LETTER
SeqSero2 (1) and its predecessor SeqSero (2) predict Salmonella serotypes from whole-genome sequencing (WGS) data by targeting genetic determinants of serotype without resorting to surrogate markers, such as multilocus sequence types (MLST). This approach maintains continuity with the well-established scheme for phenotypic serotypes but may generate incomplete prediction of an antigenic profile should a serotype determinant gene be poorly sequenced by WGS (2).
DNA libraries prepared by the Illumina Nextera XT kits are known to produce suboptimal sequencing coverage at low-GC regions; this bias has implications for subtyping and metagenomics analyses (3–6). The lipopolysaccharide O antigen determinants of Salmonella in the rfb gene cluster feature considerably lower GC content (∼30%) than the genome-wide GC average of Salmonella (∼52%). A recent evaluation of Salmonella serotype prediction tools by Uelze et al. reported a lack of O antigen prediction by SeqSero2 (7). The authors convincingly attributed such predictions to library preparation-induced low-GC sequencing bias caused by the Nextera XT kits. In contrast, genomes prepared by the newer Illumina Nextera Flex kits were free of the issue (7).
The lack of O antigen prediction in the study by Uelze et al. was alarmingly prevalent, which prompted us to reanalyze their data to investigate the cause of the reported issue. Compared to a representative set of Salmonella genomes from public health laboratories in United States and England, Nextera XT-prepared genomes in the study by Uelze et al. appeared to be disproportionately overrepresented by predictions that lacked an O antigen call (Table 1), the vast majority of which belonged to serogroup O7 (Table 2). These O7 genomes as well as Nextera XT-prepared genomes of other common serogroups (O4, O8, and O9) in the study by Uelze et al. were significantly more susceptible to GC content-associated sequencing bias against low-GC regions (Fig. 1 and 2). The biases were significant enough to affect de novo genome assembly, as measured by the L50 score (Fig. 2), and likely contributed to the uncharacteristically low antigen prediction accuracy by SISTR, another tool evaluated in that study (34.5% full match rate versus 41.9% in a previous evaluation [8]). In our benchmark data set, genomes from the U.S. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) were prepared by Illumina TruSeq kits and least affected by the sequencing bias (Fig. 1). These genomes were used in the previous evaluation of SeqSero2 (1); their bias-free nature (Fig. 3) may explain the discrepant results between the previous study and the study by Uelze et al., particularly the performance of the microassembly workflow that requires sufficient sequencing coverage of the rfb region to assemble O antigen determinant genes.
TABLE 1.
Prevalence of serotype predictions that lack an O antigen call by the microassembly workflow of SeqSero2 using Salmonella genomes from different public health laboratories
| Data set | Total no. | Library prepn | No. of predictions that lack an O antigen call | % of predictions that lack an O antigen call |
|---|---|---|---|---|
| BfRa | 578 | Nextera XT | 71 | 12.3 |
| U.S. FDAb | 3,929 | Nextera XT | 33 | 0.8 |
| U.S. PulseNetc | 196 | Nextera XT | 5 | 2.6 |
| PHEd | 202 | Nextera XT | 0 | 0 |
| U.S. NARMSe | 2,280 | TrueSeq | 5 | 0.2 |
Genomes (n = 1,263) from animal production, food, and the environment in Germany under BioProject no. PRJEB31846 were analyzed. Out of the 1,263 genomes, 578 were prepared by Nextera XT kits, of which 71 were missing an O antigen call. Another 685 were prepared by Nextera Flex kits, of which 3 were missing an O antigen call. NCBI accession numbers can be found at http://denglab.info/static/AEM_letter_datasets.xlsx.
Genomes (n = 3,929) used by FDA for an evaluation study of SeqSero2 (unpublished data). NCBI accession numbers can be found at http://denglab.info/static/AEM_letter_datasets.xlsx.
Genomes (n = 196) sequenced by state and local health departments in the United States for national surveillance of Salmonella. Genomes were randomly selected from BioProject no. PRJNA230403 to represent 16 major serotypes, including Braenderup, Infantis, Montevideo, Thompson, Agona, Heidelberg, Saintpaul, Typhimurium, Hadar, Kentucky, Muenchen, Newport, Berta, Enteritidis, Javiana, and Panama. NCBI accession numbers can be found at http://denglab.info/static/AEM_letter_datasets.xlsx.
Genomes (n = 202) were randomly selected from the Public Health England BioProject PRJNA248792 to represent 16 major serotypes as aforementioned. NCBI accession numbers can be found at http://denglab.info/static/AEM_letter_datasets.xlsx. Genomes were prepared by Nextera XT kits according to the annotation of WGS data in the depository.
Genomes (n = 2,280) from human clinical isolates submitted to the U.S. NARMS in 2015 (1). NARMS performs surveillance for antimicrobial resistance in Salmonella (https://www.cdc.gov/narms/index.html); every 20th isolate, along with serotype information, is submitted by state and local health departments in the United States. NCBI accession numbers can be found at http://denglab.info/static/AEM_letter_datasets.xlsx.
TABLE 2.
Summary of serotype predictions that lack an O antigen call by the microassembly workflow of SeqSero2 among Nextera XT-prepared genomes in the study by Uelze et al.
| Serotypea | Total no. | No. of predictions that lack an O antigen call | O group | % of O antigen-less predictions |
|---|---|---|---|---|
| Virchow | 3 | 3 | O7 | 100.0 |
| Bareilly | 4 | 3 | O7 | 75.0 |
| Infantis | 69 | 29 | O7 | 42.0 |
| Mbandaka | 58 | 17 | O7 | 29.3 |
| Paratyphi B var. Java | 55 | 5 | O4 | 9.1 |
| Agona | 41 | 3 | O4 | 7.3 |
| Typhimurium | 61 | 4 | O4 | 6.6 |
Only serotypes with at least 3 genomes that produced predictions without an O antigen call are shown. These genomes accounted for 90.1% of predictions that lacked an O antigen call from Salmonella isolates from animal production, food, and the environment in Germany (n = 1,263) under BioProject no. PRJEB31846.
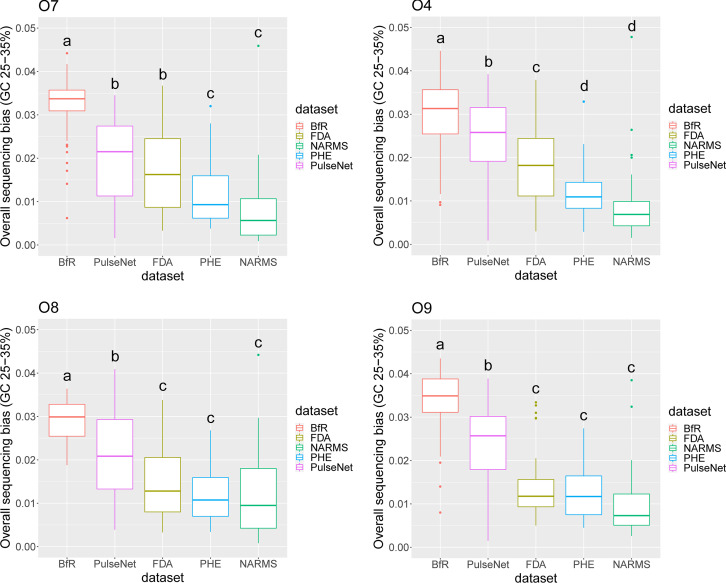
FIG 1.
Comparison of GC-associated sequencing biases of low-GC regions in genomes from four major O antigen groups. Low-GC regions are defined as regions of GC content between 25% and 35%, which is similar to the GC content of O antigen determinant genes (wzx/wzy). GC-associated sequencing bias was calculated according to methods described in reference 3. Salmonella genomes from five data sets were analyzed, including BfR (n = 415), PulseNet (n = 158), FDA (n = 145), Public Health England (PHE) (n = 160), and NARMS (n = 155). Genomes of four major O antigen groups (O7, O4, O8, and O9) in these data sets were analyzed. For the PulseNet, FDA, PHE, and NARMS data sets, the O7 group includes serotypes Braenderup, Infantis, Montevideo, and Thompson; the O4 group includes serotypes Agona, Heidelberg, Saintpaul and Typhimurium; the O8 group includes serotypes Hadar, Kentucky, Muenchen, and Newport; and the O9 group includes serotypes Berta, Enteritidis, Javiana, and Panama. Up to 10 genomes of each serotype were randomly selected from each data set. For Nextera XT-prepared genomes in the study by Uelze et al. (n = 578), all the O7, O4, O8, and O9 genomes were included except serotype Enteritidis. This serotype was overrepresented in that study (n = 115), and 20 genomes were randomly selected for this analysis. Different letters above boxes indicate that there is a significant difference (P < 0.05, analysis of variance [ANOVA]) between the values.
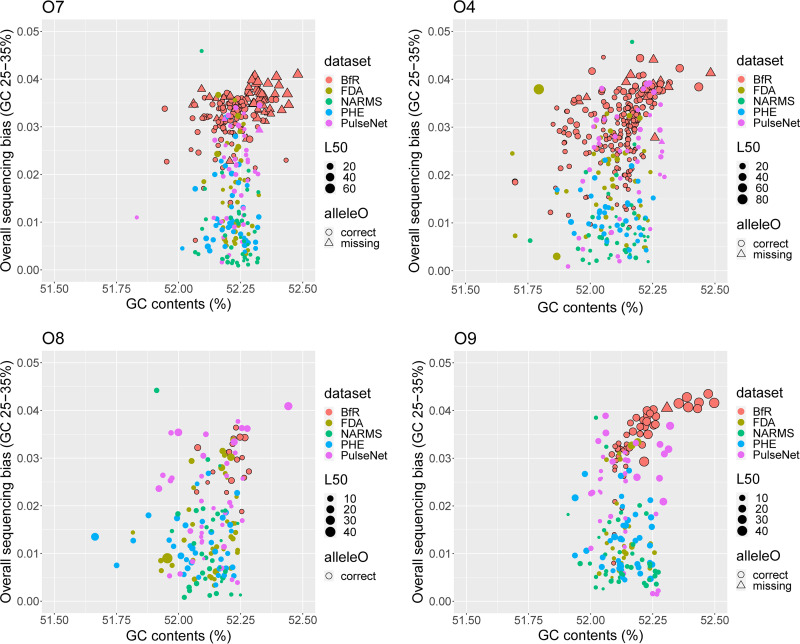
FIG 2.
Sequencing biases of low-GC regions and assembly quality of entire genomes. Low-GC regions are defined as regions of GC content between 25% and 35%, which is similar to the GC content of O antigen determinant genes (wzx/wzy). Genome assembly quality is represented by L50. GC-associated sequencing bias was calculated according to reference 3. Salmonella genomes from five data sets were analyzed, including BfR (n = 415), PulseNet (n = 158), FDA (n = 145), PHE (n = 160), and NARMS (n = 155). Serotypes of four major O antigen groups (O7, O4, O8, and O9) in these data sets were analyzed separately. The O7 group includes serotypes Braenderup, Infantis, Montevideo, and Thompson; the O4 group includes serotypes Agona, Heidelberg, Saintpaul, and Typhimurium; the O8 group includes serotypes Hadar, Kentucky, Muenchen, and Newport; and the O9 group includes serotypes Berta, Enteritidis, Javiana, and Panama. Up to 10 genomes of each serotype were randomly selected from each data set. For Nextera XT-prepared genomes in the study by Uelze et al. (n = 578), all the O7, O4, O8, and O9 genomes were included except serotype Enteritidis (n = 115). This serotype was overrepresented in that study, and 20 genomes were randomly selected for this analysis.
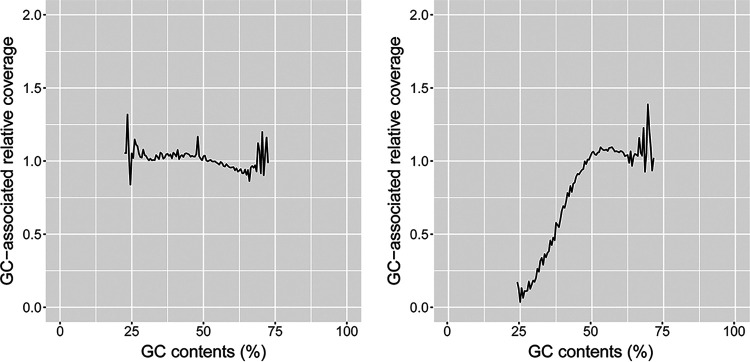
FIG 3.
An example of sequencing bias profile by TruSeq-prepared (left; Sequence Read Archive accession number SRR5740049) and Nextera XT-prepared (right; Sequence Read Archive accession number ERR3581017) genomes. The analyzed isolates were all serotype Infantis genomes. GC-associated relative coverage was calculated according to methods described in reference 3.
While unrelated to sequencing bias, the study by Uelze et al. reported misidentification of Salmonella enterica serotype Enteritidis as serotype Hillingdon, due to a misidentification of serogroup O9 as O9,46 that was specific to the k-mer workflow of SeqSero2. This issue was independently identified by multiple laboratories in the United States and addressed in later releases of SeqSero2. We note that the study by Uelze et al. described SeqSero2 workflows with obsolete terms such as “k-mer mode” and “allele-mode” and did not mention which version of SeqSero2 was evaluated. These terms were used only in the earliest test release of SeqSero2 prior to the first stable version (v.1.0.0) that was published (1).
In conclusion, the genomes used in the study by Uelze et al. were abnormally challenging for O antigen prediction because of unusually high sequencing bias that was not seen in similarly prepared genomes from other laboratories. We recommend that SeqSero2 users be mindful of the GC-related sequencing bias when analyzing Nextera XT-prepared genomes. Although it is unusual for such biases to compromise serotype prediction by SeqSero2 per our knowledge and analysis, it is unknown whether they could affect subtyping and characterization of other low-GC regions, such as Salmonella pathogenicity islands (9, 10), when genome assembly is affected by such biases (Fig. 2).
ACKNOWLEDGMENTS
We thank Patti Fields, Blake Dinsmore, Ana Lauer, and Jessica Chen of the U.S. CDC and Ruth Timme, Shaohua Zhao, and Sunee Himathongkham of the U.S. FDA for providing WGS data and/or helpful discussion.
Footnotes
For the author reply, see https://doi.org/10.1128/AEM.01260-20.
REFERENCES
- 1.Zhang S, den Bakker HC, Li S, Chen J, Dinsmore BA, Lane C, Lauer AC, Fields PI, Deng X. 2019. SeqSero2: rapid and improved Salmonella serotype determination using whole-genome sequencing data. Appl Environ Microbiol 85:e01746-19. doi: 10.1128/AEM.01746-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, Fitzgerald C, Fields PI, Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol 53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sato MP, Ogura Y, Nakamura K, Nishida R, Gotoh Y, Hayashi M, Hisatsune J, Sugai M, Takehiko I, Hayashi T. 2019. Comparison of the sequencing bias of currently available library preparation kits for Illumina sequencing of bacterial genomes and metagenomes. DNA Res 26:391–398. doi: 10.1093/dnares/dsz017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones MB, Highlander SK, Anderson EL, Li W, Dayrit M, Klitgord N, Fabani MM, Seguritan V, Green J, Pride DT, Yooseph S, Biggs W, Nelson KE, Venter JC. 2015. Library preparation methodology can influence genomic and functional predictions in human microbiome research. Proc Natl Acad Sci U S A 112:14024–14029. doi: 10.1073/pnas.1519288112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan JH, Yin Y, Reed EF, Moua K, Thomas K, Zhang Q. 2015. Impact of three Illumina library construction methods on GC bias and HLA genotype calling. Hum Immunol 76:166–175. doi: 10.1016/j.humimm.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grutzke J, Malorny B, Hammerl JA, Busch A, Tausch SH, Tomaso H, Deneke C. 2019. Fishing in the soup—pathogen detection in food safety using metabarcoding and metagenomic sequencing. Front Microbiol 10:1805. doi: 10.3389/fmicb.2019.01805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uelze L, Borowiak M, Deneke C, Szabo I, Fischer J, Tausch SH, Malorny B. 2019. Performance and accuracy of four open-source tools for in silico serotyping of Salmonella spp. based on whole-genome short-read sequencing data. Appl Environ Microbiol 86:e02265-19. doi: 10.1128/AEM.02265-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VPJ, Nash JHE, Taboada EN. 2016. The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanc-Potard AB, Solomon F, Kayser J, Groisman EA. 1999. The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol 181:998–1004. doi: 10.1128/JB.181.3.998-1004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayek N. 2013. Lateral transfer and GC content of bacterial resistant genes. Front Microbiol 4:41. doi: 10.3389/fmicb.2013.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]