Fe–S clusters function as cofactors of proteins controlling diverse biological processes, such as respiration, photosynthesis, nitrogen fixation, DNA replication, and gene regulation. The mechanism of how Actinobacteria regulate the expression of the sole Fe-S cluster assembly system in response to the various Fe–S cluster demands remains to be elucidated. In this study, we showed that SufR functions as a transcriptional repressor of the sole Fe-S cluster assembly system in the avermectin producer S. avermitilis. [4Fe-4S]-SufR binds to the promoter region of the suf operon and represses its expression. When Fe-S cluster levels are insufficient, SufR loses its [4Fe-4S] cluster and DNA-binding activity. Apo-SufR dissociates from the promoter region of suf operon, and the expression of the suf system is strongly increased by derepression to promote the synthesis of Fe-S clusters. The study clarifies how Streptomyces maintains its Fe-S cluster homeostasis through the activity of SufR to modulate the various Fe-S cluster demands.
KEYWORDS: Fe-S cluster homeostasis, SufR, [4Fe-4S] cluster, suf operon, S. avermitilis
ABSTRACT
Iron-sulfur (Fe-S) clusters are ubiquitous and versatile inorganic cofactors that are crucial for many fundamental bioprocesses in nearly all organisms. How cells maintain Fe-S cluster homeostasis is not well understood in Gram-positive bacteria. Genomic analysis showed that the Suf system, which is encoded by the sufRBDCSU operon, is the sole Fe-S cluster assembly system in the genus Streptomyces. Streptomyces avermitilis is the industrial producer of avermectins, which are widely used as agricultural pesticides and antiparasitic agents. sufR (SAV6324) encodes a putative ArsR-family transcriptional regulator, which was characterized as a repressor of the sufRBDCSU operon in this investigation. Spectroscopy and mass spectrometry demonstrated that anaerobically isolated SufR contained an oxidation-sensitive [4Fe-4S] cluster and existed as a homodimer. Electrophoretic mobility shift assays (EMSAs) and DNase I footprinting analyses revealed that [4Fe-4S]-SufR bound specifically and tightly to a 14-bp palindromic sequence (CAAC-N6-GTTG) in the promoter region of the sufR operon, repressing expression of the sufRBDCSU operon. The presence of the [4Fe-4S] cluster is critical for the DNA-binding activity of SufR. Cys182, Cys195, and Cys223 in the C-terminal region of SufR are essential for [4Fe-4S] cluster coordination, but Cys178 is not. The fourth non-Cys ligand in coordination of the [4Fe-4S] cluster for SufR remains to be identified. The findings clarify the transcriptional control of the suf operon by [4Fe-4S] SufR to satisfy the various Fe-S cluster demands. SufR senses the intracellular Fe-S cluster status and modulates the expression of the sole Fe-S cluster assembly system via its Fe-S cluster occupancy.
IMPORTANCE Fe–S clusters function as cofactors of proteins controlling diverse biological processes, such as respiration, photosynthesis, nitrogen fixation, DNA replication, and gene regulation. The mechanism of how Actinobacteria regulate the expression of the sole Fe-S cluster assembly system in response to the various Fe–S cluster demands remains to be elucidated. In this study, we showed that SufR functions as a transcriptional repressor of the sole Fe-S cluster assembly system in the avermectin producer S. avermitilis. [4Fe-4S]-SufR binds to the promoter region of the suf operon and represses its expression. When Fe-S cluster levels are insufficient, SufR loses its [4Fe-4S] cluster and DNA-binding activity. Apo-SufR dissociates from the promoter region of suf operon, and the expression of the suf system is strongly increased by derepression to promote the synthesis of Fe-S clusters. The study clarifies how Streptomyces maintains its Fe-S cluster homeostasis through the activity of SufR to modulate the various Fe-S cluster demands.
INTRODUCTION
Fe-S clusters are among the most ubiquitous and versatile cofactors of proteins in nearly all organisms. Because of their remarkable structural plasticity and intrinsic chemical/electronic properties, Fe-S clusters participate in diverse biological processes such as respiration, photosynthesis, nitrogen fixation, DNA repair, and gene regulation (1–3). The most common types of Fe-S clusters, [4Fe-4S] and [2Fe-2S], are attached to proteins primarily via cysteine ligands (4).
The formation of Fe-S clusters requires a complex biosynthetic machinery to assemble stored iron and sulfur, and the resulting clusters are transferred to the appropriate apo-protein substrates (4). Three systems for the Fe-S cluster assembly have been identified: the Isc (iron-sulfur cluster) system, the Suf (sulfur mobilization) system, and the Nif (nitrogen fixation) system (2, 4). However, the phylogenetic distribution of these systems varies substantially among species of bacteria. The Nif system was originally found in the nitrogen-fixing bacteria, in which it is specifically used for the maturation of nitrogenases (5). The Isc system is considered to be the housekeeping Fe-S cluster biogenesis pathway that mainly functions under normal growth conditions in bacteria such as Escherichia coli (6, 7). The Suf system occurs in a wide range of bacteria and archaea (8). In E. coli, the Suf system is a back-up system for Fe-S cluster assembly and is specifically activated under stress conditions, such as iron deficiency or oxidative stress (9–11). In cyanobacteria, archaea, and many Gram-positive bacteria, the Suf system seems to be the sole or the main system for Fe-S cluster assembly (12–14).
Solvent-exposed Fe-S clusters are prone to be damaged by O2, reactive oxygen species (ROS) (15, 16), and nitric oxide (NO) (17). To cope with the various demands of Fe-S cluster assembly and to maintain Fe-S cluster homeostasis, bacteria have evolved regulators that precisely control the activity of Fe-S cluster assembly systems. The [2Fe-2S] cluster-containing transcription factor IscR (Isc pathway regulator), which was first identified in the Gram-negative bacterium E. coli, is a central mediator in sensing the intracellular Fe–S status and in regulating the expression of the Isc and Suf systems (6, 7). When the intracellular Fe-S clusters are sufficient, IscR mainly exists in holo-form. [2Fe-2S]-IscR binds to the promoter region of the iscRSUA-hscBA-fdx operon (isc operon) and represses its expression (6, 18). When the cells are experiencing oxidative stress or iron limitation and the requirements for Fe-S clusters are not satisfied, IscR is mostly in apo-form. Apo-IscR loses binding activity to the promoter region of the isc operon, and the expression of isc operon is increased by derepression (6, 9). Apo-IscR can also bind to the promoter region of the sufABCDSE operon (suf operon) and activate the expression of the Suf system to maximize Fe-S assembly capacity (9, 19, 20). IscR senses the intracellular Fe–S status via its [2Fe–2S] cluster occupancy and tunes the synthesis of the Isc and Suf systems in response to the various demands for Fe–S cluster biogenesis.
Like E. coli, cyanobacteria have both Isc and Suf systems for Fe-S cluster assembly. In cyanobacteria, the Suf system rather than the Isc system is responsible for the biogenesis and maintenance of the Fe-S clusters (21). The sufBCDS genes of the Suf system in cyanobacteria are organized in an operon, which is transcribed divergently from a bidirectional promoter region with the sufR gene, while sufA and sufE are located elsewhere. The [4Fe-4S] cluster-containing SufRcy binds to the two DNA sequences in the bidirectional promoter region of sufR-sufBCDS and represses the expression of the suf operon and sufR (13, 21). In many Gram-positive bacteria such as Bacillus subtilis, mycobacteria, and Streptomyces, the Suf system is solely responsible for [Fe-S] cluster assembly and repair (12, 22). Recently, Mycobacterium tuberculosis Rv1460, a SufR homolog, was reported to repress the expression of the suf operon in Mycobacterium (23, 24).
Streptomyces spp. are Gram-positive, filamentous soil bacteria that exhibit complex morphological differentiation and that produce more than half of all known antibiotics, as well as anticancer, anthelmintic, and immunosuppressive agents (25, 26). How Streptomyces regulate the expression of the sole Fe-S cluster assembly system in response to the various Fe–S cluster demands remains to be elucidated. S. avermitilis is used for the industrial production of avermectins, a series of 16-membered macrocyclic anthelmintic agents that are widely applied in medicine, agriculture, and animal husbandry (27, 28). Like the Suf system in Bacillus subtilis and mycobacteria, the Suf system is the sole system for Fe-S cluster biogenesis in S. avermitilis. In this study, we report on the regulatory role of SufR in Fe-S cluster biogenesis in S. avermitilis.
RESULTS
The suf operon in S. avermitilis.
Although there are three distinct Fe-S assembly systems in bacteria (4), no equivalent of the nif or isc operon has been found in the S. avermitilis genome. A BLAST search identified a gene locus (SAV6324 to SAV6331) with sequence similarity to the suf operon in bacteria (Fig. 1A, Fig. S1A in the supplemental material). SAV6325 to SAV6330 are predicted to encode SufB, SufD, a putative ferredoxin subunit of phenylpropionate dioxygenase (Hcac1), SufC, SufS, and SufU (a NifU-like protein), respectively. The last gene of the locus (SAV6331) has no homology with any of the suf genes. Neither sufA nor sufE homolog genes were found in the S. avermitilis genome. It seems that the sufB, sufC, sufD, and sufS genes are highly conserved in suf operons. SufS is a cysteine desulfurase that mobilizes sulfur from free l-cysteine for Fe-S cluster biogenesis (29). SufC has ATPase activity and forms a tight complex with SufB and SufD. The SufBCD complex acts as a scaffold for the assembly and transfer of transient clusters to target proteins (30). S. avermitilis SufB, SufD, SufC, and SufS share 82%, 58%, 77%, and 52% identity with the M. tuberculosis orthologs, and 41%, 22%, 51%, and 26% identity with the E. coli orthologs, respectively. The suf operon is exceptionally conserved in all Streptomyces genomes sequenced to date, suggesting that Streptomyces species possess a functional Suf system as the sole system for Fe-S cluster assembly.
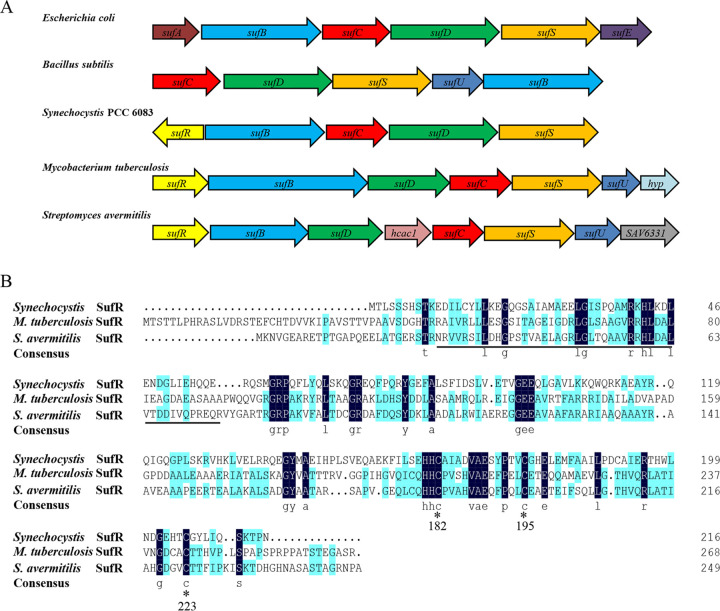
FIG 1.
Identification of a putative Fe-S cluster assembly regulator in S. avermitilis. (A) Gene organization of the suf operons from different bacterial species (Escherichia coli, Bacillus subtilis, Synechocystis PCC 6083, Mycobacterium tuberculosis, and Streptomyces avermitilis). The genes with conserved functions are indicated with the same color. (B) Multiple sequence alignment of SufR homologs. Strictly conserved amino acids are highlighted in navy blue; underline, the HTH DNA-binding domain; *, conserved cysteine.
SAV6324 (sufR), the first gene of the locus, encodes a putative ArsR-family transcriptional regulator protein. The predicted product shares 47% and 27% identity with SufR from M. tuberculosis and Synechocystis sp. PCC6803, respectively. S. avermitilis SufR contains an ArsR-family helix-turn-helix (HTH) DNA-binding domain at the N terminus and three highly conserved cysteine residues that may function in the Fe-S cluster binding motif at the C terminus (Fig. 1B). In contrast to the sufRcy gene in cyanobacteria, the sufR gene is oriented in the same direction as other suf genes in S. avermitilis, as well as in M. tuberculosis (Fig. 1A). S. avermitilis suf genes and adjacent genes are oriented in the same direction with very short intergenic regions or intein invading sequences. Reverse transcriptase PCR (RT-PCR) analysis demonstrated that these genes (from sufR to SAV6331) are organized in the same operon and are cotranscribed into a long mRNA molecule (Fig. S1). sufR and the adjacent suf genes are widely distributed among Streptomyces species, suggesting a conserved regulatory role of SufR in Fe-S cluster assembly in Streptomyces.
SufR harbors the [4Fe-4S] cluster.
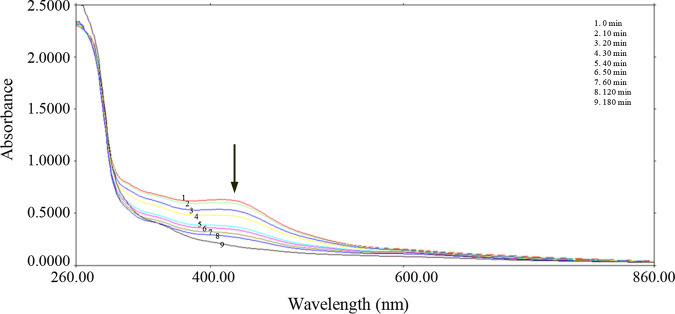
Fe-S cluster assembly regulators usually harbor Fe-S clusters in order to sense the intracellular Fe-S cluster status and to modulate their binding activity (6, 13). To investigate whether S. avermitilis SufR contains a Fe-S cluster, which is usually oxygen-sensitive, we purified the SufR protein from E. coli under strictly anaerobic conditions (O2 <8 ppm). The anaerobically purified SufR-His6 fraction was dark brown and was much darker than the aerobically purified one, suggesting the presence of Fe-S clusters (Fig. S2). The UV-visible absorbance spectrum of the anaerobically purified soluble SufR protein had a single broad absorption peak around 412 nm and no other features at longer wavelengths (Fig. 2), indicating that it is more characteristic of a [4Fe-4S] cluster rather than a [2Fe-2S] cluster (31, 32). Although the anaerobically purified [4Fe-4S]-SufR was stable and remained brown for days under anaerobic conditions, it gradually lost its color and its A412 value decreased rapidly when it was exposed to air. The A412 value decreased by approximately 35% and 75% with 30 and 60 min, respectively, of air exposure. The 412 nm peak disappeared completely after 3 h of air exposure, indicating that the [4Fe-4S] cluster is very sensitive to oxygen (Fig. 2). No characteristic [2Fe-2S] cluster absorbance in the visible region was observed with the gradual loss of [4Fe-4S] cluster signal intensity, indicating a gradual transition from holo- to apo-SufR, instead of a [4Fe-4S] to [2Fe-2S] cluster conversion.
FIG 2.
Oxygen-induced degradation of the Fe-S cluster in SufR. The SufR sample was purified under strictly anaerobic conditions (O2 <8 ppm). The results show the absorption spectra of the anaerobically purified protein SufR after exposure to air for 0 to 180 min.
Matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry was used to determine the mass of the Fe-S cluster-bound SufR. The predicted mass of C-terminal His-tagged SufR (apo-SufR) is 28.7 kDa. Two main peaks were observed in the mass spectrum; one was at approximately 56 kDa, corresponding to the dimeric form of SufR-His6, and the other was at about 28 kDa, corresponding to the monomeric protein (Fig. S3). The presence of the monomeric form was possibly due to destruction of the dimer by the ionization and vaporization processes. The findings demonstrate that SufR exists as a homodimer that contains a [4Fe-4S] cluster under anaerobic conditions.
SufR directly represses the expression of the suf operon.
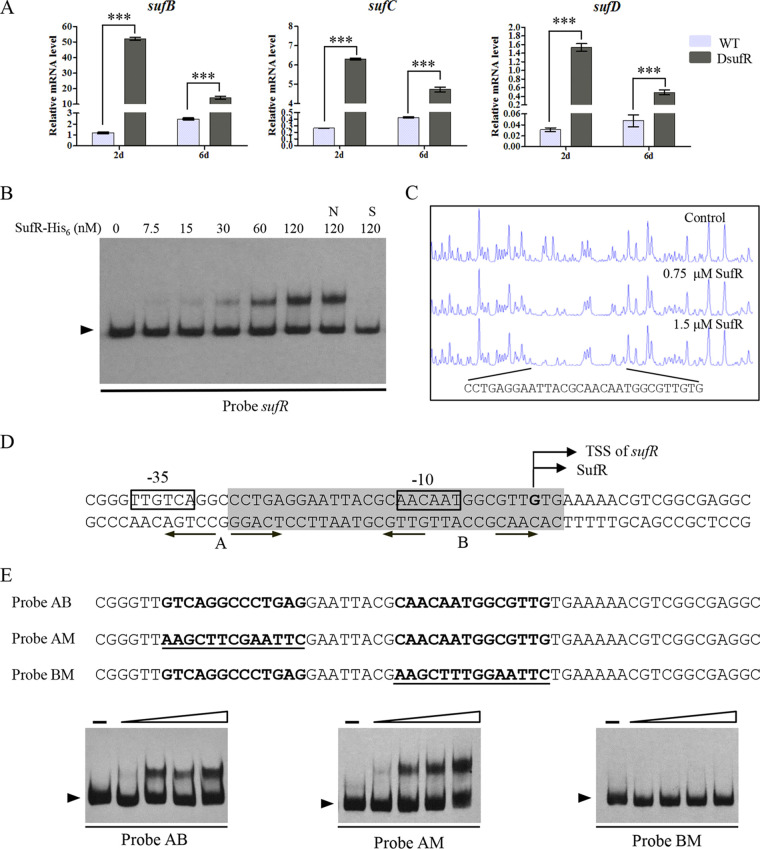
Some members of ArsR-family of transcription factors are transcriptional repressors for heavy metal ion detoxification (33). In cyanobacteria, SufR represses the expression of itself and the adjacent suf operon (13). To elucidate the function of SufR in S. avermitilis, we constructed a sufR deletion mutant (DsufR) by homologous recombination in S. avermitilis ATCC 31267. RT-qPCR analysis indicated that the transcriptional levels of sufB, sufC, and sufD were remarkably higher in DsufR than in the wild type (WT) after 2 and 6 days in soya flour-mannitol (SFM) culture, indicating that SufR represses the expression of the suf operon (Fig. 3A).
FIG 3.
SufR represses expression of the suf operon. (A) RT-qPCR analysis of the transcriptional levels of suf genes in the WT and DsufR grown on SFM for 2 or 6 days. Error bars: standard deviation of three technical replicates. ***, P < 0.001 (Student’s t test). (B) Binding of SufR-His6 to its own promoter region by EMSA. Labeled probe (0.15 nM) and various amounts of SufR-His6 were added to each reaction mixture. The 300-fold nonspecific competitor DNA (lane N) and unlabeled specific probe (lane S) were added to confirm the specificity of band shifts. Arrow: free probe. (C) DNase I footprinting assay of SufR on its own promoter region. Upper fluorogram, control reaction without protein. Protection fluorograms were acquired with increasing amounts of SufR-His6 protein. (D) Nucleotide sequences of the sufR promoter region. Shaded box, the region protected by SufR; straight arrows, inverted repeat sequences; bent arrows, sufR transcriptional and translational start sites; boxes, putative −35 and −10 regions. (E) EMSAs of probe AB and the mutated probes AM and BM to identify the SufR-binding site. Mutations were introduced into the inverted repeat sequences A and B of probe AB to generate mutated probes AM and BM. Underlining, altered nucleotides.
To determine whether SufR directly regulates the suf operon, we performed in vitro electrophoretic mobility shift assays (EMSAs) using a soluble SufR-His6 protein anaerobically purified from E. coli and the digoxigenin (DIG)-labeled probe of the sufR promoter region (probe sufR). SufR-His6 bound to the probe sufR and produced a clearly shifted band (Fig. 3B). Binding specificity was evaluated by competitive assays with excess (300-fold) unlabeled specific probe sufR, which abolished the binding of SufR-His6 to the labeled probe sufR (lane S), or with excess unlabeled nonspecific probe, which did not attenuate the retarded signal (lane N). These results indicate that SufR binds specifically to the promoter region of the suf operon and directly regulates its expression.
SufR binds to a 14-nt palindromic sequence in the sufR promoter region.
To elucidate how SufR regulates the suf operon, we determined the binding sequence of SufR in the 5′-end fluorescein-labeled promoter region of the suf operon by DNase I footprinting analysis. One 30-nt region (5′-CCTGAGGAATTACGCAACAATGGCGTTGTG-3′) in the sufR promoter region was protected by 0.75 and 1.5 μM SufR-His6 (Fig. 3C). The transcriptional start site (TSS) of sufR was determined by 5′ rapid amplification of cDNA ends (5′-RACE). The TSS was located at the G residue, which is the same position of the translational start codon of SufR (Fig. S4). The SufR-binding sequence is located between the −35 and −10 regions of the sufR promoter and contains −10 regions (Fig. 3D). Analysis of the protected region revealed two palindromic sequences that were possibly the SufR binding site: TCAGGNCCTGA (site A) and CAAC-N6-GTTG (site B). To determine which one is the recognition sequence of SufR, we introduced site-directed mutation to site A or site B of probe AB (containing both palindromic sequences) to produce probe AM or probe BM (Fig. 3E). EMSAs showed that SufR-His6 could bind equally well to probe AM, which had a mutated site A, and probe AB but could not bind to probe BM, which had a mutated site B (Fig. 3E), indicating that the palindromic sequence B (CAAC-N6-GTTG) is the SufR-binding site. The palindromic sequence is the same as the identified binding sequence of SufRcy (CAAC-N6-GTTG) in Synechocystis (13), indicating the similar regulatory role and regulatory mechanism of SufR in these bacteria. Based on these results, we conclude that SufR may directly block the transcription of the suf operon by binding to the −10 region of the sufR promoter to prevent the attachment of RNA polymerase to the promoter.
To determine whether SufR regulates genes involved in other physiological processes, we used the palindromic sequence and Virtual Footprint software to scan the S. avermitilis genome (Table S1) and verified the putative target genes with high score and annotation by EMSA using [4Fe-4S]-SufR proteins (Fig. S5). We failed to find any other genes that are regulated by SufR. Thus, SufR likely functions specifically as a transcriptional repressor of the sole Fe-S cluster assembly system in Streptomyces. The CAAC-N6-GTTG palindromic sequence is present in the promoter regions of suf operons among Streptomyces species (Fig. S6), indicating that the regulatory mechanism of SufR in Fe-S cluster assembly is conserved in Streptomyces.
[4Fe-4S] cluster is critical for the DNA-binding activity of SufR.
The DNA-binding activity of iron-sulfur cluster transcriptional regulators usually depends on the state of Fe-S clusters (34, 35). To determine whether the presence of the [4Fe-4S] cluster is required for the DNA-binding activity of SufR, we performed EMSAs using different forms of SufR with DIG-labeled sufR probe. Fresh anaerobically purified SufR resulted in a clear shifted band at a very low concentration (60 nM). When the anaerobically purified SufR was exposed to air for 1.5 h, which partially destroyed the [4Fe-4S], the binding activity decreased significantly (Fig. 4). Apo-SufR was prepared from anaerobically purified SufR using EDTA and potassium ferricyanide. Apo-SufR could not bind to the sufR probe, and no shifted band was observed even with a much larger amount of protein (480 nM). To further illustrate that the [4Fe-4S] cluster is required for DNA-binding activity, apo-SufR was reconstituted to [4Fe-4S]-SufR as described in the Materials and Methods. The ability to bind to the sufR probe was restored in Re-SufR (Fig. 4). These findings indicate that the DNA-binding activity of SufR depends on the presence of the [4Fe-4S] cluster.
FIG 4.
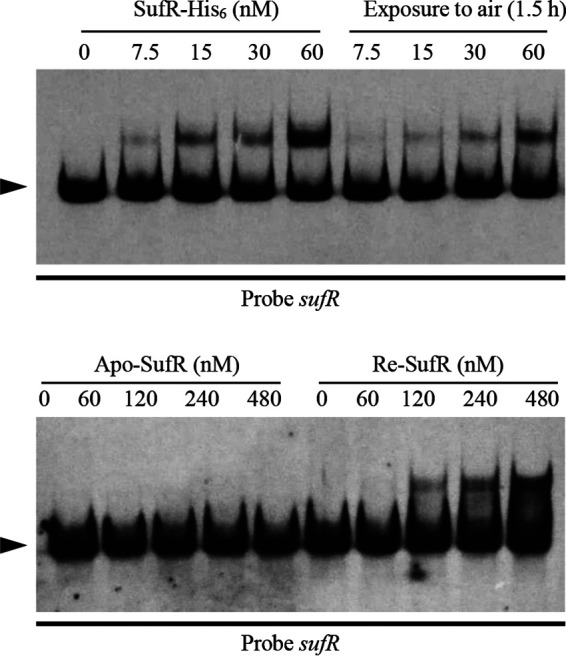
The [4Fe-4S] cluster is essential for the DNA-binding activity of SufR. EMSA analysis of the binding to the sufR promoter region using anaerobically purified SufR, air-exposed SufR (1.5 h), apo-SufR, and reconstituted SufR (Re-SufR). EMSA conditions were as described in Fig. 2. Arrow, free probe.
Cys182, Cys195, and Cys223 provide ligands to the [4Fe-4S] cluster.
The integration of Fe–S clusters into Fe-S proteins is usually coordinated by cysteine residues, and in some cases by histidine or other residues, on the proteins (35, 36). Alignment of S. avermitilis SufR with its homologs from other bacteria revealed that its C-terminal domain has three conserved cysteine residues at positions 182, 195, and 223, which are likely candidates for the coordination of the [4Fe-4S] cluster (Fig. 1B). There is a fourth cysteine residue (Cys178) in the C terminus that is absent in SufRcy, suggesting that the residue may not be essential. To investigate the role of the cysteine residues (Cys178, Cys182, Cys195, and Cys223) in SufR, the Cys residues were replaced by a Ser residue to produce variants as described in the Materials and Methods.
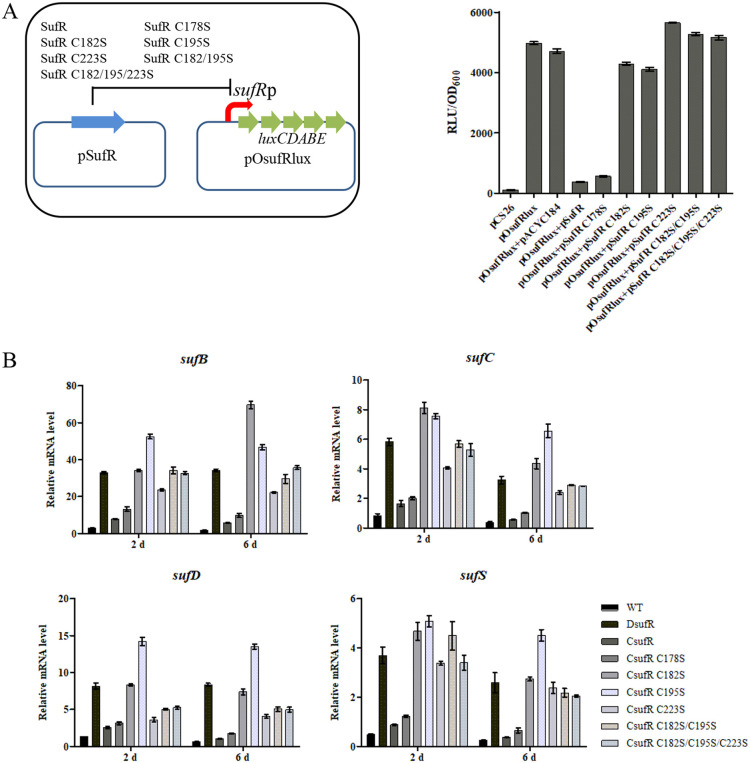
A heterologous Lux-reporter system in E. coli (37) was used to identify the effects of the cysteine residues on the DNA-binding activity of SufR. The reporter system exploited two plasmids: pSufR (based on pACYC184) was used for expressing SufR and its variants, and pOsufRlux (based on pCS26-Pac carrying the promoterless lux reporter genes) was used for expressing the lux operon under the control of the sufRp promoter (Fig. 5A). The E. coli transformant containing pOsufRlux conferred a high level of bioluminescence, while E. coli containing the control plasmid pCS26-Pac had only a low background level (Fig. 5A), indicating that the sufR promoter is able to be recognized by E. coli RNA polymerase. In comparison with the control plasmid pACYC184, the introduction of the expression plasmid pSufR dramatically reduced the bioluminescence of the transformant harboring pOsufRlux (Fig. 5A), which was consistent with the previous finding that SufR is negatively autoregulated. The bioluminescence of the pOsufRlux transformant expressing pSufRC178S was similar to that of the strain expressing pSufR, while pSufRC182S, pSufRC195S, pSufRC223S, pSufRC182S/C195S, and pSufRC182S/C195S/C223S did not repress the transcription from the sufR promoter. Considering that serine might still be able to coordinate [4Fe-4S] cluster, Cys178 was also mutated to alanine to rule out the possibility of being a ligand residue. The bioluminescence of the transformant harboring pSufRC178A was similar to the strain expressing pSufR (Fig. S7), indicating that Cys178 is not involved in the coordination of [4Fe-4S] clusters. Amino acid sequence alignment of SufR proteins from bacteria revealed that Glu198 is also highly conserved, together with Cys182, Cys195, and Cys223 (Fig. S8). However, the bioluminescence of the transformant harboring pSufRE198A was similar to the strain expressing pSufR (Fig. S7), suggesting that Glu198 is not the non-Cys ligand to coordinate the [4Fe-4S] cluster. The findings indicate that Cys182, Cys195, and Cys223, but not Cys178, are essential for the regulatory function of SufR.
FIG 5.
Three conserved cysteines are essential for SufR function in vivo. (A) The effects of Cys residue mutation on SufR repression of sufR promoter activities were investigated using an E. coli bioluminescence reporter system. A schematic representation of the reporter system (left) and the effect of SufR variants on bioluminescence level of E. coli containing reporter plasmid pOsufRlux (right). RLU (relative light units) represents bioluminescence level. (B) RT-qPCR analysis of the transcriptional levels of suf genes in S. avermitilis WT, DsufR, and complemented strain of DsufR using sufR or sufR variants. The strains were grown in FM-I for 2 or 6 days. Error bars, standard deviation of three technical replicates.
The latter conclusion was further supported by in vivo experiments in S. avermitilis. Deletion of sufR led to increased expression of sufB, sufC, sufD, and sufS genes. The transcriptional levels of the suf genes were restored to those of the WT when the complementation plasmid pSETSufR (based on the integrative plasmid pSET152) was transferred into the DsufR mutant (Fig. 5B). pSETSufRC178S (expressing the C178S SufR variant based on pSET152) also restored the expression of the suf genes of DsufR to that of the WT, while pSETSufRC182S, pSETSufRC195S, pSETSufRC223S, pSETSufRC182S/C195S, and pSETSufRC182S/C195S/C223S could not (Fig. 5B). The results further confirmed that the residues Cys182, Cys195, and Cys223 are required for proper SufR function.
The C182/195/223S triple-mutation SufR variant was purified under anaerobic conditions to assess its DNA activity by EMSA. Unlike the brown color of SufR, the C182/195/223S SufR variant was colorless (Fig. S2), suggesting the absence of the Fe-S cluster. C182/195/223S SufR could not bind to the promoter region of sufR (Fig. S9). The findings demonstrate that the [4Fe-4S] cluster is critical for the DNA-binding activity of SufR, and that the Cys182, Cys195, and Cys223 residues are essential for coordination of the [4Fe-4S] cluster of SufR.
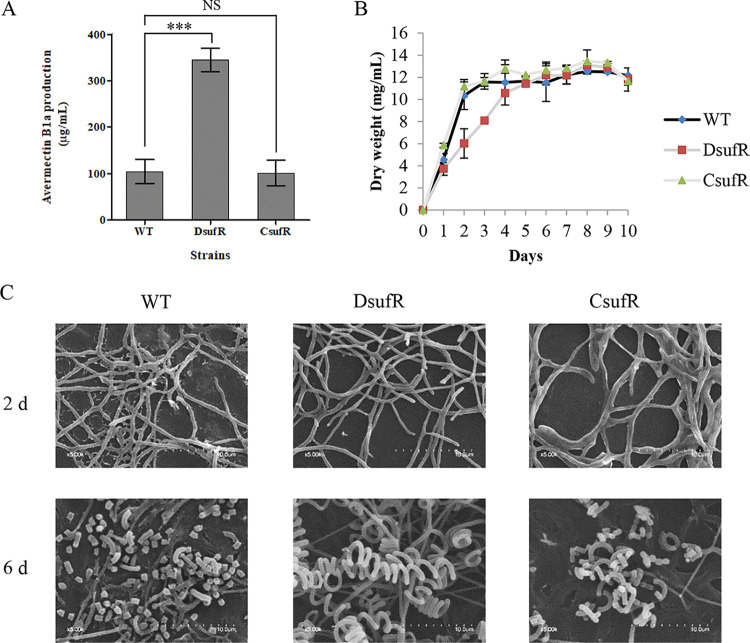
SufR represses avermectin production and promotes morphological differentiation.
In Streptomyces roseosporus, the ArsR-family transcriptional regulator DepR2 negatively regulates daptomycin production by directly binding to the promoters of the daptomycin gene cluster (38). We therefore determined whether SufR regulates secondary metabolism or morphological differentiation in S. avermitilis. Shake-flask fermentation and high-pressure liquid chromatography (HPLC) analysis showed the avermectin yield was ∼230% higher for DsufR than for the WT (Fig. 6A). The biomass was lower for DsufR than the WT in soluble fermentation medium FM-II in the exponential phase, but the biomasses were almost the same in the stationary phase (Fig. 6B). Avermectin yield and bacterial biomass were restored in CsufR by introduction of a copy of sufR into DsufR (Fig. 6A and B), indicating that the changes in avermectin yield and biomass of DsufR were due solely to the sufR deletion. Compared with the WT and CsufR, DsufR displayed obvious delays of aerial hypha formation and sporulation on SFM plates (Fig. S10), and scanning electron microscopy (SEM) showed that DsufR grown on SFM formed normal aerial hyphae and spores, although this was delayed (Fig. 6C). Therefore, the deletion of sufR delays the formation of aerial hyphae and spores. Given that no SufR-binding sites were found in the promoter regions of ave genes, and that SufR did not bind to their promoter regions (Fig. S11), we infer that SufR may affect avermectin production in an indirect manner.
FIG 6.
Avermectin production, growth, and morphological differentiation of S. avermitilis WT and sufR-related mutant strains. (A) Avermectin production of the WT, DsufR, and CsufR cultured in FM-I for 10 days. ***, P < 0.001; NS, not significant (Student’s t test). (B) Growth curves of the WT, DsufR, and CsufR in soluble FM-II. (C) SEM images showing morphological development of the WT, DsufR, and CsufR grown for 2 or 6 days on SFM media. Error bars, standard deviation of three biological replicates.
The WhiB-like (Wbl) family of regulators are small [4Fe-4S]-containing proteins that are found in actinomycetes, including the genera Mycobacterium and Corynebacterium (39, 40). Eleven and nine wbl genes were identified in S. coelicolor (41) and S. avermitilis, respectively (Table S2). Several of them (wblA, whiB, and whiD) are important in morphological differentiation (42–44). ChIP-seq analysis showed that WhiB is a transcription factor that coregulates the expression of key genes required for sporulation with WhiA (45, 46), and mutation of its cysteine residues that coordinate the WhiB [4Fe-4S] cluster abolishes the DNA binding in vivo (46). RT-qPCR analysis in the current study revealed that the transcriptional levels of most of the wbl genes were significantly decreased in DsufR (Fig. S12). Because wblA, whiB, and whiD are essential for morphological differentiation in Streptomyces, the decreased expression of wbl genes led to delayed morphological differentiation.
DISCUSSION
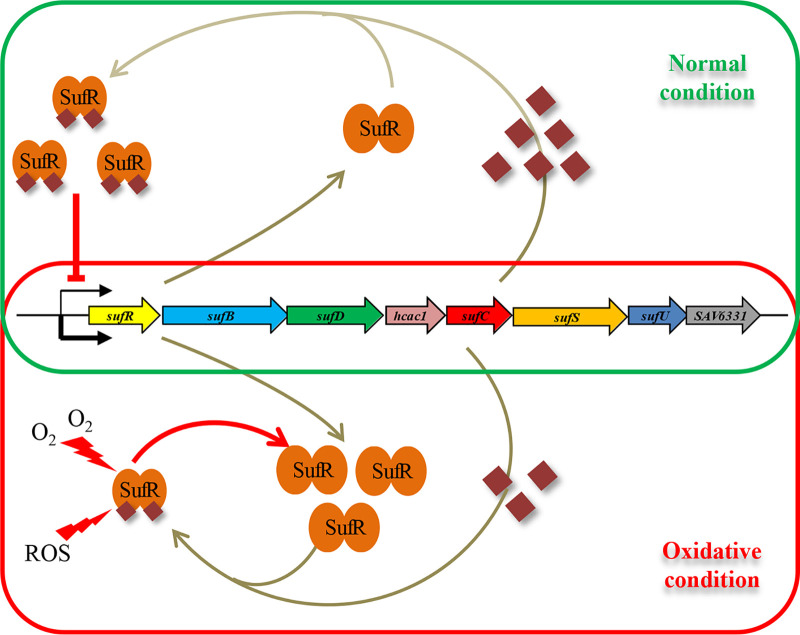
The present study elucidates the biochemical and biophysical properties and regulation function of SufR in Streptomyces. The sufRBDCSU operon is conserved among Streptomyces. Because neither Isc nor Nif systems were found in Streptomyces genomes, the Suf system is the sole biosynthetic machinery for the assembly and repair of Fe-S clusters in the genus. [4Fe-4S]-SufR exists as a homodimer and functions as a transcriptional repressor of the suf operon. Based on the findings, we propose a model for the function of SufR in maintaining Fe-S cluster homeostasis in S. avermitilis (Fig. 7). When the intracellular synthesis of Fe-S clusters is sufficient, SufR exists mainly in the form of [4Fe-4S]-SufR. Holo-SufR binds to the promoter region of the suf operon and represses its expression. Under oxidative stress, however, the Fe-S clusters are damaged by ROS, and the intracellular Fe-S cluster abundance is insufficient. SufR loses its [4Fe-4S] cluster and its DNA-binding activity. Apo-SufR dissociates from the promoter region of the suf operon, and the expression of the suf system is strongly increased by derepression in order to promote the synthesis of Fe-S clusters. Once the demand for Fe-S cluster biogenesis is met, the increased intracellular Fe-S cluster binds to SufR, and holo-SufR represses the expression of the suf operon and slows the synthesis of Fe-S clusters. Thus, SufR senses the status of intracellular Fe-S cluster via its [4Fe–4S] cluster occupancy and controls intracellular Fe-S cluster homeostasis by reversible binding to the [4Fe-4S] cluster.
FIG 7.
Proposed model of SufR-mediated Fe-S cluster biogenesis under normal and oxidative stress conditions in S. avermitilis. Under normal conditions, the Fe-S clusters (crimson rhombuses) synthesized by the Suf system are sufficient to satisfy the requirement of intracellular Fe-S cluster proteins. SufR exists mainly in the form of [4Fe-4S] SufR and represses transcription of the suf operon. Under oxidative stress, destruction of Fe-S clusters results in an increased demand for intracellular Fe-S clusters. Apo-SufR loses its ability to bind to the sufR promoter region, which leads to derepression of the suf operon and therefore to increased synthesis of Fe-S clusters.
The presence of the [4Fe-4S] cluster is critical for the DNA-binding activity of SufR. Cysteine residues or other residues coordinate the iron ions of [4Fe-4S] cluster to integrate the cluster into Fe-S proteins (35, 36). S. coelicolor WhiD is a small [4Fe-4S] cluster-containing protein with a highly conserved pattern of cysteine residues C(Xn)C(X2)C(X5)C that are essential for [4Fe-4S] cluster coordination (44). E. coli IscR, which contains a [2Fe-2S] cluster, is coordinated by three cysteine residues and one histidine residue (18). S. coelicolor NsrR contains a [4Fe-4S] cluster, which is ligated by three conserved cysteine residues and an oxygenic ligand Glu85 (34). Although the C-terminal domain of SufR has four cysteine residues (Cys178, Cys182, Cys195, and Cys223), only Cys182, Cys195, and Cys223 are essential for the DNA-binding activity of SufR in vivo, i.e., Cys178 is not required. The latter finding is consistent with the alignment of S. avermitilis SufR with its orthologs, which reveals three conserved cysteine residues at their C-terminal domains. Although a highly conserved C(X12)C(X13)C(X14)C sequence is present in the C-terminal domains of cyanobacterial SufR proteins, only the first, second, and fourth cysteine residues, which are highly conserved in all SufR proteins, are involved in ligating the [4Fe-4S] cluster; the third cysteine residue is not involved and is replaced by a threonine residue in actinomycete SufR (Fig. 1B) (13). Therefore, the fourth ligand in the coordination of the [4Fe-4S] cluster for SufR remains to be identified. Its three Cys residues and one non-Cys ligand may reduce the ability of the SufR protein to preferentially obtain Fe-S clusters rather than other substrates with all cysteinyl ligands. This results in the coexistence of apo-SufR and cluster-containing holo-SufR and thereby ensures that the sole Fe-S cluster assembly system (the Suf system) is constitutively expressed at the level required to fulfill the demands for Fe-S cluster biogenesis under normal conditions.
SufR also affects avermectin production and morphological differentiation in S. avermitilis. Because no SufR-binding sites were found in the promoter regions of ave genes or genes involved in morphological differentiation, SufR affects avermectin production and morphological differentiation in an indirect manner. The WhiB-like (Wbl) family regulators play important regulatory roles in morphological differentiation and secondary metabolism in Streptomyces. WblA is a downregulator of antibiotic production in S. coelicolor (47), S. peucetius (48), and S. ghanaensis (49). The decreased expression of wblA, a gene that downregulates antibiotic production, might result in increased avermectin production. The decreased expression of wbl genes in DsufR was possibly caused by the increased availability of [4Fe-4S] clusters due to the enhanced Suf system, which affected the DNA-binding activities of Wbl proteins to their targets and their own promoters. Therefore, SufR may affect morphological differentiation and avermectin production by influencing wbl gene expression and the DNA-binding activities of Wbl regulators.
In the Gram-negative bacterium E. coli, IscR controls two Fe-S cluster assembly systems: Isc and Suf (6, 9). IscR also directly represses hyaABCDEF, hybOABCDEFG, and napFDAGHBC, which encode anaerobic respiratory enzymes containing Fe-S clusters, and yadR and yhgl, which encode proteins related to Fe-S cluster biogenesis (20). Therefore, IscR acts as a global regulator in E. coli. The current results indicate that SufR binds to the palindromic sequence (CAACN6GTTG) in the promoter region of the suf operon and represses the transcription of the suf operon. Although several genes contain a conservative SufR binding motif in the promoter regions, they are not under the direct control of SufR (Fig. S5). It thus seems that SufR functions specifically as a repressor of the Fe-S cluster assembly system in Streptomyces. Further systematic studies, such as ChIP-seq analysis, in the future would elucidate whether SufR has additional binding sites and provide more information for the regulatory role of SufR in Streptomyces.
In summary, the current research demonstrates that SufR is a transcriptional repressor of the suf operon. The [4Fe-4S] cluster is required for the DNA-binding activity of SufR and thereby enables SufR to sense the intracellular Fe-S cluster status and to modulate the expression of the sole Fe-S cluster assembly system. Further work is needed to determine how Streptomyces maintains its Fe-S cluster homeostasis in response to diverse environmental signals, such as oxidative stress or iron deficiency.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
The strains and plasmids used in this work are listed in Table 1. The WT S. avermitilis ATCC 31267 was used as a host for gene disruption and propagation. S. avermitilis was grown at 28°C on YMS (yeast extract-malt extract-soluble starch) and SFM (soya flour-mannitol) media for sporulation, or in modified liquid YEME medium containing 25% sucrose for mycelia growth and protoplast preparation. Culture conditions and media for protoplast regeneration, phenotypic observation, and avermectin production were as described previously (50). E. coli JM109 and BL21(DE3) were used for plasmid construction and protein overexpression, respectively. E. coli ET12567 (51) was used to propagate nonmethylated plasmids for transformation into S. avermitilis. E. coli strains were grown in standard LB medium at 37°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source |
|---|---|---|
| Strains | ||
| S. avermitilis | ||
| ATCC 31267 | Wild type strain | Laboratory stock |
| DsufR | sufR deletion mutant of ATCC31267 | This study |
| CsufR | Complemented strain of DsufR with SufR | This study |
| CsufRC178S | Complemented strain of DsufR with SufRC178S | This study |
| CsufRC182S | Complemented strain of DsufR with SufRC182S | This study |
| CsufRC195S | Complemented strain of DsufR with SufRC195S | This study |
| CsufRC223S | Complemented strain of DsufR with SufRC223S | This study |
| CsufRC182S/C195S | Complemented strain of DsufR with SufRC182S/C195S | This study |
| CsufRC182S/C195S/C223S | Complemented strain of DsufR with SufRC182S/C195S/C223S | This study |
| E. coli | ||
| ET12567 | Methylation-deficient strain | 50 |
| BL21(DE3) | Host for protein overexpression | Novagen |
| Plasmids | ||
| pKC1139 | Multiple-copy, temperature-sensitive E. coli-Streptomyces shuttle vector | 51 |
| pSET152 | Integrative E. coli-Streptomyces shuttle vector | 51 |
| pET-28a(+) | Vector for His6-tagged protein overexpression in E. coli | Novagen |
| pKCDsufR | sufR deletion vector based on pKC1139 | This study |
| pSETSufR | sufR-complemented vector based on pSET152 | This study |
| pSETSufRC178S | sufR(C178S)-complemented vector based on pSET152 | This study |
| pSETSufRC182S | sufR(C182S)-complemented vector based on pSET152 | This study |
| pSETSufRC195S | sufR(C195S)-complemented vector based on pSET152 | This study |
| pSETSufRC223S | sufR(C223S)-complemented vector based on pSET152 | This study |
| pSETSufRC182S/C195S | sufR(C182S/C195S)-complemented vector based on pSET152 | This study |
| pSETSufRC182S/C195S/C223S | sufR(C182S/C195S/C223S)-complemented vector based on pSET152 | This study |
| pET28-SufR | sufR-overexpressing vector based on pET-28a(+) | This study |
| pET28-SufRC182S/C195S/C223S | sufR(C182S/C195S/C223S)-overexpressing vector based on pET-28a(+) | This study |
| pCS26-Pac | Vector containing promoterless lux reporter | 34 |
| pOsufRlux | pCS26-Pac carrying sufRp-controlled lux reporter | This study |
| pACYC184 | For protein expression in reporter system | 34 |
| pSufR | For SufR expression in reporter system | This study |
| pSufRC178S | For SufRC178S expression in reporter system | This study |
| pSufRC178A | For SufRC178A expression in reporter system | This study |
| pSufRC182S | For SufRC182S expression in reporter system | This study |
| pSufRC195S | For SufRC195S expression in reporter system | This study |
| pSufRC223S | For SufRC223S expression in reporter system | This study |
| pSufRC182S/C195S | For SufRC182S/C195S expression in reporter system | This study |
| pSufRC182S/C195S/C223S | For SufRC182S/C195S/C223S expression in reporter system | This study |
| pSufRE198A | For SufRE198A expression in reporter system | This study |
Gene deletion and complementation.
The sufR mutant (DsufR) was constructed by homologous recombination as follows. Upstream (position –519 to 16 relative to the sufR start codon) and downstream regions (position –44 to 462 relative to the sufR stop codon) of sufR were amplified by PCR with the primer sets sufR-up-FW/sufR-up-Rev and sufR-dw-FW/sufR-dw-Rev, respectively. The PCR fragments were purified, digested with EcoRI/BamHI, and BamHI/HindIII, respectively, and together inserted into EcoRI/HindIII-digested pKC1139 vector to generate the sufR deletion vector pKCDsufR. pKCDsufR was transformed into S. avermitilis WT protoplasts (52). Double-crossover recombinant strains were selected as described previously (53). The mutant was analyzed and confirmed by PCR using primer pairs listed in Table 2.
TABLE 2.
Primers used in this studya
| Purpose | Primer | Sequence (5′–3′) |
|---|---|---|
| Construction of DsufR mutant | sufR-up-Fw | CGGAATTCTCTGCATGAGCGCCAAGT (EcoRI) |
| sufR-up-Rev | CGGGATCCCGCCGACGTTTTTCACAAC (BamHI) | |
| sufR-dw-Fw | CGGGATCCGCCACAACGCATCCGCAA (BamHI) | |
| sufR-dw-Rev | CCCAAGCTTCGGTGTCGAGGAAGATGACG (HindIII) | |
| sufR-V1 | CCGCCGGACTGGAGCATCA | |
| sufR-V2 | ATGGTGACCTTGGAGCCGATGT | |
| Complementation of DsufR mutant | sufR-C-Fw | CGGGATCCGTGGCCGTCGAGCTGGGT (BamHI) |
| sufR-C-Rev | GCTCTAGACTTGAGGCGGAGCTTGGT (XbaI) | |
| sufR-CM-Fw | CGGGATCCTTCCCGACAGCCTCCTCCG (BamHI) | |
| sufR-CM-Rev | GCTCTAGAACTCCGCCTGCTTCTCCGT (XbaI) | |
| Construction of His-tagged SufR | His-sufR-Fw | CATGCCATGGAAAACGTCGGCGAGGCAC (NcoI) |
| His-sufR-Rev | CGGGATCCTTCCTCCCGGCGGTGCTTG (BamHI) | |
| EMSA | sufRp-Fw | GATGGAAGCTCGCGCACG |
| sufRp-Rev | CGGCGGCCTGGGTGAGAC | |
| sufRpAB-Fw | CGCAGGTCAGTTTAGGTATGCC | |
| sufRpAB-Rev | GGGTCTCCCGTGCCTCGC | |
| Footprinting assay | sufRp-FAM-Fw | GATGGAAGCTCGCGCACG |
| sufRp-Rev | CGGCGGCCTGGGTGAGAC | |
| Lux reporter system | CS-sufR-Fw | CCCTCGAGTCGCCGTACCGCTTCACCAG (XhoI) |
| CS-sufR-Rev | CGGGATCCCTCCCGTGCCTCGCCGAC (BamHI) | |
| sufR-P-Fw | GGGTTGTCAGGCCCTGAGGAA | |
| sufR-P-Rev | CGGGATCCTCGTTGATGCCGCGCTTGG (BamHI) | |
| Serine and alanine substitution of cysteine residues | sufRC178S-up-Fw | AGTGGGCCGGTCGGAGGG |
| sufRC178S-dw-Rev | CTCCGCCTCCGGGATGCC | |
| sufRC182S-up-Fw | GAGTCTTCCCGACAGCCTCCTC | |
| sufRC182S-dw-Rev | CGCTGCTTCTCCGCCTCCG | |
| sufRC195S-up-Fw | AGTCTTCCCGACAGCCTCCTC | |
| sufRC195S-dw-Rev | GCGGATCTGGTGGTAGACGAC | |
| sufRC223S-up-Fw | TTCCCGACAGCCTCCTCCG | |
| sufRC223S-dw-Rev | TGCTCCTTCAGGGCGGTGTC | |
| sufRC178A-up-Fw | AGTGGGCCGGTCGGAGGG | |
| sufRC178A-dw-Rev | CTCCGCCTCCGGGATGCC | |
| sufRE198A-up-Fw | AGTCTTCCCGACAGCCTCCTC | |
| sufRE198A-dw-Rev | GCGGATCTGGTGGTAGACGAC | |
| 5′RACE | Oligo(dT) anchor primer | GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTT |
| anchor primer | GACCACGCGTATCGATGTCGAC | |
| SP1sufR | CGGCGGTGCTTGCGGATG | |
| SP2sufR | GGTCTCCGCCTCGCACAGC | |
| SP3sufR | GTCAGGGCGAAGACCTTGGC |
Underlining indicates restriction enzyme sites.
For complementation of DsufR, a 1,266-bp DNA fragment containing the promoter and ORF of sufR was amplified from WT genomic DNA. The PCR product was cloned into the integrative vector pSET152 to generate a sufR-complemented vector, which was transformed into DsufR to obtain the complemented strain CsufR. The point mutation SufR complemented plasmids were also constructed by the method described above, and were transformed into DsufR to obtain corresponding complemented strains.
Anaerobic purification of SufR.
To overproduce SufR in E. coli, plasmid pETSufR was constructed as follows. The SufR coding region was amplified by PCR from S. avermitilis genomic DNA using primers His-sufR-Fw and His-sufR-Rev. The PCR product was digested with NcoI and BamHI and inserted into a His6-tagged expression vector pET28a cut with the same enzymes to generate pETSufR. pETSufRC182/195/223S was constructed using the same procedure to overproduce a C182/195/223S triple-mutation SufR variant. The resulting plasmids were transformed into E. coli BL21(DE3) for overexpression of SufR proteins with a His6-tagged C terminus.
The transformants were cultured in LB with 50 μg/ml kanamycin and 20 μM ammonium ferric citrate at 37°C and 230 rpm until an optical density at 600 nm (OD600) of 0.4 to 0.6 was attained. To facilitate the synthesis of intracellular Fe-S clusters, the cultures were placed on ice for 18 min and were induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubated at 37°C and 230 rpm. After 50 min, the cultures were supplemented with 25 μM l-methionine and 200 μM ammonium ferric citrate and incubated at 37°C for 3.5 h. The harvested cells were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole) to a total volume of 50 ml and disrupted by sonication on ice in an anaerobic workstation. The crude lysate was transferred to sealed centrifuge tubes (Beckman) and centrifuged outside the anaerobic workstation at 9,000 rpm for 50 min at 4°C to obtain a clarified SufR-containing extract. After repeated vacuuming and nitrogen filling, the extract was transferred to the anaerobic glovebox cabinet (MBRAUN Lab Star, Germany). His6-tagged protein was purified on Ni2+-NTA resin (Qiagen, Germany) according to the manufacturer’s protocol in an anaerobic glovebox cabinet (O2 <8 ppm). All solutions used for anaerobic purification underwent repeated vacuuming and nitrogen filling to remove oxygen in an anaerobic workstation.
UV-visible absorption spectroscopy and mass spectrometry of SufR.
UV-visible absorption spectra were obtained using a TU-1900 dual beam UV spectrometer (China) in the 260 to 860 nm wavelength range. Absorption spectra of SufR were immediately collected at room temperature. To study the effect of air oxidation on the [4Fe-4S] cluster of SufR, the absorption spectra of purified protein were recorded at different time intervals up to 180 min. The anaerobically purified SufR was analyzed on a Bruker Autoflex II mass spectrometer (Bruker Daltonics, Billerica, MA, USA). The mass spectrometer was operated at linear mode, 45 to 60% laser power, 337 nm nitrogen laser. Spectra were analyzed with Flex-Analysis.
Preparation of apo-SufR and reconstituted [4Fe-4S]-SufR.
Apo-SufR and reconstituted [4Fe-4S]-SufR were prepared as described previously (54). Apo-SufR was prepared from freshly purified holo-SufR solution (100 μM) using potassium ferricyanide and EDTA in a 1:50:20 molar ratio of protein:EDTA:ferricyanide at 25°C for 30 min, and then apo-SufR was exchanged into elution buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole), using Zeba spin desalting columns (Thermo Scientific).
Reconstitution of the Fe-S cluster of apo-SufR was performed in the anaerobic cabinet. An apo-protein solution (50 μM) was incubated with 500 μM FeCl3 and 500 μM Na2S at 25°C for 8 h. The reconstituted protein (Re-SufR) was immediately used for EMSAs.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays (EMSAs) were performed with a DIG Gel Shift kit (second generation; Roche, USA). The promoter region of sufR was obtained by PCR and labeled with DIG-11-ddUTP using terminal transferase. Binding reaction mixtures (20 μl) contained 0.15 nM labeled probe, binding buffer [20 mM HEPES, pH 7.0, 10 mM (NH4)2SO4, 1 mM DTT, 0.2% (wt/vol) Tween 20, 30 mM KCl], 1 μg poly(d[I-C]), and various concentrations of SufR or SufR variant. The binding reactions were performed at 25°C for 30 min and were run on 5% wt/vol native polyacrylamide gels. Gels were transferred to nylon membrane and signals were detected following the manufacturer’s protocol (Roche). The above operations were performed under aerobic conditions.
DNase I footprinting.
A fluorescent labeling procedure was used for DNase I footprinting assays. To determine the binding site of SufR in its promoter region, a 505-bp fluorescence-labeled DNA fragment corresponding to the upstream region of sufR was amplified by PCR using primer pair sufRp-FAM-Fw/sufRp-Rev and was purified from the agarose gel. A reaction mixture (25 μl) containing 400 ng of labeled probe and various amounts of SufR-His6 protein in binding buffer was incubated at 25°C for 30 min. DNase I (0.016 units) digestion was performed at 37°C for 50 s and was terminated with 50 mM EDTA. After purification, the DNA samples were analyzed with a 3730XL DNA genetic analyzer (Applied Biosystems), and the data were analyzed using GeneMarker v2.2 software.
5′-RACE.
The transcriptional start site (TSS) of sufR was identified using a 5′/3′ rapid amplification of cDNA ends (RACE) kit (second generation; Roche). Total RNA was prepared from WT culture grown in FM-I for 2 days. The cDNA was synthesized using a gene-specific primer (SP1sufR). An oligo(dA) tail was added to the purified cDNAs at the 3′ end using terminal deoxynucleotidyl transferase. The tailed cDNA was used as the template with oligo(dT)-anchor primer and another gene-specific primer (SP2sufR) to perform the first round of PCR. To obtain a single specific band, the product of first-round PCR was used as a template for the second-round PCR with the anchor primer and a nested primer (SP3sufR). The final PCR product was purified and sequenced (Invitrogen Biotechnology Corporation, China).
Serine substitution of cysteine residues in SufR.
Four cysteine residues (C178, C182, C195, and C223) were mutated to serine individually or in combination by site-directed mutagenesis using overlap extension PCR. A similar procedure was used for mutating C178 and E198 to alanine. Two overlap PCR products were amplified from WT genomic DNA using primer pairs. The purified PCR products were used as the templates in the second PCR with sufRCNS-up-Fw and sufRCNS-dw-Rev (N: 178, 182, 195, 223). Fragments (1,190-bp) were amplified using the fusion fragments as the templates with SufR-CM-Fw and sufR-CM-Rev. The products were cleaved by BamHI/XbaI and cloned into pSET152 to generate a series of constructs. DNA sequencing analysis was performed to verify the sequence of each plasmid.
Construction of biosensor strains and bioluminescence assay in E. coli.
The promoter region of sufR was amplified by PCR with primers CS-sufR-Fw/CS-sufR-Rev and ligated into the reporter vector pCS26-Pac to obtain pOsufRlux. For expression of SufR proteins, the sufR gene (WT or variants C178S, C182S, C195S, C223S, C182/195S, and C182/195/223S) containing its Shine-Dalgarno sequence and coding region was amplified with primers and ligated into pACYC184 to produce pSufR or site-mutated SufR expression plasmids. The reporter vector pOsufRlux and the SufR expression vector pSufR were cotransformed into E. coli JM109. Vectors pACYC184 and pCS26-Pac were cotransformed into E. coli JM109 as controls. The bioluminescence of E. coli reporter cultures was measured after 12 h cultivation at 37°C using a single-tube luminometer (GloMax 20/20; Promega, USA).
RNA extraction and reverse transcription quantitative PCR analysis.
Total RNAs were isolated from mycelia of S. avermitilis grown for various times in fermentation medium FM-I or on SFM solid medium, as described previously (55). RNA samples were treated with RNase-free DNase I (TaKaRa, Japan) to remove genomic DNA. Synthesis of cDNA and reverse transcription quantitative PCR (RT-qPCR) analysis were performed as described previously (55). Primer efficiencies were measured and calculated (56), and the efficiency values of all primers were between 93 and 105%. The relative expression level was quantified using the comparative threshold cycle (CT) method. Transcription of the housekeeping gene hrdB (SAV2444) was used as an internal control. Each experiment was performed in triplicate.
Scanning electron microscopy.
For specimen preparation, coverslips were embedded at a 45° angle in SFM agar inoculated with the S. avermitilis strains. The coverslips were gently removed from the agar after various periods of growth. Specimens were fixed with glutaraldehyde and osmium tetroxide, washed three times with phosphate butter, dehydrated by ethanol, dried in a LEICA critical point dryer, sputter-coated with a layer of gold, and examined with a scanning electron microscope (S3400N; Hitachi).
Production and analysis of avermectins.
Fermentation of S. avermitilis strains and HPLC analysis of avermectin yield were performed as described previously (50).
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grants from the National Natural Science Foundation of China (grants no. 31470190 and 31861143004).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Beinert H, Holm RH, Munck E. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653–659. doi: 10.1126/science.277.5326.653. [DOI] [PubMed] [Google Scholar]
- 2.Johnson DC, Dean DR, Smith AD, Johnson MK. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem 74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 3.Py B, Moreau PL, Barras F. 2011. Fe-S clusters, fragile sentinels of the cell. Curr Opin Microbiol 14:218–223. doi: 10.1016/j.mib.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 4.Ayala-Castro C, Saini A, Outten FW. 2008. Fe-S cluster assembly pathways in bacteria. Microbiol Mol Biol Rev 72:110–125. doi: 10.1128/MMBR.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Argudo I, Little R, Shearer N, Johnson P, Dixon R. 2005. Nitrogen fixation: key genetic regulatory mechanisms. Biochem Soc Trans 33:152–156. doi: 10.1042/BST0330152. [DOI] [PubMed] [Google Scholar]
- 6.Giel JL, Nesbit AD, Mettert EL, Fleischhacker AS, Wanta BT, Kiley PJ. 2013. Regulation of iron-sulphur cluster homeostasis through transcriptional control of the Isc pathway by [2Fe-2S]-IscR in Escherichia coli. Mol Microbiol 87:478–492. doi: 10.1111/mmi.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. 2001. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci U S A 98:14895–14900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi Y, Tokumoto U. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem 277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 9.Yeo WS, Lee JH, Lee KC, Roe JH. 2006. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol Microbiol 61:206–218. doi: 10.1111/j.1365-2958.2006.05220.x. [DOI] [PubMed] [Google Scholar]
- 10.Outten FW, Djaman O, Storz G. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol 52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 11.Py B, Barras F. 2010. Building Fe-S proteins: bacterial strategies. Nat Rev Microbiol 8:436–446. doi: 10.1038/nrmicro2356. [DOI] [PubMed] [Google Scholar]
- 12.Huet G, Daffe M, Saves I. 2005. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: evidence for its implication in the pathogen’s survival. J Bacteriol 187:6137–6146. doi: 10.1128/JB.187.17.6137-6146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen G, Balasubramanian R, Wang T, Wu Y, Hoffart LM, Krebs C, Bryant DA, Golbeck JH. 2007. SufR coordinates two [4Fe-4S]2+, 1+ clusters and functions as a transcriptional repressor of the sufBCDS operon and an autoregulator of sufR in cyanobacteria. J Biol Chem 282:31909–31919. doi: 10.1074/jbc.M705554200. [DOI] [PubMed] [Google Scholar]
- 14.Riboldi GP, Verli H, Frazzon J. 2009. Structural studies of the Enterococcus faecalis SufU [Fe-S] cluster protein. BMC Biochem 10:3. doi: 10.1186/1471-2091-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imlay JA. 2006. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol 59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 17.Crack JC, Green J, Thomson AJ, Le Brun NE. 2014. Iron-sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide. Acc Chem Res 47:3196–3205. doi: 10.1021/ar5002507. [DOI] [PubMed] [Google Scholar]
- 18.Fleischhacker AS, Stubna A, Hsueh KL, Guo Y, Teter SJ, Rose JC, Brunold TC, Markley JL, Munck E, Kiley PJ. 2012. Characterization of the [2Fe-2S] cluster of Escherichia coli transcription factor IscR. Biochemistry 51:4453–4462. doi: 10.1021/bi3003204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee KC, Yeo WS, Roe JH. 2008. Oxidant-responsive induction of the suf operon, encoding a Fe-S assembly system, through Fur and IscR in Escherichia coli. J Bacteriol 190:8244–8247. doi: 10.1128/JB.01161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol 60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang T, Shen G, Balasubramanian R, McIntosh L, Bryant DA, Golbeck JH. 2004. The sufR gene (sll0088 in Synechocystis sp. strain PCC 6803) functions as a repressor of the sufBCDS operon in iron-sulfur cluster biogenesis in cyanobacteria. J Bacteriol 186:956–967. doi: 10.1128/jb.186.4.956-967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albrecht AG, Netz DJ, Miethke M, Pierik AJ, Burghaus O, Peuckert F, Lill R, Marahiel MA. 2010. SufU is an essential iron-sulfur cluster scaffold protein in Bacillus subtilis. J Bacteriol 192:1643–1651. doi: 10.1128/JB.01536-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willemse D, Weber B, Masino L, Warren RM, Adinolfi S, Pastore A, Williams MJ. 2018. Rv1460, a SufR homologue, is a repressor of the suf operon in Mycobacterium tuberculosis. PLoS One 13:e0200145. doi: 10.1371/journal.pone.0200145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minch KJ, Rustad TR, Peterson EJ, Winkler J, Reiss DJ, Ma S, Hickey M, Brabant W, Morrison B, Turkarslan S, Mawhinney C, Galagan JE, Price ND, Baliga NS, Sherman DR. 2015. The DNA-binding network of Mycobacterium tuberculosis. Nat Commun 6:5829. doi: 10.1038/ncomms6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flardh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat Rev Microbiol 7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- 26.Challis GL, Hopwood DA. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci U S A 100(Suppl 2):14555–14561. doi: 10.1073/pnas.1934677100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egerton JR, Ostlind DA, Blair LS, Eary CH, Suhayda D, Cifelli S, Riek RF, Campbell WC. 1979. Avermectins, new family of potent anthelmintic agents: efficacy of the B1a component. Antimicrob Agents Chemother 15:372–378. doi: 10.1128/aac.15.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller TW, Chaiet L, Cole DJ, Cole LJ, Flor JE, Goegelman RT, Gullo VP, Joshua H, Kempf AJ, Krellwitz WR, Monaghan RL, Ormond RE, Wilson KE, Albers-Schönberg G, Putter I. 1979. Avermectins, new family of potent anthelmintic agents: isolation and chromatographic properties. Antimicrob Agents Chemother 15:368–371. doi: 10.1128/aac.15.3.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Outten FW, Wood MJ, Munoz FM, Storz G. 2003. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J Biol Chem 278:45713–45719. doi: 10.1074/jbc.M308004200. [DOI] [PubMed] [Google Scholar]
- 30.Outten FW. 2015. Recent advances in the Suf Fe-S cluster biogenesis pathway: beyond the proteobacteria. Biochim Biophys Acta 1853:1464–1469. doi: 10.1016/j.bbamcr.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duin EC, Lafferty ME, Crouse BR, Allen RM, Sanyal I, Flint DH, Johnson MK. 1997. [2Fe-2S] to [4Fe-4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry 36:11811–11820. doi: 10.1021/bi9706430. [DOI] [PubMed] [Google Scholar]
- 32.Ugulava NB, Sacanell CJ, Jarrett JT. 2001. Spectroscopic changes during a single turnover of biotin synthase: destruction of a [2Fe-2S] cluster accompanies sulfur insertion. Biochemistry 40:8352–8358. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Busenlehner LS, Pennella MA, Giedroc DP. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol Rev 27:131–143. doi: 10.1016/S0168-6445(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 34.Crack JC, Munnoch J, Dodd EL, Knowles F, Al Bassam MM, Kamali S, Holland AA, Cramer SP, Hamilton CJ, Johnson MK, Thomson AJ, Hutchings MI, Le Brun NE. 2015. NsrR from Streptomyces coelicolor is a nitric oxide-sensing [4Fe-4S] cluster protein with a specialized regulatory function. J Biol Chem 290:12689–12704. doi: 10.1074/jbc.M115.643072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reents H, Gruner I, Harmening U, Bottger LH, Layer G, Heathcote P, Trautwein AX, Jahn D, Hartig E. 2006. Bacillus subtilis Fnr senses oxygen via a [4Fe-4S] cluster coordinated by three cysteine residues without change in the oligomeric state. Mol Microbiol 60:1432–1445. doi: 10.1111/j.1365-2958.2006.05198.x. [DOI] [PubMed] [Google Scholar]
- 36.Isabella VM, Lapek JD Jr, Kennedy EM, Clark VL. 2009. Functional analysis of NsrR, a nitric oxide-sensing Rrf2 repressor in Neisseria gonorrhoeae. Mol Microbiol 71:227–239. doi: 10.1111/j.1365-2958.2008.06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tahlan K, Ahn SK, Sing A, Bodnaruk TD, Willems AR, Davidson AR, Nodwell JR. 2007. Initiation of actinorhodin export in Streptomyces coelicolor. Mol Microbiol 63:951–961. doi: 10.1111/j.1365-2958.2006.05559.x. [DOI] [PubMed] [Google Scholar]
- 38.Mao XM, Luo S, Li YQ. 2017. Negative regulation of daptomycin production by DepR2, an ArsR-family transcriptional factor. J Ind Microbiol Biotechnol 44:1653–1658. doi: 10.1007/s10295-017-1983-3. [DOI] [PubMed] [Google Scholar]
- 39.Kim JS, Lee HN, Lee HS, Kim P, Kim ES. 2013. A WblA-binding protein, SpiA, involved in Streptomyces oxidative stress response. J Microbiol Biotechnol 23:1365–1371. doi: 10.4014/jmb.1306.06032. [DOI] [PubMed] [Google Scholar]
- 40.Crack JC, den Hengst CD, Jakimowicz P, Subramanian S, Johnson MK, Buttner MJ, Thomson AJ, Le Brun NE. 2009. Characterization of [4Fe-4S]-containing and cluster-free forms of Streptomyces WhiD. Biochemistry 48:12252–12264. doi: 10.1021/bi901498v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fowler-Goldsworthy K, Gust B, Mouz S, Chandra G, Findlay KC, Chater KF. 2011. The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2). Microbiology 157:1312–1328. doi: 10.1099/mic.0.047555-0. [DOI] [PubMed] [Google Scholar]
- 42.Zheng F, Long Q, Xie J. 2012. The function and regulatory network of WhiB and WhiB-like protein from comparative genomics and systems biology perspectives. Cell Biochem Biophys 63:103–108. doi: 10.1007/s12013-012-9348-z. [DOI] [PubMed] [Google Scholar]
- 43.Liu G, Chater KF, Chandra G, Niu G, Tan H. 2013. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jakimowicz P, Cheesman MR, Bishai WR, Chater KF, Thomson AJ, Buttner MJ. 2005. Evidence that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe-4S] cluster. J Biol Chem 280:8309–8315. doi: 10.1074/jbc.M412622200. [DOI] [PubMed] [Google Scholar]
- 45.Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ. 2013. Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. mBio 4:e00684-13. doi: 10.1128/mBio.00684-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bush MJ, Chandra G, Bibb MJ, Findlay KC, Buttner MJ. 2016. Genome-wide chromatin immunoprecipitation sequencing analysis shows that WhiB is a transcription factor that cocontrols its regulon with WhiA to initiate developmental cell division in Streptomyces. mBio 7:e00523-16. doi: 10.1128/mBio.00523-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang SH, Huang J, Lee HN, Hur YA, Cohen SN, Kim ES. 2007. Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. J Bacteriol 189:4315–4319. doi: 10.1128/JB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noh JH, Kim SH, Lee HN, Lee SY, Kim ES. 2010. Isolation and genetic manipulation of the antibiotic down-regulatory gene, wblA ortholog for doxorubicin-producing Streptomyces strain improvement. Appl Microbiol Biotechnol 86:1145–1153. doi: 10.1007/s00253-009-2391-z. [DOI] [PubMed] [Google Scholar]
- 49.Rabyk M, Ostash B, Rebets Y, Walker S, Fedorenko V. 2011. Streptomyces ghanaensis pleiotropic regulatory gene wblA(gh) influences morphogenesis and moenomycin production. Biotechnol Lett 33:2481–2486. doi: 10.1007/s10529-011-0728-z. [DOI] [PubMed] [Google Scholar]
- 50.Jiang L, Liu Y, Wang P, Wen Y, Song Y, Chen Z, Li J. 2011. Inactivation of the extracytoplasmic function sigma factor Sig6 stimulates avermectin production in Streptomyces avermitilis. Biotechnol Lett 33:1955–1961. doi: 10.1007/s10529-011-0673-x. [DOI] [PubMed] [Google Scholar]
- 51.Macneil DJ, Klapko LM. 1987. Transformation of Streptomyces avermitilis by plasmid DNA. J Ind Microbiol 2:209–218. doi: 10.1007/BF01569542. [DOI] [Google Scholar]
- 52.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Yan T, Jiang L, Wen Y, Song Y, Chen Z, Li J. 2013. Characterization of SAV7471, a TetR-family transcriptional regulator involved in the regulation of coenzyme A metabolism in Streptomyces avermitilis. J Bacteriol 195:4365–4372. doi: 10.1128/JB.00716-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alam MS, Garg SK, Agrawal P. 2007. Molecular function of WhiB4/Rv3681c of Mycobacterium tuberculosis H37Rv: a [4Fe-4S] cluster co-ordinating protein disulphide reductase. Mol Microbiol 63:1414–1431. doi: 10.1111/j.1365-2958.2007.05589.x. [DOI] [PubMed] [Google Scholar]
- 55.Yang R, Liu X, Wen Y, Song Y, Chen Z, Li J. 2015. The PhoP transcription factor negatively regulates avermectin biosynthesis in Streptomyces avermitilis. Appl Microbiol Biotechnol 99:10547–10557. doi: 10.1007/s00253-015-6921-6. [DOI] [PubMed] [Google Scholar]
- 56.Ramakers C, Ruijter JM, Deprez RHL, Moorman A. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.