in zebrafish retina by immunohistochemistry and at uncovering the functions of Crb1 by comparing phenotypic differences between WT and crb1 null mutant fish. When adult fish were analyzed, 3-6 males and females were used for each experimental condition. When larval zebrafish were analyzed, sex was not considered as a biological variable because sex is undifferentiated in zebrafish at larval stages. For confocal microscopy, image stacks were collected at an interval of 0.3 μm. The numbers of animals, cells, and tissue sections used in each experiment were specified in corresponding figure legends. All experiments were not preregistered.
In addition to the above general parameters, the following considerations were applied to specific experiments: To measure the length of red cone outer segments, only confocal images that displayed perfect radial sections of cones were used. Because the outer segments of cones are cone-shaped, the longest PNA-stained outer segment section from a series of optical sections of a red cone was regarded as the length of its outer segment. To quantify photoreceptors after light damage, the DAPI-stained cell nuclei were categorized and counted according to the photoreceptor type-specific nuclear morphologies and locations as detailed in Results. To quantify the rounding process of cone nuclei after UV light damage, perfect radial retinal sections were used to measure the radial and transverse diameters of condensed/condensing nuclei with Fiji software (Schindelin et al., 2012).
Statistical analysis was performed with GraphPad software. Student's t test with two-tailed hypothesis was used to compare the differences between WT and crb1 mutant fish (see Figs. 4, 5, 8–10). One-way ANOVA followed by Tukey's post hoc test was used to evaluate the variation of a quantitative dependent variable, such as the ratios of nuclear diameters, among three or more groups of a categorical independent variable, such as time points (see Figs. 7–9). Quantitative dependent variables are reported as mean ± SEM. A p value < 0.05 was considered statistically significant. Test statistics and p values are presented in corresponding results, figures, or figure legends. Detailed statistical data are available on request.
Figure 4.
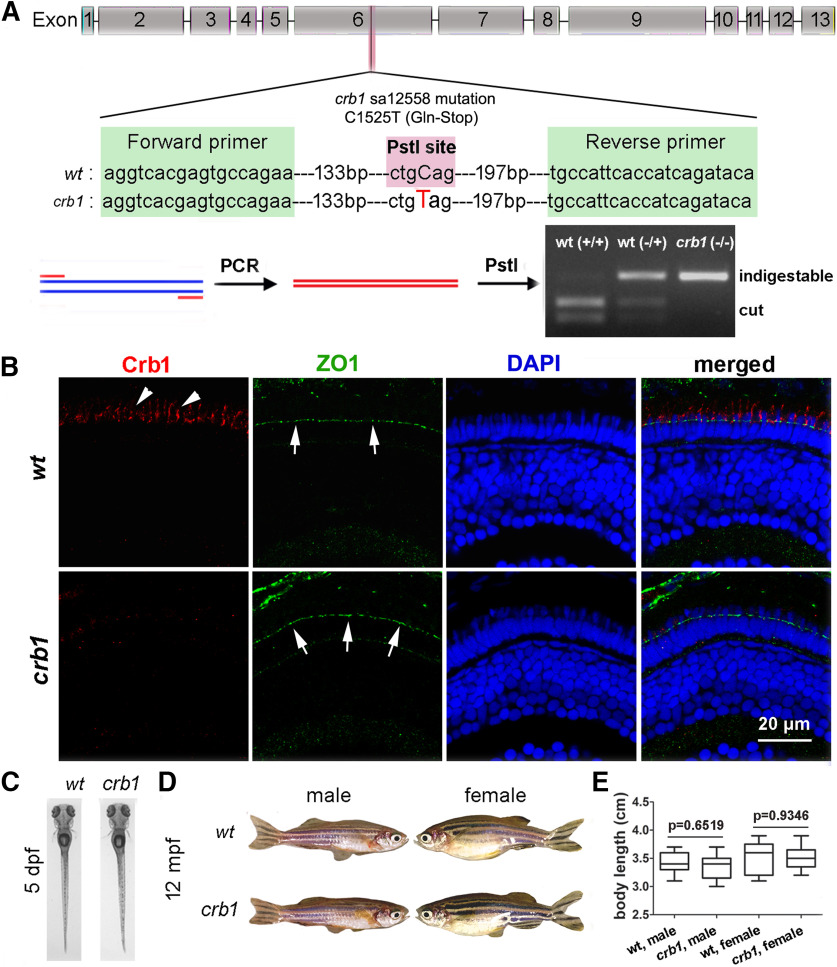
The loss of Crb1 does not compromise fish survival and development. A, The genotyping method to identify crb1 mutants: PCR amplification followed by PstI restriction digestion. B, Immunohistochemistry with anti-Crb1 antibodies revealed Crb1 signals (arrowheads) in WT (wt) but not in crb1sa12558 (crb1) mutants at 5 dpf, counterstained for ZO1 (green) of the OLM (arrows) and retinal nuclei (by DAPI, blue). Scale bar, 20 μm. C, D, The external appearances of WT fish and crb1 mutant fish at 5 dpf (C) and 12 mpf (D). E, Quantification of the body lengths of 12 mpf WT and crb1 mutant fish (6 males and 6 females). Data are mean ± SEM. p values were generated by Student's t test, two-tailed hypothesis.
Figure 5.
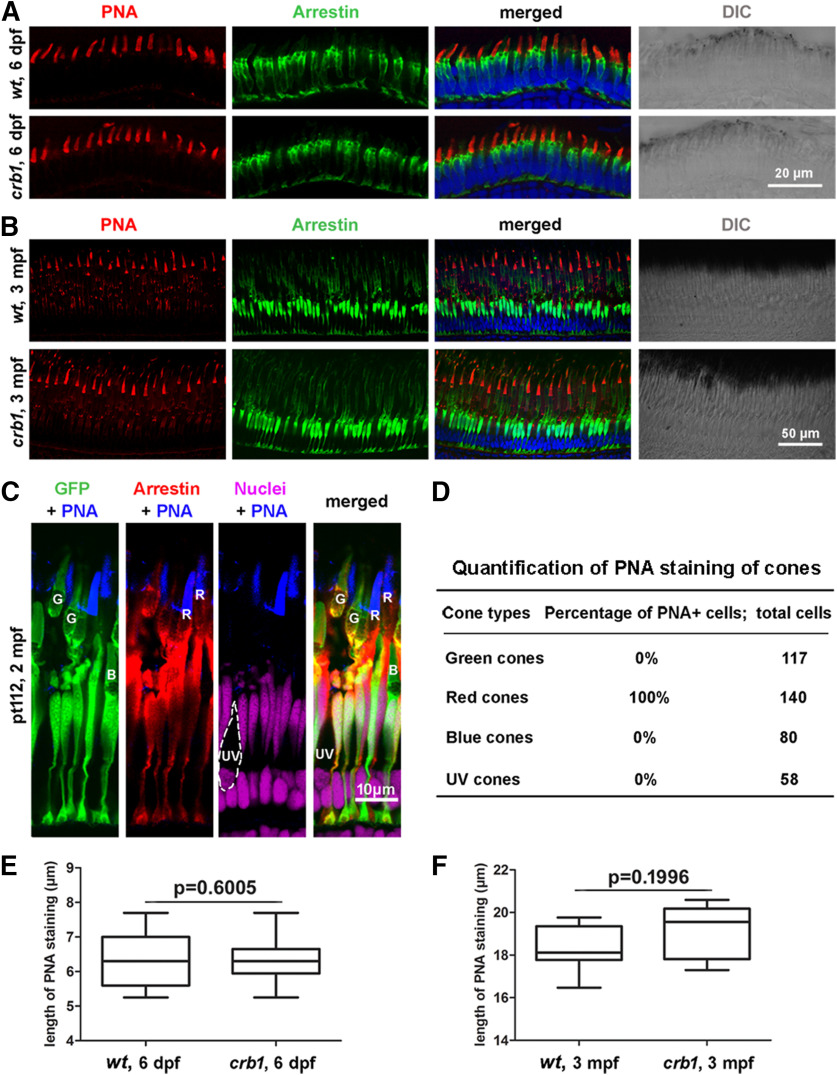
The loss of Crb1 did not compromise photoreceptor morphologies at larval and adult stages. A, B, Morphologies of WT (wt) and crb1 mutant double cones, revealed by immunostaining of arrrestin3a with the Zpr1 antibody, and red cone outer segments, stained with PNA, at 6 dpf (A, scale bar, 20 μm) and 3 mpf (B, scale bar, 50 μm). DIC, Differential interference contrast. C, PNA-stained red cone outer segments. Cone types were determined by immunostaining of arrestin3a in double cones in the Tg (LCRRH2-RH2-1:GFP)pt112 transgenic background (pt112), which expresses GFP in green and blue cones (Fang et al., 2013). The following staining and morphologic features were used to distinguish four cone types: G, green cones, positive for both GFP and arrestin3a; R, red cones, negative for GFP but positive for arrestin3a; B, blue cones, positive for GFP positive but negative for arrestin3a; UV, UV cones, shortest cones with their outer segments closest to the OLM and negative for both GFP and arrestin3a. Scale bar, 10 μm. D, Quantification of PNA staining of four cone types. E, Quantification of the lengths of red cone outer segments of dark-adapted fish at 6 dpf (phenylthiourea-treated). Thirty cones from 6 WT and 41 cones from 6 crb1 mutant were quantified. Data are mean ± SEM. p value was generated by Student's t test, two-tailed hypothesis. F, Quantification of the lengths of red cone outer segments at the central retinal region of dark-adapted 3 mpf adult fish. Eighteen cones from 3 WT and 18 cones from 3 crb1 mutants were quantified. Data are mean ± SEM. p value was generated by Student's t test, two-tailed hypothesis.
Figure 8.
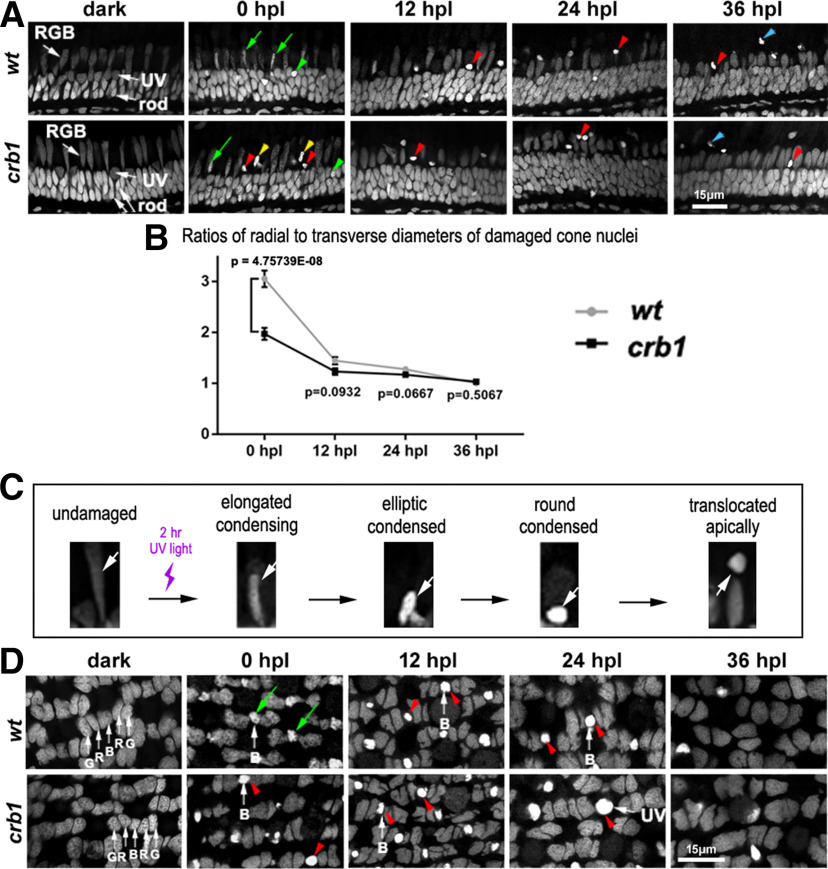
The retina of UV light-treated WT and crb1 adult fish displayed condensing/condensed chromatin in UV and blue cone nuclei. A, DAPI staining of radial sections of dorsal retina. Green arrowheads and green arrows indicate condensing UV and RGB cone nuclei, respectively. Yellow arrowheads indicate condensed elliptic nuclei. Red arrowheads indicate more condensed round nuclei. hpl, hours post a 2 h UV light irradiation. Scale bar, 15 μm. B, Comparisons between the ratios of radial to transverse diameters of condensed/condensing cone nuclei in WT and crb1 mutant fish after UV light irradiation. Data are mean ± SEM; 9 sections from 6 individuals were quantified for each group. More than 100 light-damaged cells were quantified at each time point in each group. Data are mean ± SEM. p values were generated by Student's t test, two-tailed hypothesis. C, The inferred morphologic changes and translocation of damaged cone nuclei in the RGB cone layer after UV light irradiation. The top side of images faces the retinal pigment epithelium. The bottom side of images faces the OLM. D, DAPI staining of transverse sections of the dorsal retina of UV light-treated fish. White arrows G, R, and B indicate green, red, and blue cone nuclei, respectively. Green arrows indicate condensing nuclei. Red arrowheads indicate condensed nuclei. Scale bar, 15 μm.
Figure 7.
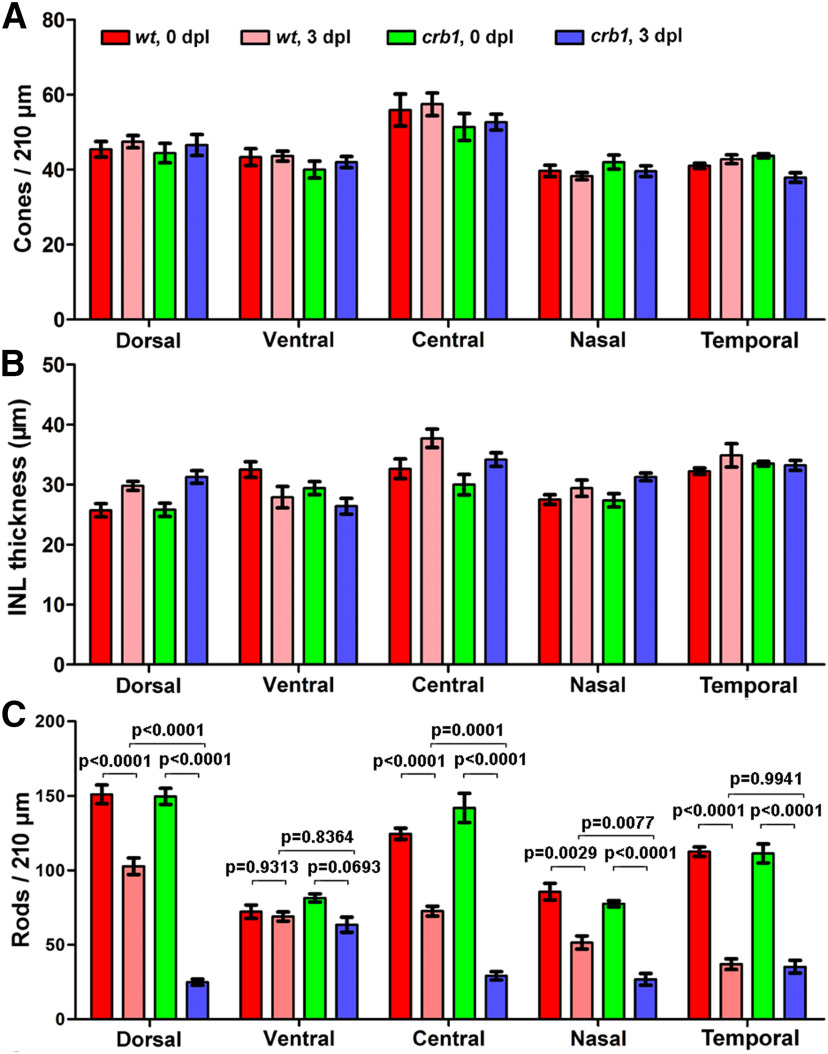
Quantification of the densities of cones and rods as well as the thicknesses of the INL in the centers of five local retinal regions (dorsal, ventral, central, nasal, and temporal) in WT and crb1 mutant fish. A, Average numbers of cones (RGB and UV cones) per 210 μm interval across retinal sections. B, Average thicknesses of the INL. C, Average numbers of rods per 210 μm interval across retinal sections. Data are mean ± SEM; 9 sections from 3 individuals were quantified for each group. Statistical significance was evaluated with one-way ANOVA and Tukey's post hoc analyses: dorsal, F(3,27) = 107.2; ventral, F(3,16) = 2.558; nasal, F(3,14) = 33.85; temporal, F(3,13) = 63.86; central, F(3,29) = 77.27.
Figure 9.
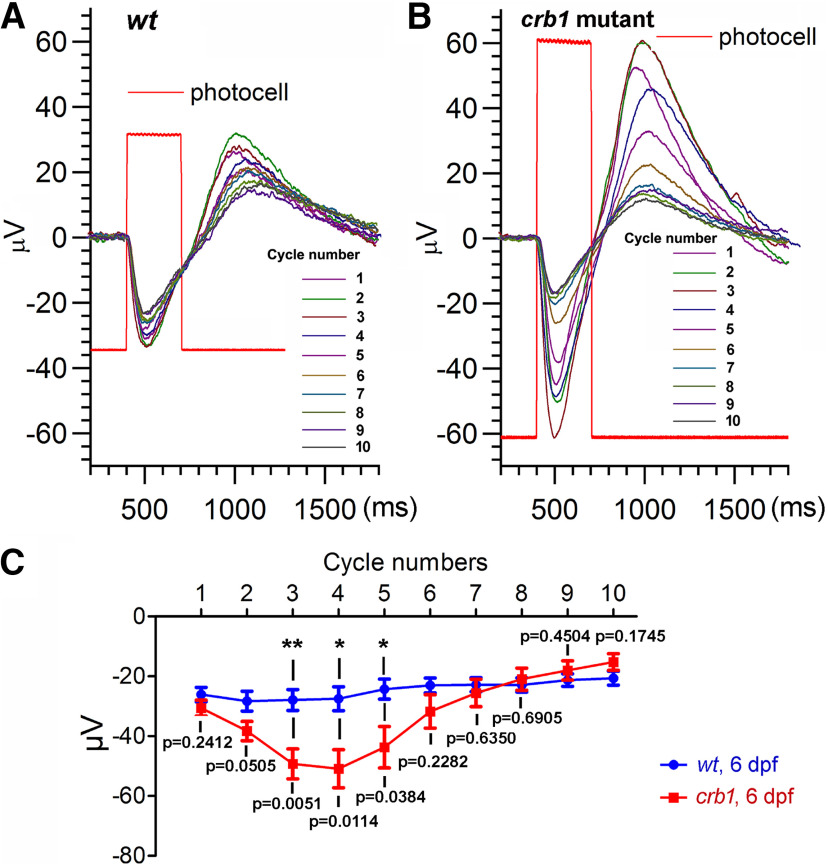
The effects of the loss of Crb1 on the ERG of cones. A, B, Representative ERG measurements of cone light responses in WT (A) and crb1 mutant larvae (B) at 6 dpf. Lines 1-10 indicate the successive cycle numbers of white light stimulation. Rectangular steps (red) represent the duration of light exposure as monitored by a photocell in the light beam. C, The average changes in the amplitudes of vitreal negativity during light stimulation over time (N = 8 eyes for wt; N = 11 eyes for crb1). Data are mean ± SEM. Statistical differences between WT and crb1 mutant fish were evaluated for each cycle of light stimulation with Student's t test, two-tailed hypothesis: *p < 0.05; **p < 0.01.
Results
Phylogenetic relationships among vertebrate Crb proteins
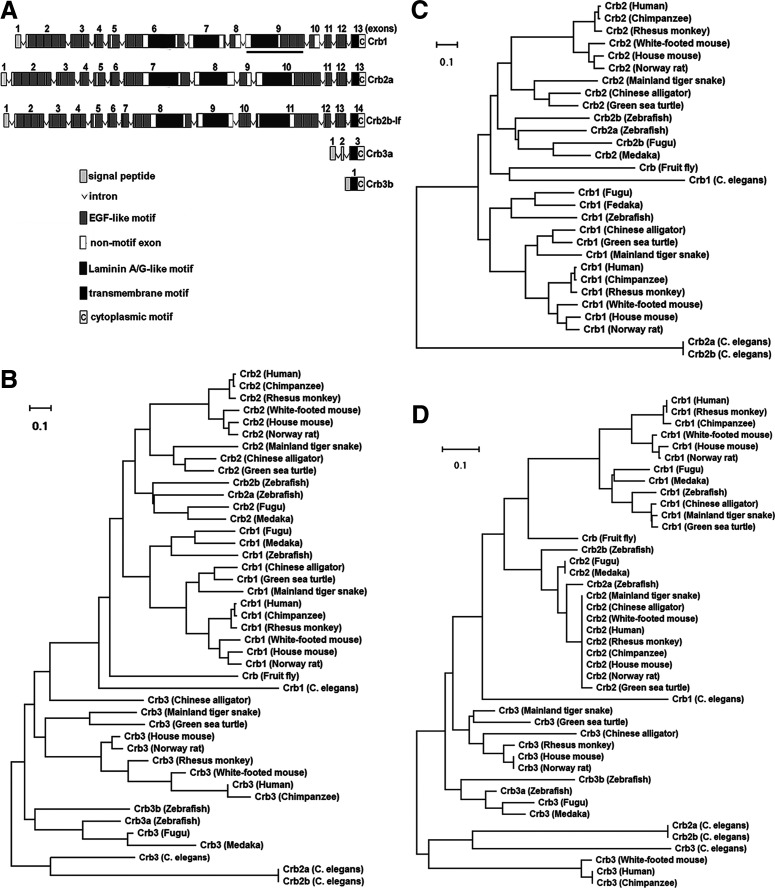
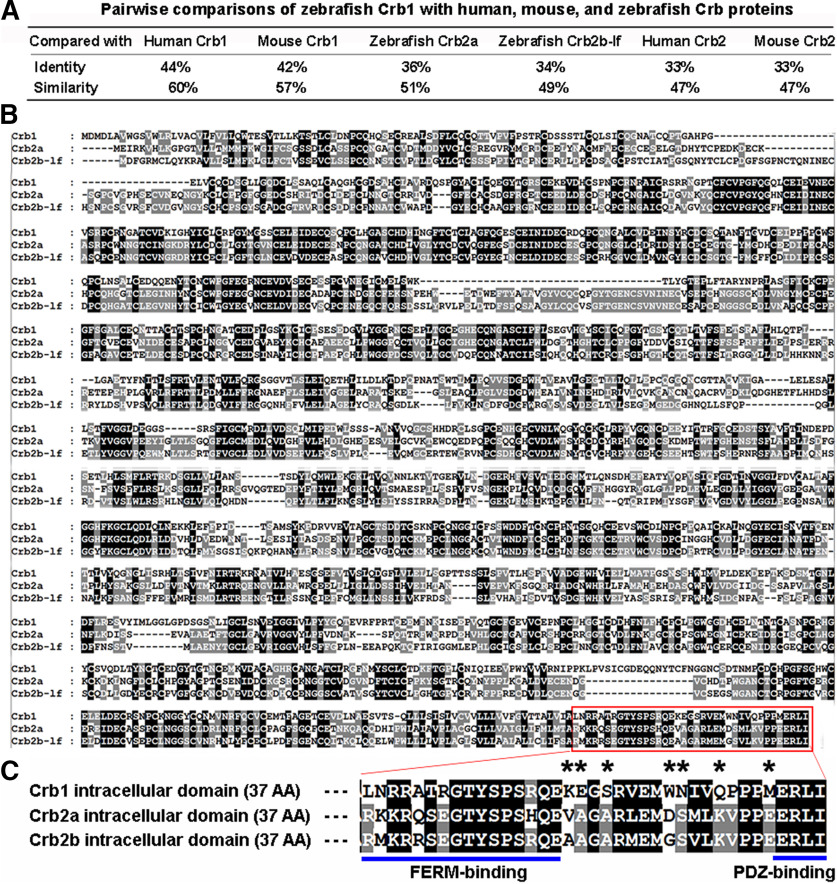
Generally speaking, proteins that are closer in phylogenetic relationships are more similar not only in structure but also in function. To gain insights into the functions of zebrafish Crb1, we evaluated its phylogenetic relationships with other Crb proteins from 14 species, including 3 teleost fish, 3 rodents, 3 primates, 3 reptiles, and 2 invertebrates. Because Crb3 proteins lack the conventional large extracellular domains (Fig. 1A), the phylogenetic analyses of Crb proteins were performed in three different ways, each using their full-length sequences (Fig. 1B), extracellular domains (Fig. 1C), or intracellular domains (Fig. 1D). All three comparisons suggest that zebrafish Crb1 is structurally more closely related to Crb1 of other vertebrate species; interestingly, the comparison of the intracellular domains suggests that zebrafish Crb1 is more closely related to reptile Crb1 proteins than to those of other 2 teleost fish. By contrast, the comparison with invertebrate Crb proteins suggests something different: While the intracellular domain of zebrafish Crb1 is more closely related to fly Crb, the extracellular domain of zebrafish Crb1 is less closely related to fly Crb and C. elegans Crb1 than zebrafish Crb2a and Crb2b are. These phylogenetic relationships are also reflected by pairwise sequence comparisons and alignments between zebrafish Crb1 and other Crb proteins (Fig. 2A,B) as well as by protein motif structures as predicted by SMART (http://smart.embl-heidelberg.de/), which revealed that zebrafish Crb1 contains 18 EGF-like motifs, compared with 19 and 21 EGF-like motifs in Crb2a and Crb2b, respectively (Fig. 1A) (Omori and Malicki, 2006). The intracellular domains of the three zebrafish Crb proteins have an identical PDZ-binding motif at the very C-terminus and similar FERM-binding motifs near the trans-membrane motif (Fig. 2C). However, the amino acids between the PDZ-binding and FERM-binding motifs of Crb1 differ significantly from those of Crb2a and Crb2b in hydrophobicity, polarity, and electric charge (Fig. 2C, asterisks).
Figure 1.
Phylogenetic relationships among invertebrate and vertebrate Crb proteins. A, Schematics of the structures of zebrafish Crb proteins. The coding exons are numbered from the 5′ end to the 3′ end. The antigenic region (amino acids 952–1258) for making anti-Crb1 antibody is underlined. B–D, The phylogenetic trees of the Crb proteins of 14 different species, generated by using full-length (B), extracellular (C), and intracellular amino acid sequences (D).
Figure 2.
Amino acid sequence comparisons of zebrafish Crb1 with human, mouse, and zebrafish Crb proteins. A, Pairwise comparisons of the sequence identities and similarities between zebrafish Crb1 and other Crb proteins. B, Alignments of the amino acid sequences of zebrafish Crb1, Crb2a, and Crb2b-lf. The last 37 amino acids in a red rectangle are the intracellular domains. C, The PDZ-binding motif and FERM-binding motif of the intracellular domains of Crb1, Crb2a, and Crb2b are underlined. Asterisks indicate the major variations of amino acids in the interval region between the PDZ-binding and FERM-binding motifs.
Together, zebrafish Crb1 is generally more like other vertebrate Crb1 proteins than Crb2 proteins, but its lesser similarity to fly Crb and C. elegans Crb1 in the extracellular domains and its more similarity to reptile Crb1 proteins than to other fish Crb1 proteins in the intracellular domains may suggest a functional diversification.
Zebrafish Crb1 localizes to the cell membranes that encircle the axonemes of cone photoreceptors
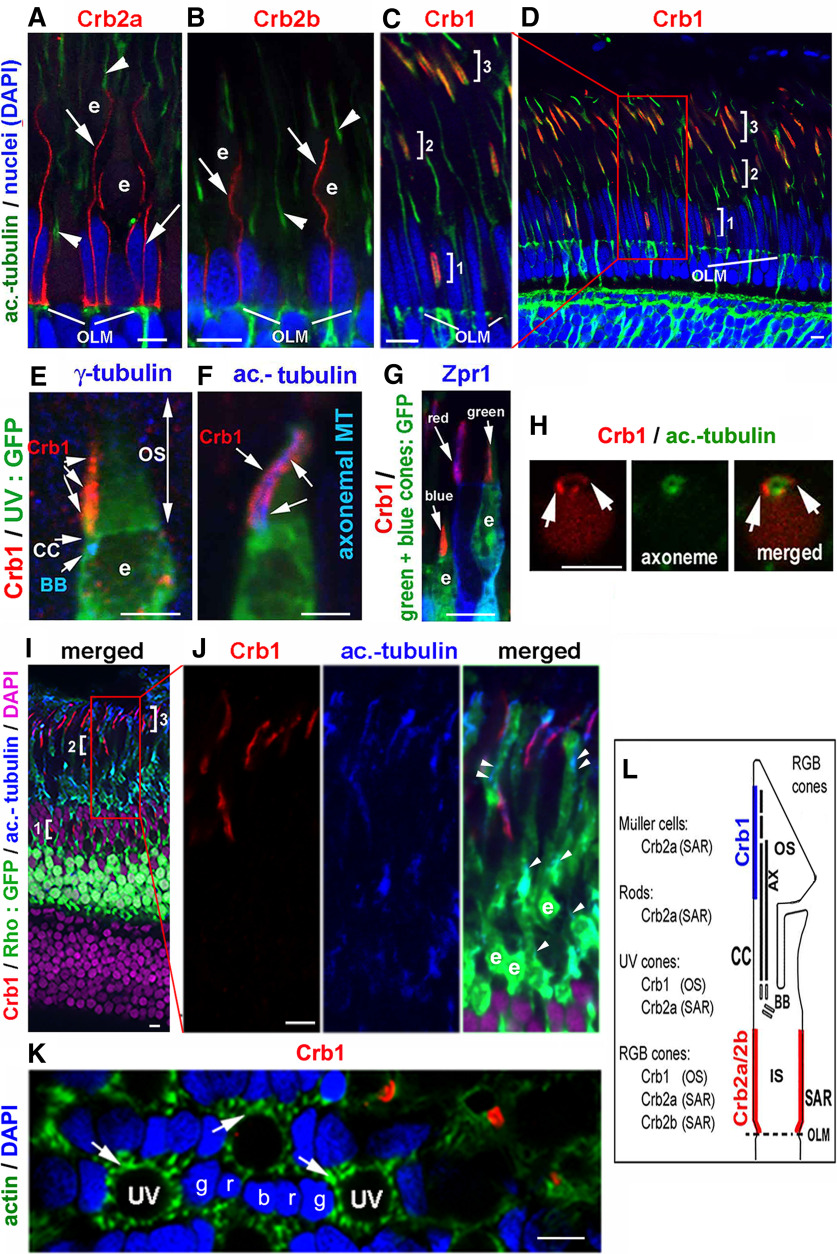
Previously, we showed that Crb1 is expressed in the neuroepithelium of the neural tube but not in the optic vesicles (Zou et al., 2013). However, Crb1's expression pattern in the adult retina was not known. Therefore, we next examined its expression in adult retina by immunohistochemistry. We found that Crb1 was expressed in the photoreceptor layer like Crb2a and Crb2b (Fig. 3A,B) (Zou et al., 2012; Fang et al., 2017). However, unlike Crb2a and Crb2b, which localized to the SARs of photoreceptors and/or the apical processes of Müller glial cells (Fig. 3A,B), at the subcellular level, Crb1 localized to a small region in the outer segments of all four types of cones (red, green, blue, and UV) (Fig. 3C–G). As a result, Crb1 signals in the photoreceptor layer were distributed in three bands (Fig. 3C,D) because the outer segments of cone subtypes are at different distances from the outer limiting membrane (OLM, which is a collection of adherens junctions and tight junctions between photoreceptors and Müller glial cells). Transverse imaging further revealed that Crb1 localized to the cell membranes that encircle the axonemes of cones (Fig. 3H). Interestingly, Crb1 is not expressed in rod photoreceptors (Fig. 3I,J), nor in Müller glial cells' apical processes, which are enriched with actin and Crb2a (Fig. 3K) (Zou et al., 2012). Thus, zebrafish Crb1 differs from Crb2a and Crb2b in both cell type-specific expression pattern and subcellular localization (Fig. 3L), suggesting that Crb1 should differ from Crb2a and Crb2b in function.
Figure 3.
Distinct localizations of Crb1, Crb2a, and Crb2b in adult zebrafish retina. In all panels, the retinal tissues were sectioned radially, except for the transverse sections in H and K. e, Ellipsoids; OS, outer segment; IS, inner segment; UV, UV cones; g, green cones; r, red cones; b, blue cones. Scale bars, 5 µm. A, B, Crb2a and Crb2b (red, arrows) localized to the SARs of photoreceptors or Müller glial cells, abutting the OLM and away from the axonemes. Arrowheads indicate anti-acetylated tubulin staining. C, D, Apical of and not abutting the OLM, Crb1 localized in three bands to the outer segment regions of UV cones (bracket 1), blue cones (bracket 2), and green/red cones (bracket 3). C, Magnified region boxed in D. E, Crb1 localized apical to the basal body (BB, blue, by anti-γ-tubulin staining) and the connecting cilium (CC) in a UV one, which was visualized by the GFP expression in the background of Tg(−5.5opn1sws1:EGFP)kj (UV:GFP) (Takechi et al., 2003). F, Crb1 (red) localized to the vicinity of the axonemal microtubules (MT, blue, by anti-acetylated tubulin staining) in a Tg(−5.5opn1sws1:EGFP)kj UV cone (green). G, Crb1 signals (red) in RGB cones in the background of Tg (LCRRH2-RH2-1:GFP)pt112(green + blue cones: GFP), which expresses GFP (green) in green and blue cones (Fang et al., 2013). Green and red cones were labeled with Zpr1 antibody (blue). H, At the transverse section of a UV cone, confirmed by its Tg(−5.5opn1sws1:EGFP)kj GFP expression (GFP signals not shown), Crb1 signals (red) surrounded the axoneme (green, by anti-acetylated-tubulin staining). I, J, Crb1 (red) distribution in the adult retina of Tg(−3.7rho:EGFP)kj2 fish (Rho:GFP), which expresses GFP (green) in rods (Hamaoka et al., 2002). J, Magnified region boxed in I. Note the lack of Crb1 signals in GFP-positive rods, whose ellipsoids (e) marked the vicinity of the basal ends of rod axonemes (arrowheads, blue, by anti-acetylated tubulin staining). Brackets 1-3 indicate the bands of Crb1 staining. K, A transverse retinal section through the region of UV cone ellipsoids showed the lack of Crb1 signals (red) in the apical processes of Müller glial cells, which were revealed by their strong actin staining (green, arrows) (Zou et al., 2012). DAPI staining (blue) showed the nuclei of RGB cones (r, g, b). L, The distinct cellular expression and subcellular localization patterns of zebrafish Crb1, Crb2a, and Crb2b in the retina are summarized with a table and depicted with a diagram.
Loss of Crb1 does not affect zebrafish survival and body development
To investigate the functions of Crb1 in zebrafish, we examined the effects of the crb1sa12558 nonsense mutation, which causes a C1525T nucleotide exchange in exon 6 and can be genotyped by a Cleaved Amplified Polymorphic Sequence analysis (Konieczny and Ausubel, 1993) (Fig. 4A). The crb1sa12558 mutation truncates Crb1 in the middle of the extracellular domain and abolishes Crb1 immunostaining with the anti-Crb1aa952-1258 antibodies at both larval stage (Fig. 4B) and adult stage (data not shown). It is reasonable to believe that any Crb1 functions that need its full-length extracellular domain, cell membrane attachment, and intracellular signaling are abolished by this mutation. Surprisingly, the development of crb1 mutants was not affected, and fish can survive to adulthood. Their external body shapes and lengths are not distinguishable from those of WT fish at both larval and adult stages (Fig. 4C–E). In addition, crb1 mutants can breed normally. These observations suggest that Crb1 is not essential for the general development and maintenance of zebrafish in captivity.
Effects of the loss of Crb1 on cone photoreceptors
Although the loss of Crb1 does not affect the overall development of zebrafish, Crb1 may still play important roles in cone maintenance because of its restrictive cone expression in the retina. Supporting this notion, Crb proteins have been shown to be required for photoreceptor morphogenesis in both invertebrates and vertebrates (Izaddoost et al., 2002; Pellikka et al., 2002; Mehalow et al., 2003; Nam and Choi, 2003; Lemmers et al., 2004). We thus next examined whether cone photoreceptors are affected in crb1 mutants. By immunohistochemistry, we found no general abnormality in retinal development. In addition, the morphologies of double cones (green and red cones, stained with the Zpr1 antibody against arrestin3a) in crb1 mutants appeared as normal as in WT at both 6 dpf and 3 mpf (Fig. 5A,B). Considering that Crb1 localizes to cone outer segments, we also evaluated the effect of the crb1sa12558 mutation on the development of cone outer segments by examining the lengths of cone outer segments stained with PNA, which was used previously as a general outer segment marker for all cone types in zebrafish (Korenbrot et al., 2013; Yu et al., 2016; Lessieur et al., 2017; Lobo et al., 2017; Shi et al., 2017; Xie et al., 2019). However, we found that PNA specifically stained the outer segments of red cones but no other cone types in zebrafish (Fig. 5C,D; see the criteria for distinguishing cone types in the figure legend). We speculate that this is because PNA has not been rigorously evaluated for cone specificity in zebrafish previously. Regardless, we found that the lengths of the red cone outer segments were not affected in crb1 mutants (Fig. 5E,F). Thus, we conclude that that Crb1 is unlikely required for the overall morphogenesis of cone photoreceptors.
Roles of zebrafish Crb1 in photoreceptor maintenance under damaging light irradiation
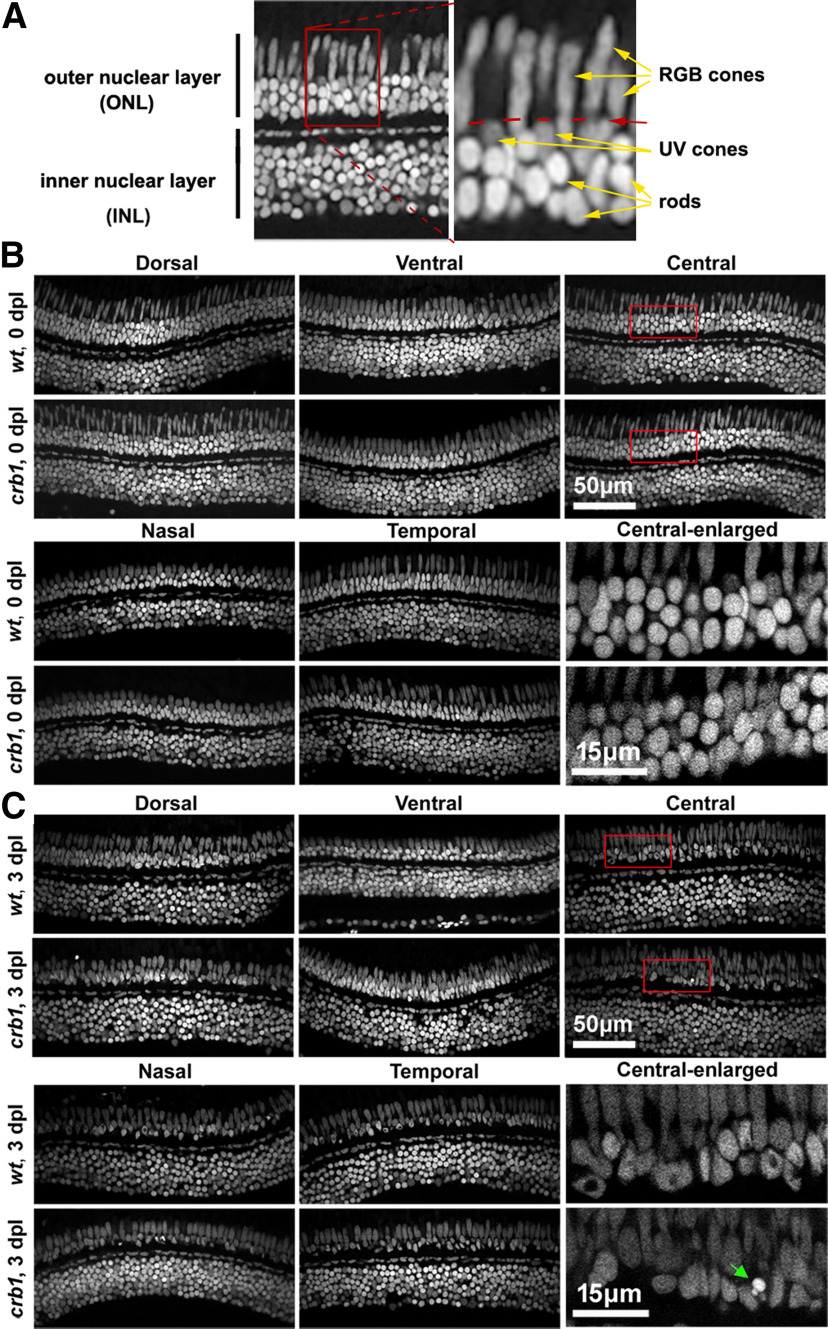
Photoreceptors are prone to damage by strong light irradiation. It has been shown that Crb proteins protect photoreceptors from light irradiation in both vertebrates and invertebrates (Johnson et al., 2002; Chartier et al., 2012; Mishra et al., 2012). Thus, we next examined whether zebrafish Crb1 protects photoreceptors in damaging light irradiation. To do so, we examined how photoreceptors in WT and crb1 mutant fish were affected by irradiation with either strong white light or UV light, which preferentially damage rods and cones, respectively (Thomas et al., 2012; Thomas and Thummel, 2013). Rods and cones were categorized according to their nuclear morphologies and locations revealed by DAPI staining: Rod nuclei are round, most intensely stained by DAPI, and reside at the basal half of the outer nuclear layer, basal to the OLM. The cone category encompasses RGB cones and UV cones. RGB cone nuclei are elongated and reside mostly apical to the OLM; and UV cones are round, less condensed than rod nuclei, and localize basal to and abutting the OLM (Fig. 6A) (Zou et al., 2012; Fang et al., 2017, 2020; Fu et al., 2018).
Figure 6.
The loss of Crb1 made rod photoreceptors prone to damage by strong white light irradiation. A, An example DAPI image of a radial section of adult zebrafish retina, showing the outer nuclear layer (ONL) and the inner nuclear layer (INL). Arrows indicate the nuclei of RGB cones, UV cones, and rods. Red dashed lines indicate the position of the OLM. Left, Boxed region is magnified as the right panel. B, C, Typical images of radial retinal sections show the nuclear morphologies (by DAPI staining) of the ONL and INL of strong white light-treated fish at 0 dpl (B) and 3 dpl (C). B, C, Bottom right, Enlarged central retinal regions boxed in red. Green arrow indicates a highly condensed nuclear residue, possibly from a dead or dying rod. Scale bars, 50 μm or 15 μm.
After strong white light irradiation, we found no differences in the densities of cones between 3 mpf WT and crb1 mutant fish at either 0 or 3 dpl (days post the start of light treatment) (Figs. 6B,C, 7A). In addition, INL and ganglion cell layers were not affected either, as exemplified by the similar thicknesses of the INL between WT and crb1 mutants (Figs. 6B,C, 7B). Conversely, the densities of rods decreased surprisingly more in crb1 mutants than in WT fish across the retina, except for the ventral and temporal retinal regions (Figs. 6B,C, 7C). Considering that Crb1 is not expressed in rods, these findings suggest that Crb1 protects rods under strong white light irradiation in a non--cell-autonomous fashion, a new mechanism that has not been observed previously in the fruit fly and mouse (Johnson et al., 2002; van de Pavert et al., 2004; Mishra et al., 2012); Pellikka et al., 2002; Chartier et al., 2012; Izaddoost et al., 2002). Conversely, the Johnson et al. study suggested an autonomous role of Crb in the morphogenesis and maintenance of the ommatidia (Johnson et al., 2002).
To evaluate whether Crb1 protects cone photoreceptors from light damage, we treated fish with UV light irradiation. Immediately after the treatment (0 hpl, hour post UV light treatment), in both WT and crb1 mutant fish, nuclei with unusual condensing chromatin were observed in radial retinal sections in the places where UV cone and RGB cone nuclei normally reside (Fig. 8A, green arrows and arrowheads). The degree of chromatin condensation became more extensive over time for both WT and crb1 mutant fish, as reflected by the decreases in the ratios of radial to transverse diameters of damaged cone nuclei for both WT and crb1 over time, particularly between 0 and 12 hpl (Fig. 8B; determined by one-way ANOVA; WT: F(3,573) = 79.32, p < 0.0001; crb1: F(3,655) = 32.58, p < 0.0001). This trend suggests an elongated-elliptic-round morphologic transition for those damaged cones in the RGB cone layer (Fig. 8C). At 24 and 36 hpl, many condensed nuclei localized apical to the bulk of normally looking RGB cone nuclei, suggesting that they might be translocating toward the retinal pigment epithelium over time (Fig. 8A, blue arrowheads; Fig. 8C). This elongated-elliptic-round morphologic transition may not apply to UV cones because UV cone nuclei are round in the first place. In addition, damaged UV cones may go through a faster condensing process, judging by their stronger DAPI staining at 0 hpl (Fig. 8A, wt, green arrowhead vs green arrows). By 36 hpl, no apparent UV cone nuclei were observed basal to the OLM where they normally reside (Fig. 8A). Although the damaging effects were observed in both WT and mutant fish (Fig. 8A), chromatin condensation progressed faster in crb1 mutants than in WT fish (Fig. 8B; 0 hpl, p = 4.75739E-08).
At 0 hpl and in radial sections, the apparent elongated-elliptic-round nuclear morphologic transition occurred in the place where RGB cones reside (Fig. 8A). However, not all RGB cone nuclei displayed such transition after UV light irradiation, suggesting that not all RGB cones were damaged. To determine whether a particular type of RGB cones were damaged by the UV light irradiation, we analyzed the retina at transverse sections, which allowed us to distinguish RGB cone types by their positions in the mirror-symmetric and pentameric alignment of RGB cones (Fig. 8D). It turned out that only blue cones of RGB cones underwent condensation (Fig. 8D). Again, the chromatin of blue cone nuclei in crb1 mutant retina appeared more condensed than that in WT, suggesting faster deterioration (Fig. 8D, 0 hpl, red arrowheads vs green arrows).
These results suggest that Crb1 plays a role in slowing down the deterioration of the blue and UV cones caused by the UV light irradiation. However, this protecting role did not last long because when examined at 12 hpl and later, the degree of damage appeared similar between WT and mutant fish (Fig. 8B).
Role of Crb1 in light responses of cones
The outer segments are the subcellular structures where photoreceptors receive light stimuli. The unique localization of zebrafish Crb1 at the outer segments made us wonder whether it plays a role in cones' responsiveness to light stimuli. We investigated the effects of Crb1 loss on cones' responsiveness to white light stimulation by recording isolated cone PIII ERG signals. Cone PIII signals were extracted from the ERG response by adding 20 mm L-aspartate to the perfusion media to block signals downstream of the cones. Cone PIII signals contain two parts, namely, transient-sustained vitreal negativity, a hyperpolarization during stimulation and vitreal positivity, a rebound depolarization after light offset. The properties of the cone responsiveness were modeled by the peak-to-peak amplitudes of both negative and positive waveforms (Nelson et al., 2019). At 6 dpf, WT and crb1 mutant larvae were stimulated with flashes of white light and their response amplitudes were recorded. We found that the vitreal negativity response amplitudes were largely not affected in WT over time (Fig. 9A,C; determined by one-way ANOVA, F(9,70) = 0.9437, p = 0.4936). By contrast, in crb1 mutants, the vitreal negativity response amplitudes increased significantly from the second round of light stimulus and reached to a peak at the fourth round (Fig. 9B,C; determined by one-way ANOVA, F(9,100) = 7.593, p < 0.0001). Then starting at the seventh round of treatment, the response amplitudes in crb1 mutants decreased to the level of WT and subsequently maintained largely steady as in WT (Fig. 9B,C). The effects of crb1sa12558 mutation on response amplitudes were significant at cycles 3-5, as revealed by the differences between the averaged responses of WT and crb1 mutant fish (Fig. 9C). Thus, these results suggest that Crb1 plays a role in modulating cones' responsiveness to bright white light over time.
Effects of the loss of Crb1 on arrestin3a translocation
Light responsiveness is related to phototransduction, which is modulated during the dark-light transition by the inner-outer-segment translocation of many phototransduction molecules, such as transducin, arrestin, and recoverin (Calvert et al., 2006). For example, the loss of arrestin3a caused a severe delay in photoresponse recovery in zebrafish (Renninger et al., 2011). To determine whether the loss of Crb1 affects arrestin translocation, which could consequently cause the increased vitreal negativity during white light stimulation, we investigated the distribution of arrestin3a during the dark/light transition by quantifying the ratios of arrestin3a intensities in the outer segments to those in the inner segments (Fig. 10A). We found that arrestin3a was similarly enriched in the inner segments of double cones in both WT and crb1 mutants in the dark. And 30 min after light exposure, arrestin3a signals translocated to the outer segments at the similar level in WT fish and crb1 mutants, although the majority of the signals still remained in the inner segments (Fig. 10B–D). As expected, loss of Crb1 did not affect the numbers of arrestin3a-positive cones (Fig. 10E). These results suggest that Crb1 is unlikely required for arrestin3a's translocation responding to dark-light transition in red and green cones. Thus, the increased vitreal negativity in crb1 mutants may be caused by a mechanism that is independent of arrestin translocation.
Discussion
In this study, we addressed the question whether zebrafish Crb1 differs from other vertebrate Crb1 and Crb2 in their expression patterns, subcellular localization, and functions in the retina. Our results suggest that zebrafish Crb1 is distinct from zebrafish Crb2a and Crb2b as well as from human and mouse Crb1 and Crb2, suggesting that zebrafish Crb1 may represent a neofunctionalization of Crb proteins during evolution.
Zebrafish Crb1's retinal expression and subcellular localization are unique
Here, we revealed the expression patterns and subcellular localization of Crb1 in the zebrafish retina. Together with the previous findings on the expression and localization patterns of Crb2a, Crb2b, Crb3a, and Crb3b (Omori and Malicki, 2006; Zou et al., 2012, 2013; Fang et al., 2017), the expression patterns of Crb proteins in the zebrafish retina have now been fully revealed: Crb1 is expressed in all types of cones but not in rods (Fig. 3); Crb2a is expressed in all photoreceptors and Müller glial cells in developed retina as well as in neuroepithelial cells in undifferentiated retina; Crb2b is restrictively expressed in RGB cones; Crb3a and Crb3b are not expressed in the retina. In other words, rods and Müller glia express only Crb2a; all cones express both Crb1 and Crb2a; RGB cones also express Crb2b, in addition to Crb1 and Crb2a; and other retinal cell types do not express any Crb proteins. The cone-specific expression pattern of zebrafish Crb1 is distinct from the expression patterns of mammalian Crb1 because, in mouse and human retina, Crb1 is also expressed in rod photoreceptors and Müller glial cells in addition to cones (Tepass et al., 1990; Pellikka et al., 2002; Mehalow et al., 2003; van de Pavert et al., 2004; Pellissier et al., 2014). Thus, the cellular expression patterns of mammalian Crb1 are similar most to zebrafish Crb2a, suggesting that the retinal functions of mammalian Crb1 are likely conducted by Crb2a rather than Crb1 in zebrafish retina.
Zebrafish Crb1 is not only distinct in its cellular expression pattern, but also in its subcellular localization because of its unique localization to the cell membranes surrounding the axonemes in cone outer segments (Fig. 3). Conventionally, Crb proteins localize to the SARs in a variety of epithelia (Tepass et al., 1990; Knust and Bossinger, 2002). For example, in mammalian retina, both Crb1 and Crb2 localize to the SARs of photoreceptors and/or Müller glia cells, immediately apical to the OLM (Mehalow et al., 2003; van de Pavert et al., 2004; van den Hurk et al., 2005; van Rossum et al., 2006; Pellissier et al., 2014). Similarly, zebrafish Crb2a and Crb2b also localize to the SARs of photoreceptor inner segments or the apical processes of Müller glial cells (Omori and Malicki, 2006; Zou et al., 2012). The non-SAR localization of zebrafish Crb1 appears to be specific for developed cone photoreceptors because we previously showed that, in the neural tube during early development, zebrafish Crb1 also localizes to the SARs, just as Crb2a does (Zou et al., 2013; Guo et al., 2018). Thus, the unique localization of Crb1 to the outer segment cell membranes that encircle the axonemes of cone photoreceptors sets itself apart from other Crb proteins.
The molecular mechanisms that underlie zebrafish Crb1's unique localization to the axonemal region of cone outer segments are yet to be determined. It is possible that Crb1 interacts with a different set of cytoplasmic partners for its outer segment localization. Supporting this hypothesis, MPP5a and MPP5b (also known as Nok and Ponli, respectively) (Wei and Malicki, 2002; Zou et al., 2010), the conventional cytoplasmic partners of Crb proteins, do not colocalize with zebrafish Crb1, but rather colocalize with Crb2a and Crb2b to the SARs (Wei et al., 2006; Zou et al., 2010). The unique axonemal cell membrane localization of Crb1 is likely not related to its PDZ-binding and FERM-binding motifs because they are identical or very similar to those of Crb2a and Crb2b, respectively. However, given the significant variations in the interval region between the FERM-binding and PDZ-binding motifs (Fig. 2C), it would be interesting to determine whether this interval region is responsible for Crb1's axonemal cell membrane localization in cones.
Roles of zebrafish Crb1 in the apicobasal polarity of the neuroepithelium
Crb proteins are normally required for the establishment and maintenance of the apicobasal polarity of a variety of epithelia (Nam and Choi, 2003; Richard et al., 2006; Pocha et al., 2011b; Letizia et al., 2013; Zou et al., 2013). However, zebrafish Crb1 is not expressed in the undifferentiated retinal neuroepithelium. In addition, the loss of Crb1 does not disrupt the apicobasal polarity of cone photoreceptors in developed retina. These observations suggest that zebrafish Crb1 is not required for the apicobasal polarity of the zebrafish retina and cone photoreceptors.
However, the nonrequirement of Crb1 for apicobasal polarity is likely because of it is functional redundancy with Crb2a in zebrafish retina, as suggested by the following two observations: First, the loss of Crb2a did not affect the epithelial apicobasal polarity of the neural tube (Malicki and Driever, 1999), where Crb1 is coexpressed with Crb2a (Zou et al., 2013). Second, the loss of Crb2a disrupted the apicobasal polarity of the undifferentiated retinal neuroepithelium, where Crb1 is not expressed (Zou et al., 2013). These observations suggest that zebrafish Crb1 still possesses the capability for regulating the apicobasal polarity of undifferentiated neuroepithelium, but this function of Crb1 is normally not necessary in the adult zebrafish retina because of the earlier, broader, and redundant expression of Crb2a.
Roles of zebrafish Crb1 in retinal and photoreceptor morphogenesis
The fact that the loss of zebrafish Crb1 did not change the general retinal patterning and cone morphology (Figs. 4, 5) is surprising because such defects were observed in other species. For example, in Drosophila, the loss of Crb reduced the length of rhabdomeres, disrupted the integrity of zonula adherens, affected the localization of adherens junctions, and interfered with the formation of the stalk membrane (Izaddoost et al., 2002; Pellikka et al., 2002). Similarly, in mice, the loss of function or malfunction of Crb1 caused displacement of photoreceptors that often formed rosettes, interruption of the OLM, upregulation of GFAP, and retinopathies, such as RP (Jacobson et al., 2003; Mehalow et al., 2003; van de Pavert et al., 2004, 2007; Aleman et al., 2011; Alves et al., 2014; Zhao et al., 2015). And in humans, the loss of CRB1 caused RP and LCA (den Hollander et al., 1999; Abouzeid et al., 2006; Vallespin et al., 2007; Bujakowska et al., 2012; Corton et al., 2013).
The noninvolvement of Crb1 in retinal development and photoreceptor morphogenesis in zebrafish may be directly related to Crb1's unique localization to cone outer segments because this localization physically segregates Crb1 from interacting with the adherens junctions and tight junctions at the OLM, which are required for the morphogenesis of photoreceptors by establishing and maintaining their apicobasal polarity. In addition, the outer segment localization also makes it unlikely for Crb1 to mediate photoreceptor patterning through trans homophilic or heterophilic physical adhesion as Crb2a and Crb2b mediate cone-cone adhesion in the formation of cone pentamers at the SARs of cone inner segments (Zou et al., 2012) because the Crb1-bearing axonemal cell membranes of cones do not cluster together in close juxtaposition, excluding the possibility of any Crb-Crb-based physical adhesion among photoreceptors. Despite Crb1's localization to cone outer segments, the length of red cones, as revealed by PNA staining (Fig. 5), was not affected by the loss of Crb1. Thus, the noninvolvement of Crb1 in retinal development and photoreceptor morphogenesis also sets it apart from other retinal Crb proteins in vertebrates.
The protecting roles of zebrafish Crb1 in cone and rod maintenance in damaging light irradiation
Previous studies showed that both Crb in Drosophila and Crb1 in mouse play a role in protecting photoreceptors from light damage (Johnson et al., 2002; van de Pavert et al., 2004; Organisciak and Vaughan, 2010; Chartier et al., 2012; Mishra et al., 2012). And a cell-autonomous mechanism was suggested for such a function of Crb in the fly (Johnson et al., 2002). Our study suggests that zebrafish Crb1 also plays protecting roles in photoreceptor maintenance under damaging light irradiation, but by different mechanisms.
For rods, we initially speculated that Crb1 should not protect rods from strong rod-damaging white light irradiation (Thomas and Thummel, 2013) because Crb1 is not expressed in rods. Surprisingly, we found that strong white light irradiation caused more reduction in rod densities in crb1 mutant fish, whereas cone survival was not affected (Figs. 6, 7). Our findings suggest that, under strong white light irradiation, Crb1 expressed in cones somehow promotes rod survival in a surprisingly non--cell-autonomous fashion.
For cones, Crb1 may play a role in cones' resistance to UV light irradiation, likely through a cell-autonomous fashion, because UV light irradiation caused faster death to blue and UV cones in crb1 mutants than in WT fish. However, this protecting role is minimal at the beginning stage after UV light irradiation because the deteriorating effects eventually become very similar between WT and mutant fish (Fig. 8).
Roles of zebrafish Crb1 in light responses
The localization of Crb1 at the outer segment cell membranes surrounding the axonemes makes it tempting to hypothesize that Crb1 regulates the trafficking of proteins that are required for phototransduction. Such function was reported in Drosophila, where Crb regulates rhodopsin transportation (Pocha et al., 2011a,b). This hypothesis is also consistent with the finding that the response amplitudes of vitreal negativity in crb1 mutants increased in the first four rounds of white light flash stimuli (Fig. 9). Such changes in light responsiveness suggest a loss of ability to appropriately adjust to light stimulation. However, these changes in light responsiveness were likely not based on arrestin3a-translocation-mediated modulation because we did not observe that the loss of Crb1 caused any change in arrestin3a translocation during dark-light transition compared with WT larvae (Fig. 10), although arrestin3a morphants displayed delayed cone response recovery (Renninger et al., 2011). Thus, how Crb1 modulates phototransduction at the outer segments remains to be discovered.
In conclusion, although the phylogenetic relationships among Crb proteins suggest that zebrafish Crb1 is more closely related to Crb1 proteins of other vertebrate species, its unique cell type-specific expression pattern, subcellular localization, and functions distinguish it from other vertebrate Crb1 and Crb2 homologs. This distinction may suggest that zebrafish Crb1 represents a neofunctionalization of Crb proteins during evolution. Our study also demonstrates the importance of considering the cell type-specific expression, subcellular localization, and animal models for a deeper understanding of the complex biology of Crb proteins.
Footnotes
The authors declare no competing financial interests.
This work was supported by EY016099 and EY025638 to X.W. The study utilized the core facility supported by National Institutes of Health Grant P30EY008098 to the Department of Ophthalmology of the University of Pittsburgh, which was funded in part by a grant from the Eye and Ear Foundation of Pittsburgh and an unrestricted grant from the Research to Prevent Blindness. This work was supported in part by National Institute of Neurological Disorders and Stroke, National Institutes of Health Intramural Research Program to R.N.
References
- Abouzeid H, Li Y, Maumenee IH, Dharmaraj S, Sundin O (2006) A G1103R mutation in CRB1 is co-inherited with high hyperopia and Leber congenital amaurosis. Ophthalmic Genet 27:15–20. 10.1080/13816810500481840 [DOI] [PubMed] [Google Scholar]
- Aleman TS, Cideciyan AV, Aguirre GK, Huang WC, Mullins CL, Roman AJ, Sumaroka A, Olivares MB, Tsai FF, Schwartz SB, Vandenberghe LH, Limberis MP, Stone EM, Bell P, Wilson JM, Jacobson SG (2011) Human CRB1-associated retinal degeneration: comparison with the rd8 Crb1-mutant mouse model. Invest Ophthalmol Vis Sci 52:6898–6910. 10.1167/iovs.11-7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves CH, Pellissier LP, Wijnholds J (2014) The CRB1 and adherens junction complex proteins in retinal development and maintenance. Prog Retin Eye Res 40:35–52. 10.1016/j.preteyeres.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Bujakowska K, Audo I, Mohand-Saïd S, Lancelot ME, Antonio A, Germain A, Léveillard T, Letexier M, Saraiva JP, Lonjou C, Carpentier W, Sahel JA, Bhattacharya SS, Zeitz C (2012) CRB1 mutations in inherited retinal dystrophies. Hum Mutat 33:306–315. 10.1002/humu.21653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgakova NA, Knust E (2009) The Crumbs complex: from epithelial-cell polarity to retinal degeneration. J Cell Sci 122:2587–2596. 10.1242/jcs.023648 [DOI] [PubMed] [Google Scholar]
- Busch-Nentwich E, Kettleborough R, Harvey S, Collins J, Ding M, Dooley C, Fenyes F, Gibbons R, Herd C, Mehroke S, Scahill C, Sealy I, Wali N, White R, Stemple DL (2012) Sanger Institute Zebrafish Mutation Project phenotype and image data submission. ZFIN Direct Data Submiss (Active Curation) [Google Scholar]
- Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Arshavsky VY (2006) Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol 16:560–568. 10.1016/j.tcb.2006.09.001 [DOI] [PubMed] [Google Scholar]
- Chartier FJ, Hardy EJ, Laprise P (2012) Crumbs limits oxidase-dependent signaling to maintain epithelial integrity and prevent photoreceptor cell death. J Cell Biol 198:991–998. 10.1083/jcb.201203083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G (2010) The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci USA 107:15810–15815. 10.1073/pnas.1004060107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corton M, Tatu SD, Avila-Fernandez A, Vallespín E, Tapias I, Cantalapiedra D, Blanco-Kelly F, Riveiro-Alvarez R, Bernal S, García-Sandoval B, Baiget M, Ayuso C (2013) High frequency of CRB1 mutations as cause of early-onset retinal dystrophies in the Spanish population. Orphanet J Rare Dis 8:20. 10.1186/1750-1172-8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander AI, Ten Brink JB, De Kok YJ, Van Soest S, Van Den Born LI, Van Driel MA, Van De Pol DJ, Payne AM, Bhattacharya SS, Kellner U, Hoyng CB, Westerveld A, Brunner HG, Bleeker-Wagemakers EM, Deutman AF, Heckenlively JR, Cremers FP, Bergen AA (1999) Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12). Nat Genet 23:217–221. 10.1038/13848 [DOI] [PubMed] [Google Scholar]
- Fang W, Bonaffini S, Zou J, Wang X, Zhang C, Tsujimura T, Kawamura S, Wei X (2013) Characterization of transgenic zebrafish lines that express GFP in the retina, pineal gland, olfactory bulb, hatching gland, and optic tectum. Gene Expr Patterns 13:150–159. 10.1016/j.gep.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Guo C, Wei X (2017) Rainbow enhancers regulate restrictive transcription in teleost green, red, and blue cones. J Neurosci 37:2834–2848. 10.1523/JNEUROSCI.3421-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Wen Y, Wei X (2020) Identification and characterization of cis-regulatory elements for photoreceptor-type-specific transcription in zebrafish. Methods Mol Biol 2092:123–145. 10.1007/978-1-0716-0175-4_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Nagashima M, Guo C, Raymond PA, Wei X (2018) Novel animal model of crumbs-dependent progressive retinal degeneration that targets specific cone subtypes. Invest Ophthalmol Vis Sci 59:505–518. 10.1167/iovs.17-22572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grawe F, Wodarz A, Lee B, Knust E, Skaer H (1996) The Drosophila genes crumbs and stardust are involved in the biogenesis of adherens junctions. Development 122:951–959. [DOI] [PubMed] [Google Scholar]
- Guo C, Zou J, Wen Y, Fang W, Stolz DB, Sun M, Wei X (2018) Apical cell-cell adhesions reconcile symmetry and asymmetry in zebrafish neurulation. iScience 3:63–85. 10.1016/j.isci.2018.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaoka T, Takechi M, Chinen A, Nishiwaki Y, Kawamura S (2002) Visualization of rod photoreceptor development using GFP-transgenic zebrafish. Genesis 34:215–220. 10.1002/gene.10155 [DOI] [PubMed] [Google Scholar]
- Izaddoost S, Nam SC, Bhat MA, Bellen HJ, Choi KW (2002) Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature 416:178–182. 10.1038/nature720 [DOI] [PubMed] [Google Scholar]
- Jacobson SG, Cideciyan AV, Aleman TS, Pianta MJ, Sumaroka A, Schwartz SB, Smilko EE, Milam AH, Sheffield VC, Stone EM (2003) Crumbs homolog 1 (CRB1) mutations result in a thick human retina with abnormal lamination. Hum Mol Genet 12:1073–1078. 10.1093/hmg/ddg117 [DOI] [PubMed] [Google Scholar]
- Johnson K, Grawe F, Grzeschik N, Knust E (2002) Drosophila crumbs is required to inhibit light-induced photoreceptor degeneration. Curr Biol 12:1675–1680. 10.1016/s0960-9822(02)01180-6 [DOI] [PubMed] [Google Scholar]
- Knust E, Bossinger O (2002) Composition and formation of intercellular junctions in epithelial cells. Science 298:1955–1959. 10.1126/science.1072161 [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4:403–410. 10.1046/j.1365-313x.1993.04020403.x [DOI] [PubMed] [Google Scholar]
- Korenbrot JI, Mehta M, Tserentsoodol N, Postlethwait JH, Rebrik TI (2013) EML1 (CNG-modulin) controls light sensitivity in darkness and under continuous illumination in zebrafish retinal cone photoreceptors. J Neurosci 33:17763–17776. 10.1523/JNEUROSCI.2659-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RE, Tan WH, Innes AM, Parboosingh JS, Schneidman-Duhovny D, Rajkovic A, Pappas J, Altschwager P, DeWard S, Fulton A, Gray KJ, Krall M, Mehta L, Rodan LH, Saller DN, Steele D, Stein D, Yatsenko SA, Bernier FP, Slavotinek AM (2016) Expansion of phenotype and genotypic data in CRB2-related syndrome. Eur J Hum Genet 24:1436–1444. 10.1038/ejhg.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi MH, Médina E, Arsanto JP, Le Bivic A (2004) CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol Biol Cell 15:1324–1333. 10.1091/mbc.e03-04-0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessieur EM, Fogerty J, Gaivin RJ, Song P, Perkins BD (2017) The ciliopathy gene ahi1 is required for zebrafish cone photoreceptor outer segment morphogenesis and survival. Invest Ophthalmol Vis Sci 58:448–460. 10.1167/iovs.16-20326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letizia A, Ricardo S, Moussian B, Martín N, Llimargas M (2013) A functional role of the extracellular domain of Crumbs in cell architecture and apicobasal polarity. J Cell Sci 126:2157–2163. 10.1242/jcs.122382 [DOI] [PubMed] [Google Scholar]
- Lobo GP, Fulmer D, Guo L, Zuo X, Dang Y, Kim SH, Su Y, George K, Obert E, Fogelgren B, Nihalani D, Norris RA, Rohrer B, Lipschutz JH (2017) The exocyst is required for photoreceptor ciliogenesis and retinal development. J Biol Chem 292:14814–14826. 10.1074/jbc.M117.795674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O, Roh MH, Liu CJ, Laurinec S, Margolis B (2003) Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1). Gene 302:21–29. 10.1016/s0378111902010843 [DOI] [PubMed] [Google Scholar]
- Malicki J, Driever W (1999) Oko Meduzy mutations affect neuronal patterning in the zebrafish retina and reveal cell-cell interactions of the retinal neuroepithelial sheet. Development 126:1235–1246. [DOI] [PubMed] [Google Scholar]
- Mehalow AK, Kameya S, Smith RS, Hawes NL, Denegre JM, Young JA, Bechtold L, Haider NB, Tepass U, Heckenlively JR, Chang B, Naggert JK, Nishina PM (2003) CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum Mol Genet 12:2179–2189. 10.1093/hmg/ddg232 [DOI] [PubMed] [Google Scholar]
- Mishra M, Rentsch M, Knust E (2012) Crumbs regulates polarity and prevents light-induced degeneration of the simple eyes of Drosophila, the ocelli. Eur J Cell Biol 91:706–716. 10.1016/j.ejcb.2012.03.006 [DOI] [PubMed] [Google Scholar]
- Nam SC, Choi KW (2003) Interaction of Par-6 and Crumbs complexes is essential for photoreceptor morphogenesis in Drosophila. Development 130:4363–4372. 10.1242/dev.00648 [DOI] [PubMed] [Google Scholar]
- Nelson RF, Balraj A, Suresh T, Torvund M, Patterson SS (2019) Strain variations in cone wavelength peaks in situ during zebrafish development. Vis Neurosci 36:E010. 10.1017/S0952523819000075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RF, Singla N (2009) A spectral model for signal elements isolated from zebrafish photopic electroretinogram. Vis Neurosci 26:349–363. 10.1017/S0952523809990113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr, Deerfield DW II (1997) GeneDoc: Analysis and Visualization of Genetic Variation. EMBNEW News 4:1–4. [Google Scholar]
- Omori Y, Malicki J (2006) oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr Biol 16:945–957. 10.1016/j.cub.2006.03.058 [DOI] [PubMed] [Google Scholar]
- Organisciak DT, Vaughan DK (2010) Retinal light damage: mechanisms and protection. Prog Retin Eye Res 29:113–134. 10.1016/j.preteyeres.2009.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. (2010) The hippo signaling pathway in development and cancer. Dev Cell 19:491–505. 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons LM, Grzeschik NA, Allott ML, Richardson HE (2010) Lgl/aPKC and Crb regulate the salvador/warts/hippo pathway. Fly (Austin) 4:288–293. 10.4161/fly.4.4.13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellikka M, Tepass U (2017) Unique cell biological profiles of retinal disease-causing missense mutations in the polarity protein Crumbs. J Cell Sci 130:2147–2158. 10.1242/jcs.197178 [DOI] [PubMed] [Google Scholar]
- Pellikka M, Tanentzapf G, Pinto M, Smith C, McGlade CJ, Ready DF, Tepass U (2002) Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature 416:143–149. 10.1038/nature721 [DOI] [PubMed] [Google Scholar]
- Pellissier LP, Lundvig DM, Tanimoto N, Klooster J, Vos RM, Richard F, Sothilingam V, Garrido MG, Bivic A, Le Seeliger MW, Wijnholds J (2014) CRB2 acts as a modifying factor of CRB1-related retinal dystrophies in mice. Hum Mol Genet 23:3759–3771. 10.1093/hmg/ddu089 [DOI] [PubMed] [Google Scholar]
- Pocha SM, Knust E (2013) Complexities of crumbs function and regulation in tissue morphogenesis. Curr Biol 23:R289–R293. 10.1016/j.cub.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Pocha SM, Shevchenko A, Knust E (2011a) Crumbs regulates rhodopsin transport by interacting with and stabilizing myosin V. J Cell Biol 195:827–838. 10.1083/jcb.201105144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocha SM, Wassmer T, Niehage C, Hoflack B, Knust E (2011b) Retromer controls epithelial cell polarity by trafficking the apical determinant crumbs. Curr Biol 21:1111–1117. 10.1016/j.cub.2011.05.007 [DOI] [PubMed] [Google Scholar]
- Quinn PM, Pellissier LP, Wijnholds J (2017) The CRB1 complex: following the trail of crumbs to a feasible gene therapy strategy. Front Neurosci 11:175. 10.3389/fnins.2017.00175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renninger SL, Gesemann M, Neuhauss SC (2011) Cone arrestin confers cone vision of high temporal resolution in zebrafish larvae. Eur J Neurosci 33:658–667. 10.1111/j.1460-9568.2010.07574.x [DOI] [PubMed] [Google Scholar]
- Richard M, Grawe F, Knust E (2006) DPATJ plays a role in retinal morphogenesis and protects against light-dependent degeneration of photoreceptor cells in the Drosophila eye. Dev Dyn 235:895–907. 10.1002/dvdy.20595 [DOI] [PubMed] [Google Scholar]
- Roh MH, Fan S, Liu CJ, Margolis B (2003) The Crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J Cell Sci 116:2895–2906. 10.1242/jcs.00500 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Obert E, Rahman B, Rohrer B, Lobo GP (2017) The retinol binding protein receptor 2 (Rbpr2) is required for photoreceptor outer segment morphogenesis and visual function in zebrafish. Sci Rep 7:16207. 10.1038/s41598-017-16498-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek AM. (2016) The family of crumbs genes and human disease. Mol Syndromol 7:274–281. 10.1159/000448109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takechi M, Hamaoka T, Kawamura S (2003) Fluorescence visualization of ultraviolet-sensitive cone photoreceptor development in living zebrafish. FEBS Lett 553:90–94. 10.1016/s0014-5793(03)00977-3 [DOI] [PubMed] [Google Scholar]
- Tepass U. (1996) Crumbs, a component of the apical membrane, is required for zonula adherens formation in primary epithelia of Drosophila. Dev Biol 177:217–225. 10.1006/dbio.1996.0157 [DOI] [PubMed] [Google Scholar]
- Tepass U, Theres C, Knust E (1990) crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell 61:787–799. 10.1016/0092-8674(90)90189-l [DOI] [PubMed] [Google Scholar]
- Thomas JL, Nelson CM, Luo X, Hyde DR, Thummel R (2012) Characterization of multiple light damage paradigms reveals regional differences in photoreceptor loss. Exp Eye Res 97:105–116. 10.1016/j.exer.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JL, Thummel R (2013) A novel light damage paradigm for use in retinal regeneration studies in adult zebrafish. J Vis Exp 80:e51017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiab L, Largueche L, Chouchane I, Derouiche K, Munier FL, Matri L, El Schorderet DF (2013) A novel homozygous R764H mutation in crumbs homolog 1 causes autosomal recessive retinitis pigmentosa. Mol Vis 19:829–834. [PMC free article] [PubMed] [Google Scholar]
- Vallespin E, Cantalapiedra D, Riveiro-Alvarez R, Wilke R, Aguirre-Lamban J, Avila-Fernandez A, Lopez-Martinez MA, Gimenez A, Trujillo-Tiebas MJ, Ramos C, Ayuso C (2007) Mutation screening of 299 Spanish families with retinal dystrophies by Leber congenital amaurosis genotyping microarray. Invest Ophthalmol Vis Sci 48:5653–5661. 10.1167/iovs.07-0007 [DOI] [PubMed] [Google Scholar]
- van de Pavert SA, Kantardzhieva A, Malysheva A, Meuleman J, Versteeg I, Levelt C, Klooster J, Geiger S, Seeliger MW, Rashbass P, Le Bivic A, Wijnholds J (2004) Crumbs homologue 1 is required for maintenance of photoreceptor cell polarization and adhesion during light exposure. J Cell Sci 117:4169–4177. 10.1242/jcs.01301 [DOI] [PubMed] [Google Scholar]
- van de Pavert SA, Meuleman J, Malysheva A, Aartsen WM, Versteeg I, Tonagel F, Kamphuis W, McCabe CJ, Seeliger MW, Wijnholds J (2007) A single amino acid substitution (Cys249Trp) in Crb1 causes retinal degeneration and deregulates expression of pituitary tumor transforming gene Pttg1. J Neurosci 27:564–573. 10.1523/JNEUROSCI.3496-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk JA, Rashbass P, Roepman R, Davis J, Voesenek KE, Arends ML, Zonneveld MN, van Roekel MH, Cameron K, Rohrschneider K, Heckenlively JR, Koenekoop RK, Hoyng CB, Cremers FP, den Hollander AI (2005) Characterization of the Crumbs homolog 2 (CRB2) gene and analysis of its role in retinitis pigmentosa and Leber congenital amaurosis. Mol Vis 11:263–273. [PubMed] [Google Scholar]
- van Rossum AG, Aartsen WM, Meuleman J, Klooster J, Malysheva A, Versteeg I, Arsanto JP, Le Bivic A, Wijnholds J (2006) Pals1/Mpp5 is required for correct localization of Crb1 at the subapical region in polarized Müller glia cells. Hum Mol Genet 15:2659–2672. 10.1093/hmg/ddl194 [DOI] [PubMed] [Google Scholar]
- Wei X, Malicki J (2002) nagie oko, encoding a MAGUK-family protein, is essential for cellular patterning of the retina. Nat Genet 31:150–157. 10.1038/ng883 [DOI] [PubMed] [Google Scholar]
- Wei X, Zou J, Takechi M, Kawamura S, Li L (2006) Nok plays an essential role in maintaining the integrity of the outer nuclear layer in the zebrafish retina. Exp Eye Res 83:31–44. 10.1016/j.exer.2005.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Han S, Qu Z, Liu F, Li J, Yu S, Reilly J, Tu J, Liu X, Lu Z, Hu X, Yimer TA, Qin Y, Huang Y, Lv Y, Jiang T, Shu X, Tang Z, Jia H, Wong F, Liu M (2019) Knockout of Nr2e3 prevents rod photoreceptor differentiation and leads to selective L-/M-cone photoreceptor degeneration in zebrafish. Biochim Biophys Acta Mol Basis Dis 1865:1273–1283. 10.1016/j.bbadis.2019.01.022 [DOI] [PubMed] [Google Scholar]
- Yu M, Liu Y, Li J, Natale BN, Cao S, Wang D, Amack JD, Hu H (2016) Eyes shut homolog is required for maintaining the ciliary pocket and survival of photoreceptors in zebrafish. Biol Open 5:1662–1673. 10.1242/bio.021584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Andrieu-Soler C, Kowalczuk L, Paz Cortés M, Berdugo M, Dernigoghossian M, Halili F, Jeanny JC, Goldenberg B, Savoldelli M, El Sanharawi M, Naud MC, van Ijcken W, Pescini-Gobert R, Martinet D, Maass A, Wijnholds J, Crisanti P, Rivolta C, Behar-Cohen F (2015) A new CRB1 rat mutation links Müller glial cells to retinal telangiectasia. J Neurosci 35:6093–6106. 10.1523/JNEUROSCI.3412-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Yang X, Wei X (2010) Restricted Localization of Ponli, a novel zebrafish MAGUK-family protein, to the inner segment interface areas between green, red, and blue cones. Invest Ophthalmol Vis Sci 51:1738–1746. 10.1167/iovs.09-4520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Wang X, Wei X (2012) Crb apical polarity proteins maintain zebrafish retinal cone mosaics via intercellular binding of their extracellular domains. Dev Cell 22:1261–1274. 10.1016/j.devcel.2012.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Wen Y, Yang X, Wei X (2013) Spatial-temporal expressions of Crumbs and Nagie oko and their interdependence in zebrafish central nervous system during early development. Int J Dev Neurosci 31:770–782. 10.1016/j.ijdevneu.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]