Establishment of the rhizobium-legume symbiosis requires a highly specific and complex signal exchange between both participants. Rhizobia perceive legume flavonoid compounds through LysR-type NodD regulators. Often, rhizobia encode multiple copies of nodD, which is one determinant of host specificity. In some species of rhizobia, the presence of multiple copies of NodD extends their symbiotic host-range. Here, we identified and characterized a second copy of nodD present in some strains of the clover microsymbiont Rhizobium leguminosarum bv. trifolii. The second nodD gene contributed to the competitive ability of the strain on white clover, an important forage legume. A screen for strains containing nodD2 could be utilized as one criterion to select strains with enhanced competitive ability for use as inoculants for pasture production.
KEYWORDS: clover, competition, Nod factor, NodD, Rhizobium leguminosarum, symbiosis
ABSTRACT
Establishment of the symbiotic relationship that develops between rhizobia and their legume hosts is contingent upon an interkingdom signal exchange. In response to host legume flavonoids, NodD proteins from compatible rhizobia activate expression of nodulation genes that produce lipochitin oligosaccharide signaling molecules known as Nod factors. Root nodule formation commences upon legume recognition of compatible Nod factor. Rhizobium leguminosarum was previously considered to contain one copy of nodD; here, we show that some strains of the Trifolium (clover) microsymbiont R. leguminosarum bv. trifolii contain a second copy designated nodD2. nodD2 genes were present in 8 out of 13 strains of R. leguminosarum bv. trifolii, but were absent from the genomes of 16 R. leguminosarum bv. viciae strains. Analysis of single and double nodD1 and nodD2 mutants in R. leguminosarum bv. trifolii strain TA1 revealed that NodD2 was functional and enhanced nodule colonization competitiveness. However, NodD1 showed significantly greater capacity to induce nod gene expression and infection thread formation. Clover species are either annual or perennial and this phenological distinction is rarely crossed by individual R. leguminosarum bv. trifolii microsymbionts for effective symbiosis. Of 13 strains with genome sequences available, 7 of the 8 effective microsymbionts of perennial hosts contained nodD2, whereas the 3 microsymbionts of annual hosts did not. We hypothesize that NodD2 inducer recognition differs from NodD1, and NodD2 functions to enhance competition and effective symbiosis, which may discriminate in favor of perennial hosts.
IMPORTANCE Establishment of the rhizobium-legume symbiosis requires a highly specific and complex signal exchange between both participants. Rhizobia perceive legume flavonoid compounds through LysR-type NodD regulators. Often, rhizobia encode multiple copies of nodD, which is one determinant of host specificity. In some species of rhizobia, the presence of multiple copies of NodD extends their symbiotic host-range. Here, we identified and characterized a second copy of nodD present in some strains of the clover microsymbiont Rhizobium leguminosarum bv. trifolii. The second nodD gene contributed to the competitive ability of the strain on white clover, an important forage legume. A screen for strains containing nodD2 could be utilized as one criterion to select strains with enhanced competitive ability for use as inoculants for pasture production.
INTRODUCTION
Rhizobia are soil bacteria that possess the ability to enter a symbiotic relationship with a legume host, culminating in the formation of nitrogen-fixing root nodules (1). Formation of the symbiosis requires a highly specific and complex signal exchange (2). The plants exude flavonoids into the rhizosphere and, in response, compatible rhizobia produce lipochitin oligosaccharide (LCO) signaling molecules known as Nod factors (NFs) (3). The biosynthesis of NFs is performed by proteins encoded by the nodulation (nod) genes, which are generally located on transmissible genetic elements (4, 5). Recognition of NFs by a compatible legume initiates a signal cascade resulting in infection thread (IT) formation and nodule organogenesis (6, 7). Once established in root nodules, the rhizobia differentiate into bacteroids that fix atmospheric nitrogen into ammonia for utilization by the host legume.
Transcriptional activation of the nod genes requires the NodD protein, a flavonoid inducer, and a cis-regulatory element upstream of nod genes known as the nod-box, where NodD binds (8–11). nodD encodes a LysR-type transcriptional regulator (12–14) that recognizes and binds flavonoid inducers (3). NodD is a transcriptional activator, but it can also act as a repressor. In R. leguminosarum, nodD is located adjacent to, and transcribed divergently from, the nodABCIJ operon (14). The nodA and nodD promoters overlap, and binding of NodD to the nodA nod-box results in repression of nodD, even in the absence of inducer (14, 15). Some species of rhizobia contain up to five isoforms of NodD (16), although R. leguminosarum is reported to contain one (11). R. leguminosarum bv. viciae and bv. trifolii nodD mutants were previously shown to be incapable of nodulation (17, 18); however, in species containing multiple copies, regulation is more complex and nodulation is not abolished by a single nodD mutation (19–22). NodD is an important determinant of host specificity (23, 24) and the presence of multiple nodD genes in some species extends host range through recognition of different signal molecules by the various NodD isoforms (19, 20, 25).
Rhizobium leguminosarum bv. trifolii is the microsymbiont of Trifolium (clover) species (26), which are valuable agricultural forage legumes (27). For the clover-rhizobium symbiosis, there are two barriers to effective nodulation for different host-strain combinations: (i) a geographical barrier (relating to the broad centers of clover diversity) and (ii) a phenological barrier (relating to growth cycle) between annual and perennial species (28). Both the biogeographic and phenological distinctions have significant implications for agronomy. Many R. leguminosarum bv. trifolii strains cannot form an effective symbiosis with clovers from a different center of diversity. Moreover, the phenology of the host is important since, although most R. leguminosarum bv. trifolii strains can form nodules on both annual and perennial clovers, they are often only effective nitrogen-fixers on one type (28). Thus, the phenological barrier has implications for selection of inoculant strains, particularly when annual and perennial clovers are grown in proximity.
Competition is an issue central to the use of rhizobial inoculant strains in agriculture. It has particular relevance to pasture production because R. leguminosarum bv. trifolii inoculum strains face significant competition from indigenous rhizobial soil populations for nodule formation (29). These indigenous populations can be highly competitive but often exhibit reduced symbiotic effectiveness relative to commercial inoculants, reducing the potential benefit of inoculants in the field (30).
The R. leguminosarum bv. trifolii strain TA1 is the commercial inoculant strain for white clover in New Zealand. In this study, we report the identification and characterization of a second copy of the nodD gene, designated nodD2, in strain TA1 and, subsequently in several other R. leguminosarum bv. trifolii strains. In the absence of NodD1, NodD2 of TA1 provided functional redundancy for nodulation, but inactivation of nodD2 did not affect nodulation kinetics and IT formation on white clover, nor induction of the nod genes in response to the major white clover flavonoid, 7,4’-dihydroxyflavone (DHF) (31) in vitro. However, NodD2 significantly enhanced competitive ability for nodule colonization, and its presence may influence effective symbiosis with perennial clover species.
RESULTS
Identification of a second copy of nodD.
Genome sequences of 13 R. leguminosarum bv. trifolii strains, which were isolated from 11 different Trifolium species (Table 1), were available within the Joint Genomes Institute Integrated Microbial Genomes (JGI/IMG) database (32) as of March 2020. BLAST analysis of these sequences indicated eight of the strains contained two copies of nodD. In contrast, none of the 16 strains of R. leguminosarum bv. viciae contained a second nodD copy.
TABLE 1.
Strains, presence of nodD2, host origin, growth cycle (habit), and center of diversity of the 13 R. leguminosarum bv. trifolii strains investigated in this section
| Strain | nodD2 | Host origin (legume typec) | Bacterial habit | Center of diversityd | Source or reference(s) |
|---|---|---|---|---|---|
| WSM1325 | No | Unknown (A) | Most effective on annual clovers | E-M | (79) |
| SRDI565 | No | T. subterraneum (A) | Primarily annual, and one perennial species of Mediterranean origin | - | (80) |
| SRDI943 | No | T. michelianum Savi cv. Paradana (A) | Primarily annual, and some perennial species of Mediterranean origin; (Fix−) nodules with the perennial clovers T. pratense and T. polymorphum | - | (81) |
| WSM1689b | No | T. uniflorum (P) | Single perennial species | E-M | (28, 39) |
| CB782 | No | T. semipilosum (P) | Perennial | A | (28, 82) |
| TA1 | Yes | T. subterraneum (A) | Annual and perennial species; broad host range on European and Mediterranean clovers | E-M | (28, 83) |
| CC275e | Yes | T. repens (P) | Range of annual and perennial species | E-M | This study/(32, 84) |
| CC283ba | Yes | T. ambiguum (P) | Highly effective on perennial species T. ambiguum | E-M | (28, 64) |
| WSM2297 | Yes | T. africanum (P) | Limited info/perennial | A | (32) |
| WSM2012a | Yes | T. rueppellianum (A) | Limited annual and perennial species | A | (28, 85) |
| WSM2304a | Yes | T. polymorphum (P) | Highly competitive on perennial species T. polymorphum | S-A | (28, 86) |
| WSM597a | Yes | T. pallidum (A) | Effective on perennial species T. polymorphum | S-A | (32, 87) |
| CC278f | Yes | T. nanum (P) | Perennial | N-A | (28) |
Known to have a narrow host range.
Forms either no nodules or ineffective nodules on a range of annual and perennial species; forms highly effective nodules on a single host, T. uniflorum.
A, annual clover species; P, perennial clover species.
E-M, Euro-Mediterranean; A, Africa; S-A, South America; N-A, North America; -, unspecified.
The designation of nodD1 or nodD2 in R. leguminosarum bv. trifolii was determined by their genetic context. In every case, nodD1 was adjacent to, and divergently transcribed from, the nodABCIJ operon (Fig. S1a in the supplemental material); the genetic context of nodD2 varied among strains, but in all cases it was located in proximity to putative transposase gene fragments (Fig. S1b). The majority of the R. leguminosarum bv. trifolii genomes are incomplete draft assemblies comprised of multiple contigs, and therefore it could not be determined whether the nodD2 homologues were located on the symbiotic plasmid. A pairwise comparison of the 13 nodD1 and 8 nodD2 sequences across the 13 R. leguminosarum bv. trifolii strains revealed they shared from 75 to 100% nucleotide identity (Table S1), with increased conservation observed toward the 5′ end (Fig. S2). Investigation into the conservation of the promoter regions upstream of nodD2 showed three distinct subgroups (Fig. S3). In all cases, canonical nod-box motifs were either absent or degenerated.
Construction of R. leguminosarum bv. trifolii nodD markerless deletion mutants.
BLASTP and BLASTN analyses of the TA1 genome sequence (NCBI Reference Sequence: CP053205 to CP053209) revealed nodD2 (IMG gene ID 2510893092) shared 89% nucleotide identity with nodD1 (IMG gene ID 2510892990), corresponding to 86% amino acid identity with NodD1. nodD2 was located ∼65 kb downstream of the nodABCIJ operon, in divergent orientation to nodD1. To investigate the functionality of nodD2, markerless nodD1, nodD2, and nodD1 nodD2 deletion mutants of TA1 were constructed and characterized (Table 2). A nodD mutant was also constructed in R. leguminosarum bv. trifolii strain WSM1325, which contains a single nodD, producing strain WSM ΔnodD. The nodD2 gene in TA1 was surrounded by DNA encoding multiple transposase gene fragments, some of which were repeated (Fig. S4). Initial experiments showed the fidelity of homologous recombination was compromised by utilizing these flanking regions for construction of the mutant, and so a region of 6 kb that encompassed nodD2 and the flanking repetitive DNA was deleted.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference(s) |
|---|---|---|
| Rhizobium species | ||
| TA1 | Wild-type strain isolated from sub clover in the 1950s in Tasmania, Australia | (83, 88) |
| TA1 ΔnodD1 | TA1 nodD1 markerless deletion | This study |
| TA1 ΔnodD2 | TA1 nodD2 markerless deletion | This study |
| TA1 ΔnodD1 ΔnodD2 | TA1 double nodD1 nodD2 markerless deletion | This study |
| WSM1325 | Wild-type strain isolated from an unidentified annual Trifolium species on the Greek Cyclades island of Serifos in 1993 | (79, 89) |
| WSM ΔnodD | WSM1325 nodD markerless deletion | This study |
| Escherichia coli | ||
| ST18 | S17 λpir ΔhemA | (90) |
| Plasmids | ||
| pFAJ1700 | Stable RK2-derived cloning vector, Apr Tcr | (52) |
| pJQ200SK | pACYC derivative, oriVp15A, sacB, Gmr | (61) |
| pSDZ | Derivative of pFAJ1700 carrying promoterless lacZ gene with downstream GFP fusion cassette, Tcr | (59) |
| pPROBE-KT | pVS1/p15A replicon containing promoterless gfp, Kmr | (53) |
| pPROBE-GT | pVS1/p15A replicon containing promoterless gfp, Gmr | (53) |
| pSFTPnodAZ | pSDZ containing TA1 369-bp nodA promoter lacZ transcriptional fusion | This study |
| pSFTPnodFZ | pSDZ containing TA1 330-bp nodF promoter lacZ transcriptional fusion | This study |
| pSFTnodD1 | pFAJ1700 containing TA1 nodD1 with a 286-bp promoter region | This study |
| pSFTnodD2 | pFAJ1700 containing TA1 nodD2 with a 292-bp promoter region | This study |
| pSFTnodD1KT | pPROBE-KT containing TA1 nodD1 with a 286-bp promoter region | This study |
| pSFTnodD2KT | pPROBE-KT containing TA1 nodD2 with a 292-bp promoter region | This study |
| pSFWnodD | pFAJ1700 containing WSM1325 nodD with a 294-bp promoter region | This study |
| pPR3 | pPROBE-KT containing a 336-bp region of the nptII promoter upstream of a promoterless gfp | (22, 52) |
| pAMNHGUSA | pFAJ1700 containing a gusA transcriptional fusion to the CC275e nifH promoter | This study |
| pAMNHCELB | pFAJ1700 containing a celB transcriptional fusion to the CC275e nifH promoter | This study |
Symbiotic phenotypes of R. leguminosarum bv. trifolii nodD single and double mutants on Trifolium repens.
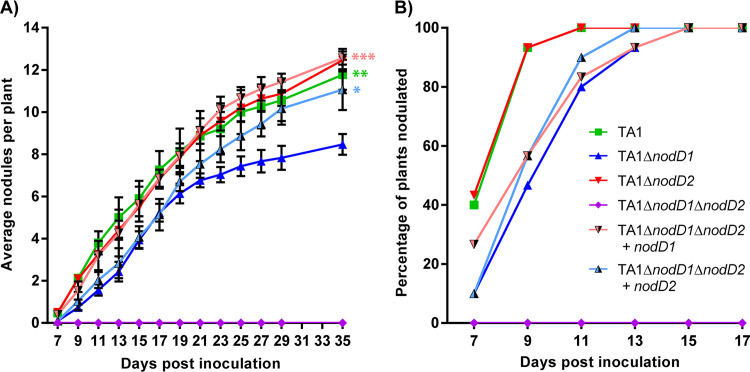
The nodulation ability of the nodD mutants was investigated on Trifolium repens (white clover) cv. Tribute (Fig. 1). Both TA1 and WSM1325 wild-type strains nodulated all plants by 13 days post-inoculation (dpi). WSM ΔnodD formed no nodules during the 42-day period, and its wild-type nodulation phenotype was restored by introduction of the complementation plasmid pSFWnodD (Fig. S5). In contrast, both nodD1 and nodD2 single mutants of strain TA1 nodulated white clover. The TA1 ΔnodD1 ΔnodD2 double mutant did not nodulate white clover, but it was complemented by introduction of either plasmid pSFTnodD1 or pSFTnodD2 containing the nodD1 or nodD2 gene of strain TA1 expressed from their native promoter regions, respectively.
FIG 1.
Nodulation kinetics of white clover plants inoculated with R. leguminosarum bv. trifolii strain TA1 or its nodD mutant derivatives. Three biological replicates were performed with 10 technical replicates in each. (A) Average number of nodules per plant. (B) Percentage of plants nodulated. Error bars represent ± standard error of the mean (SEM). Asterisks represent significant difference from TA1 ΔnodD1 determined using one-way analysis of variance (ANOVA) performed on data from day 35 with Dunnett’s multiple comparisons post hoc test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). The same statistical analysis was performed separately on data from day 11 for early infection.
TA1 ΔnodD2 showed no significant difference in nodulation kinetics compared to the wild-type strain across the course of the nodulation assay (Fig. 1). In contrast, TA1 ΔnodD1 was delayed 4 days in forming nodules on 100% of plants, and at day 35 plants inoculated with TA1 ΔnodD1 had produced significantly fewer nodules compared to plants inoculated with TA1, TA1 ΔnodD2, or TA1 ΔnodD1 ΔnodD2 complemented with either nodD1 or nodD2. In addition to the decreased nodule number, the symbiotic effectiveness of TA1 ΔnodD1 was reduced, as indicated by dry weights of plants measured at 42 dpi (Fig. S6).
By day 35, increased copy number of nodD2 correlated with a significant increase in nodule number, observed from comparing plants inoculated with TA1 ΔnodD1 (containing one copy of nodD2) and TA1 ΔnodD1 ΔnodD2 + nodD2 (containing multiple nodD2 copies). However, this difference was not observed during early infection; at day 11, TA1 ΔnodD1 had produced significantly fewer nodules than any of the strains that possessed nodD1 (TA1, P = 0.0052; TA1 ΔnodD2, P = 0.0334; TA1 ΔnodD1 ΔnodD2 + nodD1, P= 0.0420), but there was no difference compared to TA1 ΔnodD1 ΔnodD2 + nodD2 (P = 0.8916). These data suggest that while nodD1 was the preeminent nodD homologue for nodulation of white clover, nodD2 was able to partially compensate for loss of nodD1 and this was augmented by an increase in the copy number of nodD2.
In R. leguminosarum bv. trifolii strain TA1, NodD1 and NodD2 show differences in in vitro induction of the nod genes in response to flavonoids.
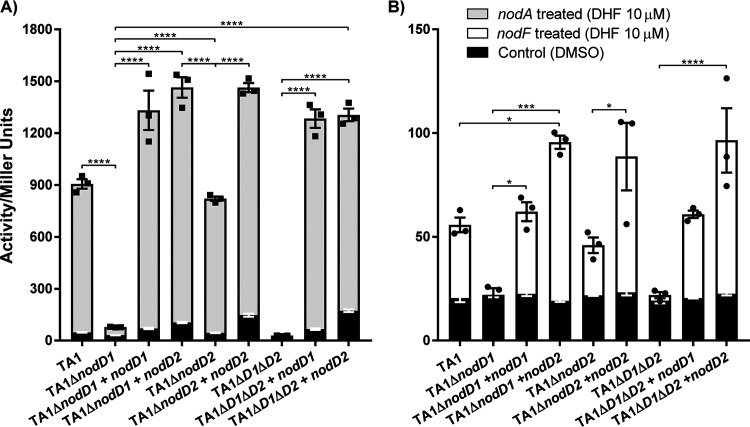
To investigate the capability of the individual copies of NodD to induce nod gene expression, β-galactosidase assays were performed on TA1 ΔnodD1 and TA1 ΔnodD2 strains containing the PnodA-lacZ and PnodF-lacZ transcriptional fusion plasmids pSFTPnodAZ and pSFTPnodFZ cultured in the presence of 10 μM DHF. The effect of adding additional copies of nodD1 or nodD2 was also tested using complementation plasmids pSFTnodD1KT and pSFTnodD2KT, respectively.
The nodA and nodF promoters were not induced in strain TA1 ΔnodD1 ΔnodD2. TA1 ΔnodD2 showed no difference in nodA or nodF induction relative to wild-type TA1 (Fig. 2A and B). TA1 ΔnodD1, on the other hand, showed a drastic reduction in activity from both promoters; relative to the control, the nodF promoter induction was negligible, while the nodA promoter was induced only 2.5-fold. However, TA1 ΔnodD2 complemented with multicopy nodD2 showed significantly higher expression of nodA compared to wild-type, as did TA1 ΔnodD1 complemented with nodD2 and TA1 ΔnodD1 ΔnodD2 complemented with either nodD1 or nodD2. Similar results were observed for the nodF promoter, where expression levels were complemented by both nodD genes; however, for this promoter, nodD2 produced higher levels of expression. Together, these results imply that in the natural genomic context, nodD2 is poorly expressed; however, when present on a complementation plasmid (∼6 to 8 copies/cell), NodD2 activity is amplified and responsive to DHF. Additionally, the nodD2 complementation plasmid induced low but significant expression of the nodA promoter in the absence of the inducer DHF (Fig. 2A; Fig. S7). In contrast, the nodF promoter showed no difference in basal promoter expression across the various strains in the presence or absence of pSFTnodD2KT.
FIG 2.
Role of NodD1 and NodD2 in activation of nodA and nodF promoters. β-galactosidase assays were conducted on TA1 nodD mutant derivatives containing nodA or nodF promoter reporter plasmids in response to 10 μM synthetic DHF. The DMSO-only controls are superimposed on the DHF-induced treatment data. (A) nodA promoter induction. (B) nodF promoter induction. Each symbol represents a biological replicate. Error bars represent ± SEM. Asterisks indicate significant difference between the treated samples determined using one-way ANOVA (Tukey’s multiple comparisons post hoc test, *, P < 0.05; ***, P < 0.001; ****, P < 0.0001).
nodD1 and nodD2 mutants show differences in infection thread formation.
To investigate the effect of the individual copies of nodD on IT formation, the plasmid pPR3 that expresses green fluorescent protein (GFP) from the constitutive nptII promoter (22) was introduced into TA1, TA1 ΔnodD1, TA1 ΔnodD2, and TA1 ΔnodD2 + pSFTnodD2 strains. The noncomplemented strains also contained plasmid pFAJ1700 as a control for the complemented strain. The numbers of ITs per plant were then enumerated at seven dpi using epifluorescence microscopy. TA1 produced on average 25.3 ITs per plant. TA1 ΔnodD2 produced an average of 32.2, and TA1ΔnodD2 complemented with nodD2 produced an average of 24.8, neither of which were significantly different from wild-type TA1 (P = 0.44 and P = 0.99, respectively). In contrast, TA1 ΔnodD1 produced significantly fewer, with only 4.3 ITs per plant (Fig. 3).
FIG 3.
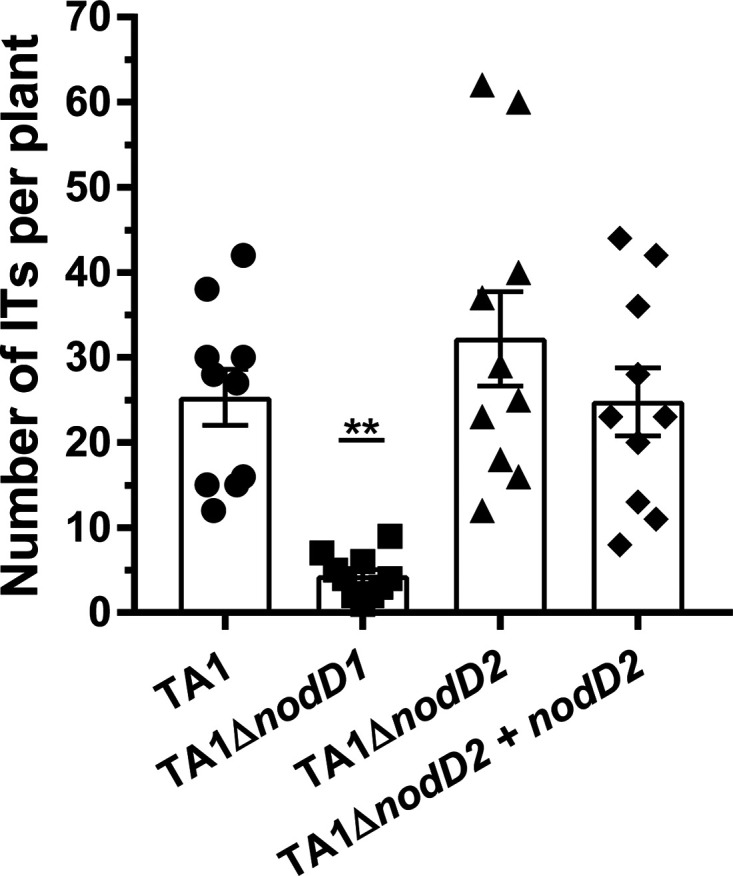
Infection thread formation on white clover by strain TA1 and its nodD mutant derivatives containing the reporter plasmid pPR3. The number of ITs per strain was observed at seven dpi on 10 plants, each represented by a symbol. The error bars represent ± the SEM. The asterisks indicate significant difference from the wild type (one-way ANOVA with Dunnett’s multiple comparisons post hoc test, **, P < 0.01).
nodD mutants are impaired in competitive nodule colonization.
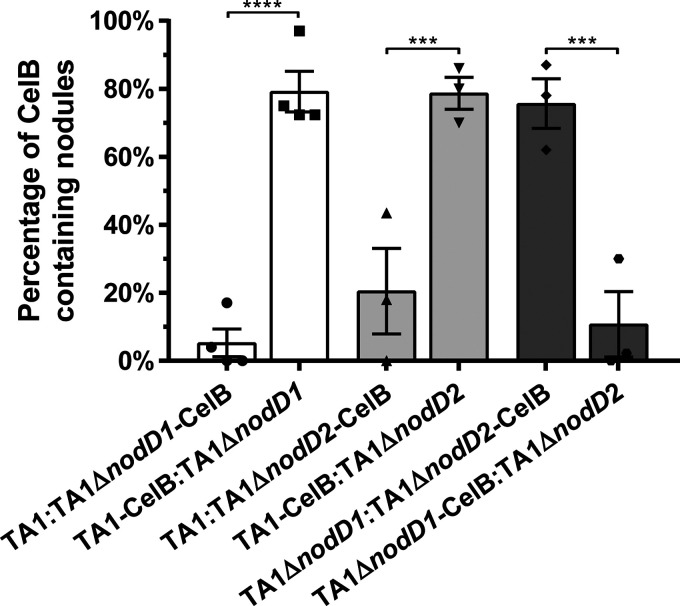
The pairwise competitive ability of TA1 and its two single nodD mutants was investigated on white clover using strains marked with plasmids pAMNHCELB and pAMNHGUSA expressing either celB or gusA, respectively. TA1 ΔnodD1 was severely disadvantaged when competed against TA1 (Fig. 4). TA1 ΔnodD2 out-competed TA1 ΔnodD1 for nodule colonization, confirming the importance of nodD1 for nodulation. However, TA1 ΔnodD2 was also significantly out-competed by TA1 for nodule colonization, despite containing functional nodD1, indicating that both genes contribute to the overall competitiveness of the strain.
FIG 4.
The relative competitive ability of TA1 wild type and nodD mutant strains. The percentage of nodules occupied by each strain was determined by the presence of blue/nonblue nodules following staining of white clover roots inoculated with pairs of strains in a 1:1 ratio (approximately 500 cells of each strain). The proportion of blue nodules from each comparison is reported as percentage. The same pairs of strains containing reciprocal reporter plasmids are linked by bar color. In each reciprocal combination, the strain lacking pAMNHCELB contained the plasmid pAMNHGUSA. The percentage indicates the proportion of blue nodules determined from the sum of the five uppermost nodules formed on 10 plants (50 nodules total where 5 nodules were present). Three to four sets of 10 plants per strain pair were examined, with each replicate set represented by a symbol. The error bars represent ± the SEM. The asterisks indicate significant difference between the reciprocal pairs of strains (one-way ANOVA with Sidak’s multiple comparisons post hoc test, ***, P < 0.001; ****, P < 0.0001).
TA1 NodD1 and NodD2 predicted protein structures suggest altered flavonoid perception.
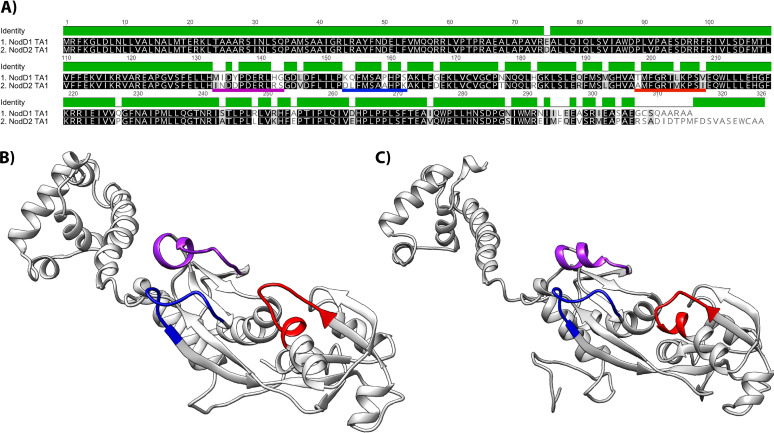
Sequence alignment of TA1 NodD1 and NodD2 revealed 86% amino acid identity (Fig. 5A). LysR-type transcriptional regulators contain an N-terminal HTH DNA binding motif (13), and TA1 NodD1 and NodD2 were identical apart from one conservative amino acid difference (E75D) in the first 120 amino acids. There were several regions of reduced conservation in the C-terminal half of the protein, three of which are underlined in Fig. 5A and highlighted in the model in Fig. 5B. These regions of reduced conservation appeared to not only alter the structures of NodD1 and NodD2 relative to each other (Fig. 5B and C), but also corresponded to the entrance of the predicted flavonoid-binding pocket (33, 34), and therefore likely alter flavonoid binding/perception of each isoform.
FIG 5.
Alignment of TA1 NodD1 and NodD2 amino acid sequences and corresponding predicted protein structures shown as ribbon models constructed using Phyre2. (A) Amino acid sequence alignment. The background tone indicates amino acid similarity, where black is identical, dark gray is similar, light gray is not similar. The green bar indicates conserved residues. Three regions corresponding to the predicted flavonoid inducer binding pocket entrance are underlined in purple, blue, and red. (B) TA1 NodD1. (C) TA1 NodD2. The location of the residues highlighted in (A) are shown on the ribbon structures with corresponding colors.
R. leguminosarum bv. trifolii NodD2 shows discrete origins and clusters according to the native clover host’s biogeography and phenology.
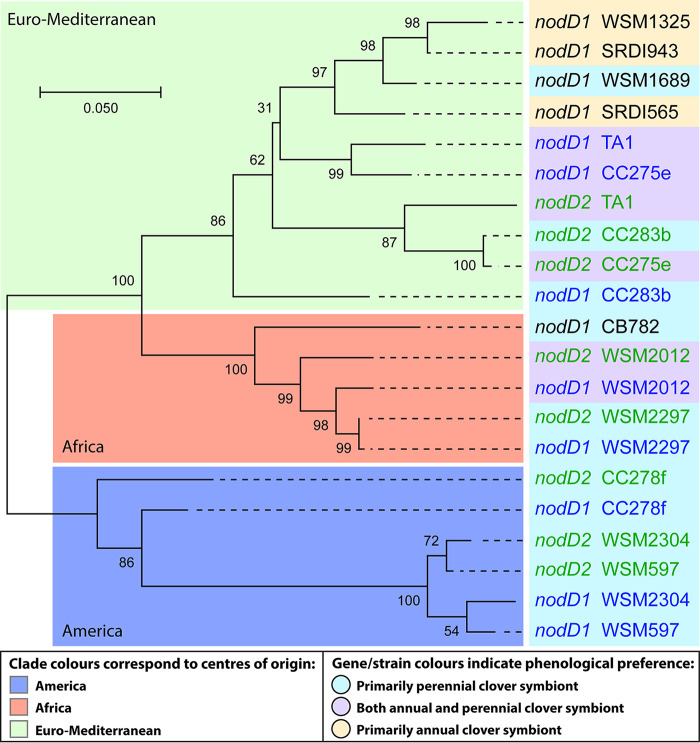
The nodD1 and nodD2 nucleotide sequence similarity of strains CC278f, WSM2304, WSM2012, WSM597, and WSM2297 ranged from 93 to 99.8% (Table S1). The two nodD homologues from each of these strains clustered in proximity (Fig. 6), and for strain WSM2297, the NodD1 and NodD2 amino acid sequences were identical (Table S2), with only two synonymous single-nucleotide polymorphisms; however, the promoter regions are not conserved (Fig. S8). Together, these results indicate that nodD2 arose via gene duplication events in these strains which, given the observation that nodD2 was located adjacent to transposon fragments in all strains examined, likely involved transposition events that are known to facilitate both gene duplications and horizontal transfer (35). In contrast, the two nodD homologues from strains TA1, CC275e, and CC283b were more divergent, with 89, 90, and 87% nucleotide identity, respectively (Table S1). The nodD2 genes from TA1, CC275e, and CC283b formed a separate clade from the other nodD2 homologues (Fig. 6), suggesting that these three strains acquired nodD2 by horizontal transfer. In particular, nodD2 from CC275e and CC283b shared 99% nucleotide identity whereas the nodD1 genes from the same strains only shared 89% identity, suggesting that the nodD2 genes were acquired recently by the two strains from a common ancestor. Taken together, these data suggest that two independent mechanisms, duplication and horizontal acquisition, were responsible for the appearance of the R. leguminosarum bv. trifolii nodD2 genes in the different strains.
FIG 6.
Evolutionary history of 21 nodD nucleotide sequences inferred by using the maximum likelihood method and Tamura-Nei model (76). For strains with one copy, nodD1 is black; for strains with both, nodD1 and nodD2 are blue and green, respectively. The phenological and geographical distributions of the strains are indicated according to the key, based on information in Table 1. The tree with the highest log likelihood (−6,198.64) is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches (77). Initial tree(s) for the heuristic search were obtained automatically by applying neighbor joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 21 nucleotide sequences. There were a total of 986 positions in the final data set. Evolutionary analyses were conducted in MEGA X (78).
The geographic centers of diversity of the hosts that form effective symbioses with the rhizobial strains examined here are shown in Fig. 6 and Table 1. To investigate the biogeography further, heat maps were constructed from the pairwise identity matrices generated from separate alignments of the entire symbiont genomes, the nodA nucleotide sequences, and nodD1/nodD2 nucleotide sequences (Fig. S9). The genomes clustered into two groups as determined by orthologous average nucleotide identity (orthoANI) (36), with the smaller group containing three distinct genospecies (defined as sharing less than 95% sequence identity) (37). These genome groups (with one exception) corresponded to the biogeographical origins of the strain, as did the nodD1, nodD2, and nodA genes. Therefore, the chromosomal lineage and symbiotic genes are grouped according to the same biogeography and nodD2 is not restricted to microsymbionts of any particular center of diversity. Strain CC278f is an exception, as its genome and nodA sequence cluster in closer proximity to the Euro Mediterranean strains, yet its nodD homologues are more similar to the South American strains, as reported previously (38).
There was also a possible association between the presence of nodD2 and enhanced ability to effectively nodulate perennial clovers (Fig. 6). nodD from four of the five strains that contain one copy cluster together, and three of those four are annual clover symbionts (Table 1). The remaining R. leguminosarum bv. trifolii strains are all capable of forming an effective symbiosis with perennial species, and some are capable of crossing the phenological barrier for effective symbiosis with both annual and perennial host types. Only strains CB782 and WSM1689 lack nodD2 but are capable of effective symbiosis with a perennial clover. Strain WSM1689 possesses a particularly narrow host range, as it is ineffective on a range of both annual and perennial hosts with the single exception of T. uniflorum (39). Strain CB782 NodD was equally similar to NodD2 and NodD1, as inferred by clustering and amino acid identity (Table S2) of 91% and 92% with WSM2012 NodD1 and NodD2, respectively.
DISCUSSION
Many rhizobial species contain multiple nodD homologues (40, 41) that perform divergent roles and extend symbiotic host range (19, 20, 42). For instance, Sinorhizobium meliloti contains three copies: nodD1, nodD2, and nodD3 (21, 43). NodD3 and SyrM, another LysR regulator, are involved in responses to different plant signals than NodD1 and NodD2 (20). In Rhizobium tropici CIAT899, a strain known for its broad host range, five copies of nodD have been identified (44). For some R. tropici CIAT899 hosts, one copy of nodD is adequate for efficient nodulation, whereas regulation is far more complex on other hosts (e.g., Lotus japonicus), where four of the five nodD homologues are required (19). R. leguminosarum was previously considered to contain one nodD (11, 18), and single nodD mutations in both R. leguminosarum bv. trifolii and R. leguminosarum bv. viciae abolished nodulation (17, 45). We did not detect a second copy of nodD in the 16 R. leguminosarum bv. viciae strain genomes present in the JGI/IMG database, yet we identified nodD2 in eight of the 13 R. leguminosarum bv. trifolii strains. In support of these findings, deletion of the single nodD of R. leguminosarum bv. trifolii strain WSM1325 abolished nodulation, whereas for strain TA1, nodulation was only abolished in the double nodD mutant.
Recently it was shown that for the Lotus symbiont Mesorhizobium japonicum (formerly M. loti) strain R7A, the two NodD isoforms (46) function at distinct stages during symbiotic infection in response to different inducer molecules (33). NodD1 promoted IT formation, whereas NodD2 was primarily involved in nod gene induction in the rhizosphere and in nodules. Likewise, for R. leguminosarum bv. trifolii there appeared to be spatially and temporally distinct roles for each NodD isoform. NodD1 and NodD2 varied in their response to DHF when at chromosomal copy number (Fig. 2), and deletion of the individual genes showed they varied in their ability to support IT initiation (Fig. 3). NodD1 was the central regulator, predominant for both activation of the nod genes in response to root signals, and for IT formation. However, in R. leguminosarum bv. trifolii strain TA1, provision of additional copies of nodD2 (present on a multicopy plasmid) conferred an advantage in nodule formation relative to a strain containing a single nodD2 copy, complementing the nodD1 mutant nodulation kinetics to near wild-type levels (Fig. 1). It also resulted in significantly increased nod promoter activation in response to DHF (Fig. 2). This increased nodD copy number and the corresponding elevated nod gene expression would therefore have augmented NF production (47, 48). These results suggest a disparity in the native expression levels of nodD1 and nodD2 in vivo, where higher nodD1 expression would result in substantial NF production, important for rhizosphere signaling and initiation of infection. Nonetheless, nodD2 conferred a significant benefit at a later stage of infection, as its absence was significantly detrimental for competitive nodule colonization (Fig. 4). Together the results suggest that R. leguminosarum bv. trifolii NodD1 and NodD2 each exert influence during temporally discrete stages of infection, and indicate that nodD2 may act later in infection.
Several regions of reduced conservation identified between NodD1 and NodD2 isoforms in M. japonicum R7A (33) were also identified between TA1 NodD1 and NodD2 (Fig. 5A). These regions are associated with important structural components, including the proposed flavonoid binding pocket entrance (33, 34) (Fig. 5B and C), and a few specific residues that are involved in NodD coinducer binding/response (49). This suggests NodD1 and NodD2 are at least partially distinct in their recognition of inducers and that NodD2 may provide a mechanism for perception of a different inducer molecule, or a similar molecule with altered affinity, relative to NodD1. In support of this hypothesis, white clover was shown previously to produce different inducers at temporally and spatially separate stages in symbiosis (50).
In M. japonicum R7A, NodD2 provides extra compatibility scrutiny, to improve selection of an appropriate rhizobial partner (33). If applied to R. leguminosarum bv. trifolii, this generalized compatibility check would presumably impose a strong selective pressure, resulting in the ubiquitous presence of nodD2, yet nodD2 is not encoded in ∼40% of R. leguminosarum bv. trifolii strains investigated. Although it is clear that NodD2 substantially enhances the competitive ability of TA1 for nodule colonization on white clover, this nodD2-mediated signaling may be unnecessary or undesirable on certain hosts. For the clover-rhizobia symbiosis, Howieson et al. (28) reported that few host-strain combinations formed effective symbioses across either the geographical or phenological barrier, with many R. leguminosarum bv. trifolii strains forming effective symbiosis with either annual or perennial clovers, but rarely both. The biogeography of the genomes of the 13 strains is congruent with the clustering of the nodD sequences, with one exception, strain CC278f. However, we found an apparent association between the preferred host phenology of R. leguminosarum bv. trifolii strains (Table 1) and the presence of a particular nodD homologue (Fig. 6). Strains with one nodD were primarily annual clover symbionts, while the majority of strains that possess nodD2 were capable of effective symbiosis with perennial clover hosts. These results suggest that nodD is linked to the specificity of R. leguminosarum bv. trifolii strains within this phenological distinction. Furthermore, Howieson et al. (28) found that within 400 cross-inoculation groups, consisting of R. leguminosarum bv. trifolii strains from the major centers of geographical diversity, along with both annual and perennial clovers, perennial clover species appeared to be much less promiscuous than annual species. This is consistent with the concept that symbiosis with perennial clovers by compatible microsymbionts is enhanced by additional regulation conferred by nodD2.
For strain WSM2297, NodD1 and NodD2 share 100% amino acid identity, but their promoter regions contain nod-boxes that are not conserved (Fig. S8). The nod-box is a 46 to 47-bp conserved motif (10, 51) containing two copies of the imperfect palindrome AT-N10-GAT, which includes the LysR motif, T-N11-A, and is the primary binding target of NodD (8, 51). It consists of two distinct halves, the distal (D) and proximal (P) halves, each containing the palinodrome that is crucial for NodD binding (51). An alignment of the WSM2297 nodD1 and nodD2 promoter regions revealed that for nodD2, the nod-box P-half contained several base pair substitutions, including a disruption to the LysR motif (Fig. S8). Furthermore, there was a base pair deletion in the region between the two nod-box motifs. Therefore, the autoregulation known to occur in the nodD1-nodA intergenic region (14, 15) would be altered if not abolished for nodD2. Similarly, upstream of TA1 nodD2, there was no apparent nod-box motif.
nodD2 in TA1 appeared to be expressed poorly in vitro, as seen with the β-galactosidase assays (Fig. 2), yet the low genomic expression of nodD2 was overcome by expression of nodD2 in trans on a multicopy vector, indicating the potential of nodD2 to be active. The possibility of expression of nodD2 from a vector-borne promoter is unlikely due to the design of the complementation plasmids; in both versions (Table 2) the multiple cloning sites containing nodD2 are flanked by transcriptional terminators (52, 53). An alternative explanation of cis-acting sequences adjacent to nodD2 causing repression of nodD2 in its chromosomal context but not in the complementation plasmid seems unlikely, as the complementation plasmids contained 292 bp of DNA upstream of nodD2. Two lines of evidence show that nodD2 is, in fact, functional in planta: first, the presence of nodD2 alone was sufficient for nodulation of white clover (Fig. 1); and second, there was a significant competitive defect in the absence of nodD2 (Fig. 4). Together these results suggest that nodD2 expression may be activated in planta, or, alternatively, it is possible that nodD2 expression may be low but constitutive. TA1 NodD2 appeared to exhibit a slight flavonoid-independent transcriptional activation (FITA) phenotype for the nodA promoter when present on the multicopy plasmid, suggesting that NodD2 may be functional in the absence of inducer (Fig. S7). Although significant expression measured using β-galactosidase assays was not observed when a single nodD2 gene was present on the genome (Fig. 2, Fig. S7), nodD2 may induce low level constitutive expression of the nodABCIJ operon, enabling NF production in the absence of inducer.
Thus, we hypothesize that the advantage conferred by NodD2 derives from NF signaling at a stage of infection that is advantageous for establishing symbiosis with particular hosts. This signaling may occur either due to NodD2 more readily recognizing a particular flavonoid profile than NodD1 or, alternatively, by providing a low level of constitutive NF production in the absence of plant signal. Therefore, the differential regulation of each nodD may account for the retention of a second copy in strains where homologues are highly conserved.
Taken together the data show that despite nodD1 being the dominant regulator, NodD2 is a functional isoform of NodD present in some strains of R. leguminosarum bv. trifolii that significantly enhances nodule colonization under conditions of competition, implicating it as an important marker for identifying competitive inoculant strains. Our data also hint at a possible link between the presence of nodD2 and the ability of an R. leguminosarum bv. trifolii strain to be an effective symbiont of perennial clover species. With an ever-increasing number of completed genome sequences becoming available, it should be possible in the future to confirm if there is a direct relationship between the presence of NodD2 in R. leguminosarum bv. trifolii and the ability to effectively nodulate perennial clover host species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and oligonucleotides.
The bacterial strains and plasmids used in this study are listed in Table 2. Oligonucleotides used in this study are listed in Table 3. R. leguminosarum bv. trifolii strains were cultured at 28°C in TY (54) or rhizobium defined medium (RDM) (55) supplemented with 0.4% (wt/vol) glucose (G/RDM). Antibiotics were used at the following concentrations: for E. coli, 25 μg/ml gentamicin, 15 μg/ml tetracycline, and 50 μg/ml kanamycin; for R. leguminosarum bv. trifolii, 50 μg/ml gentamicin, 2 μg/ml tetracycline, and 50 μg/ml neomycin. Conjugation of plasmids from E. coli to R. leguminosarum bv. trifolii was performed by biparental spot mating as described previously (56).
TABLE 3.
Primers used in this study
| Primer name | 5′ to 3′ sequence | Source |
|---|---|---|
| nodA TA1 left | TTAAAGATCTGCTCATGGCTGGTTGACTGA | This study |
| nodA TA1 right | AATTCTCGAGTCAATTAATCAGATATTTTCCACCGCACTCC | This study |
| nodF TA1 left | TTAAAGATCTTCAAAATCGCGATTCCGAGC | This study |
| nodF TA1 right | AATTCTCGAGTCAATTAATCACTGATCGGCCATCTTGTTCC | This study |
| WSM NodD LL | GCGGCCGCTCTAGAACTAGTAGATCCGGATGGTCTTTGAC | This study |
| WSM NodD LR | TTAGGTCGCTCTGGCCTCTTTCCAGGCCCTTAAAACGCAT | This study |
| WSM NodD RL | ATGCGTTTTAAGGGCCTGGAAAGAGGCCAGAGCGACCTAA | This study |
| WSM NodD RR | CGAATTGGGTACCGGGCCCCGCCGAGATAAATGCTGACCT | This study |
| TA1 NodD1 LL | GCGGCCGCTCTAGAACTAGTCGGGATGGTCTTTGACATAC | This study |
| TA1 NodD1 LR | TTAGGCCGCTCTGGCCGCTTTCCAGGCCCTTAAAACGCAT | This study |
| TA1 NodD1 RL | ATGCGTTTTAAGGGCCTGGAAAGCGGCCAGAGCGGCCTAA | This study |
| TA1 NodD1 RR | CGAATTGGGTACCGGGCCCCGAGTCACTCCGTCTAGAAGG | This study |
| LL SpeI (TA1D2) | TATAACTAGTACGTATCAGCCGGCAGTCAC | This study |
| LR badzone (TA1D2) | CTACCGCCGTTACTCCGTGACTGCACTTCAACTTCACCAA | This study |
| RL badzone (TA1D2) | TTGGTGAAGTTGAAGTGCAGTCACGGAGTAACGGCGGTAG | This study |
| RR badzone (TA1D2) | AATTGGGCCCCAGCTCAGGCGCTCAATTAG | This study |
| WSM D comp Fwd | TTAATCTAGAATGATCCGACGGTTCGAGAT | This study |
| WSM D comp Rev | AATTGAATTCTTTAAGCGACGGTAGCTCGA | This study |
| TA1 D1 comp Fwd | TTAATCTAGAATGGTCCGACGGTTCGAGAT | This study |
| TA1 D1 comp Rev | AATTGAATTCTTCGAGCTAATGCAGCTCGA | This study |
| TA1 NodD1 forward KpnI (pPROBE) | TTAAGGTACCATGGTCCGACGGTTCGAGAT | This study |
| NodD2 Complement fwd | TTAATCTAGAGGTTAACTTTACGGTGCCCT | This study |
| NodD2 Complement rev | AATTGAATTCACTGCGTATCCGCATCTTCA | This study |
| TA1 NodD2 Complement forward KpnI (pPROBE) | TTAAGGTACCGGTTAACTTTACGGTGCCCT | This study |
DNA manipulations.
Preparation of rhizobial DNA was performed as described previously (57). Extraction of plasmid DNA, cloning, agarose gel electrophoresis, and transformation by electroporation were performed using established methodology (58).
Construction of reporter plasmids.
Primer pairs nodA TA1 left plus nodA TA1 right and nodF TA1 left plus nodF TA1 right (Table 3) were used to amplify the nod-box promoter regions from the R. leguminosarum bv. trifolii strain TA1 nodABCIJ and nodFERL operons, respectively, which were cloned adjacent to the lacZ gene in the plasmid vector pSDZ (59).
Complementation of nodD1 and nodD2 mutants.
Complementation plasmids pSFTnodD1, pSFTnodD2, and pSFWnodD were constructed by cloning PCR products containing the relevant nodD gene and its native promoter region digested with XbaI and EcoRI into pFAJ1700 (52). Similarly, pSFTnodD1KT and pSFTnodD2KT were constructed by cloning the same regions contained in pSFTnodD1 and pSFTnodD2 into pPROBE-KT (53) as KpnI and XbaI fragments.
pSFTnodD1KT and pSFTnodD2KT were used for complementation of nodD mutants in β-galactosidase assays, as pPROBE-KT is compatible with the lacZ reporter plasmids. Strains contained the pSDZ and pPROBE-KT empty vectors as a control where appropriate.
Construction of the markerless deletion mutants.
The nodD mutants used in this study were constructed as markerless deletions as described by Rodpothong et al. (22), with the exception that Gibson assembly (60) was utilized to clone the desired PCR products into the suicide vector pJQ200SK (61). The constructs were confirmed by DNA sequencing, and subsequently transferred to R. leguminosarum bv. trifolii strains by biparental spot mating. Transconjugants were passaged twice on G/RDM plates containing gentamicin to select a strain which had undergone a single crossover. Sucrose-resistant, gentamicin-sensitive strains which contained the anticipated markerless deletion were then obtained by plating on RDM containing 5% sucrose and mutants were screened and confirmed by PCR.
For the TA1 nodD2 mutant, an area of 6 kb was deleted, which included nodD2 and fragments of insertion elements and transposase genes that flanked the gene.
Plant assays.
Seeds were surface-sterilized and both nodulation and IT assays were performed as described previously (62), except for IT assays in which each seedling was inoculated with 100 μl instead of 50 μl of 0.1 optical density at 600 nm (OD600) bacterial culture. The dry shoot weight of plants was recorded following oven drying at 70°C for 48 h.
Plant studies were performed using white clover (T. repens cv. Tribute). For nodulation assays, individual seedlings were planted in 18-mm diameter test tubes, or for IT assays in 10 × 10 cm square petri dishes, containing 8 or 50 ml of Jensen’s seedling agar (63), respectively. Following inoculation, plants were cultivated at 70% relative humidity with cycling from 21°C for 16 h (day) to 14°C for 8 h (night). Nodulation was scored as previously described (64).
β-galactosidase assays.
Five-milliliter TY broth cultures were inoculated with 25 μl of stationary-phase cells, supplemented with flavonoid inducer solubilized in dimethyl sulfoxide (DMSO) and grown with shaking at 180 rpm at 28°C for 18 h. Control cultures were supplemented with an equal volume of DMSO. β-galactosidase assays were performed on broths as previously described (65).
Competition assays.
A competition assay was developed based on a previously described method (66, 67). An alignment of the nifH promoters of R. leguminosarum bv. trifolii strains showed that the CC275e promoter essentially represented a conserved natural consensus sequence (data not shown). The nifH promoter was chosen because it is not expressed until the bacteria have differentiated into bacteroids inside the nodule cells (68), and would thus not be expected to confer a fitness cost during free-living growth and nodule infection. The genes gusA and celB were transcriptionally fused to the CC275e nifH promoter in pFAJ1700, resulting in the reporter constructs pAMNHGUSA and pAMNHCELB, respectively (Table 2).
The plasmids were transferred into R. leguminosarum bv. trifolii strains and mutants. Pairings of one strain containing pAMNHGUSA and the other pAMNHCELB were inoculated onto 10 white clover plants along with a further 10 plants inoculated with the same strains containing the reciprocal reporter plasmids. Coinoculation was performed by adding 100 μl of 1:1 ratio of strains giving approximately 103 cells total/plant. Seedlings were cultivated on Jensen’s seedling agar slopes in square agar plates. At 24 dpi, plant roots were harvested and immersed in 20 ml phosphate-buffered saline containing 0.1% sodium laurylsarcosine and 0.1% Triton X-100, then incubated at 70°C for 60 min to inactivate enzymes other than CelB. The roots were left to cool and then 40 μl of 50 μg/ml X-Gal was added and the roots were incubated at 37°C overnight. The five nodules closest to the cotyledons of each plant were counted and scored to determine nodule occupancy. Staining for GusA activity was not performed, as pAMNHGUSA was present only to balance the potential fitness costs conferred by carrying the plasmids.
Microscopy.
Visual investigation of IT formation by fluorescently marked R. leguminosarum bv. trifolii strains was performed using epifluorescence microscopy. Seedlings cultivated on Jensen’s seedling agar slopes in square agar plates were examined at 7 dpi using an Olympus microscope (model BX51TRF) with fluorescence illuminator (model BXRFA). Cells expressing GFP were visualized using a fluorescence mirror unit (model U-MWIB3) consisting of a 460 to 495 nm bandpass exciter, a 505 nm longpass dichroic mirror, and a 510 nm longpass emitter. The number of ITs along the entire length of the roots was recorded.
Bioinformatics.
The sequences investigated in this study (Table S3) were obtained from the IMG database (32). A range of software packages and applications were used for sequence analysis, primer design, sequence alignments, percentage identity matrices, phylogenetic trees, sequencing quality control, heat map generation, and orthoANI analysis. Software utilized included DNAStar (version 14.1.0.115, DNASTAR, Madison, WI), SnapGene Viewer (version 2.6.2, SnapGene software from GSL Biotech), Geneious (version R10) (69), ClustalX 2.1 (70), RStudio (71, 72), heatmaply (73), and OrthoANI (36). For statistical analysis of data, GraphPad Prism (version 7.02 for Windows, GraphPad Software, La Jolla, CA, USA) was used. Protein structures were predicted and modeled using the Phyre2 web portal (74) and viewed using UCSF Chimera 1.13.1 (75).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded under subcontract by the New Zealand Ministry of Business, Innovation and Employment (MBIE)/Dairy NZ, contract C10X1308 entitled “Improving forage legume-rhizobia performance,” to AgResearch.
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Oldroyd GE, Murray JD, Poole PS, Downie JA. 2011. The rules of engagement in the legume-rhizobial symbiosis. Annu Rev Genet 45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- 2.Dénarié J, Cullimore J. 1993. Lipo-oligosaccharide nodulation factors: a new class of signaling molecules mediating recognition and morphogenesis. Cell 74:951–954. doi: 10.1016/0092-8674(93)90717-5. [DOI] [PubMed] [Google Scholar]
- 3.Spaink HP. 2000. Root nodulation and infection factors produced by rhizobial bacteria. Annu Rev Microbiol 54:257–288. doi: 10.1146/annurev.micro.54.1.257. [DOI] [PubMed] [Google Scholar]
- 4.Hynes MF, Finan TM. 1998. General genetic knowledge, p 25–43, The Rhizobiaceae. Springer. [Google Scholar]
- 5.Sullivan JT, Ronson CW. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci U S A 95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gage DJ. 2004. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly S, Radutoiu S, Stougaard J. 2017. Legume LysM receptors mediate symbiotic and pathogenic signalling. Curr Opin Plant Biol 39:152–158. doi: 10.1016/j.pbi.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Goethals K, Van Montagu M, Holsters M. 1992. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc Natl Acad Sci U S A 89:1646–1650. doi: 10.1073/pnas.89.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong G-F, Burn JE, Johnston A. 1987. Evidence that DNA involved in the expression of nodulation (nod) genes in Rhizobium binds to the product of the regulatory gene nodD. Nucleic Acids Res 15:9677–9690. doi: 10.1093/nar/15.23.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rostas K, Kondorosi E, Horvath B, Simoncsits A, Kondorosi A. 1986. Conservation of extended promoter regions of nodulation genes in Rhizobium. Proc Natl Acad Sci U S A 83:1757–1761. doi: 10.1073/pnas.83.6.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlaman HR, Okker RJ, Lugtenberg BJ. 1992. Regulation of nodulation gene expression by NodD in rhizobia. J Bacteriol 174:5177–5182. doi: 10.1128/jb.174.16.5177-5182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henikoff S, Haughn GW, Calvo JM, Wallace JC. 1988. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A 85:6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddocks SE, Oyston PC. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 14.Rossen L, Shearman CA, Johnston AWB, Downie JA. 1985. The nodD gene of Rhizobium leguminosarum is autoregulatory and in the presence of plant exudate induces the nodA, B, C genes. EMBO J 4:3369–3373. doi: 10.1002/j.1460-2075.1985.tb04092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu H, Liu S, Yang Y, Chang W, Hong G. 2000. In Rhizobium leguminosarum, NodD represses its own transcription by competing with RNA polymerase for binding sites. Nucleic Acids Res 28:2784–2793. doi: 10.1093/nar/28.14.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Rhijn P, Vanderleyden J. 1995. The Rhizobium-plant symbiosis. Microbiol Rev 59:124–142. doi: 10.1128/MMBR.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Downie JA, Knight CD, Johnston AWB, Rossen L. 1985. Identification of genes and gene products involved in the nodulation of peas by Rhizobium leguminosarum. Mol Gen Genet 198:255–262. doi: 10.1007/BF00383003. [DOI] [Google Scholar]
- 18.Innes RW, Kuempel PL, Plazinski J, Canter-Cremers H, Rolfe BG, Djordjevic MA. 1985. Plant factors induce expression of nodulation and host-range genes in Rhizobium trifolii. Mol Gen Genet 201:426–432. doi: 10.1007/BF00331334. [DOI] [Google Scholar]
- 19.del Cerro P, Rolla-Santos AAP, Gomes DF, Marks BB, del R, Espuny M, Rodríguez-Carvajal MÁ, Soria-Díaz ME, Nakatani AS, Hungria M, Ollero FJ. 2015. Opening the “black box” of nodD3, nodD4 and nodD5 genes of Rhizobium tropici strain CIAT 899. BMC Genomics 16:864. doi: 10.1186/s12864-015-2033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demont N, Ardourel M, Maillet F, Prome D, Ferro M, Prome JC, Dénarié J. 1994. The Rhizobium meliloti regulatory nodD3 and syrM genes control the synthesis of a particular class of nodulation factors N-acylated by (omega-1)-hydroxylated fatty acids. EMBO J 13:2139–2149. doi: 10.1002/j.1460-2075.1994.tb06490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honma MA, Ausubel FM. 1987. Rhizobium meliloti has three functional copies of the nodD symbiotic regulatory gene. Proc Natl Acad Sci U S A 84:8558–8562. doi: 10.1073/pnas.84.23.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodpothong P, Sullivan JT, Songsrirote K, Sumpton D, Cheung K-T, Thomas-Oates J, Radutoiu S, Stougaard J, Ronson CW. 2009. Nodulation gene mutants of Mesorhizobium loti R7A-nodZ and nolL mutants have host-specific phenotypes on Lotus spp. Mol Plant Microbe Interact 22:1546–1554. doi: 10.1094/MPMI-22-12-1546. [DOI] [PubMed] [Google Scholar]
- 23.Horvath B, Bachem CW, Schell J, Kondorosi A. 1987. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant-signal, interacting with the nodD gene product. EMBO J 6:841–848. doi: 10.1002/j.1460-2075.1987.tb04829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spaink HP, Wijffelman CA, Pees E, Okker RJH, Lugtenberg B. 1987. Rhizobium nodulation gene nodD as a determinant of host specificity. Nature 328:337–340. doi: 10.1038/328337a0. [DOI] [Google Scholar]
- 25.Honma MA, Asomaning M, Ausubel FM. 1990. Rhizobium meliloti nodD genes mediate host-specific activation of nodABC. J Bacteriol 172:901–911. doi: 10.1128/jb.172.2.901-911.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dénarié J, Debellé F, Rosenberg C. 1992. Signaling and host range variation in nodulation. Annu Rev Microbiol 46:497–531. doi: 10.1146/annurev.mi.46.100192.002433. [DOI] [PubMed] [Google Scholar]
- 27.Lamont E-J, Zoghlami A, Hamilton RS, Bennett SJ. 2001. Clovers (Trifolium L.), p 79–98. In Maxted N, Bennett SJ (ed), Plant genetic resources of legumes in the Mediterranean. Springer, Dordrecht, Netherlands. [Google Scholar]
- 28.Howieson JG, Yates RJ, O'Hara GW, Ryder M, Real D. 2005. The interactions of Rhizobium leguminosarum biovar trifolii in nodulation of annual and perennial Trifolium spp. from diverse centres of origin. Aust J Exp Agric 45:199–207. doi: 10.1071/EA03167. [DOI] [Google Scholar]
- 29.Triplett EW, Sadowsky MJ. 1992. Genetics of competition for nodulation of legumes. Annu Rev Microbiol 46:399–422. doi: 10.1146/annurev.mi.46.100192.002151. [DOI] [PubMed] [Google Scholar]
- 30.Shi S, Villamizar L, Gerard E, Ronson C, Wakelin S, Ballard R, Caradus JR, O'Callaghan M. 2019. Increasing biological nitrogen fixation by white clover-rhizobia symbiosis. J NZ Grasslands 81:231–234. doi: 10.33584/jnzg.2019.81.380. [DOI] [Google Scholar]
- 31.Redmond JW, Batley M, Djordjevic MA, Innes RW, Kuempel PL, Rolfe BG. 1986. Flavones induce expression of nodulation genes in Rhizobium. Nature 323:632–635. doi: 10.1038/323632a0. [DOI] [Google Scholar]
- 32.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly S, Sullivan JT, Kawaharada Y, Radutoiu S, Ronson CW, Stougaard J. 2018. Regulation of Nod factor biosynthesis by alternative NodD proteins at distinct stages of symbiosis provides additional compatibility scrutiny. Environ Microbiol 20:97–110. doi: 10.1111/1462-2920.14006. [DOI] [PubMed] [Google Scholar]
- 34.Kostiuk NV, Belyakova MB, Leshchenko DV, Miniaev MV, Petrova MB, Kharitonova EA. 2013. Structural characterization of the NodD transcription factor. Am J Bioinf Res 3:35–41. doi: 10.5923/j.bioinformatics.20130303.01. [DOI] [Google Scholar]
- 35.Vandecraen J, Chandler M, Aertsen A, Van Houdt R. 2017. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit Rev Microbiol 43:709–730. doi: 10.1080/1040841X.2017.1303661. [DOI] [PubMed] [Google Scholar]
- 36.Lee I, Kim YO, Park S-C, Chun J. 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 37.Kumar N, Lad G, Giuntini E, Kaye ME, Udomwong P, Shamsani NJ, Young JPW, Bailly X. 2015. Bacterial genospecies that are not ecologically coherent: population genomics of Rhizobium leguminosarum. Open Biol 5:140133. doi: 10.1098/rsob.140133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mauchline TH, Hayat R, Roberts R, Powers SJ, Hirsch PR. 2014. Assessment of core and accessory genetic variation in Rhizobium leguminosarum symbiovar trifolii strains from diverse locations and host plants using PCR‐based methods. Lett Appl Microbiol 59:238–246. doi: 10.1111/lam.12270. [DOI] [PubMed] [Google Scholar]
- 39.Terpolilli J, Rui T, Yates R, Howieson J, Poole P, Munk C, Tapia R, Han C, Markowitz V, Tatiparthi R, Mavrommatis K, Ivanova N, Pati A, Goodwin L, Woyke T, Kyrpides N, Reeve W. 2014. Genome sequence of Rhizobium leguminosarum bv trifolii strain WSM1689, the microsymbiont of the one flowered clover Trifolium uniflorum. Stand Genomic Sci 9:527–539. doi: 10.4056/sigs.4988693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Göttfert M. 1993. Regulation and function of rhizobial nodulation genes. FEMS Microbiol Lett 104:39–63. doi: 10.1111/j.1574-6968.1993.tb05863.x. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez-Quinones F, Banfalvi Z, Murphy P, Kondorosi A. 1987. Interspecies homology of nodulation genes in Rhizobium. Plant Mol Biol 8:61–75. doi: 10.1007/BF00016435. [DOI] [PubMed] [Google Scholar]
- 42.Györgypal Z, Kiss GB, Kondorosi A. 1991. Transduction of plant signal molecules by the Rhizobium NodD proteins. Bioessays 13:575–581. doi: 10.1002/bies.950131106. [DOI] [PubMed] [Google Scholar]
- 43.Göttfert M, Horvath B, Kondorosi E, Putnoky P, Rodriguez-Quiñones F, Kondorosi A. 1986. At least two nodD genes are necessary for efficient nodulation of alfalfa by Rhizobium meliloti. J Mol Biol 191:411–420. doi: 10.1016/0022-2836(86)90136-1. [DOI] [PubMed] [Google Scholar]
- 44.Van Rhijn P, Feys B, Verreth C, Vanderleyden J. 1993. Multiple copies of nodD in Rhizobium tropici CIAT899 and BR816. J Bacteriol 175:438–447. doi: 10.1128/jb.175.2.438-447.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Djordjevic MA, Schofield PR, Rolfe BG. 1985. Tn5 mutagenesis of Rhizobium trifolii host-specific nodulation genes result in mutants with altered host-range ability. Mol Gen Genet 200:463–471. doi: 10.1007/BF00425732. [DOI] [Google Scholar]
- 46.Sullivan JT, Trzebiatowski JR, Cruickshank RW, Gouzy J, Brown SD, Elliot RM, Fleetwood DJ, McCallum NG, Rossbach U, Stuart GS, Weaver JE, Webby RJ, De Bruijn FJ, Ronson CW. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol 184:3086–3095. doi: 10.1128/jb.184.11.3086-3095.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castillo M, Flores M, Mavingui P, Martínez-Romero E, Palacios R, Hernández G. 1999. Increase in alfalfa nodulation, nitrogen fixation, and plant growth by specific DNA amplification in Sinorhizobium meliloti. Appl Environ Microbiol 65:2716–2722. doi: 10.1128/AEM.65.6.2716-2722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mulligan JT, Long SR. 1985. Induction of Rhizobium meliloti nodC expression by plant exudate requires nodD. Proc Natl Acad Sci U S A 82:6609–6613. doi: 10.1073/pnas.82.19.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McIver J, Djordjevic MA, Weinman JJ, Bender GL, Rolfe BG. 1989. Extension of host range of Rhizobium leguminosarum bv. trifolii caused by point mutations in nodD that result in alterations in regulatory function and recognition of inducer molecules. Mol Plant Microbe Interact 2:97–106. doi: 10.1094/mpmi-2-097. [DOI] [PubMed] [Google Scholar]
- 50.Mathesius U, Bayliss C, Weinman JJ, Schlaman HR, Spaink HP, Rolfe BG, McCully ME, Djordjevic MA. 1998. Flavonoids synthesized in cortical cells during nodule initiation are early developmental markers in white clover. Mol Plant Microbe Interact 11:1223–1232. doi: 10.1094/MPMI.1998.11.12.1223. [DOI] [Google Scholar]
- 51.Feng J, Li Q, Hu HL, Chen XC, Hong GF. 2003. Inactivation of the nod box distal half‐site allows tetrameric NodD to activate nodA transcription in an inducer‐independent manner. Nucleic Acids Res 31:3143–3156. doi: 10.1093/nar/gkg411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dombrecht B, Vanderleyden J, Michiels J. 2001. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in Gram-negative bacteria. Mol Plant Microbe Interact 14:426–430. doi: 10.1094/MPMI.2001.14.3.426. [DOI] [PubMed] [Google Scholar]
- 53.Miller WG, Leveau JHJ, Lindow SE. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol Plant Microbe Interact 13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- 54.Beringer JE. 1974. R Factor Transfer in Rhizobium leguminosarum. J Gen Microbiol 84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 55.Ronson CW, Nixon BT, Albright LM, Ausubel FM. 1987. Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functions. J Bacteriol 169:2424–2431. doi: 10.1128/jb.169.6.2424-2431.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hubber A, Vergunst AC, Sullivan JT, Hooykaas PJ, Ronson CW. 2004. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol Microbiol 54:561–574. doi: 10.1111/j.1365-2958.2004.04292.x. [DOI] [PubMed] [Google Scholar]
- 57.Sullivan JT, Patrick HN, Lowther WL, Scott DB, Ronson CW. 1995. Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc Natl Acad Sci U S A 92:8985–8989. doi: 10.1073/pnas.92.19.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 59.Ramsay JP, Tester LGL, Major AS, Sullivan JT, Edgar CD, Kleffmann T, Patterson-House JR, Hall DA, Tate WP, Hynes MF, Ronson CW. 2015. Ribosomal frameshifting and dual-target antiactivation restrict quorum-sensing–activated transfer of a mobile genetic element. Proc Natl Acad Sci U S A 112:4104–4109. doi: 10.1073/pnas.1501574112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 61.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 62.Kelly SJ, Muszyński A, Kawaharada Y, Hubber AM, Sullivan JT, Sandal N, Carlson RW, Stougaard J, Ronson CW. 2013. Conditional requirement for exopolysaccharide in the Mesorhizobium–Lotus symbiosis. Mol Plant Microbe Interact 26:319–329. doi: 10.1094/MPMI-09-12-0227-R. [DOI] [PubMed] [Google Scholar]
- 63.Vincent J. 1970. A manual for the practical study of root nodule bacteria. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 64.Miller SH, Elliot RM, Sullivan JT, Ronson CW. 2007. Host-specific regulation of symbiotic nitrogen fixation in Rhizobium leguminosarum biovar trifolii. Microbiology 153:3184–3195. doi: 10.1099/mic.0.2007/006924-0. [DOI] [PubMed] [Google Scholar]
- 65.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 66.Sánchez-Cañizares C, Palacios J. 2013. Construction of a marker system for the evaluation of competitiveness for legume nodulation in Rhizobium strains. J Microbiol Methods 92:246–249. doi: 10.1016/j.mimet.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 67.Sessitsch A, Wilson KJ, Akkermans A, De Vos W. 1996. Simultaneous detection of different Rhizobium strains marked with either the Escherichia coli gusA gene or the Pyrococcus furiosus celB gene. Appl Environ Microbiol 62:4191–4194. doi: 10.1128/AEM.62.11.4191-4194.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dixon R, Kahn D. 2004. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- 69.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 71.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 72.RStudio Team. 2015. RStudio: integrated development for R. RStudio, Inc, Boston, MA. [Google Scholar]
- 73.Galili T, O'Callaghan A, Sidi J, Sievert C. 2018. heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 34:1600–1602. doi: 10.1093/bioinformatics/btx657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 76.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 77.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 78.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reeve W, O'Hara G, Chain P, Ardley J, Bräu L, Nandesena K, Tiwari R, Copeland A, Nolan M, Han C, Brettin T, Land M, Ovchinikova G, Ivanova N, Mavromatis K, Markowitz V, Kyrpides N, Melino V, Denton M, Yates R, Howieson J. 2010. Complete genome sequence of Rhizobium leguminosarum bv. trifolii strain WSM1325, an effective microsymbiont of annual Mediterranean clovers. Stand Genomic Sci 2:347–356. doi: 10.4056/sigs.852027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reeve W, Drew E, Ballard R, Melino V, Tian R, De Meyer S, Brau L, Ninawi M, Teshima H, Goodwin L, Chain P, Liolios K, Pati A, Mavromatis K, Ivanova N, Markowitz V, Woyke T, Kyrpides N. 2013. Genome sequence of the clover-nodulating Rhizobium leguminosarum bv. trifolii strain SRDI565. Stand Genomic Sci 9:220–231. doi: 10.4056/sigs.4468250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reeve W, Drew E, Ballard R, Melino V, Tian R, De Meyer S, Brau L, Ninawi M, Daligault H, Davenport K, Erkkila T, Goodwin L, Gu W, Munk C, Teshima H, Xu Y, Chain P, Kyrpides N. 2013. Genome sequence of the clover-nodulating Rhizobium leguminosarum bv. trifolii strain SRDI943. Stand Genomic Sci 9:232–242. doi: 10.4056/sigs.4478252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roughley RJ, Date RA. 1986. The effect of strain of Rhizobium and of temperature on nodulation and early growth of Trifolium semipilosum. Ex Agric 22:123–131. doi: 10.1017/S0014479700014204. [DOI] [Google Scholar]
- 83.Reeve W, Tian R, De Meyer S, Melino V, Terpolilli J, Ardley J, Tiwari R, Howieson J, Yates R, O'Hara G, Ninawi M, Teshima H, Bruce D, Detter C, Tapia R, Han C, Wei C-L, Huntemann M, Han J, Chen I-M, Mavromatis K, Markowitz V, Ivanova N, Ovchinnikova G, Pagani I, Pati A, Goodwin L, Pitluck S, Woyke T, Kyrpides N. 2013. Genome sequence of the clover-nodulating Rhizobium leguminosarum bv. trifolii strain TA1. Stand Genomic Sci 9:243–253. doi: 10.4056/sigs.4488254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Delestre C, Laugraud A, Ridgway H, Ronson C, O’Callaghan M, Barrett B, Ballard R, Griffiths A, Young S, Blond C. 2015. Genome sequence of the clover symbiont Rhizobium leguminosarum bv. trifolii strain CC275e. Stand Genomic Sci 10:121. doi: 10.1186/s40793-015-0110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reeve W, Melino V, Ardley J, Tian R, De Meyer S, Terpolilli J, Tiwari R, Yates R, O'Hara G, Howieson J, Ninawi M, Held B, Bruce D, Detter C, Tapia R, Han C, Wei C-L, Huntemann M, Han J, Chen I-M, Mavromatis K, Markowitz V, Szeto E, Ivanova N, Mikhailova N, Pagani I, Pati A, Goodwin L, Woyke T, Kyrpides N. 2013. Genome sequence of the Trifolium rueppellianum-nodulating Rhizobium leguminosarum bv. trifolii strain WSM2012. Stand Genomic Sci 9:283–293. doi: 10.4056/sigs.4528262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reeve W, O'Hara G, Chain P, Ardley J, Bräu L, Nandesena K, Tiwari R, Malfatti S, Kiss H, Lapidus A, Copeland A, Nolan M, Land M, Ivanova N, Mavromatis K, Markowitz V, Kyrpides N, Melino V, Denton M, Yates R, Howieson J. 2010. Complete genome sequence of Rhizobium leguminosarum bv trifolii strain WSM2304, an effective microsymbiont of the South American clover Trifolium polymorphum. Stand Genomic Sci 2:66–76. doi: 10.4056/sigs.44642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reeve W, Terpolilli J, Melino V, Ardley J, Tian R, De Meyer S, Tiwari R, Yates R, O'Hara G, Howieson J, Ninawi M, Held B, Bruce D, Detter C, Tapia R, Han C, Wei C-L, Huntemann M, Han J, Chen I-M, Mavromatis K, Markowitz V, Ivanova N, Ovchinnikova G, Pagani I, Pati A, Goodwin L, Woyke T, Kyrpides N. 2013. Genome sequence of the South American clover-nodulating Rhizobium leguminosarum bv. trifolii strain WSM597. Stand Genomic Sci 9:264–272. doi: 10.4056/sigs.4508258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bullard GK, Roughley RJ, Pulsford DJ. 2005. The legume inoculant industry and inoculant quality control in Australia: 1953–2003. Aust J Exp Agric 45:127–140. doi: 10.1071/EA03159. [DOI] [Google Scholar]
- 89.Yates RJ, Howieson JG, Real D, Reeve WG, Vivas-Marfisi A, O'Hara GW. 2005. Evidence of selection for effective nodulation in the Trifolium spp. symbiosis with Rhizobium leguminosarum biovar trifolii. Aust J Exp Agric 45:189–198. doi: 10.1071/EA03168. [DOI] [Google Scholar]
- 90.Thoma S, Schobert M. 2009. An improved Escherichia coli donor strain for diparental mating. FEMS Microbiol Lett 294:127–132. doi: 10.1111/j.1574-6968.2009.01556.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.