Abstract
The current COVID‐19 pandemic constitutes a threat to the population worldwide with over 21 million infected people. There is an urgent need for the development of rapid and massive detection tools as well as the identification and isolation of infected individuals. we sought to evaluate different RT-qPCR kits and protocols to evaluate the best approach to be used omitting an RNA extraction step. We have investigated the sensitivity and performance of different commercially available RT-qPCR kits in detecting SARS-CoV-2 using 80 extracted RNA and NSS from COVID-19 diagnosed patients. We evaluated the ability of each kit to detect viral RNA from both kit-extracted or directly from a pre-boiled NSS observing that direct RNA detection is possible when Ct values are lower than 30 with the three kits tested. Since SARS-CoV-2 testing in most locations occurs once COVID-19 symptoms are evident and, therefore, viral loads are expected to be high, our protocol will be useful in supporting SARS-CoV-2 diagnosis, especially in America where COVID-19 cases have exploded in the recent weeks as well as in low- and middle-income countries, which would not have massive access to kit-based diagnosis. The information provided in this work paves the way for the development of more efficient SARS-CoV-2 detection approaches avoiding an RNA extraction step.
Keywords: SARS-CoV-2, 2019-nCoV, COVID-19, RNA extraction, RT-PCR
The ongoing COVID‐19 pandemic constitutes a threat to the population worldwide with over 21 million cases of the disease (Dong et al., 2020). The rapid spread of SARS-CoV-2 is explained by an important fraction of asymptomatic individuals that can transmit the virus (Li et al., 2020). There is an urgent need for the development of rapid and massive detection tools as well as the identification and isolation of COVID-19 cases of the disease. Both measures together with social distancing remain as the most powerful ways to avoid the appearance of new COVID-19 cases. Currently, there are several challenges associated with ramping up testing capacity, including a shortage in the chain of supplies for RNA extraction reagents. Based on this, we sought to evaluate different RT-qPCR kits and protocols to evaluate the best approach to be used omitting an RNA extraction step.
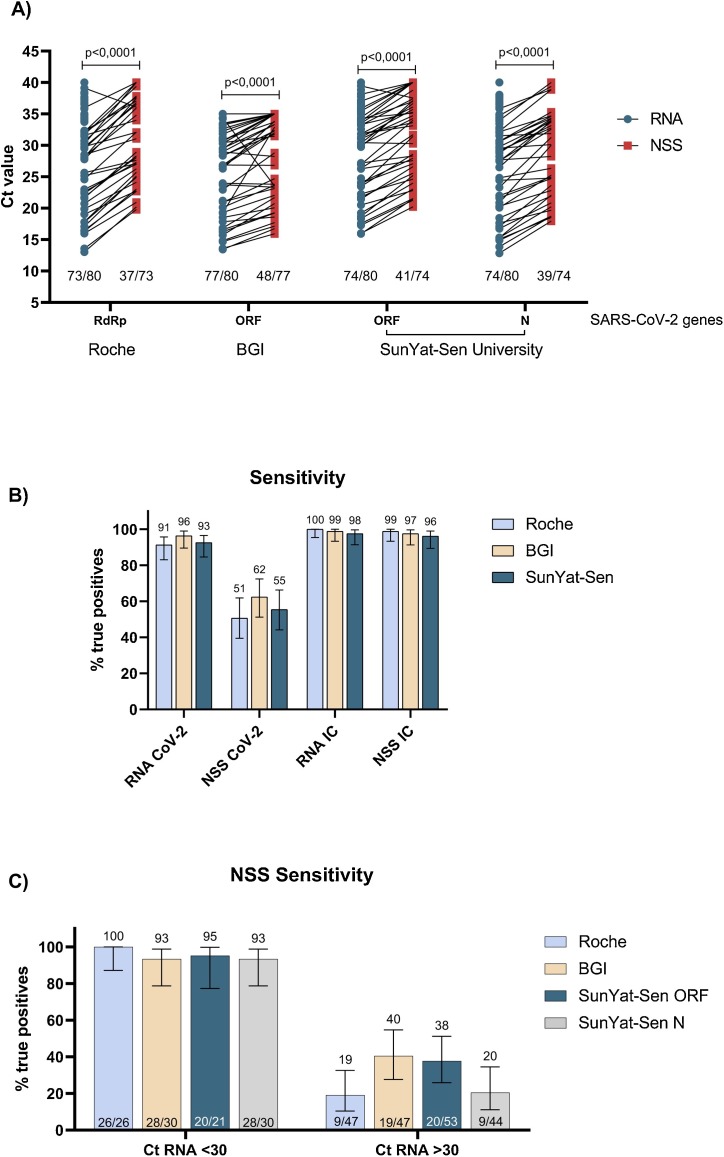
We selected eighty SARS-CoV-2 positive samples obtained from individuals visiting health care at Santiago, Chile. Nasopharyngeal swabs samples (NSS) were resuspended in 2 mL of phosphate buffer saline (PBS) and 150 μL were used to carry out RNA extraction with the Norgen Biotek CORP RNA extraction kit (cat. 24,380). A 5 μL aliquot of the extracted RNA was used to perform RT-PCR assays using the TaqMan™ 2019-nCoV Assay (cat. A15300). We confirmed the presence of viral RNA using this kit following the protocols provided by the manufacturer in a QuantStudio3 Real Time PCR System 96 wells (Thermo Fisher Scientific) showing the presence of the SARS-CoV-2 genes Orf1ab, S and N (data not shown). RNA extracted under similar conditions described above was tested with three different FDA approved and commercially available detection kits: namely SARS-CoV-2 RdRp plus EAV control (cat. 40−0777-10, Roche), Real-time fluorescent RT-PCR kit for detecting 2019-nCoV (cat. MFG030011, BGI) and Detection kit for 2019-nCoV RNA (PCR-Fluorescence Probing) (Sun Yat-sen University, cat. DA0930, Da An Gene Co., Ltd). Roche kit detected 73 out of 80 RNA samples (91 %) declared positive by the TaqMan™ 2019-nCoV Assay from ThermoFisher. Similar results were obtained with the SunYat-Sen kit (93 %). Although this kit detects 2 viral genes in the same reaction, its sensibility is similar for both genes (74 out 80). In the case of the BGI kit, we observed that 77 of 80 (96 %) were positive at the RNA level being the best results obtained in comparison with the gold standard TaqMan™ 2019-nCoV kit (Fig. 1 A).
Fig. 1.
SARS-CoV-2 RNA can be detected in direct nasopharyngeal swabs with less sensitivity in RNA simples with >30 Ct values. A. RT-qPCR amplification of SARS-CoV-2 RNA (light blue) and direct nasopharyngeal swab samples preserved in PBS (NSS, light red) using Roche, BGI and SunYat-Sen University as commercial diagnosis kits. Statistical analysis was done using a t-test with Welch's correction. Each dot corresponds to individual data points. B. A positive percentage agreement between RNA (reference) and NSS RT-PCR Bars represent the 95 % confidence intervals computed by the hybrid Wilson/Brown method. C. Total positive percentage agreement between RNA (reference) and NSS RT-qPCR results separated as Ct <30 and >30 in RNA amplification. Bars represent the 95 % confidence intervals computed by the hybrid Wilson/Brown method (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Then, we evaluated whether the different kits would be able to detect SARS-CoV-2 without a prior RNA extraction step. For this, 100 μL from each NSS were incubated 5 min at 95 °C and then centrifuged at 3000 g for 5 min, the supernatant was collected in a new tube. Finally, 5 μL of each supernatant was used to perform direct SARS CoV-2 RT-PCR detection. The results showed that a significantly lower number of positive samples was detected when the RNA extraction step was omitted (p < 00,001; Fig. 1A). The three SARS-CoV-2 detection kits evaluated in this work presented lower efficiency than the protocol including RNA extraction reaching only 37 out 73 (51 %), 48 out 77 (62 %) and 41 out 74 (45 %) positive detections for SARS-CoV-2 RdRp plus EAV control (Roche), Real-time fluorescent RT-PCR kit for detecting 2019-nCoV (BGI) and Detection kit for 2019-nCoV RNA (PCR-Fluorescence Probing) (Sun Yat-sen University), respectively (Fig. 1A). Furthermore, the clinical test showed a 51–62 % range of sensitivity which is considerably lower than the sensitivity reported by each manufacturer being over 90 % sensible (Fig. 1B). It is noteworthy that the differences in terms of sensitivity observed here could lead to a high percentage of false negatives that would affect the identification of infected individuals. The internal control of each kit does not show differences between extracted RNA or NSS, which may be explained by the high amount of cellular RNA present in the samples (Fig. 1B).
To better understand whether the amount of viral RNA present within the sample is associated with results obtained when using directly the NSS for RT-PCR, we split the samples into two positivity groups based on Ct values obtained with extracted RNA: those with Ct values under 30 (<30 Ct) and those with Ct values over 30 (>30 Ct) (Fig. 1C). A sensitivity greater than 90 % was observed with samples presenting Ct values lower than 30 with any of the 3 kits evaluated. On the other hand, the sensitivity decreases abruptly with samples having a Ct value higher than 30 with detection ranging between 19–40 %. These data strongly suggest that this technique is strongly dependent on the amount of viral RNA present in the sample, showing that it is a very efficient method at Ct values lower than 30.
Importantly, the results are also independent of the detected gene (RdRp, ORF1a or N genes) and therefore, on the kit used. Besides, the median (IQR) difference between RNA and NSS in SARS-CoV-2 Ct values (ΔCtRNA-NSS) obtained with Roche, BGI and SanYut-Sen kit were 6.2 (4.0–7.3), 2.7 (0.0–4.2) and 4.1 (2.3–5.2), respectively (Supplementary Table 1).
In summary, we extended our preliminary observations using a RNA extraction-free SARS-CoV-2 detection protocol (Beltrán-Pavez et al., 2020) using nasopharyngeal swab samples from eighty COVID-19 diagnosed individuals using three commercially available and broadly used RT-PCR kits. Our data confirm that RNA would be omitted from the protocol at least for rapid screening purposes as we observed that samples with Ct values <30 were detected independent of the Rt-PCR kit thus, allowing rapid detection of SARS-CoV-2 directly from nasopharyngeal swabs samples for screening to obtain faster results. Still, it is important to notice that a high percentage of false negatives could be envisioned using this strategy (those with lower viral loads). Nevertheless, some discrepancies in the detection of positive samples are also evident when comparing the sensitivity of detection of the different kits using the same RNA extracted samples. Therefore, this method may need improvements to increase the sensibility in all tested kits.
We have investigated the sensitivity and performance of different commercially available RT-PCR kits in detecting SARS-CoV-2 using 80 extracted RNA and NSS from COVID-19 diagnosed patients. We evaluated the ability of each kit to detect viral RNA from both kit-extracted or directly from a pre-boiled NSS observing that direct RNA detection is possible when Ct values are lower than 30 with the three kits tested. Since SARS-CoV-2 testing in most locations occurs once COVID-19 symptoms are evident and, therefore, viral loads are expected to be high (To et al., 2020), our protocol will be useful in supporting SARS-CoV-2 diagnosis, especially in America where COVID-19 cases have exploded in the recent weeks as well as in low- and middle-income countries, which would not have massive access to kit-based diagnosis. This protocol will also help to avoid the bottleneck in SARS-CoV-2 diagnosis exerted by the RNA extraction step as it reduces at least by 2 h the time required for sample processing thus, accelerating the obtention of the results. The information provided in this work paves the way for the development of more efficient SARS-CoV-2 detection approaches avoiding an RNA extraction step.
Ethical statement
The 80 nasopharyngeal samples from COVID-19 positive patients were de-identified and not considered as Human samples. However, we have a protocol approved by the Ethic Committee of the Faculty of Medicine at Universidad de Chile (Project Nº 036–2020) in order to improve SARS-CoV-2 detection strategies.
Authors contributions
CB-P, FV-E, AG, RS-R and GB participated in the study design. CB-P, LAP and GB performed the experiments. CB-P, LAP, RS-R and GPB analyzed the data. AG provided the NSS from COVID-19 diagnosed individuals. FV-E, AG, RS-R and GB wrote the manuscript. All authors approved the final version of the manuscript.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
The authors are supported by ANID Chile through Fondo Nacional de desarrollo cientifico y tecnologia (FONDECYT) grants No. 11200228 (GB-P), 1181656 (AG), 1190156 (RS-R), 1180798 (FV-E), Instituto Antártico Chileno (INACH) RT_35-19 (GB-P); Internacionalización Universidad de Chile -1566, Postdoctoral fellowship No SECTEI/138/2019 from Mexico city (LA-P). Authors would like to thank C. Joaquin Caceres (Poultry Diagnostic and Research Center, Department of Population Health, University of Georgia, USA) for critical reading of the manuscript and the Chilean Science, Technology, Knowledge and Innovation Ministry for articulating and coordinating support from the scientific community.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2020.113969.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Beltrán-Pavez C., Márquez C., Muñoz G., Valiente-Echeverría F., Gaggero A., Soto-Rifo R., Barriga G. SARS-CoV-2 detection from nasopharyngeal swab samples without RNA extraction. bioRxiv. 2020 doi: 10.1101/2020.03.28.013508. 2020.03.28.013508. [DOI] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368(80-):489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C.Y., Cai J.P., Chan J.M.C., Chik T.S.H., Lau D.P.L., Choi C.Y.C., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C.K., Poon R.W.S., Luo C.T., Cheng V.C.C., Chan J.F.W., Hung I.F.N., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.